Abstract
Background
Genetic variants of the transcription factor SIX1 and its co-factor EYA1 underlie 50% of Branchio-oto-renal syndrome (BOR) cases. BOR is characterized by craniofacial defects, including malformed middle ear ossicles leading to conductive hearing loss. In this work, we expand our knowledge of the Six1 gene regulatory network by using a Six1-null mouse line to assess gene expression profiles of E10.5 mandibular arches, which give rise to the neural crest (NC)-derived middle ear ossicles and lower jaw, via bulk RNA-sequencing.
Results
Our transcriptomic analysis led to the identification of 808 differentially expressed genes that are related to translation, NC cell differentiation, osteogenesis and chondrogenesis including components of the WNT signaling pathway. As WNT signaling is a known contributor to bone development, we demonstrated that SIX1 is required for expression of the WNT antagonist Frzb in the mandibular arch, and determined that SIX1 expression results in repression of WNT signaling.
Conclusion
Our results clarify the mechanisms by which SIX1 regulates the development of NC-derived craniofacial elements that are altered in SIX1-associated disorders. In addition, this work identifies novel genes that could be causative to this birth defect and establishes a link between SIX1 and WNT signaling during patterning of NC cells.
Keywords: SIX1, Transcriptional regulation, branchio-oto-renal syndrome, craniofacial defects, hearing loss
1. Introduction
The pharyngeal arches are transient embryonic structures that are populated by cranial neural crest cells (NCC) early in embryonic development. The first pharyngeal arch, also called the mandibular arch, contributes to the development of most elements of the vertebrate face while the other arches will contribute primarily to the development of elements of the neck.1,2 Dorsal-ventral (D-V) patterning, also referred as proximo-distal patterning, of the mandibular arch is achieved by ectodermal cues such as endothelin-1 (EDN1) and BMP that induce, in the NCC-derived mesenchyme (ectomesenchyme), nested expression of “ventral” (distal) genes such as Dlx5, Dlx3 and Hand2 while repressing “dorsal” (proximal) genes such as Dlx2 and Jag1. Rostral-caudal patterning, also referred as oral-aboral patterning, of this arch, on the other hand, is achieved by signaling cues such as FGF signaling which induces expression of “rostral” (oral) genes such as Lhx7 while repressing “caudal” (aboral) genes such as Gsc.3-7 This nested gene expression gives positional identity to the ectomesenchyme and is required for the differentiation of NCCs into different cell types such as chondrocytes, osteocytes, odontoblasts, connective tissue cells, melanocytes, glia and neurons.8,9 Several studies in mouse and zebrafish showed that NCCs in the ventral domain of the mandibular arch will give rise to lower jaw elements while those in the dorsal domain will give rise to upper jaw elements. The region between these two domains, the intermediate domain, will give rise to the jaw joint in fish. In mammals, on the other hand, the intermediate domain will give rise to middle ear ossicles (i.e., malleus and incus) while the stapes is derived from both the first and second pharyngeal arches.10-13 Thus, defects in patterning of the NCCs that populate the pharyngeal arches can lead to defects in jaw and tooth development, and/or conductive hearing loss.14-17
Branchio-oto-renal syndrome (BOR) is an autosomal dominant disorder characterized by a variable degree of craniofacial defects (e.g., ear tags and pits, branchial fistulas and cysts), hearing loss and, in some cases, kidney defects. Hearing loss in BOR patients can be caused by defects in the inner ear and/or the middle ear ossicles.18,19 Genetic variants of SIX1, a transcription factor implicated in many cellular processes during craniofacial development, and of its co-factor EYA1 underlie 50% of BOR cases. Point mutations in SIX1 will either render this protein transcriptionally inactive (e.g., R110W), block interaction with EYA1 co-activators (e.g., V17E) or create a truncated protein that likely leads to haploinsufficiency (e.g., Q22X).20-28 Interestingly, while this disorder affects men and women with a similar frequency, there is significant phenotypic variability among BOR patients, even between members of the same family that carry the same variant.19 Expression data show that, in the developing embryo face, SIX1 is expressed in NCCs within the mandibular arch, maxillary and frontonasal prominences. In addition, expression is also detected in the pharyngeal pouch endoderm in between arches and otic vesicle. Previous work in mouse demonstrated that Six1−/− (Six1-null) embryos present with severe defects in the jaws, and in the inner and middle ear at embryonic day (E) 18.5. These defects have been linked to the role of SIX1 in D-V patterning of the E10.5 mandibular arch and of the otic vesicle.29-34 It was also shown that Six1+/− (Six1-het) mice, though viable, have conductive hearing deficit which is likely caused by middle ear defects.35 Recently, a clinical study demonstrated that SIX1 function in craniofacial development goes beyond patterning of elements described above as BOR variants were linked to premature cranial suture closure or craniosynostosis.26 Therefore, SIX1 acts at different locations and time points during craniofacial development and is regulating different morphogenetic events such as NCC differentiation, placodal specification, and likely osteogenesis of cranial bones and suture patency maintenance. As around 50% of reported BOR cases are not linked to variants in SIX1 or EYA1 18,19,28, we are identifying novel putative causative genes by characterizing the transcriptome of mandibular arches from Six1-null and Six1-het embryos during the time point (E10.5) when NCCs are being patterned in the mandibular arch to give rise to elements affected in BOR patients.
Herein, we identified 808 differentially expressed genes after loss of Six1. Gene ontology analysis after clustering based on patterns of change associated these SIX1-regulated genes to processes such as translation, gene expression, extracellular matrix organization, NCC differentiation, and osteoblast and cartilage development including components of the WNT signaling pathway. As WNT signaling is a known contributor to craniofacial bone development 36-39, we performed functional validation and confirmed that SIX1 regulates this pathway using the TOP-Flash reporter vector that measures canonical WNT-specific transcriptional activity.
