Summary
Background
The true incidence of SARS-CoV-2 infection in Costa Rica was likely much higher than officially reported, because infection is often associated with mild symptoms and testing was limited by official guidelines and socio-economic factors.
Methods
Using serology to define natural infection, we developed a statistical model to estimate the true cumulative incidence of SARS-CoV-2 in Costa Rica early in the pandemic. We estimated seroprevalence from 2223 blood samples collected from November 2020 to October 2021 from 1976 population-based controls from the RESPIRA study. Samples were tested for antibodies against SARS-CoV-2 nucleocapsid and the receptor-binding-domain of the spike proteins. Using a generalized linear model, we estimated the ratio of true infections to officially reported cases. Applying these ratios to officially reported totals by age, sex, and geographic area, we estimated the true number of infections in the study area, where 70% of Costa Ricans reside. We adjusted the seroprevalence estimates for antibody decay over time, estimated from 1562 blood samples from 996 PCR-confirmed COVID-19 cases.
Findings
The estimated total proportion infected (ETPI) was 4.0 times higher than the officially reported total proportion infected (OTPI). By December 16th, 2021, the ETPI was 47% [42–52] while the OTPI was 12%. In children and adolescents, the ETPI was 11.0 times higher than the OTPI.
Interpretation
Our findings suggest that nearly half the population had been infected by the end of 2021. By the end of 2022, it is likely that a large majority of the population had been infected.
Funding
This work was sponsored and funded by the National Institute of Allergy and Infectious Diseases through the National Cancer Institute, the Science, Innovation, Technology and Telecommunications Ministry of Costa Rica, and Costa Rican Biomedical Research Agency-Fundacion INCIENSA (grant N/A).
Keywords: SARS-CoV-2, COVID-19, Cumulative incidence, Natural immunity, Sero-epidemiological survey, Costa Rica, Latin America, Middle-income country
Research in context.
Evidence before this study
In a previous study we showed that in Costa Rica, reporting of COVID-19 deaths was generally accurate during the pandemic, even if possibly slightly underestimated. On the other hand, Costa Rica reported 0.57 million case (11% of the population) by the end of 2021. The officially reported total incidence of SARS-CoV-2 infection is likely to be an underestimate of the true incidence, as has also been observed in various countries including the United States, Spain and Germany.
We searched PubMed and SciELO, using the following search terms through June 2023: COVID-19, SARS-CoV-2 incidence, Costa Rica. We did not identify any analysis of cumulative incidence in Costa Rica based on seroprevalence. The only information available regarding infection toll in Costa Rica is the official data published by the Health Ministry and based on reported cases only. In other countries, seroprevalence was used to estimate the true infection toll.
Added value of this study
To get an accurate estimate of true infection numbers, we developed a novel statistical model accounting for all the available information: the official infection toll, and the serologic COVID-19 markers (anti-spike-RBD and anti-nucleocapsid antibodies) measured in population-base controls (N = 1999) enrolled in a longitudinal COVID-19 study (RESPIRA). We also adjusted the seroprevalence to take into account the decay of antibodies over time observed in cases (N = 999) and therefore estimate the rate of false negatives in controls. Based on this model, we calculated the number of people who got infected for each reported case. The true infection toll was estimated by age, sex, wave and region.
Implications of all the available evidence
Our main finding suggests that in total 1.7 million people got infected by the end of the third wave (December 2021) in the study area. This number is 4.0 times higher than the official infection count. As a result, 47% of the population had had COVID-19 before the omicron waves in the study area. We estimate that by now the majority of the population have already been infected. In addition, most people are vaccinated at least once, likely explaining the currently low rates in disease and hospitalization. The study also showed that even if children and adolescent infections were poorly reported, their infection rate was higher than in adults. Our model allows a more accurate description and understanding of the dynamics of the pandemic, will be useful to assess the impact of future waves, and can be used in other contexts. Indeed, the future impact of COVID-19 on public health depends not only on vaccination coverage and circulating SARS-CoV-2 variants, but also on the proportion of residents with past natural infection.
Introduction
The official number of COVID-19 cases in Costa Rica published by the Health Ministry through May 2023 was 1,228,000 cases, including 9350 deaths.1 By the end of 2021, there were 573,000 cases (11% of the population), including 7350 deaths.1 In a previous study, we showed that the official reported number of deaths was accurate in 2020–2021,2 but the reported number of infections is likely to be an underestimate, as in many other countries.3, 4, 5, 6, 7 Knowing the true cumulative incidence of SARS-CoV-2 infection in a country is important to understand the dynamics of the pandemic, the decline in the case fatality rate over time, and to anticipate future developments.8 Vaccination is a proven method to achieve immunity and is highly recommended, but both vaccination and past natural infection provide strong protection against COVID-19, particularly against future hospitalization and death.9 Hybrid protection, from both a past natural infection and vaccination appears to provide better protection against new variants than vaccination alone.10,11 Thus, the future impact of COVID-19 on public health depends not only on vaccination coverage and circulating SARS-CoV-2 variants, but also on the proportion of residents with past natural infection.
The population prevalence of SARS-CoV-2 anti-nucleocapsid and anti-spike serum IgG antibodies has been used to estimate cumulative burden of infection to SARS-CoV-2.3,5,12 Infected individuals are known to mount strong antibody responses to these proteins shortly after infection that persist for months and decline slowly after recovery.13 Furthermore, whereas anti-spike antibodies are markers of either natural infection or vaccination, anti-nucleocapsid antibodies are observed only after a natural infection, and thereby identify natural infection.
