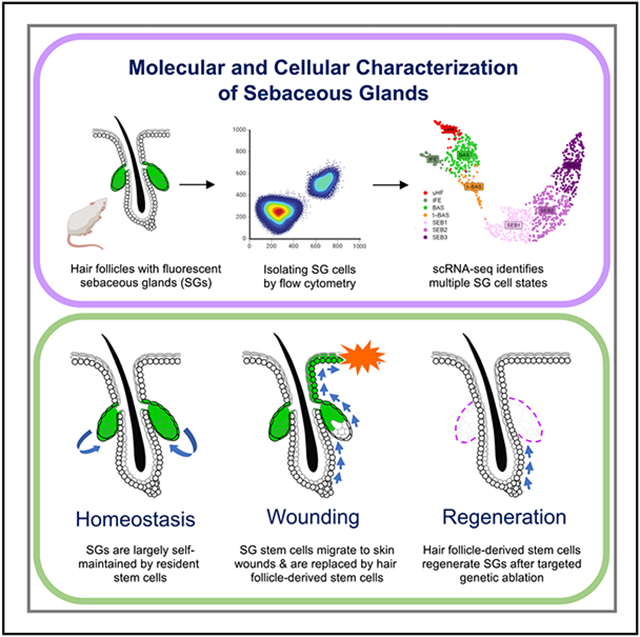
SUMMARY
Sebaceous glands (SGs) release oils that protect our skin, but how these glands respond to injury has not been previously examined. Here, we report that SGs are largely self-renewed by dedicated stem cell pools during homeostasis. Using targeted single-cell RNA sequencing, we uncovered both direct and indirect paths by which resident SG progenitors ordinarily differentiate into sebocytes, including transit through a Krt5+PPARγ+ transitional basal cell state. Upon skin injury, however, SG progenitors depart their niche, re-epithelialize the wound, and are replaced by hair-follicle-derived stem cells. Furthermore, following targeted genetic ablation of >99% of SGs from dorsal skin, these glands unexpectedly regenerate within weeks. This regenerative process is mediated by alternative stem cells originating from the hair follicle bulge, is dependent upon FGFR2 signaling, and can be accelerated by inducing hair growth. Altogether, our studies demonstrate that stem cell plasticity promotes SG durability following injury.
Graphical Abstract
In brief
Veniaminova et al. characterize the development, maintenance, and regeneration of sebaceous glands (SGs). Although SGs are largely self-maintained by dedicated stem cells during homeostasis, alternative stem cells enter and regenerate the gland following injury. This regenerative process relies on FGF signaling and can be accelerated by stimulating hair growth.
INTRODUCTION
Our skin is coated with a complex mixture of oils that serves critical roles in modulating water retention, body temperature, and the microbiome. These oily secretions, known as sebum, originate from sebaceous glands (SGs) and constitute up to 90% of the total surface lipids in the skin.1,2 Over-production of sebum by SGs can lead to “oily skin,” whereas hyposecretion of sebum is often associated with dry skin and eczematous dermatoses.3,4 Since perturbations in sebum are notably linked to common cutaneous disorders such as acne, seborrheic dermatitis, and enlarged facial pores, SGs must be exquisitely regulated in order to maintain healthy skin function and cosmetic appearance.5,6
SGs are epithelial appendages typically associated with hair follicles. These acinar structures are composed of terminally differentiated sebocytes ensheathed by a peripheral layer of undifferentiated basal progenitor cells.7,8 During maturation, sebocytes enlarge, accumulate lipids, and degrade their organelles in a specialized form of cell death known as holocrine secretion.9 This process culminates with sebocytes releasing their lipid contents through the sebaceous duct into the hair follicle infundibulum, which provides a passageway for sebum to exit the follicle and enter the skin surface.10
Since SGs are hormonally regulated, their activity varies at different stages of life.11,12 Nonetheless, the constant turnover of sebocytes necessitates that these cells be continually replenished, a process that typically takes 1–2 weeks in mice and 2–4 weeks in humans.13-16 This renewal process is made possible by stem cells, although the niche in which these cells reside has not been decisively established. Although some studies have noted that hair follicle stem cells can enter and renew the gland,17-19 other reports have indicated that SGs harbor their own dedicated stem cell pools.20-23 In addition, it remains controversial whether all basal progenitors that line the SG periphery contribute equally to sebocyte formation. Finally, whether these cellular processes become altered after injury has not been explored.
Technical challenges have posed a major hindrance to answering these questions. Because SGs are lobular structures that exhibit cellular heterogeneity along multiple axes—including proximal-distal, as well as proximity to the sebaceous duct—the spatial and molecular relationships of sebocytes at different stages of differentiation have been difficult to resolve. In addition, the lack of Cre drivers that specifically and efficiently target SGs complicates genetic fate mapping studies. Indeed, current tools for performing lineage tracing on SGs rely on mouse Cre lines that also target the hair follicle.18,22,24,25 Sebocytes are also challenging to isolate due to their complex cellular properties. Consequently, these cells typically constitute a minor sub-population in single-cell RNA sequencing (scRNA-seq) studies of skin, precluding the ability to perform deeper analyses.26-28 Finally, studies on SG function have historically relied on mouse mutants such as Asebia, which possesses impaired SGs due to a germline mutation in stearoyl-Coenzyme A desaturase-1 (Scd1).29,30
Recent reports have suggested that SGs are adaptable structures that respond to local and systemic cues, are affected by the hair cycle and immune factors, and appear to be lost in diseases such as cicatricial alopecia and psoriasis.31-37 Here, we overcome many of the technical challenges for studying SGs and perform highly targeted genetic fate mapping, scRNA-seq, and ablation studies to explore how distinct stem cell populations maintain the gland and confer resiliency in response to injury.
RESULTS
Establishing SG landmarks
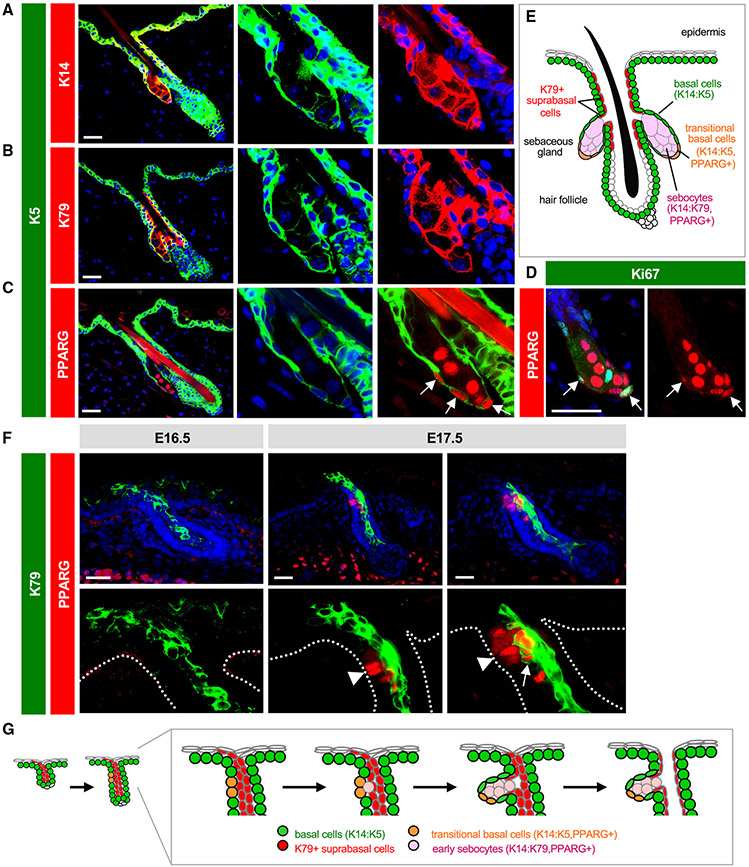
Keratins are by far the most abundant proteins in the skin, and the expression patterns of the 54 keratin family members subdivide keratinocytes by niche, function, and differentiation status.38 We previously reported that basal progenitors at the SG periphery express keratins (K) 5 and K14, which form prototypic heterodimers in multiple mouse skin stem cell compartments (Figure 1A).20 In differentiated sebocytes, however, K14 levels remain high, whereas expression of its typical binding partner K5 is reduced (Figure 1A). In its place, a different keratin, K79, becomes elevated in sebocytes and heterodimerizes with K14 (Figure 1B).20 Therefore, SG progenitors undergo a K14:K5 → K14:K79 keratin switch when they become sebocytes. Whether other keratins display similar shifts in the SG remains unclear and will be examined below.
Figure 1. Establishing SG landmarks.
(A) Co-localization of K5 (green) with K14 (red) in peripheral SG basal cells, but not in sebocytes. Middle and right panels are magnified single-channel views.
(B) Lack of co-localization of K5 with K79 (red) in sebocytes.
(C) Co-localization of K5 with PPARγ (red) in transitional basal cells of the lower SG (arrows).
(D) Co-localization of PPARγ (red) with Ki67 (green) in a subset of peripheral basal cells (arrows) in the SG.
(E) Schematic of telogen hair follicle. Note that the infundibulum and sebaceous ducts are continuously lined by differentiated K79+ cells (red).
(F) Localization of K79 (green) and PPARγ (red) in the developing hair follicle during embryonic (E) days 16.5–17.5. Middle panels, follicle with basal PPARγ+ cells (arrowhead), but minimal co-localization with K79. Right panels, follicle with early sebocytes identified by the unique co-localization of PPARγ and K79 (arrow). Dotted lines delineate the basal layer of the epidermis and hair follicle. Bottom panels are magnified views, with DAPI omitted for clarity.
(G) Schematic of SG specification. PPARγ+ basal cells (orange) initially emerge at E16.5–17.5 and give rise to early sebocytes (pink) adjacent to the K79+ cell column (red). Subsequent remodeling leads to the opening of the sebaceous duct and hair canal.39 One of two SG lobes is depicted. The second lobe may be specified later or may arise when the initial SG compartment splits into two, as has been proposed.40 Scale bar, 50 μm.
Notably, we also observed that peroxisome proliferator-activated receptor gamma (PPARγ), a master regulator of lipid metabolism and SG differentiation,41,42 is initially expressed by K5+ basal SG progenitors located at the lower (proximal) region of the gland (Figure 1C). A subset of basal PPARγ+ cells are also proliferative (Figure 1D). Because these cells express a unique combination of both basal progenitor (K5) and differentiation (PPARγ) markers, this suggests that these K5+PPARγ+ cells may behave as transitional basal cells poised to differentiate into sebocytes, a concept we revisit later (Figure 1E).
