Abstract
Screening implantable biomaterials for antifibrotic properties is constrained by the need for in vivo testing. Here, we show that the throughput of in vivo screening can be increased by cellularly barcoding a chemically modified combinatorial library of hydrogel formulations. The method involves the implantation of a mixture of alginate formulations, each barcoded with human umbilical vein endothelial cells from different donors, and the association of the identity and performance of each formulation by genotyping single nucleotide polymorphisms of the cells via next-generation sequencing. We used the method to screen 20 alginate formulations in a single mouse and 100 alginate formulations in a single non-human primate, and identified three lead hydrogel formulations with antifibrotic properties. Encapsulating human islets with one of the formulations led to long-term glycaemic control in a mouse model of diabetes, and coating medical-grade catheters with the other two formulations prevented fibrotic overgrowth. High-throughput screening of barcoded biomaterials in vivo may help identify formulations that enhance the long-term performance of medical devices and of biomaterial-encapsulated therapeutic cells.
Keywords: High-throughput screening, Next-generation sequencing, Cellular barcoding, Foreign body response, fibrosis, Type-I diabetes, Cell Therapy
One-sentence editorial summary
The throughput of the in vivo screening of hydrogels for antifibrotic properties can be increased by tagging the biomaterials with cells and reading their genotype via next-generation sequencing.
Host immune response to implanted devices involves a complex set of inflammatory cascade events, followed by cellular and collagen deposition that wall off the implanted device from the host1–7. These reactions occur over weeks post-implantation and have been difficult to adequately model in vitro4,8,9. Notably, macrophages play a crucial role in the process by adhering to large implants and secreting several cytokines and chemokines that attract fibroblast deposition to the implanted materials, leading to fibrotic overgrowth of the biomaterial1,5,6,10–12. Fibrosis has been studied and reported as a major contributing factor to the failure of many implanted devices 5–7,11,13–15. Therefore, there is an urgent need to develop materials with improved biocompatibility to mitigate foreign body responses (FBR).
One of the most investigated biomaterials is alginate-based hydrogels16,17 which have been used for cell transplantation18,19, tissue engineering20,21, and as components of medical devices22,23. However, the clinical utility of alginate hydrogels has been hindered by host immune recognition, which limits the long-term durable function of alginate-based medical implants. Pre-clinical and human trials with alginate-based hydrogels have elucidated that the FBR can be further exacerbated by the presence of encapsulated allogeneic or xenogeneic donor tissue24–30. To address this challenge, various approaches, including chemical modification of surface and adjustments of implant geometry, have been developed to mitigate fibrosis in alginate-based capsules and other implants6,18,31,32. Previously, we developed a combinatorial synthesized alginate analog library, screened a subset, and identified lead compounds containing triazole chemical modifications6. Lead formulations exhibit low levels of inflammation, higher immune protection, reduced FBR and fibrosis in rodents6,19 and non-human primates (NHP)18, and led to the launch of recent human clinical trials (NCT04541628). However, the prior studies were low-throughput as they limited the screening to 6–8 new materials per rodent. Further, there are many triazole-containing alginate analogs (~10K) that can be synthesized and tested to explore new potent hydrogel formulations that prevent FBR.
High-throughput combinatorial experiments have enormous potential for rapidly identifying small molecules, biomaterials, or biologics with superior performance, especially where a biological response remains unexplored8,33–38. Such combinatorial screens are necessary for therapeutic development for cancer, diabetes, and other diseases. Conventional in vivo material screening approaches employ one animal or one implantation site for a single material, which takes tremendous implantation efforts and animal costs to evaluate a multitude of material candidates. Therefore, efficient in vivo methods are needed to increase the throughput and reduce the number of living animals. Physical39–41 or chemical42 barcoding systems can be used for this purpose. The size or shape of biomaterials can be adjusted to distinguish among different biomaterials7. Further, strategies were used to modify the chemical backbone by attaching fluorophores to determine each material’s identity39,42. However, those parameters can modulate host immune response, and it becomes difficult to differentiate the material-elicited immune response from external factor-elicited responses. Additionally, the number of distinct barcodes achievable with these techniques is limited to less than ten and hence can’t be considered as high-throughput. Genetic barcoding is a popular tool to accomplish high-throughput screening of biomaterials or nanoparticles for various applications35,43,44 by employing DNA barcodes to differentiate biomaterials. However, external factors that induce immunogenic responses can hamper the investigation of the material’s immune properties. There are some recent reports of using next-generation sequencing (NGS) for high-throughput screening approaches45–47. However, to our best knowledge, no in vivo high-throughput screening method was developed using cellular barcoding technologies to screen hydrogels that mitigate fibrosis. Hence, a significant medical need exists to develop a reliable and effective high-throughput screening approach to identify novel biomaterials that overcome this fundamental challenge of eliminating the FBR related to biomaterial grafting.
Here, we developed a novel high-throughput in vivo screening method that utilizes xenogeneic transplantation of hydrogel encapsulated human cells into pro-fibrotic C57BL/6J rodent or NHP models for evaluating multiple hydrogel biomaterials at a single implantation site using cellular barcoding (single or dual donors) techniques. This method comprises tagging each biomaterial by encapsulating distinct barcoding cells (single or dual donors) that uniquely represent the material inside hydrogel capsules made from new alginate analogs. The cellular barcoding technique uses 20 different unique human umbilical vein endothelial cells (HUVECs) as barcoding cells for encapsulation by new alginate analogs. Moreover, each HUVEC donor possesses a distinct genotype represented by unique genetic profiles of a set of single nucleotide polymorphisms (SNPs), which enables the identification of encapsulated donor HUVECs and subsequently the material by revealing the pattern of SNP genotypes through NGS. This approach with 20 different cell populations allows the barcoding of 20 distinct materials inside a single rodent. This NGS barcoding technique was employed to screen a large library of chemically modified triazole-containing alginate analogs in a mouse model with a material screening throughput over four times greater than conventional methods. The screening throughput was further expanded in an NHP model using a dual donor barcoding strategy that enables screening up to 400 distinct materials in a single NHP, raising the material screening throughput by over 20 folds.
Additionally, the lead immune-modulating hydrogel (Z4-A10) identified from the high-throughput screening was applied to transplant xenogeneic human pancreatic islets, successfully restoring long-term glycemia in a pro-fibrotic C57BL6/J mouse model by preventing fibrosis and maintaining long-term cell viability for almost three months. In addition, methacryloyl modification of two top lead structures (Met-Z1-A3 and Met-B2-A17) demonstrated lower deposition of fibrotic tissue when applied to medical-grade catheters. This work provides the foundation for proof-of-concept of this innovative high-throughput screening of immune-modulatory alginate hydrogels that prevent fibrosis in mouse and NHP models.
