Abstract
Purpose:
We previously showed that elevated frequencies of peripheral blood CD3+CD4+CD127-GARP-CD38+CD39+ T-cells was associated with checkpoint immunotherapy resistance in metastatic melanoma patients. In the present study we sought to further investigate this population of ectoenzyme expressing T-cells (Teee).
Expeirmental Design:
Teee derived from the peripheral blood of metastatic melanoma patients were evaluated by bulk RNA-seq and flow cytometry. The presence of Teee in the tumor microenviroment was assessed using publically available single-cell RNA-seq datasets of melanoma, lung, and bladder cancer along with multispectral immunofluorescent imaging of melanoma patient FFPE specimens. Suppressive function of Teee was determined by an in vitro autologous suppression assay.
Results:
Teee had phenotypes associated with proliferation, apoptosis, exhaustion, and high expression of inhibitory molecules. Cells with a Teee gene signature were present in melanoma, lung cancer and bladder cancer patients’ tumors. CD4+ T-cells co-expressing CD38 and CD39 in the tumor microenvironment were preferentially associated with Ki67- CD8+ T-cells. Co-culture of patient Teee with autologous T-cells resulted in decreased proliferation of target T-cells. High baseline intra-tumoral frequencies of Teee were associated with checkpoint immunotherapy resistance and poor overall survival in metastatic melanoma patients.
Conclusions:
These results demonstrate that a novel population of CD4+ T-cells co-expressing CD38 and CD39 is found both in the peripheral blood and tumor of melanoma patients and is associated with checkpoint immunotherapy resistance.
Keywords: Ectoenzymes, Adenosine, Melanoma, NSCLC, Bladder Cancer, Predictive, Biomarker, PD1, CTLA4, Nivolumab, Ipilimumab
Statement of Translational Relevence:
In order to improve the efficacy of checkpoint immunotherapies, there is a critical need to determine mechanisms of resistance and predictive biomarkers. Here, we expand upon previous findings and show that a population of CD4+ T-cells co-expressing CD38 and CD39 present in the periphery and tumor microenvironment of metastatic melanoma patients is associated with treatment resistance. We demonstrate that this population of cells is present in other solid tumors, has phenotypes associated with immunosuppression, and suppresses autologous T-cell proliferation in vitro. Collectively, these data highlight a novel population of ectoenzyme expressing CD4+ T-cells in association with immunotherapy resistance and provide rationale for exploration as a potential target for augmenting the efficacy of checkpoint immunotherapies.
Introduction.
Antagonist antibodies targeting T-cell co-inhibitory receptors PD1 and CTLA4 have demonstrated efficacy in a number of advanced malignancies. The combination of nivolumab (αPD1) and ipilimumab (αCTLA4) has demonstrated objective response rates of 61% in melanoma1, 38% in non-small cell lung cancer (NSCLC)2, 38% in bladder cancer3, and 42% in renal cancer4. Despite having unprecedented efficacy, immune checkpoint inhibitor therapies fail to benefit most patients. Consequently, there is a critical need to determine mechanisms of resistance and predictive biomarkers of response, particularly actionable biomarkers that can be targeted in combination with immune checkpoint inhibition to improve treatment efficacy.
We previously reported that a population of T-cells defined by the marker set CD3+CD4+CD38+CD39+CD127-GARP- found in the peripheral blood of patients with metastatic melanoma was associated with immune checkpoint blockade resistance5,6. We observed elevated frequencies of this population of ectoenzyme expressing T-cells (Teee) in both advanced metastatic melanoma patients who progressed on nivolumab-ipilimumab or nivolumab monotherapy compared to those who responded to therapy. We also observed on-treatment expansion of Teee in high-risk resected melanoma patients that relapsed while on combination nivolumab and ipilimumab adjuvant therapy.
CD39 and CD38 are both ectoenzymes involved in the production of extracellular adenosine. These ectoenzymes and adenosine receptors are upregulated in tumor and immune cells in response to hypoxia and chronic inflammatory signaling7 in the tumor microenvironment. Extracellular ATP actively secreted by tumors8 or released from dying or stressed cells leads to inflammatory immune responses9, whereas the conversion of ATP into adenosine leads to immune suppression10. In T-cells, binding of adenosine to its receptor, A2AR, suppresses activation11,12, induces exhaustion13 , and upregulates expression of co-inhibitory receptors14. Extracellular adenosine also impairs anti-tumor immune responses by disruption of B-cell signaling15, polarization of M2 macrophages16, promotion of immunosuppressive dendritic cell phenotypes17, and recruitment of myeloid derived suppressor cells16,17. Additionally, extracellular adenosine promotes tumor proliferation and metastasis18–20.
In this study we sought to address the roles of Teee in immunotherapy resistance by characterizing their phenotype and function, assessing their presence in the tumor microenvironment of various solid malignancies, and evaluating the relationship of intra-tumor Teee to immunotherapy resistance.
Materials & Methods.
Patient samples:
Cryopreserved peripheral blood mononuclear cells (PBMCs) were obtained from previously collected baseline samples (i.e. prior to start of therapy) as part of the clinical studies S15–00906 (NCTS15–00906), MCC15400 (NCT0117646121), CA209–064 (NCT0178393822) and MCC15651 (NCT011764745). All subjects gave written informed consent for trial participation, including the research use of patient-derived materials. The studies were conducted in accordance with the Declaration of Helsinki guidelines. Patient information for the specimens used from these trials is given in Supplemental Tables 1, 2, 3 and 4. FFPE samples were obtained from the University of Colorado Anschutz Medical Campus Skin Cancer BioRepository. Patient and treatment information for the FFPE specimens is given in Supplemental Table 5. All specimens were collected under IRB-approved protocols and with informed patient consent. Helathy donor specimens were acquired from One Blood (New York, NY). All response data were based on RECIST v.1.1 criteria. Complete and partial responses were considered as “responders” and progressive disease considered as “non-responders”.
Flow cytometry and cell sorting:
Antibodies used for flow cytometric analysis and cell sorting are listed in Supplemental Table 6. Samples were acquired on an Attune Nxt 14 parameter flow cytometer (Therno-Fisher, Waltham, MA), BD Biosciences FACSymphony (Franklin Lakes, NJ) or a ID7000 spectral analyzer (Sony, New York, NY). The general gating strategy for Teee is shown in Supplemental Figure 1A. Analyses were performed using FlowJo 10.8 (Ashland, OR).
Suppression assay:
CD4+ T-cells were enriched from melanoma patient PBMC specimens (S15–00906 specimens) using an EasySep human CD4+ T-cell enrichment kit from STEMCELL Technologies (Vancouver, Canada). Isolated CD4+ T cells were sorted by flow cytometry on a BD Biosciences (Franklin Lakes, NJ) FACSAria II cytometer to obtain CD3+CD4+CD127-GARP-CD25-CD38+CD39+ cells. Autologous CD3+ T-cells (target cells) were isolated using the STEMCELL Technologies CD3 negative isolation kit and stained with Cell Trace Violet from Thermo Fisher. Target cells and Teee were plated at a ratio of 2:1 (Target:Teee) or only target cells in round-bottomed 96-well plates from Costar (Corning, NY). Co-cultures were stimulated with plate-bound anti-CD3 at 0.5ug/mL (clone OKT3) and soluble anti-CD28 at 0.5ug/mL (clone: CD28.2 (Biolegend,San Diego, CA) , and ProImmune human recombinant IL-2 (Oxford, UK) was added at a concentration of 50IU/mL. The percentage of proliferating cells was determined as the percentage of target T-cells with peaks outside of generation zero in the Cell Trace Violet channel.
RNA-Seq data analysis:
CD4+ T cells from baseline PBMC samples (S15–00906 specimens) were flow sorted into CD4+ CD127-GARP-CD38-CD39-, CD4+ CD127-GARP-CD38+CD39-, CD4+ CD127-GARP-CD38-CD39+, or CD4+ CD127-GARP-CD38+CD39+ as described under “Suppression assay”, and total RNA was immediately isolated from the sorted cells with either a Qiagen (Venlo, Netherlands) RNAEasy Plus Micro kit or a Qiagen RNAEasy Mini kit. RNA integrity numbers (RIN) were determined for each sample with an Agilent (Santa Clara, CA) 2100 Bioanalyzer. Only samples with a RIN ≥ 8.0 were used to prepare cDNA libraries with a CloneTech (Mountain View, CA) SMARTer HT kit. Sequencing was performed by the NYU Langone’s Genome Technology Center (RRID: SCR_017929) on an Illumina (San Diego, CA) NovaSeq 6000, generating approximately 50 million 100bp paired reads per sample. FastQC v0.11.9 was used to determine the quality of the reads, presence of contaminating adapter sequences and other quality control metrics. Low quality reads (Phred score < 20), reads < 20bp in length, and contaminating adapter sequences were removed with TrimGalore v0.6.7. Resulting reads were then aligned to the human genome (GRCH38, release 84) with HISAT2 v2.2.1 with the default parameters. Mapped reads were sorted and indexed with SAMtools v1.16.1 and mapped reads counted with FeatureCounts v2.0.3 with options: -p and -s 0. Subsequent analysis was performed with the R programming language. Differential gene expression analysis was conducted with DESeq2 v3.16. Genes with a count < 50 summed across all samples were removed. Next, the CD38+CD39+ samples were contrasted against the other three populations in DESeq2 accounting for the paired nature of the data with: “design = ~patient + population”. Differentially expressed genes (DEGs) were considered as those with a log2FC of > |1| and an false discovery rate (FDR) adjusted p-value of < 0.01. CSV files of the DEGs are available in the supplementary materials. Principal component analysis was performed with the DESeq2 normalized counts with the prcomp function. Volcano plots were constructed using the EnhancedVolcano package v1.11.3. Heatmaps were constructed with the DESeq2 normalized counts with the pheatmap package. Expression data from Teee populations were analyzed for enrichment within the MSigDB Hallmark Gene Set (GSEA software v4.2.3 for Mac) and normalized enrichment scores (NES) plotted as bar graphs.
