The Python image processing community has seen rapid growth from new members across many domains and with varying levels of software proficiency. Much of this growth is driven by the accessibility of the scientific Python software stack1–3. The napari application for n-dimensional image visualization and analysis can potentially further this growth by fulfilling the need for a convenient and powerful graphical interface built atop these technologies4. The plugin-based model of napari promotes extensibility, sharing, and modularity, and the rapidly growing napari community is doing an excellent job driving the development of needed features to accelerate and broaden napari’s utility.
Meanwhile, the ImageJ software ecosystem5,6, beginning with the original ImageJ and now supported by ImageJ27 and Fiji8, has developed over decades into a flourishing community for n-dimensional image processing in Java9. This maturity makes the ImageJ ecosystem a prime candidate for collaboration, and there is already demand from napari users for popular ImageJ functions. While some ImageJ features have been or will be ported to napari, this approach cannot practically scale throughout the entire ImageJ ecosystem. A direct port would significantly increase the maintenance burden on developers, distracting from work that might address new problems in the napari community. A more ideal solution would be for napari and ImageJ to integrate directly, removing the need to keep plugin ports in sync between the two.
Integrating the ImageJ ecosystem into napari presents two main challenges. The first is cross-language operation: ImageJ-based tools run on the Java platform, resulting in considerable technical barriers when attempting to integrate them into a Python program. A mechanism is needed to call Java code from Python, allowing users to access new and existing ImageJ routines simply and transparently. The second challenge is accessibility: napari users from the Python community may be unfamiliar with Java and ImageJ terminology and structure. A solution should enable these users to utilize the strengths of both ecosystems without needing to learn two separate applications.
The PyImageJ project10 provides a robust solution for Python-based ImageJ access, including its data structures and plugins. However, PyImageJ is a library for programmers, requiring explicit conversion of Python data structures such as NumPy images into equivalent Java structures before they can be passed to ImageJ routines. To make ImageJ truly accessible from napari with no additional programming, we developed another layer on top of PyImageJ, automating data conversions and enabling access to ImageJ functionality within one unified napari interface.
This new layer, called napari-imagej, provides an accessible and comprehensive solution to ImageJ ecosystem access from napari. As a napari plugin, napari-imagej is available on all operating systems supported by both napari and PyImageJ, including Linux, macOS, and Windows. Through a configuration dialog, napari-imagej users can customize their ImageJ2 installation, allowing efficient access to all ImageJ ecosystem functionality including ImageJ, ImageJ2, Fiji, and third-party plugins. The napari-imagej plugin provides two different mechanisms for accessing ImageJ ecosystem functionality, both built on the same foundation.
Much of the ImageJ ecosystem, including the ImageJ2 platform and the plugins designed for it, can be run in a headless mode without visible ImageJ components. All headless routines are discovered by the search service within ImageJ2 and integrated directly within napari within a new napari widget, as shown in Figure 1 (a). By presenting these routines via this mechanism, napari-imagej minimizes both the display footprint and the amount of Java and ImageJ terminology exposed to the user, while maintaining comparable performance to usage from within the ImageJ UI (see for benchmarking analysis). Third-party plugins and scripts written for the ImageJ2 platform are also automatically exposed in the search results, maximizing extensibility.
