Summary
The Hippo pathway is an evolutionarily conserved regulator of tissue growth that integrates inputs from both polarity and actomyosin networks. An upstream activator of the Hippo pathway, Kibra, localizes at the junctional and medial regions of the apical cortex in epithelial cells, and medial accumulation promotes Kibra activity. Here, we demonstrate that cortical Kibra distribution is controlled by a tug of war between apical polarity and actomyosin dynamics. We show that while the apical polarity network, in part via aPKC, tethers Kibra at the junctional cortex to silence its activity, medial actomyosin flows promote Kibra-mediated Hippo complex formation at the medial cortex, thereby activating the Hippo pathway. This study provides a mechanistic understanding of the relationship between the Hippo pathway, polarity, and actomyosin cytoskeleton and offers novel insights into how fundamental features of epithelial tissue architecture can serve as inputs into signaling cascades that control tissue growth, patterning, and morphogenesis.
Keywords: Kibra, Hippo pathway, polarity, actomyosin, tissue growth, epithelia
eTOC blurb
Tokamov et al. show that apical polarity and actomyosin networks control subcellular distribution of Kibra, an activator of the growth-suppressing Hippo pathway. In a tug of war, apical polarity silences Kibra via junctional tethering, while actomyosin flows promote Hippo signaling via medial Kibra accumulation.
Graphical Abstract
Introduction
Growth and morphogenesis of epithelial tissues are critical determinants of organ size. Epithelial tissues such as the Drosophila wing imaginal disc are composed of cells polarized along the apico-basal axis and tightly interconnected by intercellular junctions along the lateral domain. These characteristics feature prominently in the organization and function of the Hippo pathway, an evolutionarily conserved regulator of tissue growth.1,2 For example, an upstream activator of the Hippo pathway, Expanded (Ex), localizes at the junctional cortex (a term we use to include the marginal zone and the adherens junctions3), where it assembles and activates a signaling cascade that includes Hippo (Hpo) and Warts (Wts) kinases, thereby repressing a pro-growth transcriptional effector Yorkie (Yki).4–6 Unlike Ex, Kibra (Kib) and Merlin (Mer) accumulate at both the junctional and apicomedial cortex (referred to as the “medial” cortex in this study), where Kib assembles a Hippo signaling complex that inhibits Yki in parallel to Ex.7–9 The functional significance of these distinct subcellular Kib pools and the mechanisms that control the distribution of Kib between the junctional and medial cortex remain elusive.
Apico-basal asymmetry in epithelial cells is regulated by a mutually antagonistic relationship between apical and basolateral polarity determinants.3 Among the apical components, a transmembrane protein Crumbs (Crb) regulates polarity via associated cytoplasmic partners, including the Stardust (Sdt)/Patj and Par-6/atypical protein kinase C (aPKC) complexes. Aside from its role in polarity, however, Crb recruits Ex to the junctional cortex, thereby enabling Ex-mediated Hippo pathway activation.10,11 In the absence of Crb, Ex is diffuse throughout the cytoplasm and inactive. Previous work suggested that Crb also tethers Kib at the junctional cortex by an unknown mechanism, which suppresses Kib function.7 Additionally, ectopic aPKC activity was previously shown to inhibit Hippo signaling.12 Conversely, loss of Hippo pathway activity, at least in part via Yki-mediated transcriptional activity, promotes expansion of the apical domain in imaginal epithelial cells.13 Thus, apico-basal polarity and Hippo signaling are highly interconnected.
Another critical regulator of Hippo signaling is the actomyosin cytoskeleton. In many epithelia, the cortical actomyosin network is organized into junctional and medial pools at the apical domain.14 Junctional actomyosin cables can generate sustained forces to resist mechanical strain or drive morphogenetic events, such as junctional remodeling during convergent extension. On the other hand, the medial actomyosin network is more transient and pulsatile and is important in driving apical constriction of epithelial cells, which leads to tissue scale deformations. A large body of evidence suggests that increased F-actin polymerization and cortical tension generated by non-muscle myosin II (myosin) contractility promote the activity of Yki and its mammalian homolog, YAP.15–19 Additionally, in cultured cells cortical actin inhibits the ability of Mer to recruit and activate Wts.20 However, how actomyosin organization and dynamics influence upstream Hippo signaling components remains unknown. Specifically, the relationship between medial actomyosin pulses and Hippo signaling has not been explored. Several observations suggest that Kib is uniquely poised to mediate interactions between polarity, actomyosin, and Hippo signaling. First, Kib has a key role in recruiting and activating the Hippo kinase cascade.21–23,7 Second, previous studies have shown that Kib physically interacts with aPKC.24–26 Lastly, Kib’s localization at the junctional and medial cortex closely resembles that of actomyosin7 (Figure 1A). Here, using a combination of osmotic, pharmacological, and genetic manipulations we show that Kib distribution at the cell cortex is regulated by the apical polarity and actomyosin networks. We demonstrate that aPKC tethers Kib at the junctional cortex to inhibit Kib-mediated Hippo pathway activation, whereas medial actomyosin flows promote Kib activity by accumulating and maintaining it at the medial cortex. Our results suggest a model whereby a tug of war between the apical polarity and actomyosin networks modulates Hippo signaling and tissue growth via Kib subcellular localization.
Figure 1. Medial Kib is associated with the actomyosin network.
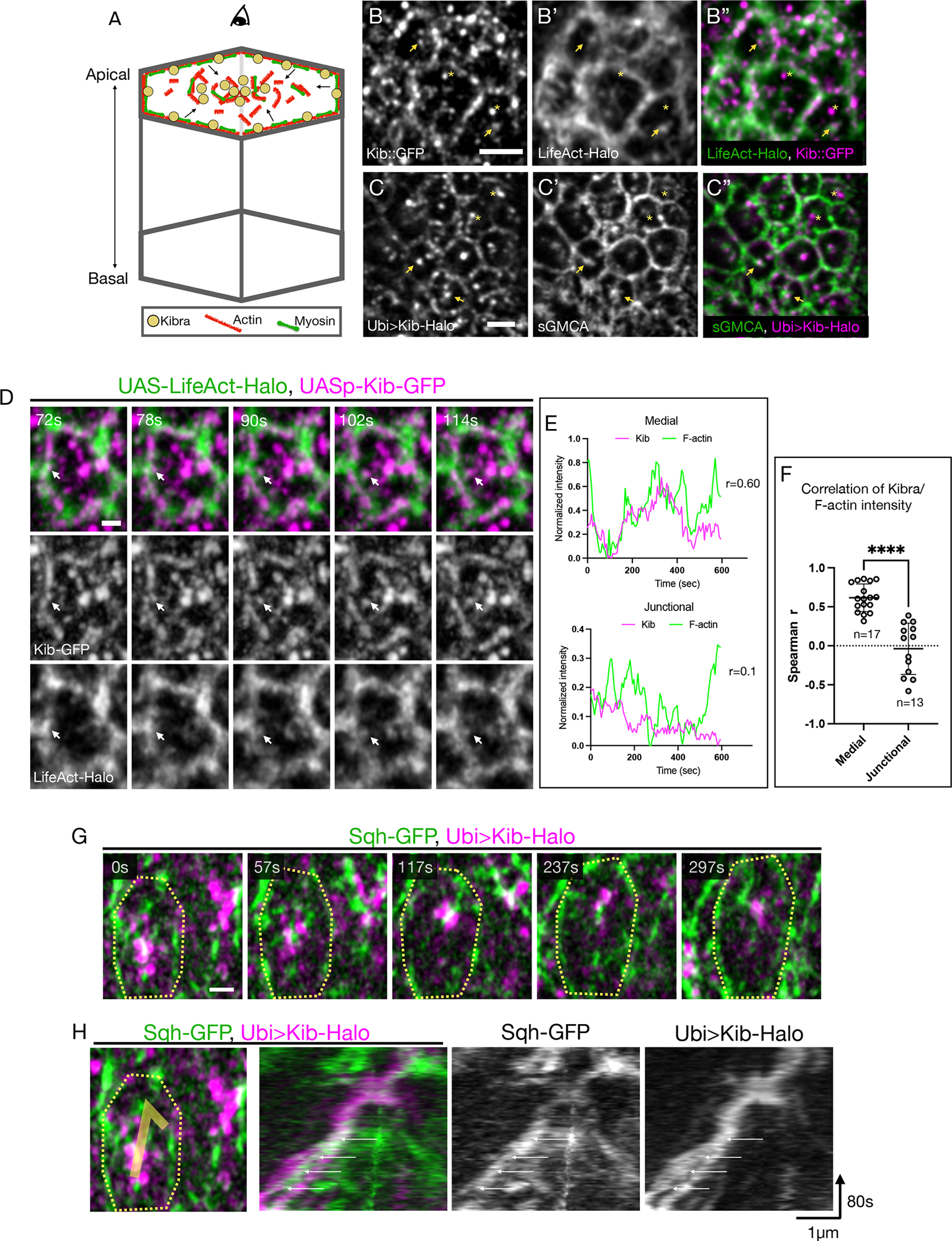
(A) A cartoon of an epithelial cell polarized along the apical-basal axis. The actomyosin network and Kib are similarly partitioned at the apical cortex into junctional and medial pools. The arrows indicate flows of actomyosin components from the junctional to the medial region. Throughout the paper, images display tissue apical projections as viewed from the top.
(B-C”) Apical projections of wing imaginal tissues expressing either endogenous Kib::GFP and LifeAct-Halo (B-B”) or Ubi>Kib-Halo and sGMCA (C-C”). Arrowheads point to medial Kib punctae that appear to decorate apical F-actin structures. Asterisks indicate medial Kib punctae that do not appear to be associated with F-actin. Scale bars = 2μm.
(D) Single frames from Video S1 of a single cell showing co-migration of a Kib cluster (magenta) with F-actin (green) from the junctional to the medial region (white arrow). Scale bar = 1μm.
(E) Representative plots of medial or junctional Kib and F-actin intensities over time and their corresponding Spearman r correlation coefficient values measured from a single cell.
(F) A plot of Spearman r coefficients from multiple cells comparing correlation of medial vs. junctional Kib and F-actin intensity over time. Statistical significance was calculated using the Mann-Whitney test; n = number of cells. Throughout the paper, unless otherwise noted data are shown as the mean ± SD and significance values are represented as follows: ****p ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05, ns = not significant.
(G) Single frames from Video S2 of a single cell showing co-migration of medial Kib (magenta) with Sqh (green). The contour line roughly outlines the cell. Scale bar = 1μm.
(H) A kymograph tracking co-migration of medial Kib and Sqh shown in E. The white arrows point to a myosin pulse increasing in intensity over time that is associated with the merger of two initially separate Kib punctae. See also Video S2.
Results
Medial Kib localization is associated with actomyosin dynamics
The actomyosin cytoskeleton is known to regulate growth via Yki, but the link between actomyosin dynamics and the upstream Hippo pathway components remains unknown (throughout the study, we use the term “dynamics” to describe general movement of actomyosin components at the cell cortex). The similarity in Kib and actomyosin subcellular distribution prompted us to investigate their localization in more detail. Using Airyscan confocal microscopy for better spatial resolution, we first examined the localization of either endogenous Kib tagged with the green fluorescent protein (Kib::GFP) or Halo-tagged Kib expressed under the ubiquitin promoter (Ubi>Kib-Halo) with respect to F-actin. To visualize F-actin, we used either UAS-driven LifeAct-Halo or a GFP-tagged Moesin actin-binding domain expressed under the spaghetti squash promoter (sGMCA). In the wing imaginal epithelium, both Kib::GFP and Ubi>Kib-Halo localized at the junctional and medial cortex, as observed previously8,27 (Figures 1B and 1C). Although both Kib and F-actin localize junctionally, Kib’s punctate pattern is distinct from the more uniformly distributed F-actin (Figures 1B–1C”). In contrast, medial Kib distribution seemed superficially similar to that of F-actin. Close examination revealed that both Kib::GFP and Ubi-Kib-Halo punctae localized adjacent to or decorated medial F-actin structures (Figures 1B–1C”), suggesting an association with the apical actomyosin network. However, we also observed medial Kib clusters that appeared to be independent of F-actin (Figures 1B–1C”). Thus, although cortical Kib organization is similar to F-actin, Kib does not appear to be tightly associated with the medial F-actin pool.
We reasoned that the lack of strong co-localization is not necessarily indicative of the absence of the relationship between Kib and F-actin. For example, in the C. elegans zygote, the distribution of a polarity regulator, Par3, is controlled by actomyosin-driven cortical flows, even though Par3 displays very little co-localization with cortical F-actin.28–30 Therefore, we examined Kib subcellular dynamics with F-actin or myosin via time lapse imaging. Because endogenous Kib is expressed at low levels in wing imaginal discs, we used UASp-Kib-GFP, which produces functional Kib with normal subcellular localization.8 We observed coordinated movement of Kib with F-actin, and Kib punctae often appeared to be carried by F-actin from the junctional to medial cortex, combining with the existing pool of Kib clusters, some of which appeared independent of F-actin (Figure 1D, Video S1). To ask if Kib medial accumulation correlates with F-actin, we measured medial Kib and F-actin intensity over time. Indeed, in the medial cortex increased Kib intensity significantly correlated with increased F-actin (Figures 1E and 1F). In contrast, junctional Kib and F-actin did not display strong correlation (Figures 1E and 1F).
