Summary
The fusion peptide of SARS-CoV-2 Spike is essential for infection. How this charged and hydrophobic domain occupies and affects membranes needs clarification. Its depth in zwitterionic, bilayered micelles at pH 5 (resembling late endosomes) was measured by paramagnetic NMR relaxation enhancements used to bias molecular dynamics simulations. Asp830 inserted deeply, along with Lys825 or Lys835. Protonation of Asp830 appeared to enhance agreement of simulated and NMR-measured depths. While the fusion peptide occupied a leaflet of the DMPC bilayer, the opposite leaflet invaginated with influx of water and choline headgroups in around Asp830 and bilayer-inserted polar sidechains. NMR-detected hydrogen exchange found corroborating hydration of the backbone of Thr827 – Phe833 inserted deeply in bicelles. Pinching of the membrane at the inserted charge and the intramembrane hydration of polar groups agree with theory. Formation of corridors of hydrated, inward-turned headgroups was accompanied by flip-flop of headgroups. Potential roles of the defects are discussed.
Graphical Abstract
Diverse experiments have implied deeper membrane insertion of the charged coronaviral fusion peptide (FP) than have published MD simulations. Van Doren et al. show compatible and complementary observations from NMR and simulations regarding FP insertion and hydration in a zwitterionic bilayer. NMR-aided MD simulations visualize provocative defects in this environment.
Introduction
Infections by coronaviruses rely upon the fusion peptide in viral-cell fusion.1–3 The importance of the fusion peptide of SARS-CoV-2 is underscored by the discovery of antibodies against it that neutralized multiple coronaviruses.1 The fusion peptide, at the N-terminus of the Spike S2 subunit, moves through changing environments during viral-cell fusion. The fusion peptide is unveiled by proteolysis of the S2’ site.4 This triggers a dramatic transformation of prefusion Spike to the rod-shaped, post-fusion S2 subunit.5,6 The rearrangement of S2 was proposed to pass through an intermediate that places the HR1 helix on the membrane of the host cell and the HR2 helix on the envelope of the coronavirus (Fig. 1A).7 Subsequent formation of a partially folded intermediate of S2 probably trimerizes about HR18 (Fig. 1B). Then HR2 folds back upon trimerized HR1 to form the hemi-fusion state of class I viral-cell fusion that uses the fusion peptide in drawing the host cell membrane close to the viral envelope9 (Fig. 1C). In virions of SARS-CoV-2, the post-fusion state of Spike appears to position the fusion peptide on the outside where it may contact a neighboring post-fusion Spike trimer.10 The SARS-CoV-2 fusion peptide was several-fold more active in membrane fusion at pH ≤ 6.5 than pH 7.4, and was not aided by calcium ions.11 This suggests the endocytic pathway could be a favored location of viral entry in some locations.11 The endocytic pathway ranges from about pH 6.5 to 59,12 and from 0.3 to 30 μM Ca2+.13,14 SARS-CoV-2 pseudoparticles entered cells efficiently through the endocytic pathway.4 The work below asks how the fusion peptide may insert in and affect a lipid bilayer under the mildly acidic and low calcium conditions of the endocytic route of infection.
Figure 1. Intermediates of coronaviral Spike S2, lipid bilayer-associated structural models, and aligned fusion peptide sequences are illustrated.
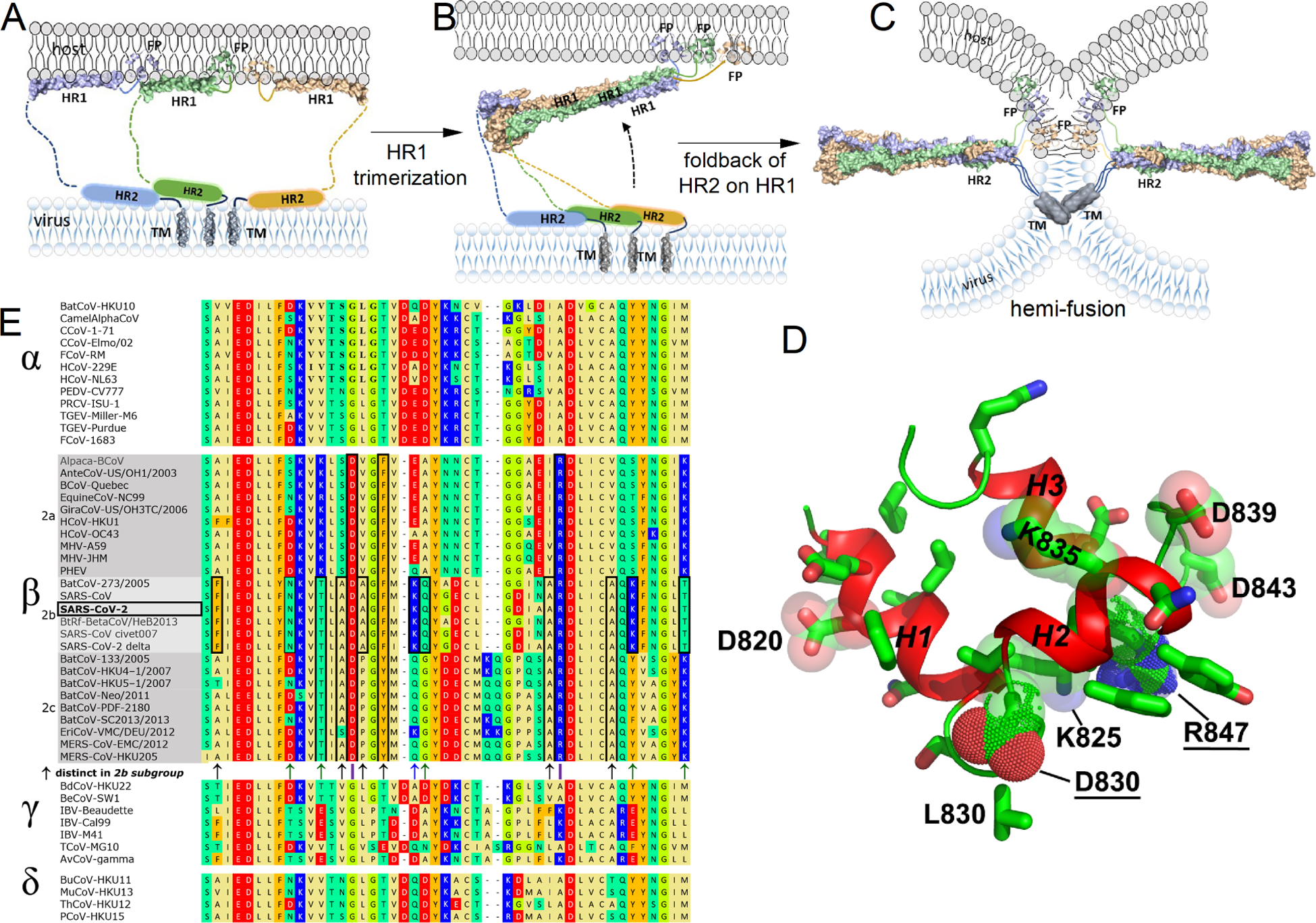
(A–C) Proposed intermediates in the maturation of the S2 subunit during viral-cell fusion have been enhanced with recent structural models. (A) The proposed intermediate placing amphipathic helices HR1 and HR2 helices upon membranes7 is depicted with the addition of the NMR structures of HR1, FP16, and transmembrane helix (TM) protomers64, each measured in bicelles. The PDB IDs of these NMR structures are: 7R95 for HR1, 7MY8 for FP, and 7LC8 for TM. (B) The intermediate prior to HR2 folding back onto HR1, imaged by cryo-ET8, is represented. (C) The six-helix bundle formed and used in illustrating S2 in the hemi-fusion state, obtained by cryo-EM5,65, has PDB ID: 6M3W. (D) The NMR structural model of a 42-residue FP associated with bicelles 16(PDB ID: 7MY8, model 1) is illustrated with sidechains. (E) 47 Spike sequences from the N-terminal end of the S2 subunit from the four Orthocoronavirinae genera15 were aligned using Kalign at EMBL-EBI.66 Sequence positions that distinguish the β-coronavirus genus are marked with purple lines. Positions characteristic of its sequences in the 2b or 2b+c groups15 are boxed and marked with arrows. Accession numbers of the sequences are listed in Table S1.
A complete fusion peptide of β-coronaviruses was mapped to the N-terminal 40-residues of the S2 subunit of Spike.15 Investigations referred to that 40-residue region as the fusion peptide or FP16–18, the usage herein, or alternatively as the fusion domain.19 The combination of a β-coronaviral FP with 1 mM Ca++ induced membrane ordering15,20,21, and depends upon anionic residues.22 Since millimolar calcium concentrations are found on the outside of plasma membranes, these results and the need for calcium and anionic residues in FP for the infectivity of β-coronaviruses2,15,21,23 suggest that FP may insert preferentially in the outside leaflet of cell membranes with the anionic residues at the surface of the membrane.22 FP was intrinsically disordered in aqueous solution, and adopted helical structure upon mixing with zwitterionic membrane mimics at pH 5 and pH 7 without calcium.16,19 When inserted in bilayered micelles (bicelles), the three helices folded into a wedge shape with a narrow tip at Leu82816 (Fig. 1D). The fusion loop connects the second and third helices and is crosslinked by the Cys840-Cys851 disulfide. The Ser816 – Tyr837 segment appeared to insert two helices in membrane-like environments, in spite of its charge.16,19 Near the hydrophobic residues lie three anionic sidechains, two cationic sidechains, and five polar, uncharged sidechains16 (Fig. 1D,E). This leads to a seeming paradox regarding the action of FP: membrane insertion simulated to be shallow due to charge, but apparently deeper insertion in membranes in diverse experiments. Unrestrained molecular dynamics (MD) simulations of unstructured chains of FP suggested early insertion events to be shallow.17,18,24 The multiple polar and charged residues interspersed among the hydrophobic sidechains16(Fig. 1D, E) seem compatible with shallow insertion. Those simulations agreed upon the tendency of Leu821 – Phe823 (the LLF motif), the helix encompassing it, and Phe833 - Ile834 to insert into lipid bilayers.17,18,24–26 Penetration of bilayers by these features is supported by NOEs between fatty acyl groups and four aromatic sidechains of helices 1 and 2.16 On the other hand, in contrast to the unbiased simulations, NMR results suggested that the 40-residue FP adopted structure in bicelles and micelles and may have inserted more deeply.16,19 Neutron reflectivity found that the Ser816-Tyr837 fragment of FP spanned an entire leaflet of a lipid bilayer and was joined by water at a measurable volume percentage in the bilayer.27 There remains a need for depth measurements and simulations to account for the sustained insertion of FP measured experimentally by membrane fusion assays, NMR, and neutron scattering.
Questions at hand regard what membrane defects might be created by FP, how they might develop, the extent of insertion of charged and polar residues, the depth and orientation of helices and sidechains, and possible relationships to membrane fusion. Alanine substitutions implicated four anionic residues of FP as important in viral-cell fusion by one or more β-coronaviruses, corresponding to E819A, D820A, D830A, and D848A in SARS-CoV-2 numbering.2,15,21,23,28 One focal point regards the influences of Asp830, because it is critical for β-coronaviral infectivity, membrane fusion, and cell-cell fusion.2,22 Asp830 and Arg847 distinguish FP of β-coronaviruses from α, γ, and δ-coronaviruses (Fig. 1E). Asp830 is located in the loop between helices 1 and 2 (Fig. 1D) thought to insert in membranes.16,19 Could other acid and basic residues such as Asp820, Asp839, Asp843, Lys825, Lys835, and Arg847 (Fig. 1D) also influence lipid bilayers? Early evidence was that the SAR-CoV equivalent of an D820A lesion (D802A in SARS-CoV) disrupted about 30% of membrane fusion between cells, while the equivalent of.the D830A lesion (D812A in SARS-CoV) disrupted about 95%.2 Lys835, Asp839, and Asp843 (SARS-CoV-2 numbering) are characteristic of the group 2b of β-coronaviruses that contains SARS-CoV and SARS-CoV-2.
The work herein investigates how a mostly folded FP of 42 residues (Ser816 – Gly857) might orient in and perturb a simple DMPC bilayer. Atomistic MD simulations have proven critical for modeling defects in lipid bilayers induced by polypeptides.29–37 Magnetic resonance measurements of proximity between lipids and polypeptides have been used to bias MD simulations.38–41 This biasing by experiments can enrich trajectories with approaches of lipids and polypeptides that may be consistent with measurements at equilibrium. Consequently, we used paramagnetic NMR to estimate dozens of lipid-protein distances for biasing MD simulations. Polypeptides were reported to insert preferentially into bilayer-like phases of bicelles,42,43 as were phospholipids.44 We measured close approaches between FP and nitroxide spin-labeled phospholipids added to DMPC:DH7PC bicelles (1:2) at the pH 5 typical of late endosomes, via carefully quantified paramagnetic NMR relaxation enhancements (PREs). The PRE-estimated proximities provided weak harmonic lipid-protein distance restraints for simulations. We conducted NMR-biased MD simulations with Asp830 deprotonated, protonated, or protonated along with three other membrane-proximal aspartate sidechains. The rationale for neutralizing one or more aspartyl carboxylate groups was to test influences of these charges and possible consequences of membrane insertion shifting an aspartate pKa upward enough for protonation. Such a rise in pKa was reported for carboxylates buried in hydrophobic protein cores45,46 and in pHLIP peptides that insert in membranes below pH 6.47,48 Regardless of neutralization of aspartyl sidechains, FP spanned the cis-leaflet, inserted Asp830 and other charged sidechains, and introduced defects of invagination and hydration internal to the DMPC bilayer around FP. This is consistent with theory49 and NMR-detected hydration of part of the backbone inserted deeply in bicelles.
Results
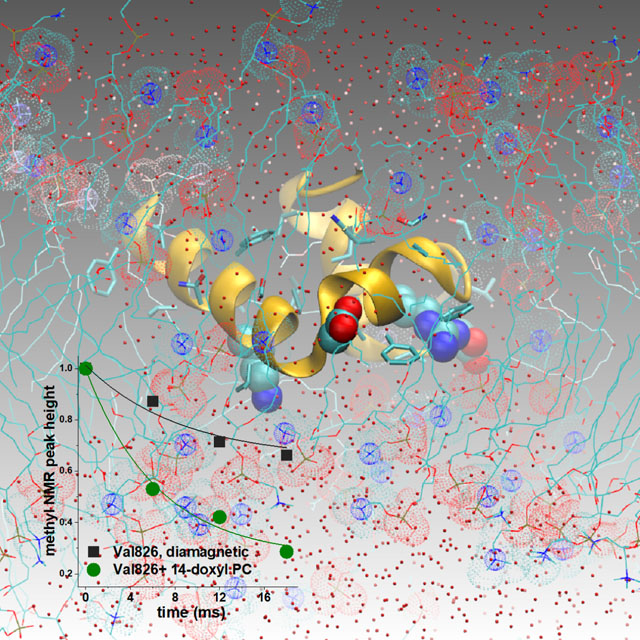
We investigated lipid bilayer insertion and disturbances by the 42-residue FP of SARS-CoV-2 using conditions to assess a mildly acidic and low [Ca2+] environment like the endocytic pathway of viral entry. The formation of the membrane mimic of bicelles composed of one DMPC per two DH7PC, was confirmed by their methyl NMR peak heights and mean diameter of 8.1 nm (Fig. S1 in Supplemental Information). NMR was used to measure lipid-protein proximities in these bicelles at pH 5. The resulting distance estimates were incorporated as gentle restraints in MD trajectories. We compared FP insertion into the simple bilayer mimic with FP in three states of aspartyl (de)protonation. We monitored the altered morphology of the lipid bilayer, fluctuations in angles and depths of FP insertion, and development of water inclusions. Paramagnetic NMR described below suggested insertion of Asp830 and other ionizable sidechains. Since the endocytic pathway of coronaviral entry ranges from pH 6.5 to pH 5, we simulated potential effects of neutralization of Asp830 that can insert in the membrane. As a control, three membrane-proximal aspartate sidechains and Asp830 were neutralized. That is, the aspartyl (de)protonation states investigated using NMR-biased simulations were: Asp830 and all aspartate residues deprotonated (Asp−1 830), only Asp830 protonated (Asp0 830), and aspartate residues 820, 830, 839, and 843 protonated (Asp0 820/830/839/843).
