Abstract
Background:
Candida spp. are opportunistic yeasts capable of forming biofilms, which contribute to resistance, increasing the urgency for new effective antifungal therapies. Repurposing existing drugs could significantly accelerate the development of novel therapies against candidiasis.
Methods:
We screened the Pandemic Response Box containing 400 diverse drug-like molecules active against bacteria, viruses or fungi, for inhibitors of C. albicans and C. auris biofilm formation. Initial hits were identified based on the demonstration of >70% inhibitory activity. Dose-response assays were used to confirm the antifungal activity of initial hits and establish their potency. The spectrum of antifungal activity of the leading compounds was determined against a panel of medically important fungi, and the in vivo activity of the leading repositionable agent was evaluated in murine models of C. albicans and C. auris systemic candidiasis.
Results:
The primary screening identified 20 hit compounds, and their antifungal activity and potency against C. albicans and C. auris were validated using dose-response measurements. From these experiments, the rapalog everolimus, emerged as the leading repositionable candidate. Everolimus displayed potent antifungal activity against different Candida spp., but more moderate levels of activity against filamentous fungi. Treatment with everolimus increased survival of mice infected with C. albicans, but not those with C. auris.
Conclusion:
The screening of the pandemic response box resulted in the identification of several drugs with novel antifungal activity, with everolimus emerging as the main repositionable candidate. Further in vitro and in vivo studies are needed to confirm its potential therapeutic use.
Keywords: Candida spp., biofilm, repurposing, screening
INTRODUCTION
Candidiasis, infections caused by Candida spp, constitutes a growing threat to human health, particularly for immune- and medically compromised individuals (1, 2). Indeed, over the last few decades progress of modern medicine has resulted in an expanding number of patients who are at increased risk for opportunistic infections, with candidiasis representing the most frequent fungal infection afflicting these patients, and now the third to fourth leading nosocomial infection in US hospitals (3). Unfortunately, invasive candidiasis carries unacceptably high morbidity and mortality rates (4, 5). Although Candida albicans represent the most frequent causative agent of candidiasis, accounting for approximately 50% of all cases, other non-albicans Candida species are on the rise (6). During the last decade since its first description in 2009 (7), C. auris has emerged as a formidable opportunistic pathogen of humans, being responsible for major outbreaks in healthcare facilities, with the aggravated circumstance that this yeast is often multi-, or even pan-fungal drug resistant (8, 9, 10).
Importantly, different manifestations of candidiasis are associated with biofilm formation both on innate and biological surfaces (11). Biofilms are highly structured communities of cells attached to a surface and enveloped within a matrix of self-produced exopolymeric materials. Fungal biofilm development has been best studied in C. albicans, where it has been demonstrated to be highly regulated and coordinated, involving processes such as adhesive interactions, morphological changes, and consortia behavior (12). Perhaps, from a clinical point of view, the most important negative consequence of Candida biofilm formation is the fact that cells within the biofilms show decreased susceptibility to antifungal therapy (13), often leading to therapeutic failure and by extension contributing to excess mortality (14). The current armamentarium of antifungal agents for the clinical therapy of invasive candidiasis is limited to three different classes, azoles, polyenes and echinocandins. However, C. albicans biofilms show decreased susceptibility (13), and C. auris biofilms have been described to be intrinsically resistant to all three classes of antifungals (15). Altogether, these point to the urgent need for the identification of novel agents with anti-Candida biofilm activity to improve or complement existing antifungal drug therapy (16).
Drug repurposing (also referred to as repositioning), which is the process of finding new therapeutic indications for current medications and abandoned or failed compounds, is gaining traction as an alternative approach to drug development (17). One of the main advantages of this approach is the fact that these drugs have already undergone several phases of clinical development, with fully characterized pharmacological and safety properties. As such, repurposing constitutes a very practical and attractive pathway to drug development, with also high potential for accelerated translation from the bench to the clinic which can rapidly bring benefits to patients (17).
Here we have screened The Pandemic Response Box® library from Medicines for Malaria Venture, a diverse library of approximately 400 anti-infective compounds assembled by MMV, in search for inhibitors of C. albicans and C. auris biofilm formation.
MATERIALS AND METHODS
Strains, media, and culture conditions
C. albicans SC5314 was used in this study, including primary screens and follow-up experiments. The C. auris panel consisting of 10 different clinical isolates was provided by the U.S. Centers for Disease Control and Prevention and Food and Drug Administration Antibiotic Resistance (AR) Bank. From this panel, C. auris strain 0390 was chosen for initial experiments, including the primary screens. Cells were grown in yeast extract-peptone-dextrose (YPD) (1% (wt/vol) yeast extract, 2% (wt/vol) peptone, 2% (wt/vol) dextrose) liquid medium in an orbital shaker (150–180 rpm) at 30 °C overnight. The cells were then washed with phosphate-buffered saline (PBS), counted with a hemocytometer, and adjusted to the desired final cell density by diluting in RPMI-1640 medium without sodium bicarbonate and supplemented with L-glutamine (Cellgro, Manassas, VA, USA), buffered with 165 mM morpholine propane sulfonic acid (Thermo-Fisher Scientific, Waltham, MA, USA) and adjusted to pH 6.9.
Chemical library
The Pandemic Response Box was kindly provided by Medicines for Malaria Venture. The library contains 400 diverse drug-like anti-infective molecules active against bacteria, viruses, or fungi (18). The list of compounds included in this box can be found at the Pandemic Response Box website (https://www.mmv.org/mmv-open/pandemicresponse-box). Compounds in the original library are provided in individual wells within 96-well microtiter plates as 10 mM solutions in dimethyl sulfoxide (DMSO). A daughter plate at a concentration of 200 μM was made by performing a 1:50 dilution from the master plate into the wells of pre-sterilized, polystyrene, flat-bottomed 96-well microtiter plates (Corning Incorporated, Corning, NY, USA), and these plates were stored at −20 °C until used.
Primary screens for inhibitors of C. albicans SC5314 and C. auris 0390 biofilm formation.
C. albicans SC5314 and C. auris 0390 strains were used in primary screens. The screens were performed following the 96-well microtiter plate model of Candida biofilm formation previously developed by our group (19, 20), and each performed in duplicate (two independent plates). Briefly, 50 μl of the fungal inoculum (2 × 106 cells/ml) were used to seed individual wells within a flat-bottom 96-well microtiter plate. The same volume of each individual library compound, prepared at a 20 μM concentration in RPMI 1640, was added to individual wells, resulting in the appropriate final volume (100 ml per well) as well as concentration of cells (1 × 106 cells/ml) and compounds (10 μM each) for screening. The plates were then incubated statically at 37 °C for 24 h to allow for biofilm formation. After the incubation, the plates were washed once with PBS to remove non-adherent cells, and the extent of biofilm formation was determined using a colorimetric assay based on the reduction of 2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide (XTT, Sigma, St. Louis, MO, USA). For both screens (C. albicans and C. auris), wells in the first column served as positive controls (no compound added), while wells in the last column served as negative controls (no cells added), respectively.
