Abstract
Artificial intelligence and machine learning techniques have progressed dramatically and become powerful tools required to solve complicated tasks, such as computer vision, speech recognition, and natural language processing. Since these techniques have provided promising and evident results in these fields, they emerged as valuable methods for applications in human physiology and healthcare. General physiological recordings are time-related expressions of bodily processes associated with health or morbidity. Sequence classification, anomaly detection, decision making, and future status prediction drive the learning algorithms to focus on the temporal pattern and model the non-stationary dynamics of the human body. These practical requirements give birth to the use of recurrent neural networks, which offer a tractable solution in dealing with physiological time series and provide a way to understand complex time variations and dependencies. The primary objective of this article is to provide an overview of current applications of recurrent neural networks in the area of human physiology for automated prediction and diagnosis within different fields. Lastly, we highlight some pathways of future recurrent neural network developments for human physiology.
Index Terms—: Deep learning, human physiology, recurrent neural network, signal processing
I. Introduction
MODERN artificial intelligence and machine learning techniques have significantly impacted a wide range of applications, and such powerful learning tools have dramatically improved results. Several ambitious goals have already been achieved: the early triumph of AlphaGo from DeepMind and a recent version of OpenAI that beat the top human players in Dota2 (a sophisticated video game)[1]. In terms of the existing achievements of machine learning, a natural question is raised: how can such an advanced technique serve human health? One answer is deep learning-assisted biomedical image processing, which can adapt Convolutional Neural Networks (CNNs) to analyze spatial information[2].
In another scenario, physiological recordings refer to sequential data rather than images. Such data sets commonly have the following characteristics: (1). They refer to collective electrical/mechanical signals representing physical variables of interest, such as electrical activity produced by the brain or skeletal muscles; (2). These data reflect the status variation of a subject/subjects in a given period of time; (3). They are naturally in the format of time-related recordings (e.g., time series), and latent causality governs two (or more) successive occurrences. In practice, detecting an event in real-time or the future is critical, and the results might be sensitive to the temporal dynamics determined by physiological conditions. Our literature survey found that most sensors used for signal acquisition were non-invasive. For example, electrocardiography (ECG) or electroencephalogram (EEG) signals were collected from electrodes attached to the skin. The data collection procedures are patient-friendly and ubiquitous for practical healthcare systems. However, interpreting these signals is not an easy task. The underlying complexity within the signals and actual physiological mechanisms are generally not visible or easy to understand. Therefore, it is challenging to predict outcomes solely based on a human expert’s experience since the physiological interactions are multidimensional, highly nonlinear, stochastic, time-variant, and patient-specific.
Artificial networks may offer solutions to the problems mentioned above. Neural networks can mathematically describe the underlying relationship. The “Universal Approximation Theorem” tells us that the neural network with one hidden layer can approximate a particular class of functions, which are large enough to capture processes of practical concern. In other words, all the members of the neural network family can approximate the nonlinear characteristics of a given system and explore the relationship from the inputs and corresponding labels, although this process is affected by many factors, such as network structures and learning algorithms. Basic feed-forward neural networks (or deep neural network, DNN) and convolutional neural networks have inherent limits in dealing with time series. DNNs cannot model the system dynamics, which describes the transitions (or time-dependencies) between states in a time sequence. Additionally, in most situations, samples always have variant lengths, which are unfeasible for DNNs to process. The CNNs are good at finding local patterns of temporal sequences, but it’s hard to discover the long-term dependency[3].
Besides the DNNs and CNNs, the Recurrent Neural Networks (RNNs), another deep learning architecture, are more suitable tools for physiological applications with sequential data or signals. RNN presents a class of artificial neural networks, which possess many of the qualities required for tackling the physiological problems: they possess both current and past features of the temporal sequences, adapt to the long-term historical changes in the data, store the past information to solve context-dependent tasks, and make predictions simultaneously with existing observations. Although RNNs were typically used to deal with sequential data like music or language, there have been attempts at applying RNNs to the area of physiology.
In computational physiology, designing a machine-learning algorithm aims to transform electrical recordings from the human cardiovascular, nervous, muscular, and other systems into computer computation in order to predict or identify events, monitor body activities, and detect anomalies[4]. For example, the ECG signal analysis focuses on classifying different types of heartbeat, thus assisting the cardiologist in achieving an accurate diagnosis for the patient; the EEG signal is a critical measure to evaluate many human functions, such as emotion and sleep qualities. It is also widely used to assess cerebral disorders, such as seizures and stroke. All these modalities carry human body information to create the solutions that transform healthcare delivery. As mentioned before, these recordings or signals are commonly presented in sequential manners, and the RNNs are thus successful paradigms in modeling complex physiological processes.
For machine learning practitioners, another goal is to improve the model performance. However, it is greatly constrained by specific conditions of physiological applications, such as feature extraction, data structure, model implementation, and subject issues. In this review, we first briefly introduce the RNN structures and highlight the model constructions according to two types of labels. Meanwhile, regarding the data collection from human subjects, we summarize the currently adopted validation strategies from a deep learning perspective, and discuss how they could affect the performance in later sections. We also present a variety of physiological applications with most representative studies and show that the RNN-based models outperform the other types of architectures, such as support vector machine (SVM) and CNN models. Furthermore, we summarize the existing issues in this field and propose possible solutions for future work.
II. RNN in general
RNN is a kind of network specifically designed for processing time dependent sequential data. Given an input sequence, , a basic RNN architecture maps the input to a target sequence with a hidden layer, as shown in Fig.1. This hidden layer aims to learn the state-wise time dependency, which is modeled as:
| (1) |
where the RNN unit is a class of functions, which will be introduced later. Based on Eq.(1), the RNN structure models the relationship between adjacent hidden states and thus has the capability to process temporal information. This is the main difference between the RNN and CNN. Moreover, the RNNs have many characteristics benefiting the physiological activities and resultant multi-channel signals: (1) For the RNN models, the sequential examples do not necessarily have the same length[5], [6]. This is another difference between the RNN and CNN, because the CNNs request all the input samples have the same dimension;(2) the mapping process keeps the time consistency between the input and output; (3) the ith element could be multi-dimensional; (4) the hidden states described by the recurrent units can be stacked as a deeper structure [7], [8]. In practice, the training process and model performance are greatly affected by the construction of the RNN unit.
Fig. 1.
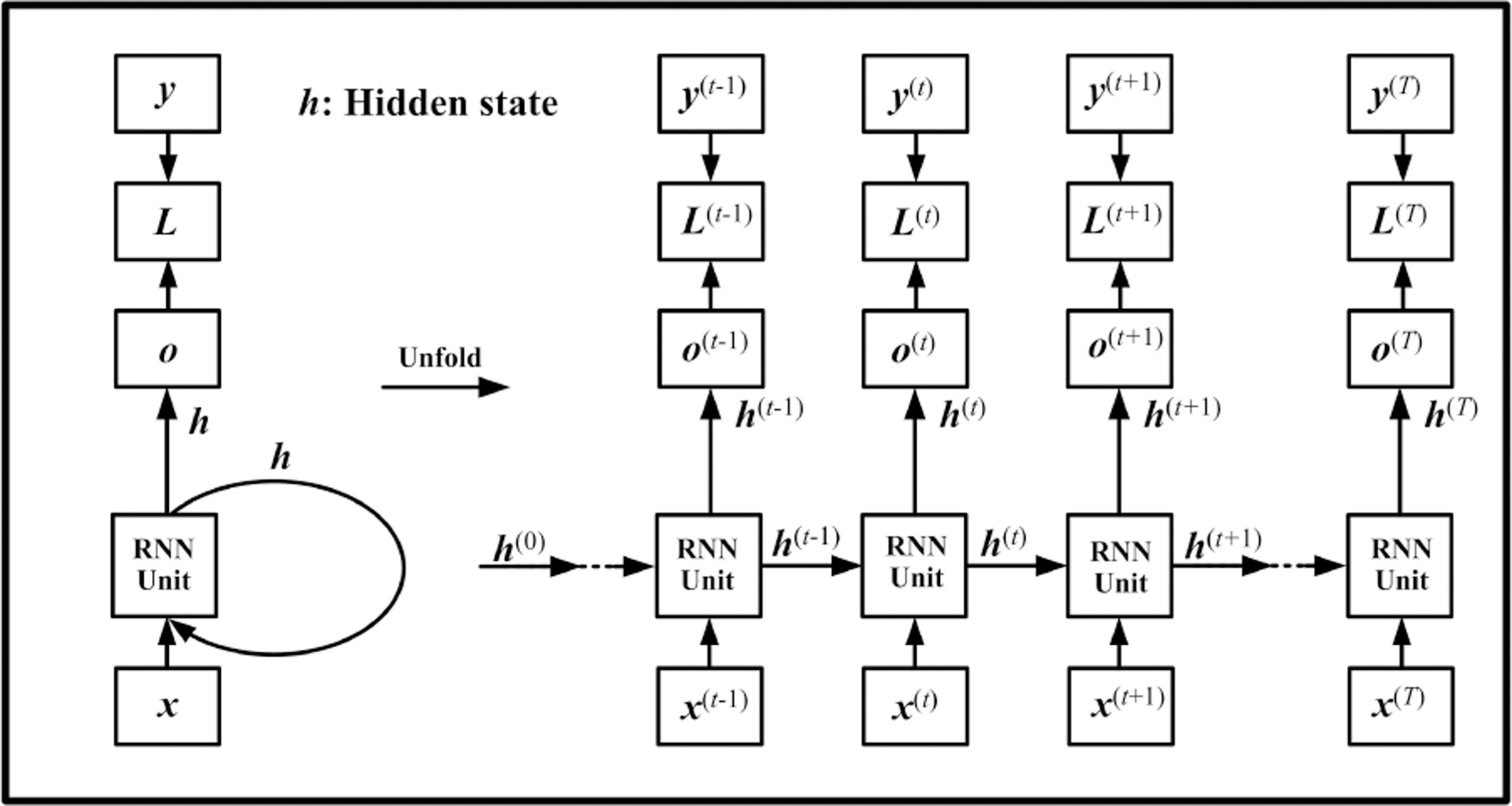
Computational graph of RNN. is the RNN output, and presents the difference between the RNN output and the desired output (target or label). is commonly used for calculating the loss function.
A. Structures of the RNN unit
1). Elman RNN:
The Elman RNN, which was named after Jeffrey Locke Elman, is the most basic RNN unit (sometimes called “Vanilla RNN”, meaning that it doesn’t have any extra features)[9]. The hidden state is calculated as:
| (2) |
where , are weights matrices, and is a bias term.
Elman RNN is one dense layer structure augmented by the inclusion of edges that span adjacent time steps[3]. The nonlinearity is introduced by using the activation function to transfer the hidden state dynamically.
2). Long-Shot Term Memory (LSTM):
Compared with Elman RNN, LSTM contains an external cell structure. It delivers the information of input states through the entire time chain and forms a shortcut connection for the hidden states, as shown in Fig. 2. Since the cell state won’t be transferred to the next layer, it is also considered as a self-loop. Moreover, in LSTM, three gate components control the information flow: input gate, output gate, and forget gate. At each time step, the cell state updates itself by two actions: 1. gathering new information from the current input and hidden state, and 2. choosing old information from the past cell state.
Fig. 2.
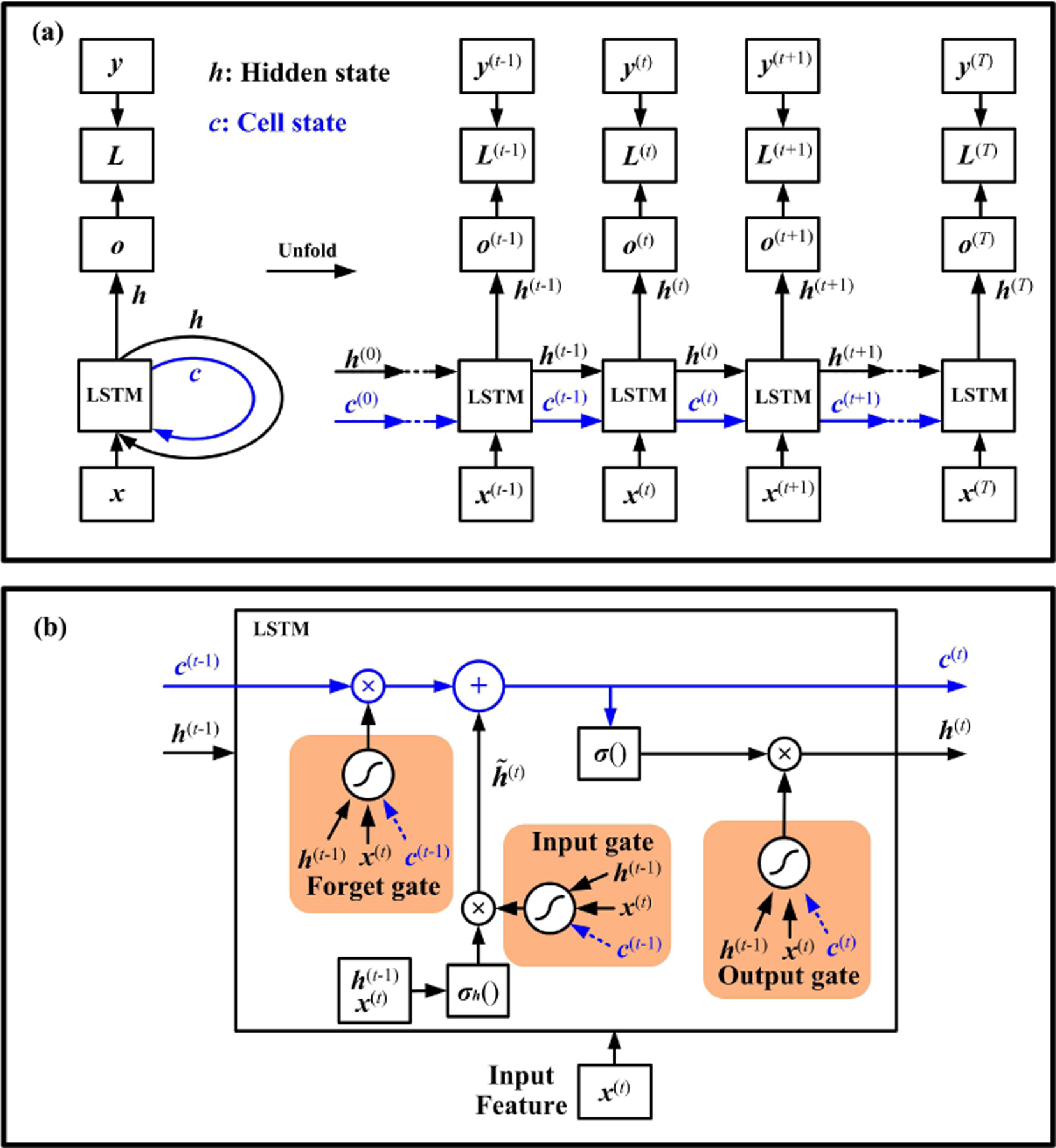
LSTM recurrent neural network. The computational graph is shown in (a). The LSTM has an extra pathway for the cell state. A recurrent unit of LSTM is shown in (b). The arrows in blue represent the internal cell state.
The input feature and the previous hidden state are used to compute an intermediate state with an activation function . This procedure is similar to that of the Elman RNN (Eq.(2)). The state can accumulate into the cell state , if the input gate allows it. The cell state is controlled by the forget gate to drop irrelevant parts of the previous cell. Meanwhile, the input gate and forget gate determine how much information is chosen from the current and past time steps for updating the cell state. Moreover, the output state can be shut off by the output gate to limit the information passed to the next hidden state. The cell state can also act as an extra input to these gating units, as shown in Fig. 2(b).
3). Gated Recurrent Unit(GRU):
GRU is another successful RNN unit design, as shown in Fig. 3 [5]. It contains two gating units: a reset gate and an update gate, as shown in Fig.3. The reset gate determines how much information is removed from the previous hidden state and generates a new state . Similar to Elman RNN (Eq.(2)), the input feature and the new state are used to compute a intermediate state with an activation function .
Fig. 3.
