Highlights
-
•
Time-restricted feeding (TRF) inhibits lung tumor growth.
-
•
TRF altered and partially restored the fecal microbiota, metabolite composition and rhythm in lung cancer mice.
-
•
Circadian expression of clock genes is affected, and correlated with fecal microbiota.
-
•
TRF mainly affects inflammatory and immune pathways.
Keywords: Time-restricted feeding, Circadian rhythm, Gut microbiota, Metabolome, Lung cancer
Abstract
The homeostasis of the gut microbiota and circadian rhythm is critical to host health, and both are inextricably intertwined with lung cancer. Although time-restricted feeding (TRF) can maintain circadian synchronization and improve metabolic disorders, the effects of TRF on the fecal microbiome, metabolome and their diurnal oscillations in lung cancer have not been discussed. We performed 16S rRNA sequencing and untargeted metabonomic sequencing of the feces prepared from models of tumor-bearing BALB/c nude mice and urethane-induced lung cancer. We demonstrated for the first time that TRF significantly delayed the growth of lung tumors. Moreover, TRF altered the abundances of the fecal microbiome, metabolome and circadian clocks, as well as their rhythmicity, in lung cancer models of tumor-bearing BALB/c nude mice and/or urethane-induced lung cancer C57BL/6J mice. The results of fecal microbiota transplantation proved that the antitumor effects of TRF occur by regulating the fecal microbiota. Notably, Lactobacillus and Bacillus were increased upon TRF and were correlated with most differential metabolites. Pathway enrichment analysis of metabolites revealed that TRF mainly affected immune and inflammatory processes, which might further explain how TRF exerted its anticancer benefits. These findings underscore the possibility that the fecal microbiome/metabolome regulates lung cancer following a TRF paradigm.
Introduction
The human gut microbiota is a huge and complex community of microorganisms, containing mainly bacteria [1]. The gut microbiota is involved in a variety of physiological processes, such as immunity and metabolism, whereas abnormalities in their composition or function are associated with disease states [2,3]. As an environmental factor in cancer susceptibility and progression, the gut microbiota is associated with multiple mechanisms, such as the regulation of inflammation, induction of DNA damage and production of metabolites responsible for tumorigenesis or tumor suppression [4], [5], [6]. Likewise, emerging studies have reported the associations of the microbiota in lung cancer, one of the deadliest cancers worldwide [7,8]. Lung cancer patients showed different gut microbiota characteri47stics, which could modulate the effectiveness of immunotherapy and chemotherapy [9], [10], [11]. Dysbiosis is associated with lung oncogenesis [8]. Thus, targeting the gut microbiota may provide new insights into the prevention and treatment of lung cancer.
In mammals, the internal biological clock system is a complicated and accurate feedback network of hierarchical oscillators [12]. The central circadian clock located in the suprachiasmatic nucleus of the hypothalamus is affected by environmental light conditions. In turn, the central clock synchronizes organismal systems by delivering signals to the peripheral clock [13], [14], [15]. The disturbance of the circadian clock is closely related to many diseases, such as obesity, diabetes and cancer [16], [17], [18]. It has been shown that circadian rhythm disturbance promotes lung tumorigenesis and circadian homeostasis has an essential tumor suppressive effect [19]. Furthermore, single cell transcriptomic analysis has shown that circadian rhythm disturbance is associated with poor prognosis and drug resistance in lung adenocarcinoma [20]. Conversely, circadian rhythm disorders have been detected in lung cancer patients. Abnormal expression of circadian rhythm genes in non-small cell lung cancer is closely associated with the clinicopathological features of patients and is involved in cancer progression in lung cancer patients [21]. This all suggests that circadian rhythm disorders have a substantial impact on lung cancer and that altering or restoring the rhythm may be capable of being an assistance in the prevention and treatment of lung cancer. Emerging evidence indicates that the gut microbiota exhibits diurnal variation, which is regulated by the host circadian clock [22,23]; reciprocally, the gut microbiota can also affect the host [24,25]. Moreover, the production of bacterial metabolites is rhythmic, and these metabolites can regulate the circadian rhythm and metabolism of the host [26]. Although the close relationships between the gut microbiota, metabolites or circadian rhythm and cancer have been explored separately, the relationship among the rhythm of gut microbiota and its metabolites and lung cancer has not yet been reported.
Rhythmic metabolism is well known to be regulated by time-restricted feeding (TRF) [27]. TRF refers to limiting daily food intake to 10 h or less without changing the quantity or quality of food intake [28]. Studies have shown that TRF alleviates metabolic diseases caused by various obesity-causing diets, and the benefits are proportional to the duration of fasting [29]. Regulating the rhythm of the gut microbiota may be a possible way for TRF to mitigate the harmful effects of a high-fat diet [30]. TRF also exerts a potential antitumor effect, such as in breast cancer or lung cancer, probably by improving insulin or metabolic phenotypes [31]. Nevertheless, no research has focused on whether TRF has an impact on the fecal microbiota, its metabolites and their rhythm in lung cancer.
In this study, we investigated the effect of TRF on the diurnal dynamics of the fecal microbiome composition and metabolites in a subcutaneous xenograft lung tumor model and urethane-administered lung tumorigenesis mice. We sought to determine the role of 6 h of TRF in lung cancer and whether the gut microbiome, its metabolites and their rhythm in lung cancer were affected. We found for the first time that in two lung tumor models, TRF significantly slowed the growth of lung tumors. Furthermore, TRF impacted the compositions of the fecal microbiome and metabolome and reprogrammed their diurnal oscillations. We also demonstrated the antitumor effects of TRF by regulating the fecal microbiota.
Materials and methods
Animals
Five-week-old male C57BL6/J mice (Vital River, Beijing, China) and BALB/c nude mice (Vital River, Beijing, China) were housed under specific pathogen-free conditions at the Experimental Animal Center of Chongqing Medical University. All mice were maintained under a 12 h light/dark cycle (lights on at 8 am), and all animal procedures were approved by the Biomedical Ethics Committee of Chongqing Medical University.
Tumor-bearing BALB/c nude mice
After one week of adaptive feeding of BALB/c nude mice, they were subcutaneously inoculated with A549 lung adenocarcinoma cells (6 × 106 cells/mouse) to generate a xenograft tumor mouse model. Once tumors became palpable, the mice were randomly divided into two groups: mice fed normal chow with ad libitum food access (NC) or normal chow with time-restricted food access (TRF). Under TRF, mice were restricted access to food for a period of 6 h during the dark (2 h after lights off to 4 h before lights on). The implanted tumor size was measured with Vernier calipers every three days and calculated by (L × W2)/2 (where L and W are the length and width of the tumor, respectively). Body weight and food intake were recorded every three days. 10:00 pm (2 h after lights off) was treated as Zeitgeber time 0 (T0). Mice were sacrificed under anesthesia and executed on Day 40, and intestine and stool samples were collected every 4 h throughout the 24 h period. Samples were collected starting at 11:00 pm as Zeitgeber time 1 (T1). Tissue and stool samples were stored at -80°C for further analysis.
Urethane-induced lung cancer mouse model
After one week of acclimatization, C57BL6/J mice were randomly separated into three groups. (1) The SN group: mice were injected intraperitoneally with saline and fed normal chow with ad libitum food access. (2) The UN group: mice were injected intraperitoneally with urethane and fed normal chow with ad libitum food access. (3) The UT group: mice were injected intraperitoneally with urethane and fed normal chow with restricted food access for a period of 6 h during the dark (2 h after lights off to 4 h before lights on). For the C57BL6/J mouse lung cancer model, lung cancer was induced by intraperitoneal injection of urethane (Sigma, USA, 10 %, 0.1 ml/10 g body weight) for 10 weeks. The urethane solution for injection was prepared with urethane powder dissolved in saline, and after a latent period of 15 weeks, the mice were sacrificed under anesthesia. Mice in the SN group were injected intraperitoneally with saline (0.1 ml/10 g body weight) for 10 weeks. TRF intervention started after grouping and continued until the end of the experiment. Body weight and food intake were recorded every three days. Tissue and stool samples were collected and stored at -80°C for further analysis.
Lung tumor enumeration
After the C57BL6/J mice were euthanized, 4 % paraformaldehyde (MACKLIN, Shanghai) was injected into the lung tissues. The lung tumors were counted under a dissecting microscope (Hitachi-7500, Japan) by three experimental researchers blinded to the groups.
Cell culture
The A549 cell line was obtained from American Type Culture Collection (ATCC). The human lung adenocarcinoma cell line A549 (ATCC, Manassas, USA) was cultivated in RPMI-1640 (Cat. #C3010-0500, VivaCell, Shanghai, China) supplemented with 10 % fetal bovine serum (Cat. #C04001-500, VivaCell, Shanghai, China) and 1 % penicillin/streptomycin (Cat. #SV30010, HyClone, USA) and maintained at 37°C in a humidified atmosphere of 95 % air and 5 % CO2. The cell line had been authenticated, and all experiments were performed with mycoplasma-free cells.
