Abstract
Purpose:
Immune checkpoint blockade (ICB) demonstrates durable clinical benefits in a minority of patients with renal cell carcinoma (RCC). We aimed to identify the molecular features that determine the response and develop approaches to enhance the response.
Experimental Design:
We investigated the effects of SET domain-containing protein 2 (SETD2) loss on the DNA damage response pathway, the cytosolic DNA sensing pathway, the tumor immune microenvironment, and the response to Ataxia telangiectasia and rad3 related (ATR) and checkpoint inhibition in RCC.
Results:
ATR inhibition activated the cyclic GMP–AMP synthase (cGAS)-Interferon regulatory factor 3 (IRF3)-dependent cytosolic DNA-sensing pathway, resulting in the concurrent expression of inflammatory cytokines and immune checkpoints. Among the common RCC genotypes, SETD2 loss is associated with preferential ATR activation and sensitizes cells to ATR inhibition. SETD2 knockdown promoted the cytosolic DNA sensing pathway in response to ATR inhibition. Treatment with the ATR inhibitor VE822 concurrently upregulated immune cell infiltration and immune checkpoint expression in Setd2 knockdown Renca tumors, providing a rationale for ATR inhibition plus ICB combination therapy. Setd2 deficient Renca tumors demonstrated greater vulnerability to ICB monotherapy or combination therapy with VE822 than Setd2 proficient tumors. Moreover, SETD2 mutations were associated with a higher response rate and prolonged overall survival in ICB-treated RCC patients but not in non-ICB-treated RCC patients.
Conclusions:
SETD2 loss and ATR inhibition synergize to promote cGAS signaling and enhance immune cell infiltration, providing a mechanistic rationale for the combination of ATR and checkpoint inhibition in RCC patients with SETD2 mutations.
Keywords: RCC, SETD2, ATR inhibitor, cGAS, Immunotherapy
Introduction
Renal cell carcinoma (RCC) is one of the ten most prevalent cancers. There will be approximately 81,800 new cases of RCC in the United States in 2023, and an estimated 14,890 patients with RCC in the United States will die of RCC (1). Immune checkpoint blockade (ICB) has revolutionized cancer treatment by providing survival benefits. However, only a minority of patients show complete response (CR) to ICB (2,3). There is an urgent clinical need to identify the genetic features that determine ICB response and to develop a mechanism-based approach to enhance the therapeutic efficacy of ICB in patients with RCC.
The tumor immune microenvironment is known to be a determinant of ICB response and can be influenced by genetic alterations and therapeutic interventions (4). Clear cell RCC (ccRCC), the most common histological subtype of RCC, is characterized by the inactivation of the von Hippel Lindau (VHL) tumor suppressor gene. Loss of other genetic components, such as Polybromo-1 (PBRM1), SET domain-containing 2 (SETD2), and BRCA-associated protein 1 (BAP1), facilitates kidney tumorigenesis driven by VHL deficiency (5). Our group and Braun et al. reported that PBRM1 loss is associated with an immune desert phenotype in RCC (6,7), and our murine Renca model revealed that Pbrm1 loss confers resistance to immunotherapy (6). Bap1 mutations are associated with a more immunogenic phenotype (8). We also reported that antiangiogenic therapy with sunitinib and bevacizumab induced an immunosuppressive tumor microenvironment associated with increased immune cell infiltration and upregulated CD274 (PD-L1) expression (9). Several pivotal phase 3 clinical trials further reported that anti-PD-1 antibody (pembrolizumab, nivolumab) plus antiangiogenic therapy (axitinib, cabozantinib, or nivolumab) led to increased overall survival (OS), progression-free survival (PFS), and objective response rates (ORR) compared to sunitinib monotherapy (10-12). A recent study showed that sitravatinib, a tyrosine kinase inhibitor, reduced immunosuppressive myeloid cells in the tumor microenvironment of patients with advanced RCC, providing a rationale for combination therapy with ICB (13).
Targeting the S-phase DNA damage repair (S-DDR) network has been reported to convert a non-immunogenic tumor microenvironment into an immunogenic tumor microenvironment by increasing immune cell infiltration, thus improving the immunotherapy response (14,15). The inhibition of Poly(ADP-Ribose) polymerase (PARP), Ataxia telangiectasia mutated (ATM)-Checkpoint kinase 2 (CHK2), or Ataxia telangiectasia and rad3 related (ATR)-Checkpoint kinase 1 (CHK1) axes promotes the accumulation of unrepaired or unprocessed deoxyribose nucleic acid (DNA) fragments, activates the cyclic GMP–AMP synthase (cGAS)-mediated cytosolic DNA-sensing pathway, and initiates the expression of immune cell-attracting factors (e.g. CCL5, CXCL10, and type I interferon (IFN) (14,15). ATM loss confers greater sensitivity to ATR inhibition in prostate cancer cells, immunocompromised pancreatic cancer models, and patients with advanced solid tumors (16-18). However, ATM mutations are rarely identified in RCC (19); thus, identifying the genetic features that influence ATM activity might provide therapeutic strategies with ATR inhibitors. Several frequently mutated genes in ccRCC are known to influence DNA damage and repair. VHL loss leads to the induction of DNA replication stress and damage accumulation, whereas PBRM1 loss rescues DNA replication stress in VHL-deficient cells (20). Loss of SETD2 impairs DNA damage repair (21,22). Thus, we hypothesized that these fundamental genetic mutations in ccRCC confer therapeutic vulnerabilities to pharmacological inhibitors targeting DNA damage repair pathways, which may further potentiate the response to immunotherapy.
In this study, we utilized RCC cell lines, controlled preclinical models, and clinical cohort validation to demonstrate SETD2 loss is associated with upregulated ATR-CHK1 activity and activation of the cytosolic DNA-sensing pathway. Setd2 deficient murine Renca tumors in mice treated with an ATR inhibitor demonstrated concurrent high immune cell infiltration and immune checkpoint expression, and upregulated responsiveness to ICB. SETD2 mutations were associated with prolonged OS in ICB-treated RCC patients but not in non-ICB-treated RCC patients. This study provides molecular guidance for the development of personalized combination therapy regimens for patients with RCC with SETD2 mutations.
Materials and Methods
Antibodies and chemical reagents
cGAS antibody (D1D3G, # 15102, RRID:AB_2732795), PBRM1 antibody (D3F7O, #91894, RRID:AB_2800173), IRF-3 antibody (D83B9, #4302, RRID:AB_1904036), phospho-Histone H2A (Ser139, 20E3, #9718, RRID:AB_2118009), phospho-ATR antibody (Ser428, #2853, RRID:AB_2290281), cleaved Caspase-3 antibody (Asp175, 5A1E, #9664, RRID:AB_2070042), CHK1 antibody (2G1D5, #2360, RRID:AB_2080320), phospho-CHK1 antibody (Ser345, 133D3, #2348, RRID:AB_331212), phospho-CHK1 (Ser296, D3O9F, #90178,RRID:AB_2800153), CD11C antibody (D1V9Y, #97585, RRID:AB_2800282), CD4 antibody (D7D2Z, 25229, RRID:AB_2798898), CD8 antibody (D4W2Z, 98941, RRID:AB_2756376), PD-1 (D7D5W, 84651, RRID:AB_2800041), PD-L1 antibody (E1L3N; 13684, RRID:AB_2687655), and PD-L1 antibody (D5V3B, 64988, RRID:AB_2799672), Rad50 antibody (3427, RRID:AB_2176936) and phospho-Rad50 antibody (Ser635, 14223, RRID:AB_2798430) were from Cell Signaling Technology. SETD2 antibody (HPA042451, RRID:AB_10806239) was purchased from Sigma-Aldrich. Phospho-IRF3 (S386, EPR2346, ab76493, RRID:AB_1523836) and phospho-ATM (S1981, EP1890Y, ab81292 , RRID:AB_1640207) were purchased from Abcam. The GAPDH antibody (6C5, sc32233, RRID:AB_627679) was purchased from Santa Cruz Biotechnology. VE822 (S7102) and Prexasertib HCl (LY2606368, S7178), were purchased from Selleck Chemicals.
Cell culture and establishment of knockdown cell lines
RCC cell lines (786O, RCC4, and Renca) were obtained from ATCC and were validated by short tandem repeat (STR) DNA fingerprinting using the Promega 16 High Sensitivity STR Kit (catalog # DC2100). Human RCC cell lines and Renca cells were maintained in DMEM containing 10% fetal bovine serum at 37 °C with 5% CO2 in a humidified incubator. 786-O (RRID:CVCL_1051), RCC4 (RRID:CVCL_0498), or Renca (RRID:CVCL_2174) cells infected with lentivirus expressing shRNA against SETD2, cGAS, or IRF3 were selected in medium containing 2 μg/ml puromycin. Lentiviral particles expressing control shRNA (SHC002V), human cGAS shRNA (TRCN0000128706 and TRCN0000148694), human IRF3 shRNA (TRCN0000005919 and TRCN0000352624), or mouse Setd2 shRNA (TRCN0000238434) were purchased from Sigma-Aldrich. Human control shRNA (V3LHS_318943) and SETD2 shRNA (V2LHS_53398 and V2LHS_53401) were purchased from Dharmacon. Cell lines were verified as mycoplasma negative by regular testing with the Mycoplasma PCR Detection Kit MycoAlert Kit (BioVision).
