Abstract
Precise arrangement of actin, myosin, and other regulatory components in a sarcomeric pattern is critical for producing contractile forces in striated muscles. Actin-interacting protein 1 (AIP1), also known as WD-repeat protein 1 (WDR1), is one of essential factors that regulate sarcomeric assembly of actin filaments. In the nematode Caenorhabditis elegans, mutation in unc-78, encoding one of the two AIP1 isoforms, causes severe disorganization of sarcomeric actin filaments and near paralysis, but mutation in sup-13 suppresses the unc-78-mutant phenotypes to restore nearly normal sarcomeric actin organization and worm motility. Here, we identified that sup-13 is a nonsense allele of arrd-15 encoding an α-arrestin. The sup-13/arrd-15 mutation suppressed the phenotypes of unc-78 null mutant but required aipl-1 that encodes a second AIP1 isoform. aipl-1 was normally expressed highly in embryos and downregulated in mature muscle. However, in the sup-13/arrd-15 mutant, the AIPL-1 protein was maintained at high levels in adult muscle to compensate for the absence of the UNC-78 protein. The sup-13/arrd-15 mutation caused accumulation of ubiquitinated AIPL-1 protein, suggesting that a normal function of sup-13/arrd-15 is to enhance degradation of ubiquitinated AIPL-1, thereby promoting transition of AIP1 isoforms from AIPL-1 to UNC-78 in developing muscle. These results suggest that α-arrestin is a novel factor to promote isoform turnover by enhancing protein degradation.
Keywords: actin, actin-interacting protein 1 (AIP1), α-arrestin, sarcomere, ubiquitination
Significance Statement.
Sarcomeric organization of contractile proteins is essential for efficient contractility of striated muscles. However, the mechanism of sarcomere assembly remains unclear. Actin-interacting protein 1 (AIP1), also known as WDR1, is a conserved protein that promotes disassembly of actin filaments and is essential for sarcomeric assembly of actin filaments. In the nematode Caenorhabditis elegans, we found that the sup-13/arrd-15 gene, encoding an α-arrestin, promotes turnover of one of the two AIP1 isoforms in striated muscle by specifically targeting one isoform for ubiquitin-dependent degradation. Our study suggests that α-arrestin is a novel factor to promote isoform transition in striated muscle.
Introduction
Dynamic assembly and disassembly of actin filaments are critical for the function of the actin cytoskeleton that plays central roles in many cell biological processes including cell migration, cell division, and morphogenesis. Because actin filaments are relatively stable, severing and depolymerization of actin filaments are often promoted for cytoskeletal reorganization and recycling of actin monomers for new rounds of polymerization (1–3). Actin depolymerizing factor (ADF)/cofilin is a key regulator of actin filament disassembly by accelerating both depolymerization and severing of actin filaments. However, ADF/cofilin by itself can only modestly sever and depolymerize actin filaments. Therefore, ADF/cofilin cooperates with other factors including coronin, actin-interacting protein 1 (AIP1), and cyclase-associated protein to promote rapid actin filament disassembly.
AIP1 (4), also known as WD-repeat protein 1 (WDR1), is an evolutionarily conserved regulator of actin filament dynamics (5, 6). AIP1 contains two seven-bladed β-propeller domains in a single molecule (7, 8) and promotes disassembly of actin filaments in the presence of ADF/cofilin (9–11). AIP1 has minimum effects on bare actin filaments but rapidly and strongly severs ADF/cofilin-decorated actin filaments (12–18). Coronin promotes binding of ADF/cofilin to actin filaments and further accelerates filament disassembly by AIP1 (16, 18, 19). In contrast, tropomyosin prevents binding of ADF/cofilin to actin filaments and inhibits filament disassembly by AIP1 (20). However, other than the regulation at the level of ADF/cofilin, how AIP1-mediated actin filament disassembly is regulated remains unknown. Since coronin, ADF/cofilin, and AIP1 are present in most eukaryotic cells (2), cells need to be equipped with a mechanism to control these strong actin filament disassembly factors in order to maintain functional actin cytoskeleton.
Mutation or knockdown of AIP1 causes abnormal actin filament dynamics and organization in vivo in most tested organisms (5, 6). Notably, point mutations in the human AIP1 gene, WDR1, cause severe blood disorders, including reduction and impaired motility of neutrophils (21–23). Therefore, WDR1 is considered a gene responsible for lazy leukocyte syndrome (24, 25). Similar WDR1 mutant phenotypes are reported in mice (26) and Zebrafish (27). AIP1 is also an essential regulator of sarcomere assembly in striated muscle. In the nematode Caenorhabditis elegans, mutations in the AIP1 gene, unc-78, cause severe disorganization of sarcomeric actin filaments and reduced muscle contractility (28–30). A cardiomyocyte-specific WDR1 knockout in mice causes similar actin disorganization and results in postnatal lethality with hypertrophic cardiomyopathy (31). Thus, genetic studies on AIP1 should provide insight in the basic in vivo function of AIP1 as well as pathogenesis of AIP1/WDR1-linked diseases.
In this study, we took advantage of C. elegans genetics to analyze an extragenic suppressor of unc-78 encoding AIP1. The sup-13 mutation was previously shown to suppress the motility defects of unc-78 mutants, but the molecular identity of sup-13 was uncharacterized (32, 33). We identified that sup-13 is a nonsense allele of arrd-15 encoding a member of the α-arrestins that are generally known as adaptors to mediate protein trafficking and degradation (34–36). We found that sup-13/arrd-15 is a regulator of a second AIP1 gene, aipl-1, that is partially redundant with unc-78 (37, 38). In wild-type muscle, UNC-78 is the predominant AIP1 isoform in adults. However, in sup-13/arrd-15 mutant muscle, AIPL-1 is maintained at high levels, without significant effects on UNC-78 levels, in adult muscle to compensate for the absence of UNC-78. Ubiquitinated AIPL-1 is accumulated in sup-13/arrd-15 mutants, suggesting that sup-13/arrd-15 mediates a process for degradation of ubiquitinated AIPL-1. Thus, our study shows that an AIP1 isoform is targeted for degradation in an α-arrestin-dependent manner in vivo and suggests a novel mechanism of isoform turnover by degradation of a selective isoform(s) in differentiated cells.
Results
sup-13 mutation suppresses sarcomeric actin disorganization of unc-78 mutants
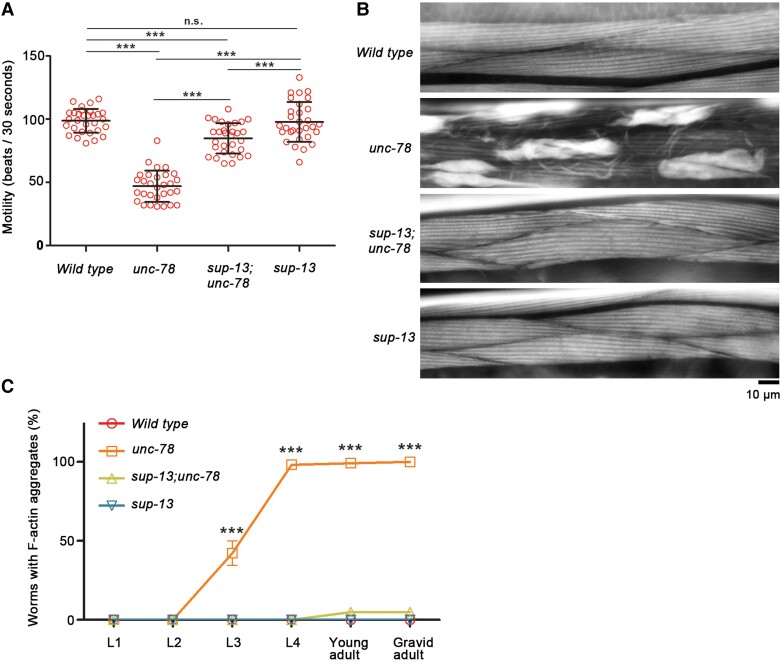
In an effort to identify genes with a functional connection to unc-78, a sup-13 mutant allele, st210, was isolated from a screen for extragenic suppressors of unc-78 and mapped to chromosome III (32, 33). The unc-78 single mutants show sluggish and slow movement (28, 39), whereas the sup-13;unc-78 double mutants show superficially normal motility (33). The original screen and analysis were performed using unc-78(e1217), a loss-of-function allele with a missense mutation (28, 30). Therefore, we tested whether the sup-13 mutation suppresses the phenotypes of unc-78(gk27), a null allele expressing no detectable UNC-78 protein (28, 30). The unc-78(gk27) single mutants moved much slower than wild type (Fig. 1A). The sup-13;unc-78(gk27) double mutants moved slightly slower than wild type but significantly faster than the unc-78(gk27) single mutants (Fig. 1A). The sup-13 single mutants moved as fast as wild type (Fig. 1A). Therefore, the sup-13 mutation suppresses the motility defects of the unc-78 mutants even in the absence of the UNC-78 protein.
