Significance
Biomolecular condensates, implicated in many cellular processes, can assemble via phase transitions of a few key driver macromolecules. Condensate formation is further modulated by the interactions with ligands such as polyubiquitin chains, a common protein posttranslational modification. Using a library of designed polyubiquitin ligand hubs with decreasing or increasing spacings between binding sites and altered binding affinities with drivers, we employ theory and experiments to determine key properties that govern how ligand hubs affect driver phase transitions. Importantly, linear (M1-linked) polyubiquitin chains are already optimized (in terms of spacing between binding sites) to maximize phase separation of driver macromolecules such as proteasomal shuttle factor UBQLN2. Polyubiquitin chains likely use their multivalent architecture to dynamically regulate condensate formation and other properties.
Keywords: ligand-induced phase transition, polyphasic linkage, biomolecular condensates, multicomponent phase separation, polyubiquitin
Abstract
Biomolecular condensates form via multivalent interactions among key macromolecules and are regulated through ligand binding and/or posttranslational modifications. One such modification is ubiquitination, the covalent addition of ubiquitin (Ub) or polyubiquitin chains to target macromolecules. Specific interactions between polyubiquitin chains and partner proteins, including hHR23B, NEMO, and UBQLN2, regulate condensate assembly or disassembly. Here, we used a library of designed polyubiquitin hubs and UBQLN2 as model systems for determining the driving forces of ligand-mediated phase transitions. Perturbations to either the UBQLN2-binding surface of Ub or the spacing between Ub units reduce the ability of hubs to modulate UBQLN2 phase behavior. By developing an analytical model based on polyphasic linkage principles that accurately described the effects of different hubs on UBQLN2 phase separation, we determined that introduction of Ub to UBQLN2 condensates incurs a significant inclusion energetic penalty. This penalty antagonizes the ability of polyUb hubs to scaffold multiple UBQLN2 molecules and cooperatively amplify phase separation. The extent to which polyubiquitin hubs promote UBQLN2 phase separation is encoded in the spacings between Ub units. This spacing is modulated by chains of different linkages and designed chains of different architectures, thus illustrating how the ubiquitin code regulates functionality via the emergent properties of the condensate. The spacing in naturally occurring linear polyubiquitin chains is already optimized to promote phase separation with UBQLN2. We expect our findings to extend to other condensates, emphasizing the importance of ligand properties, including concentration, valency, affinity, and spacing between binding sites in studies and designs of condensates.
Many membraneless organelles and cellular complexes are hypothesized to form via phase separation and contribute to multiple biological processes, such as stress response, gene expression, and protein degradation (1, 2). Biomolecular condensates comprise hundreds to thousands of unique macromolecules, but only a few of these are necessary to drive condensation in vitro (3, 4). These drivers are classified as scaffolds whereas the other nondriver components are known as clients/ligands. Scaffolds are multivalent macromolecules that can self-interact at specific sites called stickers to undergo phase separation (PS) into coexisting macromolecule-dense and macromolecule-dilute phases. Ligands, many of which do not phase separate on their own, bind to scaffolds and regulate condensate assembly/disassembly and material properties. Recent studies have uncovered several rules that dictate how different features of ligands can lead to modulation of scaffold phase behavior (5). Specifically, low- and high-valency ligands tend to inhibit and promote scaffold PS, respectively (6–13). Moreover, computational studies showed that weakly binding ligands do not affect scaffold PS as much as strongly binding ligands. Furthermore, binding of ligands to nonsticker (spacer) regions rather than sticker regions of scaffolds is more favorable for enhancing PS in a ligand:scaffold concentration-dependent manner (5–7). Importantly, these rules are best applied for systems whose scaffolds phase separate via homotypic interactions but also can be modulated by ligands; a thorough discussion regarding ligand properties and effects is presented in (5). However, an additional consideration is that ligands with the same composition but different conformations or architecture can lead to highly different scaffold phase behaviors (8, 11–13).
One prevalent set of ligands includes ubiquitin (Ub) and polyubiquitin (polyUb) chains, which are posttranslationally attached to proteins and target them to different signaling outcomes, including proteasomal degradation and the DNA damage response, among many others (14). PolyUb chains, composed of different lengths and linkages, are a regulator of biomolecular condensates in protein quality control, autophagy, and immune system activation. Condensation can be driven by ubiquitination of E3 ligases such as TRIM3 (15) or regulated by ligase-mediated ubiquitination of proteins such as Disheveled-2 in Wnt signaling for organismal development (16). Cargo proteins modified with K63-linked polyUb chains interact with p62 to form preautophagosome condensates that later recruit autophagosome machinery (12, 17); furthermore, autophagy cargo adaptor NBR1 condenses with K63- and M1-linked chains (18). NEMO condenses with K63- and M1-linked polyUb chains, but not K6-, K11-, K29-, K33- and K48-linked polyUb chains, to facilitate activation of the IKK complex leading to downstream NF-κB processing (11, 19). In contrast, hHR23B preferably binds to and condenses with K48-linked chains to form nuclear condensates and recruit the proteasome and other protein quality control components to degrade defective ribosomal proteins (13). These different polyUb chains, which differ only in the location of the isopeptide bond that connects the Ub units, signal for different cellular outcomes (the Ub code) through the regulation of condensate formation. Therefore, these studies suggest that the differential effects on the condensation of interacting proteins are a mechanism for the Ub code to be read and interpreted in the cell (14, 20). Given the growing evidence for polyUb chain involvement in condensate formation, it is essential to determine the molecular rules by which polyUb chains regulate biomolecular condensates.
We recently showed that polyUb chain conformation contributes to whether polyUb chains promote or inhibit the PS of UBQLN2, a Ub-binding shuttle protein critical to cellular protein quality control mechanisms. UBQLN2 PS is driven by homotypic interactions involving multiple folded and intrinsically disordered regions across the protein (21, 22). MonoUb drives disassembly of UBQLN2 condensates (21). However, compact tetraUb (Ub4) chains (K11 and K48) largely inhibit UBQLN2 PS, whereas extended Ub4 chains (K63 and M1) promote UBQLN2 PS. Via binding interface mapping and Kd measurement, we showed that UBQLN2 interacts similarly with these different chain types (8, 23). These collective observations suggest that the distinction between linkages manifests through the emergent property of condensation. These studies led us to hypothesize that more extended polyUb chains have greater abilities to promote UBQLN2 PS. To systematically examine the features of polyUb ligands that regulate PS, we employed a library of designed polyUb hubs where we modified either the linker length/flexibility between the Ub units on these hubs or the binding affinity between the Ub unit and UBQLN2. By combining information on the solution structures of these designed hubs and changes in UBQLN2 PS with theoretical modeling based on polyphasic linkage principles (24, 25), we determined the essential properties of polyUb ligand architecture that govern how they modulate phase separation of UBQLN2. These properties enable the prediction of how ligands of different architectures and valencies (e.g., mono- and tetraUb) modulate phase behavior and determine the designs of ligands for tighter control of specific condensate assembly as a function of ligand concentration.
Results
Decreased Binding Affinity between Ub and UBQLN2 Weakens polyUb Ability to Promote UBQLN2 Phase Separation.
We previously showed that Ub interacts with UBQLN2 at the same UBA residues that are involved in PS-driving multivalent interactions within UBQLN2 (21). These Ub:UBA interactions can either inhibit or further promote UBQLN2 PS, depending whether Ub is in monomeric or tetrameric (Ub4) states, respectively (8). To investigate how different Ub:UBA binding affinities affect UBQLN2 PS, we introduced single and double amino acid substitutions at select residues (I44 and V70) on the Ub hydrophobic binding patch (Fig. 1A) (26–28). Using NMR spectroscopy (SI Appendix, Fig. S1), we determined that all of the substitutions reduced the binding affinity between Ub and UBQLN2 to different extents in the following order: Ub > Ub V70I > V70A > I44V > V70I/I44V > V70A/I44V > I44A (SI Appendix, Table S1).
Fig. 1.
