Abstract
Motivation
Gene regulatory networks (GRNs) are a way of describing the interaction between genes, which contribute to revealing the different biological mechanisms in the cell. Reconstructing GRNs based on gene expression data has been a central computational problem in systems biology. However, due to the high dimensionality and non-linearity of large-scale GRNs, accurately and efficiently inferring GRNs is still a challenging task.
Results
In this article, we propose a new approach, iLSGRN, to reconstruct large-scale GRNs from steady-state and time-series gene expression data based on non-linear ordinary differential equations. Firstly, the regulatory gene recognition algorithm calculates the Maximal Information Coefficient between genes and excludes redundant regulatory relationships to achieve dimensionality reduction. Then, the feature fusion algorithm constructs a model leveraging the feature importance derived from XGBoost (eXtreme Gradient Boosting) and RF (Random Forest) models, which can effectively train the non-linear ordinary differential equations model of GRNs and improve the accuracy and stability of the inference algorithm. The extensive experiments on different scale datasets show that our method makes sensible improvement compared with the state-of-the-art methods. Furthermore, we perform cross-validation experiments on the real gene datasets to validate the robustness and effectiveness of the proposed method.
Availability and implementation
The proposed method is written in the Python language, and is available at: https://github.com/lab319/iLSGRN.
1 Introduction
The interaction between genes forms the dynamic biochemical network known as the gene regulatory networks (GRNs). Clarifying the biological mechanism of the cell cycle, damage repair, and apoptosis depends on understanding the information transmission in the biological GRN from a systematic perspective (Pataskar and Tiwari 2016, Jackson et al. 2020, Razaghi-Moghadam and Nikoloski 2020). In addition, GRNs can obtain the key regulatory genes related to cell diseases, which is conducive to the diagnosis and treatment of cancer and other complex diseases as well as the research and development of targeted drugs (Emmert-Streib et al. 2014, Mi et al. 2019, Oulas et al. 2019). The investigation on GRNs has great significance to the realization of precision medicine (Madhamshettiwar et al. 2012, Yan et al. 2016, Delgado-Chaves et al. 2020). With the advance of biological sequencing technology (Marbach et al. 2012, Barrett et al. 2013, Buermans and den Dunnen 2014), professionals have developed various feasible methods to reconstruct GRNs from gene expression data (Schaffter et al. 2011, Gama-Castro et al. 2016, Mercatelli et al. 2020). Some methods also incorporated different types of data (steady-state data, time-series data, etc.) in gene network inference (Bonneau et al. 2006, Werhli and Husmeier 2007, Greenfield et al. 2013, Petralia et al. 2015, Castro et al. 2019).
The methods based on information theory are widely used to infer GRNs (Margolin et al. 2006, Faith et al. 2007, Zhang et al. 2013). These methods including Pearson correlation coefficient, mutual information (MI), conditional mutual information, and Maximal Information Coefficient (MIC), apply a threshold to determine whether MI between the expression responses of two genes is sufficient to infer a connecting regulatory link (Stuart et al. 2003). The prominent advantage of MI is that it can identify the non-functional regulatory relationship between genes. Yang et al. proposed a method MICRAT (Maximal Information coefficient with Conditional Relative Average entropy and Time-series mutual information) based on MIC to infer GRN (Yang et al. 2018). The MIC provides a more general framework for measuring the dependence between two genes.
Model-based methods usually establish mathematical models to infer the regulatory relationship between genes, including Boolean network model, Bayesian network model, Neural network model, and Differential equation model. The Boolean network method adopts 0/1 value to represent gene expression level, and logic function to represent the relationship between genes (Saadatpour and Albert 2013). But the unreasonable threshold selection in the Boolean model will increase the noise, making the GRNs inaccurate (Chai et al. 2014). The Dynamic Bayesian network takes time-series data as input, which enables the Bayesian model to effectively utilize the time information in gene expression data (Kim et al. 2003, Li et al. 2014, Sanchez-Castillo et al. 2018). The Bayesian model tends to suffer from a long-running time for analyzing large-scale genomic data, which could be resolved by the Local Bayesian network model (Liu et al. 2016). A highly flexible and scalable method, the neural network model, which simulates different functional relationships can represent the dynamic relationship of genes (Vohradsky 2001, Yuan and Bar-Joseph 2019). The different neural network models could be used to process different types of gene data, such as convolutional neural networks for gene expression images (Yang et al. 2019) and recurrent neural networks for gene time-series data (Ressom et al. 2006). The differential equation model employs a differential equation to construct a model to describe the relationship between genes, which is mainly divided into the linear ordinary differential equations (ODEs) method (Matsumoto et al. 2017, Fan et al. 2019) and the non-linear ODEs method (Ma et al. 2020, Tsai et al. 2020). Ma et al. proposed a Non-linear ODEs inference algorithm to establish a dynamic gene regulatory model and utilized importance scores to effectively infer all regulatory relationships among genes (Ma et al. 2020). Henderson et al. developed a non-parametric additive differential equation model to process time-series data (Henderson and Michailidis 2014). The advantage of differential equation is that it can be exploited to analyze both steady-state and time-series data (Delgado and Gómez-Vela 2019).
