SUMMARY
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most common neuromuscular disorders and has no cure. Due to an unknown molecular mechanism, FSHD displays overlapping manifestations with the neurodegenerative disease amyotrophic lateral sclerosis (ALS). FSHD is caused by aberrant gain of expression of the transcription factor double homeobox 4 (DUX4), which triggers a pro-apoptotic transcriptional program resulting in inhibition of myogenic differentiation and muscle wasting. Regulation of DUX4 activity is poorly known. We identify Matrin 3 (MATR3), whose mutation causes ALS and dominant distal myopathy, as a cellular factor controlling DUX4 expression and activity. MATR3 binds to the DUX4 DNA-binding domain and blocks DUX4-mediated gene expression, rescuing cell viability and myogenic differentiation of FSHD muscle cells, without affecting healthy muscle cells. Finally, we characterize a shorter MATR3 fragment that is necessary and sufficient to directly block DUX4-induced toxicity to the same extent as the full-length protein. Collectively, our data suggest MATR3 as a candidate for developing a treatment for FSHD.
In brief
FSHD is one of the most common neuromuscular diseases and has no cure. It is caused by aberrant DUX4 expression, which is toxic to muscle cells. Runfola et al. identify MATR3 as a DUX4 interactor, which inhibits DUX4 expression and activity, thus opening the possibility of future therapeutic developments.
Graphical abstract
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most prevalent neuromuscular disorders1 and leads to significant lifetime morbidity, with up to 20% of patients requiring a wheelchair.2,3 Two genetic forms of FSHD have been reported. FSHD1 (MIM: 158900) accounts for 95% of the cases and is linked to the reduction in copy number of the D4Z4 microsatellite repeat at 4q35 below 10 units.3 FSHD2 (MIM: 158901) is clinically indistinguishable from FSHD14 and is associated with mutations in some of the genes encoding for regulators of D4Z4 epigenetic status.5
The common molecular outcome of the genetic and/or epigenetic alterations of the FSHD locus is chromatin relaxation and the subsequent aberrant reactivation of the double homeobox 4 (DUX4) gene.6,7 DUX4 is a transcription factor containing two homeodomains, responsible for sequence-specific DNA binding, located at the N terminus of the protein.8 Physiologically, DUX4 contributes to genome activation during the cleavage stage of early embryonic development.9–11 Subsequently, DUX4 expression is repressed in most adult somatic tissues,7 including skeletal muscle. While the pathway(s) controlling DUX4 expression and activity and leading to muscular dystrophy are not fully understood, DUX4 ectopic expression in cells as well as in skeletal muscle in vivo leads to apoptotic cell death.12–16 Increased apoptosis and its dependence on DUX4 have been documented also in FSHD muscle cells and tissues.4,15,17,18
Despite several pharmacological clinical trials,19–29 no therapeutic option is available for patients with FSHD. The activation of its direct transcriptional targets is linked to DUX4-induced muscle toxicity.12,30 Accordingly, DUX4 targets account for the majority of gene expression alterations in FSHD.13,14,16,31–42 Hence, blocking the ability of DUX4 to activate its transcriptional targets has strong therapeutic relevance.
Here, we identified Matrin 3 (MATR3) as an endogenous modulator of DUX4 expression and activity. MATR3 is a nuclear DNA- and RNA-binding protein, involved in regulation of chromosomal and genome integrity, RNA metabolism, and maintenance of the nuclear framework.43 Mutations in MATR3 have been associated with amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and distal myopathy43; however, the molecular mechanism underlying the role of MATR3 mutations in these pathologies remains poorly understood. We found that MATR3 interacts directly with the DNA-binding domain of DUX4 protein and hinders DUX4 target gene activation. Moreover, we identified a MATR3 fragment that is required and sufficient to inhibit DUX4 pathological activity in cellular models of FSHD without affecting healthy muscle cells.
RESULTS
Proteomics to identify DUX4 nuclear interactors
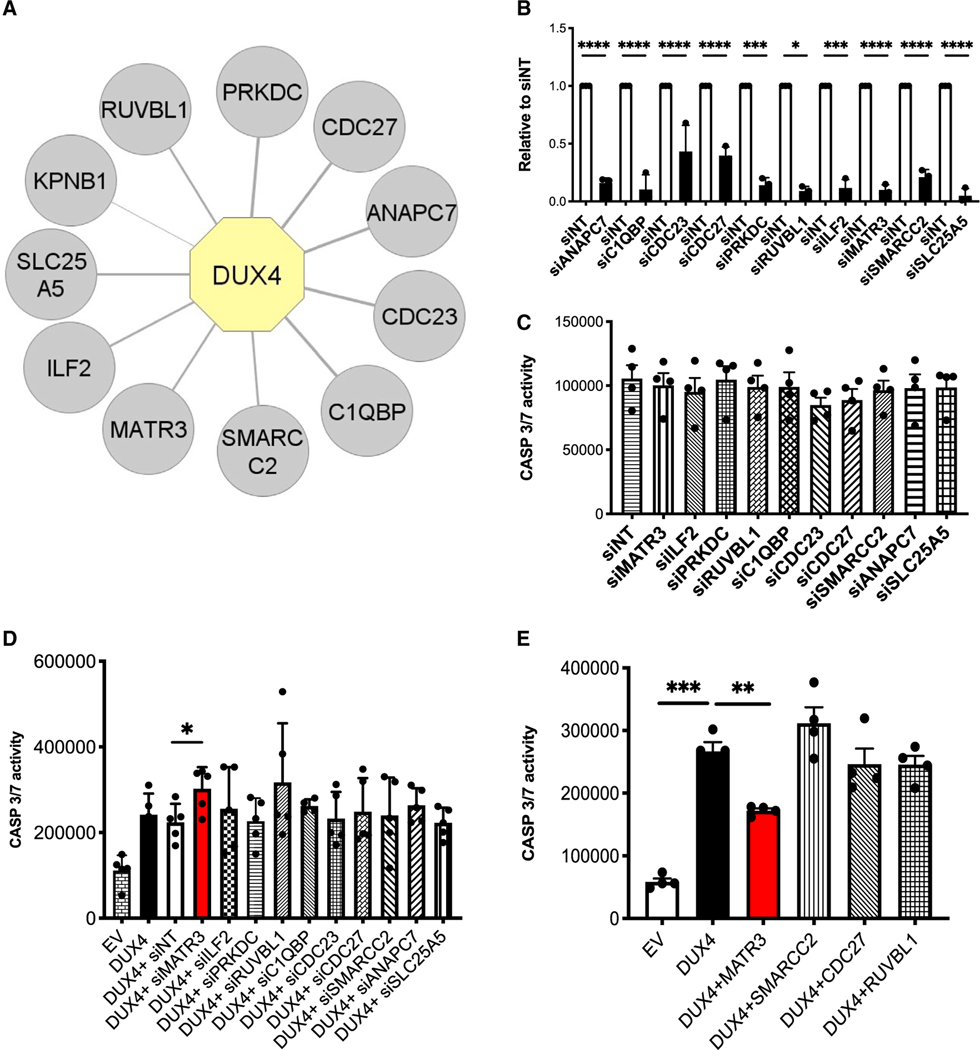
A “guilt-by-association” approach is widely used in proteomics to characterize the regulation and biological function of a protein based on the identification of its associated factors. To this aim, we generated doxycycline-inducible HEK293 cells, where DUX4 was fused to a streptavidin-binding peptide and a hemagglutinin tag (iSH-DUX4). In iSH-DUX4 cells, expression of DUX4 protein is detectable as soon as 4 h after doxycycline (dox) administration, DUX4 target genes are upregulated by 8 h, and significant apoptosis is detectable within 24 h of dox treatment (Figure S1), in line with previous results in human muscle cells.44 We performed tandem affinity purification (TAP) using nuclear extracts from dox-induced control and iSH-DUX4 cells under high stringency, followed by mass spectrometry (TAP-MS).45–48 To avoid possible side effects due to apoptosis, we collected the cells after just 8 h of dox treatment, the minimum time to observe induction of DUX4 targets but far from any detectable sign of cell death. We performed TAP purifications from three independent sets of nuclear extracts, which were nuclease treated and precleared to reduce nonspecific binding and to identify tight DUX4 interactors. Using stringent statistical evaluation, we identified 11 proteins that selectively associate with DUX4 (Figure 1A; Table S1). Beside karyopherin beta 1 (KPNB1), likely responsible for DUX4 nuclear localization, the remaining proteins have been all involved in regulation of gene expression (Table S1). Therefore, we investigated the role of each of the other 10 interactors on DUX4-mediated toxicity.
Figure 1. MATR3 protects HEK293 cells from DUX4-induced apoptosis.
(A) Graphical representation of DUX4 nuclear interactors identified by proteomics. Proteins identified in all the STREP-HA affinity purifications with a spectral count average of DUX4/EV control ratio >4 are displayed. DUX4 is highlighted in yellow, and the interactors are displayed in gray. The thickness of the edges is proportional to the spectral count average of DUX4/EV ratio.
(B) Quantitative real-time quantitative PCR showing the efficiency of knockdown for the indicated DUX4 interactors in HEK293 cells. Values are expressed relative to cells transfected with control small interfering RNAs (siRNAs; siNTs) (unpaired Student’s t test, *p ≤ 0.05; ***p ≤ 0.001; ****p ≤ 0.0001, n = 3).
(C) Caspase-3/7 activity assays performed upon knockdown of the indicated DUX4 interactors in HEK293 cells not expressing DUX4 (paired Student’s t test, n = 4).
(D) Caspase-3/7 activity assays performed in HEK293 cells collected 48 h after transfection with empty vector (EV), DUX4, or DUX4 in combination with siRNAs specific for the indicated targets (paired Student’s t test, *p < 0.05, n = 5).
(E) Caspase-3/7 activity assays performed in HEK293 cells collected 48 h after transfection with EV, DUX4, or DUX4 in combination with expression vectors for the indicated factors (paired Student’s t test, **p < 0.01, ***p < 0.001, n = 4).
See also Figures S1 and S2 and Table S1.
MATR3 inhibits DUX4-induced toxicity in HEK293 cells
To determine whether the identified interactors have a role on DUX4-mediated cell toxicity, we performed knockdown studies. Despite efficient knockdown (Figure 1B), depletion of none of the 10 selected interactors showed significant effects on cell viability in absence of DUX4 (Figure 1C). Intriguingly, silencing of MATR3 significantly increased DUX4-induced apoptosis in HEK293 cells (Figure 1D). On the other hand, MATR3 expression significantly protected cells from DUX4-induced apoptosis (Figure 1E). None of the other interactors tested were able to consistently affect DUX4 toxicity (Figures 1D and 1E). To assess the specificity of our findings, we treated HEK293 cells with the proapoptotic drug staurosporine (STS) and evaluated the effect on cell death by MATR3 (Figure S2). We found that MATR3 did not prevent STS-induced apoptosis (Figure S2), thus indicating that MATR3 per se is not a general inhibitor of apoptosis. Hence, while the other DUX4 interactors here identified might be involved in different aspects of DUX4 biology, MATR3 stood out as the only factor playing a key role in modulating DUX4-induced toxicity.
