SUMMARY
The aryl hydrocarbon receptor (AhR) regulates Th17-polarized CD4+ T cell functions, but its role in HIV-1 replication/outgrowth remains unknown. Genetic (CRISPR-Cas9) and pharmacological inhibition reveal AhR as a barrier to HIV-1 replication in T cell receptor (TCR)-activated CD4+ T cells in vitro. In single-round vesicular stomatitis virus (VSV)-G-pseudotyped HIV-1 infection, AhR blockade increases the efficacy of early/late reverse transcription and subsequently facilitated integration/translation. Moreover, AhR blockade boosts viral outgrowth in CD4+ T cells of people living with HIV-1 (PLWH) receiving antiretroviral therapy (ART). Finally, RNA sequencing reveals genes/pathways downregulated by AhR blockade in CD4+ T cells of ART-treated PLWH, including HIV-1 interactors and gut-homing molecules with AhR-responsive elements in their promoters. Among them, HIC1, a repressor of Tat-mediated HIV-1 transcription and a tissue-residency master regulator, is identified by chromatin immunoprecipitation as a direct AhR target. Thus, AhR governs a T cell transcriptional program controlling viral replication/outgrowth and tissue residency/recirculation, supporting the use of AhR inhibitors in “shock and kill” HIV-1 remission/cure strategies.
Graphical abstract
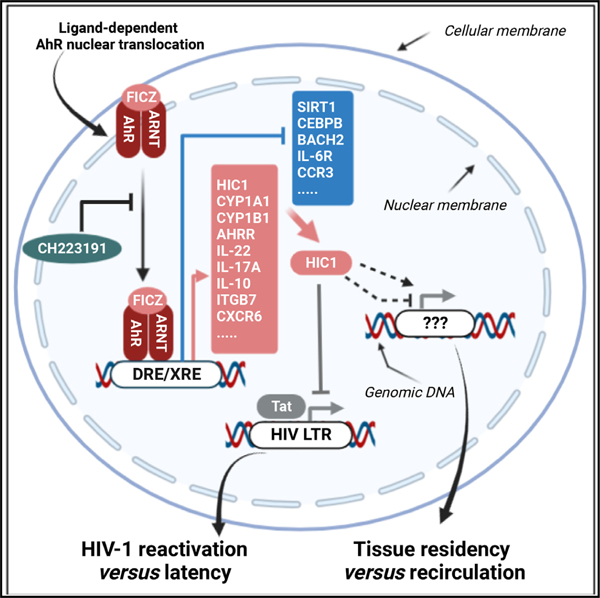
In brief
Chatterjee et al. investigated the role of AhR in controlling Th17/Th22 polarization and HIV-1 replication in CD4+ T cells. Gene editing, pharmacological blockade, and RNA sequencing demonstrated that AhR transcriptionally regulates pathogenic Th17 features and HIV-1 permissiveness/outgrowth. The authors identified the HIV-1 repressor and tissue-residency regulator HIC1 as a direct AhR target.
INTRODUCTION
Antiretroviral therapy (ART) transformed HIV-1 infection into a manageable chronic disease and increased the life expectancy of people living with HIV-1 (PLWH). However, ART does not cure HIV-1, and immune competence is not restored.1,2 Major barriers to HIV-1 remission/cure during ART include the persistence of viral reservoirs (VRs) in long-lived CD4+ T cells,3,4 residual HIV-1 transcription,5–7 and the non-restoration of mucosal CD4+ T cell-dependent intestinal barrier functions.8,9 Subsequently, ART-treated PLWH experience chronic immune activation, immune metabolism deregulation, and increased risk for non-AIDS comorbidities, such as cardiovascular disease.10,11 In this context, additional therapeutic strategies are required for HIV-1 remission/cure.
Among CD4+ T cell lineages, T helper (Th) subsets producing the hallmark cytokines interleukin-17A/F (IL-17A/F; Th17) and IL-22 (Th22) are targeted by HIV-1 for infection and subsequent depletion from mucosal sites (reviewed in Wacleche et al.,8 Planas et al.,9 and Fert et al.12). The differentiation and functionality of Th17 cells are governed by the master regulator transcription factor (TF) retinoic acid orphan receptor γt (RORγt in mice; RORC2 in humans).13,14 RORC2 represents a target for drugs designed to treat autoimmune conditions in which Th17 cells exert deleterious functions.15–17 Of particular notice, our group demonstrated the capacity of RORC2 inhibitors to block HIV outgrowth in CD4+ T cells of ART-treated PLWH,18 supporting the existence of a “RORC2-sensitive” VR.12 However, only a fraction of Th17 cells are permissive to integrative HIV-1 infection,8,9,12,19 suggesting the existence of HIV-1-resistant Th17 cells. In the context of autoimmunity, two types of Th17 cells were identified, “pathogenic” and “non-pathogenic,” based on their ability to promote experimental autoimmune encephalitis (EAE).20 The existence of “non-pathogenic” Th17 cells was demonstrated also in humans and was documented to express a unique molecular signature including the aryl hydrocarbon receptor (AhR).20,21
The AhR is a ligand-dependent nuclear receptor, initially identified as the dioxin receptor.22,23 It is a member of the basic-helix-loop-helix (bHLH)/Per-Arnt-Sim (PAS) family of proteins that interacts with a broad range of xenobiotic or natural ligands such as tryptophan metabolites, dietary products, and microbiota-derived factors.24–26 AhR resides in the cytoplasm in an inactive state due to its association with different chaperons (e.g., HSP90, XAP2, p23).26 Upon ligand binding, AhR forms a heterodimer with the AhR nuclear translocator (ARNT), which subsequently translocates into the nucleus, where it binds onto dioxin-/xenobiotic-responsive elements (DRE/XRE) on the promoter of specific genes.26 Among AhR regulated genes, the xenobiotic-metabolizing enzyme CYP1A1 (cytochrome P450 family 1 A1), AHRR (AhR repressor), and TIPARP (2,3,7,8-tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase) are involved in the negative feedback of AhR signaling.26,27 Thus, AhR is an important nuclear receptor that adapts transcriptional profiles of cells to changes in the environment.24–26
Pioneering studies demonstrated that AhR is specifically expressed by Th17-polarized CD4+ T cells from mice and humans,28 with AhR activation promoting effector functions,29–31 such as the production of IL-22 in Th22 cells and IL-10 in regulatory T cells (Tregs).28,29,32–34 Of particular notice, AhR mediates the trans-differentiation of Th17 cells into Tregs, a process dependent on AhR-mediated IL-10 production.35 Moreover, AhR was reported to transcriptionally regulate the expression of the gut-homing integrin β 7 (ITGB7) in macrophages,36 indicative of an AhR role in regulating gut tropism/residency. Furthermore, a large body of evidence identified that AhR-expressing CD4+ T cells act as important gatekeepers, mainly by producing lineage-specific cytokines (e.g., IL-17A/F, IL-22) that act on epithelial cells to strengthen their barrier functions.26 Finally, AhR activation was reported to block HIV-1 replication in macrophages,37 thus raising the question of whether AhR also restricts HIV-1 infection in CD4+ T cells.
Herein, we explored the role of AhR in modulating HIV-1 replication/outgrowth in primary CD4+ T cells isolated from the peripheral blood of ART-treated PLWH and uninfected participants. Our results provide evidence supporting a model in which AhR governs an antiviral program that, in the context of specific AhR ligands derived from microbiota, diet, and tryptophane catabolism,24–26 may reduce the susceptibility to HIV-1 acquisition during primary infection. Once the infection is established, the activation of the AhR pathway may promote HIV-1 latency in VRs of ART-treated PLWH, thus preventing VR sensing/depletion by the immune system. Finally, our results support the use of pharmacological AhR inhibitors, a class of drugs currently tested in clinically for cancer,38,39 to boost VR reactivation in “shock and kill” HIV-1 remission/cure strategies.
RESULTS
CRISPR-Cas9-mediated AhR knockout reduces IL-17A, IL-22, and IL-10 production and facilitates HIV-1 replication in memory CD4+ T cells
We explored AhR expression/activation in CD4+ T cells. Maximal AhR protein and mRNA expression in memory CD4+ T cells was observed at days 3 and 1 post-T cell receptor (TCR) triggering, respectively (Figures S1A and S1B). Interestingly, AhR expression was not induced by phytohaemagglutinin (PHA) activation (Figure S1C), likely explaining the findings by Kueck et al. that HIV-1 replication in PHA-activated CD4+ T cells was AhR independent.37 The expression of CyP1A1 mRNA, an AhR-specific target gene and a component of the cytochrome p450 family,26 culminated at day 3 post-TCR triggering (Figure S1D). Thus, TCR triggering in CD4+ T cells leads to AhR expression/activation in memory CD4+ T cells, consistent with the presence of AhR ligands in culture media, as previously reported.31
To explore the role of AhR on HIV-1 replication, CRISPR-Cas9-mediated gene editing was optimized in primary CD4+ T cells (Figure 1A) using a published protocol.40 TCR-activated memory CD4+ T cells were electroporated with two AhR-specific (AhR CRISPR RNA [crRNA]) and the non-specific (negative crRNA) guide RNA. The AhR silencing was demonstrated by western blotting visualization of AhR protein expression (Figure 1B). The AhR#1 crRNA was used for all subsequent studies. The gene editing activity of AhR#1 guide RNA was confirmed by T7 endonuclease assay (Figure 1C). Further, the efficiency of AhR#1 crRNA was tested in three other donors at the DNA level by tracking of insertions or deletions (indels) by decomposition (TIDE) analysis (Figures S2A and S2B), as reported.40,41 The editing efficacy determined by TIDE analysis showed donor-to-donor variations (i.e., 32%, 28%, and 7%), proportional with changes in AhR protein expression (Figure S2C). At day 4 post-electroporation, the CRISPR-Cas9-mediated AhR knockout (KO) resulted in decreased expression of the gut-homing molecule ITGB7 (Figures S3A and S3B), consistent with AhR-dependent ITGB7 expression in macrophages.36 The electroporation with AhR crRNA and negative crRNA did not differently affect cell viability (Figure S3C).
Figure 1. CRISPR-Cas9-mediated AhR gene editing reduces IL-22, IL-17A, and IL-10 production in memory CD4+ T cells.
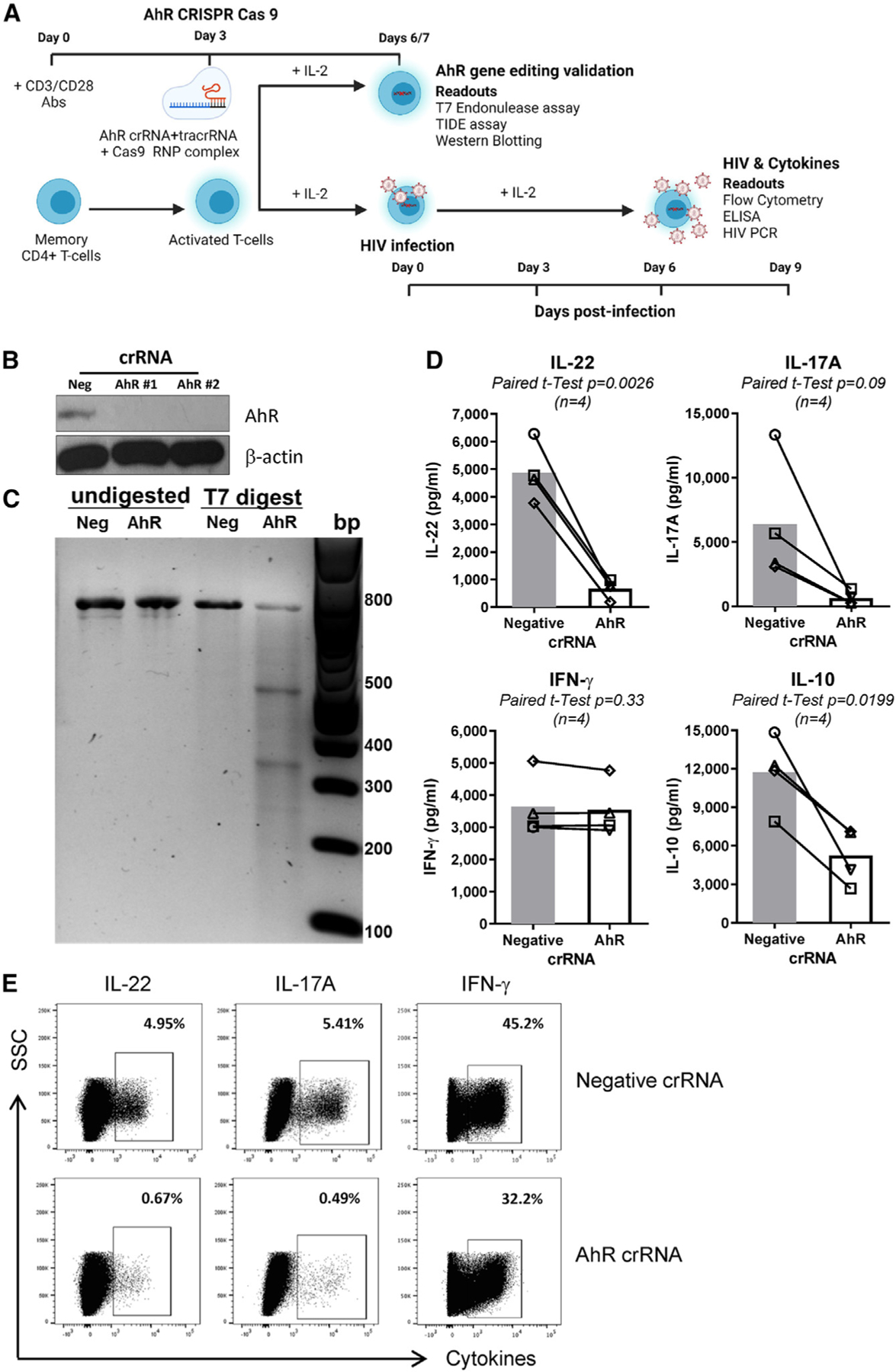
(A) Shown is the experimental flow chart of CRISPR-Cas9 ribonucleoprotein (RNP)-mediated AhR KO. Briefly, memory CD4+ T cells, isolated from peripheral blood mononuclear cells (PBMCs) of HIV-uninfected individuals, were stimulated with CD3/CD28 Abs for 3 days. Cells were electroporated with AhR-targeting (AhR crRNA) or control crRNA (negative crRNA) in the presence of the trans-activating CRISPR RNA (tracrRNA).
(B and C) 3/4 days after electroporation, cells were analyzed by western blotting (B) and the T7 endonuclease assay (C) to determine the efficacy of CRISPR-Cas9-mediated AhR gene editing at protein and DNA levels, respectively.
(D and E) 3 days after electroporation, cells were exposed to HIVTHRO for 3 h, washed, and cultured for 9 days with rhIL-2 (5 ng/mL). At day 9 post-infection, supernatants were collected, and cytokine levels were quantified by ELISA (D). In parallel, cells were stimulated with PMA (50 ng/mL) and ionomycin (1 μg/mL) for 2 h, followed by the addition of brefeldin A (2 μg/mL), and cells were incubated for another 4 h; the intracellular expression of cytokines was measured by flow cytometry (E).
Experiments in (B) and (C) and (D) and (E) were performed with cells from n = 2 and 4 HIV-uninfected individuals, respectively (donors 1–4). Paired t test p values are indicated in (D). Shown are bars that indicate mean values (D).
To explore the AhR-dependent modulation of HIV-1 permissiveness, memory CD4+ T cells from HIV-uninfected individuals were electroporated with AhR-crRNA and negative crRNA and 3 days later were exposed to the transmitted/founder HIV-1 THRO strain (HIVTHRO42). The capacity of AhR to modulate IL-22, IL-17A,28,29,31,32 and IL-1033 production is well established. Consistently, levels of IL-22, IL-17A, and IL-10, but not interferon γ (IFN-γ), were strongly downregulated in cell-culture supernatants of AhR-crRNA compared with negative crRNA electroporated CD4+ T cells at day 9 post-infection (Figure 1D). Similarly, AhR-crRNA compared with negative crRNA electroporated CD4+ T cells expressed lower levels of IL-22 and IL-17A, but similar levels of IFN-γ, as measured by intracellular staining (ICS) and flow cytometry upon PMA/ionomycin stimulation (Figure 1E). These results are indicative that CRISPR-Cas9-mediated AhR gene editing affected AhR-specific functional features of CD4+ T cells.