Similarly, time lapse imaging of Kib-Halo with GFP-tagged Spaghetti squash (myosin regulatory light chain in Drosophila, Sqh-GFP) revealed coupled movement of both proteins at the medial cortex (Figures 1G and 1H, Video S2). Together, these results suggest that actomyosin organization and dynamics might control Kib distribution between the junctional and medial cortex.
Osmotic shifts lead to coordinated changes in actomyosin and Kib distribution
To further test the relationship between actomyosin dynamics and Kib localization, we sought to acutely alter actomyosin organization and examine any potential changes in Kib localization. Changes in osmolarity have been shown to affect F-actin organization in cultured cells,31–34 so we decided to use this approach on explanted wing imaginal tissues. In wing imaginal discs incubated under isotonic conditions, both F-actin and myosin are apically enriched, where they are partitioned into junctional and medial pools (Figures 2A, 2A’, 2E, and 2E’). Strikingly, incubating wing imaginal tissues in a hypertonic solution led to medial enrichment of F-actin (Figures 2B and 2B’). Ecad localization was unaffected, indicating that this effect was not due to a general collapse of cell-cell junctions (Figure 2B). Conversely, under hypotonic conditions F-actin accumulated predominantly at the junctional cortex (Figures 2C and 2C’). Quantification of F-actin distribution revealed a decrease and increase in junctional/medial F-actin intensity under hypertonic and hypotonic conditions, respectively (Figure 2D).
Figure 2. Osmotic shifts lead to coordinated changes in actomyosin and Kib distribution.
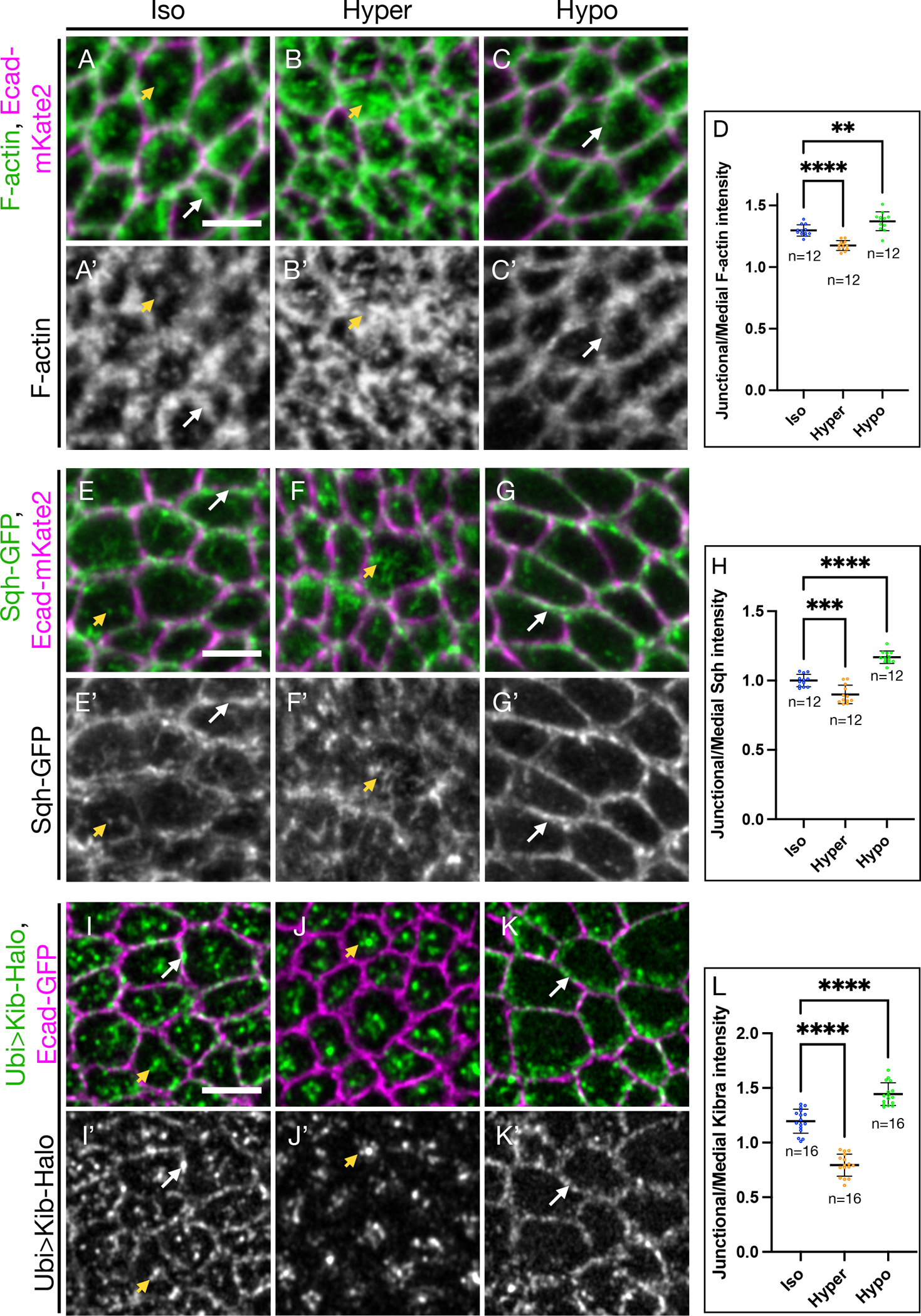
(A-C’) Apical projections of wing imaginal tissues expressing Ecad-mKate2 and an F-actin marker, sGMCA. F-actin is both junctional (white arrows) and medial (yellow arrowhead) under isotonic conditions (A and A’); under hypertonic conditions (B and B’), F-actin accumulates more medially; in contrast, under hypotonic conditions (C and C’), F-actin is more junctional.
(D) Quantification of junctional/medial F-actin intensity under osmotic shifts.
(E-G’) Apical projections of wing imaginal tissues expressing Ecad-mKate2 and Sqh-GFP. Sqh is both junctional and medial under isotonic conditions (E and E’); Sqh becomes mostly medial and is decreased at the junctions under hypertonic conditions (F and F’); in contrast, Sqh is more junctional under hypotonic conditions (G and G’).
(H) Quantification of junctional/medial Sqh intensity under osmotic shifts.
(I-K’) Under isotonic conditions (I and I’), Kib is both junctional and medial. Similar to F-actin and myosin, Kib becomes predominantly medial under hypertonic conditions (J and J’) and mostly junctional under hypotonic conditions (K and K’).
(L) Quantification of junctional/medial Kib intensity under osmotic shifts.
Scale bars = 3μm. Statistical significance was calculated using One-way ANOVA followed by Tukey’s HSD test; n = number of wing discs See also Figure S1, Video S3, and Video S4.
Similar to the changes in F-actin organization, myosin also became more medially enriched under hypertonic conditions, though it was more prominently lost from the junctions (Figures 2F and 2F’). On the other hand, hypotonic shift led to significant junctional accumulation of myosin (Figures 2G–2H). These results show that in the wing imaginal epithelium, changes in extracellular osmolarity can be used to acutely shift subcellular actomyosin distribution between junctional and medial cortical pools.
We next examined whether osmotic shifts affect Kib localization. Compared to the normal junctional and medial Kib distribution under isotonic conditions (Figures 2I and 2I’), Kib strongly accumulated medially and became almost undetectable at the junctional cortex under hypertonic conditions (Figures 2J and 2J’). In sharp contrast, Kib localized mostly at the junctional cortex under hypotonic conditions (Figures 2K and 2K’). Quantification of Kib organization revealed that as in the case of F-actin and myosin, junctional/medial Kib fluorescence decreased under hypertonic and increased under hypotonic conditions relative to isotonic controls (Figure 2L). These effects were readily reversible because Kib localization could be altered as the same tissue was sequentially subjected to hypertonic and then hypotonic conditions (Figures S1A–S1D). Additionally, time lapse imaging of Sqh-GFP with Kib-Halo revealed strong spatial and temporal correlation in Kib and myosin rearrangement upon osmotic shifts. Shift to hypertonic medium led to dynamic medial myosin flows that appeared to cluster Kib at the medial cortex (Video S3). Conversely, after a hypotonic shift, myosin and Kib simultaneously localized at the junctional cortex (Video S4). These observations suggest that changes in Kib localization are driven by changes in actomyosin organization in response to changes in cortical tension35,36 (see Discussion).
The drastic changes in Kib localization under osmotic shifts made us wonder if all cortical proteins could be similarly affected by these manipulations. We examined the localization of a spectraplakin Short stop (Shot), which links the minus-ends of apico-basal microtubule arrays to the apical actin cortex in polarized epithelia. Similar to Kib, Shot localizes apically in epithelial cells and can re-localize from the junctional to the medial cortex.37,38 However, in marked contrast to Kib, Shot seemed to enrich more junctionally under hypertonic conditions and accumulated mostly in medial clusters under hypotonic conditions (Figures S1E–S1G’). We also examined the localization of Ex and found that unlike Kib, Ex remained predominantly junctional under osmotic shifts (Figures S1H–S1H”). These observations suggest that changes in actomyosin organization could specifically modulate cortical Kib distribution.
Medial Kib localization is mediated via actomyosin dynamics
Given the strong correlation between actomyosin and Kib distribution under osmotic shifts, we next asked if changes in Kib localization were mediated by actomyosin flows. To this end, we sought to acutely stabilize F-actin using Jasplakinolide39 (Jasp) and examine the effect on Kib localization. Relative to control tissues, F-actin intensity increased dramatically in tissues treated with Jasp, confirming that Jasp potently stabilizes F-actin in the wing imaginal discs (Figures S2A–S2C). Treating tissues with Jasp under isotonic conditions was sufficient to increase junctional/medial Kib distribution (Figures 3A–3B’, 3E). Timelapse imaging of sGMCA with Ubi>Kib-Halo showed that medial Kib punctae were dynamic and moved together with F-actin in control tissues, whereas both Kib and F-actin movement was significantly impaired upon treatment with Jasp (Video S5).
Figure 3. Medial Kib accumulation is mediated via actomyosin dynamics.
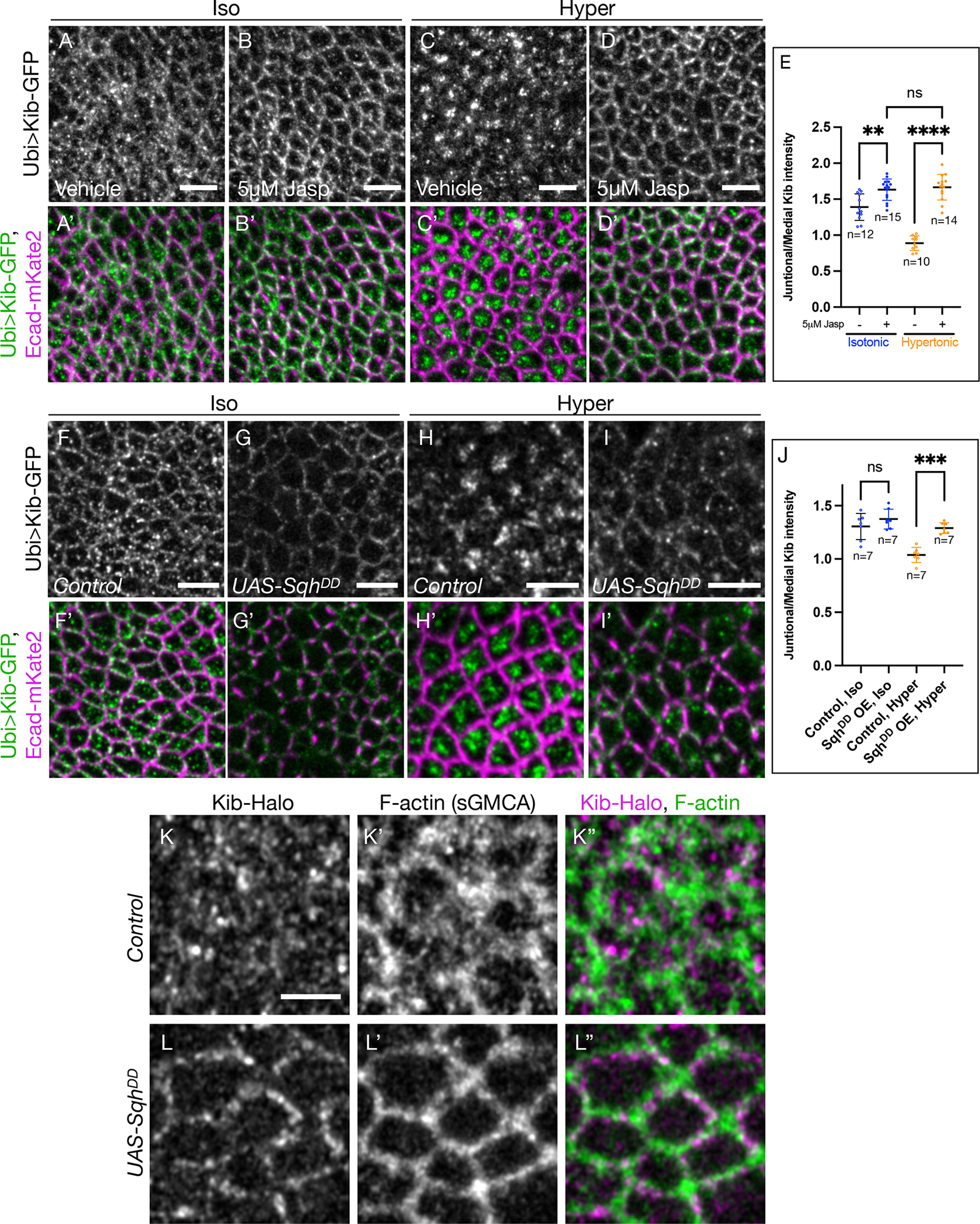
(A-E) Stabilizing F-actin prevents medial Kib localization. Compared to isotonic controls (A and A’), treating wing imaginal tissues with 5μM Jasp results in more junctional Kib (B and B’) While Kib is mostly medial under hypertonic conditions (C and C’), treatment with 5μM Jasp blocks the effect of the hypertonic shift on Kib localization (D and D’). Scale bars = 5μm. (E) Quantification of junctional/medial Kib intensity under the manipulations shown in (A-D’)
(F-I’) Dynamic myosin phosphorylation is required for medial Kib accumulation. UAS-SqhDD was expressed for 14h using hh>Gal4 driver and Gal80ts. SqhDD expression leads to slightly more junctional Kib under isotonic conditions (F-G’) and strongly inhibits medial Kib relocalization under hypertonic conditions (H-I’). Scale bars = 5μm.