Close approaches of the fusion peptide to spin-labeled lipids by NMR
The bicelles were doped with around one nitroxide spin-labeled phosphatidylcholine per leaflet, i.e., one doxyl-substituted POPC per 65 DMPC. The maximum expected radius of proton NMR line broadening by the nitroxide is expected to be 25 Å.50 The nitroxide substituent in 16:0 – 5-doxyl PC probed the shallow region below the glycerol backbone of DMPC (Fig. 2B). Deep probing of the interior of the bicelles was provided by 16:0 – 14-doxyl PC (Fig. 2C). Examples of NMR HSQC spectra of 15N-labeled FP before and after addition of the respective spin-labeled probes are shown in Fig. S2. The sharp lines and τc of 16.9 ns in the bicelles16 are consistent with monomeric character. 49 paramagnetic relaxation enhancements (PREs) of NMR spectra from each spin-labeled probe were quantified as Γ2 relaxation rate constants. This was the difference in relaxation rate constants with and without the probe, i.e., R2,paramag – R2,diamag, obtained from exponential fits with R2 > 0.97 (Fig. S3). The shallow probe introduced large NMR line broadenings (Γ2) to helix 1, helix 2, the fusion loop between helices 2 and 3, and helix 3, but not to the loop joining helices 1 and 2 (Fig. 2B). This reflects proximity of these sites to the nitroxide at the 5-position of the acyl chain. Probing with the deep 16:0 – 14-doxyl PC probe gave a much more restricted set of NMR line broadenings that is distinct: The amide peaks of Thr827 – Ala829 were broadened severely by 16:0 – 14-doxyl PC, contrasting minimal broadening by 16:0 – 5-doxyl PC (Fig. 2B,C). The spin label at the 14-position in the acyl chain also introduced large Γ2 to helix 2 (Fig. 2C). Four of the six residues buried within the polypeptide fold (Leu822, Ile834, Gly838, and Leu849) exhibit large PREs (Fig. 2), consistent with the long-range of the PREs relative to the small folding core. More paramagnetic line broadening was observed at many backbone amide peaks than at the side chain aliphatic peaks of these residues. Most of the residues with amide groups in exchange with water exhibit larger PREs to their amide groups than to their aliphatic groups, namely Ile818, Glu819, Thr827 – Ala829, Gly832, Tyr837, Gly838, Cys840, Asp843, and Leu849 (Table S2, Fig. 2). It might be possible that water molecules within or near the bicelles not only experience PREs from the spin-labeled lipid, but also exchange hydrogen with these amide groups to increase their apparent paramagnetic relaxation. Even so, the patterns of PREs suggest that Thr827 through Ala829 in the loop between helices 1 and 2 (Fig. 2) appear to insert comparatively deeply into the interior of the DH7PC-DMPC bicelles. The patterns of PREs also suggest that the N-terminus of helix 1 and the fusion loop remain shallow near the junction of acyl groups with headgroups (Fig. 2). The changes in PREs along helices 1 and 2 (Fig. 2B,C) also imply that the depth of insertion of helices 1 and 2 grew more shallow with distance from the tip at Thr827 – Ala829.
Figure 2. Paramagnetic NMR relaxation enhancements reveal locations in FP close to nitroxide spin labels deep or shallow within bicelles.
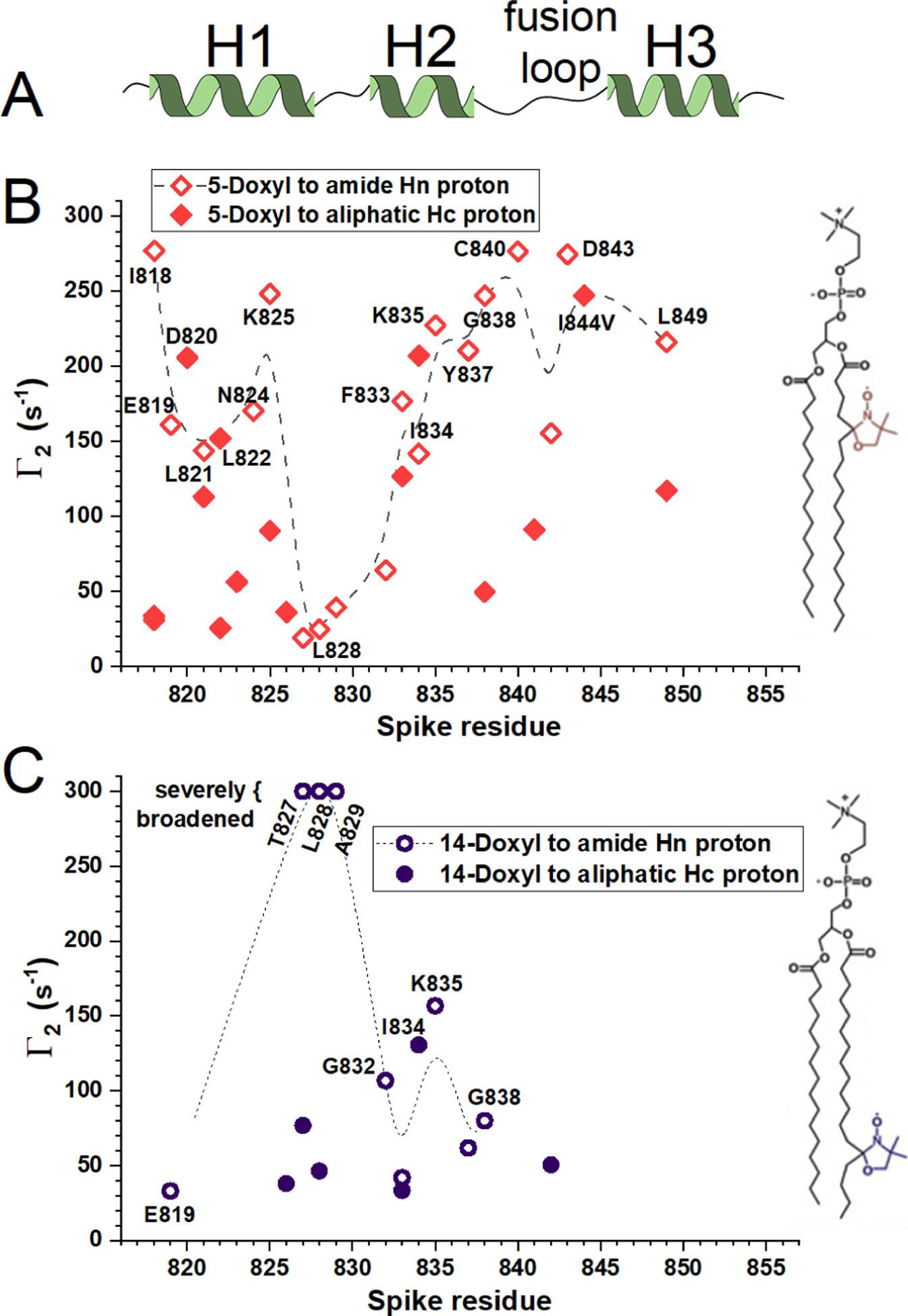
(A) The NMR-identified helices16,19 are plotted. (B,C) PREs emanating from nitroxide spin-labeled phospholipids at pH 5.0 at 303 K were detected by NMR at 800 MHz. Γ2 values are the differences between rate constants fitted to 1H T2 decays of the paramagnetic and diamagnetic samples. (B) 5-doxyl PC conferred high Γ2 values (large line broadening) to polypeptide groups that it approached. (C) 14-doxyl PC conferred high Γ2 to the most deeply inserted groups. Related to Figs. S1 – S3.
NMR biasing of MD simulations and experimental compatibility
49 close approaches of FP to spin-labeled POPC were estimated from the Γ2 measured by paramagnetic NMR. These distance estimates and six lipid-protein NOEs served as ambiguous harmonic depth restraints (Tables 1, S2), alongside 112 medium-and long-range NOEs and 12 sequential NOEs (Table S3),16 to bias a series of MD simulations using NAMD. The depth restraints were implemented as collective variables (colvars) parallel to the membrane normal along the Z-axis of the coordinate system with weak force constants (Table S2). The agreement with the colvars improved in the initial 5 to 50 ns of each production run of 300 ns, calculated with all 55 depth restraints (Figs. S4 – S12). Each simulation was replicated five times, in each of the three ionic environments. FP restrained by the 55 depth estimates remained inserted in the leaflet of the DMPC bilayer among all replicates of all three ionic environments, with very good agreement with the depth restraints (first 300 ns in Figs. S4 – S12; Table 1). Most of the replicated trajectories exhibited metastability in that one to three orientational substates were sampled by each (Figs. S4 – S12).
Table 1:
Conditions of NMR-biased MD simulationsa
| # colvars, harmonic force constants (FC), kcal/mol/Å2 | Replicates × length,μs | RMSD validation restraints | ||
|---|---|---|---|---|
| Lipid-protein PREs+NOEs,b FC | NOEs in protein,c FC | (divergent trajectories) | ||
| Asp−1 830 | 55, 0.02 | 116, 0.05 | 5 × 0.3 | 1.9 ± 1.1 |
| Asp−1 830, half of depth restraints | 28, 0.02 | 116, 0.05 | 5 × 0.2 | 5.0 ± 5.5 (2) |
| Asp−1 830, no depth restraints | 0, 0 | 116, 0.01 | 5 × 0.2 | 3.1 ± 2.5 |
| Asp0 830 | 55, 0.02 | 116, 0.05 | 5 × 0.3 | 2.2 ± 1.0 |
| Asp0 830, half of depth restraints | 28, 0.02 | 116, 0.01 | 5 × 0.2 | 2.4 ± 1.1 |
| Asp0 830, no depth restraints | 0, 0 | 116, 0.01 | 5 × 0.2 | 3.5 ± 3.0 (1) |
| Asp0 820/830/839/843 | 55, 0.02 | 116, 0.05 | 5 × 0.3 | 2.4 ± 0.8 |
| Asp0 820/830/839/843 | 28, 0.02 | 116, 0.01 | 5 × 0.2 | 7.8 ± 10.3 (1) |
| Asp0 820/830/839/843, no depth restraints | 0, 0 | 116, 0.01 | 5 × 0.2 | 3.3 ± 2.3 |
The use of loose depth restraints raises questions of the adequacy of the long restraints to the depths of the 5- and 14-postions of the acyl chains, and their compatibility with unrestrained simulations. To investigate these questions, we continued the simulations for 200 ns in each of two ways: with half of the depth restraints or with none of the depth restraints. The half of the depth restraints withheld from simulations constituted the validation set used to monitor the degree of agreement between experimental measurements and simulations (Figs. S4 – S12; Tables 1, S2). In the Asp−1 830 simulations continued with half of the depth restraints, replicates 2, 3, and 4 maintained high agreement with the depth restraints of the validation set (Fig. S4B–D). However, replicates 1 and 5 diverged out of agreement with the validating PREs because FP diffused more than 5 Å toward the surface and to shallow positioning in the DMPC bilayer (Fig. S4A,E). Fully restrained Asp−1 830 replicates 1 and 5 each had the deepest insertion of FP, including a charged loop, before withdrawal of half the depth restraints and moving to shallow insertion (Fig. S4A,E). A look at this divergence in replicate 1 finds strategic disagreement with four or five of the 27 withheld depth restraints by at least 1 or 2 Å above their estimated upper bounds (Fig. S13). In contrast, the Asp0 830 simulations continued with only half of the depth restraints never encountered this problem. They maintained almost equally good agreement with validation set of depths (Fig. S4A–E), with RMSD from the validation depths of 2.4 vs. 2.2 when all depth restraints were used (Table 1). This suggests that the protonated Asp0 830 state could be more compatible with the depth restraints measured at pH 5 than the deprotonated Asp−1 830 state. That is, Asp830 may have been at least partly protonated during the NMR measurements at pH 5.
For reference, upon withholding half of the depth restraints from continuations of Asp0 820/830/839/843 control simulations, three of the replicates maintained good agreement with the set of validation depth restraints while replicate 4 diffused 3 or 4 Å deeper into the bilayer and replicate 5 diffused across the trans-leaflet to its surface (Fig. S12). This suggests that the expected negative charge of Asp820, Asp839, and Asp843 may limit how deeply FP moves into the membrane. The results for the different ionization states together suggest that the incomplete set of depth restraints was most sufficient with only Asp830 protonated in this zwitterionic bilayer at pH 5 during NMR measurements.
For all three ionization states in the simulations, all of the trajectory replicates continued without any depth restraints remained inserted in the DMPC bilayer. In the simulations without depth restraints, the variability in depth of insertion increased in 5 of 5 replicates of Asp−1 830, 4 of 5 replicates of Asp0 830, and 4 of 5 replicates of Asp0 820/830/839/843 (Figs. S4 – S12). The more variable depths retained a surprising degree of agreement with the depth restraints: The increases of the RMS deviation from the set of validation depths were only about 60% for the Asp−1 830 and Asp0 830 states and about 40% for the Asp0 820/830/839/843 state (Table 1). This variability in simulated depths of FP insertion in the DMPC bilayer without depth restraints contrasts the steadier depths of FP insertion in the depth-restrained simulations.
Water influx into membrane mimic in silico and in NMR samples
In most of the simulations with all 55 depth restraints, water molecules and choline headgroups invaded central regions of the DMPC bilayer occupied by fatty acyl chains (Fig. 3, Movies S1 – S3). The possibility of water recruitment into the membrane was checked by NMR detection of amide hydrogen exchange with water. The backbone of helix 1 is protected from exchange with water from Asp820 through Val826 (Table S4); these are residues of helix 1 that insert into the acyl phase by NMR and simulation (Fig. 3). In contrast, nearby bicelle-inserted residues spanning from the loop joining helices 1 and 2 through the first turn of helix 2 (Figs. 2 and 3), i.e., Thr827 through Phe833, each display amide hydrogen exchange with water in NOESY spectra (Fig. S14; Table S4). The backbone of FP being exposed to water and near spin-labeled phospholipid probes (Figs. S14, 2) suggests detectable hydration of FP from Thr827 through Phe833 within the bicelles.
Figure 3. FP deformed DMPC bilayers with deep insertion of Asp830 and ingress of water and headgroups, in NMR-biased MD simulations at the three levels of protonation of aspartyl sidechains.
