Abstract
Porcine reproductive and respiratory syndrome (PRRS) is one of the most significant diseases affecting the pig industry worldwide. The PRRSV mutation rate is the highest among the RNA viruses. To date, NADC30-like PRRSV and highly pathogenic PRRSV (HP-PRRSV) are the dominant epidemic strains in China; however, commercial vaccines do not always provide sufficient cross-protection, and the reasons for insufficient protection are unclear. This study isolated a wild-type NADC30-like PRRSV, SX-YL1806, from Shaanxi Province. Vaccination challenge experiments in piglets showed that commercial modified live virus (MLV) vaccines provided good protection against HP-PRRSV. However, it could not provide sufficient protection against the novel strain SX-YL1806. To explore the reasons for this phenomenon, we compared the genomic homology between the MLV strain and HP-PRRSV or NADC30-like PRRSV and found that the MLV strain had a lower genome similarity with NADC30-like PRRSV. Serum neutralization assay showed that MLV-immune serum slightly promoted the homologous HP-PRRSV replication and significantly promoted the heterologous NADC30-like PRRSV strain replication in vitro, suggesting that antibody-dependent enhancement (ADE) might also play a role in decreasing MLV protective efficacy. These findings expand our understanding of the potential factors affecting the protective effect of PRRSV MLV vaccines against the NADC30-like strains.
Keywords: NADC30-like PRRSV, Modified live virus (MLV) vaccines, Genomic similarity, Antibody dependent enhancement (ADE), Cross protection
Highlights
-
•
The SX-YL1806 isolate is a NADC30-like PRRSV that is the dominant strain in China.
-
•
MLV vaccine provides insufficient protection against heterologous NADC30-like PRRSV.
-
•
MLV vaccine immune serum significantly promoted the heterologous NADC30-like PRRSV isolate replication in vitro.
-
•
Genomic homology and ADE of immunized serum are potential factors affecting the cross-protection of PRRSV MLV vaccines.
1. Introduction
Porcine reproductive and respiratory syndrome (PRRS) is one of the most relevant viral diseases in swine-producing countries. Since its outbreak in the 1980s, PRRS has caused enormous economic damage to the global swine industry (Wensvoort et al., 1991; Cavanagh, 1997). Porcine reproductive and respiratory syndrome virus (PRRSV) is a small, enveloped, single-stranded positive-sense RNA virus. PRRSV was newly classified into the genus Porartevirus, family Arteriviridae, and order Nidovirales according to the latest classification (King et al., 2018; Guo et al., 2021; Zhu et al., 2022). PRRSV is divided into two major genotypes based on genetic and antigenic analyses: PRRSV-1 (genotype 1, Europe) and PRRSV-2 (genotype 2, North America) (Darwich et al., 2011; Lunney et al., 2016). In 2006, a highly pathogenic PRRSV (HP-PRRSV) outbreak occurred in the Chinese mainland characterized by high fever (40–42 °C), high morbidity (50%–100%), and high mortality (20%–100%) (Tian et al., 2007, 2009; Zhou et al., 2008; Song et al., 2012). Since 2013, the NADC30-like PRRSV that likely originated from the NADC30 PRRSV strain circulating in North America around 2008, has been isolated and widely spread in China (Zhao et al., 2015; Zhou et al., 2015; Wang et al., 2017). Although the pathogenicity of NADC30-like PRRSV is lower than that of HP-PRRSV, NADC30-like PRRSV has high variability and is prone to recombination with other PRRSV to form new PRRSV strains with complex genetic backgrounds and genomic characteristics (Zhang et al., 2016; Zhou et al., 2018; Chen P. et al., 2021; Guo et al., 2021; Li Y. et al., 2021). Currently, NADC30-like PRRSV and HP-PRRSV are the dominant strains in China (Li et al., 2016; Guo et al., 2019; Li L. et al., 2021; Zhang et al., 2022).
Vaccine immunization is the primary option for controlling and preventing PRRSV infections. Generally, modified live virus (MLV) vaccines provide better protection than inactivated PRRSV vaccines (Huang and Meng, 2010; Han et al., 2017). However, because of the rapid mutation and extensive recombination of PRRSV, some commercial PRRSV MLV vaccines confer great protection against homologous strains but only limited cross-protection against heterologous PRRSV strains, such as NADC30-like PRRSV (Tian et al., 2007; Bian et al., 2017; Zhao et al., 2017; Sun et al., 2018; Rupasinghe et al., 2022). Although many studies have explored the protective effects of PRRSV MLV vaccines, the reasons for their inadequate protection against NADC30-like PRRSV remain unclear.
Antibody-dependent enhancement (ADE) is the phenomenon in which the presence of poorly neutralizing antibodies or sub-neutralizing concentrations of antibodies does not inhibit the virus but promotes its entry and replication. Some viral infections can induce ADE, such as dengue fever virus (DENV), severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome (MERS) (Wan et al., 2020; Xu et al., 2021). Interestingly, most viruses infect immune cells, particularly macrophages. The PRRSV primarily infects porcine alveolar macrophages (PAMs). When antibodies with sub-neutralizing concentrations are present, PRRSV can enhance the infection of PAMs through ADE in vitro. In addition, the viremia increases after serum transfer in vivo, and serum enhances heterologous strain infection in a strain-dependent manner (Yoon et al., 1996, 1997). Vaccine-induced enhancement has been identified as a major obstacle in developing flavivirus, coronavirus, paramyxovirus, and lentivirus vaccines (Xu et al., 2021; Soraci et al., 2022). Owing to ADE, judging whether the antibody produced after vaccine immunization is beneficial or harmful for resisting viral infection is difficult. Pigs infected with PRRSV exhibit a strong and rapid humoral response; however, the protective effect of these antibodies may change with disease development. Significant levels of non-neutralizing antibodies, particularly during the early stages of infection, may promote ADE, which can potentially make it difficult for vaccine immunization to achieve the desired effect.
In this study, we isolated a novel wild-type PRRSV strain, SX-YL1806, which belongs to the current dominant NADC30-like PRRSV in China. We found that the commercial MLV vaccine provided good protection against HP-PRRSV but could not provide sufficient protection against the novel strain SX-YL1806. To explore the potential factors affecting the poor protective effect of the MLV vaccine against NADC30-like PRRSV, we compared the genomic similarity of the MLV strain with HP-PRRSV or SX-YL1806 and evaluated whether the immune serum promotes ADE on HP-PRRSV or SX-YL1806 replication. The results showed that both genomic homology and ADE of immunized sera are potential factors affecting the cross-protection of PRRSV MLV vaccines. These findings may provide new explanations for the poor protection provided by MLV vaccines against NADC30-like PRRSV.
2. Materials and methods
2.1. Cells, viruses, and antibodies
The PRRSV strains used in this study were all stored in our laboratory and included the following: PRRSV-1 GZ11-G1 (KF001144); HP-PRRSV SD-YL1712 (MT708500); NADC30-like PRRSV SX-YL1806 (isolated in this study, OR208175); classical PRRSV VR2332 (EF536003); and CH1a (AY032626). The commercial MLV vaccine used in this study was stored in our laboratory. The African green monkey kidney cell line MARC-145 and porcine alveolar macrophage cell line CRL2843-CD163 were maintained in Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher, USA) or RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA). PAMs were obtained from 6-week-old healthy piglets, as previously described (Xiao et al., 2014), and cultured in RPMI-1640 medium supplemented with 10% FBS. A monoclonal antibody (6D10) against the PRRSV N protein was prepared and stored in our laboratory and used as the primary antibody in the indirect immunofluorescence (IFA), Western blotting (WB), and immunohistochemistry (IHC) assays. An anti-mouse IgG antibody labeled with Alexa Fluor 488 was purchased from Abways Technology (Shanghai, China).
2.2. Isolation of wild-type NADC30-like PRRSV
Lung tissues were obtained from a pig farm in Shaanxi Province, China, in 2018. Pigs on this farm displayed high fever, dyspnea, and lethargy. All lung tissues were tested using reverse transcriptase polymerase chain reaction (RT-PCR) for related pathogens, including classical swine fever virus (CSFV), porcine circovirus type 2 (PCV2), pseudorabies virus (PRV), porcine epidemic diarrhea virus (PEDV), and PRRSV. PRRSV-positive lung tissues were ground and homogenized in phosphate-buffered saline (PBS; Solarbio, China) and subjected to three freeze-thaw cycles. The supernatant was harvested after filtration through a 0.22-μm filter. The supernatant was added to the PAMs for 2 h, and the cells were cultured in RPMI-1640 medium containing 10% FBS. Subsequently, monitoring was performed daily until a cytopathic effect (CPE) was observed. IFA and WB assays were performed as described previously with minor modifications to confirm the presence of the isolated PRRSV (Feng et al., 2022). The HP-PRRSV SD-YL1712 strain was used as a positive control.
IFA: Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.25% Triton X-100 in PBS for 10 min. After blocking nonspecific binding by incubating the cells with 1% BSA in PBS for 30 min, the cells were incubated with an anti-PRRSV N monoclonal antibody (6D10), sequentially with the corresponding secondary antibodies and then visualized with a fluorescence microscope.
Western blotting: Cells were harvested and lysed in radioimmunoprecipitation assay (RIPA, Solarbio) containing protease inhibitors. The lysates were separated using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% nonfat milk in PBST for 1 h at 25 °C, and then incubated with the 6D10 at 4 °C overnight. The membranes were washed with PBST and then incubated with a secondary antibody for 1 h at 25 °C. After washing, the target proteins were detected using the enhanced chemiluminescence (ECL) kit (Beyotime Biotechnology, Shanghai, China).
2.3. Complete genome analysis
Total RNA was extracted from the isolated PRRSV SX-YL1806, as described previously (Zheng et al., 2022). A Reverse Transcription kit (Vazyme, Nanjing, China) was used for RT-PCR. The complete genome of SX-YL1806 was divided into 13 overlapping fragments and amplified by PCR using the PrimeSTAR GXL Premix (TAKARA, Dalian, China) (Supplementary Table S1). The 5′-untranslated region (UTR) and 3′UTR of SX-YL1806 were amplified by using the SMART 5′RACE and 3′RACE kits (TAKARA, Dalian, China). Purified PCR products were cloned into pMD-19T vectors (TAKARA, Dalian, China) and sequenced by a commercial service provider (Tsingke Biotechnology Co., Ltd., Beijing, China). Subsequently, the complete genome of the isolated SX-YL1806 was obtained by assembling the overlapping sequences.
Thirty-six representative PRRSV strains were collected from the GenBank database (Supplementary Table S2), and the complete genome of SX-YL1806 was aligned using the ClustalX software. Phylogenetic trees were constructed using MEGA 7.0 software based on the NSP2, ORF5, and the complete genome of SX-YL1806, respectively. Bootstrap confidence values from 1000 replicates were used to analyze the evolutionary relationship of SX-YL1806.