Quantitative real-time PCR
The total RNA, in each sample, was extracted from intestinal tissues of tumor-bearing BALB/c nude mice using TRIzol (Cat. #DP424, TIANGEN, Beijing, China), in accordance with the manufacturer's instructions. Subsequently, 1 µg RNA was reverse transcribed into cDNA, in a total 20 µl reaction system, using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Cat. #RR047A, Takara, Dalian, China) following the manufacturer's instructions. RT‒qPCR was performed using SYBR Green qPCR Master Mix (Cat. #HY-K0501, MedChemExpress, Monmouth Junction, NJ, USA). The following thermocycling conditions were used for qPCR using a CFX Maestro Real-Time PCR system (CFX Connect, Bio-Rad, USA): Initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 s; and a final extension at 72°C for 10 min. The relative expression level of mRNAs of target genes was measured with the 2−ΔΔcq method. The following primer sequences were used in this study: Actin: forward, CCACCATGTACCCAGGCATT, reverse, CAGCTCAGTAACAGTCCGCC; Bmal1: forward, CCACCTCAGAGCCATTGATACA, reverse, GAGCAGGTTTAGTTCCACTTTGTCT; Clock: forward, ATGGTGTTTACCGTAAGCTGTAG, reverse, CTCGCGTTACCAGGAAGCAT; Per1: forward, CGGATTGTCTATATTTCGGAGCA, reverse, TGGGCAGTCGAGATGGTGTA; Per2: forward, GAAAGCTGTCACCACCATAGAA, reverse, AACTCGCACTTCCTTTTCAGG; Per3: forward, AAAAGCACCACGGATACTGGC, reverse, GGGAGGCTGTAGCTTGTCA; Cry1: forward, GAGGCAAGCAGACTGAATATTG, reverse, CCCATTAGAGTTAGAAGGGACC; Cry2: forward, GAGAACTCTCACACCCTCTATG, reverse, CATAGGTGTCGTCATGGTTCTC.
Fecal microbiota transplantation (FMT)
After one week of acclimatization, five-week-old male BALB/c nude mice were treated for two weeks with an antibiotic cocktail containing ampicillin (100 mg/kg, Cat. #A8180, Solarbio, Beijing, China), neomycin sulfate (100 mg/kg, Cat. #N6386, Sigma‒Aldrich, Saint Louis, MO, USA), vancomycin hydrochloride (50 mg/kg, Cat. #T0832, Targetmol, Shanghai, China), and metronidazole (100 mg/kg, Cat. #M1547, Sigma‒Aldrich, Saint Louis, MO, USA) by gavage once a day for gut microbiota depletion. Then, all mice were subcutaneously inoculated with 5 × 106 A549 cells in 100 μl of PBS. After the tumor became palpable, the mice were randomly divided into the FMT-NC group (gavaged with gut microbiota from NC mice) and FMT-TRF group (gavaged with gut microbiota from TRF mice). From then on, the tumor volume of the mice was recorded. Stool from donor mice (NC and TRF groups) was collected and resuspended in saline at 0.1 g/ml, and then 0.2 ml of this suspension was administered to mice by oral gavage for 24 continuous days, specifically, every day for the first week and every other day from the second week until the end of the experiment. Mice were sacrificed under anesthesia, and tumor tissues were photographed on Day 24.
16S rRNA sequencing
Fresh fecal samples from BALB/c nude mice and C57BL/6J mice were collected into a sterile tube and sent to Shanghai Majorbio Company (Shanghai, China) for 16S rRNA gene V3-V4 region sequencing using the Illumina MiSeq platform. The primers used were 341F, CCTAYGGGRBGCASCAG and 806R, GGACTACNNGGGTATCTAAT. The protocol was as follows:
Genomic DNA was extracted from 0.2 g of fecal samples using the E.Z.N.A.® soil DNA Kit (Omega Biotek, Norcross, GA, USA) following the manufacturer's instructions. The V3–V4 regions of the 16S rRNA gene were amplified by PCR and paired-end sequenced on an Illumina MiSeq 2 × 250 bp platform (Illumina, San Diego, USA) using the specific primers 338F-806R in combination with barcode sequences. All PCRs were performed in triplicate. PCR amplification was performed on an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA) using the following procedure: 95°C for 3 min; 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and a final extension at 72°C for 10 min. The mixed PCR products were assessed, purified and quantified using a 2 % agarose gel, the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union, CA, USA), and Quantus™ Fluorometer (Promega, USA), respectively.
FLASH (version 1.2.7) was used to merge overlapping paired reads that overlapped longer than 10 bp into a sequence and quality-filtered with the following criteria: (I) the unassembled reads were discarded; (II) the reads of 300-bp were truncated at any site when receiving an average quality score of less than 20 over a 50-bp sliding window, and trimmed reads that were too short (< 50 bp) were discarded; and (III) the allowed mismatch base numbers for barcode and primer matching were 0 and 2, respectively. Operational taxonomic units (OTUs) were clustered with 97 % identity using UPARSE (version 7.1), and chimeric sequences were detected and removed. The taxonomy of representative OTU sequences was analyzed by the RDP Classifier (http://rdp.cme.msu.edu/) against the GreenGene Database in accordance with a confidence threshold of 70 %. Data analysis was performed at Majorbio (https://cloud.majorbio.com/).
Untargeted metabolomics analyses
Frozen stool samples from urethane-induced lung cancer C57BL/6J mice were sent to Shanghai Majorbio (Shanghai, China) for LC‒MS/MS analysis. Fifty milligrams of stool sample and 6-mm-diameter grinding beads were added to a 2-mL centrifuge tube. Metabolite extraction was performed with 400 μl of extraction solution (methanol:water=4:1 (v:v)) containing 0.02 mg/mL internal standard (L-2-chlorophenylalanine). Samples were ground using a Wonbio-96c (Shanghai Wanbo Biotechnology Co., Ltd.) frozen tissue grinder for 6 min (-10°C, 50 Hz) and then extracted by low-temperature ultrasound for 30 min (5°C, 40 kHz). The samples were stored at -20°C for 30 min and centrifuged for 15 min (4°C, 13,000 × g), and the supernatant was transferred to an injection vial for LC‒MS/MS analysis. The LC‒MS/MS analysis of the sample was conducted on a Thermo UHPLC-Q Exactive HF-X system equipped with an ACQUITY HSS T3 column (100 mm × 2.1 mm i.d., 1.8 μm; Waters, USA) at Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The mobile phases consisted of 0.1 % formic acid in water:acetonitrile (95:5, v/v) (solvent A) and 0.1 % formic acid in acetonitrile:isopropanol:water (47.5:47.5, v/v) (solvent B). Positive ion mode separation gradient: 0–20 % B for 0-3 min, 20–35 % B for 3–4.5 min, 35–100 % B for 4.5-5 min, 100 % B for 5-6.3 min, 100–0 % B for 6.3-6.4 min, 0 % B for 6.4-8 min. Separation gradient in negative ion mode: 0–5 % B for 0-1.5 min, 5–10 % B for 1.5-2 min, 10 %-30 % B for 2-4.5 min, 30–100 % B for 4.5-5 min, 100 % B for 5-6.3 min, 100 % to 0 % B for 6.3–6.4 min, 0 % B for 6.4–8 min. The flow rate was 0.40 mL/min, and the column temperature was 40°C. The mass spectrometric data were collected by a Thermo UHPLC-Q Exactive HF-X Mass Spectrometer equipped with an electrospray ionization (ESI) source operating in positive mode and negative mode. The pretreatment of raw data was performed by Progenesis QI (Waters Corporation, Milford, USA) software. The metabolites were identified by searching databases, and the main databases were the HMDB (http://www.hmdb.ca/) and Majorbio Database. The data were analyzed through the free online platform of the Majorbio cloud platform (cloud.majorbio.com).
Metabolite correlation and network statistics
R version 4.0.3 was used to calculate the Pearson's correlation coefficients between metabolites. Networks are based on metabolite-pair correlations with an estimated p value < 0.0001 and |r| > 0.6. Undirected networks were analyzed and visualized using the Compound Spring Embedder (CoSE) layout of Cytoscape (version 3.9.1).
Circadian rhythmicity analysis
Metabolome cycling was determined with the JTK_CYCLE algorithm implemented in Metacycle (p value < 0.05). The RNA expression rhythm of circadian clock genes was calculated by the CircaCompare R package (version 0.1.1). Comparison of circadian rhythm parameters between two oscillating groups was computed by the CircaCompare R package (version 0.1.1).
Statistical analysis
All statistical analyses were performed in GraphPad Prism (GraphPad Software, version 8.0.2) or R (version 4.0.3). Student's t tests or Wilcoxon rank-sum tests were used for comparisons between two groups, and one-way ANOVA or Kruskal‒Wallis H tests were used for comparisons among three groups. All data are expressed as the mean ± SEM if not otherwise stated. A p value < 0.05 was interpreted as statistically significant.
