Summary
Antibiotic resistance genes (ARGs) are emerging pollutants present in various environments. Identifying ARGs has become a growing concern in recent years. Several databases, including the Antibiotic Resistance Genes Database (ARDB), Comprehensive Antibiotic Resistance Database (CARD), and Structured Antibiotic Resistance Genes (SARG), have been applied to detect ARGs. However, these databases have limitations, which hinder the comprehensive profiling of ARGs in environmental samples. To address these issues, we constructed a non-redundant antibiotic resistance genes database (NRD) by consolidating sequences from ARDB, CARD, and SARG. We identified the homologous proteins of NRD from Non-redundant Protein Database (NR) and the Protein DataBank Database (PDB) and clustered them to establish a non-redundant comprehensive antibiotic resistance genes database (NCRD) with similarities of 100% (NCRD100) and 95% (NCRD95). To demonstrate the advantages of NCRD, we compared it with other databases by using metagenome datasets. Results revealed its strong ability in detecting potential ARGs.
Subject areas: Structural biology, Omics
Graphical abstract

Highlights
-
•
A non-redundant comprehensive database for detecting ARGs is developed
-
•
The type and number of ARGs identified are greater than those in previous databases
-
•
A user-friendly web server provides a comprehensive analysis and explanation
Structural biology; Omics
Introduction
Since the discovery of penicillin and streptomycin, antibiotics have been widely applied to provide an effective treatment for prevalent diseases.1 However, with the overuse and abuse of antibiotics, antibiotic resistance genes (ARGs) have been recognized as emerging pollutants that are widely distributed and accumulated in most natural environment niches, including aquatic water ecosystems,2 soil,3 and human feces.4 These environment niches are considered key hotspots for the spread of antimicrobial resistance.5 Additionally, with the increasing popularity and scale of metagenomic experiments, the identification of ARGs in metagenomic data with high accuracy and efficiency is essential to profile the composition of ARGs in microbial communities from different environmental niches and is indispensable in understanding the ecology and dissemination of ARGs between environment- and human-related reservoirs.6 In the last few decades, many molecular biological methods, such as traditional polymerase chain reaction (PCR) and quantitative PCR (qPCR), have been developed and applied to investigate ARGs in environmental samples.7 Compared with qPCR, the metagenomic approach is a popular tool to identify ARGs and detect new types of ARGs because of its advantages. First, the metagenomic approach has a broad coverage because it can simultaneously identify a large number of ARGs in multiple samples. By contrast, qPCR typically requires specific primers and probes to detect specific ARGs.8 Second, the metagenomic approach considerably improves detection efficiency, thereby saving time and reducing experimental costs. Metagenomic methods do not require prior knowledge or a specific primer design and can directly extract all DNA sequence information from environmental samples. This capability allows for the discovery of ARGs or unknown antibiotic resistance mechanisms. However, metagenomic analysis has specific limitations, including reduced sensitivity, need for complex data interpretation, and high costs. Therefore, the selection of the appropriate method should be based on specific research requirements and available resources.
In recent years, the application of high-throughput sequencing technology has made the analysis of ARG sequences simple and easy, and metagenomic analysis has attracted extensive attention in the identification of ARGs.9,10 It allows access to genomic data in environmental samples and does not require the isolation and culture of microorganisms before analysis.11 To date, various bioinformatic tools for metagenomic data have been developed and used to elucidate the ARGs in different environmental niches. Among the metagenomic datasets for the detection of potential ARGs, the ARG database is the most critical and should be of concern. The ARG database enables researchers to identify specific ARGs in samples and evaluate their potential contribution to the development of antibiotic resistance. Furthermore, it promotes collaboration among researchers. Building and maintaining the ARG database typically involve open collaboration, which accelerates the understanding of antibiotic resistance and facilitates the development of countermeasures. Regular updates and continuous database development should be conducted to provide up-to-date support for research on antibiotic resistance and maintain database accuracy.
