Abstract
Translation elongation factor 2 (EF2), which in Saccharomyces cerevisiae is expressed from the EFT1 and EFT2 genes, has been found to be targeted by a new family of highly specific antifungal compounds derived from the natural product sordarin. Two complementation groups of mutants resistant to the semisynthetic sordarin derivative GM193663 were found. The major one (21 members) consisted of isolates with mutations on EFT2. The minor one (four isolates) is currently being characterized but it is already known that resistance in this group is not due to mutations on EFT1, pointing to the complex structure of the functional target for these compounds. Mutations on EF2 clustered, forming a possible drug binding pocket on a three-dimensional model of EF2, and mutant cell extracts lost the capacity to bind to the inhibitors. This new family of antifungals holds the promise to be a much needed and potent addition to current antimicrobial treatments, as well as a useful tool for dissection of the elongation process in ribosomal protein synthesis.
Ribosomal protein synthesis is one of the oldest and best-conserved processes taking place in a living cell. Within this commonality, it is possible to distinguish between the eubacterial and eukaryotic translation machineries, with the members of the domain Archaea displaying an amazingly heterogeneous mixture of eubacterial and eukaryotic features (9). One of the distinguishing characteristics of the three clades is the differential sensitivity to inhibitors of the translation process (2).
Amino acid residues are added to the growing peptide chain in the ribosome by an elongation process that involves two GTP-switched elongation factors, denominated EF1 and EF2 in eukaryotes. EF1-GTP brings the aminoacyl-tRNA (as the so-called ternary complex) to the acceptor site on the ribosome. After the nascent protein chain is transpeptidated to the newly arrived tRNA, EF2 catalyzes a conformational switch of the organelle, such that the newly generated peptidyl-tRNA is moved from the acceptor site to the peptidyl site, liberating the former for a new round of elongation. EF2 is a large (more than 800-residue), probably multifunctional, and remarkable protein that apparently binds to the same ribosomal structures as the EF1-GTP–aminoacyl-tRNA complex. Observations in bacteria indicate that this can easily be accomplished, since the overall shape of the bacterial EF2 homolog mimics that of the whole ternary complex (18, 19, 25).
The main purpose of the work described here was to identify by genetic means, in Saccharomyces cerevisiae, cellular components targeted by the family of compounds denominated FPS, for fungal protein synthesis inhibitors, which are semisynthetic derivatives of the natural product sordarin (6, 13, 23). These compounds inhibit translation elongation in fungal cells with a high degree of selectivity (10, 16), despite the high degree of conservation in the translation components within eukaryotes. One of these compounds, denominated GM193663, was used to select for resistant mutants. We present genetic evidence indicating that EF2 is part of the target to which this new family of antifungals bind. While the manuscript was in preparation, a related compound was used to determine that EF2 is the target of sordarins (15). The present work independently confirms and extends those observations by investigating the mechanism of resistance and by constructing a three-dimensional model of EF2 which shows a possible binding site for the drug on the protein. Furthermore, we present evidence showing that the EF2 function does not represent the whole target for sordarins, because a second complementation group of resistant mutants in S. cerevisiae was detected.
MATERIALS AND METHODS
Strains, growth conditions, plasmids, and genomic library.
All yeast strains used in this study are derivatives of S. cerevisiae SEY6210 (MATα ura3-52 leu2-3,112 his3Δ200 trp1Δ901 lys2-81 suc2Δ9), S. cerevisiae SEY6211 (MATa ura3-52 leu2-3,112 his3Δ200 trp1Δ901 ade2-101 suc2Δ9) (S. Emr), and S. cerevisiae 373 (MATa ade2-101) (A. Jimenez).
Growth media and methods for tetrad analysis, gene disruption, and allele recovery in yeasts were as described previously (12). Yeast transformations were done by the lithium acetate method as described by Ito et al. (14). Escherichia coli DH5α [endA1 hsdR1 supE44 thi-1 recA1 gyrA9 relA1 ΔlacU169(φ80lacZΔM15)] was used for transformation and preparation of plasmid DNA. All DNA manipulations were carried out by standard procedures (3, 22).