2. Results
Workflow and preliminary analysis
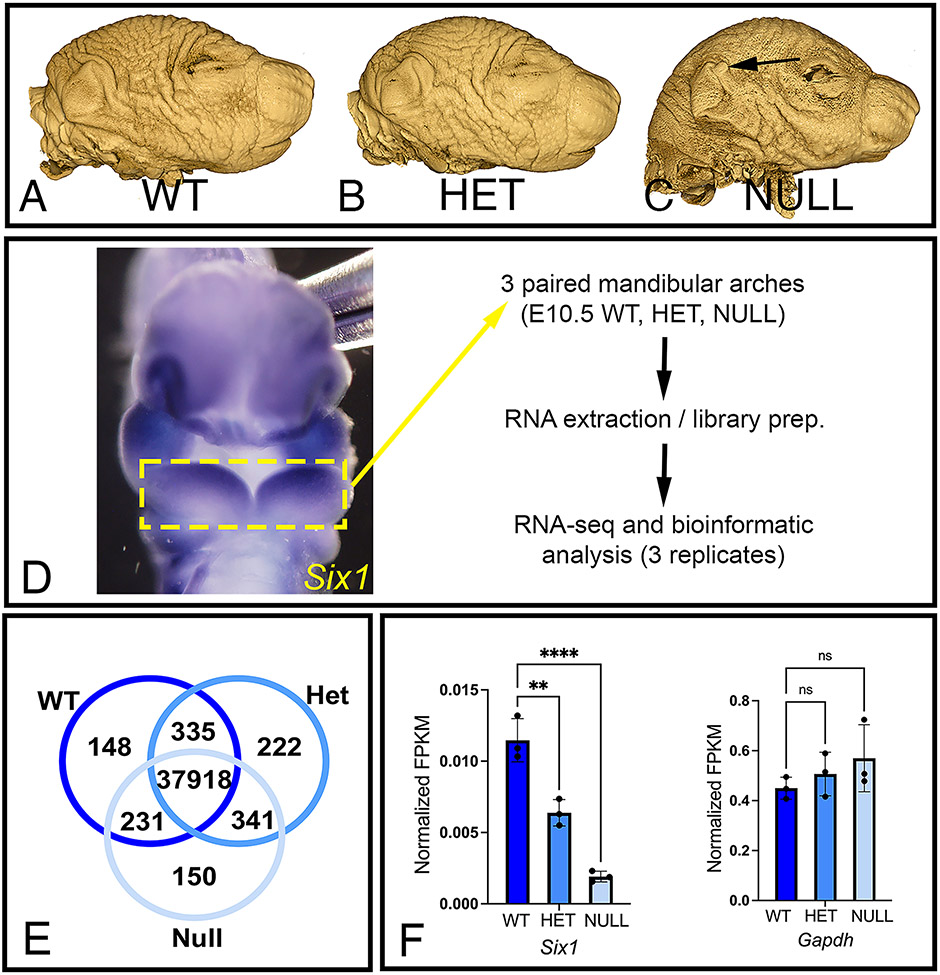
To identify genes that are regulated by SIX1 in the mandibular arch and that could be investigated as causative to BOR, we performed Illumina sequencing of RNA extracted from mandibular arches of embryos of the Six1-null line (Fig. 1A-C). We included in our analysis Six1-null embryos because OPT imaging revealed that they present with external features of BOR such as ear tags (Fig. 1C) and severe craniofacial defects including hypoplastic middle ear ossicles. 29,30,32 Six1-het embryos were also included because, even though they do not present with apparent craniofacial defects (Fig. 1B), they present with hearing deficits. 35 RNA-seq was performed using E10.5 mandibular arches because this is the time point when Six1 is localized throughout the arch mesenchyme (Fig. 1D), and when we detect changes in genes critical for D-V pattering of this arch. 29,30 We assembled a transcriptome comprising 40,841 transcripts in total. We detected 38,632 transcripts from WT with 148 transcripts that were unique to this genotype. We detected 38,816 from Six1-het embryos (222 unique genes), and 38,640 transcripts detected from Six1-null embryos (150 unique genes), although a majority of uniquely detected genes are currently unannotated (listed in Supplemental File 1) (Fig. 1E). The equally robust detection of transcripts from all three genotypes indicates that mandibular arches from Six1-het and Six1-null embryos were transcriptionally active at a level similar to WT. Expression profile of Six1 for each genotype evaluated using RNA-seq (Fig. 1F) demonstrated differences in abundance of Six1 as expected for each genotype.
Figure 1: Workflow for comparative bulk RNA-seq analysis of wild type, Six1-het and Six1-null mandibular arches.
(A–C) OPT images of wild type (WT), Six1-het (HET), and Six1-null (NULL) mouse heads reveal that, in addition to deformed skull and jaws, Six1-null mice present with BOR-like features such as ear tags (arrow in C). (D) Expression pattern for Six1 as seen with whole-mount ISH in WT E10.5 mouse embryo. Mandibular arches (dashed yellow box) were manually dissected and processed for RNA-seq. Ventral view. (E) Venn diagram showing the number of genes detected in each genotype. (F) Quantitative expression of Six1 and reference gene Gapdh in mandibular arches from each genotype. n=3 per genotype. Data was normalized with expression values for Actb. **P<0.01, ****P<0.0001, ns: not significant.
Differential expression analysis
Differential gene expression of mandibular arches from WT, Six1-het and Six1-null embryos (E10.5) resulted in 808 differentially expressed genes between WT and Six1-null arches: 317 genes were downregulated, and 491 genes were upregulated in Six1-null arches (FDR ≤ 0.25). With the exception for Six1, we noted no differentially expressed genes between WT and Six1-het arches within the FDR cutoff parameter.
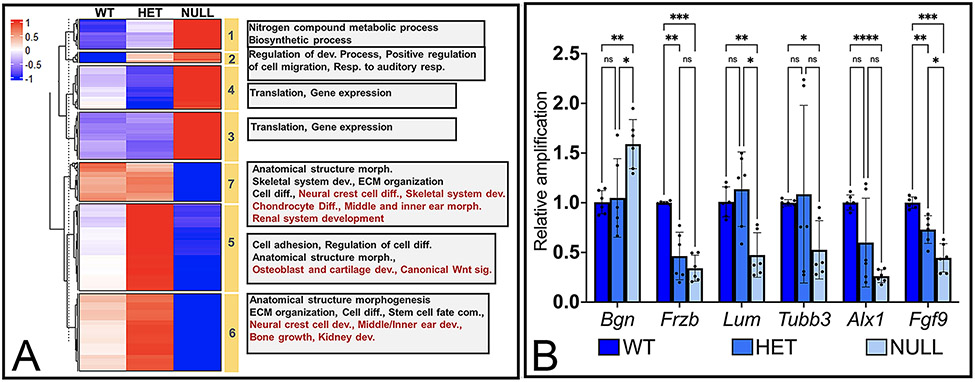
To further characterize the 808 differentially expressed genes, we performed a K-means clustering approach using averaged FPKM values for all three genotypes. Even though changes in genes in Six1-het arches did not pass the FDR cutoff parameter, we included this genotype to assist in the stratification of genes in different clusters. Using this approach, we grouped the differentially expressed genes into 7 clusters representing distinct patterns of changes in gene expression based on genotype (Fig. 2) (Supplemental File 2). Clusters 1 – 4 represent genes that were upregulated in Six1-null compared to WT arches (Fig. 2A). The major difference between these 4 clusters were changes in gene expression in Six1-het compared to expression in WT arches: genes in Clusters 1 and 2 showed an increase in expression while genes in Clusters 3 and 4 showed a decrease in expression. Gene ontology analysis of genes enriched in each cluster from Clusters 1 and 2 revealed that these primarily represented biological functions such as maintaining protein metabolism (e.g., Eef1a1, Eef2, Eif1, Eif2s3x and Eif4b) as well as ribosomal proteins (e.g., Rpl and Rps genes). Cluster 2 included genes such as Atoh8, Mdk and, Rhoc that play a role in positive regulation of cell migration. Clusters 3 and 4 primarily grouped genes involved in biosynthetic processes, translation, and maintenance of gene expression such as additional Rpl and Rps genes.