Dose-response assays for confirmation of initial hits and determination of potency
Confirmatory dose-response assays were performed to validate the inhibitory activity of the hit compounds identified during the primary screens. These assays were performed by using the same microdilution techniques for the inhibition of biofilm growth as described above, using a 10-point, 1:2 dilution dose-response format for each of the initial hits, resulting in final compound concentrations ranging from 20 to 0.04 μM. Dose response curves were generated from the normalized readings obtained from the plate reader using the positive (untreated) and negative (uninoculated) controls which were arbitrarily set as 100% and 0% growth. These data were used to calculate the IC50 values (an indication of potency), which is defined as the concentration of drug required to reduce biofilm growth by 50%, by fitting the normalized results to the variable slope Hill equation (an equation that determines the nonlinear drug dose-response relationship) using Prism 8 (GraphPad Software Inc., San Diego, CA, USA).
Cytotoxicity Assay Test
Human hepatocellular carcinoma (HepG2) cells (ATCC#HB-8065) were used to determine the cytotoxicity of selected compounds. Briefly, HepG2 cells were maintained in minimum essential medium (MEM, Gibco) enriched with 10 % fetal bovine serum (FBS), 1 mM sodium pyruvate (Gibco, Carlsbad, CA, USA), 1× MEM amino acid solution (Sigma-Aldrich), 100 IU mL penicillin, and 100 mg streptomycin (Cellgro Inc., Herndon, VA, USA). We detached the monolayer adhered cells after confluency was attained and dispersed them using 1× Trypsin/EDTA solution (Gibco, Carlsbad, CA, USA). HepG2 cells were enumerated, and 5 × 105 cells/mL were added in a total volume of 100 μL to each well of the 96-well microtiter plates containing 100 μL of serial two-fold dilutions of the compounds to be tested. The 96 well plates were incubated for 24 h at 37 °C with CO2, and the cytotoxicity was determined using the XTT cell viability assay. From these data, we derived the CC50 value, defined as the concentration of compound leading to 50 % inhibition. All experiments were performed in duplicate and were repeated two times.
Evaluation of the in vitro activity of everolimus against C. albicans SC5314 and different strains of C. auris under different growing conditions.
For follow-up experiments, pharmaceutical grade everolimus was commercially purchased from Sigma-Aldrich, after having been identified as the leading “repositionable” compound from the initial screens and confirmatory dose-response assays. For this set of experiments, we tested the activity of everolimus under three different modalities: inhibition of biofilm formation, inhibition of planktonic growth, and activity against preformed biofilms. All three different modalities used microdilution techniques in 96-well microtiter plates. The biofilm inhibitory assay was performed as described above. Antifungal susceptibility tests under planktonic conditions were performed following the Clinical and Laboratory Standards Institute (CLSI) document M27-A3 for antifungal susceptibility testing of yeasts (21), with minor modifications. Briefly, the fungal inoculum is prepared at 1 × 103 cells/ml of yeast cells and added to the wells of 96-well round bottom microtiter plates containing serial dilutions of everolimus. The plates were then incubated at 37 °C and read visually (for >50% inhibition) at 24 and 48 h. At the end of the incubation, the cells in the wells were homogenized and the absorbance determined spectrophotometrically with a microtiter plate reader to provide a more quantitative measure of inhibition. The activity against preformed biofilms was determined as previously described by our group (20). Briefly, biofilms of the different strains were formed by adding 1 × 106 cells/mL to wells of a 96-well microtiter and incubated for 24 h to allow for biofilm formation. Once mature biofilms were formed, they were washed, and serial-dilutions of everolimus were added. The plates were read using a microtiter plate reader, with both biofilm assays being read colorimetrically using the XTT-reduction assay and the planktonic assay being read as absorbance to determine the turbidity of the wells after homogenization. As described for the dose-response assay, the readings were normalized and then the IC50 values determined using Prism.
Drug combination studies with everolimus and clinically-used antifungals
We assessed the efficacy of combinations of everolimus together with fluconazole, caspofungin, micafungin, and amphotericin B against both C. albicans and C. auris by checkerboard assays, basically using a method similar to the CLSI methodology described above. For two-dimensional microplate preparation, a series of two-fold serial dilutions of the clinically-used antifungal were performed across the rows of a microtiter plate, whereas two-fold serial dilutions of everolimus were prepared across the columns. Appropriate positive (no drug) and negative (no organism) controls were also included. To assess whether each combination of drugs resulted in synergistic, indifferent, or antagonistic effects, the Fractional Inhibitory Concentration Index was used. The FICI is defined as: MICAB/MICA + MICBA/ MICB. This calculation takes the minimum inhibitory concentration (MIC) of each drug when mixed with the other and divides it by the MIC of the drug by itself. FICI values of ≤ 0.5 indicate synergy, indifference is defined as > 0.5 and ≤ 4.0, and antagonism is defined as > 4.0 (22).
Determination of the antifungal spectrum of activity of the leading repositionable candidates against a panel of medically-important fungi
Antifungal susceptibility testing was performed using standard CLSI techniques, to examine the activity of selected leading repositionable compounds against a panel of medically important fungi. All clinical fungal isolates tested form part of the collection available in the Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio. In vitro antifungal susceptibility testing was performed by broth microdilution for yeast and filamentous fungi according to the CLSI M27 and M38-A2 respectively (21, 23). Stock solutions of everolimus, MMV1633966 and MMV1593537 were prepared by dissolving the powders in DMSO. Further dilutions were prepared in RPMI medium. The concentration range used were 0.06 - 32 μg/ml for everolimus, and 0.03- 16 μg/ml for MMV1633966 and MMV1593537. Fluconazole (for yeasts), and posaconazole and voriconazole (for molds) were used for comparison purposes. The minimum inhibitory concentrations (MIC) were read visually at 50% and 100% of growth after 24 and 48 h of incubation for yeasts and filamentous fungi respectively.
Determination of the in vivo antifungal activity of everolimus in murine models of hematogenously disseminated C. albicans and C. auris infections
All animal experiments were performed following NIH guidelines and in accordance with institutional regulations (IACUC) in AAALAC-certified facilities. Animals were allowed a 1-week acclimatization period before experiments were started. Mice were randomly distributed in different cages and assigned to the different treatment arms, and persons monitoring the animals were not blinded as to the identity of different groups.
The evaluation of the in vivo efficacy of everolimus in the hematogenously disseminated model of C. albicans infections was performed following methodologies previously described by our group(24, 25). Briefly, C. albicans SC5314 strain was grown overnight in YPD broth at 25°C. Cells were harvested by centrifugation and washed three times with sterile saline. Cells were counted using a hemocytometer, and appropriate dilutions of the cells were made in sterile saline for injection. Confirmation of the number and viability of cells present in the infecting inoculum was performed via plate counts. A final volume of 200 μl containing 3.5 × 105 yeast cells was injected via the lateral tail vein into 6- to 8-week-old female BALB/c mice. Groups of mice (n = 8) were treated intraperitoneally with a dose of 2.5 mg/kg of everolimus diluted in 2% DMSO (prepared in saline for injection), starting 2 days prior to infection, with treatment continuing once daily for the observational period. A control group was on the same schedule but received vehicle-only injections.