During the pandemic, each country defined its own testing guidelines. Some countries limited testing to the most severe cases, and others attempted to test all individuals in contact with each case. Therefore, variation in testing guidelines is a key factor for understanding differences in underreporting among countries. In Costa Rica, PCR or antigen testing was only performed for symptomatic individuals, but not for asymptomatic contacts, including household members. Thus, people with mild or no symptoms might not have been diagnosed if infected. In children, who tend to have fewer symptoms,14 the underestimation likely was more considerable. Moreover, only the first case of each infected household was targeted for testing, whereas the remaining members were considered infected through epidemiological linkage only if they reported symptoms during quarantine. Being considered an epidemiological link case (18% of reported cases in Costa Rica)1 triggered quarantine, possibly leading to underreporting of symptoms, especially for uninsured workers who do not receive paid sick leave.
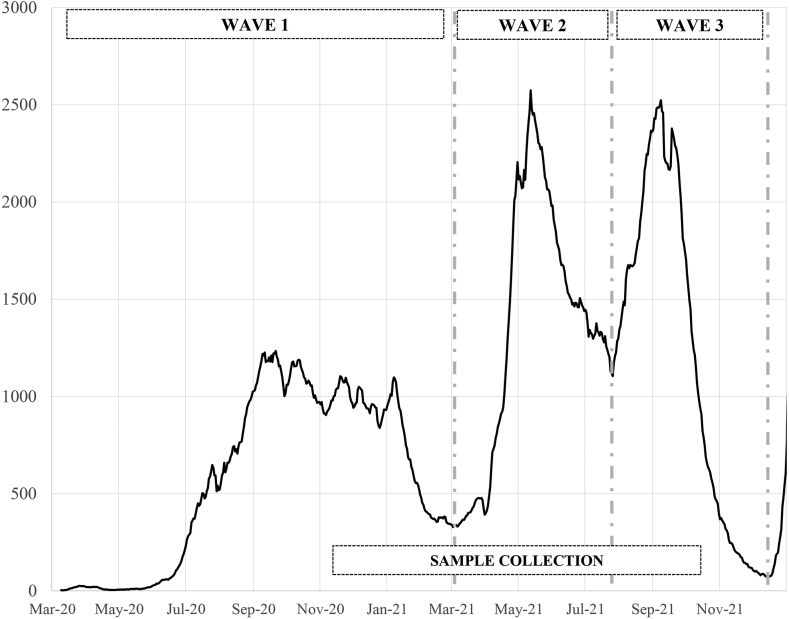
Costa Rica experienced three waves of COVID-19 in 2020 and 2021 (Fig. 1) associated with different viral variants, peaking in September 2020 (ancestral), May 2021 (Alpha and Gamma) and September 2021 (Delta). During the first wave there was no vaccination available. At the peak of the second wave, 89% of the 70+ year-olds, 52% of the 58–69-year-olds, and 4% of the 20–57-year-olds were vaccinated.15 At the peak of the third wave, 92% of the 58+ year-olds, 73% of the 20–57-year-olds, 52% of 12–19-year-olds were vaccinated, but children were not.15
Fig. 1.
Definition of the three pandemic waves, population-based samples collection period, and 14-day average number of daily cases in Costa Rica according to the Health Ministry. Sample collection in population-based controls (11/12/2020-10/15/2021).
The main objective of this study was to estimate the cumulative number of people who were infected with SARS-CoV-2 during the first three waves of the COVID-19 epidemic in Costa Rica (March 2020–December 2021) using the presence of serum antibodies as a biomarker of infection.
Methods
Data
RESPIRA cohort
RESPIRA is a population-based cohort study with active follow-up for two years.16,17 The main objective was to describe the immune response to SARS-CoV-2 infection and vaccination, its time course, epidemiologic and genetic determinants, and its protective efficacy against subsequent infection. RESPIRA was also conducted to estimate the population prevalence of infection over time and investigate determinants of acquisition, clinical presentation and household transmission.16,17 From November 2020 to October 2021, we recruited 999 PCR-confirmed COVID-19 cases selected from the national surveillance list; and 1999 matched population-based controls. The national surveillance list included all the cases diagnosed in Costa Rica in both the public and the private laboratories. By design, 30% of cases were recruited in the first two weeks after diagnosis and 70% between two weeks and 1.5 years. Cases were randomly selected within four age groups (0–19, 20–39, 40–59 and 60 or more years). Two population-based controls matched to cases on age, sex assigned at birth and minimal geostatistical unit (MGU) were randomly selected for each participating case. Population-based controls were recruited to represent the population of the area, so they were recruited regardless of self-reported prior COVID-19 infection. Thus, population-based controls are representative of the study population within strata defined by age, sex and MGU. Trained field workers visited households sequentially in defined patterns until identifying population-based controls meeting the matching criteria of the original case.16 Participation rates were 61% in cases, and 79% in population-based controls.
Study area
Cases and population-based controls were recruited from 48 of the 81 cantons of Costa Rica, where 70% (3.6 million persons) of the population reside. 68% of the participants reside in the metropolitan region known as Valle Central, i.e., the cantons around the capital. The non-metropolitan participants reside in Guanacaste, Puntarenas, and Upala.