Figure 2: Differential expression analysis of genes in WT, Six1-het and Six1-null mandibular arches.
(A) Heatmap with K-means cluster analysis of averaged, z-score scaled, FPKM values for gene expression in each genotype. Yellow bars mark cluster number. Grey boxes list gene ontology terms represented in each cluster. (B) Quantitative-PCR (qPCR) validation of expression pattern of select genes from cluster 1 (Bgn), 5 (Frzb), 6 (Lum, Tubb3), 7 (Alx1, Fgf9) in WT, Six1-het (HET) and Six1-null (NULL) arches. *P<0.05, ** P< 0.01, *** P<0.001, **** P<0.0001, ns: not significant. n=3. Actb was used as housekeeping gene.
Clusters 5 – 7 mainly grouped genes that were downregulated in Six1-null arches (Fig. 2A). Cluster 5 included genes that showed higher expression in Six1-het arches compared to those collected from WT and Six1-null mice. These included genes such as Col27a1, Dicer1, Mia3 and, Satb2 that regulate cartilage development and genes such as Hdac4 and Satb2 that regulate bone development. This cluster also contained genes that play a role in canonical WNT signaling such as Frzb and Fzd1, indicating a role for SIX1 in regulation of WNT signaling. Cluster 6 contained genes that had a slightly higher expression in Six1-het compared to WT arches while cluster 7 primarily had genes with similar expression between WT and Six1-het arches. The genes in Clusters 6 and 7 were involved in ear, craniofacial and kidney development such as Sobp, Ednra, Gbx2, Tgfb2, Foxd1 and Rdh10.
qPCR validation confirmed changes in expression of genes from Cluster 1 (Bgn), Cluster 5 (Frzb), Cluster 6 (Lum, Tubb3) and Cluster 7 (Alx1, Fgf9) in Six1-null relative to WT arches (Fig. 2B). Interestingly, while RNA-seq analysis did not detect significant changes in genes from Six1-het arches, qPCR revealed significant changes in Frzb and Fgf9 mRNA levels in Six1-het relative to WT arches (Fig. 2B).
Regulation of BOR associated features by Six1
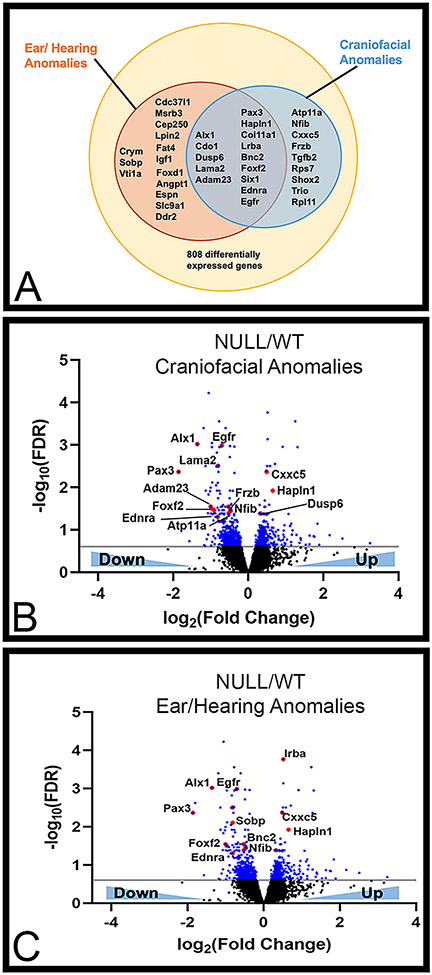
As BOR patients present with craniofacial and inner/middle ear defects, we further characterized the 808 differentially expressed genes between WT and Six1-null arches by annotating associated craniofacial and hearing phenotypes as reported on publicly available databases, Online Mendelian Inheritance in Man (OMIM) and Mouse Genome Informatics (MGI). We identified 14 genes in the two phenotypes that were differentially regulated in Six1-null arches (Fig. 3A) while 9 genes were specific to craniofacial anomalies and 14 to ear/hearing anomalies. Volcano plots were then used to depict this data based on down- or up-regulation in Six1-null compared to WT arches (Fig. 3B-C). While some genes in these groups showed upregulation in Six1-null embryos (e.g., Hapln and Dusp6), most genes were downregulated after loss of Six1, suggesting that SIX1 is primarily required for gene induction within the mandibular arch. Supplemental file 3 contains a list of up- (shades of blue) or down- (shades of red) regulated genes in Six1-null compared to WT arches and the associated possible BOR-like phenotype from Mouse Genome Informatics (MGI, https://www.informatics.jax.org/).
Figure 3: Comparison of differentially expressed genes with BOR associated phenotypes as described in OMIM and MGI databases.
(A) Venn diagram showing differentially expressed genes that have been associated with ear/hearing and craniofacial anomalies. (B–C) Volcano plots marking differentially expressed genes between WT and Six1-null (NULL) arches in blue (FDR ≤ 0.25, i.e. −0.6 on a log10 scale). Data points marked in red represent genes that have been associated with ear/hearing anomalies and craniofacial anomalies.
We next asked whether the 808 differentially expressed genes detected by RNA-seq might be potential direct regulatory targets of SIX1. We compared our dataset with previously reported putative downstream targets of SIX1 identified by ChIP-seq for SIX1 targets in mouse otic vesicles. 52 This comparison identified 322 of the 808 (~40%) differentially expressed genes as potential direct SIX1 targets. Among these, we noted several genes associated with processes that are likely to be related to features presented in BOR patients such as head development, NCC differentiation, ossification, and ear morphogenesis (Table 1). This analysis clarifies a direct role for SIX1 in regulating several morphogenetic processes required for development of the vertebrate face and skull, and highlights additional genes whose role in craniofacial development and potential link to BOR will be analyzed in future experiments.
Table 1 -.
Comparative analysis of differentially expressed genes with putative direct targets of SIX1.