Spontaneous FPS-resistant mutants were selected by plating them on yeast extract peptone dextrose (YPD) medium plates containing either 6 or 100 μg of GM193663 per ml and incubating the plates at 30°C until colonies appeared. To score the resistant phenotype after genetic crosses, agar plates containing 1 μg of the inhibitor per ml were used.
The genomic library from the resistant mutant FPR1-4 was constructed by ligating partially digested (Sau3A-I) genomic DNA into the BamHI site of the pRS316 vector (CEN6 URA3 Ampr) and transforming the ligation mixture into E. coli DH5α. Plasmid DNA from 15,000 independent primary E. coli transformants with an average insert size of 13 kb was pooled, and the pooled DNA was used to transform the yeast.
Disruption of EFT1 and EFT2 in the various wild-type and mutant strains was carried out by allele replacement with HIS3 by following standard techniques (3, 12). The disrupted loci were checked by Southern blotting.
In vitro activity and binding assays.
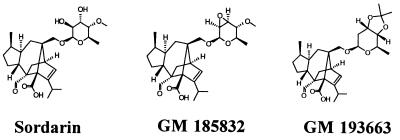
The growth inhibitory activities of FPS compounds were determined in 96-well microtiter plates by the antibiotic twofold serial dilution technique (from 125 to 0.01 μg/ml). One hundred microliters of YPD was inoculated with 105 CFU per well. The MIC was defined as the lowest concentration of compound that inhibited 95% of the control growth after 24 h of incubation at 30°C. Stock solutions of sordarin (Bioprocessing, Glaxo Wellcome, Stevenage, United Kingdom) and other FPS derivatives (Glaxo Wellcome, S.A.) were made in dimethyl sulfoxide at 5 mg/ml. The FPS compounds tested were sordarin, GR135402, GM160575, GM163420, GM165119, GM193663, and GM237354. The chemical structures of representative FPS compounds used in the main experiments described in this work are shown in Fig. 1. Commercial protein synthesis inhibitors were obtained from Sigma. Anisomycin, cycloheximide, and hygromycin were prepared in water at 10 mg/ml. Verrucarin A was dissolved at 2.5 mg/ml in water-dimethyl sulfoxide (3:1). In vitro translation assays with S50 extracts (supernatant fraction obtained by centrifugation of cell extracts at 50,000 × g for 30 min) from cells growing exponentially at 30°C (optical density at 600 nm = 2) in YPD were carried out by following the poly(U) (Sigma)-directed incorporation of [3H]phenylalanine (Amersham) into acid-insoluble material essentially as described previously (5).
FIG. 1.
Structures of the inhibitors used in this study. Sordarin is a natural product; the other two are more potent semisynthetic derivatives.
For the binding assays, it was necessary to disrupt the EFT1 gene in all mutant and parental strain pairs in order to reduce the background binding due to the EF2 protein expressed from the EFT1 locus. [3H]sordarin, labeled at the aldehyde group, was synthesized at 180 GBq/mmol by the Glaxo Wellcome Isotope Chemistry Group (Stevenage, United Kingdom). Binding assays were done with 2 mg of protein from an S50 fraction and 0.1 μg of [3H]sordarin (36 kBq) in a final volume of 500 μl under the same conditions used for poly(Phe) synthesis. After 15 min of incubation at room temperature, 400 μl from each sample was applied to a prepacked PD-10 Sephadex-G-25M column (Pharmacia) equilibrated with the binding buffer. [3H]sordarin bound to macromolecules was measured by counting the radioactivity in the excluded fractions. The radioactivity counts obtained in the presence of a 100-fold excess of cold sordarin were subtracted from all datum points. Assay points were always obtained in duplicate, and the values were averaged. Independent experiments were performed with cell extracts prepared on different days.
Molecular mapping of EFT2 mutations.
Single-strand conformation polymorphism (SSCP) mapping of the resistance mutations was carried out as described previously (20). EFT2 from the mutant and wild-type strains was amplified as six overlapping fragments of approximately 500 bp by standard PCRs with the oligonucleotide pairs listed in Table 1.
TABLE 1.