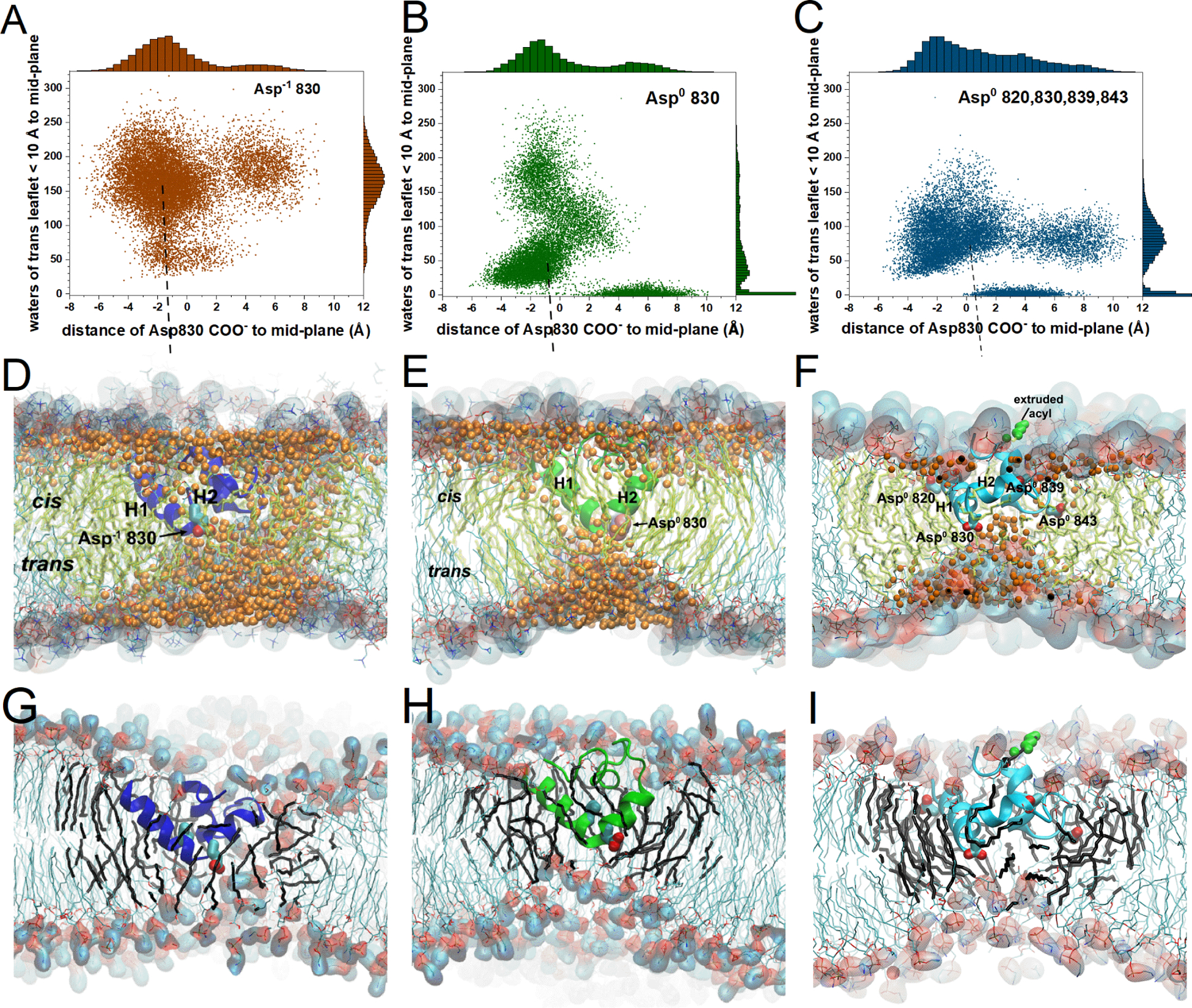
Panels A, D, G report on the fully deprotonated state, with the Asp−1 830 sidechain plotted. Panels B, E, H report on the singly protonated state, with the Asp0 830 sidechain plotted. Panels C, F, I report on the state with Asp820, Asp830, Asp839, and Asp843 all protonated and their sidechains plotted. (A–C) Each symbol represents a timepoint taken at 100 ps intervals from the simulations. The water molecules counted at each timepoint between the bilayer midplane and 10 Å into the trans-leaflet are plotted vs. the distance from the bilayer center to the Asp830 carboxyl group. The trajectories fully equilibrated with the depth restraints have combined durations of (A) 1.355 μs, (B) 1.386 μs, and (C) 1.426 μs. (D–F) Water oxygen atoms (orange) are plotted within the hydrophobic acyl phase of the DMPC bilayer. Fatty acyl chains within 20 Å of FP are tinted yellow-green. (D–I) The headgroups are highlighted with space-filling. Choline methyl carbons are cyan and phosphoryl oxygens red. (D,G) The snapshot is from a median position in (A) at 203 ns into the fifth Asp−1 830 trajectory. (E,H) The snapshot is from the median in (B) at 282 ns into the second Asp0 830 trajectory. (F,I) The snapshot is from the median in (C) at 19 ns into the trajectory 5 of Asp0 820/830/839/843. A fatty acyl sidechain pushed to the surface of the cis-leaflet over FP in an “L”-state is colored green. (G–I) Water molecules are omitted for clarity. Acyl chains within 12 Å of the protein are black. Related to Figs. S8, S15 and Movies S1 – S7.
The exceptions to water recruitment into the bilayer were a few simulations in which Asp830 was protonated (Fig. 3B,C). Water was recruited into the bilayer within the first 25 ns (Figs. S15A – S15C), especially into a dome-shaped defect in the leaflet across from FP (Fig. 3D–F). In the replicates of simulations with Asp−1 830, the water influx defect continued for another 60 to 100 ns (Fig. S15A). The numbers of water molecules drawn into the trans-leaflet across from the inserted FP tended to decrease when Asp830 was neutralized, alone or in combination with Asp820, 839, and 839 (Figs. 3A–C, 6A–C). In simulations of Asp−1 830 and Asp0 830, the carboxyl group of Asp830 was inserted deeply to just beyond the mid-plane throughout most of the trajectories, i.e. to median depths of about 1.3 and 0.7 Å, respectively, to the trans-leaflet side of the bilayer mid-plane (Fig. 3A,B,D,E). The Asp−1 830 trajectories exhibited median recruitment of 171 water molecules into the trans-leaflet opposite the inserted FP (Fig. 3A). This was well above the median recruitment of 48 or 76 water molecules in the Asp0 830 and Asp0 820/830/839/843 simulations, respectively (Fig. 3B,C). Asp−1 830 trajectories near median depth of FP and water recruitment exhibit a swath of water molecules spanning between the cis and trans leaflets of the bilayer (Fig. 3D; Movie S5). A representative Asp0 830 trajectory developed a persistent pool of water in the cis leaflet around the inserted FP (Movie S6). During the period 9 to 20 ns into trajectory 2, a “water wire”31 connected the water of the two leaflets. The water wire reformed at 47 ns and grew broader after 74 ns (Movie S6). A bowl or dome-shaped defect usually formed in the leaflet opposite FP in each protonation state, with hydrated phosphocholine headgroups drawn toward FP, and water filling the defect. However, when the -COOH group of Asp0 830 was shallower, i.e., in the cis-leaflet > 3 Å from mid-bilayer, the defect in the trans-leaflet of the DMPC bilayer usually disappeared (Fig. 3B,C).The results suggest that (i) FP was capable of recruiting water into a simple zwitterionic DMPC bilayer in monovalent cations, with or without charge on its aspartate side chains, and (ii) the anionic form of inserted Asp830 recruited more water to the interior of the bilayer.
Figure 6. One or two charges rotated into the membrane with water on either side of FP in simulations with Asp830 neutralized.
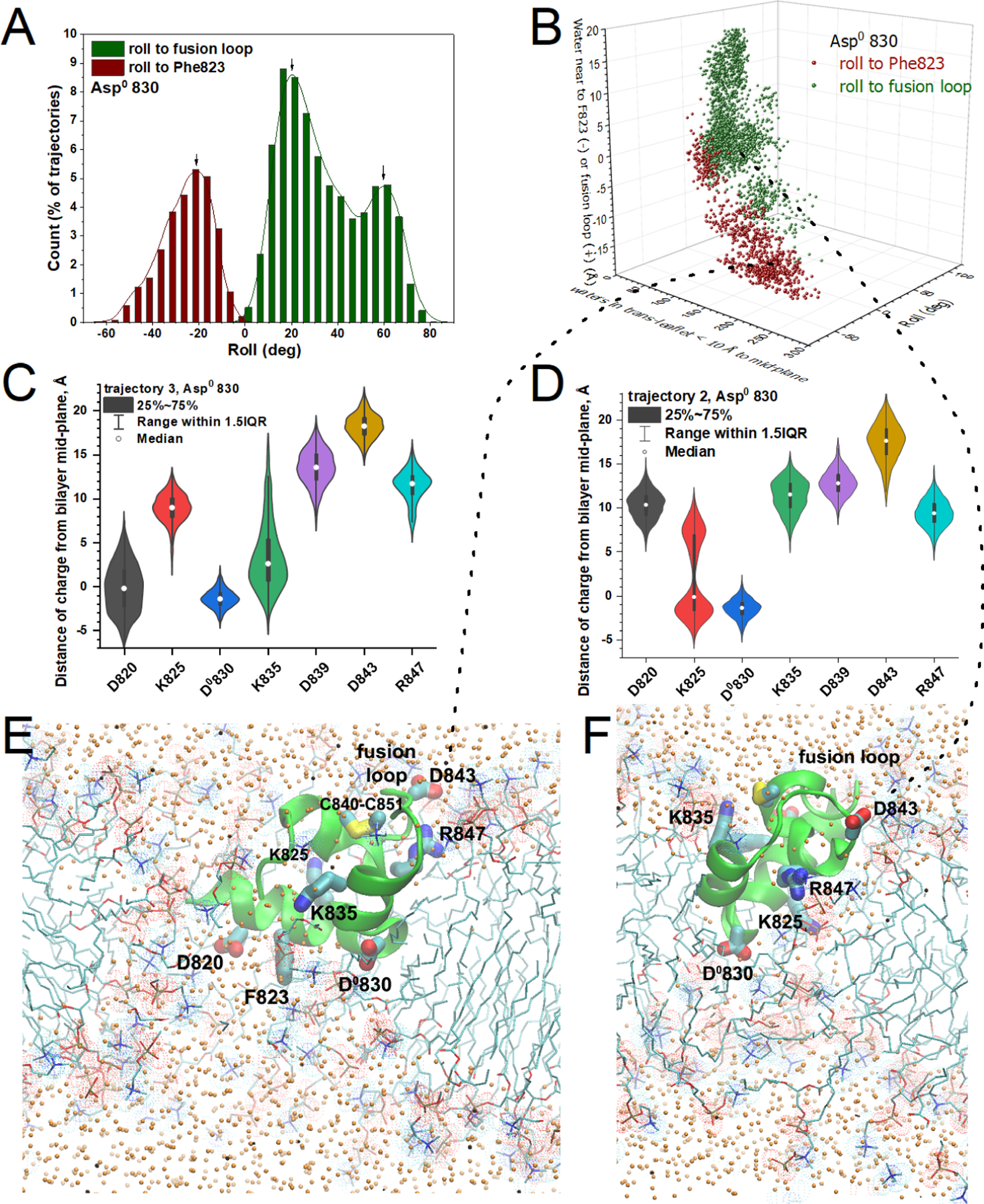
(A) The histogram plots the frequency of roll of FP toward the fusion loop or Phe823 (from the LLF motif). (B) The roll of FP correlates with the side of FP where more water accumulated in the membrane. (C,D) The distributions of distances of charged groups from the center of the bilayer (violin plots with medians and interquartile ranges as in Fig. 5) suggest those groups that inserted into the DMPC bilayer in simulations that tilted toward Phe823 (C) or toward the fusion loop (D). (E) The snapshot of FP with the side of Phe823 rolled into the membrane is drawn from 133 ns into trajectory 3. (F) The snapshot of FP partly tilting toward the fusion loop is drawn from 136 ns into trajectory 2 as lipid flip-flop was taking place. Orange dots mark water oxygen atoms. Related to Figs. S7 – S9 and S15.
Displacement of phospholipid molecules
Choline head groups entered the interior of the DMPC bilayer together with water molecules. The intruding headgroups and water molecules correlated in numbers (Figs. 3D–I, S15D). Multiple choline headgroups drew close to Asp830, suggesting attraction between the positive charge of choline and negative charge of Asp−1 830. Across from FP where the dome-shaped defect formed in the trans-leaflet, phosphatidylcholine molecules moved inward into the bilayer, with fatty acyl chains wrapping around FP and intermingling with the fatty acyl chains of the cis-leaflet. The headgroups of these phospholipids, which slid from the trans-side to the cis-side in part, collected around Asp830 and the inserted loop between helices 1 and 2, often forming the top of the water-filled, dome-shaped defect (Fig. 3G–I, Movies S1 – S3). At this defect in the trans-leaflet, the headgroups lay on the domed surface and the fatty acyl groups radiated from the curved surface, tilting them away from the overall membrane normal (Fig. 3D–I). These fatty acyl groups tilted away from the center appear to cradle FP and its aromatic and aliphatic sidechains with favorable hydrophobic interactions. In the cis-leaflet, fatty acyl chains tended to adopt a sweeping tilt around FP. Tilted fatty acyl chains often lay sandwiched between the bilayer mid-plane and helices 1 and 2 in the trajectories. Other DMPC molecules draped over FP in the cis-leaflet. In 11% of Asp0 820/830/839/843 simulations, a fatty acyl chain of a DMPC molecule was lifted to the water-exposed surface of the cis-leaflet (Figs. 3F,I and S16). With one acyl chain displayed on the surface and one acyl chain inserted, this shape of a phospholipid was named an “L”-state.51 This protrusion of a fatty acyl chain from the surface usually occurred adjacent to the disulfide bond of the fusion loop within the first 10 to 100 ns of replicates 1, 3, 4, and 5 of the Asp0 820/830/839/843 simulations (Movie S4). Headgroups bulged or protruded from the bilayer around FP in Asp−1 830 and Asp0 830 simulations, which elevated acyl chains in measurements (Fig. S16). Such bulging of headgroups from the bilayer preceded the acyl protrusion in Asp0 820/830/839/843 trajectories. The frequent tilting and translation of molecules around FP in the simulations might portend beginnings of negative Gaussian curvature (Fig. 3D–I, Movies S1–S3). The appearance of the fatty acyl chain that appeared on the cis-surface of the DMPC bilayer adjacent to the disulfide bond (Figs. 3F, S16) raises the question of this as a means of promoting the lipid mixing of membrane fusion.
Thinning of the lipid bilayer around the fusion peptide
Thinning of the bilayer at the dimple across the trans-leaflet across from FP was quantified. In the Asp−1 830 trajectories, the trans-leaflet was especially thinned near FP to a median of 9.8 Å from the mean choline center-of-mass to the mid-plane of the bilayer, i.e., to only half of the normal 20 Å thickness remote from FP (Fig. 4A). This is consistent with the large influx of water (Fig. 3A,D). All of the Asp−1 830 simulations freed of depth restraints retained thinning of the trans-leaflet. In the Asp0 830 trajectories, the trans-leaflet was thinned to around 60% of the normal thickness (median of 11.8 Å with CV = 0.2, Fig. 4B). Three of four Asp0 830 trajectories that were deeply inserted remained deeply inserted after removal of depth restraints (Figs. S7 – S9) and retained thinning of the trans-leaflet. In Asp0 820/830/839/843 simulations, most of the thinning was to 10.5 Å, but a minor population thinned only to ~15.5 Å (Fig. 4C). The Asp−1 830 trajectories tend to display a broader thinning of the bilayer in the trans-leaflet across from FP than those with protonation of aspartate sidechains (Movies S5, S6).