Fig. 1.
The sup-13 mutation suppresses motility defects and sarcomere disorganization of the unc-78 null mutant. A) Motility of young adult worms was quantified by measuring beating frequency (beats per 30 s). An unc-78 null allele, unc-78(gk27), was used in this study and simply shown as unc-78 (n = 30, data are average ± standard deviation). ***, P < 0.001. n.s., not significant. B) Adult worms were stained for F-actin by tetramethylrhodamine-phalloidin, and regions of the body wall muscle are shown. Actin was organized in a striated manner in wild type but highly disorganized with aggregates in unc-78(gk27). This phenotype was suppressed in the sup-13(st210);unc-78(gk27) double mutant to nearly an appearance of wild type. The sup-13(st210) single mutant did not show a detectable phenotype in the F-actin organization. Bar, 10 μm. C) Worms at different developmental stages were stained for F-actin by tetramethylrhodamine-phalloidin, and percentages of worms with F-actin aggregates (counted as positive when >25% of total muscle cells contained aggregates) were analyzed. For each experiment, > 500 worms were examined for each stage, and average ± standard deviation of three independent experiments are shown. ***, P < 0.001.
The sup-13 mutation also strongly suppressed disorganization of sarcomeric actin filaments in the body wall muscle of the unc-78 mutants (Fig. 1B). The body wall muscle is responsible for body movement of the worms, and actin filaments were organized in a striated pattern in wild type (Fig. 1B). In the unc-78(gk27) mutants, actin filaments were highly disorganized and accumulated in large aggregates (Fig. 1B). However, in the sup-13;unc-78(gk27) double mutants, striated organization of actin filaments were nearly indistinguishable from that in wild type (Fig. 1B). The sup-13 single mutants did not exhibit detectable defects in the actin filament organization (Fig. 1B). During development, the actin disorganization phenotype became detectable in unc-78(gk27) at the L3 larval stage and was nearly 100% penetrant after the L4 larval stage (Fig. 1C). The sup-13 mutation almost completely suppressed the actin disorganization phenotype of unc-78(gk27) at all developmental stages (Fig. 1C), and the sup-13 mutation alone did not cause actin disorganization at any stage (Fig. 1C). These cytological observations indicate that the sup-13 mutation suppresses the muscle defects of the unc-78 mutants by suppressing disorganization of sarcomeric actin filaments to restore assembly of functional contractile apparatuses.
sup-13 is an allele of arrd-15 encoding an α-arrestin
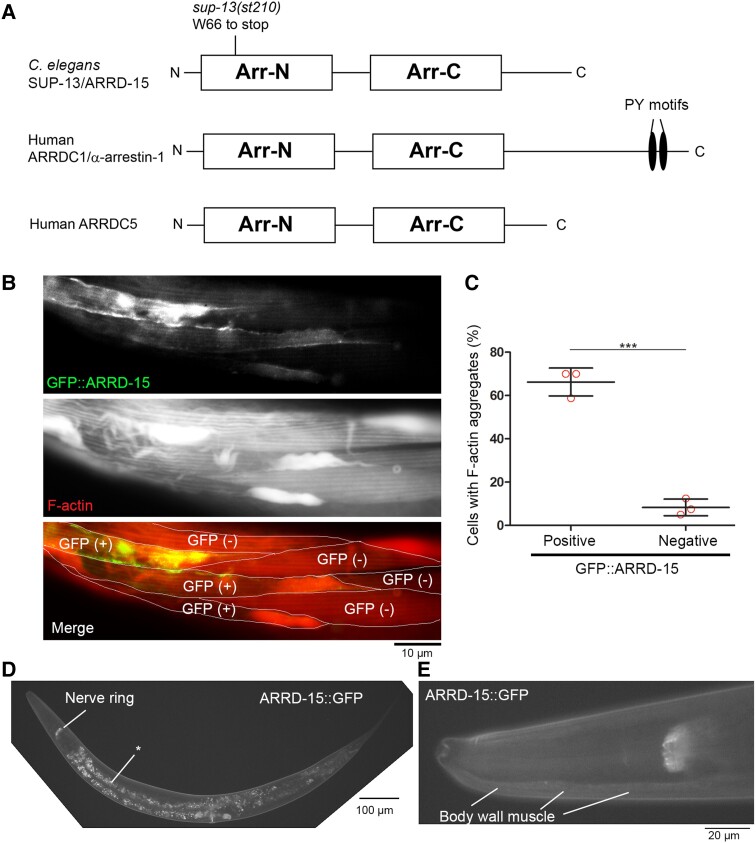
To understand the molecular nature of the sup-13 mutation, we conducted whole-genome sequencing and identified that sup-13(st210) was a nonsense mutation [W66 (TGG) to stop (TGA)] in the arrd-15 gene encoding an α-arrestin (Fig. 2A). The SUP-13/ARRD-15 protein (NCBI Reference Sequence: NP_001254970.1) is a previously uncharacterized member of α-arrestins, also known as arrestin-domain-containing proteins (ARRDCs) (Fig. 2A). These proteins commonly contain arrestin-N (Arr-N) and arrestin-C (Arr-C) domains (34–36) (Fig. 2A). In yeast, α-arrestins are also named arrestin-related trafficking adaptors (ARTs) and involved in protein trafficking and degradation as adaptors for ubiquitin (Ub) ligases (34–36). Many α-arrestins have multiple PY motifs (PP × Y or LP × Y) in their C-termini as binding sites for the WW domains of E3 Ub ligases (34–36) (Fig. 2A). However, SUP-13/ARRD-15 lacks a PY motif in the C-terminus, which is similar to mammalian ARRDC5, the least characterized member of α-arrestins (Fig. 2A).
Fig. 2.
sup-13 is a nonsense allele of arrd-15 encoding an α-arrestin. A) Domain organization of the C. elegans SUP-13/ARRD-15 protein (374 amino acids, NCBI Reference Sequence: NP_001254970.1) with a location of the sup-13(st210) mutation. Arrestin-N (Arr-N) and arrestin-C (Arr-C) domains are indicated by boxes. N- and C-termini are shown as N and C, respectively. For comparison, human ARRDC1/α-arrestin 1 (433 amino acids, NCBI Reference Sequence: NP_689498.1) and ARRDC5 (342 amino acids, NCBI Reference Sequence: NP_001073992.1) are shown. Multiple copies of PY motifs (PP × Y) are present in the C-termini of many α-arrestins but absent in SUP-13/ARRD-15 and human ARRDC5. B, C) Transgenic rescue of the sup-13 suppressor phenotype was demonstrated by expressing GFP::ARRD-15 in the body wall muscle of sup-13(st210);unc-78(gk27) using the myo-3 promoter. The worms were stained with tetramethylrhodamine-phalloidin, and sarcomeric actin filaments (F-actin) were examined in GFP::ARRD-15 positive [GFP (+)] and negative [GFP (−)] cells. In the merged image (GFP::ARRD-15 in green and F-actin in red), cell-cell boundaries are marked by white borders. B). Bar, 10 μm. The presence or absence of F-actin aggregates was examined in GFP::ARRD-15 positive and negative cells (34–40 cells per experiment) from worms with mosaic expression of GFP::ARRD-15, and percentages of cells with F-actin aggregates are shown in the graph (C). Data are average ± standard deviation of three independent experiments. ***, P < 0.001. D, E) Expression pattern of SUP-13/ARRD-15 in adult worms was examined by tagging endogenous SUP-13/ARRD-15 with GFP. Fluorescence images at low (D; bar, 100 μm) and high (E; bar 20 μm) magnifications show the entire worm and the head region, respectively. SUP-13/ARRD-15::GFP was detected in the nerve ring (D) and body wall muscle in a diffuse pattern (E). Because the fluorescence intensity of SUP-13/ARRD-15::GFP was weak, autofluorescence of the gut granules (indicated by an asterisk in D) appears relatively high.
