Abstract
Sushi, von Willebrand factor type A, epidermal growth factor, and pentraxin domain containing 1 (SVEP1) is a large extracellular matrix protein that is also detected in the circulation. Recent plasma proteomics and genomics studies have revealed a large number of associations between SVEP1 and human traits, particularly chronic disease. These include associations with cardiac death and disease, diabetes, platelet traits, glaucoma, dementia, and aging; many of which are causal. Animal models demonstrate that SVEP1 is critical in vascular development and disease, but its molecular and cellular mechanisms remain poorly defined. Future studies should aim to characterize these mechanisms and determine the diagnostic, prognostic, and therapeutic value of measuring or intervening on this enigmatic protein.
A brief history of SVEP1
SVEP1 is a poorly understood extracellular matrix (ECM) (see Glossary) protein [1,2]. The sudden rise of population plasma proteomics has revealed a profound number of associations between SVEP1 and human chronic disease. For example, among all proteins tested within each respective study, plasma SVEP1 concentration shows the highest acceleration during aging [3], among the strongest associations with heart failure hospitalization or death [4], and the strongest causal relationship with dementia [5]. In addition, Mendelian randomization (MR) analyses (Box 1) suggest SVEP1 causally relates to coronary artery disease (CAD) [6,7], hypertension [6], type 2 diabetes [6,8], platelet traits [7], and longevity [9]. Human genetic or proteomic studies identified additional SVEP1 associations, including glaucoma [10,11], pulmonary arterial hypertension [12], cardiovascular risk [13], atrial fibrillation [14], platelet reactivity [15], and aging [16,17]. Many of these diseases share certain attributes, including vascular dysfunction, chronicity, and aging, suggesting a common pathogenic mechanism may underly their association with SVEP1.
Box 1. Basics of Mendelian Randomization.
Mendelian randomization (MR) uses an experiment of nature to test for causal associations. It utilizes alleles (which are stochastically distributed across a population according to the law of independent assortment) that affect a measured variable, such as the plasma concentration of its gene product. An outcome of interest can then be regressed on the genetically encoded changes of such a variable to test causality.
This approach can estimate the causal relationship of SVEP1 to disease through regression of genetically encoded changes in plasma SVEP1 concentration with outcomes data from GWASs. One limitation in applying this approach to plasma SVEP1 is that its concentration is thought to be regulated, in part, by leakage from origin tissues [18]. This biology obscures the anatomical location of the effects of SVEP1 and may bias the magnitude of the causal estimate, though likely toward the null. Nonetheless, MR remains a powerful tool to test if the observed correlations between SVEP1 and disease reflect causal relationships.
SVEP1 was initially cloned and analyzed in 2000 by Gilgès et al., who named it Polydom for its many domains [1]. It is now referred to as SVEP1, a name derived from its constituent domains. The protein is predominately synthesized by cells of mesenchymal origin and is believed to be integrated within the matrix [2,6,7]. It also leaks from its tissues of origin [18] and circulates in plasma [19,20], likely in association with extracellular vesicles [21–23]. SVEP1 is thought to interact with several proteins, including integrins [2,24,25], angiopoietins (ANGs) [26], and platelet endothelial aggregation receptor 1 (PEAR1), a receptor tyrosine kinase-like protein [7,27].
Animal models demonstrate that SVEP1 is critical for vascular development [11,26,28,29] and mouse models support its role in vascular disease [6,24,30], consistent with its disease associations in humans. SVEP1 resides within the ECM and plasma, making it a priority candidate for disease diagnostics, prognostics, and therapeutics. Here, we provide an overview of the existing data on SVEP1, focusing on its structure, function, and role in development and disease. We also highlight key unanswered questions that, once addressed, will enhance our understanding of the protein and its impact on human biology.
Molecular composition of SVEP1
SVEP1 was first cloned using degenerative reverse transcriptase polymerase chain reaction for sequences that code for epidermal growth factor (EGF) domains [1] (Figure 1). Such domains often govern interaction with adjacent proteins and are found in many ECM proteins and transmembrane receptors [31]. SVEP1 contains a single von Willebrand factor type A (VWA) domain, a domain named after von Willebrand factor [32] and found in many ECM proteins. Many VWA domains contain metal ion-dependent adhesions sites [33] and mediate protein complex formation [32,34] or receptor–ligand interaction. Approximately half of SVEP1 is comprised of repeat complement control protein (CCP) domains, also known as Sushi domains. These domains often govern protein interactions, including with the complement family [35]. Hyalin repeat (HYR) domains are frequently found in association with CCPs [36] and are also contained within SVEP1 [37]. HYR domains are found within hyalin and are thought to govern cell adhesion and protein-protein interaction [36]. SVEP1 also contains a pentraxin domain, which is shared with the pentraxin family of pentameric proteins, including C-reactive protein and serum amyloid P component [38].
Figure 1.
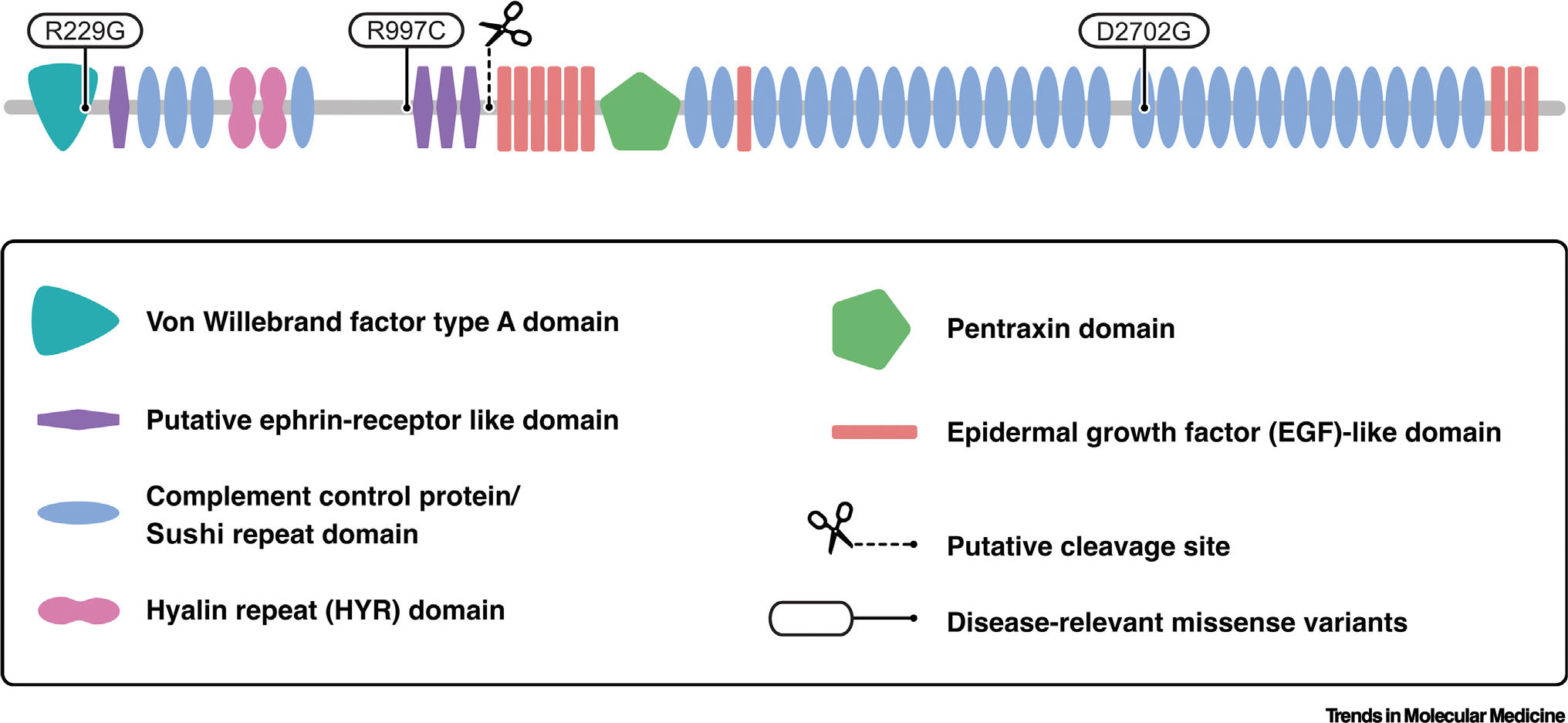
Sushi, von Willebrand factor type A, epidermal growth factor and pentraxin domain containing 1 (SVEP1) protein schematic. Simple Modular Architecture Research Tool and InterPro were used to identify and map domains [97,98]. Coding variants that associate with human disease are indicated at their corresponding peptide. Adapted, with permission, from Elenbaas et al. [7].
