Summary
Neutralizing antibodies targeting HIV-1 Env have been shown to protect from systemic infection. To explore whether these antibodies can inhibit infection of the first cells, challenge viruses based on simian immunodeficiency virus (SIV) were developed that use HIV-1 Env for entry into target cells during the first replication cycle, but then switch to SIV Env usage. Antibodies binding to Env of HIV-1, but not SIV, block HIV-1 Env-mediated infection events after rectal exposure of non-human primates to the switching challenge virus. After natural exposure to HIV-1, such a reduction of the number of first infection events should be sufficient to provide sterilizing immunity in the strictest sense in most of the exposed individuals. Since blocking infection of the first cells avoids the formation of latently infected cells and reduces the risk of emergence of antibody-resistant mutants, it may be the best mode of protection.
Keywords: HIV, AIDS, neutralizing antibody, sterilizing immunity, mucosal immunity, Fc-effector function
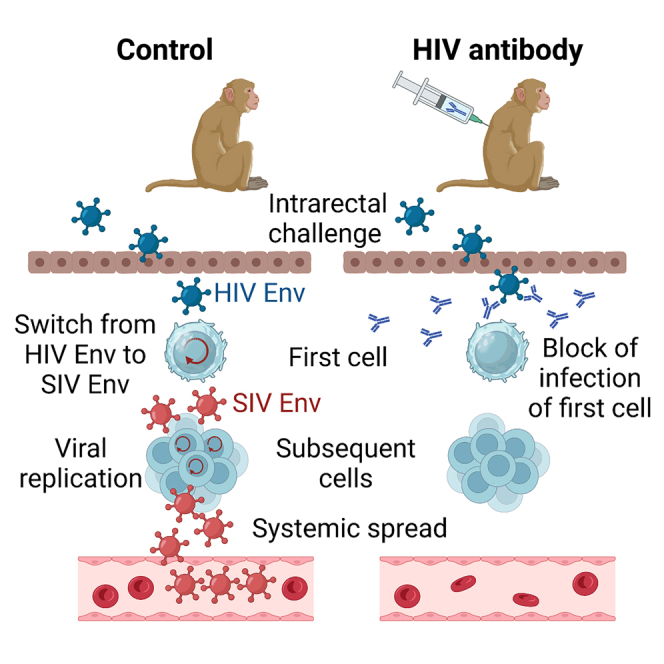
Graphical abstract
Highlights
-
•
Novel challenge virus switching from HIV-1 to SIV Env usage during the first infection cycle
-
•
HIV bnAbs block infection of first cell after mucosal exposure
-
•
Low level of protection under non-neutralizing conditions
Stab et al. show that HIV antibodies can prevent infection with viruses that use HIV-1 Env for entry into the first cell, but SIV Env for all subsequent infection cycles. Since the HIV antibodies do not bind to SIV Env, they must act prior to infection of the first cell.
Introduction
Antibodies raised after natural infection or vaccination are important mediators of protection from many viral diseases. Levels of neutralizing antibodies that bind to viral surface proteins and block entry of the virus into target cells frequently correlate with protection from disease, and passive transfer of monoclonal neutralizing antibodies has also been shown to be protective.1 Non-neutralizing effector mechanisms of the antibodies may also contribute to protection by trapping or inactivation of virions (reviewed in Excler et al.2). In addition, antibodies binding to viral proteins displayed on the surface of virus-infected cells may mediate a number of effector mechanisms, leading to destruction of infected cells, including antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis.
Protection from viral disease through antibodies may occur at different stages of infection. Sterilizing immunity in the strictest sense prevents infection of the “first cells” of the exposed host by viruses present in the inoculum. Although frequently assumed, we are not aware of any direct experimental evidence for such an antiviral sterilizing immunity in the strictest sense after mucosal exposure. After passive immunization, antibody concentrations in mucosal secretions, tissues, and surfaces tended to be 10- to >75-fold lower than in plasma,3,4,5 raising doubts about whether antibody levels sufficient for sterilizing immunity in the strictest sense are achievable. Alternatively, viruses can be contained at the portal of entry or draining lymph nodes without detectable systemic spread. In case of HIV, evidence for such abortive infections has been obtained.6,7 Transfer of neutralizing antibodies has also been shown to provide protection from systemic infection in non-human primate models of HIV infection,8 as evidenced by absence of detectable levels of viremia and lack of seroconversion. Clinical trials with the broadly neutralizing antibody VRC01 also revealed efficacy against acquisition of VRC01-sensitive HIV-1 isolates.9
In a high-dose challenge study in non-human primates, passive immunization with the broadly neutralizing antibody PGT121 provided protection from infection as defined above, although lymphatic tissues distal to the vaginal challenge site contained infectious virus.10 Detection of viral DNA in the distal tissues and transcriptomic signatures also suggested a limited degree of virus replication,10 indicating that PGT121 can contain virus replication after infection of the first cells consistent with the therapeutic efficacy of this antibody.11 However, due to the high challenge dose used, this study does not allow to exclude that PGT121 predominantly blocks infection of the first cells and that only a small percentage of challenge viruses break through this first line of defense and are then contained at subsequent rounds of infection. Whether antibodies to HIV can prevent infection of the first cells or only contain HIV replication at a later step has important implications. In the latter case, latently infected cells generated early during focal replication12 may become reactivated at a time point at which antibody concentrations have fallen beyond protective levels. In addition, low-level replication may allow HIV to acquire escape mutations leading to antibody resistance. Since highly potent monoclonal HIV antibodies are currently developed for therapeutic and prophylactic use, we aimed to explore whether PGT121 and 10–1074, two broadly neutralizing, V3 glycan-dependent antibodies,13,14 can prevent infection of the first cells after mucosal exposure of non-human primates.
Results
Development of an SIV challenge virus switching Envs
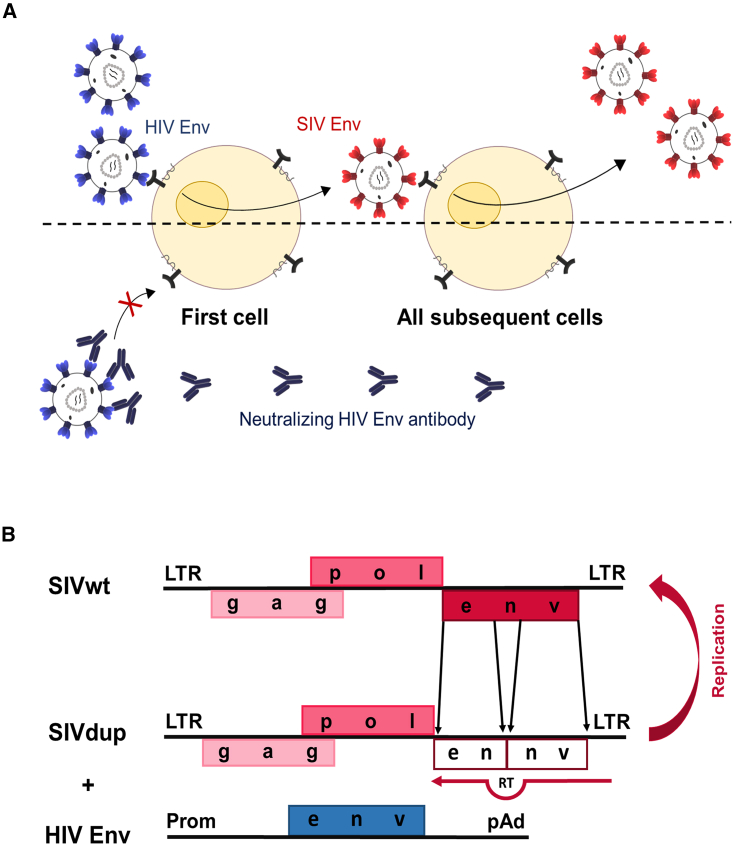
In order to determine whether antibodies can block infection of the first cells after mucosal exposure of non-human primates, the number of cells infected directly by infectious virions present in the inoculum need to be reliably quantified. This is difficult due to the low frequency of these first infection events. Although this hurdle may be overcome by a high-dose challenge and highly sensitive PCR methods,10 it is even more challenging to prove that viral nucleic acids detected at the site of inoculation are not the ones already present in the inoculum. We therefore followed an entirely different approach and generated SIV-based challenge viruses for non-human primate studies that use HIV-1 Env for entry during the first replication cycle but then switch to the use of SIV Env for all subsequent rounds (Figure 1A, upper half). If an antibody that binds to HIV-1 Env but not SIV Env prevents infection with such a challenge virus (Figure 1A, lower half), inhibition must occur at a step prior to virus entry into the first cells.
Figure 1.
SIV challenge virus switching Envs during the first round of replication
(A) Scheme of concept. Replication of an SIV challenge virus using HIV-1 Env for the first, but SIV Env for subsequent rounds of infections (upper half). If a neutralizing antibody (lower half) to HIV-1 Env prevents infection it can only do so by blocking the first infection event, since SIV Env is not targeted by the antibody.
(B) Map of SIVdup and assumed mechanism of repair during reverse transcription (RT). The inactivated SIV env with the internal direct tandem repeat (ennv) is shaded.
Retroviruses can be efficiently pseudotyped during one cycle of infection by expression of a heterologous Env protein in virus-producing cells; however, this will lead to virions containing both the heterologous and the homologous Env protein. In order to generate a challenge virus that can truly switch from HIV-1 Env during the first round of infection to SIV Env for all subsequent rounds, a direct tandem repeat was generated within env by duplicating an env fragment of the pathogenic SIVmac239 molecular clone (Figure 1B). The 3′ repeat was inserted out of frame and generated a stop codon between the duplicated env regions. Template strand switching during reverse transcription15 should lead to repair of the env open reading frame. Upon transfection of the proviral genome, designated SIVdup, a C-terminally truncated, non-functional SIV Env protein was detectable in cell lysates of transfected 293T cells (Figure 2A). Lacking the transmembrane and cytoplasmic domains, the truncated SIV Env could not be pelleted through a 35% sucrose cushion, indicating that it is not incorporated into virions. In contrast, Gag and SIV Env expressed from the parental wild-type SIV were readily detectable in the pelleted virion preparations (Figure 2A). Thus, SIVdup expresses a truncated SIV Env protein that is not incorporated into SIVdup particles.
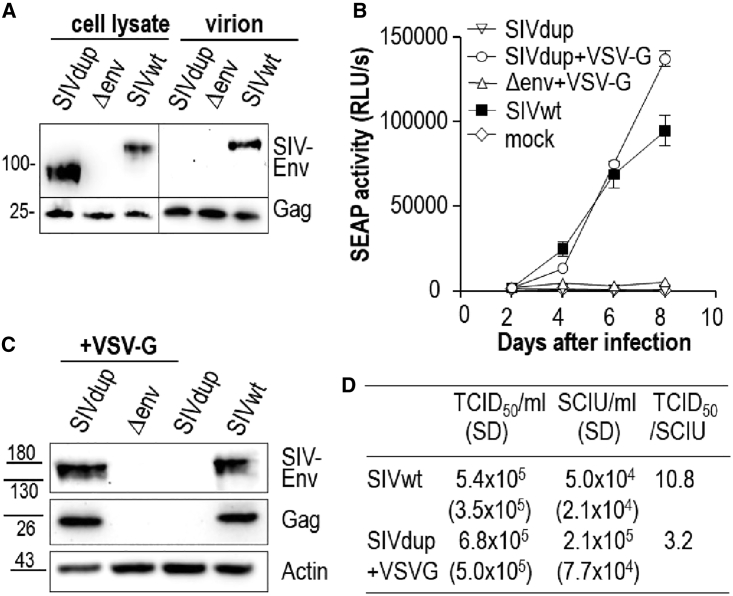
Figure 2.
