Abstract
A member of the short-chain dehydrogenase/reductase superfamily (DHRS1, SDR19C1) is a member of the short-chain dehydrogenase/reductase superfamily and a potential predictor of hepatocellular carcinoma (HCC). However, the role of DHRS1 in HCC immunity remains unclear. We systematically analyzed the association between DHRS1 and HCC immunity with transcriptional and clinical data from the Tumor Immune Estimation Resource, an integrated repository portal for tumor immune system interactions, and cBioPortal databases. Six DHRS1-associated immunomodulators strongly correlated with survival and were uncovered by exploiting univariate and multivariate Cox analyses. We created a risk score for each patient by adding the points from each immunomodulator and then classified them into high and low risk categories. Survival analysis were used to compare the overall survival between the 2 groups, and the receiver operating characteristic curve was applied to assess the accuracy of the risk score. Data from our center were adopted as the external validation set, the risk score was calculated using the risk coefficient of the 6 genes in the training cohort, and survival analysis were executed to verify the experimental group results. A nomogram was ultimately constructed with the R package. Our data revealed a correlation between the levels of immune cell infiltration and either the DHRS1 gene copy numbers or mRNA levels in HCC. Second, we generated a signature based on the 6 DHRS1-related immunomodulators (KDR, TNFRSF4, CD276, TNFSF4, SLAMF6, and SIGLEC9). We postulate that the generated risk scores would serve as an independent indicator of HCC prognosis, with an area under the receiver operating characteristic curve for the risk score of 0.743. We further established external validation sets to reconfirm the predictive validity of the risk score. Finally, a prognostic nomogram and calibration curve were created. The DHRS1 gene may exert an impact on HCC immunity. We posit that the nominated immune signature based on DHRS1-associated immunomodulators could constitute a promising prognostic biomarker in HCC.
Keywords: DHRS1, hepatocellular carcinoma, immunity, prognosis
1. Introduction
According to the most recent official cancer data, there were approximately 906,000 new primary liver cancers and 830,000 deaths in 2020, with hepatocellular carcinoma (HCC) accounting for approximately 75% to 85% of the total. The prevalence of HCC was greatest in areas of East Asia, and men died at substantially greater rates than women.[1] Because patients are asymptomatic in the early stages of hepatocellular carcinoma, HCC proceeds to an advanced stage or even metastasizes by the time it is detected, and when the greatest opportunity for localized therapy has passed (e.g., after hepatectomy, TACE, and radiofrequency ablation).[2] However, patients with HCC exhibit an appalling prognosis, with a high rate of recurrence and a poor response to chemoradiotherapy. As a result, <10% of HCC patients live for 5 years.[3] The hepatic microenvironment affects immunosurveillance, which can either hamper or foster HCC development, and the immunological state of the tumor microenvironment (TME) may thus affect the prognosis of HCC. For example, Treg infiltration in HCC abrogates the immune system of the tumor, such that the presence of abundant Treg infiltration constitutes an untoward prognosis for the tumor.[4] As immune checkpoints have been associated with tumor cell growth and escape from the immune system,[5] immune checkpoint inhibitors such as PD-1/PDL-1 have been authorized for cancer treatment to increase immune control over malignant cells. NCCN guidelines suggest the adoption of immune checkpoint inhibitors for advanced HCC because of their outstanding effectiveness.[6] Current clinical categorization, such as the tumor, node, and metastasis (TNM) staging system, however, provides no tumor immune information; therefore, it is not possible to predict the response to immune therapy.[7] Consequently, selecting a biomarker related to HCC immunity as an indicator of prognostic assessment is critical.
The short-chain dehydrogenase/reductase (SDR) family member 1 (a member of the short-chain dehydrogenase/reductase superfamily [DHRS1], also known as SDR19C1) has been localized to the nucleus, mitochondria, and endoplasmic reticulum of HeLa cells. The human gene DHRS1 was found on chromosome 14q12, with its highest level of protein expression in the liver.[8] In HCC, DHRS1 expression falls, and patients with reduced DHRS1 expression show a poorer prognosis and survival. Although it has not yet been demonstrated experimentally, some authors suggest that DHRS1 may constitute a unique and independent predictive biomarker and a potential therapeutic target for HCC.[9] Prognostic prediction models based on DHRS1-related immune genes have been established, which have proven to be both effective and reliable, suggesting a role for DHRS1 in the infiltration of immune cells in HCC. However, to our knowledge, no relevant studies on this topic have been published.
In the present investigation, we performed a search of a publicly accessible database to analyze the relationship between DHRS1 and immunity in HCC. The cancer genome atlas (TCGA)-LIHC database was used to screen for immunomodulators associated with DHRS1, and we subsequently generated a prognostic model to predict the prognosis of HCC patients. We then conducted an external validation using 62 cases of HCC at our center.
2. Materials and methods
2.1. Patients and sample tissues
Between January 2010 and May 2015, we collected fresh tissue specimens as well as clinical and prognostic data from 62 patients who were diagnosed with HCC and who underwent radical resection for HCC at Anhui Medical University’s First Affiliated Hospital. The Ethics Committee of Anhui Medical University’s First Affiliated Hospital approved our protocol, and the IRB number was 20131359. We received informed consent from participants in our study, and we adopted clinical HCC specimens in compliance with the Declaration of Helsinki.
