Highlights
-
•
ICW assay represents a reliable method to measure infection rates of PUUV.
-
•
ICW assay is suitable for quantification of replication in different cell types.
-
•
Antiviral drug testing reveals cell type specific differences for efficacy of ribavirin.
-
•
Podocytes and vero E6 cells exhibit different SI values for RBV.
Keywords: Orthohantavirus, Puumala virus, Podocytes, Infectivity, N protein, Antivirals
Abstract
Hemorrhagic fever with renal syndrome (HFRS) represents a serious zoonotic disease caused by orthohantaviruses in Eurasia. A specific antiviral therapy is not available. HFRS is characterized by acute kidney injury (AKI) with often massive proteinuria. Infection of kidney cells may contribute to the clinical picture. However, orthohantaviral replication in kidney cells is not well characterized. Therefore, we aimed to perform a reliable high-throughput assay that allows the quantification of infection rates and testing of antiviral compounds in different cell types. We quantified relative infection rates of Eurasian pathogenic Puumala virus (PUUV) by staining of nucleocapsid protein (N protein) in an in-cell Western (ICW) assay. Vero E6 cells, derived from the African green monkey and commonly used in viral cell culture studies, and the human podocyte cell line CIHP (conditionally immortalized human podocytes) were used to test the ICW assay for replication kinetics and antiviral drug testing. Quantification of infection by ICW revealed reliable results for both cell types, as shown by their correlation with immunofluorescence quantification results by counting infected cells. Evaluation of antiviral efficacy of ribavirin by ICW assay revealed differences in the toxicity (TC) and inhibitory concentrations (IC) between Vero E6 cells and podocytes. IC5O of ribavirin in podocytes is about 12-fold lower than in Vero E6 cells. In summary, ICW assay together with relevant human target cells represents an important tool for the study of hantaviral replication and drug testing.
1. Introduction
Pathogenic hantaviruses belong to the genus Orthohantavirus within the Hantaviridae family and infection is characterized by a specific organ manifestation. Members on the American continents cause hantaviral cardiopulmonary syndrome (HCPS), whereas infections with Eurasian orthohantaviruses lead to hemorrhagic fever with renal syndrome (HFRS) (Krautkrämer et al., 2013; Vaheri et al., 2013). Reservoir hosts are rodents, and humans become infected via inhalation of aerosols contaminated with excreta from infected animals. Severity of infections varies enormously between HFRS-causing hantaviruses, even between members of the same species. Dobrava-Belgrade virus (DOBV) and Sochi virus both belonging to the species Orthohantavirus dobravaense and Hantaan virus (HTNV) (species Orthohantavirus hantanense) cause severe forms of HFRS, whereas infections with Kurkino virus that is also a member of species Orthohantavirus dobravaense and Puumala virus (PUUV) (species Orthohantavirus puumalaense) display mainly as mild form of HFRS (Klempa et al., 2013). Patients with HFRS demonstrate fever, hematuria, proteinuria, an acute rise in creatinine levels and decreased levels of platelets and serum albumin (Koehler et al., 2022). Icatibant, a bradykinin receptor antagonist, was successfully tested in the treatment of two severe cases of PUUV infection but failed in two other cases (Antonen et al., 2013; Laine et al., 2015; Tuiskunen Bäck et al., 2022; Vetter et al., 2021). Early treatment of HTNV-infected patients with ribavirin has been found to increase survival and to reduce the incidence of oliguria and hemodialysis requirement, but therapeutical benefit in PUUV-infected patients has not been demonstrated (Huggins et al., 1991; Malinin and Platonov, 2017; Rusnak et al., 2009; Steininger et al., 2023). Therefore, treatment options for PUUV are limited to supportive measures, and no specific antiviral drug is yet available for therapeutic use.
Further, hantavirus research continues to be hampered by the lack of a suitable small animal model for HFRS (Perley et al., 2019; Sanada et al., 2011), thus relying mainly on cell culture models to analyze the viral replication cycle or to identify specific antiviral drugs. Vero E6 cells are widely used in viral research since they lack an intact interferon response (Emeny and Morgan, 1979). However, HFRS is characterized by its organ-specific manifestation with acute kidney injury with often massive proteinuria, and a variety of renal cell types are targeted by hantavirus infection leading to cell type-specific alterations in cellular morphology and function (Bourquain et al., 2019; Hägele et al., 2018, 2019; Krautkrämer et al., 2011; Nusshag et al., 2022; Temonen et al., 1993). Podocyte injury contributes to severe proteinuria and correlates with disease severity (Nusshag et al., 2020). Therefore, we established an in-cell Western (ICW) assay under biosafety level 2 (BSL-2) laboratory conditions for the HFRS-causing Puumala virus (PUUV) and compared the results between Vero E6 cells and a human podocyte cell line as natural target cells of hantavirus infection.
