Abstract
The Hippo kinase cascade functions as a central hub that relays input from the “outside world” of the cell and translates it into specific cellular responses by regulating the activity of Yes-associated protein 1 (YAP1). How Hippo translates input from the extracellular signals into specific intracellular responses remains unclear. Here, we show that transforming growth factor β (TGFβ)-activated TAK1 activates LATS1/2, which then phosphorylates YAP1. Phosphorylated YAP1 (p-YAP1) associates with RUNX3, but not with TEAD4, to form a TGFβ-stimulated restriction (R)-point-associated complex which activates target chromatin loci in the nucleus. Soon after, p-YAP1 is exported to the cytoplasm. Attenuation of TGFβ signaling results in re-localization of unphosphorylated YAP1 to the nucleus, where it forms a YAP1/TEAD4/SMAD3/AP1/p300 complex. The TGFβ-stimulated spatiotemporal dynamics of YAP1 are abrogated in many cancer cells. These results identify a new pathway that integrates TGFβ signals and the Hippo pathway (TGFβ→TAK1→LATS1/2→YAP1 cascade) with a novel dynamic nuclear role for p-YAP1.
Keywords: LATS1/2, restriction point, RUNX3, TAK1, TGFβ, YAP1
INTRODUCTION
Studies on cell fate determination during development have identified the classic core Hippo pathway in Drosophila (Pan, 2010). The classic human Hippo pathway kinases are MST1/2 (Drosophila Hippo/Hpo) and LATS1/2 (Drosophila Warts/Wts), which control phosphorylation of Yes-associated protein 1 (YAP1) (Drosophila Yorkie/Yki) (Pan, 2010). Considering the critical importance of the Hippo pathway in regulating cell proliferation and organ size, there is a remarkable paucity of studies that have attempted to reconcile how this pathway is mechanistically linked to the restriction (R)-point when cells make the decision to initiate cell division (Malumbres and Barbacid, 2001; Pardee, 1974; Weinberg, 2007). Importantly, this R-point regulatory mechanism is disrupted in nearly all cancer cells (Blagosklonny and Pardee, 2002; Weinberg, 2007) and renders cells independent of external growth factors to permit unrestricted growth. Defining how the Hippo pathway controls cell cycle progression in cancer cells in relation to the R-point would represent a major advance in our understanding of cell growth control.
YAP1 integrates multiple pathways that play key roles in controlling cell fate and was initially identified as a target of tyrosine protein kinases related to the SRC and YES1 oncogenes (Hansen et al., 2015; Sudol et al., 1995). Phosphorylated YAP1 at Ser-127 (p-YAP1) is generated by a signaling relay involving the human serine/threonine kinases MST1/2 and LASTS1/2, which sequesters p-YAP1 in the cytoplasm via binding to 14-3-3 proteins (Dong et al., 2007; Kanai et al., 2000; Ren et al., 2010). The requirement of MST1/2 for LATS1/2 phosphorylation is context- and cell type-dependent, and additional kinases may also be involved in LATS1/2 phosphorylation (Yin et al., 2013). The Hippo cascade can be regulated at the cell surface by G-protein-coupled receptors (Miller et al., 2012; Yu et al., 2012) and epidermal growth factor receptors (Fan et al., 2013; Reddy and Irvine, 2013). However, these signaling pathways inactivate LATS1/2. Therefore, there are substantial gaps in our understanding of the molecular mechanism that govern LATS1/2 activation by extracellular signals.
One attractive candidate for LATS1/2 activation is TGFβ. Our current understanding is that TGFβ controls cell growth and differentiation by inducing phosphorylation of SMAD2/3 (Miller et al., 2019; Nakao et al., 1997). Yet, TGFβ also activates multiple SMAD independent pathways, including the TGFβ-activated kinase 1 (TAK1) and mitogen-activated protein kinase (MAPK) pathways. Both SMAD dependent and independent pathways are rapidly attenuated by auto-feedback loops (within about 2 h post-stimulation) (Cheung et al., 2003; Denissova et al., 2000; Ishida et al., 2000; Nagarajan et al., 1999). YAP1 interacts with TGFβ-activated SMADs (Nakamura et al., 2021; Varelas et al., 2008). Importantly, the roles of TAK1 in the regulation of YAP1 have been reported (Deng et al., 2018; Onodera et al., 2019; Santoro et al., 2020). These findings establish cross-talk between the Hippo/YAP1 and TGFβ pathways.
YAP1 does not contain a DNA binding domain, but elicits transcriptional activation via interactions with sequence-specific DNA binding transcription factors. RUNX family transcription factors (RUNXs), which represent master regulators of development and are often deregulated in cancers (Ito et al., 2015), were first identified as the cognate DNA binding transcription factors of YAP1 (Yagi et al., 1999). Subsequent studies identified additional transcription factors, including TEA-domain containing transcription factors (TEADs), the p53 tumor suppressor-related p63 protein (TP63), the PAX class of homeodomain proteins, the TGFβ-activated SMAD proteins, and the FOS/JUN related AP1 transcription factor (Liu et al., 2016; Varelas, 2014; Varelas et al., 2008; Zanconato et al., 2015). TEADs are major DNA binding transcription factors that support the oncogenic activity of YAP1 (Liu-Chittenden et al., 2012), but the roles of RUNX proteins and other transcription factors capable of modulating YAP1 activity in cell growth control remain insufficiently explored.
The association of RUNX3 with the Hippo/YAP1 pathway (Jang et al., 2017; Qiao et al., 2016) is particularly interesting because RUNX3 is a key target of TGFβ signaling (Ito and Miyazono, 2003) and interacts with both YAP1 and SMAD proteins. Furthermore, RUNX3 is the core component of a novel R-point associated mitogen-stimulated protein/protein complex (Chi et al., 2017; Lee et al., 2013; 2019a; 2019b; 2020; 2023). Therefore, we examined the regulatory mechanisms that unite the Hippo pathway with the TGFβ-stimulated R-point.
Here, we show that TGFβ signaling triggers YAP1 phosphorylation through the TGFβ→TAK1→LATS→YAP1 pathway. Phosphorylated YAP1 forms a TGFβ-stimulated R-point-associated complex in the nucleus and induces expression of early target genes. Soon after, the complex disassembles and p-YAP1 is exported to the cytoplasm. When the TGFβ signal is attenuated, YAP1 re-localizes to the nucleus and associates with TEAD4. The YAP1-TEAD4 complex induces expression of late target genes. The TGFβ-stimulated spatiotemporal dynamics of YAP1 were abrogated in all cancer cell lines analyzed in our study. Collectively, our results identify a tumor suppressor pathway (TGFβ→TAK1→LATS→YAP1) that connects TGFβ signaling to the Hippo pathway and the R-point, suggesting a new role of p-YAP1 in the nucleus.
MATERIALS AND METHODS
Cell lines and culture
HEK293, NIH-3T3, PANC-1, and MKN-28 cells (ATCC, USA) were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (Gibco BRL) and 1% penicillin/streptomycin (Invitrogen, USA). WI-38 cells (Lonza, Switzerland) were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL), 1% MEM Non-Essential Amino Acids (Gibco BRL), and 1% penicillin/streptomycin (Invitrogen). H460 and A549 cells (ATCC) were maintained in RPMI 1640 medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL) and 1% penicillin/streptomycin (Invitrogen). All cell lines were incubated at 37°C under 5% CO2. Lats1/2 double KO (Lats1/2 dKO) mouse embryonic fibroblasts (MEFs) were kind gifts from Dr. Dae-Sik Lim (KAIST, Korea). Other MEFs of various genotypes were obtained from mouse embryos at 15.5 days of gestation. All MEFs were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL) and 1% penicillin/streptomycin (Invitrogen).
TGFβ stimulation
All cell lines were cultured at low cell density. Before TGFβ1 treatment, cells were cultured under serum-free conditions for 1 h and then, treated with 1 ng/ml TGFβ1 (Cat No. 100-21C; PeproTech, USA).
DNA transfection, immunoprecipitation (IP), and immunoblotting (IB)
Transient transfection of all cell lines was performed using Lipofectamine Plus reagent and Lipofectamine (Invitrogen). Cell lysates were incubated for 3 h at 4°C with appropriate mono- or polyclonal antibodies (2 µg antibody/500 µg lysate sample), followed by protein G–Sepharose beads (Amersham Pharmacia Biotech, USA) for 1 h at 4°C. For IP of endogenous proteins, lysates were incubated for 6-12 h at 4°C with the appropriate mono- or polyclonal antibodies (dilution range, 1:1,000-1:3,000), and then with protein G–Sepharose beads (Amersham Pharmacia Biotech) for 5 h at 4°C. Immunoprecipitated samples were resolved on SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to a polyvinylidene difluoride membrane (Millipore, USA). The membrane was immunoblotted with appropriate antibodies after blocking and then visualized by an AmershamTM Imager 600 (GE Healthcare, USA) after treatment with ECL solution (Amersham Pharmacia Biotech). All the western blots were reproduced at least three times.
Plasmid constructs
pcDNA3.1-Flag-hYAP and pcDNA3.0-HA-LATS1 were kind gifts from Dr. Lim Dae Sik (KAIST). The p2xFlag CMV2-YAP2-1st WW mutant (YAP-M1, Plasmid #19046), p2xFlag CMV2-YAP2-2nd WW mutant (YAP-M2, Plasmid #19047), and p2xFlag CMV2-YAP2-1st&2nd WW mutant (YAP-M1/2, Plasmid #19048) were purchased from Addgene (USA). Point mutant plasmids of RUNX3 and YAP1 were generated by site-directed mutagenesis, and all mutations were confirmed by DNA sequencing. Adeno-associated virus (AAV) expressing wild-type RUNX3 (AAV-RUNX3-WT) and RUNX3-S356A (AAV-RUNX3-S356A) were obtained from GeneCraft (Korea).
Antibodies
Antibodies targeting BRD2 (Cat No. sc-393720), BRG-1 (Cat No. sc-17796, Cat No. sc-10768), Cyclin D1 (Cat No. sc-20044), CDK4 (Cat No. sc-260), c-JUN (Cat No. sc-74543), CTGF (Cat No. sc-365970), HDAC4 (Cat No. sc-11418), Myc (Cat No. sc-40), p-c-Jun (Cat No. SC-822), p300 (Cat No. sc-584), p21 (Cat No. sc-6246), pan 14-3-3 (Cat No. sc-1657), RUNX3 (Cat No. sc-101553), SMAD3 (Cat No. sc-101154), TAF1 (Cat No. sc-735), TBP (Cat No. sc-421), TEAD4 (Cat No. sc-101184), and YAP1 (Cat No. sc-101199) were obtained from Santa Cruz Biotechnology (USA). All antibodies from Santa Cruz Biotechnology were diluted to 1:1,000. Antibodies targeting H2AK119-ub (Cat No. 8240S), H3K412-ac (Cat No. 2591S), H3K27-me3 (Cat No. 9733S), H3K4-me3 (Cat No. 9751S), c-FOS (Cat No. 2250S), RNF2 (Cat No. 5694S), EZH2 (Cat No. 5246S), p-TAK1 (Cat No. 4508), LATS1 (Cat No. 3477), LATS2 (Cat No. 5888), p-LATS (Cat No. 9157), MST1 (Cat No. 14946), and p-MST1 (Cat No. 49332) were obtained from Cell Signaling Technology (USA). All antibodies from Cell Signaling Technology were diluted to 1:1,000. Antibodies targeting RUNX3 (5G4) (Cat No. ab40278), SMAD3 (Cat No. ab40854), p-SMAD3 (Cat No. ab52903), TAK1 (Cat No. ab109526), YAP1 (Cat No. ab52771), and p-YAP1 (Cat No. ab76252) were obtained from Abcam (UK). All antibodies from Abcam were diluted to 1:3,000. Antibodies targeting HA (12CA5, dilution 1:1,000, Cat No. 11 666 606 001; Roche Applied Science, Germany), Flag (M2, dilution 1:3,000, Cat No. F1804; Sigma, USA), BRD2 (M01, dilution 1:1,000, Cat No. H00006046-M01; Abnova, Taiwan), and MLL1 (dilution 1:1,000, Cat No. A300-374A; Bethyl Laboratories Inc., USA) were used for IB and IP. Anti-RUNX3-phospho-S356 (dilution 1:1,000) polyclonal anti-serum against synthetic RUNX3 peptide phosphorylated at Ser-356 was raised in rabbits.