Results
TRF reshapes the gut microbiota composition in a urethane-induced lung cancer mouse model
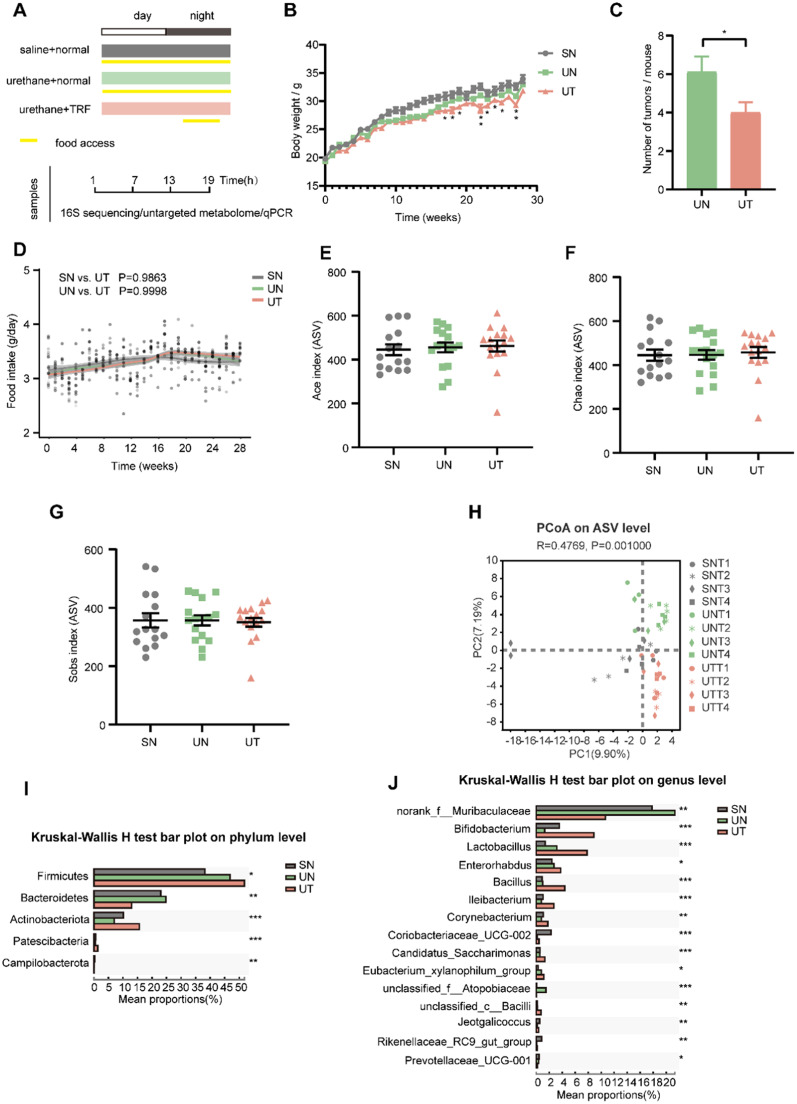
After 3 weeks of TRF intervention, C57BL/6J mice were intraperitoneally injected with urethane for 10 weeks to induce lung tumorigenesis, and TRF was continued until the end of the experiment. After an incubation period of 15 weeks, mice were anesthetized and sacrificed, and feces were collected every 6 h for 24 h for 16S rRNA sequencing (Fig. 1A). We observed that TRF reduced the weight of C57BL/6J mice with lung cancer and reduced the number of lung tumors (Fig. 1B and C). The food intakes did not differ between the three groups (Fig. 1D). This suggested that TRF significantly delayed the tumor growth.
Fig. 1.
TRF reshapes the gut microbiota composition in urethane-administrated mice. (A) Schematic diagram of experimental design and sample collection. Yellow frames indicate feeding period. (B) The average weekly weight of mice in different conditions. (C) Number of tumors per mouse in the UN and the UT groups. Data are compared between the UN and the UT groups at same timepoint using Student's t-tests. (D) Food intake trajectories. Data were analyzed by area under the curve. (E–G) Alpha diversity by Ace, Chao and Sobs index of gut microbiota in SN mice (fed a normal diet and injected intraperitoneally with saline), UN mice (fed a normal diet and injected intraperitoneally with urethane) and UT mice (fed a normal diet with time-restricted food access and injected intraperitoneally with urethane). Indexes were compared using Kruskal-Wallis H test. (H) Principal co-ordinates analysis (PCoA) plot by Binary-Euclidean dissimilarity of the gut microbiota across time points. (I, J) Kruskal-Wallis H test at the phylum level and genus level, * p < 0.05, ** p < 0.01, *** p < 0.001. Number of mice n = 15 for SN, n = 16 for UN and n = 16 for UT. Data are presented as mean ± SEM. See also Fig. S1.
Alpha diversity indices were not significantly altered in any of the three groups (Fig. 1E–G, S1A and B). Principal coordinate analysis (PCoA) of the gut microbiota demonstrated a separation between the SN group (fed a normal diet and injected intraperitoneally with saline), the UN group (fed a normal diet and injected intraperitoneally with urethane) and the UT group (fed a normal diet with time-restricted food access and injected intraperitoneally with urethane), while the UT group was relatively close to the SN group (Fig. S1C), indicating that TRF partially restores the gut microbiota composition of mice with lung tumors to the level of nonlung tumor mice. Further evaluation of the beta diversity at different time points revealed that samples of the SN group at T1 and T4 were the closest to those of the UT group at T1 and T3 (Fig. 1H).
Firmicutes and Bacteroidetes were the dominant bacterial communities at the phylum level in C57BL/6J mice (Fig. S1D). The relative abundances of Firmicutes and Actinobacteriota in the UT group were significantly increased compared to those in the UN group, while the relative abundance of Bacteroidetes was decreased (Fig. 1I). The major bacterial communities at the genus level were Akkermansia, norank_f__Muribaculaceae, Allobaculum and Staphylococcus (Fig. S1E). Among the top 15 genera with significant differences, compared to the UN group, the relative abundances of Bifidobacterium, Lactobacillus, Bacillus and Jeotgalicoccus were increased, while the relative abundances of norank_f__Muribaculaceae, unclassified_f__Atopobiaceae and Prevotellaceae_UCG-001 were decreased in the UT group, and those of norank_f__Muribaculaceae and unclassified_f__Atopobiaceae were higher than those in the SN group (Fig. 1J). LEfSe (linear discriminant analysis effect size) analysis of the gut microbiota also showed that the most representative species in the UT group included Bifidobacterium, Lactobacillus and Bacillus (Fig. S1F). Taken together, these findings indicate that TRF overly reshapes the gut microbiota diversity and composition.
TRF enhances diurnal oscillations in the gut microbiota in a urethane-induced lung cancer mouse model
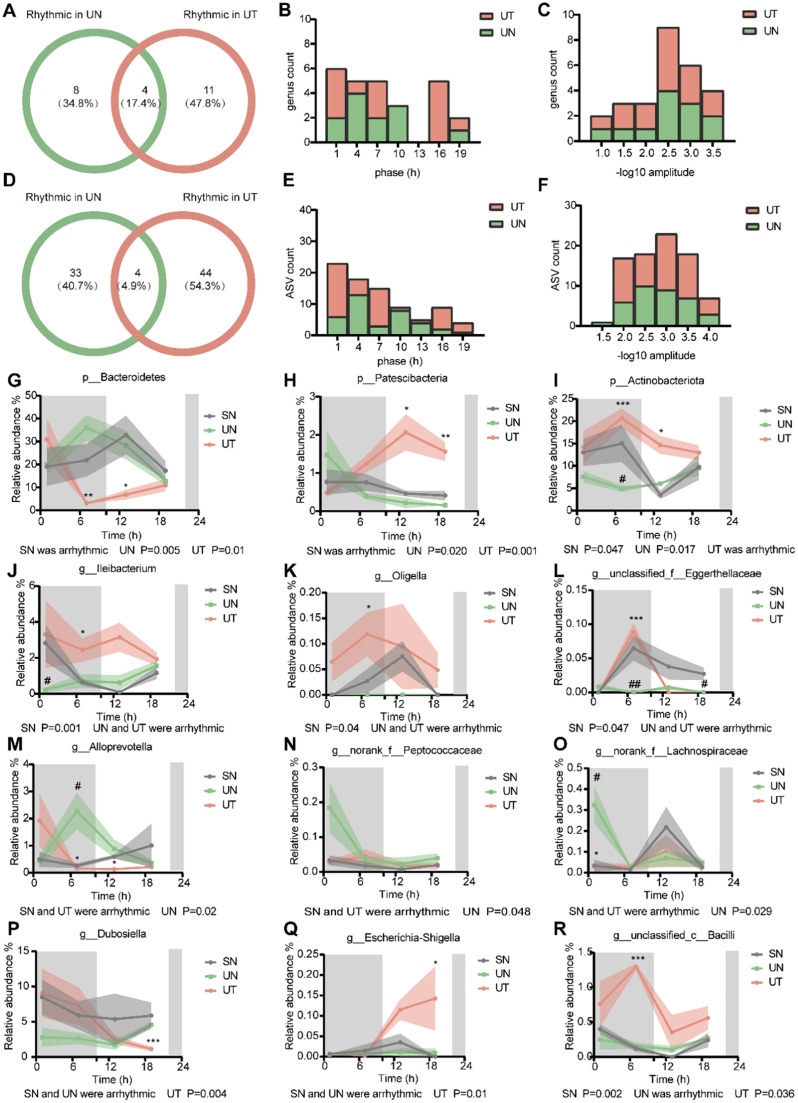
Since both the gut microbiota and circadian rhythms have been strongly linked to cancer [32], we next investigated the role of TRF on the circadian rhythm of the gut microbiota. We analyzed the diurnal dynamics of the gut microbiota using the JTK_CYCLE algorithm in MetaCycle [33,34]. At the genus level, the microbiome revealed that 8 genera (34.8 %) were exclusively oscillatory in the UN group and 11 (47.8 %) genera were only rhythmic in the UT group, while 4 genera (17.4 %) were circadian in both groups (Fig. 2A, S2A and B). The phase distribution was different between the UN group and the UT group (Fig. 2B). The UT group had more genera than the UN group at the same amplitude, indicating that the amplitude of rhythmic genera was enhanced in UT mice (Fig. 2C). At the ASV level, microbiome analysis identified 33 ASVs (40.7 %) in UN and 44 ASVs (54.3 %) in UT to be circadian, and 4 ASVs (4.9 %) were found to be commonly rhythmic in both groups (Fig. 2D, S2C and D). The phase of rhythmic ASVs was shifted in the UT group relative to the UN group (Fig. 2E). Similar to the rhythmic genera results, the amplitude of rhythmic ASVs was enhanced in UT mice compared to UN mice (Fig. 2F).