The identity of arrd-15 as sup-13 was confirmed by transgenic rescue experiments. Transgenic expression of an N-terminally tagged green fluorescent protein (GFP)::ARRD-15 fusion protein in the body wall muscle of sup-13(st210);unc-78(gk27) suppressed the suppressor phenotype and induced actin aggregate formation (Fig. 2B). GFP::ARRD-15 was expressed in the body wall muscle of sup-13(st210);unc-78(gk27) using the myo-3 promoter from extrachromosomal arrays, which often cause transgenic expression in a mosaic manner (40). The mosaic expression of GFP::ARRD-15 allowed us to compare GFP::ARRD-15-positive and negative cells within the same animals and populations. Nearly 70% of GFP::ARRD-15-positive cells had F-actin aggregates (Fig. 2B and C), whereas over 90% of GFP::ARRD-15-negative cells had striated actin organization with no aggregates (Fig. 2B and C), indicating that the suppression of the Sup-13 suppressor phenotype was caused by the transgenic expression of GFP::ARRD-15.
Expression of ARRD-15 in the body wall muscle was detected by observing localization of endogenous sup-13/arrd-15 that was C-terminally tagged with GFP (ARRD-15::GFP) by CRISPR/Cas9-mediated gene editing (Fig. 2D and E). The fluorescence levels of ARRD-15::GFP were strongest in the nerve ring (Fig. 2D) and also detected at low levels in the body wall muscle and localized in the diffuse cytoplasm (Fig. 2E). This expression pattern is consistent with the mRNA distribution data in the GExplore database (41) (Online Supplementary Fig. S1). Together, these results of muscle-specific transgenic rescue and endogenous expression of sup-13/arrd-15 in the body wall muscle indicate that sup-13/arrd-15 acts autonomously in the body wall muscle to influence sarcomeric actin organization.
The second AIP1 isoform, aipl-1, is required for sup-13/arrd-15-dependent phenotypic suppression of the unc-78 mutant phenotypes
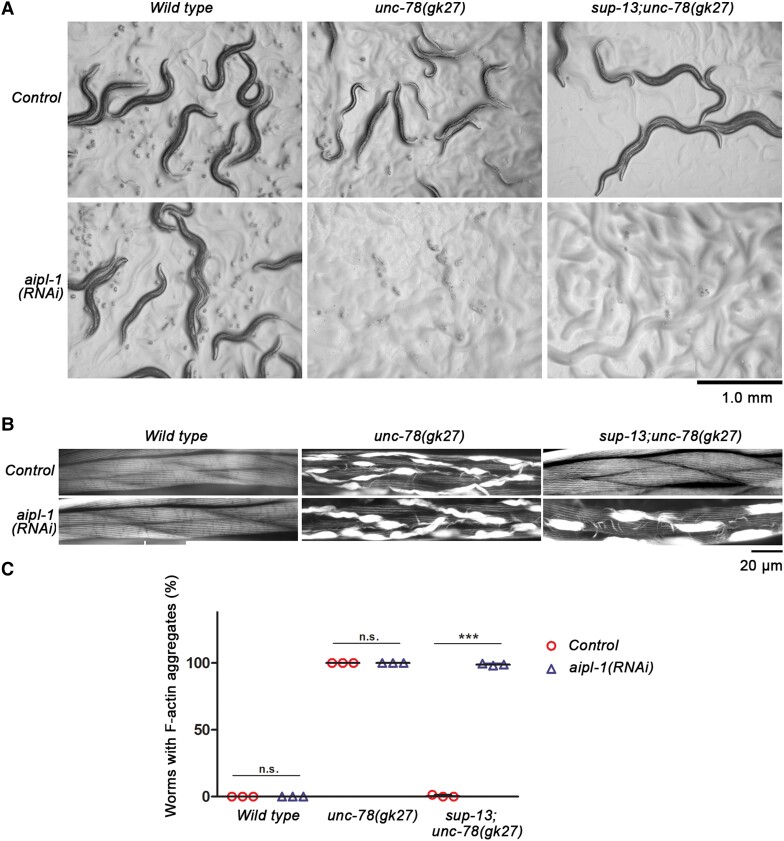
Caenorhabditis elegans has a second AIP1 gene, aipl-1, that is partially redundant with unc-78 in actin regulation (37, 38). We found that aipl-1 is essential for the phenotypic suppression by the sup-13/arrd-15 mutation in the unc-78 mutants (Fig. 3). Depletion of aipl-1 in wild type by RNA interference (RNAi) did not cause a detectable abnormality (Fig. 3A, left). However, in the unc-78(gk27) mutants, aipl-1(RNAi) caused embryonic arrest (Fig. 3A, middle), as reported previously (37), and this lethal phenotype was not suppressed by the sup-13 mutation (Fig. 3A, right). In order to observe adult phenotypes, the RNAi treatments were started from the L1 larval stage after normal embryogenesis was completed, and phenotypes analyzed in adult worms (Fig. 3B and C). Under these conditions, aipl-1(RNAi) did not cause detectable alterations in the phenotypes in wild type (Fig. 3B, left; Fig. 3C) and unc-78(gk27) (Fig. 3B, middle; Fig. 3C) but caused disorganization of actin filaments with aggregate formation in sup-13;unc-78(gk27) (Fig. 3B, right; Fig. 3C). These results and our previous reports demonstrate that unc-78 and aipl-1 have functional impacts at different developmental stages: unc-78 mutations cause stronger phenotypes in the late larval and adult stages than in the embryonic and early larval stages (28) (Fig. 1C), whereas aipl-1(RNAi) (in an unc-78 background) causes stronger phenotypes in the embryonic stages than in the adult stage (37). However, the strong adult phenotypes of aipl-1(RNAi) in sup-13;unc-78(gk27), but not in unc-78(gk27), suggest that aipl-1 became functionally significant at the adult stage in the sup-13 mutant background.
Fig. 3.
The second AIP1 gene aipl-1 is required for sup-13/arrd-15-dependent suppression of the unc-78 mutant phenotypes. A) Wild type, unc-78, or sup-13;unc-78 worms were treated with control RNAi or aipl-1(RNAi) from the L4 larval stage, and the phenotypes were examined on agar plates in the next generation. In control RNAi, all strains grew to adult worms. However, in aipl-1(RNAi), only wild type grew to adult worms, but unc-78 and sup-13;unc-78 were arrested as embryos. Bar, 1.0 mm. B) Wild type, unc-78, or sup-13;unc-78 worms were treated with control RNAi or aipl-1(RNAi) from the L1 larval stage, and stained for F-actin with tetramethylrhodamine-phalloidin when they became adults, and regions of the body wall muscle are shown. In adult muscle, aipl-1(RNAi) did not affect the F-actin organization in wild type and unc-78 but caused severe F-actin disorganization in sup-13;unc-78. Bar, 20 μm. C) Percentages of worms with F-actin aggregates (counted as positive when >25% of total muscle cells contained aggregates) were analyzed. For each experiment, >200 worms were examined for each sample, and average ± standard deviation of three independent experiments are shown (error bars are too small to be visible). ***, P < 0.001. n.s., not significant.