The C. auris infection model has been previously described (26, 27). Briefly, male ICR mice were rendered neutropenic with a single dose of pharmaceutical-grade 5-fluorouracil (5 mg/mouse) administered 24 h prior to inoculation. To prevent bacterial superinfection and deaths in the immunosuppressed mice, mice received antibacterial prophylaxis consisting of enrofloxacin at 50 ppm in their drinking water beginning 1 day prior to infection. On the day of inoculation (day 0), a clinical isolate of C. auris (DI 17-46) was used to infect mice via the lateral tail vein (0.2 ml of a yeast cell inoculum of 1 × 107 cells/mouse). Treatment groups consisted of vehicle control (2% DMSO) and everolimus at 2.5 mg/kg. Another group of animals was treated with caspofungin at 10 mg/kg as a positive control (not shown). Drugs were administered once daily by intraperitoneal injection, starting 2 days prior to infection. Ten mice were included in each study arm.
Throughout the studies, for both the C. albicans and C. auris models, mice were observed multiple times per day to prevent and minimize unnecessary pain and distress that may have occurred with infection. Any animal that appeared moribund was humanely euthanized. To determine the survival curves, days on which the mice died were recorded; for euthanized mice death was recorded as occurring the next day. Survival was plotted by Kaplan-Meier analysis and differences between groups (treated versus untreated) were analyzed using the log-rank test. Analyses were performed using Prism (GraphPad Software, Inc.).
RESULTS
Screening the Pandemic Response Box for inhibitors of C. albicans SC5314 and C. auris 0390 biofilm formation and validation of initial hits by dose-response assays.
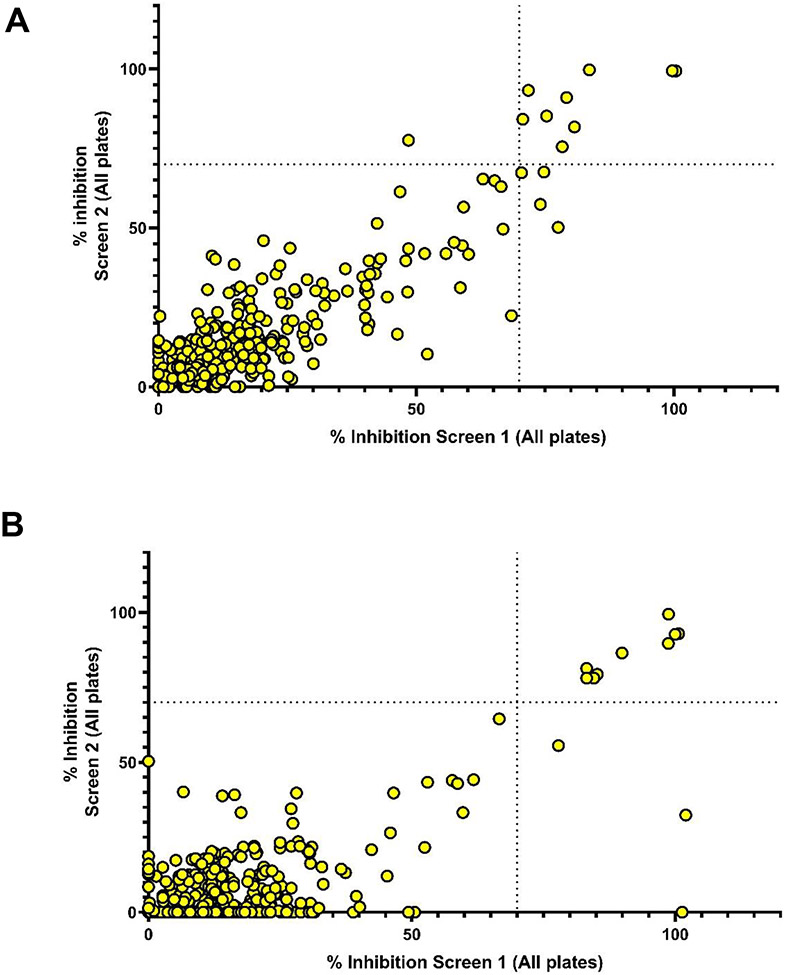
We screened the Pandemic Response Box in search of inhibitors of Candida biofilm formation. Two parallel screenings were designed against C. albicans SC5314 and C. auris 0390, each performed in duplicate. The primary screen was conducted at a concentration of 10 μM for each of the 400 compounds in the Pandemic Response Box. Initial hits were identified as compounds that inhibit biofilm growth by 70% or higher in one of the two duplicate assays. According to this criterion, a total of 14 hits were identified in the screening against C. albicans (Figure 1A, Table 1), whereas 12 initial hit compounds were found to inhibit C. auris biofilm growth (Figure 1B, Table 2). Several compounds were effective against both C. albicans and C. auris (Figure S1), resulting in the identification of a total of 20 unique hit compounds, and an overall hit rate of 5%. The compounds that inhibited biofilm formation are spread among the antifungal, antibacterial and antiviral compound sets in the Pandemic Response Box, although not surprisingly most hits fall under the antifungal category (Figure 1, Tables 1 and 2, and Figure S1).
Figure 1.
Graphical representation of data from primary screenings of the Pandemic Response Box in search for compounds with inhibitory activity against C. albicans SC5314 (Panel A) and Candida auris 0390 (Panel B) biofilm formation. For each strain, screening was performed in duplicate plates and the results, expressed as percent inhibition, are plotted on each of the two axes.
Table 1.
Identity, degree of inhibition during primary screens, and IC50 values from dose-response experiments of hit compounds in the Pandemic Response Box with inhibitory activity against C. albicans SC5314 biofilm formation.
| MMV ID |
% Inhibition Screen 1 |
% Inhibition Screen 2 |
IC50 (μM) |
Identity | Classification |
|---|---|---|---|---|---|
| 002731 | 80.68 | 81.76 | 5.475 | Ciclopirox | Antifungal |
| 1782108 | 75.29 | 85.24 | 6.275 | 4-butyl-N-(5-methyl-1,2-oxazol-3-yl)benzenesulfonamide | Antifungal |
| 1634360 | 74.77 | 67.62 | 9.050 | N-epoxymethyl-1,8-naphthalimide | Antifungal |
| 1634493 | 70.42 | 67.37 | 9.491 | Abafungin | Antifungal |
| 000043 | 74.07 | 57.44 | 18.17 | Tafenoquine | Antibacterial |
| 396785 | 100 | 99.38 | 3.295 | Alexidine | Antifungal |
| 687273 | 79.16 | 91.03 | 4.309 | SQ109 | Antibacterial |
| 1633966 | 83.57 | 99.74 | 3.101 | 1-[2-[(2,10-dichloro-5,7-dihydroindolo[2,3-b]carbazol-6-yl)oxy]ethyl]tetrazol-5-amine | Antibacterial |
| 1593537 | 99.66 | 99.47 | 5.016 | 3-[4,5-bis(4-fluorophenyl)-1H-imidazol-2-yl]-5-bromo-1H-indole | Antibacterial |
| 1634399 | 78.34 | 75.54 | 12.56 | 4-methyl-8-phenoxy-1-(2-phenylethyl)-2,3-dihydropyrrolo[3,2-c]quinoline | Antibacterial |
| 639951 | 48.50 | 77.60 | 7.403 | Everolimus | Antiviral |
| 002505 | 70.65 | 84.19 | 15.98 | Metitepine | Antiviral |
| 1006203 | 77.54 | 50.21 | 16.23 | 1,1-dioxide 1-Thioflavone | Antiviral |
| 003069 | 71.74 | 93.28 | 13.12 | Tomatidine | Antibacterial |
Table 2.