Fig. 2.
TRF enhances diurnal oscillations of gut microbiota in urethane-administrated mice. (A, D) Venn diagrams illustrate the overlap of oscillating genera and ASVs (Amplicon Sequence Variant) respectively, between UN and UT mice. (B, E) Phase distribution diagram of dual-oscillating genera and ASVs, respectively. (C, F) Amplitude distribution diagram of dual-oscillating genera and ASVs, respectively. (G-I) Relative abundance for specific bacteria at the phylum level. (J-L) Relative abundance of bacterial genera oscillating in SN the group. (M-O) Relative abundance of bacterial genera oscillating in the UN group. (P-R) Relative abundance of bacterial genera oscillating in the UT group. Samples are represented as mean ± SEM (n = 3-5 for each timepoint). Shaded areas show SEM. Data are compared between groups at same timepoint using Student's t-tests. */# p < 0.05, **/## p < 0.01, ***/### p < 0.001, * indicates UN compared to UT, # indicates UN compared to SN. Statistics of rhythmicity parameters were computed by JTK_CYCLE. See also Fig. S2.
Next, we analyzed the microbiota rhythm in the main taxa. At the phylum level, there were three phyla with rhythmic fluctuations. The abundance levels of Bacteroidetes and Patescibacteria diurnally fluctuated with opposite oscillating patterns in the UN and UT groups but were not circadian in the SN group (Fig. 2G and H). Actinobacteriota diurnally fluctuated in both the SN and UN groups but not in the UT group; however, its abundance was increased significantly in the UT group, and restored significantly to the level of the SN group (Fig. 2I). At the genus level, we show the most representative genera. Ileibacterium, Oligella and unclassified_f__Eggerthellaceae had a cyclical pattern only in the SN group (Fig. 2J–L). The abundance level of Ileibacterium in the UN group was significantly reduced compared to that in the SN group at T1, while that in the UT group was similar to that in the SN group (Fig. 2J). The abundance levels of Oligella and unclassified_f__Eggerthellaceae in the UT group were significantly increased compared to those in the UN group and were partially restored to the level in the SN group (Fig. 2K and L). Although Alloprevotella, norank_f__Peptococcaceae and norank_f__Lachnospiraceae were exclusively oscillatory in the UN group, they always maintained similar abundance levels in the SN and UT groups (Fig. 2M–O). Dubosiella and Escherichia-Shigella obtained rhythm only after TRF intervention (Fig. 2P and Q). The relative abundance of Dubosiella in the SN and UT groups decreased from the dark phase to the light phase, while in the UN group, the opposite trend was observed (Fig. 2P). Escherichia-Shigella had significant cycling in the UT group but not in the SN and UN groups, with a spike in the light phase in the SN and UT groups (Fig. 2Q). Unclassified_c__Bacilli followed the food ingestion patterns of animals under UT conditions, with their level being greatly reduced during the light phase upon SN and UT, and rhythmicity was observed in both groups (Fig. 2R). Together, our findings suggest that TRF affects diurnal oscillations of the gut microbiota in a urethane-administered mouse model of urethane-induced lung cancer.
TRF profoundly reprograms the fecal metabolites of urethane-administered mice
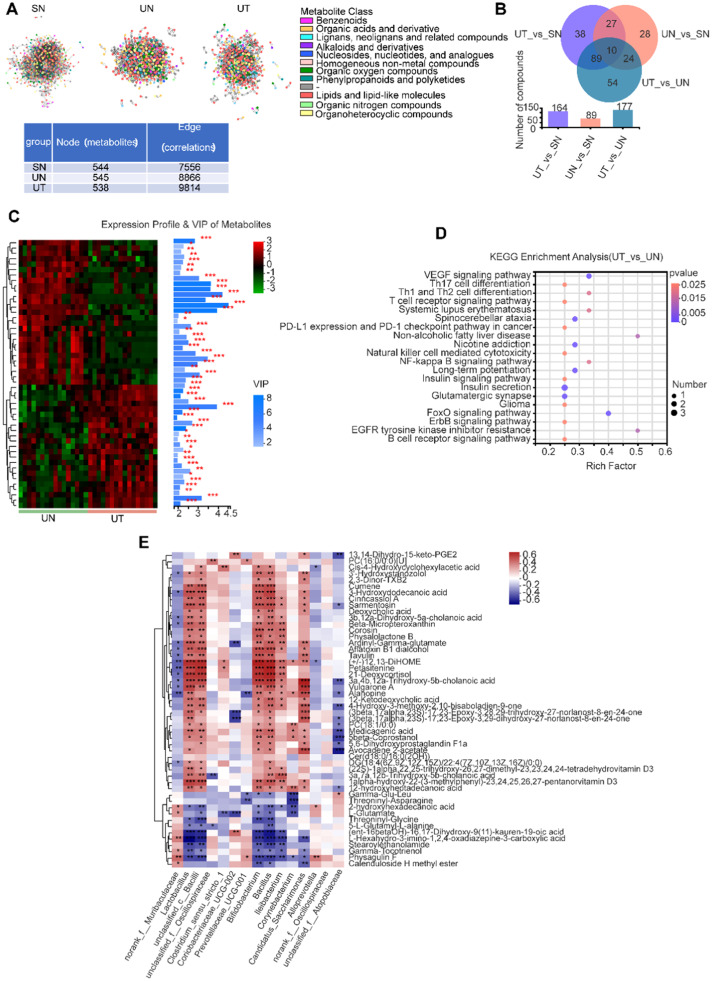
Since gut microbiota are involved in the regulation of host metabolism in part through metabolites, we further attempted to perform untargeted metabolomics analyses to examine the alterations in fecal metabolites. After TRF intervention, feces were collected from mice at 4-h intervals for 24 h for untargeted metabolomics. The metabolite expression data went through various preprocessing and quality control steps prior to statistical analyses, including peak detection, filling missing values, normalization, centering, scaling and data transformation. Thereafter, we identified a total of 590 metabolites. To reveal the underlying metabolic relationships, we constructed condition-specific correlation networks. We found that the three groups shared a similar number of nodes (∼500 metabolites), but compared with the SN or UN groups, the UT group had more edges (correlations), indicating an enhanced metabolic relationship upon TRF in lung cancer (Fig. 3A). The PLS-DA score plot showed that the samples in the three groups were significantly different from each other in both positive and negative modes, supported by permutation testing (Fig. S3A–D). The number of differential metabolites between the UN group and the UT group was 177 (Fig. 3B). The top 50 metabolites of the UT vs. UN metabolic sets were further displayed according to variable importance in projection (VIP) analysis based on the OPLS-DA model, including 27 downregulated and 23 upregulated metabolites. All metabolites had VIP scores > 1.5 (Fig. 3C). Similarly, according to the VIP analysis of the SN and UN groups, diacetone alcohol and aspartylglycosamine were significantly reduced in tumor mice but recovered after TRF intervention (Fig. S3F). KEGG pathway analysis revealed that these differential metabolites (UT vs. UN) were mainly concentrated in immune-related pathways, including the T-cell receptor signaling pathway, B-cell receptor signaling pathway, natural killer cell-mediated cytotoxicity, PD-L1 expression and PD-1 checkpoint pathway in cancer (Fig. 3D). KEGG pathway analysis of different metabolites in UN vs. SN showed significant pathways, including glycine, serine and threonine metabolism; D-arginine and D-ornithine metabolism; cysteine and methionine metabolism; arginine and proline metabolism; and ABC transporters (Fig. S3E). To better understand the relationship between fecal metabolites and the fecal microbiome, we next analyzed the correlation between the top 50 metabolites and the altered genera. Interestingly, most of the metabolites were significantly and positively correlated with six bacterial genera, namely, Lactobacillus, unclassified_c__Bacilli, Bifidobacterium, Bacillus, Ileibacterium and Candidatus_Saccharimonas (Fig. 3E), which were persistently increased upon TRF in the urethane-administered C57BL/6J mice, indicating an important role of Lactobacillus and Bacillus in response to TRF.