To determine whether these expression patterns are recapitulated during initial SG development, we examined mouse embryonic skin after hair follicle initiation but before SG formation. We have previously shown that nascent hair buds generate and extend columns of K79+ differentiated cells out into the epidermis, which subsequently undergo remodeling to form hair follicle openings.39,43 In embryonic (E) day 16.5 skin, we observed K79+ columns in developing hair buds, as expected, but no PPARγ expression (Figure 1F). By E17.5, however, we noticed basal PPARγ expression, reminiscent of the K5+PPARγ+ transitional basal cells seen in adult follicles (Figure 1F). Furthermore, we observed early sebocytes, identified by the unique co-expression of K79 and PPARγ, located immediately adjacent to the basal layer and alongside K79+ columns (Figure 1F). These findings are consistent with previous studies indicating that basal cells undergo asymmetric cell divisions to form sebocytes,40,44 and suggest a model for how the SG compartment becomes connected to the developing sebaceous duct and future hair follicle infundibulum, domains that are unified by their shared expression of K79 (Figure 1G). In total, these observations establish a set of landmarks for evaluating SGs.
SG dynamics during skin homeostasis and injury
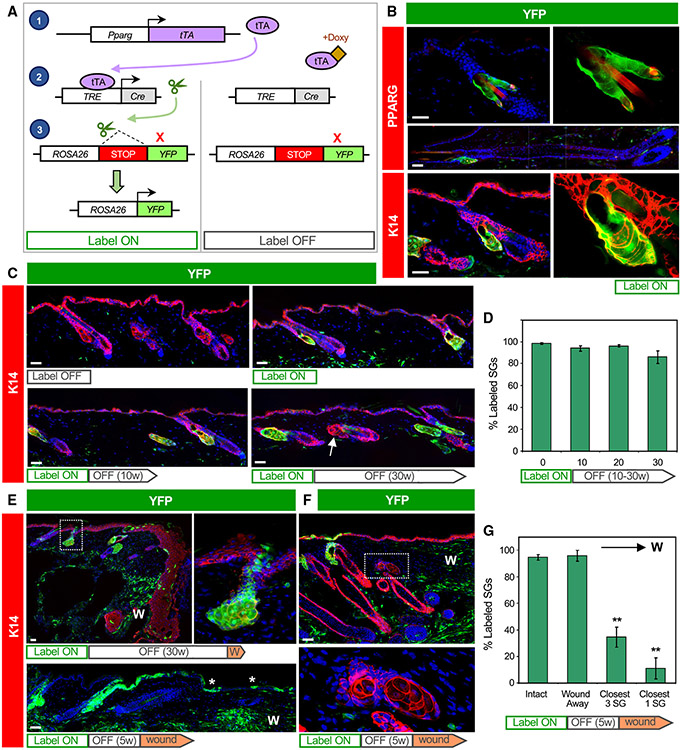
Given our observation that PPARγ is initially expressed in the SG basal layer, we next attempted to trace the fate of Pparg-expressing cells and their progeny. For this, we acquired AdipoTrak mice, in which a tetracycline-regulated transactivator (tTA) is expressed under the control of the endogenous Pparg promoter (Figure 2A).45 When coupled with a tetracycline-responsive element (TRE)-driven Cre recombinase and a Cre-inducible YFP reporter allele (PPARγ;YFP mice), these genetic elements enable PPARγ+ cells and their descendants to become permanently labeled. However, in the presence of doxycycline (doxy), tTA cannot activate Cre expression, providing temporal control over this system.
Figure 2. Tracing the SG during homeostasis and after wounding.
(A) Schematic for tracing PPARγ+ cells. Left, in the absence of doxycycline (doxy), Pparg promoter-driven tTA induces Cre expression, causing genomic recombination that activates YFP expression. Right, doxy suppresses tTA activity.
(B) Immunohistochemical localization of YFP (green) and PPARγ (red, top) or K14 (red, bottom) in label-on PPARγ;YFP mice. Basal SG cells, sebocytes, and sebaceous ducts express YFP, but other hair follicle epithelia do not, in either telogen (top, bottom) or anagen (middle). Right panels are magnified views of the left panels, with DAPI omitted.
C) Top panels, 8-week-old skin from label-off (left) or label-on (right) PPARγ;YFP mice. Bottom panels, skin from mice treated for the first time with doxy starting at 8 weeks of age, for 10–30 continuous weeks (label-on → label-off). Arrow, unlabeled SG.
(D) Quantitation of labeled SGs, following 0–30 weeks of continuous doxy treatment.
(E) Wounded skin from a label-on → label-off PPARγ;YFP mouse, examined 1 week (top) or 8 weeks (bottom) after injury. Top right panel is a magnified view of the boxed area showing labeled cells that have departed the SG and entered the epidermis. Asterisk, SG-derived YFP+ cells maintained long-term in the healed epithelium. K14 staining was omitted from the bottom panel for clarity.
(F) Wounded skin from a label-on → label-off PPARγ;YFP mouse, examined 3 weeks after injury. Bottom panel is a magnified view of the boxed area showing unlabeled, wound-proximal SGs.
(G) Quantitation of SG labeling as a function of distance from the wound site. The closest SG cluster to the wound site is designated “closest 1,” and the closest 3 SG clusters are designated “closest 3.” W, wound site. w, weeks. **p < 0.01 by one-way ANOVA and Tukey post hoc test, comparing closest 3 or closest 1 with “intact” or “wound away.” n ≥ 4 mice per time point for (D). Four mice were wounded for (G). Data are represented as mean ± SEM. Scale bar, 50 μm.
We began by analyzing 8-week-old PPARγ;YFP mice without doxy exposure (label on), and observed that >98% of all SGs were completely labeled (Figure 2B). These labeled cells included SG basal layer cells, sebocytes, and differentiated cells of the sebaceous duct but did not include the interfollicular epidermis (IFE), isthmus, or other hair follicle compartments. We also did not detect any additional epithelial cell labeling in anagen hair follicles, demonstrating the exquisite specificity for SG labeling in this system (Figure 2B).
To track the long-term fate of labeled cells in the SG, we next treated 8-week-old mice with doxy-containing chow to suppress any additional new labeling (label on → off). After 30 weeks of continuous doxy treatment, we observed that ~86% of SGs were still completely labeled (Figures 2C and 2D). To verify that induction of YFP labeling is indeed suppressed by doxy-chow, we examined adult mice that were continuously treated with doxy since gestation (label off) and observed no SG labeling, as expected (Figure 2C). These findings suggest that, under homeostatic conditions, SGs are largely self-maintained by their own dedicated stem cell pools but may receive occasional cellular input from the hair follicle.
Following skin injury, stem cells from the IFE and hair follicle migrate into the wound to promote re-epithelialization.24,46-49 To determine whether SG-derived cells exhibit similar behavior, we generated mice with labeled SGs, treated them with doxy to suppress any additional labeling (label on → off), and subsequently performed excisional wounding. One week after injury, we observed labeled cells that had moved directly out of the SG and into the migratory epithelial front (Figure 2E). These SG-derived cells contributed long term to the regenerated epidermis, since labeled cell clones were still observed at least 8 weeks after wounding (Figure 2E). Notably, after skin healing, we observed that only ~10% of SGs located closest to the wound remained labeled, whereas nearly all SGs situated away from the wound were YFP+ (Figures 2F and 2G). This suggests that injury can spur a dramatic reorganization of the SG, where resident SG progenitors depart their niche and are replaced by unlabeled hair follicle-derived stem cells. The absence of labeling in wound-proximal SGs also provides technical reassurance that de novo labeling of SGs is properly suppressed by doxy treatment.
Sebocyte isolation
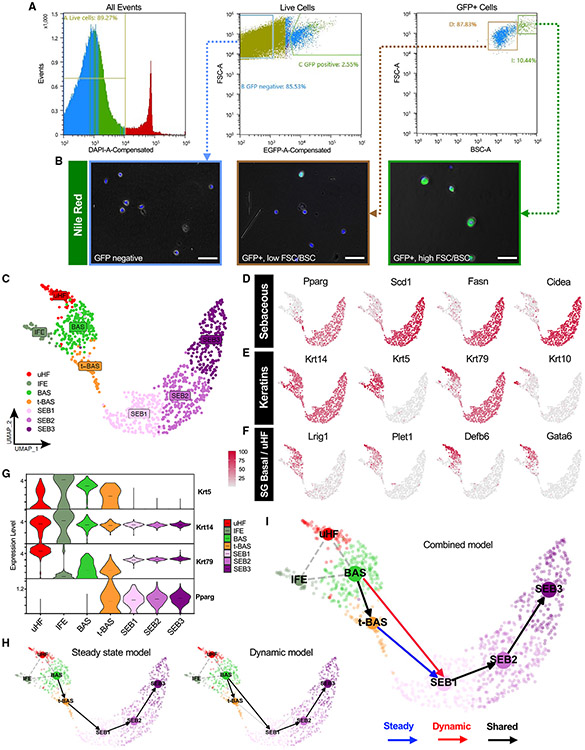
Having characterized the cell dynamics of SGs during homeostasis and injury, we next sought to investigate the molecular changes that occur during sebocyte differentiation. While previous scRNA-seq studies on mouse and human skin have included SG sub-populations, these cells are poorly represented due to challenges associated with isolating large, complex, lipid-filled sebocytes. Since PPARγ;YFP mice exhibit specific labeling of SGs, we analyzed skin epithelial cell suspensions by flow cytometry and found that YFP+ cells typically comprise 2%–4% of live cells recovered from 8-week-old label-on mice (Figure 3A). By further fractionating YFP+ cells by size and complexity (measured by forward scatter [FSC] and back scatter [BSC]), followed by staining plated cells with the lipophilic dye Nile red, we observed that the vast majority of Nile red+ sebocytes are found within the highest ~10% FSC/BSC sub-population (Figures 3A and 3B). In contrast, FSC/BSC-low YFP+ cells were only occasionally stained by Nile red and likely comprise a mix of smaller SG basal progenitors, early sebocytes, and sebaceous duct cells (Figure 3B). Overall, this approach enabled us to significantly enrich for SG cells and especially sebocytes, which accounted for <1% of all cells in our original suspension.
Figure 3. Isolating and profiling SG cells.
(A) Flow cytometry plots of isolated cell suspensions from 8-week-old PPARγ;YFP label-on skin.
(B) Nile red staining (green) of sorted keratinocyte sub-populations: bulk GFP negative (left), GFP+ with low FSC/BSC (middle), and GFP+ with high FSC/BSC (right). Note that GFP epifluorescence is not visible and does not interfere with bright Nile red staining, which was superimposed upon bright-field images.
(C) UMAP projection showing seven cell clusters isolated from YFP-sorted, 8-week-old PPARγ;YFP label-on skin.
(D) Feature plots for canonical SG genes.
(E) Feature plots for key keratin genes.
(F) Feature plots for markers of SG basal cells, sebaceous duct, isthmus, and infundibulum.
(G) Violin plots showing relative expression of key marker genes across different cell sub-populations. Note that t-BAS cells uniquely express both Krt5 and Pparg. Horizontal lines indicate median values.
(H) RNA-velocity trajectory analysis performed using scVelo with either a steady-state (left) or dynamic (right) model.