Machine-learning-based methods mainly apply machine-learning models and data structures to fit gene expression data. The GENIE3 method is model-free for inferring GRNs from steady-state series gene expression data (Huynh-Thu et al. 2010). However, it cannot be exploited for time-series data. The dynGENIE3 adapts the GENIE3 method to process time-series and steady-state series gene expression data (Huynh-Thu and Geurts 2018). Zheng et al. proposed the BiXGBoost method to solve the over-fitting problem by integrating randomization and regularization techniques (Zheng et al. 2019). The MMFGRN combines three models: a single time-series data model, a single steady-data model, and a single time-series and steady-state series data joint model (He et al. 2021). The results of MMFGRN imply that the model fusion strategy is feasible for GRN prediction. These methods are highly explanatory and can identify the regulatory direction, so the GRN obtained is often a directed network.
Currently, different inference algorithms have made great achievements, especially for small-scale GRNs. However, for large-scale networks, the high dimensionality, sparsity, and non-linearity of the network result in a significant decline in the efficiency and accuracy of inference algorithms (Fan et al. 2019, Ma et al. 2020). Here, we initially employ MIC to shrink the regulatory gene’s dimension for the given gene expression data. The next step is to infer GRNs using XGBoost and RF models. The machine-learning algorithms can efficiently capture gene regulatory relationships from both time-series data and steady-state series data. The results using the DREAM4 and Escherichia coli datasets demonstrate that our method performs better compared with state-of-the-art methods when inferring large-scale GRNs.
2 Datasets
In this article, we take the DREAM4 in silico size100 simulated data and E.coli real gene expression data as our experimental datasets. Table 1 shows the details of the datasets we used.
Table 1.
Datasets.a
| Network | DREAM4 in silico size100 |
Escherichia coli
|
||||
|---|---|---|---|---|---|---|
| Time-series data | Steady-state series | Cold stress | Heat stress | Oxidative stress | Lactose | |
| Genes | 100 | 100 | 3959 | 3959 | 3959 | 3959 |
| Samples | 10 | 200 | 3 | 3 | 3 | 3 |
| Time points | 21 | 8 | 8 | 11 | 5 | |
| TFs | 100 | 100 | 114 | 114 | 114 | 114 |
This table contains two datasets, DREAM4 and Escherichia coli, and introduces the number of genes, samples, time points, and regulatory factors in the dataset. DREAM4 contains five subnets, and the Escherichia coli dataset contains gene expression data under four different environments.
2.1 DREAM4 in silico size100 dataset
The DREAM4 in silico size100 dataset is a simulated gene expression data provided by Gene Net Weaver (GNW). The GNW is widely used for the inference of GRNs, including three public inferential GRN competitions, DREAM5, DREAM4, and DREAM3 (Schaffter et al. 2011). The in silico size100 incorporates both time-series data and steady-state series data. The time-series data contains 10 samples of 100 genes, and each sample has 21 time points. Steady-state series data includes gene expression levels under different perturbations.
2.2 Escherichia coli dataset
Real gene expression data of the E.coli dataset is collected from the GEO database (GSE20305), which records the expression levels of about 4400 genes under five different environmental perturbations (cold, heat, oxidative stress, lactose diauxie, and stationary phase) (Jozefczuk et al. 2010). The gold standard network in the RegulonDB provides experimentally validated regulatory relationships among genes (Gama-Castro et al. 2016). After matching the E.coli dataset with the gold standard network and retaining the common genes, the dataset contains 3959 genes and 114 TFs (Ma et al. 2020).