Several ARG databases have been designed, constructed, and applied to detect potential ARGs in metagenomic datasets collected from various environmental niches. The first of these databases was established in 2009 and is called the Antibiotic Resistance Genes Database (ARDB), which contains 13,293 sequences of ARGs affiliated to 257 antibiotics.12 ARGs research has been flourishing since the establishment of ARDB. Subsequently, the Comprehensive Antibiotic Resistance Database (CARD) was rigorously constructed in 2013 and continues to be updated with high frequency.13 In 2016, on the basis of the sequences of ARDB and CARD, another popular database, namely, Structured Antibiotic Resistance Genes (SARG), was designed and constructed with a hierarchical structure by integrating ARG sequences, removing the redundant ones, and re-selecting the representative query sequences.14 Similarly, on the basis of the sequences of ARDB, a total of 1,260,069 protein sequences and 1,164,479 nucleotide sequences are contained in the updated database Sequence Database of Antibiotic Resistance Genes (SDARG), which consists of 448 types of ARGs and 18 categories of antibiotics. It is used as a built-in database of an online pipeline-ARG analyzer (ARGA) to detect potential ARGs in environmental samples.7 Furthermore, DeepARG-DB, a companion database for DeepARG, was designed and constructed to enhance the quality of the built-in model.15
Although these ARGs databases have been widely applied in ARGs studies to detect potential ARGs in various environmental niches, several limitations still exist. For example, ARDB has not been updated since 2009, which means that ARGs discovered after 2009, such as NDM-116 and mcr-1,17 are not included in it. In addition, the ARDB database does not distinguish between resistant genes with deterministic resistance functions and resistant genes predicted based on homology.18 CARD and SARG are recently established and updated databases, and they contain 2,498 and 4,246 selected reference sequences, respectively. The two databases cover a limited number of high-quality sequences, which is conducive to improving the annotation speed of ARG but not enough for primer evaluation.7 Moreover, some databases, such as the Lactamase Engineering Database (LacED),19,20 Lahey Database of β-lactamases,21 β-Lactamase Alleles Initiative, and Comprehensive β-Lactamase Molecular Annotation Resource (CBMAR),22 only include specific antibiotics. These databases only contain resistance genes for β-lactams,23 and their comparison results are likely to seriously underestimate the number of existing ARGs. Moreover, we found that the identification results of potential ARGs with different ARGs databases for the same dataset present differences and inconsistencies.8 Hence, an enhanced, restyled ARGs database is urgently needed to detect potential ARGs and obtain a comprehensive profile of ARGs. Thus, in the present study, we collected the protein sequences of ARGs from three popular ARGs databases, namely, ARDB, CARD, and SARG. We eliminated the redundancy of these sequences to establish non-redundant antibiotic resistance genes databases (NRD), identified the homologous proteins of NRD from the Non-redundant Protein Database (NR) and the Protein DataBank Database (PDB), and clustered them to establish a non-redundant comprehensive antibiotic resistance genes database (NCRD) with similarities of 100% (NCRD) and 95% (NCRD95). We also assessed the identified results of the potential ARGs in seven environmental niches, and our results showed that our databases have a powerful ability to detect potential ARGs.
Results
Characteristics and features of NCRD
To obtain an enhanced, restyled database for detecting potential ARGs from metagenomic data of different environmental niches, we selected the protein sequences derived from ARDB, CARD, and SARG as fundamental sequences and removed the redundancy of these sequences to construct an initial database of ARGs, which was called NRD. The homologous proteins of NRD were identified from NR and PDB databases. The union set of the protein sequences of NRD and its homologous proteins were merged, and the redundancy of these proteins was removed to construct NCRD. Subsequently, a subdatabase called NCRD95 was constructed based on the similarity of the proteins. Three kinds of ARG databases, namely, NRD, NCRD, and NCRD95, were constructed (Figure 1). Then, the different characteristics and features of these databases were assessed and compared with those of ARDB, CARD, and SARG.
Figure 1.
Flowchart for the construction of NCRD
First, the three redesigned databases had more protein sequences than ARDB, CARD, and SARG. Specifically, we found that 23,136, 4,750, and 12,085 protein sequences were included in ARDB, CARD, and SARG, respectively, and 18,619, 710,231, and 34,008 protein sequences were included in NRD, NCRD, and NCRD95, respectively (Figure 1). The large number of protein sequences in the ARG databases meant that numerous potential ARGs, including false positive ARGs, were detected in the metagenomic dataset. The similarity and coverage of proteins were set to 90% to avoid the appearance of false positive ARGs, and the sequence length was controlled to be greater than 52 amino acids (AA) to screen reliable homologous sequences. Thus, the protein sequences of ARGs of NRD, NCRD, and NCRD95 were highly credible.
Second, the three restyled databases had abundant valuable information for exploring the profiles of ARGs from multiple perspectives. ARGs studies have focused on the gene name (subtype of ARG), antibiotic resistant type, and mechanism of resistance. ARDB, CARD, and SARG databases address only some or all of these concerns, but they still have limitations. For example, ARDB provides information from these aspects, but it is no longer updated and has been abandoned. Our three databases provided the gene name, antibiotic resistant type, and mechanism. Specifically, we observed that the numbers of subtypes of ARGs in ARDB, SARG, and CARD were 180, 225, and 338, respectively. By contrast, our databases contained a much larger number of subtypes of ARGs, which can reach 444 (Figure 1). This vast difference in numbers can be attributed to the standardization of gene names in our database. First, we retained the original names of ARGs from the three databases while standardizing their case. Second, we established a unified name based on the information provided by CARD. For instance, we retained the original names of OXA-19, OXA-20, and OXA-21, and we categorized them as OXA beta-lactamase. As a result, our databases provide two choices for users: OXA-19 and OXA beta-lactamase. Figure 2A shows that certain subtypes of ARGs found in ARDB and SARG are absent in our database, but subtypes of ARGs from CARD are included. This discrepancy is due to our merging process, where data from CARD are prioritized and placed ahead. This prioritization ensures that the sequence information from CARD is preserved when removing redundancies, resulting in the loss of some subtypes of ARGs in ARDB and SARG.
