Abstract
A recent report by Yang et al. in Cell demonstrates that faithful DNA double strand breaks and repair cycles phenocopy many aspects of aging in mice. Whether this progeroid phenotype is caused by a loss of epigenetic information remains to be conclusively determined.
Aging is a complex phenotype characterized by an array of biological changes that lead to an increase in an organism’s frailty and in the likelihood of its death. Conserved hallmarks of aging across taxa have been described1, including genomic instability, epigenetic alterations, senescent cell accumulation and exhaustion of stem cells. However, there is still much debate about which hallmarks are primary drivers, secondary mediators, or simply downstream consequences of the aging process.
A recent paper by Yang and colleagues2 attempts to test a unifying theory of aging, dubbed the “Information Theory of Aging”. Central to this theory is the notion that cellular identity is determined by a precise epigenomic landscape. As a by-product of cellular metabolism and exposure to external insults, DNA-damage inevitably accumulates with time. Repair of DNA-damage is coordinated by chromatin modifiers, which also play a key role in maintaining cellular epigenomes and, thus, cell identity. Based on this theory, the repeated relocalization of chromatin modifiers to DNA-damage sites over a lifetime leads to progressive loss of epigenomic identity, ultimately manifesting in what we know as aging at the organismal level. Crucially, the theory distinguishes itself from prior DNA-based theories of aging in that it posits that the loss of epigenomic, rather than genomic, information is the primary driver of aging2.
In order to test this theory, the authors use a mouse model expected to scramble epigenetic information while keeping genetic information intact, dubbed ICE for Inducible Changes to the Epigenome2. In this model, mice carry 2 transgenes: (i) a ubiquitously expressed Cre recombinase localizing to the nucleus upon exposure to tamoxifen [TAM], and (ii) a TAM-stabilized and nuclear-localized I-PpoI endonuclease fusion protein transgene at the Rosa26 locus, which can only be expressed once an upstream floxed stop cassette is excised by Cre. Using this system, transgenic mouse cells exposed to TAM should inducibly express nuclear I-PpoI, which can cut 20 unique canonical sites in the mouse genome. To note, one of these sites is located within the 28S rDNA sequence, of which mice carry hundreds of genomic copies3. Since using I-PpoI to induce Double-Stranded DNA Breaks [DSBs] should create “sticky” 4-basepair overhangs, these DSBs are expected to have low mutagenic potential.
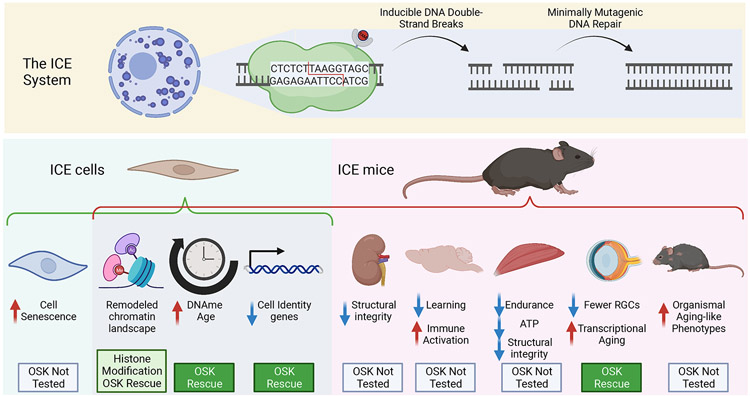
The authors first characterized the ICE system using mouse embryonic fibroblasts [MEFs]. After induction, ICE cells showed strong signs of dsDNA damage compared to controls, while displaying limited mutational load post-treatment, compatible with the notion that the ICE system increases the rate of mutation-less DSBs. ICE MEFs showed increased estimated age based on a DNA-methylation [DNAme] epigenetic clock, increased expression of senescence-associated genes, and increased frequency of markers of cell senescence. The group tested the ICE system in vivo, by inducing I-PpoI expression in young adult mice, and performed phenotypic characterization 1 and 10 months after induction. Consistent with the known progeroid impact of DNA-damage4, ICE mice exhibited visible signs of accelerated organismal aging (Figure 1). Specifically, young adult ICE mice phenocopied organismal aging phenotypes usually observed only by 24-30 months of age, such as lower body weight, reduced fat mass, decreased respiratory exchange rate, decreased activity, increased frailty, kyphosis, decreased bone density, and hair greying. System-specific age-mimicking phenotypes were also observed at the level of kidney architecture, impaired learning and increased glial cell activation in the brain, and decreased muscle function. ICE mice also exhibited accelerated DNAme epigenetic aging in multiple tissues. Thus, relatively faithful DNA damage in ICE mice seems to recapitulate many aspects of aging. To provide molecular insights into the epigenetic impact of the ICE system, the authors profiled chromatin states in ICE MEFs using mass spectrometry of histone modifications and ChIP-seq of select histone marks. Consistent with the disruptive effect of DNA damage signalling on chromatin5, the authors observed many chromatin remodelling events in ICE MEFs. For instance, there was an enrichment for neuronal-related genes among genes with decreased levels of the repressive histone mark H3K27me3 and indeed, ICE MEFs were more easily reprogrammed into neurons, which the authors hypothesize is the result of cell identity erosion in ICE cells.
