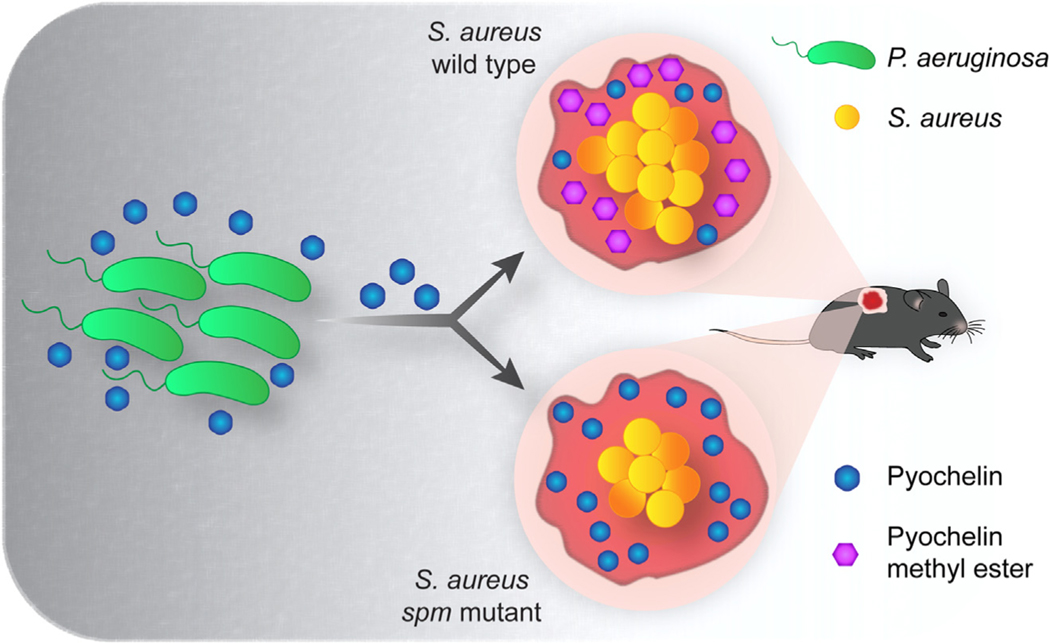
SUMMARY
Pseudomonas aeruginosa and Staphylococcus aureus are among the most frequently isolated bacterial species from polymicrobial infections of patients with cystic fibrosis and chronic wounds. We apply mass spectrometry guided interaction studies to determine how chemical interaction shapes the fitness and community structure during co-infection of these two pathogens. We demonstrate that S. aureus is equipped with an elegant mechanism to inactivate pyochelin via the yet uncharacterized methyltransferase Spm (staphylococcal pyochelin methyltransferase). Methylation of pyochelin abolishes the siderophore activity of pyochelin and significantly lowers pyochelin-mediated intracellular reactive oxygen species (ROS) production in S. aureus. In a murine wound co-infection model, an S. aureus mutant unable to methylate pyochelin shows significantly lower fitness compared with its parental strain. Thus, Spm-mediated pyochelin methylation is a mechanism to increase S. aureus survival during in vivo competition with P. aeruginosa.
In brief
Jenul et al. report that Staphylococcus aureus inactivates the Pseudomonas aeruginosa-derived siderophore pyochelin by methylation. Pyochelin methylation is dependent on the enzyme staphylococcal pyochelin methyltransferase (Spm) and increases S. aureus fitness in co-infections with P. aeruginosa.
Graphical Abstract
INTRODUCTION
Staphylococcus aureus and Pseudomonas aeruginosa are opportunistic pathogens frequently co-isolated from sites of chronic infection, including the pulmonary infections of persons with cystic fibrosis (CF)1,2 and chronic wounds.3–8 In CF pulmonary infections, co-infections with these two pathogens are associated with worsened patient outcomes due to increased inflammation, virulence factor expression, and antibiotic tolerance.1,9,10 Contrary to CF lung infections, the impact of S. aureus/P. aeruginosa co-infection in chronic wounds is not well understood. Due to the lack of comprehensive long-term clinical data on chronic wound co-infection, studies have primarily relied on in vitro, ex vivo, and animal models to better understand how co-infection with S. aureus and P. aeruginosa influences chronic wound outcomes.11–13 Despite frequent co-isolation from the infection environment, which suggests some level of co-existence, co-cultivation under standard laboratory conditions leads to antagonistic interactions between these two microbes, with P. aeruginosa outcompeting S. aureus.12,14–17 This phenomenon is highly strain dependent and influenced by the inoculum ratio of the two bacterial species as well as culturing conditions.18
P. aeruginosa isolates from initial infection of the lungs of people with CF are antagonistic toward S. aureus, while isolates from chronic infections may support co-existence.17 Research into chronic P. aeruginosa infection has shown that its prolonged persistence in the CF environment results in the accumulation of mutations, leading to increased biofilm formation and antibiotic resistance, decreased virulence, and general metabolic adaption to the CF environment, which includes a shift from siderophore-based iron acquisition to heme-based iron acquisition.19–25 Importantly, chronic wound biopsies suggest that S. aureus and P. aeruginosa occupy distinct niches and therefore may interact through chemical competition rather than physical interaction.26
S. aureus is a known early colonizer of patients with CF and the main pathogen found in young children, while P. aeruginosa is the dominant pathogen found in older patients with CF.27 In addition, P. aeruginosa is not commonly found to inhabit the skin,28 while S. aureus is a common, although transient, member of the skin microbiota of healthy individuals28 and is likely seeded from its primary niche, the anterior nares.29 Taken together, S. aureus is likely to encounter non-adapted P. aeruginosa strains during the early stages of skin wound infections as well as during the early onset of P. aeruginosa CF lung infection. The objective of this study was to determine the roles of key metabolic exchange factors governing the in vivo co-existence of non-adapted P. aeruginosa and S. aureus.
The antagonistic interactions between S. aureus and P. aeruginosa are multifactorial. P. aeruginosa produces a variety of extracellular factors that kill or inhibit growth of S. aureus, including the phenazine respiratory toxins, such as pyocyanin30; alkyl-hydroxyquinoline N-oxides, a class of respiratory toxins known to inhibit S. aureus growth31; the metalloendopeptidase LasA that lyses S. aureus via peptidoglycan cleavage32,33; rhamnolipids, which have antiadhesive effects and disperse staphylococcal biofilms34,35; and the siderophores pyoverdine36,37 and pyochelin,37,38 with the latter not only exhibiting iron chelating activity but also inducing intracellular reactive oxygen species (ROS) production in S. aureus and other bacteria.39,40 In response, S. aureus forms small-colony variants (SCVs),16 a phenotype that is selected for when S. aureus is subjected to respiratory toxins like pyocyanin and alkyl-hydroxyquinoline N-oxides.41,42
Herein, we identify an additional mechanism that S. aureus uses to respond to P. aeruginosa antagonism: inactivation of the P. aeruginosa siderophore pyochelin via methylation. Using a combination of mass spectrometry imaging (MSI), classical molecular networking, MS guided screening, genetic mutation, and in vitro and in vivo assays, we show that some S. aureus strains encode an uncharacterized methyltransferase, Spm (staphylococcal pyochelin methyltransferase), that neutralizes the siderophore- and ROS-generating functions of pyochelin. Importantly, the presence of Spm is critical for S. aureus co-existence with P. aeruginosa in a murine wound co-infection model.
RESULTS
MRSA methylates pyochelin produced by P. aeruginosa
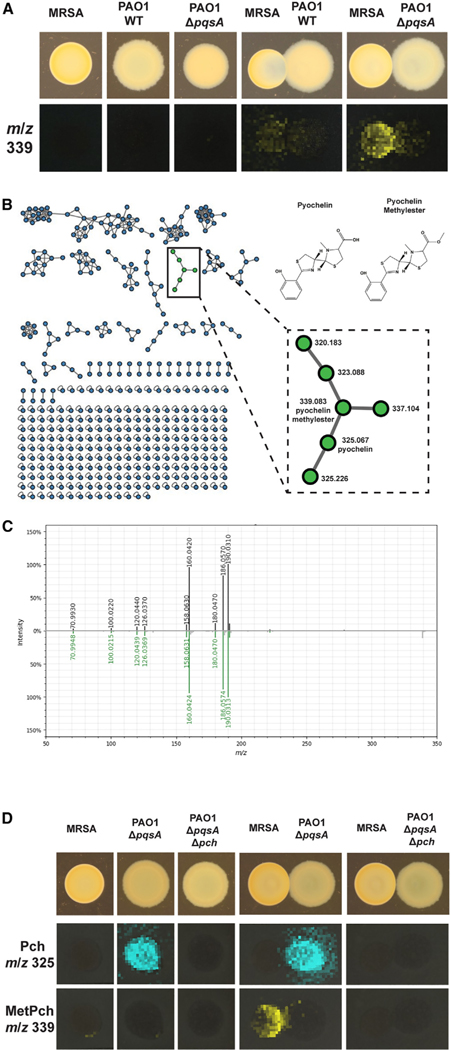
We used USA300 methicillin-resistant S. aureus strain LAC (MRSA) and P. aeruginosa strain PAO1 (PAO1) to study the natural product-mediated interaction of S. aureus and P. aeruginosa. Our rationale for choosing these two strains was that (1) both are genetically tractable, and we had the necessary expertise and tools to generate mutant and complemented strains, which are an important part of this study; (2) PAO1 is a derivative of strain PAO, which was originally isolated from a wound in Australia43; and (3) MRSA is a member of the USA300 pulsed-filed type, which is the predominant cause of community-acquired S. aureus skin and soft-tissue infections,44 making both strains suitable to study polymicrobial interaction in wound infections. To study the natural product mediated interaction of S. aureus and P. aeruginosa, USA300 MRSA and PAO1 were grown as mono-cultures or adjacent co-cultures on tryptic soy agar (TSA). After 24 h growth at 37°C, the semi-translucent hemisphere of the MRSA colony suggested a decrease in cell mass adjacent to the PAO1 colony when the two bacterial species were co-cultured (Figures 1A and S1A). A section of the agar was excised from the mono- and co-cultures and examined for the spatial distribution of the secreted metabolites by matrix-assisted laser desorption ionization MSI (MALDI-MSI).45,46 Although dozens of ions were observed, the ion at m/z 339 was exclusively detected in the co-cultures and predominantly distributed toward the MRSA colony, albeit at low intensity (Figure 1A). As this ion was not detected in PAO1 or MRSA mono-cultures, we hypothesized that it was produced by MRSA and that the observed low signal intensity was likely due to the low cell mass of the MRSA colony in the interaction due to inhibition by PAO1.
Figure 1. Pyochelin methyl ester (MetPch) is produced during interaction of S. aureus and P. aeruginosa.
(A) MALDI-MSI of S. aureus and P. aeruginosa grown as mono-cultures or interactions. Photographs of the cultures are shown on top. P. aeruginosa wild type (PAO1 WT), but not a P. aeruginosa quinolone mutant (PAO1 ΔpqsA), inhibits S. aureus WT (MRSA) when grown in co-culture. The bottom row shows the false colored m/z distribution of an unknown compound with m/z 339 that was exclusively observed during interaction of P. aeruginosa and S. aureus.
(B) The interaction specific compound (m/z 339) is annotated as MetPch, a member of the Pch molecular family, by molecular networking.
(C) Mirror plot comparing the MS2 spectrum of isolated MetPch (top; black trace) with its GNPS library match (bottom; green trace).
(D) MALDI-MSI of S. aureus and P. aeruginosa producing (ΔpqsA) or not producing Pch (ΔpqsA Δpch). Photographs of the cultures are shown on top. The middle row shows the false colored m/z distribution for Pch (m/z 325) and the bottom row for MetPch (m/z 339). Pch production can be observed by PAO1 ΔpqsA but not PAO1 ΔpqsA Δpch.
P. aeruginosa produces a variety of secreted metabolites that influence S. aureus growth directly and indirectly, including the alkyl quinolones. 2-heptyl-4(1H)-quinolone (HHQ) and 2-heptyl-3-hydroxy-4(1H)-quinolone (Pseudomonas quinolone signal [PQS]) regulate the production of antistaphylococcal factors, such as pyocyanin and rhamnolipids, while 2-heptyl-4-quinolone-N-oxide (HQNO) directly suppresses S. aureus growth by inhibiting the respiratory chain.36,45–47 The pqsABCDE biosynthetic gene cluster is required for P. aeruginosa to produce these metabolites. Therefore, the gene encoding PqsA, an anthranilate coenzyme A ligase that catalyzes the first step in alkyl quinolone biosynthesis, was deleted to render PAO1 (PAO1 ΔpqsA) deficient in the production of alkyl quinolones and pyocyanin and impair MRSA inhibition.47–50 A replicate interaction of MRSA with PAO1 ΔpqsA was inoculated and subjected to MALDI-MSI. As expected, abrogation of quinolone production by the PAO1 ΔpqsA strain (Figures 1A and S1A, top panels) led to increased MRSA cell mass of the MRSA colony adjacent to the PAO1 colony, with a concomitant increase in signal intensity for the ion at m/z 339 compared with the wild-type interaction (Figure 1A).
To facilitate the identification of the ion at m/z 339, untargeted liquid chromatography high-resolution tandem MS (LC-MS/MS) metabolomics data were collected on a chemical extraction of a duplicate interaction of MRSA and PAO1 ΔpqsA, and the data were analyzed using the classical molecular networking workflow of the GNPS platform.51 Classical molecular networking organizes compounds into molecular families by using MS/MS spectral relatedness as a proxy for structural similarity. Within the molecular network, MS/MS spectra are represented as nodes connected by edges that indicate spectral similarity. The MS/MS spectra in the data analyzed are simultaneously scored for spectral similarity to the data within the GNPS spectral libraries to provide putative annotation of compounds.
In the molecular network generated, the node representing m/z 339.0829 clustered within the pyochelin (Pch) molecular family38 ([M + H]+, calculated [calc] 325.0681, m/z 325.0671, −2.95 ppm) and was putatively annotated as Pch methyl ester (MetPch), where methylation of the Pch structure was localized to the carboxylic acid group ([M + H]+, calc: 339.0837, m/z 339.0829, −2.43 ppm). (Figures 1B and 1C). This structural annotation of Pch and MetPch was confirmed by comparing the retention time, exact mass, and MS/MS fragmentation of the microbially produced metabolites with synthesized standards (Figures S1B and S1C).