Figure 4. Thinning of both leaflets DMPC bilayer near FP.
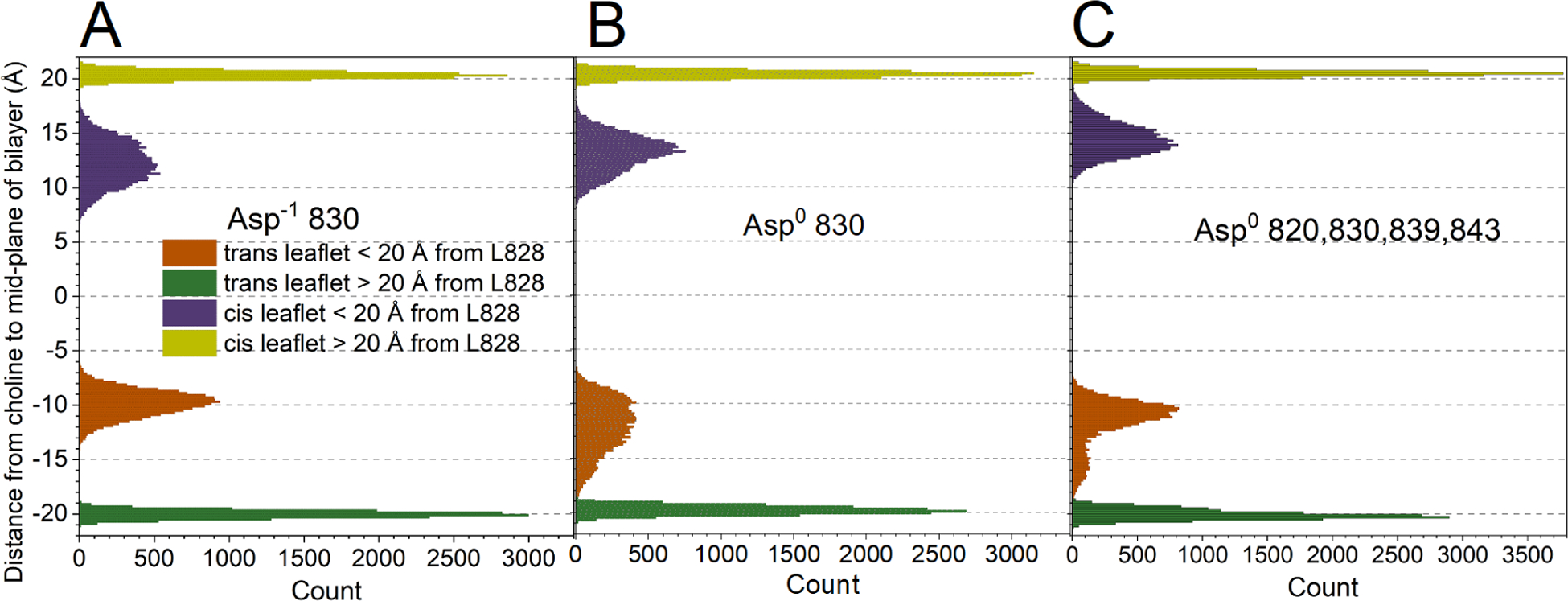
The states are (A) Asp−1 830, (B) Asp0 830, and (C) Asp0 820/830/839/843. The mean distances from the center-of-mass of the choline headgroup to the bilayer mid-plane were monitored every 100 ps, after equilibration with the NMR-derived restraints. The gold and green peaks plot the unperturbed distances remote from FP. The purple and orange peaks represent the mid-plane distances to the headgroups in the cis and trans-leaflets, respectively, perturbed by proximity to FP, i.e., within 20 Å of the Leu828 backbone at the tip of FP. Related to Fig. S15.
Thinning was also evident in the cis-leaflet as choline headgroups being recessed around the inserted FP. The recession of the choline groups was to about 60% of normal thickness in Asp−1 830 simulations, 65% in Asp0 830 simulations, and 70% in Asp0 820/830/839/843 simulations (purple in Fig. 4). The thinning of both leaflets of the DMPC bilayer around FP might resemble a very localized portion of the negative Gaussian curvature of stalk-like fusion intermediates52 (Figs. 3D–I, 5D–F).
Figure 5. Three populations of angles of membrane insertion, insertion of charges, and recruitment of water are observed in the NMR-biased MD simulations of the Asp−1 830 state of FP.
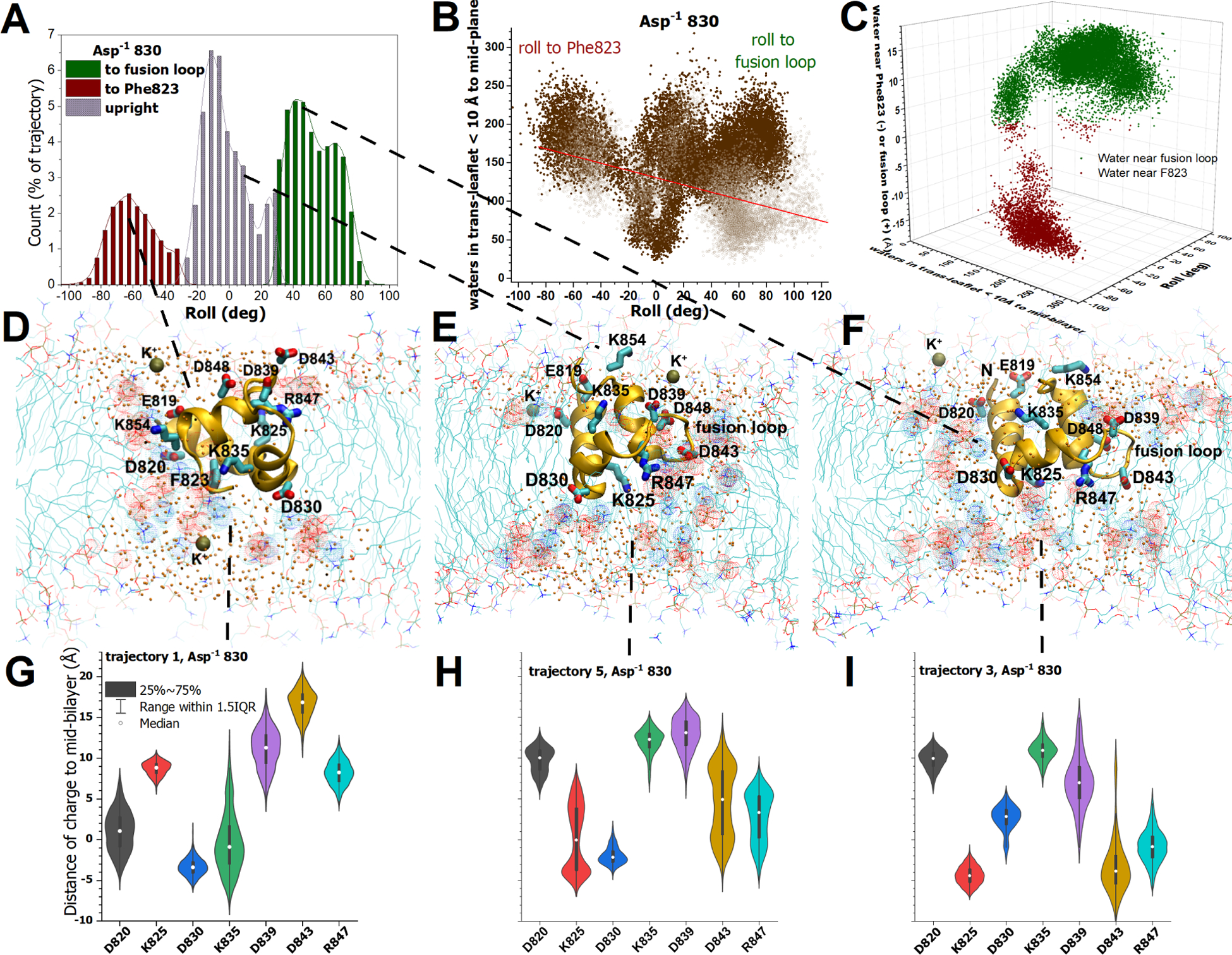
(A,B) FP was distributed in three populations of roll, relative to the initial upright position (in the equilibrated 1.355 μs of trajectories with depth restraints). Upright (0°) is defined by the vector between the backbone of deeply inserted Leu828 and Ala852 in helix 3 near the surface of the membrane. (B) The water molecules internalized between the mid-plane of the bilayer and 10 Å across the trans-leaflet were counted with 17 Å of Leu828. The numerals specify which trajectories correspond to the clusters. The open symbols are the water counts in the simulations upon withdrawal of NMR-measured depth restraints (five replicates of 200 ns each). (C) The location of the internalized water is plotted vs. the roll and the number of water molecules recruited across into the 10 Å interior slab of the trans-leaflet for the 1.355 μs of NMR-equilibrated trajectories. The location is reported as the distance of the center-of-mass of the water from the fusion loop side (green) or the Phe823 side (red). (D–F) Snapshots represent the three highly populated angles of insertion. Charged sidechains are plotted. Water oxygens are plotted as small orange spheres. Dots highlight head groups with red for phosphoryl oxygen and blue for choline. Potassium ions are plotted with bronze spheres. (D) The roll toward Phe823 is depicted in the snapshot from 115 ns into trajectory 1. (E) An upright orientation is depicted by a snapshot from 116 ns into trajectory 4, with the fusion loop in the foreground. (F) The frequent roll toward the fusion loop is drawn from trajectory 3 at 85 ns. (G–I) The variable proximity of charges in sidechains to the mid-plane of the DMPC bilayer is represented by violin plots, medians, and the 25–75% interquartile range of the distance to the charged atoms (averaged for aspartyl OD1,2 or arginine NH1,2). Related to Figs. S4 – S6 and S15.
Membrane invagination around inserted charges at varied insertion angles
In the NMR-biased simulations of the Asp−1 830 ionization state of FP, the largest protein reorientation in the simulations was roll in the membrane (Fig. 5A,B). The side facing the interior of the DMPC bilayer varied from the side displaying Phe823 (from the conserved LLF motif) to the side of the fusion loop (Fig. 5D,F). Consequently, three main populations of FP insertion angles and water recruitment into the membrane emerged in the simulations (Fig. 5A,B). The three orientations of membrane insertion of the Asp−1 830 state were similar in probability (Fig. 5A,B). A smaller, orthogonal reorientation of the N-terminus pitching forward into the membrane and back to the surface, re-exposing Glu819 and Asp820 to the aqueous phase, sometimes also occurred. In each of the three main populations of inserted orientations, 110 to 260 water molecules were typically recruited into the inner half of the leaflet across from FP, though recruitment of 30 to 100 water molecules by upright insertion was also common (Fig. 5B, Movies S5 and S7). Despite the wide range of angles of insertion, water filling of the bowl in the trans-leaflet opposite FP was shared among them (Fig. 5D–F).
Despite differences in the details of the three orientations of membrane insertion, conceptually similar physical-chemical behavior emerged in the membrane interactions. When the face displaying Phe823 rotated toward the membrane interior in trajectory 1, the Asp820 and Lys835 sidechains rotated to the middle of the bilayer, with Asp830 reaching the trans-leaflet (Fig. 5D,G). These deep charges lay among the headgroups of the invagination deep within the trans-leaflet (Fig. 5D). Upright orientations of FP consistently positioned Asp830 just to the trans-side of the mid-plane of the DMPC bilayer where it approached choline headgroups (with partial positive charge) and water (Fig. 5E,H). Lys825 and Arg847 were usually inserted deeply on the cis-side of the mid-plane near phosphoryl segments of the headgroups and water (Fig. 5E,H). Anionic Asp820 and Asp843 were also inserted (Fig. 5H) into contact with water in the bilayer, often near choline. Often, the fusion loop rolled into the acyl phase of the membrane, which could rotate the charges of Lys825 and Asp843 to the trans-side of the mid-plane, Arg847 to near mid-bilayer, and Asp830 to the cis-side (Fig. 5B,F,I). These four charged side chains tended to mingle with the headgroups and water molecules around the invagination (Fig. 5F). Though the combination of inserted charge side chains differed among the orientations of insertion, in each case the charged side chains were positioned among headgroups and water molecules in the defect turned inward. In the populations with FP inserted upright or rolled toward the fusion loop, the center-of-mass of the water that invaded the DMPC bilayer collected more often on the side of the fusion loop (Fig. 5C). The most interior water on either side of FP appears to have colocalized with the deeply inserted charged side chains and headgroups, hydrating them (Fig. 5D–F). The anionic Asp830 side chain was always inserted, and usually colocalized with choline headgroups and water. Throughout the Asp−1 830 simulations, the insertion of charged amino acid sidechains, invagination of the trans-leaflet and its headgroups, and collection of water all colocalized. Thus, the electrostatic attractions and the hydration of charge became the working, general hypothesis for deformation of the DMPC bilayer by the β-coronaviral FP at varied angles of membrane insertion.
Hydrated membrane invaginations lined with charge in the Asp0 830 state
Consequences of the charge of the Asp830 carboxyl group were investigated further by neutralizing it in silico. Protonation of Asp830 in a set of NMR-restrained simulations modified the insertion of charge and tested potential consequences should Asp830 become protonated in the late endocytic pathway (pH 5 to 5.5). Neutralization of this buried sidechain narrowed the breadth of the roll of FP in the DMPC bilayer and decreased the frequency of the roll toward the fusion loop (Fig. 6A). When the Asp0 830 state rolled toward the fusion loop, FP recruited < 125 water molecules (and often < 20) to the inner 10 Å slab of the trans-leaflet (Fig. 6B,F). The more that the Asp0 830 state rolled to the side of Phe823, the more that water tended to enter the trans-leaflet, i.e., ranging from as few as 60 water molecules to as many as 275 in the inner 10 Å slab of the trans-leaflet (Fig. 6B). Why did this recruit more water? Insertion of FP that was upright or rolled toward Phe823 often tipped Asp820 into the membrane (Fig. 6C,E), which appeared to draw additional water into the acyl region of the bilayer to solvate the negative charge of Asp820 (Fig. 6E). The roll of FP in the membrane toward its Phe823 side also moved the amino group of Lys835 deep into the bilayer where it interacted with water and phosphoryl moieties of the headgroups (Fig. 6C,E). When the Asp0 830 form of FP rolled toward the fusion loop in simulations, it rotated the charges of Lys825 and Arg847 into the acyl phase like in the Asp−1 830 state. However, protonated Asp0 830 rolled toward the fusion loop did not recruit as much water into the membrane as the Asp−1 830 state did (Figs. 5C, 6B). Thus, the emerging association of inserted charge with invagination of the trans-leaflet and ingress of water into the bilayer was corroborated. The Asp0 830 simulations suggest that anionic charge of each bilayer-inserted aspartate, e.g., Asp820 or Asp830, can enhance water movement into the bilayer.