RNA isolation and real-time PCR
RNA isolation and real-time PCR were performed as previously described (6). Total RNAs were isolated and purified using the RNeasy Mini Kit (Qiagen, 74106) and converted to cDNA using iScript™ Reverse Transcription Supermix (Bio-Rad, 1708841) mRNA expression was measured using a real-time PCR detection system (Applied Biosystems ViiA 7) in 96-well or 384-well optical plates using SsoAdvanced Universal SYBR Green supermix (Bio-Rad,1725275). GAPDH/Gapdh was used as a control. The primer sequences are listed in supplementary Table S1.
Cytosolic DNA detection
Cytosolic double-stranded DNA was detected using fluorescent PicoGreen reagent (P7581, Invitrogen). The fluorescent PicoGreen signal was imaged using a confocal microscope (Zeiss LSM880 Confocal with Airyscan). Cytosolic fluorescence intensity was analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD, USA, RRID:SCR_003070).
Cell cycle analysis
Cell samples were prepared using the Propidium Iodide Flow Cytometry Kit (ab139418, Abcam). DNA content was analyzed using a flow cytometer (Gallios Flow Cytometer, Beckman Coulter), and the cell cycle was analyzed using FlowJo software (Becton Dickinson, RRID:SCR_008520).
Measurement of colony-formation
Five hundred cells were seeded in each well of a 12-well plate and incubated with or without 50 nM of VE822. When the cells formed colonies after incubation for 5 or 6 days, they were stained with crystal violet (0.5% w/v). Images were obtained using a cell scanner (Epson Perfection V600 Photo) and colonies were counted under a microscope (LMI-3000 LAXCO).
Immunohistochemical (IHC) staining
Renca tumor FFPE (formalin-fixed paraffin-embedded) tissue sections were used for immunohistochemical staining. Briefly, slides were incubated with primary antibodies against CD11c, CD4, CD8, PD-1, or PD-L1 overnight at 4 °C, followed by incubation with SignalStain Boost IHC Detection HRP Rabbit (Cell Signaling, 8114) for 30 min at room temperature. IHC staining was performed using either the ImmPact DAB Peroxidase (HRP) Substrate (Vector Laboratories, SK-4105) for the slides stained with anti-CD11c antibody, or the ImmPact NovaRed (HRP) substrate for the remaining slides. Slides were then counterstained with Harris hematoxylin and mounted on coverslips using the Cytoseal™ mounting medium.
Quantification of Immunohistochemical Images
Images were acquired using the Vectra® 3 automated quantitative pathology imaging system. The images were then processed for quantification of immunohistochemical staining using Inform® 2.6 software from Akoya Biosciences®. The percentage positivity in the figures represents the percentage of cells in the tissue section of the tumor area that were positive for staining.
Mouse experiments
Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of The University of Texas MD Anderson Cancer Center. As previously described (6), 6-week-old BALB/c mice, half male and half female, were purchased from TACONIC. Renca cells (5 × 105) were suspended in 100 μL Matrigel matrix (Corning, 354234) diluted with PBS at a 1:1 ratio and subcutaneously injected into the backs of mice. After the tumors were palpable (i.e., tumor volume reached approximately 100 mm3), the mice were randomly divided into four groups, which were left untreated or treated with VE822 (60 mg/kg, 5 consecutive days per week) via oral gavage, PD-1 antibody (100 μg/mouse/3 days) via intraperitoneal (I.P.) injection, or both reagents concurrently. Tumors were measured daily using a caliper by a technician who was unaware of the treatment allocation. The tumor volume was calculated using the formula V = (W2 × L)/2. Mice were euthanized once the tumors reached 1000 mm3, ulceration occurred, or if they showed signs of distress. Tumors were fixed in an RNA stabilization reagent for RNA extraction or in 10% buffered formalin phosphate for IHC or Opal staining.
RCC protein expression, gene expression, and immune deconvolution analysis
Targeted RPPA proteomics data were acquired from The Cancer Genome Atlas (TCGA). Manually curated proteins associated with DNA damage response and immune signaling were utilized for supervised clustering. The ATR-CHK1 score was calculated as the average z-normalized value of ATR pS428 and CHK1 pS296. The ATM-CHK2 score was calculated as the average z-normalized value of ATM pS1981 and CHK pT68 expression levels. The ratio of the ATR-CHK1 score to the ATM-CHK2 score was calculated by z-normalizing each score and then subtracting the ATM-CHK2 score from the ATR-CHK1 score. Gene expression data from patients with ccRCC were compiled from The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas released by Braun et al. (7) and McDermott et al. (23). Signature scores were calculated as the average of the z-normalized expression values for a given pathway. The Cytosolic DNA Sensing Pathway from KEGG (24), Type I IFN Response, pDCs from Rooney et al. (25), and Dendritic Cells from Bindea et al. (26). The full gene sets for these signatures are listed in supplementary Figure S2. Meta-analyses across cohorts were performed using a fixed effects model.
RCC patient survival and response rate analysis
We curated the OS and ORR data from four primary data sources with matched SETD2 mutation data. OS data were acquired for ccRCC from TCGA pan-cancer atlas, which was predominately from-ICB-treated patients. OS and ORR data for patients with ccRCC treated with everolimus and nivolumab were acquired from Braun et al. (7). OS data for patients with ccRCC treated with ICB were acquired from Samstein et al. (27). OS and ORR to atezolizumab with or without bevacizumab were acquired from McDermott et al. (23).
Murine tumor gene expression and signature score analysis
RNA-seq reads were aligned to the mouse reference genome GRCm38 using Star RNASeq alignment software. Quality control was performed on the read data using FastQC software. HTseq was used to summarize reads per gene from the aligned BAM files. Count data were normalized using the DEseq2 R package (version 1.38.1, RRID:SCR_015687) (28). Pathway signature scores were estimated using single-sample gene set enrichment analysis (ssGSEA) implemented in the R package GSVA (version 1.42.0) (29). The abundance of T cells was estimated using the R package mMCPcounter (version 1.1.0) (30) and the abundance of dendritic cells was estimated using the digital cell quantification (DCQ) method as implemented in the R package ComICS (version 1.0.4) (31).
Mutation burden and neoantigen load analysis
The association between SETD2 mutations and tumor mutation burden or neoantigen load was analyzed using CAMOIP (32).
Data availability
RNAseq data for 786-O cells and Renca tumors were deposited to The Gene Expression Omnibus (GEO) (GSE234732, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE234732).
Results
ATR inhibition induces the expression of immune stimulatory and immune suppressive molecules via cGAS-IRF3 pathway
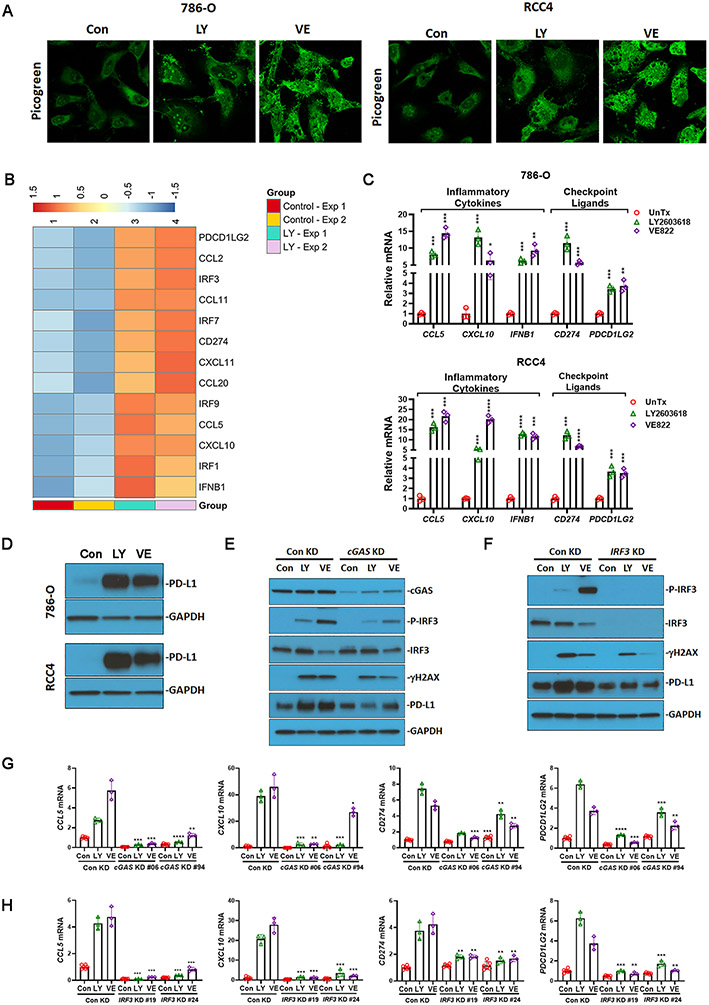
The cytosolic DNA sensing pathway connects the DNA damage response pathway to the cell-autonomous immune response (33,34). LY2603618 (rabusertib) and VE822 (berzosertib) are selective inhibitors of CHK1 and ATR, respectively, and both have been used in clinical trials. To assess the induction of cytosolic DNA following ATR-CHK1 inhibition, we treated the cells with LY2603618 or VE822, followed by staining with PicoGreen, a highly sensitive fluorescent stain that selectively binds to double-stranded DNA. We observed that both LY2603618 and VE822 increased the intensity of PicoGreen staining in the cytoplasm of 786-O and RCC4 cells (Fig. 1A), indicating the accumulation of DNA fragments after targeting ATR-CHK1-mediated DNA damage repair.
Fig. 1.