Results
Combinatorial second-generation biomaterial library development
Reports published earlier from our group demonstrated the identification of three lead triazole-containing alginate analogs from a combinatorically synthesized hydrogel library6. The first column of Extended Data Fig. 1 shows two top lead molecules (Z1-A34: hydrophilic and B1-A21: hydrophobic) reported in the previous study6. Structural analysis showed that the commonality between the lead analogs was the presence of a triazole ring (a heterocyclic five-membered ring with two carbon atoms and three nitrogen atoms). This observation suggests that triazole-containing molecules may mitigate FBR by regulating immune cell populations at the surface of these hydrogels, likely inhibiting their activation and interrupting the fibrotic processes.
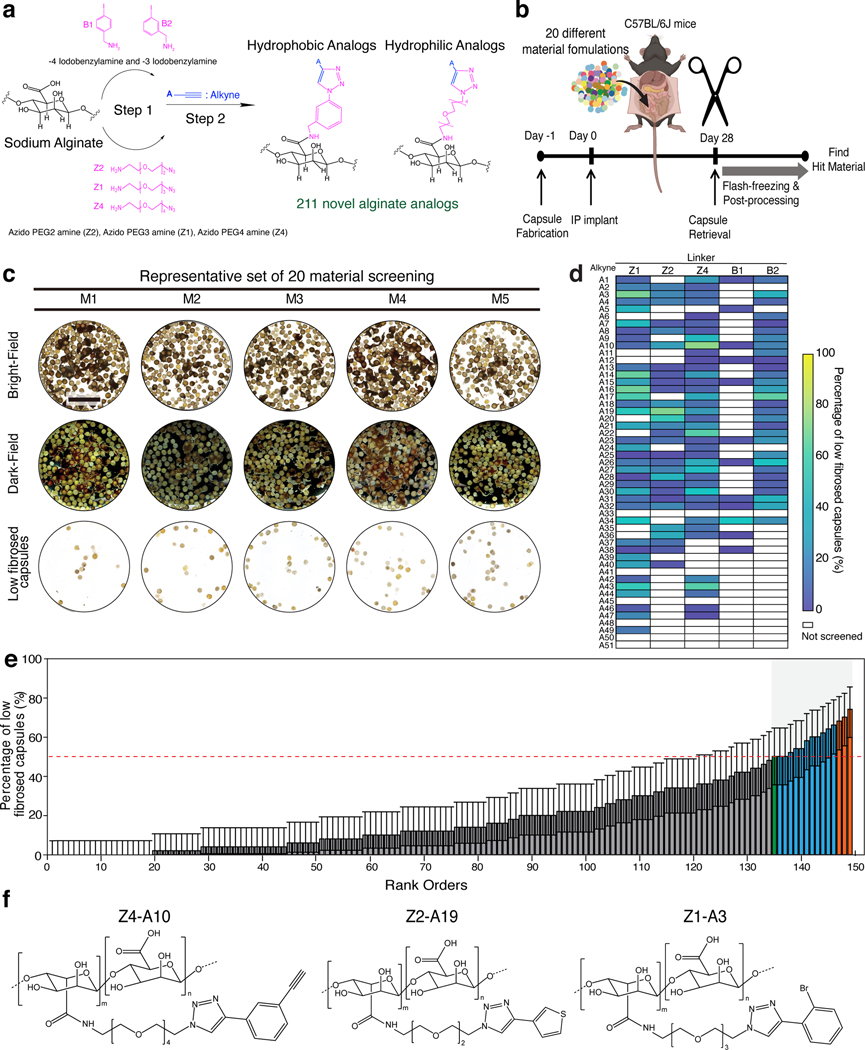
The combinatorial diversity of chemical molecular structures implies that additional alginate analogs with superior anti-fibrotic properties may exist. In this present study, a new library was designed by maintaining the structural similarity of the linkers (hydrophilic: Z1, Z2, and Z4; hydrophobic: B1 and B2) from the earlier reported leads (Z1-A34 and B1-A21)6. A total of fifty-one new alkynes (belong to the following classes: ethynyl benzene, ethynyl pyridine, ethynyl thiophene, substituted-1-yne, 1-yloxy propyne) were used. The second generation of 211 new alginate analogs (encapsulation materials) was synthesized by triazole surface modification (Extended Data Fig. 1). A combinatorial biomaterial approach was developed to generate a library of alginate-based hydrogels by covalently attaching small-molecule functionalities and keeping the triazole analogs common to all. Low molecular weight (MW), ultrapure UP-VLVG alginate with high guluronate (G) content (>60% G, ∼25 kDa MW, NovaMatrix) was used as the starting material (Supplementary Table. 1). Among the 211 unique alginate analog polymers, 150 analogs were generated from the combinatorial synthesis of three hydrophilic PEG-based linkers resembling the reported hydrophilic lead (Z1-A34) with 51 new alkynes, and 61 triazole-containing analogs from two hydrophobic linkers resembling the hydrophobic lead (B1-A21) (Extended Data Fig. 1, Supplementary Table. 2). The covalently attached triazole containing alginate analogs were characterized using elemental analysis and nuclear magnetic resonance spectroscopy (NMR) (Supplementary Fig. 1). Following confirmation of synthesis, purity, solubility, and gel-forming ability (gelation assay), 149 alginate analogs were selected for in vivo screening purposes (Extended Data Fig. 2a-c).
High-throughput materials screening using SNP genotype barcodes
We developed a high-throughput method to screen the combinatorial synthesized library of hydrogels in vivo using cellular barcoding to identify novel hydrogels that prevent fibrosis. The methods comprise (a) preparing a plurality of barcode cells derived from one or more subjects, wherein each composition of barcode cells comprises a unique profile of a plurality of SNPs that serve as a genetic barcode for each cell encapsulation material; (b) fabricating capsules using various encapsulation materials and barcode cells; (c) implanting the capsules in vivo into a test subject (mouse); (d) explanting the capsules after a set time; and (e) determining the sequence of the plurality of SNPs in the barcode cells of each explanted capsule, thereby identifying the cell encapsulation material of each capsule (Fig. 1a). Deep sequencing of 20 different HUVEC donors revealed unique genetic profiles of the selected 30 non-pathogenic SNPs: Each SNP locus could possess one of the three genotypes: homozygous for wildtype (WT), heterozygous, or homozygous for variant allele, and altogether the 30 SNPs formed unique genetic profiles that could unambiguously represent the HUVEC cell identify (Fig. 1b). Thus, by encapsulating a single donor HUVEC in each different hydrogel material, the material identity is uniquely tagged by cellular barcodes that could be read through sequencing post-implantation. The extraction method of gDNA from a single capsule was optimized (Supplementary Fig. 2) to increase DNA input for sequencing. Further, the workflow was established for NGS library preparation and material identifications (Extended Data Fig. 3-4). In our work, HUVECs are used for three main reasons: a) their intrinsically different genetic profiles are leveraged to barcode biomaterial libraries; b) the viability of encapsulated barcoding cells is an indicator of the biocompatibility of the associated biomaterial in vivo; c) the presence of xenogeneic cells in such transplantation study would induce a higher level of foreign body response and thus provide a more stringent screening environment.