To evaluate the potential recombination events in SX-YL1806, Simplot v.3.5.1 (200-bp sliding window, 20-bp step size) and RDP4.0 (RDP, GENECONV, BootScan, MaxChi, Chimaera, Siscan, and Phyloro) software were used. Subsequently, the flanking regions near the recombination breakpoints were amplified and re-sequenced to identify the natural recombination.
To compare the genomic similarity between the MLV and challenged strains, the complete genome of the MLV strain was sequenced. Each gene of the MLV strain was compared with those of HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 using MEGA and MegAlign software. The N-linked glycosylation sites of GP5 were predicted and analyzed online (http://www.cbs.dtu.dk/services/NetNGlyc/). The GP5 protein structures of the MLV strain, HP-PRRSV SD-YL1712, and NADC30-like PRRSV SX-YL1806 were predicted using Alphafold v2.3.2 software (parameters: default, uniport and uniref 90:2023–03-01, pdb-mmcif, and pdb-seqres: 2023-03-03, other database versions: default).
2.4. Animal vaccine and challenge design
Animal experiments were performed to evaluate the protective effect of the PRRSV MLV vaccine against HP-PRRSV and NADC30-like PRRSV. Twenty-three four-week-old healthy piglets (male, commercial breed) free of PRRSV, PRV, PCV2, and CSFV were selected as respiratory disease models. Among them, 20 piglets were randomly divided into four groups (n = 5); the remaining three piglets were treated as negative controls (NC). Pigs in groups NC-SD and NC-SX were injected with DMEM at 0 days post-vaccination (dpv) and inoculated intramuscularly (2 × 105 TCID50/mL) and intranasally (2 × 105 TCID50/mL) with HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806, respectively, at 42 dpv. Pigs in the MLV-SD and MLV-SX groups were intramuscularly vaccinated with a commercial MLV vaccine at a single dose at 0 dpv, and then challenged with SD-YL1712 or SX-YL1806 at 42 dpv. Pigs in the NC group were vaccinated and challenged with DMEM. All groups were fed separately and euthanized at 63 dpv.
2.5. Evaluation of the MLV vaccine immune efficacy against HP-PRRSV or NADC30-like PRRSV in piglets
The behavior of each piglet was monitored daily for unusual clinical situations such as coughing, sneezing, depression, or diarrhea. The rectal temperature of each piglet was measured daily. The growth performance of each piglet was assessed by measuring its body weight weekly (weight gain rate). Serum samples from pigs in each group were collected at 0, 4, 7, 11, 14, 21, 28, 35, 42, 49, 56, and 63 dpv to analyze the virus load and detect antibodies.
To identify the viral load in serum and lung tissues of the MLV vaccine inoculation group, viral RNA was extracted from 300 μL of serum and 1 g of lung tissue and reverse transcribed using a Reverse Transcription kit according to the manufacturer's instructions (Vazyme, Nanjing, China). Viral load was measured using absolute quantitative real-time PCR with primers targeting the ORF7 PRRSV. Standard curves were generated from serial 10-fold dilutions of the plasmid constructed using the PRRSV ORF7 (Li et al., 2022). PRRSV RNA copies were calculated using a standard curve.
The antibodies in the serum were analyzed using the Porcine Reproductive and Respiratory Syndrome Virus AB Elisa kit (JNT, JN60415, China) according to the manufacturer's instructions; the threshold for seroconversion was set at a sample-to-positive (S/P) ratio of 0.4.
All piglets were euthanized for autopsy at 63 dpv, and lung tissues were collected from each piglet. A portion of the lung tissue was used to determine the viral load by RT-qPCR, and another portion was immersed in 4% paraformaldehyde for pathological changes analysis performed by Shaanxi Yike Biotechnology Service Co. Ltd. (China). Macroscopic lesions and scores of the lungs were estimated as previously described, which were based on assigning a number to each lobe to reflect the approximate percentage of the entire lung represented by that lobe (Halbur et al., 1995; Hou et al., 2020). The microscopic lesions and scores of the lung tissues were estimated based on the extent and magnitude of interstitial pneumonia as: 0, no lesion; 1, mild/focal; 2, moderate/multifocal; 3, moderate/diffuse (alveolar wall accounting for greater than 50% of the measured section); and 4, severe/diffuse (alveolar wall accounting for greater than 75% of the measured section). Macroscopic and microscopic lung lesions were evaluated by three pathologists who were blinded to the tests.
2.6. Serum virus neutralization assay (SVN)
Sera collected at 42 dpv were heat-inactivated and subjected to a virus neutralization (SVN) test using a previously described method (Trible et al., 2015). Serum dilutions collected at 42 dpv were incubated with 200 TCID50 of viruses incorporating HP-PRRSV (SD-YL1712), NADC30-like PRRSV (SX-YL1806), classical PRRSV (CH1a, VR2332), and PRRSV-1 (GZ11-G1) in DMEM with 3% FBS for 1 h. Then, the virus-antibody mixture was transferred to a 96-well plate of confluent MARC-145 or CRL2843-CD163 cells and incubated until the CPE was noticed. IFA was performed to identify the CPE induced by PRRSV. The absence of CPE at 1:8 dilution was considered positive for PRRSV neutralization (Trible et al., 2015; Chen N. et al., 2021).
2.7. PRRSV-ADE infection assay in vitro
To study the potential ADE of the PRRSV MLV vaccine immune serum in vitro, an ADE assay was conducted, as described previously, with minor modifications (Wan et al., 2019). Briefly, serum from each piglet in the MLV-SD, MLV-SX, and NC groups was collected at 14, 21, 28, 35, and 42 dpv. Anti-PRRSV antibody-positive serum was heat-inactivated at 56 °C for 30 min and diluted at a series dilution ratio of 1:50 to 1:800 in DMEM. Serum collected from group NC was used as a negative control. Then, the diluted serum was incubated with an equal volume of HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 (104 TCID50/mL) for 1 h at 37 °C. CRL2843-CD163 cells were seeded into 24-well plates at a density of 5 × 104 cells/well 24 h before infection. The supernatant was removed, and cells were washed twice with PBS. Subsequently, 500 μL of virus-antibody mixture were inoculated in triplicate onto CRL2843-CD163 cells. Then, the 24-well plates were incubated at 37 °C for 2 h, supernatant was removed, cells were washed with PBS twice, and fresh DMEM supplemented with 10% FBS was added. The cells were harvested for RT-qPCR 48 h after the incubation.
RT-qPCR was performed as described previously with minor modifications (Zheng et al., 2022). Total RNA was isolated using the TRIzol reagent and reverse transcribed using a PrimeScript RT reagent kit (Vazyme, Nanjing, China) according to the manufacturer's instructions. RT-qPCR was performed using the ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China). The relative expression levels were calculated by the 2−ΔΔCT method. HPRT1 was used as an internal reference in CRL2843-CD163 cells.
Staphylococcal Protein A (Beyotime Biotechnology) was resuspended in sterile distilled deionized water at a concentration of 1 mg/mL. Parallel ADE assays were conducted on CRL2843-CD163 cells using a virus-antibody mixture in the presence or absence of Protein A, as previously described with slight modifications (Joseph et al., 2003). Briefly, Protein A was diluted and incubated with the MLV immune sera (sera collected at 42 dpv, 1:50 diluted) at 37 °C for 1 h, then PRRSV was added and incubated at 37 °C for a further 1 h. CRL2843-CD163 cells were infected with the PRRSV-serum complex or PRRSV-serum-protein A complex for 48 h. The infection rates of PRRSV with ADE were confirmed using IFA or WB, as described previously (Feng et al., 2022). PEDV-positive pig serum was used as an independent control.
2.8. Statistical analysis
Data analysis was performed using GraphPad Prism 6 software (La Jolla, CA, USA) with one- or two-way analysis of variance (ANOVA), followed by Tukey's t-test. Accordingly, a P-value <0.05 was considered statistically different.
3. Results
3.1. Isolation and identification of wild-type PRRSV SX-YL1806
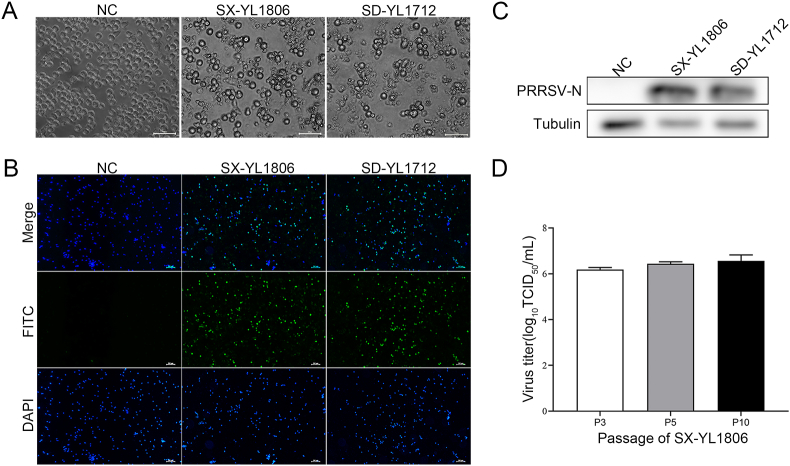
Lung tissues were obtained from a pig farm in Shaanxi Province, China, in 2018. Pigs on this farm exhibited obvious clinical signs, including coughing, sneezing, and anorexia. These samples were tested for relative pathogens, including CSFV, PCV2, PRV, PEDV, and PRRSV, by RT-PCR, and the results showed that only PRRSV was positive (data not shown). The tissue suspension was inoculated onto PAMs to isolate wild-type PRRSV. A representative PRRSV strain, SX-YL1806, was also isolated. Typical CPE induced by PRRSV SX-YL1806, such as cell disintegration, was observed in PAMs 48 h post-infection, similar to the HP-PRRSV strain SD-YL1712 (Fig. 1A). To further investigate the characteristics of PRRSV SX-YL1806, IFA and WB assays were performed. The results showed obvious positive fluorescent signals in PAMs inoculated with the SX-YL1806 and SD-YL1712 strains (Fig. 1B). PRRSV-N was detected in PAMs inoculated with PRRSV SX-YL1806 and SD-YL1712 by WB blotting on day 2 post-infection (Fig. 1C). The viral titer of SX-YL1806 from different passages was measured in PAMs. No difference in viral titers among the third, fifth, and tenth passages of SX-YL1806 was found, with the highest titer reaching 106.4 TCID50/mL (Fig. 1D). These results demonstrated that wild-type PRRSV SX-YL1806 was successfully isolated from clinical samples. Cultures from the fifth passage (SX-YL1806) were collected and subjected to genome sequencing.