2. Material and methods
2.1. Cells and viruses
Vero E6 cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10 % fetal calf serum (FCS). Stocks of Puumala virus (PUUV) strain Vranica were propagated on Vero E6 cells. To obtain virus inoculum, cell culture supernatants were harvested, clarified from cells and cell debris by centrifugation at 2000 x g for 10 min. Virus inoculum was titrated on the respective cell type. CIHP (conditionally immortalized human podocytes) (Saleem et al., 2002) were cultured in RPMI1640 supplemented with 10 % FCS and 1 % insulin-transferrin-selenium and differentiated by a shift to 37 °C for ten days. Work with virus was performed in a BSL-2 laboratory.
2.2. Immunofluorescence (IF)
For immunofluorescence, cells fixed with 3 % paraformaldehyde in PBS were stained with mouse monoclonal anti-PUUV N protein antibody (Progen, Heidelberg, Germany) and Cy3 fluorescently labeled secondary antibody. Nuclei were stained with Hoechst 33342. The percentage of infected cells was quantified by counting cells expressing N protein.
2.3. In-cell Western (ICW) assay
Cells (2 × 104 cells/well) were grown in 96 well microplates and infected with PUUV. At the indicated time points, cells were washed, fixed with 3 % paraformaldehyde in PBS, permeabilized and stained with anti-PUUV N protein (Progen) for 1 h at room temperature with shaking. Plates were washed two times for five minutes with PBS and incubated with the secondary IRDye800CW-labeled antibody and with DRAQ5/Sapphire700 cell stains (BioStatus, Leicestershire, United Kingdom) for cell number normalization for one hour at room temperature. For background control, cells were incubated with secondary antibody without first antibody. Plates were analyzed by using Odyssey infrared imaging system (LiCor Biosciences, Lincoln, NE, USA). The relative amount of N protein expression was obtained by normalizing to the cell number obtained by DRAQ5/Sapphire700 cell stains.
2.4. Viability
Cytotoxicity of drugs was determined by measuring the amount of ATP using CellTiter-Glo luminescent cell viability assay (Promega, Mannheim, Heidelberg). Viability was analyzed at day six post drug administration with a change of medium and addition of fresh drug- or solvent-containing medium after three days of incubation.
2.5. Drug treatment
Ribavirin (RBV) (Santa Cruz Biotechnology, Heidelberg, Germany) was dissolved in H2O. Cells were pretreated with the drug or solvent control at the indicated concentrations for one hour, then viral inoculum was added for one hour at a multiplicity of infection (MOI) of 1. After a triple wash with PBS, medium with the indicated drug concentration or solvent control was added to the cells. Cells were incubated for six days with a change of medium and addition of fresh drug- or solvent-containing medium after three days. Infection was determined by ICW after six days post infection.
2.6. Statistical analysis
Pearson analysis was performed using Prism 5 (GraphPad, Boston, MA, USA) to assess the correlation between quantification of infection via ICW and IF. To assess the quality of the ICW assay, the Z′-factor was calculated using the following equation, Z′=1 − (3σcinf + 3σcuninf)/(|μcinf − μcuninf|), where σcinf and σcuninf are the standard deviations of infected positive controls and uninfected negative controls, respectively. The µcinf and µcunif represent the mean of infected positive controls and uninfected negative controls, respectively.
A Z’-factor ≥ 0.5 and < 1 indicates an excellent assay with low variability of data and wide separation between signal and controls (Zhang et al., 1999).
3. Results
3.1. Determination of infectivity by in-cell Western (ICW) assay
An ICW assay was performed to detect and quantify PUUV infection. Vero E6 cells were infected at increasing amounts of viral inoculum. Infection was monitored by staining for N protein and normalization to cell number (Fig. 1A). Suitability of ICW assay was evaluated by calculation of Z´-factor. The Z′-factor of ICW assays was 0.81, which is considered to be excellent for a high-throughput assay. We compared our ICW results with the determination of infection via counting of infected cells after immunofluorescence staining for N protein and labeling of nuclei with Hoechst 33342 (Fig. 1B). The results of quantification via ICW assay were in excellent agreement with the results obtained via immunofluorescence staining (Fig. 1C) showing a highly significant linear correlation (Pearsons r = 0.9963; 95 % CI = 0.9429 to 0.9998; P = 0.0003) (Fig. 1D).
