Abstract
Objective:
Early identification of human papillomavirus associated oropharyngeal squamous cell carcinoma (HPV(+)OPSCC) is challenging and novel biomarkers are needed. We hypothesized that a panel of methylated DNA markers (MDMs) found in HPV(+) cervical squamous cell carcinoma (CSCC) will have similar discrimination in HPV(+)OPSCC tissues.
Materials and Methods:
Formalin-fixed, paraffin-embedded tissues were obtained from patients with primary HPV(+)OPSCC or HPV(+)CSCC; control tissues included normal oropharynx palatine tonsil (NOP) and cervix (NCS). Using a methylation-specific polymerase chain reaction, 21 previously validated cervical MDMs were evaluated on tissue-extracted DNA. Discrimination between case and control cervical and oropharynx tissue was assessed using area under the curve (AUC).
Results:
34 HPV(+)OPSCC, 36 HPV(+)CSCC, 26 NOP, and 24 NCS patients met inclusion criteria. Within HPV(+)CSCC, 18/21 (86%) of MDMs achieved an AUC ≥ 0.9 and all MDMs exhibited better than chance classifications relative to control cervical tissue (all p<0.001). In contrast, within HPV(+)OPSCC only 5/21 (24%) MDMs achieved an AUC ≥ 0.90 but 19/21 (90%) exhibited better than chance classifications relative to control tonsil tissue (all p<0.001). Overall, 13/21 MDMs had statistically significant lower AUCs in the oropharyngeal cohort compared to the cervical cohort, and only 1 MDM exhibited a statistically significant increase in AUC.
Conclusions:
Previously validated MDMs exhibited robust performance in independent HPV(+)CSCC patients. However, most of these MDMs exhibited higher discrimination for HPV(+)CSCC than for HPV(+)OPSCC. This suggests that each SCC subtype requires a unique set of MDMs for optimal discrimination. Future studies are necessary to establish an MDM panel for HPV(+)OPSCC.
Keywords: Oropharyngeal Cancer, Uterine Cervical Neoplasms, Human Papillomavirus Viruses, DNA Methylation, Biomarkers
INTRODUCTION
Incidence of oropharyngeal squamous cell carcinoma (OPSCC) is increasing at a faster rate than other head and neck cancers in the United States1, attributable to changes in human papillomavirus associated OPSCC (HPV(+)OPSCC) incidence2. Indeed, HPV(+)OPSCC outpaced HPV(+) cervical squamous cell carcinoma (HPV(+)CSCC) in 20152 and is now the most common HPV associated cancer in the US3. While HPV vaccination could prevent over 90% of all HPV tumors, based on current vaccination rates, cases of HPV(+)OPSCC are not expected to plateau until 20454. Identification of novel biomarkers to aid in early diagnosis or surveillance of HPV(+)OPSCC are lacking.
In general, as methods for treating cancer continue to become more sophisticated, it has become increasingly clear that earlier detection of tumors is critical. Particular emphasis has been placed on non-invasive or minimally invasive measures, which is of particular interest for HPV(+)OPSCC where no detectable premalignant lesion has been identified. Over the last 5-10 years, there has been a rapid increase in the investigation of tests which aim to identify substances shed from tumors into the blood stream. Most commonly, circulating tumor DNA (ctDNA) has been used to diagnose cancer via liquid biopsy5. Rettig et al. and Routman et al. separately investigated patients with HPV(+)OPSCC and each found an approximately 89% detection rate at diagnosis, which was associated with nodal status and lymphovascular invasion in patients who underwent surgery6, 7. However, more than half of patients without nodal disease had undetectable ctDNA at presentation. Thus, while ctDNA may be useful for treatment selection and surveillance, its performance for detecting early HPV(+)OPSCC appears limited.
Studies of multiple cancers have examined the usefulness of other techniques, such as cell-free DNA (cfDNA) methylation8-10, genetic mutations11 and DNA fragmentation12. Liu et al. targeted a panel of >100,000 key methylation regions in plasma cfDNA from 6,689 participants (2482 cancer, 4207 non-cancer)9. They found that targeted methylation analysis of cfDNA detected more than 50 cancer types across all stages, at a specificity of >99%. Tu et al. found that DNA methylation diagnostic biomarkers for CSCC could be successfully used to differentiate from normal cervical tissue13. This has similarly been found in pancreatic14, triple negative breast cancer15, prostate cancer16, and ovarian cancer17, among others. Despite the epidemic rise in the disease, this has not yet been done for HPV(+)OPSCC.