Identity, degree of inhibition during primary screens, and IC50 values from dose-response experiments of hit compounds in the Pandemic Response Box with inhibitory activity against C. auris 0390 biofilm formation.
| MMV ID |
% Inhibition Screen 1 |
% Inhibition Screen 2 |
IC50 (mM) |
Identity | Classification |
|---|---|---|---|---|---|
| 002731 | 77.83 | 55.59 | 19.43 | Ciclopirox | Antifungal |
| 1578560 | 100 | - | 12.02 | OSU-03012 | Antifungal |
| 1634494 | 89.92 | 86.48 | 2.287 | Isavuconazonium | Antifungal |
| 1634386 | 85.22 | 79.40 | 4.806 | (2R)-2-(2,4-difluorophenyl)-1,1-difluoro-3-(tetrazol-1-yl)-1-[5-[4-(2,2,2-trifluoroethoxy)phenyl]pyridin-2-yl]propan-2-ol | Antifungal |
| 637533 | 83.21 | 81.34 | 1.007 | Ketoconazole | Antifungal |
| 637528 | 84.55 | 78.12 | 3.235 | Itraconazole | Antifungal |
| 1634362 | 83.21 | 78.12 | 1.079 | Ravuconazole | Antifungal |
| 000043 | 100 | 32.42 | 11.46 | Tafenoquine | Antibacterial |
| 396785 | 100 | 92.92 | 3.411 | Alexidine | Antifungal |
| 1633966 | 100 | 92.72 | 2.923 | 1-[2-[(2,10-dichloro-5,7-dihydroindolo[2,3-b]carbazol-6-yl)oxy]ethyl]tetrazol-5-amine | Antibacterial |
| 1593537 | 98.70 | 89.69 | 3.114 | 3-[4,5-bis(4-fluorophenyl)-1H-imidazol-2-yl]-5-bromo-1H-indole | Antibacterial |
| 639951 | 98.76 | 99.41 | 0.6566 | Everolimus | Antiviral |
Next, we carried out dose-response assays to confirm the biofilm inhibitory activity of all the hit compounds and at the same time establish their potency. These assays were able to validate the inhibitory activity of all initial hits from the initial screenings against C. albicans and C. auris. From these assays, the corresponding IC50 values for each hit compound, a measure of their potency, were also calculated (Tables 1 and 2).
Since the main emphasis of these studies was on repurposing compounds as antimycotics, we focused on those hit compounds with an original classification as antibacterials or antivirals (but not antifungals). Of these and based on their activity against both Candida spp. tested, their efficacy (maximum levels of inhibition achieved) and potency (IC50 values), we selected compounds MMV1633966 and MMV1593537 as well as everolimus as our main repositionable leading compounds for further characterization of their antifungal activities. Their structure and chemical information can be found in Table S1. We note that everolimus seemed particularly effective and potent at inhibiting C. auris biofilm formation.
Further in vitro characterization of the antifungal activity of MMV1633966 and MMV1593537
Both MMV1633966 and MMV1593537 compounds showed potent biofilm-inhibitory activity against both C. albicans and C. auris, leading to almost complete inhibition of biofilm formation in primary screenings and dose-response experiments, and with calculated IC50 values in the order of 3 to 5 μM. Besides their classification as antibacterials, very little information is available for these compounds, as these chemotypes are currently in discovery and early development. To our knowledge, their antifungal activity has not yet been described. Thus, we performed a limited number of follow-up experiments with these compounds.
To preliminarily examine their antifungal spectrum of activity we performed antifungal susceptibility testing, following CLSI methodologies, of compounds MMV1633966 and MMV1593537 against a relatively small, but representative, panel of yeast and filamentous fungi, in comparison to currently available antifungal agents (fluconazole, voriconazole and posaconazole). As seen in Table S2, both compounds display activity against yeasts, including different Candida spp. and Cryptococcus neoformans, with low MIC values of 1 and 2 μg/ml detected for MMV1633966 and MMV1593537, respectively, using a 50% inhibition endpoint. Whereas MIC values for both compounds against Candida spp. were similar when read at 100% inhibition, using this endpoint MMV1593537 seemed to have more potent activity against C. neoformans than MMV1633966 (MIC 2 versus >16 μg/ml for MMV1593537 and MMV1633966 respectively). However, both compounds lacked activity against filamentous fungi, with detected MIC values of > 16 μg/ml (the highest concentration used in these experiments) at both the 50% and the 100% endpoints for all representative clinical isolates of molds tested.
To further ascertain their potential for clinical development, including for the treatment of fungal infections, in parallel experiments we examined the level of cellular cytotoxicity of these two compounds, for which we used an established assay using liver hepatocellular cells as an alternative to animal testing (Figure S2). From these experiments, the calculated CC50 values were 9.868 and 6.564 μM for compounds MMV1633966 and MMV1593537 respectively. These concentrations are similar to those at which they display antifungal activity, thereby indicating a very narrow therapeutic index, and potentially some severe limitations for their eventual development as antifungals.
Further in vitro characterization of the antifungal activity of everolimus
From results of the initial screenings and dose-response experiments everolimus, an analog of rapamycin (“rapalog”) which was categorized as an antiviral in the Pandemic Response Box associated database, emerged as the main leading repositionable compound with potent anti-biofilm inhibitory activity against both C. albicans and C. auris. Thus, we were interested in further assessing its antifungal activity and potential to be repositioned as an antimycotic for the treatment of biofilm-associated candidiasis and possibly other fungal infections.