Oligonucleotide pairs used to amplify the six overlapping fragments of EFT2
| Fragment | Oligonucleotide | Fragment size (bp) |
|---|---|---|
| A | 5′-CACAAATTATAACATAATTGC-3′ | 502 |
| 5′-CCTTGTTGATAACAACAACAGGC-3′ | ||
| B | 5′-GAAGGTGTCTGTGTCCAAACC-3′ | 487 |
| 5′-CATCTTTCTTGAAGTTCATGATAGC-3′ | ||
| C | 5′-CAACATGTTCATCTTGGACC-3′ | 518 |
| 5′-CGATTGGTTCGACAAATCTACC-3′ | ||
| D | 5′-CAAGGTCCAAACTACGTTCC-3′ | 506 |
| 5′-TTGTTTGGAGACTTGGACAAAGC-3′ | ||
| E | 5′-GGTGTTCCATTGAAGATCTCC-3′ | 515 |
| 5′-GGATCAGCCAACAAGAAACC-3′ | ||
| F | 5′-CATCCCAACCATGAGAAGAGC-3′ | 418 |
| 5′-TTCTTACAATTTGTCGTAATATTC-3′ |
Resistant alleles were rescued (12) from their chromosomal locations with pRS316 plasmids carrying a wild-type EFT2 gene gapped by the removal of either of two internal regions of the coding sequence covering either amino acids 439 to 502 or amino acids 495 to 652, as appropriate. Plasmids were recovered from Ura+ stable transformants, retransformed into a wild-type strain to check the resistant phenotype, and sequenced. Sequencing was done with an ABI Prism 310 Sequencer Analyzer according to the manufacturer’s recommendations.
Three-dimensional modeling of EF2.
Computational and structural details will be published elsewhere, but in essence, the model resulted from homology mapping of EF2 onto the crystal structure of EF-G from Thermus thermophilus (Brookhaven Protein Data Bank [PDB] entries 1dar and 1elo) (1, 7). The model was built in three main steps. First, the water of crystallization in EF-G was removed. Second, EF2 residues were substituted for the corresponding ones in EF-G by using the HOMOLOGY module of the BIOSYM package (Molecular Simulations Inc., San Diego, Calif.). Third, sequence stretches present in S. cerevisiae EF2 and absent from T. thermophilus EF-G were modeled. For this last purpose, protein fragments of known three-dimensional structure with the same or a closely related sequence were obtained from the PDB-select database, and they were fitted to the structure by using HOMOLOGY. The complete structure was refined with DISCOVER CVFF and AMBER force fields (8). The quality of modeling throughout the different steps of the process was monitored with the PROCHECK program (17). Molecular docking experiments were performed by taking into account the molecular electrostatic potential minima of GM185832, the complementarity of the atomic contact surface areas, and the chemical properties of the contact atoms by using the DOCKING and DELPHY modules of BIOSYM.
RESULTS
GM193663-resistant mutants fall into two complementation groups.
A genetic approach was used to identify targets of GM193663. Spontaneous resistant mutants in S. cerevisiae, arising at frequencies of 10−7 to 10−8, were selected at concentrations of GM193663 of 6 and 100 μg/ml on YPD medium plates. All mutants were cross resistant to the members of the FPS class tested (see Materials and Methods). No significant cross-resistance was observed with other protein synthesis inhibitors: anisomycin, cycloheximide, hygromycin, and verrucarin A.
Twenty-five independently isolated mutants were analyzed by making genetic crosses, and at least 16 spore tetrads from each cross were dissected to score the resistance phenotype of the segregants. Resistance segregated in all cases as a single mutation in a Mendelian fashion. It was observed in the backcrosses that the resistance of the heterozygous diploids always fell between those of the two parents, but the MICs for the diploids spread widely between the MIC for the sensitive parental strain (0.2 μg/ml) and the MIC for the resistant parental strain, indicating that resistance is a semidominant phenotype. All 10 mutants obtained at the lower dose fell into a single complementation group, which was named FPR1. From the 15 mutants isolated at the higher dose, 11 fell into FPR1 and the remaining 4 defined a second complementation group, FPR2. In summary, of 25 mutants analyzed, 21 fell into one complementation group and 4 fell into a second one. This defines two genetic loci capable of giving resistance to GM193663 when they are mutated.