Fig. 1.
Identification of the isolated PRRSV SX-YL1806. PRRSV-positive lung tissues were ground and homogenized in PBS, and subjected to three freeze-thaw cycles. The supernatant was harvested after filtration through a 0.22 μm filter. Then the supernatant was added to PAMs for 2 h and cultured in RPMI-1640 medium containing 10% FBS. A Cytopathic effect (CPE) induced by SX-YL1806 in PAMs was observed at day 2 post-infection. Scale bar = 400 μm. B The immunofluorescence assay (IFA) was used to detect the presence of SX-YL1806 in PAMs. At day 2 post-infection, PAMs infected with SX-YL1806 were incubated with an anti-PRRSV N monoclonal antibody (6D10) and then sequentially with corresponding secondary antibodies. Scale bar = 200 μm. C The presence of SX-YL1806 was identified by Western blotting assay in PAMs. PAMs were harvested at day 2 post-infection and lysed in RIPA containing protease inhibitor. The lysates were separated by 12% SDS-PAGE gels and transferred onto PVDF membranes. The 6D10 was used as the corresponding antibody. HP-PRRSV SD-YL1712 was treated as a positive control. D The viral titer was determined at different passages of SX-YL1806 with PAMs. The different passages of PRRSV SX-YL1806 were inoculated into PAMs at an MOI = 0.01, and cultured for 3 days. Then the viral titers were calculated according to the Manbdcs method. Data were shown as mean ± SD.
3.2. Complete genome analysis of NADC30-like PRRSV SX-YL1806
The complete genome of isolated PRRSV SX-YL1806 was 15,019 nt, excluding the poly(A) tail at the 3′ end, which contains a 190-nt 5′-UTR and a 151-nt 3′-UTR. Complete genome alignments revealed that SX-YL1806 shared 84.3% nucleotide sequence identity with JXA1 and HB-1(sh)-2002 (lineage 8), 85.4% with VR-2332 (lineage 5), 82.6% with QYYZ and GM2 (lineage 3), 84%–84.7% with LNWK130 and JS2021NADC34 (sublineage 1.5), and 89.6%–94.2% with JL580 and NADC30 (sublineage 1.8) (Table 1). Next, the homology of the nucleotide sequences of NSP2 and ORF5 between SX-YL1806 and other representative PRRSV strains was analyzed. The results revealed that NSP2 and ORF5 of SX-YL1806 shared the highest nucleotide homology with the NADC30-like PRRSV (sublineage 1.8) (Table 1). These data indicated that the isolated SX-YL1806 strain was classified as an NADC30-like PRRSV.
Table 1.
Nucleotide sequence identities of SX-YL1806 as compared to representative PRRSV strains.
| SX-YL1806 | Lineage 1 |
Lineage 8 |
Lineage 3 |
Lineage 5 |
||||
|---|---|---|---|---|---|---|---|---|
| Sublineage 1.5 |
Sublineage 1.8 |
Sublineage 8.1 |
Sublineage 8.3 |
Sublineage 8.5 |
||||
| LNWK130/JS2021NADC34 | NADC30/JL580 | CH1a | JXA1 | HB-1 (sh)-2002 | QYYZ | GM2 | VR-2332 | |
| Complete genome | 84.7/84 | 94.2/89.6 | 84.8 | 84.3 | 84.3 | 82.6 | 82.6 | 85.4 |
| Nsp2 | 76.7/75.8 | 92.6/85.2 | 75.8 | 75.5 | 75.7 | 73.4 | 73.2 | 77.8 |
| ORF5 | 87.7/86.6 | 95.4/92.7 | 87.1 | 86.1 | 85.6 | 83.4 | 83.1 | 87.2 |
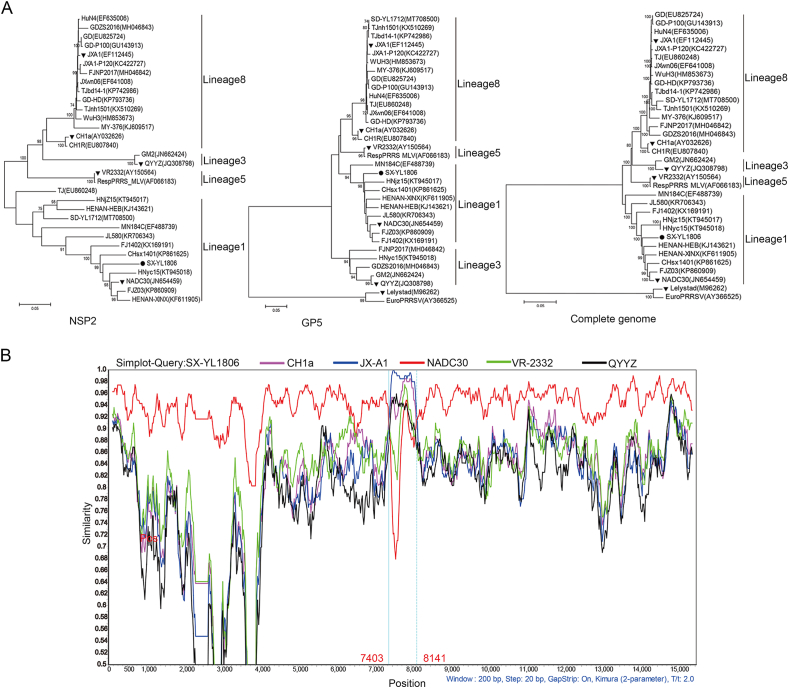
To analyze the genetic evolutionary relationship between SX-YL1806 and other references, phylogenetic trees were constructed based on the NSP2, ORF5, and the complete genome of SX-YL1806. All three phylogenetic trees showed that the isolated PRRSV SX-YL1806 was classified into lineage 1, which contained most NADC30-like PRRSV strains (Fig. 2A).
Fig. 2.
Complete genome alignment of the isolated SX-YL1806 strain. A Phylogenetic trees were constructed by MEGA 7.0 software based on the NSP2, ORF5, and complete genome of SX-YL1806, respectively. Bootstrap confidence values from 1000 replicates were used to analyze the evolutionary relationship of SX-YL1806. SX-YL1806 was highlighted with a black circle (●), while other representative strains from different lineages were marked with black triangles (▼). B Recombination analysis of SX-YL1806 were performed with Simplot v.3.5.1 software (200-bp sliding window, 20-bp step size). Five endemic clusters of circulating strains in China, CH1a-like (CH1a, AY032626), JXA1-like (JXA1, EF112445), NADC30-like (NADC30, JN654459), VR-2332-like (VR-2332, AY150564), and QYYZ-like (QYYZ, JQ308798), were chosen as representative PRRSV strains. The Y-axis showed the similarity between the SX-YL1806 and representative strains, while the X-axis indicated the genome position of the SX-YL1806.
Simplot software was used to investigate possible recombination events in the SX-YL1806 isolates. Five endemic clusters of strains circulating in China, CH1a-like (CH1a, AY032626), JXA1-like (JXA1, EF112445), NADC30-like (NADC30, JN654459), VR-2332-like (VR-2332, AY150564), and QYYZ-like (QYYZ, JQ308798), were selected as representative PRRSV strains. Strikingly, two potential recombination breakpoints were detected in the whole genomic sequence alignment of SX-YL1806, which were located at nt 7403 and nt 8141 and divided the genome of SX-YL1806 into three regions. The results showed that nt 7403–8141 was associated with the JXA1-like strain, and the other two regions were closely related to the NADC30-like strain (Fig. 2B). This analysis indicates that the SX-YL1806 isolates may have originated from a recombination event between NADC30-like PRRSV and JXA1-like PRRSV. The RDP software was used to identify the recombination of SX-YL1806, and the results showed that SX-YL1806 was a naturally recombinant virus from the NADC30-like PRRSV (major parent) and HP-PRRSV (minor parent) strains (Table 2).
Table 2.
Recombination events of SX-YL1806 detected by RDP4.0 software.
| Isolate | Breakpoints |
Major Parent | Minor Parent |
P-Value |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beginning | Ending | RDP | GENECONV | Bootscan | MaxChi | Chimera | SiScan | Phylpro | |||
| SX-YL1806 | 1 | 7403 | NADC30 | JXA1 | 6.535 × 10−72 | 3.251 × 10−69 | 2.111 × 10−73 | 1.567 × 10−14 | 4.757 × 10−19 | 1.444 × 10−18 | 3.730 × 10−14 |
| 7404 | 8141 | JXA1 | NADC30 | ||||||||
| 8142 | 15,525 | NADC30 | JXA1 | ||||||||
Collectively, the newly isolated PRRSV SX-YL1806 originated from a recombination event and was clustered with the dominant epidemic NADC30-like PRRSV.
3.3. Clinical manifestations after the MLV inoculation and PRRSV challenge
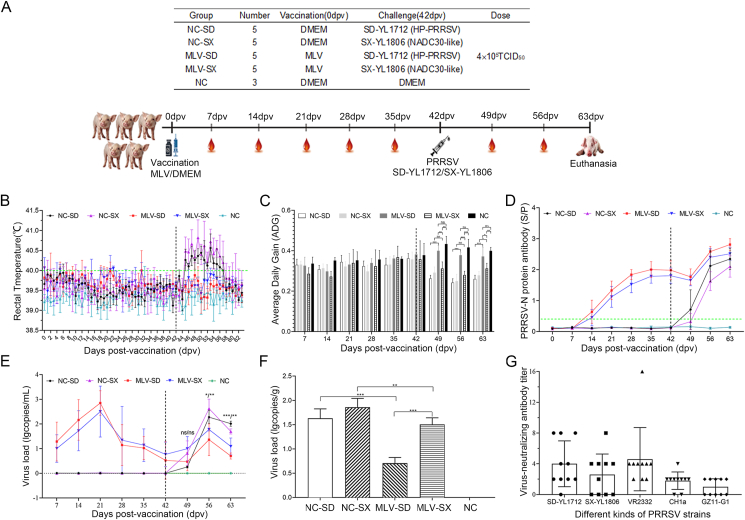
To evaluate the protective effects of the commercial PRRS-MLV vaccine against this novel strain, vaccination and challenge strategies were designed (Fig. 3A). All pigs were carefully monitored until the end of the experiment to observe the clinical signs of PRRS. Groups NC-SD and NC-SX showed obvious clinical signs, such as cough, diarrhea, anorexia, and depression, after being challenged with HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 at 42 dpv. Group NC-SD exhibited a fever since 45 dpv that lasted after 13 days, with the highest temperature reaching 40.56 °C at 53 dpv; the body temperature of group NC-SX started rising at 45 dpv and lasted until 57 dpv, and the peak temperature was 40.82 °C at 49 dpv. Interestingly, the MLV-SD and MLV-SX groups exhibited mild clinical symptoms of PRRS after immunization with MLV at 0 dpv and challenge with HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 at 42 dpv. No obvious clinical signs were observed in the NC group (Fig. 3B).