Fig. 3.
TRF profoundly reprograms fecal metabolites of urethane-administrated mice. (A) Networks of significantly correlated metabolites. Each node indicates a metabolite and color refers to metabolite class. Node and edge numbers are summarized in the table. (B) Venn diagrams of the differentially expressed metabolites (identified metabolites). (C) Variable importance in projection (VIP) analysis. The heat map on the left shows the metabolites expression profile of the UN and the UT groups, the bar chart on the right represents VIP scores of metabolites and their significance between the two groups. * p < 0.05, ** p < 0.01, *** p < 0.001. (D) Metabolic pathway enrichment analysis of differentially presented metabolites between the UN and the UT groups. (E) Heat map of correlations between altered bacterial genera and differentially expressed metabolites. The metabolite data used in this figure were aggregated at various time points within the groups. * p < 0.05, ** p < 0.01, *** p < 0.001. See also Fig. S3.
TRF alters the diurnal patterns of fecal metabolites
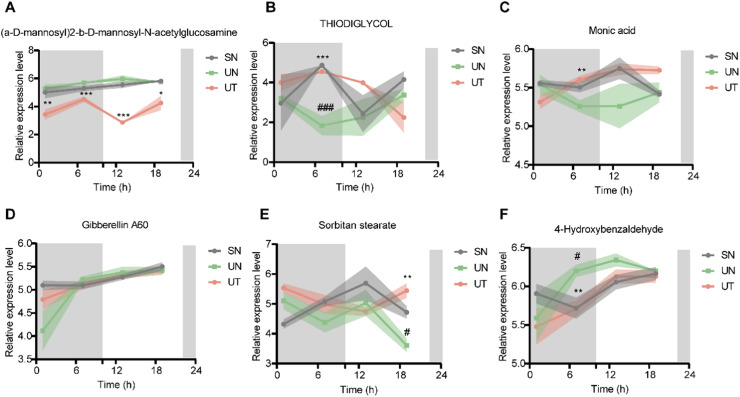
Untargeted metabolomics analysis uncovered diurnal rhythms in 146 out of 590 metabolites; among them, a-D-Mannosyl)2-b-D-mannosyl-N-acetylglucosamine, thiodiglycol and monic acid had rhythmic oscillations only in the UT group (Fig. 4A–C). The fluctuation pattern of thiodiglycol in the UT group and its level at each time point were closer to those in the SN group, reaching a peak after feeding and then dropping rapidly (Fig. 4B). The level of monic acid in the SN and UT groups peaked in the light period and maintained a similar trend (Fig. 4C). Furthermore, gibberellin A60, sorbitan stearate and 4-hydroxybenzaldehyde showed rhythmicity in the SN and UT groups but lost their rhythm in the UN group, indicating that TRF restored their rhythm (Fig. 4D–F). In brief, TRF tremendously affects the composition, level and rhythm of fecal metabolites.
Fig. 4.
24h expression profile of metabolites. (A-C) Relative expression level of a-D-mannosyl)2-b-D-mannosyl-N-acetylglucosamine, thiodiglycol and Monic acid with rhythm only in the UT group. (D-F) Relative expression level of Gibberellin A60, Sorbitan stearate and 4-Hydroxybenzaldehyde rhythmic in the SN and the UT group but arrhythmic in the UN group. Samples are represented as mean ± SEM (n = 3-5 for each timepoint). Shaded areas show SEM. Data are compared between groups at same timepoint using Student's t-tests. */# p < 0.05, **/## p < 0.01, ***/### p < 0.001, * indicate UN compared to UT, # indicate UN compared to SN. Statistics of rhythmicity parameters were computed by JTK_CYCLE.
TRF alters the fecal microbiota composition in tumor-bearing BALB/c nude mice
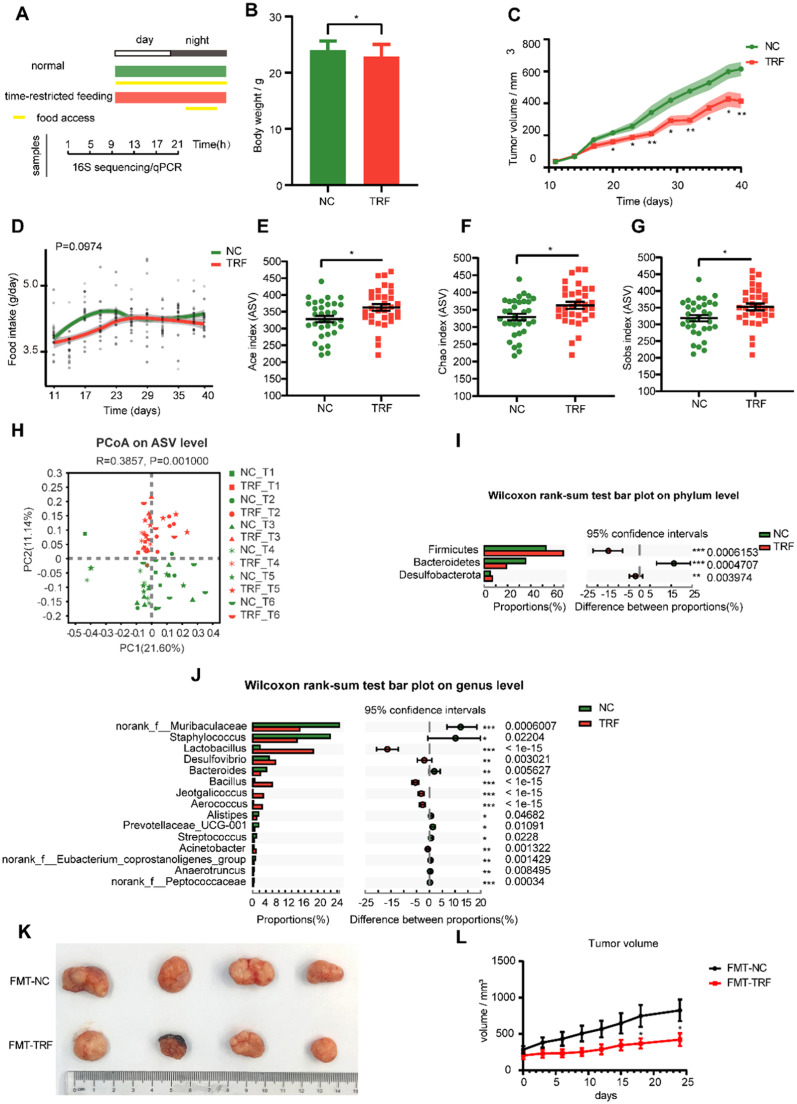
To determine whether TRF played a similar role in tumor-bearing BALB/c nude mice, five-week-old tumor-bearing BALB/c nude mice were fed normal chow with ad libitum access to food (NC) or normal chow with time-restricted feeding (TRF) for an experimental period of 40 days, and fecal samples were collected every 4 h during a 24-h period (Fig. 5A). TRF reduced the weight of tumor-bearing mice and slowed the growth of tumors (Fig. 5B and C). The food intakes did not differ between the NC and TRF groups (Fig. 5D). The gut microbiota composition was detected by 16S rRNA gene sequencing of fecal samples. Alpha diversity was compared between groups, and the Ace, Chao and Sobs indices of the TRF group were significantly higher than those of the NC group (Fig. 5E-G), indicating that TRF significantly increased the richness of the gut microbiota in the lung cancer model. There was no change in the Shannon and Shannoneven indices (Fig. S4A and S4B). Principal coordinate analysis (PCoA) of abund_jaccard beta diversity showed separation between the two groups (Fig. 5H and S4C).
Fig. 5.
Time-restricted feeding (TRF) alters the gut microbiota composition in tumor-bearing BALB/c nude mice. (A) Schematic diagram of experimental design and sample collection. Yellow frames indicate feeding period. (B) The average weight of mice in different conditions. (C) The average volume of mice in different conditions. Shaded areas show SEM. Data are compared between groups at same timepoint using Student's t-tests. (D) Food intake trajectories. Data were analyzed by area under the curve. Alpha diversity by Ace (E), Chao (F) and Sobs index (G) of gut microbiota in ad libitum normal chow (NC) and time-restricted feeding (TRF) mice. Indexes are compared using Wilcoxon rank-sum test. (H) Principal co-ordinates analysis (PCoA) plot by Abund-Jaccard dissimilarity of the gut microbiota across time points (ANOSIM, R = 0.3857, P = 0.001). Wilcoxon rank-sum test on the phylum level (I) and genus level (J), number of mice n = 33 for NC, n = 32 for TRF. (K) Photograph of dissected tumors (FMT-NC: mice received feces from the NC group; FMT-TRF: mice received feces from the TRF group; n = 6). (L) Tumor volume was measured every 3 days. * p < 0.05. * p < 0.05, ** p < 0.01, *** p < 0.001. Data are presented as mean ± SEM. See also Fig. S4.
