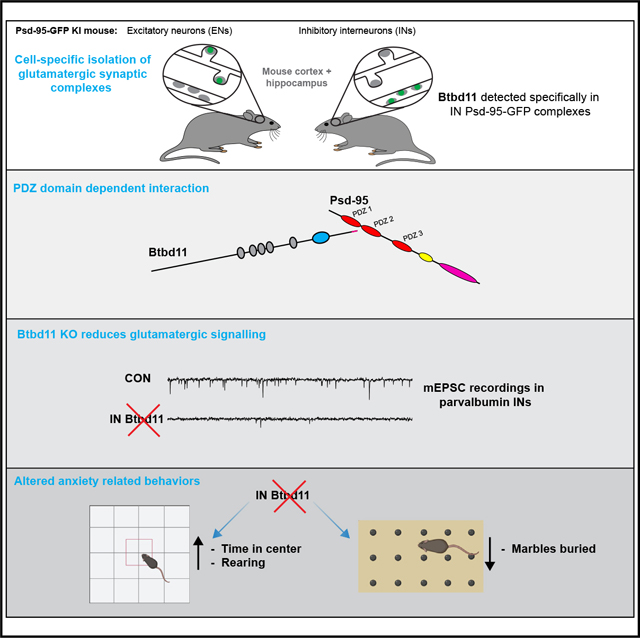
SUMMARY
Synapses in the brain exhibit cell-type-specific differences in basal synaptic transmission and plasticity. Here, we evaluated cell-type-specific specializations in the composition of glutamatergic synapses, identifying Btbd11 as an inhibitory interneuron-specific, synapse-enriched protein. Btbd11 is highly conserved across species and binds to core postsynaptic proteins, including Psd-95. Intriguingly, we show that Btbd11 can undergo liquid-liquid phase separation when expressed with Psd-95, supporting the idea that the glutamatergic postsynaptic density in synapses in inhibitory interneurons exists in a phase-separated state. Knockout of Btbd11 decreased glutamatergic signaling onto parvalbumin-positive interneurons. Further, both in vitro and in vivo, Btbd11 knockout disrupts network activity. At the behavioral level, Btbd11 knockout from interneurons alters exploratory behavior, measures of anxiety, and sensitizes mice to pharmacologically induced hyperactivity following NMDA receptor antagonist challenge. Our findings identify a cell-type-specific mechanism that supports glutamatergic synapse function in inhibitory interneurons—with implications for circuit function and animal behavior.
Graphical Abstract
In brief
Bygrave et al. identify Btbd11 as an inhibitory interneuron-specific protein localized at glutamatergic (excitatory) synapses. Conditional knockout of Btbd11 from inhibitory interneurons disrupts the function of glutamatergic synapses, neuronal circuit activity, and behaviors related to anxiety.
INTRODUCTION
The postsynaptic density (PSD) of glutamatergic synapses in the brain is a densely packed, protein-rich structure that supports excitatory synaptic transmission and synaptic plasticity. Our understanding of the PSD largely comes from studying excitatory neurons (ENs), which make up 80%–90% of neurons in cortical and hippocampal circuits.1 Impaired function of inhibitory interneurons (INs) is associated with psychiatric disease and neurologicaldisorders.2–4More specifically, impaired glutamatergic excitation of INs, particularly parvalbumin (PV)-positive INs, is linked to the pathophysiology of schizophrenia.3,4 PV-INs have been extensively studied, and their role in regulating circuit activity (i.e., controlling the precise timing of EN cell firing), supporting rhythmic neuronal activity (i.e., generation of gamma oscillations), and controlling animal behavior is well documented.5–9 Interestingly, both basal glutamatergic transmission and plasticity in INs (including PV-INs) is different from those in ENs.10–13 However, very little is known about the molecular composition of the IN glutamatergic PSD (inPSD) and whether cell-type-specific specializations exist to govern these distinct properties.
Recent evidence indicates that many components of the glutamatergic PSD undergo liquid-liquid phase separation (LLPS), suggesting that the PSD likely exists as a phase-separated structure that aids synapse function.14–17 LLPS could be particularly important for the stability and function of inPSDs, which are frequently located directly on the dendritic shaft of INs1 rather than being nested within dendritic spines as is common in ENs (but note, Sancho and Bloodgood18 and Keck et al.19). Molecular mechanisms must support PSD stability, as shaft glutamatergic synapses located within PV-INs are, on average, more stable than their counterparts in ENs when assessed with longitudinal imaging in vivo.20 Here, we use a combination of genetic tools and proteomics to identify cell-type-specific PSD proteins. We characterize Btbd11 as an inPSD that undergoes LLPS and regulates glutamatergic synapses in INs as well as neuronal circuit function. Furthermore, our data highlight how LLPS—a critical regulator of cellular function across biology—can be achieved in the same subcellular compartment (i.e., the synapse) in a cell-type-specific manner through molecular specializations.
RESULTS
Identification of Btbd11 as an inPSD protein
To gain insights into the molecular composition of inPSDs, we bred conditional Psd-95-GFP knockin mice (second-generation mice based on Fortin et al.21) with vGATCre or CaMKIICre animals, tagging a core scaffold protein at glutamatergic synapses in INs or ENs, respectively. We collected cortex and hippocampal tissue from these animals and performed GFP-immuno-isolation experiments (Figure 1A). To compensate for the lower abundance of INs, we pooled samples from 4 vGATCre:Psd-95-GFP mice to compare with 1 CaMKIICre:Psd-95-GFP mouse. Utilizing mass spectrometry and label-free quantification, we identified immuno-isolated PSD complexes preferentially originating from INs or ENs. Normalizing to the amount of Psd-95, we found proteins that were specifically enriched in INs (Figure 1B; Table S1). Such proteins were considered putative inPSD proteins. The top hit was Btbd11, an ankyrin repeat and BTB/POZ domain-containing protein.
Figure 1. Identification of Btbd11 as an inPSD protein.
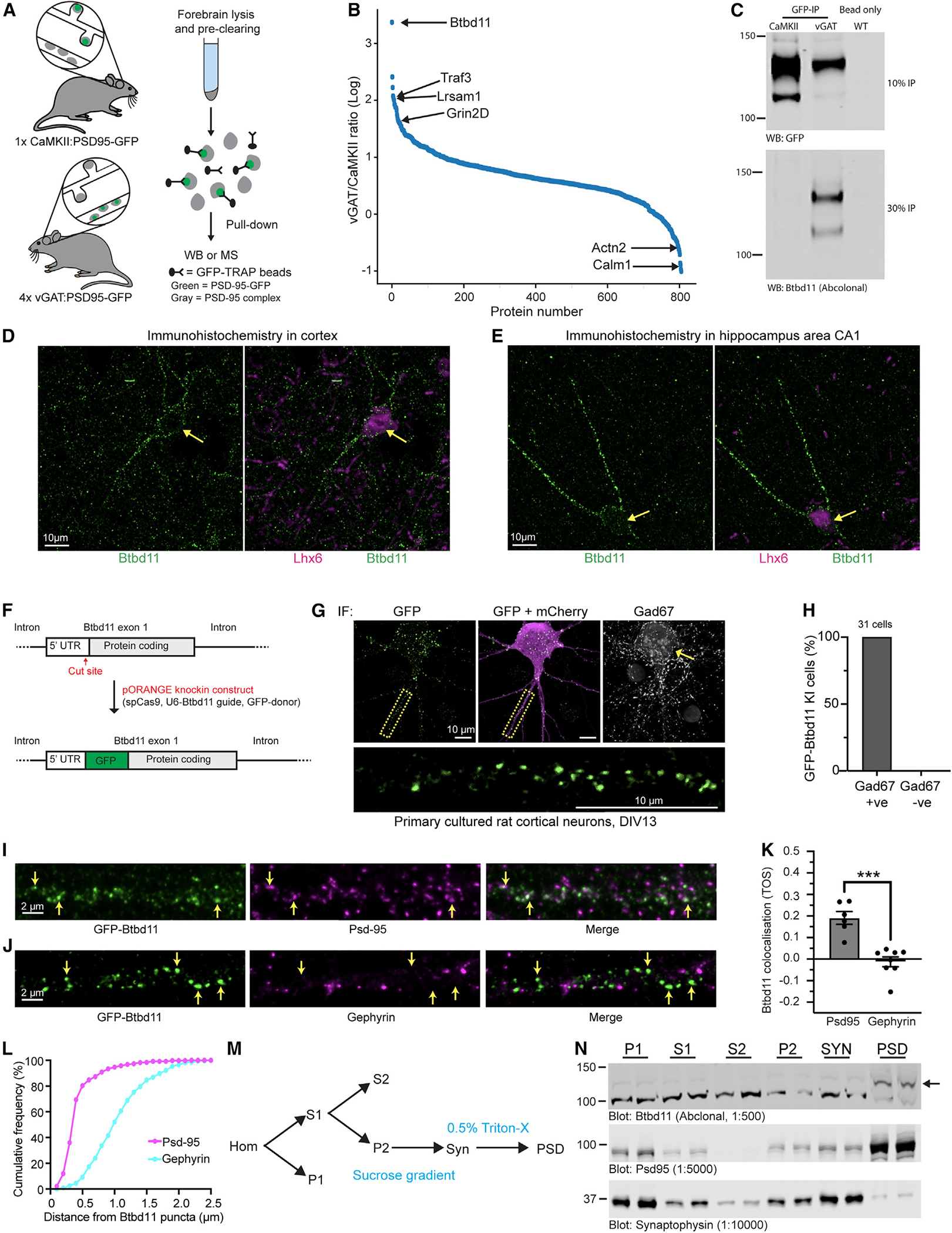
(A) Schematic of Psd-95-GFP knockin in CaMKII or vGAT positive neurons to label excitatory and inhibitory neurons, respectively.
(B) Proteins identified using semi-quantitative mass spectrometry.
(C) Psd-95-GFP immuno-isolation with CaMKII:Psd-95-GFP and 4× vGAT:Psd-95-GFP mice (pooled) or a WT animal used for a bead-only control followed by western blots for GFP (detecting Psd-95-GFP, the “bait”) from 10% of the pull-down or Btbd11 (a candidate inPSD) from 30% of the pull-down material. This is a representative example of an experiment conducted 3 times.
(D and E) Immunohistochemistry in WT mouse brain slices for Btbd11 (green) or Lhx6 (magenta) in layer 2/3 of the visual cortex (D) and area CA1 (E) of the hippocampus. Note the Btbd11-positive cells are expressing Lhx6, a marker for somatostatin and parvalbumin inhibitory interneurons.
(F) Schematic of the CRISPR knockin strategy used to label endogenous Btbd11.
(G) Example of GFP-Btbd11 knockin cell from primary cultured rat cortical neurons electroporated with knockin constructs and a mCherry cell fill (magenta). Immunofluorescence was used to boost the GFP-Btbd11 signal (green) and to identify GAD67-positive INs. Yellow arrow indicates a GAD67-positive cell. The area in the yellow dotted box is enlarged to show the GFP-Btbd11 signal. Respective scale bars indicate 10 μm.
(H) All 31 GFP-Btbd11 knockin cells were also GAD67 positive.
(I and J) Zoom in along the dendrites of GFP-Btbd11 knockin cells with immunofluorescence for Psd-95 or gephyrin, respectively. Yellow arrows indicate a subset of Btbd11 puncta. Scale bars indicate 2 μm.
(K) Co-localization of GFP-Btbd11 puncta (identified with CRISPR knockin [KI]) with Psd-95 and gephyrin (visualized with immunofluorescence) using the threshold overlap score (TOS). Bars indicate the mean, and error bars show the SEM. ***p < 0.001.
(L) Cumulative frequency plots indicating the puncta-to-puncta distance of Btbd11 with Psd-95 (magenta) or gephyrin (blue).
(M) Schematic of the fractionation protocol used to isolate the postsynaptic density (PSD) from mouse cortex. P1, crude nuclear; S1 and S2, supernatant; P2, crude synaptosomal pellet; Syn, synaptosomes; PSD, postsynaptic density.
(N) Western blot analysis of Btbd11 levels (top blot, 12 μg protein loaded) in different biochemical fractions of WT animals (1 male and 1 female, 3 months). Note the enrichment of Btbd11 in the PSD fraction. The band corresponding to Btbd11 is indicated with a black arrow (predicted weight 121.6 kDa). The fractionation efficiency was demonstrated with blots for Psd-95 (middle blot, enrichment in PSD) and synaptophysin (bottom blot, enrichment in SYN) with 2 μg protein loaded for each. See Table S2 for full statistical information on all tests.
Btbd11 was previously identified in both Psd-95 pull-down and proximity labeling experiments,22,23 but its cell-type-specific expression pattern was not explored. Furthermore, a biological function of Btbd11 remains completely unknown, in the brain or otherwise. To confirm our mass spectrometry results, we repeated GFP-immuno-isolation experiments and used western blots to confirm that Btbd11 was selectively pulled down from IN samples but not with control beads (Figures 1C and S1B). As an additional validation, we generated and validated an antibody against Btbd11 (Figure S1A) and could selectively detect Btbd11 from vGAT:Psd95-GFP samples (Figure S1C). We then explored published RNA datasets and resources and found that Btbd11 mRNA is consistent with exclusive expression in INs in the cortex and hippocampus of mice (Figure S1D).24,25
Using immunofluorescence for Btbd11 in cultured mouse hippocampal neurons, we observed punctate Btbd11 expression in GAD67-positive INs (Figures S1E and S1F). Importantly, we validated the specificity of the commercial Btbd11 antibody with Btbd11 conditional knockout (KO) mice (Figures S1E and S1F; KO mice described later). Using this Btbd11 antibody, we also detected Btbd11 in INs in the visual cortex (Figure 1D) and area CA1 of the hippocampus (Figure 1E). As in the primary neuronal cultures, the Btbd11 signal was punctate and looked to be present along dendrites. For an orthogonal validation, we designed CRISPR knockin constructs to label endogenous Btbd11 using the ORANGE method.26 We opted to tag Btbd11 at the N terminus with GFP and electroporated rat primary cultures cortical neurons with the knockin construct alongside an mCherry cell fill (Figure 1F). We observed a sparse population of cells with punctate GFP signals, which, as expected, were also positive for GAD67 (Figure 1G). Indeed, of the 31 knockin (KI) cells imaged 100% were also GAD67 positive (Figure 1H), confirming the IN-specific expression of Btbd11 in cortical neurons. We next coupled this CRISPR KI approach with immunofluorescence to evaluate if Btbd11 puncta were enriched at synapses. GFP-Btbd11 puncta overlapped with Psd-95 puncta (Figure 1I). In contrast, there was very little overlap with gephyrin, a marker of GABAergic inhibitory synapses (Figure 1J). We quantified the relative enrichment at glutamatergic synapses by calculating the co-localization of Btbd11 with Psd-95 and Gephyrin. Btbd11 was significantly more co-localized with Psd-95 compared with gephyrin (Figure 1K; see Table S1 for full statistical test details for this and all subsequent analyses). The same data were also analyzed by calculating the distances between GFP-Btbd11 puncta and Psd-95 or gephyrin puncta. We found that the puncta-to-puncta distance was considerably smaller at glutamatergic synapses, in line with the co-localization results (Figure 1L). These data indicate that Btbd11 is localized at glutamatergic synapses. We subsequently performed biochemical fractionation of wild-type (WT) mouse cortex (Figure 1M) and found that Btbd11 was enriched in the PSD fraction (Figure 1N).
Collectively, these data highlight that there are cell-type-specific differences in the composition of glutamatergic synapses and establish Btbd11 as an inPSD protein that is selectively located at glutamatergic synapses.
Btbd11 contains a PDZ binding motif that interacts with PDZ1,2 of Psd-95
Btbd11 is annotated to contain 5 ankyrin repeats, a BTB domain, and C-terminal amino acids consistent with a PDZ binding motif (PBM; Figure 2A). Utilizing protein structure prediction algorithms from AlphaFold,27 we observed that Btbd11 contains a region of potential disorder (Figure 2B; Video S1). The C-terminal region of Btbd11 is highly conserved between species, highlighting the potential importance of the PBM (Figure 2C). PBMs support interactions with PDZ domain-containing proteins, including the membrane-associated guanylate kinase (MAGUK) family of proteins such as Psd-95. We hypothesized that the PBM of Btbd11 might support a direct interaction with the PDZ domains of Psd-95.
Figure 2. Btbd11 contains a PDZ binding motif that interacts with PDZ1,2 of Psd-95.
