Significance
Positive-sense single-stranded RNA (ssRNA+) viruses, such as dengue virus (DENV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), have RNA-dependent RNA polymerase which is a promising target for broad-spectrum antivirals. We identified 2-thiouridine (s2U) as a broad-spectrum antiviral ribonucleoside analogue that exhibited antiviral activity against DENV. Furthermore, s2U inhibited SARS-CoV-2 infection including the currently circulating Omicron subvariants. In DENV2- or SARS-CoV-2-infected mice, s2U treatment significantly decreased viral load and improved survival rates. Since the efficacy of s2U against other ssRNA+ viruses including Zika virus, yellow fever virus, Japanese encephalitis virus, West Nile virus, Chikungunya virus, and other human coronaviruses was also identified, we demonstrated the potential clinical utility of s2U for Dengue, COVID-19, and other diseases caused by ssRNA+ viruses.
Keywords: dengue virus, SARS-CoV-2, positive-strand RNA viruses, antiviral, RNA-dependent RNA polymerase
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections are causing significant morbidity and mortality worldwide. Furthermore, over 1 million cases of newly emerging or re-emerging viral infections, specifically dengue virus (DENV), are known to occur annually. Because no virus-specific and fully effective treatments against these or many other viruses have been approved, there is an urgent need for novel, effective therapeutic agents. Here, we identified 2-thiouridine (s2U) as a broad-spectrum antiviral ribonucleoside analogue that exhibited antiviral activity against several positive-sense single-stranded RNA (ssRNA+) viruses, such as DENV, SARS-CoV-2, and its variants of concern, including the currently circulating Omicron subvariants. s2U inhibits RNA synthesis catalyzed by viral RNA-dependent RNA polymerase, thereby reducing viral RNA replication, which improved the survival rate of mice infected with DENV2 or SARS-CoV-2 in our animal models. Our findings demonstrate that s2U is a potential broad-spectrum antiviral agent not only against DENV and SARS-CoV-2 but other ssRNA+ viruses.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; family: Coronaviridae) is responsible for the ongoing pandemic of COVID-19, and the number of cases since December 2019 has been estimated at over 676 million (1). Likewise, many newly emerging or re-emerging viral infections have occurred in the 21st century and also continue to seriously impact human health. For instance, dengue virus (DENV; family: Flaviviridae) causes large-scale epidemics in Asia, with 390 million cases worldwide annually (2), and Chikungunya virus (CHIKV; family: Togaviridae), which is endemic in the equatorial regions, causes at least 3 million infections annually (3). The genomes of these viruses are composed of positive-sense single-stranded RNA (ssRNA+). Although extensive drug discovery research against these viruses has been undertaken, no virus-specific and fully effective treatments have been approved. With respect to SARS-CoV-2, emergency use authorization (EUA) of neutralizing antibodies has significantly contributed to suppressing disease severity; however, these are expensive and only effective for the fusion step of SARS-CoV-2 infection. Therefore, the development of novel effective therapeutic agents, particularly small-molecule compounds, against COVID-19 and many other viral diseases is essential.
Because licensed antiviral therapeutic agents are limited, broad-spectrum antiviral agents are critical for the control of emerging viral diseases. Ribavirin, which was developed in the 1970s, is a broad-spectrum antiviral agent effective against both RNA and DNA viruses (4–6), and it has been reported to be clinically beneficial in the treatment of several viral diseases (7–9). In the current COVID-19 situation, remdesivir (10) and molnupiravir (11) were originally developed to treat Ebola virus and Venezuelan equine encephalitis virus infections, respectively, and have been repurposed for SARS-CoV-2 treatment. Remdesivir is an adenosine analogue that exhibits antiviral activities against the Filoviridae, Paramyxoviridae, Pneumoviridae, Flaviviridae, and Coronaviridae families (12, 13). Molnupiravir is a cytidine analogue that exhibits antiviral activities against influenza virus (IFV), respiratory syncytial virus (RSV), and SARS-CoV-2 (14–16). The molecular target of these nucleoside analogues or their prodrugs is the viral RNA-dependent RNA polymerase (RdRp), and they inhibit viral RNA replication by inhibiting RdRp activity or by introducing a mutation into the viral genome during replication (17–19). RdRp is indispensable for the replication and transcription of viral genomes, and the core structural features of viral RdRps are functionally essential and conserved across a wide range of viruses (20–22). Thus, viral RdRp is a promising target for the development of broad-spectrum inhibitors of viruses.
Here, we report the identification of a broad-spectrum antiviral nucleoside analogue, 2-thiouridine (s2U), using phenotypic screening. Furthermore, we have successfully demonstrated the antiviral activity of s2U against several ssRNA+ viruses, including DENV serotype 2 (DENV2) and SARS-CoV-2, in vitro and in vivo.
Results
s2U Is a Broad-Spectrum Inhibitor of ssRNA+ Viruses.
We initially performed cell-based anti-DENV2 screening of 753 nucleoside analogues in a compound library from Hokkaido University. We identified a number of hit compounds and further confirmed their antiviral effects against all serotypes of DENV and flaviviruses, including Zika virus (ZIKV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and West Nile virus (WNV) (SI Appendix, Fig. S1A). We have clearly identified s2U (Fig. 1A) as a nucleoside analogue with strong antiviral activity against all eight tested flaviviruses (SI Appendix, Fig. S1B). The antiviral activity of s2U was markedly higher than that of ribavirin and favipiravir, which are known to have anti-flavivirus activity (23, 24) (SI Appendix, Fig. S1B and Table S1).
Fig. 1.
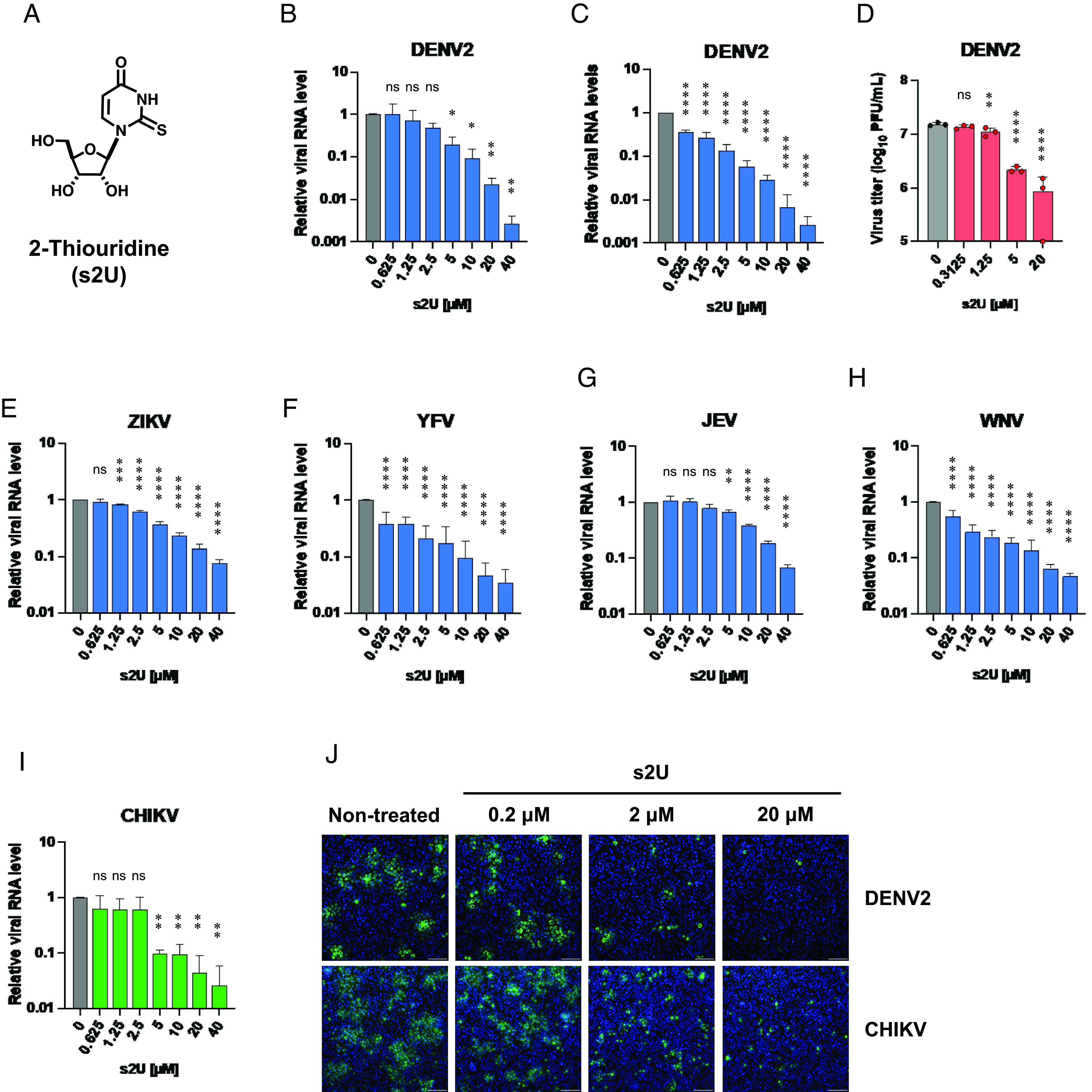
Broad-spectrum antiviral activities of s2U against Flaviviruses and CHIKV. (A) Chemical structure of 2-thiouridine (s2U). (B and C) Dose-response inhibition of DENV2 replication by s2U in VeroE6 (B) and Huh7 (C) cells. Cell lysates were collected for viral RNA determination, and viral RNA levels were determined relative to ACTB transcripts. (D) Dose-response inhibition of DENV2 propagation by s2U. Supernatants of DENV2-infected BHK-21 cells were collected at 72 hours post-infection (hpi), and dilutions were used to inoculate BHK-21 cells. Four days after inoculation, viral titers were determined by the plaque assay. (E–I) Dose-response inhibition of ZIKV (E), YFV (F), JEV (G), WNV (H), and CHIKV (I) by s2U. Cell lysates were collected for viral RNA determination, and viral RNA levels were determined relative to ACTB transcripts. (J) Dose-response inhibition of viral protein expression in the DENV2- and CHIKV- infected cells. Cells were stained with viral-specific antibodies (green, DENV2: Envelope protein, CHIKV: E1 protein) and counterstained with Hoechst 33342 nuclear dye (blue). (Scale bars indicate 200 μm.) Data are presented as mean values of biological triplicates from one of the experiments, and error bars indicate SD. Statistically significant differences were determined using a one-way ANOVA followed by Dunnett’s multiple comparisons test to compare with non-treated cells; *P < 0.01, **P < 0.005, ***P < 0.0005, and ****P < 0.0001.
