Abstract
Long ignored as a vestigial remnant of cytokinesis, the mammalian midbody (MB) is released post-abscission inside large extracellular vesicles called MB remnants (MBRs). Recent evidence suggests that MBRs can modulate cell proliferation and cell fate decisions. Here, we demonstrate that the MB matrix is the site of ribonucleoprotein assembly and is enriched in mRNAs that encode proteins involved in cell fate, oncogenesis, and pluripotency, that we are calling the MB granule. Both MBs and post-abscission MBRs are sites of spatiotemporally regulated translation, which is initiated when nascent daughter cells re-enter G1 and continues after extracellular release. MKLP1 and ARC are necessary for the localization and translation of RNA in the MB dark zone, whereas ESCRT-III was necessary to maintain translation levels in the MB. Our work reveals a unique translation event that occurs during abscission and within a large extracellular vesicle.
Keywords: midbody, MB, midbody remnant, MBR, large extracellular vesicle, EV, MBsome, mitosis, cytokinesis, abscission, intercellular bridge, RNP granule, Jun, Fos, KLF4, Arc, ESCRT-III, RNA-binding protein, RBP, RNA granule, translation, asymmetric cell division, intercellular communication
eTOC blurb
Midbodies (MBs) are released post-abscission as large extracellular vesicles called MB remnants (MBRs). Here, we demonstrate that the MB matrix is the site of ribonucleoprotein assembly and is enriched in mRNAs that encode proteins involved in cell fate, oncogenesis, and pluripotency, that we are calling the MB granule, which is unique in that it is translationally active.
Graphical Abstract
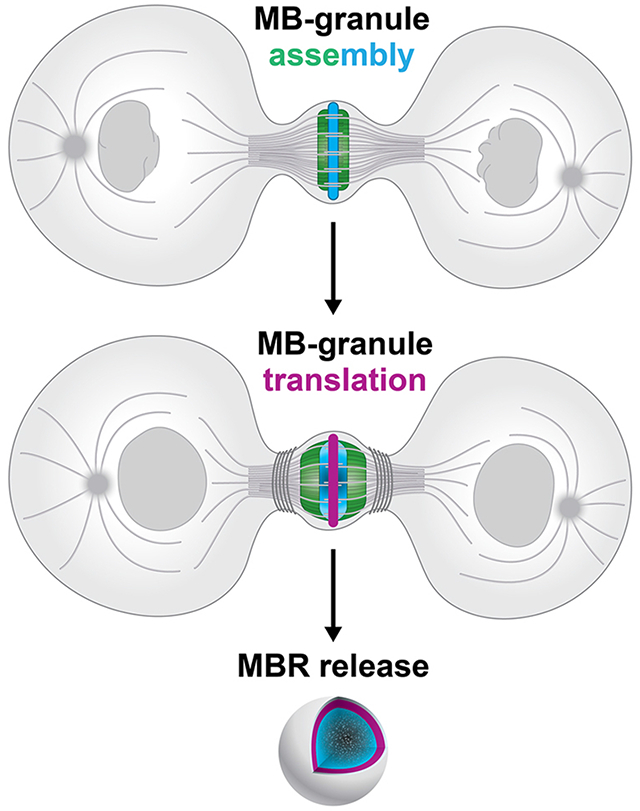
Introduction
The midbody (MB) is a protein-rich structure assembled during mitosis at the overlapping plus ends of spindle microtubules, where it recruits and positions the abscission machinery that separates dividing cells1–13. Long thought to be quickly internally degraded in daughter cell lysosomes, recent studies revealed that a majority of MBs are released extracellularly as membrane-bound particles, or extracellular vesicles, following bilateral abscission from nascent daughter cells10,14–16. Released post-abscission MB remnants (MBRs) are bound and tethered by neighboring cells, internalized, and can persist in endosomal compartments for up to 48 hours as signaling organelles (termed MB-containing endosomes or MBsomes) before being degraded by lysosomes10,12,17,18. Distinct cell types, including cancer and stem cells, exhibit differing avidities for internalizing MBRs 11,19, and exogenous addition of MBRs correlates with increased proliferation and tumorigenic behavior11,12,20. MBRs have been implicated in specifying apicobasal polarity and lumenogenesis in epithelia21; specifying primary cilium formation22, neurite formation23, and dorsoventral axis formation in Caenorhabditis elegans embryos24; and specifying stem cell pluripotency25. The functional importance of MBR signaling in the regulation of cell behavior, architecture, and fate is an emerging field, but the signaling mechanisms are only beginning to be understood.
MB structure and composition suggest mechanistic insights. Proteomic analyses of mitotic MBs and MBRs revealed enriched levels of large numbers (approximately 100) of RNA-binding proteins, ribosomal and translational regulators, and RNA-processing proteins1,12,20,26,27, several of which have been implicated in phase-separated condensate formation, but the functional significance was unclear. Given these data, we hypothesized that RNA and ribonucleoprotein (RNP) complexes may play unappreciated structural and/or functional roles in MB biology. Supporting this, a population of polypurine-repeat-containing long non-coding RNAs were localized to the MB28, but the identities and functions of these RNAs remain unknown. In the central core of the MB lies the MB matrix4,29–31, a structure of unknown composition. It appears as a prominent electron-dense stripe in electron micrographs30,32, similar to other membrane-less organelles33–36; under polarized light it is birefringent37, that is, with a refractive index sharply distinct from the surrounding cytoplasm. Whether RNA plays any role in MB structure or function remains unknown and is the subject of this study.
Here, we further define the structural components, organization, and behavior of MBs throughout their uniquely complex life cycle. Using a quantitative transcriptomic approach, we identified a population of mRNAs enriched in mitotic MBs and confirmed their presence in MBRs released by abscission. We demonstrate that the MB is the assembly site of an RNP granule. We show the biochemical activities of MBs are temporally coupled to cell cycle status: MBs initiate translation of stored mRNAs in late telophase as pre-abscission daughter cells re-enter G1 of the cell cycle and continue translation following abscission. Last, we found that MKLP1 and ARC play a role in promoting the assembly and maintenance of RNA aggregates and active translation at the MB. In contrast, ESCRT-III is necessary for the modulation of translation levels. We present a model in which the assembly and transfer of RNP complexes are central to post-mitotic MBR function and suggest a unique mode of intercellular communication via extracellular vesicles with defined biogenesis that is coupled to abscission and inherently links cell division status with signaling capacity.
Results
Midbodies (MBs) and midbody remnants (MBRs) are sites of RNA storage.
An MB-enriched transcriptome was identified using a comparative genomics approach. Note that these pre-abscission MBs are distinct from isolated post-abscission MBRs, whose proteome has been recently reported26. Here, three cell cycle-specific RNA-Seq libraries were prepared from synchronized Chinese Hamster Ovary (CHO) cell populations in interphase, metaphase, and MB/intercellular bridge stage (Fig. 1A), similar methods to our cell cycle proteome published earlier38,39. Specifically, whole-cell tubulin, metaphase spindles, and MB spindle microtubules were harvested from CHO cells at interphase, metaphase, and MB stage, respectively, and mRNAs associated with these structures were isolated and sequenced using our previously published methods1. Comparative analysis identified 22 transcripts enriched in the MB stage relative to total mRNAs associated with metaphase microtubules, with enrichment defined as reads per kilobase million (RPKM) values greater than 1.0 and defined enrichment as 2-fold more reads in the midbody compared to metaphase spindles (Fig. 1B; Supp. Tables 1–4). Gene ontology analysis identified that the 22 transcripts encoded factors that function in cell fate, cell cycle, RNA processing, and signal transduction (Fig. 1C). Remarkably, a majority of these 22 RNAs encodes proteins expressed during late telophase and in the MBR in HeLa cells (Fig. S1A). To our surprise, one transcript encoded a critical regulator of cytokinesis, the Centralspindlin component of kinesin Kif23/Mklp1/CHO129,40. Another transcript encoded a member of the TIS11 family of RNA-binding proteins, Zfp36/TIS11, which has been implicated in regulating RNA stability and RNP granule function in multiple contexts41–43. Perhaps more surprising were the 10-transcription factor-encoding mRNAs identified that are implicated in proliferation, pluripotency, cell fate, cell death, and oncogenesis and that did not have a reported role in cytokinesis (Fig. 1C; Supp. Table 1). In addition, re-evaluation of 99 previously identified RNA binding proteins identified in the midbody proteome 1, suggest that these RBPs might perform multiple functions in nucleic acid binding, post-mitotic cell fate functions, cell division, proliferation, and development (Fig. S1B–C).
Figure 1: Midbodies and midbody remnants are sites of RNA storage. MKLP1 and ARC are necessary for mRNA localization and maintenance in the dark zone.
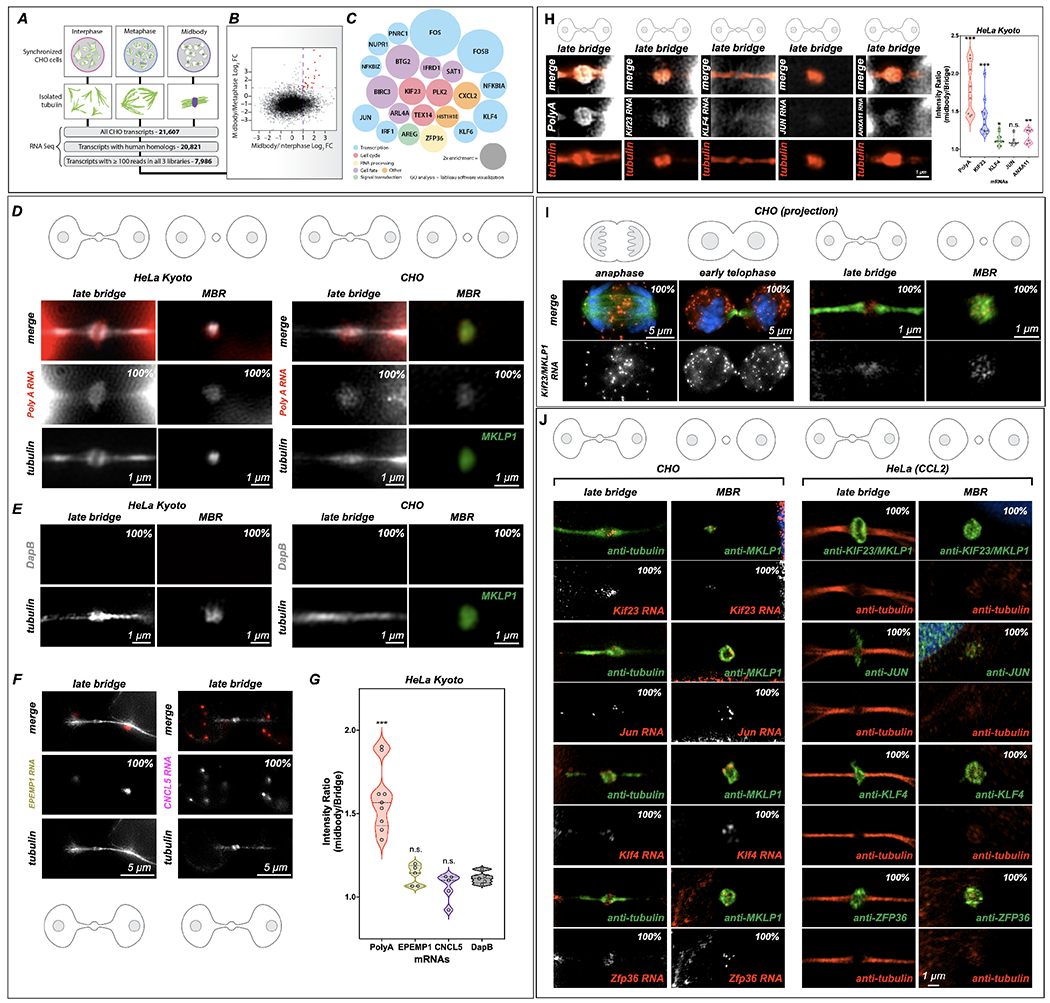
(A-C) RNA-Seq analysis of the MB transcriptome. mRNA was sequenced from three stages of the cell cycle: interphase, metaphase, and late cytokinesis (or “MB stage”). Tubulin structures were purified, and associated RNAs were isolated and analyzed by RNA-Seq. Of 21,607 distinct CHO transcripts identified, 20,821 could be annotated by gene ontology. Of those, 7,986 had ≥100 reads in all cell cycle stages and were further analyzed as plotted in (B). Raw data can be found in Supp.Tables 1–4.
(B) Transcripts with ≥100 reads in all three populations were compared and plotted based on their log2 enrichment scores (RPKM/RPKM). Dotted lines at x = 1 and y = 1 indicate minimum values for 2-fold enrichment. The 22 transcripts enriched in the MB relative to both interphase and metaphase are highlighted in red.
(C) Enrichment score (relative diameter) and gene ontology groups of the 22 MB-enriched transcripts; colors correspond to gene ontology biological process terms (Fig. 1; see also Supp. Tables 1–4).
(D) Single-molecule RNAscope (RNA in situ) hybridization revealed mRNA enrichment in the MB and released MBRs. PolyA-positive mRNAs (red) localized to mitotic MBs and post-mitotic MBRs in both CHO and HeLa Kyoto cells, in contrast with the bacterial DapB negative control (E).
(E) The bacterial DapE was not found at the midbody in HeLa Kyoto or CHO cells midbodies or MBRs.
(F-G) Two mRNAs. EPEMP1 and CNCL5, identified from interphase enriched RNAseq data (see Table 4) were not enriched in the midbody. *Significance was determined and denoted by * or n.s. n.s denotes not significant.
(H) Localization of PolyA, MKLP1, KLF4, JUN and ANXA11 to the midbody in HeLa Kyoto cells. Here we observed an enrichment of PolyA and MKLP1 RNAs but less so the other transcription factors, KLF4 and JUN, and ANXA11 (plot). *Significance was determined by comparing data to DapB 1E. n.s. denotes not significant. See S2C for CHO cell RNAscope quantification for similar probes.
(I) mRNA encoding Kif23, an MB-resident kinesin required for abscission, localized to the spindle overlap from anaphase through abscission; however, in early telophase, Kif23/MKLP1 was also found in the cytoplasm in distinct puncta as well as at the MB dark zone. In late telophase (or G1), puncta were found in the dark zone but were also highly enriched in cell bodies; the released MBR contained Kif23/MKLP1 NA molecules; tubulin is shown in green.
(J) mRNAs identified as MB-enriched by RNA-Seq co-localized to the MB and MBR in CHO cells. In HeLa cells, their complementary proteins were localized to the dark zone and the MBR. RNAscope experiments demonstrated that four mRNAs (Kif23, Jun, Klf4, and Zfp36) localized within the MB matrix, or alpha-tubulin-free zone, of the mitotic MB during G1, and post-mitotically in the MBRs. See S2C for CHO cell RNAscope quantification for similar probes. *Significance was determined by comparing data to DapB 1E. Proteins encoded by these transcripts similarly localized to mitotic MBs and post-mitotic MBRs in HeLa cells.
All data were done in triplicate and quantifications are noted on each figure at a minimum of n=10 for each stage.