In vitro characterization of SIVdup
(A) Western blot analyses of cells transfected with SIVdup, an env deletion mutant of SIV (Δenv), or wild-type SIV (SIVwt) and virions released from the transfected cells with antibodies to SIV Env and Gag.
(B) Replication kinetics of SIVdup, VSV-G pseudotyped SIVdup, VSV-G pseudotyped Δenv and SIVwt on CEMxSEAP indicator cells exposed to 1 × 104 infectious units of virus. Mean and standard deviation of the secreted embryonic alkaline phosphatase (SEAP) activity of triplicate cultures are shown.
(C) Western blot analyses of cell lysates from day 7 cultures infected as shown in (B) with antibodies to SIV Env, Gag, and actin.
(D) Titers of SIVwt and VSV-G pseudotyped SIVdup in a viral outgrowth assay (TCID50) and a single cycle of infection assay (SCIU). Mean and standard deviation (SD) of three independent experiments and the ratio of the titers are shown. See also Figure S1.
To test whether the tandem repeat could be removed during the initial replication cycle, SIVdup was pseudotyped by cotransfection with a VSV-G expression plasmid. The pseudotyped virus was then used to infect CEMxSEAP indicator cells supporting replication of SIV. The pseudotyped SIVdup led to an increase in reporter gene activity from day 2 to day 8 (Figure 2B). In contrast, reporter gene activity remained at background levels after infection with a replication-deficient env-deletion mutant of SIV also pseudotyped with VSV-G. Baseline levels of reporter gene activity after incubation with non-pseudotyped SIVdup particles confirmed that the tandem repeat destroys Env function (Figure 2B). Western blot analyses of cell lysates from CEMxSEAP indicator cells infected with VSV-G pseudotyped SIVdup and wild-type SIV also revealed SIV Env proteins co-migrating at the same size (Figure 2C). In addition, characteristic syncytia were also readily detectable 2 to 4 days after infection with VSV-G pseudotyped SIVdup, indicating repair of the interrupted SIV env sequences (data not shown). Day 7 supernatants of CEMxSEAP indicator cells infected with VSV-G pseudotyped SIVdup or wild-type SIV also contained infectious virions (3.3⋅103 infectious units [IU]/mL and 3.9⋅103 IU/mL, respectively) as determined by titration on TZMbl cells, while no infectious virions could be detected after inoculation of SIVdup or a VSV-G pseudotyped env deletion mutant of SIV. To further confirm reversion of the pseudotyped SIVdup to wild-type SIV after replication, RNA from the infected indicator cells on day 7 was amplified by RT-PCR with primers spanning the duplication site in env (Figure S1). Sequence analysis revealed that amplicons from indicator cells infected with pseudotyped SIVdup or wild-type SIV harbored the identical wild-type SIVmac239 sequence confirming accurate repair of the SIV env sequence. No amplicons were obtained from CEMxSEAP indicator cells exposed to non-pseudotyped SIVdup confirming its replication deficiency.
To investigate how frequently pseudotyped SIVdup is repaired during the first replication cycle, we compared the titer of the pseudotyped SIVdup on CEMxSEAP indicator cells in a single round of replication with the titer of emerging replication competent viruses in a limiting dilution viral outgrowth assay on the same cell line. As the sensitivity of the two assays may differ, we used wild-type SIV that did not require repair as a benchmark. Titers of wild-type SIV were 5.4 ∗105 tissue culture infectious doses 50% (TCID50)/mL in the viral outgrowth assay and 5.0∗104 single-cycle infectious units (SCIU)/mL, respectively (Figure 2D). Thus, the viral outgrowth assay is 10.8-fold more sensitive than the titration in the single-cycle assay. If the repair efficiency of the pseudotyped SIVdup was 100%, we would also expect a ratio of 10.8 of the TCID50/mL to the SCIU/mL. However, the ratio of the TCID50/mL to the SCIU/mL of the pseudotyped SIVdup was only 3.2 (Figure 2D), indicating that the efficacy of viral outgrowth of SIVdup is only 30% (3.2 divided by 10.8) of the viral outgrowth efficacy of wild-type SIV. Thus, three in 10 infection events should lead to repair of the env gene of SIVdup.
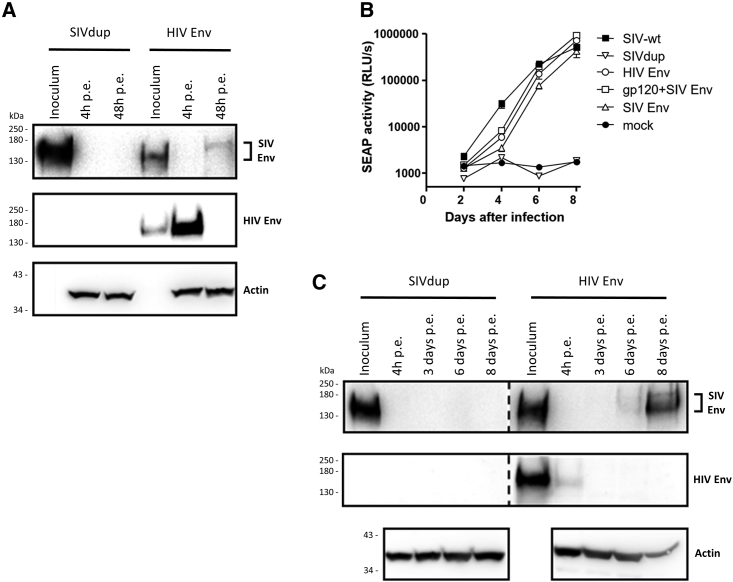
The SIVdup could also be pseudotyped with the HIV-1 Env of the SF162P3Nc8 clone (HIV Env) of an SIV/HIV-1 hybrid virus previously used for mucosal challenges in non-human primates,16 resulting in titers in the range of 1 × 105 IU per mL. Two days after infection of TZMbl cells, the repaired SIV Env could be detected by western blot analysis, while the Env protein from the inoculum was detectable 4 h but not 2 days after infection (Figure 3A). The HIV Env pseudotyped SIVdup, but not non-pseudotyped SIVdup initiated a spreading infection in CEMxSEAP indicator cells (Figure 3B). Western blot analyses indicated replication via the repaired SIV Env, and not via HIV Env (Figure 3C).
Figure 3.
Characterization of SIVdups pseudotyped with different lentiviral Env proteins
(A) Western blot analyses of TZMbl cell lysates 4 h and 48 h post exposure (p.e.) to 2 × 105 IU of HIV Env pseudotyped SIVdup (SIVdup+HIV Env) or the same volume of SIVdup particles. Viral inoculums were included as controls. Proteins detected by the antibodies used are indicated.
(B) Secreted embryonic alkaline phosphatase (SEAP) activity in cultures exposed to 2.8 × 103 IU of wild-type SIV (SIV-wt) or SIVdup pseudotyped with the indicated Env proteins. Non-pseudotyped SIVdup and mock infections were included as controls. Mean and standard deviation of reporter gene activity (SEAP) of triplicate cultures are shown.
(C) Western blot analysis of cell lysates of CEMxSEAP cells at the indicated days post exposure to SIVdup+HIV Env or non-pseudotyped SIVdup.
Protection after low-dose challenge
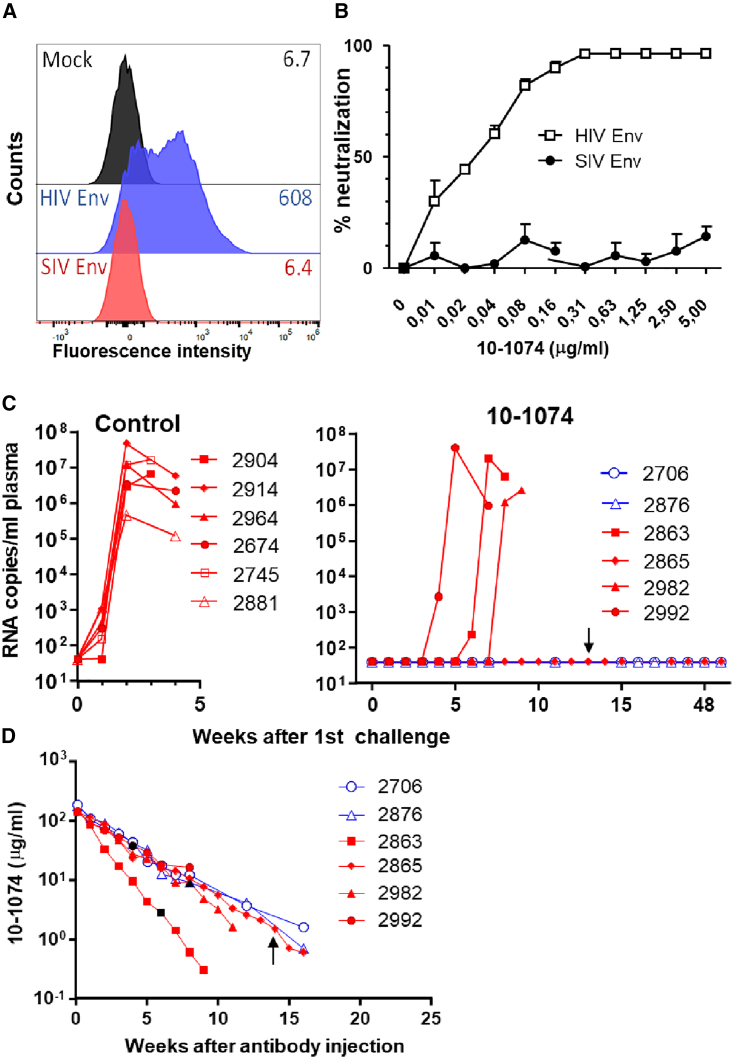
To prepare for challenge studies we first confirmed that PGT121, a broadly neutralizing antibody repeatedly used in non-human primate studies, could bind to SF162P3N HIV Env, but not SIV Env (Figure 4A). In contrast to SIV Env pseudotypes, SIVdup pseudotyped with HIV Env was also efficiently neutralized by PGT121 (Figure 4B).
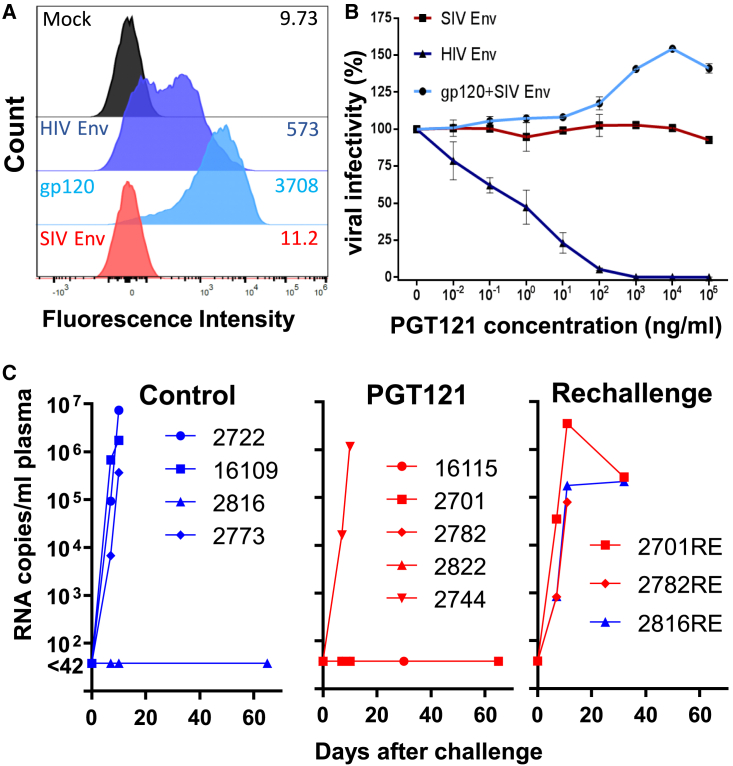
Figure 4.