Next, we performed cell-based antiviral assays (MTT assay or resazurin assay) using several RNA viruses to assess whether the antiviral activity of s2U was effective across a broad spectrum of viruses. s2U exhibited sub-micromolar to micromolar antiviral activity against ssRNA+ viruses, including viruses in the Flaviviridae, Togaviridae, and Coronaviridae families (Table 1). The cell-based assay revealed that the 50% cytotoxic concentration (CC50) value of s2U was high (e.g., >400 μM for BHK-21, VeroE6, and MRC5 cells) (SI Appendix, Fig. S1C). On the other hand, s2U did not show antiviral activity against negative-sense single-stranded RNA (ssRNA−) viruses, including rabies virus (RABV), Rift Valley fever virus (RVFV), and IFV (Table 1).
Table 1.
Antiviral activity of s2U against various RNA viruses (CPE-based assay)
| Virus | Strain | Cell | EC50 (μM)* | Virus | Strain | Cell | EC50 (μM)† |
|---|---|---|---|---|---|---|---|
| MTT/resazurin assay | |||||||
| DENV2 | D2/hu/INDIA/09-74 | BHK-21 | 1.5 ± 0.10 | RABV | HEP | BHK-21 | >100 |
| AG-P10 (Mouse-adapted) | BHK-21 | 1.3 ± 0.12 | LACV | ATCC VR-1834 | MDBK | >100 | |
| ZIKV | MR766 | BHK-21 | 3.8 ± 0.96 | LPHV | 11SB17 | KB | >100 |
| YFV | 17D-204 | BHK-21 | 3.2 ± 1.4 | LCMV | Armstrong | KB | >100 |
| JEV | Beijing-1 | VeroE6 | 7.6 ± 1.3 | JUNV | Candid #1 | 293T | >100 |
| WNV | NY99 | BHK-21 | 8.6 ± 1.8 | SFTSV | ArtLN/2017 | MDCK | >100 |
| VeroE6 | 4.0 ± 0.24 | RVFV | MP12 | MDCK | >100 | ||
| CHIKV | SL10571 | BHK-21 | 3.8 ± 0.12 | TPMV | VRC-66412 | VeroE6 | >100 |
| VeroE6 | 2.2 ± 0.23 | IAV H5N1 | A/Hong Kong/483/97 | A549 | >100 | ||
| HCoV | 229E | MRC5 | 0.94 ± 0.16 | IAV H7N9 | A/Anhui/1/2013 | MA104/TMPRSS2 | >100 |
| HCoV | OC43 | MRC5 | 4.8 ± 0.25 | Virus | Strain | Cell | EC50 (μM)* |
| SARS-CoV-2 | WK-521 (Ancestral) | VeroE6/TMPRSS2 | 2.3 ± 0.43 | SARS-CoV-2 | TY38-873 (Omicron BA.1) | VeroE6/TMPRSS2 | 2.0 ± 0.16 |
| QK002 (Alpha) | VeroE6/TMPRSS2 | 2.2 ± 0.56 | TY40-385 (Omicron BA.2) | VeroE6/TMPRSS2 | 1.8 ± 0.094 | ||
| TY8-612 (Beta) | VeroE6/TMPRSS2 | 2.2 ± 0.23 | TY41-703 (Omicron BA.4) | VeroE6/TMPRSS2 | 1.8 ± 0.013 | ||
| TY7-501 (Gamma) | VeroE6/TMPRSS2 | 1.8 ± 0.37 | TY41-702 (Omicron BA.5) | VeroE6/TMPRSS2 | 2.9 ± 0.18 | ||
| TY11-927 (Delta) | VeroE6/TMPRSS2 | 1.9 ± 0.22 | TY41-796 (Omicron BQ.1.1) | VeroE6/TMPRSS2 | 2.0 ± 0.44 | ||
| MA-P10 (Mouse-adapted) | VeroE6/TMPRSS2 | 2.5 ± 0.34 | TY41-795 (Omicron XBB.1) | VeroE6/TMPRSS2 | 1.9 ± 0.18 | ||
| Virus | Strain | Cell | EC50 (μM)* | Fold change‡ | |||
| Resazurin assay | |||||||
| rgDENV2 | Wild type | BHK-21 | 0.86 ± 0.12 | ||||
| rgDENV2 | NS5-G605V | BHK-21 | 5.1 ± 0.69 | 6.1 | |||
Antiviral assays were carried out as described in SI Appendix, Table S3. EC50: 50% effective concentration.
*EC50 values represent mean values ± SEM from at least three independently performed experiments (n = 2).
‡Fold change is calculated from the ratio of rgNS5-G605V/rgWT.
†EC50 values represent mean values from at least three independently performed experiments (n = 2).
qRT-PCR analysis confirmed that s2U inhibited the replication of the ssRNA+ viruses [DENV2, ZIKV, YFV, JEV, WNV, CHIKV, human coronavirus [HCoV]-229E, HCoV-OC43, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV)] in a dose-dependent manner (Figs. 1 B, C, E–I and 2 A–D, Tables 1 and 2, and SI Appendix, Fig. S2). However, s2U did not inhibit viral genome replication of ssRNA− viruses, such as RABV and RVFV, and the DNA virus herpes simplex virus-1 (HSV-1) (SI Appendix, Fig. S3 F–H and Table 2). Next, we performed immunofluorescence assays (IFA) and observed dose-dependent inhibition of viral protein expression in DENV2−, CHIKV−, and HCoV-OC43-infected cells (Fig. 1J and SI Appendix, Fig. S3A). Furthermore, we confirmed that s2U inhibited the progeny virus production of DENV2 in a dose-dependent manner (Fig. 1D).
Fig. 2.
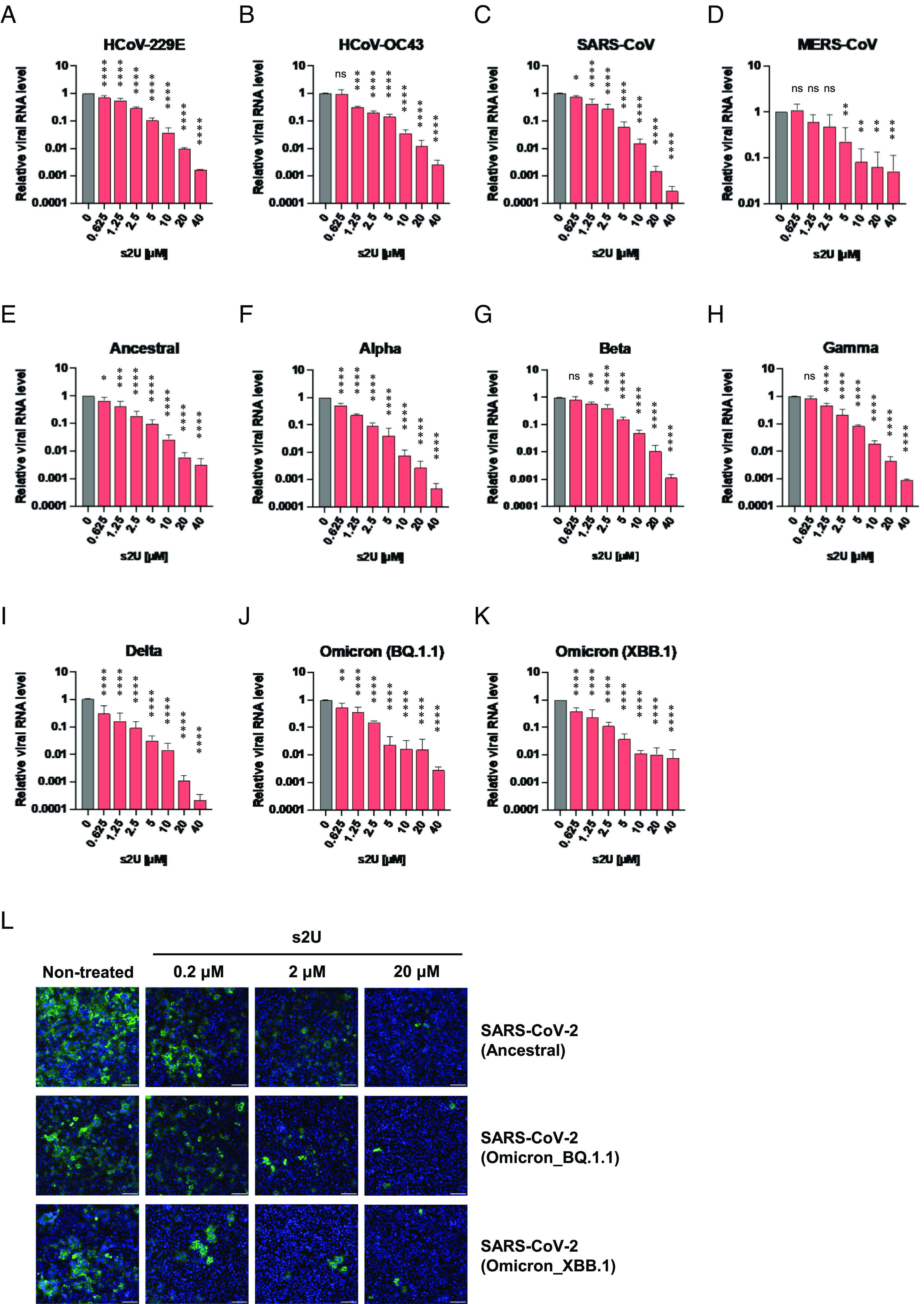
Broad-spectrum antiviral activities of s2U against human coronaviruses, including SARS-CoV-2. (A–K) Dose-response inhibition of HCoV-229E (A), HCoV-OC43 (B), SARS-CoV (C), MERS-CoV (D), and several SARS-CoV-2 variants (E–K) by s2U. Cell lysates were collected for viral RNA determination, and viral RNA levels were determined relative to ACTB transcripts. (L) Dose-response inhibition of viral protein expression in the SARS-CoV-2-infected cells. Cells were stained with viral-specific antibodies (green, Nucleocapsid) and counterstained with Hoechst 33342 nuclear dye (blue). (Scale bars indicate 200 μm.) Data are presented as mean values of biological triplicates from one of the experiments, and error bars indicate the SD. Statistically significant differences were determined using a one-way ANOVA followed by Dunnett’s multiple comparisons test to compare with non-treated cells; *P < 0.01, **P < 0.005, ***P < 0.0005, and ****P < 0.0001.