Poly(Phe) synthesis is resistant to GM193663 in mutant cell extracts.
One of the most common mechanisms of resistance to antimicrobials arises through changes in transport systems for the inhibitory compounds. Since that is uninformative regarding the mode of action of the inhibitor, the sensitivity of poly(Phe) synthesis to GM193663 was tested with cell extracts from mutant strains. The 50S supernatants were prepared from total cell homogenates obtained from one of the most dominant mutants in each group (FPR1-5 and FPR2-6). Poly(U)-directed synthesis of poly(Phe) was assayed in the presence of increasing concentrations of GM193663. The inhibition curves showed that resistance is also manifested in cell extracts (Fig. 2), thus excluding transport mechanisms and indicating that the mutations affect elements involved in the mode of action of the inhibitor. It is also apparent that the curve for the extract from the FPR2 mutant has a complex shape. It could be interpreted to mean that there are at least two cellular components with different sensitivities to GM193663. Clarification of this issue will be facilitated by the identification of the gene involved. Cloning and characterization of the gene mutated in complementation group FPR1 are presented below. Group FPR2 is currently being analyzed and will be described elsewhere.
FIG. 2.
Inhibition of poly(Phe) synthesis by GM193663 in extracts from wild-type and resistant strains. Cell extracts from mutants belonging to complementation groups 1 (FPR1) and 2 (FPR2) and from their corresponding parental strains were programmed with poly(U) to synthesize poly(Phe) in the presence of various concentrations of GM193663, as described in Materials and Methods. Data from two independent experiments are represented together. •, resistant mutant; ○, wild-type parental strain.
The gene mutated in complementation group 1 is EFT2.
Attempts to clone FPR1 by transforming wild-type libraries into the least dominant mutant strain failed. Therefore, a gene library on a centromeric vector was constructed from one of the most dominant alleles, FPR1-4 (see Materials and Methods). Screening of 23,000 S. cerevisiae colonies transformed with the library DNA yielded six GM193663-resistant (GM193663r) isolates. Plasmid DNA recovered from the resistant transformants was analyzed to identify the common regions and the minimum sequence capable of conferring resistance. This was found to be a 3.5-kb EcoRI fragment, which was then partially sequenced. The sequences obtained matched that of EFT2, one of the two genes which encode the 842-residue EF2 in S. cerevisiae (21). As a further check, EFT2 was disrupted in the FPR1-4 haploid strain. The disruptant was viable due to the presence of an intact EFT1 locus, but resistance was lost, confirming that the mutation giving GM193663 resistance was on EFT2. In agreement with this result, independent work with the pathogenic yeast Candida albicans has shown that EF2 is the primary sordarin-binding protein in cell extracts from this organism (11). GM193663 can therefore inhibit the growth of yeast cells by interacting with EF2.
Most resistance mutations cluster on a 50-amino-acid segment of EF2.
S. cerevisiae EF2 has 53% homology to bacterial EF-G, and it presumably folds into the same structural and functional domains (see below) (1, 7). Localization of resistance mutations within the EF2 protein could give clues to the mechanism of resistance and hence to the mode of action of GM193663. To avoid sequencing this large gene from all mutants, EFT2 from several mutant strains was amplified as a series of six overlapping PCR fragments. SSCP techniques were used to map the mutations to individual fragments. Fragments from 11 mutants displayed altered mobility compared with that of the corresponding fragment from the parental strain. In all cases, the mobility change affected fragment D or E only (Fig. 3A), indicating that changes in just a small area of the protein can confer GM193663 resistance.
FIG. 3.
Mapping of the GM193663r mutations on the primary structure of EF2. (A) Overlapping fragments of the EFT2 gene used in the mapping by SSCP. Base pair 1 corresponds to the A in the first ATG codon. (B) Position and nature of the amino acid change in the sequenced mutant alleles of EFT2. (C) Protein sequence comparison of the region in EF2 from different organisms corresponding to the region where most GM193663 resistance mutations were found in S. cerevisiae. The mutated positions are boxed, and the amino acid residue found in the resistant mutant is shown on top of each box. Fu, fungi; An, animals; Pl, plants; Pr, protists.