Of particular importance, CRISPR-Cas9-mediated AhR KO significantly increased replication of HIVTHRO (Figure 2), as observed by measuring levels of HIV-DNA integration at day 3 post-infection (Figure 2A) and HIV-p24 levels in cell-culture supernatants at days 3, 6, and 9 post-infection (Figures 2B and 2C). Also, there was a significant increase in intracellular HIV-p24 expression (Figures 2D and 2E), despite a slight decrease in cell viability at day 9 post-infection (Figure 2F). A similar increase in HIV-DNA integration was observed when AhR silencing was induced in TCR-activated CD4+ T cells with AhR (siAhR) versus non-targeting small interfering RNA (siNT) upon exposure to the replication-competent CCR5-tropic HIVNL4.3BaL strain (Figure S4). Thus, AhR acts as a negative regulator of HIV-1 replication in CD4+ T cells.
Figure 2. CRISPR-Cas9-mediated AhR gene editing boosts HIV-1 replication in memory CD4+ T cells.
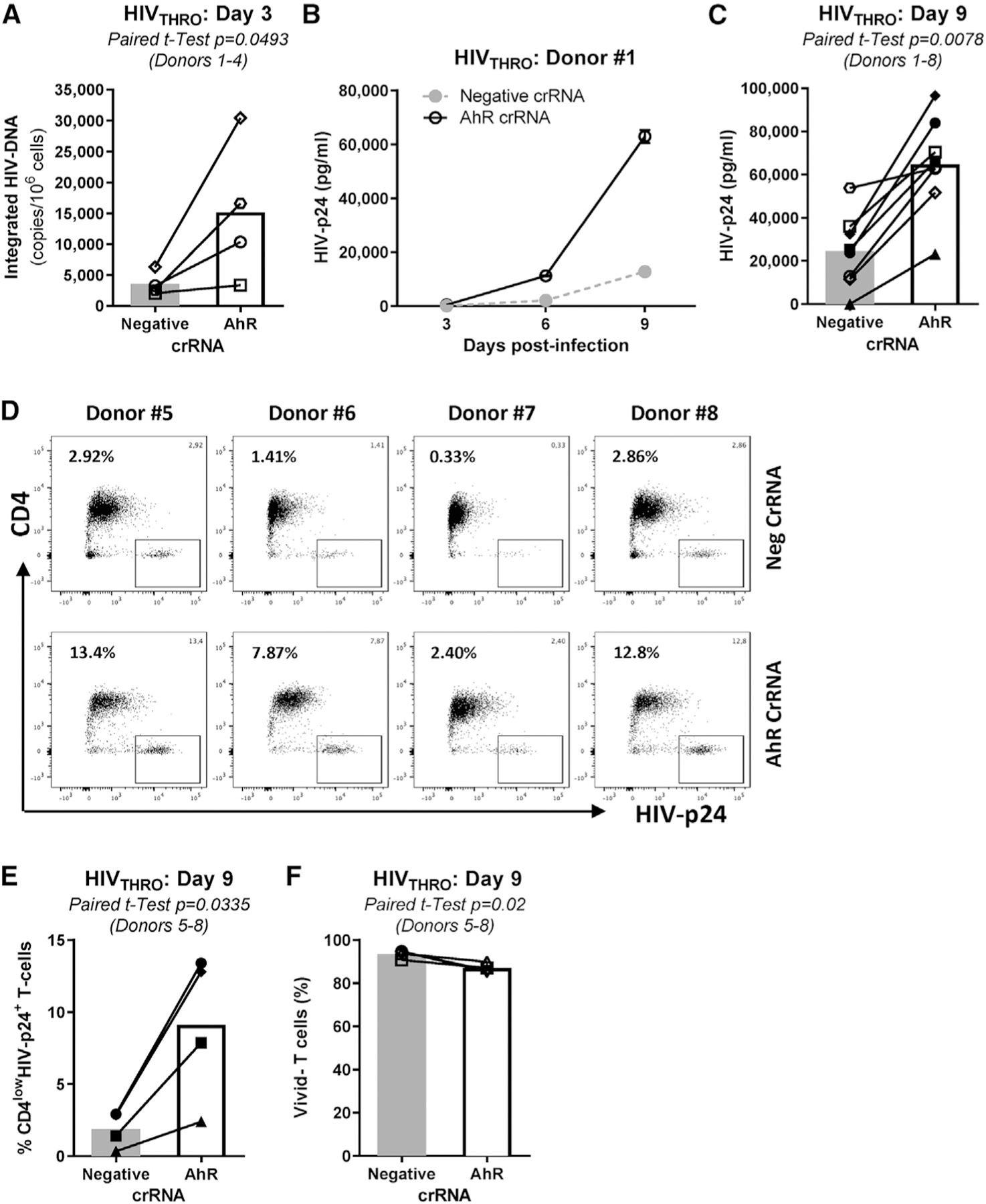
The experimental flow chart in Figure 1A was used to investigate the effect of CRISPR-Cas9-mediated AhR gene editing on HIV-1 replication. Briefly, TCR-activated memory CD4+ T cells were electroporated with AhR crRNA or negative crRNA in the presence of the tracrRNA. 3 days after electroporation, cells were exposed to HIVTHRO for 3 h, washed, and cultured for 9 days in the presence of rhIL-2 (5 ng/mL).
(A–C) Shown are cell-associated integrated HIV-DNA levels at day 3 post-infection (n = 4, donors 1–4) (A) and HIV-p24 levels in cell-culture supernatants at days 3, 6, and 9 post-infection in one representative donor (B) and statistical analysis of HIV-1 replication at day 9 post-infection in donors 1–8 (C). Finally, cells were harvested at day 9 post-infection and stained on the surface with CD4 Abs and intracellularly with HIV-p24 Abs.
(D and E) Shown are the effects of AhR gene editing on the frequency of CD4lowHIV-p24+ T cells in each individual donor (D) and statistical analysis in donors 5–8 (E).
(F) The viability dye Aqua Vivid was used to determine cell viability (Vivid−) at day 9 post-infection in donors 5–8.
Paired t test p values are indicated in (A), (C), (E), and (F). Shown are bars that indicate mean values (A), (C), (E), and (F).
Pharmacological AhR triggering/blockade modulates IL-22, IL-17A, and IL-10 production in memory CD4+ T cells
To explore the effects of ligand-mediated AhR triggering/blockade on Th lineage cytokine production, we used the AhR agonist 6-formylindolo [3,2-b] carbazole (FICZ), a photooxidation product of tryptophan,43 and the AhR antagonist CH223191, a chemical inhibitor that blocks binding of ligands to AhR.44 As expected, the activation of memory CD4+ T cells via the TCR in the presence of FICZ led to a >10-fold increase in CYP1A1 mRNA expression compared with DMSO, while exposure to CH223191 decreased CYP1A1 mRNA expression (Figure S5). Subsequently, dose-response experiments demonstrated that CH223191 decreased IL-22, IL-17A, and IL-10, but not IFN-γ, production (Figures S6A–S6D, left panels) at 5 and 10 μM. At the opposite end, FICZ increased IL-22 and IL-10, but not IL-17A and IFN-γ, production at 100 nM (Figures S6A–S6D, right panels). At the concentrations tested, CH223191 and FICZ did not affect cell viability nor cell proliferation estimated by the expression of Ki67 (Figures S7A and S7B), a surrogate marker of the cell cycle S phase, thus excluding the cytotoxic effects of these drugs.
Pharmacological AhR blockade facilitates HIV-1 replication at post-entry steps, between reverse transcription and integration
To explore the effects of ligand-mediated AhR triggering/blockade on HIV-1 replication, TCR-activated memory CD4+ T cells were cultured in the presence/absence of optimal CH223191 (10 μM) and FICZ (100 nM) doses. Results revealed that AhR blockade strongly promoted HIV-1 replication compared with DMSO, while minor changes were observed upon exposure to FICZ (Figures 3A and 3B). Of note, a CH223191 dose-response increase in HIV-DNA levels was observed upon exposure to HIVTHRO, but no changes were observed with FICZ (Figures S8A and S8B). Biologically active doses of CH223191 and FICZ did not affect the expression of the HIV-1 receptor CD4, nor the coreceptors CCR5/CXCR4 (Figures S9A–S9D), indicative of post-entry mechanisms. Nevertheless, CH223191 significantly downregulated ITGB7 expression (Figures S9A and S9E). Consistent with the documented antiviral role of IL-2245 and IL-10,46,47 recombinant IL-22 and/or IL-10 partially or completely counteracted the effect of CH223191 on HIV-1 replication in CD4+ T cells (Figure S10). Together, these results provide evidence that AhR blockade boosts HIV-1 replication in CD4+ T cells via mechanisms independent of CD4/CCR5/CXCR4 expression but likely linked to the reduced production of the antiviral cytokines IL-22 and IL-10.
Figure 3. The AhR antagonism increases HIV-1 replication in CD4+ T cells at post-entry levels, between reverse transcription and integration.
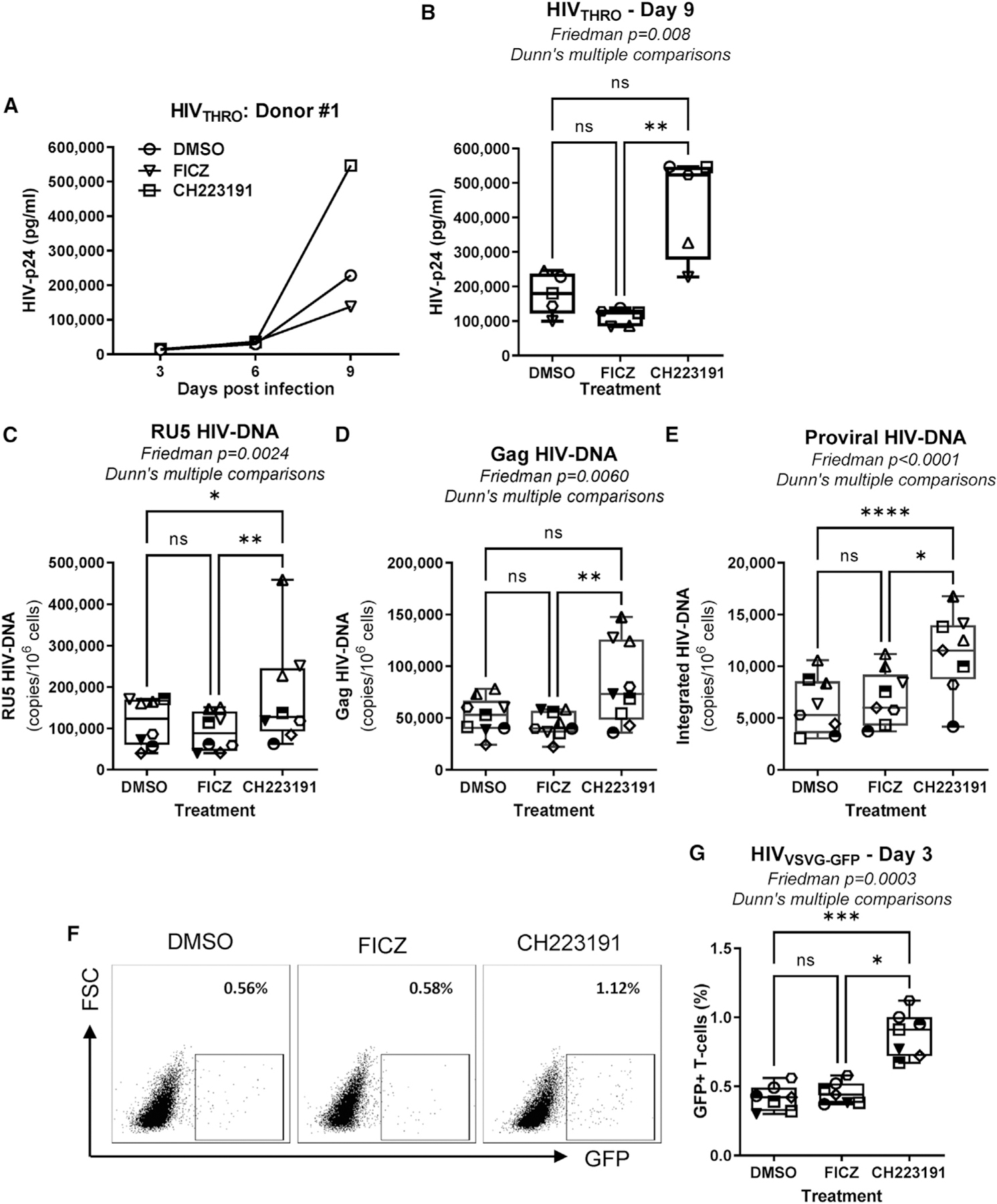
Memory CD4+ T cells isolated from PBMCs of HIV-uninfected individuals were activated via CD3/CD28 and cultured in the presence/absence of the AhR antagonist CH223191 (10 μM) or agonist FICZ (100 nM) for 3 days. Cells were exposed to transmitted founder (T/F) HIVTHRO (50 ng HIV-p24/106 cells) and cultured for up to 9 days with rhIL-2 in the presence/absence of CH223191 or FICZ.
(A and B) Shown are HIV-p24 levels quantified by ELISA in cell-culture supernatants collected at days 3, 6, and 9 post-infection in one representative donor (A) and statistical analysis of results obtained using cells from n = 5 donors at day 9 post-infection (B). In parallel, cells were exposed to single-round VSV-G-pseudotyped HIV-1 encoding gfp in place of nef (HIVVSVG-GFP) (50 ng HIV-p24/106 cells) and cultured for 3 days in the presence/absence of CH223191 or FICZ.
(C–E) Levels of early (RU5) (C) and late (Gag) reverse transcripts (D), as well as integrated HIV-DNA levels (E), were quantified by real-time nested PCR at day 3 post-infection.
(F and G) The GFP expression was measured by flow cytometry as an indicator of HIV-1 transcription/translation. Shown is the frequency of GFP+ T cells in one representative donor (F) and statistical analysis of GFP expression in T cells from n = 7 donors (G). Each symbol represents one donor.
(B–E and G) The Friedman test p values and Dunn’s multiple comparison significance are indicated on the graphs (*p < 0.05; **p < 0.01; ***p < 0.001). Shown are boxes and whisker plots, with minimum to maximum values.
To determine at which step of the viral replication cycle AhR ligands acted, TCR-activated memory CD4+ T cells were exposed to single-round vesicular stomatitis virus (VSV)-G-pseudotyped GFP-expressing HIV-1 (HIVVSVG-GFP), entering cells via the low-density lipoprotein (LDL) receptor,48 independently of CD4 and coreceptors. CH223191 significantly increased cell-associated levels of early (RU5) and late (Gag) reverse transcripts and integrated HIV-DNA when compared with DMSO or FICZ (Figures 3C–3E). Consistently, increased levels of GFP expression, indicative of efficient HIV-1 transcription, were observed upon AhR blockade (Figures 3F and 3G), while FICZ did not exert a significant effect on HIV-DNA levels or HIV-1 transcription when compared with DMSO (Figures 3C–3G), likely due to the activation of the AhR pathway by ligands present in the culture media.31 Thus, pharmacological AhR blockade facilitates HIV-1 replication at early/late post-entry reverse transcription levels, subsequently leading to efficient viral integration, transcription, and translation.
