Abstract
Modular proteins are regulatory proteins that carry out more than one function. These proteins upregulate or downregulate a biochemical cascade to establish homeostasis in cells. To switch the function or alter the efficiency (based on cellular needs), these proteins require different facilitators that bind to a site different from the catalytic (active/orthosteric) site, aka ‘allosteric site’, and fine-tune their function. These facilitators (or effectors) are allosteric modulators. In this Review, we have discussed the allostery, characterized them based on their mechanisms, and discussed how allostery plays an important role in the activity modulation and function fine-tuning of proteins. Recently there is an emergence in the discovery of allosteric drugs. We have also emphasized the role, significance, and future of allostery in therapeutic applications.
Keywords: Allostery, Multi-domain proteins, Drug design, Enzyme kinetics, Protein function regulation, Therapeutics
Graphical Abstract
1. Introduction
Proteins are versatile biomacromolecules that are involved in almost every cellular process: genome repair and maintenance, molecules transportation, energy generation, cellular movement, cell division, host immunity and defense, and so on. The cellular functions operate in a pathway fashion or a cascade of reaction for which the protein requires more than one binding partner that either (i) binds to the same site at a different time to confer different functions or (ii) bind to different sites at the same time to regulate a function. For instance, proteins involved in genomic regulations - transcription, replication, repair, and maintenance, have multiple regulators that bind to the same protein to switch and fine-tune its function. An excellent example of such regulation is DNA helicase, which binds ATP (adenosine triphosphate) and DNA at two different sites and utilizes ATP-driven energy to unwind the DNA [1], [2]. Similarly, there are pathways that are controlled by the end product through the allosteric regulation of proteins involved in it. One such example is the regulation of tryptophan (trp)-repressor by tryptophan in the trp-operon, where tryptophan, if present in excess, binds to trp-repressor protein and holds it on the operator region on the DNA and stops the further expression of trp-genes required for trp-biosynthesis [3], [4]. These regulations are due to the presence of more than one binding site in protein and such regulations are called “allosteric” regulations. The term “allostery”, first coined by Jacques Monod and Francois Jacob in 1961 [5], is a combination of two Greek words: allos meaning “other” and stereos meaning “solid”, which in the biological context, translates to regulation by other sites. The allosteric sites can either be in close proximity to the orthosteric (active/substrate binding) site or at a distance (Fig. 1A). A timeline of evolution of allostery has been nicely reviewed by Liu and Nussinov [6].
Fig. 1.
(A) Proximity of allosteric and orthosteric sites in proteins. (B) Effect of different allosteric effectors (PAM: Positive allosteric modulator; NAM: Negative allosteric modulator) on enzyme activity.
Allostery is a phenomenon where the binding of a molecule at one site of a protein affects the activity of a distinct functional site. Allostery is quite common in proteins involved in, but not limited to, signal transduction, epigenetics regulation, enzymatic activity, etc. In protein biochemistry, allostery can be observed in two major scenarios: (i) protein has a single domain that contains more than one binding pocket or site, and (ii) protein contains more than one domain (Fig. 1A). Modular proteins contain multiple domains which work in synergy for their efficient productivity and fine-tuning the activity. In multi-domain proteins, every domain may have its distinct role or binding partner (effectors) which can efficiently tune the role of the same protein in multiple cellular processes. These effectors are called “allosteric regulators/ligands”.
Allostery is, however, not restricted to regulation by small molecule effectors (substrate, co-factors. inhibitors, etc.). Macromolecules, such as DNA, RNA, and proteins also act as allosteric regulators. For example, in the case of DNA repair, one protein senses the DNA damage through a distinct domain and relays the signal to other regulatory or repair proteins through its other domain, via inter/intra-domain contact or spatial domain rearrangement or crosstalk. Similarly, the protein involved in chromatin methylation maintenance recognizes methyl-cytosine on one DNA strand and methylates specific lysine on histone through its other domain [7], [8], [9], [10], [11]. The activity of these proteins depends on the presence/absence of a specific effector molecule(s) like damaged DNA is an effector for DNA damage response (DDR) proteins.
Structural studies of complex biomacromolecular assemblies have paved the way to study the mechanism of allosteric regulation and explore the potential of allostery [12], which has been harnessed for quite some time for target-specific drug designing and industry-scale custom biosynthesis of small molecules, mostly active pharmaceutical ingredients (APIs) [13], [14], [15], [16]. Allostery can also be exploited to treat diseases [17]. For example, allosteric drugs against G-protein coupled receptors (GPCRs) are widely used to treat neurological diseases [18]. To date, several allosteric drugs have been approved by different regulatory agencies such as FDA and are being used to treat some major diseases like epilepsy, hypertension, neuropathic pain, dementia, HIV, acute leukemia, melanoma, insomnia, and dyslexia, among many others [18], [19], [20].
In this review, we discuss the allostery-mediated crosstalk in modular proteins with an emphasis on the role of allostery in (i) the accurate functionality of modular proteins and (ii) drug design.
2. Allosteric effectors
Allosteric regulators have a functional outcome that is observed as a change in: (i) enzyme-catalyzed reaction (for enzymes), (ii) recognition of a binding partner (for non-enzymatic proteins), and (iii) regulation of ion flux (neither enzymatic nor substrate-binding). While biochemical assays help address the allosteric regulation of enzymes, biophysical studies assist in unraveling the allosteric regulation of non-enzymatic proteins. Macromolecular structural studies not only complement biochemical and biophysical studies but also provide mechanistic insights, such as local and global conformational changes and dynamics, into the allosteric regulation of proteins at atomic resolution.
For the proteins with catalytic activity, the effectors affect (i) their rate of reaction, either favorably or unfavorably, in terms of product formation, or (ii) alter their substrate/product specificity. Biochemical kinetics assays (enzyme assays) give a detailed insight into how much the effector affects the rate of an enzymatic reaction, and whether they increase or decrease the reaction. These effects can be monitored by measuring rate constants and apparent rate constants, such as Vmax (maximum velocity of the reaction), KM (Michaelis constant), and kcat/KM (catalytic efficiency) for the reaction in the presence and absence of the effector. Based on how the reaction is affected, the allostery can be classified into two categories:
-
(a)
Positive Allostery: It is when the binding of a molecule at an allosteric site enhances the enzyme or receptor activity at the orthosteric site. Positive allostery is widely known as “allosteric activation” and the effectors are called “positive allosteric modulators (PAMs)”. The agonists or partial agonists induce positive allostery (Fig. 1B).
-
(i)
Agonist - An effector molecule that binds to a protein at a specific site (other than the catalytic/active site) and stimulates/increases the protein activity at the catalytic site to the highest possible reaction rate. It can also be called an ‘activator’ or ‘full agonist’.
-
(ii)
Partial agonist - An effector molecule that activates the enzyme but not to its maximum efficiency.
-
(b)
Negative Allostery: Also known as allosteric inhibition, it is when the binding of a molecule at the allosteric site restricts or inhibits the enzyme activity at the orthosteric site. Such effectors are called “negative allosteric modulators (NAMs)” which include antagonists or neutral antagonists, partial antagonists, and inverse agonists (Fig. 1B).
-
(i)
Inverse agonist - The effector that shuts even the basal or constitutive activity of the enzyme or receptor. These antagonists preferentially stabilize the inactive conformational state, resulting in a reduction in basal receptor activity.
-
(ii)
Neutral antagonist – These antagonists can inhibit the effects of both agonist and inverse agonist, thereby reducing the agonist-activated receptor signaling or increasing the receptor signaling in the concurrent presence of an inverse agonist. These antagonists are believed to bind both active and inactive conformations of the receptors or enzymes
-
(iii)
Partial antagonist - The effector that reduces the enzyme activity, but not to the basal level.
Below, we have discussed, with examples, how allosteric regulators affect the functional outcomes of proteins.
Enzymes involved in signal transduction are best suited to describe the functional outcomes of allostery, as they are dependent on signaling molecules (effectors) for their further course of action. For example, the proteins involved in DNA damage repair are highly regulated by allosteric regulators. The sensor proteins in these systems have multiple domains - (i) the sensory/regulatory domains that recognize the DNA damage at the site; (ii) the catalytic domain that catalyzes the formation of signaling molecules and (iii) the flag-bearer of the signaling molecule that invites the repair proteins to the affected site. One such example is the Poly(ADP-ribose) polymerase (PARP) family of proteins. The founding member of the PARP family is the PARP1 which senses the DNA breaks through the zinc-finger (ZnF) domains at the N-terminal and relays a signal to the C-terminal catalytic domain to synthesize poly(ADP-ribose) (PAR) on the centrally positioned auto-modification domain (AD). PAR acts as a signaling molecule to recruit repair proteins such as XRCC1, Ligase 4, etc. [21], [22] In this case, the damaged DNA, which has no direct contact with the catalytic domain, yet elicits its catalytic activity, serves as an allosteric agonist or PAM (Fig. 2A). Recently, our group has shown that PAR also independently activates the catalytic activity of PARP1, but not as much as DNA [23], [24], which makes PAR a PAM (partial agonist) (Fig. 2A). PARP1 recognizes DNA and PAR through a distinct set of domains, which further indicates that PAR and DNA could simultaneously affect PARP1’s activity [23], [24]. In the presence of PAR, the DNA-dependent activity of PARP1 gets reduced, which further makes PAR a NAM or partial antagonist [23], [24]. The study revealed a mechanism that showed that the same compound has both enhancing and diminishing effects based on the other effector's presence (Fig. 2A).
Fig. 2.
Effect of allosteric regulators on the functional outcome of proteins. (A) Signal (DNA damage)-induced PARP1 recruits repair proteins to initiate the DNA repair pathway. Schematic representation of allosteric regulation of PARP1 by DNA (agonist) and PAR (both partial agonist and partial antagonist). (B) Schematic representation of RNA degradation cascade by type III CRISPR-Cas system, where Cas10 and Csm6 are allosterically regulated by RNA and cOAs, respectively. The operon system and the genes associated with the type III CRISPR-Cas system is shown at the bottom of the panel. The left side of the panel represents the recognition of the viral RNA by the type III CRISPR-Cas system. (C) Schematic representation of DNA-induced allosteric activation of cGAS protein in cGAS-STING pathway.
