Abstract
In Streptomyces ambofaciens, the producer of the macrolide antibiotic spiramycin, an open reading frame (ORF) was found downstream of srmA, a gene conferring resistance to spiramycin. The deduced product of this ORF had high degrees of similarity to Streptomyces lividans glycosyl transferase, which inactivates macrolides, and this ORF was called gimA. The cloned gimA gene was expressed in a susceptible host mutant of S. lividans devoid of any background macrolide-inactivating glycosyl transferase activity. In the presence of UDP-glucose, cell extracts from this strain could inactivate various macrolides by glycosylation. Spiramycin was not inactivated but forocidin, a spiramycin precursor, was modified. In vivo studies showed that gimA could confer low levels of resistance to some macrolides. The spectrum of this resistance differs from the one conferred by a rRNA monomethylase, such as SrmA. In S. ambofaciens, gimA was inactivated by gene replacement, without any deleterious effect on the survival of the strain, even under spiramycin-producing conditions. But the overexpression of gimA led to a marked decrease in spiramycin production. Studies with extracts from wild-type and gimA-null mutant strains revealed the existence of another macrolide-inactivating glycosyl transferase activity with a different substrate specificity. This activity might compensate for the effect of gimA inactivation.
Macrolides inhibit protein synthesis by binding to the large ribosomal subunit (14). Several mechanisms of resistance to macrolides have been described. The most widespread consists of target modification by methylation of 23S rRNA (for a review, see reference 45). Resistance can also be the result of antibiotic export (23; reference 41 and references included) or inactivation. Various chemical modifications leading to macrolide inactivation have been described phosphorylation (24, 25), esterification of the lactone ring (2, 26), and glycosylation (11, 21, 33, 38). All these resistance mechanisms have been identified in pathogens and in Streptomyces spp., both those producing and those not producing macrolide antibiotics. Often, macrolide producers have several genes involved in self-resistance. For instance, in Streptomyces fradiae, the tylosin producer, four resistance genes have been cloned (4, 9, 35, 47).
Streptomyces ambofaciens produces the macrolide antibiotic spiramycin (31). Spiramycin is composed of a 16-atom lactone ring with two amino sugars and one neutral sugar attached. The mycelium is sensitive to spiramycin during exponential growth phase and then becomes resistant, with the concomitant production of spiramycin, during stationary phase. In S. ambofaciens, at least two resistance mechanisms are present (29). Several resistance determinants have been cloned (34, 39). One of them, srmA, encodes a monomethyltransferase acting at position A2058 of 23S rRNA (28), thereby conferring resistance to macrolides-lincosamides-streptogramin B (type I resistance phenotype [30]). Here we report the cloning, sequencing, and characterization of gimA, a new macrolide resistance gene located downstream of srmA.
MATERIALS AND METHODS
Strains and culture conditions.
Strains and plasmids used in this study are listed in Table 1. Streptomyces strains were maintained on Hickey-Tressner (HT) medium at 30°C (32). For spiramycin production, S. ambofaciens strains were grown in MP5 liquid medium (29) at 27°C. Streptomyces lividans strains were maintained on R2YE medium (17) at 30°C. AS1 medium (3) was used for conjugation experiments, and cultures were grown at 37°C (see below). S. lividans strains were grown in tryptic soy broth medium (TSB; Difco) at 30°C for preparation of S30 cellular extracts. To ensure maintenance of plasmids carrying the thiostrepton resistance gene (tsr), nosiheptide (NO) was added to the solid medium at 40 μg/ml and to the liquid medium at 10 μg/ml for S. lividans cultures, and at 200 μg/ml to the solid medium for S. ambofaciens cultures.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. ambofaciens ATCC 23877 | Wild-type strain | 31 |
| S. ambofaciens OS81 | Non-spiramycin-producing mutant of ATCC 23877 | 29 |
| S. ambofaciens OS41.99 | Hmr; derivative of ATCC 23877, with gimA inactivated by gene replacement | This study |
| S. ambofaciens OS41.99NP | Hmr; derivative of OS81, with gimA inactivated by gene replacement | |
| S. lividans OS456 | Hmr; derivative of TK21 (18), with lrm and mgt inactivated by gene replacement; highly sensitive to macrolides | 30 |
| M. luteus DSM1790 | Indicator strain, highly sensitive to macrolides | |
| M. luteus Cgr | Mutant strains of M. luteus DSM1790, resistant to CG | This study |
| E. coli DH5α | Strain used for DNA cloning | Clontech |
| E. coli S17.1 | Donor strain used for E. coli/Streptomyces conjugation | 40 |
| Plasmids | ||
| pHP45 Ωhyg | Apr Hmr; source of the Ωhyg cassette | 10 |
| pWED1 | Apr; cosmid vector derived from pWE15 (44) by deletion of the 4.1-kb HpaI-HpaI fragment, removing the mammalian expression module | This study |