Figure 2.
Characteristics and features of NRD in comparison with ARDB, CARD, SARG, and NCRD95
(A) Venn plot showing the subtypes of ARGs existing in different databases.
(B) Venn plot showing the antibiotic-resistant types in different databases.
Third, the three databases have more antibiotic-resistant types than the other databases. We found that the same antibiotic-resistant types (categories) are present in NRD, NCRD, and NCRD95. Hence, we chose NRD as an example and conducted a comparison with ARDB, CARD, and SARG (Figure 2B). The results showed that 14, 23, and 17 antibiotic-resistant types are present in ARDB, CARD, and SARG, respectively, and 29 antibiotic resistant types are included in NRD (Figure 2B). Among these antibiotic resistant types, 14 are present in all databases; 12 are common to CARD and NRD; 2 are common to SARG, CARD and NCRD; and 1 is common to SARG and NRD (Figure 2B). In the process of unifying the antibiotic resistant types, the small class was classified into a large class by performing searches. For instance, carbapenem, cephalosporin, and cephamycin belong to the beta-lactam class. In the annotation information, we retained the broad class and the specific subclass, such as |beta-lactam|carbapenem; cephalosporin; cephamycin|. Additional information on the comparison of the six database subtypes is given in Figure 3. Given that multidrug and beta-lactam contain a larger number of sequences, we singled them out for comparison (Figure 3). Overall, our databases contain the most information numerically and categorically.
Figure 3.
Detailed information on antibiotic-resistant types in six databases
(A) Comparison of 27 antibiotic-resistant types in the six databases. From left to right are ARDB, SARG, CARD, NRD, NCRD95, and NCRD.
(B) Comparison of the number of beta-lactams in the six databases.
(C) Comparison of the number of multidrugs in the six databases.
With regard to the potential mechanism of ARGs, on the basis of the information on the mechanism of ARGs in CARD, including antibiotic target alternation, antibiotic target protection, antibiotic target replacement, antibiotic efflux, antibiotic inactivation, antibiotic efflux, and reduced permeability to antibiotics, we provided the mechanism information of ARGs in the three enhanced databases.
The protein sequences with highly credible and valuable information, including the subtypes of ARGs, antibiotic resistant types, and mechanisms, were found in our three redesigned ARGs databases.
Profiles of the ARGs of various environmental niches annotated with NCRD were estimated and compared with those annotated with other ARGs databases
The metagenomic datasets from seven environmental niches were assembled, the potential genes were predicted, and the ARGs candidates in ARDB, CARD, SARG, NRD, and NCRD were identified and compared to further verify the universality and application of NCRD. The results showed that the numbers of ARGs identified were in the order of NCRD > NRD > CARD > SARG > ARDB with the same filtered parameters for the same sample (Figure S1). Overall, the number of ARGs identified by ARDB, CARD, SARG, NRD, NCRD95, and NCRD was 20,198, 41,935, 28,787, 53,008, 66,519, and 66,067, respectively. Then, the average number of ARGs in each sample identified by each database was calculated, and the numbers were 288.5, 599.1, 411.2, 757.3, 950.3, and 943.8 for ARDB, CARD, SARG, NRD, NCRD95, and NCRD, respectively. The results of the databases were about 1.26–3.29 times those of the previously established databases. In addition, the average numbers of ARGs identified by NCRD95 and NCRD were compared with those identified by the other databases. The results also showed that the number of ARGs detected by NCRD95 was 3.29, 1.59, and 2.31 times that detected by ARDB, CARD, and SARG, respectively, indicating that our databases have great potential in detecting potential ARGs. At the same time, paired sample t-tests were performed to compare the differences in the alignment results of NRD, NCRD, and NCRD95 for all samples (Figure S2). The results showed considerable differences among NRD, NCRD, and NCRD95 in terms of the total number of identified ARGs and the subtypes of ARGs. However, no remarkable difference in the number of ARGs by type was observed between NCRD and NCRD95.
We selected an urban drinking water source (Chaohu Lake) as an example to visualize and provide a brief explanation (Figure 4A). The number of potential ARGs, the subtypes of ARGs, and the antibiotic-resistant types identified with NCRD, NCRD95, and NRD were higher than those identified with ARDB, CARD, and SARG (Figure 4A). In particular, the profiles of ARGs identified by NCRD and NCRD95 were relatively similar (Figure 4A). These results suggest that comprehensive profiles of ARGs in various environmental niches can be obtained by aligning sequences to NCRD, NCRD95, and NRD; among them, NCRD95 is the most suitable for detecting potential ARGs from the perspective of time.