Another important example comes from the bacterial anti-phage system, CRISPR-cas (clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas)). In the type III CRISPR system, the dynamics of cas10 and csm6 proteins play an important role in providing innate immunity to the bacteria against infective phage [25], [26]. Upon recognition of the target RNA by CRISPR-RNA-guided effector complex (also includes cas10), the PALM domain of cas10 synthesizes a second messenger molecule, cyclic-oligoadenylates (cOAs) using ATP. Here, the RNA acts as an allosteric regulator (agonist or PAM) that induces cOA formation. The CRISPR-cas type III system is further regulated by cOAs. cOAs consist of a different number of adenylates, ranging from two (cyclic diadenylate; cA2) to six (cyclic hexa-adenylate; cA6) (Fig. 2B). Further, cOAs bind to the CRISPR-associated Rossmann fold (CARF) domain of the csm6 protein and activate its HEPN nuclease domain. In the absence of cOA, the HEPN domain of csm6 remains in an inactive conformation. The binding of cOA to CARF induces a globular active conformational change in the csm6 so that HEPN domains come into functional conformation and degrade the RNA [25], [27]. Here, cOAs can be regarded as PAM (agonist) (Fig. 2B).
In eukaryotes, the presence of cytosolic nucleic acid indicates pathogen invasion. The Chen group discovered that to clear the cytosolic DNA, eukaryotes use the cGAS-STING (cyclic GMP-AMP synthase-Stimulator of interferon genes) pathway [28], [29], [30], [31]. The detection of cytoplasmic dsDNA induces global conformational change in cGAS leading to its dimerization and subsequent activation to synthesize cyclic GMP-AMP (cGAMP) from GTP and ATP substrates (Fig. 2C) [31], [32], [33], [34]. The cGAS possess three DNA binding sites - site A, site B and site C. The dsDNA binding to site A and site B leads to cGAS dimerization (Fig. 2C) [33]. The DNA bound to the ‘site C’ causes the clustering of cGAS-DNA complex, leading to the formation of the molecular condensate, which further activates the innate immune signaling (Fig. 2C) [33], [35]. The active site, including the ‘helix spine’ and ‘activation loop’ of cGAS which remains disordered in the absence of dsDNA, becomes ordered upon DNA binding and facilitates cGAMP synthesis [36]. cGAMP then binds to STING and causes the release of C-terminal tails (CTT) and STING polymerization. Upon polymerization, STING translocate from the endoplasmic reticulum to Golgi, where it initiates the autophagy, for the clearance of cytosolic dsDNA and pathogens. The mechanism of the cGAS-STING pathway has been extensively reviewed by Hopfner and Hornung [35].
3. Agonist, neutral antagonist and inverse agonist: Role in therapeutics
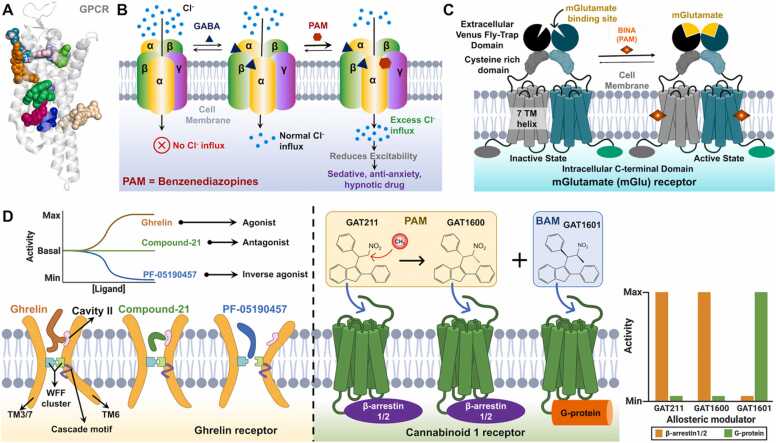
Apart from the standard functional aspects, PAMs have extended their branches to pharmaceuticals, where they are designed for treatment. Being a major class of sensory proteins, G-protein coupled receptors (GPCRs), are highly potent drug targets. GPCRs are one of the major membrane proteins involved in cellular environmental sensing and downstream signaling. Robert J. Lefkowitz and Brian K. Kobilka shared the Nobel Prize in Chemistry (2012) for their studies on the GPCRs. GPCRs are highly regulated by allosteric effectors as they possess multiple allosteric sites (Fig. 3A). One such PAM is a class of drugs called benzodiazepines that act as a GABA (Gamma-aminobutyric acid; an inhibitory neurotransmitter) agonist. GABA binds to the GABA receptor, a class C G-protein coupled receptor (GPCR) protein embedded in the synaptic membrane, which regulates the chloride flux [37], [38], [39], [40]. Benzodiazepines bind to the allosteric site of the GABAA receptor and promote the chloride influx (thus, an agonist) causing reduced neuron excitability, thus are used as a sedative, hypnotic (sleep-inducing), and anxiolytic (anti-anxiety) drug (Fig. 3B). Regulation of GABAA receptor by effector molecule to alter the ion flux is an excellent example of allosteric regulation of non-enzymatic and non-substrate-binding protein with high therapeutical importance. Another such allosterically regulated GPCR is the metabotropic glutamate (mGlu) receptor that has a modulatory activity to L-glutamate, a major excitatory neurotransmitter in the central nervous system and is therefore essential in the fine-tuning of synapses [41]. mGlu receptor is highly dynamic in nature and rapidly oscillates between active and inactive states. Structural and functional studies by Pin and Margeat groups have shown that BINA, a PAM of mGlu receptor binds to its allosteric site in the transmembrane region and stabilizes the mGlu receptor in the active state [42]. Allosteric activation of mGlu receptors is linked to the treatment of neuro-disorders such as autism, Parkinson’s disease, schizophrenia, and Fragile X disease, among others and thus PAMs for these receptors have a wide range of therapeutic interest (Fig. 3C).
Fig. 3.
(A) Representative GPCR with ligands bound at different experimentally validated allosteric sites. Schematic representation of allosteric regulation of different GPCRs - (B) GABAA receptor, (C) mGlu receptor, (D) ghrelin and cannabinoid 1 receptors. The effect of agonist, antagonist, and inverse agonist on the ghrelin receptor is shown by a representative kinetic plot. The effect of PAM and BAM on the cannabinoid receptor is shown by a representative histogram.
(Panel A is adapted from Wakefield et al. [55]).
The effect of agonist, antagonist, and inverse agonist has been nicely reported in the study from the Shao group, where they have studied the allosteric activation and inhibition of the ghrelin receptor, another GPCR. The role of the ghrelin receptor has a vast range, including appetite regulation, alcohol consumption, adipocyte metabolism, and glucose homeostasis [43], [44], [45], [46], due to its broad distribution and multiple signaling pathways through divergent G-protein coupling or β-arrestin recruitment [47]. The ghrelin receptor has a basal constitutive activity of 50%, which increases to 100% in the presence of acetylated-ghrelin, thus acetylated-ghrelin acts as an agonist for the ghrelin receptor. Structural studies have shown that the constitutive activity of the ghrelin receptor is due to the interaction between the WFF (Trp-Phe-Phe) cluster. Ghrelin induces favorable conformational displacement in the WFF cluster resulting in an increase in its activity. This makes ghrelin a potent agonist for the ghrelin receptor compared to compound 21, which acts as a NAM or neutral antagonist and brings its activity to the basal level, i.e., this inhibitor neither increases nor decreases the basal activity of the ghrelin receptor [48], [49]. On the other hand, the conformational change imposed by another NAM, PF-05190457, which pushes apart the WFF cluster resulting in the inhibition of its basal activity [49], makes it an inverse agonist (Fig. 3D). Inhibition of the ghrelin–ghrelin receptor signaling axis and deacylation of ligands or deletion of receptors could potentially prevent obesity and type 2 diabetes. Thus, blockade of the ghrelin receptor by NAMs has been proven to be a great therapeutic approach for the treatment of related diseases [50], [51], [52], [53], [54].
4. Biased allosteric modulators for therapeutics
Recently, a novel class of allosteric modulators called biased allosteric modulators (BAM), has been discovered that bias the GPCRs towards specific functions [56], [57], [58], [59]. It is also referred to as “biased agonism” for its feature of functional selectivity of multi-function proteins. A ligand-activated GPCR can selectively couple to different transducers (i.e., G protein or β-arrestin) and regulate diverse processes in the nervous system. Thakur and Laprairie's groups first discovered the BAM for cannabinoid 1 receptor (CB1R) as a result of the “magic-methyl effect” on a known agonist PAM, GAT211 [56]. Methylation at the alpha position of the nitro-group, yielded two diastereomers, (-)-(S,R)−13 conformer (GAT1600) and (+)-(R,S)−14 conformer (GAT1601). Out of the two, GAT1600 exhibited enhanced PAM potency, whereas GAT1601 exhibited biasing towards G-proteins and drastically reduced CB1R coupling to ꞵ-arrestin1/2 (Fig. 3D). Another such BAM is PNR-4–20 which biases the G-protein coupling to CB1R [60]. Cannabinoid signaling has been considered the therapeutic target for treating glaucoma, pain, addiction, obesity, inflammation, and other diseases. G-protein-biased modulation of CB1R offers a safe therapeutic candidate for these diseases [56]. The cannabinoid receptor signaling mediated by allosteric PAM and BAMs has been extensively reviewed by the Zhou group [61]. Besides G-protein coupling BAMs, ꞵ-arrestin1/2 coupling BAMs have also been identified for another set of GPCRs, β-adrenergic receptors (ꞵ-ARs), primary regulators of cardiac function and a potent drug target for cardiac disease [57]. Lefkowitz and Rockman's group has identified that a molecule called Cmpd-6, which was primarily discovered as a PAM for ꞵ2-ARs, selectively enhances the affinity and cellular signaling of carvedilol, a known β-arrestin–biased β-blocker for β1-ARs [57]. In vivo, Cmpd-6 provides an enhanced β-arrestin–dependent cardioprotective effect of carvedilol during ischemia/reperfusion injury–induced apoptosis. Their study identifies the potential therapeutic application of Cmpd-6 for enhanced clinical benefits of carvedilol in cardiac disease treatment [57]. Therefore, exploring the biased allostery of the therapeutically relevant receptors has tremendous potential in tuning physiology to develop safer and targeted therapeutics. Regulation of GPCRs by allosteric modulators has been extensively reviewed by several groups [42], [55], [62], [63], [64].