MATR3 interacts with the DNA-binding domain of DUX4
MATR3 was originally identified as a major component of the nu clear matrix.49 However, it also associates with nucleic acids through zinc-finger domains (ZFs) and RNA recognition motifs (RRMs) and contributes to the regulation of gene expression, chromatin accessibility, DNA repair, and RNA metabolism, via still unclear mechanism.43 The rest of the protein consists of two large intrinsically disordered regions.50
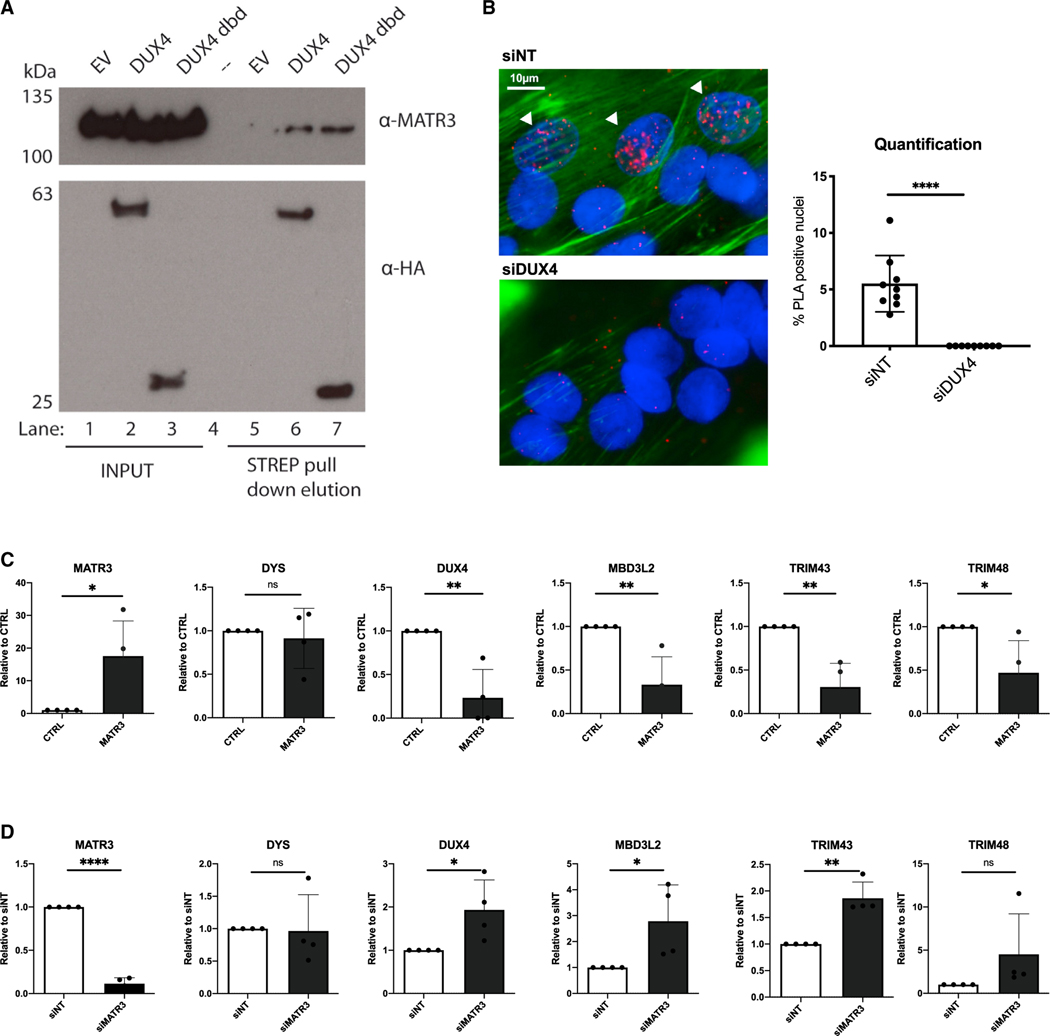
To validate our findings, we initially performed semi-endogenous Strep-Tactin pull-down experiments using nuclease-treated nuclear extracts from HEK293 cells ectopically expressing DUX4. As shown in Figure 2A, DUX4 protein specifically interacts with the endogenous MATR3, validating our proteomics. Intriguingly, endogenous MATR3 was equally able to interact with a DUX4 fragment retaining just the DNA-binding domain (DUX4 dbd) (Figure 2A), suggesting that MATR3 interacts with this region. We then wanted to assess whether the interaction observed in HEK293 cells was conserved in FSHD muscle cells. Since the endogenous DUX4 protein is expressed only in a minority of FSHD myonuclei (Figure S3),51,52 co-immunoprecipitation experiments in FSHD muscle cells are not feasible. Therefore, to investigate the interaction between the endogenous DUX4 and MATR3 proteins, we took advantage of in situ proximity ligation assay (PLA), which allows us to visualize protein-protein interactions at a single-cell level.53 We performed PLA in human primary FSHD muscle cells using antibodies specific for DUX4 and MATR3. As shown in Figures 2B, a positive PLA signal was detectable in a fraction of FSHD myonuclei, as expected from the DUX4-expression pattern (Figure S3). Importantly, the PLA signal disappeared upon depletion of DUX4 (Figure 2B), as well as when omitting DUX4 and/or MATR3 antibodies, or when using antibodies for DUX4 and WDR5, a chromatin remodeling factor involved in the regulation of the FSHD locus54 that does not interact with DUX4, as negative controls (Figure S4). Collectively, these results demonstrate that the endogenous DUX4 and MATR3 proteins interact in FSHD muscle cells and that MATR3 interaction occurs with the DUX4 dbd.
Figure 2. MATR3 interacts with DUX4 dbd and blocks DUX4 transcriptional activity in primary FSHD muscle cells.
(A) Strep-Tactin pull-down in HEK293 cells transfected with EV, DUX4 full-length, or DUX4 dbd. Nuclear proteins were incubated with Strep-Tactin beads, pull-down complexes were eluted with D-Biotin, and immunoblotting was performed with antibodies against MATR3 (detecting endogenous MATR3) or HA (detecting DUX4 and DUX4 dbd, lane 6 and 7, respectively).
(B) Proximity ligation assay (PLA) performed in terminally differentiated FSHD muscle cells treated with control (siNT) or DUX4 siRNAs. Positive PLA signals (white arrows) are present in nuclei of FSHD cells treated with siNT (top left), while they are absent in cells treated with siDUX4 (bottom left). The bar graph on the right shows quantification of the percentage of PLA-positive nuclei (unpaired Student’s t test, ****p ≤ 0.0001, n = 9). Scale bar: 10 μm.
(C) Quantitative real-time quantitative PCR for the indicated genes performed on RNA from differentiated primary FSHD muscle cells transduced with a control (CTRL) or MATR3 lentiviruses. Data are represented as relative to CTRL (unpaired Student’s t test, *p < 0.05, **p < 0.01, n = 4).
(D) Quantitative real-time quantitative PCR for the indicated genes performed on RNA from primary FSHD muscle cells transfected with control (siNT) or MATR3 (siMATR3) siRNAs. Data are represented as relative to siNT (unpaired Student’s t test, *p < 0.05, **p < 0.01, ****p ≤ 0.0001, n = 4).
See also Figures S3 and S4.
MATR3 inhibits the expression of DUX4 and DUX4 targets in FSHD muscle cells
Since DUX4’s ability to activate gene expression is linked to its toxicity,12–14,30,34,55 we analyzed the expression of DUX4 targets upon MATR3 loss- and gain-of-function in primary FSHD muscle cells. DUX4 target genes were significantly downregulated in cells overexpressing MATR3 (Figure 2C). On the contrary, MATR3 knockdown caused increased expression of DUX4 targets (Figure 2D). Surprisingly, we found that the expression of the endogenous DUX4 gene was significantly decreased by MATR3 gain of function, while MATR3 loss of function led to a significant increase of DUX4 expression in FSHD muscle cells (Figures 2C and 2D). MATR3 manipulation did not cause any significant alteration in the expression of the control muscle gene Dystrophin (Figures 2C and 2D). Thus, MATR3 delivery inhibits DUX4 expression and its transcriptional activity in a relevant FSHD cellular model.
MATR3 rescues cell death of FSHD muscle cells
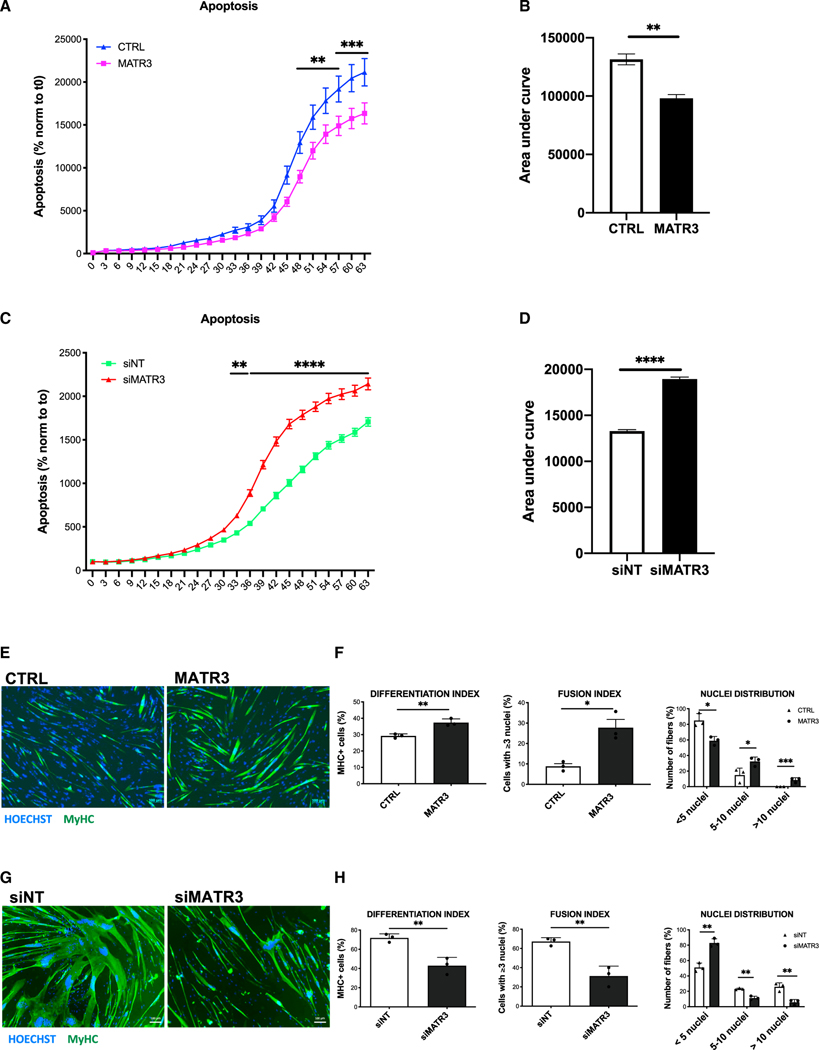
Apoptosis has been documented in FSHD muscle cells and tissues as a functional outcome of aberrant DUX4 expression.16,35,39 Since DUX4 is expressed by a minority of FSHD muscle cells nuclei52,56 (Figure S3), only a fraction of FSHD muscle cells undergo DUX4-induced cell death, making it difficult to monitor the efficacy of possible therapeutic treatments. To address this issue, we took advantage of live, real-time, single-cell apoptotic assays, which allows us to correlate apoptotic signals with high-definition phase contrast images in an automated and unbiased manner. Importantly, we used primary FSHD muscle cells, which have been reported to display higher than 10% DUX4-positive myonuclei,18 to help with the detection of cell death. To test the ability of MATR3 to protect from endogenous DUX4-induced cell death, we transduced primary FSHD muscle cells from two different FSHD donors with a control lentivirus or a lentivirus expressing MATR3 and monitored cell death over time. As shown in Figures 3A, 3B, S5A, and S5B, MATR3 delivery leads to a significant decrease of cell death with respect to control-infected FSHD muscle cells. Conversely, MATR3 knockdown in primary muscle cells from two different FSHD donors caused a significant increase of cell death with respect to control knockdown (Figures 3C, 3D, S5C, and S5D).
Figure 3. MATR3 overexpression rescues viability and myogenic differentiation of FSHD muscle cells.
(A) Live-cell, real-time, caspase-3/7 apoptotic assays on primary FSHD muscle cells transduced with CTRL or MATR3 lentiviruses. Live-cell imaging was performed by IncuCyte S3 Imager system and quantified using the IncuCyte software. Data are reported as a percentage (%) of green-fluorescent apoptotic cells normalized to time 0 (two-way ANOVA, **p < 0.01, ***p ≤ 0.001, n = 3).
(B) Quantification of the area under the curve in (A) (unpaired Student’s t test, **p < 0.01).
(C) Live-cell, real-time, caspase-3/7 apoptotic assays on primary FSHD muscle cells transfected with control (siNT) or MATR3 (siMATR3) siRNAs. Live-cell imaging was performed by IncuCyte S3 Imager system and quantified using the IncuCyte software. Data are reported as a percentage (%) of green-fluorescent apoptotic cells normalized to time 0 (two-way ANOVA, **p < 0.01, ****p ≤ 0.0001, n = 3).
(D) Quantification of the area under the curve in (C) (unpaired Student’s t test, ****p ≤ 0.0001).
(E) Representative images of myosin heavy-chain (MyHC; green) and nuclei (Hoechst 33342, blue) staining performed on differentiated primary FSHD muscle cells transduced with CTRL or MATR3 lentiviruses. Scale bar: 100 μm.
(F) Quantification of differentiation index, fusion index, and nuclei distribution in primary FSHD muscle cells treated as in (E) (unpaired Student’s t test, *p < 0.05, **p < 0.01, ***p ≤ 0.001, n = 3).
(G) Representative images of MyHC (green) and nuclei (Hoechst 33342, blue) staining performed on differentiated primary FSHD muscle cells transfected with control (siNT) or MATR3 (siMATR3) siRNAs. Scale bar: 100 μm.
(H) Quantification of differentiation index, fusion index, and nuclei distribution in primary FSHD muscle cells treated as in (G) (unpaired Student’s t test, **p < 0.01, n = 3).
See also Figures S5 and S6.
MATR3 rescues myogenic differentiation of FSHD muscle cells
DUX4 expression interferes with muscle differentiation,35 and muscle cells from patients with FSHD display impaired myogenesis.38,57 We wondered if MATR3, by allowing survival of DUX4-expressing cells, can rescue the myogenic defects of FSHD muscle cells. To test this, we transduced primary muscle cells from two different FSHD donors with control or MATR3 lentiviruses and measured their ability to differentiate into myotubes. As shown in Figures 3E, 3F, S5E, and S5F, MATR3 significantly rescues the myogenic and fusion indexes of FSHD muscle cells, allowing them to produce myotubes with a significantly increased number of myonuclei with respect to control-infected muscle cells. On the other hand, we scored a significant worsening of myogenic differentiation when we silenced MATR3 in FSHD primary myotubes (Figures 3G, 3H, S5G, and S5H). Notably, myogenesis was unaffected by MATR3 modulation in primary muscle cells from healthy donors (Figures S6A–S6D), thus showing that the MATR3 effect on FSHD muscle cells is DUX4 dependent.
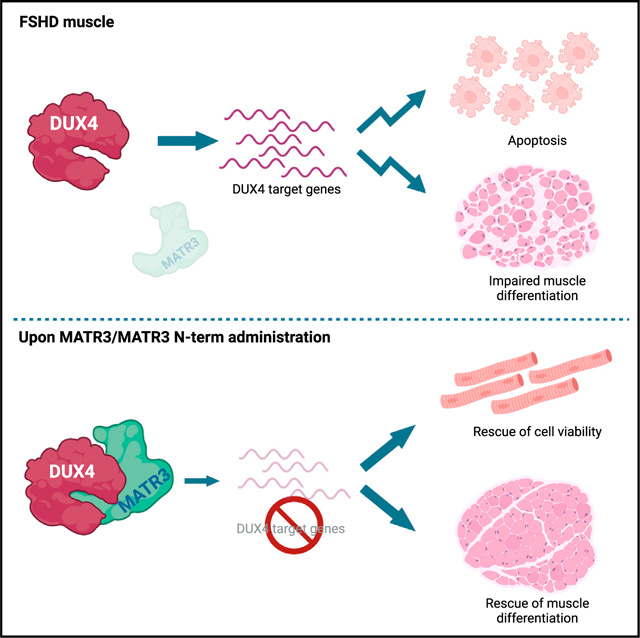
Collectively, our results strongly indicate that MATR3 is a natural, physiological inhibitor of DUX4, whose delivery to FSHD muscle cells prevents activation of DUX4 target genes and induction of apoptosis and improves myogenic differentiation.