Glycoproteomic studies [20,39] identify SVEP1 as a likely glycoprotein that contains a number of potential glycosylation sites [40]. Recombinant murine SVEP1 is efficiently secreted from cells and is cleaved in the process [2]. This generates fragments of approximately 125 and 300 kDa under reducing and denaturing conditions. We have confirmed this cleavage event and its approximate end products in a similar expression system [6]. SVEP1 may exist as a multimer, like many other ECM proteins, though this has not been formally tested.
Tissue expression and localization of SVEP1
SVEP1 is expressed in vasculature [6,7,26,28], adipose [7,30,41], and bone marrow [1,42,43] tissues. The vascular human placenta expresses extremely high levels of SVEP1 [44] and maternal plasma levels of the protein increase ≥20-fold over the course of gestation [45]. Lung, stomach, and intestinal tissue also express the transcript [2]. The cells that express SVEP1 include bone-marrow-derived mesenchymal stem cells [46], preosteoblastic cells [47], activated skeletal muscle satellite cells [48], and vascular cells including adventitial fibroblasts [49] and vascular smooth muscle cells [6,24].
Precisely where SVEP1 exists within the ECM remains unclear. Previous studies reporting in situ SVEP1 staining relied on unvalidated antibodies and did not include necessary negative controls, such as genetic knockouts [50]. This is of particular importance when using antibodies against SVEP1 since it shares many domains with other ECM proteins, resulting in a high potential for undiscernible crossreactivity. Poor antibody validation has led to significant confusion about where SVEP1 is localized within tissue, though SVEP1 pulldown assays suggest it may integrate with the basement membrane after secretion [7]. The generation and rigorous validation of anti-SVEP1 antibodies, or similar reagents, is necessary to characterize the location of SVEP1 during development and pathology.
SVEP1 binding partners
Several SVEP1 binding partners have been proposed, but it is unclear which of these, if any, are responsible for mediating the developmental and disease phenotypes associated with SVEP1. The first protein reported to bind with SVEP1 was integrin α9β1 [2]. Integrin α9 heterodimerizes only with integrin β1 to form integrin α9β1 [51], which binds with an estimated affinity of 32.4 nM to residues 2636–2644, which are conserved in humans and contained within a CCP domain in the C-terminus of murine SVEP1 [2]. Integrin α9β1 is expressed on a wide variety of cell types, particularly muscle and epithelia [52] and is known for its role in various developmental processes, including lymphangiogenesis [53] and hematopoiesis [54] through the regulation of cell growth, migration, differentiation, and other behaviors.
PEAR1, a receptor tyrosine kinase-like protein [27], also interacts with SVEP1 [7]. Plasma SVEP1 levels are influenced by PEAR1 levels in humans [7], suggesting endothelial PEAR1 binds and sequesters SVEP1 from the plasma. Indeed, the recombinant proteins bind to each other with an affinity of 0.7–8.8 nM in label-free binding assays. PEAR1 tissue expression is highly correlated with SVEP1 [7] and the two genes share numerous disease associations, including platelet reactivity [15,55] and cardiovascular disease [56,57]. PEAR1 is also thought to play a role in neoangiogenesis [58] and hematopoiesis [59].
SVEP1 binds to tyrosine-protein kinase receptor Tie-1 (TIE) [60,61], and potentially ANG1 and ANG2 [26]. These data suggest that SVEP1 modulates ANG–TIE signaling. Such regulation of growth factors is a common function of ECM proteins [62]. The ANG–TIE signaling pathway is involved in many of the same developmental and disease processes as SVEP1, including angiogenesis [63], atherosclerosis [64], and glaucoma [11].
Additional binding partners to SVEP1 likely exist. Several partners have been proposed, but their characterization remains limited. These include integrin α4 [24,65] and members of the complement [66] and Notch family [6]. Further investigation is necessary to validate these interactions and determine how they contribute to the biological functions of SVEP1. Given its large size and interaction with multiple signaling proteins, SVEP1 may serve as a signaling nidus and coordinate its effects through multiple interactions.
Cellular response to SVEP1
Various cell types adhere to SVEP1 in a dose-dependent manner [2,6,7,67], consistent with the functions of the domains that comprise the protein. SVEP1-induced cell adhesion depends on integrin α9β1 in rhabdomyosarcoma cells [2], and integrin signaling is activated in various cells exposed to SVEP1 [6]. Certain cells also proliferate rapidly [6] and migrate [61] in response to SVEP1. SVEP1-induced cell proliferation has been shown to depend on integrin α9β1 and Notch signaling [6]; however, the specificity of these effects is unclear. Consistent with its effects on proliferation, exposure of endothelial cells and human coronary artery smooth muscle cells to SVEP1 results in robust AKT/mTOR activation [7,61]. This signaling is dependent on PEAR1 [7] and is of particular interest, since AKT/mTOR signaling plays a key role in the development and disease processes associated with SVEP1 [68–76].
