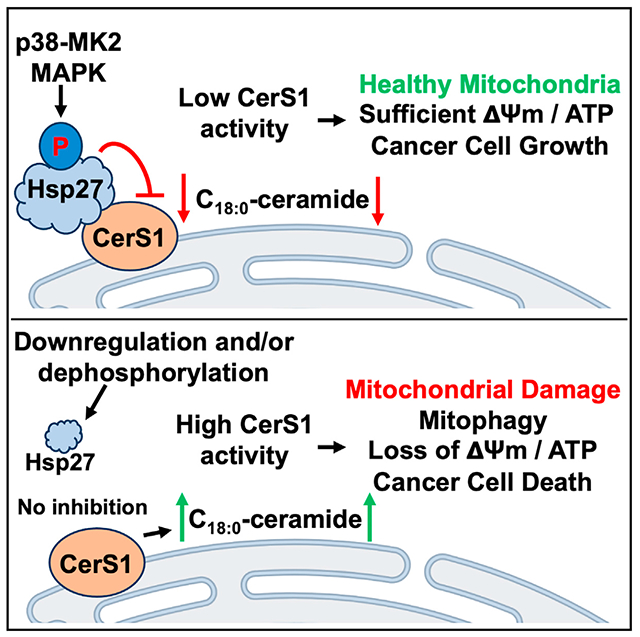
SUMMARY
Sphingolipids have key functions in membrane structure and cellular signaling. Ceramide is the central molecule of the sphingolipid metabolism and is generated by ceramide synthases (CerS) in the de novo pathway. Despite their critical function, mechanisms regulating CerS remain largely unknown. Using an unbiased proteomics approach, we find that the small heat shock protein 27 (Hsp27) interacts specifically with CerS1 but not other CerS. Functionally, our data show that Hsp27 acts as an endogenous inhibitor of CerS1. Wild-type Hsp27, but not a mutant deficient in CerS1 binding, inhibits CerS1 activity. Additionally, silencing of Hsp27 enhances CerS1-generated ceramide accumulation in cells. Moreover, phosphorylation of Hsp27 modulates Hsp27-CerS1 interaction and CerS1 activity in acute stress-response conditions. Biologically, we show that Hsp27 knockdown impedes mitochondrial function and induces lethal mitophagy in a CerS1-dependent manner. Overall, we identify an important mode of CerS1 regulation and CerS1-mediated mitophagy through protein-protein interaction with Hsp27.
Graphical abstract
In brief
Boyd et al. report that Hsp27 associates with CerS1 and inhibits its activity via direct interaction in vitro and in cells. Hsp27-CerS1 interaction can be modulated by phosphorylation of Hsp27, and downregulation of Hsp27 induces CerS1-dependent lethal mitophagy.
INTRODUCTION
Sphingolipids are important structural components of biological membranes, and some of the sphingolipid family members also have signaling properties modulating important downstream biological processes.1,2 Ceramide is the central molecule in the sphingolipid (SL) pathway, and it is composed of a sphingosine backbone that is N-acylated with fatty acids of varying lengths. De novo ceramide production takes place in the endoplasmic reticulum (ER). Once the sphingoid backbone is formed by the sequential activities of serine palmitoyltransferase (SPT) and 3-ketosphinganine reductase, dihydroceramide is formed by the ceramide synthases (CerS), where the dihydrosphingosine backbone is N-acylated using fatty acyl-coenzymes (FA-CoAs).1,2 Ceramide synthases also acylate sphingosine to directly generate ceramide within the salvage/recycling pathway.1,2 Six CerS isoforms (CerS1–CerS6), encoded by distinct genes, have been identified and shown to preferentially utilize fatty acyl-CoAs with different chain lengths to generate distinct ceramide species.3,4 Functionally, CerS-generated ceramides have critical and discrete roles in multiple biologies including apoptosis,5–7 response to chemotherapy and radiation,8–11 epithelial-mesenchymal transition,12 autophagy,13 mitophagy,14,15 ER stress response,16 anti-inflammatory therapy,17 graft-versus-host disease,18,19 insulin resistance,20,21 cerebral ischemia/reperfusion injury,22,23 and neuronal development.24,25 Hence, it is extremely important to identify the molecular mechanisms regulating individual CerS enzymes to have a better understanding of the biological functions of ceramide.
The human genome contains at least ten small heat shock proteins (sHSPs) that are characterized by their small molecular weight, ranging from 15 to 40 kDa, and by having a highly conserved α-crystallin domain.26 Belonging to sHSPs, human Hsp27 (also known as Hsp25 in mouse) is encoded by the Hspb1 gene and has important functions in multiple physiological processes such as ATP-independent chaperoning,27,28 cell death,29,30 and cancer metastasis.31,32 Hsp27 is also implicated in Alzheimer’s disease through Tau-protein-binding activity.33–35 With respect to cancer therapy, elevated Hsp27 levels in breast, ovarian, pancreatic, colon, and bladder cancers are associated with poor prognosis.36–43 In addition, Hsp27 overexpression has been linked to resistance to chemotherapeutic agents in lung, prostate, breast, pancreas, and colon cancers.44–52
One of the well-established pathways leading to Hsp27 phosphorylation is activation of the p38-MK2 (MAPKAPK2) mitogen-activated protein kinase (MAPK) pathway.53–55 p38-MK2-induced phosphorylation of Hsp27 is implicated in regulation of actin cytoskeleton dynamics such that phosphorylated Hsp27 is involved in actin-rich stress fiber reorganization and increased motility.56,57 In addition, increased phosphorylation status of Hsp27 is associated with poor response to anti-cancer therapy.38,46,50
In this study, to uncover possible allosteric regulation of CerS by other proteins, we used a proteomics approach and discovered that CerS1, but not the other CerS tested, specifically interacts with Hsp27. Our results indicate that interaction of Hsp27 with CerS1 decreases CerS1 enzymatic activity. We have identified the domains of Hsp27 that are required for association with CerS1 and have found that phosphorylation status of Hsp27 influences its interaction with CerS1. Biologically, downregulation of Hsp27 induced CerS1-generated C18:0-ceramide accumulation and in turn caused CerS1-dependent mitochondrial dysfunction and cancer cell death. Our study identifies a fundamental mechanism of CerS1 regulation that depends on protein-protein interaction which can be regulated by phosphorylation of Hsp27.
RESULTS
Hsp27 interacts with CerS1
CerS-generated ceramides are signaling molecules controlling a multitude of biological responses.58 However, there is limited information on how CerS enzymes are regulated. To identify proteins that interact with CerS and can potentially regulate CerS activity, after immunoprecipitation (IP) of CerS proteins, we used proteomics59 and found that Hsp27, encoded by HSPB1 gene, interacts with CerS1 but not with other CerS enzymes tested in HCT116 colorectal carcinoma cells (Figures 1A and S1A). Next, we tested the interaction of hemagglutinin (HA)-tagged Hsp27 with CerS1. HA-Hsp27 was detected after IP of CerS1 using either anti-FLAG antibody or an antibody that recognizes endogenous CerS1 (Figure 1B). Moreover, the interaction of endogenous CerS1 with endogenous Hsp27 was also confirmed (Figure 1C). We also established the interaction of Hsp27 with CerS1 using reverse IP, where HA-tagged Hsp27 is immunoprecipitated (Figure 1D). Likewise, when HA-tagged Hsp27 was immunoprecipitated in the presence of FLAG-tagged CerS proteins, only FLAG-CerS1 was detected in the IP complexes (Figure S1B). Further, using proximity ligation assays (PLA), which do not rely on cell lysis, we showed the interaction of Hsp27 and CerS1 in intact cells (Figure 1E). We did not detect significant PLA signal between CerS1 and PYGL (glycogen phosphorylase, liver), another cytosolic protein (Figure S1C). Although not a measure of interaction, we also observed significant co-localization between CerS1 and Hsp27 when analyzed using immunofluorescence confocal microscopy (Figure 1F), but not between CerS1 and PYGL (Figure S1D). Since CerS enzymes are localized to the ER and Hsp27 is a cytosolic protein, their interaction likely occurs on the cytosolic surface of the ER. Taken together, these data suggest the presence of a previously unidentified specific interaction between Hsp27 and CerS1 occurring on the ER-cytosol interface.
Figure 1. Determination of CerS1-Hsp27 association.
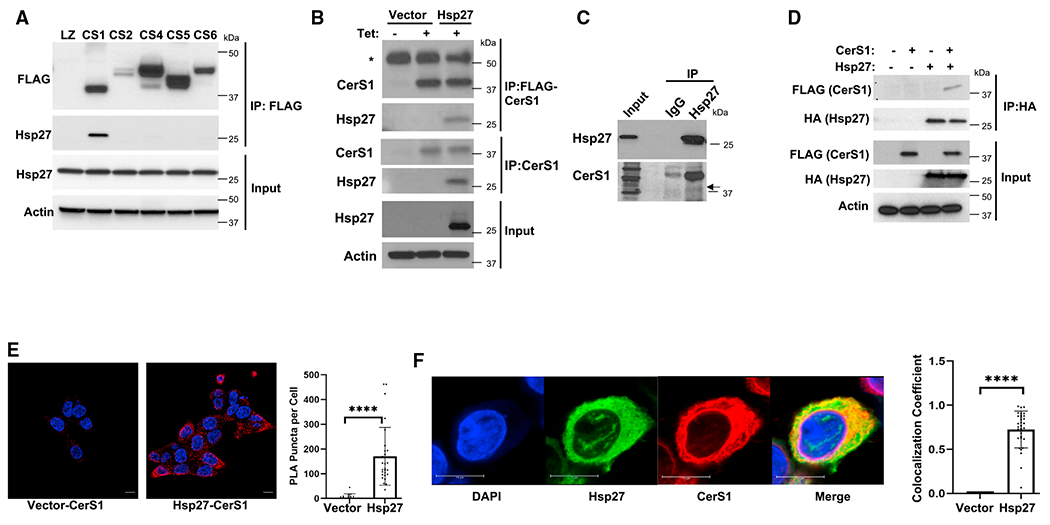
(A) FLAG-tagged CerS1, CerS2, CerS4, CerS5, and CerS6 were immunoprecipitated, and the interaction of Hsp27 with CerS proteins was determined by western blotting in HCT116 cells.
(B) HA-tagged Hsp27 in pcDNA4 plasmid was expressed in HCT116 cells stably transfected with Tet-inducible FLAG-CerS1. pcDNA4 plasmid without insert (Vector) was used as control. CerS1-Hsp27 interaction was determined by western blotting after immunoprecipitation (IP) with anti-FLAG or anti-CerS1 antibodies. Asterisk indicates heavy-chain immunoglobulin G.
(C) Interaction of endogenous Hsp27 with CerS1 was detected using IP with anti-Hsp27 antibody followed by western blotting using anti-CerS1 antibody.
(D) HA-tagged Hsp27 was expressed as in (B), and CerS1-Hsp27 interaction was determined by western blotting after IP with anti-HA antibody.
(E) Interaction of FLAG-tagged CerS1 and HA-Hsp27 was identified using proximity ligation assay (PLA) as described in STAR Methods (scale bar, 10 μm) (left). PLA signal was quantitated (right). n ≥ 3 for all samples from three independent experiments. Data represent mean ± SD. Statistical analysis by two-tailed unpaired t test, ****p < 0.0001.
(F) Cellular co-localization of HA-Hsp27 with FLAG-CerS1 proteins was determined by immunofluorescence confocal laser scanning microscopy (scale bar, 10 μm) (left). Co-localization of Hsp27 and CerS1 was quantitated as Manders’ coefficient (right). n = 31 for each group. Data represent mean ± SD. Statistical analysis by two-tailed unpaired t test, ****p < 0.0001.
Hsp27 inhibits CerS1 activity through interaction via its N terminus
HSPs generally function to restore the folding of their client proteins to re-establish a functional client in adaptive responses. On the other hand, maladaptive changes of HSP proteins have been documented in pathological conditions. Notably, elevated Hsp27 mRNA abundance is associated with resistance to chemotherapy, increased metastasis potential, and poor overall prognosis for cancer patients.60–62 Importantly, reduced CerS1 expression and/or C18:0-ceramide generation is also associated with similar poor prognostic factors in cancer.63–65 Therefore, we hypothesized that Hsp27 acts as an endogenous inhibitor of CerS1 through physical association in cancer. To test this hypothesis, we first investigated the biochemical relevance of Hsp27-CerS1 interaction. Ectopic expression of Hsp27 (Figure 2A, upper panel) significantly inhibited the production of NBD-C18:0-ceramide, a CerS1-specific ceramide product, but not the production of NBD-C16:0-ceramide (CerS5/6 product) in in vitro activity assays (Figure 2A, lower panel). In an independent project, while screening cell lines for differences in CerS activities, we identified HepG2 cells to have elevated endogenous CerS1 activity compared to HCT116 cells. Whereas CerS1 protein abundance is slightly lower in HCT116 cells compared to HepG2 cells (Figure S2A), coincidentally, HepG2 cells have lower Hsp27 protein abundance compared to HCT116 cells (Figure 2B, upper panel, lanes 1 and 2). While elevated CerS1 activity in HepG2 cells may be due to differences in the CerS1 levels in the basal conditions, more importantly, ectopic Hsp27 expression in HepG2 cells still caused a decrease in the endogenous CerS1 activity when measured in vitro (Figure 2B, lower panel). Since ectopic expression of proteins can cause other unforeseen changes in the cells that can affect enzymatic activities, we expressed and purified recombinant Hsp27 (Figures S2B and 2C) and tested its effects on CerS1 in in vitro enzyme activity assays. Remarkably, purified Hsp27 caused a concentration-dependent decrease in CerS1 activity (Figure 2D) when included in the enzyme activity assay reaction, suggesting that Hsp27 directly inhibits CerS1 enzymatic activity.