Fragments displaying an altered mobility were cloned, and both strands were completely sequenced, as was the corresponding region of the original library clone, FPR1-4. A single base change was detected in each case, leading to an amino acid substitution in the EF2 protein. Two mutants were found to have the same mutation. Two others had different substitutions at the same position in the protein, reducing to nine the total number of mutated positions. The nature of the amino acid changes is shown in Fig. 3B. Six of the altered positions clustered on a 50-amino-acid segment which maps to domain 3 of EF-G (see below); the remaining three changes map to positions flanking this region. Although the clustering of mutations around the one found in the genomic clone made it unlikely that they were PCR artifacts, the possibility was checked by rescuing the mutations from the genomes of several resistant isolates by using “gap repair” techniques for allele rescue (see Materials and Methods). The repaired plasmids were transformed into a Δeft1 Δeft2 S. cerevisiae 6210, and the region of interest was sequenced. They were found to confer resistance and to have the expected base changes, confirming that the observed amino acid replacements were responsible for the resistance.
Figure 3C shows the six more tightly clustered mutations in the context of the sequences of other eukaryotic EF2 proteins from widely divergent groups. The nature of the amino acid substitutions does not give obvious clues to the mechanism of resistance, but the sequence alignment shows that they are located in a region bracketing a highly conserved and hence potentially important region. It is therefore surprising how radical an amino acid change EF2 can tolerate and yet remain functional. The Q490E and Y521D mutations introduce an acidic residue at a position where none is found naturally. Another “unnatural” change is introduced by the A562P mutation. Yet, the resistant mutants grow at a rate not significantly different from that of the wild type in the absence of the drug (data not shown).
Resistant EF2 molecules have reduced affinity for GM193663.
Clustering of GM193663 mutations within a small area of the EF2 protein strongly suggested that they might change the structure of a binding site for the drug. The hypothesis was tested by preparing cell extracts from the resistant mutants and measuring the level of binding of [3H]sordarin to macromolecules by gel filtration. This is an available labeled analog of GM193663 which competes for binding and shows cross-resistance (data not shown). A background of binding activity was expected, and indeed was observed, due to the presence of the EF2 protein expressed from the EFT1 gene, the second gene encoding EF2 in S. cerevisiae (21, 24). To simplify the system, EFT1 was interrupted in eight resistant strains with unique mutations in EF2. Cell extracts containing cytosol and ribosomes were prepared from the deletants, and the level of [3H]sordarin binding was measured. It was observed that GM193663 resistance mutations in EF2 eliminate the binding of the labeled analog to cellular macromolecules, thus explaining the resistance. Table 2 shows the results from two independent binding experiments. It can be seen that when all EF2 molecules in the cell carry a mutation for resistance to GM193663, there is no significant sordarin binding to macromolecules.
TABLE 2.
Specific binding of [3H]sordarin to macromolecules in cell extracts from wild-type and GM193663r mutant strainsa
| Strain | Mutation | dpmb
|
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| 373 | None | 85,640 | 71,265 |
| FPR1-3 | Ala562Pro | 125 | 0 |
| FPR1-5 | Phe612Leu | 172 | 0 |
| SEY6210 | None | 23,184 | 188,515 |
| FPR1-18 | Ile529Ser | 229 | 76 |
| FPR1-19 | Ser523Thr | 0 | 191 |
| FPR1-21 | Tyr521Asp | 0 | 87 |
| FPR1-24 | Val561Phe | 4,853 | 1,040 |
| FPR1-26 | Ala475Val | 0 | 1,770 |
| FPR1-30 | Pro559Ser | 99 | 7,053 |
Resistant isolates are listed below their corresponding parental strain.
3H radioactivity associated with macromolecules, measured in two independent experiments (experiments 1 and 2), as described in Materials and Methods.
Mutated positions define a putative binding pocket in a three-dimensional model of EF2.
Completely independent of the work described above, a three-dimensional model of S. cerevisiae EF2 was constructed by using as a scaffold the crystal structure of EF-G from T. thermophilus (PDB entries 1dar and 1elo) (1, 7) (see Materials and Methods). The values from the Ramachandran plots obtained for the modeled EF2 structure (Table 3) allow a high degree of confidence in the overall correctness of the model.