sup-13/arrd-15 controls the protein level of AIPL-1, but not UNC-78, possibly in a Ub-dependent manner
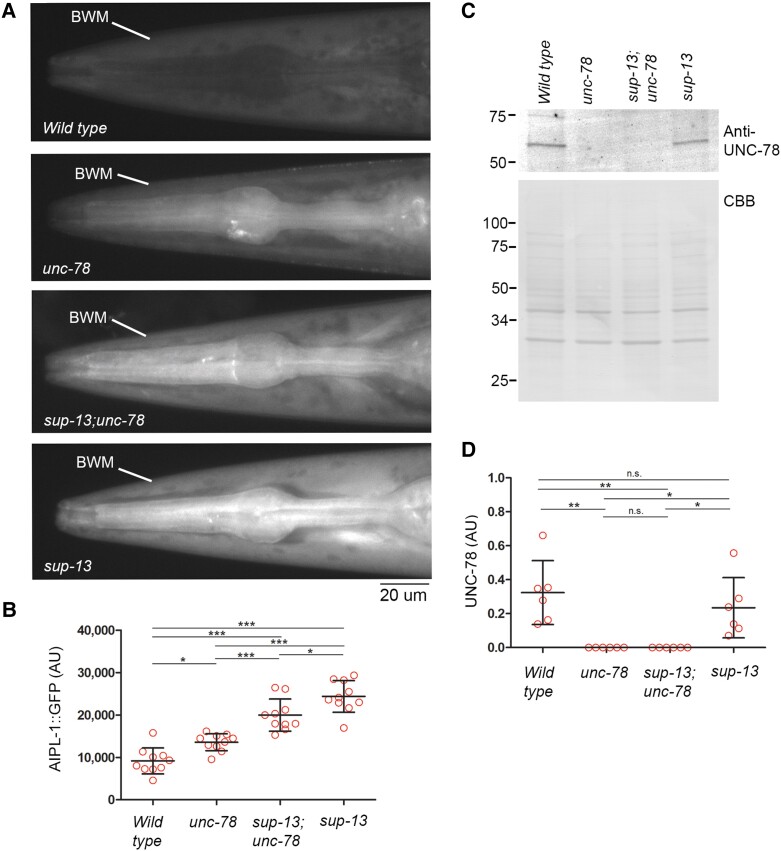
Examination of AIPL-1 protein levels revealed that the sup-13/arrd-15 mutation altered the AIPL-1 protein level in the adult body wall muscle (Fig. 4). Since AIPL-1 is expressed in many tissues (37), the endogenous aipl-1 gene was C-terminally tagged with GFP by CRISPR/Cas9-mediated gene editing, and its expression in the body wall muscle was examined by fluorescence microscopy. The AIPL-1::GFP protein levels were determined as fluorescence intensity. In wild type, AIPL-1::GFP was very low in the body wall muscle (Fig. 4A and B) and slightly increased in unc-78(gk27) (Fig. 4A and B). However, in sup-13;unc-78(gk27) and sup-13, AIPL-1::GFP was significantly increased in the body wall muscle as compared with wild type and unc-78(gk27) (Fig. 4A and B), indicating that the sup-13/arrd-15 mutation was responsible for the high levels of AIPL-1::GFP in the body wall muscle. In contrast, the UNC-78 protein levels were not altered by the sup-13/arrd-15 mutation (Fig. 4C and D). The UNC-78 protein was examined by Western blot using whole adult worm lysates because UNC-78 is predominantly expressed in the body wall muscle in adults (28, 30). Indistinguishable levels of UNC-78 were detected in wild type and sup-13, and the absence of UNC-78 was confirmed in unc-78(gk27) and sup-13;unc-78(gk27) (Fig. 4C and D). These results indicate that the sup-13/arrd-15 mutation specifically increases the level of AIPL-1, but not UNC-78, in the adult body wall muscle. Therefore, a normal function of sup-13/arrd-15 is to decrease the level of AIPL-1 in the adult muscle.
Fig. 4.
The sup-13/arrd-15 mutation increases the protein levels of AIPL-1 in the body wall muscle. A, B) The AIPL-1::GFP protein levels in the body wall muscle were determined in the head region where the autofluorescence of the gut granules is absent. Fluorescence images of AIPL-1::GFP were captured using the same exposure settings for the four strains (A; bar, 20 μm), and the AIPL-1::GFP protein levels were determined as fluorescence intensity (arbitrary units) within the body wall muscle (BWM in A) in B. Data are average ± standard deviation (n = 10). *, 0.01 < P < 0.05. ***, P < 0.001. C, D) The UNC-78 protein levels in whole worm lysates were determined by Western blot using anti-UNC-78 antibody. Whole worm lysates of an equal number of young adults (30 worms) were loaded for each strain and probed with anti-UNC-78 antibody (C, top). Total proteins were stained with Coomassie Brilliant Blue (C, bottom) and used as a loading control to normalize the UNC-78 signals (arbitrary units) in D. Positions of molecular weight markers in kDa are shown on the left. Data are average ± standard deviation (n = 6). *, 0.01 < P < 0.05. **, 0.001 < P < 0.01. n.s., not significant.
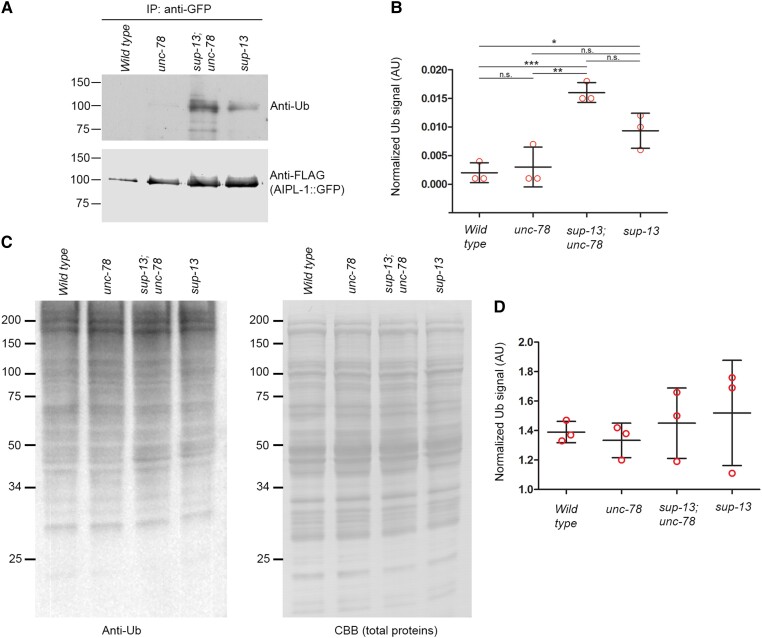
To determine the regulatory mechanism of AIPL-1 protein levels by sup-13/arrd-15, we examined the ubiquitination levels of AIPL-1 and found that ubiquitinated AIPL-1 was accumulated in the sup-13/arrd-15 mutants (Fig. 5A and B). AIPL-1::GFP was immunoprecipitated, and probed with anti-Ub antibody in Western blot (Fig. 5A). Ubiquitinated AIPL-1::GFP was nearly undetectable in wild type and unc-78(gk27) but detected at high levels in sup-13 and sup-13;unc-78(gk27) (Fig. 5A and B). Therefore, in the sup-13/arrd-15 mutants, AIPL-1 protein might be maintained at high levels because ubiquitinated AIPL-1 was accumulated without being processed by the downstream protein degradation pathway. However, levels of ubiquitinated proteins in total worm lysates were not significantly different among the four strains (Fig. 5C and D), suggesting that sup-13/arrd-15 regulates degradation of only specific target proteins rather than systemic degradation of ubiquitinated proteins. These results suggest that sup-13/arrd-15 acts after the ubiquitination process and promotes degradation of ubiquitinated AIPL-1.
Fig. 5.
Ubiquitinated AIPL-1 is accumulated in the sup-13/arrd-15 mutants. A, B) AIPL-1::GFP was immunoprecipitated with anti-GFP magnetic beads and probed with anti-Ub (Ub) antibody (A, top) and anti-DYKDDDDK (FLAG epitopes in AIPL-1::GFP) antibody (A, bottom). The anti-Ub signals (arbitrary units) were normalized to anti-DYKDDDDK signals and shown in the graph (B). Data are average ± standard deviation (n = 3). *, 0.01 < P < 0.05. **, 0.001 < P < 0.01. ***, P < 0.001. n.s., not significant. C, D) Whole lysates of adult worms were examined by Western blot using anti-Ub antibody (C, left). Total proteins on the blot were stained with Coomassie Brilliant Blue (CBB) (C, right) and used as a loading control to normalize the anti-Ub signals (arbitrary units) in D. Positions of molecular weight markers in kDa are shown on the left in A and C. Data are average ± standard deviation (n = 3). There were no significant differences among the data.