Fig. 3.
Vaccination and challenge strategies. A At 0 dpv, 4-week-old healthy piglets were inoculated with the commercial MLV-PRRSV vaccine at the indicated dose according to the manufacturer's instructions or with DMEM as a mock-vaccination control. Then, a HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 challenge was performed in the piglets immunized with MLV vaccine or DMEM at 42 dpv. Sera was collected at the indicated time points. All piglets were euthanized at 63 dpv. B Rectal temperature of piglets was recorded daily. C Average daily gain rate of piglets was measured weekly. D PRRSV-N protein special antibodies were determined weekly by a commercial Porcine Reproductive and Respiratory Syndrome Virus AB Elisa kit (JNT, JN60415, China) according to the manufacturer's instructions, and the threshold for seroconversion was set at a S/P (sample-to-positive) ratio of 0.4. The virus load in serums (E) and lung tissue (F) were detected by real-time qPCR. A recombinant plasmid containing the ORF7 gene of PRRSV was applied to construct a standard curve. The RNA copies of PRRSV were calculated according to the standard curve. G Virus neutralization properties of immune serums from 42 dpv. Sera dilutions collected at 42 dpv were incubated with 200 TCID50 of viruses incorporating HP-PRRSV (SD-YL1712), NADC30-like PRRSV (SX-YL1806), Classical PRRSV (CH1a, VR2332), and PRRSV-1 (GZ11-G1) in DMEM medium with 3% FBS for 1 h. Then, the virus–antibody mixture was transferred to a 96-well plate of confluent MARC-145 cells or CRL2843-CD163 cells and incubated until the CPE was noticed. Then the immunofluorescence assay was carried out to identify the CPE induced by PRRSV. The absence of CPE at 1:8 dilution was considered positive for the presence of PRRSV neutralization. Data were shown as mean ± SD ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
The average daily gain (ADG) of the different groups showed no significant differences before the challenge. However, the ADG of the MLV-SD group was significantly higher than that of the NC-SD group after the challenge with SD-YL1712 at 42 dpv. The ADG of the MLV-SX group was higher than that of the NC-SX group at 63 dpv, whereas there was no significant difference between these two groups within two weeks after the challenge with SX-YL1806 (49 dpv and 56 dpv). The ADG of the MLV-SD group was significantly higher than that of the MLV-SX group at 42 dpv. In addition, no significant differences between the MLV-SD and NC groups were observed, whereas the ADG of the MLV-SX group was significantly lower than that of the NC group at 42, 56, and 63 dpv (Fig. 3C).
3.4. Antibody responses to HP-PRRSV and NADC30-like PRRSV in pigs
From 14 to 42 dpv, pigs in the MLV-SD and MLV-SX groups had high levels of antibodies against the PRRSV N protein. Following the challenge with SD-YL1712 or SX-YL1806 at day 42 post-vaccination, PRRSV-positive antibodies in the MLV-SD and MLV-SX groups decreased slightly at 49 dpv, then increased significantly and were higher than those in the NC-SD and NC-SX groups at 56–63 dpv. In addition, the pigs in the NC-SD group were seroconverted at 49 dpv. In contrast, those in the NC-SX group were seroconverted at 56 dpv. As expected, no antibodies were detected in the NC group (Fig. 3D).
3.5. Viral load in serum and lung tissues of the MLV vaccine inoculation pigs
The viral load in the serum of all groups was determined from day 7–63 post-vaccination with a seven-day interval. Viremia in groups MLV-SD and MLV-SX gradually increased as the MLV vaccine was injected; the highest point appeared at 21 dpv and then gradually declined. After challenge with HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 on day 42 post-vaccination, the viremia of the NC-SD, NC-SX, MLV-SD, and MLV-SX groups increased. No significant difference in viremia between the NC-SD and MLV-SD groups at 49 dpv was found. However, the serum viral load in the MLV-SD group was significantly lower than that in the NC-SD group at 56 and 63 dpv. Similarly, no significant difference in viremia between the NC-SX and MLV-SX groups at 49 dpv was observed, whereas the serum viral load of the MLV-SX group was significantly lower than that of the NC-SX group at 56 and 63 dpv. In contrast, no viremia was detected in the NC group (Fig. 3E).
The viral load in the lung tissues was measured on day 63 post-vaccination. Among the two groups challenged with SD-YL1712, the viral load of the lung tissues in the MLV-SD group was significantly lower than that in the NC-SD group. Similarly, among the two groups challenged with SX-YL1806, the viral load of lung tissues in the MLV-SX group was significantly lower than that in the NC-SX group. Moreover, the viral load of the lung tissues in the MLV-SX group was significantly higher than that in the MLV-SD group. In contrast, PRRSV was not detected in the lung tissues of the NC group (Fig. 3F).
3.6. PRRSV-neutralizing antibodies
To explore the role of viral-neutralizing antibodies in conferring cross-protection, a virus-neutralizing test was performed for the 42-dpv sera of the MLV groups. Surprisingly, only three out of ten piglets immunized with the MLV vaccine developed homologous neutralizing activity against HP-PRRSV SD-YL1712 (1:8). Likewise, only one in ten piglets exhibited high neutralizing activity against NADC30-like PRRSV SX-YL1806 (1:8) or classical PRRSV VR2332 (1:16). Furthermore, the neutralizing activity against the CH1a and PRRSV-1 GZ11-G1 strains was not detected in any of the piglets infected with the MLV vaccine (<1:8) (Fig. 3G).
3.7. Macroscopic and histopathological lesions in the lungs
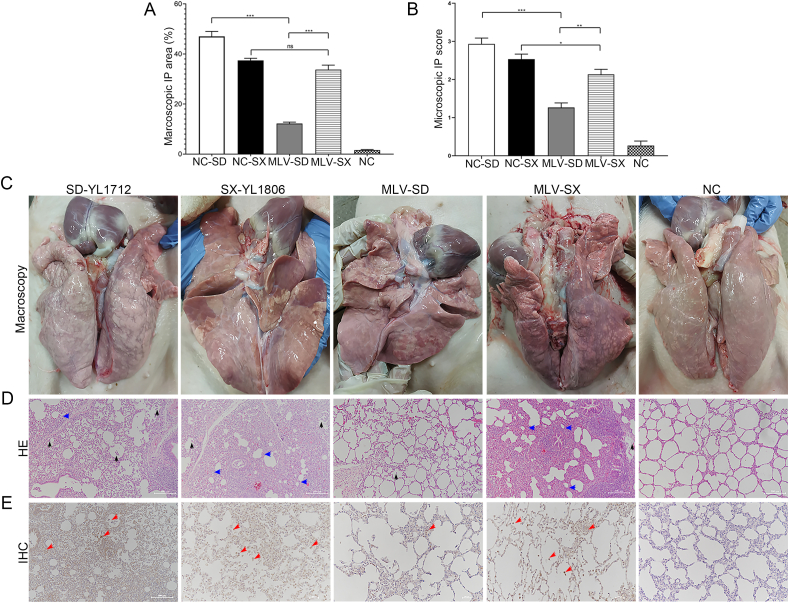
Pigs in the different groups infected with PRRSV showed different degrees of interstitial pneumonia. Severe macroscopic lesions of the lungs were observed in the NC-SD group, including pulmonary consolidation, edema, and hemorrhage, which were significantly more severe than those observed in the MLV-SD group. Similarly, the macroscopic lesions of the lungs in the MLV-SX group were significantly more severe than those in the MLV-SD group. Interestingly, the macroscopic lung lesions in the MLV-SX group were still severe, similar to those in the NC-SX group. No obvious macroscopic lesions were observed in the lungs of the NC group (Fig. 4A and C).
Fig. 4.
Macroscopic and microscopic tissue lesions and immunohistochemistry assay of infected pigs. All piglets were euthanized for autopsy at 63 dpv, and lung tissues were collected and detected. A, C The macroscopic lesion and scores of lungs were estimated based on assigning a number to each lobe to reflect the approximate percentage of the entire lung represented by that lobe. B, D Lung sections of piglets stained with haematoxylin and eosin (HE). The microscopic lesion and score of lung tissues were estimated based on the extent and magnitude of interstitial pneumonia: 0, no lesion; 1, mild/focal; 2, moderate/multifocal; 3, moderate/diffuse (alveolar wall accounting for greater than 50% of the measuring section); 4, severe/diffuse (alveolar wall accounting for greater than 75% of the measuring section). Both macroscopic and microscopic lung lesions were evaluated by three pathologists under blind tests. E Detection of PRRSV-N protein in lung tissue of piglets with immunohistochemistry (IHC) assay, the 6D10 was used as the corresponding antibody. Thickening of alveolar septum and shrinkage of alveolar space were shown by blue arrows; inflammatory cell infiltration was indicated by black arrows; positive signals of PRRSV were shown by red arrows. Scale bar = 500 μm. Data were shown as mean ± SD ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
Obvious microscopic lesions in the NC-SD, NC-SX, and MLV-SD groups were observed, including thickening of the alveolar septum, shrinkage of the alveolar space (blue arrows), and inflammatory cell infiltration (black arrows). Compared to the NC-SD group, the microscopic lesions of the lungs from the MLV-SD group were significantly reduced. Similarly, the microscopic lesions of the lungs in the MLV-SX group were significantly more severe than those in the MLV-SD group. Moreover, the MLV-SX group showed slightly milder microscopic lesions than the NC-SX group but still had high microscopic lesion scores. No obvious microscopic lesions were observed in the lungs of the NC group (Fig. 4B and D).
IHC staining was performed to identify the presence of PRRSV in lung tissues. Positive PRRSV signals were observed in challenged lung tissues. Compared with the NC-SD group, the positive signals in the MLV-SD group were reduced. Similar to the NC-SX group, PRRSV-positive signals were observed in the MLV-SX group. No obvious PRRSV-positive signals were observed in the NC group (Fig. 4E).
3.8. The genomic similarity between the MVL strain and SD-YL1712 or SX-YL1806
To compare the genomic similarities between the MVL strain and the HP-PRRSV SD-YL1712 or the NADC30-like PRRSV SX-YL1806, the complete genome of the MLV strain was sequenced. The genomic homology of each gene of the MLV strain was compared with that of SD-YL1712 or SX-YL1806, and the results showed that almost all non-structural protein (NSP) genes of the MLV strain shared higher nucleotide sequence (84.2%–99.1% identity) and amino acid sequence (79.4%–100% identity) similarity with SD-YL1712 compared to SX-YL1806. Moreover, the genomic homology between open reading frames 2–7 gene (ORF2–ORF7) of the MLV strain and SD-YL1812 (97.8%–99.4%, 96.1%–99.1% identity) was higher than that between the MLV strain and SX-YL1806 (82.9%–89.5%, 81.1%–94.3% identity), including some immune genes such as ORF5 (Table 3).