MATR3 N-terminal fragment binds directly to DUX4 dbd
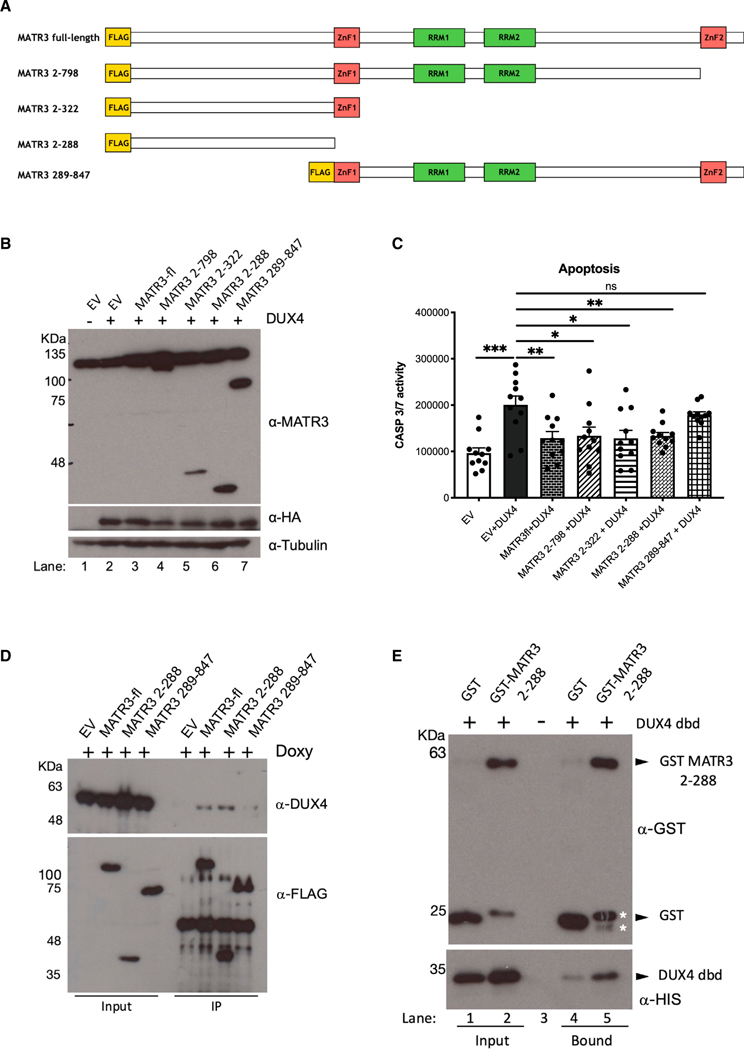
To gain insights into the interaction between DUX4 and MATR3 proteins, we expressed four MATR3 deletion mutants in HEK293 cells (Figure 4A), we assessed their nuclear localization (Figure S7), and we tested their effect on DUX4-mediated apoptosis in comparison with full-length MATR3 (Figures 4B and 4C). Intriguingly, we found that an N-terminal fragment of MATR3 devoid of any known functional domain (MATR32_288) was sufficient to inhibit DUX4-induced cell death to the same extent of full-length MATR3 (Figure 4C). On the other hand, a MATR3 fragment lacking the above-mentioned N-terminal part (MATR3289_847) was unable to significantly protect from DUX4-induced apoptosis (Figure 4C). Accordingly, MATR32_288 interacts with DUX4 protein similarly to full-length MATR3, while its counterpart MATR3289_847 is not able to do so (Figure 4D). Notably, by performing pull-down experiments using purified, recombinant versions of the DUX4 dbd and MATR32_288 (Figure S8), we found that MATR32_288 interacts directly with the DUX4 dbd (Figure 4E). Overall, our data indicate that the N-terminal fragment of MATR3 is required and sufficient to directly bind to the DUX4 dbd.
Figure 4. MATR32–288 is sufficient to block DUX4-induced apoptosis and directly bind DUX4 dbd.
(A) Schematic representation of MATR3 full length (MATR3-fl) and MATR3 deletion mutants MATR32–798, MATR32–322, MATR32–288, and MATR3289–847. The N-terminal yellow box indicates the FLAG tag.
(B) Immunoblotting on total protein extracts from HEK293 cells transfected with EV, DUX4, or DUX4 in combination with the indicated MATR3 constructs, incubated with anti-MATR3 (recognizing endogenous as well as transfected MATR3), anti-HA (recognizing transfected DUX4), and anti-tubulin (as loading control).
(C) Caspase-3/7 assay in HEK293 cells transfected with EV, DUX4, and DUX4 in combination with the indicated MATR3 constructs (unpaired Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001, n = 10).
(D) FLAG-immunoprecipitation (IP) on total protein extracts from HEK293 cells transfected with EV, MATR3-fl, or the indicated MATR3 constructs in combination with DUX4, incubated with anti-DUX4 and anti-FLAG (recognizing transfected MATR3 constructs).
(E) Pull-down assay with recombinant, purified His-DUX4 dbd plus purified GST-MATR32–288 or GST (as negative control), analyzed by immunoblotting with antibodies against GST or His tag. Asterisks (*) indicate the position of two degradation products of GST-MATR32–288.
See also Figures S7 and S8.
MATR3 N-terminal fragment blocks DUX4 transcriptional activity in FSHD muscle cells
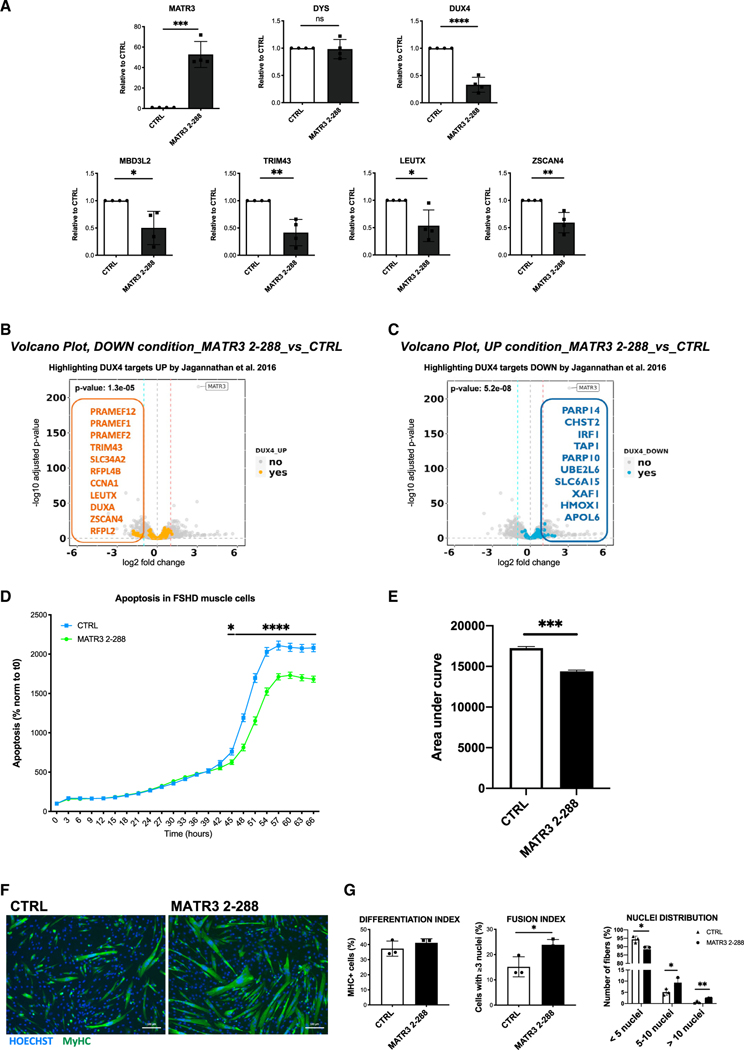
To test the ability of MATR32_288 to impair DUX4 transcriptional activity, we transduced primary muscle cells isolated from two independent patients with FSHD with lentiviruses carrying either MATR32_288 or a control lentivirus and evaluated the expression of DUX4 target genes. As shown in Figures 5A and S9A, MATR32_288 significantly reduced the expression of DUX4 targets compared to the control. Moreover, as observed for full-length MATR3, MATR32_288 led to a significant decrease of DUX4, without impairing the expression of Dystrophin, used as control (Figure 5A). To broadly evaluate the effect of MATR32_288 expression, we performed RNA sequencing in FSHD primary muscle cells transduced with MATR32_288 or its control (Table S5). We found that MATR32_288 caused a significant impact on the DUX4-dependent gene expression, which is the major molecular signature of FSHD.14,34 In particular, we found that genes upregulated by DUX4 were significantly downregulated by MATR32_288 expression, and vice versa (Figures 5B and 5C). Instead, no significant correlation was found between genes upregulated or downregulated by both DUX4 and MATR32_288 (Figures S9B and S9C).
Figure 5. MATR32–288 blocks DUX4-dependent gene expression and rescues viability and muscle differentiation in primary FSHD muscle cells.
(A) Quantitative real-time quantitative PCR for the indicated genes performed on RNA from differentiated primary FSHD muscle cells transduced with a CTRL or with MATR32–288 lentiviruses. Data are represented as relative to CTRL (unpaired Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n = 3).
(B) Volcano plot showing the significantly downregulated genes by MATR32–288 overexpression among the lists of DUX4 upregulated targets from Jagannathan et al.,34 filtered for absolute values of |log2 fold change (FC)| >1.
(C) Volcano plot showing the significantly upregulated genes by MATR32–288 overexpression among the lists of DUX4 downregulated targets from Jagannathan et al.,34 filtered for absolute values of |log2 FC| >1.
(D) Live-cell, real-time, caspase-3/7 apoptotic assays on primary FSHD muscle cells transduced with CTRL or MATR32–288 lentiviruses. Live-cell imaging was performed by IncuCyte S3 Imager system and quantified using the IncuCyte software. Data are reported as a percentage (%) of green-fluorescent apoptotic cells normalized to time 0 (two-way ANOVA, *p < 0.05, ****p ≤ 0.0001, n = 3).
(E) Quantification of the area under the curve in (D) (unpaired Student’s t test, ***p ≤ 0.001).
(F) Representative images of MyHC (green) and nuclei (Hoechst 33342, blue) staining performed on differentiated primary FSHD muscle cells transduced with CTRL or MATR32–288 lentiviruses. Scale bar: 100 μm.
(G) Quantification of differentiation index, fusion index, and nuclei distribution in primary FSHD muscle cells treated as in (F) (unpaired Student’s t test, *p < 0.05, **p < 0.01, n = 3).
See also Figures S9 and S10 and Table S5.
MATR3 N-terminal fragment rescues viability and muscle differentiation of FSHD muscle cells
To evaluate the phenotypic effect of MATR32_288 delivery in a relevant cellular system, we performed live-imaging apoptotic assays in primary FSHD myotubes transduced with a lentivirus carrying MATR32_288 or a control lentivirus. As shown in Figures 5D and 5E, MATR32_288 was sufficient to significantly block DUX4-induced toxicity. Notably, no effect on apoptosis was observed upon expression of MATR32_288 in healthy primary muscle cells (Figures S10A and S10B), thus indicating a DUX4-specific effect by MATR32_288. Upon expression of MATR32_288 in FSHD primary muscle cells, we observed a significant rescue of muscle differentiation (Figures 5F and 5G) that was scored by an increased differentiation index, improvement of the fusion index, and distribution of nuclei in the myotubes (Figure 5G). Overall, our data indicate that MATR32_288 is required and sufficient to directly bind the DUX4 protein and inhibit its toxic activities in FSHD muscle cells.
DISCUSSION
Although the genetics underlying FSHD has been studied since decades, there is still an incomplete understanding of FSHD pathophysiology, and no therapeutic option is currently available for patients with FSHD.8 Two subtypes of the disease have been described, FSHD1 and FSHD2, which have different genetic causes but are clinically indistinguishable.8 Importantly, both have as common outcome loss of repression at the FSHD locus and the consequent aberrant expression of DUX4, which is the main driver of the disease.8 Therefore, the current therapeutic emphasis in the FSHD field is on blocking DUX4 at different levels, including by modulating DUX4-regulating pathways, targeting DUX4 mRNA, DUX4 protein, or cellular pathways regulated by DUX4.33 However, results in patients with FSHD thus far have been inconclusive, because of poor specificity and/or inefficient in vivo delivery, or inconsistent, due to small sample size and high variability.33,58
MATR3 is a physiological inhibitor of DUX4
In this study, we hypothesized that identifying DUX4 nuclear interactors and acting on their availability might be a route to specifically interfere with DUX4 toxicity. While we expected to find a DUX4 co-factor required for apoptosis, we instead identified MATR3 as a physiological inhibitor of DUX4. We propose that MATR3 protects from DUX4-induced toxicity by preventing transcriptional activation of DUX4 target genes, which are toxic to muscle cells. As a result, MATR3 not only promotes survival of FSHD muscle cells but also ameliorates their myogenic differentiation. Thus, MATR3 rescues two key features of DUX4-induced toxicity.