(J) Quantification of junctional/medial Kib intensity under the manipulations shown in (F-I’). Statistical significance in (E) and (J) was calculated using One-way ANOVA followed by Tukey’s HSD test; n = number of wing discs.
(K-L”) Compared to control cells (K-K”), cells expressing SqhDD (L-L”) display significantly more junctional Kib and F-actin. Scale bar = 3μm. See also Figure S2, Figure S3, Video S5, and Video S6.
We then asked if stabilizing F-actin could prevent medial Kib enrichment induced by the hypertonic shift. Indeed, while Kib accumulated medially in a control hypertonic solution, this relocalization was effectively blocked by the addition of Jasp (Figures 3C–3E). To test if F-actin dynamics were required to maintain the medial Kib pool, we first concentrated Kib medially by incubating tissues in a hypertonic solution and then transferred them into a hypertonic solution with or without Jasp. Treatment with Jasp reversed hypertonically induced medial Kib accumulation (Figures S2D–S2E’), suggesting that F-actin dynamics not only accumulate but also maintain Kib at the medial cortex.
We also wondered if inhibiting medial myosin dynamics could have a similar effect on Kib localization. Dynamic phosphorylation-dephosphorylation cycling of the myosin regulatory light chain is required for medial pulsatile myosin activity during morphogenesis.40 Therefore, we examined Kib localization upon ectopic expression of a phosphomimmetic version of Sqh (SqhDD).41 As with F-actin stabilization under Jasp treatment, ectopic SqhDD expression resulted in significant stabilization of myosin, as evidenced by the increased intensity of GFP-tagged myosin heavy chain (Zip-GFP, Figures S2F–S2H). Ectopic SqhDD expression under isotonic conditions also resulted in a more junctional appearance of Kib (Figures 3F–3G’), though the effect was not quantitatively significant (Figure 3J), likely because the transgene was expressed transiently in the background of the endogenous Sqh. Timelapse imaging of Ubi>Kib-Halo and sGMCA strongly suggested that SqhDD dramatically inhibited medial F-actin flows, as both F-actin and Kib appeared significantly more junctional over time in SqhDD-expressing cells compared to control cells (Video S6, Figures 3J–3K”). SqhDD expression also significantly blocked medial Kib localization under hypertonic conditions (Figures 3H–3J). Similarly, transient depletion of Sqh blocked medial Kib accumulation both under isotonic and hypertonic conditions (Figures S2I–S2J’). Collectively, our data suggest that Kib accumulation at the medial cortex is promoted and actively maintained by medial actomyosin flows.
Actomyosin-driven medial Kib assembles a Hippo signaling complex
Previous work using genetic manipulations has shown that Kib is a key protein that recruits other Hippo pathway components into a signaling complex at the medial cortex, and that medially-localized Kib is more active in restricting growth27. However, we wondered if Hippo complex assembly could also occur when Kib is acutely accumulated at the medial cortex upon hypertonic shift. Activation of Hippo signaling by Kib requires the recruitment of Mer, and Mer helps recruit the core kinase cassette to the medial cortex.27 Under isotonic conditions, we observed some colocalization between Kib and Mer at the medial and junctional cortex (Figures 4A–4A”). Under hypertonic conditions, both proteins strongly accumulated at the medial cortex and their colocalization was more pronounced (Figures 4B–4B”), suggesting that actomyosin-driven medial Kib can rapidly recruit Mer into a signaling complex. Conversely, under hypotonic conditions Kib and Mer were both at the junctional cortex where their colocalization was less apparent (Figures 4C–4C”).
Figure 4. Actomyosin-driven medial Kib assembles a Hippo signaling complex.
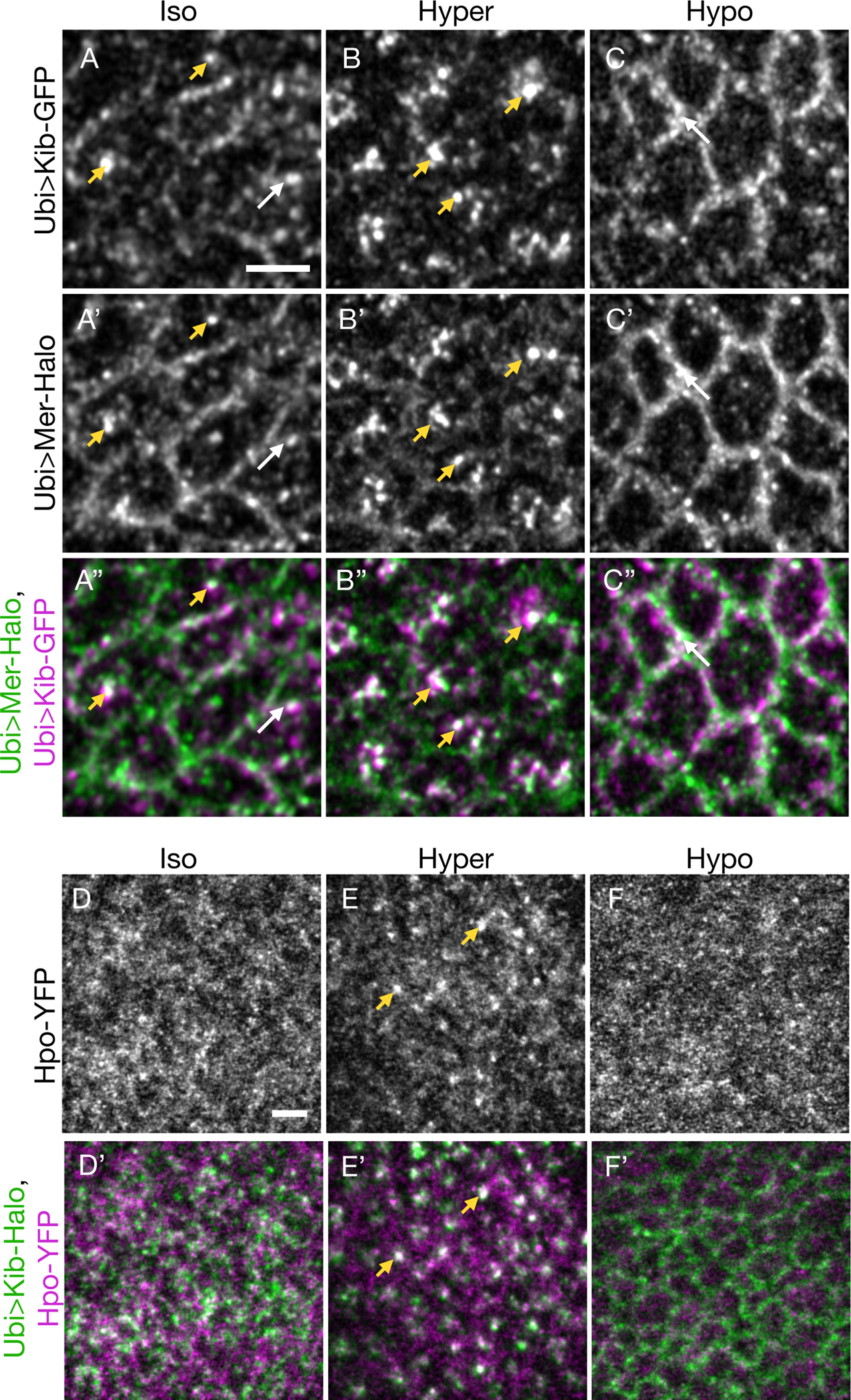
(A-C”) Mer displays some co-localization with Kib at the junctional (white arrows) and medial (yellow arrowheads) cortex under isotonic conditions (A-A”). Under hypertonic conditions, Mer is strongly recruited to medial foci with Kib (B-B”). Conversely, under hypotonic conditions, both Mer and Kib localize mainly at the junctional cortex (C-C”).
(D-F’) Hpo is normally diffuse under isotonic conditions (D and D’). However, under the hypertonic shift, Hpo is recruited to the medial cortex with Kib (E and E’). In contrast, Hpo remains diffuse under hypotonic conditions (F and F’). Scale bars = 3μm.
Next, we examined the effect of Kib relocalization under osmotic shifts on Hpo recruitment using endogenously expressed Hpo-YFP. Under isotonic conditions, Hpo-YFP appeared mostly diffuse, with no discernable localization pattern (Figures 4D and 4D’). However, as was observed with Mer, distinct Hpo foci formed and colocalized with medial Kib upon hypertonic shift (Figures 4E and 4E’). Conversely, shift to hypotonic conditions led to predominantly diffuse Hpo-YFP, with no detectible junctional localization (Figures 4F and 4F’). Collectively, these data suggest that acute medial Kib accumulation via actomyosin flows promotes Hippo complex assembly.
Crb controls Kib localization via actomyosin organization and cortical tethering
To this point, our results demonstrate that actomyosin flows promote medial Kib accumulation and Hippo complex formation. However, our observations that Kib accumulates junctionally when actomyosin dynamics are inhibited suggests that Kib could be tethered at the junctional cortex. We previously showed that Crb is required for junctional Kib localization,27 prompting us to further investigate the relationship between Crb and Kib.
Since loss of Crb results in similar medial Kib accumulation as observed under the hypertonic shift (Figures S3A–S3B’), we wondered if the actomyosin network was also altered in cells lacking Crb. Previous studies reported that loss of Crb leads to increased medial actomyosin organization.42,43 Consistent with those reports, we found that F-actin organization in crb-null clones was significantly more medial (Figures S3C and S3C’). We also found that similar to hypertonic conditions, loss of Crb disrupted junctional myosin organization and led to increased medial myosin accumulation (Figures S3D–S3F, Video S7). These observations suggest that medial accumulation of Kib in cells lacking Crb could be caused by increased medial actomyosin flows. Indeed, stabilizing F-actin via Jasp treatment blocked medial Kib accumulation in crb-depleted cells (Figures S3G–S3H’). Interestingly, while Jasp treatment resulted in more junctional Kib localization in control cells, it failed to restore junctional Kib in cells depleted of Crb (Figures S3G–S3H’). These results suggest that Crb can regulate Kib via two simultaneous mechanisms: 1) by inhibiting medial actomyosin flows and 2) by providing a tether to maintain Kib at the junctional cortex.
How could Crb tether Kib junctionally? Crb has a short intracellular region containing only two known functional motifs. The FERM-binding motif, which interacts with Ex, Moesin, and Yurt,3 is not required for Kib junctional localization.27 Consistent with this, while Crb depletion led to more medial Kib, loss of Ex had no effect on Kib localization (Figures S4A–S4B”). On the other hand, Crb’s PDZ-binding motif interacts with Sdt/Patj and aPKC/Par6 complexes, which in turn could recruit Kib since Kib itself does not contain a PDZ domain. Interestingly, loss of Sdt or Patj had no effect on Kib localization (Figures S4C–S4D”), suggesting that Crb could tether Kib at the junctional cortex via aPKC/Par6.
aPKC tethers Kib at the cell cortex in cultured cells
Several lines of evidence suggest that aPKC could tether Kib cortically downstream of Crb. First, Kib contains an aPKC binding motif and is known to physically interact with aPKC in Drosophila and mammalian cells.24–26 Second, aPKC is highly enriched at the junctional cortex, where it partially colocalizes with Kib punctae (Figures S4E–S4E”). Third, we found that in the wing imaginal disc, loss of Crb leads to a significant decrease in cortical aPKC (Figures S4F–S4F”), consistent with previous observations and reports that Crb forms a complex with Par6 and aPKC and promotes aPKC activity.13,44–46
To test the tethering role of aPKC, we first used cultured Schneider’s 2 (S2) cells to ask if aPKC can recruit Kib to the cell cortex. When expressed by itself, Kib normally aggregates in cytosolic foci and almost never appears at the cortex23 (Figures 5A and 5E). In contrast, co-expression of Par6 and aPKC, both of which localize cortically in S2 cells, led to recruitment of Kib to the cell cortex in ~40% of cells, as evidenced by the appearance of a bright rim around the cell optically sectioned through the equatorial plane (Figures 5B–B”, E). To ask if Crb can modify cortical Kib recruitment via Par6/aPKC, we expressed the intracellular region of Crb (Crbi) alone or together with Par6 and aPKC and examined Kib localization. Crbi by itself did not recruit Kib to the cortex (Figures 5C and 5C’, 5E). However, Kib localized cortically in over 60% of cells when Crbi, Par6, and aPKC were co-expressed (Figures 5D–5E).
Figure 5. aPKC tethers Kib at the cell cortex in S2 cells.