Based upon the spatial distribution of MetPch in the MSI data, we hypothesized that S. aureus methylates Pch produced by P. aeruginosa by an unknown mechanism. This hypothesis is supported by the fact that P. aeruginosa is a known producer of Pch,52 while S. aureus does not harbor biosynthetic genes for the production of Pch or related molecules. To verify that production of MetPch was dependent upon Pch biosynthesis by P. aeruginosa, MRSA was co-cultured with the PAO1 pqsA mutant impaired in Pch biosynthesis (PAO1 ΔpqsA Δpch) on TSA. Indeed, the absence of MetPch in the co-culture was associated with the loss of Pch production by PAO1 ΔpqsA Δpch (Figure 1D).
Although the spatial distribution of MetPch favored localization to the MRSA colony, several CF lung P. aeruginosa isolates produce a methylated Pch.53,54 To confirm that MRSA was the biosynthetic origin of MetPch during interaction with PAO1, MRSA cultures were incubated with cell-free supernatant from PAO1 ΔpqsA, which contains Pch (Figure S2A). While low levels of MetPch were detected in PAO1 ΔpqsA cell-free supernatant, incubation of the supernatant with MRSA increased the levels of MetPch over 190-fold (Figure S2B). These results indicate that the PAO1 strain used in our study produces low levels of MetPch, which is in accordance with the observed MetPch production by P. aeruginosa CF isolates.54 Nonetheless, the sharp increase in MetPch production from co-incubation of PAO1 ΔpqsA with MRSA wild type (WT) demonstrated that MRSA produces an enzyme that methylates the carboxylic acid of Pch to generate MetPch. It is also important to note that the steep increase of MetPch levels in PAO1 ΔpqsA supernatant treated with MRSA compared with the untreated PAO1 ΔpqsA supernatant control (Figure S2B) did not correspond to an equivalent decrease in Pch signal intensity in the MRSA-treated PAO1 ΔpqsA supernatant (Figure S2A). Quantification of synthetic Pch and MetPch under the same conditions resulted in an approximately 10-fold higher signal intensity of MetPch compared with Pch at equimolar concentrations (Figure S2C), suggesting a difference in the ionization efficiency of the two compounds during electrospray ionization (ESI).
Pch is methylated by Spm
To identify the S. aureus gene encoding the enzyme responsible for biotransformation of Pch to MetPch, an LC-MS guided screen of the Nebraska Transposon Mutant Library,55 a fully sequenced and sorted library of transposon insertions in the USA300 S. aureus LAC derivative JE2, was employed to identify mutants that did not produce MetPch when incubated with cell-free PAO1 ΔpqsA supernatant containing Pch. Of the 576 mutants tested, transposon mutant NE502 (transposon inserted in gene SAUSA300_1977) was the only one unable to produce MetPch (Data S1). In silico analysis of the SAUSA300_1977 gene sequence identified it as encoding a putative methyltransferase (TcmP-like; IPR016874), which we termed Spm (staphylococcal pyochelin methyltransferase).
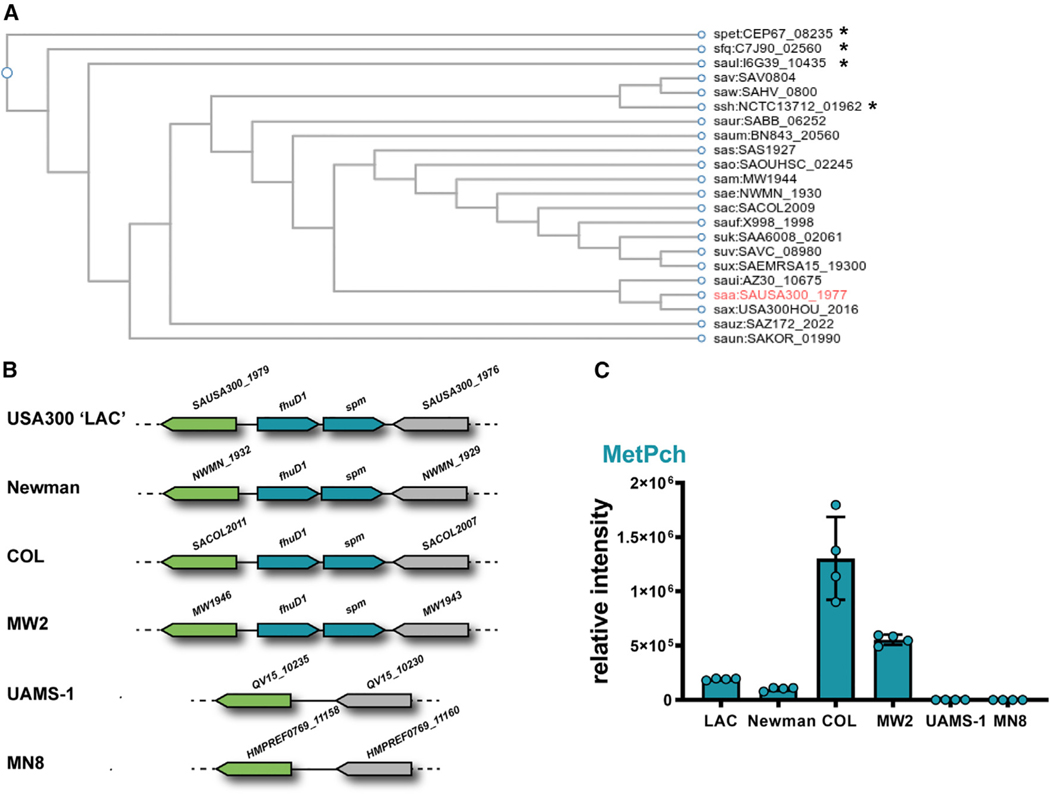
To assess whether Spm homologs were present within other staphylococcal genomes, the sequence of the spm gene was searched against the genomes available in the KEGG database (Figure 2A). This analysis revealed that Spm is conserved in 17 out of 53 S. aureus strains (32%) and in 4 out of 35 coagulase-negative staphylococci (11%) represented in the database. To substantiate that strains that encode spm in their genomes were capable of converting Pch to MetPch, six S. aureus strains with sequenced genomes (including LAC USA300) were incubated with Pch and evaluated for MetPch production using LC-MS. The four S. aureus strains harboring an spm homolog, including USA300 LAC, Newman, COL, and MW2, produced MetPch, while the two S. aureus strains lacking spm (UAMS-1 and MN8) did not (Figures 2B and 2C).
Figure 2. MetPch is only produced by S. aureus strains harboring spm.
(A) Dendrogram of Spm (SAUSA300_1977) orthologs in staphylococcal species. Dendrogram was constructed based on KEGG database entries. *Coagulase-negative staphylococcal species.
(B) Genetic environment of spm (SAUSA300_1977) in six sequenced S. aureus strains. The gene spm is encoded in a putative operon with fhuD1.
(C) MetPch was detected from cultures of S. aureus strains encoding the spm gene but not by spm-negative strains (UAMS-1, MN8) when incubated with PAO1 ΔpqsA supernatant. n = 4 biological replicates. Values are mean ± SDs.
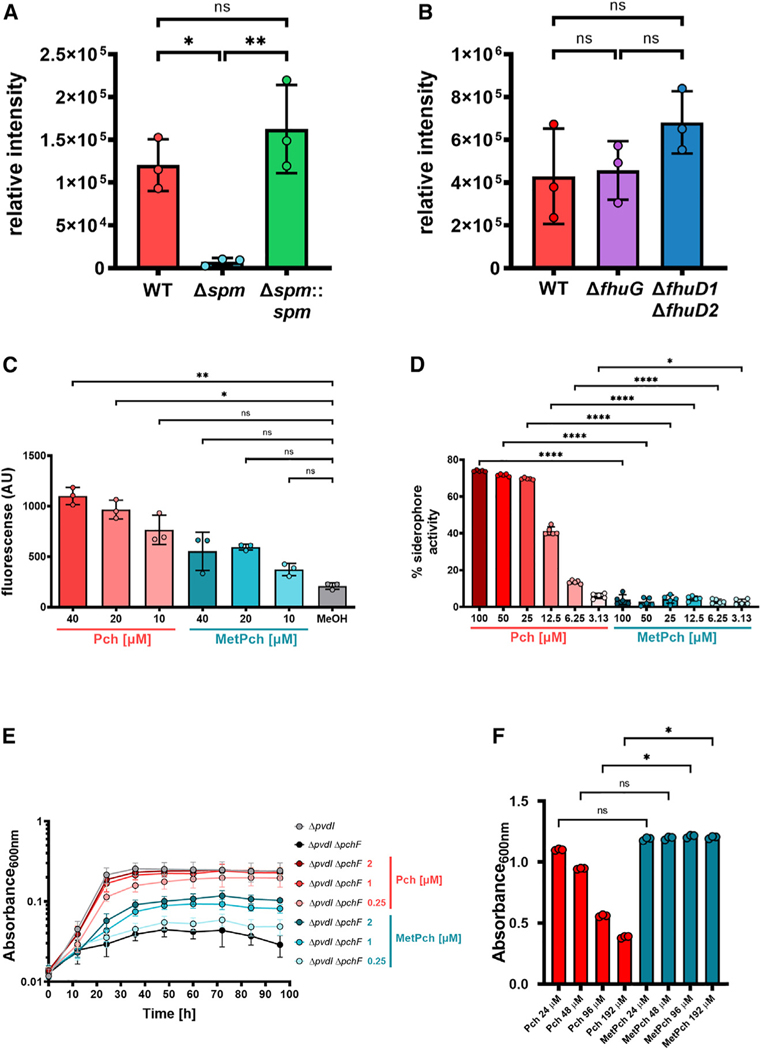
To further establish that Spm is responsible for the methylation of Pch, a markerless in-frame deletion of spm in the MRSA background (MRSA Δspm) was generated, and this mutant was incubated with cell-free supernatant from PAO1 ΔpqsA, which contains Pch. Compared with MRSA WT, MRSA Δspm produced significantly lower levels of MetPch, while genetic complementation of MRSA Δspm with spm (MRSA Δspm::spm) recovered production of MetPch to WT levels (Figure 3A). PAO1 ΔpqsA is capable of very low intrinsic MetPch production (Figure S2B), and thus we conclude that the low amount of MetPch observed from the co-incubation of cell-free PAO1 ΔpqsA with MRSA Δspm stems from the PAO1 ΔpqsA supernatant. These results confirm that Spm is necessary for the efficient biotransformation of Pch to MetPch by S. aureus.
Figure 3. Spm-mediated methylation leads to Pch inactivation.
(A) Incubation of PAO1 ΔpqsA cell-free supernatant with overnight cultures of MRSA WT, an spm mutant strain (Δspm), and its corresponding complementation strain (Δspm::spm) results in high amounts of MetPch production by WT and Δspm::spm but not by Δspm. n = 3 biological replicates. One-way ANOVA with Tukey’s multiple comparison.
(B) The Fhu xenosiderophore uptake machinery does not promote MetPch production. Mutation of the two ferric-siderophore binding proteins fhuD1 and fhuD2 (ΔfhuD1 ΔfhuD2), as well as fhuG (ΔfhuG), which is essential for the uptake of several hydroxamate xenosiderophores, does not result in significantly lowered MetPch production. n = 3 biological replicates. One-way ANOVA with Tukey’s multiple comparison.
(C) Exposure to Pch, but not MetPch, results in significantly elevated ROS production in MRSA WT compared with the carrier control (MeOH). n = 3 biological replicates. Kruskal-Wallis with Dunn’s multiple comparison.
(D) Siderophore activity of Pch and MetPch was evaluated by liquid Chrome Azural S assay. Siderophore activity of MetPch is significantly reduced compared with Pch across all tested concentrations. n = 5 biological replicates. One-way ANOVA and Šidák’s multiple comparison.
(E) Pch, but not MetPch, supports P. aeruginosa growth under low iron conditions. Shown are 96 h growth curves of a PAO1 pyoverdine mutant (ΔpvdI) and a pyoverdine/Pch double mutant (ΔpvdI ΔpchF) supplemented with different concentrations of synthetic Pch or MetPch. To create a low iron environment, 500 µM 2,2′-bipyridine was added to the growth medium. n = 5 biological replicates from 4 independent experiments.
(F) Exposure to Pch, but not MetPch, results in growth inhibition of MRSA under low iron conditions (chelex-treated TSB). Growth was significantly reduced in samples treated with high concentrations of Pch (96 and 192 µM) compared with samples treated with equimolar amounts of MetPch. n = 3 biological replicates. Kruskal-Wallis with Dunn’s multiple comparison.
(A–F) Values are mean ± SDs. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns, not significant.
Spm is encoded in a putative operon downstream of the ferric-siderophore binding protein FhuD1.56 The genomes of S. aureus strains that do not encode the spm gene lack the whole fhuD1-spm locus (Figure 2B). FhuD1 and FhuD2 are ferric-siderophore binding proteins that bind iron (III)-hydroxamate siderophore complexes, with a partially overlapping substrate range, and are part of the ferrichrome uptake (Fhu) system.56 The xenosiderophores bound by these two proteins are transported into S. aureus cells via the siderophore-permeases and ATPase transporter machinery encoded by the fhuCBG operon.57 Although Pch is a phenolate siderophore and the Fhu xenosiderophore acquisition system is used by S. aureus to acquire iron from exogenous hydroxamate siderophores, the proximity of spm to fhuD1 within the MRSA genome suggested that the Fhu system may directly or indirectly influence methylation of Pch. To test the influence of the Fhu system on the production of MetPch, markerless in-frame deletions of fhuD1/fhuD2 and fhuG in the MRSA background were generated (MRSA ΔfhuD1 ΔfhuD2 and MRSA ΔfhuG, respectively), and these mutants were incubated with PAO1 ΔpqsA cell-free supernatant containing Pch. Although MRSA ΔfhuD1 ΔfhuD2 produced, on average, higher levels of MetPch than MRSA WT and MRSA ΔfhuG, the difference in production levels between the strains was not statistically significant (Figure 3B).