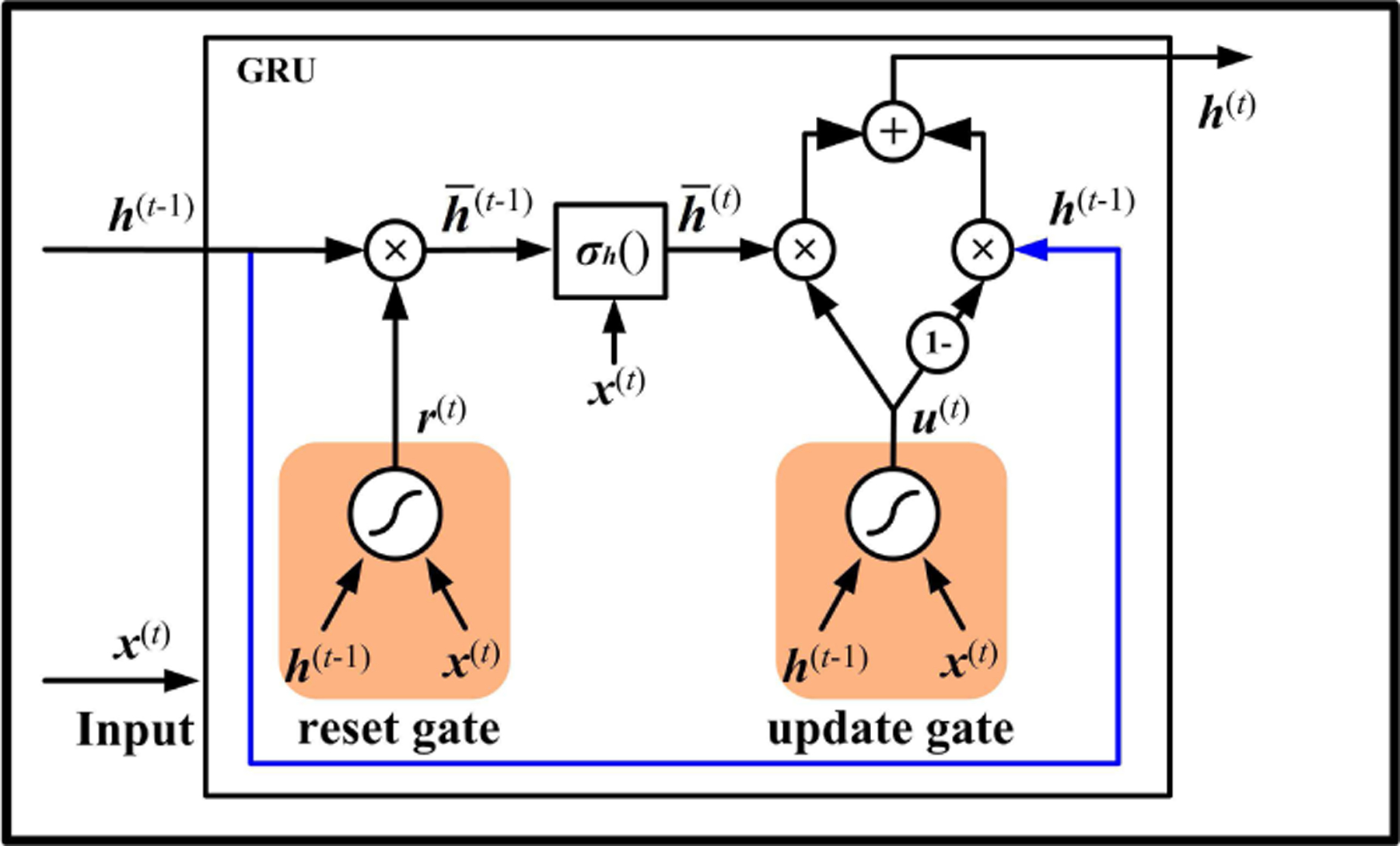
The unit structure of GRU.
If the reset gate outputs zero, the unit only involves the information of current input . To calculate the hidden state , the unit needs to select the meaningful information from the intermediate state by an update gate. Meanwhile, to make the output quickly react to the previous state , GRU also includes a shortcut connection, as delineated with blue lines in Fig. 3. The useful part of can also be controlled by the update gate, which acts like a two-way switch and simultaneously controls both forget and output information.
In these three RNN units, the Elman RNN employs the simplest structure, and it was thus widely applied in various physiological studies. However, training Elman RNN would raise issues known as exploding gradient and vanishing gradient with the first-order optimization method (like Gradient Descent)[10]. Compared to the Elman unit, both LSTM and GRU establish shortcut paths with gate components. The primary advantage of these two units is that they efficiently address the gradient vanishing problem when the time lag is extremely long[3]. Therefore, they have the capability of learning long-term dependency for time sequences. Besides, the GRU uses fewer parameters than the LSTM and thus reduces the computational time in the training and inference processes. In physiology, which kind of RNN unit is the most appropriate for a given task is still an open question. We will offer more discussion with physiological studies in Section V-B.
B. Bidirectional RNN
The previously discussed RNN computational graph (shown in Fig. 1) learns information from the previous time steps, meaning that RNNs are causal systems from a control theory perspective. In some cases, the RNN also needs to model the temporal dependency from the future to get better representations of the entire signal. Such a function can be conducted by simply adding an extra backward path to form a bidirectional RNN, as shown in Fig.4. The bidirectional RNN performs well on language-related tasks because the semantic analysis requires the previous and future words or sounds. Intuitively, bidirectional RNNs are not suitable for the physiological signal analysis, especially when online detection or classification is desired, because the future input is unobserved. A feasible solution is to specify a fixed size window (or buffer) around the current time step when the classification results are highly sensitive to the upcoming input. In abnormal heartbeat detection and emotion classification tasks, some studies attempted to use bidirectional RNNs, since the future observation on physiological signals may give us more evidence for decision-making at the current time point[11], [12], [7], [13].
Fig. 4.
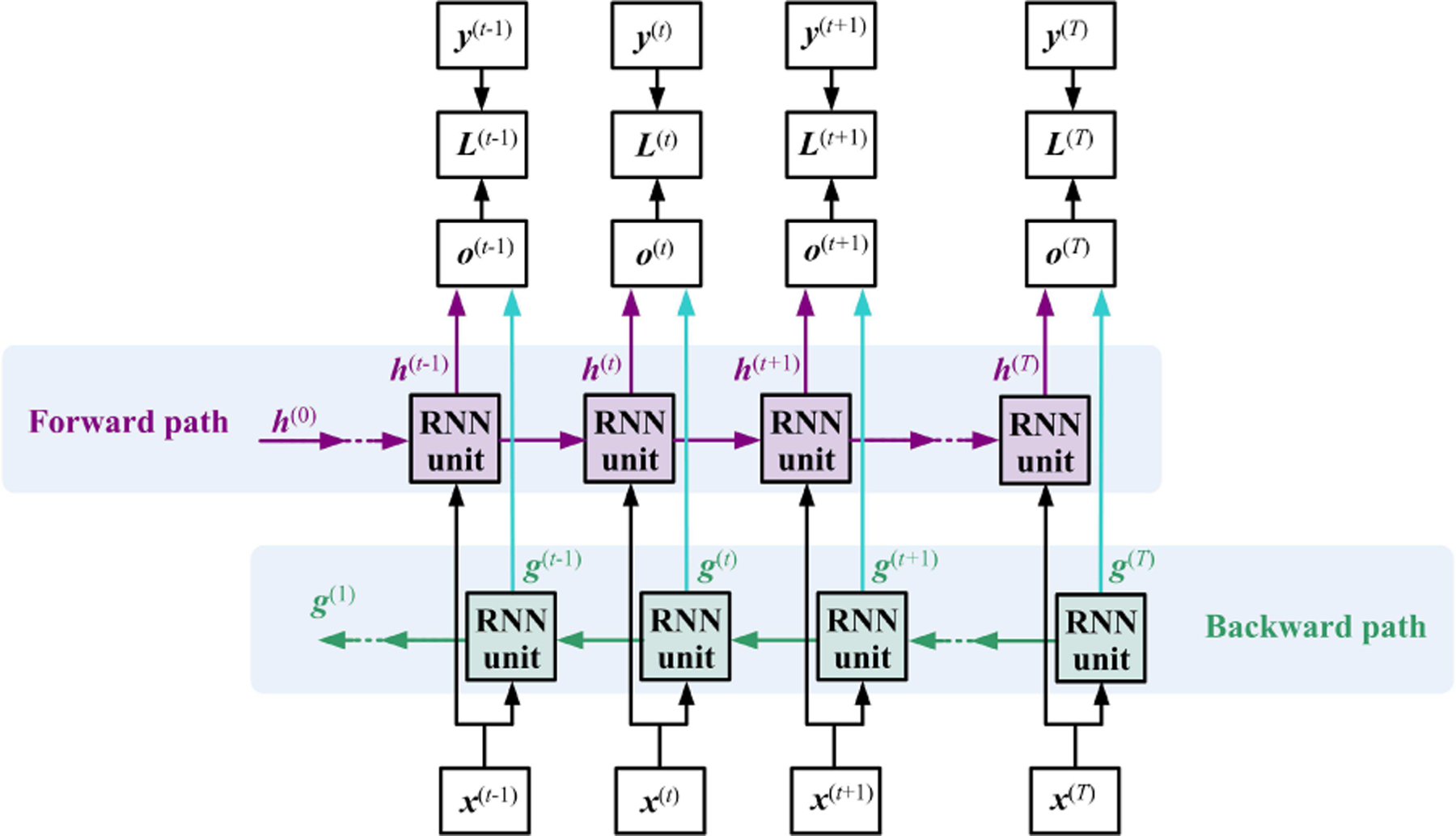
The general bidirectional RNN has two time flow paths. The variables and present the hidden state for the sub-RNN moving forward and backward, respectively.
III. Experiment design with RNN
In physiological applications, Supervised learning is commonly used because the expected output (label) always exists. Besides, some Unsupervised learning techniques, such as autoencoder and clustering, may also help the physiological studies, which will be introduced in Section IV with examples. The experimental design depends on practical requirements and special considerations, which are sometimes beyond the scope of computer science or engineering. We will discuss the experiment design from two levels of view.
A. Model implementation
For model implementation, one should first analyze the data structure on hand. The physiological signals, which are always used as input data, are time sequences. As shown in Fig. 5(a) and (b), the annotated label structure would lead to two scenarios. Scenario I is that a sequence sample is annotated with a sequential label, framed as a ‘Many-to-Many’ problem, such as the study of sleep stage classification[14]. The model output was also a sequence with the same length as the label sequence. Another scenario II is that a sequence sample is annotated with a single label. For example, the ECG signal in a segment has only one label[15], [16]. This scenario is also described as a ‘Many-to-One’ problem since the considered output is a scalar value for each sequential input.
Fig. 5.
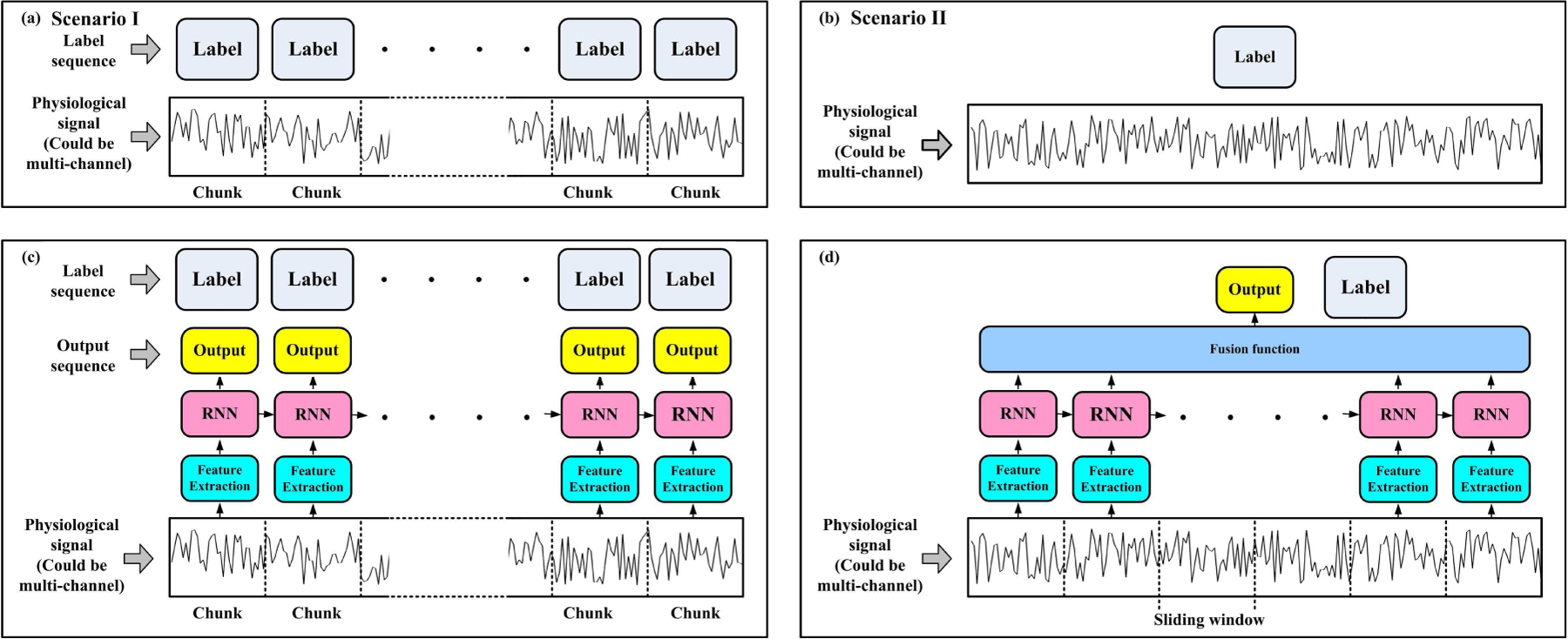
The implementations of RNN models are determined by the label structure of each signal sample. (a) shows a signal sequence with a sequential label. The general applied RNN could be designed in (c). Sometimes a signal sequence could only have one annotated label, as shown in (b), and the RNN could be designed in the form of (d). Although (c) and (d) show one-layer unidirectional RNN, multiple stacked layers or bidirectional RNN are also adoptable.
1). Model output construction:
The RNN model structures have slight differences at the final output part in these two scenarios. For scenario I, one can use the structure shown in Fig.1, Fig.2(a), and Fig.4, since the output in these figures is also sequential. For scenario II, there would be several ways to construct a fusion function (layer) to obtain the output, as shown in Fig.5(d).
A. The easiest way to construct the output is to use the hidden state at the last step with one or more dense layers. For example, to diagnose arrhythmia based on ECG signal, Oh et al. fed the LSTM’s hidden state at last time step to 3-dense layers for final output[17]. Chang et al. and Hofmann et al. also applied a similar configuration [18], [19]. For the bidirectional RNNs, one can concatenate the last hidden states of both forward and backward paths into a signal vector, and then feed it into dense layers for final output, as introduced in the studies of Lynn et al. and Supratak et al.[20], [21].
B. Another way of designing a fusion function is to employ an attention layer and then a dense layer as the final output. [22], [11], [23], [14]. The attention layer is calculated by a weighted sum of all the hidden state vectors from the RNN. A study from Shashikumar et al. suggested that the attention mechanism could improve the accuracy of classifying paroxysmal atrial fibrillation[11].
C. The third way of constructing the output also uses the information of all the hidden states. They could be flattened first and concatenated into a 1-D vector, then fed to dense layer(s) for the final output. For example, the studies reported by Yildirim and Liu et al. applied such a way for arrhythmia classification[24], [25], [26], [27]; Xing et al. also designed one dense layer with all hidden states as input for emotion recognition from EEG[28].
There could be other designs for the fusion function, such as sparse projection on hidden states[29] or averaging all the hidden states[30]. Method B and C may not be compatible with variant length samples since the concatenated vector should have a uniform length. Padding zeros might be a solution. However, more studies are needed to discuss the padding effects.
2). Input construction:
TThe input constructions of the two scenarios shown in Fig.5(a) and (b) are also slightly different. In scenario I, the signals are manually divided into consecutive chunks (or slices, epochs, segments) with clinical or other practical purposes, as shown in Fig.5(a). To construct an input sequence, a feature extraction module might be needed, which is also called ‘epoch processing block’ in the study of Phan et al.[31]. This module forms a vector representing each chunk’s information, and then vectors from all the chunks connect into an input sequence, as shown in Fig.5(c).
In each chunk, there are several ways to design a feature extraction module: 1. Directly flatten the chunk-wise data into a 1-D vector. The data lengths in all the chunks should be the same. 2. Knowledge-based features. Such as the R-R interval of the ECG signal[32], [33], [34]. 3. Handcraft features with engineering methods, including the statistic features. For example, mean value, standard deviation, frequency-domain features, and spectral features [35], [36], [27]. 4. Deep learning method to form a end-to-end system. For instance, CNN-1D, autoencoder, or even lower level RNN[21], [14], [22].
The sequential data in scenario II can also be sliced into chunks with sliding windows, and then the designer can apply the feature extraction module in each window to form the input sequence for RNN. The most straightforward design is that the window size is one and skip the feature extraction. In this case, the raw signal or recording is directly fed into the RNN as an input sequence, as introduced in [37]. Similar to scenario I, designers can also use the handcrafted features with overlapped windows[38], [11]. One typical design applies the Short-Time Fourier Transform (STFT) or Continuous Wavelet Transform (CWT) on the signals to form a spectral image, as introduced in [18], [39], [38]. Such an image composes a sequence of feature vectors. The CNN-1D layers can also be treated as a sliding window, in which the filter size determines the window size and stride determines how large two adjacent windows are overlapped[40], [41], [25], [17], [21]. More sophisticated designs could combine the two ways: calculate the spectral image and then apply CNN-1D on each frequency vector or CNN-2D on the spectral image[30], [42], [43].
Sliding windows can also serve the model design in scenario I. The chunks at a time could use as an input sequence for the label at , as suggested in [14]. The feature extraction module is critical for constructing the input sequence for RNN model. It can employ very flexible and complicated structures. The later sections will introduce more about this module.
B. Subject issue
Unlike other prevalent deep learning tasks, such as image classification or natural language processing, the physiological application has a very peculiar issue related to the subjects (persons, patients, participants, or users). One subject could offer more than one training pair in data collection, and all the data samples may not be independent of each other. Assigning of training, validation, and testing sets must consider the subject effect. Based on our survey, Cross-subject (Inter-subject) prediction and Within-subject (Intra-subject) prediction are commonly used strategies in physiological applications.
1). Cross-subject prediction:
Based on the samples acquired from certain subjects, it is desired for an RNN to model the universal pattern and predict the event of interest on the unseen subjects. The common practice of examining the model generalization is leave-one-subject-out, or k-fold subjects cross-validation, as shown in Fig 6(a). For example, Joseph Futoma et al. trained an RNN to detect sepsis on an unseen participant[44]. Chang et al., Hou et al., and Shashikumar et al. also conducted cross-subject experiments for the ECG classification studies[18], [45], [11]. The basic assumption of such a design is that the physiological information shared in the training group can benefit the testing group. Compared to the within-subject prediction, the cross-subject prediction is substantially challenging because human characteristics are subject-specific, and inter-subject variability can seriously degrade the performance[46].
Fig. 6.