Proximity ligation assay (PLA)
The PLA was performed using the Duolink® In Situ PLA® Kit (Sigma). Briefly, cells were grown, fixed, and permeabilized. The samples were then incubated overnight at 4°C with primary antibodies against the two proteins to be examined, washed (buffer A: 0.01 M Tris-HCl [pH 7.4], 0.15 M NaCl, and 0.05% Tween 20), incubated at 37°C for 60 min with specific probes, stained for F-actin to visualize the cytoplasm, and washed with buffer B (0.2 M Tris-HCl [pH 7.5] and 0.1 M NaCl). Signals were visualized as distinct fluorescent spots under a fluorescence microscope (Carl Zeiss AXIO Zoom.V16 and ApoTome.2; Carl Zeiss, Germany). Background correction, contrast adjustment of raw images, and quantification of fluorescence signals were performed using the Zen 2012 Blue Edition software (Carl Zeiss).
Immunofluorescence staining
HEK293 cells were grown to the appropriate density on 22 mm cover slips (Thermo Fisher Scientific, UK). Cells were serum-starved for 1 h and then stimulated with TGFβ1, washed with phosphate-buffered saline, and then fixed in a solution of 4% formaldehyde for 15 min at room temperature. Cells were incubated for 45 min in a solution of 10% fetal bovine serum/phosphate-buffered saline/0.1% Triton X-100. Cells were incubated with the indicated primary antibody for 6-12 h at 4°C, followed by Alexa Fluor 488 anti-mouse and Alexa 594 anti-rabbit antibodies for 1 h at 25°C. The cells were then stained with DAPI for 7 min. Signals were visualized as distinct fluorescent spots under a fluorescence microscope (Carl Zeiss AXIO Zoom.V16 and ApoTome.2). Background correction, contrast adjustment of raw images, and quantification of fluorescence signals were performed using the Zen 2012 Blue Edition software (Carl Zeiss).
Inhibitors and siRNA treatment
The TGFβRI kinase inhibitor (SB431542) and TAK1 kinase inhibitor (5Z-7) were purchased from Sigma-Aldrich (USA). Cells were treated with a TGFβRI kinase inhibitor (SB431542, 1 µM) or a Tak1 kinase inhibitor (5Z-7, 1 µM), serum-starved for 1 h, and then stimulated with TGFβ1. Cells were harvested at the indicated time points after TGFβ1 stimulation. SiRNAs targeting TAK1 (6885-1), YAP1 (10413-1), and c-JUN (3725-3) were purchased from Bioneer (Korea). Knockdown analysis was performed by transfecting HEK293 cells with 50 nM siRNA using RNAiMAX (Invitrogen) before serum starvation. Cells were harvested at the indicated time points after TGFβ1 stimulation.
Chromatin immunoprecipitation (ChIP) and Re-ChIP assays
ChIP assays were performed using the ChIP assay kit (Cat No. 17-295; Millipore). HEK293 cells were serum-starved for 1 h and then stimulated with TGFβ1. Cells were harvested at the indicated time points and cross-linked with formaldehyde (1% [v/v], 10 min, 37°C). Chromatin was immunoprecipitated with the indicated antibodies. Re-ChIP assays were performed using the ChIP assay kit (Cat No. 17-295). Briefly, the eluant of the primary immunocomplex was obtained from the first ChIP and then subjected to further IP with the indicated second antibodies. The p21 or CTGF promoter region was amplified by PCR using the following primers, as reported previously (Chi et al., 2017; Qiao et al., 2016):
(p21-forward: CACCAGACTTCTCTGAGCCCCAG)
(p21-reverse: GCACTGTTAGAATGAGCCCCCTTTC)
(CTGF-forward: ATATGAATCAGGAGTGGTGCGA)
(CTGF-reverse: CAACTCACACCGGATTGATCC)
RNA sequencing
Total RNA was isolated using Trizol reagent (Invitrogen). RNA quality was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, The Netherlands) and RNA quantification was performed using a ND-2000 Spectrophotometer (Thermo Fisher Scientific). Libraries were prepared from total RNA using the NEBNext Ultra II Directional RNA-Seq Kit (New England Biolabs, UK). Isolation of mRNA was performed using the Poly(A) RNA Selection Kit (Lexogen, Austria). The isolated mRNAs were used for cDNA synthesis and shearing in accordance with the manufacturer’s instructions. Indexing was performed using the Illumina indexes 1-12. The enrichment step was carried out using PCR. Subsequently, libraries were checked using the TapeStation HS D1000 Screen Tape (Agilent Technologies) to evaluate the mean fragment size. Quantification was performed using the library quantification kit and a StepOne Real-Time PCR System (Life Technologies, USA). High-throughput sequencing was performed as paired-end 100 sequencing using NovaSeq 6000 (Illumina, USA).
Quantification and statistical analysis
Quality control of raw sequencing data was performed using FastQC (Simon, 2010). Adapters and low-quality reads (<Q20) were removed using FASTX_Trimmer (Hannon Lab, 2014) and BBMap (Bushnell, 2014). Then, the trimmed reads were mapped to the reference genome using TopHat (Trapnell et al., 2009). The Read Count data were processed based on a FPKM + Geometric normalization method using EdgeR within R (R Core Team, 2020). FPKM (fragments per kb per million reads) values were estimated using Cufflinks (Roberts et al., 2011).
Data and software availability
Data mining and graphic visualization were performed using ExDEGA (Ebiogen Inc., Korea). The RNA sequencing data are available under accession number GSE226590 at Gene Expression Omnibus (GEO).
RESULTS
TGFβ signaling phosphorylates YAP1 at Ser-127 and triggers nuclear-cytoplasmic shuttling of the protein
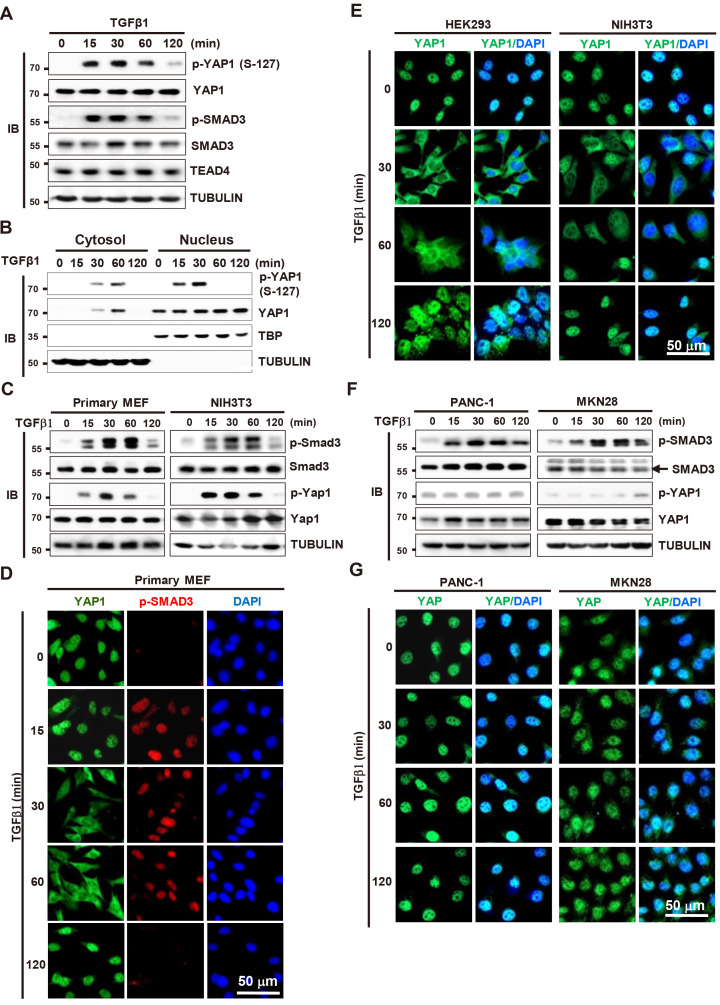
SMAD3 is phosphorylated at S-423 and -425 (p-SMAD3) 15 min after exposure to TGFβ (Shi and Massague, 2003). In this study, we asked whether YAP1 is phosphorylated by TGFβ stimulation. IB analysis revealed that YAP1 was also phosphorylated at Ser-127 (p-YAP1) at the same time as SMAD3 in HEK293 cells (human embryonic kidney cells) (Fig. 1A). Both p-SMAD3 and p-YAP1 levels were maintained until 60 min after stimulation and returned to base line at 120 min (Fig. 1A). SMAD3 phosphorylation occurs at the cytoplasm and the p-SMAD3 moves into the nucleus (Shi and Massague, 2003). By contrast, the p-YAP1 was first detected at the nucleus (15-30 min after TGFβ stimulation) and then detected at the cytoplasm (30-60 min) (Fig. 1B). These results suggest that YAP1 is phosphorylated at the nucleus and the p-YAP1 moves to the cytoplasm in response to TGFβ stimulation.
Fig. 1. TGFβ signaling phosphorylates YAP1 at Ser-127 and triggers nuclear-cytoplasmic shuttling of the protein.
(A) HEK293 cells were stimulated with TGFβ1. Time-dependent phosphorylation of SMAD3 and YAP1 was analyzed by immunoblotting (IB) at the indicated time points. (B) HEK293 cells cultured at low cell density were stimulated with TGFβ1 (1 ng/ml). Cells were harvested at the indicated time points. The levels of p-YAP1, YAP1, and RUNX3 were measured by IB in the cytoplasmic and nuclear fractions of the samples. (C) Primary MEF and immortalized MEF (NIH3T3) were stimulated with TGFβ1. Time-dependent phosphorylation of SMAD3 and YAP1 was analyzed by IB at the indicated time points. (D) Primary MEFs were stimulated with TGFβ1. Time-dependent subcellular localization of YAP1 and p-SMAD3 was analyzed by triple immunofluorescence staining at the indicated time points (green, YAP1; red, p-SMAD3; blue, DAPI). (E) Immortalized normal cell lines (HEK293 [human embryonic kidney] and NIH3T3 [MEF]) were stimulated with TGFβ1. Time-dependent subcellular localization of YAP1 was analyzed by double immunofluorescence staining (green, YAP1; blue, DAPI). (F) Cancer cell lines (MKN28 [human gastric cancer] and PANC-1 [human pancreatic cancer]) were stimulated with TGFβ1. Time-dependent phosphorylation of SMAD3 and YAP1 was analyzed by IB at the indicated time points. (G) MKN28 and PANC-1 cell lines were stimulated with TGFβ1, and time-dependent subcellular localization of YAP1 was analyzed by double immunofluorescence staining (green, YAP1; blue, DAPI). TGFβ, transforming growth factor β; YAP1, Yes-associated protein 1; MEF, mouse embryonic fibroblast.
We further confirmed whether YAP1 is exported to the cytoplasm in response to TGFβ-stimulation. In primary embryonic fibroblast cells (MEFs), SMAD3 and YAP1 were phosphorylated at the same time points as in HEK293 cells (Fig. 1C). Triple immunofluorescence staining of MEFs revealed that p-SMAD3 localized to the nucleus 15 min after TGFβ stimulation and remained there until 60 min post-stimulation (Fig. 1D). Notably, YAP1 was exported to the cytoplasm at 30 min post-stimulation and remained there until 60 min (Fig. 1D). When the TGFβ signal was attenuated (120 min after stimulation), p-SMAD3 disappeared and YAP1 re-localized to the nucleus (Fig. 1D). These results, together with the results shown in Fig. 1B, demonstrate that TGFβ signaling phosphorylates YAP1 at Ser-127 and triggers nuclear-cytoplasmic shuttling of the protein.
We then asked whether TGFβ-stimulated YAP1 phosphorylation and its nuclear-cytoplasmic shuttling are general phenomena in various cell lines. In primary MEFs and all four analyzed immortalized normal cell lines (HEK293, NIH3T3, WI-38, and HaCaT), p-YAP1, as well as p-SMAD3, was detected at the same time points (15-60 min) after TGFβ stimulation (Fig. 1C, Supplementary Fig. S1A). In all these cell lines, YAP1 was exported to the cytoplasm 30-60 min after TGFβ stimulation, after which it re-localized to the nucleus after 120 min, as in primary MEFs (Fig. 1E, Supplementary Fig. S1B). These results demonstrate that YAP1 phosphorylation/dephosphorylation and subsequent nuclear-cytoplasmic shuttling of YAP1 in response to TGFβ stimulation is a general phenomenon in MEFs and immortalized normal cell lines.