Table 3.
The genomic homology between the MLV strain and the challenged strains.
| MLV | HP-PRRSV SD-YL1712 |
NADC30-like PRRSV SX-YL1806 |
||
|---|---|---|---|---|
| Genomic region | nt% | aa% | nt% | aa% |
| Full-length | 95.3 | - | 85.9 | - |
| 5′UTR | 100 | - | 90.4 | - |
| ORF1a | 91.7 | 89.6 | 80.8 | 81.6 |
| ORF1b | 98.5 | 99 | 87.6 | 95.8 |
| NSP1 | 97.6 | 98.4 | 83.2 | 90.1 |
| NSP2 | 84.2 | 79.4 | 74.7 | 70 |
| NSP3 | 97.4 | 98.3 | 82.5 | 91.8 |
| NSP4 | 97.5 | 97.5 | 84.8 | 93.1 |
| NSP5 | 98 | 97.6 | 85.5 | 92.4 |
| NSP6 | 95.8 | 100 | 95.8 | 100 |
| NSP7 | 98.1 | 97.7 | 89.6 | 93.1 |
| NSP8 | 98.5 | 100 | 98.5 | 100 |
| NSP9 | 98.4 | 99.2 | 89.7 | 97 |
| NSP10 | 98.7 | 99.3 | 85.6 | 94.3 |
| NSP11 | 98.2 | 97.8 | 89.8 | 96 |
| NSP12 | 99.1 | 99.3 | 88.7 | 94.1 |
| ORF2 | 98.4 | 99.1 | 85.6 | 90.1 |
| ORF3 | 98.4 | 98.8 | 82.9 | 81.1 |
| ORF4 | 97.8 | 96.1 | 85.7 | 87.6 |
| ORF5 | 99 | 98.5 | 85.6 | 87 |
| ORF6 | 99.4 | 98.3 | 89.5 | 94.3 |
| ORF7 | 98.7 | 98.4 | 89 | 88.6 |
| 3′UTR | 99.3 | - | 89.3 | - |
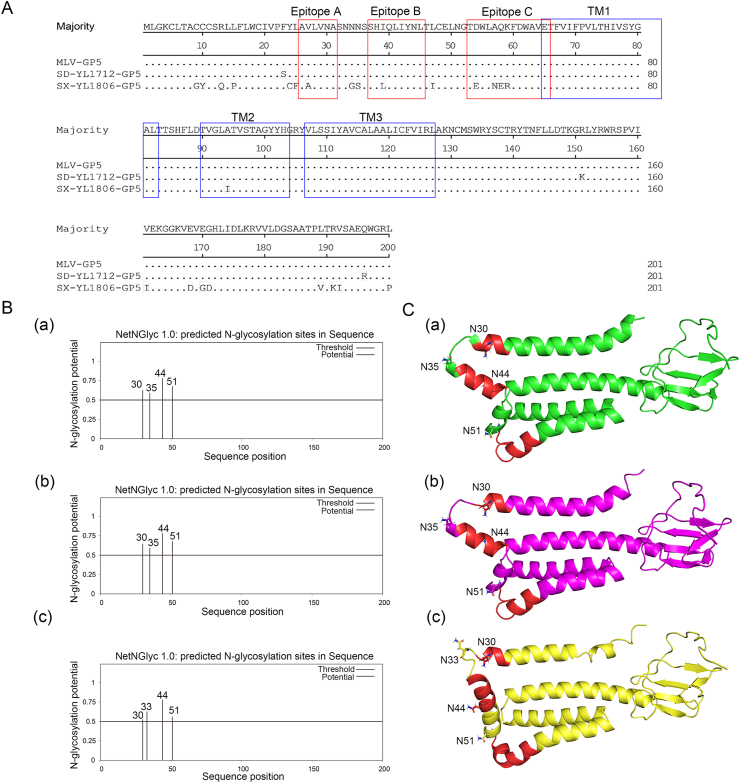
GP5 is the major glycoprotein with the highest variability in PRRSV, and the amino acid homology of GP5 among different PRRSV strains is low. The hypervariability of GP5 among different PRRSV strains may be responsible for its lack of cross-protection. Three linear epitopes of PRRSV were identified, located in GP5. Epitope A is hypervariable and immunodominant and can induce many antibodies against GP5 but not neutralizing antibodies. Epitope B is conserved and functions as a broadly neutralizing epitope. However, it is not immunodominant. Epitope C appears to be a homologous neutralizing epitope but is not conserved (Ostrowski et al., 2002). In this study, three linear epitopes in GP5 of MLV and challenged PRRSV strains were compared. The epitopes A, B, and C were all conserved between MLV and SD-YL1712. Conversely, epitopes A, B, and C of SX-YL1806 were different from those of the MLV strain, especially amino acids at positions 54, 57, 58, and 59 of epitope C were mutated, which may affect the cross-protective effect of the MLV vaccine on SX-YL1806. The three transmembrane (TM) regions of GP5 were similar in the three PRRSV strains (Fig. 5A).
Fig. 5.
The genomic similarity of GP5 between the MLV strain and the challenged strains HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806. A GP5 amino acid analysis of three PRRSV strains by MegAlign software. Three transmembrane (TM) regions of GP5 were shown in blue boxes. Three linear antigenic epitopes of GP5 were shown in red boxes. B The N-glycosylation sites (NGS) of PRRSV GP5 protein were predicted and analyzed online (http://www.cbs.dtu.dk/services/NetNGlyc/). (B-a), (B-b) and (B-c) indicated the MLV strain, HP-PRRSV SD-YL1712 and NADC30-like PRRSV SX-YL1806, respectively. C The GP5 protein structures of these three PRRSV strains were predicted by Alphafold v2.3.2 software (The parameters: default parameters, uniport and uniref 90: 2023-03-01, pdb-mmcif and pdb-seqres: 2023-03-03, other database versions: default). (C-a), (C-b) and (C-c) indicated the MLV strain, HP-PRRSV SD-YL1712 and NADC30-like PRRSV SX-YL1806, respectively. N30, N33, N35, N44, and N51 referred to the corresponding NGS.
In addition, the N-glycosylation sites (NGS) of MLV, SD-YL1712, and SX-YL1806 were predicted and compared, and were likely related to the immune evasion and persistence of viruses by shielding neutralizing antibodies. The results showed that there were four NGSs in each PRRSV strain. The NGSs of the MLV strain and SD-YL1712 had the same pattern (30, 35, 44, and 51), whereas the NGSs of SX-YL1806 were mutated (30, 33, 44, and 51) (Fig. 5B).
Furthermore, the structures of the GP5 proteins of these three strains were analyzed by homologous modeling using Alphafold v2.3.2. The results showed that the MLV strain and HP-PRRSV SD-YL1712 both formed the α helixes at 35–45, 47–52, and 60–80 aa, while NADC30-like PRRSV SX-YL1806 formed the α helixes at 37–52 and 54–80 aa, resulting in significant differences in the structure of GP5. Because the primary neutralizing epitope B (36–45 aa) was located here, this structural difference may have affected the protective effect of the MLV vaccine against the heterologous strain SX-YL1806 (Fig. 5C).
3.9. The immune serums could slightly promote the replication of HP-PRRSV SD-YL1712 in vitro
We further explored whether porcine sera after immunization with the MLV vaccine could promote PRRSV replication in vitro. Pig sera were collected from the MLV-SD group at different time points after immunization with the MLV vaccine and serially diluted (1:50–1:800). The immune sera were co-incubated with SD-YL1712 and then transferred to CRL2843-CD163 cells to analyze PRRSV replication using RT-qPCR, IFA, and WB. Unimmunized and PEDV-positive pig serum served as controls.
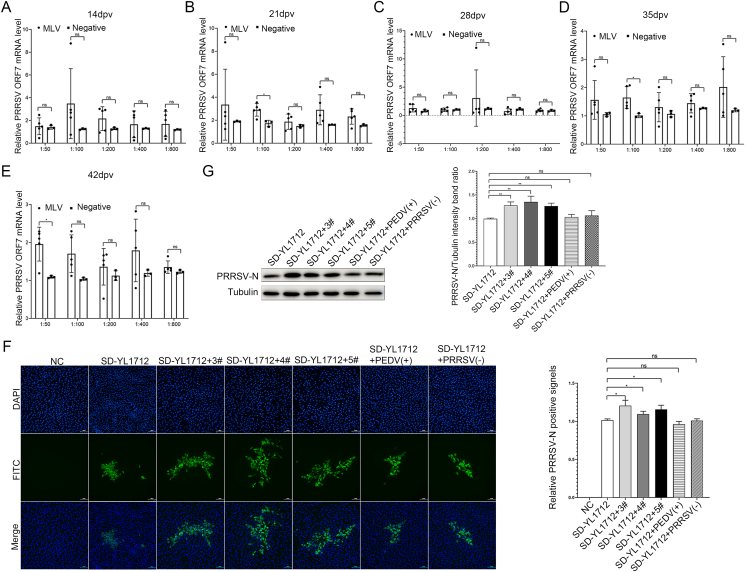
When co-incubated with HP-PRRSV SD-YL1712, the immune sera mildly promoted PRRSV replication. Only one serum sample from the MLV-SD group collected at 14 dpv increased PRRSV replication by 8.7-fold after 1:100 dilution, whereas the other serum sample did not significantly promote PRRSV replication compared to the negative control (Fig. 6A). Serum collected at 21 dpv significantly promoted PRRSV replication by 2.9-fold after a 1:100 dilution (Fig. 6B). Furthermore, one serum sample collected at 28 dpv promoted PRRSV replication by up to 12-fold after a 1:200 dilution, but there was no significant difference between the mean values of these two groups (Fig. 6C). Serum collected at 35 dpv significantly promoted the replication of HP-PRRSV SD-YL1712 by 1.6-fold after a 1:100 dilution (Fig. 6D), and serum collected at 42 dpv mildly promoted replication by up to 2.0-fold after a 1:50 dilution (Fig. 6E).
Fig. 6.