Descriptions of several additional cell responses to SVEP1 have been published, but reproduction of these effects is lacking. Many of these studies utilized unpurified recombinant SVEP1 in solution, which is not a physiological approach to interrogating an ECM protein that is immobilized in vivo. In fact, circulating SVEP1 may also be functionally immobilized given its association with extracellular vesicles [21–23]. The generation of force between a ligand in the ECM and its receptor is often critical for signal transduction; soluble ligands are free in solution and may not be able to transmit sufficient force for receptor activation [77]. We believe this biology suggests SVEP1 immobilization should be the standard approach for cellular assays. The use of purified recombinant protein also reduces the risk of confounded data. Modified cellular expression of SVEP1 in vitro is also not ideal, since SVEP1 produced by cultured cells (which typically lack a robust matrix) is readily excreted into the media as a soluble protein. Rigorous and physiologic assay design is necessary to improve reproducibility and dissect the precise mechanisms of the effects of SVEP1 on cell behavior.
SVEP1 and vascular development
Murine embryos lacking SVEP1 exhibit marked edema by mid-gestation and die immediately after birth [26,28]; this is attributed to severe disruptions in lymphatic vessels and capillaries [11,26,28]. Zebrafish lacking Svep1 also have lymphatic defects, suggesting that the role of SVEP1 in lymphatic development is evolutionarily conserved [28,60]. Zebrafish studies also support a role of Svep1 in blood vessel anastomosis [29]. Similarly, mice lacking SVEP1 in neural crest-derived cells develop a hypomorphic Schlemm’s canal and disrupted vasculature within the eye [11]. Consistent with these developmental findings, analysis of genetic variation within SVEP1 in humans strongly suggests that loss of SVEP1 is not developmentally tolerated [78].
The specific mechanisms by which SVEP1 contributes to development are not well understood, but current understanding is that adjacent endothelial cells respond to SVEP1 produced by nearby mesenchymal cells [6,26,28]. Integrin α9β1 is involved in lymphangiogenesis [53], but the phenotype of Itga9−/− mice is notably milder than Svep1−/− mice [28,79]. Surprisingly, the zebrafish Svep1 lacks the integrin α9β1 binding domain altogether [28], signifying that other proteins likely have a more prominent role. Mice with constitutive loss of Pear1 also fail to phenocopy the developmental defects observed in Svep1−/− mice, despite the role of PEAR1 in neoangiogenesis [29,58]. The potential regulation of ANG-TIE signaling by SVEP1 is a promising explanation for these developmental effects. Interaction with ANG1 and ANG2 has been proposed as a mechanism [26], but in vivo evidence is lacking. Direct interaction between SVEP1 and the tyrosine-protein kinase receptor TIE1 [60,61] has also recently been proposed by independent groups. Further research is needed to characterize how these pathways mediate the critical role of SVEP1 in vascular development.
SVEP1 and human disease
Our understanding of SVEP1’s role in disease has been greatly influenced by its human genomic and proteomic associations. Genome-wide association studies (GWASs) have revealed several associations between SVEP1 and human traits and diseases. Aptamer-based proteomics have rapidly expanded these associations, with many of these studies identifying SVEP1 as the protein most strongly associated with the outcome of interest. Analyses of these data by MR has yielded additional causal associations. Table 1 outlines the SVEP1 disease and trait associations with the strongest supporting data (Table 1, Figure 2), and categories of SVEP1 disease associations are discussed in more detail below. A summary of their supporting data and proposed mechanisms is included; however, the mechanisms underlying the associations remain poorly understood.
Table 1.
SVEP1 human disease and trait associations
| Associated disease or trait | Associated allele(rs number) | Refs | |
|---|---|---|---|
| Genetic associations | Coronary artery disease | rs111245230 | [80] |
| Blood pressure | rs111245230 | [80] | |
| Type 2 diabetes | rs111245230 | [80] | |
| Primary congenital glaucoma | rs761025824 | [90] | |
| Open-angle glaucoma | rs61751937 | [10] | |
| Platelet reactivity | rs61751937 | [15] | |
| Red cell distribution width | rs189173017 | [85] | |
| Associated disease or trait | Direction of association | ||
| Protein associations | Coronary artery disease | Positive | [8] |
| Systolic blood pressure | Positive | [8] | |
| Type 2 diabetes | Positive | [8] | |
| Heart failure | Positive | [4] | |
| Cardiovascular risk | Positive | [13,99] | |
| Atrial fibrillation | Positive | [14] | |
| Pulmonary arterial hypertension | Positive | [12] | |
| Cirrhosis | Positive | [100] | |
| Cognitive decline | Positive | [92] | |
| Dementia | Positive | [5] | |
| Longevity | Undefined | [16] | |
| Age | Positive | [3] | |
| Gestational age | Positive | [45] | |
| Causal associations | Coronary artery disease | Positive | [6,7] |
| Hypertension | Positive | [6] | |
| Type 2 diabetes | Positive | [6,8] | |
| Mean platelet volume | Positive | [7] | |
| Platelet count | Negative | [7] | |
| Dementia | Positive | [5] | |
| Parental lifespan | Negative | [9] |
Figure 2.
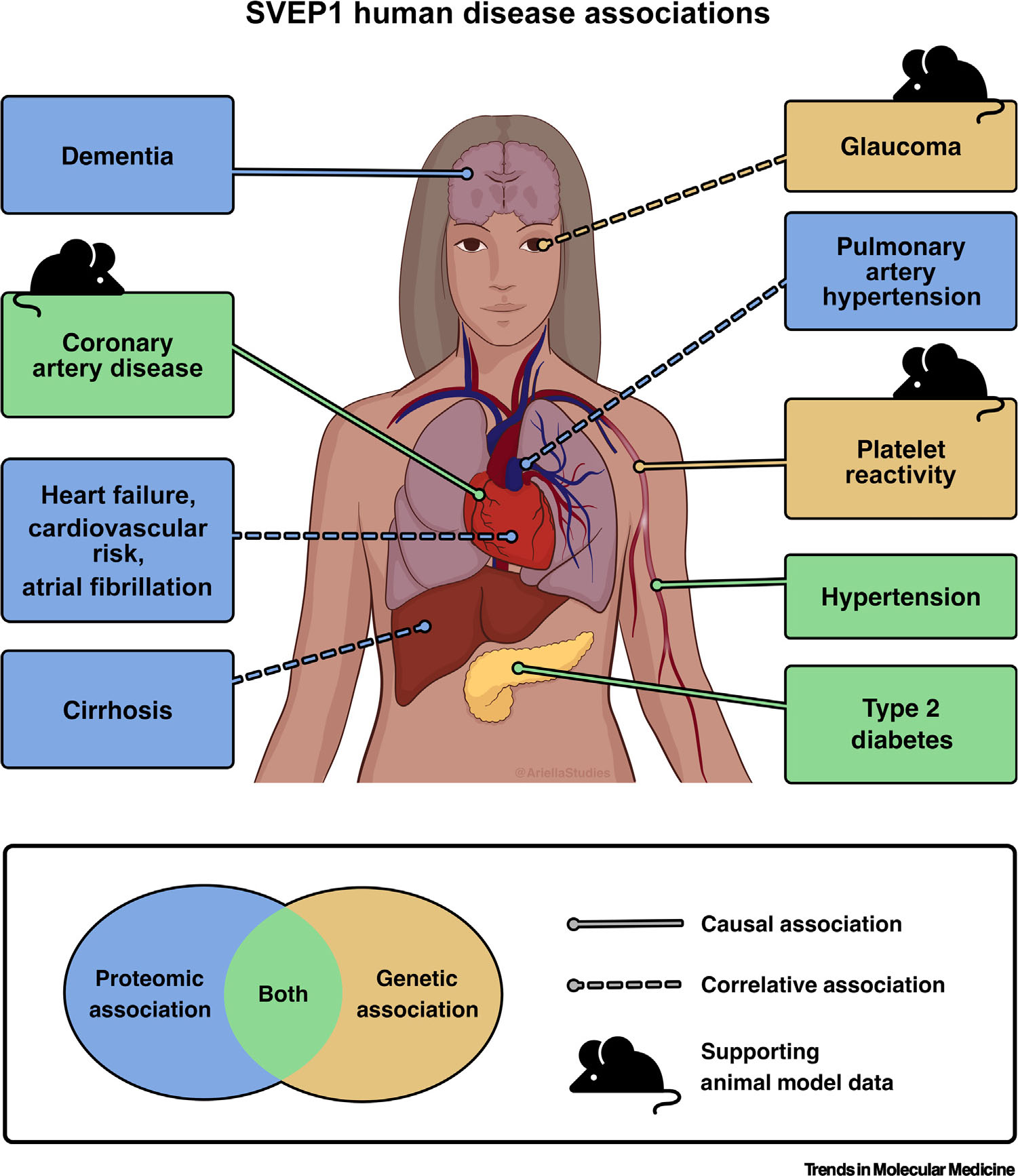
Sushi, von Willebrand factor type A, epidermal growth factor and pentraxin domain containing 1 (SVEP1) human disease associations. A list of the SVEP1 human disease and trait associations with the strongest supporting data. The traits and diseases with a causal association with SVEP1 are indicated with solid lines. Dashed lines represented correlative associations. Pointers indicate organs affected by the associated trait or disease, not necessarily the physiological location of the effects of SVEP1. Of note, many of the associated diseases are chronic conditions that relate to cardiometabolic or vascular dysfunction. See Table 1 for the associated references.