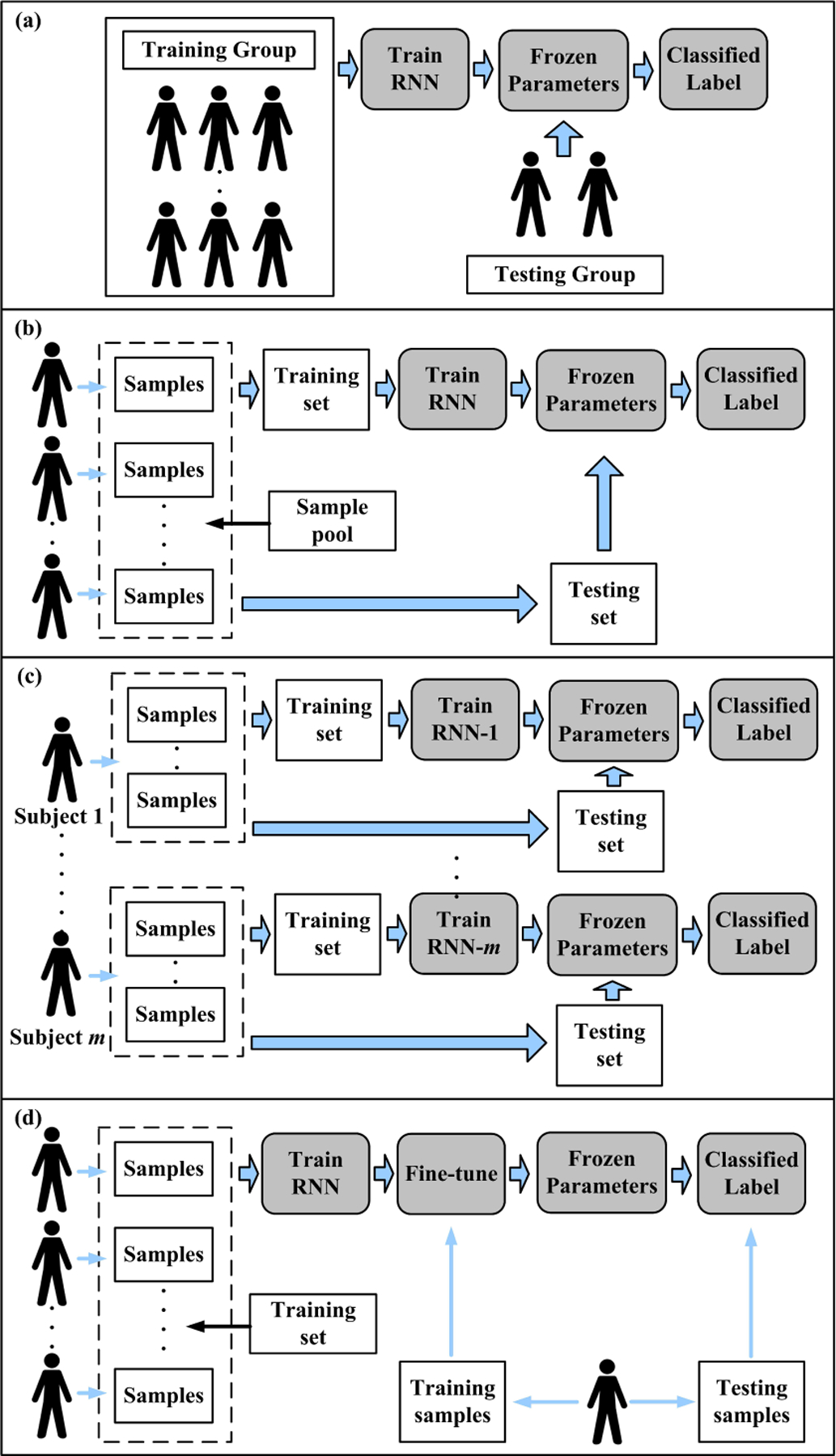
The experiment designs in computational physiology. (a)Cross-subject prediction; (b), (c), and (d) describe three different strategies of within-subject prediction. (b): the mixed manner; (c) patient-specific manner; (d) fine-tuning manner
Diagnosing an unseen patient from the experience of diagnosing former patients is attractive for both clinical and technical investigators. To obtain robust RNNs, gathering data from more subjects is helpful, although it is costly or sometimes infeasible. We will discuss this issue in Section V-C.
2). Within-subject prediction:
In some studies, the RNN-based models were trained by some samples (or clinical attempts) collected from one/some subject(s) and validated/tested by the samples collected from the same subject(s). The validation/testing pairs were unseen for the model, but the subject(s) information was partially seen in the model. In practice, there are mainly three ways to implement within-subject studies.
1. Mixed manner.
Each subject provided several samples, and the whole sample pool was collected from multiple subjects. All the samples were mixed and randomly divided into training, validation, and testing sets, as shown in Fig. 6(b). This manner assumed that all the samples were independent and identically distributed regardless of the subject effect. Some ECG classification studies adopted this manner[37], [24], [17], [26]. In the applications of emotion recognition with EEG signals, some studies also applied ”trial-oriented” recognition, which was similar to mixed manner[30], [47], [28].
2. Subject-specific (subject-dependent) manner.
This manner tended to train a specific model for just one subject due to the variability among the subjects[32]. The training and validation sets were collected from the same subject for just one model training, and the participant’s group thus required multiple models, as shown in Fig6(c). This manner assumed that only the samples collected from the same subject share an identical pattern. Some epileptic seizures prediction studies preferred to use this manner, such as the studies reported in [27], [48].
3. Fine-tuning manner.
This manner attempted to balance the information of other subjects and the testing subject. The model could be first trained by the data collected from other subjects, and then fine-tuned by partitions data of the tested subject (also known as target domain). Such a manner believed that the training set collected from training group helped model the common patterns, but it has insufficient personalized information of unseen subjects due to the interuser differences. To build up the blood glucose prediction model, Dong et al. used this manner to train a model on multiple patients and then fine-tuned the model for one patient [49]. Similarly, Phan et al. fine-tuned SeqSleepNet[14] and DeepSleepNet[21] models, which are well-developed RNN-based models in sleep stage classification[31].
The strategy choice is greatly determined by the study purposes, practical requirement, data structure, and physiological considerations. However, different strategies on the same dataset could lead to significantly different results. We will discuss this issue in Section V-C.
IV. Applications in physiology
The main machine learning task in physiology is to develop an automatic diagnostic or patient status monitoring system. Analyzing the disorder, predicting the onset of a seizure, classifying a subjects’ state, and even forecasting a possible disease from the time-related signals are all desired tasks. RNNs are the top options for dealing with temporal information and learning the relationship between the signals and the symptoms. As mentioned before, such relationships are generally not well understood and are hard to evaluate with existing human knowledge. The RNN framework endows physiological data analysis with a highly flexible, inductive, nonlinear modeling ability. Based on our survey, we found that the RNNs have already served human physiology from the top (brain) to the bottom (gait), as shown in Fig.7.
Fig. 7.
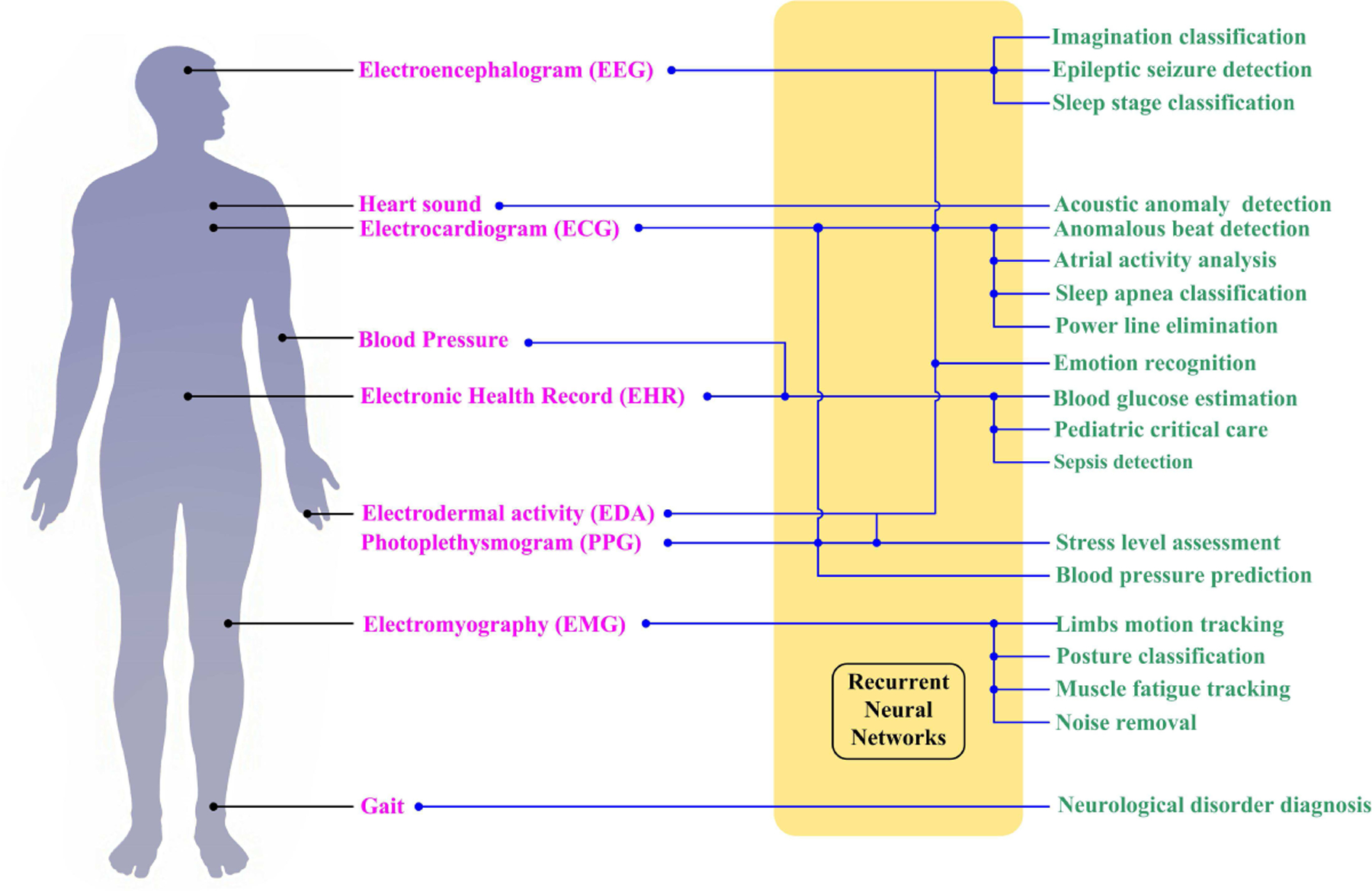
Representative applications of RNN in the human body for diagnosis and event detection.
The works cited in this review were found by use of aggregate research databases including PubMed (MEDLINE), Springer Link, Google Scholar, and IEEE Xplore. Keyword searches were conducted through these databases with search terms such as ‘recurrent neural network’, ‘long short-term memory’ and ‘gated recurrent unit’ (and their acronyms) with the combination of ‘physiology’, ‘electrocardiogram’, ‘electromyography’, ‘electroencephalogram’, ‘photoplethysmogram’, ‘epileptic seizure’, ‘emotion recognition’, ‘sleep stage’, ‘blood glucose’ and ‘gait’ (and their acronyms). In the following sections, we will summarize the majority of studies in ECG classification, emotion recognition, epileptic seizure detection, sleep stage classification, and blood glucose level prediction with RNNs. To present up-to-date studies in these applications, we focus on the papers published after 2015. Besides, we also summarize the studies in other physiological fields from the most representative studies published after 2010. We mainly cover the human-subject studies that applied RNN for analyzing the physiological time sequence. Additional studies, such as biomedical image processing and document/tabular analysis, will not be included.
A. Electrocardiographic signal analysis
The ECG plays a significant role in the diagnosis of cardiovascular status[32]. It has become a focus of investigations since it consists of unobtrusive, effective, non-invasive, low cost, and widely available procedures using sensors (or electrodes). As a physiological measure, the ECG represents the sequential cardiac electrical activities, such as depolarization and repolarization of the cardiac muscle. When the deep learning models are applied, The ECG signal could be directly fed into the network without any elaborate preprocessing[50]. Alternatively, knowledge-based features, such as morphology (or shape) features, heartbeat interval (R-R interval), and heart rate, could also be used as input[45]. It is worth noticing that all of these features are also variant over time, which offers us a way to detect events of interest with RNNs.
The tasks of ECG classification are usually ‘Many to One’ problems, namely classifying the clinical label based on the signal segment in each beat. Therefore, most studies would consider segment-level classification. Some studies also attempted to model the dependency between successive heartbeats, such as the studies reported by Shashikumar et al., Chang et al., and Mousavi et al[18], [11], [54]. The detailed applications of RNNs in diagnosing abnormalities or analyzing signals are listed in Table I.
TABLE I.
Summary of works carried out using RNN structure with ECG Signals
| Author | Heartbeat typesa | Model architecture | Performance | Datasetb |
|---|---|---|---|---|
| Zhang et al. 2017[51] | N, VEB, SVEB and F. | The raw ECG signal segment was fed into a 2-layer stacked LSTM. | The accuracies were 99.6% and 99.0% for detecting SVEB and VEB, respectively. | MITDB |
| Maknickas et al. 2017[52] | N, AF, O, and noise. | • 23 handcraft features were extracted from each ECG segment and then connected in the time order to form an input sequence. • The model was composed of 3-layer LSTM and followed by a dense layer. |
Macro-averaging F1-score was 0.78. | CinC |
| Schwab et al. 2017[22] | N, AF, O, and noise. | • Handcrafted and autoencoder calculated features were extracted from beat-wise ECG segments and then connected in the time order to form an input sequence. • An ensemble model contained 15 RNNs and 4 Hidden Semi-Markov models. The outputs of sub-models were concatenated as an input for a dense layer to obtain the final result. |
Macro-averaging F1-score was 0.79. | CinC |
| Warrick et al. 2017[40] | N, AF, O, and noise. | The ECG signal segment was firstly fed into one CNN-1D layer. Three layers of LSTM were then applied at the top of the CNN layer | Macro-averaging F1-score was 0.80. | CinC |
| Zihlmann et al. 2017[39] | N, AF, O, and noise. | • Proposed an ensemble model with 5 deep networks, either CNN only models or CNN and RNN models. • The authors first calculated the logarithmic time-frequency spectrogram as an input image. • The RNN part was composed of a 3-layer bidirectional LSTM. |
Macro-averaging F1-score was 0.821 | CinC |
| Xiong et al. 2018[41] | N, AF, O, and noise. | • The ECG signal segment was first fed into a CNN part composed of 16 residual blocks with CNN-1D layers. • Three layers of Elman RNN were then applied at top of CNN parts. |
Macro-averaging F1-score was 0.864. | CinC |
| Singh et al. 2018[37] | N and A. | The authors applied three kinds of RNNs with Elman, LSTM, and GRU. Each RNN had three stacked layers, and the ECG signal segment was used as an input sequence. | LSTM showed the best accuracy of 88.1%. | MITDB |
| Shashikumar et al. 2018[11] | AF and O. | • The wavelet power spectrum was calculated in 30s non-overlapping windows. 5-layer CNN was applied on each spectrum to extract local features and thus form a feature sequence in 10 mins. • The feature sequence was fed into a bidirectional Elman RNN with a soft attention layer, followed by a dense layer for decision-making. |
• Testing accuracy was 96%. • Transfer learning was conducted on PPG recordings as input data and obtained 97% accuracy. |
Collected from 2850 patients |
| Yildirim. 2018[24] | N, LBBB, RBBB, PB and VPC | • The detailed coefficients were calculated from 4 levels of the discrete wavelet transform. These coefficients with original signals were connected into a 5-channel time sequence as input. • The sequence was fed into 2-layer unidirectional and bidirectional LSTMs separately. |
• For the unidirectional LSTM, the accuracy was 99.25%. • For the bidirectional LSTM, the accuracy was 99.39% |
MITDB |
| Oh et al. 2018[17] | N, LBBB, RBBB, APB and VPC | • The ECG signal segment was first fed into the CNN part, composed of 3 CNN-1D layers. • The CNN part was followed by 1-layer LSTM. The last time step’s output was fed to 3 dense layers for final classification. |
The accuracy was 98.10% | MITDB |
| Chang et al. 2018[18] | N and AF | • The author calculated the time-frequency spectrum on each ECG recording, which contained multiple heartbeats. • Fed the spectrum into the LSTM model according to the time order. |
The accuracies were 98.3% and 87.0% for within- and cross-subject experiments | MITDB, CinC and self recorded data set |
| Tan et al. 2018[12] | N and CAD | • Each ECG segment was sliced into multiple shorter segments with overlapping windows. Then they were put together into a 2D matrix as an input. • The input was first fed into 2-layer CNN, and 3-layer LSTM was designed on the top of CNN. |
• For non-patient specific task, the accuracy was 99.85%. • For patient specific task, the accuracy was 95.76%. |
INCARTDB |
| Yildirim et al. 2019[25] | N, LBBB, RBBB, APB and VPC | • The authors applied a CNN-based autoencoder to extract the feature from each ECG segment. • The feature sequence was fed into 1-layer LSTM. |
The accuracy was 99.11% | MITDB |
| Hou et al. 2019[45] | Two tasks: 1.N, LBBB, RBBB, APB and VPC; 2. AAMI standardc |
• The authors applied an RNN-based autoencoder to extract the features from each ECG signal segment. • Both encoder and decoder were designed with 1-layer LSTM. • An SVM was used for the final decision-making based on the features. |
• For task 1, the accuracy was 99.74%. • For task 2, the accuracy was 99.45% |
MITDB |
| Wang et al. 2019[32] | N, VEB, SVEB and F | • The ECG signal segment was fed into the RNN as a morphological vector. • The RR interval features were fed into a dense layer as temporal input with an extra clinical label. • RNN and dense layer outputs were combined and fed to another dense layer for final decision-making. |
• Accuracies of detection SVEB on three datasets were: 99.7%, 99.0% and 99.9%. • Accuracies of detection VEB on three datasets were: 99.7%, 99.8% and 99.4% |
MITDB, SVDB and INCARTDB |
| Liu et al. 2019[26] | N, VPC, RBBB and APB | • An ensemble model with 3 CNNs and 3 RNNs was proposed. • The signal was decomposed into 6 levels of intrinsic mode functions (IMF)with empirical mode decomposition. • Three low IMFs were fed into three bidirectional LSTMs, and three high IMFs were fed into three CNNs • The sub-models were first trained separately, and an SVM was used as a fusion layer. |
The accuracy was 99.1% | INCARTDB |
| Saadatnejad et al. 2019[53] | VEB vs. non-VEB and SVEB vs. non-SVEB | • The authors proposed an ensemble model with two sub-RNN models. • The original signal, RR-interval features, and wavelet features were combined as pairs for the inputs of sub-RNN models. |
Evaluated the model with three different sub-set of MITDB. The average accuracies for those two tasks were 99.4% and 98.6% | MITDB |
| Mousavi et al. 2019[54] | NA, SA, VA, FA, and QA | • A Seq2seq model was designed. The input was a sequence of multiple beats, and the output was sequential labels. • The ECG signals were first fed into CNN-1D layers to extract feature sequences, which were then fed into an encoder. • Both encoder and decoder were bidirectional Elman RNN. |
Intra-patient (within-subject) paradigm achieved 99.92% accuracy. Inter-patient (cross-subject) paradigm achieved 99.53% accuracy. | MITDB |
N: Normal rhythm; A: Arrhythmia; AF: Atrial fibrillation rhythm; APB: Atrial premature beats or atrial premature complex; CAD: Coronary artery disease; F: Fusion of a ventricular ectopic beat and a normal beat; LBBB: Bundle branch block; PB: Paced beat; RBBB: Right bundle branch block; VEB: Ventricular ectopic beat; VPC: Ventricular premature contraction; SVPC: Supraventricular premature contraction; SVEB: Supraventricular ectopic beat; O: Other types of rhythms.