Pharmacological AhR triggering/blockade modulates IL-22/IL-17A production and HIV-1 replication in memory CCR6+CD4+ T cells
Among memory CD4+ T cells, IL-17A and IL-22 are mainly produced by cells expressing the surface marker CCR6.28,34,49 Our group and others reported that CCR6+ T cells are major targets for HIV-1 infection.8,9,12,19 Therefore, we sought to determine whether AhR modulates HIV-1 replication in CCR6+ T cells. Fluorescence-activating cell sorted (FACS) memory CCR6+ and CCR6− T cells (Figure S11) were stimulated via CD3/CD28 in the presence/absence of FICZ or CH223191 and exposed to HIVTHRO. At day 3 post-TCR triggering, CCR6+ versus CCR6− T cells preferentially produced IL-22 and IL-17A (Figures 4A and 4B). CH223191 significantly decreased IL-22 and IL-17A production in CCR6+ T cells (Figures 4C and 4D). Finally, the highest levels of HIV replication occurred in CCR6+ versus CCR6− T cells, with CH223191 robustly increasing HIVTHRO replication in CCR6+, but not CCR6−, T cells (Figures 4E and 4F). Collectively, these results demonstrate that the pharmacological AhR blockade facilitates HIV-1 replication in CCR6+ T cells while downregulating IL-22 and IL-17A production.
Figure 4. The AhR blockade increases HIV-1 replication in CCR6+CD4+ T cells.
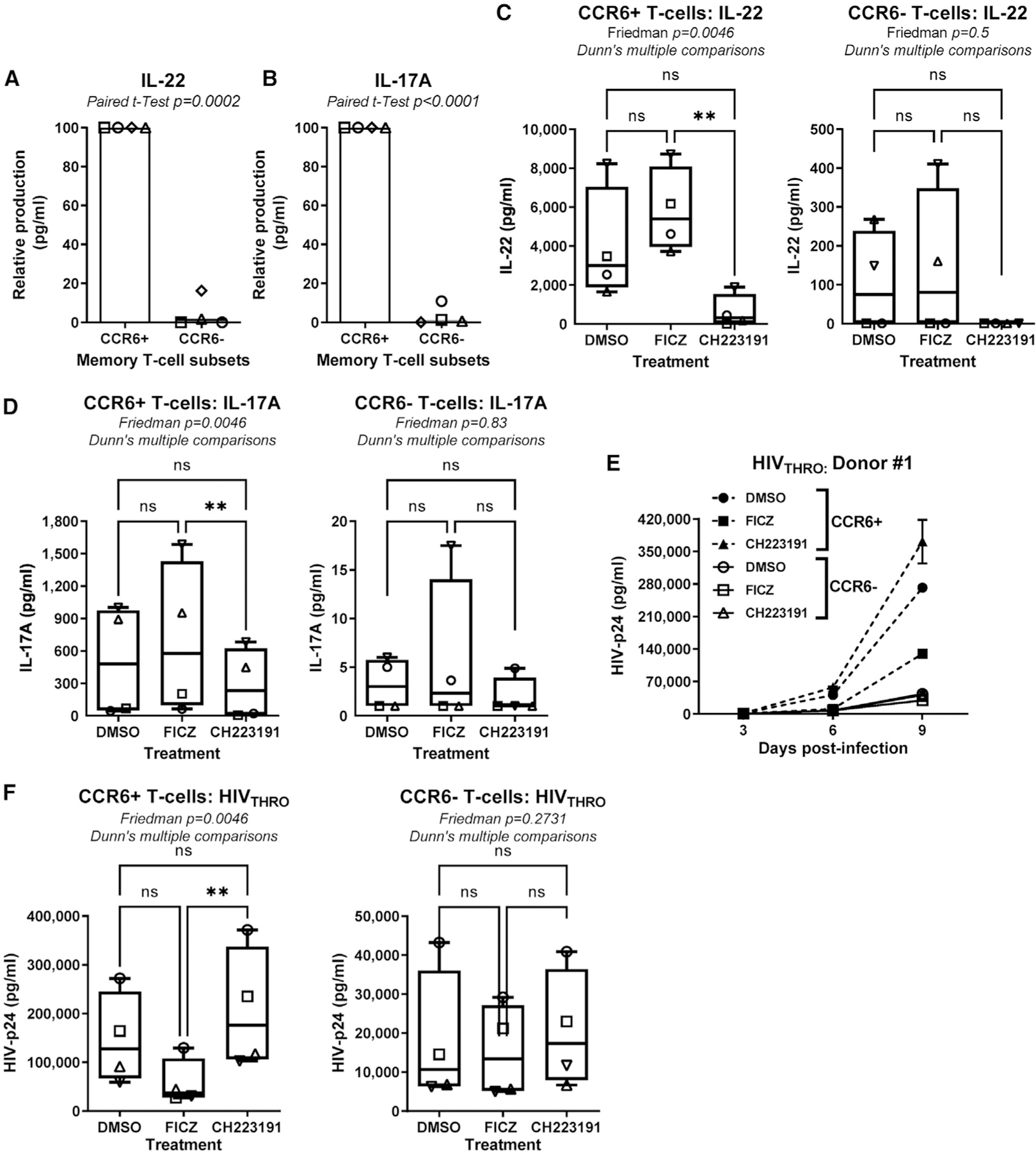
Flow cytometry-sorted memory CCR6+ and CCR6−CD4+ T cells isolated from PBMCs of HIV-uninfected individuals were stimulated via CD3/CD28 Abs in the presence/absence of the FICZ (100 nM) or CH223191 (10 μM) for 3 days. Cells were exposed to T/F HIVTHRO (50 ng HIV-p24/106 cells) and cultured with rhIL-2 in the presence/absence of AhR drugs.
(A–D) Levels of IL-22 (A and C) and IL-17A (B and D) were measured by ELISA in cell-culture supernatants of CCR6+ versus CCR6− T cells (A and B; relative cytokine production) treated or not with FICZ or CH223191 (C and D; absolute cytokine levels) at day 3 post-TCR triggering (n = 4).
(E and F) Shown are HIV-p24 levels quantified by ELISA in cell-culture supernatant collected at day 3, 6, and 9 post-infection in one representative donor (E) and statistical analysis of results obtained using cells from n = 4 different HIV-uninfected donors (F, CCR6+ T cells, left panel; CCR6− T cells, right panel). Each symbol represents one donor.
Paired t test p values (A and B) and Friedman test p values and Dunn’s multiple comparison significance (C, D, and F) are indicated on the graphs (*p < 0.05; **p < 0.01; ***p < 0.001). Shown are bars that indicate mean values (A–B) and boxes and whisker plots, with minimum to maximum values (C–D ) and (F).
Pharmacological AhR blockade boosts HIV-1 outgrowth in memory CD4+ T cells of ART-treated PLWH
To explore the effect of AhR pathway on HIV-1 outgrowth, we performed a simplified viral outgrowth assay (VOA) previously set up by our group18,50 using the experimental procedure depicted in Figure 5A. In preliminary experiments, we demonstrated that TCR triggering in memory CD4+ T cells of ART-treated PLWH (Table S1) resulted in the upregulation of CYP1A1 mRNA (Figure S12), indicative of AhR pathway activation. The AhR antagonist CH223191 significantly increased the frequency of HIV-p24+ T cells (Figures 5B and 5C) and soluble HIV-p24 levels (Figures 5D and S13) without differently affecting cell viability (Figure 5E). At the opposite end, the AhR agonist FICZ showed only moderate effects on blocking HIV-1 outgrowth (Figures 5B–5D), in line with the possibility that the AhR pathway is already activated in cells from ART-treated PLWH. These findings show that AhR blockade facilitates viral outgrowth from CD4+ T cells of ART-treated PLWH.
Figure 5. The AhR blockade boosts HIV-1 outgrowth in CD4+ T cells of ART-treated PLWH.
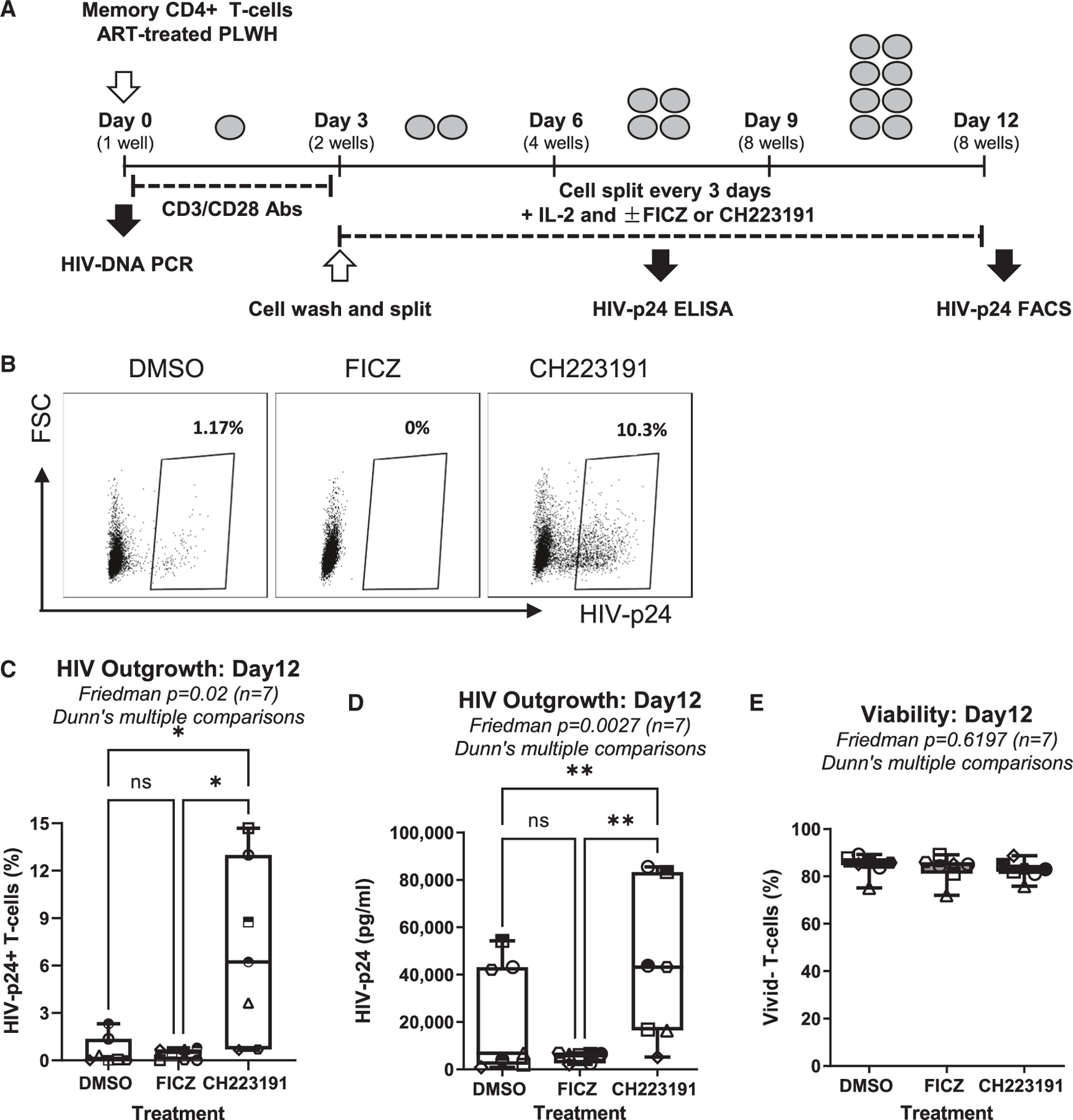
(A) Shown is the flow chart of the viral outgrowth assay (VOA). Briefly, memory CD4+ T cells isolated from the PBMCs of ART-treated PLWH were activated with CD3/CD28 Abs and cultured with rhIL-2 in the presence/absence of FICZ (100 nM) or CH223191 (10 μM). At day 12 post-infection, HIV-1 replication and cell viability were measured.
(B–E) Shown are the percentages of HIV-p24+ T cells (8 splitting replicates pooled per condition) from one representative donor (ART #2; Table S1) (B), the statistical analysis of the percentage of HIV-p24+ cells (C), the HIV-p24 levels in cell-culture supernatants (median values of HIV-p24 levels from 8 splitting replicates per condition) (D), and the viability of cells harvested at day 12 of VOA (E) in experiments performed with cells from n = 7 donors. Friedman test p values with Dunn’s multiple comparisons indicated on the graphs (*p < 0.05; **p < 0.01; ***p < 0.001).
Shown are boxes and whisker plots, with minimum to maximum values (C–E).
Genome-wide transcriptional profiling identifies molecular mechanisms by which AhR blockade boosts HIV-1 outgrowth in CD4+ T cells of ART-treated PLWH
To get insights into AhR-dependent regulatory mechanisms, RNA sequencing using the Illumina technology was performed on TCR-activated memory CD4+ T cells of ART-treated PLWH cultured in the presence/absence of CH223191 (Figure 6A). Differentially expressed genes (DEGs) were identified based on p values, adjusted p values, and fold change (FC) ratios (Data S1 and S2) and illustrated in the volcano plot (Figure 6B). Upregulated DEGs (n = 204) included transcripts encoding for cell surface markers, cytokines, and transcriptional regulators previously linked to T cell activation and HIV permissiveness (e.g., CCR3, IL6, CD38, CD276/B7-H3, BACH2, CD274/B7-H1/PD-L1, LAG3, IL6R, CXCL10, LEF1, TBX21). Downregulated DEGs (n = 258) included known AhR transcriptional targets/regulators (e.g., CYP1A1, CYP1B1, AHRR, TIPARP), Th17/Th22 cytokines (e.g., IL22, IL17F, IL26, IL24, IL1A), cell residency markers (e.g., ITGB7, S1PR1, CXCR3, CXCR5, CXCR6), and the HIV repressor HIC1 (hypermethylated in cancer 1)51 (Data S1 and S2). The downregulation of CYP1A1, CYP1B1, and AHRR (AhR repressors) as well as ITGB7 by CH223191 validates the AhR specificity of this antagonist (Data S2).
Figure 6. Transcriptional reprogramming of CD4+ T cells upon pharmacological AhR inhibition.
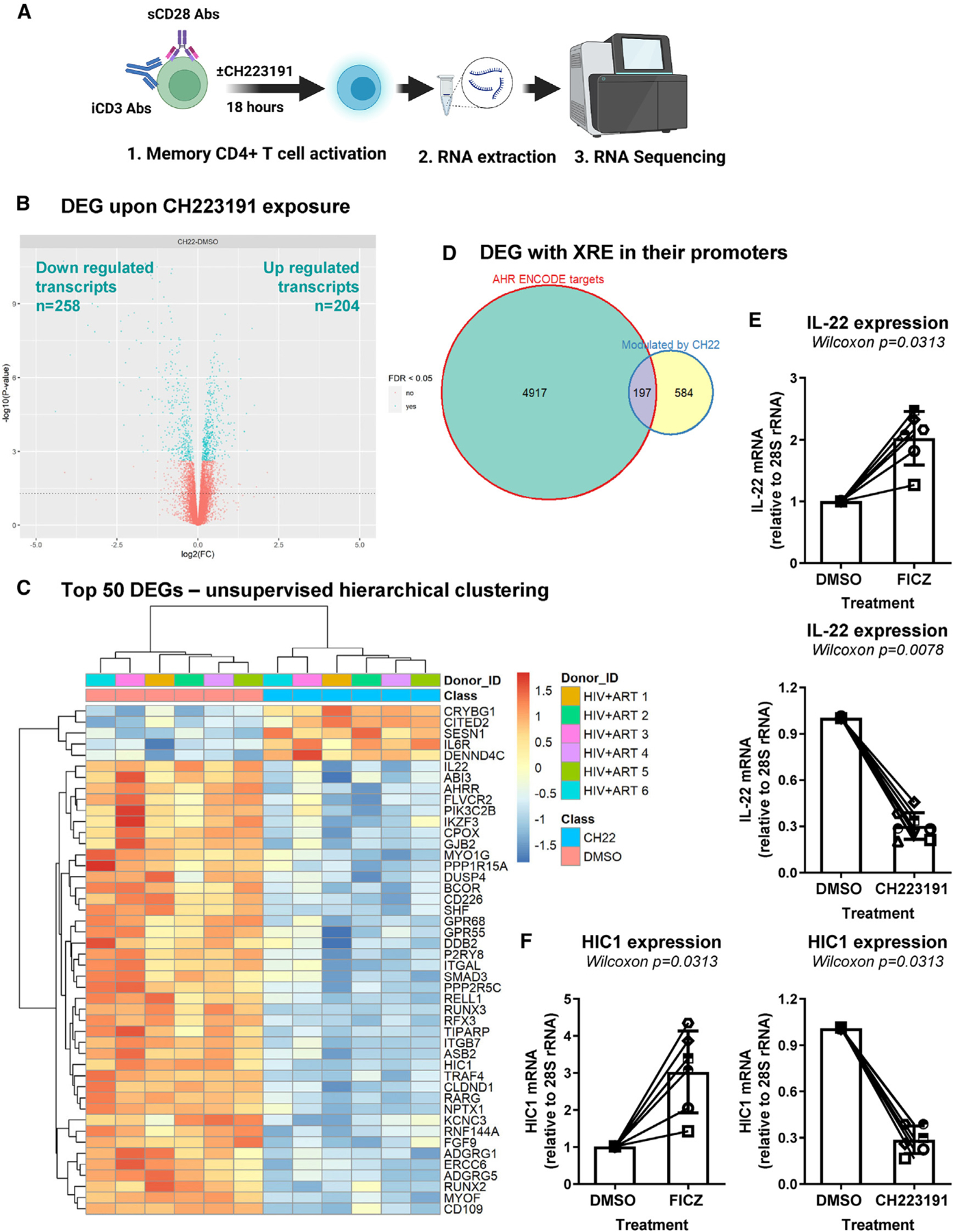
(A) Shown is the flow chart of RNA sequencing. Briefly, memory CD4+ T cells of ART-treated PLWH (n = 6) were isolated and stimulated with CD3/CD28 Abs and cultured in the presence/absence of CH223191 (10 μM) for 18 h. Total RNA was extracted and used for RNA sequencing (Illumina Technology).