Next, we examined the effect of TGFβ stimulation on YAP1 phosphorylation in four cancer cell lines (PANC-1, MKN28, H460, and A549). In these cell lines, p-SMAD3 was detected 15 min after TGFβ stimulation (Fig. 1F, Supplementary Fig. S1C). However, in all four cancer cell lines, p-YAP1 levels were base line and were unchanged by TGFβ stimulation (Fig. 1F, Supplementary Fig. S1C). In these cell lines, YAP1 localized to the nucleus, and its subcellular localization was not changed by TGFβ1 stimulation (Fig. 1G, Supplementary Fig. S1D). These results demonstrate that TGFβ-dependent YAP1 phosphorylation and subsequent cytoplasmic export are mediated through a SMAD-independent pathway, and that this pathway is disrupted in many cancer cell lines.
Studies show that YAP1 retains TGFβ-activated SMAD3 in the nucleus (Varelas et al., 2008; 2010). However, a recent report shows that nuclear localization of TGFβ-activated SMAD2/3 is not affected by the presence or absence of YAP1/TAZ (Labibi et al., 2020). Our results show that p-SMAD3 localized to the nucleus 15 min after TGFβ stimulation and remained there, even when YAP1 was exported to the cytoplasm (30-60 min after stimulation) (Fig. 1D). When the TGFβ signal was attenuated (120 min after), p-SMAD3 disappeared (Fig. 1C), but dephosphorylated SMAD3 was still detected in the nucleus (Supplementary Fig. S2). These results, together with those of previous reports, suggest that p-SMAD3 localizes to the nucleus without the aid of YAP1, but dephosphorylated SMAD3 remains in the nucleus via interaction with unphosphorylated YAP1, which re-localizes to the nucleus after the TGFβ signal is attenuated.
TGFβ signaling facilitates YAP1 phosphorylation through TAK1→LATS pathway activation
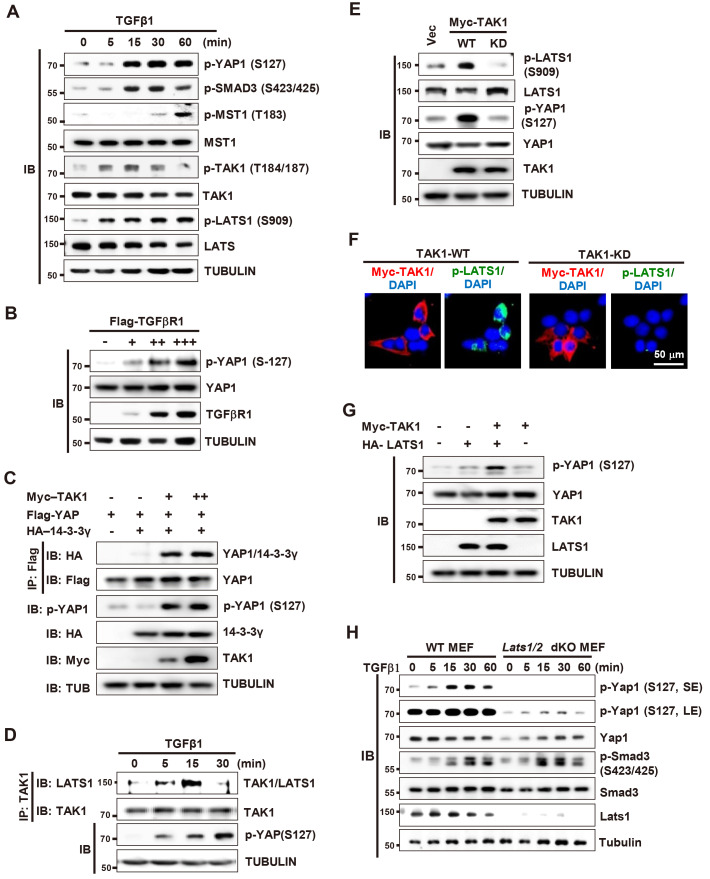
We then investigated how the TGFβ signal is transduced to YAP1. Analysis of the time-dependent activation of TGFβ signaling pathways and the Hippo pathway in HEK293 cells revealed that TAK1 was activated by phosphorylation 5 min after TGFβ1 stimulation, which is a little bit earlier than that of YAP1 and SMAD3 (Fig. 2A). MST1 was also activated, but at the later time points than YAP1 phosphorylation (60 min after) (Fig. 2A), suggesting a possibility that TAK1 is involved in TGFβ-stimulated phosphorylation of YAP1. To investigate this possibility, we analyzed the effect of TGFβ receptor I or TAK1 overexpression. Overexpression of Flag-tagged TGFβ receptor I (Flag-TGFβRI) increased p-YAP1 levels (Fig. 2B). Overexpression of Myc-tagged TAK1 (Myc-TAK1) also increased p-YAP1 levels and facilitated interaction between YAP1 and 14-3-3 (Fig. 2C). These results demonstrate that TAK1 is involved in TGFβ-stimulated YAP1 phosphorylation.
Fig. 2. TGFβ-activated TAK1 activates LATS1/2, which phosphorylates YAP1 at S127.
(A) HEK293 cells were stimulated with TGFβ1. Cells were harvested at the indicated time points and expression of the indicated proteins was measured by immunoblotting (IB). (B) HEK293 cells were transfected with increasing amounts of Flag-TGFβRI. Cells were harvested and the level of p-YAP1-S127 was measured by IB. (C) HEK293 cells were transfected with Flag-YAP1, HA-14-3-3-γ, and increasing amounts of Myc-TAK1. Cells were harvested and p-YAP1-S127 and YAP1-14-3-3-γ interactions were measured by immunoprecipitation (IP) and IB. (D) HEK293 cells were stimulated with TGFβ1. Cells were harvested at the indicated time points, and the interaction between TAK1 and LATS1 was measured by IP and IB. (E) HEK293 cells were transfected with wild-type (WT) or a kinase dead mutant (KD) of Myc-TAK1. The level of expressed Myc-TAK1 and activated LATS1 (p-LATS1-S909) was analyzed by IB. (F) HEK293 cells were transfected with Myc-TAK1-WT or Myc-TAK1-WT. The level of expressed Myc-TAK1 and activated LATS1 (p-LATS1-S909) was analyzed by triple immunofluorescence staining (red, Myc-TAK1; green, p-LATS1; blue, DAPI). (G) HEK293 cells were transfected with Myc-TAK1-WT and HA-LATS1. The level of p-YAP (S127) was analyzed by IB. (H) WT MEFs and Lats1/2 double knockout MEFs (Lats1/2 dKO MEFs) were stimulated with TGFβ1. Cells were harvested at the indicated time points and expression of the indicated proteins was measured by IB. TGFβ, transforming growth factor β; TAK1, TGFβ-activated kinase 1; Vec, vector; dKO, double KO; MEF, mouse embryonic fibroblast; SE, short exposure; LE, long exposure.
Notably, LATS1, a core Hippo pathway kinase that phosphorylates YAP1 directly, was also activated at the same time point as TAK1 after TGFβ-stimulation (Fig. 2A). IP/IB analysis revealed that TAK1 associated with LATS1 at 5-15 min post-TGFβ1 stimulation, and then dissociated (Fig. 2D). Overexpression of Myc-TAK1-WT (wild-type), but not Myc-TAK1-KD (kinase dead TAK1 mutant at Lys-63 to Trp), activated LATS1 (Figs. 2E and 2F). Consistent with this, co-expression of Myc-TAK1 and HA-LATS1 acted synergistically to increase phosphorylation of YAP1 (Fig. 2G). In addition, TGFβ-stimulated YAP1 phosphorylation was markedly reduced by knockout of Lats1/2 in MEFs (Lats1/2 dKO MEF, p-Yap1) (Fig. 2H). These results demonstrate that TGFβ signaling facilitates YAP1 phosphorylation mainly through TAK1→LATS pathway activation.
The TGFβ→TAK1→LATS→YAP1 pathway regulates nuclear-cytoplasmic shuttling of YAP1
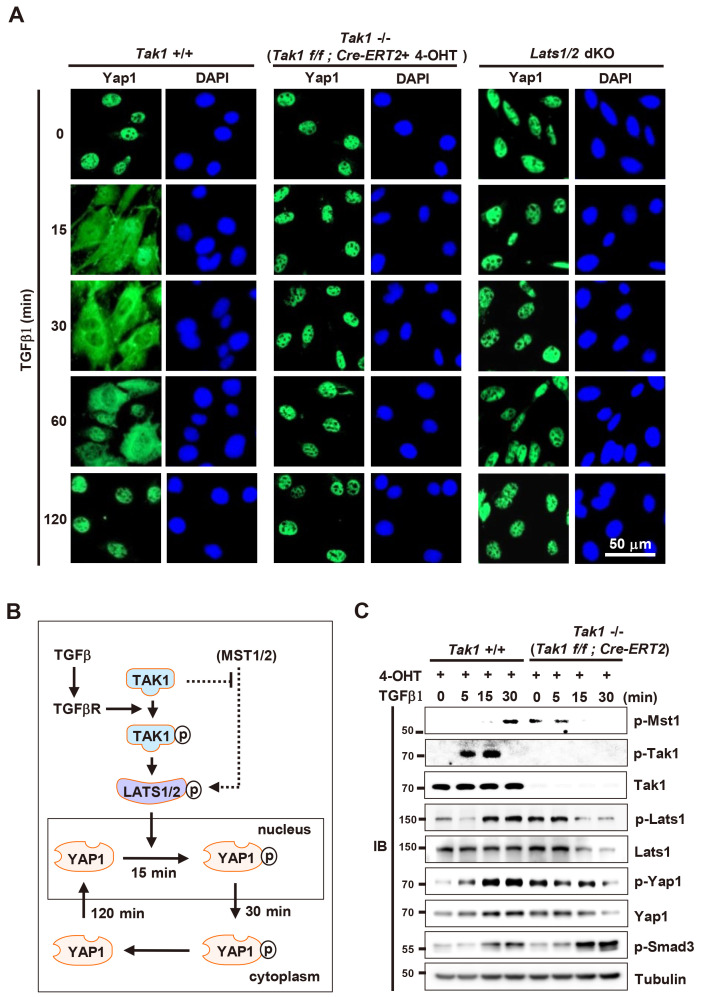
Next, we examined whether TAK1 and LATS1/2 are essential for TGFβ-stimulated nuclear-cytoplasmic shuttling of YAP1. For this purpose, we obtained MEFs from Tak1f/f;Cre-ERT2 mouse embryos. In these MEFs, Tak1 was targeted completely 24 h after treatment with 4-hydroxytamoxifen (4-OHT) (Supplementary Fig. S3A). The 4-OHT treated Tak1f/f;Cre-ERT2 MEFs proliferated normally up until 24 h after treatment, and their viability was reduced then after (Supplementary Fig. S3B). Therefore, we used Tak1f/f MEFs and Tak1f/f;Cre-ERT2 MEFs treated with 4-OHT for 24 h as WT and Tak1 KO MEFs, respectively. We then compared TGFβ-stimulated nuclear-cytoplasmic shuttling of YAP1 in WT, Tak1 KO, and Lats1/2 dKO MEFs. Under low cell density culture conditions without TGFβ1, YAP1 localized mainly to the nucleus in all three cell lines (Fig. 3A). When WT MEFs were stimulated with TGFβ1, YAP1 began to be exported to the cytoplasm 15 min later, but still localized mainly to the nucleus (Fig. 3A). YAP1 localized mainly in the cytoplasm 30-60 min after stimulation and then re-localized to the nucleus 120 min after (Fig. 3A), as seen for HEK293 cells and other untransformed cell lines. However, in Tak1 KO and Lats1/2 dKO MEFs, YAP1 was not exported to the cytoplasm until 120 min after TGFβ stimulation (Fig. 3A). These results demonstrate that the TGFβ→TAK1→LATS→YAP1 pathway regulates nuclear-cytoplasmic shuttling of YAP1 (Fig. 3B).