2.2. Data sources and processing
Gene expression and clinical data of patients with HCC were gathered from the TCGA-LIHC database (http://portal.gdc.cancer.gov/repository).[10] The limma package in R was applied to conduct analyses on the RNA expression data.[11] The Kaplan–Meier plotter method was employed to generate survival curves and confirm survival differences across groups.
2.3. Tumor immune cell infiltration
The tumor immune estimation resource (TIMER) (http://timer.cistrome.org/) is an interactive web tool that allows for the comprehensive and flexible analysis and visualization of tumor-infiltrating immune cells. TIMER deduces the abundance of tumor-infiltrating immune cells (B cells, CD4 + T cells, CD8 + T cells, macrophages, neutrophils, and dendritic cells) from gene expression profiles of different cancer types in the TCGA.[12] In this study, the correlations between DHRS1 expression levels/copy number alterations and 6 types of immune cells were estimated using the TIMER “immune association” modules.
2.4. Gene set enrichment analysis
To classify patients into high and low-expression groups, we measured the median DHRS1 expression using LIHC RNA-seq. The gene set enrichment analysis (GSEA) software’s reference molecular signature database (MSigDB) (version 4.1.0) was then applied to investigate the potential regulatory pathways in which DHRS1-related genes participated.
2.5. Immunomodulators and related genes
Tumor immune system interactions (TISIDB) is a website in which data from a variety of sources are collected and integrated, including PubMed, high-throughput screening, TCGA, and other public databases such as Uniprot, GO, and DrugBank. We downloaded information on tumor-infiltrating lymphocytes (TILs) and immunomodulators related to DHRS1 from the TISIDB database for tumor immune interactions and sought correlations between them. (http://cis.hku.hk/TISIDB/).[13] Then, DHRS1-associated immunomodulators were used to construct a protein-protein interaction network using the search tool for the retrieval of interaction gene/proteins database (https://string-db.org/). Afterward, DHRS1-associated immunomodulators were subjected to GO and KEGG analysis using the consensus online tool with FDR < 0.05 (http://cpdb.molgen.mpg.de/).
2.6. Construction of the prognostic signature and nomogram
The association of DHRS1-associated immunomodulators and prognosis were analyzed by stepwise Cox regression analysis using the R package “survmine”. Using DHRS1-associated immunomodulators, we constructed a prognostic multiple immune gene signature model,[14] and our risk score was created by screening the associated immune genes using multivariate Cox regression to establish a prognostic index: risk score = β1 × 1 + β2 × 2 +. + βixi.[15] We accounted for each gene’s expression level (xi) in this formula and designated Cox regression coefficients for each gene by βi. A stepwise Cox analysis was used to identify independent prognostic factors. The Kaplan–Meier survival curves were ultimately generated to evaluate the relationship between the risk score and overall survival (OS),[16] and time-dependent receiver operating characteristic (ROC) curves[17] were used to determine the prognostic accuracy of the risk scores.
2.7. RNA extraction and qPCR
Total RNA was isolated from HCC tissues using TRIzol reagent as directed by the manufacturer, and then reverse-transcribed into cDNA using a Prime ScriptTM RT Reagent Kit (AG, Changsha, China). Using GAPDH as an internal reference, the Ct method was used to determine the relative changes in mRNA.[18] (see Table S1, Supplemental Digital Content, http://links.lww.com/MD/K398, which demonstrates the primer sequences).
2.8. Construction of nomogram T
A nomogram was specifically developed to visualize our statistical prediction models, and we routinely used it to present results from logistic regression or COX regression analyses. The nomogram was created for R software via the rms package. The bootstrap method was utilized in conjunction with a calibration curve (1000 replicates) to visualize the difference between predicted and actual probabilities. The forecasting precision of a nomogram was measured using the concordance index. Based on the results of the regression, we produced multiple lines at a specific scale, and the graphs were used to conveniently deduce the risk or probability of an individual’s survival. Each of the characteristics was rated separately, including age, sex, TNM staging, and risk score. Then these variables were combined to generate a total score and to allow estimation of the calibration curve for 1-, 3-, and 5-year OS.[19]
2.9. Statistical analysis
All statistical analyses were performed by R version 4.0.3 and the GSEA analysis software (GSEA Desktop Application v4.1, Broad Institute, Inc, Cambridge, MA). Outcomes were implemented using SPSS (version 22.0), GraphPad Prism (version 8.0), and the aforementioned network tools. We generated Kaplan–Meier survival curves and time-dependent ROC curves with the R packages“survival” and “survival ROC,” respectively. Relevant immunological genes were identified using Spearman correlation analysis, and the threshold for statistical significance was set at P < .05.
3. Results
3.1. Association between DHRS1 and immune infiltration cells in HCC
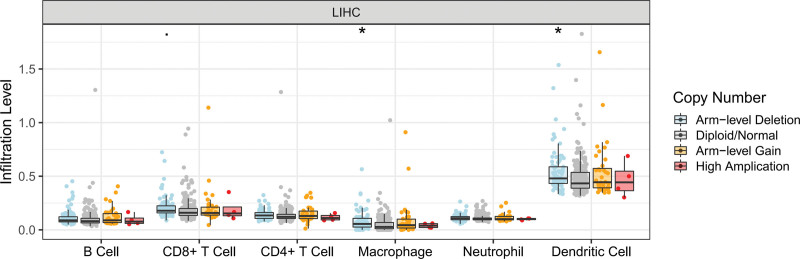
Infiltrated immune cells – key components of the TME – exert an impact on the prognosis of HCC patients. With limited evidence, Li et al reported that DHRS1 might contribute to the immune response in HCC.[20] Here, we further investigated the relationship between DHRS1 and HCC immunity. In TIMER, several types of immune cell infiltration were found in LIHC, including macrophages and dendritic cells, and they appeared to be associated with changes in DHRS1 gene copy levels (Fig. 1). The expression of DHRS1 mRNA in LIHC was also positively correlated with some immune types and negatively correlated with others (Figure S1, Supplemental Digital Content, http://links.lww.com/MD/K397).
