Abstract
With the increasing acreage of genetically modified crops worldwide, rapid and efficient detection technologies have become very important for the regulation and screening of GM organisms. We constructed a method based on loop-mediated isothermal amplification (LAMP), CRISPR-Cas12a and lateral flow assay (LAMP-CRISPR-Cas12a-LFA). It is an intuitive, sensitive and specific fluorescence detection and test strip system to detect CP4-EPSPS and Cry1Ab/Ac genes in field screening. The LAMP-CRISPR-Cas12a-LFA method has a limit of detection (LOD) of 100 copies based on lateral flow test strips after optimization of the conditions with screened specific primers, and the entire detection process can be completed within 1 h at 61 °C. The system was used to evaluate field test samples and showed high reproducibility after testing products containing CP4-EPSPS and Cry1Ab/Ac genes, and both were detectable. The LAMP-CRISPR-Cas12a-LFA method established in this paper functions as a rapid field detection method. It requires only one portable thermostatic instrument, which renders it compatible with the rapid detection of field samples and useable at experimental workstations, in law enforcement field work, and in local inspection and quarantine departments.
Keywords: GM organisms, CRISPR-Cas12a, LAMP, Rapid detection
Highlights
-
•
An accurate, sensitive, time-saving and portable on-site screening system for the detection of Genetically Modified Products developed using LAMP -CRISPR Cas12a-LFA for test strip detection.
-
•
The limit of detection of lateral flow biosensor method was as low as 100 copies/μL within 1h.
-
•
The LAMP -CRISPR Cas12a-LFA system is fit for on-site rapid molecular identification screening of Genetically Modified Products.
1. Introduction
Since their first commercial rollout in 1996, the acreage of GM crops has expanded significantly (Lin and Pan, 2016). The global area dedicated to GM organisms has increased ∼112-fold, from 1.7 million hectares in 1996 to 190.4 million hectares in 2019 (ISAAA, 2021). Over the past 26 years, insect-resistant (Cry1Ab/Ac gene) and herbicide-resistant (CP4-EPSPS gene) crops have become the most widely planted GM crops, and these transgenic traits have resulted in major economic benefits. In 2019, the area dedicated to herbicide-tolerant crops reached 43% of all worldwide agricultural areas, and about 12% of these transgenic crops also have insect-resistant traits. The study of transgenic crops with CP4-EPSPS and Cry1Ab/Ac genes has far-reaching implications for food safety testing, environmental and monitoring. Due to the widespread application of genetically modified organism (GMO) technology and the large-scale cultivation of GM crops, the safety of GM food has gradually aroused people's concern (Ponti, 2005). At present, countries have enacted corresponding laws and regulations for standardized management, and most countries have established mandatory labeling systems for the detection and monitoring of GM foods (Devos et al., 2022; Singh, 2021). The United States is one of the world's largest growers of GMOs. Its government has been supportive of the development of genetic modification technology. The U.S. Food and Drug Administration (FDA) requires that GM foods undergo rigorous safety assessment and be labeled on food packaging. The EU also has a strict labeling system for GM products that mandates clear labeling for products with more than 0.9% GM content. The Chinese government supports the development of genetic modification technology, but it has stricter controls on genetically modified foods. No GM staple crop seeds have been approved for commercial cultivation in the country. A labeling system established under these regulations requires the addition of easier and more sensitive testing methods for GMOs detection (Huang et al., 2022). Various approaches to GM detection have been reported (Kamle et al., 2017), including PCR-based detecting methods (Chen et al., 2019; Holst-Jensen, 2009; Shang et al., 2020), fluorescent quantitative PCR (Akiyama et al., 2010; Cho et al., 2013; Kageyama et al., 2003), enzyme-linked immunosorbent assay (ELISA) (Emslie et al., 2007; Morenkov, 2000; Zeng et al., 2021), protein blotting (Mishra et al., 2017) (western blotting), and protein microarray (Ranek et al., 2020). However, these methods also have significant disadvantages (Qian et al., 2018). For example, the detection rate of GM crops is low because the proteins become denatured during treatment, and detection requires expensive instruments and complex procedures, which are costly (Deng et al., 2013; Wu et al., 2014; Xu et al., 2017).
The current demand for the detection of transgenic products is increasingly inclined toward rapid visual field detection, and the rapidly developing isothermal amplification technique loop-mediated isothermal amplification (LAMP) has been widely used in transgenic detection (Prasannakumar et al., 2021; Takabatake et al., 2018; Wu et al., 2020). The clustered regularly interspaced short palindromic repeats (CRISPR) system's trans-cleavage effect detection technology has high sensitivity, strong targeting and rapid detection, and its use has become popular in research and development in related fields. CRISPR-related protein Cas12a, a class II V CRISPR system RNA-guided nucleic acid endonuclease with incidental cleavage activity, has been combined with amplification technology to develop new DNA detection methods, such as DETECTR and HOLMES, which enable rapid and sensitive detection of the test target (Chen et al., 2018; Du et al., 2021; Murugan et al., 2020; Wang et al., 2020). Amplification techniques such as loop-mediated isothermal amplification are relatively mature isothermal (Tsugunori et al., 2000) amplification technologies. They are known for their high efficiency, high specificity, and low cost (Lee et al., 2019), and are widely used in nucleic acid detection, such as virus detection and transgenic detection (Joshi et al., 2021; Pirc et al., 2021). The combination of CRISPR and LAMP can realize the detection of low copy molecules. The lateral flow test strip is combined with CRISPR and LAMP detection to visualize the results, which is conducive to large-scale screening and detection in the field.