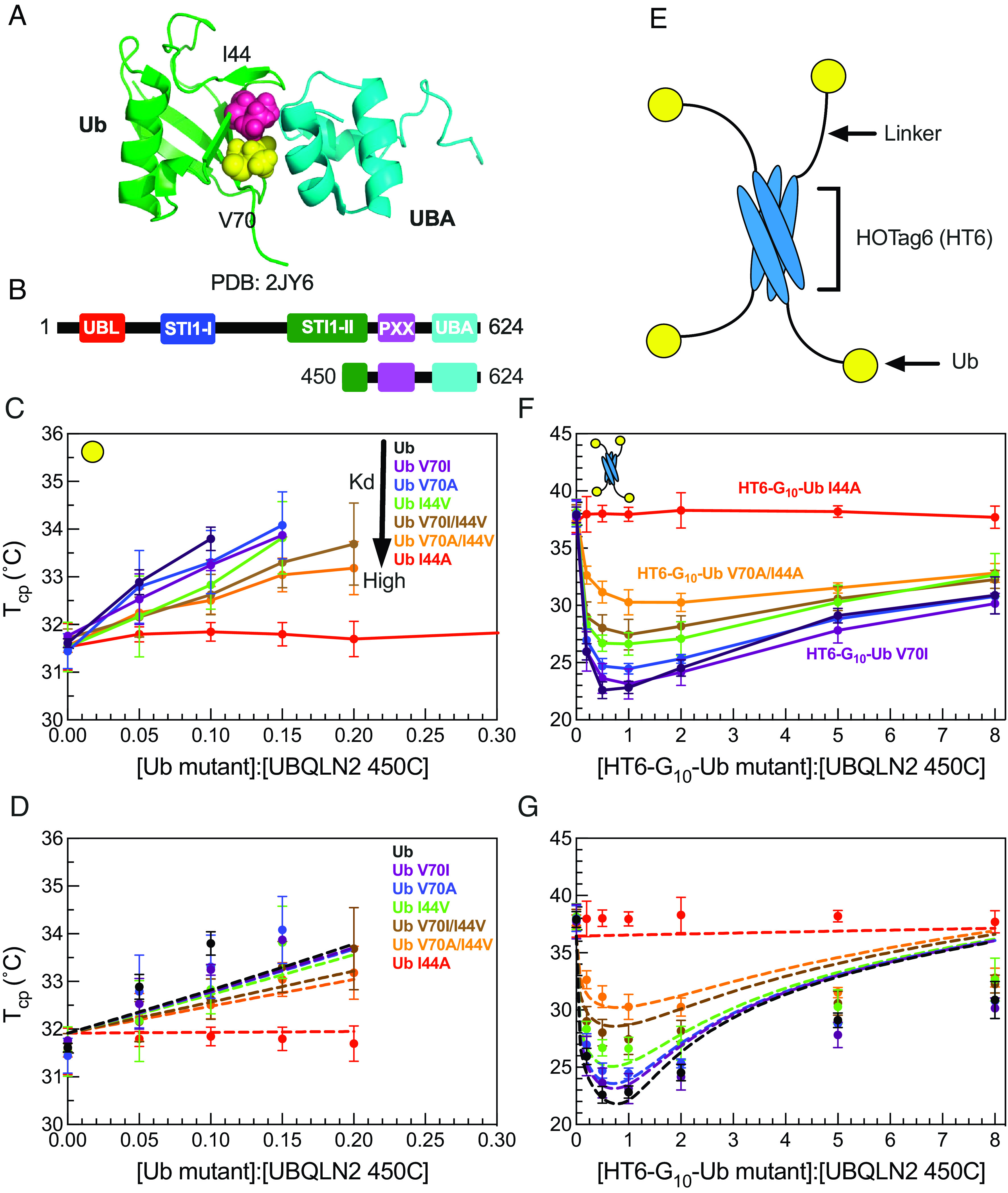
HT6-Ub variants with weakened binding affinity do not efficiently promote phase separation of UBQLN2/HT6-Ub. (A) Structure of the Ub/UBA complex with the hydrophobic patch residues I44 and V70 of Ub represented in spheres. (B) Domain structure of full-length UBQLN2 and UBQLN2 450-624 construct (450C) used in this study. (C) Cloud point temperature curves of 450C PS in the presence of monoUb variants. PS occurs above phase boundary. (D) Comparison of theory (dotted lines) to experiments (points) for the monoUb variants in panel C. (E) Architecture of HOTag6-G10-Ub (HT6-Ub) ligand. (F) Cloud point temperature curves of 450C PS in the presence of HT6-Ub variants. (G) Theory fit (dotted lines) to HT6-Ub variants using an inclusion energy of 9.5 kT. Data points and error bars in panels C and F reflect averages and SDs across at least n = 6 experiments using two protein stocks.
We first quantified the effects of the substitutions in monoUb on the PS of a UBQLN2 C-terminal construct 450–624 (450C, Fig. 1B), which exhibits similar PS behavior to full-length UBQLN2 (21). We performed temperature-ramp turbidity assay experiments that monitor the change in A600 values (as a proxy for PS) at different Ub:450C ratios, but fixed 450C concentration and buffer composition. We used these turbidity data to build cloud point temperature (Tcp) curves (SI Appendix) that are a proxy for the low-concentration arm of temperature-component phase diagrams (29). At temperatures above the cloud point curves, the UBQLN2:Ub solutions are phase-separated, whereas the solutions are not phase-separated below the curves. As the binding affinity between UBQLN2 and Ub variants decreased, Ub variants were less effective at binding to UBA and inhibiting 450C PS, which occurred at lower temperatures and higher Ub:450C ratios (Fig. 1C). The I44A substitution completely disrupted Ub:UBA binding; hence, Ub I44A had little or no effect on 450C phase separation (Fig. 1C).
We next quantified how substitutions in the designed tetrameric HOTag6-(G)10-Ub (HT6-Ub, Fig. 1E) affect 450C PS (8). HT6-Ub, a structural mimic of a multi-monoubiquitinated substrate, can significantly promote UBQLN2 PS over a wide range of HT6-Ub:UBQLN2 ratios (8) (Fig. 1F). Cloud point temperature curves showed that as the binding affinity between UBQLN2 and HT6-Ub variants decreased, HT6-Ub variants were less effective at enhancing 450C PS (Fig. 1F). Together, data for both Ub and HT6-Ub variants demonstrated that binding affinities are positively correlated with the ability of the ligand Ub/HT6-Ub to modulate the phase behavior of the PS-driver 450C. These trends are consistent with results from a previous theoretical study using a generic PS-driver and its mono- and divalent ligands (6).
Theory Development for How Ub Ligands Inhibit and Promote Phase Separation.
We developed a theory to reconcile the observation that monoUb and polyUb inhibit and promote PS, respectively. Our formalism is equivalent to the general polyphasic linkage framework of Wyman and Gill (24, 25). In our treatment, various quantities take specific meanings in the context of polyUb/UBQLN2 condensates. The key elements of the theory (SI) include the enforcements of the chemical equilibrium of polyUb-UBQLN2 binding and the equilibrium of molecular partitioning between phases (dilute/dense). These two equilibria are linked, and this serves as the basis for polyphasic linkage concepts (5, 24, 25). Here, we consider UBQLN2 as a PS “driver,” which is a macromolecule capable of PS on its own. The Ub or polyUb ligand is termed a “hub,” by which we refer to the ligand’s ability to interact with multiple UBQLN2 molecules, depending on the number of Ub units present on the hub. We assume that a driver has a single binding site for the hub, in accord with our experimental data where UBQLN2 binds to a single Ub molecule. Chemical equilibrium is enforced via the equilibrium constants for the reaction: :
| [1] |
where is the dissociation constant for a driver/hub complex (oligomer) containing one hub molecule and n drivers (henceforth referred to as an n-mer). , , and are the concentrations of monomer (unbound) drivers, monomer (unbound) hubs, and n-mers, respectively, and the i-index indicates either the dilute phase (V = vapor) or the dense phase (L = liquid). Partitioning equilibrium is enforced by introducing transfer free energies and :
| [2a] |
| [2b] |
where indicates unfavorable transfer for the hub to the dense phase (the hub is unable to phase separate independently), and indicates favorable transfer for the driver to the dense phase. The transfer energies, and all other energies, are given in units of kBT, where T is the absolute temperature and kB is the Boltzmann constant. The favorable interactions captured by will be temperature-dependent, which can be incorporated in the theory by noting that in the absence of hub molecules
| [3] |
where is the saturation concentration of the driver at a particular temperature (SI), and is the total concentration of driver in the liquid (henceforth, capital H and D indices indicate total concentrations of hubs and drivers, while lowercase h and d indicate concentration of hubs and drivers in the unbound state). Combining Eqs. 2 and 3, we find
| [4] |
Eq. 4 is the essence of the polyphasic linkage concept (5, 24, 25). This equation says that PS can occur with below the if the concentration of unbound driver in the dense phase is less than the concentration of driver in the homogenous driver fluid (UBQLN2 PS alone). This condition is easy to attain in the presence of hubs because n-mers will occupy a significant fraction of the dense phase volume such that . In other words, n-mers dilute the concentration of unbound drivers in the dense state, which lowers their chemical potential and allows PS to occur at lower concentration. To determine a condition for the onset of PS, we use the fact that when polymers phase separate the mass concentration in the dense phase is insensitive to the molecular weight (or equivalently, polymerization number) of the individual molecules (5). This is because the microscopic interactions and mesh structure of the dense phase are both much smaller than the molecules. In agreement with this expectation, the concentration of UBQLN2 molecules in the dense phase is nearly unchanged as polyUb is added (8). The approximation of constant driver concentration in the dense phase is empirically valid for polyUb/UBQLN2 condensates (8), but not generally applicable to polyphasic linkage systems since the ligand/hub molecule would be expected to displace driver molecules (SI). With this approximation, our criterion for PS is that the total concentration of drivers in the dense phase is equal to the concentration of the homogeneous fluid
| [5] |
With the use of Eqs 1–4, this condition can be cast in terms of the dilute phase concentrations (SI).
| [6] |
We refer to the quantity as the “inclusion energy.” It accounts for two detrimental effects from transferring an n-mer to the dense phase. The first, described by , is the disruptive effect of inserting the hub within the network of attractive interactions formed by the drivers. In our model, this contribution depends only on the number of Ub modules in a hub. The second effect, captured by , comes from the fact that drivers bound to the hub are constrained in their interactions with the surrounding fluid so that the driving force for an n-mer to enter the dense state may differ from . We expect that these constraints will depend on the specific linkage and/or geometry of the hub. The total inclusion energy, , is the only free parameter in Eq. 6. In summary, this theoretical treatment is best applied for systems whose PS is driven by homotypic interactions (e.g., the UBQLN2 driver), where the ligand/hub (e.g., polyUb) is multivalent in design and does not phase separate on its own.