| Stem Cell Development |
Neural Crest Cell Differentiation |
Ossification | Bone Development | ||||
|---|---|---|---|---|---|---|---|
| Up* | Down* | Up* | Down* | Up* | Down* | Up* | Down* |
| Dicer1 | Dicer1 | Rpl38 | Ddr2 | Has2 | Bnc2 | ||
| Ednra | Ednra | Fat4 | Tmem107 | Ddr2 | |||
| Frzb | Frzb | Fhl2 | Fat4 | ||||
| Fzd1 | Pax3 | Fzd1 | Igf1 | ||||
| Nrg1 | Rdh10 | Igf1 | Osr2 | ||||
| Pax3 | Rspo2 | Pdgfc | |||||
| Rdh10 | Satb2 | Thbs1 | |||||
| Tgfb2 | |||||||
| Chd3 | |||||||
| Cartilage Development |
Head Development | Ear Morphogenesis |
WNT Pathway | ||||
| Up* | Down* | Up* | Down* | Up* | Down* | Up* | Down* |
| Prrx2 | Dicer1 | Ddit4 | Dclk2 | Myc | Edrna | Cav1 | Ednra |
| Nfib | Dkk1 | Dicer1 | Prrx2 | Frzb | Dkk1 | Foxd1 | |
| Osr2 | Mgarp | Ednra | Rpl38 | Lrig1 | Frzb | ||
| Rspo2 | Tiparp | Egfr | Osr2 | Fzd1 | |||
| Satb2 | Fat4 | Sobp | Chd24 | ||||
| Tgfb2 | Nfib | Whrn | Fat1 | ||||
| Thbs1 | Nrg1 | Fat3 | |||||
| Osr2 | Pcdh9 | ||||||
| Pcdh19 | Pcdh19 | ||||||
| Ptch1 | Wnt7b | ||||||
| Rere | Tcf4 | ||||||
| Robo1 | |||||||
| Satb2 | |||||||
| Syne2 | |||||||
| Tgfb2 | |||||||
| Whrn | |||||||
Increased (Up) or decreased (Down) expression in Six1−/− (NULL) embryos relative to control (WT) embryos
Six1 induces expression of the WNT repressor Frzb
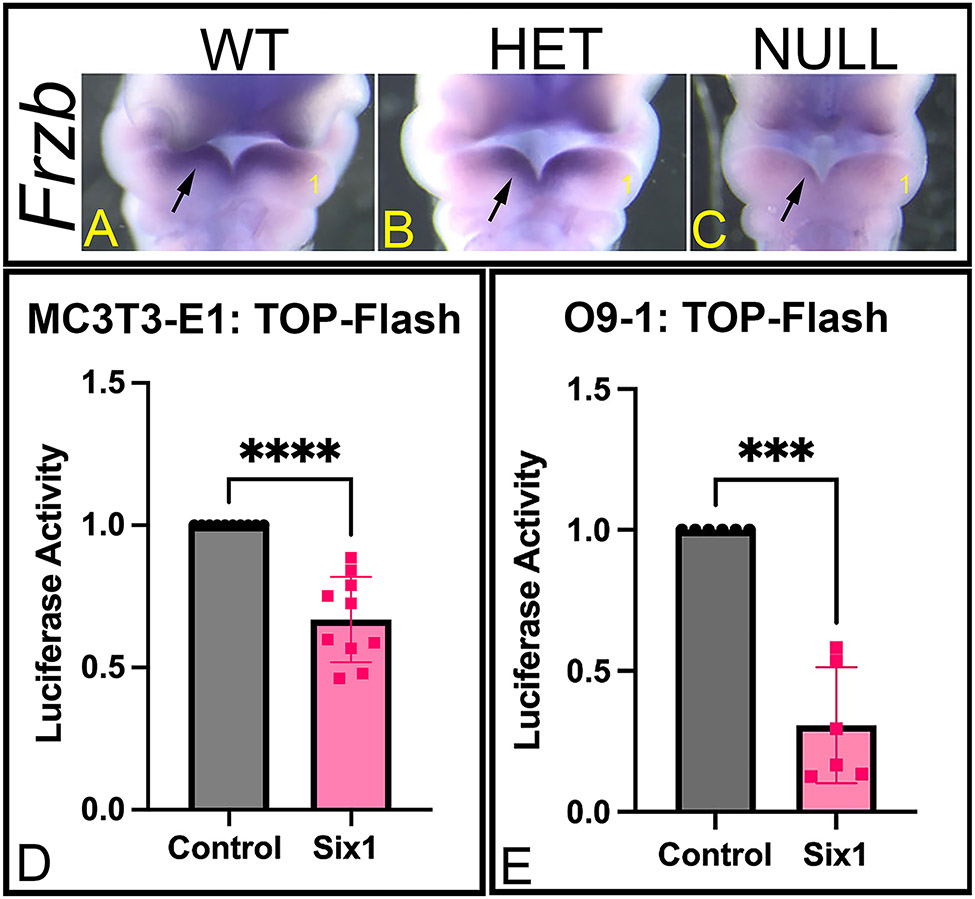
We detected a number of WNT pathway genes to be differentially regulated in Six1-null arches (e.g., Frzb, Frz1 in Cluster 5) and these were also found to be putative regulatory targets of SIX1 (Table 1). Because WNT signaling is a known contributor to craniofacial bone development 36-39, we further evaluated how SIX1 regulates this pathway. In our RNA-seq (Fig. 2A) and qPCR (Fig. 2B) analyses, we noted that Frzb (Cluster 5), a known WNT antagonist, was downregulated in Six1-het and Six1-null compared to WT arches; thus, we decided to further characterize changes in Frzb in mutant embryos by whole-mount ISH. Spatial expression analysis showed that in WT mandibular arches, Frzb is expressed within the Six1 expression domain (compare Fig. 1D with Fig. 4A); ISH analysis demonstrated that Frzb expression was severely downregulated in Six1-null embryos (Fig. 4C). Additionally, we noted a small decrease in the expression domain for Frzb in Six1-het embryos (Fig. 4B) confirming findings from qPCR assay. Finally, to determine whether SIX1 regulates WNT signaling in the mandibular arch, we performed luciferase assays using calvarial pre-osteoblastic cells (MC3T3-E1) or cranial NCCs (O9-1), and the TOP-Flash reporter vector. SIX1 overexpression in these cells caused a significant reduction in activity of the luciferase reporter (~33% reduction in MC3T3-E1 cells, P<0.0001, Fig. 4D; ~70% reduction in O9-1 cells, P<0.001, Fig. 4E), indicating that SIX1 represses WNT signaling in cells derived from the developing face and skull.
Figure 4: Regulation of WNT signaling by SIX1.
(A-C) Whole-mount ISH for Frzb in E10.5 WT (A), Six1-het (HET) (B), and Six1-null (NULL) (C) mouse embryos. Ventral view. The heart was removed to aid visualization. 1. Mandibular arch. Loss of Six1 disrupts expression of Frzb in the mandibular arch (arrow). (D-E) Luciferase activity of the TOP-Flash reporter vector is significantly decreased in MC3T3-E1 (D) and O9-1 (E) cells after Six1 overexpression indicating that SIX1 represses canonical WNT-specific transcriptional activity. ***P<0.001, ****P<0.0001. n=3.
3. Discussion
Branchio-oto-renal syndrome (BOR) is a birth defect that affects derivatives of the otic placode and mandibular arches leading to variable degrees of craniofacial defects and sensorineural and/or conductive hearing loss. As the genetic causes of this disorder are known in only 50% of affected individuals 18,19,28, we used bulk RNA-seq of mandibular arches from embryos of a Six1-null mouse line to identify genes associated with SIX1 that could be causative for BOR and serve as targets for future studies.