Figure 1.
Immune relevance of DHRS1 in HCC based on tumor immune estimation resource (TIMER). Comparison of tumor infiltration levels among tumors with different DHRS1 copy number alterations*P < .05, **P < .01, ***P < .005. DHRS1 = a member of the short-chain dehydrogenase/reductase superfamily, HCC = hepatocellular carcinoma.
3.2. DHRS1 is associated with immune-related pathways in LIHC
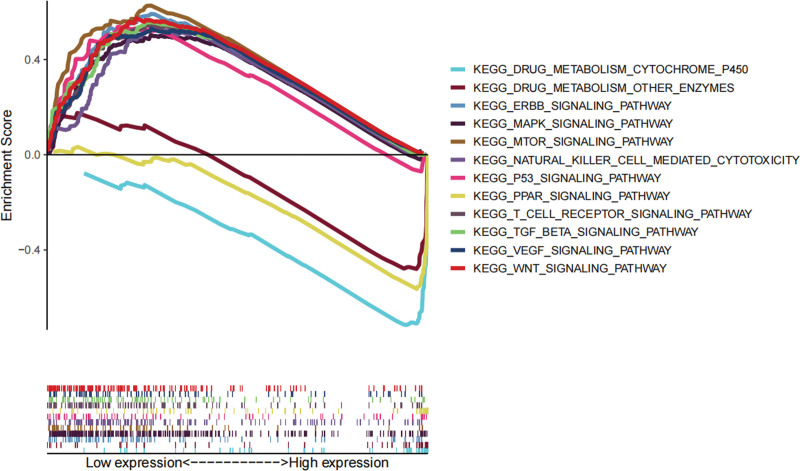
In addition, we investigated potential signaling pathways regulated by DHRS1 in LIHC that may be responsible for modulating the immunological response. Several immune-related signaling pathways (including the MAPK signaling network, the mTOR-signaling pathway, and the P53-signaling system) were found to be associated with DHRS1 by gene set enrichment analysis (GSEA, Fig. 2), which validated that DHRS1 may affect tumor progression through immune-related pathways in HCC.
Figure 2.
Representative signaling pathways for DHRS1 single gene GSEA analysis. DHRS1 = a member of the short-chain dehydrogenase/reductase superfamily, GSEA = gene set enrichment analysis.
3.3. DHRS1 is associated with immunomodulators in HCC
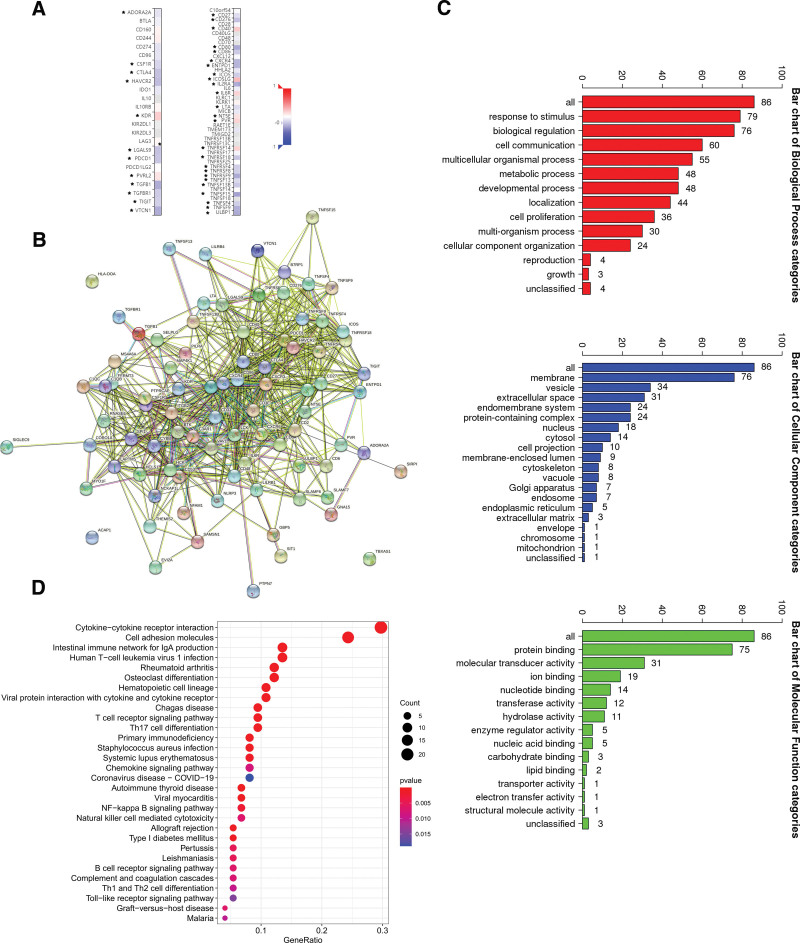
We next aimed to explore the relationship between DHRS1 and immunomodulators. We obtained 25 immunostimulators (CD27, CD276, CD40, CD80, CD86, CXCR4, ENTPD1, ICOS, ICOSLG, IL2RA, IL6R, LTA, PVR, NT5E, TNFRSF14, TNFRSF18, TNFRSF4, TNFRSF8, TNFRSF9, TNFSF13, TNFSF13B, TNFSF15, TNFSF4, TNFSF9, and ULBP1) and 12 immunoinhibitors (ADORA2A, CSF1R, CTLA4, HAVCR2, KDR, LGALS9, PDCD1, PVRL2, TGFB1, TGFBR1, TIGIT, and VTCN1) as agents that were significantly associated with DHRS1 in the LIHC (Fig. 3A). By constructing a protein-protein interaction network of the 37 immunomodulators, we obtain a close interaction relationship among DHRS1-related 50 immunoregulatory genes. (Fig. 3B). These genes were then annotated using GO terms, and subsequent KEGG analysis revealed that they played roles in immunological signaling pathways, such as T cell receptor signaling route and B cell receptor signaling pathway. (Fig. 3C–D)
Figure 3.