Oncogenic viral infections alter DNA methylation18 making this a particularly attractive candidate biomarker class for HPV(+)OPSCC19. In HPV mediated cancer, HPV protein E6 increases DNA methylation by inactivation of p53, a DNA methyltransferase 1 (DNMT1) inhibitor20. Consequently, there has been increased interest in evaluating epigenetic differences between HPV-driven and non-HPV-driven cancers rather than differences between cancer and non-cancer20. It is possible that HPV-mediated SCC may have similarities across subsites. As HPV mediates not only OPSCC, but also CSCC, we hypothesized that methylated DNA markers of HPV(+)CSCC would provide strong discrimination between HPV(+)OPSCC and NOP. We have previously validated a panel of methylated DNA markers (MDM) that discriminated between HPV(+)CSCC and benign cervical tissues with individual AUCs >0.9021. Here we sought to assess classification accuracy of these MDMs in HPV(+)OPSCC.
MATERIALS AND METHODS
Study population and patient selection
After institutional review board approval (IRB: 19-005075) we identified patients with HPV(+)CSCC, HPV(+)OPSCC, normal oropharyngeal palatine tonsil tissue (NOP), and normal uterine cervical tissue (NCS) treated at Mayo Clinic, Rochester, MN. HPV(+)OPSCC patients were identified through the Department of Otolaryngology Oropharynx REDCap Cancer database (1990-2019 at time of data retrieval). All tissues were formalin-fixed, paraffin-embedded (FFPE) and identified from an archive of residual clinical specimens in the Mayo Clinic Tissue Registry. Clinical history was obtained from the medical record and included age at diagnosis, sex at birth (male vs female), race (white vs non-white), tobacco and alcohol use, cervical Papanicolaou (Pap) test history (if relevant), and comorbidity status as defined using the Adult Comorbidity Evaluation-27 (ACE-27)22.
Patients were included if they had a primary (non-recurrent) tumor (OPSCC; CSCC) and normal histology (controls; NOP, NCS), no prior history of neoplastic or malignant disease of the target site (head and neck; uterine cervix), no exposure to chemotherapeutics within 1 year, no prior therapeutic radiation to the target region, no prior transplant, had clinical history available for review, sufficient target tissue available (>5mm), and were > 18 years of age. Normal cervical patients were excluded for any history of an abnormal Pap smear. Patients were considered HPV(+) if they had positive DNA in-situ hybridization for high-risk HPV (16, 18, 31, 33, or 51) or positive results on p16 staining (>70% diffuse nuclear and cytoplasmic staining). Patients were excluded if they declined Minnesota Research Authorization, if they had metastatic disease at presentation, if they presented with a synchronous primary tumor, or had a second primary active malignancy.
Tissue procurement and quantitative methylation specific PCR
2 mm FFPE tissue cores (marked by JJG) between 10 and 12 mm deep were macrodissected by the Biospecimens Accession and Processing Core lab from the area of interest within each block received from the Tissue Registry. The panel of methylated DNA markers (MDMs) was previously validated for HPV(+)CSCC21. These markers were then evaluated using DNA from independent FFPE HPV(+)OPSCC, HPV(+)CSCC, NOP, and NCS tissue using methylation-specific polymerase chain reaction (PCR). White blood cells (WBCs) were used as a background control.
Candidate MDMs were assayed by quantitative methylation specific PCR assays (qMSP), as previously published17. DNA was purified using the Qiagen (Valencia, CA) QIAmp FFPE DNA Tissue Kit (part#56404). Ten ng of sample DNA (per MDM) was bisulfite converted using the EZ-96 DNA Methylation kit (Zymo Research, Irvine CA) and amplified with primers recognizing the converted sequence using SYBR Green detection on Roche 480 LightCyclers (Roche, Basel Switzerland). Serially diluted universally methylated DNA samples were used as positive control standards, and negative controls included bisulfite converted and unconverted leukocyte-derived genomic DNA, and converted whole genome amplified (unmethylated) DNA. All laboratory staff were blinded to sample group information (cancer versus normal).
Statistical analysis
A target accrual of at least 31 patients per group was set to give 80% power for detecting an AUC of 0.85 given a null AUC of 0.7 while fixing alpha to 0.05. Up to 36 patients per group were included, where possible, to protect against potential assay failure or other sample dropout. Continuous variables were summarized as median (interquartile range), and discrete variables were summarized as frequency (percent). Each MDM was normalized to β-actin using generalized additive models (GAM) fit to the respective control group (NCS for HPV(+)CSCC; NOP for HPV(+)OPSCC). Discrimination between groups was assessed using AUC with corresponding 95% confidence intervals. A standard z-test was used to compute (unpaired) p-values for comparing performance between cervical and oropharyngeal tissue. Stratified AUCs were summarized by sex in oropharyngeal patients. The β-actin adjusted mean marker level across all 21 MDMs was used as a global measure of background signal in control patients and summarized for NCS and NOP separately. All analyses were performed in R (version 4.1; R Core Team). Two-sided P < 0.05 was considered statistically significant.