Table 2.
Antiviral activity of s2U against various RNA and DNA viruses (qPCR assay)
| Virus | Strain | Cell | EC50 (μM) | EC90 (μM) | Virus | Strain | Cell | EC50 (μM) | EC90 (μM) |
|---|---|---|---|---|---|---|---|---|---|
| qPCR Assay | |||||||||
| DENV2 | D2/hu/INDIA/09-74 | VeroE6 | 2.4 | 7.4 | SARS-CoV-2 | WK-521 (Ancestral) | VeroE6/TMPRSS2 | 1.0 | 4.3 |
| Huh7 | <0.63 | 3.4 | QK002 (Alpha) | VeroE6/TMPRSS2 | 0.64 | 2.3 | |||
| ZIKV | MR766 | VeroE6 | 3.7 | 23 | TY8-612 (Beta) | VeroE6/TMPRSS2 | 1.7 | 7.6 | |
| YFV | 17D-204 | VeroE6 | <0.63 | 11 | TY7-501 (Gamma) | VeroE6/TMPRSS2 | 1.3 | 3.6 | |
| JEV | Beijing-1 | VeroE6 | 7.5 | 29 | TY11-927 (Delta) | VeroE6/TMPRSS2 | <0.63 | 2.1 | |
| WNV | NY99 | VeroE6 | <0.63 | 10 | TY41-796 (Omicron_BQ.1.1) | VeroE6/TMPRSS2 | 0.74 | 3.6 | |
| CHIKV | SL10571 | VeroE6 | 1.7 | 13 | TY41-795 (Omicron_XBB.1) | VeroE6/TMPRSS2 | <0.63 | 2.7 | |
| HCoV | 229E | MRC5 | 1.3 | 6.2 | RABV | HEP | BHK-21 | >40 | >40 |
| HCoV | OC43 | VeroE6 | 1.1 | 4.2 | RVFV | MP12 | VeroE6 | >40 | >40 |
| SARS-CoV | Hanoi | VeroE6 | 1.2 | 4.5 | HSV-1 | F | VeroE6 | >40 | >40 |
| MERS-CoV | EMC2012 | VeroE6/TMPRSS2 | 2.3 | 8.2 | |||||
Antiviral assays were carried out as described in SI Appendix, Tables S4 and S5. EC50: 50% effective concentration. EC90: 90% effective concentration. EC50 and EC90 values represent mean values from a single experiment with biological triplicates (SI Appendix, Fig. S2).
Notably, s2U also inhibited viral RNA replication of the SARS-CoV-2 ancestral strain and each variant of concern (VOC), including the Delta variant (B.1.617.2 lineage) and Omicron subvariants (BA.1, BA.2, BA.4, BA.5, BQ.1.1, and XBB.1 lineages) in a dose-dependent manner (Fig. 2 E–K, Tables 1 and 2, and SI Appendix, Fig. S2). Viral protein expression and progeny virus production in SARS-CoV-2-infected cells were also inhibited by s2U in a dose-dependent manner (Fig. 2L and SI Appendix, Fig. S3 B–E).
s2U Blocks RNA Synthesis by Stalling of Viral RdRp.
We performed antiviral assays with simultaneous treatment with various doses of the four ribonucleosides (adenosine, guanosine, uridine, and cytidine) to confirm whether s2U acts as a nucleoside analogue. The antiviral activity of s2U was reduced following the addition of an excess of exogenous pyrimidine ribonucleosides (uridine and cytidine) to the infected cells but not purine ribonucleosides (adenosine and guanosine) (Fig. 3A and SI Appendix, Fig. S4A), suggesting that s2U inhibits viral RNA replication by acting as a pyrimidine ribonucleoside analogue.
Fig. 3.
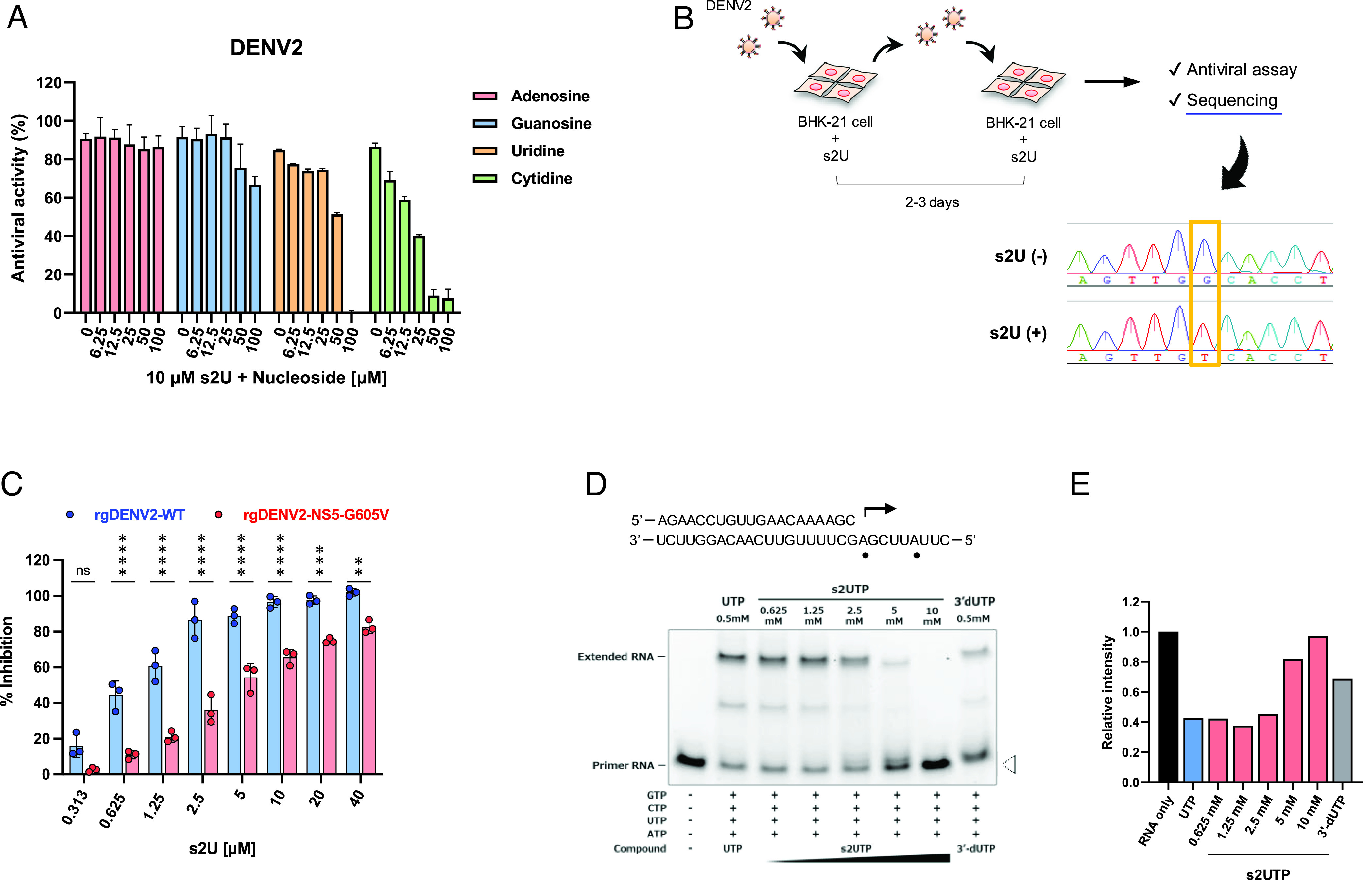
Molecular target and mechanism of action of s2U. (A) Ribonucleoside competition of DENV2 inhibition by s2U. DENV2 [multiplicity of infection (MOI) = 0.01]-infected BHK-21 cells were treated with 10 μM of s2U and serial dilutions of exogenous nucleosides. A resazurin reduction assay was performed at 4 days post-infection (dpi). (B) Schematic presentation of the experimental design for drug-escape mutant selection and sequence result. BHK-21 cells were infected with DENV2 in the presence of s2U. The passage of infected cells or culture supernatant was performed every 2 to 3 d. Base substitution was detected using Sanger sequencing. (C) Effect of s2U resistance mutation on anti-DENV2 activity of s2U. BHK-21 cells were infected with rgDENV2-WT or rgDENV2-NS5-G605V (MOI = 0.1) containing a serially diluted compound. A resazurin reduction assay was performed at 4 dpi. (D) Analysis of viral RdRp stalling by s2U 5′-triphosphate (s2UTP). Denaturing polyacrylamide gel electrophoresis fraction of RNA transcripts produced through primer extension by ZIKV RdRp in the presence of the indicated nucleotides. The RNA primer/template sequence used in this assay is indicated at the top (small black circles indicate the incorporation sites of UTP). (E) Relative band intensities of fluorescently labeled RNA primers. Relative fluorescence intensities of each RNA primer (white arrowhead in Fig. 3D) were normalized by the RNA sample without UTP or s2UTP (black bar, RNA only). Anti-DENV2 activities (%; A and C) are expressed relative to the values for the DMSO-treated, infected samples and non-infected samples. Data are presented as mean values, and error bars indicate SD. Statistically significant differences between wildtype and G605V viruses (C) were determined using a two-way ANOVA followed by Bonferroni’s multiple comparisons tests; *P < 0.01, **P < 0.005, ***P < 0.0005, and ****P < 0.0001.