Fig. 1 |. Cell barcoding strategy enables high-throughput materials screening.
a, Overall schematic diagram for the workflow of biomaterials screening: 20 different HUVEC donors were encapsulated with corresponding materials and implanted into mice for evaluating the anti-fibrotic property and biocompatibility for four weeks using NGS assay (created with BioRender.com). b, Twenty unique HUVECs have been sequenced via NGS to identify their specific SNPs, which can be used as a barcode to tag and identify different encapsulation materials in vivo. c-d, To prove all donors (H1-H20) can be deconvoluted by NGS, a mixture of 20 different capsules encapsulating 20 different HUVECs donors was implanted in NSG mice for four weeks. c, Bright-field and dark-field images of the pre-implant and post-implant; capsules were retrieved with minimal cell deposition, indicating no fibrosis, similar to pre-implant capsules. Encapsulated cells are still alive (live cells: green, dead cells: red) after 4weeks of implantation. Scale bar, 2 mm. d, The capsules from one mouse were analyzed with NGS, and 195 out of 200 capsules were successfully identified. Across the donors, the identified capsules percent are evenly distributed from donor 1 to donor 20.
To prove all donors (H1-H20, Supplementary Table. 3) can be deconvoluted by NGS, a mixture of 20 different hydrogel capsules (made from the library of 149 novel hydrogel analogs) encapsulating 20 unique HUVECs (10 capsules per material) were implanted in the intraperitoneal (IP) space of NSG (NOD SCID gamma) mice for four weeks. The NSG mouse lacks mature T cells, B cells, and natural killer cells to induce normal innate immunity function48,49. Screening in immunodeficient mice allows us to assess this cellular barcoding strategy with minimal host immune responses against implanted materials. Imaging of post-retrieval capsules showed minimal cell deposition, indicating a lack of fibrosis and high viability of encapsulated cells (Fig. 1c). The capsules from one mouse were analyzed with the NGS technique, and 195 out of 200 capsules (~96.5%) were successfully identified based on their unique cellular barcoding (Fig. 1d). The identified percentages were evenly distributed across donors. These results indicate that cellular barcoding using different HUVEC donors and NGS genotyping can be leveraged as a suitable strategy for determining biomaterial identity without altering material properties.
Chemically modified alginates reduce FBRs in immune-competent mice in mixed settings
The ability of the newly synthesized alginate analogs to mitigate fibrosis was evaluated (Fig. 2a). A total of 149 novel hydrogel formulations were screened using 20 different HUVEC donors. After 28 days of in vivo implantation in mice, the capsules were retrieved from the IP space, and post processes were performed to identify immune-protective materials (Fig. 2b). All explanted capsules were separated into three groups (L: Low, M: Medium, H: High fibrosed capsules) based on surface fibrosis levels under the microscope. Capsules in the low fibrosed (L) groups showed a clear surface with low fibrosis deposition. In contrast, the high fibrosed (H) group showed higher deposition of fibrotic tissues on capsule surfaces, resulting in the formation of aggregate due to fibrotic overgrowth between capsules. Capsules between two groups were assigned as medium fibrosed group (M). Only the L group capsules were selected for materials identification using NGS assay (Fig. 2c).
Fig. 2 |. High-throughput screening of combinatorially synthesized chemically modified alginates using unique cellular barcoding facilitates identifying new hydrogels with reduced fibrosis in immune-competent mice.
a, A library of immunomodulatory biomaterials; a total of 211 novel alginate analogs were synthesized. b, The mixtures of different materials were implanted in the same implantation site to increase screening throughput. NGS assay was used to determine material identity by demultiplexing the SNP genotype of encapsulated HUVECs. Typical implantation of 200 alginate capsules (~1.5 mm diameter, 10 capsules/material) allows the simultaneous evaluation of 20 different implanted materials, with sufficient independent capsules per material to enable statistical analysis. c, Representative results from one of the rounds. After four weeks of implantation, capsules were explanted. Clear capsules with low fibrosis (bottom row), similar to pre-implant capsules, were separated for further analysis. Scale bar, 10 mm. d, Heat map summarizing material screening for the entire alginate analogs. e, 149 new materials were screened, and corresponding lead materials were identified via NGS assay (Corresponding materials of rank order in the x-axis are listed in Supplementary Table 4). Error bars represent 95% confidence intervals from a binomial distribution (n=5 biological replicates). The average level of Z1-A34 (previous positive material, labeled as a green bar) was marked as a red dotted line. Top materials over Z1-A34 were labeled with a blue bar. f, Representative structures of top three lead alginate analogs (orange bars in e).
The percentage of low fibrosed capsules represented the material’s anti-fibrosis performance (Fig. 2e, Supplementary Table. 4). A total of 15 alginate analogs (blue and orange color bars in Fig. 2e), including one previously known material (Z1-A34, marked as the green bar in Fig. 2e), had above 50% of low fibrosed capsules, suggesting five or more capsules appeared clear with low fibrosis from 10 implanted capsules. The screening result was also presented as a heat map (Fig. 2d) to visualize the fibrosis outcomes of different combinations of alkynes and linkers. These results show that some of the alkynes (A3, A17, A19, A30, A43) displayed low fibrosis levels across the different linkers. These alkynes contain phenyl bromine, pyridine, thiophene, ethoxy, and cyclopropyl functionality, respectively (Supplementary Table. 2). Some similarities of these alkynes include a lack of branched structures and longer carbon chains, which makes these alkynes relatively compact. Additionally, these alkynes all contain a carbon ring structure, and most have an electronegative atom (O, N, S, Br) in or around the carbon ring.
Fig. 2f shows the chemical structures of the top three identified lead alginate analogs (orange bars in Fig. 2e) with the lowest FBR or fibrosis (Z4-A10, Z2-A19, and Z1-A3). All three new hydrogels contain triazole moiety in the backbone, which further supports the role of triazole in prohibiting fibrosis. Interestingly all top three lead materials have hydrophilic PEG linkers attached with the alginate, in addition to the functionalization of bromobenzene (Z1-A3), thiophene (Z2-A19), and ethynylbenzene (Z4-A10) to the triazole (Fig. 2f). Notably, in the top 15 leads, only two hydrophobic analogs were identified (B2-A17 and B1-A34), one of which was later used for catheter coating application.