Fig. 3. The TGFβ→TAK1→LATS→ YAP1 pathway regulates the spatiotemporal dynamics of YAP1.
(A) WT, Tak1 KO, and Lats1/2 dKO MEFs were stimulated with TGFβ1. Time-dependent subcellular localization of YAP1 was measured by double immunofluorescence staining at the indicated time points (green, YAP1; blue, DAPI). (B) Schematic illustration of predicted signaling pathways regulating YAP1 phosphorylation regulating subcellular localization of YAP1. (C) WT and Tak1 KO MEFs were stimulated with TGFβ1. Cells were harvested at the indicated time points and time-dependent phosphorylation of Mst1, Tak1, Lats1, YAP1, and Smad3 was analyzed by immunoblotting (IB). TGFβ, transforming growth factor β; TAK1, TGFβ- activated kinase 1; WT, wild-type; dKO, double KO; MEF, mouse embryonic fibroblast.
Unexpectedly, the levels of p-Lats1 and p-YAP1 in Tak1 KO MEFs in the absence of TGFβ were as high as those in TGFβ-stimulated WT MEFs (15 min after) (Fig. 3C). Interestingly, the level of Mst1 phosphorylation was increased when Tak1 was dephosphorylated in WT MEFs (Fig. 3C). Consistently, the level of p-Mst1 was increased by Tak1 deletion in the absence of TGFβ (Fig. 3C). Although it is unclear how the levels of p-Lats1 and p-YAP1 are increased in Tak1 KO MEFs in the absence of TGFβ, our results suggest a possibility that Tak1 may inhibit Mst1 activity under normal cell culture conditions (Fig. 3B).
Biphasic role of YAP1 in response to TGFβ stimulation
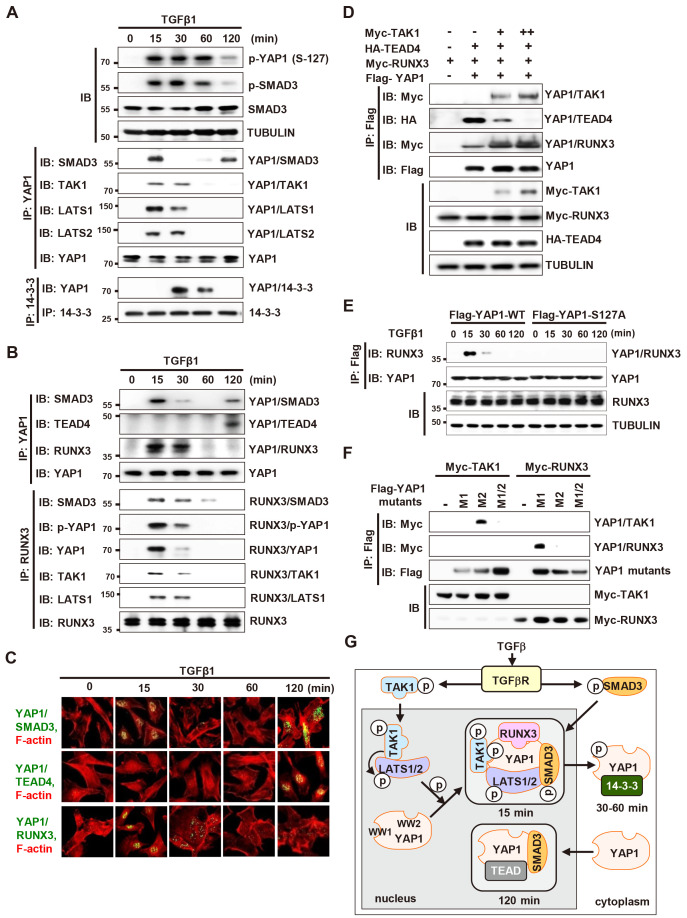
It has been reported that 2 h after TGFβ stimulation, SMADs interact with YAP1 (Fujii et al., 2012; Nakamura et al., 2021; Varelas et al., 2008). At the time point, TGFβ signal is attenuated. Therefore, we examined whether YAP1-SMAD3 interaction occurs while TGFβ signal is activated. TGFβ stimulation followed by IP/IB analysis at various time points revealed that YAP1 interacted with SMAD3 15 min after stimulation (Fig. 4A). YAP1 also interacted with TAK1 and LATS1 at the time point (Fig. 4A). Then, the complex was destroyed, and p-YAP1 associates with 14-3-3 (30-60 min) (Fig. 4A). When the TGFβ signal was attenuated, unphosphorylated YAP1 formed a complex with SMAD3 again, but not with TAK1 and LATS1 (Fig. 4A). These results demonstrate that YAP1-SMAD3 interaction is biphasic: YAP1 associated with SMAD3 15 min after TGFβ-stimulation, dissociated 30 min after, and re-associated 120 min after.
Fig. 4. Biphasic role of YAP1 in TGFβ signaling.
(A and B) HEK293 cells cultured at low cell density were stimulated with TGFβ1 (1 ng/ml). Cells were harvested at the indicated time points and expression of the indicated proteins, as well as interactions among proteins, were measured by immunoprecipitation (IP) and immunoblotting (IB). (C) PLA showing YAP1-SMAD3, YAP1-TEAD4, and YAP1-RUNX3 interactions at the indicated time points after TGFβ1 stimulation. Green or yellow fluorescence indicates associations between indicated proteins. F-actin was stained (red) to visualize the cytoplasmic compartment. Quantification of YAP1-SMAD3, YAP1-TEAD, and YAP1-RUNX3 PLA signals (n = 20) is shown in (C). (D) HEK293 cells were transfected with Flag-YAP1, HA-TEAD4, Myc-RUNX3, and increasing amounts of Myc-TAK1. Cells were harvested and the interactions between proteins were measured by IP and IB. (E) HEK293 cells were transfected with Flag-YAP1-WT or Flag-YAP1-S127A. Cells were harvested and the interaction between YAP and RUNX3 was measured by IP and IB. (F) HEK293 cells were transfected with Myc-RUNX3 or Myc-TAK1, and Flag-YAP1-M1 (a WW1 domain-disrupted YAP1 mutant; W199A and P202A), Flag-YAP1-M2 (a WW2 domain-disrupted YAP1 mutant; W258A and P261A), or Flag-YAP1-M1/2 (a mutant in which the YAP1 WW1 and WW2 domains are disrupted). Cells were harvested and the interactions between proteins were measured by IP and IB. TAK1 and RUNX3 bound to the WW1 and WW2 domain of YAP1, respectively. (G) Schematic illustration of sequential TGFβ-stimulated molecular interactions occurring on YAP1. TGFβ-activated TAK1 activates LATS1/2. The activated LATS1/2 phosphorylates YAP1. p-YAP1 forms a complex with p-TAK1, p-LATS, p-SMAD3, and RUNX3 in the nucleus at 15 min after TGFβ1 stimulation. At 30-60 min after stimulation, the complex dissociates and p-YAP1 associates with 14-3-3 and is exported from the nucleus to the cytoplasm. When the TGFβ signal is attenuated by auto-feedback inhibitory mechanisms at 120 min, YAP1 re-localizes to the nucleus and associates with TEAD4 and SMAD3. TGFβ, transforming growth factor β.
TGFβ-activated SMADs interacts with YAP1, which interacts with TEADs (Fujii et al., 2012; Nakamura et al., 2021; Varelas et al., 2008). TGFβ-activated SMADs also interact with RUNX3 (Ito and Miyazono, 2003), which interacts with YAP1 (Chi et al., 2005; Ito and Miyazono, 2003; Jang et al., 2017; Li et al., 2002; Qiao et al., 2016). Therefore, we investigated whether TGFβ-stimulation affects interactions among YAP1, SMAD3, TEAD4, and RUNX3. Interestingly, YAP1 interacted with TEAD4 and RUNX3 at distinct time points after TGFβ stimulation. YAP1 interacted with RUNX3 as well as SMAD3, 15 min after TGFβ-stimulation (Fig. 4B). The YAP1-TEAD4 interaction occurred 120 min after TGFβ-stimulation, when the TGFβ signal was attenuated and YAP1 was unphosphorylated (Fig. 4B). The TGFβ-stimulated time-dependent change in YAP1 partners was further confirmed in a PLA (Fig. 4C). The results demonstrated that soon after TGFβ-stimulation, both YAP1 and SMAD3 were phosphorylated; p-YAP1 and p-SMAD3 then formed a complex with RUNX3 (p-YAP1/p-SMAD3/RUNX3). When the TGFβ signal was attenuated, unphosphorylated YAP1 and SMAD3 formed a complex with TEAD4 (YAP1/SMAD3/TEAD4).
To understand the mechanism for the changing partners of YAP1, we transfected HEK293 cells with Myc-TAK1, Flag-YAP1, HA-TEAD4, and Myc-RUNX3. IP/IB analysis showed that TAK1 associated with YAP1, and reduced the YAP1-TEAD4 interaction and enhanced YAP1-RUNX3 interaction markedly (Fig. 4D). In addition, a phosphorylation-defective YAP1 mutant, YAP1-S127A (Ser-127 to Ala), failed to interact with RUNX3 after TGFβ-stimulation (Fig. 4E). This YAP1 mutant interacted with TEAD4 15 min after TGFβ-stimulation and the complex was maintained for long time (see below). These results suggest that TGFβ-activated TAK1 facilitates the YAP1-RUNX3 interaction and inhibits the YAP1-TEAD4 interaction through YAP1 phosphorylation at Ser-127.
Since YAP1 interacted with TAK1 and RUNX3 simultaneously (Fig. 4D), we mapped the regions of YAP1 that are involved in each interaction. For the purpose, we constructed Flag-YAP1-M1 (a WW1 domain-disrupted YAP1 point mutant; W199A and P202A), Flag-YAP1-M2 (a WW2 domain-disrupted YAP1 point mutant; W258A and P261A), or Flag-YAP1-M1/2 (a mutant in which the YAP1 WW1 and WW2 domains are disrupted) (Supplementary Fig. S4). Transfection of the constructs with Myc-TAK1 or Myc-RUNX3 followed by IP/IB analysis showed that YAP1 interacts with TAK1 and RUNX3 through its WW1 and WW2 domains, respectively (Fig. 4F).
YAP1 interacted with TAK1, LATS1, and SMAD3 15-30 min after stimulation (Fig. 4A). RUNX3 also interacted with TAK1 and LATS1 at the same time point (Fig. 4B), demonstrating that YAP1, TAK1, LATS1, SMAD3, and RUNX3 form a single complex. These results suggest that early after TGFβ1 stimulation, SMAD3 is phosphorylated by TGFβR kinase, and YAP1 is phosphorylated through the TGFβ→TAK1→LATS pathway; p-YAP1 then forms a complex with p-TAK1, p-LATS, p-SMAD3, and RUNX3 in the nucleus. Then, the complex is destroyed, and p-YAP1 associates with 14-3-3 and is exported to the cytoplasm. After the TGFβ signal is attenuated, unphosphorylated YAP1 relocalizes to the nucleus and forms a complex with SMAD3 and TEAD4 (Fig. 4G). These results identify a new signaling pathway (TGFβ→TAK1→LATS→YAP1) connecting the TGFβ and Hippo pathways.
