Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease caused by distinctive mutations in individual patients; therefore, each patient may display different cell-type compositions. Although most patients with AML achieve complete remission (CR) through intensive chemotherapy, the likelihood of relapse remains high. Several studies have attempted to characterize the genetic and cellular heterogeneity of AML; however, our understanding of the cellular heterogeneity of AML remains limited. In this study, we performed single-cell RNA sequencing (scRNAseq) of bone marrow-derived mononuclear cells obtained from same patients at different AML stages (diagnosis, CR, and relapse). We found that hematopoietic stem cells (HSCs) at diagnosis were abnormal compared to normal HSCs. By improving the detection of the DNMT3A R882 mutation with targeted scRNAseq, we identified that DNMT3A-mutant cells that mainly remained were granulocyte-monocyte progenitors (GMPs) or lymphoid-primed multipotential progenitors (LMPPs) from CR to relapse and that DNMT3A-mutant cells have gene signatures related to AML and leukemic cells. Copy number variation analysis at the single-cell level indicated that the cell type that possesses DNMT3A mutations is an important factor in AML relapse and that GMP and LMPP cells can affect relapse in patients with AML. This study advances our understanding of the role of DNMT3A in AML relapse and our approach can be applied to predict treatment outcomes.
Keywords: acute myeloid leukemia, cancer genomics, copy number variation, DNMT3A R882, relapse, single-cell RNA sequencing
INTRODUCTION
Acute myeloid leukemia (AML) is a type of blood cancer in which abnormal immature cells accumulate in the bone marrow (BM) and interfere with normal function (Khwaja et al., 2016). Although 60%-70% of patients with AML achieve complete remission (CR) through intensive chemotherapy, the likelihood of relapse remains high (Estey and Döhner, 2006). Various studies have attempted to determine the causes of relapse (Shlush et al., 2017; Vosberg and Greif, 2019; Yilmaz et al., 2019); however, further in-depth research is required as each patient responds differently to treatment. AML is caused by DNA mutations in hematopoietic stem cells (HSCs) or progenitor cells during the early stages of hematopoiesis (DiNardo and Cortes, 2016; Yu et al., 2020). Although genome and transcriptome analyses have identified causes of drug resistance and therapeutic targets (Alanazi et al., 2020; Arindrarto et al., 2021; Tyner et al., 2018), our understanding of the cellular heterogeneity of AML remains poor.
Recent studies have used single-cell RNA sequencing (scRNAseq) (Choi and Kim, 2019) to determine the cell-type composition in patients with AML and to identify cell-type specific gene expression changes (Petti et al., 2019; van Galen et al., 2019). Although several single cell mutation analyses of AML have successfully measured clonal complexity and the order of mutations during disease progression (Ediriwickrema et al., 2020; Li et al., 2023; Miles et al., 2020; Paguirigan et al., 2015; Pellegrino et al., 2018; Stetson et al., 2021; Zhai et al., 2022), single-cell transcriptome analysis on sequential samples obtained at diagnosis (Dx), CR, and relapse (Rel) is required to distinguish cell-types harboring the mutations and to determine its effect on gene expression profiling in the cells.
DNMT3A (DNA methyltransferase 3 alpha) encodes methyltransferase that methylates DNA (Okano et al., 1999) and forms a tetramer, regulating expression of genes involved in cell differentiation and development (Jia et al., 2007; Wu et al., 2012). The most common DNMT3A mutation, R882, blocks tetramer formation and inhibits enzymatic activity (Emperle et al., 2019; Russler-Germain et al., 2014; Yamashita et al., 2010). DNMT3A-mutant clones are considered as pre-leukemic (Dillon et al., 2021), as DNMT3A mutations that induce clonal hematopoiesis (CH) do not significantly affect the hematological phenotypes of normal individuals (Buscarlet et al., 2017) and DNMT3A mutations can persist after treatment during CR in AML patients (Garg et al., 2015; Jongen-Lavrencic et al., 2018). Nonetheless, CH with DNMT3A mutations is significantly associated with increased risk of hematologic malignancy (Genovese et al., 2014; Jaiswal et al., 2014), indicating that DNMT3A mutations occur in early stages of AML and eventually lead AML progression. In addition, studies showed that DNMT3A mutations are undetected during CR and reappears at Rel in some patients (Chien et al., 2017; Park et al., 2020), suggesting that DNMT3A-mutant cells can behave like leukemic clones. Indeed, studies demonstrated that Dnmt3a deficiency in mouse HSC/progenitor cells lose their differentiation ability but displayed enhanced self-renewal, thereby affecting AML pathogenesis (Challen et al., 2012; Mayle et al., 2015). DNMT3A mutation also showed proliferation advantage in HSCs (Shlush et al., 2014). Thus, further studies are required to elucidate the specific role of DNMT3A in AML relapse.
Here, we analyzed the single-cell transcriptome data of 29 BM samples from healthy donors and AML patients with different disease stages (Dx, CR, and Rel) to continuously observe transcriptional and mutational changes. In addition, targeted scRNAseq was performed to enhance detection of DNMT3A mutations. Our findings demonstrate that single-cell technology can dissect heterogeneity of HSCs and leukemic cells (LC) in AML and help elucidate the connection between AML relapse and DNMT3A mutation persistence in CR.
MATERIALS AND METHODS
Sample selection and cell preparation
Previously, we selected patients with DNMT3A R882 at the time of AML diagnosis with available BM samples (Ahn et al., 2018) (informed consent obtained). This study was approved by the Institutional Ethics Review Board at Chonnam National University Hwasun Hospital (No. CNUHH-2014-083). The treatments and sampling times of each patient are detailed in Supplementary Table S1. BM-derived mononuclear cells (BM-MNCs) were isolated from BM using density gradient centrifugation in Ficoll.
Targeted DNA sequencing
Genomic DNA was extracted from cryopreserved BM samples using a QIAamp DNA blood mini-kit (Qiagen, USA) according to the manufacturer’s protocol. Targeted deep sequencing was performed on 83 genes with previously reported recurrent driver mutations (Ley et al., 2013; Papaemmanuil et al., 2016). Agilent custom probes were designed to cover exons of the targeted genes and were sequenced using HiSeq 2000 (Illumina, USA). Variants were called as reported previously (Ahn et al., 2018).
Single-cell RNA sequencing
Cells were prepared using a LUNA-FLTM Automated Fluorescence Cell Counter (Logos Biosystems, Korea) according to the 10× Genomics Single Cell Protocols Cell Preparation Guide and the Guidelines for Optimal Sample Preparation flowchart (Documents CG00053 and CG000126, respectively). Libraries were prepared using Chromium controller according to the 10× Chromium Next GEM Single Cell 3’ v3.1 protocol (CG000315). Briefly, cell suspensions were diluted in nuclease-free water to achieve a target cell count of 10,000, mixed with master mix, and loaded into a Chromium Next GEM chip G with Single Cell 3’ v3.1 Gel Beads and Partitioning Oil. RNA transcripts from single cells were uniquely barcoded and reverse-transcribed within droplets. cDNAs were pooled and subjected to end repair, single ‘A’ base addition, and adapter ligation, before being purified and enriched using PCR to create the final cDNA library which was quantified according to the qPCR Quantification Protocol Guide (KAPA; Roche, Switzerland) and qualified using an 4200 TapeStation (Agilent Technologies, USA). Libraries were sequenced using a Hiseq platform (Illumina) according to the user guide.
Sequence alignment, quality control, clustering, and cell type annotation
FASTQ files were aligned to the human reference sequence (hg19) using CellRanger count pipeline (10× Genomics, ver. 3.0.2). The output gene-barcode matrices were imported into Seurat R package (v3.2.1) for scRNAseq data analysis and visualization. Cells with nFeature_RNA < 200 or > 25% mitochondrial genes were filtered out. After low quality cells were removed, data were normalized using the NormalizedData function with “LogNormalize” and scale factor 10,000. Highly variable features were identified using the FindVariableFeatures function with selection.method “vst”. Data were scaled using the ScaleData function. Once the Seurat objects of each sample were merged, batch effects were removed using the RunFastMNN function. Cells were clustered using FindNeighbors (reduction = “mnn”, dims = 1:30) with the FindClusters function (resolution = 1.8) based on the shared nearest neighbor. Datasets were visualized through non-linear dimensional reduction using Uniform Manifold Approximation and Projection (UMAP). To merge clusters with similar gene expression patterns, clusters were compared using the bimod test with the FindMarkers function (p_val_adj < 0.01, avg_logFC ≥ 1). Then, cluster pairs with < 10 differentially expressed genes (DEGs) were merged. Cluster cell types were annotated based on marker genes detected using the FindAllMarkers function (min.diff.pct = 0.3, log_fold change > 0.25).
Identification of cell type composition
To compare cell type composition by sample or disease stage (control [CON], Dx, CR, and Rel), 34 cell types were classified into ten major groups: HSC; LC; CD36+ LC; GMP1 and GMP2 (GMP, granulocyte-monocyte progenitor); Promono, monocytes, macrophages and dendritic cells (DCs) (mononuclear phagocyte system, MPS); EPC, pro-E to late ortho-E, and megakaryocyte (erythrocytes); lymphoid-primed multipotential progenitor (LMPP); pro B, B cells, and plasma cells (B cells); lymphoid cells, T cells, CD8 T, NK T cells, NK cells (T/NK cells); and Unassigned. The number of cells per group was subdivided by the number of cells per sample or disease stage.
Gene module analysis
To compare the expression of gene modules related to HSC and erythroid differentiation at each disease stage, the total number of unique molecular identifiers (UMIs) of gene module at each disease stage was normalized and log2 transformed. Gene modules were determined using previously identified gene sets (Addya et al., 2004; Alanazi et al., 2020).
Differential expression analysis
DEGs between HSCs, LCs, mutant cells, and wild-type (WT) cells were identified using MAST, which uses a hurdle model, through the FindMarkers function of Seurat. DEGs (|log_fold change| > 0.1, P < 0.05) were visualized using a volcano plot. HSC and LC clusters were compared to identify cell type-specific DEGs; next, a new set of DEGs was obtained by comparing each cluster to the remaining 33 clusters. The DEGs that overlapped in both DEG sets were displayed using a volcano plot. DNMT3A mutant and WT cells from AML samples (Dx, CR, and Rel) were also compared.