Dual donor cells barcoding expands the high-throughput potential
To translate this novel barcoding strategy to screen a larger batch of anti-fibrotic biomaterials, a dual donor barcoding strategy was utilized, and its feasibility was tested in an NHP model. Only 20 different materials were implanted in one mouse. In contrast, in the NHP model, due to the larger capacity of implantable space, the material batch size can be increased to 100, resulting in a 5-fold increase in throughput. With single donor encapsulation, its throughput is limited by the number of available unique donors. Here, a new method using a dual donor barcoding strategy was devised to increase the screening throughput without purchasing and validating new cell donors. By mixing two different HUVECs at a ratio of 1:2 (Fig. 3a), 400 unique permutations of barcoding compositions were created with only 20 HUVEC donors (Fig. 3b), which significantly expands existing donors’ barcoding capacity. With this ratio, the convoluted variant allele frequencies (VAFs) now include seven possibilities (0, 1/6, 1/3, 1/2, 2/3, 5/6, and 1). Hence, the donor identification strategy is modified from direct comparison with individual donors to log-likelihood analysis (Extended Data Fig. 4). The likelihoods of all possible combinations were assessed from Gaussian distribution to determine the mixed donor identity. The mixing ratio of two donors was optimized (Supplementary Fig. 3), and small cohort screening (comparison of three materials) was performed to confirm the feasibility of a dual donor barcoding strategy (Extended Data Fig. 5).
Fig. 3 |. Dual-donors barcoding enables to scale up the materials screening in the NHP model.

a, Two HUVEC donors were mixed at a 1:2 ratio and encapsulated in various materials. b, With 20 HUVEC donors mixed at the ratio of 1:2, 20×20=400 distinct SNP profiles were created. c, 100 donor pairs were encapsulated with corresponding materials and implanted into IP space in a NHP for four weeks. Thirty capsules per material were used, and a total of 3000 capsules were implanted. d, The representative images of preimplant capsules. e, After four weeks, all the free-floating capsules in IP space were collected and used for material identification (white arrow: capsules with fibrotic tissue aggregation, yellow arrow: free-floating capsules). f, Summary of donor pair identification. Among a total of 503 selected capsules, 466 (92.6%) were identified with high confidence, 32 (6.36%) with lower confidence, and 5 (0.99%) capsules failed to be identified. g, Distribution of confidence level of analyzed capsules. Goodness is the difference between the log-likelihood of the most likely pair and the second most likely donor pair, and thus high goodness indicates a low chance of misidentification. Capsules at the upper right corner have higher confidence, and those with log-likelihood below −200 or goodness less than 10 are considered “less confident”. h, The chemical structure of the top 4 identified leads.
Following the development of 400 novel dual barcodes, a set of 100 unique triazole containing alginate analogs (randomly selected from the 211 newly synthesized analogs library (represented in Extended Data Fig. 1) encapsulated with the unique combinations of two different HUVECs (1:2 ratio) were implanted (30 capsules per materials) in the IP space of an NHP for four weeks (Fig. 3c). The representative images of pre-implant capsules showed the homogenous distribution of 1.5 mm capsules (Fig. 3d). Live/dead staining of pre-implant capsules confirms that cells were viable before implantation to NHP (Fig. 3d). After four weeks, all the free-floating capsules in IP space were retrieved and used for material identification (white arrow: tissue aggregated capsules, yellow arrow: free-floating capsules, Fig. 3e) using NGS assay. From 3000 implanted capsules, a total of 503 free-floating capsules were retrieved and used for NGS analysis, resulting in ~92.6% of sampled capsules being successfully identified with high confidence (Fig. 3f). The remaining 7.4% capsules were not confidently identified either because of insufficient sequencing depth (N=5, failed) or low barcode matching likelihood (N=32, less confident). The distribution of capsules in confidence space was plotted in Fig. 3g. This study successfully demonstrates the in vivo screening applicability and feasibility of 100 new materials in the NHP model with dual barcoding strategies. Four lead hydrogels were identified from 100’s that showed minimum fibrosis in NHP (Z1-A2, Z4-A11, Z1-A27, and Z2-A27, Fig. 3h). All these four lead hydrogels contain hydrophilic linkers. Interestingly, these four leads from NHP screening were not identified as the top 15 leads in the mouse study and hence were not utilized for further applications in this study.
Chemically modified alginates reduce FBRs in immune-competent mice in individual settings
Top 10 lead hydrogels from mouse screening were implanted individually in the IP space in a pro-fibrotic C57BL/6J mouse model to validate whether materials identified from mixed settings still outperformed individually. Since smaller capsules can provoke more rapid immune responses, this study used microcapsules with 300~400μm in size to test the selected lead hydrogels in difficult conditions7. Further, a negative control hydrogel (SLG20) and one positive control hydrogel reported earlier (Z1-A34)6 were used in this study. After two weeks of implantation, dark-field images showed less fibrosis deposition in lead materials than controls (Fig. 4a). The SLG20 control had more elevated immune responses, and most microcapsules were aggregated within fibrosis tissues, in contrast to selected lead materials, including Z4-A10 and Z1-A3. Surface fibrosis levels were determined by imaging capsules with macrophage and fibroblast markers and RT-qPCR analysis (Fig. 4b-c)6,50. From immunofluorescence imaging (Fig. 4b), Z4-A10 and Z1-A3 showed the least intensity of macrophage (CD68, green) and myofibroblast (α-SMA, red) markers compared with SLG20 control, indicating low fibrosis levels. Reverse transcription PCR analysis of fibrotic markers (α-SMA and Col1a1) revealed that lead materials have significantly lower expression of both markers, indicating lower fibrosis and reduced collagen deposition on capsule surface compared to SLG20 (Fig. 4c). Z4-A10 and Z1-A3 looked most promising in preventing the FBRs among all top leads and hence were considered for further applications, including delivery of xenogeneic human islets (Z4-A10) in diabetic rodents and coating of medical-grade catheters (Z1-A3). These results confirmed the anti-fibrosis effect of screened lead materials in individual settings, providing various immunomodulatory application opportunities.
Fig. 4 |. Lead hydrogels show low fibrosis intraperitoneally in C57BL/6J mice.

a, Representative dark-field images of preimplant and explanted microcapsules (300 ~ 400μm size) retrieved from IP space after two weeks. Scale bar, 2mm. b, Representative confocal images of explanted microcapsules; Capsules were stained with CD68 (green), DAPI (blue), and α-SMA (red) markers. c, RT-qPCR analysis to compare RNA expression in different materials. Expression of fibrotic markers (α-SMA and Col1a1) were normalized to SLG20 (control). All bar graphs are mean±s.e.m. of biological replicates (n=5). Two-way ANOVA with Bonferroni correction was used for statistical analysis (****P < 0.0001, SLG20 control vs. others).
Lead hydrogel restores long-term glycemia using xenogeneic human islets in an immunocompetent animal model
Our newly discovered anti-fibrotic alginate (Z4-A10, Supplementary Scheme. 5-6) hydrogels were used to encapsulate xenogeneic human islets. These formulations provide a highly porous and anti-fibrotic hydrogel outer membrane to enable long-term nutrient diffusion, high islet viability, and low fibrosis in vivo. Our high-throughput screening in mice used a high density of cell loading, ~30,000 HUVECs per capsule, to provide enhanced selection pressure for identifying materials that can protect densely packed encapsulated xenogeneic cells from rejection. For our diabetes correction studies, we maintained a similar cell density per capsule (15K-60K cells per capsule) to assess the efficacy of our lead formulation in enabling long-lasting viability and protection of pancreatic islets in STZ-induced C57BL/6J mice.