Targeting the DNA damage pathway induced the expression of immune stimulatory and immune suppressive molecules via cGAS- and IRF3-dependent cytosolic DNA response. (A) LY2603618 and VE822 induced cytosolic DNA accumulation in 786-O and RCC4 cells. Double-stranded DNA was detected with the fluorescent Picogreen reagent. (B) Heatmap showing LY2603618 induced gene expression that both positively and negatively modulate tumor immune microenvironment. 786-O cells were untreated (Control) or treated with 25nM LY2603618 for 48 hrs. Samples from two independent experiments (Exp1 and Exp2) were subjected to RNAseq. (C) LY2603618 and VE822 induced the expression of inflammatory cytokines (CCL5, CXCL10, and IFNB1) and immune checkpoint ligands (CD274 and PDCD1LG2) in 786-O and RCC4 cells. (D) LY2603618 and VE822 induced PD-L1 protein expression in 786-O and RCC4 cells. (E) cGAS knockdown reduced IRF3 phosphorylation, γH2AX, and PD-L1 expression in response to LY2603618 and VE822. cGAS knockdown 786-O stable cell line #06 was used. (F) IRF3 knockdown reduced γH2AX and PD-L1 expression in response to LY2603618 and VE822. IRF3 knockdown 786-O stable cell line #19 was used. (G) cGAS knockdown reduced CCL5, CXCL10, CD274, and PDCD1LG2 mRNA expression in response to LY2603618 and VE822. (H) IRF3 knockdown reduced CCL5, CXCL10, CD274, and PDCD1LG2 mRNA expression in response to LY2603618 and VE822. RCC parental cells (786-O cells, RCC4 cells), 786-O cells stably expressing control shRNA, cGAS shRNA, or IRF3 shRNA were treated with 25nM LY2603618 or 2.5μM VE822 for 48 hrs. Con, Untreated control; LY, LY2603618; VE, VE822. Protein expression was analyzed by immunoblot using antibodies against cGAS, IRF3, P-IRF3, γH2AX, PD-L1, and GAPDH. CCL5, CXCL10, IFNB1, CD274, and PDCD1LG2 mRNA levels were detected using real-time PCR. Data represent mean±s.d., n= 3. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001, compared with untreated control in C, with control knockdown cells in each treatment condition in G and H.
The tumor immune microenvironment is modulated by tumor-autonomous expression of chemokines and immune checkpoint molecules (35). Here, we observed that the CHK1 inhibitor LY2603618 induced the expression of multiple tumor landscape-modulating genes, including CCL and CXCL chemokines, interferon and interferon regulatory factors (IRFs), and immune checkpoint ligands (Fig. 1B). To validate these results using real-time PCR, we found that pharmacological inhibition of the ATR-CHK1 axis with LY2603618 or VE822 induced the expression of inflammatory cytokines (CCL5, CXCL10, and IFNB1) and immune checkpoint ligands (CD274 or PD-L1, PDCD1LG2 or PD-L2) in both the 786-O and RCC4 cell lines (Fig. 1C). Consistently, LY2603618 and VE822 increased PD-L1 protein levels in both 786-O and RCC4 cells (Fig. 1D).
It is widely accepted that targeting the DNA damage response network promotes an innate immune response via the cGAS-mediated DNA sensing pathway. To further study the dependence of cGAS and IRF3 on the activation of the cytosolic DNA-sensing pathway in response to ATR or CHK1 inhibition, we examined the effects of cGAS and IRF3 knockdown on the activity of the signaling pathway. In cGAS knockdown cells, as validated by real-time PCR and immunoblotting (Fig. S1A), IRF3 phosphorylation and target gene expression (CCL5, CXCL10, CD274, and PDCD1LG2) were largely suppressed in response to LY2603618 or VE822 treatment (Fig 1E,1G). Similarly, IRF3 knockdown substantially reduced the expression of both inflammatory cytokines and immune checkpoint ligands (Fig. 1F, 1H, Fig. S1B). These results indicated that cGAS and IRF3 are indispensable for the expression of both immunostimulatory and immunosuppressive molecules. Interestingly, the accumulation of γH2AX in the presence of LY2603618 and VE822 was reduced in cGAS or IRF3 knockdown cells (Fig. 1E, 1F). It is likely that the absence of cGAS or IRF3 phosphorylation upregulates DNA damage repair as previously reported (36,37).
SETD2 loss is associated with preferential ATR activation and sensitivity to ATR inhibition
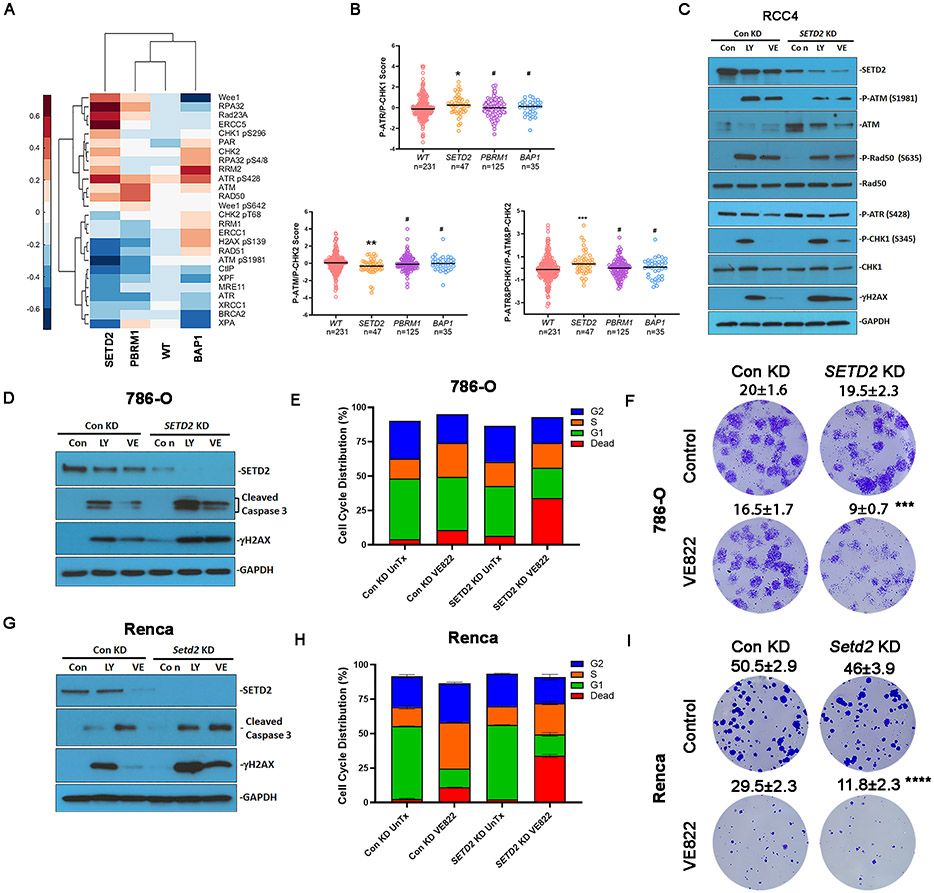
ATM activity is associated with autophosphorylation at S1981, and ATM activates multiple substrates via phosphorylation, such as CHK2 at T68 and Rad50 at S635 (38). ATR phosphorylates CHK1 at S317 and S345, which promotes autophosphorylation at S296, and the phosphorylation of all these residues is required for CHK1-mediated DNA damage response (39). We performed unsupervised clustering of DDR proteins based on the mutational status of tumors in The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) (Fig. 2A), and observed that ATR-CHK1 activity increased and ATM/CHK2 activity decreased in SETD2 mutant tumors. We then specifically compared the RPPA activation scores as indicated by pATR (S428)-pCHK1 (S296) and pATM (S1981)-pCHK2 (T68) phosphorylation levels across ccRCC genotypes and found a significant difference in SETD2 mutants (n=47), but not in PBRM1 (n=125) or BAP1 mutants (n=35), compared to wild-type samples (n=231) (Fig. 2B). SETD2 mutated tumors were associated with increased phosphorylation levels of ATR-CHK1 and decreased phosphorylation levels of ATM-CHK2 (Fig. 2B). Therefore, the ratio of pATR-pCHK1 to pATM-pCHK2 was significantly higher in SETD2 mutated samples than that in SETD2 wild type samples (Fig. 2B). These results indicate that SETD2 deficient tumors may rely more on the ATR-CHK1 axis for DNA damage response, which may confer sensitivity to ATR or CHK1 inhibitors.
Fig. 2.
SETD2 loss was associated with preferential ATR activation and vulnerability to ATR inhibition. (A) Heatmap showing clustering of DDR proteins based on the mutational status of tumors in TCGA KIRC. Each column represents the average value for tumors with respective mutations or wild type (WT) for PBRM1, SETD2, and BAP1. (B) RPPA activation scores for P-ATR&P-CHK1 signaling, P-ATM&P-CHK2 signaling, or ratio of P-ATR&P-CHK1 to P-ATM&P-CHK2 signaling. Rank-sum test. (C) SETD2 knockdown influenced ATM and ATR signaling in response to LY2603618 or VE822. RCC4 cells expressing control shRNA or SETD2 shRNA were treated with 25nM LY2603618 or 2.5μM VE822 for 48 hrs. Protein expression was analyzed by immunoblotting using antibodies against SETD2, P-ATM, ATM, P-Rad50, Rad50, P-ATR, P-CHK1, CHK1, γH2AX, and GAPDH. (D, G) SETD2 or Setd2 knockdown increased caspase 3 cleavage in response to LY2603618 and VE822 in (D) 786-O cells and (G) Renca cells. Protein expression was analyzed by immunoblot using antibodies against SETD2, cleaved caspase 3, γH2AX, and GAPDH. (E, H) SETD2/Setd2 knockdown increased cell death in response to VE822 treatment in (E) 786-O cells and (H) Renca cells. 786-O cells and Renca cells were treated with or without 2.5 μM VE822 for 24 hrs. The cell cycle distribution results represent the combined results of two independent experiments. (F, I) SETD2/Setd2 knockdown reduced the clonogenicity of (F) 786-O cells and (I) Renca cells. 786-O cells and Renca cells were treated with or without 50 nM VE822 for 5 days. Data represeFnt mean ± s.d., n= 4. ***P<0.001, and ****P<0.0001. Compared with control knockdown treated with VE822. Unpaired t test.