(B) Volcano plots for all probes in each linear model presented with the log2 FC on the x axis and the negative logarithm of the adjusted p values for false discovery rate (FDR) on the y axis. The red/blue color code is used based on the 5% FDR threshold.
(C–F) Shown are the representation of AhR direct target genes modulated by CH223191, as identified using the ENCODE Bioinformatic tool (https://www.encodeproject.org) (C); top differentially expressed genes (DEGs; identified based on p values) in CD4+ T cells exposed or not to CH223191 (D); IL-22 mRNA levels quantified by qPCR in CD4+ T cells exposed or not to FICZ or CH223191 (n = 6) (E); and HIC1 mRNA levels in CD4+ T cells exposed or not to FICZ or CH223191 (F).
(E and F) Each symbol represents an individual donor; bars indicate median values. Wilcoxon matched-pairs signed rank test p values are indicated on the graphs. Shown are bars that indicate mean ± SD values.
Encyclopedia of DNA elements (ENCODE)
The top 50 modulated transcripts (e.g., IL22, AHRR, ITGB7, HIC1) are depicted in Figure 6C (Data S1 and S2). Among genes modulated by CH223191, a fraction of them (197 out of 584 transcripts) represent putative AhR transcriptional targets, as suggested by in silico analyses of the transcriptional promoter of these genes for the presence of putative DRE/XRE using the Bioinformatics tool ENCODE (https://www.encodeproject.org) (Figure 6D). The CH223191-modulated genes with DRE/XRE in their promoters include transcripts upregulated (e.g., OASL, CCND1/cyclin D1, IL6R, BATF3, SGK1, RORA, IFI35, LGALS3BP, CEBPB, GADD45A, SIRT1) and downregulated (e.g., CYP1A1, SMAD3, TRAF4, IKZF3, IL1R1, RPTOR, IL32) (Data S3). The downregulation of IL-22 mRNA upon CH223191 exposure (Figure 6C) and its increase by FICZ were confirmed by RT-PCR (Figure 6E), thus validating the functional activity of AhR drugs in these experimental settings.
The zinc-finger transcriptional repressor HIC1,52 reported as an HIV transcription repressor,51 was one of the top downregulated cellular transcripts upon exposure to CH223191 (Figure 6C; Data S2). Consistent with the RNA sequencing (RNA-seq) data, real-time RT-PCR quantification of HIC1 mRNA demonstrated that exposure to CH223191 significantly decreased, while exposure to FICZ robustly upregulated HIC1 mRNA expression (Figure 6F).
Gene set variation analysis (GSVA)
To extract additional meaning from these RNA-seq results, GSVA was performed using the UC San Diego/Broad Institute Molecular Signature Database (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb). This allowed the identification of top modulated canonical pathways in response to CH223191-exposed cells, including pathways upregulated (e.g., Th1 differentiation, IFN-α response, IFN-γ response, Toll-like receptor 3 signaling, Toll-like receptor 4 signaling, negative regulation of IL-2 production, Toll-like receptor 9 signaling, positive regulation of IL-17 production, αβ T cell proliferation, and positive regulation of T cell proliferation) and downregulated (e.g., zinc ion homeostasis) (Figures S14A and S14B). The modulation of these pathways may be either the direct effect of AhR-mediated transcriptional regulation and/or the consequence of HIV-1 sensing in the VOA.
NCBI HIV-1 interactor
Further, data mining was performed by interrogating the NCBI HIV-1 interactor database (Figure S15). Among top HIV-1 interactors, upregulated transcripts included LGALS1, CD4, CCR3, DPP4, LGALS3BP, CEBPB, CD38, NFKBIA, PTK2B, HERC5, BACH2, SGK1, IL6, NOTCH2, SIRT1, CD28, and LDLR, while top downregulated transcripts included LCK, ITGB7, CD226, ITGAL, IKZF3, IL1A, GPR15, ITGA4, CXCR6, and CXCR3 (Figure S15). Real-time validations were performed for SIRT1, CEBPB (CCAAT/enhancer-binding protein β), and BACH2 mRNA, transcripts included on the list of putative direct AhR targets (Data S3). While CH223191 exerted minor effects, FICZ significantly and slightly decreased SIRT1 and CEBPB mRNA, respectively (Figures S16A and S16B). Finally, BACH2 mRNA expression significantly increased and decreased upon exposure to CH223191 and FICZ, respectively (Figure S16C).
Together, these results revealed the transcriptional reprogramming associated with AhR blockade in CD4+ T cells of ART-treated PLWH and identified HIC1, SIRT1, CEBPB, and BACH2 as putative AhR transcriptional targets and HIV interactors.
Identification of HIC-1 as a direct target of the AhR pathway
We previously reported that HIC-1 inhibits Tat-dependent HIV-1 gene transcription.51 To determine whether AhR directly regulates HIC1 transcription, an in silico search was performed with three other databases (i.e., GeneCards, a human genome database, the Eukaryotic Promoter Database, and Jaspar) in an effort to identify putative DREs/XREs in the HIC1 promoter. Precisely, we used in our Jaspar in silico analysis the AhR position frequency matrix (PFM) of the Jaspar Vertebrate 2022 library (MA0006.1) on both strands of the HIC1 promoter.53 The presence of four putative AhR-binding sites were identified, located at positions (nt −792 to −798), (nt −232 to −238), (nt +785 to +791), and (nt +902 to 908) (where nt +1 corresponds to the first nucleotide of the HIC1 transcription start site [TSS]) and were referred to as AhR#1, AhR#2, AhR#3, and AhR#4, respectively (Figure 7A; Data S4). To determine whether the AhR, together with its partner ARNT, could be recruited in vivo to the HIC1 promoter, chromatin immunoprecipitation followed by quantitative PCR assay (ChIP-qPCR) was performed using two cell lines (STAR Methods). Importantly, our results showed the in vivo recruitment of both AhR and ARNT using three primer sets, covering the chromatin regions comprising the four potential AhR-binding sites (AhR#1, AhR#2, AhR#3, and AhR#4) of the HIC1 promoter (Figures 7B and 7C). The mmp9 cellular gene, known to recruit in vivo AhR/ARNT to its promoter,54 was used as a positive control (Figures 7B and 7C). Altogether, our results demonstrate that the HIC1 promoter is a direct target for AhR/ARNT in vivo and support that the HIC1 promoter contains four distinct binding sites for the AhR/ARNT complex. Thus, HIC1 may be added to the list of direct AhR gene targets.
Figure 7. The AhR/ARNT complex is recruited in vivo to the HIC1 promoter.
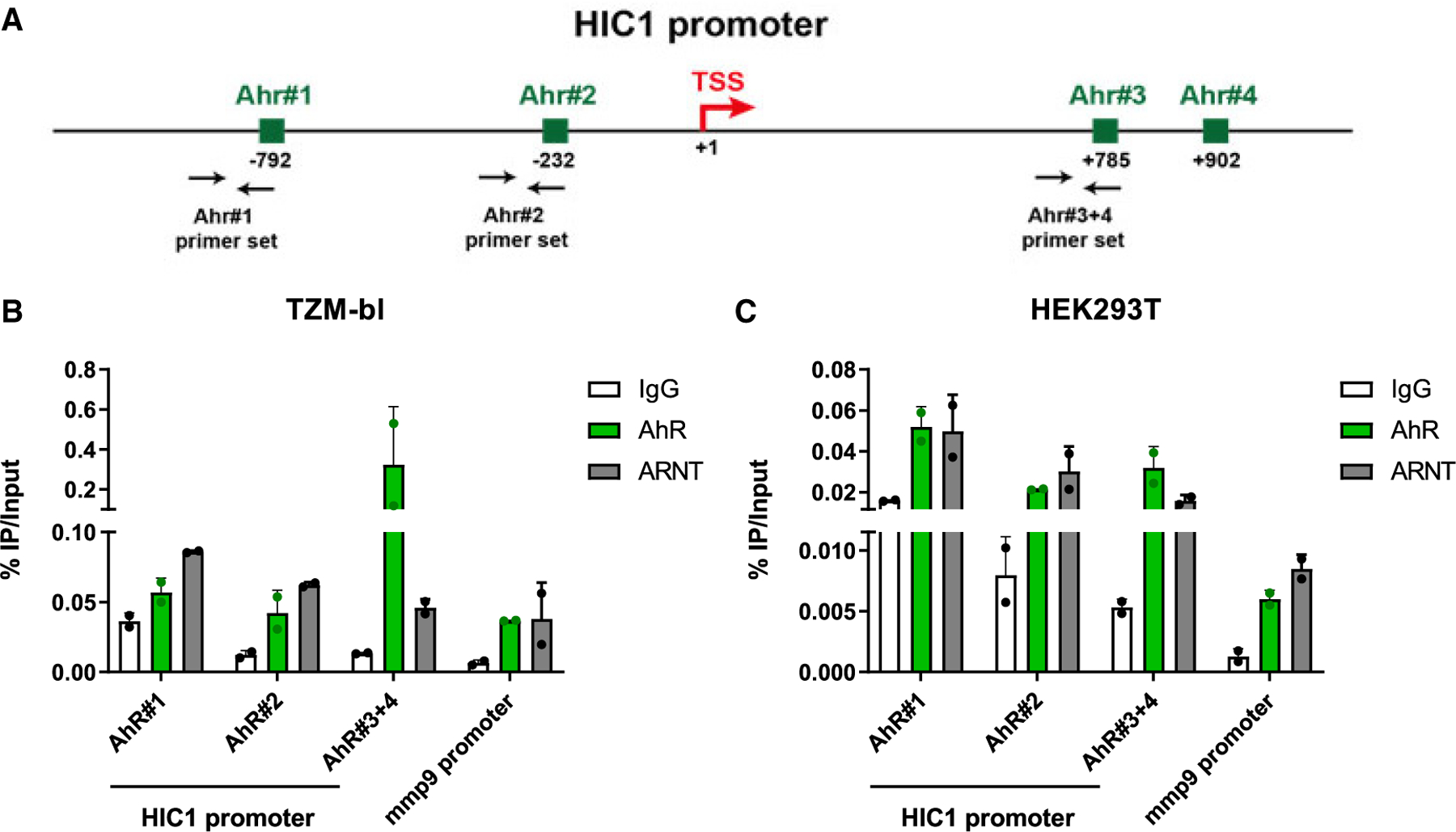
(A) Schematic representation of the HIC1 promoter, with the localization of the potential AhR-/ARNT-binding sites identified by in silico analysis (using the Jaspar 2022 consensus motif) and with the localization of the ChIP-qPCR primer sets.
(B and C) Chromatin prepared from TZM-bl (B) or HEK293T (C) cells was immunoprecipitated using specific Abs directed against AhR and ARNT or using an immunoglobulin G (IgG) as background measurement. Results are presented as histograms indicating percentages of immunoprecipitated DNA compared with the input DNA (% IP/input). Data are the means ± SD from one experiment representative of four independent experiments. Shown are bars that indicate mean ± SD values.
DISCUSSION
In this article, we provide evidence that AhR expression/activation induced by TCR triggering limits HIV-1 infection/outgrowth in CD4+ T cells. Briefly, the genetic/pharmacological inhibition of AhR increased HIV-1 replication in vitro and enhanced viral outgrowth from CD4+ T cells of ART-treated PLWH. RNA-seq revealed the transcriptional reprogramming associated with increased viral outgrowth prompted by AhR blockade, with in silico analyses identifying putative AhR target genes listed on the NCBI HIV-1 Interactor Database. Among AhR-modulated genes, ChIP experiments identified HIC1, an HIV-1 transcriptional repressor51 and modulator of intestinal homeostasis and T cell tissue residency,55,56 as a direct AhR target. These results support a model in which AhR activation favors the gut homing/residency via the induction of ITGB7 and CXCR6 expression and fuels the persistence of “silent” HIV reservoirs in CD4+ T cells of ART-treated PLWH. At the opposite end, pharmacological AhR blockade facilitates viral outgrowth and, by interfering with tissue residency, likely promotes the recirculation of “reactivated” reservoir cells from deep tissues into the blood stream.
One important finding of our study is the demonstration that AhR expression at RNA/protein levels, along with its activity (CYP1A1 transcription), are upregulated upon TCR triggering. We demonstrated that the AhR pathway controls HIV-1 replication at post-entry levels, with no changes in the expression of the HIV-1 receptor CD4 and coreceptors CCR5 and CXCR4 upon AhR activation/blockade. In fact, CH223191-mediated AhR blockade increased the efficacy of early/late HIV-1 reverse transcription, subsequently favoring HIV-DNA integration and de novo viral protein expression. Finally, CH223191 strongly boosted HIV-1 outgrowth from CD4+ T cells of ART-treated PLWH. Our results are consistent with previous studies by Kueck et al. reporting an antiviral role for AhR in macrophages.37 In the latter study, AhR activation restricted productive HIV-1 replication via the regulation of cyclin-dependent kinase (CDK)1/2 expression and SAMHD1-mediated restriction of reverse transcription.37 Although our RNA-seq results did not reveal the modulation of CDK1/2 nor SAMHD1 mRNA expression in CD4+ T cells, transcripts encoding for cyclin D1 (CCND1) were upregulated by CH223191. The involvement of AhR in controlling the dNTP levels via SAMHD1 phosphorylation/dephosphorylation in CD4+ T cells remains a possibility to be tested.