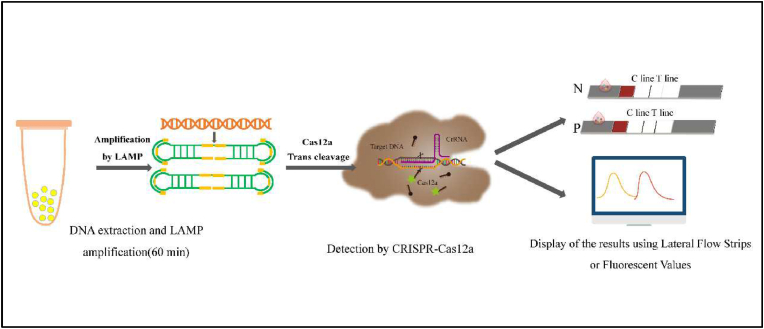
In this paper, the CRISPR-Cas12a system was combined with the established LAMP and lateral flow test strips to establish an intuitive, sensitive, and specific detection method for CP4-EPSPS and Cry1Ab/Ac. The entire detection process requires only a portable thermostat, and can be performed quickly and onsite. This method is simple to operate and is highly sensitive. It is thus convenient for the detection of targets present at low concentrations. It can be used at experimental workstations, in law enforcement field work, and in territorial inspection and quarantine departments. Fig. 1 shows the detailed detection mode and how the results are displayed. The advantages of this assay are as follows: (1) visualization of the assay results by combining with lateral flow test strips; (2) ability to recognize low concentration targets generating strong fluorescent signals; and (3) differentiation of specific and non-specific amplification products by fluorescence detection. LAMP-CRISPR-Cas12a is less enzyme-dependent, requires less manual labor, and is more efficient in both money and time. It facilitates timely monitoring and rapid development of transgenic control strategies.
Fig. 1.
Detection of GMO products by the LAMP-CRISPR/Cas12a system.
Genomic DNA was extracted from GMO products. LAMP amplification was performed at 60 °C for 12–15 min, followed by the Cas12a trans-cleavage reaction. The reaction mixture was directly loaded on the lateral flow strip. This portable CRISPR/Cas12a-based lateral flow platform was suitable for sensitive detection of GMOs. In the presence of the target gene, the ssDNA reporter complex of Bioten-15NT-FAM was captured by the capture probe and pre-modified on the test line. The test line and the control line are displayed. The results are visible to the naked eye. In high-throughput detection, the ssDNA reporter group of HEX-12NT-BHQ1 is used to directly utilize a fluorescence reader for fluorescence measurement.
2. Materials and methods
2.1. Materials
2.1.1. Reagents
2.1.1.1. Sample preparation
Genomic DNA from the seeds of transgenic material was extracted using a DP350 extraction kit (Tiangen Biotechnology, Beijing, China). The transgenic products used in this paper were maize, soybean, cotton, canola, alfalfa, and sugar beet containing the CP4-EPSPS gene and maize, soybean, cotton, and rice containing the Cry1Ab/Ac gene (Table 1). The various GMO detection samples were provided by the Crop Ecological Environment Safety Monitoring and Test Center, Ministry of Agriculture, People's Republic of China. First, samples were crushed in a grinder, then plant genomic DNA was extracted and purified from 0.1 g of powder made from various transgenic crops using a genomic extraction kit. The quality and concentration of DNA samples were evaluated using a Nano Drop 1000 UV/Vis spectrophotometer (Thermo Scientific) and 1% agarose gel electrophoresis for subsequent amplification experiments.
Table 1.
Experimental materials.
| GMO events | ID | CP4 EPSPS | Cry1Ab/Ac |
|---|---|---|---|
| Transgenic maize | NK603 | + | – |
| Transgenic soybean | RRS MON89788 | + | – |
| Transgenic cotton | MON1445 MON88913 | + | – |
| Transgenic rapeseed | GT73 | + | – |
| Transgenic alfalfa | J101 | + | – |
| Transgenic beet | H7-1 | + | – |
| Transgenic maize | Bt11 | – | + |
| Transgenic soybean | MON87701 | – | + |
| Transgenic cotton | M0N531 | – | + |
| Transgenic rice | KMD | – | + |
| Transgenic rice | KF | – | + |
| Negative control | NON-GMO | – | – |
2.2. LAMP primer design
Primer design is the key to LAMP reactions. LAMP primer sets were pre-designed at https://primerexplorer.jp/e/ based on the genome sequences of CP4-EPSPS and Cry1Ab/Ac. The LAMP primers include two external primers F3 and B3, and two internal primers FIP and BIP. Based on the principle of LAMP, five sets of primer regions were designed for different regions of template DNA as shown in Table 2. The specificity of the primer sets was examined using NCBI BLAST, while the secondary structure and primer dimerization were examined using DNAMAN.
Table 2.
LAMP primers, sgRNA and ssDNA in this study.
| Primer Name | Sequences | |
|---|---|---|
| LAMP-CP4-1 | LAMP-CP4-F1 | GTGAAGTCCGCTGTTCTG |
| LAMP-CP4-B1 | GAAAGCAGTAGAGGATGGAT | |
| LAMP-CP4-FIP-1 | TCAGTGTGGTCACGAGTCATTCTCAACACCCCAGGTATC | |
| LAMP-CP4-BIP-1 | GTGTGCGTACCATCCGTCTTCACCTGGAACATCAATCACT | |
| LAMP-CP4-2 | LAMP-CP4-F2 | CTTGCTGGTCTCAACACC |
| LAMP-CP4-B2 | TGGGAAAGCAGTAGAGGAT | |
| LAMP-CP4-FIP-2 | CAAAACCTTGAAGCATCTTTTCAGTAGGTATCACCACTGTTATCGA | |
| LAMP-CP4-BIP-2 | GTGTGCGTACCATCCGTCTTGGATCACCTGGAACATCAA | |
| LAMP-CP4-3 | LAMP-CP4-F3 | AGGGTACCTATGGCTTCC |
| LAMP-CP4-B3 | AATCACTTGACCGGTGAG | |
| LAMP-CP4-FIP-3 | TCGATAACAGTGGTGATACCTGGTCAAGTGAAGTCCGCTGT | |
| LAMP-CP4-BIP-3 | CACTGAAAAGATGCTTCAAGGTTTTACCTTCAAGACGGATGGT | |
| LAMP-CP4-4 | LAMP-CP4-F4 | CTCAACACCCCAGGTATC |
| LAMP-CP4-B4 | GAAAGCAGTAGAGGATGGAT | |
| LAMP-CP4-FIP-4 | CACCAAAACCTTGAAGCATCTTTTACCACTGTTATCGAGCCA | |
| LAMP-CP4-BIP-4 | GTGTGCGTACCATCCGTCTTCACCTGGAACATCAATCACTT | |