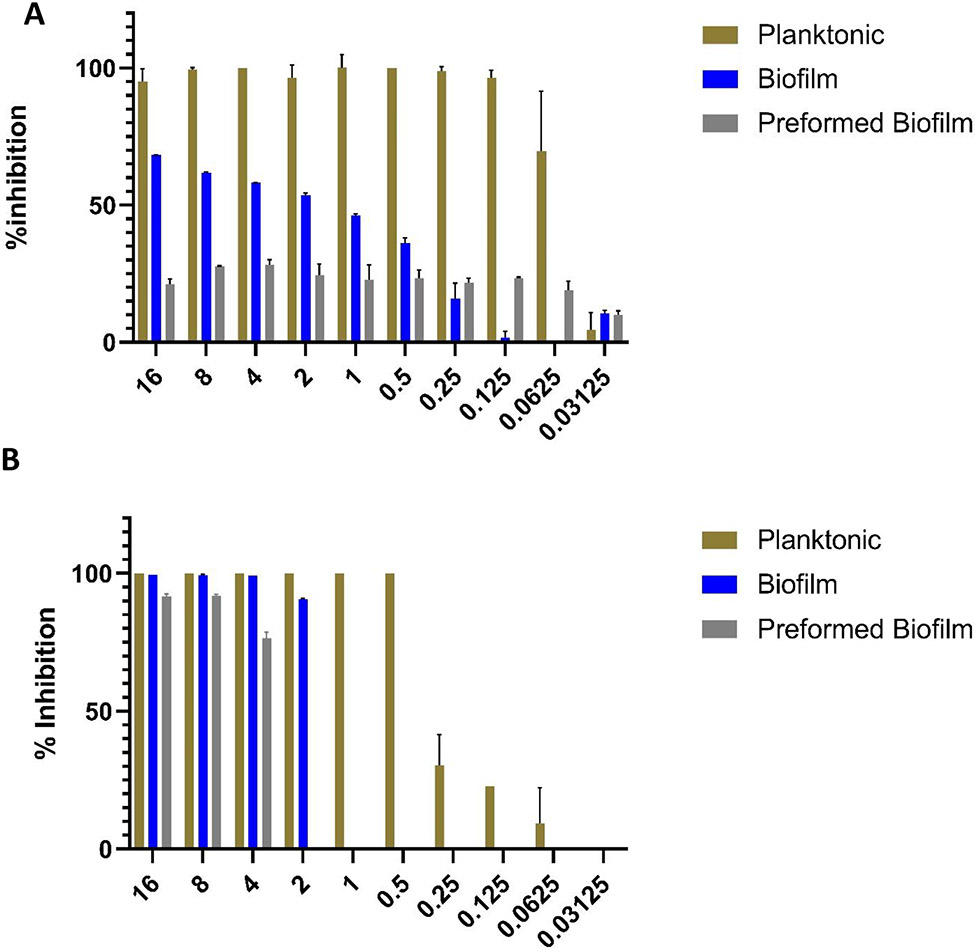
In a first series of experiments, we determined the inhibitory effect of commercially purchased, pharmaceutical grade everolimus against the same strains of C. albicans SC314 and C. auris 0390, under three different growth conditions and treatment modalities: inhibition of biofilm formation, inhibition of planktonic growth, and activity against preformed biofilms. As seen in Figure 2, treatment with everolimus completely or almost completely abolished planktonic growth at concentrations as low as 0.125 μg/ml (for C. albicans) and 0.5 μg/ml (for C. auris). In agreement with results from the primary screening and confirmatory dose-response measurements, everolimus showed increased biofilm inhibitory activity against C. auris as compared to C. albicans; although as expected the concentrations required to inhibit biofilm formation were slightly elevated as compared to those required to inhibit planktonic growth. Lastly, as anticipated, fully formed biofilms of both species displayed the lowest levels of susceptibility against everolimus as compared to the other two growth modalities. In the case of C. albicans, preformed biofilms inhibition levels were relatively low, even at the highest concentrations of everolimus tested. In contrast, almost complete inhibition of C. auris preformed biofilms was observed at everolimus concentrations of 4 - 8 μg/ml.
Figure 2.
Activity of everolimus against (A) C. albicans SC5314 and (B) C. auris 0390, under the three different growing conditions: inhibition of planktonic growth, biofilm inhibition, and preformed biofilm.
Next, we determined the activity of everolimus against all other nine C. auris strains in the CDC FDA AR Bank panel, also under the same three different treatment modalities (planktonic, inhibition of biofilm and activity against preformed biofilms). Results of this set of experiments are shown in Figures S3, S4 and S5. Overall, the susceptibility patterns of all clinical isolates tested were similar to those observed for C. auris strain 0390, with the most variability observed in the treatment of preformed biofilms.
We examined the activity of everolimus in combination with four clinically used antifungals, fluconazole, caspofungin, micafungin, and amphotericin B, against both C. albicans and C. auris, mostly to evaluate its potential as a potentiator of current therapeutic options. To assess the interactions of combinations of drugs, we calculated the fractional inhibitory concentration index (FICI) as described above in Materials and Methods. Results of this set of experiments indicated that all combinations between everolimus and all four antifungal agents, and against both Candida species, resulted in “indifference”, with FICI values ranging from 1 - 3.
We extended our observations on the in vitro antifungal activity of everolimus by testing its activity against an extended panel of medically important fungi. Results of these experiments for yeasts and filamentous fungi respectively are shown in Tables 3 and 4. Regarding yeasts, results confirmed the antifungal activity of everolimus against all Candida spp. tested; however, Cryptococcus neoformans displayed decreased susceptibility, particularly when the 100% reading end point was used. Likewise, when the 100% endpoint was used, all species of filamentous fungi tested showed intrinsic resistance to everolimus (MIC > 32 μg/ml, the highest concentration tested; Table 4). However, we note that using the less demanding 50% reading endpoint we detected some limited and highly variable levels of antifungal activity against different clinical isolates from the Mucorales, Scedosporium spp. and Fusarium spp.
Table 3.
MIC values of everolimus against multiple clinical isolates belonging to different species of yeast, in comparison to fluconazole. Values are in μg/ml.
| Species | Isolate | Everolimus | Fluconazole | |
|---|---|---|---|---|
| 50% | 100% | 50% | ||
| C. parapsilosis QC | ATCC22019 | 0.5 | 1 | 1 |
| C. krusei QC | ATCC6258 | 1 | >32 | 16 |
| Candida albicans | ATCC 90028 | 0.5 | 1 | ≤0.125 |
| SC5314 | 0.5 | 1 | ≤0.125 | |
| Ca-1 | 0.25 | 1 | 0.5 | |
| Candida auris | Cau-1 | 1 | 1 | >64 |
| Cau-2 | 0.5 | 1 | >64 | |
| Cau-3 | 1 | 1 | 2 | |
| Candida glabrata | Cg-1 | 0.5 | 1 | 64 |
| Cg-2 | 0.5 | 1 | 4 | |
| Cg-3 | 0.5 | 1 | 0.5 | |
| Candida parapsilosis | Cp-1 | 0.5 | 1 | 0.5 |
| Cp-2 | 0.5 | 1 | 0.25 | |
| Cp-3 | 0.5 | 1 | 0.5 | |
| Cryptococcus neoformans | Crn-1 | >32 | >32 | 64 |
| Crn-2 | 1 | >32 | 4 | |
| H99 | 1 | >32 | 16 | |
Table 4.
MIC values of everolimus against multiple clinical isolates belonging to different species of filamentous fungi, in comparison to voriconazole and/or posaconazole. Values are in μg/ml.
| Species | Isolate | Everolimus | Voriconazole | Posaconazole | |
|---|---|---|---|---|---|
| 50% | 100% | 100% | 100% | ||
| P. variotii QC | MYA-3630 | >32 | >32 | 0.125 | ≤0.03 |
| Rhizopus arrhizus | Rh-1 | ≤0.06 | >32 | --- | 1 |
| Rh-2 | >32 | >32 | --- | 0.5 | |
| Rh-3 | >32 | >32 | --- | 0.5 | |
| Mucor spp. | Mc-1 | ≤0.06 | >32 | --- | 2 |
| Mc-2 | 16 | >32 | --- | 1 | |
| Mc-3 | ≤0.06 | >32 | --- | 2 | |
| Aspergillus flavus | ATCC204304 | >32 | >32 | 1 | --- |
| Af-1 | >32 | >32 | 1 | --- | |
| Af-2 | >32 | >32 | 1 | --- | |
| Aspergillus fumigatus | AF293 | >32 | >32 | 0.5 | --- |
| Af-1 | >32 | >32 | >16 | --- | |
| Af-2 | >32 | >32 | 4 | --- | |
| Fusarium spp. | Fs-1 | 4 | >32 | >16 | --- |
| Fs-2 | 4 | >32 | >16 | --- | |
| Fs-3 | 2 | >32 | >16 | --- | |
| Scedosporium spp. | Sc-1 | >32 | >32 | >16 | --- |
| Sc-2 | ≤0.06 | >32 | 2 | --- | |
| Sc-3 | 0.25 | >32 | 1 | --- | |
| Altenaria spp. | Al-1 | 32 | >32 | 1 | --- |
| Curvularia spp. | Cu-1 | >32 | >32 | 0.5 | --- |
| Exserohilum spp. | Ex-1 | 32 | >32 | 2 | --- |
In vivo efficacy of everolimus in the murine models of hematogenously disseminated C. albicans and C. auris infections.