Figure 1: The ICE mouse model recapitulates many aspects of aging.
The ICE system creates inducible double strand breaks at 20 unique sites in the mouse nuclear genome, that are repaired with minimal mutagenesis. ICE cells show age-related signatures in vitro and ICE mice phenocopy many aspects of aging in vivo. Aspects of ICE-induced phenotypes can be rescued with OSK (Oct4-Sox2-Klf4) transient overexpression. Created with BioRender.com
Finally, the authors attempted to rescue the ICE phenotypes using partial reprogramming with OSK factors, which they previously showed can rescue aged phenotypes in mouse retinal ganglion cells [RGCs]6. After OSK treatment, select phenotypes induced in ICE cells and tissues appeared rescued (e.g. marker gene expression, H3K9me3 levels, DNAme age acceleration). In addition, OSK expression rescued RGC gene expression in aged ICE mouse retina tissue, consistent with transcriptional rejuvenation.
This study makes a convincing case that the ICE model leads to accelerated organismal aging, suggesting that high mutational load after DSBs is not a necessary intermediate for progeroid-effects of genomic instability1,4. However, the notion that progeroid phenotypes are only directly mediated by observed changes to the epigenome (the so-called “Information Theory of Aging”) is not directly tested in this study. Importantly, the type of system used in this study (inducible nuclear localization of a restriction enzyme like I-PpoI or SacI) robustly induces the DNA-damage response and P53 signalling7-9, which can lead to cell senescence. Importantly, senescent cells secrete a panoply of pro-inflammatory factors, leading to a deleterious state of chronic sterile inflammation which can drive aspects of aging1. This study identified increased senescence markers in ICE cells2, suggesting that the secretory impact of senescent cells in the ICE model may need to be accounted for. Interestingly, transient nuclear localization of I-PpoI in mouse epidermal stem cells was recently found to promote selective elimination of damaged stem cells through differentiation10. Although stem cell niches were not directly queried in this study, loss of adult stem cell populations by ICE-activation mediated attrition could explain a number of age-related phenotypes in ICE mice, and can be addressed in future studies with single cell technologies. In summary, additional evidence will be required to prove or disprove the “Information Theory of Aging”. However, this study provided the field with a new model, whereby a short organism-wide burst of DSBs in adulthood can have a long-term detrimental impact on mice that resembles aging.
Acknowledgements
Figure created with BioRender.com. The authors thank Dr. Minhoo Kim for feedback on the manuscript. B.A.B. is supported by NIGMS R35 GM142395, NIA R01 AG076433, Simons Foundation award SF811217 and Pew Biomedical Scholar award #00034120.
Footnotes
Declaration of Interests
The authors have no conflict of interest.
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2023). Hallmarks of aging: An expanding universe. Cell 186, 243–278. 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Yang JH, Hayano M, Griffin PT, Amorim JA, Bonkowski MS, Apostolides JK, Salfati EL, Blanchette M, Munding EM, Bhakta M, et al. (2023). Loss of epigenetic information as a cause of mammalian aging. Cell 186, 305–326 e327. 10.1016/j.cell.2022.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watada E, Li S, Hori Y, Fujiki K, Shirahige K, Inada T, and Kobayashi T (2020). Age-Dependent Ribosomal DNA Variations in Mice. Mol Cell Biol 40. 10.1128/MCB.00368-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieckher M, Garinis GA, and Schumacher B (2021). Molecular pathology of rare progeroid diseases. Trends Mol Med 27, 907–922. 10.1016/j.molmed.2021.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Hauer MH, and Gasser SM (2017). Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev 31, 2204–2221. 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Brommer B, Tian X, Krishnan A, Meer M, Wang C, Vera DL, Zeng Q, Yu D, Bonkowski MS, et al. (2020). Reprogramming to recover youthful epigenetic information and restore vision. Nature 588, 124–129. 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkovich E, Monnat RJ Jr., and Kastan MB (2007). Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9, 683–690. 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Sturgill D, Tran AD, Sinclair DA, and Oberdoerffer P (2016). Controlled DNA double-strand break induction in mice reveals post-damage transcriptome stability. Nucleic Acids Res 44, e64. 10.1093/nar/gkv1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White RR, Milholland B, de Bruin A, Curran S, Laberge RM, van Steeg H, Campisi J, Maslov AY, and Vijg J (2015). Controlled induction of DNA double-strand breaks in the mouse liver induces features of tissue ageing. Nat Commun 6, 6790. 10.1038/ncomms7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato T, Liu N, Morinaga H, Asakawa K, Muraguchi T, Muroyama Y, Shimokawa M, Matsumura H, Nishimori Y, Tan LJ, et al. (2021). Dynamic stem cell selection safeguards the genomic integrity of the epidermis. Dev Cell 56, 3309–3320 e3305. 10.1016/j.devcel.2021.11.018. [DOI] [PubMed] [Google Scholar]