Figure 2. Hsp27 inhibits CerS1 activity in cells and in vitro.
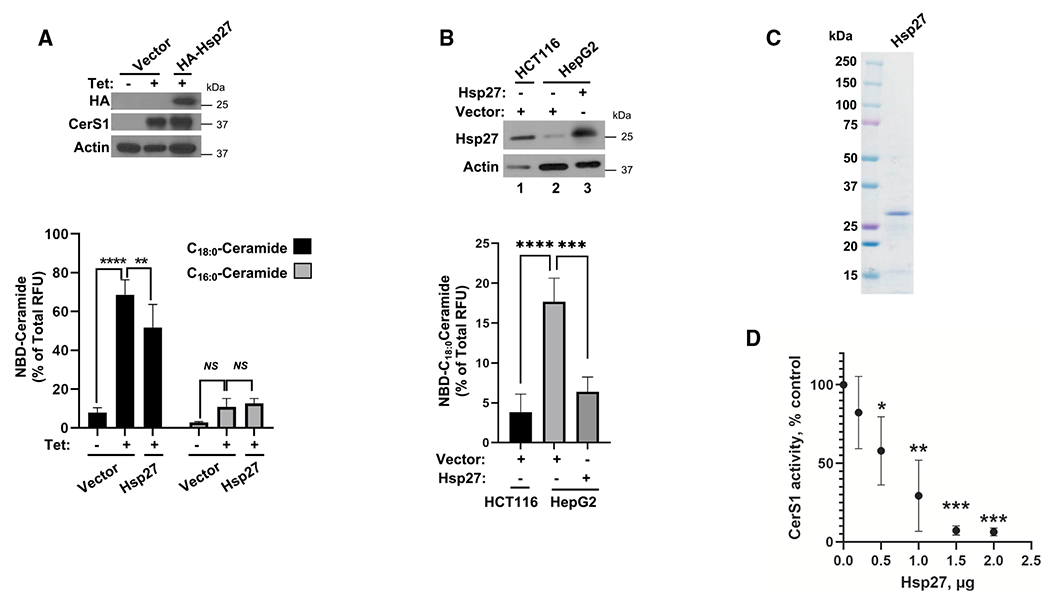
(A) Ectopic expression of Hsp27 in pcDNA4 plasmid in HCT116 cells that stably express Tet-inducible CerS1 is confirmed by western blotting (top). pcDNA4 plasmid without insert (Vector) was used as control. In vitro ceramide synthase activities of CerS1 and CerS5/6 were measured after Hsp27 expression using C18-CoA and C16-CoA substrates, respectively (bottom). n ≥ 4 for all samples from independent experiments. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, ****p < 0.0001, **p < 0.005, NSp > 0.05.
(B) Ectopic expression of Hsp27 was confirmed by western blotting (top). Effect of Hsp27 expression on CerS1 in vitro activity was measured in microsomes (bottom). n ≥ 3 for all samples from at least three independent experiments. Data represent mean ± SD. Statistical analysis by one-way ANOVA with Tukey’s multiple comparison test, ****p < 0.0001.
(C) Purity of recombinantly produced Hsp27 was confirmed using Coomassie blue staining.
(D) Effect of recombinant Hsp27 on microsomal CerS1 enzymatic activity was measured as described in STAR Methods. n ≥ 3 independent experiments for each group with 2–3 technical replicates. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, *p < 0.05, **p < 0.01, ***p < 0.001.
To gain a more detailed understanding of how CerS1 and Hsp27 interact, we investigated the peptide regions, which are important for the Hsp27-CerS1 association. We generated multiple deletion mutants of Hsp27 (Figure 3A, upper panel) and tested their interaction with CerS1 using IP. Deletion of the first 84 amino acids of Hsp27 (ΔN) greatly reduced its association with CerS1, while deletion of C-terminal amino acids (ΔC) did not have a significant effect on CerS1 interaction (Figure 3A, lower panel). In addition, the α-crystallin domain of Hsp27 alone, lacking both the N and C termini (core), had a significantly reduced association with CerS1 (Figure 3A, lower panel), indicating that the α-crystallin domain of Hsp27 is also important for the binding but that this core on its own is not sufficient to support CerS1 interaction, since the N terminus is also required. As our data indicated the importance of the N terminus of Hsp27 for CerS1 interaction, we next purified recombinant ΔN-Hsp27 (Figure 3B, left panel) and tested its effects on CerS1 activity in vitro. Consistent with its limited interaction with CerS1, recombinant ΔN-Hsp27 failed to inhibit CerS1 activity compared to wild-type Hsp27 (Figure 3B, right panel). These results collectively suggest that the N terminus of Hsp27 is required for Hsp27-CerS1 association and possible steric inhibition of CerS1. To test whether the N terminus of Hsp27 alone can support these functions, the amino acids 1–84 of Hsp27 were cloned adjacent to the N terminus of enhanced GFP (eGFP), and association of this chimera (N-term-eGFP) (Figure 3C, upper panel) with CerS1 was tested. N-term-eGFP association with CerS1 was modestly detected but significantly reduced compared to the wild-type Hsp27 (Figure 3C, lower panel). Concomitantly, ectopic expression of N-term-eGFP did not cause a significant change in CerS1 activity in the in vitro settings (Figure 3D), implicating that the N terminus of Hsp27 alone cannot efficiently interact with or inhibit CerS1 activity. Taken together, Hsp27 can inhibit CerS1 activity sterically by interacting with CerS1, which requires the N-terminal amino acids (1–84) of Hsp27 in the presence of the α-crystallin domain.
Figure 3. Identification of domains in Hsp27 that are required for CerS1-Hsp27 association.
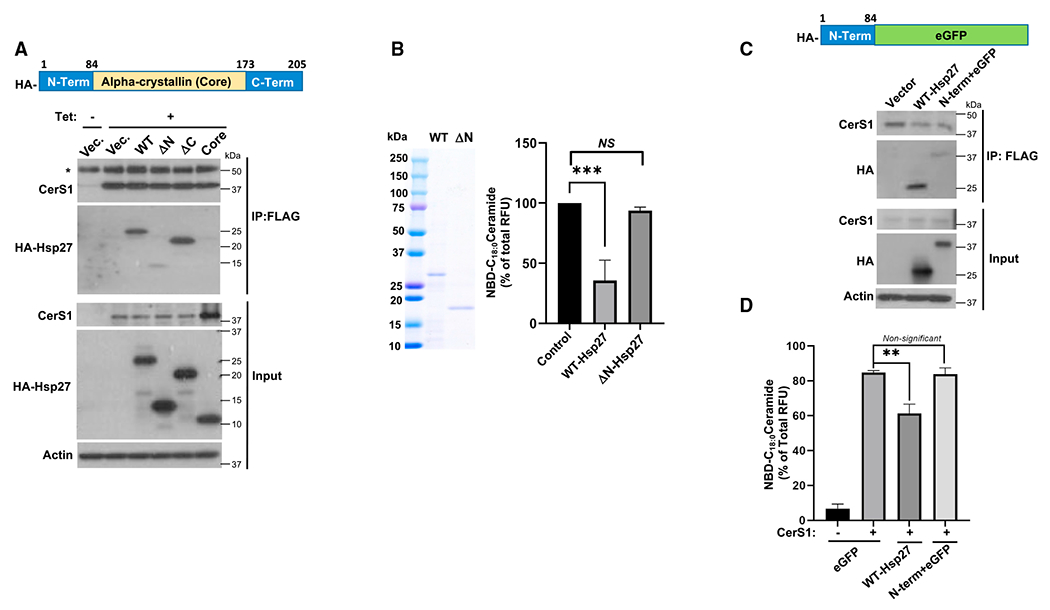
(A) Top: schematic representation of Hsp27 protein with domains that were deleted or mutated. N-Term, N-terminal amino acids 1–84; Alpha-crystallin (Core), amino acids 84–173; C-Term, C-terminal amino acids 173–205. Bottom: association of wild type (WT) and deletion mutants of HA-tagged Hsp27 in pcDNA4 plasmid with FLAG-tagged CerS1 was determined in HCT116 cells by anti-FLAG IP followed by western blotting using an anti-HA antibody. pcDNA4 plasmid without insert (Vector) was used as control. Heavy chain of the immunoprecipitating antibody is indicated by an asterisk.
(B) Purity of recombinantly produced WT and N terminus deleted mutant of Hsp27 was confirmed using Coomassie blue staining (left), and their effects on microsomal CerS1 enzymatic activity were determined (right). n = 4 independent experiments for each group with 2–3 technical replicates. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, ***p <0.001.
(C) Top: schematic representation of the HA-tagged chimeric protein (N-Term + eGFP) containing the first 84 amino acids of Hsp27 (N-Term) upstream of enhanced green fluorescent protein (eGFP). Interaction of HA-tagged WT and N-Term + eGFP with FLAG-tagged CerS1 was identified by anti-FLAG IP followed by western blotting using anti-HA antibody (bottom).
(D) In vitro CerS1 activity was measured in microsomes from cells expressing vector (eGFP), WT Hsp27, or chimeric protein (N-Term + eGFP). n = 3 independent experiments for each group with 2–3 technical replicates. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, **p < 0.05, NSp > 0.05.
Phosphorylation of Hsp27 dictates its interaction with CerS1
Hsp27 can be phosphorylated by the p38-MK2 MAPKAP kinase pathway at its N-terminal residues S15, S78, and S82,53–56,66 resulting in changes in its biological functions. Given that we have identified the N terminus of Hsp27 to be crucial for the binding and inhibition of CerS1, we investigated whether the phosphorylation of Hsp27 modulates Hsp27-CerS1 association. Osmotic stress is a known activator of p38, an upstream kinase in Hsp27 phosphorylation.67 Therefore, we first tested the effect of sorbitol-induced osmotic stress on p38 activation and Hsp27 phosphorylation in HCT116 cells. Sorbitol treatment induced p38 and Hsp27 phosphorylation within 15 min (Figure 4A). More importantly, osmotic-stress-induced phosphorylation of Hsp27 caused an increase in the association of Hsp27 with CerS1 (Figure 4B). Additionally, UV irradiation, another known activator of p38 MAPK,68 induced phosphorylation of Hsp27 (Figure S3A) and caused an increase in the association of Hsp27 with CerS1 (Figure 4C).
Figure 4. Phosphorylation of Hsp27 enhances Hsp27-CerS1 interaction and amplifies inhibition of CerS1 activity.
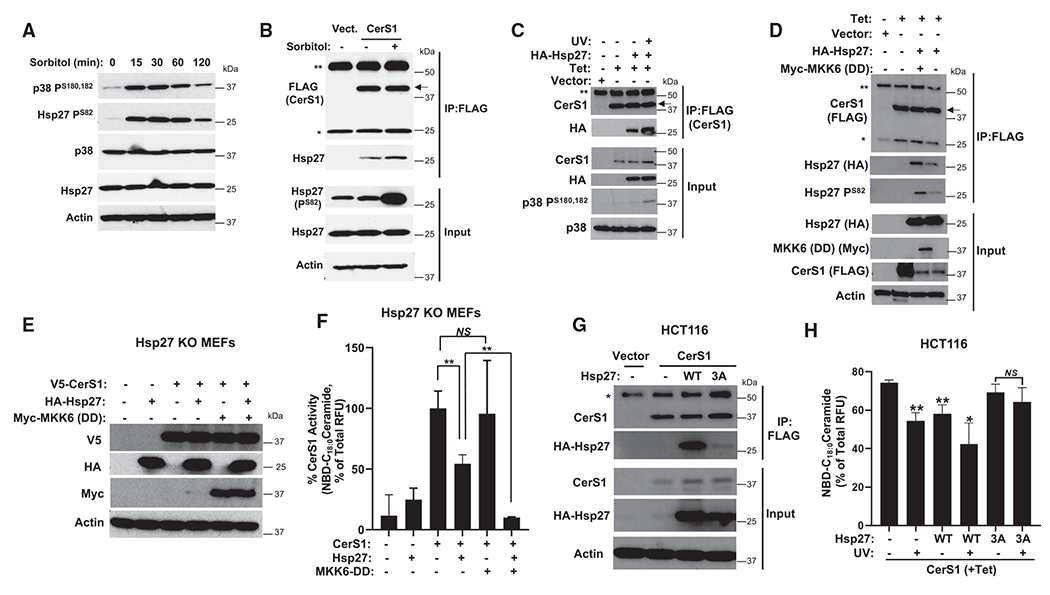
(A) Phosphorylation of Hsp27 and p38 MAPK was detected after sorbitol-induced osmotic stress by western blotting in HCT116 cells.