Identification and analysis of immunomodulators associated with the DHRS1 gene. (A) Immunostimulators (left panel) and immunoinhibitors (right panel) agents are significantly associated with DHRS1 in the LIHC. (B) Protein-protein network of 37 DHRS1-associated immunomodulators and 50 closely related genes in LIHC, produced by the STRING online server. (C) GO annotation of 37 DHRS1-associated immunomodulators and 50 closely connected genes in LIHC. (D) KEGG enrichment analysis of 37 DHRS1-associated immunomodulators and 50 closely related genes. *P < .05, **P < .01, ***P < .001. DHRS1 = a member of the short-chain dehydrogenase/reductase superfamily, STRING = the retrieval of interaction gene/proteins.
3.4. Prognostic implications of DHRS1-associated immunomodulators in LIHC
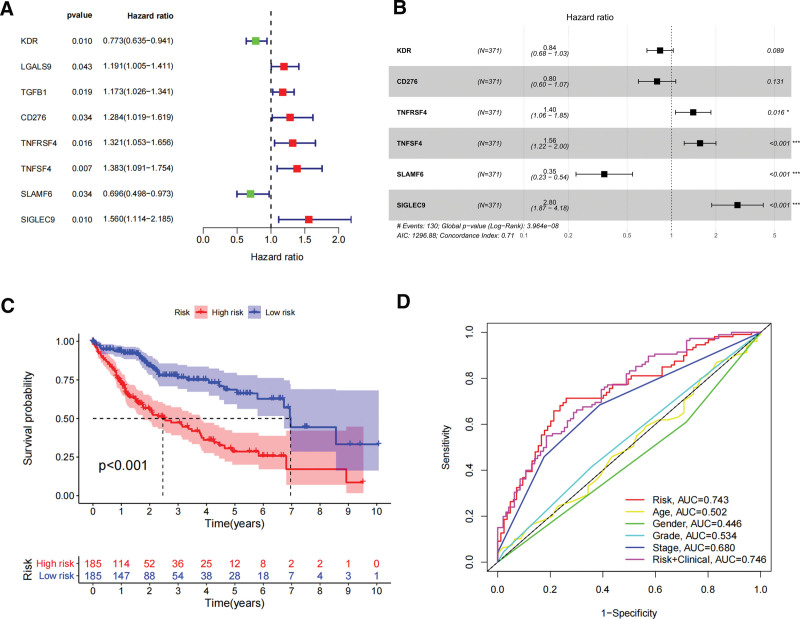
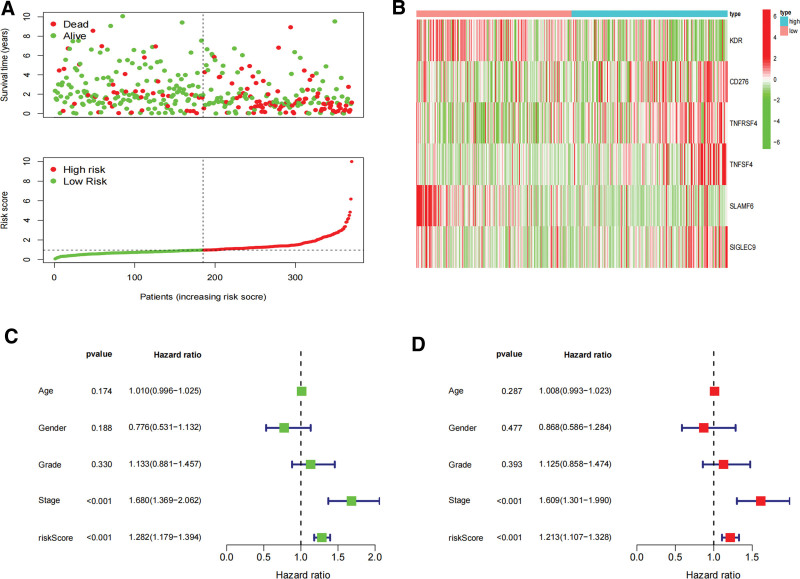
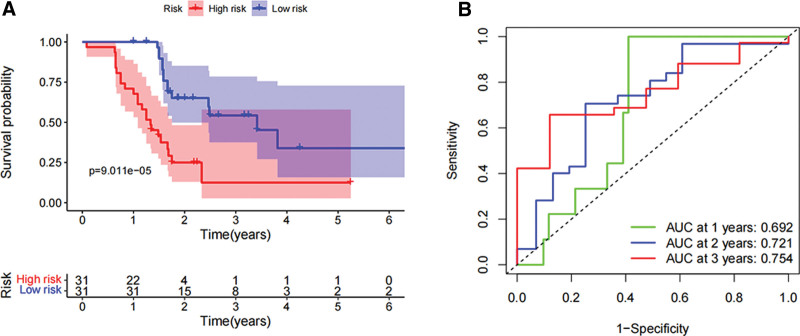
In order to assess the prognostic value of 87 DHRS1-related immunomodulators, firstly, we selected 8 genes involved in prognosis using univariate Cox regression analysis (Fig. 4A), and then we identified 6 prognostic genes by multivariate Cox regression analysis (Fig. 4B). The expression coefficients of these 6 genes were as follows: KDR = −0.178, CD276 = −0.225, TNFRSF4 = 0.3388, TNFSF4 = 0.4457, SLAMF6 = −1.049, and SIGLEC9 = 1.0294. The risk scores were (−0.178 * X1) + (−0.225 * X2) + (0.3388 * X3) + (0.4457 * X4) + (−1.049 * X5) + (1.0294 * X6), with Xi representing each gene’s expression levels. The risk score was calculated by accumulating the product of these 6 genes expression and its coefficient in each sample. We downloaded the mRNA expression data of patients with TCGA-LIHC, and we noted that those patients with lower risk ratings fared much better than those with higher risk ratings using the Kaplan–Meier survival curve (log-rank test, P < .001) (Fig. 4C). The areas under the ROC curve (AUC) for risk score and staging were 0.743 and 0.680, respectively, and the AUC for the risk score and clinical score combined was 0.746 (Fig. 4D). The distribution of risk scores, survival status, and characteristic gene expression patterns of the HCC patients are illustrated in (Fig. 5A–B). Figure 5C displays the results of the univariate Cox regression analysis of the TCGA-LIHC, showing a statistically significant association between risk score and supplemental cont survival (hazard ratio = 1.282, 95% confidence interval = 1.179–1.394, P < .001). Our multivariate Cox regression analysis subsequently revealed