Gene set enrichment analysis (GSEA)
Enriched gene sets between two groups were identified using curated (c2) or ontology (c5) gene sets through GSEA (http://www.broadinstitute.org/gsea/index.jsp) with 1000 permutations and the Signal2Noise metric for ranking genes. Gene sets > 500 or < 5 were excluded from analysis. Enriched gene sets were filtered using P < 0.05 and false discovery rate < 0.25.
Single cell gene regulatory network analysis
Transcription factor activity and gene regulatory networks were identified using SCENIC (v1.1.2-01) (Aibar et al., 2017) with HSC, LC, and CD36+ LC raw count matrices extracted from the Seurat object. Cells within each cell type were classified according to disease stage (CON and CR, or Dx and Rel). For motif ranking, 10 kb around the transcription start site (TSS) and 500 bp upstream of the TSS were selected. Potential transcription factor targets and gene regulatory networks were inferred using GENIE3: genes were filtered using the default geneFiltering function and the activity of each regulon group was scored using the AUCell package. Cell type-specific regulators were identified based on the regulon specificity score and visualized using pheatmap. A scaled regulon activity score was used to compare the log2 fold-change of regulon activity between cell types.
VirtualKO
Virtual knockout analysis was performed using scTenifoldKnk (Osorio et al., 2022). Ribosomal and mitochondrial genes were excluded from the raw count matrix of LC cluster, and only genes expressed in more than 5% of total cells were used for analysis. Gene functional annotation and enrichment tests were performed using ‘Enrichr’ package (Kuleshov et al., 2016) with virtual KO perturbed genes. The results were visualized using ‘igraph’ package (https://github.com/igraph/rigraph).
Targeted DNMT3A sequencing for scRNAseq libraries
To improve detection of the DNMT3A R882 mutation, PCR was performed on cDNAs with a cell barcode (CB) and a UMI, including those from all CR samples and the AML02-Rel sample. We used primers that bound upstream of the mutation site (chr2:25457242 and 25457243) and to the 10× Read1 sequence for CB and UMI (Supplementary Table S2). During amplification (29 cycles), different index sequences were added to the primers near the mutation site to distinguish samples. Amplified cDNAs from each sample were pooled to generate a sequencing library for MiSeq.
DNMT3A-mutant cell identification
To detect DNMT3A-mutant cells (chr2:25457242 C>T or 25457243 G>T [for AML03]), sequencing reads with a mapping quality < 30 were filtered out and reads with both a CB and a Chromium molecular barcode were analyzed. For each read, DNMT3A-mutant cells were determined by identifying the mutation site and CB. Mutations were only identified in sequences in which the CIGAR string was M (alignment match). Cells with at least one read with mutations were labeled mutant cells. Those containing only reads without mutations were labeled WT cells.
Additional steps were required to identify mutant cells in the targeted sequencing data using only reads with both the 10× Read1 sequence and the sequence of the mutation site target primers. Reads with sequencing quality < 20 and those without a sample identification index sequence were filtered out. Only reads with a CB sequence matching the cells from the Seurat object were extracted. WT cells had a ‘G’ (chr2:25457242 C>T) or ‘C’ (AML03, chr2:25457243 G>A; CC47 chr2: 25457243 G>T) at the mutation site, whereas mutant cells had ‘A’ or ‘T’. Reads were discarded if any other base was found. Reads with the same UMI but different mutation site bases were also discarded. Cells with DNMT3A mutant reads were identified using their CB and sample index sequence. Because the number of mutation cover reads per cell was greatly improved in targeted sequencing compared to scRNAseq, the criteria for discriminating mutant cells were different from scRNAseq. Cells were labeled as mutant cells if they had more mutant reads than WT reads. We also labeled cells as mutant cells if the number of WT reads was less than 1.5-fold the number of mutant reads.
Copy number variation (CNV) analysis
To determine the cell type of clonal origin from CR to Rel, chromosomal copy number alterations were identified using InferCNV (https://github.com/broadinstitute/inferCNV) (Patel et al., 2014) with the raw count matrix of gene-cell containing read counts extracted from the Seurat object. All cells from three control samples were used as a normal reference. Mitochondrial genes were excluded from gene ordering files. In AML01, stem/progenitor cell-types from CR and Rel cells were analyzed. In AML02, stem/progenitor cell types from CR and LC and LMPP from Rel were analyzed. Parameters: cutoff = 0.1, denoise = TRUE, HMM = TRUE, and cluster_by_grpups = TRUE or FALSE. Cell information of inferCNV outputs was identified using infercnvNGCHM.
Nanopore sequencing
To determine the DNMT3A mutation in three relapse-origin cells in AML02, we performed long-read Nanopore sequencing on the scRNAseq library. cDNAs from these three cells were amplified independently using primers designed for the 10× Read1 sequence and CB (forward) and the DNMT3A mutation site (chr2:25457242 and 25457243; reverse). The MinION Kit protocol (SQK-LSK109; Oxford Nanopore Technologies, UK) was run for 43 m on a MinION sequencing device. Data were generated using MinKNOW 20.10.3 software. Bases were called using Guppy (v.4.2.2) and sequenced reads were aligned to the human reference genome (hg19) using minimap2.
To call DNMT3A variants in relapse-origin cells in AML02, we used a custom python script in the pysam Python package with biobookshelf (https://github.com/ahs2202/biobookshelf). Reads with an average mapping quality < 60 or mutation site base quality < 20 were filtered out. The mutation site base was identified in reads with a 10× Read1 sequence with a maximum allowed error rate of 0.2 (including substitutions, insertions, and deletions) and a CB for three cells with a maximum allowed error rate of 0.3. Reads with a mutation site base other than WT or mutation were filtered out. If both WT and mutant bases were identified in reads with the same CB and UMI, all reads with the same UMI were filtered out.
Trajectory analysis
Cells from relapsed patients were pseudotemporally ordered from CR to Rel using the Monocle R package (v2.14.0) (Trapnell et al., 2014): in AML01, GMP1 at CR and GMP1 and LC at Rel were analyzed, whereas in AML02, LMPP and HSC at CR and LMPP and LC at Rel were analyzed. CellDataSet objects for Monocle were created from raw count data using the newCellDataSet function (expressionFamily = negbinomial.size). The estimateSizeFactors and estimateDispersions functions were used to normalize mRNA differences across cells and analyze DEGs. Cells were ordered along a pseudotime trajectory using the reduceDimension and orderCells functions. Genes expressed in at least ten cells were used to identify DEGs that changed with pseudotime. Genes clustered based on pseudotemporal expression were visualized using the plot_pseudotime_heatmap function. Gene ontology analysis of biological processes was performed for highly variable genes at each stage using DAVID (Huang et al., 2009).
Statistics and reproducibility
When comparing cell type composition by disease stage, the P value was determined by two proportion test using prop.test function in R. In the Gene module analysis, P values were determined using the Wilcoxon rank-sum test, two-sided. All analyses are reproducible using raw data with Materials and Methods section.
Data sharing statement
The raw data was deposited in Korean Nucleotide Archive (KoNA; https://kobic.re.kr/kona) with the accession ID, PRJKA220155.
RESULTS
Identification of cell compositions in BM samples from patients with AML
To explore the cellular heterogeneity in AML, BM-MNCs were obtained from three healthy donors and six patients with AML (Fig. 1A). During long-term follow-up (median, 9.2 years), three patients with AML relapsed (AML01, AML02, and AML05) and 3 (AML03, AML04, and AML06) maintained CR after intensive chemotherapy. Samples were obtained at the Dx, CR, and Rel stages from two patients who relapsed (AML01 and AML02). Dx and CR samples were obtained from two non-relapse patients (AML03 and AML04). Only CR samples were obtained from the other patients (AML05 and AML06; Supplementary Table S1).
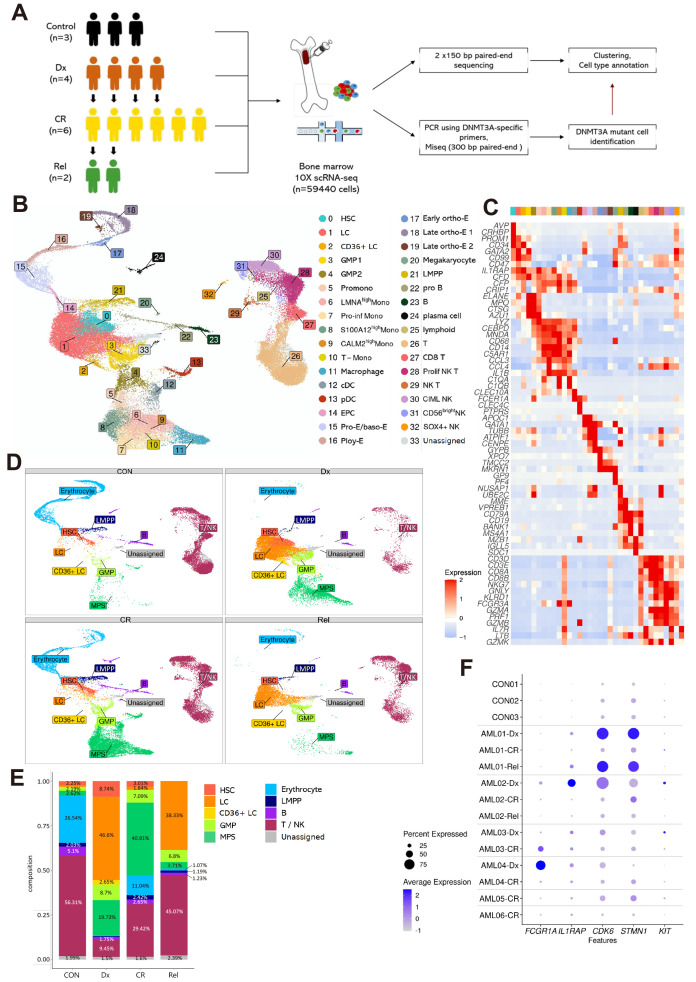
Fig. 1. Single-cell RNA sequencing (scRNAseq) analysis of patients with acute myeloid leukemia (AML) reveals differences in cell-type proportions with disease stage.