With aspartates neutralized, basic and polar groups distorted the membrane
To probe more shallowly inserting carboxylate groups, aspartate residues 820, 830, 839, and 843 were all protonated in silico in another set of NMR-biased MD simulations. These Asp0 820/830/839/843 trajectories largely rolled toward the fusion loop (Fig. 7A,B), as no negative charges remained to resist immersion of the loop in the acyl phase. The trans-leaflet curved inward opposite FP and took in a moderate amount of water in four of five of the simulation replicates (Figs. 3C and 7B,F). The variability in the main cluster of roll and hydration (Fig. 7B) is suggested by frames with roll toward the fusion loop of 60° and 120 waters hydrating the interior of the trans-leaflet and of 83° and 81 water molecules in this region (Fig. 7C,D). Characteristic of the roll toward the fusion loop was deep insertion of the Lys825 and Arg825 charges among the hydrated phospholipid head groups lining the invagination (Fig. 7C–E). These basic side chains were joined by neutralized Asp843 and sometimes neutralized Asp830. The number of water molecules that entered the interior of the leaflet in trans correlated negatively with the mean distance of the basic and aspartyl side chains (Lys825, 835; Arg847; Asp0 820, 830, 839, 843) from the bilayer mid-plane (Pearson’s r of −0.65; Fig. 7F). The mean position of Lys825 and Arg847 charges correlate negatively with the internalized water with Pearson’s r of −0.55 (Fig. S18). These suggest the sufficiency of the inserted cations for electrostriction of the membrane and water recruitment. It also suggests that protonated, polar aspartyl side chains may also enhance the hydrated defect.
Figure 7. Simulations with neutralized aspartates, Asp0 820/830/839/843, rolled toward the fusion loop, rotating Lys825 and Arg847 into the DMPC bilayer with recruitment of water toward them.
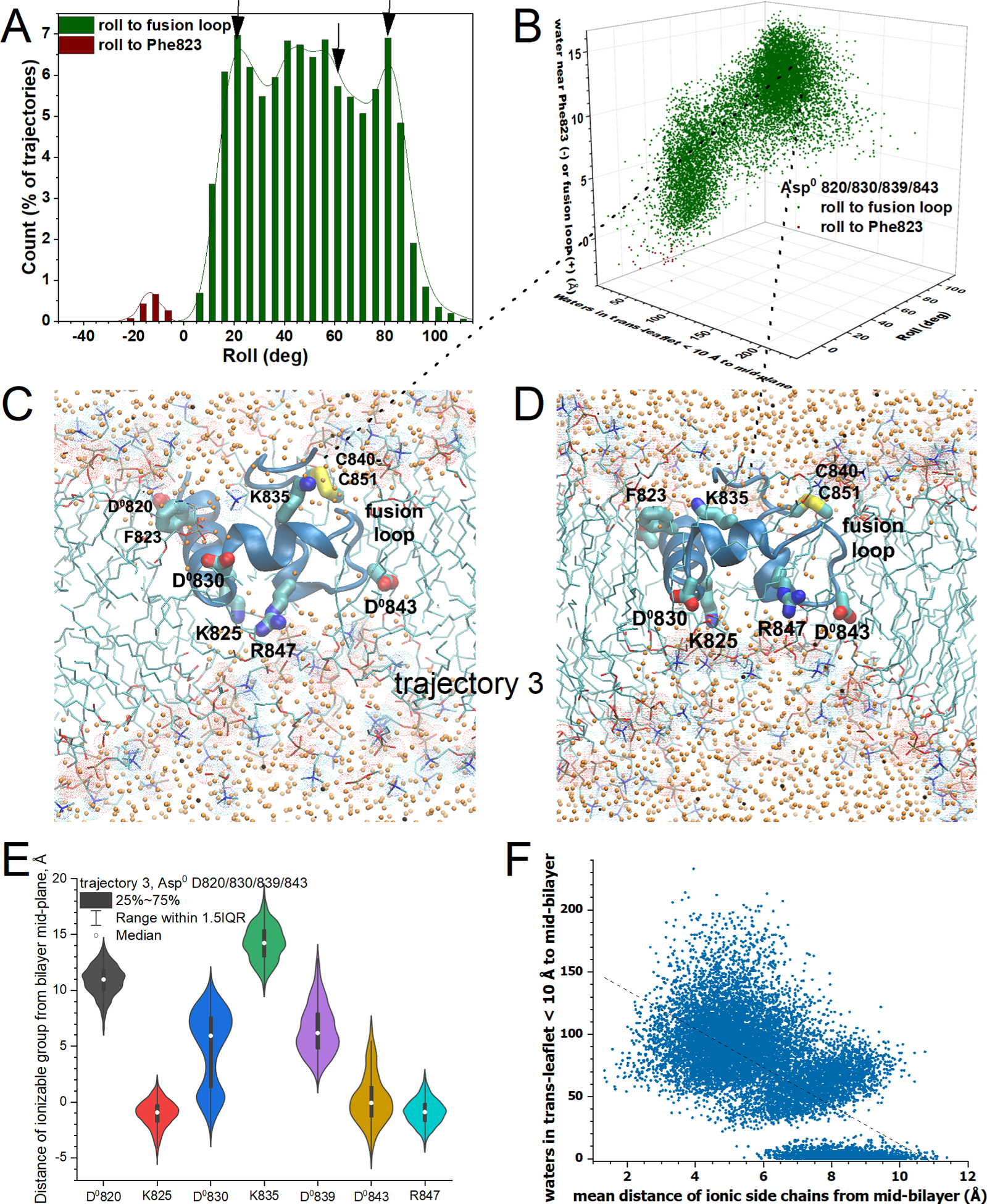
(A) The histogram of frequency of roll orientations is plotted as in Figs. 5A, 6A. (B) Roll and location of center-of-mass of water recruited into the DMPC bilayer are plotted vs. waters recruited into the internal half of the trans-leaflet, as in Figs. 5C, 6B. (C, D) Snapshots are illustrated from the third Asp0 820/830/839/843 trajectory replicate, with rotations by 82° and 60° toward the fusion loop and recruitment of 82 and 120 waters, respectively (C,D), to the 10 Å interior slab of trans-leaflet. Sidechains of inserted basic and neutralized aspartic residues are labeled. (E) Violin plots show the distributions of the proximity of positive charges and neutralized carboxylate residues to the center of the bilayer for trajectory 3, including medians and interquartile ranges. (F) The number of water molecules counted in the trans-leaflet within 10 Å of the mid-plane of the DMPC bilayer is plotted against the mean distance to the mid-plane of the acidic or basic groups of the sidechains listed in panel (E). Related to Figs. S10 – S12 and S15.
Flip-flop of headgroups through watery corridors around fusion peptide
In all five simulations with Asp830 ionized (Asp−1 830), some choline headgroups diffused to the opposite leaflet of the DMPC bilayer. DMPC molecules positioned on the flanks of the defect in the trans-leaflet, with their acyl chains radiating away from the membrane normal (Figs. 3G, 8A–D), were close enough for the choline head groups to diffuse between leaflets of the membrane. An example of flip-flop of several phospholipids early in a simulation is drawn from trajectory 3 (Fig. 8A–D; Movie S8). DMPC molecules flip-flopped in both directions across the bilayer, with the traffic in the cis to trans direction averaging about 70% the volume in the trans to cis direction in the Asp−1 830 trajectories (Fig. 8E,F). In the example, the cis-to-trans flip-flop preferred to pass by helix 1 or 2 (past Asp820, Asp830, or Lys835) while the trans-to-cis flip-flop was near the fusion loop between helices 2 and 3 (Fig. 8A–D and Movie S8). The flip-flop of DMPC molecules correlated with the simulated accumulation of waters of hydration in the membrane near FP. As the waters’ collective radius of gyration in the middle of the membrane grew beyond about 8 Å, flip-flop of choline head groups commenced (Fig. 8G). These dimensions match and exceed the breadth of a choline headgroup. This suggests the need for a water-rich portal large enough for the headgroup to diffuse through.
Figure 8. Flip-flop of DMPC proceeded bidirectionally through the transmembrane water jacket around FP.
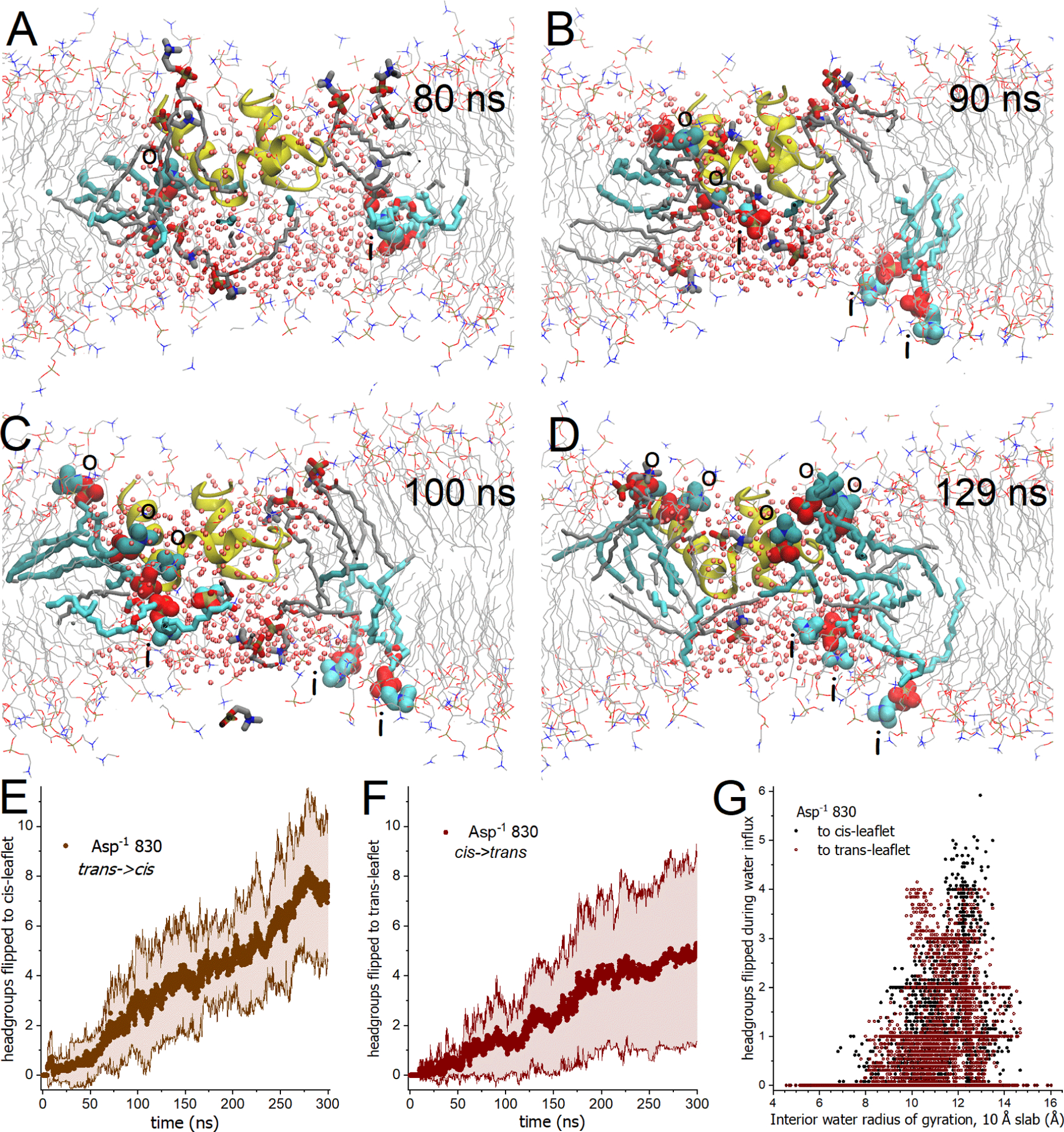
(A–D) Snapshots of the Asp−1 830 state depict the progress of the initial flip-flops of DMPC from the trans to cis-leaflet (darker cyan, labeled “o”) and from the cis to trans-leaflet (lighter cyan, labeled “i”) during the 80 to 130 ns segment of trajectory 3. Affected DMPC molecules are plotted with heavy gray sticks prior to their flip-flop. Small pink spheres mark water oxygen atoms. FP (yellow) tilted toward the fusion loop. Movie S8 portrays the same events. (A) The first headgroup to emerge into the opposite leaflet (from both directions) is plotted. (B) Two DMPC molecules had flip-flopped from trans to cis, two from cis to trans, and another midway through a cis to trans flip-flop. (C) Three flip-flops had occurred in both directions. (D) Three DMPC had flip-flopped from cis to trans and five from trans to cis. (E–G) Headgroup atoms were counted as they crossed the mid-plane from one leaflet into the other leaflet. Counts of headgroup atoms are normalized by 13 atoms per choline headgroup. (E,F) Solid symbols mark represent the average of the five replicates of gently restrained simulations of the Asp−1 830 state for flip-flop trans to cis (E) and cis to trans (F). Confidence bands of ± 1σ are marked by shading thin lines. (G) Flip-flop of DMPC is plotted against the radius of gyration of the water within the central region of the bilayer (within 5 Å from the mid-plane) during initial influx of water into the bilayer and initial flip-flops, i.e., 0 – 85 ns of trajectories 1 to 4 and 0 – 180 ns of trajectory 5. Related to Movie S8 and Fig. S17.
Flip-flop of the choline headgroups became less frequent in silico upon protonation of Asp830, and infrequent with protonation of Asp820, 830, 839, and 843 (Fig. S17A–D). In the case of Asp0 830 trajectories, flip-flop of choline headgroups ranged from none in replicates 1 and 5 up to seven in the trans to cis direction and up to six in in the cis to trans direction of trajectory 3. In Asp0 830 simulations, slowing of flip-flop (Fig. S17A,B) paralleled overall slowing of influx of water into the trans-leaflet for Asp0 830 (Fig. S17B). Asp0 820/830/839/843 trajectories ranged from no flip-flop in trajectories 1 and 4 up to three lipids flip-flopped trans to cis in trajectory 5 and two cis to trans in trajectory 2. In trajectory 5, two trans to cis flip-flops passed by Arg847 and Asp0 843 of the fusion loop and one trans to cis flip-flop passed Asp0 830 (Fig. S17H). The flip-flop in Asp0 820/830/839/843 trajectories occurred when the influx of water molecules into the trans-leaflet exceeded 140 molecules into the 10 Å slab nearest the mid-plane (Fig. S17F). In all states of protonation of Asp sidechains in the simulations, cis to trans flip-flop awaited movement of at least 60 water molecules into the interior of the trans-leaflet while trans to cis flip-flop awaited influx of at least 80 water molecules into the interior of the trans-leaflet (Fig. S17E–G). These results support the working hypotheses that (i) flip-flop required watery corridors wide enough for the headgroup to diffuse through and (ii) such water-containing passages were more likely to form upon deep insertion of a charge, especially Asp−1 830, but also Asp−1 820, Lys825, Asp−1 843, or Arg847.
Discussion
Limitations of the study
The choice of lipids was constrained by the need of solution NMR for a membrane mimic that tumbles quickly enough for sharp NMR lines. Small bicelles fulfill that need, provide a small bilayer region, and are well-characterized, but are limited to simple lipid compositions.53–56 The bicelles lacked the physiologically important and abundant cholesterol, anionic lipids, curvature-inducing headgroups such as ethanolamine, and C16 and C18 acyl chains. POPC and cholesterol influence fluidity. POPC, anionic lipids, and cholesterol have been favored for assays of membrane fusion by coronaviral fusion peptides.2 More work using lipid compositions that are more physiological and diverse will be needed in mechanistic studies of the membrane insertion of viral fusion peptides and proteins. A trimeric context for positioning FP10 could influence lipid mixing and water penetration. However, there is an experimental gap in sample preparation between viral fusion peptides and intact viral glycoproteins used in membrane fusion assays. The lipid measurements in this study were limited to their proximity to FP. Solid-state NMR can provide additional strategic measurement strategies for larger proteins and more complex mixtures of lipids in bilayers.57–59
How may a charged coronaviral FP insert into a zwitterionic bilayer?