To identify the molecular target of s2U, we selected a drug-escape mutant by passaging DENV2 in the presence of gradually increasing s2U concentrations. We observed an almost complete decrease in s2U susceptibility at passage 19 and identified a single base substitution (G1814T) within the viral non-structural protein 5 (NS5) in this escape mutant virus (Fig. 3B). This single mutation resulted in a single amino acid substitution (G605V) in the RdRp coding region, which was not observed in the in-parallel-passaged viruses in the absence of s2U.
Next, we constructed recombinant DENV2 possessing NS5-G605V (rgDENV2-WT and rgDENV2-NS5-G605V) using a reverse genetics system (25) to evaluate replication fitness and s2U resistance caused by the G605V mutation. The mutation affected viral replication in different cell lines (SI Appendix, Fig. S4 B and C). Although the G605V mutation slightly affected the sensitivity of ribavirin and favipiravir in the antiviral assay (2.3- and 2.2-fold decrease, respectively) (SI Appendix, Table S1), it reduced the sensitivity to s2U by 6.1-fold compared with recombinant rgDENV2-WT (Table 1 and Fig. 3C).
Nucleoside analogues can sometimes serve as substrates for the mitochondrial RNA polymerase (POLRMT), resulting in mitochondrial toxicity and other side effects (26). To assess the impact of s2U on POLRMT and RNA polymerase II (RNA pol II) activity, we performed mitochondrial protein synthesis assays using an In-Cell ELISA method (26). s2U did not affect the steady-state levels of mitochondrial-encoded [cytochrome c oxidase I (COX-I)] and nuclear-encoded [succinate dehydrogenase (SDH)-A] proteins, suggesting that POLRMT and RNA pol II activities were unaffected (SI Appendix, Fig. S5).
To characterize the molecular mechanism of action of s2U, we performed an in vitro primer extension assay (27) using ZIKV NS5 protein and s2UTP. Although UTP was incorporated into the RNA template and extended this, the incorporation of s2UTP instead of UTP might likely block RNA extension in the presence of the next correct ribonucleotide (Fig. 3 D and E). These results suggest that s2UTP acts on viral RdRp and inhibits viral RNA synthesis.
In Vivo Efficacy of s2U against DENV2.
Subsequently, we employed an animal model to evaluate the in vivo antiviral activity of s2U. A mouse-adapted DENV2 model has been established using AG129 [interferon (IFN)-α/β and IFN-γ receptor-deficient 129/Sv] mice (28), and this model has been utilized to develop of anti-DENV drugs and vaccines (29–31). Thus, we established a mouse-adapted DENV2 strain (DENV2 AG-P10) based on this model (28) to evaluate the in vivo efficacy of the compound. This mouse-adapted DENV2 strain had two amino acid substitutions (NS4B-A119T and NS5-E802Q) that developed during viral passages and demonstrated higher pathogenicity in AG129 mice compared to the parental clinical isolate (SI Appendix, Fig. S6).
AG129 mice were intraperitoneally inoculated with DENV2 AG-P10 and treated twice daily with s2U [50 or 150 mg/kg of body weight (BW)] by oral gavage starting immediately after infection and also at 8- or 24- h after infection (Fig. 4A). After five consecutive days of treatment, the survival rate of virus-inoculated AG129 mice was significantly increased in the mice that received the s2U treatment starting immediately after infection (median survival: 12.5 d at 50 mg/kg and >16 d at 150 mg/kg) compared with the vehicle-treated mice (median survival: 9 d) (Fig. 4B). The survival rate of virus-inoculated AG129 mice was also significantly increased in the mice that received the s2U treatment (150 mg/kg) starting 8- or 24- h after infection [median survival: 9 d (vehicle), 12 d (8 h delay), and 13 d (24 h delay)] (Fig. 4C). A dose-dependent decrease in viral RNA load at 3 days post-infection (dpi) was also observed in serum, spleen, kidney, and liver samples (Fig. 4D and SI Appendix, Fig. S8 A–D). These data suggest that s2U protects against DENV2-induced mortality by decreasing viral propagation in vivo in AG129 mice.
Fig. 4.
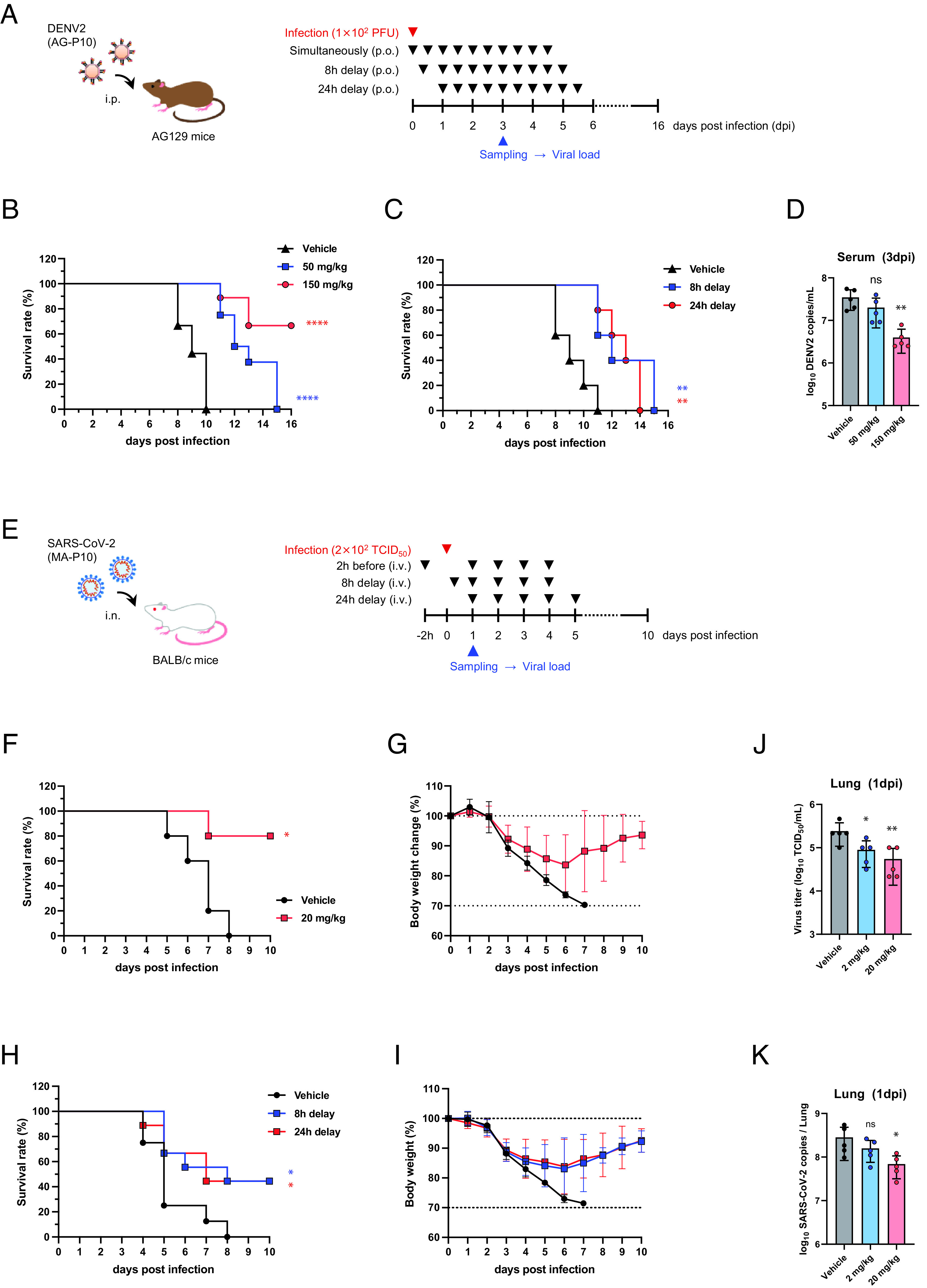
In vivo efficacy of s2U in the DENV2 and SARS-CoV-2 mouse model. (A) Schematic representation of the survival and viremia studies using AG129 mice and strain DENV2 AG-P10. (B) Effect of s2U on survival of DENV2 AG-P10-infected (1 × 102 plaque-forming units (PFU)] mice (7-wk-old, female) orally treated twice daily with s2U (50 or 150 mg/kg) compared with vehicle-treated mice. Treatment started immediately after infection. Data are from two independent studies with 9 (in total, vehicle and 150 mg/kg) or 8 (in total, 50 mg/kg) mice per group. (C) Effect of s2U on survival of DENV2 AG-P10-infected (1 × 102 PFU) mice (7-wk-old, female) orally treated twice daily with s2U (150 mg/kg) compared with vehicle-treated mice. Treatment started 8 or 24 h after infection (n = 5 per group). (D) Effect of s2U on viremia at 3 dpi in mice treated twice daily with s2U (50 or 150 mg/kg) compared with vehicle-treated mice (n = 5 per group, 7-wk-old, male). Viral RNA copies/mL of serum samples were quantified using qRT-PCR. (E) Schematic representation of the survival and viremia studies using BALB/c mice and SARS-CoV-2 MA-P10. (F–I) Effect of s2U on survival and body weight change in SARS-CoV-2 MA-P10-infected [2 × 102 50% tissue culture infection dose (TCID50)] mice intravenously administered 20 mg/kg s2U daily compared with vehicle-treated mice [n = 5 (F and G), n = 8 (H and I, vehicle), and n = 9 (H and I, s2U) per group, 30- to 50-wk-old, female]. Treatment was started 2 h before (F) and 8 or 24 h after (H) infection. Survival (F and H) and body weight (G and I) of the mice were monitored daily. (J and K) Effect of s2U on viremia at 1 dpi in mice intravenously administered 2 or 20 mg/kg s2U daily compared with vehicle-treated mice (n = 5 per group, 5-wk-old, female). Virus titers in lung samples (J) were quantified by a standard TCID50 assay using VeroE6/TMPRSS2 cells. Viral RNA copies/mL in lung samples (K) were quantified using qRT-PCR. Data are presented as mean values, and error bars indicate SD. Statistically significant differences between the s2U-treated and vehicle-treated groups were determined using a log-rank (Mantel–Cox) test (B, F, and H) and one-way ANOVA followed by Dunnett’s multiple comparisons tests (D, J, and K); *P < 0.01, **P < 0.005, and ****P < 0.0001.