| pIJ4070 | Apr; pIJ2925 (19) containing the ermE-up promoter mutant ermEp* (6) | 5 |
| pIJ903 | Apr Nor; low-copy-number Streptomyces/E. coli vector | 22 |
| pOJ260 | Gnr; replicative vector in E. coli, nonreplicative in Streptomyces; used for conjugation experiments | 7 |
| pOS41.78 | Apr; cosmid from the S. ambofaciens ATCC 23877 gene library in pWED1 hybridizing with srmA | This study |
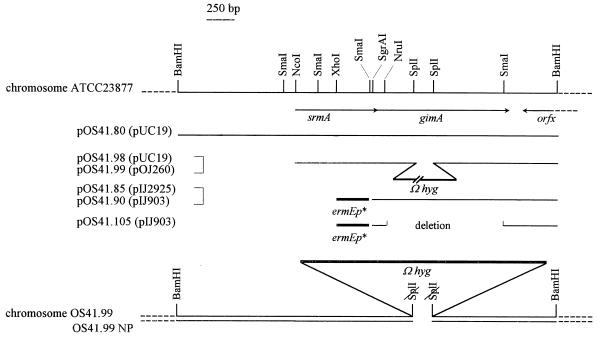
| pOS41.80 | Apr; derived from pOS41.78; 3.6-kb BamHI fragment hybridizing with the end of srmA cloned into pUC19 (Fig. 1) | This study |
| pOS41.85 | Apr; derived from pOS41.80; SgrAI-BamHI fragment cloned into pIJ4070 BamHI (Fig. 1) | This study |
| pOS41.90 | Apr Nor; derived from pOS41.85; fragment containing ermEp* and gimA cloned into pIJ903 (Fig. 1) | This study |
| pOS41.105 | Apr Nor; derived from pOS41.90; in-frame deletion in gimA from NruI to SmaI (Fig. 1) | This study |
| pOS41.98 | Apr Hmr; derived from pOS41.80; NcoI-BamHI fragment with the internal SplI-SplI fragment replaced by Ωhyg (Fig. 1) | This study |
| pOS41.99 | Gnr Hmr; insert from pOS41.98 cloned into pOJ260 (Fig. 1) | This study |
Micrococcus luteus cells grown at 37°C in Luria-Bertani (LB) medium (Gibco BRL) were added to soft agar overlay on LB plates. Observation of growth inhibition zones was done after incubation of petri plates for 24 to 48 h at 37°C. The M. luteus congocidin (CG)-resistant mutant was grown in LB medium in the presence of CG at 5 μg/ml.
Antibiotics.
The antibiotics were obtained from the following sources: hygromycin B (HM), Boehringer (Mannheim, Germany); ampicillin, Appligene (Illkirch, France); chalcomycin, tylosin, geneticin (GN), erythromycin, oleandomycin, and nalidixic acid, Sigma (St. Louis, Mo.); carbomycin and azithromycin, Pfizer (Orsay, France); josamycin, Pharmuka (Genevilliers, France); rosaramicin, Schering-Plough Corporation (Union, N.J.); neospiramycin, forocidin, spiramycin, NO, and CG, Rhône-Poulenc Rorer (Paris, France); angolamycin, lankamycin, methymycin, and pikromycin, E. Cundliffe (Leicester University, Leicester, United Kingdom).
DNA manipulations and bacterial transformation.
DNA extraction and manipulation and transformations of Escherichia coli and Streptomyces were performed according to standard protocols (17, 36).
DNA sequencing.
The DNA sequencing protocol was as described by Sanger et al. (37), with modifications as described by Biggin et al. (8). A Deaza T7 sequencing kit was obtained from Pharmacia, and α-35S-dATP was obtained from Amersham. Sequences were determined for both strands.
Computer-assisted sequence analysis.
Sequence comparisons with databases were performed with the FASTA (27) and BLAST programs (1).
Preparation of crude extracts.
This preparation was performed essentially as described by Cundliffe (11), except that cell breakage was obtained by ultrasonic treatment. Spores were used to inoculate TSB. Cultures were grown with vigorous shaking for 24 h. Mycelia were harvested by filtration, washed twice with HS buffer (10 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 1 M NH4Cl, 5 mM β-mercaptoethanol) and once with HRS buffer (10 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol). Mycelia were resuspended in HRS buffer prior to cell breakage by ultrasonication, followed by centrifugation at 30,000 × g for 30 min. The supernatant was then dialyzed against HM buffer (10 mM HEPES-KOH [pH 7.5], 5 mM MgCl2, 5 mM β-mercaptoethanol, 10% glycerol), resulting in S30 extracts, and stored as aliquots at −70°C. For S. ambofaciens, cultures were grown in TSB containing spiramycin at 10 μg/ml for 20 h, and 10 μg of spiramycin per ml was again added 90 min before harvesting. Spiramycin was added because the cultivation time was too short to allow production and appearance of the resistance. As the expression of some resistance genes (29), including srmA and probably gimA (15), could be induced by spiramycin, this treatment ensured their early expression.
In vitro inactivation of macrolides.
Complete reaction mixtures (total volume, 100 μl) contained dialyzed S30 extract (90 μl), UDP-glucose (1 mM), and macrolide (100 μg/ml) and were incubated at 30°C. At time zero and after various times of incubation, samples were removed and frozen at −20°C to stop the reaction. They were then applied to Whatman AA paper discs (6 mm in diameter), and residual antibiotic activity was examined by bioassay with M. luteus.
Glycosyl transferase assays.
The experiments were performed according to the method developed by Cundliffe (11) except that S30 extracts were used instead of S100*. S30 extract was incubated (total volume, 110 μl) with 0.125 μCi of UDP-[14C]glucose (specific activity, 335 mCi/mmol or 12.4 GBq/mmol) and 50 μg of macrolide antibiotic. The extent of 14C-glycosylation of the drug was calculated by the subtraction of background values obtained from drug-free control assays. For S. lividans harboring the cloned gene, 10 μl of S30 extracts was used, while for S. ambofaciens, 100 μl was needed for the assays.
Resistance conferred by gimA.