5. Allostery based on the direction
As discussed in the above examples, the binding of a molecule at a distal site alters the active site dynamics. This is also possible in the reverse order, that is, the changes at the active site, due to the presence/absence of a binding partner, can affect the allosteric site binding dynamics. Thus, based on the direction of cause-and-effect, the allostery can be classified into - (i) forward allostery, (ii) reverse allostery, and (iii) bi-directional allostery.
Forward allostery: It is when the binding of an effector at the allosteric site alters the catalytic activity of the enzyme.
Reverse allostery: It is when the binding of an effector at the active site regulates the effectiveness of the non-catalytic binding site.
Bidirectional Allostery: Some proteins show both forward and reverse allostery. This is referred to as bidirectional allostery.
As we have seen in the example of PARP1, the binding of DNA by N-terminal ZnF domains increases the catalytic activity of the catalytic domain at the C-terminal, which is a necessary step for the initiation of DNA repair. This is the case, where the effector (DNA) binding at the non-catalytic site regulates the functional dynamics of the catalytic sub-unit, it is a favorable “forward allostery” [21], [22], [23], [24] (Fig. 4A). PARP1 has great clinical significance as it is overexpressed in several types of cancer, including breast cancer, ovarian cancer, pancreatic cancer, prostate cancer, etc., which makes it a very potent drug target. PARP inhibitors (PARPis) block the DNA-repair function of PARP in cancer cells and kill them. Most of the clinical PARPis are NAD analogs that target the catalytic site in PARP1. Studies from the Pommier group identified that these clinical PARPis not only inhibit the PARP1 activity but also impose an effect on DNA binding [65], [66], suggesting a PARPi-induced alteration in the interdomain crosstalk in PARP1. A further investigation carried out by the Pascal group has shown that the PARPis causes either (i) strong allosteric pro-retention of PARP1 and PARP2 on DNA, (ii) no or mild allosteric pro-retention of PARPs on DNA, or (iii) allosteric pro-release of PARP from DNA [67], [68]. Such regulation by inhibitors that bind to the catalytic site and affect the allosteric roles of the proteins are examples of “reverse allostery” (Fig. 4A). PARPis-mediated PARP trapping on DNA is an example of bidirectional allostery. Structural studies from the Pascal group have shown that DNA binding at the N-terminal ZnF domains activates the leucine switch at the C-terminal which favors the NAD+ binding at the C-terminal CAT domain [22]. Activated leucine switch, in the presence of DNA, can also facilitate high-affinity binding of the orthosteric inhibitors (NAD+ analogs) to the CAT domain. Based on the inhibitor category, PARPi can further modulate the DNA retention at the N-terminal region of PARP1. These two activities: (i) DNA binding-mediated PARPi binding (forward allostery), and (ii) PARPi-mediated DNA retention (reverse allostery), are co-dependent phenomena, thus this mechanism can be classified as “bi-directional allostery” (Fig. 4A). Due to this co-dependency, such PARPis are identified as more effective drug molecules in cancer treatment [65], [66], [69]. However, there are non-NAD analog PARPis, which do not bind to the CAT domain, yet inhibit the PARP activity. The Tulin and Skorski groups have identified one such non-NAD analog allosteric PARPi, (called 5F02) that prevents the H4-induced PARP1 activation and has a significant effect in the treatment of BRCA-deficient leukemia [70], [71], [72]. This type of inhibition can be classified under unfavorable “forward allostery”.
Fig. 4.
Mechanism of regulation of different proteins by different types of allosteric effectors. (A) Schematic representation of forward and reverse allostery in PARP1 (panel-i) and bidirectional allostery in phosphofructokinase (PFK) (panel-ii). (B) Schematic representation of active and inactive states of KRAS GTPase with G12C mutation, and its allosteric inhibition by suicidal (covalent) inhibitor.
Protein kinases are another abundant protein that is highly regulated at both allosteric and orthosteric (active) sites. Phosphoinositide-dependent protein kinase 1 (PDK1) is another example in which bidirectional allostery has been reported. PDK1 consists of an active site that binds ATP and an allosteric site called PDK1-interacting fragment (PIF) pocket that binds PIFtide, a 24 residues polypeptide sequence. The binding of PIFtide activates PDK1. An interesting mechanism of PDK1 inhibitor has been revealed by the Biondi group [73], where an orthosteric inhibitor (PS653) locks the active site in an open conformation, rigidifies the allosteric PIF pocket, and inhibits PIFtide binding. This can be classified as unfavorable “reverse allostery”. Another inhibitor (adenosine), then binds to the active site and relaxes the PIF pocket facilitating the binding of PIFtide, thus exhibiting a favorable “reverse allosteric” mechanism (Fig. 4A).
6. Reversible and irreversible effectors
The retention time of an effector at the binding site is decided by the binding affinity, aka molar equilibrium dissociation constant (KD), which is the ratio of the dissociation (koff) and association (kon) rate constants of the effector for the protein. Most potent effectors have high kon and low koff rate constants, indicating a higher rate of complex formation than their dissociation. These effectors are reversible effectors which bind to the protein with non-bonded interactions, such as hydrogen bonds, electrostatic interactions, salt bridges, water bridges, hydrophobic, and van der Waal interactions and are called non-covalent inhibitors. Some allosteric effectors, on the other hand, can permanently lock the protein in a certain conformation by binding irreversibly through a covalent bond, along with other non-bonded interactions, which provided selectivity and target site specificity. Such effectors are called ‘covalent inhibitors’ or ‘suicidal inhibitors’. Most of the allosteric drugs available in the market are non-covalent effectors, development of covalent allosteric inhibitors is in the nascent stage.
Cee and group used a structure-based drug design approach to identify a covalent allosteric inhibitor, AMG510 or Sotorasib, against KRAS GTPase, a proto-oncogene with G12C mutation, which has been observed in 13% of non-small cell lung cancer (NSCLC) patients [74]. The KRAS protein regulates cell proliferation by switching between the active (GTP-bound) and inactive (GDP-bound) forms [75]. Sotorasib targets the cysteine to bind irreversibly to the KRAS G12C variant and ceases it in its inactive form leading to the inactivation of KRAS and eventually cell death (Fig. 4B). Sotorasib is the first KRAS granted accelerated approval by FDA for the treatment of advanced NSCLC in adults with a KRAS G12C mutation.
7. Feedback regulation by end-product
A number of cascades/pathways are regulated by their end-products that bind to an allosteric site of an enzyme and either activate or inhibit its activity. Such a type of regulation is also known as feedback allosteric regulation. These feedback regulations are required for the enzyme’s selective activation/inhibition based on its demand under a given physiological condition. For example, the trp operon is only activated under tryptophan-deficient conditions in the cell. Based on the impact of the end products on the activity of the enzymes, feedback regulations are classified into “positive” and “negative”.
Positive feedback: Also referred to as “feedback activation”, it is when the binding of product/by-product enhances the activity of the enzyme.
Negative feedback: The binding of the product shuts/slows down the enzymatic activity or further product formation. It is also known as “feedback inhibition”.
Glycolysis is a pathway highly regulated by the end product, ATP. In glycolysis, the enzyme phosphofructokinase (PFK) that utilizes ATP to convert fructose-6-phosphate (F6P) to fructose 1,6-bis-phosphate (FBP) and produce ADP as a by-product, is allosterically regulated by both ATP and ADP (Fig. 5A). PFK exists in two conformational states - R and T, at equilibrium. PFK contains an active site that binds ATP and F6P and an allosteric site [76]. When the ATP concentration in the cell is high and the cell no longer needs to synthesize it more, ATP binds to the allosteric site of PFK and traps it in the T state which has a reduced affinity for F6P, thus halts/slows the reaction [77] (Fig. 5A). On the other hand, when the concentration of ATP drops in cells, the ATP in the allosteric pocket of PFK is replaced by ADP which causes a shift in the equilibrium to R conformation and accelerates the reaction for FBP production [78]. Apart from ADP, AMP also acts as an allosteric activator of PFK (Fig. 5A). Such regulation by the end product that leads to the accelerated activity of the enzyme is a positive feedback allosteric mechanism.
Fig. 5.
Allosteric regulation by end-products. (A) Allosteric regulation of PKF by substrate (ATP) and end by-product (ADP). (B) Allosteric regulation of RNR enzymes by their end-products, where the end-products provide substrate specificity to RNR, whereas the last end-product (dATP) acts as an inhibitor of RNR. Schematic representation of allosteric regulation of active (S-dimer) and inactive (I-dimer) states of RNR by ATP and dATP. The panel on the right shows the mechanism of allosteric activation and inhibition of RNR, through a conformational change in loop 2, by different effectors and substrates. (C) Crosstalk-mediated allosteric regulation of SUVH and CMT proteins by their respective end-products for maintenance of epigenetic marks in the genome.
Panel C is adapted from Abhishek et al. FEBS J [9].