Cardiometabolic disease
In 2016, we reported a robust association between a coding variant in SVEP1 (rs111245230, encoding p.D2702G) and coronary artery disease (CAD) [80]. This variant also associated with blood pressure and type 2 diabetes, but not with lipids, suggesting it may influence vascular disease pathogenesis in a lipid-independent manner. Individuals with increased plasma levels of SVEP1 were subsequently found to be at greater risk for incident CAD and type 2 diabetes [8]. Increased plasma SVEP1 also associates with increased systolic blood pressure and poor survival after incident CAD [8]. MR analyses suggest SVEP1 is deleterious, since genetic variation which associate with elevated levels of plasma SVEP1 also positively associate with CAD [6,7], hypertension [6], and type 2 diabetes [6,8].
A recent study of patients with heart failure found that plasma SVEP1 was as strongly associated with heart-failure-associated hospitalization or cardiovascular death as N-terminal pro-B-type natriuretic peptide (NT-proBNP) [4]. Increased plasma SVEP1 was also shown to portend a poor prognosis in patients with pulmonary arterial hypertension [12]. A machine learning proteomic surrogate for cardiovascular outcomes identified SVEP1 as one of the top plasma proteins predictive of myocardial infarction, stroke, heart failure, or death [13].
Consistent with these observations, SVEP1-deficient mouse models were found to have lower plaque burden and complexity when compared to controls [6]. Winkler et al. reported the opposite phenotype [30] in a murine study confounded by differing proportions of male and female animals [81,82]. Integrin signaling [25], Notch signaling [25], and differential CXC motif chemokine ligand (CXCL)1 expression [30] have all been proposed as the molecular mechanisms behind these animal phenotypes; however, Itga9−/− mouse models fail to phenocopy the atherogenesis phenotypes of Svep1−/− mice [25] and changes in CXCL1 expression failed to replicate in other models of SVEP1 depletion [6,7]. Interaction with PEAR1 is an intriguing but untested hypothesis related to how SVEP1 influences atherosclerosis, since PEAR1 also appears to causally relate to CAD [7,56,57,83] through unclear mechanisms.
Additional cardiometabolic phenotypes have also been studied in mice with experimentally reduced SVEP1. Ex vivo aortic contraction studies from Svep1+/− mice suggest SVEP1 may regulate vascular contraction through integrins α9 and α4 [24]. Despite this finding, mice lacking SVEP1 post development do not appear to have a clear vascular reactivity phenotype when assessed by in vivo catheterization [7]. These mice similarly lack a glucose metabolism phenotype, as assessed by glucose and insulin tolerance tests, as well as indirect calorimetry [7]. Future studies are needed to clarify the role and mechanisms of SVEP1 in murine cardiometabolic disease, given the inconsistencies in vascular and atherosclerotic phenotypes reported for murine SVEP1-depletion models. The association between increased plasma SVEP1 and poor cardiovascular outcomes in humans is nonetheless stunning. Additional plasma proteomic studies are ongoing and are likely to reveal additional associations between SVEP1 cardiometabolic disease.
Platelet biology and hematopoiesis
Recent data points to an intriguing role of SVEP1 in platelet biology and hematopoiesis. A missense polymorphism (rs61751937 encoding SVEP1 p.R229G) strongly associates with platelet reactivity in response to ADP [15]; SVEP1 R229G, along with other SVEP1 variants, also associate with additional platelet traits [84], and MR suggests these SVEP1 trait associations are causal [7]. Mice lacking SVEP1 postdevelopment had similar platelet counts to controls, but their platelets had fewer platelet preactivation markers in response to ADP. Exogenous SVEP1 induced adhesion, activation, and agglutination of platelets ex vivo, supporting the role of SVEP1 in promoting platelet activation. Importantly, the activation of platelets by SVEP1 was shown to depend on PEAR1 [7].
Variation within the locus containing SVEP1 is associated with numerous additional hematological parameters, including red blood cell traits [85] and white blood cell counts [84]. Mice lacking SVEP1 had increased lymphocytes and white blood cell counts, in addition to increased red blood cell counts [7]. These findings overlap with the hematological associations of SVEP1 in humans [84] and suggest the protein may regulate hematopoiesis, as has been described in animal models [86] and ex vivo [87].
These findings support the human disease associations of SVEP1 and implicate the protein in platelet biology and hematopoiesis. SVEP1 also associates with death from septic shock [88], perhaps reflecting its association with vascular and hematological traits. The mechanisms by which SVEP1 impacts hematopoiesis remain unclear; however, integrin α9β1 [89] and PEAR1 [59] are also thought to regulate hematopoiesis. How SVEP1 influences platelet biology and hematopoiesis is an important area of future study that may relate to the other SVEP1 disease associations.
