Summary
Thyroid nodules are a common disease, and fine needle aspiration cytology (FNAC) is the primary method to assess their malignancy. For the diagnosis of follicular thyroid nodules, however, FNAC has limitations. FNAC can classify them only as Bethesda IV nodules, leaving their exact malignant status and pathological type undetermined. This imprecise diagnosis creates difficulties in selecting the follow-up treatment. In this retrospective study, we collected ultrasound (US) image data of Bethesda IV thyroid nodules from 2006 to 2022 from five hospitals. Then, US image-based artificial intelligence (AI) models were trained to identify the specific category of Bethesda IV thyroid nodules. We tested the models using two independent datasets, and the best AI model achieved an area under the curve (AUC) between 0.90 and 0.95, demonstrating its potential value for clinical application. Our research findings indicate that AI could change the diagnosis and management process of Bethesda IV thyroid nodules.
Subject area: medical imaging, neural networks, cancer
Graphical abstract

Highlights
-
•
Individuals with Bethesda category IV thyroid nodules are subtyped using an AI model
-
•
AI-based method can serve as a supplement to fine needle aspiration cytology (FNAC)
-
•
The management of Bethesda IV thyroid nodules could be changed by AI technology
Medical imaging; Neural networks; Cancer
Introduction
Thyroid nodules have a high incidence and may be present in 50% of individuals. Although most of these nodules are benign, about 8% of them are malignant tumors.1 Fine needle aspiration cytology (FNAC) is the standard diagnostic tool for evaluating the malignancy of thyroid nodules.2,3,4,5 However, FNAC also has limitations. For example, benign and malignant follicular thyroid tumors share similar cellular characteristics, making it difficult to differentiate them based on FNAC.3 A more extensive analysis of the nodule structure, coupled with an evaluation of a potential invasion of blood vessels, is essential to provide further diagnostic insights. FNAC, which relies on a limited and fragmented cell sample, can indicate only the presence of follicular nodules and classify them as Bethesda IV. Histopathological examination following surgical resection of the nodules remains the gold standard for diagnosing such nodules.4,6 Because 10%–30% of follicular thyroid nodules are malignant tumors, a vague diagnosis of Bethesda IV makes it difficult to formulate an accurate follow-up treatment and management plan.5
It is known that Bethesda IV thyroid nodules mainly include follicular thyroid carcinoma (FTC); the follicular variant of papillary thyroid carcinoma (FV-PTC); and benign nodules (BNs) such as follicular thyroid adenoma (FTA) or adenomatoid hyperplastic nodule (AHN).3,6 Among these, FTC has a high degree of malignancy and is prone to hematogenous or distant metastasis; FV-PTC has a low degree of malignancy, slow development, and rare metastasis; and AHN and FTA are benign lesions.7,8,9 Thus, the degree of malignancy and the prognosis of these Bethesda IV thyroid nodules vary greatly. However, due to the lack of effective methods to distinguish them accurately, many patients with benign Bethesda IV thyroid nodules have received excessive treatment, including surgical operation and radioactive I131 treatment.9,10,11 Meanwhile, some FTC patients miss the optimal treatment opportunity because their pathological type cannot be confirmed in time.
To address this problem, early methods tended to identify the follicular thyroid nodules by analyzing the statistical significance of serum indicators or ultrasound (US) image characteristics, such as serum thyroid-stimulating hormone, microcalcification, hypoechogenicity, or halo signs.12,13,14 However, these statistics-based methods cannot yield stable results. Furthermore, the sample size of the relevant studies was small. To improve performance, later studies tried to address the problem by applying genomic sequence methods.15,16 For example, an RNA and machine learning method were used to evaluate the malignant risk of follicular thyroid nodules.15 The model classified thyroid nodules into “benign” and “suspicious” based on their gene expression patterns. According to the study, the previous RNA and machine learning classifier has a sensitivity of 91% and a specificity of 68% and therefore, it can help to reduce unnecessary surgeries to some extent. Other studies have used gene mutation-related methods to evaluate Bethesda IV thyroid nodules. Typical representatives include ThyroSeq V3, which can detect 112 thyroid cancer-related gene mutations.17,18 Compared with the RNA methods, ThyroSeq V3 can provide more genomic information, including point mutation, gene fusion, and gene expression variation. Recent studies indicated that this method has increased the specificity to a range of 77%–82%.18
The previous genetics-based methods have made some progress, obtaining better sensitivity and specificity than traditional statistical methods. However, they also have some shortcomings. First, most genetics-based methods require the use of an extra fine needle aspiration (FNA) to collect thyroid nodule samples.16,17 Because of the heterogeneity of the tumor, the accuracy of sampling depends on the experience of physicians, and extra FNA may also lead to complications, including bleeding, infection, or pseudocysts, for example. Second, even if hundreds of genes or mutations are evaluated, the previous genetics-based methods still have non-negligible rates of false negatives and false positives. Moreover, these methods are expensive and require a laboratory environment.
Recently, artificial intelligence (AI) has advanced rapidly. New-generation AI models such as swin-transformer (ST) and ChatGPT,19,20 which employ transformer technology, have demonstrated performance capabilities superior to the deep convolutional neural networks (DCNNs) used previously.21,22,23,24,25,26 These new AI models offer novel possibilities for an accurate diagnosis, treatment, and management of many different cancers.26,27,28 In the field of thyroid nodule diagnosis and evaluation, computer-aided diagnosis (CAD) models based on US images and AI technologies have made many breakthroughs.4,5,27 However, the use of new-generation transformer-based AI technologies to assist in the diagnosis of common Bethesda IV thyroid nodules has not been deeply explored. Given the strong feature-perception ability of AI models, in this study, we retrospectively collected the US image data of Bethesda category IV thyroid nodules from 2006 to 2022 from five hospitals and undertook the project of training the AI models to identify the specific types of Bethesda category IV thyroid nodules.