Low-dose challenge of PGT121 treated rhesus monkeys
(A) Binding of PGT121 to 293T cells transfected with an empty vector control (Mock) or expression plasmids encoding SIV Env, HIV Env, or a membrane-anchored HIV gp120. One representative experiment out of two is shown.
(B) Infectivity of SIVdup pseudotyped with HIV-1 Env (HIV Env), SIV Env, or HIV-1 gp120 and SIV Env (gp120+SIV Env) on TZMbl cells in the presence of the indicated concentrations of PGT121. Mean and standard deviation of three replicates of one representative experiment out of two is shown.
(C) Viral RNA load after low dose challenge. Rhesus macaques were treated with 1 mg/kg body weight of control antibody (n = 4) or PGT121 (n = 5) 24 h prior to rectal inoculation with 2,800 infectious units of HIV Env pseudotyped SIVdup. The uninfected control monkey (2816) and two of the uninfected monkeys from the PGT121 treatment group (2701, 2782) were re-challenged 70 days after the first challenge. Plasma viral RNA levels at the indicated time points are shown. Four- and five-digit numbers are monkey designations. Animals that were re-challenged 70 days after the first challenge are given the suffix “RE.” See also Table S1 for PGT121 serum concentrations.
To determine whether PGT121 could block infection of the first cells after rectal exposure, four monkeys received an isotype-matched control antibody, while five monkeys were treated intravenously with 1 mg/kg body weight of PGT121. One day later, monkeys were challenged rectally with a low dose (2,800 IU) of HIV Env pseudotyped SIVdup. Assuming a conversion rate of pseudotyped SIVdup to replication competent SIV of 30% this corresponds to an inoculation dose of approximately 800 IU of replication competent SIV. At the time of challenge, serum concentrations of PGT121 ranged from 3.3 to 4.8 μg/mL (Table S1). Three of the four control animals were infected with SIVdup as evidenced by detection of viral RNA in the plasma 7 and 10 days after inoculation (Figure 4C). In contrast, only one of five PGT121-treated animals became viremic and seroconverted. The uninfected control animal (2816) and two of the four uninfected PGT121 animals (2701, 2782) were re-challenged on day 70 after the first challenge. The previously challenged animals had obtained a single intrarectal injection of 56 ng of SIV-p27 CA and 47 ng HIV gp120, a dose not expected to raise any immune response to the challenge virus. As expected, the animals were seronegative on the day of re-challenge and antibodies to HIV Env could not be detected by ELISA either (Table S1). This also implies that PGT121 levels of animals 2701 and 2782 had fallen below detectable levels, consistent with the reported half-life of PGT121 of 5.8 days.17
SIV RNA could not be detected in the plasma of the three animals at the time of re-challenge, but all three became viremic 7 days after the re-challenge (Figure 4C). Thus, six out of seven challenges resulted in infection in the absence of PGT121, while only one out of five monkeys got infected in its presence (p = 0.046; one-tailed Fisher's exact test). Despite this breakthrough infection in one of the animals, the results indicate that PGT121 prevented acquisition of infection prior to challenge virus entry into the first cell and thus provided sterilizing immunity in the strictest sense in the majority of the PGT121-treated monkeys.
Protection after repeated low-dose challenge
To confirm this result with the broadly neutralizing monoclonal antibody 10–1074 in an independent repeated low-dose challenge experiment, a second stock of the HIV Env pseudotyped SIVdup challenge virus was generated. The 10–1074 bound to HIV Env, but not SIV Env and its IC50 for HIV Env pseudotyped SIVdup was 0.023 μg/mL (Figures 5A and 5B). For the repeated low-dose challenge experiment, six monkeys were treated intravenously with 10 mg/kg body weight of 10–1074, while six control animals were mock-treated. Starting 1 week after the antibody administration, monkeys were challenged at weekly intervals by the intrarectal route with 6,000 IUs of the second SIVdup challenge stock. Prior to each challenge, blood samples were taken for viral RNA analysis. The weekly challenges were stopped 1 week after the first detection of viremia. Five out of six mock-treated control monkeys had detectable viral RNA levels in the blood 1 week after the first challenge, the other became positive for viral RNA after the second challenge. In contrast, none of the 10–1074 animals became viral RNA positive even after three challenges (p = 0.001; one-tailed Fisher's exact test). The challenges were continued in four of the 10–1074-treated monkeys to determine the 10–1074 concentration at breakthrough infection. Breakthrough infections were observed after four, six, and eight inoculations, respectively (Figure 5C). 10–1074 serum concentrations at the time point of breakthrough infection in these three monkeys varied from 2.8 to 38 μg/mL with a median concentration of 9 μg/mL (Figure 5D). One animal remained uninfected even after the last of 14 inoculations, a time point at which the 10–1074 plasma concentration was 1.5 μg/mL. Consistently, all viral RNA-positive monkeys became highly viremic and seroconverted. Median peak viremia at week 2 after the assumed take of the challenge virus did not differ significantly (two-tailed Mann-Whitney test) between the control group and the 10–1074 group consistent with reversion of the SIVdup challenge virus after one round of infection to SIVmac239. For two monkeys that were viral RNA negative 1 week after the third challenge, the repeated low-dose challenge was discontinued after the fourth challenge. These two and the monkey resistant to a total of 14 challenges remained viral RNA negative until autopsy 50–51 weeks after the first challenge (Figure 5C). Attempts to isolate infectious virus from peripheral blood mononuclear cells (PBMCs) and single mononuclear cell suspensions of the spleen, bone marrow, tonsils, and lymph nodes by a sensitive C8166 co-culture assay at the time of autopsy were not successful, although a total of 5–6 × 107 cells were analyzed from each animal (data not shown). In addition, none of the three monkeys seroconverted during the observation period. Therefore, the 10–1074 antibody provided sterilizing immunity in the repeated low-dose challenge experiment.
Figure 5.
Repeated low-dose challenge of 10–1074 treated rhesus monkeys
(A) Binding of 10–1074 to 293T cells transfected with an empty vector control (Mock) or expression plasmids encoding SIV Env and HIV Env. One representative experiment out of two is shown.
(B) Neutralization of SIVdup pseudotyped with HIV-1 Env (HIV Env) or SIV Env on TZMbl cells by 10–1074. Mean and standard deviation of three replicates of one experiment are shown.
(C) Viral RNA load after repeated low-dose challenge. Rhesus monkeys were intravenously injected with 10 mg/kg body weight of 10–1074 (n = 6) or were mock treated (n = 6) prior to weekly challenges with 6,000 infectious units of HIV Env pseudotyped SIVdup. In monkeys 2706 and 2876 (blue symbols), the repeated challenges were stopped after four inoculations. All other monkeys were challenged until 1 week after the first detection of SIV RNA in the plasma. Plasma viral RNA levels at the indicated time points are shown.
(D) Concentration of 10–1074 in serum at the indicated time points after injection of the antibody. Black symbols indicate the antibody concentrations at the time points of breakthrough infection. Monkey 2865 was exposed to challenge virus 14 times with the last challenge marked by a black arrow.
Development of a switching challenge virus targeted in a non-neutralizing manner
To explore whether antibodies may also block infection of the first cell by non-neutralizing mechanisms, we aimed to develop an SIVdup challenge virus to which PGT121 can bind without neutralizing it. Cotransfection of SIVdup with plasmids encoding SIV Env and a membrane-anchored gp120 of HIV Env should lead to SIVdup particles that simultaneously contain SIV Env for entry into target cells and gp120 as a binding site for PGT121. As expected and in contrast to HIV Env pseudotyped SIVdup, PGT121 did not neutralize SIVdup particles pseudotyped with SIV Env+gp120 (Figures 4A and 4B), although binding of PGT121 to gp120 expressed on the surface of transfected 293T cells exceeded its binding to HIV Env (Figure 4A). To further characterize SIVdup virions pseudotyped with HIV Env, SIV Env, or gp120+SIV Env, particles were purified from the supernatant of transfected 293T cells through a 35% sucrose cushion. Western blot analyses confirmed the presence of HIV gp120, HIV Env, SIV Env, and SIV Gag in the pelleted particles (Figure S2). The molar ratio of HIV Env and HIV gp120 to SIV Gag in these pelleted virion preparations was determined by ELISA to be 0.31 and 1.78, respectively. The higher ratio of HIV gp120 to SIV Gag in the gp120+SIV Env pseudotypes may be due to higher expression levels of gp120 due to codon optimization and more efficient incorporation into virions or exosomes co-purified with the virions. To further confirm that gp120 is indeed incorporated into SIVdup particles, virion preparations obtained after sucrose density centrifugation were immunoprecipitated by PGT121. SIV Gag and SIV Env could be co-precipitated with gp120 from the gp120+SIV Env pseudotypes, but not from the SIV Env pseudotype where the SIV proteins were only detectable in the washing fraction (Figure S2). This indicates that cotransfection of SIVdup with expression plasmids for gp120 and SIV Env results in virion that contain gp120 and SIV Env. Replication kinetics of the HIV Env pseudotyped SIVdup and the SIVdup pseudotyped with gp120+SIV Env were also very similar, consistent with the reversion to the same wild-type SIV after a single round of replication (Figure 3B).
Protection after simultaneous high-dose challenge
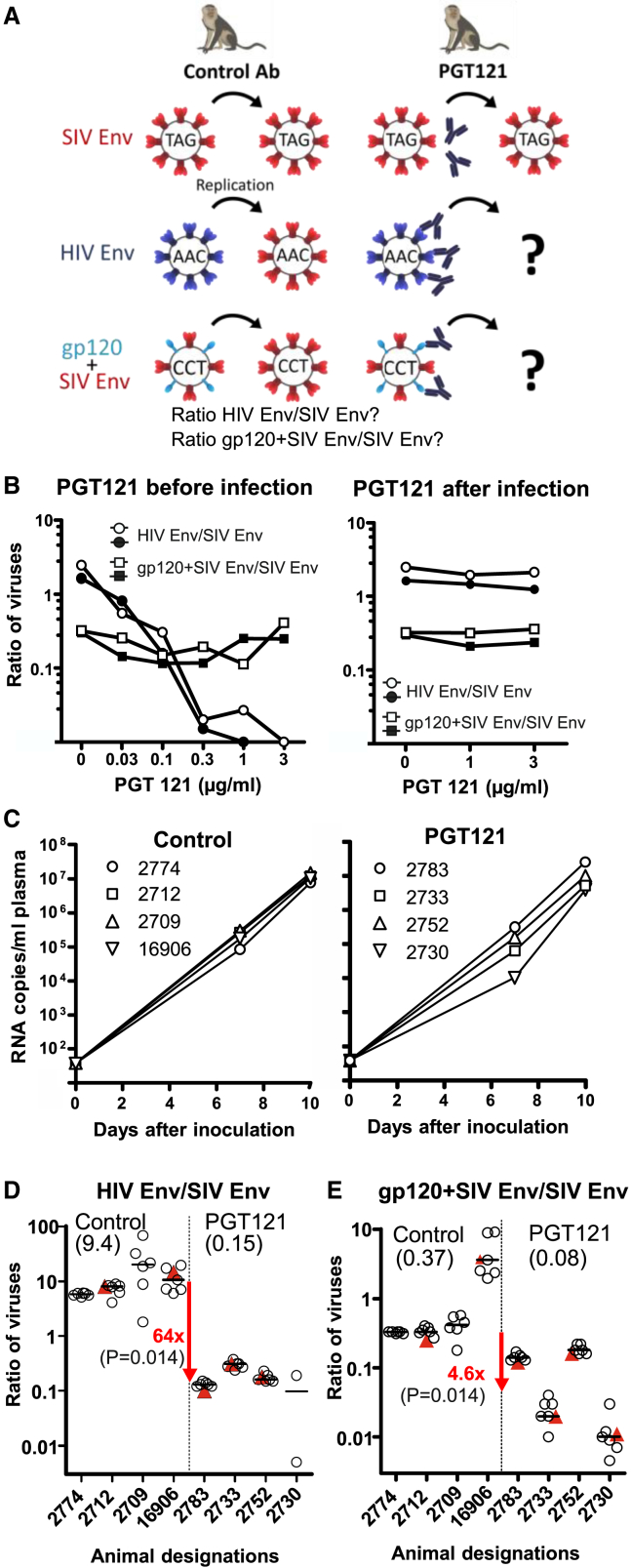
The susceptibility of non-human primates to acquisition of challenge viruses after mucosal exposure may differ substantially from one individual to the other. To control for these potential variations and to reduce the number of animals needed, a simultaneous challenge strategy was designed in which non-human primates are co-exposed to the SIV Env, HIV Env, and gp120+SIV Env pseudotypes (Figure 6A). Since SIVdup pseudotyped with SIV Env was not inhibited by PGT121 (Figure 4B), treatment with this antibody prior to a simultaneous challenge with the three pseudotypes should reduce the ratio of the HIV Env pseudotype to the SIV Env pseudotype. Similarly, if PGT121 inhibits infection by non-neutralizing mechanisms, the ratio of SIVdup pseudotyped with gp120+SIV Env to SIV Env pseudotypes should be reduced. The different SIVdup pseudotypes should revert to the same SIV during the first replication cycle. To discriminate between the different pseudotypes after simultaneous challenge, we introduced genetic tags into SIVdup that allowed for determining the ratio of the emerging SIVs by next generation sequencing as described in method details and Figure S3. Since gp120 and HIV Env are only present during the first infection cycle, differences in the ratio of SIVs derived from different SIVdup pseudotypes between controls and PGT121-treated monkeys must be due to the inhibitory activity of PGT121 during the first infection cycle (Figure 6A).