Period
During the study period (March 2020–December 2021), three COVID-19 waves were observed. The end of each wave (and thus the beginning of the following wave) was defined using the nadir of infections following a peak according to the Health Ministry data. Using this definition, the first wave was from March 1st, 2020 to March 3rd, 2021, the second wave from March 4th, 2021 to July 26th, 2021 and the third wave from July 27th, 2021 to December 16th, 2021 (Fig. 1).
RESPIRA sampling stopped in mid-October 2021, but we extrapolated the number of people who got infected for each detected case based on our model and the Health Ministry data to the end of the third wave.
Informed consent
The study was approved by the ethical committee of the Central Social Security (CCSS) IRB (Protocol number: R020- SABI-00261). The study is observational and was considered of minimal risk to participants. All participants or their legal representatives signed informed consent or assent (for children 12–17 years old) in the presence of a witness, as mandated by Costa Rican law.
Antibody testing
3785 blood samples from 2972 individuals (1976 population-based controls, 996 PCR-confirmed cases) were tested for antibodies against SARS-CoV-2 nucleocapsid and the receptor-binding-domain (RBD) of the spike proteins. 2961 blood samples were collected at enrollment and 824 during follow-up. The 2223 samples from 1976 controls (up to 2 samples per participant) were used to estimate SARS-CoV-2 seroprevalence in the study area. The 1562 samples from 996 PCR-confirmed cases were used to estimate the decay of seropositivity after SARS-CoV-2 infection.
Analysis of antibody responses to SARS-CoV-2 was performed at the German Cancer Research Center (DKFZ) Heidelberg as described by Butt et al.18 (Supplementary Materials, Appendix 1). A blood sample was considered seropositive (SP) if anti-nucleocapsid (anti-N) antibodies were above 4000 MFI (Median Fluorescence Intensity) and anti-spike-RBD (anti-S1-RBD) antibodies were above 513 MFI. These thresholds were defined using 96 pre-pandemic samples (Supplementary Materials, Appendix 2). Using both anti-N and anti-S1-RBD produces a low rate of false positives.
Health Ministry data
Cumulative number of infections since the beginning of the pandemic, according to public anonymized Health Ministry data,19 were computed by day within 80 categories defined by sex (men/women), age-group (0–9, 10–19,…, 60–69, 70+), and socioeconomic characteristics of the canton (metropolitan region: low/intermediate/high income cantons; non-metropolitan region: low/intermediate income cantons). After September 5, 2021, sex data were not available in the Health Ministry database. Thus, we assumed a similar age-sex distribution before and after this date.
Official daily number of infections (ond) and official total proportion infected (OTPI)
We used the Health Ministry database to calculate the official number of cases on day d (ond), and the cumulative proportion of people who were identified as COVID-19 cases on or before day d (OTPId). OTPI was calculated for each stratum defined by age-group a, sex s, canton group c, and day of the sample collection d and was associated to each population-based control according to his/her characteristics (a, s, c, d) as:
| (1) |
where popa,s,c is the population total in the age-sex-canton group from the National Institute of Statistics and Census (INEC).20 ona,s,c,j is the daily official number of cases on day j in (a,s,c).
We assumed no repeat infections when defining OTPI because reinfections represented only 0.2% of all infections in the official data during waves 2 and 3.21
The model
The main objective was to estimate the SARS-CoV-2 infection total in Costa Rica by modelling the number of people who got infected for each case reported to the official national registry. Multiplying the officially reported prevalence of infection by this number yielded an estimate of the total number of people infected in the study population. To achieve this objective, we estimated the relation between the seropositivity to anti-N and anti-S1-RBD antibodies in the RESPIRA population-based controls and the official infection total (Health Ministry database) the day of sample collection. In order to take into account that some people do not have detectable antibodies despite having been infected in the past, the decay of antibodies over time after infection, and therefore of seropositivity, was included in the model. The antibody decay curve was estimated using the sub-set of RESPIRA PCR-confirmed cases.
Estimation of the number of people who got infected for each detected case
The probability for a cohort member i to have a positive serology on day di of the epidemic depends on the probabilities of being infected in the days d preceding and including day di, and the probability that a previously infected person is seropositive t days after infection (with t = di−d). Indeed, because the probability of seropositivity increases in the first days after infection and then decreases with time since infection, the prevalence of seropositivity on a given day does not capture all individuals previously infected. To be precise,
| (2) |
where is the true probability of infection on day d for someone with age a, sex s, and canton group c.
We made the key assumption that:
| (3) |
namely that the actual probability of infection on day d equals the officially reported proportion infected (Health Ministry database) times a factor that depends on age, sex, and canton group. We assumed this factor to be constant over time and checked the sensitivity of our results to this assumption (Supplementary Materials, Appendix 3A). The factor exp() represents the number of people who got infected for each officially reported case.
As a result:
| (4) |
We used logistic regression on PCR-confirmed cases to estimate the probability wa,s,t that someone in age group a and of gender s would be seropositive t days after infection:
| (5) |
where , , are the coefficients associated respectively with age a, sex s and time since infection t categorized into six-time intervals (0–14 days, 15–29, 30–89, 90–179, 180–364, 365+). Vaccination status and comorbidities were not included in the model because they were not associated with a higher probability across time of being seropositive among PCR-confirmed cases, given our definition of seropositivity including the presence of anti-N antibodies. Results from other models including interactions were not shown as they led to similar estimates of numbers infected as the main effects model in Equation (5).