To the best of our knowledge, this was the first study to utilize a new generation of transformer-based AI technology to diagnose Bethesda IV thyroid nodules. It also employed the most extensive dataset that has ever been used to investigate this sub-class. Four AI models, including DCNNs and the new-generation ST model, were used to address this diagnostic challenge. We first tested all the models on two independent datasets from different hospitals. Subsequently, the 10-fold cross-validation method was employed to further assess the models’ robustness and stability. The best AI model obtained an area under curve (AUC) exceeding 0.90, indicating its potential clinical value. Our study demonstrates that an AI-based CAD model not only may serve as an important supplement to FNAC but can also be expected to change the diagnosis, treatment, and management of Bethesda category IV thyroid nodules.
Results
Study design and participant characteristics
Initially, a total of 1,875 cases with FNAC results classified as Bethesda IV between May 2006 and April 2022 were collected from five hospitals, yielding a total of 10,746 US images. All patients underwent total thyroidectomy or lobectomy in subsequent treatment, and the pathological results were finally obtained. Based on our exclusion criteria, we rejected (1) cases containing unqualified images (including cases with missing US images or nodules too large to be displayed in a single US image); (2) cases with preoperative treatment (including patients who had undergone radiotherapy or other neck surgery); and (3) cases having incomplete clinical information (including missing information about previous treatments). There were no restrictions on the size of the previous Bethesda IV nodules except for nodules too large to be displayed in a single image. When patients had multiple nodules that met the inclusion criteria, we randomly selected one nodule for the analysis. Finally, we retained 1690 qualified cases and 7566 US images. Figure 1 shows the specific enrollment process of the training set and independent test sets. These cases were then divided into one training set and two independent test sets. The training set comprised 1349 (79.9%) cases from three centers. The two independent testing sets from two separate centers comprised 163 (9.6%) and 178 (10.5%) cases, respectively. Table 1 shows the statistical data that were collected from multiple centers. Table S1 in the supplementary materials provide the equipment information and details of the data acquisition.
Figure 1.
Enrollment of the training set and independent test sets
Table 1.
Data distributions of multiple centers
| Category | Center 1 |
Center 2 |
Center 3 |
Center 4 |
Center 5 |
|
|---|---|---|---|---|---|---|
| Training Set | Test set 1 | Test set 2 | ||||
| Number | 555 | 552 | 242 | 163 | 178 | |
| Age, mean (SD) | 57.4 (13.7) | 59.0 (12.6) | 55.6 (11.8) | 55.2 (13.3) | 54.6 (12.4) | |
| <50, number (Mean, SD) | 178 (41.1, 6.9) | 114 (41.3, 7.0) | 84 (42.2, 6.2) | 52 (39.2, 7.1) | 63 (41, 6.2) | |
| ≥50, number (Mean, SD) | 377 (62.6, 8.0) | 438 (63.0, 8.3) | 158 (60.7, 6.8) | 111 (62.5, 7.3) | 115 (60.8, 6.6) | |
| Sex | ||||||
| Female | 396 | 415 | 170 | 113 | 131 | |
| Male | 159 | 137 | 72 | 50 | 47 | |
| FTC (Mean size, SD) | 113 (1.71, 0.74) | 102 (1.95, 0.53) | 31 (1.86, 0.69) | 45 (1.60, 0.92) | 53 (1.73, 0.77) | |
| FV-PTC (Mean size, SD) | 195 (1.73, 0.70) | 156 (1.79, 0.60) | 92 (1.61, 0.75) | 57 (1.68, 0.66) | 59 (1.71, 0.59) | |
| BN | ||||||
| AHN (Mean size, SD) | 114 (1.66, 0.51) | 173 (1.73, 0.59) | 76 (1.53, 0.81) | 30 (1.55, 0.57) | 39 (1.49, 0.73) | |
| FTA (Mean size, SD) | 133 (1.63, 0.50) | 121 (1.54, 0.62) | 43 (1.79, 0.65) | 31 (1.75, 0.76) | 27 (1.59, 0.63) | |
SD: standard deviation; The unit of mean size is centimeters (cm); Center 1: Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital); Centre 2: Taizhou Cancer Hospital; Center 3: Affiliated Hangzhou First People’s Hospital of Zhejiang University’s School of Medicine. Center 4: Sun Yat-sen University Cancer Center; Center 5: Zhejiang Provincial People’s Hospital.
The AI model and data labeling
Four representative AI models were used in the experiment, including the ST model, ThyNet, RadImageNet, and ResNet50, all of which were trained by the transfer-learning method.4,19,29,30 Figure 2 shows the general training and test procedure. All of the deep-learning models employed a three-class design, and a one-hot method was used to label the training samples. As is shown in Figure 2A, the FTC, FV-PTC, and BN samples are labeled as “1, 0, 0”, “0, 1, 0”, and “0, 0, 1”, respectively. The labeled values “0” and “1” represent the probabilities for each category. Finally, the model outputs three predicted probabilities corresponding to the three categories.
Figure 2.
Graphical summary of the training and testing procedure
(A) The image ROI and one-hot labels; (B) A transformer-based AI model; (C) The test US images; (D) The output feature heat maps and prediction results. MLP: multi-layer perceptron, W-MSA: multi-head self-attention modules with window, SW-MSA multi-head self-attention modules with shifted window, LN: layer normalization.
The image training and testing flow of the ST model is shown in Figures 2B and 2C.The region of interest (ROI) of the nodule is extracted from the US images and normalized to 224 × 224 pixels. The details of image normalization are presented in Figure S1 of the supplementary information. As seen in Figure 2B, the model first performs downsampling on the input US image. This operation uses three scales, namely 4 times, 8 times, and 16 times. The downsampled image is then sent to the Transformer block after patch merging. These blocks learn image features in receptive fields of different scales through W-MSA and SW-MSA. Once the training is completed, the model will record the parameters and test the independent test set.