Glaucoma
Glaucoma is also associated with variation within the SVEP1 locus. The same coding variant that associates with platelet reactivity (p.R229G) was found to associate with open angle glaucoma in a GWAS of adults [10]. Glycine at 229 correlates with increased plasma SVEP1 levels (through unclear mechanisms), suggesting elevated SVEP1 may contribute to open angle glaucoma. Another missense allele within SVEP1 (rs761025824 encoding p.R997C) was found in four of five affected members of a family of patients with primary congenital glaucoma and who also harbored a mutation in TEK (the gene that encodes TIE2 which is a receptor for ANG1) [90]. Exposure of human umbilical vein endothelial cells to SVEP1 R997C resulted in less TEK expression than wild-type SVEP1, suggesting that SVEP1 modulates TEK-related primary congenital glaucoma [90]. Deletion of Svep1 in neural crest cells of mice resulted in defects within Schlemm’s canal and increased intraocular pressure, suggesting insufficient SVEP1 may lead to primary congenital glaucoma [11].
Future studies are necessary to determine if chronic excess SVEP1 is also sufficient to disrupt Schlemm’s canal and promote glaucoma, as one might hypothesize from the human GWAS. It is unclear how SVEP1 governs the development of Schlemm’s canal; however, its potential interaction with the ANG–TIE signaling pathway [26,60,90] is a leading hypothesis. Both Pear1 and Itga9 are also expressed within Schlemm’s canal and abundant PEAR1 is observed in trabecular meshwork cell–ECM adhesion complexes [91], raising questions about whether these receptors also interact with SVEP1 to govern development or homeostasis of Schlemm’s canal.
Aging and cognitive function
The disease associations of SVEP1 are broad, but themes related to the pathophysiology of these diseases provide clues to the function of the protein. Relation to aging and chronicity are characteristics of many of its associated diseases. Additional studies have identified genetic and plasma protein associations between SVEP1 and measures of aging or cognitive decline [13,16,92]. One such analysis found SVEP1 to be the plasma protein with the strongest causal association with dementia among 4877 proteins analyzed [5]. A causal association between plasma SVEP1 and decreased parental lifespan has also been reported [9].
Although increased SVEP1 appears to constitute a health risk during aging, it is also necessary for embryonic viability and vascular development (Figure 3). This biological phenomenon, termed antagonistic pleiotropy, has long been described [93]. According to this hypothesis, selective pressure across evolution heavily favors alleles necessary for survival up to the peak reproductive capacity of an organism, even if such alleles have a negative effect on survivorship after reproductive capacity wanes [94,95]. The effects of SVEP1 on human biology appear to closely follow this paradigm, given its causal association with age-related disease.
Figure 3.
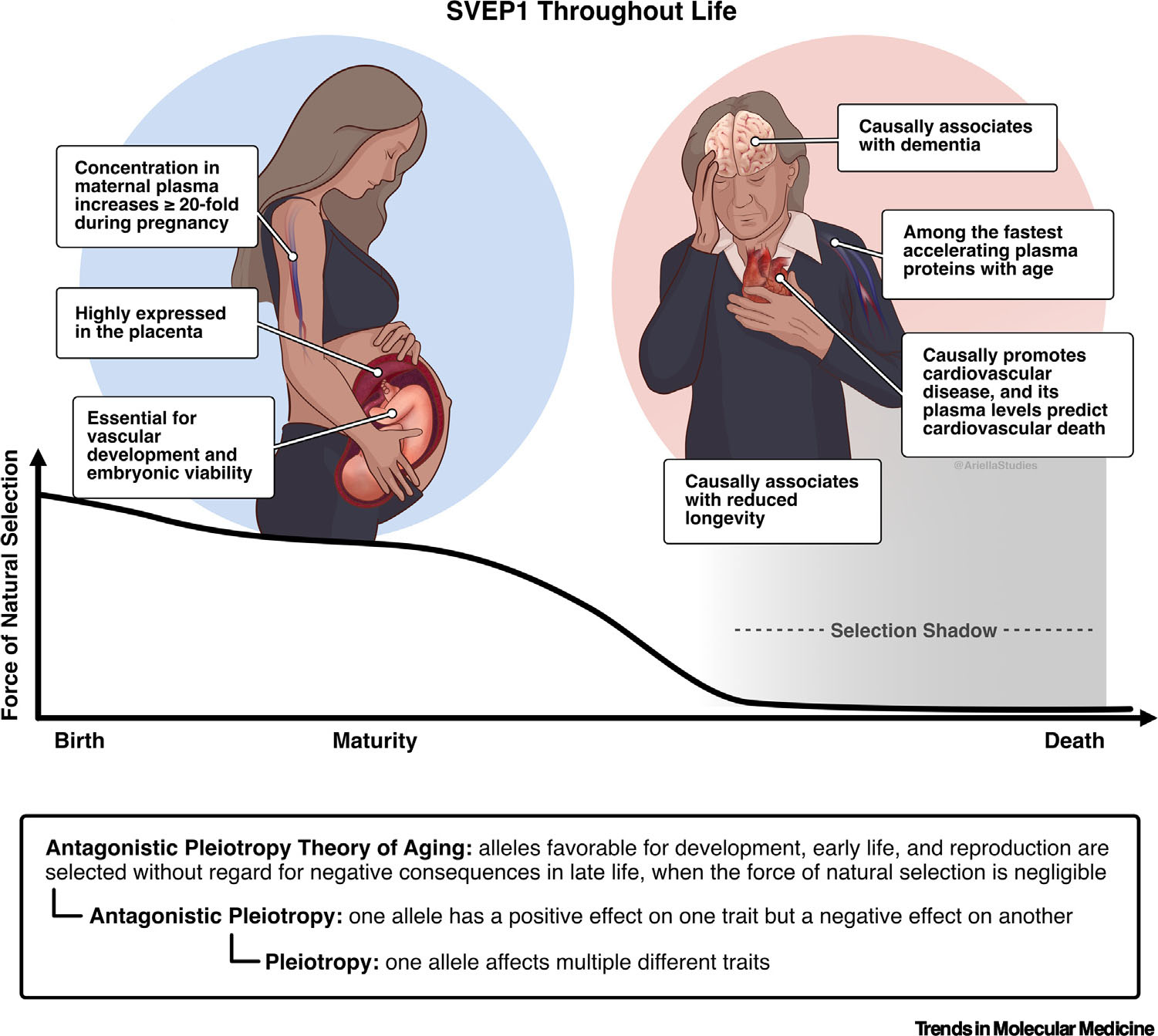
Sushi, von Willebrand factor type A, epidermal growth factor and pentraxin domain containing 1 (SVEP1) throughout life. SVEP1 and the antagonistic pleiotropy theory of aging. SVEP1 is essential for embryonic viability and appears to promote age-related disease later in life. This can be explained by the theory of antagonistic pleiotropy, which states that alleles necessary for development and reproduction are favored without regard for their effects in later life. The loss of selective pressure on alleles during the period proceeding max reproductive capability is referred to as the selection shadow.
The ECM is thought to play a critical role in aging [96], but the specific effects of SVEP1 remain unclear. The robust activation of mTOR signaling in endothelial and smooth muscle cells [7] provides a reasonable hypothesis, since mTOR is among the most recognized regulators of longevity [72,74]. Further addressing this intriguing question may shed light on the biology underlying the disease associations of SVEP1 and will be an important direction for future investigation.