Finally, based on Equations (4), (5), and assuming can be represented by main effects, we fit the Bernoulli seropositivity status of each control with a generalized linear model with a log-link, a binomial distribution, and an offset :
| (6) |
Estimates of and are in Supplementary Material, Table S1. Here, the probability that control subject i with characteristics age ai, sex si, and canton group ci and whose blood sample was collected on day is seropositive is where represents the officially reported weighted proportion infected (Equation (4)).
Estimated total proportion infected (ETPI)
The estimated total proportion infected is the proportion of the population which has been infected at or before day d. It can be estimated from the daily official number of infections (Health Ministry data) and the number of people who got infected for each detected case:
| (7) |
where .
Estimation of the confidence intervals
The confidence intervals were estimated using empirical bootstrap quantiles (1000 replications), with a bootstrap resampling at the participant level. At each replication, both the cases used to estimate the probability to be seropositive as a function of time since infection, and the population-based controls used to estimate the number of people who got infected for each detected case, were resampled.
Software
Statistical analyses were computed using STATA@18.
Role of the funding source
The funding sources did not have any role in the study design, data collection, data analysis, interpretation, writing of the report.
Results
Table 1 presents the characteristics of the population-based controls who provided the 2223 blood samples evaluated. 414 of these samples were collected during the first wave, 889 during the second wave, and 920 during the third wave.
Table 1.
Characteristics of the population-based controls associated with each blood sample (N = 2223).
| N (%) | Time of sample collection |
|||
|---|---|---|---|---|
| Wave 1 |
Wave 2 |
Wave 3 |
||
| N (%) | N (%) | N (%) | ||
| Total | 2223 | 414 (19) | 889 (40) | 920 (41) |
| Age | ||||
| <20y | 392 (18) | 39 (10) | 172 (44) | 181 (46) |
| 20–39y | 658 (30) | 137 (21) | 248 (38) | 273 (41) |
| 40–59y | 671 (30) | 116 (17) | 266 (40) | 289 (43) |
| 60y+ | 502 (23) | 122 (24) | 203 (40) | 177 (35) |
| Sex | ||||
| Men | 1037 (47) | 191 (18) | 401 (39) | 445 (43) |
| Women | 1186 (53) | 223 (19) | 448 (41) | 475 (40) |
| Region | ||||
| Non-metropolitan region | 714 (32) | 95 (13) | 284 (40) | 335 (47) |
| Metropolitan region | 1509 (68) | 319 (21) | 605 (40) | 585 (39) |
20% of the blood samples from population-based controls were seropositive (Table 2). According to the characteristics of the population-based controls, by the day of the blood sample collection, the official total proportion infected was 7%. As a result, crude seroprevalence was 2.7-fold higher [95% Confidence Interval: 2.5–3.0] than expected according to the official reports.
Table 2.
Seroprevalence, official total proportion infected, and their ratio overall and by characteristics of population-based controls (N = 2223).
| Seroprevalence (SP) N (%) | Official Total Proportion Infected (OTPI) % | Ratio SP/OTPI [95% CI] | |
|---|---|---|---|
| Total | 442 (20) | 7 | 2.7 [2.5–3.0] |
| Period of the sample collection | p < 0.01 | ||
| Wave 1 | 57 (14) | 4 | 3.2 [2.5–4.1] |
| Wave 2 | 136 (15) | 6 | 2.6 [2.2–3.0] |
| Wave 3 | 249 (27) | 10 | 2.7 [2.4–3.0] |
| Age | p < 0.01 | ||
| <20y | 86 (22) | 3 | 6.7 [5.5–8.0] |
| 20–39y | 129 (20) | 10 | 2.0 [1.7–2.3] |
| 40–59y | 157 (23) | 9 | 2.6 [2.2–2.9] |
| 60y+ | 70 (14) | 5 | 2.9 [2.3–3.5] |
| Sex | p = 0.99 | ||
| Men | 206 (20) | 7 | 2.7 [2.4–3.1] |
| Women | 236 (20) | 7 | 2.7 [2.4–3.0] |
| Region | p = 0.04 | ||
| Non-metropolitan region | 160 (22) | 7 | 3.2 [2.8–3.7] |
| Metropolitan region | 282 (19) | 7 | 2.5 [2.2–2.8] |
OTPI: mean official total proportion infected in the study population by the day of the sample collection (expected proportion of seropositivity based on day of sample collection, age, sex and region, according to the cumulative proportion of people infected according to the Health Ministry data). SP: crude seroprevalence in population-based controls (without correction for antibody decay). p, chi-square test between the variable and seroprevalence. Twenty percent of the blood samples from population-based controls were seropositive. The official total proportion infected for a given age, sex and canton group, OTPIa,s,c,d, was averaged over the distribution of (a,s,c,d) in controls, where d is the day of sampling the control, to produce the estimate OTPI = 7%. Other values of OTPI were obtained similarly for selected values of age, sex, canton group and wave of the epidemic.