First, we confirmed that the MB is a site of RNA storage by verifying that polyA-containing mRNAs were enriched in the MBs and MBRs of HeLa Kyoto (Fig. 1D, 1G) and CHO cells (Fig. 1D). Similar results were seen in HeLa (CCL2) cells, suggesting that RNA targeting, and storage are likely to be a general property of MBs (Fig. 2A, 2C). We used DapB, a bacterial (Bacilus subtilis) RNA as a control (Fig 1E), which in both CHO and HeLa Kyoto cells do not localize to the bridge or MBRs (Fig. 1E. S2A–B). Next, we chose two other RNAs that were not enriched in our midbody RNAseq data, EPEMP1 and CNCL5, to determine if they were found in the MB (Fig 1F–G). Here, all three RNAs EPEMP1, CNCL5 and DapB were not enriched at the MB, in contrast to PolyA (Fig. 1G; S2A–B). PolyA localization in whole cells with regard to the midbody PolyA RNA localization was also determined (Fig. S2A). Here, both the cell bodies and the midbody dark zone have PolyA RNA signal, suggesting that RNA is localizing to this discrete spot at the end of mitosis, in addition to the PolyA signal in the cell bodies (Fig. S2A, Zoom). Next, we determined how enriched Poly A, KIF23, and KLF4, Jun and ANXA11 were in the midbody in HeLa Kyoto cells (Fig 1H). Here, we quantified RNAscope probe localization in the midbody versus the bridge and determine that PolyA and MKLP1 were over-enriched when compared to these three other RNAs (Fig 1H). Of special interest was the dynamic cell cycle-localization pattern observed for transcripts of Kif23/Mklp1 (Kif23 is used to probe CHO cells and MKLP1 is used to probe human cells), which encode an atypical (non-processive) kinesin motor that is widely used as an MB marker and that critically regulates cytokinesis and abscission27,44–47. In CHO cells, Kif23 transcripts were localized to the site of spindle microtubule overlap from early anaphase through late telophase (Fig. 1I; S2C), coincident with the localization of KIF23 protein (S1A)48. However, the early telophase pattern was unusual. KIF23 protein is normally found at the MB ring44, but we observed that, in early telophase, Kif23 RNA expression occurred as small puncta found throughout the cell bodies and in two distinct spots adjacent to the dark zone (Fig. 1I, early telophase). Following abscission, Kif23 transcripts were found in released MBRs, confirming that these transcripts are present in MBs throughout their life cycle (Fig 1I, MBR).
Fig. 2. MKLP1 and Arc are important for PolyA localization or translation at the MB.
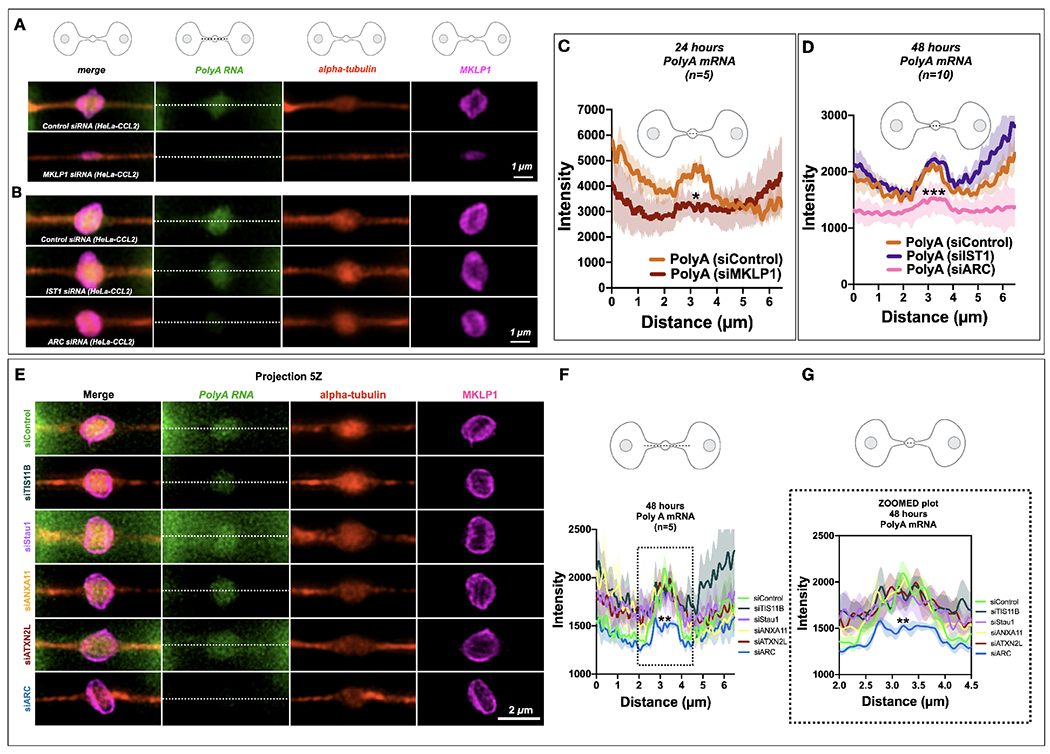
(A-B) PolyA signals (green) localized to the MB matrix surrounded by MKLP1 signal (magenta) in HeLa cells. RNAscope fixation techniques led to loss of the MB dark zone as seen by the tubulin bulge along the intercellular canal (red). Scale bars are 1 μm unless noted.
(C) Quantification of the line scans revealed that loss of MKLP1 by siRNA knockdown led to a decrease in polyA mRNA in MKLP1 siRNA-treated cells. *denotes significance
(D) Loss of ESCRT-III/IST1 did not affect RNA levels, but loss of ARC led to decreased levels of polyA mRNA in the MB. *denotes significance
(E-G) The RBP Arc leads to a decrease of PolyA RNA localization or maintenance at the MB, whereas loss of TIS11B, Stau1, ANXA11 or ATXN2L do not lead to a decrease in PolyA RNA signal. Note there is a slight insignificant decrease in siTIS11B treated cells. (F-G) Line scans across the bridge are shown (F) and a zoomed portion (G)(dotted line) shows the area of the dark zone. *denotes significance
Next, we selected for further testing three mRNAs from our RNA-Seq data that encode distinct classes of proteins (Fig. 1J): the oncogenic transcription factor Jun, the pluripotency-regulating transcription factor Klf4, and the RNA-binding/RNP granule constituent Zfp36/TIS11. We confirmed that all four mRNAs, KIF23, KLF4, JUN, ZFP36 localized to the MBs and MBRs by RNAscope analysis in CHO cells and were significantly enriched when compared to DapB (control) alone (Fig 1J, CHO; S2C for Violin plots). As in CHO cells, proteins encoded by each of these mRNAs were also observed in MBs and released MBRs in HeLa cells (Fig. 1J, HeLa (CCL2).
Last, we identified genes necessary to target or maintain RNA localization to the MB. Using a PolyA RNAscope probe in HeLa cells, we found that PolyA was enriched in the dark zone of the MB in HeLa (CCL2) cells (Fig. 2A–B). PolyA RNA enrichment may be dependent on KIF23/MKLP1 (Fig. 2A), a kinesin44,47,49, and ARC (Fig. 2B), a repurposed viral-like capsid protein involved in synaptic plasticity and memory50. ARC was identified in preliminary RNAseq data of isolated MBRs from different cell types, and since ARC is a protein that is involved in moving RNAs between neurons50, we surmised that ARC might also move RNAs between all cells. In contrast, ESCRT-III subunit IST1, a protein complex necessary for abscission51,52 which additionally functions as an RNA-binding protein53,54, was not required for the localization of PolyA RNA to the dark zone of the MB in HeLa cells (Figure 2B). We also tested other midbody RBPs that might be necessary to target RNA to the MB including TIS11B, Stau1, ANXA11, and ATXN2L, and ARC. Loss of ARC led to a loss of the PolyA signal (Fig 2E–G).
These RNAscope data confirmed our transcriptomic findings that MB-enriched mRNA populations were localized to MB structures assembled during mitosis, and that factors necessary for RNA movement and cellular transport were required for their localization to the MB. Equally, these RNAs are stored and released as MBRs. Combined, our data suggest three testable mechanistic hypotheses: specific mRNAs may be physically sequestered at the MB in RNP granules; MB-targeted mRNAs may be locally translated; these mRNAs and proteins may play an important role in MBR function.
Midbodies are assembly sites of ribonucleoprotein granules.
Several lines of evidence suggest the MB may harbor a phase-separated RNP condensate, given that the MB stores RNA (Fig. 1), is highly enriched in RNA-binding proteins (e.g., Staufen, eIF3e, Ataxin-2L, PABP, and the 40S and 60S ribosomal proteins)1,26,55,56 is enriched in known RNP granule components, including AnnexinA11(ANXA11)57, and exhibits birefringence. RNA granules are heterogeneous in composition and function but generally contain solid-like, mobility-restricted structural core components and more labile, liquid-like components that control mRNA flux and translational availability. Yet, it remains unclear how RNA granules are dynamically regulated, assembled, maintained, and disassembled.
First, we investigated if midbodies are bona-fide RNA aggregates are reversibly disruptable by challenge with the aliphatic alcohol 1,6-hexanediol, which distinguishes liquid-like assemblies from solid-like assemblies by rapidly dissolving only the former58–60. A 90-second treatment with 7.5% hexanediol was sufficient to disrupt MB matrix in dividing HeLa cells, affecting noticeable but incomplete dispersion of the kinesin KIF23 protein from its native MB localization (Fig. 3A). When hexanediol challenge was followed by recovery in normal medium in a 0- to 30-minute timed series, KIF23 exhibited progressively wider spatial dispersion, accompanied by reaggregation of increasingly larger assemblies that were usually physically continuous with the native MB (Fig. 3A, T=30). Importantly, the main structural component of MBs—bundled microtubules—was unaffected by hexanediol treatment, suggesting the MB matrix exhibits material properties consistent with a liquid-like assembly, whereas other structural components, such as microtubules, do not. In parallel with our KIF23 results, PolyA mRNA also exhibited hexanediol-sensitive dispersion from its normal midzone domain and remained detectable in association with KIF23-positive aggregates, but in complementary domains (Fig. 3B). KIF23 has traditionally been attributed to a structural role in MB-bundling spindle microtubules at the midzone and in assembling abscission machinery at the MB 44. However, our data suggest that KIF23/MKLP1 may have an additional role in the positional assembly or tethering of RNA aggregates at the antiparallel microtubule overlap of the spindle midzone, which we observed after short interfering RNA (siRNA) knockdown of MKLP1 (Fig. 2k). To determine if the 1,6-hexanediol sensitive behavior is unique to KIF23/MKLP1, we performed live imaging on GFP-MKLP1 and GFP-MKLP2/KIF20A, a related kinesin-6 family member61–63. Here, we found that only GFP-MKLP1 was sensitive to 1,6-hexanediol, suggesting that this behavior is unique to this kinesin-6 family member (Fig. 3C). We then used FRAP to determine that KIF23/MKLP1 behaved as a non-mobile component within the native MB granule, as there was very little recovery of MKLP1-GFP fluorescence during the very late stages of cytokinesis (Fig. 3D). This suggests that KIF23 may serve as an immobile kinesin scaffold for the MB RNP granule or MB granule. Our FRAP data were gathered in the context of a native MB within an established RNA granule anchored to microtubules. We interpret these data to suggest that KIF23 behavior exhibits solid-like behavior in intact, native MBs and liquid-like behaviors when weakly hydrophobic bonds are disrupted with 1,6-hexanediol. This is consistent with a functional role for KIF23 in tethering liquid-like RNP aggregates to the microtubule component of the cytoskeleton.
Figure 3: Midbody proteins and RNAs behave as ribonucleoprotein granules.
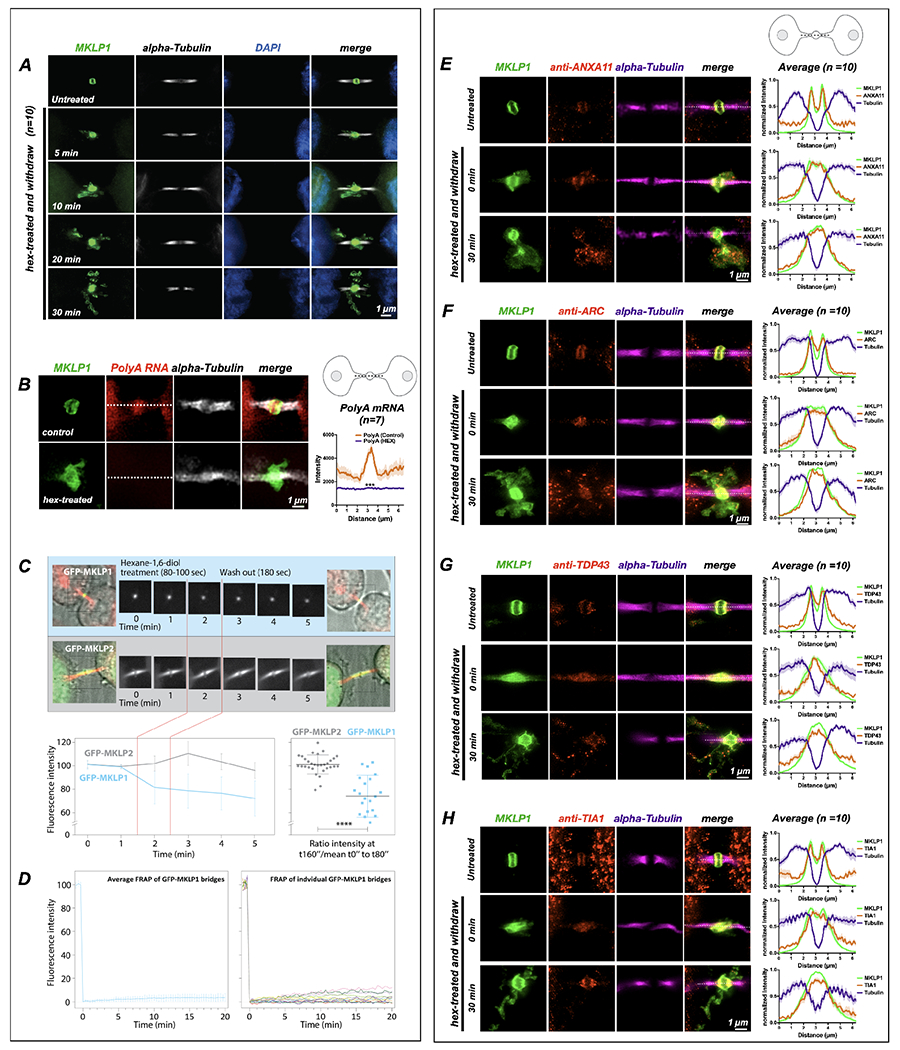
(A) Synchronized HeLa cells (n=10) were treated at the MB stage for 90 seconds with 1,6-hexanediol and then were allowed to recover in normal medium for specified times (T = minutes post-hexanediol). The MB kinesin MKLP1 protein dispersed upon hexanediol addition, reforming spatially disseminated aggregates over time that surrounded the bridge in projected Z-series images. The MB structural component alpha-tubulin was unaffected by hexanediol treatment.