Figure 4.
Benchmark of the ARGs databases and sequence alignment tools for rapidly detecting potential ARGs in metagenomic datasets of various environmental niches
(A) Benchmark of the number of potential ARGs, subtypes of ARGs, and number of antibiotic-resistant types identified in Chaohu Lake by using ARDB, CARD, SARG, NRD, NCRD95, and NCRD.
(B) Total number of potential ARGs, number of subtypes of potential ARGs, and number of antibiotic-resistant types in the 10 samples from Chaohu Lake identified by BLAST and DIAMOND against the six databases.
(C) Time required to profile the composition of ARGs for several samples from Chaohu Lake by using BLAST and DIAMOND. The tests were performed on an Ubuntu server with 4×Intel(R) Xeon(R) CPU E5-2680 v3 at 2.50GHz and with 64 GB of memory.
Selection of sequence aligners to rapidly detect potential ARGs
The ARG profiles in 10 Chaohu Lake samples (CH01–CH10) identified by BLAST and DIAMOND against the six databases were compared to rapidly detect the profiles of ARGs and ensure the consistency of ARGs. The results were visualized in a boxplot, and the same connection was applied to the same sample (Figure 4B). The resulting connections were almost parallel regardless of the total number of identifications, number of gene names, or number of classifications, indicating that the results of DIAMOND and BLAST were similar.
The time required to detect the potential ARGs by DIAMOND and BLAST was summarized and compared (Figure 4C). The results showed that BLAST was much slower than DIAMOND in ARG identification. The time required by NRD and NCRD95 did not differ considerably from that required by ARDB, SARG, and CARD databases regardless of whether BLAST or DIAMOND was used (Figure 4C). Considering that the results of potential ARGs identified by BLAST and DIAMOND showed no notable differences, we recommend DIAMOND as the first choice in profiling the composition of ARGs because it can greatly decrease the time required. Furthermore, we found that the time required to identify potential ARGs against NCRD was much greater than that for NCRD95 (Figure 4C). Given that the results profiled by NCRD and NCRD95 were similar, NCRD95 was recommended as the database for detecting ARGs in metagenomic datasets.
Web server
The related files of the databases are provided in the GitHub of our laboratory (https://github.com/LabHanmz/NCRD) to broaden the application our ARG databases. A web server (http://ncrd.single-cell.cn/index/) for the identification of potential ARGs was constructed to provide a user-friendly graphical interface for accessing the NCRD databases. The primary use case supported by the web interface is described as follows.
-
(1)
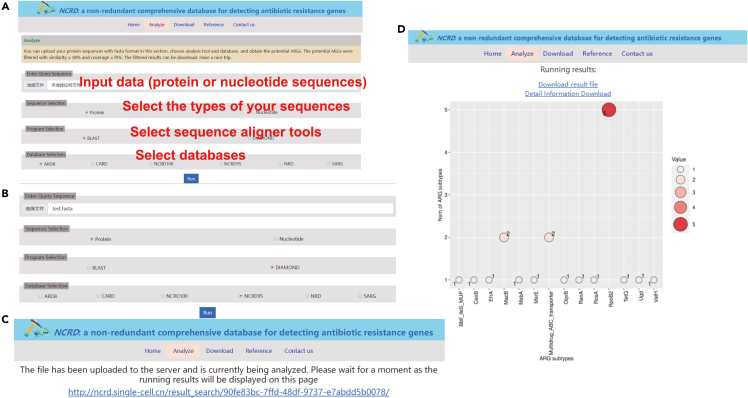
Users can upload their protein or nucleotide sequences in the FASTA format in the Analyze section (Figure 5), choose an analysis tool and database, and obtain the potential ARGs. The potential ARGs are filtered with similarity ≥80% and coverage ≥70%, and the filtered results can be downloaded after the completion of analysis (Figure 5C).
-
(2)
The default align sequence tool is BLAST, and the default database is NCRD95.
-
(3)
Users can obtain the download links of the databases, including ARDB, CARD, SARG, NCRD95, and NCRD.
-
(4)
The analysis result is shown in a bubble diagram (Figure 5D).
Figure 5.
Representative screenshot of the web interface
(A) Analysis interface introduction.
(B) Sample data analysis selection.
(C) Analysis results page.
(D) Bubble diagram showing the analysis results.