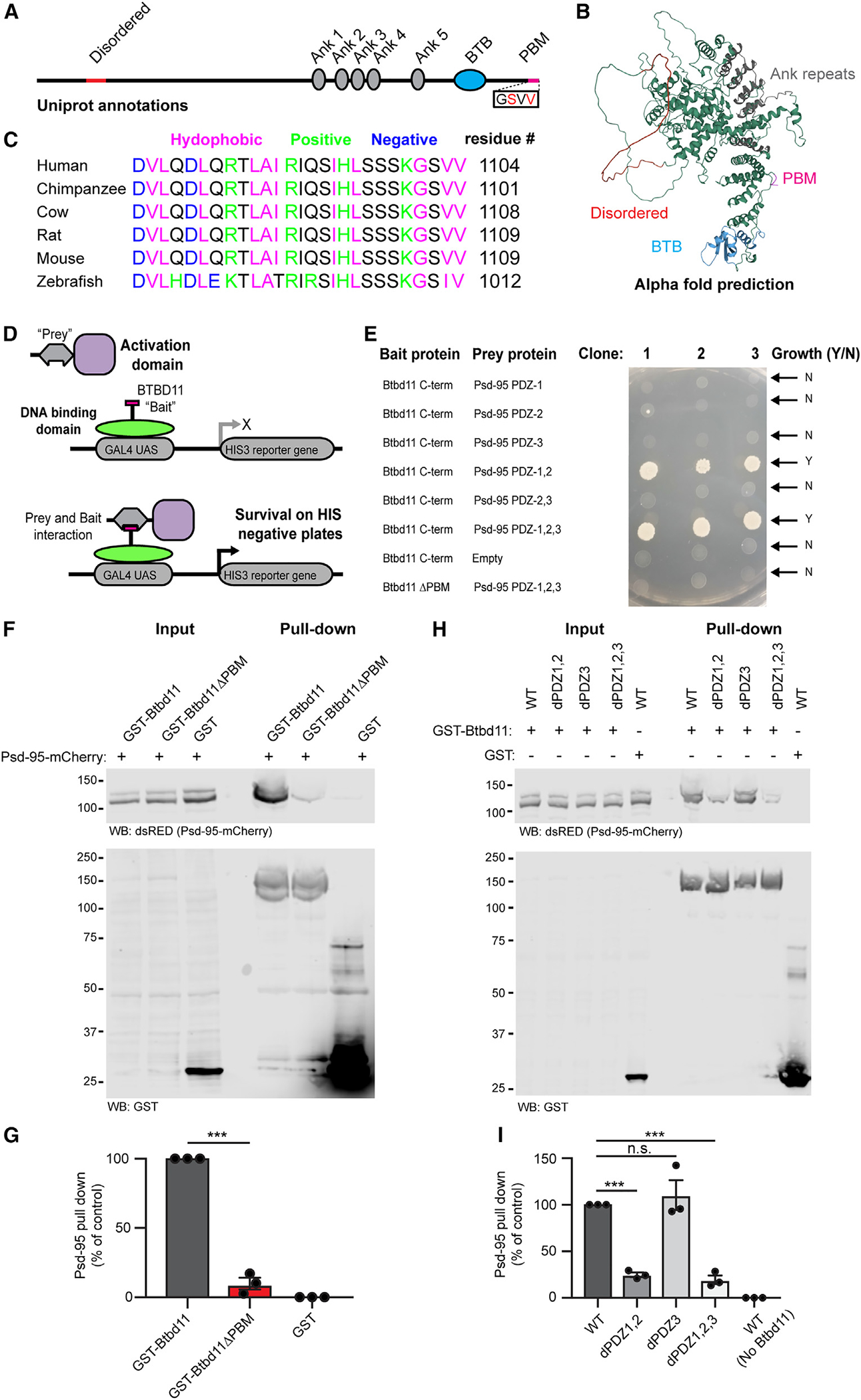
(A) Schematic depiction of Btbd11 with annotations from Uniprot. Red: disordered, gray: ankyrin repeats (Ank), blue: BTB domain (BTB), magenta: PDZ binding motif (PDM).
(B) Predicted structure of Btbd11 from AlphaFold with domains shaded as in (A).
(C) C-terminal region of Btbd11 in different species showing the conservation of the PBM.
(D) Schematic of targeted yeast 2-hybrid experiment to assess binding of Btbd11 with different PDZ domains of Psd-95.
(E) Results from targeted yeast 2-hybrid experiment with growth indicating an interaction between Btbd11 and Psd-95.
(F) GST pull-down experiments evaluating the ability of GST-Btbd11 or a mutant lacking the PBM (GST-Btbd11ΔPBM) to interact with Psd-95-mCherry in HEK cells. GST only was included as a negative control.
(G) Qualification of Psd-95-mCherry pulled down by GST-Btbd11 or GST-Btbd11ΔPBM.
(H) GST pull-down experiments in HEK cells evaluating the ability of GST-Btbd11 to interact with Psd-95-mCherry point mutants designed to disrupt PDZ domain binding.
(I) Quantification of Psd-95-mCherry mutants pulled down by GST-Btbd11. Bars display the mean, and error bars display SEM. ***p < 0.001.
To test an interaction between Btbd11 and Psd-95 through PBM-PDZ domain interactions, we used a yeast 2-hybrid approach. The C terminus of Btbd11 ± the PBM was used as “bait,” and different combinations of the 3 PDZ domains in Psd-95 were used as “prey” (Figure 2D). We only observed yeast growth, a readout of a protein-protein interaction, when PDZ domains 1,2 or 1,2,3 were present (Figure 2E). Btbd11 lacking the PBM did not interact with PDZ 1,2,3 of Psd-95, highlighting the necessity of the PBM for binding.
As a complementary approach, we adopted a GST pull-down strategy in mammalian cells. We cloned full-length GST-Btbd11 ± the PBM and expressed these GST fusion proteins (or GST only) in HEK cells alongside Psd-95-mCherry, then lysed the cells and performed a GST pull-down. Btbd11 could pull down Psd-95-mCherry but only when the PBM was present (Figures 2F and 2G). To confirm the binding was mediated by PDZ1,2 of Psd-95, we cloned mutants of Psd-95-mCherry in which the individual PDZ domain function was disrupted (dPDZ1–3) based on previously generated mutants.28 Confirming our observations in yeast, we found that diminished binding to Btbd11 with Psd-95 harbored mutations in PDZ1,2 or PDZ1,2,3 (Figures 2H and 2I). Mutations to PDZ3 alone had no discernible effect on Btbd11 binding, highlighting the specificity of the interaction.
Having established a direct interaction with Psd-95, we sought to identify other potential interaction partners. We conducted a yeast 2-hybrid screen using either the BTB domain or the C-terminal region containing the PBM of Btbd11 as bait. From the screen with the C terminus of Btbd11, we identified well-known PDZ-containing synaptic proteins including Psd-93, Sap-102, and Pick1 as putative interactors (Figure S2). Using the BTB domain, we pulled out Ataxin1 and Ataxin-1-like as potential interactors (Figure S2). Although not yet tested, we expect that the tandem ankyrin repeats of Btbd11 facilitate other protein-protein interactions, as is common for proteins with ankyrin repeats.29
LLPS of Btbd11 when expressed with Psd-95
We next sought to explore the properties of exogenously expressed Btbd11. To do this, we cloned GFP- and mCherry-tagged Btbd11 with or without the PBM necessary for an interaction with Psd-95. In HEK cells, expression of GFP-Btbd11 alone led to the formation of striking fibril-like structures (Figure 3A). Remarkably, when Psd-95-mCherry was co-expressed with GFP-Btbd11, the fibril structures were replaced with large spherical intracellular droplets with a high degree of Btbd11 and Psd-95 co-localization (Figures 3B and 3C). Droplet formation was critically dependent upon an interaction between Btbd11 and Psd-95 since deletion of the PBM of Btbd11 abolished their formation (Figures 3D and 3E). By contrast, formation of fibrils was not dependent upon the PBM (see Figure S3 for quantification of fibril and droplet formation under different conditions).
Figure 3. Liquid-liquid phase separation of Btbd11 when expressed with Psd-95.
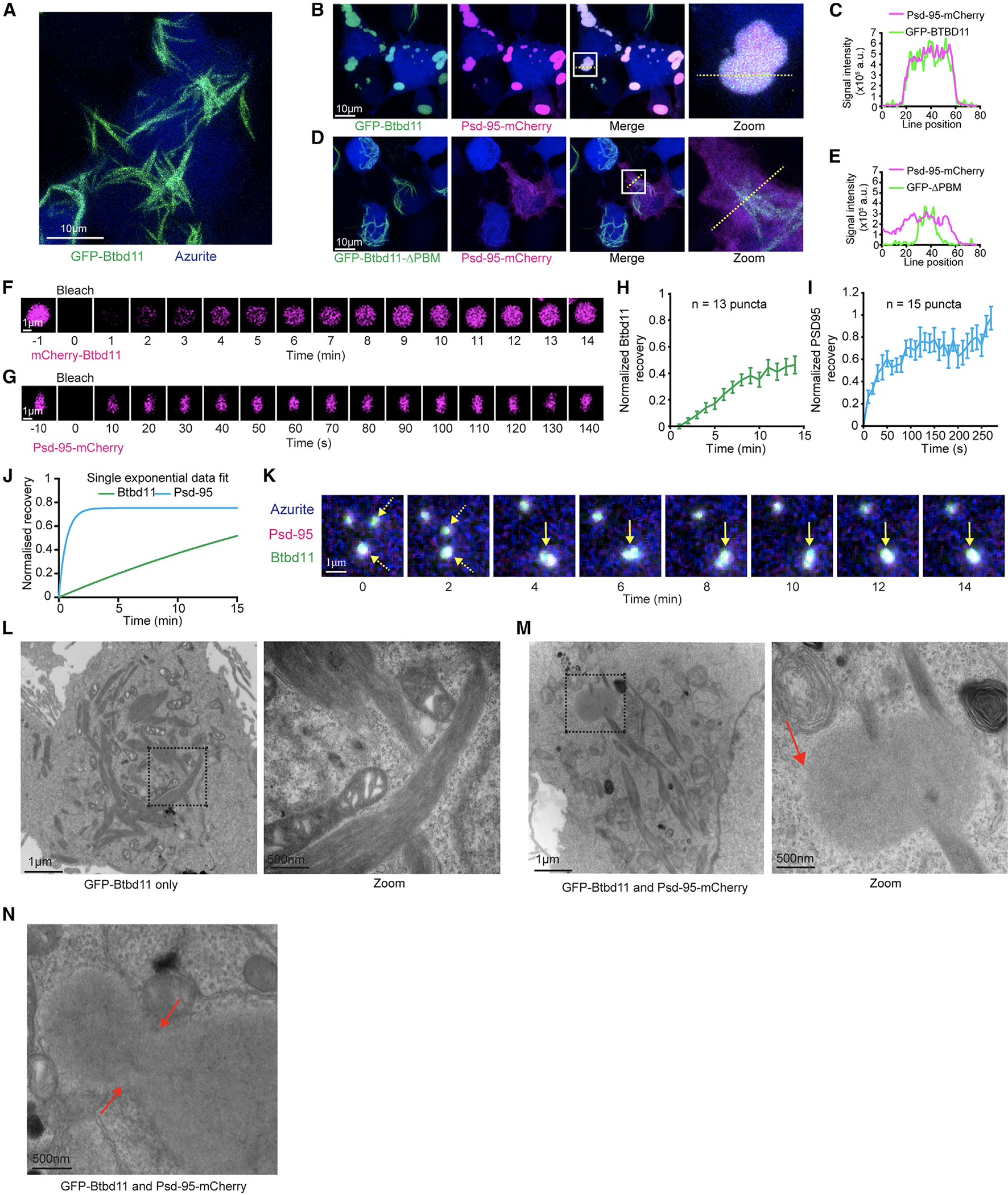
(A) Expression of GFP-Btbd11 (green) and an azurite cell fill (blue) in HEK cells led to the formation of large fibril-like assemblies. Scale bar: 10 μm.
(B and C) Co-expression of GFP-Btbd11 (green) with Psd-95-mCherry (magenta) and an azurite cell fill (blue). The region within the white box is enlarged on the right (zoom), and the yellow dotted line indicates where a line scan (C) is shown for Psd-95-mCherry and GFP-Btbd11.
(D and E) Co-expression of GFP-Btbd11ΔPBM (green) with Psd-95-mCherry (magenta) and an azurite cell fill (blue). The region within the white box is enlarged on the right (zoom), and the yellow dotted line indicates where a line scan (E) is shown for Psd-95-mCherry and GFP-Btbd11ΔPBM.
(F and G) Fluorescence recovery after photobleaching (FRAP) of mCherry-Btbd11 and Psd-95-mCherry co-expressed with Psd-95-GFP and GFP-Btbd11, respectively. Note the difference in timescales for Btbd11 and Psd-95. Scale bar: 1 μm.
(H and I) Quantification of fluorescence recovery for Btbd11 (n = 13) and Psd-95 (n = 15), respectively. Error bars indicate SEM.
(J) Plot of the exponential fit of FRAP data for Btbd11 (green) and Psd-95 (blue).
(K) Longitudinal confocal imaging of a HEK cell transfected with GFP-Btbd11 (green), Psd-95-mCherry (magenta), and an azurite cell fill (blue). Yellow dotted arrows indicate two puncta that come together and form a single droplet (solid yellow arrow). Scale bar: 1 μm.
(L and M) Electron microscope images of HEK cells transfected with GFP-Btbd11 or GFP-Btbd11 and Psd-95-mCherry, respectively. The black dotted box is enlarged on the right of each panel. Respective scale bars are indicted at the bottom left of each image. In (M), the red arrow indicates a putative droplet observed with Btbd11 and Psd-95 co-expression but not with Btbd11 expression alone.
(N) An example of a putative Btbd11 and Psd-95 droplet that looks to be in the process of fusion or fission with red arrows indicating the neck. Respective scale bars are indicated at the bottom left of each image.
The spherical puncta formed by Btbd11 and Psd-95 were reminiscent of biomolecular condensates arising from LLPS of abundant PSD proteins including SynGAP and Psd-95.16 A hallmark of LLPS is fluorescent recovery after photobleaching (FRAP), indicating a dynamic exchange of biomolecules between putative condensates and the surrounding cytosol—a phenomenon that is not expected if puncta are formed from protein aggregates locked into a solid state. To test this, we transiently expressed mCherry-Btbd11 with Psd-95-GFP (Figure 3F) or Psd-95-mCherry with GFP-Btbd11 (Figure 3G) and photobleached individual puncta in HEK cells. We observed signal recovery for both Btbd11 (Figure 3H) and Psd-95 (Figure 3I), with Psd-95 recovering at a faster rate (Figure 3J). It is important to note that FRAP alone does not definitively confirm LLPS.30
LLPS can be facilitated by proteins that contain regions of intrinsic disorder.31 Btbd11 contains predicted regions of disorder within its N-terminal region (Figures 2A and 2B) that we speculated could promote LLPS. To test this, we cloned a GFP- and mCherry-Btbd11 that lacked a large portion of the N terminus upstream of the ankyrin repeats (5xANK-BTB). When expressed with Psd-95, large aggregates formed in which there was also a high degree of co-localization between 5xANK-BTB and Psd-95 (Figure S3). However, in addition to lacking the spherical droplets observed with full-length Btbd11, the 5xANK-BTB assemblies did not show FRAP, indicating the structures were unlikely to be liquid-like assemblies (Figure S3).
Another hallmark of LLPS is the coalescence of puncta over time to form larger droplets. Through time-lapse imaging of HEK cells expressing GFP-Btbd11 and Psd-95-mCherry, we observed fusion of puncta over time (Figure 3K; Video S2). Frequently purified proteins—or portions of proteins—are mixed in vitro to validate LLPS. Our attempts to purify Btbd11 to conduct such experiments have so far been unsuccessful, likely because Btbd11 readily forms fibrils, making protein purification challenging. Therefore, to get an orthogonal validation that the puncta were true “membrane-less organelles,” we performed electron microscopy on HEK cells expressing either GFP-Btbd11 alone or in combination with Psd-95-mCherry. In the Btbd11-only condition, we found cells with fibril-like structures (Figure 3L). We observed fibril-like structures when Btbd11 was expressed with Psd-95 but also identified droplet-like assemblies in the cytoplasm, which occasionally even appeared to be impaled by the fibrils (Figure 3M). Critically, these droplets appeared electron dense but clearly lacked any kind of lipid bilayer. Furthermore, we found examples of droplets that looked like they could be in the process of fusion (Figure 3N).
Exogenous expression of Btbd11 stabilizes Psd-95 at glutamatergic synapses
We next tested exogenous expression of GFP-Btbd11 in rat hippocampal neurons. Large fibril-like structures formed in the cell body and dendrites of transfected cells (Figure 4A). These fibril structures were stable, as observed by a lack of FRAP when a portion of the fibril was bleached (Video S3). GFP-Btbd11 was also observed at glutamatergic synapses when imaged with higher magnification, as indicated by co-localization with Psd-95, but not gephyrin (Figure 4B), confirming our previous observations with endogenously tagged Btbd11. We speculated that Btbd11’s PBM would be necessary for proper synaptic targeting and confirmed this by showing that there was no synapse localization of Btbd11 that lacked a PBM (Figures 4C and 4D).
Figure 4. Exogenous expression of Btbd11 stabilizes Psd-95 at glutamatergic synapses.
