Abstract
Cause of Kawasaki disease (KD) is unknown. KD is often resistant to treatment with intravenous immunoglobulin (IVIG). Sano’s score, which is derived from total bilirubin (TBIL), aspartate aminotransferase (AST) and C-reactive protein (CRP), is predictive of IVIG resistance in Japan. A recent study reported that Torquetenovirus (TTV), especially TTV7, was present at a high viral load in the patients with KD. We used PCR to quantify TTV load and amplicon next generation sequencing to detect individual TTV species. We used serum samples that were collected between 2002 and 2005 from 57 Japanese KD patients before IVIG treatment. Correlations between TTV load and Sano’s score, the biomarkers that constitute this score, and IVIG resistance were examined. TTV load was positively correlated with Sano’s score (P = 0.0248), TBIL (P = 0.0004), and AST (P = 0.0385), but not with CRP (P = 0.6178). TTV load was marginally correlated with IVIG resistance (P = 0.1544). Presence of TTV7 was correlated with total TTV load significantly (P = 0.0231). The correlations between biomarkers for KD and TTV load suggested that TTV may play a role in the pathophysiology of KD. We hypothesize that TTV7 may be associated with a higher total viral load in KD.
Subject terms: Microbiology, Diseases, Medical research, Pathogenesis, Risk factors
Introduction
Kawasaki disease (KD) is a febrile illness characterized by mucocutaneous manifestations (e.g., conjunctivitis, cheilitis, rash, cervical lymphadenopathy, and/or oedematous extremities)1,2. Unless the disease is diagnosed and treated promptly, 40% of children with KD may develop coronary aneurysms3, which can form thromboses or stenoses4. In Japan, 17,000 children are affected by KD annually. Nearly 400,000 Japanese children have suffered from this potentially lethal illness5. Increasing numbers of cases have been reported in other countries and regions.
Developed in the 1980s–1990s, intravenous immunoglobulin (IVIG) is the mainstay of treatment of KD6,7. However, even with IVIG treatment, 20–40% of patients with KD relapse and require additional immunomodulatory therapy8,9. Multiple scoring systems have been developed to predict this “IVIG resistance”. The scores developed by Sano10, Kobayashi11, and Egami12 are frequently used in Japan, although these are less sensitive when used in non-Japanese populations13. A patient with KD who is predicted to have a high risk of IVIG resistance will be given immunomodulatory therapy in conjunction with the first IVIG treatment. For example, using Sano’s scoring method, either total bilirubin (TBIL) ≥ 0.9 mg/dL, aspartate aminotransferase (AST) ≥ 200 IU/L, or C-reactive protein (CRP) ≥ 7 mg/dL constitutes one score. A summed score ≥ 2 indicates a high-risk patient to whom an intravenous methylprednisolone pulse dose would be given10. Even with this augmented treatment, a substantial proportion of those patients develop IVIG resistance. Therefore, more specific modes of diagnostic tests and treatments have been needed. However, the cause of KD remains unknown 60 years after its discovery14, thus hindering development of specific diagnostic tests or treatments.
Epidemiological characteristics of KD, such as the presence of seasonality or spatiotemporal clustering15–17, suggest that KD is triggered by an infectious agent. It is likely that a genetically predisposed child can develop KD when they are infected with a causal microbe18–21. Epidemiological comparisons between KD and paediatric infectious diseases support presence of a viral agent for KD22,23.
A prospective metagenomic study identified high viral loads of Torquetenovirus (TTV) 7 in some patients with KD, while this virus was not identified in control subjects24. Although the result was not statistically significant due to the small sample size, this finding warrants further study to examine the possible association between KD and TTV. Use of quantitative PCR technology has been increasingly common in the study of TTV kinetics25. Using this technology, we quantified the viral loads of TTV in serum samples from patients with KD. In addition, we used newly developed amplicon next generation sequence (NGS) to identify individual species of TTV. These virological data were correlated to biomarkers that constitute Sano’s score and to IVIG resistance.
Results
Participants
Between 2002 and 2005, 236 patients with KD were enrolled from four hospitals in Osaka prefecture (i.e. Itami city hospital, Ikeda municipal hospital, Toyonaka municipal hospital and Minoh city hospital). Per the study protocol, we stored the serum only after written informed consent was obtained from the patients’ guardians. We collected pre-treatment serum samples from a total of 61 patients. Four of these pre-treatment samples had insufficient volume (< 200 μL). Therefore, the analysis included serum samples collected from 57 patients (Supplementary File 1). Of these patients, pre-treatment serum samples were collected from six patients one day before the first IVIG treatment and from 51 patients on the day of the first IVIG treatment. We also analysed longitudinal samples collected from these patients between day − 1 and day 15. Of the 57 patients studied, 30 were male (53%). The mean age of the patients was 3.0 years (median: 2.7 years; range: 0.2–9.3 years). The first IVIG was given to all patients. The median timepoint of the first IVIG was day 4 of the course of the illness (range: days 2–11), assuming that the onset of KD is day 1. In seven patients, whether the patient was IVIG-resistant was not recorded. Among the remaining 50 patients, 8 (16%) were resistant to the first IVIG treatment. On the day of treatment initiation, Sano’s score indicated a high risk of IVIG resistance (i.e., ≥ 2 score value) in 17 patients (30%). These patients were given an intravenous methylprednisolone pulse on the first day of IVIG treatment.