(B) Treatment with 1,6-hexanediol (hex) also affected polyA localization at the dark zone (n=7). We observed a loss of polyA and dissolution of the MKLP1 signal in the intercellular bridge.
(C) Live imaging of hexanediol-treated HeLa cells expressing a GFP-MKLP1 fusion protein and incubated with fluorescent SiR-tubulin (red) revealed a rapid and sustained partial loss (30% decrease) of MKLP1 levels at the native MB location; in contrast, the closely related mitotic kinesin MKLP2 fused to GFP exhibited no change in intensity after hexanediol treatment. The 30% loss of MKLP1-GFP after hexanediol treatments reveals that this kinesin is specifically sensitive to 1,6-hexanediol.
(D) FRAP analysis of GFP-MKLP1 MBs showed no recovery after photobleaching, suggesting little mobility of GFP-MKLP1 within the MB granule in native MBs.
(E-H) A functional range of MB matrix proteins (ANXA11, ARC, TDP-43, and TIA1) dispersed and reaggregated in apposition to MKLP1 upon hexanediol treatment (T=0 seconds) and after a long recovery time (T=30 seconds)(n=10 for E-H). Interestingly, all hexanediol-sensitive components tested reaggregated in domains complementary, but tightly apposed, to MKLP1. Of note, we often observed that only a portion of MB factors moves farther away from their original location in the intercellular bridge after hexanediol treatment. For example, the bulk of TIA1 remained diffuse in the dark zone immediately after treatment, but TIA1 quickly assembled back to its normal localization pattern after 30 minutes. MB expression in untreated controls was similar to MKLP1 for all hexanediol-sensitive MB factors (Fig. 4A, B). See also Fig. 4 for a timed series of hexanediol-mediated dissolution and reaggregation of RacGAP, TIA1, ANXA11, and ARC.
Scale bars are 1 μm.
Next, we determined whether hexanediol altered the localization of other MB proteins known to function in cytokinesis, as well as putative RNP granule components identified in MBs (Fig. 3E–H; Fig. 4A–B). In non-treated cells, ANXA11, ARC, TDP-43, and TIA1 all localized to the MB (Fig. 4A, controls). After hexanediol treatment and washout, all of the factors tested, were sensitive to hexanediol treatment (Fig. 3E–H, Fig. 4B). Additionally, other MB factors and RNA-binding proteins, including the citron rho-interacting kinase CIT-K, the GTPase RacGAP, and the polyA-binding protein PABP, all of which localized to the MB and MBRs in control cells (Fig. 4A), were hexanediol-sensitive (Fig. 4B). RacGAP, which comprises the Centralspindlin complex with KIF23, formed discrete puncta complementary to KIF23 that resided in KIF23-free pockets directly abutting KIF23 domains (Fig. 4B). Similar patterns of dissolution and reaggregation were observed for two other MB proteins required for cytokinesis, namely CIT-K (Fig. 4B), which directly binds KIF23 and organizes late-stage MB structure, and the phospholipid-binding protein ANXA11 (Fig. 3E), which can tether RNA granules to organellar membranes57. Other midbody factors and RNA-binding proteins also exhibited the same hexanediol-sensitive behaviors (Fig. 3F–H; Fig. 4B). TIA1 localized to the MB matrix and did not appreciably disperse after hexanediol treatment, suggesting it might be an immobile component of the MB structure (Fig. 4A–B). Three of these RNA-binding proteins, TIA1, PABP, and TDP-43, function in the assembly and dynamic regulation of stress granules, which are reversible membrane-less organelles that execute cytoprotective defense against environmental stressors by sequestering and translationally silencing mRNAs64,65. We also determined that double-stranded RNA, a known extracellular vesicle marker66,67, was also located in the MB and MBRs. After hexanediol treatment, double-stranded RNA was found in the cloud of MKLP1 (Fig. 4B, zoomed image). In combination, our data suggest that RNAs targeted to MBs are assembled into phase-separated RNP granules containing mRNA and RNA-binding proteins.
Fig. 4. Hexanediol-sensitive proteins and double-stranded RNAs localize to the midbody matrix and are sensitive to hexanediol.
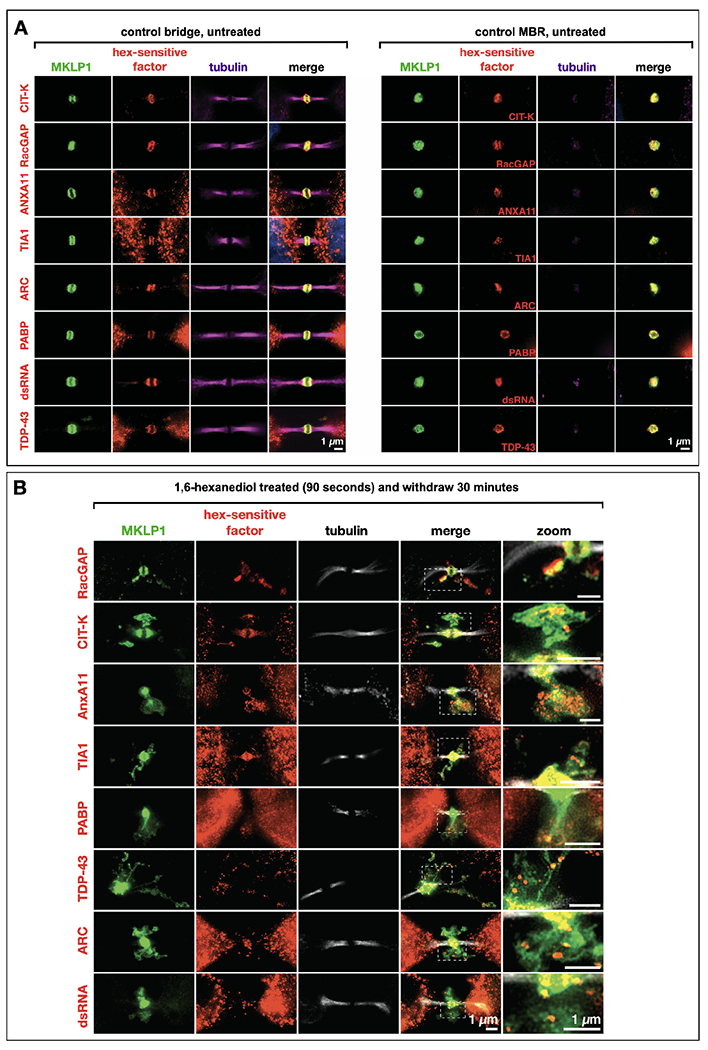
(A) A range of MB-localized proteins and double-stranded RNAs exhibited sensitivity to 1-6’ hexanediol (hex) treatment, causing their dispersal and progressive reaggregation over time. These factors localized to the MB matrix (red) in mitotic MBs. MKLP1 (green) was used as a marker of the MB matrix, and alpha-tubulin staining (magenta) was used to visualize the dark zone interruption. All assays are done in triplicate and a minimum of n=5 for each.
(B) The factors in (A) all remained co-localized with MKLP1 following abscission and release of the MB as an MBR. All assays are done in triplicate and a minimum of n=5 for each.
Scale bars: 1 μm.
Midbodies and midbody remnants are sites of localized translation.
To determine whether MB mRNAs are translationally activated or silenced, we used two methods to quantify translation. We used the puromycin-based SUnSET technique to label nascent peptides and visualize sites of recent translation using anti-puromycin antibodies68,69, and OPP-ClickIT and HPG-ClickIT68, to determine whether active translation occurs in the MB and MBRs. OPP-and HPG-ClickIT are different from puromycin-based assay because when incubated with live cells, OPP or HPG react with translating ribosomes and become covalently attached to elongating peptides68,70,71. Synchronized HeLa cells pulsed with puromycin for 4 minutes in early telophase (ET) showed little evidence of MB translation (~39% (n=15/38) of the MB dark zones displayed puromycin labeling at the ET stage)(Fig. 5A, 6A–B). Parallel cells pulsed just 15 minutes later, in late telophase (LT), showed sharply demarcated toroidal domains of translation encircling the spindle midzone and MB matrix (100% of the MB dark zones displayed puromycin rings at the LT stage) (Fig. 5B; 6B). The ring-like localization pattern was similar to the localization of both the large and small ribosomal subunits (Fig. 5C), suggesting perhaps the ribosomes and translation events occur in a particular ring-like compartment surrounding the RNA in MB dark zone. High levels of translation continued in singly abscised MBs and in doubly abscised and released MBRs (100% of the MBRs displayed puromycin labeling) (Fig. 6B). The puromycin localization in the MB was also observed in different cell types, including CHO, Retinal Pigmented Epithelial (RPE) cells, Neural Stem/Progenitor cells (NSPCs) (Fig. 6C), We confirmed that this translation signal was indeed active using OPP-ClickIT and HPG-ClickIT68. We observed that the HPG-ClickIT signal gave a hazy disk in the MB in early G1(Fig. 5B). In both singly abscised and doubly abscised MBRs, we saw two distinct regions of HPG-ClickIT signal: a central core and a faint ring of translation around the MBRs that we called the G1 ring (Fig. 5A). Staining with anti-puromycin antibodies revealed a more distinct ring, perhaps owing to the diffusion barrier created by ESCRT 72 in the MB, which is located in the same compartment where the bulk of the MB ribosomes and translation regulators are found (Fig. 5C, 40S and 60S)31
Figure 5: The midbody is a translation which is regulated by IST1, MKLP1 and ARC.
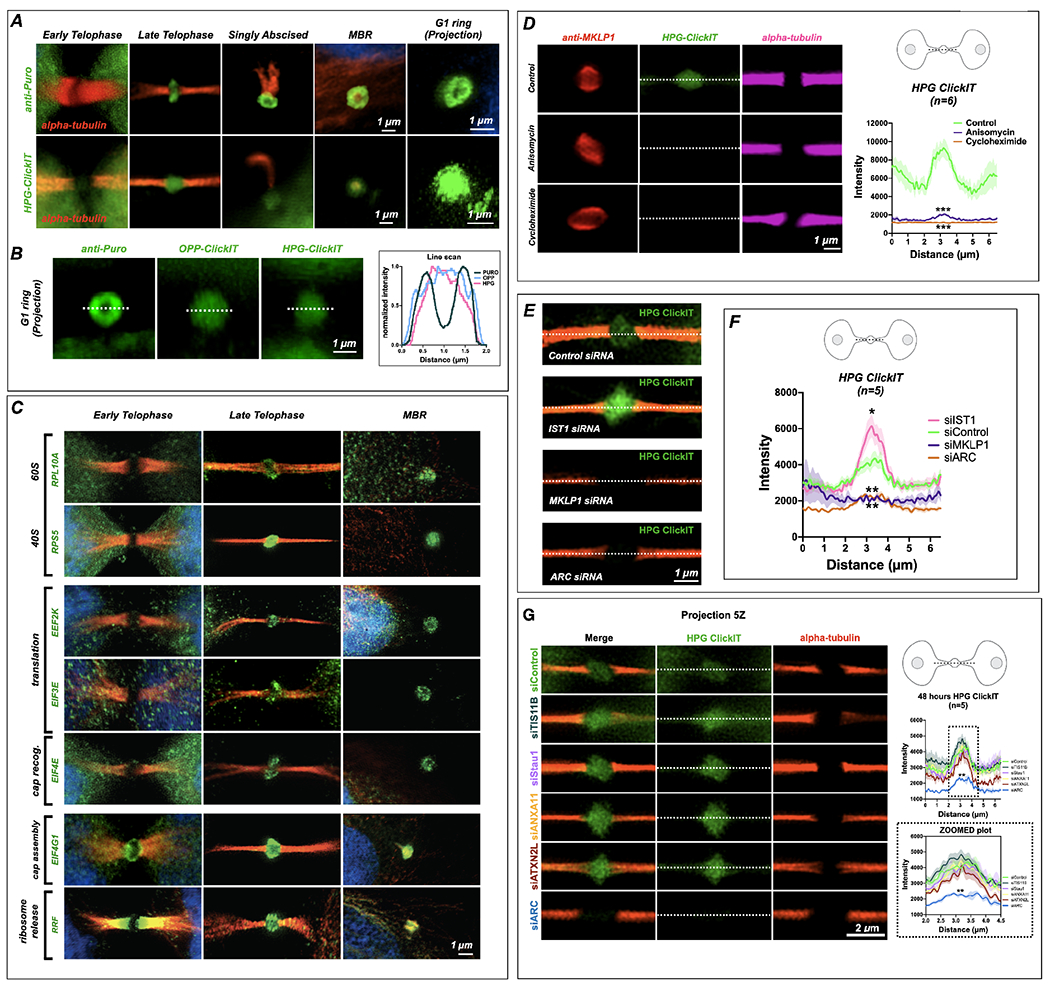
(A) SUnSET labeling (α-Puro) revealed that the MB is a translation platform during abscission. Translation was undetected in early MBs (early telophase) but observed at high levels in late MBs (late telophase/G1), in abscising midbodies, and in released extracellular MBRs. Projection revealed that translation occurred in a toroid shape encircling the MB matrix or dark zone. (See Fig. 6B for quantification of α-Puro rings per stage) HPG-ClickIT analysis revealed a similar pattern, which suggests that active translation occurred in the dark zone. The HPG-ClickIT pattern appeared as a hazy disk surrounded by a faint ring or cloud.
(B) The images show the translation patterns from α-Puro (ring) and the OPP-ClickIT and HPG-ClickIT reagents (hazy disk), which indicate a site of recent translation. The graph shows the normalized intensity of the ring (puro) and disk (OPP-ClickIT or HPG-ClickIT) patterns.
(C) Coincident with the puromycin rings, rings were observed for all translation factors previously identified by midbody proteomics (Skop, 2004). Here, 40S and 60S ribosomal subunits (RPL10A and RPS5), translation elongation factors (EEF2K and EIF3E), a cap recognition factor (EIF4E), and a cap assembly regulator (EIF4G1) were first robustly detected in late-stage MBs (abscission/G1) and remained detectable in MBRs. The translational regulators EIF4G1 (cap assembly) and RRF (ribosome release) were present in lateral MB domains in early telophase but re-localized to the translation/ribosome ring at the abscission/G1 transition.
(D) The robust control HPG-ClickIT signal in the MB dark zone was significantly reduced after treatment with the translation inhibitors anisomycin and cycloheximide. Asterix denote significance.
(E) Several candidate MB markers were tested, and ESCRT-III/IST1 was found to regulate the levels of active translation in the MB. Here, ESCRT-III/IST1 loss leads to significantly increased levels of HPG-ClickIT (green). MKLP1 and ARC both lead to a loss of HPG-ClickIT (green) signal. Con: control.
(F) Quantification of the HPG-ClickIT signals in control, IST1, MKLP1, and ARC siRNA knockdown cells (n=5). Con: control. Asterix denote significance.
(G) Arc siRNA treatment leads to a decrease of translation activity in the midbody as visualized by HPG-ClickIT in HeLa (CCL2). Knockdown of TIS11B, Stau1, ANXA11, ATXN2L did not lead to a loss of HPG-ClickIT signal. Line scans across the bridge are shown (top graph) and a zoomed portion (dotted line in top corresponds to zoomed part in bottom graph) shows the area of the dark zone (n=5 for each assay). Asterix denote significance for ARC.