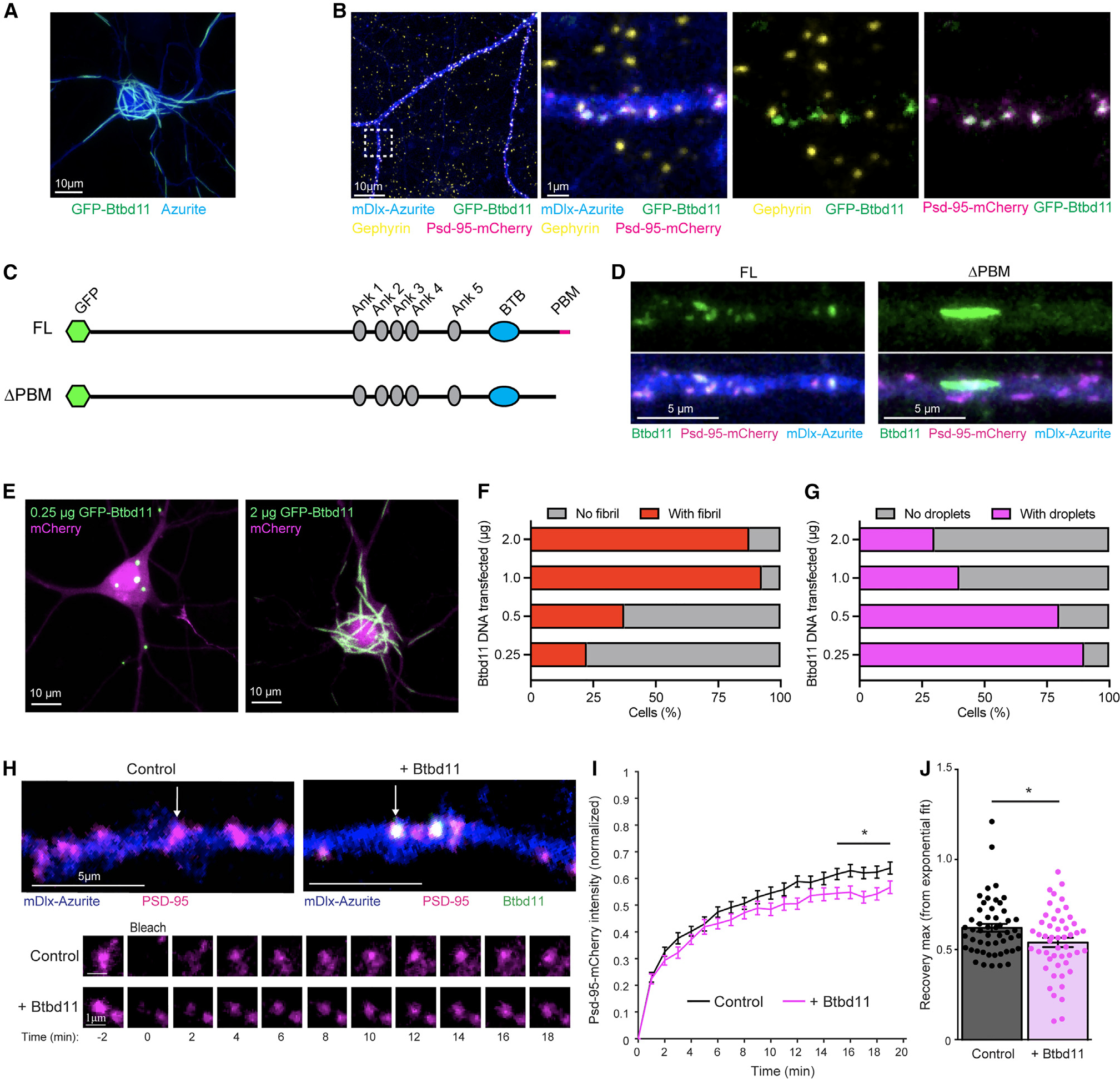
(A) Confocal image from a primary cultured rat hippocampal neuron transfected with GFP-Btbd11 (green) and azurite as a cell fill (blue). Scale bar: 10 μm.
(B) Confocal image from a primary cultured putative IN transfected with GFP-Btbd11 (green), Psd-95-mCherry (magenta), and mDlx-azurite (blue) with immunohistochemistry for gephyrin (yellow). The region within the white dotted box is enlarged on the right with different combinations of channels. Scale bars are in the bottom left of the images.
(C) Schematic of GFP-Btbd11 and GFP-Btbd11ΔPBM constructs.
(D) Confocal image of putative primary cultured hippocampal interneurons, identified with an azurite cell fill (blue) under the control of the mDlx enhancer to drive expression in INs. Full-length (FL) GFP-Btbd11 (left) and GFP-Btbd11ΔPBM (ΔPBM, right) shown in green were co-expressed with Psd-95-mCherry (magenta). Scale bar: 5 μm.
(E) Confocal image from transfected primary cultured rat hippocampal neurons with varying amounts of GFP-Btbd11 (green) and mCherry as a cell fill (magenta). Scale bar: 10 μm.
(F and G) Quantification of the proportion of cells with fibrils (red bars) or droplets (magenta bars), respectively, when GFP-Btbd11 is transfected in different quantities.
(H) Live-cell confocal imaging and FRAP experiments in putative INs (identified with mDlx-azurite) in which Psd-95-mCherry (magenta) is bleached when expressed alone or in the presence of GFP-Btbd11 (green). The bottom panels show FRAP of the individual puncta labeled in the top panels with a white arrow.
(I) Quantification of Psd-95-mCherry FRAP under control conditions (black) or with overexpression of Btbd11 (magenta). *p < 0.05.
(J) Quantification of the estimated recovery maximum from an exponential fit of the FRAP data for each bleached punctum. Lines or bars show the mean, and error bars display SEM.
The formation of fibril-like structures is common to proteins that also undergo LLPS, with liquid-to-solid phase transitions occurring at saturating concentrations.30,32 We tested if fibril formation was dose dependent by expressing increasing amounts of GFP-Btbd11 DNA (0.25 – 2 μg/well) alongside an mCherry cell fill (Figure 4E). While there was variability in the amount of DNA transfected within each transfection dose, clear patterns emerged whereby at high doses, almost all mCherry-positive cells contained Btbd11 fibrils (Figure 4F). Interestingly, at the lower doses, we observed many cells that contained small droplet-like assemblies reminiscent of the LLPS condensates observed in HEK cells (Figure 4G, and see Figure 3). We speculate that these droplets form when Btbd11 interacts with endogenous Psd-95 and that the fibrils form when a concentration threshold is surpassed. As fibril structures were not observed with our endogenous CRISPR labeling of Btbd11 (at least under the basal conditions evaluated), we imagine that fibril formation is unlikely to be physiologically relevant to cellular function—but does highlight intriguing properties of the protein.
The stability of Psd-95 is the same in INs as in ENs, despite INs generally lacking dendritic spines.21 We independently confirmed these observations with FRAP of overexpressed Psd-95-mCherry in putative INs or ENs (Figures S4A–S4C). It has been postulated that biomolecular condensation through LLPS at the synapse might promote the stability of densely packed PSD proteins against the forces of Brownian motion.14,17 We suspect that inPSDs have protein specializations to promote their stability and function. Could Btbd11 promote the stability of Psd-95 at the synapse, possibly by driving LLPS? To explore this possibility, we expressed Psd-95-mCherry with or without GFP-Btbd11 in putative INs in hippocampal cultures identified with an Azurite cell fill under the control of the mDlx enhancer.33 Psd-95 stability was monitored by FRAP of Psd-95-mCherry puncta and the recovery curves compared with or without the addition of GFP-Btbd11 (Figure 4H). Psd-95-mCherry FRAP was slower in the presence of overexpressed Btbd11, indicating a larger immobile fraction—or a stabilizing effect on Psd-95 at the synapse (Figures 4I and 4J). Intriguingly, when Btbd11 was overexpressed in ENs (which normally lack Btbd11), no stabilization of Btbd11 was observed (Figures S4D and 4E). It is possible that the stabilizing effect on Psd-95 results from Btbd11 promoting LLPS at the PSD.
Btbd11 KO reduces glutamatergic signaling in PV-INs
We next explored the effects of Btbd11 loss of function through genetic KO by generating Btbd11 gene-trap mice using in vitro fertilization and frozen sperm from the European Mutant Mouse Archive (https://www.infrafrontier.eu). The gene-trap mice did not show reduced levels of Btbd11 mRNA or protein (Figures S5A and S5B), probably due to a truncation in the gene-trap cassette of this line.34 By crossing the gene-trap mice with a constitutive Flp recombinase line, we converted the gene-trap mice into conditional KO animals (Btbd11F/F). This flanks the critical exon 5 with loxP sites (Figure S5C). We validated Btbd11 KO using primary cultured cortical neurons from Btbd11F/F mice with adeno-associated viruses (AAVs) delivering GFP or GFP-Cre. There was reliable loss of Btbd11 mRNA and protein in the presence of GFP-Cre, observed with northern blots and western blots, respectively (Figures 5A and 5B).
Figure 5. Btbd11 KO reduces glutamatergic signaling in PV-INs.
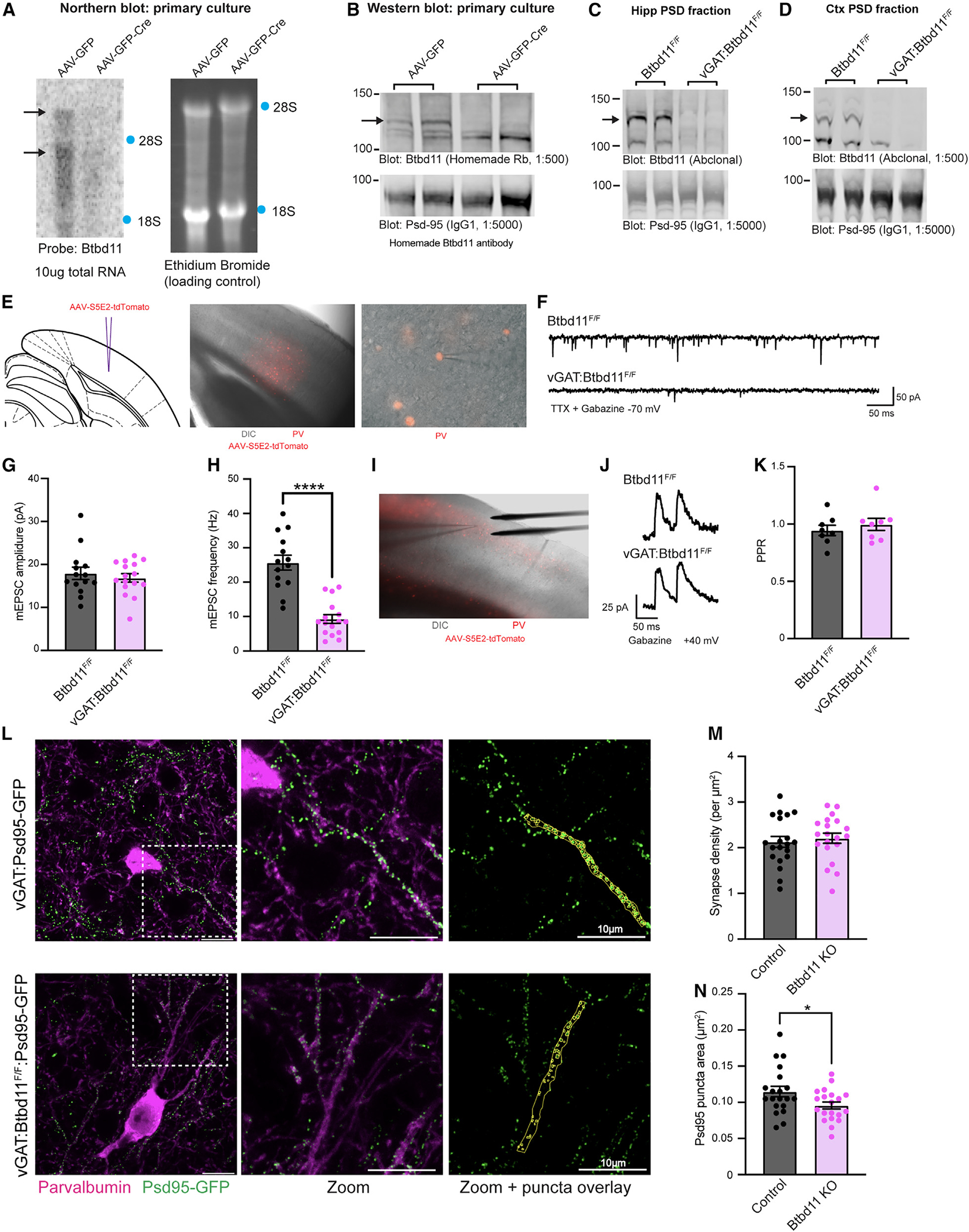
(A) Northern blot to evaluate levels of Btbd11 mRNA in Btbd11F/F cultures transduced with AAV-GFP (left lane) or AAV-GFP-Cre (right lane) and harvested at DIV14. 10 μg RNA was loaded onto a gel, and ethidium bromide staining was used to confirm equal loading of RNA. Locations of 18S and 28S ribosomal RNA are indicated with blue dots.
(B) Western blots characterizing the levels of Btbd11 (homemade antibody, top blot) and Psd-95 (bottom blot, as a loading control) in primary cortical Btbd11F/F cultures transduced with AAV-GFP (left lanes) or AAV-GFP-Cre (right lanes). 12 μg protein was loaded from the PSD fraction. Cells harvested at DIV12 and DIV14 (one sample per condition per time point). A black arrow indicates the band corresponding to Btbd11.
(C and D) Western blots evaluating the levels of Btbd11 (Abclonal antibody, top blot) and Psd-95 (bottom blot, as a loading control) from the (C) hippocampal and (D) cortical PSD fractions of female Btbd11F/F or vGAT:Btbd11F/F mice aged 3 months. 20 μg protein was loaded. Note the hippocampal PSD material was not generated with a sucrose gradient and so serves as a proxy for the PSD fraction. A black arrow indicates the band corresponding to Btbd11.
(E) Schematic depicting the site of AAV-S5E2-tdTomato injection, used to visualize PV-INs in the V1 (left), with zoomed-out (middle) and zoomed-in (right) merged images of differential interference contrast (DIC) and mCherry fluorescence.
(F) Example mEPSC traces recorded from V1 PV-INs in Btbd11F/F or vGAT:Btbd11F/F mice recorded at −70 mV in the presence of TTX and gabazine.
(G and H) The mEPSC amplitude and frequency, respectively, of mEPSCs recorded from PV-INs from Btbd11F/F (black) animals or vGAT:Btbd11F/F (magenta) mice. ****p < 0.0001.
(I) DIC image showing the placement of the electrical stimulating electrode for recordings in the V1.
(J) Example traces used to calculate the PPR in Btbd11F/F or vGAT:Btbd11F/F mice with cells held at +40 mV (to allow for the AMPA/NMDA ratio to also be calculated; see Figure S5G).
(K) Summary PPR data recorded from PV-INs from Btbd11F/F (black) or vGAT:Btbd11F/F (magenta) mice.
(L) Representative confocal images (maximum intensity projections) from the V1 of vGAT:Psd-95-GFP (control) or vGAT:Btbd11F/F:Psd-95-GFP (Btbd11 KO) slices. Endogenous Psd-95-GFP (green) shows putative glutamatergic synapses and is visualized alongside PV detected with immunohistochemistry (magenta). Psd-95-GFP puncta in PV neuron dendrites were manually identified (yellow regions of interest [ROIs], right panel). Scale bar: 10 μm.
(M) Glutamatergic synapse density, calculated by the density of Psd-95-GFP puncta, in PV-INs in control and Btbd11 KO animals.
(N) Median Psd-95-GFP puncta area per dendritic region in PV-INs of control and Btbd11 KO animals. *p < 0.05. Bars display the mean and error bars show S.E.M. For further details, see Figure S5.
To create IN-specific Btbd11KO mice, we bredvGATCre/Wtmice with Btbd11F/F animals to generate litters of vGATWt/Wt:Btbd11F/F (control [CON]) and vGATCre/Wt:Btbd11F/F (vGAT-Btbd11 KO) animals. Confirming our previous data that Btbd11 was specific to INs, we observed a total loss of Btbd11 in the hippocampal (Figure 5C) and cortical (Figure 5D) PSD fractions of vGAT-Btbd11 KO mice. As PV-INs displayed the highest level of Btbd11 expression (based on RNA datasets; see Figure S1D), we decided to assess glutamatergic synapse function within PV-INs from CON or vGAT-Btbd11 KO mice by measuring miniature excitatory postsynaptic currents (mEPSCs) with whole-cell patch-clamp recordings. We expressed tdTomato in PN-INs of the visual cortex in CON or vGAT-Btbd11 KO littermates using an AAV with a PV-specific enhancer35 to enable fluorescently guided recordings (Figures 5E and 5F). While the amplitude remained unchanged between the conditions (Figure 5G), we observed a dramatic decrease in the frequency of mEPSCs (Figure 5H). The mEPSC rise and decay time was equivalent in PV-INs from CON and vGAT-Btbd11 KO mice (Figures S5D–S5F). A frequency decrease could reflect fewer functional glutamatergic synapses in PV-INs of vGAT-Btbd11 KO mice or decreased presynaptic release probability. We measured the paired-pulse ratio (PPR) using electrical stimulation to explore presynaptic release probabilities (Figures 5I and 5J). The PPR was comparable between groups, suggesting that presynaptic release probabilities were unchanged with Btbd11 KO from INs(Figure 5K). We also assessed the AMPA/NMDA ratio in PV-INs and found no difference between CON and vGAT-Btbd11 KO mice (Figure S5G). We next aimed to establish if the density of glutamatergic synapses was decreased in PV-INs in vGAT-Btbd11 KO mice. It can be challenging to conduct quantitative cell-specific immunohistochemistry (IHC) for synaptic proteins in dense neural tissue. Therefore, we used a genetic approach generating vGAT:Psd-95-GFP (CON) and vGAT:Btbd11F/F:Psd-95-GFP (Btbd11 KO) animals wherein all INs have Psd-95 labeled with GFP with or without Btbd11 present in INs. This enabled unambiguous counting of Psd-95-GFP puncta specifically in the dendrites of PV-INs (Figure 5L). Surprisingly, we saw no decrease in Psd-95-GFP puncta density in PV-INs, used as a proxy of glutamatergic synapse density (Figure 5M). However, we did observe that the median Psd-95-GFP puncta size was smaller in PV-INs from Btbd11 KO animals (Figure 5N).
Loss of Btbd11 impacts circuit function in vitro and in vivo
INs are well known to play a critical role in regulating the activity of neuronal circuits. Having found that vGAT-Btbd11 KO mice have reduced glutamatergic synaptic activity in PV-INs, we speculated that network properties might be abnormal following Btbd11 KO. To test this, we first returned to our in vitro cell culture system and prepared primary hippocampal cultures from postnatal day 0 (P0) Btbd11F/F pups. We delivered GFP (CON) or GFP-Cre (KO) and jRGEGCO1 to cultures through AAV transduction at day in vitro (DIV) 1–2 (Figure 6A). Spontaneous activity of the cultures was measured with live-cell imaging of the jRGECO1 signal to track Ca2+ dynamics. Large, and synchronous, Ca2+ transients were observed throughout the cultures (Videos S4 and S5). We quantify these activity patterns in CON and KO cultures (Figure 6B). As expected for a disinhibited network, there was an increased frequency of Ca2+ transients in KO cultures (Figure 6C). Interestingly, the expression of PV protein was dramatically upregulated both in KO hippocampal cultures (observed with immunofluorescence; Figures 6D and 6E) and KO cortical cultures (observed with western blots; Figures 6F and 6G), potentially reflecting elevated activity in the cultures since PV expression is dynamically regulated by activity of PV neurons.8
Figure 6. Loss of Btbd11 impacts circuit function in vitro and in vivo.
