Abstract
Mutations in the rifampin resistance-determining (Rif) regions of the rpoB gene of Staphylococcus aureus mutants obtained during therapy or in vitro were analyzed by gene amplification and sequencing. Each of the resistant clinical isolates, including five nonrelated clones and two strains isolated from the same patient, and of the 10 in vitro mutants had a single base pair change that resulted in an amino acid substitution in the β subunit of RNA polymerase. Eight mutational changes at seven positions were found in cluster I of the central Rif region. Certain substitutions (His481/Tyr and Asp471/Tyr [S. aureus coordinates]) were present in several mutants. Substitutions Gln468/Arg, His481/Tyr, and Arg484/His, which conferred high-level rifampin resistance, were identical or in the same codon as those described in other bacterial genera, whereas Asp550/Gly has not been reported previously. Substitutions at codon 477 conferred high- or low-level resistance, depending on the nature of the new amino acid. The levels of resistance of in vivo and one-step in vitro mutants carrying identical mutations were similar, suggesting that no other resistance mechanism was present in the clinical isolates. On the basis of these data and the population distribution of more than 4,000 clinical S. aureus isolates, we propose ≤0.5 and ≥8 μg/ml as new breakpoints for the clinical categorization of this species relative to rifampin.
Staphylococcus is the bacterial genus most frequently responsible for infections of prosthetic devices, osteomyelitis, and endocarditis (11, 15, 23). During the last 10 years, the proportion of methicillin-resistant Staphylococcus aureus (MRSA) isolates in hospitals has reached nearly 29% in the United States (19) and 40% in Europe (27). Most MRSA isolates are resistant to multiple antibiotics (4, 16) and more than 50% are resistant to rifampin, whereas 1% of methicillin-susceptible S. aureus (MSSA) isolates are resistant to rifampin (27). Vancomycin is the therapy of choice against S. aureus when β-lactams are inappropriate. However, because of poor tissue diffusion and moderate bactericidal activity (2), vancomycin is often combined with rifampin for deep-seated infections (6, 10). Fluoroquinolones in association with rifampin also represent an alternative strategy for the prevention of the emergence of resistant mutants in the treatment of serious MSSA infections (7).
Rifampin acts by interacting specifically with the β subunit of the bacterial RNA polymerase encoded by the rpoB gene (1). Rifampin resistance in Escherichia coli (13, 22) and S. aureus is due to alterations in the target leading to a reduced affinity of the enzyme for the antibiotic (17). Alignment of the predicted amino acid sequence of the S. aureus RNA polymerase β subunit with those of other bacterial genera identified conserved domains in all the sequences (1). One of them, between amino acids 486 and 717 (E. coli coordinates), includes the loci responsible for rifampin resistance in E. coli (13), Mycobacterium tuberculosis (24), Streptococcus pneumoniae (9), and Neisseria meningitidis (5). We have thus amplified and sequenced portions of rpoB from rifampin-susceptible and -resistant S. aureus isolates from the same patient. These portions correspond to the mutated regions in rifampin-resistant mutants of the previously studied bacterial genera. Mutants obtained in vitro were also studied. Attempts to correlate the various levels of rifampin resistance and the position of the rpoB mutations were made.
MATERIALS AND METHODS
Bacterial strains.
Rifampin-susceptible and -resistant matched clinical strains of MSSA (three pairs and three strains from the same patient) or MRSA (two pairs) were used in the study. Each pair and the three strains from the same patient were isolated between 1992 and 1997 before and during therapy with rifampin either alone or in combination (Table 1). Plasmid-free S. aureus RN4220, which was susceptible to antibiotics, was used for the in vitro selection of rifampin-resistant mutants (14).
TABLE 1.