Figure 6.
Simultaneous high-dose challenge experiment
(A) Scheme of concept. SIVdups genetically tagged (TAG, AAC, CCT) and pseudotyped with the indicated Env proteins are co-injected into non-human primates treated with control antibody or PGT121. A change in the ratio of the genetically tagged SIVdup pseudotypes by PGT121 reveals inhibition of infection of the first cells by neutralization (ratio of HIV Env/SIV Env) and/or non-neutralizing mechanisms (ratio gp120+SIV Env/SIV Env).
(B) Ratio of derivatives of SIVdup pseudotypes in monkey PBMCs infected with a mixture of SIVdups pseudotyped with SIV Env, HIV Env, or gp120+SIV Env. PGT121 was either added before or 4 h after infection. Results of each of two independent experiments are shown.
(C–E) Rhesus macaques were treated with control antibody (n = 4) or PGT121 (n = 4) 24 h prior to rectal inoculation with a 4:1:0.5 mixture of genetically tagged SIVdups pseudotyped with HIV Env, SIV Env, or gp120+SIV Env. (C) Viral RNA load after challenge. (D) Ratio of tagged SIVs derived from HIV and SIV Env pseudotyped SIVdup in the plasma (red triangle) or lymphatic tissues (open circle) of control and PGT121-treated macaques 10 days after exposure. (E) Ratio of SIVs derived from SIVdup pseudotyped with gp120+SIV Env and SIV Env. The median of the ratios of each animal is marked by a horizontal bar. The medians of the individual median ratios of all animals of each of the two groups are provided in brackets under the group designation. The fold reduction in the group medians by PGT121 are indicated in red with the p values of one-tailed Mann-Whitney tests given in brackets. See also Figures S2 and S3 and Tables S1–S3.
To model the simultaneous challenge concept in vitro, rhesus monkey PBMCs were infected with a 4:1:0.5 mixture (based on infectious titers on TZMbl) of three differentially tagged SIVdups that were pseudotyped with HIV Env, SIV Env, or gp120+SIV Env, respectively. The PGT121 antibody was either pre-incubated with the SIVdup inoculum or added to the PBMC culture after infection with the SIVdup pseudotypes. After a 7-day culture period, the ratios of the SIVs derived from the HIV Env to SIV Env pseudotypes were determined. When adding PGT121 prior to infection, a dose-dependent decrease of the ratio of HIV Env to SIV Env from 2.1 to <0.02 (Figure 6B) indicated potent neutralization of the HIV Env pseudotyped SIVdup during the first infection cycle. In contrast, this ratio was only marginally affected if PGT121 was added after the initial infection. Differences in susceptibility of TZMbl cells used for the titration of the stocks and rhesus PBMCs to infection with the HIV Env and SIV Env pseudotyped SIVdup may explain why the ratio of progeny viruses derived from the pseudotypes in the PBMCs differs slightly from the ratio of the TZMbl titers of the inoculum even in the absence of PGT121.
The ratio of derivatives of the gp120+SIV Env pseudotype to derivatives of the SIV Env pseudotype was not reduced in a dose-dependent manner even if PGT121 was added prior to infection (Figure 6B), indicating that PGT121 did not efficiently inhibit the first infection cycle in PBMCs under non-neutralizing conditions.
For the simultaneous in vivo challenge experiment, rhesus monkeys were treated with PGT121 or a control antibody. One day later, monkeys were intrarectally inoculated with a mixture of high doses of three SIVdup challenge viruses pseudotyped either with HIV Env (dose: 2 × 105 IU, corresponding to at least 86 monkey infectious doses), SIV Env (dose: 5 × 104 IU), or gp120+SIV Env (dose: 2.3 × 104 IU). Since the SIV Env pseudotype is not targeted by PGT121 and was used at a high dose, all animals should get infected. Indeed, viral RNA could be detected in the plasma 7 and 10 days after inoculation in all animals and the viral loads did not differ between the two groups (Figure 6C). Ten days after inoculation, the ratio of SIVs derived from the three different challenge viruses was determined in plasma and different lymphatic tissues (Tables S2 and S3). The median ratio of SIV derived from the HIV Env pseudotype to SIV derived from the SIV Env pseudotype was reduced from 9.35 in the control group to 0.146 in the PGT121-treated animals (Figure 6D). Since PGT121 cannot affect infection events with the SIV Env pseudotype, the 64-fold decrease in the ratio of the HIV Env pseudotype to the SIV Env pseudotype by PGT121 must be due to a 64-fold reduction of infection events with the HIV Env pseudotypes. The median ratio of SIV derived from the gp120+SIV Env pseudotype to SIV derived from the SIV Env pseudotype was reduced by PGT121 from 0.37 to 0.08 indicating a 4.6-fold reduction of infection events by a non-neutralizing mechanism (Figure 6E). Thus, the simultaneous challenge experiment clearly confirmed the results of the low-dose challenge experiments and provided a quantitative measure of the efficacy of prevention of the first infection events under neutralizing and non-neutralizing conditions.
Discussion
Although neutralizing antibodies against HIV-1 have been repeatedly shown to protect from systemic infection in non-human primate studies,8 our results now provide direct evidence that neutralizing antibodies to HIV Env can efficiently block infection of the first cells in a low-dose, a repeated low-dose, and a simultaneous high-dose mucosal transmission model. This implies that sufficient levels of antibody are reached at the mucosal surface, and/or in the interstitial fluid surrounding the first target cells, presumably in the gut lamina propria, or in draining lymph nodes. Our study does not allow determination of whether these first target cells protected from infection are located at the site of inoculation or at distal sites. However, a number of non-human primate studies indicate that the first cells infected are CD4+ T cells forming local foci at the basolateral surface of the mucosal epithelium (reviewed in Lewis18). Also considering the results of Liu et al.,10 the following scenario emerges: Neutralizing antibodies efficiently reduce the number of infection events of the first cells at the site of inoculation. At a high-dose challenge, a few challenge viruses may overwhelm this first line of defense, but then can be controlled by neutralizing antibodies at later stages even at distal sites. However, under more limiting conditions involving exposure to lower infectious doses, as observed in most heterosexual transmissions,19 decreasing the number of infection events may substantially reduce the probability of infection and thus provide solid sterilizing immunity at the level of the first cells. Therefore, containment of virus replication at later stages may only become relevant under conditions in which infections are caused by a larger number of founder viruses.
Regarding the mechanisms mediating protection from infection of the first cell, neutralization seems to play the dominant role. PGT121 was at least 10-fold more efficient in preventing infection by HIV Env pseudotyped SIVdup than by the gp120+SIV Env pseudotype, which is bound, but not neutralized by PGT121. However, PGT121 reduced the number of first infection events even under non-neutralizing conditions by a factor of 4.6 suggesting that non-neutralizing effector mechanisms may contribute to protection. The 64-fold reduction of infection events under neutralizing conditions may therefore be due to synergistic effects of classical neutralization and Fc-dependent effector functions such as trapping of the virion in the mucus and blocking of epithelial transcytosis or DC-mediated trans-infection of CD4+ T cells.2 Our results suggesting that neutralizing and non-neutralizing mechanisms can block infection of the first cell are consistent with previous studies observing reduced protection from systemic SHIV infection by a neutralizing antibody (b12) with an impaired Fcγ receptor binding activity20,21 due to the LALA mutation in the Fc region. However, LALA mutants of PGT121 were shown to protect from intravenous challenge with cell-associated SHIV as efficiently as the parental antibody.22 Similarly, an LALAPG mutant of PGT121, which further reduces the residual Fcγ receptor binding activity of LALA mutants, also protected from intravaginal SHIV challenge as efficiently as the parental PGT121.23 Since a mouse model for passive immunization against HIV provided evidence that the effect of Fc-effector functions on protection might be masked by saturating neutralization activity,24 Hangartner et al. also applied PGT121 and its LALAPG mutant at semiprotective doses and observed similar levels of protection from intravaginal SHIV challenge for both antibodies. Therefore, the precise binding properties of the antibody used may also affect the efficacy of antibody Fc-effector mechanisms.
The observation by Liu et al., that PGT121 increased the frequency at which challenge virus RNA can be detected at distal sites, also raised the possibility that PGT121 facilitated virus translocation across the mucosal barrier to distal sites, potentially via immune complex capture on Fc-receptor bearing cells.10 If virions were opsonized by non-neutralizing antibodies, this could lead to enhancement of virus acquisition. The slight enhancement of viral infectivity observed at high concentrations of PGT121 (Figure 4B) in vitro would be consistent with this hypothesis. However, our observation that PGT121 reduces the number of first infection events under non-neutralizing conditions rather than enhancing infection clearly argues against this potential concern.
From a theoretical point of view, blocking infection of the first cell would be the best way to protect, since this avoids the establishment of latently infected cells that may reactivate the latent provirus once antibody levels have fallen beyond protective levels. However, given the difficulties in raising high enough levels of protective antibodies by vaccination, it may be more important to identify the effector mechanism requiring the lowest concentration of Ab levels for protection. This could then allow to optimize passive and active immunization for this particular effector mechanisms. Comparison of serum concentrations of 10–1074 at breakthrough infection with SF162P3N pseudotyped SIVdup and SHIVAD8-EO25 suggests that higher levels are needed for blocking infection of the first cell. However, differences in the two challenge viruses and the higher SIVdup challenge dose limit the strength of this conclusion. Indirect evidence that protection from systemic infection is easier to achieve by blockage of the first infection event comes from studies on post-exposure prophylaxis. If antibodies were equally efficient in blocking infection of the first cell and controlling early virus replication at subsequent steps, the efficiency of post-exposure prophylaxis in preventing systemic infection should be similar whether the antibodies are administered at the time of challenge or 1 or 2 days after challenge. However, the non-human primate studies indicate that efficacy is rapidly lost,26,27 suggesting that blocking infection of the first cells may be easier to achieve than control of virus spread after the first round of infection.