Data availability
The data generated in this study are available upon request from the corresponding author, pending approval from the Mayo Clinic IRB and Exact Sciences.
RESULTS
Patient demographics
36 HPV(+)CSCC, 24 NCS, 34 HPV(+)OPSCC, and 26 NOP patients met inclusion criteria. As shown in Table 1, HPV(+)OPSCC patients were older (median 57, IQR=48-62 years) than NOP patients (median 45, IQR=33-66 years) and had slightly increased alcohol and tobacco exposure. ACE-27 comorbidity scores were similar between the two groups. Males represented 88% of the HPV(+)OPSCC and 58% of NOP patients. When compared to HPV(+)CSCC, HPV(+)OPSCC patients were older, more often male, more likely to use alcohol and less likely to use tobacco. Tumors were identified earlier in HPV(+)CSCC, with 83% at stage I compared to 47% at stage I in HPV(+)OPSCC. More stage II tumors were found in HPV(+)OPSCC (44% vs 11% in HPV(+)CSCC), and stage III tumors were comparably represented (9% and 6% for HPV(+)OPSCC and HPV(+)CSCC, respectively).
Table 1:
Clinical characteristics of case and control patients providing tissues for methylation analysis.
| CSCC (N=36) | NCS (N=24) | OPSCC (N=34) | NOP (N=26) | |
|---|---|---|---|---|
| Age | ||||
| Median (Q1, Q3) | 43 (37, 53) | 44 (38, 55) | 57 (48, 62) | 45 (33, 66) |
| Female | 36 (100.0%) | 24 (100.0%) | 4 (11.8%) | 11 (42.3%) |
| Caucasian | 26 (72.2%) | 24 (100.0%) | 33 (97.1%) | 25 (96.2%) |
| Alcohol | ||||
| N-Miss | 16 | 1 | 1 | 2 |
| No | 11 (55.0%) | 7 (30.4%) | 5 (15.2%) | 6 (25.0%) |
| Yes | 9 (45.0%) | 16 (69.6%) | 28 (84.8%) | 18 (75.0%) |
| Ever Tobacco Use | ||||
| N-Miss | 2 | 1 | 0 | 0 |
| No | 11 (32.4%) | 11 (47.8%) | 19 (55.9%) | 17 (65.4%) |
| Yes | 23 (67.6%) | 12 (52.2%) | 15 (44.1%) | 9 (34.6%) |
| ACE-27 | ||||
| 1 | 19 (52.8%) | 5 (20.8%) | 17 (50.0%) | 8 (30.8%) |
| 2 | 9 (25.0%) | 15 (62.5%) | 8 (23.5%) | 8 (30.8%) |
| 3 | 6 (16.7%) | 3 (12.5%) | 7 (20.6%) | 8 (30.8%) |
| 4 | 2 (5.6%) | 1 (4.2%) | 2 (5.9%) | 2 (7.7%) |
| PAP Test Results | ||||
| N-Miss | 35 | 15 | 34 | 21 |
| ASCUS | 0 (0.0%) | 0 (0.0%) | 0 | 1 (20.0%) |
| Negative | 1 (100.0%) | 9 (100.0%) | 0 | 4 (80.0%) |
| Cancer Stage | ||||
| N-Miss | 0 | 24 | 0 | 26 |
| I | 30 (83.3%) | 0 | 16 (47.1%) | 0 |
| II | 4 (11.1%) | 0 | 15 (44.1%) | 0 |
| III | 2 (5.6%) | 0 | 3 (8.8%) | 0 |
| Cancer Tissue Purity | ||||
| N-Miss | 0 | 24 | 0 | 26 |
| Median (Q1, Q3) | 70 (60, 70) | NA | 80 (80, 80) | NA |
CSCC: HPV(+) Cervical Squamous Cell Carcinoma; NCS: Normal Cervical Tissue; OPSCC: HPV(+) Oropharyngeal Squamous Cell Carcinoma; NOP: Normal Oropharyngeal Palatine Tonsil; ACE-27: Adult Comorbidity Evaluation-27; PAP: Papanicolaou; ASCUS: Atypical Squamous Cells of Undetermined Significance; NA: non-applicable