Capsules with three different densities (4K IEQ/mL of alginate, 8K IEQ/mL of alginate, and 16K IEQ/mL of alginates, Extended Data Fig. 6) of human islets were prepared using Z4-A10 alginates, the lead triazole containing alginate identified through the high-throughput screening (Fig. 2e-f). Previous lead Z1-A34 and control SLG20 capsules were prepared for comparison. The pre-implant dithizone staining and live/dead imaging of the capsule groups (Z4-A10 and control SLG20) demonstrate the viability of the islets (Fig. 5a). The Z4-A10 capsules at a density of 4K IEQ/mL demonstrate long-term restoration of euglycemia and maintain glycemic correction until 80 days of data recording with the average blood glucose (BG) levels below 250, considered as the BG level of a healthy mouse, at a fasting condition (Fig. 5b). However, the control SLG20 alginate at the same dose (IEQ density of 4K/mL) failed to maintain glycemic correction for more than four weeks. Intravenous glucose tolerance test (IVGTT) was performed after four hours of fasting on day 75, showing encapsulated islet cells restored normoglycemia to a rate comparable with healthy C57BL/6J mice (Fig. 5c). Further, the post-retrieval capsule images (dark-field) display minimal fibrotic overgrowth on the surface of Z4-A10 capsules compared with SLG20 (Fig. 5d). Dithizone staining also supports the long-term islet viability after 80 days of implantation (Fig. 5e). The concentration of human c-peptide, a surrogate biomarker for insulin production, was measured from the serum separated from mouse blood 80 days post-transplantation. Higher levels of c-peptide secretion were observed in the Z4-A10 group compared to SLG20, suggesting better improved long-term viability (Fig. 5f).
Fig. 5 |. Lead hydrogel encapsulating xenogeneic human islets demonstrates a diabetic reversal in immunocompetent C57BL/6J mice.

a, Representative images of pre-implant capsules. Z4-A10 capsules containing human islets at a density of 10 IEQ/capsule, 20 IEQ/capsule, and 40 IEQ/capsule, respectively. Z1-A34 and SLG20 capsule was used as control material. Dithizone staining indicates viable islets within the capsule matrix. After encapsulation, islets show good viability (live: green, dead: red). b, Blood glucose levels for both Z4-A10 and SLG20 groups (4,000 IEQ/mL density) were monitored until mice were euthanized (Two-way ANOVA with Bonferroni multiple comparisons, ****P < 0.0001 (SLG20 vs. Z4-A10)). c, IVGTT test with Z4-A10 capsule (4,000 IEQ/mL) implant group in diabetic mouse, and non-implant group in diabetic mice and non-diabetic mice (Two-way ANOVA with Bonferroni multiple comparisons, ns; not significant, ****P < 0.0001 (all comparisons)). d-e, Representative dark-field (d), and dithizone staining (red, e) images of explanted Z4-A10 and SLG20 capsules (4,000 IEQ/mL). f, Human c-peptide measurements at 80 days post-transplantation (SLG20 vs. Z4-A10). g-i, Blood glucose monitoring with high islets density groups: Z4-A10 capsules (g), Z1-A34 capsules (h), and SLG20 capsules (i). All error bars denote mean ± s.e.m of biological replicates; Two-way ANOVA with Bonferroni multiple-comparison correction; Z4-A10 (n=8), Z1-A34 (n=6), and SLG20 (n=6).
In high-density encapsulation groups, Z4-A10 capsules with 8K and 16K IEQ/mL concentration could maintain long-term glycemic control >50 days of function (Fig. 5g). In contrast, the previous lead Z1-A34 and control SLG20 group failed to maintain glycemic control for no more than 10 days of implantation (Fig. 5h-i). However, the previous lead (Z1-A34, Fig 5h) failed early with higher density groups (at 8K and 16K IEQ/mL), even though it maintained glucose control for 50 days with the lowest density group. Our results suggest that Z4-A10 is enhanced in protecting encapsulated islets from foreign body response, which maintains longer viability and function of grafts.
Lead anti-fibrotic materials show low fibrosis when coated with catheters
Towards testing the effectiveness of newly developed small molecules against preventing fibrosis in the context of other medical devices, medical-grade silicone catheters were plasma treated and coated with methacryloyl modified Z1-A3 (one new hydrophilic lead) and B2-A17 (one new hydrophobic lead) (Fig. 6a, Supplementary Scheme. 7-10). Silicone catheters are inherently hydrophobic in nature. Hence, we selected one top hydrophobic lead (B2-A17) in addition to one hydrophilic lead (Z1-A3) to find out if hydrophilicity/hydrophobicity plays a role in mitigating fibrosis. Additionally, we coated catheters with the previous hydrophilic lead Z1-A34 and a previously explored alternative anti-fibrotic coating using the zwitterionic MPC31. ToF-SIMS, XPS, and SEM were used to analyze the surface chemistry of the uncoated and coated catheters that confirmed the successful coating of the catheters (Fig. 6a-b, Extended Data Fig. 7a-d).
Fig. 6 |. Lead small molecules can be translated for medical applications, including catheter coating.
a, Chemical structures of the unmodified or coated with either Met-Z1-A3, Met-B2-A17, Met-Z1-A34, or MPC catheter. b, ToF-SIMS data for unmodified, Met-Z1A3, Met-B2-A17, Met-Z1-A34, and MPC modified catheters showing the area with normalized intensity (a.u.) by total ion intensity for the main peaks (CN-, Br-), indicating successful coating. c-d, Representative histology images of the measured fibrotic capsule for unmodified and coated catheters after 4 weeks (c) and 8 weeks (d). The thinner, less dense purple band of cells at the tissue-catheter interface of the coated catheters indicates a milder immune response. e-f, Quantification of fibrotic capsule thickness for unmodified and coated catheters. One-way ANOVA with Bonferroni correction was used for statistical analysis; after 4 weeks (e) **P = 0.002 (Z1-A3, n=5), ***P = 0.0002 (B2-A17, n=5), **P = 0.0011 (Z1-A34, n=4), **P = 0.0025 (MPC, n=4) for comparisons with unmodified control (n=5); after 8 weeks (f) ***P = 0.0489, ns; not significant for comparisons with unmodified control. All error bars denote mean ± s.e.m of biological replicates.