To confirm these findings in RCC cell lines, we generated stable SETD2 knockdown RCC4 and 786-O cell lines. The knockdown efficiency was validated by immunoblotting, which showed reduced SETD2 protein level in SETD2 knockdown cells (Fig. 2C, S1C). LY2603618 and VE822 decreased SETD2 protein levels (Fig. 2C, 2D, S1C); however, neither inhibitor decreased SETD2 mRNA expression (Fig. S1E), indicating that targeting the ATR-CHK1 axis downregulated SETD2 protein levels at the post-translational level. In SETD2 knockdown cells, the induced phosphorylation of ATM was reduced in the presence of LY2603618 and VE822 (Fig. 2C). The ATM-mediated phosphorylation of Rad50 (S635) was also reduced (Fig. 2C, S1C). In contrast, the phosphorylation levels of ATR (S428) and ATR-mediated CHK1 phosphorylation at S345 were slightly higher than those in the control knockdown cells (Fig. 2C, S1C, S1D). These results were consistent with the proteomic analysis of the TCGA-KIRC dataset, indicating that SETD2 deficiency was associated with preferential ATR activation relative to ATM.
Since SETD2 loss is associated with preferential ATR activation, we hypothesized that SETD2 deficiency confers sensitivity to ATR-CHK1 inhibition. As expected, γH2AX accumulation was further promoted in SETD2 knockdown cells in the presence of LY2603618 or VE822 (Fig. 2C, 2D). Fig. 2D shows that LY2603618 and VE822 induced the cleavage of Caspase 3 in 786-O cells, an indicator of apoptosis. SETD2 knockdown increased Caspase 3 cleavage (Fig. 2D), indicating that apoptosis was promoted in SETD2 knockdown cells. Flow cytometry results showed that, in control knockdown cells, VE822 treatment increased the fraction of cells in the S phase, an indication of S-phase arrest, and increased the fraction of dead cells from approximately 3% to 10% (Fig. 2E, Fig. S1F). In contrast, in SETD2 knockdown cells, VE822-induced S phase arrest was not as obvious, but VE822 treatment increased the number of dead cells to more than 30% (Fig. 2E, Fig. S1F). Although RCC4 cells were relatively more resistant to VE822 treatment (2.5 μM VE822, 24 hr treatment), SETD2 knockdown cells also demonstrated vulnerability to high dose and prolonged treatment (5 μM, 48 hr) (Fig. S1F, S1G). We further validated that SETD2 knockdown cells were preferentially dying by performing a colony formation assay and found that VE822 reduced the colony formation of SETD2 deficient cells to a greater extent than that in control knockdown cells (Fig. 2F). We confirmed these results with the widely used murine RCC cell line Renca and found that depletion of Setd2 mirrored the results found in human 786-O cells following treatment with LY2603618 and VE822, including exacerbated induction of γH2AX, caspase 3 cleavage, and increased cell death (Fig. 2G-2I, Fig. S1F). These results indicated that SETD2 loss enhanced DNA damage in the presence of an ATR inhibitor, and a large fraction of cells arrested in the G1 phase died because of the failure of DNA damage repair.
SETD2 loss is associated with upregulated cytosolic DNA sensing pathway
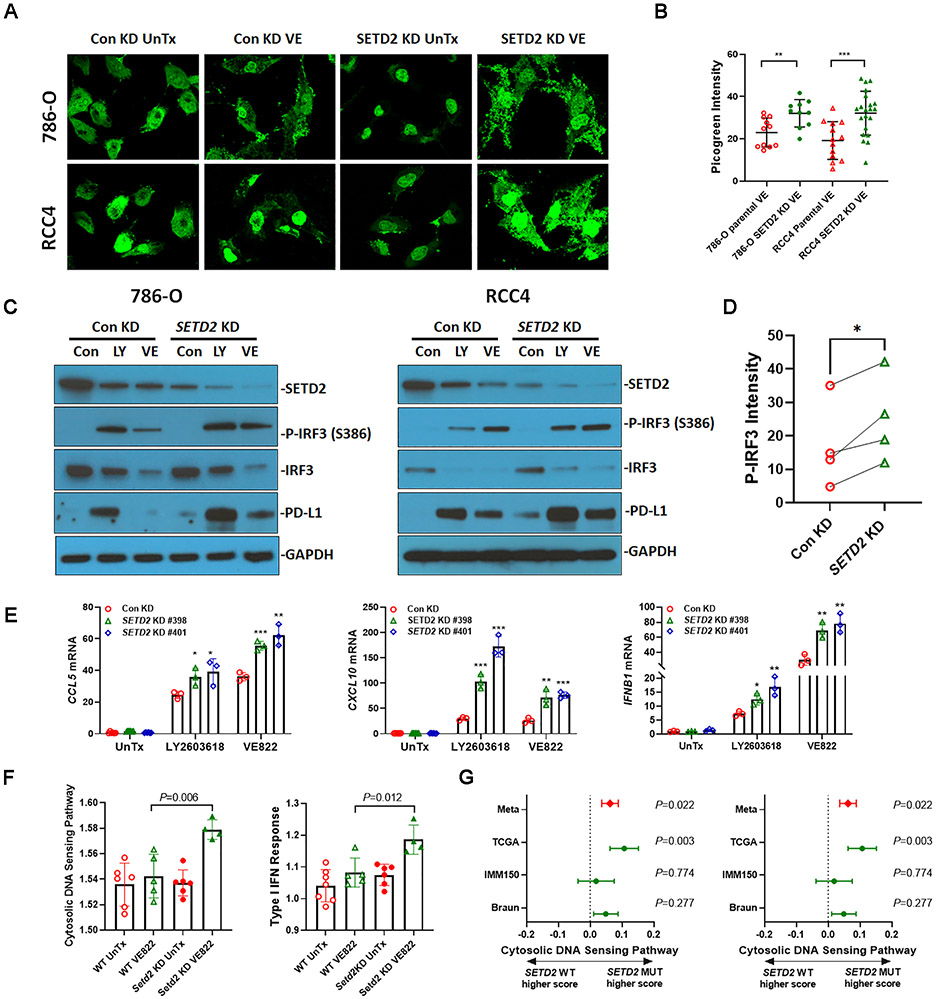
Since SETD2 knockdown enhanced DNA damage following ATR-CHK1 inhibition, it is conceivable that SETD2 loss promotes the cytosolic DNA-sensing pathway. First, we studied its effects on cytosolic DNA accumulation. As indicated by the enhanced intensity of PicoGreen staining, VE822 treatment led to greater cytosolic DNA accumulation in SETD2 deficient 786-O cells and RCC4 cells than in control knockdown cells (Fig. 3A, 3B). Next, we studied the DNA sensing pathway induced by targeting the DNA damage response in SETD2 deficient 786-O cells and RCC4 cells. Both LY2603618 and VE822 induced IRF3 phosphorylation, which was further elevated in SETD2 deficient cells (Fig. 3C, 3D). LY2603618 and VE822 treatment also induced PD-L1 protein expression to a greater extent in SETD2 deficient cells (Fig. 3C). Consistently, SETD2 deficiency promoted the expression of downstream target genes including CCL5, CXCL10, and IFNB1 in response to LY2603618 or VE822 treatment (Fig. 3E, S1H).
Fig. 3.
SETD2 loss was associated with increased cytosolic DNA response. (A) SETD2 knockdown increased cytosolic DNA accumulation in response to VE822 treatment in 786-O and RCC4 cells. Double-stranded DNA was detected with the fluorescent Picogreen reagent. (B) The quantification of cytosolic Picogreen intensity. The intensity of Picogreen in the cytoplasm was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Data represent mean±s.d. **, P<0.01, ***, P<0.001. Unpaired t test. (C) SETD2 knockdown increased IRF3 phosphorylation and PD-L1 expression in response to LY2603618 and VE822 treatment. Protein expression was analyzed by immunoblotting using antibodies against SETD2, IRF3, P-IRF3, PD-L1, and GAPDH. (D) Quantification of P-IRF3 relative amount in 786-O and RCC4 cells. The intensity of P-IRF3 immunoblot bands in the presence of LY2603618 and VE822 was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). ***, P<0.001. Paired t test. (E) SETD2 knockdown increased the expression of CCL5, CXCL10, and IFNB1 in response to LY2603618 and VE822 treatment in 786-O cells. Unpaired t test comparing SETD2 knockdowns to control knockdown in each treatment condition. In experiments A-E, 786-O and RCC4 cells expressing control shRNA or SETD2 shRNA were treated with 25 nM LY2603618 or 2.5 μM VE822 for 48 hrs. (F) Setd2 knockdown increased signature scores of cytosolic DNA sensing pathway and Type I IFN response pathway in Renca tumors treated with VE822. (G) SETD2 mutated RCC tumors were associated with increased signature scores of cytosolic DNA sensing pathway and Type I IFN response pathway in RCC samples from The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas release (SETD2 WT, n=428; SETD2 mutants n=47), Braun et al. (SETD2 WT, n=134; SETD2 mutants n=43) (7), and McDermott et al. (SETD2 WT, n=138; SETD2 mutants n=55) (23). Difference with 95% confidence interval shown; fixed effects model used for meta-analysis value (Meta).