3 Materials and methods
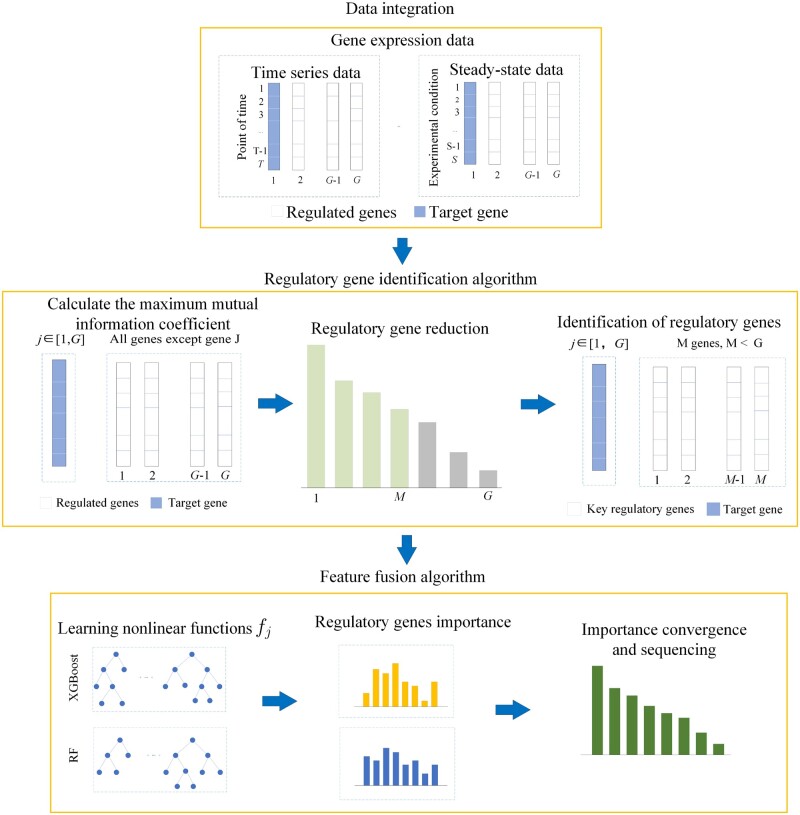
The overall flow of our method is shown in Fig. 1. Assuming that there are G genes in total, we first combined time-series data with steady-state series data. Then, we removed the redundant gene regulatory links using the regulatory gene identification algorithm to reduce the dimension of large-scale gene data. The MIC has a good ability to identify regulatory relationships between genes, so we applied MIC to construct the regulatory gene identification algorithm (Yang et al. 2018). After shrinking the candidate regulatory genes of target gene j (j = 1,2,…,G), we obtained M regulatory genes (M≪G). In the feature fusion algorithm, we applied a non-linear ODEs model to describe gene regulatory relationship behaviors. The non-linear ODEs are more suitable for simulating the dynamic characteristics of genes and processing the time-series data and steady-state series data. Then, the XGBoost and RF are used to train the non-linear ODEs model, respectively, and the importance scores derived from the above two models are fused to obtain the final gene regulatory relationship.
Figure 1.
Overall flow chart of the iLSGRN. The overall process mainly consists of three parts: data integration, regulatory gene identification algorithm, and feature fusion algorithm
3.1 Non-linear ODE model
We employ a non-linear ODEs to construct gene networks. We define the non-linear ODEs model for time-series data as follows:
| (1) |
where is the expression value of target gene j at time t, is decay rate, is a vector containing all regulatory gene expression levels at time t, and represents the non-linear function of M regulatory genes with respect to target gene j. Furthermore, for discrete time-series data, we utilize the difference equation to approximate the differential equation and simplify Formula 1 to:
| (2) |
where b is the time step, the default value of b is 1. For steady-state series data, we define . Then, the non-linear ODEs model for steady-state series data is simplified to Equation (3). Where e denotes different experimental conditions.
| (3) |
3.2 Regulatory gene identification algorithm
According to the sparsity of large-scale GRNs, we proposed a regulatory gene identification algorithm. The approach calculates the correlation between the target gene and all candidate regulatory genes using MIC. The prediction accuracy of the proposed approach can be improved by identifying the most important regulatory genes. Moreover, our algorithm can effectively shrink potential candidate genes and avoid massive computational costs in the following training step of the machine-learning model.
We define the MI between gene x and gene y as:
| (4) |
| (5) |
where X and Y represent the expression sequences for gene x and y, respectively. is the joint probability distribution function of X and Y, and and denote the marginal probability distribution functions of X and Y. Generally, the plane including x and y is divided into grids to calculate MIC, are the numbers of horizontal and vertical grids, respectively, , and is used to normalize the MIC (Yang et al. 2018).
For G genes in the whole gene expression dataset, gene j is selected as the target gene, and the remaining genes are chosen as candidate regulatory genes. We calculate the MIC between a given target gene j and a candidate regulatory gene, exclude redundant regulatory genes according to the threshold of MIC, and attain the regulatory gene sets for the target gene j. After repeating the above steps to obtain the regulatory gene set for all target genes, our algorithm achieves the goal of regulatory gene identification.
3.3 Feature fusion algorithm
The feature fusion method employs XGBoost and RF models to learn the function . At present, these two advanced machine-learning algorithms support parallel computing and can substantially enhance the efficiency of the fusion algorithm. After obtaining the regulatory gene for target gene j, we adopt machine-learning algorithms to learn the non-linear function respectively, and calculate the importance scores between regulatory genes and target gene j. The regulatory gene importance list for target gene j is calculated based on geometric mean method as follows:
| (6) |
where is the regulatory gene importance list of target gene j derived by the XGBoost model. is the regulatory gene importance list of target gene j derived by the RF model. We multiply these two vectors to generate the final regulatory gene importance list for target gene j. After repeating the above steps to obtain the importance lists for all target genes, we finally integrate the importance lists into the importance matrix, which shows the regulatory relationship between any two genes.