The ferric uptake regulator (Fur) is an iron-dependent transcriptional repressor that regulates genes involved in iron import and acquisition.58 When iron is abundant, Fur binds ferrous iron and blocks transcription of iron acquisition genes. The promoter region of the fhuD1/spm operon contains a Fur box,56 a conserved sequence for Fur binding, indicating that this operon may be regulated by Fur activity. To measure transcriptional activation of the fhuD1/spm operon, sGFP was fused to the fhuD1/spm promoter, and fluorescence was measured in both WT MRSA and fur mutant (MRSA fur:tet). Promoter activity was significantly increased in MRSA fur:tet compared with WT (Figure S2D). Similar results were observed when transcription of spm and fhuD1 was measured by qRT-PCR and compared between MRSA WT and MRSA fur:tet (Figures S2E and S2F). Collectively, these results suggest that spm expression is iron dependent and that its expression is controlled by Fur in S. aureus.
Methylation of Pch neutralizes its biological functions
Pch is a phenolate siderophore produced by Pseudomonas and Burkholderia species.38,59 In addition to its iron chelating activity,52 exposure to Pch induces intracellular ROS production by an unknown mechanism in several bacterial species, including S. aureus.39,40 We hypothesized that methylation of the carboxylic acid of Pch by S. aureus could disrupt Pch-driven biological activities.
To test if MetPch had reduced antibacterial activity against S. aureus compared with Pch, MRSA was treated with equimolar concentrations of synthetic Pch and MetPch (10, 20, and 40 µM), and ROS levels were assessed by measuring the production of 2′,7′-dichlorofluorescein (DCF) from 2′,7′-dichlorodihydrofluorescein (H2DCF) by fluorescence (Figure 3C). While MRSA treated with Pch had a concentration-dependent increase in ROS production, exposure to MetPch did not result in significant increase in ROS production for all tested concentrations compared with carrier control (Figure 3C).
P. aeruginosa produces two siderophores, Pyoverdine (Pvd) and Pch. Pvd is considered the primary siderophore of P. aeruginosa due to its higher affinity for iron.60 However, Pch is produced at moderately low iron concentrations, and Pvd is only produced at extremely low iron concentrations,61 likely due to the metabolic burden of its production.37 Importantly, the carboxylic acid moiety of Pch is required for iron binding as well as its interactions with its transporter (FptA) located on the outer membrane of P. aeruginosa.62 To determine if MetPch had reduced iron binding capacity compared with Pch, iron binding of synthetic Pch and MetPch was measured via Chrom Azurol S (CAS) assay. Iron binding by MetPch was significantly lower than iron binding by Pch for all concentrations tested, ranging from 3.13 to 100 µM (Figure 3D).
To ascertain if the reduced iron binding of MetPch observed in the CAS assay reflected reduced support for P. aeruginosa growth under iron-limited conditions, a PAO1 mutant strain deficient in the production of both Pvd and Pch (PAO1 ΔpchF ΔpvdI) was generated and chemically complemented with synthetic Pch or MetPch at equimolar concentrations in low iron medium (M9 minimal medium supplemented with 500 µM 2,2′-bipyridine) (Figure 3E). Only PAO1 ΔpchF ΔpvdI is unable to grow robustly under iron-limited conditions, as the production of either siderophore is sufficient to support P. aeruginosa growth (Figure S3). While chemical complementation of PAO1 ΔpchF ΔpvdI with Pch was sufficient to recover P. aeruginosa growth to PAO1 ΔpvdI levels, MetPch was not (Figure 3E). Conversely, when S. aureus was grown in low iron medium (chelex-treated TSB) and subjected to Pch or MetPch, high concentrations of Pch inhibited its growth, while MetPch had no effect on S. aureus growth at any of the concentrations tested (Figure 3F). These results indicate that methylation of the carboxylic acid of Pch by S. aureus abrogates iron binding and ROS generation by Pch, providing a protective effect from this compound.
Spm increases S. aureus fitness in co-infection with PAO1
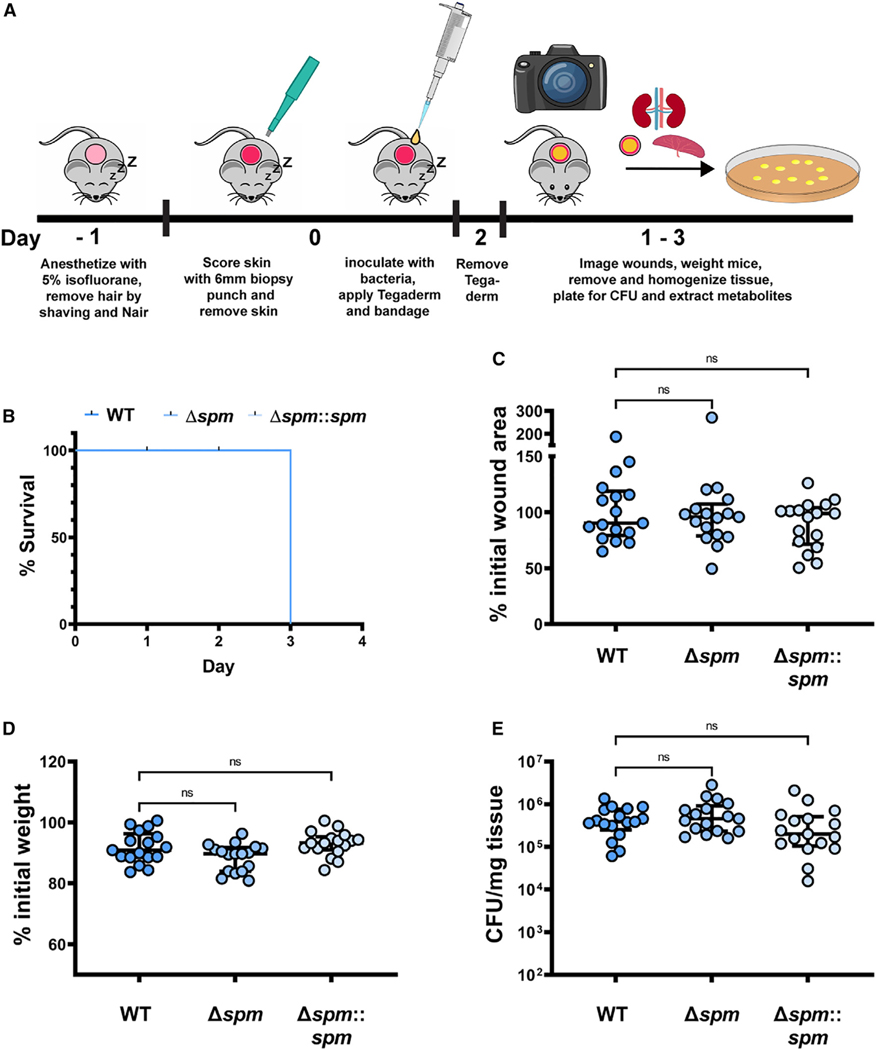
Taken together, our in vitro results show that S. aureus produces Spm to methylate the carboxylic acid moiety of Pch to protect itself from ROS production and iron limitation during interaction with P. aeruginosa. This ability of S. aureus to diminish Pch bioactivity may represent a viable competition strategy for S. aureus to co-exist with P. aeruginosa in skin wounds, one of their primary interaction environments. To test this hypothesis, a murine skin wound model was used to evaluate PAO1 and MRSA virulence and fitness as well as microbial competition between PAO1 and MRSA, MRSA Δspm, or MRSA Δspm::spm (Figure 4A).
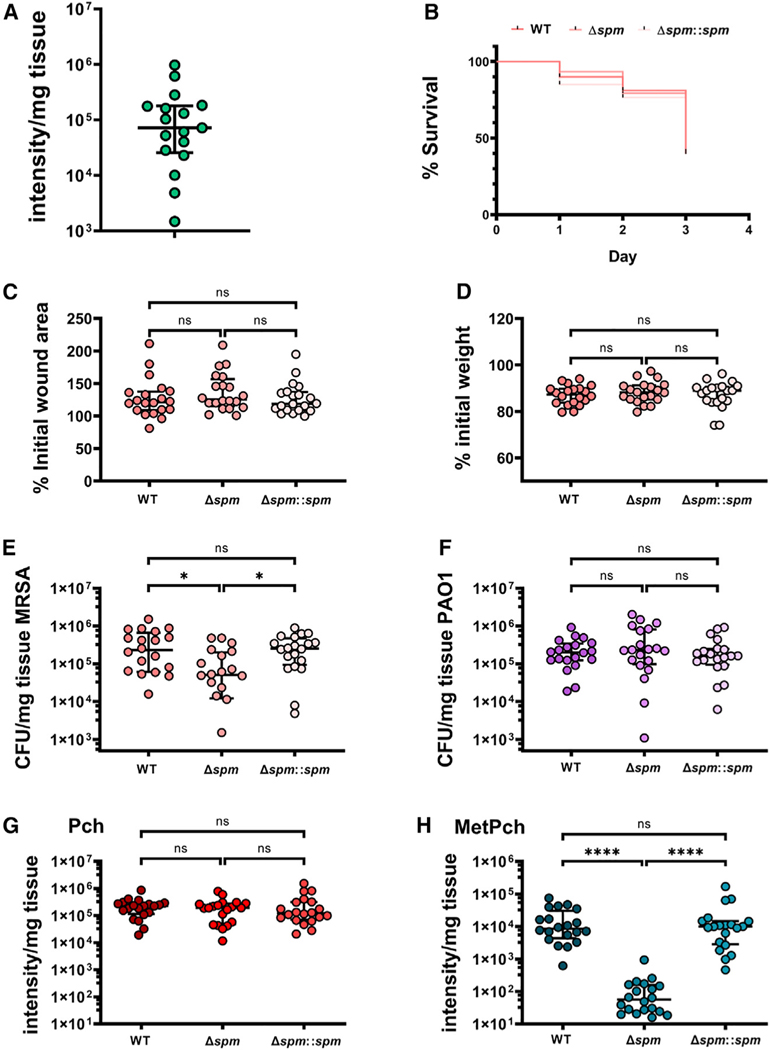
Figure 4. Spm does not change fitness or virulence of MRSA.
(A) Schematic of our murine wound skin infection model.
(B) Murine survival after skin wound infection with MRSA WT, the spm mutant strain (Δspm), and the spm complemented mutant (Δspm::spm).
(C) The change in murine skin wound size area between start and end of the experiment. Changes were calculated from the measured wound size area before bacterial infection and at the endpoint of the murine skin wound model. The changes are expressed as a percentage (%) of the initial wound size area at the endpoint of the experiment.
(D) The change in murine weight between start and end of the experiment. Changes were calculated from the murine weight before bacterial infection and at the endpoint of the murine skin wound model. The changes are expressed as a percentage (%) of the initial murine weight at the endpoint of the experiment.
(E) MRSA colony-forming units (CFUs) recovered from mouse tissue at the endpoint of the experiment.
(B–E) n = 17 biological replicates from 3 independent experiments for each bacterial strain. (C–E) Kruskal-Wallis with Dunn’s multiple comparison. Values are median ± interquartile range. Each dot represents values from a single mouse. ns, not significant.
Spm is an uncharacterized enzyme. Therefore, we first measured its influence on MRSA wound infections. Single infections with MRSA WT, MRSA Δspm, and MRSA Δspm::spm were performed to determine if Spm influences MRSA fitness or infection progression. Murine survival as well as changes to wound size and murine weight between the day of infection and the day of sacrifice were assessed as indicators of infection severity. All mice from the three groups (MRSA WT, MRSA Δspm, and MRSA Δspm::spm) survived over the course of the 3 day experiment, and no significant changes in wound size, weight, or MRSA fitness, as determined by colony-forming unit (CFU) recovery of the three MRSA strains from mouse wound tissue, were observed (Figures 4B–4E). This demonstrates that Spm does not affect the ability of MRSA to cause infection or to persist in our in vivo murine skin wound model.
Pch biosynthesis by PAO1 in the wound environment is a prerequisite to assess the role of Spm during bacterial wound co-infection. Therefore, we established a protocol for Pch extraction from PAO1 wound infections and successfully quantified Pch produced by PAO1 in our murine skin wound infection model (Figure 5A). No quantifiable MetPch amounts were detected in PAO1 mono-infections, but examination of our raw data showed that several samples contained trace amounts of MetPch that fell below the limit of quantitation threshold for our data analysis workflow.
Figure 5. Mutation of spm impairs the fitness of MRSA during co-infection with PAO1.
(A) Quantification of Pch recovered from mouse tissue infected with PAO1.
(B) Murine survival after skin wound co-infection with PAO1 and MRSA.
(C) The change in murine skin wound size area between start and end of the experiment. Changes were calculated from the measured wound size area before bacterial infection and at the endpoint of the murine skin wound model. The changes are expressed as a percentage (%) of the initial wound size area at the endpoint of the experiment.
(D) The change in murine weight between start and end of the experiment. Changes were calculated from the murine weight before bacterial infection and at the endpoint of the murine skin wound model. The changes are expressed as a percentage (%) of the initial murine weight at the endpoint of the experiment.
(E and F) MRSA (E) and PAO1 (F) CFUs recovered from mouse tissue at the endpoint of the experiment.
(G and H) Quantification of (G) Pch and (H) MetPch recovered from mouse tissue co-infected with PAO1 and MRSA. All co-infections were performed with PAO1 WT and either MRSA WT, the Spm mutant strain (Δspm), or the Spm complemented strain (Δspm::spm).
The co-infecting MRSA strain is indicated in the figure legend (B) or below the x axis (C–H). (A) n = 17 biological replicates from 3 independent experiments. (B–H) n = 20 biological replicates from 4 independent experiments. (C–H) Kruskal-Wallis with Dunn’s multiple comparison. Values are median ± interquartile range. Each dot represents values from a single mouse. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns, not significant.
Next, our murine wound co-infection model was employed to determine the role of Spm during MRSA/PAO1 wound co-infection. Infection severity was assessed by the same indicators as used for the MRSA single infections (murine survival, wound size progression, weight loss). Mice co-infected with MRSA WT and PAO1 WT showed a lowered, although not significant, probability to survive until the end of the experiment (day 3) compared with mono-infections with MRSA WT or PAO1 WT (Figure S4A). In addition, a significant increase in wound size progression in mice co-infected with MRSA WT and PAO1 WT was observed compared with mono-infections (Figure S4B) and significantly less weight loss in mice infected with MRSA WT compared with PAO1 WT but not compared with co-infections (Figure S4C).