Seroprevalence was 27% in blood samples collected during wave 3, compared to 15% and 14% in blood samples collected in wave 2 and 1 respectively. Seroprevalence in children and adolescents was 6.7-fold higher [5.5–8.0] than the OTPI. This ratio is higher than in older age groups: 2.0 [1.7–2.3] in 20–39-year-olds, 2.6 [2.2–2.9] in 40–59-year-olds, and 2.9 [2.3–3.5] in 60+ year-olds. Seroprevalence was lower in 60+ year-olds compared to 20–59-year-olds, in agreement with the OTPI. However, seroprevalence was similar in children/adolescents and in adults, whereas the official total proportions infected were significantly lower in children and adolescents (3%) compared to adults (9–10%). Seroprevalence was similar in men and in women (20%), in agreement with the OTPI. Seroprevalence was slightly higher in the non-metropolitan region (22%) compared to the metropolitan region (19%), whereas the OTPI were similar in both regions (7%).
The proportion seropositive among PCR-confirmed cases peaks at 30–89 days after a PCR-positive test and declines slowly thereafter, both in men and women and at all ages of PCR-positive diagnosis (Supplementary Materials, Table S2). Based on this pattern of seropositivity following infection in cases, we estimated that 74% of the population-based controls who got infected were seropositive when the blood sample was collected (Fig. 2). This proportion varied by sample collection wave because the mean time between infection and blood collection was lower in wave 1 compared to waves 2 and 3 (Supplementary Materials, Table S3). Thus, 83% of samples from wave 1 compared to 73% and 72% of samples from waves 2 and 3, respectively, were still seropositive when the blood sample was collected (Supplementary Materials, Figure S1). Seropositivity after infection was significantly lower in children and adolescents compared to adults and the elderly (Fig. 2). Thus, the seroprevalence underestimated the cumulative incidence in children and adolescents more than in adults.
Fig. 2.
Estimated probability to be seropositive the day of the sample collection among those previously infected by sex and age (N = 2223). Example: On average, the probability to be seropositive on collection day for a previously infected 60–69y woman is 77%. Estimated probability of seropositivity was calculated using W in Equation S1 (Supplementary Material, Appendix 4).
By the end of the first and second waves, the estimated infection total (ETPI) in our study population (representing 70% of the population of Costa Rica) was 16% [14–17] and 32% [29–35] respectively, corresponding to 0.57 [0.51–0.63] and 1.15 [1.03–1.28] million cases, respectively (Table 3). ETPIs were similar in men and in women; in the metropolitan region and in the non-metropolitan region and in children/adolescents and adults. ETPIs were lower in people 60 years and older.
Table 3.
Official (OTPI) and estimated (ETPI) total proportion infected at the end of each pandemic wave and participant characteristics.
| March 3rd, 2021 |
July 26th, 2021 |
December 16th, 2021 |
Ratioa ETPI/OTPI [95% CI] | ||||
|---|---|---|---|---|---|---|---|
| (End of wave 1) |
(End of wave 2) |
(End of wave 3) |
|||||
| OTPI (%) | ETPI % [95% CI] | OTPI (%) | ETPI % [95% CI] | OTPI (%) | ETPI % [95% CI] | ||
| Total | 4 | 16 [14–17] | 8 | 32 [29–35] | 12 | 47 [42–52] | 4.0 [3.6–4.5] |
| Men | 4 | 15 [13–18] | 8 | 31 [26–35] | 12 | 45 [38–53] | 3.9 [3.3–4.5] |
| Women | 4 | 16 [14–19] | 8 | 33 [28–38] | 12 | 48 [42–56] | 4.1 [3.6–4.8] |
| <20y | 2 | 16 [13–20] | 3 | 36 [28–45] | 5 | 60 [47–75] | 11.0 [8.6–13.8] |
| 20–39y | 6 | 16 [13–19] | 12 | 31 [26–38] | 16 | 43 [36–52] | 2.7 [2.2–3.2] |
| 40–59y | 5 | 17 [14–19] | 11 | 34 [29–39] | 15 | 46 [40–54] | 3.2 [2.7–3.7] |
| 60y+ | 3 | 12 [9–15] | 6 | 20 [15–25] | 8 | 29 [22–37] | 3.6 [2.7–4.6] |
| Non-metropolitan region | 3 | 15 [13–17] | 7 | 32 [28–37] | 11 | 54 [46–63] | 4.9 [4.2–5.7] |
| Metropolitan region | 4 | 16 [14–18] | 9 | 32 [28–36] | 12 | 45 [40–51] | 3.8 [3.4–4.3] |
By the end of the third wave, the ETPI was 4.0 times higher than the OTPI. We estimated that 1.69 [1.51–1.89] million people, representing 47% [42–52] of the study population, had been infected, whereas the OTPI in the study area was 12% (0.42 million). ETPI was similar in men and in women. ETPI was higher in the non-metropolitan region (54% [46–63]) compared to the metropolitan region (45% [40–51], p = 0.04). ETPI was 60% [47–75] in children and adolescents, 11.0 times the official total proportion infected. In age groups 20–39y, 40–59y, and 60+ y, the ETPI was 43% [36–52], 46% [40–54], and 29% [22–37], representing 2.7, 3.2 and 3.6 times the OTPI, respectively.
Discussion
The cumulative incidence of COVID-19 reported to the surveillance systems does not reflect the true total COVID-19 infections in Costa Rica. We developed a model based on antibody response that calculated the number of people who got infected for each detected case, taking into account the decay of the antibodies over time. We estimated that in the study area (70% of the population of Costa Rica), 1.7 million people got infected by the end of the third wave (December 2021), 4.0 times more than the official infection total. As a result, 47% of the population had COVID-19 before the Omicron waves in the study area.