Scale bars are 1 μm unless noted.
Figure 6. The midbody is a site of spatiotemporally regulated translation which also occurs in different cell types.
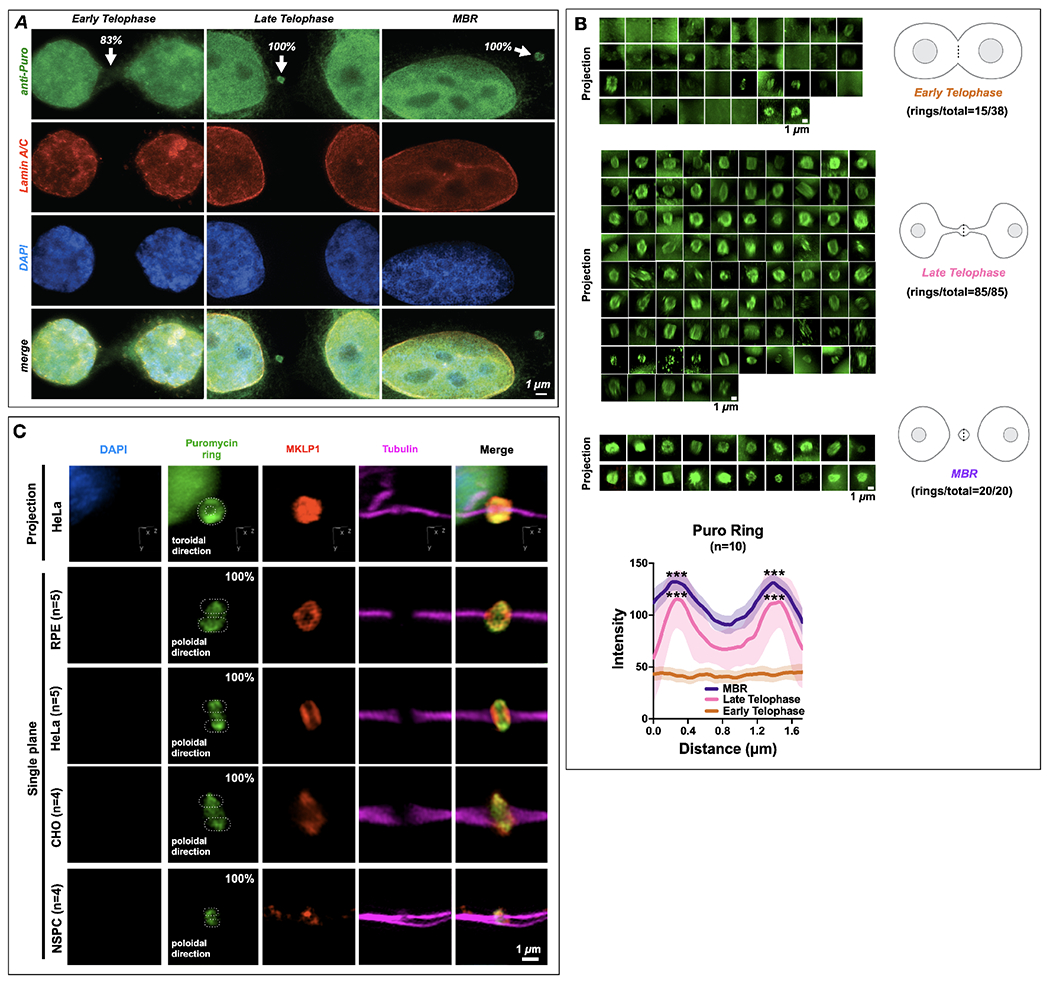
(A) Translational onset (α-Puro; arrowheads at MB) occurred precisely as cells formally exited mitosis at the G1 transition, coincident with the mature reformation of the nuclear envelope (detected by lamin A/C) and the de-condensation of chromatin (DNA detected by DAPI staining). DAPI: 4’,6-diamidino-2-phenylindole. Quantification is noted at each stage in figure; 83% Early Telophase (n =10/12), 100% Late Telophase (n =3/3) and 100% MBR (n =3/3).
(B) Quantification of the number of distinct puromycin rings observed at different points during the late stages of mitosis, namely early telophase (ET), late telophase (LT), and MBR. The α-Puro label was primarily found in late telophase/G1 and continued in the MBR stage after MBR release. Quantification is noted next to each stage. Line scans denoted by the dotted line in each schematic was quantified for data sets and plotted (n=10). Here, the puromycin ring is seen prominently during Late Telophase and MBR stages. Asterix denotes significance.
(C) Retinal pigment epithelium cells (RPE)(n=5), HeLa CCL2 cells(n=5), CHO cells (n=4), and neural stem/progenitor cells (NSPCs)(n=4) all had puromycin rings labeled with MKLP1 within the bridge (tubulin). DAPI: 4’,6-diamidino-2-phenylindole. Quantification is noted next to each cell type, which are 100% for each cell type.
Scale bars are 1 μm unless noted.
We treated MBs with anisomycin or cycloheximide to inhibit translation68. We observed that the HPG-ClickIT signal was abolished after these drug treatments and the MKLP1 localization was often distorted (Fig. 5D), suggesting that active translation during late telophase might be necessary for the proper maintenance of MB structure.
ARC, ESCRT-III and MKLP1 regulate translation activity in midbodies.
To determine which genes might be necessary for the unique translation event that occurs in MBs, we knocked down ESCRT-III/IST1, MKLP1, and ARC using siRNAs in the HeLa (CCL2) cell line (Fig. 5E–G). Surprisingly, depletion of ESCRT-III/IST1 led to a sharp increase in active translation in the MB (Fig. 5E–F, S3A). Conversely, in MKLP1 and ARC siRNA-treated cells translation was entirely abolished at the MB (Fig. 5E–F, S3A), suggesting either that these genes or proteins are required for translation or that they are required to target or maintain MB RNA. However, because we observed thinner tubulin bundles in the siMKLP1-treated cell midbodies, the lack of translation could also be due to a failure to properly assemble the midbody or a failure to target mRNAs to be translated. In both parental HeLa (CCL2) and MKLP1-GFP HeLa Kyoto expressing cell lines, we only observed failure of cytokinesis (bi-nucleate daughter cells) in MKLP1 siRNA-treated cells (Fig. S3B), whereas ESCRT-III/IST1 led to delays in abscission (% MB bridge, Fig. S3C). We favor the latter suggestion, as loss of MKLP1 and ARC led to a loss of RNA signal in the MB (Fig. 2A–B, E). Additionally, of the targets we knocked down, only the loss of MKLP1 led to a thinning of the microtubules in the midbody (Fig. 2A), suggesting that in MKLP1 siRNA-treated cells there may be a limited ability to target RNAs.
We made an unexpected finding that MKLP1 may promotes global translation events, as HPG-ClickIT levels were increased at the MB dark zone in the MKLP1-GFP Kyoto HeLa cell line when compared with the HeLa (CCL2) cell line (Fig. S4A). However, when we compared the dark zone region in HeLa Kyoto cell line to the MKLP1-GFP HeLa Kyoto cell line we did not observe any statistically significant differences (Fig. S4B). Next, we quantified significant translation activity throughout the cell bodies, and the MB dark zone in the MKLP1-GFP HeLa Kyoto cell lines (Fig. S5A). These data suggest that MKLP1 may promote translation in distinct cellular sites (in the cell body), and these data represent a caution to others that use of this MKLP1-GFP cell line could confound their results. We found that knockdown of ESCRT-III, MKLP1, and ARC by siRNAs had similar effects on translation in the MKLP1-GFP HeLa Kyoto cell line. Loss of ESCRT-III led to increased levels of translation, and MKLP1 and ARC appeared to be required for translation (Fig. S3A–B). Overall, our finding that localized translation in the MB initiated prior to daughter cell separation raises the possibility that assembly of the MB -granule and translation of its RNA contents may be a necessary step during the late steps of abscission. In addition, we have discovered an autonomous extracellular vesicle with active translation activity, and may reflect a transition in the life stage of the MB RNA granule that is critical to post-mitotic MBR function.
A primary function of many RNP membrane-less compartments such as stress granules is to regulate the translational availability of mRNAs by reversible partitioning into translationally silenced condensates33,73–75. Although it is accepted that global translation is severely restricted during mitosis, MB and MBR RNA interference screening and proteome analysis suggest the presence of large complements of both 40S and 60S ribosomal subunit proteins1,12,26 and translation initiation and elongation factors1,12,26,76,77; we confirmed MB and MBR localization of these proteins in representative samples (Fig. 5C).
Translation starts at the M/G1 transition.
Dividing daughter cells exit mitosis while still joined by the intercellular bridge containing the MB and undergo abscission only after re-entering the G1 phase of the cell cycle when they resume global protein synthesis78–80. We used SUnSET staining69 to determine the relative timing of MB translation initiation with three hallmarks of the M/G1 transition: re-initiation of global translation, nuclear envelope reassembly, and chromatin decondensation. In late telophase, newly segregated chromosomes are fully condensed, the nuclear envelope is beginning to reform, and translation in the MB and daughter cell body was almost undetectable (Fig. 6A, Early Telophase; 83%, n = 10/12). As daughter cells progress into G1, chromatin de-condensation initiates as the nuclear envelope becomes continuous, and active MB translation was observable in the intercellular bridge (Fig. 6A, Late Telophase; 100%, n = 3/3). Following abscission, the euchromatin of interphase daughter cells was observable within fully formed nuclear envelopes, and actively translating extracellular MBRs were visible on plasma membrane surfaces (Fig. 6A, MBR, 100%, n = 3/3). We, therefore, hypothesize that the G1 transition triggers a burst of translation in a juxta-granular compartment of the MB.
Supporting this hypothesis, we found that most proteins encoded by MB-enriched mRNAs (identified in Fig. 1C) were first detectable in the MB only after the G1 transition (n=10/12; Fig. S1A, C) In telophase, the two cytokinesis factors, KIF23 and TEX14 are seen in the MB matrix and flanking arms of the intercellular bridge, respectively. In contrast, the remaining 10 proteins, which have no reported role in cytokinesis, were undetectable in early telophase, except KLF4, despite being readily seen in late telophase (Fig. S1A); these factors included five transcription factors (JUN, cFOS, FOSB, KLF6, and IRF1), the transcriptional inhibitor IKBalpha, the RNA granule component ZFP36/TIS11, histone HISTH1, and the multifunctional BIRC3 protein. In contrast, all 12 factors were readily detected in the MB at later stages following transition into G1 and remained detectable in post-abscission MBRs. These data strongly suggest that mRNAs targeted to the MB RNA granule become translationally available coincident with the M/G1 transition and may reflect a critical life cycle transition as the mitotic MB matures toward release as an extracellular MBR with post-mitotic functions.
ARC leads to a decrease in RNA and translation at the midbody.
To identify which MB RBP might be responsible for the assembly or maintenance of RNA and the translation activity in the MB, we took a close look at our previously published MB proteome (Fig. S1B). Here, we identify several candidates that might be important for this function, which include TIS11B, Staufen/Stau, Annexin all, Ataxin 2L and ARC, all of which localized to the MB during G1/late telophase of the cell cycle (Fig. S1C). Using siRNA knockdown, we observed that the PolyA signal at the MB was found in all of our knockdowns except ARC (Fig 2A, 2E; Fig. S6A). There was a slight decrease in PolyA signal in TIS11b siRNA-treated cells (Fig. 2E). However, ARC was the only factor whose depletion led to a decrease the HPG-ClickIT signal (Fig. 2B, 2D, 2E–G), suggesting that ARC is critical for RNA maintenance and translational activity in the MB.
DISCUSSION
Until recently, the MB was thought to regulate assembly of the abscission machinery during cytokinesis and then be immediately degraded following cell separation. However, studies in the last decade have demonstrated that post-mitotic MBRs are released by abscission as extracellular vesicles, are internalized to form signaling MBsomes in target cells, and may contribute to driving highly proliferative fates such as tumor and stem cells6,11,12,19,26,29,81–83. In support of this idea, MBRs are preferentially accumulated in tumor and stem cells, and exogenous MBRs can upregulate proliferation-promoting genes, the proliferative index, and anchorage-independent invasiveness12,20,84. Although the functional importance of post-mitotic MB signaling has been established, the underlying mechanisms remain poorly understood. Recent advances identify a requirement for integrins and EGFR receptor tyrosine kinase signaling in MBsome function; however, this simple model does not sufficiently account for the strikingly large size of MB derivatives nor their structural complexity and multi-stage life cycles. In this study, we characterized the structural components of MB derivatives to gain insight into post-mitotic MBR signaling mechanisms. Importantly, we demonstrated that the MB is the assembly site of an RNP granule that is packaged and released within a large 1- to 2-μm extracellular vesicle following the terminal stages of cell division. We used a transcriptomic approach to characterize specific mRNA populations that are enriched at the MB in a translationally quiescent granule called the MB granule and demonstrated that local translation of MB granule mRNAs was initiated as cells exit mitosis and MBRs are released by abscission. By identifying ongoing translation in MBRs, implies that dynamic translational availability of MB granule mRNAs may play an active role in subsequent target-cell binding and/or signaling by MBRs.
The reversible formation of RNA granules is the primary mechanism by which cells control translational availability and localization of RNAs to rapidly respond to changing cellular demands85–87. During mitosis, RNA and ribosomal protein sequestration in condensates facilitates global shutdown of protein synthesis and regulates cytoplasmic partitioning88. This study identified a unique subtype of mitotic RNA granule with localized assembly at the overlapping spindle microtubules that define MB positioning; thus, we called it the MB granule. The locations of the MB granule and MB matrix precisely correlate. We report that MB granules are also exquisitely sensitive to hexanediol, a behavior typical of liquid-like assemblies; MB granule-associated RNA-binding proteins and RNA dispersed almost immediately upon hexanediol treatment and then progressively reaggregated in heterotopic puncta and broad clouds continuous with the native MB after hexanediol removal. Interestingly, RNA and RNA-binding proteins reaggregated in domains complementary to reaggregating KIF23, revealing organization within the reforming liquid-like assembly. KIF23 has previously only been suggested to function in microtubule bundling and vesicular trafficking to the MB44–46,81,89,90, so its liquid-like behavior was not predicted, especially as spindle microtubules were unaffected by hexanediol treatment. FRAP analysis indicated that MKLP1-GFP was an immotile component of MB granules. We interpret the data to suggest that KIF23 performs a tethering function in MB granules by binding microtubules with its N-terminal motor domains and by binding RNA or RNA-binding protein assemblies with the predicted intrinsically disordered regions near its C-terminus. Equally, we hypothesize that ARC may play a role to protect MB RNAs from degradation given ARC assembles viral-like capsids in cells50. It remains possible that the heterotopic material observed reflects formation of hexanediol-induced stress granules58 or other aberrant granules caused by prolonged hexanediol exposure91. We think this scenario is unlikely, as we could never detect the stress granule marker G3BP with any of several antibodies tested, and our 90-second hexanediol treatments were far below the 50-minute threshold reported for cytotoxic granule induction91.