End products of enzymes also act as allosteric inhibitors of the same enzyme as seen in the PARP case discussed in Section 4 (Fig. 4A). In some cases, the end product is both an allosteric inhibitor and an activator. An interesting case is ribonucleotide reductase (RNR), a two-subunit tetrameric enzyme, that reduces ribonucleotide to deoxyribonucleotide. In association with partner proteins, RNR fulfills the dNTPs requirement for DNA maintenance in cells. RNR is tightly regulated by dNTPs. RNR has non-specificity for substrate, as all four ribonucleotides are its substrate. However, for the conversion of a specific substrate, RNR requires a specific allosteric effector (Fig. 5B). Structural studies by the Ando group showed that the allosteric effectors of class 1b RNR cause distinct spatial reorganization of RNR subunits, making them either catalytically active or inactive [79]. Their studies showed that the binding of dATP, a negative effector, at the allosteric site gives RNR an inactive I-shape conformation which further results in the filamentation of RNR [79]. On the other hand, the binding of positive effectors gives it an active S-shape conformation [80] (Fig. 5B). The extensive structural studies by the Drennan group have revealed the allosteric mechanism of class Ia RNR functioning [81]. They showed that each allosteric effector modulates the active site loop conformation in such a way that the catalytic pocket can bind to a specific substrate only. Different effector changes the loop conformation differently (Fig. 5B). The flexibility of the active site loop gives substrate promiscuity to the RNR [81]. Mechanism of RNR is an excellent example of allosteric regulation of an enzyme towards substrate specificity.
The allosteric feedback mechanism is also applicable in the dual-enzyme system, where the product of one enzyme regulates the activity of the other. One such system in the SUVH-CMT enzyme system for epigenetic regulation (Fig. 5C). SUVH (Su-(var) 3–9 homolog) is a histone lysine methyltransferase (HKMT) that recognizes 5-methylcytosine (5mC) on the hemi-methylated DNA strand through the SRA domain and subsequently catalyzes dimethylation of lysine 9 on histone H3 (H3K9me2) through SET domain. The H3K9me2 mark allosterically activates the CMT (chromomethylase) that recognizes the H3K9me2 mark through the chromodomain and methylates the cytosine at 5th carbon (5mC) on the unmethylated DNA strand through MTase (methyltransferase) domain. In such cases, we see that both enzymes are not only allosterically regulated by the end-product of one another but have allosteric regulation within themselves too. In SUVH protein, 5mC-containing DNA acts as an allosteric regulator for the SET domain to catalyze H3K9me2, and simultaneously, H3K9me2 acts as an allosteric activator of the MTase domain of CMT [9], [11], [82], [83], [84], [85]. These allosteric regulations are co-dependent and work in a cyclic fashion, thus are known as the “reinforcement feedback loop” (Fig. 5C).
8. Regulation via multiple allosteric sites for function fine-tuning
Proteins involved in more than one pathway require different effectors to tune their function. These effectors either bind to the same site at different times or at different sites. Structural studies of signaling proteins like GPCRs have revealed multiple effector binding sites which regulate the signaling processes. GPCRs have 7 allosteric regulatory sites (microswitches), which include PIF motif, CWxP motif, DRY motif, hydrophobic patches, NPXXY motif, and Na+ binding site located within the transmembrane region, and an intracellularly located transducer binding site (Fig. 3A,D). The allosteric regulation of GPCRs has been reviewed in detail by the Christopoulos group [63].
PARP is another multi-domain protein that is regulated by multiple effectors that bind to PARP1 at different sites. Apart from DNA that binds to the N-terminal ZnF domains (as discussed in Section 1; Fig. 2A) and elicit self-PARylation of PARP1, there are other effectors, such as HMGN1 (High mobility group - nucleosome binding protein 1), HPF1 (Histone PARylation factor 1), YB-1 (Y-box-binding protein 1), etc. that either affects the self-PARylation of PARP1 or alters its substrate specificity. Bustin group has shown that HMGN1 binds to the catalytic domain of PARP1 (but not at the NAD+ binding site) and increases its DNA-dependent self-PARYlation activity by 10 folds [86]. HPF1 also interacts with the catalytic domain of PARP1 and redirects its self-PARylation activity to trans-PARylation of histone, specifically targeting serine [87], [88], [89], [90]. YB-1, on the other hand, competes with DNA to bind to PARP1 and restricts its self-PARylation activity [91], [92].
Advancements in computational approaches have revolutionized protein pocketome prediction. Programs such as SiteMap [93], FTMap (http://ftmap.bu.edu/), FTSite (http://ftsite.bu.edu/), molecular probing and docking, and others have revealed the previously not known allosteric sites, some of which have been experimentally validated in due course of time [94].
Multiple allosteric sites in a target protein provides an opportunity to design multivalent ligand switch that can help in designing “programmable effector” to tune the protein’s activity. In recent studies, using nucleic acids as a substrate, several groups have successfully outlined the framework of developing programmable molecular switches [95], [96]. This approach can have a tremendous application in designing “specialized/programmable” drug to monitor and regulate the active-inactive states of the target protein. This approach can also be used in designing a programmable machinery with applications in synthetic biology, biosensors development, and so on.
9. Kinetics of allostery
The kinetics of allostery is measured in two aspects: (i) enzyme/receptor activity, and (ii) substrate/ligand binding.
Enzymes with one substrate follow Michaelis-Menten kinetics, which describes the change in the rate of reaction (v) with respect to the substrate concentration ([S]) through the equation: , where is the rate of reaction, i.e, rate of substrate depletion (d[S]/dt) or rate of product (P) formation (d[P]/dt); Vmax is the maximum achievable rate of reaction; and KM is the Michaelis-Menten (MM) constant that denotes the substrate concentration at which the rate of reaction is half of the Vmax. This equation represents a non-linear rectangular hyperbolic function. The allosteric modulator, based on its positive or negative effect, can help the reaction attain equilibrium faster or slower. Either way, it makes the enzyme disobey the Michaelis-Menten kinetics and transforms the function to sigmoidal. The mathematics behind the kinetics of allosteric enzymes has been nicely reviewed by Einav et al. [97]. In terms of inhibition, non-competitive and uncompetitive inhibition are the types of allosteric inhibition, where inhibitor [I] binds to the allosteric site in an enzyme [E] either (i) in the presence of the substrate [S] (uncompetitive inhibition: [I] binds to [ES] complex), or (ii) irrespective of the presence of substrate (non-competitive inhibition: [I] binds to either [E] or [ES] complex). A non-competitive inhibitor doesn’t affect the KM, as it is not dependent on the substrate, but reduces the Vmax. On the other hand, an uncompetitive inhibitor reduces both KM and Vmax of the reaction.
Apart from the non-competitive and uncompetitive allosteric inhibition mechanisms, we propose a new type of inhibition, called ‘allosteric competition’, which has a mechanism of ‘competitive inhibition’, but the kinetics of ‘uncompetitive inhibition’. This would be the case in multi-domain enzymes that possess more than one allosteric site or have allosteric sites across the domains. In this case, a molecule binding to one allosteric site would affect the binding of an activator on another allosteric site. Though there would be no direct competition between the two allosteric molecules as both molecules bind to distinct sites, the binding of one would cause the dissociation of another, possibly due to spatial rearrangement of domains (Fig. 6A). Based on the effect on the enzyme activity, the allosteric competitor can be a PAM or NAM sub-type. Multi-domain proteins involved in multiple pathways have numerous regulators that bind to different domains and regulate the overall function of the protein. We observed this typical behavior in PARP1. Multiple studies have shown that PARP1 is activated by DNA, which binds to the N-terminal Zn-finger domains of PARP1 [21], [22], [23], [24]. Whereas, PAR, the catalytic product of PARP1, binds to the central domains of PARP1 (feedback mechanism) and causes dissociation on the activator DNA bound to the N-terminal domains [23], [24]. Though PAR and DNA are recognized by different sets of domains, the binding of PAR affects the DNA binding. This phenomenon can be explained as the allosteric competition (not for the same site of binding) between PAR and DNA to bind to PARP1. Since the DNA serves as the activator of PARP1, dislodging of DNA reduces the catalytic activity of PARP1. Due to this, both KM and Vmax would be affected, which is a characteristic of uncompetitive inhibition. At this point, mathematical modeling of such ‘allosteric competitive enzyme kinetics’ would be of great interest as that would help develop better allosteric drugs/inhibitors.
Fig. 6.
(A) Schematic representation of inhibition by ‘allosteric competition’. (B) Approaches to determine the effect of the allosteric modulator on substrate binding and catalytic activity of proteins.
Direct or indirect biochemical assays are routinely employed to measure enzyme kinetics by exploiting the spectroscopic signatures like absorbance or fluorescence of the compounds (Fig. 6B). Direct assays utilize the distinguished spectroscopic properties of the substrate used and/or the product formed during enzyme catalysis. Direct measurement depends on the native properties of the substrate and/or product, which may not be present in non-native compounds and thus cannot be used. In such cases, the indirect kinetics measurement can be taken advantage of, by converting the substrate and/or product to some other compound using a secondary enzyme or chemicals which can be quantified by its characteristic properties (Fig. 6B).
Isothermal titration calorimetry (ITC) is one of the direct methods to measure enzyme kinetics [98]. ITC measures the heat change (produced or absorbed) during the reaction in real time. Single injection and multiple injection assays can be performed to characterize enzyme allosteric interactions. In a single injection ITC kinetic assay, the heat change upon enzyme-substrate titration is measured till it reaches back to the baseline (indicating exhaustion of the substrate during the reaction). The substrate concentration is taken several folds more than KM and the enzyme concentration is chosen so that the reaction ends within a few minutes.
Winzor group used the ITC to measure the kinetics of pyruvate kinase (PK) that catalyzes the formation of pyruvate from the phosphoenolpyruvate, releasing an ATP [99]. It is believed that the allosteric binding of osmolytes, like phenylalanine, inactivates PK, causing cellular damage in phenylketonuria [100], [101]. To understand the active/inactive transition of PK in the presence of osmolytes, they performed ITC kinetics measurement of PK with the native substrate phosphoenolpyruvate in the absence and presence of phenylalanine and observed a sigmoidal shape right shift in the standard MM curve in the absence of phenylalanine indicating allosteric regulation. Further, with the addition of proline, the curve shifted back to the MM curve, indicating restored active enzyme conformation.