Effector mechanisms preventing infection of the first cell may also differ from those required for control of virus replication at subsequent steps. For cell-free transmissions, ADCC, for example, can be excluded as an effector mechanism of sterilizing immunity in the strictest sense, since virus infection is blocked prior to viral entry into the first cells. However, ADCC may allow control of virus replication at subsequent replication cycles.28 When aiming for sterilizing immunity in the strictest sense, the potency with which active or passive immunization blocks infection of the first cell may therefore also be considered as a criterion for selection of candidates for clinical development. The quantitative readout of the simultaneous non-human primate challenge model described allows one to optimize active and passive immunization approaches for blocking infection of the first cells and to reduce the number of animals required, thereby facilitating development of antibody-based HIV-1 prevention strategies.
Sterilizing immunity in the strictest sense also deprives the virus of the chance of diversifying from a small number of founder viruses to a large swarm of viruses and should thus lower the risk of emergence of neutralization-resistant viral variants. This may be of particular importance for prevention of other quickly adapting human pathogenic viruses, such as influenza viruses or SARS-CoV-2.
Limitations of the study
In contrast to other non-human primate challenge models of HIV infection, but similar to the most widely used in vitro neutralization assay,29 our challenge viruses are pseudotypes. We cannot exclude that this difference enhances the susceptibility of the challenge virus to antibody mediated Fc-effector functions in vivo. In particular, a higher Env content may increase the density of antibodies bound to the virion and thus the degree of opsonization. A higher degree of opsonization by polyclonal sera rather than monoclonal antibodies may also explain the weak protection induced by passive transfer of polyclonal sera without measurable neutralizing activity against the SIV challenge virus.30 However, further studies directly assessing the effect of the degree of opsonization on protection from HIV infection are needed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti HIV-1 p-24 (183-H12-5C) | NIH AIDS Research and Reagent Program | Cat# 1513 |
| Mouse monoclonal anti SIVmac251 gp120 (KK8) | NIH AIDS Research and Reagent Program | Cat# 1403 |
| Mouse monoclonal anti-β Actin antibody (AC-15) | Sigma Aldrich | Cat# A5441; RRID:AB_476744 |
| Goat polyclonal anti HIV-1 gp120 | Acris Antibodies | Cat# BP1035 |
| Rat Alexa Fluor 647 anti-mouse IgG1 | BioLegend | Cat# 406618; RRID:AB_2563477 |
| HIV-1 broadly neutralizing IgG1 (PGT121) | Laboratory of Dennis Burton | Cat# bNAberID_24 RRID:AB_2491041 |
| anti-Dengue NS1 IgG1 (DEN3) | Laboratory of Dennis Burton | N/A |
| HIV-1 broadly neutralizing IgG1 (10–1074) | Laboratories of Michel Nussenzweig and Florian Klein | N/A |
| peroxidase-conjugated F(ab')₂ fragment of goat anti-human IgG | Jackson ImmunoResearch Labs | Cat# 109-036-088, RRID:AB_2337594 |
| Rabbit anti-human IgG antibody, FITC-conjugated | Dako | Cat# F0202, RRID:AB_2335710 |
| Bacterial and virus strains | ||
| DH5α E. coli strain, competent | New England Biolabs | Cat# C2987 |
| SIVdup (pseudotyped with SIV Env, HIV Env, gp120+SIV Env, VSV-G) | This paper | N/A |
| SIVmac239 | This paper | N/A |
| Biological samples | ||
| Rhesus monkey PBMCs, plasma, lymphatic tissues | German Primate Center | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Staphylococcal enterotoxin B (SEB) | Sigma Aldrich | ⋅S4881; CAS: 11100-45-1 |
| Staphylococcal enterotoxin A (SEA) | Sigma Aldrich | S9399; CAS: 642595-84-4 |
| HIV-1 ConB gp120 | This paper | N/A |
| Critical commercial assays | ||
| QIAamp DNA Mini Kit | Qiagen | Cat#/ID: 51304 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat#/ID: 52906 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat# Q32854 |
| MiSeq Reagent Kit v3 | Illumina | MS-102-3003 |
| Bright-Glo™ Luciferase Assay System | Promega | Cat# E2610 |
| NucleoBond® Xtra Maxi EF Kit | Macherey-Nagel | Ref. 740424.10 |
| Agencourt AMPure XP system | Beckman Coulter | Cat# A63881 |
| In-Fusion® HD EcoDry™ Cloning Kit | TaKaRa | Cat# 638912 |
| One Step PrimeScript RT-PCR Kit | TaKaRa | Cat#: RR064B |
| Deposited data | ||
| Next generation sequencing data | This paper | SRA, BioProject: PRJNA958152 |
| Experimental models: Cell lines | ||
| HEK-293T/17 | ATCC | ATCC® CRL-11268™ |
| CEMx174 | NIH AIDS Research and Reagent Program | Cat# 272 |
| CEMxSEAP | (Means et al., 1997)31 | N/A |
| CEM-M7-CCR5 | (Brandt et al., 2002)32 | N/A |
| TZMbl | NIH AIDS Research and Reagent Program | Cat# 8129 |
| Experimental models: Organisms/strains | ||
| M.mulatta of Indian origin | German Primate Center | N/A |
| Oligonucleotides | ||
| Cloning of SIVdup: envdup500s: GAGGAGGAGATCCGGATTAGAT TTAGGTATTGTGCACCTC |
This paper | N/A |
| Cloning of SIVdup: envdupanti: GAAGGTAACTTCCGGATCTCC TCCTCCAGGA |
This paper | N/A |
| Introduction of genetic tags: dupFlag-s: GTACAGACAACAGA ACCCCATACCNGTNGGNAACA TTTACAGGAGATG |
This paper | N/A |
| Introduction of genetic tags: dupFlag-a: GATCCATCTCCTGT AAATGTTNCCNACNGGTATGG GGTTCTGTTGTCT |
This paper | N/A |
| Primers flanking the genetically tagged region: NGS-2dupGag-s: TCGTCGGC AGCGTCAGATGTGTATAAGAG ACAGCAACCAGCTCCACAACAAGG |
This paper | N/A |
| Primers flanking the genetically tagged region: NGS-2dupGag-a: GTCTCGTGGG CTCGGAGATGTGTATAAGAGACA GCTGTCTACATAGCTCTGAAATGGC |
This paper | N/A |
| Viral quantification: gag forward primer: ACCCAGTACAACAAATAGGTGGTAACT |
Sigma-Aldrich | N/A |
| Viral quantification: gag reverse primer: TCAATTTTACCCAGGCATTTAATGT |
Sigma-Aldrich | N/A |
| Viral quantification: gag probe: 5′-6FAM-TGTCCACCTGCCATTAA GCCCGAG-TAMRA-3′ |
Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| pBRmac239 | (Schnell et al., 2000)33 | N/A |
| SIVdupAAC, SIVdupTAG, SIVdupCCT expression constructs | This paper | N/A |
| pCAGGS-SF162P3Nc8 | C. Cheng-Mayer (Ho et al., 2007)16 | N/A |
| pcdSenv251co | This paper | N/A |
| gp120-162P3N-GTM/CD | This paper | N/A |
| pHIT-G | (Fouchier et al., 1997)34 | N/A |
| pΔenv-GFP | (Schnell et al., 2000)33 | N/A |
| pcD-HIVgp120ΔKR-His | (Storcksdieck genannt Bonsmann, 2014)35 | N/A |
| Software and algorithms | ||
| FlowJo Analysis Software | TreeStar | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 5.00 for Windows | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| QIAGEN CLC Genomics Workbench 9 | Qiagen Aarhus | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Klaus Überla (klaus.ueberla@fau.de).
Materials availability
All unique and stable reagents generated in this study are available from the lead contact with a completed Material Transfer Agreement.
Experimental model and study participant details
Animals
The rhesus monkeys (Macaca mulatta) used in this study were cared for by experienced personnel at the German Primate Center (DPZ) and kept according to the German Animal Welfare Act which complies with the European Union guidelines on the use of non-human primates for biomedical research and the Weatherall report. The study was approved by the Lower Saxony State Office for Consumer Protection and Food Safety and carried out with the project licenses 33.19-42502-04-13/1348 and 33.19-42502-04-18/3039. Conforming to § 11 of the German Animal Welfare act, the DPZ has the permission to breed and house non-human primates under license number 392001/7 granted by the local veterinary office. Monkeys were kept in groups of two if socially compatible with each other or, if not, in single cages. Animals which had to be individually caged had continual visual, olfactory and acoustic contact to their roommates and were still able to groom their neighbors through small mash inserts in the separating side walls. Each cage was equipped with a perch. The animals had unlimited water access and were fed with dry monkey biscuits containing adequate carbohydrate, energy, fat, fiber (10%), mineral, protein, and vitamin content twice daily. The feed was enriched by fresh fruit or vegetables and varying treats like nuts, cereal pulp and different seeds to make foraging more attractive. Moreover, for environmental enrichment monkeys were offered feeding puzzles, alternating toys and wood sticks for gnawing. During the study animals were assessed by experienced animal care attendants twice a day for any signs of distress, pain or sickness by checking water and feed intake, feces consistency and the general condition. In case of any abnormal presentation animals were attended by veterinarians.
Cells
HEK-293T/17 and TZMbl cell lines were cultured in DMEM (Life Technologies, Carlsbad, USA) supplemented with 10% FCS and 1% penicillin/streptomycin. CEMx174, CEMxSEAP, and CEM-M7-CCR5 cells were cultured in RPMI 1640 (Life Technologies) also supplemented with 10% FCS and 1% penicillin/streptomycin.
SIVdup challenge viruses
Pseudotyped SIVdup virus particles were generated by transient co-transfection of HEK-293T/17 cells with genetically tagged SIVdup (SIVdupAAC, SIVdupTAG, SIVdupCCT) and corresponding Env expression constructs (HIV Env, SIV Env or gp120+SIV Env) by the polyethylenimine method (PEI) essentially as described before.36 Briefly, the cells were seeded in 175 cm2 cell culture flasks at 2⋅107 cells per flask 24 h before transfection. Then, 80 μg DNA (40 μg of the genetically tagged SIVdup plasmid, 20 μg of HIV or SIV Env encoding plasmids, 20 μg of the gp120 expression plasmid or carrier DNA) were added to 5 mL of the serum-free DMEM containing 1% PenStrep and were thoroughly mixed with 100 μg PEI. After 10 min incubation at RT, the transfection mixture was combined with 15 mL of DMEM containing 1.5% FCS and 1% PenStrep and was given to the cells. The cells were cultivated at 37°C, 5% CO2. 16 h after transfection the medium was changed to remove all traces of PEI and the cells were washed twice with 1xPBS. After additional 28 h virus containing supernatants were collected, centrifuged at 1500xg for 3 min, filtered through a 0.45 μm filter unit, and stored in 1 mL aliquots at −80°C. These aliquots were used for the in vitro infection experiments with rhesus PBMCs and the low- and high-dose challenge experiments. Wild type SIVmac239 (SIVwt), the VSV-G pseudotyped SIVdup (SIVdup+VSV-G), non-pseudotyped SIVdup (SIVdup), and the VSV-G pseudotyped SIV env deletion mutant (Δenv+VSV-G) were produced by the same protocol using the plasmids described in the section Other plasmids.
Method details
Generation of SIVdup expression constructs
To generate SIVdup, a fragment spanning nucleotides 7541–8019 (numbering according to GenBank entry M33262.1) of the SIVmac239 proviral DNA was amplified by PCR using primers envdup500s (5′ GAG GAG GAG ATC CGG ATT AGA TTT AGG TAT TGT GCA CCT C) and envdupanti (5′ GAA GGT AAC TTC CGG ATC TCC TCC TCC AGG A). The PCR product was inserted out of frame into BspeEI digested and dephosphorylated pBRmac239 proviral plasmid using the In-Fusion HD EcoDry Cloning Kit (Takara Clontech, Mountain View, CA, USA). The resulting construct contained direct repeats of 478 bp separated by a stop codon and encoded a truncated Env ectodomain of 387 amino acids. To introduce silent mutations as genetic tags into gag of SIVdup the oligonucleotides dupFlag-s (5′GTA CAG ACA ACA GAA CCC CAT ACC NGT NGG NAA CAT TTA CAG GAG ATG) and dupFlag-a (5′GAT CCA TCT CCT GTA AAT GTT NCC NAC NGG TAT GGG GTT CTG TTG TCT) were phosphorylated and annealed prior to ligation into BsrGI/BamHI digested SIVdup. Two mutants (SIVdupTAG, SIVdupCCT) differing by at least two positions at the randomized sequence from each other and the wild type sequence (SIVdupAAC) were selected for subsequent experiments.