Correlation between biomarkers for KD and TTV viral load
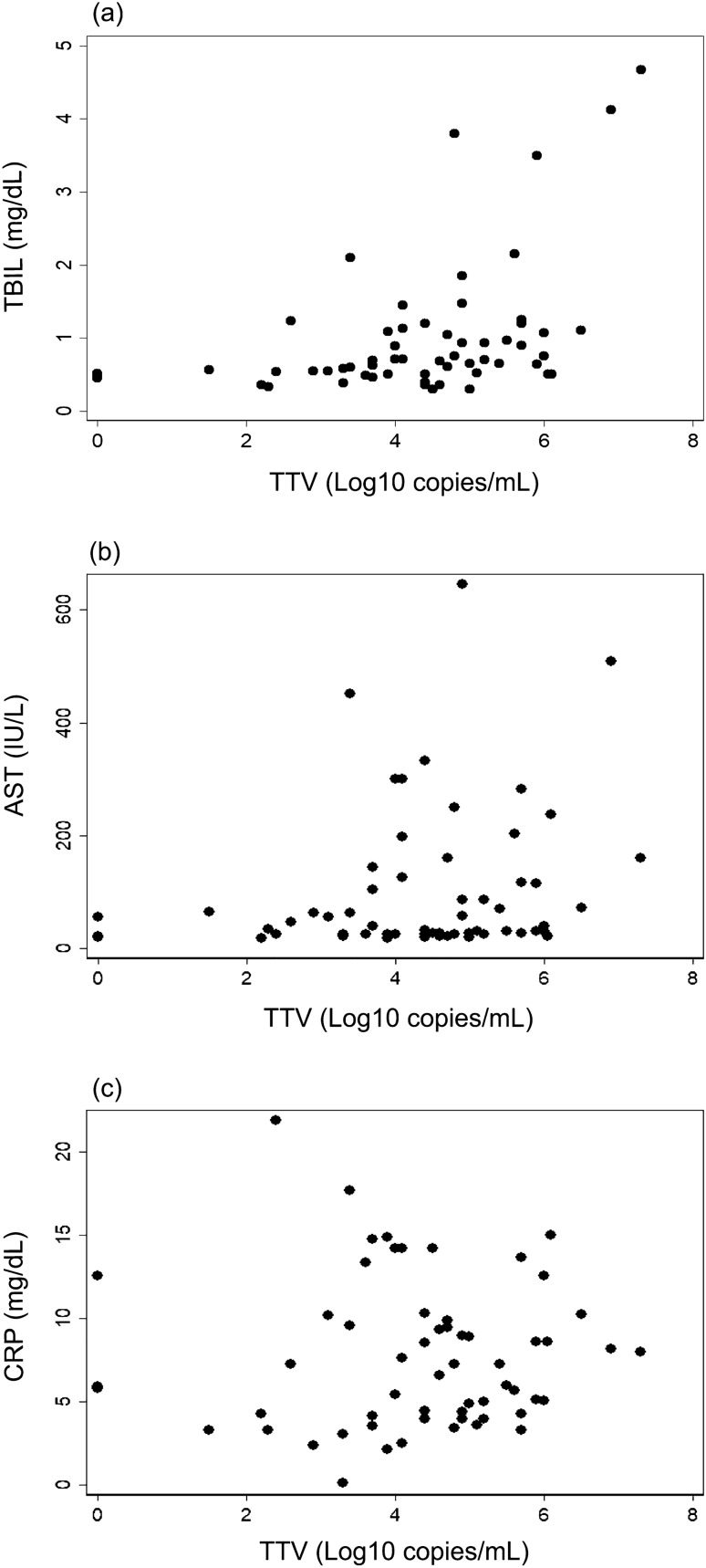
The TTV viral load was positively correlated with TBIL and AST, but not with CRP (Fig. 1). The highly skewed distributions of these biomarkers were normalised26, in subsequent statistical analyses (Supplementary File 2). Spearman’s rank correlation analysis (Table 1a), univariate linear regression analysis (Table 1b), and multivariate linear regression analysis (Table 1c) indicated that the correlations between TTV load and TBIL/AST were statistically significant (P < 0.05).
Figure 1.
The biomarkers that constitute Sano’s score, and viral loads of Torquetenovirus (TTV). TTV viral loads were plotted against TBIL (a), AST (b), and CRP (c), which constitute Sano’s score that predicts IVIG resistance. TBIL and AST were positively correlated with TTV viral load; the result was statistically significant.
Table 1.
Correlations between Torquetenovirus viral loads and biomarkers for Kawasaki disease.
| n = 57 | TBIL | AST | CRP |
|---|---|---|---|
| (a) Spearman’ rank correlation analysis | |||
| TTV | R = 0.4476 (P = 0.0005) | R = 0.2743 (P = 0.0389) | R = 0.1114 (P = 0.4092) |
| (b) Univariate regression analysis† | Coefficients (P value) | Coefficients (P value) | Coefficients (P value) |
|---|---|---|---|
| TTV | 0.192 (P = 0.0007) | 0.0170 (P = 0.0412) | 0.0604 (P = 0.6449) |
| Adjusted R2 | 0.176 (P = 0.0007) | 0.0568 (P = 0.0412) | − 0.0142 (P = 0.6449) |
| (c) Multivariate regression analysis† | Coefficients (P value) | Coefficients (P value) | Coefficients (P value) |
|---|---|---|---|
| TTV | 0.203 (P = 0.0004) | 0.0174 (P = 0.0385) | 0.0671 (P = 0.6178) |
| Sex (male 1; female: 0) | − 0.286 (P = 0.0748) | − 0.0262 (P = 0.3123) | 0.0138 (P = 0.9738) |
| Age (years) | − 0.017 (P = 0.6958) | − 0.0065 (P = 0.3303) | 0.0721 (P = 0.5068) |
| Adjusted R2 | 0.1973 (P = 0.0021) | 0.0564 (P = 0.1069) | − 0.0437 (P = 0.8830) |
†For regression analyses, the dependent variables (i.e., TBIL, AST, CRP) were normalized using the Box and Cox method.
Correlation between Sano’s score for IVIG resistance and TTV viral load
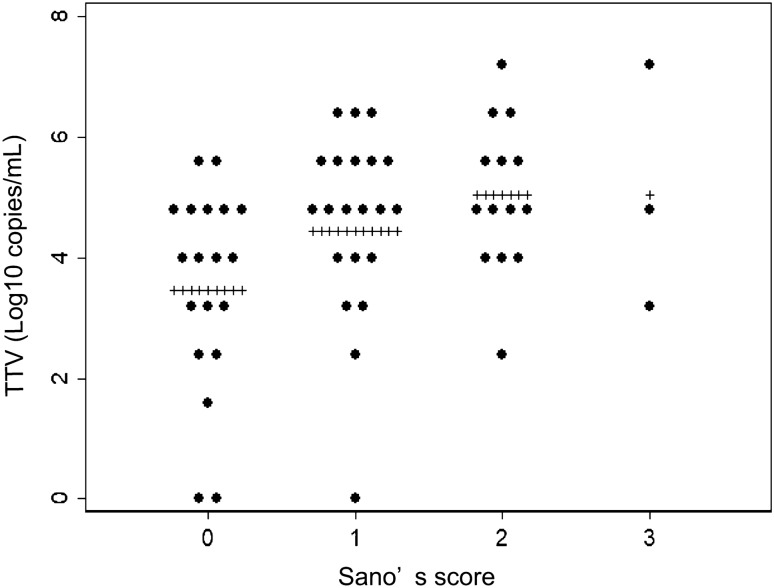
Sano’s scores were positively correlated with the TTV viral loads (Fig. 2). The statistical analyses indicated that Sano’s score was positively correlated with TTV viral load in Spearman’s rank correlation (R = 0.3645, P = 0.0045), a univariate logistic regression analysis (odds ratio [OR] 1.777, P = 0.0262), and a multivariate analysis that considered sex and age (OR 1.772, P = 0.0248) (Table 2).