MDM panel shows better discrimination between benign and malignant tissue in the cervical setting
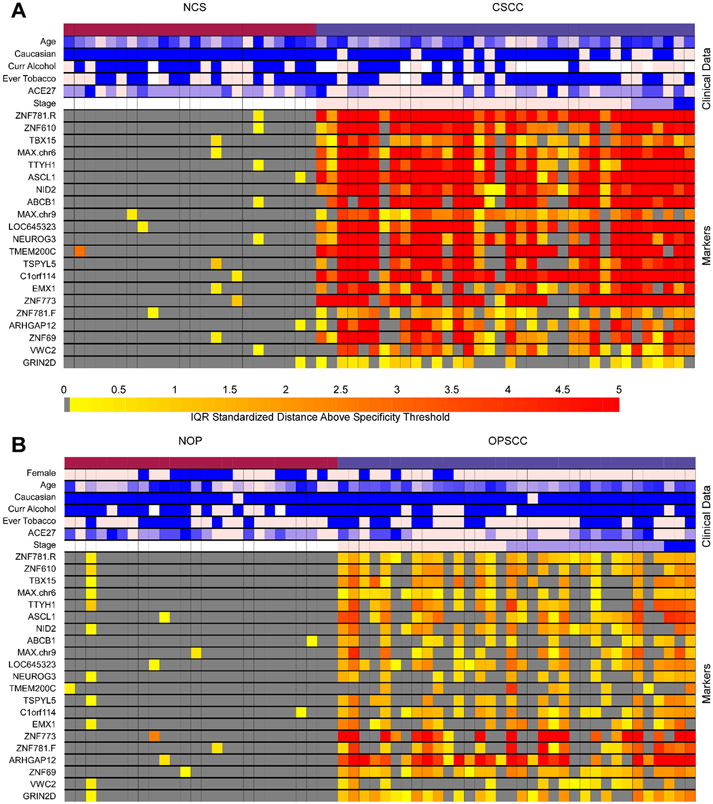
The discrimination of 21 MDMs was evaluated in HPV(+)CSCC and HPV(+)OPSCC compared to their respective controls. AUCs and corresponding 95% confidence intervals are displayed in Table 2, and MDM intensity is visualized in Figure 1. Within HPV(+)CSCC, 18/21 (86%) MDMs achieved an AUC ≥ 0.90 and all MDMs exhibited better than chance classifications relative to control cervical tissue (all p<0.001). Within HPV(+)OPSCC, 5/21 (24%) achieved an AUC ≥ 0.90, 15/21 (71%) achieved an AUC ≥ 0.8, and 19/21 (90%) exhibited better than chance classifications relative to control tonsil tissue (all p<0.001). AUCs of 7 MDMs (LOC645323, TSPYL5, C1orf114, ZNF773, ZNF781.F, ARHGAP12 and ZNF69) were not significantly different between HPV(+)OPSCC and HPV(+)CSCC. The AUC for one MDM (GRIN2D) was significantly higher in HPV(+)OPSCC. The remaining 13 MDMs had lower AUCs in HPV(+)OPSCC compared to HPV(+)CSCC. Distributions of individual β-actin -adjusted MDMs by group are presented in Figure 2.
Table 2:
Discrimination between HPV(+) cancers and their respective control groups
| CSCC vs. NCS AUC (95% CI) |
OPSCC vs. NOP AUC (95% CI) |
Cervical vs. Oropharyngeal P-valuea |
|
|---|---|---|---|
| ZNF781.R | 1 (1-1) | 0.92 (0.86-0.99) | 0.020 |
| ZNF610 | 0.99 (0.98-1) | 0.89 (0.8-0.97) | 0.016 |
| TBX15 | 0.99 (0.96-1) | 0.82 (0.72-0.93) | 0.003 |
| MAX.chr6 | 0.99 (0.97-1) | 0.86 (0.76-0.96) | 0.010 |
| TTYH1 | 0.99 (0.98-1) | 0.82 (0.71-0.93) | 0.002 |
| ASCL1 | 0.98 (0.93-1) | 0.86 (0.76-0.95) | 0.019 |
| NID2 | 0.98 (0.95-1) | 0.86 (0.77-0.95) | 0.021 |
| ABCB1 | 0.97 (0.93-1) | 0.74 (0.61-0.86) | 0.001 |
| MAX.chr9 | 0.97 (0.92-1) | 0.82 (0.72-0.93) | 0.011 |
| LOC645323 | 0.95 (0.89-1) | 0.88 (0.8-0.97) | 0.195 |
| NEUROG3 | 0.95 (0.89-1) | 0.7 (0.57-0.83) | 0.001 |
| TMEM200C | 0.95 (0.89-1) | 0.54 (0.39-0.69) | < 0.001 |
| TSPYL5 | 0.94 (0.87-1) | 0.82 (0.72-0.93) | 0.058 |
| C1orf114 | 0.94 (0.86-1) | 0.88 (0.8-0.97) | 0.337 |
| EMX1 | 0.93 (0.86-0.99) | 0.77 (0.65-0.89) | 0.019 |
| ZNF773 | 0.93 (0.86-1) | 0.9 (0.82-0.98) | 0.570 |
| ZNF781.F | 0.92 (0.86-0.99) | 0.79 (0.67-0.91) | 0.058 |
| ARHGAP12 | 0.91 (0.83-0.99) | 0.9 (0.81-1) | 0.922 |
| ZNF69 | 0.89 (0.8-0.97) | 0.95 (0.9-1) | 0.202 |
| VWC2 | 0.88 (0.79-0.97) | 0.57 (0.42-0.72) | 0.001 |
| GRIN2D | 0.72 (0.59-0.85) | 0.91 (0.83-0.99) | 0.019 |
Two-sided
CSCC: HPV(+) Cervical Squamous Cell Carcinoma; NCS: Normal Cervical Tissue; OPSCC: HPV(+) Oropharyngeal Squamous Cell Carcinoma; NOP: Normal Oropharyngeal Palatine Tonsil; AUC: Area under the curve; CI: confidence interval
Figure 1. MDM values were elevated in HPV(+) tumor tissue compared to corresponding control tissue.