In multiple injections kinetic assay, low enzyme concentration but sufficient to provide a good signal, is used. Substrate concentration is adjusted such that during initial injections [S] < < Km and towards final injections [S] > > Km. With each injection, a drift in the baseline is observed which is directly proportional to the velocity of the reaction. Rolfsson's group used multiple injections approach to characterize the kinetics of phosphorylation of gluconate by gluconokinase [102]. It was observed that at higher gluconate concentration injected into the enzyme at a constant ATP concentration, the enzyme activity reduced indicating substrate inhibition. Also, with varying ATP concentration inhibition the change noted in the Hanes-Woolf plot confirmed the substrate inhibition through ternary complex formation Gluconokinase-ADP-gluconate.
ITC multiple injections assays are advantageous over single injection assays: (i) less enzyme is required; (ii) the amount of substrate consumed in multiple injections assays is very low (< 5%), therefore the product formed is less which is good for assessing the kinetics of enzymes having feedback inhibition. There are certain disadvantages also (i) signal-to-noise ratio is low since less enzyme concentration is used; (ii) multiple injections assay is relatively time-consuming.
10. Binding kinetics in drug discovery
Apart from kinetics studies, allosteric modulation can also be explained in terms of the effect of effector binding on substrate binding by using biophysical techniques like ITC, biolayer interferometry (BLI), surface plasmon resonance (SPR), and fluorescence polarization (FP) or anisotropy (FA), Microscale thermophoresis (MST) (Fig. 6B). These techniques can be employed to understand the effect of allosteric effectors on the binding parameters of the other binding partner or substrate. The effect can be monitored by analyzing the apparent molar equilibrium dissociation constant (appKD) in the presence of an effector. Techniques like BLI and SPR which calculate the rate constants of association (kon) and dissociation (koff) would provide further detailed insight into the mode of allostery. Effector molecules can affect the appKD value by altering the apparent kon (appkon) and/or koff (appkoff) (Fig. 4C).
Allosteric effector primarily locks the enzyme into either active or inactive conformation, thus behaving as either an activator or inhibitor. The Heitman group studied the effect of PAM, NAM effectors, and an antagonist on the substrate (metabotropic glutamate; mGlu) binding kinetics of the mGlu receptor 2 (mGluR2) [103]. Their studies showed that allosteric modulators, PAM and NAM, affect the binding kinetics parameters (kon and koff) of the orthosteric ligands (both agonist and antagonist) binding, however, the binding affinity may or may not be affected [103]. Apart from in vitro studies, the effect of allosteric modulators on GPCRs has also been demonstrated in cells, by live-cell imaging [104].
The binding kinetics parameters have emerged as a model to improve the therapeutic profile of the drugs [105]. Recently, contrary to the belief that the side effects of antipsychotic drugs (APDs) are due to their high dissociation rate, the Charlton group identified that it is otherwise. They discovered that the association rate constants of APDs to dopamine receptor 2 were responsible for the side effects [106]. They report that the optimization of association kinetics could deliver better results. For a drug molecule to be effective, it should have a very low koff value, i.e., high residence time (1/koff) in the receptor binding pocket [107]. Since the allosteric effectors have a significant effect on the active site dynamics, these can be used to optimize the binding kinetics of drugs at the active site. An allosteric effector that can significantly increase the appkon and reduce the appkoff, of the drug molecule at the active site could provide a promising effective way of treatment.
11. Allostery in drug design
There are several ways to understand allosteric mechanisms, which can be further translated to either direct medical applications or green chemistry for the biosynthesis of compounds with agricultural or healthcare applications. Apart from the approaches discussed above, several computational and experimental approaches to studying allostery have been discussed in detail by the Sattin group [108].
The advancements in structure biology, X-ray crystallography, cryo-EM, and in silico methods have facilitated an in-depth understanding of global and local conformational dynamics of protein allostery (Fig. 4B), leveraged for the development of novel allosteric drugs to treat various human diseases. Despite the field of allosteric drugs is continually growing, there are a few tens of FDA-approved allosteric drugs [109], [110], [111], as opposed to thousands of approved orthosteric drugs.
Ganaxolone is one such recently approved (2022) allosteric drug to treat seizures in people with cyclin-dependent kinase-like 5 (CDKL5) deficiency disorder (CDD). Ganaxolone is a PAM of GABA receptor which boosts the action of GABA resulting in a hyperpolarization of the neuron making it more resistant to excitation [112]. Unlike benzodiazepines which are also PAMs of synaptic GABAA as mentioned earlier, Ganaxolone binds to extra-synaptic GABAA receptors further strengthening the excitability resistance of neurons.
Kinase inhibitors comprise a promising class of drugs for the treatment of cancers. There are both orthosteric (categorized as type-I and type-II) drugs that target the ATP binding site and allosteric drugs that bind to an allosteric site, either adjacent to ATP binding region (classified as type-III) or distant from the ATP binding pocket (type-IV). MEK1 and MEK2 are threonine/tyrosine kinases of the mitogen-activated protein kinase kinase (MAPKK) involved in the RAS-RAF-MEK-ERK signal transduction pathway which regulates cell proliferation, differentiation, apoptosis, etc. A kinase, BRAF, activates MEK by phosphorylating it, and the BRAF V600E/K variant makes it constitutively active. Cobimetinib and trametinib are the FDA-approved type-III allosteric drugs against MEK [113], [114]. Both these inhibitors are used to treat melanoma carrying the BRAF V600E/K mutation. Trametinib specifically binds to un-phosphorylated MEK1 and MEK2, whereas cobimetinib binds to phosphorylated MEK1 with higher potency, thus inhibiting downstream proliferation signaling.
Combinations of antiretroviral drugs are used to treat HIV (Human Immunodeficiency Virus) infection. One of the components of these antiretrovirals is non-nucleoside reverse transcriptase inhibitors (NNRTI) that inhibit the replication of HIV-1 by binding directly to the reverse transcriptase in a non-competitive fashion. Currently, there are five NNRTIs (nevirapine, delavirdine, efavirenz, etravirine, and rilpivirine) approved by FDA. Rilpivirine is another such second-generation NNRTI that brings conformational changes in the reverse transcriptase by binding to an allosteric pocket adjacent to the DNA polymerizing processing site and thus hampering its functions [115], [116].
The discovery and development of drugs is an expensive and time-consuming process. The development of in silico interventions aids to accelerate the screening and cut down on extravagant costs. Enasidenib is an example of a drug that was discovered entirely using in silico approaches against mutated isocitrate dehydrogenase-2 (IDH2) for the treatment of adult patients with relapsed or refractory acute myeloid leukemia (AML) [117], [118]. IDH2 is a homodimer mitochondrial enzyme that converts isocitrate to ⍺-ketoglutarate (αKG) in Kreb’s cycle. The mutated IDH2 further converts αKG to 2-hydroxyglutarate, an oncogenic compound that inhibits αKG-dependent dioxygenases, causing aberrant chromatin modifications that subsequently lead to leukemia. Enasidenib binds to the allosteric site and locks the homodimer a conformation that prevents the oncogenic 2-hydroxyglutarate formation and thus helps to treat AML.
Many orthosteric and a few allosteric drugs, independently have proven to be effective to treat a large number of diseases. But sometimes the therapeutic effectiveness of drugs reduces due to off-target effects and drug resistance. Therefore, a novel approach to use a combination of both allosteric and orthosteric drugs has been proposed and is under clinical trials to overcome the shortcomings [119]. One such combination therapy is approved for treating human epidermal growth factor receptor 2 (HER2) positive breast cancer. Orthosteric drugs, pertuzumab and trastuzumab, target the HER2 receptor which inhibits the PI3K-Akt signaling pathway and the allosteric drug docetaxel prevents microtubule polymerization, which together prevents cell proliferation and thus treats cancer [120], [121].
For non-small-cell lung cancer (NSCLC), gefitinib, erlotinib, and lapatinib are widely used first-generation orthosteric EGFR (epidermal growth factor receptor) drugs. However, eventually, patients have developed drug resistance through EGFR T790M mutation which increases the binding affinity of ATP outcompeting the binding of drugs. A third-generation, covalent drug, Osimertinib, was approved to overcome the drawbacks of first-generation drugs. Jane's group identified a mutant selective (EGFR L858R/T790M) allosteric inhibitor JBJ-04–125–02, which along with Osimertinib proved to be more effective in killing cancer cells than individual drugs [122]. Binding of JBJ-04–125–02 causes conformational changes in the EGFR which resensitizes the cells to Osimertinib.
Allosteric drugs are more promising than conventional orthosteric drugs, either standalone or in combination with orthosteric drugs. Owing to a marginal probability of the existence of the same allosteric binding site in other proteins, allosteric drugs are highly specialized in terms of their target molecule. In order to expand the perimeter of allosteric drugs, more structural studies are required. With the advancement in computational approaches, such as molecular dynamics simulation, free energy simulation, quantum-mechanics/molecule mechanics (QM/MM), and the integration of artificial intelligence (AI; programs like AlphaFold, RosettaFold, RFDiffusion, etc.) [123], [124], [125], [126], novel targets with allosteric sites can be identified and therapeutics can be developed for specialized treatment with minimum side effects. These computational tools, especially simulations, are invaluable in understanding the mechanism of protein function, modelling enzymatic activity, screening molecules for effective drugs and so on. Since the implement of AI in biological studies is in its nascent stage, there are some drawbacks with AI-based programs that might result in biased outcomes in molecular modeling and docking, but it is still phenomenal in structure prediction [127], [128]. Further advancements in this field will help in better understanding of biological phenomena and simultaneously set a step forward in precision medicine.