Discussion
The existence of ARGs has attracted increasing attention and concern, and the identification of ARGs in current environmental microbiome studies is a key step in exploring the distribution and migration of ARGs.24,25 Although several ARG databases have been constructed, limitations still exist in these databases because of their preferences. A comprehensive ARGs database and two associated ARGs databases (NRD, NCRD, and NCRD95, respectively) were constructed to facilitate the detection and understanding of antibiotic resistance. The protein sequences of NRD were merged with those from ARDB, CARD, and SARG, and the subtypes of ARGs, antibiotic-resistant types, and mechanism of antibiotic resistance were re-unified. Next, the homologous proteins of NRD were identified from NR and PDB databases to construct NCRD, which contained too many protein sequences. Thus, we removed the redundancy of the protein sequences of NCRD to construct NCRD95.
The constituent of the three databases was summarized and compared with ARDB, CARD, and SARG. We found that the three redesigned ARG databases had more protein sequences with high credibility and more valuable information on ARGs, including subtypes of ARGs, antibiotic-resistant types, and mechanisms, compared with ARDB, CARD, and SARG. Afterward, we applied the ARGs databases to profile the ARGs compositions in different environmental samples. The results showed that the universality of NCRD, NCRD95, and NRD and their identification results were better than those of other databases, indicating that our databases demonstrate superiority in different ecological environments.
In consideration of the identification results and the time required, we recommend the use of NRD and NCRD95 databases to detect the profiles of ARGs. Specifically, we recommend NRD, the smallest database, because of its remarkable advantages in accuracy and speed. Its results are based on protein sequences that have been identified as ARGs because it is the combination of three existing databases. Furthermore, additional categories and mechanisms have been appended to this database, and abundant useful information can be obtained when it is used to identify ARGs. Meanwhile, NCRD95 was generated from NCRD on the basis of the similarities of proteins to reduce the time required for detecting potential ARGs. Many ARGs candidates can be rapidly detected when NCRD95 is used with DIAMOND.
In conclusion, despite the progress made in establishing comprehensive ARGs databases, the potential of these databases is limited due to infrequent updates. Our database will be updated in sync with the CARD database to address this issue. We have uploaded corresponding data to our website (http://ncrd.single-cell.cn/index/).
Limitations of the study
Our databases still have limitations. For instance, although we used stringent screening parameters to filter out homologous ARG sequences, all ARGs identified as other genes in microbiome studies should still undergo experimental verification. Moreover, although our databases contain information on the mechanisms of a series of ARGs, numerous mechanisms in the annotation information remain unidentified and require further supplementation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Chaohu Lake | Han et al.26 | GenBank: PRJNA593890 |
| Freshwater lake sediments | Wang et al.27 | N/A |
| Wastewater | Martínez Arbas et al.28 | GenBank: PRJNA230567 |
| Seawater | N/A | GenBank: PRJEB1787 and PRJNA398459 |
| Mouse feces | Mitchell et al.29 | GenBank: PRJEB40312 |
| Feces of patients with RA | Tisza and Buck30 | GenBank: PRJEB6997 |
| Feces of patients with CRC | Gupta et al.31 | GenBank: PRJNA531273 |
| Software and algorithms | ||
| CD-HIT | Fu et al.32 | v4.8.1 |
| Trimmomatic | Bolger et al.33 | v0.32 |
| MEGAHIT | Li et al.34 | v1.2.9 |
| Prodigal | Hyatt et al.35 | v2.6.3 |
| BLAST | Boratyn et al.36 | v2.5.0+ |
| DIAMOND | Buchfink et al.37 | v2.1.8 |
| Other | ||
| NCRD | This Paper | v1.2; http://ncrd.single-cell.cn/index/ |
| ARDB | Liu and Pop12 | v1.1 |
| CARD | McArthur et al.13 | v3.2.6 |
| SARG | Yin et al.14 | v2.2 |
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact Zhi Wang (zwang@apm.ac.cn).
Materials availability
This study did not generate new unique data.
Experimental model and study participant details
This study is a computational science research and does not use experimental models in the life sciences.
Method details
Data processing and establishment of NCRD
To establish a comprehensive and complete database, we collected and cleaned resistance gene sequences from three published ARG databases, namely, ARDB (v1.1),12 CARD (v3.2.6),13 and SARG (v2.2).14 First, we downloaded ARDB from the website http://ardb.cbcb.umd.edu/, selected the document named arbg.tab to extract the accession number of ARGs, and downloaded 23,136 protein sequences with the downloading protein sequence function in “ape” and “rentrez” packages on the R platform (v4.0.2). Second, the protein sequences contained in the document (named protein_fasta_protein_homolog_model.fasta) of the CARD database (https://card.mcmaster.ca/) were downloaded. A total of 4,750 protein sequences belonging to 23 ARG types were obtained from CARD. Last, we chose the protein sequences of SARG, which included 12,085 protein sequences belonging to 17 ARG types (Figure 1). The NR and PDB databases were downloaded on May 6, 2023, and May 29, 2023, respectively.