(A) Schematic workflow of the study. BM-MNCs (bone marrow-derived mononuclear cells) collected from three healthy donors and six patients with AML at different disease stages (Dx, CR, and Rel) were subjected to scRNAseq. Control (CON), healthy donor; Dx, diagnosis; CR, complete remission; Rel, relapse. (B) UMAP visualization of total cells. Colors indicate 34 cell-types. HSC, hematopoietic stem cell; LC, leukemic cell; GMP, granulocyte-monocyte progenitor; Promono, promonocyte; Mono, monocyte; cDC, conventional dendritic cell; pDC, plasmacytoid dendritic cell; EPC, erythrocyte precursor cell; Pro-E, proerythroblast; baso-E, basophilic erythroblast; Poly-E, polychromatophilic erythroblast; ortho-E, orthochromatic erythroblast; LMPP, lymphoid-primed multipotential progenitor. (C) Heatmap showing the expression of cell-type specific markers used for annotation. Each column represents a cluster (0-33). (D) UMAP visualization showing the composition of ten cell-type groups according to disease stage (CON, Dx, CR, and Rel). GMP, cluster 3-4; MPS (mononuclear phagocyte system), cluster 5-13; Erythrocyte, cluster 14-20; B, cluster 22-24; T/NK, cluster 25-32. (E) Relative cell-type proportion according to disease stage. Samples were grouped by disease stage, not by patient. The number above the graph indicates the percentage of the proportion. The Proportion percentage, which was less than 1%, were not indicated. (F) Dot plot showing expression of known AML marker genes (FCGR1A, IL1RAP, CDK6, STMN1, and KIT) between samples. Dot size represents the percentage of cells expressing the genes within the samples. Blue and white represent high and low expression, respectively.
After removing low-quality cells, the remaining 59,440 cells were grouped into 34 clusters, annotated with marker genes (Figs. 1B and 1C, Supplementary Table S3). As some clusters were not shared between samples (Supplementary Fig. S1A), the 34 cell-types were divided into 10 larger groups for overall comparison of cell-type composition (Supplementary Fig. S1B). BM-MNCs from healthy donors (controls) had similar cell-type proportions; however, the cell-type composition differed significantly among patients with AML and among disease stages in the same patient. For instance, AML01 and AML02 had higher proportions of LCs and lower proportions of T/NK cells at Dx than at CR (nominal P < 2.2e-16 by two proportion test). At Rel, the proportion of LCs was higher than that at CR in both AML01 and AML02, but was similar to that at Dx in AML01 and remarkably lower than that at Dx in AML02 (LCs in AML02 CR: 0.4%, nominal P < 2.2e-16 by two proportion test). In addition, the cell compositions at Dx and Rel differed significantly from those at CON and CR (Figs. 1D and 1E). The proportion of LCs was remarkably higher at Dx and Rel than at CON and CR, whereas the proportion of erythrocytes was lower at Dx and Rel (LCs at CON: 0.93%, erythrocytes in Dx: 0.46%, nominal P < 2.2e-16 by two proportion test). The proportion of HSCs was the lowest at Rel (CON: 2.2, Dx: 8.7, CR: 3.0, Rel: 0.23%, nominal P < 2.2e-16 by two proportion test). These results highlight the heterogeneity of AML at the cellular level and show the temporal dynamics of cell populations during AML progression.
We investigated whether disease stages could be represented by genes that are reported to be highly expressed in myeloid leukemia (Fig. 1F) (Handschuh et al., 2018; Landberg et al., 2016; Liu et al., 2021; Scolnik et al., 2002; Xu and Guo, 2020). These genes were identified via bulk RNAseq of BM and blood (STMN1, KIT, and CDK6), The Cancer Genome Atlas Program (FCGR1A and CDK6), or RNAseq of leukemic stem cells (KIT). The expression of IL1RAP, CDK6, and STMN1 was upregulated at Dx and Rel, and downregulated at CR in AML01, but this pattern was not observed in other relapsed sample, AML02. FCGR1A expression was high at Dx and low at CR in AML04, but it displayed the opposite trend in AML03. Although KIT expression was slightly higher at Dx in AML02 and AML03, a small number of cells expressed KIT in other patients and at Rel in AML02, similar to the controls. The cell types commonly detected in all samples were HSCs, LCs, GMP2 cells, LMPPs, T cells, CD8 T cells, proliferating NK T cells, CIML (cytokine-induced memory-like) NK cells, and CD56 bright NK cells. We identified the expression of markers of these cell types in each sample (Supplementary Fig. S2A). The expression of genes in HSCs and LCs showed distinctive patterns at Dx and Rel in some patients. However, it was difficult to distinguish their expression between Dx and CR in most patients. This result showed that these genes known to be overexpressed in myeloid leukemia cannot distinguish disease stages with our data. We found some genes with consistently higher expression at Dx and Rel than at CR. The expression of CLEC11A, HOMER3, FAM101B, and ATP8B4 was lower at CR than at Dx and Rel in all patients, although their expression was also low at Rel in AML02, probably because of the heterogeneity of patients with AML (Supplementary Fig. S2B). Analyses of the expression of these genes in LCs revealed that the expression of HOMER3, FAM101B, and ATP8B4 was also high at Rel in AML02 (Supplementary Fig. S2C). Although more data analysis and further experiments are needed for validation, we suggest these genes as markers that distinguish CR from Dx and Rel.
Transcriptional landscape of LC and HSC heterogeneity
HSCs and LCs were not only closely located in the UMAP projection, but also initially clustered together based on similar transcriptome patterns before being subclustered into HSCs and LCs. Although HSC and LC populations were close to each other, they exclusively expressed some of their cell-type specific markers (HSC markers: AVP and CRHBP; LC marker: CD99) (Figs. 2A and 2B). Interestingly, the proportion of HSCs was higher at Dx than at CON, whereas only a few HSCs were found at Rel. To determine whether HSCs at Dx differed from those at CON or CR, we compared the expression of HSC markers at each disease stage (Fig. 2C). HSCs showed the highest expression of HSC markers at CON; whereas at Dx, HSCs expressed CD34 but rarely expressed AVP and CRHBP. Furthermore, HSCs at Dx displayed decreased expression of genes related to erythrocyte development (GYPC, HBA1, HBA2, and HBB) compared to the CON and CR (Fig. 2D, Supplementary Fig. S3A). These results suggest that despite their abundance, HSCs at Dx in AML are abnormal.
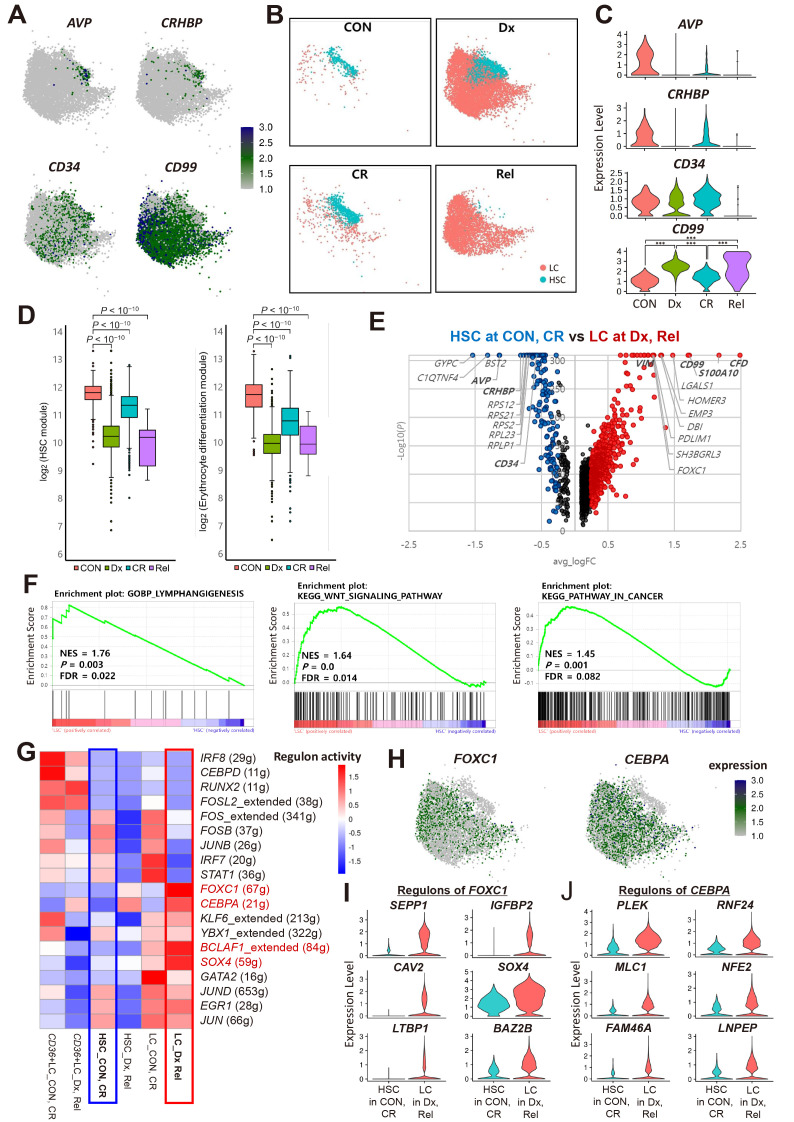
Fig. 2. Transcriptional landscape of leukemic cells (LCs) and hematopoietic stem cells (HSCs).
(A) Expression of known HSC and LC marker genes in HSC and LC. AVP, CRHBP, and CD34 for HSC, CD99 for LC. CON, healthy donor; Dx, diagnosis; CR, complete remission; Rel, relapse. (B) UMAP visualization showing the distribution of HSCs and LCs by disease stage. (C) Violin plot showing the expression of known HSC (AVP, CRHBP, CD34) and LC (CD99) marker genes in HSCs by disease stage. ***P < 0.001 by Wilcoxon rank-sum test using FindMarkers function. (D) Expression of HSC and erythrocyte differentiation gene modules in HSCs by disease stage. P values were determined using the Wilcoxon rank-sum test, two-sided. (E) DEGs (differentially expressed genes) in LCs at Dx and Rel vs HSCs at CON and CR. Red dots represent upregulated genes in LCs at Dx and Rel. Blue dots represent upregulated genes in HSCs at CON and CR. Black dots represent genes with a log2 fold change < 0.25. Known markers of each cell-type are shown in bold. (F) GSEA (gene set enrichment analysis) plots showing enrichment of cancer or stem cell associated gene sets in LCs at Dx and Rel compared to HSCs at CON and CR. NES, normalized enrichment score; P, P value; FDR, false discovery rate. (G) Heatmap showing the regulon activities of transcription factors (TFs) in HSCs, LCs, and CD36+ LCs divided by disease stage (Dx and Rel; CON and CR). Parentheses next to transcription factors indicate the number of target genes of the transcription factors. Red represents higher regulon activity. Red text indicates TFs that were overexpressed in LCs at Dx and Rel compared to HSCs at CON and CR. (H) Expression of the top two TFs in LCs at Dx and Rel compared to HSCs at CON and CR. (I and J) Violin plots showing the expression of the top six target genes of FOXC1 (I) or CEBPA (J) with higher expression in LCs at Dx and Rel compared to HSCs at CON and CR.