Firmicutes and Bacteroidetes were still the major bacterial communities at the phylum level (Fig. S4D). At the genus level, the dominant bacterial communities in the observed genera were norank_f__Muribaculaceae, Staphylococcus and Lactobacillus (Fig. S4E). Notably, consistent with our above results in C57BL/6J mice, TRF mice had a higher relative abundance of Firmicutes and a lower relative abundance of Bacteroidetes than NC mice (Fig. 5I). The relative abundances of the top 15 genera are also shown, the relative abundances of Lactobacillus, Bacillus and Jeotgalicoccus were significantly increased after TRF, conversely, the relative abundances of norank_f__Muribaculaceae and Prevotellaceae_UCG-001 were decreased in the TRF group compared to the NC group (Fig. 5J), which was consistent with the results observed in the urethane-induced lung cancer mouse model. Linear discriminant analysis (LDA) showed that Lactobacillus, Bacillus and Jeotgalicoccus were the important characteristic genera in the TRF group compared with the NC group (Fig. S4F). Together, consistent with the results in the C57BL/6J mice, TRF alters the gut microbiota compositions in tumor-bearing BALB/c nude mice, such as Lactobacillus and Bacillus at the genus level, as well as Firmicutes and Bacteroidetes at the phylum level.
To further confirm the effect of TRF on tumor growth by altering the gut microbiota in mice, BALB/c nude mice were gavaged with a cocktail of antibiotics every day for 14 days to deplete the gut microbiota of the mice. Then, A549 lung cancer cells were injected subcutaneously into BALB/c nude mice. The mice were randomly divided into two groups, the FMT-NC group and the FMT-TRF group, and the prepared fecal microbiota from the NC group and TRF group were gavaged to tumor-bearing BALB/c nude mice for 24 days. After intervention, we found that the tumor growth of mice receiving time-restricted feeding mouse feces (FMT-NC) was slower than that of mice receiving NC mouse feces (FMT-TRF) (Fig. 5K and L). This response fully depicts that the alterations in the gut microbiota caused by TRF are sufficient to slow the growth of tumors.
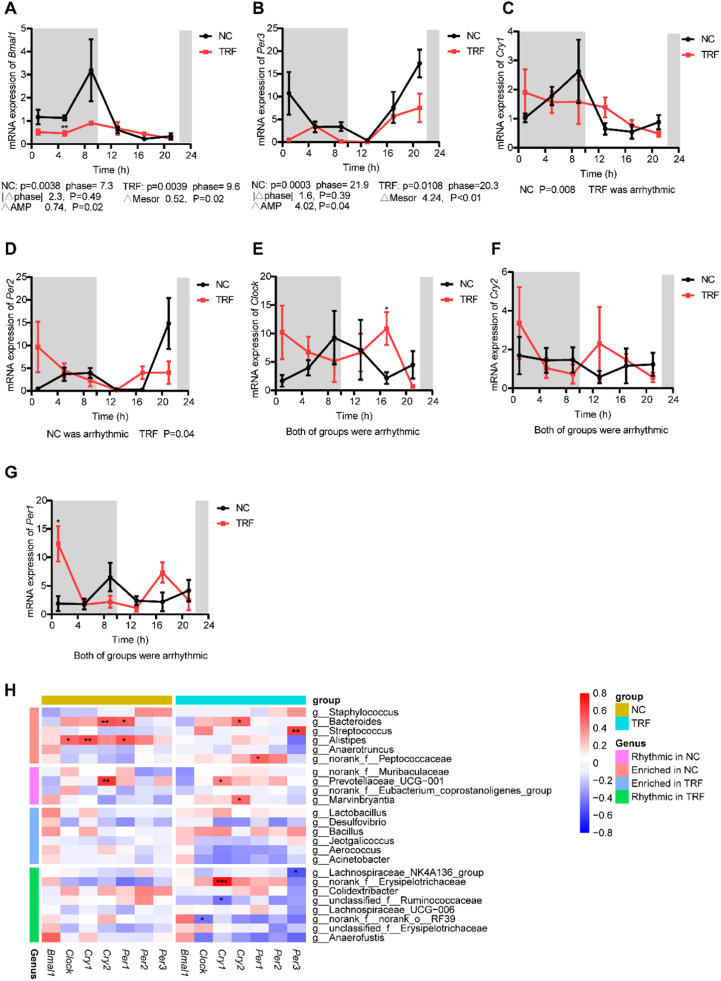
TRF alters the circadian expression of core clock genes in the intestine
To investigate whether TRF also affected the expression of endogenous clock genes, we measured the mRNA expression levels of brain and muscle ARNT-like 1 (Bmal1), circadian locomotor output cycle protein kaput (Clock), cryptochrome circadian regulator 1 (Cry1), cryptochrome circadian regulator 2 (Cry2), period circadian regulator 1 (Per1), period circadian regulator 2 (Per2) and period circadian regulator 3 (Per3) in the intestine (Fig. 6A-G). The expression of Bmal1 and Per3 exhibited clear circadian rhythm in both the NC and TRF groups (Fig. 6A and B). Diurnal rhythms of Bmal1 and Per3 showed modest phase shifts by 2 h and exhibited a decrease in amplitude (Fig. 6A and B). The 24-h expression profile of Cry1 became arrhythmic under TRF (Fig. 6C), Per2 levels followed the feeding pattern of mice under TRF conditions, and rhythmicity was observed only in the TRF group (Fig. 6D). The expression of Per2 peaked during the dark period in the TRF group, but peaked during the light period in the NC group (Fig. 6D). The expression of Clock, Cry2 and Per1 did not show circadian rhythm in either group; however, they tended to have the opposite pattern of fluctuations over 24 h (Fig. 6E–G). Although a spike was observed in Per1 in the NC and TRF mice during the dark phase, the abundance of Per1 agreed with the feeding pattern of animals under TRF conditions (Fig. 6G). These results suggested that TRF alters the expression pattern of some clock genes in the intestine. Next, to determine whether the gut microbiota was associated with the intestinal expression of clock genes, we performed correlation analysis among altered genera and circadian clock genes (Fig. 6H). Interestingly, the number of correlations was greater in the TRF group than in the NC group (Fig. 6H). In the NC group, all the genera with a significant correlation with clock genes were enriched (Fig. 6H). In conclusion, TRF altered the circadian expression of intestinal clock genes and enhanced the correlation between clock genes and the gut microbiota to some extent.
Fig. 6.
TRF alters the circadian expression of core clock genes in intestines. (A-G) Bmal1, Clock, Cry1, Cry2, Per1, Per2, Per3 mRNA expression in intestines of NC and TRF mice (n = 3-4 mice per time point). Gray shaded areas represent periods of darkness, while the rest represent periods of light. (H) Heatmap of the correlation coefficient between the core clock genes and altered genera. Data are presented as mean ± SEM. Data are compared between groups at same timepoint using Student's t-tests. * p < 0.05, ** p < 0.01, *** p < 0.001. Statistics and comparison of rhythmicity parameters were computed by CircaCompare.
Discussion
The circadian oscillators of mammals exist in many organs, tissues and cells. In addition to being regulated by the central clock located in the suprachiasmatic nucleus of the hypothalamus, the peripheral clock is also influenced by diet [35,36]. Time-restricted feeding (TRF) is a dietary pattern that limits the intake of all nutrients to certain hours of the day without restricting the quantity or quality of nutrients [37] and has been reported to reprogram circadian metabolism in contexts of high-fat diet-induced obesity and metabolic disorders [38]. For example, TRF alleviated the side effects of a high-fat diet by modulating the circadian rhythm of the gut microbiota [30]. However, it is unknown whether TRF could modulate the fecal microbiota, metabolites and their rhythm, contributing to other host phenotypes and physiologies, particularly lung cancer, which is intertwined with rhythmic disruption, the gut microbiota and metabolites [19,39]. In this study, we found for the first time that TRF delayed the growth of lung tumors, probably by regulating the fecal microbiota, and TRF altered the fecal microbiome metabolome and its diurnal oscillations in lung cancer mice.
We performed 16S rRNA sequencing of feces prepared from two lung cancer models and found that TRF effectively reshaped the fecal microbiota composition. Microbial diversity was reported to be increased in response to improved metabolic health [3,40]. Time-restricted eating was found to increase microbial richness and diversity in healthy adults [41]. Consistent with these studies, we found that the beta diversity of the fecal microbiota was changed upon TRF in two models, and the alpha diversity was increased in tumor-bearing BALB/c nude mice. Furthermore, urethane-induced lung cancer mice revealed that TRF partially restored the fecal microbiota compositions to normal levels, as reflected by the fecal microbiota composition of the UT group being closer to that of the SN group than that of the UN group. Moreover, at the phylum level, TRF resulted in an increase in Firmicutes abundance and a decrease in Bacteroidetes abundance in models of tumor-bearing BALB/c nude mice and urethane-induced lung cancer. The results were supported by a series of previous works [9,42,43]. For example, Firmicutes in the gut microbiota were reduced in subjects with chronic liver diseases and hepatocellular carcinoma patients [42]. We also found that at the genus level, Lactobacillus and Bacillus were persistently increased significantly in the TRF group, which have been proven to have effective antitumor activity [44,45]. Lactobacillus administration conferred a beneficial role in lung cancer and colon cancer [46,47]. Moreover, the relative abundances of Bifidobacterium and Corynebacterium decreased in lung cancer mice and were recovered after TRF intervention. Bifidobacterium and Corynebacterium were reported as beneficial bacteria in a study of lung cancer [48,49]. These results indicate that TRF reshapes the abundance of the gut microbiota in lung cancer. In addition, fecal microbiota transplantation experiments on nude mice confirmed that the altered gut microbiota delayed tumor growth, this result indicate that the gut microbiota played an important role in the inhibitory effect of TRF on tumors.