Comparison of the infection severity outcomes of the three co-infections of PAO1 with either MRSA WT, MRSA Δspm, or MRSA Δspm::spm showed no difference in murine survival, wound size progression, or weight loss (Figures 5B–5D). However, MRSA fitness was significantly altered between Spm-positive and Spm-negative MRSA as evidenced by the significantly reduced recovery of CFUs of MRSA Δspm compared with MRSA WT and MRSA Δspm::spm from the wounds (Figure 5E). P. aeruginosa fitness was similar for all co-infections (Figure 5F). Collectively, our data indicate that Spm is not involved in MRSA virulence but is required for optimal fitness of MRSA in polymicrobial skin wound infection with P. aeruginosa.
To determine if MRSA produced MetPch from Pch synthesized by PAO1 in vivo, Pch and MetPch were quantified from wound tissues of the murine skin wound co-infections. Both Pch and MetPch were detected in the wound tissue. While comparable amounts of Pch were measured in all three co-infections (Figure 5G), MetPch was produced at more than 100-fold lower in MRSA Δspm co-infections with PAO1 compared with co-infections with MRSA WT, and these levels were recovered with genetic complementation of the spm mutant strain (Δspm::spm) (Figure 5H). Importantly, levels of MetPch positively correlated with recovered MRSA CFUs in the polymicrobial skin wound infection model (Spearman’s rank correlation coefficient; ρ = 0.5186), further supporting that S. aureus deactivates Pch through Spm-mediated methylation to enhance fitness in the presence of P. aeruginosa (Figure 5E), which likely contributes to co-existence in these dual-species infections.
DISCUSSION
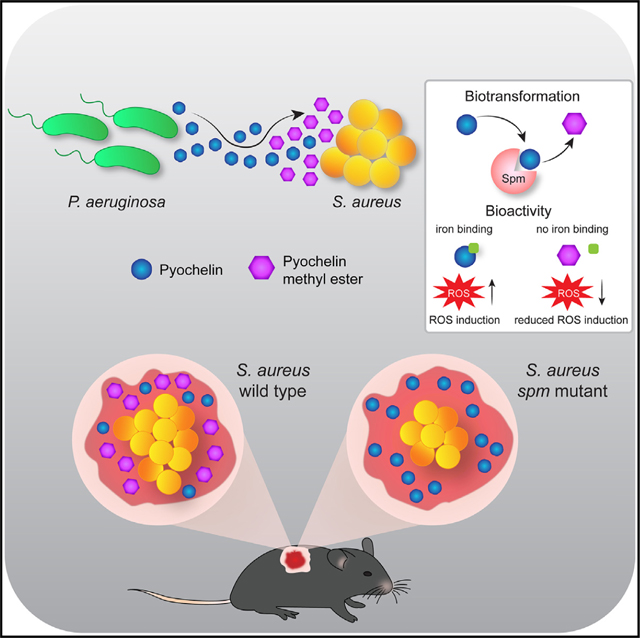
Using a MS-directed approach, we investigated the interaction of S. aureus and P. aeruginosa in vitro and in an in vivo murine skin wound model that reflects the early stages of wound coinfection. We demonstrate an underappreciated role for the P. aeruginosa-derived siderophore Pch during in vivo competition of PAO1 and MRSA in skin wound co-infection. In addition, our data clearly show that S. aureus inactivates Pch by methylation via the previously uncharacterized enzyme Spm, and this translates to enhanced fitness of MRSA in a wound co-infection model with PAO1 (Figure 6). We validated that the methylation of Pch by Spm occurs on the carboxylic acid group by comparing the experimentally derived compounds with chemically synthesized authentic standards.
Figure 6. Model of S. aureus/P. aeruginosa chemical interaction.
P. aeruginosa produces and secretes Pch during infection. S. aureus WT cells inactivate Pch through enzymatic methylation. S. aureus mutant cells lacking the Spm methyltransferase are unable to methylate Pch. Pch methylation leads to the loss of siderophore activity and reduced intracellular ROS production in S. aureus cells, resulting in a higher fitness of S. aureus during competition with P. aeruginosa in vivo.
The spm gene is encoded in an operon with the fhuD1 gene, which encodes a ferric-siderophore binding protein involved in the uptake of the two xenosiderophores, ferrichrome and ferrioxamine B,56 suggesting a possible role of the Fhu xenosiderophore uptake machinery in Pch methylation. Simultaneous mutation of fhuD1 and fhuD2 (MRSA ΔfhuD1 ΔfhuD2) or fhuG (MRSA ΔfhuG), an essential component of the Fhu xenosiderophore import machinery, did not result in significant changes of measured MetPch levels compared with MRSA WT. These data indicate that the Fhu system is most likely not involved in Pch or MetPch transportation. The slight, yet not significant, increase of measured MetPch levels observed in the MRSA ΔfhuD1 ΔfhuD2 background could be explained by FhuD1 and FhuD2 binding of Pch and excluding it from Spm-mediated methylation. Alternatively, FhuD1 and FhuD2 might bind MetPch, preventing its diffusion into the cell-free supernatant and excluding it from our metabolite extracts.
P. aeruginosa CF isolates are capable of producing low levels of MetPch.53,54 Our data show that the P. aeruginosa model strain PAO1, a derivative of P. aeruginosa strain 1 isolated from a wound in Melbourne, Australia, in 1954,43 is also capable of producing trace amounts of MetPch, which is over 190-fold lower than Spm-mediated MetPch production by MRSA. This demonstrates that MetPch production is not unique to CF isolates. Based on our own results, MetPch is an inactive version of Pch, and thus we expect its biosynthesis does not yield any advantages for P. aeruginosa. Therefore, we suggest that MetPch is a byproduct of Pch biosynthesis by P. aeruginosa, likely due to mis-methylation of the carboxy group by the stuffed methyltransferase present in the PchF enzyme.63 This is supported by the low concentrations of MetPch compared with Pch observed in P. aeruginosa cultures. In addition, future work could focus on screening of co-isolated pairs of S. aureus and P. aeruginosa to determine the effect of co-evolution of these two pathogens in regard to Pch methylation.
We show that methylation of Pch by Spm contributes to enhanced MRSA fitness in a skin wound co-infection model with P. aeruginosa. Based upon the similarity of the infection outcomes, including survival, wound size progression, and weight loss, in the mono-species infection model with WT MRSA and a deletion mutant of Spm, we determined that Spm does not contribute to MRSA virulence in our murine skin wound infection model. However, in the co-infection model with PAO1, Spm was required to maintain WT population levels of MRSA. From the two-dimensional distribution of MetPch in the MALDI-MSI data, we postulate that methylation of Pch is localized to S. aureus growth in vivo. From our in vitro studies, local methylation of Pch by S. aureus would result in increased survival by impeding iron binding by Pch, thereby improving its ability to compete for environmental iron and decreasing induction of its own ROS by detoxifying Pch.
Methylation of Pch by S. aureus did not affect P. aeruginosa fitness in our murine wound co-infection model. This is best explained by the fact that MetPch is inactive regarding its siderophore bioactivity and therefore cannot exert a negative effect on P. aeruginosa WT. This could be further explored by investigating the possibility of competitive binding of MetPch to the Pch transporter FptA, which was not in the scope of our study. In this hypothetical scenario, diffusion of MetPch through the wound tissue, first as Pch from P. aeruginosa to S. aureus and afterward as MetPch from S. aureus to P. aeruginosa, would likely limit the effectiveness of MetPch to compete with Pch for FptA binding.
Intriguingly, deactivation of Pch by esterification of the carboxylic acid moiety appears to be a conserved mechanism between a variety of microorganisms, including the fungus Phellinus noxius64 and soil bacterium Bacillus amyloliquefaciens.65 The apparent wide-spread mechanism, s, to deactivate Pch via chemical modification suggests that Pch in microbial organisms from different kingdom may have a larger impact on microbial ecology than is currently appreciated. The herein presented study demonstrates the molecular and genetic mechanism behind Pch methylation and its importance for S. aureus fitness in polymicrobial infection.
Limitations of the study
Our study does not allow us to conclude if Spm has evolved for the main purpose of Pch detoxification, but the fact that spm is repressed by Fur, similar to Pch repression,52 might indicate this to be the case. Further work will be necessary to determine if spm expression is also Fur and iron dependent in other S. aureus strains and how differences in expression strength might influence Pch detoxification and overall S. aureus fitness in polymicrobial infections with P. aeruginosa. In addition, we hypothesize that MetPch is primarily localized close to S. aureus cells in our murine wound infection model. To shed further light on this hypothesis, the spatial distribution of S. aureus, P. aeruginosa, Pch, and MetPch could be analyzed by a combination of high-resolution microscopy and MALDI-MSI of co-infected mouse wound tissue sections.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Alexander R. Horswill (alexander.horswill@cuanschhutz.edu).
Materials availability
All generated bacterial mutant and complemented strains and reporter strains generated in this study are available from the lead contact.
Data and code availability
All mass spectrometry data, including the raw files, mzXML files, metadata tables and MZmine settings have been deposited at MassIVE (MassIVE: MSV000089230; P. aeruginosa/S. aureus WT, spm mutant, spm complementation interaction, MassIVE: MSV000089231; P. aeruginosa/S. aureus WT, fhuG mutant, fhuD1/D2 double mutant interaction, MassIVE: MSV000089233, Pyochelin methylation by S. aureus transposon library mutants, MassIVE: MSV000089234; Pyochelin methyl ester quantification murine skin wound model S. aureus/P. aeruginosa co-infection) and are publicly available as of the date of publication. Password for all datasets: MetPch2022.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Murine strain
Mouse strain C57BL/6J was obtained from Jackson Laboratories (000664). All mice used for experiments were female and aged 7 weeks upon arrival and were maintained and housed in the ABSL-AHSB at the University of Colorado Anschutz Medical Campus accredited by the Association for Assessment and Accreditation of Laboratory Care International (AAALAC). All animal studies described herein were performed in accordance with best practices outlined by the Office of Laboratory Animal Resources (OLAR) and Institutional Animal Care and Use Committee (IACUC) at the University of Colorado (protocol #00987).
METHOD DETAILS
Media and growth conditions
Staphylococcus aureus, Pseudomonas aeruginosa, and Escherichia coli strains and plasmids used in this work are listed in STAR Methods. S. aureus was regularly cultured in Bacto Tryptic Soy Broth (TSB; BD) and E. coli and P. aeruginosa were cultured in Lennox Lysogeny Broth (LB; RPI) at 37°C with 200 rpm shaking. To create environments with low iron availability, media were treated with Chelex-100 (chelex: BioRad) as follows: 0.5% (w/v) Bacto Casamino Acids Technical (CAA; BD) was incubated for 5 h with 5 g/L chelex, filtered with a Nalgene Rapid-flow Filter Unit (0.2 μm aPES membrane; Thermo Scientific) to remove the chelex resin, adjusted to pH 7.4, autoclaved, and supplemented with MgCl2 (0.4 mM final concentration). TSB was incubated with 2 g/L chelex for 3 h, filtered with a Nalgene Rapid-flow Filter Unit (0.2 µm aPES membrane; Thermo Scientific) to remove the chelex resin, and autoclaved. Experiments to determine P. aeruginosa growth support by pyochelin and pyochelin methyl ester were performed in M9 minimal medium supplemented with 500 µM 2,2′-Bipyridine (Fisher Scientific). M9 minimal medium was prepared as follows: 5x M9 salts: 64 g/L Na2HPO4*7H2O, 15 g/L KH2PO4, 2.5 g/L NaCl, and 5 g/L NH4Cl. 5x M9 salts were diluted in H2O containing MgSO4 (2 mM final concentration), CaCl2 (0.1 mM final concentration), and glycerol (0.5% (v/v) final concentration). Antibiotics were added to the media where necessary at the following concentrations: chloramphenicol (Cam), 10 µg/mL; erythromycin (Erm), 5 µg/mL; and tetracycline (Tet), 1 µg/mL. E. coli strains with plasmids were maintained on media supplemented with ampicillin (Amp) at 100 µg/mL; kanamycin (Kan), 50 µg/mL; gentamicin (Gen) 20 µg/mL or spectinomycin (Spec) at 50 µg/mL.
Construction of gene deletion mutants
Markerless gene mutations were introduced using the temperature-sensitive plasmid pJB38 for S. aureus or pEX2G for P. aeruginosa carrying DNA fragments (~1000 bp in size) flanking the region targeted for deletion. The flanking regions were amplified by Phusion High-Fidelity DNA polymerase (New England BioLabs) with the primers listed in Table S1.
S. aureus
DNA fragments (~1000 bp in size) flanking the region targeted for deletion and pJB38 were digested with restriction enzymes, as indicated in Table S1, and subsequently purified with the QIAquick PCR purification kit (Qiagen). After triple ligation of the flanking region pairs with pJB38, the resulting plasmid was electroporated into E. coli DC10b. E. coli cells carrying the plasmid were selected on LB plates containing 100 µg/mL Amp, single colonies were picked, patched on new LB plates containing 100 µg/mL Amp, and the presence of the plasmid containing the flanking regions was confirmed by PCR using the primers listed in Table S1. The plasmid was recovered from overnight cultures of positive clones with a QIAquick Spin miniprep kit (Qiagen) and the sequence of the flanking regions was confirmed by in-house sequencing with their respective construction primers and sequencing primers listed in Table S1. The plasmid was electroporated into the S. aureus target strain, positive clones carrying pJB38 with the desired flanking regions were selected on TSA plates containing 10 µg/mL Cam at 30°C. For homologous recombination, positive clones were streaked on TSA-Cam and incubated at 42°C for 24 h. Cam-resistant colonies were restreaked on TSA-Cam and incubated at 42°C overnight. Single colonies were picked and incubated in 5 mL TSB at 30°C with 200 rpm shaking overnight. The resulting overnight cultures were diluted 1:1000 in TSB and incubated at 30°C with 200 rpm shaking overnight. This was repeated for six consecutive days, and subsequently, dilutions (10−6, 10−7, and 10−8) were plated on TSA plates containing anhydrotetracycline (ahTet) 200 ng/mL for counterselection and incubated at 30°C overnight. Single colonies were then patched on TSA and TSA-Cam plates and grown at 30°C overnight. Plasmid loss was indicated by growth on TSA but not TSA-Cam. Colonies were screened for the desired mutation by PCR with primers (Table S1) located on the S. aureus chromosome outside of the homology arm regions.