A basic function of RNA granules is to translationally silence phase-separated mRNAs, and MB granule transcripts are subject to cell cycle-entrained translational control. Proteomic and immunofluorescence analyses revealed that both mitotic MBs and post-mitotic MBRs harbored large quantities of ribosomal proteins and translational regulators. As translation is largely silenced during mitosis, it is perhaps expected that no translation was detectable in telophase-stage MBs or intercellular bridges. Concomitant with the nascent daughter cells re-entering G1 of the cell cycle and reinitiating global translation, a hazy disk and ring of translation was observed within the MB granule but not appreciably in flanking regions. Translational onset temporally prefigures abscission, so it is tempting to speculate that local translation is required for terminal cell separation; however, we have been unable to generate evidence supporting this hypothesis. The loss of ESCRT-III/IST-1, which leads of increased levels of translation, suggested that ESCRT-III proteins may also function as a RBP, perhaps by maintaining mRNAs release from the dense core of the MB-granule, in addition to its role in abscission92. Remarkably, active translation persists as post-abscission MBRs are released as extracellular vesicles, strongly implying a post-abscission function for ongoing protein synthesis. MBRs have been shown to enrich phosphatidylserine in their outer membrane leaflets only subsequent to abscission, as they mature toward their ultimate fate of engulfment and MBsome signaling. We suggest that active translation may play a parallel role in maturation of MBRs that potentiates recognition and engulfment by target cells and/or mediates MBsome signaling10,12,20.
A central problem in the field of MB biology is that the molecular mechanisms underlying MBR and MBsome signaling remain poorly understood. One possible impediment is that MBRs had simply not been conceptualized as extracellular vesicles until more recently20 and did not benefit from the intense research interest focused on extracellular vesicle-mediated intercellular communication, including by direct RNA transfer. In this work, we newly identify MBRs as a unique subtype of extracellular vesicles with several distinguishing features: biogenesis in mitosis that inherently links cell division status with intercellular signaling; a complex life cycle with both membrane-less and membrane-bound stages; a cargo comprised of a selectively loaded RNA granule; active translation of mRNA cargo; and an extremely large carrying capacity that is greater than 1000-fold more than exosomes on average. We showed that mRNAs were assembled into an MB granule in association with KIF23 and were lost following hexanediol-induced disruption. Our data strongly support the hypothesis that MBRs and internalized MBsomes can signal, at least in part, by the direct transfer of MB granule components. It is also possible that MBR-mediated transfer of RNP complexes is specific to HeLa cells. However, we observed that CHO cells, retinal pigment epithelium cells, and neural stem cells also harbored MKLP1-positive MBs with puromycin-labeled rings that appeared in G1 (Fig. 6C). We favor the hypothesis that MB granule-mediated RNA transfer is a signaling mechanism fundamental to all cells that divide using an MB, and that MBRs are selectively loaded with distinct transcriptomes in a cell type-specific manner and use ARC, a viral-like capsid50, to maintain RNA stability and facilitate the mechanism of cell-cell communication in all cell types, not just neurons. These hypotheses are readily testable.
We propose a model of the MB life cycle that frames its complex structural dynamism in terms of a distinct post-mitotic signaling function that has been suggested previously8,10,12,25,84: intercellular communication via extracellular vesicle-mediated transfer of RNA (Fig. 7, model). During anaphase of mitosis, selected mRNAs and MKLP1 coacervate and are tethered to overlapping regions of the antiparallel spindle microtubules by a process involving KIF23/MKLP1. As spindle microtubules constrict into an intercellular bridge during early telophase, individual coacervates coalesce into a single large RNP granule at the midzone that we called the MB granule. As nascent daughter cells transition to G1, peri-granular translation initiates throughout the MB and outward toward the ribosome-rich ring, presaging the bilateral assembly of the abscission machinery. Following scission and MBR release as a membrane-bound extracellular vesicle, severed microtubules depolymerize, and domains of translation are radialized around the MB granule. Bound and internalized MBRs evade degradation and persist as MBsomes, releasing MB granule constituents into the recipient cell’s cytoplasm. We suggest that liberated MB granule RNAs are a critical functional component of MBsome signaling in recipient cells that act as templates for direct translation of effector proteins or as templates for epigenetic silencing or as a combination of these two mechanisms.
Figure 7. Model of the unique life cycle of the midbody granule and biogenesis of the midbody remnant, a unique actively translating extracellular vesicle with RNA cargo.
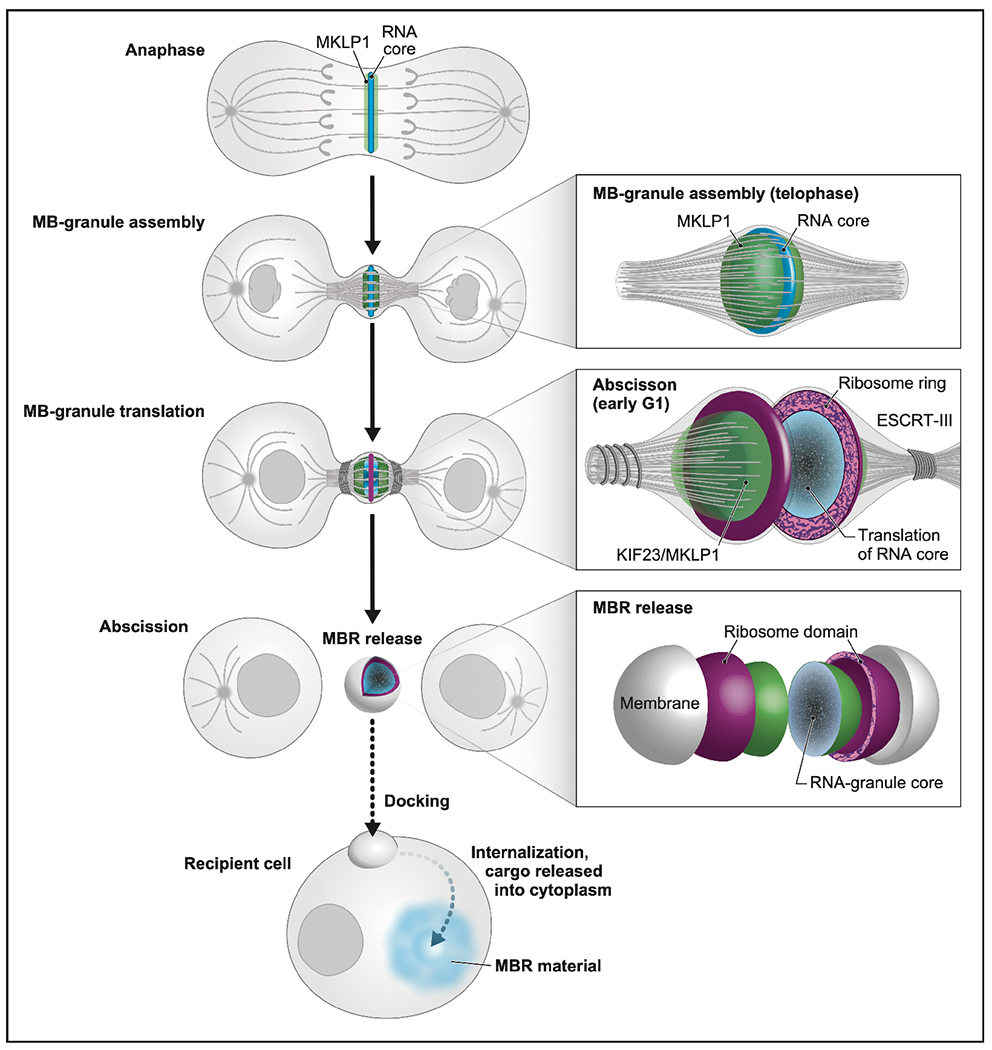
We present a model in which the MB not only plays its traditionally considered role in abscission, but also mediates a form of intercellular communication reported previously by Crowell et al10, Peterman et al12, and Chaigne et al.25, via a RNA cargo. In anaphase, MB-targeted RNAs and associated RNA-binding proteins, such as MKLP1/KIF23 and ARC (both green), begin to form small phase-separated RNP condensates (blue) at the spindle microtubule overlap. Actomyosin ring constriction drives intercellular bridge formation and accretion of a single large MB granule in telophase. At the abscission/G1 transition, ribosomes (magenta) and translation factors surround the RNA core (blue). Translation is active throughout the entire MB granule (blue) and is followed by assembly of the abscission machinery and scission. The MBR is released, which harbors an MB granule core surrounded by a shell of active translation. We propose that MBRs dock to and are internalized by recipient cells (dotted arrow), and this process is followed by the transfer of MB granule cargo, including RNA, across endo-lysosomal membranes into the cytoplasm (dotted arrow in cell) as suggested previously by Crowell et al10, Peterman et al12,84, and Chaigne et al11,25. We hypothesize that the instructional information resides in the MB granule RNA and serves as templates for either direct translation or epigenetic modulation.
Limitations of the study
We recognize that this work lacks functional insights into the nature of the RNAs found in the MB and MBRs. In addition, although we took multiple approaches to study midbody RNAs and active translation, we were unable to determine if MKLP1 directly regulates translation. This limitation is primarily due to the technical challenges of imaging a small population of cells (20%) that have not failed cytokinesis after MKLP1 siRNA treatments which have very thin microtubules in the intercellular bridge. These aspects would provide a clearer understanding of the assembly of RNA at the midbody and subsequent translation.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ahna Skop (skop@wisc.edu)
Materials Availability
Raw imaging files will be shared by the lead contact upon request.
Data and code availability
RNA sequences associated with this study have been deposited into the National Institutes of Health Sequence Read Archive (Bioprojects: PRJNA555245 and SRA:SRP215214).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell culture:
Chinese Hamster Ovary (CHO) cells (ATCC® CCI-61 ™) were maintained at 37°C and 5% CO2 in DMEM/F-12 (Thermo Fisher, Cat# 11330057) with 10% FBS (Thermo Fisher, Cat# 26140079) and 1% Penicillin-Streptomycin (Thermo Fisher, Cat# 15140-122). “Interphase” CHO cells were asynchronous populations cultured for 48 hours before RNA isolation. Synchronized CHO cell populations were grown as described (Skop, 2004)1: cells were blocked in S phase by two rounds of growth for 16 hours in medium supplemented with 2 mM thymidine (Sigma, Cat# T1895-5G) interrupted with 8 hours incubation with DMEM/F-12 medium. Following the second thymidine block, cells were released into DMEM/F-12 medium for 5 hours and then treated with 100 ng/ml nocodazole (Sigma, Cat# M1404) in DMEM/F-12 medium for 4 hours to arrest cells in metaphase. Mitotic cells were isolated by mechanical shake-off and transferred to DMEM/F-12 medium. “Metaphase” samples were incubated for 15 minutes to allow mitotic spindles to reform, and spindle associated RNAs were isolated. Following nocodazole wash-out, “MB” samples were incubated for 30–45 minutes until contractile rings were apparent (late telophase/G1), and MB-associated RNAs were harvested. To harvest stage-specific RNAs, interphase microtubules, metaphase spindles, and MBs were isolated as described (Skop, 2004)1. HeLa cells (CCL-2; ATCC) were cultured at 37°C and 5% CO2 in DMEM/high-glucose/GlutaMAX medium (10564029; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum Thermo Fisher, Cat# 26140079 and 1% penicillin-streptomycin (Thermo Fisher, Cat# 15140-122). HeLa cells were synchronized using a similar double thymidine-block procedure, as previously described 93. HeLa cells were synchronized to arrest in prophase by culture in 50 ng/ml nocodazole in DMEM/high-glucose/GlutaMAX medium for 16 hours. The mitotic cells were harvested by shake-off, centrifugation (200g/1000rpm by Eppendorf centrifuge 5702, 1min), and release from high-precision cover glasses (Zeiss, Germany, Cat# REF# 0109030091) with pre-warmed DMEM/high-glucose/GlutaMAX medium (90 min, early midbody during telophase; 4 hours, late midbody during G1). The cells were treated with 91 μM puromycin (Sigma-Aldrich, Cat# P8833) in DMEM/high-glucose/GlutaMAX medium for 4 minutes before fixation. Primary hippocampal mouse neural stem progenitor cells (NSPCs) were isolated by extracting and dissociating hippocampi from 3-5 mice roughly 6 weeks of age, as described previously in Moore et al,94. GFP-MKLP1 and GFP MKLP2 HeLa cells95were cultured at 37°C and 5% CO2 in DMEM/Glutamax (#31966; Gibco, Invitrogen Life Technologies) supplemented with 10% FCS, 1% penicillin-Streptomycin and kept under G418 (40 μg/mL, Gibco). NSPCs were cultured at 37°C/5% CO2 in serum-free media: DMEM/F12 GlutaMax (10565018; Invitrogen) with B27 (1:50, 17504044;Invitrogen), penicillin-streptomycin-fungi-zone (1:100, 15140122; Invitrogen), 20 ng/mL FGF-2 (100-18B; PeproTech), EGF (AF-100-15; PeproTech) and 5ug/mL Heparin (H3149; Sigma), as previously described (Morrow et al., 2020). RPE-1 (ATCC® CRL-4000™) was cultured in DMEM/F12 (Thermo Fisher) supplemented with 10% fetal bovine serum and penicillin/streptomycin at 37°C in an atmosphere of 5% CO2.
METHOD DETAILS
CHO midbody RNA purification and Illumina library preparation:
CHO microtubule pellets (interphase, metaphase, and MB stage) were resuspended in approximately 100 μl phosphate-buffered saline (PBS). RNA was purified from each sample using a Qiagen RNeasy kit. PolyA RNA was purified from 1 μg RNA from each sample using an Exiqon LNA dT purification kit in accordance with the manufacturer’s instructions. PolyA RNA at the final purification step was eluted using Illumina Elute/Prime/Fragment buffer. Illumina RNA libraries were constructed using the Illumina TruSeq RNA Sample Preparation Kit v2 in accordance with the manufacturer’s instructions. Each library was barcoded and sequenced on an Illumina HiSeq 2500 system.
Annotation assignment and RNA-Seq data filtering:
RNA-Seq reads were collapsed into unique reads using a custom Perl script96. Unique reads were aligned to the RefSeq sequences for the Chinese hamster (Cricetulus griseus) using Bowtie 297. Reads mapping to transcripts were quantified using a custom Perl script96 or HTSeq98. For comparison analyses, we only considered genes with at least 100 reads in all three libraries. After alignment, hamster orthologs were identified using BLASTx (National Center for Biotechnology Information); annotations were automatically assigned using DAVID (https://david.ncifcrf.gov/) and PANTHER (www.pantherdb.org) and then manually curated using gene ontology terms and the UCSC Genome Browser database. Enrichment scores were defined as the ratio of normalized read counts (in RPKM) between libraries; all comparative quantitative analyses of RNA levels were performed using RPKM values or reads per million values. RNA-Seq resulted in 21,607 transcripts, 20,821 of which had human orthologs, with at least one read in any of the three libraries (interphase, metaphase, and MB), resulting in 15,636 transcripts in the interphase library, 17,813 transcripts in the metaphase library, and 16,528 transcripts in the MB library. After low-abundance reads were discarded, 10,424 entries remained in the interphase library, 9,336 entries in the metaphase library, and 8,139 entries in the MB library. These groups overlapped, giving 7,986 entries with at least 100 reads in all three libraries.