| LAMP-CP4-5 | LAMP-CP4-F5 | TACCTATGGCTTCCGCTC |
| LAMP-CP4-B5 | TTCAAGACGGATGGTACG | |
| LAMP-CP4-FIP-5 | GCTCGATAACAGTGGTGATACCAGTGAAGTCCGCTGTTCT | |
| LAMP-CP4-BIP-5 | AATCATGACTCGTGACCACACTGACCGTCAGCATCAGTCTC | |
| LAMP- CRY-1 | LAMP-CRY-F1 | GTAAGGTCGTTGTAACGGCT |
| LAMP-CRY-B1 | ACATGAACAGCGCCTTGAC | |
| LAMP-CRY-FIP-1 | CACCTCAGCGTGCTTCGAGATGCAGCATCGAATCCCCA | |
| LAMP-CRY-BIP-1 | AGCTGCTTGAACGTACACGGACCAGCTATCCCATTGTTCGCA | |
| LAMP- CRY-2 | LAMP-CRY-F2 | GCCAGTGTTGTACCAACGAA |
| LAMP-CRY-B2 | ACATGAACAGCGCCTTGAC | |
| LAMP-CRY-FIP-2 | TGGGGATTCGATGCTGCAACCGTGGTCGGTGTAGTTTCCAA | |
| LAMP-CRY-BIP-2 | CGAAGCACGCTGAGGTGAAGAATTGTTCGCAGTCCAGAACT | |
| LAMP- CRY-3 | LAMP-CRY-F3 | GTGTTGTACCAACGAACAG |
| LAMP-CRY-B3 | GCCTTGACCACAGCTATC | |
| LAMP-CRY-FIP-3 | GATTCGATGCTGCAACCATCATAGTTTCCAATCAGCCTAGT | |
| LAMP-CRY-BIP-3 | GAAGCACGCTGAGGTGAAGACAGTCCAGAACTACCAAGTT | |
| LAMP- CRY-4 | LAMP-CRY-F4 | GTACCAACGAACAGCGTG |
| LAMP-CRY-B4 | CAGTTCAACGACATGAACAG | |
| LAMP-CRY-FIP-4 | TGGGGATTCGATGCTGCAACTCCAATCAGCCTAGTAAGGT | |
| LAMP-CRY-BIP-4 | GGTGAAGATTAGCTGCTTGAACGACCACAGCTATCCCATTG | |
| LAMP- CRY-5 | LAMP-CRY-F5 | GTAAGGTCGTTGTAACGGCT |
| LAMP-CRY-B5 | ACATGAACAGCGCCTTGAC | |
| LAMP-CRY-FIP-5 | TCTTCACCTCAGCGTGCTTCGAGCATCGAATCCCCACCT | |
| LAMP-CRY-BIP-5 | AGCTGCTTGAACGTACACGGACACAGCTATCCCATTGTTCGC | |
| CP4-SgRNA-1 | AAUUUCUACUGUUGUAGAUGGUGCUAACCUUACCGUUGAGACU | |
| Cry-SgRNA -2 | AAUUUCUACUGUUGUAGAUCCCAAACACGCUAACGUCUCGAAG | |
| HEX-12NT-BHQ1 | HEX-TTTTTTTTTTTT-BHQ1 | |
| FITC-8NT-Biotin | FITC-TTTTTTTT-Biotin |
2.3. LAMP condition optimization
The LAMP reaction targeting CP4-EPSPS and Cry1Ab/Ac genes contained 12.5 μL 2 × LAMP buffer, 10 × LAMP primer mixture (16 μM FIP, 16 μM BIP, 2 μM F3, 2 μM B3) and 8.5 μL distilled water. The above reagents were mixed thoroughly with 2.0 μL of extracted DNA, and the reaction was incubated at a constant temperature of 55–65 °C for 30–60 min, and finally at 80 °C for 5 min to inactivate the enzymes in the reaction system to avoid affecting the subsequent digestion reaction. To optimize the LAMP detection system, the LAMP reaction temperature range (55–65 °C) and LAMP reaction time (0–60 min) were analyzed. The LAMP amplification products were analyzed and compared by electrophoresis and relative fluorescence intensity measurements as well as by test strip assay. Distilled water without DNase/RNase was used as a negative control to exclude possible false positive results in the LAMP reaction.
2.4. LAMP-CRISPR-Cas12a
A Cas12a cleavage experiment was used in all detection experiments. This Cas12a cleavage system was purchased from Tolo Biotech (Tolo Biotech, Anhui, China). The in vitro cleavage reaction of the CRISPR-Cas12a assay was optimized in a total volume of 20 μL containing 2 μL CRISPR-Cas12a enzyme, 2 μL buffer, 1 μL 10 μmol/L ssDNA reporter gene, 2 μL 10 μmol/L sgRNA, 0.5 μL 5.0 U/μL ribonuclease inhibitor and 12.5 μL ribonuclease-free water. The mixture was first kept at 37 °C for 30 min, and then reacted at 85 °C for 2 min to inactivate Cas12a enzyme. Finally, 20 μL of the above reaction mixture was mixed with 180 μL of distilled water and transferred to a black opaque enzyme labeling plate, after which the fluorescence value was read from a multifunctional enzyme labeling instrument. The excitation wavelength was 480 nm and the spectra were recorded between 500 and 650 nm.
2.5. LAMP –CRISPR-Cas12a-LFA
To assess the visual presentation of results for portable applications, LAMP-CRISPR-Cas12a was combined with a lateral flow assay (LAMP-CRISPR-Cas12a-LFA). An example of test strips prepared in the laboratory, biotin ligand and goat anti-rabbit IgG were dropped onto a nitrocellulose filter membrane at a flow rate of 1 μL/cm to form a control line and a test line, respectively. The glass fiber sample pads, conjugate pads and absorbent pads were sequentially assembled along the nitrocellulose membrane on an adhesive backing pad with an overlap of 2 mm and then cut into 0.3 cm wide strips. The conjugate pad of the test strips was laid with colloidal gold-labeled FITC antibody complex. Biotin ligand and goat anti-rabbit antibody were immobilized on the NC membrane at the positions corresponding to the C and T lines, respectively. The chromogenic agent for the strips was colloidal gold, and an increase or appearance of signal was correlated with a positive result. In the LAMP-Cas12a-LFA system, trans-cleavage was activated only upon binding to an activator (experimental target) that had complementary base-pairing with crRNA. Positive samples produce two bands in the test and control capture bands, while negative samples produce only one band in the control line. First, 95 μL of buffer was mixed in a tube, and 3 μL of gilt was added to the binding pad on the test strip with 5 μL of CRISPR-Cas12a reaction product as described above. The strips were then placed upright in each tube for 5 min at room temperature. Subsequently, the strips were removed and placed on a horizontal surface, and the results were visualized using the naked eye. This experimental test strip assay was repeated three times for each experiment.