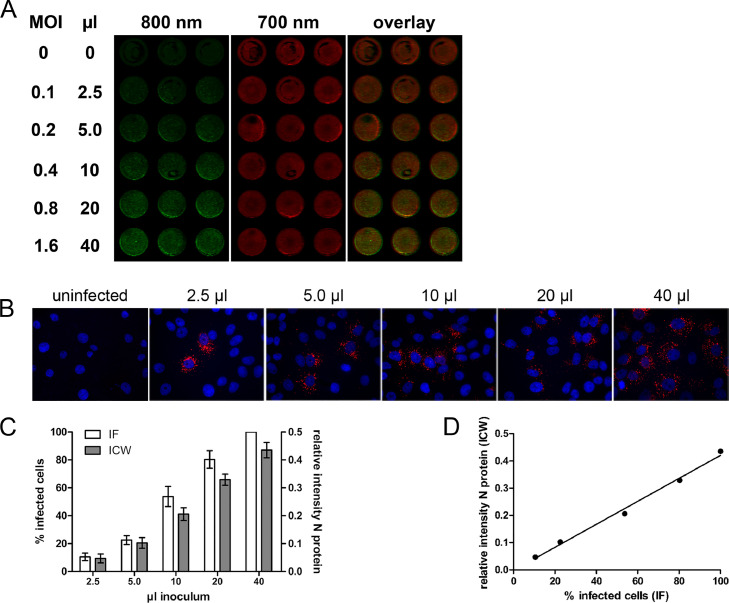
Fig. 1.
Determination of infectivity of PUUV by ICW assay. A) Vero E6 cells were infected in triplicates with increasing amounts of PUUV or were left uninfected. At 2 days post infection, cells were fixed and subjected to ICW assay. Infection was monitored by staining against PUUV N protein. N protein was detected on the 800 nm channel and cell number was determined by DRAQ5 and Sapphire700 cell stains on the 700 nm channel. B) For IF, infected Vero E6 cells were stained for N protein with Cy3 fluorescently labeled antibody (red) and nuclei were stained with Hoechst 33342 (blue). C) Quantification of infection by IF and ICW assay. In IF assay, percentage of infected cells was calculated by counting of cells positive for N protein normalized to the number of cells visualized by Hoechst 33342. In ICW assay, relative infectivity was calculated as N protein levels detected in Vero E6 cells normalized to relative cell number as described in A). Three independent experiments were performed for each assay. Data are shown as mean with standard deviation (SD). D) Relationship between quantification of infection by IF and ICW was analyzed using the Pearson correlation.
In a next step, we compared the quantification by ICW and immunofluorescence in a replication kinetic experiment (Fig. 2). Cells were infected at an MOI of 0.05. The viral spread was detectable by ICW and immunofluorescence, and both methods provide comparable results in terms of increases in infection rates at the indicated time points.
Fig. 2.
Determination of PUUV replication kinetics in Vero E6 cells by ICW and IF. Vero E6 cells were inoculated with PUUV at an MOI of 0.05 and infection cells was quantified by detection of N protein via ICW or IF at the indicated time points, dpi: days post infection. Three independent experiments were performed for each assay. Mean ± SD were shown.
Together, ICW assay is a useful and reliable method to measure relative infection rates by orthohantavirus PUUV.
3.2. Testing of antiviral activity
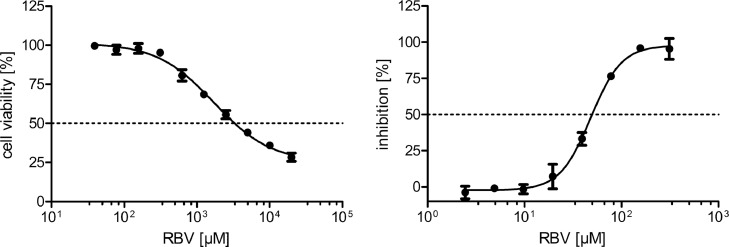
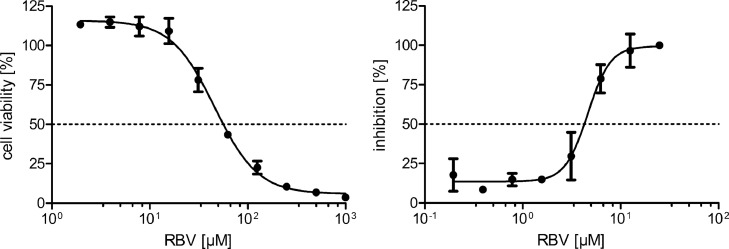
To evaluate the suitability of ICW assay as high-throughput assay for antiviral drug testing, we performed infection assays with the nucleoside analogue ribavirin (RBV), which is described to inhibit hantaviral infection in vitro (Buys et al., 2011; Murphy et al., 2000; Sun et al., 2007). We analyzed the viability of cells in the presence of different concentrations of RBV (Fig. 3). The TC50 toxic concentration for RBV in Vero E6 cells is 3308.86 µM. The inhibitory concentration (IC) of RBV for infection of Vero E6 cells with PUUV was assessed by ICW and revealed an efficient inhibition with an IC50 of 49.83 µM (Fig. 3).
Fig. 3.