CinC: A database provided by Computing in Cardiology 2017 challenge[15]; MITDB: MIT-BIH arrhythmia database[16]; SVDB: MIT-BIH Supraventricular Arrhythmia Database; INCARTDB: St. Petersburg Institute of Cardiological Technics 12-lead Arrhythmia Database[15]
AAMI standard includes 5 classes: NA: Normal beat, LBBB, RBBB, atrial escape beat, Nodal escape beat; SA: APB, SVPC, Aberrated atrial premature beat, Nodal premature beat; VA: VPC, VEB; FA: Fusion of normal and ventricular beat; QA: PB, Fusion of paced and normal beat, unclassified beat.
The LSTM is becoming more popular in recent years for ECG-based heart disorder diagnosis, although the Elman RNN is still competitive due to its computational efficiency[41]. Besides the RNN method, many studies attempted to apply CNN solely for the ECG classification problems. Moreover, the CNN models were much deeper than the proposed RNN models, as listed in Table I. Acharya et al. proposed an 11-layer deep structure with one-dimensional CNN to detect coronary artery disease and achieved 95% accuracy[55]. Kachuee et al. also attempted residual blocks with CNN-1D layers in a 13-layer deep learning model, which obtained 93.4% accuracy on the MIT-BIH database[56]. Jun et al. designed also designed an 11-layer deep structure with 2-dimensional CNN layers for arrhythmia classification on the same database, and the beat waveform was treated as a gray-scale image for input construction[57]. They achieved 99.05% accuracy, which was slightly lower than RNN-based models, such as the study presented by Hou et al[45].
B. Emotion Recognition via RNN
Human emotions influence all aspects of our diurnal experience. Automatic emotion recognition has been an active research field for decades from psychology, cognitive science, and engineering. Physiological signals are strongly correlated with emotion and offer more objective evidence, since emotion leaded physiological reactions are involuntary. Among all the modalities, such as EEG, ECG, electrooculography (EOG), temperature, blood volume pressure, electromyography (EMG), electrodermal activity(EDA), the EEG signal has drawn particular attention as it comes directly from the human brain[58], [59], [47]. EEG signals are recorded by electrodes placed on the participants’ scalp and reflect the electrical activity of the brain signals. The EEG-based assessment method is superior in the neuroscience domain due to its non-invasive ability to detect deep brain structures. The recent studies of emotion recognition with EEG and RNN are listed in Table II.
TABLE II.
A summary of contributions dealing with the applications of RNNS to emotion recognition with EEG signals.
| Author | Emotion classesa | Model architecture | Performance | Datasetb |
|---|---|---|---|---|
| Li, Xiang et al. 2016[30] | Low/high arousal and low/high valence. | • The authors calculated a scalogram (a time-frequency representation) in each channel by CWTc, then stacked the scalograms from all the channels to form a frame sequence. • Each frame was fed into 2 CNN-1D layers, followed by one LSTM layer. • The average values of all hidden states were fed to a softmax layer for final output. |
Mean accuracies were 72.06% and 74.12% on valence and arousal classification, respectively. | DEAP |
| Li,Youjun et al. 2017 [47] | Low/high arousal and low/high valence. | • The 32 leads conduction was first mapped to a 9*9 matrix. The PSD features from the raw EEG signal were filled into this matrixc. • The matrix was then interpolated into a 200*200 image at each time step. • The images were first fed into 2layer CNN with max pooling and one dense layer. • An LSTM and a dense layer were used for final decision-making with the feature sequence obtained from the last time step. |
The average accuracy was 75.21% | DEAP |
| Alhagry et al. 2017[64] | Low/high arousal, low/high valence, and low/high liking. | • Raw signal in each segment was used as input for two-layer LSTM. | For the arousal, valence, and liking classes, average accuracies were 85.65%, 85.45%, and 87.99% . | DEAP |
| Yang et al. 2018[65] | Low/high arousal and low/high valence. | • The authors designed a parallel model constructed with RNN and CNN. • The signal was first fed into one dense layer and two stacked LSTM layers. The last hidden state was fed to another dense layer. • Single vector at single time step was transferred into 2D frame according to the position of the electrodes, and the frame sequences were fed into 3 CNN layers. • Both CNN and RNN outputs were concatenated for final decision-making. |
Mean accuracies were 90.80% and 91.03% on valence and arousal classification tasks, respectively | DEAP |
| Hofmann et al. 2018[19] | Low/high arousal. | • Raw signals were decomposed with either Spatio-Spectral decomposition or Source Power Comodulation. • In the 1s segment, the decomposed signal with 250 samples was fed to 2-layer LSTM, and 2 dense layers were used for final classification. |
• Model with Spatio -Spectral decomposition got 63.4% accuracy. • Model with Source Power Comodulation got 62.3% accuracy. |
Self-collected EEG data from 45 participants. |
| Li, Yang et al. 2018[62] | Positive, Neutral and Negative emotion. | • Both left and right hemispheres had 31 channels of EEG signals. • The authors calculated DEcin 9s signals with 1s window and 5 frequency bands to form a time sequence. • LSTM was applied as a feature extractor for sequence. • Domain Adversarial Neural Networks was used based on the LSTM. |
• When conducted subject-specific experiment, the accuracy was 92.38% • When leave-one-subject-out cross-validation was considered, the accuracy was 83.28% |
SEED |
| Li, Yang et al. 2018[63] | Positive, Neutral and Negative emotion. | Proposed an extension version of [62] by adding a subject discriminator. | Achieved 84.14% accuracy. | SEED |
| Zhang et al. 2018[29] | Positive, Neutral and Negative emotion. | • The authors extracted DEcfeatures in 1s for all 62 electrodes. • In a single time step, Elman RNN was applied to model the spatial dependency from 4 directions. • The outputs of the spatial model at each time step were formed as sequences and fed into bi-directional Elman RNN. |
Achieved 89.5% accuracy. | SEED |
| Xing et al. 2019[28] | Low/high arousal and low/high valence. | • A stacked DNN autoencoder was applied for obtaining the sequence representation with the raw signal as input. • A Hanning window is used for segmenting the sequence representation, and frequency band power features and correlation coefficients were extracted in each window. • The feature sequence was fed to 1layer LSTM, and all the hidden states were fed into a dense layer for final decision-making. |
Mean accuracies were 81.10% and 74.38% on valence and arousal classification tasks, respectively. | DEAP |
| Li, Xiang et al. 2020[66] | Positive and Negative for SEED. Low/high arousal and low/high valence for DEAP | • Different types of autoencoder (Traditional, Restricted Boltzmann machine, and variational autoencoder) were first applied to obtain the latent sequences. • The latent sequence was fed into LSTM as an input, and 2 dense layers were used for final decision-making. |
• For DEAP, mean accuracies were 76.23% and 79.89% on valence and arousal classification tasks, respectively. • For SEED, the accuracy was 85.81%. |
DEAP and SEED |
Valence, arousal, and dominance space is a dimensional representation of emotions. Valence ranges from unpleasant to pleasant; arousal ranges from calm to activated and can describe the emotional intensity[67];
SEED: Shanghai Jiao Tong University Emotion EEG Dataset, which contains 15 subjects’ EEG data with 62 channels sorted according to 10–20 systems. In SEED, there are three categories of emotions (positive, neutral, and negative)[61]. DEAP: a Database for Emotion Analysis using Physiological signals, which contains EEG signals from 32 subjects, and the EEG signals were collected with 32 channels according to 10–20 system. Each participant rated the emotion in terms of arousal, valence, like/dislike, dominance, and familiarity[58].
DE: differential entropy; PSD: power spectral density; CWT: continuous wavelet transform.
Most analyses focused on the time-frequency representation for features extraction from the EEG signals since some studies suggested the correlation between valence/arousal and the frequency bands. For example, pictures and music-induced higher arousal associates decreased alpha oscillatory power[60]. Considering these characteristics of EEG signals, the power spectrum densities, continuous wavelet transformation analysis, or differential entropy features offers promising features for investigating the patterns of brain activities for specific emotion [61], [29], [62], [63], [47]. For the dataset SEED, DEAP, and other 10–20 system constructed EEG data, an additional concern is how to represent not only the temporal dependency but also the spatial connection of the multi-channel signals. The EEG components from different brain regions may also correlate with emotions. Collaborating with RNN, some studies listed in Table. II attempted to model the spatially-adjacent dependency according to the positions of electrodes. For example, the studies reported in [47] mapped the multi-channel signals to 2-dimensional image sequences and further extracted deep features by CNN layers, thus improving the accuracy compared with the study in [30].
Although various approaches have been proposed for EEG-based emotion recognition, most experimental results cannot be compared directly for different setups of experiments. Recently, we have several publicly available emotional EEG datasets, but there is still a lack of standard protocol for evaluating the performance. For example, the studies of Li, Youjun et al., and Li, Xiang et al. employed the trial-oriented 5-fold cross-validation (mixed manner) to implement their model[30], [47]. In contrast, Alhagry et al. and Yang et al. trained the model in a subject-specific manner. Nevertheless, some studies compared their performances with non-RNN methods under similar experiment setups. Li, Xiang et al. proposed C-RNN architecture achieved better performance than the random forest and support vector machine[30]. On both DEAP and SEED datasets, Li, Xiang et al. also suggested RNN with variational autoencoder outperformed support vector machine, random forest, k-nearest neighbors, sample logistic regression, naive Bayes classifier, and DNN[66].
C. Epileptic Seizure Detection
Epilepsy is a chronic neurological disorder caused by abnormal excessive or synchronous neuronal activities in the patient’s brain[68]. Based on the EEG recording, the study of the brain activity and the neurodynamic behavior of epileptic seizures provides required clinical diagnostic information. However, EEG analysis is time-consuming for the neurologist through qualitative visual inspection of raw data. Current studies in automatically detecting epileptic seizures have already utilized the merit of RNN, which helps to explore the characteristics of EEG, as summarized in Table. III.
TABLE III.
A summary of contributions describing the application of RNNS for epileptic seizure detection.
| Author | Learning taska | Model architecture | Performance | Datasetb |
|---|---|---|---|---|
| Thodoroff et al. 2016[69] | Whether a 30-second segment of EEG signal contains a seizure or not. | • In each 30s segment, the multi-channel signal was first coded into 3-channel images with 1s windows by spatial 2D projection and fast Fourier transform. • The image sequences were fed into CNN layers to extract local features. • The feature sequence was fed into a bidirectional LSTM, and a dense layer was applied on the RNN output through all the time steps for final decision-making. |
Average sensitivity was 85% and positive rate was 0.8/hour. | CHB-MIT |
| Raghu et al. 2017 [70] | Three tasks: 1. Normal vs. pre-ictal; 2. normal vs. epileptic; 3. pre-ictal vs. epileptic. | • A Weiner filter first removed the 50Hz power line noise. • In the 1s segment, log energy entropy, wavelet packet log energy entropy, and wavelet packet norm entropy were calculated as input features. • Elman RNN with two hidden layers was used as a classifier. |
The accuracies of three tasks were: 99.70%, 99.70% and 99.85%. | Uni Bonn |
| Ahmedt-Aristizabal et al. 2018 [71] | normal vs. inter-ictal vs. ictal | The raw EEG signals were directly fed to 2-layer LSTM, in which the output at the last step was fed into a dense layer for final decision-making. | The accuracy and average AUC were 95.54% and 0.9582. | Uni Bonn |
| Tsiouris et al. 2018[27] | pre-ictal vs. inter-ictal. | • First extracted 643 features in 5s chunk and then formed an input feature sequence for each signal segment. • Fed the sequence into 2-layer LSTM, followed by fully connected layers for final output. |
The sensitivity and specificity were 99.28% and 99.60%. | CHB-MIT |
| Daoud et al. 2019[48] | pre-ictal vs. inter-ictal. | • Multi-channel raw signals were fed into the CNN layers or a pre-trained encoder to extract features. • The feature sequences were then fed into bidirectional LSTM, and the outputs at last step were concatenated for final decision-making. |
Both models obtained 99.66% accuracy, 99.72% sensitivity, and 99.60% specificity. | CHB-MIT |
| Huang et al. 2019[72] | Seizure or not seizure. | • A channel dropout layer was first applied to the multi-channel EEG signals. The signals were then fed to the multi-scale CNN layers to extract multi-scale features, followed by an attention model. • The feature sequences were fed into bidirectional LSTM and GRU as two streams. The outputs of the RNN were concatenated as inputs for a dense layer at each time step. Then, the dense layer’s output sequences were fed into a global average pooling layer for final output. |
Specificity and Sensitivity were 93.94% and 92.88%, respectively. | CHB-MIT |
| Abbasi et al. 2019[73] | Four tasks: 1. pre-ictal vs. ictal; 2. pre-ictal vs. inter-ictal; 3. inter-ictal vs. ictal; 4. pre-ictal vs. inter-ictal vs. ictal. | • The raw EEG signal was divided into five components by discrete cosine transform. • Hurst exponent and auto-regressive–moving-average features were extracted for each component. • The features were used as input of 2-layer LSTM for final output. |
The accuracies for the four tasks were 99.17%, 97.78%, 97.78% and 94.81%. | Uni Bonn |
| Hussein et al. 2019[74] | Four tasks: 1. normal vs. ictal; 2. non-ictal vs. ictal; 3. normal vs. inter-ictal vs. ictal; 4. 5 type ictal states classificationc. | • The 4096*1 raw signal was first reshaped into 2048*2, and then fed into an LSTM with 2048 steps. • A dense layer was applied to the RNN output at all the time steps. • The output of the dense layer was fed into an averaging pooling layer, and a softmax layer was used for final decision-making. |
All tasks reached 100% accuracy | Uni Bonn |
For the epileptic seizure studies, there are several states: normal state, which describes healthy subjects’ EEG signal without any seizure history; pre-ictal state, which is defined by the period just before the seizure; ictal state, which is during the seizure occurrence, post-ictal state, that is assigned to the period after the seizure took place; and post-ictal state, that is assigned to the period after the seizure took place.[75].
CHB-MIT: CHB( Children’s Hospital Boston)-MIT(Massachusetts Institute of Technology) Scalp EEG Database, contains 23 patients divided among 24 cases (a patient has two recordings). The dataset consists of 969 Hours of scalp EEG recordings with 173 seizures[15]. Uni Bonn: data samples were collected in the Department of Epileptology at Bonn University[76]. This database was divided into five sets named A, B, C, D, and E. Sets A and B included surface EEG signals collected from five healthy participants. Set A was recorded from the five participants when they were awake and rested with their eyes open, while set B was recorded when their eyes were closed. Sets C, D, and E included signals collected from the cerebral cortex of five epileptic patients. Set E was taken from those patients while experiencing active seizures, and sets C and D were recorded throughout the seizure-free interims. The electrodes of set D and set C were implanted within the brain epileptogenic zone and the hippocampal formation of the obverse cerebral hemisphere, respectively.
The 5 types were A∼E sets provided by Uni Bonn dataset.
Almost all the studies in Table. III adopted the with-in subject strategy, leading to relatively fair comparisons. Only the study carried out by Thodoroff et al. considered cross-subject detection[69]. With a similar reason to the EEG-based emotion classification tasks, some studies constructed handcraft features from time-frequency representations[69], [70], [27]. Alternatively, deep models, such as CNN and autoencoder, were also applied to learn the lower-level features[48], [72].
On the CHB-MIT dataset, the model designed by Daoud et al. gave the state-of-art performance (99.72% accuracy) when the CNN layers and pre-trained encoder were used to extract the features[48]. Their model outperformed the CNN and DNN models. On the Uni Bonn dataset, a state-of-art performance was achieved by directly feeding the reshaped raw signal into RNN. The accuracy reached 100%[74], suggesting that the handcraft features were not robust enough. Other studies also applied non-RNN-based methods on the same dataset, but they did not exceed the best performance. For example, Acharya et al. conducted a 13-layer deep CNN, and obtained 88.7% accuracy[77]. Lu and Triesch proposed a deep CNN with residual structure and the system gave 99.0% accuracy[78].