Discussion
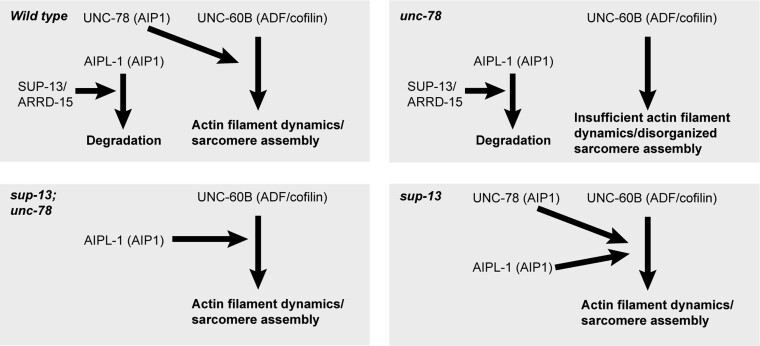
In this study, we identified the molecular nature of the sup-13/arrd-15 gene and characterized how the sup-13/arrd-15 mutation suppresses the unc-78 mutant phenotypes. Based on the current and previous data, we propose that the isoform turnover of AIP1 occurs at two levels in C. elegans muscle. First, expression of unc-78 and aipl-1 are transcriptionally regulated in an opposite manner as determined by the promoter analyses: the unc-78 promoter is more active in adult muscle than embryonic muscle (30), whereas the aipl-1 promoter is more active in embryonic muscle than in adult muscle (37). Therefore, during muscle development, expression of unc-78 is increased, but that of aipl-1 decreased. Second, the AIPL-1 protein is specifically cleared in adult muscle by sup-13/arrd-15 in a Ub-dependent manner. In the sup-13/arrd-15 mutants, AIPL-1 protein is maintained at high levels in adult muscle and is capable of compensating for the absence of UNC-78 in the unc-78-null mutants (Fig. 6). This is supported by our previous observations that forced expression of GFP::AIPL-1 in adult muscle of the unc-78-null mutants rescues the sarcomeric actin organization (37), indicating that AIPL-1 is sufficient for suppressing the unc-78-mutant phenotype. In addition, knockdown of aipl-1 in sup-13;unc-78(gk27) suppressed the Sup-13 suppressor phenotype (Fig. 3), indicating that aipl-1 is necessary for suppressing the unc-78-mutant phenotype in the sup-13/arrd-15 mutant background. Thus, the transition of AIP1 isoforms in C. elegans muscle appears to be a highly regulated process.
Fig. 6.
Model for the regulatory mechanism of AIP1 isoform turnover in the body wall muscle. The current study suggests that SUP-13/ARRD-15 specifically targets AIPL-1 for degradation in the body wall muscle. Therefore, in wild type, UNC-78 is the predominant AIP1 isoform that cooperates with UNC-60B (ADF/cofilin) to promote actin filament dynamics. In unc-78 single mutants, there is not enough AIP1 proteins to promote actin filament dynamics, causing sarcomere defects. However, in sup-13;unc-78 double mutants, AIPL-1 is not degraded and compensates for the absence of UNC-78. In sup-13 single mutants, both UNC-78 and AIPL-1 promotes actin filament dynamics and do not cause apparent sarcomere defects.
Multiple AIP1 isoforms are present only in a limited number of species, and protein levels of AIP1 may be regulated in a species-specific manner. In Arabidopsis thaliana, two AIP1 isoforms are differentially expressed in reproductive and vegetative tissues and regulate actin organization in respective tissues (42–44). However, most species have only one gene encoding AIP1 (5, 6, 45). Mammals have a single AIP1 gene, known as WDR1 (46). In the mouse heart, the WDR1 protein level is high in embryos and then gradually decreased by unknown mechanism during postembryonic development (31). AIP1 can strongly sever ADF/cofilin-bound actin filaments at very low concentrations in vitro (12–16). Therefore, high AIP1/WDR1 levels may be maintained when actin cytoskeletal reorganization is highly active in embryonic heart, but AIP1/WDR1 may be decreased when mature myofibrils are assembled and stabilized in postembryonic heart. However, it is still not clear why turnover of the AIP1 isoforms occurs in C. elegans muscle. Biochemically, UNC-78 and AIPL-1 are qualitatively very similar in promoting disassembly of ADF/cofilin-bound actin filaments (37), preference for ADF/cofilin isoforms (29, 37), and pH sensitivity (45). Genetically, unc-78 and aipl-1 are partially redundant in assembly of contractile apparatuses in striated and nonstriated muscles (37, 38). Therefore, there may be unknown differences in the function and/or regulation of the two AIP1 isoforms, which might be beneficial for C. elegans.
Generally, α-arrestins are known to promote ubiquitination of target proteins by recruiting E3 Ub ligases to the substrates (34–36). However, in our study, the AIPL-1 protein levels were increased with accumulation of ubiquitinated AIPL-1 protein in the sup-13/arrd-15 mutants. These results suggest that SUP-13/ARRD-15 is involved in processing ubiquitinated AIPL-1 for degradation rather than promoting ubiquitination of AIPL-1. Alternatively, SUP-13/ARRD-15 may indirectly regulate degradation of ubiquitinated AIPL-1. If there is a negative regulator of AIPL-1 degradation, promoting Ub-dependent degradation of this regulator by SUP-13/ARRD-15 can accelerate AIPL-1 degradation. Therefore, additional investigation is needed to identify other factors involved in the SUP-13/ARRD-15-mediated regulation of AIPL-1.
Most of the previously characterized α-arrestins have multiple PY motifs (PP × Y or LP × Y) in their C-termini as binding sites for the WW domains of Rsp5/Nedd4-like E3 Ub ligases (34–36). However, SUP-13/ARRD-15 lacks this motif (Fig. 2A), and how it is involved in the degradation of a target protein is currently unknown. In mammals, arrestin-domain-containing 5 (ARRDC5) is the only α-arrestin lacking a PY motif, and its biological function is poorly understood. One recent study has shown that ARRDC5 is enriched in testis and required for normal spermatogenesis in mice (47), although whether ARRDC5 is involved in protein degradation is unknown. C. elegans has one β-arrestin gene, arr-1 (48), and 28 α-arrestin genes. The arr-1 β-arrestin gene is involved in G-protein-coupled receptor signaling (49). However, only two α-arrestin genes, cnp-1/arrd-17 and ttm-2, have been previously characterized in C. elegans. cnp-1/arrd-17 is required for chemosensation (50). ttm-2 is required for defense against bacterial toxins (51). Either cnp-1/arrd-17 or ttm-2 does not have a consensus PY motif and has not been implicated in protein degradation. Therefore, metazoan α-arrestins lacking a PY motif may recruit a Ub ligase using an alternative mechanism such as phosphor-Ser/Thr dependent binding to a WW domain (52) or be involved in unknown biological processes.
Our study suggests that an α-arrestin is involved in isoform switching of muscle proteins during development. Many sarcomeric proteins undergo isoform transition during muscle differentiation, development, and under pathological conditions (53). Although isoform switching at levels of transcription and alternative splicing has been extensively studied, the clearance mechanism for old isoforms is still poorly understood. Since misexpression of inappropriate isoforms can cause pathological conditions in muscle, clearing old or misexpressed isoforms will be important to complete normal isoform transition. For example, abnormal expression of troponin T isoforms due to misregulated alternative splicing is implicated in the pathogenesis of myotonic dystrophy (54). Expression of a nonmuscle tropomyosin isoform in skeletal muscle can cause dystrophic phenotypes (55, 56). In addition to these structural proteins, cofilin isoforms also transition from a nonmuscle isoform (cofilin-1) to a muscle isoform (cofilin-2) during muscle differentiation (57, 58). Cofilin-1 is normally cleared by the Ub-proteasomal system after muscle differentiation, but failure to clear cofilin-1 from mature muscle causes sarcomere disorganization and impaired force generation thereby enhancing phenotypes in Emery-Dreifuss muscular dystrophy (59). The Ub-proteasomal system has been shown to be important for normal sarcomere assembly (60–62), and muscle atrophy, and hypertrophy (63–65). To date, Ub ligases, MuRF-1, MAFbx-1/atrogin-1, and Cbl-b have been associated with muscle atrophy (66–68), and UBR4 with hypertrophy (64), but function of an α-arrestin in a Ub-dependent pathway has not been determined in vertebrate muscle. Therefore, our study suggests a novel role of an α-arrestin in protein turnover in normal muscle development or under pathological conditions.
Materials and methods
Caenorhabditis elegans strains and culture
The worms were cultured following standard methods (69). N2 wild type, RW2337 sup-13(st210);unc-78(e1217), and CB1217 unc-78(e1217) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). ON5 unc-78(gk27) was reported previously (28). ON320 sup-13(st210);unc-78(gk27) was generated by crossing RW2337 and ON5 unc-78(gk27) and selecting sup-13(st210);unc-78(gk27) homozygotes in the F2 generation. ON341 sup-13(st210) was generated by crossing ON320 and N2 and selecting sup-13(st210) homozygotes in the F2 generation. Other transgenic strains are described below. All strains used in this study are listed in Online Supplementary Table S1.