TGFβ-stimulated phosphorylation of YAP1 triggers formation of an R-point-associated activator complex
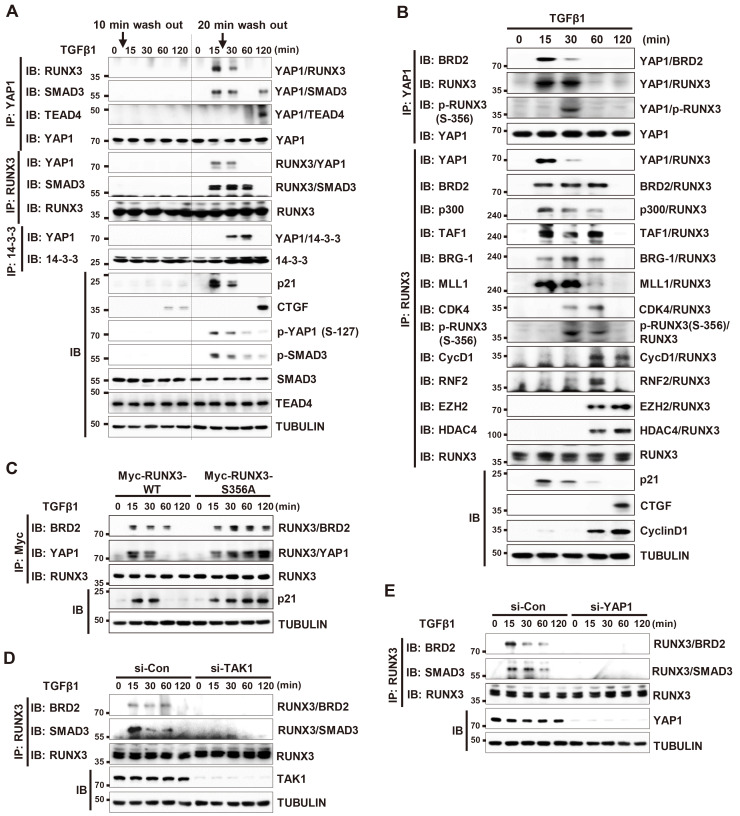
In response to extracellular signals, the cell makes a critical decision for cell fate (commitment) at R-point. Once the R-point decision is made, extracellular signals are no longer required to execute subsequent cell-autonomous programs (Malumbres and Barbacid, 2001; Pardee, 1974; Weinberg, 2007). To understand when the TGFβ-stimulated R-point decision is made, we treated HEK293 cells with TGFβ1 for only a short time (10 or 20 min) and measured time-dependent interactions among YAP1, RUNX3, SMAD3, and TEAD4. The results revealed that 10 min of TGFβ1 treatment was insufficient to induce interaction among the proteins (Fig. 5A, left). However, a 20 min treatment triggered all the sequential protein-protein interactions, just like continuous TGFβ1 stimulation (Fig. 5A, right). These results suggest that the TGFβ-stimulated commitment is made between 10 and 20 min after stimulation; therefore, the complex formed 15 min after TGFβ stimulation might be associated with the R-point.
Fig. 5. p-YAP1 and RUNX3 form a TGFβ-dependent R-point-associated complex.
(A) HEK293 cells were stimulated with TGFβ1 for 10 or 20 min. Then, the medium was replaced with TGFβ1-free medium. Cells were harvested at the indicated time points and the interactions among YAP1, RUNX3, SMAD3, TEAD4, and 14-3-3 were measured by immunoprecipitation (IP) and immunoblotting (IB). (B) HEK293 cells were stimulated with TGFβ1. Cells were harvested at the indicated time points and time-dependent interactions among the indicated proteins were measured by IP and IB. (C) HEK293 cells were transfected with Myc-RUNX3-WT or Myc-RUNX3-S356A (a CDK4-mediated phosphorylation-defective RUNX3 mutant) and then stimulated with TGFβ1. Cells were harvested and the time-dependent interactions between the proteins were measured by IP and IB. (D) HEK293 cells were transfected with control siRNA (si-Con) or TAK1-specific siRNA (si-TAK1), and then stimulated with TGFβ1 for the indicated times. Cells were harvested and the time-dependent interactions between proteins were measured by IP and IB. (E) HEK293 cells were transfected with control siRNA (si-Con) or YAP1-specific siRNA (si-YAP1) and then stimulated with TGFβ1 for the indicated times. Cells were harvested and the time-dependent interactions between proteins were measured by IP and IB. TGFβ, transforming growth factor β.
Previously, we identified a mitogen-stimulated R-point-associated RUNX3 containing activator (RPA-RX3-AC) complex that activates R-point-associated gene expression (Lee et al., 2019a). Interaction between p300, RUNX3, and BRD2 play key roles in formation of the RPA-RX3-AC complex. p300 acetylates RUNX3 and histone 4. Next, BRD2 binds simultaneously to RUNX3 and histone 4 through its two bromodomains. Then, BRD2 recruits SWI-SNF (chromatin remodeling complex), MLL1 (activating chromatin by methylating histone, H3K4), and TFIID (basal transcriptional machinery) to trigger expression of R-point associated genes, including p21 (Lee et al., 2019a). p21 plays a key role in R-point regulation by activating Cyclin D-CDK4/6 and inhibiting Cyclin E-CDK (Chi et al., 2017; Malumbres and Barbacid, 2001; Pardee, 1974; Weinberg, 2007). When the mitogen-stimulated R-point decision is made, the RPA-RX3-AC complex is destroyed by CDK4-mediated RUNX3 phosphorylation at Ser-356.
Therefore, we investigated whether the complex formed 15 min after TGFβ stimulation is similar to the mitogen-stimulated R-point-associated complex (RPA-RX3-AC). IP/IB analysis revealed that YAP1 and RUNX3 associated with BRD2 15-30 min after TGFβ1 stimulation (Fig. 5B). At these time points, RUNX3 interacted with SMAD3, p300, MLL1, BRG-1 (a component of SWI-SNF), and TAF1 (a component of TFIID) (Fig. 5B). All of these RUNX3-interacting proteins are components of the RPA-RX3-AC complex (Lee et al., 2019a). At 30 min after TGFβ stimulation, CDK4 associated with the complex and phosphorylated RUNX3 at Ser-356 (Fig. 5B). Then, the complex was destroyed (Fig. 5B). Destruction of the complex was inhibited by expression of Myc-RUNX3-S356A, a CDK4-mediated phosphorylation-defective RUNX3 mutant (Fig. 5C). As the result, p21 expression was maintained for long time (Fig. 5C). These results suggest that the structure and mechanism responsible for dissociation of the complex are very similar to those of the RPA-RX3-AC complex. Therefore, we named the complex formed 15 min after TGFβ stimulation the “TGFβ-stimulated R-point associated activator (TGFβ-RPA-AC) complex”.
To understand the role of the TGFβ-stimulated R-point-associated activator (TGFβ-RPA-AC) complex in cell cycle regulation, we obtained an AAV expressing wild-type RUNX3 (AAV-RUNX3-WT) or RUNX3-S356A (AAV-RUNX3-S356A). HEK293 cells were infected with AAV-RUNX3-WT, AAV- RUNX3-S356A, or control AAV-empty and the proliferation rates of the cells were measured. The results revealed that the proliferation rate of the RUNX3-S356A expressing cells was significantly lower than that of the control and RUNX3-WT expressing cells (Supplementary Fig. S5). This result may suggest that the TGFβ-stimulated R-point-associated complex inhibits cell cycle progression by inducing p21 as long as the integrity of the complex is maintained.
SiRNA-mediated knockdown of TAK1 or YAP1 abolished the interactions between RUNX3, BRD2, and SMAD3 (Figs. 5D and 5E). These results demonstrate that the TGFβ→TAK1→LATS1/2→YAP1 pathway is essential for formation of the TGFβ-RPA-AC complex.
YAP1 and RUNX3 play key roles in TGFβ-stimulated gene expression regulation
CDKN1A (cyclin-dependent kinase inhibitor 1, referred to hereafter as p21) is one of the major targets of TGFβ signaling, and RUNX3 plays a key role in TGFβ-stimulated induction of p21 (Chi et al., 2005). CTGF is a major target of the YAP1/TEAD complex (Zhao et al., 2008), and its expression is induced by TGFβ stimulation (Kothapalli et al., 1998). Notably, p21 was induced 15 min after TGFβ stimulation (when the TGFβ-RPA-AC complex was formed), and CTGF was induced 120 min after stimulation (when the YAP1/SMAD3/TEAD4 complex was formed) (Figs. 5A and 5B).
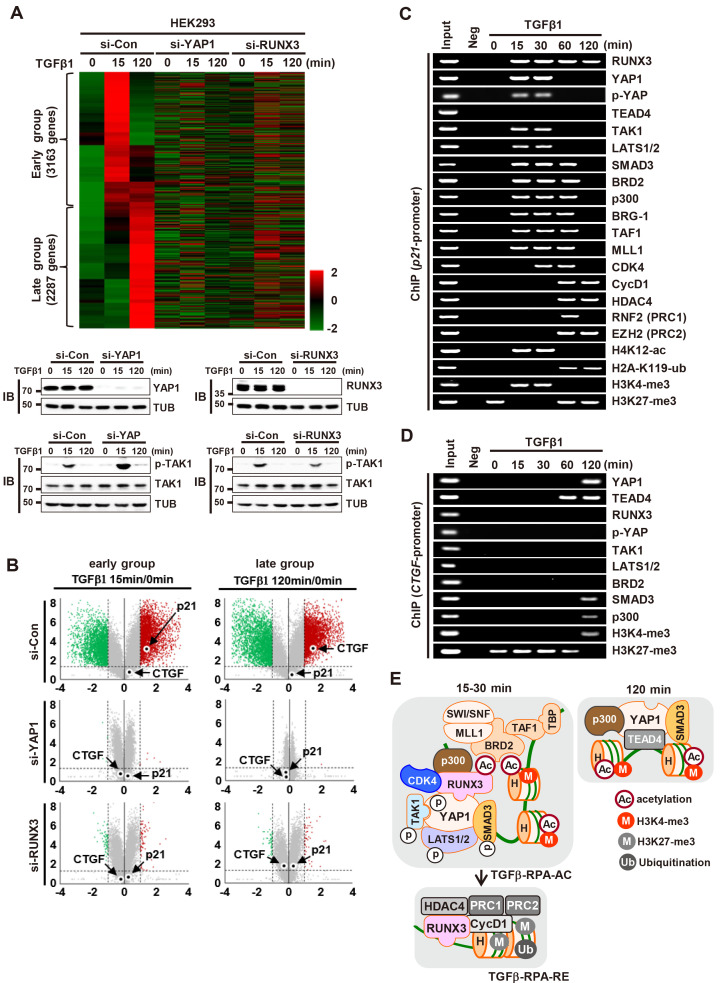
Next, we investigated the roles of YAP1 and RUNX3 in TGFβ-stimulated gene expression regulation by mRNA sequencing analysis. The results revealed that TGFβ-induced genes were categorized into two groups, early (3,163 genes) and late (2,287 genes), which are induced at 15 min after and 120 min after stimulation, respectively (Fig. 6A). Notably, siRNA-mediated knockdown of YAP1 led to significant disruption with respect to induction of both early and late genes (Figs. 6A and 6B). Consistent with the IB analysis shown in Figs. 5A and 5B, p21 was included in the early group, and CTGF was included in the late group (Fig. 6B). These results suggest that YAP1 forms distinct complexes (TGFβ-RPA-AC and YAP1/SMAD3/TEAD4), and that these complexes play roles in time-dependent induction of distinct target genes after TGFβ signaling. In this study, we used p21 and CTGF as representative early and late targets, respectively, induced by TGFβ-activated YAP1.
Fig. 6. YAP1 and RUNX3 play key roles in TGFβ-stimulated gene expression regulation.
(A) HEK293 cells were treated with control siRNA (si-con), RUNX3-specific siRNA (si-RUNX3), or YAP1-specific siRNA (si-YAP1), and then stimulated with TGFβ1. RNA was extracted from the cells at the indicated time points, and gene expression patterns were analyzed by mRNA sequencing (n = 3; biological replicates). Heatmap showing genes (5,450 genes) upregulated 15 min or 120 min after TGFβ1 stimulation, and expression of the genes in si-RUNX3 or si-YAP1-treated cells. (B) Volcano plot showing fold changes (log2FC > 1) in gene expression in the indicated samples (15 min or 120 min), along with statistical significance (P < 0.05, -log10[P value]). Significantly up- (red) or downregulated (green) genes are highlighted. (C) HEK293 cells were stimulated with TGFβ1. The binding of RUNX3, YAP1, TEAD4, TAK1, LATS1/2, SMAD3, BRD2, p300, BRG-1, TAF1, MLL1, CDK4, Cyclin D1, HDAC4, RNF2, and EZH2 to the p21 promoter and histone markers (H4K12-ac, H2A-K119-ub, H3K4-tri-met, and H3K27-tri-met) at the locus was measured by ChIP at the indicated time points. (D) HEK293 cells were stimulated with TGFβ1. Binding of RUNX3, YAP1, TEAD4, TAK1, LATS1/2, SMAD3, BRD2, and p300 to the CTGF promoter and histone markers (H3K4-tri-met and H3K27-tri-met) at the locus was measured by ChIP at the indicated time points. (E) Schematic illustration of the complexes formed after TGFβ stimulation. YAP1 forms a TGFβ-RPA-AC complex 15-30 min after TGFβ stimulation. Subsequently, TGFβ-RPA-AC is destroyed and RUNX3 forms a RPA-RX3-RE complex 60-120 min after stimulation. YAP1 forms a YAP1/TEAD4/SMAD3/p300 complex 120 min after stimulation. TGFβ, transforming growth factor β; IB, immunoblotting; ChIP, chromatin immunoprecipitation.