Single-cell RNA-seq data analysis:
Publicly available scRNA-seq datasets GSE11597823, GSE17999424, GSE14965225, and GSE12057526 were retrieved from the Gene Expression Omnibus (GEO). Normalized count matrices (i.e., TPM or log2(+1) normalized to 10,000 counts per cell) or non-normalized count matrices were imported into Seurat (v4.3.0). The analysis pipeline consisted of normalizing (if not normalized prior) and scaling/centering. CD3+CD4+ T-cells were subsetted from the data based on strategies tailored for each dataset. Broadly these strategies consisted of removing cells with low expression of CD3E and/or high expression of CD8A and/or low expression of CD4 and/or low mitochondrial contamination. Principal components (PCs) were produced and the appropriate number of PCs to use for clustering identified with elbow plots. Clusters and uniform manifold approximation and projection (UMAPs) were created with default settings, except for the number of dimensions. All datasets were analyzed for the presence of patient bias, and where necessary, this bias was removed with canonical correlation analysis in Seurat. Cells were analyzed for enriched expression of the 49 upregulated genes identified in the Teee gene signature obtained from the flow-sorted bulk RNA-seq, plus ENTPD1 and CD4, with the Seurat AddModuleScore function.
In comparisons of Teee frequencies between responding and non-responding patients and in survival analysis, Teee cells were defined as subset CD4+ cells with greater than 0.1 counts (this cutoff was directed by results from violin plots) for both CD38 and ENTPD1 and the number of cells satisfying this criterion counted for each sample. Teee counts were then divided by the total number of subset CD4+ cells for each patient to calculate the percentage of Teee cells amongst the CD4+ TIL.
For correlation and differential gene expression of Teee and Tregs, the normalized/scaled/centered counts from cells that were enriched for the Teee signature score were retrieved and classified as Teee cells. The normalized/scaled/centered counts from cells that comprised clusters with high expression of FOXP3 and IL2RA were retrieved and classified as Tregs. Pearson correlations between Tregs and Teee cells were calculated using the R cor function and hierarchical clustered/plotted with the R corrplot function. DEGs between Tregs and Teee cells were identified with the Seurat FindMarkers function with default settings. Statistical significance was determined by the Wilcoxon Rank Sum test and p-values were Bonferroni corrected. Significantly differentially expressed genes were those meeting the criteria of log2FC > |0.5| and FDR adjusted P-value < 0.1. DEGs were plotted with the R EnhancedVolcano package and key phenotype-related genes labeled. While the same list was used to label key genes, not all genes were plotted for each comparison as some genes were filtered by the FindMarkers function.
Multiplex immunofluorescence:
Multiplexed immunofluorescence (mIF) analysis was performed with an Akoya Bioscience’s (Marlborough, MA) Vectra Polaris platform on FFPE pre-treatment resected metastatic melanoma tumor samples using the protocol described in Johnson A. et al27. A nine-parameter immunofluorescence panel was optimized by the CU Anschutz HIMSR core according to best practices. Panel antibody information is given in Supplemental Table 7. Briefly, the slides were deparaffinized by baking for 3 hours at 65 degrees (F) in a laboratory oven, heat treated in antigen retrieval buffer using TBS, blocked using Akoya blocking buffer for 5 minutes, and incubated with primary antibodies for 30 minutes each, followed by horseradish peroxidase (HRP)-conjugated secondary antibody polymer, and HRP-reactive OPAL fluorescent reagents that use TSA chemistry to deposit dyes on the tissue immediately surrounding each HRP molecule for 10 min. To prevent further deposition of fluorescent dyes in subsequent staining steps, the slides were stripped in between each stain with heat treatment in antigen retrieval buffer. Repeat the blocking, antibody staining through wash steps for each individual antibody per the staining order and dilution factor indicated on the table. Whole tissue was stained and imaged at 10x with the standard Akoya 6-Plex Manual Detection Kit for the purpose of region selection and annotation. Up to 15 regions of tumor-stroma border per specimen were identified with the aid of H&E-stained consecutive tissue slices. Region selection and stamping for acquisition was done after review of H&E staining of consecutive tissue slices to identify the ROIs along the tumor border containing at least some lymphocytes. Two pathologists were consulted to help identify suitable tumor/border regions that met this criteria. The slides were then re-stained with the optimized nine-parameter panel (Akoya 8-Plex plus DAPI Kit) using their Opal immunohistochemistry Multiplex Assay on a Leica (Wetzlar, Germany) autostainer and regions of interest individually captured with the Akoya Bioscience’s Vectra Polaris at 40x resolution.
The resulting images were spectrally unmixed and Akoya’s InForm software v.2.5.1 used to identify the tumor-stroma interface; only tumor-stroma interface regions were analyzed. A subset of regions was used for the training process which consists of: 1) using InForm to demarcate the various tumor and stroma, 2) performing cell segmentation training by identifying nucleus (DAPI, Ki67), cytoplasm (tumor: SOX10/S100; T-cells: CD3, CD4, CD8), 3) adjusting intensity (starting with DAPI on a 3X blurred image to prevent simultaneous detection of two nuclei) for each for each marker, 4) training InForm (for morphology and melanoma markers (i.e., SOX10 and S100)) and reviewing additional regions with representative samples of various tumor and stroma patterns to ensure a robust training algorithm, and 5) retraining as needed. Phenotype training consisted of: 1) tagging at least 30 representative cells, 2) reviewing approximately a dozen images for accuracy and 3) batch applying the trained model. Cell itemization including tissue segmentation, cell segmentation, and phenotyping were exported. Akoya phenoptrReports software (version 0.3.2) and R packages tidyverse (1.3.2), and readxl (1.4.1) were used in combination to consolidate the exported cell data and perform spatial analysis. The count_phenotypes and count_touching_cells functions were used to count the number of cells identified as each phenotype and to quantify the number of pairs of touching cells. A correction was used for the spatial analysis that involved calculating the origin of each ROI, the offset of X and Y from the origin and then multiplying by 3 to account for the 3-factor blurring.
Statistics:
Statistical significance of comparisons between the frequency of peripheral blood Teee in melanoma patients and healthy donors were determined by a Welch’s t-test to account for unequal variance. Contrasts of flow cytometry phenotyping between the four CD38/CD39 quadrants were performed with repeated measures ANOVAs and Dunnett’s post hoc tests comparing the CD38+CD39+ population against the other populations. For mIF analysis, comparison of the proportion of CD3+CD8+Ki67- vs. CD3+CD8+Ki67+ adjacent to CD3+CD4+CD38+CD39+ cells was performed using a paired t-test. To adjust for base rate differences, the frequencies were calculated by dividing the number of CD3+CD8+Ki67- cells in contact with Teee by the total number of CD3+CD8+Ki67- cells, and the same for Ki67+ cells. Comparison of the frequency of CD3+CD8+ adjacent to CD3+CD4+CD38+CD39+ cells in progressing vs. responding patients was performed with a Student’s t-test. Statistical significance of suppression assays was determined by paired t-tests in cases of two groups and by a repeated measures ANOVA with Dunnett’s posthoc tests in cases of more than two groups. Comparisons of the frequency of Teee in the melanoma scRNA-seq data in responders vs non-responders were performed with a Welch’s t-test. For survival analysis, patients were categorized as Teee high or Teee low based on being above or below the median percentage of Teee cells (as a percentage of CD4+). Analysis was performed with the R suvminer (0.4.9) package and statistical significance was evaluated by a Cox proportional hazards regression. For relationships between Teee frequency and CD8 gene expression in the scRNA-seq dataset, a linear regression was performed and R2 and p-values assessed by ANOVAs.
Data Availability:
Bulk RNA-seq reads generated for this project have been deposited at NCBI Gene Expression Omnibus (GEO) at accession number GSE224099. Code related to the analysis of the RNA-seq data is available at https://github.com/SciOmicsLab/Ectoenzyme_Tcells. Supplementary files containing differentially expressed genes and the Teee gene signature accompany this manuscript.
Results.
Peripheral blood Teee frequencies are elevated in melanoma patients.
We first determined if the Teee phenotype (CD3+CD4+CD38+CD39+CD127-GARP-), previously identified as being associated with immunotherapy resistance in metastatic melanoma patients, was similarly elevated in the peripheral blood of melanoma patients compared to healthy individuals. The frequency of Teee as a percent of the parent CD3+CD4+ population was assessed by flow cytometry using three clinical datasets (Supplemental Figure 2). The gating strategy and a representative plot of the Teee population is shown in Supplemental Figure 1A. Increased circulating frequencies of Teee were observed at baseline in stage II/IV metastatic melanoma patients (=2.44%) compared to healthy donors (=1.37%) in clinical trial CA209–064 that compared two sequences of ipilimumab and nivolumab (Supplemental Figure 2A; 95% CI difference of means = −0.041 to 2.19, p=0.0586, n=42 & n=11), in MCC15400, a study of nivolumab with a peptide vaccine in melanoma patients that progressed after receiving ipilimumab (Supplemental Figure 2B; 95% CI difference of means = 0.290 to 0.759, p<0.0001, n= 76 & n=17), and at baseline in stage III/IV resected melanoma patients in clinical trial MCC15651 in which they received adjuvant nivolumab and a peptide vaccine. (Supplemental Figure 2C; 95% CI difference of means = −0.029 to 0.733, p=0.0679, n= 18 & n=8). We did not observe a relationship between age and Teee frequency in any of the datasets (Supplemental Figure 7). Sample level information of patient characteristics and Teee frequencies can be found in Supplemental Tables 2–4.