Viability and efficacy of RBV in PUUV-infected Vero E6 cells. Shown is viability relative to the solvent control, which is set at 100 %. Infection of cells pretreated with RBV for 1 h and infected at an MOI of 0.05 was analyzed by ICW. Inhibition of infection was calculated relative to control cells incubated with solvent control that were set at 100 %. Data of three independent experiments were presented as mean ± SD.
3.3. In-cell Western assay in human podocytes
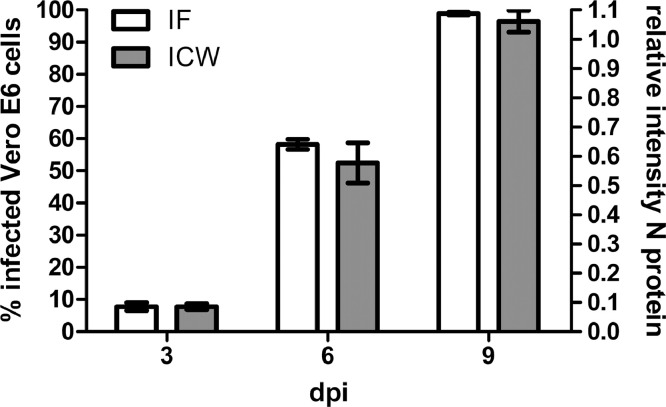
To confirm the results from Vero E6 cells in relevant hantaviral human target cells, we repeated the experiments in the podocyte cell line CIHP. First, we tested the suitability of ICW to determine relative infection rates in podocytes by comparison of the replication kinetics measured by ICW and IF (Fig. 4). Quantification of infection by IF and ICW assay in PUUV-infected podocytes revealed similar results for the increase of infection over time as in Vero E6 cells.
Fig. 4.
Determination of PUUV replication kinetics in podocytes by ICW and IF. Cells were inoculated with PUUV at an MOI of 0.05 and infection was quantified by detection of N protein via ICW or IF at the indicated time points, dpi: days post infection. Three independent experiments were performed for each assay. Mean ± SD were shown.
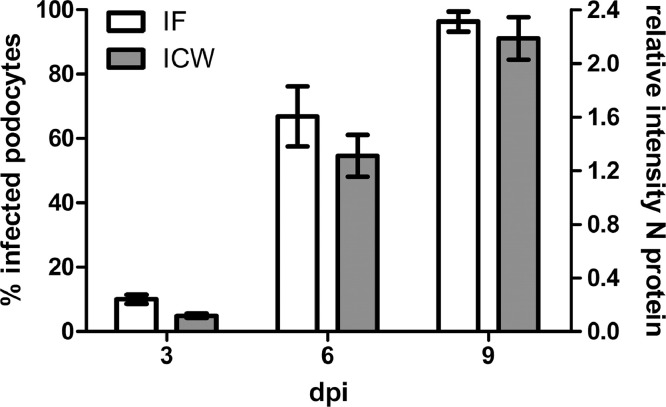
In addition, we tested the efficacy of RBV to inhibit PUUV infection in human podocytes (Fig. 5). Podocytes demonstrate a TC50 of 56.26 µM for RBV and an IC50 of 4.25 µM for RBV in PUUV infection. We observed a massive difference in the cytotoxic and inhibitory concentration of RBV for Vero E6 cells and podocytes (Table 1). Compared to Vero E6 cells, the TC50 and IC50 of RBV for podocytes is about 60-fold and 12-fold lower, respectively. These differences result in a selectivity index (SI), which is 5-fold lower than in Vero E6 cells.
Fig. 5.
Viability and efficacy of RBV in PUUV-infected podocytes. Shown is viability relative to the solvent control, which is set at 100 %. Infection of cells pretreated with RBV for 1 h and infected at an MOI of 0.05 was analyzed by ICW. Percentage of inhibition was calculated relative to infection of control cells incubated with solvent control. Data of three independent experiments were presented as mean ± SD.
Table 1.
Toxic concentration (TC50) of RBV in Vero E6 cells and podocytes and inhibitory concentration (IC50) and selectivity index (SI) of RBV for PUUV infection.
| TC50 (µM) | IC50 (µM) | SI (TC50/IC50) | |
|---|---|---|---|
| Vero E6 | 3308.86 | 49.83 | 66.40 |
| Podocytes | 56.26 | 4.25 | 13.24 |
4. Discussion
Zoonotic diseases such as hantavirus infections bear enormous pathogenic risk for humans. Therefore, identification of effective drugs is an important step in the development of antiviral therapies. The quest for antiviral compounds often begins in cell culture models by quantifying infected cells. In vitro infection assays for hantaviruses mostly rely on the detection of the viral genome via quantitative RT-PCR or N protein expression via immunofluorescence and Western blot.