(B and C) (B) Association of Hsp27 with CerS1 in control or sorbitol-treated cells was determined using IP followed by western blotting. pcDNA4 plasmid without insert (Vector) was used as control. Double asterisk (**) and single asterisk (*) denote heavy and light chain of immunoprecipitating antibody, respectively. Arrow points to specific band. (C) Association of Hsp27 with CerS1 was detected in cells after induction of Hsp27 phosphorylation with UV treatment in Tet-inducible CerS1-expressing cells.
(D) Tet-inducible CerS1-expressing HCT116 cells were transfected with indicated plasmids, and Hsp27-CerS1 association was determined.
(E) Hsp27 KO MEFs were transduced with indicated viral particles, and their expression was confirmed by western blotting.
(F) Hsp27 KO MEFs were transduced as in (E), and CerS1 microsomal activity was determined. n = 3 independent experiments for each group with 2–3 technical replicates. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, **p < 0.05, NSp > 0.05.
(G) Association of wild type (WT) or S15A, S78A, and S82A (3A) mutant of Hsp27 with CerS1 was detected in HCT116 cells.
(H) CerS1 microsomal activity was determined from HCT116 cells expressing WT or S15A, S78A, and S82A (3A) point mutant of Hsp27 after UV treatment. n = 8–10 for each group. Data represent mean ± SD. Statistical analysis by one-way ANOVA with Sidak’s multiple comparison test, **p < 0.05, ***p < 0.01.
External stimuli such as osmotic stress and UV can activate a multitude of signaling pathways in addition to the p38 MAPK pathway. Therefore, we induced p38 activation using ectopic expression of a constitutively active upstream kinase, MKK6, with point mutations at S207D and T211D (DD),69,70 and found that MKK6-DD induced phosphorylation of Hsp27 and increased its association with CerS1 (Figure 4D). To investigate the biochemical relevance without the possible interference of endogenous Hsp27, we used Hsp27-knockout mouse embryonic fibroblasts (Hsp27 KO MEFs)71 to express Hsp27 and MKK6-DD (Figure 4E) and measured the CerS1 enzyme activity. Hsp27 expression inhibited CerS1 activity, and this inhibition was greatly enhanced when MKK6-DD was co-expressed (Figure 4F), suggesting that phosphorylation of Hsp27 enhances the ability of Hsp27 to inhibit CerS1 activity by regulating Hsp27-CerS1 association. Importantly, mutagenesis of the three phosphorylation sites to alanine (S15A, S78A, S82A) in Hsp27 (3A) greatly reduced its interaction with CerS1 compared to wild type (Figure 4G). Moreover, while expression of wild-type Hsp27 inhibited CerS1 activity alone and UV treatment amplified this inhibition, the 3A mutant of Hsp27 did not affect CerS1 activity alone or after UV treatment even in the presence of endogenous Hsp27 in HCT116 cells (Figure 4H). We did not observe the UV-induced decrease in CerS1 activity when 3A mutant was expressed in the cells, implying that the ectopically expressed mutant Hsp27 can hinder the activity of endogenous protein, possibly acting as a dominant-negative mutant.26 Likewise, the differential effects of wild-type and 3A mutant Hsp27 on CerS1 activity were also observed in HepG2 cells (Figures S3B and S3C). Taken together, these data suggest that Hsp27 phosphorylation at its N terminus is crucial in modulating the inhibition of CerS1 by Hsp27 by regulating Hsp27 interaction with CerS1.
Hsp27 downregulation induces CerS1 activation in situ and potentiates the effects of CerS1-induced mitophagy
After investigating the effects of Hsp27 on CerS1 activity in vitro, we explored the possible regulation of CerS1 by Hsp27 in cells. To achieve this, we determined the effects of Hsp27 downregulation on CerS activity in situ, using a 17C-sphingosine-labeling mass spectrometry approach. Our data showed increased CerS1 activity upon downregulation of Hsp27, as evident by the increased generation of 17C-C18:0-ceramide and 17C-C18:1-ceramide generation (Figures 5A and 5B), which is consistent with our observations of elevated CerS1 activity in vitro.
Figure 5. Downregulation of Hsp27 induces CerS1 activity in cells.
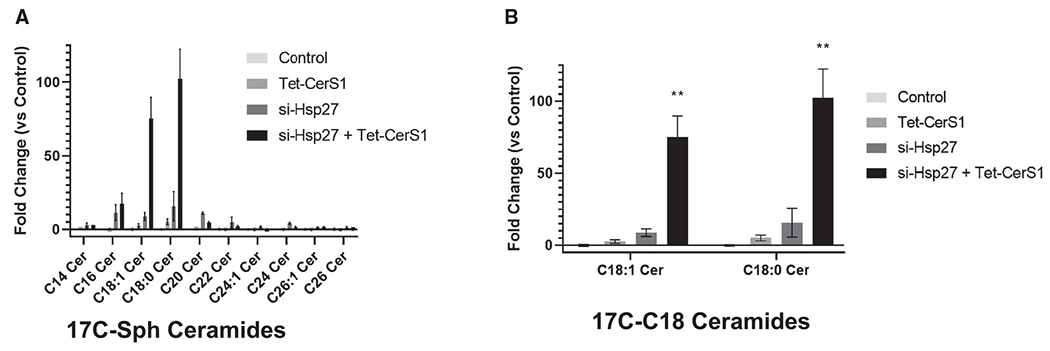
(A) In situ CerS activity was measured by liquid chromatography-mass spectrometry using 17C-sphingosine (Sph) labeling of cells that express Tet-inducible CerS1 without or with Hsp27 knockdown in HCT116 cells. Ceramide species with 17C-Sph backbone are shown.
(B) C18:1 and C18:0 ceramides with 17C-Sph backbone are shown. n = 3 independent experiments for each group. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, **p < 0.01.
Next, we examined the biological significance of Hsp27-CerS1 association in the cells and focused on lethal mitophagy, one of the established biologies shown to be regulated by CerS1.14,15,72,73 Mitophagic vesicle formation, defined by LysoTracker and MitoTracker co-staining, was observed in Tet-induced CerS1-expressing cells compared to the controls (Figure 6A, upper panels), consistent with the previously published reports.14,15,72,73 More importantly, downregulation of Hsp27 alone induced mitophagy and further significantly enhanced CerS1-induced mitophagic vesicle formation (Figure 6A, lower right panel). Moreover, we found enhanced LC3B-II formation and decreased Aco2 protein abundance, which are known molecular markers of autophagy/mitophagy, in cells with Tet induction and Hsp27 small interfering RNA (siRNA) transfections (Figure 6B). Similarly, the effects of the pharmacological mitophagy inducer, sodium selenite,72,73 on cell viability were augmented in the presence of Hsp27 downregulation (Figure S4A). Of note, sodium selenite-induced mitophagy is dependent on CerS1 (Figure S4B). Furthermore, in parallel with increased mitophagy, mitochondrial function further declined upon Hsp27 downregulation. Mitochondrial membrane potential as determined by JC1 staining measured by flow cytometry (Figure 6C), cellular total ATP levels (Figure 6D), and mitochondrial bioenergetic activity measured by Mito Stress assays using a Seahorse analyzer (Figures 6E–6G) were further reduced upon Hsp27 downregulation. Additionally, the loss of mitochondrial function was reflected in decreased cellular viability, consistent with an increase in CerS1-regulated lethal mitophagy (Figures S4C and S4D). Overall, these data support the notion that Hsp27-CerS1 interaction modulates the activity of CerS1 in cells and, hence, modifies C18:0-ceramide generation and its downstream biological functions.
Figure 6. Downregulation of Hsp27 potentiates CerS1-induced mitophagy.
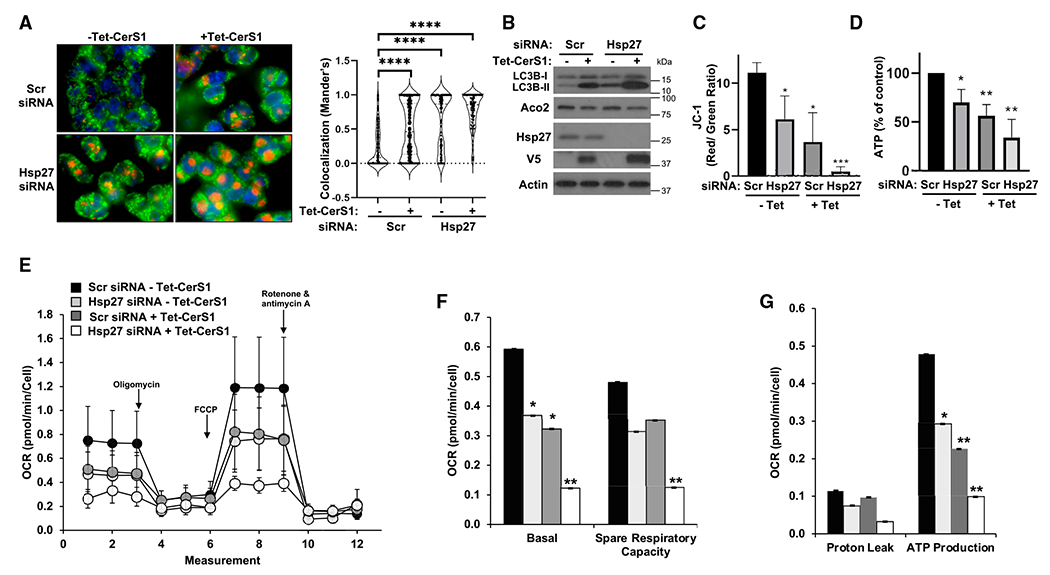
(A) Lysosomal clearance of mitochondria (mitophagic vesicle formation) was detected by co-staining of cells with MitoTracker green and LysoTracker red dye colocalization (left) in HCT116 cells. Co-localization was quantitated as described in STAR Methods (right). n ≥ 99 for each group. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Tukey’s multiple comparison test, ****p < 0.0001.
(B) LC3B-II formation and decrease in mitochondrial Aco2 were detected by western blotting.
(C) Changes in mitochondrial membrane potential was determined by JC-1 staining and flow cytometry in control (–Tet) or CerS1-expressing cells (+Tet) after transfections with control, non-targeting (Scr), or Hsp27 siRNA. n = 3 independent experiments for each group with 2–3 technical replicates. Data represent mean ± SD. Statistical analysis by one-way ANOVA with Dunnett’s multiple comparison test, *p < 0.05, ***p < 0.001.
(D) Cellular total ATP levels were quantitated as described in STAR Methods.
(E) Oxygen consumption rate was measured using Mito Stress Assay Kit in a XFe24 Seahorse bioanalyzer instrument.
(F and G) (F) Basal and spare respiratory capacity, and (G) proton leak and ATP production were calculated from data obtained in (E) as described by the manufacturer. n = 15 for each group from three independent experiments of five replicates. Data represent mean ± SD. Statistical analysis by two-way ANOVA with Dunnett’s multiple comparison test, *p < 0.05, ***p < 0.001.
To investigate whether CerS1 is required for the effects of Hsp27 knockdown on mitochondria and cell survival, we down-regulated CerS1 in the presence of Hsp27 siRNA in HCT116 cells (Figure S4E). While Hsp27 knockdown induced mitochondrial fission, a known CerS1-induced phenotype,14,15,72,73 mitochondrial fission was prevented when cells were co-transfected with CerS1 siRNA (Figure 7A, left panel). Quantitation of mitochondrial branching via image analysis confirmed these results. Reduction of mitochondrial branching, an indicator of mitochondrial fission, upon Hsp27 knockdown was prevented when CerS1 was downregulated (Figure 7A, right panel). In addition, loss of mitochondrial membrane potential due to Hsp27 knockdown was significantly prevented by silencing of CerS1 as measured by accumulation of tetramethylrhodamine methyl ester (TMRM), a cell-permeable dye that accumulates in active mitochondria (Figure 7B). Consistent with the prevention of mitochondrial defects, silencing of CerS1 expression also prevented Hsp27-knockdown-induced cell death (Figure 7C). Taken together, these data suggest that CerS1 is a direct downstream target of Hsp27 that is required for the induction of lethal mitophagy upon Hsp27 knockdown.
Figure 7. Mitochondrial defects induced by Hsp27 knockdown are CerS1 dependent.