TABLE 3.
Quality tests of T. thermophilus EF-G crystal structure and S. cerevisiae EF2 model by using the PROCHECK programa
| Elongation factor | Percent residues in the following locations:
|
SD of ω angles | No. of bad contacts | |||
|---|---|---|---|---|---|---|
| Most favored region | Additional allowed zones | Generously allowed regions | Disallowed regions | |||
| T. thermophilus EF-G (691 amino acids) | 90.1 | 8.9 | 0.5 | 0.5 | 6.0 | 1 |
| S. cerevisiae EF2 (842 amino acids) | 80.2 | 17.0 | 1.6 | 1.2 | 10.6 | 7 |
Ramachandran plots were generated with the PROCHECK program by using the coordinates from PDB entries 1dar and 1elo for EF-G and from the theoretical model presented here for EF2 (see Materials and Methods).
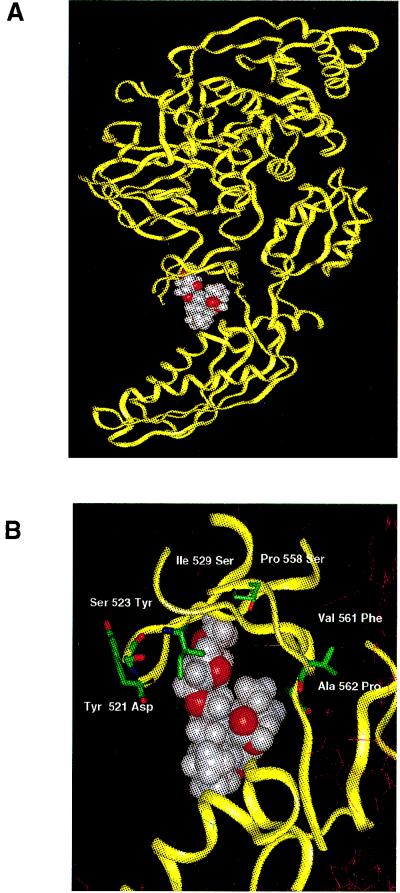
Next, a molecular docking experiment was performed with EF2 and another member of the FPS family of compounds, GM185832 (see Materials and Methods). The most favorable binding site that emerged was the one shown in Fig. 4A, at a cleft in what would be domain 3 in EF-G. Remarkably, this predicted binding site coincided with the location on the model of the sequenced GM193663 resistance mutations. Seven of the 10 mutations which block binding fall exactly on or around this predicted binding site (Fig. 4B). This area of the protein corresponds to a very flexible domain of EF-G, presumed to be a hinge connecting the G-protein portion of the molecule to putative rRNA-binding domains (1, 7). According to this model, FPS inhibitors could act as a molecular wedge, reducing the flexibility of the factor around domain 3. It can clearly be seen in Fig. 4B that the mutations would block access to or change the structure of the proposed binding pocket.
FIG. 4.
Calculated three-dimensional model of S. cerevisiae EF2. (A) Site of most favorable interaction of GM185832 on EF2, obtained from molecular docking experiments (see Materials and Methods for details). (B) Detail of the predicted contact region between the protein and the inhibitor, with the addition of the side chains of the amino acid residues present at the positions found to be mutated in the different resistant alleles.
The gene mutated in complementation group 2 is not EFT1.
Since mutations in EFT2, encoding EF2, confer resistance to FPS compounds, EFT1, the second expressed gene encoding this factor in S. cerevisiae (21), was a natural candidate as an explanation for the second complementation group of resistant mutants. Disruption of EFT1 in a mutant from this group did not abolish resistance, however, negating the hypothesis. The result was confirmed by genetic crosses and segregation analysis between FPR2-6 and an EFT1 locus genetically tagged with HIS3. Thus, there is at least one more cellular component, apart from EF2, involved in the mode of action of FPS compounds.