Before integrating the three databases, ARDB, CARD, and SARG databases were compared, and we found that CARD contained the most complete information, including gene name, category, and mechanism of a certain ARG. Hence, on the basis of the strategy used in CARD, we unified the gene names of ARGs for ARDB and SARG. Given that the gene name of ARG was not given in CARD, we named it based on the gene name of SARG. Additionally, because the gene name of ARG did not appear in CARD and SARG, we renamed it based on the gene name of ARDB. We added valuable information, such as categories and mechanisms, to ARGs that lacked essential information on the basis of the types of ARGs. With these strategies, the information of each ARG gene, including initial gene name, unified gene names, ARG types, ARG categories, and mechanisms, was updated and implemented, and three formal databases with unified gene names were obtained. Notably, several problems were still encountered in this step. For instance, we had to manually check and calibrate the information of each ARG. Specifically, several genes that only had accession numbers (no gene names) were called N/A in CARD. We searched their gene names and ARG types in the National Center for Biotechnology Information (NCBI) depending on their accession numbers and added the information to our unified database.
After obtaining unified ARDB, CARD, and SARG databases, we reduced the redundancy of the protein sequences in the three databases by using CD-HIT (v4.8.1)32 with the following parameters: -c 1, -aL 1, -aS 1, and -M 0. We retained 6,828, 4,750, and 12,085 protein sequences for a downstream analysis. Then, we merged these protein sequences, removed their redundancy, and chose representative protein sequences. During the removal of redundancy, the order of retention was CARD > SARG > ARDB, and sequences with abundant information were preferentially retained. A total of 18,619 protein sequences were obtained, and the protein set of these sequences was called the nonredundant antibiotic resistance genes database (NRD), which contained 29 ARG types (Figure 1). In this database, the entries from ARDB, CARD, and SARG are divided into 3,461, 4,750, and 10,408. To determine the newest ARGs, the homologous proteins of the 18,619 proteins were identified from NR and PDB databases by using DIAMOND with the following parameters: E-value ≤ 1×10-5, qcovhsp ≥ 90%, and ppos ≥ 90%. The shortest sequence lengths of ARDB, CARD, and SARG databases were determined to be 17, 53, and 19 AA, respectively. Only sequences that were longer than 52 AA were retained to enhance alignment reliability. Subsequently, the screened homologous proteins and the 18,619 protein sequences were merged, and their redundancy was eliminated using CD-HIT to create NCRD. In accordance with the conditions used in CD-HIT, a subset database, namely, NCRD95 (CD-HIT: c = 0.95), was constructed. A total of 710,231 and 34,008 protein sequences belonged to 29 ARG types in NCRD and NCRD95, respectively (Figure 1).
Acquisition and processing of metagenomic datasets
Seven different environments, namely, urban drinking water source (Chaohu Lake),26 freshwater lake sediments,27 wastewater of a wastewater treatment plant,28 seawater, mouse feces,29 feces of patients with rheumatoid arthritis (RA),30 and feces of patients with colorectal cancer (CRC),31 were selected to verify the application of NCRD in different environmental niches. The metagenomic datasets of seawater (GenBank: PRJEB1787 and PRJNA398459), wastewater (GenBank: PRJNA230567), the RA patient’s feces (GenBank: PRJEB6997), the CRC patient’s feces (GenBank: PRJNA531273), and mouse feces (GenBank: PRJEB40312) were downloaded from the sequence read archive of NCBI. The Chaohu Lake samples were from our previous project.26 The lake sediment samples were from our ongoing research project. In particular, 10 metagenomic datasets were collected for each of the seven types of environmental niches. The quality of the 70 metagenomic datasets was controlled with Trimmomatic (v0.32)33 by using the same parameters as those described in our previous study.8 Subsequently, the high-quality reads were assembled by MEGAHIT (v1.2.9)34 with the following parameters: --meta-large and k-mer ranged from 27 to 127 with a step of 10. The assembled contigs with length > 500 bp were retained, and the potential genes and their corresponding proteins were predicted with Prodigal (v2.6.3)35 under default settings. Then, the ARG profiles of the 70 metagenomic datasets were detected against ARDB, CARD, SARG, NRD, NCRD, and NCRD95 by DIAMOND, and the results were filtered based on the following settings: E value ≤ 1×10-5, qcovhsp ≥ 70%, and ppos ≥ 80%.26 The numbers and types of ARGs in the different environmental niches and different databases were estimated and compared.