Figure 2.
Sano’s score and TTV viral load. TTV viral loads in pre-treatment serum samples classified based on Sano’s score. Plus (+) indicates mean value in each group.
Table 2.
Logistic regression model to predict Sano’s score based upon Torquetenovirus viral loads and covariates.
| n = 57 | |
|---|---|
| (a) Univariate logistic regression analysis | Odds ratio (P value) |
| TTV | 1.777 (P = 0.0262) |
| (b) Multivariate logistic regression analysis | Odds ratio (P value) |
|---|---|
| TTV | 1.772 (P = 0.0248) |
| Sex (male: 1; female: 0) | 0.622 (P = 0.4428) |
| Age (years) | 0.929 (P = 0.6826) |
Correlation between Sano’s risk score and IVIG resistance
Resistance to the first IVIG treatment was correlated with Sano’s scores (Supplementary File 3). The correlation was statistically significant in a univariate analysis (OR 2.72, P = 0.0405. n = 50), and a multivariate regression analysis that considered age and sex (OR 3.05, P = 0.0366. n = 50).
Correlation between TTV load and resistance to IVIG
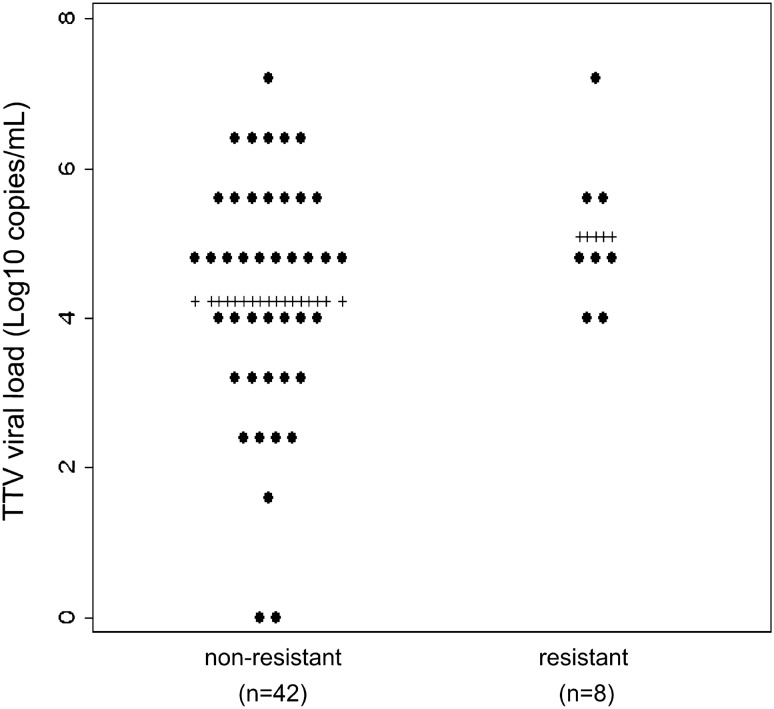
In our participants, IVIG resistance had only a marginal correlation with TTV viral load in both the univariate and multivariate analyses (Fig. 3, Table 3). We used Monte Carlo simulation to estimate the sample size necessary to identify a statistically significant correlation (i.e., α = 0.05) between TTV load and IVIG resistance (Supplementary File 4). We considered ORs of 1 or 3 for male sex, and 1 or 0.7 for age (in years). Depending on the scenario, a sample size of 150–250 participants would achieve a statistical power of 80%.
Figure 3.
Resistance to intravenous immunoglobulin (IVIG) and TTV viral load. TTV viral loads in pre-treatment serum samples classified based on IVIG resistance. Plus (+) indicates mean value in each group.
Table 3.
Logistic regression model to predict resistance to the first intravenous immunoglobulin, based on Torquetenovirus viral loads and covariates.
| n = 50 | |
|---|---|
| (a) Univariate logistic regression analysis | Odds ratio (P value) |
| TTV | 1.569(P = 0.1567) |
| (b) Multivariate logistic regression analysis | Odds ratio (P value) |
|---|---|
| TTV | 1.666 (P = 0.1544) |
| Sex (male: 1; female: 0) | 3.652 (P = 0.1500) |
| Age (years) | 1.135 (P = 0.5404) |
Temporal change in TTV viral load and IVIG resistance
The TTV viral load decreased over time (Fig. 4). In the entire population of participants, the viral load (log10 copies/mL) decreased by 0.042 per day (Table 4, column (a)). This result corresponded to a daily decrease of only 9.2%. This small downward trend was statistically significant (P = 0.0002) in the subgroup without IVIG resistance (Table 4, column (b)), but non-significant (P = 0.1304) in the group with IVIG resistance (Table 4, column (c)). However, the latter result may have been due to the small sample size available for this subgroup. We incorporated the presence of individual TTV species into this longitudinal analysis. However, due to the small sample size, none of the species showed any statistically significant contributions to the temporal trend of total TTV load. The dataset used for this longitudinal analysis is available in Supplementary File 5.
Figure 4.