Results for independent test set 1
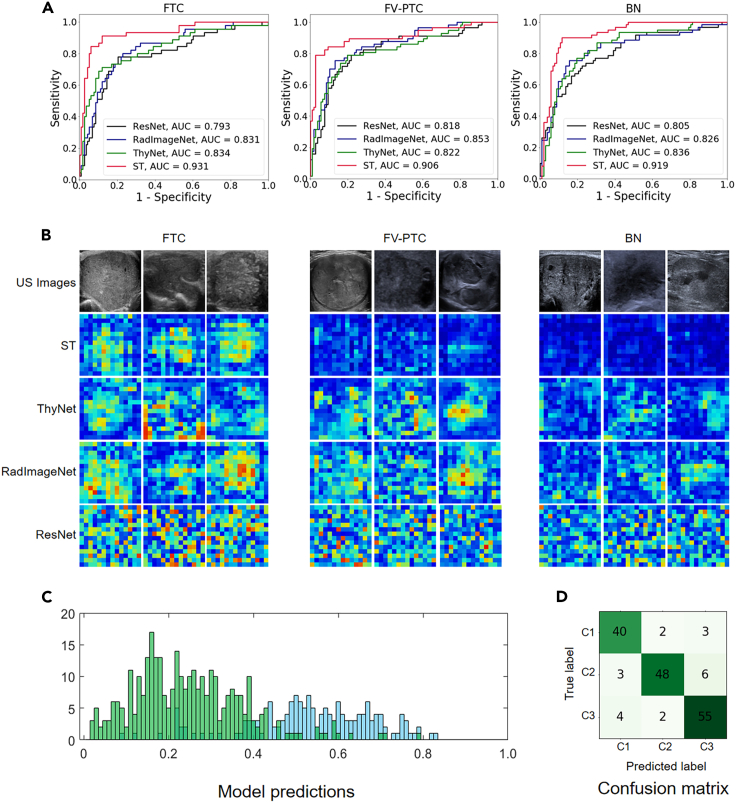
The receiver operating characteristic (ROC) curve of the identification results generated by the different AI models in independent test set 1 is shown in Figure 3. The test set included 167 cases in total, of which 45 were FTC, 57 were FV-PTC, and 61 were BN. As shown in Figure 3A, the new-generation ST model (red curves) achieved better performance than DCNN models, and its AUC value range was 0.906–0.931. In comparison, the AUC of the three deep-learning models was between 0.793 and 0.853.
Figure 3.
The test results of the independent test set 1
(A) The ROC curves of different AI models; (B) The original US images (row 1) and the output feature heat maps of the models (rows 2 to 5); (C) The distribution of the prediction values. The blue bars represent the output that should be predicted as a positive sample, the green bars represent the output that should be predicted as a negative sample, the horizontal axis represents probability, and the vertical axis represents the number of samples; (D) The confusion matrix and C1 to C3 represent FTC, FV-PTC, and BN, respectively.
Figure 3B shows the original US images together with the feature heat maps of the ST model; the first row is the original US images of FTC, FV-PTC, and BN while the second to fifth rows are the corresponding feature heat maps. In general, FTC with higher malignancy obtained significantly higher energy than the others. The heat maps of BNs had the lowest energy. The energy of FV-PTC with low malignancy was between the other two types. This pattern is more obvious in the ST model’s feature maps, which exhibit higher accuracy. The representation of the feature heat maps was found to be highly consistent with the conclusion of the malignant degree of follicular thyroid nodules.
The statistical histogram and confusion matrix are shown in Figures 3C and 3D, respectively. Figure 3C displays the statistical distribution of the predicted probability output by the model. The horizontal axis represents the predicted probability. The blue bars represent the distribution of samples that should be predicted as positive, while green represents the distribution of samples that should be predicted as negative. This means that ideally, the blue bars should all be concentrated around probability 1, while the green bars should all be concentrated around probability 0. It is evident that while the model falls short of the ideal scenario, samples with different labels have begun to cluster and become distinguishable. Table 2 compares the sensitivity and specificity indicators of several methods, among which the ST model achieved the best results. The sensitivity of the ST model for FTC was 88.9% (40/45, 95% CI: 75.2%-0.95.8%); the specificity was 94.1% (111/118, 95% CI: 87.7%–97.4%); and the AUC was 0.931 (p < 0.001). The sensitivity for FV-PTC was 84.2% (48/57, 95% CI: 71.6%–92.1%); the specificity was 96.2% (102/106, 95% CI: 90.1%–98.8%); and the AUC was 0.906 (p < 0.001). For FTC plus FV-PTC versus BN, the sensitivity was 91.2% (93/102, 95% CI: 83.5%–95.6%); the specificity was 90.2% (55/61, 95% CI: 79.1%–95.9%); and the AUC was 0.919 (p < 0.001). Other indicators, such as positive predictive value (PPV), negative predictive value (NPV), accuracy (ACC), and F1 score are shown in Table S2.
Table 2.
Identification results of different models in independent test 1
| Model | FTC vs. (FV-PTC + BN) |
FV-PTC vs. (FTC + BN) |
(FTC + FV-PTC) vs. BN |
|||
|---|---|---|---|---|---|---|
| SEN. | SPE. | SEN. | SPE. | SEN. | SPE. | |
| ResNet50 | 0.667 | 0.881 | 0.702 | 0.821 | 0.824 | 0.689 |
| RadImageNet | 0.689 | 0.873 | 0.754 | 0.840 | 0.873 | 0.721 |
| ThyNet | 0.733 | 0.890 | 0.719 | 0.858 | 0.843 | 0.738 |
| Swin-transformer | 0.889 | 0.941 | 0.842 | 0.962 | 0.912 | 0.902 |
Results for independent test set 2
Independent test set 2 comprised 178 cases, including 53 cases of FTC, 59 cases of FV-PTC, and 66 cases of BN. Figure 4 shows the test results for this dataset. As seen in Figure 4A, the AUC of the DCNN models was 0.784–0.846, while the ST-based method performed significantly better than the DCNNs, with the AUC between 0.917 and 0.945. Figure 4B displays the original US images together with the feature heat maps, where the first row includes the original US images of FTC, FV-PTC, and BN while the second to fifth rows give the corresponding feature heat maps. As may be seen, the characteristics were found to be very similar to those of independent test set 1. That is, the energy of FTC was significantly higher than that of the other two nodules, the heatmap of BNs resulted from significantly lower energy, and the energy of FV-PTC with lower malignancy was somewhere between the two. The highly consistent performance of the two test sets’ feature heat maps indicates that the model successfully learned the pattern of convolutional features of different follicular thyroid nodules.