To test the fibrotic response on uncoated and small molecule coated catheters in a pro-fibrotic C57BL/6J mice model, these catheters were implanted in mice subcutaneous space with explants at four and eight weeks. Histology and imaging of the explanted catheters after four weeks showed that the unmodified catheter explant led to thicker dark purple tissue deposition (which is collagen-rich and indicative of fibrotic overgrowth23) between the catheter and the skin tissue compared to the small molecule-coated catheters (Met-Z1-A3 and Met-B2-A17, as well as the coated controls) (Fig. 6c-f). The results indicate that the small molecule coated catheters are generally more capable of preventing FBR than the unmodified catheters at earlier timepoints even though the effects are almost comparable with the earlier published leads and MPC. However, only the Met-Z1-A3 catheter maintained a significantly better anti-fibrotic performance than the unmodified catheter by the 8 weeks explant compared to other groups. It is also noticeable that the fibrotic capsule thicknesses at 8 weeks were much thinner than those at 4 weeks, which is reflected by previous findings in literature51. This suggests that the 4-week timepoint was a good initial test of the anti-fibrotic coatings, but differences between the small molecules’ anti-fibrotic properties become more apparent with the progression of the immune response. Raman spectroscopy was also performed on explanted catheters to assess the longevity of the small molecule coatings after extended in vivo implantation (Extended Data Fig. 7e-g). The presence of a peak at 1400 cm−1 wavelength shift when using a 785 nm laser was previously established as an indicator of a triazole-containing compound31. All catheter groups coated with a triazole-containing small molecule (Met-Z1-A3, Met-B2-A17, Met-Z1-A34) showed a noticeable 1400 cm−1 wavelength shift both pre-implant and post-explant for both timepoints. These results indicate that these three tested small molecules, including a hydrophilic lead, a hydrophobic lead, and a previously established lead molecule, all remain coated on the catheter implants over an extended period in vivo.
Discussion
Combined our results show the development of a novel in vivo high-throughput screening method using cellular barcoding to screen an extensive library of anti-fibrotic alginate analogs that mitigates FBR. Using chemical structural similarity and a combinatorial approach, a large library of alginate hydrogels was synthesized by keeping the triazole in all of them. We demonstrate that our cell barcoding approach coupled with NGS can screen up to 20 alginate materials in a single rodent and greater than 100 materials in a non-human primate. Given the vast diversity of HUVEC donors from the population, our only constraint is the volume of materials implanted within the given site in an animal.
Previous low-throughput studies could only screen 6–8 distinctive materials at a time to identify hydrogels with low levels of inflammation6. Additionally, hydrogels in those studies contained no immunologic cues from allogenic or xenogeneic cells. In our present work, using cellular barcoding and NGS, the screening ability can be expanded to 20 different materials per rodent, enabling a 4-fold increase in material screening throughput and a 96-fold increase in sequencing throughput by amending unique position barcodes to each capsule. To further raise the screening selection pressure, densely packed HUVECs were encapsulated with chemically modified alginates to form microspheres, then tested in the pro-fibrotic C57BL/6J murine model. This approach allows for a higher chance to discover materials with even better immune-protection abilities that could be proven to enable long-term protection of allogeneic or xenogeneic cells in larger animals and, ultimately, clinically translatable to human patients. With this approach, a total of 149 newly synthesized second-generation triazole-containing alginate analogs were screened, and several leads that mitigated FBRs were identified. Further, a dual donor barcoding strategy by mixing two different HUVECs was developed to expand the barcoding library to 400 different codes that were further successfully tested in murine and NHP models to scale up the material screening throughput. It is important to highlight that the materials screening can be scaled up based on the maximum implantable volume of capsules in mouse/NHP IP space. Accordingly, 200 capsules or 20 different materials can be screened per round, keeping at least 10 capsules per material to ensure enough independent capsules for statistical significance. This can be further enhanced to 40 materials if we keep just 5 capsules per material. In the case of NHP, based on a maximum 40 ml IP implantable volume, a maximum total of 400 materials can be implanted per round with at least 30 capsules per material. Hence, the screening of materials can be scaled up at least 6–7 fold in a mouse and over 50 fold in an NHP model.
Additionally, the lead hydrogels were tested individually using smaller microcapsules, which supported their ability to reduce fibrosis. Two of the identified leads, Z1-A3 and B2-A17, were used to coat medical-grade silicone catheters for 4 and 8 weeks to demonstrate their utility in preventing fibrosis in a mouse model. Moreover, previously published molecules (Z1-A34, zwitterionic MPC polymer) were used to benchmark the study against established materials preventing fibrosis. Results demonstrated the newly discovered lead (Z1-A3) performed better than earlier published molecules in preventing fibrosis over 8 weeks in a subcutaneous mouse model.
Transplantation of islet cells represents a promising therapy for type 1 diabetes, affecting approximately 1.6 million Americans, including nearly 200,000 children and adolescents - triggers insulin-producing pancreatic islet cells to be destroyed by the body’s immune system18,19,22,31. However, it presents many challenges, including delivering islets, maintaining viability, and minimizing fibrosis to protect the construct. Many research groups, including ours, demonstrated the successful transplantation of human stem cell-derived β (SC-β) cells to the immunocompetent as well as immunocompromised mice and NHP models18,19. Additionally, there are reports that use xenogeneic rat islet cells in an immunocompetent animal model18,19,22,31. However, human SC-β cells or xenogeneic rat islets are far less immunogenic than human pancreatic islets. In this present work, transplantation of xenogeneic human islets to the immunocompetent pro-fibrotic C57BL/6J models was demonstrated using anti-fibrotic alginate (Z4-A10) capsules with highly porous and anti-fibrotic protections to enable better nutrient diffusion, improved islet viability, and low fibrosis. The top lead hydrogel (Z4-A10) was employed to transplant xenogeneic human islet cells, demonstrating long-term restoration of glycemia in the diabetic mouse model even at a higher density than the current standard. Notably, this is among a few published reports that showed the implantation of high-density xenogeneic human islets in an immunocompetent animal model with the successful restoration of euglycemia, with long-lasting islets cell viability and minimum fibrosis on the surface of the implant construct52. Moreover, the previously reported lead molecule failed to maintain normal glycemia with high cell density beyond 10 days, indicating the utility of our newly identified lead molecules in preventing FBR and protecting cell viability. These results further support the development of an implantable cell therapeutic construct that eliminates the use of steroids or other immunomodulatory drugs to suppress the immune system from attacking islets.
Unlike previous studies that directly used synthetic DNA strands as barcoding material, our genetic barcodes used barcoding donor cells as a tool for high-throughput screening. The barcoding donor cells serve three purposes: 1) their intrinsically different genetic profiles that are leveraged to barcode biomaterial libraries; 2) the viability of encapsulated barcoding cells is an indicator of the biocompatibility of the associated biomaterial in vivo; 3) the presence of xenogeneic cells in such transplantation study would induce a higher level of foreign body response and thus provide a more stringent screening environment. This is the first report using cellular barcoding to perform high-throughput screening to identify materials that prevent foreign body response.