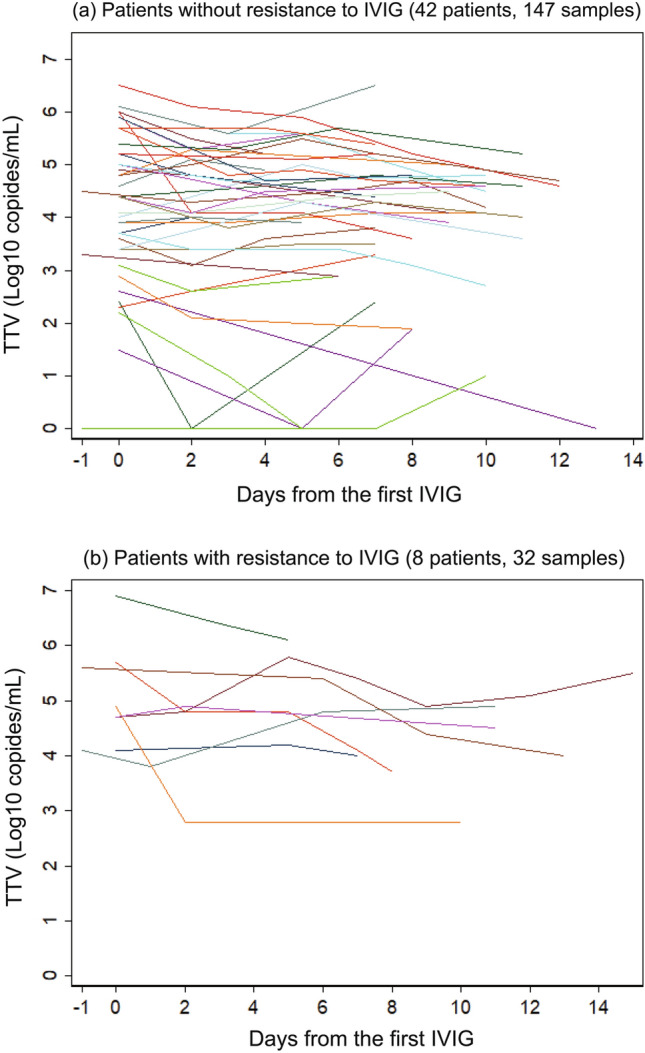
Temporal change in TTV viral load. TTV viral load versus days elapsed since day of first IVIG treatment for 42 patients (147 samples) who were not resistant (a), and 8 patients (32 samples) who were resistant (b) to IVIG.
Table 4.
Random effect regression analysis to explain Torquetenovirus viral loads.
| (a) All patients 50 patients 179 samples |
(b) Patients without resistance 42 patients 147 samples |
(c) Patients with resistance 8 patients 32 samples |
|
|---|---|---|---|
| Univariate analysis | |||
| Days after 1st IVIG | − 0.0420 (P = 0.0001) | − 0.0437 (P = 0.0002) | − 0.0376 (P = 0.1304) |
| Multivariate analysis | |||
| Days after 1st IVIG | − 0.0417 (P = 0.0001) | − 0.0433 (P = 0.0003) | − 0.0383 (P = 0.1241) |
| Sex (male: 1; female: 0) | 0.455 (P = 0.3072) | 0.243 (P = 0.6398) | 1.038 (P = 0.1546) |
| Age (years) | − 0.0746 (P = 0.4988) | − 0.0852 (P = 0.5223) | − 0.0583 (P = 0.6637) |
Presence of individual TTV species
The DNA extracted from the pre-treatment samples (n = 57) were sequenced by the amplicon NGS technology (Accession number: PRJDB15714). Each of all 22 TTV species, which are defined in the update taxonomic classification27, were examined bioinformatically. Consequently, 17 TTV species were present in at least one pre-treatment samples (Supplementary File 6). Interestingly, the patient who presented the highest total TTV load and the largest TBIL value was positive only with TTV7 (Sample 402 in Supplementary File 1). The presence of TTV7 was marginally correlated with the normalised TBIL (Pearson’s R = 0.2172, P = 0.1046) as is shown in Supplementary File 6. The presence of TTV7 was positively correlated with the total TTV load (Pearson’s R = 0.3005, P = 0.0231, Supplementary File 6). TTV7 was positive in three KD patients (Supplementary File 7).
Discussion
In this study, we found that the total viral load of TTV was associated with the hepatobiliary biomarkers that constitute Sano’s score10. A recent study in Japan indicated that TBIL has sensitivity and specificity which were superior to other biomarkers in predicting resistance to IVIG augmented by corticosteroid treatment28. Of note, all scores recommended in Japanese guidelines as being predictive for IVIG resistance (i.e., Kobayashi, Egami, and Sano) use liver enzyme(s)10–12. It has also long been assumed that KD severity is associated with hepatobiliary system damage29,30. Therefore, the significant correlation between TTV load and TBIL/AST identified in the present study suggested that TTV was involved in the hepatobiliary dysfunction which is crucial to the pathophysiology of KD. However, we are aware that the correlations between TTV load and the indicators of KD severity (i.e., Sano’s score, TBIL, and AST) identified in our present study do not necessarily demonstrate that there is a causal link between TTV and KD.
The correlation between TTV load and IVIG resistance was only marginally significant in the present study. This result might be due to the small sample size used. We estimated that 150–250 KD patients would be necessary to identify the presence of a statistically significant correlation between TTV load and IVIG resistance. However, the methylprednisolone pulse which was given to the high-risk patients (i.e. with ≥ 2 Sano scores) may have effectively prevented IVIG-resistance in some patients, and hence blunted the statistical power of our analysis. Therefore, a smaller sample size may be sufficient in a future study if the treatment to high-risk patients is not augmented with adjunct therapies including methylprednisolone pulse.
It is widely assumed that KD has an infectious trigger. Diverse microorganisms have been proposed as candidate causative agent(s) of KD31,32. Recently, it was reported that SARS-CoV-2 infection can result in an illness that mimics the symptoms and cardiac complications of KD33. Finding this illness, named paediatric inflammatory multisystem syndrome temporally associated with COVID-1934 or multisystem inflammatory syndrome in children (MIS-C)35,36, revived interest in a possible viral involvement in the pathophysiology of KD. However, to our knowledge, no study has found that a specific virus is associated with the development of KD or with illness severity.
In 1997, Nishizawa et al. first discovered TTV as a possible agent for hepatitis of unknown origin37. Subsequently, TTV was assigned to a new family, Anelloviridae38. During their lifetime, almost all human beings are infected with at least one TTV species that establishes a persistent infection39. TTV is associated with febrile episodes40, or respiratory illnesses41,42, in children. There are numerous reports of TTV involvement in liver diseases43,44. These results were consistent with our finding that a higher viral load of TTV was correlated with more severe hepatobiliary damage in patients with KD. The correlation between TTV load and hepatobiliary biomarkers for KD found in our study suggested that TTV plays a role in the pathophysiology of KD. Development of an experimental animal model would be crucial in understanding whether (and how) TTV is involved in the development of vasculitis, and in elucidating the relationship between TTV and KD.