Other plasmids
The construct pCAGGS-SF162P3Nc816 encoding for the SHIV SF162P3N Env was a kind gift from Dr. C. Cheng-Mayer. The construct pcdSenv251co contains a codon-optimized sequence of env of an early transmitted SIVmac251 founder virus (clone K11 in37; GenBank: FJ578048.1) in the pcDNA3.1(+) vector. To express membrane-anchored gp120 of SF162P3Nc8, a codon-optimized sequence encoding amino acid 1 to 496 of P3N-838 with a isoleucine to leucine mutation at amino acid 21 was fused via a G4S linker (amino acids SGGGGSGGGGS) to the sequence encoding the membrane-spanning and cytoplasmic domains of VSV-G (amino acid 425 to 495 of Gene bank: NP_955548.1) and cloned into the pcDNA3.1(+) resulting in gp120-162P3N-GTM/CD. The proviral wild type SIVmac239 expression construct, pBRmac239, the pHIT-G construct encoding the G protein of VSV and pΔenv-GFP, designated Δenv in the current work and containing a deletion in env (188 bp) and nef (325 bp) of pBRmac239, have been previously described.33,34 pcD-HIVgp120ΔKR-His was kindly provided by Storcksdieck genannt Bonsmann (Storcksdieck genannt Bonsmann, 2014). All plasmids were purified from overnight bacterial cultures using NucleoBond Xtra Maxi EF Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions.
In vitro replication studies
Titration on TZMbl cells
The infectious units (IU) of the SIVdup pseudotypes were determined using TZMbl indicator cell line susceptible to infection with R5-and X4-tropic viruses and containing Tat-inducible β-galactosidase and firefly luciferase expression cassettes. In brief, cells were seeded in a 24-well plate at a density of 1 × 105 cells per well. After 24 h, triplicates of at least three serial 10-fold dilutions of virus were used to infect cells. 4 h after infection, 1 mL of culture medium was added to each well and cells were incubated for 48 h at 37°C, 5%CO2. Subsequently, β-galactosidase staining of the infected cells was carried out. The number of β-galactosidase expressing cells per well was determined and used for the calculation of the IU per mL supernatant.
Replication assay in CEMxSEAP cells
To determine whether SIVdup pseudotypes undergo more than one infection cycle, 1 × 105 CEMx174 cells were incubated in 24-well plates in triplicates for 4 h with 1 × 104 IU of HEK-293T/17 supernatants containing VSV-G pseudotyped SIVdup, wild type SIV, or VSV-G pseudotyped SIVdeltaEnv corresponding to 79 ng, 59 ng, and 70 ng p27 CA, respectively. CEMx174 cells were also exposed to 362 ng p27 CA of non-pseudotyped SIVdup. Subsequently, cells were washed three times with PBS and co-cultured with 1 × 106 CEMxSEAP reporter cells in 6 mL culture medium for 8 days. CEMxSEAP reporter cells contain a Tat-inducible secreted embryonic alkaline phosphatase (SEAP) expression cassette.31 SEAP levels were measured in supernatants at different time points using Phospha-Light SEAP Reporter Gene Assay System (Life Technologies) according to manufacturer’s instructions. Due to inefficient entry of HIV-1 Env pseudotypes of SIVdup into CEMx174 cells, replication assays with HIV Env and gp120+SIV Env pseudotyped SIVdups were performed by replacing the CEMx174 cells during the initial 4 h incubation period by CEM-M7-CCR5 cells32 and infecting them with 2.8 × 103 IU of the respective SIVdup pseudotypes. Thereafter, 1 × 106 CEMxSEAP cells were added and the SEAP activity measured during an eight day culture period.
TCID50 assay
The 50% tissue culture infectious dose (TCID50) of replication competent revertants emerging from VSV-G pseudotyped SIVdup and the TCID50 of wild type SIVmac239 was determined in parallel on CEMxSEAP cells in 96-well plates by a limiting dilution assay essentially as previously described.39 Therefore, virus containing supernatants were serially diluted and mixed with the cells. Every 4 to 5 days, cells were split by transferring of 50 μL infected cell suspension to 150 μL fresh CEMxSEAP cells. SEAP levels in the supernatants were determined using Phospha-Light SEAP Reporter Gene Assay System (Life Technologies) to identify wells with replication-competent SIV. Wells becoming virus positive within 3 weeks of cultivation formed the data basis for calculation of the TCID50 titer.
Determination of SCIU
To determine the single-cycle infectious units (SCIU) of SIVdup pseudotypes and wild type SIV on CEMxSEAP cells, cells were incubated with the viruses for 4 h. After washing, the RT inhibitor Lamivudine was added at a final concentration of 100 μM to block subsequent rounds of infection. Two days after infection, cells were fixed and permeabilized and stained for intracellular Gag expression with an anti-Gag-antibody (183-H12-5C; 30 μg/mL) and an anti-mouse Alexa 647-labelled secondary antibody (BioLegend). Based on the number of cells at the start of the infection experiment and the percentage of Gag-positive cells after 48 h, the SCIU were calculated.
Experiments with PBMCs
Rhesus monkey PBMCs were stimulated with a mixture of SEB and SEA (Sigma Aldrich, München, Germany) at 10 ng/mL each for 24 h prior to simultaneous infection with 2.2 × 104 IU (234 ng p27 CA) of the HIV Env SIVdup pseudotype, 5.5 × 103 IU (13 ng p27 CA) of the SIV Env SIVdup pseudotype, and 2.7 × 103 IU (7.6 ng p27 CA) of gp120+SIV Env pseudotype of SIVdup. The virus mix was either incubated with different concentrations of PGT121 for 1 h prior to incubation with stimulated PBMCs for another 4 h or PGT121 was added after the 4-h incubation period of the virus mix with the PBMCs. RNA was extracted from the supernatant of the PBMC cultures on day 7 for subsequent NGS analyses.
Western blot analyses
Western blot analyses were performed using monoclonal mouse anti p24 antibody (183-H12-5C, obtained through the NIH AIDS Research and Reagent Program from Dr. B. Chesebro and Dr. H. Chen), monoclonal mouse anti-SIVmac251 gp120 antibody (KK8, obtained through the NIH AIDS Research and Reagent Program from Dr. K. Kent) and polyclonal goat anti HIV-1 gp120 IgG (BP1035, Acris, Herford, Germany). TZMbl, CEM-M7-CCR5, or CEMxSEAP cells were incubated and cultured with SIVdup or its pseudotypes essentially as described in the section “In vitro replication studies” above. After washing of the cells at the indicated culture periods, cells were lysed in lysis buffer and proteins of the cell lysates were separated by SDS Page gel electrophoresis and blotted onto nitrocellulose membranes. SIVdup particles and its pseudotypes were also pelleted from the supernatant of transfected 293T cells by ultracentrifugation through a 35% sucrose cushion. The pellets containing the virions were resuspended for Western blot analyses. To verify the incorporation of Env proteins into SIVdup particles, the resuspended virions were also immunoprecipited with PGT121 using Protein G coated Dynabeads (Life Technologies) according to the manufacturer’s instructions. In brief, Dynabeads loaded with PGT121 were incubated with the resuspended SIVdup virions for 30 min at room temperature and washed. The beads were resuspended in SDS sample buffer for subsequent Western blot analyses.
Quantification of gag and Env content
The SIV p27 CA concentrations of viral stocks were determined with the SIV p27 ELISA assay kit (Cat# SK845, XpressBio, Frederic, US) or by coating of plates with serial dilutions of the viral stocks and using recombinant HIVp24 (Rec. HIV-1-p24, Aalto Bio Reagents, AG6054, Dublin, Ireland) as standard and the HIV-1 p24 specific antibody 183-H12-5C cross-reacting with SIV Gag and an anti-mouse IgG-HRP (Dianova 115-035-062; 1:5000) as secondary antibody. The Gag content of SIVdup virions pelleted through a 35% sucrose cushion was determined by coating ELISA plates with dilutions of the virion preparations and a serial dilutions of recombinant HIV-1 Gag40 as standard. The HIV-1 p24 specific antibody 183-H12-5C cross-reacting with SIV Gag and an anti-mouse IgG-HRP (Cat# P0260, Dako) were used to detect the plate-bound Gag proteins. The HIV-1 gp120 concentrations of viral stocks and SIVdup virions pelleted through the sucrose cushion were determined by coating the ELISA plates with the viral stocks or the virions and a serial dilution of recombinant gp120 of ConB with a C-terminal His-tag35 as a standard. The amount of coated HIV Env was then determined using the HIV Env antibody 2G12 and an anti-human IgG-HRP (Cat# P0214, Dako or Cat# 109-036-088, Jackson ImmunoResearch, West Grove, PA, USA).
PGT121 and 10–1074 binding assay
To determine PGT121 and 10–1074 binding to HIV Env, a membrane anchored gp120 or SIV Env, HEK293T17 cells were co-transfected with an expression plasmid for Blue Fluorescent Protein (BFP) and an empty vector or with BFP and expression plasmids encoding for the respective envelope proteins. 48 h post transfection, 2 × 105 cells were stained with 15 μg/ml of the respective antibody in FACS buffer (2% BSA, 2 mM NaN3 in PBS) for 30 min at RT. After 3 washing steps with FACS buffer, the cells were incubated for 20 min at RT in the dark with a 1:400 dilution of a mouse anti-human IgG coupled to Alexa Fluor 647 antibody (HP6017, Biolegend, USA). Subsequent to washes with FACS buffer, the cells were fixed with 2% paraformaldehyde in PBS and analyzed on an AttuneNxt (Thermofisher) flow cytometer. PGT121 and 10–1074 binding to the respective transfected cells were finally displayed using FlowJo software (Tree Star Inc.)
Neutralization assay
To determine an adequate virus dilution for the neutralization assay, 2-fold serial dilutions of the virus were prepared using neutralization medium (DMEM and 10% FCS with Penicillin (100 Units/ml) and Streptomycin (100 μg/mL) or 50 μg/mL Gentamicin and 25 mM HEPES) and 100 μL of each dilution were added to 11 vertical rows of a 96-well flat-bottom microtiter plate. Afterward, TZMbl cells were adjusted to the cell count of 1⋅105 cells per mL of the neutralization medium and 25 μg/mL DEAE-Dextran was added to the cell suspension. At last, 100 μL of the cell suspension were mixed with the diluted virus. After incubation for 72 h at 37°C, 5% CO2 the plate was examined under the microscope and wells with the clear cytopathic effect were excluded from the analysis. The luciferase activity was measured using Bright-Glo Luciferase Assay System (Promega, Fitchburg, USA).
In vitro neutralization assays were carried out essentially as described.41 Therefore, serial dilutions of PGT121 and 10–1074 were added to each well of a 96-well flat-bottom microtiter plate. Subsequently, the appropriate virus dilution resulting in approximately 100.000 RLUs luciferase activity was added to each well. TZMbl cells were adjusted to cell counts of 2⋅105/mL and after addition of 25 μg/mL DEAE-Dextran the cell suspension was added to the antibody-virus mix or to the virus only controls. The plate was incubated at 37°C, 5% CO2. After 48 h, luciferase activity was determined using Bright-Glo Luciferase Assay System (Promega, Fitchburg, USA) and IC50 values were calculated using GraphPad Prism 5.00 for Windows (San Diego, California, USA).