HT6-Ub Imparts a Significant Inclusion Energy that Is Offset by Ub Binding Affinity to Promote UBQLN2 Phase Separation.
We tested our theoretical model on our 450C data for monoUb and HT6-Ub variants (Fig. 1 D and G). First, we performed a global fit to the cloud-point temperature curves for HT6-Ub variants of different Ub:UBA binding affinities. The global fit was performed using the experimentally obtained binding affinities between monoUb and UBA (SI Appendix, Fig. S1 and Table S1). We assumed that each Ub in HT6-Ub interacted with the UBA of a single 450C with the same Kd as the isolated Ub:UBA interaction; previous data suggested that this is the case for HT6-Ub and UBQLN2 (8). Thus, an HT6-Ub molecule can maximally have four 450C molecules bound. Importantly, the theory-derived cloud-point temperature curves mirrored the experimental ones for HT6-Ub variants of different Ub:UBA binding affinities (Fig. 1G). Stronger binding between Ub and UBA stabilized HT6-Ub:450C PS at low Ub:UBQLN2 ratios with the lowest Tcp generally near 1:1 stoichiometry of Ub:450C. The best fit value of the inclusion energy was 9.5 kT, suggesting that it costs almost 2.5 kT to transfer each ubiquitin unit to the dense phase (Fig. 1G). This value is very close to the energy of inserting RFP tags into SUMO-SIM condensates (30) and is also consistent with the entropic repulsion with computer models showing that biomolecular condensates tend to exclude foreign particles (31). The Ub inclusion penalty is offset by the favorable binding between the multiple Ub units on the hub and the 450C molecules. Thus, the reentrant behavior at stoichiometries greater than 1:1 is due to hubs that incur the full 9.5 kT inclusion energy but, having less than four drivers bound, have a reduced driving force for phase separation. We next considered the monoUb ligand. Using the same parameters, and an inclusion energy that is one-fourth of the HT6-Ub inclusion energy, we also recapitulated the cloud point temperature curves for the monoUb variants (Fig. 1D). These results suggest that a single Ub unit is unfavorable to enter the dense phase, thus driving destabilization of UBQLN2 PS.
We note that the theory curves do not capture the phase boundary at higher Ub:UBQLN2 ratios; the theory predicts a lower degree of PS (higher Tcp) than what we observe (lower Tcp). We believe that this discrepancy is due to cooperativity in the binding of drivers to the hub. We speculate that the same favorable driver–driver interactions that promote PS will make it more likely for drivers to bind to hubs that already have drivers bound. Thus, when there is an insufficient amount of drivers to saturate the hubs, the hubs will prefer fully bound or fully empty states, rather than the randomly occupied states assumed by the theory. Because of the unfavorable hub inclusion energy, a smaller number of fully bound hubs is more favorable to PS than a larger number of partially bound hubs. Due to the systematic overestimation of the phase boundary at high stoichiometries, we only used Ub:UBQLN2 ratios less than or equal to 2.0 to fit the model parameters.
HT6-Ub with Longer Linkers Are more Extended in Solution and Less Effective at Enhancing UBQLN2 Phase Separation.
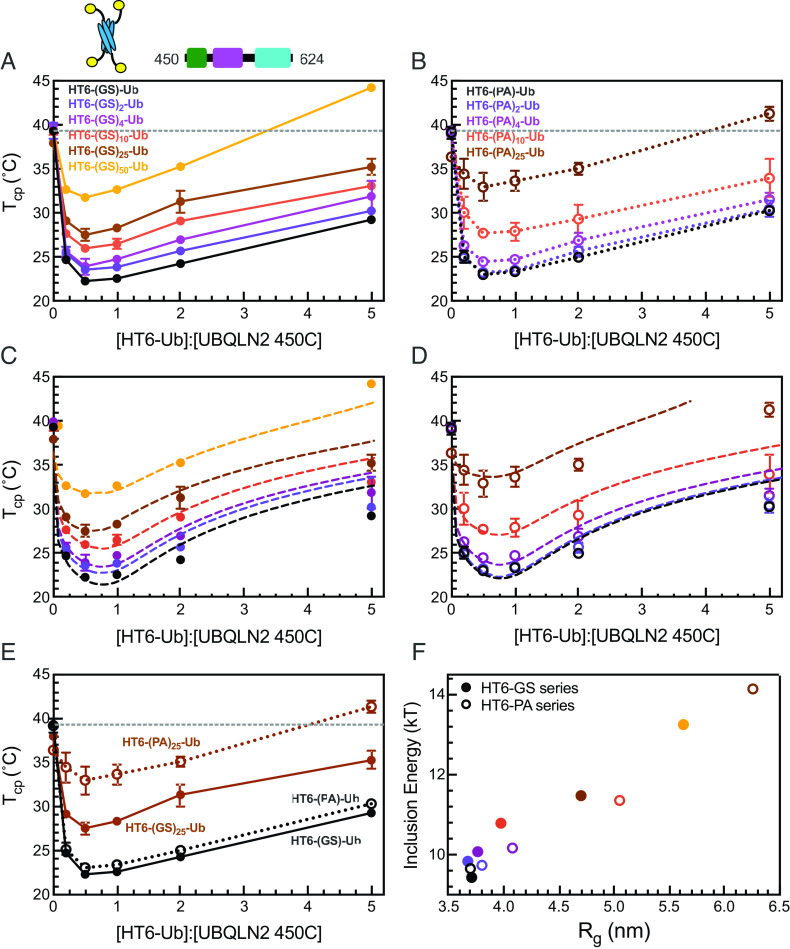
Our recent work demonstrated that more extended polyUb chains promoted UBQLN2 PS, with the most extended HT6-G10-Ub and the most compact K48-linked Ub4 exhibiting the most and least PS-enhancing ability, respectively (8). However, HT6-G10-Ub and polyUb chains of different linkages differ not only in conformation but also in overall topology, steric hindrance within the polyUb chains, accessibility to the Ub hydrophobic binding surface, and orientation of the Ub units, stymieing clear interpretation of the observed correlation between ligand conformations and ligand ability to modulate PS. Moreover, we hypothesize that there is a limit to the positive correlation between the extendedness of polyUb hubs and the enhancement of UBQLN2 phase separation. Beyond this limit, the Ub units in more extended polyUb hubs would be too far apart to bring the driver molecules close together to enhance phase separation. Here, we aimed to systematically investigate how changes in polyUb hub conformation modulate UBQLN2 PS by using the simplicity of our designed tetrameric HT6-Ub, the architecture of which enabled accommodations for linkers of varying lengths and flexibility without interfering with the Ub units. We created a library of linkers comprising multiple GS (Gly-Ser) or PA (Pro-Ala) blocks (Fig. 2A and SI Appendix, Fig. S2), which are flexible and rigid in conformation, respectively (32). These different HT6-Ub constructs potentially mimic multi-monoubiquitinated substrates of different tertiary structures and sizes, resulting in varying distances between ubiquitination sites.
Fig. 2.