While MATR3 is ubiquitously expressed, its levels vary in different tissues. Given that skeletal muscle is one of the tissues displaying the lowest MATR3 levels,43 it is tempting to speculate that the normal free pool of endogenous MATR3 in skeletal muscle is not sufficient to fully counteract DUX4 function when it is aberrantly expressed in FSHD. Accordingly, we found that MATR3 knockdown increases the expression of DUX4 targets and worsens the pathogenic signs of FSHD muscle cells. On the contrary, overexpressed MATR3 interacts with DUX4 and inhibits its toxicity.
MATR3 is a bifunctional DUX4 inhibitor
In addition to directly blocking DUX4-dependent transcription, we found that MATR3 is also able to inhibit DUX4 expression. Recently, a positive feedforward mechanism has been reported.59 It involves the DUX4 target MBD3L2, which is selectively expressed in FSHD muscle cells and is required for the full induction of DUX4 transcription in FSHD muscle cells. Indeed, MBD3L2 counteracts DUX4 transcriptional repression mediated by the nucleosome remodeling deacetylase (NuRD) complex. Importantly, MDB3L2 knockdown significantly decreases DUX4 expression in FSHD muscle cells, suggesting that MBD3L2 contributes to the amplification of DUX4 transcription in FSHD muscle cells.59 Since we found that MATR3 expression is associated with MBD3L2 downregulation, it is tempting to speculate that MATR3 decreases DUX4 expression in FSHD muscle cells indirectly, by blocking the induction of MDB3L2 by DUX4.
Identification of a MATR3 fragment devoid of possible toxic domains
Since MATR3 is a protein involved in many biological processes,43,50 administration of full-length MATR3 could lead to side effects. Indeed, previous reports showed toxicity associated with full-length MATR3 overexpression.60–62 For this reason, we characterized the portion of MATR3 involved in the interaction with DUX4 and identified an N-terminal fragment of MATR3 (MATR32_288) that is required and sufficient to directly bind the DUX4 dbd and inhibit its activity. Importantly, we have shown that MATR32_288 blocks DUX4 transcriptional activity comparably to full-length MATR3, with the functional consequence of promoting survival and myogenic differentiation of FSHD muscle cells. Intriguingly, MATR3 toxicity was linked to its RRMs,60–62 which are absent in MATR32_288. Accordingly, MATR32_288 does not affect cell proliferation and survival of healthy donor muscle cells. Importantly, all MATR3 interactors identified thus far bind MATR3 regions located away from the region interacting with DUX4. Moreover, unlike DUX4, most of the other MATR3 interactors bind MATR3 indirectly through nucleic acid bridges.43 Thus, while further work is required, we are cautiously optimistic that a safe MATR3-based DUX4 inhibitor can be developed.
MATR3 could contribute to FSHD heterogeneity
Despite having distinct clinical definitions, FSHD shares intriguing molecular features with ALS, the most common motor neuron disease. Overlapping aspects include altered proteostasis, aberrant RNA metabolism, activation of human endogenous retroviruses, increased oxidative stress, aggregates of TDP-43, and cell death.18,30,35,37,57,63–72 A molecular explanation for these similarities is still lacking. Intriguingly, evidence in the literature indicates that MATR3 dysfunction is integrally linked to ALS pathogenesis, since MATR3 interacts and forms aggregates with TDP-43 in ALS,43 and that MATR3 mutations cause ALS.43 ALS is linked to different forms of muscular disorders. In this regard, a recurrent mutation in MATR3 causes asymmetric progressive autosomal-dominant distal myopathy,73 which also shows clinical manifestations overlapping with FSHD. While MATR32_288 does not display any known functional domain, the hotspot of ALS-causing mutations74 and the S85C mutation causing distal myopathy73 fall within this region. Given these considerations, it is tempting to think about these disorders as different phenotypes of the same spectrum, which could help to identify common pathological pathways and therapeutic targets.
FSHD is characterized by an extensive intrafamilial variability in clinical severity and disease progression75,76 that is only partially explained by currently known FSHD disease modifiers.8,77 Hence, it is plausible to think that MATR3 may act as an additional modifier of disease severity in families with FSHD. For example, a MATR3 mutation decreasing its ability to bind DUX4 could be associated with a more severe FSHD phenotype. On the contrary, a MATR3 mutation strengthening its binding to DUX4 would be protective. In the future, it would be interesting to investigate if mutations in MATR32_288, which is responsible for DUX4 binding, affect its ability to inhibit DUX4 and segregate with the variability of clinical phenotype encountered in FSHD families.
In summary, our results show that MATR3 is a natural inhibitor of DUX4 activity that binds to the DUX4 dbd and inhibits activation of its targets and induction of apoptosis. Moreover, we identified an N-terminal fragment of MATR3 that is sufficient to control DUX4 expression and toxic activity, without affecting healthy muscle cells. Our results offer a perspective for the development of therapeutic strategies to effectively treat FSHD.
Limitations of the study
Our data are in line with the possibility that MATR3, by binding to the DUX4 dbd, inhibits DUX4 association to its genomic targets. To demonstrate our hypothesis, chromatin immunoprecipitation (ChIP) for DUX4 in FSHD muscle cells overexpressing MATR3 would be required. Nevertheless, ChIP for the endogenous DUX4 is not feasible since it is expressed only in a small percentage of FSHD muscle nuclei. Future work will be required to determine if this issue could be overcome by using single-nucleus ChIP. Another limitation concerns the fact that the MATR32_288 fragment identified in the present study is still too big to be efficiently developed into a drug-like molecule. Therefore, in a therapeutic perspective, further studies are required to identify the minimal MATR3 region sufficient to inhibit DUX4.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Davide Gabellini (gabellini.davide@hsr.it).
Materials availability
Plasmids generated in this study are available upon request to the lead contact.
Data and code availability
Proteomics data have been deposited at ProteomeXchange database via the PRIDE partner repository (ProteomeXchange: PXD011073 and https://doi.org/10.6019/PXD011073) and RNA-seq data have been deposited at GEO database (GEO: GSE210008). They are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat# F1804, RRID: AB_262044 |
| Mouse monoclonal anti-HA.11 (Clone 16B12) | Covance | Cat# MMS-101R, RRID: AB_291262 |
| Rabbit polyclonal anti MATR3 | Thermo Fisher Scientific | Cat# PA5-57720, RRID: AB_2643782 |
| Mouse monoclonal anti-6xHis | Clontech | Cat# 631212, RRID: AB_2721905 |
| Mouse monoclonal anti-GST | Sigma-Aldrich | Cat# G1160, RRID: AB_259845 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026, RRID: AB_477593 |
| Anti-mouse IgG-HRP | Jackson ImmunoResearch | Cat # 715-035-150, RRID: AB_2340770 |
| Anti-rabbit IgG-HRP | Jackson ImmunoResearch | Cat # 711-035-152, RRID: AB_10015282 |
| Rabbit monoclonal anti-DUX4 E5-5 | Abcam | Cat# ab124699, RRID: AB_10973363 |
| Mouse monoclonal anti-DUX4 P2B1 (for PLA) | Sigma-Aldrich | Cat# SAB5200019, RRID: AB_2612761 |
| Mouse monoclonal anti-myosin | DSHB | Cat# MF20, RRID: AB_2147781 |
| Rabbit monoclonal anti-WDR5 (clone D9E1I) | Cell Signaling Technology | Cat# 13105, RRID:AB_2620133 |
| Alexa 555 goat anti-rabbit | Molecular Probes | Cat# A-27039, RRID: AB_2536100 |
| Alexa 555 goat anti-mouse | Molecular Probes | Cat # A-21422, RRID: AB_141822 |
| Alexa Fluor 488 goat anti-mouse | Molecular Probes | Cat# A-32723, RRID: AB_2633275 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Rosetta2 (DE3) pLysS competent cells | Novagen | Cat #71403 |
|
| ||
| Biological samples | ||
|
| ||
| FSHD primary muscle cells | Richard Fields Center for FSHD Research Biobank | Table S6 of this paper |
| Healthy primary muscle cells | Richard Fields Center for FSHD Research Biobank | Table S6 of this paper |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Staurosporine | Sigma-Aldrich | 19123 |
| Doxycycline | Sigma-Aldrich | D9891 |
| D-Biotin | Sigma-Aldrich | 2031 |
| DL-Dithiothreitol | Sigma-Aldrich | D9779 |
| Trypsin-EDTA (0,5%) | Gibco | 15400054 |
| Dimethyl sulfoxide | Sigma-Aldrich | D2650 |
| L-Glutamine | Thermo Fisher Scientific | 25030081 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Paraformaldehyde | Electron Microscopy Sciences | 157–8 |
| GST recombinant protein | This paper | N/A |
| GST-MATR3 2–288 recombinant protein | This paper | N/A |
| HIS-DUX4 dbd recombinant protein | This paper | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| Caspase-Glo 3/7 luminescent assay | Promega | G8091 |
| CellTiter-Glo luminescent assay | Promega | G7570 |
| CASP3/7 assay GREEN reagent | Essen BioScience | 4440 |
| Duolink in situ Red kit | Sigma-Aldrich | DUO92101 |
| QuikChange XL Site-Directed Mutagenesis Kit | Agilent Technologies | 200517 |
| Gateway LR Clonase Enzyme mix | Invitrogen | 11791019 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | L3000001 |
| Lipofectamine LTX Reagent with PLUS Reagent | Thermo Fisher Scientific | 15338-100 |
| PureLink RNA Mini Kit | Thermo Fisher Scientific | 12183025 |
| DNAseI (PureLink DNase Set) | Thermo Fisher Scientific | 12185010 |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | 18080051 |
| iTaq Universal SYBR Green Supermix | Biorad | 1725122 |
| Polyfect Transfection Reagent | QIAGEN | 301107 |
| Slide-A-Lyzer dialysis cassettes | Thermo Fisher Scientific | 66380 |
| Protein G Sepharose beads | GE Healthcare | 71708300 |
| Strep-Tactin Sepharose beads | IBA Lifesciences | 2-1201-002 |
| anti-HA-agarose beads | Sigma-Aldrich | A2095 |
| Glutathione-Sepharose beads | GE Healthcare | 17513201 |
| anti-FLAG M2 Affinity Gel | Sigma-Aldrich | A2220 |
| His-Select Nickel Affinity gel beads | Sigma-Aldrich | H0537 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE210008 and Table S5 |
| Raw and analyzed proteomics data | This paper | ProteomeXchange: PXD011073, https://doi.org/10.6019/PXD011073 and Table S1 |
| DUX4 core target gene list | Table S4 (Jagannathan S et al.)34 | https://doi.org/10.1093/hmg/ddw271 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T cells | ATCC | CRL-3216 |
| Flp-In T-REx 293 cells | Thermo Fisher Scientific | R78007 |
| DUX4 Flp-In T-Rex cells (HEK-iSH-DUX4) | This manuscript | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S2 for a list of primers for cloning | This paper | N/A |
| See Table S3 for a list of siRNAs | N/A | N/A |
| See Table S4 for a list of primers for RT-qPCR | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV_FLAG-MATR3 | (Salton et al.)78 | RRID: Addgene_32880 |
| pCMV_FLAG-SMARCC2 | (Xi et al.)79 | RRID: Addgene_19142 |
| pECE-M2-CDC27 | (Banko et al.)80 | RRID: Addgene_31661 |
| pCDNA-3xFLAG-RUVBL1 | (Venteicher et al.)81 | RRID: Addgene_51635 |
| pCMV_FLAG-MATR3 2_288 | This paper | N/A |
| pCMV_FLAG-MATR3 2_322 | This paper | N/A |
| pCMV_FLAG-MATR3 2_798 | This paper | N/A |
| pCMV-FLAG-MATR3 289_end | This paper | N/A |
| pCS2-mkgDUX4 vector | (Snider et al.)52 | RRID: Addgene_21156 |
| pET-GB1-DUX4 dbd | This paper | N/A |
| pGEX-2tk- MATR3 2_288 | This paper | N/A |
| pCDNA5/FRT/TO STREP-HA | Dr. Superti-Furga G, CeMM Research Center for Molecular Medicine | Austrian Academy of Sciences, 1090 Vienna, Austria |
| pCMV-dR8.91 | Broad Institute | http://www.broadinstitute.org/rnai/trc |
| VSV-G/pMD2G | Broad Institute | http://www.broadinstitute.org/rnai/trc |
| pFUGW:GFP | Dr. Shanahan CM, King’s College London, London, UK | https://doi.org/10.1091/mbc.E16-06-0346 |
| pFUGW:GFP- MATR3-full length | This paper | N/A |
| pFUGW:GFP- MATR3 2_288 | This paper | N/A |
| pLBC2-BS-RFCA-BCVIII | Dr. Mullighan CG, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA | https://doi.org/10.1038/ng.3691 |
| pLBC2-BS-RFCA-BCVIN-att-MATR3-full length | This paper | N/A |
| pLBC2-BS-RFCA-BCVIII-att-MATR3 2_288 | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad PRISM 8 | GraphPad Software | https://www.graphpad.com/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| CFX Manager Software 3.1 | Biorad | https://www.bio-rad.com/it-it/sku/1845000-cfx-manager-software?ID=1845000 |
| MaxQuant (v 1.5.2.8) | (Cox J. et al.)82 | https://doi.org/10.1021/pr101065j |
| STAR aligner (v 2.5.3a) | (Dobin A et al.)83 | https://doi.org/10.1093/bioinformatics/bts635 |
| CRAN package pheatmap (v.1.0.12) | CRAN package | https://cran.r-project.org/package=pheatmap |
| CRAN package “venn” (v1.11). | CRAN package | https://CRAN.R-project.org/package=venn |
| FeatureCounts (v 1.6.4) | (Liao Y et al.)84 | https://doi.org/10.1093/bioinformatics/btt656 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell culture
HEK293T cells were originally obtained from American Type Culture Collection (ATCC). Flp-In T-REx 293 cells were originally obtained by Thermo Fisher Scientific. Cell lines were authenticated by morphology, growth characteristics, tested mycoplasma-free, and frozen. Cells were cultured for <3 months after thawing.