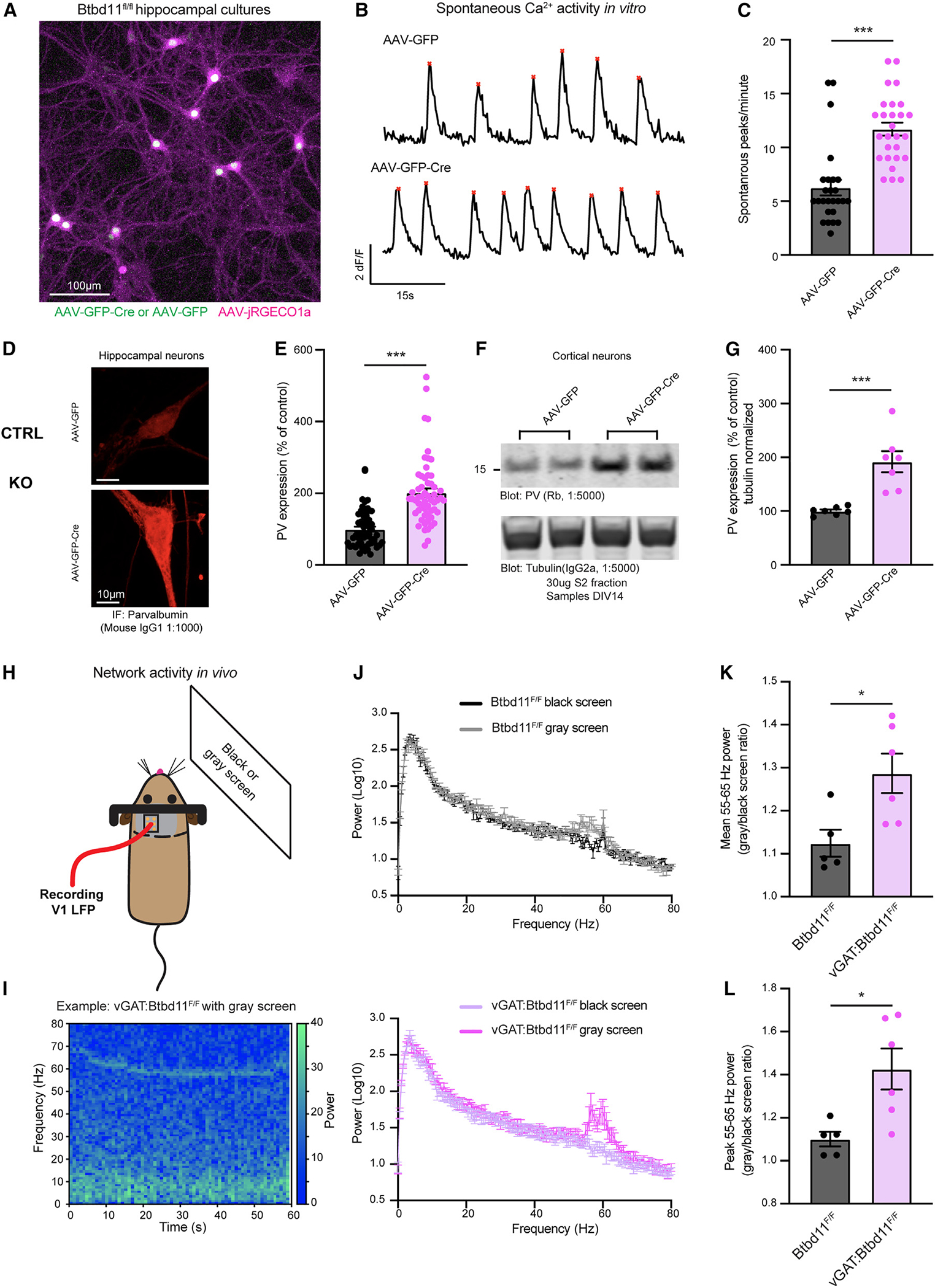
(A) Confocal live-cell imaging of primary cultured hippocampal neurons from Btbd11F/F mice transduced with AAV-jRGECO1a (magenta) and AAV-GFP or AAV-GFP-Cre (green). The example image is with AAV-GFP-Cre. Scale bar: 100 μm.
(B) Example traces for control (AAV-GFP, top) and KO (AAV-GFP-Cre, bottom) cultures showing the average Ca2+ activity across multiple neurons in one field of view over a 60 s period. Red stars indicate automatically identified peaks.
(C) Quantification of large Ca2+ transients across multiple ROIs and coverslips from 3 independent batches of neurons. Control (AAV-GFP) data are shown in black and KO (AAV-GFP-Cre) data in magenta. ***p < 0.001.
(D) Confocal image showing immunofluorescence of PV in DIV14 primary culture hippocampal Btbd11F/F neurons transduced with AAV-GFP (control, top) or AAV-GFP-Cre (KO, bottom). Scale bar: 10 μm.
(E) Quantification of PV immunofluorescence data with control data in black and KO data in magenta.
(F) Western blot from the cytosolic S2 fraction of DIV14 primary cultured cortical Btbd11F/F neurons transduced with AAV-GFP (control) or AAV-GFP-Cre (KO). Top blot shows levels of PV, and the bottom blot shows alpha-tubulin used as a loading control. 30 μg lysate was run.
(G) Quantification of western blot data evaluating levels of PV (normalized to alpha-tubulin levels) with control data in black and KO data in magenta.
(H) Schematic of in vivo setup used to assess narrowband gamma oscillations in the V1 with presentation of a gray screen.
(I) Example spectrogram showing the power over time in the 0–80 Hz range from the V1 of a vGAT:Btbd11F/F mouse presented with a gray screen.
(J) Power spectra for Btbd11F/F mice (n = 5, top; dark gray: dark screen, light gray: gray screen) and vGAT:Btbd11F/F mice (n = 6, bottom; light magenta: dark screen, dark magenta gray: gray screen).
(K and L) Quantification of the mean or peak 55–65 Hz activity, respectively, presented as a ratio of gray screen/black screen. *p < 0.05. Bars display mean, and error bars show SEM.
Next, we assessed circuit function in vivo using CON and vGAT-Btbd11 KO mice. PV-IN function has been closely tied to the regulation of fast local field potential (LFP) oscillations in the 30–100 Hz “gamma” frequency.5,6,9 A narrowband gamma oscillation ~55–65 Hz can be induced in the visual cortex through presentation of a gray screen alone.36 We exploited this straightforward assay to explore visually evoked gamma oscillations in the primary visual cortex (V1) of CON and vGAT-Btbd11 KO animals (Figures 6H and 6I). We implanted a tungsten electrode targeted to layer 4 of the V1 as in Cooke et al.37 and attached a metal bar to enable head fixation of animals. After recovery from surgery, mice were handled and habituated to head restraint, then presented with either a black or gray screen (Figure 6J). As previously reported, we observed a narrowband gamma oscillation in the 55–65 Hz range. Interestingly, we found that the gray/black screen ratio of both mean (Figure 6K) and peak (Figures 6L) 55–65 Hz activity was elevated in vGAT-Btbd11 KO animals compared with CONs, indicating increased power in the narrowband gamma frequency.
Btbd11 KO mice display altered exploratory behavior, reduced anxiety, and sensitization to an NMDA receptor antagonist
We assessed the behavior of CON and vGAT-Btbd11 KO animals in an open field to evaluate locomotor activity. Animals were run in a brightly illuminated open field, with infrared beam breaks used as a readout of activity. No differences were observed in beam breaks, indicating normal locomotor activity in vGAT-Btbd11 KO mice (Figure 7A). In a separate cohort of mice run in the dark, we also found no differences in locomotor activity (Figures S7A and S7B). Interestingly, vGAT-Btbd11 KO animals made significantly more rearing events during the 30 min of exploration, a measure of exploratory drive (Figure 7B). The open field task is frequently used as an assay for anxiety in rodents. Exploration of the center region of the arena is indicative of a lower anxiety state. We evaluated the beam breaks in the center of the apparatus during the first 5 min of open field exploration (the period where the arena is most novel and anxiogenic) and found that vGAT-Btbd11 KO mice made significantly more beam breaks in the center region, suggesting that vGAT-Btbd11 KO mice might be less anxious than CONs (Figure 7C). As a secondary test of anxiety levels, we ran animals on the elevated plus maze (EPM) test of anxiety in which animals choose between entering open (anxiogenic) vs. closed arms. There was no difference between groups in the total distance traveled the maze (Figure S7C) or the total entries into the open arm vs. the closed arm (expressed as a ratio; Figure S7D). However, during the test, we noticed that some animals spent time at the extreme ends of the open arm, presumably the most anxiogenic portion of the maze (Figure S7E). When analyzing the entries to the extreme end of the open arm vs. entries into the closed arm (expressed as a ratio), we found a significantly higher ratio for vGAT-Btbd11 KO mice compared with CONs (Figure S7F). Furthermore, using automated freezing detection software, we detected the time each animal spends freezing on the maze, a measure commonly used as a readout of fear. We found that vGAT-Btbd11 KO mice spent less time freezing compared with CONs (Figure S7G). Together, these data indicate that vGAT-Btbd11 KO mice are less anxious on the EPM compared with CONs, though the phenotype was subtle and not observed in a crude analysis of the data. To evaluate anxiety behavior further, we conducted the hole-board test and counted the number of nose pokes made during a 3 min session. Anxious animals generally make fewer nose pokes, or “head dips.”38 We found that male (Figure 7D), but not female (Figure S7H), vGAT-Btbd11 KO mice made significantly more nose pokes compared with CONs. As an additional test of anxiety, we ran a cohort of mice through a marble burying task. Mice are thought to bury marbles when anxious, and anxiolytic drugs lead to animals burying fewer marbles.39 We observed no difference in marble burying in female animals (Figure S7I); however, in male mice, vGAT-Btbd11 KO animals buried significantly fewer marbles than CONs (Figure 7F), consistent with vGAT-Btbd11 KO animals having reduced anxiety, albeit in a sex-specific manner.
Figure 7. Btbd11 KO mice display altered exploratory behavior, reduced anxiety, and sensitization to a NMDA receptor antagonist.
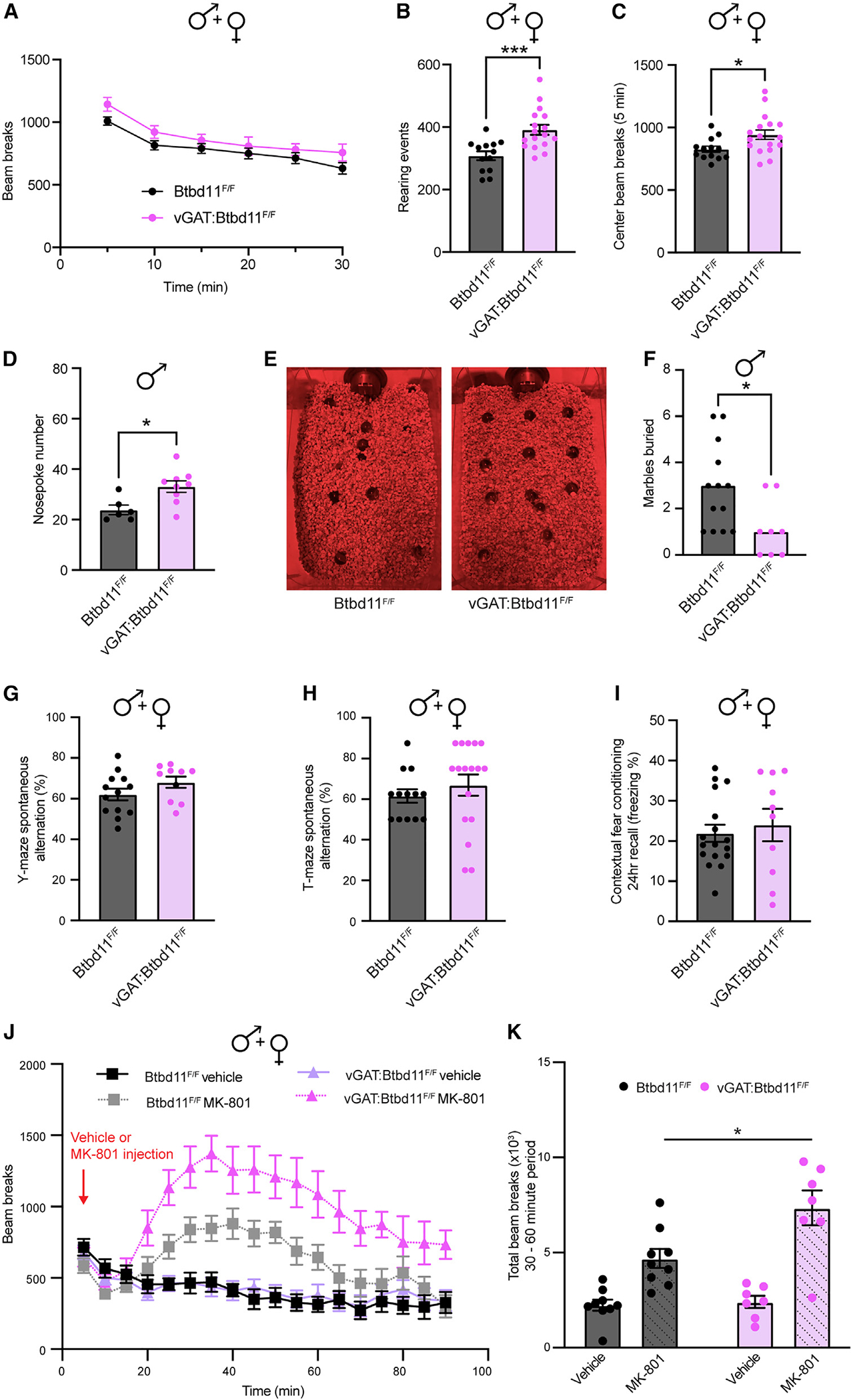
(A) Locomotor activity of male and female mice exploring a novel environment.
(B) Rearing events during 30 min of open field exploration in male and female animals. ***p < 0.001.
(C) Beam breaks in the center of the open field arena during the first 5 min of exploration in male and female animals. *p < 0.05.
(D) Number of nose pokes made by male animals.
(E) Representative photos showing the number of marbles buried by Btbd11F/F or vGAT:Btbd11F/F mice.
(F) Quantification of marble burying data. Bars indicate the median.
(G) Spontaneous alternation of male and female animals in a Y-maze test of short-term spatial memory.
(H) T-maze spontaneous alternation performance in male and female animals.
(I) Contextual fear conditioning recall data 24 h after conditioning. Freezing was used as a proxy for memory performance and compared between male and female animals.
(J) Locomotor activity of mice in an open field arena following injection with either saline or MK-801 (0.2 mg/kg).
(K) Quantification of the total infrared beam breaks in the 30–60 min period after injection. Unless specified, line or bars display mean, and error bars show SEM. Btbd11F/F animals displayed in black and vGAT:Btbd11F/F displayed in magenta throughout.
For further details, see Figure S7.
We subsequently assessed short-term spatial memory using a spontaneous alternation version of the Y-maze. We observed no difference between CON and vGAT-Btbd11 KO animals, indicating that under basal conditions, spatial short-term memory was intact (Figure 7G). As a second test of short-term spatial memory, we ran a cohort of mice through a spontaneous T-maze alternation task.40 There was no difference between the groups in spontaneous alternation behavior (Figure 7H). To evaluate emotional learning, we conducted a contextual fear conditioning assay evaluating freezing behavior 24 h after conditioning. There was no difference between CON and vGAT-Btbd11 KO mice, indicating normal emotional learning (Figure 7I).
Genetic manipulations that impact glutamatergic synapses in INs often display altered sensitivity to NMDA receptor antagonist challenge. For example, mice that lack NMDA receptors (NMDARs) in PV-INs are more sensitive to the effects of the NMDAR antagonist MK-801.41 Since vGAT-Btbd11-KO mice had reduced glutamatergic recruitment of PV-INs, we speculated that they might be predisposed to MK-801 challenge. Therefore, we administered CON and vGAT-Btbd11 KO mice with MK-801 (0.2 mg kg−1) or saline (as a vehicle) and measured their locomotor response in the open field apparatus (Figure 7J). MK-801 administration led to a dramatic increase in locomotor activity; however, this was particularly prominent in vGAT-Btbd11 KO mice, indicating an increased sensitivity to the drug (Figure 7K).
DISCUSSION
In this study, we identify Btbd11 as a cell-type-specific synaptic protein and characterize its basic properties and function using biochemistry, imaging, electrophysiology, and behavior.
Glutamatergic SSPSDs show cell-type-specific protein specializations
Using immuno-isolation and proteomics, we demonstrate that it is possible to identify inPSD-specific proteins. A similar method was used to examine differences in PSDs from different subpopulations of ENs,42 highlighting the usefulness of the approach. Initially, we focused our effort on a thorough characterization of Btbd11, but in future studies, it will be interesting to explore the other putative inPSD proteins (Table S1). For example, we identify Traf3 and Lrsam1 as putative inPSD proteins, with links to ischemia43 and neurodegeneration,44 respectively. Furthermore, by combining proteomic with new methods such as in vivo proximity labeling,23,45 it should be possible to uncover yet more PSD protein specializations, including those that do not form complexes with Psd-95 or that form only transient or weak interactions with Psd-95.
Btbd11 is localized exclusively to inPSDs in cortex and hippocampus
Converging evidence shows that Btbd11 is expressed exclusively in INs within cortical and hippocampal tissue; however, it is possible that in other brain regions, this cell-type specificity will be lost. It is noteworthy that Btbd11 was previously identified as a reliable marker of cortical interneurons, although no function or properties of Btbd11 were explored.46 The subcellular distribution of Btbd11 is highly targeted to glutamatergic synapses—via interactions with Btbd11’s PBM. Our yeast 2-hybrid screen indicates putative interacting proteins, including known synaptic proteins that impact synapse function. It will be important to further characterize the interactome of Btbd11 to gain further mechanistic insights into how it regulates glutamatergic synapses in INs.