3.4 Evaluation method
Usually, a confusion matrix is applied to evaluate the accuracy of the inference algorithm, including the number of true positive (TP) samples, false negative (FN) samples, false positive (FP) samples, and true negative (TN) samples. Based on the confusion matrix, four evaluation indicators including recall, precision, true positive rate (TPR), and false positive rate (FPR), could be further calculated.
| (7) |
| (8) |
| (9) |
| (10) |
This study applies two evaluation metrics, area under precision–recall curves (AUPR) and area under receiver operating characteristic (AUROC), to assess the performance of the inference methods for GRNs. The AUROC and AUPR scores can better reflect the comprehensive performance of the classification algorithms. And we also define an overall score to evaluate the performance of the different methods.
| (11) |
We also consider the early precision ratio (EPR) to evaluate the accuracy of the top k edges of the inferred network. And k denotes the number of edges with the label “1” in the gold standard network. Early precision (EP) refers to the fraction of TPs in the top k edges, and the EPR represents the ratio of the EP of the proposed model to a random predictor, where a random predictor’s EP is the edge density. The edge density is defined as the ratio of edges with the label “1” to all potential edges in the gold standard network, and the network of n genes has potential edges (Pratapa et al. 2020).
| (12) |
| (13) |
3.5 Pseudocode flow chart
Algorithm 1 is the pseudo code flow chart of our method. Our algorithm takes the gene expression time-series data and steady-state series data as input, and the regulatory importance list for all genes as output.
Algorithm 1: The proposed algorithm for solving GRNs inference
Input: time-series data , including G genes and T time sampling points, steady sequence data , including G genes and S experimental conditions, threshold .
Output: gene regulatory network .
1 Initialize .
2 Calculate MIC coefficient.
3 for j = 1; j ≤ G; j + + do
4 for i ≠ j; i ≤ G; i++ do
5 Combining time-series samples with steady-state series samples, the input and output pairs of training samples are obtained.
.
.
6 Dimensionality reduction based on MIC
if < MIC (, )
Retain gene i to list .
else
skip.
Obtain all regulatory genes of target gene j from list .
7 Build a non-linear ODEs model . Using XGBoost and RF machine-learning model to calculate importance score and .
8 Model importance score fusion.
.
9 All importance scores are obtained after ranking all importance.
10 Return .
3.6 Related methods for inference of GRN
In this article, we make a comparison experiment of iLSGRN with other mainstream methods, including GENIE3, dynGENIE3, BiXGBoost, Non-linear ODEs, and MMFGRN. GENIE3 and dynGENIE3 utilize importance scores from RF to identify regulators for each target gene, and dynGENIE3 is an adapted version of the GENIE3 to deal with time-series and steady-state series data. BiXGBoost is based on the bidirectional model, and considers both candidate regulatory genes and target genes for a specific gene and uses XGBoost to calculate feature importance as regulatory gene relationships. Non-linear ODEs method combines non-linear ODE models and XGBoost to infer GRNs. MMFGRN fuses three models to infer GRNs through a weighted fusion strategy, including a time-series data model based on LightGBM and a steady-state data model based on LightGBM, as well as a time-series and steady-state data joint model based on XGBoost.
3.7 Parameter setting and cross-validation experiment
When setting the MIC thresholds in our method, we referred to the results in a previous study, which also used MIC to infer GRNs. In the study (Yang et al. 2018), a suitable MIC threshold (0.15) on the DREAM4 in silico size100 dataset was recommended. We set several values within a small neighboring range (0.1–0.2) of this threshold to infer the GRNs and chose the one that corresponded to the optimal result. Supplementary Figs S1 and S4 show that the MIC distributions on the E.coli dataset are significantly wider than those on the DREAM4 dataset, and the previous study has reported that the GRN of E.coli is basically very sparse (Fan et al. 2019), thus, we searched for the optimal threshold in a relatively large range (0.2–0.7) to exclude more redundant regulatory relationships on the E.coli dataset. Since this parameter setting method may cause overfitting, we further conducted cross-validation experiments to validate the robustness of the proposed method.
In the cross-validation experiment for the iLSGRN, we divided the genes into 2-folds, the training set and the test set. Here, we mainly discuss the effect of threshold and learning rate in XGBoost, and use the grid search method to adjust the parameters. Specifically, we set 10 different learning rates of XGBoost and 30 different thresholds of MIC on one training set, conducted a total of 300 experiments and recorded the overall score for each experiment. Next, the proposed model with the optimal parameters was applied on the test set. Finally, we switched the training set and test set and repeated the above-mentioned steps. The performance of the prediction algorithm is estimated by averaging the accuracy results of the two test sets.
In addition, we used the grid search method to search the optimal parameters for other inference methods in comparison experiments. For GENIE3 and dynGENIE3, the parameters “max_depth” and “n_estimators” were selected to optimize. For Non-linear ODEs and BiXGBoost, the optimized parameters include: “learning_rate, max_depth, n_estimators.” In MMFGRN, we optimized the parameters “learning_rate” in XGBoost and “learning_rate” in LigthGBM.