Various dilutions of spore suspensions of S. lividans OS456(pOS41.90) or -(pIJ903) (control) were plated on HT medium supplemented with increasing concentrations of various macrolides or without antibiotics. The cultures were then incubated for 48 h before counting was performed.
Spiramycin production assays.
For spiramycin production, MP5 was inoculated with spores at a concentration of 2.5 × 106 spores/ml. No pregermination treatment was done before inoculation. The cultures (70 ml in 500-ml baffled flasks) were incubated at 27°C in an orbital shaker at 250 rpm. In order to take in account the variability of individual cultures, six identical cultures of each strain were included in the study. At 0, 32, 45, 54, 72, and 94 h, samples were withdrawn, and supernatants were independently frozen. The presence of spiramycin in the supernatant of each sample was detected and quantified by bioassay against M. luteus and by high-pressure liquid chromatography (HPLC).
For bioassays, 70 μl of supernatant was applied to Whatman AA paper discs (12 mm in diameter) in parallel with known quantities of spiramycin. Discs were laid on plates containing M. luteus. A CG-resistant mutant (Table 1) obtained for this purpose was used because S. ambofaciens produces CG in addition to spiramycin. Plates were incubated at 4°C for 2 h to allow antibiotic diffusion and then were incubated at 37°C. Diameters of growth inhibition zones were measured after 48 h. Analytic HPLC was carried out as described by Dary et al. (12). Mixtures of spiramycin I, II, and III were used as standards.
Conjugal transfer from E. coli to S. ambofaciens.
Conjugal transfer was performed essentially as described by Bierman et al. (7). Spores of S. ambofaciens ATCC 23877 or OS81 (Table 1) pregerminated as described by Hopwood et al. (17) were used. The optimal conditions were found to be 106 S. ambofaciens CFU and 6 × 107 E. coli CFU per plate. Plates were incubated at 37°C. After 18 h, they were washed with LB and gently scraped to remove the E. coli layer. Then they were covered with an overlay containing HM (at a final concentration of 50 μg/ml) to select transconjugants and nalidixic acid (at a final concentration of 50 μg/ml) to inhibit E. coli growth. Incubation was continued for 5 days.
Nucleotide sequence accession number.
The nucleotide sequence obtained in this study has been deposited in the GenBank/EMBL database under accession no. AJ223970.
RESULTS
Cloning and sequencing of gimA.
Downstream of macrolide resistance gene srmA (28), sequence analysis revealed a truncated open reading frame (ORF). The putative GTG start codon of that ORF overlapped the srmA stop codon (GTGA). To investigate a possible role of this second ORF in resistance, the cloning of the complete ORF was undertaken. An S. ambofaciens gene library constructed in cosmid pWED1 (Table 1) was probed by colony hybridization with a DNA fragment containing srmA and the truncated ORF. This led to the isolation of cosmid pOS41.78, from which a 3.6-kb BamHI fragment containing srmA and 1.7 kb of downstream DNA sequence was subcloned into pUC19 to yield pOS41.80. The sequence of the region downstream of srmA was determined. It revealed the presence of a 1,254-bp ORF which could encode a 45-kDa protein (data not shown). Its GTG start codon was preceded, 8 bp upstream, by the sequence GAGGAG, which could constitute a good Shine-Dalgarno sequence. This ORF showed a typical Streptomyces codon usage (46). The analysis of the deduced protein sequence revealed high degrees of similarity to proteins deduced from mgt of S. lividans (82.5% identity) (20) and oleD of Streptomyces antibioticus (80.9% identity) (16). Mgt from S. lividans is a glycosyl transferase inactivating macrolides by the addition of a glucose residue (11, 20). The gene mgt is also cited as mgtA in the literature. S. lividans is not known to produce any macrolide. In S. antibioticus, producer of the macrolide oleandomycin, OleD is a glycosyl transferase inactivating several macrolides including oleandomycin (33). Because of the strong similarity of the product of the ORF downstream of srmA to Mgt from S. lividans, we hypothesized that this product might also be a glycosyl transferase inactivating macrolides. Therefore, the ORF was called gimA.
The gimA structural gene was cloned downstream from the strong, constitutive ermE-up promoter mutant ermEp* (6), yielding plasmid pOS41.90 (Table 1; Fig. 1). This plasmid was used for most of the experiments described below.
FIG. 1.
Map of the chromosome of S. ambofaciens in the region of srmA and gimA. The inserts found in the plasmid constructions used for this study are indicated. The names of the corresponding vectors are in parentheses. The srmA-gimA chromosomal regions of S. ambofaciens strains, in which gimA has been inactivated by gene replacement, are also presented.
Activity of the gimA product.
In a first approach, experiments were performed to test if the gimA product could inactivate macrolides. As S. ambofaciens possesses several other resistance genes, this was done with the cloned gimA gene. pOS41.90 was introduced into S. lividans OS456 (Table 1), a mutant strain of S. lividans in which the resistance genes lrm and mgt have been inactivated, providing a host strain highly sensitive to macrolides and with no background Mgt activity (30). Crude S30 extracts were prepared from OS456(pOS41.90) and from OS456(pIJ903) (control strain containing the vector without insert) and used for various tests: inactivation of macrolides followed by biological assay of residual antibiotic activity with M. luteus as the indicator strain and glycosylation tests using UDP-[14C]glucose.