DISCUSSION
The ribosomal protein synthesizing machinery is thought to be highly conserved among eukaryotic organisms, perhaps with the exception of fungi, which have an additional soluble elongation factor (EF3) not present in other analyzed eukaryotes (4). It was therefore widely believed that protein synthesis inhibitors could make good antibacterial compounds but were unlikely to be selective inhibitors of eukaryotic microbes, again with the possible exception of drugs aimed at fungal EF3. It therefore came as a surprise when our genetic analysis showed that S. cerevisiae EF2, one of the most ancient and conserved proteins throughout evolution, was a target of Glaxo Wellcome’s FPS family of selective antifungals, as evidenced by the fact that single point mutations on the protein conferred resistance to six different members of the class.
Eighty percent of the resistance mutations affected EF2. The fact that 7 of 10 alleles sequenced had changes clustered on a 50-amino-acid segment of the EF2 protein, the same one predicted to fold into an FPS-binding pocket in our modeling experiments, plus the fact that mutants displayed negligible binding to FPS compounds, strongly suggests that the mutated positions define the binding site for the drug. Mutations outside of this pocket could reduce the binding indirectly by affecting the folding of the binding site. Preliminary results from cross-linking and protease digestion experiments are in agreement with the hypothesis presented above. A detailed analysis by mass spectroscopy will be undertaken to establish this point.
The proposed FPS binding site is located on what would be domain 3 of EF2 by comparison with the X-ray structure of EF-G. Figure 3C shows that some of these positions are highly conserved in EF2 proteins from different organisms. Yet all these substitutions allow mutated EF2 to catalyze translation elongation. This agrees with the notion, deduced from the EF-G structure, that a precise conformation may not be essential for the role of this protein domain, which may only require some global physicochemical property, such as flexibility, to function (1, 7). Impediment of EF-G’s flexibility has been suggested to underlie fusidic acid’s mode of action (7). FPS inhibitors could be doing exactly the same thing, acting also on this hinge region, but from the side of the factor opposite the putative binding site for fusidic acid. Recently communicated results, obtained with cloned EF2 and a different sordarin derivative, found resistant mutants with changes in parts of the protein not affected by the mutations described here but corresponding to areas found to be mutated in EF-G proteins resistant to fusidic acid. The mechanism of resistance brought about by those mutations is unknown, but in agreement with the hypothesis presented above, some mutants were found to have cross-resistance to fusidic acid (15).
It is noteworthy that 21 spontaneous mutants with changes on EFT2 were found but that none had mutations on EFT1. This may be due to the partial dominance of the resistant phenotype and the fact that the EFT2 promoter seems to be 2.5-fold more active than that of EFT1 (24). A highly dominant phenotype may be needed to detect EFT1 mutants when EFT2 is being expressed at such high levels.
The existence of a second complementation group of FPS-resistant mutants indicates that the cellular function inhibited by FPS compounds is not carried out by EF2 alone. The involvement of more than one component in defining the functional target for sordarins could contribute to the selectivity of these compounds, despite the high degrees of homology between individual molecular components of the target. Our genetic data show that the gene mutated in the second complementation group does not encode EF2. Preliminary biochemical experiments indicate that resistance in this group is associated with ribosomes (data not shown), and in C. albicans, high-affinity binding to EF2 requires the presence of ribosomes (11), supporting a role for ribosomal components in the interaction between the inhibitors and EF2. Given the number of isolates analyzed so far, the mutant screen cannot be considered saturated. Thus, the pathway inhibited by sordarins could still contain additional components, all presumably involved in the EF2 function and hence ribosomal translocation.
New selective antifungal agents are sorely needed in the clinic. Compounds from this family are very good candidates that could be used to close some important gaps in the existing antifungal drug portfolio. FPS inhibitors can also be useful tools in dissecting the mechanism of the elongation cycle in eukaryotic ribosomes, including identification of the ribosomal components involved, something that we hope to start achieving once cloning of the FPR2 gene is accomplished.
ACKNOWLEDGMENTS
We are indebted to M. J. Serramía for expert technical assistance, M. Gómez for performing gene disruptions, J. M. Domínguez for measuring poly(Phe) synthesis, and our colleagues in the Isotope Chemistry Group (Glaxo Wellcome) for providing tritiated sordarin. The many scientific discussions and the critical reading of the manuscript by J. P. G. Ballesta are also gratefully acknowledged.