The proviral effects of CH223191 were mainly observed in Th17/Th22-polarized CCR6+ T cells, consistent the preferential expression of AhR in Th17 cells.28,31 AhR regulates the expression of multiple cytochrome P450 transcripts like CYP1A1 and CYP1B1,26,27 as well as cytokines, including IL-22, IL-17A, and IL-10 in CD4+ T cells.20,21,28,31,35 While IL-17A exhibits proviral features,57 the antiviral effects of IL-22 and IL-10 are documented,45,46 with IL-10 contributing to VR persistence in simian immunodeficiency virus (SIV) infection models.47 The downregulation of IL-22 and IL-10 by CH223191 may explain the increased viral outgrowth observed in CD4+ T cells of ART-treated PLWH. Consistent with AhR expression by “non-pathogenic” Th17 cells, our RNA-seq results revealed that the pharmacological inhibition of AhR increased the expression of the “pathogenic” Th17 markers IL23R,20 SGK1,58 and RORA.59
Of particular importance, our results revealed that, similar to its effect in macrophages,36 AhR upregulates the expression of the gut-homing molecule ITGB7 in CD4+ T cells. Thus, the transcriptional program governed by AhR may imprint CD4+ T cells with a gut-homing tropism. In addition to ITGB7, other transcripts for chemokines, chemokine receptors, and adhesion molecules involved in tissue-specific trafficking were upregulated (CCR3, CXCL13, CXCL11, CXCL10) or downregulated (GPR15, ITGA1, CD226, ITGA2B, S1PR1, ITGAL, ITGA4, CXCR6, CXCR3, CXCR5) by CH223191. GPR15 mediates gut homing under the control of AhR,60 and S1PR1 is a master regulator of T cell egress from the lymph nodes,61 while CCR3 was previously documented by our group as preferentially expressed on HIV-infected cells in vitro.62 Of particular notice, HIC1, induced by FICZ and decreased by CH223191 in CD4+ T cells of ART-treated PLWH, represents a key regulator in intestinal homeostasis55 and was recently reported to promote T cell tissue residency.56 Importantly, by in silico analysis, we here identified four putative AhR-/ARNT-binding sites in the HIC1 promoter, and ChIP-qPCR experiments demonstrated that the AhR/ARNT complex was recruited in vivo to the HIC1 promoter in both TZM-bl and HEK293T cell lines. These results originally demonstrate that HIC1 is a direct AhR target. Future studies should explore the role of the AhR-HIC1 interplay in controlling the tissue residency in the gut, mainly during HIV-1 infection, a condition associated with increased expression of AhR ligands.63,64 Based on our results, one may anticipate that AhR blockade promotes the recirculation of tissue-residence CD4+ T cells carrying VRs via HIC1-dependent mechanisms.
RNA-seq revealed that CH223191 downregulated the expression of CYP1A1, AHRR, and TIPARP, involved in the negative feedback regulation of AhR signaling.26,27 In addition, we observed the modulation of lineage cytokines important for HIV-1 pathogenesis, including the downregulation of IL1A, IL1R1, IL24, and IL26 and the upregulation of IL27, IL6, IL6R, IL7R, IL18RAP, and IL12RB1 expression. Among these transcripts, the ENCODE search points to the possibility that IL6R and IL1R1 genes are directly regulated by AhR, as they express DREs/XREs in their promoters. Thus, AhR activation may facilitate T cell responsiveness to IL-1α, to the detriment of IL-6. Of particular importance, we report here that genes encoding for immune checkpoint activators/inhibitors are modulated by AhR, as reflected by the upregulation of CD276/B-H3, CD38, CD274/PD-L1, LAG3, and CD59 and the downregulation of CD109, CD160, TIGIT, CD96, and CD27 in the presence of CH223191. Whether these AhR-modulated transcripts represent surface markers for T cells carrying replication-competent VRs remains to be evaluated using single-cell analysis techniques.
In TCR-activated CD4+ T cells of ART-treated PLWH, CH223191 modulated the expression of various sets of genes previously identified as HIV interactors. Among them, HIC1 was documented to inhibit Tat-dependent HIV-1 transcription,51 raising the possibility that HIC1 downregulation is one mechanism by which viral outgrowth was boosted by CH223191. This is consistent with our in silico search and ChIP experiments supporting the fact that HIC1 transcription is directly regulated by AhR. Of note, downregulation of HIC1 coincided with the upregulation of IL-6, a proinflammatory cytokine associated with HIV-1 disease progression65 and HIV-1 replication46 and a negative regulator of HIC1 expression.66 In addition, SIRT1, CEBPB, and BACH2 were also identified as HIV interactors included on the list of putative AhR gene targets. Of note, SIRT1 is a histone deacetylase repressed at the transcriptional level by HIC1 that, in turn, regulates HIC1 activity.51,52 Also, SIRT1 facilitates Tat-dependent HIV-1 transcription67 and represents a therapeutic target also in HIV-1 infection.68 SIRT1 is used as a therapeutic target in cancer.69 CEBPB, is involved in the transcriptional regulation of multiple genes, including HIV-1.70 Finally, BACH2 caught our attention based on reports demonstrating preferential HIV-1 integration in this gene.71–73 These results raise the possibility that CH223191-mediated BACH2 expression may facilitate HIV-1 integration into this gene; alternatively, since HIV-1 preferentially integrates in BACH2,71–73 CH223191-mediated BACH2 transcription may explain the increased viral outgrowth.
The evidence in this article that AhR acts as a barrier to HIV-1 infection/outgrowth in TCR-activated CD4+ T cells is in line with studies by Kueck et al. performed in macrophages.37 In contrast, a report by Zhou et al. demonstrated that exposure to AhR ligands (e.g., kynurenine, FICZ) promoted viral transcription in resting CD4+ T cells of ART-treated PLWH and reported the presence of AhR-binding DREs in the HIV-1 promoter.74 These controversial results raise further questions on potentially different AhR-mediated mechanisms of action in resting versus TCR-activated CD4+ T cells. Also, the capacity of AhR to bind directly to the HIV-1 promoter as an activator versus a repressor may be dependent on the cell activation status and the expression of other HIV-1-dependency factors. These aspects remain to be further investigated.
Altogether, results included in this article support a model in which AhR acts as a barrier to HIV-1 infection/outgrowth in CD4+ T cells and a master regulator of cell trafficking/tissue residency, at least in part by modulating HIC1 expression. AhR activation may play a dual role during HIV-1 infection. On one side, AhR activation may limit HIV-1 acquisition during primary infection in CD4+ T cells. On the other side, AhR triggering potentially maintains “silent” VRs in ART-treated PLWH. This is consistent with the state of deep latency observed in CD4+ T cells residing in the gut.75–77 This model is consistent with the documented abundance of AhR ligands in the GALT,24–26 the abnormal expression of IDO-1 and Trp catabolites in ART-treated PLWH,63,64 and the maintenance of a state of deep HIV-1 latency at the intestinal level.75–78 Finally, in the context where AhR antagonists are currently tested in clinical trials for cancer,38,39 their ability to reactivate/purge VRs in ART-treated PLWH deserves investigations in “shock and kill” strategies.
Limitations of the study
Our study has limitations that we highlight here. Single-round infection with VSV-G-pseudotyped HIV demonstrated a direct action of AhR on the initial infection/outgrowth. However, our study does not distinguish between the effects of AhR on the initial infection/reactivation versus cell-to-cell spreading of infection in vitro. Although we provide evidence that AhR directly modulates HIC1 expression, it remains unclear whether HIC1 represses HIV transcription in CD4+ T cells as we demonstrated for myeloid cells.51 Moreover, the fact that FICZ had limited effects on cytokine production and HIV replication/outgrowth was considered to be due to the presence of other AhR ligands in cell culture media, as previously reported.31 However, natural AhR ligands may be expressed in cells upon TCR triggering and/or HIV exposure; these possibilities require future investigations. Furthermore, our study used only FICZ and CH223191 for pharmacological studies. Different natural and synthetic AhR ligands differ in their biological activities,26 thus prompting the need for further studies to assess the effect of various ligands on HIV replication/outgrowth. Finally, in addition to its role as a ligand-activated TF, AhR exhibits an intrinsic E3 ubiquitin ligase activity that targets the degradation of steroid receptors (e.g., estrogen receptors).79 In our study, we did not differentiate between genomic and non-genomic actions of AhR-mediated functions in CD4+ T cells; also, we did not perform experiments on cells from males and females. Future studies should address these issues in an effort to explain controversies on the role of AhR in antiviral immunity.80
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Petronela Ancuta (petronela.ancuta@umontreal.ca).
Materials availability
This study did not generate unique reagents.
Data and code availability
RNA-seq data have been deposited at Gene Expression Omnibus (GEO) database under accession GSE198078 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE198078) and are publicly available as of the date of publication, as listed in the key resources table. Original Western blot images have been deposited at Mendeley database and are publicly available as of the date of publication (https://data.mendeley.com/datasets/4ry9gxv7tp/1).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Purified NA/LE Mouse Anti-Human CD3 (Clone UCHT1) | BD | Cat# 555329; RRID: AB_395736 |
| Purified NA/LE Mouse Anti-Human CD28 (Clone CD28.2) | BD | Cat# 555725; RRID: AB_396068 |
| HIV-1 core (p24) antigen-FITC (Clone KC57) | Beckman Coulter | Cat# 6604665; RRID: AB_1575987 |
| Mouse anti-human CD4 Alexa Fluor 700 (Clone RPAT4) | BD | Cat# 557922; RRID: AB_396943 |
| Mouse anti-human CD4 PerCP-Cy5.5 (Clone RPAT4) | BioLegend | Cat# 300530; RRID: AB_893328 |
| Mouse anti-human CD3 Pacific Blue (Clone UCHT1) | BD | Cat# 558117; RRID: AB_397038 |
| Mouse anti-human CCR6 PE (Clone 11A9) | BD | Cat# 559562; RRID: AB_397273 |
| Mouse anti-human CCR5 PE (Clone 2D7/CCR5) | BD | Cat# 555993; RRID: AB_396279 |
| Mouse anti human IFN-γ Alexa Fluor 700 (Clone B27) | BD | Cat# 557995; RRID: AB_396977 |
| Mouse anti human CXCR4 APC (Clone 12G5) | Thermo Fisher | Cat# 17–9999–42; RRID: AB_1724113 |
| Mouse anti human Ki67 BV421 (Clone B56) | BD | Cat# 562899; RRID: AB_2686897 |
| Rat anti human Integrinβ7 PE-Cy 5 (Clone FIB504) | BD | Cat# 551059; RRID: AB_394026 |
| Mouse anti human CD56 FITC (Clone HCD56) | eBioscience | Cat# 318304; RRID: AB_604100 |
| Mouse anti human IL-17A PE (Clone eBio64DEC17) | eBioscience | Cat# 12–7179–42; RRID: AB_1724136 |
| Mouse anti human IL-22 PE-Cyanine7 (Clone 22URTI) | eBioscience | Cat# 25–7229–42; RRID: AB_10853346 |
| Mouse anti human CD8 FITC (Clone BW135/80) | Miltenyi Biotech | Cat# 130–080–601; RRID: AB_244336 |
| Mouse anti human CD19 FITC (Clone LT19) | Miltenyi Biotech | Cat# 130–091–328; RRID: AB_244222 |
| Mouse anti human CD45RA APCeFluor780 (Clone HI100) | eBioscience | Cat# 47–0458–42; RRID: AB_10853641 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (405 nm excitation) | Invitrogen | Cat# L34957 |
| Rabbit anti human AhR | Abcam | Cat# ab190797; RRID: AB_2894707 |
| Rabbit anti human AhR | Cell Signaling | Cat# 83200; RRID: AB_2800011 |
| Rabbit anti human ARNT | Cell Signaling | Cat# 5537; RRID: AB_10694232 |
| Normal Rabbit IgG | Cell Signaling | Cat# 2729; RRID: AB_1031062 |
| Monoclonal anti-βActin (clone AC15) | Sigma | Cat# A5441; RRID: AB_476744 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| HIV-1 THRO plasmid (pTHRO.c/2626), subtype B | NIH AIDS Reagent Program (Contribution of Dr. John Kappes and Dr. Christina Ochsenbauer) | Cat#11745 |
| NL4.3BaL HIV plasmid | From Dr Roger J Pomerantz (Thomas Jefferson University, Philadelphia, Pennsylvania, USA) and Dr. Michel J. Tremblay (Université Laval, Québec City, QC, Canada) | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Leukaphereses of ART treated and untreated people living with HIV | Recruited at the Montreal Chest Institute, McGill University Health Center and Center Hospitalier de l’Université de Montréal via the collaboration of Dr Jean-Pierre Routy’s group | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Dulbecco’s Modified Eagle’s medium (DMEM) | Thermo Fisher | Cat# 11995065 |
| Roswell Park Memorial Institute (RPMI) | Thermo Fisher | Cat# 11875–093 |
| Penicillin/Streptomycin | Gibco | Cat# 15140–122 |
| Fetal Bovine Serum (FBS) | Sigma | Cat# F2442 |
| Bovine Serum Albumin (BSA) | BioShop | Cat# ALB001 |
| Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat# 20012050 |
| Tween 20 | Thermo Fisher | Cat# BP337–500 |
| PVDF membrane | Millipore Sigma | Cat# ISEQ00010 |
| RIPA Buffer 10X | NEB | Cat# 9806S |
| rhIL-2 | R&D Systems | Cat# 202–IL–050 |
| rhIL-10 | R&D Systems | Cat# 217–IL |
| rhIL-22 | R&D Systems | Cat# 782–IL–010 |
| CH-223191 | Sigma | Cat# C8124 |
| FICZ | Sigma | Cat# SML1489 |
| DNAzol™ Reagent | Invitrogen | Cat# 10503027 |
| T7 Endonuclease I - 250 units | NEB | Cat# M0302S |
| Phusion® High-Fidelity DNA Polymerase | NEB | Cat# M0530S |
| UltraComp ebeads | eBioscience | Cat# 01–2222–42 |
| Formaldehyde 37% | Sigma | Cat#F79500 |
| LC480 probe master mix | Roche | Cat# 4707494001 |
| Amersham ECL Prime Western Blotting System | Cytiva | Cat# RPN2232 |
| Brefeldin A | Sigma | Cat# B7651 |
| PMA | Sigma | Cat# P1585 |
| Ionomycin | Sigma | Cat# I0634 |
| Luna Universal qPCR Master Mix | NEB | Cat# M3003E |
| TMB Peroxidase Substrate | Fitzgerald | Cat# 42R–TB102 |
| Streptavidin Poly-HRP40 | Fitzgerald | Cat# 65R–S104PHRP–200UG |
|
| ||
| Critical commercial assays | ||
|
| ||
| Memory CD4+ T cell Isolation Kit, human | Miltenyi | Cat#130–091–893 |
| Fixation/Permeabilization Solution Kit (Cytofix/Cytoperm) | BD | Cat# 554714 |
| Human IL-10 DuoSet ELISA | R&D Systems | Cat# DY217B |
| Human IL-17A (homodimer) ELISA Ready-SET-Go | Thermo Fisher | Cat# 88–7176–86 |
| Human IL-22 DuoSet ELISA | R&D Systems | Cat# DY782 |
| Human IFN-γ ELISA | Thermo Fisher | Cat# 88–7316–86 |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat# 204245 |
| Human T cell Nucleofector® Kit | Lonza | Cat# VPA–1002 |
| Rneasy Plus Mini Kit | Qiagen | Cat# 74136 |
| CHIP assay kit | EMD Millipore | Cat# 17–295 |
| Luna® Universal qPCR Master Mix | NEB | Cat# M3003S |
| MyTaq Extract PCR kit | Bioline | Cat# BIO–21126 |
| High-fidelity enzyme Q5 | NEB | Cat# M0530L |
| QIAEX II Gel extraction kit | Qiagen | Cat# 20021 |
| GeneJET PCR Purification Kit | Thermo Fisher | Cat# K0702 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-Sequencing dataset | This paper | GEO: GSE198078 |
| Original western blot images relative to Figures 1, S1 and S2. | This paper; Mendeley Data | https://data.mendeley.com/datasets/4ry9gxv7tp/1 |
| Gene Ontology: Biological Processes database | Broad Institute | https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=BP |
| Homo Sapiens database GRCh 37 version75 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| ACH-2 cell line | NIH HIV Reagent Program | Cat# ARP–349 |
| 293T cells | ATCC | Cat# CRL–3216 |
| TZM-bl cells | NIH HIV Reagent Program | Cat# ARP–8129 |
| HIV-p24 Abs producing hybridome | From Dr. Michel J. Tremblay (Université Laval, Québec City, QC, Canada) | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S3: Oligonucleotides | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Kallisto software version 0.44.0 | Pachter Lab | https://pachterlab.github.io/kallisto/ |
| FlowJo version 10 | BD | https://www.flowjo.com/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Snapgene software | Snapgene | https://www.snapgene.com/ |
| TIDE software | Tide | https://tide.nki.nl/ |
| ICE software | Synthego | https://ice.synthego.com/ |
| QuantStudio™ Design & Analysis Software | Thermo Fisher | https://www.thermofisher.com/ |
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Ethics statement
In this study, leukapheresis collection from ART-treated PLWH and HIV-uninfected study participants was performed in compliance with the principles included in the Declaration of Helsinki. This study is approved by Institutional Review Board of the McGill University Health Centre and the CHUM-Research Center, Montreal, QC, Canada. All study participants have provided signed informed consents and agreed to publish the results in Journal with their samples.