An enrichment threshold of ≥2 was used to identify MB-specific and MB-enriched transcripts. MB-specific transcripts had an RPKM score of ≥2 for both the MB/metaphase and MB/interphase ratios. MB-enriched transcripts had a score of ≥2 in either the MB/metaphase or MB/interphase ratios. The log2 enrichment score of MB/metaphase transcripts was plotted against the log2 enrichment score of MB/interphase transcripts using the R programming language and package ggplot2 (R Core Team, 2018; https://www.R-project.org/)99
Gene ontology:
Gene ontology analysis was performed using human ortholog UniProt IDs as input for PANTHER100. Biological process terms (transcription, cell cycle, RNA processing, cell fate, signal transduction, and DNA processing) were assigned through a combination of PANTHER/UniProt analysis and manual annotation and were assembled into Fig. 1 and Supp. Tables 1, 2, and 3.
Tableau visualization:
We delivered our annotated data for the 22 MB-enriched RNAs into Tableau (https://www.tableau.com) to create Fig. 1C. Each color represents an association with a particular gene ontology term, and the size of each circle correlates to the enrichment score.
Immunofluorescence:
MBs and MBRs from synchronized and asynchronized cells (HeLa or CHO), respectively, were fixed. Cells were cultured on high-precision cover glasses, fixed in 3% paraformaldehyde (Electron Microscopy Sciences, Cat# 15735-85) with 0.3% Triton® X-100 (Sigma-Aldrich, Cat# T9284) in PHEM buffer (60 mM PIPES, 27 mM HEPES, 10 mM EGTA, 4 mM MgSO4, pH 7.0) for 10 min at room temperature, blocked for 60 min in blocking solution (PHEM with 3% bovine serum albumin(BSA) (Sigma-Aldrich, Cat# A2153)), and incubated with primary or secondary antibodies in blocking solution (PHEM with 3% BSA). Cover glasses were mounted on slides using Fluoro-Gel mounting medium (Electron Microscopy Sciences, Cat# 17985-03) for SIM microscopy.
RNAscope/Fluorescent in situ hybridization:
RNA in situ hybridization was performed using the RNAscope Multiplex Fluorescent kit (Cat# 323100; Advanced Cell Diagnostics, Inc.) in accordance with the manufacturer’s instructions. Briefly, CHO or HeLa cells were fixed for 30 minutes with 4% paraformaldehyde in 0.1 M PBS (15735-85; Electron Microscopy Sciences) on cover glasses coated in poly-L-lysine (P4832; Sigma), dehydrated through a graded ethanol series (50%, 70%, 100%, 100%), and stored overnight at 4°C. Cells were rehydrated through a graded ethanol series (100%, 70%, 50%, PBS, PBS) at room temperature and pretreated with hydrogen peroxide and then protease III for 10 minutes each prior to hybridization. Cells were hybridized using custom RNAscope probe sets designed against Klf4, Jun, PolyA, Zfp36, Kif23, and DapB (control) mRNA sequences (Cat# 563611; 563621; 318631; 563631; 558051; 310043; Advanced Cell Diagnostics, Inc., respectively).The preamplifier, amplifier and HRP-labeled probes were then hybridized sequentially, followed by immunofluorescence labeling with Alexa488 or Alexa568 conjugated tyramide (AAT Bioquest, Cat# 11070; Thermo Fisher Scientific Inc, Cat# B40956, respectively). Subsequent immunofluorescent stainings were performed using anti-α-tubulin and/or anti-MKLP1 antibodies in Dulbecco’s PBS (DPBS) with 3% bovine serum albumin and 0.1% saponin (AAA1882014; Thermo Fisher Scientific). Cover glasses were mounted on microscope slides using Fluoro Gel mounting medium.
Structured illumination microscopy imaging:
Structured illumination microscopy was performed on a motorized inverted Eclipse Ti-E structured illumination microscope (Nikon) at the University of Wisconsin–Madison Biochemistry Optical Core. Images were captured on an Andor iXon 897 electron-multiplying charge-coupled device camera (Andor Technology). Images were captured and processed using NIS-Elements AR with N-SIM software (Nikon).
Hexanediol treatments:
HeLa cells were cultured and synchronized as described above. HeLa cells were released from the S phase block by transfer to a normal medium for 8.25 to 8.5 hours, and contractile ring-mediated early MB stages were visually confirmed. Cells were treated with medium supplemented with 7.5% 1,6-hexanediol (240117; Sigma-Aldrich) for 90 seconds, washed with PBS, and incubated in normal medium. Cells were fixed and processed for immunofluorescence as described above. GFP-MKLP1 expressing HeLa cells were treated with hexanediol as described above, and then processed for time-lapse imaging.
FRAP experiment:
GFP-MKLP1 expressing cells were imaged using an inverted NickonEclipse TiE microscope equipped with a CSU-X1 spinning disk confocal scanning unit (Yokogawa) and a EMCCD Camera (Evolve 512 Delta, Photometrics). Bleaching was performed by scanning 3 iterations of 488 nm excitation throughout the bleaching ROI. Images were acquired every 20 seconds with a x100 1.4 NA PL-APO VC objective lens and MetaMorph software (MDS).
Puromycin labeling to visualize translation in midbodies:
HeLa and CHO cells were cultured and synchronized as described above. MB-stage cells or asynchronous cell populations were treated with medium supplemented with 91 μM puromycin for 4 minutes, washed twice in DPBS, and immediately fixed in 3% paraformaldehyde with 0.3% Triton® X-100 in PHEM buffer on poly-L-lysine-coated cover glasses for 10 minutes. Translation was visualized using anti-puromycin primary antibodies (Millipore Sigma, Cat# MABE343), co-incubated with anti-MKLP1 antibodies (Novus Biologicals, Cat# NBP2-56923) as a marker for midbodies and midbody remnants and processed as described above. We quantified then the number of puro rings we observed at different stages and plotted this using Excel and GraphPad Prism.
HPG-ClickIT and OPP-ClickIT experiments:
For analysis of newly synthetized proteins, HeLa cells were washed and grown in methionine-free RPMI media (Thermo Fisher Scientific, Cat# A1451701) for 2 hours containing HPG (400 μM; manufacturer’s guideline is 50μM) with/without 9.4μM anisomycin or 335μM cycleheximide (A9789; C1988; Sigma, respectively). After incubation, cells were fixed with 4% paraformaldehyde for 15min, washed with DPBS containing 3% BSA and then 0.25% Triton-X-100 was incubated to the cells for 5min. For the detection of Click-IT HPG, Click-IT reaction cocktail containing the Alexa Fluor® 488 azide or BP Fluor 555 azide (BroadPharm, Cat# BP-25564) was incubated for 30min in dark. Additionally Click-IT® Plus OPP Alexa Fluor® 488 protein synthesis assay kit was used for another type of detection of newly synthetized proteins. HeLa cells were incubated in growth media with 20μM Click-IT® OPP (O-propargyl-puromycin) working solution for 4 min, and then fixed by the same methods with HPG Click-IT®. The fixed cells were then incubated with Click-IT® OPP reaction cocktail for 30 min at room temperature in dark.
siRNA experiments and genes:
HeLa cells were seeded in 6-well plates and cultured in DMEM/high-glucose/GlutaMAX at 37 °C under a 5% CO2 atmosphere before transfection. After one-day growth, siRNA transfection (at a final concentration of 80pmols) was performed using lipofectamine™ RNAiMAX. 8μl siRNAs and 6μl siRNA transfection reagent were diluted in each 100μl Opti-MEM™ media (Thermo Fisher, Cat# 31985062), then mixed and incubated for 5 min at room temperature. Subsequently, the mixtures totally 214μl were added to each well containing cells and 800μl growth medium. The mixture was then incubated in HeLa cells for 15 hours. Following that, new media were replaced to reduce toxicity of transfected reagent and then the transfected cells were cultured up to 24h or 48h since the transfection. For a matured midbody synchronization, the siRNA transfected cells were blocked by nocodazole (25ng/ml) for 5 hours, then mitotically rounded cells were physically shake with new culture media and the floating cells were transferred on poly-L-lysine coated cover slip. And then the prophase cells were released to the midbody-stage for 4 hours. The synchronization was performed before 9 hours from a fixed time point (24h or 48h).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of siRNA experiments:
To determine the number of bi-nucleates or multi-nucleates, we visualized DAPI, MKLP1, Phalloidin, and α-tubulin in the control and siRNA treated samples both using 20x objective of an ECHO Revolve Microscope (Echo Laboratories, San Diego, CA, USA). For IST1 siRNA experiments, we determined the number of cells stuck at the midbody bridge stage versus non-dividing cells. All visualized images were analyzed from at least 100 nuclei per each group.
Quantification of fixed midbody bridges and midbody remnants:
To quantify MKLP1, RBP, midbody factors, and tubulin signals in the intercellular bridges or MBRs, we performed line scans using profile plot analysis in ImageJ/FIJI. Fluorescence intensity values represent the average fluorescence intensity measured from a 6.5μm wide line along the axis of the midbody bridge. Line scan data were analyzed using unpaired two-tailed Student’s t-test. P-values below 0.05 were considered significant and reported in figures as *p<0.05; **p<0.01; ***p<0.001. p value above 0.05 were considered not significant and were not reported in figures. Statistical analyses were performed using Excel and Graphpad Prism software. All line scan results are shown as mean ± SEM of at least five images. *denotes significance, n.s. denotes not significant.
Quantification of translation signals in the midbody:
To quantify the α-Puro, OPP Click-IT® and HPG(L-Homopropargylglycine) Click-IT® signals we performed line scans. Fluorescence intensity values represent the average fluorescence intensity measured from a 6.5μm wide line along the axis of the midbody bridge. Graphs were assembled using GraphPad Prism. All line scan results are shown as mean ± SEM of at least five images.
Quantification of FRAP images:
To quantify MKLP1 and MKLP2 dynamics in HeLa cells, the mean intensity values for two different regions of identical areas were obtained for each image frame, as photobleached (Fp) and not photobleached (Fo). An empty region of the frame was used to measure the background (Fb). The pre-photobleaching value was normalized to 1 for each sample. The fraction of fluorescent recovery for each frame was calculated as follows: (Fp−Fb)/(Fo−Fb) and plotted as a function of time.
Supplementary Material
Table S1: Midbody-enriched transcripts related to Figure 1.
MB-enriched transcripts were identified as the 22 transcripts that had an enrichment score (RPKM/RPKM) of ≥2 when compared with both interphase (“midbody/interphase enrichment score,” highlighted in yellow) and metaphase (“midbody/metaphase enrichment score,” highlighted in peach). Colors and gene ontology terms correspond to those in Fig. 1C and 1D.
Table S2: Midbody-enriched transcripts in metaphase related to Figure 1.
MB-enriched transcripts in metaphase were identified as the 86 transcripts that had an enrichment score (RPKM/RPKM) of ≥2 when compared with metaphase (“midbody/metaphase enrichment score,” highlighted in peach).
Table S3: Midbody-enriched transcripts in interphase related to Figure 1. MB-enriched transcripts in interphase were identified as the 1051 transcripts that had an enrichment score (RPKM/RPKM) of ≥2 when compared with interphase (“midbody/interphase enrichment score,” highlighted in yellow).
Table S4: Raw data from CHO cell RNA-Seq related to Figure 1.
