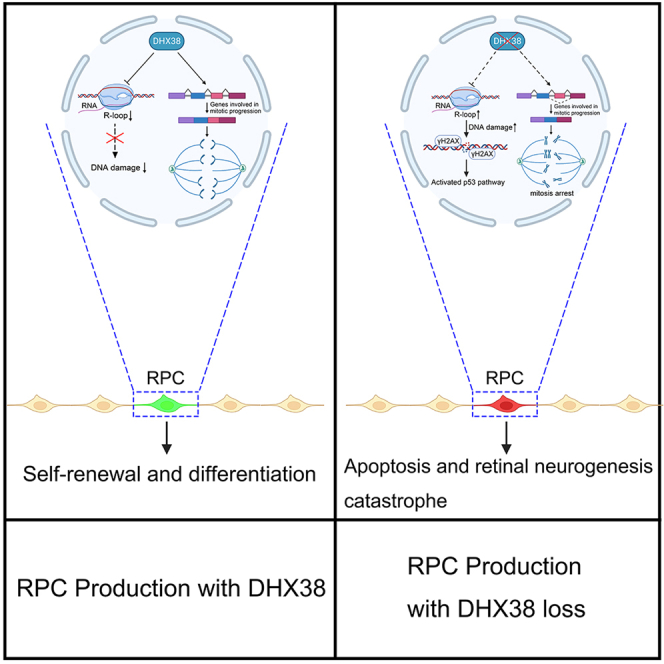
Summary
DEAH-Box Helicase 38 (DHX38) is a pre-mRNA splicing factor and also a disease-causing gene of autosomal recessive retinitis pigmentosa (arRP). The role of DHX38 in the development and maintenance of the retina remains largely unknown. In this study, by using the dhx38 knockout zebrafish model, we demonstrated that Dhx38 deficiency causes severe differentiation defects and apoptosis of retinal progenitor cells (RPCs) through disrupted mitosis and increased DNA damage. Furthermore, we found a significant accumulation of R-loops in the dhx38-deficient RPCs and human cell lines. Finally, we found that DNA replication stress is the prerequisite for R-loop-induced DNA damage in the DHX38 knockdown cells. Taken together, our study demonstrates a necessary role of DHX38 in the development of retina and reveals a DHX38/R-loop/replication stress/DNA damage regulatory axis that is relatively independent of the known functions of DHX38 in mitosis control.
Subject areas: Molecular physiology, Developmental genetics
Graphical abstract
Highlights
-
•
Dhx38 is essential for RPCs survival and retinal development in zebrafish
-
•
Dhx38 deletion triggers R-loop-dependent genome instability in vivo and in vitro
-
•
Replication stress caused by R-loop accumulation is the main cause of DNA damage
Molecular physiology; Developmental genetics
Introduction
The vertebrate retina is an integrated, extensive, and highly ordered collection of multiple cell types. Retinal neurons and glia are generated in a conserved order across all vertebrates during development. In zebrafish, ganglion cells generated first, followed by the production of cone photoreceptors, horizontal cells, and most of the amacrine neurons, and rods, bipolar cells and glia produced last, and this birth order is similar in mice.1,2,3 Retinal progenitor cells (RPCs) are the origin of all retinal neurons and glia. A precise spatiotemporal regulation of the survival, proliferation, and differentiation of RPCs is critical for retinal development.
Notably, retinal tissue has unexpectedly high levels of the expression of splicing factors, and mutations in eight splicing factors (PRPF3, PRPF4, PRPF6, PRPF8, PRPF31, SNRNP200, RP9, and DHX38) have been associated with retinitis pigmentosa (RP), a common subtype of retinal degeneration in humans.4,5 Deletion of the splicing factor prpf31 in zebrafish causes severe retinal developmental defects characterized by abnormal differentiation of RPCs,6 suggesting the important roles of splicing factors in retinal development and maintenance. However, it remains unknown whether other RP-related splicing factors play a role in retinal development. Therefore, the specific function of RP-related splicing factors in the retina and their role in retinal development merit further study.
DHX38 (DEAH-box Helicase 38) is a component of the spliceosome and is essential for the catalytic step II in the pre-mRNA splicing process.7,8 Mutations in DHX38 can cause autosomal recessive RP (arRP), and the role of DHX38 in the retinal development and maintenance remains unknown.9,10 Our previous study has reported that DHX38 indirectly regulates mitosis by modulating the alternative splicing of cell cycle-related genes in zebrafish hematopoietic tissues.11 Additionally, in vitro study has shown that DHX38 participates in chromosome segregation during mitosis in human cells independent of its splicing function.12 However, in this regard, the physiological function of DHX38 in retinal tissue remains unclear.
R-loop structures comprise a complementary RNA/DNA hybrid and a displaced single-stranded DNA (ssDNA) often formed during the coordinated transcription.13 Although essential for several biological processes, increase of R-loops can also induce DNA damage and genomic instability.14,15 Several studies have shown that mutations in splicing factors can increase R-loops and induce genomic instability.16,17 For example, SLU7, a protein involved in catalytic step II during pre-mRNA splicing process, can suppress the unscheduled R-loops formation and maintain genomic integrity in vitro.7,18 Besides, ATP-dependent RNA helicases, such as DDX19, DDX23, DDX1, and DDX9, have also been reported to induce R-loop accumulation when deleted.19,20,21,22,23,24,25 Although DHX38 has been reported to interact with RNA/DNA hybrids in vitro,26 whether DHX38 is involved in the R-loop regulation and genome stability maintenance, especially in the retina, remains unclear.
Here, we showed that dhx38 depletion in zebrafish caused severe defects in retinal development. Loss of dhx38 triggers the R-loop-dependent DNA damage, which ultimately results in p53-mediated apoptosis of RPCs. Similarly, results were observed in human cells deleted for DHX38, suggesting that the molecular mechanism of DHX38 is probably highly conserved in human cells and zebrafish. Furthermore, we found that DNA replication stress is the main cause of R-loop induced DNA damage in DHX38-deficient cells. Altogether, our study, for the first time, revealed the necessary roles of DHX38 in regulating R-loop homeostasis and its role in retinal development.