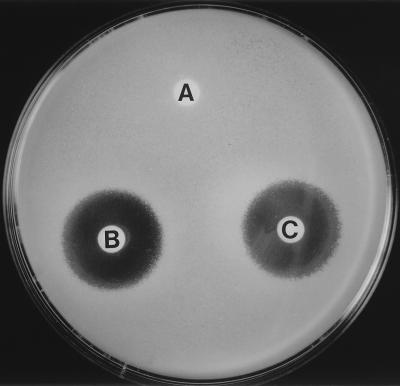
Extracts from OS456(pOS41.90) were able to inactivate some macrolides in the presence of UDP-glucose. In particular, chalcomycin and rosaramicin were completely inactivated after 1 h at 30°C, while no spiramycin inactivation could be detected. In the absence of UDP-glucose, no inactivation was observed (Fig. 2). This UDP-glucose-dependent inactivation was not detected with extracts from OS456(pIJ903). Inactivation was also observed with UDP-galactose as a cofactor (data not shown).
FIG. 2.
In vitro inactivation of chalcomycin by S30 extracts from S. lividans OS456(pOS41.90). Paper disks impregnated with samples from inactivation assays were used to test residual antibiotic activity on M. luteus. Samples came from complete reaction mixture: S30 extract, chalcomycin, and UDP-glucose (A); reaction mixture without UDP-glucose (B); and reaction mixture without S30 (C).
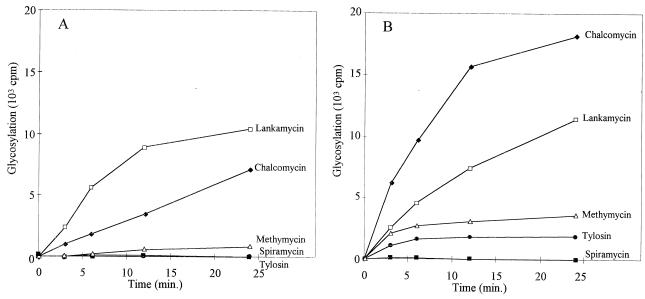
Extracts from OS456(pOS41.90) were used, with UDP-[14C]glucose, to test and quantify the transfer of glucose to macrolides. Chalcomycin, which was rapidly inactivated, was used in this assay. [14C]glucose was transferred to chalcomycin, and almost all the [14C]glucose available in the reaction mixture was transferred within 6 min (see below and Fig. 3).
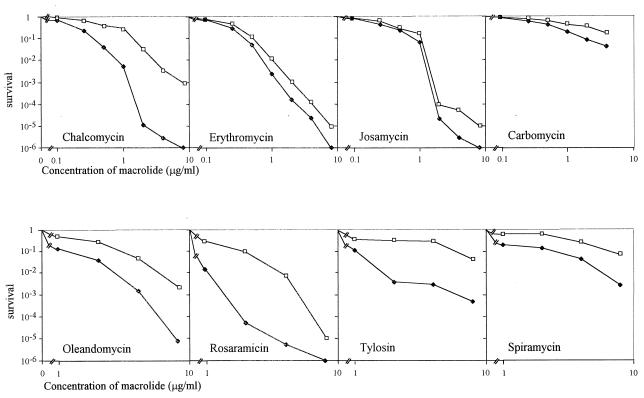
FIG. 3.
Glycosylation of macrolide antibiotics with extracts from S. lividans OS456(pOS41.90). Reaction mixtures contained crude GimA together with 5 μl of UDP-[14C]glucose and 5 μl of macrolide solution in dimethyl sulfoxide (at 10 mg/ml) as described in Materials and Methods. (A) Glycosylation of various macrolides. The same results were obtained with erythromycin, angolamycin, and lankamycin; only those for lankamycin are shown. (B) Glycosylation of spiramycin and its precursors compared with glycosylation of chalcomycin.
These results demonstrated that the gimA product could inactivate macrolide antibiotics by transfer of a glucose residue. UDP-galactose could replace UDP-glucose, but as discussed by Cundliffe (11) it is unknown if UDP-galactose is used per se or if it must be converted to UDP-glucose (or vice versa). Therefore, GimA activity was called glycosyl transferase rather than glucosyl transferase.
Substrate specificity of GimA.
The radiotransfer assay allows quantification of the GimA activity. This assay was used to study the substrate specificity of GimA, in order to compare it with those of other known macrolide-inactivating glycosyl transferases from S. lividans (11), S. antibioticus (33), and Streptomyces hygroscopicus (38). Moreover, the radiotransfer assay should indicate if GimA could glycosylate spiramycin precursors in the biosynthetic pathway. This last point could not be studied by inactivation, because some of the precursors of spiramycin lacked antibiotic activity.
With UDP-[14C]glucose as the cofactor, crude S30 extracts from OS456(pOS41.90) were tested on various macrolides (Fig. 3A). Among those, chalcomycin was the most active substrate. Methymycin, tylosin, pikromycin, and rosaramicin were four of the best substrates. Oleandomycin, josamycin, and carbomycin were glycosylated to a lesser extent. Macrolides that were found to be as poor substrates of GimA as lankamycin were erythromycin and angolamycin. Spiramycin was also a very poor substrate (Fig. 3B). As gimA originates from a spiramycin producer, it was interesting to study the glycosylation of spiramycin precursors. Forocidin consists of the 16-atom lactone ring with only mycaminose attached. Neospiramycin possesses two amino sugars, mycaminose and forosamine. The biosynthetic pathway ends with spiramycin with three sugars attached, the mycarose being attached to mycaminose. The results presented in Fig. 3B demonstrate that neospiramycin was as poorly modified as spiramycin but, interestingly, that forocidin could be glycosylated.
Resistance profile conferred by gimA.