Teee have phenotypes associated with enhanced proliferation, apoptosis, and immune checkpoint expression.
To investigate genes differentially expressed by the Teee population, baseline PBMC samples from three metastatic melanoma patients were flow sorted into: 1) CD3+CD4+CD127-GARP-CD38-CD39-, 2) CD3+CD4+CD127-GARP-CD38+CD39-, 3) CD3+CD4+CD127-GARP-CD38-CD39+, and 4) CD3+CD4+CD127-GARP-CD38+CD39+ (Teee) cells, and subsequently analyzed by bulk RNA-seq (Figure 1). Examination of the first two principal components (PC) showed that the Teee populations from all three patients were resolved from the other populations along PC1 (Supplemental Figure 3A). Differential expression analysis of Teee resulted in 809 differentially expressed genes (DEGs) (log2FC > |1| and FDR adjusted p-value < 0.01) compared to the CD38-CD39- population (Supplemental Figure 4A), 604 DEGs compared to the CD38+CD39- population (Supplemental Figure 4C), and 168 DEGs compared to the CD38-CD39+ population (Supplemental Figure 4E). Enrichment analysis of hallmark gene sets for these comparisons was also performed (Supplemental Figures 4B, 4D, 4E). In all three comparisons, proliferation-associated gene sets (e.g. G2M checkpoint) displayed the greatest enrichment in the Teee population. Other noteworthy enriched genes sets present across comparisons included apoptosis/DNA repair/p53 signaling and apical junction/epithelial mesenchymal transition. Ninety overlapping DEGs were obtained between the three comparisons (Figure 1A, 1B).
Figure 1. Peripheral blood Teee have a gene expression profile associated with proliferation, apoptosis and inhibitory checkpoints.
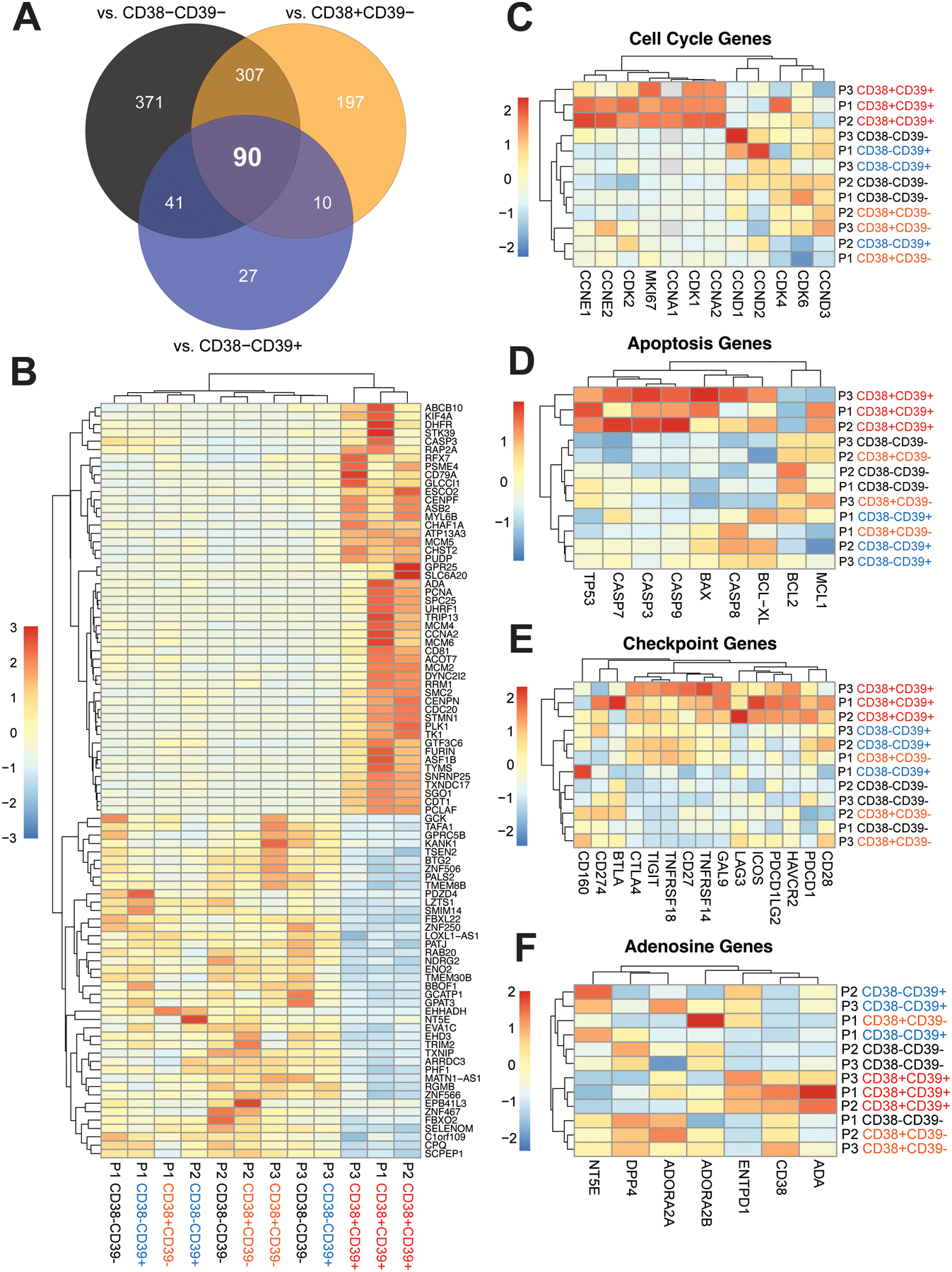
Baseline peripheral blood specimens from melanoma patients were flow sorted into CD3+CD4+CD127-GARP-CD38-CD39-, CD3+CD4+CD127-GARP-CD38+CD39-, CD3+CD4+CD127-GARP-CD38-CD39+, and CD3+CD4+CD127-GARP-CD38+CD39+ (Teee) populations and evaluated by bulk RNA-sequencing. (A) A Venn diagram is shown representing the overlap of the identified differentially expressed genes (DEGs) based on comparison of the Teee population against the other three CD38/CD39 quadrants. The 90 DEGs in the middle are shared between all comparisons. (B) The overlapping 90 DEGs are plotted as a heatmap of normalized counts scaled by row. Genes with relative increased expression in Teee are plotted in shades of red and those with relative decreased expression in shades of blue. Dendrograms represent hierarchical clustering of samples (columns) and genes (rows) by Euclidean distance. (C-F) Heatmaps of normalized expression values of canonical genes associated with (C) cell cycle, (D) apoptosis, (E) T-cell co-stimulation/inhibition, and (F) adenosine generation/signaling are plotted.
Based on the observed DEGs and hallmark gene set analysis, we hypothesized that Teee were highly proliferative, apoptotic, and upregulated genes indicative of an exhausted phenotype. To address this, we looked at sets of select genes canonically associated with these phenotypes. Teee samples clustered together for expression of cell proliferation-associated genes, with upregulation of G1/S phase associated cyclins and cyclin-dependent kinases (Figure 1C). Teee samples also clustered together for expression of apoptosis related genes, including upregulation of TP53 (p53 gene) and caspases (Figure 1D) with downregulation of the anti-apoptotic gene BCL2. Teee samples also clustered together with high expression of checkpoint inhibitor genes, including PDCD1 (PD1), CTLA4, HAVCR2 (TIM3), LAG3, and PDCD1LG2 (PDL2) (Figure 1E). Teee samples also clustered together for expression of adenosine pathway related genes, including high expression of ADA, and low expression of DPP4 (CD26) and NT5E (CD73) (Figure 1F). We also investigated the expression of genes canonically associated with CD4+ T-cell polarization phenotypes (Supplemental Figure 3B). No clear trends in expression patterns associated with known polarization phenotypes were observed.
We next used flow cytometry to determine if Teee had a proliferative, apoptotic, and exhausted phenotype as observed in the bulk RNA-seq analysis. Additional baseline PBMC samples from advanced melanoma patients were gated into CD38/CD39 quadrants of the CD3+CD4+CD127-GARP- parent population as shown in Supplemental Figure 1A.The Teee population showed the highest expression of the proliferation marker Ki67 (Fig. 2). Teee also had high frequencies of cells positive for surface expression of the apoptosis marker annexin V, as did the CD38-CD39+ subset. Teee and the CD38-CD39+ population both had relatively low expression of TCF1, with a higher expression of TOX in the Teee subset, a phenotype associated with exhausted T-cells28,29. TOX and TCF1 are shown as geometric mean fluorescence intensity (gMFI) readouts in leiu of percent expressing cells as very high percentages of cells were positive for these markers. Representative histograms for TOX and TCF1 are shown in Supplemental Figure 1B. Teee had the highest frequency of expression of the co-inhibitory markers CTLA4 and TIM3 and were PD1high. The gating strategy defining PD1neg/low/high is shown in Supplemental Figure 1C. The CD38-CD39+ population had the highest frequency of PD1low, with Teee having a slightly lower frequency. Teee also had relatively high expression of PDL1. Additionally, Teee had the second lowest expression of CD73, a key enzyme in extracellular adenosine production, and the lowest expression of CD26, a protein associated with adenosine breakdown.