Origin and properties of the S. aureus strains
| Strain | Date of isolation (mo/yr) | Origin | Patient’s therapy | Resistance phenotypea | Rifampin MIC (μg/ml) |
|---|---|---|---|---|---|
| BM4626 | 4/1992 | Bone | Rifampin plus pefloxacin | Tc | 0.008 |
| BM4627 | 4/1992 | Pef Rif Tc | 128–256 | ||
| BM4364 | 2/1996 | Bone | Vancomycin, followed by rifampin plus pristinamycin | Emc Fos Fus Km Met Pc Pef Tm | 0.008 |
| BM4364-R | 3/1996 | Emc Fos Fus Km Met Pc Pef Rif Tm | 256 | ||
| BM4365 | 6/1996 | Blood | Rifampin | Pc | 0.008 |
| BM4365-R | 10/1996 | Pc Rif | 256 | ||
| BM4366 | 1/1997 | Skin | Rifampin | Pc Emc | 0.008 |
| BM4366-R | 1/1997 | Pc Emc Rif | 128–256 | ||
| BM4367 | 3/1997 | Urine | Rifampin plus vancomycin | Emc Fos Km Met Pc Pef Tm | 0.016 |
| BM4367-R | 4/1997 | Urine and pulmonary tree | Emc Fos Km Met Pc Pef Rif Tm | 4 | |
| BM4368 | 3/1997 | Urine | Rifampin plus vancomycin | Pc Tc | 0.016 |
| BM4368-R1 | 3/1997 | Pulmonary tree | Rifampin plus vancomycin | Pc Rif Tc | 128 |
| BM4368-R2 | 4/1997 | Pulmonary tree | Pc Rif Tc | 4 |
Emc, constitutive resistance to macrolide, lincosamide, and streptogramin β-type antibiotics; Fos, resistance to fosfomycin; Fus, resistance to fusidic acid; Km, resistance to kanamycin; Met, resistance to methicillin; Pc, resistance to penicillin G; Pef, resistance to pefloxacin; Rif, resistance to rifampin; Tm, resistance to tobramycin.
Isolation of rifampin-resistant mutants of S. aureus RN4220 and of clinical isolate MSSA BM4368.
Approximately 108 CFU of exponentially growing bacteria was plated onto Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) containing concentrations of rifampin (provided by Merrell-Dow Research, Milan, Italy) of 0.016 to 64 μg/ml. After 24 h of incubation at 37°C, the numbers of colonies on agar with rifampin concentrations of ≥0.032 μg/ml were counted and the mutation frequencies were determined relative to the total count of viable organisms plated.
Antibiotic susceptibility testing.
S. aureus strains were screened for rifampin resistance by the disk-agar diffusion method (Sanofi Diagnostics Pasteur), and resistance was confirmed by determination of the MICs by dilution in Mueller-Hinton agar (18) with an inoculum of 104 CFU per spot.
Analysis of total DNA by pulsed-field gel electrophoresis.
Contour-clamped homogeneous electric field electrophoresis of SmaI restriction endonuclease (United States Biochemicals, Cleveland, Ohio) digests of genomic DNA was performed with a CHEF-DR II system (Bio-Rad Laboratories, Nazareth, Belgium) as described previously (3). Strains were assigned to the same macrorestriction genotype when they shared electrophoretic restriction patterns that differed by three or fewer fragments (26) and displayed a coefficient of similarity (CS) equal to or greater than 0.85 (8). The CS was calculated as follows: CS = 2 × number of matching bands/total number of bands in both strains.
Detection of mutations in the rpoB gene.
Total DNA from S. aureus was purified (29) and was used as a template for amplification by PCR. Two portions of the rpoB gene from S. aureus were amplified: a 702-bp fragment from nucleotide positions 441 to 673 (S. aureus coordinates) corresponding to the so-called rifampin resistance-determining (Rif) region in the center of the E. coli rpoB gene (13) and a 158-bp fragment from nucleotide positions 94 to 144 in which a substitution (Val143/Phe) conferring rifampin resistance has also been reported in E. coli (22). The 20-mer oligodeoxyribonucleotides used as primers were F3 (5′-AGTCTATCACACCTCAACAA) and F4 (5′-TAATAGCCGCACCAGAATCA) for the larger fragment and D1 (5′-GTGTAAAAGTGCGTCTAATC) and D2 (5′-ATAAACGGATGGTGAACGAA) for the smaller fragment. Amplification was carried out in a 100-μl volume containing 40 pmol of each oligonucleotide primer, each 2′-deoxynucleoside 5′-triphosphate at a concentration of 100 mM, reaction buffer (United States Biochemicals), 2 μl of a template DNA sample containing 100 ng of DNA, and 1 U of Taq DNA polymerase (United States Biochemicals). The reactions were performed in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) for 35 cycles. The conditions were 4 min at 94°C for denaturation, 3 min at 54°C for preannealing, 45 s at 52°C for annealing, and 45 s at 72°C for polymerization for the 702-bp fragment. The PCR conditions for the 158-bp fragment differed by the lack of preannealing and by the use of an annealing temperature of 55°C. The amplification products were purified on Microspin S-400 HR columns (Pharmacia LKB Biotechnology, Uppsala, Sweden), cloned into pCRII vector (Original TA Cloning Kit; Invitrogen), and sequenced by the dideoxy chain termination method (20) with T7 DNA polymerase (T7 Sequencing kit; Pharmacia) and [α-35S]dATP (Amersham Radiochemical Center, Amersham, England).