It was unknown whether a macrolide-inactivating glycosyl transferase could confer a resistant phenotype or not. To answer this question, the resistances of S. lividans OS456(pOS41.90) to various macrolides were compared to those of S. lividans OS456(pIJ903) (control strain). This study could only be performed with the cloned gimA gene and not with S. ambofaciens, which possesses several resistance mechanisms.
The results are presented in Fig. 4. Some of the macrolides tested in the radiotransfer assay were not available in sufficient amounts for this experiment. Among the ones tested, those for which the difference in survival between OS456(pOS41.90) and the control strain was the greatest were chalcomycin, rosaramicin, tylosin, and oleandomycin. In these cases, the presence of gimA increased the survival by at least a factor of 100 at several concentrations of the antibiotics. These four antibiotics were among the best substrates for GimA in vitro. However, for other macrolides tested, no simple correlation between the level of resistance in vivo and the in vitro activity could be established. For instance, with spiramycin, a clear difference in survival between the strain harboring gimA and the control strain was observed, even though spiramycin was one of the poorest substrates. These results might indicate that different macrolides are taken up and/or accumulated by S. lividans with different efficiencies.
FIG. 4.
Resistance conferred by gimA. Spores from S. lividans OS456(pOS41.90) and OS456(pIJ903) (control) were plated on HT medium without or with different macrolides at various concentrations. The survival rates at various macrolide concentrations, each calculated as the ratio of the number of CFU on the medium with the macrolide to the number of CFU on the same medium without the antibiotic (log scale), are presented for OS456(pOS41.90) (white squares) and OS456(pIJ903) (black squares).
Gene replacement inactivating gimA in S. ambofaciens.
In order to determine the role played by gimA in the self-protection of S. ambofaciens from spiramycin, its inactivation was attempted both in the ATCC 23877 wild-type strain and in the non-spiramycin-producing mutant OS81 (Table 1). As shown in Fig. 1, an SplI fragment internal to gimA was deleted and replaced by the Ωhyg cassette, generating pOS41.98 (in pUC19) and then pOS41.99 (in conjugative vector pOJ260; conferring GN resistance) (Table 1). Then, pOS41.99 was introduced into S. ambofaciens via conjugal transfer from E. coli S17.1 (40) and HM selection was applied. Five days after conjugation, Hmr transconjugants were picked and examined for their GN resistance. Hmr Gns clones were obtained from both strains. Such clones had presumably lost the pOJ260 part by a double-crossover recombination event. The replacement of gimA by its disrupted counterpart from pOS41.99 was confirmed by Southern blot analysis (data not shown). For instance, the gimA probe revealed that the hybridizing BamHI fragment had increased from 3.6 kb in the wild type to 6.1 kb in the mutant strains. Hybridization with Ωhyg confirmed its presence in the mutants and its absence in the wild-type and OS81 strains. One mutant strain with gimA interrupted derived from the wild-type strain was designated OS41.99. One equivalent mutant derived from OS81 was designated OS41.99NP. These two strains were not affected in their growth or sporulation.
Effect of gimA on spiramycin production.
The issue investigated was the ability of strain OS41.99 to produce spiramycin, and the possible effect of gimA disruption or overexpression on the level of spiramycin production. OS41.99 was first grown in MP5 production medium, and the supernatant activity was tested by a bioassay using M. luteus Cgr (Table 1) and further characterized by HPLC. It was found that OS41.99 could still produce spiramycin, indicating that gimA is not a resistance gene essential for the survival of the strain, even under antibiotic production conditions.
Therefore, as GimA could glycosylate some spiramycin precursors, and as GimA could confer resistance towards spiramycin in vivo, it was of interest to investigate whether or not the inactivation or the constitutive expression of gimA had an effect on the spiramycin production level. The production of spiramycin by the following four S. ambofaciens strains was studied: wild-type strain ATCC 23877, OS41.99 where gimA is inactivated, OS41.99(pOS41.90) (where gimA expression is governed by the strong and constitutive ermE-up promoter mutant ermEp*), and, as a control, OS41.99(pOS41.105). The last plasmid was derived from pOS41.90 by an in-frame deletion removing most of the gimA gene (Fig. 1). This control was done to ensure that a possible effect observed with pOS41.90 was due to the presence of gimA, and not to the presence of the vector carrying ermEp*. As has been reported, introduction of plasmid vectors could affect antibiotic production in some strains (42).
All strains were grown under the same conditions, with no selective pressure. The presence of the low-copy-number replicative plasmids pOS41.90 and pOS41.105 was checked at different times by plating the dispersed mycelia on medium without antibiotics, followed by replica plating on medium with NO. After 94 h of growth in MP5, the plasmids were still present in 100% of the CFU. The growth rates of the different strains in MP5 were similar (data not shown). The levels of spiramycin production were determined by HPLC.
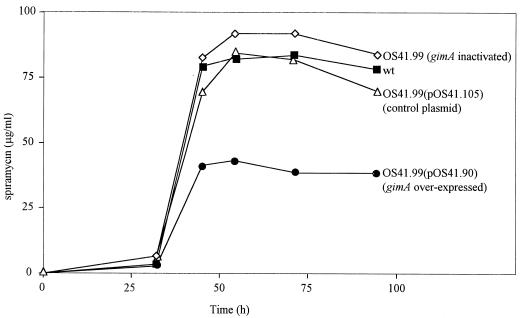
The mean values for spiramycin production are presented in Fig. 5. The observed slight increase in spiramycin production between the gimA mutant OS41.99 and the wild-type strain is not significant (data not shown). However, for strain OS41.99(pOS41.90), where gimA is constitutively expressed, the level of spiramycin production was significantly decreased, by a factor of 2, in comparison with that of OS41.99 and wild-type strains. This effect could be attributed to the expression of gimA, carried by pOS41.90, as in the same strain the presence of control plasmid pOS41.105 caused a nonsignificant decrease.