4 Results
In this section, we extensively compared our method with five state-of-the-art methods: Non-linear ODEs (Ma et al. 2020), GENIE3 (Huynh-Thu et al. 2010), dynGENIE3 (Huynh-Thu and Geurts 2018), BiXGBoost (Zheng et al. 2019), and MMFGRN (He et al. 2021). A comparative experiment was first conducted on the DREAM4 in silico size100 and the E.coli gene expression dataset. Then, we performed cross-validation and ablation experiments to verify the robustness and stability of the method. There is no denying that the decay rate is still an important factor. However, it is difficult to set an exact decay rate value for all genes, and we consider the decay rate as a constant.
4.1 Performance evaluation on DREAM4 in silico size100 dataset
In this part, we performed experiments of the above-mentioned approaches on the five sub-datasets of the DREAM4 dataset containing time-series data and steady-state series data. The main parameters of the iLSGRN on DREAM4 in silico size100 dataset are shown in Supplementary Table S1. Table 2 summarizes the results of these GRN inference methods on the DREAM4 in silico size100 dataset. The distribution of the MIC is provided by Supplementary Fig. S1. The AUROC curves of iLSGRN are provided by Supplementary Fig. S2. The iLSGRN achieves the highest AUROC in Networks 1 and 5 and the highest AUPR in all networks. Although the AUROC scores of the iLSGRN do not reach the best in Networks 2–4, our method is still more competitive than other inference methods in most subnets. The Cluster Bar Chart of the results for various methods on each sub-dataset of the DREAM4 in silico size100 is shown in Supplementary Fig. S3. The iLSGRN achieves the best overall score on all sub-datasets in Supplementary Fig. S3. The EPR results on DREAM4 dataset for each method are shown in Supplementary Table S2. The EPR values of iLSGRN on the five sub-networks are 29.08, 13.57, 20.83, 18.01, and 19.40, respectively, which are higher than other inference methods.
Table 2.
The results of various methods on DREAM4 in silico size 100 dataset.
| Methods | Net1 | Net2 | Net3 | Net4 | Net5 | |
|---|---|---|---|---|---|---|
| GENIE3 | AUROC | 0.825 | 0.783 | 0.813 | 0.826 | 0.791 |
| AUPR | 0.129 | 0.119 | 0.147 | 0.150 | 0.113 | |
| dynGENIE3 | AUROC | 0.843 | 0.746 | 0.803 | 0.793 | 0.824 |
| AUPR | 0.236 | 0.144 | 0.227 | 0.221 | 0.158 | |
| BiXGBoost | AUROC | 0.806 | 0.730 | 0.765 | 0.735 | 0.769 |
| AUPR | 0.235 | 0.152 | 0.261 | 0.204 | 0.214 | |
| Non-linear ODEs | AUROC | 0.857 | 0.758 | 0.795 | 0.787 | 0.798 |
| AUPR | 0.370 | 0.213 | 0.322 | 0.318 | 0.294 | |
| MMFGRN | AUROC | 0.854 | 0.772 | 0.792 | 0.701 | 0.747 |
| AUPR | 0.371 | 0.215 | 0.328 | 0.319 | 0.337 | |
| iLSGRN | AUROC | 0.859 | 0.771 | 0.799 | 0.800 | 0.839 |
| AUPR | 0.462 | 0.262 | 0.389 | 0.357 | 0.342 |
The best results for AUROC and AUPR scores are in bold.
4.2 Performance evaluation on E.coli dataset
Here, we apply the iLSGRN and other inference methods on the E.coli dataset. The main parameters of the iLSGRN on E.coli dataset are shown in Supplementary Table S3. Table 3 summarizes the performance of these methods on the E.coli dataset. As seen in Table 3, the iLSGRN obtains the best AUROC scores on Cold Stress, Heat Stress, and Oxidative stress sub-datasets, and the best AUPR scores on Cold Stress, Oxidative stress, and Lactose sub-datasets. MMFGRN achieves the best AUPR score on the Heat Stress subset and the best AUROC score on the Lactose subset. Overall, our method still maintains a high prediction accuracy and is competitive for reconstructing large-scale GRNs. Supplementary Fig. S4 shows the distribution of the MIC on the E.coli dataset and Supplementary Fig. S5 shows the AUROC curves of iLSGRN for each subset of E.coli data. Supplementary Fig. S6 is the cluster bar chart of the results for each method in comparison experiment. The EPR results on E.coli dataset for each method are shown in Supplementary Table S4. On the E.coli dataset, the iLSGRN obtains the highest EPR values on Cold stress, Oxidative stress, and the same EPR value as MMFGRN on Lactose.
Table 3.
The results of various methods on E.coli.
| Methods | Cold stress |
Heat stress |
Oxidative stress |
Lactose |
||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | |
| GENIE3 | 0.522 | 0.016 | 0.547 | 0.020 | 0.579 | 0.035 | 0.528 | 0.019 |
| BiXGBoost | 0.470 | 0.013 | 0.523 | 0.015 | 0.503 | 0.012 | 0.505 | 0.011 |
| Non-linear ODEs | 0.552 | 0.019 | 0.558 | 0.017 | 0.605 | 0.027 | 0.583 | 0.018 |
| MMFGRN | 0.689 | 0.023 | 0.812 | 0.044 | 0.762 | 0.038 | 0.833 | 0.035 |
| iLSGRN | 0.784 | 0.026 | 0.878 | 0.032 | 0.803 | 0.059 | 0.753 | 0.036 |
The best results for AUROC and AUPR scores are in bold.