12. Conclusion
Allostery plays an important role in regulating and/or fine-tuning enzyme/receptor activity as per cellular requirements. It has specific significance in modular proteins as they have multiple binding sites, specific for different binding partners or regulators. Each binding partner of these proteins affects the regulation by another binding partner, in either a positive or negative way. Understanding allosteric regulation in modular proteins not only provides mechanistic insights but is also useful in discovering its previously unknown association with different pathways. Apart from fundamental research, allostery is gaining a high significance in healthcare. Allosteric drugs are showing promising results in disease treatment. In this Review, we have summarized different mechanisms of allosteric regulation of enzyme functions along with their role in drug design.
CRediT authorship contribution statement
Suman Abhishek: Investigation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Waghela Deeksha: Investigation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Krishnapura R Nethravathi: Formal analysis, Visualization. Mehdi D Davari: Funding acquisition, Conceptualization, Writing - review & editing. Eerappa Rajakumara: Investigation, Supervision, Funding acquisition, Conceptualization, Writing - review & editing.
Declaration of Competing Interest
Authors declare no competing interest.
Acknowledgements
E.R. thanks the Department of Biotechnology (DBT) and the Science and Engineering Research Board, Department of Science and Technology, Government of India, for the Ramalingaswami re-entry fellowship and the Early Career Research Award, respectively. S.A. thanks the Ministry of Education, Government of India, for the fellowship. W.D. thanks the University Grants Commission, Government of India for the fellowship. M.D.D. is supported through funds from IPB Halle.
Contributor Information
Suman Abhishek, Email: abhisheks.iith@gmail.com.
Mehdi D. Davari, Email: mehdi.davari@ipb-halle.de.
Eerappa Rajakumara, Email: eraj@bt.iith.ac.in.
References
- 1.Wong I., Lohman T.M. Allosteric effects of nucleotide cofactors on escherichia coli rep helicase&dna binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 2.Davidson R.B., Hendrix J., Geiss B.J., McCullagh M. Allostery in the dengue virus NS3 helicase: insights into the NTPase cycle from molecular simulations. PLOS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn M.F. Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch Biochem Biophys. 2012;519:154–166. doi: 10.1016/j.abb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niks D., Hilario E., Dierkers A., Ngo H., Borchardt D., Neubauer T.J., et al. Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states. Biochemistry. 2013;52:6396–6411. doi: 10.1021/bi400795e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monod J., Jacob F. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 6.Liu J., Nussinov R. Allostery: an overview of its history, concepts, methods, and applications. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Yuan J., Lin J., Chen L., You L.-Y., Chen S., et al. Molecular basis of locus-specific H3K9 methylation catalyzed by SUVH6 in plants. Proc Natl Acad Sci. 2023;120 doi: 10.1073/pnas.2211155120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., Harris C.J., Zhong Z., Chen W., Liu R., Jia B., et al. Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc Natl Acad Sci USA. 2018;115:E8793–E8802. doi: 10.1073/pnas.1809841115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abhishek S., Deeksha W., Rajakumara E. Mechanistic insights into allosteric regulation of methylated DNA and histone H3 recognition by SRA and SET domains of SUVH5 and the basis for di-methylation of lysine residue. FEBS J. 2023;290:1060–1077. doi: 10.1111/febs.16633. [DOI] [PubMed] [Google Scholar]
- 10.Rajakumara E., Law J.A., Simanshu D.K., Voigt P., Johnson L.M., Reinberg D., et al. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satish M., Nivya M.A., Abhishek S., Nakarakanti N.K., Shivani D., Vani M.V., et al. Computational characterization of substrate and product specificities, and functionality of S-adenosylmethionine binding pocket in histone lysine methyltransferases from Arabidopsis, rice and maize. Protein: Struct, Funct, Bioinforma. 2018;86:21–34. doi: 10.1002/prot.25399. [DOI] [PubMed] [Google Scholar]
- 12.Tsai C.-J., Del Sol A., Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajakumara E., Abhishek S., Nitin K., Saniya D., Bajaj P., Schwaneberg U., et al. Structure and cooperativity in substrate–enzyme interactions: perspectives on enzyme engineering and inhibitor design. ACS Chem Biol. 2022;17:266–280. doi: 10.1021/acschembio.1c00500. [DOI] [PubMed] [Google Scholar]
- 14.Choi J.-M., Han S.-S., Kim H.-S. Industrial applications of enzyme biocatalysis: current status and future aspects. Biotechnol Adv. 2015;33:1443–1454. doi: 10.1016/j.biotechadv.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Wells A.S., Finch G.L., Michels P.C., Wong J.W. Use of enzymes in the manufacture of active pharmaceutical ingredients—a science and safety-based approach to ensure patient safety and drug quality. Org Process Res Dev. 2012;16:1986–1993. doi: 10.1021/op300153b. [DOI] [Google Scholar]
- 16.Robinson P.K. Enzymes: principles and biotechnological applications. Essays Biochem. 2015;59:1–41. doi: 10.1042/bse0590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nussinov R., Tsai C.-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 18.Wong T.-S., Li G., Li S., Gao W., Chen G., Gan S., et al. G protein-coupled receptors in neurodegenerative diseases and psychiatric disorders. Sig Transduct Target Ther. 2023;8:1–57. doi: 10.1038/s41392-023-01427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y., Todd N., Thathiah A. The role of GPCRs in neurodegenerative diseases: avenues for therapeutic intervention. Curr Opin Pharm. 2017;32:96–110. doi: 10.1016/j.coph.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Azam S., Haque M.E., Jakaria M., Jo S.-H., Kim I.-S., Choi D.-K. G-protein-coupled receptors in CNS: a potential therapeutic target for intervention in neurodegenerative disorders and associated cognitive deficits. Cells. 2020;9:506. doi: 10.3390/cells9020506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eustermann S., Wu W.-F., Langelier M.-F., Yang J.-C., Easton L.E., Riccio A.A., et al. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol Cell. 2015;60:742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langelier M.-F., Planck J.L., Roy S., Pascal J.M. Structural basis for DNA damage–dependent Poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deeksha W., Abhishek S., Rajakumara E. PAR recognition by PARP1 regulates DNA-dependent activities and independently stimulates catalytic activity of PARP1. FEBS J. 2023 doi: 10.1111/febs.16907. [DOI] [PubMed] [Google Scholar]
- 24.Deeksha W., Abhishek S., Giri J., Rajakumara E. Regulation of PARP1 and its apoptotic variant activity by single-stranded DNA. FEBS J. 2023 doi: 10.1111/febs.16875. [DOI] [PubMed] [Google Scholar]
- 25.Huang F., Zhu B. The cyclic oligoadenylate signaling pathway of type III CRISPR-Cas systems. Front Microbiol. 2021;11 doi: 10.3389/fmicb.2020.602789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J., Feng M., Zhang H., She Q. Characterization of a novel type III CRISPR-Cas effector provides new insights into the allosteric activation and suppression of the Cas10 DNase. Cell Discov. 2020;6:1–16. doi: 10.1038/s41421-020-0160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia N., Jones R., Yang G., Ouerfelli O., Patel D.J. CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol Cell. 2019;75:944–956. doi: 10.1016/j.molcel.2019.06.014. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao D., Wu J., Wu Y.-T., Du F., Aroh C., Yan N., et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X.-D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Wu J., Du F., Xu H., Sun L., Chen Z., et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Shu C., Yi G., Chaton C.T., Shelton C.L., Diao J., et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie W., Lama L., Adura C., Tomita D., Glickman J.F., Tuschl T., et al. Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc Natl Acad Sci. 2019;116:11946–11955. doi: 10.1073/pnas.1905013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooy R.M., Sohn J. The allosteric activation of cGAS underpins its dynamic signaling landscape. eLife. 2018;7 doi: 10.7554/eLife.39984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopfner K.-P., Hornung V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat Rev Mol Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 36.Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G., et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu S., Sridhar A., Teng J., Howard R.J., Lindahl E., Hibbs R.E. Structural and dynamic mechanisms of GABAA receptor modulators with opposing activities. Nat Commun. 2022;13:4582. doi: 10.1038/s41467-022-32212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter L., de Graaf C., Sieghart W., Varagic Z., Mörzinger M., de Esch I.J.P., et al. Diazepam-bound GABAA receptor models identify new benzodiazepine binding-site ligands. Nat Chem Biol. 2012;8:455–464. doi: 10.1038/nchembio.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baur R., Tan K.R., Lüscher B.P., Gonthier A., Goeldner M., Sigel E. Covalent modification of GABAA receptor isoforms by a diazepam analogue provides evidence for a novel benzodiazepine binding site that prevents modulation by these drugs. J Neurochem. 2008;106:2353–2363. doi: 10.1111/j.1471-4159.2008.05574.x. [DOI] [PubMed] [Google Scholar]