As observed for HTNV in HUVECs (primary human umbilical vein endothelial cells) and A549 cells (lung carcinoma epithelial cell line), the detection of N protein by in-cell Western assay provides a useful large-scale method for the relative quantification of hantavirus infection (Ma et al., 2017). Using PUUV instead of HTNV as HFRS-causing hantavirus allows testing of anti-hantaviral drugs under BSL-2 conditions. For RBV, an inhibitory effect on the replication of the pathogenic Eurasian hantaviruses HTNV and DOBV in Vero E6 cells was observed in several studies by FFU (focus-forming unit) assay and detection of viral genome (Buys et al., 2011; Mayor et al., 2021; Murphy et al., 2000). The IC50 values of RBV range from 2.65 µM to 72 µM for HTNV and DOBV (Buys et al., 2011; Mayor et al., 2021; Sun et al., 2007). To the best of our knowledge, the antiviral activity of RBV against PUUV has not been tested in vitro, but our results demonstrate that PUUV infection in vitro is reduced by RBV. An IC50 value of 49.83 µM for PUUV in Vero E6 cells is within the range noticed for the inhibition of other HFRS-causing hantaviruses in Vero E6 cells. We also observed an inhibition of PUUV in podocytes. In line with these in vitro results, a decrease in viral load was observed in patients with HFRS caused by PUUV after treatment with RBV (Steininger et al., 2023; Vetter et al., 2021). In contrast, another study could not identify reduction of viral load by RBV treatment (Malinin and Platonov, 2017) and the reasons for these conflicting results remain speculative. However, our results now demonstrate concentration-dependent and cell-type specific characteristics of RBV in vitro that may contribute to the ineffectiveness of RBV in certain contexts in vivo to eradicate the virus. This is of particular importance since hantavirus infection is known to manifest in different organs and in certain organ-specific cell types in the lung or kidneys of patients with HCPS or HFRS, respectively. Thus, our results for varying TC50 and IC50 concentrations for RBV in Vero E6 cells and podocytes are of high clinical interest as they indicate that drug target levels differ in terms of toxicity and antiviral efficacy in a cell type-specific manner.
The antiviral effect of RBV relies on different mechanisms and for hantavirus the inhibition by lowering levels of intracellular levels of GTP via inhibition of IMPDH (inosine-5′-monophosphate dehydrogenase), by interaction with viral polymerase as well as by enhancement of viral mutagenesis has been described (Chung et al., 2007; Severson et al., 2003; Sun et al., 2007). It may be possible that these mechanisms exert cell type specific differences. In addition, uptake of RBV is complex and varies between cell types due to expression of nucleoside transporter proteins which mediate intracellular uptake of RBV (Ibarra and Pfeiffer, 2009; Jarvis et al., 1998; Podgorska et al., 2005). For example, podocytes express transporter proteins allowing a concentration gradient-independent transport of RBV into the cells (Nakajo et al., 2007). Cell-type specific differences in the efficacy of RBV with higher IC50 concentration for Vero cells were also observed for orthopoxviruses, vesicular stomatitis virus and Sendai virus (Shah et al., 2010; Smee et al., 2001). Vero cells show a lower intracellular accumulation of RBV compared to other cell types resulting in partial resistance to antiviral activity of RBV (Shah et al., 2010).
These data, together with our findings, underline, that the choice of an appropriate cell culture model in antiviral research is of special importance.
Further, to identify new broad-range anti-hantaviral drugs, different pathogenic members have to be tested and other pathogenic hantavirus besides HTNV and PUUV should be integrated. In this regard, the ICW assay is a suitable method to gain a better understanding of cell type-specific infection rates and efficacy of drugs. Compared to determination of infection by immunofluorescence assay and manual counting, ICW assay allows greater throughput and is time and cost saving. The list of orthohantaviruses is growing and newly identified members may pose a pathogenic risk to humans in the future. Orthohantaviruses share high homology but differ enormously in their pathogenicity and the replicative fitness of viruses plays often a key role in the clinical course. Viral load and disease severity correlate in hantavirus infections (Korva et al., 2013; Tuiskunen Bäck et al., 2022; Yi et al., 2014). Thus, the ICW assay has large clinical implications since it represents an uncomplicated and fast method to test and identify antiviral drug candidates which may help to prevent severe disease courses.
5. Conclusions
We quantified relative infection of Vero E6 cells and podocytes infected with PUUV by ICW assay. We could show that results obtained by staining for N protein and counting of infected cells correspond to the results of the ICW assay. Further, we identified cell-specific drug level targets for RBV with potential context-dependent clinical implications. These findings may be transferable to other antiviral drugs and may assist in fighting against zoonoses in the future since the ICW assay is a suitable high-throughput method for the analysis of viral infectivity.
Together, the analysis of hantavirus replication, pathogenesis, and the search for specific anti-hantaviral drugs will be facilitated by the large-scale determination of relative infectivity by ICW assay.