4.3 Performance of the cross-validation experiments on E.coli dataset
To evaluate the robustness of the proposed method, we conducted 2-fold cross-validation experiments on the E.coli dataset. The main parameters of iLSGRN in the cross-validation experiment are shown in Supplementary Table S5. The cross-validation experimental results are shown in Table 4 and Supplementary Figs S7 and S8. In Table 4, iLSGRN has the highest scores except for the AUROC on the Cold Stress sub-dataset. Supplementary Fig. S7 shows the boxplot of overall score and threshold for E.coli dataset. Supplementary Fig. S8 is the cluster bar chart of the cross-validation results on the E.coli dataset. Although our method does not achieve the best scores in certain sub-datasets, our method still outperforms other methods, indicating the great potential of iLSGRN for inferring large-scale GRNs.
Table 4.
The results of the 2-fold cross-validation experiments on E.coli.
| Method | Cold stress |
Heat stress |
Oxidative stress |
Lactose |
||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | |
| GENIE3 | 0.441 | 0.020 | 0.531 | 0.015 | 0.409 | 0.028 | 0.515 | 0.017 |
| BiXGBoost | 0.442 | 0.011 | 0.510 | 0.013 | 0.517 | 0.005 | 0.467 | 0.012 |
| Non-linear ODEs | 0.491 | 0.016 | 0.512 | 0.017 | 0.557 | 0.021 | 0.569 | 0.014 |
| MMFGRN | 0.572 | 0.018 | 0.425 | 0.020 | 0.541 | 0.033 | 0.463 | 0.015 |
| iLSGRN | 0.571 | 0.023 | 0.635 | 0.065 | 0.694 | 0.082 | 0.563 | 0.019 |
The best results for AUROC and AUPR scores are in bold.
4.4 Results of the ablation experiment
We performed ablation experiments on the DREAM4 in silico size100 and E.coli datasets to validate the effectiveness of each module in the proposed method. In the ablation experiments, the effects of our regulatory gene recognition algorithm and feature fusion algorithm can be evaluated systematically, as shown in Fig. 2. The “RF” represents only applying the RF algorithm to calculate the importance score. The “XGBoost” indicates only applying the XGBoost algorithm to calculate the importance score. The “XGBoost+RF” means combining the XGBoost with RF without the regulatory gene recognition algorithm.
Figure 2.
Cluster Bar Chart of the ablation experiment’s overall score on the DREAM4 in silico size100 and E.coli datasets
The iLSGRN achieves the highest overall scores on all sub-datasets compared to the single-model approaches. From Fig. 2, we observed that the overall score gradually increases with the integration of the three single models. The results of the ablation experiments show that the regulatory gene recognition algorithm and the feature fusion algorithm have great advantages in gene network prediction.
4.5 Computational complexity
The iLSGRN algorithm is implemented based on XGBoost and RF. The computational complexity of XGBoost is on the order of , where is the number of trees, indicates the number of genes, represents the maximum depth of trees, and means the total number of samples. And the computational complexity of RF is on the order of , where is the maximum depth of trees and denotes the number of trees. We further compared the computational complexity of iLSGRN with GENIE3, BiXGBoost, Non-linear ODEs, and MMFGRN. Among these methods, BiXGBoost utilizes the XGBoost to construct two regression equations. GENIE3 is implemented by the RF and Non-linear ODEs inference algorithm is based on the XGBoost. MMFGRN constructs one XGBoost model and two LightGBM models to process steady-state and time-series gene expression data for model fusion.
The running time of the different methods on the DREAM4 in silico size100 and E.coli datasets is shown in Supplementary Table S6. Longer running time indicates higher computational complexity of the model. And these measurements are conducted on a computer with Intel Core i7-9750H CPU, clocked at 2.60 GHz and 16 GB memory. Our proposed iLSGRN algorithm spends considerable running time while achieving more accurate prediction performance.
5 Discussion
Currently, many computational methods for inference of GRNs have been proposed. In this article, we propose a scalable method based on model fusion to infer GRNs from time-series and steady-state series gene expression data. Our method first calculates the MIC between genes and sets a suitable threshold to exclude the redundant and enough weak gene regulatory relationships. Then, we combine XGBoost and RF to learn the non-linear ODEs model and fuse the importance scores. The experimental results on the DREAM4 and E.coli datasets show that our method effectively eliminates large redundant gene expression information and improves the accuracy of inferring regulatory relationships.