Results
Dhx38 mutant zebrafish exhibits severe retinal developmental defects
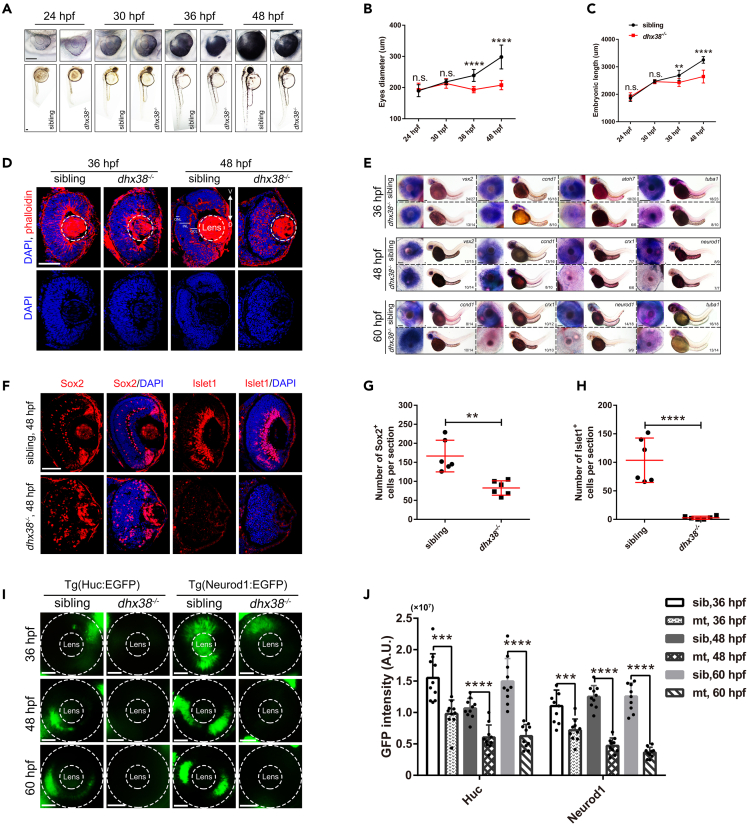
To examine the temporal and spatial expression of Dhx38 in zebrafish, we first performed whole-mount in situ hybridization (WISH) on zebrafish embryos at different developmental stages. Across the fish body, we found that the expression pattern of Dhx38 was abundant in the central nervous system, especially in the eyes, suggesting that Dhx38 may play a critical role in eye development (Figure S1A). Next, to determine whether dhx38 deletion has an impact on the development of the zebrafish retina, we explored this question with a zebrafish dhx38 loss-of-function mutant.11 We found that dhx38 homozygous mutants showed pronounced developmental defects at 36 h postfertilization (hpf) and 48 hpf, including a smaller head, abnormal eye development, and a curled-tail (Figures 1A–1C), and died at 3–4 days post fertilization (dpf). The detailed retinal phenotypes of dhx38 knockout zebrafish were observed using immunofluorescence and WISH assays with the corresponding antibodies and probes. Compared with the normal retinal layers of siblings, dhx38 mutants exhibited disordered cell arrangement and condensed nuclear morphology at 48 hpf (Figure 1D). The RPC markers (vxs2 and ccnd1) were expressed normally at 36 hpf, but up-regulated significantly at 48 hpf and 60 hpf in the dhx38 mutants (Figure 1E). Contrastly, the markers of neuronal precursors (atoh7, and crx1) and mature neurons (tuba1, and neurod1) were all down-regulated in the dhx38 mutants at 36–60 hpf, compared with the siblings (Figure 1E). To further confirm these results, an immunofluorescence assay was used to examine the expression patterns of Sox2 (RPCs marker), Zpr1 and Zpr3 (photoreceptor cell markers), Islet1 (inner retinal cells markers), and Gfap (glial cells markers) in the dhx38 mutants. Similarly, the irregular distribution of RPCs marker (Sox2) and the dramatically reduced expression of markers in mature retinal cells suggested that RPC differentiation into mature retinal cells may be interrupted in the dhx38 mutants (Figures 1F–1H and S1B). Using the Tg(Neurod1:EGFP) and Tg(Huc:EGFP) transgenic zebrafish lines, in which the specialized and post-mitotic neurons were labeled with EGFP,6 it was also found that the numbers of EGFP positive cells decreased significantly in the dhx38 mutants (Figures 1I and 1J). Altogether, our results indicate that RPC differentiation in the dhx38 mutant zebrafish was severely impaired compared with siblings during retinal neurogenesis.
Figure 1.
Dhx38 deletion impaired retinal morphology and RPC differentiation
(A) The morphology of bodies and eyes in siblings and dhx38−/− embryos at 24, 30, 36, and 48 hpf. Scale bar, 100 μm.
(B, C) Quantification of the embryonic length and eye size in siblings and dhx38−/− embryos shown in A. n = 10 for each panel. Data was shown as mean ± SD. n.s., no significance; ∗∗p < 0.01, ∗∗∗∗p < 0.0001 as indicated.
(D) Retinal sections of siblings and dhx38 mutants were stained with phalloidin and DAPI at 36 and 48 hpf. V, ventral side, D, dorsal side. n = 9 for each panel. Scale bar, 50 μm.
(E) Whole-mount in situ hybridization for RPCs marker (ccnd1 and vsx2), for neural precursors (atoh7 and crx1), and for specialized neurons (neurod1) and mature neurons (tuba1) at 36, 48 and 60 hpf. Scale bar, 100 μm.
(F) Retinal sections of siblings and dhx38−/− embryos were immunostained using Sox2 (a marker for RPCs), and Islet1 (a marker for neuron cells) antibodies at 48 hpf.
(G and H) Quantification of the Sox2+ and Islet1+ cells in the retinas of siblings and dhx38−/− embryos shown in F. n = 6 per panel. Scale bar, 50 μm. Data was shown as mean ± SD. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 as indicated.
(I) Distribution of Neurod1:EGFP (specialized neurons) and Huc:EGFP (post mitotic neurons) labeled cells in the whole retina of siblings and dhx38−/− transgenic zebrafish at 36, 48 and 60 hpf. The dashed circles indicate the eye and lens, respectively. Scale bar, 50 μm.
(J) Quantification of the GFP intensity in the eyes of siblings and dhx38−/− embryos at different stages shown in I. n = 10 per panel. Data was shown as mean ± SD. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 as indicated.
Dhx38 deficiency causes severe apoptosis and abnormal mitosis of RPCs in zebrafish
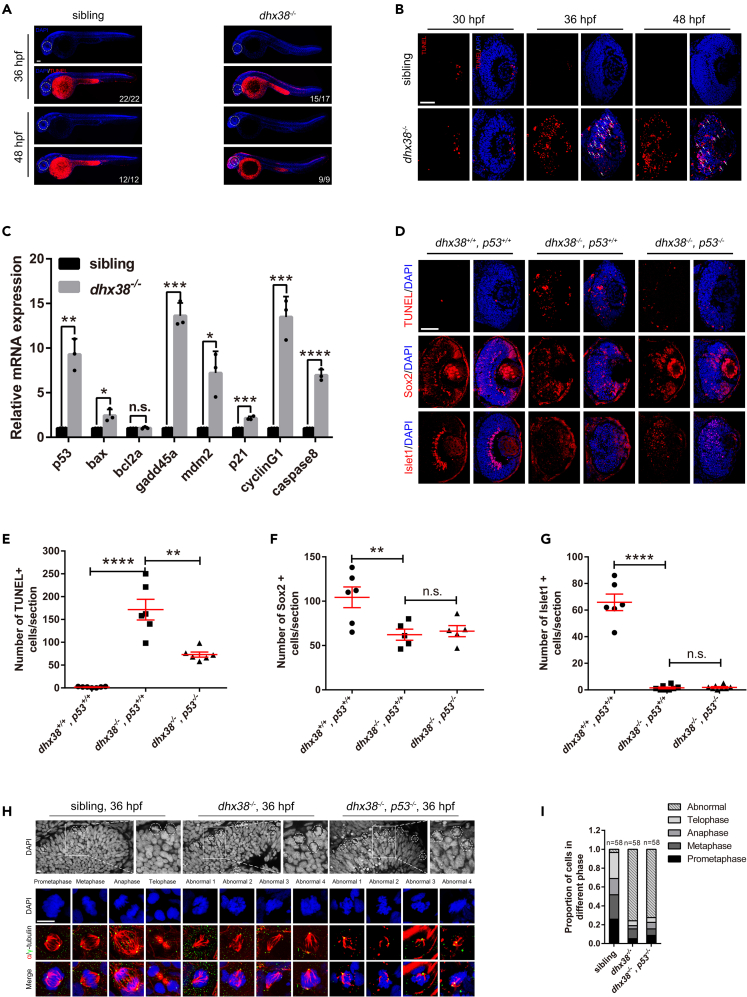
To further investigate the cause of retinal developmental defects in dhx38 mutant zebrafish, we first performed TUNEL assay to detect whether the dhx38 knockout zebrafish retinas were undergoing apoptosis. We observed extensive apoptotic signals in RPCs of the dhx38 knockout zebrafish retinas at different stages, and this apoptosis was partially p53-dependent (Figures 2A–2E, S2A, and S2D). Next, co-staining of TUNEL+ and retinal cell-type-specific markers showed that almost all TUNEL positive cells were Sox2 positive (Figures S2B and S2C), suggesting that the apoptotic cells were RPCs. However, we next found that the differentiation defects of RPCs were not alleviated by p53 deletion (Figures 2D, 2F and 2G), suggesting that the activation of p53 pathway is the direct cause of RPCs’ apoptosis, but is likely not the primary cause of the RPCs differentiation defects in the dhx38 mutants.
Figure 2.
Dhx38 deletion leads to p53-dependent apoptosis and abnormal mitosis in zebrafish retinas
(A) Whole embryo TUNEL staining showed many apoptotic cells in the retina, brain and spinal cord of dhx38 mutants at 36 and 48 hpf. White arrows, apoptotic signals. Scale bar, 100 μm.
(B) Retinal sections showed many apoptotic signals at 30, 36 and 48 hpf in dhx38 mutant zebrafish retinas White arrows, apoptotic RPCs. n = 9 per panel; Scale bar, 50 μm.
(C) qPCR showed activation of the p53 pathway in the dhx38 mutant at 36 hpf. Data was shown as mean ± SD. n.s., no significance; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 as indicated.