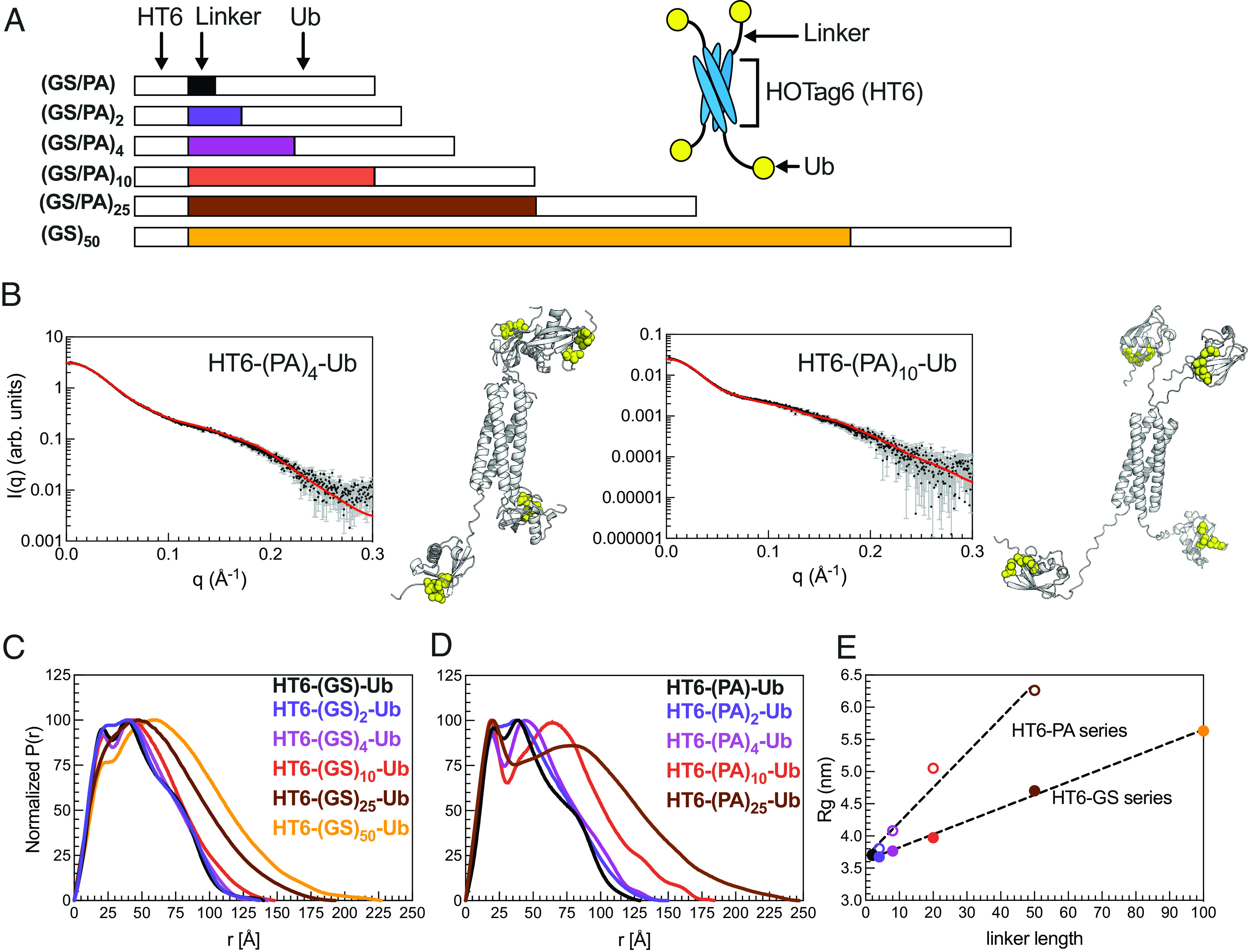
HT6-Ub constructs with longer linkers are larger in size and more extended in overall conformation. (A) Library of HT6-Ub constructs with different linkers with a cartoon representation of the spider-like HT6-Ub geometric architecture. (B) Experimental X-ray scattering curves (black) for designed HT6-(PA)x-Ub constructs overlaid with scattering curve predicted (red) from single best structure (shown) derived from SASSIE conformational ensembles. Hydrophobic patch residues L8, I44, and V70 are represented as yellow spheres. (C and D) P(r) profiles from SAXS experiments on HT6-Ub ligands of different linker lengths. (E) Radius of gyration (Rg) as determined from Guinier fits to SAXS experiments for HT6-Ub constructs (closed circles: GS series and open circles: PA series) as a function of linker length (# of amino acids). Black dotted lines represent simple linear regression of each dataset.
We performed size-exclusion chromatography coupled with multiangle light scattering and small-angle X-ray scattering (SEC-MALS-SAXS) on the HT6-Ub constructs with different GS and PA linkers to gain insights on how these linkers affected the overall conformations and structures of the HT6-Ub hubs. SEC-MALS experiments confirmed that each HT6-Ub construct examined herein existed as a tetramer in solution and that elution time, hence overall size of the HT6-Ub complex, increased with linker length (SI Appendix, Fig. S3 and Table S2). Representative structures of these HT6-Ub constructs were determined by refining AlphaFold-predicted starting structures (33) against our scattering data using conformational ensembles constructed with SASSIE (34) (Fig. 2B). Pair distance distribution function (P(r)) analysis showed that the HT6-Ub constructs with longer linkers adopt more extended structures (Fig. 2C). Guinier analysis of collected SAXS data also displayed a positive linear relationship between linker length and radius of gyration (Rg), with Rg for constructs with (PA)x linkers increasing more than with (GS)x linkers as a function of linker length (Fig. 2D and SI Appendix, Fig. S4 and Table S2). HT6-Ub with PA linkers were much more extended than those containing GS linkers of the same length, consistent with (PA)x being more rigid than (GS)x. For all HT6-Ub constructs used, the Ub units do not exhibit significant changes to structure or dynamics compared to monoUb, as observed by NMR spectroscopy experiments (SI Appendix, Fig. S5 A–E).
To compare flexibility of Ub tethered to HT6 to that of monoUb on a residue-by-residue basis, we performed NMR spin relaxation experiments on backbone amide 15N resonances. 15N R1 & R2 relaxation rates monitor backbone dynamics on millisecond to nanosecond timescales. MonoUb has the highest R1 rate of 1.73 s−1 while Ub tethered to HT6 by (PA)2 and (GS)2 linkers showed the lowest R1 rate of 0.83 s−1 (SI Appendix, Fig. S5). 15N R1 rates for Ub decreased while 15N R2 rates increased in the order of decreasing linker length: (Ub > HT6(GS)50Ub > HT6(PA)10Ub > HT6(GS)10Ub > HT6(G)10Ub > HT6(PA)2Ub ~ HT6(GS)2Ub). Decreases in R1 rate with corresponding increases in R2 rate indicate an overall slower rotational tumbling rate for the protein; hence Ub tethered to HT6 with a shorter linker tumbled more slowly than Ub tethered with a longer linker. Collective observations from spin-relaxation and SEC-MALS-SAXS experiments indicated that Ub tethered to HT6 with a longer linker exhibited a faster tumbling rate (has a higher degree of freedom to move) and adopted more extended structures.
We next performed turbidity assays of 450C in the presence of HT6-Ub constructs containing different linkers. The resulting cloud point temperature curves significantly shifted as a function of linker length (Fig. 3). Importantly, the overall shape of these curves (minima and slopes with increasing Ub:450C ratios) remained unchanged as linker length was modulated, unlike that of the binding variant HT6-Ub/450C cloud point temperature curves (Fig. 1F), indicating that the Ub:450C binding surface was unperturbed. Indeed, NMR titration experiments reported similar Kd values and chemical shift perturbations (CSPs) between UBA and the Ub units across several HT6-Ub constructs (SI Appendix, Table S1 and Fig. S6). These results are consistent with those from our prior work which showed that differences in UBQLN2 phase separation behavior do not correlate with changes in Kd between polyUb of different linkages and UBQLN2 (8). Therefore, these results suggest that the differences in the cloud point temperature curves for HT6-(GS/PA)x-Ub and 450C stemmed solely from changes to the HT6-Ub linkers.
Fig. 3.
HT6-Ub constructs with longer linkers are less effective at enhancing phase separation of UBQLN2. (A and B) Cloud point temperature curves of 450C PS in the presence of HT6-Ub variants containing variable GS (A) or PA (B) linker length. Data points and error bars in panels A and B reflect averages and SDs across at least n = 6 experiments using two protein stocks. Cartoon above (A) denotes use of HT6-Ub ligands with 450C (fixed concentration of 50 µM) for these experiments. PS of 450C and HT6-Ub occurs at temperatures above (but not below) phase boundaries. (C and D) Theory fit (dotted lines) to HT6-variants. (E) Comparison of 450C cloud point temperature curves with HT6-GS-Ub (filled circles, black), HT6-PA-Ub (open circles, black), HT6-(GS)25-Ub (filled circles, brown), and HT6-(PA)25-Ub (open circles, brown). (F) Inclusion energy (kT) as determined from theory compared to the Rg of each HT6-Ub ligand used (as color-coded in panels C and D).
Our results show that the HT6-Ub constructs with the shortest linkers (HT6-GS-Ub & HT6-PA-Ub) were the most effective at enhancing 450C PS (lower Tcp). Increasing either GS or PA repeat length diminished the ability of HT6-Ub to promote 450C phase separation over a wide Ub:450C range (Fig. 3 A and B). Indeed, HT6-Ub constructs with longer (PA)x linkers, which are much more extended than those with longer (GS)x linkers of the same length (Fig. 3E), were less effective at enhancing 450C PS compared to their (GS)x counterparts. These results appear to contradict our previous finding that more extended polyUb chains can more effectively enhance UBQLN2 PS (8). To understand this contradiction, we used our theoretical model to determine the inclusion energy. We assume that these inclusion energies are the sum of the linkage-independent effect of inserting the hubs into the condensate (sh) and a linkage-dependent effect on driver–driver interactions (ΔS) (SI). This analysis revealed that the inclusion energy of introducing the HT6-Ub hub into the dense phase was lowest (most favorable) for hubs with the shortest linker and increased for hubs with longer linkers (Fig. 3 B, D, and F). Together, our experimental and modeling results suggest that the HT6-Ub hubs with shorter linkers are better accommodated within the condensate of drivers, hence lowering the inclusion energy for hub incorporation and enhancing PS. Our data suggest that the HT6-Ub constructs with the shortest linkers are already optimized to maximize phase separation with UBQLN2. Our previous work (8) included more compact chains (K48-, K63-, and M1-Ub4) and a more extended HT6-Ub construct (with a 10-residue linker), but not the most optimal HT6-Ub constructs (with a 2-residue linker) studied here.