Because TTV load is correlated with immunosuppression45,46, quantitative PCR of TTV has been established as a highly reliable and useful clinical methodology25,47–49. This methodology has been used in longitudinal studies50,51. Quantitative and longitudinal relationships between viral load and the course of illness are important in future pathophysiological studies of KD. Our study found that the TTV viral load decreased only slightly after the KD was treated with IVIG. However, it is unclear whether the temporal decrease in TTV viral load was due to the effect of IVIG because all patients received at least one IVIG dose. The temporal relationship between TTV viral load and IVIG resistance would be an interesting topic for future large-scale studies.
A newly developed TTV amplicon NGS showed that presence of TTV7 was marginally correlated with TBIL (P = 0.1046). This may be because TTV7 was significantly associated with larger total TTV loads (P = 0.0231). This finding is consistent with a previous report that the load of TTV7 in KD patients were high24. This hypothesis that TTV7 has a propensity of proliferating into a high viral load and hence of developing severe KD should be tested in a future study.
Mathematical studies predict that at least one agent for KD persistently infects the host22,52. Consistent with these predictions, TTV remains persistently in the host39. It was also predicted mathematically that the causative microbe for KD infects most children and that only a small proportion of these infections develop KD22. Almost all human beings are infected with at least one TTV species43. Therefore, TTV possesses many of the characteristics predicted for the agent(s) of KD. Although a previous study did not identify an association between TTV and KD53, we found that the PCR primer sets used in that study did not detect many species of TTV.
Our study had limitations, most importantly its small sample size and the absence of information for cardiac complications. In addition, our study lacked control subjects (i.e. children without KD). Although the fundamental pathophysiology of KD is systemic vasculitis, biomarkers for vasculitis (e.g. D-Dimer and Fibrin Degradation Products) were not recorded in our study. The objective of the original study, approved in 2001, was to elucidate the relationship between cytokine profiles and the severity of KD. Therefore, control subjects were not enrolled in this study. To address these issues, a future study should compare prevalence and viral load between patients with KD and control subjects. In this new study, indicators of vasculitis (e.g., D-Dimer and fibrin degradation products) should also be correlated to the TTV load.
In the Japanese diagnostic criteria for KD, the diagnosis of the complete form of KD is entertained when at least 5 signs/symptoms (e.g., fever, conjunctivitis, cheilitis, cervical lymphadenopathy, erythema and oedema of the fingers/toes) are present, regardless of the presence of overlapping infections54. Therefore, in the present study, KD was not diagnostically excluded solely based on the presence of infections. However, it would be interesting to consider not only TTV but many other microbes in a future study.
An advantage of our study was that our results were not interfered with COVID outbreak and MIS-C. A future large-scale study using updated methodologies is warranted to examine the relationship between TTV and KD.
Conclusions
In 57 patients with KD, the viral load of TTV was positively correlated with the levels of hepatobiliary biomarkers constituting Sano’s score, which predicts resistance to IVIG in Japanese population. A larger study is necessary to identify a positive correlation between TTV and KD.
Methods
Ethical approval and informed consent
This study was approved by the ethics committee of the Osaka University Faculty of Medicine (references: 128-1 and 128-2) in November 2001. The guardians of the participants provided written informed consent to participate in the study. The samples were not used for the original objective and were stored at − 20 ℃. The samples were used for the present analysis in accordance with Japanese ethical guidelines for medical and biological sciences55.
Diagnosis and treatment
KD was diagnosed based on Japanese diagnostic criteria, version 554. All patients diagnosed as having KD were treated with IVIG (2 g/kg) and aspirin (30 mg/kg/day). An intravenous methylprednisolone pulse (30 mg/kg) was given to patients with KD who had values of 2 or 3 for Sano’s score, in parallel with the first IVIG. A patient who remained febrile (> 37.5 °C) 48 h after initiation of the first IVIG was classified as IVIG-resistant. A second IVIG treatment was given to all IVIG-resistant patients. When defervescence occurred, the dose of aspirin was decreased to 5 mg/kg/day. Blood tests were conducted before each IVIG, and at least once before discharge. A patient was discharged when the fever subsided for more than 48 h, all the biomarkers normalised and the coronary arteries did not show progressive dilatation.
DNA extraction from serum
DNA was extracted from 200 μL serum using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) following the manufacturer’s protocol. Extracted DNA was eluted into 200 μL buffer AE, which was then aliquoted in equal amounts into three tubes. The tubes were stored at – 80 °C until use.
Quantification of total TTV DNA in individual samples
Extracted viral DNA was amplified using real-time PCR amplification performed on a 7500 Fast Instrument® (Applied Biosystems, MA, USA). The TTV load was determined using a single-step TaqMan real-time PCR assay that targeted the highly conserved 5’UTR of the TTV genome. Copy numbers were quantified as previously described56,57. Briefly, the method amplified a 63-nucleotide UTR fragment with a detection limit of < 10 viral genomes per ml plasma/serum.