MDM intensity and clinical covariates for normal cervical tissue (NCS) and HPV(+)CSCC (Panel A) and normal oropharyngeal palatine tonsil (NOP) and HPV(+)OPSCC (Panel B). Each row is a clinical covariate/MDM, and each column is a unique patient sample. MDM intensities were calculated separately for Panel A and Panel B. Marker levels below the 95th percentile in each control group were set to dark gray; increasing color intensity from yellow to red reflects increasing IQR standardized distance above this threshold. For continuous and ordinal clinical covariates, light pink reflects the lowest value and blue the highest. For binary clinical covariates, blue represents “yes,” and light pink represents “no.” Missing data is reflected with white boxes.
Figure 2. Most methylated DNA markers (MDMs) show better separation between benign and malignant tissues in cervical compared to oropharyngeal tissues.
DNA was extracted from FFPE tumor tissues (OPSCC, n=34; CSCC, n=36), normal oropharyngeal palatine tonsil (NOP, n=26) and normal cervical tissue (NCS, n=24). Methylation of selected DNA markers was determined using quantitative methylation specific PCR assays, β-actin adjusted marker levels among the groups are depicted by violin plots with embedded box plots representing minimum, Q1, median, Q3 and maximum values).
Sex differences in the cohorts had no obvious effect on marker performance
As these MDMs were discovered and previously validated in cervical tissue (all female) and oropharyngeal tissues were obtained primarily from males (Table 1), we performed stratified analyses to assess sex differences in MDM performance (Table 3). For most markers, we observed similar performance across sex, although statistical precision among females was very poor. Marker levels differed between NCS and NOP (Figure 3), with NOP patients exhibiting higher median methylation levels and greater variance compared to NCS, which may be explained by the origin of the panel or may indicate less underlying variability in cervical tissue methylation compared to tonsil.
Table 3:
Sex-stratified and overall AUCs with corresponding 95% confidence intervals.
| OPSCC Males vs. NOP Males AUC (n=45) |
OPSCC Females vs. NOP Females AUC (n=15) |
OPSCC vs. NOP Overall AUC |
|
|---|---|---|---|
| ZNF781.R | 0.91 (0.83-0.99) | 0.95 (0.85-1) | 0.92 (0.86-0.99) |
| ZNF610 | 0.88 (0.78-0.98) | 0.91 (0.72-1) | 0.89 (0.8-0.97) |
| TBX15 | 0.86 (0.75-0.96) | 0.7 (0.23-1) | 0.82 (0.72-0.93) |
| MAX.chr6 | 0.85 (0.74-0.96) | 0.91 (0.75-1) | 0.86 (0.76-0.96) |
| TTYH1 | 0.83 (0.71-0.95) | 0.61 (0.17-1) | 0.82 (0.71-0.93) |
| ASCL1 | 0.86 (0.76-0.97) | 0.73 (0.41-1) | 0.86 (0.76-0.95) |
| NID2 | 0.89 (0.79-0.98) | 0.73 (0.44-1) | 0.86 (0.77-0.95) |
| ABCB1 | 0.74 (0.6-0.88) | 0.64 (0.27-1) | 0.74 (0.61-0.86) |
| MAX.chr9 | 0.87 (0.77-0.97) | 0.61 (0.25-0.98) | 0.82 (0.72-0.93) |
| LOC645323 | 0.87 (0.77-0.97) | 0.86 (0.65-1) | 0.88 (0.8-0.97) |
| NEUROG3 | 0.7 (0.55-0.85) | 0.57 (0.17-0.96) | 0.7 (0.57-0.83) |
| TMEM200C | 0.52 (0.35-0.7) | 0.73 (0.45-1) | 0.54 (0.39-0.69) |
| TSPYL5 | 0.83 (0.71-0.95) | 0.75 (0.5-1) | 0.82 (0.72-0.93) |
| C1orf114 | 0.88 (0.79-0.98) | 0.82 (0.54-1) | 0.88 (0.8-0.97) |
| EMX1 | 0.8 (0.68-0.93) | 0.59 (0.23-0.95) | 0.77 (0.65-0.89) |
| ZNF773 | 0.87 (0.77-0.98) | 1 (1-1) | 0.9 (0.82-0.98) |
| ZNF781.F | 0.8 (0.67-0.93) | 0.77 (0.38-1) | 0.79 (0.67-0.91) |
| ARHGAP12 | 0.9 (0.79-1) | 0.93 (0.78-1) | 0.9 (0.81-1) |
| ZNF69 | 0.98 (0.94-1) | 0.77 (0.42-1) | 0.95 (0.9-1) |
| VWC2 | 0.59 (0.42-0.75) | 0.39 (0.06-0.71) | 0.57 (0.42-0.72) |
| GRIN2D | 0.92 (0.83-1) | 0.89 (0.65-1) | 0.91 (0.83-0.99) |
CSCC: HPV(+) Cervical Squamous Cell Carcinoma; NCS: Normal Cervical Tissue; OPSCC: HPV(+) Oropharyngeal Squamous Cell Carcinoma; NOP: Normal Oropharyngeal Palatine Tonsil; AUC: Area under the curve; CI: confidence interval
Figure 3. Mean methylation levels were elevated and had greater variance in NOP compared to NCS tissues.
