Summary
Background
Accurate tumour response prediction to targeted therapy allows for personalised conversion therapy for patients with unresectable colorectal cancer liver metastases (CRLM). In this study, we aimed to develop and validate a multi-modal deep learning model to predict the efficacy of bevacizumab in patients with initially unresectable CRLM using baseline PET/CT, clinical data, and colonoscopy biopsy specimens.
Methods
In this multicentre cohort study, we retrospectively collected data of 307 patients with CRLM from the BECOME study (NCT01972490) (Zhongshan Hospital of Fudan University, Shanghai) and two independent Chinese cohorts (internal validation cohort from January 1, 2018 to December 31, 2018 at Zhongshan Hospital of Fudan University; external validation cohort from January 1, 2020 to December 31, 2020 at Zhongshan Hospital—Xiamen, Shanghai, and the First Hospital of Wenzhou Medical University, Wenzhou). The main inclusion criteria were that patients with CRLM had pre-treatment PET/CT images as well as colonoscopy specimens. After extracting PET/CT features with deep neural networks (DNN) and selecting related clinical factors using LASSO analysis, a random forest classifier was built as the Deep Radiomics Bevacizumab efficacy predicting model (DERBY). Furthermore, by combining histopathological biomarkers into DERBY, we established DERBY+. The performance of model was evaluated using area under the curve (AUC), sensitivity, specificity, positive predictive value, and negative predictive value.
Findings
DERBY achieved promising performance in predicting bevacizumab sensitivity with an AUC of 0.77 and 95% confidence interval (CI) [0.67–0.87]. After combining histopathological features, we developed DERBY+, which had more robust accuracy for predicting tumour response in external validation cohort (AUC 0.83 and 95% CI [0.75–0.92], sensitivity 80.4%, specificity 76.8%). DERBY+ also had prognostic value: the responders had longer progression-free survival (median progression-free survival: 9.6 vs 6.3 months, p = 0.002) and overall survival (median overall survival: 27.6 vs 18.5 months, p = 0.010) than non-responders.
Interpretation
This multi-modal deep radiomics model, using PET/CT, clinical data and histopathological data, was able to identify patients with bevacizumab-sensitive CRLM, providing a favourable approach for precise patient treatment. To further validate and explore the clinical impact of this work, future prospective studies with larger patient cohorts are warranted.
Funding
The National Natural Science Foundation of China; Fujian Provincial Health Commission Project; Xiamen Science and Technology Agency Program; Clinical Research Plan of SHDC; Shanghai Science and Technology Committee Project; Clinical Research Plan of SHDC; Zhejiang Provincial Natural Science Foundation of China; and National Science Foundation of Xiamen.
Keywords: Colorectal cancer, Liver metastases, Multi-modal, Deep learning, Treatment efficacy
Research in context.
Evidence before this study
We searched PubMed with the term “(colorectal cancer liver metastases) AND (bevacizumab)) AND (predict)” published from database inception up to 01 May 2023 with no language restrictions. We found that most studies primarily use tumour morphological modifications of enhanced computed tomography imaging to predict the efficacy of bevacizumab. Only three studies about bevacizumab efficacy prediction using PET/CT, and they mainly used metabolic parameter such as SUVmax, which may lose a large amount of key information of PET/CT images and lead to limited prediction accuracy. We believe that individual biomarkers alone cannot adequately predict the therapeutic response to treatment, given the complexity of this clinical issue. Therefore, we have employed a deep learning approach to integrate PET/CT images, clinical data, and histological protein biomarkers with the aim of constructing a predictive model for bevacizumab efficacy in patients with colorectal cancer liver metastases (CRLM).
Added value of this study
In this multicentre study, we developed and validated a multi-modal model for predicting the response to bevacizumab in patients with CRLM. The model was constructed using baseline PET/CT images, clinical data, and histological protein biomarkers, employing a deep learning approach. The proposed model (DERBY), showing its robust prediction ability, performed accurately predicted the response to bevacizumab, with an area under the curve (AUC) of 0.83 and a hazard ratio of 13.60 (95% confidence interval [5.44–37.28]). In addition, significant differences in progression-free survival (median survival: 9.6 vs 6.3 months) and overall survival (median survival: 27.6 vs 18.5 months) were noted between DERBY+-predicted sensitive patients with CRLM with insensitive ones.
Implications of all the available evidence
Our developed model, compared with the current bevacizumab administration strategy, not only improves the treatment response rate by about 20% but also reduces the use of bevacizumab in almost 50% of the patients with Ras-mutated CRLM. This change is beneficial for improving their prognosis, reducing unnecessary complications, and facilitating the relocating of societal healthcare resources. This study represents a step forward in using artificial intelligence for personalised treatment and prognosis prediction in patients with CRLM. Further validation, exploration of clinical impact, and prospective studies are needed to establish the reliability, generalisability, and utility of these models in the clinical management of patients with unresectable CRLM.
Introduction
Colorectal cancer (CRC) is currently the third most common cancer in the world, and its mortality rate ranks second among all cancers.1 More than 50% of patients with CRC develop liver metastases, and colorectal cancer liver metastases (CRLM) is the leading cause of CRC-related death.2 Liver resection, when clinically possible, is the best therapy for individuals with CRLM, and can increase the 5-year survival rate from 5% to over 50%.3 However, 70–80% of patients with CRLM can be considered initially unresectable.4 Hence, conversion therapy has a pivotal role in this initially unresectable group for improving clinical outcomes.5
Bevacizumab, an anti-angiogenic drug, has been recommended as a first-line regimen for CRLM conversion therapy by the NCCN Clinical Practice Guidelines in oncology6 and shown to be effective for improving overall survival (OS) of these patients.7,8 In the BECOME study (NCT01972490), we demonstrated that bevacizumab, when combined with FOLFOX, improved the objective response rate (ORR, 54.5%) of patients with initially unresectable CRLM.9 In other words, nearly half of patients with CRLM were insensitive to bevacizumab. Therefore, identification of patients who are sensitive to bevacizumab is crucial to improve response rates and reduce adverse events caused by ineffective treatment.5,10
During the past decade, radiomics has advanced quickly into clinical applications.11 Technically, radiomics extracts quantitative data from medical images from PET, MRI, and enhanced CT with a high throughput, then transforms imaging information into a high-dimensional, mineable form to support model decision-making, which can significantly enhance the use of conventional imaging techniques.12, 13, 14, 15 For instance, Dohan et al. developed a radiomic signature at baseline and 2-month CT from the PRODIGE9 study among metastatic CRC (mCRC) patients undergoing first-line treatment with FOLFIRI and bevacizumab, which was able to predict OS and identify responders better than the RECIST 1.1 criteria.16 Additionally, as tumour metabolism–a fundamental principle of PET/CT imaging–is linked to angiogenesis,17 PET/CT result can be a predictor of bevacizumab efficacy. As a matter of fact, there have been studies showing the value of PET/CT in predicting bevacizumab therapy efficacy.18, 19, 20 Although the metabolic parameter (SUVmax) of PET/CT could help predict and evaluate the effectiveness of bevacizumab therapy for patients with CRLM,18,19 the current models lose a large amount of key information of PET/CT images, and may lead to limited prediction accuracy. In contrast to the metabolic parameters, in a cohort of 79 breast cancer patients, the radiomic signature constructed by baseline PET/CT was shown to be able to predict the pathological complete response rate of neoadjuvant chemotherapy (NAC).20 Moreover, a recent study showed that a multi-modal machine learning predictor outperformed single-scale models for therapy response of breast cancer.21 However, the PET/CT imaging signature-based multi-modal model for predicting efficacy of bevacizumab therapy in patients with CRLM has not been investigated.
In this study (Fig. 1), we proposed a multi-modal deep radiomics framework using baseline PET/CT images and clinical data of patients with CRLM from the BECOME study,9 and developed the DERBY model for predicting bevacizumab efficacy in RAS-mutant patients with CRLM. Furthermore, considering that histopathology coupled with machine learning might help in cancer treatment selection,10,22 a multi-modal model (DERBY+) that combined PET/CT images, clinical data, and histological protein biomarkers, was developed and evaluated for synergy in improving prediction in both internal and external validation cohorts.