P. aeruginosa
DNA fragments (~1000 bp in size) flanking the region targeted for deletion and pEX2G were digested with restriction enzymes, as indicated in Table S1, and subsequently purified with the QIAquick PCR purification kit (Qiagen). After triple ligation of the flanking region pairs with pEX2G, the resulting plasmid was electroporated into E. coli Top10. E. coli cells carrying the plasmid were selected on LB plates containing 20 µg/mL Gen, single colonies were picked, patched on a new LB plate containing 20 µg/mL Gen, and the presence of the plasmid containing the flanking regions was confirmed by PCR using the primers listed in Table S1. The plasmids were recovered from overnight cultures of positive clones with a QIAquick Spin miniprep kit (Qiagen), and the sequence of the flanking regions was confirmed by in-house sequencing with their respective construction primers and sequencing primers listed in Table S1. The plasmids were transferred to the Pseudomonas aeruginosa PAO1 target strain by conjugation. Single recombinants were selected on Vogel-Bonner minimal medium (VBMM)74 supplemented with 160 µg/mL Gen to create merodiploid strains. The merodiploid strains were resolved by an overnight outgrowth in LB followed by plating on VBMM supplemented with 7.5% sucrose for counterselection. Single colonies were picked, patched on VBMM agar plates containing 7.5% sucrose and gene deletions were confirmed by PCR with primers located on the chromosome outside of the homology arm regions as listed in Table S1.
Construction of complemented S. aureus mutant strains
The spm mutation in the MRSA Δspm background was repaired by introducing the intact spm gene at its original location on the chromosome. The complemented spm strain (MRSA Δspm::spm) was constructed by amplifying the spm gene and flanking regions from the MRSA wild type genome. First, the spm gene and a region covering 508 bp upstream of spm were amplified with primers listed in Table S1, digested with KpnI and SalI and ligated into pJB38 digested with the same restriction enzymes. The resulting plasmid was electroporated into E. coli DC10B. After propagation in the E. coli strain, the plasmid was digested with NcoI and SalI and ligated with a DNA fragment spanning 559 bp downstream of the spm coding sequence which was amplified with primers listed in Table S1 digested with NcoI and SalI. The resulting plasmid (pJB38spmKI) carries the wild type spm gene and flanking regions for homologues recombination and an NcoI restriction site located 110 bp downstream of the spm stop codon that serves as a watermark for the repaired strain. Introduction of the plasmid into E. coli DC10B, transfer to MRSA Δspm, selection for repaired strains and plasmid sequencing was carried out as described for gene deletion mutant construction.
Construction of S. aureus LAC fur mutant
The furtet cassette from AH2367 was transduced into S. aureus USA300 LAC using phage, and deletions were confirmed by PCR with primers HC479 and HC480 (Table S1)
Construction of promoter fusion vector
A transcriptional fusion of the fhuD1/spm promoter to sGFP was constructed using plasmid pHC48. A region containing the putative promoter region of the fhuD1/spm operon was amplified by Phusion High-Fidelity DNA polymerase (New England BioLabs) with the primers listed in Table S1. The PCR products and pHC48 were digested with restriction enzymes, as indicated in Table S1, and subsequently purified with the QIAquick PCR purification kit (Qiagen). The resulting plasmid, pHC48-spmprom, was electroporated into E. coli DC10b. E. coli cells carrying the plasmid were selected on LB plates containing 10 µg/mL Cam, single colonies were picked, patched on a new LB plate containing 10 µg/mL Cam, and the presence of the plasmid containing the promoter region was confirmed by PCR using the primers listed in Table S1. The plasmid was recovered from overnight cultures of positive clones with a QIAquick Spin miniprep kit (Qiagen), and the sequence of the flanking regions was confirmed by in-house sequencing with their respective construction primers and sequencing primers listed in Table S1.
Promoter fusion activity
S. aureus cells harboring the spm promoter fusion vector pHC48-spmprom were grown overnight in TSB containing Cam 10 µg/mL or Cam 10 µg/mL and Tet 2 µg/mL for the S. aureus LAC fur:tet background. Cells were washed with PBS and resuspended in fresh media. Cultures were set to an OD600 of 0.05 and 200 µL of each sample were transferred to 96-well black plates with clear bottom (Corning Incorporated) and incubated at 37°C with 1,000 rpm (Microtiter Plate Shaker Incubator SI505; Stuart) for 24 h. Promoter activities were measured with an Infinite M PLEX plate reader (Tecan) with the following settings: excitation wavelength 549 nm, emission wavelength 588 nm, number of flashes 10, settle time 10 ms, gain 80, integration time 40 µs, Z-position 20,000 µm. To assure similar growth between different strain backgrounds bacterial growth was measured with the following settings: wavelength 600 nm, number of flashes 10.
qRT-PCR
Bacteria were grown in high- (TSB) and low-iron media (TSB treated with 2 g/L chelex for 3h) at 37°C with shaking. Overnight cultures were washed and resuspended to an OD600 of 0.05 in the respective medium. Bacterial cultures were grown to mid-log phase, 1 mL aliquots were harvested by centrifugation and stored at −80°C before RNA extraction. Thawed bacterial cell pellets were resuspended in 100 µL TE buffer and transferred to bead-beating tubes.
Thawed pellets were re-suspended in 100 µL TE buffer and suspension was transferred to bead-beating tube. Samples were bead-beated for 60 s to lyse cells. Then spun quickly at full speed for 1 s to burst any bubbles. 650µL of RLT + β-mercaptoethanol was added to each sample and the beat-beating step was repeated. Samples were then spun down for 30 s at full speed. Next, 600 µL of sample supernatant was transferred to a clean 2 mL tube containing 900 µL EtOH and mixed by pipetting up and down 7 times. 750µL of the mixture was immediately loaded onto an RNeasy column. Columns were spun at full speed for 30 s and flow-through discarded. The remaining lysate/EtOH mixture was loaded onto the same column and the spin step was repeated. 700µL of RW1 buffer was added to column. Samples were spun down at full speed for 30 s and the flow-through was discarded. Columns were washed twice with 500µL fresh RPE buffer and samples were again spun down at full speed for 30 s and the flow-through was discarded.
To dry columns, samples were again spun down at full speed for 2 min 53 µL of nuclease-free ddH2O was added to the membrane and incubated for 1 min. Columns were again spun down at full speed for 1 min to elute RNA.
Samples were immediately treated with DNAse. Reactions contained 53 µL RNA sample, 6 µL 10X DNAse buffer, 1 µL DNase. Samples were incubated for 1h at 37°C after which 7 µL DNAse deactivation buffer was mixed with each sample to stop DNAse treatment and samples were centrifuged at 10.0000 g for 3 min 42 µL of RNA sample was transferred to a new 1.5mL tube and immediately stored at −80°C. The remaining 5–10µL RNA sample was transferred to a separate tube for immediate quantification of RNA concentration.
cDNA generation was performed with the iScript RT master mix kit. Aliquots of RNA were normalized to a concentration of 250 ng/µL and reactions were setup as follows: 2 µL RNA sample, 4 µL of iScript master mix and 14 µL of DEPC treated H2O. Samples were resuspended to a final volume of 200 µL and stored at −20°C.
Primer used for RT-qPCR are listed in Table S1.
Pyochelin and pyochelin methyl ester extractions from agar plates
P. aeruginosa and S. aureus cells were grown on agar plates as described for MALDI MSI. Agar pieces containing either P. aeruginosa or S. aureus colonies from single incubations or co-incubations were excised from agar plates. Agar pieces from 5 plates were pooled and extracted with 5 mL of acidified EtOAc. 2 mL of the acidified EtOAc phase were removed, centrifuged at 5,000 rpm (Biofuge pico, Heraeus) to remove nonsoluble particulates and evaporated to dryness in a SpeedVac vacuum concentrator (SPD131DDA SpeedVac Concentrator; Thermo Scientific) and stored at – 20° C.
Pyochelin and pyochelin methyl ester extractions from liquid cultures
Ethyl acetate (EtOAc; J.T. Baker and Fisher Scientific) and Optima-grade methanol (MeOH; Fisher Scientific) were used. P. aeruginosa PAO1 wild type or ΔpqsA cells were grown overnight in LB (RPI) at 37°C with 200 rpm shaking. Bacterial cells were centrifuged at 5,000 rpm (Biofuge pico, Heraeus) and washed with 0.5% chelex treated CAA. New cultures were seeded in 0.5% chelex treated CAA at a starting OD600 of 0.05 and incubated at 37°C with 200 rpm shaking. After 48 h the cultures were centrifuged at 3,800 rpm (Eppendorf Centrifuge 5810 R) for 30 min at room temperature. The supernatant was decanted and filtered using a Nalgene Rapid-flow Filter Unit (0.2 µm aPES membrane; Thermo Scientific). The cell free supernatant was acidified to pH 1.8–2 with 6 M HCl. Pyochelin and pyochelin methyl ester were extracted with 3 volumes EtOAc and evaporated to dryness in a Rotavapor (Büchi; Rotavapor R-300) or a SpeedVac vacuum concentrator (SPD131DDA SpeedVac Concentrator; Thermo Scientific). Dried samples were resuspended in MeOH and stored at – 20° C. Samples were centrifuged for 5 min at 10,000 rpm (Thermo; Sorvall ST 40R) to remove nonsoluble particulates and diluted as needed in MeOH containing 1 µM glycocholic acid.
Micro plug solvent screen
Extraction of the unknown metabolite at m/z 339 was tested with 15 different solvents (Table S2). Briefly, P. aeruginosa ΔpqsA and S. aureus cells were grown in co-cultures on agar plates as described for MALDI-TOF MSI. Agar plugs from the interaction zone (highest intensity of m/z 339 in MALDI-TOF MSI) were harvested with the bottom end of a 100 µL pipette tip. For each solvent, plugs from 4 interaction plates were pooled, homogenized with a pipette tip and incubated with 200 µL solvent for 1 h at room temperature. Tubes were centrifuged at 5,000 rpm (Biofuge pico, Heraeus), 150 µL of clean solvent were transferred in a fresh tube, evaporated to dryness in a SpeedVac vacuum concentrator (SPD131DDA SpeedVac Concentrator; Thermo Scientific) and stored at – 20° C.
LC-MS guided transposon mutant screen
P. aeruginosa ΔpqsA was grown in 50 mL of 0.5% chelex treated CAA supplemented with 0.4 mM MgCl2 at pH 7.4 for 48 h at 37°C with shaking. The P. aeruginosa culture was centrifuged at 3,800 rpm in an Eppendorf Centrifuge 5810 R at room temperature for 20 min. The supernatant was filtered with a Nalgene Rapid-Flow Filter Unit (0.2 µm aPES membrane; Thermo Scientific). The cell free supernatant was used for co-incubation with the S. aureus transposon mutants.
S. aureus transposon mutants were grown in 96-well plates in 220 µL TSB at 700 rpm at 37°C (Microtiter Plate Shaker Incubator SI505; Stuart) for 18 h. 200 µL of each grown culture were transferred to a 96-deep well plate, mixed with 400 µL cell free P. aeruginosa ΔpqsA supernatant. The plates were sealed with Gas permeable sealing membranes (Breathe-Easy; Diversified Biotech; 9123–6100) and incubated at 37°C with 800 rpm shaking at 37°C (Microtiter Plate Shaker Incubator SI505; Stuart). S. aureus train JE2, the parental strain of the transposon library was used as a control. After incubation, proteins were crashed out by addition of 300 µL ethyl acetate and all samples were evaporated to dryness in a SpeedVac vacuum concentrator (SPD131DDA SpeedVac Concentrator; Thermo Scientific). Dried samples were stored at −20C until further processing. Samples were resuspended in 200 µL Optima-grade MeOH and incubated in a sonicator bath (Branson; 5800) at room temperature for 40 min and an additional 30 min in the dark at room temperature. Plates were centrifuged at 10,000 rpm (Biofuge pico, Heraeus) and clear MeOH extracts were diluted 1:10 v/v with MeOH containing 1 µM glycocholic acid for subsequent analysis by LC-MS.
Murine model of skin infection
P. aeruginosa strain PAO1 was grown overnight in 5 mL of LB medium at 37°C shaking at 200 rpm. S. aureus strains of LAC USA300 (MRSA WT, Δspm and Δspm::spm) were all grown overnight in 5 mL of TSB medium at 37°C shaking at 200 rpm. For mouse infections, overnight cultures of MRSA and PAO1 were diluted 1:100 into 35 mL TSB and LB, respectively, and grown shaking in flasks at 200 rpm to an OD600 of 0.5 (≈2 h for MRSA and ≈3 h for PAO1). Subcultured bacteria were then pelleted and resuspended in sterile saline, so that 10 µL of each culture was normalized to ≈5×105 CFU. 1 mL of each strain suspension was aliquoted into 1.5 mL Eppendorf tubes and kept on ice for murine infection inocula.
All in vivo infections were on female C57BL/6 WT mice obtained from Jackson Laboratories (Bar Harbor, ME). All animals were housed and maintained at the University of Colorado Anschutz Medical Campus Animal Care Facility accredited by the Association for Assessment and Accreditation of Laboratory Care International (AAALAC) and mice were allowed to acclimate to the BSL-2 level facility for at least seven days prior to their inclusion in this study’s in vivo murine model. All animal studies described herein were performed in accordance with best practices outlined by the Office of Laboratory Animal Resources (OLAR) and Institutional Animal Care and Use Committee (IACUC) at the University of Colorado (protocol #00987).
A murine model of wound infection was used to assess chemical interactions between MRSA and PAO1 during polymicrobial infection. One day prior to infection, mice were anesthetized (inhalation of isoflurane, 2–3%), and the fur on the dorsal surface was carefully shaved and Nair applied to completely expose the skin. On day 0, mice were anesthetized, and exposed dorsal skin was sterilized with a PVP iodine prep pad (PDI Healthcare, Woodcliff Lake, NJ). Bupivacaine hydrochloride was injected subcutaneously with a 30-gauge insulin syringe at a dose of 1–2 mg/kg in the medial thoracic region as an analgesic for the area to be wounded. A disposable 6 mm biopsy punch (Integra Miltex) was used alongside dissection scissors and forceps to excise a circular section of skin 6 mm in diameter to create a wound. For single infections, 10 µL (5 × 105 CFU) of MRSA inoculum in sterile saline was inoculated into the open wound by pipette. For co-infections, 5 µL (2.5 × 105 CFU) of both MRSA and PAO1 were inoculated into the open wound, for a final inoculating dose of 5 × 105 CFU.