Compared with other model fusion methods, such as MMFGRN, our method utilizes MIC in the regulatory gene recognition algorithm to remove massive redundant regulatory links in large-scale GRNs. It fundamentally reduces the influence of redundant and enough weak genes on identifying GRNs. For the coexistence of time-series data and steady-state series data, the non-linear ODEs model can more precisely simulate the dynamic characteristics between genes and retain the information in steady-state series data. Moreover, the non-linear ODEs model can comprehensively deal with the non-linear relationships in GRNs. Meanwhile, we integrate XGBoost with RF machine-learning model, i.e. Boost and bagging, to obtain more accurate GRN.
Although our method has improved the prediction accuracy compared to other state-of-the-art methods, there are still some limitations. We only tested the validity of the proposed method on the simulated gene expression dataset of DREAM4 and the real gene expression dataset of E.coli. The inference methods obtained low AUPR results on the E.coli dataset, which may be due to the sparsity of the GRNs. The real biological datasets only contain steady-state series gene expression data and lack time-series data, which may affect the performance of the developed method. We will further extend the proposed approach on more comprehensive real gene expression datasets in the future. Moreover, this study did not consider the batch effects of gene expression data, which may affect the calculation of the MIC or feature importance assessment. The feature fusion algorithm will increase the computational complexity and time cost of the proposed method due to the training process of XGBoost and RF models. Therefore, developing a faster and more robust algorithm is one of the future directions.
Supplementary Material
Contributor Information
Yiming Wu, School of Information Science and Technology, Dalian Maritime University, Dalian 116026, China.
Bing Qian, School of Information Science and Technology, Dalian Maritime University, Dalian 116026, China.
Anqi Wang, Department of Statistics and Actuarial Science, The University of Hong Kong, Hong Kong 999077, China.
Heng Dong, School of Information Science and Technology, Dalian Maritime University, Dalian 116026, China.
Enqiang Zhu, Institution of Computing Science and Technology, Guangzhou University, Guangzhou 510006, China.
Baoshan Ma, School of Information Science and Technology, Dalian Maritime University, Dalian 116026, China.
Author contributions
Yiming Wu (Conceptualization [equal], Investigation [equal], Methodology [equal], Software [equal], Validation [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Bing Qian (Conceptualization [equal], Investigation [equal], Writing—review & editing [equal]), Anqi Wang (Writing—review & editing [equal]), Heng Dong (Conceptualization [equal], Investigation [equal], Writing—review & editing [equal]), Enqiang Zhu (Supervision [equal], Writing—review & editing [equal]), and Baoshan Ma (Conceptualization [equal], Methodology [equal], Supervision [equal], Writing—review & editing [equal])
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
The authors have declared no conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China [62272115, 61471078].
References
- Barrett T, Wilhite SE, Ledoux P et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Reiss DJ, Shannon P et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol 2006;7:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans HP, den Dunnen JT. Next generation sequencing technology: advances and applications. Biochim Biophys Acta 2014;1842:1932–41. [DOI] [PubMed] [Google Scholar]
- Castro DM, de Veaux NR, Miraldi ER et al. Multi-study inference of regulatory networks for more accurate models of gene regulation. PLoS Comput Biol 2019;15:e1006591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai LE, Loh SK, Low ST et al. A review on the computational approaches for gene regulatory network construction. Comput Biol Med 2014;48:55–65. [DOI] [PubMed] [Google Scholar]
- Delgado-Chaves FM, Gómez-Vela F, Divina F et al. Computational analysis of the global effects of Ly6E in the immune response to coronavirus infection using gene networks. Genes (Basel) 2020;11:831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado FM, Gómez-Vela F. Computational methods for gene regulatory networks reconstruction and analysis: a review. Artif Intell Med 2019;95:133–45. [DOI] [PubMed] [Google Scholar]
- Emmert-Streib F, Dehmer M, Haibe-Kains B. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Front Cell Dev Biol 2014;2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 2007;5:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan A, Wang H, Xiang H et al. Inferring large-scale gene regulatory networks using a randomized algorithm based on singular value decomposition. IEEE/ACM Trans Comput Biol Bioinform 2019;16:1997–2008. [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Santos-Zavaleta A et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 2016;44:D133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield A, Hafemeister C, Bonneau R. Robust data-driven incorporation of prior knowledge into the inference of dynamic regulatory networks. Bioinformatics 2013;29:1060–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Tang J, Zou Q et al. MMFGRN: a multi-source multi-model fusion method for gene regulatory network reconstruction. Brief Bioinform 2021;22:bbab166. [DOI] [PubMed] [Google Scholar]