RESULTS AND DISCUSSION
Population distribution of S. aureus.
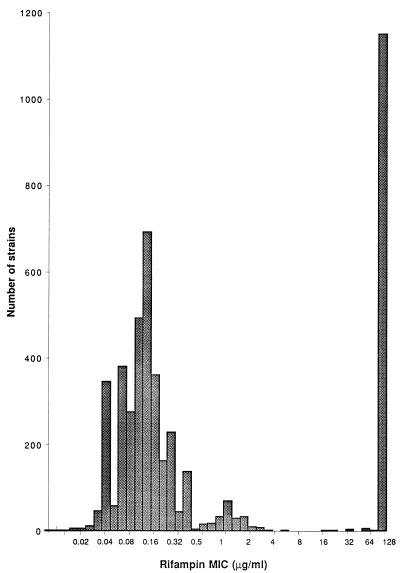
The MICs of rifampin for 4,644 clinical S. aureus strains isolated at Hopital Henri Mondor between 1993 and 1996 are presented in Fig. 1. On the basis of this multimodal distribution, the strains were categorized into categories of susceptible (MICs, ≤0.5 μg/ml), low-level resistant (MICs, 1 to 4 μg/ml), and high-level resistant (MICs, ≥8 μg/ml) according to the indicated breakpoints.
FIG. 1.
Distribution of rifampin MICs for 4,644 S. aureus strains isolated at Henri Mondor Hospital between 1993 and 1996.
Typing of clinical isolates.
The members of each of the five pairs and of the three isolates from the same patient had similar resistance phenotypes except for that for rifampin (Table 1). Pulsed-field gel electrophoresis of total DNA digested with SmaI is suitable for discrimination of S. aureus clones (25). The two members of each pair and the three strains from the same patient had indistinguishable DNA profiles (data not shown), indicating that the resistant mutants were selected in vivo under therapy with rifampin. The five pairs of clinical strains and the three isolates from the same patient displayed six SmaI patterns (CS range, 0.5 to 0.7) and were therefore not clonally related.
Selection of rifampin-resistant mutants.
Rifampin-resistant mutants were obtained at frequencies of 10−7 to 10−8 by plating S. aureus RN4220 (MIC, 0.008 μg/ml) and clinical isolate MSSA BM4368 (MIC, 0.016 μg/ml) onto solid medium containing rifampin at various concentrations. The mutation frequencies to low- and high-level rifampin resistance were similar and were not affected by the concentration of the selecting agent, which was less than 2 μg/ml. This result suggests that resistance to high levels of rifampin does not arise by sequential independent events but, rather, arises in a single-step fashion. The relative ease with which rifampin-resistant S. aureus mutants were obtained confirms that this antibiotic must not be prescribed alone and that considerable attention must be paid to interactions between rifampin and other antimicrobial agents. In fact, rifampin-resistant S. aureus BM4627 and BM4364-R mutants emerged in vivo under combination therapy. Ten mutants of S. aureus RN4220 obtained in vitro in two independent experiments were selected for further studies.
DNA sequence analysis and susceptibility to rifampin.