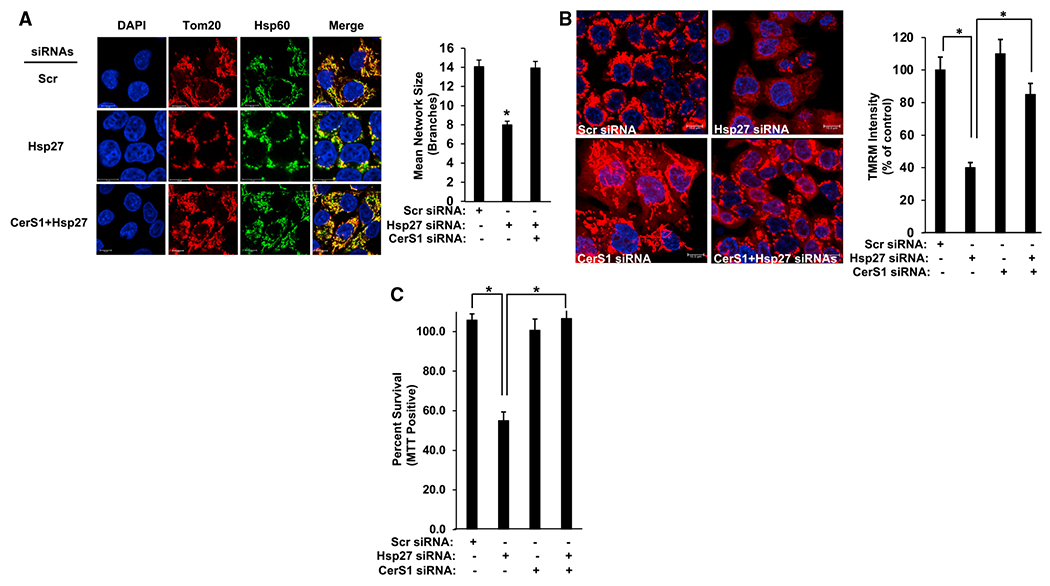
(A) Left: mitochondria are observed by Tom20 and Hsp60 co-staining in HCT116 cells transfected with control (Scr), Hsp27, or CerS1 and Hsp27 siRNAs, as described in STAR Methods. Scale bars, 10 μm. Right: mitochondrial mean network size (branching) was determined by MiNA (mitochondrial network) plug-in in ImageJ as described. n = 3 for each group. Data represent mean ± SD. Statistical analysis by one-way ANOVA with Tukey’s multiple comparison test, *p < 0.05.
(B) Left: mitochondrial membrane potential was visualized using TMRM in cells after transfections with control (Scr), Hsp27, CerS1, or CerS1 and Hsp27 siRNAs. Scale bars, 10 μm. Right: TMRM intensity was measured in the images as described in STAR Methods. n = 3 for each group. Data represent mean ± SD. Statistical analysis by one-way ANOVA with Tukey’s multiple comparison test, *p < 0.05.
(C) Cell viability was determined using MTT positivity in cells transfected with control (Scr), Hsp27, CerS1, or CerS1 and Hsp27 siRNAs. n = 3 independent experiments for each group with two technical replicates. Data represent mean ± SD. Statistical analysis by one-way ANOVA with Tukey’s multiple comparison test, *p < 0.05.
DISCUSSION
In this study, we identified Hsp27 as an important regulator of CerS1. More importantly, our data show that through the protein-protein interaction, Hsp27 can inhibit CerS1 enzymatic activity in vitro and in cells. This interaction appears to depend on the phosphorylation of Hsp27 on its N terminus, as the deletion mutant of Hsp27 lacking this domain neither interacts with CerS1 nor can inhibit CerS1 activity. Moreover, phosphodeficient mutant (S15A, S78A, S82A) of Hsp27 does not interact or inhibit CerS1. Mechanistically, p38-MK2 kinase cascade-induced phosphorylation of Hsp27 on these sites can influence the affinity of Hsp27 to CerS1, pointing to a fine-tuning ability of cells in regulating CerS1 activity in stress-response conditions. Biologically, downregulation of Hsp27 increased CerS1 activity in cells and induced CerS1-dependent mitophagy and cell death, implicating that Hsp27 keeps CerS1 in an inactive state as an anti-cell-death measure in cancers. Collectively, this report identifies a crucial role for Hsp27 in regulating CerS1 activity and downstream CerS1-generated ceramide-induced mitophagy and cell death.
The CerS enzymes preferentially use fatty acyl-CoAs with different chain lengths to generate distinct ceramide species.3,58,74 The specificity of CerS for particular fatty acyl-CoA consumption is attributed to the differences of CerS enzymes in a short segment of amino acids located in a predicted ER luminal loop.75,76 In the current study, Hsp27 preferentially interacted with CerS1 but not with the other CerS enzymes tested. At this point, determinants of this specific affinity of Hsp27 toward CerS1 are unknown. One possible mechanism might involve the engagement of the substrate-selecting domain of CerS1 in Hsp27 recognition, and future studies will help define the mechanisms of apparent specificity. In addition, CerS1 is the only known CerS homolog that lacks the hox-like domain in its sequence.77 The hox-like domain might be actively preventing the interaction between Hsp27 and the other CerS homologs. Another possibility is that the C-18:0 ceramide that is generated by CerS1 facilitates the interaction between CerS1 and Hsp27, as CerS1 is highly specific for the generation of this ceramide species.
The activity of CerS proteins is considered to be regulated by homo- and heterodimerization,78–80 phosphorylation,81 and acetylation.82 We observed a slight increase in the CerS in vitro activity toward NBD-C16:0-ceramide generation after CerS1 expression, which was not affected by Hsp27, while C18:0-ceramide generation was inhibited (Figure 2A). This suggests that CerS1 might be dimerizing with CerS5/6 to influence their activity which is not modulated by Hsp27. Although not as robust as CerS1, CerS4 can also generate C18:0 ceramide in vitro83 and when overexpressed in cells.84 Recently, fatty acid transport protein 2 (FATP2) and CerS2 interaction has been documented.85 FATP2 might deliver the FA-CoA substrate for the CerS2 catalytic activity via direct interaction in cells.85 At this point, we do not know whether Hsp27 is involved in the supplementation of C18:0-FA-CoA to CerS1. Association between the ribosomal protein RPL29P31 (Permit) and CerS1 was shown to direct the localization of newly translated CerS1 from the ER to the outer mitochondrial membrane (OMM) and facilitate mitophagy.73 Our confocal microscopy data suggest that the interaction of Hsp27 and CerS1 is occurring on the ER/cytoplasm interface. Currently, we do not know whether Hsp27-CerS1 interaction can prevent the transport of CerS1 to the OMM via Permit. Moreover, it is possible that the effects we observe on mitochondria upon Hsp27 downregulation and, hence, CerS1 activation is due to accumulation of C18:0 ceramide in the mitochondria-associated membrane extension of ER without the involvement of Permit. Interestingly, our previous studies showed that Charcot-Marie-Tooth disease-associated mutants of Hsp27 can cause decreased mitochondrial ceramide generation.86 A role for Permit in the Hsp27-mediated regulation of CerS1 activity is still a possibility.
Mechanistically, our results suggest that the N terminus of Hsp27 is required for its interaction with CerS1. From the structural point of view, sHSPs can exist in the form of monomers and dimers but can also assemble into large multimeric complexes that vary in size and contain up to 24–40 subunits through interactions via the α-crystallin domain.87 The N terminus of Hsp27 is identified as a disordered domain that can influence Hsp27 binding affinity toward Tau protein.34 In addition, the phosphorylation sites of Hsp27 that we have identified to be critical for determining CerS1 interaction also lie in this region. Furthermore, the α-crystallin domain itself did not support the interaction with CerS1. These findings raise the possibility that the N terminus and phosphorylation of Hsp27 might structurally change the oligomerization status and/or affinity to binding partners in coordination with the α-crystallin domain.
Multiple roles of sphingolipid signaling have been well documented in cellular response to stress conditions. In the current study, activation of the p38-MK2 MAPK pathway via osmotic stress, UV exposure, or upstream kinase (MKK6) activation induced phosphorylation of Hsp27, which in turn resulted in amplified Hsp27-CerS1 interaction and further modulation of CerS1 enzymatic activity. Involvement of p38 in degradation of CerS1 in response to cisplatin was previously implicated.88 However, importantly, our study is one of the first examples of acute regulation of CerS enzymes that is dependent on kinase cascade activation.
Hsp27 has been recognized as a valid target for anti-cancer therapy. Notably, a second-generation anti-sense oligonucleotide gene therapy targeting Hsp27 (OGX-427) is being tested in clinical trials for prostate, bladder, ovarian, pancreas, and lung cancers.89–92 Moreover, Hsp27-regulated downstream pathways can also be therapeutic targets for cancer.93–96 Recently, a crucial role for Hsp27 in Apc-driven transformation and colorectal cancer formation was also established.97 Furthermore, a role for MK2-mediated phosphorylation of Hsp27 in mesenchymal cells was demonstrated in intestinal carcinogenesis.94 Additionally, the MK2/Hsp27 axis acts as a survival mechanism for pancreatic cancers.98 Along these lines, inhibitors of MK2 are proposed as a viable approach to the treatment of colitis and colitis-associated colon cancers.99–101 In our study, downregulation of Hsp27 induced CerS1-dependent mitophagy and cell death in colon cancer cells, bringing the possibility that CerS1 inactivation via Hsp27 is a critical step in cellular transformation and tumor formation. In addition, data show that the phosphorylated form of Hsp27 had an increased affinity to CerS1 and further suppressed CerS1 activity in comparison with the unphosphorylated counterpart. Our study, along with the mentioned previous work, raises the possibility of using possible changes in ceramide concentrations in the sera of cancer patients as a biomarker to determine the patient population that can/will benefit from anti-Hsp27 or anti-MK2 modalities.
In summary, we describe a substantial mechanism of CerS1 regulation via direct interaction with Hsp27, shed light on the molecular underpinnings of this interaction, and show its biological significance in controlling mitochondrial energy metabolism and mitophagy. In addition, we provide mechanistic information on how Hsp27 abundance in cancer can be an anti-cell-death factor. Further studies will define the functions of this interaction in diabetes, aging, and neuropathies in which a role for CerS1 has already been implicated.86,102–104
Limitations of the study
One limitation of this study is that we still do not know whether Hsp27 can respond to changes in the cellular C18:0-ceramide levels and act to inhibit CerS1. This type of regulatory mechanism might exist in non-pathological settings as an extension of a more complex regulatory circuit. Addressing this point would be a future challenge and contribute to our current understanding of the mechanisms that maintain the sphingolipid homeostasis. Another question that remained unanswered is whether there is crosstalk between CerS1 and other CerS enzymes upon Hsp27-mediated inhibition of CerS1. In the current study we did not observe any change in the in vitro activity toward C16:0-ceramide generation; however, we did not determine the activities toward other ceramides, e.g., C24:0 ceramide for CerS2 activity. In addition, although not as robust as CerS1, CerS4 can also generate C18:0 ceramide in vitro83 and when overexpressed in cells.84 We did not detect any Hsp27-CerS4 interaction. However, there is a possibility that in cell lines with elevated CerS4 expression, changes in the cellular C18:0 ceramide may not be attributed only to CerS1.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Can E. Senkal (Can.Senkal@vcuhealth.org).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
Original, uncropped western blot image files have been deposited at Mendeley Data and are publicly available as of the date of publication, Mendeley Data, V1, https://doi.org/10.17632/jd96sv7fts.1.
All the statistical data is available in Table S1.
This study does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell lines and growth conditions
HCT116 colorectal carcinoma cells were originally provided by Dr. Richard J. Youle (NINDS, NIH, USA). Cells were grown in DMEM (cat# 10-017-CV, Corning) supplemented with 10% FBS (Cat# S12495, lot:K20158, Bio-Techne) without antibiotics at 37°C humidified incubator with 5% CO2. Stable expression of tetracycline inducible CerS1 in HCT116 cells was performed as described previously.59 Hsp27 knock-out MEFs were kindly provided by Dr. Jonathan Dean.71 Possible mycoplasma contaminations were monitored regularly by MycoAlert mycoplasma detection kit (Lonza).
METHOD DETAILS
Plasmid constructs and siRNAs
cDNA encoding FLAG-tagged CerS1 in pLenti6.3/TO/DEST plasmid was generated as described.59 cDNA encoding HA-tagged Hsp27 was amplified by PCR and cloned between XhoI and EcoRI sites in pcDNA4 plasmid. Deletion mutants of Hsp27 was generated using PCR and cloned into the same sites in pcDNA4 plasmid. Expression plasmid encoding myc-tagged MKK6 (DD) was a kind gift from Cindy Miranti and was obtained from Addgene (plasmid #86094). Plasmid encoding Hsp27 mutant with S15A, S78A, and S82A triple mutation (3A) was obtained from Addgene (plasmid #84997) and 3A Hsp27 was subcloned into pcDNA4 plasmid. Plasmid transfections were conducted using X-tremeGENE 9 DNA transfection reagent (Roche) transfection reagent as instructed by the manufacturer. Control, non-targeting (Scr) siRNA was obtained from Qiagen (cat# 1027281). SMARTpool siRNAs targeting Hsp27 (cat# L-005269) and CerS1 (cat# L-010275) were from Dharmacon. siRNA transfections were conducted using Lipofectamine RNAiMAX (ThermoFisher) as described by the manufacturer using 20 nM siRNA final concentration for Hsp27 and 5 nM siRNA for CerS1.