(I) Trajectory analysis incorporating results from both steady-state and dynamic models, suggesting that BAS cells enter the transitional t-BAS state before differentiating into SEB-1 sebocytes (blue arrow) or can differentiate directly into SEB-1 sebocytes (red arrow). Black arrows, lineage relationships identified by both models. Gray dotted lines indicate statistical connectivity between clusters. Trajectories predicted by scVelo originating from the IFE were removed for clarity. Scale bar, 50 μm. See also Figures S1 and S2, Data S1 and S2.
Characterizing initial sebocyte differentiation
After devising a strategy to isolate SG cells, we performed targeted scRNA-seq on YFP+ cells sorted from 8-week-old skin and visualized these data in two-dimensional space by Uniform Manifold Approximation and Projection (UMAP) using Seurat. We identified seven cell clusters, including three sebocyte clusters (SEB1–3) that exhibit expression of established SG biomarkers (Pparg, Scd1, Fasn, Cidea) (Figures 3C and 3D). We also identified one cluster representing SG basal cells (BAS) that expresses high level Krt5, Krt14, and Lrig1, which encode markers of SG stem cells24 (Figures 3C, 3E, and 3F). Flanking the BAS cluster, one minor cluster likely comprises mixed upper hair follicle (uHF) cells of the infundibulum and sebaceous duct, as assessed by markers Krt79, Krt17, Krt10, Cst6, Plet1, Defb6, and Gata6, cataloged previously by us and others (Figures 3C, 3E, 3F, S1A, and S1B).43,50-53 A second minor cluster consists of blended Krt5+ basal and Krt1+ suprabasal cells of the IFE, likely originating from SG cells that had departed their niche following mild skin agitation such as scratching (Figures 3C and S1A).
Notably, a final cell cluster extended out from the BAS cluster toward the SEB sub-populations. Cells in this cluster uniquely express a combination of basal markers (Krt14, Krt5) as well as sebocyte differentiation markers (Pparg), strongly suggesting that these are the K5+PPARγ+ transitional basal cells (t-BAS) identified above (Figures 1C, 3C, and 3G). Also consistent with our above findings, we observed that downstream of the t-BAS state, all differentiated SEB clusters express Krt14 and Krt79— but not Krt5—reinforcing the notion that SG progenitors undergo a K14:K5 → K14:K79 keratin shift during sebocyte differentiation (Figure 3G). Indeed, aside from Krt14 and Krt79, no other keratins were expressed at appreciable levels in the 3 SEB clusters (Figure S1A), consistent with our previous observation that K79 serves a non-redundant structural role in the SG.20
To infer cell-state transitions between clusters, we next performed RNA-velocity analysis using scVelo and visualized trajectories using partition-based graph abstraction (PAGA). A steady-state model of transcriptional dynamics predicted a trajectory whereby BAS cells pass through the t-BAS transitional state to become SEB1 cells (Figure 3H, left). However, a dynamic model also predicted that a subset of BAS cells can bypass the t-BAS state to directly become SEB1 cells (Figure 3H, right). Overall, our findings suggest that, during homeostasis, resident SG basal progenitors can take either an indirect or direct path to differentiate into SEB1 sebocytes (Figure 3I).
Characterizing sebocyte cell states
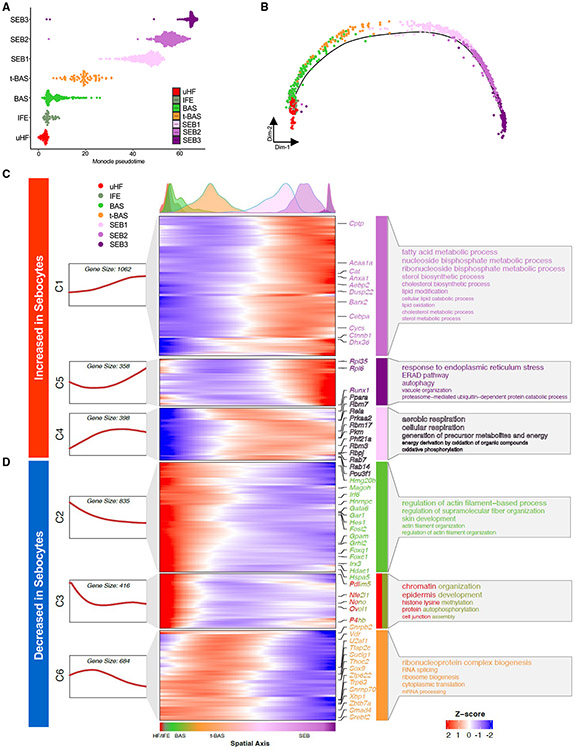
Once specified, sebocytes accumulate lipids and undergo a specialized degradative process to release sebum. To better understand the cell-state transitions that occur during sebocyte maturation, we visualized the pseudotemporal dynamics of SG cells using Monocle 2, which ordered the cells in a linear trajectory without significant branching. Although the minor uHF and IFE cell states were inter-mixed by this analysis, a single trajectory pointed from BAS to t-BAS, and then sequentially through SEB-1, -2, and -3 terminal states, consistent with results by RNA-velocity analysis (Figures 4A and 4B).
Figure 4. Pseudotemporal dynamics of gene expression during sebocyte differentiation.
(A and B) Pseudotemporal ordering of seven cell sub-populations isolated from YFP-sorted, PPARγ;YFP label-on skin using Monocle 2.
(C and D) Rolling-wave plot and smoothed expression pattern of pseudotime-dependent genes (n = 3,753) that cluster into six gene modules (C1–C6). Peak positions of the cell populations were visualized by kernel density estimation (top), along the pseudospatial axis (bottom). Also shown are the corresponding expression curve (left) and representative enriched GO terms (right) for each gene module, with larger font size corresponding to increased statistical significance. Transcription factors from each module are indicated. See also Data S3 and S4.
We next identified pseudotime-dependent differentially expressed genes (DEGs) and performed Gene Ontology (GO) analysis to identify cellular processes that become altered during sebocyte maturation. Across the pseudotime trajectory, 3,753 DEGs were identified and grouped by K-medoid clustering into six gene modules with distinct expression patterns and biological functions. Notably, three modules (C1, C5, C4) of gene expression changes were increased in sebocytes relative to the other cell populations. These modules included genes associated with lipid metabolism, endoplasmic reticulum (ER) stress response, autophagy, and aerobic respiration (Figure 4C). By contrast, three modules (C2, C3, C6) were decreased in sebocytes and were associated with cell functions such as mRNA processing, translation, chromatin organization, and cytoskeletal processes (Figure 4D). Finally, expression of androgen receptor and androgen response genes was increased during sebocyte differentiation (Figures S1B and S1C).8,37,54,55 Taken together, these changes indicate that sebocytes comprise a terminally differentiating, hormone-responsive cell lineage characterized by the shutdown of core cellular processes, the degradation of key structural components, and finally autophagic cell death.
Spatially mapping sebocyte cell states
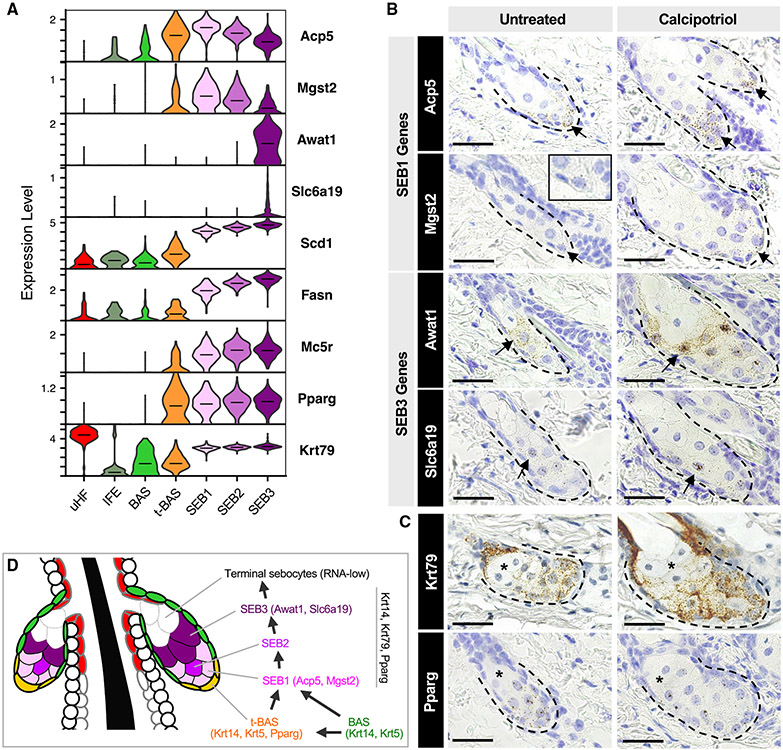
Our RNA-velocity and pseudotime analyses both suggest that sebocytes undertake a unidirectional SEB-1 → SEB-2 → SEB-3 trajectory. This path is further supported by a stepwise elevation in expression of canonical SG markers, such as Scd1, Fasn, and Mc5r (Figure 5A). To spatially resolve the three SEB clusters in the SG, we identified DEGs that define each cell state and found that SEB-1 cells are enriched for Acp5 and Mgst2 expression (Figures 5A and S2). Although the SEB-2 cluster appears to represent an intermediate state with no unique markers, SEB-3 cells display increased Awat1 and Slc6a19 mRNA (Figures 5A and S2).
Figure 5. Spatial mapping of different sebocyte cell states.
(A) Violin plots showing relative expression of key marker genes in the SG. Horizontal lines indicate median values.
(B) RNAscope in situ staining for genes enriched in SEB-1 (Acp5, Mgst2), and genes enriched in SEB-3 (Awat1, Slc6a19). Arrow, region where gene is highly expressed. Inset, magnified view of Mgst2 staining.
(C) RNAscope staining for Krt79 and Pparg in the SG. Asterisk, RNA-low terminal sebocytes. Left column, untreated wild-type skin. Right column, calcipotriol-treated skin.
(D) Schematic summarizing both direct and indirect paths for differentiation of SG basal cells into sebocytes. Scale bar, 50 μm.
By RNAscope in situ staining, we next confirmed that expression of Acp5 and Mgst2 (SEB-1) is predominantly localized to the lower SG (Figure 5B). On the other hand, expression of Awat1 and Slc6a19 (SEB-3) is enriched in sebocytes occupying a more central position in the gland (Figure 5B). For all four genes, we further observed that their expression patterns are recapitulated in skin treated with calcipotriol (MC903), a vitamin D analog that causes SG enlargement, facilitating the visualization of lower-abundance transcripts (Mgst2, Slc6a19) (Figure 5B).
Finally, we observed an additional sebocyte population that is rarely stained by any RNAscope probes, including probes targeted against pan-sebocyte markers such as Pparg and Krt79 (Figures 5A and 5C). These sebocytes, located at the distal end of the gland, closest to the sebaceous duct, comprise roughly 20%–50% of the total SG volume, and likely represent the most terminal cell state downstream of SEB-3. Because these terminal sebocytes are RNA-low, they are likely not represented in our scRNA-seq dataset. Overall, our findings suggest that resident basal progenitors differentiate into SEB1 sebocytes primarily in the lower SG and that these sebocytes transition unidirectionally along multiple cell states as they move toward the sebaceous duct, as summarized in Figure 5D.