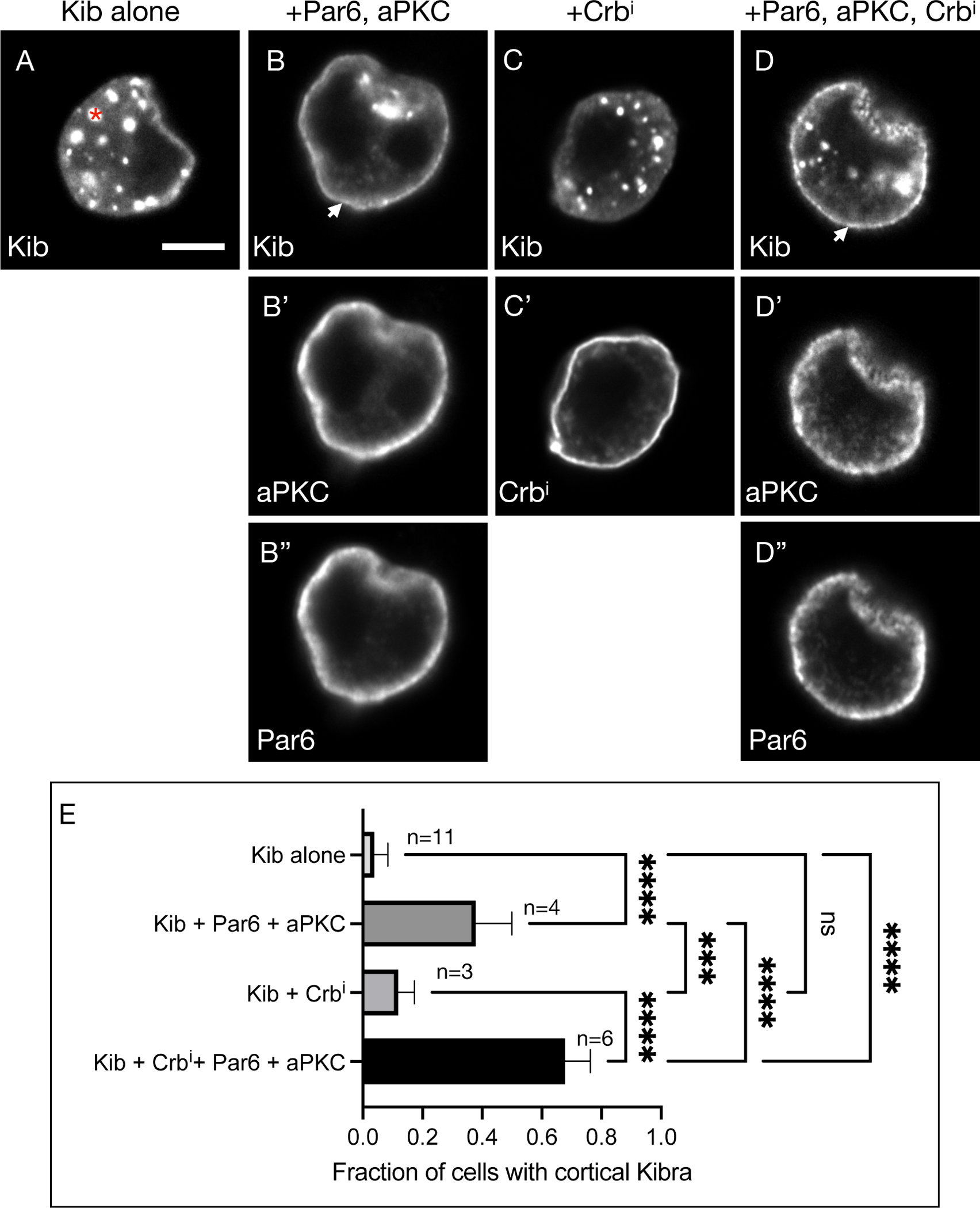
(A) Expressed by itself, Kib often aggregates in cytoplasmic foci (asterisk) in cultured S2 cells. Scale bar = 5μm.
(B-B”) Co-expression of aPKC and Par6 leads to cortical Kib recruitment (arrowhead in B).
(C-C’) Expression of Crbi alone does not result in cortical Kib recruitment.
(D-D”) Addition of Crbi enhances Par6/aPKC-mediated cortical Kib recruitment.
(E) Quantification of the fraction of cells displaying cortical Kib under the conditions shown in (A-D”). Statistical significance was calculated using One-way ANOVA followed by Tukey’s HSD test; n = number of biological replicates (≥ 50 cells counted per replicate). See also Figure S4.
If Crb functions to stabilize aPKC, then cortically anchoring aPKC by other means should also lead to robust cortical Kib recruitment. To test this idea, we adapted a previously published method of inducing polarity in S2 cells that expresses aPKC fused to the cytoplasmic terminus of the homophilic adhesion protein, Echinoid47 (Ed). Expressing this fusion construct (Ed:GFP:aPKC) in S2 cells leads to cell clustering (via the Ed extracellular domain) and enrichment of aPKC at cell-cell contacts (Figure 6A). As a control, we used an analogous construct lacking aPKC (Ed:GFP). Using this approach in combination with Kib truncations, we asked if Ed:GFP:aPKC could recruit Kib to cell-cell contacts and if so, whether the aPKC-binding region of Kib is necessary and sufficient for this interaction (Figures 6A and 6B). As expected, wild-type Kib (KibWT) appeared largely cytoplasmic in cells clustered via Ed:GFP, indicating that formation of cell-cell contacts per se does not lead to cortical Kib localization (Figures 6C–6C”, 6G). In sharp contrast, Kib was recruited to cell-cell adhesion sites upon Ed:GFP:aPKC expression (Figures 6D–6D”, 6G). Furthermore, a Kib variant lacking the aPKC-binding region (KibΔaPKC) failed to localize at the cell-cell contacts (Figures 6E–6E”, 6G), whereas a C-terminal fragment of Kib containing the aPKC-binding region (Kib858-1288) was robustly recruited by Ed:GFP:aPKC (Figures 6F–6G). Together, these results suggest that aPKC recruits Kib to the cell cortex and that the aPKC-binding motif of Kib is both necessary and sufficient for this interaction.
Figure 6. The aPKC-binding domain in Kib is necessary and sufficient for its cortical recruitment by aPKC.
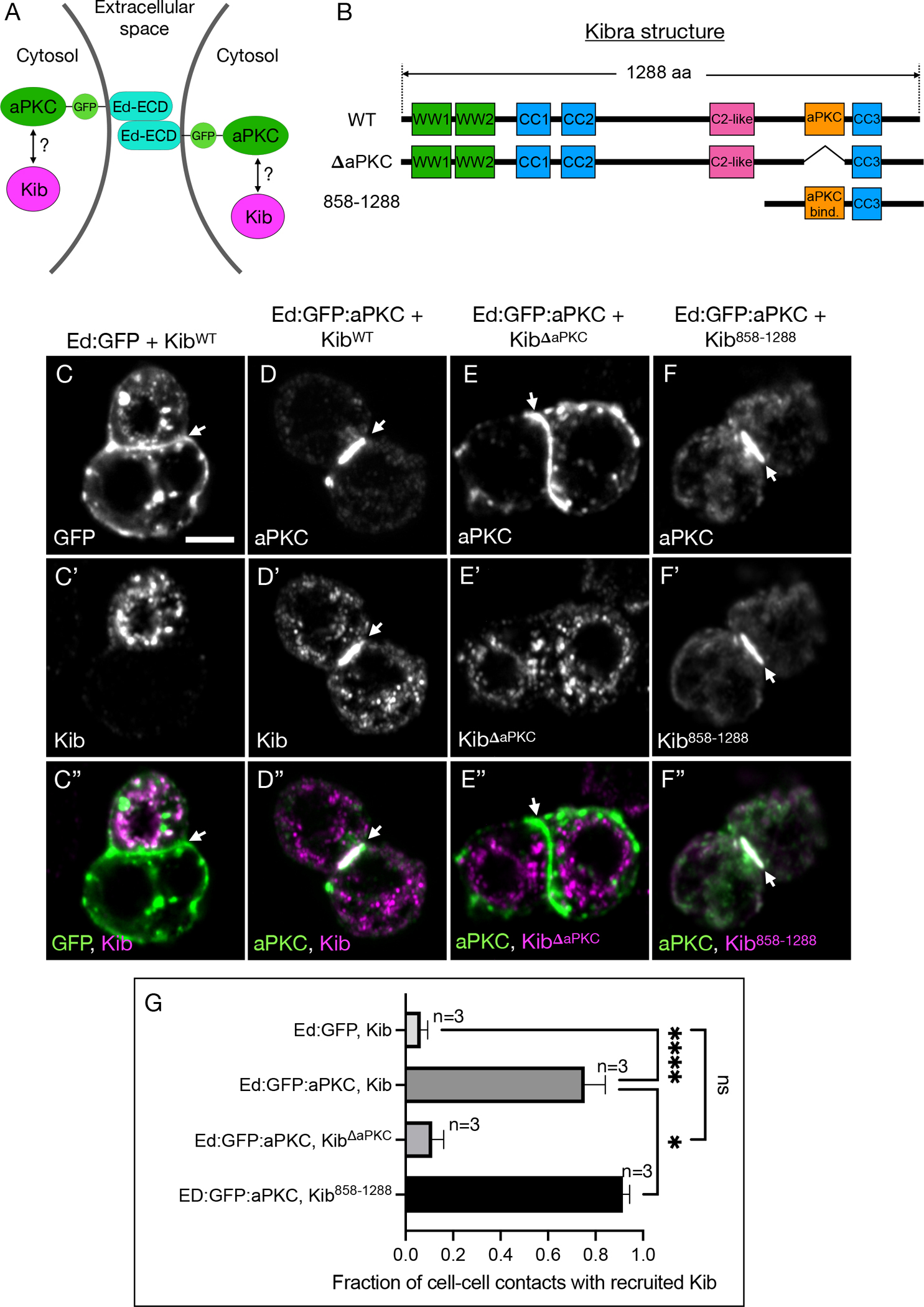
(A) A schematic of two cells clustered via Ed extracellular domain (Ed-ECD) fused to aPKC on the intracellular side.
(B) Protein structure cartoons of wild type (WT) Kib, Kib lacking the aPKC-binding motif (ΔaPKC), or Kib containing the last 431 amino acids (858–1288).
(C-C”) In control cell clusters generated via Ed:GFP, Kib aggregates predominantly in cytoplasmic foci and is absent from cell-cell contacts (arrowhead). Scale bar = 5μm.
(D-D”) When cells are clustered via Ed:GFP:aPKC, Kib is recruited to cell-cell contacts.
(E-E”) Ed:GFP:aPKC fails to recruit KibΔaPKC to cell-cell contacts.
(F-F”) Ed:GFP:aPKC robustly recruits Kib858-1288 to cell-cell contacts.
(G) Quantification of the fraction of cell-cell contacts with recruited Kib under the coanditions shown in (C-F”). Statistical significance was calculated using One-way ANOVA followed by Tukey’s HSD test; n = number of biological replicates ((≥ 50 cell-cell contacts couanted per replicate). See also Figure S4.
aPKC tethers Kib at the junctional cortex to limit Kib-mediated Hippo signaling
We next tested if aPKC tethers Kib at the junctional cortex in the wing imaginal discs and the potential functional significance of this interaction. Loss of aPKC activity disrupts cell polarity and constitutive depletion of aPKC in the wing imaginal discs leads to cell death,48 making it difficult to assess the role of aPKC in localizing Kib using genetic manipulations. Therefore, we used a previously published analog-sensitive allele of aPKC, aPKCas4, which produces a functional kinase that can be acutely inhibited by a small molecule 1NA-PP1.49 Treatment of wing imaginal discs homozygous for aPKCas4 with 1NA-PP1 for only 15 min severely disrupted normal cortical aPKC localization (Figures S5A and S5B), similar to what was previously observed in Drosophila larval neuroblasts.49 Importantly, 1NA-PP1 had no effect on aPKC or Kib-GFP distribution in the background of wild-type aPKC (Figures S5C–S5E’), indicating that the effect of 1NA-PP1 is specific to aPKCas4.
To test the effect of aPKC inhibition on Kib localization, we treated wing imaginal explants expressing Ubi>Kib-GFP in aPKCas4 background with 1NA-PP1 for 15 min. Strikingly, this resulted in Kib accumulation almost exclusively at the medial cortex (Figures 7A–7B’), supporting the role of aPKC as a junctional tether for Kib. Importantly, Mer also accumulated medially under 1NA-PP1 treatment whereas Ex remained junctional (Figures S6A–S6D’), suggesting that aPKC inhibition affects Kib-mediated signaling specifically and consistent with the previous observation that acute aPKC inhibition does not affect apical Crb localization.50
Figure 7. Tethering by aPKC limits Kib-mediated Hippo apathway activation.