FIG. 5.
Effect of gimA expression on spiramycin production in S. ambofaciens. Six identical independent cultures of each strain were grown in MP5 medium. Samples were withdrawn at 0, 32, 45, 54, 71, and 94 h. Spiramycin was detected and quantified by HPLC. For each strain, the curve was drawn from the averages of the six independent values obtained for each time. wt, wild type.
Evidence for the existence of another glycosyl transferase inactivating macrolides in S. ambofaciens.
S30 extracts were prepared from the S. ambofaciens wild-type strain and the OS41.99 mutant. Surprisingly, extracts from both strains inactivated chalcomycin in the presence of UDP-glucose (data not shown). Moreover, these extracts were used in radiotransfer assays, and a comparison of the results presented in Fig. 3A and 6A shows that the substrate specificity of GimA was clearly different from that of the activity detected in OS41.99. For instance, lankamycin, which was a very poor substrate for GimA (Fig. 3A), was found to be a very active substrate for the activity detected in both S. ambofaciens strains (Fig. 6). The activity observed in the wild-type strain (Fig. 6B) could result from the addition of both activities. A comparison of results presented in Fig. 6A and 6B shows that lankamycin was glycosylated to comparable levels with extracts from both strains but that gimA inactivation led to a decrease in the glycosylation levels of chalcomycin, tylosin, and methymycin, which were good substrates for GimA. These observations indicated the existence of another macrolide-inactivating glycosyl transferase activity, with a substrate specificity different from that of GimA.
FIG. 6.
Glycosylation of macrolides with S30 extracts from S. ambofaciens. Reaction mixtures contained crude extract together with 5 μl of UDP-[14C]glucose and 5 μl of macrolide solution in dimethyl sulfoxide (at 10 mg/ml) as described in Materials and Methods. (A) Extracts from OS41.99 (gimA-null mutant); (B) extract from the wild-type strain.
DISCUSSION
The mechanism of macrolide inactivation by glycosylation at the C-2′ position has been most often described for Streptomyces: this glycosyl transferase activity was first described for Streptomyces vendargensis acting on erythromycin A (21) and seems widespread among Streptomyces strains (38). It was studied for strains not producing macrolides such as S. lividans (11), and S. hygroscopicus (38). In S. antibioticus, producer of the macrolide oleandomycin, two glycosyl transferases (OleD and OleI) and a glycosidase (OleR) are involved in oleandomycin modification during its biosynthesis (33).
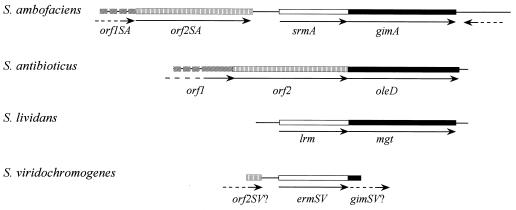
In S. ambofaciens, the gimA gene was found immediately downstream of srmA (28), a gene encoding a methyltransferase conferring macrolide resistance by target modification. As shown in Fig. 7, macrolide-inactivating glycosyl transferase genes are associated with methyltransferase genes in S. ambofaciens, in S. lividans, and probably in Streptomyces viridochromogenes. The last two strains are not known to produce any macrolide antibiotics. For S. antibioticus, no methyltransferase gene is found upstream of oleD or oleI. This is in agreement with the observation that ribosomes from S. antibioticus are sensitive to oleandomycin, even during production (13). Interestingly, the two ORFs upstream of oleD have high degrees of similarity to those upstream of srmA. This could indicate that some deletion or insertion events occurred at the corresponding loci in S. antibioticus or in S. ambofaciens. In S. lividans, the region located upstream of lrm is completely different from those found in the three other strains, indicating transfer may have occurred in this region.
FIG. 7.
Comparison of the genetic environments around macrolide resistance genes in various Streptomyces species. gimA, oleD, and mgt gene products have about 80% identity. srmA, lrm, and ermSV encode monomethyltransferases with about 80% identity. The deduced products of orf1SA and orf2SA from S. ambofaciens and of orf1 and orf2 from S. antibioticus have, respectively, 75 and 76% identity. The deduced product of the end of an ORF upstream of ermSV has 69% identity with the deduced products of orf2 and orf2SA. The deduced product of the short sequence downstream of ermSV is similar to glycosyl transferases inactivating macrolides. The GenBank/EMBL accession numbers are AJ223970 for gimA, Z22577 for oleD, M74717 for mgt, and U59450 for ermSV.
The substrate specificities of macrolide-inactivating glycosyl transferases from S. lividans, S. antibioticus, and S. hygroscopicus and that of GimA from S. ambofaciens are different. The size of the lactone ring does not seem to be a factor discriminating the poor and good substrates as discussed by Cundliffe (11). As in other cases, the sugar glycosylated by GimA is probably the one attached to the C-5 position of the 16- and 14-member lactone rings (21) (corresponding to C-3 in the 12-member lactone ring of methymycin). On this sugar, the position modified is probably that of the 2′-OH group. As expected in this case, angolamycin, which lacks this hydroxyl group, was revealed to be a very poor substrate of GimA. Our results, with the exception of those for tylosin, are in agreement with the suggestion that macrolides monosubstituted at position C-5 are better substrates than those disubstituted (43).