Human subjects
Leukapheresis were collected from ART-treated PLWH (n = 7; Table S1) and HIV-uninfected study participants (n = 17; Table S2) recruited at the McGill University Health Centre and Centre Hospitalier de l’Université de Montréal (CHUM, Montreal, QC, Canada). Peripheral blood mononuclear cells (PBMCs) (109–1010) were isolated from leukapheresis and cryopreserved as previously described.81 The clinical characteristics of study participants are listed in Tables S1 and S2.
METHOD DETAILS
Flow cytometry analysis
For flow cytometry analysis antibodies (Abs) were listed in the key resources table. Intracellular staining was performed with BD Cytofix/Cytoperm kit (BD Biosciences, Franklin Lakes, NJ, USA), as we previously described.57,82 The viability dye LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Vivid, Life Technologies, CA, USA) was used to exclude dead cells. Flow cytometry analysis was performed using an LSRII cytometer (BD Pharmingen San Diego, CA, USA). The positivity gates were placed using the fluorescence minus one (FMO) strategy. All flow cytometry data were analyzed with the FlowJo software version 10.6.1 (Tree Star, Inc).
Cell sorting and TCR activation
Memory CD4+ T cells with (>95% purity) were enriched from PBMCs by negative selection using magnetic beads (Miltenyi, Bergisch Gladbach, Germany).57,81,82 In other experiments, CCR6+ and CCR6− memory CD4+ T-cells were sorted by flow cytometry (BDAria III), as we previously described.81,82 Cells were activated using immobilized CD3 and soluble CD28 antibodies (1 μg/mL; BD Pharmingen San Diego, CA, USA) for 3 days and used for subsequent experiments.
AhR ligands
The following AhR drugs were used in this study: the antagonist CH-223191 (Sigma, St. Louis, MO, USA; concentrations: 1.25, 2.5, 5, and 10 μM) and the agonist FICZ (Sigma, St. Louis, MO, USA; concentrations: 10 and 100 nM). Stock solutions of these ligands were prepared in dimethylsulfoxide (DMSO), aliquoted and stored according to the manufacturer’s instructions.
HIV-1 infection in vitro
The following HIV-1 strains were used (key resources table): replication-competent CCR5-tropic Transmitted Founder T/F THRO (HIVTHRO) and NL4.3BaL (HIVNL4.3BaL), and single-round VSVG-HIV-GFP (HIVVSVG). The HIVTHRO molecular clone plasmid (Catalog No. 11919) was obtained from Dr. John Kappes and Dr. Christina Ochsenbauer through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.83 HIVNL4.3BaL is an NL4.3-based provirus expressing the BaL envelope, provided by Michel Tremblay (U Laval, Québec) and originating from Dr Roger J Pomerantz (Thomas Jefferson University, Philadelphia, Pennsylvania, USA). The HIVVSVG stock was generated using a plasmid encoding for an env- NL4.3-based provirus, with gfp in place of nef, and another plasmid encoding for VSV-G, as previously described.81,84 HIV-1 stocks were generated by Fugene-mediated transfection, as we previously described.81,84 Memory CD4+ T-cells activated via CD3/CD28 for 3 days were exposed to HIV-1 (25 ng HIV-p24/106 cells) for 3 h at 37°C. Cells were washed to remove the unbound virions. Then, cells were cultured in the presence of IL-2 (5 ng/mL; R&D Systems, Minneapolis, MN, USA), and in the presence or the absence of CH-223191 or FICZ, at the indicated concentrations up to 9 days, with media being refreshed every 3 days. Viral replication was monitored by flow cytometry upon HIV-p24 intracellular staining and by ELISA soluble HIV-p24 quantification, as previously described.57,81,84 In parallel, HIV-infected cells were harvested at day 3 post-infection for PCR-based HIV-DNA quantification as described below.
Quantification of Gag and integrated HIV-DNA
RU5 and Gag reverse transcripts and integrated HIV-DNA levels were quantified by nested real-time PCR (Light Cycler 480, Roche) (Table S3; key resources table). Experiments were performed in triplicates on 0.5–1×105 T-cells/PCR. HIV-1 copy numbers were normalized to CD3 copy numbers (two CD3 copies per cell), as previously described.81,84
Viral outgrowth assay (VOA)
The VOA was performed, as we previously described50,82 (Table S3; key resources table). Briefly, memory CD4+ T-cells (1 × 106 cells/mL RPMI, 10% FBS, 1% Penicillin/Streptomycin) were isolated by MACS from PBMCs of ART-treated PLWH and cultured in 48-well plates in the presence of CD3/CD28 Abs (1 μg/mL). At day 3 post-TCR triggering, cells were washed and split into two clean 48-well plate wells and cultured with IL-2 (5 ng/mL; R&D Systems), in the presence or the absence of CH-223191 (10 μM) and FICZ (100 nM). Cells were further split at day 6 and 9 post-stimulation and half of the media was replenished with IL-2 containing or not AhR drugs. Cells were harvested at day 12 and the intracellular HIV-p24 expression was measured by flow cytometry. In parallel, soluble HIV-p24 levels were quantified by ELISA in the collected supernatant of days 3, 6, 9 and 12, as previously described.81,84
CRISPR/Cas9-mediated AhR KO
For AhR gene editing, we used the CRISPR/Cas9 technology, and a published protocol adapted for primary CD4+ T-cells40 (Figure 1A). Briefly, memory CD4+ T-cells were stimulated via CD3/CD28 for 3 days prior to electroporation. Electroporation was performed using the Amaxa P3 Primary Cell 96-well Nucleofector kit and 4D-Nucleofecter (Lonza, Walkersville, MD, USA). crRNAs were selected from CRISPR sgRNA database of (Genscript, USA). crRNA, tracrRNA and Cas9 were purchased from IDT (IDT, Newark, NJ, USA). crRNA and trans-activating crRNA (tracrRNA) were mixed at equimolar concentrations in a sterile PCR tube. Oligos were annealed by heating at 95°C for 5 min in PCR thermocycler and the mix was slowly cooled to room temperature. An equimolar concentration of Cas9-NLS was slowly added to the crRNA:tracrRNA and incubated for 10 min at room temperature to generate Cas9 RNPs complex. For each reaction, 1 × 106 T-cells were pelleted and re-suspended in 100 μL nucleofection solution (Lonza, Walkersville, MD, USA). T cell suspensions were mixed with 5 μL RNP complex and cell/RNP mix transferred to Nucleofector cuvette. Cells were electroporated using program T-020 on the Amaxa 4D-Nucleofector (Lonza, Walkersville, MD, USA). After nucleofection, pre-warmed T cell media was used to transfer transfected cells in 48-well plates and cells were allowed to culture in the presence of IL-2 (5 ng/mL) for 3–5 days. The AhR gene KO was confirmed by Western Blot (Figure 1B). crRNA sequences were as follows: AhR#1: 5′- AAGTCGGTCTCTATGCCGCT-3′ and AhR#2: 5′- TTGCTGCTCTACAGTTATCC-3′ (Table S3; key resources table).
T7 endonuclease assay
The AhR gene KO was also confirmed by T7 endonuclease assay, as previously described.85 Cells were lysed with DNAzol (Invitrogen, Waltham, Massachusetts, USA) and genomic DNA was extracted. AhR target regions were PCR-amplified using the forward primer 5′-GGGAATCACTGTGCTACAAATGC-3′′and the reverse primer 5′-CAGAAAATCCAGCAAGATGGTGT-3′. The amplification was performed using the PFU HiFi DNA polymerase (NEB, Germany) following the manufacturer’s instructions. The PCR products were gel-purified using the QIAEX II Gel extraction kit (Qiagen, Germany). After purification, the PCR products were denatured and slowly re annealed to form hetero-duplex DNA and digested with T7 endonuclease (NEB, Germany) for 30 min at 37°C. The resulting DNA fragments were visualized by 1.5% agarose gel electrophoresis (Table S3; key resources table).
TIDE assay
The TIDE assay was performed to determine the efficacy of silencing, as previously reported.40,41 Briefly, genomic DNA was extracted using MyTaq Extract PCR kit according to the manufacturer (Bioline, Cat number BIO-21126). PCR amplification was performed using the high-fidelity enzyme Q5 from New England Biolab (NEB, Cat number M0530L). Amplification was performed with the following locus specific primers: Ahr_1 (5′- TTCGGAAGAATTTAACCCATTC-3′), Ahr_2 (5′-AACTGACGCTGAGCCTAAGAAC). The PCR products obtained were sent for sequencing at the Center d’expertise et de service de génome Québec (CHU-Ste-Justine, Montré al, Canada). Sequence alignments were visualized using the Snapgene software (https://www.snapgene.com/) and cleavage activity was assessed using both TIDE (https://tide.nki.nl/) and ICE software (https://ice.synthego.com/#/) (Table S3; key resources table).
AhR siRNA
AhR RNA interference were performed in primary CD4+ T cells, as described previously.86 Briefly, memory CD4+ T-cells were stimulated by CD3/CD28 Abs for 2 days. Activated cells were nucleofected with 100 μM AhR or non-targeting (NT1) siRNA (ON-TARGETplus SMART pool, Dharmacon) using the Amaxa Human T cell Nucleofector Kit (Lonza, Walkersville, MD, USA), according to the manufacturer’s protocol. Nucleofected cells (2 × 106) were cultured for 24 h at 37°C in the presence of IL-2 (5 ng/mL). Cells were exposed to HIV-1 and cultured up to 3 days. To check the efficacy of KO, the AhR gene expression level was assessed by SYBR Green real-time RT-PCR 24h post-nucleofection. At day 3 post-infection, cells were stained with LIVE/DEAD Fixable Dead Cell Stain Kit (Vivid, Life Technologies,CA) were analyzed by FACS (BD LSRII) (Table S3; key resources table).
SYBR Green quantitative real time RT-PCR
Total RNA was isolated using the RNAeasy mini kit (Qiagen, Hilden, Germany). The AhR mRNA was quantified by one step SYBR Green real-time RT-PCR using the AhR Quantitect primer (Qiagen, Hilden, Germany). The RT-PCR reactions were carried out using the Light Cycler 480 II (Roche, Basel, Switzerland). The relative expression of AhR was normalized to the housekeeping gene 28S rRNA (Table S3; key resources table), as we previously reported.19,82,84,86 Each reaction was performed in triplicates.
Western Blot
Cells were lysed with the RIPA buffer (Cell Signaling) supplemented with phosphatase inhibitors (PhosSTOP) and protease inhibitor cocktail (Complete Mini EDTA-free, Roche) for 10 min at 4°C and centrifuged at 14,000 g for 10 min. Proteins were quantified by Bradford assay (Bio-Rad, Hercules, CA, USA). Samples (15–30 μg protein/well) were loaded on SDS PAGE gel (90 min, 100 V), transferred to PVDF membrane (Millipore) and blocked with 5% BSA. Subsequently membranes were incubated with primary AhR antibodies (Clone ab190797, Abcam, Cambridge, UK) and β-actin (Sigma, USA). Immunoreactive bands were detected using HRP-conjugated secondary antibodies and revealed with ECL substrate (GE Healthcare, Chicago, IL, USA) (Table S3; key resources table).
ELISA
HIV-p24 levels were quantified in cell culture supernatants using a homemade ELISA, with a hybridome provided by Dr Michel Tremblay,87 as described previously.81 IL-17A, IL-10 and IL-22 (DuoSet ELISA R&D Systems, Minneapolis, MN, USA) IFN-γ (Invitrogen, Waltham, Massachusetts, USA) cytokine levels in cell culture supernatants were measured, as per manufacturer’s instructions (Table S3; key resources table).
Illumina RNA sequencing and analysis
CD4+ memory T-cells were isolated from ART-treated PLWH, activated with CD3/CD28 Abs, and cultured in the presence or the absence of the AHR antagonist CH223191 (10 μM) for 18h.Total RNA was extracted using RNeasy Plus mini kit (Qiagen, Germantown, Maryland, USA). Genome-wide transcriptional profiling was performed by Genome Québec (Montreal, QC, Canada) using the Illumina RNA-Sequencing technology (NovaSeq6000 S4 PE 100bp 25M reads). Briefly, the paired-end sequencing reads were aligned to coding and non-coding transcripts from Homo Sapiens database GRCh 37 version75 and quantified with the Kallisto software version 0.44.0. The entire RNA-Sequencing dataset and the technical information requested by Minimum Information About a Microarray Experiment (MIAME) are available at the GEO database under accession GSE198078.
Differentially expressed genes (DEG) were identified based on p values (p < 0.05), adjusted p values (adj. p < 0.05) and fold-change (FC, cutoff 1.3). Statistical analyses were performed using R version 4.21. Differential expression analysis was performed using the limma Bioconductor R package (version 3.52.2) on the log2-counts per million (logCPM) transformed transcript-level and gene-level data. Gene set enrichment analysis (GSEA; C2, C3, C5, C7, C8, and Hallmark databases) was performed using the GSVA method (package version 1.344.2) on the logCPM data using a Gaussian cumulative distribution function. Finally, ChIP-seq peaks annotation of AhR targets (ENCODE dataset ENCFF763XLN; reference genome HG19) was done using packages ChIPseeker (version 1.32.1) and ENCODExplorerData (version 0.99.5).
Chromatin immunoprecipitation assays
ChIP assays were performed following the ChIP assay kit from EMD Millipore on two cell lines: 1) a HeLa-derivative cell line that expresses high levels of the HIV-1 receptor CD4 and both coreceptors CXCR4 and CCR5 and contains beta-galactosidase and luciferase reporter genes under the control of the HIV-1 LTR (TZM-bl); and 2) the human embryonic kidney cell line HEK293T. Briefly, cells were cross-linked for 10 min at room temperature with 1% formaldehyde before lysis followed by chromatin sonication (Bioruptor Plus, Diagenode) to obtain DNA fragments of 200–400 bp. Chromatin immunoprecipitations were performed with chromatin from 5×106 cells and 5 μg of antibodies against either AhR (Cell Signaling 83200, RRID:AB_2800011) or ARNT (Cell Signaling 5537, RRID:AB_10694232). A Normal Rabbit IgG (Cell Signaling 2729, RRID:AB_1031062) was used as a negative control. Quantitative real-time PCR reactions were performed using 1/60 of the immunoprecipitated DNA and the Luna Universal qPCR Master Mix (NEB). Several regions were studied using oligonucleotide primer pairs to cover the different potential AhR/ARNT binding sites on the HIC1 promoter including AhR#1 (FW: CACCCACACAAACCTGACCT, RV: GGCCCTAGAATCCCCCTGTA), AhR#2 (FW: TTTTCCGAACTGGGGCTGTG, RV: TTCGCCCTCCCACCTGTA) and AhR#3 + 4 (FW: AGTTCCGGGGGAGAAGGG, RV: GCGGGGAGGGCGTTATATC), the mmp9 promoter used as positive control for the recruitment of AhR (FW: TCTCATGCTGGTGCTGCC, RV: CTTTAAGGAGGCGCTCCTGTG) (Table S3; key resources table). Relative quantification using the standard curve method was performed for each primer pair and 96-well Optical Reaction plates were read in a QuantStudio3 PCR instrument (Applied Biosystem). Fold enrichments were calculated as percentages of input values. Primer sequences used for quantification were designed using the software Primer 3.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analyses were performed using the Prism 7 (GraphPad, Inc., La Joya, CA, USA) software. In each graph and Figure legend, specific statistical tests were applied. p-values are indicated on the graphs with statistical significance as follows: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Supplementary Material
Highlights.