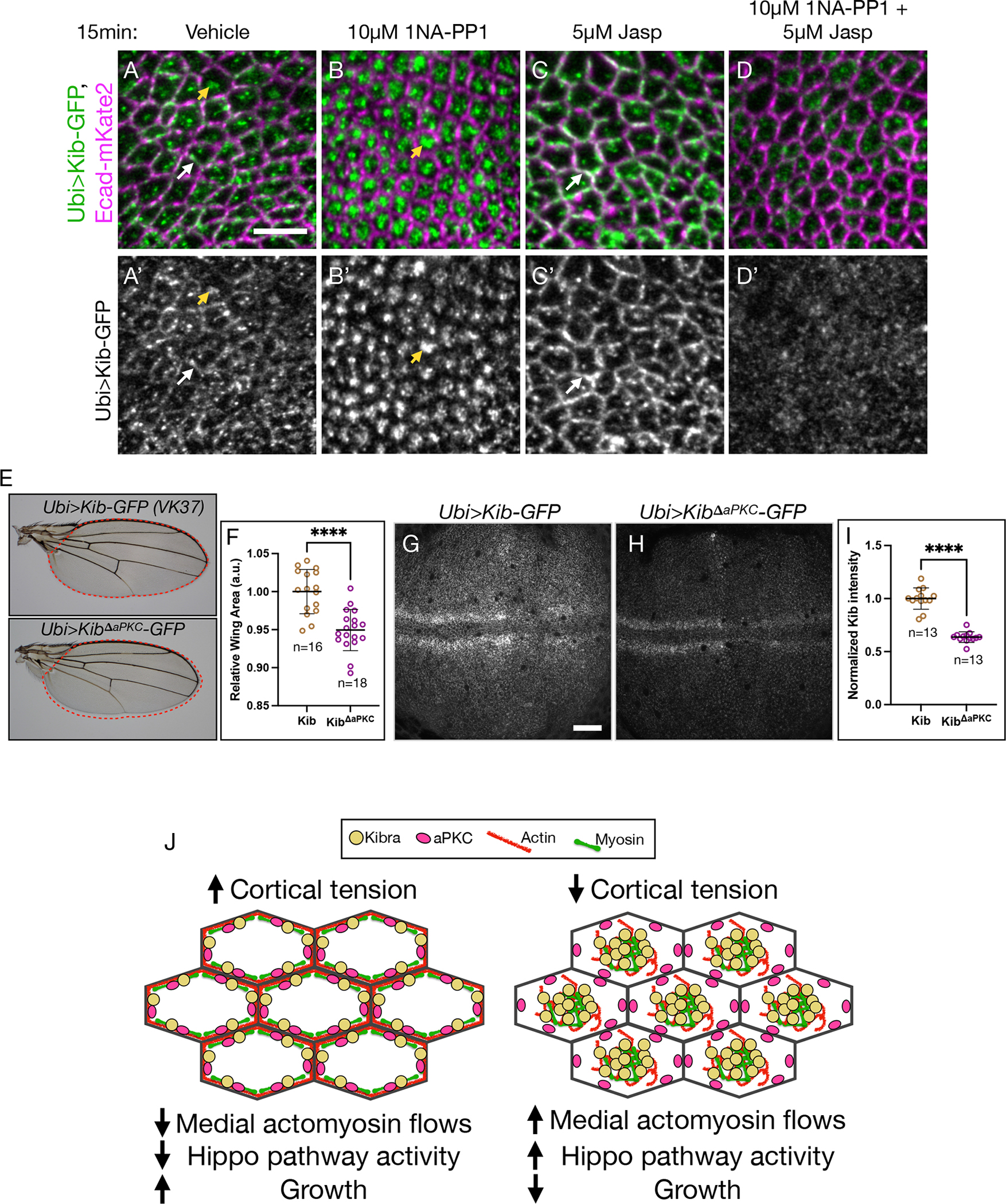
(A-D’) In tissues homozygous for aPKCas4 allele, Kib localizes at the junctional (white arrows) and medial (yellow arrowheads) cortex under control conditions (A and A’) but is predominantly medial under aPKC inhibition with 1NA-PP1 (B and B’). Treatment with Jasp leads to more junctional Kib accumulation (C and C’). Under simultaneous treatment with 1NA-PP1 and Jasp, Kib fails to accumulate medially or junctionally (D and D’). Scale bar = 5μm.
(E and F) Wings ectopically expressing KibΔaPKC (Ubi>KibΔaPKC-GFP) are slightly smaller the ones expressing wild-type Kib (Ubi>Kib-GFP); n = number of wings.
(G-I) Wild-type Kib (G and I) is more stable than KibΔaPKC (H and I); n = number of wing dics. Scale bar = 20μm. Transgenes in (E-H) are identically expressed.
Statistical significance in F and I was calculated using Mann-Whitney test.
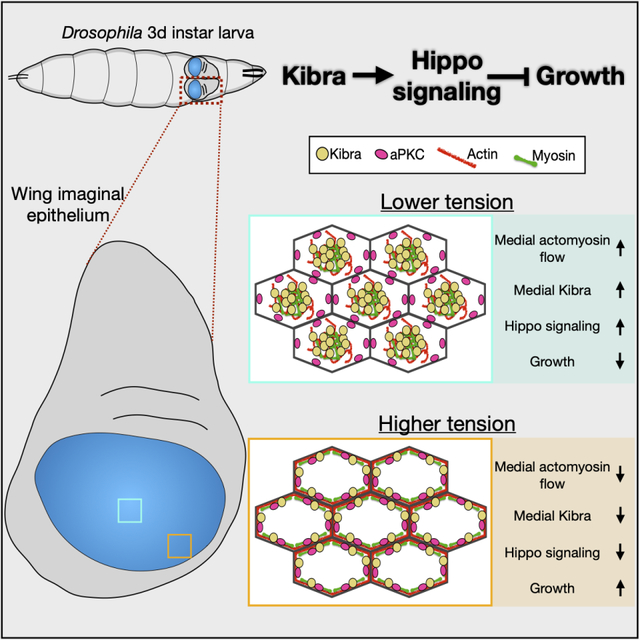
(J) A cartoon model of Kib regulation via apical polarity and actomyosin flows. Under high cortical tension medial actomyosin flow decreases, allowing aPKC to tether and inhibit Kib at the junctional cortex, resulting in more growth. Under low cortical tension, stronger medial actomyosin flows untether Kib from the junctional cortex and accumulate it medially, where it recruits associated components to promote Hippo signaling and inhibit growth. See also Figure S5 and Figure S6.
As with loss of Crb, aPKC inhibition affected actomyosin organization. Recent studies in Drosophila reported that acute aPKC inactivation leads to increased medial myosin activity and apical constriction in embryonic and the follicular epithelial cells.49,51,50 In agreement with these reports, we observed increased medial myosin and F-actin accumulation in the wing imaginal disc cells under aPKC inhibition (Figures S6E–S6H’), suggesting that changes in actomyosin organization could be responsible for medial Kib accumulation under aPKC inhibition. Consistent with this hypothesis, inhibition of aPKC in the presence of Jasp resulted in diffuse Kib that failed to accumulate at the junctional or medial cortex (Figures 7C–7D’). Collectively, these results strongly suggest that Kib cortical distribution is controlled by the opposing action of aPKC, which tethers Kib at the junctional cortex, and actomyosin flows, which accumulate Kib medially.
What could be the potential function of junctional Kib recruitment by aPKC? Previous work suggested that loss of Crb enhances Kib’s ability to activate the Hippo pathway and suppress tissue growth.27 These observations suggest that aPKC could inhibit Kib via junctional tethering. To test this idea, we compared adult wing size from flies expressing either wild-type Kib or KibΔaPKC under the ubiquitin promoter and from identical genomic locations. Flies expressing KibΔaPKC had mildly undergrown wings compared to those expressing wild-type Kib (Figures 7E and 7F). Additionally, despite its heightened ability to suppress growth, KibΔaPKC abundance was significantly lower than that of wild-type Kib (Figures 7G–7I), suggesting that KibΔaPKC is intrinsically more active than wild-type Kib. We observed similar differences in adult wing growth and protein abundance when wild-type Kib and KibΔaPKC were expressed under UAS control using a wing-specific driver Nubbin>Gal4 (Figures S5F–S5J), indicating that these differences were not due to the ubiquitous expression of the transgenes. Together, these results suggest that aPKC-mediated tethering of Kib at the junctional cortex limits Kib-mediated Hippo pathway activation.
Discussion
Proper spatial organization of subcellular signaling components is critical for controlling the specificity and amplitude of signaling pathways.52–54 Despite our long-standing knowledge that upstream Hippo pathway regulators are enriched at the apical cortex of epithelial cells, our understanding of how these proteins are organized and the importance of apical distribution in regulating their signaling output is far from complete. In this study, we focused on the cortical distribution of Kib, a key upstream Hippo pathway activator that is organized not only along the apico-basal axis, with strong enrichment apically, but also across the apical cortex into junctional and medial pools. We demonstrate that such distribution of Kib is achieved by opposing influences from the apical polarity and actomyosin networks. While the apical polarity network, at least in part via aPKC, tethers Kib at the junctional cortex to silence its activity, centripetal actomyosin flows promote Hippo signaling via accumulation of Kib and the associated Hippo signaling components at the medial cortex (Figure 7J).
Our study identifies a previously unrecognized role of the medial actomyosin network in regulating the Hippo pathway. Actomyosin-generated cortical tension is a conserved stimulus that inhibits Hippo signaling and promotes Yki activity and growth. However, while cortical tension inhibits Hippo signaling at the kinase level via Ajuba-mediated sequestration of Wts,18,19 how the actomyosin cytoskeleton might affect upstream Hippo pathway regulators is unknown. A striking observation in our study is that manipulations that affect actomyosin organization and cortical tension, such as osmotic shifts or genetic manipulation of myosin function, can drive dramatic changes in Kib localization (Figures 2, 3, S3, S6 and 7; Videos S3 and S4). Given that myosin organization and dynamics are known to be influenced by cortical tension,35,36 we propose that cortical tension could regulate medial Kib localization via modulation of actomyosin flows. Specifically, we propose that higher cortical tension promotes junctional actomyosin enrichment and stabilization, resulting in decreased medial flows, greater aPKC-dependent tethering of Kib at the junctions (less medial Kib), and attenuated Hippo signaling (Figure 7J). The opposite occurs under lower cortical tension. This mechanism could be important in the developing wing imaginal disc, where higher tension at the tissue periphery is thought to inhibit the Hippo pathway to promote growth via Yki.55–60 Consistent with this prediction, Kib is less medial in cells at the periphery than at the center of the tissue (Figures S6I–S6L). More generally, this mechanism could also function during tissue morphogenesis driven by actomyosin dynamics, such as during the apical constriction that drives furrow formation in a range of epithelia. Tension-driven actomyosin flows have also been reported to transport ZO-1 clusters to tight junctions during zebrafish epiboly,61 suggesting that this could be a general mechanism to control cortical protein distribution during tissue growth and morphogenesis. The relative importance of Mer/Kib-mediated and Ajuba/Wts-mediated tension sensing in regulating overall pathway output remains to be determined, but these tension sensing mechanisms could have distinct functions in different developmental contexts.
How could actomyosin-mediated medial Kib accumulation promote Kib activity? We observed that actomyosin flows drive translocation of Kib punctae from the junctional cortex, where aPKC is enriched, to the medial cortex, where Kib punctae appear to form larger assemblies (Videos S1 and S4). Furthermore, the formation of such medial assemblies is enhanced by treatment with hypertonic medium, Crb depletion, and acute aPKC inhibition, all of which also increase medial actomyosin organization, and inhibited when the actomyosin network is stabilized (Figures 2, 3, S3, and 7). We propose that medial actomyosin flows promote Hippo signaling by driving Kib coalescence and Hippo signaling complex formation at the medial cortex, away from Kib’s junctional inhibitors, including aPKC (Figure 7J). A recent study also argued that medial Kib (but not junctional) undergoes phase separation, thereby promoting its ability to assemble pathway components.9 Our results are consistent with this possibility, though they do not directly confirm it. The kinetics of Kib cluster formation and its regulation by actomyosin flows will have to be resolved by further studies.
Several lines of evidence suggest that medial accumulation promotes Kib activity. First, loss of Crb, which results in predominantly medial Kib, enhances Kib-mediated signaling.7 Second, the size and dynamics of medial Kib assemblies are similar to biomolecular condensates formed by Kib in mammalian cells, and phase transition was suggested to promote Kib activity at the medial cortex.9 Third, Hpo co-accumulates with Kib at the medial cortex but not at the junctional cortex (Figures 4D–4F’).7 Because Hpo dimerizes and trans-autoactivates,62 it is likely that medial Kib accumulation provides the means of increasing Hpo activation via local concentration, as has been suggested for their mammalian counterparts.9 Fourth, Merlin, which promotes Kib’s ability to assemble pathway components, also accumulates with Kib medially.7 Additionally, given the evidence that medial Kib assemblies are significantly more stable than the junctional Kib pool,9 it is possible that medial accumulation promotes Hippo signaling by increasing the cortical dwell time of Kib complexes.63,64
One of the surprising aspects of our findings is the specificity of actomyosin-mediated regulation of Kib localization. Ex and Kib are similar in size (~154kDa and 144kDa, respectively), assemble similar signaling complexes, colocalize junctionally, and are known to physically interact in cultured S2 cells.22,21,65,7 However, while Kib rapidly relocalizes from the junctional to the medial cortex under hypertonic conditions or aPKC inhibition, Ex remains junctional (Figures 2, 7, S1D–S1D”, and S6C–S6D’). This difference in behavior cannot be explained solely by the difference in tethering strengths because loss of Crb untethers both Ex and Kib from the junctional cortex, but whereas Ex becomes cytoplasmically diffuse, Kib accumulates medially (Robinson et al., 2010; Su et al., 2017, Figures S3A–S3B’). These observations are consistent with previous reports indicating that Ex and Kib function in parallel to form separate signaling modules,23,22,7,8 and they also raise the question of how Kib localization could be specifically regulated by the actomyosin flows. Kib lacks an actin-binding domain and our observations of Kib localization with respect to F-actin and myosin suggest that Kib is unlikely to associate tightly with the actomyosin network (Figure 1). Three non-mutually exclusive possibilities could explain why actomyosin flows accumulate Kib but not Ex at the medial cortex. First, the multivalency of Kib could enable weak interactions with the actomyosin cytoskeleton and/or its regulators, thereby facilitating advective transport by actomyosin flows, as has been shown for Par3 in the C. elegans zygote.28,29 Second, the interaction between Kib and aPKC could be modulated by factors that also regulate actomyosin dynamics. Third, in addition to its junctional tethering, Kib could be stabilized by a yet unknown anchor at the medial cortex. The tools and framework developed in this study provide a strong foundation for future investigation of these possibilities.