Immunoprecipitation, determination of proteins by LC/MS-MS, and western blotting
Anti-FLAG immunoprecipitations were carried out using Anti-FLAG M2 Affinity gel (Sigma, St. Louis, MO) as described by the manufacturer. LC/MS-MS was performed as described previously.59 Anti-HA, anti-CerS1, and anti-Hsp27 immunoprecipitations were carried out using anti-HA antibody (cat# 3724S, Cell Signaling Technology, Danvers, MA), anti-CerS1 antibody (cat# 65096, Santa Cruz Biotechnology, Santa Cruz, CA), and anti-Hsp27 antibody (cat# 2402, Cell Signaling Technology, Danvers, MA), respectively. Cell lysates were incubated with the antibodies overnight at 4°C, and immune complexes were sedimented using A/G plus agarose beads (cat# sc-2003, Santa Cruz Biotechnology, Santa Cruz, CA). Total cell lysates or immunoprecipitated complexes were mixed with 4X sample buffer (200 mM Tris-HCl pH 6.8, 40% glycerol, 8% SDS, 400 mM DTT, and 0.05% bromophenol blue). The samples were boiled for 5 min and resolved in 4–20% Tris-HCl polyacrylamide gels (Thermo Fisher Scientific, Waltham, MA). After transferring the proteins to PVDF membranes (Bio-Rad, Hercules, CA), the blots were blocked with 5% non-fat milk in TBS-T (1X Tris-buffered Saline with 0.3% Tween 20). The blots were incubated with the primary antibodies overnight at 4°C on a platform rocker. After washes with TBS-T and incubation with HRP-labeled secondary antibodies, the blots were washed for a final time and developed using ECL (Thermo Fisher Scientific, Waltham, MA). The primary antibodies used in the study were as follows: Anti-FLAG (#F1804, Sigma-Aldrich), anti-actin (#A3853, Sigma-Aldrich), anti-Hsp27 (cat# 95357, Cell Signaling Technology), anti-HA (cat# 3724S, Cell Signaling Technology) anti-V5 (cat# R960-25, Thermo Fisher Scientific). anti-p38 (cat# 8690S, Cell Signaling Technology), anti-phospho-p38 (cat# 4511, Cell Signaling Technology), anti-phospho Hsp27 (cat# 9709, Cell Signaling Technology). anti-myc (cat# 2276, Cell Signaling Technology), anti-LC3B (cat# 12741, Cell Signaling Technology), anti-Aco2 (cat# 6571T, Cell Signaling Technology). HRP conjugated secondary antibodies (cat# 111-035-003, cat# 115-035-003, and cat# 705-035-147, all 1:5000) were from Jackson ImmunoResearch (West Grove, PA).
Proximity ligation assay and immunofluorescence confocal microscopy
Proximity ligation assays (PLA) were performed using Duolink in situ orange PLA starter kit (# DUO92102, Sigma-Aldrich) with anti-HA and anti-FLAG antibodies as described by the manufacturer. Immunostaining of FLAG-CerS1 and HA-Hsp27 was performed as described previously.59 The PLA spots and CerS1-Hsp27 cellular localization were detected using a Zeiss LSM880 confocal microscope equipped with 63x PlanApo oil immersion lens (NA 1.4). Images were analyzed by isolating each cell in each image using Fiji (ImageJ), and the Coloc 2 or JACoP plugins were used to quantitate the colocalization using Mander’s M1 coefficient as the readout.
In vitro and in situ ceramide synthase activity assay
In vitro ceramide synthase activity assays were performed as described previously105 with minor modifications. Briefly, cells were mechanically homogenized in 25 mM Tris-HCl pH 7.4, 25 mM KCl, 2 mM MgCl2, and 250 mM sucrose supplemented with protease inhibitor cocktail (K1007, ApexBio). Total microsomes were isolated using centrifugation at 100,000 xg for 1h at 4°C. Microsomes were resuspended in homogenization buffer and incubated with 15 μM NBD-Sphingosine (cat# 25348, Cayman Chemical), 20 μM defatted BSA, and 50 μM fatty acyl-CoA (cat# P9716 (palmitoyl-CoA) and S0802 (stearoyl-CoA), Sigma-Aldrich) in a 100 μL reaction at 37°C. Reactions were terminated by addition of chloroform/methanol (2:1 v/v). The phases were separated by centrifugation at 3000 xg for 10 min at 4°C. The bottom phase was transferred into a new glass vial and the lipids were dried under N2 stream. Lipids were resuspended in methanol and transferred into HPLC vials for injection. The fluorescent NBD-Sph and NBD-ceramide from the reactions were detected and measured as described105 using a Thermo-Fisher Vanquish HPLC coupled with a fluorescence detector. In situ ceramide synthase activity was measured by 17C-Sphingosine labeling as described.106 Briefly, cells were incubated with 1 μM 17C-Sphingosine for 15 min and its incorporation into ceramide in the cells was determined by LC/MS.
Production and purification of recombinant Hsp27
Doubly tagged (HA and His) wild-type and ΔN-Hsp27 were produced by overexpression in E. coli BL21 strain using pET28a plasmid. Briefly, single colonies were grown O/N at 37°C. Fresh LB media was inoculated the next day and induced with 100 μM IPTG when the OD600 reached between 0.5 and 0.6. Cells were grown for another 4h at 37°C before collection by centrifugation at 6000 rpm for 30 min at 4°C. Pellets were resuspended in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 2 μg/mL of leupeptin, 10 μM aprotinin, 1 mM PMSF and 5 mM β-mercaptoethanol). After a freeze-thaw cycle the lysate was centrifuged at 12,000 rpm for 45 min at 4°C and the recombinant proteins were purified using Ni-NTA (cat# 30210, Qiagen) agarose beads. The beads were washed extensively in wash buffer (35 mM Tris-HCl pH 7.5, 0.5 M KCl, 10% glycerol, 15 mM imidazole, 2 μg/mL of leupeptin, 10 μM aprotinin, 1 mM PMSF, and 5 mM β-mercaptoethanol) for 4 times. The proteins were eluted from the Ni-NTA beads using elution buffer (35 mM Tris-HCl pH 7.5, 0.5 M KCl, 10% glycerol, 200 mM imidazole, 2 μg/mL of leupeptin, 10 μM aprotinin, 1 mM PMSF, and 5 mM β-mercaptoethanol). Protein fractions were then separated on SDS-PAGE, stained with Coomassie Brilliant Blue and transferred onto PVDF membranes for western blotting using anti-HA primary antibody.
Mitochondria and lysosome imaging and immunofluorescence microscopy
HCT116 Tet-CerS1 cells were plated in 35mm confocal dishes (cat# P35GC-1.5-10-C, MatTek Corp.) with center wells. The following day the cells were transfected with siRNAs as indicated. 5 h later, CerS1 expression was induced by tetracycline (Tet) treatment. 72 h later, after a 1X PBS wash, cells were incubated in cell culture incubator for 15 min with fresh media with 100 nM of LysoTracker red (LTR) (cat# L12492, Thermofisher Scientific) and 75 nM of MitoTracker Green (MTG) (cat# M7514, Thermo Fisher Scientific). Hoechst dye was added and incubated for an additional 5 min. After incubation, the cells were rinsed three times with media and imaged immediately on a Zeiss 710 confocal microscope. Images were taken with a 63X oil lens using 405 nm, 488 nm, and 647 nm for Hoechst, MTG, and LTR, respectively. Images were analyzed by isolating each cell in each image using Fiji (ImageJ), and the Coloc 2 plugin was used to measure colocalization using Mander’s M1 coefficient as the readout. Tom20 and Hsp60 immunofluorescence confocal imaging was carried out as described59 using anti-Tom20 (cat# FL-145, Santa Cruz Biotechnology) and anti-Hsp60 (cat# 1052, Santa Cruz Biotechnology) antibodies. Mitochondrial mean network branching is measured using ImageJ (FIJI) with MiNA macro extension as described.107 Staining of cells with TMRM (cat# T668, Thermofisher Scientific) was conducted as described by the manufacturer and image intensities were measured using ImageJ (FIJI).
Determination of mitochondrial membrane potential
Cells were plated in 60 mm dishes and allowed to attach overnight. siRNA transfections were conducted as described above. 5 h later, CerS1 expression was induced with tetracycline (Tet, 1 μg/mL) treatment. 72 h later, cells collected and pelleted. To stain, cells were resuspended in 2 μM JC-1 (cat# 15003, Cayman Chemicals) in 2 mL DMEM. After an incubation for 30 min in the cell culture incubator, cells were pelleted and resuspended in PBS. Flow cytometric analysis was done using a Fortessa flow cytometer with the 488 and 561 nm lasers. Filter set used was 488–530/30-A and 561–585/15-A. Single cell population was gated and used for analysis. The ratio of the number of red (561 nm laser) and green (488 nm laser) events were used to represent mitochondrial polarization.
Determination of cellular ATP levels
Cells were plated in a white 96-well clear bottom plate (cat# 3610, Corning) in 100 μL DMEM. Cells were allowed to attach overnight and were then transfected with indicated siRNAs. 5 h later, CerS1 expression was induced with tetracycline. 72 h after induction, the Luminescent ATP Detection Assay Kit (cat# ab113849, Abcam) was used to measure cellular ATP levels. Briefly, 50 μL of detergent was added to the 150 μL of volume already in the wells. The plate was rocked for 5 min at room temperature. Next, 50 μL of substrate from the kit was added and the plate was rocked another 5 min in the dark. The plate was allowed to sit for 10 min in the dark. Luminescence from wells were measured by reading on a luminometer.
Live cell oxygen consumption
A Seahorse XFe24 analyzer (Agilent Technologies) was used to measure oxygen consumption rate (OCR). Briefly, HCT116 cells were plated on Agilent Seahorse XF24 cell culture plates and incubated overnight to ensure cell attachment. The next morning, cells were incubated in a humidified non-CO2 incubator at 37°C for 1 h in assay medium (Seahorse XF Base Media supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose, pH 7.4) and then oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured using the Agilent XF Cell Mito Stress Test Kit (Agilent Technologies) with Wave software. 1.5 μM of oligomycin, 1.5 μM Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin A were used. Cells were treated with 0.025% trypsin-EDTA solution, neutralized with DMEM with 10% FBS, stained with trypan blue then counted. Cell number was used to normalize the measurements from the analyzer.