Figure 2. Teee have increased expression of proliferation, apoptosis, exhaustion, and co-inhibitory markers.
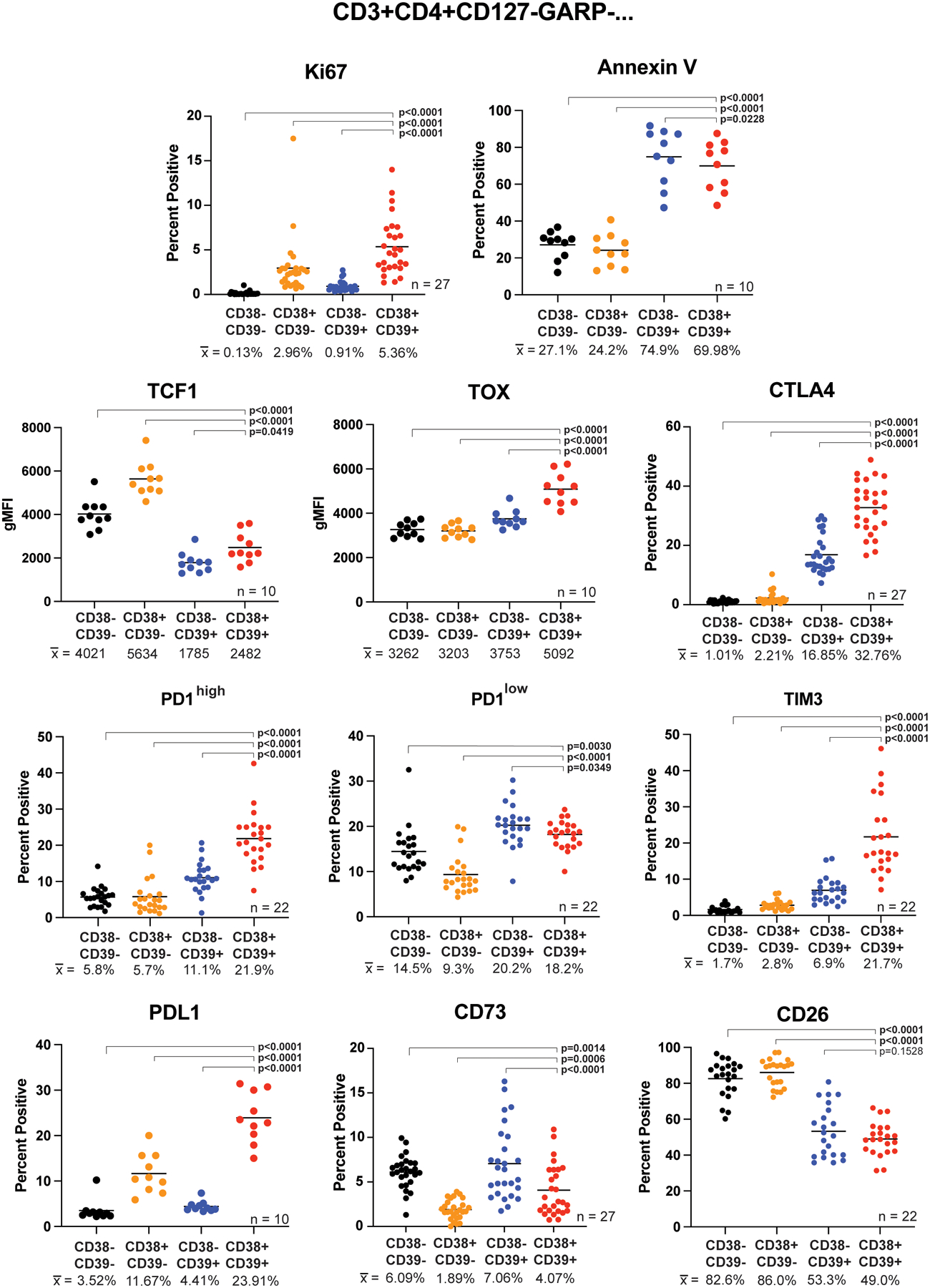
Baseline peripheral blood cells from melanoma patients were stained and analyzed by flow cytometry. The CD3+CD4+CD127-GARP- parent population was gated and the CD38/CD39 quadrants assessed for expression of the indicated markers. The CD38-CD39- population is shown in black, CD38+CD39- in orange, CD38-CD39+ in blue, and CD38+CD39+ (Teee) in red. Corresponding sample sizes assessed are indicated in the lower right of each panel. Population sample mean values are given below each group and represented by horizontal bars in the graphs. Significance was determined by repeated measure ANOVAs with Dunnett’s multiple comparison tests of the Teee population against the other CD38/CD39 quadrants.
Teee are present in melanoma patient tumors.
We next assessed the presence of Teee within tumor infiltrating lymphocytes (TIL) by flow cytometry of two freshly resected melanoma tumors. A high frequency of CD38+CD39+ cells (Teee) were present in the parent CD3+CD4+CD127-GARP- population (Supplemental Figure 5). As observed for peripheral blood-derived Teee, tumor infiltrating Teee expressed high levels of TOX and low levels of TCF1 relative to the other CD38/CD39 populations in both samples.
A population of cells with a Teee gene signature is found in TIL from melanoma, NSCLC, and bladder cancer.
To further investigate tumor-infiltrating Teee, we utilized publicly available scRNA-seq datasets to determine if cells with a Teee gene signature were found in patient tumors. We used the 49 upregulated genes from Figure 1B in addition to CD4 and ENTPD1 to create a Teee gene signature. CD38 was included in the 49 overlapping genes and consequently included in the signature. The gene signature is available in the supplemental files accompanying this manuscript.
Figure 3A shows a UMAP projection of the CD4+ TIL concatenated from 31 baseline patients from the published melanoma dataset GSE115978, with coloring by expression of the Teee gene signature. Cells with a high Teee signature score predominantly composed cluster 9 (Figure 3B). Phenotypic analysis of the clusters showed that cells in cluster 9 had the highest expression of the proliferation associated genes MKI67 (Ki67), PCNA and TOP2A (Figure 3C). This cluster also had high levels of expression of CTLA4, HAVCR2 (LAG3), PDCD1 (PD1), TOX and CASP3 along with relatively low levels of TCF7 (TCF1). These phenotypes align with the Teee phenotypes observed in the RNA-seq and flow cytometry data of peripheral blood Teee.
Figure 3. Cells with a Teee gene signature are present in TIL from melanoma, NSCLC, and bladder cancer.
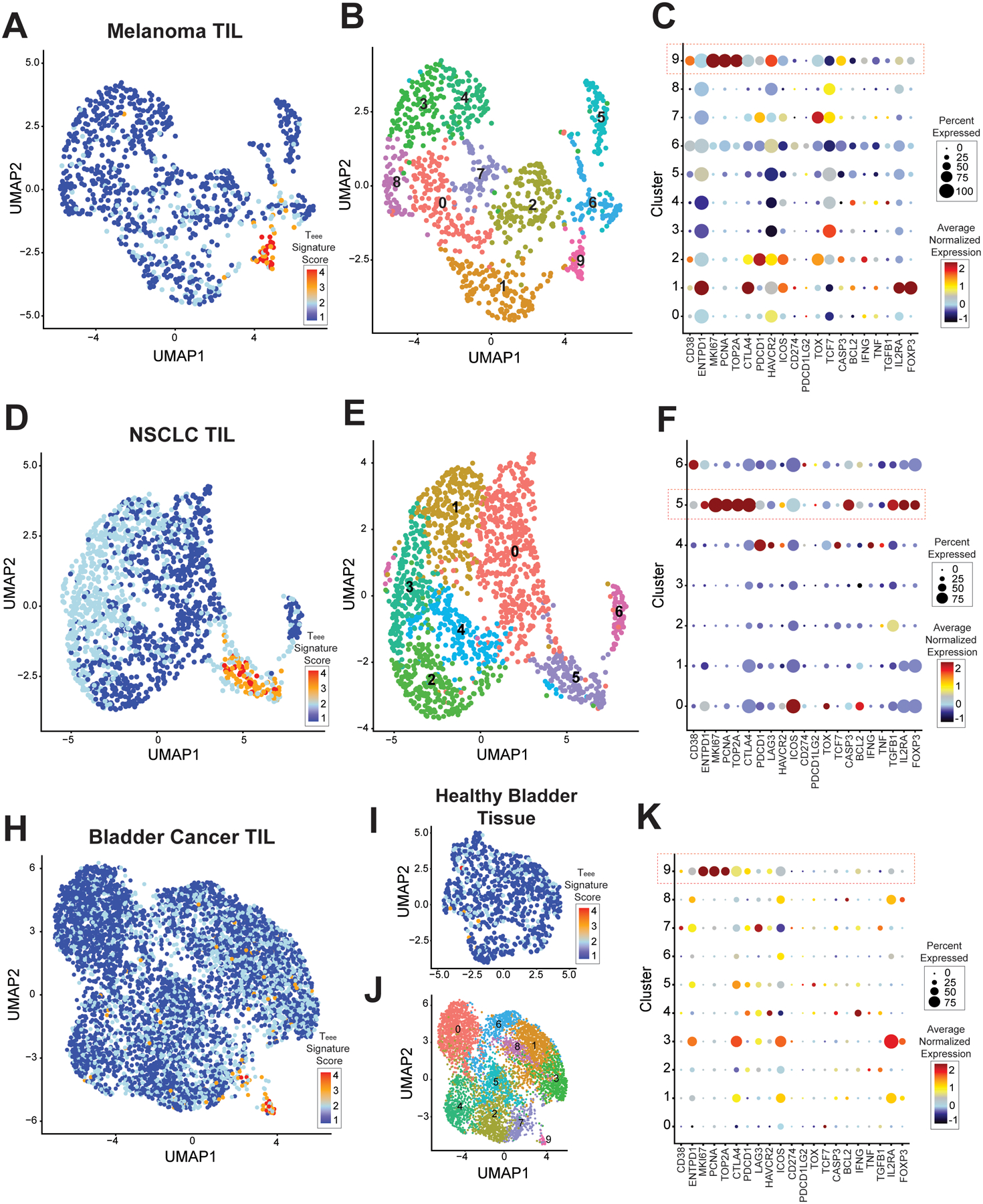
Publicly available single-cell RNA-seq datasets were analyzed for the presence of Teee cells. (A-C) Melanoma dataset GSE115978. (A) A UMAP plot of is shown of the CD3+CD4+ TIL concatenated across samples. Cells are colored by expression of the Teee signature genes, with higher expression scores shown in red. (B) The same UMAP plot is shown, colored by clusters. (C) A dot plot is shown with each row showing a cluster and the columns showing select genes. Genes with higher average expression in a cluster are colored red, genes with lower expression are colored blue, and genes with intermediate expression are colored yellow. Larger circle size denotes expression by higher proportion of cells within that cluster. (D-F) Non-small cell lung cancer dataset GSE179994. UMAP plots of CD3+CD4+ TIL concatenated across samples colored by (D) Teee signature and (E) clustering, (F) and a dot plot as above are shown. (H-K) Bladder cancer dataset GSE149652. UMAP plots of CD3+CD4+ TIL concatenated across samples colored by (H) Teee signature and (J) clustering, (K) and a dot plot as above are shown. (I) A UMAP plot of CD3+CD4+ cells from adjacent healthy bladder tissue is shown. Cells are colored based on Teee signature expression.