Motility assays
Worm motility was quantified as described previously (70). Briefly, adult worms were placed in M9 buffer. Then, one beat was counted when a worm swung its head to either right or left. The total number of beats in 30 s was recorded.
Genomic DNA isolation and whole-genome sequencing
We sequenced RW2337 sup-13(st210) III;unc-78(e1217) X and CB1217 unc-78(e1217) X as described previously (71). Genomic DNA was submitted to the British Columbia Cancer Agency Canada’s Michael Smith Genome Sciences Centre for whole-genome sequencing using the Illumina PET HiSeq technology to produce 150 bp long paired end reads. Burrows-Wheeler Alignment (BWA) (72) was used to align the sequencing reads against the C. elegans reference genome (version WS249). SAMtools (73) was applied to remove duplicate reads. Integrative Genomics Viewer (74, 75) was employed to identify the breakpoints of large deletions, medium sized insertions, and translocations. Finally, CooVar (76) was used to examine the effect of the variations on the coding sequences. sup-13 was previously mapped near the center of chromosome III by three-factor mapping using dpy-17 and lon-1 (32, 77). The sequences of RW2337 and CB1217 were compared, and a G > A mutation at III:8934387 was the only nonsense mutation (W66 to stop in arrd-15) within a 5-Mb interval in a protein-coding gene in the sup-13 mutant (RW2337), which is an expected mutation frequency for ethyl methanesulfonate mutagenesis in C. elegans as reported in the Million Mutation Project (78). To determine whether this change is the sup-13 mutation, RW2337 was outcrossed with ON5 unc-78(gk27), and mutations in the F2 sup-13;unc-27(gk27) double homozygotes were analyzed. As a result, the nonsense mutation in the arrd-15 gene was linked with the Sup-13 suppressor phenotype.
Transgenic expression of GFP::ARRD-15 in the body wall muscle
The cDNA for ARRD-15 (NCBI Reference Sequence: NP_001254970.1) was cloned in-frame at the 3′ end of the GFP sequence of pPD-118.20 (provided by Dr. Andrew Fire, Stanford University) containing the myo-3 promoter that is active in the body wall muscle (79), and injected into the syncytial gonad of ON320 sup-13(st210) III;unc-78(gk27) X to generate transgenic animals with extrachromosomal arrays as described (30). The generated strain is ON369 sup-13(st210) III;unc-78(gk27) X;ktEx252[myo-3p::gfp::ARRD-15a, pRF4].
CRISPR/Cas9-mediated genome editing
For the aipl-1::gfp strain, the GFP sequence was inserted in the genome in-frame at the 3′-end of the aipl-1 coding region by CRISPR/Cas9-mediated genome editing using vectors and protocols described by Dickinson et al. (80). A single guide (sg) RNA target sequence (TATAAACATGAAAACTAGAG) was cloned into pDD162 (Addgene Plasmid # 47549) for expression of both sgRNA and Cas9 nuclease. Homology arms of ∼500 bp each were fused with the GFP::self-excising cassette region (also containing three copies of FLAG/DYKDDDDK epitopes) from pDD282 (Addgene Plasmid #66823) by fusion PCR using Q5 High-Fidelity DNA Polymerase (New England Biolabs) and used as a homologous repair template as described previously (81). Hygromycin-resistant worms with a roller phenotype were isolated as knock-in worms. They were treated at 34°C for 4 h to induce excision of the self-excising cassette (80). In the next generation, nonroller GFP-positive worms were isolated and outcrossed three times, and ON355 aipl-1(kt4[aipl-1::3xFLAG::gfp]) was established.
For the arrd-15::gfp strain, the GFP sequence was inserted in the genome in-frame at the 3′-end of the arrd-15 coding region by CRISPR/Cas9-mediated genome editing at SunyBiotech (Fujian, China), and PHX4137 arrd-15(syb4137[arrd-15::gfp]) was isolated.
RNAi experiments
RNAi experiments were performed by feeding with Escherichia coli expressing double-stranded RNA as described previously (82). An RNAi clone for aipl-1 (V-9L16) was obtained from MRC Geneservice (Cambridge, United Kingdom) (83). An empty vector L4440 (provided by Dr. Andrew Fire, Stanford University) was used as a control. When L4 larvae were treated with RNAi, phenotypes were characterized in the F1 generation (82). When L1 larvae were treated with RNAi, phenotypes were characterized when the treated worms became adults (84).
Immunoprecipitation of AIPL-1::GFP
Age-synchronized young adults were mixed with ice cold immunoprecipitation buffer (25 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5% IGEPAL CA-630, 1 mM phenylmethylsulfonyl fluoride, 1 μM leupeptin and 1 μM pepstatin) and mechanically lysed on ice with 20 passages through a Balch cell homogenizer with a 12 μm tungsten carbide ball bearing (Isobiotec, Heidelberg Germany) (85). The lysates were centrifuged at 16,100 × g for 10 min at 4°C. Protein concentrations were determined using a Pierce BCA Protein Assay Kit (catalog # 23225, Thermo Fisher Scientific, Waltham, MA), and an equal amount of proteins (> 1 mg) was used for immunoprecipitation. The lysates were incubated for 1 h at 4°C with 10 μl suspension (binding capacity for 0.8–1.1 μg of GFP) of GFP-Trap Magnetic Particles M-270 (catalog # gtd, ChromoTek, Planegg Germany), which was pre-equilibrated with the immunoprecipitation buffer. The magnetic particles were washed three times with the immunoprecipitation buffer, and bound proteins were eluted by adding 15 μl of sodium dodecyl sulfate (SDS) lysis buffer (2% SDS, 80 mM Tris-HCl, 5% β-mercaptoethanol, 15% glycerol, 0.05% bromophenol blue, pH 6.8) and heating at 95°C for 5 min. Magnetic particles were separated from solution and the supernatant stored at −20°C for downstream application.
Western blotting
Whole worm lysates were prepared from an equal number of young adults (30 worms for Fig. 4C) by directly harvesting worms in 15 μl of SDS lysis buffer or harvesting and washing age-synchronized worms in M9 buffer (Fig. 5C) and lysis in SDS lysis buffer, which were heated at 95°C for 2 min followed by sonication, and another heating at 95°C for 2 min. Protein concentrations of the latter were determined by a filter-paper dye binding assay using Coomassie Brilliant Blue G (86), and 10 μg proteins were used for each sample. The lysates were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (10 or 12% polyacrylamide gel), and the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with Bullet Blocking One (catalog # 13779-56, Nacalai USA, San Diego, CA) for 5 min at room temperature. For detection of UNC-78 (Fig. 4C), the blots were probed with rabbit anti-UNC-78 polyclonal antibody (1:500 dilution) (29), diluted in Signal Enhancer HIKARI solution (Nacalai USA, San Diego, CA). After washing with phosphate-buffered saline containing 0.1% Tween 20 (PBS-T), the membranes were treated with IRDye 680RD goat anti-rabbit IgG polyclonal secondary antibody (1:7,500 dilution) (catalog # 926-68071, Li-Cor, Lincoln, NE), washed with PBS-T, and scanned with the Odyssey Classic Infrared Imaging System (Li-Cor). For detection of ubiquitinated proteins in whole worm lysates (Fig. 5C), the blots were probed with anti-Ub mouse monoclonal (P4D1) antibody (1:1,000 dilution) (Cat # 3739, Cell Signaling, Danvers, MA), followed by horseradish peroxidase-conjugated goat anti-mouse IgG antibody (1:2,000 dilution) (catalog # 31431, Thermo Fisher Scientific, Waltham, MA). The signals were detected by incubating with a SuperSignal West Pico Chemiluminescent Substrate (catalog # 34080, Thermo Fisher Scientific), and images acquired by an Azure Biosystems c600 imaging system. The band intensity was quantified using Image Studio Lite (Li-Cor, NE USA) for Fig. 4C or ImageJ (87) for Fig. 5C, and normalized to total protein amounts as determined by staining membranes with Coomassie Brilliant Blue R-250 and densitometry using ImageJ as described previously (88).