D. Sleep Stage Classification
Sleep plays a vital role in human health. Abnormalities in sleep timing and circadian rhythm are common comorbidities in numerous disorders, such as apnea, insomnia, and narcolepsy[79]. Automatically monitoring the sleep stage would significantly benefit the clinical research and practice for evaluating a subject’s neurocognitive performance. Many studies have been trying to automate sleep stage scoring based on multi-channel signals from electrodes. These signals are generally called polysomnogram (PSG), typically consisting of EEG, EOG (electrooculogram), EMG, and ECG. Most sleep stage classification problems could be described as ‘Many-to-Many’ problems1, since the labels were commonly in the sequential form synchronized with the PSG signals. Meanwhile, as described in Fig.5(a), each signal chunk had one corresponding label annotated by the human expert2.
The current studies for sleep staging are summarized in TABLE.IV. The RNN was indispensable for this task when deep learning methods were used. Similar to the task of emotion recognition, some studies for sleep staging used the frequency domain features for constructing the input, such as the log-power spectrum, based on the frequency bands of the rhythms of EEG signals. Meanwhile, according to the American Academy of Sleep Medicine (AASM) standard, the five sleeping stages are highly characterized by the frequency bands[86], [81]. Most of the studies applied a cross-subject strategy. For example, Suparatak et al. applied k-fold subjects cross-validation, and Phan et al. used leave-one-subject-out cross-validation[21], [23]. The DeepSleepNet developed by Supratak et al. obtained higher overall accuracy than the non-RNN sparse autoencoder[87] and CNN-based method[88]. Based on LSTM, Phan et al. further improved the accuracy with SeqSleepNet with learned features. They suggested that such a model worked better than the CNN-only[88], [89], DNN-only, and regular machine learning methods, such as SVM and random forest[38].
TABLE IV.
A summary of contributions describing the application of RNNS for sleep stage classification.
| Author | Model architecture | Dataseta,b | Performance |
|---|---|---|---|
| Supratak et al. 2017[21] | • The authors proposed a hierarchical structure called DeepSleepNet with CNN and RNN parts. • Each signal chunk was first fed into a CNN part composed of two parallel CNN-1Ds with different filter sizes, and the two outputs were then concatenated as a chunk-level feature. • All chunk features were connected as temporal sequences and fed into a 2-layer bidirectional LSTM with a residual connection. |
MASS and Sleep-EDF | For MASS, the accuracy and macro F1-score were 86.2% and 0.817; for Sleep-EDF, the accuracy and macro F1-score were 82.0% and 0.769. |
| Dong et al. 2017[38] | • In each signal chunk, engineering features were extracted.. • Each feature was first fed into 2 dense-layers. The outputs of all the chunks in each recording were then connected into a feature sequence. • The feature sequence was fed into an LSTM as an input vector. |
MASS | The accuracy and macro F1-score were 85.92% and 0.805 |
| Phan et al. 2018[23] | • The authors first calculated the log-power spectral coefficients in each chunk as model input. • A DNN model was used as a filter bank for feature extraction and dimension reduction. • A two-layer bidirectional GRU with an attention layer was then applied on the top of the DNN part. The attention vector was fed to a softmax layer for final output. • After the model training, the softmax layer was replaced by an SVM for final decision-making. |
Sleep-EDF Expanded | When in-bed part was considered only, the accuracy and macro F1-score were 79.1% and 0.698; when before and after sleep periods were also considered, the accuracy and macro F1-score were 82.5% and 0.72. |
| Michielli et al. 2018[81] | • Each 30s signal chunk was divided into 30 smaller chunks with 1s length, and 55 features were extracted in each chunk. • The authors designed a 2-level LSTM-based classifier. After feature selection, the feature sequences were fed into the first-level classifier. It classified 4-classes, in which the stages N1 and REM were merged into a single class. • The samples identified as N1 stage/REM were then fed to the second binary classifier. |
Sleep-EDF | The accuracy was 86.7% |
| Phan et al. 2019[14] | • The authors proposed a hierarchical structure called SeqSleepNet with filter bank layers and two levels of RNNs. • Raw data had 3 channels (EEG, EOG and EMG). In each chunk, power spectrums image was calculated in each channel. The filter bank layer was applied to these images. • Connected the time point features as chunk-level sequence, which was fed into first level RNN (bidirectional GRU) with attention layer. • The authors connected the features as a chunk-level sequence, which was fed into bidirectional GRU with an attention layer. |
MASS | The accuracy and macro F1-score were 87.1% and 0.833 |
| Phan et al. 2019[31] | • The authors proposed a transfer learning strategy. • The author conducted either the CNN part from DeepSleepNet or the RNN part from SeqSleepNet to extract chunk-level features in each chunk. • The chunk-level features were connected as feature sequences and fed into a 2-layer bidirectional LSTM with residual connection. |
The model was trained on MASS, and fine- turned on Sleep-EDF-SC, Sleep-EDF-ST, Surrey-cEEGGrid, and Surrey-PSG | The accuracy and macro F1-score obtained from transfer learning outperfomed directly training on all the four datasets. |
| Phan et al. 2019[82] | The authors proposed a Fusion model, which was composed of DeepSleepNet and SeqSleepNet with slight modifications. | MASS | The accuracy and macro F1-score were 88.0% and 0.843. |
| Mousavi et al. 2019[83] | • The authors proposed a Seq2seq model called SleepEEGNet. • The signal was first fed into CNN layers for extracting the feature sequence, which was used as input for an encoder.. • Both encoder and decoder were constructed with bidirectional LSTM and attention mechanism. |
Sleep-EDF Expanded (version 1 and 2) | The accuracy and macro F1-score were 84.26% and 0.7966 for version 1; 80.03% and 0.7355 for version 2. |
MASS: Montreal Archive of Sleep Studies[84]; Sleep-EDF: a database in European Data Format[15]; Sleep-EDF Expanded: An expanded version of Sleep-EDF[15]; Sleep-EDF-SC: the Sleep Cassette, a subset of the Sleep-EDF Expanded dataset; Sleep-EDF-ST: the Sleep Telemetry, a subset of the Sleep-EDF Expanded dataset; Surrey-cEEGGrid and Surrey-PSG: collected at the University of Surrey using the behind-the-ear electrodes and PSG electrodes respectively [85].
Sleeping stages are commonly categorized into five classes: rapid eye movement, three sleep stages corresponding to different depths of sleep (N1∼N3), and wakefulness.
E. Blood glucose level prediction
Diabetes mellitus is a common public health issue, and the prevalence of diabetes diagnoses has increased substantially over the past 30 years among adults in the U.S[90]. The culprit of this metabolic disorder is insulin release or action, which leads to hyperglycemia. Managing the blood glucose (BG) levels of a diabetic patient can benefit glycaemic control and reduce costly complications[91].
Continuous subcutaneous glucose monitoring is becoming the most popular tool with a micro-invasive sensor to measure BG, such as an adhesive patch. One primary task for machine learning algorithms is to forecast abnormal changes in glucose concentration to take preventive action in time and avoid life-threatening risks[92]. To implement this, there are two types of inputs: using the past BG concentration only, or using the past BG concentration and external factors, such as food, drug, insulin intake, and activity, as shown in Table.V. The features are commonly extracted from physiological models when the external factors were included as input modalities[91], [92]. All these features describe the effects of carbohydrate, insulin, exercise, sleep, and glucose dynamics, and they are characterized as glucose-related variables.
TABLE V.
A summary of existing studies applying RNNS for BG level prediction.
| Author | Dataset | Input modalities | Model architecture | Performance |
|---|---|---|---|---|
| Gu et al. 2017[91] | 35 non-diabetic subjects, 38 type I and 39 type II diabetic patients | The past BG data, meal, drug, insulin intake, physical, activity, and sleep quality. | • The features were conducted from physiological and temporal perspectives based on the external factors. • This study proposed an Md3RNN model, which had three divisions: 1. A grouped input layer with three different sets of weights corresponding to non-diabetic, type I, and type II diabetic patients; 2. shared staked LSTM; 3. personalized output layers assigned for individual users. |
The average accuracy was 82.14% |
| Fox et al. 2018[95] | 40 patients with type 1 diabetes over three years. | The past BG data only. | The authors proposed four models with Seq2seq structures. All the encoders used GRUs, and only the decoder parts were different: • DeepMO: Fully connected layers were used for prediction. • SeqMO: GRU was used for decoder. • PolyMO: Multiple fully connected layers were used to learn the coefficients of a polynomial model. • PolySeqMO: the latent vector was first fed into a GRU, and the hidden states were used to learn the coefficients of a polynomial model. |
PolySeqMO achieved the lowest absolute percentage error, which was 4.87. |
| Dong et al. 2019[49] | 40 type I diabetic patients. | The past BG data only. | • Raw data sequence was directly fed into a GRU layer. The output at the last step was fed to 2 dense layers for final output. • Train a model on multiple patients and then fine tune for one patient. |
The mean square error at 30 and 45 min were 0.419 and 0.594 (MMOL/L)2, respectively. |
| Dong et al. 2019[93] | 40 type I and 40 type II diabetic patients.. | The past BG data only. | • The 1-day sequential BG data were first assigned into different clusters by a k-mean method. • The authors designed parallel dense layers for different clusters. • Each sequence was first fed into the dense layer according to the corresponding cluster, and the outputs of all parallel layers were fed into a shared LSTM for final output. |
For type I, the mean square errors were 0.104, 0.318, and 0.556 (MMOL/L)2 at 30 min, 45 min, and 60 min prediction, respectively; for type II, the mean square errors were 0.060, 0.143 and 0.306 (MMOL/L)2 at 30 min, 45 min, and 60 min prediction, respectively. |
| He et al. 2020[92] | 112 subjects, including non-diabetic people, type I and type II diabetic patients. | The past BG data, food, drug intake, activity, sleep quality, time, and other personal configurations. | • The feature sequence was conducted from physiological models and auto-correlation encoder. • The input sequence was first fed into a personal characteristic dense layer, in which the weights and bias were uniquely assigned for each subject. • All the subject-specific dense layers shared a GRU layer for final output. |
The root mean square errors were 0.29, 0.47, and 0.91 (MMOL/L) at 15 min, 30 min, and 60 min prediction, respectively |
| Zhu et al. 2020[94] | OhioT1DM [96] and silicon dataset from the UVA-Padova simulator[97]. | The past BG recording, insulin bolus, meal intake and time index. | • The electronic health record in a sliding window was directly fed into the dilated RNN. • The dilated RNN was structured by 3-layer Elman RNN. The second and third layers had skipped connection through time. The last step was used for final decision-making. • The model was first trained on both datasets and then fine-tuned for the specific subject. |
The root-mean-square error was 18.9 mg/dL at 30 min prediction. |
Predicting the value at a future step is traditionally an auto-regressive problem. When RNN is applied, this problem could be re-formulated as a ”Many-to-One” scenario. For the prediction with multiple steps, such as predicting the BL in 15, 30, and 60 min, one can construct the model output as a multi-dimensional vector[93], [94]. Most RNN-based BL prediction studies have proven that the RNN models offered better performance (lower errors) than the other methods, such as the autoregressive model and support vector regression[92], [95], [94], indicating that RNN was more robust in searching the historical information with sufficient non-linearities. Besides, one innovative design structured this prediction problem into the seq2seq model adapting RNN-based autoencoder, as introduced in the study of Foxet al[95]. Their comparative studies suggested that the ‘Many-to-One’ structure (DeepMO) could not offer the best performance. Instead, the PolySeqMO outperformed all other structures. More details could be found in Table.V.
A critical challenge for the BG studies is modeling the typical pattern among all the subjects while considering the subject-specific characteristic. To address this issue, the practitioners in BG prediction generally design the model with 2 (or more) divisions: one learned the personalized pattern with different weights and biases, and the other one learned the common dynamics by shared RNN[93], [92], [91]. Other studies also attempt to apply the idea of fine-tuning manner, as shown in Fig. 6(c)[49], [94]. Glucose metabolism in the human body is a long-term process. For example, the effect of insulin intake can be present for more than ten hours, and measuring BGL variation demands days. Therefore, it is challenging to obtain a large number of samples. Meanwhile, within-subject experiment is the main strategy in the existing studies since the inter-patient variability makes it hard to find a generic model. We will discuss more in Section V-C.
F. Other applications
Besides the applications mentioned above, the RNN also exhibited its power in other physiological fields. Singh et al. attempted to classify automotive drivers’ stress levels based on the Galvanic Skin Response and Photoplethysmography signals[98]. Their study compared the traditional DNN and the Elman RNN, and pointed out that the RNN was the most optimal structure for stress level detection. Futoma et al. applied an LSTM combined with the Multitask Gaussian Process to detect the onset of sepsis[44]. Mastorocostas et al. designed a block-diagonal RNN, a modified version of the Elman RNN, to analyze lung sounds[99]. Cheng et al. used a deep LSTM to detect the obstruction of sleep apnea based on ECG signals[100]. Su et al. used ECG and PPG to predict blood pressure[34]. Their study applied a res-RNN architecture with a residual connection similar to the ResNet based on the CNNs[101]. Liu et al. involved historical blood pressure records, heart rate, and temperature in predicting future blood pressure with LSTMs[102]. More importantly, they used the subject’s profile as an extra input vector to address the cross-subject issue, and we will discuss it in Section V-C3. Hussain et al investigated the preterm prediction for pregnant women using Electrohysterography (EHG) technique [36]. Bahrami Rad et al. also used PSG to detect non-apneic and non-hypopneic arousals with 3-layers bidirectional LSTM[103]. Yang et al. detected heartbeat anomalies based on heart sounds with a two-layer GRU[35].
The EEG signals are also valuable measures for stroke detection and rehabilitation, and an increasing number of studies attempted to analyze such a disease with RNNs. Choi et al. designed a hybrid model with CNN and bidirectional LSTM for early stroke detection[104]. Fawaz et al. proposed a learnable Fast Fourier Transform method collaborating with LSTM RNN to classify stroke/non-stroke patients[105]. To identify post-stroke patients based on EEG, Sansiagi et al. applied a 3-layer LSTM with discrete wavelet representations[106].
Another physiological application of RNN is the pain assessment, which is challenging to complete in clinical practice. One method to objectively assess the pain level is to detect the protective behaviors by wearable motion sensors and the surface EMG signals. Based on these modalities, Wang et al. applied a 3-layer LSTM model to identify the patients with chronic lower back pain[107]; Li et al. proposed a similar structure with extra dense layers[108]; Yuan et al. constructed an LSTM-based autoencoder to extract the latent features, and then applied attention mechanism to the feature sequences[109]. Another physiological modality is functional near-infrared spectroscopy, which measures the hemodynamic response in the brain. Rojas et al. employed this modality and bidirectional LSTM to classify thermal-induced pain perceptions[110].
Some studies have also attempted to use RNN in EMG signals analysis. These signals reflect the muscular electrical activities and offer a widely adopted method for evaluating the neuromuscular status and identifying body movement. Xia et al. proposed an EMG-based forearm movement estimation system with LSTM [43]. By analyzing the EMG collected from seven upper body muscles, Bengoetxea et al. identified different figure-eight movements[111]. Wang et al. classified the left-hand postures using LSTM [42]. Li et al. built the relationship between EMG and stimulated muscular torque with a NARX strategy [46].
Some physiological signals can also use for identification with the help of RNNs. Salloum et al. and Lynn et al. used ECG signals to conduct biometric identification with RNN, and the accuracies were more than 98% [20], [112]. Moreover, Zhang et al. used ballistocardiogram as input for a similar task[113]. The above studies all compared the accuracies between GRU and LSTM and reported that there were no significant differences between these two units. More discussion will be presented in Section V-B.
Recently, some studies are attempting to measure the swallowing-induced events with on-neck sensors signals and RNN models. Mao et al. applied a multi-layer Elman RNN to track the hyoid bone movement during swallowing[114]. They then combined CNN-1D and GRUs to identify the laryngeal vestibule status (opening or closure)[115]. Importantly, they pointed out that the RNN-based model performed better than the CNN model. Khalifa et al. proposed a hybrid CNN-RNN model to detect the upper esophageal sphincter opening with swallowing acceleration signals. The proposed model used a GRU-based RNN for modeling time dependencies after time-localized feature extraction from raw signals using CNN[116], [117].
Another critical physiological task is human gait analysis, as the data can hold information about medical and neurodegenerative disorders. Zhao et al. applied LSTMs on force-sensitive resistors signals to identify neurodegenerative diseases, such as Parkinson’s disease, Huntington’s disease, and amyotrophic lateral sclerosis[118]. Zhen et al. used LSTM with accelerometer signals collected from the thigh, calf, and foot to identify swing and stance phases in the gait circles[119]. GAO et al. proposed a structure that combined LSTM and 1D CNN to classify the abnormal gait with wearable inertial measurement units[120]. Tortora et al. attempted to decode the gait patterns from EEG signals using LSTM[121]. In this case, the input was the sequential features of EEG, and the labels are swing or stance phase. The application of RNNs in human gait analysis still has a long way to go in physiology since the related studies are relatively limited.
V. Existing issues and future work
Although many studies have reported using RNNs to solve a wide range of problems, as introduced above, there remain several issues facing the further development of RNNs in physiological applications.
A. Finding features for RNN models
1). Knowledge-based feature engineering:
The principal idea of feature extraction is selecting the meaningful components of sequential data to predict events of interest. These features are supposed to be related to these events, and feature extraction requires external knowledge. Involving this knowledge in the RNN model design is a natural methodology, especially when some typical applications’ signal features have been previously explored. For example, in the epilepsy detection studies, wavelet-based methods were prevalent in constructing features from EEG signals, because wavelet transforms were extensively studied and well established to analyze brain activity[70], [122]. The study by Schwab et al. aimed to classify cardiac arrhythmias based on ECG and manually extracted features from engineering and clinical perspectives, such as the amplitude of R point and QRS duration in the ECG waveform[22]. All these features were widely studied biomarkers for cardiac disorders. They designed a 5-layer GRU or bidirectional LSTM with a Markov Model and attention mechanism. Although it was considerably complicated, such a sophisticated structure indeed provided state-of-the-art performance. All the previously reported studies in features extraction will help the RNNs’ design, especially in ECG and EEG-related tasks. However, seeking features might be intractable when the domain knowledge is insufficient[123].