In Vivo Efficacy of s2U against SARS-CoV-2.
SARS-CoV-2 (Ancestral strain) does not show infectivity and pathogenicity in normal mice. Thus, human angiotensin-converting enzyme 2 transgenic mice are widely used as a small-animal model of SARS-CoV-2 infection (32, 33). A mouse-adapted SARS-CoV-2 model was recently established, which has promoted research on SARS-CoV-2 pathogenicity and the development of drugs and vaccines against COVID-19 (34, 35). Therefore, to evaluate the in vivo efficacy of s2U, we also established a mouse-adapted SARS-CoV-2 strain (SARS-CoV-2 MA-P10) by following previous reports (36, 37) (SI Appendix, Fig. S7 A and B). The resultant mouse-adapted virus harbors a G498H substitution in the spike protein and can efficiently replicate in normal mice (SI Appendix, Fig. S7 C–G). These results were consistent with reports of other mouse models (36–39).
The effect of s2U on virus-induced mortality was assessed by daily intravenous administration (20 mg/kg) for five consecutive days starting 2 h before infection and 8 or 24 h after infection (Fig. 4E). BALB/c mice (30 to 50 wk old) were intranasally infected and then monitored for a maximum of 10 d. We observed a statistically significant increase in survival rate (80% survival) after 5 d of s2U treatment compared with the vehicle-treated mice (Fig. 4F). The peak BW loss in mice treated with s2U was approximately 17%, and the BW was recovered following the administration of s2U, while the BW of vehicle-treated mice continued to decrease daily (Fig. 4G). The survival rate of virus-inoculated mice was also significantly increased (44% survival) in the mice that received the s2U treatment (20 mg/kg) starting 8 or 24 h after infection (Fig. 4H). The peak BW loss in mice treated with s2U was approximately 17%, and the BW was also recovered following the administration of s2U (Fig. 4I).
The mouse-adapted SARS-CoV-2 replicates rapidly, and the viral load in the lungs peaks at 1 dpi (SI Appendix, Fig. S7 D and E) (37). Therefore, we next assessed the effect of s2U on the viral load at 1 dpi following intravenous administration (2 or 20 mg/kg) to 5-wk-old infected BALB/c mice. A dose-dependent decrease in viral titer and viral RNA load was observed in the lungs of s2U-treated mice compared with vehicle-treated mice (Fig. 4 J and K). We also observed decreased viral titers in the lungs following twice-daily oral administration of s2U (300 mg/kg) starting immediately after infection (SI Appendix, Fig. S8 I and J). Finally, we detected lower levels of several pro-inflammatory cytokines (IFN-β, IL-6, and CXCL10) in lung homogenates prepared from mice treated with s2U compared with those of vehicle-treated mice (SI Appendix, Fig. S8 E–H). These data suggest that s2U protects against SARS-CoV-2-induced lung inflammation and mortality by decreasing viral propagation.
In Vivo Pharmacokinetic Analysis of s2U.
Five-week-old female BALB/c mice were treated with s2U using oral (150 mg/kg) or intravenous (20 mg/kg) administration. Plasma samples were analyzed by LC-MS/MS, and the standard non-compartmental analysis was conducted to determine the pharmacokinetic parameters. The simulation of the repeated dose concentration-time profiles by nonparametric superposition showed that once-daily intravenous and twice-daily oral administration of s2U maintains the validated concentration of more than 50% effective concentration (EC50) or 90% effective concentration (EC90) against DENV2 and SARS-CoV-2 (SI Appendix, Fig. S10).
Discussion
This study identified the antiviral ribonucleoside analogue s2U as a strong inhibitor of viral replication of different clinically relevant ssRNA+ virus families, including Flaviviridae, Togaviridae, and Coronaviridae, without significant cytotoxicity. Additionally, Alam and colleagues reported that s2U inhibits the replication of murine norovirus-1 (MNV-1) (40), which belongs to the Caliciviridae family. Collectively, our findings indicate that s2U inhibits the replication of ssRNA+ viruses but not ssRNA− and DNA viruses. These findings suggest that the broad-spectrum antiviral activity of s2U against ssRNA+ viruses is due to a unique mechanism of action.
Our studies demonstrated an s2U-escape mutant with a G605V substitution in the NS5 protein of the DENV2 D2/hu/INDIA/09-74 strain, which is located at motif B of DENV2 RdRp. Moreover, s2UTP inhibited RNA extension catalyzed by the ZIKV RdRp. s2UTP has been reported to be incorporated into the RNA template of feline calicivirus and human norovirus by ProPol (a precursor comprised of both the proteinase and polymerase), resulting in the inhibition of norovirus and calicivirus polymerase activity, respectively (41). Another study provided structural evidence of s2U binding to MNV-1 RdRp and showed that s2U interacted with Thr309 and other amino acids that form the active site pocket (40). Thr309 is positioned in a motif B of MNV-1 RdRp, which corresponds to the same region including Gly605 of the DENV2 RdRp (42) and Ala688 of the SARS-CoV-2 RdRp (43) (SI Appendix, Fig. S9). These findings suggest that the molecular target of s2U is the active site of ssRNA+ viral RdRp. In addition, with regard to two catalytic aspartic acids, Asp346 and Asp250, while Asp346 is well conserved among a wide range of viruses, Asp250 is not conserved in ssRNA− viruses, which may also be the only reason for s2U efficacy solely for ssRNA+ viruses. Sofosbuvir (SOF) (44) and 4′-fluorouridine (4′-FIU) (45) are potent uridine analogues against hepatitis C virus and RSV, respectively. 4′-FIU also inhibits SARS-CoV-2 replication in vitro and in vivo (45). SOF and 4′-FIU 5′-triphosphates (TP) are incorporated into the RNA template by mimicking UTP, inducing stalling of viral RdRp (45, 46). SOF-TP is also incorporated into the RNA template by ZIKV RdRp, resulting in inhibition of viral RNA synthesis (27). Our study demonstrated s2U as another uridine analogue, and s2UTP induced stalling of ZIKV RdRp. These findings suggest that s2U inhibits viral RNA synthesis by blocking the activity of the viral RdRp, while further investigation including a comprehensive in vitro RNA extension assay and s2U complex structure of DENV and/or SARS-CoV-2 RdRp is necessary to conclude the detailed mechanism of action of s2U.
Once-daily intravenous and twice-daily oral administration of s2U to mice significantly reduced the viral load of DENV2 and SARS-CoV-2, respectively. Moreover, continuous administration of s2U to mice significantly increased the survival rate of both DENV2- and SARS-CoV-2-infected mice at prophylactic and therapeutic time points. In our pharmacokinetic analysis, simulation of the repeated dose concentration-time profile revealed that once-daily intravenous and twice-daily oral administration of s2U maintains the effective concentration against DENV2 and SARS-CoV-2 (SI Appendix, Fig. S10). s2U exhibited broad anti-coronaviral efficacy with equally strong activity against SARS-CoV-2 VOCs in vitro, including the currently circulating Omicron variants. Thus, we can conclude that the s2U should retain its efficacy against future variants of SARS-CoV-2 which may have substantial resistance to spike-targeting antibody therapeutics or vaccines. Furthermore, s2U also exhibited broad anti-flavivirus efficacy with equally strong activity in vitro. These findings suggest the high potential of s2U as an efficacious broad-spectrum oral and/or intravenous agent against ssRNA+ viruses, making it a promising therapeutic option for Dengue, COVID-19, and other diseases caused by different ssRNA+ viruses.
With regard to the safety (toxicity and mutagenicity) of s2U, we have performed the in vivo toxicity assays using BALB/c mice (SI Appendix, Fig. S11). This revealed that no BW loss was observed when mice were administrated at high concentrations of s2U via oral (300, 150 mg/kg) and intravenous (50, 25 mg/kg) routes. We also performed the following two sets of experiments, with regard to mutagenicity. First, we could not obtain any mutation in other viral proteins during the isolation of the drug escape mutant. Second, the mutagenicity tests [AMES (Bacterial reverse mutation test) and HPRT (in vitro gene mutation test using the Hprt genes in V79 cells)] showed that s2U does not have sufficient ability to introduce mutations (SI Appendix, Table. S2). These data clearly suggest that s2U does not work as a mutagen.