- 40.Maldifassi M.C., Baur R., Pierce D., Nourmahnad A., Forman S.A., Sigel E. Novel positive allosteric modulators of GABAA receptors with anesthetic activity. Sci Rep. 2016;6:25943. doi: 10.1038/srep25943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory K.J., Goudet C. International union of basic and clinical pharmacology. CXI. Pharmacology, signaling, and physiology of metabotropic glutamate receptors. Pharmacol Rev. 2021;73:521–569. doi: 10.1124/pr.119.019133. [DOI] [PubMed] [Google Scholar]
- 42.Cao A.-M., Quast R.B., Fatemi F., Rondard P., Pin J.-P., Margeat E. Allosteric modulators enhance agonist efficacy by increasing the residence time of a GPCR in the active state. Nat Commun. 2021;12:5426. doi: 10.1038/s41467-021-25620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theander-Carrillo C., Wiedmer P., Cettour-Rose P., Nogueiras R., Perez-Tilve D., Pfluger P., et al. Ghrelin action in the brain controls adipocyte metabolism. J Clin Invest. 2006;116:1983–1993. doi: 10.1172/JCI25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanagi S., Sato T., Kangawa K., Nakazato M. The homeostatic force of ghrelin. Cell Metab. 2018;27:786–804. doi: 10.1016/j.cmet.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Davis J. Hunger, ghrelin and the gut. Brain Res. 2018;1693:154–158. doi: 10.1016/j.brainres.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 46.Kojima M., Kangawa K. Ghrelin: more than endogenous growth hormone secretagogue. Ann N Y Acad Sci. 2010;1200:140–148. doi: 10.1111/j.1749-6632.2010.05516.x. [DOI] [PubMed] [Google Scholar]
- 47.Evron T., Peterson S.M., Urs N.M., Bai Y., Rochelle L.K., Caron M.G., et al. G Protein and β-Arrestin Signaling Bias at the Ghrelin Receptor*. J Biol Chem. 2014;289:33442–33455. doi: 10.1074/jbc.M114.581397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiimura Y., Horita S., Hamamoto A., Asada H., Hirata K., Tanaka M., et al. Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode. Nat Commun. 2020;11:4160. doi: 10.1038/s41467-020-17554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin J., Cai Y., Xu Z., Ming Q., Ji S.-Y., Wu C., et al. Molecular mechanism of agonism and inverse agonism in ghrelin receptor. Nat Commun. 2022;13:300. doi: 10.1038/s41467-022-27975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J., Zhao T.-J., Goldstein J.L., Brown M.S. Inhibition of ghrelin O-acyltransferase (GOAT) by octanoylated pentapeptides. Proc Natl Acad Sci. 2008;105:10750–10755. doi: 10.1073/pnas.0805353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu S.-C., Xu J., Chinookoswong N., Liu S., Steavenson S., Gegg C., et al. An acyl-ghrelin-specific neutralizing antibody inhibits the acute ghrelin-mediated orexigenic effects in mice. Mol Pharm. 2009;75:901–907. doi: 10.1124/mol.108.052852. [DOI] [PubMed] [Google Scholar]
- 52.Wortley K.E., Rincon J.-P., del, Murray J.D., Garcia K., Iida K., Thorner M.O., et al. Absence of ghrelin protects against early-onset obesity. J Clin Invest. 2005;115:3573–3578. doi: 10.1172/JCI26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y., Butte N.F., Garcia J.M., Smith R.G. Characterization of adult ghrelin and ghrelin receptor knockout mice under positive and negative energy balance. Endocrinology. 2008;149:843–850. doi: 10.1210/en.2007-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colldén G., Tschöp M.H., Müller T.D. Therapeutic potential of targeting the ghrelin pathway. Int J Mol Sci. 2017;18:798. doi: 10.3390/ijms18040798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wakefield A.E., Mason J.S., Vajda S., Keserű G.M. Analysis of tractable allosteric sites in G protein-coupled receptors. Sci Rep. 2019;9:6180. doi: 10.1038/s41598-019-42618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garai S., Leo L.M., Szczesniak A.-M., Hurst D.P., Schaffer P.C., Zagzoog A., et al. Discovery of a biased allosteric modulator for cannabinoid 1 receptor: preclinical anti-glaucoma efficacy. J Med Chem. 2021;64:8104–8126. doi: 10.1021/acs.jmedchem.1c00040. [DOI] [PubMed] [Google Scholar]
- 57.Wang J., Pani B., Gokhan I., Xiong X., Kahsai A.W., Jiang H., et al. β-Arrestin–biased allosteric modulator potentiates carvedilol-stimulated β adrenergic receptor cardioprotection. Mol Pharm. 2021;100:568–579. doi: 10.1124/molpharm.121.000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slosky L.M., Caron M.G., Barak L.S. Biased allosteric modulators: new frontiers in GPCR drug discovery. Trends Pharmacol Sci. 2021;42:283–299. doi: 10.1016/j.tips.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcaraz A.R., Bond R.A., McConnell B.K. Biased allosteric modulators as a novel approach to induce biased signalling in G-protein coupled receptors: The case of beta 2 adrenergic receptor in cardiovascular diseases. FASEB J. 2020;34 doi: 10.1096/fasebj.2020.34.s1.06759. 1–1. [DOI] [Google Scholar]
- 60.Ford B.M., Franks L.N., Tai S., Fantegrossi W.E., Stahl E.L., Berquist M.D., et al. Characterization of structurally novel G protein biased CB1 agonists: Implications for drug development. Pharm Res. 2017;125:161–177. doi: 10.1016/j.phrs.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye L., Cao Z., Wang W., Zhou N. New Insights in Cannabinoid Receptor Structure and Signaling. Current Molecular Pharmacology n.d.;12:239–248. [DOI] [PMC free article] [PubMed]
- 62.Shen S., Zhao C., Wu C., Sun S., Li Z., Yan W., et al. Allosteric modulation of G protein-coupled receptor signaling. Front Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1137604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thal D.M., Glukhova A., Sexton P.M., Christopoulos A. Structural insights into G-protein-coupled receptor allostery. Nature. 2018;559:45–53. doi: 10.1038/s41586-018-0259-z. [DOI] [PubMed] [Google Scholar]
- 64.Wakefield A.E., Bajusz D., Kozakov D., Keserű G.M., Vajda S. Conservation of allosteric ligand binding sites in G-protein coupled receptors. J Chem Inf Model. 2022;62:4937–4954. doi: 10.1021/acs.jcim.2c00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murai J., Huang S.N., Das B.B., Renaud A., Zhang Y., Doroshow J.H., et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pommier Y., O’Connor M.J., de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aaf9246. 362ps17-362ps17. [DOI] [PubMed] [Google Scholar]
- 67.Zandarashvili L., Langelier M.-F., Velagapudi U.K., Hancock M.A., Steffen J.D., Billur R., et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science. 2020;368:eaax6367. doi: 10.1126/science.aax6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langelier M.-F., Lin X., Zha S., Pascal J.M. Clinical PARP inhibitors allosterically induce PARP2 retention on DNA. Sci Adv. 2023;9:eadf7175. doi: 10.1126/sciadv.adf7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murai J., Huang S.-Y.N., Renaud A., Zhang Y., Ji J., Takeda S., et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas C., Ji Y., Lodhi N., Kotova E., Pinnola A.D., Golovine K., et al. Non-NAD-Like poly(ADP-Ribose) Polymerase-1 inhibitors effectively eliminate cancer in vivo. EBioMedicine. 2016;13:90–98. doi: 10.1016/j.ebiom.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nieborowska-Skorska M., Maifrede S., Ye M., Toma M., Hewlett E., Gordon J., et al. Non-NAD-like PARP1 inhibitor enhanced synthetic lethal effect of NAD-like PARP inhibitors against BRCA1-deficient leukemia. Leuk Lymphoma. 2019;60:1098–1101. doi: 10.1080/10428194.2018.1520988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hewlett E., Toma M., Sullivan-Reed K., Gordon J., Sliwinski T., Tulin A., et al. Novel allosteric PARP1 inhibitors for the treatment of BRCA-deficient leukemia. Med Chem Res. 2020;29:962–978. doi: 10.1007/s00044-020-02537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulze J.O., Saladino G., Busschots K., Neimanis S., Süß E., Odadzic D., et al. Bidirectional allosteric communication between the ATP-binding site and the regulatory PIF pocket in PDK1 protein kinase. Cell Chem Biol. 2016;23:1193–1205. doi: 10.1016/j.chembiol.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 74.Lanman B.A., Allen J.R., Allen J.G., Amegadzie A.K., Ashton K.S., Booker S.K., et al. Discovery of a Covalent Inhibitor of KRASG12C (AMG 510) for the Treatment of Solid Tumors. J Med Chem. 2020;63:52–65. doi: 10.1021/acs.jmedchem.9b01180. [DOI] [PubMed] [Google Scholar]
- 75.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans P.R., Farrants G.W., Hudson P.J. Phosphofructokinase: structure and control. Philos Trans R Soc Lond B Biol Sci. 1981;293:53–62. doi: 10.1098/rstb.1981.0059. [DOI] [PubMed] [Google Scholar]
- 77.Schirmer T., Evans P.R. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]
- 78.Li Y., Rivera D., Ru W., Gunasekera D., Kemp R.G. Identification of allosteric sites in rabbit phosphofructo-1-kinase. Biochemistry. 1999;38:16407–16412. doi: 10.1021/bi991761l. [DOI] [PubMed] [Google Scholar]