Concluding remarks
SVEP1 is a complex and intriguing ECM protein. Recent data implicate the protein in vascular development, chronic disease, and aging. Despite this, there is no consensus on how SVEP1 regulates cells, tissues, or disease. Future studies are necessary to characterize the location of SVEP1 within tissue and its molecular interactions. Elucidating these mechanisms will broaden our understanding of vascular development and disease pathogenesis. Rigorous antibody validation and the use of more physiologic cellular assays are important steps to accomplishing these goals.
Measurements of plasma SVEP1 concentration may have diagnostic and prognostic value across many fields in medicine. The development of a targeted assay to measure plasma SVEP1 concentration is a critical next step to evaluating its clinical utility, since the disease associations of SVEP1 were established using untargeted proteomic techniques. Such an assay can also address important questions related to the protein’s apparent ingress into plasma, stability, and other variables that influence its concentration (see Outstanding questions).
Outstanding questions.
What is the precise location of SVEP1 within the ECM? How does SVEP1 enter circulation and in what form does it circulate?
Which proteins interact with SVEP1 in vivo, and how do these interactions contribute to its role in development and disease?
What is the diagnostic or prognostic value of measuring circulating SVEP1? Does this approximate gestational age, cardiovascular risk, platelet reactivity, risk of developing glaucoma, and/or biological age?
Is it possible to safely target SVEP1 for the treatment or prevention of human chronic disease? What are the most effective strategies to modulate its activity?
Variation of plasma SVEP1 concentration is, in part, genetically determined, suggesting a therapeutic window exists to safely reduce SVEP1 activity (see Clinician’s corner). Mouse models of chronic SVEP1 depletion and the theory of antagonistic pleiotropy also hint that functional levels of plasma SVEP1 could be safely reduced in adults. These characteristics, combined with its existence within the extracellular space, make it a priority candidate for intervention. The development of therapies related to SVEP1 require a deeper understanding of the molecular mechanisms by which it contributes to disease, emphasizing the need for continued research on this fascinating protein.
Clinician’s corner.
SVEP1 is an ECM protein with important roles in vascular development, chronic disease, and aging.
Measuring plasma SVEP1 concentration may have significant diagnostic and prognostic value across various fields of medicine.
SVEP1 appears to associate as strongly as NT-proBNP with heart failure hospitalization or cardiovascular death in patients with heart failure.
Investigating the molecular mechanisms of SVEP1 could lead to new therapeutic approaches to treat or prevent disease.
Highlights.
Human genomic and proteomic data implicate Sushi, von Willebrand factor type A, epidermal growth factor, and pentraxin domain containing 1 (SVEP1) in the pathogenesis of human chronic disease, including cardiovascular disease, glaucoma, and dementia.
SVEP1 is critical for vascular development and appears to promote aging, consistent with the antagonistic pleiotropy theory of aging.
Many cell types adhere and/or proliferate in response to SVEP1, but the mechanisms underlying its role in development and disease are unclear.
SVEP1 is contained within the extracellular matrix and circulates in plasma, making it a priority candidate for disease diagnostics, prognostics, and therapeutics.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (NIH) to JSE (T32GM007200, T32HL134635, and F30HL152521), ACR (T32AR060791 and the Musculoskeletal Research Center Fund), NOS (R01HL159171, R01HL131961, UM1HG008853, P01HL151328), by the Longer Life Foundation: A RGA/Washington University Collaboration (LLF 2021-007 to NOS), and by the Foundation for Barnes-Jewish Hospital (NOS).
Glossary
- AKT
serine/threonine kinase that plays a crucial role in cell survival, growth, and metabolism. It is involved in various cellular processes, including cell proliferation, apoptosis, and protein synthesis.
- ANG–TIE
The signaling pathway involving the interaction between ANGs and TIE receptors. This pathway plays a significant role in angiogenesis, the formation of new vessels.
- Antagonistic pleiotropy
description of a situation where a single gene or genetic variant has multiple effects on different traits or functions. These effects can be beneficial for certain traits at one stage of life but detrimental to other traits at a different stage, leading to trade-offs in evolutionary fitness.
- Aptamer-based proteomics
A technique that uses DNA or RNA oligomers called aptamers that bind to known target proteins. This method enables the identification and quantification of proteins in complex biological samples at a greater scale than traditional methods.
- Cardiometabolic disease
cluster of common chronic medical conditions which share interrelated risk factors such as high blood pressure, elevated blood sugar, and elevated lipids. This term encompasses cardiovascular conditions such as heart disease and stroke along with metabolic disorders such as type 2 diabetes and obesity.
- Extracellular matrix (ECM)
complex network of proteins and carbohydrates that provides structural support and biochemical cues to cells in tissues and organs. It influences various cellular processes, including cell adhesion, migration, and differentiation.
- Genome-wide association studies (GWASs)
large studies that investigate the association between genetic variants across the genome and the prevalence of specific traits or diseases. GWASs help identify genetic markers associated with various conditions and provide insights into the genetic basis of complex traits.
- Integrins
family of cell surface receptors that mediate cell–cell and cell–ECM interactions. They play a crucial role in cell adhesion, migration, signaling, and tissue organization.
- Label-free binding assay
technique used to measure the interaction between molecules without the need for labeling or modification of the molecules. It enables the quantitative analysis of binding affinities and kinetics between proteins, small molecules, or other biomolecules.
- Lymphangiogenesis
process of formation and remodeling of lymphatic vessels. It involves the growth and branching of lymphatic endothelial cells and plays a critical role in immune responses and tissue fluid homeostasis.
- Mendelian Randomization
statistical method that uses genetic variants as instrumental variables to assess causal relationships between exposures and disease outcomes. It leverages the principles of Mendelian inheritance to infer causal effects in observational studies.
- mTOR
mechanistic target of rapamycin (mTOR) is a protein kinase that regulates cell growth, metabolism, and survival in response to various environmental signals. It integrates inputs from growth factors, nutrients, and cellular energy status to control protein synthesis, aging, and other important processes.
- Platelet endothelial aggregation receptor 1 (PEAR1)
receptor tyrosine kinase-like protein that strongly associates with platelet reactivity in humans and is activated by SVEP1.
- Schlemm’s canal
lymphatic-like vessel located near the anterior chamber angle of the eye. It plays a crucial role in regulating intraocular pressure by maintaining the proper drainage of aqueous humor.
Footnotes
Declaration of interests
NOS has received investigator-initiated research funds from Regeneron Pharmaceuticals unrelated to the content of this study. JSE, IHJ, AA, and NOS are co-inventors on US Patent App. 17/978,128 assigned to Washington University focused on SVEP1 and PEAR1. The other authors have declared no conflicts.