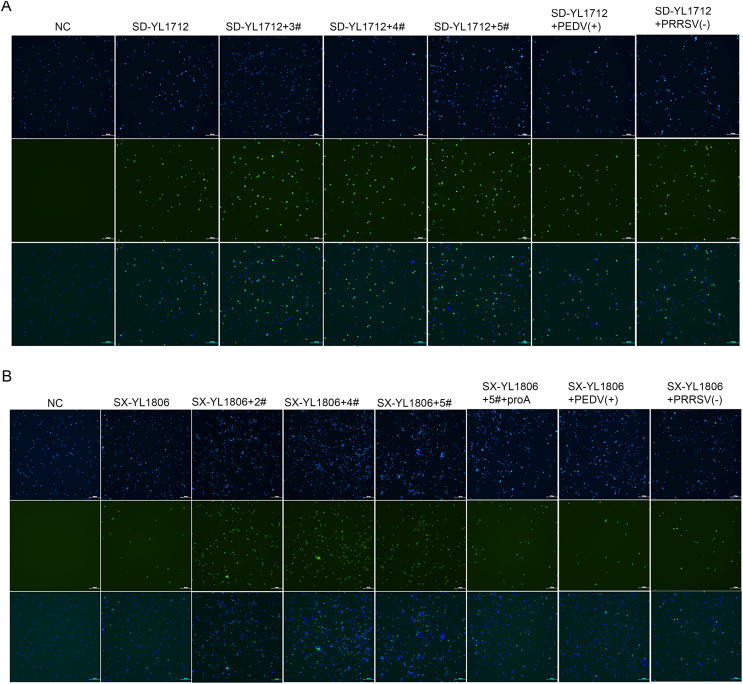
Promoting effect of immunized serum on HP-PRRSV SD-YL1712 replication in vitro. A–E CRL2843-CD163 cells were infected with virus–immunized serums mixture (HP-PRRSV SD-YL1712) for 48 h, then the cells were harvested for the relative quantitative RT-qPCR assay. The MLV-immunized serums were collected from group MLV-SD at 14 dpv, 21 dpv, 28 dpv, 35 dpv, and 42 dpv, respectively; serum collected from group NC was treated as a negative control. Serums were diluted by 1:50, 1:100, 1:200, 1:400, and 1:800. F–G The CRL2843-CD163 cells were infected with the PRRSV-serum complex for 48 h (HP-PRRSV SD-YL1712), then the cells were harvested for immunofluorescence assay (F) or Western blotting assay (G). The MLV-immunized serums were collected from group MLV-SD at 42 dpv and diluted by 1:50. 3#, 4#, and 5# represented the three piglets in group MLV-SD. The 6D10 was used as the corresponding antibody. Unimmunized pig serums [PRRSV (−)] and PEDV-positive [PEDV (+)] pig serums served as controls. Scale bar = 200 μm. Data were shown as mean ± SD ∗P < 0.05; ns, not significant.
Furthermore, the infection rates of PRRSV with ADE were also confirmed by IFA and WB assays, which showed that when co-incubated with HP-PRRSV SD-YL1712, immune sera collected at 42 dpv from the MLV-SD group mildly promoted PRRSV replication and slightly increased positive signals of PRRSV-N protein (approximately 1.3 times) (Fig. 6F and G). The same conclusion was reached for the PAMs (Supplementary Fig. S1A).
3.10. The immune serums could significantly promote the replication of NADC30-like PRRSV SX-YL1806 in vitro
Similarly, we collected pig sera from the MLV-SX group at different times after immunization with the MLV vaccine and serially diluted them (1:50–1:800). The immune sera were co-incubated with NADC30-like PRRSV SX-YL1806 and then transferred to CRL2843-CD163 cells to analyze PRRSV replication using RT-qPCR, IFA, and WB. Unimmunized and PEDV-positive pig serum served as controls.
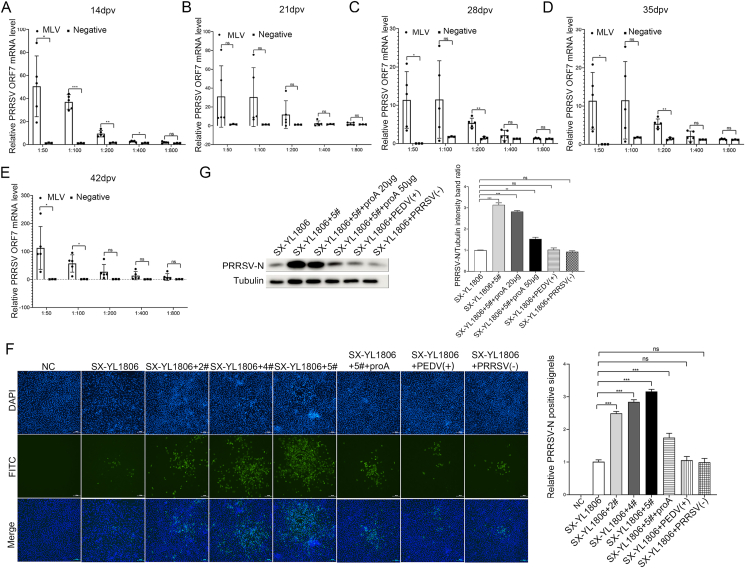
When co-incubated with NADC30-like PRRSV SX-YL1806, almost all immune sera collected at different times significantly promoted PRRSV replication, and sera diluted at 1:50 and 1:100 seemed to have the most obvious promotion effect. Serum collected at 14 dpv significantly promoted PRRSV replication after 1:50-fold and 1:100-fold dilutions, with the highest fold being 88.3-fold and the mean value being 50.5-fold (Fig. 7A). Serum collected at 28 dpv also significantly promoted PRRSV replication after a 1:50-fold dilution, with the highest fold being 20.8-fold, and the mean value being 11.4-fold (Fig. 7C). Similarly, sera collected at 35 dpv significantly promoted PRRSV replication after 1:100-fold dilution, with the highest fold being 54.1-fold and the mean value being 35.5-fold (Fig. 7D). Moreover, serum collected at 42 dpv also significantly promoted PRRSV replication after 1:50-or 1:100-fold dilutions, with the highest fold being 235.9-fold and the mean value being 112.8-fold (Fig. 7E). Serum collected at 21 dpv did not significantly promote the replication of PRRSV after 1:50-fold or 1:100 dilutions because of the large individual differences within the group, which affected the calculation of the mean. However, all serum samples had higher relative PRRSV ORF7 mRNA levels than the negative group after 1:50-fold or 1:100-fold dilutions (Fig. 7B).
Fig. 7.
Promoting effect of immunized serum on NADC30-like PRRSV SX-YL1806 replication in vitro. A–E CRL2843-CD163 cells were infected with virus–immunized serums mixture (NADC30-like PRRSV SX-YL1806) for 48 h, then the cells were harvested for the relative quantitative RT-qPCR assay. The MLV-immunized serums were collected from group MLV-SX at 14 dpv, 21 dpv, 28 dpv, 35 dpv, and 42 dpv, respectively; serum collected from group NC was treated as a negative control. Serums were diluted by 1:50, 1:100, 1:200, 1:400, and 1:800. F–G The CRL2843-CD163 cells were infected with the PRRSV-serum complex or PRRSV-serum-Protein A complex for 48 h (NADC30-like PRRSV SX-YL1806), then the cells were harvested for immunofluorescence assay (F) or Western blotting assay (G). The MLV-immunized serums were collected from group MLV-SX at 42 dpv and diluted by 1:50. 2#, 4#, and 5# represented the three piglets in group MLV-SX. The 6D10 was used as the corresponding antibody. Unimmunized pig serums [PRRSV (−)] and PEDV-positive pig [PEDV (+)] serums served as controls. Scale bar = 200 μm. Data were shown as mean ± SD ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
In addition, the IFA and WB results showed that when co-incubated with NADC30-like PRRSV SX-YL1806, immune sera collected at 42 dpv from group MLV-SX significantly promoted PRRSV replication and significantly increased positive signals of PRRSV-N protein (about three times). In contrast, PEDV-positive serum or negative porcine serum did not significantly promote the replication of NADC30-like PRRSV (Fig. 7F and G). The same conclusion was reached for the PAMs (Supplementary Fig. S1B).
To investigate whether the phenomenon of MLV-immune serum-promoting NADC30-like PRRSV replication was mediated by the formation of antigen (PRRSV)-antibody (IgG) complexes, Protein A known to bind nonspecifically to IgG, was used in this study. The results showed that the replication-promoting effect of MLV-immune serum on NADC30-like PRRSV was inhibited by Protein A (Fig. 7F and G).
Collectively, the immune sera collected from the MLV-SD group slightly promoted the replication of the homologous strain HP-PRRSV SD-YL1712. In contrast, immune sera collected from the MLV-SX group significantly promoted replication of the heterologous strain NADC30-like PRRSV SX-YL1806, and this replication-promoting effect was inhibited by Protein A.
4. Discussion
Although African swine fever is prevalent worldwide, PRRS is still one of the most important animal infectious diseases endangering the global pig industry (Lunney et al., 2010). Vaccination is important to prevent and control PRRSV and to eliminate PRRSV (Chen N. et al., 2021; Jeong et al., 2021). Although the PRRSV vaccine has a good protective effect against homologous strains, its protective effect against heterologous strains is incomplete (Chen X. et al., 2021; Ding et al., 2021; Li Y. et al., 2021). In this study, immunization with an MLV vaccine provided some protection against NADC30-like PRRSV SX-YL1806 in piglets, including an improvement in temperature, ADG level, viremia, and lung tissue virus load (Fig. 3), whereas piglets in the MLV-SX group still had a high lung tissue virus load and lung tissue injury (Fig. 4). These results indicate that MLV immunization did not provide adequate protection against NADC30-like PRRSV SX-YL 1806 in piglets.
Immune vaccine-induced neutralizing antibodies are considered an important means of PRRSV clearance (Vanhee et al., 2009). However, the neutralizing antibodies against PRRSV are often generated late, which appears to be an important reason for the unsatisfactory immune effects of the PRRSV vaccine (Osorio et al., 2002; Lopez et al., 2007). In this study, 3/10 of the serum from pigs immunized with MLV vaccine for 42 days showed neutralization properties against HP-PRRSV (≥1:8), and only 1/10 showed neutralization properties against NADC30 like PRRSV (≥1:8) (Fig. 3G), indicating that the piglets did not generate sufficient neutralizing antibodies even 42 days after immunization with the MLV vaccine.
Interestingly, although no neutralization properties of pig serum were observed at 42 dpv, the immunized MLV vaccine still protected piglets in the MLV-SD group against the HP-PRRSV SD-YL1712 challenge (Fig. 3, Fig. 4), which indicates that factors other than neutralizing antibodies, such as cellular immunity, may play key roles in this protection. In addition, the protection observed may be associated with the type of PRRSV strain since it was not produced in the NADC30-like PRRSV group (MLV-SX group).