2.6. LAMP-CRISPR-Cas12a-LFA specificity
To verify the specificity of the system, a total of 7 transgenic crops and non-transgenic crops were tested. LAMP amplified for 30–35 min at 61 °C. The amplified products were first analyzed using 1.5% agarose gel electrophoresis. Then the amplified products were mixed with CRISPR-Cas12a system and incubated at 37 °C for 30 min to observe the fluorescence results with a multi-functional enzyme marker, and finally the incubation products were incubated in lateral flow test strips in buffer at room temperature for 5 min with a reaction system containing CP4-EPSPS and Cry1Ab/Ac, sgRNA, Cas12a and LAMP products, and the visualization results were observed directly by the naked eye.
2.7. LAMP-CRISPR-Cas12a-LFA sensitivity
Calculation of the copy number of pMD18T plasmids containing CP4-EPSPS and Cry1Ab/Ac sequences based on plasmid and insert molecular weight. The pMD18T plasmid DNA standard was used as a template for LAMP-CRISPR-Cas12a fluorometric assay at 10-fold dilution to determine the detection limit, with a plasmid concentration ranging from 3.65 × 104 to 10 copies/mL. The diluted sequences were then amplified and the amplified products were subjected to gel electrophoresis analysis and fluorescence value determination. Finally, the incubation product was added to the test paper, and the product was assayed and analyzed to determine the T-line.
2.8. LAMP-CRISPR-Cas12a-LFA transgenic products
Suitable LAMP primers and sgRNAs were selected based on the assay results to validate the performance of the system in detecting different transgenic products containing CP4-EPSPS and Cry1Ab/Ac genes, respectively. Eight transgenic strains containing CP4-EPSPS and five transgenic strains containing Cry1Ab/Ac were tested. After LAMP amplification, the CRISPR-Cas12a digestion system was added to observe the fluorescence values, and finally the test strips were tested to observe the detection results and compare them to the fluorescence detection results for analysis.
2.9. Statistical analysis
Each experiment was performed in triplicate. The fluorescence intensity measurement was performed using a multi-purpose microplate reader (Tecan, Mannedorf, Switzerland). Data analysis was performed using SPSS 23.0 multivariate analysis software (IBM Crop, Chicago, USA). Data are expressed as the mean ± standard deviation (SD). For parametric data, comparisons of different groups were performed by one-way analysis of variance followed by the Tukey–Kramer HSD test (α = 0.05) for multiple comparisons. Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software Inc., Chicago, USA). P < 0.05 was considered statistically significant.
3. Results
In this paper, using transgenic products as research objects, through the analysis of CP4-EPSPS and Cry1Ab/Ac genes specific fragments, we optimized the specific primers, amplification reaction time and temperature of LAMP reaction, and established a method of LAMP, CRISPR-Cas12a and lateral flow detection (LAMP-CRISPR-Cas12a), which can provide technological support for the screening of transgenic products.
3.1. LAMP reaction primer screening
Five sets of LAMP primers were designed for the relatively conserved sequences of CP4-EPSPS and Cry1Ab/Ac genes (Table 2), and the LAMP products generated by different primers were subjected to gel electrophoresis. In the LAMP reaction of CP4-EPSPS gene, only primer 1 and primer 5 produced amplified fragments, and the amplified fragment of primer 1 was clearer and more complete than that of primer 5, as shown in Fig. 2A. In the Cry1Ab/Ac gene LAMP reaction, only primer 5 produced bright and clear amplification fragment, as shown in Fig. 2B.
Fig. 2.
LAMP primer screening. A: CP4-EPSPS gene primer screening, 1–5: LAMP-CP4-F/R, B: Cry1Ab/Ac gene primer screening, 1–5: LAMP-Cry-F/R. NTC: system without added template. M: DL2000 Marker. In Figure A, only LAMP-CP4-1 and LAMP-CP4-5 primers produced amplification fragments, and LAMP-CP4-1 amplification fragment was clearer and more complete than LAMP-CP4-5, so LAMP-CP4-1 was selected as the primer for subsequent LAMP detection of CP4-EPSPS gene. In Figure B, in the Cry1Ab/Ac gene detection LAMP reaction, only LAMP- CRY-5 produced a bright and clear amplicon, which can be used as the primer for subsequent Cry1Ab/Ac gene LAMP detection.
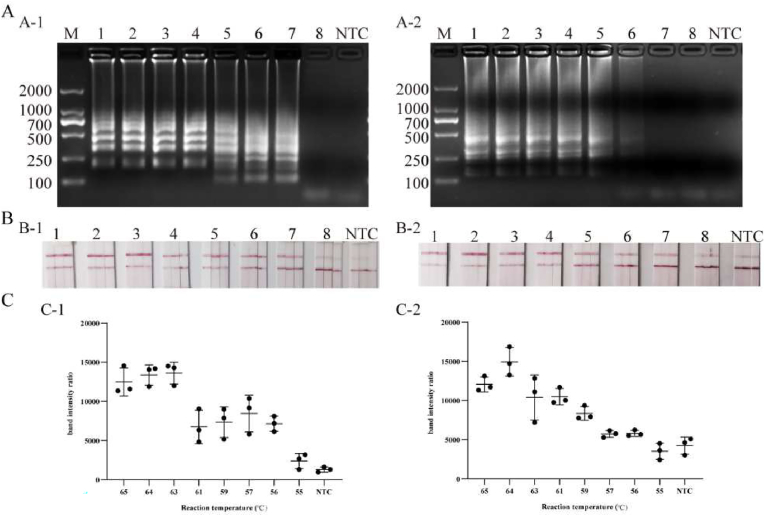
3.2. LAMP reaction temperature optimization
To determine the optimal temperature for LAMP, the reaction was carried out between 55 and 65 °C for 60 min. The LAMP amplification product of CP4-EPSPS could be detected at 56–65 °C (Fig. 3), and the LAMP amplification product of Cry1Ab/Ac could be detected at 57–65 °C (Fig. 3). As the temperature increased, the amplified bands became denser, brighter, and clearer. When the temperature reached 61 °C, the ladder-like bands could be clearly observed, and the test strips showed more obvious T lines. Therefore, the amplification temperature of 61 °C was used for subsequent amplification.