In conclusion, we have demonstrated that s2U exhibits strong and safe, broad-spectrum antiviral activity against ssRNA+ viruses by inhibiting viral RdRp activity. Viral RdRp represents a promising target for the development of broad-spectrum inhibitors because it is functionally and structurally conserved among a wide range of viruses. Thus, the use of s2U (and s2U derivatives) could be extended for the development of new drugs against related or novel viruses, and these compounds may possibly contribute to our progress in the area of pandemic preparedness.
Materials and Methods
Cell Lines.
BHK-21 (ATCC, CCL-10), VeroE6 (ATCC, CRL-1586), MA104 (RIKEN BRC, RCB0994), 293T (RIKEN BRC, RCB2202), HepG2 (ATCC, HB-8065), Huh7 (RIKEN BRC, RBRC-RCB1942), A549 (RIKEN BRC, RCB0098), KB (ATCC, CCL17), MDBK (ATCC, CCL-22), and MDCK (ATCC, CCL-34) cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and penicillin–streptomycin (P/S, Wako) at 37 °C. MRC5 (ATCC, CCL-171) cells were also maintained in Minimum Essential Medium GlutaMAX Supplement (Gibco) supplemented with 10% FBS, nonessential amino acids (NEAA, Wako), sodium pyruvate (Wako), and P/S at 37 °C. THP-1 (ATCC, TIB-202) and MOLT4 (JCRB, JCRB9031) cells were maintained in Roswell Park Memorial Institute 1640 (RPMI-1640, Gibco) supplemented with 10% FBS and P/S at 37 °C. C6/36 (ATCC, CRL-1660) cells were maintained in Minimum Essential Medium (MEM, Nissui) supplemented with 10% FBS, NEAA, and L-Alanyl-L-glutamine (Wako) at 28 °C.
Generation of TMPRSS2- and ACE2-Expressing Cells.
VeroE6 cells stably expressing human TMPRSS2 (VeroE6/TMPRSS2) were generated by lentiviral transduction with CSII-CMV-TMPRSS2-IRES2-Bsd and blasticidin-based selection as described in a previous report (47). MA104 cells stably expressing human TMPRSS2 (MA104/TMPRSS2) were also generated using the same method. VeroE6 and A549 cells stably expressing human ACE2 and TMPRSS2 (VeroE6/ACE2/TMPRSS2 and A549/ACE2/TMPRSS2) were generated by lentiviral transduction with pLVSIN-CMV-ACE2-Puro and puromycin-based selection as described in a previous report (48, 49). For the lentiviral vector preparation, 293T cells were co-transfected with the aforementioned lentiviral vector plasmid and Lentiviral High Titer Packaging Mix (Takara Bio).
Viruses.
DENV1 (D1/hu/PHL/10–07 strain), DENV2 (D2/hu/INDIA/09–74 strain, GenBank: LC367234), DENV3 (D3/hu/Thailand/00–40 strain), and DENV4 (D4/hu/Solomon/09–11 strain) were amplified in C6/36 cells.
ZIKV (MR766 strain, GenBank: LC002520), YFV (17D–204 strain), JEV (Beijing–1 strain), and WNV (NY99 strain) were amplified in BHK-21 cells.
CHIKV strain SL10571 was propagated as described in a previous report (50).
Human coronavirus strain OC43 (HCoV-OC43) and 229E (HCoV-229E) were purchased from ATCC (VR-1558 and VR-740, respectively) and amplified as described in a previous report (51).
SARS-CoV strain Hanoi was kindly provided by Nagasaki University and was amplified in VeroE6 cells (52).
MERS-CoV strain EMC2012 was kindly provided by Erasmus University Medical Center and amplified in VeroE6/TMPRSS2 cells (53).
SARS-CoV-2 strain WK-521 (Ancestral, Pango Lineage: A, GISAID: EPI_ISL_408667), QK002 (Alpha, Pango Lineage: B.1.1.7, GISAID: EPI_ISL_768526), TY8-612 (Beta, Pango Lineage: B.1.351, GISAID: EPI_ISL_1123289), TY7-501 (Gamma, Pango Lineage: P.1, GISAID: EPI_ISL_833366), TY11-927 (Delta, Pango Lineage: B.1.617.2, GISAID: EPI_ISL_2158617), TY38-873 (Omicron, Pango Lineage: BA.1, GISAID: EPI_ISL_7418017), TY40-873 (Omicron, Pango Lineage: BA.2, GISAID: EPI_ISL_9595859), TY41-703 (Omicron, Pango Lineage: BA.4, GISAID: EPI_ISL_13278440), TY41-702 (Omicron, Pango Lineage: BA.5, GISAID: EPI_ISL_13241867), TY41-796 (Omicron, Pango Lineage: BQ.1.1, GISAID: EPI_ISL_15579783), and TY41-795 (Omicron, Pango Lineage: XBB.1, GISAID: EPI_ISL_15669344), a clinical isolate from a patient with COVID-19, were kindly provided by National Institute of Infectious Diseases (NIID); the original stock of these virus strains were prepared by inoculation of VeroE6/TMPRSS2 cells.
RABV high egg passage Flury (HEP) strain was propagated as described in the previous report (54).
La Cross virus (LACV) was purchased from ATCC (VR-1834) and amplified on BHK-21 cells.
Leopards Hill virus (LPHV, 11SB17 strain) was amplified in KB cells as previously described (55).
Lymphocytic choriomeningitis virus (LCMV, Armstrong strain) and Junin virus (JUNV, Candid #1 strain) were kindly provided by NIID and were amplified in BHK-21 cells.
Severe fever with thrombocytopenia syndrome virus (SFTSV, ArtLN/2017 strain) was propagated as described in a previous report (56).
Recombinant Rift Valley fever virus (RVFV, MP12 strain) was rescued as described previously (57) and was amplified in BHK-21 cells.
Thottapalayam thottimvirus (TPMV, VRC-66412 strain) was kindly provided by Hokkaido University and was amplified in VeroE6 cells (58).
Avian influenza A virus H5N1 (IAV-H5N1, A/Hong Kong/483/97 strain) was kindly provided by University of Hong Kong and was propagated in embryonated chicken eggs and harvested from virus-containing allantoic fluids.
Avian influenza A virus H7N9 (IAV-H7N9, A/Anhui/1/2013 strain) was kindly provided by NIID and was propagated in embryonated chicken eggs and harvested from virus-containing allantoic fluids.
Herpes simplex virus (HSV-1, F strain) was kindly provided by NIID and was amplified in VeroE6 cells.
WNV, CHIKV, SARS-CoV, MERS-CoV, SARS-CoV-2, SFTSV, RVFV, IAV-H5N1, and IAV-H7N9 were propagated in a biosafety level-3 (BSL-3) facility at the International Institute for Zoonosis Control, Hokkaido University.
Compounds.
All compounds that were screened, including 2-thiouridine, were synthesized at the Faculty of Pharmaceutical Sciences, Hokkaido University, and their chemical identity and purity were determined by high-performance liquid chromatography and mass spectrometry analysis. Ribavirin, Favipiravir, GS-5734 (Remdesivir), GS-441524, 2-thiouridine, and Chloramphenicol were purchased from Sigma-Aldrich, PharmaBlock Sciences, Inc., MedChemExpress, Carbosynth Limited, Cayman Chemical Company, and Calbiochem, respectively. For in vitro studies, all compounds were solubilized in 100% dimethyl sulfoxide (DMSO; Sigma-Aldrich) and were diluted in 2% FBS/MEM. For in vivo studies, 2-thiouridine was dissolved in DMSO and diluted with 0.5% methylcellulose (MC) aqueous solution to prepare 50 or 150 mg/mL solutions for oral administration. For intravenous administration, 2-thiouridine was also dissolved in OTSUKA NORMAL SALINE (Otsuka Pharmaceutical Co., Ltd.).
Cell-Based Antiviral and Cytotoxicity Assays.
The MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) and resazurin reduction assays were carried out as previously described (25, 51, 59). These were performed to calculate cell viability following viral-induced cytopathic effects (CPE) and cytotoxicity. Assay conditions for all viruses are as described in SI Appendix, Table S3. Cells and viruses were incubated in 96-well plates with the twofold serially diluted compound (n = 2) in all assays. The EC50 value was defined in GraphPad Prism versions 8.4.3 and 9.5.1 (GraphPad Software) with a variable slope (four parameters). Non-infected cells were used as a control for 100% inhibition, whereas for infected cells, DMSO alone was used as a control for 0% inhibition. The CC50 value for each cell line was also measured using the CellTiter-Glo (Plomega) or resazurin reduction assays. Cell-free samples were used as 100% cytotoxicity control, and DMSO-treated cells were used as 0% cytotoxicity control.
Quantification of Viral RNA with qRT–PCR.
Assay conditions for all viruses are described in SI Appendix, Table S4. Briefly, cells were seeded onto 48-well plates the previous day and infected with the virus containing the serially diluted compound. After a certain period of time, total RNA was isolated with PureLink RNA Mini Kit (Ambion; Thermo Fischer Scientific). Viral RNA from all samples was quantified using real-time RT-PCR analysis with EXPRESS One-step Superscript qRT-PCR kit (Invitrogen; Thermo Fischer Scientific) and QuantStudio 7 Flex Real-Time PCR system (Applied Biosystems; Thermo Fischer Scientific). The primers and probe sequences were designed in previous reports and are described in SI Appendix, Table S5, with primers and probe for ACTB (Hs01060665_g1, Applied Biosystems) and 18S rRNA (Hs99999901_s1, Applied Biosystems) transcripts used as internal controls. The EC90 value was defined in GraphPad Prism versions 8.4.3 and 9.5.1 with a variable slope (Find ECanything, F = 90).