Following inoculation, the wounds were covered with the transparent dressing Tegaderm, to allow for observation and prevent contamination, followed by 2 bandages to protect the wounded area. Groups of at least 5 mice were used for each test condition per experiment. Animals were euthanized by CO2 inhalation followed by cervical dislocation as a secondary method of euthanasia at the completion of the experiment. However, some mice were sacrificed early to minimize distress and prevent spontaneous mortality according to the University of Colorado Guidelines for Establishing Humane Endpoints in Animal Study Proposals. Each animal’s physical condition was monitored daily to evaluate signs of morbidity (including body weight, physical appearance, clinical signs, unprovoked behavior, or response to external stimuli) that may constitute an early endpoint as the most humane option. Therefore, if animals exhibited signs of distress according to these guidelines, such as weakness, inability to drink water or eat, signs of systemic infection, or severe weight loss, mice were euthanized.
At daily timepoints over the course of 3 days, animals were monitored, weights measured, and wounds photographed adjacent to a ruler. Baseline body weights of mice were measured before infection, and everyday thereafter for 3 days or until animals were sacrificed before the third day of infection using a laboratory scale. Wound sizes were analyzed from photos taken daily with a Canon Rebel Powershot, and measured by scaling and determining wound area using ImageJ software (National Institutes of Health, NIH) calculated using the following equation: (A0-At)/A0·100.
Pyochelin and pyochelin methyl ester extractions from mouse tissue
Homogenized mouse skin tissue was acidified with 3 drops of 6 M HCl and transferred to a scintillation vial. To remove remaining homogenate, tubes were washed twice with 500 µL EtOAc, the solvent was transferred and pooled with the homogenate into the same scintillation vial. An additional 1 mL of EtOAc was added to the homogenate and the samples were mixed by pipetting. Phases were allowed to separate for 5 min and 1 mL of the EtOAc phase was transferred to a fresh tube. Homogenate was extracted three times, yielding a total of 3 mL of the EtOAc phase, which was concentrated to dryness in a SpeedVac vacuum concentrator (SPD131DDA SpeedVac Concentrator; Thermo Scientific). Samples were stored at – 20° C until further use.
To prepare mouse skin tissue extracts for LC-MS analysis, samples were thawed on room temperature, resuspended in 120 µL MeOH, incubated for 40 min in a sonicator bath (5800; Branson) at room temperature and an additional 30 min in the dark at room temperature. Samples were mixed by pipetting, centrifuged for 2 min at 10,000 rpm (Sorvall ST 40R; Thermo) to remove non-soluble particulates and 100 µL of clean extract were transferred to a fresh tube. Samples were stored at – 20° C or directly diluted 1:10 (v/v) in MeOH containing 1 µM glycocholic acid.
The tissues were then homogenized in three 30 s increments (for a total of 90 s) using a bead beater, serially diluted, and selectively plated for CFU counts. All tissue samples were plated on mannitol salt agar (MSA) with cefoxitin to select for MRSA and Pseudomonas Isolation Agar to select for PAO1. Tissue samples were then frozen at −80° C to preserve for later chemical analysis.
MALDI-TOF MSI
Overnight cultures of MRSA or PAO1 (WT, ΔpqsA or ΔpqsA Δpch) were diluted to OD600 of 0.2 and 5 µL of the bacterial solution were spotted on tryptic soy agar plates (TSA; Remel) supplemented with agar (2% (w/v) final concentration). For bacterial co-incubations, the bacterial solutions were spotted 5 mm apart. Samples were prepared for MALDI-MSI as previously described.45,46 Briefly, for each sample a region of agar including the bacterial colonies was excised from the culture, laid on top of a MALDI MSP 96 anchor plate (Bruker Daltonics). Universal matrix (1:1 2,5-dihydroxybenzoic acid (DHB): α-cyano-4-hydroxycinnamic acid (CHCA); (Sigma-Aldrich)) was applied manually using a 53 µm molecular sieve. Samples were dried at 37°C overnight and a photograph was taken. All colonies were subjected to MALDI-TOF MSI in positive reflectron mode using 500 µm spatial resolution in both X and Y dimensions by a Bruker Daltonics Autoflex Speed. Two dimensional metabolite distribution was visualized using Bruker FlexImaging version 4.0. False colored images were optimized for visualization of ion distribution.
Metabolomics data collection
Mass spectrometry data acquisition was performed using a Bruker Daltonics Maxis II HD quadrupole time of flight (qTOF) mass spectrometer with a standard electrospray ionization (ESI) source. Mass spectrometer tuning was performed by infusion with tuning mix ESITOF (Agilent Technologies) at a 3-µL/min flow rate. As a lock mass internal calibrant, hexakis (1H,1H,2H-difluoroethoxy)phosphazene ions (Apollo Scientific,m/z 622.1978) located on a wick within the source was used. Samples were introduced by an Agilent 1290 ultra-performance liquid chromatography (UPLC) system using a 10-µL injection volume. The solvent system for UPLC separation consisted of Optima-grade water (Fisher Scientific) and acetonitrile (ACN, Fisher Scientific) (Buffer A: 98:2; Buffer B: 2:98) with 0.1% Formic acid (FA, Fisher Scientific). Extracts were separated using a Phenomenex Kinetex 2.6-µm C18 column (2.1 mm by 50 mm) or a Phenomenex Kinetex 1.7-µm C18 column (2.1 mm by 50 mm) using a 9 min, linear water-ACN gradient at a flow rate of 0.5 mL/min. To compare the retention times of isolated and synthetic Pch and MetPch isomers, samples were separated using a Phenomenex Luna 5 µm C18(2) column (250 mm by 2 mm) using a 20 min, linear water-ACN gradient at a flow rate of 0.5 mL/min. The mass spectrometer was operated in data-dependent positive ion mode, automatically switching between full-scan MS and MS/MS acquisitions. Full-scan MS spectra (m/z 50 to 1,500) were acquired in the TOF-MS, and the top five most intense ions in a particular scan were fragmented via collision-induced dissociation (CID) using the stepping function in the collision cell.
Molecular networking
A classical molecular network was created from the mzXML files using the online workflow (version release 30) (https://ccms-ucsd.github.io/GNPSDocumentation/) on the GNPS website (http://gnps.ucsd.edu).51 Briefly, the data was filtered by removing all MS/MS fragment ions within +/− 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top 6 fragment ions in the +/− 50Da window throughout the spectrum. The precursor ion mass tolerance was set to 0.05 Da and an MS/MS fragment ion tolerance of 0.1 Da. A network was then created where edges were filtered to have a cosine score above 0.7 and more than 6 matched peaks. Further, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. Finally, the maximum size of a molecular family was set to 100, and the lowest scoring edges were removed from molecular families until the molecular family size was below this threshold. The spectra in the network were then searched against GNPS’ spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks. The molecular network (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=b225a233163545b7b8eba55c53fff754) was visualized in Cytoscape (version 3.8.2).75
Total synthesis of pyochelin and pyochelin methyl ester
All reactions were carried out in flame dried glassware under argon unless water was used in the reaction. Reagents and anhydrous solvents were purchased from Sigma-Aldrich, Acros Organics or TCI and used without further purification unless otherwise stated. Reactions were monitored using silica gel 60 F254 TLC plates. Plates were visualized with 254 nm UV light or when appropriate a basic KMnO4 solution or 5% solution of FeCl3 in methanol. Flash chromatography was performed using 60 µm mesh standard grade silica gel from Millipore Sigma. NMR solvents were obtained from Cambridge Isotope Labs and used as is. All 1H NMR (400 MHz) were recorded at 25°C on a Bruker Avance spectrometer. All 13C NMR (101 MHz) spectra were also recorded at 25°C on Bruker Avance spectrometers. Chemical shifts (δ) are given in parts per million (ppm) relative to the respective NMR solvent; coupling constants (J) are in hertz (Hz). Abbreviations used are s, singlet; d, doublet; dd, doublet of doublets; td, triplet of doublets; m, multiplet. All high-resolution mass spectrometry measurements were made in the Mass Spectrometry and Proteomics Facility at the University of Notre Dame. Preparatory HPLC purification was performed on a Preparatory HPLC purification was performed on an Agilent 1260 Infinity HPLC fitted with a Jupiter 4 µm Proteo 90 Å (250 × 21.2 mm) LC column running a 45 min method with a gradient of 20–95% MeCN in Water with 0.1% TFA running a gradient of 20–95% MeCN in Water with 0.1% TFA.
(S)-2-(2-hydroxyphenyl)-N-methoxy-N-methyl-4,5-dihydrothiazole-4-carboxamide
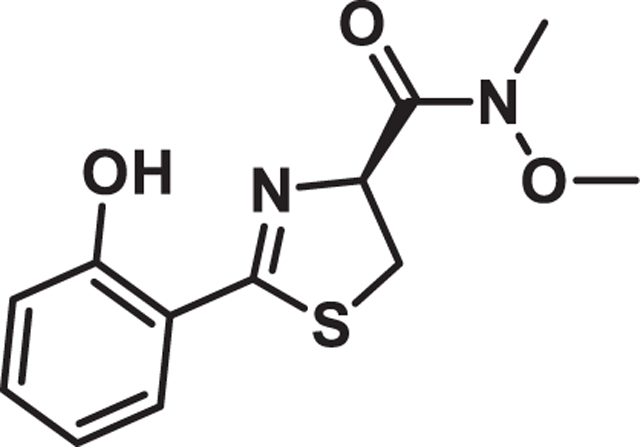
To a solution of 2-hydroxybenzonitrile 8 (2.5 g, 21 mmol) in methanol (93 mL) was added D-Cysteine HCl hydrate (3.7 g, 21 mmol) and phosphate buffer 0.1 M (pH 6.4) (75 mL). The solution was adjusted to pH 6.4 by addition of solid K2CO3 and the reaction mixture was stirred at 60°C overnight. The mixture was concentrated under reduced pressure, and the yellow crude material was diluted with water (100 mL). The solution was adjusted to pH 2.0 by addition of solid citric acid. After extraction with DCM (3 × 100 mL), the organic layers were collected, dried over Na2SO4 and filtered. The solvent was removed under reduced pressure to afford the as a yellow powder (4.2 g, 90%).
To a flamed dried reaction flask charged with carboxylic acid (3.8 g, 17 mmol) under argon was added anhydrous DMF (95 mL). Then, EDC (13 g, 68 mmol), HOBt (13 g, 68 mmol), N,O-dimethyl hydroxylamine hydrochloride (3.3 g, 34 mmol) were added. Triethylamine (9.5 mL, 68 mmol) was added dropwise by syringe. The reaction mixture was stirred overnight at room temperature. The reaction was then diluted with water and the mixture was brought to pH 2 by addition of 1 M HCl and extracted with ethyl acetate (3 × 100 mL). The combined organic layers were washed with brine, dried over anhydrous sodium sulfate, filtered, and concentrated. The crude material was purified by silica gel chromatography (10%–100% EtOAc in hexanes) to give an off-white solid (2.5 g, 55% yield). NMR values matched previously reported spectra.76
N-methyl-L-cysteine hydrochloride
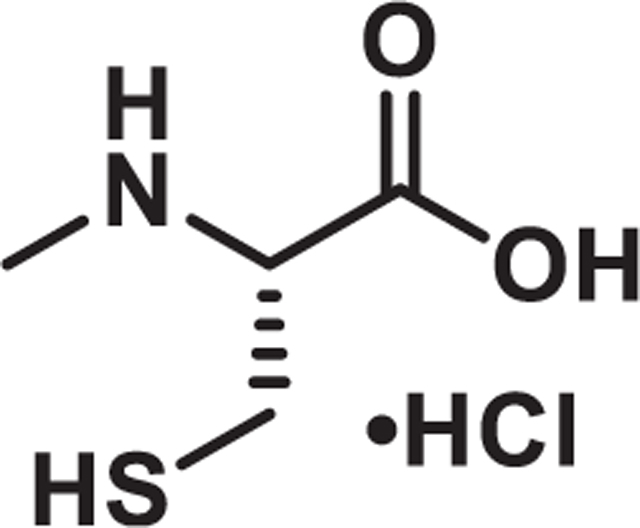
A solution of L-cysteine hydrochloride (16.0 g, 102 mmol) in water (10 mL) and NaOH (1 M, 10.2 mL, 10.2 mmol) was added aqueous formaldehyde (11.3 mL, 152 mmol, 37% by weight). After stirring at room temperature for 24 h, the reaction was cooled to 0°C and ethanol (25 mL) and pyridine (12 mL) were added. The resulting white precipitate was filtered and dried under vacuum (12.5 g, 93%). Used in next step without further purification.
The carboxylic acid (5.0 g, 38 mmol) from the previous step was dissolved in liquid ammonia (100 mL) condensed at −78°C. Water (0.68 mL, 38 mmol) was added. Sodium metal was added until the solution retained a deep blue color. The ammonia was allowed to evaporate, and the reaction was quenched with sat. ammonium chloride. After drying, the residue was dissolved in water and acidified to pH 1 with 6N HCl and water was removed under vacuum. The remaining residue was washed multiple times with hot ethanol and the organic layer was concentrated under vacuum to afford the product as a sticky white solid (2.4 g, 37%). NMR values matched previously reported spectra.77 DMF was used as internal standard to calculate purity of amino acid by weight to ensure no inorganic salts were present.
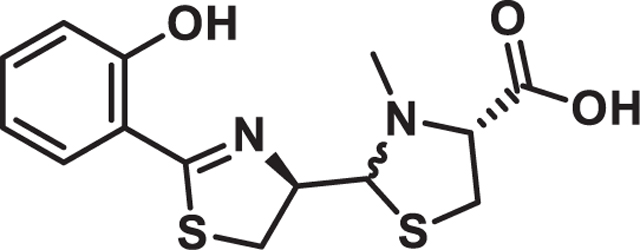
Pyochelin I and II
Weinreb amide 13 (2.5 g, 9.4 mmol) was added to a flame-dried flask under argon and dissolved in dry THF (190 mL). The reaction mixture was brought to −78°C in a bath of dry ice and acetone. Lithium aluminum hydride (4M, 2.6 mL, 10 mmol) was added dropwise by syringe down the side of the reaction flask into the reaction mixture. The reaction was stirred at between −50 and −30° C and monitored by TLC. After 2 h, the reaction was quenched by slow addition of saturated NH4Cl and the reaction was allowed to warm to room temperature while stirring vigorously until two phases formed. The phases were separated, and the aqueous layer was extracted with EtOAc (3 × 25 mL). The combined organic layers were washed with brine, dried over anhydrous sodium sulfate, filtered, and concentrated. Due to instability the aldehyde was used as quickly as possible in the cyclization without any purification.