AhR activation, induced upon TCR triggering, boosts non-pathogenic Th17 functions
AhR gene editing decreases IL-22/IL-10 production and increases HIV-1 replication
AhR pharmacological blockade promotes HIV outgrowth in cells of ART-treated PLWH
AhR directly regulates HIC1, an HIV-1 repressor and tissue-residency regulator
ACKNOWLEDGMENTS
The authors thank Dr. Dominique Gauchat and Philippe St Onge (Flow Cytometry Core Facility, CHUM-Research Center, Montré al, QC, Canada) for expert technical support with polychromatic flow cytometry sorting; Olfa Debbeche (Biosafety Level 3 Core Facility CHUM-Research Center, Montré al, QC, Canada) and Mario Legault (FRQ-S/AIDS and Infectious Diseases Network; Montréal, QC, Canada) for help with ethical approvals and informed consents; and Josée Girouard and Angie Massicotte (McGill University Health Centre, Montréal, QC, Canada) for their key contribution to blood collection and clinical information from PLWH and uninfected study participants. The authors also thank Dr. Roger J. Pomerantz (Thomas Jefferson University, Philadelphia, PA, USA) and Dr. Michel Tremblay (Université Laval, Quebec, QC, Canada) for providing the NL4.3BaL HIV plasmid and the HIV-p24 Abs producing hybridome, respectively and Dr. Jean-Franç ois Schmouth (CRCHUM, Transgenesis and Animal Modeling Platform) for technical assistance in performing the TIDE assay. The authors also thank Dr. Ariberto Fassati (UCL, London, UK) and Edwin Caballero for the critical reading of the manuscript and Dr. Nicolas Cermakian (Douglas Institute; McGill University) for valuable discussions on the project. Finally, the authors acknowledge the key contributions of all study participants for their precious gift of leukapheresis essential for this study. P.A. was supported by a grant from the Canadian HIV Cure Enterprise Team Grant (CanCURE 1.0) funded by the Canadian Institutes of Health (CIHR) in partnership with CANFAR and IAS (CanCURE 1.0; # HIG-133050 to P.A.); the Canadian HIV Cure Enterprise Team Grant (CanCURE 2.0) funded by the CIHR (#HB2–164064); and CIHR project grants to P.A. (PJT #153052; PJT 178127). Core facilities and PLWH cohorts were supported by the Fondation du CHUM and the FRQS/AIDS and Infectious Diseases Network. The funding institutions played no role in the design, collection, analysis, and interpretation of data. T.R.W.S. received doctoral fellowships from the Université de Montréal and Fonds de Recherche Québec Santé (FRQ-S). J.-P.R. holds the Louis Lowenstein Chair in Hematology and Oncology, McGill University. C.V.L. acknowledges funding from the Belgian National Fund for Scientific Research (CDR J.0181.21; F.R.S-FNRS, Belgium); the French INSERM agency “ANRS/Maladies infectieuses émergentes” (grant ECTZ189331); the “Télévie” program of the F.R.S.-FNRS; ViiV Healthcare (grant no. 21199 RHIVIERA WP1); the “Fondation Roi Baudouin” (A20/TT/0176 (2019-J1820640–213631)); the Internationale Brachet Stiftung (IBS; grant 22–5); the Walloon Region (“Fonds de Maturation”); The “Amis des Instituts Pasteur à Bruxelles” (Amis IPB-Bourses - Echéance 01.04.2022); Université Libre de Bruxelles (ULB - Action de Recherche Concertée [ARC] grant); the NEAT (European AIDS Treatment Network) program; the European Union’s Marie Sklodowska Curie COFUND action; and the US National Institutes of Health (NIH) (MDC grant UM1AI164562 cofunded by the National Heart, Lung, and Blood Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, the National Institute on Drug Abuse, and the National Institute of Allergy and Infectious Diseases). A.D. is funded by an “Aspirant” fellowship from the F.R.S.-FNRS (Contract A 2/5 – MCF/BIC - AR 181 FC 33943). O.H. is funded by fellowships from the Belgian “Fonds pour la formation à la Recherche dans l’Industrie et dans l’Agriculture (FRIA) (F.R.S.-FNRS)” and from “Les Amis des Instituts Pasteur à Bruxelles” (Contract F 3/5/5 - FRIA/FC - 6035 FC 31697). C.V.L. is “Directrice de Recherches” of the F.R.S-FNRS (Contract A 11/5 – XH/JA FC 51320). The laboratory of C.V.L. is part of the ULB-Cancer Research Center (U-CRC) (Faculty of Medicine, ULB).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112634.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Davenport MP, Khoury DS, Cromer D, Lewin SR, Kelleher AD, and Kent SJ (2019). Functional cure of HIV: the scale of the challenge. Nat. Rev. Immunol 19, 45–54. 10.1038/s41577-018-0085-4. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong MAWP, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, et al. (2021). Research priorities for an HIV cure: international AIDS society global scientific strategy 2021. Nat. Med 27, 2085–2098. 10.1038/s41591-021-01590-5. [DOI] [PubMed] [Google Scholar]
- 3.Cohn LB, Chomont N, and Deeks SG (2020). The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe 27, 519–530. 10.1016/j.chom.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sengupta S, and Siliciano RF (2018). Targeting the latent reservoir for HIV-1. Immunity 48, 872–895. 10.1016/j.immuni.2018.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siliciano JD, and Siliciano RF (2020). Nonsuppressible HIV-1 viremia: a reflection of how the reservoir persists. J. Clin. Invest 130, 5665–5667. 10.1172/JCI141497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siliciano JD, and Siliciano RF (2022). In vivo dynamics of the latent reservoir for HIV-1: New insights and implications for cure. Annu. Rev. Pathol 17, 271–294. 10.1146/annurev-pathol-050520-112001. [DOI] [PubMed] [Google Scholar]
- 7.Mediouni S, Lyu S, Schader SM, and Valente ST (2022). Forging a functional cure for HIV: transcription regulators and inhibitors. Viruses 14, 1980. 10.3390/v14091980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wacleche VS, Landay A, Routy JP, and Ancuta P (2017). The Th17 lineage: from barrier surfaces homeostasis to autoimmunity, cancer, and HIV-1 pathogenesis. Viruses 9, 303. 10.3390/v9100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Planas D, Routy JP, and Ancuta P (2019). New Th17-specific therapeutic strategies for HIV remission. Curr. Opin. HIV AIDS 14, 85–92. 10.1097/COH.0000000000000522. [DOI] [PubMed] [Google Scholar]
- 10.Butterfield TR, Landay AL, and Anzinger JJ (2020). Dysfunctional immunometabolism in HIV infection: contributing factors and implications for age-related comorbid diseases. Curr. HIV AIDS Rep 17, 125–137. 10.1007/s11904-020-00484-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagle A, Goerlich E, Post WS, Woldu B, Wu KC, and Hays AG (2022). HIV and global cardiovascular Health. Curr. Cardiol. Rep 24, 1149–1157. 10.1007/s11886-022-01741-1. [DOI] [PubMed] [Google Scholar]
- 12.Fert A, Raymond Marchand L, Wiche Salinas TR, and Ancuta P (2022). Targeting Th17 cells in HIV-1 remission/cure interventions. Trends Immunol 43, 580–594. 10.1016/j.it.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, and Littman DR (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. [DOI] [PubMed] [Google Scholar]
- 14.Unutmaz D (2009). RORC2: the master of human Th17 cell programming. Eur. J. Immunol 39, 1452–1455. 10.1002/eji.200939540. [DOI] [PubMed] [Google Scholar]
- 15.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidović D, et al. (2011). Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472, 491–494. 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, et al. (2014). Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity 40, 477–489. 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Withers DR, Hepworth MR, Wang X, Mackley EC, Halford EE, Dutton EE, Marriott CL, Brucklacher-Waldert V, Veldhoen M, Kelsen J, et al. (2016). Transient inhibition of ROR-gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat. Med 22, 319–323. 10.1038/nm.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiche Salinas TR, Zhang Y, Sarnello D, Zhyvoloup A, Marchand LR, Fert A, Planas D, Lodha M, Chatterjee D, Karwacz K, et al. (2021). Th17 cell master transcription factor RORC2 regulates HIV-1 gene expression and viral outgrowth. Proc. Natl. Acad. Sci. USA 118, e2105927118. 10.1073/pnas.2105927118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wacleche VS, Goulet JP, Gosselin A, Monteiro P, Soudeyns H, Fromentin R, Jenabian MA, Vartanian S, Deeks SG, Chomont N, et al. (2016). New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology 13, 59. 10.1186/s12977-016-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol 13, 991–999. 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramesh R, Kozhaya L, McKevitt K, Djuretic IM, Carlson TJ, Quintero MA, McCauley JL, Abreu MT, Unutmaz D, and Sundrud MS (2014). Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med 211, 89–104. 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burbach KM, Poland A, and Bradfield CA (1992). Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc. Natl. Acad. Sci. USA 89, 8185–8189. 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukunaga BN, Probst MR, Reisz-Porszasz S, and Hankinson O (1995). Identification of functional domains of the aryl hydrocarbon receptor. J. Biol. Chem 270, 29270–29278. 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 24.Hooper LV (2011). You AhR what you eat: linking diet and immunity. Cell 147, 489–491. 10.1016/j.cell.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Lamas B, Natividad JM, and Sokol H (2018). Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol 11, 1024–1038. 10.1038/s41385-018-0019-2. [DOI] [PubMed] [Google Scholar]
- 26.Stockinger B, Shah K, and Wincent E (2021). AHR in the intestinal microenvironment: safeguarding barrier function. Nat. Rev. Gastroenterol. Hepatol 18, 559–570. 10.1038/s41575-021-00430-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutiérrez-Vázquez C, and Quintana FJ (2018). Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 48, 19–33. 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, and Stockinger B (2008). The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109. 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 29.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, and Weiner HL (2008). Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71. [DOI] [PubMed] [Google Scholar]
- 30.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, and Kishimoto T (2008). Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc. Natl. Acad. Sci. USA 105, 9721–9726. 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veldhoen M, Hirota K, Christensen J, O’Garra A, and Stockinger B (2009). Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J. Exp. Med 206, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, Kozoriz D, Weiner HL, and Quintana FJ (2010). Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat. Immunol 11, 846–853. 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, and Kuchroo VK (2010). The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol 11, 854–861. 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trifari S, Kaplan CD, Tran EH, Crellin NK, and Spits H (2009). Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol 10, 864–871. [DOI] [PubMed] [Google Scholar]
- 35.Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limón P, Paiva RS, Ching T, et al. (2015). Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225. 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monteiro P, Gilot D, Le Ferrec E, Lecureur V, N’Diaye M, Le Vee M, Podechard N, Pouponnot C, and Fardel O (2007). AhR- and c-maf-dependent induction of beta7-integrin expression in human macrophages in response to environmental polycyclic aromatic hydrocarbons. Biochem. Biophys. Res. Commun 358, 442–448. [DOI] [PubMed] [Google Scholar]
- 37.Kueck T, Cassella E, Holler J, Kim B, and Bieniasz PD (2018). The aryl hydrocarbon receptor and interferon gamma generate antiviral states via transcriptional repression. Elife 7, e38867. 10.7554/eLife.38867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray IA, Patterson AD, and Perdew GH (2014). Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer 14, 801–814. 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean JW, and Zhou L (2022). Cell-intrinsic view of the aryl hydrocarbon receptor in tumor immunity. Trends Immunol 43, 245–258. 10.1016/j.it.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, and Marson A (2016). A Cas9 ribonucleoprotein Platform for functional genetic studies of HIV-host interactions in primary human T cells. Cell Rep 17, 1438–1452. 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brinkman EK, Chen T, Amendola M, and van Steensel B (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42, e168. 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, et al. (2013). Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. USA 110, 6626–6633. 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rannug A, and Rannug U (2018). The tryptophan derivative 6-formylindolo[3,2-b]carbazole, FICZ, a dynamic mediator of endogenous aryl hydrocarbon receptor signaling, balances cell growth and differentiation. Crit. Rev. Toxicol 48, 555–574. 10.1080/10408444.2018.1493086. [DOI] [PubMed] [Google Scholar]
- 44.Zhao B, Degroot DE, Hayashi A, He G, and Denison MS (2010). CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol. Sci 117, 393–403. 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macedo AB, Novis CL, De Assis CM, Sorensen ES, Moszczynski P, Huang SH, Ren Y, Spivak AM, Jones RB, Planelles V, and Bosque A (2018). Dual TLR2 and TLR7 agonists as HIV latency-reversing agents. JCI Insight 3, e122673. 10.1172/jci.insight.122673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kedzierska K, and Crowe SM (2001). Cytokines and HIV-1: interactions and clinical implications. Antivir. Chem. Chemother 12, 133–150. 10.1177/095632020101200301. [DOI] [PubMed] [Google Scholar]
- 47.Harper J, Ribeiro SP, Chan CN, Aid M, Deleage C, Micci L, Pino M, Cervasi B, Raghunathan G, Rimmer E, et al. (2022). Interleukin-10 contributes to reservoir establishment and persistence in SIV-infected macaques treated with antiretroviral therapy. J. Clin. Invest 132, e155251. 10.1172/JCI155251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkelshtein D, Werman A, Novick D, Barak S, and Rubinstein M (2013). LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 110, 7306–7311. 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan Q, Kozhaya L, ElHed A, Ramesh R, Carlson TJ, Djuretic IM, Sundrud MS, and Unutmaz D (2011). Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. J. Exp. Med 208, 1875–1887. 10.1084/jem.20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Planas D, Raymond Marchand L, Massanella M, Chen H, Wacleche VS, Gosselin A, Goulet JP, Filion M, Routy JP, et al. (2020). Improving HIV outgrowth by optimizing cell-culture conditions and supplementing with all-trans retinoic acid. Front. Microbiol 11, 902. 10.3389/fmicb.2020.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Douce V, Forouzanfar F, Eilebrecht S, Van Driessche B, Ait-Ammar A, Verdikt R, Kurashige Y, Marban C, Gautier V, Candolfi E, et al. (2016). HIC1 controls cellular- and HIV-1- gene transcription via interactions with CTIP2 and HMGA1. Sci. Rep 6, 34920. 10.1038/srep34920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen WY, Wang DH, Yen RC, Luo J, Gu W, and Baylin SB (2005). Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123, 437–448. 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 53.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, et al. (2022). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res 50, D165–D173. 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim DJ, Iwasaki A, Chien AL, and Kang S (2022). UVB-mediated DNA damage induces matrix metalloproteinases to promote photoaging in an AhR- and SP1-dependent manner. JCI Insight 7, e156344. 10.1172/jci.insight.156344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burrows K, Antignano F, Bramhall M, Chenery A, Scheer S, Korinek V, Underhill TM, and Zaph C (2017). The transcriptional repressor HIC1 regulates intestinal immune homeostasis. Mucosal Immunol 10, 1518–1528. 10.1038/mi.2017.17. [DOI] [PubMed] [Google Scholar]