Quantification of Viral DNA with qPCR.
VeroE6 cells were seeded the previous day and infected with HSV-1 at a MOI of 0.1 containing the serially diluted compound. At 24 hpi, total DNA was isolated with DNeasy Blood & Tissue Kit (Qiagen). Viral DNA from samples was quantified using real-time PCR analysis with TaqMan Fast Advanced Master Mix (Applied Biosystems) and QuantStudio 7 Flex Real-Time PCR system. The primers and probe sequences were designed in previous reports and are described in SI Appendix, Table S5, with primers and probe for ACTB transcripts used as internal controls.
Indirect IFA.
Assay conditions for all viruses are as described in SI Appendix, Table S4. Briefly, cells were seeded onto 48-well plates the previous day and infected with the virus containing the serially diluted compound. After a certain period of time, cells were fixed with the buffered formalin (Masked Form A, Japan Tanner Co.), permeabilized with 0.5% Triton X-100 in PBS or ice-cold methanol. Cells were then stained with the 4G2 (supernatant from D1-4G2-4-15 cells, ATCC HB-122), Anti-Chikungunya virus E1 mAb, clone 1B6 (MAB12424, Abnova), anti-coronavirus antibody (MAB9013, Merck Millipore), SARS-CoV-2 nucleocapsid antibody (HL344; GTX635679, GeneTex), and Alexa Fluor Plus 488-conjugated anti-mouse or anti-rabbit IgG antibody (Invitrogen). Cell nuclei were counterstained with Hoechst 33342 (Molecular Probes). Cells were then evaluated using fluorescence microscopy (IX73, Olympus). Images were processed with cellSens Standard 1.16 (Olympus).
Plaque Assays.
For DENV2, BHK-21 cells were inoculated with DENV2 at an MOI of 0.01. After 1 h of incubation, the cells were washed and fed with fresh culture medium containing the serially diluted compound. The culture supernatants were harvested at 72 hpi. Monolayers of BHK-21 cells were inoculated with serial dilutions of supernatants for 1 h at 37 °C. The cells were then overlaid with MEM containing 2% FBS, and 1% Methylcellulose. At 4 dpi, cells were fixed with buffered formalin and stained with 1% crystal violet.
For SARS-CoV-2, VeroE6/TMPRSS2 (Ancestral) and VeroE6/ACE2/TMPRSS2 (Omicron_XBB.1), and A549/ACE2/TMPRSS2 (Ancestral and Omicron_XBB.1) cells were inoculated with SARS-CoV-2 at an MOI of 0.01. After 1 h of incubation, the cells were washed and fed with fresh culture medium containing the serially diluted compound. The culture supernatants were harvested at 24 hpi from VeroE6/TMPRSS2 cells and 48 hpi from other cells. Monolayers of VeroE6/ACE2/TMPRSS2 cells were inoculated with serial dilutions of supernatants for 1 h at 37 °C. The cells were then overlaid with DMEM containing 2% FBS, 0.5% Bacto Agar (Becton Dickinson), and gentamicin (25 μg/mL, Wako). At 2 dpi, cells were fixed with buffered formalin and stained with 1% crystal violet.
Ribonucleoside Competition of DENV2 and HCoV Inhibition.
BHK-21 or MRC5 cells were infected with DENV2 (MOI = 0.01) or HCoV-229E (MOI = 0.005), and assay media was supplemented with s2U at 10 or 15 μM alone or in combination with 6.25 to 100 μM exogenous ribonucleosides (Adenosine, Guanosine, Uridine, Cytidine, Sigma-Aldrich). Resazurin reduction assays were carried out at 96 or 72 hpi, respectively. Antiviral activities (%) are expressed relative to the values for the DMSO-treated, infected samples and non-infected samples.
Isolation of Drug-Escape Mutants.
BHK-21 cells were seeded onto 12-well plates and were infected with DENV2 at an MOI of 0.01 containing 2 μM or 6 μM of s2U. The passage of infected cells was performed every 2 to 3 d. When a CPE was observed, culture supernatants were transferred to non-infected BHK-21 cells in the presence of the compound. After continuous culture of the viruses for 42 d (passage 19), the viral RNA was isolated and amplified by RT-PCR using the PrimeScript II High Fidelity One Step RT-PCR Kit (Takara Bio) and specific primers. Viral genomes were then examined by Sanger sequencing using specific primers and the 3500xL Genetic Analyzer (Life Technologies). The sequence was compared to that of wild-type virus.
Construction of Recombinant and Mutant DENV2 Infectious Clones.
Recombinant DENV2 (D2/hu/INDIA/09-74, rgDENV2-WT) was rescued and amplified as described previously (25). A point mutation fragment was amplified using PCR-based site-directed mutagenesis method and inserted into a plasmid containing the whole DENV2 genome. The T3 promoter and DENV2 genomic region of the plasmid was amplified by PCR using KOD One PCR Master Mix -Blue- (Toyobo), and DENV2 genomic RNA was synthesized using a mMessage mMachine T3 Kit (Thermo Fisher Scientific). The genomic RNA was transfected into BHK-21 cells with TransIT-mRNA reagent (Mirus), and culture supernatants were collected when CPEs were observed.
Primer Extension Polymerase Activity Assay.
The primer extension assay was performed according to a previous report (27, 51). For analysis of the competitive inhibition ability of s2UTP, 200 nM of recombinant ZIKV NS5 (40546-V08B, Sino Biological) and 50 nM RNA primer-template complexes (RNA P/T) were incubated 30 °C for 15 min in reaction buffer [10 mM Tris-HCl (pH 7.5), 10 mM DTT, 5 mM MgCl2, 5% glycerol, 0.05% Triton-X 100, and 0.02 U/μL RNasein (Promega)], and then UTP analogues [500 μM UTP (Thermo Fischer Scientific), 500 μM 3′-dUTP (TriLink), or serially diluted s2UTP (0.625 to 10 mM)] were added and incubated at 30 °C for 10 min. Primer extension reactions were initiated by the addition of ATP, GTP, CTP, and UTP (100 μM each). The reactions were performed at 30 °C for 2 h and stopped by the addition of quenching buffer (7M Urea, 1× TBE buffer, and 50 mM EDTA). The quenched samples were denatured at 95 °C for 5 min, and the primer extension products were separated on a 15% denaturing polyacrylamide gel (Invitrogen). After electrophoresis, the gels were scanned using an Amersham ImageQuant 800 Fluor system (Cytiva). The band intensities were analyzed by ImageQuant TL version 8.2.0 (Cytiva).
Ethical Statement.
All the animal experiments were performed in accordance with the National University Corporation, Hokkaido University Regulations on Animal Experimentation (https://www.global.hokudai.ac.jp/rules-and-regulations-for-check/). The protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Hokkaido University (approval no. 18-0046, 18-0149, and 20-0060).
Determination of Antiviral Activity In Vivo.
In the DENV2 infection model, SPF, sex-matched 7-wk-old AG129 mice were inoculated intraperitoneally with 100 μL of 2 × 102 PFU/mouse of DENV2 AG-P10 strain (as described in SI Appendix, SI Text). After inoculation, mice were treated with s2U twice daily by oral administration (50 or 150 mg per kg) for five consecutive days. Mice were monitored daily and body weight was also determined daily. To evaluate the viral load, the infected mice were euthanized at 3 dpi, and serum, spleen, kidney, and liver were collected and homogenized in PBS with TissueRuptor.
In SARS-CoV-2 infection model, SPF, 5-wk-old (for viral load) or 30 to 50-wk-old (for survival study) female BALB/c mice were inoculated intranasally with 50 μL of 2 × 102 TCID50/mouse of SARS-CoV-2 MA-P10 strain (as described in SI Appendix, SI Text) under anesthesia. Mice were treated with s2U by intravenous (2 or 20 mg/kg, once daily) or oral (300 mg/kg, twice daily) administration. Treatment was continued for five consecutive days for the survival study. On day 1 after inoculation, the infected mice were euthanized, and whole lung tissues were harvested and homogenized in PBS with a TissueRuptor. The homogenates were centrifuged for 10 min at 3,000 rpm to pellet tissue debris and the supernatants were subjected to standard TCID50 assay using VeroE6/TMPRSS2 cells for virus titration.
Viral RNA isolation from serum or tissue samples was performed using QIAamp Viral RNA Mini Kit (Qiagen) or PureLink RNA Mini Kit, respectively. Viral RNA level was quantified by qRT-PCR analysis as described above with 18S rRNA transcripts used as internal controls.
Statistical Analysis.