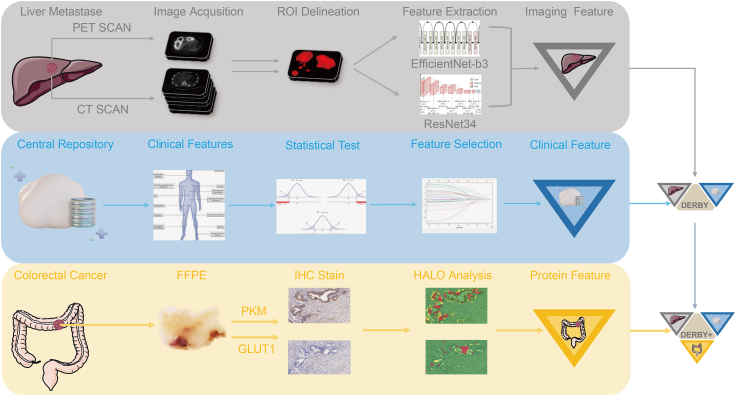
Fig. 1.
Overall study strategy. Baseline PET/CT images of liver metastases, clinical data, and colonoscopy biopsy specimens were retrospectively selected for feature extraction. After feature evaluating, three sets of signatures were generated (Clinical, Imaging, Histological signature, respectively). Clinical and imaging signatures were combined to build the DERBY model, and the DERBY+ model was further constructed by combining the DERBY with histological signatures. Abbreviations: ROI, region of interest; FFPE, formalin-fixed and parrffin-embedded; IHC, immunohistochemistry.
Methods
Ethics
The study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University (No. B2021-172). Written consent and institutional approval were obtained from all study participants.
Study design and patient eligibility
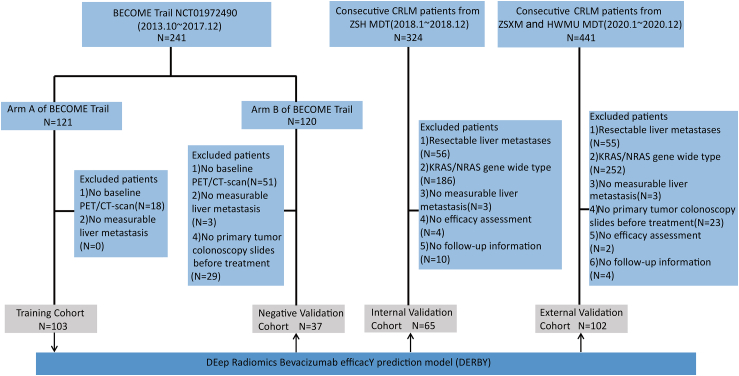
In this multicentre cohort study, we collected 307 patients with CRLM (Fig. 2). The training cohort and negative validation cohort were derived from the BECOME study (NCT01972490),9 for whom baseline PET/CT images were available. In the training cohort (n = 103), each patient received mFOLFOX6 (day 1: oxaliplatin 85 mg/m2, folinic acid 400 mg/m2, and fluorouracil 400 mg/m2 intravenous bolus and then 2400 mg/m2 over 46 h continuous infusion) combined with bevacizumab (Arm A of the BECOME study). In contrast, all patients were treated with mFOLFOX6 alone in the negative validation cohort (n = 37) (Arm B of the BECOME study).
Fig. 2.
Study design and participants. Patients from Arm A of the BECOME study were treated with mFOLFOX6 with bevacizumab, mFOLFOX6 in Arm B. Abbreviations: ZSH, Zhongshan Hospital; ZSHX, Zhongshan Hospital of Xiamen; HWMU, the First Affiliated Hospital of Wenzhou Medical University; MDT, multi-disciplinary team.
The internal validation cohort was derived from consecutive mCRC patients of the multi-disciplinary team (MDT) at Zhongshan Hospital (ZSH), share the same MDT, surgical team, and PET/CT imaging equipment with training cohort, from 01 January 2018 to 31 December 2018. Inclusion criteria were as follows: (1) Patients were histologically confirmed for colorectal adenocarcinoma with unresectable liver-limited or liver-dominant metastases23; (2) PET/CT at baseline were available; and (3) First line treated with FOLFOX + bevacizumab. Exclusion criteria were as follows: (1) Resectable liver metastases; (2) Wide-type KRAS/NRAS; (3) No measurable liver metastasis; (4) No efficacy assessment; and (5) No follow-up information. The external validation cohort came from the MDT of Zhongshan Hospital–Xiamen (ZSHX) and the First Hospital of Wenzhou Medical University (HWMU), from 01 January 2020 to 31 December 2020, and these patients come from different geographical regions and were subjected to diverse PET/CT imaging equipment, with data collection conducted by the respective doctors at their respective hospitals. Additional exclusion criteria compared to the ZSH cohort included no primary tumour colonoscopy slides before treatment. This study was approved by the Ethics Committee of ZSH in accordance with the Declaration of Helsinki. All study participants have provided informed consent.
Efficacy assessment and follow-up
We assessed bevacizumab efficacy with tumour response, which was provided by the MDT and based on RECIST version 1.1.24 Overall response rate (ORR) was defined as the sum of complete response (CR) and partial response (PR). Disease control rate (DCR) contained CR, PR, and stable disease (SD). Progression-free survival (PFS) was defined as the period from the start of treatment with bevacizumab plus chemotherapy, or chemotherapy alone to the date of disease progression or death.9 OS was calculated as the interval between the start of chemotherapy and the date of the last follow-up, or until death from any cause, at which point the data was censored.
PET/CT imaging
All patients were asked to fast at least 6 h (until reaching a serum glucose level of <150 mg/dL) and was injected with approximately 3.7 MBq/kg body weight of 18F-FDG. 18F-FDG PET/CT scans were performed using a uMI 510/uMI 550/uMI 780 scanner (United Imaging Healthcare, Shanghai, China) or Discovery VCT scanner (GE Healthcare, Milwaukee, Wisconsin, USA) for the ZSH and ZSHX cohort, or a Gemini TF 64 (Philips Healthcare, Best, the Netherlands) for the HWMU cohort. Images were obtained approximately 60 min after the 18F-FDG injection, from the skull base to the mid-thigh. The PET acquisition time per bed position was 2 min for the ZSH and ZSHX cohort, and 1.5 min for the HWMU cohort. PET images were acquired in three dimensions. The PET images were reconstructed onto a 128 × 128 matrix with CT-based attenuation using the ordered subset expectation maximisation algorithm.25 The diagnostic CT scan parameters were as follows: Discovery VCT (140 mAs; 120 kV; pitch, 0.516; slice thickness, 1.25 mm), uMI 510/uMI 550/uMI 780 (140 mAs; 120 kV; pitch, 0.9875; slice thickness, 1.0 mm), or Gemini TF 64 (150 mAs; 120 kV; pitch, 0.83; 5.0 mm).
Segmentation of PET/CT images
PET images were segmented using a uWS-MI workstation (United Imaging Healthcare, Shanghai, China). Our study nuclear physician manually enclosed the regions of interest (ROI) of PET images in a cropping box. Each lesion was segmented using an adaptive threshold segmentation method.26 This segmentation method combines the threshold algorithm and the region-growing algorithm. The former was determined with lesion segmentation through iteration with the seed pixels and those pixels with intensity values greater than the optimal to generate the lesion region. Compared to the frequently employed relative or absolute fixed SUV thresholds, this method utilises all the information from the signal to the background, resulting in superior lesion delineation. Moreover, it is independent of image properties, scanner types, reconstruction, and imaging noise. The CT image ROIs were delineated according to their corresponding ROIs in the PET images.
Feature extraction of PET/CT images
Prior to feature extraction, we first reformulated the PET/CT image serials into PET/CT image pairs through an image preprocessing pipeline (See online Supplementary methods). Due to the relatively small size of our training dataset, it is difficult to train a classification network in a three-dimensional (3D) form. However, using a purely two-dimensional (2D) training approach would result in the loss of continuity information between different slices. To address this challenge, we made a compromise between 2D and 3D and designed a 2.5D training approach based on our understanding of this data collection. Specifically, we reformulated the PET/CT image sequences into PET/CT image pairs by selecting three geometrically correspondent CT slices for each PET slice. For each image pair, CT and PET features were computed in parallel using a Resnet-34 network27 and EfficientNet-b3 network,28 respectively. The PET feature extraction was conducted in basic 2D, while the extracted 2D feature maps of 3 consecutive CT slices were fused using 1 × 1 convolution. The extracted features were then concatenated into a Gaussian Mixture Model (GMM),29 the output of which was a fused representation of PET and CT image features and saved as the final image features for the current PET/CT image pair, denoted as . Network training details and GMM definition can be seen in online Supplementary methods.