Violin plots (with embedded box plots displaying minimum, Q1, median, Q3 and maximum) show adjusted mean marker level across 21 MDMs in control patients (normal cervical tissue (NCS) and normal oropharyngeal palatine tonsil (NOP)).
DISCUSSION
In this study, we observed that methylation markers discovered and validated in cervical cancer achieved some success in discriminating between HPV(+)OPSCC and NOP. 90% of markers examined provided better than chance classification between these groups. Additionally, 5/21 (24%) markers exhibited excellent discriminatory potential (AUC estimates > 0.9). Although there were fewer females in the OPSCC group, stratified analysis revealed similar discrimination in males and females (Table 3). While some markers performed well, discriminating each cancer type from its respective control tissue, overall, the markers did not achieve the same degree of discrimination in HPV(+)OPSCC as in HPV(+)CSCC.
It has been suggested that HPV infection results in an early change in the carcinogenesis timeline for epithelial cells, leading to key alterations in the tumor microenvironment (TME)23. Sartor et al demonstrated that HPV(+) cell lines had higher DNA methylation in genic and LINE-1 regions than HPV(−) SCC cell lines19. This is consistent with our observation that several MDMs (ZNF781.R, ZNF773, ARHGAP12, ZNF69, GRIN2D) from our HPV(+)CSCC panel achieved excellent discrimination in HPV(+)OPSCC. However, most markers still performed better in HPV(+)CSCC, motivating ongoing targeted discovery and validation testing for HPV(+)OPSCC. As stated above, the HPV infection event is the key factor in tumorigenesis for these subsites, however, differences in the epithelial structure (reticulated vs non), tumor microenvironment (immune privileged vs non), microbiome, carcinogen exposure, and other factors likely impact the choice of methylation targets between these subsites.
Demographics and other risk factors have been shown to be associated with altered levels of methylation. Using methylation sensitive digital PCR assessments, Lei et al. found that heavy alcohol consumption was strongly correlated with accelerated epigenetic aging24. In addition, consistent with trends in SEER age-adjusted incidence rates for oropharynx and tonsil cancer by sex, which in 2019 were 5 per 100,000 (95% CI 4.9-5.2) for males and 0.9 per 100,000 for females (95% CI 0.9-1.0)25, we had significantly more male patients in our HPV(+)OPSCC cohort than female. It is known that sex hormones, aging and smoking can influence the epigenetics seen in different cancer populations26-28. Our study lacked power to assess the impact of these other modulators on the methylation patterns we observed in our cohorts, underscoring the need for further investigation into methylation differences between HPV(+)OPSCC and NOP tissues.
The majority of the MDMs evaluated in this study reside in genes that have been previously associated with other solid cancers by our group (Supplemental Table 1). The most common cancer association was esophageal squamous cell carcinoma (ESCC) where over half of the 21 current MDMs resided in genes that were identified as very discriminative hypermethylation events29. Additionally, 13 of the 21 MDMs we identified were found in differentially methylated genes of multiple cancers including pancreatic, colorectal, gastric, lung, ovarian, and lymphoma. These cancers have widely different histologies, microenvironments and little or no HPV involvement. AUCs for OPSCC with the multiple-cancer MDMs ranged from 0.57 to 0.92. Conversely, 8 of the 21 MDMs were unique to CSCC and the performance of these in OPSCC ranged from AUC of 0.54 to 0.95. Generally, there does not appear to be a strong relationship between the site specificity or universality of these MDMs and their ability to discriminate between HPV(+)OPSCC and NOP.