Raw data from RNA-Seq experiments on all three CHO cell libraries (interphase, metaphase, and MB). The CHO RefSeq accession number, the read count, and the RPKM for each transcript in each library are displayed. The human UniProt ID corresponding to each transcript is displayed together with the e-value for each ID. MB/interphase and MB/metaphase enrichment scores and the enrichment scores after log2 transformation are given. Ints2 (UniProt ID: Q9H0H0) and Cse1l (UniProt ID: P55060) were used as controls in the qPCR experiments and are highlighted in yellow at the top.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Annexin XI (mouse monoclonal) | Santa Cruz | Cat#sc-46686 |
| Anti-ARC (rabbit polyclonal) | Custom antibody provided from Jason Shepherd | University of Utah |
| Anti-ARC (mouse monoclonal) | Santa Cruz | Cat#sc-17839 |
| Anti-ATXN2L (rabbit polyclonal) | Thermo Scientific | Cat#PA5-59601 |
| Anti-BIRC3 (rabbit polyclonal) | Sigma | Cat#HPA002317 |
| Anti-cFOS (mouse monoclonal) | Santa Cruz | Cat#sc-8047 |
| Anti-CRIK (mouse monoclonal) | Santa Cruz | Cat#sc-390437 |
| Anti-dsRNA (mouse monoclonal) | EMD Millipore | Cat#MABE1134 |
| Anti-EEF2K [EP881Y] (rabbit monoclonal) | Abcam | Cat#ab45168 |
| Anti-EIF3E (rabbit polyclonal) | Sigma | Cat#HPA023973 |
| Anti-EIF4E [Y448] (rabbit monoclonal) | Abcam | Cat#ab33766 |
| Anti-EIF4G1 (rabbit polyclonal) | Abcam | Cat#ab47625 |
| Anti-FOSB (mouse monoclonal) | Santa Cruz | Cat#sc-398595 |
| Anti-Histone H1 (mouse monoclonal) | Santa Cruz | Cat#sc-8030 |
| Anti-IRF1 (mouse monoclonal) | Santa Cruz | Cat#sc-74530 |
| Anti-IκBα (mouse monoclonal) | Santa Cruz | Cat#sc-1643 |
| Anti-JUN (rabbit polyclonal) | Sigma | Cat#HPA059474 |
| Anti-KLF4 (rabbit polyclonal) | Sigma | Cat#HPA002926 |
| Anti-KLF6 (mouse monoclonal) | Santa Cruz | Cat#sc-365633 |
| Anti-Lamin A/C (mouse monoclonal) | Cell Signaling | Cat#4777S |
| Anti-MKLP1; Discontinued (rabbit polyclonal) | Santa Cruz | Cat#sc-867 |
| Anti-MKLP1 (rabbit polyclonal) | Novus | Cat#NBP2-56923 |
| Anti-MRRF (rabbit polyclonal) | Novus | Cat#NBP2-33586 |
| Anti-PABP (mouse monoclonal) | Santa Cruz | Cat#sc-166027 |
| Anti-Puromycin, clone 12D10 (mouse monoclonal) | EMD Millipore | Cat#MABE343 |
| Anti-Puromycin, clone 12D10, Alexa Fluor® 488 Conjugate (mouse monoclonal) | EMD Millipore | Cat#MABE343-AF488 |
| Anti-Puromycin, clone 12D10, Alexa Fluor® 647 Conjugate (mouse monoclonal) | EMD Millipore | Cat#MABE343-AF647 |
| Anti-RacGAP1 (mouse monoclonal) | Santa Cruz | Cat#sc-166477 |
| Anti-RPL10A (rabbit polyclonal) | Sigma | Cat#HPA053803 |
| Anti-RPS5 (rabbit polyclonal) | Genetex | Cat#GTX32851 |
| Anti-dsRNA (mouse monoclonal) | Fisher Scientific | Cat#MABE1134100 |
| Anti-Stau1 (mouse monoclonal) | Santa Cruz | Cat#sc-390820 |
| Anti-TARDBP/TDP43 (mouse monoclonal) | Santa Cruz | Cat#sc-376311 |
| Anti-TEX14 (rabbit polyclonal) | Thermo Fisher | Cat#PA5-44140 |
| Anti-TIA1 (mouse monoclonal) | Santa Cruz | Cat#sc-166247 |
| Anti-TIS11B (mouse monoclonal) | Santa Cruz | Cat#sc-293267 |
| Anti-ZFP36 (rabbit polyclonal) | Thermo Fisher | Cat#PA5-40876 |
| Anti-α-tubulin (DM1A) (mouse monoclonal) | Thermo Scientific | Cat#62204 |
| Anti-α-tubulin, clone DM1A, Alexa Fluor® 488 conjugate (mouse monoclonal) | EMD Millipore | Cat#16-232 |
| Anti-α-tubulin, clone DM1A, Alexa Fluor® 555 conjugate (mouse monoclonal) | EMD Millipore | Cat#05-829X-555 |
| Anti-α-tubulin, clone DM1A, Alexa Fluor® 647 conjugate (mouse monoclonal) | EMD Millipore | Cat#05-829-AF647 |
| Alexa Fluor® 488-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat#111-545-003 |
| Alexa Fluor 568 Dnk Anti-Mouse IgG | Abcam | Cat#ab175472 |
| Alexa Fluor® 594-conjugated AffiniPure Sheep Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat#515-585-003 |
| Alexa Fluor® 647-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat#111-605-003 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher | Cat#A11004 |
| Chemicals, peptides, and recombinant proteins | ||
| Nocodazole (25ng/ml or 50ng/ml for HeLa; 100ng/ml for CHO) | Sigma | Cat#M1404 |
| Puromycin dihydrochloride (final concentration 91μM) | Sigma | Cat#P8833 |
| Thymidine (final concentration 2mM) | Sigma | Cat#T1895 |
| 1,6-Hexanediol, 99% (final concentration 7.5%; Dissolved by media) | Sigma | Cat#240117 |
| Cycloheximide (final concentration 335μM) | Sigma | Cat#C1988 |
| L-Homopropargylglycine hydrochloride (400μM; Water soluble; Extra amount HPG required for Click-iT™ HPG Alexa Fluor™ 488 Protein Synthesis Assay Kit) | Sigma | Cat#900893 |
| Anisomycin (final concentration 9.4μM) | Sigma | Cat#A9789 |
| Fluoro-Gel Mounting Medium with TES Buffer | Electron Microscopy Sciences | Cat#17985-30 |
| BP Fluor 555 Azide | BroadPharm | Cat#BP-25564 |
| AF568 tyramide reagent | Thermo Scientific | Cat#B40956 |
| AF488 tyramide reagent | AAT Bioquest | Cat#11070 |
| Phalloidin CruzFluor 555 | Santa Cruz | Cat#sc-363794 |
| Critical commercial assays | ||
| Life Technologies click It Plus Opp Af488 Kit 50 (OPP; 20μM; similar amount of puromycin 91μM) | Thermo Scientific | Cat#C10456 |
| Click-iT™ HPG Alexa Fluor™ 488 Protein Synthesis Assay Kit Invitrogen [HPG (400μM) eight times higher concentration than manufaturer’s recommendation] | Thermo Scientific | Cat#C10428 |
| RPMI, no methionine (for HPG reaction) | Thermo Scientific | Cat#A1451701 |
| RNAScope Multiplex Fluorescent Detection Reagent Kit V2 | Advanced Cell Diagnostics | Cat#323100 |
| PolyA | Advanced Cell Diagnostics | Cat#318631 |
| dapB | Advanced Cell Diagnostics | Cat#310043 |
| Cricetulus griseus JUN (targeting 120-1471 of XM_007643818.1) | Advanced Cell Diagnostics | Cat#563621 |
| Cricetulus griseus KLF4 (targeting 447-1862 of XM_003511916.2) | Advanced Cell Diagnostics | Cat#563611 |
| Cricetulus griseus KIF23 (targeting 834-1819 of NM_001243981.1) | Advanced Cell Diagnostics | Cat#558051 |
| Cricetulus griseus ZFP36 (targeting 671-1622 of XM_007644391.1) | Advanced Cell Diagnostics | Cat#563631 |
| Hs-Kif23 | Advanced Cell Diagnostics | Cat#1159491-C1 |
| Hs-KLF4 | Advanced Cell Diagnostics | Cat#457461 |
| Hs-JUN | Advanced Cell Diagnostics | Cat#470541 |
| Hs-ANXA11 | Advanced Cell Diagnostics | Cat#1159481-C1 |
| Hs-EPEMP1 | Advanced Cell Diagnostics | Cat#493721 |
| Hs-CNCL5 | Advanced Cell Diagnostics | Cat#433161 |
| Cg-PPIB | Advanced Cell Diagnostics | Cat#450461 |
| Lipofectamine RNAiMAX Transfection Reagent [(1:35.66) dilution 6μl/Opti-MEM 200μl] | Thermo Scientific | Cat#13-778-075 |
| Opti-MEM™ media (for siRNAs transfection) | Thermo Scientific | Cat#31985062 |
| Experimental models: Cell lines | ||
| Hamster: CHO-K1 cells | ATCC | CCL-61 |
| Human: HeLa cells | ATCC | CCL-2 |
| Human: HeLa Kyoto cells | Capalbo, et al 201927 | University of Cambridge, UK |
| Human: MKLP1-GFP expressing HeLa Kyoto cells | Douglas, et al 201048 | University of Warwick, UK |
| Human: hTERT RPE-1 cells | ATCC | CRL-4000 |
| Mouse: AtT-20/D16v-F2 cells | ATCC | CRL-1795 |
| Mouse: Primary hippocampal mouse neural stem cells (NSPC cells) | Moore et al., 201594 | University of Wisconsin-Madison |
| Oligonucleotides | ||
| control A siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-37007 |
| ARC siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-29721 |
| MKLP1 siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-35936 |
| IST1 siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-93481 |
| TIS11B siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-76672 |
| Annexin XI siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-29694 |
| Stau1 siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-76586 |
| ATXN2L siRNA (374nM; dilution 10μM stock 8ul/Opti-MEM 200μl) | Santa Cruz | Cat#sc-93060 |
| Software and algorithms | ||
| ImageJ/FiJI | https://fiji.sc | |
Highlights.
The midbody is the assembly site of a RNP granule, we call the MB granule
Distinct oncogenic and pluripotent transcription factor RNAs are packaged in MBs and MBRs
The MB and MBR are sites of active translation
Multiple cell types including cancer, stem, neural stem, have actively translating MBRs
Acknowledgments
The authors thank Judith Kimble, Robert Singer, Michael Sussman, Harmit Malik, Diana Chu, Abby Dernburg, Karen Schindler, Daniel Jung, Maureen Barr, Francisco Pelegri, Dana Miller, Elizabeth Torr and Smit Patel, and Premal Shah, for sharing their expertise, support, and advice. ARS is particularly grateful to John G. White for his mentorship, friendship, and discussions about the midbody over the years. We are indebted to the work performed by Jessica Shivas, Jennifer Gilbert, and Lan Qin during the initial stages of this project. Special thanks to Chris Morrow and Darcie Moore for the NSPC cells; Sakae Ikeda and Akihiro Ikeda for the RPE cells. Special thanks to Kurt Weiss, Elle Grevstad and Peter Favreau for their technical assistance with structured illumination microscopy at the University of Wisconsin–Madison Biochemistry Optical Core. We can’t thank Adam Steinberg enough (artforscience.com) for pushing the science further by showing gaps in our knowledge throughout the process of our research in our model. ARS is supported by the National Institutes of Health (R01 GM139695-01A1). Imaging was performed at the University of Wisconsin–Madison Biochemistry Optical Core, WI, United States, which was established with support from the University of Wisconsin–Madison Department of Biochemistry Endowment. JS is funded by a NIH Transformative R01 NS115716 and a Chan Zuckerberg Initiative Ben Barres Early Career Acceleration Award. MDB was supported by the National Institutes of Health R01 GM122893 and GM144352. Part of this work has been supported by Institut Pasteur, CNRS, and ANR (Cytosign, SeptScort) to AE, AP received a fellowship from the Doctoral School Complexité du Vivant ED515, contrat n°2611 bis/2016 and Fondation ARC pour la recherche sur le cancer (DOC20190508876).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
References
- 1.Skop AR, Liu H, Yates J, Meyer BJ, and Heald R (2004). Dissection of the Mammalian Midbody Proteome Reveals Conserved Cytokinesis Mechanisms. Science 305, 61–66. 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mierzwa BE, Chiaruttini N, Redondo-Morata L, Filseck J.M. von, König J, Larios J, Poser I, Müller-Reichert T, Scheuring S, Roux A, et al. (2017). Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 19, 787–798. 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guizetti J, and Gerlich DW (2010). Cytokinetic abscission in animal cells. Semin Cell Dev Biol 21, 909–916. 10.1016/j.semcdb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Matuliene J, and Kuriyama R (2004). Role of the Midbody Matrix in Cytokinesis: RNAi and Genetic Rescue Analysis of the Mammalian Motor Protein CHO1. Mol Biol Cell 15, 3083–3094. 10.1091/mbc.e03-12-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steigemann P, and Gerlich DW (2009). Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol 19, 606–616. 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Schink KO, and Stenmark H (2011). Cell differentiation: midbody remnants - junk or fate factors? Curr Biology Cb 21, R958–60. 10.1016/j.cub.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Sagona AP, and Stenmark H (2010). Cytokinesis and cancer. Febs Lett 584, 2652–2661. 10.1016/j.febslet.2010.03.044. [DOI] [PubMed] [Google Scholar]
- 8.Crowell EF, Tinevez J-Y, and Echard A (2013). A simple model for the fate of the cytokinesis midbody remnant: implications for remnant degradation by autophagy. Bioessays News Rev Mol Cell Dev Biology 35, 472–481. 10.1002/bies.201200132. [DOI] [PubMed] [Google Scholar]
- 9.Addi C, Bai J, and Echard A (2018). Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr Opin Cell Biol 50, 27–34. 10.1016/j.ceb.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Crowell EF, Gaffuri A-L, Gayraud-Morel B, Tajbakhsh S, and Echard A (2014). Engulfment of the midbody remnant after cytokinesis in mammalian cells. J Cell Sci 127, 3840–3851. 10.1242/jcs.154732. [DOI] [PubMed] [Google Scholar]
- 11.Chaigne A, and Brunet T (2022). Incomplete abscission and cytoplasmic bridges in the evolution of eukaryotic multicellularity. Curr Biol 32, R385–R397. 10.1016/j.cub.2022.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Peterman E, Gibieža P, Schafer J, Skeberdis VA, Kaupinis A, Valius M, Heiligenstein X, Hurbain I, Raposo G, and Prekeris R (2019). The post-abscission midbody is an intracellular signaling organelle that regulates cell proliferation. Nat Commun 10, 3181. 10.1038/s41467-019-10871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frémont S, and Echard A (2018). Membrane Traffic in the Late Steps of Cytokinesis. Curr Biology Cb 28, R458–R470. 10.1016/j.cub.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 14.Chen C-T, Ettinger AW, Huttner WB, and Doxsey SJ (2012). Resurrecting remnants: the lives of post-mitotic midbodies. Trends Cell Biol 23, 118–128. 10.1016/j.tcb.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ettinger AW, Wilsch-Bräuninger M, Marzesco A-M, Bickle M, Lohmann A, Maliga Z, Karbanová J, Corbeil D, Hyman AA, and Huttner WB (2011). Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat Commun 2, 503. 10.1038/ncomms1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arai Y, Sampaio JL, Wilsch-Bräuninger M, Ettinger AW, Haffner C, and Huttner WB (2015). Lipidome of midbody released from neural stem and progenitor cells during mammalian cortical neurogenesis. Front Cell Neurosci 9, 325. 10.3389/fncel.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou G, Gentili C, and Gönczy P (2014). Stereotyped distribution of midbody remnants in early C. elegans embryos requires cell death genes and is dispensable for development. Cell Res 24, 251–253. 10.1038/cr.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Presle A, Frémont S, Salles A, Commere P-H, Sassoon N, Berlioz-Torrent C, Gupta-Rossi N, and Echard A (2021). The viral restriction factor tetherin/BST2 tethers cytokinetic midbody remnants to the cell surface. Curr Biol. 10.1016/j.cub.2021.02.039. [DOI] [PubMed] [Google Scholar]
- 19.Kuo T-C, Chen C-T, Baron D, Onder TT, Loewer S, Almeida S, Weismann CM, Xu P, Houghton J-M, Gao F-B, et al. (2011). Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol 13, 1214–1223. 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai A, Greening DW, Xu R, Chen M, Suwakulsiri W, and Simpson RJ (2021). Secreted midbody remnants are a class of extracellular vesicles molecularly distinct from exosomes and microparticles. Commun Biology 4, 400. 10.1038/s42003-021-01882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangan AJ, Sietsema DV, Li D, Moore JK, Citi S, and Prekeris R (2016). Cingulin and actin mediate midbody-dependent apical lumen formation during polarization of epithelial cells. Nat Commun 7, 12426. 10.1038/ncomms12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernabé-Rubio M, Andrés G, Casares-Arias J, Fernández-Barrera J, Rangel L, Reglero-Real N, Gershlick DC, Fernández JJ, Millán J, Correas I, et al. (2016). Novel role for the midbody in primary ciliogenesis by polarized epithelial cells. J Cell Biology 214, 259–273. 10.1083/jcb.201601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding BN, Moccia A, Drunat S, Soukarieh O, Tubeuf H, Chitty LS, Verloes A, Gressens P, Ghouzzi VE, Joriot S, et al. (2016). Mutations in Citron Kinase Cause Recessive Microlissencephaly with Multinucleated Neurons. Am J Hum Genet 99, 511–520. 10.1016/j.ajhg.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh D, and Pohl C (2014). Coupling of Rotational Cortical Flow, Asymmetric Midbody Positioning, and Spindle Rotation Mediates Dorsoventral Axis Formation in C. elegans. Dev Cell 28, 253–267. 10.1016/j.devcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Chaigne A, Labouesse C, White IJ, Agnew M, Hannezo E, Chalut KJ, and Paluch EK (2020). Abscission Couples Cell Division to Embryonic Stem Cell Fate. Dev Cell. 10.1016/j.devcel.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Addi C, Presle A, Frémont S, Cuvelier F, Rocancourt M, Milin F, Schmutz S, Chamot-Rooke J, Douché T, Duchateau M, et al. (2020). The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat Commun 11, 1941. 10.1038/S41467-020-15205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capalbo L, Bassi ZI, Geymonat M, Todesca S, Copoiu L, Enright AJ, Callaini G, Riparbelli MG, Yu L, Choudhary JS, et al. (2019). The midbody interactome reveals unexpected roles for PP1 phosphatases in cytokinesis. Nat Commun 10, 4513. 10.1038/s41467-019-12507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng R, Shen Z, Tripathi V, Xuan Z, Freier SM, Bennett CF, Prasanth SG, and Prasanth KV (2010). Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J Cell Sci 123, 3734–3744. 10.1242/jcs.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellitto C, and Kuriyama R (1988). Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J Cell Biology 106, 431–439. 10.1083/jcb.106.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullins JM, and McIntosh JR (1982). Isolation and initial characterization of the mammalian midbody. J Cell Biology 94, 654–661. 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullins J, and Biesele J (1977). Terminal phase of cytokinesis in D-98S cells. J Cell Biology 73, 672–684. 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullins JM, and Biesele JJ (1973). Cytokinetic activities in a human cell line: the midbody and intracellular bridge. Tissue Cell 5, 47–61. 10.1016/s0040-8166(73)80005-9. [DOI] [PubMed] [Google Scholar]