Determination of cell growth
Cellular growth after treatments or transfections was determined by 3-(4,5-dimethyl-2-thizolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, as described previously.8
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as means ± SD of at least three independent studies (n ≥ 3). Group comparisons were performed with twotailed unpaired t-tests for comparison of two groups, and with one-way or two-way ANOVA for comparison of more than two groups with Tukey’s or Dunnett’s multiple comparison tests using GraphPad Prism Software, version 9.4.1., p < 0.05 was considered significant. All statistical information is provided in Table S1.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-HA | Cell Signaling Technology | Cat# 3724S (RRID:AB_1549585) |
| Goat polyclonal anti-CerS1 | Santa Cruz Biotechnology | Cat# sc-65096 (RRID:AB_2132952) |
| Mouse monoclonal anti-Hsp27 | Cell Signaling Technology | Cat# 2402 (RRID:AB_331761) |
| Rabbit polyclonal Anti-FLAG | Sigma-Aldrich | Cat# F7425 (RRID:AB_439687) |
| Mouse monoclonal Anti-FLAG | Sigma-Aldrich | Cat# F1804 (RRID:AB_262044) |
| Mouse monoclonal anti-Actin | Sigma-Aldrich | Cat# A3853 (RRID:AB_262137) |
| Rabbit monoclonal anti-Hsp27 | Cell Signaling Technology | Cat# 95357 (RRID:AB_2800246) |
| Mouse monoclonal anti-V5 | Thermo Fisher Scientific | Cat# R960-25 (RRID:AB_2556564) |
| Rabbit monoclonal anti-p38 | Cell Signaling Technology | Cat# 8690S (RRID:AB_10999090) |
| Rabbit monoclonal anti-phospho-p38 | Cell Signaling Technology | Cat# 4511 (RRID:AB_2139682) |
| Rabbit monoclonal anti-phospho Hsp27 | Cell Signaling Technology | Cat# 9709 (RRID:AB_11217429) |
| Mouse monoclonal anti-myc | Cell Signaling Technology | Cat# 2276 (RRID:AB_331783) |
| Rabbit monoclonal anti-LC3B | Cell Signaling Technology | Cat# 12741 (RRID:AB_2617131) |
| Rabbit monoclonal anti-Aco2 | Cell Signaling Technology | Cat# 6571 (RRID:AB_2797630) |
| Rabbit polyclonal anti-Tom20 | Santa Cruz Biotechnology | Cat# FL-145 (RRID:AB_2207533) |
| Goat polyclonal anti-Hsp60 | Santa Cruz Biotechnology | Cat# sc-1052 (RRID:AB_631683) |
| Peroxidase AffiniPure Goat Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch | Cat# 111-035-003 (RRID:AB_2313567) |
| Peroxidase AffiniPure Goat Anti-Mouse IgG (H + L) | Jackson ImmunoResearch | Cat# 115-035-003 (RRID:AB_10015289) |
| Peroxidase AffiniPure Donkey Anti-Goat IgG (H + L) | Jackson ImmunoResearch | Cat# 705-035-147 (RRID:AB_2313587) |
| Alexa Fluor 488-Donkey anti-Goat IgG (H + L) | Thermo Fisher Scientific | Cat# A11055 (RRID:AB_2534102) |
| Alexa Fluor 488-Donkey anti-Rabbit IgG (H + L) | Thermo Fisher Scientific | Cat# A21206 (RRID:AB_2535792) |
| Alexa Fluor 647-Donkey anti-Mouse IgG (H + L) | Thermo Fisher Scientific | Cat# A31571 (RRID:AB_162542) |
| Alexa Fluor 647-Donkey anti-Rabbit IgG (H + L) | Thermo Fisher Scientific | Cat# A31573 (RRID:AB_2536183) |
| Bacterial strains | ||
| E. coli BL21 | New England Biolabs | Cat# C2530H |
| Chemicals, peptides and recombinant proteins | ||
| Tween 20 | RPI | Cat# P20370–1.0 |
| NBD-Sphingosine | Cayman Chemical | Cat# 25348 |
| Defatted BSA | GeminiBio | Cat# 700-100P |
| Protease inhibitor cocktail | ApexBio | Cat# K1007 |
| Palmitoyl-CoA | Sigma-Aldrich | Cat# P9716 |
| Stearoyl-CoA | Sigma-Aldrich | Cat# S0802 |
| TMRM | Thermo Fisher Scientific | Cat# T668 |
| JC-1 | Cayman Chemicals | Cat# 15003 |
| LysoTracker red | Thermo Fisher Scientific | Cat# L12492 |
| MitoTracker Green | Thermo Fisher Scientific | Cat# M7514 |
| 17C-Sphingosine | Avanti Polar Lipids | Cat# 860640P-5mg |
| Purified wild type Hsp27 | This study | N/A |
| Purified DN-Hsp27 | This study | N/A |
| Chloroform | Sigma-Aldrich | Cat# C2432-2.5L |
| Methanol | Fisher | Cat# A452-4 |
| Tetracycline | Goldbio | Cat# 64-75-5 |
| Imidazole | RPI | Cat# I52000–100.0 |
| Glycerol | Acros organics | Cat# 15892-0025 |
| IPTG | RPI | Cat# I56100–5.0 |
| Leupeptin | ApexBio | Cat# A2570 |
| Aprotinin | ApexBio | Cat# A2574 |
| PMSF | Sigma-Aldrich | Cat# 93482-50ML-F |
| β-mercaptoethanol | Sigma-Aldrich | Cat# M3148-100ML |
| Oligomycin | Cell Signaling Technology | Cat# 9996L |
| FCCP | Sigma-Aldrich | Cat# C2920-10MG |
| Rotenone | Sigma-Aldrich | Cat# R8875 |
| Antimycin A | Sigma-Aldrich | Cat# A8674-25MG |
| MTT | RPI | Cat# M92050–1.0 |
| Hoechst dye | Thermo Fisher Scientific | Cat# 62249 |
| Trypan blue | GIBCO | Cat# 15250-061 |
| Critical commercial assays | ||
| anti-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat# A2220 (RRID:AB_10063035) |
| MycoAlert mycoplasma detection kit | Lonza | Cat# 75870-454 |
| Roche X-tremeGENE HP DNA Transfection Reagent | Sigma-Aldrich | Cat# 6365787001 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | Cat# 13778-150 |
| ECL | Thermo Fisher Scientific | Cat# 32106 |
| Agilent XF Cell Mito Stress Test Kit | Agilent Technologies | Cat# 103015-100 |
| Duolink in situ orange PLA starter kit | Sigma-Aldrich | Cat# DUO92102 |
| Luminescent ATP Detection Assay Kit | Abcam | Cat# ab113849 |
| Experimental models: Cell lines | ||
| Human: HCT116 | Laboratory of Dr. Richard J. Youle | N/A |
| Mouse: Hsp27 knock-out MEFs | Laboratory of Dr. Jonathan Dean | N/A |
| Recombinant DNA | ||
| pLenti6.3/TO/DEST-FLAG-CerS1 | Senkal et al., 2017 | N/A |
| pcDNA4 -HA-Hsp27 | This study | N/A |
| pcDNA4 -3A-HA-Hsp27 | This study | N/A |
| Expression plasmid encoding myc-MKK6(DD) | Laboratory of Dr. Cindy Miranti | Addgene (plasmid #86094) |
| pET28a-HA-Hsp27-6xHis | This study | N/A |
| pET28a-HA-DNHsp27-6xHis | This study | N/A |
| Oligonucleotides | ||
| Control siRNA | Qiagen | Cat# 1027281 |
| SMARTpool Hsp27 siRNA | Dharmacon | Cat# L-005269 |
| SMARTpool CerS1 siRNA | Dharmacon | Cat# L-010275 |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| ImageJ | NIH, USA | https://imagej.nih.gov/ij/ |
| FlowJo | BDBIOSCIENCES | https://www.flowjo.com/ |
| Wave software | Agilent Technologies | https://www.agilent.com/ |
| Other | ||
| DMEM | Corning | Cat# 10-017-CV |
| Trypsin-EDTA | Corning | Cat# 25-051-CI |
| FBS | Bio-Techne | Cat# S12495, lot:K20158 |
| Mounting Medium with DAPI | Abcam | Cat# ab104139 |
| Ni-NTA agarose | Qiagen | Cat# 30210 |
| A/G plus agarose beads | Santa Cruz Biotechnology | Cat# sc-2003 |
| Non-fat milk | RPI | Cat# M17200–500.0 |
| 4-20% Tris-HCl pre-cast polyacrylamide gels | Thermo Fisher Scientific | Cat# WXP42020BOX |
| PVDF membranes | Bio-Rad Laboratories | Cat# 1620177 |
Highlights.
Ceramide synthase 1 (CerS1), but not the other CerS tested, associates with Hsp27
Hsp27 inhibits CerS1 activity in vitro and in situ via direct interaction
Hsp27-CerS1 interaction is tuned by p38-MK2 MAPK-induced phosphorylation of Hsp27
Knockdown of Hsp27 induces CerS1-dependent mitochondrial defects and mitophagy
ACKNOWLEDGMENTS
This study was supported by NIH/NCI research grant R37 CA239532 and Harrison Scholar funds from Massey Comprehensive Cancer Center to C.E.S. J.S. was supported by a Fulbright Visiting Scholar fellowship and sabbatical support from Birzeit University. We thank the Department of Medicine at Stony Brook University for the pilot project funds. Lipidomics, microscopy, and flow cytometry shared resources at the Massey Comprehensive Cancer Center are funded by NIH/NCI Cancer Center Support grant P30 CA016059.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113081.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Hannun YA, and Obeid LM (2018). Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol 19, 175–191. 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogretmen B. (2018). Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 18, 33–50. 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JW, Park WJ, and Futerman AH (2014). Ceramide synthases as potential targets for therapeutic intervention in human diseases. Biochim. Biophys. Acta 1841, 671–681. 10.1016/j.bbalip.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Zelnik ID, Ventura AE, Kim JL, Silva LC, and Futerman AH (2020). The role of ceramide in regulating endoplasmic reticulum function. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 1865, 158489. 10.1016/j.bbalip.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Fekry B, Jeffries KA, Esmaeilniakooshkghazi A, Ogretmen B, Krupenko SA, and Krupenko NI (2016). CerS6 Is a Novel Transcriptional Target of p53 Protein Activated by Non-genotoxic Stress. J. Biol. Chem 291, 16586–16596. 10.1074/jbc.M116.716902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernández-Corbacho MJ, Canals D, Adada MM, Liu M, Senkal CE, Yi JK, Mao C, Luberto C, Hannun YA, and Obeid LM (2015). Tumor Necrosis Factor-alpha (TNFalpha)-induced Ceramide Generation via Ceramide Synthases Regulates Loss of Focal Adhesion Kinase (FAK) and Programmed Cell Death. J. Biol. Chem 290, 25356–25373. 10.1074/jbc.M115.658658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotolo JA, Mesicek J, Maj J, Truman JP, Haimovitz-Friedman A, Kolesnick R, and Fuks Z (2010). Regulation of ceramide synthase-mediated crypt epithelium apoptosis by DNA damage repair enzymes. Cancer Res. 70, 957–967. 10.1158/0008-5472.CAN-09-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, and Ogretmen B (2007). Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther 6, 712–722. 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 9.Saddoughi SA, Garrett-Mayer E, Chaudhary U, O’Brien PE, Afrin LB, Day TA, Gillespie MB, Sharma AK, Wilhoit CS, Bostick R, et al. (2011). Results of a phase II trial of gemcitabine plus doxorubicin in patients with recurrent head and neck cancers: serum C(1)(8)-ceramide as a novel biomarker for monitoring response. Clin. Cancer Res 17, 6097–6105. 10.1158/1078-0432.CCR-11-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Yin X, Allan R, Lu DD, Maurer CW, Haimovitz-Friedman A, Fuks Z, Shaham S, and Kolesnick R (2008). Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science 322, 110–115. 10.1126/science.1158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, Haimovitz-Friedman A, Fuks Z, and Kolesnick R (2010). Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell. Signal 22, 1300–1307. 10.1016/j.cellsig.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmond V, Dufour F, Poiroux G, Shoji K, Malleter M, Fouqué A, Tauzin S, Rimokh R, Sergent O, Penna A, et al. (2015). Downregulation of ceramide synthase-6 during epithelial-to-mesenchymal transition reduces plasma membrane fluidity and cancer cell motility. Oncogene 34, 996–1005. 10.1038/onc.2014.55. [DOI] [PubMed] [Google Scholar]
- 13.Spassieva SD, Mullen TD, Townsend DM, and Obeid LM (2009). Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J 424, 273–283. 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, et al. (2012). Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat. Chem. Biol 8, 831–838. 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dany M, Gencer S, Nganga R, Thomas RJ, Oleinik N, Baron KD, Szulc ZM, Ruvolo P, Kornblau S, Andreeff M, and Ogretmen B (2016). Targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML. Blood 128, 1944–1958. 10.1182/blood-2016-04-708750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senkal CE, Ponnusamy S, Manevich Y, Meyers-Needham M, Saddoughi SA, Mukhopadyay A, Dent P, Bielawski J, and Ogretmen B (2011). Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network. J. Biol. Chem 286, 42446–42458. 10.1074/jbc.M111.287383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffmann S, Ferreiros N, Birod K, Eberle M, Schreiber Y, Pfeilschifter W, Ziemann U, Pierre S, Scholich K, Grösch S, and Geisslinger G (2012). Ceramide synthase 6 plays a critical role in the development of experimental autoimmune encephalomyelitis. J. Immunol 188, 5723–5733. 10.4049/jimmunol.1103109. [DOI] [PubMed] [Google Scholar]
- 18.Sofi MH, Heinrichs J, Dany M, Nguyen H, Dai M, Bastian D, Schutt S, Wu Y, Daenthanasanmak A, Gencer S, et al. (2017). Ceramide synthesis regulates T cell activity and GVHD development. JCI Insight 2, e91701. 10.1172/jci.insight.91701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albeituni S, and Stiban J (2019). Roles of Ceramides and Other Sphingolipids in Immune Cell Function and Inflammation. Adv. Exp. Med. Biol 1161, 169–191. 10.1007/978-3-030-21735-8_15. [DOI] [PubMed] [Google Scholar]