The PRRSV genome mutates rapidly, and genomic homology among different PRRSV strains is low (Guo et al., 2018; Li et al., 2022). In this study, the MLV vaccine provided good protection against HP-PRRSV but did not provide sufficient protection against the NADC30-like PRRSV SX-YL1806 (Fig. 3, Fig. 4). We then compared the genomic homology between the MLV strain and HP-PRRSV SD-YL1712 or NADC30-like PRRSV SX-YL1806 and found that the MLV strain had higher genomic similarity with SD-YL1712 than with SX-YL1806, especially in the NSP2 and ORF5 genes (Table 3). Open reading frame (ORF) genes, such as ORF5, are the major immune genes of PRRSV, and the difference in genomic homology between the MLV strain and HP-PRRSV or NADC30-like PRRSV may be a potential reason for the difference in MLV vaccine immune efficacy. We performed a detailed analysis of the ORF5 gene between the MLV strain and challenged PRRSV strains and found that MLV and HP-PRRSV SD-YL1712 had three similar linear epitopes, A, B, and C, whereas epitopes A, B, and C of SX-YL1806 were mutated (Fig. 5A). In addition, the NGSs and GP5 structures of SX-YL1806 were different from those of the MLV strain and HP-PRRSV SD-YL1712, which may have influenced the protective effects of the MLV vaccine against SX-YL1806 (Fig. 5B and C). There are at least nine types of vaccines used in China, but almost all of them originated from HP-PRRSV or classical PRRSV, and none of them were derived from NADC30-like PRRSV (Li et al., 2022). Thus, improving the genomic homology between the vaccine strain and the dominant epidemic PRRSV may be a good strategy.
Antibodies triggered by vaccines can block infections via antibody neutralization. However, antibodies induced by vaccines may enhance viral infection in ADE (Anderson et al., 2013; Khurana et al., 2013). In this study, MLV immunization did not adequately protect against NADC30-like PRRSV attacks, although no significantly enhanced clinical manifestations were observed in the MLV-SD group compared with the NC-SD group. We collected porcine serum at 14, 21, 28, 35, and 42 days after immunization with the MLV vaccine and evaluated its effect on the homologous strain HP-PRRSV SD-YL1712 and the heterologous strain NADC30 like PRRSV SX-YL1806 replication in vitro using CRL2843-CD163 cells. Immunized serum diluted at 1:50 or 1:100 significantly promoted the replication of NADC30-like PRRSV SX-YL1806 (average 11.4–112.8 times) (Fig. 7A–E). In contrast, it had a slightly mildly promoted effect on the ADE of HP-PRRSV SD-YL1712 (average 1.6–2.9 times) (Fig. 6A–E). The replication-promoting effect of the NADC30-like PRRSV SX-YL1806 was inhibited by Protein A (Fig. 7F and G). These results indicate the potential risk of ADE in MLV-immunized pig serum against homologous and heterologous strains in vitro.
Notably, the replication-promoting effect against HP-PRRSV or NADC30-like PRRSV caused by MLV-immunized serum discussed in this study was evaluated in vitro, and no more severe clinical enhancement in the MLV-SX group compared to the NC-SX group was observed in vivo. One possible explanation is that the immunized serum can promote PRRSV replication in vivo at some concentrations; however, it may not be significantly enhanced by other immune reactions, such as cellular immunity. Considering the fluctuating rule of antibodies following PRRSV MLV immunization, we speculate that the replication-promoting effect of MLV-immunized serum on heterologous PRRSV may aggravate PRRS or interfere with the efficacy of the vaccine against NADC30-like PRRSV. However, further studies are required to confirm this hypothesis. Avoiding the replication-promoting effect of MLV-immune serum may be an important measure to improve the effectiveness of the PRRSV vaccines against heterologous PRRSV.
5. Conclusions
In conclusion, this study systematically evaluated the protective effects of a commercial MLV vaccine against the isolated field-circulating strains NADC30-like PRRSV SX-YL1806 and HP-PRRSV SD-YL1712. The MLV vaccine did not provide sufficient cross-protection against SX-YL1806. Furthermore, this study found that the MLV strain shared higher genomic similarity with HP-PRRSV SD-YL1712 than with NADC30-like PRRSV SX-YL1806. Compared to the homologous strain SD-YL1712, the immunized serum significantly promoted the heterologous NADC30-like PRRSV SX-YL1806 replication in vitro. This study suggests that the genomic similarity between the MLV and challenged PRRSV strains, together with the replication-promoting effect to NADC30-like PRRSV caused by MLV-immune serum, may be two important factors affecting the protective effect of PRRSV MLV vaccines against heterologous PRRSV, which has not received sufficient attention until now and requires more in-depth studies.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
All experimental programs involving pigs were carried out in accordance with the guidelines of the Universal Declaration on Animal Welfare from the World Organization for Animal Health, and the Ethical Committee of Lanzhou Veterinary Research Institute of Chinese Academy of Agricultural Science (CAAS) approved by the Ministry of Agriculture and Rural Affairs. All pigs used in this study were carefully fed and suffering of animals was minimized.
Author contributions
Yang Li: conceptualization, data curation, formal analysis, investigation, methodology, visualization, writing-original draft, writing-review & editing. Lele Xu: conceptualization, data curation, formal analysis, investigation, methodology, visualization, writing-original draft, writing-review & editing. Dian Jiao: data curation, formal analysis, investigation, methodology, software, validation. Zifang Zheng: data curation, investigation, methodology, validation. Zhihao Chen: investigation, methodology. Yang Jing: methodology. Zhiwei Li: methodology. Zhiqian Ma: conceptualization, resources. Yingtong Feng: conceptualization. Xuyang Guo: software validation. Yuan He: software validation. Yumiao Wang: conceptualization. Haixue Zheng: conceptualization, funding acquisition, project administration. Shuqi Xiao: conceptualization, funding acquisition, project administration, supervision, visualization, writing-review & editing.
Conflict of interest
All authors declare they have no conflicts of interest.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (32172846), the Earmarked Fund for CARS-35, the Science and Technology Major Project of Gansu Province (22ZD6NA001), the Science Foundation for Distinguished Young Scholars of Shaanxi Province (2021JC-18), the Natural Science Foundation of Gansu Province (23JRRA1153), the Science and Technology Plan Project of Gansu Province (23JRRA561), the Chinese Academy of Agricultural Science and Technology Innovation Project (CAAS-ASTIP-JBGS-20210602), and the Strategic Priority Research Program of the National Center of Technology Innovation for Pigs (NCTIP-XD/C03).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2023.08.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figs. 1
References
- Anderson L.J., Dormitzer P.R., Nokes D.J., Rappuoli R., Roca A., Graham B.S. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine. 2013;31(Suppl. 2):B209–B215. doi: 10.1016/j.vaccine.2012.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian T., Sun Y., Hao M., Zhou L., Ge X., Guo X., Han J., Yang H. A recombinant type 2 porcine reproductive and respiratory syndrome virus between NADC30-like and a MLV-like: genetic characterization and pathogenicity for piglets. Infect. Genet. Evol. 2017;54:279–286. doi: 10.1016/j.meegid.2017.07.016. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Chen N., Li S., Tian Y., Li X., Li S., Li J., Qiu M., Sun Z., Xiao Y., Yan X., Lin H., Yu X., Tian K., Shang S., Zhu J. Chimeric HP-PRRSV2 containing an ORF2-6 consensus sequence induces antibodies with broadly neutralizing activity and confers cross protection against virulent NADC30-like isolate. Vet. Res. 2021;52:74. doi: 10.1186/s13567-021-00944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Tan X., Lao M., Wu X., Zhao X., Zhou S., Yu J., Zhu J., Yu L., Tong W., Gao F., Yu H., Liu C., Jiang Y., Tong G., Zhou Y. The novel PRRSV strain HBap4-2018 with a unique recombinant pattern is highly pathogenic to piglets. Virol. Sin. 2021;36:1611–1625. doi: 10.1007/s12250-021-00453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.X., Zhou X., Guo T., Qiao S., Guo Z., Li R., Jin Q., Hu X., Xing G., Deng R., Wan B., Zhang G. Efficacy of a live attenuated highly pathogenic PRRSV vaccine against a NADC30-like strain challenge: implications for ADE of PRRSV. BMC Vet. Res. 2021;17:260. doi: 10.1186/s12917-021-02957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwich L., Gimeno M., Sibila M., Diaz I., de la Torre E., Dotti S., Kuzemtseva L., Martin M., Pujols J., Mateu E. Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet. Microbiol. 2011;150:49–62. doi: 10.1016/j.vetmic.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Ding Y., Wubshet A.K., Ding X., Zhang Z., Li Q., Dai J., Hou Q., Hu Y., Zhang J. Evaluation of four commercial vaccines for the protection of piglets against the highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) QH-08 strain. Vaccines (Basel) 2021;9:1020. doi: 10.3390/vaccines9091020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Guo X., Tian H., He Y., Li Y., Jiang X., Zheng H., Xiao S. Induction of HOXA3 by porcine reproductive and respiratory syndrome virus inhibits type I interferon response through negative regulation of HO-1 transcription. J. Virol. 2022;96 doi: 10.1128/JVI.01863-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Liu Z., Tong X., Wang Z., Xu S., Chen Q., Zhou J., Fang L., Wang D., Xiao S. Evolutionary dynamics of type 2 porcine reproductive and respiratory syndrome virus by whole-genome analysis. Viruses. 2021;13:2469. doi: 10.3390/v13122469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Chen X.X., Li R., Qiao S., Zhang G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol. J. 2018;15:2. doi: 10.1186/s12985-017-0910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Chen X.X., Li X., Qiao S., Deng R., Zhang G. Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in central China during 2016-2017: NADC30-like PRRSVs are predominant. Microb. Pathog. 2019;135 doi: 10.1016/j.micpath.2019.103657. [DOI] [PubMed] [Google Scholar]
- Halbur P.G., Paul P.S., Frey M.L., Landgraf J., Eernisse K., Meng X.J., Lum M.A., Andrews J.J., Rathje J.A. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the lelystad virus. Vet. Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- Han J., Zhou L., Ge X., Guo X., Yang H. Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2017;209:30–47. doi: 10.1016/j.vetmic.2017.02.020. [DOI] [PubMed] [Google Scholar]