The underestimation of the infection total observed in Costa Rica was far less than in Mexico through October 2020, where the true number of cases might have been more than 30 times higher than the official infection total.6 Nevertheless, the underestimation in Costa Rica was more severe than observed in various high-income countries. In the United States, in May 2021, for every COVID-19 case reported since the beginning of the pandemic, there were 2.1 people with detectable SARS-CoV-2 antibodies.5 In Germany by the end of 2020,7 and in Navarre (Spain) in May 2022,3 around half of the infections had been detected and reported.
The ratio of estimated infection total to official infection total varied by age group: 11.0 times higher in 0–19y, 2.7 times higher in 20–39y, 3.2 times higher in 40–59y, and 3.6 times higher in 60+ y people. The ETPI was lower in 60+ y people compared to the rest of the population after each wave, in concordance with studies in various other countries.3,5, 6, 7 The ETPI was similar in children/adolescents and in adults during the first two waves, suggesting that even if children and adolescents were mantained isolated due to school closure during the pandemic, they were infected, possibly in the household. After the third wave, the ETPI was higher in children/adolescents compared to adults. This might be due to vaccination,22 as adults were vaccinated on a large scale when the third wave peaked, but children were not. Similar results were found in the United States.23 The ETPI was similar in men and in women, consistent with the expectation that men and women are equally susceptible to SARS-CoV-2, and with international studies.23 The ETPI was higher in the non-metropolitan compared to the metropolitan region, particularly during the third wave, and so was the underestimation of the OTPI. It suggests that the mitigation measures to avoid transmission were more successful in the urban than in the more rural and lower income areas.
The model we used was based on two assumptions, which we aimed to address in sensitivity analyses. The first assumption is that the underestimation of the official infection total is constant across time. This assumption was needed because of the relatively small sample sizes compared to the large time span of blood collection but might be false if the percentage of detection by the official registry varied across time. Nevertheless, the official guidelines for access to testing or for epidemiological linkage did not vary in Costa Rica during the period studied. The detection rate might have changed with the Omicron variant, but our study was based on pre-Omicron waves. As a result, the fatality rate after infection by age did not vary substantially during the period studied.1 Moreover, sensitivity analyses using only blood samples from specific waves led to similar results (Supplementary Materials, Appendix 3A) and supported the hypothesis that the number of people who got infected for each detected case was constant in Costa Rica in 2020–2021. Our methods can be adapted to contexts where this assumption is incorrect by including time in Equation (6). The second assumption is that the decay of antibodies of PCR-confirmed cases is similar to that of undetected cases. This assumption appears to be valid given that our sensitivity analysis using only mild cases produced similar results as the main analysis (Supplementary Materials, Appendix 3B). Moreover, the analysis of the positivity using only anti-S1-RBD antibodies in unvaccinated people also led to similar results (Supplementary Materials, Appendix 3C). The analysis of crude seropositivity based on anti-N and anti-S1-RBD, without adjustment for decaying seropositivity over time, also led to the conclusion that the official infection total seriously underestimated total infections; by the end of the third wave, 33% of the population was seropositive to COVID-19 antibodies, 2.8 times more than the OTPI (Supplementary Materials, Appendix 3D). We believe this is an underestimate, however, because crude seroprevalences do not account for loss of seropositivity with increasing time from infection. Another limitation of our work is the use of the same threshold to define seropositivity, regardless of age and sex. This issue was addressed by estimating seropositivity after infection by age and sex. We assumed that the probability that a previously infected individual would be seropositive, wa,s,t, depends only on age, sex and time since infection. Other covariates we tested, including certain comorbidities, did not improve the prediction significantly, and therefore, were not further considered. Nevertheless, better models based on individualized estimations of the probability to be seropositive after infection would be useful. Moreover, antibody dynamics could also change over calendar time, in particular with variants that cause more severe disease and possibly higher antibody responses. However, in sensitivity analyses we found similar antibody dynamics when we excluded cases with moderate disease and hospitalized cases (Supplementary Materials, Appendix 3B). A final limitation is that this work was restricted to selected cantons representing 70% of the population of Costa Rica. The results might be different in the rest of the country.
This study also had strengths. The control sample was large and population-based, although matched to the age, sex and minimal geostatistical unit distribution of the cases. The distribution of the education level in the population-based controls was also consistent with national data (ENAHO study)24 in groups defined by age, sex, and region. In particular, 54% of the population-based participants aged 20 years and older did not complete high school, and 25% went to college. These numbers agreed well with expected proportions based on national age-, sex-, and geographic area-specific rates of 56% [54–58] and 26% [24–28] respectively. By estimating ka,s,c, the age-, sex-, and canton group-specific number of people who got infected for each case reported, and by applying ka,s,c to the official number of cases for these groups, we accommodated sociodemographic differences between the RESPIRA population-based controls and the source population in the study area. The proportion of population-based controls found in the surveillance lists of reported cases before the end of each wave agreed with what would be expected based on the age-, sex-, and canton group-specific rates reported in the source population (OTPI). In particular, 5.0%, 9.3% and 13.4% of the population-based controls were reported to the surveillance system before the end of the waves 1, 2 and 3 respectively; and these numbers agreed well with the expected proportions based on the OTPI of 4.2% [3.4–5.2], 8.4% [7.2–9.7], and 12.0% [10.5–13.5] respectively. This is evidence that the population-based controls have reported COVID-19 infection rates consistent with those of the source population within categories defined by age, sex, and canton group, leading to unbiased estimation of k in the model. Requiring both anti-N and anti-S1-RBD to be positive made the assay more specific than anti-N alone. We obtained precise estimates of the cumulative infection burden at the end of each wave, despite the fact that there were few blood samples on those specific days, and the rest of the blood samples were collected over a large time interval (one year) when SARS-CoV-2 was spreading rapidly in the population. This methodology can be adapted to other populations and contexts where the dynamics of the infection may be different. Finally, a key novel contribution of our paper is the correction for antibody dynamics in population-based participants using data from a sample of COVID-19 PCR-confirmed cases. The difference between the estimates based on corrected and the uncorrected approaches was substantial, as we estimated that 26% of the previously infected people were antibody negative when their sample was collected.