Selection of clinical factors
We collected eight clinical factors associated with therapy efficacy and recurrence of patients with CRLM as previously reported,30, 31, 32, 33, 34 including SUVmax of PET/CT imaging, clinical risk score, pre-operative CEA (carcinoembryonic antigen), pre-operative CA19-9, the site of primary tumour, the numbers of liver metastases, maximum diameter of liver metastases, and the distribution of liver metastases. To identify correlated clinical predictors of bevacizumab efficacy, we conducted the least-absolute-shrinkage-and-selection-operator (LASSO) regression with different regularisation terms (Supplementary Figure S1). The factors selected with LASSO were used as clinical features, , in the final integrated predictive model.
Immunohistochemical (IHC) staining and HALO analysis
To identify primary tumour-related features associated with bevacizumab efficacy, we conducted the literature search35,36 and selected two protein markers (GLUT1 and PKM) through the following experiments (Supplementary Figure S2). We performed IHC staining in specimens from colonoscopy and analysed with the HALO platform (v3.3.2541.420) (Indica Labs, NM, USA) to display the metabolic status of primary CRC (Supplementary Figure S3). We derived IHC scores of these two protein markers, and used theses scores as biopsy features, denoted as , in the final predictive model (See online Supplementary methods).
Establishment of single-scale models
Before building the fused model, we trained three distinct models to assess the individual predictive value of images, clinic features and biopsy features. For image features, we directly used the feature extraction network to predict, and the classification results of a patient were determined by majority voting of the prediction from all PET/CT image pairs. While for clinic and biopsy features, we used the Python package “scikit-learn, version 0.2.2”,37 to evaluate the individual predictive value of each predictor type.
Establishment of the DERBY and DERBY+ models
Before combining features from different cohorts, we firstly addressed the huge dimensional gap between deep features learned from networks (1024-dimensions) and the selected as well as by constructing a GMM. The GMM is a probabilistic model which can effectively projected the high-dimensional deep feature vectors onto a lower-dimensional space while still retain the most important information within the data.38 The detailed explanations of our GMM definition and feature transformation procedure are shown in Supplementary materials. The GMM-transformed features are denoted as and served as image features in the final predictive model.
After comparing multiple usually used classification algorithms (See online Supplementary methods, Supplementary Figure S4), we decided to build the DERBY and DERBY+ model using random forest classifier.39
The DERBY model was built using a random forest classifier, which took and as input to give a prediction for each PET/CT image pair (Fig. 1). The final decision for a patient was made through a weighted voting among all image pairs from that patient, where the weight of each PET/CT pair was determined according to the ROI size on the corresponding PET image. The DERBY model was built on the training cohort (See online Supplementary methods) from the BECOME study, then validated in internal and external validation cohorts. We further proposed the DERBY+ model by incorporating histological features into the DERBY (Fig. 1). The DERBY+ model was trained on a histology cohort (Supplementary Figure S5) and validated in the same external validation cohort. The output of the DERBY model was a binary prediction of tumour response to bevacizumab as either responders or non-responders. Specifically, the therapy response score (TRS) was acquired by aggregating the model's probability output for each image pair, which was evaluated for the likelihood of a patient who may benefit from bevacizumab therapy. Patients with TRS ≥ 0.5 were defined as responders, while patients with TRS<0.5 were non-responders (See online Supplementary methods).
Model evaluation and statistical analysis
All processing and analysis steps were conducted in Python 3.7.2 and the R software 4.1.3 (https://www.r-project.org). The retrospective power calculation for the external validation cohort for OS, PFS and ORR, and the corresponding power were 87%, 90% and 93%, respectively. Detailed process for power calculation was provided in online Supplementary methods. The linearity of continuous variables was tested before model constructed. The performance of the random forest classifier was evaluated in the internal and external validation cohorts. Performance was measured based on predictive accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and the receiver operating characteristic (ROC) analysis. The calibration of model was assessed using brier loss as well as a post hoc Hosmer–Lemeshow test. A smaller value of brier loss indicates better calibration of the model, while a p-value greater than 0.05 for the Hosmer–Lemeshow test suggests that the model is consistently calibrated. The contribution of each feature in the final DERBY+ model is illustrated with partial plot analysis. The Kaplan–Meier method and log-rank test were applied in PFS and OS analyses to assess the clinical outcomes. Following the assessment of the proportional hazard assumption (Supplementary Figure S6), we applied the Cox proportional hazards regression model to ascertain the independent prognostic predictors in the univariable analysis. The association between the risk factors and tumour response was analysed using unconditional logistic regression models. The R package “car” (version 3.1-1) was adapted to test the linearity of continuous variables. The “survminer” package (version 0.4.8) was adapted to plot a survival curve. The “pROC” package (version 1.18.0) was adapted to plot a ROC curve. The “rmda” package (version 1.6) was adapted to decision curve analysis. The Mann–Whitney U test and the chi-square test were performed for continuous variables and categorical variables, respectively. A p-value of less than 0.05 was considered statistically significant for analysis.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or the writing of this report.
Results
Study cohort and patient characteristics
A total of 307 patients with CRLM were enrolled in this study from multiple centres (Fig. 2). The training cohort (n = 103) for the DERBY model consisted of patients of arm A (mFOLFOX6 plus bevacizumab) from the BECOME study. The internal validation cohort (n = 65) were collected from consecutive patients with CRLM from ZSH cohort, while external validation cohort (n = 102) was derived from ZSHX and HWMU cohort. As for DERBY+, it was trained in histology cohort (n = 82), which included patients eligible for baseline colonoscopy specimens in training and internal validation cohorts and evaluated in external validation cohort. To exclude the interference of chemotherapy alone, negative validation cohort (n = 37) enrolled members of arm B (mFOLFOX6) from the BECOME study.
The clinical characteristics of patients were represented in Table 1. All these patients received conversion therapy with RAS mutated type. The efficacy of treatment was assessed by ORR, DCR, PFS, OS, clinical benefit rate and surgery for liver metastases in Supplementary Table S1. The number (percentage) of patients with PR, who we defined as responders, were 49 (47.6%), 27 (41.5%), 39 (47.6%), 46 (45.1%), 11 (29.8%) in training, internal validation, histology, external validation cohort, and negative validation cohort, respectively. Notably, patients in the negative validation cohort who received chemotherapy alone exhibited the poorest performance across all efficacy measures.
Table 1.