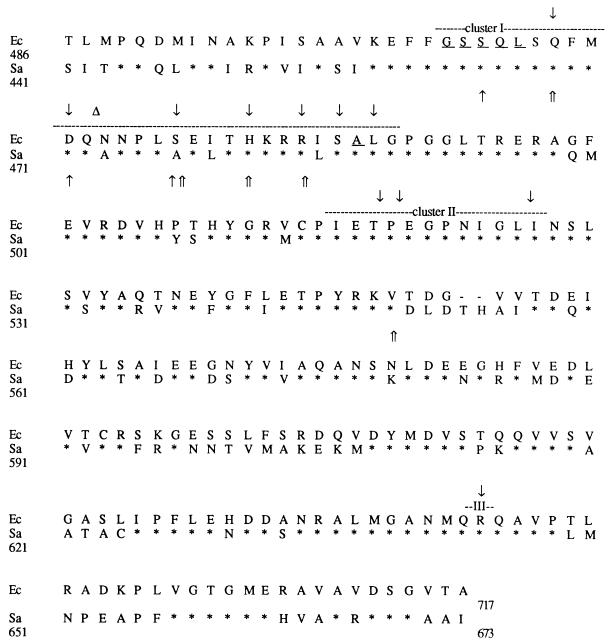
The sequences of the rpoB Rif gene region from nucleotides 441 to 673 (S. aureus coordinates) of seven high-level-resistant clinical isolates and of their susceptible counterparts, two in vitro low-level-resistant mutants (RN4220-R1 and RN4220-R2), and eight high-level-resistant mutants were determined (Fig. 2). Relative to their susceptible counterparts, the 17 in vitro and in vivo mutants had a single base pair change in the Rif region of the rpoB gene that resulted in an amino acid substitution, although the occurrence of other mutations in nonsequenced regions of the rpoB gene cannot be excluded. Eight mutational changes were found at seven positions: six clustered from nucleotide positions 464 to 484 and one was at position 550 (Table 2). The mutations in the E. coli rpoB gene involved in rifampin resistance can be assigned to three clusters, clusters I, II, and III (13). Since no strain had mutations in clusters II and III, we do not know if these regions are involved in rifampin resistance in S. aureus, as has been described for high- and low-level (cluster II) or very low level (cluster III) resistance in M. tuberculosis (24) and E. coli (13) (Fig. 2). Replacement of aspartate 550 by a glycine was a new substitution found outside the clusters at a position where E. coli and M. tuberculosis have a threonine and a valine, respectively.
FIG. 2.
Amino acid sequence comparison of the Rif regions of E. coli (Ec) and S. aureus (Sa). Clusters I, II, and III are indicated by dashed lines. Identical amino acids are indicated by asterisks, and gaps are indicated by dashes. Positions known to be involved in rifampin resistance in E. coli are indicated by downward-pointing arrows (mutations) or triangles (insertions) or are underlined (deletions). Mutations leading to low- and high-level rifampin resistance in S. aureus are indicated by single and double upward-pointing arrows, respectively.
TABLE 2.
Resistance to rifampin of S. aureus strains and mutations in the rpoB gene
| Strain | Rifampin concn (μg/ml) used for selection | Rifampin MIC (μg/ml) | Mutation (amino acid substitution)a | Corresponding mutation in the E. coli β subunit |
|---|---|---|---|---|
| RN4220-R5 | 2 | 4 | TCT→CCT (Ser464→Pro) | Δ[Gly507-Leu511]b |
| RN4220-R6 | 2 | 4 | TCT→CCT (Ser464→Pro) | Δ[Gly507-Leu511]b |
| RN4220-R10 | 32 | 256 | CAA→CGA (Gln468→Arg) | Gln513→Pro/Leu |
| RN4220-R2 | 0.016 | 2 | GAC→TAC (Asp471→Tyr) | Asp516→Val/Asn |
| BM4368-R2 | In vivo | 4 | GAC→TAC (Asp471→Tyr) | Asp516→Val/Asn |
| BM4367-R | In vivo | 4 | GAC→TAC (Asp471→Tyr) | Asp516→Val/Asn |
| RN4220-R1 | 0.016 | 1 | GCT→GTT (Ala477→Val) | Ser522→Phe |
| RN4220-R3 | 0.016 | 256 | GCT→GAT (Ala477→Asp) | Ser522→Phe |
| BM4368-R1 | In vivo | 128 | GCT→GAT (Ala477→Asp) | Ser522→Phe |
| RN4220-R4 | 0.032 | 256 | CAT→TAT (His481→Tyr) | His526→Tyr |
| RN4220-R7 | 2 | 256 | CAT→TAT (His481→Tyr) | His526→Tyr |
| RN4220-R8 | 2 | 256 | CAT→TAT (His481→Tyr) | His526→Tyr |
| RN4220-R9 | 16 | 256 | CAT→TAT (His481→Tyr) | His526→Tyr |
| BM4365-R | In vivo | 256 | CAT→TAT (His481→Tyr) | His526→Tyr |
| BM4364-R | In vivo | 256 | CAT→TAT (His481→Tyr) | His526→Tyr |
| BM4366-R | In vivo | 128 | CGT→CAT (Arg484→His) | Arg529→Cys/Ser |
| BM4627 | In vivo | 128 | GAT→GGT (Asp550→Gly) | NDc |
The underscores indicate the base changes.
Deletion.
ND, not described.