We next assessed CD4+ TIL concatenated from 8 baseline and 14 post-treatment patient samples in a NSCLC dataset (GSE179994). Cells with a high Teee signature score predominantly composed cluster 5 (Figure 3D, and 3E). As in the melanoma dataset, this cluster had high frequencies and expression levels of proliferation associated genes (MKI67, PCNA, TOP2A), co-inhibitory genes (CTLA4, PDCD1), and CASP3 (Figure 3F).
We also assessed CD4+ TIL concatenated from 7 baseline patients in a bladder cancer dataset (GSE149652). Figure 3H shows a UMAP of with CD4+ TIL shown with coloring by expression of the Teee gene signature. Cells with a high Teee signature score predominantly composed cluster 9 (Figure 3J). As in the melanoma and NSCLC datasets, the Teee signature cluster (cluster 9) of the TIL had high expression of proliferation associated genes (MKI67, PCNA, TOP2A), co-inhibitory genes (CTLA4, PDCD1), and CASP3 (Figure 3K). In addition to cells infiltrating tumor, this dataset included adjacent healthy bladder tissue. Assessment of the Teee signature in the CD4+ T-cells of the healthy bladder tissue did not demonstrate any clustering of cells with expression of the Teee signature (Figure 3I).
Tumor infiltrating Teee are distinct from Tregs.
We previously showed that peripheral blood Teee were distinct from regulatory T-cells (Tregs), with Tregs being predominantly CD38-CD39+ and Teee having limited expression of FOXP3 and CD255. To determine if intra-tumor Teee were also distinct from Tregs, we compared the gene expression of Teee clusters to Treg clusters (those with the highest expression of FOXP3 and IL2RA). While positive correlations in gene expression were observed between Teee clusters in the different datasets, negative correlations of Teee with Tregs were observed within and between datasets (Supplemental Figure 6A). Differential gene expression analysis comparing Teee and Treg clusters in each dataset showed numerous differences (Supplemental Figure 6B-E). Supplemental files containing the DEGs for these comparisons accompany this manuscript.
CD3+CD4+CD38+CD39+ cells are present in melanoma tumor tissue and are adjacent to Ki67- CD8+ TIL.
Having identified Teee in melanoma tumors by flow cytometry and cells with a Teee gene signature in various solid tumors in scRNA-seq datasets, we next sought to identify Teee in situ within tumors and determine their relation to CD8+ TIL. To accomplish this, we utilized multispectral immunofluorescence imaging to evaluate FFPE baseline tumor tissue from metastatic melanoma patients treated with checkpoint inhibition. The marker set CD3+CD4+CD38+CD39+ was used to define Teee. CD127 and GARP were not included in the panel due to technical limitations on the number of markers. A representative image of single stains and the merged set is shown in Figure 4A. Teee were identifiable in melanoma tumors as shown by the magenta-colored cells in Figure 4B. Sample information on patient demographics, tumor, and Teee frequencies can be found in Supplemental Table 5. We were also able to identify CD3+CD8+ T-cells with or without expression of the proliferation marker Ki67, illustrated with cyan and orange coloring, respectively, in the representative image. CD8+ T-cells had overall high expression of Ki67 (Figure 4C), but CD3+CD8+ T-cells in contact with Teee comprised a higher percentage of the Ki67- CD8+ population than in the Ki67+ population (Figure 4D; 95% CI −12.25% to −2.811%, p=0.0033, n=22).
Figure 4. CD3+CD4+CD38+CD39+ cells found in the melanoma tumor microenvironment are associated with Ki67- CD8+ T-cells.
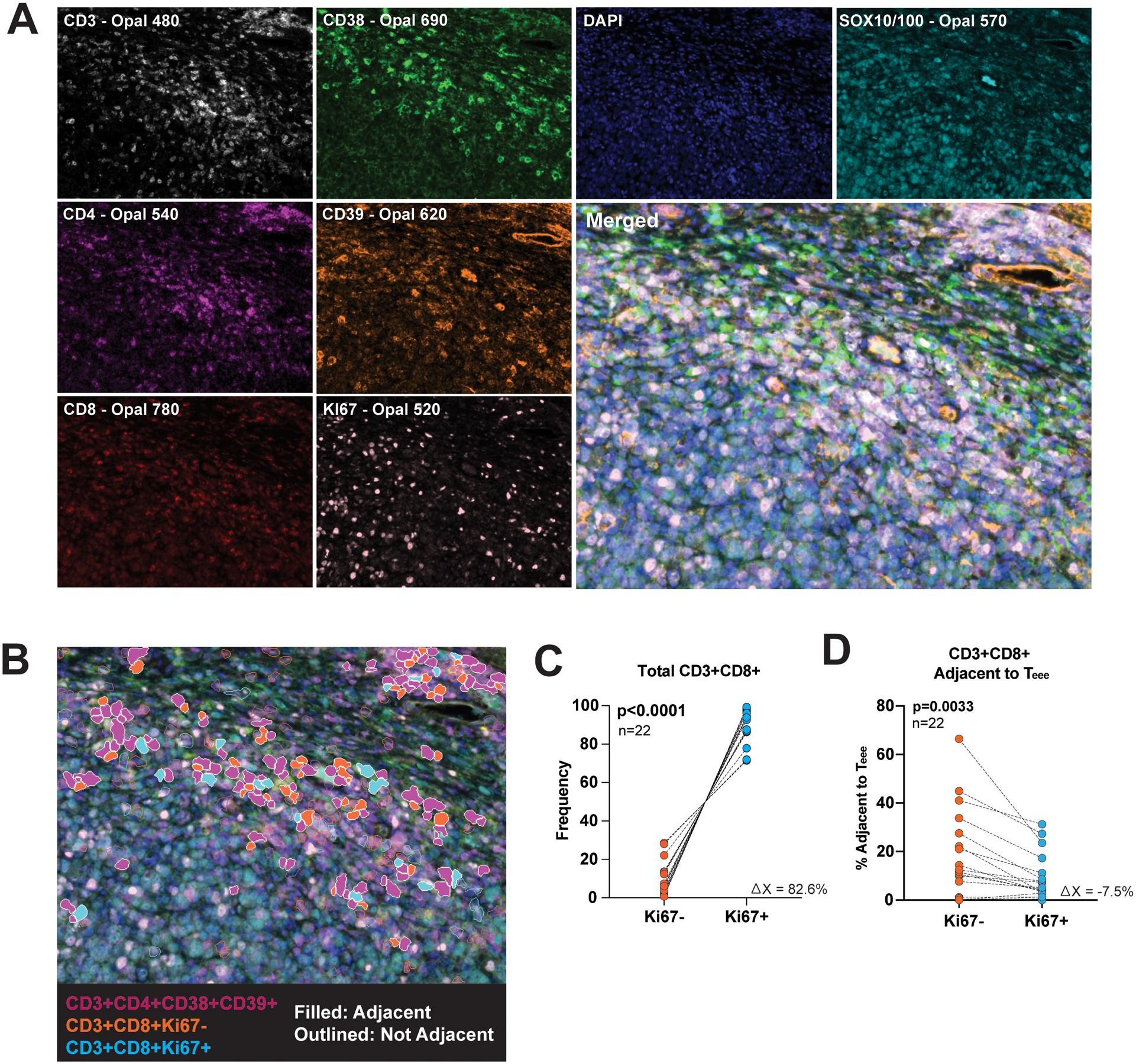
Baseline tumor tissue FFPE samples from metastatic melanoma patients treated with checkpoint inhibition were evaluated by multiplex immunofluorescent imaging. (A) Representative images of CD3 (white), CD38 (green), DAPI (blue), SOX10/100 (cyan), CD4 (purple), CD39 (orange), CD8 (red), and Ki67 (pink) staining, along with the merged image are shown. (B) Cells expressing the marker set CD3+CD4+CD38+CD39+ are artificially colored purple, CD3+CD8+Ki67- cells colored orange and CD3+CD8+Ki67+ colored blue. CD3+CD4+CD38+CD39+ and CD3+CD8+ cells in contact with one another are filled in by color and those not in contact are only outlined. (C) The frequency of Ki67- and Ki67+ CD3+CD8+ cells in each specimen are plotted. Each dot pair represents one patient. (D) The frequency of CD3+CD8+Ki67- and CD3+CD8+Ki67+ adjacent to CD3+CD4+CD38+CD39+ cells as a percent of the parent Ki67−/+ populations are plotted. Each dot pair represents one patient. The corresponding p-value was determined by a paired t-test.