Funding
This project was partially funded by grants from the German Center for Infection Research (DZIF), (TTU 01.806 Broad-spectrum Antivirals). The sponsor is not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
CRediT authorship contribution statement
Christian Nusshag: Methodology, Validation, Formal analysis, Writing – review & editing. Pamela Schreiber: Writing – review & editing, Visualization. Josephine Uhrig: Investigation, Visualization. Martin Zeier: Writing – review & editing, Project administration. Ellen Krautkrämer: Methodology, Investigation, Formal analysis, Writing – original draft, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Vanessa Bollinger for excellent technical assistance. Stefan Hägele and Alexander Müller for initially measurement of viability and inhibitory concentrations.
Data availability
Data will be made available on request.
References
- Antonen J., Leppanen I., Tenhunen J., Arvola P., Makela S., Vaheri A., Mustonen J. A severe case of Puumala hantavirus infection successfully treated with bradykinin receptor antagonist icatibant. Scand. J. Infect. Dis. 2013;45(6):494–496. doi: 10.3109/00365548.2012.755268. [DOI] [PubMed] [Google Scholar]
- Bourquain D., Bodenstein C., Schurer S., Schaade L. Puumala and tula virus differ in replication kinetics and innate immune stimulation in human endothelial cells and macrophages. Viruses. 2019;11(9) doi: 10.3390/v11090855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buys K.K., Jung K.H., Smee D.F., Furuta Y., Gowen B.B. Maporal virus as a surrogate for pathogenic New World hantaviruses and its inhibition by favipiravir. Antivir. Chem. Chemother. 2011;21(5):193–200. doi: 10.3851/IMP1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D.H., Sun Y., Parker W.B., Arterburn J.B., Bartolucci A., Jonsson C.B. Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus. J. Virol. 2007;81(21):11722–11729. doi: 10.1128/JVI.00874-07. doi JVI.00874-07 [pii]0874-07 [pii]10.1128/JVI.00874-07 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny J.M., Morgan M.J. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979;43(1):247–252. doi: 10.1099/0022-1317-43-1-247. doi. [DOI] [PubMed] [Google Scholar]
- Hägele S., Müller A., Nusshag C., Reiser J., Zeier M., Krautkrämer E. Motility of human renal cells is disturbed by infection with pathogenic hantaviruses. BMC Infect. Dis. 2018;18(1):645. doi: 10.1186/s12879-018-3583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägele S., Müller A., Nusshag C., Reiser J., Zeier M., Krautkrämer E. Virus- and cell type-specific effects in orthohantavirus infection. Virus Res. 2019;260:102–113. doi: 10.1016/j.virusres.2018.11.015. [DOI] [PubMed] [Google Scholar]
- Huggins J.W., Hsiang C.M., Cosgriff T.M., Guang M.Y., Smith J.I., Wu Z.O., LeDuc J.W., Zheng Z.M., Meegan J.M., Wang Q.N., et al. Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J. Infect. Dis. 1991;164(6):1119–1127. doi: 10.1093/infdis/164.6.1119. doi. [DOI] [PubMed] [Google Scholar]
- Ibarra K.D., Pfeiffer J.K. Reduced ribavirin antiviral efficacy via nucleoside transporter-mediated drug resistance. J. Virol. 2009;83(9):4538–4547. doi: 10.1128/JVI.02280-08. doi JVI.02280-08 [pii]2280-08 [pii]10.1128/JVI.02280-08 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis S.M., Thorn J.A., Glue P. Ribavirin uptake by human erythrocytes and the involvement of nitrobenzylthioinosine-sensitive (es)-nucleoside transporters. Br. J. Pharmacol. 1998;123(8):1587–1592. doi: 10.1038/sj.bjp.0701775. doi 0701775 [pii]10.1038/sj.bjp.0701775 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B., Avsic-Zupanc T., Clement J., Dzagurova T.K., Henttonen H., Heyman P., Jakab F., Krüger D.H., Maes P., Papa A., Tkachenko E.A., Ulrich R.G., Vapalahti O., Vaheri A. Complex evolution and epidemiology of Dobrava-Belgrade hantavirus: definition of genotypes and their characteristics. Arch. Virol. 2013;158(3):521–529. doi: 10.1007/s00705-012-1514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler F.C., Di Cristanziano V., Spath M.R., Hoyer-Allo K.J.R., Wanken M., Muller R.U., Burst V. The kidney in hantavirus infection-epidemiology, virology, pathophysiology, clinical presentation, diagnosis and management. Clin. Kidney J. 2022;15(7):1231–1252. doi: 10.1093/ckj/sfac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korva M., Saksida A., Kejzar N., Schmaljohn C., Avsic-Zupanc T. Viral load and immune response dynamics in patients with haemorrhagic fever with renal syndrome. Clin. Microbiol. Infect. 2013;19(8):E358–E366. doi: 10.1111/1469-0691.12218. [DOI] [PubMed] [Google Scholar]