Compared to the class I fusion peptides of influenza hemagglutinin and HIV GP41, the membrane-inserting surfaces of FP from SARS-CoV-2 Spike display more charged and polar sidechains alongside the hydrophobic sidechains16 (Fig. 1D). The high energetic costs of inserting proteins with charged sidechains into the low dielectric core of membranes are well-known.49,60 Why does coronaviral FP insert into zwitterionic membranes in monovalent salt nonetheless? Two considerations of the theory of insertion of charge and polar groups into the hydrophobic phase of a membrane regard the deformation of the membrane by electrostriction and hydration.49 Parsegian considered potential mechanisms of ion transport through membranes in terms of ion – low dielectric interactions.49 He proposed that the mechanism most capable of lowering the energy of ion entry into the membrane should be a “pore” filled with very polarizable water or protein.49 Solvation of the ion with a very polarizable, neutral molecule like water minimizes the energy of ion entry next best. Furthermore, an ion in the membrane pinches (thins) the low dielectric membrane along the membrane normal drawn through the charge.49 Protonation of Asp830 decreased the electrostatic recruitment of headgroups from the trans-leaflet. Yet neighboring Lys825 or Lys835 remained inserted in the bilayer in simulations. Asp830 protonation decreased the pinching and invagination in the trans-leaflet, and decreased the water influx into this invagination (Fig. 3B,E,H). Parsegian indicated that hydration of an ion in a “pore” would make the thinned region of the pore from a bridge through the membrane. These principles and behaviors are evident in NMR-biased simulations of FP in bilayers of DMPC in which combinations of the inserted charges of Asp820/Asp830/Lys835, Lys825/Asp830, and Lys825/Asp830/Asp843/Arg847 lay near the apex of the largest water-filled pits observed in the trans-leaflet. The amount of water that entered the membrane appeared to correlate with the combined proximity of Asp, Lys, and Arg sidechains to the mid-plane of the bilayer (Fig. 7F). A path of water tended to cross through the cis-leaflet (Figs. 3D,G,J, 5E–G, 8G–H), as Parsegian’s theoretical models predicted. Uncharged, polar sidechains probably also attracted water for solvation into the membrane. Insertion of FP deep enough to place an ion near the middle of the membrane retained electrostriction and water bridging in the simulations (Fig. 7C,D). Parsegian pointed out that the electrostrictive pressure is greatest through the charge inserted in the membrane.49 In accordance with this prediction, the region of thinning and water pooling tended to shift to the most deeply inserted charges; see Figs. 5D–F, 6E, 7C,D for examples. The pressure of electrostriction may have done the work of turning headgroups into the membrane (intrusion), aimed around FP in the cis-leaflet and in the invagination in the trans-leaflet. The headgroup intrusion seems was accompanied by lateral tilting of the fatty acyl chain (Figs. 3G–I, 8A–D). The repositioning of the phospholipids laterally within the membrane, combined with the water bridging across the bilayer, predicted by Parsegian, appear to have fostered the simulated lipid flip-flop (Fig. 8 and Movie S8).
More subtle membrane invagination and thinning by the influenza HA fusion peptide may also be promoted by electrostriction. Simulated bilayer invagination by a single fusion peptide was shallow and localized to the N-terminus.36 The simulated invagination for a trimer seemed to be deeper and attributable to the charge of its protonated alpha-amino group.61 Clustering of free influenza fusion peptides displaced lipids and thinned the hydrophobic phase.37
Could membrane perturbation by fusion peptide aid membrane fusion?
Schaefer et al. simulated the affinity between FP and a membrane to be more than enough to bring host and viral membranes into apposition.18 The study herein, albeit limited by the simplicity of the lipid composition, suggests the possibility of electromechanical distortions of phospholipid bilayers by coronaviral fusion peptides. If such distortions carry over into more physiological lipid compositions, they might participate in coronaviral-cell fusion. For juxtaposed membranes to proceed to fusion, the repulsive forces between them need to be overcome by attractive forces.62 Once the fusion stalk forms, it needs to expand for membrane fusion to occur.
The repulsive forces between juxtaposed membranes arise from hydration, steric clashes of dynamic headgroups, and electrostatic repulsion between their respective headgroups.62 Simulated submersion or intrusion of phospho-linkages of POPC by the influenza HA2 fusion peptide caused shallow invagination of the headgroups near the N-terminus of the influenza fusion peptide.36 The authors proposed that this would withdraw the headgroups out of steric overlap and decrease electrostatic repulsion between the headgroups of the two membranes. The invagination in the cis-leaflet by the SARS-CoV-2 fusion peptide is deeper and broader, with the headgroups submerged more deeply (Figs. 3 – 8). Retraction of headgroups from contact by insertion of β-coronaviral fusion peptide could be postulated to help decrease repulsion between apposed membranes in fusion. Calcium interactions with aspartate sidechains22 also appear likely to diminish electrostatic repulsion at the plasma membrane where calcium exceeds 2 mM.
The simulated invagination of the trans-leaflet (Figs. 3 – 8) looks similar in depth to that in coarse-grained simulations of the junctional complex with full-length influenza hemagglutinin.61 The invagination in the trans-leaflet of the hemagglutinin simulations enabled the trans-leaflet to move into the fusion junction where headgroups of the trans-leaflet and cis-leaflet appeared to mix, and meet the viral membrane mimic.61 The lipid flip-flop simulated in the presence of SARS-CoV-2 FP (Figs. 8 and S17) suggests the possibility of analogous traffic between trans- and cis-leaflets that could be conducive to the lipid mixing of membrane fusion. Based on neutron reflectivity and scattering, Santamaria et al. also hypothesized mixing of lipids at sites of fusion initiation by SARS-CoV-2 FP.27 Pabis et al. proposed that distortion of the trans-leaflet by influenza hemagglutinin was pivotal in the simulated fusion process and dependent upon the deep insertion of the N-terminus of the influenza fusion peptide into contact with a phosphoryl group of the trans-leaflet.61 In the SARS-CoV-2 FP simulations, electrostatically favorable contacts fluctuate between basic choline headgroups of the invaginated trans-leaflet and acidic Asp830, Asp820, and Asp843 (Figs. 3, 5 – 8). Lys825, Lys835, and Arg847 also approached the phosphoryl portions of the invaginated headgroups. The simulated behaviors raise the question of FP also distorting the trans-leaflet and mixing lipids of more physiological composition.
The NMR evidence for hydration of the FP backbone while inserted in bicelles (Figs. S8, 2C) is intriguing. It appears related to the provocative lipid flip-flop through hydrated portals suggested by NMR-biased simulations. If the coronaviral FP could promote a concurrent transmembrane flux of lipids and water, that might favor fusion pore models over proximity models.63 Neutron reflectivity measured water introduced to a membrane by a 22-residue FP peptide from SARS-CoV-2 Spike, supporting a fusion pore model.27 The trajectories herein portray the opening in the trans leaflet as a fluctuating, water-filled invagination that connects to “wires” or “sheets” of water to establish a fluctuating, hydrated corridor across the membrane (Figs. 3, 5 – 8 and Movies S1 – S3, S5 – S8).
Concluding remarks
In MD simulations of SARS-CoV-2 FP in a zwitterionic bilayer, constrained by depth estimates by NMR, FP and part of its acidic and basic sidechains inserted into a leaflet of the bilayer in monovalent cations. Deep insertion of FP charge into the DMPC bilayer was accompanied by phosphocholine headgroups and water molecules migrating together into the membrane in the simulations, consistent with hydration of part of the bicelle-inserted backbone detected by NMR. The simulated influx of polar groups occurred around FP in the cis-leaflet and across in the trans-leaflet. This was the case in the three major angles of insertion of FP in the membrane and in most of the simulations performed with Asp830 neutralized alone or in combination with Asp820, Asp839, and Asp843. Upon protonation of the four insertable aspartate sidechains, protrusion of a hydrophobic lipid tail at the fusion loop occurred. The simulated membrane pinching and water conduits past hydrophilic groups of FP follows the theory of electrostriction and intramembrane hydration described by Parsegian.49 The pinching and water filling of the defect in the trans-leaflet consistently accompanied deep insertion of charged sidechains to near the mid-plane of the DMPC bilayer, and enlarged with deep insertion of additional charges. When fluctuating watery passages opened wide enough for a choline headgroup, flip-flop of DMPC between leaflets proceeded in both directions past FP, suggesting a simulated avenue of lipid mixing between leaflets. Whether the simulated membrane manipulation and lipid flip-flop by FP occur in coronaviral-cell fusion is unknown.
Star Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Steven Van Doren (vandorens@missouri.edu).
Materials availability
All unique/stable reagents generated in the study are available from the lead contact.
Data and code availability
The NMR-biased MD trajectories, collective variables, and protein structure files have been deposited in osf.io in project “SARS2_fusion_peptide_in_membrane: NMR-biased_MD” at https://osf.io/95tre/
The analysis scripts written in Tcl or PLUMED have been deposited in osf.io in the project “SARS2_fusion_peptide_in_membrane: NMR-biased_MD” at https://osf.io/95tre/
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Bacterial host strain E. coli BL21-Gold (DE3) RIL was used to express FP.
Method Details
Preparation of FP
A protein fusion of the protein G B1 domain with the 42-residue fusion peptide of Spike from SARS-CoV-2 with a stabilizing I844V mutation was expressed in E. coli BL21(DE3)67 (Gold RIL strain) using PG minimal medium containing 15NH4 and 13C6 D-glucose for uniform labeling.16,68 The sequence after completion of the preparative steps was: 816-SFIED LLFNKVTLAD AGFIKQYGDC LGDVAARDLI CAQKFNG-857. The bacterial cell pellets were lysed by sonication and the insoluble protein recovered in inclusion bodies by centrifugation. The insoluble pellet was dissolved in 6 M urea, 20 mM Tris (pH 8.0). The supernatant was loaded onto a column of nitriloacetic acid resin (GenScript), washed with several column volumes of 20 mM imidazole (20 mM Tris, pH 8.0), and eluted with 300 mM imidazole (pH 8.0). Disulfide bonds were reduced with 10 mM dithiothreitol. To fold the protein and form the disulfide bond, GB1-FP was diluted to 0.1 mg/ml by dialysis overnight against 3 mM cysteine, 0.3 mM cystine, 20 mM Tris (pH 8.0), followed by dialysis against 20 mM Tris (pH 8.0) without thiol redox reagents. GB1-FP was treated with TEV protease and the GB1 domain removed by using chromatography on nitriloacetic acid resin (see above). Pure FP was captured on Q-Sepharose resin, eluted with 1 M NaCl in 20 mM Tris (pH 8.0), and dialyzed into 20 mM Tris-acetate (pH 5.0) for NMR.16
Preparation of bicelles
DMPC vesicles were prepared using three freeze-thaw cycles, followed by bath sonication until the milky white suspension became translucent with a blue tint at 42 °C.69,70 The large unilamellar vesicles and multilamellar vesicles were collected by microcentrifugation for 30 min. A concentrated suspension of these DMPC vesicles was dissolved in DH7PC and incubated at 42 °C for 1 to 2 h until formation of a homogeneous suspension of bicelles.8,59 The stock solution of bicelles was a mixture of 50 mM DMPC with 100 mM DH7PC. This was estimated to correspond to 0.385 mM bicelles, based on the estimate of 130 DMPC per bicelle and 65 DMPC per leaflet, based on diameters of bicelles of 8 to 9 nm. The ratio of one DMPC to two DH7PC (q=0.5) was verified from the 1:2 ratio of their respective methyl peak areas in the 1H NMR spectrum (Fig. S1A).71 The integrity of the bicelles was also evident from their mean diameter of 8.1 nm by dynamic light scattering (Fig. S1B).
Measurement and interpretation of PREs
NMR samples contained 0.4 mM 15N/13C FP (I844V) and 0.24 mM DMPC/DH7PC bicelles (q=0.5). Spin-labeled 16:0–5-doxyl PC or 16:0–14-doxyl PC was added to 0.48 mM. This resulted in an average of one spin label per 65 DMPC molecules, that is, about one spin-label POPC per leaflet of the bicelles. The nitroxide substituents in the 2-acyl chain lay at the 5-position closer to the ester linkage or the 14-position near the methyl terminus. The NMR peak assignments from BMRB entry 30909 were used. A PROJECT-CPMG train that suppresses 1JHH couplings72 was inserted in 15N HSQC and 13C HMQC pulse sequences for measuring exponential relaxation decays without J-modulation from the diamagnetic and paramagnetic samples.69 CPMG periods of 0, 6, 12, 18 ms were measured. The peak heights, y, from the exponential relaxation decays were fitted with the expression:
Eq. 1: , where R2 is the apparent relaxation rate constant, x the length of the CPMG period, and y0 a constant offset fitted. The PREs were estimated quantitatively from the apparent relaxation rate constants: Γ2 = R2,paramag − R2,diamag. The close approaches, r, between the spin-labeled lipid and protein groups that underwent line broadening from the spin label were estimated from the relationship:
eq. 2: , where κ =1.23e-44 m6 s−2 and τc is the rotational correlation time. The τc in the bicelles, estimated from the 15N R2/R1 ratio,16 was 16.2 ns.
PREs and NOEs as ambiguous harmonic restraints in MD simulations
PREs.
Lipid-protein proximities estimated from quantitative fits of PREs (eq. 1; Fig. 2) were satisfied in the restrained simulations by close approaches to the 5- or 14-position of the central fatty acyl chain of any of several DMPC molecules. These ambiguous distance restraints were implemented as collective variables (colvars). The simulations evaluated the energy of colvars for PREs emanating from 16:0–5-doxyl PC to the 5R and S hydrogen atoms of twelve DMPC molecules of the cis-leaflet surrounding FP (DMPC residues 128 – 139). The simulations evaluated the colvars for PREs from 16:0–14-doxyl PC to C14 of 29 DMPC molecules (DMPC residues 128 – 139 and 17 more DMPC molecules randomly and widely dispersed through the cis-leaflet). There was no energy penalty from a colvar with a close approach within radius r of one of the C14 atoms in a colvar from the 14-doxyl label, or within r x 1.15 of a 5R or S hydrogen atom in a colvar from the 5-doxyl label. The harmonic force constant used with the PRE-based colvars was 0.02 kcal/mol/Å2. 33 colvars were used to bias simulations toward agreement with PREs from the 5-doxyl spin label and 16 colvars for PREs from the 14-doxyl spin label.
NOEs.
The six lipid-protein NOEs assigned16 were implemented similarly as colvars to bias MD simulations gently using force constants of 0.02 kcal/mol/Å2. Four of the NOEs were ambiguously defined to H7 methylene protons of both fatty acyl chains and two of the NOEs to H2 protons of both fatty acyl chains among twelve DMPC molecules surrounding the inserted FP (DMPC molecules 128 – 139) in the cis-leaflet. The conformation of FP measured in bicelles16 was maintained during the simulations using 116 NOE-based distances, implemented as colvars. The NOEs comprised 24 long-range NOEs (≥ 5 residues apart in sequence), 80 medium-range NOEs (two to four residues apart), and 12 sequential NOEs. Harmonic force constants of 0.05 kcal/mol/Å2 were used in these colvars.