Certain mutations in cluster I were frequently found: the His481/Tyr substitution occurred in two clinical isolates and in four high-level-resistant mutants obtained independently in vitro; the Asp471/Tyr substitution was present in two in vivo low-level-resistant mutants, BM4368-R2 and BM4367-R, and in the in vitro low-level-resistant mutant RN4220-R2. The MICs of rifampin for mutants that were obtained in vivo and in vitro and that had identical mutations were similar and were within 1 dilution, suggesting that reduced affinity of the enzyme for the antibiotic following the occurrence of a mutation in the Rif region of the rpoB gene is the major mechanism of resistance in S. aureus.
Comparative analysis of the level of resistance to rifampin in S. aureus and of the mutation sites indicated that high-level resistance correlated with mutations at codons 468 and 481 and that low-level resistance was associated with a mutation at codon 471 (S. aureus coordinates). These codons correspond to those associated with respective similar levels of resistance in E. coli (13), N. meningitidis (5), M. tuberculosis (24), and Mycobacterium leprae (12). However, replacement of alanine at position 477 by a valine led to low-level resistance (mutant RN4220-R1), and replacement of alamine at position 477 by an aspartate led to high-level resistance (mutants RN4220-R3 and BM4368-R1). This is not surprising since alanine and valine are both hydrophobic amino acids, whereas aspartate is acidic. It thus appears that the level of rifampin resistance in S. aureus depends not only on the position of the mutation but also on the nature of the new amino acid.
Relative to the corresponding susceptible strain, BM4368, resistant clinical isolates BM4368-R1, selected under rifampin therapy (1,800 mg per day), and BM4368-R2, obtained 3 days after the end of the treatment, had single base pair changes that resulted in a different level of rifampin resistance. They shared the same mutation as in vitro low-level-resistant mutant RN4220-R2 and high-level-resistant mutant RN4220-R3, respectively (Table 2). The low- and high-level-resistant mutants characterized in the present study are adequately categorized by using the breakpoints based on the population distribution of a large number of clinical isolates (Fig. 1).
Mutations at positions 468 and 481 led to resistance to very high concentrations of rifampin and might point to amino acid changes in the β subunit that prevent the binding of rifampin to RNA polymerase. Indeed, it has been shown in E. coli that residues 516 to 540 (positions 471 to 495 in Rif region cluster I; S. aureus coordinates) are part of the target of rifampin (28) and participate, along with residues 1065 and 1237 (E. coli coordinates), in the formation of the initiation site when the β subunit is assembled in the RNA polymerase complex (21).
We also sequenced the rpoB region, from codons 94 to 144, which corresponds to the region containing a Rif mutation (Val146/Phe) in E. coli (22). None of the 17 S. aureus mutants examined in the present study had substitutions in their N-terminal cluster, which is distant, by almost 400 amino acids, from the central Rif region. This mutation, not detected in subsequent studies, is probably rare but suggests that the amino acid residue at position 146 and the central Rif region might jointly form the rifampin binding site (21).
In conclusion, using gene amplification and sequencing, we have established that rifampin resistance in S. aureus is probably due to mutations in the Rif region of the rpoB gene and that the resistance levels are dependent on both the location and the nature of the amino acid substitution. With one exception, the resistance mutations detected in S. aureus were identical to or were in the same codon as those in other eubacteria, confirming that the regions implicated in the interaction with rifampin are conserved among procaryotes.
ACKNOWLEDGMENTS
We thank R. Leclercq for having incited us to initiate this study and B. Périchon and G. Gerbaud for constant technical advice to H. Aubry-Damon.
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases.