We then characterized the transcriptome of LCs and HSCs. LCs are disease-specific cells that originate from progenitor cells. Studies comparing HSCs and LCs have been conducted previously to identify LC characteristics and find treatment targets (Eppert et al., 2011; Sachs et al., 2020; Schuurhuis et al., 2013; Thakral et al., 2023). We also aimed to identify differences in the transcriptome between HSCs and LCs. The DEGs between them included known markers (CD34 and AVP for HSCs; S100A10, CD99, and VIM for LCs; Supplementary Fig. S3B). Meanwhile, the GSEA revealed that gene sets related to lymphangiogenesis, Hedgehog and WNT signaling, and dedifferentiation were enriched in LCs (Supplementary Fig. S3C). As the HSCs at Dx were abnormal, LCs at Dx and Rel were further compared with HSCs at CON and CR. In the DEG analysis, similar genes were identified with DEGs in all HSCs and LCs, but the log2 fold-change value increased (Fig. 2E). In the GSEA, gene sets related to lymphangiogenesis, WNT signaling pathway, and pathway in cancer were enriched in LCs at Dx and Rel (Fig. 2F). In addition, gene sets related to NOTCH signaling, self-renewal, and leukemia were identified in addition to the terms obtained when comparing all HSCs and LCs (Supplementary Fig. S3D). Gene sets related to the differentiation of T cells and macrophage-derived foam cells and reversible differentiation were enriched in HSCs (Supplementary Figs. S3E and S3F).
Next, single-cell regulatory network inference and clustering was used to identify gene regulatory networks in LCs compared with those in normal HSCs. The regulon activity of transcription factors (TFs) FOXC1 and CEBPA was higher in LCs at Dx and Rel than HSCs at CON and CR (Fig. 2G). CEBPA and FOXC1 also showed a higher regulon activity in HSCs at Dx and Rel than at CON and CR, suggesting that they may affect HSCs in patients with AML (Fig. 2H). Additionally, representative target genes of FOXC1 and CEBPA showed increased expression in LCs at Dx and Rel compared with those in HSCs at CON and CR (Figs. 2I and 2J). We then performed virtual KO analysis of FOXC1 and CEBPA to assess their potential effect on LCs (Supplementary Fig. S4). The results indicated that they negatively regulate the expression of CD34 and GYPC, which is involved in erythrocyte development, whereas they upregulate the expression of CD99 and malignant tumor-related TF HOXB3 (Sauvageau et al., 1997). In addition, both FOXC1 and CEBPA regulate genes involved in the BDNF signaling pathway and prostaglandin biosynthesis, which are related to cancer cell migration, proliferation, and adhesion (Meng et al., 2019; Menter and DuBois, 2012).
To verify the presence of abnormal HSCs at Dx and to compare the transcriptomes between HSCs and LCs, we analyzed a public scRNAseq dataset (GSE227903). As it is challenging to find only normal HSCs in the samples of patients with AML, we analyzed these data together with our data of three healthy controls. First, HSCs and LCs were identified and changes in their cell proportion were examined at different disease stages (Supplementary Figs. S5A-S5C). Similar to results in our data, there were many HSCs and LCs at Dx, but HSCs at Dx rarely expressed AVP and CRHBP (Supplementary Fig. S5D). There were only a few HSCs at MRD (Supplementary Fig. S5C). These HSCs at MRD rarely expressed AVP and CRHBP, similar to HSCs from Dx (Supplementary Fig. S5D). DEGs between HSCs at CON and LCs at Dx and Rel included their marker genes, consistent with our results (Supplementary Fig. S5E). The GSEA results showed that gene sets related to the regulation of lymphangiogenesis, WNT signaling pathway, and pathways in cancer were enriched in LCs at Dx and Rel compared with those in HSCs at CON (Supplementary Fig. S5F). In addition, FOXC1 and CEBPA were exclusively expressed in LCs at Dx and Rel (Supplementary Fig. S5G).
Identification of DNMT3A-mutant cell-types and signatures
Along with scRNAseq, targeted DNA sequencing was performed to identify the DNMT3A variant allele frequency (VAF) at CR (Supplementary Table S4). Two relapsed patients (AML01 and AML05) and one non-relapsed patient (AML03) had low DNMT3A mutation frequencies at CR (Fig. 3A), whereas one relapsed patient (AML02) and two non-relapsed patients (AML04 and AML06) retained high DNMT3A mutation frequencies at CR (>20%). To determine the difference between patients at CR with low and high VAF at the cellular level, DNMT3A mutations were called and cell-types were identified. As scRNAseq has dropout events in which a transcript is technically undetected despite its presence in a cell, this analysis has limitations when focused on a specific gene. We enhanced variant detection in individual cells by performing targeted sequencing on scRNAseq libraries targeting DNMT3A R882 (Fig. 3B, Supplementary Table S2). Targeted scRNAseq data dramatically improved the number of reads covering DNMT3A R882 per cell (Fig. 3C, Supplementary Fig. S6A). Moreover, the number of cells covering the mutation site increased by 2-4 times (Supplementary Fig. S6B). DNMT3A VAF obtained from scRNAseq data were overall similar to those obtained from targeted DNA sequencing (Supplementary Fig. S6C).
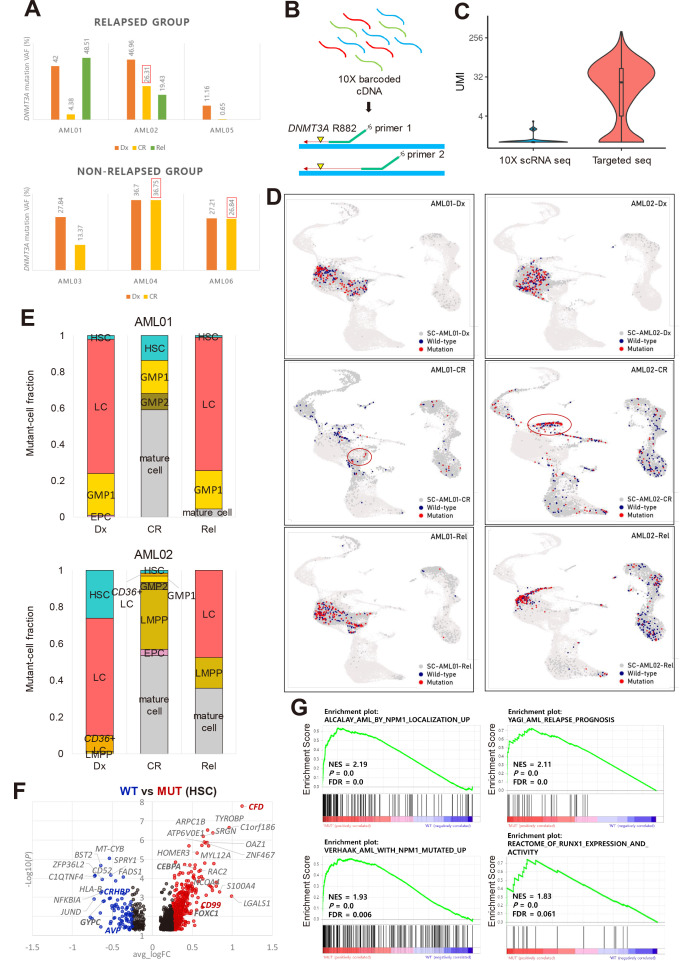
Fig. 3. Identification of DNMT3A-mutant cell-type and signatures.
(A) Targeted DNA sequencing results showing that AML02 (relapsed) and AML04 and AML06 (non-relapsed) have a high DNMT3A VAF at CR. AML, acute myeloid leukemia; VAF, variant allele frequency; CR, complete remission. (B) Simple description of targeted sequencing process. Single cell barcoded cDNAs were amplified using mutation site-specific primers and a custom Python script was used to identify DNMT3A-mutant cells with at least one mutant read. (C) Unique molecular identifiers (UMIs) per cell at the DNMT3A mutation locus from all CR samples and Rel samples from AML02. (D) Plot showing DNMT3A-mutant and wild-type (WT) cells projected onto the UMAP cluster from AML01 and AML02 according to disease stages. Red, DNMT3A-mutant cell; navy, DNMT3A WT cell; dark gray, no DNMT3A coverage. Red circle indicates the cell-type with the highest mutant-cell fraction at CR: GMP1 for AML01, LMPP for AML02. (E) Fraction of DNMT3A-mutant cells in each cluster at different disease stages in AML01 and AML02. Mutant cell fraction: number of mutant cells in each cell-type divided by the total number of mutant cells in each sample. Mature cell-types excluding stem/progenitor cell-types were merged. HSC, hematopoietic stem cell; LC, leukemic cell; GMP, granulocyte-monocyte progenitor; EPC, erythrocyte precursor cell; Dx, diagnosis; Rel, relapse; LMPP, lymphoid-primed multipotential progenitor. (F) DEGs (differentially expressed genes) between DNMT3A-mutant and WT cells in HSC of AML samples. Red dots indicate upregulated genes in DNMT3A-mutant cells. Blue dots indicate downregulated genes. Black dots indicate genes with a log2 fold change < 0.25. Genes of interest are marked in bold. (G) GSEA (gene set enrichment analysis) plots of unregulated gene sets in DNMT3A-mutant cells compared to WT cells. NES, normalized enrichment score; P, P value; FDR, false discovery rate.