Fig. 3.
LAMP reaction temperature optimization. A, B, and C are the results of agarose gel electrophoresis analysis, lateral flow test strip detection, and band intensity ratio, respectively. A-1, A-1, A-1: LAMP reaction temperature optimization for CP4-EPSPS gene. B-2, B-2, B-2: LAMP reaction temperature optimization for Cry1Ab/Ac gene. 1–8: 65 °C, 64 °C, 63 °C, 61 °C, 59 °C, 57 °C, 56 °C, 55 °C. NTC: system without the addition of template. The optimal temperature of the reaction was finally determined to be 61 °C.
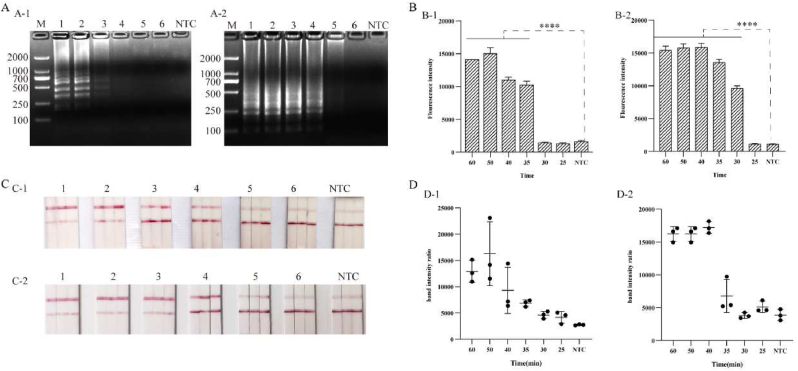
3.3. LAMP reaction time optimization
To optimize the reaction time, LAMP reactions were performed for durations of 25, 30, 35, 40, 45, 50, and 60 min, respectively. The results showed that the CP4-EPSPS gene produced amplicons at 35 min by LAMP reaction, and the fluorescence value decreased continuously with the shortening of amplification time. Furthermore, the Cry1Ab/Ac gene could detect the amplification product by 30 min of LAMP reaction and produced strong fluorescence values. The test strip detection results were consistent with the fluorescence values. Therefore, the target gene could be detected by 30–35 min of LAMP reaction (Fig. 4).
Fig. 4.
LAMP reaction time optimization. a, b, c, and d are the results of agarose gel analysis, fluorescence detection, and lateral flow test strip detection of band intensity ratio, respectively. a-1, b-1, c-1, and d-1 are the time optimization of CP4-EPSPS gene. The results show that 35 min is the optimal time. A-2, B-2, C-2, D-2 are the time optimization for Cry1Ab/Ac gene. The results show the optimal time is 30 min. 1–6 are: 60, 50, 40, 35, 30, 25 min, respectively. NTC: the system without template addition. Note: **** is significantly different (p < 0.0001).
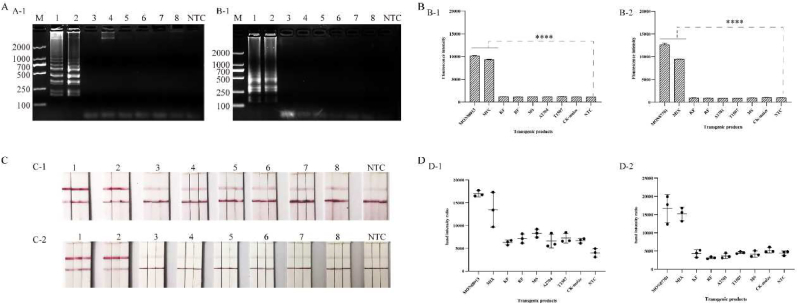
3.4. LAMP-CRISPR-Cas12a test strip specificity assay
The samples containing the PAT gene, CP4-EPSPS gene, Cry1Ab/Ac gene, and nucleic acid samples of the blank control were processed to evaluate the LAMP-CRISPR-Cas12a system specificity assay. The experimental results showed that the sgRNA of CRISPR-Cas12a system was highly specific to the genes being tested. Only the GMs containing CP4-EPSPS and Cry1Ab/Ac genes initiated the subsequent LAMP reaction to produce amplification fragments with clear fluorescent signals, while no strong positive fluorescent signals were detected in the reactions of other transgenic and non-transgenic crops. Next, using lateral flow test strips for specific visualization, a clear band was only observed at the T-line of the samples containing CP4-EPSPS and Cry1Ab/Ac and the transgenic mixture among the eight samples (Fig. 5). In addition, the T-line intensity was stronger than the C-line and consistent with the fluorescence intensity.
Fig. 5.
LAMP-CRISPR-Cas12a specificity assay. A, B, C, and D are the results of agarose gel analysis, fluorescence detection and lateral flow test strip detection and band intensity ratio, respectively. A-1, B-1, C-1, and D-1 are the results of LAMP-CRISPR-Cas12a specificity assay for CP4-EPSPS gene. 1–8 are the results of transgenic soybean MON88913, DNA mixed samples (MON88913: BT11: TC1507 = 1:1:1), transgenic rice KF, transgenic oilseed rape RF, transgenic oilseed rape MS, transgenic maize TC1507, A2704, non-transgenic maize. A-2, B-2, C-2, D-2 are the results of LAMP-CRISPR-Cas12a specificity assay for Cry1Ab/Ac gene. 1–8 are transgenic soybean MON87701, DNA mixed samples (MON88913: BT11: TC1507 = 1:1:1), transgenic soybean RRS, transgenic rape RF, transgenic maize A2704, TC1507, transgenic rape MS, and non-transgenic maize, respectively. NTC: No template was added to the system. Note: **** is significantly different (P < 0.0001).