To examine AIPL-1 ubiquitination in the immunoprecipitated samples, the samples were subjected to SDS-PAGE and transferring to PVDF membranes, as described above. The membranes were probed with anti-Ub mouse monoclonal (P4D1) antibody (1:1,000 dilution) (Cat # 3739, Cell Signaling, Massachusetts USA), followed by horseradish peroxidase-conjugated goat anti-mouse IgG antibody (1:2,000 dilution) (catalog # 31431, Thermo Fisher Scientific, Waltham, MA). The signals were detected by incubating with a SuperSignal West Pico Chemiluminescent Substrate (catalog # 34080, Thermo Fisher Scientific) and exposing to X-ray films. The membranes were blocked again, and probed with rabbit anti-DYKDDDDK epitope polyclonal antibody (1:5,000 dilution) to detect AIPL-1::GFP with three copies of the FLAG epitope (DYKDDDDK), followed by incubation with IRDye 680RD goat anti-rabbit IgG polyclonal secondary antibody (1:7500). After washing with PBS-T, the signals were detected by scanning with an Odyssey Classic Infrared Imaging System. The signals for anti-Ub and anti-DYKDDDDK were quantified with ImageJ and Image Studio Lite, respectively, and the anti-Ub signals were normalized to the anti-DYKDDDDK signals.
Fluorescence microscopy
Staining of worms with tetramethylrhodamine-phalloidin (catalog # P1951, MilliporeSigma, St. Louis, MO) was performed as described (28, 89). Samples were observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope (Nikon Instruments, Tokyo, Japan) with a CFI Plan Fluor ELWD 40 × (NA 0.60) objective. Images were captured by a Hamamatsu ORCA Flash 4.0 LT sCMOS camera (Hamamatsu Photonics, Shizuoka, Japan) and processed by NIS-Elements AR V5.02.01 (Nikon Instruments) and Adobe Photoshop CS3.
To measure fluorescence intensity of AIPL-1::GFP, live worms were mounted and immobilized with 25% Pluronic F-127 (catalog # 2730-50G, Biovision, Milpitas, CA) in M9 buffer containing 0.5 mM levamisole and 0.1% tricaine methanesulfonate. Fluorescence images were captured with the same settings for all samples in NIS-Elements using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40 × (NA 0.60) objective. Fluorescence intensity of each worm was determined using ImageJ as average fluorescence intensity at 10 randomly selected points within the cytoplasm of the body wall muscle in the head region.
Statistics
The following methods were used for statistical tests in GraphPad Prism 5 (Version 5.04): one-way analysis of variance (ANOVA) and Tukey post hoc test (Figs. 1A; 4B and D; and 5B and D); two-way ANOVA and Bonferonni post hoc test (Fig. 1C); unpaired t-test (Figs. 2C and 3C).
Supplementary Material
Contributor Information
Mario Lewis, Department of Pathology, Emory University School of Medicine, Atlanta, GA 30322, USA; Department of Cell Biology, Emory University School of Medicine, Atlanta, GA 30322, USA.
Kanako Ono, Department of Pathology, Emory University School of Medicine, Atlanta, GA 30322, USA; Department of Cell Biology, Emory University School of Medicine, Atlanta, GA 30322, USA.
Zhaozhao Qin, Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
Robert C Johnsen, Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
David L Baillie, Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
Shoichiro Ono, Department of Pathology, Emory University School of Medicine, Atlanta, GA 30322, USA; Department of Cell Biology, Emory University School of Medicine, Atlanta, GA 30322, USA; Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Supplementary material
Supplementary material is available at PNAS Nexus online.
Funding
Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (P40 OD010440). This work was supported by grants from the National Institutes of Health (AR048615) to S.O. and National Science and Engineering Research Council of Canada (9975-2011) to D.L.B.
Author contributions
M.L. and K.O. performed experiments, analyzed data, and generated figures. Z.Q. and R.C.J. performed experiments and analyzed data. D.L.B. designed experiments and analyzed data. S.O. designed experiments, analyzed data, and wrote the manuscript.
Data availability
All data are contained in the article.
References
- 1. De La Cruz EM, Gardel ML. 2015. Actin mechanics and fragmentation. J Biol Chem. 290:17137–17144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ono S. 2007. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 258:1–82. [DOI] [PubMed] [Google Scholar]
- 3. Brieher W. 2013. Mechanisms of actin disassembly. Mol Biol Cell. 24:2299–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amberg DC, Basart E, Botstein D. 1995. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 2:28–35. [DOI] [PubMed] [Google Scholar]
- 5. Ono S. 2003. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry. 42:13363–13370. [DOI] [PubMed] [Google Scholar]
- 6. Ono S. 2018. Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation. Biochem Biophys Res Commun. 506:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voegtli WC, Madrona AY, Wilson DK. 2003. The structure of Aip1p, a WD repeat protein that regulates cofilin-mediated actin depolymerization. J Biol Chem. 278:34373–34379. [DOI] [PubMed] [Google Scholar]
- 8. Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. 2004. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 279:31697–31707. [DOI] [PubMed] [Google Scholar]
- 9. Okada K, Obinata T, Abe H. 1999. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J Cell Sci. 112:1553–1565. [DOI] [PubMed] [Google Scholar]
- 10. Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. 1999. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 145:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aizawa H, et al. 1999. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 4:311–324. [DOI] [PubMed] [Google Scholar]
- 12. Ono S, Mohri K, Ono K. 2004. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 279:14207–14212. [DOI] [PubMed] [Google Scholar]
- 13. Nadkarni AV, Brieher WM. 2014. Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr Biol. 24:2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Q, Courtemanche N, Pollard TD. 2015. Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. J Biol Chem. 290:2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gressin L, Guillotin A, Guerin C, Blanchoin L, Michelot A. 2015. Architecture dependence of actin filament network disassembly. Curr Biol. 25:1437–1447. [DOI] [PubMed] [Google Scholar]
- 16. Jansen S, et al. 2015. Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nat Comm. 6:7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayakawa K, Sekiguchi C, Sokabe M, Ono S, Tatsumi H. 2019. Real-time single-molecule kinetic analyses of AIP1-enhanced actin filament severing in the presence of cofilin. J Mol Biol. 431:308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang VW, Nadkarni AV, Brieher WM. 2020. Catastrophic actin filament bursting by cofilin, Aip1, and coronin. J Biol Chem. 295:13299–13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kueh HY, Charras GT, Mitchison TJ, Brieher WM. 2008. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 182:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu R, Ono S. 2006. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton. 63:659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Standing AS, et al. 2017. Autoinflammatory periodic fever, immunodeficiency, and thrombocytopenia (PFIT) caused by mutation in actin-regulatory gene WDR1. J Exp Med. 214:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhns DB, et al. 2016. Cytoskeletal abnormalities and neutrophil dysfunction in WDR1 deficiency. Blood. 128:2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Papadopoulou C, et al. 2019. Monogenic mimics of Behcet’s Disease in the young. Rheumatology. 58:1227–1238. [DOI] [PubMed] [Google Scholar]
- 24. Southwick FS. 2016. The lazy leukocyte syndrome revisited. Blood. 128:2112–2113. [DOI] [PubMed] [Google Scholar]
- 25. Tur-Gracia S, Martinez-Quiles N. 2021. Emerging functions of cytoskeletal proteins in immune diseases. J Cell Sci. 134:jcs253534. [DOI] [PubMed] [Google Scholar]
- 26. Kile BT, et al. 2007. Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia. Blood. 110:2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bowes C, et al. 2019. Coronin 1A depletion restores the nuclear stability and viability of Aip1/Wdr1-deficient neutrophils. J Cell Biol. 218:3258–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ono S. 2001. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J Cell Biol. 152:1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mohri K, Ono S. 2003. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J Cell Sci. 116:4107–4118. [DOI] [PubMed] [Google Scholar]
- 30. Mohri K, Ono K, Yu R, Yamashiro S, Ono S. 2006. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol Biol Cell. 17:2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan B, et al. 2014. A cardiomyocyte-specific Wdr1 knockout demonstrates essential functional roles for actin disassembly during myocardial growth and maintenance in mice. Am J Pathol. 184:1967–1980. [DOI] [PubMed] [Google Scholar]
- 32. Waterston RH, Francis GR. 1985. Genetic analysis of muscle development in Caenorhabditis elegans. Trends Neurosci. 8:270–276. [Google Scholar]