LLPS of Btbd11 with Psd-95
We show that exogenously expressed Btbd11 undergoes LLPS with Psd-95. LLPS is highly sensitive to protein concentration, but since the PSD concentrates synaptic proteins, we expect the concentration of Btbd11 and Psd-95 to surpass a threshold to support biomolecular condensation at glutamatergic synapses in INs. Thus far, difficulties in purifying full-length Btbd11 (which we find is necessary for LLPS properties) have prevented in vitro studies to calculate the precise concentration of Btbd11 and Psd-95 required to trigger LLPS. Phase separation at the PSD is likely ubiquitous among neuronal cell types and has been argued to underscore the assembly and stability of the PSD.14,16,17 It is tempting to speculate that Btbd11 can stabilize Psd-95 at the synapse through promoting LLPS; however, we note that our LLPS data come from overexpressed Btbd11 and might not faithfully reflect what happens with endogenous Btbd11. If this was indeed the case, it likely reflects a generalizable phenomenon, for which we have identified a particular cell-type-specific specialization involving Btbd11 utilized by INs. LLPS of the inPSD may play an even more critical role than it does at the PSD in excitatory neurons, as the inPSD is exposed on the dendritic shaft and is not compartmentalized in spine-like protrusions thought to limit diffusion to and from the synapse.
Reduced glutamatergic recruitment in PV neurons of Btbd11 KO mice
With whole-cell patch-clamp recordings, we observed a decrease in the mEPSC frequency in PV-INs when Btbd11 was knocked out from INs. This phenotype is like that of IN-specific deletion of ErbB4 or global KO of Brevican, two proteins that are also enriched at glutamatergic synapses in PV-INs.47–49 Because a measure of presynaptic release (PPR) was unchanged with Btbd11 KO, we expect that this mEPSC phenotype is a result of decreased glutamatergic synapses within PV-INs. We expect this is a consequence of a destabilizing effect on Psd-95 due to Btbd11 KO, as we found that Btbd11 overexpression was able to stabilize Psd-95 at the PSD of cultured INs. In support of this idea is the observation that Psd-95-GFP puncta were smaller in PV-INs that lack Btbd11, potentially highlighting a decreased stability of Psd-95 without Btbd11 present as an interaction partner and scaffolding protein. It will be interesting to test if other IN subtypes, such as those expressing somatostatin, also receive reduced glutamatergic input following Btbd11 KO. Furthermore, in subsequent studies, it will be informative to elucidate if there is a time dependence of Btbd11 deletion or if this phenotype is dependent on gene deletion early in development (Cre switches on early in the vGATCre line).
Altered network properties, behavior, and sensitivity to NMDAR challenge in Btbd11 KO mice
In cultured Btbd11 KO neurons, we observed an increase in spontaneous Ca2+ transients, consistent with a lack of inhibition in the cultures. Furthermore, we observed an exaggerated induced gamma frequency oscillation in vivo when mice were presented with a gray screen. Because of the role PV-INs have in regulating the firing of other neurons and supporting synchronous activity, we suspect that altered activity of PV-INs—through loss of glutamatergic signaling—is responsible for these changes in network properties. Behaviorally, we observed that IN-specific Btbd11 KO mice display measures of increased exploratory behavior (rearing events) and decreased anxiety that were particularly pronounced in male animals. In tests of short-term spatial memory and contextual fear conditioning, there was no difference between CON and IN-specific Btbd11 KO animals, in contrast to other manipulations of PV-INs.50 Interestingly, IN-specific Btbd11 KO animals are sensitized to NMDAR antagonism with MK-801. Previous work has shown that INs could be preferentially sensitive to NMDAR antagonism at certain doses.51 Furthermore, deletion of NMDARs from different populations of INs results in differential sensitivity to MK-801 challenge.41,52–54 The increased sensitivity of vGAT-Btbd11 KO mice to MK-801 likely manifests because of their compromised glutamatergic synapses in PV-INs. In future studies, it will be interesting to test if other stressors, such as post-weaning isolation54,55 or reduced environmental enrichment,53 have exaggerated effects on vGAT-Btbd11 KO mice compared with CON animals.
Limitations of the study
In this study, we have uncovered behavioral and physiological alterations when Btbd11 is deleted from vGAT-positive INs. As vGAT expression occurs early in development, Btbd11 KO will also happen early. Therefore, we cannot rule out whether Btbd11 plays a developmental role in regulating glutamatergic synapses. Future work will be required to see if adult KO of Btbd11 recapitulates the same phenotypes. Furthermore, in the present study, we restricted our analysis to cortical and hippocampal INs. Based on in situ hybridization data from the Allen Institute, both vGAT and Btbd11 are expressed outside of the cortex and hippocampus. Therefore, we do not know if the specificity of Btbd11 for INs in maintained in other brain regions.
In summary, we reveal Btbd11 as an IN-specific glutamatergic synaptic protein and show that it plays an important role in regulating glutamatergic synapses using biochemistry and in vitro live-cell imaging experiments through to in vivo physiology and behavior. It will be important to uncover if Btbd11—or other inPSD proteins—could be potential therapeutic targets for neurological disorders in which GABAergic signaling is disrupted.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Richard Huganir (rhuganir@jhmi.edu).
Materials availability
Plasmids generated in this study are available upon request. Requests for plasmids should be directed to and will be fulfilled by the lead contact.
Data and code availability
Proteomic data is available from the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD041635.
No unique code is described in this study.
All other data and any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal care
All animals were treated in accordance with the Johns Hopkins University Animal Care and Use Committee guidelines. Transgenic mice (outlined below) and timed pregnant Sprague-Dawley rats were used in this study. Mice were housed in a temperature and humidity-controlled vivarium with ad libitum access to drinking water and food.
GFP-immuno-isolation
Cortex and hippocampus from 1 CaMKII:Psd-95-GFP and 4 vGAT:Psd-95-GFP animals were collected from male mice aged 11–12 weeks. Brain tissue was homogenized in 8 mL of ice-cold lysis buffer (1% DOC, 50mM Tris (pH9), 50mM NaF, 20μM ZnCl2, 1mM Na3VO4) with the addition of Pefabloc SC (Roche), Okadaic acid (200nM) and a protease inhibitor cocktail (Roche Complete) with a Dounce homogenizer and solubilized for 1 h rotating at 4°C; as previously reported.22 Samples were then clarified with spins at 17,000g and 50,000g each for 30 min and at 4°C. Further lysis buffer was added so that GFP-immuno-isolation experiments were set up in 10 mL total volume. For each sample 50μL of pre-washed GFP-TRAP agarose beads (Chromotek) was added and incubated with rotation overnight at 4°C. Beads were washed x6 with ice-cold lysis buffer (with the same inhibitors as before) using spin-filter columns (spin 1500g for 1 min). Samples were eluted with 1% SDS containing 2.5% beta-mercaptoethanol. The vGAT samples were serially eluted to increase the concentration of eluant (since the abundance of Psd-95-GFP is much lower in the vGAT:Psd-95-GFP mice). Samples were then subject to mass spectrometry.
Note: the same pull-down procedure was followed to get samples for western blots (Figure 1C), except 100μL of bead slurry was used for the CaMKII:Psd-95 GFP and each of the 4x vGAT:Psd95-GFP animals that were later were pooled. Additionally, a wildtype animal was included with 400μL of control beads (Chromotek) to check that Btbd11 was not just binding non-specifically to the beads. After the IP and the beads were washed samples were eluted in Laemmli Buffer and used for western blots. In Figure S1B the same procedure was followed except 5x vGAT:Psd95-GFP animals were pooled for the vGAT condition and 50μL of bead slurry was used per animal (and there was no bead-only control.
Mass spectrometry
GFP-immuno-isolation samples (see above) were reduced with DTT, alkylated with IAA and TCA/Acetone precipitated with x8 volume overnight at 37°C. Samples were then reconstituted in 12uL 500mM TEAB, 48uL water and 25 ng/μL trypsin/LysC added. Samples were proteolyzed at 37°C overnight. Peptides were desalted on Qasis U-HLB plates, eluted with 60% acetonitrile/0.1% TFA, and dried. Desalted peptides, 10% and 20% were analyzed by nano-LC/MS/MS on QExactive Plus at resolution 140K on precursor and 35K on fragment, using 85 min gradient from 2% acetonitrile/0.1% formic acid to 98% acetonitrile/0.1% formic acid. Injected peptides showed abundant base peak chromatographs. Database search: FilesRC option was applied and MSMS spectra were searched with Mascot 6.1 via Proteome Discoverer 2.3 against RefSeq2017_83_mouse and a small database with added enzymes, bovine serum albumin (our standard). Mass tolerance 4ppm for precursor, 0.01Da for fragment variable modifications: carbamidomethylation on Cys, oxidation on M, deamidation NQ. Peptides validated with Percolator. Mascot.dat files compiled in Scaffold. Note: just the 20% samples were used for analysis. Label-free quantification was performed, and protein abundances were normalized to the abundance of Psd-95. The ratio of vGAT/CaMKII abundance was then calculated to identify putative inPSD proteins (plotted in Figure 1B). From the ratio data, proteins with ratios greater than the mean + 2X the standard deviation were classified as putative inPSD candidates (Table S1).
Western blots
Samples in Laemmli Buffer were heated to 60°C for 15 min prior to running western blots. Depending on target protein Mw, precast 4–12% or 8% Bis-Tris gels were used (Thermo Fisher Scientific) with NuPAGE MOPS SDS running buffer (Thermo Fisher Scientific). Proteins were transferred to nitrocellulose membrane (GE Healthcare) and blocked for 1 h with Odyssey Blocking Buffer (Li-COR bioscience) before probing with primary antibodies (see below) overnight at 4°C or at room temperature for 1–2 h. Antibodies were made up in TBS supplemented with 3% BSA and 0.1% Tween 20. Blots were washes with TBS with 0.1% Tween 20 (TBST) 4x and incubated with secondary antibodies (all fluorescently conjugated with 680 or 800nm dyes and used at 1:10000 and from Li-COR bioscience) made up in TBS supplemented with 3% BSA and 0.1% Tween 20 and 0.02% SDS for 1 h at room temp. Blots were washed 4x in TBST and imaged on a Li-COR scanner. Bands were quantified in Image Studio (Li-COR bioscience). Primary antibodies used: Tanaka anti dsRed (Living colors, rabbit polyclonal, 1:1000), Santa Cruz anti-GFP (B2, IgG2a, 1:2000), homemade anti-Btbd11 (rabbit polyclonal, 1:500), Abclonal Btbd11 (Rabbit, 1:500), Neuromab anti-Psd-95 (K28/74 mouse IgG1, 1:5000), Sigma anti-GST (GST-2, mouse IgG2B, 1:5000), Swant anti-Parvalbumin (PV27, rabbit polyclonal, 1:5000), Santa Cruz anti alpha-tubulin (B-7, mouse IgG2a, 1:5000) and Sigma Synaptophysin (SVP-38 mouse, 1:10000).
Immunohistochemistry for Btbd11
WT mice (3 months, male) were transcardially perfused with ice-cold PBS followed by 4% PFA (in PBS) then subject to 2 h of continued post fixation in PFA. After washing with PBS, the brains were sliced on a vibratome (60mm thick). Slices were washed x3 with PBS, then incubated with 90% methanol (in 10mM MES buffer) at RT for 30 min. Slices were permeabilized in PBST (0.5% Triton-X-100) for 20 min, and then blocked for 1 h at RT in 5% NGS, 3% BSA and 0.15% Triton X-100. The same blocking buffer was used for the primary antibody incubation (rabbit Btbd11, Abclonal 1:500 and mouse Lhx6, Santa Cruz A-9 1:200) overnight at RT. The next day slices were washed 4x with PBS, then incubated with secondary antibodies (Thermo Fisher Alexa Fluor goat anti rabbit 568 and Alexa Fluor goat anti mouse IgG2a 488, both at 1:500) in PBS containing 2.5% NGS, 1.5% BSA, and 0.15% Triton X-100 at RT for 2 h. Slices were then washed 4x with PBS, and then mounted with PermaFluor media (Thermo Fisher Scientific) and stored at 4C in the dark. Images were acquired on a confocal microscope (Zeiss LSM 880) at optimal resolution with a 633 objective from area CA1 of the hippocampus or layer 2/3 of the visual cortex.
Immunofluorescence
Neurons were rinsed 1X with PBS at room temperature and then fixed for 15 min at room temperature with 4% paraformaldehyde (Electron Microscopy Sciences) made up in PBS and supplemented with 4% sucrose. For certain experiments (Figure 1G) this fixation step was replaced with a 20-min fixation in methanol at −20°C. In other instances, to promote GAD67 and Psd-95 immunofluorescence, cells were subject to a 90s methanol wash (−20°C) following the paraformaldehyde fixation described above. Cells were washed 4x with PBS and incubated in primary antibodies (see below) in a pH 7.4 gelatin buffer (30 mM phosphate buffer, 0.2% gelatin, 0.3% Triton X-100, and 0.25M NaCl) overnight at 4°C. Cells were washed 4x with PBS and then incubated with secondary antibodies in the same GDB buffer for 1 h at room temperature. All 488, 568 and 647 secondary antibodies were raised in goat, Alexa Fluor conjugated, used at 1:500 and purchased from Thermo Fisher Scientific. All 405 conjugated antibodies were raised in goats, DyLight 405 conjugated, used at 1:500, and purchased from Jackson Immuno Research Laboratories. Cells were washed 4x in PBS, briefly rinsed in distilled water and mounted on slides with PermaFluor mounting media (Thermo Fisher Scientific). Slides were stored in the dark at 4°C.
Primary antibodies used:
Abcam anti-GFP (ab13970, chicken, 1:2000), Neuromab anti-Psd-95 (K28/43, mouse IgG2a, 1:500), homemade anti-Btbd11 (rabbit polyclonal, 1:100), Tanaka anti dsRed (Living colors, rabbit polyclonal, 1:1000), Santa Cruz anti-Gad-67 (F-6, mouse IgG3, 1:100), Swant anti-parvalbumin (Clone 235, mouse, IgG1, 1:1000), Synaptic Systems anti-Gephyrin (147 011, mouse, IgG1, 1:500).
Analysis of Btbd11 colocalization with Psd95 and gephyrin
The EzColocalization ImageJ plugin61 was used to evaluate the colocalization of endogenously tagged GFP-Btbd11 with Psd95 and Gephyrin labeled with immunofluorescence. In FIJI, maximum intensity projections were generated from confocal Z-stacks, background was subtracted (rolling ball radius, 10 pixels) and the images were median filtered (1 pixel). For each KI cell, ROIs were drawn and dendritic segments, in which the colocalization analysis was to be conducted. The EzColocalization plugin was used, and the threshold overlap score (TOS) was computed. For each channel, the threshold (FT) was set to 10% to identify the top 10% of pixels as being over the threshold. With this analysis a TOS of −1 equates to perfect anticorrelation, 0 for non-colocalization and 1 for complete colocalization. After calculating the TOS values for each dendritic region, they were averaged for each KI cell. The GFP-Btbd11 colocalization with Psd95 or gephyrin was compared with a T test.
Yeast two-hybrid experiments
Different combinations of pDBLeu-Btbd11-C-terminus (consisting of the terminal 58 or 54 amino acids of Btbd11 for Btbd11 and Btbd11ΔPBM, respectively) and pPC86-PSD95 PDZ constructs were co-transformed into PJ69 yeast cells56 and bait/prey pairs were selected for by growth on -Leu, Trp media at 30°C. Btbd11-PSD95 interactions were then tested by the ability of the different yeast bait/prey combinations to grow on -Leu, Trp, His media.
To identify putative Btbd11 interaction partners the BTB domain of Btbd11 or the C-terminal region of Btbd11 were cloned into pDBLeu and used as baits in yeast 2-hybrid screens with a rat hippocampal library. Plasmids from yeast clones that could grow on -Leu, Trp, His, Ade media were rescued and analyzed by sequencing.
Cell-culture
HEK cells
HEK cells were grown on 10cm plates (for biochemistry) or on collagen (Advanced Biomatrix) coated glass coverslips in 12-well plates (for immunofluorescence and live-cell imaging) in DMEM (Gibco) supplemented with 10% Fetal Bovine Serum (Hyclone) and Penicillin-Streptomycin antibiotics (100 U/ml, Thermo Fisher Scientific). Cells were maintained at 37C with 5% CO2. For biochemistry (GST-Pulldown experiments) HEK cells were transfected via Calcium Phosphate precipitation. For immunofluorescence and live-cell imaging experiments HEK cells were transfected with Lipofectamine 2000 (Invitrogen).
Primary cultured rat neurons
Timed pregnant Sprague-Dawley rats were purchased (ENVIGO) and dissected at embryonic day 18. Dissection media (to make 1L) consists of 100 mL 10x HBSS (Gibco), 10 mL Penicillin-Streptomycin (Thermo Fisher Scientific), 10 mL pyruvate (Gibco), 10 mL HEPES (Gibco) 10 mM final, 12 mL of 2.5M Glucose (Sigma) stock 30 mM final and 860 mL Milli-Q water. Cortical cells were plated onto glass coverslips coated with 1 mg/ml Poly-L-Lysine hydrobromide (Sigma) at a density of 250K/well (of a 12-well plate) and grown in Neurobasal Plus Medium (Gibco) supplemented with 2% B-27 Plus (Gibco), 2mM Glutamax (Thermo Fisher Scientific), Penicillin-Streptomycin (100U/ml, Thermo Fisher Scientific), and 5% horse serum (Hyclone). Hippocampal cells were plated at a density of 100K/well (of a 12-well plate) in the same manner except with standard Neurobasal Medium and standard B27. For hippocampal cultures, cells were swapped to serum-free media (as above but lacking horse serum) at DIV1 and subsequently fed once a week with serum-free media. For cortical neurons, cells were fed with media containing 1% horse serum and FdU (Sigma) at DIV4, then subsequently fed twice per week with serum free media (Neurobasal Plus Medium and B27 Plus). Cells were maintained at 37C with 5% CO2.