(D) p53 deletion in dhx38 mutants significantly reduces apoptosis of RPCs, but could not rescue retinal differentiation defects. 36 hpf for TUNEL; 48 hpf for Sox2 and Islet1. Scale bar, 50 μm.
(E‒G) The quantitative analysis of TUNEL-positive cells, Sox2-positive cells and Islet1-positive cells is shown in D. n ≥ 5 per panel. Data was shown as mean ± SD. n.s., no significance; ∗∗p < 0.01, ∗∗∗∗p < 0.0001 as indicated.
(H) The spindle and nuclei of RPCs in siblings, dhx38−/− and dhx38−/−/p53−/− zebrafish were stained using anti-α-tubulin (red) and γ-tubulin (green) antibodies and DAPI (blue), respectively. The different types of spindle abnormalities are shown in the panels (abnormal 1–4). Hexagon, mitotic karyotype. Scale bar, 10 μm.
(I) Quantitative analysis of the number of RPCs at each stage of mitosis in siblings and dhx38 mutant embryos shown in (H).
The accumulation of undifferentiated RPCs in dhx38 mutant zebrafish suggested that RPCs were either hyper-proliferative or arrested in the cell cycle. We then examined the cell cycle status in sibling and dhx38 mutant zebrafish. As expected, we found that dhx38 mutant RPCs had abnormal cell cycle and were arrested at mitotic phase using immunostaining labeling of RPCs with EdU, pH3 and alpha/gamma-tubulin antibodies (Figures 2H, 2I, S2E, and S2F). This result was further reinforced by the observation of DAPI staining under a high-resolution microscope, where many abnormal nuclear structures resembling chromosome nondisjunction during mitosis27 could be observed in the dhx38 mutant retinas at 36 hpf (Figure 2H). Next, we also found that the mitotic defect phenotype could not be rescued by p53 deletion (Figures 2H and 2I), suggesting that the elevated p53 levels are unlikely to be involved in the mitotic defects caused by dhx38 knockout. Furthermore, the pH3 positive cells partially overlapped with the TUNEL signals in the dhx38 mutants at both 36 hpf and 48 hpf using the co-staining assay (Figures S2G and S2H), suggesting that the cell cycle arrested RPCs eventually undergo apoptosis. Based on these results, we speculated that such an abnormal chromosome segregation process and mitotic arrest possibly causes the differentiation defect of RPCs in the dhx38 mutants.
Accumulation of R-loops is the main cause of DNA damage and subsequent apoptosis of RPCs in dhx38 mutants
Mutations in many splicing factors can lead to the accumulation of DNA damage in vivo and in vitro.28,29,30,31 We firstly detected DNA damage in the dhx38 mutants by immunostaining using the γH2AX antibody, which labels the DNA double-strand breaks. As expected, the percentages of γH2AX-labeled cells in the dhx38 mutant retinas (averagely 25%) were apparently higher than those in the siblings at 36 hpf (Figures 3A and 3B, white arrows). Meanwhile, the protein levels of γH2AX were also significantly up-regulated in the dhx38 mutant zebrafish (Figures 3C and 3D). In addition, we performed alkaline comet assay to measure the DNA damage levels in individual cells.32 The significant increase in tail moment (Figures 3E and 3F) confirmed that there were more DNA breaks in the dhx38 mutants.
Figure 3.
Dhx38 constraint of R-loop levels is critical for RPCs homeostasis
(A) Immunofluorescence analysis of dhx38−/− and sibling retinas at 30 hpf and 36 hpf. Scale bar, 50 μm.
(B) Quantitative analysis of the percentage of γH2AX-positive cells in each condition is shown in A. n = 8 per panel. Data was shown as mean ± SD. ∗p < 0.05, ∗∗∗∗p < 0.0001 as indicated.
(C and D) The protein levels of γH2AX in siblings and dhx38 mutants at different time points were detected using Western blotting. GAPDH was used to normalize protein loading. The red arrows indicated the corresponding protein bands. Data was shown as mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001 as indicated.
(E) Alkaline comet assay showed increased DNA damage in dhx38 mutants at 36 hpf and 48 hpf. White arrows, DNA with single or double-strand breaks. Scale bar, 100 μm.
(F) Quantitative analysis of the DNA damaged cells shown in D. At least 100 cells from 15 embryos were quantified per group. Data was shown as mean ± SD. ∗∗∗∗p < 0.0001 as indicated.
(G) Schematic of the RNH1 overexpression experiments.
(H and J) Confocal images showing the immunofluorescence of R-loops (H) and γH2AX (J) levels in cells isolated from siblings and dhx38 mutants that were either (hsp70:M27RNASEH1-mCherry) negative (left) or (hsp70:M27RNASEH1-mCherry) positive (right). Scale bar, 10 μm.
(I and K) Quantification of R-loops (I) and γH2AX (K) levels in 15 embryos with at least 100 cells per group. Data was shown as mean ± SD. ∗∗∗∗p < 0.0001 as indicated.
R-loops are considered as a source of DNA damage that triggers genomic instability.33 As DHX38 can bind to RNA/DNA hybrids,26 the basic structure of R-loops in vitro, we speculated that DHX38 might play a role in preventing the formation of R-loops. To verify this hypothesis, we performed single-cell immunofluorescence using the S9.6 antibody34 to detect R-loop levels in zebrafish. We found that R-loop level was significantly increased in the dhx38 mutants compared with the siblings (Figure S3A). Next, to determine whether DNA damage was induced by the elevated R-loops, rescue experiments were performed by over-expressing RNase H (RNH1), which can degrade R-loops in cells, in the dhx38 mutants (Figure 3G). We found that R-loop and DNA damage levels were significantly reduced when RNH1 was overexpressed (Figures 3H–3K), suggesting that DNA damage depends on the accumulation of R-loops in the dhx38 mutants. More importantly, the apoptosis of RPCs was also significantly rescued by the overexpression of RNH1 (Figures S3C and S3D), suggesting that R-loop-dependent DNA damage is the main cause of RPC apoptosis in the dhx38 mutant zebrafish. However, we found that overexpression of RNH1 could not rescue the mitotic defects of RPCs (Figures S3G‒S3I), suggesting that the unscheduled R-loops accumulation is likely not involved in the mitotic defects caused by dhx38 knockout. Importantly, we next found that these retinal phenotypes could be fully rescued by overexpressing wild-type DHX38 (dhx38-WT), rather than a helicase-dead DHX38 mutant (dhx38-ΔH) (Figures S3J‒S3M), suggesting that the retinal developmental defects observed in dhx38 mutant zebrafish are indeed caused by dhx38 deletion, and the helicase activity of DHX38 is essential for the function of DHX38 in R-loop formation and DNA damage.
Conserved roles of DHX38 in R-loop accumulation, DNA damage and genomic instability in human cells
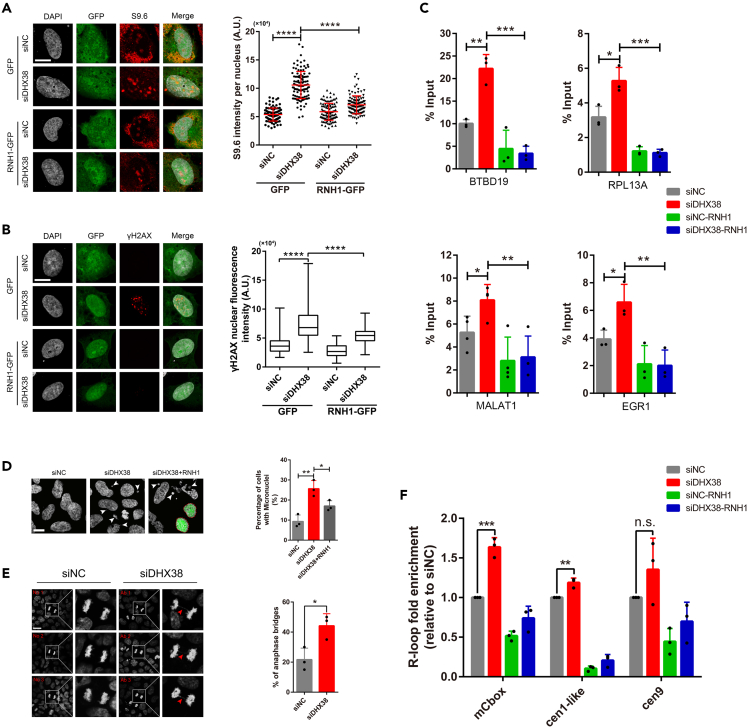
The DHX38 sequence is highly conserved between humans and zebrafish. The human DHX38 shows 80% overall identity and 89% homology to the zebrafish Dhx38. To verify if their functions are also evolutionary conserved, we knocked down the DHX38 expression using small interfering RNAs (siRNAs) in cultured human cell lines HEK293 and ARPE-19 (Figures S4A–S4D). Then, we discovered that the R-loop and DNA damage levels were increased significantly in HEK293 and ARPE-19 cells after interference with DHX38, which could be eliminated by RNase H1 overexpression (Figures 4A, 4B, and S4E–S4H). The alkaline comet assay also revealed an accumulation of DNA breaks in the siDHX38 cells (Figure S4I). Therefore, it was consider that DHX38 is required to maintain genomic integrity in human cells, primarily by preventing DNA breaks caused by R-loop accumulation. Next, we performed DRIP-qPCR using the S9.6 antibody to quantify R-loop accumulation at various actively transcribed genes (BTBD19, RPL13A, MALAT1 and EGR1) previously reported to have a propensity to form these structures.35 DHX38 knockdown induced R-loop accumulation at all four analyzed gene loci compared with the control, which were sensitive to RNase H1 treatment (Figure 4C). These results suggest that DHX38-depleted cells showed an accumulation of R-loops.