Figure 4.
The test results of the independent test set 2
(A) The ROC curves of different AI models; (B) The original US images (row 1) and the output feature heat maps of the models (rows 2 to 5); (C) The distribution of the prediction values. The blue bars represent the output that should be predicted as a positive sample, the green bars represent the output that should be predicted as a negative sample, the horizontal axis represents probability, and the vertical axis represents the number of samples; (D) The confusion matrix and C1 to C3 represent FTC, FV-PTC, and BN, respectively.
The statistical distribution of the model’s predicted probability output is depicted in Figure 4C. The horizontal axis represents the predicted probability output by the model. Ideally, the predicted probabilities for these samples should be 0 and 1, respectively. It is evident that samples with different labels have started to cluster and become distinguishable. The confusion matrix of model recognition results is reported in Figure 4D. Table 3 shows the sensitivity and specificity of several AI models, among which the new-generation AI ST method achieved the best results. Specifically, its recognition sensitivity for FTC was 86.8% (45/53, 95% CI: 74.0–94.1%), its specificity was 92.0% (115/125, 95% CI: 85.4%–95.9%), and the AUC was 0.934 (p < 0.001). The recognition sensitivity for FV-PTC was 83.1% (49/59, 95% CI: 76.9%–86.3%), the specificity was 89.9% (107/119, 95% CI: 70.6%–91.1%), and the AUC was 0.914 (p < 0.001). For benign follicular nodules, the sensitivity was 83.3% (55/66, 95% CI: 71.7%–91.0%) and the specificity was 93.8% (105/112, 95% CI: 81.7%-91. Other indicators, such as PPV, NPV, ACC, and the F1 score are shown in Table S3.
Table 3.
Identification results of different models in independent test 2
| Model | FTC vs. (FV-PTC + BN) |
FV-PTC vs. (FTC + BN) |
(FTC + FV-PTC) vs. BN |
|||
|---|---|---|---|---|---|---|
| SEN. | SPE. | SEN. | SPE. | SEN. | SPE. | |
| ResNet50 | 0.717 | 0.864 | 0.763 | 0.882 | 0.875 | 0.758 |
| RadImageNet | 0.755 | 0.920 | 0.780 | 0.874 | 0.857 | 0.773 |
| ThyNet | 0.811 | 0.912 | 0.797 | 0.933 | 0.857 | 0.803 |
| Swin-transformer | 0.868 | 0.976 | 0.831 | 0.941 | 0.893 | 0.924 |
Results of the 10-fold cross-validation
To further validate the robustness and reliability of the model, we employed 10-fold cross-validation on the dataset. The multi-center data were evenly divided into 10 subsets, with each subset containing 167 cases. And the ratio of the test set to the training set was 2 : 8. In each round, we sequentially selected two subsets to form a test set, while the remaining eight subsets were used as training sets (see Figure S2 of the supplementary information for details). Therefore, the test set in each round of testing contained 334 cases, with 68 cases of FTC, 110 cases of FV-PTC, and 156 cases of BN. And the distribution of ROC curves from 10 rounds of independent testing are presented in Figure 5. The average sensitivity, specificity, and AUC values are reported in Table 4.
Figure 5.
The distribution of ROC curves of the 10-fold cross-validation where R1 to R10 represent the ten rounds of testing
Table 4.
Results of average AUC, sensitivity, and specificity in the 10-fold cross-validation
| Model | FTC vs. (FV-PTC + BN) |
FV-PTC vs. (FTC + BN) |
(FTC + FV-PTC) vs. BN |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC (SD) | SEN. (SD) | SPE. (SD) | AUC (SD) | SEN. (SD) | SPE. (SD) | AUC (SD) | SEN. (SD) | SPE. (SD) | |
| ResNet50 | 0.790 (0.015) | 0.684 (0.032) | 0.830 (0.013) | 0.783 (0.017) | 0.683 (0.031) | 0.819 (0.010) | 0.803 (0.014) | 0.865 (0.012) | 0.658 (0.022) |
| RadImageNet | 0.830 (0.012) | 0.713 (0.032) | 0.857 (0.014) | 0.816 (0.013) | 0.662 (0.035) | 0.834 (0.014) | 0.836 (0.010) | 0.854 (0.016) | 0.693 (0.025) |
| ThyNet | 0.838 (0.011) | 0.716 (0.030) | 0.859 (0.014) | 0.822 (0.010) | 0.735 (0.024) | 0.832 (0.012) | 0.840 (0.009) | 0.887 (0.014) | 0.699 (0.017) |
| Swin-transformer | 0.933 (0.015) | 0.860 (0.054) | 0.935 (0.009) | 0.920 (0.014) | 0.831 (0.021) | 0.938 (0.016) | 0.935 (0.010) | 0.927 (0.016) | 0.896 (0.021) |
As seen in Figure 5, although the ROC curves of each method exhibit fluctuations, they generally converge within a certain range. Among them, the ST method achieved the best performance in the 10-fold cross-validation, with AUC and standard deviation of 0.933 ± 0.015, 0.920 ± 0.014, and 0.935 ± 0.010 (see Table 4, row 4 of the data), respectively. The ThyNet and RadImageNet, which were used for medical image processing, performed lower than ST but better than the standard ResNet. Their average AUC, sensitivity, and specificity values are detailed in Table 4. In general, the results of independent test set 1 and 2 and the 10-fold cross-validation demonstrate a high level of consistency. This provides additional evidence for the AI models’ stability and robustness.