The ability of glycosyl transferases inactivating macrolides to confer resistance in vivo has never been investigated until now. This was done with the cloned gene gimA, introduced into a strain susceptible to macrolides, S. lividans OS456. Resistance was indeed observed, but the levels of resistance were very low, in particular, much lower than those conferred by target modifying enzymes: those encoded by either tlrD, ermE (30), or srmA (28). The low level of resistance was probably not due to a low level of expression of gimA. Indeed, cellular extracts prepared from OS456(pOS41.90), the strain used for the whole-cell resistance study, showed greater glycosyl transferase activity than the ones prepared from the S. ambofaciens wild-type strain. Although the level of resistance conferred by GimA was low, GimA nevertheless conferred resistance to chalcomycin and tylosin, while SrmA was unable to confer any resistance to these two macrolides.
In a strain producing a macrolide antibiotic, a gene encoding a glycosyl transferase inactivating macrolides might play a role in the self-protection of the strain, as oleI does in S. antibioticus. Nevertheless, gene gimA could be inactivated in S. ambofaciens without any deleterious effect, even during production of spiramycin. But S. ambofaciens possesses additional macrolide resistance mechanisms (including a second macrolide-inactivating glycosyl transferase), which might provide protection against the produced antibiotic.
In S. antibioticus, oleI is located among oleandomycin biosynthetic genes and its product is supposed to be the enzyme responsible for oleandomycin glycosylation during biosynthesis. oleD is not located in the oleandomycin cluster and might play a more general role, one not directly involved in oleandomycin biosynthesis (33). In S. ambofaciens, gimA is dispensable; it is probably not located in the spiramycin cluster, and its product is not very active on spiramycin. A comparison of genetic organization and protein sequences indicated that gimA is more related to mgt and oleD than to oleI. Therefore, gimA could be involved in the resistance to exogenous macrolides rather than in self-protection against spiramycin during biosynthesis. However, the constitutive overexpression of gimA led to a decrease in spiramycin production. This effect could be due to the action of GimA on spiramycin itself, or more probably on spiramycin precursors such as forocidin. These glycosylated precursors might not be substrates for later steps of the biosynthetic pathway, or the glycosylated spiramycin produced might not be reactivated.
ACKNOWLEDGMENTS
We are very grateful to E. Cundliffe for introducing us to the use of the radiotransfer assay for the measurement of macrolide-inactivating glycosyl transferase activity, for the kind gift of various macrolides, and for helpful discussions. We thank P. Lacroix for the kind gift of spiramycin precursors and CG, J. Hugueville for HPLC analysis, P. Mazodier for providing E. coli S17-1, and M. Bibb for providing pIJ4070. We thank M. Guérineau for support and critical reading of the manuscript.
This work was supported by CNRS and the Alliance Programme. A.G. received a PhD fellowship from the Ministère de l’Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Autissier D, Courvalin P. Analysis of the nucleotide sequence of the ereB gene encoding the erythromycin esterase type II. Nucleic Acids Res. 1986;14:4987–4999. doi: 10.1093/nar/14.12.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltz R H. Genetic recombination by protoplast fusion in Streptomyces. Dev Ind Microbiol. 1980;21:43–54. [Google Scholar]
- 4.Baltz R H, Seno E T. Genetics of Streptomyces fradiae and tylosin biosynthesis. Annu Rev Microbiol. 1988;42:547–574. doi: 10.1146/annurev.mi.42.100188.002555. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, M. J. Personal communication.
- 6.Bibb M J, White J, Ward J M, Janssen G R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol Microbiol. 1994;14:533–545. doi: 10.1111/j.1365-2958.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- 7.Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 8.Biggin M D, Gibson T J, Hong G F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci USA. 1983;80:3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birmingham V A, Cox K L, Larson J L, Fishman S E, Hershberger C L, Seno E T. Cloning and expression of a tylosin resistance gene from a tylosin-producing strain of Streptomyces fradiae. Mol Gen Genet. 1986;204:532–539. doi: 10.1007/BF00331036. [DOI] [PubMed] [Google Scholar]
- 10.Blondelet-Rouault M H, Weiser J, Lebrihi A, Branny P, Pernodet J L. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 11.Cundliffe E. Glycosylation of macrolide antibiotics in extracts of Streptomyces lividans. Antimicrob Agents Chemother. 1992;36:348–352. doi: 10.1128/aac.36.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dary A, Bourget N, Girard N, Simonet J M, Decaris B. Amplification of a particular DNA sequence in Streptomyces ambofaciens RP181110 reversibly prevents spiramycin production. Res Microbiol. 1992;143:99–112. doi: 10.1016/0923-2508(92)90039-q. [DOI] [PubMed] [Google Scholar]
- 13.Fierro J F, Hardisson C, Salas J A. Resistance to oleandomycin in Streptomyces antibioticus, the producer organism. J Gen Microbiol. 1987;133:1931–1939. doi: 10.1099/00221287-133-7-1931. [DOI] [PubMed] [Google Scholar]
- 14.Gale E F, Cundliffe E, Reynolds P E, Richmond M H, Waring M J. The molecular basis of antibiotic action. London, United Kingdom: Wiley; 1981. [Google Scholar]
- 15.Gourmelen, A. Unpublished data.