These results indicate that SETD2/Setd2 loss is associated with the upregulation of the cytosolic DNA sensing pathway in response to ATR-CHK1 inhibition in human and murine RCC cell lines. To confirm our findings in a preclinical model, we grew both control knockdown and Setd2 knockdown Renca tumors subcutaneously in immune-competent BALB/c mice, and mice were left untreated or treated with VE822. We then analyzed the signature scores of the cytosolic DNA sensing pathway and the type I IFN response in Renca tumors. Setd2 knockdown Renca tumors treated with VE822 showed higher signature scores than those of control knockdown tumors (Fig. 3F). We further investigated the effects of SETD2 mutations on cytosolic DNA-sensing pathways in human ccRCC tumor datasets, including TCGA-KIRC (40), IMmotion 150 (23), and Braun ccRCC (41). We specifically focused on transcriptional signatures representing the cytosolic DNA sensing pathway (24) and type I IFN response (25). Meta-analysis showed that SETD2 mutated tumors were associated with the upregulation of both signature scores (Fig. 3G). These in vivo and in vitro results collectively indicate that SETD2 loss promotes the cytosolic DNA-sensing pathway, resulting in upregulation of cytosolic DNA accumulation, inflammatory cytokine expression, and PD-L1 expression.
ATR inhibition concurrently upregulates immune cell infiltration and immune checkpoint expression in Setd2 deficient tumors
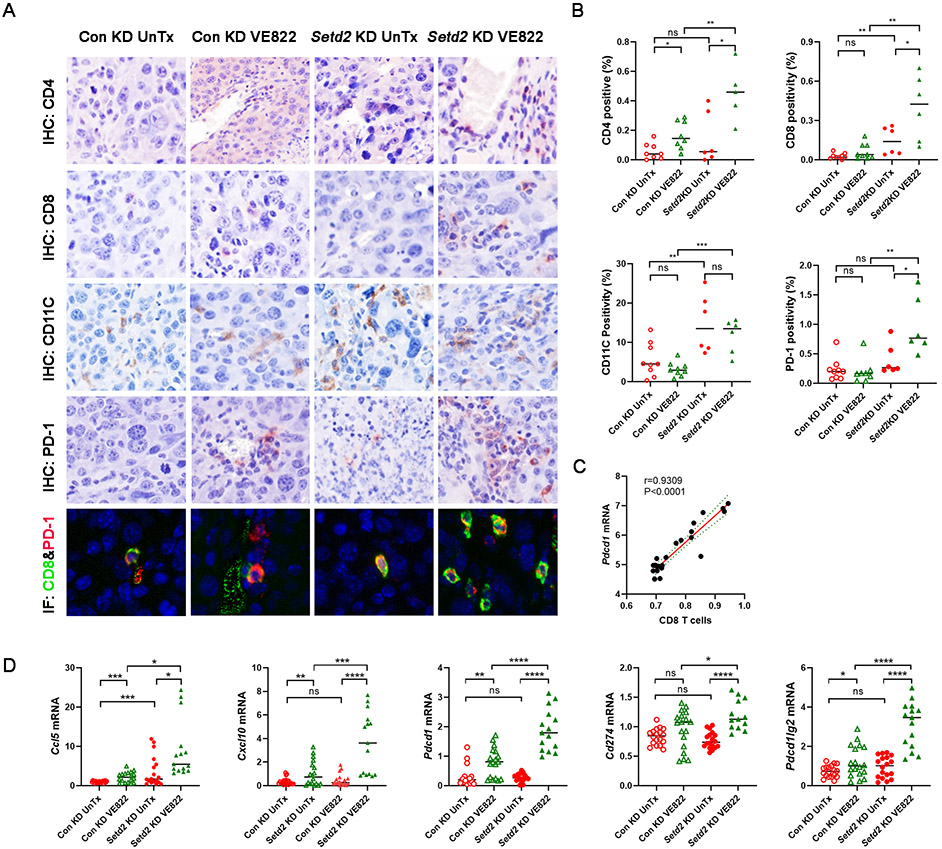
CCL5, CXCL10, and type I interferon are cytokines involved in T cell attraction, dendritic cell migration, and T cell-dendritic cell interactions (14). Based on the signaling pathway signature scores and immune cell scores derived from gene expression-based inference, Fig. S2A shows that the cytosolic DNA sensing pathway was positively correlated with the type I IFN response pathway, CD4 T cells, CD8 T cells, and dendritic cells in pooled Renca tumors. Bulk RNA-seq deconvolution revealed that VE822 treatment increased the abundance of CD4 T cells, CD8 T cells, and dendritic cells in both control knockdown and Setd2 knockdown Renca tumors, and that Setd2 knockdown tumors undergoing VE822 treatment demonstrated higher T cell and dendritic cell abundance than control knockdown tumors (Fig. S2B). We validated these observations by immunohistochemical (IHC) staining, confirming that Setd2 knockdown tumors demonstrated more abundant CD4, CD8, and CD11C positive cells after receiving VE822 than control knockdown tumors (Fig. 4A, 4B). Importantly, Setd2 knockdown and VE822 synergistically promoted the expression of PD-1 in Renca tumors (Fig. 4A, 4B). Multiplex Opal immunofluorescence staining further confirmed the colocalization of CD8 and PD-1 (Fig. 4A). Consistent with these findings, Pdcd1 mRNA levels were positively correlated with the abundance of CD8 positive T cells in Renca tumors (Fig. 4C). Real-time PCR assay of the Renca tumor samples revealed that VE822 treatment induced the expression of Ccl5, Cxcl10, Cd274 (PD-L1), Pdcd1lg2 (PD-L2), and Pdcd1 (PD-1) in both Setd2 proficient and Setd2 deficient tumors, but the induction was greater in Setd2 deficient tumors (Fig. 4D). These results demonstrated that Setd2 loss and VE822 treatment synergistically promoted immune cell infiltration and immune checkpoint expression, providing a rationale for ATR inhibition plus ICB combination therapy.
Fig. 4.
VE822 treatment concurrently upregulated immune cell infiltration and immune checkpoint expression in Setd2 knockdown Renca tumors. (A-B) VE822 increased immune cell infiltration and PD-1 expression in Setd2 knockdown tumors based on IHC staining. (A) Representative images. (B) Quantification of immunohistochemical positivity of CD4, CD8, CD11C, and PD-1 in Renca tumors. Murine Renca tumors were immunohistochemically stained with antibodies against CD4, CD8, CD11C, and PD-1 or co-stained with CD8 (green) and PD-1 (red) with immunofluorescence. The percentages of positively stained cells were analyzed using inForm software. Control knockdown and Setd2 knockdown Renca tumors were treated with VE822 or left untreated (UnTx) as a control group. Unpaired t test. (C) The correlation between CD8 T cells and Pdcd1 expression. Renca tumors were subjected to RNAseq, and the expression level of Pdcd1 was derived and the relative CD8 T cell level was analyzed using gene expression-based inference. (D) Setd2 knockdown and VE822 synergized to induce the expression of inflammatory cytokines and immune checkpoints in Renca tumors. The mRNA levels of Ccl5, Cxcl10, Pdcd1, Cd274, and Pdcd1lg2 were analyzed using real-time PCR.