Figure 4.
DHX38 depletion led to R-loop-dependent genome instability in human cells
(A) Representative images and quantification of S9.6 staining per nucleus in ARPE-19 cells treated with siNC or siDHX38. Cells were transfected with either a control (GFP) or a vector expressing RNH1-GFP. At least 100 cells were calculated from three independent experiments. Scale bar, 10 μm. Data was shown as mean ± SD. ∗∗∗∗p < 0.0001 as indicated.
(B) Immunofluorescence of γH2AX in ARPE-19 cells after siNC and siDHX38 transfection with or without RNH1-GFP overexpression. At least 100 cells were calculated from three independent experiments. Scale bar, 10 μm. Data was shown as mean ± SD. ∗∗∗∗p < 0.0001 as indicated.
(C) DRIP-qPCR using the S9.6 antibody for the genes BTBD19, RPL13A, MALAT1 and EGR1 in HEK293 cells after being transfected with the indicated siRNAs. The relative abundance of RNA: DNA hybrids immunoprecipitated was represented as a percentage of the input material. Data was shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as indicated.
(D) Representative images of DHX38-depleted HEK293 cells showing micronuclei (white arrows). Dotted line, cells transfected with RNH1-GFP. Scale bar, 10 μm. Data was shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01 as indicated.
(E) Anaphase bridges (red arrows) in siNC and siDHX38 transfected HEK293 cells. Microphotographs showing anaphase bridges in DHX38-depleted cells. The percentages of anaphase cells with anaphase bridges were plotted. No, normal; Ab, anaphase bridges. Scale bar, 20 μm. Data was shown as mean ± SD. ∗p < 0.05 as indicated.
(F) Detected R-loop levels using DRIP-qPCR assay on specific centromeric α-SAT arrays (mCbox, cen1-like, and cen9) in siNC and siDHX38 cells. Graph shows the R-loop fold enrichment normalized to control (siNC) conditions. Data was shown as mean ± SD. n.s., no significance; ∗∗p < 0.01, ∗∗∗p < 0.001 as indicated.
Furthermore, the siDHX38 HEK293 cells exhibited an increased frequency of genomic aberrations, including anaphase bridges and micronuclei (MN), and the increased MN events could be partially abolished by RNase H1 overexpression (Figures 4D and 4E). Chromosome fragments lacking functional centromeres could form MN and R-loop plays a role in maintaining homeostasis of centromeric function.36,37 To verify whether DHX38 deficiency altered R-loop homeostasis in the centromeric region, we performed DRIP-qPCR assay on specific centromeric α-SAT arrays as reported previously.37 Interestingly, we found that DHX38 deletion increased R-loop levels on two α-SAT repeats (mCbox and cen1-like) when compared to control cells (Figure 4F). These results suggested that MN formation in DHX38-deficient cells is associated with altered R-loop homeostasis in the centromere region. In summary, our results suggest that the DHX38 function is well conserved between humans and zebrafish, and the DHX38 deficiency causes R-loop-dependent DNA damage and genomic instability.
DNA replication stress mediates the DNA damage caused by R-loop accumulation in DHX38-deficient cells
The formation of R-loops is an important cause of replication fork stalling, resulting in ATR-dependent replication stress and DNA damage.38 To test whether R-loop accumulation causes DNA damage through the induction of replication fork collapse in DHX38-knockdown cells, we performed FACS analysis and found that the siDHX38 cells showed an increased proportion of cells in the S-phase (Figure 5A). The EdU and pH3 staining showed a significantly reduced number of actively replicating cells and a modest increase in the number of G2/M-phase cells in the siDHX38 cells (Figures S5A–S5D).
Figure 5.
DHX38 inhibition slowed down ongoing DNA synthesis and caused cell-cycle arrest
(A) FACS analysis of PI (propidium iodide) staining in HEK293 cells treated with either control siNC or siDHX38. N = 4 separate experiments. Data was shown as mean ± SD. n.s., no significance; ∗∗p < 0.01 as indicated.
(B and C) The experimental scheme of DNA fiber assay (left) and representative DNA fibers from the identified conditions (middle), quantified by CldU track length (right). For each condition the lengths of the CIdU tracks of n ≥ 100 fibers were quantified. Scale bar, 3 μm. Data was shown as mean ± SD. ∗∗∗∗p < 0.0001 as indicated.
(D and E) Left: Western blotting of total HEK293 cell extracts upon asynchronously growing (D) or serum starvation (E) after DHX38 knockdown. Cell cycle distribution was verified using FACS analysis (right panel).
(F and G) Immunofluorescence images of γH2AX fluorescence in asynchronously growing or serum starvation HEK293 cells after DHX38 knockdown. Quantification of at least 100 cells from three independent experiments. Scale bar, 30 μm. Data was shown as mean ± SD. ∗∗∗∗p < 0.0001 as indicated.
Then, we performed DNA fiber stretching experiments to directly measure the dynamics of DNA replication.39 Cells were labeled with the IdU (red) to identify newly replicated DNA regions and then with the CIdU (green) to visualize the progression of replication forks during the second labeling (Figure 5B). We found that DHX38 knockdown significantly reduced DNA synthesis (Figure 5B). Importantly, the replisome impairment was partially rescued by RNase H1 overexpression (Figure 5C), suggesting that it is R-loops dependent. These results demonstrate that the DHX38-deficient cells have reduced DNA synthesis and increased replication stress due to the R-loop formation.
To further understand the role of DNA replication stress in generating DNA damage, we synchronized the cells in the G0 phase via serum starvation to minimize DNA replication. Under this condition, DHX38 knockdown could no longer induce DNA damage (Figures 5D–5G). Next, pulse labeling of cells with the uridine analog 5-ethyluridine (EU), which was incorporated into newly synthesized RNA,40 revealed no significant difference in the transcriptional activity between the siDHX38 and siNC cells (Figures S5E and S5F), indicating that the transcription rate was not affected by DHX38 knockdown. Altogether, these results suggest that the R-loop-mediated replication stress is the direct cause of DNA damage observed in the DHX38-knockdown cells.
Discussion
In this study, we identify Dhx38 as a key regulator of RPCs homeostasis and retinal development. Deletion of Dhx38 in zebrafish causes excessive R-loop accumulation, DNA replication stress and DNA damage, triggering the p53 signaling pathway and ultimately causing apoptosis of RPCs. Overall, we reveal the important roles of DHX38 and the involved pathways in the survival and differentiation of RPCs, and the physiological function of Dhx38 in the retina and its role in retinal development and maintenance.