This study highlights the relationship between Kib and aPKC. Our results suggest that junctional tethering by aPKC attenuates Kib’s ability to activate the Hippo pathway. We show that aPKC recruits Kib to the cortex in S2 cells (Figures 5 and 6), and Kib lacking the ability to bind aPKC is intrinsically more active in vivo (Figure 7). Because Crb is necessary for aPKC recruitment to the junctional cortex, these observations are consistent with previous data showing that loss of Crb potentiates Kib-mediated Hippo pathway activation.7 Similar to our observations of the relationship between Kib and aPKC in the wing, previous work in the Drosophila fat body suggested that aPKC inhibits Kib’s ability to promote starvation-induced autophagy,26 although it is unclear if Kib-mediated regulation of autophagy involves Hippo signaling. Nonetheless, multiple lines of evidence suggest that aPKC regulates Kib function through direct interaction.
Our results also suggest that aPKC likely is not the only component that tethers Kib because ectopically expressed KibΔaPKC is still able to localize junctionally (Figures S5K and S5L). However, the additional Kib tether must, catalytically and/or structurally, depend on aPKC, because simultaneous inhibition of both aPKC and actomyosin dynamics prevents junctional accumulation of full-length Kib normally observed when actomyosin dynamics alone are perturbed. Both Par6 and the Drosophila homolog of Par3, Bazooka (Baz), mislocalize under acute aPKC inhibition,66,67 suggesting that they could assist aPKC in stabilizing Kib at the junctional cortex. On one hand, the involvement of Baz in aPKC-mediated Kib regulation seems unlikely since aPKC was shown to exclude Baz from the marginal zone to the adherens junctions in the follicular epithelium.44 On the other hand, this exclusion requires aPKC phosphorylation of Baz on S980, as non-phosphorylated Baz (S980A) associates with Par6/aPKC at the apical membrane.44,68 Given that Kib is believed to bind to aPKC’s kinase domain and inhibit aPKC’s kinase activity,25,69 it is unlikely that aPKC bound to Kib would be able to exclude Par3 from binding Par6. In addition, mammalian Kib also directly interacts with Patj.70 Although we find that depletion of Patj has no effect on junctional Kib localization, the existence of multiple interactions stabilizing Kib could obscure the role of individual components, especially under long-term genetic perturbations. Regardless of the molecular details, however, the existence of multiple tethers also suggests that Kib could be associated with a broader network of polarity organizers.
Finally, our work highlights the importance of understanding the role and regulation of Hippo signaling in a broader developmental context. Proper organ development involves robust control of both tissue size and shape, suggesting that these processes must be regulated in a concerted fashion. Cell polarity and actomyosin networks play central roles in the regulation of growth and morphogenesis,3,71–73 but our understanding of how the interaction between epithelial architecture and mechanical forces is biochemically translated into intracellular signaling events is currently limited. Our findings that Kib subcellular organization and activity are regulated by the tug of war between apical polarity and actomyosin networks suggest that Kib could serve as a nexus in coordinating growth and morphogenesis. Further investigation of how Kib and other Hippo signaling components fit into the molecular network governing epithelial cell organization will provide valuable insight into the broader functions of the Hippo pathway in development.
Limitations of the Study
A limitation in this study (and the field at large) is the lack of a biosensor that directly reports Kib-mediated Hippo pathway activation in living cells. Thus, although the results in this paper and previous studies suggest that medial Kib accumulation promotes Hippo pathway activation, determining the state of Kib-mediated signaling with accuracy at the subcellular level remains a significant challenge.
RESOURCE AVAILABILITY
Lead contact
Please contact the Lead Contact, Richard Fehon (rfehon@uchicago.edu), for reagents and resources generated in this study.
Materials availability
Materials such as plasmids and Drosophila stocks generated for this study are available upon request.
Data and code availability
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Drosophila husbandry
Drosophila melanogaster was cultured using standard techniques at 25°C (unless otherwise noted). For Gal80ts experiments, crosses were maintained at 18°C and moved to 29°C for the duration specified in each experiment. For wing disc dissections, both male and female larvae were used. For adult wing growth comparison, only females were used.
Drosophila genotypes used for data collection
Drosophila stocks used in this study are listed in the Key Resource Table.
KEY RESOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-aPKC | Santa Cruz Biotechnology | Lot#: G2304 |
| Mouse monoclonal anti-Crb | DSHB | Cq4 |
| Mouse monoclonal anti-Myc | Cell Signaling | Product #2276 |
| Mouse monoclonal anti-FLAG | Sigma Aldrich | Cat# F1804; RRID:AB_262044 |
| Chemicals, peptides, and recombinant proteins | ||
| 1-NA-PP1 | Cayman | Item # 10954 |
| Jasplakinolide | Cayman | Item # 11705 |
| Janelia Fluor® Halo Tag® Ligand | Promega | Cat# GA1120 |
| Experimental models: Cell lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Peter Cherbas | RRID:CVCL_TZ72 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| D. melanogaster: Kib::GFP | Su et al.27 | |
| D. melanogaster: UAS-LifeAct-Halo2 | Bloomington Drosophila Stock Center | BDSC:67625; FlyBase: FBti0186932 |
| D. melanogaster: Ubi>Kib-Halo | This paper | N/A |
| D. melanogaster: sGMCA | Kiehart et al.79 | N/A |
| D. melanogaster: Sqh-GFP | Bloomington Drosophila Stock Center | BDSC:57145; FlyBase: FBti0150058 |
| D. melanogaster: Ecad::GFP | Huang et al.80 | N/A |
| D. melanogaster: Ubi>Kib-GFP | Tokamov et al.8 | N/A |
| D. melanogaster: Ubi>Kib𝚫aPKC-GFP | This paper | N/A |
| D. melanogaster: Zip-GFP | Rauskolb et al.18 | N/A |
| D. melanogaster: UAS-SqhDD | Mitonaka et al.41 | N/A |
| D. melanogaster: Ecad-3XmKate2 | Pinheiro et al.81 | N/A |
| D. melanogaster: Ex-YFP | Su et al.27 | N/A |
| D. melanogaster: Ubi-Mer-Halo | This paper | N/A |
| D. melanogaster: Hpo-YFP | Su et al.27 | N/A |
| D. melanogaster: Halo-Snap-aPKC | Erdmann et al.82 | N/A |
| D. melanogaster: Mer-YFP | Su et al.27 | N/A |
| D. melanogaster: RNAi of Crb: GD14463 | Vienna Drosophila RNAi Center | VDRC: 39177 |
| D. melanogaster: RNAi of Sqh: GD1695 | Vienna Drosophila RNAi Center | VDRC: 7916 |
| D. melanogaster: RNAi of Patj: y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01489}attP2 | Bloomington Drosophila Stock Center | BDSC: 35747; FlyBase: FBti0145012 |
| D. melanogaster: RNAi of Sdt: y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00851}attP2 | Bloomington Drosophila Stock Center | BDSC: 33909; FlyBase: FBti0140562 |
| D. melanogaster: RNAi of Ex: KK100573 | Vienna Drosophila RNAi Center | VDRC: 109281 |
| D. melanogaster: UAS-Utrophin-ABD-GFP | Rauzi et al.83 | N/A |
| D. melanogaster: aPKCas4 | Hannaford et al.49 | N/A |
| Recombinant DNA | ||
| pMT-Kib-GFP | This paper | N/A |
| pMT-Par-6-Myc | This paper | N/A |
| pMT-aPKC | This paper | N/A |
| pAc.5.1-Crbi-3xFLAG | This paper | N/A |
| pMT-Ed:GFP | Johnston et al.47 | N/A |
| pMT-Ed:GFP:aPKC | Johnston et al.47 | N/A |
| Ubi>Kib-GFP-FLAG | This paper | N/A |
| Ubi>Kib𝚫aPKC-GFP-FLAG | This paper | N/A |
| Ubi>Kib858–1288-GFP-FLAG | This paper | N/A |
| Software and algorithms | ||
| Fiji (ImageJ) | Schneider et al.84 | https://imagej.nih.gov/ij/ |
| Cellpose | Stringer et al.76 | https://www.cellpose.org |
| Scikit | van der Walt et al.75 | https://scikit-image.org |
| GraphPad Prism | GraphPad Software | N/A |
Figure 1B: Nub>Gal4, UAS-LifeAct-Halo; Kib::GFP
Figure 1C: Ubi>Kib-Halo; sGMCA
Figure 1D: Nub>Gal4, UAS-LifeAct-Halo/+; UASp-Kib-GFP
Figure 1G: Ubi>Kib-Halo; Sqh-GFP
Figure 2A–C’: Ecad-3x-mKate2; sGMCA
Figure 2E–G’: Ecad-3x-mKate2; Sqh-GFP
Figure 2I–K’: Ubi>Kib-Halo Ecad::GFP
Figure 3A–D: Ubi>Kib-GFP Ecad-3x-mKate2
Figure 3F–I’: Ecad-3x-mKate2, tub>Gal80ts/UAS-SqhDD ; Ubi>Kib-GFP/+
Figure 3K–L”: Ubi>Kib-Halo/UAS-SqhDD; sGMCA, hh>Gal4/tub>Gal80ts
Figure 4A–C”: Ubi>Kib-GFP/Ubi>Mer-Halo
Figure 4D–F’: Ubi>Kib-Halo/+; Hpo-YFP/+
Figure 7A–D’: aPKCas4 Ecad-3x-mKate2; Ubi>Kib-GFP/+
Figure 7E–I: Ubi>Kib-GFP/+ or Ubi>KibΔaPKC-GFP/+
Figure S1A–C’: Ubi>Kib-GFP Ecad-3x-mKate2
Figure S1E–G’: Shot-GFP/Ecad-3x-mKate2
Figure S1H–H”: Ex-YFP
Figure S2A–C: sGMCA
Figure S2D–E’: Ubi>Kib-GFP Ecad-3x-mKate2
Figure S2F–H: Zip:GFP/UAS-SqhDD ; hh>Gal4/tub>Gal80ts
Figure S2I–J’: Ecad-3x-mKate2, tub>Gal80ts/UAS-Sqh RNAi; Ubi>Kib-GFP/+
Figure S3A–B’: Ubi>Kib-GFP Ecad-3x-mKate2/UAS-Crb RNAi; hh>Gal4/+
Figure S3C–C’: y w hsFlp; Nub>Gal4, UAS-Utr-GFP/+; 82BFRT crb1/82BFRT Ubi>RFP
Figure S3D–F: Ecad-3x-mKate2/UAS-Crb RNAi; Sqh-GFP/hh>Gal4
Figure S3G–H’: Ubi>Kib-GFP Ecad-3x-mKate2/UAS-Crb RNAi; hh>Gal4/+
Figure S4A–A’: Ecad-3x-mKate2/UAS-Crb RNAi; Ubi>Kib-GFP hh>Gal4/+
Figure S4B–B’: Ecad-3x-mKate2/UAS-Ex RNAi; Ubi>Kib-GFP hh>Gal4/+
Figure S4C–C’: Ecad-3x-mKate2/+; Ubi>Kib-GFP hh>Gal4/UAS-Sdt RNAi
Figure S4D–D’: Ecad-3x-mKate2/+; Ubi>Kib-GFP hh>Gal4/UAS-Patj RNAi
Figure S4E–E”: Halo-Snap-aPKC; Ubi>Kib-GFP
Figure S4F–F”: UAS-Crb RNAi/+; hh>Gal4/+
Figure S5A–B: aPKCas4
Figure S5C: w1118
Figure S5D–E’: Ubi>Kib-GFP Ecad-3x-mKate2
Figure S5F–L: Nub>Gal4/+; UASt-Kib-GFP or UASt-KibΔaPKC-GFP/+
Figure S6A–B’: aPKCas4 Ecad-3x-mKate2; Mer-YFP
Figure S6C–D’: aPKCas4 Ecad-3x-mKate2; Ex-YFP
Figure S6E–F’: aPKCas4 Ecad-3x-mKate2; Sqh-GFP
Figure S6G–H’: aPKCas4 Ecad-3x-mKate2; sGMCA
METHOD DETAILS
Data collection
All in vivo data was collected from wing imaginal disc explants from wandering third instar larvae. Unless otherwise noted, images displaying subcellular localization of Kib, F-actin, or myosin were acquired from the wing disc pouch, avoiding the dorsal-ventral boundary and the pouch periphery.
Osmotic and pharmacological treatments
All osmotic solutions were prepared fresh per experiment. Schneider’s Drosophila Medium (Sigma) supplemented with 10% Fetal Bovine Serum (Thermo Fisher Scientific) was used as isotonic medium (~360mOsm). To make a hypertonic solution, the osmolarity of the isotonic solution was increased to ~460mOsm using 1M NaCl. To make a hypotonic solution, the isotonic medium was diluted with deionized water to 216mOsm.