DUX4 Flp-In T-REx 293 cells were generated by transfection of Flp-In T-REx 293 cells with pCDNA5/FRT/TO STREP-HA_DUX4 and the pOG44 Flp-Recombinase Expression vector (Thermo Fisher Scientific), according to manufacturer’s instructions. Resistant cells were selected using Blasticidin and Hygromycin B (Thermo Fisher Scientific, #R210-01, #10687010). The parental Flp-In T-REx 293 cells were grown in parallel and used as negative control.
HEK293T cells were grown in DMEM high glucose medium with L-Glutamine and Sodium Pyruvate (EuroClone, #ECM0728L) supplemented with 10% FBS (Thermo Fisher Scientific, #A4766801) and 1% penicillin/streptomycin (Pen/Strep, Thermo Fisher Scientific, #15140122). Flp-In T-REx 293 and DUX4-Flp-In T-REx 293 cells were grown in DMEM high glucose medium with L-Glutamine and Sodium Pyruvate supplemented with 10% Tetracycline-negative FBS (EuroClone, #ECS0182L) and 1% Pen/Strep and STREP-HA_DUX4 expression was induced upon doxycycline (Sigma-Aldrich, #D9891) administration. All the above cell lines were maintained at 37°C in a humidified incubator with 5% CO2.
Human primary cells
Human primary muscle cells derived from FSHD patients and healthy donors were kindly provided by Dr. Rabi Tawil, Richard Fields Center for FSHD Research Biobank (Department of Neurology, University of Rochester, NY, USA). Information about muscle cells is available in Table S6. Myoblasts were authenticated by morphology, growth characteristics, PCR for tissue-specific gene expression and differentiation markers, tested mycoplasma-free, and frozen. Cells were cultured for <2 months after thawing and were maintained at 37°C in a humidified incubator with 5% O2 and 5% CO2.
Myoblasts were grown in Ham’s F-10 medium (Euroclone, #ECB7503L), supplemented with L-Glutamine (Thermo Fisher Scientific, #25030081), 20% FBS, 1% Pen/Strep, 10 ng/ml bFGF (Tebu-bio, #100-18B), and 1 mM dexamethasone (Sigma-Aldrich, #D4902). To induce differentiation, growth medium was replaced by DMEM:F12 (1:1, Sigma-Aldrich, #D9785) supplemented with 20% knockout serum replacement (KOSR, Thermo Fisher Scientific, #10828028), 3.151 g/L glucose, 10 mM MEM non-essential amino acids (Thermo Fisher Scientific, #11140050), 1 mM sodium pyruvate (Thermo Fisher Scientific, #11360070). Differentiation was carried out for 96h.
METHOD DETAILS
Constructs and cloning procedures
Expression pCMV vectors carrying FLAG-tagged full-length MATR3, SMARCC2, CDC27 and RUVBL1 were obtained by Addgene (see “Recombinant DNA” section in the “key resources table”).78–81 FLAG-MATR3 2_288, 2_322 and 2_798 mutants were generated by the introduction of termination codons into the pCMV vector carrying full-length MATR3 through mutagenic PCR, using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, #200517). FLAG-MATR3 289_end coding sequence was amplified by PCR (GoTaq flexi DNA polymerase, Promega) and cloned into a pCMV-Tag2B vector by restriction digest cloning.
DUX4 full-length and DUX4 dbd inserts were generated by PCR using pCS2-mkgDUX4 vector (see “Recombinant DNA” section in the “key resources table”) as template and cloned into the pCDNA5/FRT/TO STREP-HA destination vector, kindly provided by Dr. Giulio Superti-Furga (CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, 1090 Vienna, Austria), by gateway technology (Thermo Fisher Scientific).
For recombinant protein purification, DUX4 dbd was cloned into the bacterial expression vector pET-GB1, carrying GB1 and 6xHIS tags,85 and MATR3 2_288 was cloned into pGEX-2tk vector by restriction digest (see “Recombinant DNA” section in the “Key resources table”). Primers used for cloning are listed in Table S2.
Total proteins extraction and immunoblotting
HEK293 cells were harvested and lysed in IP buffer (50mM Tris-HCl pH 7,5; 150mM NaCl; 1% NP-40; 5mM EDTA; 5mM EGTA; protease inhibitors). Cell extracts were resolved on a 10% or 6% SDS-PAGE acrylamide gels, transferred to nitrocellulose blotting membrane (GE Healthcare Life Sciences) and incubated with the following primary antibodies: anti-FLAG M2 (Sigma-Aldrich, #F1804), anti-HA.11 (Covance, #MMS-101R), anti-MATR3 (Thermo Fisher Scientific, #PA5-57720), anti-6xHis (Clontech, #631212), anti-GST (Sigma-Aldrich, #G1160), anti-tubulin (Sigma-Aldrich, #T9026). Anti-mouse IgG-HRP #715-035-150 or anti-rabbit IgG-HRP #711-035-152 (Jackson ImmunoResearch) were used as secondary antibodies.
Affinity purification from nuclear extracts
For the STREP-HA affinity purifications of parental and iSH-DUX4 Flp-In T-REx 293 cells, 1μg/mL of doxycycline was added to the cell culture media for 8h prior to cell harvesting. Nuclear protein extraction and STREP-HA affinity purification was conducted as previously described.86 Briefly, cells were lysed in buffer N (300 mM sucrose, 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1 mM DTT, 0.75 mM spermidine, 0.15 mM spermine, 0.1% Nonidet P-40, 50 mM NaF, protease inhibitors) for 5 min on ice and then centrifuged (500g for 5min) to separate the nuclear pellet from the supernatant containing the cytoplasmic fraction. The nuclear pellet was then washed with buffer N and resuspended in buffer C420 (20 mM HEPES pH 7.9, 420 mM NaCl, 25% glycerol, 1 mM EDTA, 1 mM EGTA, 0.1 mM DTT, 50 mM NaF, protease inhibitors), vortexed briefly, and shaken vigorously for 30 min. Samples were then centrifuged at 100000g for 1 h at 4°C and the supernatant containing the soluble nuclear proteins were collected and quantified by Bradford assay. 100 mg of nuclear extracts were used for the two-step affinity purifications. Prior to purification, nuclear extracts were adjusted to 150 mM NaCl with HEPES buffer (20 mM HEPES, 50 mM NaF, protease inhibitors) and brought to the final volume of 7.5 mL with TNN-HS buffer (50 mM HEPES pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, protease inhibitors). Samples were then incubated for 20 min at 4°C on a rotating wheel with RNase A, benzonase and avidin to remove nucleic acids and saturate endogenously biotinylated proteins, respectively. Nuclear extracts were then precleared with Protein G Sepharose beads (GE Healthcare, #71708300) for 1h at 4°C on rotation, and then incubated with Strep-Tactin Sepharose beads (IBA Lifesciences, #2-1201-002) overnight at 4°C on rotation. The day after the flow-through was removed, beads were washed 3 times with TNN-HS buffer and proteins bound to the beads were eluted with 3 consecutive incubations with 300 μl of 2.5 mM D-Biotin (Sigma-Aldrich, #2031) in TNN-HS buffer. The biotin eluate was subsequently incubated with anti-HA-agarose beads (Sigma-Aldrich, #A2095) for 2 h at 4°C on a rotating wheel. Samples were centrifuged for 3 min at 300 g and beads were washed 3 times with TNN-HS buffer. Another two washing steps with TNN-HS buffer without detergent and inhibitors were performed to remove traces of detergent that are detrimental to LC—MS analysis. Finally, proteins were eluted in 50 μl 2% SDS buffer, boiled 5 min at 95°C and centrifuged 3 min at 300g. The supernatant containing the eluted proteins was processed according to the Filter Aided Sample Preparation (FASP) protocol87 to remove the SDS prior to trypsin digestion, using EMD Millipore Amicon Ultra-0.5 Centrifugal Filter Units (Thermo Fisher Scientific, #UFC500324). Within the procedure, samples were reduced with Dithiothreitol (DTT), alkylated with Iodoacetamide and digested with Trypsin sequencing grade (Sigma-Aldrich), as previously described.88
MS analysis and protein identification
Tryptic peptides were desalted using StageTip C18 (Thermo Fisher Scientific, #87782) and analyzed by nLC-MS/MS using a Q-Exactive mass spectrometer (Thermo Fisher Scientific) equipped with a nano-electrospray ion source and a nanoUPLC Easy nLC 1000 (Proxeon Biosystems). Peptide separations occurred on a homemade (75 μm i.d., 12 cm long) reverse phase silica capillary column, packed with 1.9-μm ReproSil-Pur 120 C18-AQ (Dr. Maisch GmbH, Germany). A gradient of eluents A (distilled water with 0.1% v/v formic acid) and B (acetonitrile with 0.1% v/v formic acid) was used to achieve separation (300 nl/min flow rate), from 5% B to 50% B in 88 minutes. Full scan spectra were acquired with the lock-mass option, resolution set to 70,000 and mass range from m/z 300 to 2000 Da. The ten most intense doubly and triply charged ions were selected and fragmented.
To quantify proteins, the raw data were loaded into the MaxQuant82 software version 1.5.2.8 to search the human_proteome 20180425 (93,606 sequences; 37,037,628 residues). Searches were performed with the following settings: trypsin as proteolytic enzyme; 3 missed cleavages allowed; carbamidomethylation on cysteine as fixed modification; protein N-terminus-acetylation, methionine oxidation as variable modifications. Peptides and proteins were accepted with an FDR less than 1%. Label-free protein quantification was based on the spectral counts considering only proteins identified with minimum two peptides in any SH-DUX4 purification. The following filtering criterion was used to discriminate the specific interactors of SH-DUX4: the protein must be detected in all the three STREP-HA DUX4 biological replicates with spectral counts fold enrichment > 4 with respect of the control affinity purifications performed on parental cells.
Transfection of siRNA and plasmids
siRNA transfection was performed using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, #L3000001), following manufacturer’s instructions. For FSHD muscle cells, siRNAs were delivered 24 h after induction of differentiation and myotubes were harvested 96 h after induction of differentiation. The list of siRNAs used in this study is provided in Table S3.
Plasmids were delivered by using Lipofectamine LTX Reagent with PLUS Reagent (Thermo Fisher Scientific, #15338-100), following manufacturer’s instructions.
When transfection of both siRNAs and plasmids was required, HEK293 cells were reverse-transfected with siRNAs by Lipofectamine 3000 Transfection Reagent and the day after they were transfected with plasmids using Lipofectamine LTX Reagent with PLUS Reagent, following manufacturer’s instructions. Cells were harvested 48h after the last transfection.
Lentiviral production and transduction
For MATR3 overexpression experiments in FSHD muscle cells, lentiviral particles were produced in HEK293T cells. Briefly, HEK293T cells at 90% confluence were transfected in 10 cm dish with 6.5 μg of lentiviral vectors, 6 μg of pCMV-dR8.91 plasmid and 0.65 μg of pCMV-VSV-G plasmid. Lentiviral constructs used for gene expression and differentiation experiments carry either GFP alone as control (pFUGW:GFP, a kind gift from Shanahan CM lab), or MATR3-full length or MATR3 2_288 cDNA fused to GFP. Lentiviral vectors used for IncuCyte assays carry BFP alone (pLBC2-BS-RFCA-BCVIII, a kind gift from Dr. Mullighan CG lab, used as negative control), or MATR3-full length or MATR3 2_288 cDNA. After three collections of viral suspension (48 h, 72 h and 96 h post-transfection), viral preparations were concentrated of 100-fold by ultra-centrifuging at 20,000 rpm for 2h at 4°C and then resuspended in Opti-MEM Reduced Serum Medium (Thermo Fisher Scientific, #31985070) and stored at −80°C. FSHD muscle cells were transduced 24h after induction of differentiation and harvested 72h after infection.
RNA extraction, reverse transcription, and quantitative real-time PCR
Total RNA was extracted using PureLink RNA Mini Kit (Thermo Fisher Scientific), following manufacturer’s instructions. Briefly, cells were lysed in Lysis buffer supplemented with 2-mercaptoethanol and homogenized through a 21-gauge syringe needle. After adding one volume of 70% ethanol, lysates were loaded onto the spin cartridges provided by the kit, washed, treated with DNAseI (PureLink DNase Set, Thermo Fisher Scientific), and eluted in RNAse-free water.
cDNA synthesis was performed using SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific), following manufacturer’s instructions.
Quantitative real-time PCR (qPCR) was performed with iTaq Universal SYBR Green Supermix (Biorad) using CFX96 Real-Time PCR Detection System (Bio-Rad). Primers used for RT-qPCR are listed in Table S4. Relative quantification was performed using the ΔΔCt method. Specific details of each data set are provided in the Figure legends.
Immunofluorescence
Immunofluorescence of DUX4 was performed on FSHD muscle cells were plated on coverslips and differentiated for 96h. Myotubes were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. After 3 washes in PBS, cells were permeabilized in 1% Triton X-100 (Sigma-Aldrich) in PBS for 15 min at room temperature and blocked in 2% goat serum, 2% horse serum, 2% BSA, 0.1% Triton-X100 in PBS, for 45 min at room temperature. Cells were then incubated with 1:100 anti-DUX4 E5-5 antibody (Abcam) at 37°C in a humid chamber overnight. After 3 washes in PBS, cells were incubated with fluorescent-conjugated Alexa 555 goat anti-rabbit secondary antibody (Molecular Probes) for 45 min at RT and rinsed again in PBS. Counterstaining with Hoechst 33342 was performed for 10 min at RT and after 3 washes in PBS coverslips were mounted and imaged by a fluorescence microscope.