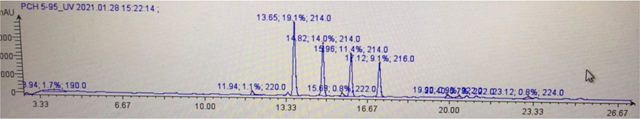
Potassium acetate (4.1 g, 42 mmol) and N-methyl-L-cysteine hydrochloride (2.4 g, 14 mmol) were added to a solution of the crude aldehyde in EtOH (250 mL) and H2O (62.5 mL). The mixture was stirred at 25°C overnight and then diluted with H20, adjusted to pH 4.5 with 6 N HCl then extracted with EtOAc. The organic layers were worked up to yield crude pyochelin as a mixture of 4 isomers. HPLC (15 cm, C12 column) analysis showed the four separate isomers eluting at 13.65, 14.82, 15.96 and 17.12 min on a 30 min 5–95% ACN (0.1% FA) LC method (LC trace below). Each of the four major peaks has the correct mass of [M + H]+ 325 m/z. The natural pyochelin isomers were determined to be the second and third peaks on LC based on the 1H NMR shifts of their methyl esters. Peaks one and four are neopyochelin. Purification of 125 mg of crude material on prep HPLC (20–95 ACN in aq. 0.1% TFA) led to 40 mg each of pyochelin and neopyochelin as their bis-trifluoro acetate salts. 15 mg of each was put aside then the rest was dissolved in a pH 4.5 phosphate buffer (680 mg potassium dihydrogen phosphate in 100 mL DI water) then extracted with ethyl acetate and dried under high vacuum to afford a mixture of pyochelin I and II which could not be purified further due to their rapid interconversion.
Copy of LC trace (30 min, 5–95% MeCN in 0.1% Formic acid)
Pyochelin Methylesters
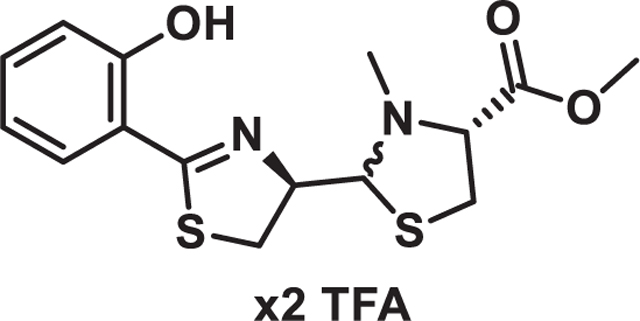
To a mixture of the free bases of pyochelin I and II (100 mg, 0.3 mmol) in benzene (5 mL) and MeOH (1.25 mL) was added TMSCHN2 (2M, 200 µL, 0.4 mmol) at room temperature. The reaction was allowed to stir for 30 min before being concentrated under vacuum. The residue was then purified via prep HPLC (20–95 ACN in aq. 0.1% TFA) to afford the pyochelin methyl esters in a 4:1 ratio as their bis-trifluoro acetate salts. NMR values for pyochelin I methyl ester and pyochelin II methyl ester matched those previously reported (Figure S5).78
CAS assay
CAS reagent was prepared as follows: 25 mL of 2 mM CAS reagent (Chem-Impex International) in water were mixed with 5 mL of 1 mM FeCl3 in 10 mM HCl, giving CAS reagent stock solution. For a 10 mL stock solution of the final CAS solution, 0.6 mL of CAS reagent stock solution were mixed with 0.4 mL of a 5 mM HDTMA solution in water and 9 mL of a 111 mM PIPES solution in water (pH 6.8). 4 mM stock solutions of synthetic pyochelin and pyochelin methyl ester in MeOH were serially diluted 1:2 (v/v) with MeOH, giving stock solutions of pyochelin and pyochelin methyl ester at 4 mM, 2 mM 1 mM, 0.5 mM, 0.25 mM and 0.125 mM concentrations. 5 µL of each stock solution were mixed with 195 µL of the final CAS solution in 96-black well plates (Corning Incorporated) and incubated at room temperate in the dark. CAS solution incubated with MeOH was used as reference. After 5 h of incubation, the fluorescence of the samples was recorded on an Infinite M PLEX plate reader (Tecan) with the following settings: Excitation 425 nm, emission 630 nm, Bandwidth 9 nm, Number of flashes 25. Siderophore activity of Pch and MetPch was calculated according to the following formula: (Ar-As)/Ar x 100% (Ar = absorbance of reference and As = absorbance of sample)
ROS assay
Stock solutions of 2′,7′-dichlorofluorescin diacetate (DCF-DA; Sigma) were prepared by dissolving DCF-DA at a final concentration of 5 mM in absolute ethanol (Fisher Scientific) and stored at – 20°C. The DCF-DA stock solution was then hydrolyzed to the non-fluorescent 2′,7′-dichlorodihydrofluorescein (DCF-H) by mixing 0.5 mL DCF-DA (5 mM) with 2 mL of 0.1 N NaOH at room temperate for 30 min. The reaction was stopped by adding 7.5 mL of 10x PBS pH 7.4 (0.1 M, without calcium and magnesium, Gibco). S. aureus cultures grown in TSB overnight were used to inoculate fresh TSB cultures at a starting OD600 of 0.05 and incubated at 37°C with shaking (200 rpm). After 2.5 h, the OD600 of the S. aureus cultures was measured and all cultures were set to an OD600 of 1. Cultures were incubated with either Pch or MetPch at final concentrations of 10 µM, 20 µM and 40 µM or an equal volume of methanol (control) for 90 min at 37°C with 800 rpm shaking (Microtiter Plate Shaker Incubator SI505; Stuart). Bacterial cultures containing either Pch, MetPch or methanol were adjusted to OD600 of 1 and centrifuged at 5,000 g (Biofuge pico, Heraeus) for 10 min. Bacterial cell pellets were resuspended in 100 µL DCF-H (50 µM) solution and incubated for 40 min 100 µL were added to a 96-well black plate (Corning Incorporated). DCF-H incubated with sterile medium was used as control. ROS production was determined as a function of oxidation of the non-fluorescent DCF-H to 2′,7′-dichloroofluorescein (DCF) in an Infinite M PLEX plate reader (Tecan) with the following settings: Excitation 488 nm (bandwidth 9 nm); emission 515 nm (bandwidth 20 nm).
P. aeruginosa growth support assay
P. aeruginosa strains were pre-grown for 36 h in M9 minimal medium supplemented with 0.5% (v/v) glycerol as carbon source, spun down and resuspended in either M9 glycerol or M9 glycerol containing 500 µM 2,2′-Bipyridine (Fisher Scientific). The bacterial stock solutions were used to inoculate M9 glycerol and M9 glycerol medium containing 500 µM 2,2′-Bipyridine at a starting OD600 of 0.05. Bacterial growth was followed over the course of 96 h in the presence of three different concentrations (0.25 µM, 1 µM and 2 µM) of pyochelin or pyochelin methyl ester or an equivalent volume of methanol. OD600 measurements were taken at 0 h and every 12 h thereafter in an Infinite M PLEX plate reader (Tecan).
S. aureus growth inhibition assay
S. aureus LAC wild type was grown overnight in chelex treated TSB. Cultures were centrifuged and resuspended in chelex treated TSB. The bacterial stock solutions were used to inoculate chelex treated TSB medium at a starting OD600 of 0.05 and 198 µL of bacterial cultures were transferred to 96-well plates. 4 mM stock solutions of synthetic pyochelin and pyochelin methyl ester in MeOH were diluted to yield working solutions of 1.92 mM, 0.96 mM, 0.48 mM and 0.24 mM. 198 µL of S. aureus cultures at OD600 of 0.05 were incubated with 2 µL of Pch and MetPch working solutions to yield final concentrations of 19.2 µM, 9.6 mM, 4.8 µM and 2.4 µM of synthetic pyochelin or pyochelin methyl ester. 2 µL MeOh was used for control cultures. The bacterial cultures were incubated at 37°C with 1000 rpm shaking (Microtiter Plate Shaker Incubator SI505; Stuart) for 24 h. Bacterial growth was determined by OD600 measurements with an Infinite M PLEX plate reader (Tecan).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed with GraphPad Prism version 9.4.1. Statistical tests performed and the p values of datasets are indicated for each individual experiment in the corresponding figure legend.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| S. aureus USA300 LAC/MRSA WT | Boles et al.66 | AH1263 |
| S. aureus USA300 LAC Δspm/MRSA Δspm | This study | AH6121 |
| S. aureus USA300 LAC Δspm::spm/MRSA Δspm::spm | This study | AH6080 |
| S. aureus USA 300 LAC fur::tet | This study | AH4449 |
| S. aureus 8325-4 fur::tet | Horsburgh et al.67 | AH2367 |
| P. aeruginosa PAO1 | Holloway et al.68 | AH5602 |
| P. aeruginosa PAO1 ΔpqsA | Toyofuku et al.50 | AH5136 |
| P. aeruginosa PAO1 ΔpvdI | This study | AH6075 |
| P. aeruginosa PAO1 ΔpchF | This study | AH6197 |
| P. aeruginosa PAO1 ΔpvdI ΔpchF | This study | AH6076 |
| P. aeruginosa PAO1 Δpch | This study | AH6148 |
| E. coli DC10b | Monk et al.69 | AH2811 |
| E. coli Top10 | Invitrogen | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Methanol | Fisher Chemical | A456-212 |
| Ethyl Acetate | J.T. Baker | 02-002-839 |
| Acetonitrile | Fisher Chemical | A955-212 |
| Formic Acid | Fisher Chemical | A117-50 |
| 2-Cyano phenol | Chem-Impex Inc. | 26803 |
| D-cysteine hydrochloride hydrate | Sigma-Aldrich | C8005 |
| TMS-diazomethane | Sigma-Aldrich | 527254 |
| Formaldehyde (37% by weight in water) | Sigma-Aldrich | 252549 |
| N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide HCl | Sigma-Aldrich | 17750 |
| Hydroxy benzotriazole | Sigma-Aldrich | 157260 |
| Lithium aluminum hydride | Sigma-Aldrich | 593702 |
| Hexadecyltrimethylammonium bromide (HDTMA) |
Sigma-Aldrich | H9151 |
| 2′,7′-dichlorofluorescin diacetate | Sigma-Aldrich | D6883-250MG |
| 2,2′-Bipyridine | Thermo-Scientific Chemicals | A15782-14 |
| Chrome azurole S | Honeywell | 72687-25G |
|
| ||
| Critical commercial assays | ||
|
| ||
| QIAprep Spin Miniprep Kit | Qiagen | 27104 |
| QIAquick Gel Extraction Kit | Qiagen | 28704 |
| QIAquick PCR Purification Kit | Qiagen | 28104 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | 1725121 |
| iScript RT Supermix | Bio-Rad | 1708841 |
|
| ||
| Deposited data | ||
|
| ||
| mass spectrometry data | This study | MassIVE: MSV000089230, MassIVE: MSV000089231, MassIVE: MSV000089233, MassIVE: MSV000089234 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | Jackson Laboratories | 000664 |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 | N/A | |
|
| ||
| Recombinant DNA | ||
|
| ||
| pJB38; Mutation generation vector | Bose et al.70 | pJB38 |
| pEX2G; Mutation generation vector | Rietsch et al.71 | pEX2G |
| pHC48; S. aureus constitutive dsRed expression vector | Ibberson et al.72 | pHC48 |
| pJBspm; pJB38-spm; construct for unmarked deletion of spm | This study | pJBspm |
| pJBspmKI; pJB38-spmKI; construct to repair Δspm mutant strain; water mark: NcoI restriction site |
This study | pJBspmKI |
| pJBfhuD1; pJB38-fhuD1; construct for unmarked deletion of fhuD1 | This study | pJBfhuD1 |
| pJBfhuD2; pJB38-fhuD2; construct for unmarked deletion of fhuD2 | This study | pJBfhuD2 |
| pJBfhuG; pJB38-fhuG; construct for unmarked deletion of fhuG | This study | pJBfhuG |
| pEXpch; pEX2G-pch; construct for unmarked deletion of the entire pyochelin biosynthesis cluster | This study | pEXpch |
| pEXpchF; pEX2G-pchF; construct for unmarked deletion of pchF | This study | pEXpchF |
| pEXpvdI; pEX2G-pvdI; construct for unmarked deletion of pvdI | This study | pEXpvdI |
| pHCspmprom; pHC48-spmprom, promoter region of the spm/fhuD1 operon with dsRed on the pHC48 backbone. | This study | pHCspmprom |
|
| ||
| Software and algorithms | ||
|
| ||
| Prism, V9.4.1 | GraphPad Software | https://graphpad.com/scientificsoftware/prism |
| CFX Manager, V3.1 | Bio-RAD | https://www.bio-rad.com/en-us/sku/1845000-cfx-manager-software?ID=1845000 |
| MZmine, V2.53 | Pluskal et al.73 | http://mzmine.github.io |
| GNPS V30 | Wang et al.51 | https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp |
| FlexImaging, V4.1 | Bruker Daltonics | https://www.bruker.com |
| DataAnalysis, V4.2 | Bruker Daltonics | https://www.bruker.com |
| Cytoscape, V3.8.2 | Cytoscape Consortium | https://cytoscape.org |
Highlights.