Non-human primate studies
Twenty-nine purpose-bred Indian-ancestry rhesus monkeys (Macaca mulatta) of either sex were taken from the breeding colony of the German Primate Center. They were 2.8–8 years old with a body weight between 3.8 and 8 kg. All monkeys were seronegative for simian immunodeficiency virus, simian retrovirus type D, and simian T-lymphotropic virus type 1 and randomly assigned to the different study groups, either receiving the anti-HIV-1 Env antibodies or the anti-Dengue antibody (see below). Blood was collected from the femoral vein using the vacutainer system (BD). For this purpose, for virus inoculation, and as premedication for euthanasia animals were anesthetized by intramuscular (i.m.) injection of a mixture of 5mg ketamine, 1mg xylazine and 0.01mg atropine per kg body weight (BW). Rectal challenges were performed in a volume of 3 mL as described.42 Following virus administration animals were kept in ventral recumbency with their hips elevated for 20 min. At necropsy animals were euthanized by an overdose of 200 mg sodium pentobarbital per kg BW injected into the bloodstream. Tissue sampling at necropsy comprised mesenteric, inguinal, axillary, retropharyngeal, submandibulary and rectal lymph nodes, spleen, tonsil and bone marrow.
Low-dose challenge experiment
HIV-1 broadly neutralizing IgG1 antibodies PGT121 or anti-Dengue NS1 IgG1 (DEN3) control antibody were diluted in saline and administered into the saphenous vein at a dose of 1 mg/kg body weight in a total volume of 10 mL 24 h before challenge. The challenge contained a total of 2800 IU of HIV Env pseudotyped SIVdup consisting of 2000 IU (44 ng p27 CA) of the pseudotype with the AAC tag, 600 IU (9.7 ng p27 CA) of that with the TAG tag and 200 IU (2.4 ng p27 CA) of that with the CCT tag. Different amounts of HIV Env pseudotyped SIVdups were used to determine the order of magnitude of the initial infection events in each animal. Blood samples were taken on day 7 and 10 or 11 after inoculation for determination of viral RNA levels. Infected animals were sacrificed on day 10/11 or on day 30 to determine the presence of the differentially tagged SIVdups. Uninfected macaques were monitored for viral RNA levels for at least 30 days. Three of the uninfected animals were re-challenged on day 70 after the first challenge in exactly the same way as for the first challenge, but without any additional antibody administrations.
Repeated low-dose challenge experiment
For the repeated low-dose challenge experiment monkeys were intravenously injected with 10–1074 antibody at a dose of 10 mg/kg body weight or mock-treated with 0.9% NaCl. Starting seven days later, monkeys were challenged weekly by the intrarectal route with 6000 IU (16 ng of p27 CA) of a second AAC-tagged HIV Env pseudotyped SIVdup challenge stock. Blood samples were taken weekly just prior to the subsequent challenge. Challenge virus exposure was stopped one week after animals became viral RNA positive in plasma.
Simultaneous high-dose challenge experiment
PGT121 and DEN3 antibodies were administered as described for the low-dose challenge experiment. In the simultaneous high-dose challenge experiment, monkeys were exposed once to 2 × 105 IU (2132 ng of p27 CA) of the HIV Env (AAC), 0.5 × 105 IU (119 ng of p27 CA) of the SIV Env (TAG), and 0.23 × 105 IU (69 ng of p27 CA) of gp120+SIV Env (CCT) pseudotypes. Expecting a strong reduction of infection events of the HIV Env pseudotyped SIVdup by PGT121, we used a higher dose for this challenge virus than for the SIV Env pseudotyped SIVdup virus in order to be able to also detect larger degrees of inhibition. Since the IU obtained for the stock of the gp120+SIV Env pseudotype was lower, the highest possible dose was used for this challenge virus. Based on the low-dose challenge experiment that resulted in infection of 6 out of 7 control animals at a dose of 2000 IU of the AAC-tagged HIV Env pseudotyped SIVdup, the inoculation dose of the same HIV Env pseudotype in the high-dose challenge experiment corresponded to at least 86 (= 2 × 105/2000 x 6/7) monkey infectious doses (MID). Since the median ratio of the HIV Env pseudotype to the SIV Env pseudotype in the control group of the high dose challenge experiment was 9.4, the MID of the SIV Env pseudotype was at least 9.2 (=86/9.4). Based on a ratio of HIV Env pseudotype to gp120+SIV Env pseudotype of 20.9 in the control group, the MID of the latter was calculated to be at least 4.1 (=86/20.9).
Viral RNA quantification and isolation of genomic DNA
Viral RNA copies in plasma were quantified using the TaKaRa One-step PrimeScript RT-PCR kit, gag forward primer (5′-ACCCAGTACAACAAATAGGTGGTAACT-3′), gag reverse primer (5′-TCAATTTTACCCAGGCATTTAATGT-3′) and a fluorescently labeled probe (5′-6FAM(6-carboxyfluorescein)-TGTCCACCTGCCATTAAGCCCGAG-TAMRA(6-carboxytetramethylrhodamine-3′) along with an RNA standard. Briefly, RT-PCR reactions contained 8.5μL eluted RNA, 0.4 μM SIV gag forward, 0.4 μM gag reverse primer, 0.2 μM probe, 0.5 μL reverse transcriptase, 0.5 μL HS-mix, and 12.5 μL mastermix in a reaction volume of 25 μL. PCR conditions were 30 min at 45°C and for 10 s at 95°C for reverse transcription and activation, followed by 45 cycles at 95°C for 5 s and at 60°C for 30 s in a Rotor-Gene Q apparatus (Qiagen). Quantification was performed by two independent PCR reactions. In the repeated low-dose challenge experiment plasma samples were considered positive, if both reactions were positive. A no template control, a negative plasma control and a SIV-positive control sample that were processed along with the other plasma samples were included in each run. The detection limit for viral RNA was 42 copies per mL plasma. Genomic DNA was isolated from cell pellets obtained from PBMCs and lymph node cells of infected animals using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Amplicons spanning the tagged region were generated and analyzed by NGS.
Analysis of blood and tissue samples for infectious virus
Cell-associated infectious virus was determined in blood, lymph nodes from different regions, spleen, tonsil, and bone marrow by a limiting dilution co-culture assay. Peripheral blood mononuclear cells (PBMC) were separated and mononuclear cells (MNC) from tissue purified as described.43,44 The complete MNC yield from each organ was kept in RPMI supplemented with 20% FCS and antibiotics at 2 x 106 cells/ml and stimulated with SEB/SEA at 10 ng/ml each for 24 h. Thereafter, recombinant human IL-2 (TEBU) was added at 100 I.U./mL for another 48 h without washing. Next, the primary cells were washed, counted and for each organ up to five aliquots containing 0.5–2 x 106 MNC were co-cultured with 3 × 106 C81-66 indicator cells except PBMC for which only a single co-culture with 3 x 106 primary cells was set up. All cultures were tested for intracellular viral antigen by an immunoperoxidase assay44 at weeks 2, 3 and 4 after start of cultivation.
Serum analyses
Seropositivity was determined by the INNO-LIA HIV I/II Score kit (Fujirebio, Tokyo, Japan), which also readily cross-reacts with SIV Gag and –Env specific antibodies due to the phylogenetic proximity of HIV-1 Gag/HIV-2 Env and SIVmac239. Serum PGT121 concentrations were determined on plates coated with 100 ng HIV-1 ConB gp120 per well. ConB gp120 was purified from the supernatant of 293T cells transfected with pcD-HIVgp120ΔKR-His using Lentil Lectin Sepharose 4B affinity chromatography. We used serial dilutions of PGT121 as standard, 1:40 dilutions of serum samples from PGT121-treated monkeys, a peroxidase-conjugated F(ab')₂ fragment of goat anti-human IgG (Jackson ImmunoResearch Cat.#109-036-088) at a 1:3000 dilution as secondary antibody, and a chemiluminescent substrate. Serum concentrations of 10–1074 were determined accordingly using serial dilutions of 10–1074 as standard.
NGS analysis
A gag fragment spanning the genetic tag was amplified by PCR from extracted RNA or DNA with the primers NGS-2dupGag-s (5′TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CAA CCA GCT CCA CAA CAA GG) and NGS-2dupGag-a (5′GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GCT GTC TAC ATA GCT CTG AAA TGG C). The PCR products were purified using Agencourt AMPure XP system (Beckman Coulter). Illumina Nextera-compatible dual indices were added to both ends of the above amplicons using 8 cycles of PCR with 200 nM of unique index primer pairs for each amplicon. Index-attachment to amplicons was verified by gel electrophoresis, and the indexed amplicon libraries were purified with Agencourt AMPure XP beads. NGS libraries were then quantified by the Qubit dsDNA HS (High Sensitivity) Assay Kit (Thermo Fisher Scientific, Eugene, USA), diluted to 4 nM in 1xTE, denatured by addition of 1 vol 0.2 N NaOH for 5 min, and neutralized by dilution to 20 p.m. in 5xSSC/0.05% Tween 20. The 20 p.m. libraries were sequenced using 600 cycle v3 reagent kits on an Illumina MiSeq instrument. The frequency of the different tags was determined by aligning quality trimmed NGS reads (total average 83.7 k reads per sample) with high stringency to SIV reference genomes containing the three SIVdup tags with the Genome workbench 9 (CLCbio/Qiagen). Sequencing errors and the demultiplexing process of parallel Illumina sequencing may lead to misclassification of identifiers45 resulting in assignment of sequences present in the same run to other samples. To get an estimate of these background counts for the SIVdup tags, non-SIV samples sequenced on the same run were analyzed for the presence of SIVdup tags. Number of reads of misassigned SIVdup tags were in the range of 0–463. Therefore, samples were only considered positive for each of the SIVdup tags, if the number of reads exceeded the maximal number of misassigned reads for the same SIVdup tag on the same run by a factor of two. The cut-offs and the number of reads of the different SIVdup tags used to calculate the ratio of the SIVdup tags are listed in Table S2. For each sample analyzed, the number of reads of the AAC, CCA, and TAG tag was the primary readout. Dividing the number of reads for the AAC tag by the number of reads for the TAG tag provided the ratio of viruses derived from the HIV Env and the SIV Env pseudotypes (Table S3). Similarly, the ratio of the derivatives of gp120+SIV Env and SIV Env pseudotypes was calculated by dividing the number or reads for the CCA tag by the number of reads for the TAG tag. Minority species may have escaped detection in samples with low viral load levels as they may not be represented in the sample added to the PCR. This may explain why low minority species were detected in some but not all tissues of the same animal. Therefore, only samples positive for both SIVdup tags were used to calculate the median of the ratio of the two SIVdup tags for each individual animal. From these individual median ratios of the SIVdup tags the medians of the ratio of the SIVdup tags of the control and the PGT121 group were determined and the fold reduction in the ratio by PGT121 treatment was calculated.
Quantification and statistical analysis
Statistical analysis
Fisher's exact test was performed to determine whether PGT121 or 10-1074 administration leads to a significant reduction in the frequency of infections after low-dose challenge virus exposure. The significance of reduced ratios of HIV Env to SIV Env pseudotypes and gp120+SIV Env to SIV Env pseudotypes after PGT121 treatment was analyzed by the non-parametric Mann-Whitney test using the GraphPad Prism 5.01 program. Since PGT121 and 10–1074 have previously been shown to be potent inhibitors of HIV-1 Env-mediated infections in cell culture and non-human primates, one-tailed statistical tests were applied. Statistical parameters including the p values, mean and SD measures are indicated in the Figures and Figure Legends.