For immunofluorescence of FLAG-tagged MATR3 mutants, HEK293 cells were plated on coverslips, transfected as described in the “transfection” section above and fixed in 4% PFA 48 h after transfection. Cells were then permeabilized with 1% TritonX-100 in PBS and immunostained with anti-FLAG M2 antibody (Sigma-Aldrich, dilution 1:1000) followed by Alexa Fluor 488 (Molecular Probes, dilution 1:500) and DAPI. Cells were imaged using an upright fluorescence microscope (Observer.Z1, Zeiss).
Myotube morphology analysis
For myotube morphology analysis, cells were fixed in 4% PFA, permeabilized with 1% TritonX-100 in PBS and immunostained with mouse MF20 antibody (Developmental Studies Hybridoma Bank; dilution 1:2) followed by Alexa Fluor 488 or 555 goat anti-mouse (Molecular Probes, dilution 1:500) and Hoechst 33342. Cells were imaged using a fluorescence microscope (Observer.Z1, Zeiss). The differentiation index was scored by determining the percentage of myosin heavy chain-positive (MHC+) nuclei. Fusion index analysis was performed with ImageJ software by determining the percentage of nuclei included or not into myotubes (myosin positive syncytia containing at least 3 nuclei). The nuclei distribution was quantified by determining the percentage of myofibers carrying the indicated number of nuclei. For each treatment, three differentiation experiments were performed and at least 4 fields per well were analyzed.
Cell viability and apoptotic assay
Cell viability was measured using the CellTiter-Glo luminescent assay (Promega), following manufacturer’s instructions. Apoptosis was measured through Caspase-Glo 3/7 luminescent assay (Promega) following manufacturer’s instructions, and luminescence was quantified by Wallace 1420 multilabel Victor3 microplate reader (Perkin Elmer). These assays were performed in HEK-iSH-DUX4 cells 24h after doxycycline administration, and in HEK293 cells 48h after transfection.
For Staurosporine treatment, HEK293 cells were transfected with pCMV-FLAG empty vector or carrying MATR3 full-length, and then treated with 4 μM DMSO or 4 μM Staurosporine (Sigma-Aldrich) for 6 hours. Cells were then collected and the Caspase 3/7 Glo assay was performed.
Apoptotic levels in primary human myotubes from FSHD patients or healthy individuals transduced with lentiviral vectors carrying BFP-MATR3 full-length, BFP-MATR3 2_288 or BFP only as control were determined using the IncuCyte live-cell imaging system (Essen BioScience). Briefly, 24 h post-transduction, differentiation medium was replaced with fresh medium containing 1ml/ml of IncuCyte CASP3/7 assay GREEN reagent (Essen BioScience) and the green fluorescence signal was acquired by the IncuCyte S3 Imager system as follows: 36 scans/well every 3 h for 72 h. The signal was analyzed using the IncuCyte software. Results are shown as average of the fluorescent signal from three replicates normalized to the fluorescence detected at time 0.
Strep-Tactin pull-down assays
HEK293T cells were transfected with pCDNA5/FRT/TO STREP-HA empty vector, or carrying DUX4 full-length or DUX4 dbd using PolyFect Transfection Reagent (QIAGEN), according to manufacturer’s instructions. 24 hours after transfection, cells were harvested, and nuclear extracts were prepared as described above. Pull-down was performed using 600 μg of nuclear proteins, by adding 2 volumes of HEPES buffer (20mM Hepes pH 8; 50mM NaF; protease inhibitors) and TNN buffer (50mM Hepes pH 8.0; 150mM NaCl; 5mM EDTA; 0.5% NP-40 substitute; 50mM NaF; protease inhibitors) to reduce the NaCl concentration. Nuclear extracts were incubated for 1h at 4°C with Avidin, Benzonase and RNase A and precleared with Protein G Sepharose beads for 1h at 4°C with rotation. Protein complexes were obtained by incubation of nuclear extracts with 40 μl of Strep-Tactin Sepharose beads overnight at 4°C in gentle rotation. After 3 washes with IP-buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 1% NP-40, 1mM EDTA, 0.5mM EGTA), proteins were eluted adding 2.5mM D-Biotin. Input (10% or 0,5%) and bound fractions (10%) of the pull down were analyzed by immunoblotting.
Proximity ligation assay
Proximity ligation assay of MATR3 and DUX4 or WDR5 and DUX4 (negative control) was performed in muscle cells plated on coverslips and differentiated for 96h, by using Duolink in situ Red kit (Sigma-Aldrich). Myotubes were fixed in 4% paraformaldehyde in PBS for 10 min at 4°C. After 3 washes in PBS, cells were permeabilized in 1% TritonX-100 in PBS for 15 min at room temperature and blocked in 2% goat serum, 2% horse serum, 2% BSA, 0.1% Triton-X100 in PBS, for 45 min at room temperature. Coverslips were then incubated with primary antibodies and PLA was carried out following manufacturer’s instructions and imaged using a fluorescence microscope. Primary antibodies used are α-MATR3 (1:200, Thermo Fisher Scientific), α-DUX4 (1:50, P2B1, Sigma-Aldrich) and α-WDR5 (1:200, Cell Signaling Technology).
FLAG-immunoprecipitation (FLAG-IP) assay
HEK-iSH-DUX4 cells were transfected with pCMV-FLAG empty vector, or carrying MATR3 full-length, MATR3 2–288 or MATR3 289–847, using Lipofectamine LTX Reagent with PLUS Reagent, following manufacturer’s instructions. 24 hours after transfection, DUX4 expression was induced by 1 μg/ml doxycycline administration. Cells were harvested 8 h post-induction and lysed in IP buffer (50mM Tris-HCl pH 7,5; 150mM NaCl; 1% NP-40; 5mM EDTA; 5mM EGTA; protease inhibitors). Protein extracts were treated with Avidin, Benzonase and RNase A for 1h at 4°C and then incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) for 3h at 4°C with rotation. After 3 washes with IP buffer, proteins were eluted in Laemmli buffer and Input (10%), and IP fractions were analyzed by immunoblotting with anti-FLAG M2 and anti-DUX4 E5-5 antibodies.
Recombinant protein purification
6×His-DUX4 dbd, GST and GST-MATR3 2_288 constructs were transferred to Rosetta2(DE3) pLys E. coli (Novagen) following manufacturer’s instructions, and selected on kanamycin or ampicillin plates, respectively. A single bacterial colony was grown in LB medium supplemented with antibiotics at 37°C until they reached an OD600nm= 0.6–0.7. The induction was made with 1 mM IPTG (Biosciences) for 3 hours at 37°C for GST-MATR3 2_288 and GST or 20 hours at 18°C for 6×His-DUX4 dbd. Bacterial pellets were resuspended in Lysis Buffer 1 (PBS; 1mM PMSF; 5mM 2-mercaptoethanol) or Lysis Buffer 2 (50 mM NaH2PO4, 1M NaCl, pH 8.0, plus protease inhibitors) for GST- and His-tagged proteins, respectively. Bacteria were then sonicated using a Bandelin-sonoplus HD3100 (probe MS73) sonicator (10 cycles of 30sec on and 30sec off 80% amplitude), incubated by gentle rotation for 15 minutes at 4°C after adding Triton X-100 (1%; Sigma-Aldrich), and centrifuged at 15000 rpm at 4°C for 20 minutes. Cleared lysates were incubated for 1 hour at 4°C with Glutathione-Sepharose beads (GE Healthcare) or His-Select Nickel Affinity gel beads (Sigma-Aldrich), for GST- and His-tagged proteins, respectively. Beads were washed with Lysis Buffer 1 or Lysis Buffer 2 plus 10 mM imidazole (Fluka), for GST- and His-tagged proteins respectively. Proteins were eluted with elution solution 1 (20mM glutathione, 100mM Tris-HCl pH8.0, 120mM NaCl) for GST-tagged protein and with elution solution 2 (50 mM NaH2PO4, 1M NaCl, 250 mM imidazole pH 8.0) for His-tagged proteins.
Proteins were dialyzed overnight at 4°C in Slide-A-Lyzer dialysis cassettes (Thermo Fisher Scientific) in Lysis solution. The purification steps and the obtained proteins were analyzed by Coomassie Blue staining loading samples on 10% polyacrylamide gels. Purified proteins were quantified at Nanodrop and on gel using Bovine Serum Albumin (BSA) as protein standard.
GST pulldown assay
For GST pulldown assays, 40 pmol of purified GST or GST-MATR3 2_288 were immobilized on Glutathione-Sepharose beads (GE Healthcare) and incubated with 40 pmol of purified soluble His-DUX4 dbd in 1 ml of IPP100 buffer (10 mM Tris–HCl, pH 8, 100 mM NaCl, 0.1% NP-40) supplemented with 2 mM DTT and 1mM PMSF for 2 hours at 4°C. Beads were washed three times with IPP100 buffer and boiled in Laemmli buffer. Bound fractions (10%) were loaded on 10% acrylamide gel and immunoblotted using α-GST (Sigma-Aldrich) and α−6xHis antibody (Clontech).
RNA-sequencing
NGS libraries were generated with the mRNA Stranded TruSeq protocol. Sequencing was performed on an Illumina Nova-Seq 6000 (Illumina, San Diego, CA), obtaining an average of 50 million pair-end reads, 100nt long per sample. Reads were trimmed using Trimmomatic, version 0.39, to remove adapters and exclude low-quality reads from the analysis. The remaining reads were then aligned to the reference genome GRCh38, GENCODE v31, using STAR aligner, version 2.5.3a.83 FeatureCounts (v 1.6.4) software84 was used to assign reads to the corresponding genes. Only genes with a CPM (Counts per million) value higher than 1 in at least four samples were retained. Gene expression read counts were exported and analyzed in the R environment (v. 4.0.5) to identify differentially expressed genes (DEGs, Table S5) using the DESeq Bioconductor library.89 To control differences due to library preparation batches, we included this variable in the DGE design (~condition+batch). P-values were adjusted using a threshold for false discovery rate (FDR) ≤ 0.05.90 Using the 500 most variable genes in terms of RPKM (counts per million reads normalized on library sizes and gene lengths), we perform Principal Component Analysis (prcomp function in R) and clustering analysis via heatmap (CRAN package pheatmap - v.1.0.12). Volcano plot analysis was performed in R [using ggplot2 R library (v3.3.5)], labelling the 20 upregulated and downregulated genes with the most significant adjusted p-value. The lists of genes induced or repressed by more than two-fold by iDUX4, vDUX4 and enDUX4 proposed by Jagannathan et al. (Table S4)34 were compared to the modulated genes of this study, filtered for absolute values of |log2 FC| > 1. Annotation differences between this dataset and our annotation (GENCODE v31 basic annotation) were corrected by updating the gene names from Jagannathan et al. to their most recent gene alias in GENCODE v31. This conversion was performed using the Ensembl IDs, which remain comparable across different annotation versions. To test the statistical significance of the overlap between gene lists, we used the Bioconductor package GeneOverlap (v3.15) using the number of expressed features in our experiment as genome size. List intersections were drawn with the CRAN package “venn” (v1.11).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, San Diego, USA). Quantitative data are presented as mean ± standard deviation (SD). Statistical significance was calculated on at least three independent experiments. P-value: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Details of statistical test used for each dataset are provided in the corresponding figure legend.
Supplementary Material
Highlights.