- Krautkrämer E., Grouls S., Stein N., Reiser J., Zeier M. Pathogenic old world hantaviruses infect renal glomerular and tubular cells and induce disassembling of cell-to-cell contacts. J. Virol. 2011;85(19):9811–9823. doi: 10.1128/JVI.00568-11. doi JVI.00568-11 [pii]10.1128/JVI.00568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkrämer E., Zeier M., Plyusnin A. Hantavirus infection: an emerging infectious disease causing acute renal failure. Kidney Int. 2013;83(1):23–27. doi: 10.1038/ki.2012.360. [DOI] [PubMed] [Google Scholar]
- Laine O., Leppanen I., Koskela S., Antonen J., Makela S., Sinisalo M., Vaheri A., Mustonen J. Severe Puumala virus infection in a patient with a lymphoproliferative disease treated with icatibant. Infect. Dis. 2015;47(2):107–111. doi: 10.3109/00365548.2014.969304. [DOI] [PubMed] [Google Scholar]
- Ma H.W., Ye W., Chen H.S., Nie T.J., Cheng L.F., Zhang L., Han P.J., Wu X.A., Xu Z.K., Lei Y.F., Zhang F.L. In-cell western assays to evaluate hantaan virus replication as a novel approach to screen antiviral molecules and detect neutralizing antibody titers. Front. Cell. Infect. Microbiol. 2017;7:269. doi: 10.3389/fcimb.2017.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinin O.V., Platonov A.E. Insufficient efficacy and safety of intravenous ribavirin in treatment of haemorrhagic fever with renal syndrome caused by Puumala virus. Infect. Dis. 2017;49(7):514–520. doi: 10.1080/23744235.2017.1293841. [DOI] [PubMed] [Google Scholar]
- Mayor J., Engler O., Rothenberger S. Antiviral efficacy of ribavirin and favipiravir against hantaan virus. Microorganisms. 2021;9(6) doi: 10.3390/microorganisms9061306. doi microorganisms9061306 [pii]microorganisms-09-01306 [pii]10.3390/microorganisms9061306 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.E., Kariwa H., Mizutani T., Yoshimatsu K., Arikawa J., Takashima I. I antiviral activity of lactoferrin and ribavirin upon hantavirus. Arch. Virol. 2000;145(8):1571–1582. doi: 10.1007/s007050070077. [doi] [DOI] [PubMed] [Google Scholar]
- Nakajo A., Khoshnoodi J., Takenaka H., Hagiwara E., Watanabe T., Kawakami H., Kurayama R., Sekine Y., Bessho F., Takahashi S., Swiatecka-Urban A., Tryggvason K., Yan K. Mizoribine corrects defective nephrin biogenesis by restoring intracellular energy balance. J. Am. Soc. Nephrol. 2007;18(9):2554–2564. doi: 10.1681/ASN.2006070732. doi ASN.2006070732 [pii]10.1681/ASN.2006070732 [doi] [DOI] [PubMed] [Google Scholar]
- Nusshag C., Boegelein L., Schreiber P., Essbauer S., Osberghaus A., Zeier M., Krautkrämer E. Expression profile of human renal mesangial cells is altered by infection with pathogenic puumala orthohantavirus. Viruses. 2022;14(4) doi: 10.3390/v14040823. doi v14040823 [pii]viruses-14-00823 [pii]10.3390/v14040823 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusshag C., Stütz A., Hägele S., Speer C., Kälble F., Eckert C., Brenner T., Weigand M.A., Morath C., Reiser J., Zeier M., Krautkrämer E. Glomerular filtration barrier dysfunction in a self-limiting, RNA virus-induced glomerulopathy resembles findings in idiopathic nephrotic syndromes. Sci. Rep. 2020;10(1):19117. doi: 10.1038/s41598-020-76050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perley C.C., Brocato R.L., Kwilas S.A., Daye S., Moreau A., Nichols D.K., Wetzel K.S., Shamblin J., Hooper J.W. Three asymptomatic animal infection models of hemorrhagic fever with renal syndrome caused by hantaviruses. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0216700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgorska M., Kocbuch K., Pawelczyk T. Recent advances in studies on biochemical and structural properties of equilibrative and concentrative nucleoside transporters. Acta Biochim. Pol. 2005;52(4):749–758. doi 20051165 [pii] [PubMed] [Google Scholar]