REFERENCES
- 1.AEvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al Karadaghi S, Svensson L A, Liljas A. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amils R, Ramirez L, Sanz J L, Marin I, Pisabarro A G, Sanchez E, Ureña D. Phylogeny of antibiotic action. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 645–654. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 4.Belfield G P, Tuite M F. Translation elongation factor 3: a fungus-specific translation factor? Mol Microbiol. 1993;9:411–418. doi: 10.1111/j.1365-2958.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 5.Colthurst D R, Chalk P, Hayes M, Tuite M F. Efficient translation of synthetic and natural mRNAs in an mRNA-dependent cell-free system from the dimorphic fungus Candida albicans. J Gen Microbiol. 1991;137:851–857. doi: 10.1099/00221287-137-4-851. [DOI] [PubMed] [Google Scholar]
- 6.Coval S J, Puar M S, Phife D W, Terracciano J S, Patel M. SCH57404, an antifungal agent possessing the rare sordaricin skeleton and a tricyclic sugar moiety. J Antibiot. 1995;48:1171–1172. doi: 10.7164/antibiotics.48.1171. [DOI] [PubMed] [Google Scholar]
- 7.Czworkowski J, Wang J, Steitz T A, Moore P B. The crystal structure of elongation factor g complexed with GDP, at 2.7 angstrom resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dauber-Osguthorpe P, Roberst V A, Wolff J. Structure and energetics of ligand binding to proteins: E. coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1992;4:409–417. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- 9.Dennis P P. Ancient ciphers—translation in Archaea. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez J M, Kelly V A, Kinsman O S, Marriott M S, Gomez de las Heras F, Martin J J. Sordarins: a new class of antifungals with selective inhibition of the protein synthesis elongation cycle in yeast. Antimicrob Agents Chemother. 1998;42:2274–2278. doi: 10.1128/aac.42.9.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez J M, Martin J J. Identification of elongation factor 2 as the essential protein targeted by sordarins in Candida albicans. Antimicrob Agents Chemother. 1998;42:2279–2283. doi: 10.1128/aac.42.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guthrie C, Fink G R. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press Inc.; 1991. [PubMed] [Google Scholar]
- 13.Hauser D, Sigg H P. Isolierung und Abbau von Sordarin. [Isolation and decomposition of sordarin.] Helv. Chim Acta. 1971;54:1178–1190. doi: 10.1002/hlca.19710540427. [DOI] [PubMed] [Google Scholar]
- 14.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Justice M C, Hsu M J, Tse B, Ku T, Balkovec J, Schmatz D, Nielsen J. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J Biol Chem. 1998;273:3148–3151. doi: 10.1074/jbc.273.6.3148. [DOI] [PubMed] [Google Scholar]
- 16.Kinsman O S, Chalk P A, Jackson H C, Middleton R F, Shuttleworth A, Rudd B A M, Jones C A, Noble H M, Wildman H G, Dawson M J, Stylli C, Sidebottom P J, Lamont B, Lynn S, Hayes M V. Isolation and characterisation of an antifungal antibiotic ( GR135402) with protein synthesis inhibition. J Antibiot. 1998;51:41–49. doi: 10.7164/antibiotics.51.41. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 19.Nyborg J, Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Clark B F C, Reshetnikova L. Structure of the ternary complex of EF-Tu: macromolecular mimicry in translation. Trends Biochem Sci. 1996;21:81–82. [PubMed] [Google Scholar]
- 20.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perentesis J P, Phan L D, Gleason W B, LaPorte D C, Livingston D M, Bodley J W. Saccharomyces cerevisiae elongation factor 2. Genetic cloning, characterization of expression, and G-domain modeling. J Biol Chem. 1992;267:1190–1197. [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schneider G, Anke H, Sterner O. Xylarin, an antifungal Xylaria metabolite with an unusual tricyclic uronic acid moiety. Nat Prod Lett. 1995;7:309–316. [Google Scholar]
- 24.Veldman S, Rao S, Bodley J W. Differential transcription of the two Saccharomyces cerevisiae genes encoding elongation factor 2. Gene. 1994;148:143–147. doi: 10.1016/0378-1119(94)90248-8. [DOI] [PubMed] [Google Scholar]
- 25.Wilson K S, Noller H F. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]