Designed Linear(M1-linked) polyUb Chains with Longer Linkers Are Less Effective at Enhancing UBQLN2 Phase Separation.
Ligand hub architecture could affect PS of drivers (e.g., UBQLN2) via differences in hub geometry, binding constraints, and the proximity of bound drivers. The HT6-Ub hub comprises Ub units that can rotate independently from one another. In contrast, the Ub units in polyUb chains are covalently attached to one another and thus experience more constraints. Therefore, the trends observed for HT6-Ub with varying linker lengths might differ from those observed for polyUb chains with varying linker lengths. Since conjugation of polyUb chains requires specific recognition by particular Ub-conjugating enzymes, most chains are not amenable to manipulation. One exception is the linear M1-linked chain in which the C terminus of one Ub is covalently linked to the N-terminal amine of the next, and so forth; this chain type adopts extended conformations in solution (8, 35). Moreover, structural and NMR dynamics studies (36, 37) revealed that the last four residues of Ub (residues 73–76) are unstructured and very flexible. Therefore, this linear architecture enabled us to create a large set of M1-Ub4 constructs that contained both shorter and longer linkers than the naturally occurring M1-Ub4 to examine the effects of changing the spacing between Ub units on UBQLN2 phase separation (Fig. 4).
Fig. 4.
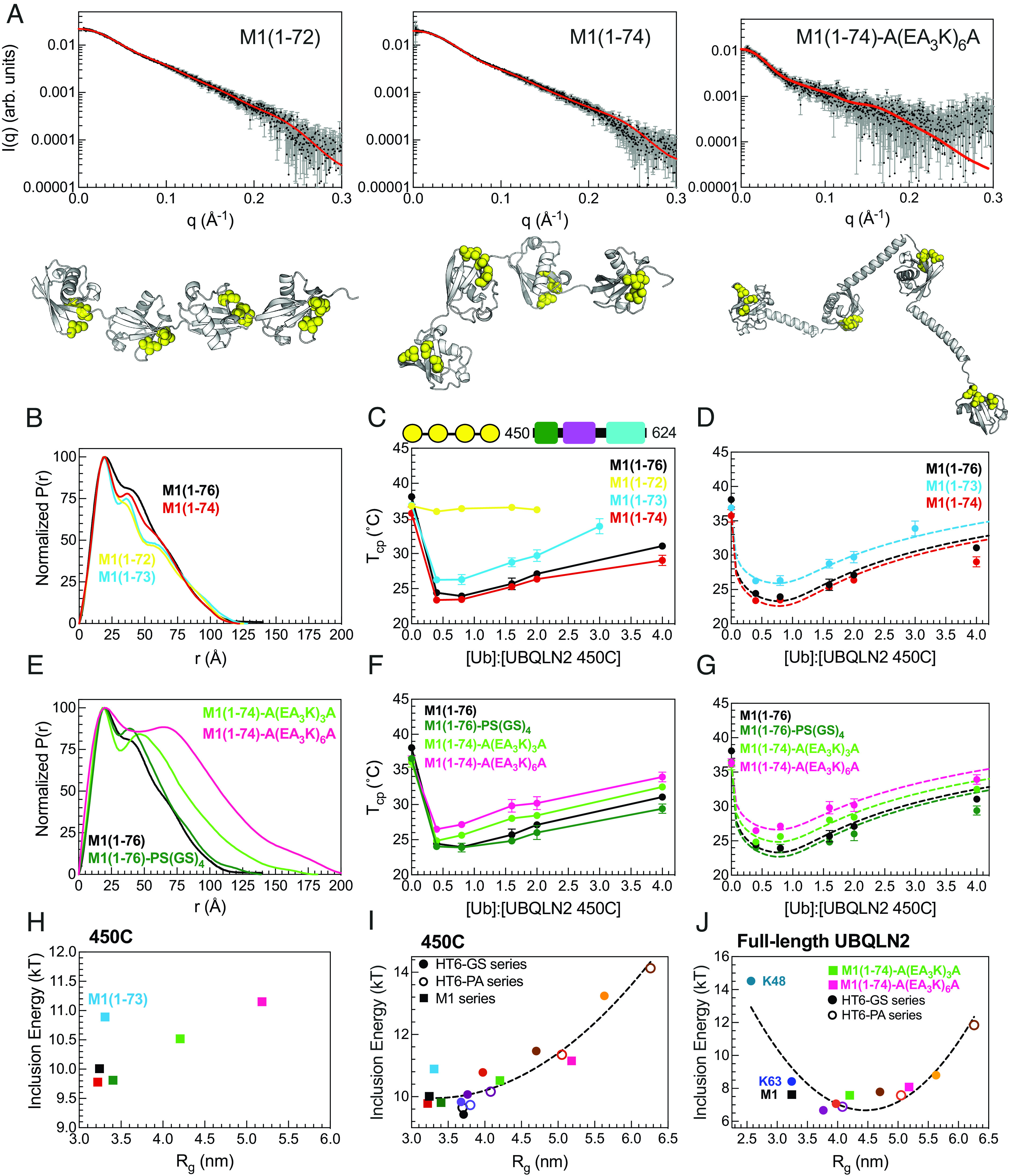
Designed linear polyUb chains with either short or long Ub-Ub linkers are less effective at enhancing UBQLN2 phase separation. (A) Experimental X-ray scattering curves (black) for designed M1-Ub4 constructs overlaid with scattering curve predicted (red) from single best structure (shown) derived from SASSIE conformational ensembles. Hydrophobic patch residues L8, I44, and V70 are represented as yellow spheres. (B) P(r) profiles of designed M1-Ub4 constructs with shorter Ub-Ub linkers. (C) Cloud point temperature curves of 450C with designed M1-Ub4 constructs. PS of 450C and M1-Ub4 constructs occurs at temperatures above (but not below) phase boundaries. (D) Theory-derived curves overlaid on experimental data. (E) P(r) profiles of designed M1-Ub4 constructs with longer Ub-Ub linkers. (F) Cloud point temperature curves of 450C (fixed concentration of 50 µM) with designed M1-Ub4 constructs. (G) Theory-derived curves overlaid on experimental data. (H) Theory-derived hub inclusion energy for M1-Ub4 constructs of various Ub-Ub linker lengths (as color-coded in panels D and G). Label is added for M1-Ub4 (1-73) as this construct binds less effectively to 450C. (I) For 450C PS, theory-derived hub inclusion energy for all designed polyUb hubs in this study. Data points are color-coded as in panels D and G, and from Fig. 3F. Inclusion energies for M1-Ub4 and HT6-Ub constructs are represented as squares and circles, respectively. (J) For full-length UBQLN2 PS, theory-derived hub inclusion energy for naturally occurring Ub4 polyUb chains, two designed M1-Ub4 constructs of longer Ub-Ub linker lengths, HT6-(GS)x-Ub hubs (filled circles), and HT6-(PA)x-Ub hubs (open circles). Parabolic trendlines (dotted lines) in panels I and J are drawn to guide the eye. Rg values in panels H, I, and J are of the different ligand hubs used.
First, we examined shorter linkers, i.e., by deleting residues from the C terminus of Ub for all three Ub-Ub linkers in an M1-Ub4 construct [(1-72, 1-73, 1-74, and 1-76 (WT)]. Although the scattering profiles and Rg values of these different constructs were relatively unchanged (Fig. 4A and SI Appendix, Fig. S7), distinct peaks appeared in the M1-Ub4 (1-72) and M1-Ub4 (1-73) P(r) profiles (Fig. 4B). We next compared SASSIE-constructed conformational ensembles for each construct against the experimental SAXS scattering curves (Fig. 4A). This analysis revealed that the Ub units in M1-Ub4 (1-72) exhibited restricted mobility due to the short linkers between Ub units. NMR spectra of these chains were similar to that of monoUb, indicating that the Ub units remained well-structured (SI Appendix, Fig. S8). However, CSPs between M1-Ub4 (1-72) or M1-Ub4 (1-73) and monoUb suggested that the hydrophobic patch residues (8,44,70) made Ub-Ub contacts not present in naturally occurring M1-Ub4 or the HT6-Ub hubs (SI Appendix, Fig. S8 A–D). NMR titration experiments revealed that binding between M1-Ub4 (1-72) and UBQLN2 UBA is significantly attenuated (SI Appendix, Fig. S8G). Modeling of the UBA to SASSIE structures of M1-Ub4(1-72) suggested that multiple UBA domains cannot be accommodated on this shortened Ub4. In line with our structural data, cloud point temperature curves of 450C with M1-Ub4 (1-74) and M1-Ub4 (WT) were very similar and almost within error of each other (Fig. 4 C and D). However, M1-Ub4 (1-73) was less effective at enhancing 450C PS than M1-Ub4 (1-74), and M1-Ub4 (1-72) had no effect on 450C PS. These results agree with our previous data on the compact K48-Ub4 chains, the Ub units of which interact with one another and impede the ability of the chain to promote UBQLN2 PS.