This study has several important limitations. First, while cervical cancer and oropharyngeal cancer comprise the majority of HPV mediated squamous cell carcinoma, our study did not investigate other HPV mediated subsites. In addition, we used tonsillectomy specimens from adult patients with non-malignant disease to serve as our normal oropharyngeal controls. However, these tissue specimens may not represent normal tonsil as these tonsils were likely removed for infectious or inflammatory reasons and they may not be representative of base of tongue tonsillar tissue, as this subsite is less likely removed for benign disease. Our ability to distinguish between tissues of origin was limited as we used a validated panel of MDMs for cervical cancer across all sites (normal cervix, normal tonsil, HPV(+)OPSCC, HPV(+)CSCC) without a validated panel in HPV(+)OPSCC for comparison. Finally, we were not able to accrue 31 patients to each group, thus slightly decreasing our power to detect differences between case and control tissues.
CONCLUSION
Using a panel of MDMs previously validated for HPV(+)CSCC, we found excellent discrimination between independent HPV(+)CSCC and NCS and only moderate discrimination between HPV(+)OPSCC and NOP. One criterion for the selection of the markers in HPV(+)CSCC tissue was low background levels in normal cervical tissues. The lower discrimination observed in HPV(+)OPSCC is likely reflective of the increased background levels in methylation in normal tissue, rather than decreased methylation of these markers in cancerous tissue (Figures 2 and 3). Differences in methylation levels may be dependent on demographics, such as alcohol or tobacco exposure, age, or sex, among others. Despite known differences in the rates of HPV(+)OPSCC in males and females, we did not identify a difference in methylation markers based on sex for HPV(+)OPSCC in our analysis, although statistical precision among females was poor. Future studies investigating a validated MDM panel for HPV(+)OPSCC and HPV driven disease across sites is needed.
Supplementary Material
Highlights.
Methylated DNA markers distinguished cervical cancer from normal cervical tissue.
Cervical panel performed well but not optimally in HPV-associated oropharynx cancer.
A panel targeted to oropharynx cancer may be predictive of cancer and site-specific
FINANCIAL SUPPORT:
This work was supported by internal funding (KMVA). Additional support was provided by Exact Sciences under a sponsored research agreement with Mayo Clinic. Additional support was provided by the National Cancer Institute (CA214679) to JBK. This work used shared resources/cores supported by the Mayo Clinic Comprehensive Cancer Center Support Grant (P30 CA015083); its contents are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health. Funding sources had no role in study design; data collection, analysis and interpretation; manuscript preparation or the decision to publish.
ABBREVIATIONS:
- ACE-27
Adult Comorbidity Evaluation-27
- AUC
Area under the curve
- CSCC
Cervical squamous cell carcinoma
- cfDNA
Cell-free DNA
- ctDNA
Circulating tumor DNA
- FFPE
Formalin-fixed, paraffin-embedded
- HPV
Human papillomavirus
- HPV(+)OPSCC
HPV-associated oropharyngeal squamous cell carcinoma
- MDM
Methylated DNA marker
- NCS
Normal cervical tissue
- NOP
Normal oropharynx palatine tonsil
- OPSCC
Oropharyngeal squamous cell carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
DISCLOSURES: Messrs. Taylor and Mahoney, and Drs. Van Abel, Routman, Ma, and Kisiel have disclosed inventions of Mayo Clinic intellectual property that are licensed to Exact Sciences (Madison, WI) for which they could earn royalties, paid to Mayo Clinic.