Deficiency of certain splicing factors could induce DNA damage and various stages of mitotic defects.41,42 Our previous study reported that deletion Dhx38 in zebrafish hematopoietic tissues results in DNA damage and mitotic defects due to the abnormal alternative splicing of specific genes.11 Here, we found that DNA damage caused by DHX38 deletion in human cells and zebrafish RPCs is mainly due to the replication stress caused by R-loop accumulation. Interestingly, we also found that the abnormal R-loop accumulation was not involved in mitotic defects, suggesting that there may be other mechanisms regulating mitosis. Here, to identify whether alternative splicing of mitosis-related genes in retinal tissue is similarly affected by dhx38 deletion, we performed RT-PCR assay using the retinal tissue. We found that alternative splicing of mitosis-associated genes was also significantly affected in retinal tissue, and was more evident when compared to hematopoietic tissues (Figure S6), suggesting that the mitotic defects of RPCs may be caused by abnormal alternative splicing of genes involved in mitosis. However, studies from other groups have reported that DHX38 directly regulates chromosome segregation by affecting the function of Aurora B.12 Overall, we conclude that whether DHX38 regulates mitosis indirectly through pre-mRNA splicing or directly interacts with specific proteins is the main issue to be addressed in this field.
Although we observed DNA damage and p53-dependent apoptosis in both retinal and hematopoietic tissues, we found that the phenotype of the DNA damage levels, p53 pathway and apoptosis was more evident in retinal tissues compared to hematopoietic tissues. Indeed, we found that retinal tissues show obvious DNA damage levels, apoptosis and increased p53 protein levels at 30, 36, and 48 hpf (Figures 2B, 3A–3F, and S2D). At that time point, other tissues (such as inner ear cells, hematopoietic progenitor cells) are not or only mildly affected as detected by immunostaining (data not shown here), suggesting that the retinal tissue is more sensitive to the insufficiency of Dhx38 than other tissues. Importantly, at the molecular level, the splicing defects of pre-mRNAs are more striking in the retinal tissues than hematopoietic tissues (Figures S6F‒S6H, see PSI value, e.g., mis18a, scrib, cep131).11 This result is consistent with previous studies on Prpf31, another pathogenic gene of RP.6,43 These results suggest that retinal tissues may have a higher requirement for Dhx38 compared to other tissues. As a conclusion, we emphasize that retinal tissue is the first and most severely affected during the decreasing of Dhx38 levels with age at the early embryonic stage.
In the dhx38 knockout zebrafish, some of the RPCs are apoptotic, while others are arrested in the mitotic phase, characterized by the abnormal structures of spindles and defects in chromosome segregation. Meanwhile, in the DHX38 knockdown human cells, the cell cycle is arrested in the S-phase, probably due to the reduced DNA replication speed and elevated DNA damage level. The different DHX38 protein levels and cellular states in the RPCs and cultured cells may be responsible for these phenotypic differences. For example, the maternal mRNAs and proteins in zebrafish embryos will decrease rapidly with time until complete evaporation during embryonic development.44 At earlier stages, the maternal Dhx38 protein may support the RPCs to enter the mitotic phase in the dhx38 knockout zebrafish. However, at later stages, the remaining Dhx38 protein may be insufficient for the normal splicing of spindle assembly related genes, causing the arrest of cell division. Conversely, RNA-interfering of DHX38 in human cells would cause a rapid decline of the protein level, rather than the complete elimination of DHX38. These methodological differences may cause similar but not exactly the identical phenotypes in the in vivo and in vitro models.
RP is a cluster of inherited retinal dystrophies primarily characterized by progressive loss of photoreceptor cells.45,46 Whether or not the role we determined for Dhx38 in R-loop metabolism is related to the pathogenic mechanism of Dhx38-associated RP remains a question to be addressed. Although several studies suggested that the R-loop is involved in the molecular mechanisms of multiple neurological diseases.47,48 the relationship between the R-loops and RP remains unknown. Unfortunately, dhx38 homozygous mutants are embryonically lethal and so are unsuitable as RP animal models to detect whether the alteration of these RPCs also occurs in degenerating photoreceptors. However, the outer photoreceptor segments are continuously renewed (10% of rod discs daily) and have a more vigorous metabolic activity than other cell types, requiring photoreceptor cells to be more transcriptionally efficient in synthesizing the required substances to maintain these activities, making them more dependent on transcription-related factors, such as splicing factors, than other cell types.49,50 Collectively, our study improves our understanding of the specific biological function of Dhx38 in retinal neurogenesis and provides mechanistic insights into the study of Dhx38 function in retinal tissues and the potential pathogenesis of RP caused by splicing factor mutations.
Limitations of the study
Although our findings shed light on the specific function of DHX38 in retinal development in zebrafish, we did not elucidate the pathogenic mechanism of RP caused by DHX38 mutations due to homozygous lethality of dhx38 embryos. It is necessary to conduct RP-associated mutation model to explore the specific molecular mechanism of RP caused by DHX38 mutation. Another limitation of this study is that we only revealed the role of DHX38 in the regulation of R-loop, and further investigation is required to elucidate how DHX38 maintains R-loop homeostasis in vivo and in vitro.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Zpr-1 | Zebrafish International Resource Center | N/A |
| Zpr-3 | Zebrafish International Resource Center | N/A |
| Gfap | Genetex | Cat# GTX128741; RRID:AB_2814877 |
| Sox2 | Genetex | Cat# GTX124477; RRID:AB_11178063 |
| Islet1 | Genetex | Cat# GTX102807; RRID:AB_11179180 |
| γH2AX | Cell Signaling | Cat# 9718; RRID:AB_2118009 |
| p53 | Genetex | Cat# GTX128135: RRID:AB_2864277 |
| α-tubulin | Genetex | Cat# GTX628802; RRID:AB_2716636 |
| γ-tubulin | Genetex | Cat# GTX636480; RRID:AB_2909990 |
| phosphorylated histone H3 | Affinity | Cat# AF3358; RRID:AB_2834773 |
| S9.6 | Millipore | Cat# MABE1095; RRID:AB_2861387 |
| DHX38 | Proteintech | Cat# 10098–2; RRID:AB_2092294 |
| GAPDH | Proteintech | Cat# 60004–1; RRID:AB_2107436 |
| mouse anti-BrdU | Becton Dickinson | Cat# 347580; RRID:AB_400326 |
| rat anti-BrdU antibody | Abcam | Cat# Ab6326; RRID:AB_305426 |
| Critical commercial assays | ||
| Alexa Fluor 594 Phalloidin | Thermo Scientific | A12381 |
| Cell-Light EdU | Ribobio | C10310-1 |
| Cell-Light EU Apollo567 RNA Imaging Kit | Ribobio | C10316-1 |
| TUNEL BrightRed Apoptosis Detection Kit | Vazyme Biotech | A11 |
| Comet Assay Kit | Trevigen | 4252-040-K |
| Experimental models: Cell lines | ||
| HEK-293 cell lines | This paper | N/A |
| ARPE-19 cell lines | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Zebrafish: dhx3812_21del | This paper | N/A |
| Zebrafish: Tg(Neurod1:EGFP) | China Zebrafish Resource Center (CZRC) | N/A |
| Zebrafish: Tg(Huc:EGFP) | China Zebrafish Resource Center (CZRC) | N/A |
| Software and algorithms | ||
| GraphPad Prism 5 | GraphPad Software, Inc | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | National Institutes of Health(NIH) | https://imagej.nih.gov/ij/ |
| Quantity One | Bio-rad | https://www.bio-rad.com/en-us/product/quantity-one-1-danalysis-software?ID=1de9eb3a-1eb5-4edb-82d2-68b91bf360fb |
| Step One Software v2.3 | Thermo Fisher | https://www.thermofisher.com/us/en/home/technical-resources/softwaredownloads/StepOne-and-StepOnePlusReal-Time-PCR-System.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mugen Liu (lium@mail.hust.edu.cn).
Materials availability
All unique materials generated in this study are available from the lead contact upon completion of materials transfer agreement with Huazhong University of Science and Technology.
Experimental model and study participant details
Zebrafish lines
Zebrafish (Danio rerio), AB strain, were maintained in a circulating water system (pH 6.6-7.4,26°C-28.5°C) with a cycle of 14h of light and 10h of dark. The complete knockout of dhx38 results in embryonic lethality at about 3–4 dpf in zebrafish. As a result, the dhx38 homozygotes were generated by crossing the dhx38 heterozygotes with each other. The Tg (neurod1: Egfp) and Tg (huc: Egfp) lines were purchased from China Zebrafish Resource Center (CZRC). All zebrafish were maintained in accordance with guidelines approved by the Ethics Committee of the College of Life Science and Technology, Huazhong University of Science and Technology.