- Hou F.H., Lee W.C., Liao J.W., Chien M.S., Kuo C.J., Chung H.P., Chia M.Y. Evaluation of a type 2 modified live porcine reproductive and respiratory syndrome vaccine against heterologous challenge of a lineage 3 highly virulent isolate in pigs. PeerJ. 2020;8 doi: 10.7717/peerj.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Meng X.J. Novel strategies and approaches to develop the next generation of vaccines against porcine reproductive and respiratory syndrome virus (PRRSV) Virus Res. 2010;154:141–149. doi: 10.1016/j.virusres.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C.G., Khatun A., Nazki S., Kim S.C., Noh Y.H., Kang S.C., Lee D.U., Yang M.S., Shabir N., Yoon I.J., Kim B., Kim W.I. Evaluation of the cross-protective efficacy of a chimeric PRRSV vaccine against two genetically diverse PRRSV2 field strains in a reproductive model. Vaccines (Basel) 2021;9:1258. doi: 10.3390/vaccines9111258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph T., Kibenge M.T., Kibenge F.S.B. Antibody-mediated growth of infectious salmon anaemia virus in macrophage-like fish cell lines. J. Gen. Virol. 2003;84:1701–1710. doi: 10.1099/vir.0.19087-0. [DOI] [PubMed] [Google Scholar]
- Khurana S., Loving C.L., Manischewitz J., King L.R., Gauger P.C., Henningson J., Vincent A.L., Golding H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- King A.M.Q., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M.L., Rubino L., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2018) Arch. Virol. 2018;163:2601–2631. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
- Li C., Zhuang J., Wang J., Han L., Sun Z., Xiao Y., Ji G., Li Y., Tan F., Li X., Tian K. Outbreak investigation of NADC30-like PRRSV in south-east China. Transbound Emerg Dis. 2016;63:474–479. doi: 10.1111/tbed.12530. [DOI] [PubMed] [Google Scholar]
- Li L., Chen J., Cao Z., Cao Y., Guo Z., Tong W., Zhou Y., Li G., Jiang Y., Liu C., Yu L., Qiao S., Liu J., Tong G., Gao F. Recombinant bivalent live vectored vaccine against classical swine fever and hp-prrs revealed adequate heterogeneous protection against NADC30-like strain. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.822749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Jiao D., Jing Y., He Y., Han W., Li Z., Ma Z., Feng Y., Xiao S. Genetic characterization and pathogenicity of a novel recombinant PRRSV from lineage 1, 8 and 3 in China failed to infect MARC-145 cells. Microb. Pathog. 2022;165 doi: 10.1016/j.micpath.2022.105469. [DOI] [PubMed] [Google Scholar]
- Li Y., Xu G.X., Du X.Q., Xu L.L., Ma Z.Q., Li Z.W., Feng Y.T., Jiao D., Guo W.P., Xiao S.Q. Genomic characteristics and pathogenicity of a new recombinant strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 2021;166:389–402. doi: 10.1007/s00705-020-04917-8. [DOI] [PubMed] [Google Scholar]
- Lopez O.J., Oliveira M.F., Garcia E.A., Kwon B.J., Doster A., Osorio F.A. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clin. Vaccine Immunol. 2007;14:269–275. doi: 10.1128/CVI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney J.K., Benfield D.A., Rowland R.R. Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 2010;154:1–6. doi: 10.1016/j.virusres.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney J.K., Fang Y., Ladinig A., Chen N., Li Y., Rowland B., Renukaradhya G.J. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. 2016;4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- Osorio F.A., Galeota J.A., Nelson E., Brodersen B., Doster A., Wills R., Zuckermann F., Laegreid W.W. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology. 2002;302:9–20. doi: 10.1006/viro.2002.1612. [DOI] [PubMed] [Google Scholar]
- Ostrowski M., Galeota J.A., Jar A.M., Platt K.B., Osorio F.A., Lopez O.J. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 2002;76:4241–4250. doi: 10.1128/JVI.76.9.4241-4250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupasinghe R., Lee K., Liu X., Gauger P.C., Zhang J., Martinez-Lopez B. Microbiol Spectr; 2022. Molecular Evolution of Porcine Reproductive and Respiratory Syndrome Virus Field Strains from Two Swine Production Systems in the Midwestern united states from 2001 to 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Fang L., Zeng S., Li B., Chen H., Xiao S. Complete genome sequence of porcine reproductive and respiratory syndrome virus isolated from piglet stool samples. J. Virol. 2012;86:4040–4041. doi: 10.1128/JVI.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soraci L., Lattanzio F., Soraci G., Gambuzza M.E., Pulvirenti C., Cozza A., Corsonello A., Luciani F., Rezza G. COVID-19 vaccines: current and future perspectives. Vaccines (Basel) 2022;10:608. doi: 10.3390/vaccines10040608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.F., Zhou L., Bian T., Tian X.X., Ren W.K., Lu C., Zhang L., Li X.L., Cui M.S., Yang H.C., Yu H. Efficacy evaluation of two commercial modified-live virus vaccines against a novel recombinant type 2 porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2018;216:176–182. doi: 10.1016/j.vetmic.2018.02.016. [DOI] [PubMed] [Google Scholar]
- Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C., Hu Y., Chen X., Hu D., Tian X., Liu D., Zhang S., Deng X., Ding Y., Yang L., Zhang Y., Xiao H., Qiao M., Wang B., Hou L., Wang X., Yang X., Kang L., Sun M., Jin P., Wang S., Kitamura Y., Yan J., Gao G.F. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z.J., An T.Q., Zhou Y.J., Peng J.M., Hu S.P., Wei T.C., Jiang Y.F., Xiao Y., Tong G.Z. An attenuated live vaccine based on highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) protects piglets against hp-prrs. Vet. Microbiol. 2009;138:34–40. doi: 10.1016/j.vetmic.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Trible B.R., Popescu L.N., Monday N., Calvert J.G., Rowland R.R.R. A single amino acid deletion in the matrix protein of porcine reproductive and respiratory syndrome virus confers resistance to a polyclonal swine antibody with broadly neutralizing activity. J. Virol. 2015;89:6515–6520. doi: 10.1128/JVI.03287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee M., Delputte P.L., Delrue I., Geldhof M.F., Nauwynck H.J. Development of an experimental inactivated PRRSV vaccine that induces virus-neutralizing antibodies. Vet. Res. 2009;40:63. doi: 10.1051/vetres/2009046. [DOI] [PubMed] [Google Scholar]
- Wan B., Chen X., Li Y., Pang M., Chen H., Nie X., Pan Y., Qiao S., Bao D. Porcine FcγRIIb mediated PRRSV ADE infection through inhibiting IFN-β by cytoplasmic inhibitory signal transduction. Int. J. Biol. Macromol. 2019;138:198–206. doi: 10.1016/j.ijbiomac.2019.07.005. [DOI] [PubMed] [Google Scholar]
- Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., Zhou Y., Du L., Li F. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94:e02015–e02019. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.J., Xie W., Chen X.X., Qiao S., Zhao M., Gu Y., Zhao B.L., Zhang G. Molecular epidemiology of porcine reproductive and respiratory syndrome virus in central China since 2014: the prevalence of NADC30-like PRRSVs. Microb. Pathog. 2017;109:20–28. doi: 10.1016/j.micpath.2017.05.021. [DOI] [PubMed] [Google Scholar]
- Wensvoort G., Terpstra C., Pol J.M., ter Laak E.A., Bloemraad M., de Kluyver E.P., Kragten C., van Buiten L., den Besten A., Wagenaar F., et al. Mystery swine disease in The Netherlands: the isolation of lelystad virus. Vet. Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- Xiao S.Q., Zhang A.K., Zhang C., Ni H.B., Gao J.M., Wang C.B., Zhao Q., Wang X.P., Wang X., Ma C., Liu H.L., Li N., Mu Y., Sun Y.N., Zhang G.P., Hiscox J.A., Hsu W.H., Zhou E.M. Heme oxygenase-1 acts as an antiviral factor for porcine reproductive and respiratory syndrome virus infection and over-expression inhibits virus replication in vitro. Antivir. Res. 2014;110:60–69. doi: 10.1016/j.antiviral.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Xu L., Ma Z., Li Y., Pang Z., Xiao S. Antibody dependent enhancement: unavoidable problems in vaccine development. Adv. Immunol. 2021;151:99–133. doi: 10.1016/bs.ai.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.J., Wu L.L., Zimmerman J.J., Platt K.B. Field isolates of porcine reproductive and respiratory syndrome virus (PRRSV) vary in their susceptibility to antibody dependent enhancement (ADE) of infection. Vet. Microbiol. 1997;55:277–287. doi: 10.1016/s0378-1135(96)01338-7. [DOI] [PubMed] [Google Scholar]
- Yoon K.J., Wu L.L., Zimmerman J.J., Hill H.T., Platt K.B. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol. 1996;9:51–63. doi: 10.1089/vim.1996.9.51. [DOI] [PubMed] [Google Scholar]
- Zhang J., Bai J., Sun Y., Liu X., Gao Y., Wang X., Yang Y., Jiang P. Comparison of pathogenicity of different subgenotype porcine reproductive and respiratory syndrome viruses isolated in China. Microb. Pathog. 2022;168 doi: 10.1016/j.micpath.2022.105607. [DOI] [PubMed] [Google Scholar]
- Zhang Q.Y., Jiang P., Song Z.B., Lv L., Li L., Bai J. Pathogenicity and antigenicity of a novel NADC30-like strain of porcine reproductive and respiratory syndrome virus emerged in China. Vet. Microbiol. 2016;197:93–101. doi: 10.1016/j.vetmic.2016.11.010. [DOI] [PubMed] [Google Scholar]
- Zhao H., Han Q., Zhang L., Zhang Z., Wu Y., Shen H., Jiang P. Emergence of mosaic recombinant strains potentially associated with vaccine JXA1-R and predominant circulating strains of porcine reproductive and respiratory syndrome virus in different provinces of China. Virol. J. 2017;14:67. doi: 10.1186/s12985-017-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Ye C., Chang X.B., Jiang C.G., Wang S.J., Cai X.H., Tong G.Z., Tian Z.J., Shi M., An T.Q. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J. Virol. 2015;89:10712–10716. doi: 10.1128/JVI.01446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Fu X., Ling X., Sun H., Li Y., Ma Z., Wei B., Zheng H., Xiao S. Host cells actively resist porcine reproductive and respiratory syndrome virus infection via the IRF8-Microrna-10a-SRP14 regulatory pathway. J. Virol. 2022;96 doi: 10.1128/jvi.00003-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Wang Z., Ding Y., Ge X., Guo X., Yang H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 2015;21:2256–2257. doi: 10.3201/eid2112.150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Kang R.M., Yu J.F., Xie B., Chen C.Y., Li X.Y., Xie J., Ye Y.G., Xiao L., Zhang J.L., Yang X., Wang H.N. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among NADC30-like, JXA1-like, and MLV-like strains. Viruses-Basel. 2018;10:551. doi: 10.3390/v10100551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.J., Hao X.F., Tian Z.J., Tong G.Z., Yoo D., An T.Q., Zhou T., Li G.X., Qiu H.J., Wei T.C., Yuan X.F. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound Emerg Dis. 2008;55:152–164. doi: 10.1111/j.1865-1682.2008.01020.x. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Yuan L., Hu D., Lian Z., Yao X., Liu P., Li X. Isolation and genomic characterization of a Chinese NADC34-like PRRSV isolated from Jiangsu Province. Transbound Emerg Dis. 2022;69:e1015–e1027. doi: 10.1111/tbed.14392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.