This work was presented as a poster presentation at the AACR in New Orleans, LA on April 13, 2022
REFERENCES
- 1.Fakhry C, Krapcho M, Eisele DW, D'Souza G. Head and neck squamous cell cancers in the United States are rare and the risk now is higher among white individuals compared with black individuals. Cancer. 2018;124:2125–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Dyne EA, Henly SJ, Saraiya M, Thomas CC, Markowitz LE, Benard VB. Trends in Human Papillomavirus–Associated Cancers — United States, 1999–2015. MMWR Morb Mortal Wkly Rep 2018;67:918–924. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Cancers Associated with Human Papillomavirus USUDB, no. 32. Atlanta, GA: Centers for Disease Control and Prevention, US Department of Health and Human Services; 2022. [Google Scholar]
- 4.Schuman A, Anderson KS, Day AT, Ferrell J, Sturgis EM, Dahlstrom KR. Is 2045 the best we can do? Mitigating the HPV-related oropharyngeal cancer epidemic. Expert Rev Anticancer Ther. 2022;22:751–761. [DOI] [PubMed] [Google Scholar]
- 5.Klein EA, Richards D, Cohn A, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. 2021;32:1167–1177. [DOI] [PubMed] [Google Scholar]
- 6.Rettig EM, Wang AA, Tran NA, et al. Association of Pretreatment Circulating Tumor Tissue-Modified Viral HPV DNA With Clinicopathologic Factors in HPV-Positive Oropharyngeal Cancer. JAMA Otolaryngol Head Neck Surg. 2022;148:1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Routman DM, Kumar S, Chera BS, et al. Detectable Postoperative Circulating Tumor Human Papillomavirus DNA and Association with Recurrence in Patients With HPV-Associated Oropharyngeal Squamous Cell Carcinoma. Int J Radiat Oncol Biol Phys. 2022;113:530–538. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Gole J, Gore A, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11:3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583. [DOI] [PubMed] [Google Scholar]
- 11.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu J, Chen S, Wu S, Wu T, Fan R, Kuang Z. Tumor DNA Methylation Profiles Enable Diagnosis, Prognosis Prediction, and Screening for Cervical Cancer. Int J Gen Med. 2022;15:5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Guo S, Liu X, et al. Noninvasive detection of pancreatic ductal adenocarcinoma using the methylation signature of circulating tumour DNA. BMC Med. 2022;20:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manoochehri M, Borhani N, Gerhauser C, et al. DNA methylation biomarkers for noninvasive detection of triple-negative breast cancer using liquid biopsy. Int J Cancer. 2023;152:1025–1035. [DOI] [PubMed] [Google Scholar]
- 16.Krausewitz P, Kluemper N, Richter AP, et al. Early Dynamics of Quantitative SEPT9 and SHOX2 Methylation in Circulating Cell-Free Plasma DNA during Prostate Biopsy for Prostate Cancer Diagnosis. Cancers (Basel). 2022;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marinelli LM, Kisiel JB, Slettedahl SW, et al. Methylated DNA markers for plasma detection of ovarian cancer: Discovery, validation, and clinical feasibility. Gynecol Oncol. 2022;165:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneda A, Matsusaka K, Aburatani H, Fukayama M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445–3450. [DOI] [PubMed] [Google Scholar]
- 19.Sartor MA, Dolinoy DC, Jones TR, et al. Genome-wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekanayake Weeramange C, Tang KD, Vasani S, Langton-Lockton J, Kenny L, Punyadeera C. DNA Methylation Changes in Human Papillomavirus-Driven Head and Neck Cancers. Cells. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakkum-Gamez JN, Graham RP, Broderick BT, et al. Discovery and validation of novel methylated DNA markers of cervical cancer. Journal of Clinical Oncology 2021. 39:15_suppl, 5526–5526. [Google Scholar]
- 22.Monteiro AR, Garcia AR, Pereira TC, et al. ACE-27 as a prognostic tool of severe acute toxicities in patients with head and neck cancer treated with chemoradiotherapy: a real-world, prospective, observational study. Support Care Cancer. 2021;29:1863–1871. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Yuan O, Cui H, et al. Bioinformatic analysis identifies HPV-related tumor microenvironment remodeling prognostic biomarkers in head and neck squamous cell carcinoma. Front Cell Infect Microbiol. 2022;12:1007950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei MK, Gibbons FX, Gerrard M, Beach SRH, Dawes K, Philibert R. Digital methylation assessments of alcohol and cigarette consumption account for common variance in accelerated epigenetic ageing. Epigenetics. 2022;17:1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oropharynx & Tonsil Recent Trends in SEER Age-Adjusted Incidence Rates, 2000-2019. Vol 20222019. [Google Scholar]
- 26.Lopes-Ramos CM, Quackenbush J, DeMeo DL. Genome-Wide Sex and Gender Differences in Cancer. Front Oncol. 2020;10:597788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett KL, Lee W, Lamarre E, et al. HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Gene Chromosome Cancer. 2010;49:319–325. [DOI] [PubMed] [Google Scholar]
- 28.Johnson AA, Akman K, Calimport SRG, Wuttke D, Stolzing A, de Magalhães JP. The Role of DNA Methylation in Aging, Rejuvenation, and Age-Related Disease. Rejuvenation Research 2012;15:483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Y, Taylor W, Bamlet WR, et al. Methylated DNA Markers of Esophageal Squamous Cancer and Dysplasia: An International Study. Cancer Epidemiol Biomarkers Prev. 2020;29:2642–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author, pending approval from the Mayo Clinic IRB and Exact Sciences.