Teee suppress autologous T-cell proliferation in vitro.
Given elevated peripheral blood frequencies of Teee were associated with immunotherapy resistance and we observed that CD8+ TIL adjacent to Teee were more likely to be Ki67-, we hypothesized that Teee may suppress T-cell proliferation. To address this, we co-cultured Teee with Cell Trace Violet labeled autologous CD3+CD25- T-cells in the presence of αCD3/CD28 stimulation. Teee suppressed the proliferation of both the CD4+ (Figures 5A & 5B; 95% CI difference of means = −15.16% to −0.38%, p=0.0432, n=5) and CD8+ (Figure 5C & 5D; 95% CI difference of means = −6.44% to −1.76%, p=0.0082, n=5) populations in all patient specimens evaluated. We also compared the proliferation of autologous target T-cells in the presence of the other CD3+CD4+CD127-GARP-CD38/CD39 populations. The presence of Teee significantly reduced expression of Ki67 in target T-cells compared to cultures with the CD38-CD39- (95% CI difference of means = −15.64% to −5.59%, p=0.0005) and CD38+CD39- (95% CI difference of means = −14.28% to −3.22%, p=0.0020) populations (Figure 5E). No significant difference was observed in comparisons of Teee co-cultures against those with CD38-CD39+ population (95% CI difference of means = −6.25% to 4.80%, p=0.9889).
Figure 5. Teee suppress proliferation of autologous T-cells in vitro.
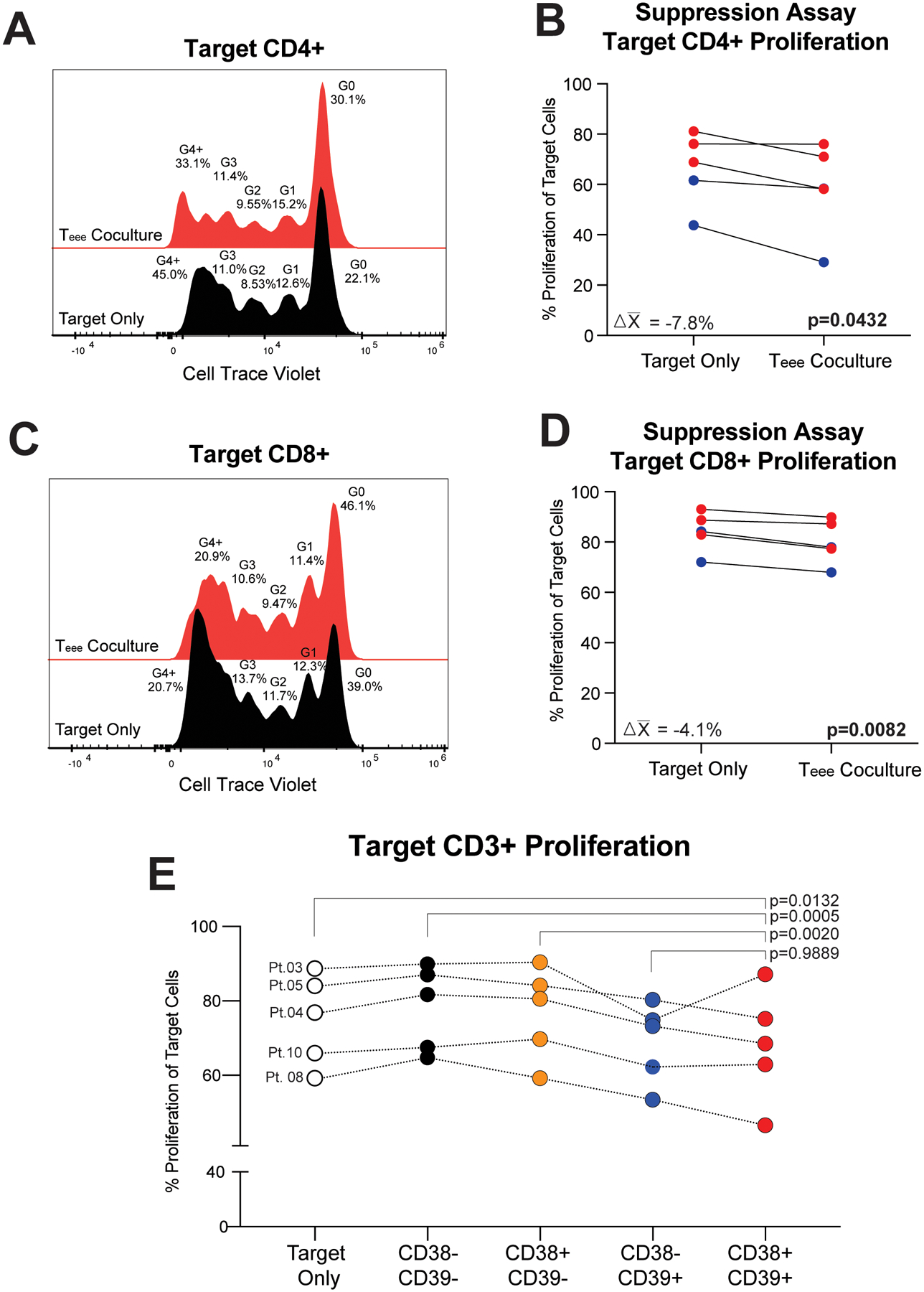
Baseline peripheral blood cells from metastatic melanoma patients were flow sorted for Teee. Autologous bulk CD3+ T-cells as targets were stained with Cell Trace Violet, activated by plate bound αCD3 and soluble αCD28, and cultured alone or in the presence of Teee at a target to Teee ratio of 2:1. (A) A representative plot of Cell Trace Violet is shown for target CD4+ T-cells alone (black histogram) or in the presence of Teee (red histogram). (B) The percentages of proliferating target CD4+ T-cells for five samples evaluated are shown. Lines connect paired patient samples. P-values were determined by paired t-tests. Mean changes in the percentage of proliferating target cells are given in the corresponding panel. Red coloring indicates specimens from non-responding patients and blue coloring represents responding patients. (C,D) Target CD8+ T-cells are likewise shown. (E) Target CD3+ T-cells were cultured alone or in the presence of autologous CD3+CD4+CD127-GARP-CD38-CD39- (black dots), CD3+CD4+CD127-GARP-CD38+CD39- (orange dots), CD3+CD4+CD127-GARP-CD38-CD39+ (blue dots), or CD3+CD4+CD127-GARP-CD38+CD39+ (Teee, red dots). The percentages of proliferating target CD3+ T-cells for five samples evaluated are shown. Lines connect paired patient samples. P-values were determined by repeated measures ANOVA and Dunnett’s posthoc tests.
Elevated frequencies of tumor-infiltrating cells with the Teee gene signature are associated with immunotherapy resistance in metastatic melanoma.
As Teee from the peripheral blood of metastatic melanoma patients were associated with checkpoint inhibitor resistance and suppressed autologous T-cell proliferation, we addressed if Teee present in the tumor microenvironment were also associated with checkpoint inhibitor resistance. We analyzed a publicly available, clinically annotated scRNA-seq dataset of TIL from 19 baseline metastatic melanoma patients treated with checkpoint inhibitor immunotherapy (GSE120575). Patient demographics and treatment information is given in Supplemental Table 8. Figures 6A and 6B show that a discrete cluster of cells, cluster 6, had high expression of the Teee gene signature. Analysis of gene expression in this cluster showed the phenotype observed previously with Teee, including high expression of proliferation-associated genes (MKI67, PCNA, TOP2A), co-inhibitory markers (CTLA4, PD1), and CASP3 (Figure 6C). These cells also had high expression of TOX and low expression of TCF7. We next colored cells by patient response, with non-responding patients in red and responding patients in blue, and found that cluster 6 was predominantly populated by cells from non-responding patients (Figure 6D). Non-responding patients had higher mean frequencies of Teee relative to responding patients (Figure 6E; 95% CI of difference between means −19.8% to −4.2%, p=0.0064, n=10 & n=9). Patients with greater than median frequencies of Teee positive cells likewise had shorter overall survival compared to patients frequencies below the median (Figure 6F; p=0.039).
Figure 6. Tumor-infiltrating Teee cells are associated with immunotherapy resistance in metastatic melanoma.