MATR3 is a nuclear interactor of DUX4 in FSHD muscular dystrophy
MATR3 is a bifunctional DUX4 inhibitor that blocks its expression and pathogenic activity
MATR32_288 binds directly to DUX4 DNA-binding domain
MATR32_288 is required and sufficient to inhibit DUX4
ACKNOWLEDGMENTS
We thank Dr. R. Tawil and the University of Rochester, Fields Center for FSHD, for providing the human primary muscle cells from healthy donors and patients with FSHD used in this work. We thank Dr. G. Superti-Furga for the pcDNA/FRT/TO/STREP-HA, Dr. C.M. Shanahan for the pFUGW:GFP, and Dr. C.G. Mullighan for the pLBC2-BS-RFCA-BCVIII constructs. The MF20 monoclonal antibody developed by Dr. D.A. Fischman was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242, USA. Research in D.G.’s laboratory is funded by the Italian Ministry of Health (grant number RF-2018-12366631), the European Joint Programme on Rare Diseases (grant number EJPRD20-191), the Italian Association for Cancer Research (grant number IG 2017-ID 19919), and the National Institutes of Health-National Cancer Institute (grant number 1R21CA249378-01). R.G. was supported by a co-funded fellowship between Vita-Salute San Raffaele University and the European Union Seventh Framework Marie Curie Program (COFUND-INVEST, GA-2010-267264). The graphical abstract was created with BioRender (https://www.biorender.com).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113120.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research. Received: March 28, 2023
DECLARATION OF INTERESTS
D.G., R.G., V.R., and C.C. are inventors of the patent “Inhibitor of DUX4 and uses thereof” WO/2020/152367. This had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
REFERENCES
- 1.Deenen JCW, Arnts H, van der Maarel SM, Padberg GW, Verschuuren JJGM, Bakker E, Weinreich SS, Verbeek ALM, and van Engelen BGM. (2014). Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 83, 1056–1059. 10.1212/WNL.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Statland JM, and Tawil R. (2014). Risk of functional impairment infacioscapulohumeral muscular dystrophy. Muscle Nerve 49, 520–527. 10.1002/mus.23949. [DOI] [PubMed] [Google Scholar]
- 3.Padberg GW, Frants RR, Brouwer OF, Wijmenga C, Bakker E, and Sandkuijl LA. (1995). Facioscapulohumeral muscular dystrophy in the dutch population. Muscle Nerve 18, 81–84. 10.1002/mus.880181315. [DOI] [PubMed] [Google Scholar]
- 4.Statland JM, Odrzywolski KJ, Shah B, Henderson D, Fricke AF, van der Maarel SM, Tapscott SJ, and Tawil R. (2015). Immunohistochemical Characterization of Facioscapulohumeral Muscular Dystrophy Muscle Biopsies. J. Neuromuscul. Dis. 2, 291–299. 10.3233/JND-150077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia FF, Drew AP, Nicholson GA, Corbett A, and Kumar KR. (2021). Facioscapulohumeral muscular dystrophy type 2: an update on the clinical, genetic, and molecular findings. Neuromuscul. Disord. 31, 1101–1112. 10.1016/j.nmd.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Sacconi S, Briand-Suleau A, Gros M, Baudoin C, Lemmers RJLF, Rondeau S, Lagha N, Nigumann P, Cambieri C, Puma A, et al. (2019). FSHD1 and FSHD2 form a disease continuum. Neurology 92, e2273–e2285. 10.1212/WNL.0000000000007456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemmers RJLF, Van Der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, Van Ommen GJ, Padberg GW, et al. (2010). A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653. 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mocciaro E, Runfola V, Ghezzi P, Pannese M, and Gabellini D. (2021). DUX4 role in normal physiology and in FSHD muscular dystrophy. Cells 10, 3322. 10.3390/cells10123322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934. 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ. (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet. 49, 935–940. 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941–945. 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, and Harper SQ. (2011). DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 69, 540–552. 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, vanderMaarel SM, Ruzzo WL, Gentleman RC, Tawil R, and Tapscott SJ. (2012). DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Dev. Cell 22, 38–51. 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Z, Snider L, Balog J, Lemmers RJLF, van der Maarel SM, Tawil R, and Tapscott SJ. (2014). DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 23, 5342–5352. 10.1093/hmg/ddu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickard AM, Petek LM, and Miller DG. (2015). Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 24, 5901–5914. 10.1093/hmg/ddv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñ ez NG, Leo O, et al. (2007). The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 17, 611–623. 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Sandri M, El Meslemani AH, Sandri C, Schjerling P, Vissing K, Andersen JL, Rossini K, Carraro U, and Angelini C. (2001). Caspase 3 expression correlates with skeletal muscle apoptosis in Duchenne and facioscapulo human muscular dystrophy. A potential target for pharmacological treatment? J. Neuropathol. Exp. Neurol. 60, 302–312. 10.1093/jnen/60.3.302. [DOI] [PubMed] [Google Scholar]
- 18.Block GJ, Narayanan D, Amell AM, Petek LM, Davidson KC, Bird TD, Tawil R, Moon RT, and Miller DG. (2013). Wnt/β-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet. 22, 4661–4672. 10.1093/hmg/ddt314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tawil R, McDermott MP, Pandya S, King W, Kissel J, Mendell JR, and Griggs RC. (1997). A pilot trial of prednisone in facioscapulohumeral muscular dystrophy. FSH-DY Group. Neurology 48, 46–49. 10.1212/wnl.48.1.46. [DOI] [PubMed] [Google Scholar]
- 20.Passerieux E, Hayot M, Jaussent A, Carnac G, Gouzi F, Pillard F, Picot M-C, Böcker K, Hugon G, Pincemail J, et al. (2015). Effects of vitamin C, vitamin E, zinc gluconate, and selenomethionine supplementation on muscle function and oxidative stress biomarkers in patients with facioscapulohumeral dystrophy: a double-blind randomized controlled clinical trial. Free Radic. Biol. Med. 81, 158–169. 10.1016/j.freeradbiomed.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Kissel JT, McDermott MP, Mendell JR, King WM, Pandya S, Griggs RC, and Tawil R; FSH-DY Group (2001). Randomized, double-blind, placebo-controlled trial of albuterol in facioscapulohumeral dystrophy. Neurology 57, 1434–1440. 10.1212/wnl.57.8.1434. [DOI] [PubMed] [Google Scholar]
- 22.Elsheikh BH, Bollman E, Peruggia M, King W, Galloway G, and Kissel JT. (2007). Pilot trial of diltiazem in facioscapulohumeral muscular dystrophy. Neurology 68, 1428–1429. 10.1212/01.wnl.0000264017.08217.39. [DOI] [PubMed] [Google Scholar]
- 23.Wagner KR, Fleckenstein JL, Amato AA, Barohn RJ, Bushby K, Escolar DM, Flanigan KM, Pestronk A, Tawil R, Wolfe GI, et al. (2008). A phase I/IItrial of MYO-029 in adult subjects with muscular dystrophy. Ann. Neurol. 63, 561–571. 10.1002/ana.21338. [DOI] [PubMed] [Google Scholar]
- 24.Matsumura T, Yokoe M, Nakamori M, Hattori N, Saito T, Nozaki S, Fujimura H, and Shinno S. (2004). A clinical trial of creatine monohydrate in muscular dystrophy patients. Clin. Neurol. 44, 661–666. [PubMed] [Google Scholar]
- 25.van der Kooi EL, Kalkman JS, Lindeman E, Hendriks JCM, van Engelen BGM, Bleijenberg G, and Padberg GW. (2007). Effects of training and albuterol on pain and fatigue in facioscapulohumeral muscular dystrophy. J. Neurol. 254, 931–940. 10.1007/s00415-006-0432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payan CA, Hogrel JY, Hammouda EH, Lacomblez L, Ollivier G, Doppler V, Eymard B, Attarian S, Pouget J, Desnuelle C, and Laforêt P. (2009). Periodic Salbutamol in Facioscapulohumeral Muscular Dystrophy: A Randomized Controlled Trial. Arch. Phys. Med. Rehabil. 90, 1094–1101. 10.1016/j.apmr.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 27.Mellion ML, Ronco L, Berends CL, Pagan L, Brooks S, van Esdonk MJ, van Brummelen EMJ, Odueyungbo A, Thompson LA, Hage M, et al. (2021). Phase 1 clinical trial of losmapimod in facioscapulohumeral dystrophy: Safety, tolerability, pharmacokinetics, and target engagement. Br. J. Clin. Pharmacol. 87, 4658–4669. 10.1111/bcp.14884. [DOI] [PubMed] [Google Scholar]
- 28.Sitzia C, Meregalli M, Belicchi M, Farini A, Arosio M, Bestetti D, Villa C, Valenti L, Brambilla P, and Torrente Y. (2019). Preliminary evidences of safety and efficacy of flavonoids- And omega 3-based compound for muscular dystrophies treatment: A randomized double-blind placebo controlled pilot clinical trial. Front. Neurol. 10, 755. 10.3389/fneur.2019.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glasser CE, Gartner MR, Wilson D, Miller B, Sherman ML, and Attie KM. (2018). Locally acting ACE-083 increases muscle volume in healthy volunteers. Muscle Nerve 57, 921–926. 10.1002/mus.26113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Homma S, Beermann ML, Boyce FM, and Miller JB. (2015). Expression of FSHD-related DUX4-FL alters proteostasis and induces TDP-43 aggregation. Ann. Clin. Transl. Neurol. 2, 151–166. 10.1002/acn3.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerji CRS, and Zammit PS. (2021). Pathomechanisms and biomarkers in facioscapulohumeral muscular dystrophy: roles of DUX4 and PAX7. EMBO Mol. Med. 13, e13695. 10.15252/emmm.202013695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim KRQ, Nguyen Q, and Yokota T. (2020). Dux4 signalling in the pathogenesis of facioscapulohumeral muscular dystrophy. Int. J. Mol. Sci. 21, 729. 10.3390/ijms21030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schätzl T, Kaiser L, and Deigner HP. (2021). Facioscapulohumeral muscular dystrophy: genetics, gene activation and downstream signalling with regard to recent therapeutic approaches: an update. Orphanet J. Rare Dis. 16, 129. 10.1186/s13023-021-01760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagannathan S, Shadle SC, Resnick R, Snider L, Tawil RN, van der Maarel SM, Bradley RK, and Tapscott SJ. (2016). Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet. 25, 4419–4431. 10.1093/hmg/ddw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppée F, et al. (2008). An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779. 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dmitriev P, Bou Saada Y, Dib C, Ansseau E, Barat A, Hamade A, Dessen P, Robert T, Lazar V, Louzada RAN, et al. (2016). DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic. Biol. Med. 99, 244–258. 10.1016/j.freeradbiomed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Barro M, Carnac G, Flavier S, Mercier J, Vassetzky Y, and Laoudj-Chenivesse D. (2010). Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J. Cell Mol. Med. 14, 275–289. 10.1111/j.1582-4934.2008.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanderplanck C, Ansseau E, Charron S, Stricwant N, Tassin A, Laoudj-Chenivesse D, Wilton SD, Coppée F, and Belayew A. (2011). The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One 6, e26820. 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shadle SC, Zhong JW, Campbell AE, Conerly ML, Jagannathan S, Wong CJ, Morello TD, van der Maarel SM, and Tapscott SJ. (2017). DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet. 13, e1006658. 10.1371/journal.pgen.1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Wang Z, Jin S, Hao H, Zheng L, Zhou B, Zhang W, Lv H, and Yuan Y. (2014). Dux4 induces cell cycle arrest at G1 phase through upregulation of p21 expression. Biochem. Biophys. Res. Commun. 446, 235–240. 10.1016/j.bbrc.2014.02.105. [DOI] [PubMed] [Google Scholar]
- 41.Feng Q, Snider L, Jagannathan S, Tawil R, van der Maarel SM, Tapscott SJ, and Bradley RK. (2015). A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. Elife 4, e04996. 10.7554/eLife.04996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shadle SC, Bennett SR, Wong CJ, Karreman NA, Campbell AE, van der Maarel SM, Bass BL, and Tapscott SJ. (2019). DUX4-induced bidirectional HSATII satellite repeat transcripts form intranuclear double-stranded RNA foci in human cell models of FSHD. Hum. Mol. Genet. 28, 3997–4011. 10.1093/hmg/ddz242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malik AM, and Barmada SJ. (2021). Matrin 3 in neuromuscular disease: physiology and pathophysiology. JCI Insight 6, e143948. 10.1172/jci.insight.143948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, and Kyba M. (2016). DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44, 5161–5173. 10.1093/nar/gkw141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, and Séraphin B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 46.Köcher T, and Superti-Furga G. (2007). Mass spectrometry–based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815. 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 47.Glatter T, Wepf A, Aebersold R, and Gstaiger M. (2009). An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237. 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varjosalo M, Sacco R, Stukalov A, van Drogen A, Planyavsky M, Hauri S, Aebersold R, Bennett KL, Colinge J, Gstaiger M, and Superti-Furga G. (2013). Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods 10, 307–314. 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 49.Belgrader P, Dey R, and Berezney R. (1991). Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J. Biol. Chem. 266, 9893–9899. [PubMed] [Google Scholar]