2). Finding features through deep RNNs:
Besides extracting the features by human knowledge, scholars were also aware of the merit in deep learning: it is possible to seek features via the deep architecture itself. In computer vision, deep CNN architecture has been historically successful by generating “feature maps” in intermediate layers. However, the situations were more complicated in dealing with physiological data. If seeking features from the raw data is desirable, increasing the model capacity may be needed. Chauhan and Vig first attempted to feed raw electrocardiographic signals into a three-layer LSTM RNN to conduct anomaly detection[124]. It was quite a deep structure in processing the physiological temporal data. Qiu et al. proposed a three-layer LSTM to remove the power line interference in ECG, in which the input was also raw data[125].
The drawback of the raw signal input is the number of time steps through which the error signal of RNNs has to propagate[22]. The LSTM and GRU are specifically designed to solve the long time dependency problems, and they are not hardware friendly due to the difficulties in parallelized computation[126]. Therefore, the design of hardware accelerators is a path for future work. An alternative way is the modification of the entire deep RNN structures to reduce the calculation of back-propagation through time, as introduced in the next section.
3). Finding features through deep structures:
From 2018, there was a tendency to combine convolutional networks with RNNs (C-RNN) for physiological application[11], [12], [41], [43]. Although the purpose of each network in these studies was different, the deep structures were similar: the CNN layers aimed to extract the local features, and the RNN connected the temporal relationship among these features. Shashikumar et al. treated the 1-D ECG signals as 2-D pictures by calculating the wavelet power spectrum[11]. Based on the spectral “image”, they implemented a 5-layers CNN, at the top of which was a one-layer bidirectional Elman RNN. Unlike Shashikumar’s work, Tan et al. and Andrea et al. used a 1-D CNN as the bottom layer to extract the features of 1-D signals[12]. Andrea et al. also proposed a “siamese architecture” besides the C-RNN to improve accuracy. The structure reported by Xiong et al. was more advanced: they applied the residual block and the batch normalization techniques to cardiac arrhythmias detection with an Elman RNN[41]. These ideas are prevalent in image-related tasks and have been transferred to physiological studies. In the above studies, the CNN layers provide short-term local features and are easy to parallelize in computation. Additionally, when a convolutional layer is introduced, the pooling technique is also applicable to reduce the signal length or time steps, and the computation is therefore simplified. The studies of hybrid structures just started in physiological applications, but they created new ideas in future studies.
B. Choice of RNN unit
Section II-A introduced the Elman, LSTM, and GRU, and they are widely used units in most physiological studies. The LSTM and GRU are famous structures to address the problem of ‘gradient vanishing’, typically associated with the long-term training of Elman RNN. Based on the existing studies shown in Section IV, the best choices for building up the RNN models are LSTM and GRU. However, we should explore more in future studies.
1). LSTM vs. GRU:
Choosing between LSTM and GRU might be a hard question for the model designer. The detailed comparison between these two units was presented in [5]. Also, based on polyphonic music modeling and speech signal modeling, this study suggested no concrete conclusion on which of the two units was better. In terms of physiological studies, we can get similar results. Zhang et al. conducted ballistocardiogram-based biometric identification with two types of units and reported that LSTM and GRU were not significantly different in accuracy[113]. Lynn suggested that GRU was slightly better than LSTM with ECG signals[20]. Dong et al. reported that LSTM and GRU achieved similar performance for sleep stage classification[38]. Latif et al. conducted RNN-based abnormal heartbeat detection with phonocardiography and reported that the accuracy difference of GRU and LSTM was smaller than 1% [127].
Although GRU gave similar results with LSTM, it employs fewer parameters and thus is computationally efficient compared to LSTM. Latif et al. also reported that GRU took 35% less run-time than Bi-directional LSTM while achieving a comparable result, suggesting that GRU was more suitable for deploying mobile or wearable devices with limited hardware resources[127]. From the perspective of algorithm deployment, the GRU is more competitive and hardware-friendly than LSTM.
2). The use of Elman RNN:
Elman RNN may still be valuable for future model design. Although training Elman RNN is suffered by ‘gradient vanishing’ problem, we didn’t see an absolute disappearance in current studies from Section IV. Elman RNN is simple and computationally efficient, and we need to figure out how to address the ‘gradient vanishing’ problem. One way is reducing the length of input sequence with deep models (such as CNN) at a lower level, as suggested by Xiong et al., Shashikumar et al., and Zhang et al.[11], [41], [29]. The model proposed by Xiong et al. even outperformed other LSTM-based models with the same dataset, as shown in Table. I. Mousavi also conducted a Seq2seq model with CNN layers and bidirectional Elman RNN, and they achieved better performance for ECG classification than the LSTM ones[54].
Compared to Elman RNN, LSTM and GRU have additive gating components. According to the analysis carried out by Chung et al., these additions effectively create shortcut paths that bypass multiple temporal steps[5]. For effectively training the Elman RNN, an alternative way is to create the shortcut paths outside the recurrent loops. Several studies attempted to investigate this idea. In atrial fibrillation detection, Shashikumar et al. added a soft attention layer on the top of Elman RNN for the final output[11]. Another profound study was proposed by Zhu et al. in 2020. They designed a dilated Elman RNN structure with skipped time-step connections on each successive layer and reduced the long-term dependency[94]. They also compared the Elman with LSTM and GRU, and suggested that the Elman gave the best performance with significantly reduced parameters number. For the ‘Many-to-One’ scenario, methods B and C (Section III-A) create the shortcuts outside recurrent units. These structures may help the Elman RNN solve the vanished gradient problem since the backpropagation-through-time is not the only way for weight updating.
3). Other choices:
Although LSTM and GRU are widely used RNN units, they are not the only choices for constructing the RNN models. There were several other unit types, which were modified versions of existing units and showed promising results.
Quasi-Recurrent Neural Networks (Q-RNN, 2016)[128].
Q-RNN is a hybrid structure inspired by LSTM. It combines CNN and LSTM, and enables parallel computation across time-steps. Q-RNN achieved comparable results with LSTM on language modeling tasks.
Simple Recurrent Units (SRU, 2017) [129].
SRU holds the idea of cell states while only using forget and reset gates. One improvement is replacing the matrix multiplication of cell state with point-wise multiplication, making the unit computation parallelizable. SRU achieved more robust performance than LSTM and Q-RNN but used less computational time on various language processing tasks.
Independently Recurrent Neural Network (IndRNN, 2018) [130].
IndRNN is similar to the Elman unit. In Eq.(2, the hidden state is updated by matrix multiplication, while IndRNN replaces matrix multiplication with point-wise multiplication (Hadamard product). IndRNN got a higher accuracy on both image and language tasks.
Just Another NETwork (JANET, 2018) [131].
JANET only keeps the forget gate of LSTM and removes all other gates. JANET outperformed the LSTM on MNIST and pM-NIST databases. JANET was also applied to the MIT-BIH ECG database and achieved 89.4% accuracy under cross-subject classification. This performance is higher than the study reported by Hou et al., who also conducted cross-subject classification with the same dataset[45].
All these currently proposed units have shown promising performances with simplified structures. They have not been broadly investigated in physiological applications, and comparative results are still absent.
C. Subject effects
Section III-B introduced the experiments that could employ either within-subject or cross-subject strategies for model training. Meanwhile, in Section IV, the comparisons of existing methods are under the same strategy to avoid the impact of the subject issue. Although this issue exists for all kinds of machine learning designs (e.g., DNN and CNN), it causes obstacles for RNN development.
1). Performance comparison:
Using inconsistent strategies stagnated side-by-side comparisons among the studies. It is hard to design a better model structure if we cannot measure the performance fairly. With the same datasets, performances can get changed by the innovative model designs, and the strategies in the experiments. Some studies attempted to compare the performance discrepancy under different strategies with RNN. Tan et al. conducted both a mixed manner and an equivalent way of cross-subject prediction for ECG classification, and accuracies were 99.85% and 95.76%, respectively[12]. For the same task, Hou et al. also compared beats-based cross-validation (mixed manner) and record-based cross-validation (cross-subject prediction), and the accuracies were 99.74% and 85.20%, respectively[45]. In the EEG emotion recognition task, Li et al. used both mixed manner and leave-one-subject-out cross-validation, and the accuracies were 92.38% and 83.28%, respectively[62]. Thodoroff et al. employed both patient-specific and cross-patient settings for seizure detection, and the sensitivities were 95–100% and 85%, respectively[69].
For even comparison, an ideal way is using a standard protocol for all the practitioners in a specific field. Building up universal standard test datasets is challenging and requires collaboration across organizations and disciplines. Fortunately, the effort is currently ongoing, such as the dataset provided by Computing in Cardiology Challenge, in which the testing set was strictly defined for all the participating groups[15], as shown in Table I. Some research groups would also spontaneously use the same protocol, such as the protocol proposed by Zheng et al. for EEG emotion recognition[61], and leave-one-subject-out cross-validation in pain level assessment[109]. With the evenly comparative results, we can objectively evaluate the development and have a clear vision for future model design.
Considering the subject effect in computational physiology, the deep learning practitioners should clearly describe how they conduct the validation process. Our survey found that it is challenging to compare some performances among published studies since sometimes the types of validation were not clearly stated. Moreover, the comparative analysis should be carefully carried out. Picking up the reported values from other studies is arbitrary because they may not use the same strategy. Fortunately, some studies conducted comparison by re-performing the methods proposed from the other ones under the same strategy. Based on these investigations, we could see the advantages of the RNN model, as introduced in Section IV.
2). Practicability:
Most pilot studies were in the prototype stage, and adopting a within-subject strategy was for the proof of concept purpose. Although they suggested that the RNN could achieve better results, we also need to consider whether the within-subject study is practically feasible.The models trained under within-subject strategies must be re-trained (or fine-tuned) with new training set for the unseen subjects. This is not typically an issue if the devices have sufficient computational power. The problem is whether human experts are indispensable for labeling new data. If the labels for unseen subjects can be automatically obtained without human experts, personalized models are practically feasible. One example is blood glucose prediction, in which monitoring devices could measure the glucose level continuously. Therefore, it is efficient for the pre-trained models to capture the training pairs for a new subject, and the inter-person variability issue could be addressed in practice.
If the labels are not easy to acquire and human experts are necessary for the labeling process, off-line fine-tuning for a personalized model is the only choice. For an unseen subject, the model tuning procedures will be constrained by many factors, such as the availability of the human raters. Since within-subject studies have already achieved very high performance, more studies should focus on the scalability of the personalized models. In the future, we would like to see investigations that practically implement the within-subject model for the unseen subjects at the inference stage, especially when the recordings of unseen subjects were not collected or labeled before training the pre-trained model. These studies would enlighten the way to make protocols of data collection, human experts scheduling, inter/intra-rater reliability analysis, off-line tuning for the personalized model, and model deployment for the unseen subjects.
3). Implementation of cross-subject prediction:
Developing the physiological system for unseen patients is the most nature circumstance. As introduced in Section IV and V-C1, the accuracies of some studies under within-subjects classification tasks have already achieved more than 90%, even approaching 100%. However, cross-subject prediction accuracies stayed 80% ~ 90%. In the future, we should consider the hypothesis that it is possible to predict one person’s status by other persons’ examples via RNN models. The inter-patient variability will seriously affect the results when the cross-subject prediction is conducted, but it does not suggest the impossibility of capturing the personalized pattern based on training groups. For example, the state-of-the-art cross-subject performance has reached 99.53% for ECG classification by using the Seq2seq model[54].
We can imagine whether the human experts(raters) are ‘personalized’ or ‘user specific’ when labeling the data. However, this expert-level performance is only obtainable when the studied cohort is large, i.e., hundreds of subjects or more [31], [91]. Although some datasets were manually annotated and publicly available, such as MIT-BIH of ECG signals[16], DEAP of EEG signals[58], more data is still needed to improve the model’s cross-subject prediction generalization. Collecting more datasets for training is a solution, but it is challenging due to practical constraints, such as time cost for labeling, monetary expense, ethical review, and privacy problem.
The individual characteristics impede the model’s generalization across the patients, and the samples may be conditionally identical distributed under different subjects. However, rigorous mathematical or numerical analysis is absent, and it is unclear how the inter-subject variability impacts the model’s generalization error bound. Meanwhile, we need to investigate whether the input signals contain the class information and represent the individual characteristics. Unsupervised learning methods, such as the autoencoder model, may help us answer those questions by analyzing the latent space. A typical structure involving unsupervised learning was proposed by Dong et al. for blood glucose prediction, in which a combined model with the K-mean method and RNN model was proposed (Clu-RNN)[93]. The idea was that the input vectors collected from some subgroups might share similar patterns. Moreover, Li et al. attempted to use the unsupervised learning method first to search the sequence of the features for emotion recognition[66]. This method achieved the best cross-subject accuracy for the SEED dataset compared with other studies.
In physiological applications, each patient could be characterized by external factors, such as gender, age, weight, body mass index, medical history, and personal profile. Suppose the model cannot capture the complete personalized information based on the input signals. In that case, we can encode the external factors as an auxiliary input vector and involve them in the model design. Liu et al. adopted such an idea in blood pressure prediction by embedding a contextual information cue (personal profile), including age, gender, body mass index, height, weight, and temperature[102]. In practice, the personal information is easy to achieve and may help to improve the model’s performance for cross-subject prediction.
Data augmentation is a widely used way for improving the model generalization. Augmentation is not readily accessible for sequential physiological input since there is no way to augment the inputs from unseen subjects. If the latent factors, some mathematical descriptions of training subjects, follow some distribution, would it be possible to model this and sample it to get more training data? Generative models are good options. Such ideas have already been successfully implemented in other fields, but more effort is needed in processing the physiological sequences.
Another way to obtain more data is multi-task learning. We introduced that the emotion recognition, seizure detection, and sleep stage classification tasks adopted EEG(PSG) signals as inputs. Although they were collected from different domains and varied setups, they may share common features. Joint datasets and multi-task learning could extract these features. This method increases the number of subjects and achieves more robust features. Additionally, some unsupervised learning methods, such as RNN-based autoencoder, allow involving more EEG datasets without any annotation. Multi-task learning may offer us new solutions to improve the cross-subject prediction performance for the task of sub-domain and ideas for other big-data-related physiological problems, such as transfer learning.
D. Other opportunities
-
‐
Ensemble model. There are so many ways of constructing RNN models, as introduced in Section IV. Also, there are many choices for feature extraction, hyperparameters setup, and the types of RNN units. By including different deep learning architectures, the ensemble model partially addresses the problem of searching for the optimal structure while improving robustness. This technique is prevalent in many fields and has shown encouraging results. In physiological tasks, only several studies attempted to implement this technique with RNNs, such as the studies carried out by Schwab et al. and Zihlmann et al. for ECG classification[22], [39].
-
‐
Seq2seq model. In Section III-A, we described the ”Many-to-Many” scenario, in which both the input and output are sequences, and we also discussed the typical way of model construction. Alternatively, the Seq2seq model is also suitable for the ”Many-to-Many” scenario[80]. This model is well-developed in natural language processing, but it is not drawing enough attention in the physiological area. Fox et al. borrowed the idea of the Seq2seq model and designed the PolySeqMO to predict the blood glucose[95]. Mousavi et al. adopted the Seq2seq model to classify the sleep stages[83]. They also proposed a similar structure for ECG classification and reached the best performance (shown in Table. I)[54]. Moreover, some advanced techniques accompanied by Seq2seq, such as attention mechanism[132], transformer-based architectures[133], can also be transferred to physiological applications.
-
‐
Generative model. As stated in Section V-C3, generative models may help for data augmentation. Generative adversarial network (GAN) has mainly been developed and applied to images or artificial audio generation[134], [135]. Recent studies have already attempted to generate EEG and ECG signals with advanced techniques like Wasserstein GANs with gradient penalty [136], [137]. Besides data augmentation, GANs can also serve other physiological tasks, such as anomaly detection with well-trained discriminators, signal denoising, and signal synthesis/restoration for missing channel(s) of multiple-sensor systems[138], [139]. Variational Autoencoder (VAE) is another kind of generative model, which offers an alternative manner for describing the distribution of given data in latent space[140]. This model could also generate more physiological sequences, such as ECG generation reported by Kuznetsov et al.[141]. VAE may enlighten new studies in computational physiology, but more investigations are still needed.
VI. Conclusion
This review provided a comprehensive overview of existing studies attempting to apply RNNs in the field of human physiology. The RNN is particularly amenable for monitoring and detecting various physiological states in real-time due to its capability of processing time-dependent sequential data. Our survey revealed that RNNs have already been widely studied in diverse healthcare applications. The modern neural networks and computational power techniques have facilitated addressing health issues.
Acknowledgment
This review work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R01HD092239, R01HD074819. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies

Shitong Mao received the BSc and MSc degree from Harbin Institute of Technology, China, in 2008 and 2010, respectively. He is working toward the Ph.D. degree at the University of Pittsburgh, Swanson school of engineering, Department of Electrical and Computer Engineering. His current research interests include pattern recognition, machine learning, biomedical signal processing, computer vision, electrical devices development, and their applications in healthcare systems.

Ervin Sejdić (S’00-M’08-SM’16) received the B.E.Sc. and Ph.D. degrees in electrical engineering from the University of Western Ontario, London, ON, Canada, in 2002 and 2008, respectively. From 2008 to 2010, he was a Post-Doctoral Fellow with the University of Toronto, Toronto, ON, Canada, with a cross-appointment with Bloorview Kids Rehab, Toronto, ON, Canada, Canada’s largest children’s rehabilitation teaching hospital. From 2010 to 2011, he was a Research Fellow with the Harvard Medical School, Boston, MA, USA, with a cross-appointment with the Beth Israel Deaconess Medical Center. In 2011, he joined the Department of Electrical and Computer Engineering, University of Pittsburgh, Pittsburgh, PA, USA. as a tenure-track Assistant Professor. In 2017, he was promoted to a tenured Associate Professor. He holds secondary appointments with the Department of Bioengineering, Swanson School of Engineering, with the Department of Biomedical Informatics, School of Medicine, and also with the Intelligent Systems Program, School of Computing and Information, University of Pittsburgh. He joined the University of Toronto and North York General Hospital in 2021. His current research interests include biomedical signal processing, gait analyses, swallowing difficulties, advanced information systems in medicine, rehabilitation engineering, assistive technologies, and anticipatory medical devices.