Having identified cells with DNMT3A R882, we investigated their distribution in each patient. As undifferentiated cells accumulate in AML, differentiated cell-types were grouped as mature cells to focus on stem/progenitor cell-types. As only CR samples were obtained from AML05 and AML06, changes in mutant cell distribution could not be identified in these patients. First, we analyzed changes in the distribution of DNMT3A-mutant cells from Dx to Rel in relapsed patients AML01 and AML02. In both patients at Dx, the fraction of DNMT3A-mutant cells was the highest in LCs (Figs. 3D and 3E); however, the mutant LCs almost disappeared at CR and reappeared at Rel (Figs. 3D and 3E). In AML01, the proportion of mutant GMP1s at Dx was also high and persisted at CR and Rel although the actual number of mutant GMP1s was low at CR (Figs. 3D and 3E). In AML01, the DNMT3A VAF decreased at CR and increased again at Rel, following a dynamic pattern similar to that of leukemic burden suggesting that mutant GMP1s remaining during CR may represent residual LCs. In AML02, the total number of DNMT3A-mutant cells at CR was similar to that at Dx and Rel (Fig. 3D). AML02 had a high proportion of mutant LMPPs at CR compared to other patients, whereas mutant LMPPs were rarely present at Dx (Figs. 3D and 3E, Supplementary Figs. S7A and S7B) and persisted until Rel while spreading to adjacent LCs on UMAP (Fig. 3D). Thus, in a patient with persistent DNMT3A mutation post-treatment, the DNMT3A mutation persisted mainly in a pre-leukemic state in LMPPs and mature cells.
Next, we examined the distribution of DNMT3A-mutant cells at Dx and CR in non-relapsed patients (AML03, AML04, and AML06). In AML03, there were similar proportions of mutant HSCs, LCs, and GMP1s at Dx; whereas at CR, mutant cells were mainly HSCs and monocytes, not GMP1s (Supplementary Figs. S7A and S7B). Although AML04 and AML06 had mutant LMPPs at CR, no mutant LMPPs were detected at Dx in AML04 as in AML02 (Supplementary Figs. S7A and S7B), suggesting that patients with a high DNMT3A VAF at CR (AML02, AML04, and AML06) may have a high proportion of mutant cell-types not found at Dx, especially LMPPs.
Having distinguished mutant and WT cells through single-cell sequencing, we investigated the specific cellular effects of DNMT3A mutation. The expression of LC markers (CD99 and CFD) and TFs with high activity in LCs (CEBPA and FOXC1) was upregulated in DNMT3A-mutant cells (Fig. 3F). Furthermore, the GSEA revealed that the mutant cells were enriched for gene sets related to AML- and NPM1-mutated signatures (Fig. 3G, Supplementary Fig. S7C). DNMT3A-mutant cells were also enriched for AML relapse-related gene sets, such as AML relapse prognosis and reactome of RUNX1 expression and activity, and showed higher expression of RUNX1 (Figs. 3F and 3G). These results suggest that DNMT3A mutant cells have gene signatures related to AML and LC maintenance and may therefore be involved in AML relapse.
We then explored the distribution of DNMT3A-mutant GMP1s in more detail (Supplementary Fig. S8A). GMP1s were divided into three subclusters: progenitor marker (GATA2) high, myeloblast to promyelocyte stage gene (PRTN3) high, and proliferation marker (TOP2A) high (Supplementary Figs. S8A and S8B). In AML01, three of the four DNMT3A-mutant GMP1s found at CR were in the progenitor marker high subcluster (Supplementary Fig. S8C); however, no mutant GMP1s were found at CR in AML03, a non-relapsed patient with a low DNMT3A VAF at CR. To further explore the distribution of mutant LMPPs in patients with a high DNMT3A VAF at CR, we subclustered LMPPs into progenitor B cell marker (VPREB1) high and myeloid progenitor marker high (FLT3 and MPO) clusters (Supplementary Fig. S8D and S8E). Overall, the number of DNMT3A-mutant cells was the highest at CR in AML02 (relapsed) compared with that in AML04 and AML06, and DNMT3A-mutant cells were evenly distributed in LMPP subclusters (Supplementary Fig. S8F).
Identification of relapse-related clones in patients with relapsed AML
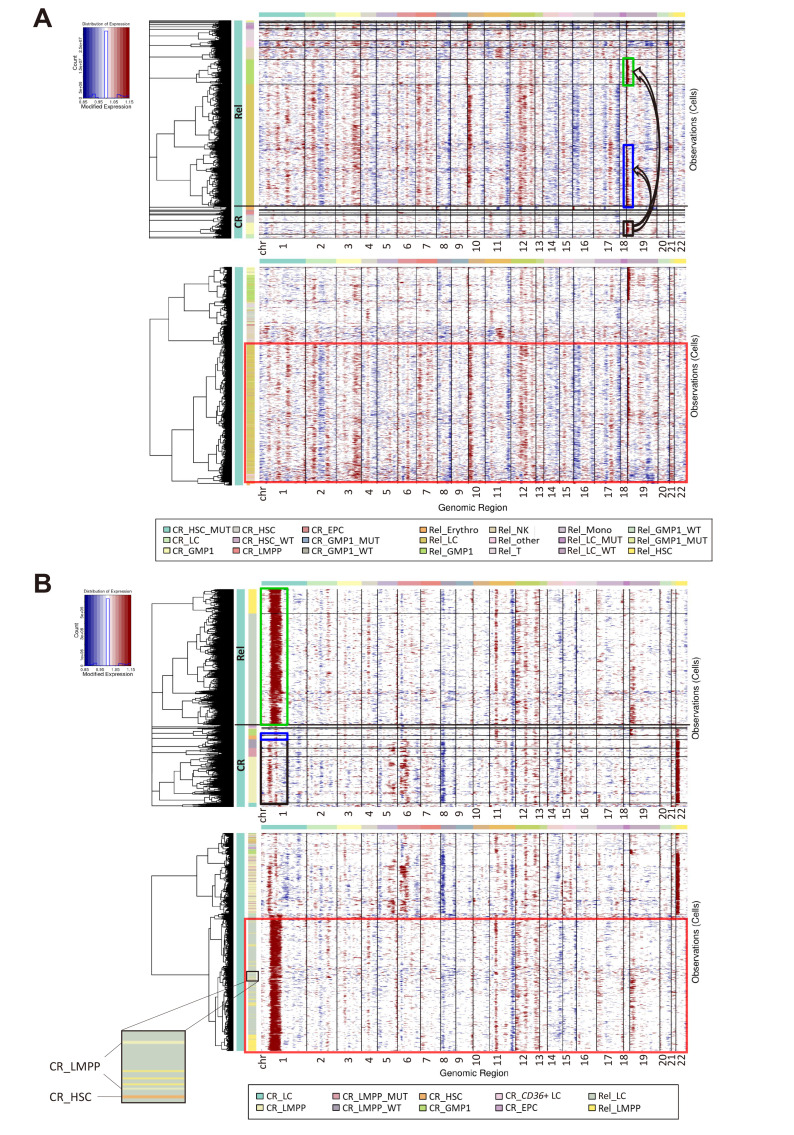
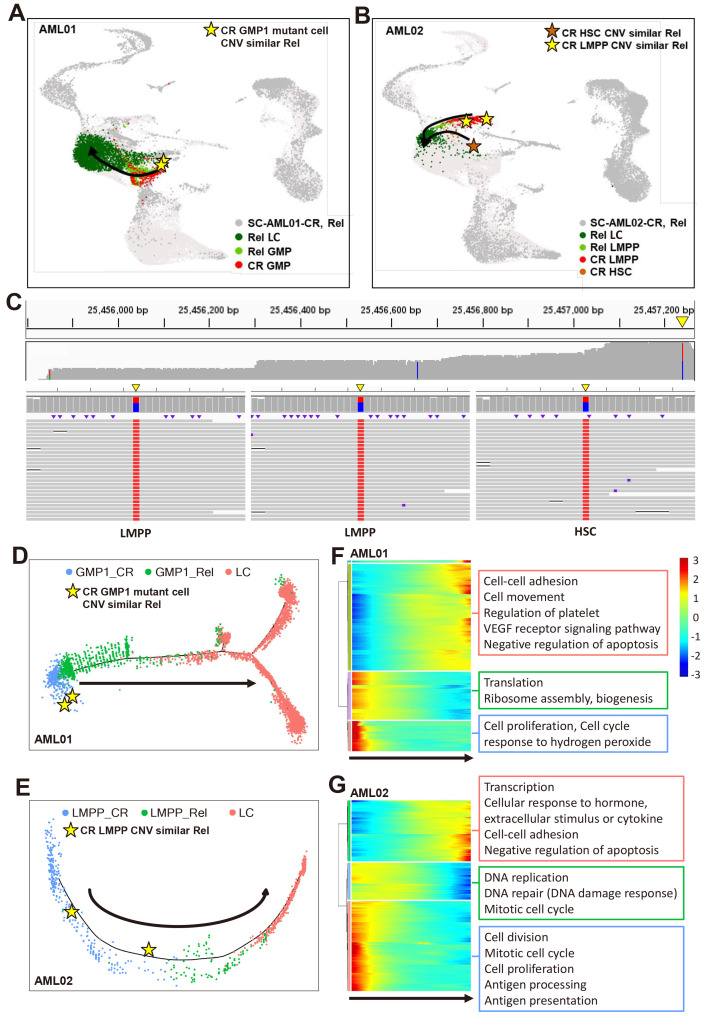
As we identified the progenitor cell-types in which DNMT3A mutations persisted at CR in AML, we examined whether these cell-types lead to relapse by performing single-cell CNV analysis. An amplified region in chr19 was identified in GMP1s at CR in AML01 (Fig. 4A, top, black box), and GMP1s showed similar CNV patterns at Rel (Fig. 4A, top, green box). Some LCs were also amplified in a specific chr19 region at Rel similar to GMP1s at CR (Fig. 4A, top, blue box). As clones originated from the same cell have similar CNV patterns (Erickson et al., 2022), we clustered cells based only on the CNV patterns to determine whether the LCs at relapse originated from cells at remission. Clustering according to CNV patterns revealed that some CR cells clustered with LCs at Rel (Fig. 4A, bottom, red box), including GMP1s, LCs, LMPPs, and CD36+ LCs (number of cells clustered with LCs at Rel, GMP1: 14, LC: 7, LMPP: 3, CD36+ LC: 1). In particular, GMP1s had a similar CNV pattern at CR to LCs at Rel and clustered at the highest ratio, including some DNMT3A-mutant cells (2/14 GMP1). Thus, the DNMT3A-mutant LCs and GMP1s detected at Rel in AML01 may originate from GMP1s at CR (Fig. 5A).
Fig. 4. Identification of clones involved in relapse in patients with relapsed acute myeloid leukemia (AML).