- 50.Salem A, Wilson CJ, Rutledge BS, Dilliott A, Farhan S, Choy WY, and Duennwald ML. (2021). Matrin3: Disorder and ALS Pathogenesis. Front. Mol. Biosci. 8, 794646. 10.3389/fmolb.2021.794646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tassin A, Laoudj-Chenivesse D, Vanderplanck C, Barro M, Charron S, Ansseau E, Chen YW, Mercier J, Coppée F, and Belayew A. (2013). DUX4 expression in FSHD muscle cells: How could such a rare protein cause a myopathy? J. Cell Mol. Med. 17, 76–89. 10.1111/j.1582-4934.2012.01647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snider L, Geng LN, Lemmers RJLF, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, and Miller DG. (2010). Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181. 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Söderberg O, Leuchowius K-J, Gullberg M, Jarvius M, Weibrecht I, Larsson L-G, and Landegren U. (2008). Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232. 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Mocciaro E, Giambruno R, Micheloni S, Cernilogar FM, Andolfo A, Consonni C, Pannese M, Ferri G, Runfola V, Schotta G, and Gabellini D. (2023). WDR5 is required for DUX4 expression and its pathological effects in FSHD muscular dystrophy. Nucleic Acids Res. 51, 5144–5161. 10.1093/nar/gkad230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corona ED, Jacquelin D, Gatica L, and Rosa AL. (2013). Multiple Protein Domains Contribute to Nuclear Import and Cell Toxicity of DUX4, a Candidate Pathogenic Protein for Facioscapulohumeral Muscular Dystrophy. PLoS One 8, e75614. 10.1371/journal.pone.0075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones TI, Chen JCJ, Rahimov F, Homma S, Arashiro P, Beermann ML, King OD, Miller JB, Kunkel LM, Emerson CP, et al. (2012). Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 21, 4419–4430. 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caron L, Kher D, Lee KL, McKernan R, Dumevska B, Hidalgo A, Li J, Yang H, Main H, Ferri G, et al. (2016). A human pluripotent stem cell model of facioscapulohumeral muscular dystrophy-affected skeletal muscles. Stem Cells Transl. Med. 5, 1145–1161. 10.5966/sctm.2015-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen J, DeSimone A, Lek M, and Lek A. (2021). Therapeutic Approaches in Facioscapulohumeral Muscular Dystrophy. Trends Mol. Med. 27, 123–137. 10.1016/j.molmed.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell AE, Shadle SC, Jagannathan S, Lim J-W, Resnick R, Tawil R, van der Maarel SM, and Tapscott SJ. (2018). NuRD and CAF-1-mediated silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins. Elife 7, e31023. 10.7554/eLife.31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malik AM, Miguez RA, Li X, Ho Y-S, Feldman EL, and Barmada SJ. (2018). Matrin 3-dependent neurotoxicity is modified by nucleic acid binding and nucleocytoplasmic localization. Elife 7, e35977. 10.7554/eLife.35977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramesh N, Kour S, Anderson EN, Rajasundaram D, and Pandey UB. (2020). RNA-recognition motif in Matrin-3 mediates neurodegeneration through interaction with hnRNPM. Acta Neuropathol. Commun. 8, 138. 10.1186/s40478-020-01021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sprunger ML, Lee K, Sohn BS, and Jackrel ME. (2022). Molecular determinants and modifiers of Matrin-3 toxicity, condensate dynamics, and droplet morphology. iScience 25, 103900. 10.1016/j.isci.2022.103900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackenzie IR, Rademakers R, and Neumann M. (2010). TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9, 995–1007. 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 64.Douville R, Liu J, Rothstein J, and Nath A. (2011). Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 69, 141–151. 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, Lee M-H, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K, et al. (2015). Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 7, 307ra153. 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowen LN, Tyagi R, Li W, Alfahad T, Smith B, Wright M, Singer EJ, and Nath A. (2016). HIV-associated motor neuron disease: HERV-K activation and response to antiretroviral therapy. Neurology 87, 1756–1762. 10.1212/WNL.0000000000003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krug L, Chatterjee N, Borges-Monroy R, Hearn S, Liao W-W, Morrill K, Prazak L, Rozhkov N, Theodorou D, Hammell M, and Dubnau J. (2017). Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 13, e1006635. 10.1371/journal.pgen.1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor JP, Brown RH, and Cleveland DW. (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, and Tapscott SJ. (2013). DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis. PLoS Genet. 9, e1003947. 10.1371/journal.pgen.1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, Tawil R, Chung S-A, Masny PS, and Figlewicz DA. (2003). Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul. Disord. 13, 322–333. 10.1016/s0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 71.Turki A, Hayot M, Carnac G, Pillard F, Passerieux E, Bommart S, Raynaud de Mauverger E, Hugon G, Pincemail J, Pietri S, et al. (2012). Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free Radic. Biol. Med. 53, 1068–1079. 10.1016/j.free-radbiomed.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 72.Dandapat A, Bosnakovski D, Hartweck LM, Arpke RW, Baltgalvis KA, Vang D, Baik J, Darabi R, Perlingeiro RCR, Hamra FK, et al. (2014). Dominant Lethal Pathologies in Male Mice Engineered to Contain an X-Linked DUX4 Transgene. Cell Rep. 8, 1484–1496. 10.1016/j.celrep.2014.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Senderek J, Garvey SM, Krieger M, Guergueltcheva V, Urtizberea A, Roos A, Elbracht M, Stendel C, Tournev I, Mihailova V, et al. (2009). Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am. J. Hum. Genet. 84, 511–518. 10.1016/j.ajhg.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boehringer A, Garcia-Mansfield K, Singh G, Bakkar N, Pirrotte P, and Bowser R. (2017). ALS Associated Mutations in Matrin 3 Alter Protein-Protein Interactions and Impede mRNA Nuclear Export. Sci. Rep. 7, 14529. 10.1038/s41598-017-14924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neguembor MV, Previtali S, and Gabellini D. (2015). Facioscapulohumeral Dystrophy. In TheOnline Metabolic and Molecular Bases of Inherited Disease, Valle DL, Antonarakis SE, Ballabio A, Beaudet AL, and Mitchell GA, eds. (McGraw-Hill Medical; ). 10.1036/ommbid.216.1. [DOI] [Google Scholar]
- 76.Wang LH, and Tawil R. (2016). Facioscapulohumeral Dystrophy. Curr. Neurol. Neurosci. Rep. 16, 66. 10.1007/s11910-016-0667-0. [DOI] [PubMed] [Google Scholar]
- 77.Mul K, Voermans NC, Lemmers RJLF, Jonker MA, van der Vliet PJ, Padberg GW, van Engelen BGM, van der Maarel SM, and Horlings CGC. (2018). Phenotype-genotype relations in facioscapulohumeral muscular dystrophy type 1. Clin. Genet. 94, 521–527. 10.1111/cge.13446. [DOI] [PubMed] [Google Scholar]
- 78.Salton M, Elkon R, Borodina T, Davydov A, Yaspo ML, Halperin E, and Shiloh Y. (2011). Matrin 3 binds and stabilizes mRNA. PLoS One 6, e23882. 10.1371/JOURNAL.PONE.0023882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xi Q, He W, Zhang XHF, Le HV, and Massagué J. (2008). Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J. Biol. Chem. 283, 1146– 1155. 10.1074/JBC.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, Villén J, Wang B, Kim SR, Sakamoto K, et al. (2011). Chemical genetic screen for AMPKα2 substrates uncovers a network of proteins involved in mitosis. Mol. Cell 44, 878–892. 10.1016/J.MOL-CEL.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, and Artandi SE. (2008). Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–957. 10.1016/J.CELL.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 83.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/BIOINFORMATICS/BTS635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/BIOINFORMATICS/BTT656. [DOI] [PubMed] [Google Scholar]
- 85.Zhou P, and Wagner G. (2010). Overcoming the solubility limit with solubility-enhancement tags: successful applications in biomolecular NMR studies. J. Biomol. NMR 46, 23–31. 10.1007/s10858-009-9371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giambruno R, Grebien F, Stukalov A, Knoll C, Planyavsky M, Rudashevskaya EL, Colinge J, Superti-Furga G, and Bennett KL. (2013). Affinity purification strategies for proteomic analysis of transcription factor complexes. J. Proteome Res. 12, 4018–4027. 10.1021/pr4003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wiśniewski JR, Zougman A, Nagaraj N, and Mann M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 88.Huber ML, Sacco R, Parapatics K, Skucha A, Khamina K, Müller AC, Rudashevskaya EL, and Bennett KL. (2014). abFASP-MS: affinity-based filter-aided sample preparation mass spectrometry for quantitative analysis of chemically labeled protein complexes. J. Proteome Res. 13, 1147–1155. 10.1021/pr4009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benjamini Y, and Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc. B 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited at ProteomeXchange database via the PRIDE partner repository (ProteomeXchange: PXD011073 and https://doi.org/10.6019/PXD011073) and RNA-seq data have been deposited at GEO database (GEO: GSE210008). They are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat# F1804, RRID: AB_262044 |
| Mouse monoclonal anti-HA.11 (Clone 16B12) | Covance | Cat# MMS-101R, RRID: AB_291262 |
| Rabbit polyclonal anti MATR3 | Thermo Fisher Scientific | Cat# PA5-57720, RRID: AB_2643782 |
| Mouse monoclonal anti-6xHis | Clontech | Cat# 631212, RRID: AB_2721905 |
| Mouse monoclonal anti-GST | Sigma-Aldrich | Cat# G1160, RRID: AB_259845 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026, RRID: AB_477593 |
| Anti-mouse IgG-HRP | Jackson ImmunoResearch | Cat # 715-035-150, RRID: AB_2340770 |
| Anti-rabbit IgG-HRP | Jackson ImmunoResearch | Cat # 711-035-152, RRID: AB_10015282 |
| Rabbit monoclonal anti-DUX4 E5-5 | Abcam | Cat# ab124699, RRID: AB_10973363 |
| Mouse monoclonal anti-DUX4 P2B1 (for PLA) | Sigma-Aldrich | Cat# SAB5200019, RRID: AB_2612761 |
| Mouse monoclonal anti-myosin | DSHB | Cat# MF20, RRID: AB_2147781 |
| Rabbit monoclonal anti-WDR5 (clone D9E1I) | Cell Signaling Technology | Cat# 13105, RRID:AB_2620133 |
| Alexa 555 goat anti-rabbit | Molecular Probes | Cat# A-27039, RRID: AB_2536100 |
| Alexa 555 goat anti-mouse | Molecular Probes | Cat # A-21422, RRID: AB_141822 |
| Alexa Fluor 488 goat anti-mouse | Molecular Probes | Cat# A-32723, RRID: AB_2633275 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Rosetta2 (DE3) pLysS competent cells | Novagen | Cat #71403 |
|
| ||
| Biological samples | ||
|
| ||
| FSHD primary muscle cells | Richard Fields Center for FSHD Research Biobank | Table S6 of this paper |
| Healthy primary muscle cells | Richard Fields Center for FSHD Research Biobank | Table S6 of this paper |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Staurosporine | Sigma-Aldrich | 19123 |
| Doxycycline | Sigma-Aldrich | D9891 |
| D-Biotin | Sigma-Aldrich | 2031 |
| DL-Dithiothreitol | Sigma-Aldrich | D9779 |
| Trypsin-EDTA (0,5%) | Gibco | 15400054 |
| Dimethyl sulfoxide | Sigma-Aldrich | D2650 |
| L-Glutamine | Thermo Fisher Scientific | 25030081 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Paraformaldehyde | Electron Microscopy Sciences | 157–8 |
| GST recombinant protein | This paper | N/A |
| GST-MATR3 2–288 recombinant protein | This paper | N/A |
| HIS-DUX4 dbd recombinant protein | This paper | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| Caspase-Glo 3/7 luminescent assay | Promega | G8091 |
| CellTiter-Glo luminescent assay | Promega | G7570 |
| CASP3/7 assay GREEN reagent | Essen BioScience | 4440 |
| Duolink in situ Red kit | Sigma-Aldrich | DUO92101 |
| QuikChange XL Site-Directed Mutagenesis Kit | Agilent Technologies | 200517 |
| Gateway LR Clonase Enzyme mix | Invitrogen | 11791019 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | L3000001 |
| Lipofectamine LTX Reagent with PLUS Reagent | Thermo Fisher Scientific | 15338-100 |
| PureLink RNA Mini Kit | Thermo Fisher Scientific | 12183025 |
| DNAseI (PureLink DNase Set) | Thermo Fisher Scientific | 12185010 |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | 18080051 |
| iTaq Universal SYBR Green Supermix | Biorad | 1725122 |
| Polyfect Transfection Reagent | QIAGEN | 301107 |
| Slide-A-Lyzer dialysis cassettes | Thermo Fisher Scientific | 66380 |
| Protein G Sepharose beads | GE Healthcare | 71708300 |
| Strep-Tactin Sepharose beads | IBA Lifesciences | 2-1201-002 |
| anti-HA-agarose beads | Sigma-Aldrich | A2095 |
| Glutathione-Sepharose beads | GE Healthcare | 17513201 |
| anti-FLAG M2 Affinity Gel | Sigma-Aldrich | A2220 |
| His-Select Nickel Affinity gel beads | Sigma-Aldrich | H0537 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE210008 and Table S5 |
| Raw and analyzed proteomics data | This paper | ProteomeXchange: PXD011073, https://doi.org/10.6019/PXD011073 and Table S1 |
| DUX4 core target gene list | Table S4 (Jagannathan S et al.)34 | https://doi.org/10.1093/hmg/ddw271 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T cells | ATCC | CRL-3216 |
| Flp-In T-REx 293 cells | Thermo Fisher Scientific | R78007 |
| DUX4 Flp-In T-Rex cells (HEK-iSH-DUX4) | This manuscript | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S2 for a list of primers for cloning | This paper | N/A |
| See Table S3 for a list of siRNAs | N/A | N/A |
| See Table S4 for a list of primers for RT-qPCR | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV_FLAG-MATR3 | (Salton et al.)78 | RRID: Addgene_32880 |
| pCMV_FLAG-SMARCC2 | (Xi et al.)79 | RRID: Addgene_19142 |
| pECE-M2-CDC27 | (Banko et al.)80 | RRID: Addgene_31661 |
| pCDNA-3xFLAG-RUVBL1 | (Venteicher et al.)81 | RRID: Addgene_51635 |
| pCMV_FLAG-MATR3 2_288 | This paper | N/A |
| pCMV_FLAG-MATR3 2_322 | This paper | N/A |
| pCMV_FLAG-MATR3 2_798 | This paper | N/A |
| pCMV-FLAG-MATR3 289_end | This paper | N/A |
| pCS2-mkgDUX4 vector | (Snider et al.)52 | RRID: Addgene_21156 |
| pET-GB1-DUX4 dbd | This paper | N/A |
| pGEX-2tk- MATR3 2_288 | This paper | N/A |
| pCDNA5/FRT/TO STREP-HA | Dr. Superti-Furga G, CeMM Research Center for Molecular Medicine | Austrian Academy of Sciences, 1090 Vienna, Austria |
| pCMV-dR8.91 | Broad Institute | http://www.broadinstitute.org/rnai/trc |
| VSV-G/pMD2G | Broad Institute | http://www.broadinstitute.org/rnai/trc |
| pFUGW:GFP | Dr. Shanahan CM, King’s College London, London, UK | https://doi.org/10.1091/mbc.E16-06-0346 |
| pFUGW:GFP- MATR3-full length | This paper | N/A |
| pFUGW:GFP- MATR3 2_288 | This paper | N/A |
| pLBC2-BS-RFCA-BCVIII | Dr. Mullighan CG, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA | https://doi.org/10.1038/ng.3691 |
| pLBC2-BS-RFCA-BCVIN-att-MATR3-full length | This paper | N/A |
| pLBC2-BS-RFCA-BCVIII-att-MATR3 2_288 | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad PRISM 8 | GraphPad Software | https://www.graphpad.com/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| CFX Manager Software 3.1 | Biorad | https://www.bio-rad.com/it-it/sku/1845000-cfx-manager-software?ID=1845000 |
| MaxQuant (v 1.5.2.8) | (Cox J. et al.)82 | https://doi.org/10.1021/pr101065j |
| STAR aligner (v 2.5.3a) | (Dobin A et al.)83 | https://doi.org/10.1093/bioinformatics/bts635 |
| CRAN package pheatmap (v.1.0.12) | CRAN package | https://cran.r-project.org/package=pheatmap |
| CRAN package “venn” (v1.11). | CRAN package | https://CRAN.R-project.org/package=venn |
| FeatureCounts (v 1.6.4) | (Liao Y et al.)84 | https://doi.org/10.1093/bioinformatics/btt656 |