References
- 1.Gilges D et al. (2000) Polydom: a secreted protein with pentraxin, complement control protein, epidermal growth factor and von Willebrand factor A domains. Biochem. J. 352, 49–59 [PMC free article] [PubMed] [Google Scholar]
- 2.Sato-Nishiuchi R et al. (2012) Polydom/SVEP1 is a ligand for integrin α9β1. J. Biol. Chem. 287, 25615–25630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehallier B et al. (2019) Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 25, 1843–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L et al. (2022) Aptamer proteomics for biomarker discovery in heart failure with reduced ejection fraction. Circulation 146, 1411–1414 [DOI] [PubMed] [Google Scholar]
- 5.Walker KA et al. (2021) Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat. Aging 1, 473–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung I-H et al. (2021) SVEP1 is a human coronary artery disease locus that promotes atherosclerosis. Sci. Transl. Med. 13, eabe0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elenbaas JS et al. (2023) SVEP1 is an endogenous ligand for the orphan receptor PEAR1. Nat. Commun. 14, 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emilsson V et al. (2022) Coding and regulatory variants are associated with serum protein levels and disease. Nat. Commun. 13, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrot N et al. (2021) A trans-omic Mendelian randomization study of parental lifespan uncovers novel aging biology and therapeutic candidates for chronic diseases. Aging Cell 20, e13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharahkhani P et al. (2021) Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 12, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson BR et al. (2021) Cellular crosstalk regulates the aqueous humor outflow pathway and provides new targets for glaucoma therapies. Nat. Commun. 12, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes CJ et al. (2022) Using the plasma proteome for risk stratifying patients with pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 205, 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams SA et al. (2022) A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci. Transl. Med. 14, eabj9625. [DOI] [PubMed] [Google Scholar]
- 14.Norby FL et al. (2021) Proteomics and risk of atrial fibrillation in older adults (from the Atherosclerosis Risk in Communities [ARIC] Study). Am. J. Cardiol. 161, 42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keramati AR et al. (2021) Genome sequencing unveils a regulatory landscape of platelet reactivity. Nat. Commun. 12, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebastiani P et al. (2021) Protein signatures of centenarians and their offspring suggest centenarians age slower than other humans. Aging Cell 20, e13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yashin AI et al. (2010) Joint influence of small-effect genetic variants on human longevity. Aging 2, 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pernemalm M et al. (2019) In-depth human plasma proteome analysis captures tissue proteins and transfer of protein variants across the placenta. Elife 8, e41608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun BB et al. (2018) Genomic atlas of the human plasma proteome. Nature 558, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y et al. (2020) Glyco-CPLL: an integrated method for in-depth and comprehensive N-glycoproteome profiling of human plasma. J. Proteome Res. 19, 655–666 [DOI] [PubMed] [Google Scholar]
- 21.Ye B et al. (2022) Differential proteomic analysis of plasma-derived exosomes as diagnostic biomarkers for chronic HBV-related liver disease. Sci. Rep. 12, 14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navajas R et al. (2022) Quantitative proteomic analysis of serum-purified exosomes identifies putative pre-eclampsia-associated biomarkers. Clin. Proteomics 19, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan KH et al. (2014) Plasma biomarker discovery in pre-eclampsia using a novel differential isolation technology for circulating extracellular vesicles. Am. J. Obstet. Gynecol. 211 380.e1–13 [DOI] [PubMed] [Google Scholar]
- 24.Morris GE et al. (2022) The integrin ligand SVEP1 regulates GPCR-mediated vasoconstriction via integrins α9β1 and α4β1. Br. J. Pharmacol. 179, 4958–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung I-H et al. (2022) Vascular smooth muscle- and myeloid cell-derived integrin α9β1 does not directly mediate the development of atherosclerosis in mice. Atherosclerosis 360, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morooka N et al. (2017) Polydom is an extracellular matrix protein involved in lymphatic vessel remodeling. Circ. Res. 120, 1276–1288 [DOI] [PubMed] [Google Scholar]
- 27.Kauskot A et al. (2012) A novel mechanism of sustained platelet αIIbβ3 activation via PEAR1. Blood 119, 4056–4065 [DOI] [PubMed] [Google Scholar]
- 28.Karpanen T et al. (2017) An evolutionarily conserved role for Polydom/Svep1 during lymphatic vessel formation. Circ. Res. 120, 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coxam B et al. (2022) Svep1 stabilises developmental vascular anastomosis in reduced flow conditions. Development 149, dev199858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler MJ et al. (2020) Functional investigation of the coronary artery disease gene SVEP1. Basic Res. Cardiol. 115, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wouters MA et al. (2005) Evolution of distinct EGF domains with specific functions. Protein Sci. 14, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittaker CA and Hynes RO (2002) Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum M et al. (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuckwell D (1999) Evolution of von Willebrand factor A (VWA) domains. Biochem. Soc. Trans. 27, 835–840 [DOI] [PubMed] [Google Scholar]
- 35.Norman DG et al. (1991) Three-dimensional structure of a complement control protein module in solution. J. Mol. Biol. 219, 717–725 [DOI] [PubMed] [Google Scholar]
- 36.Callebaut I et al. (2000) HYR, an extracellular module involved in cellular adhesion and related to the immunoglobulin-like fold. Protein Sci. 9, 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigrist CJA et al. (2013) New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Clos TW (2013) Pentraxins: structure, function, and role in inflammation. ISRN Inflamm. 2013, 379040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halim A et al. (2012) Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell. Proteomics 11 M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The UniProt C (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benrick A et al. (2017) Adiponectin protects against development of metabolic disturbances in a PCOS mouse model. Proc. Natl. Acad. Sci. 114, E7187–E7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shur I et al. (2007) SVEP1 expression is regulated in estrogen-dependent manner. J. Cell. Physiol. 210, 732–739 [DOI] [PubMed] [Google Scholar]
- 43.Tikhonova AN et al. (2019) The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong DL et al. (2017) The core transcriptome of mammalian placentas and the divergence of expression with placental shape. Placenta 57, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarca AL et al. (2022) Human plasma proteome during normal pregnancy. J. Proteome Res. 21, 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shur I et al. (2006) Molecular and cellular characterization of SEL-OB/SVEP1 in osteogenic cells in vivo and in vitro. J. Cell. Physiol. 206, 420–427 [DOI] [PubMed] [Google Scholar]
- 47.Glait-Santar C et al. (2012) Expression pattern of SVEP1 alternatively-spliced forms. Gene 505, 137–145 [DOI] [PubMed] [Google Scholar]
- 48.Shefer G and Benayahu D (2010) SVEP1 is a novel marker of activated pre-determined skeletal muscle satellite cells. Stem Cell Rev. Rep. 6, 42–49 [DOI] [PubMed] [Google Scholar]