By the end of the third wave (December 2021), when 12% had been identified in the official surveillance reports as infected in the study area, we estimated that 47% of the population had been infected by SARS-CoV-2, 4.0 times more than reported. The official infection total severely underestimated infections in children and adolescents. The underestimation of true infection rates was similar in men and in women. With the emergence of the Omicron variant, the total number of reported cases was twice as high in December 2022 as in December 2021, despite a likely lower detection rate. As a result, and given our estimate that nearly half the population had already been infected by the end of 2021, it is likely that a large majority of the population has been infected at least once. In addition, 96% of the adults had received at least one dose of vaccine by the end of 2022. Thus, most inhabitants of Costa Rica are now likely to have some hybrid immunity against future COVID-19 infections.
The proportions of the population with previous natural infections and vaccination varied across countries, especially during 2020–2021. Previous infections and vaccinations may have modified the rates of hospitalization and deaths from later Omicron variants. Inter-country comparisons of the lethality of infections and prevalence of post-covid symptoms in relation to past exposure to infection and vaccination may indicate whether such sources of immunity protect against future COVID-19 variants. This underscores the necessity of continued monitoring to promote effective prevention strategies suited to the current state of the pandemic.
Contributors
Conceptualization (RF, NA, RP, AH, MG, RH), Data curation (RF, NA), Formal analysis (RF, NA, RP, MG, RH), Investigation (RF, NA, RP, TW, JB, JF, KR, RP, CP, VL, MG, RH), Methodology (RF, NA, RP, TW, Aab, JB, AH, MG, RH), Funding acquisition (AAp, CP, RH), Project administration (AAp, CP, VL), Resources (AAp, TW, JB, RH), Validation (RF, NA), Supervision (RH), Visualization (RF, NA), Writing—original draft (RF, NA), Writing—review & editing (RF, NA, AAp, RP, TW, Aab, JB, JF, KR, RP, CP, AH, VL, MG, RH). RF and NA have directly accessed and verified the underlying data reported in the manuscript.
Data sharing statement
Sharing of deidentified data can be discussed on a case-by-case basis upon request to the corresponding author.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgements
This work was partially funded by the Divisions of Intramural Research, National Institute of Allergy and Infectious Diseases, and the National Cancer Institute (contract HHSN261201700012I/75N91020F00001), the Science, Innovation, Technology and Telecommunications Ministry of Costa Rica (grant N°FI-005-20), and Costa Rican Biomedical Research Agency-Fundacion INCIENSA (grant N/A). The work of Tim Waterboer was generously supported by the Dieter Morszeck Foundation (Project “High-Throughput Serolomics Open Lab”). Mitchell H. Gail, Neha Agarwala and Ruth M. Pfeiffer were supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
Members of the RESPIRA Study Group
Caja Costarricense de Seguro Social: Alejandro Calderón, Karla Moreno, Melvin Morera, Roy Wong.
Ministerio de Salud, Costa Rica: Roberto Castro.
Agencia Costarricense de Investigaciones Biomédicas—Fundación INCIENSA, Costa Rica: Bernal Cortés, Rebecca Ocampo, Michael Zúñiga, Juan Carlos Vanegas.
Fogarty International Center, National Institutes of Health: Kaiyuan Sun.
Universidad de Costa Rica: Cristina Barboza-Solís.
German Cancer Research Center, Heidelberg, Germany: Marco Binder.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lana.2023.100616.
Contributor Information
Romain Fantin, Email: rclement@acibfunin.com.
Neha Agarwala, Email: agarwalan2@mail.nih.gov.