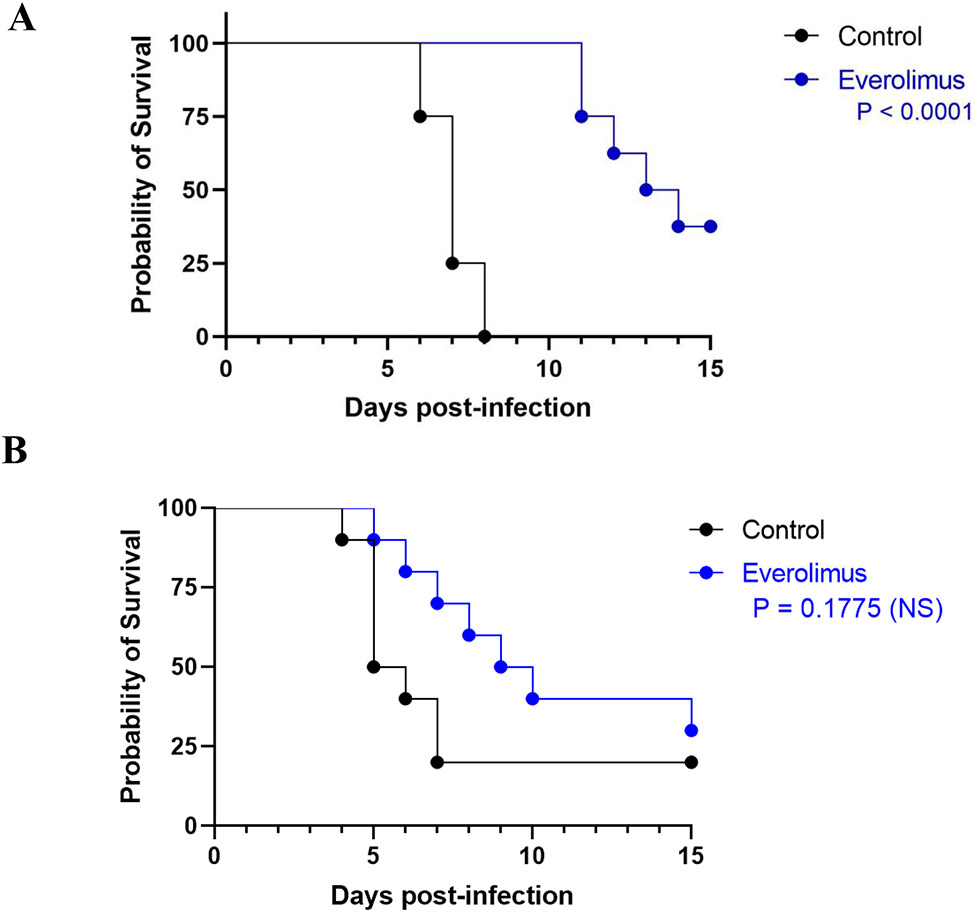
We proceeded to examine the efficacy of everolimus in the clinically relevant murine models of hematogenously disseminated candidiasis caused by C. albicans and C. auris (Figure 3). Treatment with everolimus increased the survival of animals infected with C. albicans compared the untreated control group, and these differences were statistically significant (P < 0.0001). Everolimus-treated mice infected with C. auris also showed an increase in median survival as compared to animals in the control/untreated group (9.5 versus 5.5 days); however, the overall differences in the resulting survival curves did not achieve statistical significance (P = 0.1775).
Figure 3.
In vivo activity of everolimus in the murine models of hematogenously disseminated invasive infections by C. albicans (A) and C. auris (B).
DISCUSSION
Candidiasis remains the most common fungal infection in hospitalized patients and other clinical settings, affecting an expanding population of at-risk patients (1, 2, 5, 6). C. albicans is the main cause of candidiasis, but in the last decade infections caused by the emergent species C. auris represent a serious concern (8). In fact, the Centers for Disease Control and Prevention in the US (CDC) has identified C. auris as an “urgent threat”, and most recently the World Health Organization (WHO) in its first ever global fungal priority pathogen list named both C. albicans and C. auris as “critical priority” (28, 29, 30). Both C. albicans and C. auris are fully capable of forming biofilms, which greatly contributes to their pathogenic potential (11, 15, 31). Cells within these biofilms show decreased susceptibility (in the case of C. albicans) or intrinsic resistance/tolerance (in the case of C. auris) against all clinically-used antifungal agents for the treatment of candidiasis, currently the azoles, amphotericin B and the echinocandins (13, 15, 32); and biofilm resistance represents a major contributor factor to poor patient outcomes (14, 33). These facts underline the need for the identification of novel antifungals with anti-Candida biofilm activity (31).
The development of new drugs, including antifungals, is a costly and time-consuming process, with high attrition rates (17). Repurposing existing drugs as antifungals represents a potentially much faster and economical alternative, which has been gaining traction in the last decade (17). Most recently, these repurposing efforts have been greatly facilitated by the availability of chemical libraries of existing drugs, and others at different stages of clinical development (34). Some of these are provided as open-source compound collections by a variety of organizations. One such example is the Pandemic Response Box, launched in 2019 by MMV, which contains 400 diverse drug-like molecules consisting mostly of chemotypes with in vitro activity against viral, bacterial or fungal microorganisms (18). In the antifungal space, this collection has already been screened for molecules with activity against eumycetoma, Scedosporium and Lomentospora species, and the Mucorales (35, 36, 37). Here we have screened the Pandemic Response Box® to identify compounds with inhibitory activity against C. albicans and C. auris biofilm formation. The screening technique is based on the 96-well microtiter plate model of Candida biofilm formation and susceptibility testing originally developed by our group, which is easy, rapid, cost-effective and highly reproducible (19). We have previously successfully used these techniques for screening other repurposing chemical libraries against C. albicans and C. auris, also with emphasis on the identification of biofilm inhibitors (34, 38, 39, 40).
Besides several known antifungals, our initial screens resulted in the identification of several compounds not previously classified as antifungals with inhibitory activity against Candida biofilms, which were interesting from a repurposing point of view. These compounds were subsequently validated in dose-response confirmatory experiments (see Tables 1 and 2). From these results, we identified compounds MMV1633966 and MMV1593537 and everolimus to be among the leading repositionable compounds, and we selected these compounds for a more in-depth characterization of their in vitro antifungal activity in follow-up experiments. In the Pandemic Response Box database, both MMV1633966 and MMV1593537 compounds are classified as antibacterials, although little additional information overall is available for both compounds; and to our knowledge, their antifungal activity has not yet been described. These compounds showed potent biofilm-inhibitory activity in vitro against both C. albicans and C. auris, with IC50 values in the low micromolar range, and are highly effective, almost completely abolishing biofilm formation in C. albicans and resulting in approximately 90% maximum inhibition of C. auris biofilm formation. In follow-up in vitro experiments, both MMV1633966 and MMV1593537 displayed a somewhat narrow spectrum of antifungal activity, with no activity against filamentous fungi. However, determination of in vitro cytotoxicity levels reveal that these compounds may be toxic at similar concentrations to those at which they exert their antifungal activity, which may compromise their further development an antifungal. Clearly, their inclusion in this collection indicates that these molecules have potential as anti-infectives, but that they have not yet been completely optimized, and as such these molecules would still need to undergo preclinical optimization (and eventually human testing), which makes them less attractive from the repurposing point of view given the pressing need for short-term solutions.