- 56.Crowl JT, Heeg M, Ferry A, Milner JJ, Omilusik KD, Toma C, He Z, Chang JT, and Goldrath AW (2022). Tissue-resident memory CD8(+) T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat. Immunol 23, 1121–1131. 10.1038/s41590-022-01229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiche Salinas TR, Gosselin A, Raymond Marchand L, Moreira Gabriel E, Tastet O, Goulet JP, Zhang Y, Vlad D, Touil H, Routy JP, et al. (2021). IL-17A reprograms intestinal epithelial cells to facilitate HIV-1 replication and outgrowth in CD4+ T cells. iScience 24, 103225. 10.1016/j.isci.2021.103225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, and Kuchroo VK (2013). Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517. 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang R, Campbell S, Amir M, Mosure SA, Bassette MA, Eliason A, Sundrud MS, Kamenecka TM, and Solt LA (2021). Genetic and pharmacological inhibition of the nuclear receptor RORalpha regulates TH17 driven inflammatory disorders. Nat. Commun 12, 76. 10.1038/s41467-020-20385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swaminathan G, Nguyen LP, Namkoong H, Pan J, Haileselassie Y, Patel A, Ji AR, Mikhail DM, Dinh TT, Singh H, et al. (2021). The aryl hydrocarbon receptor regulates expression of mucosal trafficking receptor GPR15. Mucosal Immunol 14, 852–861. 10.1038/s41385-021-00390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benechet AP, Menon M, Xu D, Samji T, Maher L, Murooka TT, Mempel TR, Sheridan BS, Lemoine FM, and Khanna KM (2016). T cell-intrinsic S1PR1 regulates endogenous effector T-cell egress dynamics from lymph nodes during infection. Proc. Natl. Acad. Sci. USA 113, 2182–2187. 10.1073/pnas.1516485113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ancuta P, Autissier P, Wurcel A, Zaman T, Stone D, and Gabuzda D (2006). CD16+ monocyte-derived macrophages activate resting T cells for HIV infection by producing CCR3 and CCR4 ligands. J. Immunol 176, 5760–5771. [DOI] [PubMed] [Google Scholar]
- 63.Jenabian MA, El-Far M, Vyboh K, Kema I, Costiniuk CT, Thomas R, Baril JG, LeBlanc R, Kanagaratham C, Radzioch D, et al. (2015). Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. J. Infect. Dis 212, 355–366. 10.1093/infdis/jiv037. [DOI] [PubMed] [Google Scholar]
- 64.Chen J, Xun J, Yang J, Ji Y, Liu L, Qi T, Wang Z, Zhang R, Shen Y, Ponte R, et al. (2019). Plasma indoleamine 2,3-dioxygenase activity is associated with the size of the human immunodeficiency virus reservoir in patients receiving antiretroviral therapy. Clin. Infect. Dis 68, 1274–1281. 10.1093/cid/ciy676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lederman MM, Funderburg NT, Sekaly RP, Klatt NR, and Hunt PW (2013). Residual immune dysregulation syndrome in treated HIV infection. Adv. Immunol 119, 51–83. 10.1016/B978-0-12-407707-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun X, Qu Q, Lao Y, Zhang M, Yin X, Zhu H, Wang Y, Yang J, Yi J, and Hao M (2019). Tumor suppressor HIC1 is synergistically compromised by cancer-associated fibroblasts and tumor cells through the IL-6/pSTAT3 axis in breast cancer. BMC Cancer 19, 1180. 10.1186/s12885-019-6333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, et al. (2005). SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol 3, e41. 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinzone MR, Cacopardo B, Condorelli F, Di Rosa M, and Nunnari G (2013). Sirtuin-1 and HIV-1: an overview. Curr. Drug Targets 14, 648–652. 10.2174/1389450111314060005. [DOI] [PubMed] [Google Scholar]
- 69.Limagne E, Thibaudin M, Euvrard R, Berger H, Chalons P, Végan F, Humblin E, Boidot R, Rébé C, Derangère V, et al. (2017). Sirtuin-1 activation controls tumor growth by impeding Th17 differentiation via STAT3 deacetylation. Cell Rep 19, 746–759. 10.1016/j.celrep.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Mameli G, Deshmane SL, Ghafouri M, Cui J, Simbiri K, Khalili K, Mukerjee R, Dolei A, Amini S, and Sawaya BE (2007). C/EBPbeta regulates human immunodeficiency virus 1 gene expression through its association with cdk9. J. Gen. Virol 88, 631–640. 10.1099/vir.0.82487-0. [DOI] [PubMed] [Google Scholar]
- 71.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, and Frenkel LM (2014). HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573. 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. (2014). HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183. 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cesana D, Santoni de Sio FR, Rudilosso L, Gallina P, Calabria A, Beretta S, Merelli I, Bruzzesi E, Passerini L, Nozza S, et al. (2017). HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat. Commun 8, 498. 10.1038/s41467-017-00609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou YH, Sun L, Chen J, Sun WW, Ma L, Han Y, Jin X, Zhao QX, Li T, Lu H, et al. (2019). Tryptophan metabolism activates aryl hydrocarbon receptor-mediated pathway to promote HIV-1 infection and reactivation. mBio 10, 02591–19. 10.1128/mBio.02591-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsiao F, Frouard J, Gramatica A, Xie G, Telwatte S, Lee GQ, Roychoudhury P, Schwarzer R, Luo X, Yukl SA, et al. (2020). Tissue memory CD4+ T cells expressing IL-7 receptor-alpha (CD127) preferentially support latent HIV-1 infection. PLoS Pathog 16, e1008450. 10.1371/journal.ppat.1008450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, Kim P, Chen TH, Milush J, Hunt PW, et al. (2018). Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog 14, e1007357. 10.1371/journal.ppat.1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, and Wong JK (2018). HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med 10, eaap9927. 10.1126/sci-translmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Busman-Sahay K, Starke CE, Nekorchuk MD, and Estes JD (2021). Eliminating HIV reservoirs for a cure: the issue is in the tissue. Curr. Opin. HIV AIDS 16, 200–208. 10.1097/COH.0000000000000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, et al. (2007). Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446, 562–566. [DOI] [PubMed] [Google Scholar]
- 80.Torti MF, Giovannoni F, Quintana FJ, and García CC (2021). The aryl hydrocarbon receptor as a modulator of anti-viral immunity. Front. Immunol 12, 624293. 10.3389/fimmu.2021.624293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gosselin A, Wiche Salinas TR, Planas D, Wacleche VS, Zhang Y, Fromentin R, Chomont N, Cohen ÉA, Shacklett B, Mehraj V, et al. (2017). HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS 31, 35–48. 10.1097/QAD.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Planas D, Fert A, Zhang Y, Goulet JP, Richard J, Finzi A, Ruiz MJ, Marchand LR, Chatterjee D, Chen H, et al. (2020). Pharmacological Inhibition of PPARy Boosts HIV Reactivation and Th17 Effector Functions, While Preventing Progeny Virion Release and de novo Infection. Pathog. Immun 5, 177–239. 10.20411/pai.v5i1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, et al. (2012). Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol 86, 2715–2728. 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Planas D, Zhang Y, Monteiro P, Goulet JP, Gosselin A, Grandvaux N, Hope TJ, Fassati A, Routy JP, and Ancuta P (2017). HIV-1 selectively targets gut-homing CCR6+CD4+ T cells via mTOR-dependent mechanisms. JCI Insight 2, e93230. 10.1172/jci.insight.93230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grajkowska LT, Ceribelli M, Lau CM, Warren ME, Tiniakou I, Nakandakari Higa S, Bunin A, Haecker H, Mirny LA, Staudt LM, and Reizis B (2017). Isoform-specific expression and feedback regulation of E protein TCF4 control dendritic cell lineage specification. Immunity 46, 65–77. 10.1016/j.immuni.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cleret-Buhot A, Zhang Y, Planas D, Goulet JP, Monteiro P, Gosselin A, Wacleche VS, Tremblay CL, Jenabian MA, Routy JP, et al. (2015). Identification of novel HIV-1 dependency factors in primary CCR4(+)CCR6(+)Th17 cells via a genome-wide transcriptional approach. Retrovirology 12, 102. 10.1186/s12977-015-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bounou S, Leclerc JE, and Tremblay MJ (2002). Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J. Virol 76, 1004–1014. 10.1128/jvi.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at Gene Expression Omnibus (GEO) database under accession GSE198078 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE198078) and are publicly available as of the date of publication, as listed in the key resources table. Original Western blot images have been deposited at Mendeley database and are publicly available as of the date of publication (https://data.mendeley.com/datasets/4ry9gxv7tp/1).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Purified NA/LE Mouse Anti-Human CD3 (Clone UCHT1) | BD | Cat# 555329; RRID: AB_395736 |
| Purified NA/LE Mouse Anti-Human CD28 (Clone CD28.2) | BD | Cat# 555725; RRID: AB_396068 |
| HIV-1 core (p24) antigen-FITC (Clone KC57) | Beckman Coulter | Cat# 6604665; RRID: AB_1575987 |
| Mouse anti-human CD4 Alexa Fluor 700 (Clone RPAT4) | BD | Cat# 557922; RRID: AB_396943 |
| Mouse anti-human CD4 PerCP-Cy5.5 (Clone RPAT4) | BioLegend | Cat# 300530; RRID: AB_893328 |
| Mouse anti-human CD3 Pacific Blue (Clone UCHT1) | BD | Cat# 558117; RRID: AB_397038 |
| Mouse anti-human CCR6 PE (Clone 11A9) | BD | Cat# 559562; RRID: AB_397273 |
| Mouse anti-human CCR5 PE (Clone 2D7/CCR5) | BD | Cat# 555993; RRID: AB_396279 |
| Mouse anti human IFN-γ Alexa Fluor 700 (Clone B27) | BD | Cat# 557995; RRID: AB_396977 |
| Mouse anti human CXCR4 APC (Clone 12G5) | Thermo Fisher | Cat# 17–9999–42; RRID: AB_1724113 |
| Mouse anti human Ki67 BV421 (Clone B56) | BD | Cat# 562899; RRID: AB_2686897 |
| Rat anti human Integrinβ7 PE-Cy 5 (Clone FIB504) | BD | Cat# 551059; RRID: AB_394026 |
| Mouse anti human CD56 FITC (Clone HCD56) | eBioscience | Cat# 318304; RRID: AB_604100 |
| Mouse anti human IL-17A PE (Clone eBio64DEC17) | eBioscience | Cat# 12–7179–42; RRID: AB_1724136 |
| Mouse anti human IL-22 PE-Cyanine7 (Clone 22URTI) | eBioscience | Cat# 25–7229–42; RRID: AB_10853346 |
| Mouse anti human CD8 FITC (Clone BW135/80) | Miltenyi Biotech | Cat# 130–080–601; RRID: AB_244336 |
| Mouse anti human CD19 FITC (Clone LT19) | Miltenyi Biotech | Cat# 130–091–328; RRID: AB_244222 |
| Mouse anti human CD45RA APCeFluor780 (Clone HI100) | eBioscience | Cat# 47–0458–42; RRID: AB_10853641 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (405 nm excitation) | Invitrogen | Cat# L34957 |
| Rabbit anti human AhR | Abcam | Cat# ab190797; RRID: AB_2894707 |
| Rabbit anti human AhR | Cell Signaling | Cat# 83200; RRID: AB_2800011 |
| Rabbit anti human ARNT | Cell Signaling | Cat# 5537; RRID: AB_10694232 |
| Normal Rabbit IgG | Cell Signaling | Cat# 2729; RRID: AB_1031062 |
| Monoclonal anti-βActin (clone AC15) | Sigma | Cat# A5441; RRID: AB_476744 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| HIV-1 THRO plasmid (pTHRO.c/2626), subtype B | NIH AIDS Reagent Program (Contribution of Dr. John Kappes and Dr. Christina Ochsenbauer) | Cat#11745 |
| NL4.3BaL HIV plasmid | From Dr Roger J Pomerantz (Thomas Jefferson University, Philadelphia, Pennsylvania, USA) and Dr. Michel J. Tremblay (Université Laval, Québec City, QC, Canada) | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Leukaphereses of ART treated and untreated people living with HIV | Recruited at the Montreal Chest Institute, McGill University Health Center and Center Hospitalier de l’Université de Montréal via the collaboration of Dr Jean-Pierre Routy’s group | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Dulbecco’s Modified Eagle’s medium (DMEM) | Thermo Fisher | Cat# 11995065 |
| Roswell Park Memorial Institute (RPMI) | Thermo Fisher | Cat# 11875–093 |
| Penicillin/Streptomycin | Gibco | Cat# 15140–122 |
| Fetal Bovine Serum (FBS) | Sigma | Cat# F2442 |
| Bovine Serum Albumin (BSA) | BioShop | Cat# ALB001 |
| Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat# 20012050 |
| Tween 20 | Thermo Fisher | Cat# BP337–500 |
| PVDF membrane | Millipore Sigma | Cat# ISEQ00010 |
| RIPA Buffer 10X | NEB | Cat# 9806S |
| rhIL-2 | R&D Systems | Cat# 202–IL–050 |
| rhIL-10 | R&D Systems | Cat# 217–IL |
| rhIL-22 | R&D Systems | Cat# 782–IL–010 |
| CH-223191 | Sigma | Cat# C8124 |
| FICZ | Sigma | Cat# SML1489 |
| DNAzol™ Reagent | Invitrogen | Cat# 10503027 |
| T7 Endonuclease I - 250 units | NEB | Cat# M0302S |
| Phusion® High-Fidelity DNA Polymerase | NEB | Cat# M0530S |
| UltraComp ebeads | eBioscience | Cat# 01–2222–42 |
| Formaldehyde 37% | Sigma | Cat#F79500 |
| LC480 probe master mix | Roche | Cat# 4707494001 |
| Amersham ECL Prime Western Blotting System | Cytiva | Cat# RPN2232 |
| Brefeldin A | Sigma | Cat# B7651 |
| PMA | Sigma | Cat# P1585 |
| Ionomycin | Sigma | Cat# I0634 |
| Luna Universal qPCR Master Mix | NEB | Cat# M3003E |
| TMB Peroxidase Substrate | Fitzgerald | Cat# 42R–TB102 |
| Streptavidin Poly-HRP40 | Fitzgerald | Cat# 65R–S104PHRP–200UG |
|
| ||
| Critical commercial assays | ||
|
| ||
| Memory CD4+ T cell Isolation Kit, human | Miltenyi | Cat#130–091–893 |
| Fixation/Permeabilization Solution Kit (Cytofix/Cytoperm) | BD | Cat# 554714 |
| Human IL-10 DuoSet ELISA | R&D Systems | Cat# DY217B |
| Human IL-17A (homodimer) ELISA Ready-SET-Go | Thermo Fisher | Cat# 88–7176–86 |
| Human IL-22 DuoSet ELISA | R&D Systems | Cat# DY782 |
| Human IFN-γ ELISA | Thermo Fisher | Cat# 88–7316–86 |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat# 204245 |
| Human T cell Nucleofector® Kit | Lonza | Cat# VPA–1002 |
| Rneasy Plus Mini Kit | Qiagen | Cat# 74136 |
| CHIP assay kit | EMD Millipore | Cat# 17–295 |
| Luna® Universal qPCR Master Mix | NEB | Cat# M3003S |
| MyTaq Extract PCR kit | Bioline | Cat# BIO–21126 |
| High-fidelity enzyme Q5 | NEB | Cat# M0530L |
| QIAEX II Gel extraction kit | Qiagen | Cat# 20021 |
| GeneJET PCR Purification Kit | Thermo Fisher | Cat# K0702 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-Sequencing dataset | This paper | GEO: GSE198078 |
| Original western blot images relative to Figures 1, S1 and S2. | This paper; Mendeley Data | https://data.mendeley.com/datasets/4ry9gxv7tp/1 |
| Gene Ontology: Biological Processes database | Broad Institute | https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=BP |
| Homo Sapiens database GRCh 37 version75 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| ACH-2 cell line | NIH HIV Reagent Program | Cat# ARP–349 |
| 293T cells | ATCC | Cat# CRL–3216 |
| TZM-bl cells | NIH HIV Reagent Program | Cat# ARP–8129 |
| HIV-p24 Abs producing hybridome | From Dr. Michel J. Tremblay (Université Laval, Québec City, QC, Canada) | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S3: Oligonucleotides | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Kallisto software version 0.44.0 | Pachter Lab | https://pachterlab.github.io/kallisto/ |
| FlowJo version 10 | BD | https://www.flowjo.com/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Snapgene software | Snapgene | https://www.snapgene.com/ |
| TIDE software | Tide | https://tide.nki.nl/ |
| ICE software | Synthego | https://ice.synthego.com/ |
| QuantStudio™ Design & Analysis Software | Thermo Fisher | https://www.thermofisher.com/ |
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.