Interestingly, knockdown of RUNX3 disrupted induction of genes in both groups (Fig. 6A), although RUNX3 formed a TGFβ-RPA-AC complex only at 15 min after TGFβ-stimulation. These results suggest that early group genes induced by the complex affect induction of late group genes by regulating late group genes. The TGFβ-induced YAP1-dependent genes involved in the TGFβ pathway, Hippo pathway, cell cycle regulation, and epithelial-mesenchymal transition (EMT) are listed in Supplementary Fig. S6. Detailed RNA sequencing data is provided in the Excel file (Supplementary Data 1).
Dynamic regulation of target chromatin loci by the TGFβ- stimulated complexes
ChIP analysis revealed that all components of the TGFβ-RPA-AC complex (YAP1, RUNX3, TAK1, LATS1/2, SMAD3, p300, BRD2, CDK4, MLL1, SWI/SNF, and TFIID) were recruited to the p21 locus 15-30 min after TGFβ1 stimulation (Fig. 6C). At these time points, the p21 locus was marked with activating histone modifications (H4K12-ac and H3K4-tri-met) (Fig. 6C) and p21 expression was induced (Figs. 5A, 5B, and 6B). Thus, p300 and MLL1 of the TGFβ-RPA-AC complex might contribute to H4K12 acetylation and H3K4 tri-methylation, respectively.
Polycomb group (PcG) complexes are classified into two categories: Polycome repressor 1 and 2 (PRC1 and PRC2). Ring finger protein 2 (RNF2), which ubiquitinates H2A at lysine 119 (H2A-K119-ub), is a component of PRC1 (Wang et al., 2004) and enhancer of zeste homologs (EZH2), which trimethylates histone 3 at lysine 27 (H3K27-tri-met), is a component of PRC2 (Bracken et al., 2003). H2A-K119-ub and H3K27-tri-met histone marking by PRC1 and PRC2 are characteristics of inactive chromatin (Cao et al., 2002). Sixty minutes after TGFβ1 stimulation, RUNX3 associated with Cyclin D1, RNF2, EZH2, and HDAC4 (Fig. 5B). These results demonstrate that the RUNX3-Cyclin D1-PRC1-PRC2-HDAC4 complex (hereafter named the TGFβ-stimulated R-point associated repressor [TGFβ-RPA-RE] complex) is formed after the TGFβ-RPA-AC complex is destroyed. The TGFβ-RPA-RE complex is very similar to the RPA-RX3-RE complex, which is formed after the mitogen-stimulated R-point (Lee et al., 2019a).
ChIP analysis revealed that the TGFβ-RPA-RE complex was recruited to the p21 locus 60 min after TGFβ1 stimulation (Fig. 6C). At that time point, H4K12-ac and H3K4-tri-met (activatory histone marks) were erased, and H2A-K119-ub and H3K27-tri-met (inhibitory histone marks) were enriched, at the locus (Fig. 6C). Consistent with this, p21 expression was suppressed markedly 60 min after stimulation (Figs. 5A, 5B, and 6B). These results suggest that the p21 locus is activated by the TGFβ-RPA-AC complex early after TGFβ1 stimulation, and that the locus is subsequently inactivated by RPA-RX3-RE. HDAC4, PRC1, and PRC2 of TGFβ-RPA-RE might contribute to inactivation of the locus by deacetylating H4K12-ac, ubiquitinating H2A-K119-ub, and methylating H3K4-tri-met, respectively.
As TEAD4 was not included in the TGFβ-RPA-AC complex, the protein was not recruited to the p21 locus at any time point (Fig. 6C). Rather, TEAD4 was recruited to the CTGF locus 60-120 min after TGFβ stimulation (Fig. 6D). Then, YAP1, SMAD3, and p300 were recruited to the locus 120 min after stimulation (Fig. 6D). At that time point, the histone markers of the CTGF locus changed from inhibitory modifications (H3K27-tri-met) to activating modifications (H3K4-tri-met) (Fig. 6D). Consistent with this, CTGF expression was induced 120 min after TGFβ stimulation (Figs. 5A, 5B, and 6B). However, RUNX3, TAK1, LATS1/2, and BRD2 were not recruited to the CTGF locus at any time point, demonstrating that the TGFβ-RPA-AC complex is not recruited to the locus (Fig. 6D). These results suggest that the TGFβ signal activates a distinct set of target genes through formation of different complexes at different time points. When the TGFβ→TAK1→LATS1/2→YAP1 pathway was activated (15 min after stimulation), YAP1 formed a TGFβ-RPA-AC complex and activated early group genes, including p21. Then, the complex was destroyed and the RPA-RX3-RE complex was formed, which suppressed early group genes (Fig. 6E). When the TGFβ signal was attenuated (120 min after stimulation), YAP1 formed a different complex with TEADs, SMADs, and p300, and then activated late group genes, including CTGF (Fig. 6E).
AP1 is an essential component of the TGFβ-RPA-AC complex
TGFβ activates AP1 (c-JUN/c-FOS heterodimeric transcription factor) through the MAPK pathway, after which activated AP1 associates with SMADs to mediate TGFβ-induced transcription (Zhang et al., 1998). AP1 also associates with YAP1 (Zanconato et al., 2015). Therefore, we investigated whether AP1 contributes to TGFβ-RPA-AC complex formation. TGFβ stimulation followed by IP/IB analysis revealed that YAP1 and AP1 associated 15-30 min after stimulation, dissociated 60 min after stimulation, and re-associated 120 min after stimulation (Fig. 7A). By contrast, RUNX3 and AP1 associated 15-60 min after stimulation and then dissociated (Fig. 7A). ChIP analysis revealed that AP1 was recruited to the p21 locus 15-60 min after stimulation, and to the CTGF locus 120 min after stimulation (Supplementary Fig. S7A). These results suggest that AP1 is recruited to the p21 locus as a component of the TGFβ-RPA-AC complex, and to the CTGF locus through interaction with YAP1/TEAD.
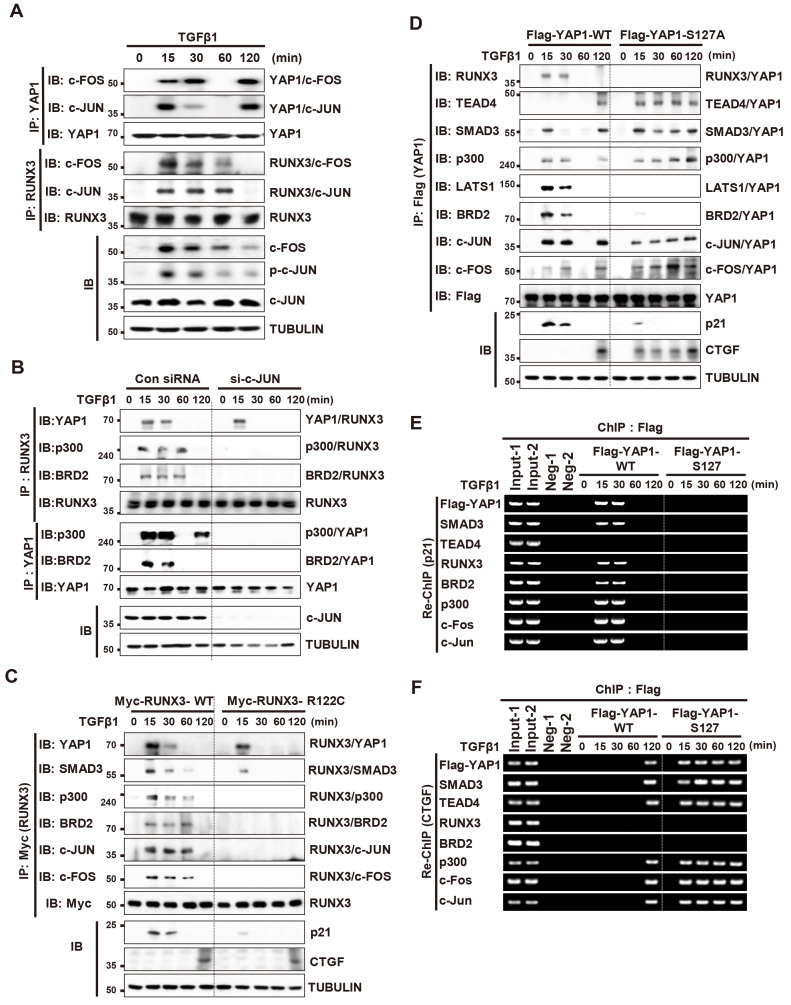
Fig. 7. AP1 is an essential component of the TGFβ-RPA-AC complex.
(A) HEK293 cells were stimulated with TGFβ1. Cells were harvested at the indicated time points and expression of the indicated proteins and time-dependent interactions among proteins were measured by immunoprecipitation (IP) and immunoblotting (IB). (B) HEK293 cells were transfected with control siRNA (si-Con) or c-Jun-specific siRNA (si-c-JUN), and then stimulated with TGFβ1. Cells were harvested and the time-dependent interactions between proteins was measured by IP and IB. (C) HEK293 cells were transfected with Myc-RUNX3-WT or Myc-RUNX3-R122C (an inactive form of RUNX3 mutant) and then stimulated with TGFβ1. Cells were harvested and the time-dependent interactions between proteins were measured by IP and IB. (D) HEK293 cells were transfected with Flag-YAP1-WT or Flag-YAP1-S127A (a phosphorylation-defective YAP1 mutant), and then stimulated with TGFβ1. Cells were harvested at the indicated time points and the interactions between proteins were measured by IP and IB. (E and F) HEK293 cells cultured at low cell density were transfected with Flag-YAP1-WT or Flag-YAP1-S127A, and then stimulated with TGFβ1. ChIP was performed using an anti-Flag antibody, followed by sequential ChIP (Re-ChIP) with anti-SMAD3, -TEAD4, -RUNX3, -BRD2, -p300, -c-FOS, and -c-JUN antibodies. Binding of SMAD3, TEAD4, RUNX3, BRD2, p300, c-FOS, and c-JUN Flag-YAP1 (WT or S127A) to the p21 (B) or CTGF (C) promoter at the locus was measured by Re-ChIP at the indicated time points. TGFβ, transforming growth factor β; ChIP, chromatin immunoprecipitation.
We then examined the role of AP1 in TGFβ-RPA-AC complex formation. SiRNA-mediated knockdown of c-JUN did not affect the YAP1-RUNX3 interaction, but it abolished the RUNX3-p300, RUNX3-BRD2, YAP1-p300, and YAP1-BRD2 interactions, which are essential for TGFβ-RPA-AC complex formation (Fig. 7B). p300 is essential for recruitment of BRD2 to the R-point-associated complex (Lee et al., 2019a). p300 also associates with AP1 (Goodman and Smolik, 2000). These results suggest that AP1 associates with the YAP1/RUNX3 complex and plays a role in recruiting p300, which enables subsequent association of BRD2.
RUNX3-R122C (Arg-122 to Cys) is an inactive RUNX3 mutant found in a gastric cancer patient (Li et al., 2002). Exogenously-expressed Myc-RUNX3-R122C interacted with YAP1, but failed to interact with AP1, p300, and BRD2 (Fig. 7C). These results suggest that RUNX3-R122C fails to form a TGFβ-RPA-AC complex due to failure of the interaction with AP1. These results together suggest that the p-TAK1/p-LATS/p-YAP1/RUNX3/p-SMAD3 complex is formed 15 min after TGFβ stimulation, and then associates with MAPK pathway-activated AP1. p300 is recruited to the complex through interaction with AP1 and then facilitates recruitment of BRD2 by acetylating RUNX3 and histone 4. Subsequently, BRD2 recruits MLL1, SWI/SNF, and TFIID to form the TGFβ-RPA-AC complex (Supplementary Fig. S7B).