One-way ANOVA followed by Dunnett’s multiple comparisons tests, two-way ANOVA followed by Bonferroni’s multiple comparisons tests, or the unpaired t-test for the in vitro and in vivo antiviral assay and the log–rank (Mantel–Cox) test for the in vivo survival test was performed to determine the statistical significance using GraphPad Prism versions 8.4.3 and 9.5.1.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Tomohiko Takasaki (Kanagawa Prefectural Institute of Public Health, Japan) for providing DENV1-4 and CHIKV, Dr. Koichi Morita (Nagasaki University, Japan) for providing SARS-CoV Hanoi strain, Dr. Bart Haagmans (Erasmus University Medical Center, Netherlands) for providing MERS-CoV EMC2012 strain, Drs. Masayuki Saijyo, Masayuki Shimojima, Mutsuyo Ito, and Ken Maeda [National Institute of Infectious Diseases (NIID), Japan] for providing SARS-CoV-2 variants, Dr. Chang-Kweng Lim (NIID, Japan) for providing RABV HEP strain, Dr. Masayuki Saijo (NIID, Japan) for providing LCMV Armstrong strain, Dr. Akihiro Ishii (Hokkaido University, Japan) for providing LPHV 11SB17 strain, Dr. Shigeru Morikawa (NIID) for providing Junin virus Candid #1 strain, Dr. Shinji Makino (University of Texas Medical Branch, USA) for providing RVFV plasmid, Dr. Karl-Klaus Conzelmann (Max von Pettenkofer-Institute, Germany) for providing BSR-T7/5 cells, Dr. Kumiko Yoshimatsu (Hokkaido University, Japan) for providing TPMV VRC-66412 strain, Dr. Kennedy F. Shortridge (University of Hong Kong, Hong Kong) for providing IAV-H5N1 A/Hong Kong/483/97 strain, and Dr. Hiroyuki Miyoshi (RIKEN BRC, Japan) for providing lentiviral vector plasmid CSII-CMV-MCS-IRES2-Bsd. We also thank Takao Sanaki, Yuki Maruyama, Masaaki Izawa, Takao Shishido, Akira Naito, Haruka Maeda, Etsuko Hayashi, Mai Kishimoto, Yukari Itakura, Satoko Otsuguro, and Tatsuya Zenko for their excellent assistance. We would like to thank Enago (www.enago.jp) for the English language review. Scientific Research on Innovative Areas and International Group from the MEXT/JSPS KAKENHI [JP20H05873 (K. Maenaka)]. Japan Agency for Medical Research and Development [JP21wm0225018 (Y.O.), JP22wm0225017 (H.S.), JP223fa627005 (A.S., H.S., and K. Maenaka), JP20ae0101047, JP21fk0108463, JP17am0101093, JP22ama121037 (K. Maenaka)]. Japan Science and Technology Agency Moonshot R&D [JPMJMS2025 (Y.O. and Y.M.)]. Takeda Science Foundation (K. Maenaka). Atlantic Philanthropies Director (W.W.H., partially supported).
Author contributions
K.U., A.S., Y.O., H.S., A.M., and K. Maenaka designed research; K.U., H.N., A.S., S.T., S. Kusakabe, M.S., N.M., S.I., M.I., K.K., A.M., and K. Maenaka performed research; M.S., K.T., and K. Matsuno contributed new reagents/analytic tools; K.U., H.N., A.S., S.T., S. Kusakabe, S.I., M.T., Y.A., S. Kita, K.K., Y.M., H.K., A.M., and K. Maenaka analyzed data; Y.O. funding acquisition; Y.M., W.W.H., H.S., and K. Maenaka funding acquisition supervision; H.K. and A.M. supervision; and K.U., A.S., K.K., W.W.H., H.S., and K. Maenaka wrote the paper.
Competing interests
The authors K.U., H.N., A.S., S.T., and S. Kusakabe are employees of Shionogi & Co., Ltd. The other authors declared no conflict of interest. We have filed an application with the Japanese patent office.
Footnotes
This article is a PNAS Direct Submission. M.S.D. is a guest editor invited by the Editorial Board.
Preprint servers: bioRxiv, https://doi.org/10.1101/2022.12.14.520006.
Contributor Information
Akihiko Sato, Email: akihiko.sato@shionogi.co.jp.
Akira Matsuda, Email: matuda@pharm.hokudai.ac.jp.
Katsumi Maenaka, Email: maenaka@pharm.hokudai.ac.jp.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.COVID-19 Dashboard by the Center for Systems Science and Engineering at Johns Hopkins University. https://coronavirus.jhu.edu/map.html. Accessed 13 March 2023.
- 2.Pierson T. C., Diamond M. S., The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaid A., et al. , Arthritogenic alphaviruses: Epidemiological and clinical perspective on emerging arboviruses. Lancet Infect. Dis. 21, e123–e133 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Witkowski J. T., Robins R. K., Sidwell R. W., Simon L. N., Design, synthesis, and broad spectrum antiviral activity of 1-(β-D-ribofuranosyl-1,2,4-triazole-3-carboxamidef and related nucleosides. J. Med. Chem. 15, 1150–1154 (1972). [DOI] [PubMed] [Google Scholar]
- 5.Huggins J. W., Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Rev. Infect. Dis. 11, 750–761 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Graci J. D., Cameron C. E., Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 16, 37–48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick J. B., et al. , Lassa fever. N. Engl. J. Med. 314, 20–26 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Smith D. W., et al. , A controlled trial of aerosolized ribavirin in infants receiving mechanical ventilation for severe respiratory syncytial virus infection. N. Engl. J. Med. 325, 24–29 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Fried M. W., et al. , Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347, 975–982 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Beigel J. H., et al. , Remdesivir for the treatment of Covid-19—Final report. N. Engl. J. Med. 383, 1813–1826 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayk Bernal A., et al. , Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 386, 509–520 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo M. K., et al. , GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 7, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheahan T. P., et al. , Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, eaal3653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon J. J., et al. , Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 62, e00766-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toots M., et al. , Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 11, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahl A., et al. , SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 591, 451–457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruijssers A. J., Denison M. R., Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 35, 57–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon C. J., Tchesnokov E. P., Feng J. Y., Porter D. P., Götte M., The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295, 4773–4779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheahan T. P., et al. , An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12, eabb5883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkataraman S., Prasad B. V. L. S., Selvarajan R., RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses 10, 1–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Te Velthuis A. J. W., Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 71, 4403–4420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L., et al. , RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 213, 113201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crance J. M., Scaramozzino N., Jouan A., Garin D., Interferon, ribavirin, 6-azauridine and glycyrrhizin: Antiviral compounds active against pathogenic flaviviruses. Antiviral Res. 58, 73–79 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Delang L., Abdelnabi R., Neyts J., Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 153, 85–94 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Nobori H., et al. , Identification of Compound-B, a novel anti-dengue virus agent targeting the non-structural protein 4A. Antiviral Res. 155, 60–66 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Jin Z., et al. , Structure-activity relationship analysis of mitochondrial toxicity caused by antiviral ribonucleoside analogs. Antiviral Res. 143, 151–161 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Lu G., et al. , Analysis of ribonucleotide 5′-triphosphate analogs as potential inhibitors of Zika virus RNA-dependent RNA polymerase by using nonradioactive polymerase assays. Antimicrob. Agents Chemother. 61, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shresta S., Sharar K. L., Prigozhin D. M., Beatty P. R., Harris E., Murine model for dengue virus-induced lethal disease with increased vascular permeability. J. Virol. 80, 10208–10217 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin Z., et al. , An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. U.S.A. 106, 20435–20439 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaptein S. J. F., et al. , A pan-serotype dengue virus inhibitor targeting the NS3–NS4B interaction. Nature 598, 504–509 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Brewoo J. N., et al. , Immunogenicity and efficacy of chimeric dengue vaccine (DENVax) formulations in interferon-deficient AG129 mice. Vaccine 30, 1513–1520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Jiang R., et al. , Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182, 50–58.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oladunni F. S., et al. , Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 11, 6122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen D. R., et al. , An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021). [DOI] [PubMed] [Google Scholar]
- 35.DiPiazza A. T., et al. , COVID-19 vaccine mRNA-1273 elicits a protective immune profile in mice that is not associated with vaccine-enhanced disease upon SARS-CoV-2 challenge. Immunity 54, 1869–1882.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H., et al. , Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leist S. R., et al. , A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang K., et al. , Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 67, 103381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., et al. , SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J. Virol. 95, e02477-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam I., et al. , Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin. Virology 426, 143–151 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Belliot G., et al. , Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase. J. Virol. 79, 2393–2403 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong P., Peersen O. B., Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bravo J. P. K., Dangerfield T. L., Taylor D. W., Johnson K. A., Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell 81, 1548–1552.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sofia M. J., et al. , Discovery of a fluoro-2′-β- C -methyluridine Nucleotide Prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 53, 7202–7218 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Sourimant J., et al. , 4′-Fluorouridine is an oral antiviral that blocks respiratory syncytial virus and SARS-CoV-2 replication. Science 375, 161–167 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fung A., et al. , Efficiency of incorporation and chain termination determines the inhibition potency of 2′-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob. Agents Chemother. 58, 3636–3645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasaki M., et al. , SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 17, e1009233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasaki M., et al. , Air-liquid interphase culture confers SARS-CoV-2 susceptibility to A549 alveolar epithelial cells. Biochem. Biophys. Res. Commun. 577, 146–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki M., et al. , S-217622, a SARS-CoV-2 main protease inhibitor, decreases viral load and ameliorates COVID-19 severity in hamsters. Sci. Transl. Med. 15, eabq4064 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wada Y., et al. , Discovery of a novel antiviral agent targeting the nonstructural protein 4 (nsP4) of chikungunya virus. Virology 505, 102–112 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Uemura K., et al. , 5-Hydroxymethyltubercidin exhibits potent antiviral activity against flaviviruses and coronaviruses, including SARS-CoV-2. iScience 24, 103120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thai H. T. C., et al. , Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42, 1956–1961 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raj V. S., et al. , Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki M., et al. , The role of heparan sulfate proteoglycans as an attachment factor for rabies virus entry and infection. J. Infect. Dis. 217, 1740–1749 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Ishii A., et al. , A nairovirus isolated from African bats causes haemorrhagic gastroenteritis and severe hepatic disease in mice. Nat. Commun. 5, 5651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuno K., et al. , Fatal tickborne phlebovirus infection in captive Cheetahs, Japan. Emerg. Infect. Dis. 24, 1726–1729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikegami T., Won S., Peters C. J., Makino S., Rescue of infectious rift valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80, 2933–2940 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okumura M., et al. , Development of serological assays for Thottapalayam virus, an insectivore-borne hantavirus. Clin. Vaccine Immunol. 14, 173–181 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanaki T., et al. , Inhibition of dengue virus infection by 1-stearoyl-2-arachidonoyl-phosphatidylinositol in vitro. FASEB J. 33, 13866–13881 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.