SGs regenerate following genetic ablation
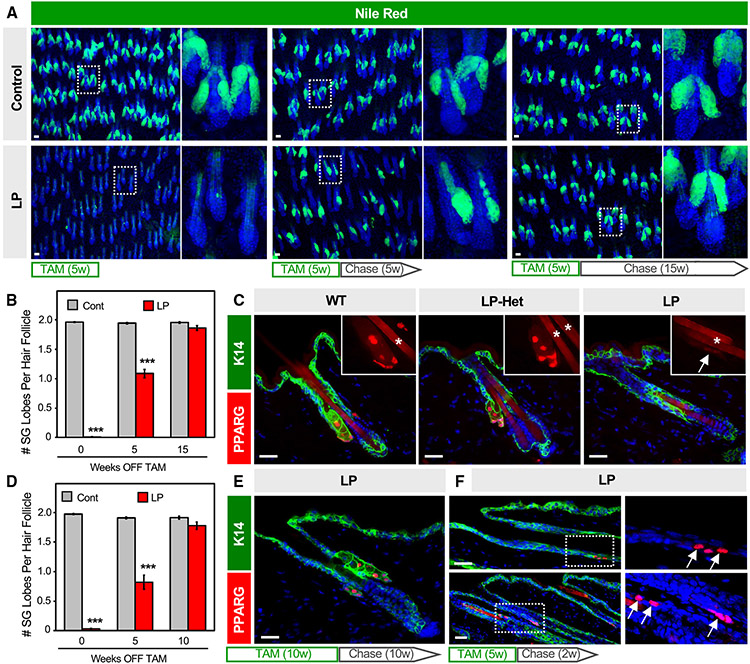
Given that Pparg is expressed in both the t-BAS and SEB1-3 cell states (Figure 5A), we next tested its requirement for SG homeostasis in adult skin. We therefore generated mice expressing tamoxifen-inducible Lrig1-CreERT2 coupled with homozygous conditional alleles for Pparg (LP mice), which enables targeted deletion of Pparg in SG stem cells.56,57 When 8-week-old LP mice were treated with tamoxifen (TAM)-containing chow for five continuous weeks, 99% of SGs were ablated from dorsal skin, confirming the absolute requirement for PPARγ in SG maintenance (Figure 6A).58 Surprisingly, however, when these LP mice were subsequently removed from TAM treatment (“chase”), roughly half of all SGs reappeared within 5 weeks, with full recovery seen after 15 weeks’ chase (Figures 6A and 6B). Regenerated SGs expressed PPARγ, indicating that they were derived from cells that had not undergone Cre-mediated recombination (Figure S3A). Since SG regeneration has not been previously documented, these observations propelled our studies in an unexpected direction.
Figure 6. SGs regenerate following genetic ablation.
(A) Nile red staining (green) of skin whole mounts from control or LP mice treated with tamoxifen (TAM)-containing chow for five continuous weeks, then moved onto normal chow (“chase”) for an additional 0 (left), 5 (middle), or 15 (right) weeks. Right panels are magnified views of the boxed areas.
(B) Quantitation for (A).
(C) Localization of PPARγ (red) in wild-type (left), Lrig1-CreERT2;Pparg-fiox/+ (LP-Het, middle) or LP mice (right) following 5 weeks of TAM-chow. Insets, magnified views of PPARγ staining. Arrow, faint PPARγ staining at the hair follicle isthmus in LP skin. Asterisk, hair shaft autofluorescence.
(D) Quantitation of SGs similar to (B) but for mice treated with 10 continuous weeks of TAM-chow, followed by 0–10 weeks’ chase.
(E) Regenerated SGs express PPARγ (red).
(F) Expression of PPARγ (red, arrows) in basal K14+ cells (green) of the upper anagen ORS (top panels) and isthmus (bottom panels), after 5 weeks of TAM-chow and 2 weeks’ chase. Right panels are magnified single-channel views of the boxed areas. w, weeks. ***p < 0.001 by unpaired t test comparing control (cont) and LP skin from the same time point. n ≥ 7 mice, per genotype, per time point for (B) and (D). Data are represented as mean ± SEM. Scale bar, 50 μm. See also Figure S3.
Cellular mechanisms for SG regeneration
To better understand how SGs regenerate, we checked whether PPARγ is fully ablated from the hair follicle. In LP mice treated with TAM-chow for 5 weeks (no chase), we observed that PPARγ is almost completely abolished from the isthmus/junctional zone, as expected, leaving behind residual “nubs” of K5+PPARγ-negative cells (Figures 6C and S3B). However, we also occasionally observed very faint PPARγ staining, at an intensity level far lower than what is seen in skin when only one copy of Pparg is intentionally deleted (Pparg-flox/+, or LP-Het) (Figure 6C). Thus, trace PPARγ staining in LP follicles is unlikely to be caused by incomplete recombination within the Lrig1+ domain. Rather, this may reflect cells that had newly entered the isthmus and had either recently begun expressing PPARγ or had recently deleted PPARγ. Faint PPARγ staining was seen even in LP mice that were treated with TAM-chow for 10 continuous weeks (not shown), and here again PPARγ+ SGs regenerated with similar kinetics after TAM removal (Figures 6D and 6E).
If non-recombined cells enter the isthmus following SG ablation, where are they coming from? To address this, we examined LP mice at earlier time points after ceasing TAM treatment. Interestingly, in LP mice treated with TAM-chow for 5 weeks, followed by a shorter 2 weeks’ chase, we observed ectopic PPARγ expression in basal cells within the upper outer root sheath of anagen follicles (Figures 6F and S3C). This domain has previously been shown to be derived from bulge cells,59 which we confirmed are not targeted by Lrig1-CreERT2 (Figure S3D). In addition, we observed high-level PPARγ expression reappearing in basal cells at the isthmus, which can also be derived from upper bulge cells over time (Figure 6F).60 Altogether, our findings suggest that non-recombined bulge-derived cells rapidly migrate into the isthmus/junctional zone to regenerate SGs following genetic ablation. In contrast to homeostatic self-renewal, this regenerative process is likely akin to the recruitment of replacement SG progenitors after skin wounding (Figures 2E and 2F).
Modulation of SG regeneration by hair cycle and fibroblast growth factor signaling
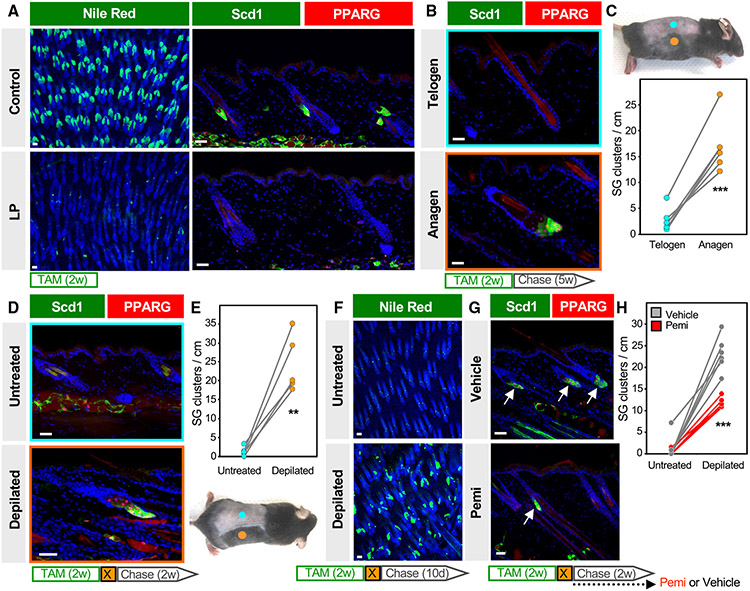
As a final question, we asked what signals instruct progenitor cells to regenerate SGs. For this, we shortened the experimental window and treated 6-week-old LP mice with TAM-chow for 2 weeks (no chase), which caused a 97% reduction in PPARγ+/Scd1+ SGs (Figures 7A and S3E). At this point, hair follicles have uniformly entered the telogen resting phase at 8 weeks of age. Since subsequent re-entry to anagen growth is asynchronous in adult skin, this provided us the opportunity to assess SG regeneration in anagen and telogen skin from the same animal (Figures 7B and 7C). Indeed, we observed that, 5 weeks after TAM withdrawal, anagen skin exhibited a >6-fold increase in SGs compared to adjacent telogen skin from the same animal (Figure 7C).
Figure 7. SG regeneration is modulated by hair cycling and FGFR signaling.
(A) Left, Nile red (green) staining of skin whole mounts from control (top) or LP (bottom) mice treated with TAM-chow for two continuous weeks (no chase). Right, confirmation of SG loss by staining for Scd1 (green) and PPARγ (red).
(B) Scd1/PPARγ staining in telogen (top) or anagen (bottom) skin from the same animal, following 2 weeks of TAM-chow and 5 weeks’ chase.
(C) Top, example of LP mouse treated with TAM-chow for 2 weeks, followed by 5 weeks’ chase. Sites of natural anagen (orange) or telogen (blue) are denoted. Bottom, SG quantitation for (B). Paired samples are connected by lines.
(D) Identification of regenerated SGs by Scd1/PPARγ staining in mice treated with TAM-chow for two continuous weeks, then depilated (X) and chased for two additional weeks.
(E) Bottom, example of LP mouse used in (D). Sites of depilation (orange) or no treatment (blue) are denoted. Top, quantitation of SG abundance for (D). Paired samples are connected by lines.
(F). Nile red (green) staining of whole mounts from untreated (top) or depilated (bottom) LP skin, where mice were treated with TAM-chow for 2 weeks, depilated, and chased for 10 days.
(G) Identification of regenerated SGs by Scd1/PPARγ staining (arrows), with similar treatment protocol as in (D) but with additional daily treatment with FGFR inhibitor (pemi) or vehicle during the 2-week chase period.
(H) Quantitation for (G) in LP mice treated with vehicle (gray) or pemi (red). Samples from the same mouse are connected by lines. w, weeks; d, days. **p < 0.01, ***p < 0.001. Paired t test for (C) and (E); unpaired t test comparing only depilated samples for (H). n = 6 mice for (C), n = 5 mice for (E), and n = 11 mice for (H). Scale bar, 50 μm. See also Figures S4 and S5.
To better explore the connection between hair growth and SG regeneration, we next depilated 8-week-old LP mice immediately after completing a 2-week course of TAM treatment. Depilation-induced anagen skin similarly exhibited a >10-fold increase in PPARγ+/Scd1+ SGs compared to non-depilated skin from the same animal (Figures 7D and 7E). These effects were quantitated 2 weeks after depilation/TAM removal, but differences in SG regeneration were apparent even after just 10 days, when most follicles in depilated skin were in early anagen (Figure 7F).