Oncogenic mutation of YAP1 disrupts the TGFβ-stimulated gene expression program.
We then examined whether oncogenic mutation of YAP1 affects the TGFβ-stimulated gene expression regulation. YAP1-S127A is an oncogenic mutant of YAP1 (Schlegelmilch et al., 2011; Zhang et al., 2009; 2011; Zhao et al., 2007). Exogenously-expressed Flag-YAP1-WT formed a TGFβ-RPA-AC complex 15-30 min after TGFβ1 stimulation, dissociated 60 min after, and formed the YAP1/TEAD4/SMAD3/p300/AP1 complex 120 min after (Fig. 7D). Re-ChIP analysis revealed that the TGFβ-RPA-AC complex was recruited to the p21 locus 15-30 min after stimulation (Fig. 7E), and that the YAP1/TEAD4/SMAD3/p300/AP1 complex was recruited to the CTGF locus 120 min after TGFβ stimulation (Fig. 7F). p21 and CTGF were induced when the TGFβ-RPA-AC or YAP1/TEAD4/SMAD3/p300/AP1 complex was recruited to each locus, respectively (Fig. 7D). However, exogenously-expressed Flag-YAP1-S127A failed to form a TGFβ-RPA-AC complex at any time point after TGFβ1 stimulation (Fig. 7D), nor was it recruited to the p21 locus (Fig. 7E). Instead, Flag-YAP1-S127A formed a YAP1/TEAD4/SMAD3/p300/AP1 complex 15 min after stimulation and the complex was maintained until 120 min after (Fig. 7D). The complex was recruited to the CTGF locus (Fig. 7F), where it induced CTGF expression at the time points (15-120 min after) (Fig. 7D). These results demonstrate that the oncogenic YAP1 mutation disrupts the TGFβ-stimulated regulatory program by inhibiting TGFβ-RPA-AC complex formation and facilitating YAP1/TEAD4/SMAD3/p300/AP1 complex formation.
The TGFβ-stimulated regulatory program is disrupted in cancer cell lines
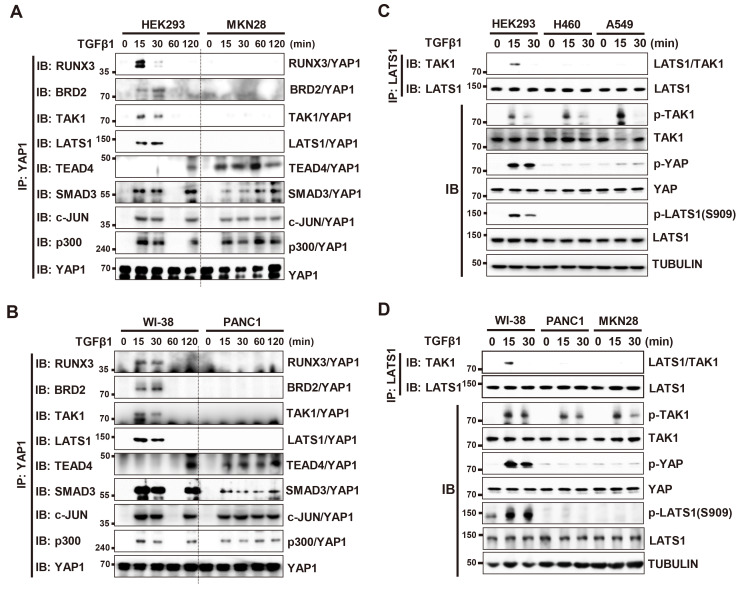
In HEK293 and WI-38 cells (immortalized normal cell lines), the TGFβ-RPA-AC complex was formed 15 min after TGFβ stimulation and the YAP1/TEAD4/SMAD3/AP1/p300 complex was formed 120 min after (Figs. 8A and 8B). However, in the MKN28 and PANC1 cell lines (cancer cell lines), the TGFβ-RPA-AC complex was not formed at any time point, and the YAP1/TEAD4/SMAD3/AP1/p300 complex was formed 15 min after TGFβ stimulation and maintained for a long time (Figs. 8A and 8B). This failure of the TGFβ-RPA-AC complex formation in cancer cells is essentially the same as those observed for Flag-YAP1-S127A-expressed HEK293 cells (Fig. 7D). Notably, in all four cancer cell lines (H460, A549, PANC1 and MKN28), TAK1 was normally activated, but LATS1 was not activated by TGFβ stimulation (Figs. 8C and 8D). These results demonstrate that abrogation of the TGFβ→TAK1→LATS1/2→YAP1 pathway leads to failure of TGFβ-RPA-AC complex formation, which is associated with malignant transformation. The TGFβ-stimulated regulatory program in immortalized normal cell lines, and abrogation of the program in cancer cell lines, are summarized in Figs. 9A and 9B, respectively.
Fig. 8. The TGFβ→TAK1→LATS→YAP1 pathway is disrupted in transformed cell lines.
(A) HEK293 (immortalized normal cell line) and MKN28 (cancer cell line) cells were stimulated with TGFβ1. Cells were harvested at the indicated time points, and expression of the indicated proteins and time-dependent interactions among the proteins were measured by immunoprecipitation (IP) and immunoblotting (IB). (B) WI-38 (immortalized normal cell line) and PANC1 (cancer cell line) cells were stimulated with TGFβ1 at low cell density. Cells were harvested at the indicated time points, and the levels of the indicated proteins and time-dependent interactions among the proteins were measured by IP and IB. (C and D) Immortalized normal cells (HEK293 and WI-38) and cancer cells (H460, A549, PANC1, and MKN28) were stimulated with TGFβ1. Time-dependent phosphorylation of LATS1 was analyzed by IB at the indicated time points. TGFβ, transforming growth factor β.
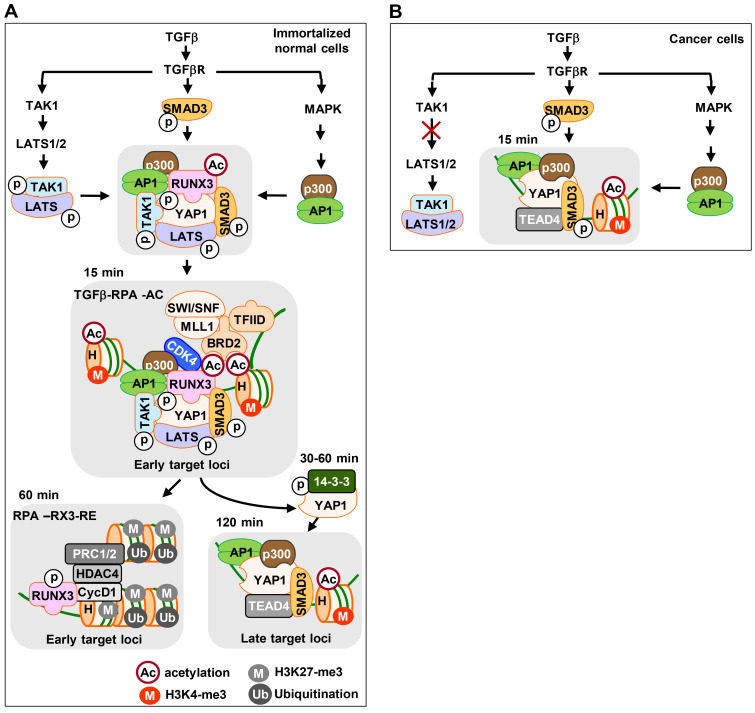
Fig. 9. Schematic illustration of sequential molecular interactions in response to TGFβ stimulation.
(A) The TGFβ→TAK1→LATS→YAP1 pathway facilitates YAP1 phosphorylation in immortalized normal cell lines. p-YAP1, p-SMAD3 (activated by the SMAD pathway), and AP1 (activated by the MAPK pathway) form a TGFβ-RPA-AC complex 15 min after TGFβ stimulation. The TGFβ-RPA-AC complex induces expression of early target genes. Then, the complex dissociates and p-YAP1 is exported from the nucleus to the cytoplasm via interaction with 14-3-3 (30-60 min after stimulation). When the TGFβ signal is attenuated (120 min after stimulation), unphosphorylated YAP1 re-localizes to the nucleus and forms the YAP1/TEAD4/SMAD3/AP1/p300 complex. The complex then activates late target genes. (B) In cancer cell lines, the TGFβ→TAK1→LATS→YAP1 pathway is abrogated due to the failure of TAK1-mediated LATS1/2 phosphorylation. As the result, the TGFβ-RPA-AC complex is not formed. Instead, a YAP1/TEAD4/SMAD3/AP1/p300 complex is formed 15 min after TGFβ stimulation and is maintained for a long time. The complex activates late target genes from early time points. TGFβ, transforming growth factor β.
DISCUSSION
The key unknowns about the Hippo pathway are which extracellular signals activate LATS1/2, and what are the molecular mechanisms underlying commitment to specific cellular responses? In this study, we show that TGFβ-activated TAK1 activates LATS1/2, which phosphorylates YAP1; p-YAP1 then forms a TGFβ-RPA-AC complex, which plays a role in gene expression regulation in the nucleus. These results identify a new pathway (TGFβ→TAK1→LATS→YAP1) that connects TGFβ signals, the Hippo pathway.
It is thought that YAP1 phosphorylation inactivates its nuclear activity. However, our results demonstrate that YAP1 phosphorylation in fact activates its nuclear activity by triggering the p-YAP1-RUNX3 interaction, leading to TGFβ-RPA-AC complex formation. To modulate chromatin accessibility and regulate gene transcription, a special transcription factor is required to activate a complex network of other proteins, including coactivators, histone-modifying complexes, chromatin remodeling complexes, and the basal transcription machinery. Such special transcription factors that have the capacity to associate with condensed chromatin and modulate chromatin accessibility are known as pioneer factors (Jozwik and Carroll, 2012). RUNX3 is a pioneer factor of the mitogen-stimulated R-point (Lee et al., 2019a). Our results show that the p-YAP1-RUNX3 interaction is essential for formation of the TGFβ-RPA-AC complex, which opens closed target chromatin loci and activates target gene expression. These results identify a new role for p-YAP1 in the nucleus: an essential partner of pioneer factor RUNX3 for regulation of the TGFβ-stimulated gene expression regulation.
Although RUNX proteins were the first identified DNA binding partners of YAP1, the biological meaning of the YAP1-RUNX interaction was insufficiently studied in relation to the YAP1-TEADs interaction. One explanation is because the p-YAP1 interaction with RUNX3 is transient and occurs only for a limited time after TGFβ stimulation in normal cells, while this interaction does not occur in cancer cells. In contrast, unphosphorylated YAP1 interacts with TEADs and the resulting complex persists for a long time in cancer cells.
We show that the TGFβ-TAK1-LATS-YAP1 pathway triggers a precise sequence of molecular events. First, YAP1 phosphorylation in the nucleus and supports formation of a transient TGFβ-induced RPA-AC complex and subsequent export of p-YAP1 to the cytoplasm. After dephosphorylation or new synthesis in the absence of a TGFβ signal, unphosphorylated YAP1 relocalizes to the nucleus to form a stable YAP1/TEAD4/SMAD3/p300/AP1 complex. These spatiotemporal dynamics of YAP1 regulation appears to be associated with the R-point program that determines cell proliferation versus alternative cell fates (e.g., differentiation, and apoptosis). The R-point program is disrupted in nearly all cancer cells (Malumbres and Barbacid, 2001; Pardee, 1974; Weinberg, 2007). Indeed, the TGFβ-stimulated spatiotemporal dynamics of YAP1 were disturbed in all cancer cell lines we analyzed due to failure of TGFβ–TAK1-LATS-YAP1 pathway activation. The spatiotemporal dynamics of YAP1 were also disturbed by the oncogenic YAP1 mutation (YAP1-S127A). These results suggest that the TGFβ–TAK1-LATS-YAP1 pathway is a tumor suppressor pathway.
It is worth mentioning that SWI/SNF, a chromatin remodeling complex that functions in tumor suppression (Kadoch and Crabtree, 2015), forms a complex with YAP1 and inhibits the pro-oncogenic activity of YAP1 (Chang et al., 2018). The complex does not contain TEADs (Chang et al., 2018). These results are consistent with our observations that the TGFβ-RPA-AC complex, which contains YAP1, RUNX3, and SWI/SNF, but not TEAD, functions as a tumor suppressor. Therefore, SWI/SNF may inhibit (at least partly) the pro-oncogenic activity of YAP1 and function as a tumor suppressor by forming the TGFβ-RPA-AC complex.