REFERENCES
- 1.Aboshkiwa M, Rowland G, Coleman G. Nucleotide sequence of the Staphylococcus aureus RNA polymerase rpoB gene and comparison of its predicted amino acid sequence with those of other bacteria. Biochem Biophys Acta. 1995;1262:73–78. doi: 10.1016/0167-4781(95)00054-k. [DOI] [PubMed] [Google Scholar]
- 2.Ackerman B H, Vannier A M, Eudy E B. Analysis of vancomycin time-kill studies with Staphylococcus species by using a curve stripping program to describe the relationship between concentration and pharmacodynamic response. Antimicrob Agents Chemother. 1992;36:1766–1769. doi: 10.1128/aac.36.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubry-Damon H, Legrand P, Brun-Buisson C, Astier A, Soussy C-J, Leclercq R. Reemergence of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus: roles of an infection control program and changes in aminoglycoside use. Clin Infect Dis. 1997;25:647–653. doi: 10.1086/513749. [DOI] [PubMed] [Google Scholar]
- 4.Brumfitt W, Hamilton-Miller J. Methicillin-resistant Staphylococcus aureus. N Engl J Med. 1989;320:1188–1196. doi: 10.1056/NEJM198905043201806. [DOI] [PubMed] [Google Scholar]
- 5.Carter P E, Abadi F J R, Yakubu D E, Pennington T H. Molecular characterization of rifampin-resistant Neisseria meningitidis. Antimicrob Agents Chemother. 1994;38:1256–1261. doi: 10.1128/aac.38.6.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers H F. Methicillin-resistant staphylococci. Clin Microbiol Rev. 1988;1:173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desplaces N, Acar J F. New quinolones in the treatment of bone and joint infections. Rev Infect Dis. 1988;10:179–183. doi: 10.1093/clinids/10.supplement_1.s179. [DOI] [PubMed] [Google Scholar]
- 8.Dice L R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 9.Enright M, Zawadski P, Pickerill P, Dowson C G. Molecular evolution of rifampicin resistance in Streptococcus pneumoniae. Microb Drug Resist. 1998;4:65–70. doi: 10.1089/mdr.1998.4.65. [DOI] [PubMed] [Google Scholar]
- 10.Graziani A L, Lawson L A, Gibson G A, Steinberg M A, MacGregor R R. Vancomycin concentrations in infected and noninfected bone. Antimicrob Agents Chemother. 1988;32:1320–1322. doi: 10.1128/aac.32.9.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas D W, MacAndrew M P. Bacterial osteomyelitis in adults: evolving considerations in diagnosis and treatment. Am J Med. 1996;101:550–560. doi: 10.1016/s0002-9343(96)00260-4. [DOI] [PubMed] [Google Scholar]
- 12.Honoré N, Cole S T. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother. 1993;37:414–418. doi: 10.1128/aac.37.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 14.Kreiswirth B A, Löfdahl S, Betley M J, O’Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 15.Lew D P, Waldvogel F A. Osteomyelitis. N Engl J Med. 1997;336:999–1007. doi: 10.1056/NEJM199704033361406. [DOI] [PubMed] [Google Scholar]
- 16.Maple P A C, Hamilton-Miller J, Brumfitt W. Worldwide antibiotic resistance in methicillin-resistant Staphylococcus aureus. Lancet. 1989;ii:537–540. doi: 10.1016/s0140-6736(89)90076-7. [DOI] [PubMed] [Google Scholar]
- 17.Morrow T O, Harmon S A. Genetic analysis of Staphylococcus aureus RNA polymerase mutants. J Bacteriol. 1979;137:374–383. doi: 10.1128/jb.137.1.374-383.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 19.Panlilio A L, Culver D H, Gaynes R P, Banerjee S, Henderson T S, Tolson J S, Martone W J. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975–1991. Infect Control Hosp Epidemiol. 1992;13:582–586. doi: 10.1086/646432. [DOI] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severinov K, Mustaev A, Severinova E, Kozlov M, Darst S A, Goldfarb A. The beta subunit Rif-cluster I is only angstroms away from the active center of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:29428–29432. doi: 10.1074/jbc.270.49.29428. [DOI] [PubMed] [Google Scholar]
- 22.Severinov K, Soushko M, Goldfarb A, Nikiforov V. RifR mutations in the beginning of the Escherichia coli rpoB gene. Mol Gen Genet. 1994;244:120–126. doi: 10.1007/BF00283512. [DOI] [PubMed] [Google Scholar]
- 23.Sorrell T C, Packham D R, Shaker S, Foldes M, Munro R. Vancomycin therapy for methicillin-resistant Staphylococcus aureus. Ann Intern Med. 1982;97:344–350. doi: 10.7326/0003-4819-97-3-344. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi H, Aramaki H, Nikaido Y, Mizuguchi Y, Nakamura M, Koga T, Yoshida S. Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1996;144:103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x. [DOI] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voss A, Milatovic D, Wallrauch-Schwarz C, Rosdahl V T, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]
- 28.Wehrli W. Rifampin: mechanisms of action and resistance. Rev Infect Dis. 1983;5:S407–S411. doi: 10.1093/clinids/5.supplement_3.s407. [DOI] [PubMed] [Google Scholar]
- 29.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [DOI] [PubMed] [Google Scholar]