Cell culture
HEK293 (human embryonic kidney) cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% Fetal Bovine Serum (FBS), and 1% peniciliin/streptomycin (Invitrogen) in 5% CO2 atmosphere at 37°C. ARPE-19 (American Type Culture Collection, CRL-2302) were cultured at 37°C in DMEM/F12 (Gibco) supplemented with 10% FBS, and 1% peniciliin/streptomycin.
Method details
siRNAs, plasmids and transfections
Transient transfection of siRNA and plasmids was conducted using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. siRNA duplexes were purchased from RiboBio (Guangzhou, China). Assays were performed 48 or 72 h after siRNA transfection and 24 or 48 h after plasmid transfection. The siRNAs and plasmids used to transfect cells in this study are listed in Table S1.
Generation of dhx38-knockout zebrafish by CRISPR/Cas9 and genotyping
The dhx38 mutant zebrafish strain was generated by CRISPR/Cas9 technology as described previously.11 In brief, guide RNAs (gRNAs) and Cas9 plasmids were purchased from CZRC. The site-specific target was designed using CHOPCHOP (http://chopchop.cbu.uib.no/). The gRNAs and Cas9 mRNAs were synthesized using the mMESSAGE mMACHINE T7 Transcription Kit (#AM1344, Thermo Fisher Scientific) and mMESSAGE mMACHINE T3 Transcription Kit (AM1348, Thermo Fisher Scientific), respectively. For microinjection, gRNAs (100 ng/μL) and Cas9 mRNAs (300 ng/μL) were mixed and co-injected into wild-type zebrafish embryos at the one-cell stage. For genotyping, embryos were harvested and incubated in NaOH and lysed at 95°C for 10 min, then Tris-HCl (pH = 8.0) with 1% NaOH volume was added and centrifuged for 2 min and used as template for subsequent PCR and enzyme digestion. The primers identify the zebrafish embryo genotypes are listed in Table S1.
Immunofluorescence
The zebrafish cryo-sectioning and immunofluorescence assay were conducted as described previously.6 For cell immunofluorescence, cells were grown on coverslips overnight before siRNA and plasmid transfection. 48 h-post-siRNA and 24 h-post-plasmid transfection, cells were washed twice using ice-cold PBS, and then fixed with 4% PFA for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 20 min. After washing twice with PBS, cells were blocked in 10% goat serum for 1 h at room temperature (RT), and incubated with primary antibodies diluted in PBS-1% bovine serum albumin (BSA) overnight at 4°C. Then cells were washed thrice in PBS. Incubated mouse or rabbit Alexa Fluor 488, 594, or 647-conjugated secondary antibody (1:1000 diluted in PBS-1% BSA) for 1 h at RT, washed thrice again in PBS and stained with DAPI for 10 min at RT. Cells and sections were imaged under a confocal microscope (FV3000, Olympus) at ×100 or ×60. ImageJ was used for quantifying the fluorescence intensity.
Zebrafish single-cell immunofluorescence
Zebrafish single-cell immunofluorescence was conducted as described previously with minor modifications.51 Briefly, to prepare single cell suspensions, embryos were placed in D-PBS after genotyping, and then incubated in Liberase (Roche, 1:65 in D-PBS) for 20 min at 37°C. 5% FBS was added to terminate the reaction. The cell suspensions were filtered using a 45-μm cell strainer and pelleted via centrifugation at 900g for 5 min. Cells were resuspended and spotted onto the poly-L-lysine treated slides to dry at RT for 1 h. After drying, slides were fixed with 4% PFA for 10 min and permeabilized with ice-cold methanol at −20°C for 5 min. After washing thrice with 0.1% Tween 20, slides were blocked with blocking solution (5% BSA/0.2% milk in PBS) for 1 h at RT. Primary antibodies of S9.6 (1:50 dilution) and γH2AX (1:400 dilution) were added to the slides and incubated at 4°C overnight. After washing thrice with 1% Tween 20, the secondary antibody (1:1000 dilution) was added and incubated for 1 h at 37°C. After three final washes, cells were stained with DAPI and covered with a glass coverslip. Images were observed under a confocal microscope (FV3000, Olympus) using a ×100 objective lens. ImageJ was used for quantifying the fluorescence intensity.
Western blotting
Zebrafish embryos and cell lysates were prepared using RIPA buffer containing protease inhibitor cocktail (Sigma). Western blotting was conducted as described previously.52 The following primary antibodies were used in this study: anti-DHX38 (Proteintech, 10098-2; 1:1000), anti-p53 (Genetex, GTX128135; 1:1000), anti-γH2AX (Cell Signaling, 9718; 1:1000), and anti-GAPDH (Proteintech, 60004-1; 1:1000). Protein images were obtained using a ChemiDoc XRS+ gel imaging system (Bio-Rad).
RNA-extraction and real-time quantitative RT-PCR
Total RNA was extracted from 36 hpf zebrafish embryos and whole-cell lysates using Trizol reagent (Life Technologies) following the manufacturer’s instructions. The cDNA was synthesized using the TransScript All-in-One First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). The RT-PCR amplification was performed using the AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Ltd) in a StepOnePlus real-time PCR machine (Life Technologies) and analyzed using the GraphPad 5.1 software. The sequences of all RT-PCR primers used in this study are listed in Table S1.
Whole-mount in situ hybridization
Whole-mount in situ hybridization (WISH) was conducted as described previously.52 The cDNA of wild-type embryos was used as a template to design RNA probes confirmed by Sanger sequencing. The digoxigenin-labeled RNA probes were synthesized using MAXIscript SP6 Transcription Kit (Invitrogen, United States). The images were captured under an optical microscope (BX53, Olympus). After imaging, genomic DNA was extracted from each embryo and used for genotyping.
Comet assay
The omet assay was conducted using the CometAssay Reagent kit for Single Cell Gel Electrophoresis Assay (Trevigen, Cat#4252-040-K) following the manufacturer’s instructions. Briefly, cells were mixed with LMAgarose at a ratio of 1:10 (cell:agarose) at 37°C, and spread onto the CometSlide. After being kept in darkness for 10 min at 4°C, the slides were immersed in the Lysis Solution at 4°C overnight. After removing the lysis buffer, slides were immersed in 50 mL 1× neutral electrophoresis buffer at 4°C for 30 min. Then, the slides were placed in an electrophoresis slide tray covered with Slide Tray Overlay, and run at 21 V for 30 min at 4°C. Then, slides were immersed in DNA Precipitation Solution for 30 min followed by 70% ethanol for 30 min at RT and dried at 37°C for 20 min. Finally, slides were stained with 50 μL diluted SYBR Gold in darkness for 30 min and imaged using fluorescence microscopy (ECLIPSE 80i, Nikon). Comet tail events were analyzed using CASP software version 1.2.2 and at least 100 cells per sample were analyzed from each independent experiment.
DNA/RNA immunoprecipitation (DRIP) assay
DRIP-qPCR was performed as described previously.35,37 Briefly, genomic DNA was extracted from HEK293 cells using a genomic DNA Kit (OMEGA) following the manufacturer’s instructions. Then, the harvested genomic DNA was digested with a restriction enzyme cocktail (HindIII, EcoRI, XbaI, SspI, BsrGI) using the CutSmart buffer r2.1 (New England Biolabs) with 2 mM Spermidine at 37°C overnight. Digested DNA was purified via phenol/chloroform extraction followed by RNase H treatment or no treatment in RNase H buffer (New England Biolabs) overnight at 37°C. After setting aside 2% for input DNA, 4 μg digested DNA (treated or non-treated with RNase H) was bound to 10 μL S9.6 antibody in 400 μL DRIP binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, 0.05% Triton X-100) at 4°C overnight gently inverting on a rotative shaker. Then, DNA/antibody complexes were immunoprecipitated to 50 μL prewashed Dynabeads Protein A (Thermofisher Scientific) at 4°C overnight on a rotative shaker. After washing four times with the binding buffer, the beads were eluted using 200 μL elution buffer (50 mM Tris–HCl, pH8.0, 10 mM EDTA, 0.5% SDS) containing 2 μL proteinase K (20 mg/mL) in a thermomixer at 55°C, 800 rpm for 1 h. Then, the eluted DNA was purified using the Qiagen MinElute Kit following the manufacturer’s conditions. qPCR was conducted to calculate the enrichment of DNA/RNA hybrids based on the input ratio53 at actively transcribed genes, and the primers used in this study are listed in Table S1.