The top three lead small molecules (Z4-A10, Z2-A19, and Z1-A3) found through this high-throughput screening approach share similar structural features. For instance, all three lead molecules contain a hydrophilic PEG linker and a triazole motif connecting the linker and alkyne. Additional similarities exist between the alkynes themselves, namely that all three include a carbon ring structure, often with an electronegative atom (O, N, S, Br) in or around the carbon ring, and alkynes lack branched structures and longer carbon chains. These structural features also appear in anti-fibrotic small molecules reported by previous combinatorial screenings, indicating a possible pattern for identifying additional formulations6. The only hydrophobic lead identified was B2-A17, which contains hydrophobic phenyl methanamine as the linker and pyridine as the alkyne. It can also be observed that the hydrophilic PEG linkers (Z1, Z2, and Z4) are better compared to the hydrophobic linkers (B1 and B2), as 9 out of the top 10 leads were hydrophilic PEG linkers alginates. Moreover, the PEG3 linker (Z1) was most frequently identified in the top leads (5 times in the top 10 leads) compared to other PEG linkers (PEG2: Z2 and PEG4: Z4). With respect to material interaction, hydrophilic PEG linkers-based small molecule leads (Z4-A10, Z1-A3, Z2-A19) work much better when attached to the hydrophilic alginates for cell encapsulation. On the other hand, hydrophobic lead (B2-A17) showed better improvement in mitigating fibrosis when used to coat hydrophobic silicone catheters than hydrophilic lead (Z1-A3). These observations may support the direct correlation of the performance of the anti-fibrotic small molecules with the compatibility of the different materials, surfaces, and hydrophilicity/hydrophobicity. Some of the alkynes (A17, A34, and A43) were repetitive among the hits and may have higher potential than others. However, more structural and computational analyses are required to understand specific structural features’ exact role in their anti-fibrotic abilities.
In addition, our results show that the newly discovered chemically modified alginate formulations (Z1-A3, Z4-A10, and Z2-A19) were not significantly different in terms of mechanical properties in comparison to one another, nor in comparison to unmodified alginate. We do observe some changes in the permeability to molecules around 150kDa. However, these molecules were also coated onto non-porous catheters and were shown to be effective in reducing foreign responses. This result indicates that the in vivo responses are mainly driven by their chemical properties due to surface modification using small molecules.
One limitation of the study is due to genetic homology between humans and primates, DNA from attaching primate cells could be amplified with human SNP targets and thus skew the signals. Design of SNP targets that differentiate species or additional bioinformatic species distinction is required to remove cross-species reads. Another limitation of this study is the relatively low DNA yield of the desired cells (HUVECs) from the capsules with higher fibrosed ones. In this study, we considered only low fibrosed capsules to find lead molecules. However, this can cause a temporary barrier if we target identifying all the single capsules, even with highly fibrosed ones that go into a mouse or primate. To solve this, more work is currently undergoing to increase the DNA output, which will enable our ability to identify every single material being implanted. Lastly, the nature of this rapid screening procedure does not leave time for investigating the long-term stability, toxicity, or biocompatibility of the lead small molecules. These unexplored aspects can be evaluated either with an additional mouse study or during future primate studies to collect more impactful results.
In summary, we developed a new high-throughput in vivo screening method in rodents’ models for testing multiple biomaterials at a single implantation site using cellular barcoding and NGS analysis. A total of 211 newly synthesized chemically modified triazole-containing alginate analogs were screened using single and dual barcoding strategies to identify several novel lead molecules that mitigate FBR. One of the molecules (Z4-A10) determined from the mouse screening study was applied to transplant xenogeneic human pancreatic islets, successfully restoring long-term glycemia and preventing fibrosis in the C57BL/6J mouse model. Further, methacryoyl modifications of two other lead molecules (Z1-A3 and B2-A17) were employed to coat medical-grade catheters, demonstrating reduced fibrotic tissue deposition. Additionally, the dual barcoding strategy was successfully validated to screen hundreds of combinatorial synthesized hydrogels in a single primate, exponentially reducing the number of living subjects and the time of the entire study. Overall, using this robust cellular barcoding strategy, a novel high-throughput screening method was established to identify new materials that mitigate the foreign body response and improve the long-sustaining performance of implanted products. The clinic-focused applications of these novel materials support their possible future utilization in various biomedical applications, including cell therapies for cancer and diabetes.
Methods
Materials/Reagents
All chemicals were from Sigma Aldrich (unless mentioned otherwise), USA, and used without further purification. Alginates used in this study are described in Supplementary Table. 1.
Generic strategy for the synthesis and characterization of alginate
To generate high-resolution structure-function relationships, we synthesized a library of 211 next-generation analogs (containing the crucial triazole ring) that incorporate additional structural changes, such as chain extensions and differential substitution.
Amide coupling reactions were performed using one equivalent of UP-VLVG alginate and one equivalent of amine linkers (5 different amine linkers) in the presence of a coupling agent of 0.5 equivalent of 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride resulting in 5 distinct amine linker conjugated alginate polymers. These linker-modified alginate derivatives were purified by dialysis for three days (in saline and water) using a 10–12 kDa dialysis membrane followed by lyophilization. These five molecules were characterized by NMR and elemental analysis for their purity and % modification of the starting alginates. In the following step, one equivalent of alkynes (51: alkynes) was conjugated with the appropriately modified alginates by copper-catalyzed click reactions. A total of 211 triazole-containing alginate derivatives were generated from the synthesis. All 211 different alginates were purified by dialysis, followed by lyophilization, and chemically characterized by NMR.
Synthesis of small molecules
Synthesis of Z4-A10 amine:
1,3-diethynylbenzene (1 equiv., 4g, 32 mmol) was added in a 1000 mL round bottom flask containing 450 mL of 5:1 methanol: water (375 mL methanol and 75 mL water) followed by dropwise addition of tris((1-benzyl-4-triazolyl)methyl)amine (0.25 equiv., 2.932 gm, 5.52 mmol) dissolved in 30 mL of 5:1 methanol: water (25 mL methanol and 5 mL water) and stirring for 15 minutes. This was followed by addition of triethylamine (0.25 equiv., 0.77 mL, 5.52 mmol) and copper iodide (0.1 equiv., 422 mg, 2.22 mmol). The reaction flask was cooled to 0°C for 15 minutes while being purged with argon before adding 11-azido-peg-4-amine (1 equiv., 6.92 mL, 32 mmol). The reaction was stirred for 5 mins at room temperature, followed by overnight stirring at 55 °C. The reaction mixture was filtered over Celite, and the solvent was removed using a rotavap. The crude reaction was then purified by liquid chromatography with dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% on a 120 gm ISCO silica column.