Next, we examined M1-Ub4 constructs with increased linker length between Ub units (Fig. 4 E–G). The overall SAXS scattering profile and Rg for an M1-Ub4 chain with a 10-residue linker [PS(GS)4] between each Ub unit were largely unchanged compared to M1-Ub4 (WT) despite the addition of these 10 residues (Fig. 4E). In line with these solution properties, the overall cloud point temperature curve of 450C with M1-Ub4 [PS(GS)4] is similar to M1-Ub4. We hypothesized that the [PS(GS)4] linker following G75G76 in each Ub is too flexible to elicit change in conformation of M1-Ub4. Therefore, we installed sequences A(EA3K)3A or A(EA3K)6A which are predicted to be helical (32) between the Ub units in a M1 (1-74) construct. SAXS data suggested that the structures of these M1-Ub4 constructs are significantly more extended compared to M1-Ub4 (1-76) as Rg values increased by nearly 10 Å (SI Appendix, Table S2). Experimental cloud point temperature curves of 450C with these polyUb hubs of longer linkers showed that these hubs were less effective at enhancing 450C PS (Fig. 4H). Using our theory to disentangle the architecture-dependent contributions to the inclusion energy, we found that the inclusion energy is minimized for hubs containing linkers of optimal lengths (Fig. 4I). Our theory is unable to determine the inclusion energy for M1-Ub4 (1-72) as the hub is unable to bind to 450C molecules, thus no longer meeting a critical assumption of our model (SI Appendix, Fig. S8G). Furthermore, our data suggest that wild-type M1 (1-76) polyUb chains are well optimized to enhance 450C PS at low Ub:450C concentrations, compared to other M1 polyUb chains where the linker is shortened or lengthened (Fig. 4).
Our data from the structurally distinct sets of M1-Ub4 and HT6-Ub constructs establish a general trend that compact hubs with unhindered binding ability are favored for inducing 450C PS (Fig. 4I and SI Appendix, Table S3). The inclusion energies for the M1-Ub4 and HT6-Ub constructs nearly overlap when plotted as a function of Rg. From these results, we can conclude that the geometry of the ligand hub is not as important as density/spacing of Ub units.
To determine whether full-length UBQLN2 responds to polyUb hubs in a similar manner as the shorter UBQLN2 450C construct, we obtained UBQLN2 cloud point temperature curves in the presence of linear M1 polyUb chains with longer linkers (A(EA3K)3A and A(EA3K)6A) as well as HT6-Ub hubs with different GS or PA linker lengths and combined these results with recent work on polyUb of different linkages (K48, K63, M1) (8) (SI Appendix, Fig. S9). Consistent with our 450C data above, hubs with linkers that are either too short or too long (have higher inclusion energy) do not enhance UBQLN2 PS compared to hubs with optimal linkers [e.g., HT6-(GS)4-Ub] (Fig. 4J and SI Appendix, Table S3). Our results reveal that the free energy cost of inserting hubs into the dense phase is dependent on the spacing of Ub binding sites. We propose that this Ub-Ub spacing is part of the Ub code that regulates PS via polyUb chains of different lengths and linkages, as well as by ubiquitination at multiple sites of target proteins (represented by the HT6-Ub constructs in this study). The different linkages used in the Ub code deliver biochemical information via Ub-Ub spacing.
Discussion
Using a combination of experiment and analytical theory, we determined that two properties of the multivalent polyUb hub contribute significantly to the mechanisms underlying ligand-mediated phase transitions (38) for polyUb-UBQLN2 condensates: 1) effective linker length between binding sites within the hub and 2) binding affinity between the hub and phase-separating driver molecules. We find that the specific geometry of the ligand hub matters less than the binding site spacing in hubs. These tunable features of multivalent ligand hubs modulate multicomponent PS as demonstrated for the polyUb-UBQLN2 system. Furthermore, the natural linker length between Ub units in M1-linked polyUb chains appears optimized to maximize PS, with the other naturally occurring polyUb chains reducing both effective Ub-Ub distance and PS due to their compact conformations (e.g., K48-Ub4).
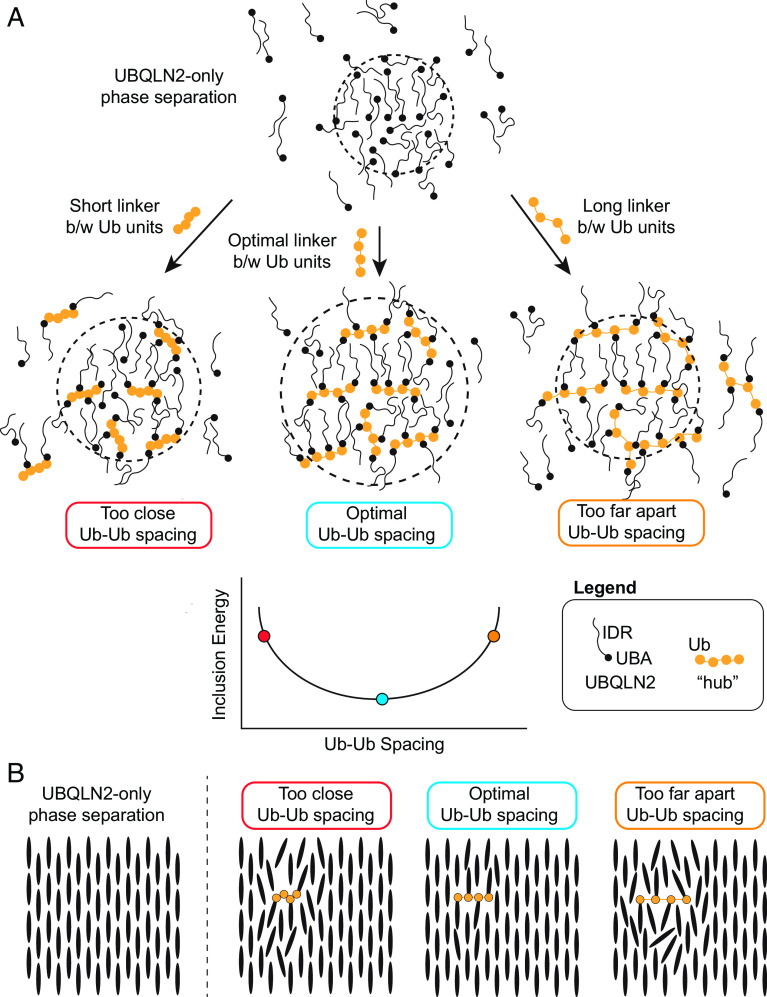
The analytical theory developed here uses the concepts of polyphasic linkage (5, 24, 25) to determine how ligand hubs impact PS of driver molecules. The theory explains preferential inclusion/exclusion of ligand hubs in condensates. For ligand incorporation into the dense phase of driver condensates, there is an energetic penalty from the ligand disrupting the driver-only interactions in the dense phase (Fig. 5). This energetic penalty is tuned by the physicochemical properties of the ligand hub, including valency, binding site spacing, and binding affinity. Despite the energetic penalty, introducing multivalent polyUb ligand hubs into UBQLN2 condensates still promotes UBQLN2 PS relative to UBQLN2 PS in the absence of a polyUb hub ligand. This is because the multivalency of the polyUb hub enables the accumulation of multiple sets of favorable binding energies between each Ub unit and UBQLN2. However, when spacing of the Ub units are too close or too far, polyUb hubs and UBQLN2 molecules are not in a favorable arrangement, thus destabilizing PS (Fig. 5B). This argument suggests that nonideal polyUb hubs (i.e., hubs with nonoptimal Ub-Ub spacing) may introduce voids and/or steric clashes among the UBQLN2 molecules such that they are not in their ideal arrangement for both a) interaction with the polyUb ligand, and b) maintaining the condensate state.
Fig. 5.
Introducing a ligand into condensates incurs an energetic penalty (inclusion energy) that can be reduced by optimizing the ligand architecture. (A) Adding a ligand to the condensate disrupts the optimal binding of the surrounding driver (UBQLN2) molecules. This disruption can be minimized if the binding sites on the ligand are commensurate with the spacing of the surroundings. This optimal spacing minimizes the inclusion energy. In the presence of any ligand hub, PS of the driver molecules is enhanced (depicted here as a larger droplet size) and less UBQLN2 is found in the dilute phase. This represents the experimental observation that all of these multivalent hubs still promote PS of the driver molecules. (B) This schematic illustrates only the dense phase of the condensates in (A). The coarse-grained depiction of UBQLN2 molecules as rigid rods omits the complication of polymer disorder to highlight the disruptions in the density and bonding network.