DNA fiber assay
DNA fiber assay was performed following standard protocols with slight modifications.54 Briefly, exponentially growing cells were first incubated with 25 μM IdU (iododeoxyuridine, Sigma) for 20 min. After washing twice with warmed PBS, cells were labeled with 250 μM CldU (chlorodeoxyuridine, Sigma) for 30 min. Then, the cells were collected and scraped into ice-cold PBS, and diluted to 200–300 cells per microliter. Next, 2 μL cell suspension was pipetted onto one end of the glass slide and lysed in 15 μL lysis buffer (50 mM EDTA and 0.5% SDS in 200 mM Tris-HCl, pH 7.5) for 10 min. Slides were tilted to 25° to allow the fibers to spread along the bottom of the slide and then dried horizontally. After drying, slides were immersed in methanol/acetic acid (3:1) for 10 min and then incubated with 2.5 M HCl for 80 min followed by washing thrice with PBST (PBS +0.1% Tween 20), and blocked with 5% BSA in PBS for 60 min. IdU and CIdU were detected using the following primary antibodies: mouse anti-BrdU antibody (Becton Dickinson, Cat#347580) and rat anti-BrdU antibody (Abcam Cat#Ab6326), and then diluted in blocking solution and applied at 4°C overnight. Then, slides were washed twice with PBS and stringency buffer (10 mM Tris-HCl pH 7.4, 40 mM NaCl, 0.02% Tween 20, 0.02% NP-40) for 10 min, and blocked again in blocking solution for 60 min at RT. After washing twice with PBS, secondary fluorescence-conjugated antibodies were applied for 1 h at RT in darkness. Then, slides were washed twice and mounted with Fluoromount-G. DNA fibers were detected using an Olympus FV3000 confocal microscope.
Flow cytometry
To analyze cell cycle profiles, cells were harvested and washed twice using ice-cold PBS. After fixing in 70% ethanol, cells were resuspended in PBS, treated with RNase A (20 μg/mL), and stained with PI (20 μg/mL) for 30 min at RT. The flow cytometry data were obtained using CytoFlex (Beckman Coulter), and analyzed using FlowJo software.
Anaphase bridges and micronuclei assay
To count anaphase bridges and micronuclei, cells were cultured on glass coverslips and transfected with siRNA for 48 h. To increase the events of mitosis, cells were treated with 50 ng/mL nocodazole (Sigma) in DMEM/10% FBS at 37°C for 4 h to synchronize the cell cycle. Then, nocodazole was removed, and the cells were washed twice with PBS and incubated with DMEM/10% FBS at 37°C for 1h before harvesting. Then cells were fixed with 4% PFA for 15 min and permeabilized with 0.5% Triton X-100 for 20 min. After washing twice in PBS, cells were stained with DAPI for 10 min at RT. The percentage of cells with micronuclei and anaphase bridges in mitosis anaphase was quantified. At least 100 cells per experiment were analyzed.
TUNEL assay
TUNEL assay was conducted using the TUNEL BrightRed Apoptosis Detection Kit (Vazyme Biotech Co., Ltd, Cat#A11) following the manufacturer’s instructions. Briefly, retinal sections were dried for 20 min and fixed in 4% PFA for 30 min at RT. After washing twice in PBS, the sections were treated with 20 μg/mL proteinase K for 10 min and washed thrice in PBS. After incubation in 1×Equilibration Buffer for 20 min at RT, sections were incubated with TdT buffer for 1 h at 37°C and then washed twice in PBS. The apoptotic cells were labeled using red fluorescence. The images were captured under a confocal microscope (FV1000, Olympus).
EdU cell proliferation assay
For the EdU cell proliferation assay, zebrafish embryos were incubated in 2 mM 5-ethynyl-2′-deoxyuridine (EdU) with 10% DMSO in E3 medium for 30 min at 4°C, and then fixed in 4% paraformaldehyde at 4°C overnight. After genotyping, the heads of the sibling and dhx38−/− embryos were cryo-protected in 30% sucrose at 4°C overnight and embedded in OCT (SAKURA) for cryo-sectioning. The proliferating cells were visualized using the Cell-Light EdU Apollo567 in vitro Kit (RiboBio, Cat#C10310-1) following the manufacturer’s instructions. The images were captured under a confocal microscope (FV1000, Olympus).
Quantification and statistical analysis
All data were represented as mean ± SD and analyzed with a two-tailed t-test by using the GraphPad Prism software. The number of samples used in each experiment is shown in the figure or figure legends. Statistical significance was as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. n.s., no significance, p > 0.05. All experiments were independently repeated at least thrice.
Acknowledgments
The authors thank the China Zebrafish Resource Center (CZRC) and the construction laboratories (Nechiporuk, Alex. OHSU School of medicine, Department of cell and developmental biology) for the Tg(huc:EGFP) and Tg(neurod1:EGFP) transgenic zebrafish lines. This work was supported by grants from the Ministry of Science and Technology of China (No.2018YFA0801000) and the National Natural Science Foundation of China (No. 82071010, 32270646, and 32370830).
Author contributions
M.L., F.L., and Z.T. developed the concept of this study. K.S., Y.H., and J.L. designed the study. S.Y., Y.H., and Y.Z. performed most of the experiments in this study. J.T., P.G., and D.J. performed the generation of dhx38−/− zebrafish by CRISPR/Cas9 technology. X.C. and H.H. contributed in vector construction and Western blot. M.R., P.L., and J.L. helped in real-time PCR assays and analyzed the data. K.S., J.L., X.R., R.J., X Z., X.S., F.L., and M.L. prepared the draft and final version of the manuscript. All authors reviewed the results and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 30, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108103.
Contributor Information
Fei Liu, Email: liufei2018@ihb.ac.cn.
Mugen Liu, Email: lium@mail.hust.edu.cn.