Lastly, to identify factors that modulate SG regeneration, we turned back to our scRNA-seq data and found that Fgfbp3, which encodes a potentiator of fibroblast growth factor (FGF) signaling,61 is among only a handful of genes for secreted factors whose expression is enriched in the SEB lineage (Figure S4A). Furthermore, we noted that, among the four major FGF receptors (FGFRs) in mice, only Fgfr2 is expressed in the SG (Figures S4A and S4B), consistent with enriched FGFR2 localization seen in SG basal and transitional basal layer cells (Figure S4C). We therefore treated LP mice with the FGFR2 inhibitor pemigatinib (pemi)62 and examined SG regeneration after depilation. Although FGFR2 inhibition did not prevent hair follicles from reentering anagen (Figure S4D), significantly fewer SGs regenerated in pemi-treated mice compared to vehicle-treated controls (Figures 7G, 7H, and S4E). This effect was associated with fewer phosphorylated-p44/42 (pErk1/2) mature sebocytes in pemi-treated mice, while overall levels of FGFR2 appeared unchanged (Figures S4C, S4F, and S4G). Altogether, these findings identify a robust and previously unrecognized process for regenerating SGs that can be modulated by hair growth and FGFR2 signaling.
DISCUSSION
Numerous technical challenges have long hindered the study of SGs. In particular, Cre-mediated approaches for manipulating these appendages typically drive genetic recombination in multiple skin compartments, complicating the interpretation of results. Although mice expressing a sebocyte-specific, Scd3 promoter-driven Cre have been reported, this system likely does not cause recombination in SG basal layer cells, and recombination efficiency in sebocytes remains unclear.63 Another long-standing challenge has been the inability to purify SG cells for molecular profiling. Indeed, we observed that sebocytes constituted <1% of all cells prior to enrichment, consistent with their relative paucity in published scRNA-seq atlases of mouse and human skin.26-28
By overcoming multiple technical hurdles, our study paints a vibrant portrait of the cellular and molecular architecture of SGs during development, homeostasis, wounding, and regeneration. Several themes have emerged. First, SGs are largely self-renewed by resident stem cell pools during homeostasis, although cells originating from outside the gland can also occasionally contribute. Second, when the SG stem cell niche is perturbed, either by wounding or genetic ablation, alternative stem cells rapidly enter the SG domain to repopulate the gland. These findings are consistent with the view that stem cells within discrete hair follicle niches serve largely compartmentalized roles during homeostasis but become highly plastic following injury.19,48,64
A third theme is that, while PPARγ is essential for sebocyte differentiation, this transcription factor is initially expressed in SG basal cells. This is seen during development, homeostasis, and regeneration. We should emphasize that these t-BAS transitional basal cells—which represent the earliest cells in the SG to express Pparg but also the latest cells to express high-level Krt5 (Figures 1C and 3G)—are unlikely to be SG stem cells in adult skin. Similar to transitional basal cells in the IFE that express differentiation markers such as K10, these K5+PPARγ+ cells likely possess limited replication potential and are poised to differentiate.65-68 While we cannot formally rule out the possibility that t-BAS cells can revert back to PPARγ-negative basal (BAS) cells, which are likely the stem cells that maintain the SG during homeostasis, such a path is not supported by our scRNA-seq trajectory analysis (Figure 3I).
If expression of PPARγ indeed predisposes basal cells to differentiate into sebocytes, this raises the question of how the entire SG, including PPARγ-negative basal cells, becomes labeled in adult PPARγ;YFP label-on mice. Unfortunately, examining newborn skin provided little clarity, as early labeling can be seen in both PPARγ+ and PPARγ-negative cells dispersed around the developing upper follicle prior to formation of the mature SG (Figure S5A). Additional studies are needed to clarify how these patterns resolve over time to achieve specific labeling of the entire adult SG. Related to this, we were also unable to acutely switch on labeling of PPARγ+ cells in adult mice that were maintained on doxy-containing chow and subsequently moved onto normal chow (label off → on) (Figure S5B). The reason for this remains unclear; nonetheless, this technical limitation prevented us from tracing the fate of adult PPARγ+ cells.
Previous studies using multi-color lineage tracing have reported that basal cells located along the entire SG periphery can give rise to differentiated sebocytes.21 While our trajectory analyses suggest that BAS progenitors can directly form sebocytes without transitioning through the t-BAS intermediate state, both the direct and indirect paths for sebocyte formation invariably funnel through the SEB-1 state, before moving unidirectionally along progressively more differentiated SEB-2 and SEB-3 lineages. A final, terminal cell state—defined not by scRNA-seq but instead by low-level RNA in situ staining-juxtaposes the sebaceous duct (Figures 5B and 5C). Since SEB-1 sebocytes are located near the proximal end of the SG, this implies that new sebocyte formation also primarily occurs within the lower SG. Why our observations differ from those of previous reports remains unclear, but it may have to do with the complex geometry of the gland, as well as differences in experimental timing.
Unexpectedly, we observed that SGs regenerate following genetic ablation of PPARγ and that non-recombined, bulge-derived cells likely give rise to regenerated glands. Although we detected ectopic PPARγ expression in the upper outer root sheath (ORS) of mutant follicles (Figure 6F), SGs reappeared at the original sites from where they were lost. These findings demonstrate that bulge-derived cells—which can either move upward after wounding or downward during hair growth—have the potential to express PPARγ upon departing their niche. At the same time, the factors that specify the exact site of SG development and regeneration remain elusive. Some of these factors likely involve gradients of Wnt and Hedgehog signaling, as well as AP-1 transcription factor activity, since perturbation of any of these components can drive ectopic SG formation.18,69-71 These gradients may potentially specify both permissive sites for SG formation, as well as non-permissive zones, such as the ORS, which does not form SGs in spite of ectopic PPARγ expression in LP mutants.
Our hair cycle studies also revealed that anagen hair growth, a process associated with increased cell proliferation and movement, greatly accelerates SG regeneration.19 In contrast, SGs hardly regenerate in telogen skin, indicating that follicles do not automatically regenerate SGs by default. Rather, microenvironmental factors in the skin are likely also critical. At least one of these factors may be Fgfbp3, which binds and liberates FGFs from the extracellular matrix to activate FGFRs.61 Although Fgfbp3 null mice do not possess obvious skin defects,72 mutant mice lacking FGFR2 have smaller SGs in tail skin.73 Concordantly, acute genetic deletion of Fgfr2 causes atrophy of eyelid meibomian glands, which are highly related to SGs, and these glands can also partially recover over time.74,75 Other glandular epithelia, such as mammary and prostate glands, can similarly regenerate after experimental injury in a manner that recapitulates embryonic development.76
While SG regeneration has not been previously reported, SG loss or hypoplasia has been associated with several skin pathologies, including cicatricial alopecia, psoriasis, and atopic dermatitis.4,33,34,36 Chemotherapy can also induce SG atrophy,77 while lymphocytic attack of SGs has been observed in a mouse model of acute graft-versus-host disease.78 Isotretinoin, which is used to treat severe acne, reduces SG size by up to 90%.79,80 Even in normal skin, SG size and activity increase and diminish at different stages throughout life.11,81 Whether SGs undergo regeneration in these varied contexts remains unclear but is conceivable in light of our findings. In summary, our work identifies distinct mechanisms for SG maintenance and regeneration, which may ultimately enable these appendages to be preserved following challenges to the skin.
Limitations of the study
Because PPARγ+ t-BAS cells are unlikely to be stem cells, it remains unclear why the entire SG, including PPARγ-negative BAS cells, are labeled in PPARγ;YFP label-on mice. In addition, the direct and indirect pathways for sebocyte differentiation require further characterization, including identifying molecular mediators that govern these cell fate transitions. Similarly, it will be important to clarify the cellular and molecular mechanisms of FGFR2 signaling during SG regeneration and maintenance, especially regarding the identity and source of FGF ligands, which were not expressed by cells in our scRNA-seq dataset. At this time, we cannot formally exclude the possibility that terminal sebocytes appeared low for RNA by in situ staining due to artifacts of tissue processing. Finally, future scRNA-seq studies of SGs should incorporate larger cell counts to potentially profile changes in the gland during disease or aging.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sunny Wong (sunnyw@umich.edu).
Materials availability
All reagents generated in this study are available from the lead contact.
Data and code availability
Single cell RNA sequencing data generated for this study have been deposited in the GEO database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225252.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
For labeling studies, PPARγ;YFP mice were fed doxycycline-containing chow (1 g/kg, BioServ Inc, F3949) ad libitum to suppress tTA activity, starting at 8 weeks of age, unless otherwise indicated in the text. For SG ablation and regeneration studies, LP mice and Cre-negative littermate controls were fed irradiated TAM-containing chow (400 mg/kg, Envigo TD.130860) starting at either 6 or 8 weeks of age, as indicated in the text. Pemigatinib (INCB054828, SelleckChem) was dissolved in DMSO to a stock concentration of 4 mg/mL, then subsequently diluted in PEG 400/5% dextrose in water (75:25 v/v). Mice were treated daily at a dose of 1 mg/kg body weight by oral gavage for 14 consecutive days after depilation during the chase period. To assess Lrig1-CreERT2-mediated recombination, Lrig1-CreERT2;ROSA-YFP mice were fed TAM-containing chow starting at 8 weeks of age for 5 continuous weeks. PPARγ;YFP, LP, Cre-negative littermate control and Lrig1-CreERT2;ROSA-YFP mice were of a mixed genetic background, and both genders were analyzed in roughly equal numbers for experiments. Calcipotriol (C4369, Sigma) was dissolved in 100% ethanol and 5.3 nmols were applied topically onto shaved skin for 9 consecutive days at a volume of 200 μL, then harvested 1 day after the final treatment. For calcipotriol, IHC and RNAscope characterization studies, staining was performed on skin from C57BL/6 mice of both genders, 8-10 week of age, unless otherwise indicated in the text.
METHOD DETAILS
Whole mount analysis
Whole mounts of telogen dorsal skin were performed as previously described.20 Briefly, skin was shaved, excised, stretched on a paper towel, covered with Elmer’s No-Wrinkle rubber cement and overlayed with cellophane tape. Following incubation for 6 hours in 5 mM EDTA/PBS at 37°C, the epidermis was separated from the dermis and fixed in formalin for 30 minutes at room temperature. Finally, the samples were incubated with Nile Red (4 μg/ml) and DAPI (1 μg/ml) for 30 min in PBS with gentle agitation at room temperature, then mounted with Vectashield on a microscope slide and imaged.