The publicly available single-cell RNA-seq melanoma dataset GSE120575 was assessed for the association of tumor-infiltrating Teee and patient outcomes. (A) A UMAP plot is shown of the CD3+CD4+ TIL concatenated across patient samples. Cells are colored by expression of the Teee signature genes, with higher expression scores shown in red. (B) The same UMAP plot is shown, colored by clusters. (C) A dot plot is shown with each row showing a cluster and the columns showing select genes. Genes with higher average expression in a cluster are colored red, genes with lower expression are colored blue, and genes with intermediate expression are colored yellow. Larger circle size denotes expression by higher proportion of cells within that cluster. (D) A UMAP plot is shown as before. Cells from non-responding patients are colored red and cells from responding patients are colored blue. (E) The frequency of Teee (CD38+CD39+) cells as a percentage of the CD4+ T-cell population for each baseline patient sample are plotted, comparing non-responding and responding patients. The corresponding p-value was determined by a Welch’s t-test. (F) Patients were divided based on median frequency of tumor infiltrating Teee at baseline and survival assessed. Patients with lower than median frequency of Teee are shown in blue and patients with higher than median frequency in red. The corresponding p-value was determined by Cox proportional hazards regression. (G-I) The frequency of Teee as a percentage of the CD4+ T-cell population is plotted against the frequency of CD8+ T-cells expressing (G) MKI67, (H) TOX, and (I) TCF7. Non-responding patients are represented with red circles and responding patients with blue triangles. Unadjusted R2 and p-values were determined by linear regression ANOVAs.
We also assessed the frequency of tumor infiltrating Teee in relation to tumor infiltrating CD8+ cells. While we did not observe a significant relationship between Teee and the frequency of CD8+ T-cells expressing MKI67 (Figure 6G), all responding patients had relatively low frequencies of both CD8+MKI67+ cells and Teee. There was a significant positive association of Teee frequency and the percentage of CD8+ T-cells expressing TOX (Figure 6H) (R2=0.2232 , p=0.0411) and a negative association of Teee frequency and the percentage of CD8+ T-cells expressing TCF7 (Figure 6I) (R2=0.501 , p=0.0007).
Discussion.
In this study we characterized a population of CD3+CD4+CD127-GARP-CD38+CD39+ cells and demonstrated that Teee were present in the tumor microenvironment and increased frequencies of intratumoral Teee were predictive of shorter overall survival in metastatic melanoma patients. We found that distinct clusters of cells with a Teee gene signature were present in various solid tumors and that the gene expression patterns of these clusters was highly correlated across datasets.
Teee expressed high levels of markers associated with proliferation, co-inhibitory receptors/ligands, and exhaustion markers. These phenotypes were consistent across Teee characterized in the PBMC and Teee detected in the scRNA-Seq datasets of various tumors. Although Teee cells had low TCF1 and high TOX expression, a phenotype consistent with T-cell exhaustion30, they had upregulation of proliferation markers and functioned to suppress autologous T-cells in vitro. Teee also expressed high frequencies of CTLA4, TIM3, LAG3 and PD1. Teee had a predominately PD1high phenotype, which has been shown to correlate with reduced effector function31. Upregulation of an exhaustion phenotype and co-inhibitory receptors have been associated with adenosine signaling in T-cells13,14, and the observed phenotype may be a downstream consequence of enhanced adenosine generation through CD38 and CD39, which are defining markers of Teee. In addition to expressing co-inhibitory receptors, Teee expressed PDL1. While canonically expressed by innate immune cells and tumor cells, we previously showed that T-cell expression of PDL1 was associated with immunotherapy resistance32,33 and that PDL1 expressed by T-cells suppressed autologous T-cell effector functions33.
Teee did not display gene expression patterns associated with known CD4+ T-cell polarizations. In the bulk RNA-seq data, Teee and the other CD38/CD39 populations had greater variability in expression of transcription factor genes (i.e. TBX21, RORC, GATA3, FOXP3, BCL6, IKZF2) and polarization associated cytokines (i.e. IL2, TNF, IL23A, IL17B, TGFB1, IL17C, IFNG, IL10, IL21, IL4) between patient samples than between the CD38/CD39 populations. We previously showed the Teee were distinct from Tregs, with the majority of Tregs having a CD38-CD39+ phenotype and the majority of Teee being FOXP3- and/or CD25-. The data in this study provide additional evidence of Teee being distinct from Tregs. Interestingly, while Teee had relatively lower expression of IL2RA and FOXP3 in both melanoma scRNA-seq datasets and the bladder cancer dataset, the Teee cluster in the NSCLC dataset had the highest level of expression of both these genes. One possible explanation is that this cluster contained a mix of both Teee and Tregs. Additionally, the data presented do not address how Teee originate; it is possible that Teee develop from a population of Tregs.
We also observed phenotypes associated with apoptosis in the Teee population. In bulk RNA-Seq data, Teee had significant upregulation of CASP3 (caspase 3 gene). We found similar increases in other caspase genes, but not CASP8. This suggests that Teee had activation of the intrinsic but not the extrinsic death pathway. Teee from the peripheral blood also downregulated the anti-apoptotic gene BCL2. Upregulation of CASP3 and decreased BCL2 were likewise observed in the Teee clusters from the scRNA-Seq tumor datasets. High frequencies of Teee expressing annexin V were seen in the flow cytometry data, consistent with the apoptotic gene expression profile seen. In the analysis of Teee in the bulk RNA-Seq data, the gene CDT1 had high expression in the Teee population. CDT1 regulates the early stage of DNA replication, and deviations in its expression causes genome instability34. This may be a contributor to the observed apoptosis. Reconciling the observed high proliferation observed in Teee, it is also possible that Teee are experiencing activation induced death35.
In comparisons of single positive populations (i.e. CD38+CD39- and CD38-CD39+) to double positive (Teee), Teee most closely resembled the CD38-CD39+ population, with the fewest differentially expressed genes between the two populations. However, Teee clustered discretely and had distinct differences in expression of many markers including increased Ki67, TOX, CTLA4, PD1high, TIM3 and PDL1. The phenotype of the CD38+CD39- population was distinct from Teee, with low levels of annexin V, co-inhibitory receptors and TOX, and high levels of TCF1. CD38+CD39- cells also had intermediate expression of Ki67. The CD38-CD39- population had the greatest number of differentially expressed genes when compared to Teee. This population had low expression of most markers evaluated except for TCF1, CD73 and CD26. Collectively, the analysis of the CD38/CD39 populations suggest that the four populations have distinct phenotypes. These observed differences in phenotypes across the CD38/CD39 populations raises the question of the role(s) each population plays in the anti-tumor immune responses and the plasticity of each of these populations.
The phenotype of Teee includes co-expression of CD38 and CD39, suggesting a phenotype associated with adenosine generation. We also observed low levels of CD26 expression by Teee. CD26 is a surface enzyme that, when bound to adenosine deaminase (ADA), degrades adenosine to inosine36. Interestingly, high gene expression of ADA was seen in Teee in the overlapping list of DEGs. Low levels of CD73, the enzyme at the intersection of the CD38 and CD39 pathways producing adenosine, was also observed in the Teee population. These two phenotypes, increased ADA and decreased CD73, are contrary to what would be expected in association with adenosine production. However, CD73 may be present on tumor and/or other immune cells within the tumor microenvironment37,34, contributing to adenosine generation.
Supporting a possible mechanistic link of Teee with immunotherapy resistance, Teee derived from melanoma patient PBMCs were able to suppress autologous T-cell proliferation in vitro. The suppression occurred without a source of adenosine precursors, which may have limited suppression by Teee. This suggests that suppression may be mediated by mechanisms other than or in addition to adenosine generation (e.g. PDL1). In FFPE specimens we also observed a preferential association of Teee in contact with Ki67- CD8+ TIL. Additionally, while no correlation of intra-tumor Teee frequency with CD8+ TIL MKI67 expression was observed, correlations were observed with CD8+ TIL exhaustion phenotypes (i.e. TOXhigh and TCF7low). Interestingly, in this scRNA-seq dataset, all responding patients had both low frequencies of MKI67 expressing CD8+ TIL and low Teee frequencies. It’s possible that high levels of CD8 proliferation may be indicative of exhaustion. It is important to note that the observed correlations with Teee frequency and CD8+ T-cell exhaustion phenotypes may be consequence of a shared cause and not directly mechanistically related. However, taken together, the data in this study suggest a mechanistic role of Teee in immunotherapy resistance.
While the data presented herein suggest a mechanistic role of Teee as a predictive biomarker of response to checkpoint inhibition therapy, there are limitations that should be considered. In the current study, patient specimens and public datasets were analyzed retrospectively. To confirm Teee as predictive of patient outcomes, a prospective analysis of a large set of baseline samples from patients treated with checkpoint inhibitors should be employed. Patients with malignancies other than melanoma and patients treated with other therapies should also be evaluated. This will determine if Teee frequencies in the periphery and/or in the tumor microenvironment are predictive or prognostic and extend the findings of the study beyond metastatic melanoma. Additionally, in the present study, paired healthy tissue was only available/assessed in the bladder cancer dataset. While these data suggest the absence of Teee from healthy tissue, assessment of additional healthy tissue is needed to address this hypothesis. Finally, further studies assessing the mechanism of suppression need to be performed.
In this study we built upon our previous findings that elevated peripheral blood frequencies of Teee were associated with checkpoint inhibitor resistance in metastatic melanoma patients by characterizing the phenotype, function and location of those cells. We found that high intra-tumor frequencies of cells with a Teee phenotype were associated with checkpoint inhibitor resistance and poor overall survival. We also observed that Teee were present in NSCLC and bladder cancer. Teee had phenotypes known to be associated with therapeutic resistance and directly suppressed autologous T-cells. These results highlight Teee as a potential rational target to improve the efficacy of checkpoint inhibition.