Transcriptomic analysis identified a link between Six1 and translation, neural crest cell differentiation, chondrogenesis and osteogenesis
RNA-sequencing of mandibular arches collected from WT, Six1-het and Six1-null embryos detected a similar number of transcripts between the three genotypes. This finding confirms past assertions that loss of Six1 rather than leading to increased cell death or decreased cell proliferation is controlling gene expression and enhancer availability thus leading to changes in patterning and differentiation of NCCs. 30,53 Comparison between the transcriptome of the three genotypes identified 808 genes differentially regulated in Six1-null embryos while we were unable to identify significant changes in Six1-het embryos relative to WT embryos. The lack of significant changes in Six1-het embryos is likely caused by increased transcriptome variability after loss of one allele for Six1. BOR patients carrying the same variant allele in a family present with a highly variable phenotype. 18,19 It has been proposed that phenotypic variability in BOR is caused by a dosage effect in which the amount of available protein determines development of an embryonic structure as a certain threshold needs to be exceeded for expression of different genes. 54,55 In addition, as SIX1 function relies on co-factor interaction 33,56-58, association with different co-factors could vary based on the amount of SIX1 protein available which in turn would lead to different patterns of gene expression. Two reported SIX1 variants (Q11X and Q22X) generate a truncated protein that is degraded (data not shown) what would match the Six1-het genotype. Comparison of features between patients carrying the Q11X or Q22X variants confirms that SIX1 haploinsufficiency caused by the truncation disrupts craniofacial development differently. 26 Environmental factors and genetic modifiers are another possible explanation to the phenotypic and transcriptomic variabilities. 54 Interestingly, the severity and frequency of the upper jaw phenotype in Six1-null embryos are different between mouse strains which suggests the role of modifier genes. 30,59 It will be highly informative to compare in future studies the phenotype of embryos from the Six1-null line with that of embryos from a mouse line carrying a BOR variant (e.g., R110W).
Despite this negative finding regarding Six1-het mouse embryos, Six1-null embryos present with severe craniofacial defects including features of BOR such as ear tags, malformed middle ear ossicle and inner ear defects. 29-32,35 Thus Six1-null embryos can be used instead of Six1-het embryos for the identification of SIX1-regulated genes that can later be studied as potential causative genes in unresolved BOR cases. Clustering and gene ontology analysis of genes differentially expressed in Six1-null compared to WT mandibular arches demonstrated that SIX1 has an important function regulating translation, NCC differentiation, chondrogenesis and osteogenesis. Interestingly, genes associated with translation were upregulated after loss of Six1 indicating that this transcription factor represses metabolic processes within the mandibular arch. Conversely, genes associated with differentiation events were downregulated in Six1-null arches indicating that SIX1 induces differentiation of NCCs, particularly towards cartilage and bone. Previous work showed that SIX1 is required for metabolic processes and patterning of NCCs within the mandibular arch by controlling expression of Edn-1 in the arch endoderm. Loss of Six1 leads to changes in genes that pattern the ventral (Dlx5) and dorsal (Dlx2) domains of the mandibular arch. In addition, there is an expansion of Prrx1 and Prrx2, and repression of Pou3f3 in the intermediate domain. 30,60,61 This function of SIX1 in D-V patterning of the mandibular arch, particularly the intermediate domain that will give rise to the middle ear 62,63, is likely the reason why Six1-null embryos and BOR patients present with defects in the middle ear ossicles leading to conductive hearing loss. 29,35 Alternatively, as our data also indicated a role for SIX1 in chondrogenesis and osteogenesis, middle ear defects could be caused by later changes in cartilage and bone development. Six1 is required for craniofacial cartilage development in Xenopus, and it has been reported that variants in SIX1 cause premature cranial suture closure or craniosynostosis, indicating that SIX1 may be regulating cranial bone development and/or suture homeostasis. 26,30,64 This work is the first to identify a link between SIX1 and genes crucial for development of cartilage and bone that could account for craniofacial defects in patients with BOR. Further studies are required to clarify whether defects in BOR patients are caused by changes in NCC patterning or disruption of cartilage/bone development, and whether different SIX1 variants would affect either patterning or chondrogenesis/osteogenesis.
Six1 regulates WNT signaling in the mandibular arch
The WNT signaling pathway is crucial for craniofacial and bone development as it is required for NCC and later, osteoblast differentiation. It is also critical during adulthood for maintenance of bone mass, osteoclast formation and control of bone fracture healing. 36-39 Our analysis indicated a link between SIX1 and genes that control osteogenesis, including components of the WNT signaling pathway. While two genes were upregulated (Cav1 and Dkk1), the majority of genes identified in this study linked to the WNT signaling pathway were downregulated after loss of SIX1. This finding suggests that SIX1 is required either directly or indirectly for the expression of genes that regulate this pathway. Our in vitro and in vivo data at E10.5, and previous work at later stages during odontogenesis in the mouse 65, show that despite the fact that SIX1 regulates both, inducers and repressors of WNT, the final outcome is the repression of WNT signaling in the oral region of the mandibular arch where SIX1 is expressed. Based on our work, it seems that SIX1 is accomplishing this repression by several mechanisms that may or may not require induction of Frzb expression in NCCs. In addition, while in mouse Frzb knockout animals are viable and do not have a described craniofacial phenotype, frzb is required for ethmoid plate extension and lower jaw convergence in zebrafish. Of note, frzb is required for bapx1 (nkx3.2) expression, a gene that patterns the intermediate domain in zebrafish from which the fish jaw joint is originated. This structure is homologous to the mammalian middle ear and suggests a role for Frzb in patterning of the middle ear. 62,63,66,67 It will be interesting to verify in future studies if mice lacking Frzb present with mild defects in craniofacial elements such as the middle ear that were not reported in previous studies and would indicate a potential link to hearing disorders such as BOR.
4. Conclusion
Our bulk RNA-seq analysis and comparison of differentially expressed genes with previously available data informed us of several SIX1-regulated genes that are associated with craniofacial and ear anomalies and therefore could be linked to BOR. However, these genes may be directly or indirectly regulated by SIX1 and further work using ChIP-seq is required. In addition, SIX1 is known to function as a transcriptional activator or repressor depending on co-factor binding. 33 We are currently investigating which co-factors regulate SIX1 transcriptional activity in the mandibular arch. Findings from these studies will help elucidate the complex function of SIX1 during craniofacial development and the pathogenesis of birth defects such as BOR.
5. Experimental Procedures
Mice
Wild type (WT) 129S1/SvlmJ mice were purchased from The Jackson Laboratory and Six1-null line was kindly provided by Dr. Kiyoshi Kawakami (Jichi Medical School, Japan). Genotyping of embryos from the Six1-null line was performed as previously described. 29 The sex of the embryos was not determined. All experiments using mice were approved by the Institutional Animal Care and Use Committee (IACUC) at the George Washington University School of Medicine and Health Sciences (A2022-019 and A2022-020). The George Washington University Animal Research Facility is certified by the Association for Assessment and Accreditation of Laboratory Animal Care.