- 20.Raichur S, Wang ST, Chan PW, Li Y, Ching J, Chaurasia B, Dogra S, Öhman MK, Takeda K, Sugii S, et al. (2014). CerS2 Haploinsufficiency Inhibits beta-Oxidation and Confers Susceptibility to Diet-Induced Steatohepatitis and Insulin Resistance. Cell Metab. 20, 687–695. 10.1016/j.cmet.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Turpin SM, Nicholls HT, Willmes DM, Mourier A, Brodesser S, Wunderlich CM, Mauer J, Xu E, Hammerschmidt P, Brönneke HS, et al. (2014). Obesity-Induced CerS6-Dependent C16:0 Ceramide Production Promotes Weight Gain and Glucose Intolerance. Cell Metab. 20, 678–686. 10.1016/j.cmet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Novgorodov SA, Chudakova D, Zhu H, Bielawska A, Bielawski J, Obeid LM, Kindy MS, and Gudz TI (2007). JNK3 signaling pathway activates ceramide synthase leading to mitochondrial dysfunction. J. Biol. Chem 282, 25940–25949. 10.1074/jbc.M701812200. [DOI] [PubMed] [Google Scholar]
- 23.Novgorodov SA, and Gudz TI (2011). Ceramide and mitochondria in ischemic brain injury. Int J. Biochem. Mol. Biol 2, 347–361. [PMC free article] [PubMed] [Google Scholar]
- 24.Novgorodov SA, Chudakova DA, Wheeler BW, Bielawski J, Kindy MS, Obeid LM, and Gudz TI (2011). Developmentally regulated ceramide synthase 6 increases mitochondrial Ca2+ loading capacity and promotes apoptosis. J. Biol. Chem 286, 4644–4658. 10.1074/jbc.M110.164392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L, Spassieva SD, Jucius TJ, Shultz LD, Shick HE, Macklin WB, Hannun YA, Obeid LM, and Ackerman SL (2011). A deficiency of ceramide biosynthesis causes cerebellar purkinje cell neurodegeneration and lipofuscin accumulation. PLoS Genet. 7, e1002063. 10.1371/journal.pgen.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carra S, Alberti S, Arrigo PA, Benesch JL, Benjamin IJ, Boelens W, Bartelt-Kirbach B, Brundel BJJM, Buchner J, Bukau B, et al. (2017). The growing world of small heat shock proteins: from structure to functions. Cell Stress Chaperones 22, 601–611. 10.1007/s12192-017-0787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kampinga HH, and Garrido C (2012). HSPBs: small proteins with big implications in human disease. Int. J. Biochem. Cell Biol 44,1706–1710. 10.1016/j.biocel.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Yang C, Wang H, Zhu D, Hong CS, Dmitriev P, Zhang C, Li Y, Ikejiri B, Brady RO, and Zhuang Z (2015). Mutant glucocerebrosidase in Gaucher disease recruits Hsp27 to the Hsp90 chaperone complex for proteasomal degradation. Proc. Natl. Acad. Sci. USA 112, 1137–1142. 10.1073/pnas.1424288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellier G, Liu D, Bellot G, Holme AL, and Pervaiz S (2013). Small molecule sensitization to TRAIL is mediated via nuclear localization, phosphorylation and inhibition of chaperone activity of Hsp27. Cell Death Dis. 4, e890. 10.1038/cddis.2013.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X, Wang H, Cao L, and Tang D (2015). HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 34, 5617–5625. 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Xu X, Yu Y, Graham M, Prince ME, Carey TE, and Sun D (2010). Silencing heat shock protein 27 decreases metastatic behavior of human head and neck squamous cell cancer cells in vitro. Mol. Pharm 7, 1283–1290. 10.1021/mp100073s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiota M, Bishop JL, Nip KM, Zardan A, Takeuchi A, Cordonnier T, Beraldi E, Bazov J, Fazli L, Chi K, et al. (2013). Hsp27 regulates epithelial mesenchymal transition, metastasis, and circulating tumor cells in prostate cancer. Cancer Res. 73, 3109–3119. 10.1158/0008-5472.CAN-12-3979. [DOI] [PubMed] [Google Scholar]
- 33.Baughman HER, Clouser AF, Klevit RE, and Nath A (2018). HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation. J. Biol. Chem 293, 2687–2700. 10.1074/jbc.M117.803411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baughman HER, Pham THT, Adams CS, Nath A, and Klevit RE (2020). Release of a disordered domain enhances HspB1 chaperone activity toward tau. Proc. Natl. Acad. Sci. USA 117, 2923–2929. 10.1073/pnas.1915099117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Zhu Y, Lu J, Liu Z, Lobato AG, Zeng W, Liu J, Qiang J, Zeng S, Zhang Y, et al. (2022). Specific binding of Hsp27 and phosphorylated Tau mitigates abnormal Tau aggregation-induced pathology. Elife 11, e79898. 10.7554/eLife.79898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Feng C, Liu J, Zhao L, Liu J, Zhang W, Liu N, and Niu Y (2016). Heat shock protein 27 and gross cystic disease fluid protein 15 play critical roles in molecular apocrine breast cancer. Tumour Biol. 37, 8027–8036. 10.1007/s13277-015-4712-4. [DOI] [PubMed] [Google Scholar]
- 37.Vahid S, Thaper D, Gibson KF, Bishop JL, and Zoubeidi A (2016). Molecular chaperone Hsp27 regulates the Hippo tumor suppressor pathway in cancer. Sci. Rep 6, 31842. 10.1038/srep31842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baylot V, Andrieu C, Katsogiannou M, Taieb D, Garcia S, Giusiano S, Acunzo J, Iovanna J, Gleave M, Garrido C, and Rocchi P (2011). OGX-427 inhibits tumor progression and enhances gemcitabine chemotherapy in pancreatic cancer. Cell Death Dis. 2, e221. 10.1038/cddis.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melle C, Ernst G, Escher N, Hartmann D, Schimmel B, Bleul A, Thieme H, Kaufmann R, Felix K, Friess HM, et al. (2007). Protein profiling of microdissected pancreas carcinoma and identification of HSP27 as a potential serum marker. Clin. Chem 53, 629–635. 10.1373/clinchem.2006.079194. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai T, Kashida H, Komeda Y, Nagai T, Hagiwara S, Watanabe T, Kitano M, Nishida N, Fujita J, and Kudo M (2017). Stress Response Protein RBM3 Promotes the Development of Colitis-associated Cancer. Inflamm. Bowel Dis 23, 57–65. 10.1097/MIB.0000000000000968. [DOI] [PubMed] [Google Scholar]
- 41.Lebret T, Watson RWG, Molinié V, O’Neill A, Gabriel C, Fitzpatrick JM, and Botto H (2003). Heat shock proteins HSP27, HSP60, HSP70, and HSP90: expression in bladder carcinoma. Cancer 98, 970–977. 10.1002/cncr.11594. [DOI] [PubMed] [Google Scholar]
- 42.Lee MS, Lee J, Lee S, Yoo SM, Kim JH, Kim WT, Kim WJ, and Park J (2018). Clinical, prognostic, and therapeutic significance of heat shock protein 27 in bladder cancer. Oncotarget 9, 7961–7974. 10.18632/oncotarget.24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saini J, and Sharma PK (2018). Clinical, Prognostic and Therapeutic Significance of Heat Shock Proteins in Cancer. Curr. Drug Targets 19, 1478–1490. 10.2174/1389450118666170823121248. [DOI] [PubMed] [Google Scholar]
- 44.Lelj-Garolla B, Kumano M, Beraldi E, Nappi L, Rocchi P, Ionescu DN, Fazli L, Zoubeidi A, and Gleave ME (2015). Hsp27 Inhibition with OGX-427 Sensitizes Non-Small Cell Lung Cancer Cells to Erlotinib and Chemotherapy. Mol. Cancer Ther 14, 1107–1116. 10.1158/1535-7163.MCT-14-0866. [DOI] [PubMed] [Google Scholar]
- 45.Konda JD, Olivero M, Musiani D, Lamba S, and Di Renzo MF (2017). Heat-shock protein 27 (HSP27, HSPB1) is synthetic lethal to cells with oncogenic activation of MET, EGFR and BRAF. Mol. Oncol 11, 599–611. 10.1002/1878-0261.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu CL, Chen SF, Wu MZ, Jao SW, Lin YS, Yang CY, Lee TY, Wen LW, Lan GL, and Nieh S (2016). The molecular and clinical verification of therapeutic resistance via the p38 MAPK-Hsp27 axis in lung cancer. Oncotarget 7, 14279–14290. 10.18632/oncotar-get.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui Y, Sun Y, Hu S, Luo J, Li L, Li X, Yeh S, Jin J, and Chang C (2016). Neuroendocrine prostate cancer (NEPCa) increased the neighboring PCa chemoresistance via altering the PTHrP/p38/Hsp27/androgen receptor (AR)/p21 signals. Oncogene 35, 6065–6076. 10.1038/onc.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CH, Wu YT, Hsieh HC, Yu Y, Yu AL, and Chang WW (2014). Epidermal growth factor/heat shock protein 27 pathway regulates vasculogenic mimicry activity of breast cancer stem/progenitor cells. Biochimie 104, 117–126. 10.1016/j.biochi.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 49.Zhang S, Zhang XQ, Huang SL, Chen M, Shen SS, Ding XW, Lv Y, and Zou XP (2015). The Effects of HSP27 on Gemcitabine-Resistant Pancreatic Cancer Cell Line Through Snail. Pancreas 44, 1121–1129. 10.1097/MPA.0000000000000418. [DOI] [PubMed] [Google Scholar]
- 50.Kang D, Choi HJ, Kang S, Kim SY, Hwang YS, Je S, Han Z, Kim JH, and Song JJ (2015). Ratio of phosphorylated HSP27 to non-phosphorylated HSP27 biphasically acts as a determinant of cellular fate in gemcitabine-resistant pancreatic cancer cells. Cell. Signal 27, 807–817. 10.1016/j.cellsig.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Hayashi R, Ishii Y, Ochiai H, Matsunaga A, Endo T, Hasegawa H, and Kitagawa Y (2012). Suppression of heat shock protein 27 expression promotes 5-fluorouracil sensitivity in colon cancer cells in a xenograft model. Oncol. Rep 28, 1269–1274. 10.3892/or.2012.1935. [DOI] [PubMed] [Google Scholar]
- 52.Shimada T, Tsuruta M, Hasegawa H, Okabayashi K, Shigeta K, Ishida T, Asada Y, Suzumura H, Koishikawa K, Akimoto S, and Kitagawa Y (2018). Heat shock protein 27 knockdown using nucleotide-based therapies enhances sensitivity to 5-FU chemotherapy in SW480 human colon cancer cells. Oncol. Rep 39, 1119–1124. 10.3892/or.2018.6180. [DOI] [PubMed] [Google Scholar]
- 53.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, and Nebreda AR (1994). A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell 78, 1027–1037. [DOI] [PubMed] [Google Scholar]
- 54.Kostenko S, and Moens U (2009). Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell. Mol. Life Sci 66, 3289–3307. 10.1007/s00018-009-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gurgis FMS, Ziaziaris W, and Munoz L (2014). Mitogen-activated protein kinase-activated protein kinase 2 in neuroinflammation, heat shock protein 27 phosphorylation, and cell cycle: role and targeting. Mol. Pharmacol 85, 345–356. 10.1124/mol.113.090365. [DOI] [PubMed] [Google Scholar]
- 56.Hoffman L, Jensen CC, Yoshigi M, and Beckerle M (2017). Mechanical signals activate p38 MAPK pathway-dependent reinforcement of actin via mechanosensitive HspB1. Mol. Biol. Cell 28, 2661–2675. 10.1091/mbc.E17-02-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tormos AM, Rius-Pérez S, Jorques M, Rada P, Ramirez L, Valverde ÁM, Nebreda ÁR, Sastre J, and Taléns-Visconti R (2017). p38alpha regulates actin cytoskeleton and cytokinesis in hepatocytes during development and aging. PLoS One 12, e0171738. 10.1371/journal.pone.0171738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JL, Mestre B, Shin SH, and Futerman AH (2021). Ceramide synthases: Reflections on the impact of Dr. Cell. Signal 82, 109958. 10.1016/j.cellsig.2021.109958. [DOI] [PubMed] [Google Scholar]
- 59.Senkal CE, Salama MF, Snider AJ, Allopenna JJ, Rana NA, Koller A, Hannun YA, and Obeid LM (2017). Ceramide Is Metabolized to Acylceramide and Stored in Lipid Droplets. Cell Metab. 25, 686–697. 10.1016/j.cmet.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lampros M, Vlachos N, Voulgaris S, and Alexiou GA (2022). The Role of Hsp27 in Chemotherapy Resistance. Biomedicines 10, 897. 10.3390/biomedicines10040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zoubeidi A, and Gleave M (2012). Small heat shock proteins in cancer therapy and prognosis. Int. J. Biochem. Cell Biol 44, 1646–1656. 10.1016/j.biocel.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Yang S, Xiao H, and Cao L (2021). Recent advances in heat shock proteins in cancer diagnosis, prognosis, metabolism and treatment. Biomed. Pharmacother 142, 112074. 10.1016/j.biopha.2021.112074. [DOI] [PubMed] [Google Scholar]