Flow cytometry
Label-on PPARγ;YFP mice were euthanized at 8 weeks of age, and dorsal skin was shaved and removed. The epidermis was separated from the dermis and cell suspensions were obtained by overnight trypsinization (0.25% trypsin, Invitrogen) at 4°C, as previously described.43 Single cells were resuspended in 2% BSA/HBSS, stained with DAPI to exclude dead cells, and sorted using a SH800 cell sorter (Sony). For scRNA-seq, 60,000 YFP+ cells from an 8 week-old PPARγ;YFP label-on male mouse were sorted into 300 μL of 2% BSA/HBSS buffer, at a ratio of 3:1 FSC/BSC-high:FSC/BSC-low, where “high” cells represented the largest ~10% of cells by FSC/BSC, and “low” cells comprised the remaining 90% by FSC/BSC (Figure 3A). For visualizing sebocytes, cells were sorted into PBS, stained with Nile Red and DAPI without fixation, and imaged.
Single cell library preparation
Single cell suspensions were subjected to counting on the LUNA Fx7 Automated Cell Counter (Logos Biosystems) and diluted to a concentration of 300 cells/μL. Single nuclei 3′ Gene Expression LT libraries were generated using the 10x Genomics Chromium instrument following the manufacturer’s protocol (Chromium Next GEM Single Cell 3′ LT Kit v3.1). In brief, suspensions were loaded onto the 10x chip along with reverse transcription (RT) master mix and appropriate gel beads. Following generation of single-cell gel bead-in-emulsions (GEMs), reverse transcription was performed, and the resulting Post GEM-RT product was cleaned up and the cDNA was amplified. cDNA was subjected to enzymatic fragmentation and size selection to optimize the cDNA size prior to final library construction following the manufacturer’s protocol (10x Genomics). Final library quality was assessed using the LabChip GX (PerkinElmer). Libraries were then subjected to paired-end sequencing according to manufacturer’s protocol (Illumina NovaSeq 6000). Four LT reactions were run in parallel (LT1-4) from the same animal.
Single cell data processing and analysis
Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed fastq files, and the CellRanger Pipeline (10x Genomics) was used to align reads and generate count matrices against the mouse genome GRCm38/mm10. For downstream analysis, the Seurat (v4.3.0) R package83 was used to combine the 4 cell libraries and a merged Seurat object was generated. Genes detected in <3 cells were removed. Low-quality cells were further filtered on the basis of total UMI counts per cell (>900 and <80,000), number of detected genes (>200 and <7,000) and mitochondrial genes fraction (<15%). Applying these filters resulted in a final dataset of 1,066 single cell transcriptomes (Figures S6A and S6B).
To account for batch effects, the merged Seurat object was normalized using the NormalizeData() function with a scale factor of 10,000, and variable features were identified using FindVariableFeatures() with 2,000 genes. Principal component analysis (PCA) was used and the first 30 principal components (PCs) were further summarized using UMAP dimensionality reduction. We chose to use 30 PCs based on results from analyses using Elbow plots. Clustering was conducted using the FindNeighbors() and FindClusters() functions using 30 PCA components and a resolution parameter set to 0.7. A library-split UMAP plot was generated by DimPlot() function to evaluate inter-sample differences. For batch effect detection across different libraries, the distribution of the first principal component (PC1) obtained after PCA analysis was visualized by VlnPlot(). As no obvious batch effect was observed between samples (Figures S6C-S6E), we utilized the processed merged Seurat object for subsequent analysis.
For potential doublet detection, we identified doublets with DoubletFinder (v2.0).82 The doublets were predicted using the cleaned pre-processed merged Seurat data. We did not filter doublets because no discrete doublet-enriched cluster was identified, and only few doublets were observed in the dataset.
Cluster markers were interpreted and assigned using established cell type annotations: Krt5/Krt1(+), Lrig1(−) and Pparg(−) for blended interfollicular epidermis (IFE); Defb6(+), Cst6(+), Krt17(+) and Krt79(+) for mixed upper hair follicle cells (uHF); Krt5(+), Krt14(+), Lrig1(+) and Pparg(−) for SG basal cells (BAS); Krt5(+) and Pparg(+) for transitional basal cells (t-BAS); and Cidea(+), Scd1 (+) and Fasn(+) for differentiated sebocytes (SEB1/2/3). Absence of non-epithelial cell lineages was confirmed by assessing canonical markers, including Pecam1, Cdh5 (endothelial); Pdgfra, Col1a1, Col3a1 (fibroblast); Ptprc, Cd52 (immune); Adipoq (adipocyte); Pmel, Mlana (melanocyte); and others.
To assess the effects of cell cycle heterogeneity on cell clustering, cell cycle phase scores were estimated using Seurat’s CellCycleScoring function with mouse homologs of the cell cycle gene sets provided by Seurat. No obvious clustering differences were found between G2M and S phases within differentiating cells (Figures S7A and S7B). The signals separating non-cycling cells and cycling cells were also checked by combined G2M and S phase gene scoring (cycling cell scoring) and showed high correlation between cycling cell score and corresponding cell states (Figure S7C).
To identify DEGs in each cell cluster, we used the Seurat FindAllMarkers function and the COSG (v0.9.0) R package84 (Figure S2). The COSG-identified top genes were used to establish the cell identity of each cluster, along with markers described in the literature for assigned cell states. Gene signature scores were calculated on the basis of the scRNA-seq data. For each gene signature, individual cells were scored using UCell (v2.2.0) R package89 and projected onto UMAP plots (Figure S7D).
scVelo (v.0.2.5)85 and Monocle 286,87 were used for trajectory analysis. For scVelo, reads that passed quality control after clustering were used as input for the velocyto command line. The mouse expressed repeat annotation file was retrieved from UCSC genome browser. The genome annotation file was provided by CellRanger. The output loom file was used as input to estimate velocity. Velocity embedding was estimated using either the steady-state or likelihood-based dynamical model. PAGA was performed using the sc.tl.paga function in scVelo. For Monocle 2, we built a new CellDataSet object from the cluster-annotated Seurat object using the newCellDataSet function. We used the differentialGeneTest function to derive DEGs from each cluster, and genes with q < 1 × 10−4 were used to order cells in pseudotime. Dimension reduction was performed using the DDRTree algorithm and cells were ordered along the trajectory.
Gene Ontology enrichment analysis was performed using clusterProfiler.88 bitr() was first employed to map gene symbols to Entrez IDs using org.Mm.eg.db (v3.16.0)90 as the reference database, and then the enrichGO function was used with “ont = “BP”, pAdjust-Method = “BH”, pvalueCutoff = 0.01, and qvalueCutoff = 0.05”.
Immunohistochemistry and RNAscope
Frozen sections were probed with antibodies against the following antigens: FGFR2 (1:1000), GFP (1:1000), Ki67 (1:300), K14 (1:1000), K5 (1:1000), K79 (1:400), p44/p42 (1:100), PPARγ (1:300) and Scd1 (1:300). In some cases, fluorescent images were processed using the Auto-Blend feature of Adobe Photoshop CS6 to automatically maximize image sharpness across multiple focal planes. RNAscope 2.5 Brown kit (ACD Bio) was used for RNA in situ staining according to manufacture’s protocol. After deparaffinization, 5 μm sections were boiled for 15 minutes in RNAscope retrieval buffer, treated with protease for 30 minutes and incubated with target probes for 2 hours at 40°C. Probe detection was performed according to manufacturer’s instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
SG quantitation
All analyses utilized a minimum of 4 mice per genotype (≥ 2 per gender) and timepoint. Experiments utilized matched mutant and control litter-mate animals, whenever possible. To quantitate SGs in whole mounts, 2 representative fields at 5x magnification were photographed for DAPI and Nile Red staining, and subsequently all images were divided into thirds by drawing guide lines. SG presence or absence was scored for every third hair follicle that intersected these guide lines, yielding 18-25 randomly selected follicles per field. To quantitate SGs from sections, frozen skin sections (8 μm) were stained with antibodies against PPARγ and Scd1. The number of PPARγ/Scd1 double-positive SG clusters was then counted and normalized to the length of the skin section.
Statistics
SG quantitation data are depicted as means from independent biological replicates. Unpaired t tests were performed in most cases to determine statistical significance. For matched samples harvested from the same animal, paired t tests were used for comparisons between groups. Error bars are depicted as SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-FGFR2 | Cell Signaling | Cat # 23328S |
| Chicken anti-K14 | Biolegend | Cat # 906004 |
| Chicken anti-K5 | Biolegend | Cat # 905903 |
| Rat anti-Ki67 | eBioscience | Cat # 14-5698-80 |
| Goat anti-K79 | Santa Cruz | Cat # sc-243156 |
| Rabbit anti-PPARγ | Cell Signaling | Cat # 2443S |
| Rabbit anti-p44/42 (pErk1/2) | Cell Signaling | Cat # 4370 |
| Chicken anti-GFP | Abcam | Cat # ab13970 |
| Goat anti-Scd1 | Santa Cruz | Cat # sc-14719 |
| Biological samples | ||
| Mouse tissue samples, obtained in accordance with guidelines established by the University of Michigan Unit for Laboratory Animal Medicine | This manuscript | Study protocol # PRO00010041 |
| Chemicals, peptides, and recombinant proteins | ||
| Doxycycline chow (1 g/kg) | BioServ Inc | Cat # F3949 |
| Tamoxifen chow (400 mg/kg, irradiated) | Envigo | Cat # TD.130860 |
| 0.25% Trypsin (no EDTA) | Invitrogen | Cat # 15050065 |
| Albumin, Bovine Fraction V (BSA) | Research Prod. International | Cat # A30075 |
| Hank’s balanced salt solution (HBSS) | Gibco | Cat # 14025092 |
| Nair hair removal lotion | Nair | Cat # B001E6OAM8 |
| Hematoxylin | Sigma | Cat # HHS16 |
| DAPI | Sigma | Cat # 32670 |
| Nile Red | Sigma | Cat # N3013 |
| Pemigatinib (INCB054828) | SelleckChem | Cat # S0088 |
| Calcipotriol (MC903) | Sigma | Cat # C4369 |
| Critical commercial assays | ||
| RNAscope 2.5 HD Reagent Kit-BROWN | ACD (RNAscope) | Cat # 322310 |
| RNAscope 2.5 Pretreat Reagents-H202 and Protease Plus | ACD (RNAscope) | Cat # 322330 |
| RNAscope Target Retrieval | ACD (RNAscope) | Cat # 322000 |
| RNAscope Wash Buffer | ACD (RNAscope) | Cat # 310091 |
| Deposited data | ||
| Data files for single-cell RNA sequencing | This study | GEO: GSE225252 |
| Experimental models: Organisms/strains | ||
| Mouse: Lrig1tm1.1(cre/ERT2)Rjc (Lrig1-CreERT2) | The Jackson Laboratory | Cat # 018418 |
| Mouse: B6.129-Ppargtm2Rev/J (Pparg-flox) | The Jackson Laboratory (by way of Dr. Y. Eugene Chen) | Cat # 004584 |
| Mouse: B6;129-Ppargtm1.1(tTA)/Jmgr/J (AdipoTrak) | The Jackson Laboratory | Cat # 024755 |
| Mouse: B6.Cg-Tg(tetO-cre)1Jaw/J (TRE-Cre) | The Jackson Laboratory (by way of Dr. A. Dlugosz) | Cat # 006234 |
| Mouse: Gt(ROSA)26Sortm1(EYFP)Cos (YFP reporter) | The Jackson Laboratory | Cat # 006148 |
| Mouse: C57BL/6J | The Jackson Laboratory | Cat # 000664 |
| Oligonucleotides | ||
| In situ probe: mouse Acp5 | ACD (RNAscope) | Cat # 465001 |
| In situ probe: mouse Mgst2 | ACD (RNAscope) | Cat # 819931 |
| In situ probe: mouse Awat1 | ACD (RNAscope) | Cat # 1172821-C1 |
| In situ probe: mouse Slc6a19 | ACD (RNAscope) | Cat # 897821 |
| In situ probe: mouse Pparg | ACD (RNAscope) | Cat # 418821 |
| In situ probe: mouse Krt79 | ACD (RNAscope) | Cat # 436201 |
| Software and algorithms | ||
| Cell Ranger v6.1.2 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| DoubletFinder v2.0 | McGinnis et al.82 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Seurat v4.3.0 | Hao et al.83 | https://github.com/satijalab/seurat |
| COSG v0.9.0 | Dai et al.84 | https://github.com/genecell/COSGR |
| scVelo v0.2.5 | Bergen et al.85 | https://github.com/theislab/scvelo |
| Monocle 2 | Qiu et al.86,87 | https://github.com/cole-trapnell-lab/monocle2-rge-paper |
| clusterProfiler v4.6.0 | Yu et al.88 | https://github.com/YuLab-SMU/clusterProfiler |
| UCell 2.2.0 | Andreatta and Carmona89 | https://github.com/carmonalab/UCell |
| R | R Core | https://www.r-project.org/ |
| Python | Python Software Foundation | https://www.python.org/ |
| org.Mm.eg.db | Carlson90 | http://bioconductor.org/packages/org.Mm.eg.db/ |
Highlights.