that risk score was an independent predictor of HCC survival (hazard ratio = 1.213, 95% confidence interval of total = 1.107–1.328, P < .001) (Fig. 5D). Sixty-two HCC tissue samples were chosen for external validation to evaluate the validity of the risk score (see Table S2, Supplemental Digital Content, http://links.lww.com/MD/K399, which demonstrates the patient characteristics between high and low risk group). Moreover, the risk score of the 62 HCC patients at our center was calculated using the risk coefficients of the 6 genes in the experimental group, and the survival analysis was applied to verify the experimental group’s results. We thereby discovered that patients with lower risk scores possessed a significantly longer survival time and survival rate, consistent with the results from the training set (Fig. 6A). The AUCs for the biomarker-based prognostic model at 1, 2, and 3 years were 0.692, 0.721, and 0.754, respectively (Fig. 6B).
Figure 4.
Develop prognostic gene signatures based on 87 DHRS1-related immune genes. (A) Univariate COX regression of DHRS1-associated immunomodulators. (B) Multivariate Cox regression analysis of DHRS1-associated immunomodulators. (C) Survival curves of the high and low risk groups of the TCGA set. (D) Time-dependent ROC curves of DHRS1-associated prognostic model of the TCGA set. DHRS1 = a member of the short-chain dehydrogenase/reductase superfamily, ROC = the receiver operating characteristic, TCGA = the cancer genome atlas.
Figure 5.
Characteristics of the DHRS1-associated prognostic signature and value in the TCGA dataset. (A) The distribution of risk scores and the survival status of HCC patients. (B) Heatmap of the DHRS1-associated prognostic signature expression profiles between the high and low risk groups. (C) Univariate Cox regression analyzes the relationship between risk score and overall survival. (D) Multivariate Cox regression analyzes the relationship between risk score and overall survival. *P < .05, **P < .01, ***P < .001. DHRS1 = a member of the short-chain dehydrogenase/reductase superfamily, HCC = hepatocellular carcinoma, TCGA = the cancer genome atlas.
Figure 6.
The identification of risk scores correlated with survival in patients with LIHC in the external validation set. (A) Survival analysis of the risk score in the external validation set. (B) ROC curves and area under the curve (AUC) for 1-, 2-, and 3-year survival in the external validation set. *P < .05, **P < .01, ***P < .001. ROC = the receiver operating characteristic.