3.5. LAMP-CRISPR-Cas12a test strip sensitivity assay
Diluted CP4-EPSPS and Cry1Ab/Ac plasmid standards were used to determine the limit of detection, and the results indicated that the limit of detection for the reaction was 100 copies (Fig. 6). Amplification and fluorescence occurred in CP4-EPSPS and Cry1Ab/Ac plasmid templates at plasmid concentrations of 3.89 × 104∼1 × 102 copies (Fig. 6C), and the amplified bands gradually became darker and the fluorescence intensity gradually decreased. Below 1 × 102 copies, no band and no fluorescence signal were observed. Meanwhile, the detection using the lateral flow method was 100 copies (Fig. 6D), and the results were consistent with the fluorescence detection results.
Fig. 6.
LAMP-CRISPR-Cas12a sensitivity assay. A, B, C, D are the results of agarose gel analysis, fluorescence assay, lateral flow test strip assay and band intensity ratio. A-1, B-1, C-1, D-1 are the sensitivity assay results of CP4-EPSPS gene. G1: 3.89 × 104 copies, G2: 3.89 × 103 copies, G3: 3.89 × 102 copies, G4: 200 copies, G5: 100 copies, G6: 50 copies, G7: 10 copies. G3: 3.89 × 102 copies, G4: 200 copies, G5: 100 copies, G6: 50 copies, G7: 10 copies. A-2, B-2, C-2, D-2 are the sensitivity results of Cry1Ab/Ac gene, G1: 3.81 × 104 copies, G2: 3.81 × 103 copies, G3: 3.81 × 102 copies, G4: 200 copies, G5: 100 copies, G6: 50 copies, G7: 10 copies. NTC: no template was added to the system. The final detection limit for CP4-EPSPS and Cry1Ab/Ac genes was determined to be 100 copies. Note: **** is significantly different (p < 0.0001).
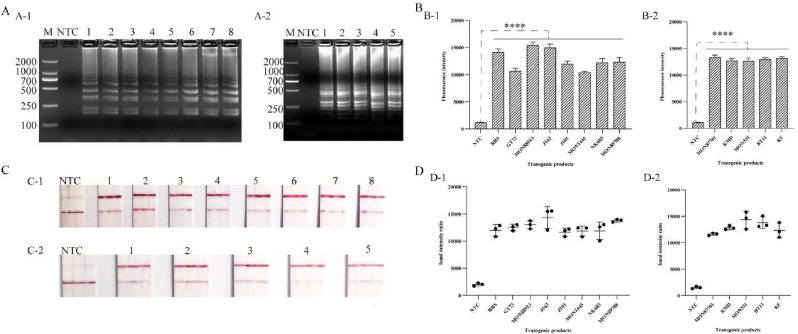
3.6. LAMP-CRISPR-Cas12a test strip transgenic product detection
To test the stability of this reaction system, transgenic products containing CP4-EPSPS and Cry1Ab/Ac genes were selected for validation. The results showed that all of them produced fluorescence signals except the blank control, indicating that the reaction system is stable and feasible. Then, LAMP amplification products and CRISPR-Cas12a were combined with sgRNA targeting CP4-EPSPS and Cry1Ab/Ac genes to form a test strip detection system. The results showed that all transgenic positive lines showed the appearance of T and C lines, and both T and C line bands were homogeneous, while the negative samples showed only C lines, which was consistent with the fluorescence assay results shown in Fig. 7.
Fig. 7.
LAMP-CRISPR-Cas12a transgenic product assays. a, b, c, and d are the results of agarose gel analysis, fluorescence assay, lateral flow test strip assay and band intensity ratio results. a-1, b-1, c-1, d-1 is CP4-EPSPS gene transgenic product assays, 1–8 are transgenic soybean RRS, transgenic oilseed rape GT73, transgenic soybean MON88913, transgenic alfalfa J163, J101, transgenic cotton MON1445, transgenic maize NK603, transgenic soybean MON89788, respectively. B is Cry1Ab/Ac transgenic product testing, A-2, B-2, C-2, D-2 is Cry1Ab/Ac transgenic product testing, 1–5 transgenic soybean MON87701, transgenic rice KMD, transgenic cotton MON531, transgenic maize BT11, transgenic rice KF. M: DL2000 Marker, NTC: no template added to the system. Note: **** is significantly different (P < 0.0001).
4. Discussion
Recent studies have indicated that sequence-specific nucleic acid detection is still the most practical detection method. LAMP is a relatively mature isothermal amplification technique, and it is widely used in nucleic acid detection, including in virus detection and transgenic detection, because of its efficient amplification, high specificity, and low cost. The LAMP reaction requires four primers that recognize six different regions on the target (Lee et al., 2019), which can greatly increase specificity, and the reaction process involves continuous cycles of target synthesis, leading to rapid and efficient amplification. Combining CRISPR with LAMP enables the detection of low-copy molecules. The RNA-mediated DNA-targeted CRISPR effector protein Cas12a side-branch cleavage proposed in this study uses Cas12a non-specific targeting of ssDNA for lateral targeting cleavage as a starting point (Shi et al., 2022). The CRISPR-Cas12a system is a new nucleic acid detection technology. Its trans-cleavage activity provides a new idea for nucleic acid detection. In this study, we established a low-cost, highly sensitive, convenient LAMP-CRISPR-Cas12a mobile test strip system that combines LAMP and a CRISPR-Cas12a assay. The results are visualized using mobile test strips, which can be used for the rapid detection of transgenes in the field. The assay established in this study is simple, relatively low-cost, rapid, specific, and highly sensitive, and it requires minimal equipment to detect the presence of target DNA fragments in a given sample. In combination with the lateral flow test strip technique, the test results are visible to the naked eye, which is of great practical value in the clinical testing of nucleic acids in China. By changing the type of primers used, the target of detection can also be changed. Combining LAMP isothermal amplification with the Cas12a enzyme at a At room temperature (Pang et al., 2020) enables a true single-tube assay mixture (Li et al., 2021). The LAMP and CRISPR-Cas12a reagents in the LAMP-CRISPR-Cas12a assay can be lyophilized and stored at room temperature, thus reducing the cost of cold storage and of the transportation of reagents. CRISPR-Cas12a could also have great potential in the development of next-generation nucleic acid detection biosensors due to the trans-cleavage ability of Cas effector proteins.