Optical projection tomography imaging
Paraformaldehyde-fixed heads from E18.5 mouse embryos (three heads per genotype) were rinsed briefly in phosphate buffered saline (PBS) then embedded in analytical grade low melting point agarose (Promega) (1.1% in Milli-Q water). The agarose blocks were glued to magnetic stubs using Loctite-454 Prism surface-insensitive adhesive gel. After mounting, the agarose blocks were trimmed and hung, immersed in 100% methanol, in glass jars (magnetic stub to lid) overnight. The methanol was replaced twice (each for a further 24 hours), to ensure complete dehydration. Following methanol dehydration, the blocks were placed in BABB clearing agent (1 part benzyl alcohol to 2 parts benzyl benzoate) for 24 hours and the solution again replaced twice for 24 hours each time to ensure complete optical clearing of the tissue. Cleared samples were imaged in a Bioptonics 3001M Optical Projection Tomograph at ~13.2 microns resolution using a 1024 x 1024 image format and a 0.9 degree rotation step. Imaging was conducted under UV light using a GFP1 filter (425nm excitation, 475nm emission) to detect tissue autofluorescence. Raw imaging data (TIF format) were reconstructed into multidimensional slice data (BMP format) using NRecon v1.6.9 software (Skyscan, Belgium). Reconstructed data were then imported into Drishti v2.6.5 Volume Exploration software for 3D rendering, visual assessment and image generation. 40
RNA extraction
Six1+/+ (WT), Six1+/− (Six1-het) and Six1−/− (Six1-null) embryos at embryonic day (E) 10.5 41 were harvested in PBS and stored in RNAlater solution (Thermo). The ventral and intermediate (v+i) domains of the first pharyngeal arch (mandibular arch) were dissected in RNAlater solution then transferred to TRI-Reagent (Zymo). RNA was extracted from three pooled dissected arches of the same genotype using the Direct-zol RNA Microprep kit with DNase-I treatment (Zymo). After RNA Bioanalyzer QC control, 1mg of RNA was sent for Illumina sequencing. The unused RNA was stored at −80°C. Library was prepared using the TruSeq Stranded Total RNA Library Prep Human/Mouse/Rat kit (Illumina) and, after library QC, sequenced using the Illumina NextSeq 500 (Mid Output 2x75bp).
Data processing pipeline
Fastq files containing RNA-Seq reads were transferred to GWU's high performance computing cluster, Pegasus. FastQC was used to generate a quality report for each file. The resulting report files were aggregated into a single report using multiQC. Because reads had sufficient quality, no trimming was deemed necessary. Paired-end reads were aligned to the GRCm39 mouse genome (RefSeq assembly version GCF_000001635.27) using STAR. The resulting SAM files were used as input for featureCounts to count reads to exons. 42 Reads across different technical replicates were summed in R. Any NA values were changed to zeros. The R package HTSFilter was used to remove genes with low signal. 43 The R package DESeq2 was used for differential expression analysis between each pair of conditions. 44 The alpha value used was 0.05 and the Benjamini-Hochberg (BH) method was used to estimate p-adjusted (FDR) values.
Clustering analysis
Genes identified as significantly altered in their expression values (FDR ≤ 0.25) were used for clustering analysis based on their averaged expression values (FPKM) for each genotype. The K-means clustering was supplemented with hierarchical clustering and a heatmap was generated based on these gene expression change using the ComplexHeatmap R-package (https://github.com/jokergoo/ComplexHeatmap). 45
Gene ontology analysis
Gene function and phenotype analysis were performed based on reports generated using PantherDB and Visual Annotation Display (VLAD) gene list analysis and visualization tool available on the mouse genome informatics database (MGI, http://www.informatics.jax.org). 46-48 Further phenotypic annotation for craniofacial and hearing anomalies were based on information available on Online Mendelian Inheritance in Man (OMIM, https://omim.org/) and Mouse Genome Informatics (MGI, https://www.informatics.jax.org/).
Quantitative real time PCR (qPCR)
RNA that was not sent for sequencing was used for cDNA synthesis using the iScript Advanced cDNA Syntehsis kit (Bio-rad). qPCR was performed using 5ng of cDNA and the Sso Advanced Universal SYBR Green Supermix (Bio-Rad) in a Bio-Rad CFX-384 Real Time Thermal Cycler. QuantiTect Assay primers for Actb (housekeeping gene), Six1, Bgn, Lum, Tubb3, Alx1, Fgf9 and Frzb were purchased from Qiagen. All assays were performed in duplicates at least three times. Statistical analysis between WT, HET and NULL embryos was performed with Prism and significance calculated using a paired 2-way ANOVA with Tukey’s multiple comparisons test.
Whole mount in situ hybridization (ISH)
Whole mount ISH was performed as previously described. 49 Digoxigenin-labeled probe for Six1 was synthesized using the pZErO-2-Six1 (kindly provided by Dr. Heide Ford). Digoxigenin-labeled probe for Frzb was synthesized using the pCRII-Frzb plasmid (generated by PCR and TOPO TA cloning, Thermo). E10.5 embryos were incubated in BM-Purple (Roche) to detect Six1 probe, or with 4-nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) (Sigma) to detect Frzb probe. All ISH experiments were performed on a minimum of three embryos from different litters.
Cell culture and Luciferase assay
O9-1 (Sigma) and MC3T3-E1 (ATCC) cells were kindly provided by Dr. David Clouthier (University of Colorado Anschutz Medical Campus). O9-1 cells were culture with Complete ES Cell Medium with 15% FBS (Gibco) and LIF plus bFGF and penicillin/streptomycin in plates coated with Matrigel (Corning). 50 MC3T3-E1 cells were cultured with MEM α with GlutaMAX Supplement (Thermo) with 20% FBS (Gibco) and penicillin/streptomycin (Gibco). 51 Cells were passed every 2-3 days with TrypLE Express (Gibco). For luciferase assay experiments, cells were plated in a 24-well plate (O9-1 cells - 25,000 cells per well; MC3T3-E1 cells - 200,000 cells per well). 24 hours after plating, cells were transfected using Lipofectamine 3000 (Thermo) with 200ng of the TCF firefly lucirefase reporter vector (pTOP-Flash) and 20ng of Renilla luciferase reporter (pTK-Renilla). In addition, cells were either transfected with control vector (pCMV) or pCMV2-3’Flag-Six1. 48 hours after transfection, cells were harvested in 1X Passive Lysis Buffer and stored at −80°C overnight to further facilitate cell lysis. 20 μL of lysate was used in the analysis with the Dual Luciferase Assay kit (Promega). Experiments were repeated at least five independent times and read on a Varioskan Flash Plate Reader (Thermo). Statistical analysis between cells transfected with control vector or the Six1 overexpression vector was performed with GraphPad Prism and significance calculated using a paired two-tailed t-test.
Supplementary Material
Acknowledgments
We thank Dr. Sally Moody (The George Washington University School of Medicine and Health Sciences) for helpful discussion and valuable feedback on this study and manuscript. We thank Dr. Heide Ford (University of Colorado School of Medicine) for generously providing the pCMV2-3’Flag-Six1 and Dr. Kiyoshi Kawakami (Jichi Medical School, Japan) for kindly providing the Six1-null mice. We thank Dr. Keith Crandall, Tyson Dawson and Castle Raley (The Genomics Core at The George Washington University) for performing the Illumina sequencing and bioinformatic analysis. We also thank Dr. Steve Klein for their helpful discussions during this study.