Here, we used two sets of designed polyUb hubs (HT6-Ub and M1-Ub4 chains of different Ub-Ub spacings) to dissect hierarchical contributions (39) to PS of driver/hub systems. We have not perturbed UBQLN2 (driver) molecules but focus exclusively on the effects of the ligand/hub. Despite the very different architecture of how the Ub units are arranged on the hubs (spider-like vs. beads on a string for HT6-Ub and M1-Ub4, respectively), several themes emerged from our data. First, this work and our prior report (8) showed that crowding the Ub units too close together disfavors UBQLN2 PS. Second, increased distance between Ub units also disfavors UBQLN2 PS (Fig. 4). Third, because of these competing trends, there is an optimal spacing of Ub units that maximizes the propensity for PS (Fig. 4). Fourth, the optimal ligand hub size and Ub-Ub spacing is similar for both full-length UBQLN2 and 450C constructs (Fig. 4 I and J). These data demonstrate the versatility of different ligand hub designs on effecting similar outcomes on PS of a scaffold like UBQLN2. Specifically, a multi-monoubiquitinated substrate (e.g., HT6-Ub) could take the place of a polyubiquitin chain and promote UBQLN2 PS over a wide range of Ub:UBQLN2 ratios. Increasing the GS/PA linker length in HT6-Ub mimics the effect of longer intrinsically disordered regions in ubiquitinated substrates on the ability to modulate shuttle protein PS.
In the UBQLN2 and polyUb multicomponent system, both macromolecules colocalize into the same condensates (8). However, this need not be the case for other multicomponent phase-separating systems. Additional complexity in phase behaviors can also arise from ligand-mediated phase transitions that are not explicitly modeled here. For example, multiphase condensates could form and/or condensates could exhibit different material and rheological properties as seen for nucleic acid/protein condensates (40–42).
An important caveat is that our results do not exclude the possibility that UBQLN2 or polyUb undergo structural/conformational changes during the phase transition to the condensed state. Indeed, UBQLN2 may undergo a conformational change upon binding Ub, as we recently demonstrated in ref. 43. These conformational changes may be subsumed within the energetic differences of the inclusion energy values for full-length UBQLN2 and 450C (Fig. 4).
Conceptually, our work shows that ligand hubs provide a sensitive mechanism to regulate condensate formation: 1) small differences in spacing between binding sites (e.g., Ub-Ub) can be magnified through driver PS (e.g., UBQLN2), and 2) PS of a driver can be driven to lower concentration with a ligand hub. Similarly, recent work on Speckle-type POZ protein (SPOP), a tumor suppressor that self-oligomerizes, has shown that SPOP cancer mutations can potentiate phase separation of SPOP with the DAXX substrate (44). DAXX has a low intrinsic tendency to phase separate, but SPOP can act as a hub that brings multiple DAXX molecules together to promote DAXX PS (45, 46). Wild-type SPOP requires a high DAXX:SPOP ratio to form the liquid-like condensates associated with DAXX ubiquitination. In contrast, cancer-associated SPOP mutations reduce SPOP oligomerization leading to more favorable arrangements between SPOP and DAXX to promote liquid-like SPOP/DAXX condensates at lower concentrations than for wild-type SPOP. In other words, cancer mutations optimize the SPOP hub to provide conditions that optimally promote DAXX PS. In a similar way, the polyUb hub optimizes UBQLN2 interactions to favor PS, but less optimal spacing between Ub units on the hub will disfavor UBQLN2 PS.
Interestingly, the Ub-Ub spacing in naturally occurring M1-linked polyUb chains is generally optimal at promoting UBQLN2 PS (Fig. 4 C and F and SI Appendix, Fig. S9). This observation suggests that the evolutionarily conserved length and sequence of the Ub C-terminal tail (27, 47), aside from known roles in Ub conjugation to proteins, is also well-poised for multivalent interactions with Ub-binding proteins. This insight is important given recent evidence that M1-linked polyUb chains drive multicomponent PS of other Ub-binding proteins such as NEMO and p62 (11, 12, 17, 19). Linker shortening or lengthening by the removal or addition of multiple residues on the Ub C-terminal tail (Ub residues 73–76) reduced the ability of the polyUb hub to maximally promote UBQLN2 PS. These results suggest that the Ub code takes advantage of the optimal Ub-Ub spacing and the sensitivity of the high density (short spacer) side of the inclusion energy curve (Fig. 4 I and J) in regulating condensate formation. The spacing within natural M1-linked polyUb chains is optimized for Ub-binding domains of highly diverse architectures, specifically a compact three-helix UBA domain located at one end of p62 and UBQLN2 in contrast to a long coiled-coil region in NEMO (48, 49). Moreover, M1-linked polyUb chains are optimized to both enhance the PS of systems that can phase separate on their own (UBQLN2) and drive the PS of systems that cannot (p62, NEMO). The heterotypic interactions lower the intrinsic csat (saturation concentration) of the Ub-binding partner, permitting regulation of condensate assembly/disassembly (5, 50). These data suggest that the architecture of M1-linked polyUb chains has favorable properties (flexibility and spacing of Ub binding sites) and may have been evolutionarily tuned to promote PS of systems driven by either homotypic or heterotypic interactions.
Aside from Ub, other Ub-like modifiers of proteins, such as SUMO, UFM1, and NEDD8, form chains of different linkages or hybrid chains (i.e., Ub-SUMO or Ub-NEDD8 chains) (51, 52). SUMO and UFM1 contain unstructured regions of different lengths on either or both of their termini. Subsequently, the spacings between SUMO units on a chain are larger than those between Ub units. We showed that the UBA domain of UBQLN2 is unable to interact with M1-Ub4 (1-72) or M1-Ub4 (1-73) due to insufficient spacing between the Ub units. Just like the naturally occurring M1-Ub4 (1-76) is optimized to accommodate several UBQLN2 molecules and enhance PS, the spacings between the units of SUMO chains, depending on linkage types, might have been optimized for binding to their partners (e.g., proteins with multiple SIM modules). Protein polySUMOylation has been shown to regulate the assembly and dissolution of various biomolecular condensates, such as PML bodies, nucleoli, and stress granules (53). These multivalent Ub, UFM1, or SUMO chains could communicate their cellular code through modulating condensate assembly/disassembly. Therefore, knowledge of how changes in the spacing between Ub units affect UBQLN2 phase separation is also relevant to these Ub-like modifiers and their respective binding partners. This framework can be further extended to quantify the effects of other posttranslational modifications on phase-separating systems (54, 55).
In summary, ligand hubs offer a tunable knob for modulating condensate assembly and disassembly. The interplay between ligand hub architecture and driver molecule arrangement leverages the sensitivity of phase separation in amplifying small differences, such as the distance between binding sites on a ligand hub. As a result, the total effect on condensates cannot be considered a sum of individual parts (e.g., specific interactions including driver–driver and ligand–driver) but rather an emergent property of the condensates (38). We expect that polyUb chains (homotypic linkages, mixed linkages, and branched) and other polyUb-like systems (e.g., polyPARylation) use their multivalent architecture to dynamically regulate condensate properties and functions. For Ub and Ub-like proteins, we believe that the effect of these ligand hubs is the mechanistic underpinning of the Ub code.
Materials and Methods
All protein constructs (SI Appendix, Table S4) used for this study were expressed in Escherichia coli bacteria cells. Details for cloning, expression, and purification of all protein constructs are provided in SI Appendix. Full-length UBQLN2 or UBQLN2 450C were mixed with different Ub hubs and subjected to phase separation experiments using temperature-based spectrometry experiments. Protocols for phase separation turbidity measurements, cloud point temperature determination, and dilute/dense phase concentration measurements are fully described in SI Appendix. Designed Ub hubs were characterized structurally and dynamically using NMR experiments for chemical shifts and relaxation, respectively. Additional experiments including SEC-MALS-SAXS (Dataset S1), and computational modeling (SI Appendix, Table S5) were used to determine the structures of designed ubiquitin ligand hubs. Analytical theory for inclusion energy determination of turbidity curves is fully described in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
This work was supported by NIH R01GM136946 (all protein purifications, turbidity assays, structural modeling, and NMR experiments) to C.A.C. and NIH R01GM141235 (theoretical modeling) to J.D.S. NMR data were acquired on an 800 MHz NMR spectrometer funded by NIH-shared instrumentation grant 1S10OD012254. This research used resources of the Advanced Photon Source (APS), a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was supported by NIH grant P30 GM138395. Use of the Pilatus 3 1 M detector was provided by grant 1S10OD018090-01 from NIGMS. We acknowledge support and assistance from Dr. Jesse Hopkins and Dr. Maxwell Watkins on collecting SEC-MALS-SAXS data at APS. Suzanne Enos and Antara Chaudhuri were supported by NSF REU grant CHE1950802. We acknowledge Yiran Yang for assistance in purifying some of the proteins for this project. We thank Susan Krueger, Tanja Mittag, and the Condensate Colloquium series for scientific discussions on this project.