Discussion
Identifying the specific category of follicular thyroid nodules is a difficult challenge for accurately diagnosing thyroid cancer. FNAC can provide only the vague diagnosis of Bethesda category IV, which leaves its malignancy and prognosis clinically uncertain. Among all types of follicular thyroid nodules, FTC is the most malignant, and more aggressive treatment methods such as surgery are usually necessary.2,4 In contrast, the FV-PTC type can be treated relatively conservatively due to its low degree of malignancy, and for benign follicular nodules, an active surveillance strategy can be considered. Because different types of Bethesda IV thyroid nodules correspond to different prognoses and treatment strategies, it is crucial to develop a method that can accurately identify their specific category.
In this study, we retrospectively collected 7566 US images of Bethesda IV thyroid nodules from 2006 to 2022 from five hospitals. Then, we trained the AI models to identify specific categories of Bethesda IV thyroid nodules. To mitigate potential central effects, we evaluated our model using two independent test sets, resulting in a sensitivity of 89%–91% and specificity of 90%–92% for diagnosing malignant Bethesda IV thyroid nodules. Compared to genetics-based methods, AI not only offers non-invasiveness but also demonstrates superior specificity. Thus, our AI-based diagnostic model can be easily integrated into the existing diagnostic process to facilitate the auxiliary diagnosis of Bethesda IV follicular nodules. Specifically, when patients with thyroid nodules receive FNAC and the examination results are Bethesda category IV, the AI model can promptly be used to perform a further identification. If the AI model identifies the nodules as FTC, more aggressive treatment methods such as total thyroidectomy should be considered.3,31,32 If the AI model identifies them as low malignant FV-PTC or benign, additional examinations such as ThyroSep V3 or RNA-based classifier can be performed to further rule out the possibility of FTC.15,16,17 Physicians can combine the results of multimodal examinations to make a comprehensive judgment.
Although our method can be well integrated into the clinical diagnostic workflow, there are still challenges facing its practical clinical implementation. Firstly, combining US imaging with state-of-the-art deep-learning techniques for distinguishing Bethesda IV nodules represents a macro-level radiomics approach. Yet, technologies like ThyroSep V3 or RNA-based classifiers employ a micro-level genomics approach. Both approaches have made progress, but they still produce a certain proportion of false positive and false negative results. In actual clinical scenarios, where physicians need to perform comprehensive assessments of patients with Bethesda IV thyroid nodules, they may encounter a multitude of information from radiomics and genomics. Effectively integrating and analyzing this information poses a significant challenge. Currently, research on an auxiliary diagnosis of Bethesda IV thyroid nodules is still limited within individual omics domains. There is still no research that can construct a unified model from both macro and micro levels to thereby achieve a truly multimodal and cross-omics auxiliary diagnosis. In addition, the implementation of AI models in clinical practice raises ethical and legal concerns. Issues such as patient privacy, data security, and liability for diagnostic decisions need to be addressed to ensure the responsible and ethical use of AI technology. Addressing these challenges will be essential for the successful implementation and widespread adoption of an auxiliary diagnostic model for Bethesda IV thyroid nodules in clinical practice.
Limitations of the study
This study has some limitations. First, because our study included only Bethesda IV patients who underwent surgery and had surgical pathology (because we needed a gold standard to determine tumor characteristics), the proportion of malignancy was higher than expected. In the overall dataset, BN is 46.6% (787 cases), FV-PTC is 33.1% (559 cases), and the proportion of FTC is 20.3% (344 cases), which represents a certain deviation in the data proportions. This phenomenon occurred because, after an evaluation by physicians, some patients diagnosed with Bethesda IV thyroid nodules concluded that their risk of malignancy was low and chose a conservative treatment instead of surgery. This phenomenon is also commonly observed in the surgical treatment of other types of tumors. In the future, we plan to address this issue through prospective studies, which will not only increase the quantity of data but also provide a more balanced dataset. Second, as a branch of FV-PTC, the encapsulated follicular variant of papillary thyroid carcinoma (EFV-PTC) has been independently named, “non-invasive follicular thyroid neoplasm with papillary-like nuclear features” (NIFTP).33 It is generally believed that this tumor has a low degree of malignancy. However, we did not treat NIFTP as a separate category, because many hospitals identify only FV-PTC, and NIFTP has not been further subdivided. Establishing a prospective research queue and setting NIFTP as a separate sub-queue will help explore the recognition performance of AI for NIFTP categories. This is also one of the key issues for our prospective research. Third, in this retrospective study, we have recorded the sex information (see Table 1), but the gender information is missing. This is because the patients are not required to provide gender information in the studied multi-center hospital, but the registration information includes sex information. Generally speaking, factors such as sex, age, and race have been found to have certain correlations with the risk of thyroid tumors, although they are not determinative factors.2,6,7 Given that our study primarily focuses on US-based thyroid tumor diagnosis research, we did not incorporate these non-imaging factors as a primary focus in our model.