Characteristics of patients in each study cohorts.a
| Characteristic | Training Cohort (n = 103) |
Internal Validation Cohort (n = 65) |
Histology Cohort (n = 82) |
External Validation Cohort (n = 102) |
Negative Validation Cohort (n = 37) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | |
| Sex | ||||||||||
| Female | 33 | 32.0 | 22 | 33.9 | 27 | 32.9 | 32 | 31.4 | 10 | 27.0 |
| Male | 70 | 68.0 | 43 | 66.1 | 55 | 67.1 | 70 | 68.6 | 27 | 73.0 |
| Age, years | ||||||||||
| Median (range) | 58 (30–75) | 58 (31–81) | 59 (30–82) | 60 (29–78) | 61 (29–72) | |||||
| >65 | 22 | 21.4 | 14 | 21.5 | 19 | 23.2 | 34 | 33.3 | 10 | 27.0 |
| Preop. CEA, ng/ml | ||||||||||
| <5 | 15 | 14.6 | 14 | 21.5 | 17 | 20.7 | 19 | 18.6 | 5 | 13.5 |
| 5–199 | 54 | 52.4 | 35 | 53.9 | 48 | 58.6 | 66 | 64.6 | 20 | 54.1 |
| ≥200 | 34 | 33.0 | 16 | 24.6 | 17 | 20.7 | 17 | 9.8 | 12 | 32.4 |
| Preop. CA19–9, U/ml | ||||||||||
| <200 | 55 | 53.4 | 34 | 52.3 | 53 | 64.6 | 56 | 54.9 | 20 | 54.1 |
| ≥200 | 48 | 46.6 | 31 | 47.7 | 29 | 35.4 | 46 | 45.1 | 17 | 45.9 |
| Clinical risk scoreb | ||||||||||
| 0–2 | 14 | 13.6 | 11 | 16.9 | 12 | 14.6 | 15 | 14.7 | 6 | 16.2 |
| 3–5 | 89 | 86.4 | 54 | 83.1 | 70 | 85.4 | 87 | 85.3 | 31 | 83.8 |
| Primary tumor sitec | ||||||||||
| Right-sided | 37 | 35.9 | 20 | 30.8 | 25 | 30.5 | 31 | 30.4 | 13 | 35.1 |
| Left-sided | 66 | 64.1 | 45 | 69.2 | 57 | 69.5 | 71 | 69.6 | 24 | 64.9 |
| No. of liver metastases | ||||||||||
| <3 | 10 | 9.7 | 11 | 16.9 | 7 | 8.5 | 25 | 24.5 | 6 | 16.2 |
| ≥3 | 93 | 90.3 | 54 | 83.1 | 75 | 91.5 | 77 | 75.5 | 31 | 83.8 |
| Maximum size of metastases, cm | ||||||||||
| <5 | 69 | 67.0 | 41 | 63.1 | 54 | 65.9 | 69 | 67.6 | 23 | 62.2 |
| ≥5 | 34 | 33.0 | 24 | 36.9 | 28 | 34.1 | 33 | 32.4 | 14 | 37.8 |
| Distribution of liver metastasis | ||||||||||
| Unilobar | 16 | 15.5 | 10 | 15.4 | 13 | 15.9 | 28 | 27.5 | 9 | 24.3 |
| Bilobar | 87 | 84.5 | 55 | 84.6 | 69 | 84.1 | 74 | 72.5 | 28 | 75.7 |
| SUVmax | ||||||||||
| Mean ± SD | 11.9 ± 7.8 | 11.8 ± 5.2 | 12.3 ± 8.1 | 11.1 ± 5.2 | 12.3 ± 5.0 | |||||
| PKM Score | ||||||||||
| Mean ± SD | – | – | 11.7 ± 4.9 | 12.9 ± 5.2 | 12.3 ± 8.6 | |||||
| GLUT1 Score | ||||||||||
| Mean ± SD | – | – | 11.2 ± 3.7 | 10.6 ± 4.3 | 11.6 ± 5.0 | |||||
NOTE. Data presented as %.
Abbreviations: CEA, carcinoembryonic antigen; CA19-9, Carbohydrate antigen199; SUV, standardized uptake value; SD, standard deviation.
All enrolled patients developed synchronous liver metastases. Histology cohort was consisting of patients with colonoscopy biopsy specimens in training and validation cohorts, thus, there was a overlap of patients in three cohorts.
Clinical risk factors included lymphatic spread of primary cancer, simultaneous metastases, or interval <12 months from primary tumor resection to metastasis, CEA>200 ng/mL, no. of liver metastasis >1, and largest size of liver metastasis >5 cm. Each risk factor was 1 point.
Right-sided included tumors from cecal to two thirds of proximal transverse colon; left-sided represented tumors from one third of distal transverse colon to rectum.
Visualisation of multi-modal imaging features and selection of clinical features
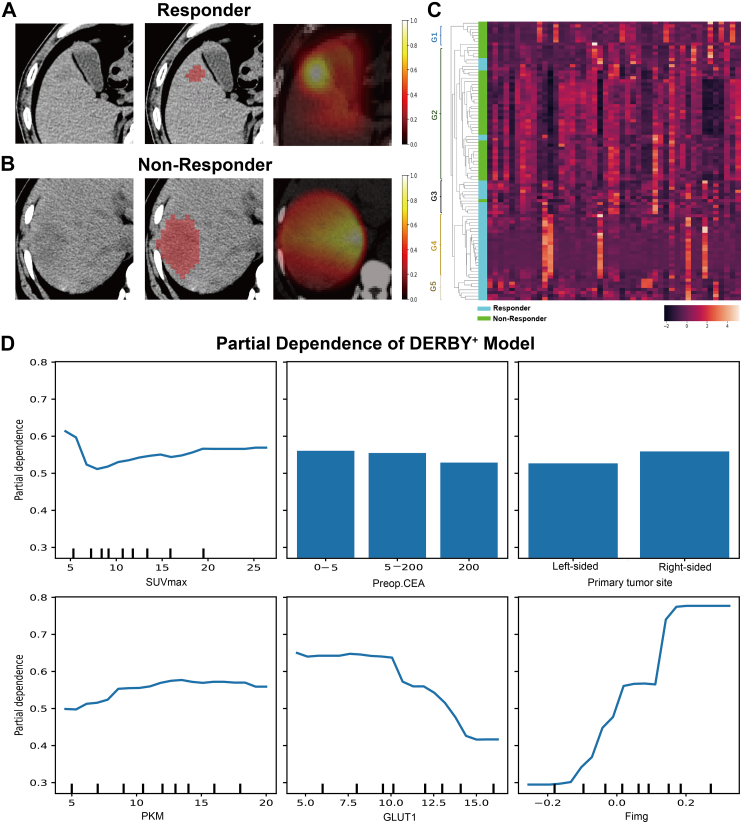
The focusing areas of DNN on PET and CT images were showed in Fig. 3 using Grad-CAM.29 As shown in Fig. 3A, the DNN automatically focused on the most crucial areas within the tumour region for bevacizumab responders. In contrast, for non-responders, the DNN's attention was relatively even within the region of interest, and the decisive areas could not be identified (Fig. 3B). Additionally, the result of unsupervised hierarchical clustering performed on the extracted deep features was presented in Fig. 3C. Of note, there were five distinct subgroups, and the treatment outcomes within each subgroup were largely consistent, indicating that the features learned by the DNN captured characteristics that helped distinguish patients with different bevacizumab efficacy. The individual contribution of each feature in the final classification model is presented in Fig. 3D, where we can conclude that the image feature plays a primary role in prediction.
Fig. 3.
Visualisation of PET/CT features. A, B. The figures from left to right are patients' CT image, segmented tumour areas and our model response which indicates the self-learned important areas for predicting bevacizumab efficacy. C. The heatmap plotted according to the unsupervised hierarchical clustering of deep features learned by our model. There are five instinct subgroups, denoted as G1-G5 in C. Feature representation within each subgroup shows similar pattern. Patients within G1 and G2 mostly got negative outcome from bevacizumab treatment, while most patients within G3, G4 and G5 showed positive bevacizumab efficacy. D. Partial dependency graph for DERBY+ model. Continuous features are presented in curves and discrete features in bars. The curves and bars reflect how our model tends towards a positive or negative prediction as the corresponding feature value changes. As is shown in the graph, all involved features show correlation with the prediction, and the image features and immune marker GLUT1 have more clear and large contribution to model decision.
Supplementary Fig. 1A depicts the results of the LASSO regression, which identified PET/CT SUVmax, pre-operative CEA, and primary tumour site as the most relevant predictors for bevacizumab efficacy. Supplementary Figure S1B further shows the correlation heatmap across these clinical features, indicating that none of these features exhibited high correlation with each other. Particularly, the correlation across the three selected predictors was less than 0.1, thus being largely independent.
Evaluation and predictive performance of the DERBY+ model
The calibration consistency of the model's probability outputs was evaluated using Hosmer–Lemeshow test. The p-values were consistently above 0.05 (Supplementary Table S2), indicating that both models exhibit strong probability consistency.