Setd2 loss and ATR inhibitor synergize to promote immunotherapy response in Renca tumors
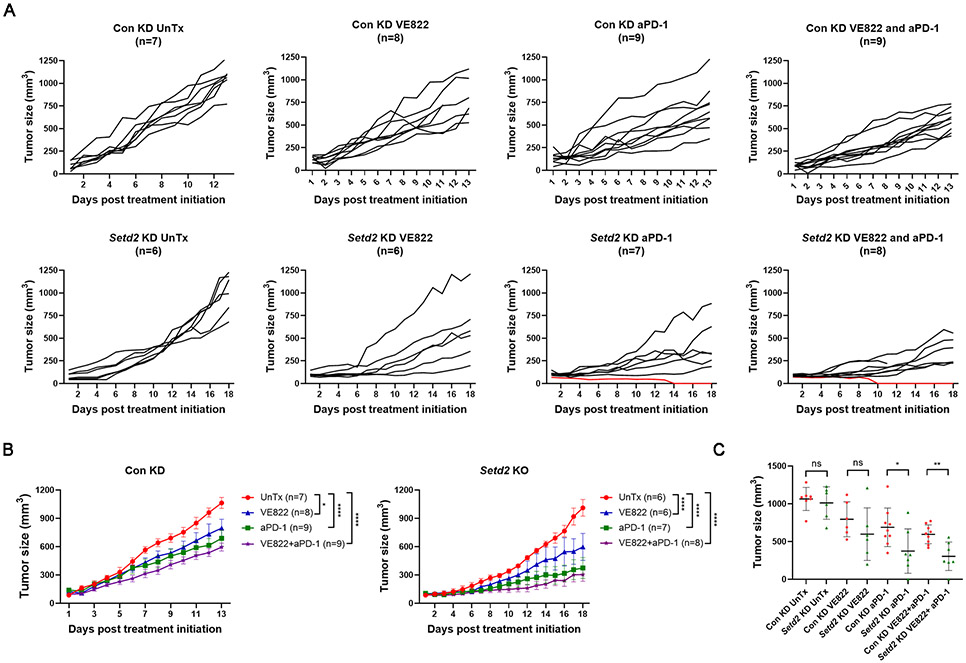
These results described above indicate that VE822 treatment increased T cell and dendritic cell infiltration in the tumor microenvironment, especially in Setd2 deficient tumors. On the other hand, VE822 also upregulated immune checkpoint proteins. We hypothesized that combination therapy with VE822 and ICB would activate antitumor immunity and provide more therapeutic benefit by counteracting immune checkpoint upregulation. When Renca tumors reached 100-200 mm3, BALB/c mice received VE822 or PD-1 antibody as monotherapy or both agents as combination therapy. The control knockdown Renca tumors grew rapidly, and by day 13, most untreated mice were sacrificed because the tumor sizes reached 1000 mm3. Monotherapy with either VE822 or PD-1 antibody reduced tumor growth (Fig. 5A, 5B). Combination therapy with both drugs provided better control of tumor growth, and none of the tumors reached 1000 mm3 in 13 days (Fig. 5A, 5B). Setd2 knockdown Renca tumors grew slowly at the beginning, and once they reached approximately 500 mm3, they started to grow quickly (Fig. 5A, 5B). Treatment with VE822 or PD-1 antibody alone decreased their growth, and the combination reduced their growth to a much greater extent (Fig. 5A, 5B). Notably, two Setd2 knockdown tumors exhibited CR: one received PD-1 antibody monotherapy, and the other received PD-1 antibody plus VE822 (Fig. 5A, red lines). Therapeutic CR has not been previously observed in the Renca model with other tumor genotypes and other treatments. Regardless of the various responses in the same group, these observations suggest that Setd2 deficient tumors are more responsive to ICB. We further compared the tumor size on the day of sacrifice when both untreated groups exhibited comparable sizes (Fig. 5C). After treatment with PD-1 blocking antibodies with or without VE822, Setd2 knockdown tumors were significantly smaller than control knockdown tumors (Fig. 5C). Although it did not reach the significance threshold, VE822-treated Setd2 knockout tumors were smaller (Fig. 5C). These results indicate that Setd2 loss confers greater sensitivity to immune checkpoint blockade with or without ATR inhibitor treatment in RCC Renca tumors.
Fig. 5.
Setd2 loss and VE822 synergized to promote Renca tumor immune responsiveness. (A) Individual tumor growth curves. (B) Aggregate tumor growth curves. Monotherapy with VE822 or anti-PD-1 antibody, or combination treatment suppressed both control knockdown and Setd2 knockdown Renca tumor growth. *P<0.05, and ****P<0.0001. Two-way ANOVA analysis. (C) Setd2 knockdown tumors demonstrated greater sensitivity to anti-PD-1 antibody with or without VE822. The final tumor volume in each treatment group was displayed on a per-mouse basis with mean ± SD. ns, p>0.05, *P<0.05, and **P<0.01. Unpaired T test. Renca cells were subcutaneously injected into the back of Balb/c mice. Once the tumors reached 100-200 mm3, mice bearing Renca tumors received monotherapy with anti-PD-1 antibody (200 μg/mouse, every 3 days, I.P.), VE822 (60mg/kg, 5 consecutive days per week, oral gavage), or combination therapy with both reagents.
SETD2 loss is associated with increased response to immunotherapy in RCC patients
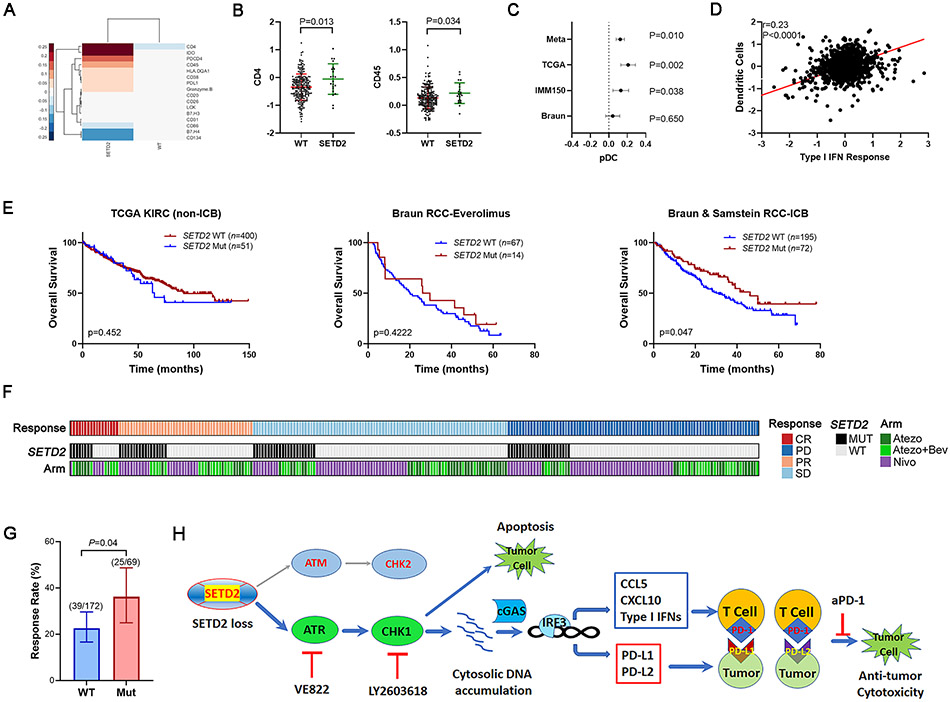
To identify the influence of SETD2 mutations on the human RCC tumor immune microenvironment, we first analyzed TCGA RPPA data and found that SETD2 mutated tumors were associated with altered expression of immune proteins (Fig. 6A). Although the expression of CD8 and CD11C was not available, we found the expression of CD45 and CD4 was significantly higher in SETD2 mutated samples (Fig. 6B). The SETD2 mutated samples from Clark et al. (42) also demonstrated higher CD11C expression, although the difference is not significant (P=0.42) probably due to limited patient number and heterogeneity among patients (Fig. S2C). Further transcriptional deconvolution analysis indicated that plasmacytoid dendritic cells (pDCs), a subtype of dendritic cells that secrete high levels of type I interferons, were elevated in SETD2 mutated tumors (Fig. 6C). We also observed a positive correlation between dendritic cells and the type I IFN response (Fig. 6D). These results indicated that SETD2 loss is associated with higher T cell and dendritic cell infiltration in human RCC.
Fig. 6.
SETD2 loss promoted response to immunotherapy in ccRCC patients. (A) Clustergram of immune proteins from RPPA targeted proteomics. A value of 0 represents the average of the entire cohort. (B) ccRCC samples with SETD2 mutations demonstrated higher CD4 and CD45 expression. (C) ccRCC samples with SETD2 mutations demonstrated higher pDC infiltration. Difference with 95% confidence interval for pDC content inferred from RNAseq in RCC cohorts; fixed effects model used for meta-analysis value (Meta). (D) Correlation between dendritic cells and type I IFN response pathway expression signatures determined from RNAseq. Data from patients with ccRCC in C and D was compiled from The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas release, Braun et al. (7), and McDermott et al. (23). (E) SETD2 mutations were associated with prolonged overall survival in ICB-treated patients but not non-ICB-treated patients. Kaplan-Meier plots were derived from non-ICB-treated RCC patients from TCGA-KIRC, everolimus-treated RCC patients from Braun cohorts (7), ICB-treated RCC patients in Braun and Samstein cohorts (7,27). Log-rank test. (F-G) SETD2 mutations were associated with increased response rate in ICB-treated ccRCC patients. Objective response in the Braun cohort following Nivolumab treatment (Nivo) and the IMmotion150 cohort following treatment with either atezolizumab (Atezo) or atezolizumab in combination with bevacizumab (Atezo+ Bev). Merged response rate (PR/CR) to ICB from Braun and IMmotion150 cohorts. Error bars indicate a 95% confidence interval determined by the Clopper Pearson method. Mantel-Haenszel test. (H) Working model. SETD2 loss is associated with preferential activation of the ATR/CHK1 axis of the DNA damage response pathway. The inhibition of this axis led to DNA damage-induced cell death, and the accumulation of cytosolic DNA, which activated cGAS and consequently IRF3 phosphorylation. As a transcription factor, the phosphorylated IRF3 initiated the expression of inflammatory cytokines (CCL5, CXCL10, and Type I IFNs), which promoted the immune cell infiltration into the tumor immune microenvironment. IRF3 also initiated the transcription of immune checkpoint ligands (PD-L1 and PD-L2), and thus inhibited the T cell activity via their interaction with PD-1. The immune checkpoint blockade with anti-PD-1 antibody disrupted the interaction between PD-1 and PD-L1 or PD-L2 and conferred anti-tumor immunity.