Zhaohui Tang, Email: zh_tang@hust.edu.cn.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Seritrakul P., Gross J.M. Genetic and epigenetic control of retinal development in zebrafish. Curr. Opin. Neurobiol. 2019;59:120–127. doi: 10.1016/j.conb.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson R., Tracey-White D., Webster A., Moosajee M. The zebrafish eye-a paradigm for investigating human ocular genetics. Eye (Lond) 2017;31:68–86. doi: 10.1038/eye.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassett E.A., Wallace V.A. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Růžičková Š., Staněk D. Mutations in spliceosomal proteins and retina degeneration. RNA Biol. 2017;14:544–552. doi: 10.1080/15476286.2016.1191735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farkas M.H., Grant G.R., White J.A., Sousa M.E., Consugar M.B., Pierce E.A. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genom. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Liu F., Lv Y., Sun K., Zhao Y., Reilly J., Zhang Y., Tu J., Yu S., Liu X., et al. Prpf31 is essential for the survival and differentiation of retinal progenitor cells by modulating alternative splicing. Nucleic Acids Res. 2021;49:2027–2043. doi: 10.1093/nar/gkab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z., Reed R. Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 9.Latif Z., Chakchouk I., Schrauwen I., Lee K., Santos-Cortez R.L.P., Abbe I., Acharya A., Jarral A., Ali I., Ullah E., et al. Confirmation of the Role of DHX38 in the Etiology of Early-Onset Retinitis Pigmentosa. Invest. Ophthalmol. Vis. Sci. 2018;59:4552–4557. doi: 10.1167/iovs.18-23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajmal M., Khan M.I., Neveling K., Khan Y.M., Azam M., Waheed N.K., Hamel C.P., Ben-Yosef T., De Baere E., Koenekoop R.K., et al. A missense mutation in the splicing factor gene DHX38 is associated with early-onset retinitis pigmentosa with macular coloboma. J. Med. Genet. 2014;51:444–448. doi: 10.1136/jmedgenet-2014-102316. [DOI] [PubMed] [Google Scholar]
- 11.Tu J., Yu S., Li J., Ren M., Zhang Y., Luo J., Sun K., Lv Y., Han Y., Huang Y., et al. Dhx38 is required for the maintenance and differentiation of erythro-myeloid progenitors and hematopoietic stem cells by alternative splicing. Development. 2022;149 doi: 10.1242/dev.200450. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura K., Cho Y., Tokunaga K., Nakao M., Tani T., Ideue T. DEAH box RNA helicase DHX38 associates with satellite I noncoding RNA involved in chromosome segregation. Gene Cell. 2019;24:585–590. doi: 10.1111/gtc.12707. [DOI] [PubMed] [Google Scholar]
- 13.Crossley M.P., Bocek M., Cimprich K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang E.Y.C., Tsai S., Aristizabal M.J., Wells J.P., Coulombe Y., Busatto F.F., Chan Y.A., Kumar A., Dan Zhu Y., Wang A.Y.H., et al. MRE11-RAD50-NBS1 promotes Fanconi Anemia R-loop suppression at transcription-replication conflicts. Nat. Commun. 2019;10:4265. doi: 10.1038/s41467-019-12271-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prendergast L., McClurg U.L., Hristova R., Berlinguer-Palmini R., Greener S., Veitch K., Hernandez I., Pasero P., Rico D., Higgins J.M.G., et al. Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability. Nat. Commun. 2020;11:4534. doi: 10.1038/s41467-020-18306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Bonnet A., Grosso A.R., Elkaoutari A., Coleno E., Presle A., Sridhara S.C., Janbon G., Géli V., de Almeida S.F., Palancade B. Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability. Mol. Cell. 2017;67:608–621.e6. doi: 10.1016/j.molcel.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez M., Urtasun R., Elizalde M., Azkona M., Latasa M.U., Uriarte I., Arechederra M., Alignani D., Bárcena-Varela M., Álvarez-Sola G., et al. Splicing events in the control of genome integrity: role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res. 2019;47:3450–3466. doi: 10.1093/nar/gkz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodroj D., Recolin B., Serhal K., Martinez S., Tsanov N., Abou Merhi R., Maiorano D. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017;36:1182–1198. doi: 10.15252/embj.201695131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cristini A., Groh M., Kristiansen M.S., Gromak N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018;23:1891–1905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Germain D.R., Poon H.Y., Hildebrandt M.R., Monckton E.A., McDonald D., Hendzel M.J., Godbout R. DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Mol. Cell Biol. 2016;36:2794–2810. doi: 10.1128/MCB.00415-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeiro de Almeida C., Dhir S., Dhir A., Moghaddam A.E., Sattentau Q., Meinhart A., Proudfoot N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell. 2018;70:650–662.e8. doi: 10.1016/j.molcel.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sridhara S.C., Carvalho S., Grosso A.R., Gallego-Paez L.M., Carmo-Fonseca M., de Almeida S.F. Transcription Dynamics Prevent RNA-Mediated Genomic Instability through SRPK2-Dependent DDX23 Phosphorylation. Cell Rep. 2017;18:334–343. doi: 10.1016/j.celrep.2016.12.050. [DOI] [PubMed] [Google Scholar]
- 24.Tedeschi F.A., Cloutier S.C., Tran E.J., Jankowsky E. The DEAD-box protein Dbp2p is linked to noncoding RNAs, the helicase Sen1p, and R-loops. RNA. 2018;24:1693–1705. doi: 10.1261/rna.067249.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saponaro M., Kantidakis T., Mitter R., Kelly G.P., Heron M., Williams H., Söding J., Stewart A., Svejstrup J.Q. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang I.X., Grunseich C., Fox J., Burdick J., Zhu Z., Ravazian N., Hafner M., Cheung V.G. Human proteins that interact with RNA/DNA hybrids. Genome Res. 2018;28:1405–1414. doi: 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uribe R.A., Gross J.M. Id2a influences neuron and glia formation in the zebrafish retina by modulating retinoblast cell cycle kinetics. Development. 2010;137:3763–3774. doi: 10.1242/dev.050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Q., Wang F., Xiang Y., Zhang X., Zhao Z.A., Gao Z., Liu W., Lu X., Liu Y., Yu X.J., et al. Maternal BCAS2 protects genomic integrity in mouse early embryonic development. Development. 2015;142:3943–3953. doi: 10.1242/dev.129841. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Trépanier V., Beaujois R., Viranaicken W., Drobetsky E., DesGroseillers L. The downregulation of the RNA-binding protein Staufen2 in response to DNA damage promotes apoptosis. Nucleic Acids Res. 2016;44:3695–3712. doi: 10.1093/nar/gkw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajesh C., Baker D.K., Pierce A.J., Pittman D.L. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickramasinghe V.O., Venkitaraman A.R. RNA Processing and Genome Stability: Cause and Consequence. Mol. Cell. 2016;61:496–505. doi: 10.1016/j.molcel.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olive P.L., Banáth J.P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 33.Brambati A., Zardoni L., Nardini E., Pellicioli A., Liberi G. The dark side of RNA:DNA hybrids. Mutat. Res. Rev. Mutat. Res. 2020;784 doi: 10.1016/j.mrrev.2020.108300. [DOI] [PubMed] [Google Scholar]
- 34.Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 35.Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenech M., Knasmueller S., Bolognesi C., Holland N., Bonassi S., Kirsch-Volders M. Micronuclei as biomarkers of DNA damage, aneuploidy, inducers of chromosomal hypermutation and as sources of pro-inflammatory DNA in humans. Mutat. Res. Rev. Mutat. Res. 2020;786 doi: 10.1016/j.mrrev.2020.108342. [DOI] [PubMed] [Google Scholar]
- 37.Racca C., Britton S., Hédouin S., Francastel C., Calsou P., Larminat F. BRCA1 prevents R-loop-associated centromeric instability. Cell Death Dis. 2021;12:896. doi: 10.1038/s41419-021-04189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Muse T., Aguilera A. Transcription-replication conflicts: how they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016;17:553–563. doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- 39.Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jao C.Y., Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maslon M.M., Heras S.R., Bellora N., Eyras E., Cáceres J.F. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife. 2014;3 doi: 10.7554/eLife.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin J., Brocher J., Fischer U., Winkler C. Mutant Prpf31 causes pre-mRNA splicing defects and rod photoreceptor cell degeneration in a zebrafish model for Retinitis pigmentosa. Mol. Neurodegener. 2011;6:56. doi: 10.1186/1750-1326-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miccoli A., Dalla Valle L., Carnevali O. The maternal control in the embryonic development of zebrafish. Gen. Comp. Endocrinol. 2017;245:55–68. doi: 10.1016/j.ygcen.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Pagon R.A. Retinitis pigmentosa. Surv. Ophthalmol. 1988;33:137–177. doi: 10.1016/0039-6257(88)90085-9. [DOI] [PubMed] [Google Scholar]
- 46.Daiger S.P., Sullivan L.S., Bowne S.J. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013;84:132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groh M., Lufino M.M.P., Wade-Martins R., Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richard P., Manley J.L. R Loops and Links to Human Disease. J. Mol. Biol. 2017;429:3168–3180. doi: 10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 50.Zhao C., Bellur D.L., Lu S., Zhao F., Grassi M.A., Bowne S.J., Sullivan L.S., Daiger S.P., Chen L.J., Pang C.P., et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am. J. Hum. Genet. 2009;85:617–627. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorrells S., Nik S., Casey M.J., Cameron R.C., Truong H., Toruno C., Gulfo M., Lowe A., Jette C., Stewart R.A., Bowman T.V. Spliceosomal components protect embryonic neurons from R-loop-mediated DNA damage and apoptosis. Dis. Model. Mech. 2018;11 doi: 10.1242/dmm.031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu S., Jiang T., Jia D., Han Y., Liu F., Huang Y., Qu Z., Zhao Y., Tu J., Lv Y., et al. BCAS2 is essential for hematopoietic stem and progenitor cell maintenance during zebrafish embryogenesis. Blood. 2019;133:805–815. doi: 10.1182/blood-2018-09-876599. [DOI] [PubMed] [Google Scholar]
- 53.El Hage A., Tollervey D. Immunoprecipitation of RNA:DNA Hybrids from Budding Yeast. Methods Mol. Biol. 2018;1703:109–129. doi: 10.1007/978-1-4939-7459-7_8. [DOI] [PubMed] [Google Scholar]
- 54.Schwab R.A.V., Niedzwiedz W. Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J. Vis. Exp. 2011:e3255. doi: 10.3791/3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.