- 63.Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, Day TA, Jiang JC, Jazwinski SM, Hannun YA, et al. (2004). Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem 279, 44311–44319. 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- 64.Abuhusain HJ, Matin A, Qiao Q, Shen H, Kain N, Day BW, Stringer BW, Daniels B, Laaksonen MA, Teo C, et al. (2013). A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis. J. Biol. Chem 288, 37355–37364. 10.1074/jbc.M113.494740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, Bielawski J, Day TA, Gillespie MB, Sinha D, et al. (2007). Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 256, 101–111. 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka T, Iino M, and Goto K (2018). Sec6 enhances cell migration and suppresses apoptosis by elevating the phosphorylation of p38 MAPK, MK2, and HSP27. Cell. Signal 49, 1–16. 10.1016/j.cellsig.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 67.Nishida T, Hattori K, and Watanabe K (2017). The regulatory and signaling mechanisms of the ASK family. Adv. Biol. Regul 66, 2–22. 10.1016/j.jbior.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 68.Hotamisligil GS, and Davis RJ (2016). Cell Signaling and Stress Responses. Cold Spring Harb. Perspect. Biol 8, a006072. 10.1101/cshperspect.a006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haar ET, Prabakhar P, Liu X, and Lepre C (2007). Crystal structure of the p38 alpha-MAPKAP kinase 2 heterodimer. J. Biol. Chem 282, 9733–9739. 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- 70.Frank SB, Berger PL, Ljungman M, and Miranti CK (2017). Human prostate luminal cell differentiation requires NOTCH3 induction by p38-MAPK and MYC. J. Cell Sci 130, 1952–1964. 10.1242/jcs.197152. [DOI] [PubMed] [Google Scholar]
- 71.Crowe J, Aubareda A, McNamee K, Przybycien PM, Lu X, Williams RO, Bou-Gharios G, Saklatvala J, and Dean JLE (2013). Heat shock protein B1-deficient mice display impaired wound healing. PLoS One 8, e77383. 10.1371/journal.pone.0077383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas RJ, Oleinik N, Panneer Selvam S, Vaena SG, Dany M, Nganga RN, Depalma R, Baron KD, Kim J, Szulc ZM, and Ogretmen B (2017). HPV/E7 induces chemotherapy-mediated tumor suppression by ceramide-dependent mitophagy. EMBO Mol. Med 9, 1030–1051. 10.15252/emmm.201607088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oleinik N, Kim J, Roth BM, Selvam SP, Gooz M, Johnson RH, Lemasters JJ, and Ogretmen B (2019). Mitochondrial protein import is regulated by p17/PERMIT to mediate lipid metabolism and cellular stress. Sci. Adv 5, eaax1978. 10.1126/sciadv.aax1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stiban J, Tidhar R, and Futerman AH (2010). Ceramide synthases: roles in cell physiology and signaling. Adv. Exp. Med. Biol 688, 60–71. 10.1007/978-1-4419-6741-1_4. [DOI] [PubMed] [Google Scholar]
- 75.Tidhar R, Ben-Dor S, Wang E, Kelly S, Merrill AH Jr., and Futerman AH (2012). Acyl chain specificity of ceramide synthases is determined within a region of 150 residues in the Tram-Lag-CLN8 (TLC) domain. J. Biol. Chem 287, 3197–3206. 10.1074/jbc.M111.280271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tidhar R, Zelnik ID, Volpert G, Ben-Dor S, Kelly S, Merrill AH Jr., and Futerman AH (2018). Eleven residues determine the acyl chain specificity of ceramide synthases. J. Biol. Chem 293, 9912–9921. 10.1074/jbc.RA118.001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zelnik ID, Rozman B, Rosenfeld-Gur E, Ben-Dor S, and Futerman AH (2019). A Stroll Down the CerS Lane. Adv. Exp. Med. Biol 1159, 49–63. 10.1007/978-3-030-21162-2_4. [DOI] [PubMed] [Google Scholar]
- 78.Laviad EL, Kelly S, Merrill AH Jr., and Futerman AH (2012). Modulation of ceramide synthase activity via dimerization. J. Biol. Chem 287, 21025–21033. 10.1074/jbc.M112.363580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jensen SA, Calvert AE, Volpert G, Kouri FM, Hurley LA, Luciano JP, Wu Y, Chalastanis A, Futerman AH, and Stegh AH (2014). Bcl2L13 is a ceramide synthase inhibitor in glioblastoma. Proc. Natl. Acad. Sci. USA 111, 5682–5687. 10.1073/pnas1316700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JL, Ben-Dor S, Rosenfeld-Gur E, and Futerman AH (2022). A novel C-terminal DxRSDxE motif in ceramide synthases involved in dimer formation. J. Biol. Chem 298, 101517. 10.1016/j.jbc.2021.101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sassa T, Hirayama T, and Kihara A (2016). Enzyme Activities of the Ceramide Synthases CERS2-6 Are Regulated by Phosphorylation in the C-terminal Region. J. Biol. Chem 291, 7477–7487. 10.1074/jbc.M115.695858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Novgorodov SA, Riley CL, Keffler JA, Yu J, Kindy MS, Macklin WB, Lombard DB, and Gudz TI (2016). SIRT3 Deacetylates Ceramide Synthases: IMPLICATIONS FOR MITOCHONDRIAL DYSFUNCTION AND BRAIN INJURY. J. Biol. Chem 291, 1957–1973. 10.1074/jbc.M115.668228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mizutani Y, Kihara A, and Igarashi Y (2005). Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J 390, 263–271. 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gencer S, Oleinik N, Kim J, Panneer Selvam S, De Palma R, Dany M, Nganga R, Thomas RJ, Senkal CE, Howe PH, and Ogretmen B (2017). TGF-beta receptor I/II trafficking and signaling at primary cilia are inhibited by ceramide to attenuate cell migration and tumor metastasis. Sci. Signal 10, eaam7464. 10.1126/scisignal.aam7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim JL, Mestre B, Malitsky S, Itkin M, Kupervaser M, and Futerman AH (2022). Fatty acid transport protein 2 interacts with ceramide synthase 2 to promote ceramide synthesis. J. Biol. Chem 298, 101735. 10.1016/j.jbc.2022.101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwartz NU, Linzer RW, Truman JP, Gurevich M, Hannun YA, Senkal CE, and Obeid LM (2018). Decreased ceramide underlies mitochondrial dysfunction in Charcot-Marie-Tooth 2F. Faseb. J 32, 1716–1728. 10.1096/fj.201701067R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arrigo AP (2017). Mammalian HspB1 (Hsp27) is a molecular sensor linked to the physiology and environment of the cell. Cell Stress Chaperones 22, 517–529. 10.1007/s12192-017-0765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sridevi P, Alexander H, Laviad EL, Pewzner-Jung Y, Hannink M, Futerman AH, and Alexander S (2009). Ceramide synthase 1 is regulated by proteasomal mediated turnover. Biochim. Biophys. Acta 1793, 1218–1227. 10.1016/j.bbamcr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosenberg JE, Hahn NM, Regan MM, Werner L, Alva A, George S, Picus J, Alter R, Balar A, Hoffman-Censits J, et al. (2018). Apatorsen plus docetaxel versus docetaxel alone in platinum-resistant metastatic urothelial carcinoma (Borealis-2). Br. J. Cancer 118, 1434–1441. 10.1038/s41416-018-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu EY, Ellard SL, Hotte SJ, Gingerich JR, Joshua AM, Gleave ME, and Chi KN (2018). A randomized phase 2 study of a HSP27 targeting antisense, apatorsen with prednisone versus prednisone alone, in patients with metastatic castration resistant prostate cancer. Invest. New Drugs 36, 278–287. 10.1007/s10637-017-0553-x. [DOI] [PubMed] [Google Scholar]
- 91.Hendriks LEL, and Dingemans AMC (2017). Heat shock protein antagonists in early stage clinical trials for NSCLC. Expert Opin. Investig. Drugs 26, 541–550. 10.1080/13543784.2017.1302428. [DOI] [PubMed] [Google Scholar]
- 92.Ko AH, Murphy PB, Peyton JD, Shipley DL, Al-Hazzouri A, Rodriguez FA, Womack MS 4th, Xiong HQ, Waterhouse DM, Tempero MA, et al. (2017). A Randomized, Double-Blinded, Phase II Trial of Gemcitabine and Nab-Paclitaxel Plus Apatorsen or Placebo in Patients with Metastatic Pancreatic Cancer: The RAINIER Trial. Oncol 22, 1427–e129. 10.1634/theoncologist.2017-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Le TK, Cherif C, Omabe K, Paris C, Lannes F, Audebert S, Baudelet E, Hamimed M, Barbolosi D, Finetti P, et al. (2023). DDX5 mRNA-targeting antisense oligonucleotide as a new promising therapeutic in combating castration-resistant prostate cancer. Mol. Ther 31, 471–486. 10.1016/j.ymthe.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nappi L, Aguda AH, Nakouzi NA, Lelj-Garolla B, Beraldi E, Lallous N,Thi M, Moore S, Fazli L, Battsogt D, et al. (2020). Ivermectin inhibits HSP27 and potentiates efficacy of oncogene targeting in tumor models. J. Clin. Invest 130, 699–714. 10.1172/JCI130819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sevin M, Kubovcakova L, Pernet N, Causse S, Vitte F, Villeval JL, Lacout C, Cordonnier M, Rodrigues-Lima F, Chanteloup G, et al. (2018). HSP27 is a partner of JAK2-STAT5 and a potential therapeutic target in myelofibrosis. Nat. Commun 9, 1431. 10.1038/s41467-018-03627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cherif C, Nguyen DT, Paris C, Le TK, Sefiane T, Carbuccia N, Finetti P, Chaffanet M, Kaoutari AE, Vernerey J, et al. (2022). Menin inhibition suppresses castration-resistant prostate cancer and enhances chemosensitivity. Oncogene 41, 125–137. 10.1038/s41388-021-02039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Neerven SM, Smit WL, van Driel MS, Kakkar V, de Groot NE, Nijman LE, Elbers CC, Léveillé N, Heijmans J, and Vermeulen L (2022). Intestinal Apc-inactivation induces HSP25 dependency. EMBO Mol. Med 14, e16194. 10.15252/emmm.202216194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grierson PM, Dodhiawala PB, Cheng Y, Chen THP, Khawar IA, Wei Q, Zhang D, Li L, Herndon J, Monahan JB, et al. (2021). The MK2/Hsp27 axis is a major survival mechanism for pancreatic ductal adenocarcinoma under genotoxic stress. Sci. Transl. Med. 13, eabb5445. 10.1126/scitranslmed.abb5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Henriques A, Koliaraki V, and Kollias G (2018). Mesenchymal MAPKAPK2/HSP27 drives intestinal carcinogenesis. Proc. Natl. Acad. Sci. USA 115, E5546–E5555. 10.1073/pnas.1805683115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strasser SD, Ghazi PC, Starchenko A, Boukhali M, Edwards A, Suarez-Lopez L, Lyons J, Changelian PS, Monahan JB, Jacobsen J, et al. (2019). Substrate-based kinase activity inference identifies MK2 as driver of colitis. Integr. Biol 11, 301–314. 10.1093/in-tbio/zyz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suarez-Lopez L, Kong YW, Sriram G, Patterson JC, Rosenberg S, Morandell S, Haigis KM, and Yaffe MB (2020). MAPKAP Kinase-2 Drives Expression of Angiogenic Factors by Tumor-Associated Macrophages in a Model of Inflammation-Induced Colon Cancer. Front. Immunol 11, 607891. 10.3389/fimmu.2020.607891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turner N, Lim XY, Toop HD, Osborne B, Brandon AE, Taylor EN, Fiveash CE, Govindaraju H, Teo JD, McEwen HP, et al. (2018). A selective inhibitor of ceramide synthase 1 reveals a novel role in fat metabolism. Nat. Commun. 9, 3165. 10.1038/s41467-018-05613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turpin-Nolan SM, Hammerschmidt P, Chen W, Jais A, Timper K, Awazawa M, Brodesser S, and Brüning JC (2019). CerS1-Derived C18:0 Ceramide in Skeletal Muscle Promotes Obesity-Induced Insulin Resistance. Cell Rep. 26, 1–10.e7. 10.1016/j.celrep.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 104.Tosetti B, Brodesser S, Brunn A, Deckert M, Blüher M, Doehner W, Anker SD, Wenzel D, Fleischmann B, Pongratz C, et al. (2020). A tissue-specific screen of ceramide expression in aged mice identifies ceramide synthase-1 and ceramide synthase-5 as potential regulators of fiber size and strength in skeletal muscle. Aging Cell 19, e13049. 10.1111/acel.13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Couttas TA, and Don AS (2016). Fluorescent Assays for Ceramide Synthase Activity. Methods Mol. Biol 1376, 23–33. 10.1007/978-1-4939-3170-5_3. [DOI] [PubMed] [Google Scholar]
- 106.Spassieva S, Bielawski J, Anelli V, and Obeid LM (2007). Combination of C(17) sphingoid base homologues and mass spectrometry analysis as a new approach to study sphingolipid metabolism. Methods Enzymol. 434, 233–241. 10.1016/S0076-6879(07)34012-3. [DOI] [PubMed] [Google Scholar]
- 107.Valente AJ, Maddalena LA, Robb EL, Moradi F, and Stuart JA (2017). A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 119, 315–326. 10.1016/jacthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original, uncropped western blot image files have been deposited at Mendeley Data and are publicly available as of the date of publication, Mendeley Data, V1, https://doi.org/10.17632/jd96sv7fts.1.
All the statistical data is available in Table S1.
This study does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.