Everolimus is a second generation “rapalog” (analog of rapamycin) and was classified in the Pandemic Response Box database as an antiviral. Interestingly, rapamycin was originally discovered because of its antifungal activity (41, 42), but then found to exert immunosuppressive and anti-proliferative properties (43). These effects are due to its inhibition of the mammalian target of rapamycin (mTOR), which is involved in cancer. Everolimus has been FDA-approved for the treatment of different types of cancer and is also used in solid organ transplantation (44). Everolimus was specifically designed structurally to have improved pharmacokinetic properties over sirolimus. More specifically, everolimus is the 40-O-(2-hydroxyethyl) derivative of sirolimus, with this modification leading to improved pharmacological properties as well as differences in drug-target protein interactions, which altogether result in overall higher potency as compared to sirolimus (44). In the field of antifungal research, another rapalog, FK 506 (tacrolimus), has been described to potentiate the antifungal activity of azoles and overcome resistance in vitro (45), but its usefulness is limited due to its immunosuppressive properties. Everolimus displays much more potent antifungal activity than tacrolimus (over 50 – 100 times more potent, results not shown) and lower immunosuppressive effects (46), making it a potentially viable candidate for being repurposed as an antifungal. In our primary screen and subsequent dose-response assays, everolimus showed potent biofilm-inhibitory activity, particularly against C. auris, with IC50 values approaching the picomolar range (Tables 1 and 2). Of note, these concentrations are achievable in patients (44). A series of follow-up in vitro experiments confirmed the activity of everolimus under two different growing modalities (planktonic growth and inhibition of biofilm formation) against C. albicans and a panel of C. auris clinical isolates; but, as expected, preformed biofilms formed by these strains were mostly resistant against this drug. Our experiments to examine the antifungal spectrum of activity of everolimus indicated that, among yeasts, this drug is active against all Candida species tested but not against C. neoformans (Table 3). Likewise, everolimus did not exhibit significant levels of antifungal activity against filamentous fungi (Table 4). Finally, treatment with everolimus increased survival of mice infected with C. albicans, but no statistically significant survival differences were observed in animals infected with C. auris, which we attribute in part to the fact that the C. auris model uses immunosuppressed mice.
CONCLUSION
The screening of the Pandemic Response Box and subsequent characterization of initial hit compounds resulted in the identification of several leading repositionable drugs with inhibitory activity against C. albicans and C. auris biofilm formation, for which there is a dire need to develop effective therapies. From these results and subsequent in vitro studies, the rapalog everolimus emerged as the most attractive leading candidate, with very potent antifungal activity also against other yeast species, but not filamentous fungi. Although our results on the activity of everolimus may represent an excellent starting point towards its repurposing for the treatment of fungal infections, further preclinical studies are required on both its in vitro and in vivo antifungal properties, followed by subsequent clinical studies.
Supplementary Material
Figure S1. (A) Initial hits were classified into three classes: antifungals, antibacterials, and antivirals. (B) Venn diagram showing initial hits active against either or both C. albicans SC5314 and C. auris 0390.
Table S1. Structure and chemical information of the leading repositionable compounds everolimus, MMV1633966 and MMV1593537.
Table S2. MIC values of MMV1633966 and MMV1593537 after exposure to planktonic forms of medically important yeast and filamentous fungi. FLZ, fluconazole; VRZ, voriconazole; PSZ, Posaconazole.
Figure S2. Cytotoxicities of the MMV1633966 (MMV1) and MMV1593537 (MMV2) compounds. Cytotoxicity was measured using a human hepatocellular carcinoma (HepG2) cell line. For each compound, the concentration that reduced cell viability by 50% (CC50) was calculated based on these assays.
Figure S3. Activity of everolimus against different clinical isolates of C. auris under planktonic growth conditions.
Figure S4. Inhibition of biofilm formation by different clinical isolates of C. auris by everolimus.
Figure S5. Activity of everolimus against mature biofilms formed by different clinical isolates of C. auris.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R33AI140823 and R21AI156100 from the National Institute of Allergy and Infectious Diseases. Additional support was provided by the Margaret Batts Tobin Foundation, San Antonio, TX. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript, and the content is solely the responsibility of the authors. We thank the Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention (CDC), Clinical and Environmental Microbiology Branch for providing the Candida auris panel (recipient JL-R.). The authors are indebted to Medicines for Malaria Venture (MMV, Switzerland) for providing the Pandemic Response Box library.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Thomas-Ruddel DO, Schlattmann P, Pletz M, Kurzai O, Bloos F. Risk Factors for Invasive Candida Infection in Critically Ill Patients: A Systematic Review and Meta-analysis. Chest. 2022;161(2):345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsay SV, Mu Y, Williams S, Epson E, Nadle J, Bamberg WM, et al. Burden of Candidemia in the United States, 2017. Clin Infect Dis. 2020;71(9):e449–e53. [DOI] [PubMed] [Google Scholar]
- 3.Magill SS, O'Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, et al. Changes in Prevalence of Health Care-Associated Infections in U.S. Hospitals. N Engl J Med. 2018;379(18):1732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37(9):1172–7. [DOI] [PubMed] [Google Scholar]
- 5.Koehler P, Stecher M, Cornely OA, Koehler D, Vehreschild M, Bohlius J, et al. Morbidity and mortality of candidaemia in Europe: an epidemiologic meta-analysis. Clin Microbiol Infect. 2019;25(10):1200–12. [DOI] [PubMed] [Google Scholar]
- 6.Quindos G, Marcos-Arias C, San-Millan R, Mateo E, Eraso E. The continuous changes in the aetiology and epidemiology of invasive candidiasis: from familiar Candida albicans to multiresistant Candida auris. Int Microbiol. 2018;21(3):107–19. [DOI] [PubMed] [Google Scholar]
- 7.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–4. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarti A, Singh S. Multidrug-resistant Candida auris: an epidemiological review. Expert Rev Anti Infect Ther. 2020;18(6):551–62. [DOI] [PubMed] [Google Scholar]
- 9.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, et al. Candida auris: a Review of the Literature. Clin Microbiol Rev. 2018;31(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kean R, Brown J, Gulmez D, Ware A, Ramage G. Candida auris: A Decade of Understanding of an Enigmatic Pathogenic Yeast. J Fungi (Basel). 2020;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramage G, Martinez JP, Lopez-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006;6(7):979–86. [DOI] [PubMed] [Google Scholar]