NMR biasing of MD simulations
The three variants of the system (Asp−1 830, Asp−0 830, and Asp0 820/830/839/843) were generated using CHARMM-GUI and Membrane Builder.73–76 The CHARMM36 all-atom forcefield (validated for DMPC)77,78 was employed using NAMD 2.13, its Colvars module, and compilation for GPUs.79 TIP3P water molecules were used.80,81 The assemblages were built with 127 DMPC molecules in the trans-leaflet, 119 DMPC molecules in the cis-leaflet, and surface areas of each leaflet of about 8150 Å2. The structure of FP adopted in bicelles (PDB: 7MY8)16 was inserted in the cis-leaflet with the vector between Cα atoms of Leu828 and Asp848 aligned along the Z-axis. Each simulation box had dimensions of approximately 90.3 × 90.3 × 85.9 Å. Layers of water around 23.4 Å thick sandwiched the zwitterionic membrane with FP largely inserted. The ionic strength of all simulations was set to 150 mM using primarily KCl. Among the Asp−1 830 trajectories (deprotonated), five were run in 150 mM KCl and another five in 120 mM KCl plus 12 mM CaCl2 (two calcium ions in the simulation box) for ionic strength of 150 mM.
The simulations were performed at 303.15 K where the bicelle-inserted fold of FP is stable16 and DMPC is fluid. The isobaric-isothermal (NPT) ensemble maintained temperature control by the Nosé-Hoover chain method82 and pressure control by the Langevin piston method.83 Semi-isotropic pressure coupling were used by setting useFlexibleCell and useConstantRatio each to yes. Periodic boundary conditions were used. Non-bonded interactions were evaluated with switching from 10 to 12 Å. Electrostatics were evaluated by the particle mesh Ewald method.84 Using rigid bonds to hydrogen, time steps were 1 fs in the first three steps of equilibration, 2 fs in the next three steps of equilibration, and 2 fs during production runs. The NMR-based colvars were active throughout. During the six steps of initial equilibration, the force constants of the protein positional restraints decreased through steps of 10, 5.0, 2.5, 1.0, 0.5, and 0.1 kcal/mol, to none during production runs. The production runs were not considered fully equilibrated, however, until after the energies of the NMR-based lipid-protein colvars had eased and stabilized (marked in Figs. S4—S12). For each of the conditions,1.5 μs of trajectories (five replicates of 300 ns each) were calculated with the gentle biasing by colvars representing PRE and NOE measurements mentioned above.
Withdrawal of depth restraints.
The Asp−1 830 and Asp0 830 trajectories were extended 200 ns per replicate with only half of the lipid-protein depth restraints (Table S2) or without any lipid-protein depth restraints. These extensions decreased the force constants for NOEs within the polypeptide by five-fold to 0.01 kcal/mol/Å2. In the dynamic C-terminal segment from Ala845 to Gly857, NOE force constants were decreased five-fold further to 0.002 kcal/mol/Å2.
Analyses of MD trajectories
Snapshots at 100 ps intervals were used to illustrate and analyze the trajectories in VMD 1.9.3 using Tcl and PLUMED scripts.85,86 The trajectories were preprocessed with unwrapping (protein or lipids), centering of the bilayer mid-plane at Z=0, and wrapping for centering laterally. Unwrapping used a Tcl script of Jerome Hennin. Centering and wrapping used a Tcl script of Jeff Comer. We wrote Tcl scripts to count and locate water molecules and choline headgroups, monitor flip-flop of choline headgroups, monitor radii of gyration of ensembles of interior water, monitor distances of calcium ions to carboxylate groups, and monitor energies of violations of the colvars for PREs and NOEs. We wrote scripts for use the PLUMED-GUI for VMD86 to measure the depths and tilts of insertion of FP in the lipid bilayer. Essential dynamics analyses (EDA or PCA) of trajectories were calculated using ProDy and depicted using the NMWiz plugin for VMD.87
Quantification and statistical analysis
Measurements were quantified using the Tcl and PLUMED analysis scripts posted. NMR peak heights were measured using NMRFAM-Sparky or Bruker Topspin 3.5. Data were fitted or plotted using Origin, versions 2020–2023. Statistical parameters were calculated using Origin or Microsoft Excel. Centers of distributions were defined by mean or median as indicated in Figures and Tables. Dispersion of points of key distributions was illustrated in the figures and summarized by SD, coefficient of variation, interquartile range, and root-mean-squared deviation given in Tables and Figures.
Supplementary Material
Movie S1. Rotation through a median Asp−1 830 frame shows the distortion of the membrane and fatty acyl chains near the fusion peptide. Related to Figs. 3 to 5. The snapshot is drawn at 203 ns into trajectory 5. Waters are omitted for clarity.
Movie S2. Rotation through a median Asp0 830 frame shows the distortion of the membrane and fatty acyl chains. Related to Figs. 3, 4, and 6. The frame is from trajectory 2 at 282 ns.
Movie S3. Rotation through a median Asp0 820/830/839/843 frame shows the distortion of the membrane and fatty acyl chains. Related to Figs. 3, 4, and 7. The frame is from trajectory 5 at 19 ns.
Movie S4. Protrusion of an acyl chain (green) over the fusion loop in Asp0 820/830/839/843 trajectory. Related to Fig. 3. The disulfide and protonated Asp839 and Asp843 in the fusion loop are plotted in this segment running through 34 ns.
Movie S5. Water spans the DMPC bilayer in a representative Asp−1 830 trajectory. Related to Figs. 3 to 5. Trajectory 5 illustrated from 140 to 300 ns. Orange spheres mark water oxygens in a 25 Å slab and within 15 Å of the fusion peptide. Fatty acyl chains within 20 Å of the fusion peptide are yellow. Charged sidechains in the membrane are plotted. K+ ions are plotted with light blue spheres.
Movie S6. Formation of a narrow water bridge across the DMPC bilayer a representative Asp0 830 trajectory. Related to Figs. 3, 4, and 6. Trajectory 2 from 0 to 120 ns is illustrated as in Movie S5.
Movie S7. Top-down view of the cis-leaflet of the Asp−1 830 points out the dense pool of water near the fusion peptide. Related to Figs. 3 to 5. Small spheres indicate water molecules (pink: O; white: H) in near the bilayer mid-plane.
Movie S8. Bidirectional lipid flip-flop through aqueous passages flanking the fusion peptide. Related to Fig. 8A–D. The flip-flop of DMPC molecules during Asp−1 830 trajectory 3 is captured from 80 – 130 ns. Choline headgroups are plotted with spheres. DMPC molecules are colored white upon flip-flop from the trans- to cis-leaflet. DMPC molecules are colored cyan upon flip-flop from the cis- to trans-leaflet.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21-Gold (DE3) RIL | Agilent | 230245 |
| Chemicals, peptides, and recombinant proteins | ||
| Ni-NTA resin | GenScript | L00250 |
| TEV protease | GenScript | Z03030 |
| DMPC | Avanti Polar Lipids | 850345 |
| DH7PC | Avanti Polar Lipids | 850306 |
| 16:0–5 Doxyl PC | Avanti Polar Lipids | 810601 |
| 16:0–14 Doxyl PC | Avanti Polar Lipids | 810605C discontinued; use 810604 |
| Recombinant FP prepared from GB1-FP | Koppisetti et al. (2021) | N/A |
| Deposited data | ||
| NMR-biased MD trajectories in .DCD format | This paper | DOI 10.17605/OSF.IO/95TRE |
| collective variables,.psf files, and .pdb files used for MD simulations | This paper | DOI 10.17605/OSF.IO/95TRE |
| Oligonucleotides | ||
| GB1-FP_seq with GGGKKKK tag prior to deletion: gat ata cat atg cac cac cat cac cat cac ggc ggc acg tat aaa ctt att ctc aat ggt aag aca ctt aaa ggg gag act acg aca gag gcg gta gac gca gca acg gca gag aag gtg ttc aaa caa tac gca aat gat aac ggg gtg gac ggg gaa tgg aca tac gat gat gcg acc aaa aca ttt act gtc act gaa ggg ggt gag aat tta tat ttt caa agt ttt att gaa gac ttg ctt ttc aac aaa gtg acg ctg gca gac gcg ggt ttc att aag cag tat ggg gac tgc ttg ggt gat att gca gcg cgg gat ttg att tgt gct caa aag ttc aat ggc ggt ggt ggt aaa aag aaa aaa tga gaa ttc gag ctc | Koppisetti et al. (2021) | N/A |
| Recombinant DNA | ||
| pET27b(+) with GB1-FP insert and deletion of C-terminal GGGKKKK tag. Hosted in E. coli BL21-Gold (DE3) RIL. | Koppisetti et al. (2021) | N/A |
| Software and algorithms | ||
| Tcl analysis scripts and PLUMED-VMD analysis scripts | This paper | DOI 10.17605/OSF.IO/95TRE |
Highlights.
Fusion peptide insertion in a DMPC bilayer reconciled NMR and simulations.
Hydrogen exchange and simulations find hydration of the fusion peptide in the bilayer.
Simulated defects feature membrane pinching at inserted charge and lipid flip-flop.
Similar distortions were hypothesized to promote influenza viral-cell fusion.
Acknowledgments
National Science Foundation RAPID award 2030473 supported this work. The 800 MHz NMR spectrometer was purchased in part using NIH grant S10RR022341. The MD simulations utilized the HPC resources of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource (supported by NIH grant P41GM111135), and the Extreme Science and Engineering Discovery Environment (XSEDE, supported by National Science Foundation grant ACI-1548562). Tanner Roy and Catherine Cummings measured the dynamic light scattering of the bicelles. We thank Richard Pastor and Amy Rice for discussion and Ioan Kosztin and Zachary Berndsen for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
References
- 1.Dacon C, Tucker C, Peng L, Lee C-CD, Lin T-H, Yuan M, Cong Y, Wang L, Purser L, Williams JK, et al. (2022). Broadly neutralizing antibodies target the coronavirus fusion peptide. Science (80-.) 377, 728–735. 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madu IG, Roth SL, Belouzard S, and Whittaker GR (2009). Characterization of a Highly Conserved Domain within the Severe Acute Respiratory Syndrome Coronavirus Spike Protein S2 Domain with Characteristics of a Viral Fusion Peptide. J. Virol 83, 7411–7421. 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang T, Bidon M, Jaimes JA, Whittaker GR, and Daniel S (2020). Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res 178, 104792. 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch B-J, Rey FA, and Veesler D (2017). Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci 114, 11157–11162. 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM, Rawson S, Rits-Volloch S, and Chen B (2020). Distinct conformational states of SARS-CoV-2 spike protein. Science (80-.) 369, 1586–1592. 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaitanya CS, M. LJ, Rodolfo G, and Ad B (2021). Transient lipid-bound states of spike protein heptad repeats provide insights into SARS-CoV-2 membrane fusion. Sci. Adv 7, eabk2226. 10.1126/sciadv.abk2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcink TC, Kicmal T, Armbruster E, Zhang Z, Zipursky G, Golub KL, Idris M, Khao J, Drew-Bear J, McGill G, et al. (2022). Intermediates in SARS-CoV-2 spike–mediated cell entry. Sci. Adv 8, eabo3153. 10.1126/sciadv.abo3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JM, and Whittaker GR (2016). Fusion of Enveloped Viruses in Endosomes. Traffic 17, 593–614. 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai L, Zhu G, Yang M, Cao L, Xing X, Yin G, Chan C, Qin C, Rao Z, Wang X, et al. (2021). Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein. Proc. Natl. Acad. Sci 118, e2112703118. 10.1073/pnas.2112703118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birtles D, Oh AE, and Lee J (2022). Exploring the pH dependence of the SARS-CoV-2 complete fusion domain and the role of its unique structural features. Protein Sci 31, e4390. 10.1002/pro.4390. [DOI] [Google Scholar]
- 12.Mellman I, Fuchs R, and Helenius A (1986). ACIDIFICATION OF THE ENDOCYTIC AND EXOCYTIC PATHWAYS. Annu. Rev. Biochem 55, 663–700. 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 13.Narayanaswamy N, Chakraborty K, Saminathan A, Zeichner E, Leung K, Devany J, and Krishnan Y (2019). A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat. Methods 16, 95–102. 10.1038/s41592-018-0232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerasimenko JV, Tepikin AV, Petersen OH, and Gerasimenko OV (1998). Calcium uptake via endocytosis with rapid release from acidifying endosomes. Curr. Biol 8, 1335–1338. 10.1016/S0960-9822(07)00565-9. [DOI] [PubMed] [Google Scholar]
- 15.Lai AL, Millet JK, Daniel S, Freed JH, and Whittaker GR (2017). The SARS-CoV Fusion Peptide Forms an Extended Bipartite Fusion Platform that Perturbs Membrane Order in a Calcium-Dependent Manner. J. Mol. Biol 429, 3875–3892. 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koppisetti RK, Fulcher YG, and Van Doren SR (2021). Fusion Peptide of SARS-CoV-2 Spike Rearranges into a Wedge Inserted in Bilayered Micelles. J. Am. Chem. Soc 143, 13205–13211. 10.1021/jacs.1c05435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khelashvili G, Plante A, Doktorova M, and Weinstein H (2021). Ca2+-dependent mechanism of membrane insertion and destabilization by the SARS-CoV-2 fusion peptide. Biophys. J 120, 1105–1119. 10.1016/j.bpj.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer SL, Jung H, and Hummer G (2021). Binding of SARS-CoV-2 Fusion Peptide to Host Endosome and Plasma Membrane. J. Phys. Chem. B 125, 7732–7741. 10.1021/acs.jpcb.1c04176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birtles D, and Lee J (2021). Identifying Distinct Structural Features of the SARS-CoV-2 Spike Protein Fusion Domain Essential for Membrane Interaction. Biochemistry 60, 2978–2986. 10.1021/acs.biochem.1c00543. [DOI] [PubMed] [Google Scholar]
- 20.Lai AL, and Freed JH (2021). SARS-CoV-2 Fusion Peptide has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca2+. J. Mol. Biol 433, 166946. 10.1016/j.jmb.2021.166946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straus MR, Tang T, Lai AL, Flegel A, Bidon M, Freed JH, Daniel S, and Whittaker GR (2020). Ca2+ Ions Promote Fusion of Middle East Respiratory Syndrome Coronavirus with Host Cells and Increase Infectivity. J. Virol 94, 10.1128/jvi.00426-20. 10.1128/jvi.00426–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai AL, and Freed JH (2022). Negatively charged residues in the membrane ordering activity of SARS-CoV-1 and -2 fusion peptides. Biophys. J 121, 207–227. 10.1016/j.bpj.2021.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carten JD, Khelashvili G, Bidon MK, Straus MR, Tang T, Jaimes JA, Weinstein H, Whittaker GR, and Daniel S (2023). A mechanistic understanding of the modes of Ca ion binding to the SARS-CoV-1 fusion peptide and their role in the dynamics of host membrane penetration. bioRxiv 10.1101/2022.03.03.482731. [DOI] [PubMed] [Google Scholar]
- 24.Gorgun D, Lihan M, Kapoor K, and Tajkhorshid E (2021). Binding mode of SARS-CoV-2 fusion peptide to human cellular membrane. Biophys. J 120, 2914–2926. 10.1016/j.bpj.2021.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen H, Wu Z, and Chen L (2022). Different Binding Modes of SARS-CoV-1 and SARS-CoV-2 Fusion Peptides to Cell Membranes: The Influence of Peptide Helix Length. J. Phys. Chem. B 126, 4261–4271. 10.1021/acs.jpcb.2c01295. [DOI] [PubMed] [Google Scholar]