To inhibit F-actin dynamics, tissues were incubated in an isotonic or hypertonic solution with DMSO (0.5% v/v) or 5μM Jasp before mounting. To inhibit aPKC, tissues homozygous for aPKCas4 allele were incubated in an isotonic solution with DMSO (0.1% v/v) or 10μM 1NA-PP1 before mounting. Unless indicated otherwise, all incubations were done for 15 min in a humid chamber, and tissues were imaged live immediately after incubation.
Live imaging
Except for Fig. S4F–F”, tissues were imaged live. Live imaging of the wing imaginal discs was adapted from Restrepo et al.74 Briefly, wing imaginal tissues were first dissected in Schneider’s Drosophila Medium (Sigma) supplemented with 10% Fetal Bovine Serum (Thermo Fisher Scientific) on a siliconized glass slide. Tissues were then transferred with a pipette in 5–10μl of medium to a glass bottom microwell dish (MatTek, 35mm petri dish, 14mm microwell) with No. 1.5 coverglass. The discs were oriented so that the apical side of the disc proper faced the coverglass. A Millicell culture insert (Sigma, 12mm diameter, 8μm membrane pore size) was prepared in advance by cutting off the bottom legs with a razor blade and removing any excess membrane material around the rim of the insert. The insert was carefully placed into the 14mm microwell space, directly on top of the drop containing the tissues. The space between the insert and the microwell was sealed with ~15μl of mineral oil and 200μl of isotonic medium was added into the insert chamber. For osmotic shift movies with Sqh-GFP and Ubi>Kib-Halo, isotonic medium was removed and 200μl of media with indicated osmolarity was added into the chamber of the insert immediately before scanning.
For Halo labeling, wing imaginal tissues were incubated in a humid chamber in 150μl of 100nM Halo dye (see Key Resource Table) diluted in 1X PBS for 15min. Tissues were then rinsed twice in the culture medium and mounted as described above. For osmotic shift experiments, tissues were subjected to indicated osmotic solutions after Halo labeling.
An inverted Zeiss LSM880 laser scanning confocal microscope equipped with a GaAsP spectral detector and the Airyscan module was used for all imaging. All timelapse images and still images in Figs. 1, 2I–K’, S1D–D”, 4, S4E–E”, and S6E–H’ were taken using the Airyscan in superresolution mode. Timelapse acquisition was performed using a PIFOC P-737 Piezo Z nanopositioner stage insert.
Image analysis and quantification
ImageJ was used for basic image processing, including generation of maximum projections, background subtraction, and Gaussian blur application.
To quantify the correlation between Kib and F-actin dynamics in Fig. 1E–F, Kib and F-actin intensity were measured in the medial or junctional regions of cells. For medial Kib/F-actin measurements, cells were selected solely based on the appearance of Kib coalescence at the medial cortex (i.e., cells displaying medial Kib accumulation were predicted to have medial F-actin accumulation as well). A region of interest (ROI) was drawn for individual cells using the polygon selection tool in ImageJ, such that the ROI did not contact cell junctions across all timeframes. Plot Z Profile function in ImageJ was then used to extract the mean intensity values for Kib and F-actin over time. For junctional intensity, measurements were taken from random junctions. The intensity values were plotted and the Spearman’s correlation coefficient (r) was calculated using GraphPad Prism software.
To quantify junctional/medial intensity, maximum projections were segmented via Cellpose (using Ecad-mKate2 as the junctional marker) and a standard Scikit watershed algorithm.75,76 The junctional mask generated by segmentation was then applied to quantify mean junctional Kib intensity, while remaining non-junctional signal was classified as medial. The ratio of mean junctional to medial intensity was then calculated.
Immunostaining of imaginal tissues
In Fig. S4F–F”, the wing imaginal discs from wandering late third instar larvae were fixed in 2% paraformaldehyde (PFA)/PBS and stained as previously described.77 Primary antibodies listed in Key Resource Table were used at the following concentrations: anti-Crb (1:1000), anti-aPKC (1:1000). Secondary antibodies (diluted 1:1000) were from Jackson ImmunoResearch Laboratories. Tissues were mounted in Prolong Diamond Antifade Mountant (Thermo Fisher Scientific).
Cortical Kib recruitment in S2 cells
Drosophila S2 cells were maintained in Schneider’s Insect Medium (Sigma) containing 10% Insect Medium Supplement (Sigma). For experiments in Figure 5, the following constructs were used: pMT-Kib-GFP, pMT-Par-6-Myc, pMT-aPKC, and pAc.5.1-Crbi-3xFLAG. Briefly, 2.7 × 106 S2 cells (S2-DGRC) were transfected with total of 500ng of the indicated DNA constructs using dimethyldioctadecylammonium bromide (DDAB, Sigma) at 250μg/ml in 6-well plates.78 To induce expression of pMT constructs, 350μM CuSO4 was added to the wells 48h after transfection. Cells were collected 72h after transfection, fixed with 2% PFA/PBS for 15min, stained in PBS/0.1% Saponin/1% Normal goat serum (PSN) for 1h with primary (anti-Myc [1:10,000] and anti-aPKC [1:1000], see also Key Resource Table) and 1h with secondary antibodies, with 30 min washes. Cells were mounted in Prolong Diamond Antifade Mountant (Thermo Fisher Scientific) and cortical Kib localization was scored blind (at least 50 cells per condition) via widefield fluorescence using a Zeiss Axioplan 2ie microscope. Representative images in Figure 5 were acquired using Zeiss LSM880 confocal microscope.
For experiments in Fig. 6, the following constructs were used: pMT-Ed:GFP or pMT-Ed:GFP:aPKC, Ubi>Kib-GFP-FLAG, Ubi>KibΔaPKC-GFP-FLAG, and Ubi>Kib858-1288-GFP-FLAG. Cells were transfected and expression of pMT constructs was induced as described above. Cells were resuspended 72h after transfection and gently mixed (in a 6-well plate) on an orbital shaker at 70 rpm for 30min to promote cell-cell clustering. After mixing, cells were collected, stained with primary (anti-aPKC and anti-FLAG [1:20,000], see also Key Resource Table) and secondary antibodies, mounted, and scored blind as described above.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests and plots were generated in GraphPad Prism software. The type of statistical test, representation of data, and significance are reported within plots and/or figure legends.
Supplementary Material
Video S1. Kib movement from the junctional to the medial cortex is coordinated with F-actin flow. Related to Figure 1.
Video S2. Medial Kib movement across the apical cortex is coordinated with medial myosin flow. Related to Figure 1.
Video S3. Hypertonic shift leads to increased medial myosin flow which appear to cluster Kib medially. Related to Figure 2.
Video S4. Hypotonic shift leads to decreased medial myosin flow and concomitant accumulation of myosin and Kib at the junctional cortex. Related to Figure 2.
Video S5. Treatment with Jasp stabilizes F-actin and leads to more junctional Kib accumulation. Related to Figure 3.
Video S6. Ectopic expression of SqhDD reduces medial F-actin flow and leads to more junctional Kib localization. Related to Figure 3.
Video S7. Depletion of crb disrupts myosin organization. Related to Figure 3.
Highlights.
Medial actomyosin flows promote Kibra accumulation at the medial cortex
Actomyosin-driven medial Kibra assembles a Hippo signaling complex
aPKC tethers Kibra at the junctional cortex and represses its activity
A tug of war between apical polarity and actomyosin flows controls Hippo signaling
Acknowledgements
We thank K. Irvine, Y. BellaÏche, D. St Johnston, E. Morais de Sá, C. Doe, J. Knoblich, A. Wodarz, E. Knust, S. Horne-Badovinac, B. Glick, the Developmental Studies Hybridoma Bank, the Bloomington stock center, and the VDRC stock center for fly stocks and other reagents. We thank A. Feil for technical help and A. Williams for helpful discussions and help with quantification. We thank S. Horne-Badovinac for helpful comments on the manuscript and E. Munro and the members of the Munro lab for helpful discussion and feedback. SAT was supported by NIH T32 GM007183 and a NSF-GRF. AR was supported by NIH T32 GM007183 and a NSF GRF. MG was supported by R35GM127091. This work was supported by a grant from the National Institute of Health to RGF (R01NS034783).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Boggiano JC, Vanderzalm PJ, and Fehon RG (2011). Tao-1 Phosphorylates Hippo/MST Kinases to Regulate the Hippo-Salvador-Warts Tumor Suppressor Pathway. Developmental Cell 21, 888–895. 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genevet A, and Tapon N (2011). The Hippo pathway and apico–basal cell polarity. Biochemical Journal 436, 213–224. 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 3.Tepass U (2012). The Apical Polarity Protein Network in Drosophila Epithelial Cells: Regulation of Polarity, Junctions, Morphogenesis, Cell Growth, and Survival. Annual Review of Cell and Developmental Biology 28, 655–685. 10.1146/annurev-cellbio-092910-154033. [DOI] [PubMed] [Google Scholar]
- 4.Boedigheimer MJ, Nguyen KP, and Bryant PJ (1997). Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Developmental Genetics 20, 103–110. . [DOI] [PubMed] [Google Scholar]
- 5.McCartney BM, Kulikauskas RM, LaJeunesse DR, and Fehon RG (2000). The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development 127, 1315–1324. [DOI] [PubMed] [Google Scholar]
- 6.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, and Halder G (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nature Cell Biology 8, 27–36. 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 7.Su T, Ludwig MZ, Xu J, and Fehon RG (2017). Kibra and Merlin Activate the Hippo Pathway Spatially Distinct from and Independent of Expanded. Developmental Cell 40, 478–490.e3. 10.1016/j.devcel.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokamov SA, Su T, Ullyot A, and Fehon RG (2021). Negative feedback couples Hippo pathway activation with Kibra degradation independent of Yorkie-mediated transcription. eLife 10, e62326. 10.7554/eLife.62326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Choi K, Su T, Li B, Wu X, Zhang R, Driskill JH, Li H, Lei H, Guo P, et al. (2022). Multiphase coalescence mediates Hippo pathway activation. Cell 185, 4376–4393.e18. 10.1016/j.cell.2022.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, and Pan D (2010). The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proceedings of the National Academy of Sciences 107, 10532–10537. 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson BS, Huang J, Hong Y, and Moberg KH (2010). Crumbs Regulates Salvador/Warts/Hippo Signaling in Drosophila via the FERM-Domain Protein Expanded. Current Biology 20, 582–590. 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, and Richardson HE (2010). Lgl, aPKC, and Crumbs Regulate the Salvador/Warts/Hippo Pathway through Two Distinct Mechanisms. Current Biology 20, 573–581. 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 13.Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, and Tapon N (2009). The Hippo pathway regulates apical-domain size independently of its growth-control function. Journal of Cell Science 122, 2360–2370. 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao H, and Blankenship JT (2020). The pulse of morphogenesis: actomyosin dynamics and regulation in epithelia. Development 147, dev186502. 10.1242/dev.186502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, and Janody F (2011). Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337–2346. 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 16.Sansores-Garcia L, Bossuyt W, Wada K-I, Yonemura S, Tao C, Sasaki H, and Halder G (2011). Modulating F-actin organization induces organ growth by affecting the Hippo pathway. The EMBO journal 30, 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, and Piccolo S (2013). A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 154, 1047–1059. 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Rauskolb C, Sun S, Sun G, Pan Y, and Irvine KD (2014). Cytoskeletal Tension Inhibits Hippo Signaling through an Ajuba-Warts Complex. Cell 158, 143–156. 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibar C, Kirichenko E, Keepers B, Enners E, Fleisch K, and Irvine KD (2018). Tension-dependent regulation of mammalian Hippo signaling through LIMD1. J Cell Sci 131, jcs214700. 10.1242/jcs.214700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin F, Yu J, Zheng Y, Chen Q, Zhang N, and Pan D (2013). Spatial Organization of Hippo Signaling at the Plasma Membrane Mediated by the Tumor Suppressor Merlin/NF2. Cell 154, 1342–1355. 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genevet A, Wehr MC, Brain R, Thompson BJ, and Tapon N (2010). Kibra Is a Regulator of the Salvador/Warts/Hippo Signaling Network. Developmental Cell 18, 300–308. 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Zheng Y, Dong J, Klusza S, Deng W-M, and Pan D (2010). Kibra Functions as a Tumor Suppressor Protein that Regulates Hippo Signaling in Conjunction with Merlin and Expanded. Developmental Cell 18, 288–299. 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumgartner R, Poernbacher I, Buser N, Hafen E, and Stocker H (2010). The WW Domain Protein Kibra Acts Upstream of Hippo in Drosophila. Developmental Cell 18, 309–316. 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Büther K, Plaas C, Barnekow A, and Kremerskothen J (2004). KIBRA is a novel substrate for protein kinase Cζ. Biochemical and Biophysical Research Communications 317, 703–707. 10.1016/j.bbrc.2004.03.107. [DOI] [PubMed] [Google Scholar]
- 25.Yoshihama Y, Sasaki K, Horikoshi Y, Suzuki A, Ohtsuka T, Hakuno F, Takahashi S-I, Ohno S, and Chida K (2011). KIBRA Suppresses Apical Exocytosis through Inhibition of aPKC Kinase Activity in Epithelial Cells. Current Biology 21, 705–711. 10.1016/j.cub.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 26.Jin A, Neufeld TP, and Choe J (2015). Kibra and aPKC regulate starvation-induced autophagy in Drosophila. Biochemical and Biophysical Research Communications 468, 1–7. 10.1016/j.bbrc.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Su T, Ludwig MZ, Xu J, and Fehon RG (2017). Kibra and Merlin Activate the Hippo Pathway Spatially Distinct from and Independent of Expanded. Developmental Cell 40, 478–490.e3. 10.1016/j.devcel.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro E, Nance J, and Priess JR (2004). Cortical Flows Powered by Asymmetrical Contraction Transport PAR Proteins to Establish and Maintain Anterior-Posterior Polarity in the Early C. elegans Embryo. Developmental Cell 7, 413–424. 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Goehring NW, Trong PK, Bois JS, Chowdhury D, Nicola EM, Hyman AA, and Grill SW (2011). Polarization of PAR Proteins by Advective Triggering of a Pattern-Forming System. Science 334, 1137–1141. 10.1126/science.1208619. [DOI] [PubMed] [Google Scholar]