Although YAP1 plays essential roles in initiation or progression of various cancers (Zanconato et al., 2016), it was recognized recently as a tumor suppressor as well (Cottini et al., 2014; Pearson et al., 2021). TGFβ also elicits completely opposite responses under different physiological conditions. At the early stage of tumorigenesis, TGFβ has tumor suppressive functions, primarily through cell cycle arrest and apoptosis. At the late stage, TGFβ acts as a driver of tumor progression and metastasis (Ikushima and Miyazono, 2010; Massague, 2008). Our results suggest that the opposite roles of YAP1 and TGFβ may occur because their roles are not determined by their own activity, but determined in a context- and/or cell type-dependent manner at the R-point.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Funding Statement
ACKNOWLEDGMENTS S.-C.B. is supported by a Creative Research Grant (NRF-2014R1A3A2030690) through the National Research Foundation (NRF) of Korea. J.-W.L. is supported by Basic Science Research Program grant (NRF-2021R1I1A1A01060610) of Korea. M.-K.K. is supported by Basic Science Research Program grant (NRF-2017R1A6A3A11028050) of Korea. S.-H.S. is supported by Basic Science Research Program grant (NRF-2021R1I1A1A01059185). E.-G.K. is supported by Medical Research Center (MRC-2020R1A5A2017476) of Korea. D.-S.L. is supported by the National Creative Research Initiatives (NRF-2020-2079551) of Korea.
Footnotes
AUTHOR CONTRIBUTIONS
M.-K.K., S.-H.H., T.-G.P., S.-H.S., and J.-W.J. analyzed assembly and disassembly of the R-point–associated complexes after TGFβ-stimulation. J.-Y.L., Y.-S.L., S.-Y.Y., and X.-Z.C. maintained knockout mice, prepared primary MEFs and performed ChIP assay. J.-W.L. analyzed RNA sequencing data. E.-G.K., D.S.L., A.J.W., J.-W.L. and S.-C.B. designed experiments and interpreted the results. J.-W.L. and S.-C.B. planed the project and wrote the manuscript. All authors contributed to the editing of the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Blagosklonny M.V., Pardee A.B. The restriction point of the cell cycle. Cell Cycle. 2002;1:103–110. doi: 10.4161/cc.1.2.108. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. BBMap. 2014. [Retrieved August 19, 2021]. from https://sourceforge.net/projects/bbmap/
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chang L., Azzolin L., Di Biagio D., Zanconato F., Battilana G., Lucon Xiccato R., Aragona M., Giulitti S., Panciera T., Gandin A., et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018;563:265–269. doi: 10.1038/s41586-018-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P.C., Campbell D.G., Nebreda A.R., Cohen P. Feedback control of the protein kinase TAK1 by SAPK2a/p38alpha. EMBO J. 2003;22:5793–5805. doi: 10.1093/emboj/cdg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X.Z., Lee J.W., Lee Y.S., Park I.Y., Ito Y., Bae S.C. Runx3 plays a critical role in restriction-point and defense against cellular transformation. Oncogene. 2017;36:6884–6894. doi: 10.1038/onc.2017.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X.Z., Yang J.O., Lee K.Y., Ito K., Sakakura C., Li Q.L., Kim H.R., Cha E.J., Lee Y.H., Kaneda A., et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol. Cell. Biol. 2005;25:8097–8107. doi: 10.1128/MCB.25.18.8097-8107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottini F., Hideshima T., Xu C., Sattler M., Dori M., Agnelli L., ten Hacken E., Bertilaccio M.T., Antonini E., Neri A., et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat. Med. 2014;20:599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Lu J., Li W., Wu A., Zhang X., Tong W., Ho K.K., Qin L., Song H., Mak K.K. Reciprocal inhibition of YAP/TAZ and NF-kappaB regulates osteoarthritic cartilage degradation. Nat. Commun. 2018;9:4564. doi: 10.1038/s41467-018-07022-2.d52048cf054c48528cb5a67044142ce7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissova N.G., Pouponnot C., Long J., He D., Liu F. Transforming growth factor beta -inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6397–6402. doi: 10.1073/pnas.090099297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R., Kim N.G., Gumbiner B.M. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2569–2574. doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Toyoda T., Nakanishi H., Yatabe Y., Sato A., Matsudaira Y., Ito H., Murakami H., Kondo Y., Kondo E., et al. TGF-beta synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. J. Exp. Med. 2012;209:479–494. doi: 10.1084/jem.20111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. doi: 10.1101/gad.14.13.1553. [DOI] [PubMed] [Google Scholar]
- Hannon Lab., author FASTX toolkit. 2014. [Retrieved August 19, 2021]. from http://hannonlab.cshl.edu/fastx_toolkit/
- Hansen C.G., Moroishi T., Guan K.L. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25:499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H., Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat. Rev. Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Ishida W., Hamamoto T., Kusanagi K., Yagi K., Kawabata M., Takehara K., Sampath T.K., Kato M., Miyazono K. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem. 2000;275:6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Ito Y., Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr. Opin. Genet. Dev. 2003;13:43–47. doi: 10.1016/S0959-437X(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Jang J.W., Kim M.K., Lee Y.S., Lee J.W., Kim D.M., Song S.H., Lee J.Y., Choi B.Y., Min B., Chi X.Z., et al. RAC-LATS1/2 signaling regulates YAP activity by switching between the YAP-binding partners TEAD4 and RUNX3. Oncogene. 2017;36:999–1011. doi: 10.1038/onc.2016.266. [DOI] [PubMed] [Google Scholar]
- Jozwik K.M., Carroll J.S. Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer. 2012;12:381–385. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- Kadoch C., Crabtree G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F., Marignani P.A., Sarbassova D., Yagi R., Hall R.A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L.C., et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli D., Hayashi N., Grotendorst G.R. Inhibition of TGF-beta-stimulated CTGF gene expression and anchorage-independent growth by cAMP identifies a CTGF-dependent restriction point in the cell cycle. FASEB J. 1998;12:1151–1161. doi: 10.1096/fasebj.12.12.1151. [DOI] [PubMed] [Google Scholar]
- Labibi B., Bashkurov M., Wrana J.L., Attisano L. Modeling the control of TGF-beta/Smad nuclear accumulation by the Hippo pathway effectors, Taz/Yap. iScience. 2020;23:101416. doi: 10.1016/j.isci.2020.101416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Kim D.M., Jang J.W., Park T.G., Song S.H., Lee Y.S., Chi X.Z., Park I.Y., Hyun J.W., Ito Y., et al. RUNX3 regulates cell cycle-dependent chromatin dynamics by functioning as a pioneer factor of the restriction-point. Nat. Commun. 2019a;10:1897. doi: 10.1038/s41467-019-09810-w.ce5cd98dd35b4a8b97f2416472079307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Lee Y.S., Kim M.K., Chi X.Z., Kim D., Bae S.C. Role of RUNX3 in restriction point regulation. Cells. 2023;12:708. doi: 10.3390/cells12050708.dd1719ce589445a88288d836a578d16d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Park T.G., Bae S.C. Involvement of RUNX3 and BRD family members in restriction point. Mol. Cells. 2019b;42:836–839. doi: 10.14348/molcells.2019.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Lee J.W., Jang J.W., Chi X.Z., Kim J.H., Li Y.H., Kim M.K., Kim D.M., Choi B.S., Kim E.G., et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell. 2013;24:603–616. doi: 10.1016/j.ccr.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Lee J.W., Somg S.H., Kim D.M., Lee J.W., Chi X.Z., Ito Y., Bae S.C. K-Ras-activated cells can develop into lung tumors when Runx3-mediated tumor suppressor pathways are abrogated. Mol. Cells. 2020;43:889–897. doi: 10.14348/molcells.2020.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.L., Ito K., Sakakura C., Fukamachi H., Inoue K., Chi X.Z., Lee K.Y., Nomura S., Lee C.W., Han S.B., et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/S0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Liu X., Li H., Rajurkar M., Li Q., Cotton J.L., Ou J., Zhu L.J., Goel H.L., Mercurio A.M., Park J.S., et al. Tead and AP1 coordinate transcription and motility. Cell Rep. 2016;14:1169–1180. doi: 10.1016/j.celrep.2015.12.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y., Huang B., Shim J.S., Chen Q., Lee S.J., Anders R.A., Liu J.O., Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.S.J., Schmierer B., Hill C.S. TGF-beta family ligands exhibit distinct signalling dynamics that are driven by receptor localisation. J. Cell Sci. 2019;132:jcs234039. doi: 10.1242/jcs.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E., Yang J., DeRan M., Wu C., Su A.I., Bonamy G.M., Liu J., Peters E.C., Wu X. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 2012;19:955–962. doi: 10.1016/j.chembiol.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Nagarajan R.P., Zhang J., Li W., Chen Y. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J. Biol. Chem. 1999;274:33412–33418. doi: 10.1074/jbc.274.47.33412. [DOI] [PubMed] [Google Scholar]
- Nakamura R., Hiwatashi N., Bing R., Doyle C.P., Branski R.C. Concurrent YAP/TAZ and SMAD signaling mediate vocal fold fibrosis. Sci. Rep. 2021;11:13484. doi: 10.1038/s41598-021-92871-z.dbec92e0fd3d4248a6d8406d315a86f9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E., Tamaki K., Hanai J., Heldin C.H., Miyazono K., et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Teramura T., Takehara T., Fukuda K. Transforming growth factor beta-activated kinase 1 regulates mesenchymal stem cell proliferation through stabilization of Yap1/Taz proteins. Stem Cells. 2019;37:1595–1605. doi: 10.1002/stem.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee A.B. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.D., Huang K., Pacal M., McCurdy S.R., Lu S., Aubry A., Yu T., Wadosky K.M., Zhang L., Wang T., et al. Binary pan-cancer classes with distinct vulnerabilities defined by pro- or anti-cancer YAP/TEAD activity. Cancer Cell. 2021;39:1115–1134.e12. doi: 10.1016/j.ccell.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Lin S.J., Chen Y., Voon D.C., Zhu F., Chuang L.S., Wang T., Tan P., Lee S.C., Yeoh K.G., et al. RUNX3 is a novel negative regulator of oncogenic TEAD-YAP complex in gastric cancer. Oncogene. 2016;35:2664–2674. doi: 10.1038/onc.2015.338. [DOI] [PubMed] [Google Scholar]
- R Core Team, author. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [Google Scholar]
- Reddy B.V., Irvine K.D. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell. 2013;24:459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Zhang L., Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev. Biol. 2010;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R., Zanotto M., Simionato F., Zecchetto C., Merz V., Cavallini C., Piro G., Sabbadini F., Boschi F., Scarpa A., et al. Modulating TAK1 expression inhibits YAP and TAZ oncogenic functions in pancreatic cancer. Mol. Cancer Ther. 2020;19:247–257. doi: 10.1158/1535-7163.MCT-19-0270. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J.R., Zhou D., Kreger B.T., Vasioukhin V., Avruch J., Brummelkamp T.R., et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- Simon A. FastQC. 2010. [Retrieved August 19, 2021]. from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Sudol M., Bork P., Einbond A., Kastury K., Druck T., Negrini M., Huebner K., Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- Varelas X., Miller B.W., Sopko R., Song S., Gregorieff A., Fellouse F.A., Sakuma R., Pawson T., Hunziker W., McNeill H., et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B.M., Dembowy J., Yaffe M.B., Zandstra P.W., Wrana J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Weinberg R.A. pRb and control of the cell cycle clock. In: Weinberg R.A., editor. The Biology of Cancer. Garland Sciences; New York: 2007. pp. 275–329. [Google Scholar]
- Yagi R., Chen L.F., Shigesada K., Murakami Y., Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Yu J., Zheng Y., Chen Q., Zhang N., Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J., Yuan H., Tumaneng K., Li H., et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M., Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Pasolli H.A., Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Milton C.C., Humbert P.O., Harvey K.F. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng X.H., Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M., et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data mining and graphic visualization were performed using ExDEGA (Ebiogen Inc., Korea). The RNA sequencing data are available under accession number GSE226590 at Gene Expression Omnibus (GEO).