In addition, the black box feature of deep learning itself will also be the main challenge for clinical applications in the future. Although deep learning has achieved exciting diagnostic results in many diseases, physicians still prefer transparent, understandable, and interpretable diagnostic models to better gain the confidence of their supervisors and patients.34,35 In a future study, we expect to introduce explainable AI (XAI) and other technologies to improve the model’s interpretability.37 For example, we can use XAI and uother technologies to analyze the change rules of the feature map and thereby obtain a more transparent and interpretable model.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| Python 3.9 Python | Python | https://www.python.org |
| PyTorch 1.7.1 PyTorch | PyTorch | https://pytorch.org |
| SPSS 25.0 SPSS | SPSS | https://spss.en.softonic.com/ |
| Swin-Transformer | This paper | https://github.com/dumzj/ai_bethesdaiv |
| ThyNet | Peng et al.4 | https://github.com/sprint2200/ThyNet |
| RadImageNet | Mei et al.30 | https://github.com/BMEII-AI/RadImageNet |
| ResNet-50 | PyTorch | https://github.com/pytorch/vision/blob/main/torchvision/models/resnet.py |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Dong Xu (xudong@zjcc.org.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The Ultrasound data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at github and is publicly available as of the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
AI models
We experimented with four AI models for US image processing: ST model, ThyNet, RadImageNet and ResNet50. Among them, the ST model uses a transformer framework with multi-head self-attention and technology, while the other three use traditional DCNN structures. Compared with the DCNN, the ST model divides the input image into multiple windows of different scales and applies transformer modules on each scale. To improve the association between the receptive field and the surrounding area, it uses a shifted window mechanism on each scale, making the attention computation depend only on neighboring windows instead of the entire image. Additionally, the ST model uses a cyclic shift operation in each transformer module, rearranging the feature vectors within each window so that adjacent features can be covered by the next shifted window, enhancing the information flow and representation ability. The specific implementation of the ST model is listed in row 6 of the key resources table.
The ThyNet, RadImageNet, and ResNet50 utilize convolutional layers, pooling layers, activation functions, and fully connected layers for image processing. All of them incorporate skip connections to facilitate the flow of information across layers without degradation. Specifically, ThyNet adopts a hybrid structure that integrates sparsity, group convolution and feed-forward technologies into a unified model, thereby enhancing network authentication capabilities. The detailed implementation of the ThyNet model can be found in row 7 of the key resources table. The RadImageNet employs Inception-ResNet-v2, DenseNet121, InceptionV3 and other models as the backbone. It utilizes global average pooling and dropout techniques with a rate of 0.5 to centralize feature processing and enhance model robustness. On the other hand, the ResNet50 employs a residual structure with residual blocks that add the outputs of previous layers to the inputs of subsequent layers. The specific implementations of these three models are listed in rows 8 and 9 of the key resources table.
Participant details
This study involved human subjects, and the inclusion and exclusion criteria are shown in Figure 1. Initially, a total of 10746 US images from 1875 cases from May 2006 to April 2022 were collected from five hospitals. After screening by the exclusion criteria, 1690 qualified cases and 7566 US images remained. All participants were Asian descent yellow race individuals from China, among whom 1636 (96.8%) were of Han ethnicity. The remaining 54 (3.2%) invividuals belonged to ethnic minorities groups, with 52 being Zhuang ethnicity and 2 being Miao ethnicity. The study was reviewed and approved by the institutional review boards of the Medical Ethics Committee of Zhejiang Cancer Hospital (IRB-2020-287); Affiliated Hangzhou First People’s Hospital of Zhejiang University’s School of Medicine (IRB-2019-010-01); Sun Yat-sen University Cancer Center (IRB-B2021-021-02); Taizhou Cancer Hospital (IRB-2023-001) and Zhejiang Provincial People’s Hospital (IRB-KT2022-0140). The requirement of informed consent was waived due to the study’s retrospective nature.
Method details
Study design
First, we divided the collected Bethesda IV thyroid nodules into three categories, including FTC, FV-PTC, and BN. Then, we divided the training set and the test set by an overall ratio of approximately 8:2. Specifically, the training set comprised 1349 (79.9%) cases from centers 1 to 3; the independent testing set 1 was from center 4, which comprised 163 (9.6%) cases; and the independent testing set 2 was from center 5, which comprised 178 (10.5%) cases. We extracted the lesion’s ROI from the US images of the thyroid nodules and labeled them using one-hot encoding based on the final pathology results. Subsequently, we fed the ROI and corresponding labels to the AI models to perform a transfer-learning procedure. The overall training and testing process is shown in Figure 2.
All the AI models were pre-trained on ImageNet-1000. During transfer learning, we froze all layers except the output layer to retain the common features learned by the pre-training operation, updating only the output layer to accommodate new tasks. The loss function of all models was cross-entropy loss. For the optimization algorithm, a commonly used Adam method was selected, and the learning rate was set to 0.001. The data augmentation methods were also applied to expand the training set to enhance the robustness of the model. The specific methods included random scaling and brightening by 0.9-1.1 times, −10° to +10° random rotations, horizontal or vertical reversals, adding white Gaussian noise to the US images, and random clipping.
Data acquisition
During image collection, the patients were examined in the supine position with the neck straight. Both sides of the neck were fully exposed, and the thyroid gland was scanned in the transverse and longitudinal axes. Quality control for all data was ensured by a senior ultrasound radiologist. The ultrasound images of the patients were collected retrospectively using 26 different pieces of equipment manufactured by the General Electric Company, Philips, Esaote, Siemens, and Toshiba. Table S1 shows the detailed list.
Quantification and statistical analysis
ROC curves and AUC values were adopted to demonstrate the ability of AI models to identify Bethesda IV thyroid nodules. Based on the model’s prediction values, we have separately given ROC curves for each category and calculated the corresponding AUC values. In addition, the comprehensive performance of the networks was evaluated on the basis of sensitivity, specificity, positive predict value (PPV), negative predict value (NPV), accuracy (ACC), F1 score, P-value, and 95% CI. These indicators were calculated using SPSS 25.0 as well as Python-based matplotlib and sklearn packages. To better display the experimental results, the feature heat map, confusion matrix, and distribution of the predicted values were calculated, as shown in Figures 3 and 4.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 82071946), the Natural Science Foundation of Zhejiang Province (No. LZY21F030001), the Pioneer and Leading Goose R&D Program of Zhejiang (No. 2023C04039), the National Key Research and Development Program of China (2022YFF0608403), the Research Program of National Health Commission Capacity Building and Continuing Education Center (CSJRZC2021JJSJ001), the Research Program of Zhejiang Provincial Department of Health (No. 2021KY099, 2022KY110, 2023KY066 and 2019RC108), and the Qiantang Cross Fund of Institute of Basic Medicine and Cancer of Chinese Academy of Sciences.