Amplicon NGS of TTV species
Targeted enrichment is often necessary to generate complete viral genomes in clinical samples58. For this aim, we developed an amplicon NGS method59, to identify individual TTV species. The sensitivity and specificity of this methodology will be reported elsewhere. We enriched the circular DNAs of anelloviruses, using Rolling Circle Amplification (RCA) technology which is based on Phi29 DNA polymerase60. RCA reaction was performed on a mixture of a DNA sample (< 10 ng), 25 µM of exonuclease-resistant random primer (Thermo Fisher Scientific, CA, USA), 4 mM of deoxynucleotides (dNTPs) (Solis BioDyne, Tartu, Estonia) and 10 U of φ29 DNA polymerase (Thermo Fisher Scientific, CA, USA). Amplification was performed at 30 °C for 18 h, followed by inactivation of Phi 29 DNA polymerase at 65 °C for 10 min. The resulting linear double-stranded DNA product was spectrophotometrically quantified by NanoDrop Lite instrument (Thermo Fisher Scientific, CA, USA) and tenfold diluted to be used as the template in the Universal Anelloviruses inverse-PCR61. The Universal Anellovirus inverse-PCR was applied to a mixture of 50 μL consisting of 1 × PrimeSTAR® GXL Buffer (Takara, Shiga, Japan), 200 μM dNTP mixture (Takara, Shiga, Japan), 0.3 μM forward primer, 0.3 μM reverse primer, 1.25 U of PrimeSTAR® GXL DNA polymerase (Takara, Shiga, Japan) and 500 to 1000 ng of template. Primers are as follows: 206INV_TTVFor, 5′-CAA GGG GCA ATT CGG GCT C-3′; 205INV_TTVRev, 5′-ACT NCG GTG TGT AAA CTC ACC T-3′. Positions of the 5′-end nucleotide of primers in the reference TTV1 genome (Accession: NC_002076) are used as primer prefix names with either forward (For) or reverse (Rev) orientation. The PCR was performed in 35 cycles of 98 °C for 10 s, 60 °C for 15 s, and 68 °C for 4 min. To verify successful amplification, 10 µL of product was analysed by electrophoresis on 1% w/v agarose gel. If the expected band was present on ultraviolet transilluminator, the remaining PCR product (about 40 µL) was purified by NucleoSpin Gel and PCR Clean-up (Macherey–Nagel, Düren, Germany). The purified PCR products were subjected to fluorimetric analysis using Qubit (Thermo Fisher Scientific, CA, USA) to determine the concentration of the recovered nucleic acids. Libraries were prepared using the Illumina DNA Prep kit (Illumina, San Diego, CA, USA), starting with an initial DNA input of 150 ng. The pool of libraries was quantified using Qubit, and the sizes of fragments were estimated using the BioAnalyzer (Agilent Technologies, Inc. CA, USA). The denatured libraries were then loaded in 12 pM concentration on Miseq V3 cartridge (Illumina, CA, USA) to be sequenced with Paired-end 2 × 250 bp.
Bioinformatical analysis
We developed a bioinformatical pipeline which consists of reference-based approach and de-novo assembly. Briefly, low-quality reads were filtered and adapters were trimmed with Fastp programme62. Reads of human origin (Accession: GRCh38.p13) were eliminated using Kraken 263. Using BBmap64, we aligned the remaining reads against a database of species-level anellovirus genomes27. The mapped reads reconstructed the full or near-full-length genomic sequences by consensus calling or de novo assembly. Subsequently, Blastn programme classified the reconstructed sequence into the anellovirus genomes at the species level. The reconstructed viral genomes were mapped with the non-human reads using BWA-MEM software version 0.7.1765. Then, the number of reads assigned to each individual species was reported in the log10 of reads mapped per million (Log RPM) (Supplementary File 1). We assumed that a TTV species was present in a sample when Log RPM was positive.
Statistical analysis
Stata SE 13 (Stata, TX, USA) was used for the statistical analyses. In the linear regression analyses, the independent variables (i.e., TBIL, AST, and CRP) had highly skewed distributions. Therefore, these variables were normalised using the method proposed by Box and Cox26, as in the following equation:
“λ” was − 0.619589 for TBIL, − 0.5450522 for AST, and 0.4735706 for CRP. The results indicated that the normalised values had symmetrical distributions (Supplementary File 2).
Sample size calculations
We estimated the sample size necessary to identify a statistically significant correlation between TTV viral load and IVIG resistance. We used Monte Carlo simulations66,67, and included sex and age as covariates in the multivariate logistic regression model. The distributions of viral loads and ages in our study participants were reproduced in the simulations. Age (years) was assumed to follow a two-parameter gamma distribution, with a shape parameter of 1.9 and a scale parameter of 1.5. Viral load (log10 copies per mL) was simulated as a normal distribution with a mean of 4.4 and a standard deviation of 1.55; the negative values were set to zero. The proportions of male and female participants were assumed to be equal. The OR for TTV viral load was fixed to 1.6, based on the results of the analysis (Table 3). The sensitivity analysis included an OR value for male sex of 1 or 3; the OR value for age (years) was 1 or 0.7, the latter of which was translated from a previously reported OR value (i.e., 0.974 for age in months)11. We conducted 10,000 simulations, and power was computed as the proportion of simulations in which the coefficient for viral load was statistically significant (i.e., two-sided P-value < 0.05). The Stata script for the simulation is available in Supplementary File 8.
Supplementary Information
Acknowledgements
We are grateful to Tiago Pereira, Noriko Tokuda, Kazunori Miki, Ichiro Maki, Taro Matsuoka and Takehisa Yamamoto for assistance with this study.
Author contributions
P.G.S., K.M., F.M., F.F. and T.I. conducted the experiments. Y.N., M.P. and J.H. conceived the study. T.M., T.S. and K.O. assumed responsibility for management of clinical data and samples. All authors contributed to manuscript preparation and agreed with submission of the manuscript in its present form.
Data availability
All the data used in the present analysis are available as supplementary information. Nucleotide sequences obtained in the present study is registered in DNA Databank of Japan with a BioProject accession code of PRJDB15714 (BioSample: SAMD00598640-SAMD00598696).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pietro Giorgio Spezia, Kazunari Matsudaira, Fabrizio Maggi and Junichi Hara.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-45327-5.