- 30.Chang Y, and Dickinson DJ (2022). A particle size threshold governs diffusion and segregation of PAR-3 during cell polarization. Cell Reports 39, 110652. 10.1016/j.celrep.2022.110652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornet M, Isobe Y, and Lemanski LF (1994). Effects of anisosmotic conditions on the cytoskeletal architecture of cultured PC12 cells. J. Morphol. 222, 269–286. 10.1002/jmor.1052220305. [DOI] [PubMed] [Google Scholar]
- 32.Di Ciano C, Nie Z, Szászi K, Lewis A, Uruno T, Zhan X, Rotstein OD, Mak A, and Kapus A (2002). Osmotic stress-induced remodeling of the cortical cytoskeleton. American Journal of Physiology-Cell Physiology 283, C850–C865. 10.1152/ajpcell.00018.2002. [DOI] [PubMed] [Google Scholar]
- 33.Pietuch A, Brückner BR, and Janshoff A (2013). Membrane tension homeostasis of epithelial cells through surface area regulation in response to osmotic stress. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1833, 712–722. 10.1016/j.bbamcr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 34.van Loon AP, Erofeev IS, Maryshev IV, Goryachev AB, and Sagasti A (2020). Cortical contraction drives the 3D patterning of epithelial cell surfaces. Journal of Cell Biology 219, e201904144. 10.1083/jcb.201904144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Gonzalez R, Simoes SDM, Röper J-C, Eaton S, and Zallen JA (2009). Myosin II Dynamics Are Regulated by Tension in Intercalating Cells. Developmental Cell 17, 736–743. 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duda M, Kirkland NJ, Khalilgharibi N, Tozluoglu M, Yuen AC, Carpi N, Bove A, Piel M, Charras G, Baum B, et al. (2019). Polarization of Myosin II Refines Tissue Material Properties to Buffer Mechanical Stress. Developmental Cell 48, 245–260.e7. rauzi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Röper K, and Brown NH (2003). Maintaining epithelial integrity: a function for gigantic spectraplakin isoforms in adherens junctions. The Journal of Cell Biology 162, 1305–1315. 10.1083/jcb.200307089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booth AJR, Blanchard GB, Adams RJ, and Röper K (2014). A Dynamic Microtubule Cytoskeleton Directs Medial Actomyosin Function during Tube Formation. Developmental Cell 29, 562–576. 10.1016/j.devcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bubb MR, Senderowicz AM, Sausville EA, Duncan KL, and Korn ED (1994). Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. Journal of Biological Chemistry 269, 14869–14871. 10.1016/S0021-9258(17)36545-6. [DOI] [PubMed] [Google Scholar]
- 40.Vasquez CG, Tworoger M, and Martin AC (2014). Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. Journal of Cell Biology 206, 435–450. 10.1083/jcb.201402004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitonaka T, Muramatsu Y, Sugiyama S, Mizuno T, and Nishida Y (2007). Essential roles of myosin phosphatase in the maintenance of epithelial cell integrity of Drosophila imaginal disc cells. Developmental Biology 309, 78–86. 10.1016/j.ydbio.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 42.Flores-Benitez D, and Knust E (2015). Crumbs is an essential regulator of cytoskeletal dynamics and cell-cell adhesion during dorsal closure in Drosophila. eLife 4, e07398. 10.7554/eLife.07398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salis P, Payre F, Valenti P, Bazellieres E, Le Bivic A, and Mottola G (2017). Crumbs, Moesin and Yurt regulate junctional stability and dynamics for a proper morphogenesis of the Drosophila pupal wing epithelium. Sci Rep 7, 16778. 10.1038/s41598-017-15272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morais-de-Sá E, Mirouse V, and St Johnston D (2010). aPKC Phosphorylation of Bazooka Defines the Apical/Lateral Border in Drosophila Epithelial Cells. Cell 141, 509–523. 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walther RF, and Pichaud F (2010). Crumbs/DaPKC-Dependent Apical Exclusion of Bazooka Promotes Photoreceptor Polarity Remodeling. Current Biology 20, 1065–1074. 10.1016/j.cub.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 46.Dong W, Lu J, Zhang X, Wu Y, Lettieri K, Hammond GR, and Hong Y (2020). A polybasic domain in aPKC mediates Par6-dependent control of membrane targeting and kinase activity. Journal of Cell Biology 219, e201903031. 10.1083/jcb.201903031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston CA, Hirono K, Prehoda KE, and Doe CQ (2009). Identification of an Aurora-A/PinsLINKER/ Dlg Spindle Orientation Pathway using Induced Cell Polarity in S2 Cells. Cell 138, 1150–1163. 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sotillos S, Díaz-Meco MT, Caminero E, Moscat J, and Campuzano S (2004). DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. The Journal of Cell Biology 166, 549–557. 10.1083/jcb.200311031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hannaford M, Loyer N, Tonelli F, Zoltner M, and Januschke J (2019). A chemical-genetics approach to study the role of atypical protein kinase C in Drosophila. Development, dev.170589. 10.1242/dev.170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osswald M, Barros-Carvalho A, Carmo AM, Loyer N, Gracio PC, Sunkel CE, Homem CCF, Januschke J, and Morais-de-Sá E (2022). aPKC regulates apical constriction to prevent tissue rupture in the Drosophila follicular epithelium. Current Biology, S0960982222013859. 10.1016/j.cub.2022.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biehler C, Rothenberg KE, Jette A, Gaude H-M, Fernandez-Gonzalez R, and Laprise P (2021). Pak1 and PP2A antagonize aPKC function to support cortical tension induced by the Crumbs-Yurt complex. eLife 10, e67999. 10.7554/eLife.67999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DYR, and Reiter JF (2005). Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021. 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 53.Lancaster MA, Schroth J, and Gleeson JG (2011). Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nat Cell Biol 13, 700–707. 10.1038/ncb2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rys JP, DuFort CC, Monteiro DA, Baird MA, Oses-Prieto JA, Chand S, Burlingame AL, Davidson MW, and Alliston TN (2015). Discrete spatial organization of TGFβ receptors couples receptor multimerization and signaling to cellular tension. eLife 4, e09300. 10.7554/eLife.09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aegerter-Wilmsen T, Aegerter CM, Hafen E, and Basler K (2007). Model for the regulation of size in the wing imaginal disc of Drosophila. Mechanisms of Development 124, 318–326. 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Aegerter-Wilmsen T, Heimlicher MB, Smith AC, de Reuille PB, Smith RS, Aegerter CM, and Basler K (2012). Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size. Development 139, 3221–3231. 10.1242/dev.082800. [DOI] [PubMed] [Google Scholar]
- 57.LeGoff L, Rouault H, and Lecuit T (2013). A global pattern of mechanical stress polarizes cell divisions and cell shape in the growing Drosophila wing disc. Development 140, 4051–4059. 10.1242/dev.090878. [DOI] [PubMed] [Google Scholar]
- 58.Mao Y, Tournier AL, Hoppe A, Kester L, Thompson BJ, and Tapon N (2013). Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J 32, 2790–2803. 10.1038/emboj.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hariharan IK (2015). Organ Size Control: Lessons from Drosophila. Developmental Cell 34, 255–265. 10.1016/j.devcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan Y, Heemskerk I, Ibar C, Shraiman BI, and Irvine KD (2016). Differential growth triggers mechanical feedback that elevates Hippo signaling. Proceedings of the National Academy of Sciences 113, E6974–E6983. oskar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwayer C, Shamipour S, Pranjic-Ferscha K, Schauer A, Balda M, Tada M, Matter K, and Heisenberg C-P (2019). Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell 179, 937–952.e18. 10.1016/j.cell.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Jin Y, Dong L, Lu Y, Wu W, Hao Q, Zhou Z, Jiang J, Zhao Y, and Zhang L (2012). Dimerization and Cytoplasmic Localization Regulate Hippo Kinase Signaling Activity in Organ Size Control. Journal of Biological Chemistry 287, 5784–5796. 10.1074/jbc.M111.310334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Case LB, Zhang X, Ditlev JA, and Rosen MK (2019). Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097. 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM, Biswas KH, Kuriyan J, and Groves JT (2019). A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103. 10.1126/science.aau5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun S, Reddy BVVG, and Irvine KD (2015). Localization of Hippo signalling complexes and Warts activation in vivo. Nature Communications 6. 10.1038/ncomms9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aguilar-Aragon M, Elbediwy A, Foglizzo V, Fletcher GC, Li VSW, and Thompson BJ (2018). Pak1 Kinase Maintains Apical Membrane Identity in Epithelia. Cell Reports 22, 1639–1646. 10.1016/j.celrep.2018.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hannaford M, Loyer N, Tonelli F, Zoltner M, and Januschke J (2019). A chemical-genetics approach to study the role of atypical protein kinase C in Drosophila. Development, dev.170589. 10.1242/dev.170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benton R, and St Johnston D (2003). Drosophila PAR-1 and 14–3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115, 691–704. [DOI] [PubMed] [Google Scholar]
- 69.Yoshihama Y, Izumisawa Y, Akimoto K, Satoh Y, Mizushima T, Satoh K, Chida K, Takagawa R, Akiyama H, Ichikawa Y, et al. (2013). High expression of KIBRA in low atypical protein kinase C-expressing gastric cancer correlates with lymphatic invasion and poor prognosis. Cancer Science 104, 259–265. 10.1111/cas.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duning K, Schurek E-M, Schluter M, Bayer M, Reinhardt H-C, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem MA, et al. (2008). KIBRA Modulates Directional Migration of Podocytes. Journal of the American Society of Nephrology 19, 1891–1903. 10.1681/ASN.2007080916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.LeGoff L, and Lecuit T (2016). Mechanical Forces and Growth in Animal Tissues. Cold Spring Harb Perspect Biol 8, a019232. 10.1101/cshperspect.a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Irvine KD, and Shraiman BI (2017). Mechanical control of growth: ideas, facts and challenges. Development 144, 4238–4248. 10.1242/dev.151902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fomicheva M, Tross EM, and Macara IG (2020). Polarity proteins in oncogenesis. Current Opinion in Cell Biology 62, 26–30. 10.1016/j.ceb.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Restrepo S, Zartman JJ, and Basler K (2016). Cultivation and Live Imaging of Drosophila Imaginal Discs. In Drosophila Methods in Molecular Biology., Dahmann C, ed. (Springer; New York: ), pp. 203–213. 10.1007/978-1-4939-6371-3_11. [DOI] [PubMed] [Google Scholar]
- 75.van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, and Yu T (2014). scikit-image: image processing in Python. PeerJ 2, e453. 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stringer C, Wang T, Michaelos M, and Pachitariu M (2021). Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106. 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 77.McCartney BM, and Fehon RG (1996). Distinct cellular and subcellular patterns of expression imply distinct functions for the Drosophila homologues of moesin and the neurofibromatosis 2 tumor suppressor, merlin. The Journal of Cell Biology 133, 843–852. 10.1083/jcb.133.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han K (1996). An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Research 24, 4362–4363. 10.1093/nar/24.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, and Montague RA (2000). Multiple Forces Contribute to Cell Sheet Morphogenesis for Dorsal Closure in Drosophila. Journal of Cell Biology 149, 471–490. short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang J, Zhou W, Dong W, Watson AM, and Hong Y (2009). Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. U.S.A. 106, 8284–8289. 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pinheiro D, Hannezo E, Herszterg S, Bosveld F, Gaugue I, Balakireva M, Wang Z, Cristo I, Rigaud SU, Markova O, et al. (2017). Transmission of cytokinesis forces via E-cadherin dilution and actomyosin flows. Nature 545, 103–107. 10.1038/nature22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Erdmann RS, Baguley SW, Richens JH, Wissner RF, Xi Z, Allgeyer ES, Zhong S, Thompson AD, Lowe N, Butler R, et al. (2019). Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags. Cell Chemical Biology 26, 584–592.e6. 10.1016/j.chembiol.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rauzi M, Lenne P-F, and Lecuit T (2010). Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468, 1110–1114. 10.1038/nature09566. [DOI] [PubMed] [Google Scholar]
- 84.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Kib movement from the junctional to the medial cortex is coordinated with F-actin flow. Related to Figure 1.
Video S2. Medial Kib movement across the apical cortex is coordinated with medial myosin flow. Related to Figure 1.
Video S3. Hypertonic shift leads to increased medial myosin flow which appear to cluster Kib medially. Related to Figure 2.
Video S4. Hypotonic shift leads to decreased medial myosin flow and concomitant accumulation of myosin and Kib at the junctional cortex. Related to Figure 2.
Video S5. Treatment with Jasp stabilizes F-actin and leads to more junctional Kib accumulation. Related to Figure 3.
Video S6. Ectopic expression of SqhDD reduces medial F-actin flow and leads to more junctional Kib localization. Related to Figure 3.
Video S7. Depletion of crb disrupts myosin organization. Related to Figure 3.
Data Availability Statement
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