Funding
This work would not have been possible without the support of the National Institute of Health/National Institute of Dental and Craniofacial Research (R03 DE028964 to AT) and The George Washington University School of Medicine and Health Sciences.
Footnotes
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data Availability Statement
All RNA-seq datasets generated in this study are available through the FaceBase data repository (http://www.facebase.org, Record ID 1-YNZC, DOI 10.25550/1-YNZC).
7. References
- 1.Abe M, Ruest LB, Clouthier DE. Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor-deficient mice. Int J Dev Biol. 2007;51(2):97–105. https://doi.org 10.1387/ijdb.062237ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. Apr 2000;127(8):1671–9. [DOI] [PubMed] [Google Scholar]
- 3.Clouthier DE, Garcia E, Schilling TF. Regulation of facial morphogenesis by endothelin signaling: insights from mice and fish. Am J Med Genet A. Dec 2010;152a(12):2962–73. https://doi.org 10.1002/ajmg.a.33568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. Jan 1999;126(1):51–61. https://doi.org 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- 5.Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol. Nov 15 2012;371(2):121–35. https://doi.org 10.1016/j.ydbio.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. Jun 2010;137(11):1843–52. https://doi.org 10.1242/dev.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander C, Zuniga E, Blitz IL, et al. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. Dec 2011;138(23):5135–46. https://doi.org 10.1242/dev.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. Oct 11 2002;298(5592):381–5. https://doi.org 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- 9.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. Nov 2005;207(5):501–61. https://doi.org 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. Jan 1 2000;217(1):10–24. https://doi.org 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- 11.Ruest LB, Kedzierski R, Yanagisawa M, Clouthier DE. Deletion of the endothelin-A receptor gene within the developing mandible. Cell Tissue Res. Mar 2005;319(3):447–53. https://doi.org 10.1007/s00441-004-0988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavares AL, Garcia EL, Kuhn K, Woods CM, Williams T, Clouthier DE. Ectodermal-derived Endothelin1 is required for patterning the distal and intermediate domains of the mouse mandibular arch. Dev Biol. Nov 1 2012;371(1):47–56. https://doi.org 10.1016/j.ydbio.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavares AL, Clouthier DE. Cre recombinase-regulated Endothelin1 transgenic mouse lines: novel tools for analysis of embryonic and adult disorders. Dev Biol. Apr 15 2015;400(2):191–201. https://doi.org 10.1016/j.ydbio.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin S, Matalova E, Simpson C, Yoshida H, Tucker AS. Incudomalleal joint formation: the roles of apoptosis, migration and downregulation. BMC Dev Biol. Dec 5 2007;7:134. https://doi.org 10.1186/1471-213X-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon CT, Weaver KN, Zechi-Ceide RM, et al. Mutations in the endothelin receptor type A cause mandibulofacial dysostosis with alopecia. Am J Hum Genet. Apr 2 2015;96(4):519–31. https://doi.org 10.1016/j.ajhg.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard AB, Kanai SM, Krock B, et al. Loss-of-function of Endothelin receptor type A results in Oro-Oto-Cardiac syndrome. Am J Med Genet A. May 2020;182(5):1104–1116. https://doi.org 10.1002/ajmg.a.61531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanai SM, Heffner C, Cox TC, et al. Auriculocondylar syndrome 2 results from the dominant-negative action of PLCB4 variants. Dis Model Mech. Apr 1 2022;15(4). https://doi.org 10.1242/dmm.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochhar A, Fischer SM, Kimberling WJ, Smith RJ. Branchio-oto-renal syndrome. Am J Med Genet A. Jul 15 2007;143A(14):1671–8. https://doi.org 10.1002/ajmg.a.31561. [DOI] [PubMed] [Google Scholar]
- 19.Smith RJH. Branchiootorenal Spectrum Disorders. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, eds. GeneReviews((R)). Seattle (WA): 2018. [Google Scholar]
- 20.Kochhar A, Orten DJ, Sorensen JL, et al. SIX1 mutation screening in 247 branchio-oto-renal syndrome families: a recurrent missense mutation associated with BOR. Hum Mutat. Apr 2008;29(4):565. https://doi.org 10.1002/humu.20714. [DOI] [PubMed] [Google Scholar]
- 21.Lee KY, Kim S, Kim UK, Ki CS, Lee SH. Novel EYA1 mutation in a Korean branchio-oto-renal syndrome family. Int J Pediatr Otorhinolaryngol. Jan 2007;71(1):169–74. https://doi.org 10.1016/j.ijporl.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 22.Mehdizadeh T, Majumdar HD, Ahsan S, Tavares ALP, Moody SA. Mutations in SIX1 Associated with Branchio-oto-Renal Syndrome (BOR) Differentially Affect Otic Expression of Putative Target Genes. J Dev Biol. Jun 30 2021;9(3). https://doi.org 10.3390/jdb9030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah AM, Krohn P, Baxi AB, et al. Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis Model Mech. Mar 3 2020;13(3). https://doi.org 10.1242/dmm.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orten DJ, Fischer SM, Sorensen JL, et al. Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum Mutat. Apr 2008;29(4):537–44. https://doi.org 10.1002/humu.20691. [DOI] [PubMed] [Google Scholar]
- 25.Sanggaard KM, Rendtorff ND, Kjaer KW, et al. Branchio-oto-renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur J Hum Genet. Nov 2007;15(11):1121–31. https://doi.org 10.1038/sj.ejhg.5201900. [DOI] [PubMed] [Google Scholar]
- 26.Calpena E, Wurmser M, McGowan SJ, et al. Unexpected role of SIX1 variants in craniosynostosis: expanding the phenotype of SIX1-related disorders. J Med Genet. Jan 12 2021. https://doi.org 10.1136/jmedgenet-2020-107459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization of six SIX1 Branchio-oto-renal syndrome mutations. J Biol Chem. Jul 31 2009;284(31):20781–90. https://doi.org 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Valle A, Wang X, Potocki L, et al. HERV-mediated genomic rearrangement of EYA1 in an individual with branchio-oto-renal syndrome. Am J Med Genet A. Nov 2010;152A(11):2854–60. https://doi.org 10.1002/ajmg.a.33686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozaki H, Nakamura K, Funahashi J, et al. Six1 controls patterning of the mouse otic vesicle. Development. Feb 2004;131(3):551–62. https://doi.org 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- 30.Tavares ALP, Cox TC, Maxson RM, Ford HL, Clouthier DE. Negative regulation of endothelin signaling by SIX1 is required for proper maxillary development. Development. Jun 1 2017;144(11):2021–2031. https://doi.org 10.1242/dev.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. Jun 2003;120(6):669–79. [DOI] [PubMed] [Google Scholar]
- 32.Guo C, Sun Y, Zhou B, et al. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J Clin Invest. Apr 2011;121(4):1585–95. https://doi.org 10.1172/jci44630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. Dec 2004;131(23):5871–81. https://doi.org 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- 34.Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. Sep 2000;96(2):253–7. [DOI] [PubMed] [Google Scholar]