TRF partially restored the diurnal rhythm of the ileal or gut microbiome that was damaged by a high-fat diet [23,50]. Host feeding rhythms can synchronize with the intestinal microbiota to enhance the rhythms of intestinal innate immunity [51]. Our results were consistent with previous observations that TRF enhances rhythmic metabolism [23,50]. Our rhythm analysis showed that the gut microbiota was very dynamic under TRF conditions, with more cycling genera. In the urethane-induced lung cancer mouse model, 8 (34.8 %) genera and 33 (40.7 %) ASVs were exclusively oscillatory in the UN group, and 11 (47.8 %) genera and 44 (50.3 %) ASVs were rhythmic only in the UT group. Additionally, the amplitude of rhythmic genera and ASVs was enhanced in UT mice. These results demonstrated that TRF shapes a unique rhythm fluctuation pattern. However, additional studies are warranted to detect the diurnal dynamics of the gut microbiota in other lung cancer models after TRF intervention. In conclusion, TRF alters and restores the rhythm of the gut microbiota.
It has been pointed out that metabolites may be related to tumorigenesis and progression, and metabolites also have rhythmic fluctuations, which can regulate host metabolism [4,26]. We collected feces from C57BL/6J mice for nontargeted metabonomic sequencing. The correlation network analysis highlighted the powerful metabolite crosstalk under TRF. Metabolite enrichment analysis revealed pathways that are mostly related to immunity and inflammation. The implications of findings related to TRF and the host's immune and inflammatory response have been discussed previously [52], [53], [54]. Moreover, tumors effectively inhibit the immune response by activating a negative regulatory pathway (also called a checkpoint) related to immune homeostasis [55,56]. This situation may explain the antitumor effect of TRF on lung cancer mice. Interestingly, we further found that most of the differential metabolites were significantly positively correlated with beneficial genera such as Lactobacillus and Bacillus. This may indicate that beneficial genera play a role in the antitumor effect of TRF through some specific metabolites. We also found that TRF impacted the rhythm of fecal metabolites. (a-D-Mannosyl)2-b-D-mannosyl-N-acetylglucosamine, thiodiglycol and monic acid only had rhythm fluctuations in the UT group, while gibberellin A60, sorbitan stearate and 4-hydroxybenzaldehyde in tumor-bearing mice recovered the lost rhythm after TRF intervention. Our results are consistent with those of previous studies. TRF changed the rhythm of serum and muscle metabolites [57]. TRF reduced the deleterious effects of a high-fat diet by regulating the circadian rhythm of liver lipid metabolism and the gut microbiota [30]. The above results indicate that the level and rhythm alterations of fecal metabolites caused by lung cancer can be restored by TRF to a certain extent. However, the roles of these metabolites in cancer in response to TRF are unknown, and additional studies are warranted to explore the underlying mechanism.
The central and peripheral clocks are believed to rely on a molecular clock mechanism, which is generated by two transcription/translation feedback loops that work together to produce a strong 24 h gene expression rhythm [58]. The core feedback loop is driven by four complete clock proteins: two activators (Clock and Bmal1) and two repressors (Per and Cry) [59]. Then, we detected the circadian expression of the core circadian clock genes in the intestines. Bmal1 and Per3 had a slight phase shift between the two groups. Clock, Cry2 and Per1 had opposite fluctuation patterns in the absence of rhythm, while Cry1 and Per2 lost and gained rhythm after TRF, respectively. TRF was reported to affect the circadian clock under different conditions [60], [61], [62]. Time-restricted feeding prevented the destruction of the peripheral circadian rhythm during chronic jet lag [60]. The TRF regimen brought the circadian clock into a fixed feeding time and prevented the interruption of normal cell metabolic processes caused by a high-fat diet [62]. This finding fully proves that the peripheral clock can be affected by TRF. Clock, Cry2 and Per1 with opposite fluctuation patterns and Per2 with rhythm only in the TRF group might play an important role in the antitumor effect of TRF. The gut microbiota was proven to regulate host circadian rhythms and metabolism in response to dietary cues [63]. Low light exposure boosted the circadian rhythm of the host clock through interactions with the cecal microbial community [22]. The gut microbiota regulates energy storage and body fat accumulation through the circadian clock [25]. Likewise, our correlation analysis results confirmed the relationship between the gut microbiota and the circadian clock. We found more correlations between the genera and the core circadian clock genes in the TRF group than in the NC group. However, whether TRF could regulate the rhythm of the gut microbiota by the circadian clock in lung cancer is a subject that needs to be explored in future studies.
Conclusions
In conclusion, our study was the first to show that TRF significantly delayed lung tumor growth, and confirmed the antitumor effects of TRF through the regulation of the fecal microbiota. In addition, we revealed that TRF altered the abundance levels of the gut microbiota and its metabolites, as well as their rhythmicity in lung cancer mice. This result suggests that targeting the fecal microbiota, its metabolites and circadian rhythm by the TRF regimen could be a potential nonpharmacological intervention for cancer treatment.
Funding
This research was funded by the National Natural Science Foundation of China (No. 8210120502), China Postdoctoral Science Foundation (2021M693755), and CQMU Program for Youth Innovation in Future Medicine (No. W0085).
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Animal Care and Use Committee, Chongqing Medical University (protocol code IACUC-CQMU-2023-0006).
CRediT authorship contribution statement
Gaofeng Fang: Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Visualization. Shengquan Wang: Validation, Investigation, Resources, Data curation. Qianyao Chen: Investigation, Resources, Data curation. Han Luo: Investigation, Resources, Data curation. Xuemei Lian: Writing – review & editing, Supervision, Project administration, Funding acquisition. Dan Shi: Conceptualization, Methodology, Software, Formal analysis, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the online platform of Majorbio Cloud Platform (www.majorbio.com) for support from data analysis in this study. We would like to thank the technicians and staffs at the Department of Nutrition and Food Hygiene, School of Public Health, Chongqing Medical University for performing experiments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100943.
Contributor Information
Xuemei Lian, Email: xuemeilian@cqmu.edu.cn.
Dan Shi, Email: danshi@cqmu.edu.cn.
Appendix. Supplementary materials
Data availability
Original raw data in this manuscript were deposited to Sequence Read Archive (SRA) database of NCBI (Accession number: SRP425124). The data generated in the present study may be requested from the corresponding author.