SGs are largely self-renewed by resident stem cells during homeostasis
Alternative hair follicle stem cells regenerate the gland after ablation
scRNA-seq identifies direct and indirect paths for sebocyte differentiation
Transitional basal cells in the SG co-express Keratin 5 and PPARγ
ACKNOWLEDGMENTS
We are grateful to the Dlugosz lab (University of Michigan) for helpful discussions and sharing reagents; Dr. Y. Eugene Chen (University of Michigan) for sharing mice; Ann Marie Deslauriers-Cox for flow cytometry; Dr. Allison C. Billi (University of Michigan) for advice on single-cell preparation; and Tricia Tamsen, Olivia Koues, and the Advanced Genomics Core (University of Michigan) for single-cell sequencing. S.Y.W. acknowledges the support of the LEO Foundation (LF18017) and the NIH (R01AR065409 and R01AR080654). S.X.A. acknowledges the support of the National Science Foundation (CBET2134916). S.Y.W. and S.X.A. were jointly funded by the American Cancer Society (TLC-21-161-01-TLC). S.N. was partly supported by a Japan Society for the Promotion of Science (JSPS) research fellowship. The authors also acknowledge support from the UM Skin Biology and Disease Resource-based Center (P30AR075043) and NCI Cancer Center Support Grant (P30CA046592).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113121.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Zouboulis CC, Coenye T, He L, Kabashima K, Kobayashi T, Niemann C, Nomura T, Oláh A, Picardo M, Quist SR, et al. (2022). Sebaceous immunobiology - skin homeostasis, pathophysiology, coordination of innate immunity and inflammatory response and disease associations. Front. Immunol 13, 1029818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niemann C, and Horsley V (2012). Development and homeostasis of the sebaceous gland. Semin. Cell Dev. Biol 23, 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovászi M, Szegedi A, Zouboulis CC, and Törőcsik D (2017). Sebaceous-immunobiology is orchestrated by sebum lipids. Dermatoendocrinol. 9, e1375636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi VY, Leo M, Hassoun L, Chahal DS, Maibach HI, and Sivamani RK (2015). Role of sebaceous glands in inflammatory dermatoses. J. Am. Acad. Dermatol 73, 856–863. [DOI] [PubMed] [Google Scholar]
- 5.Smith KR, and Thiboutot DM (2008). Thematic review series: skin lipids. Sebaceous gland lipids: friend or foe? J. Lipid Res 49, 271–281. [DOI] [PubMed] [Google Scholar]
- 6.Lee SJ, Seok J, Jeong SY, Park KY, Li K, and Seo SJ (2016). Facial Pores: Definition, Causes, and Treatment Options. Dermatol. Surg 42, 277–285. [DOI] [PubMed] [Google Scholar]
- 7.Hinde E, Haslam IS, Schneider MR, Langan EA, Kloepper JE, Schramm C, Zouboulis CC, and Paus R (2013). A practical guide for the study of human and murine sebaceous glands in situ. Exp. Dermatol 22, 631–637. [DOI] [PubMed] [Google Scholar]
- 8.Cottle DL, Kretzschmar K, Schweiger PJ, Quist SR, Gollnick HP, Natsuga K, Aoyagi S, and Watt FM (2013). c-MYC-induced sebaceous gland differentiation is controlled by an androgen receptor/p53 axis. Cell Rep. 3, 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer H, Fumicz J, Rossiter H, Napirei M, Buchberger M, Tschachler E, and Eckhart L (2017). Holocrine secretion of sebum is a unique DNase2-dependent mode of programmed cell death. J. Invest. Dermatol 137, 587–594. [DOI] [PubMed] [Google Scholar]
- 10.Schneider MR, and Paus R (2014). Deciphering the functions of the hair follicle infundibulum in skin physiology and disease. Cell Tissue Res. 358, 697–704. [DOI] [PubMed] [Google Scholar]
- 11.Zouboulis CC, and Boschnakow A (2001). Chronological ageing and photoageing of the human sebaceous gland. Clin. Exp. Dermatol 26, 600–607. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Shui G, Wang G, Wang C, Sun S, Zouboulis CC, Xiao R, Ye J, Li W, and Li P (2014). Cidea control of lipid storage and secretion in mouse and human sebaceous glands. Mol. Cell Biol 34, 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung Y, Tam J, Ray Jalian H, Rox Anderson R, and Evans CL (2015). Longitudinal, 3D in vivo imaging of sebaceous glands by coherent anti-stokes Raman scattering microscopy: normal function and response to cryotherapy. J. Invest. Dermatol 135, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein EH, and Epstein WL (1966). New cell formation in human sebaceous glands. J. Invest. Dermatol 46, 453–458. [PubMed] [Google Scholar]
- 15.Plewig G, and Christophers E (1974). Renewal rate of human sebaceous glands. Acta Derm. Venereol 54, 177–182. [PubMed] [Google Scholar]
- 16.Weinstein GD (1974). Cell kinetics of human sebaceous glands. J. Invest. Dermatol 62, 144–146. [DOI] [PubMed] [Google Scholar]
- 17.Panteleyev AA, Rosenbach T, Paus R, and Christiano AM (2000). The bulge is the source of cellular renewal in the sebaceous gland of mouse skin. Arch. Dermatol. Res 292, 573–576. [DOI] [PubMed] [Google Scholar]
- 18.Petersson M, Brylka H, Kraus A, John S, Rappl G, Schettina P, and Niemann C (2011). TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J. 30, 3004–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lin K, Choo H, Chen Y, Zhang X, Xu RH, Wang X, and Wu Y (2023). Distinct bulge stem cell populations maintain the pilosebaceous unit in a β-catenin-dependent manner. iScience 26, 105805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veniaminova NA, Grachtchouk M, Doane OJ, Peterson JK, Quigley DA, Lull MV, Pyrozhenko DV, Nair RR, Patrick MT, Balmain A, et al. (2019). Niche-specific factors dynamically regulate sebaceous gland stem cells in the skin. Dev. Cell 51, 326–340.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen MS, Hannezo E, Ulyanchenko S, Estrach S, Antoku Y, Pisano S, Boonekamp KE, Sendrup S, Maimets M, Pedersen MT, et al. (2019). Tracing the cellular dynamics of sebaceous gland development in normal and perturbed states. Nat. Cell Biol 21, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Füllgrabe A, Joost S, Are A, Jacob T, Sivan U, Haegebarth A, Linnarsson S, Simons BD, Clevers H, Toftgård R, and Kasper M (2015). Dynamics of Lgr6+ Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem Cell Rep. 5, 843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghazizadeh S, and Taichman LB (2001). Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. EMBO J. 20, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page ME, Lombard P, Ng F, Göttgens B, and Jensen KB (2013). The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretzschmar K, Cottle DL, Donati G, Chiang MF, Quist SR, Gollnick HP, Natsuga K, Lin KI, and Watt FM (2014). BLIMP1 is required for postnatal epidermal homeostasis but does not define a sebaceous gland progenitor under steady-state conditions. Stem Cell Rep. 3, 620–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, Sequeira I, Sandberg R, and Kasper M (2020). The molecular anatomy of mouse skin during hair growth and rest. Cell Stem Cell 26, 441–457.e7. [DOI] [PubMed] [Google Scholar]
- 27.Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, and Kasper M (2016). Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 3, 221–237.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, Fassett MS, Golovato J, Gray M, Ghadially R, et al. (2018). Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep. 25, 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Eilertsen KJ, Ge L, Zhang L, Sundberg JP, Prouty SM, Stenn KS, and Parimoo S (1999). Scd1 is expressed in sebaceous glands and is disrupted in the asebia mouse. Nat. Genet 23, 268–270. [DOI] [PubMed] [Google Scholar]
- 30.Sundberg JP, Boggess D, Sundberg BA, Eilertsen K, Parimoo S, Filippi M, and Stenn K (2000). Asebia-2J (Scd1(ab2J)): a new allele and a model for scarring alopecia. Am. J. Pathol 156, 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, Truong A, Doebel T, Sakamoto K, Cui CY, et al. (2019). Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell 176, 982–997.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choa R, Tohyama J, Wada S, Meng H, Hu J, Okumura M, May RM, Robertson TF, Pai RAL, Nace A, et al. (2021). Thymic stromal lymphopoietin induces adipose loss through sebum hypersecretion. Science 373, eabd2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rittié L, Tejasvi T, Harms PW, Xing X, Nair RP, Gudjonsson JE, Swindell WR, and Elder JT (2016). Sebaceous Gland Atrophy in Psoriasis: An Explanation for Psoriatic Alopecia? J. Invest. Dermatol 136, 1792–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karnik P, Tekeste Z, McCormick TS, Gilliam AC, Price VH, Cooper KD, and Mirmirani P (2009). Hair follicle stem cell-specific PPARgamma deletion causes scarring alopecia. J. Invest. Dermatol 129, 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichenbach B, Classon J, Aida T, Tanaka K, Genander M, and Göritz C (2018). Glutamate transporter Slc1a3 mediates inter-niche stem cell activation during skin growth. EMBO J. 37, e98280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stenn KS, Sundberg JP, and Sperling LC (1999). Hair follicle biology, the sebaceous gland, and scarring alopecias. Arch. Dermatol 135, 973–974. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Chinnappan M, Prestwood CA, Edwards M, Artami M, Thompson BM, Eckert KM, Vale G, Zouboulis CC, McDonald JG, and Harris-Tryon TA (2021). Interleukins 4 and 13 drive lipid abnormalities in skin cells through regulation of sex steroid hormone synthesis. Proc. Natl. Acad. Sci. USA 118, e2100749118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X, Hobbs RP, and Coulombe PA (2013). The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol 25, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mesler AL, Benedeck RE, and Wong SY (2021). Preparing the hair follicle canal for hair shaft emergence. Exp. Dermatol 30, 472–478. 10.1111/exd.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frances D, and Niemann C (2012). Stem cell dynamics in sebaceous gland morphogenesis in mouse skin. Dev. Biol 363, 138–146. [DOI] [PubMed] [Google Scholar]
- 41.Ramot Y, Mastrofrancesco A, Camera E, Desreumaux P, Paus R, and Picardo M (2015). The role of PPARγ-mediated signalling in skin biology and pathology: new targets and opportunities for clinical dermatology. Exp. Dermatol 24, 245–251. [DOI] [PubMed] [Google Scholar]