Dr. Sejdić was a recipient of many awards. As a graduate student, he was awarded two prestigious awards from the Natural Sciences and Engineering Research Council of Canada. In 2010, he was the recipient of the Melvin First Young Investigator’s Award from the Institute for Aging Research at Hebrew Senior Life, Boston, MA, USA. In 2016, President Obama named Prof. Sejdić as a recipient of the Presidential Early Career Award for Scientists and Engineers, the highest honor bestowed by the U.S. Government on science and engineering professionals in the early stages of their independent research careers. In 2017, he was the recipient of the National Science Foundation CAREER Award, which is the National Science Foundation’s most prestigious award in support of career-development activities of those scholars who most effectively integrate research and education within the context of the mission of their organization.
Footnotes
Some studies in this field also named such a scenario as the ‘Sequence-to-Sequence’ sleep staging problem, which has a similar name with the well-known Seq2seq model[80]. For disambiguation, we use the term ‘Many-to-many’ to describe the scenario in Fig.5(a)
Some EEG or PSG studies named a signal slice in a specific window as ‘epoch’. However, in deep learning studies, ‘epoch’ usually refers to the iterations that an entire dataset is used for training a model. For disambiguation, we use ‘chunk’ instead of ‘epoch’ to represent the signal slice.
Contributor Information
Shitong Mao, Department of Electrical and Computer Engineering, University of Pittsburgh, Pittsburgh, PA, 15213, USA..
Ervin Sejdić, Edward S. Rogers Department of Electrical and Computer Engineering, Faculty of Applied Science and Engineering, University of Toronto, Toronto, Ontario, Canada, and North York General Hospital, Toronto, Ontario, Canada..
References
- [1].“Openai five benchmark: Results,” https://blog.openai.com/openai-five-benchmark-results/, 2018-August-06.
- [2].Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, and Thrun S, “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, p. 115, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lipton ZC, Berkowitz J, and Elkan C, “A critical review of recurrent neural networks for sequence learning,” arXiv preprint arXiv:1506.00019, 2015.
- [4].Khalifa Y, Mandic D, and Sejdić E, “A review of hidden markov models and recurrent neural networks for event detection and localization in biomedical signals,” Information Fusion, vol. 69, pp. 52–72, 2021. [Google Scholar]
- [5].Chung J, Gulcehre C, Cho K, and Bengio Y, “Empirical evaluation of gated recurrent neural networks on sequence modeling,” arXiv preprint arXiv:1412.3555, 2014.
- [6].Graves A, Supervised Sequence Labelling Berlin, Heidelberg: Springer, 2012, pp. 5–13. [Google Scholar]
- [7].He L, Jiang D, Yang L, Pei E, Wu P, and Sahli H, “Multimodal affective dimension prediction using deep bidirectional long short-term memory recurrent neural networks,” in Proceedings of the 5th International Workshop on Audio/Visual Emotion Challenge. ACM, 2015, pp. 73–80.
- [8].Goodfellow I, Bengio Y, and Courville A, Deep Learning. MIT Press, 2016.
- [9].Elman JL, “Finding structure in time,” Cognitive Science, vol. 14, no. 2, pp. 179–211, 1990. [Google Scholar]
- [10].Pascanu R, Mikolov T, and Bengio Y, “On the difficulty of training recurrent neural networks,” in Proceedings of the 30th International Conference on Machine Learning, 2013, pp. 1310–1318.
- [11].Shashikumar SP, Shah AJ, Clifford GD, and Nemati S, “Detection of paroxysmal atrial fibrillation using attention-based bidirectional recurrent neural networks,” in Proceedings of the 24th ACM Special Interest Group on Knowledge Discovery & Data Mining(SIGKDD), 2018, pp. 715–723.
- [12].Tan JH, Hagiwara Y, Pang W, Lim I, Oh SL, Adam M, San Tan R, Chen M, and Acharya UR, “Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals,” Computers in Biology and Medicine, vol. 94, pp. 19–26, 2018. [DOI] [PubMed] [Google Scholar]
- [13].Brady K, Gwon Y, Khorrami P, Godoy E, Campbell W, Dagli C, and Huang TS, “Multi-modal audio, video and physiological sensor learning for continuous emotion prediction,” in Proceedings of the 6th International Workshop on Audio/Visual Emotion Challenge. ACM, 2016, pp. 97–104.
- [14].Phan H, Andreotti F, Cooray N, Chén OY, and De Vos M, “SeqSleepNet: End-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging,” IEEE Transactions on Neural Systems and Rehabilitation Engineering, vol. 27, no. 3, pp. 400–410, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng C-K, and Stanley HE, “Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals,” circulation, vol. 101, no. 23, pp. e215–e220, 2003. [DOI] [PubMed] [Google Scholar]
- [16].Moody GB and Mark RG, “The impact of the MIT-BIH arrhythmia database,” IEEE Engineering in Medicine and Biology Magazine, vol. 20, no. 3, pp. 45–50, 2001. [DOI] [PubMed] [Google Scholar]
- [17].Oh SL, Ng EY, San Tan R, and Acharya UR, “Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats,” Computers in Biology and Medicine, vol. 102, pp. 278–287, 2018. [DOI] [PubMed] [Google Scholar]
- [18].Chang Y-C, Wu S-H, Tseng L-M, Chao H-L, and Ko C-H, “AF detection by exploiting the spectral and temporal characteristics of ECG signals with the LSTM model,” in 2018 Computing in Cardiology Conference (CinC), vol. 45. IEEE, 2018, pp. 1–4. [Google Scholar]
- [19].Hofmann SM, Klotzsche F, Mariola A, Nikulin VV, Villringer A, and Gaebler M, “Decoding subjective emotional arousal during a naturalistic VR experience from EEG using LSTMs,” in 2018 IEEE International Conference on Artificial Intelligence and Virtual Reality (AIVR) IEEE, 2018, pp. 128–131. [Google Scholar]
- [20].Lynn HM, Pan SB, and Kim P, “A deep bidirectional GRU network model for biometric electrocardiogram classification based on recurrent neural networks,” IEEE Access, vol. 7, pp. 145 395–145 405, 2019. [Google Scholar]
- [21].Supratak A, Dong H, Wu C, and Guo Y, “DeepSleepNet: a model for automatic sleep stage scoring based on raw single-channel EEG,” IEEE Transactions on Neural Systems and Rehabilitation Engineering, vol. 25, no. 11, pp. 1998–2008, 2017. [DOI] [PubMed] [Google Scholar]
- [22].Schwab P, Scebba GC, Zhang J, Delai M, and Karlen W, “Beat by beat: Classifying cardiac arrhythmias with recurrent neural networks,” 2017 Computing in Cardiology (CinC), pp. 1–4, 2017.
- [23].Phan H, Andreotti F, Cooray N, Chén OY, and De Vos M, “Automatic sleep stage classification using single-channel EEG: Learning sequential features with attention-based recurrent neural networks,” in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE, 2018, pp. 1452–1455. [DOI] [PubMed] [Google Scholar]
- [24].Yildirim Ö, “A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification,” Computers in Biology and Medicine, vol. 96, pp. 189–202, 2018. [DOI] [PubMed] [Google Scholar]
- [25].Yildirim O, Baloglu UB, Tan R-S, Ciaccio EJ, and Acharya UR, “A new approach for arrhythmia classification using deep coded features and LSTM networks,” Computer Methods and Programs in Biomedicine, vol. 176, pp. 121–133, 2019. [DOI] [PubMed] [Google Scholar]
- [26].Liu F, Zhou X, Cao J, Wang Z, Wang H, and Zhang Y, “A LSTM and CNN based assemble neural network framework for arrhythmias classification,” in 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE, 2019, pp. 1303–1307. [Google Scholar]
- [27].Tsiouris KM, Pezoulas VC, Zervakis M, Konitsiotis S, Koutsouris DD, and Fotiadis DI, “A long short-term memory deep learning network for the prediction of epileptic seizures using EEG signals,” Computers in Biology and Medicine, vol. 99, pp. 24–37, 2018. [DOI] [PubMed] [Google Scholar]
- [28].Xing X, Li Z, Xu T, Shu L, Hu B, and Xu X, “SAE+ LSTM: A new framework for emotion recognition from multi-channel EEG,” Frontiers in Neurorobotics, vol. 13, p. 37, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang T, Zheng W, Cui Z, Zong Y, and Li Y, “Spatial–temporal recurrent neural network for emotion recognition,” IEEE Transactions on Cybernetics, vol. 49, no. 3, pp. 839–847, 2018. [DOI] [PubMed] [Google Scholar]
- [30].Li X, Song D, Zhang P, Yu G, Hou Y, and Hu B, “Emotion recognition from multi-channel EEG data through convolutional recurrent neural network,” in 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE, 2016, pp. 352–359. [Google Scholar]
- [31].Phan H, Chén OY, Koch P, Lu Z, McLoughlin I, Mertins A, and De Vos M, “Towards more accurate automatic sleep staging via deep transfer learning,” arXiv preprint arXiv:1907.13177, 2019. [DOI] [PubMed]
- [32].Wang G, Zhang C, Liu Y, Yang H, Fu D, Wang H, and Zhang P, “A global and updatable ECG beat classification system based on recurrent neural networks and active learning,” Information Sciences, vol. 501, pp. 523–542, 2019. [Google Scholar]
- [33].Ringeval F, Eyben F, Kroupi E, Yuce A, Thiran J-P, Ebrahimi T, Lalanne D, and Schuller B, “Prediction of asynchronous dimensional emotion ratings from audiovisual and physiological data,” Pattern Recognition Letters, vol. 66, pp. 22–30, 2015. [Google Scholar]
- [34].Su P, Ding XR, Zhang YT, Liu J, Miao F, and Zhao N, “Long-term blood pressure prediction with deep recurrent neural networks,” in IEEE Engineering in Medicine and Biology Society International Conference on Biomedical & Health Informatics (BHI) IEEE, 2018, pp. 323–328. [Google Scholar]
- [35].Yang TCI and Hsieh H, “Classification of acoustic physiological signals based on deep learning neural networks with augmented features,” in Computing in Cardiology Conference (CinC) IEEE, 2016, pp. 569–572. [Google Scholar]
- [36].Hussain AJ, Fergus P, Al-Askar H, Al-Jumeily D, and Jager F, “Dynamic neural network architecture inspired by the immune algorithm to predict preterm deliveries in pregnant women,” Neurocomputing, vol. 151, pp. 963–974, 2015. [Google Scholar]
- [37].Singh S, Pandey SK, Pawar U, and Janghel RR, “Classification of ECG arrhythmia using recurrent neural networks,” Procedia Computer Science, vol. 132, pp. 1290–1297, 2018. [Google Scholar]
- [38].Dong H, Supratak A, Pan W, Wu C, Matthews PM, and Guo Y, “Mixed neural network approach for temporal sleep stage classification,” IEEE Transactions on Neural Systems and Rehabilitation Engineering, vol. 26, no. 2, pp. 324–333, 2018. [DOI] [PubMed] [Google Scholar]
- [39].Zihlmann M, Perekrestenko D, and Tschannen M, “Convolutional recurrent neural networks for electrocardiogram classification,” in 2017 Computing in Cardiology (CinC) IEEE, 2017, pp. 1–4. [Google Scholar]
- [40].Warrick P and Homsi MN, “Cardiac arrhythmia detection from ECG combining convolutional and long short-term memory networks,” in 2017 Computing in Cardiology (CinC) IEEE, 2017, pp. 1–4. [Google Scholar]
- [41].Xiong Z, Nash MP, Cheng E, Fedorov VV, Stiles MK, and Zhao J, “ECG signal classification for the detection of cardiac arrhythmias using a convolutional recurrent neural network,” Physiological Measurement, vol. 39, no. 9, p. 094006, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang W, Chen B, Xia P, Hu J, and Peng Y, “Sensor fusion for myoelectric control based on deep learning with recurrent convolutional neural networks,” Artificial Organs, vol. 42, no. 9, pp. E272–E282, 2018. [DOI] [PubMed] [Google Scholar]
- [43].Xia P, Hu J, and Peng Y, “EMG-based estimation of limb movement using deep learning with recurrent convolutional neural networks,” Artificial Organs, vol. 42, no. 5, pp. E67–E77, 2018. [DOI] [PubMed] [Google Scholar]
- [44].Futoma J, Hariharan S, and Heller K, “Learning to detect sepsis with a multitask Gaussian process RNN classifier,” arXiv preprint arXiv:1706.04152, 2017.
- [45].Hou B, Yang J, Wang P, and Yan R, “LSTM based auto-encoder model for ECG arrhythmias classification,” IEEE Transactions on Instrumentation and Measurement, 2019.
- [46].Li Z, Hayashibe M, Fattal C, and Guiraud D, “Muscle fatigue tracking with evoked EMG via recurrent neural network: Toward personalized neuroprosthetics,” IEEE Computational Intelligence Magazine, vol. 9, no. 2, pp. 38–46, 2014. [Google Scholar]
- [47].Li Y, Huang J, Zhou H, and Zhong N, “Human emotion recognition with electroencephalographic multidimensional features by hybrid deep neural networks,” Applied Sciences, vol. 7, no. 10, p. 1060, 2017. [Google Scholar]
- [48].Daoud H and Bayoumi MA, “Efficient epileptic seizure prediction based on deep learning,” IEEE Transactions on Biomedical Circuits and Systems, vol. 13, no. 5, pp. 804–813, 2019. [DOI] [PubMed] [Google Scholar]
- [49].Dong Y, Wen R, Zhang K, and Zhang L, “A novel RNN-based blood glucose prediction approach using population and individual characteristics,” in 2019 IEEE 7th International Conference on Bioinformatics and Computational Biology (ICBCB) IEEE, 2019, pp. 145–149. [Google Scholar]
- [50].Chauhan S, Vig L, and Ahmad S, “ECG anomaly class identification using LSTM and error profile modeling,” Computers in Biology and Medicine, vol. 109, pp. 14–21, 2019. [DOI] [PubMed] [Google Scholar]
- [51].Zhang C, Wang G, Zhao J, Gao P, Lin J, and Yang H, “Patient-specific ECG classification based on recurrent neural networks and clustering technique,” in 13th International Association of Science and Technology for Development (IASTED) Conference on Biomedical Engineering (BioMed) IEEE, 2017, pp. 63–67. [Google Scholar]
- [52].Maknickas V and Maknickas A, “Atrial fibrillation classification using QRS complex features and LSTM,” in 2017 Computing in Cardiology (CinC) IEEE, 2017, pp. 1–4. [Google Scholar]
- [53].Saadatnejad S, Oveisi M, and Hashemi M, “LSTM-based ECG classification for continuous monitoring on personal wearable devices,” IEEE Journal of Biomedical and Health Informatics, 2019. [DOI] [PubMed]
- [54].Mousavi S and Afghah F, “Inter-and intra-patient ECG heartbeat classification for arrhythmia detection: a sequence to sequence deep learning approach,” in 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE, 2019, pp. 1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Acharya UR, Fujita H, Lih OS, Adam M, Tan JH, and Chua CK, “Automated detection of coronary artery disease using different durations of ECG segments with convolutional neural network,” Knowledge-Based Systems, vol. 132, pp. 62–71, 2017. [Google Scholar]
- [56].Kachuee M, Fazeli S, and Sarrafzadeh M, “ECG heartbeat classification: A deep transferable representation,” in 2018 IEEE International Conference on Healthcare Informatics (ICHI) IEEE, 2018, pp. 443–444. [Google Scholar]
- [57].Jun TJ, Nguyen HM, Kang D, Kim D, Kim D, and Kim Y-H, “ECG arrhythmia classification using a 2-d convolutional neural network,” arXiv preprint arXiv:1804.06812, 2018.
- [58].Koelstra S, Muhl C, Soleymani M, Lee J-S, Yazdani A, Ebrahimi T, Pun T, Nijholt A, and Patras I, “DEAP: A database for emotion analysis; using physiological signals,” IEEE Transactions on Affective Computing, vol. 3, no. 1, pp. 18–31, 2012. [Google Scholar]
- [59].Shu L, Xie J, Yang M, Li Z, Li Z, Liao D, Xu X, and Yang X, “A review of emotion recognition using physiological signals,” Sensors, vol. 18, no. 7, p. 2074, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Luft CDB and Bhattacharya J, “Aroused with heart: modulation of heartbeat evoked potential by arousal induction and its oscillatory correlates,” Scientific Reports, vol. 5, p. 15717, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zheng W-L and Lu B-L, “Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks,” IEEE Transactions on Autonomous Mental Development, vol. 7, no. 3, pp. 162–175, 2015. [Google Scholar]
- [62].Li Y, Zheng W, Cui Z, Zhang T, and Zong Y, “A novel neural network model based on cerebral hemispheric asymmetry for EEG emotion recognition.” in International Joint Conferences on Artificial Intelligence(IJCAI), 2018, pp. 1561–1567.
- [63].Li Y, Zheng W, Zong Y, Cui Z, Zhang T, and Zhou X, “A bi-hemisphere domain adversarial neural network model for EEG emotion recognition,” IEEE Transactions on Affective Computing, 2018.