- 35.Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. Sep 2003;130(17):3989–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodnough LH, Dinuoscio GJ, Atit RP. Twist1 contributes to cranial bone initiation and dermal condensation by maintaining Wnt signaling responsiveness. Dev Dyn. Feb 2016;245(2):144–56. https://doi.org 10.1002/dvdy.24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodnough LH, Dinuoscio GJ, Ferguson JW, Williams T, Lang RA, Atit RP. Distinct requirements for cranial ectoderm and mesenchyme-derived wnts in specification and differentiation of osteoblast and dermal progenitors. PLoS Genet. Feb 2014;10(2):e1004152. https://doi.org 10.1371/journal.pgen.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibarra BA, Machen C, Atit RP. Wnt-Dependent Activation of ERK Mediates Repression of Chondrocyte Fate during Calvarial Development. J Dev Biol. Jun 27 2021;9(3). https://doi.org 10.3390/jdb9030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda K, Kobayashi Y, Koide M, et al. The Regulation of Bone Metabolism and Disorders by Wnt Signaling. Int J Mol Sci. Nov 6 2019;20(22). https://doi.org 10.3390/ijms20225525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Limaye A. Drishti, A Volume Exploration and Presentation Tool. Developments in X-Ray Tomography Viii. 2012;8506. https://doi.org 10.1117/12.935640. [DOI] [Google Scholar]
- 41.Kaufman MH. The atlas of mouse development. London ; San Diego: Academic Press; 1992:xvi, 512 p. [Google Scholar]
- 42.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features [Article]. Bioinformatics. Apr 2014;30(7):923–930. https://doi.org 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 43.Rau A, Gallopin M, Celeux G, Jaffrezic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments [Article]. Bioinformatics. Sep 2013;29(17):2146–2152. https://doi.org 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 [Article]. Genome Biol. 2014;15(12):38. https://doi.org 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu ZG, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data [Article]. Bioinformatics. Sep 2016;32(18):2847–2849. https://doi.org 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 46.Mi HY, Ebert D, Muruganujan A, et al. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive api. Nucleic Acids Res. Jan 2021;49(D1):D394–D403. https://doi.org 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson JE, Bult CJ. Visual annotation display (VLAD): a tool for finding functional themes in lists of genes [Article]. Mamm Genome. Oct 2015;26(9-10):567–573. https://doi.org 10.1007/s00335-015-9570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blake JA, Baldarelli R, Kadin JA, et al. Mouse Genome Database (MGD): Knowledgebase for mouse-human comparative biology [Article]. Nucleic Acids Res. Jan 2021;49(D1):D981–D987. https://doi.org 10.1093/nar/gkaa1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clouthier DE, Hosoda K, Richardson JA, et al. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. Mar 1998;125(5):813–24. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen BH, Ishii M, Maxson RE, Wang J. Culturing and Manipulation of O9-1 Neural Crest Cells. J Vis Exp. Oct 9 2018;(140). https://doi.org 10.3791/58346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang PW, Horton JA. Variable osteogenic performance of MC3T3-E1 subclones impacts their utility as models of osteoblast biology. Sci Rep. Jun 5 2019;9(1):8299. https://doi.org 10.1038/s41598-019-44575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Zhang T, Ramakrishnan A, et al. Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. Apr 6 2020;48(6):2880–2896. https://doi.org 10.1093/nar/gkaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu JY, Danis EP, Nance S, et al. SIX1 reprograms myogenic transcription factors to maintain the rhabdomyosarcoma undifferentiated state. Cell Rep. Feb 1 2022;38(5):110323. https://doi.org 10.1016/j.celrep.2022.110323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng H, Xu H, Chen B, et al. Genetic and Phenotypic Variability in Chinese Patients With Branchio-Oto-Renal or Branchio-Oto Syndrome. Front Genet. 2021;12:765433. https://doi.org 10.3389/fgene.2021.765433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang YG, Sun SP, Qiu YL, Xing QH, Lu W. A novel mutation in EYA1 in a Chinese family with Branchio-oto-renal syndrome. BMC Med Genet. Aug 7 2018;19(1):139. https://doi.org 10.1186/s12881-018-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tavares ALP, Jourdeuil K, Neilson KM, Majumdar HD, Moody SA. Sobp modulates Six1 transcriptional activation and is required during craniofacial development. bioRxiv. 2021:2021.04.05.438472. https://doi.org 10.1101/2021.04.05.438472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neilson KM, Keer S, Bousquet N, et al. Mcrs1 interacts with Six1 to influence early craniofacial and otic development. Dev Biol. Nov 1 2020;467(1-2):39–50. https://doi.org 10.1016/j.ydbio.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Oghi KA, Zhang J, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. Nov 20 2003;426(6964):247–54. https://doi.org 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 59.Liu Z, Li C, Xu J, et al. Crucial and Overlapping Roles of Six1 and Six2 in Craniofacial Development. J Dent Res. May 2019;98(5):572–579. https://doi.org 10.1177/0022034519835204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kingsbury TJ, Kim M, Civin CI. Regulation of cancer stem cell properties by SIX1, a member of the PAX-SIX-EYA-DACH network. Adv Cancer Res. 2019;141:1–42. https://doi.org 10.1016/bs.acr.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays. Jul 2000;22(7):616–26. https://doi.org. [DOI] [PubMed] [Google Scholar]
- 62.Tucker AS, Watson RP, Lettice LA, Yamada G, Hill RE. Bapx1 regulates patterning in the middle ear: altered regulatory role in the transition from the proximal jaw during vertebrate evolution. Development. Mar 2004;131(6):1235–45. https://doi.org 10.1242/dev.01017. [DOI] [PubMed] [Google Scholar]
- 63.Anthwal N, Joshi L, Tucker AS. Evolution of the mammalian middle ear and jaw: adaptations and novel structures. J Anat. Jan 2013;222(1):147–60. https://doi.org 10.1111/j.1469-7580.2012.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coppenrath K, Tavares ALP, Shaidani NI, Wlizla M, Moody SA, Horb M. Generation of a new six1-null line in Xenopus tropicalis for study of development and congenital disease. Genesis. Dec 2021;59(12):e23453. https://doi.org 10.1002/dvg.23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takahashi M, Ikeda K, Ohmuraya M, et al. Six1 is required for signaling center formation and labial-lingual asymmetry in developing lower incisors. Dev Dyn. Sep 2020;249(9):1098–1116. https://doi.org 10.1002/dvdy.174. [DOI] [PubMed] [Google Scholar]
- 66.Kamel G, Hoyos T, Rochard L, et al. Requirement for frzb and fzd7a in cranial neural crest convergence and extension mechanisms during zebrafish palate and jaw morphogenesis. Dev Biol. Sep 15 2013;381(2):423–33. https://doi.org 10.1016/j.ydbio.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Lories RJ, Peeters J, Bakker A, et al. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice. Arthritis Rheum. Dec 2007;56(12):4095–103. https://doi.org 10.1002/art.23137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq datasets generated in this study are available through the FaceBase data repository (http://www.facebase.org, Record ID 1-YNZC, DOI 10.25550/1-YNZC).