Author contributions
S.K.K.G., T.P.D., and C.A.C. designed research; S.K.K.G., T.P.D., S.E.E., A.C., and C.A.C. performed research; J.D.S. contributed new reagents/analytic tools; S.K.K.G., T.P.D., J.D.S., and C.A.C. analyzed data; and S.K.K.G., T.P.D., J.D.S., and C.A.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. N.W. is a guest editor invited by the Editorial Board.
Contributor Information
Jeremy D. Schmit, Email: schmit@phys.ksu.edu.
Carlos A. Castañeda, Email: cacastan@syr.edu.
Data, Materials, and Software Availability
SAXS data for HT6-Ub & M1-Ub4 ligand hubs are deposited in SASBDB: https://www.sasbdb.org/project/1831/ (56). Raw data for figures and scripts are deposited at https://doi.org/10.5281/zenodo.8199908 (57). All other data supporting the findings of this study are included in this article and/or supporting information.
Supporting Information
References
- 1.Alberti S., Hyman A. A., Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Holehouse A. S., Pappu R. V., Functional implications of intracellular phase transitions. Biochemistry 57, 2415–2423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ditlev J. A., Case L. B., Rosen M. K., Who’s in and Who’s Out—Compositional control of biomolecular condensates. J. Mol. Biol. 430, 4666–4684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruff K. M., Dar F., Pappu R. V., Polyphasic linkage and the impact of ligand binding on the regulation of biomolecular condensates. Biophys. Rev. 2, 021302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruff K. M., Dar F., Pappu R. V., Ligand effects on phase separation of multivalent macromolecules. Proc. Natl. Acad. Sci. U.S.A. 118, e2017184118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa J. R., et al. , Liquid network connectivity regulates the stability and composition of biomolecular condensates with many components. Proc. Natl. Acad. Sci. U.S.A. 117, 13238–13247 (2020). 10.1073/pnas.1917569117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dao T. P., et al. , Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains. EMBO Rep. 23, e55056 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders D. W., et al. , Competing protein–RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang P., et al. , G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du M., Ea C.-K., Fang Y., Chen Z. J., Liquid phase separation of NEMO induced by polyubiquitin chains activates NF-κB. Mol. Cell 82, 2415–2426.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun D., Wu R., Zheng J., Li P., Yu L., Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda S., et al. , Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Komander D., Rape M., The Ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Tozawa T., et al. , Ubiquitination-coupled liquid phase separation regulates the accumulation of the TRIM family of ubiquitin ligases into cytoplasmic bodies. PLoS ONE 17, e0272700 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vamadevan V., Chaudhary N., Maddika S., Ubiquitin assisted phase separation of Dishevelled-2 promotes Wnt signaling. J. Cell Sci. 135, jcs260284 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Zaffagnini G., et al. , p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, e98308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turco E., et al. , Reconstitution defines the roles of p62, NBR1 and TAX1BP1 in ubiquitin condensate formation and autophagy initiation. Nat. Commun. 12, 5212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goel S., et al. , Linear ubiquitination induces NEMO phase separation to activate NF-κB signaling. Life Sci. Alliance 6, e202201607 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dao T. P., Castañeda C. A., Ubiquitin-modulated phase separation of shuttle proteins: Does condensate formation promote protein degradation? BioEssays 42, 2000036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dao T. P., et al. , Ubiquitin modulates liquid–liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng T., Galagedera S. K. K., Castañeda C. A., Previously uncharacterized interactions between the folded and intrinsically disordered domains impart asymmetric effects on UBQLN2 phase separation. Protein Sci. 30, 1467–1481 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D., Raasi S., Fushman D., Affinity makes the difference: Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 377, 162–180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyman J., Gill S. J., Ligand-linked phase changes in a biological system: Applications to sickle cell hemoglobin. Proc. Natl. Acad. Sci. U.S.A. 77, 5239–5242 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyman J., Binding and Linkage: Functional Chemistry of Biological Macromolecules (University Science Books, 1990). [Google Scholar]
- 26.Pickart C. M., Fushman D., Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Vijay-Kumar S., Bugg C. E., Cook W. J., Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 (1987). [DOI] [PubMed] [Google Scholar]
- 28.Beal R. E., Toscano-Cantaffa D., Young P., Rechsteiner M., Pickart C. M., The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry 37, 2925–2934 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Jones H. B., Dao T. P., Castañeda C. A., Single amino acid substitutions in stickers, but not spacers, substantially alter UBQLN2 phase transitions and dense phase material properties. J. Phys. Chem. B 123, 3618–3629 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Bhandari K., Cotten M. A., Kim J., Rosen M. K., Schmit J. D., Structure-function properties in disordered condensates. J. Phys. Chem. B 125, 467–476 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigorev V., Wingreen N. S., Zhang Y., Conformational entropy of intrinsically disordered proteins bars intruders from biomolecular condensates. bioRxiv [Preprint] (2023). 10.1101/2023.03.03.531005 (Accessed 15 July 2023). [DOI]
- 32.Chen X., Zaro J. L., Shen W.-C., Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 65, 1357–1369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirdita M., et al. , ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis J. E., Raghunandan S., Nanda H., Krueger S., SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput. Phys. Commun. 183, 382–389 (2012). [Google Scholar]
- 35.Jussupow A., et al. , The dynamics of linear polyubiquitin. Sci. Adv. 6, eabc3786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fushman D., Varadan R., Assfalg M., Walker O., Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Prog. Nucl. Magn. Reson. Spectrosc. 44, 189–214 (2004). [Google Scholar]
- 37.Castañeda C. A., et al. , Linkage-specific conformational ensembles of non-canonical polyubiquitin chains. Phys. Chem. Chem. Phys. 18, 5771–5788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pappu R. V., Cohen S. R., Dar F., Farag M., Kar M., Phase transitions of associative biomacromolecules. Chem. Rev. 123, 8945–8987 (2023), 10.1021/acs.chemrev.2c00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmit J. D., Feric M., Dundr M., How hierarchical interactions make membraneless organelles tick like clockwork. Trends Biochem. Sci. 46, 525–534 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee P. R., Milin A. N., Moosa M. M., Onuchic P. L., Deniz A. A., Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. 56, 11354–11359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alshareedah I., Moosa M. M., Raju M., Potoyan D. A., Banerjee P. R., Phase transition of RNA–protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. U.S.A. 117, 15650–15658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu T., Spruijt E., Multiphase complex coacervate droplets. J. Am. Chem. Soc. 142, 2905–2914 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robb C. G., Dao T. P., Ujma J., Castañeda C. A., Beveridge R., Ion Mobility mass spectrometry unveils global protein conformations in response to conditions that promote and reverse liquid–liquid phase separation. J. Am. Chem. Soc. 145, 12541–12549 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabri N., et al. , Reduction of oligomer size modulates the competition between cluster formation and phase separation of the tumor suppressor SPOP. bioRxiv [Preprint] (2023). 10.1101/2023.02.11.528154 (Accessed 15 April 2023). [DOI] [PMC free article] [PubMed]
- 45.Bouchard J. J., et al. , Cancer mutations of the tumor suppressor SPOP disrupt the formation of active. Phase-separated compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmit J. D., Bouchard J. J., Martin E. W., Mittag T., Protein network structure enables switching between liquid and gel states. J. Am. Chem. Soc. 142, 874–883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickart C. M., Eddins M. J., Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta BBA Mol. Cell Res. 1695, 55–72 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Lo Y.-C., et al. , Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshikawa A., et al. , Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett. 583, 3317–3322 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Saha S., et al. , Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cappadocia L., Lima C. D., Ubiquitin-like protein conjugation: Structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez Berrocal D. A., Witting K. F., Ovaa H., Mulder M. P. C., Hybrid chains: A collaboration of ubiquitin and ubiquitin-like modifiers introducing cross-functionality to the ubiquitin code. Front. Chem. 7, 931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keiten-Schmitz J., Röder L., Hornstein E., Müller-McNicoll M., Müller S., SUMO: Glue or solvent for phase-separated ribonucleoprotein complexes and molecular condensates? Front. Mol. Biosci. 8, 673038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hofweber M., Dormann D., Friend or foe—Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brady J. P., et al. , Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. U.S.A. 114, E8194–E8203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galagedera S. K. K., Castañeda C. A., Decoding optimal ligand design for multicomponent condensates. SASBDB. https://www.sasbdb.org/project/1831/. Deposited 17 March 2023.
- 57.Galagedera S. K. K., Castañeda C. A., Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units. Zenodo. 10.5281/zenodo.8199908. Deposited 25 September 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
SAXS data for HT6-Ub & M1-Ub4 ligand hubs are deposited in SASBDB: https://www.sasbdb.org/project/1831/ (56). Raw data for figures and scripts are deposited at https://doi.org/10.5281/zenodo.8199908 (57). All other data supporting the findings of this study are included in this article and/or supporting information.