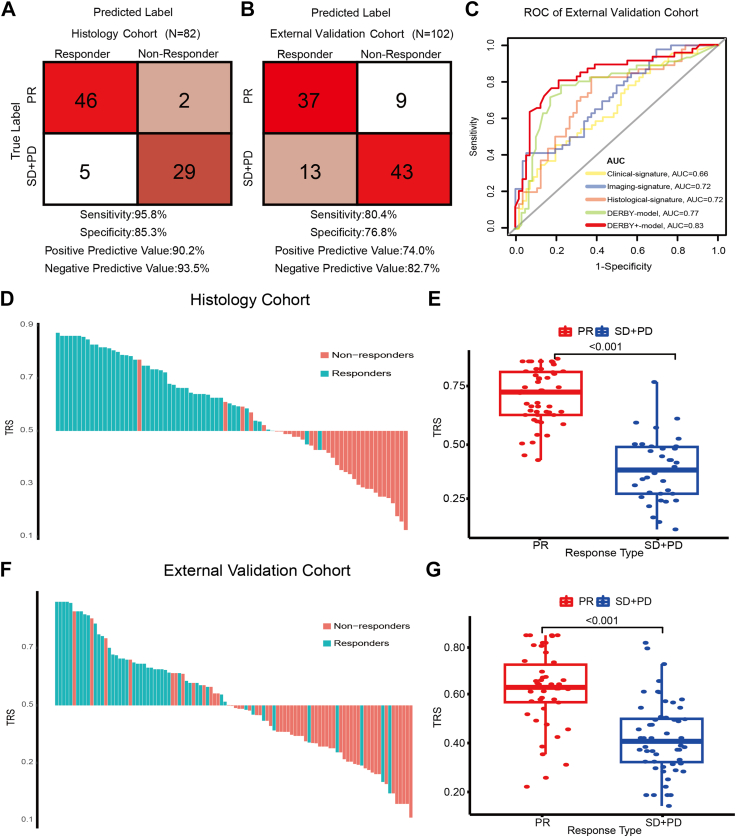
The DERBY+ showed satisfied performance in the independent histology cohort with an AUC of 0.95 (95% CI: 0.91–1.00), sensitivity of 95.8%, specificity of 85.3% (Fig. 4A). Notable, the DERBY+ model (AUC: 0.83) presented higher accuracy than individual predictor types (AUC: Clinical-signature:0.66, Imaging-signature:0.72, Histological-signature:0.72, DERBY: 0.77), in the external validation cohort (Fig. 4B and C). As shown in Supplementary Figure S7, the result of calibration analysis indicates that the DERBY+ model provides better probability calibration in the external test than DERBY with a lower brier loss (0.166 vs 0.200). Logistic regression analysis also confirmed that the DERBY+ was a significant predictor for bevacizumab therapy response (OR [Odds Ratio]: 13.60, 95% CI: 5.44–37.28, p < 0.001, Supplementary Table S3). Furthermore, the TRS of DERBY+ was significantly different between the PR group and the SD+PD group in both the histology cohort and the external validation cohort (both p-values <0.001, Fig. 4D–G). As expected, the DERBY+ failed with chemotherapy alone in the negative controls, with an AUC of 0.58 (95% CI: 0.39–0.76), sensitivity of 40.0%, and specificity of 73.0% (Table 2), demonstrating the specificity of this model for bevacizumab. These results showed that the DERBY+ demonstrated the capacity of predicting bevacizumab efficacy and showed robust performance in independent samples.
Fig. 4.
Prediction performance of the DERBY+and therapy response scores. A, B. Sensitivity, specificity, positive predictive value, and negative predictive value of DERBY+ in the histology cohort and external validation cohort. C. ROC curve of clinical, radiomic, histological signatures and DERBY, DERBY+ models in external validation cohort. D, F. The therapy response score (TRS) for each patient in the histology (n = 82) and external validation cohorts (n = 102). E, G. TRS of the PR and SD + PD groups in histology cohort and external validation cohort. The statistical analysis of TRS was done with z-test. Abbreviations: TRS, therapy response score; ROC, receiver operating characteristic curve.
Table 2.
Prediction performance of DERBY and DERBY+ model.
| Training cohort (N = 103) | Internal validation cohort (N = 65) | Histology cohort (N = 82) | External validation cohort (N = 102) | Negative validation cohort (N = 37) | |
|---|---|---|---|---|---|
| DERBY model | |||||
| AUC (95% CI) | 0.92 (0.87–0.97) | 0.84 (0.74–0.93) | – | 0.77 (0.67–0.87) | 0.53 (0.34–0.73) |
| Sensitivity (95% CI) | 0.84 (0.69–0.93) | 0.67 (0.48–0.81) | – | 0.68 (0.54–0.80) | 0.31 (0.12–0.59) |
| Specificity (95% CI) | 0.80 (0.67–0.88) | 0.84 (0.66–0.94) | – | 0.80 (0.65–0.89) | 0.67 (0.43–0.85) |
| PPV (95% CI) | 0.75 (0.60–0.86) | 0.81 (0.61–0.93) | – | 0.78 (0.63–0.89) | 0.42 (0.16–0.71) |
| NPV (95% CI) | 0.87 (0.75–0.94) | 0.71 (0.54–0.84) | – | 0.70 (0.56–0.81) | 0.56 (0.35–0.75) |
| DERBY+ model | |||||
| AUC (95% CI) | – | – | 0.95 (0.91–1.00) | 0.83 (0.75–0.92) | 0.58 (0.39–0.76) |
| Sensitivity (95% CI) | – | – | 0.96 (0.85–0.99) | 0.80 (0.66–0.90) | 0.40 (0.17–0.67) |
| Specificity (95% CI) | – | – | 0.85 (0.68–0.94) | 0.77 (0.63–0.87) | 0.73 (0.50–0.88) |
| PPV (95% CI) | – | – | 0.90 (0.78–0.96) | 0.74 (0.59–0.85) | 0.50 (0.22–0.78) |
| NPV (95% CI) | – | – | 0.94 (0.77–0.99) | 0.83 (0.69–0.91) | 0.64 (0.42–0.81) |
Abbreviation: AUC, area under curve; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
Prognosis value of DERBY+ model
Interestingly, Kaplan–Meier analysis showed that the DERBY+-predicted responders achieved a favourable PFS and OS in the histology cohort (Fig. 5A and B). In the external validation cohort, the DERBY+ displayed consistent performance in predicting survivals. Specifically, the DERBY+ predicted responders had longer PFS (median: 9.6 vs 6.3 months, HR [hazard Ratio]: 0.52, 95% CI: 0.35–0.79, p-value = 0.002) and OS (median: 27.6 vs 18.5 months, HR: 0.50, 95% CI: 0.29–0.86, p-value = 0.010) than non-responders (Fig. 5C and D).
Fig. 5.
Kaplan–Meier survival analysis based on the DERBY+model. A, B. Kaplan–Meier estimates of PFS and OS stratified by the DERBY+ in histology cohort. C, D. Kaplan–Meier estimates of PFS and OS stratified by the DERBY+ in external validation cohort. The statistical analysis of PFS and OS was done with log-rank test. Hazard ratio and 95% confidence interval was calculated with cox-regression analysis. Abbreviations: PFS, progression free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
Clinical utility of DERBY+ model
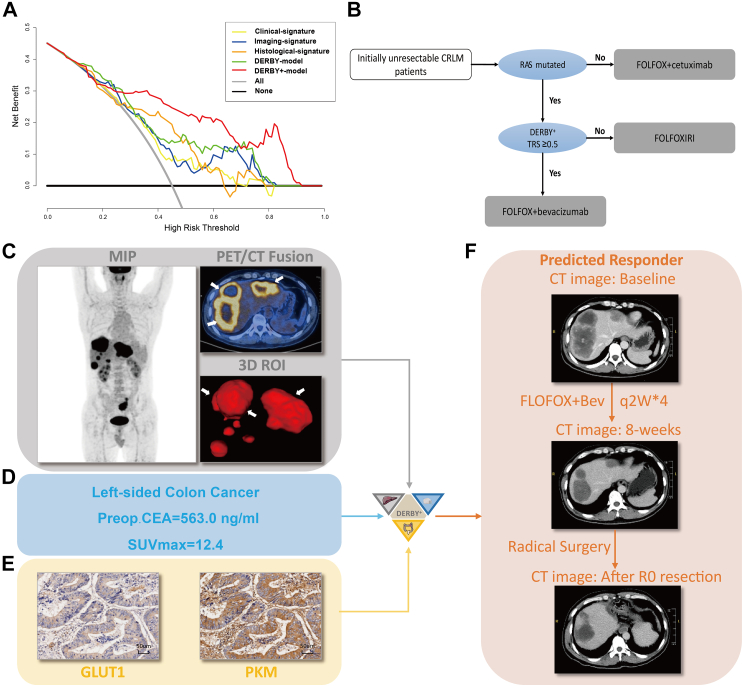
The clinical utility of the DERBY+ model to identify patients sensitive to bevacizumab was examined using decision curve analysis. Regardless of all bevacizumab strategy and no bevacizumab strategy, the clinical, imaging, histological signatures and the DERBY model, and the DERBY+ model consistently showed a positive and larger net benefit across a wide range of risk thresholds (Fig. 6A). Consequently, we proposed an alternative guideline could be employed to guide treatment of patients with CRLM (Fig. 6B). To display the straightforward procedure and encouraging discriminatory ability of the DERBY+ model for bevacizumab sensitivity, a representative case was provided in Fig. 6C–F.