Supplementary Material
Acknowledgements:
We extend our appreciation to the NYU Langone Cytometry and Cell Sorting Laboratory, the NYU Langone Genomics Core, the University of Colorado Anschutz Medical Campus Skin Cancer BioReposityory, and the University of Colorado Anschutz Medical Campus Human Immune Monitoring Shared Resource (HIMSR) for their services and assistance.
Funding:
NCI R00CA230201, NCI P50CA225450, NCI P30CA016087
Conflicts of Interest Disclosures:
Ankita Mitra: None
Brian Thompson: None
Ann Strange: None
Carol M. Amato: None
Igor Dolgalev: None.
Melinda Vassallo: None.
Jonathan Hester-McCullough: None.
Tomoaki Muramatsu: None.
Diana Kimono: None.
Amrutesh Puranik: None.
Jeffrey S. Weber: Received less than $10,000 dollars per annum from Merck, Genentech, Astra Zeneca, GSK, Novartis, Nektar, Celldex, Incyte, Biond, ImCheck, Sellas, Evaxion and EMD Serono and $10-25,000 dollars from BMS for membership on Advisory Boards; Equity in Biond, Evaxion, OncoC4, Instil Bio and Neximmune; on scientific advisory boards for CytoMx, Incyte, ImCheck, Biond, Sellas, Instil Bio OncoC4 and Neximmune and was remunerated between $10,000-$75,000 dollars; NYU, but not me personally, received research support from BMS, Merck, GSK, Moderna, Pfizer, Novartis and Astra Zeneca; Moffitt Cancer Center filed a patent on an IPILIMUMAB biomarker and on TIL preparation that I am named on, and Biodesix filed a PD-1 patent that I was named on; I receive less $6000 yearly in royalties
David Woods: Owns stock in BMS (less than $1000).
References.
- 1.Postow MA et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 372, 2006–2017 (2015). https://doi.org: 10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paz-Ares L et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol 22, 198–211 (2021). https://doi.org: 10.1016/S1470-2045(20)30641-0 [DOI] [PubMed] [Google Scholar]
- 3.Sharma P et al. Nivolumab Alone and With Ipilimumab in Previously Treated Metastatic Urothelial Carcinoma: CheckMate 032 Nivolumab 1 mg/kg Plus Ipilimumab 3 mg/kg Expansion Cohort Results. J Clin Oncol 37, 1608–1616 (2019). https://doi.org: 10.1200/JCO.19.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motzer RJ et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N Engl J Med 378, 1277–1290 (2018). https://doi.org: 10.1056/NEJMoa1712126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khushalani NI et al. Phase II clinical and immune correlate study of adjuvant nivolumab plus ipilimumab for high-risk resected melanoma. J Immunother Cancer 10 (2022). https://doi.org: 10.1136/jitc-2022-005684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods DM et al. Nivolumab and ipilimumab are associated with distinct immune landscape changes and response-associated immunophenotypes. JCI Insight 5 (2020). https://doi.org: 10.1172/jci.insight.137066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takenaka MC et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 22, 729–740 (2019). https://doi.org: 10.1038/s41593-019-0370-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Virgilio F, Sarti AC, Falzoni S, De Marchi E & Adinolfi E Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat Rev Cancer 18, 601–618 (2018). https://doi.org: 10.1038/s41568-018-0037-0 [DOI] [PubMed] [Google Scholar]
- 9.Elliott MR et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461, 282–286 (2009). https://doi.org: 10.1038/nature08296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allard B, Beavis PA, Darcy PK & Stagg J Immunosuppressive activities of adenosine in cancer. Curr Opin Pharmacol 29, 7–16 (2016). https://doi.org: 10.1016/j.coph.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 11.Skalhegg BS et al. Cyclic AMP-dependent protein kinase type I mediates the inhibitory effects of 3’,5’-cyclic adenosine monophosphate on cell replication in human T lymphocytes. J Biol Chem 267, 15707–15714 (1992). [PubMed] [Google Scholar]
- 12.Vang T et al. Activation of the COOH-terminal Src kinase (Csk) by cAMP-dependent protein kinase inhibits signaling through the T cell receptor. J Exp Med 193, 497–507 (2001). https://doi.org: 10.1084/jem.193.4.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mastelic-Gavillet B et al. Adenosine mediates functional and metabolic suppression of peripheral and tumor-infiltrating CD8(+) T cells. J Immunother Cancer 7, 257 (2019). https://doi.org: 10.1186/s40425-019-0719-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horenstein AL, Bracci C, Morandi F & Malavasi F CD38 in Adenosinergic Pathways and Metabolic Re-programming in Human Multiple Myeloma Cells: In-tandem Insights From Basic Science to Therapy. Front Immunol 10, 760 (2019). https://doi.org: 10.3389/fimmu.2019.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minguet S et al. Adenosine and cAMP are potent inhibitors of the NF-kappa B pathway downstream of immunoreceptors. Eur J Immunol 35, 31–41 (2005). https://doi.org: 10.1002/eji.200425524 [DOI] [PubMed] [Google Scholar]
- 16.Ryzhov S et al. Adenosinergic regulation of the expansion and immunosuppressive activity of CD11b+Gr1+ cells. J Immunol 187, 6120–6129 (2011). https://doi.org: 10.4049/jimmunol.1101225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorrentino C, Miele L, Porta A, Pinto A & Morello S Myeloid-derived suppressor cells contribute to A2B adenosine receptor-induced VEGF production and angiogenesis in a mouse melanoma model. Oncotarget 6, 27478–27489 (2015). https://doi.org: 10.18632/oncotarget.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beavis PA et al. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc Natl Acad Sci U S A 110, 14711–14716 (2013). https://doi.org: 10.1073/pnas.1308209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou JZ et al. Differential impact of adenosine nucleotides released by osteocytes on breast cancer growth and bone metastasis. Oncogene 34, 1831–1842 (2015). https://doi.org: 10.1038/onc.2014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi Y et al. Blockade of Adenosine A2b Receptor Reduces Tumor Growth and Migration in Renal Cell Carcinoma. J Cancer 11, 421–431 (2020). https://doi.org: 10.7150/jca.31245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber JS et al. Safety, efficacy, and biomarkers of nivolumab with vaccine in ipilimumab-refractory or -naive melanoma. J Clin Oncol 31, 4311–4318 (2013). https://doi.org: 10.1200/JCO.2013.51.4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber JS et al. Sequential administration of nivolumab and ipilimumab with a planned switch in patients with advanced melanoma (CheckMate 064): an open-label, randomised, phase 2 trial. Lancet Oncol 17, 943–955 (2016). https://doi.org: 10.1016/S1470-2045(16)30126-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerby-Arnon L et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 175, 984–997 e924 (2018). https://doi.org: 10.1016/j.cell.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat Cancer 3, 108–121 (2022). https://doi.org: 10.1038/s43018-021-00292-8 [DOI] [PubMed] [Google Scholar]
- 25.Oh DY et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 181, 1612–1625 e1613 (2020). https://doi.org: 10.1016/j.cell.2020.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sade-Feldman M et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020 (2018). https://doi.org: 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson AM et al. Cancer Cell-Specific Major Histocompatibility Complex II Expression as a Determinant of the Immune Infiltrate Organization and Function in the NSCLC Tumor Microenvironment. J Thorac Oncol 16, 1694–1704 (2021). https://doi.org: 10.1016/j.jtho.2021.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan O et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218 (2019). https://doi.org: 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1 (2016). https://doi.org: 10.1126/sciimmunol.aai8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blank CU et al. Defining ‘T cell exhaustion’. Nat Rev Immunol 19, 665–674 (2019). https://doi.org: 10.1038/s41577-019-0221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thommen DS et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 24, 994–1004 (2018). https://doi.org: 10.1038/s41591-018-0057-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacquelot N et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat Commun 8, 592 (2017). https://doi.org: 10.1038/s41467-017-00608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woods DM et al. Decreased Suppression and Increased Phosphorylated STAT3 in Regulatory T Cells are Associated with Benefit from Adjuvant PD-1 Blockade in Resected Metastatic Melanoma. Clin Cancer Res 24, 6236–6247 (2018). https://doi.org: 10.1158/1078-0432.CCR-18-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pozo PN & Cook JG Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes (Basel) 8 (2016). https://doi.org: 10.3390/genes8010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green DR, Droin N & Pinkoski M Activation-induced cell death in T cells. Immunol Rev 193, 70–81 (2003). https://doi.org: 10.1034/j.1600-065x.2003.00051.x [DOI] [PubMed] [Google Scholar]
- 36.Dong RP et al. Determination of adenosine deaminase binding domain on CD26 and its immunoregulatory effect on T cell activation. J Immunol 159, 6070–6076 (1997). [PubMed] [Google Scholar]
- 37.Chen S et al. CD73: an emerging checkpoint for cancer immunotherapy. Immunotherapy 11, 983–997 (2019). https://doi.org: 10.2217/imt-2018-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA-seq reads generated for this project have been deposited at NCBI Gene Expression Omnibus (GEO) at accession number GSE224099. Code related to the analysis of the RNA-seq data is available at https://github.com/SciOmicsLab/Ectoenzyme_Tcells. Supplementary files containing differentially expressed genes and the Teee gene signature accompany this manuscript.