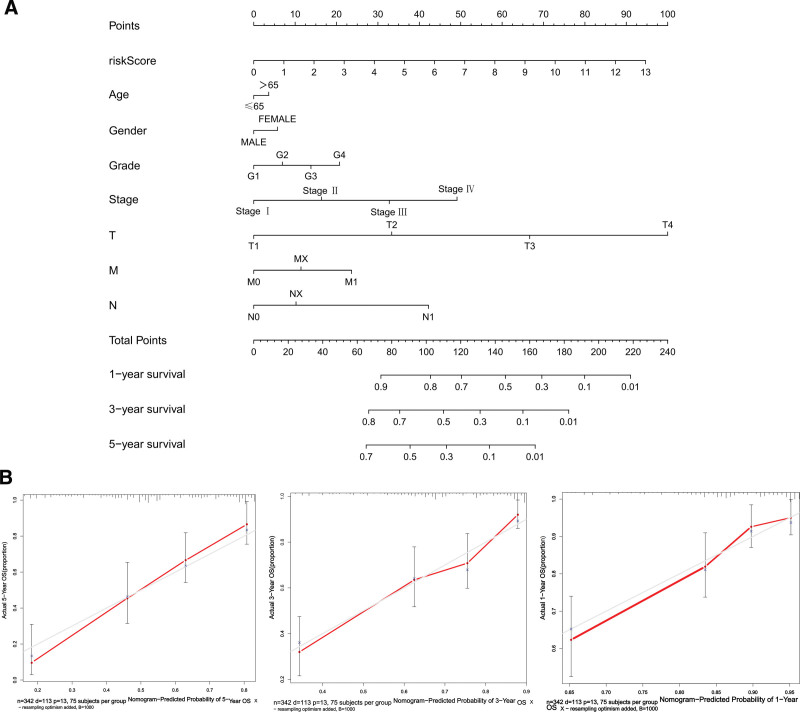
3.5. Construction of nomogram
To estimate the probability of survival for HCC patients, we constructed a nomogram that allowed for risk score, stage, TNM, age, and sex (Fig. 7A). We calibrated the nomogram for 1-, 3-, and 5-year survival, and the calibration curve indicated a satisfactory match between the nomogram-predicted probability and the ideal reference line (Fig. 7B). Additionally, the concordance index was 0.71, which suggested good predictive power.
Figure 7.
Establishment of the prognostic nomogram with the inclusion of the risk score. (A) A nomogram for predicting the overall survival of TCGA-LIHC patients. (B) Calibration curves of nomogram-predicted 1-, 3-, and 5-year survival of HCC patients. Red line: nomogram-predicted survival curve. Gray line: ideal survival reference curve. HCC = hepatocellular carcinoma, TCGA = the cancer genome atlas.
4. Discussion
In this study, we demonstrated that DHRS1 was closely associated with immunity in HCC, with DHRS1 expression linked to infiltrating immune cells and immunomodulators. We constructed multiple-gene risk-prediction signatures from the DHRS1-associated immunomodulators using stepwise COX regression and validated the prognostic model using clinical samples from our center. Finally, we used our prognostic model and clinical characteristics to generate an HCC prognostic nomogram for each patient that could then be used to guide HCC treatment after surgery.
Many members of the SDR family have been shown to display diverse functions in different illnesses; for example, DHRS12 has been used as an independent predictor of prognosis in cervical cancer patients.[21] Furthermore, the activation of DHRS2 reduced the development of nasopharyngeal cancer by regulating lipid metabolism.[22] By increasing Nrf2 ubiquitination, BDH2 (DHRS6) may augment endoplasmic reticulum stress and trigger cell death in gastric cancer.[23] Liang et al[24] ascertained that BDH2 reduced Bcl-2 expression, leading to elevated Bax levels and preventing HCC cells from proliferating. These findings collectively show that DHRS1 gene homologs are involved in cancer regulation.
Li et al[25] reported low-level expression of DHRS1 in HCC tumor tissues and that DHRS1 mRNA and protein levels were associated with OS. DHRS1 levels drop as the tumor stage and grade rise, indicating the potential involvement of DHRS1 in HCC. However, how DHRS1 contributed to HCC was not previously understood. The TME is important in HCC development and therapeutic response, wherein cancer cells interact with multiple immune components to form a suppressive immune microenvironment that promotes immune escape, proliferation, invasion, and metastasis of HCC cells and that mediates drug resistance.[26] We identified a correlation between DHRS1 gene copy number and infiltrating immune cells in pan-cancer via the TIMER database and demonstrated that the abundance of most immune subsets changed with the levels of DHRS1 mRNA using the TISIDB online tool. Evaluation of DHRS1-associated immunomodulators using TCGA-LIHC data with KEGG analysis suggests that the MAPK signaling pathway, the mTOR-signaling pathway, and the P53 signaling route are all involved in DHRS1-mediated immune responses.[27] The roles of MAPK signaling in immunity and immunotherapy have been extensively studied. Activation of mTOR regulates tumor growth, metastasis, and immunity;[28] and essential components of tumor immunology and homeostatic regulation of immune responses involve the p53 tumor suppressor pathway.[29] Thus, the aforementioned results indicated a link between DHRS1 and immunity.
The liver is a powerful immune organ in the body, serving immune-defensive and immune-regulatory functions.[30] TILs, including T cells and NK cells, are the immune cells that produce the greatest effects against tumors, and the impact of TILs on antitumor efficacy is proportional to their abundance and activity.[31] TIL therapy was therefore developed to assess prognosis by directly identifying the targets of cancer cells. MITD1 gene expression was shown to be linked to immune infiltration in HCC, and studies by Shen et al[32] have shown that it may serve as a predictive biomarker for Cox regression analysis. In the current study, we analyzed DHRS1 in a similar manner. We determined that multiple TILs and immunomodulators were abundantly associated with DHRS1, and using gene co-expression analysis, we observed potential mechanisms linking the high levels of TILs and immunomodulators to DHRS1. Multiple immunomodulator genes, such as KDR, TNFRSF4, CD276, TNFSF4, SLAMF6, and SIGLEC9, were shown to be co-expressed with DHRS1. Cox regression analysis revealed that KDR, CD276, and SLAMF6 were protective for HCC patients, while TNFRSF4, TNFSF4, and SIGLEC9 were risk factors.[33] found SLAMF6 to be a predictive gene by adding the stromal and immune scores in HCC. Yoshiji et al[34] depicted monotherapy with KDR/Flk-1 as boosting apoptosis in tumor cells – specifically in HCC cells. According to Cheng et al,[35] patients with HCC who expressed the CD276 protein and developed vasculogenic mimicry reflected a poorer prognosis. Hong et al[36] constructed an immune-related risk score model and uncovered a significant correlation between TNFSF4 and patient risk score. In a study of potential immunotherapeutic targets in HCC patients, Cai et al[37] showed that the immune gene TNFRSF4 might comprise a promising prognostic biomarker for HCC. Ren et al[38] also found that SIGLEC9 signaling was greatly reduced in HCC, suggesting that it might constitute an anti-oncogenic gene. Using DHRS1-related immunomodulators, we herein developed a predictive model that categorized HCC patients into high and low risk categories and produced a risk score that was substantially linked to survival.