- Rusnak J.M., Byrne W.R., Chung K.N., Gibbs P.H., Kim T.T., Boudreau E.F., Cosgriff T., Pittman P., Kim K.Y., Erlichman M.S., Rezvani D.F., Huggins J.W. Experience with intravenous ribavirin in the treatment of hemorrhagic fever with renal syndrome in Korea. Antiviral Res. 2009;81(1):68–76. doi: 10.1016/j.antiviral.2008.09.007. doi S0166-3542(08)00421-X [pii]10.1016/j.antiviral.2008.09.007 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M.A., O'Hare M.J., Reiser J., Coward R.J., Inward C.D., Farren T., Xing C.Y., Ni L., Mathieson P.W., Mundel P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002;13(3):630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- Sanada T., Kariwa H., Nagata N., Tanikawa Y., Seto T., Yoshimatsu K., Arikawa J., Yoshii K., Takashima I. Puumala virus infection in Syrian hamsters (Mesocricetus auratus) resembling hantavirus infection in natural rodent hosts. Virus Res. 2011;160(1–2):108–119. doi: 10.1016/j.virusres.2011.05.021. doi S0168-1702(11)00221-8[pii]10.1016/j.virusres.2011.05.021. [DOI] [PubMed] [Google Scholar]
- Severson W.E., Schmaljohn C.S., Javadian A., Jonsson C.B. Ribavirin causes error catastrophe during hantaan virus replication. J. Virol. 2003;77(1):481–488. doi: 10.1128/JVI.77.1.481-488.2003. doi 1381 [pii]10.1128/jvi.77.1.481-488.2003 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N.R., Sunderland A., Grdzelishvili V.Z. Cell type mediated resistance of vesicular stomatitis virus and Sendai virus to ribavirin. PLoS One. 2010;5(6):e11265. doi: 10.1371/journal.pone.0011265. doi 10-PONE-RA-16972R1 [pii]10.1371/journal.pone.0011265 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smee D.F., Bray M., Huggins J.W. Antiviral activity and mode of action studies of ribavirin and mycophenolic acid against orthopoxviruses in vitro. Antivir. Chem. Chemother. 2001;12(6):327–335. doi: 10.1177/095632020101200602. [doi] [DOI] [PubMed] [Google Scholar]
- Steininger P., Herbst L., Bihlmaier K., Willam C., Körper S., Schrezenmeier H., Klüter H., Pfister F., Amann K., Weiss S., Krüger D.H., Zimmermann R., Korn K., Hofmann J., Harrer T. Fatal puumala hantavirus infection in a patient with common variable immunodeficiency (CVID) Microorganisms. 2023;11(2) doi: 10.3390/microorganisms11020283. doi microorganisms11020283 [pii]microorganisms-11-00283 [pii]10.3390/microorganisms11020283 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Chung D.H., Chu Y.K., Jonsson C.B., Parker W.B. Activity of ribavirin against Hantaan virus correlates with production of ribavirin-5′-triphosphate, not with inhibition of IMP dehydrogenase. Antimicrob. Agents Chemother. 2007;51(1):84–88. doi: 10.1128/AAC.00790-06. doi AAC.00790-06 [pii]0790-06 [pii]10.1128/AAC.00790-06 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temonen M., Vapalahti O., Holthofer H., Brummer-Korvenkontio M., Vaheri A., Lankinen H. Susceptibility of human cells to Puumala virus infection. J. Gen. Virol. 1993;74(Pt 3):515–518. doi: 10.1099/0022-1317-74-3-515. [DOI] [PubMed] [Google Scholar]
- Tuiskunen Bäck A., Rasmuson J., Thunberg T., Rankin G., Wigren Byström J., Andersson C., Sjödin A., Forsell M., Ahlm C. Clinical and genomic characterisation of a fatal Puumala orthohantavirus case with low levels of neutralising antibodies. Infect. Dis. 2022;54(10):766–772. doi: 10.1080/23744235.2022.2076904. (Lond)[doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaheri A., Strandin T., Hepojoki J., Sironen T., Henttonen H., Makela S., Mustonen J. Uncovering the mysteries of hantavirus infections. Nat. Rev. Microbiol. 2013;11(8):539–550. doi: 10.1038/nrmicro3066. doi. [DOI] [PubMed] [Google Scholar]
- Vetter P., L'Huillier A.G., Montalbano M.F., Pigny F., Eckerle I., Torriani G., Rothenberger S., Laubscher F., Cordey S., Kaiser L., Schibler M. Puumala virus infection in family, Switzerland. Emerg. Infect. Dis. 2021;27(2):658–660. doi: 10.3201/eid2702.203770. doi 20-3770[pii]10.3201/eid2702.203770 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Zhang Y., Zhang Y., Ma Y., Zhang C., Li Q., Liu B., Liu Z., Liu J., Zhang X., Zhuang R., Jin B. Increased plasma cell-free DNA level during HTNV infection: correlation with disease severity and virus load. Viruses. 2014;6(7):2723–2734. doi: 10.3390/v6072723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [doi] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.