- 16.Hernandez C, Olano C, Mendez C, Salas J A. Characterization of a Streptomyces antibioticus gene cluster encoding a glycosyltransferase involved in oleandomycin inactivation. Gene. 1993;134:139–140. doi: 10.1016/0378-1119(93)90189-a. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 18.Hopwood D A, Kieser T, Wright H M, Bibb M J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983;129:2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- 19.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins G, Cundliffe E. Cloning and characterization of two genes from Streptomyces lividans that confer inducible resistance to lincomycin and macrolide antibiotics. Gene. 1991;108:55–62. doi: 10.1016/0378-1119(91)90487-v. [DOI] [PubMed] [Google Scholar]
- 21.Kuo M S, Chirby D G, Argoudelis A D, Cialdella J I, Coats J H, Marshall V P. Microbial glycosylation of erythromycin A. Antimicrob Agents Chemother. 1989;33:2089–2091. doi: 10.1128/aac.33.12.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lydiate D J, Malpartida F, Hopwood D A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35:223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 23.Mendez C, Salas J A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol Lett. 1998;158:1–8. doi: 10.1111/j.1574-6968.1998.tb12792.x. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi N, Emura A, Matsuyama H, O’Hara K, Sasatsu M, Kono M. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2359–2363. doi: 10.1128/aac.39.10.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi N, Katayama J, O’Hara K. Cloning and nucleotide sequence of the mphB gene for macrolide 2′-phosphotransferase II in Escherichia coli. FEMS Microbiol Lett. 1996;144:197–202. doi: 10.1111/j.1574-6968.1996.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 26.Ounissi H, Courvalin P. Nucleotide sequence of the gene ereA encoding the erythromycin esterase in Escherichia coli. Gene. 1985;35:271–278. doi: 10.1016/0378-1119(85)90005-8. [DOI] [PubMed] [Google Scholar]
- 27.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pernodet, J. L. Unpublished data.
- 29.Pernodet J L, Alegre M T, Blondelet-Rouault M H, Guerineau M. Resistance to spiramycin in Streptomyces ambofaciens, the producer organism, involves at least two different mechanisms. J Gen Microbiol. 1993;139:1003–1011. doi: 10.1099/00221287-139-5-1003. [DOI] [PubMed] [Google Scholar]
- 30.Pernodet J L, Fish S, Blondelet-Rouault M H, Cundliffe E. The macrolide-lincosamide-streptogramin B resistance phenotypes characterized by using a specifically deleted, antibiotic-sensitive strain of Streptomyces lividans. Antimicrob Agents Chemother. 1996;40:581–585. doi: 10.1128/aac.40.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinnert-Sindico S. Une nouvelle espèce de Streptomyces productrice d’antibiotiques: Streptomyces ambofaciens n. sp. caractères culturaux. Ann Inst Pasteur (Paris) 1954;87:702–707. [PubMed] [Google Scholar]
- 32.Pridham T G, Anderson P, Foley C, Lindenfelser L A, Hesseltine C W, Benetdict R C. A selection of media for maintenance and taxonomic study of Streptomyces. Antibiot Annu. 1957;1956–1957:947–953. [PubMed] [Google Scholar]
- 33.Quiros L M, Aguirrezabalaga I, Olano C, Mendez C, Salas J A. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol. 1998;28:1177–1185. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 34.Richardson M A, Kuhstoss S, Solenberg P, Schaus N A, Rao R N. A new shuttle cosmid vector, pKC505, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene. 1987;61:231–241. doi: 10.1016/0378-1119(87)90187-9. [DOI] [PubMed] [Google Scholar]
- 35.Rosteck P R, Jr, Reynolds P A, Hershberger C L. Homology between proteins controlling Streptomyces fradiae tylosin resistance and ATP-binding transport. Gene. 1991;102:27–32. doi: 10.1016/0378-1119(91)90533-h. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki J, Mizoue K, Morimoto S, Omura S. Microbial glycosylation of macrolide antibiotics by Streptomyces hygroscopicus ATCC 31080 and distribution of a macrolide glycosyl transferase in several Streptomyces strains. J Antibiot (Tokyo) 1996;49:1110–1118. doi: 10.7164/antibiotics.49.1110. [DOI] [PubMed] [Google Scholar]
- 39.Schoner B, Geistlich M, Rosteck P, Jr, Rao R N, Seno E, Reynolds P, Cox K, Burgett S, Hershberger C. Sequence similarity between macrolide-resistance determinants and ATP-binding transport proteins. Gene. 1992;115:93–96. doi: 10.1016/0378-1119(92)90545-z. [DOI] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Pühler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 41.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–24. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas D I, Cove J H, Baumberg S, Jones C A, Rudd B A. Plasmid effects on secondary metabolite production by a streptomycete synthesizing an anthelminthic macrolide. J Gen Microbiol. 1991;137:2331–2337. doi: 10.1099/00221287-137-10-2331. [DOI] [PubMed] [Google Scholar]
- 43.Vilches C, Hernandez C, Mendez C, Salas J A. Role of glycosylation and deglycosylation in biosynthesis of and resistance to oleandomycin in the producer organism, Streptomyces antibioticus. J Bacteriol. 1992;174:161–165. doi: 10.1128/jb.174.1.161-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahl G M, Lewis K A, Ruiz J C, Rothenberg B, Zhao J, Evans G A. Cosmid vectors for rapid genomic walking, restriction mapping, and gene transfer. Proc Natl Acad Sci USA. 1987;84:2160–2164. doi: 10.1073/pnas.84.8.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright F, Bibb M J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 47.Zalacain M, Cundliffe E. Cloning of tlrD, a fourth resistance gene, from the tylosin producer, Streptomyces fradiae. Gene. 1991;97:137–142. doi: 10.1016/0378-1119(91)90021-3. [DOI] [PubMed] [Google Scholar]