Primary cultured mouse neurons
P0 Btbd11 F/F pups were dissected and plated as above except 5% fetal bovine serum was used in place of horse serum, and that Neurobasal Plus Medium and B27 Plus supplement was used for both hippocampal and cortical cultures. Hippocampal neurons were plated onto Poly-L-Lysine coated (1 mg/ml) coverslips at a density of 200K/well (of a 12-well plate) and cortical neurons were plated at a density of 6.5–7M cells per 10cm plate (also Poly-L-Lysine coated. Cells were fed twice a week with serum free media. AAVs were added to cultures at DIV1 or DIV2. AAV.CMV.PI.EGFP.WPRE.bGH (Serotype 2/9; titer ≥1×1013 vg/mL, was a gift from James M. Wilson), AAV.CMV.HI.eGFP-Cre.WPRE.SV40 (Serotype 2/9; titer ≥1×1013 vg/mL, was a gift from James M. Wilson) and AAV.Syn.NES-jRGECO1a.WPRE.SV40 (Serotype 2/9; titer ≥1×1013 vg/mL, a gift from Douglas Kim & GENIE Project) viruses were purchased from Addgene. For Figure S1D mixed cultures were prepared as above, except with mixed litters of Btbd11 F/F and vGAT:Btbd11 F/F P0 pups and AAV-CAG-DIO-tdTomato (Serotype 2/9; Addgene 28306-AAV9, a gift from Edward Boyden) was added to cultures. This led to inhibitory neurons being labeled with tdTomato, but only those that also were lacking Btbd11. Cells were maintained at 37C with 5% CO2.
PSD preparation of hippocampus and cultured neurons
Btbd11 F/F primary cultured cortical neurons (1 × 10cm plate per condition) were collected by plate scraping, or one hippocampus (per Gene Trap, vGAT:Btbd11 F/F or Btbd11 F/F control animal) was dissected, and then homogenized by passage through a 26g needle, 12 times, in homogenization buffer (320mM sucrose, 5mM sodium pyrophosphate, 1mM EDTA, 10mM HEPES pH 7.4, 200nM okadaic acid, protease inhibitor cocktail (Roche)). The homogenate was centrifuged at 800g for 10 min at 4°C to yield the P1 and S1 fractions. S1 was further centrifuged at 17,000g for 20 min at 4°C to yield P2 and S2 fractions. P2 was resuspended in milliQ water, adjusted to 4mM HEPES pH 7.4 from a 1M HEPES stock solution, and incubated with agitation at 4°C for 30 min. The resuspended P2 was centrifuged at 25,000g for 20 min at 4C to yield LP1 and LS2. LP1 was resuspended in 50mM HEPES pH 7.4, then mixed with an equal volume of 1% Triton X-100 and incubated with agitation at 4°C for 15 min. The PSD fraction was generated by centrifugation at 32,000×g for 20 min at 4°C. The final PSD pellet was resuspended in 1x RIPA buffer. Protein quantification was performed via Bradford assay and samples made up in Laemmli Buffer containing 5% Beta-mercaptoethanol and frozen.
PSD preparation from cortex samples
Mouse cortex from wildtype (Figure 1N; 1x male and 1x female 3 months) or Btbd11 F/F and vGAT:Btbd11 F/F animals (Figure 5D) were homogenized in ice-cold buffer (320mM sucrose, 5mM sodium pyrophosphate, 1mM EDTA, 10mM HEPES pH 7.4, 200nM okadaic acid, protease inhibitor cocktail (Roche)) using 12–15 strokes of a Dounce homogenizer. The homogenate was centrifuged at 1000g for 10 min at 4C to generate the S1 and P1 (Figure 1M). The S1 was further centrifuged at 17,000g for 30 min at 4C to obtain the S2 and P2 fractions. The P2 fraction was resuspended in 3 mL of homogenization buffer (as above) and loaded onto a discontinuous sucrose gradient consisting of 2.5mL of 1.2, 1 and 0.8M sucrose (10mM HEPES, 5mM NaPPI, 1mM EDTA and protease and phosphatase inhibitors). The fraction was then subject to ultracentrifugation at 82,500g for 2 h at 4C, and 1 mL of the resulting fraction at the 1M and 2M interface was collected. The Synaptosomes were diluted with 10mM HEPES to restore the sucrose concentration back to 320mM and then pelleted with centrifugation at 100,000g for 30 min at 4C. Synaptosomes were resuspended in 1.5mL of 50mM HEPES (pH7.4 with inhibitors added) and then mixed with an equal part of 1% Triton X-100 and incubated for 15 min at 4C. The PSD material was then pelleted with centrifugation at 50,000g for 2 min at 4C. The PSD pellet was resuspended in RIPA buffer.
GST-pulldown experiments
HEK cells (transfected via Ca2+ phosphate precipitation) grown to confluence on a 10cm plate were washed 1X with PBS at room temp and then lysed in ice-cold lysis buffer (1X Tris-Buffered Saline, 1% NP-40, 10mM NaPPi, 10mM NaF, 200nM okadaic acid, 1mM Na3VO4 and a home-made protease inhibitor cocktail). Cells were scraped from the plate and lysed by rotation for 20 min at 4°C. Lysates were centrifuged at 17,000g for 15 min and the supernatant retained. 20μl of supernatant was kept as 2% INPUT. 30μL of pre-washed Glutathione Sepharose 4B bead slurry (GE Healthcare) was added to each sample and incubated with rotation for 2-h at 4°C. Beads were washed in spin columns 5X with lysis buffer and then eluted with 30–40μL of Laemmli Buffer containing 5% Beta-mercaptoethanol and frozen or run immediately on western blots.
Confocal microscopy and image analysis
All confocal images were acquired with a Zeiss LSM 880 microscope. Neurons and HEK cells were imaged in 37°C pH 7.4 ACSF (120mM NaCl, 5 mM KCl, 10mM HEPES, 10mM glucose, 2mM CaCl2 and 1mM MgCl2) in a humidity-controlled chamber. For FRAP experiments (neuron and HEK), the laser power and repetitions needed for successful bleaching was optimized for each experiment, and then kept consistent. Images were collected with oil-immersive 40x or 63× objectives, except for the Ca2+ imaging experiments in mouse neurons, which was collected using a 10× air objective.
Analysis of FRAP data
For FRAP experiments in neurons time-series Z-stacks were acquired with two timepoints acquired before photobleaching. For each dendritic region at least 5 putative synaptic puncta (Psd-95-mCherry puncta) were bleached, ensuring that there were several puncta in the field of view that were not bleached. Images were analyzed in FIJI. Maximum image projections were generated for the timeseries, and a median filter was applied to all channels (1-pixel) and the Azurite cell-fill channel was used to correct for movement during imaging with a rigid-body transformation (MultiStackReg plugin). Regions of interest (ROIs) were drawn around bleached and unbleached puncta as well as regions of background (away from the dendritic signal). Background signal was subtracted from the puncta signal, which was then normalized to the average intensity of the unbleached puncta at each time point (to account for the low levels of acquisition bleach over time). The signal in the bleached ROIs was then normalized such that the average baseline was centered on 1 and the post-bleach time point was 0. For each puncta the estimated maximum recovery was estimated with a one phase exponential fit (in GraphPad Prism 9). For comparisons +/− GFP-Btbd11 analysis was performed on decoded data with just the motion-corrected Psd-95-mCherry signal, so the investigator had no knowledge of the experimental condition.
Analysis of puncta-to-puncta distance
Imaris 9 (Bitplane) was used to estimate the puncta-to-puncta distance shown in Figure 1K. The GFP channel (GFP-Btbd11 knockin) with smoothing (1μm) was used to generate a surface along the dendrite of the knockin cell. This surface was used as a mask for the other channel (i.e., Psd-95 or Gephyrin) for spot detection and analysis. The spot-detection feature was then used to detect GFP-Btbd11 and Psd-95 or Gephyrin puncta. We then calculated the distance of the nearest Psd-95 or Gephyrin puncta to each GFP-Btbd11 puncta (using the center of the detected spot as the center point of each punctum).
Analysis of PV levels with immunofluorescence
Imaris 9 (Bitplane) was used to generate a surface around the cell body of PV-IN using PV immunofluorescence signal with smoothing (2–3μm) and an estimated diameter of 5–10μm. This surface was then used to calculate the average intensity of PV for each cell.
Cloning and molecular biology
All constructs were generated using HiFi assembly (New England Biolabs). mDlx-Azurite was generated by replacing EGFP with Azurite from the pAAV-mDlx-GFP-Fishell-1 plasmid (Addgene number: 83900, a gift from Gordon Fishell).
Generation of pORANGE Btbd11 constructs were generated using the pORANGE Cloning template vector (Addgene number: 131471, a gift from Harold MacGillavry). The guide sequence to target the N terminus of Btbd11 was: 5-ACGGCG GCTGCAGCATGAAG-3′. cDNA for mouse Btbd11 was purchased with a C-terminal Myc tag (Origene catalog number: MR217199). Using this cDNA as template pCAG-GFP-Btbd11 was generated, removing the C-terminal Myc tag (exposing the PBM of Btbd11). A linker (GGGGSGGGGTR) was added between EGFP and Btbd11. The 5xANK-BTB mutant consisted of the last 512 amino acids of Btbd11 (i.e., a large N-terminal deletion). pCMV-Psd-95-mCherry point mutants were generated based on the mutations described previously.28 pCIS-GST-Btbd11 was generated by subcloning Btbd11 into a pCIS-GST expression vector. The sequence of all constructs was confirmed with DNA sequencing.
Electron microscopy
Cells grown in 35 mm tissue culture dishes (Falcon 3001) were briefly rinsed with 37 C PBS, then fixed with 2.5% glutaraldehyde in 100 mM phosphate buffer (Sorenson’s) containing 5 mM MgCl2 pH 7.4, for 2.5 h at room temperature on a slow rocker. After a 30 min buffer rinse (100 mM phosphate buffer with 3% sucrose and 5mM MgCl2), cells were post-fixed in 1% osmium tetroxide in 100 mM phosphate buffer with 5 mM MgCl2 at 4°C for 1 h in the dark. Samples were then rinsed 100 mM maleate buffer containing 3% sucrose pH 6.2 and en-bloc stained with 2% uranyl acetate (0.22 μm filtered) in the same buffer for 1 h in the dark. Plates were dehydrated in a graded series of ethanols then infiltrated in Eponate 12 (Pella) overnight without catalyst. The next day cells were further embedded with fresh epon containing 1.5% DMP-30 (catalyst). Culture dishes were cured at 37°C for three days, and further polymerized at 60°C overnight. Cured discs were removed from the plastic dish and 3 mm circles punched out and glued to epon blanks for sectioning. 80 nm thin compression free sections were obtained with a Diatome diamond knife (35°). Sections were picked up onto 1 × 2 mm formvar coated copper slot grids (Polysciences), and further stained with uranyl acetate followed by lead citrate. Grids were examined on a Hitachi H-7600 TEM operating at 80 Kv. Images were digitally captured with an XR-50, 5-megapixel CCD camera (AMT).
Generation of Btbd11 conditional KO mice
In vitro fertilization of C57Blk/6J mice was performed by the Johns Hopkins Transgenic Core using frozen sperm obtained from the European Mutant Mouse Archive (Btbd11tm1a(EUCOMM)Wtsi, https://www.infrafrontier.eu/). Offspring were genotyped as recommended with a common forward primer (5′−3’: TCCTGTCTTAATGCCCCCTG), a wildtype reverse primer (5′−3’: TTCTGGCG GTTCTAAATCCTG) and a mutant reverse primer (5′−3’: TCGTGGTATCGTTATGCGCC). Btbd11 Gene Trap mice were backcrossed with C57Blk/6J animals. To generate Btbd11 conditional knockout mice, Btbd11 Gene Trap mice were crossed with constitutive FLPe-expressing animals. Correct conversion was confirmed with PCR as described by the European Mutant Mouse Archive. Conditional animals were bred to homozygosity. To generate IN specific knockout animals, Btbd11 conditional mice (Btbd11 F/F) were crossed with vGATCre animals (Jackson lab stock: 028862). Mice that were homozygous for floxed Btbd11 (Btbd11 F/F) were used as controls and animals that were heterozygous for vGATcre and homozygous for floxed Btbd11 were used as conditional interneuron-specific KOs (denoted as vGAT:Btbd11 F/F).
Stereotaxic surgery
Virus injection: pAAV-S5E2-dTom-nlsdTom virus (Addgene number: 135630, a gift from Jordane Dimidschstein) was packaged by the Janelia Vector core with AAV2/9 serotype (virus titer after 1:1 dilution: 2 × 1013 GC/ml). Stereotaxic surgery was conducted as previously described.57 Male and female Btbd11 F/F or vGAT:Btbd11 F/F mice aged 4–5 weeks were anesthetized with isoflurane, placed into a stereotaxic frame (Kopf) with their body temperature monitored and maintained at 37C with a closed-loop temperature control system (Kent Instruments). Animals were injected subcutaneously with sterile saline (VetOne; 0.5mL) to maintain hydration and buprenorphine (ZooPharm; 1 mg/kg) and lidocaine (VetOne; 2%) to provide analgesia, with lidocaine injected locally under the skin over the skull. An incision was made to expose the skull with a scalpel, and a craniotomy performed to expose the brain surface (see below for coordinates). Glass pipettes (Drummond Science Company; Wiretrol II) were pulled (Sutter Instruments) and sharpened to a 30° angle (Medical Systems Corp) and used for controlled virus injection with a pneumatic injector (Narishige) at a rate of 100 nl/min. After each injection the pipette was kept in place for 5 min before being raised to the next injection depth or being removed slowly from the brain (to prevent backflow of virus). To target the visual cortex the following stereotaxic coordinates and injection volumes were followed, relative to bregma and pia. AP: −3.8, ML: +/− 2.6, Z: −450 (200nL) and −300 (200nL). After injection the skin was sutured (Ethicon) and sealed with glue (VetBond). Animals were placed in a heated cage to recover with access to soft-ened food and monitored closely. Animals were left to recover from surgery (and to provide time for virus expression) for at least 12 days before being used for slice electrophysiology experiments.
Electrode implant:
The initial surgery was performed as described above. Male Btbd11 F/F or vGAT:Btbd11 F/F mice aged 2–5 months were used for in vivo electrophysiology experiments. A craniotomy was made above the V1 in the left hemisphere (AP: level with lambda, ML: 3.2) where a 50μm polyimide-insulated tungsten wire was implanted at a depth of −0.45mm relative to pia. An additional craniotomy was made above the cerebellum just behind lambda and just to the right of the midline for a ground screw. A final craniotomy was made in the right hemisphere (AP: +1.5, ML: 0.5–0.6) for a reference electrode consisting of 125μm stainless-steel coated with PTFE. Wires were connected to a mill-max adaptor with metal pins and the implant secured with light-curable dental cement (3M RelyX). To enable head fixation, a custom-made metal head bar was secured to the skull. Mice were left to recover for 1 week before habituation to handling began. After habituation to handling, mice were habituated to brief periods of head fixation prior to recording sessions. Electrode placements were determined by making electrolytic lesions at the conclusion of experiments which were imaged on DAPI stained brain sections on a fluorescent microscope.
Slice physiology
Mice were deeply anesthetized with isoflurane. Animals irresponsive to toe pinches were decapitated, and brains were rapidly extracted and sectioned using a vibratome (Leica VT-1200). The cutting ACSF contained (in mM) 85 NaCl, 65 sucrose, 25 NaHCO3, 10 glucose, 4 MgCl2, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2 (pH 7.35, ~308mOsm). 300 μm coronal slices containing the visual cortex were collected and recovered for 10min in cutting ACSF at 32°C, after which slices were transferred to ACSF solution containing (in mM) 130 NaCl, 24 NaHCO3, 10 glucose, 3.5 KCl, 2.5 CaCl2, 1.5 MgSO4, 1.25 NaH2PO4 (pH 7.35, ~303mOsm). Slices were left to recover for 1h at room temperature before recordings. All ACSF solutions were saturated with 95% O2 and 5% CO2. For recording, a single slice was transferred to a heated chamber (34–35°C) and perfused with ACSF using a peristaltic pump (WPI). tdTomato-expressing PV-INs in layer 2/3 the visual cortex were identified on an upright microscope equipped for differential interference contrast (DIC) microscopy (Olympus BX51WI) and LED fluorescence (X-Cite, 120 LED). Whole-cell patch-clamp recordings were made using a MultiClamp 700B amplifier (12.5kHz digitization) with pClamp 10.3 software (Molecular Devices). Voltage-clamp recordings were made using borosilicate glass pipets (King Precision Glass Inc., KG-33 ID1.00 OD 1.50) with resistance 2.0–3.0MΩ, filled with internal solution containing (in mM) 117 Cs-methanesulphonate, 20 HEPES, 5 QX-314, 5 TEA-Cl, 4 ATP-Mg, 2.8 NaCl, 1 Na2-phosphocreatine, 0.4 EGTA, 0.4 GTP-Na (pH 7.30, ~290 mOsm). Access resistance was continually monitored throughout recording; cells in which access resistance rose above 20MΩ were excluded from analysis. Membrane potentials were not corrected for junction potentials.
For mEPSC analysis, recordings were conducted at −70 mV in the presence of TTX (1μm) and the GABA receptor antagonist SR-95531 (5 μM) to isolate quantal glutamatergic currents. Data were analyzed offline using Clampfit (v 10.7). Data were low pass filtered offline at 1 kHz using a Bessel filter. Using a template-matching approach, 200 events were analyzed per cell. The average amplitude and frequency of the mEPSCs was calculated, as well as the half-maximal rise and decay times.