References
- 1.McCrindle BW, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A scientific statement for health professionals from the American heart association. Circulation. 2017;135:e927–e999. doi: 10.1161/cir.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T, et al. Revision of diagnostic guidelines for Kawasaki disease (6th revised edition) Pediatr. Int. 2020;62:1135–1138. doi: 10.1111/ped.14326. [DOI] [PubMed] [Google Scholar]
- 3.Ogata S, et al. Coronary artery outcomes among children with Kawasaki disease in the United States and Japan. Int. J. Cardiol. 2013;168:3825–3828. doi: 10.1016/j.ijcard.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukazawa R, et al. JCS/JSCS 2020 guideline on diagnosis and management of cardiovascular sequelae in Kawasaki disease. Circ. J. 2020;84:1348–1407. doi: 10.1253/circj.CJ-19-1094. [DOI] [PubMed] [Google Scholar]
- 5.Makino N, et al. Nationwide epidemiologic survey of Kawasaki disease in Japan, 2015–2016. Pediatr. Int. 2019;61:397–403. doi: 10.1111/ped.13809. [DOI] [PubMed] [Google Scholar]
- 6.Furusho K, et al. High-dose intravenous gammaglobulin for Kawasaki disease. Lancet. 1984;324:1055–1058. doi: 10.1016/S0140-6736(84)91504-6. [DOI] [PubMed] [Google Scholar]
- 7.Newburger JW, et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N. Engl. J. Med. 1991;324:1633–1639. doi: 10.1056/nejm199106063242305. [DOI] [PubMed] [Google Scholar]
- 8.Tremoulet AH, et al. Resistance to intravenous immunoglobulin in children with Kawasaki disease. J. Pediatr. 2008;153:117–121. doi: 10.1016/j.jpeds.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam JY, et al. Intravenous immunoglobulin resistance in Kawasaki disease patients: Prediction using clinical data. Pediatr. Res. 2023 doi: 10.1038/s41390-023-02519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano T, et al. Prediction of non-responsiveness to standard high-dose gamma-globulin therapy in patients with acute Kawasaki disease before starting initial treatment. Eur. J. Pediatr. 2007;166:131–137. doi: 10.1007/s00431-006-0223-z. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T, et al. Prediction of intravenous immunoglobulin unresponsiveness in patients with Kawasaki disease. Circulation. 2006;113:2606–2612. doi: 10.1161/circulationaha.105.592865. [DOI] [PubMed] [Google Scholar]
- 12.Egami K, et al. Prediction of resistance to intravenous immunoglobulin treatment in patients with Kawasaki disease. J. Pediatr. 2006;149:237–240. doi: 10.1016/j.jpeds.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 13.Piram M, et al. Defining the risk of first intravenous immunoglobulin unresponsiveness in non-Asian patients with Kawasaki disease. Sci. Rep. 2020;10:3125. doi: 10.1038/s41598-020-59972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawasaki T. Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children [In Japanese] Arerugi. 1967;16:178–222. [PubMed] [Google Scholar]
- 15.Nakamura Y, Yanagawa I, Kawasaki T. Temporal and geographical clustering of Kawasaki disease in Japan. Prog. Clin. Biol. Res. 1987;250:19–32. [PubMed] [Google Scholar]
- 16.Hearn J, et al. Spatiotemporal clustering of cases of Kawasaki disease and associated coronary artery aneurysms in Canada. Sci. Rep. 2018;8:17682. doi: 10.1038/s41598-018-35848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burney JA, et al. Temporal clustering of Kawasaki disease cases around the world. Sci. Rep. 2021;11:22584. doi: 10.1038/s41598-021-01961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onouchi Y, et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat. Genet. 2008;40:35–42. doi: 10.1038/ng.2007.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khor CC, et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet. 2011;43:1241–1246. doi: 10.1038/ng.981. [DOI] [PubMed] [Google Scholar]
- 20.Onouchi Y, et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat. Genet. 2012;44:517–521. doi: 10.1038/ng.2220. [DOI] [PubMed] [Google Scholar]
- 21.Lee YC, et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat. Genet. 2012;44:522–525. doi: 10.1038/ng.2227. [DOI] [PubMed] [Google Scholar]
- 22.Nagao Y, Urabe C, Nakamura H, Hatano N. Predicting the characteristics of the aetiologic agent for Kawasaki disease from other paediatric infectious diseases in Japan. Epidemiol. Infect. 2016;144:478–492. doi: 10.1017/S0950268815001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim JH, et al. Seasonal trends of viral prevalence and incidence of Kawasaki disease: A Korea public health data analysis. J. Clin. Med. 2021 doi: 10.3390/jcm10153301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thissen JB, et al. A novel variant of torque teno virus 7 identified in patients with Kawasaki Disease. PLoS ONE. 2018 doi: 10.1371/journal.pone.0209683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doberer K, et al. Torque teno virus load is associated with subclinical alloreactivity in kidney transplant recipients: A prospective observational trial. Transplantation. 2021;105:2112–2118. doi: 10.1097/tp.0000000000003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Box GEP, Cox DR. An analysis of transformations. J. R. Stat. Soc. B. 1964;26:211–243. [Google Scholar]
- 27.Varsani A, et al. Taxonomic update for mammalian anelloviruses (family Anelloviridae) Arch. Virol. 2021;166:2943–2953. doi: 10.1007/s00705-021-05192-x. [DOI] [PubMed] [Google Scholar]
- 28.Miyata K, et al. Risk factors of coronary artery abnormalities and resistance to intravenous immunoglobulin plus corticosteroid therapy in severe Kawasaki disease: An analysis of post RAISE. Circ. Cardiovasc. Qual. Outcomes. 2021;14:e007191. doi: 10.1161/circoutcomes.120.007191. [DOI] [PubMed] [Google Scholar]
- 29.Cheng F, et al. Analysis of hyperbilirubinemia in patients with Kawasaki disease. Medicine. 2020;99:e21974. doi: 10.1097/md.0000000000021974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi DY, Kim JY, Choi EY, Choi JY, Yang HR. Hepatobiliary risk factors for clinical outcome of Kawasaki disease in children. BMC Pediatr. 2014;14:51. doi: 10.1186/1471-2431-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nigro G, et al. Active or recent parvovirus B19 infection in children with Kawasaki disease. Lancet. 1994;343:1260–1261. doi: 10.1016/s0140-6736(94)92154-7. [DOI] [PubMed] [Google Scholar]
- 32.Rowley AH, et al. RNA-containing cytoplasmic inclusion bodies in ciliated bronchial epithelium months to years after acute Kawasaki disease. PLoS ONE. 2008;3:e1582. doi: 10.1371/journal.pone.0001582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdoni L, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: An observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/s0140-6736(20)31103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Royal College of Paediatrics and Child Health. Paediatric Multisystem Inflammatory Syndrome Temprally Associated with COVID-19 (PIMS): Guidance for Clinicians. https://www.rcpch.ac.uk/resources/guidance-paediatric-multisystem-inflammatory-syndrome-temporally-associated-covid-19-pims (2020).
- 35.Centers for Disease Control and Prevention. Multisystem Inflammatory Syndrome in Children (MIS-C) Associated with Coronavirus Disease 2019 (COVID-19). https://emergency.cdc.gov/han/2020/han00432.asp (2020).
- 36.World Health Organization. Multisystem Inflammatory Syndrome in Children and Adolescents Temporally Related to COVID-19. https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19 (2020).