- [64].Alhagry S, Fahmy AA, and El-Khoribi RA, “Emotion recognition based on EEG using LSTM recurrent neural network,” Emotion, vol. 8, no. 10, pp. 355–358, 2017. [Google Scholar]
- [65].Yang Y, Wu Q, Qiu M, Wang Y, and Chen X, “Emotion recognition from multi-channel EEG through parallel convolutional recurrent neural network,” in 2018 International Joint Conference on Neural Networks (IJCNN) IEEE, 2018, pp. 1–7. [Google Scholar]
- [66].Li X, Zhao Z, Song D, Zhang Y, Pan J, Wu L, Huo J, Niu C, and Wang D, “Latent factor decoding of multi-channel EEG for emotion recognition through autoencoder-like neural networks,” Frontiers in Neuroscience, vol. 14, p. 87, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Soleymani M, Asghari-Esfeden S, Fu Y, and Pantic M, “Analysis of EEG signals and facial expressions for continuous emotion detection,” IEEE Transactions on Affective Computing, vol. 7, no. 1, pp. 17–28, 2015. [Google Scholar]
- [68].Pippa E, Zacharaki EI, Mporas I, Tsirka V, Richardson MP, Koutroumanidis M, and Megalooikonomou V, “Improving classification of epileptic and non-epileptic EEG events by feature selection,” Neurocomputing, vol. 171, pp. 576–585, 2016. [Google Scholar]
- [69].Thodoroff P, Pineau J, and Lim A, “Learning robust features using deep learning for automatic seizure detection,” in Machine Learning for Healthcare Conference, 2016, pp. 178–190.
- [70].Raghu S, Sriraam N, and Kumar GP, “Classification of epileptic seizures using wavelet packet log energy and norm entropies with recurrent Elman neural network classifier,” Cognitive Neurodynamics, vol. 11, no. 1, pp. 51–66, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ahmedt-Aristizabal D, Fookes C, Nguyen K, and Sridharan S, “Deep classification of epileptic signals,” in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE, 2018, pp. 332–335. [DOI] [PubMed] [Google Scholar]
- [72].Huang C, Chen W, and Cao G, “Automatic epileptic seizure detection via attention-based CNN-BiRNN,” in 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE, 2019, pp. 660–663. [Google Scholar]
- [73].Abbasi MU, Rashad A, Basalamah A, and Tariq M, “Detection of epilepsy seizures in neo-natal EEG using LSTM architecture,” IEEE Access, vol. 7, pp. 179 074–179 085, 2019. [Google Scholar]
- [74].Hussein R, Palangi H, Ward RK, and Wang ZJ, “Optimized deep neural network architecture for robust detection of epileptic seizures using EEG signals,” Clinical Neurophysiology, vol. 130, no. 1, pp. 25–37, 2019. [DOI] [PubMed] [Google Scholar]
- [75].Chiang C-Y, Chang N-F, Chen T-C, Chen H-H, and Chen L-G, “Seizure prediction based on classification of EEG synchronization patterns with on-line retraining and post-processing scheme,” in 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE, 2011, pp. 7564–7569. [DOI] [PubMed] [Google Scholar]
- [76].Andrzejak RG, Lehnertz K, Mormann F, Rieke C, David P, and Elger CE, “Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state,” Physical Review E, vol. 64, no. 6, p. 061907, 2001. [DOI] [PubMed] [Google Scholar]
- [77].Acharya UR, Oh SL, Hagiwara Y, Tan JH, and Adeli H, “Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals,” Computers in Biology and Medicine, vol. 100, pp. 270–278, 2018. [DOI] [PubMed] [Google Scholar]
- [78].Lu D and Triesch J, “Residual deep convolutional neural network for EEG signal classification in epilepsy,” arXiv preprint arXiv:1903.08100, 2019.
- [79].Wulff K, Gatti S, Wettstein JG, and Foster RG, “Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease,” Nature Reviews Neuroscience, vol. 11, no. 8, pp. 589–599, 2010. [DOI] [PubMed] [Google Scholar]
- [80].Sutskever I, Vinyals O, and Le QV, “Sequence to sequence learning with neural networks,” in Advances in Neural Information Processing Systems, 2014, pp. 3104–3112.
- [81].Michielli N, Acharya UR, and Molinari F, “Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals,” Computers in Biology and Medicine, vol. 106, pp. 71–81, 2019. [DOI] [PubMed] [Google Scholar]
- [82].Phan H, Chén OY, Koch P, Mertins A, and De Vos M, “Fusion of end-to-end deep learning models for sequence-to-sequence sleep staging,” in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE, 2019, pp. 1829–1833. [DOI] [PubMed] [Google Scholar]
- [83].Mousavi S, Afghah F, and Acharya UR, “SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach,” PloS one, vol. 14, no. 5, p. e0216456, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].O’reilly C, Gosselin N, Carrier J, and Nielsen T, “Montreal archive of sleep studies: an open-access resource for instrument benchmarking and exploratory research,” Journal of Sleep Research, vol. 23, no. 6, pp. 628–635, 2014. [DOI] [PubMed] [Google Scholar]
- [85].Sterr A, Ebajemito JK, Mikkelsen KB, Bonmati-Carrion MA, Santhi N, Della Monica C, Grainger L, Atzori G, Revell V, Debener S et al. , “Sleep EEG derived from behind-the-ear electrodes (cEEGrid) compared to standard polysomnography: a proof of concept study,” Frontiers in Human Neuroscience, vol. 12, p. 452, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rosenberg RS and Van Hout S, “The American academy of sleep medicine inter-scorer reliability program: sleep stage scoring,” Journal of clinical sleep medicine, vol. 9, no. 01, pp. 81–87, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tsinalis O, Matthews PM, Guo Y, and Zafeiriou S, “Automatic sleep stage scoring with single-channel EEG using convolutional neural networks,” arXiv preprint arXiv:1610.01683, 2016.
- [88].Tsinalis O, Matthews PM, and Guo Y, “Automatic sleep stage scoring using time-frequency analysis and stacked sparse autoencoders,” Annals of Biomedical Engineering, vol. 44, no. 5, pp. 1587–1597, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Phan H, Andreotti F, Cooray N, Chén OY, and De Vos M, “Joint classification and prediction CNN framework for automatic sleep stage classification,” IEEE Transactions on Biomedical Engineering, vol. 66, no. 5, pp. 1285–1296, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mendola N, Chen T, Gu Q, Eberhardt M, and Saydah S, “Prevalence of total, diagnosed, and undiagnosed diabetes among adults: United States, 2013–2016.” National Center for Health Statistics (NCHS) Data Brief, no. 319, pp. 1–8, 2018. [PubMed] [Google Scholar]
- [91].Gu W, Zhou Y, Zhou Z, Liu X, Zou H, Zhang P, Spanos CJ, and Zhang L, “Sugarmate: Non-intrusive blood glucose monitoring with smartphones,” Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies, vol. 1, no. 3, pp. 1–27, 2017. [Google Scholar]
- [92].He M, Gu W, Kong Y, Zhang L, Spanos CJ, and Mosalam KM, “CausalBG: Causal recurrent neural network for the blood glucose inference with IoT platform,” IEEE Internet of Things Journal, vol. 7, no. 1, pp. 598–610, 2019. [Google Scholar]
- [93].Dong Y, Wen R, Li Z, Zhang K, and Zhang L, “Clu-RNN: A new RNN based approach to diabetic blood glucose prediction,” in 2019 IEEE 7th International Conference on Bioinformatics and Computational Biology (ICBCB) IEEE, 2019, pp. 50–55. [Google Scholar]
- [94].Zhu T, Li K, Chen J, Herrero P, and Georgiou P, “Dilated recurrent neural networks for glucose forecasting in type 1 diabetes,” Journal of Healthcare Informatics Research, pp. 1–17, 2020. [DOI] [PMC free article] [PubMed]
- [95].Fox I, Ang L, Jaiswal M, Pop-Busui R, and Wiens J, “Deep multi-output forecasting: Learning to accurately predict blood glucose trajectories,” in Proceedings of the 24th International Conference on Knowledge Discovery & Data Mining, 2018, pp. 1387–1395.
- [96].Marling C and Bunescu RC, “The OhioT1DM dataset for blood glucose level prediction.” in Knowledge Discovery in Healthcare Data co-located with the 27th International Joint Conference on Artificial Intelligence, 2018, pp. 60–63.
- [97].Pesl P, Herrero P, Reddy M, Xenou M, Oliver N, Johnston D, Toumazou C, and Georgiou P, “An advanced bolus calculator for type 1 diabetes: system architecture and usability results,” IEEE Journal of Biomedical and Health Informatics, vol. 20, no. 1, pp. 11–17, 2015. [DOI] [PubMed] [Google Scholar]
- [98].Singh RR, Conjeti S, and Banerjee R, “A comparative evaluation of neural network classifiers for stress level analysis of automotive drivers using physiological signals,” Biomedical Signal Processing and Control, vol. 8, no. 6, pp. 740–754, 2013. [Google Scholar]
- [99].Mastorocostas PA and Theocharis JB, “A stable learning algorithm for block-diagonal recurrent neural networks: Application to the analysis of lung sounds,” IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), vol. 36, no. 2, pp. 242–254, 2006. [DOI] [PubMed] [Google Scholar]
- [100].Cheng M, Sori WJ, Jiang F, Khan A, and Liu S, “Recurrent neural network based classification of ECG signal features for obstruction of sleep apnea detection,” in IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC) IEEE, 2017, pp. 199–202. [Google Scholar]
- [101].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778.
- [102].Liu J, Wu Y, Yuan Z, and Sun X, “Blood pressure prediction with multi-cue based rbf and LSTM model,” in 2018 9th International Conference on Information Technology in Medicine and Education (ITME) IEEE, 2018, pp. 72–76. [Google Scholar]
- [103].Bahrami Rad A, Zabihi M, Zhao Z, Gabbouj M, Katsaggelos AK, and Särkkä S, “Automated polysomnography analysis for detection of non-apneic and non-hypopneic arousals using feature engineering and a bidirectional LSTM network,” arXiv, pp. arXiv–1909, 2019.
- [104].Choi Y-A, Park S-J, Jun J-A, Pyo C-S, Cho K-H, Lee H-S, and Yu J-H, “Deep learning-based stroke disease prediction system using real-time bio signals,” Sensors, vol. 21, no. 13, p. 4269, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fawaz S, Sim KS, and Tan SC, “Encoding rich frequencies for classification of stroke patients eeg signals,” IEEE Access, vol. 8, pp. 135 811–135 820, 2020. [Google Scholar]
- [106].Sansiagi W, Djamal EC, Djajasasmita D, and Wulandari A, “Post-stroke identification of eeg signals using recurrent neural networks and long short-term memory,” International Journal of Advances in Intelligent Informatics, vol. 7, no. 2, pp. 137–150, 2021. [Google Scholar]
- [107].Wang C, Olugbade TA, Mathur A, Williams A. C. De C., Lane ND, and Bianchi-Berthouze N, “Recurrent network based automatic detection of chronic pain protective behavior using mocap and semg data,” in Proceedings of the 23rd international symposium on wearable computers, 2019, pp. 225–230.
- [108].Li Y, Ghosh S, Joshi J, and Oviatt S, “Lstm-dnn based approach for pain intensity and protective behaviour prediction,” in 2020 15th IEEE International Conference on Automatic Face and Gesture Recognition (FG 2020) IEEE, 2020, pp. 819–823. [Google Scholar]
- [109].Yuan X and Mahmoud M, “Alanet: Autoencoder-lstm for pain and protective behaviour detection,” in 2020 15th IEEE International Conference on Automatic Face and Gesture Recognition (FG 2020) IEEE, 2020, pp. 824–828. [Google Scholar]
- [110].Rojas RF, Romero J, Lopez-Aparicio J, and Ou K-L, “Pain assessment based on fnirs using bi-lstm rnns,” in 2021 10th International IEEE/EMBS Conference on Neural Engineering (NER) IEEE, 2021, pp. 399–402. [Google Scholar]
- [111].Bengoetxea A, Leurs F, Hoellinger T, Cebolla AM, Dan B, McIntyre J, and Cheron G, “Physiological modules for generating discrete and rhythmic movements: Action identification by a dynamic recurrent neural network,” Frontiers in Computational Neuroscience, vol. 8, p. 100, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Salloum R and Kuo C-CJ, “ECG-based biometrics using recurrent neural networks,” in 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE, 2017, pp. 2062–2066. [Google Scholar]
- [113].Zhang X, Zhang Y, Zhang L, Wang H, and Tang J, “Ballistocardiogram based person identification and authentication using recurrent neural networks,” in 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI) IEEE, 2018, pp. 1–5. [Google Scholar]
- [114].Mao S, Zhang Z, Khalifa Y, Donohue C, Coyle JL, and Sejdic E, “Neck sensor-supported hyoid bone movement tracking during swallowing,” Royal Society open science, vol. 6, no. 7, p. 181982, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Mao S, Sabry A, Khalifa Y, Coyle JL, and Sejdic E, “Estimation of laryngeal closure duration during swallowing without invasive x-rays,” Future Generation Computer Systems, vol. 115, pp. 610–618, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Khalifa Y, Donohue C, Coyle JL, and Sejdić E, “Upper esophageal sphincter opening segmentation with convolutional recurrent neural networks in high resolution cervical auscultation,” IEEE journal of biomedical and health informatics, vol. 25, no. 2, pp. 493–503, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].——, “On the robustness of high-resolution cervical auscultation-based detection of upper esophageal sphincter opening duration in diverse populations,” in Big Data III: Learning, Analytics, and Applications, vol. 11730. International Society for Optics and Photonics, 2021, p. 117300M. [Google Scholar]
- [118].Zhao A, Qi L, Dong J, and Yu H, “Dual channel LSTM based multifeature extraction in gait for diagnosis of neurodegenerative diseases,” Knowledge-Based Systems, vol. 145, pp. 91–97, 2018. [Google Scholar]
- [119].Zhen T, Yan L, and Yuan P, “Walking gait phase detection based on acceleration signals using LSTM-DNN algorithm,” Algorithms, vol. 12, no. 12, p. 253, 2019. [Google Scholar]
- [120].Gao J, Gu P, Ren Q, Zhang J, and Song X, “Abnormal gait recognition algorithm based on LSTM-CNN fusion network,” IEEE Access, vol. 7, pp. 163 180–163 190, 2019. [Google Scholar]
- [121].Tortora S, Ghidoni S, Chisari C, Micera S, and Artoni F, “Deep learning-based bci for gait decoding from EEG with LSTM recurrent neural network,” Journal of Neural Engineering, 2020. [DOI] [PubMed]
- [122].Gandhi T, Panigrahi BK, and Anand S, “A comparative study of wavelet families for EEG signal classification,” Neurocomputing, vol. 74, no. 17, pp. 3051–3057, 2011. [Google Scholar]
- [123].Xu Y, Hong K, Tsujii J, and Chang EIC, “Feature engineering combined with machine learning and rule-based methods for structured information extraction from narrative clinical discharge summaries,” Journal of the American Medical Informatics Association, vol. 19, no. 5, pp. 824–832, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Chauhan S and Vig L, “Anomaly detection in ECG time signals via deep long short-term memory networks,” in International Conference on Data Science and Advanced Analytics (DSAA) IEEE, 2015, pp. 1–7. [Google Scholar]
- [125].Qiu Y, Xiao F, and Shen H, “Elimination of power line interference from ECG signals using recurrent neural networks,” in Engineering in Medicine and Biology Society (EMBS), 39th Annual International Conference of the IEEE IEEE, 2017, pp. 2296–2299. [DOI] [PubMed] [Google Scholar]
- [126].Stollenga MF, Byeon W, Liwicki M, and Schmidhuber J, “Parallel multi-dimensional LSTM, with application to fast biomedical volumetric image segmentation,” in Advances in Neural Information Processing Systems, 2015, pp. 2998–3006.
- [127].Latif S, Usman M, Rana R, and Qadir J, “Phonocardiographic sensing using deep learning for abnormal heartbeat detection,” IEEE Sensors Journal, vol. 18, no. 22, pp. 9393–9400, 2018. [Google Scholar]
- [128].Bradbury J, Merity S, Xiong C, and Socher R, “Quasi-recurrent neural networks,” arXiv preprint arXiv:1611.01576, 2016.
- [129].Lei T, Zhang Y, Wang SI, Dai H, and Artzi Y, “Simple recurrent units for highly parallelizable recurrence,” arXiv preprint arXiv:1709.02755, 2017.
- [130].Li S, Li W, Cook C, Zhu C, and Gao Y, “Independently recurrent neural network (IndRNN): Building a longer and deeper RNN,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 5457–5466.
- [131].Van Der Westhuizen J and Lasenby J, “The unreasonable effectiveness of the forget gate,” arXiv preprint arXiv:1804.04849, 2018.
- [132].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, and Polosukhin I, “Attention is all you need,” in Advances in Neural Information Processing Systems, 2017, pp. 5998–6008.
- [133].Wang Z, Ma Y, Liu Z, and Tang J, “R-transformer: Recurrent neural network enhanced transformer,” arXiv preprint arXiv:1907.05572, 2019.
- [134].Radford A, Metz L, and Chintala S, “Unsupervised representation learning with deep convolutional generative adversarial networks,” arXiv preprint arXiv:1511.06434, 2015.
- [135].Donahue C, McAuley J, and Puckette M, “Adversarial audio synthesis,” arXiv preprint arXiv:1802.04208, 2018.
- [136].Golany T and Radinsky K, “Pgans: Personalized generative adversarial networks for ECG synthesis to improve patient-specific deep ECG classification,” in Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, 2019, pp. 557–564. [Google Scholar]
- [137].Gulrajani I, Ahmed F, Arjovsky M, Dumoulin V, and Courville AC, “Improved training of Wasserstein GANs,” in Advances in neural information processing systems, 2017, pp. 5767–5777.
- [138].Singh P and Pradhan G, “A new ECG denoising framework using generative adversarial network,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2020. [DOI] [PubMed]
- [139].Rajan D and Thiagarajan JJ, “A generative modeling approach to limited channel ECG classification,” in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE, 2018, pp. 2571–2574. [DOI] [PubMed] [Google Scholar]
- [140].Kingma DP and Welling M, “Auto-encoding variational bayes,” arXiv preprint arXiv:1312.6114, 2013.
- [141].Kuznetsov V, Moskalenko V, and Zolotykh NY, “Electrocardiogram generation and feature extraction using a variational autoencoder,” arXiv preprint arXiv:2002.00254, 2020.