- 12.Desai JV, Mitchell AP. Candida albicans Biofilm Development and Its Genetic Control. Microbiol Spectr. 2015;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierce CG, Srinivasan A, Uppuluri P, Ramasubramanian AK, Lopez-Ribot JL. Antifungal therapy with an emphasis on biofilms. Curr Opin Pharmacol. 2013;13(5):726–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tumbarello M, Posteraro B, Trecarichi EM, Fiori B, Rossi M, Porta R, et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J Clin Microbiol. 2007;45(6):1843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherry L, Ramage G, Kean R, Borman A, Johnson EM, Richardson MD, et al. Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida auris. Emerg Infect Dis. 2017;23(2):328–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Moraes DC. Current scenario of the search for new antifungal agents to treat Candida auris infections: An integrative review. J Mycol Med. 2022;32(1):101232. [DOI] [PubMed] [Google Scholar]
- 17.Farha MA, Brown ED. Drug repurposing for antimicrobial discovery. Nat Microbiol. 2019;4(4):565–77. [DOI] [PubMed] [Google Scholar]
- 18.Samby K, Besson D, Dutta A, Patra B, Doy A, Glossop P, et al. The Pandemic Response Box horizontal line Accelerating Drug Discovery Efforts after Disease Outbreaks. ACS Infect Dis. 2022;8(4):713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce CG, Uppuluri P, Tristan AR, Wormley FL Jr., Mowat E, Ramage G, et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc. 2008;3(9):1494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45(9):2475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CLSI. Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—2nd ed. CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. 2008. [Google Scholar]
- 22.Meletiadis J, Verweij PE, TeDorsthorst DT, Meis JF, Mouton JW. Assessing in vitro combinations of antifungal drugs against yeasts and filamentous fungi: comparison of different drug interaction models. Med Mycol. 2005;43(2):133–52. [DOI] [PubMed] [Google Scholar]
- 23.CLSI. Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard—2nd ed. CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. 2008. [Google Scholar]
- 24.Pierce CG, Chaturvedi AK, Lazzell AL, Powell AT, Saville SP, McHardy SF, et al. A Novel Small Molecule Inhibitor of Candida albicans Biofilm Formation, Filamentation and Virulence with Low Potential for the Development of Resistance. NPJ Biofilms Microbiomes. 2015;1:15012-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romo JA, Pierce CG, Chaturvedi AK, Lazzell AL, McHardy SF, Saville SP, et al. Development of Anti-Virulence Approaches for Candidiasis via a Novel Series of Small-Molecule Inhibitors of Candida albicans Filamentation. mBio. 2017;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiederhold NP, Lockhart SR, Najvar LK, Berkow EL, Jaramillo R, Olivo M, et al. The Fungal Cyp51-Specific Inhibitor VT-1598 Demonstrates In Vitro and In Vivo Activity against Candida auris. Antimicrob Agents Chemother. 2019;63(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiederhold NP, Najvar LK, Jaramillo R, Olivo M, Patterson H, Connell A, et al. The Novel Arylamidine T-2307 Demonstrates In Vitro and In Vivo Activity against Candida auris. Antimicrob Agents Chemother. 2020;64(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(CDC) CfDCaP. Antibiotic Resistance Threats Report. 2019. p. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- 29.Fisher MC, Denning DW. The WHO fungal priority pathogens list as a game-changer. Nat Rev Microbiol. 2023;21(4):211–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Organization WH. WHO fungal priority pathogens list to guide research, development and public health action. . 2022. p. https://www.who.int/publications/i/item/9789240060241. [Google Scholar]
- 31.Wall G, Lopez-Ribot JL. Current Antimycotics, New Prospects, and Future Approaches to Antifungal Therapy. Antibiotics (Basel). 2020;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taff HT, Mitchell KF, Edward JA, Andes DR. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013;8(10):1325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall G, Montelongo-Jauregui D, Vidal Bonifacio B, Lopez-Ribot JL, Uppuluri P. Candida albicans biofilm growth and dispersal: contributions to pathogenesis. Curr Opin Microbiol. 2019;52:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wall G, Lopez-Ribot JL. Screening Repurposing Libraries for Identification of Drugs with Novel Antifungal Activity. Antimicrob Agents Chemother. 2020;64(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim W, Nyuykonge B, Eadie K, Konings M, Smeets J, Fahal A, et al. Screening the pandemic response box identified benzimidazole carbamates, Olorofim and ravuconazole as promising drug candidates for the treatment of eumycetoma. PLoS Negl Trop Dis. 2022;16(2):e0010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rollin-Pinheiro R, Xisto M, de Castro-Almeida Y, Rochetti VP, Borba-Santos LP, Fontes YDS, et al. Pandemic Response Box(R) library as a source of antifungal drugs against Scedosporium and Lomentospora species. PLoS One. 2023;18(2):e0280964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xisto M, Rollin-Pinheiro R, de Castro-Almeida Y, Dos Santos-Freitas GMP, Rochetti VP, Borba-Santos LP, et al. Promising Antifungal Molecules against Mucormycosis Agents Identified from Pandemic Response Box((R)): In Vitro and In Silico Analyses. J Fungi (Basel). 2023;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vila T, Lopez-Ribot JL. Screening the Pathogen Box for Identification of Candida albicans Biofilm Inhibitors. Antimicrob Agents Chemother. 2017;61(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wall G, Chaturvedi AK, Wormley FL Jr., Wiederhold NP, Patterson HP, Patterson TF, et al. Screening a Repurposing Library for Inhibitors of Multidrug-Resistant Candida auris Identifies Ebselen as a Repositionable Candidate for Antifungal Drug Development. Antimicrob Agents Chemother. 2018;62(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wall G, Chen E, Hull MV, Lopez-Ribot JL. Screening the CALIBR ReFRAME Library in Search for Inhibitors of Candida auris Biofilm Formation. Front Cell Infect Microbiol. 2020;10:597931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sehgal SN, Baker H, Vezina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J Antibiot (Tokyo). 1975;28(10):727–32. [DOI] [PubMed] [Google Scholar]
- 42.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo). 1975;28(10):721–6. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klawitter J, Nashan B, Christians U. Everolimus and sirolimus in transplantation-related but different. Expert Opin Drug Saf. 2015;14(7):1055–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y, Puumala E, Robbins N, Cowen LE. Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and Beyond. Chem Rev. 2021;121(6):3390–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bravo-San Pedro JM, Senovilla L. Immunostimulatory activity of lifespan-extending agents. Aging (Albany NY). 2013;5(11):793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Initial hits were classified into three classes: antifungals, antibacterials, and antivirals. (B) Venn diagram showing initial hits active against either or both C. albicans SC5314 and C. auris 0390.
Table S1. Structure and chemical information of the leading repositionable compounds everolimus, MMV1633966 and MMV1593537.
Table S2. MIC values of MMV1633966 and MMV1593537 after exposure to planktonic forms of medically important yeast and filamentous fungi. FLZ, fluconazole; VRZ, voriconazole; PSZ, Posaconazole.
Figure S2. Cytotoxicities of the MMV1633966 (MMV1) and MMV1593537 (MMV2) compounds. Cytotoxicity was measured using a human hepatocellular carcinoma (HepG2) cell line. For each compound, the concentration that reduced cell viability by 50% (CC50) was calculated based on these assays.
Figure S3. Activity of everolimus against different clinical isolates of C. auris under planktonic growth conditions.
Figure S4. Inhibition of biofilm formation by different clinical isolates of C. auris by everolimus.
Figure S5. Activity of everolimus against mature biofilms formed by different clinical isolates of C. auris.