In the analysis of ARG prediction, sequence alignment and searching against a reference database are key steps and consume much time. At present, two popular tools, namely, BLAST (v2.5.0+)36 and DIAMOND (v2.1.8),37 are adopted as sequence aligners to predict and annotate the function of genes and proteins. Several benchmark analyses of the speed and resource requirements of the two tools have been conducted, and they suggest that DIAMOND has better performance and sensitivity than BLAST.37 However, comparisons of ARGs predicted by DIAMOND and BLAST remain lacking. In this study, 10 metagenomic datasets of Chaohu Lake were selected as input datasets to estimate the profiles of ARGs aligned with DIAMOND and BLAST and select a suitable tool for predicting ARGs in vast microbiome sequence data. The profiles of ARGs detected by DIAMOND and BLAST were compared with those in different databases. The time required to detect the ARG composition was estimated.
Quantification and statistical analysis
In this study, the paired t-test of the “rstatix” package in R (v4.0.2) was used to detect the difference in the comparison results of NRD, NCRD and NCRD95 for all samples. Moreover, we use the ‘add_significance’ function to convert the P value into a significant symbol. Specifically, “∗”: p ≤ 0.05; “∗∗”: p ≤ 0.01; “∗∗∗”: p ≤ 0.001; “∗∗∗∗”: p ≤ 0.0001; ns: not significant.
Acknowledgments
This work was partially supported by grants for Scientific Research of BSKY (XJ201916) from Anhui Medical University, the Natural Science Foundation in Higher Education of Anhui (KJ2021A0244), the Young Foundation of Anhui Medical University (2020xkj015), the Key Project of Hubei Province Natural Science Foundation (2020CFA110), and the Youth Innovation Promotion Association, Chinese Academy of Sciences (2018369).
Author contributions
Y.M. performed the data analyses and was a major contributor in writing the manuscript. X.H. has made a huge contribution in website building. N.Z. collected relevant data and supervised the database development. M.H. and Z.W. designed the study, supervised and complemented the writing. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 5, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108141.
Contributor Information
Zhi Wang, Email: zwang@apm.ac.cn.
Maozhen Han, Email: hanmz@ahmu.edu.cn.
Supplemental information
Data and code availability
-
•
The accession number of the samples used in the analysis are listed in the key resources table. All metagenome datasets used in this work are available from public sources as cited in the manuscript. NCRD is a one-stop and user-friendly interface and freely available at http://ncrd.single-cell.cn/index/, and the workflow can be downloaded from https://github.com/LabHanmz/NCRD.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- 1.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., Mueller A., Schäberle T.F., Hughes D.E., Epstein S., et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S., Zhang C., Li F., Hua T., Zhou Q., Ho S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review. J. Hazard Mater. 2021;411:125148. doi: 10.1016/j.jhazmat.2021.125148. [DOI] [PubMed] [Google Scholar]
- 3.Chen B., He R., Yuan K., Chen E., Lin L., Chen X., Sha S., Zhong J., Lin L., Yang L., et al. Polycyclic Aromatic Hydrocarbons (PAHs) Enriching Antibiotic Resistance Genes (ARGs) in the Soils. Environ. Pollut. 2017;220:1005–1013. doi: 10.1016/j.envpol.2016.11.047. [DOI] [PubMed] [Google Scholar]
- 4.Ma L., Xia Y., Li B., Yang Y., Li L.G., Tiedje J.M., Zhang T. Metagenomic Assembly Reveals Hosts of Antibiotic Resistance Genes and the Shared Resistome in Pig, Chicken, and Human Feces. Environ. Sci. Technol. 2016;50:420–427. doi: 10.1021/acs.est.5b03522. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S., Abbas M., Rehman M.U., Huang Y., Zhou R., Gong S., Yang H., Chen S., Wang M., Cheng A. Dissemination of antibiotic resistance genes (ARGs) via integrons in Escherichia coli: A risk to human health. Environ. Pollut. 2020;266:115260. doi: 10.1016/j.envpol.2020.115260. [DOI] [PubMed] [Google Scholar]
- 6.Li Y., Xu Z., Han W., Cao H., Umarov R., Yan A., Fan M., Chen H., Duarte C.M., Li L., et al. HMD-ARG: hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome. 2021;9:40. doi: 10.1186/s40168-021-01002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Z., Wu Y., Feng K., Yang M., Zhang Y., Tu Q., Wang J., Deng Y. ARGA, a pipeline for primer evaluation on antibiotic resistance genes. Environ. Int. 2019;128:137–145. doi: 10.1016/j.envint.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 8.Peng Z., Mao Y., Zhang N., Zhang L., Wang Z., Han M. Utilizing Metagenomic Data and Bioinformatic Tools for Elucidating Antibiotic Resistance Genes in Environment. Front. Environ. Sci. 2021;9 doi: 10.3389/fenvs.2021.757365. [DOI] [Google Scholar]
- 9.Zhao R., Yu K., Zhang J., Zhang G., Huang J., Ma L., Deng C., Li X., Li B. Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches. Water Res. 2020;186:116318. doi: 10.1016/j.watres.2020.116318. [DOI] [PubMed] [Google Scholar]