- 26.Villalaín J (2023). SARS-CoV-2 Protein S Fusion Peptide Is Capable of Wrapping Negatively-Charged Phospholipids. Membranes (Basel) 13. 10.3390/membranes13030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santamaria A, Batchu KC, Matsarskaia O, Prévost SF, Russo D, Natali F, Seydel T, Hoffmann I, Laux V, Haertlein M, et al. (2022). Strikingly Different Roles of SARS-CoV-2 Fusion Peptides Uncovered by Neutron Scattering. J. Am. Chem. Soc 144, 2968–2979. 10.1021/jacs.1c09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bidon MK, Khelashvili G, Straus MR, Tang T, Carten JD, Weinstein H, Whittaker GR, and Daniel S (2022). The interaction of calcium ions with specific residues in the SARS-CoV fusion peptide and the regulation of viral infectivity. bioRxiv 10.1101/2022.03.03.482731. [DOI] [Google Scholar]
- 29.Lagüe P, Roux B, and Pastor RW (2005). Molecular Dynamics Simulations of the Influenza Hemagglutinin Fusion Peptide in Micelles and Bilayers: Conformational Analysis of Peptide and Lipids. J. Mol. Biol 354, 1129–1141. 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 30.Alfredo FJ, J. TD, Gunnar von H., and H. WS (2005). Interface connections of a transmembrane voltage sensor. Proc. Natl. Acad. Sci 102, 15059–15064. 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett WFD, and Tieleman DP (2014). The Importance of Membrane Defects—Lessons from Simulations. Acc. Chem. Res 47, 2244–2251. 10.1021/ar4002729. [DOI] [PubMed] [Google Scholar]
- 32.Perrin BS, and Pastor RW (2016). Simulations of Membrane-Disrupting Peptides I: Alamethicin Pore Stability and Spontaneous Insertion. Biophys. J 111, 1248–1257. 10.1016/j.bpj.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrin BS, Fu R, Cotten ML, and Pastor RW (2016). Simulations of Membrane-Disrupting Peptides II: AMP Piscidin 1 Favors Surface Defects over Pores. Biophys. J 111, 1258–1266. 10.1016/j.bpj.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihailescu M, Sorci M, Seckute J, Silin VI, Hammer J, Perrin BS, Hernandez JI, Smajic N, Shrestha A, Bogardus KA, et al. (2019). Structure and Function in Antimicrobial Piscidins: Histidine Position, Directionality of Membrane Insertion, and pH-Dependent Permeabilization. J. Am. Chem. Soc 141, 9837–9853. 10.1021/jacs.9b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsson P, and Kasson PM (2013). Lipid Tail Protrusion in Simulations Predicts Fusogenic Activity of Influenza Fusion Peptide Mutants and Conformational Models. PLOS Comput. Biol 9, 1–9. 10.1371/journal.pcbi.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Légaré S, and Lagüe P (2014). The influenza fusion peptide promotes lipid polar head intrusion through hydrogen bonding with phosphates and N-terminal membrane insertion depth. Proteins Struct. Funct. Bioinforma 82, 2118–2127. 10.1002/prot.24568. [DOI] [PubMed] [Google Scholar]
- 37.Rice A, Haldar S, Wang E, Blank PS, Akimov SA, Galimzyanov TR, Pastor RW, and Zimmerberg J (2022). Planar aggregation of the influenza viral fusion peptide alters membrane structure and hydration, promoting poration. Nat. Commun 13, 7336. / 10.1038/s41467-022-34576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marinelli F, and Faraldo-Gómez JD (2015). Ensemble-Biased Metadynamics: A Molecular Simulation Method to Sample Experimental Distributions. Biophys. J 108, 2779–2782. 10.1016/j.bpj.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hustedt EJ, Marinelli F, Stein RA, Faraldo-Gómez JD, and Mchaourab HS (2018). Confidence Analysis of DEER Data and Its Structural Interpretation with Ensemble-Biased Metadynamics. Biophys. J 115, 1200–1216. 10.1016/j.bpj.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prior SH, Fulcher YG, Koppisetti RK, Jurkevich A, and Van Doren SR (2015). Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors. Structure 23, 2099–2110. 10.1016/J.STR.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcink TC, Simoncic JA, An B, Knapinska AM, Fulcher YG, Akkaladevi N, Fields GB, and Van Doren SR (2019). MT1-MMP Binds Membranes by Opposite Tips of Its β Propeller to Position It for Pericellular Proteolysis. Structure 27. 10.1016/j.str.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee D, Walter KFA, Brückner A-K, Hilty C, Becker S, and Griesinger C (2008). Bilayer in Small Bicelles Revealed by Lipid–Protein Interactions Using NMR Spectroscopy. J. Am. Chem. Soc 130, 13822–13823. 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- 43.Lindberg M, Biverståhl H, Gräslund A, and Mäler L (2003). Structure and positioning comparison of two variants of penetratin in two different membrane mimicking systems by NMR. Eur. J. Biochem 270, 3055–3063. 10.1046/j.1432-1033.2003.03685.x. [DOI] [PubMed] [Google Scholar]
- 44.Caldwell TA, Baoukina S, Brock AT, Oliver RC, Root KT, Krueger JK, Glover KJ, Tieleman DP, and Columbus L (2018). Low-q Bicelles Are Mixed Micelles. J. Phys. Chem. Lett 9, 4469–4473. 10.1021/acs.jpclett.8b02079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehler EL, Fuxreiter M, Simon I, and Garcia-Moreno E B (2002). The role of hydrophobic microenvironments in modulating pKa shifts in proteins. Proteins Struct. Funct. Bioinforma 48, 283–292. 10.1002/prot.10153. [DOI] [PubMed] [Google Scholar]
- 46.Isom DG, Castañeda CA, Cannon BR, Velu PD, and García-Moreno E, B. (2010). Charges in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci 107, 16096–16100. 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vila-Viçosa D, Silva TFD, Slaybaugh G, Reshetnyak YK, Andreev OA, and Machuqueiro M (2018). Membrane-Induced pKa Shifts in wt-pHLIP and Its L16H Variant. J. Chem. Theory Comput 14, 3289–3297. 10.1021/acs.jctc.8b00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva TFD, Visca H, Klumpp C, Andreev OA, Reshetnyak YK, and Machuqueiro M (2023). Arginine Residues Modulate the Membrane Interactions of pHLIP Peptides. J. Chem. Inf. Model 10.1021/acs.jcim.3c00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.PARSEGIAN A (1969). Energy of an Ion crossing a Low Dielectric Membrane: Solutions to Four Relevant Electrostatic Problems. Nature 221, 844–846. 10.1038/221844a0. [DOI] [PubMed] [Google Scholar]
- 50.Battiste JL, and Wagner G (2000). Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39, 5355–5365. [DOI] [PubMed] [Google Scholar]
- 51.Smirnova YG, Marrink S-J, Lipowsky R, and Knecht V (2010). Solvent-Exposed Tails as Prestalk Transition States for Membrane Fusion at Low Hydration. J. Am. Chem. Soc 132, 6710–6718. 10.1021/ja910050x. [DOI] [PubMed] [Google Scholar]
- 52.Kuzmin PI, Zimmerberg J, Chizmadzhev YA, and Cohen FS (2001). A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci 98, 7235–7240. 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prosser RS, Evanics F, Kitevski JL, and Al-Abdul-Wahid MS (2006). Current Applications of Bicelles in NMR Studies of Membrane-Associated Amphiphiles and Proteins,. Biochemistry 45, 8453–8465. 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 54.Durr UH, Soong R, and Ramamoorthy A (2013). When detergent meets bilayer: birth and coming of age of lipid bicelles. Prog Nucl Magn Reson Spectrosc 69, 1–22. 10.1016/j.pnmrs.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dufourc EJ (2021). Bicelles and nanodiscs for biophysical chemistry11A tribute to Prof Michèle Auger, University Laval, Canada. Biochim. Biophys. Acta - Biomembr 1863, 183478. 10.1016/j.bbamem.2020.183478. [DOI] [PubMed] [Google Scholar]
- 56.Lu Z, Van Horn WD, Chen J, Mathew S, Zent R, and Sanders CR (2012). Bicelles at low concentrations. Mol Pharm 9, 752–761. 10.1021/mp2004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ladizhansky V (2017). Applications of solid-state NMR to membrane proteins. Biochim. Biophys. Acta - Proteins Proteomics 1865, 1577–1586. 10.1016/j.bbapap.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 58.Mandala VS, Williams JK, and Hong M (2018). Structure and Dynamics of Membrane Proteins from Solid-State NMR. Annu. Rev. Biophys 47, 201–222. 10.1146/annurev-biophys-070816-033712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray DT, Das N, and Cross TA (2013). Solid State NMR Strategy for Characterizing Native Membrane Protein Structures. Acc. Chem. Res 46, 2172–2181. 10.1021/ar3003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hessa T, White SH, and von Heijne G (2005). Membrane Insertion of a Potassium-Channel Voltage Sensor. Science (80-.) 307, 1427. 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 61.Pabis A, Rawle RJ, and Kasson PM (2020). Influenza hemagglutinin drives viral entry via two sequential intramembrane mechanisms. Proc. Natl. Acad. Sci 117, 7200–7207. 10.1073/pnas.1914188117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Israelachvili JN (2011). Interactions of Biological Membranes and Structures. In Intermolecular and Surface Forces (Academic Press; ), pp. 577–613. [Google Scholar]
- 63.Lindau M, and Almers W (1995). Structure and function of fusion pores in exocytosis and ectoplasmic membrane fusion. Curr. Opin. Cell Biol 7, 509–517. 10.1016/0955-0674(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 64.Fu Q, and Chou JJ (2021). A trimeric hydrophobic zipper mediates the intramembrane assembly of SARS-CoV-2 spike. J. Am. Chem. Soc 143, 8543–8546. 10.1021/jacs.1c02394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan X, Cao D, Kong L, and Zhang X (2020). Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun 11, 3618. 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lassmann T (2020). Kalign 3: multiple sequence alignment of large datasets. Bioinformatics 36, 1928–1929. 10.1093/bioinformatics/btz795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Studier FW, and Moffatt BA (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 68.Studier FW (2005). Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif 41, 207–234. 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Koppisetti RK, Fulcher YG, Jurkevich A, Prior SH, Xu J, Lenoir M, Overduin M, and Van Doren SR (2014). Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12. Nat. Commun 5, 5552. 10.1038/ncomms6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marcink TC, Koppisetti RK, Fulcher YG, and Van Doren SR (2017). Mapping lipid bilayer recognition sites of metalloproteinases and other prospective peripheral membrane proteins 10.1007/978-1-4939-6863-3_5. [DOI] [PubMed]
- 71.Piai A, Fu Q, Dev J, and Chou JJ (2017). Optimal Bicelle Size q for Solution NMR Studies of the Protein Transmembrane Partition. Chem. – A Eur. J 23, 1361–1367. 10.1002/chem.201604206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aguilar JA, Nilsson M, Bodenhausen G, and Morris GA (2012). Spin echo NMR spectra without J modulation. Chem. Commun 48, 811–813. [DOI] [PubMed] [Google Scholar]
- 73.Jo S, Kim T, Iyer V, and Im W (2008). CHARMM-GUI: a web-based graphical user interface for CHARMM [DOI] [PubMed]
- 74.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, et al. (2016). CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput 12, 405–413. 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu EL, Cheng X, Jo S, Rui H, Song KC, Dávila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, et al. (2014). CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem 35, 1997–2004. 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jo S, Lim JB, Klauda JB, and Im W (2009). CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J 97, 50–58. 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, and Pastor RW (2010). Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 114, 7830–7843. 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang J, and MacKerell AD Jr. (2013). CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34, 2135–2145. 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Hénin J, Jiang W, et al. (2020). Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys 153, 44130. 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- 81.DURELL SR, BROOKS BR, and BEN-NAIM A (1994). Solvent-induced forces between two hydrophilic groups. J. Phys. Chem 98, 2198–2202. [Google Scholar]
- 82.Martyna GJ, Tobias DJ, and Klein ML (1994). Constant pressure molecular dynamics algorithms. J. Chem. Phys 101, 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
- 83.Feller SE, Zhang Y, Pastor RW, and Brooks BR (1995). Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys 103, 4613–4621. 10.1063/1.470648. [DOI] [Google Scholar]
- 84.Darden T, York D, and Pedersen L (1993). Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- 85.Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 27–28,33–38. 0263785596000185 [pii]. [DOI] [PubMed] [Google Scholar]
- 86.Giorgino T (2014). PLUMED-GUI: An environment for the interactive development of molecular dynamics analysis and biasing scripts. Comput. Phys. Commun 185, 1109–1114. 10.1016/j.cpc.2013.11.019. [DOI] [Google Scholar]
- 87.Bakan A, Meireles LM, and Bahar I (2011). ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27. 10.1093/bioinformatics/btr168 LB - Bakan2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Rotation through a median Asp−1 830 frame shows the distortion of the membrane and fatty acyl chains near the fusion peptide. Related to Figs. 3 to 5. The snapshot is drawn at 203 ns into trajectory 5. Waters are omitted for clarity.
Movie S2. Rotation through a median Asp0 830 frame shows the distortion of the membrane and fatty acyl chains. Related to Figs. 3, 4, and 6. The frame is from trajectory 2 at 282 ns.
Movie S3. Rotation through a median Asp0 820/830/839/843 frame shows the distortion of the membrane and fatty acyl chains. Related to Figs. 3, 4, and 7. The frame is from trajectory 5 at 19 ns.
Movie S4. Protrusion of an acyl chain (green) over the fusion loop in Asp0 820/830/839/843 trajectory. Related to Fig. 3. The disulfide and protonated Asp839 and Asp843 in the fusion loop are plotted in this segment running through 34 ns.
Movie S5. Water spans the DMPC bilayer in a representative Asp−1 830 trajectory. Related to Figs. 3 to 5. Trajectory 5 illustrated from 140 to 300 ns. Orange spheres mark water oxygens in a 25 Å slab and within 15 Å of the fusion peptide. Fatty acyl chains within 20 Å of the fusion peptide are yellow. Charged sidechains in the membrane are plotted. K+ ions are plotted with light blue spheres.
Movie S6. Formation of a narrow water bridge across the DMPC bilayer a representative Asp0 830 trajectory. Related to Figs. 3, 4, and 6. Trajectory 2 from 0 to 120 ns is illustrated as in Movie S5.
Movie S7. Top-down view of the cis-leaflet of the Asp−1 830 points out the dense pool of water near the fusion peptide. Related to Figs. 3 to 5. Small spheres indicate water molecules (pink: O; white: H) in near the bilayer mid-plane.
Movie S8. Bidirectional lipid flip-flop through aqueous passages flanking the fusion peptide. Related to Fig. 8A–D. The flip-flop of DMPC molecules during Asp−1 830 trajectory 3 is captured from 80 – 130 ns. Choline headgroups are plotted with spheres. DMPC molecules are colored white upon flip-flop from the trans- to cis-leaflet. DMPC molecules are colored cyan upon flip-flop from the cis- to trans-leaflet.
Data Availability Statement
The NMR-biased MD trajectories, collective variables, and protein structure files have been deposited in osf.io in project “SARS2_fusion_peptide_in_membrane: NMR-biased_MD” at https://osf.io/95tre/
The analysis scripts written in Tcl or PLUMED have been deposited in osf.io in the project “SARS2_fusion_peptide_in_membrane: NMR-biased_MD” at https://osf.io/95tre/
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