- 42.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, and Evans RM (2013). PPARgamma signaling and metabolism: the good, the bad and the future. Nat. Med 19, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veniaminova NA, Vagnozzi AN, Kopinke D, Do TT, Murtaugh LC, Maillard I, Dlugosz AA, Reiter JF, and Wong SY (2013). Keratin 79 identifies a novel population of migratory epithelial cells that initiates hair canal morphogenesis and regeneration. Development 140, 4870–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldman A, Mukha D, Maor II, Sedov E, Koren E, Yosefzon Y, Shlomi T, and Fuchs Y (2019). Blimp1+ cells generate functional mouse sebaceous gland organoids in vitro. Nat. Commun 10, 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, and Graff JM (2008). White fat progenitor cells reside in the adipose vasculature. Science 322, 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, and Cotsarelis G (2005). Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med 17, 1351–1354. [DOI] [PubMed] [Google Scholar]
- 47.Vagnozzi AN, Reiter JF, and Wong SY (2015). Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration. Cell Cycle 14, 3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang S, Kuri P, Aubert Y, Brewster M, Li N, Farrelly O, Rice G, Bae H, Prouty S, Dentchev T, et al. (2021). Lgr6 marks epidermal stem cells with a nerve-dependent role in wound re-epithelialization. Cell Stem Cell 28, 1582–1596.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haensel D, Jin S, Sun P, Cinco R, Dragan M, Nguyen Q, Cang Z, Gong Y, Vu R, MacLean AL, et al. (2020). Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics. Cell Rep. 30, 3932–3947.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raymond K, Richter A, Kreft M, Frijns E, Janssen H, Slijper M, Praetzel-Wunder S, Langbein L, and Sonnenberg A (2010). Expression of the orphan protein Plet-1 during trichilemmal differentiation of anagen hair follicles. J. Invest. Dermatol 130, 1500–1513. [DOI] [PubMed] [Google Scholar]
- 51.Panteleyev AA, Paus R, Wanner R, Nürnberg W, Eichmüller S, Thiel R, Zhang J, Henz BM, and Rosenbach T (1997). Keratin 17 gene expression during the murine hair cycle. J. Invest. Dermatol 108, 324–329. [DOI] [PubMed] [Google Scholar]
- 52.Zeeuwen PLJM, van Vlijmen-Willems IMJJ, Hendriks W, Merkx GFM, and Schalkwijk J (2002). A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum. Mol. Genet 11, 2867–2875. [DOI] [PubMed] [Google Scholar]
- 53.Donati G, Rognoni E, Hiratsuka T, Liakath-Ali K, Hoste E, Kar G, Kayikci M, Russell R, Kretzschmar K, Mulder KW, et al. (2017). Wounding induces dedifferentiation of epidermal Gata6+ cells and acquisition of stem cell properties. Nat. Cell Biol 19, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayer-Garner IB, Givens V, and Smoller B (1999). Immunohistochemical staining for androgen receptors: a sensitive marker of sebaceous differentiation. Am. J. Dermatopathol 21, 426–431. [DOI] [PubMed] [Google Scholar]
- 55.Zouboulis CC, Chen WC, Thornton MJ, Qin K, and Rosenfield R (2007). Sexual hormones in human skin. Horm. Metab. Res 39, 85–95. [DOI] [PubMed] [Google Scholar]
- 56.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. (2012). The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, and Evans RM (2003). Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 100, 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sardella C, Winkler C, Quignodon L, Hardman JA, Toffoli B, Giordano Attianese GMP, Hundt JE, Michalik L, Vinson CR, Paus R, et al. (2018). Delayed hair follicle morphogenesis and hair follicle dystrophy in a lipoatrophy mouse model of Pparg total deletion. J. Invest. Dermatol 138, 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu YC, Pasolli HA, and Fuchs E (2011). Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brownell I, Guevara E, Bai CB, Loomis CA, and Joyner AL (2011). Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 8, 552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taetzsch T, Brayman VL, and Valdez G (2018). FGF binding proteins (FGFBPs): Modulators of FGF signaling in the developing, adult, and stressed nervous system. Biochim. Biophys. Acta, Mol. Basis Dis 1864, 2983–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu PCC, Koblish H, Wu L, Bowman K, Diamond S, DiMatteo D, Zhang Y, Hansbury M, Rupar M, Wen X, et al. (2020). INCB054828 (pemigatinib), a potent and selective inhibitor of fibroblast growth factor receptors 1, 2, and 3, displays activity against genetically defined tumor models. PLoS One 15, e0231877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dahlhoff M, Camera E, Schäfer M, Emrich D, Riethmacher D, Foster A, Paus R, and Schneider MR (2016). Sebaceous lipids are essential for water repulsion, protection against UVB-induced apoptosis, and ocular integrity in mice. Development 143, 1823–1831. [DOI] [PubMed] [Google Scholar]
- 64.Schepeler T, Page ME, and Jensen KB (2014). Heterogeneity and plasticity of epidermal stem cells. Development 141, 2559–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cockburn K, Annusver K, Gonzalez DG, Ganesan S, May DP, Mesa KR, Kawaguchi K, Kasper M, and Greco V (2022). Gradual differentiation uncoupled from cell cycle exit generates heterogeneity in the epidermal stem cell layer. Nat. Cell Biol 24, 1692–1700. 10.1038/s41556-41022-01021-41558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen E, Johnson C, Redmond CJ, Nair RR, and Coulombe PA (2022). Revisiting the significance of keratin expression in complex epithelia. J. Cell Sci 135, jcs260594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aragona M, Sifrim A, Malfait M, Song Y, Van Herck J, Dekoninck S, Gargouri S, Lapouge G, Swedlund B, Dubois C, et al. (2020). Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature 584, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S, Drummond ML, Guerrero-Juarez CF, Tarapore E, MacLean AL, Stabell AR, Wu SC, Gutierrez G, That BT, Benavente CA, et al. (2020). Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat. Commun 11, 4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allen M, Grachtchouk M, Sheng H, Grachtchouk V, Wang A, Wei L, Liu J, Ramirez A, Metzger D, Chambon P, et al. (2003). Hedgehog signaling regulates sebaceous gland development. Am. J. Pathol 163, 2173–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh K, Camera E, Krug L, Basu A, Pandey RK, Munir S, Wlaschek M, Kochanek S, Schorpp-Kistner M, Picardo M, et al. (2018). JunB defines functional and structural integrity of the epidermo-pilosebaceous unit in the skin. Nat. Commun 9, 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gu LH, and Coulombe PA (2008). Hedgehog signaling, Keratin 6 induction, and sebaceous gland morphogenesis. Am. J. Pathol 173, 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tassi E, Garman KA, Schmidt MO, Ma X, Kabbara KW, Uren A, Tomita Y, Goetz R, Mohammadi M, Wilcox CS, et al. (2018). Fibroblast Growth Factor Binding Protein 3 (FGFBP3) impacts carbohydrate and lipid metabolism. Sci. Rep 8, 15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grose R, Fantl V, Werner S, Chioni AM, Jarosz M, Rudling R, Cross B, Hart IR, and Dickson C (2007). The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 26, 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang X, Zhong X, Huang AJ, and Reneker LW (2022). Spontaneous acinar and ductal regrowth after meibomian gland atrophy induced by deletion of FGFR2 in a mouse model. Ocul. Surf 26, 300–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parfitt GJ, Lewis PN, Young RD, Richardson A, Lyons JG, Di Girolamo N, and Jester JV (2016). Renewal of the Holocrine Meibomian Glands by Label-Retaining, Unipotent Epithelial Progenitors. Stem Cell Rep. 1, 399^–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Centonze A, Lin S, Tika E, Sifrim A, Fioramonti M, Malfait M, Song Y, Wuidart A, Van Herck J, Dannau A, et al. (2020). Heterotypic cell-cell communication regulates glandular stem cell multipotency. Nature 584, 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selleri S, Seltmann H, Gariboldi S, Shirai YF, Balsari A, Zouboulis CC, and Rumio C (2006). Doxorubicin-induced alopecia is associated with sebaceous gland degeneration. J. Invest. Dermatol 126, 711–720. [DOI] [PubMed] [Google Scholar]
- 78.Murphy GF, Lavker RM, Whitaker D, and Korngold R (1991). Cytotoxic folliculitis in GvHD. Evidence of follicular stem cell injury and recovery. J. Cutan. Pathol 18, 309–314. [DOI] [PubMed] [Google Scholar]
- 79.Landthaler M, Kummermehr J, Wagner A, and Plewig G (1980). Inhibitory effects of 13-cis-retinoic acid on human sebaceous glands. Arch. Dermatol. Res 269, 297–309. [DOI] [PubMed] [Google Scholar]
- 80.Goldstein JA, Comite H, Mescon H, and Pochi PE (1982). Isotretinoin in the treatment of acne: histologic changes, sebum production, and clinical observations. Arch. Dermatol 118, 555–558. [PubMed] [Google Scholar]
- 81.Plewig G, and Kligman AM (1978). Proliferative activity of the sebaceous glands of the aged. J. Invest. Dermatol 10, 314–317. [DOI] [PubMed] [Google Scholar]
- 82.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 8, 329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dai M, Pei X, and Wang XJ (2022). Accurate and fast cell marker gene identification with COSG. Brief. Bioinform 23, bbab579. [DOI] [PubMed] [Google Scholar]
- 85.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. [DOI] [PubMed] [Google Scholar]
- 86.Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andreatta M, and Carmona SJ (2021). UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J 19, 3796–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carlson M. (2015). Org. Mm. Eg. Db: Genome Wide Annotation for Mouse (Bioconductor). http://bioconductor.org/packages/org.Mm.eg.db/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA sequencing data generated for this study have been deposited in the GEO database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225252.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.