- 10.Costa P.S., Reis M.P., Ávila M.P., Leite L.R., de Araújo F.M.G., Salim A.C.M., Oliveira G., Barbosa F., Chartone-Souza E., Nascimento A.M.A. Metagenome of a microbial community inhabiting a metal-rich tropical stream sediment. PLoS One. 2015;10:e0119465. doi: 10.1371/journal.pone.0119465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Abreu V.A.C., Perdigão J., Almeida S. Metagenomic Approaches to Analyze Antimicrobial Resistance: An Overview. Front. Genet. 2020;11:575592. doi: 10.3389/fgene.2020.575592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B., Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., De Pascale G., Ejim L., et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/aac.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin X., Jiang X.T., Chai B., Li L., Yang Y., Cole J.R., Tiedje J.M., Zhang T. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics. 2018;34:2263–2270. doi: 10.1093/bioinformatics/bty053. [DOI] [PubMed] [Google Scholar]
- 15.Arango-Argoty G., Garner E., Pruden A., Heath L.S., Vikesland P., Zhang L. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018;6:23. doi: 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann P., Dortet L., Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm. Trends Mol. Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/s1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 18.Bengtsson-Palme J., Larsson D.G.J., Kristiansson E. Using metagenomics to investigate human and environmental resistomes. J. Antimicrob. Chemother. 2017;72:2690–2703. doi: 10.1093/jac/dkx199. [DOI] [PubMed] [Google Scholar]
- 19.Thai Q.K., Pleiss J. SHV Lactamase Engineering Database: a reconciliation tool for SHV β-lactamases in public databases. BMC Genom. 2010;11:563. doi: 10.1186/1471-2164-11-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thai Q.K., Bös F., Pleiss J. The Lactamase Engineering Database: a critical survey of TEM sequences in public databases. BMC Genom. 2009;10:390. doi: 10.1186/1471-2164-10-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush K., Jacoby G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/aac.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava A., Singhal N., Goel M., Virdi J.S., Kumar M. CBMAR: a comprehensive β-lactamase molecular annotation resource. Database. 2014;2014 doi: 10.1093/database/bau111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boolchandani M., D'Souza A.W., Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019;20:356–370. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanganyado E., Gwenzi W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019;669:785–797. doi: 10.1016/j.scitotenv.2019.03.162. [DOI] [PubMed] [Google Scholar]
- 25.Buckner M.M.C., Ciusa M.L., Piddock L.J.V. Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. FEMS Microbiol. Rev. 2018;42:781–804. doi: 10.1093/femsre/fuy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han M., Zhang L., Zhang N., Mao Y., Peng Z., Huang B., Zhang Y., Wang Z. Antibiotic resistome in a large urban-lake drinking water source in middle China: Dissemination mechanisms and risk assessment. J. Hazard Mater. 2022;424:127745. doi: 10.1016/j.jhazmat.2021.127745. [DOI] [PubMed] [Google Scholar]
- 27.Wang C., Mao Y., Zhou W., Li Y., Zou G., Chen B., Wang Z. Inhomogeneous antibiotic distribution in sediment profiles in anthropogenically impacted lakes: Source apportionment, fate drivers, and risk assessment. J. Environ. Manag. 2023;341:118048. doi: 10.1016/j.jenvman.2023.118048. [DOI] [PubMed] [Google Scholar]
- 28.Martínez Arbas S., Narayanasamy S., Herold M., Lebrun L.A., Hoopmann M.R., Li S., Lam T.J., Kunath B.J., Hicks N.D., Liu C.M., et al. Roles of bacteriophages, plasmids and CRISPR immunity in microbial community dynamics revealed using time-series integrated meta-omics. Nat. Microbiol. 2021;6:123–135. doi: 10.1038/s41564-020-00794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell A.L., Scheremetjew M., Denise H., Potter S., Tarkowska A., Qureshi M., Salazar G.A., Pesseat S., Boland M.A., Hunter F.M.I., et al. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018;46:D726–D735. doi: 10.1093/nar/gkx967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisza M.J., Buck C.B. A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023202118. e2023202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta A., Dhakan D.B., Maji A., Saxena R., Pk V.P., Mahajan S., Pulikkan J., Kurian J., Gomez A.M., Scaria J., et al. Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India. mSystems. 2019;4:e00438-19. doi: 10.1128/mSystems.00438-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D., Luo R., Liu C.M., Leung C.M., Ting H.F., Sadakane K., Yamashita H., Lam T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y., et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The accession number of the samples used in the analysis are listed in the key resources table. All metagenome datasets used in this work are available from public sources as cited in the manuscript. NCRD is a one-stop and user-friendly interface and freely available at http://ncrd.single-cell.cn/index/, and the workflow can be downloaded from https://github.com/LabHanmz/NCRD.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.