To conduct external verification of risk score accuracy, we selected 62 HCC patients with tumor tissues for the present study. The mRNA expression levels of 6 immunomodulators were determined by qPCR, and these values were entered into the formula for the internal validation set. The survival curve was generated in combination with the clinical data, and the results of the external validation set were identical to the internal validation set. Furthermore, in the external validation set, we confirmed a correlation between DHRS1 expression and immunomodulators by qPCR. The above results suggest that the model established by DHRS1-related immunomodulators can be employed as an independent factor in predicting patient prognosis.
However, there were still limitations to the present study. First, this study was based on bioinformatics analysis of the TCGA-LIHC databases, and ascertaining whether DHRS1 actually regulates the immunomodulator functions requires further experimental research. Additionally, the number of samples used for external validation (62) was inadequate. Therefore, additional external studies are needed to confirm the results of our study.
5. Conclusion
DHRS1 may be involved in the regulatory process of the HCC immune microenvironment. Through analysis, we identified 6 prognostic immune genes associated with the immune microenvironment and HCC prognosis. The prognosis-predicted model generated by the 6 DHRS1-related immunomodulators could be adopted to anticipate the prognosis of HCC patients.
Author contributions
Conceptualization: Yuxian Shen.
Funding acquisition: Hongchuan Zhao, Yuxian Shen.
Methodology: Wei Wang.
Resources: Sa Xu, Tao Meng, Fuyan Wang, Guoxing Wang, Fan Huang, Guobin Wang, Xiaojun Yu, Ruolin Wu, Liujin Hou, Xinghua Zhang.
Software: Sa Xu, Tao Meng, Fuyan Wang, Guoxing Wang, Fan Huang, Guobin Wang, Xiaojun Yu, Ruolin Wu, Liujin Hou, Zhenghui Ye, Xinghua Zhang, Wei Wang.
Supervision: Hongchuan Zhao, Yuxian Shen.
Validation: Hongchuan Zhao, Yuxian Shen.
Visualization: Sa Xu, Tao Meng, Fuyan Wang, Guoxing Wang, Fan Huang, Guobin Wang, Xiaojun Yu, Ruolin Wu, Liujin Hou, Zhenghui Ye, Xinghua Zhang.
Writing – original draft: Yuxian Shen.
Writing – review & editing: Guoxing Wang, Fan Huang, Guobin Wang, Xiaojun Yu, Ruolin Wu, Liujin Hou, Zhenghui Ye, Xinghua Zhang.
Supplementary Material
Abbreviations:
- AUC
- areas under the ROC curve
- DHRS1
- a member of the short-chain dehydrogenase/reductase superfamily
- GSEA
- gene set enrichment analysis
- HCC
- hepatocellular carcinoma
- OS
- overall survival
- ROC
- the receiver operating characteristic
- SDR
- short-chain dehydrogenase/reductase
- TCGA
- the cancer genome atlas
- TILs
- tumor-infiltrating lymphocytes
- TIMER
- the tumor immune estimation resource
- TISIDB
- tumor immune system interactions
- TME
- the tumor microenvironment
- TNM
- tumor, node, and metastasis
Supplemental Digital Content is available for this article.
The datasets generated during and/or analyzed during the current study are available from the first author on reasonable request.
SX and TM contributed equally to this work.
This work was supported by grants from the National Natural Science Foundation of China (No. 81973336, 81902003, and 31800702), Anhui Medical University Research Fund (No.2021xkj147) and Anhui Provincial Higher Education Research Fund (No.2022AH051189). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. We also thank Let Pub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
The authors have no conflicts of interest to disclose.
How to cite this article: Xu S, Wang W, Meng T, Wang F, Wang G, Huang F, Wang G, Yu X, Wu R, Hou L, Ye Z, Zhang X, Zhao H, Shen Y. Construction and validation of a immune-related prognostic gene DHRS1 in hepatocellular carcinoma based on bioinformatic analysis. Medicine 2023;102:42(e35268).
Contributor Information
Sa Xu, Email: xusa4262@163.com.
Wei Wang, Email: wwok68@hotmail.com.
Tao Meng, Email: joe_mt@163.com.
Fuyan Wang, Email: wwok68@hotmail.com.
Guoxing Wang, Email: wwok68@hotmail.com.
Fan Huang, Email: huang_f@vip.126.com.
Guobin Wang, Email: wwok68@hotmail.com.
Xiaojun Yu, Email: yuxiaojunhefei@163.com.
Ruolin Wu, Email: wuruolin0685@126.com.
Liujin Hou, Email: 592490328@qq.com.