To assess the paired pulse ratio (PPR), electrical stimuli were delivered using a bipolar stimulating electrode (FHC, MX21AES) placed laterally to the recorded cell. To allow for the concurrent calculation of AMPA/NMDA ratios, recordings were conducted at +40 mV, and in the presence of SR-95531 (5 μM) to isolate glutamatergic currents. Cells were then additionally perfused with D-AP5 (50 μM) to isolate AMPA currents. NMDA currents were calculated via digital subtraction offline. Analysis was performed offline using Clampfit.
Ca2+ imaging and analysis
Mouse Btbd11 F/F hippocampal cultures transduced with AAV-GFP or AAV-GFP-Cre and AAV-jRGECO1a were transferred to ACSF (recipe as above) and allowed to equilibrate for 10–15 min before being imaged in a temperature and humidity-controlled chamber. Images (512×512 pixels) were acquired with a 10× objective at 4Hz for 60s from multiple regions of interest for each coverslip. In FIJI, each time series was projected to a single plane (maximum intensity projection) to aid identifying the soma of neurons. A square/rectangle was drawn at the cell body of neurons and assigned as ROIs. ROIs were then opened on the original time series which was median filtered (0.5 pixels). The mean signal intensity was extracted for each neuron in the field of view. Data were then processed with custom written Python scripts using Jupyter notebooks to: 1) convert the signal to represent dF/F assigning F0 as the 10th percentile of the signal, 2) calculate the average dF/F of all neurons in each imaging region to capture the synchronous activity, 3) convert the data to a Z score, 4) subtract the minimum value from each time point to center the baseline around 0, and 5) use find_peaks and peak_prominences (from scipy library) with parameters [height threshold = 2, distance threshold = 8, prominence threshold = 1] to automatically identify spontaneous activity in the cultures.
In vivo recordings
Following habituation to handling and head fixation, mice (5 control and 6 knockout) were head restrained and presented with a monitor positioned at 45° with respect to their right eye. As a visual stimulation, mice were presented with a black screen or a gray screen for 90s. Recordings were made with a 32-channel RHD2132 head stage (Intan Technologies, CA, US) via a custom-built adaptor and an Open Ephys acquisition board via an SPI-cable (Intan Technologies). Data were amplified and digitized by the RHD2132 headstage, sampled at 30kHz, and digitally bandpass filtered between 0.1 and 300 Hz before further processing in MATLAB (Mathworks) and custom written scripts in Jupyter notebooks. Data were down sampled to 1kHz and power analyzed with compute_spectrum and specgram from the Neurodsp and matplotlib libraries, respectively. The mean and peak 55Hz–65Hz power was calculated with presentation of the black or gray screen and converted to a gray/black screen ratio.
Analysis of Psd95 puncta size and density
To evaluate the glutamatergic synapse density in control and IN-specific Btbd11 KO mice we generated vGAT+/−:Btbd11 F/F:Psd95-GFP+/− animals and vGAT+/−:Psd95-GFP+/− to serve as Btbd11 KO and control mice, respectively. Animals aged 6 weeks, were perfused with ice-cold PBS followed by ice-cold 4% PFA in PBS. IHC was then conducted on free-floating 40μm thick slices prepared on a cryostat (Leica). Slices from 4 control and 4 KO mice (6 weeks) were washed with PBS x3 for 10 min, then PBST (0.3% Triton X-100) for 20 min, all at RT. Slices were then blocked for 1 h in PBST (0.15% Triton X-100) with 5% Goat serum at RT, before incubation with a PV primary antibody (Swant, Rb 1:2000) in PBST (0.15% Triton X-100) with 5% normal goat serum (Vector labs) at 4C overnight. Slices were then washed 4x in PBS for 10 min at RT before being incubated with a secondary antibody (Goat anti rabbit Alexa Fluor 568, Thermo, 1:500) in the same blocking buffer for 2 h at RT. Slices were then washed 4x with PBS, with the penultimate wash containing the nuclear stain DAPI. Slices were then mounted with PermaFluor media (Thermo Fisher Scientific) and stored at 4C in the dark. Slides were imaged on a Zeiss LSM 880. Stacks were acquired from layer 2/3 of the visual cortex at optimal resolution with a 63× objective and identical laser settings for each group. The endogenous Psd95-GFP signal was acquired alongside the PV and DAPI signal. Images were analyzed in FIJI. All inhibitory INs had Psd95-GFP puncta, but we restricted the analysis to dendrites of PV neurons (to complement mEPSC recordings in Figure 5). All analysis was performed by an experimenter blind to the experimental condition. To aid puncta detection the Psd95-GFP channel was background subtracted (rolling ball = 10) and then Gaussian blurred (1-pixel). Centered on a region of primary PV dendrite a maximum intensity projection was made from 5 optical z-planes. A region of PV-IN primary dendrite was used to generate a region of interest (ROI), then Psd95-GFP puncta were manually identified within the dendritic region. Freehand ROIs were drawn around each puncta to enable both the density of puncta, but also the puncta size to be calculated for each dendritic region. For each cell one dendrite was analyzed, and 5 cells were analyzed per animal (20 cells/dendrites per group in total). For analysis, the puncta density was calculated per dendrite area (μm2) and then compared between groups with a T-test. The distribution of puncta size was skewed, so for each dendrite the median size was used. The median puncta size was then compared per group with a T-test.
Behavioral testing
Animals were housed in a holding room on a reverse light cycle. Testing was conducted during the dark phase after animals were habituated to handling. The number, sex and age of animals used for each experiment are outlined in Table S2.
Locomotor activity
Locomotor activity was assessed by placing animals in an open field arena (40 × 40cm; San Diego Instruments Inc.) and measuring the number of infrared beam breaks during either for 90min in the dark (Figures S7A and S7B) or for 30 min in the light (Figure 7A) session. Data were binned into 5 min periods for analysis. Information was extracted for rearing events and beam breaks in the central portion of the arena using the Photobeam Activity System software (San Diego Instruments Inc.).
Elevated plus maze
To assess anxiety an elevated plus maze (66 cm long and 5 cm wide; San Diego Instruments Inc), consisting of two closed arms and two open arms suspended 54 cm above the ground was used. The test was conducted in a well-lit room and the maze was thoroughly cleaned between animals with disinfectant (MB-10). Animals were placed onto the center of the elevated plus maze facing an open arm and allowed to explore for 5 min, wherein animal position was tracked using ANYmaze software (Stoelting, IL). Zones were drawn for the open and closed arms, with an additional zone drawn at the end of the open arm presumably in the most anxiogenic portion of the maze). Zone entry settings were set such that an entry required 70% of the animal to be in the new zone. A zone exit occurred when less than 30% of the animals’ body area remained in the zone. Freezing was detected with Anymaze software (threshold 2s).
Hole board assay
The hole board assay was used as a measure of exploratory behavior. A hole board (40 cm × 40 cm) with 16 holes (diameter 3cm) was used (ANY-maze). Animals were tested under red-light with gentle illumination from two lamps. Mice were placed into the arena for 3 min, and nose pokes were manually scored for the entire duration of the test. The apparatus was thoroughly cleaned between animals with disinfectant (MB-10).
Marble burying assay
The marble burying test was conducted in a clean cage (regular home cage) filled 1/3 full of clean sawdust. 12 marbles were arranged in 4 rows of 3 marbles. Animals were placed into the cage for 30 min in the dark (under red light). After 30 min the animals were carefully removed (making sure not to disturb any marbles) and the number of buried marbles counted. A marble needed to be 2/3 covered with sawdust to be considered buried.
Y-maze spontaneous alternation
Spatial short-term memory was assessed in the Y-maze spontaneous alternation task. Mice were placed at the end of one arm of a Y-maze consisting of three 38cm long arms (San Diego Instruments Inc) and allowed to freely explore the maze for 5 min. Animal location was automatically recorded and tracked using Anymaze Tracking Software (Stoelting). The percent of spontaneous alternation was calculated using the equation: % alternation = (total alternations/(total arm entries −2))×100.
T-maze spontaneous alternation
The T-maze was used as a test of spatial short-term memory. A clear Perspex T-maze was used and cleaned between animals with disinfectant (MB-10). Animals were tested in a dimly lit room and two trials were collected per day per animal for 4 subsequent days. For each trial an animal was placed in the start arm and allowed to run down the maze and make a choice of one of the goal arms (sample trial). A choice was made when all four paws were inside one of the goal arms. A plastic divider was then used to keep the mouse in the chosen arm for 20s. The animal was then gently removed from the maze and immediately returned to the start arm (with the goal arm divider removed) where it was allowed to make a choice of either goal arm (choice trial). As before, all four paws in an arm was considered to be a decision. After the animal reached the end of the chosen arm it was removed from the maze and placed into a temporary holding cage before being given a second trial with an inter-trial interval of 45–60s. A correct decision was scored when the animal alternated arms between the sample and choice trials. The average alternation was calculated for all 8 trials run over 4 days.
Contextual fear conditioning
Contextual fear conditioning was conducted as described previously.50 On day 1, mice were placed in Med Associates chambers (light setting 1) and after 2 min they were subject to a single foot shock 0.7 mA lasting 2s. Animals remained in the chamber for a further 2 min. Between animals the chambers were thoroughly cleaned with disinfectant (MB-10). 24hrs later animals were returned to the identical chamber for 5 min, and freezing behavior was automatically recorded (detection threshold = 15, minimum of 6 frames). The degree of freezing was used as a proxy for contextual fear memory.
Locomotor activity with MK-801 challenge:
Activity was assessed as above, except the area was illuminated animals received an intraperitoneal injection of saline or MK-801 maleate (0.2 mg/kg; Tocris). A within-subject design was followed, and the test was repeated 2-days later with drug assignment reversed. The order of saline vs. MK-801 was counterbalanced for genotype.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 9 was used for all statistical analyses. Figures were plotted in Prism 9 or using Python and Jupyter Notebooks. Images were processed in FIJI60 and often presented as maximum intensity projections. Images were frequently median filtered to reduce noise. Any adjustment of brightness and contrast was performed uniformly across the image. Figures were assembled in Adobe Illustrator. Details of all statistical tests (n numbers, age, and sex of animals) can be found in Table S2. To reduce the chance of bias, FRAP, mEPSC analyses, PPR, AMPA/NMDAR ratio, and Psd-95-GFP puncta were performed on decoded files and all behavioral experiments were conducted by an investigator that was unaware of the genotype of the animals being tested. Except for the marble burying and hole-board behavioral assays, male and female animals were analyzed together. Data were assumed to follow the normal distribution, but no formal tests of normality were conducted. Non-parametric tests were used for the marble burying test and the freezing time on the elevated plus maze.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| ChromoTek GFP-Trap agarose beads | ChromoTek | Cat# Gta |
| ChromoTek Binding Control Agarose Beads | ChromoTek | Cat# bab |
| Rabbit Living Colors dsRed Polyclonal Antibody | Takara | Cat# 632496, RRID:AB_10013483 |
| Mouse anti-GFP | Santa Cruz Biotechnology | Cat# sc-9996, RRID:AB_627695 |
| Rabbit anti Btbd11 | This paper | N/A |
| Rabbit anti Btbd11 | Abclonal | Cat# A20192 |
| Mouse anti Psd95 (clone K28/74) | NeuroMab | Cat# K28/74, RRID:AB_2877478) |
| Mouse anti GST | Sigma-Aldrich | Cat# SAB4200692, RRID:AB_259845 |
| Rabbit anti Parvalbumin | Swant | Cat# PV27, RRID:AB_2631173 |
| Mouse anti Alpha-tubulin | Santa Cruz Biotechnology | Cat# sc-5286, RRID:AB_628411 |
| Mouse anti Synaptophysin | Sigma-Aldrich | Cat# S5768, RRID:AB_477523 |
| Mouse anti Lhx6 | Santa Cruz Biotechnology | Cat# sc-271433, RRID:AB_10649856 |
| Chicken anti GFP | Abcam | Cat# ab13970, RRID:AB_300798 |
| Mouse anti Psd95 (clone K28/43) | NeuroMab | Cat# K28/43, RRID:AB_2877189 |
| Mouse anti GAD67 | Santa Cruz Biotechnology | Cat# sc-28376, RRID:AB_627650 |
| Mouse anti parvalbumin | Swant | Cat# 235, RRID:AB_10000343 |
| Mouse anti Gephyrin | Synaptic Systems | Cat# 147011, RRID:AB_887717 |
| IRDye 680RD Goat anti-Mouse IgG Secondary Antibody | LI-COR Biosciences | Cat# 926–68070, RRID:AB_10956588 |
| IRDye 800CW Goat anti-Rabbit IgG Secondary Antibody | LI-COR Biosciences | Cat# 926–32211, RRID:AB_621843 |
| IRDye 680LT Goat anti-Mouse IgG1-Specific Secondary Antibody | LI-COR Biosciences | Cat# 926–68050, RRID:AB_2783642 |
| IRDye 800CW Goat anti-Mouse IgG2a-Specific Secondary Antibody | LI-COR Biosciences | Cat# 926–32351, RRID:AB_2782998 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A-11011, RRID:AB_143157 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A-21131, RRID:AB_2535771 |
| Goat anti-Chicken IgY (H + L) Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A-11039, RRID:AB_142924 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A-21134, RRID:AB_2535773 |
| DyLight 405-AffiniPure Goat Anti-Mouse IgG, Fc_ Subclass 3 Specific | Jackson Immuno Research | Cat# 115-475-209, RRID:AB_2338803 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A-21124, RRID:AB_2535766 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A-21240, RRID:AB_2535809 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Escherichia coli: Stbl3 | ThermoFisher | Cat #C737303 |
| Escherichia coli: Top10 | ThermoFisher | Cat #C404010 |
| PJ69-4A yeast cells | (James et al., 1996)56 | N/A |
| AAV.CMV.PI.EGFP.WPRE.bGH (serotype 2/9) | Addgene | Cat# 105530-AAV9 |
| AAV.CMV.FII.eGFP-Cre.WPRE.SV40 (serotype 2/9) | Addgene | Cat# 105545-AAV9 |
| AAV-FLEX-tdTomato | Addgene | Cat# 28306-AAV9 |
| AAV-S5E2-dTom-nlsdTom (serotype 2/9) | Janelia Farm | N/A |
| AAV.Syn.NES-jRGECO1a.WPRE.SV40 (serotype 2/9) | Addgene | Cat# 100854-AAV9 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Sodium pyrophosphate decahydrate | Sigma-Aldrich | Cat# 221368; CAS: 13472-36-1 |
| Sodium orthovanadate | Sigma-Aldrich | Cat# S6508; CAS: 13721-39-6 |
| Sodium Fluoride | Sigma-Aldrich | Cat# S7920, CAS: 7681-49-4 |
| Sodium Chloride | Sigma-Aldrich | Cat# S7653, CAS: 7647-14-5 |
| Potassium Chloride | Sigma-Aldrich | Cat# P9333, CAS: 7447-40-7 |
| Sodium deoxycholate | Sigma-Aldrich | Cat# 30970, CAS: 302-95-4 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787, CAS: 9036-19-5 |
| IGEPAL CA-630 (NP-40) | Sigma-Aldrich | Cat# I8896, CAS: 9002-93-1 |
| β-Mercaptoethanol | Millipore | Cat# 444203, CAS: 60-24-2 |
| MES | Fisher | Cat# 50–188-3347 |
| HEPES, ≥99.5% (titration) | Sigma-Aldrich | Cat# H3375; CAS: 7365-45-9 |
| Sucrose | Sigma-Aldrich | Cat# S7903; CAS: 57-50-1 |
| cOmplete, EDTA-free | Roche | Cat# 04 693 132 001 |
| Pefabloc SC (AEBSF) | Roche | Cat# 11427393103 |
| PhosSTOP™ | Roche | Cat# 04 906 837 001 |
| Okadaic acid, Free acid, >98% | LC Laboratories | Cat# O-2220 CAS: 78111-17-8 |
| Ethylenediaminetetraacetic acid disodium salt dihydrate | Sigma-Adlrich | Cat# ED2SS; CAS: 6381-92-6 |
| Glycine | Fisher Scientific | Cat# BP381-5 CAS: 56-40-6 |
| Sodium dodecyl sulfate | Sigma-Aldrich | Cat# 436143, CAS: 151-21-3 |
| Normal Goat Serum | Vector Laboratories | Cat# S-1000-20 |
| Neurobasal Medium | Gibco | Cat# 21103049 |
| Neurobasal Plus Medium | Gibco | Cat# A3582901 |
| B-27 Supplement | Gibco | Cat# 17504044 |
| B-27 Plus Supplement | Gibco | Cat# A3582801 |
| Glutamax Supplement | Gibco | Cat# 35050061 |
| Penicillin-Streptomycin | Gibco | Cat# 15070063 |
| Fetal Bovine Serum | HyClone | Cat# SH30070.03 |
| Poly-L-lysine hydrobromide | Sigma-Aldrich | Cat# P2636 |
| Paraformaldehyde (16%) | Electron Microscopy Sciences | Cat# 15711 |
| Tween 20 | Fisher Scientific | Cat# BP337-500; CAS: 9005-64-5 |
| Lipofectamine 2000 | Invitrogen | Cat# 11668019 |
| SR 95531 hydrobromide | Tocris | Cat# 1262 |
| Tetrodotoxin citrate | Tocris | Cat# 1069 |
| D-AP5 | Tocris | Cat# 0106 |
| MK-801 maleate | Tocris | Cat# 0924 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T | ATCC | Cat # CRL-3216 |
|
| ||
| Deposited data | ||
|
| ||
| Raw Mass Spectrometry Data Files | This paper | ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD041635 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Btbd11 (rederived from frozen sperm) | European Mouse Mutant Archive | Btbd11tm1a(EUCOMM)Wtsi |
| Mouse: vGAT-cre | The Jackson Laboratory | Strain #:028862 |
| Mouse: CaMKII-cre | The Jackson Laboratory | Strain #:005359 |
| Mouse: Psd95-GFP-ENABLED v2 | From Haining Zhong, OHSU | N/A |
| Rat: Sprague-Dawley (timed pregnant E18) | Envigo | Order code: 002-US |
|
| ||
| Oligonucleotides | ||
|
| ||
| Guide sequence to target the N terminus | This paper | N/A |
| of Btbd11: 5-ACGGCGGCTGCAGCATGAAG-3’ | ||
|
| ||
| Recombinant DNA | ||
|
| ||
| pAAV-S5E2-dTom-nlsdTom | Addgene | Cat# 135630 |
| pAAV-mDlx-GFP-Fishell-1 | Addgene | Cat# 83900 |
| pAAV-mDIx-Azurite | This paper | N/A |
| pORANGE Cloning template vector | Addgene | Cat# 131471 |
| Btbd11 (Myc-DDK-tagged) - Mouse BTB (POZ) domain containing 11 (Btbd11), transcript variant 1 | Origene | Cat# MR217199 |
| pCAG-Azurite | (Fangetal., 2021)57 | N/A |
| pCAG-GFP-Btbd11 | This paper | N/A |
| pCAG-mCherry-Btbd11 | This paper | N/A |
| pCAG-GFP-5xANK BTB (truncation) | This paper | N/A |
| pCAG-mCherry-5xANK BTB (truncation) | This paper | N/A |
| pCAG-GFP-Btbd11 Δ PBM | This paper | N/A |
| pCIS-GST-Btbd11 | This paper | N/A |
| pCIS-GST-Btbd11 Δ PBM | This paper | N/A |
| pDBLeu-Btbd11-C-terminus | This paper | N/A |
| pDBLeu-Btbd11-C-terminus Δ PBM | This paper | N/A |
| pDBLeu-Btbd11 (BTB domain only) | This paper | N/A |
| pPC86-PSD95 PDZ1 | (Kim etal., 1998)58 | N/A |
| pPC86-PSD95 PDZ2 | (Kim etal., 1998)58 | N/A |
| pPC86-PSD95 PDZ3 | (Kim etal., 1998)58 | N/A |
| pPC86-PSD95 PDZ1,2 | (Kim etal., 1998)58 | N/A |
| pPC86-PSD95 PDZ2,3 | (Kim etal., 1998)58 | N/A |
| pPC86-PSD95 PDZ1,2,3 | (Kim etal., 1998)58 | N/A |
| pCMV-Psd-95-mCherry | (Araki et al., 2020)59 | N/A |
| pCMV-Psd-95-mCherry Δ PDZ1,2 | This paper | N/A |
| pCMV-Psd-95-mCherry Δ PDZ3 | This paper | N/A |
| pCMV-Psd-95-mCherry Δ PDZ1,2,3 | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| LI-COR Image Studio | LI-COR Biosciences | N/A |
| ANY-maze behavior tracking software | Stoelting | N/A |
| ImageJ | (Schneider et al. 2012)60 | https://imagej.nih.gov/ij/ |
| IMARIS | Oxford Instruments | Version 9 |
| Illustrator | Adobe | Version 2023 |
| Prism | Graphpad | Version 9 |
| MATLAB | Mathworks | https://www.mathworks.com/ |
| Python 3.7 | Jupyter notebooks | Ver 3.7 |
|
| ||
| Other | ||
|
| ||
| Nanodrop One C | Thermo Fisher | Cat# 840–317500 |
| LSM 880 microscope | Zeiss | N/A |
Highlights.