- Henderson J, Michailidis G. Network reconstruction using nonparametric additive ODE models. PLoS One 2014;9:e94003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu VA, Geurts P. dynGENIE3: dynamical GENIE3 for the inference of gene networks from time series expression data. Sci Rep 2018;8:3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu VA, Irrthum A, Wehenkel L et al. Inferring regulatory networks from expression data using tree-based methods. PLoS One 2010;5:e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CA, Castro DM, Saldi G-A et al. Gene regulatory network reconstruction using single-cell RNA sequencing of barcoded genotypes in diverse environments. Elife 2020;9:e51254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefczuk S, Klie S, Catchpole G et al. Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol 2010;6:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Imoto S, Miyano S. Inferring gene networks from time series microarray data using dynamic Bayesian networks. Brief Bioinform 2003;4:228–35. [DOI] [PubMed] [Google Scholar]
- Li P, Gong P, Li H et al. Gene regulatory network inference and validation using relative change ratio analysis and time-delayed dynamic Bayesian network. EURASIP J Bioinform Syst Biol 2014;2014:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Zhang S-W, Guo W-F et al. Inference of gene regulatory network based on local Bayesian networks. PLoS Comput Biol 2016;12:e1005024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Fang M, Jiao X. Inference of gene regulatory networks based on nonlinear ordinary differential equations. Bioinformatics 2020;36:4885–93. [DOI] [PubMed] [Google Scholar]
- Madhamshettiwar PB, Maetschke SR, Davis MJ et al. Gene regulatory network inference: evaluation and application to ovarian cancer allows the prioritization of drug targets. Genome Med 2012;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Costello JC, Küffner R et al. ; DREAM5 Consortium. Wisdom of crowds for robust gene network inference. Nat Methods 2012;9:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 2006;7 (Suppl. 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Kiryu H, Furusawa C et al. SCODE: an efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Bioinformatics 2017;33:2314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D, Scalambra L, Triboli L et al. Gene regulatory network inference resources: a practical overview. Biochim Biophys Acta Gene Regul Mech 2020;1863:194430. [DOI] [PubMed] [Google Scholar]
- Mi Z, Guo B, Yin Z et al. Disease classification via gene network integrating modules and pathways. R Soc Open Sci 2019;6:190214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulas A, Minadakis G, Zachariou M et al. Systems bioinformatics: increasing precision of computational diagnostics and therapeutics through network-based approaches. Brief Bioinform 2019;20:806–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataskar A, Tiwari VK. Computational challenges in modeling gene regulatory events. Transcription 2016;7:188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia F, Wang P, Yang J et al. Integrative random forest for gene regulatory network inference. Bioinformatics 2015;31:i197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratapa A, Jalihal AP, Law JN et al. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods 2020;17:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razaghi-Moghadam Z, Nikoloski Z. Supervised learning of gene-regulatory networks based on graph distance profiles of transcriptomics data. NPJ Syst Biol Appl 2020;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressom HW, Zhang Y, Xuan J et al. Inference of gene regulatory networks from time course gene expression data using neural networks and swarm intelligence. In: IEEE Symposium on Computational Intelligence and Bioinformatics and Computational Biology, Toronto, ON, Canada, 28-29 September. 1–8. IEEE, 2006.
- Saadatpour A, Albert R. Boolean modeling of biological regulatory networks: a methodology tutorial. Methods 2013;62:3–12. [DOI] [PubMed] [Google Scholar]
- Sanchez-Castillo M, Blanco D, Tienda-Luna IM et al. A Bayesian framework for the inference of gene regulatory networks from time and pseudo-time series data. Bioinformatics 2018;34:964–70. [DOI] [PubMed] [Google Scholar]
- Schaffter T, Marbach D, Floreano D. GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics 2011;27:2263–70. [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D et al. A gene-coexpression network for global discovery of conserved genetic modules. Science 2003;302:249–55. [DOI] [PubMed] [Google Scholar]
- Tsai M-J, Wang J-R, Ho S-J et al. GREMA: modelling of emulated gene regulatory networks with confidence levels based on evolutionary intelligence to cope with the underdetermined problem. Bioinformatics 2020;36:3833–40. [DOI] [PubMed] [Google Scholar]
- Vohradsky J. Neural model of the genetic network. J Biol Chem 2001;276:36168–73. [DOI] [PubMed] [Google Scholar]
- Werhli AV, Husmeier D. Reconstructing gene regulatory networks with Bayesian networks by combining expression data with multiple sources of prior knowledge. Stat Appl Genet Mol Biol 2007;6:Article15. [DOI] [PubMed] [Google Scholar]
- Yan W, Xue W, Chen J et al. Biological networks for cancer candidate biomarkers discovery. Cancer Inform 2016;15:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Xu Y, Maxwell A et al. MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data. BMC Syst Biol 2018;12:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fang Q, Shen HB. Predicting gene regulatory interactions based on spatial gene expression data and deep learning. PLoS Comput Biol 2019;15:e1007324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Bar-Joseph Z. Deep learning for inferring gene relationships from single-cell expression data. Proc Natl Acad Sci USA 2019;116:27151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu K, Liu Z-P et al. NARROMI: a noise and redundancy reduction technique improves accuracy of gene regulatory network inference. Bioinformatics 2013;29:106–13. [DOI] [PubMed] [Google Scholar]
- Zheng R, Li M, Chen X et al. BiXGBoost: a scalable, flexible boosting-based method for reconstructing gene regulatory networks. Bioinformatics 2019;35:1893–900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.