References
- 1.Collins S.M., Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology. 2009;136:2003–2014. doi: 10.1053/j.gastro.2009.01.075. [DOI] [PubMed] [Google Scholar]
- 2.Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 3.Sonnenburg J.L., Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt A.P., Redinbo M.R., Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 2017;67:326–344. doi: 10.3322/caac.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill C.I.R., Rowland IR. Diet and cancer: assessing the risk. Br J Nutr. 2002;88(Suppl 1):S73–S87. doi: 10.1079/BJN2002632. [DOI] [PubMed] [Google Scholar]
- 6.Shin N.R., Whon T.W., Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends. Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Pizzo F., Maroccia Z., Hammarberg Ferri I., Fiorentini C. Role of the microbiota in lung cancer: insights on prevention and treatment. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23116138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto T. Microbiota and lung cancer. Semin. Cancer Biol. 2022;86 doi: 10.1016/j.semcancer.2022.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W.Q., Zhao S.K., Luo J.W., Dong X.P., Hao Y.T., Li H., et al. Alterations of fecal bacterial communities in patients with lung cancer. Am. J. Transl. Res. 2018;10:3171–3185. [PMC free article] [PubMed] [Google Scholar]
- 10.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 11.Daillère R., Vétizou M., Waldschmitt N., Yamazaki T., Isnard C., Poirier-Colame V., et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi J.S., Hong H.K., Ko C.H., McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 14.Mohawk J.A., Green C.B., Takahashi JS. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogenesch J.B., Ueda HR. Understanding systems-level properties: timely stories from the study of clocks. Nat. Rev. Genet. 2011;12:407–416. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 16.Archer S.N., Laing E.E., Möller-Levet C.S., van der Veen D.R., Bucca G., Lazar A.S., et al. Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc. Natl. Acad. Sci. U S A. 2014;111:E682–EE91. doi: 10.1073/pnas.1316335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buxton O.M., Cain S.W., O'Connor S.P., Porter J.H., Duffy J.F., Wang W., et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci. Transl. Med. 2012;4:129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schernhammer E.S., Laden F., Speizer F.E., Willett W.C., Hunter D.J., Kawachi I., et al. Night-shift work and risk of colorectal cancer in the nurses' health study. J. Natl. Cancer Inst. 2003;95:825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 19.Papagiannakopoulos T., Bauer M.R., Davidson S.M., Heimann M., Subbaraj L., Bhutkar A., et al. Circadian rhythm disruption promotes lung tumorigenesis. Cell. Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He L., Fan Y., Zhang Y., Tu T., Zhang Q., Yuan F., et al. Single-cell transcriptomic analysis reveals circadian rhythm disruption associated with poor prognosis and drug-resistance in lung adenocarcinoma. J. Pineal. Res. 2022;73:e12803. doi: 10.1111/jpi.12803. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Liu R., Zhang B., Huo H., Song Z. Advances in the study of circadian genes in non-small cell lung cancer. Integr. Cancer. Ther. 2022;21 doi: 10.1177/15347354221096080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Li Y., Yuan Y., Wang J., Zhang S., Zhu R., et al. Reducing light exposure enhances the circadian rhythm of the biological clock through interactions with the gut microbiota. Sci. Total Environ. 2023;858 doi: 10.1016/j.scitotenv.2022.160041. [DOI] [PubMed] [Google Scholar]
- 23.Zarrinpar A., Chaix A., Yooseph S., Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thaiss C.A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A.C., et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Kuang Z., Yu X., Ruhn K.A., Kubo M., Hooper LV. The intestinal microbiota regulates body composition through NFIL and the circadian clock. Science. 2017;357:912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leone V., Gibbons S.M., Martinez K., Hutchison A.L., Huang E.Y., Cham C.M., et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoogian E.N.C., Panda S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev. 2017;39:59–67. doi: 10.1016/j.arr.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung H., Chou W., Sears D.D., Patterson R.E., Webster N.J.G., Ellies LG. Time-restricted feeding improves insulin resistance and hepatic steatosis in a mouse model of postmenopausal obesity. Metabolism. 2016;65:1743–1754. doi: 10.1016/j.metabol.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaix A., Zarrinpar A., Miu P., Panda S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20 doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y., Xu H., Xie Z., Wang L., Sun Y., Yang H., et al. Time-restricted feeding reduces the detrimental effects of a high-fat diet, possibly by modulating the circadian rhythm of hepatic lipid metabolism and gut microbiota. Front Nutr. 2020;7 doi: 10.3389/fnut.2020.596285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das M., Ellies L.G., Kumar D., Sauceda C., Oberg A., Gross E., et al. Time-restricted feeding normalizes hyperinsulinemia to inhibit breast cancer in obese postmenopausal mouse models. Nat. Commun. 2021;12:565. doi: 10.1038/s41467-020-20743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishehsari F., Voigt R.M., Keshavarzian A. Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat. Rev. Endocrinol. 2020;16:731–739. doi: 10.1038/s41574-020-00427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G., Anafi R.C., Hughes M.E., Kornacker K., Hogenesch JB. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu A.C., Lewis W.G., Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat. Chem. Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- 36.Yasumoto Y., Hashimoto C., Nakao R., Yamazaki H., Hiroyama H., Nemoto T., et al. Short-term feeding at the wrong time is sufficient to desynchronize peripheral clocks and induce obesity with hyperphagia, physical inactivity and metabolic disorders in mice. Metabolism. 2016;65:714–727. doi: 10.1016/j.metabol.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Sutton E.F., Beyl R., Early K.S., Cefalu W.T., Ravussin E., Peterson CM. Early time-restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018;27 doi: 10.1016/j.cmet.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman H., Genzer Y., Cohen R., Chapnik N., Madar Z., Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J. 2012;26:3493–3502. doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- 39.Vernocchi P., Gili T., Conte F., Del Chierico F., Conta G., Miccheli A., et al. Network analysis of gut microbiome and metabolome to discover microbiota-linked biomarkers in patients affected by non-small cell lung cancer. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21228730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 41.Zeb F., Wu X., Chen L., Fatima S., Haq I.U., Chen A., et al. Effect of time-restricted feeding on metabolic risk and circadian rhythm associated with gut microbiome in healthy males. Br. J. Nutr. 2020;123:1216–1226. doi: 10.1017/S0007114519003428. [DOI] [PubMed] [Google Scholar]
- 42.Chen T., Ding R., Chen X., Lu Y., Shi J., Lü Y., et al. Firmicutes and Blautia in gut microbiota lessened in chronic liver diseases and hepatocellular carcinoma patients: a pilot study. Bioengineered. 2021;12:8233–8246. doi: 10.1080/21655979.2021.1982273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almalki M.A., Khalifa AYZ. Silver nanoparticles synthesis from Bacillus sp KFU36 and its anticancer effect in breast cancer MCF-7 cells via induction of apoptotic mechanism. J. Photochem. Photobiol. B. 2020;204 doi: 10.1016/j.jphotobiol.2020.111786. [DOI] [PubMed] [Google Scholar]
- 45.Badgeley A., Anwar H., Modi K., Murphy P., Lakshmikuttyamma A. Effect of probiotics and gut microbiota on anti-cancer drugs: mechanistic perspectives. Biochim Biophys. Acta Rev. Cancer. 2021;1875 doi: 10.1016/j.bbcan.2020.188494. [DOI] [PubMed] [Google Scholar]
- 46.Le Noci V., Bernardo G., Manenti G., Infante G., Khaleghi Hashemian D., Minoli L., et al. Live or heat-killed lactobacillus rhamnosus aerosolization decreases adenomatous lung cancer development in a mouse carcinogen-induced tumor model. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budu O., Banciu C., Pinzaru I., Sarău C., Lighezan D., Șoica C., et al. A combination of two probiotics, lactobacillus sporogenes and clostridium butyricum, inhibits colon cancer development: an in vitro study. Microorganisms. 2022;10 doi: 10.3390/microorganisms10091692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu F., Li J., Guan Y., Lou Y., Chen H., Xu M., et al. Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung cancer. Int. J. Biol. Sci. 2019;15:2381–2392. doi: 10.7150/ijbs.35980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadler T.E., Castro JE. The effects of Corynebacterium parvum and surgery on the Lewis lung carcinoma and its metastases. Br. J. Surg. 1976;63:292–296. doi: 10.1002/bjs.1800630410. [DOI] [PubMed] [Google Scholar]
- 50.Dantas Machado A.C., Brown S.D., Lingaraju A., Sivaganesh V., Martino C., Chaix A., et al. Diet and feeding pattern modulate diurnal dynamics of the ileal microbiome and transcriptome. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks J.F., Behrendt C.L., Ruhn K.A., Lee S., Raj P., Takahashi J.S., et al. The microbiota coordinates diurnal rhythms in innate immunity with the circadian clock. Cell. 2021;184 doi: 10.1016/j.cell.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson R.B., Zhang R., Chen Y.J., Peters K.M., Sawyez C.G., Sutherland B.G., et al. Two-week isocaloric time-restricted feeding decreases liver inflammation without significant weight loss in obese mice with non-alcoholic fatty liver disease. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21239156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cissé Y.M., Borniger J.C., Lemanski E., Walker W.H., Nelson RJ. Time-restricted feeding alters the innate immune response to bacterial endotoxin. J. Immunol. 2018;200:681–687. doi: 10.4049/jimmunol.1701136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okawa T., Nagai M., Hase K. Dietary intervention impacts immune cell functions and dynamics by inducing metabolic rewiring. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.623989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topalian S.L., Drake C.G., Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 57.Lundell L.S., Parr E.B., Devlin B.L., Ingerslev L.R., Altıntaş A., Sato S., et al. Time-restricted feeding alters lipid and amino acid metabolite rhythmicity without perturbing clock gene expression. Nat. Commun. 2020;11:4643. doi: 10.1038/s41467-020-18412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manella G., Sabath E., Aviram R., Dandavate V., Ezagouri S., Golik M., et al. The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nat. Metab. 2021;3:829–842. doi: 10.1038/s42255-021-00395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Partch C.L., Green C.B., Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desmet L., Thijs T., Mas R., Verbeke K., Depoortere I. Time-restricted feeding in mice prevents the disruption of the peripheral circadian clocks and its metabolic impact during chronic jetlag. Nutrients. 2021;13 doi: 10.3390/nu13113846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jamshed H., Beyl R.A., Della Manna D.L., Yang E.S., Ravussin E., Peterson C.M. Early time-restricted feeding improves 24-hour glucose levels and affects markers of the circadian clock, aging, and autophagy in humans. Nutrients. 2019;11 doi: 10.3390/nu11061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E.A., Gill S., et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi H., Rao M.C., Chang EB. Gut microbiota as a transducer of dietary cues to regulate host circadian rhythms and metabolism. Nat. Rev. Gastroenterol. Hepatol. 2021;18:679–689. doi: 10.1038/s41575-021-00452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original raw data in this manuscript were deposited to Sequence Read Archive (SRA) database of NCBI (Accession number: SRP425124). The data generated in the present study may be requested from the corresponding author.