- 37.Nishizawa T, et al. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 38.Biagini P, et al. IXth Report of the International Committee for the Taxonomy of Viruses. Elsevier Academic Press; 2012. pp. 331–341. [Google Scholar]
- 39.Zehender G, et al. Molecular epidemiology of TT virus in Italy and phylogenesis of viral isolates from subjects at different risk for parenteral exposure. J. Med. Virol. 2001;63:76–84. doi: 10.1002/1096-9071(200101)63:1<76::AID-JMV1012>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 40.McElvania TeKippe E, et al. Increased prevalence of anellovirus in pediatric patients with fever. PLoS ONE. 2012;7:e50937. doi: 10.1371/journal.pone.0050937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Rosal T, et al. Torque teno virus in nasopharyngeal aspirate of children with viral respiratory infections. Pediatr. Infect. Dis. J. 2023;42:184–188. doi: 10.1097/inf.0000000000003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pifferi M, et al. Associations between nasal Torquetenovirus load and spirometric indices in children with asthma. J. Infect. Dis. 2005;192:1141–1148. doi: 10.1086/444389. [DOI] [PubMed] [Google Scholar]
- 43.Spandole S, Cimponeriu D, Berca LM, Mihăescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch. Virol. 2015;160:893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 44.Nobili V, et al. Is Torquetenovirus a potential cause of liver damage in children? J. Infect. 2005;50:368–369. doi: 10.1016/j.jinf.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Touinssi M, et al. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. J. Clin. Virol. 2001;21:135–141. doi: 10.1016/s1386-6532(01)00157-3. [DOI] [PubMed] [Google Scholar]
- 46.Maggi F, et al. Early post-transplant Torquetenovirus viremia predicts cytomegalovirus reactivations in solid organ transplant recipients. Sci. Rep. 2018;8:15490. doi: 10.1038/s41598-018-33909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studenic P, et al. Torque teno virus quantification for monitoring of immunomodulation with biological compounds in the treatment of rheumatoid arthritis. Rheumatology. 2021 doi: 10.1093/rheumatology/keab839. [DOI] [PubMed] [Google Scholar]
- 48.Uhl P, et al. Torque teno viral load reflects immunosuppression in paediatric kidney-transplanted patients: A pilot study. Pediatr. Nephrol. 2021;36:153–162. doi: 10.1007/s00467-020-04606-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernández-Ruiz M, et al. Monitoring of alphatorquevirus DNA levels for the prediction of immunosuppression-related complications after kidney transplantation. Am. J. Transplant. 2019;19:1139–1149. doi: 10.1111/ajt.15145. [DOI] [PubMed] [Google Scholar]
- 50.Bueno F, et al. Kinetics of torque teno virus DNA in stools may predict occurrence of acute intestinal graft versus host disease early after allogeneic hematopoietic stem cell transplantation. Transpl. Infect. Dis. 2021;23:e13507. doi: 10.1111/tid.13507. [DOI] [PubMed] [Google Scholar]
- 51.Albert E, et al. Kinetics of Alphatorquevirus plasma DNAemia at late times after allogeneic hematopoietic stem cell transplantation. Med. Microbiol. Immunol. 2019;208:253–258. doi: 10.1007/s00430-019-00586-w. [DOI] [PubMed] [Google Scholar]
- 52.Pitzer VE, et al. Modelling seasonal variations in the age and incidence of Kawasaki disease to explore possible infectious aetiologies. Proc. Biol. Sci. 2012;279:2736–2743. doi: 10.1098/rspb.2011.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chua PK, et al. Lack of association between Kawasaki syndrome and infection with parvovirus B19, human herpesvirus 8, TT virus, GB virus C/hepatitis G virus or Chlamydia pneumoniae. Pediatr. Infect. Dis. J. 2000;19:477–479. doi: 10.1097/00006454-200005000-00019. [DOI] [PubMed] [Google Scholar]
- 54.Committee for Japanese diagnostic guideline of Kawasaki Disease. Japanese Diagnostic Guideline of Kawasaki Disease. Version 5 [In Japanese]. https://jspccs.jp/wp-content/uploads/Kawasakitebiki_5.pdf (2002).
- 55.Japan's Ministry of education and Ministry of Health Labour and Welfare. Ethical Guideline for the Medical Researches Involving Human Individuals [In Japanese]. https://www.mhlw.go.jp/file/06-Seisakujouhou-10600000-Daijinkanboukouseikagakuka/0000153339.pdf (2014).
- 56.Maggi F, et al. Low prevalence of TT virus in the cerebrospinal fluid of viremic patients with central nervous system disorders. J. Med. Virol. 2001;65:418–422. doi: 10.1002/jmv.2051. [DOI] [PubMed] [Google Scholar]
- 57.Maggi F, et al. TT virus in the nasal secretions of children with acute respiratory diseases: Relations to viremia and disease severity. J. Virol. 2003;77:2418–2425. doi: 10.1128/JVI.77.4.2418-2425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quick J, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isaacs SR, et al. Amplification and next generation sequencing of near full-length human enteroviruses for identification and characterisation from clinical samples. Sci. Rep. 2018;8:11889. doi: 10.1038/s41598-018-30322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Focosi D, et al. Molecular validation of pathogen-reduction technologies using rolling-circle amplification coupled with real-time PCR for Torquetenovirus DNA quantification. Transfus. Med. 2021;31:371–376. doi: 10.1111/tme.12807. [DOI] [PubMed] [Google Scholar]
- 62.Chen S, Zhou Y, Chen Y, Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joint Genome Institute. BBMap Guide. https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbmap-guide/ (2023).
- 65.Li, H. Burrow-Wheeler Aligner for Short-Read Alignment (see minimap2 for Long-Read Alignment). https://github.com/lh3/bwa (2017).
- 66.Kontopantelis E, Springate DA, Parisi R, Reeves D. Simulation-based power calculations for mixed effects modeling: Ipdpower in Stata. J. Stat. Softw. 2016;74:1–25. doi: 10.18637/jss.v074.i12. [DOI] [Google Scholar]
- 67.Arnold BF, Hogan DR, Colford JM, Jr, Hubbard AE. Simulation methods to estimate design power: An overview for applied research. BMC Med. Res. Methodol. 2011;11:94. doi: 10.1186/1471-2288-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in the present analysis are available as supplementary information. Nucleotide sequences obtained in the present study is registered in DNA Databank of Japan with a BioProject accession code of PRJDB15714 (BioSample: SAMD00598640-SAMD00598696).