Zhenghui Ye, Email: 767725431@qq.com.
Xinghua Zhang, Email: zhangxh8888@qq.com.
Hongchuan Zhao, Email: zhc0117@sina.com.
References
- [1].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- [2].Reig M, Forner A, Rimola J, et al. BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J Hepatol. 2022;76:681–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:293–313. [DOI] [PubMed] [Google Scholar]
- [4].Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li X, Ramadori P, Pfister D, et al. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer. 2021;21:541–57. [DOI] [PubMed] [Google Scholar]
- [6].Li B, Chan HL, Chen P. Immune checkpoint inhibitors: basics and challenges. Curr Med Chem. 2019;26:3009–25. [DOI] [PubMed] [Google Scholar]
- [7].Vaddepally RK, Kharel P, Pandey R, et al. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers (Basel). 2020;12:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang X, Wang F, Zhong M, et al. The biomarkers of hyperprogressive disease in PD-1/PD-L1 blockage therapy. Mol Cancer. 2020;19:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the “Immunoscore” in the classification of malignant tumours. J Pathol. 2014;232:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zemanova L, Navratilova H, Andrys R, et al. Initial characterization of human DHRS1 (SDR19C1), a member of the short-chain dehydrogenase/reductase superfamily. J Steroid Biochem Mol Biol. 2019;185:80–9. [DOI] [PubMed] [Google Scholar]
- [11].Li L, Situ HJ, Ma WC, et al. Decreased DHRS1 expression is a novel predictor of poor survival in patients with hepatocellular carcinoma. Biomark Med. 2021;15:1319–31. [DOI] [PubMed] [Google Scholar]
- [12].Wei L, Jin Z, Yang S, et al. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34:1615–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li T, Fu J, Zeng Z, et al. TIMER20 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–2. [DOI] [PubMed] [Google Scholar]
- [16].George B, Seals S, Aban I. Survival analysis and regression models. J Nucl Cardiol. 2014;21:686–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rich JT, Neely JG, Paniello RC, et al. A practical guide to understanding Kaplan–Meier curves. Otolaryngol Head Neck Surg. 2010;143:331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44. [DOI] [PubMed] [Google Scholar]
- [20].Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125. [DOI] [PubMed] [Google Scholar]
- [21].Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364–70. [DOI] [PubMed] [Google Scholar]
- [22].Zhang Y, Wu J, Jing H, et al. Increased DHRS12 expression independently predicts poor survival in patients with high-grade serous ovarian cancer. Future Oncol. 2018;14:2579–88. [DOI] [PubMed] [Google Scholar]
- [23].Luo X, Li N, Zhao X, et al. DHRS2 mediates cell growth inhibition induced by Trichothecin in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2019;38:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu JZ, Hu YL, Feng Y, et al. BDH2 triggers ROS-induced cell death and autophagy by promoting Nrf2 ubiquitination in gastric cancer. J Exp Clin Cancer Res. 2020;39:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liang H, Xiong Z, Li R, et al. BDH2 is downregulated in hepatocellular carcinoma and acts as a tumor suppressor regulating cell apoptosis and autophagy. J Cancer. 2019;10:3735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lu C, Rong D, Zhang B, et al. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer. 2019;18:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang Y, Liu S, Yang Z, et al. Anti-PD-1/L1 lead-in before MAPK inhibitor combination maximizes antitumor immunity and efficacy. Cancer Cell. 2021;39:1375–1387.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hua H, Kong Q, Zhang H, et al. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ghosh M, Saha S, Bettke J, et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell. 2021;39:494–508.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43(2 Suppl 1):S54–62. [DOI] [PubMed] [Google Scholar]
- [31].Weber EW, Maus MV, Mackall CL. The emerging landscape of immune cell therapies. Cell. 2020;181:46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shen H, Wang Z, Ren S, et al. Prognostic biomarker MITD1 and its correlation with immune infiltrates in hepatocellular carcinoma (HCC). Int Immunopharmacol. 2020;81:106222. [DOI] [PubMed] [Google Scholar]
- [33].Liu B, Zeng L, Shao Y, et al. Expression and function of SLAMF6 in CD8(+) T lymphocytes of patients with severe aplastic anemia. Cell Immunol. 2021;364:104343. [DOI] [PubMed] [Google Scholar]
- [34].Yoshiji H, Kuriyama S, Hicklin DJ, et al. KDR/Flk-1 is a major regulator of vascular endothelial growth factor-induced tumor development and angiogenesis in murine hepatocellular carcinoma cells. Hepatology. 1999;30:1179–86. [DOI] [PubMed] [Google Scholar]
- [35].Cheng R, Wang B, Cai XR, et al. CD276 promotes vasculogenic mimicry formation in hepatocellular carcinoma via the PI3K/AKT/MMPs pathway. Onco Targets Ther. 2020;13:11485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hong WF, Gu YJ, Wang N, et al. Integrative characterization of immune-relevant genes in hepatocellular carcinoma. J Clin Transl Hepatol. 2021;9:301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cai Q, Zhu M, Duan J, et al. Comprehensive analysis of immune-related prognosis of TK1 in hepatocellular carcinoma. Front Oncol. 2021;11:786873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ren X, Ji Y, Jiang X, et al. Down-regulation of siglec-2 (CD22) predicts worse overall survival from HBV-related early-stage hepatocellular carcinoma: a preliminary analysis from Gene Expression Omnibus. Biosci Rep. 2018;38:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.