Next, we evaluated the effect of SETD2 loss on immune checkpoint blockade responses in patients with RCC using publicly available databases. First, SETD2 mutations did not demonstrate a prognostic value in the largely untreated patients with RCC from the TCGA-KIRC cohort (SETD2 WT, n=400; SETD2 mutants n=51) or in patients from Braun et al. (7) who received the mTOR inhibitor everolimus (SETD2 WT, n=67; SETD2 mutants n=14) (Fig. 6E). In contrast, in patients who received anti-PD-1 (nivolumab) in Braun et al. (7) and in patients who received ICB from Samstein et al. (27) (SETD2 WT, n=195; SETD2 mutants n=72), SETD2 mutations were associated with favorable OS (Fig. 6E, S2D). These results indicate that prolonged OS in patients with SETD2 mutated tumors is more likely related to a better response to ICB treatment, but not due to a generally better prognosis. To further confirm that we were not observing prognostic effects, we analyzed the ORR of patients treated with atezolizumab (anti-PD-L1) with or without bevacizumab in the IMmotion150 study, as well as in patients who received nivolumab in Braun cohorts (7,23) as a function of SETD2 mutation status (Fig. 6F, 6G). We found that patients with SETD2 mutant tumors exhibited a higher ORR when treated with ICB (25/69) compared with patients with wild-type SETD2 tumors (39/172) (Fig. 6G). Taken together, SETD2 mutations enhanced the response to immunotherapy in multiple patient cohorts.
Discussion
Although immunotherapy with ICB monotherapy revolutionized RCC treatment by increasing OS, the ORR was only 25% (43). The development of ICB combination therapy, both with ICB doublets and ICB/TKIs, has increased the ORR, PFS, and CR rates (3), but the ability to identify those most likely to derive profound benefits from ICB-based therapy is still limited. Here, we used RCC cell lines, murine RCC Renca tumors, and multiple RCC patient cohorts to demonstrate that SETD2 loss and ATR inhibition synergistically promote the cGAS-mediated cytosolic DNA sensing pathway and enhance immune responsiveness in RCC. Targeting the ATR-CHK1 axis with pharmacological inhibitors activated the cGAS-IRF3-mediated cytosolic DNA-sensing pathway, leading to the expression of immune cell-attracting factors (CCL5, CXCL10, and Type I IFN) and immune checkpoint molecules (PD-1, PD-L1, and PD-L2). SETD2 loss is associated with preferential ATR-CHK1 activity over ATM-CHK2 activity and sensitizes cells to ATR inhibition. ATR inhibition induced a more immunoinhibitory tumor microenvironment in Setd2 deficient tumors, providing a rationale for ATR inhibition plus ICB combination therapy (Fig. 6H). ATR pharmacological inhibitors can induce immune-independent cell death (apoptosis) and promote immune-dependent tumor killing when administered in combination with immune checkpoint inhibitors.
Both the ATR-CHK1 and ATM-CHK2 axes are critical pathways that mediate the DNA damage response, and there are functional interactions between both axes. Previous studies have shown that loss of ATM function confers greater sensitivity to ATR inhibitors (16-18). ATM mutations are rarely found in RCC, and we compared the effect of SETD2, PBRM1 and BAP1 mutations on ATR and ATM activity and found that SETD2 knockdown in RCC cells suppressed ATM-CHK2 activity and increased ATR-CHK1 activity, implying that SETD2 loss engenders greater dependence on ATR activity to compensate for the suppressed ATM pathway and thus confers greater sensitivity to ATR inhibition. Here, we found that SETD2 loss sensitized cells to ATR inhibition in vitro and in vivo with increased cell death, cGAS signaling, and immunotherapy response. SETD2 plays a critical role in maintaining genomic integrity, suppressing replication stress, and enhancing double-stranded DNA repair (21,44). SETD2 interacts with the mismatch recognition protein MutSα and is co-enriched at DNA damage sites in response to oxidative stress, which in turn recruits ATM and activates the ATM-CHK2 pathway (45). SETD2 mediates trimethylation of H3K36, and H3K36me3 physically interacts with MutSα and recruits it to the chromatin (46). It is conceivable that SETD2 mutated tumors are less effective at transducing DNA damage signals from MutSα to ATM due to its conformational change, accelerated degradation, or reduced methyltransferase activity.
SETD2 is the third most commonly mutated gene in ccRCC, with a prevalence of approximately 15% in TCGA KIRC dataset, and its association with immune responsiveness remains unclear. One possible reason for this is that there are generally insufficient samples in most clinical trials to make robust observations. Another possible reason is that SETD2-mutated patients also harbor other genetic mutations that influence the response to immunotherapy. Our collective analysis of multiple RCC cohorts revealed that SETD2 mutations were associated with an improved response rate and prolonged OS in ICB-treated RCC patients. Pan-cancer analysis has shown that SETD2 mutations are associated with a higher tumor mutation burden and favorable clinical outcomes (47). However, individual analysis of TCGA-KIRC did not reveal a higher tumor mutation burden or neoantigen load associated with SETD2 mutations (Fig. S2E). More importantly, in ccRCC cohorts, there is not a single cohort in which high TMB is associated with a better response rate in ccRCC (48). Defects in DNA damage repair can induce antitumor immunity via neoantigen production and the activation of the cGAS pathway (49). Our results indicate that SETD2 mutations may influence tumor immunogenicity mainly via cGAS pathway activation in ccRCC but not via neoantigen load upregulation.
Our analysis of the patient datasets was retrospective and had several inherent limitations. First, due to the limited number of SETD2 mutated patients in each cohort, it was difficult to gain statistical significance in individual cohorts, and we combined the cohorts to perform a collective analysis. We do appreciate that combining cohorts across treatments is potentially problematic, because different cohorts might use different immunotherapy agents, and we cannot exclude the possibility that SETD2 mutations interact differently with these immune checkpoint inhibitors. However, all patients from Braun cohort received the same IBC treatment, nivolumab, and SETD2 mutated patients did show prolonged OS with a P=0.0885. A similar trend was obtained with patients from the Samstein cohort although they did not annotate which patients received which specific agent. Furthermore, the results of the combination analysis of RCC cohorts are congruent with results obtained from animal experiments, and our data demonstrate general internal consistency and directionality. It potentially provides important information for future clinical trials and personalized treatment. Second, all the available data sets used in this study consisted of RCC patients who did not receive an ATR inhibitor, and SETD2 loss may have less influence on cGAS signaling and the tumor immune microenvironment under unstimulated conditions, although SETD2 mutated samples still demonstrated increased activity of the cytosolic DNA sensing pathway and response to ICB. Since SETD2 is required for ATM activity and DNA damage repair (45) and SETD2-mutant RCC cells demonstrate impaired DNA damage signaling (21), the long-term inactivation of SETD2 in RCC tumors is likely sufficient to induce DNA damage accumulation and activate the cytosolic DNA sensing pathway. However, such a chronic change is not expected to be as effective as that induced by the pharmacological inhibitors of ATR. Immune-competent animal models are required to isolate and clarify the influence of SETD2 loss on immunotherapy response, with or without combination therapy with an ATR inhibitor. In this study, Setd2 deficient Renca cells and tumors demonstrated vulnerability to ATR kinase inhibition, upregulated the cytosolic DNA sensing pathway, and enhanced immune cell infiltration, which mirrored the features of human RCC cell lines and RCC patient tumors. Comparing isogenically paired Renca tumors, Setd2 knockdown Renca tumors develop a more immunogenic TME and are more responsive to ICB after receiving ATR inhibitors. Although our current study and animal experiments in small cell lung cancer and ovarian cancer show the benefit of simultaneously targeting DNA damage response and immune checkpoint (34,50), recent clinical trial data further revealed that sequential immunotherapy and targeted therapy showed OS benefit in BRAF+ melanoma (51), and it may be interesting for the future animal studies to consider sequencing treatment with the ATR inhibition and PD-1 blocking antibodies in addition to testing combination strategies. Considering tumor heterogeneity (52), private mutations in SETD2 in the evolutionary trajectories of tumor progression may limit the clinical impact of targeting this pathway. In addition, increasing T-cell infiltration is not always predictive of immunotherapy response, and we cannot completely exclude the possibility that ATR inhibition will not clinically improve immunotherapy response in patients with RCC.
In the future, clinical studies that compare ICB monotherapy, combination, or sequential therapy with an ATR inhibitor will further identify the influence of fundamental gene mutations on therapeutic response in RCC patients. Taken together, this study utilized both isogenic preclinical models and clinical cohort validation to demonstrate that SETD2 loss promotes sensitivity to ICB, which may be further potentiated by ATR-CHK1 inhibition, thus providing mechanistic evidence for a combination therapy regimen for RCC patients with SETD2 mutations.
Supplementary Material
Translational relevance.
Immune checkpoint blocking agents (ICB) have become a mainstay for the treatment of patients with renal cell carcinoma (RCC); however, only a minority of patients show complete responses to ICB. A recent clinical trial has revealed that Ataxia telangiectasia and rad3 related (ATR) inhibitor provides benefit in patients with advanced solid tumors with Ataxia telangiectasia mutated (ATM) mutations. However, ATM mutations themselves rarely occur in RCC patients. Our study demonstrates that SET domain-containing protein 2 (SETD2) loss is associated with decreased ATM activity and sensitizes RCC cells to ATR inhibition. ATR inhibition concurrently upregulates inflammatory cytokine expression, immune cell infiltration, and immune checkpoint expression to a greater extent in Setd2 deficient tumors, and consequently potentiates sensitivity to ICB. These findings can guide the development of combination therapy for RCC patients with SETD2 mutations and further provide a strategy to screen patients who will benefit from treatment with ATR inhibitors, particularly in combination with ICB, by identifying complementary genetic lesions that reduce ATM activity.
Acknowledgments
This work was supported by DOD grant W81XWH-17.1.0307, Kidney Cancer Association Grant 13653766, Koch Center Award, Philip Guentert Memorial Fund, and Adopt-a-Scientist Foundation to E. Jonasch, NCI grant R00CA240689 to D.J. McGrail, NIH/NCI U24 CA 264006 to R. Akbani, NIH R50 Grant R50CA221675 to Y. Lu, UT MD Anderson Cancer Center CCSG grant 5 P30 CA016672 (including Biostatistics Shared Resource) and NIH 1S10OD024977-01. The RPPA Core was supported by NCI Grant CA16672 to P. Pisters.