- 49.Wirka RC et al. (2019) Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlen M et al. (2016) A proposal for validation of antibodies. Nat. Methods 13, 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takada Y et al. (2007) The integrins. Genome Biol. 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmer EL et al. (1993) Sequence and tissue distribution of the integrin alpha 9 subunit, a novel partner of beta 1 that is widely distributed in epithelia and muscle. J. Cell Biol. 123, 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bazigou E et al. (2009) Integrin-α9 is required for fibronectin matrix assembly during lymphatic valve morphogenesis. Dev. Cell 17, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas DS et al. (2009) The integrin α9β1 on hematopoietic stem and progenitor cells: involvement in cell adhesion, proliferation and differentiation. Haematologica 94, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson AD et al. (2010) Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 42, 608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ansari N et al. (2021) PEAR1 polymorphisms as a prognostic factor in hemostasis and cardiovascular diseases. J. Thromb. Thrombolysis 51, 89–95 [DOI] [PubMed] [Google Scholar]
- 57.Lewis JP et al. (2013) Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ. Cardiovasc. Genet. 6, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandenbriele C et al. (2015) Platelet endothelial aggregation receptor-1: a novel modifier of neoangiogenesis. Cardiovasc. Res. 108, 124–138 [DOI] [PubMed] [Google Scholar]
- 59.Zhang S et al. (2023) PEAR1 is a potential regulator of early hematopoiesis of human pluripotent stem cells. J. Cell. Physiol. 238, 179–194 [DOI] [PubMed] [Google Scholar]
- 60.Hußmann M et al. (2023) Svep1 is a binding ligand of Tie1 and affects specific aspects of facial lymphatic development in a Vegfc-independent manner. eLife 12, e82969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato-Nishiuchi R et al. (2023) Polydom/SVEP1 binds to Tie1 and promotes migration of lymphatic endothelial cells. J. Cell Biol. 222, e202208047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schultz GS and Wysocki A (2009) Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 17, 153–162 [DOI] [PubMed] [Google Scholar]
- 63.Suri C et al. (1996) Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell 87, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 64.Woo KV et al. (2011) Tie1 attenuation reduces murine atherosclerosis in a dose-dependent and shear stress–specific manner. J. Clin. Invest. 121, 1624–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrews SL et al. (2023) SVEP1 influences monocyte to macrophage differentiation via integrin α4β1/α9β1 and Rho/Rac signalling. Biochimica et Biophysica Acta (BBA) - Molecular. Cell Res. 1870, 119479. [DOI] [PubMed] [Google Scholar]
- 66.Ojha H et al. (2019) Spatially conserved motifs in complement control protein domains determine functionality in regulators of complement activation-family proteins. Commun. Biol. 2, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lengsfeld AM et al. (1974) Interaction of phalloidin with actin. Proc. Natl. Acad. Sci. 71, 2803–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woulfe DS (2010) Akt signaling in platelets and thrombosis. Expert. Rev. Hematol. 3, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X et al. (2018) The PI3K/AKT pathway in obesity and type 2 diabetes. J. Biol. Sci. 14, 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Y et al. (2021) Role of PI3K in the progression and regression of atherosclerosis. Front. Pharmacol. 12, 632378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aslan JE et al. (2011) S6K1 and mTOR regulate Rac1-driven platelet activation and aggregation. Blood 118, 3129–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saxton RA and Sabatini DM (2017) mTOR signaling ingrowth, metabolism, and disease. Cell 168, 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sciarretta S et al. (2018) New insights into the role of mTOR signaling in the cardiovascular system. Circ. Res. 122, 489–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weichhart T (2018) mTOR as regulator of lifespan, aging, and mTOR as regulator of lifespan, aging, and cellular senescence: a mini-review. Gerontology 64, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar V et al. (2019) Therapeutic suppression of mTOR (mammalian target of rapamycin) signaling prevents and reverses salt-induced hypertension and kidney injury in Dahl salt-sensitive rats. Hypertension 73, 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huber S et al. (2007) Inhibition of the mammalian target of rapamycin impedes lymphangiogenesis. Kidney Int. 71, 771–777 [DOI] [PubMed] [Google Scholar]
- 77.Chen Y et al. (2017) Receptor-mediated cell mechanosensing. Mol. Biol. Cell 28, 3134–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karczewski KJ et al. (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang XZ et al. (2000) Fatal bilateral chylothorax in mice lacking the integrin alpha9beta1. Mol. Cell. Biol. 20, 5208–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stitziel NO et al. (2016) Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N. Engl. J. Med. 374, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maeda N et al. (2007) Anatomical differences and atherosclerosis in apolipoprotein E-deficient mice with 129/SvEv and C57BL/6 genetic backgrounds. Atherosclerosis 195, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Man JJ et al. (2020) Sex as a biological variable in atherosclerosis. Circ. Res. 126, 1297–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nie XY et al. (2018) Genetic mutations in PEAR1 associated with cardiovascular outcomes in Chinese patients with acute coronary syndrome. Thromb. Res. 163, 77–82 [DOI] [PubMed] [Google Scholar]
- 84.Chen MH et al. (2020) Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213.e1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuchenbaecker K et al. (2022) Insights into the genetic architecture of haematological traits from deep phenotyping and whole-genome sequencing for two Mediterranean isolated populations. Sci. Rep. 12, 1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yvernogeau L et al. (2020) Multispecies RNA tomography reveals regulators of hematopoietic stem cell birth in the embryonic aorta. Blood 136, 831–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tran V and O’Neill HC (2022) Role of SVEP1 in stroma-dependent hematopoiesis in vitro. Front. Cell Dev. Biol. 9, 760480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakada T. a. et al. (2015) Identification of a nonsynonymous polymorphism in the SVEP1 gene associated with altered clinical outcomes in septic shock. Crit. Care Med. 43, 101–108 [DOI] [PubMed] [Google Scholar]
- 89.Schreiber TD et al. (2009) The integrin alpha9beta1 on hematopoietic stem and progenitor cells: involvement in cell adhesion, proliferation and differentiation. Haematologica 94, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young TL et al. (2020) SVEP1 as a genetic modifier of TEK-related primary congenital glaucoma. Invest. Ophthalmol. Vis. Sci. 61, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maddala R and Rao PV (2020) Global phosphotyrosinylated protein profile of cell-matrix adhesion complexes of trabecular meshwork cells. Am. J. Phys. Cell Phys. 319, C288–C299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindbohm JV et al. (2022) Plasma proteins, cognitive decline, and 20-year risk of dementia in the Whitehall II and Atherosclerosis Risk in Communities studies. Alzheimers Dement. 18, 612–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams GC (1957) Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 [Google Scholar]
- 94.Kirkwood TBL and Austad SN (2000) Why do we age? Nature 408, 233–238 [DOI] [PubMed] [Google Scholar]
- 95.Statzer C et al. (2023) Extracellular matrix dynamics as an emerging yet understudied hallmark of aging and longevity. Aging Dis. 14, 670–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park JYC et al. (2023) Strategic outline of interventions targeting extracellular matrix for promoting healthy longevity. Am. J. Phys. Cell Phys. 325, C90–C128 [DOI] [PubMed] [Google Scholar]
- 97.Letunic I et al. (2020) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Paysan-Lafosse T et al. (2023) InterPro in 2022. Nucleic Acids Res. 51, D418–D427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deo R et al. (2023) Proteomic cardiovascular risk assessment in chronic kidney disease. Eur. Heart J. 44, 2095–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sveinbjornsson G et al. (2022) Multiomics study of nonalcoholic fatty liver disease. Nat. Genet. 54, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]