Rolando Herrero, Email: rherrero@acibfunin.com.
the RESPIRA Study Group:
Alejandro Calderón, Karla Moreno, Melvin Morera, Roy Wong, Roberto Castro, Bernal Cortés, Rebecca Ocampo, Michael Zúñiga, Juan Carlos Vanegas, Kaiyuan Sun, Cristina Barboza-Solís, and Marco Binder
Appendix A. Supplementary data
References
- 1.Ministerio de Salud . 2020. Situación nacional COVID-19.https://oges.ministeriodesalud.go.cr/ [Google Scholar]
- 2.Fantin R., Barboza-Solís C., Hildesheim A., et al. Excess mortality from COVID 19 in Costa Rica: a registry based study using Poisson regression. Lancet Reg Health Am. 2023;20 doi: 10.1016/j.lana.2023.100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castilla J., Lecea Ó., Martín Salas C., et al. Seroprevalence of antibodies against SARS-CoV-2 and risk of COVID-19 in Navarre, Spain, May to July 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.33.2200619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergeri I., Whelan M.G., Ware H., et al. Global SARS-CoV-2 seroprevalence from January 2020 to April 2022: A systematic review and meta-analysis of standardized population-based studies. PLoS Med. 2022;19 doi: 10.1371/journal.pmed.1004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones J.M., Stone M., Sulaeman H., et al. Estimated US infection- and vaccine-induced SARS-CoV-2 seroprevalence based on blood donations, July 2020-May 2021. JAMA. 2021;326:1400–1409. doi: 10.1001/jama.2021.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres-Ibarra L., Basto-Abreu A., Carnalla M., et al. SARS-CoV-2 infection fatality rate after the first epidemic wave in Mexico. Int J Epidemiol. 2022;51:429–439. doi: 10.1093/ije/dyac015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuhauser H., Rosario A.S., Butschalowsky H., et al. Nationally representative results on SARS-CoV-2 seroprevalence and testing in Germany at the end of 2020. Sci Rep. 2022;12 doi: 10.1038/s41598-022-23821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sigal A. Milder disease with Omicron: is it the virus or the pre-existing immunity? Nat Rev Immunol. 2022;22:69–71. doi: 10.1038/s41577-022-00678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein C., Nassereldine H., Sorensen R.J.D., et al. Past SARS-CoV-2 infection protection against re-infection: a systematic review and meta-analysis. Lancet. 2023;401:833. doi: 10.1016/S0140-6736(22)02465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ntziora F., Kostaki E.G., Karapanou A., et al. Protection of vaccination versus hybrid immunity against infection with COVID-19 Omicron variants among Health-Care Workers. Vaccine. 2022;40:7195–7200. doi: 10.1016/j.vaccine.2022.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Lai F.T.T., Yan V.K.C., et al. Comparing hybrid and regular COVID-19 vaccine-induced immunity against the Omicron epidemic. NPJ Vaccines. 2022;7:1–9. doi: 10.1038/s41541-022-00594-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demonbreun A.R., McDade T.W., Pesce L., et al. Patterns and persistence of SARS-CoV-2 IgG antibodies in Chicago to monitor COVID-19 exposure. JCI Insight. 2021;6 doi: 10.1172/jci.insight.146148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedele G., Stefanelli P., Bella A., et al. Anti-SARS-CoV-2 antibodies persistence after natural infection: a repeated serosurvey in Northern Italy. Ann Ist Super Sanita. 2021;57:265–271. doi: 10.4415/ANN_21_04_01. [DOI] [PubMed] [Google Scholar]
- 14.Nikolopoulou G.B., Maltezou H.C. COVID-19 in Children: where do we Stand? Arch Med Res. 2022;53:1–8. doi: 10.1016/j.arcmed.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CCSS–Subárea de Vigilancia Epidemiológica . 2021. Vacunación contra COVID-19.https://www.ccss.sa.cr/web/coronavirus/vacunacion [Google Scholar]
- 16.Loria V. Evaluation of immune response and household transmission of SARS-CoV-2 in Costa Rica: the RESPIRA Study. BMJ Open; [in press]. [DOI] [PMC free article] [PubMed]
- 17.Sun K., Loria V., Aparicio A., et al. Behavioral factors and SARS-CoV-2 transmission heterogeneity within a household cohort in Costa Rica. Commun Med. 2023;3:102. doi: 10.1038/s43856-023-00325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butt J., Murugan R., Hippchen T., et al. From multiplex serology to serolomics-A novel approach to the antibody response against the SARS-CoV-2 proteome. Viruses. 2021;13:749. doi: 10.3390/v13050749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ministerio de Salud de Costa Rica Base anonimizada de casos confirmados COVID 19. https://oges.ministeriodesalud.go.cr/evolucioncovid.html
- 20.INEC Estadísticas demográficas. 2011–2050. Proyecciones nacionales. Población por años calendario, según sexo y grupos quinquenales de edades. https://www.inec.cr/poblacion/estimaciones-y-proyecciones-de-poblacion
- 21.Gómez-Meléndez A. Análisis y simulación espacial de la pandemia COVID-19 a nivel cantonal, para el caso de Costa Rica. CIOdD-UCR; 2022. Reporte epidemiológico 22. [Google Scholar]
- 22.Fantin R., Herrero R., Hildesheim A., et al. Estimating vaccine effectiveness against SARS-CoV-2 infection, hospitalization and death from ecologic data in Costa Rica. BMC Infect Dis. 2022;22:767. doi: 10.1186/s12879-022-07740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiegand R.E., Deng Y., Deng X., et al. Estimated SARS-CoV-2 antibody seroprevalence trends and relationship to reported case prevalence from a repeated, cross-sectional study in the 50 states and the District of Columbia, United States—October 25, 2020–February 26, 2022. Lancet Reg Health Am. 2023;18 doi: 10.1016/j.lana.2022.100403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.INEC . 2021. Encuesta Nacional de Hogares 2021–sistema de consultas.http://sistemas.inec.cr:8080/bininec/RpWebEngine.exe/Portal?BASE=ENAHO2021&lang=esp [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.