(A and B) InferCNV heatmap of CR and Rel in AML01 (A) or AML02 (B). Top: cells grouped by cell-type labeled in different colors. Bottom: cells grouped according to CNV (copy number variation) patterns. Chromosomal copy number alterations (red, amplification; blue, deletion) inferred in each chromosomal location (columns) across cells (rows). Datasets from three healthy donors were used as a control. Partial enlarged view in bottom left shows LMPPs and HSCs at CR clustered with LCs at Rel in AML02. CR, complete remission; Rel, relapse; HSC, hematopoietic stem cell; LC, leukemic cell; GMP, granulocyte-monocyte progenitor; EPC, erythrocyte precursor cell; LMPP, lymphoid-primed multipotential progenitor; WT, wild type.
Fig. 5. Identification of transcriptional changes along the cell trajectory towards leukemic cells (LCs).
(A and B) UMAP visualization of mutant cell-types in AML01 (A) or AML02 (B) showing the expected direction from CR to Rel. Star-shaped dots represent cells at CR with a similar CNV to LCs at Rel. AML, acute myeloid leukemia; CR, complete remission; GMP, granulocyte-monocyte progenitor; CNV, copy number variation; Rel, relapse; LC, leukemic cell; LMPP, lymphoid-primed multipotential progenitor; HSC, hematopoietic stem cell. (C) Integrative genome viewer of Nanopore sequencing reads for relapse-origin cells in AML02. Top: coverage of Nanopore sequencing for DNMT3A. Bottom: raw Nanopore sequencing reads mapped to mutation site in two LMPPs and one HSC. Yellow triangle indicates the position of DNMT3A 24257242. Wild type (WT) or mutation sites (DNMT3A 25457242) are indicated by color. Blue, WT (base: C); red, mutation (base: T); black, deletion; purple, insertion; white, non-covered region. (D and E) Trajectory analysis of LCs and cell-types, including relapse-origin cells in AML01 (D) and AML02 (E). GMP1 for AML01, LMPP for AML02. Arrow indicates the direction from CR cells to LCs at Rel. Star-shaped dots represent cells at CR with a similar CNV to LCs and LMPPs at Rel. (F and G) Heatmap of top 500 highly variable genes (rows) clustered based on the pseudotemporal expression pattern in AML01 (F) or AML02 (G). Bottom arrow indicates the direction from CR cells to LCs at Rel.
In AML02, the CNV analysis identified a strongly amplified region in chr1 in LCs and LMPPs at Rel (Fig. 4B, top, green box) and in a few stem/progenitor cells at CR (Fig. 4B, top, blue and black box). However, most cells at CR had an amplified region in chr22 that was not found at Rel, suggesting that only some cells without chr22 amplification at CR could be relapse-origin cells. Therefore, we clustered cells based on CNV patterns to identify the CR cells that persisted and expanded at Rel (Fig. 4B, bottom, red box). Almost all LCs and LMPPs at Rel clustered together with a few HSCs and LMPPs at CR owing to a similar CNV pattern (Fig. 4B, bottom, partial enlarged view), but did not include other CR stem/progenitor cells. Thus, the LCs and LMPPs at Rel in AML02 appear to originate from LMPPs and HSCs at CR (Fig. 5B).
As even targeted scRNAseq did not cover the DNMT3A R882 site in these cells, we performed additional targeted sequencing to confirm whether these cells had the DNMT3A mutation. The LMPPs and HSCs at CR with a CNV pattern similar to those of cells at Rel had both WT and mutant reads, confirming that they were DNMT3A-mutant cells (Fig. 5C). Thus, DNMT3A mutations were identified in LMPPs and HSCs in AML02 at CR (relapsed, high DNMT3A VAF in CR) that were not originally discovered at Dx, with CNV analysis confirming that these LMPPs and HSCs likely produce clones at Rel.
Identification of transcriptional changes from CR to Rel
From our results, we predicted that cells at relapse may originate from GMP1s and LMPPs at CR. Despite several AML studies, the progression from CR to Rel has not been fully elucidated. Therefore, our aim was to identify the transcriptional changes that occur in cells as the disease progress from CR to cells at Rel, particularly LCs. In AML01, a relapsed patient with a low DNMT3A VAF at CR, transcriptional changes were observed when GMP1s (containing relapse-origin cells) became LCs (Fig. 5D). As expected, GMP1s were ordered in trajectory from CR to Rel and finally to LCs. We selected CR cells with an earlier actual time as the starting point. At the trajectory starting point, DNMT3A-mutant GMP1s (relapse-origin) were identified. In AML02, a relapsed patient with a high DNMT3A VAF at CR, the trajectory started with LMPPs, which are involved in relapse, at CR, and ended with LCs (Fig. 5E). In both cases, we identified highly variable genes along the trajectory and associated biological pathways. In AML01, the expression of genes associated with cell proliferation and translation decreased from GMP1s at CR compared with those at Rel and in LCs (Fig. 5F). Conversely, the expression of cell–cell adhesion and VEGF receptor signaling pathway-related genes was upregulated as GMP1s proceeded to develop into LCs. In AML02, the expression of genes related to cell division and antigen presentation decreased, whereas that of genes related to transcription and cell–cell adhesion increased from LMPPs at CR to LMPPs at Rel and in LCs (Fig. 5G). In both relapsed patients, the expression of proliferation and cell cycle-related genes decreased during the early stages of progression from CR to Rel, whereas the expression of genes related to cell–cell adhesion and negative apoptotic regulation increased towards LCs, suggesting these biological pathways influence LC formation.
Finally, we analyzed scRNAseq data from six additional patients (Dx 6, CR 6, and Rel 2) to cross validate our results (Supplementary Fig. S9A). A comparison of LCs and HSCs in the additional data showed DEGs and gene sets similar to those identified in Fig. 2 such as Notch signaling and T cell differentiation (Supplementary Figs. S9B-S9D). Furthermore, a comparison between DNMT3A-mutant and WT cells confirmed that gene sets “AML relapse prognosis” and “AML with NPM1 mutated up” were enriched in DNMT3A-mutant cells (Supplementary Fig. S9E). Among the additional patients, two relapsed patients (CC47 and CC70) continued to show high in DNMT3A VAF at CR, similar to AML02. At CR, DNMT3A mutations in these patients were highly enriched in mature cells, but also detected in HSCs, LCs, progenitors, GMPs and LMPPs (Supplementary Figs. S9F and S9G). Through CNV analysis, we confirmed that some progenitor cells in CC47 (number of cells clustered with LCs at Rel, progenitor: 8, HSC: 1, EPC: 3, GMP1: 1, LMPP: 2, LC: 5) and HSCs and EPCs at CR in CC70 (HSC: 4, EPC: 6, progenitor: 3, LMPP: 8, LC: 9) had CNV patterns similar to those of LCs at Rel (Supplementary Fig. S10). Therefore, we expected that DNMT3A-mutant cells remaining in these cell-types during remission might be involved in relapse. This indicates that AML is highly heterogeneous among individuals.
DISCUSSION
AML is a heterogeneous disease caused by distinctive mutations in each patient that may result in different cell-type compositions. In this study, using scRNAseq, we characterized the heterogeneity of HSCs and LCs in AML, examined their proportion changes in different AML stages, and identified the TFs that may govern the cell-type identity of HSCs and LCs. Moreover, we characterized the effect of the DNMT3A mutation in AML and its association with relapse. Targeted sequencing and CNV analysis at the single-cell level indicated that DNMT3A-mutant GMPs and LMPPs at CR could preclude relapse in different ways.
Advances in single-cell technology have improved our understanding of heterogeneous AML at the cellular level (Ediriwickrema et al., 2020; Paguirigan et al., 2015; Pellegrino et al., 2018; Petti et al., 2019; Povinelli et al., 2018; van Galen et al., 2019). Droplet-based scRNAseq, can only sequence the 3' end of transcripts and produces a sparse gene expression matrix, limiting the detailed analysis of mutation-harboring cells. To improve detection of the DNMT3A mutation, we performed targeted scRNAseq for DNMT3A. Several studies have noted that DNMT3A mutations increase with aging but has only limited biological impact in normal individuals (Buscarlet et al., 2017). However, we found that DNMT3A-mutant cells were enriched for “AML relapse prognosis” and “AML with NPM1 mutated up”, consistent with a previous study that reported the frequent co-occurrence of DNMT3A and NPM mutations (Loghavi et al., 2014), as observed in the four patients in our study. Although it may not be surprising that mutant cells are rich in disease-associated gene sets, we could accurately identify the effects of DNMT3A mutations by comparing mutant cells to WT cells rather than comparing bulk levels in patients with and without DNMT3A mutations. In addition, DNMT3A-mutant cells had higher expression of RUNX1, which is involved in LC maintenance and negatively correlated with survival of AML patients, than WT cells (Na et al., 2020; Wesely et al., 2020).
Previous studies defined LCs based on several surface markers and compared LCs and HSCs to identify LC gene signatures (Chung et al., 2017; Heo et al., 2020; Jan et al., 2011); however, we characterized LCs and HSCs using transcriptome patterns. Although these known markers distinguished LCs from HSCs, there was another abnormal HSC subpopulation that expressed CD34 but not AVP and CRHBP, and genes related to erythropoiesis at Dx, which may explain the low proportion of red blood cells in AML. In addition, almost no HSCs were observed at Rel (Figs. 1E and 2B), consistent with a previous report that HSCs decrease at Rel (Wang et al., 2017).
By comparing LCs with normal HSCs, we identified master TFs and their regulons in LCs including FOXC1 and CEBPA. FOXC1 is involved in the induction of cancer stem cell traits and tumor development (Cao et al., 2018; Han et al., 2017), while CEBPA is a critical TF for myeloid lineage differentiation (McKnight, 2001). Stem/progenitor cells with CEBPA downregulation display impaired myeloid differentiation (Lin et al., 2007; Pabst and Mueller, 2009; Radomska et al., 1998; Zhang et al., 2004). Meanwhile, the overexpression of CEBPA may exert oncogenic effects in precursor B acute lymphoblastic leukemia (Chapiro et al., 2006). Based on our data, FOXC1 and CEBPA overexpression in AML may also affect LC maintenance or development.