- 79.Thomas W.C., Brooks F.P., Burnim A.A., Bacik J.-P., Stubbe J., Kaelber J.T., et al. Convergent allostery in ribonucleotide reductase. Nat Commun. 2019;10:2653. doi: 10.1038/s41467-019-10568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eriksson M., Uhlin U., Ramaswamy S., Ekberg M., Regnström K., Sjöberg B.-M., et al. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/S0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 81.Zimanyi C.M., Chen P.Y.-T., Kang G., Funk M.A., Drennan C.L. Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli. eLife. 2016;5 doi: 10.7554/eLife.07141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson L.M., Bostick M., Zhang X., Kraft E., Henderson I., Callis J., et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du J., Johnson L.M., Groth M., Feng S., Hale C.J., Li S., et al. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol Cell. 2014;55:495–504. doi: 10.1016/j.molcel.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 85.Jackson J.P., Johnson L., Jasencakova Z., Zhang X., PerezBurgos L., Singh P.B., et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 86.Masaoka A., Gassman N.R., Kedar P.S., Prasad R., Hou E.W., Horton J.K., et al. HMGN1 protein regulates poly(ADP-ribose) polymerase-1 (PARP-1) self-PARylation in mouse fibroblasts. J Biol Chem. 2012;287:27648–27658. doi: 10.1074/jbc.M112.370759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibbs-Seymour I., Fontana P., Rack J.G.M., Ahel I. HPF1/C4orf27 Is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol Cell. 2016;62:432–442. doi: 10.1016/j.molcel.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kurgina T.A., Moor N.A., Kutuzov M.M., Naumenko K.N., Ukraintsev A.A., Lavrik O.I. Dual function of HPF1 in the modulation of PARP1 and PARP2 activities. Commun Biol. 2021;4:1259. doi: 10.1038/s42003-021-02780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langelier M.-F., Billur R., Sverzhinsky A., Black B.E., Pascal J.M. HPF1 dynamically controls the PARP1/2 balance between initiating and elongating ADP-ribose modifications. Nat Commun. 2021;12:6675. doi: 10.1038/s41467-021-27043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suskiewicz M.J., Zobel F., Ogden T.E.H., Fontana P., Ariza A., Yang J.-C., et al. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature. 2020;579:598–602. doi: 10.1038/s41586-020-2013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alemasova E.E., Pestryakov P.E., Sukhanova M.V., Kretov D.A., Moor N.A., Curmi P.A., et al. Poly(ADP-ribosyl)ation as a new posttranslational modification of YB-1. Biochimie. 2015;119:36–44. doi: 10.1016/j.biochi.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 92.Alemasova E.E., Moor N.A., Naumenko K.N., Kutuzov M.M., Sukhanova M.V., Pestryakov P.E., et al. Y-box-binding protein 1 as a non-canonical factor of base excision repair. Biochim Biophys Acta. 2016;1864:1631–1640. doi: 10.1016/j.bbapap.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 93.Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 94.Hedderich J.B., Persechino M., Becker K., Heydenreich F.M., Gutermuth T., Bouvier M., et al. The pocketome of G-protein-coupled receptors reveals previously untargeted allosteric sites. Nat Commun. 2022;13:2567. doi: 10.1038/s41467-022-29609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu L., Yuan M., Jin Y., Zhou G., Li T., Li L., et al. Tunable dual-effector allostery system for nucleic acid analysis with enhanced sensitivity and an extended dynamic range. Anal Chem. 2021;93:8170–8177. doi: 10.1021/acs.analchem.1c00055. [DOI] [PubMed] [Google Scholar]
- 96.Lauzon D., Vallée-Bélisle A. Programing chemical communication: allostery vs multivalent mechanism. J Am Chem Soc. 2023;145:18846–18854. doi: 10.1021/jacs.3c04045. [DOI] [PubMed] [Google Scholar]
- 97.Einav T., Mazutis L., Phillips R. Statistical mechanics of allosteric enzymes. J Phys Chem B. 2016;120:6021–6037. doi: 10.1021/acs.jpcb.6b01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y., Wang G., Moitessier N., Mittermaier A.K. Enzyme kinetics by isothermal titration calorimetry: allostery, inhibition, and dynamics. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.583826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lonhienne T.G.A., Winzor D.J. Calorimetric demonstration of the potential of molecular crowding to emulate the effect of an allosteric activator on pyruvate kinase kinetics. Biochemistry. 2002;41:6897–6901. doi: 10.1021/bi020064h. [DOI] [PubMed] [Google Scholar]
- 100.Hörster F., Schwab M.A., Sauer S.W., Pietz J., Hoffmann G.F., Okun J.G., et al. Phenylalanine reduces synaptic density in mixed cortical cultures from mice. Pedia Res. 2006;59:544–548. doi: 10.1203/01.pdr.0000203091.45988.8d. [DOI] [PubMed] [Google Scholar]
- 101.van Spronsen F.J., Hoeksma M., Reijngoud D.-J. Brain dysfunction in phenylketonuria: Is phenylalanine toxicity the only possible cause. J Inherit Metab Dis. 2009;32:46–51. doi: 10.1007/s10545-008-0946-2. [DOI] [PubMed] [Google Scholar]
- 102.Rohatgi N., Guðmundsson S., Rolfsson Ó. Kinetic analysis of gluconate phosphorylation by human gluconokinase using isothermal titration calorimetry. FEBS Lett. 2015;589:3548–3555. doi: 10.1016/j.febslet.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 103.Doornbos M.L.J., Vermond S.C., Lavreysen H., Tresadern G., IJzerman A.P., Heitman L.H. Impact of allosteric modulation: Exploring the binding kinetics of glutamate and other orthosteric ligands of the metabotropic glutamate receptor 2. Biochem Pharmacol. 2018;155:356–365. doi: 10.1016/j.bcp.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 104.May L.T., Self T.J., Briddon S.J., Hill S.J. The effect of allosteric modulators on the kinetics of agonist-G protein-coupled receptor interactions in single living cells. Mol Pharm. 2010;78:511–523. doi: 10.1124/mol.110.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tonge P.J. Drug–Target Kinetics in Drug Discovery. ACS Chem Neurosci. 2018;9:29–39. doi: 10.1021/acschemneuro.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sykes D.A., Moore H., Stott L., Holliday N., Javitch J.A., Lane J.R., et al. Extrapyramidal side effects of antipsychotics are linked to their association kinetics at dopamine D2 receptors. Nat Commun. 2017;8:763. doi: 10.1038/s41467-017-00716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Knockenhauer K.E., Copeland R.A. The importance of binding kinetics and drug–target residence time in pharmacology. British Journal of Pharmacology n.d.;n/a. 10.1111/bph.16104. [DOI] [PubMed]
- 108.Civera M., Moroni E., Sorrentino L., Vasile F., Sattin S. Chemical and biophysical approaches to allosteric modulation. Eur J Org Chem. 2021;2021:4245–4259. doi: 10.1002/ejoc.202100506. [DOI] [Google Scholar]
- 109.Liu X., Lu S., Song K., Shen Q., Ni D., Li Q., et al. Unraveling allosteric landscapes of allosterome with ASD. Nucleic Acids Res. 2020;48:D394–D401. doi: 10.1093/nar/gkz958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaczor A.A., Wróbel T.M., Bartuzi D. Allosteric modulators of dopamine D2 receptors for fine-tuning of dopaminergic neurotransmission in cns diseases: overview, pharmacology, structural aspects and synthesis. Molecules. 2023;28:178. doi: 10.3390/molecules28010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mullard A. 2022 FDA approvals. Nat Rev Drug Discov. 2023;22:83–88. doi: 10.1038/d41573-023-00001-3. [DOI] [PubMed] [Google Scholar]
- 112.Yasmen N., Sluter M.N., Yu Y., Jiang J. Ganaxolone for management of seizures associated with CDKL5 deficiency disorder. Trends Pharmacol Sci. 2023;44:128–129. doi: 10.1016/j.tips.2022.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Unsworth A.J., Bye A.P., Kriek N., Sage T., Osborne A.A., Donaghy D., et al. Cobimetinib and trametinib inhibit platelet MEK but do not cause platelet dysfunction. Platelets. 2019;30:762–772. doi: 10.1080/09537104.2018.1514107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu P., Clausen M.H., Nielsen T.E. Allosteric small-molecule kinase inhibitors. Pharmacol Ther. 2015;156:59–68. doi: 10.1016/j.pharmthera.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 115.Usach I., Melis V., Peris J.-E. Non-nucleoside reverse transcriptase inhibitors: a review on pharmacokinetics, pharmacodynamics, safety and tolerability. J Int AIDS Soc. 2013;16:18567. doi: 10.7448/IAS.16.1.18567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ferretti F., Boffito M. Rilpivirine long-acting for the prevention and treatment of HIV infection. Curr Opin HIV AIDS. 2018;13:300–307. doi: 10.1097/COH.0000000000000474. [DOI] [PubMed] [Google Scholar]
- 117.Abou Dalle I., DiNardo C.D. The role of enasidenib in the treatment of mutant IDH2 acute myeloid leukemia. Ther Adv Hematol. 2018;9:163–173. doi: 10.1177/2040620718777467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reed D.R., Elsarrag R.Z., Morris A.L., Keng M.K. Enasidenib in acute myeloid leukemia: clinical development and perspectives on treatment. CMAR. 2019;11:8073–8080. doi: 10.2147/CMAR.S162784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ni D., Li Y., Qiu Y., Pu J., Lu S., Zhang J. Combining allosteric and orthosteric drugs to overcome drug resistance. Trends Pharmacol Sci. 2020;41:336–348. doi: 10.1016/j.tips.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 120.Swain S.M., Baselga J., Kim S.-B., Ro J., Semiglazov V., Campone M., et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Swain S.M., Miles D., Kim S.-B., Im Y.-H., Im S.-A., Semiglazov V., et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA): end-of-study results from a double-blind, randomised, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:519–530. doi: 10.1016/S1470-2045(19)30863-0. [DOI] [PubMed] [Google Scholar]
- 122.To C., Jang J., Chen T., Park E., Mushajiang M., De Clercq D.J.H., et al. Single and dual targeting of mutant EGFR with an allosteric inhibitor. Cancer Discov. 2019;9:926–943. doi: 10.1158/2159-8290.CD-18-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Watson J.L., Juergens D., Bennett N.R., Trippe B.L., Yim J., Eisenach H.E., et al. De novo design of protein structure and function with RFdiffusion. Nature. 2023:1–3. doi: 10.1038/s41586-023-06415-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sheik Amamuddy O., Veldman W., Manyumwa C., Khairallah A., Agajanian S., Oluyemi O., et al. Integrated computational approaches and tools forallosteric drug discovery. Int J Mol Sci. 2020;21:847. doi: 10.3390/ijms21030847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nussinov Ruth, Mingzhen Zhang, Yonglan Liu, Hyunbum Jang. AlphaFold, allosteric, and orthosteric drug discovery: ways forward. Drug Discov Today. 2023 doi: 10.1016/j.drudis.2023.103551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nussinov Ruth, Mingzhen Zhang, Yonglan Liu, Hyunbum Jang. AlphaFold, artificial intelligence (AI), and allostery. J Phys Chem B. 2022;126(34):6372–6383. doi: 10.1021/acs.jpcb.2c04346. [DOI] [PMC free article] [PubMed] [Google Scholar]