Author contributions
Conceptualization: J.Y., Y.Z., J.S., J.Z., P.L., and D.X.; methodology: J.Y., Z.L., J.X., B.F., X.L., W.L., D.O., Y.L., and N.F.; writing – original draft: J.Y., Y.Z., and J.S.; writing – review & editing: J.X., M.Y., J.C., L.C., C.Y., L.W. and K.W.; sample collection: J.Y., Y.Z., J.S., Z.L., X.L., and M.Y. All authors reviewed the whole work and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108114.
Contributor Information
Jianhua Zhou, Email: zhoujh@sysucc.org.cn.
Ping Liang, Email: liangping301@126.com.
Dong Xu, Email: xudong@zjcc.org.cn.
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA. Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Rossi E.D., Baloch Z. The Impact of the 2022 WHO Classification of Thyroid Neoplasms on Everyday Practice of Cytopathology. Endocr. Pathol. 2023;34:23–33. doi: 10.1007/s12022-023-09756-2. [DOI] [PubMed] [Google Scholar]
- 3.Cibas E.S., Ali S.Z. The 2017 Bethesda system for reporting thyroid cytopathology. Thyroid. 2017;27:1341–1346. doi: 10.1089/thy.2017.0500. [DOI] [PubMed] [Google Scholar]
- 4.Peng S., Liu Y., Lv W., Liu L., Zhou Q., Yang H., Ren J., Liu G., Wang X., Zhang X., et al. Deep learning-based artificial intelligence model to assist thyroid nodule diagnosis and management: a multicentre diagnostic study. Lancet. Digit. Health. 2021;3:e250–e259. doi: 10.1016/S2589-7500(21)00041-8. [DOI] [PubMed] [Google Scholar]
- 5.Yu J., Deng Y., Liu T., Zhou J., Jia X., Xiao T., Zhou S., Li J., Guo Y., Wang Y., et al. Lymph node metastasis prediction of papillary thyroid carcinoma based on transfer learning radiomics. Nat. Commun. 2020;11:4807. doi: 10.1038/s41467-020-18497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christofer Juhlin C., Mete O., Baloch Z.W. The 2022 WHO classification of thyroid tumors: novel concepts in nomenclature and grading. Endocr. Relat. Cancer. 2023;30:e220293. doi: 10.1530/ERC-22-0293. [DOI] [PubMed] [Google Scholar]
- 7.Yip L., Gooding W.E., Nikitski A., Wald A.I., Carty S.E., Karslioglu-French E., Seethala R.R., Zandberg D.P., Ferris R.L., Nikiforova M.N., Nikiforov Y.E. Risk assessment for distant metastasis in differentiated thyroid cancer using molecular profiling: A matched case-control study. Cancer-Am Cancer Soc. 2021;127:1779–1787. doi: 10.1002/cncr.33421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baloch Z.W., Asa S.L., Barletta J.A., Ghossein R.A., Juhlin C.C., Jung C.K., LiVolsi V.A., Papotti M.G., Sobrinho-Simões M., Tallini G., Mete O. Overview of the 2022 WHO Classification of Thyroid Neoplasms. Endocr. Pathol. 2022;33:27–63. doi: 10.1007/s12022-022-09707-3. [DOI] [PubMed] [Google Scholar]
- 9.Kotani T., Asada Y., Aratake Y., Umeki K., Yamamoto I., Tokudome R., Hirai K., Kuma K., Konoe K., Araki Y. Diagnostic usefulness of dipeptidyl aminopeptidase IV monoclonal antibody in paraffin-embedded thyroid follicular tumours. J. Pathol. 1992;168:41–45. doi: 10.1002/path.1711680108. [DOI] [PubMed] [Google Scholar]
- 10.Haugen B.R., Alexander E.K., Bible K.C., Doherty G.M., Mandel S.J., Nikiforov Y.E., Pacini F., Randolph G.W., Sawka A.M., Schlumberger M., et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim P.H., Suh C.H., Baek J.H., Chung S.R., Choi Y.J., Lee J.H. Unnecessary thyroid nodule biopsy rates under four ultrasound risk stratification systems: a systematic review and meta-analysis. Eur. Radiol. 2021;31:2877–2885. doi: 10.1007/s00330-020-07384-6. [DOI] [PubMed] [Google Scholar]
- 12.Kuo T.C., Wu M.H., Chen K.Y., Hsieh M.S., Chen A., Chen C.N. Ultrasonographic features for differentiating follicular thyroid carcinoma and follicular adenoma. Asian J. Surg. 2020;43:339–346. doi: 10.1016/j.asjsur.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Maia F.F.R., Matos P.S., Pavin E.J., Vassallo J., Zantut-Wittmann D.E. Value of ultrasound and cytological classification system to predict the malignancy of thyroid nodules with indeterminate cytology. Endocr. Pathol. 2011;22:66–73. doi: 10.1007/s12022-011-9159-6. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S.Y., Shin J.H., Oh Y.L., Kim T.H., Lim Y., Choi J.S. Role of Ultrasound in Predicting Tumor Invasiveness in Follicular Variant of Papillary Thyroid Carcinoma. Thyroid. 2017;27:1177–1184. doi: 10.1089/thy.2016.0677. [DOI] [PubMed] [Google Scholar]
- 15.Patel K.N., Angell T.E., Babiarz J., Barth N.M., Blevins T., Duh Q.Y., Ghossein R.A., Harrell R.M., Huang J., Kennedy G.C., et al. Performance of a Genomic Sequencing Classifier for the Preoperative Diagnosis of Cytologically Indeterminate Thyroid Nodules. JAMA Surg. 2018;153:817–824. doi: 10.1001/jamasurg.2018.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L., Smola B., Lew M., Pang J., Cantley R., Pantanowitz L., Heider A., Jing X. Performance of Afirma genomic sequencing classifier vs gene expression classifier in Bethesda category III thyroid nodules: An institutional experience. Diagn. Cytopathol. 2021;49:921–927. doi: 10.1002/dc.24765. [DOI] [PubMed] [Google Scholar]