Some studies have suggested that patients with DNMT3A R882 have a high probability of relapse (Yuan et al., 2016), while others have reported that DNMT3A R882 does not affect patient outcomes, even if the VAF remains high during CR (Bhatnagar et al., 2016). Here, we also had a case of relapse with a very low DNMT3A VAF at CR (AML05) and cases with no relapse but a high VAF at CR (AML04, AML06; Fig. 3A). Therefore, the specific mutant cell-type that persists at CR may play an important role in relapse. By analyzing mutations and transcriptomes together at the single-cell level, we observed DNMT3A-mutant cell-types and their distribution according to AML disease stage. Notably, the changes in the distribution of mutant cells differed according to the DNMT3A VAF at CR. In the relapsed patient with a low DNMT3A VAF at CR, DNMT3A mutations were mainly found in LC and GMP1s at Dx and mutant GMP1s persisted until CR and Rel, although the frequency was lower at CR, indicating that DNMT3A-mutant GMP1s may be leukemic clones. Conversely, the relapsed patient with a high DNMT3A VAF at CR mainly had DNMT3A mutations in LMPPs that were not present at Dx but persisted until Rel, suggesting that DNMT3A-mutant LMPPs have pre-leukemic features at CR and eventually become LCs at Rel. GMP1s and LMPPs are known to display functional similarities with leukemic stem cells (Goardon et al., 2011) and we confirmed that these mutant GMP1s and LMPPs contain clones that progress to Rel and that some GMP1s and LMPPs containing the DNMT3A mutation at CR can transform into LCs at Rel.
Together, our findings provide further insights into the heterogeneity of AML between individuals and during the clinical course of disease. Single-cell analysis revealed that DNMT3A-mutant GMP1s and LMPPs can affect relapse in patients with AML. Although the highly heterogeneous nature of AML requires analysis of more patient samples to generalize the results, our novel approach that identified critical cell-types involved in relapse can contribute to finding the cause of AML relapse.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Funding Statement
ACKNOWLEDGMENTS This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future Planning (NRF-2015R1A2A1A10054579) and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1720160), Chonnam National University Hwasun Hospital Institute for Biomedical Science (HCRI21006), an NRF grant funded by the Korean government (MSIT) (No. 2019R1C1C1005403, 2021M3H9A2097520, 2019R1A5A8083404, and 2018R1A2A1A05078480), and GIST-MIT Research Collaboration grant funded by the GIST.
Footnotes
AUTHOR CONTRIBUTIONS
J.S.A., H.J.K., and D.D.H.K. designed the study. J.S.A., H.J.K., and M.Y.K. collected samples and data. S.I.S. and S.G.B. performed experiments. J.P. and S.G.B. interpreted data. S.G.B. analyzed the data. S.G.B., J.S.A., and J.P. wrote the manuscript. J.P., J.S.A., and H.J.K. reviewed the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Addya S., Keller M.A., Delgrosso K., Ponte C.M., Vadigepalli R., Gonye G.E., Surrey S. Erythroid-induced commitment of K562 cells results in clusters of differentially expressed genes enriched for specific transcription regulatory elements. Physiol. Genomics. 2004;19:117–130. doi: 10.1152/physiolgenomics.00028.2004. [DOI] [PubMed] [Google Scholar]
- Ahn J.S., Kim H.J., Kim Y.K., Lee S.S., Ahn S.Y., Jung S.H., Yang D.H., Lee J.J., Park H.J., Lee J.Y., et al. Assessment of a new genomic classification system in acute myeloid leukemia with a normal karyotype. Oncotarget. 2018;9:4961–4968. doi: 10.18632/oncotarget.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanazi B., Munje C.R., Rastogi N., Williamson A.J., Taylor S., Hole P.S., Hodges M., Doyle M., Baker S., Gilkes A.F., et al. Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia. Leukemia. 2020;34:427–440. doi: 10.1038/s41375-019-0596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arindrarto W., Borràs D.M., de Groen R.A., van den Berg R.R., Locher I.J., van Diessen S.A., van der Holst R., van der Meijden E.D., Honders M.W., de Leeuw R.H., et al. Comprehensive diagnostics of acute myeloid leukemia by whole transcriptome RNA sequencing. Leukemia. 2021;35:47–61. doi: 10.1038/s41375-020-0762-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar B., Eisfeld A.K., Nicolet D., Mrózek K., Blachly J.S., Orwick S., Lucas D.M., Kohlschmidt J., Blum W., Kolitz J.E., et al. Persistence of DNMT 3A R882 mutations during remission does not adversely affect outcomes of patients with acute myeloid leukaemia. Br. J. Haematol. 2016;175:226–236. doi: 10.1111/bjh.14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M., Provost S., Zada Y.F., Barhdadi A., Bourgoin V., Lépine G., Mollica L., Szuber N., Dubé M.P., Busque L. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–762. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- Cao S., Wang Z., Gao X., He W., Cai Y., Chen H., Xu R. FOXC1 induces cancer stem cell-like properties through upregulation of beta-catenin in NSCLC. J. Exp. Clin. Cancer Res. 2018;37:220. doi: 10.1186/s13046-018-0894-0.97c8dd2aa3eb4cbb986ebc1122ff038c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapiro E., Russell L., Radford-Weiss I., Bastard C., Lessard M., Struski S., Cave H., Fert-Ferrer S., Barin C., Maarek O., et al. Overexpression of CEBPA resulting from the translocation t (14; 19)(q32; q13) of human precursor B acute lymphoblastic leukemia. Blood. 2006;108:3560–3563. doi: 10.1182/blood-2006-03-010835. [DOI] [PubMed] [Google Scholar]
- Chien W., Sun Q.Y., Ding L.W., Mayakonda A., Takao S., Liu L., Lim S.L., Tan K.T., Garg M., De Sousa Maria Varela A., et al. Diagnosis and relapse: cytogenetically normal acute myelogenous leukemia without FLT3-ITD or MLL-PTD. Leukemia. 2017;31:762–766. doi: 10.1038/leu.2016.343. [DOI] [PubMed] [Google Scholar]
- Choi Y.H., Kim J.K. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol. Cells. 2019;42:189–199. doi: 10.14348/molcells.2019.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.S., Eng W.S., Hu W., Khalaj M., Garrett-Bakelman F.E., Tavakkoli M., Levine R.L., Carroll M., Klimek V.M., Melnick A.M., et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci. Transl. Med. 2017;9:eaaj2025. doi: 10.1126/scitranslmed.aaj2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon L.W., Ghannam J., Nosiri C., Gui G., Goswami M., Calvo K.R., Lindblad K.E., Oetjen K.A., Wilkerson M., Soltis A.R., et al. Personalized single-cell proteogenomics to distinguish acute myeloid leukemia from non-malignant clonal hematopoiesis. Blood Cancer Discov. 2021;2:319–325. doi: 10.1158/2643-3230.BCD-21-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo C.D., Cortes J.E. Mutations in AML: prognostic and therapeutic implications. Hematology Am. Soc. Hematol. Educ. Program. 2016;2016:348–355. doi: 10.1182/asheducation-2016.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediriwickrema A., Aleshin A., Reiter J.G., Corces M.R., Köhnke T., Stafford M., Liedtke M., Medeiros B.C., Majeti R. Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv. 2020;4:943–952. doi: 10.1182/bloodadvances.2019001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperle M., Adam S., Kunert S., Dukatz M., Baude A., Plass C., Rathert P., Bashtrykov P., Jeltsch A. Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns. Nucleic Acids Res. 2019;47:11355–11367. doi: 10.1093/nar/gkz911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., Van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Erickson A., He M., Berglund E., Marklund M., Mirzazadeh R., Schultz N., Kvastad L., Andersson A., Bergenstråhle L., Bergenstråhle J., et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature. 2022;608:360–367. doi: 10.1038/s41586-022-05023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey E., Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- Garg M., Nagata Y., Kanojia D., Mayakonda A., Yoshida K., Haridas Keloth S., Zang Z.J., Okuno Y., Shiraishi Y., Chiba K., et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015;126:2491–2501. doi: 10.1182/blood-2015-05-646240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goardon N., Marchi E., Atzberger A., Quek L., Schuh A., Soneji S., Woll P., Mead A., Alford K.A., Rout R., et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Han B., Bhowmick N., Qu Y., Chung S., Giuliano A.E., Cui X. FOXC1: an emerging marker and therapeutic target for cancer. Oncogene. 2017;36:3957–3963. doi: 10.1038/onc.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschuh L., Kaźmierczak M., Milewski M.C., Góralski M., Łuczak M., Wojtaszewska M., Uszczyńska-Ratajczak B., Lewandowski K., Komarnicki M., Figlerowicz M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and-M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int. J. Oncol. 2018;52:656–678. doi: 10.3892/ijo.2017.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S.K., Noh E.K., Ju L.J., Sung J.Y., Jeong Y.K., Cheon J., Koh S.J., Min Y.J., Choi Y., Jo J.C. CD45 dim CD34+ CD38− CD133+ cells have the potential as leukemic stem cells in acute myeloid leukemia. BMC Cancer. 2020;20:285. doi: 10.1186/s12885-020-06760-1.eda3cd9af6494bd38b568743b89a2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M., Chao M.P., Cha A.C., Alizadeh A.A., Gentles A.J., Weissman I.L., Majeti R. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5009–5014. doi: 10.1073/pnas.1100551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongen-Lavrencic M., Grob T., Hanekamp D., Kavelaars F.G., Al Hinai A., Zeilemaker A., Erpelinck-Verschueren C.A., Gradowska P.L., Meijer R., Cloos J., et al. Molecular minimal residual disease in acute myeloid leukemia. N. Engl. J. Med. 2018;378:1189–1199. doi: 10.1056/NEJMoa1716863. [DOI] [PubMed] [Google Scholar]
- Khwaja A., Bjorkholm M., Gale R.E., Levine R.L., Jordan C.T., Ehninger G., Bloomfield C.D., Estey E., Burnett A., Cornelissen J.J., et al. Acute myeloid leukaemia. Nat. Rev. Dis. Primers. 2016;2:16010. doi: 10.1038/nrdp.2016.10. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landberg N., Hansen N., Askmyr M., Ågerstam H., Lassen C., Rissler M., Hjorth-Hansen H., Mustjoki S., Järås M., Richter J., et al. IL1RAP expression as a measure of leukemic stem cell burden at diagnosis of chronic myeloid leukemia predicts therapy outcome. Leukemia. 2016;30:255–258. doi: 10.1038/leu.2015.135. [DOI] [PubMed] [Google Scholar]
- Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., Jr., Baty J.D., Jr., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Du Y., Cai Y., Liu W., Lv Y., Huang B., Zhang L., Wang Z., Liu P., Sun Q., et al. Single-cell analysis reveals the chemotherapy-induced cellular reprogramming and novel therapeutic targets in relapsed/refractory acute myeloid leukemia. Leukemia. 2023;37:308–325. doi: 10.1038/s41375-022-01789-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.C., Lee C.Y., Tien H.F., Hu C.Y., Tang J.L., Lin L.I. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in acute myeloid leukemia. Blood. 2007;110:2113. doi: 10.1182/blood.V110.11.2113.2113. [DOI] [Google Scholar]
- Liu W., Yi J.M., Liu Y., Chen C., Zhang K.X., Zhou C., Zhan H.E., Zhao L., Morales S., Zhao X.L., et al. CDK6 is a potential prognostic biomarker in acute myeloid leukemia. Front. Genet. 2021;11:600227. doi: 10.3389/fgene.2020.600227.3364977b4b9c4497bc4054b28e69a84a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loghavi S., Zuo Z., Ravandi F., Kantarjian H.M., Bueso-Ramos C., Zhang L., Singh R.R., Patel K.P., Medeiros L.J., Stingo F., et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J. Hematol. Oncol. 2014;7:74. doi: 10.1186/s13045-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A., Yang L., Rodriguez B., Zhou T., Chang E., Curry C.V., Challen G.A., Li W., Wheeler D., Rebel V.I., et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S.L. McBindall-a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–261. doi: 10.1016/S0092-8674(01)00543-8. [DOI] [PubMed] [Google Scholar]
- Meng L., Liu B., Ji R., Jiang X., Yan X., Xin Y. Targeting the BDNF/TrkB pathway for the treatment of tumors. Oncol. Lett. 2019;17:2031–2039. doi: 10.3892/ol.2018.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menter D.G., DuBois R.N. Prostaglandins in cancer cell adhesion, migration, and invasion. Int. J. Cell Biol. 2012;2012:723419. doi: 10.1155/2012/723419.02520ef4ed9a468e82138ca53577c5bb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L.A., Bowman R.L., Merlinsky T.R., Csete I.S., Ooi A.T., Durruthy-Durruthy R., Bowman M., Famulare C., Patel M.A., Mendez P., et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020;587:477–482. doi: 10.1038/s41586-020-2864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Y., Huang G., Wu J. The role of RUNX1 in NF1-related tumors and blood disorders. Mol. Cells. 2020;43:153–159. doi: 10.14348/molcells.2019.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Osorio D., Zhong Y., Li G., Xu Q., Yang Y., Tian Y., Chapkin R.S., Huang J.Z., Cai J.J. scTenifoldKnk: an efficient virtual knockout tool for gene function predictions via single-cell gene regulatory network perturbation. Patterns (N. Y.) 2022;3:100434. doi: 10.1016/j.patter.2022.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst T., Mueller B.U. Complexity of CEBPA dysregulation in human acute myeloid leukemia. Clin. Cancer Res. 2009;15:5303–5307. doi: 10.1158/1078-0432.CCR-08-2941. [DOI] [PubMed] [Google Scholar]
- Paguirigan A.L., Smith J., Meshinchi S., Carroll M., Maley C., Radich J.P. Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci. Transl. Med. 2015;7:281re2. doi: 10.1126/scitranslmed.aaa0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.J., Kwon A., Cho B.S., Kim H.J., Hwang K.A., Kim M., Kim Y. Characteristics of DNMT3A mutations in acute myeloid leukemia. Blood Res. 2020;55:17–26. doi: 10.5045/br.2020.55.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M., Sciambi A., Treusch S., Durruthy-Durruthy R., Gokhale K., Jacob J., Chen T.X., Geis J.A., Oldham W., Matthews J., et al. High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res. 2018;28:1345–1352. doi: 10.1101/gr.232272.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti A.A., Williams S.R., Miller C.A., Fiddes I.T., Srivatsan S.N., Chen D.Y., Fronick C.C., Fulton R.S., Church D.M., Ley T.J. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat. Commun. 2019;10:3660. doi: 10.1038/s41467-019-11591-1.e7387f8418fd4439a8a668e9564cc12b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povinelli B.J., Rodriguez-Meira A., Mead A.J. Single cell analysis of normal and leukemic hematopoiesis. Mol. Aspects Med. 2018;59:85–94. doi: 10.1016/j.mam.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska H.S., Huettner C.S., Zhang P., Cheng T., Scadden D.T., Tenen D.G. CCAAT/enhancer binding protein α is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 1998;18:4301–4314. doi: 10.1128/MCB.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russler-Germain D.A., Spencer D.H., Young M.A., Lamprecht T.L., Miller C.A., Fulton R., Meyer M.R., Erdmann-Gilmore P., Townsend R.R., Wilson R.K., et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K., Sarver A.L., Noble-Orcutt K.E., LaRue R.S., Antony M.L., Chang D., Lee Y., Navis C.M., Hillesheim A.L., Nykaza I.R., et al. Single-cell gene expression analyses reveal distinct self-renewing and proliferating subsets in the leukemia stem cell compartment in acute myeloid leukemia. Cancer Res. 2020;80:458–470. doi: 10.1158/0008-5472.CAN-18-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau G., Thorsteinsdottir U., Hough M.R., Hugo P., Lawrence H.J., Largman C., Humphries R.K. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;6:13–22. doi: 10.1016/S1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- Schuurhuis G.J., Meel M.H., Wouters F., Min L.A., Terwijn M., de Jonge N.A., Kelder A., Snel A.N., Zweegman S., Ossenkoppele G.J., et al. Normal hematopoietic stem cells within the AML bone marrow have a distinct and higher ALDH activity level than co-existing leukemic stem cells. PLoS One. 2013;8:e78897. doi: 10.1371/journal.pone.0078897.afdfbec2f3a140adb7a2c8c49ec3e1ce [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik M.P., Morilla R., de Bracco M.M., Catovsky D., Matutes E. CD34 and CD117 are overexpressed in AML and may be valuable to detect minimal residual disease. Leuk. Res. 2002;26:615–619. doi: 10.1016/S0145-2126(01)00182-5. [DOI] [PubMed] [Google Scholar]
- Shlush L.I., Mitchell A., Heisler L., Abelson S., Ng S.W., Trotman-Grant A., Medeiros J.J., Rao-Bhatia A., Jaciw-Zurakowsky I., Marke R., et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547:104–108. doi: 10.1038/nature22993. [DOI] [PubMed] [Google Scholar]
- Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson L., Balasubramanian D., Ribeiro S.P., Stefan T., Gupta K., Xu X., Fourati S., Roe A., Jackson Z., Schauner R., et al. Single cell RNA sequencing of AML initiating cells reveals RNA-based evolution during disease progression. Leukemia. 2021;35:2799–2812. doi: 10.1038/s41375-021-01338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakral D., Singh V.K., Gupta R., Jha N., Khan A., Kaur G., Rai S., Kumar V., Supriya M., Bakhshi S., et al. Integrated single-cell transcriptome analysis of CD34+ enriched leukemic stem cells revealed intra-and inter-patient transcriptional heterogeneity in pediatric acute myeloid leukemia. Ann. Hematol. 2023;102:73–87. doi: 10.1007/s00277-022-05021-4. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P., Hovestadt V., Wadsworth M.H., 2nd, Hughes T.K., 2nd, Griffin G.K., 2nd, Battaglia S., 2nd, Verga J.A., 2nd, Stephansky J., 2nd, Pastika T.J., 2nd, Story J.L., 2nd, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–1281.e24. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg S., Greif P.A. Clonal evolution of acute myeloid leukemia from diagnosis to relapse. Genes Chromosomes Cancer. 2019;58:839–849. doi: 10.1002/gcc.22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Stiehl T., Raffel S., Hoang V.T., Hoffmann I., Poisa-Beiro L., Saeed B.R., Blume R., Manta L., Eckstein V., et al. Reduced hematopoietic stem cell frequency predicts outcome in acute myeloid leukemia. Haematologica. 2017;102:1567–1577. doi: 10.3324/haematol.2016.163584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesely J., Kotini A.G., Izzo F., Luo H., Yuan H., Sun J., Georgomanoli M., Zviran A., Deslauriers A.G., Dusaj N., et al. Acute myeloid leukemia iPSCs reveal a role for RUNX1 in the maintenance of human leukemia stem cells. Cell Rep. 2020;31:107688. doi: 10.1016/j.celrep.2020.107688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Huang K., Yu J., Le T., Namihira M., Liu Y., Zhang J., Xue Z., Cheng L., Fan G. Dnmt3a regulates both proliferation and differentiation of mouse neural stem cells. J. Neurosci. Res. 2012;90:1883–1891. doi: 10.1002/jnr.23077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.l., Guo Y. FCGR1A serves as a novel biomarker and correlates with immune infiltration in four cancer types. Front. Mol. Biosci. 2020;7:581615. doi: 10.3389/fmolb.2020.581615.e09b97ac64eb4887a85549324b5a62bd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y., Yuan J., Suetake I., Suzuki H., Ishikawa Y., Choi Y., Ueno T., Soda M., Hamada T., Haruta H., et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Yilmaz M., Wang F., Loghavi S., Bueso-Ramos C., Gumbs C., Little L., Song X., Zhang J., Kadia T., Borthakur G., et al. Late relapse in acute myeloid leukemia (AML): clonal evolution or therapy-related leukemia? Blood Cancer J. 2019;9:7. doi: 10.1038/s41408-019-0170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Li Y., Zhang D., Wan D., Jiang Z. Clinical implications of recurrent gene mutations in acute myeloid leukemia. Exp. Hematol. Oncol. 2020;9:4. doi: 10.1186/s40164-020-00161-7.eaab548edef04a4da03d33c1928473db [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.Q., Peng L., Zeng W.J., Jiang B.Y., Li G.C., Chen X.P. DNMT3A R882 mutations predict a poor prognosis in AML: a meta-analysis from 4474 patients. Medicine (Baltimore) 2016;95:e3519. doi: 10.1097/MD.0000000000003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Singh P., Dolnik A., Brazda P., Atlasy N., Del Gaudio N., Döhner K., Döhner H., Minucci S., Martens J., et al. Longitudinal single-cell transcriptomics reveals distinct patterns of recurrence in acute myeloid leukemia. Mol. Cancer. 2022;21:166. doi: 10.1186/s12943-022-01635-4.cdc5c6225a8845a29d882f5c207da3a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M.L., Dayaram T., Owens B.M., Shigematsu H., Levantini E., Huettner C.S., Lekstrom-Himes J.A., et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBPα. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.