5. Conclusion
LAMP amplification has been widely used in transgenic detection because of its low cost, low enzyme dependence and high efficiency. In this paper, we designed five sets of LAMP primers for the CP4-EPSPS and Cry1Ab/Ac genes. The amplification system of LAMP was then optimized, along with the ultra-sensitive recognition system of CRISPR-Cas12a. Finally, a group of highly specific LAMP primers were selected. The optimized reaction time was 30–35 min and the reaction temperature was 61 °C. The whole detection process could be completed within 1 h. The LAMP-CRISPR-Cas12a-based fluorescence detection system and test strip detection system established in this study are easy to operate, highly sensitive and specific, and only need constant temperature equipment to achieve large-scale detection.
Author contributions
Hua Liu: Conceptualization, methodology, writing, reviewing, and editing of the manuscript. Xiuwen Hu and Fang Cheng: Writing the original draft, funding acquisition, data curation. Haijuan Zeng and Chuan He: Formal analysis, validation, and investigation. Xueming Tang: Validation, investigation, and supervision. Jinbin Wang: Funding acquisition, supervision, writing, reviewing, and editing the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Shanghai Agriculture Applied Technology Development Program (Grant No. 2020-2-1), the Shanghai Agricultural Science and Technology Innovation Action Plan (No. 21N31900800), Shanghai Key Laboratory of Agricultural Genetics and Breeding (No.21DZ2271900), 2023 SAAS Project on Agricultural Science and Technology Innovation Supporting Area [SAAS Application Basic Study 2023(08)], and the Shanghai Science and Technology Commission The belt and road project (20310750500).
Biographies
Dr. Hua Liu is an associate professor at the Institute of Biotechnology, Shanghai Academy of Agricultural Sciences in the field of rapid food safety testing. Her main research is focus on LAMP-Cas12a technologies for traceability of genetically modified crops and rapid detection.
Xiuwen Hu is a student of food science at Institute of Biotechnology Research, Shanghai Academy of Agricultural Sciences. Her research interests include nucleic and fast testing technologies based CRISPR-Cas12a.
Haijuan Zeng is an associate professor of food analysis in Institute of Biotechnology Research, Shanghai Academy of Agricultural Sciences. Her main research is focus on accurate, fast and multi-target testing technologies for quality and safety of agro-product.
Chuan He is an associant professor of food analysis in Institute of Biotechnology RT-PCR Research, Shanghai Academy of Agricultural Sciences. Her main research is focus on RPA technologies for traceability of GMOs and its products.
Fang Cheng is a associant professor of food pre-processing at Institute of Natural Sciences, Shanghai Jiao Tong University. Her research interests include nucleic and fast testing technologies for GMOs and its products.
Dr. Xueming Tang is a professor of GMOs safety at School of Agriculture and Biology, Shanghai Jiao Tong University. He works on technologies for molecular detection of GMOs safety.
Dr. Jinbin Wang is an associate professor of food safety in Institute of Biotechnology Research, Shanghai Academy of Agricultural Sciences, He works on technologies for molecular detection of pathogenic microorganism.
Handling Editor: Dr. Quancai Sun
Contributor Information
Xueming Tang, Email: xueming.tang@sjtu.edu.cn.
Jinbin Wang, Email: wangjinbin@saas.sh.cn.
Data availability
No data was used for the research described in the article.
References
- Akiyama H., Makiyama D., Nakamura K., Sasaki N., Minegishi Y., Mano J., Kitta K., Ozeki Y., Teshima R. A novel detection system for the genetically modified canola (Brassica rapa) line RT73. Anal. Chem. 2010;82:9909–9916. doi: 10.1021/ac102434q. [DOI] [PubMed] [Google Scholar]
- Chen D., Zhang M., Ma M., Hai H., Li J., Shan Y. A novel electrochemical DNA biosensor for transgenic soybean detection based on triple signal amplification. Anal. Chim. Acta. 2019;1078:24–31. doi: 10.1016/j.aca.2019.05.074. [DOI] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Costa M.D., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.-Y., Jeong R.-D., Yoon Y.-N., Lee S.-H., Shin D.B., Kang H.-W., Lee B.C. One-step multiplex reverse transcription-polymerase chain reaction for the simultaneous detection of three rice viruses. J. Virol Methods. 2013;193:674–678. doi: 10.1016/j.jviromet.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Deng J., Li S., Hong J., Ji Y., Zhou Y. Investigation on subcellular localization of Rice stripe virus in its vector small brown planthopper by electron microscopy. Virol. J. 2013;10:310. doi: 10.1186/1743-422X-10-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos Y., Oberkofler L., Glandorf D.C.M. Reference Module in Biomedical Sciences. Elsevier; 2022. Genetically modified plants and food/feed: risk assessment considerations☆⁎. [Google Scholar]
- Du Y.-C., Wang S.-Y., Wang Y.-X., Ma J.-Y., Wang D.-X., Tang A.-N., Kong D.-M. Terminal deoxynucleotidyl transferase combined CRISPR-Cas12a amplification strategy for ultrasensitive detection of uracil-DNA glycosylase with zero background. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112734. [DOI] [PubMed] [Google Scholar]
- Emslie K.R., Whaites L., Griffiths K.R., Murby E.J. Sampling plan and test protocol for the semiquantitative detection of genetically modified canola (Brassica napus) seed in bulk canola seed. J. Agric. Food Chem. 2007;55:4414–4421. doi: 10.1021/jf070267i. [DOI] [PubMed] [Google Scholar]
- Holst-Jensen A. Testing for genetically modified organisms (GMOs): past, present and future perspectives. Biotechnol. Adv. 2009;27:1071–1082. doi: 10.1016/j.biotechadv.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Huang Y.H., Fan D.F., Jiao Y., Wu X.Z., Ye J.M. Enlightenment of GMO labeling system in other countries to China. Curr. Biotechnol. 2022;12(4):516–522. [Google Scholar]
- ISAAA Global biotechnology/GM crop commercialization development in 2019. China Biotechnol. 2021;41:114–119. [Google Scholar]
- Joshi S., Sharma V., Ramesh V., Singh R., Salotra P. Development of a novel loop-mediated isothermal amplification assay for rapid detection of Mycobacterium leprae in clinical samples. Indian J. Dermatol. Venereol. Leprol. 2021:1–7. doi: 10.25259/IJDVL_248_19. [DOI] [PubMed] [Google Scholar]
- Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B., Takeda N., Katayama K. Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamle M., Kumar P., Patra J.K., Bajpai V.K. Current perspectives on genetically modified crops and detection methods. 3 Biotech. 2017;7:219. doi: 10.1007/s13205-017-0809-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Kang L.H., Kim J.O., Paik S.Y., Lee H.K., Kim S.Y., Park Y.-J. Rapid detection of carbapenemase-producing Enterobacteriaceae (CPE) using a simple DNA extraction method and Loop-mediated isothermal amplification (LAMP) J. Microbiol. Methods. 2019;167 doi: 10.1016/j.mimet.2019.105746. [DOI] [PubMed] [Google Scholar]
- Li S., Huang J., Ren L., Jiang W., Wang M., Zhuang L., Zheng Q., Yang R., Zeng Y., Luu L.D.W., Wang Y., Tai J. A one-step, one-pot CRISPR nucleic acid detection platform (CRISPR-top): application for the diagnosis of COVID-19. Talanta. 2021;233 doi: 10.1016/j.talanta.2021.122591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., Pan T.M. Perspectives on genetically modified crops and food detection. J. Food Drug Anal. 2016;24:1–8. doi: 10.1016/j.jfda.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Tiwari S., Gomes A.V. Protein purification and analysis: next generation Western blotting techniques. Expet Rev. Proteonomics. 2017;14:1037–1053. doi: 10.1080/14789450.2017.1388167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morenkov O.S. Development of immunoenzyme methods for detecting antibodies to Aujeszky's disease virus gB glycoprotein in swine serum. Vopr. Virusol. 2000;45:45–48. [PubMed] [Google Scholar]
- Murugan K., Seetharam A.S., Severin A.J., Sashital D.G. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J. Biol. Chem. 2020;295:5538–5553. doi: 10.1074/jbc.RA120.012933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., Xu J., Liu Y., Peng H., Le X.C. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology. Anal. Chem. 2020;92 doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Pirc M., Alic S., Dreo T. Rapid loop-mediated isothermal amplification for detection of the ralstonia solanacearum species complex bacteria in symptomatic potato tubers and plants. Methods Mol. Biol. 2021;2354:401–413. doi: 10.1007/978-1-0716-1609-3_20. [DOI] [PubMed] [Google Scholar]
- Ponti L. Transgenic crops and sustainable agriculture in the European context. Bull. Sci. Technol. Soc. 2005;25:289–305. [Google Scholar]
- Prasannakumar M.K., Parivallal P.B., Pramesh D., Mahesh H.B., Raj E. LAMP-based foldable microdevice platform for the rapid detection of Magnaporthe oryzae and Sarocladium oryzae in rice seed. Sci. Rep. 2021;11:178. doi: 10.1038/s41598-020-80644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C., Wang R., Wu H., Ping J., Wu J. Recent advances in emerging DNA-based methods for genetically modified organisms (GMOs) rapid detection. TrAC, Trends Anal. Chem. 2018;109:19–31. [Google Scholar]
- Ranek M.J., Oeing C., Sanchez-Hodge R., Kokkonen-Simon K.M., Dillard D., Aslam M.I., Rainer P.P., Mishra S., Dunkerly-Eyring B., Holewinski R.J., Virus C., Zhang H., Mannion M.M., Agrawal V., Hahn V., Lee D.I., Sasaki M., Van Eyk J.E., Willis M.S., Page R.C., Schisler J.C., Kass D.A. CHIP phosphorylation by protein kinase G enhances protein quality control and attenuates cardiac ischemic injury. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Xu Y., Huang K., Luo Y., Xu W. Multiplex pyrosequencing quantitative detection combined with universal primer-multiplex-PCR for genetically modified organisms. Food Chem. 2020;320 doi: 10.1016/j.foodchem.2020.126634. [DOI] [PubMed] [Google Scholar]
- Shi Y., Kang L., Mu R., Xu M., Duan X., Li Y., Yang C., Ding J.-W., Wang Q., Li S. CRISPR/Cas12a-Enhanced loop-mediated isothermal amplification for the visual detection of Shigella flexneri. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.845688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. In: Policy Issues in Genetically Modified Crops. Singh P., Borthakur A., Singh A.A., Kumar A., Singh K.K., editors. Academic Press; 2021. Chapter 4 - genetically modified crops in India: politics, policies, and political economy; pp. 75–96. [Google Scholar]
- Takabatake R., Kagiya Y., Minegishi Y., Yeasmin S., Futo S., Noguchi A., Kondo K., Mano J., Kitta K. Development and evaluation of rapid screening detection methods for genetically modified crops using loop-mediated isothermal amplification. Food Chem. 2018;252:390–396. doi: 10.1016/j.foodchem.2017.12.036. [DOI] [PubMed] [Google Scholar]
- Tsugunori N., Hiroto O., Harumi M., Toshihiro Y., Keiko W., Nobuyuki A., Tetsu H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;E63 doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ke Y., Liu W., Sun Y., Ding X. A one-pot toolbox based on Cas12a/crRNA enables rapid foodborne pathogen detection at attomolar level. ACS Sens. 2020;5:1427–1435. doi: 10.1021/acssensors.0c00320. [DOI] [PubMed] [Google Scholar]
- Wu H., Zhang X., Wu B., Qian C., Zhang F., Wang L., Ye Z., Wu J. Rapid on-site detection of genetically modified soybean products by real-time loop-mediated isothermal amplification coupled with a designed portable amplifier. Food Chem. 2020;323 doi: 10.1016/j.foodchem.2020.126819. [DOI] [PubMed] [Google Scholar]
- Wu J., Ni Y., Liu H., Ding M., Zhou X. Monoclonal antibody-based serological assays and immunocapture-RT-PCR for detecting Rice dwarf virus in field rice plants and leafhopper vectors. J. Virol Methods. 2014;195:134–140. doi: 10.1016/j.jviromet.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Xu Q., Liu H., Yuan P., Zhang X., Chen, et al. Development of a simplified RT-PCR without RNA isolation for rapid detection of RNA viruses in a single small brown planthopper (Laodelphax striatellus Fallén) Virol. J. 2017;14 doi: 10.1186/s12985-017-0732-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Wang J., Jia J., Wu G., Yang Q., Liu X., Tang X. Development of a lateral flow test strip for simultaneous detection of BT-Cry1Ab, BT-Cry1Ac and CP4 EPSPS proteins in genetically modified crops. Food Chem. 2021;335 doi: 10.1016/j.foodchem.2020.127627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.