- 33.Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Bio 18, 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M-T, Kriwacki RW, and Brangwynne CP (2020). Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214. 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, and Hyman AA (2009). Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science 324, 1729–1732. 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 36.Lafontaine DLJ, Riback JA, Bascetin R, and Brangwynne CP (2020). The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Bio, 1–18. 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 37.Mullins JM, and McIntosh JR (1979). Birefringence of the mammalian midbody. Exp Cell Res 121, 395–399. 10.1016/0014-4827(79)90019-3. [DOI] [PubMed] [Google Scholar]
- 38.Bonner MK, Poole DS, Xu T, Sarkeshik A, Yates JR, and Skop AR (2011). Mitotic spindle proteomics in Chinese hamster ovary cells. Plos One 6, e20489. 10.1371/journal.pone.0020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonner MK, Han BH, and Skop A (2013). Profiling of the Mammalian Mitotic Spindle Proteome Reveals an ER Protein, OSTD-1, as Being Necessary for Cell Division and ER Morphology. Plos One 8, e77051. 10.1371/journal.pone.0077051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuriyama R, Gustus C, Terada Y, Uetake Y, and Matuliene J (2002). CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J Cell Biology 156, 783–790. 10.1083/jcb.200109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma W, and Mayr C (2018). A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3’UTR-Mediated Protein-Protein Interactions. Cell 175, 1492–1506.e19. 10.1016/j.cell.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanduja S, Blanco FF, and Dixon DA (2011). The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev Rna 2, 42–57. 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciais D, Cherradi N, and Feige J-J (2013). Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cell Mol Life Sci 70, 2031–2044. 10.1007/s00018-012-1150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu C-Κ, Coughlin M, and Mitchison TJ (2012). Midbody assembly and its regulation during cytokinesis. Mol Biol Cell 23, 1024–1034. 10.1091/mbc.e11-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishima M, Pavicic V, Grüneberg U, Nigg EA, and Glotzer M (2004). Cell cycle regulation of central spindle assembly. Nature 430, 908–913. 10.1038/nature02767. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura M, Verboon JM, Prentiss CL, and Parkhurst SM (2020). The kinesin-like protein Pavarotti functions noncanonically to regulate actin dynamics. J Cell Biol 219. 10.1083/jcb.201912117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi S, Fusaki N, Ohta S, Iwahori Y, lizuka Y, Inagawa K, Kawakami Y, Yoshida K, and Toda M (2012). Downregulation of KIF23 suppresses glioma proliferation. J Neuro-oncol 106, 519–529. 10.1007/s11060-011-0706-2. [DOI] [PubMed] [Google Scholar]
- 48.Douglas ME, Davies T, Joseph N, and Mishima M (2010). Aurora B and 14-3-3 Coordinately Regulate Clustering of Centralspindlin during Cytokinesis. Curr Biol 20, 927–933. 10.1016/j.cub.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matuliene J, and Kuriyama R (2002). Kinesin-like Protein CHO1 Is Required for the Formation of Midbody Matrix and the Completion of Cytokinesis in Mammalian Cells. Mol Biol Cell 13, 1832–1845. 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, Belnap DM, Erlendsson S, Morado DR, et al. (2018). The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 172, 275–288.e18. 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mierzwa B, and Gerlich DW (2014). Cytokinetic Abscission: Molecular Mechanisms and Temporal Control. Dev Cell 31, 525–538. 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Renvoisé B, Parker RL, Yang D, Bakowska JC, Hurley JH, and Blackstone C (2010). SPG20 Protein Spartin Is Recruited to Midbodies by ESCRT-III Protein Ist1 and Participates in Cytokinesis. Mol Biol Cell 21, 3293–3303. 10.1091/mbc.e09-10-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talledge N, McCullough J, Wenzel D, Nguyen HC, Lalonde MS, Bajorek M, Skalicky J, Frost A, and Sundqust WI (2018). The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids. Biorxiv, 386532. 10.1101/386532. [DOI] [Google Scholar]
- 54.Vietri M, Radulovic M, and Stenmark H (2020). The many functions of ESCRTs. Nat Rev Mol Cell Bio 21, 25–42. 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 55.Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, et al. (2018). RNP-Granule Assembly via Ataxin-2 Disordered Domains Is Required for Long-Term Memory and Neurodegeneration. Neuron 98, 754–766.e4. 10.1016/j.neuron.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 56.Gnazzo MM, Uhlemann E-ME, Villarreal AR, Shirayama M, Dominguez EG, and Skop AR (2016). The RNA-binding protein ATX-2 regulates cytokinesis through PAR-5 and ZEN-4. Mol Biol Cell 27, 3052–3064. 10.1091/mbc.e16-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y-C, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon-Abell J, Shen Y, et al. (2019). RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell 179, 147–164.e20. 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler JR, Matheny T, Jain S, Abrisch R, and Parker R (2016). Distinct stages in stress granule assembly and disassembly. Elife 5, e18413. 10.7554/elife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain A, and Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saha S, and Hyman AA (2017). RNA gets in phase. J Cell Biol 216, 2235–2237. 10.1083/jcb.201706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Echard A, Jollivet F, Martinez O, Lacapère J-J, Rousselet A, Janoueix-Lerosey I, and Goud B (1998). Interaction of a Golgi-Associated Kinesin-Like Protein with Rab6. Science 279, 580–585. 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 62.Fontijn RD, Goud B, Echard A, Jollivet F, Marie J. van, Pannekoek H, and Horrevoets AJG (2001). The Human Kinesin-Like Protein RB6K Is under Tight Cell Cycle Control and Is Essential for Cytokinesis. Mol. Cell. Biology 21, 2944–2955. 10.1128/mcb.21.8.2944-2955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hill E, Clarke M, and Barr FA (2000). The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. Embo J 19, 5711–5719. 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogler TO, Wheeler JR, Nguyen ED, Hughes MP, Britson KA, Lester E, Rao B, Betta ND, Whitney ON, Ewachiw TE, et al. (2018). TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563, 508–513. 10.1038/s41586-018-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zbinden A, Pérez-Berlanga M, Rossi PD, and Polymenidou M (2020). Phase Separation and Neurodegenerative Diseases: A Disturbance in the Force. Dev Cell 55, 45–68. 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 66.Kalluri R, and LeBleu VS (2020). The biology, function, and biomedical applications of exosomes. Sci New York N Y 367, eaau6977. 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bart G, Fischer D, Samoylenko A, Zhyvolozhnyi A, Stehantsev P, Miinalainen I, Kaakinen M, Nurmi T, Singh P, Kosamo S, et al. (2021). Characterization of nucleic acids from extracellular vesicle-enriched human sweat. Bmc Genomics 22, 425. 10.1186/s12864-021-07733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Enam SU, Zinshteyn B, Goldman DH, Cassani M, Livingston NM, Seydoux G, and Green R (2020). Puromycin reactivity does not accurately localize translation at the subcellular level. Elife 9, e60303. 10.7554/elife.60303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J, Xu Y, Stoleru D, and Salic A (2012). Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc National Acad Sci 109, 413–418. 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chao JA, Yoon YJ, and Singer RH (2012). Imaging Translation in Single Cells Using Fluorescent Microscopy. Csh Perspect Biol 4, a012310. 10.1101/cshperspect.a012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samo TJ, Smriga S, Malfatti F, Sherwood BP, and Azam F (2014). Broad distribution and high proportion of protein synthesis active marine bacteria revealed by click chemistry at the single cell level. Frontiers Mar Sci 1, 48. 10.3389/fmars.2014.00048. [DOI] [Google Scholar]
- 72.Franceschi ND, Alqabandi M, Miguet N, Caillat C, Mangenot S, Weissenhorn W, and Bassereau P (2018). The ESCRT protein CHMP2B acts as a diffusion barrier on reconstituted membrane necks. J Cell Sci 132, jcs217968. 10.1242/jcs.217968. [DOI] [PubMed] [Google Scholar]
- 73.Sabari BR (2020). Biomolecular Condensates and Gene Activation in Development and Disease. Dev Cell 55, 84–96. 10.1016/j.devcel.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 74.Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 75.Ouyang JPT, Folkmann A, Bernard L, Lee C-Y, Seroussi U, Charlesworth AG, Claycomb JM, and Seydoux G (2019). P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev Cell. 10.1016/j.devcel.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Echard A, Hickson GRX, Foley E, and O’Farrell PH (2004). Terminal Cytokinesis Events Uncovered after an RNAi Screen. Curr Biol 14, 1685–1693. 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilker EW, Vugt M.A.T.M. van, Artim SC, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. (2007). 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature 446, 329–332. 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 78.Gershony O, Pe’er T, Noach-Hirsh M, Elia N, and Tzur A (2014). Cytokinetic abscission is an acute G1 event. Cell Cycle GeorgetTex 13, 3436–3441. 10.4161/15384101.2014.956486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pyronnet S, Dostie J, and Sonenberg N (2001). Suppression of cap-dependent translation in mitosis. Gene Dev 15, 2083–2093. 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanenbaum ME, Stern-Ginossar N, Weissman JS, and Vale RD (2015). Regulation of mRNA translation during mitosis. Elite 4, e07957. 10.7554/elife.07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lie-Jensen A, Ivanauskiene K, Malerød L, Jain A, Tan KW, Laerdahl JK, Liestøl K, Stenmark H, and Haglund K (2019). Centralspindlin Recruits ALIX to the Midbody during Cytokinetic Abscission in Drosophila via a Mechanism Analogous to Virus Budding. Curr Biol 29, 3538–3548.e7. 10.1016/j.cub.2019.09.025. [DOI] [PubMed] [Google Scholar]
- 82.McNeely KC, and Dwyer ND (2021). Cytokinetic Abscission Regulation in Neural Stem Cells and Tissue Development. Curr Stem Cell Reports, 1–13. 10.1007/s40778-021-00193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pohl C, and Jentsch S (2008). Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell 132, 832–845. 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 84.Peterman E, and Prekeris R (2019). The postmitotic midbody: Regulating polarity, stemness, and proliferation. J Cell Biology 218, 3903–3911. 10.1083/jcb.201906148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bauer KE, Bargenda N, Schieweck R, Illig C, Segura I, Harner M, and Kiebler MA (2022). RNA supply drives physiological granule assembly in neurons. Nat. Commun 13, 2781. 10.1038/S41467-022-30067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krause LJ, Herrera MG, and Winklhofer KF (2022). The Role of Ubiquitin in Regulating Stress Granule Dynamics. Front. Physiol 13, 910759. 10.3389/fphys.2022.910759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schisa JA, and Elaswad MT (2021). An Emerging Role for Post-translational Modifications in Regulating RNP Condensates in the Germ Line. Front. Mol. Biosci 8, 658020. 10.3389/fmolb.2021.658020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharp JA, Perea-Resa C, Wang W, and Blower MD (2020). Cell division requires RNA eviction from condensing chromosomes. J Cell Biol 219. 10.1083/jcb.201910148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raich WB, Moran AN, Rothman JH, and Hardin J (1998). Cytokinesis and Midzone Microtubule Organization in Caenorhabditis elegans Require the Kinesin-like Protein ZEN-4. Mol Biol Cell 9, 2037–2049. 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.White EA, and Glotzer M (2012). Centralspindlin: at the heart of cytokinesis. Cytoskelet Hoboken N J 69, 882–892. 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kroschwald S, Maharana S, and Simon A (2017). Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters. 10.19185/matters.201702000010. [DOI] [Google Scholar]
- 92.Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, and Gerlich DW (2011). Cortical Constriction During Abscission Involves Helices of ESCRT-III–Dependent Filaments. Science 331, 1616–1620. 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 93.Ma HT, and Poon RYC (2016). Cell Cycle Synchronization, Methods and Protocols. Methods Mol Biology 1524, 189–201. 10.1007/978-1-4939-6603-5. [DOI] [PubMed] [Google Scholar]
- 94.Moore DL, Pilz GA, Araúzo-Bravo MJ, Barral Y, and Jessberger S (2015). A mechanism for the segregation of age in mammalian neural stem cells. Science 349, 1334–1338. 10.1126/science.aac9868. [DOI] [PubMed] [Google Scholar]
- 95.Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermúdez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, et al. (2013). A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol 15, 325–334. 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- 96.Schwarz DS, and Blower MD (2014). The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradationThe RNase XendoU regulates ER structure. J Cell Biology 207, 41–57. 10.1083/jcb.201406037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anders S, Pyl PT, and Huber W (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wickham H (2016). ggplot2, Elegant Graphics for Data Analysis. R 10.1007/978-3-319-24277-4. [DOI] [Google Scholar]
- 100.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, and Thomas PD (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189. 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Midbody-enriched transcripts related to Figure 1.
MB-enriched transcripts were identified as the 22 transcripts that had an enrichment score (RPKM/RPKM) of ≥2 when compared with both interphase (“midbody/interphase enrichment score,” highlighted in yellow) and metaphase (“midbody/metaphase enrichment score,” highlighted in peach). Colors and gene ontology terms correspond to those in Fig. 1C and 1D.
Table S2: Midbody-enriched transcripts in metaphase related to Figure 1.
MB-enriched transcripts in metaphase were identified as the 86 transcripts that had an enrichment score (RPKM/RPKM) of ≥2 when compared with metaphase (“midbody/metaphase enrichment score,” highlighted in peach).
Table S3: Midbody-enriched transcripts in interphase related to Figure 1. MB-enriched transcripts in interphase were identified as the 1051 transcripts that had an enrichment score (RPKM/RPKM) of ≥2 when compared with interphase (“midbody/interphase enrichment score,” highlighted in yellow).
Table S4: Raw data from CHO cell RNA-Seq related to Figure 1.
Raw data from RNA-Seq experiments on all three CHO cell libraries (interphase, metaphase, and MB). The CHO RefSeq accession number, the read count, and the RPKM for each transcript in each library are displayed. The human UniProt ID corresponding to each transcript is displayed together with the e-value for each ID. MB/interphase and MB/metaphase enrichment scores and the enrichment scores after log2 transformation are given. Ints2 (UniProt ID: Q9H0H0) and Cse1l (UniProt ID: P55060) were used as controls in the qPCR experiments and are highlighted in yellow at the top.
Data Availability Statement
RNA sequences associated with this study have been deposited into the National Institutes of Health Sequence Read Archive (Bioprojects: PRJNA555245 and SRA:SRP215214).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