Fig. 6.
Clinical utility of the DERBY+ model. A. Decision curve analysis was performed to compare the efficacy of DERBY+ model with other models: applying bevacizumab based on clinical signature, applying bevacizumab based on imaging signature, applying bevacizumab based on histological signature, applying bevacizumab based on DERBY model and applying bevacizumab for all, not applying bevacizumab. B. Proposed alternative guideline to use DERBY+ for decision support for patients with initially unresectable CRLM. C. A 48-year-old man was initially diagnosed with liver metastases and pelvic lymph node metastases of left-sided colon cancer. Imaging features: MIP showed the systematic tumour burden, a PET/CT fusion image showed multiple liver metastases distributed in two lobes with the largest metastasis being 84 mm in diameter, and there was a 3-dimensional view of ROI. D. Clinical features: left-sided colon cancer, high Preop.CAE level (563.0 ng/mL), and SUVmax = 12.4. E. Protein features: IHC staining of pre-treatment colonoscopy specimens showed low GLUT1 expression and high PKM expression. F. After inputting the trimodal information, DERBY+ showed the patient to be bevacizumab-sensitive. After four cycles of FOLFOX + bevacizumab treatment, we observed from the CT images before and after treatment that the liver metastases were significantly less numerous and smaller, achieving a partial response status. The patient then underwent a successful radical resection with a progression-free survival of 15.5 months. Abbreviations: MIP, maximal intensity projection; Bev, bevacizumab; CRLM, colorectal cancer liver metastases; TRS, therapy response score.
Discussion
Identifying surrogates to predict efficacy of bevacizumab in conversion therapy for unresectable patients with CRLM is urgently needed in the clinic. It would offer opportunities to reduce tumour progression from inappropriate drug administration and potentially help identify more patients who may benefit from surgery. To the best of our knowledge, our work represents the pioneering effort in constructing a multi-modal model empowered by deep learning techniques to predict the response to bevacizumab in patients with CRLM.
Specifically, we presented the DERBY+ model that incorporated several innovational features for improving predictive accuracy in recruited individuals from multiple sites. Firstly, we designed a multi-modal framework to jointly extract features from PET/CT images, combining anatomical and metabolism information. We proposed a novel 2.5D training scheme on this particular data collection to address the insufficiency of training samples. Secondly, to reduce the dimension gap between high-dimensional deep features and non-imaging feature types such as the clinical factors, a GMM was employed to convert the extracted imaging features into a compact form, so that the high-dimensional imaging features would not be over-dominant in the final predictive model. Thirdly, the DERBY+ model was established by incorporating the deep learning baseline PET/CT imaging, clinical data, and colonoscopy biopsy specimens, thus enabling the prediction of bevacizumab efficacy before treatment initiation, with the potential to reduce excessive treatment-related adverse events and costs. Importantly, the discovery phase of the current study was developed upon a high-quality clinical trial, the BECOME study, and two independent cohorts were used for validation. Taken together, we proposed a promising tool for predicting bevacizumab response before initiation of treatment. We expect this innovative approach would help clinicians stratify patients with CRLM and identify those who can benefit from bevacizumab treatment.
Currently, the RESIST 1.1 criteria or early tumour shrinkage are clinically used to assess tumour response and differentiate the outcomes of patients.24,40 However, these methods are limited by hysteresis, by which they would be applicable at least 2 months after the initiation of conversion therapy. To overcome this constraint, previous studies have explored several baseline biomarkers and/or features that are relevant to bevacizumab treatment, including miRNAs,41,42 proteins,43 and SUVmax of PET/CT.19 Nevertheless, previous studies were merely correlation analysis, with no capacity to directly output prediction of bevacizumab treatment efficacy. In addition, a single biomarker or predictor type likely cannot represent the complex tumour properties of those initially unresectable CRLM. Our comparison experiment in Fig. 4C also shows these plain features are inadequate for constructing a reliable prediction model. During the past decade, radiomics-based models have been proposed to predict bevacizumab efficacy, however with critical limitations. For example, Dohan et al.16 developed a CT radiomics model that was shown to outperform RECIST1·1 and morphological criteria, but this model required images at both baseline and 2-month after treatment. While Wei et al.44 proposed a CT radiomics model to identify histopathologic growth pattern and showed that these patterns could be helpful in predicting early bevacizumab response, the AUC of response prediction was around 0.70,16,44 leading significant room for further improvement. Moreover, these methods relied on hand-crafted image descriptors for image feature extraction,16,44 thus with the possibility that optimal features for predicting bevacizumab efficacy are not captured. In recent years, DNNs have been widely adopted in radiomic research to automatically extract image features. For example, Wei et al.45 developed a deep radiomics model to predict chemotherapy response in patients with CRLM and achieved better performance than using hand-crafted features. In the current study, the DERBY+ was developed to address these technical gaps and limitations as well as clinical needs by presenting a semi-automatic delineating deep learning model, which can predict bevacizumab efficacy at baseline.
Two previous multi-modal models performed well in predicting targeted therapy response in lung and breast cancer.21,46 In these studies, each omics type played a unique role in efficacy prediction in terms of drug mechanism of action. When selecting modal types, considering that bevacizumab is an anti-angiogenic drug that acts by blocking angiogenesis and can induce metabolic disorders,47 we selected PET/CT images of liver lesions to reveal the status of micro vessels and metabolism of metastases. Furthermore, a consensus has been reached that clinical factors can improve the predictive capability of radiomics models.48, 49, 50 In this study, we applied clinical factors to help the DNN understand systemic tumour profiles. We selected SUVmax of PET/CT,19,51 pre-operative CEA,52,53 and primary tumour site54,55 as clinical factors using LASSO regression, which were proven to correlate with bevacizumab efficacy. In fact, combing PET/CT images and clinical factors, the DERBY model achieved an AUC of 0.77 in the external validation cohort, suggesting the validity of these clinical factors. Moreover, we added IHC staining of endoscopic biopsies to represent the metabolic condition of primary tumour. By incorporating endoscopic biopsy information, the DERBY+ model further improved the performance in the external validation cohort to an AUC of 0.83. The partial graph presented in Fig. 3D also demonstrates the effect of PKM and GLUT1 for bevacizumab therapy response, which consistent with our results of IHC (Supplementary Figure S2A and B). The ORR of patients predicted as responders by the model was significantly higher than that of patients predicted as non-responders (74.0% vs 17.3% p < 0.001), and the PFS was also significantly longer (9.6 months vs 6.3 months, p = 0.002). Traditional radiomics models were usually competent at predicting either prognosis or efficacy.16,20 Our DERBY + model, being designed for efficacy prediction, also showed a predictive ability in OS (responder vs non-responder: 27.6 months vs 18.5 months, p = 0.010). The improved performance of the DERBY+ model indicated that these multi-modal features for the action mechanism of bevacizumab were relevant.