Btbd11 is exclusively expressed in cortical and hippocampal inhibitory interneurons
Btbd11 binds to Psd-95 and localizes to glutamatergic synapses
Knockout of Btbd11 decreases mEPSC frequency in PV interneurons
Btbd11 KO alters neuronal circuit activity and anxiety-related behaviors
ACKNOWLEDGMENTS
We would like to thank members of the Huganir laboratory for helpful discussions. We would like to thank Sarah Rodriguez and Ashley Irving for help with mouse colony management; Chip Hawkins for conducting IVF to generate Btbd11 mice; Ingie Hong and Elena Lopez-Ortega for help with in vivo electrophysiology recordings and analysis pipelines; William Hale for help with primary cultured mouse neurons; Mike Delannoy for processing of samples for electron microscopy; Paul Worley for useful discussions and input on experimental design; and Tatiana Boronina and Robert Cole for running and analyzing mass spectrometry data. BioRender was used to make the graphical abstract. This work was supported by K99/R00 award K99MH124920 (A.M.B.), R01NS036715 (R.L.H.), and RF1MH120119 (H.Z.).
Footnotes
DECLARATION OF INTERESTS
R.L.H. is scientific co-founder and SAB member of Neumora Therapeutics and SAB member of MAZE Therapeutics. M.S. is scientific co-founder and SAB member of Neumora Therapeutics and SAB member of Biogen, Cerevel, and Vanqua.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112591.
REFERENCES
- 1.Hu H, Gan J, and Jonas P (2014). Interneurons. Fast-spiking, parvalbumin+ GABAergic interneurons: from cellular design to microcircuit function. Science 345, 1255263. 10.1126/science.1255263. [DOI] [PubMed] [Google Scholar]
- 2.Marín O (2012). Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 13, 107–120. 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 3.Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S, and Grace AA (2008). Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 31, 234–242. 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis DA, Hashimoto T, and Volk DW (2005). Cortical inhibitory neurons and schizophrenia. Nat. Rev. Neurosci. 6, 312–324. 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 5.Sohal VS, Zhang F, Yizhar O, and Deisseroth K (2009). Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698–702. 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann EO, Suckling JM, Hajos N, Greenfield SA, and Paulsen O (2005). Perisomatic feedback inhibition underlies cholinergically induced fast network oscillations in the rat hippocampus in vitro. Neuron 45, 105–117. 10.1016/j.neuron.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Kuhlman SJ, Olivas ND, Tring E, Ikrar T, Xu X, and Trachtenberg JT (2013). A disinhibitory microcircuit initiates critical-period plasticity in the visual cortex. Nature 501, 543–546. 10.1038/nature12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donato F, Rompani SB, and Caroni P (2013). Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature 504, 272–276. 10.1038/nature12866. [DOI] [PubMed] [Google Scholar]
- 9.Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, and Moore CI (2009). Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 459, 663–667. 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang MC, Park JM, Pelkey KA, Grabenstatter HL, Xu D, Linden DJ, Sutula TP, McBain CJ, and Worley PF (2010). Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat. Neurosci. 13, 1090–1097. 10.1038/nn.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matta JA, Pelkey KA, Craig MT, Chittajallu R, Jeffries BW, and McBain CJ (2013). Developmental origin dictates interneuron AMPA and NMDA receptor subunit composition and plasticity. Nat. Neurosci. 16, 1032–1041. 10.1038/nn.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamsa KP, Heeroma JH, Somogyi P, Rusakov DA, and Kullmann DM (2007). Anti-Hebbian long-term potentiation in the hippocampal feedback inhibitory circuit. Science 315, 1262–1266. 10.1126/science.1137450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, and Monyer H (1995). Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron 15, 193–204. 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Wu X, Wu H, and Zhang M (2020). Phase separation at the synapse. Nat. Neurosci. 23, 301–310. 10.1038/s41593-019-0579-9. [DOI] [PubMed] [Google Scholar]
- 15.Zeng M, Díaz-Alonso J, Ye F, Chen X, Xu J, Ji Z, Nicoll RA, and Zhang M (2019). Phase separation-mediated TARP/MAGUK complex condensation and AMPA receptor synaptic transmission. Neuron 104, 529–543. 10.1016/j.neuron.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng M, Shang Y, Araki Y, Guo T, Huganir RL, and Zhang M (2016). Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 166, 1163–1175. 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Z, Chen X, Zeng M, and Zhang M (2019). Phase separation as a mechanism for assembling dynamic postsynaptic density signalling complexes. Curr. Opin. Neurobiol. 57, 1–8. 10.1016/j.conb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Sancho L, and Bloodgood BL (2018). Functional distinctions between spine and dendritic synapses made onto parvalbumin-positive interneurons in mouse cortex. Cell Rep. 24, 2075–2087. 10.1016/j.celrep.2018.07.070. [DOI] [PubMed] [Google Scholar]
- 19.Keck T, Scheuss V, Jacobsen RI, Wierenga CJ, Eysel UT, Bonhoeffer T, and Hübener M (2011). Loss of sensory input causes rapid structural changes of inhibitory neurons in adult mouse visual cortex. Neuron 71, 869–882. 10.1016/j.neuron.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Melander JB, Nayebi A, Jongbloets BC, Fortin DA, Qin M, Ganguli S, Mao T, and Zhong H (2021). Distinct in vivo dynamics of excitatory synapses onto cortical pyramidal neurons and parvalbumin-positive interneurons. Cell Rep. 37, 109972. 10.1016/j.celrep.2021.109972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortin DA, Tillo SE, Yang G, Rah JC, Melander JB, Bai S, SolerCedeñ o O, Qin M, Zemelman BV, Guo C, et al. (2014). Live imaging of endogenous PSD-95 using ENABLED: a conditional strategy to fluorescently label endogenous proteins. J. Neurosci. 34, 16698–16712. 10.1523/JNEUROSCI.3888-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández E, Collins MO, Uren RT, Kopanitsa MV, Komiyama NH, Croning MDR, Zografos L, Armstrong JD, Choudhary JS, and Grant SGN (2009). Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 5, 269. 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uezu A, Kanak DJ, Bradshaw TWA, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, and Soderling SH (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129. 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furlanis E, Traunmüller L, Fucile G, and Scheiffele P (2019). Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat. Neurosci. 22, 1709–1717. 10.1038/s41593-019-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346. 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willems J, de Jong APH, Scheefhals N, Mertens E, Catsburg LAE, Poorthuis RB, de Winter F, Verhaagen J, Meye FJ, and MacGillavry HD (2020). Orange: a CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18, e3000665. 10.1371/journal.pbio.3000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, et al. (2020). Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710. 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 28.Imamura F, Maeda S, Doi T, and Fujiyoshi Y (2002). Ligand binding of the second PDZ domain regulates clustering of PSD-95 with the Kv1.4 potassium channel. J. Biol. Chem. 277, 3640–3646. 10.1074/jbc.M106940200. [DOI] [PubMed] [Google Scholar]
- 29.Mosavi LK, Cammett TJ, Desrosiers DC, and Peng ZY (2004). The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448. 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alberti S, and Dormann D (2019). Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194. 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 31.Uversky VN (2017). Protein intrinsic disorder-based liquid–liquid phase transitions in biological systems: complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 239, 97–114. 10.1016/j.cis.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological Fibrillization. Cell 163, 123–133. 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi G-A, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 19, 1743–1749. 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryder E, Gleeson D, Sethi D, Vyas S, Miklejewska E, Dalvi P, Habib B, Cook R, Hardy M, Jhaveri K, et al. (2013). Molecular characterization of mutant mouse strains generated from the EUCOMM/KOMP-CSD ES cell resource. Mamm. Genome 24, 286–294. 10.1007/s00335-013-9467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vormstein-Schneider D, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias-Garcia MA, Allaway K, Sakopoulos S, Schneider G, Stevenson O, et al. (2020). Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci. 23, 1629–1636. 10.1038/s41593-020-0692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleem AB, Lien AD, Krumin M, Haider B, Rosón MR, Ayaz A, Reinhold K, Busse L, Carandini M, Harris KD, et al. (2017). Subcortical source and modulation of the narrowband gamma oscillation in mouse visual cortex. Neuron 93, 315–322. 10.1016/j.neuron.2016.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooke SF, Komorowski RW, Kaplan ES, Gavornik JP, and Bear MF (2015). Visual recognition memory, manifested as long-term habituation, requires synaptic plasticity in V1. Nat. Neurosci. 18, 262–271. 10.1038/nn.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda H, Tsuji M, and Matsumiya T (1998). Changes in head-dipping behavior in the hole-board test reflect the anxiogenic and/or anxiolytic state in mice. Eur. J. Pharmacol. 350, 21–29. 10.1016/s0014-2999(98)00223-4. [DOI] [PubMed] [Google Scholar]
- 39.Deacon RMJ (2006). Digging and marble burying in mice: simple methods for in vivo identification of biological impacts. Nat. Protoc. 1, 122–124. 10.1038/nprot.2006.20. [DOI] [PubMed] [Google Scholar]
- 40.Deacon RMJ, and Rawlins JNP (2006). T-maze alternation in the rodent. Nat. Protoc. 1, 7–12. 10.1038/nprot.2006.2. [DOI] [PubMed] [Google Scholar]
- 41.Bygrave AM, Masiulis S, Nicholson E, Berkemann M, Barkus C,Sprengel R, Harrison PJ, Kullmann DM, Bannerman DM, and Kätzel D (2016). Knockout of NMDA-receptors from parvalbumin interneurons sensitizes to schizophrenia-related deficits induced by MK-801. Transl. Psychiatry 6, e778. 10.1038/tp.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu F, Collins MO, Harmse J, Choudhary JS, Grant SGN, and Komiyama NH (2020). Cell-type-specific visualisation and biochemical isolation of endogenous synaptic proteins in mice. Eur. J. Neurosci. 51, 793–805. 10.1111/ejn.14597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Tao T, Liu G, Gao X, Gao Y, Zhuang Z, Lu Y, Wang H, Li W, Wu L, et al. (2021). TRAF3 mediates neuronal apoptosis in early brain injury following subarachnoid hemorrhage via targeting TAK1-dependent MAPKs and NF-kB pathways. Cell Death Dis. 12, 10. 10.1038/s41419-020-03278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra R, Upadhyay A, Prajapati VK, Dhiman R, Poluri KM, Jana NR, and Mishra A (2019). LRSAM1 E3 ubiquitin ligase: molecular neurobiological perspectives linked with brain diseases. Cell. Mol. Life Sci. 76, 2093–2110. 10.1007/s00018-019-03055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, and Ting AY (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887. 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossier J, Bernard A, Cabungcal JH, Perrenoud Q, Savoye A, Gallopin T, Hawrylycz M, Cuénod M, Do K, Urban A, et al. (2015). Cortical fast-spiking parvalbumin interneurons enwrapped in the perineuronal net express the metallopeptidases Adamts8, Adamts15 and Neprilysin. Mol. Psychiatry 20, 154–161. 10.1038/mp.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Favuzzi E, Marques-Smith A, Deogracias R, Winterflood CM, Sánchez-Aguilera A, Mantoan L, Maeso P, Fernandes C, Ewers H, and Rico B (2017). Activity-dependent gating of parvalbumin interneuron function by the perineuronal net protein Brevican. Neuron 95, 639–655. 10.1016/j.neuron.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 48.Tan Z, Robinson HL, Yin DM, Liu Y, Liu F, Wang H, Lin TW, Xing G, Gan L, Xiong WC, et al. (2018). Dynamic ErbB4 activity in hippocampal-prefrontal synchrony and top-down attention in rodents. Neuron 98, 380–393. 10.1016/j.neuron.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.delPino I, García-Frigola C, Dehorter N, Brotons-Mas JR, Alvarez-Salvado E, Martínez de Lagrán M, Ciceri G, Gabaldón MV, Moratal D, Dierssen M, et al. (2013). Erbb4 deletion from fast-spiking interneurons causes schizophrenia-like phenotypes. Neuron 79, 1152–1168. 10.1016/j.neuron.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 50.He X, Li J, Zhou G, Yang J, McKenzie S, Li Y, Li W, Yu J, Wang Y, Qu J, et al. (2021). Gating of hippocampal rhythms and memory by synaptic plasticity in inhibitory interneurons. Neuron 109, 1013–1028. 10.1016/j.neuron.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Homayoun H, and Moghaddam B (2007). NMDA receptor hypofunction produces opposite effects on prefrontal cortex interneurons and pyramidal neurons. J. Neurosci. 27, 11496–11500. 10.1523/JNEUROSCI.2213-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlén M, Meletis K, Siegle JH, Cardin JA, Futai K, Vierling-Claassen D, Ruhlmann C, Jones SR, Deisseroth K, Sheng M, et al. (2012). A critical role for NMDA receptors in parvalbumin interneurons for gamma rhythm induction and behavior. Mol. Psychiatry 17, 537–548. 10.1038/mp.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bygrave AM, Masiulis S, Kullmann DM, Bannerman DM, and Kätzel D (2018). Gene-environment interaction in a conditional NMDAR-knockout model of schizophrenia. Front. Behav. Neurosci. 12, 332. 10.3389/fnbeh.2018.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, Quinlan EM, and Nakazawa K (2010). Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat. Neurosci. 13, 76–83. 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Z, Rompala GR, Zhang S, Cowell RM, and Nakazawa K (2013). Social isolation exacerbates schizophrenia-like phenotypes via oxidative stress in cortical interneurons. Biol. Psychiatry 73, 1024–1034. 10.1016/j.biopsych.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.James P, Halladay J, and Craig EA (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang H, Bygrave AM, Roth RH, Johnson RC, and Huganir RL (2021). An optimized crispr/cas9 approach for precise genome editing in neurons. Elife 10, e65202. 10.7554/eLife.65202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JH, Liao D, Lau LF, and Huganir RL (1998). SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20, 683–691. 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 59.Araki Y, Hong I, Gamache TR, Ju S, Collado-Torres L, Shin JH, and Huganir RL (2020). Syngap isoforms differentially regulate synaptic plasticity and dendritic development. Elife 9, 562733–e56328. 10.7554/eLife.56273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stauffer W, Sheng H, and Lim HN (2018). EzColocalization: an ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Sci. Rep. 8, 15764. 10.1038/s41598-018-33592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data is available from the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD041635.
No unique code is described in this study.
All other data and any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