- 33. Waterston RH. 1988. Muscle. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Woodbury (NY): Cold Spring Harbor Laboratory Press. p. 281–335. [Google Scholar]
- 34. Puca L, Brou C. 2014. Alpha-arrestins—new players in notch and GPCR signaling pathways in mammals. J Cell Sci. 127:1359–1367. [DOI] [PubMed] [Google Scholar]
- 35. Kahlhofer J, Leon S, Teis D, Schmidt O. 2021. The alpha-arrestin family of ubiquitin ligase adaptors links metabolism with selective endocytosis. Biol Cell. 113:183–219. [DOI] [PubMed] [Google Scholar]
- 36. Zbieralski K, Wawrzycka D. 2022. Alpha-arrestins and their functions: from yeast to human health. Int J Mol Sci. 23:4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ono S, et al. 2011. The two actin-interacting protein 1 genes have overlapping and essential function for embryonic development in Caenorhabditis elegans. Mol Biol Cell. 22:2258–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ono K, Ono S. 2014. Two actin-interacting protein 1 isoforms function redundantly in the somatic gonad and are essential for reproduction in Caenorhabditis elegans. Cytoskeleton. 71:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mello C, Fire A. 1995. DNA transformation. Methods Cell Biol. 48:451–482. [PubMed] [Google Scholar]
- 41. Hutter H, Ng MP, Chen N. 2009. GExplore: a web server for integrated queries of protein domains, gene expression and mutant phenotypes. BMC Genomics. 10:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allwood EG, et al. 2002. Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell. 14:2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kiefer CS, et al. 2015. Arabidopsis AIP1-2 restricted by WER-mediated patterning modulates planar polarity. Development. 142:151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diao M, Li X, Huang S. 2020. Arabidopsis AIP1-1 regulates the organization of apical actin filaments by promoting their turnover in pollen tubes. Sci China Life Sci. 63:239–250. [DOI] [PubMed] [Google Scholar]
- 45. Nomura K, Hayakawa K, Tatsumi H, Ono S. 2016. Actin-interacting protein 1 promotes disassembly of actin-depolymerizing factor/cofilin-bound actin filaments in a pH-dependent manner. J Biol Chem. 291:5146–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adler HJ, Winnicki RS, Gong TW, Lomax MI. 1999. A gene upregulated in the acoustically damaged chick basilar papilla encodes a novel WD40 repeat protein. Genomics. 56:59–69. [DOI] [PubMed] [Google Scholar]
- 47. Giassetti MI, et al. 2023. ARRDC5 Expression is conserved in mammalian testes and required for normal sperm morphogenesis. Nat Comm. 14:2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fukuto HS, et al. 2004. G protein-coupled receptor kinase function is essential for chemosensation in C. elegans. Neuron. 42:581–593. [DOI] [PubMed] [Google Scholar]
- 49. Palmitessa A, et al. 2005. Caenorhabditus elegans arrestin regulates neural G protein signaling and olfactory adaptation and recovery. J Biol Chem. 280:24649–24662. [DOI] [PubMed] [Google Scholar]
- 50. Jee C, et al. 2012. CNP-1 (ARRD-17), a novel substrate of calcineurin, is critical for modulation of egg-laying and locomotion in response to food and lysine sensation in Caenorhabditis elegans. J Mol Biol. 417:165–178. [DOI] [PubMed] [Google Scholar]
- 51. Huffman DL, et al. 2004. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A. 101:10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reinhardt HC, Yaffe MB. 2013. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol. 14:563–580. [DOI] [PubMed] [Google Scholar]
- 53. Ahmed RE, Tokuyama T, Anzai T, Chanthra N, Uosaki H. 2022. Sarcomere maturation: function acquisition, molecular mechanism, and interplay with other organelles. Philos Trans R Soc Lond B Biol Sci. 377:20210325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ho TH, et al. 2004. Muscleblind proteins regulate alternative splicing. EMBO J. 23:3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kee AJ, Gunning PW, Hardeman EC. 2009. A cytoskeletal tropomyosin can compromise the structural integrity of skeletal muscle. Cell Motil Cytoskeleton. 66:710–720. [DOI] [PubMed] [Google Scholar]
- 56. Kee AJ, et al. 2004. Sorting of a nonmuscle tropomyosin to a novel cytoskeletal compartment in skeletal muscle results in muscular dystrophy. J Cell Biol. 166:685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ono S, Minami N, Abe H, Obinata T. 1994. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J Biol Chem. 269:15280–15286. [PubMed] [Google Scholar]
- 58. Vartiainen MK, et al. 2002. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell. 13:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vignier N, et al. 2021. The non-muscle ADF/cofilin-1 controls sarcomeric actin filament integrity and force production in striated muscle laminopathies. Cell Rep. 36:109601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang J, et al. 2020. Myofibril assembly and the roles of the ubiquitin proteasome system. Cytoskeleton (Hoboken). 77:456–479. [DOI] [PubMed] [Google Scholar]
- 61. Wang J, et al. 2021. Inhibitors of the ubiquitin proteasome system block myofibril assembly in cardiomyocytes derived from chick embryos and human pluripotent stem cells. Cytoskeleton (Hoboken). 78:461–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carlisle C, Prill K, Pilgrim D. 2017. Chaperones and the proteasome system: regulating the construction and demolition of striated muscle. Int J Mol Sci. 19:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bonaldo P, Sandri M. 2013. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech. 6:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hunt LC, et al. 2019. A key role for the ubiquitin ligase UBR4 in myofiber hypertrophy in Drosophila and mice. Cell Rep. 28:1268–1281.e1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rodriguez J, et al. 2014. Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cell Mol Life Sci. 71:4361–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mayans O, Labeit S. 2012. MuRFs: specialized members of the TRIM/RBCC family with roles in the regulation of the trophic state of muscle and its metabolism. Adv Exp Med Biol. 770:119–129. [DOI] [PubMed] [Google Scholar]
- 67. Nikawa T, Ishidoh K. 2020. Ubiquitin ligase Cbl-b and inhibitory Cblin peptides. Biochim Biophys Acta Proteins Proteom. 1868:140495. [DOI] [PubMed] [Google Scholar]
- 68. Rom O, Reznick AZ. 2016. The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radic Biol Med. 98:218–230. [DOI] [PubMed] [Google Scholar]
- 69. T. Stiernagle. 2006. Maintenance of C. elegans. WormBook 10.1895/wormbook.1.101.1, 201-11. [DOI]
- 70. Epstein HF, Thomson JN. 1974. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 250:579–580. [DOI] [PubMed] [Google Scholar]
- 71. Ono K, Qin Z, Johnsen RC, Baillie DL, Ono S. 2020. Kettin, the large actin-binding protein with multiple immunoglobulin domains, is essential for sarcomeric actin assembly and larval development in Caenorhabditis elegans. FEBS J. 287:659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotech. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vergara IA, Frech C, Chen N. 2012. Coovar: co-occurring variant analyzer. BMC Res Notes. 5:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Davis P, et al. 2022. Wormbase in 2022-data, processes, and tools for analyzing Caenorhabditis elegans. Genetics. 220:iyac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thompson O, et al. 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 135:385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200:1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ono S, Ono K. 2020. Two Caenorhabditis elegans calponin-related proteins have overlapping functions that maintain cytoskeletal integrity and are essential for reproduction. J Biol Chem. 295:12014–12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ono S, Ono K. 2002. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 156:1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kamath RS, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. [DOI] [PubMed] [Google Scholar]
- 84. Ono K, Yu R, Ono S. 2007. Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev Dyn. 236:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bhaskaran S, et al. 2011. Breaking Caenorhabditis elegans the easy way using the Balch homogenizer: an old tool for a new application. Anal Biochem. 413:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Minamide LS, Bamburg JR. 1990. A filter paper dye-binding assay for quantitative determination of protein without interference from reducing agents or detergents. Anal Biochem. 190:66–70. [DOI] [PubMed] [Google Scholar]
- 87. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Welinder C, Ekblad L. 2011. Coomassie staining as loading control in Western blot analysis. J Proteome Res. 10:1416–1419. [DOI] [PubMed] [Google Scholar]
- 89. Ono S. 2022. Imaging of actin cytoskeleton in the nematode Caenorhabditis elegans. Methods Mol Biol. 2364:149–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the article.