Acknowledgments
This work was supported by grants from the German Research Foundation to K.Ü. and C.S.-H. (Ue45/13-1, STA447/6-1, Ue45/13-2). We would like to thank Dr M. Storcksdieck genannt Bonsmann, Dr G. Nabi, and Dr H. Elsayed for help with experiments; Regina Bütermann, Klaus Sure, Doris Jungnickl, Sandra Heine, and Nicole Leuchte for excellent technical assistance; and Antonina Klippert, Maria Daskalaki, and Matthias Mietsch for veterinary help in performing the animal experiments; and Dr. Michel Nussenzweig for providing the 10–1074 antibody.
Author contributions
Conceptualization, C.S.-H. and K.Ü.; Methodology and investigation, V.S., C.S.-H., E.R., A.E., E.R., K.F., U.S., B.T., and M.T.; Writing original draft, V.S. and K.Ü.; Writing review and editing, V.S., C.S.-H., A.E., D.R.B., M.T., and K.Ü.; Resources, D.R.B. and F.K.; Funding acquisition, C.S.-H. and K.Ü. All authors reviewed the final version.
Declaration of interests
D.R.B. is a paid consultant of IAVI. B.T.’s current affiliation is Department of Biochemistry and Pathobiochemistry, Systems Biochemistry, Ruhr-University Bochum, Bochum, Germany.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 6, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101201.
Supplemental information
Data and code availability
Standardized datatypes have been deposited at the NCBI Sequence Read Archive (SRA) and are publicly available as of the date of publication. Accession numbers are listed in the key resource table. Other data reported in the paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.
References
- 1.Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Excler J.-L., Ake J., Robb M.L., Kim J.H., Plotkin S.A. Nonneutralizing functional antibodies: a new “old” paradigm for HIV vaccines. Clin. Vaccine Immunol. 2014;21:1023–1036. doi: 10.1128/CVI.00230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shingai M., Donau O.K., Plishka R.J., Buckler-White A., Mascola J.R., Nabel G.J., Nason M.C., Montefiori D., Moldt B., Poignard P., et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein K., Veazey R.S., Warrier R., Hraber P., Doyle-Meyers L.A., Buffa V., Liao H.-X., Haynes B.F., Shaw G.M., Shattock R.J. Neutralizing IgG at the portal of infection mediates protection against vaginal simian/human immunodeficiency virus challenge. J. Virol. 2013;87:11604–11616. doi: 10.1128/JVI.01361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parren P.W., Marx P.A., Hessell A.J., Luckay A., Harouse J., Cheng-Mayer C., Moore J.P., Burton D.R. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulkarni P.S., Butera S.T., Duerr A.C. Resistance to HIV-1 infection: lessons learned from studies of highly exposed persistently seronegative (HEPS) individuals. AIDS Rev. 2003;5:87–103. [PubMed] [Google Scholar]
- 7.McChesney M.B., Collins J.R., Lu D., Lu X., Torten J., Ashley R.L., Cloyd M.W., Miller C.J. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4(+)-T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 1998;72:10029–10035. doi: 10.1128/jvi.72.12.10029-10035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegu A., Borate B., Huang Y., Pauthner M.G., Hessell A.J., Julg B., Doria-Rose N.A., Schmidt S.D., Carpp L.N., Cully M.D., et al. A Meta-analysis of Passive Immunization Studies Shows that Serum-Neutralizing Antibody Titer Associates with Protection against SHIV Challenge. Cell Host Microbe. 2019;26:336–346.e3. doi: 10.1016/j.chom.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corey L., Gilbert P.B., Juraska M., Montefiori D.C., Morris L., Karuna S.T., Edupuganti S., Mgodi N.M., deCamp A.C., Rudnicki E., et al. Two Randomized Trials of Neutralizing Antibodies to Prevent HIV-1 Acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/nejmoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Ghneim K., Sok D., Bosche W.J., Li Y., Chipriano E., Berkemeier B., Oswald K., Borducchi E., Cabral C., et al. Antibody-mediated protection against SHIV challenge includes systemic clearance of distal virus. Science. 2016;353:1045–1049. doi: 10.1126/science.aag0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.-W., Shekhar K., Gupta S., et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavez L., Calvanese V., Verdin E. HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.-P., Wang S.-K., Ramos A., Chan-Hui P.-Y., Moyle M., et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J.F., Halper-Stromberg A., Gnanapragasam P.N.P., Spencer D.I.R., Seaman M.S., et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhodes T.D., Nikolaitchik O., Chen J., Powell D., Hu W.-S. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 2005;79:1666–1677. doi: 10.1128/JVI.79.3.1666-1677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho S.h., Tasca S., Shek L., Li A., Gettie A., Blanchard J., Boden D., Cheng-Mayer C. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 2007;81:8621–8633. doi: 10.1128/JVI.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moldt B., Rakasz E.G., Schultz N., Chan-Hui P.-Y., Swiderek K., Weisgrau K.L., Piaskowski S.M., Bergman Z., Watkins D.I., Poignard P., Burton D.R. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis G.K. The first 24 h. Curr. Opin. HIV AIDS. 2016;11:561–568. doi: 10.1097/COH.0000000000000319. [DOI] [PubMed] [Google Scholar]
- 19.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H., et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hessell A.J., Hangartner L., Hunter M., Havenith C.E.G., Beurskens F.J., Bakker J.M., Lanigan C.M.S., Landucci G., Forthal D.N., Parren P.W.H.I., et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 21.Hessell A.J., Poignard P., Hunter M., Hangartner L., Tehrani D.M., Bleeker W.K., Parren P.W.H.I., Marx P.A., Burton D.R. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons M.S., Lloyd S.B., Lee W.S., Kristensen A.B., Amarasena T., Center R.J., Keele B.F., Lifson J.D., La Branche C.C., Montefiori D., et al. Partial efficacy of a broadly neutralizing antibody against cell-associated SHIV infection. Sci. Transl. Med. 2017;9:eaaf1483. doi: 10.1126/scitranslmed.aaf1483. [DOI] [PubMed] [Google Scholar]
- 23.Hangartner L., Beauparlant D., Rakasz E., Nedellec R., Hozé N., McKenney K., Martins M.A., Seabright G.E., Allen J.D., Weiler A.M., et al. Effector function does not contribute to protection from virus challenge by a highly potent HIV broadly neutralizing antibody in nonhuman primates. Sci. Transl. Med. 2021;13 doi: 10.5281/zenodo.4474217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bournazos S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., Ravetch J.V. Broadly Neutralizing Anti-HIV-1 Antibodies Require Fc Effector Functions for In Vivo Activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautam R., Nishimura Y., Gaughan N., Gazumyan A., Schoofs T., Buckler-White A., Seaman M.S., Swihart B.J., Follmann D.A., Nussenzweig M.C., Martin M.A. A single injection of crystallizable fragment domain-modified antibodies elicits durable protection from SHIV infection. Nat. Med. 2018;24:610–616. doi: 10.1038/s41591-018-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrantelli F., Buckley K.A., Rasmussen R.A., Chalmers A., Wang T., Li P.-L., Williams A.L., Hofmann-Lehmann R., Montefiori D.C., Cavacini L.A., et al. Time dependence of protective post-exposure prophylaxis with human monoclonal antibodies against pathogenic SHIV challenge in newborn macaques. Virology. 2007;358:69–78. doi: 10.1016/j.virol.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura Y., Igarashi T., Haigwood N.L., Sadjadpour R., Donau O.K., Buckler C., Plishka R.J., Buckler-White A., Martin M.A. Transfer of neutralizing IgG to macaques 6 h but not 24 h after SHIV infection confers sterilizing protection: Implications for HIV-1 vaccine development. Proc. Natl. Acad. Sci. USA. 2003;100:15131–15136. doi: 10.1073/pnas.2436476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz J.A., Bar-On Y., Lu C.-L., Fera D., Lockhart A.A.K., Lorenzi J.C.C., Nogueira L., Golijanin J., Scheid J.F., Seaman M.S., et al. Non-neutralizing Antibodies Alter the Course of HIV-1 Infection In Vivo. Cell. 2017;170:637–648.e10. doi: 10.1016/j.cell.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarzotti-Kelsoe M., Bailer R.T., Turk E., Lin C.l., Bilska M., Greene K.M., Gao H., Todd C.A., Ozaki D.A., Seaman M.S., et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alter G., Yu W.H., Chandrashekar A., Borducchi E.N., Ghneim K., Sharma A., Nedellec R., McKenney K.R., Linde C., Broge T., et al. Passive Transfer of Vaccine-Elicited Antibodies Protects against SIV in Rhesus Macaques. Cell. 2020;183:185–196.e14. doi: 10.1016/j.cell.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Means R.E., Greenough T., Desrosiers R.C. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 1997;71:7895–7902. doi: 10.1128/jvi.71.10.7895-7902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandt S.M., Mariani R., Holland A.U., Hope T.J., Landau N.R. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 2002;277:17291–17299. doi: 10.1074/jbc.M108232200. [DOI] [PubMed] [Google Scholar]
- 33.Schnell T., Foley P., Wirth M., Münch J., Uberla K. Development of a self-inactivating, minimal lentivirus vector based on simian immunodeficiency virus. Hum. Gene Ther. 2000;11:439–447. doi: 10.1089/10430340050015905. [DOI] [PubMed] [Google Scholar]
- 34.Fouchier R.A., Meyer B.E., Simon J.H., Fischer U., Malim M.H. HIV-1 infection of non-dividing cells: Evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storcksdieck Genannt Bonsmann M. Ruhr-University Bochum; 2014. Induction and Modulation of Cellular and Humoral Immune Responses against HIV-1 by Immunization with DNA and Virus-like Particle Vaccines. Dissertation for Degree of Doctor of Natural Sciences. [Google Scholar]
- 36.Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 37.Keele B.F., Li H., Learn G.H., Hraber P., Giorgi E.E., Grayson T., Sun C., Chen Y., Yeh W.W., Letvin N.L., et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren W., Mumbauer A., Zhuang K., Harbison C., Knight H., Westmoreland S., Gettie A., Blanchard J., Cheng-Mayer C. Mucosal transmissibility, disease induction and coreceptor switching of R5 SHIVSF162P3N molecular clones in rhesus macaques. Retrovirology. 2013;10:9. doi: 10.1186/1742-4690-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson V.A., Byington R.E. In: Techniques in HIV-Research. Aldovini A., Walker B.D., editors. 1990. Quantitative assays for virus infectivity; pp. 71–85. [Google Scholar]
- 40.McKinstry W.J., Hijnen M., Tanwar H.S., Sparrow L.G., Nagarajan S., Pham S.T., Mak J. Expression and purification of soluble recombinant full length HIV-1 Pr55Gagprotein in Escherichia coli. Protein Expr. Purif. 2014;100:10–18. doi: 10.1016/j.pep.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Montefiori D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 42.Tenbusch M., Ignatius R., Temchura V., Nabi G., Tippler B., Stewart-Jones G., Salazar A.M., Sauermann U., Stahl-Hennig C., Uberla K. Risk of immunodeficiency virus infection may increase with vaccine-induced immune response. J. Virol. 2012;86:10533–10539. doi: 10.1128/JVI.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl-Hennig C., Dittmer U., Nißlein T., Petry H., Jurkiewicz E., Fuchs D., Wachter H., Mätz-Rensing K., Kuhn E.M., Kaup F.J., et al. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J. Gen. Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- 44.Stahl-Hennig C., Steinman R.M., Tenner-Racz K., Pope M., Stolte N., Mätz-Rensing K., Grobschupff G., Raschdorff B., Hunsmann G., Racz P. Rapid infection of oral mucosal-associated lymphoid tissue with simian immunodeficiency virus. Science. 1999;285:1261–1265. doi: 10.1126/science.285.5431.1261. [DOI] [PubMed] [Google Scholar]
- 45.Kircher M., Sawyer S., Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Standardized datatypes have been deposited at the NCBI Sequence Read Archive (SRA) and are publicly available as of the date of publication. Accession numbers are listed in the key resource table. Other data reported in the paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.