Clinically, the DERBY+ model has the potential to assist oncologists in making personalised medicine decisions for patients with CRLM. The DERBY+ model could output an unambiguous prediction result, in terms of bevacizumab responder or bevacizumab non-responder, based on the multi-modal features. For those patients predicted insensitive to bevacizumab, it would be worthwhile to administer intense chemotherapy regimens without bevacizumab (FOLOFOXIRI regimen), since this subgroup of patients is unlikely to benefit greatly from bevacizumab (Fig. 6B). Such a strategy can reduce unnecessary adverse events and may increase the conversional resection rate of them. Especially for patients with RAS-mutant or right-sided CRC, almost all of whom received chemotherapy plus bevacizumab as conversion therapy, whether to choose bevacizumab is directly associated with their prognosis and cost. Notably, the DERBY+ performed well in terms of NPV, achieving 82.7%, suggesting that the model has the potential to reliably identify individuals who are insensitive to bevacizumab. Among those DERBY+-predicted insensitive patients with CRLM, only 5.8% (3 of 52) of patients were successfully converted by chemotherapy combined with bevacizumab and accepted R0 resection surgery. The conversion resection rate of patients predicted by the DERBY+ to be sensitive to bevacizumab was five-fold higher (30.0%, 15 of 50) than non-responders. These resulted demonstrated that DERBY+ can significantly reduce the utilisation of bevacizumab, while ensuring a high conversion resection rate, in nearly half of patients with RAS-mutant CRLM and held immense significance in effectively reallocating societal healthcare resources. The DERBY+ showed a PPV of 74.0%, which means that 74.0% of predicted positive patients obtained ORR, almost 20% higher than Arm A of the BECOME study.9 In summary, our results illustrated that formulating treatment plans based on the “second opinion” provided by DERBY+ can not only enhance patients' treatment response rates but also facilitate the rational allocation of societal resources, thereby presenting promising prospects for clinical translation.
There are some limitations in the current study. Firstly, considering the retrospective nature of our study and we were unable to enroll all patients from the BECOME study and the MDT group due to unavailable baseline PET/CT data and colonoscopy specimens, thus potentially causing selection bias and confounding. Secondly, the proposed DERBY+ model used a semi-automatic segmentation pipeline for tumour ROI, which still required manual initialisation and expert post-processing. Thirdly, the protein markers selected for this study were derived from the literature, and optimal candidates may need to be screened using proteomic data from tumour specimens of patients with CRLM treated before and after bevacizumab. Finally, this is a retrospective designed study, the conclusion needs to be validated by a prospective trial. Therefore, we have registered prospective clinical trial (NCT05354674) to validate the robustness of DERBY+ model.
In conclusion, we constructed the DERBY+, a novel deep radiomics based multi-modal model to predict bevacizumab efficacy with prognostic value as well for patients with CRLM. The robust performance of the DERBY+ model presented the promise for developing tailored regimens before conversion therapy administration, with the ultimate goal of reducing mortality.
Contributors
WCh, LW, HS, YJ and JX was responsible for overall study design. SZ, DS, WM and WCe accessed and verified the underlying data. SZ, DS and FL was responsible for analysis and interpretation of data. SZ, DS, WM, YL, FL, LW and WCh was responsible for writing, review, and/or revision of the manuscript. LY, JX and WCh were study supervisions and guarantors. All the authors have read, discussed, and unanimously approved the final version of the manuscript. All authors had full access to the data and had the final responsibility for the decision to submit for publication.
Data sharing statement
The datasets used or analysed in the current study are available from the corresponding authors upon reasonable request.
Declaration of interests
We declare no competing interests.
Acknowledgements
This trial was funded by the National Natural Science Foundation of China (82072653, 81602035, 82172816); Fujian Provincial Health Commission Project (2021GGB032); Xiamen Science and Technology Agency Program (3502Z20224ZD1067); Clinical Research Plan of SHDC (No. SHDC2020CR5006); Shanghai Science and Technology Committee Project (19511121300); Clinical Research Plan of SHDC (No. SHDC2020CR3037B); Zhejiang Provincial Natural Science Foundation of China (LY22H160021); National Science Foundation of Xiamen (2023J06057). We thank collaborators at Zhongshan Hospital and Zhongshan Hospital Xiamen of Fudan University, the First Affiliated Hospital of Wenzhou Medical University for assistance with histopathology.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102271.
Contributor Information
Jianmin Xu, Email: xujmin@aliyun.com.
Hongcheng Shi, Email: shi.hongcheng@zs-hospital.sh.cn.
Yuan Ji, Email: ji.yuan@zs-hospital.sh.cn.
Lisheng Wang, Email: lswang@sjtu.edu.cn.
Wenju Chang, Email: chang.wenju@zs-hospital.sh.cn.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E., Cervantes A., Nordlinger B., Arnold D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3) doi: 10.1093/annonc/mdu260. iii1-9. [DOI] [PubMed] [Google Scholar]
- 3.House M.G., Ito H., Gönen M., et al. Survival after hepatic resection for metastatic colorectal cancer: trends in outcomes for 1,600 patients during two decades at a single institution. J Am Coll Surg. 2010;210(5):744–752. doi: 10.1016/j.jamcollsurg.2009.12.040. [DOI] [PubMed] [Google Scholar]
- 4.Tsilimigras D.I., Brodt P., Clavien P.A., et al. Liver metastases. Nat Rev Dis Primers. 2021;7(1):27. doi: 10.1038/s41572-021-00261-6. [DOI] [PubMed] [Google Scholar]
- 5.Raphael M.J., Karanicolas P.J. Regional therapy for colorectal cancer liver metastases: which modality and when? J Clin Oncol. 2022;40(24):2806–2817. doi: 10.1200/JCO.21.02505. [DOI] [PubMed] [Google Scholar]
- 6.Benson A.B., Venook A.P., Al-Hawary M.M., et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19(3):329–359. doi: 10.6004/jnccn.2021.0012. [DOI] [PubMed] [Google Scholar]
- 7.Rosen L.S., Jacobs I.A., Burkes R.L. Bevacizumab in colorectal cancer: current role in treatment and the potential of biosimilars. Target Oncol. 2017;12(5):599–610. doi: 10.1007/s11523-017-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limited R.R. 2009. Avastin (bevacizumab) summary of product characteristics. last update 10/19/2016. [Google Scholar]
- 9.Tang W., Ren L., Liu T., et al. Bevacizumab plus mFOLFOX6 versus mFOLFOX6 alone as first-line treatment for RAS mutant unresectable colorectal liver-limited metastases: the BECOME randomized controlled trial. J Clin Oncol. 2020;38(27):3175–3184. doi: 10.1200/JCO.20.00174. [DOI] [PubMed] [Google Scholar]
- 10.Tran K.A., Kondrashova O., Bradley A., Williams E.D., Pearson J.V., Waddell N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021;13(1):152. doi: 10.1186/s13073-021-00968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambin P., Rios-Velazquez E., Leijenaar R., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayerhoefer M.E., Materka A., Langs G., et al. Introduction to radiomics. J Nucl Med. 2020;61(4):488–495. doi: 10.2967/jnumed.118.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambin P., Leijenaar R.T.H., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 14.Lam L.H.T., Do D.T., Diep D.T.N., et al. Molecular subtype classification of low-grade gliomas using magnetic resonance imaging-based radiomics and machine learning. NMR Biomed. 2022;35(11) doi: 10.1002/nbm.4792. [DOI] [PubMed] [Google Scholar]
- 15.Le V.H., Kha Q.H., Minh T.N.T., Nguyen V.H., Le V.L., Le N.Q.K. Development and validation of CT-based radiomics signature for overall survival prediction in multi-organ cancer. J Digit Imaging. 2023;36(3):911–922. doi: 10.1007/s10278-023-00778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dohan A., Gallix B., Guiu B., et al. Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut. 2020;69(3):531–539. doi: 10.1136/gutjnl-2018-316407. [DOI] [PubMed] [Google Scholar]
- 17.De Bock K., Georgiadou M., Schoors S., et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154(3):651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Lastoria S., Piccirillo M.C., Caracò C., et al. Early PET/CT scan is more effective than RECIST in predicting outcome of patients with liver metastases from colorectal cancer treated with preoperative chemotherapy plus bevacizumab. J Nucl Med. 2013;54(12):2062–2069. doi: 10.2967/jnumed.113.119909. [DOI] [PubMed] [Google Scholar]