Figure 1:
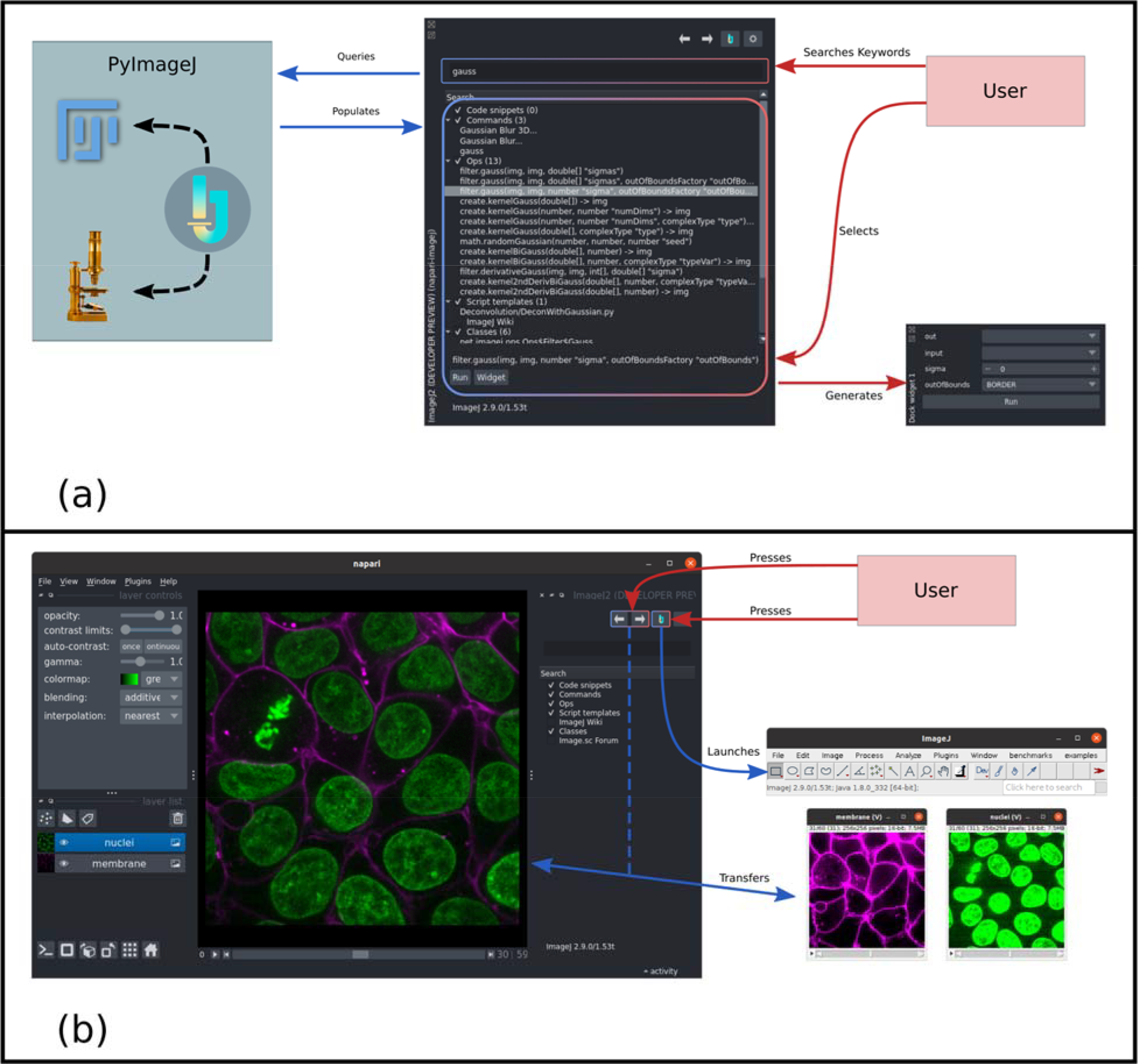
(a) Headless ImageJ ecosystem routines are executable directly from the napari interface by typing search terms into the napari-imagej search bar. A napari widget for executing a routine can then be generated by selecting any of the corresponding results shown in the panel beneath. (b) The ImageJ button in the napari-imagej toolbar launches the ImageJ user interface. From this interface, any ImageJ ecosystem routine can be executed, including third-party plugins. Multidimensional image data can be passed between the napari and ImageJ interfaces using the transfer buttons, also located in the napari-imagej toolbar.
Many plugins and macros written for the original ImageJ were not designed to run without the ImageJ graphical interface visible. To enable workflows including these routines, we also provide additional controls to launch the ImageJ interface and to explicitly transfer napari layers to and from ImageJ, as shown in Figure 1 (b). While the headless widget-based approach described above is promoted for its flexibility and cleaner integration within napari, all ImageJ ecosystem functionality can be run using this graphical method, including both ImageJ2 and original ImageJ plugins.
To run headless ImageJ routines, napari-imagej transparently converts inputs from napari to their ImageJ ecosystem equivalents. Input types supported by napari-imagej include napari image, shapes, and points layers, napari labels, napari surfaces, and Python built-in types such as numeric values and strings of text. By performing these conversions on behalf of the user, napari-imagej minimizes the burden of utilizing ImageJ routines written in other languages.
The napari-imagej plugin builds on the foundation of PyImageJ to make the ImageJ ecosystem accessible within the Python ecosystem. Users can utilize ImageJ and Fiji directly in napari, without explicit data conversion and in tandem with other napari plugins, opening the door to more expressive and interoperable workflows.
Acknowledgements
The napari-imagej developers would like to thank T. Lambert, G. Bokota, D.D. Pop, J. Nunez-Iglesias, and all of the napari core developers for their assistance on the integration with the napari application; T. Burke for collaboration on Python-based image labelings; and N. Chiaruttini, and J. Chacko for early user testing and feedback. This work has been supported by the National Institutes of Health (P41GM135019) to K.W.E.; Chan Zuckerberg Initiative funding to C.T.R. and K.W.E.; and additional internal funding from the Laboratory for Optical and Computational Instrumentation and the Morgridge Institute for Research.
Footnotes
Competing Interests
The authors declare no competing interests.
Code availability
The source code, documentation, tutorials and use cases for napari-imagej, which is made available under the open-source BSD 2-clause license, can be found online at https://github.com/imagej/napari-imagej
Data availability
All data used for napari-imagej use cases are available via https://napari-imagej.readthedocs.io/
References
- 1.Harris CR et al. Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Virtanen P et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Walt S et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sofroniew Nicholas et al. napari: a multi-dimensional image viewer for Python. (2022) doi: 10.5281/ZENODO.3555620. [DOI] [Google Scholar]
- 5.Schindelin J, Rueden CT, Hiner MC & Eliceiri KW The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder AB et al. The ImageJ ecosystem: Open source software for image visualization, processing, and analysis. Protein Sci. 30, 234–249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rueden CT et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rueden CT et al. PyImageJ: A library for integrating ImageJ and Python. Nat. Methods 19, 1326–1327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used for napari-imagej use cases are available via https://napari-imagej.readthedocs.io/