Footnotes
Conflict of interest
R.A., a bioinformatics consultant for the University of Houston. No potential conflicts of interest were disclosed by the other authors.
Reference:
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73(1):17–48 doi 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Santoni M, Massari F, Di Nunno V, Conti A, Cimadamore A, Scarpelli M, et al. Immunotherapy in renal cell carcinoma: latest evidence and clinical implications. Drugs Context 2018;7:212528 doi 10.7573/dic.212528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasanov E, Gao J, Tannir NM. The Immunotherapy Revolution in Kidney Cancer Treatment: Scientific Rationale and First-Generation Results. Cancer J 2020;26(5):419–31 doi 10.1097/PPO.0000000000000471. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen KB, Spranger S. Modulation of the immune microenvironment by tumor-intrinsic oncogenic signaling. J Cell Biol 2020;219(1) doi 10.1083/jcb.201908224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell TJ, Turajlic S, Rowan A, Nicol D, Farmery JHR, O'Brien T, et al. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell 2018;173(3):611–23 e17 doi 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu XD, Kong W, Peterson CB, McGrail DJ, Hoang A, Zhang X, et al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nat Commun 2020;11(1):2135 doi 10.1038/s41467-020-15959-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun DA, Hou Y, Bakouny Z, Ficial M, Sant' Angelo M, Forman J, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med 2020;26(6):909–18 doi 10.1038/s41591-020-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T, Lu R, Kapur P, Jaiswal BS, Hannan R, Zhang Z, et al. An Empirical Approach Leveraging Tumorgrafts to Dissect the Tumor Microenvironment in Renal Cell Carcinoma Identifies Missing Link to Prognostic Inflammatory Factors. Cancer Discov 2018;8(9):1142–55 doi 10.1158/2159-8290.CD-17-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XD, Hoang A, Zhou L, Kalra S, Yetil A, Sun M, et al. Resistance to Antiangiogenic Therapy Is Associated with an Immunosuppressive Tumor Microenvironment in Metastatic Renal Cell Carcinoma. Cancer Immunol Res 2015;3(9):1017–29 doi 10.1158/2326-6066.CIR-14-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov D, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380(12):1116–27 doi 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 11.Choueiri TK, Powles T, Burotto M, Escudier B, Bourlon MT, Zurawski B, et al. Nivolumab plus Cabozantinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2021;384(9):829–41 doi 10.1056/NEJMoa2026982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motzer R, Alekseev B, Rha SY, Porta C, Eto M, Powles T, et al. Lenvatinib plus Pembrolizumab or Everolimus for Advanced Renal Cell Carcinoma. N Engl J Med 2021;384(14):1289–300 doi 10.1056/NEJMoa2035716. [DOI] [PubMed] [Google Scholar]
- 13.Msaouel P, Goswami S, Thall PF, Wang X, Yuan Y, Jonasch E, et al. A phase 1-2 trial of sitravatinib and nivolumab in clear cell renal cell carcinoma following progression on antiangiogenic therapy. Sci Transl Med 2022;14(641):eabm6420 doi 10.1126/scitranslmed.abm6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabanon RM, Rouanne M, Lord CJ, Soria JC, Pasero P, Postel-Vinay S. Targeting the DNA damage response in immuno-oncology: developments and opportunities. Nat Rev Cancer 2021;21(11):701–17 doi 10.1038/s41568-021-00386-6. [DOI] [PubMed] [Google Scholar]
- 15.Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat Rev Cancer 2023;23(2):78–94 doi 10.1038/s41568-022-00535-5. [DOI] [PubMed] [Google Scholar]
- 16.Rafiei S, Fitzpatrick K, Liu D, Cai MY, Elmarakeby HA, Park J, et al. ATM Loss Confers Greater Sensitivity to ATR Inhibition Than PARP Inhibition in Prostate Cancer. Cancer Res 2020;80(11):2094–100 doi 10.1158/0008-5472.CAN-19-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlop CR, Wallez Y, Johnson TI, Bernaldo de Quiros Fernandez S, Durant ST, Cadogan EB, et al. Complete loss of ATM function augments replication catastrophe induced by ATR inhibition and gemcitabine in pancreatic cancer models. Br J Cancer 2020;123(9):1424–36 doi 10.1038/s41416-020-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yap TA, Tan DSP, Terbuch A, Caldwell R, Guo C, Goh BC, et al. First-in-Human Trial of the Oral Ataxia Telangiectasia and RAD3-Related (ATR) Inhibitor BAY 1895344 in Patients with Advanced Solid Tumors. Cancer Discov 2021;11(1):80–91 doi 10.1158/2159-8290.CD-20-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng H, Jiang X, Cui J, Yin G, Shi B, Liu Q, et al. Genomic Analysis Reveals Novel Specific Metastatic Mutations in Chinese Clear Cell Renal Cell Carcinoma. Biomed Res Int 2020;2020:2495157 doi 10.1155/2020/2495157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espana-Agusti J, Warren A, Chew SK, Adams DJ, Matakidou A. Loss of PBRM1 rescues VHL dependent replication stress to promote renal carcinogenesis. Nat Commun 2017;8(1):2026 doi 10.1038/s41467-017-02245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM, et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 2014;3:e02482 doi 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014;7(6):2006–18 doi 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24(6):749–57 doi 10.1038/s41591-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353–D61 doi 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160(1-2):48–61 doi 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39(4):782–95 doi 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51(2):202–6 doi 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7 doi 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petitprez F, Levy S, Sun CM, Meylan M, Linhard C, Becht E, et al. The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med 2020;12(1):86 doi 10.1186/s13073-020-00783-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altboum Z, Steuerman Y, David E, Barnett-Itzhaki Z, Valadarsky L, Keren-Shaul H, et al. Digital cell quantification identifies global immune cell dynamics during influenza infection. Mol Syst Biol 2014;10(2):720 doi 10.1002/msb.134947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin A, Qi C, Wei T, Li M, Cheng Q, Liu Z, et al. CAMOIP: a web server for comprehensive analysis on multi-omics of immunotherapy in pan-cancer. Brief Bioinform 2022;23(3) doi 10.1093/bib/bbac129. [DOI] [PubMed] [Google Scholar]
- 33.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst 2017;109(1) doi 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov 2019;9(5):646–61 doi 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilgelm AE, Richmond A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front Immunol 2019;10:333 doi 10.3389/fimmu.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018;563(7729):131–6 doi 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 37.Zierhut C, Yamaguchi N, Paredes M, Luo JD, Carroll T, Funabiki H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019;178(2):302–15 e23 doi 10.1016/j.cell.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatei M, Jakob B, Chen P, Kijas AW, Becherel OJ, Gueven N, et al. ATM protein-dependent phosphorylation of Rad50 protein regulates DNA repair and cell cycle control. J Biol Chem 2011;286(36):31542–56 doi 10.1074/jbc.M111.258152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci U S A 2008;105(52):20752–7 doi 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499(7456):43–9 doi 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun DA, Street K, Burke KP, Cookmeyer DL, Denize T, Pedersen CB, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 2021;39(5):632–48 e8 doi 10.1016/j.ccell.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark DJ, Dhanasekaran SM, Petralia F, Pan J, Song X, Hu Y, et al. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 2019;179(4):964–83 e31 doi 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 2015;373(19):1803–13 doi 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanu N, Gronroos E, Martinez P, Burrell RA, Yi Goh X, Bartkova J, et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 2015;34(46):5699–708 doi 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo S, Fang J, Xu W, Ortega J, Liu CY, Gu L, et al. Interplay between H3K36me3, methyltransferase SETD2, and mismatch recognition protein MutSalpha facilitates processing of oxidative DNA damage in human cells. J Biol Chem 2022;298(7):102102 doi 10.1016/j.jbc.2022.102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013;153(3):590–600 doi 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu M, Zhao B, Liu M, Wu L, Li Y, Zhai Y, et al. Pan-cancer analysis of SETD2 mutation and its association with the efficacy of immunotherapy. NPJ Precis Oncol 2021;5(1):51 doi 10.1038/s41698-021-00193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 2021;32(5):661–72 doi 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Shih DJH, Lin SY. Role of DNA repair defects in predicting immunotherapy response. Biomark Res 2020;8:23 doi 10.1186/s40364-020-00202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Yang L, Wang C, Zhao W, Ju Z, Zhang W, et al. Inhibition of the ATM/Chk2 axis promotes cGAS/STING signaling in ARID1A-deficient tumors. J Clin Invest 2020;130(11):5951–66 doi 10.1172/JCI130445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ascierto PA, Mandala M, Ferrucci PF, Guidoboni M, Rutkowski P, Ferraresi V, et al. Sequencing of Ipilimumab Plus Nivolumab and Encorafenib Plus Binimetinib for Untreated BRAF-Mutated Metastatic Melanoma (SECOMBIT): A Randomized, Three-Arm, Open-Label Phase II Trial. J Clin Oncol 2023;41(2):212–21 doi 10.1200/JCO.21.02961. [DOI] [PubMed] [Google Scholar]
- 52.Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 2018;173(3):581–94 e12 doi 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data for 786-O cells and Renca tumors were deposited to The Gene Expression Omnibus (GEO) (GSE234732, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE234732).