Staphylococcus aureus methylates pyochelin produced by Pseudomonas aeruginosa
Pyochelin is methylated by the staphylococcal pyochelin methyltransferase (Spm)
Pyochelin loses its biological activity upon Spm-mediated methylation
Pyochelin methylation increases S. aureus fitness in co-infections with P. aeruginosa
ACKNOWLEDGMENTS
We thank Prof. Masanori Toyofuku, University of Tsukuba, for sharing P. aeruginosa strains and Dr. Heidi Crosby for strain S. aureus LAC fur::tet. This work was supported by Switzerland National Science Foundation Early Postdoc Mobility Fellowship P2ZHP3_168585 (C.J.); by American Heart Association (AHA) postdoctoral fellowship 20POST35220011 (K.S.); by NIH public health service grants AI083211, AI153185, and AI166805 (A.R.H.) and DE022350 (C.M.); and by NIH NIGMS R35 GM128690 (V.V.P.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112540.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hubert D, Réglier-Poupet H, Sermet-Gaudelus I, Ferroni A, Le Bourgeois M, Burgel PR, Serreau R, Dusser D, Poyart C, and Coste J. (2013). Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J. Cyst. Fibros. 12, 497–503. 10.1016/j.jcf.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Yung DBY, Sircombe KJ, and Pletzer D. (2021). Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol. 116, 1–15. 10.1111/mmi.14699. [DOI] [PubMed] [Google Scholar]
- 3.Rahim K, Saleha S, Zhu X, Huo L, Basit A, and Franco OL. (2017). Bacterial contribution in chronicity of wounds. Microb. Ecol. 73, 710–721. 10.1007/s00248-016-0867-9. [DOI] [PubMed] [Google Scholar]
- 4.Registry, C.F.F.P. (2019). 2019 Annual Data Report (Cystic Fibrosis Foundation; ). [Google Scholar]
- 5.Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen B, Klein BM, and Krogfelt KA. (2006). Multiple bacterial species reside in chronic wounds: a longitudinal study. Int. Wound J. 3, 225–231. 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibberson CB, and Whiteley M. (2020). The social life of microbes in chronic infection. Curr. Opin. Microbiol. 53, 44–50. 10.1016/j.mib.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Körber A, Schmid EN, Buer J, Klode J, Schadendorf D, and Dissemond J. (2010). Bacterial colonization of chronic leg ulcers: current results compared with data 5 years ago in a specialized dermatology department. J. Eur. Acad. Dermatol. Venereol. 24, 1017–1025. 10.1111/j.1468-3083.2010.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.Bessa LJ, Fazii P, Di Giulio M, and Cellini L. (2015). Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: some remarks about wound infection. Int. Wound J. 12, 47–52. 10.1111/iwj.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, and Goldberg JB. (2016). Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 35, 947–953. 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 10.Sagel SD, Gibson RL, Emerson J, McNamara S, Burns JL, Wagener JS, Ramsey BW, Inhaled Tobramycin in Young Children Study Group, Cystic Fibrosis Foundation Therapeutics Development Network. Inhaled tobramycin in young children study, G., and cystic fibrosis foundation therapeutics development, N. (2009). impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J. Pediatr. 154, 183–188. 10.1016/j.jpeds.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, and Rumbaugh KP. (2011). An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6, e27317. 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, and Rumbaugh KP. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728. 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, and Davis SC. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8, e56846. 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lightbown JW, and Jackson FL. (1956). Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem. J. 63, 130–137. 10.1042/bj0630130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machan ZA, Pitt TL, White W, Watson D, Taylor GW, Cole PJ, and Wilson R. (1991). Interaction between Pseudomonas aeruginosa and Staphylococcus aureus: description of an anti-staphylococcal substance. J. Med. Microbiol. 34, 213–217. 10.1099/00222615-34-4-213. [DOI] [PubMed] [Google Scholar]
- 16.Biswas L, Biswas R, Schlag M, Bertram R, and Götz F. (2009). Smallcolony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75, 6910–6912. 10.1128/AEM.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldan R, Cigana C, Testa F, Bianconi I, De Simone M, Pellin D, Di Serio C, Bragonzi A, and Cirillo DM. (2014). Adaptation of Pseudomonas aeruginosa in Cystic Fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 9, e89614. 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niggli S, and Kümmerli R. (2020). Strain background, species frequency, and environmental conditions are important in determining Pseudomonas aeruginosa and Staphylococcus aureus population dynamics and species coexistence. Appl. Environ. Microbiol. 86, e00962–20. 10.1128/AEM.00962-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Vos D, De Chial M, Cochez C, Jansen S, Tümmler B, Meyer JM, and Cornelis P. (2001). Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch. Microbiol. 175, 384–388. 10.1007/s002030100278. [DOI] [PubMed] [Google Scholar]
- 20.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103, 8487–8492. 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marvig RL, Damkiær S, Khademi SMH, Markussen TM, Molin S, and Jelsbak L. (2014). Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5, e00966–e00914. 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marvig RL, Dolce D, Sommer LM, Petersen B, Ciofu O, Campana S, Molin S, Taccetti G, and Johansen HK. (2015). Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol. 15, 218. 10.1186/s12866-015-0563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khademi SMH, Sazinas P, and Jelsbak L. (2019). Within-Host adaptation mediated by intergenic evolution in Pseudomonas aeruginosa. Genome Biol. Evol. 11, 1385–1397. 10.1093/gbe/evz083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camus L, Briaud P, Vandenesch F, and Moreau K. (2021). How bacterial adaptation to cystic fibrosis environment shapes interactions between Pseudomonas aeruginosa and Staphylococcus aureus. Front. Microbiol. 12, 617784. 10.3389/fmicb.2021.617784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camus L, Vandenesch F, and Moreau K. (2021). From genotype to phenotype: adaptations of Pseudomonas aeruginosa to the cystic fibrosis environment. Microb. Genom. 7, mgen000513. 10.1099/mgen.0.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen B, Andersen AS, Krogfelt KA, Givskov M, and Tolker-Nielsen T. (2009). Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47, 4084–4089. 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Registry, C.F.F.P. (2021). 2020 Annual Data Report (Cystic Fibrosis Foundation; ). [Google Scholar]
- 28.Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams RE. (1963). Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol. Rev. 27, 56–71. 10.1128/br.27.1.56-71.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welsh MA, Eibergen NR, Moore JD, and Blackwell HE. (2015). Small molecule disruption of quorum sensing cross-regulation in Pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes. J. Am. Chem. Soc. 137, 1510–1519. 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machan ZA, Taylor GW, Pitt TL, Cole PJ, and Wilson R. (1992). 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30, 615–623. 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 32.Lache M, Hearn WR, Zyskind JW, Tipper DJ, and Strominger JL. (1969). Specificity of a bacteriolytic enzyme from Pseudomonas aeruginosa. J. Bacteriol. 100, 254–259. 10.1128/jb.100.1.254-259.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessler E, Safrin M, Olson JC, and Ohman DE. (1993). Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268, 7503–7508. [PubMed] [Google Scholar]
- 34.Pamp SJ, and Tolker-Nielsen T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189, 2531–2539. 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood TL, Gong T, Zhu L, Miller J, Miller DS, Yin B, and Wood TK. (2018). Rhamnolipids from Pseudomonas aeruginosa disperse the biofilms of sulfate-reducing bacteria. NPJ Biofilms Microbiomes 4, 22. 10.1038/s41522-018-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdallah MA, Pfestorf M, and Döring G. (1989). Pseudomonas aeruginosa pyoverdin: structure and function. Antibiot. Chemother. 42, 8–14. 10.1159/000417597. [DOI] [PubMed] [Google Scholar]
- 37.Cornelis P. (2010). Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86, 1637–1645. 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 38.Youard ZA, Wenner N, and Reimmann C. (2011). Iron acquisition with the natural siderophore enantiomers pyochelin and enantio-pyochelin in Pseudomonas species. Biometals 24, 513–522. 10.1007/s10534-010-9399-9. [DOI] [PubMed] [Google Scholar]
- 39.Adler C, Corbalán NS, Seyedsayamdost MR, Pomares MF, de Cristóbal RE, Clardy J, Kolter R, and Vincent PA. (2012). Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS One 7, e46754. 10.1371/journal.pone.0046754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong KS, Cheow YL, and Lee SM. (2017). The role of reactive oxygen species in the antimicrobial activity of pyochelin. J. Adv. Res. 8, 393–398. 10.1016/j.jare.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noto MJ, Burns WJ, Beavers WN, and Skaar EP. (2017). Mechanisms of pyocyanin toxicity and genetic determinants of resistance in Staphylococcus aureus. J. Bacteriol. 199, e00221–17. 10.1128/JB.00221-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szamosvári D, and Böttcher T. (2017). An unsaturated quinolone N-oxide of Pseudomonas aeruginosa modulates growth and virulence of Staphylococcus aureus. Angew. Chem., Int. Ed. Engl. 56, 7271–7275. 10.1002/anie.201702944. [DOI] [PubMed] [Google Scholar]
- 43.Holloway BW. (1955). Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13, 572–581. 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 44.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, and Blumberg HM. (2006). Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144, 309–317. 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 45.Yang YL, Xu Y, Straight P, and Dorrestein PC. (2009). Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 5, 885–887. 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, Fleming TC, Wenter R, Moore BS, Golden SS, Pogliano K, and Dorrestein PC. (2012). Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 194, 6023–6028. 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, and Rahme LG. (2004). Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 101, 1339–1344. 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman JP, Hudson LL, McKnight SL, Farrow JM 3rd, Calfee MW, Lindsey CA, and Pesci EC. (2008). Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J. Bacteriol. 190, 1247– 1255. 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, and Manoil C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184, 6472–6480. 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, and Uchiyama H. (2008). Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J. Bacteriol. 190, 7947–7956. 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox CD, and Graham R. (1979). Isolation of an iron-binding compound from Pseudomonas aeruginosa. J. Bacteriol. 137, 357–364. 10.1128/jb.137.1.357-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lybbert AC, Williams JL, Raghuvanshi R, Jones AD, and Quinn RA. (2020). Mining public mass spectrometry data to characterize the diversity and ubiquity of P. aeruginosa specialized metabolites. Metabolites 10, 445. 10.3390/metabo10110445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinn RA, Adem S, Mills RH, Comstock W, DeRight Goldasich L, Humphrey G, Aksenov AA, Melnik AV, da Silva R, Ackermann G, et al. (2019). Neutrophilic proteolysis in the cystic fibrosis lung correlates with a pathogenic microbiome. Microbiome 7, 23. 10.1186/s40168-019-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, and Bayles KW. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537–e00512. 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebulsky MT, and Heinrichs DE. (2001). Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183, 4994–5000. 10.1128/JB.183.17.4994-5000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sebulsky MT, Hohnstein D, Hunter MD, and Heinrichs DE. (2000). Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182, 4394–4400. 10.1128/JB.182.16.4394-4400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price EE, and Boyd JM. (2020). Genetic regulation of metal ion homeostasis in Staphylococcus aureus. Trends Microbiol. 28, 821–831. 10.1016/j.tim.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas MS. (2007). Iron acquisition mechanisms of the Burkholderia cepacia complex. Biometals 20, 431–452. 10.1007/s10534-006-9065-4. [DOI] [PubMed] [Google Scholar]
- 60.Cornelis P, and Dingemans J. (2013). Pseudomonas aeruginosa adaptsits iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 3, 75. 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dumas Z, Ross-Gillespie A, and Kümmerli R. (2013). Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc. Biol. Sci. 280, 20131055. 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobessi D, Celia H, and Pattus F. (2005). Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol. 352, 893–904. 10.1016/j.jmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Ronnebaum TA, McFarlane JS, Prisinzano TE, Booker SJ, and Lamb AL. (2019). Stuffed methyltransferase catalyzes the penultimate step of pyochelin biosynthesis. Biochemistry 58, 665–678. 10.1021/acs.biochem.8b00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho YN, Hoo SY, Wang BW, Hsieh CT, Lin CC, Sun CH, Peng CC, Lin C, and Yang YL. (2021). Specific inactivation of an antifungal bacterial siderophore by a fungal plant pathogen. ISME J. 15, 1858–1861. 10.1038/s41396-020-00871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molina-Santiago C, Vela-Corcía D, Petras D, Díaz-Martínez L, Pérez-Lorente AI, Sopeña-Torres S, Pearson J, Caraballo-Rodríguez AM, Dorrestein PC, de Vicente A, and Romero D. (2021). Chemical interplay and complementary adaptative strategies toggle bacterial antagonism and co-existence. Cell Rep. 36, 109449. 10.1016/j.celrep.2021.109449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boles BR, Thoendel M, Roth AJ, and Horswill AR. (2010). Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5, e10146. 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horsburgh MJ, Ingham E, and Foster SJ. (2001). In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475. 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holloway BW, Krishnapillai V, and Morgan AF. (1979). Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43, 73–102. 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monk IR, and Foster TJ. (2012). Genetic manipulation of Staphylococci-breaking through the barrier. Front. Cell. Infect. Microbiol. 2, 49. 10.3389/fcimb.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bose JL, Fey PD, and Bayles KW. (2013). Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 79, 2218–2224. 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rietsch A, Vallet-Gely I, Dove SL, and Mekalanos JJ. (2005). ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 102, 8006–8011. 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ibberson CB, Parlet CP, Kwiecinski J, Crosby HA, Meyerholz DK, and Horswill AR. (2016). Hyaluronan modulation impacts Staphylococcus aureus biofilm infection. Infect. Immun. 84, 1917–1929. 10.1128/IAI.01418-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pluskal T, Castillo S, Villar-Briones A, and Oresic M. (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 11, 395. 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogel HJ, and Bonner DM. (1956). Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218, 97–106. [PubMed] [Google Scholar]
- 75.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Noël S, Guillon L, Schalk IJ, and Mislin GLA. (2011). Synthesis of fluorescent probes based on the pyochelin siderophore scaffold. Org. Lett. 13, 844–847. 10.1021/ol1028173. [DOI] [PubMed] [Google Scholar]
- 77.Park JD, and Kim DH. (2002). Cysteine derivatives as inhibitors for carboxypeptidase A: synthesis and structure-activity relationships. J. Med. Chem. 45, 911–918. 10.1021/jm010272s. [DOI] [PubMed] [Google Scholar]
- 78.Rinehart KL, Staley AL, Wilson SR, Ankenbauer RG, and Cox CD. (1995). Stereochemical assignment of the pyochelins. J. Org. Chem. 60, 2786–2791. 10.1021/jo00114a029. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry data, including the raw files, mzXML files, metadata tables and MZmine settings have been deposited at MassIVE (MassIVE: MSV000089230; P. aeruginosa/S. aureus WT, spm mutant, spm complementation interaction, MassIVE: MSV000089231; P. aeruginosa/S. aureus WT, fhuG mutant, fhuD1/D2 double mutant interaction, MassIVE: MSV000089233, Pyochelin methylation by S. aureus transposon library mutants, MassIVE: MSV000089234; Pyochelin methyl ester quantification murine skin wound model S. aureus/P. aeruginosa co-infection) and are publicly available as of the date of publication. Password for all datasets: MetPch2022.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.