- 17.Skaugen J.M., Taneja C., Liu J.B., Wald A.I., Nikitski A.V., Chiosea S.I., Seethala R.R., Ohori N.P., Karslioglu-French E., Carty S.E., et al. Performance of a Multigene Genomic Classifier in Thyroid Nodules with Suspicious for Malignancy Cytology. Thyroid. 2022;32:1500–1508. doi: 10.1089/thy.2022.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steward D.L., Carty S.E., Sippel R.S., Yang S.P., Sosa J.A., Sipos J.A., Figge J.J., Mandel S., Haugen B.R., Burman K.D., et al. Performance of a Multigene Genomic Classifier in Thyroid Nodules With Indeterminate Cytology: A Prospective Blinded Multicenter Study. JAMA Oncol. 2019;5:204–212. doi: 10.1001/jamaoncol.2018.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Lin Y., Cao Y., Hu H., Wei Y., Zhang Z., Lin S., Guo B. Swin transformer: hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021;1:10012–10022. [Google Scholar]
- 20.Korngiebel D.M., Mooney S.D. Considering the possibilities and pitfalls of Generative Pre-trained Transformer 3 (GPT-3) in healthcare delivery. NPJ Digit. Med. 2021;4:93. doi: 10.1038/s41746-021-00464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiani L. MRI-based deep learning for TLE diagnosis. Nat. Rev. Neurol. 2023;19:197. doi: 10.1038/s41582-023-00798-y. [DOI] [PubMed] [Google Scholar]
- 22.Thieme A.H., Zheng Y., Machiraju G., Sadee C., Mittermaier M., Gertler M., Salinas J.L., Srinivasan K., Gyawali P., Carrillo-Perez F., et al. A deep-learning algorithm to classify skin lesions from mpox virus infection. Nat. Med. 2023;29:738–747. doi: 10.1038/s41591-023-02225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh A.H.W., Norn C., Kipnis Y., Tischer D., Pellock S.J., Evans D., Ma P., Lee G.R., Zhang J.Z., Anishchenko I., et al. De novo design of luciferases using deep learning. Nature. 2023;614:774–780. doi: 10.1038/s41586-023-05696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., Wu X., Lin D., Zhao L., Li M., Yun D., Lin Z., Pang J., Li L., Wu Y., et al. DeepFundus: A flow-cytometry-like image quality classifier for boosting the whole life cycle of medical artificial intelligence. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2022.100912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan C., Schoppe O., Parra-Damas A., Cai R., Todorov M.I., Gondi G., von Neubeck B., Böğürcü-Seidel N., Seidel S., Sleiman K., et al. Deep Learning Reveals Cancer Metastasis and Therapeutic Antibody Targeting in the Entire Body. Cell. 2019;179:1661–1676.e19. doi: 10.1016/j.cell.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazard T., Bataillon G., Naylor P., Popova T., Bidard F.C., Stoppa-Lyonnet D., Stern M.H., Decencière E., Walter T., Vincent-Salomon A. Deep learning identifies morphological patterns of homologous recombination deficiency in luminal breast cancers from whole slide images. Cell Rep. Med. 2022;3:100872. doi: 10.1016/j.xcrm.2022.100872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao J., Lei Z., Yue W., Feng B., Li W., Ou D., Feng N., Lu Y., Xu J., Chen W., et al. DeepThy-Net: A multimodal deep learning method for predicting cervical lymph node metastasis in papillary thyroid cancer. Adv. Intell. Syst. 2022;4 [Google Scholar]
- 28.Hong R., Liu W., DeLair D., Razavian N., Fenyö D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021;2:100400. doi: 10.1016/j.xcrm.2021.100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA. 2016. Deep Residual Learning for Image Recognition; pp. 770–778. ResNet: [Google Scholar]
- 30.Mei X., Liu Z., Robson P.M., Marinelli B., Huang M., Doshi A., Jacobi A., Cao C., Link K.E., Yang T., et al. RadImageNet: An Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning. Radiol. Artif. Intell. 2022;4 doi: 10.1148/ryai.210315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tessler F.N., Middleton W.D., Grant E.G., Hoang J.K., Berland L.L., Teefey S.A., Cronan J.J., Beland M.D., Desser T.S., Frates M.C., et al. ACR Thyroid Imaging, Reporting and Data System (TI-RADS): White Paper of the ACR TI-RADS Committee. J. Am. Coll. Radiol. 2017;14:587–595. doi: 10.1016/j.jacr.2017.01.046. [DOI] [PubMed] [Google Scholar]
- 32.Cibas E.S., Ali S.Z. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid. 2017;27:1341–1346. doi: 10.1089/thy.2017.0500. [DOI] [PubMed] [Google Scholar]
- 33.Nikiforov Y.E., Seethala R.R., Tallini G., Baloch Z.W., Basolo F., Thompson L.D.R., Barletta J.A., Wenig B.M., Al Ghuzlan A., Kakudo K., et al. Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol. 2016;2:1023–1029. doi: 10.1001/jamaoncol.2016.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauritsen S.M., Kristensen M., Olsen M.V., Larsen M.S., Lauritsen K.M., Jørgensen M.J., Lange J., Thiesson B. Explainable artificial intelligence model to predict acute critical illness from electronic health records. Nat. Commun. 2020;11:3852. doi: 10.1038/s41467-020-17431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatia S., He L. Machine-generated theories of human decision-making. Science. 2021;372:1150–1151. doi: 10.1126/science.abi7668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ultrasound data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at github and is publicly available as of the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.