- 19.Mertens J., De Bruyne S., Van Damme N., et al. Standardized added metabolic activity (SAM) IN 18F-FDG PET assessment of treatment response in colorectal liver metastases. Eur J Nucl Med Mol Imaging. 2013;40(8):1214–1222. doi: 10.1007/s00259-013-2421-z. [DOI] [PubMed] [Google Scholar]
- 20.Antunovic L., De Sanctis R., Cozzi L., et al. PET/CT radiomics in breast cancer: promising tool for prediction of pathological response to neoadjuvant chemotherapy. Eur J Nucl Med Mol Imaging. 2019;46(7):1468–1477. doi: 10.1007/s00259-019-04313-8. [DOI] [PubMed] [Google Scholar]
- 21.Sammut S.J., Crispin-Ortuzar M., Chin S.F., et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature. 2022;601(7894):623–629. doi: 10.1038/s41586-021-04278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Laak J., Litjens G., Ciompi F. Deep learning in histopathology: the path to the clinic. Nat Med. 2021;27(5):775–784. doi: 10.1038/s41591-021-01343-4. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., Chang W., Zhou B., et al. Conventional transarterial chemoembolization combined with systemic therapy versus systemic therapy alone as second-line treatment for unresectable colorectal liver metastases: randomized clinical trial. Br J Surg. 2021;108(4):373–379. doi: 10.1093/bjs/znaa155. [DOI] [PubMed] [Google Scholar]
- 24.Eisenhauer E.A., Therasse P., Bogaerts J., et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Chilcott A.K., Bradley K.M., McGowan D.R. Effect of a Bayesian penalized likelihood PET reconstruction compared with ordered subset expectation maximization on clinical image quality over a wide range of patient weights. AJR Am J Roentgenol. 2018;210(1):153–157. doi: 10.2214/AJR.17.18060. [DOI] [PubMed] [Google Scholar]
- 26.Boudraa A.-O., Zaidi H. Quantitative analysis in nuclear medicine imaging. 2006. Image segmentation techniques in nuclear medicine imaging; pp. 308–357. [Google Scholar]
- 27.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Deep residual learning for image recognition. [Google Scholar]
- 28.Tan M., Le Q. International conference on machine learning; 2019. Efficientnet: rethinking model scaling for convolutional neural networks. PMLR. [Google Scholar]
- 29.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE international conference on computer vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization. [Google Scholar]
- 30.Shim J.R., Lee S.D., Han S.S., et al. Prognostic significance of (18)F-FDG PET/CT in patients with colorectal cancer liver metastases after hepatectomy. Eur J Surg Oncol. 2018;44(5):670–676. doi: 10.1016/j.ejso.2018.01.243. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto M., Kobayashi T., Ishiyama K., et al. Efficacy of repeat hepatectomy for recurrence following curative hepatectomy for colorectal liver metastases: a Retrospective Cohort Study of 128 patients. Int J Surg. 2016;36(Pt A):96–103. doi: 10.1016/j.ijsu.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Wang H.W., Yan X.L., Wang L.J., et al. Characterization of genomic alterations in Chinese colorectal cancer patients with liver metastases. J Transl Med. 2021;19(1):313. doi: 10.1186/s12967-021-02986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong Y., Fortner J., Sun R.L., Brennan M.F., Blumgart L.H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230(3):309–318. doi: 10.1097/00000658-199909000-00004. discussion 18-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai S., Ye Y., Kong X., Li J., Ding K. A predictive model for early recurrence of colorectal-cancer liver metastases based on clinical parameters. Gastroenterol Rep (Oxf) 2021;9(3):241–251. doi: 10.1093/gastro/goaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai W., Xu Y., Mo S., et al. GLUT3 induced by AMPK/CREB1 axis is key for withstanding energy stress and augments the efficacy of current colorectal cancer therapies. Signal Transduct Target Ther. 2020;5(1):177. doi: 10.1038/s41392-020-00220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graziano F., Ruzzo A., Giacomini E., et al. Glycolysis gene expression analysis and selective metabolic advantage in the clinical progression of colorectal cancer. Pharmacogenomics J. 2017;17(3):258–264. doi: 10.1038/tpj.2016.13. [DOI] [PubMed] [Google Scholar]
- 37.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12(null):2825–2830. [Google Scholar]
- 38.Chaddad A., Hassan L., Desrosiers C. Deep radiomic analysis for predicting coronavirus disease 2019 in computerized tomography and X-ray images. IEEE Trans Neural Netw Learn Syst. 2022;33(1):3–11. doi: 10.1109/TNNLS.2021.3119071. [DOI] [PubMed] [Google Scholar]
- 39.Biau G., Scornet E. A random forest guided tour. Test. 2016;25(2):197–227. [Google Scholar]
- 40.Heinemann V., Stintzing S., Modest D.P., Giessen-Jung C., Michl M., Mansmann U.R. Early tumour shrinkage (ETS) and depth of response (DpR) in the treatment of patients with metastatic colorectal cancer (mCRC) Eur J Cancer. 2015;51(14):1927–1936. doi: 10.1016/j.ejca.2015.06.116. [DOI] [PubMed] [Google Scholar]
- 41.Vincenzi B., Zoccoli A., Schiavon G., et al. Dicer and Drosha expression and response to Bevacizumab-based therapy in advanced colorectal cancer patients. Eur J Cancer. 2013;49(6):1501–1508. doi: 10.1016/j.ejca.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Ulivi P., Canale M., Passardi A., Marisi G., Valgiusti M., Frassineti G.L., et al. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of Bevacizumab efficacy in patients with metastatic colorectal cancer. Int J Mol Sci. 2018;19(1) doi: 10.3390/ijms19010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger M.D., Stintzing S., Heinemann V., et al. A polymorphism within the vitamin D transporter gene predicts outcome in metastatic colorectal cancer patients treated with FOLFIRI/bevacizumab or FOLFIRI/cetuximab. Clin Cancer Res. 2018;24(4):784–793. doi: 10.1158/1078-0432.CCR-17-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei S., Han Y., Zeng H., et al. Radiomics diagnosed histopathological growth pattern in prediction of response and 1-year progression free survival for colorectal liver metastases patients treated with bevacizumab containing chemotherapy. Eur J Radiol. 2021;142 doi: 10.1016/j.ejrad.2021.109863. [DOI] [PubMed] [Google Scholar]
- 45.Li H., Boimel P., Janopaul-Naylor J., et al. Deep convolutional neural networks for imaging data based survival analysis of rectal cancer. Proc IEEE Int Symp Biomed Imaging. 2019;2019:846–849. doi: 10.1109/ISBI.2019.8759301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S., Yu H., Gan Y., et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. 2022;4(5):e309–e319. doi: 10.1016/S2589-7500(22)00024-3. [DOI] [PubMed] [Google Scholar]
- 47.Limited R.R. 2009. Avastin (bevacizumab) summary of product characteristics.https://www.ema.europa.eu/en/documents/product-information/avastin-epar-product-information_en.pdf Available from: [Google Scholar]
- 48.Wu S., Zheng J., Li Y., et al. A radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin Cancer Res. 2017;23(22):6904–6911. doi: 10.1158/1078-0432.CCR-17-1510. [DOI] [PubMed] [Google Scholar]
- 49.Li M., Zhang J., Dan Y., et al. A clinical-radiomics nomogram for the preoperative prediction of lymph node metastasis in colorectal cancer. J Transl Med. 2020;18(1):46. doi: 10.1186/s12967-020-02215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F., Liu D., Wang K., et al. Deep learning radiomics based on contrast-enhanced ultrasound might optimize curative treatments for very-early or early-stage hepatocellular carcinoma patients. Liver Cancer. 2020;9(4):397–413. doi: 10.1159/000505694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coudert B., Pierga J.Y., Mouret-Reynier M.A., et al. Long-term outcomes in patients with PET-predicted poor-responsive HER2-positive breast cancer treated with neoadjuvant bevacizumab added to trastuzumab and docetaxel: 5-year follow-up of the randomised Avataxher study. eClinicalMedicine. 2020;28 doi: 10.1016/j.eclinm.2020.100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prager G.W., Braemswig K.H., Martel A., et al. Baseline carcinoembryonic antigen (CEA) serum levels predict bevacizumab-based treatment response in metastatic colorectal cancer. Cancer Sci. 2014;105(8):996–1001. doi: 10.1111/cas.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jürgensmeier J.M., Schmoll H.J., Robertson J.D., et al. Prognostic and predictive value of VEGF, sVEGFR-2 and CEA in mCRC studies comparing cediranib, bevacizumab and chemotherapy. Br J Cancer. 2013;108(6):1316–1323. doi: 10.1038/bjc.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yaeger R., Chatila W.K., Lipsyc M.D., et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33(1):125–136.e3. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holch J.W., Ricard I., Stintzing S., Modest D.P., Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: a meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. doi: 10.1016/j.ejca.2016.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.