Synthesis of Z1-A3 amine:
1-bromo-2-ethynylbenzene (1 equiv., 2g, 11 mmol) was added in a 250 mL round bottom flask containing 180 mL of 5:1 methanol: water (150 mL methanol and 30 mL water) (5:1 methanol: water) followed by dropwise addition of tris((1-benzyl-4-triazolyl)methyl)amine (0.25 equiv., 1.466 gm, 2.76 mmol) dissolved in 24 mL of 5:1 methanol: water (20 mL methanol and 4 mL water) and stirring for 15 minutes. This was followed by addition of triethylamine (0.25 equiv., 0.385 mL, 2.76 mmol) and copper iodide (0.1 equiv., 211 mg, 1.11 mmol). The reaction flask was cooled to 0°C for 15 minutes while being purged with argon before adding 11-azido-peg-3-amine (1 equiv., 2.193 mL, 11.05 mmol). The reaction was stirred for 5 mins at room temperature, followed by overnight stirring at 55 °C. The reaction mixture was filtered over Celite, and the solvent was removed using a rotavap. The crude reaction was then purified by liquid chromatography with dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% on a 120 gm ISCO silica column.
Synthesis of B2-A17 amine:
3-iodobenzylamide (1 equiv., 4 g, 14.8 mmol) was added in a 50 mL round bottom flask containing 24 mL of methanol followed by sequential addition of triethylamine (2.4 equiv., 3.6 gm, 35.62 mmol), sodium azide (2 equiv., 1.93 g, 29.68 mmol), water (6 mL), copper iodide (0.15 equiv., 423.89 mg, 2.22 mmol), sodium ascorbate (0.1 equiv., 293.95, 0.1 eq, 2.97 mmol), 1.83 ml trans-N-N′-dimethylcyclohexene-1,2-diamine (0.2 equiv., 422.12 mg, 2.97 mmol). The mixture was evacuated and flashed with argon three times, and (4-(4-(pyridine-2-yl)-1H-1,2,3-triazol-1-yl)phenyl)methanamine(1 equiv., 3.72 g, 14.8 mmol) was added. The reaction was stirred for 5 mins at room temperature, followed by overnight stirring at 55°C. The reaction mixture was filtered over Celite, and the solvent was removed using a rotavap. The crude reaction was then purified by liquid chromatography with dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% on a 120 gm ISCO silica column.
Synthesis of B1-A51 amine:
(4-iodophenyl)methanamine (1 equiv., 2g, 15.13 mmol) was added to 90 mL of methanol. Triethylamine (2.4 equiv., 3.67 g, 36.3 mmol) and sodium azide (2 equiv., 1.97 g, 30.3 mmol) were added to the reaction flask. 40 mL of ultrapure water was added to the flask after everything previously added was dissolved. Sodium ascorbate (0.1 equiv., 300 mg, 1.5 mmol) and copper iodide (0.15 equiv., 432 mg, 2.27 mmol) were added to the flask. The flask was argon purged by bubbling argon through the mixture for 15 minutes. Trans-N,N’-dimethylcyclohexane-1,2-diamine (0.2 equiv., 430 mg, 3.03 mmol) and 1-ethynyl-2-methoxybenzene (1 equiv., 2 g, 15.13 mmol) were added to the flask. The reaction was stirred for 5 mins at room temperature, followed by overnight stirring at 55 °C under argon. The reaction mixture was filtered over Celite, and the solvent was removed using a rotavap. The crude reaction was then purified by liquid chromatography with dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% on a 120 gm ISCO silica column.
Synthesis of Z1-A34 amine:
4-Propargylthiomorpholine 1,1-Dioxide (1 eq.) was added to a 250 mL round bottom flask and dissolved in methanol: water mixture (5:1). Consequently, Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (0.25 eq.), Triethylamine (0.25 eq.), and copper iodide (0.1 eq.) were added. The reaction mixture was purged with argon for 15 mins and cooled to 0°C, after which 11-azido-3,6,9-trioxaundecan-1-amine (1 eq., 6.30 g, 28.86 mmol) was added. The reaction mixture was stirred at room temperature for 15 mins and afterward heated to 55°C for overnight. The reactions were cooled to room temperature and filtered through Celite to remove any insoluble parts. The filtrate was dried using a rotavap under reduced pressure with silica. The crude reaction was then purified by liquid chromatography with dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% on a 120 gm ISCO silica column and further characterized with ESI mass and NMR mass spectroscopy.
Synthesis of methacryloyl Z1-A3 (Met-Z1-A3):
2-(2-(2-(2-(4-(2-bromophenyl)-1H-1,2,3-triazol-1-yl)ethoxy)ethoxy)ethoxy)ethan-1-amine (1 equiv., 1.4 gm, 3.51 mmol) was added to a 50 mL round bottom flask and purged with argon for 3 times (10 mins each). 25 mL of anhydrous dichloromethane was added, followed by triethylamine (1.5 equiv., 734 μL, 5.265 mmol) and methacryloyl chloride (1.5 equiv., 509.6 μL, 5.265 mmol) in a dropwise manner under argon atmosphere. The reaction was stirred for 5–6 hours, followed by purification on a 40 gm ISCO silica column using dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% to obtain ~1 gm of colorless solid (Met-Z1-A3). The compound was characterized by mass spectroscopy and NMR to confirm the mass and purity.
Synthesis of methacryloyl B2-A17 (Met-B2-A17):
(3-(4-(puridin-2-yl)-1H-1,2,3-triazol-1-yl)phenyl)methanamine) (1.2 gm, 4.78 mmol, B2-A17) was added to a 50 mL round bottom flask and purged with argon three times (10 mins each). 25 mL of anhydrous dichloromethane (30 mL) was added, followed by triethylamine (1.5 equiv., 7.17 mmol, 999.6 μL) and methacryloyl chloride (1.5 equiv., 7.17 mmol, 693.99 μL) in a dropwise manner under argon atmosphere. The reaction was stirred for 5–6 hours, followed by purification on a 40 gm ISCO silica column using hexane/ethyl acetate (1:0 to 0:1) to obtain ~1 gm of colorless solid (Met-B2-A17). The compound was characterized by mass spectroscopy and NMR to confirm the mass and purity.
Synthesis of methacryloyl Z1-A34 (Met-Z1-A34):
4-((1-(2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)-1H-1,2,3-triazol-4-yl)methyl)thiomorpholine 1,1-dioxide (1 equiv., 1.3 gm, 3.32 mmol) was added to a 50 mL round bottom flask and purged with argon for 3 times (10 mins each). 25 mL of anhydrous dichloromethane was added, followed by triethylamine (1.5 equiv., 692 μL, 4.98 mmol) and methacryloyl chloride (1.5 equiv., 480.7 μL, 4.98 mmol) in a dropwise manner under argon atmosphere. The reaction was stirred for 5–6 hours, followed by purification on a 40 gm ISCO silica column using dichloromethane: ultra (22% MeOH in DCM with 3% NH4OH) mixture 0% to 40% to obtain ~1.2 gm of colorless solid (Met-Z1-A34). The compound was characterized by mass spectroscopy and NMR to confirm the mass and purity.
Functionalization of alginates with small molecules
Functionalization of alginate with Z4-A10 amine:
1.5 g of UP-VLVG (1 equiv) was dissolved in 45 mL of water. Then 7.65 mmol of the Z4-A10 (1 eq) amine was dissolved in 22.5 mL acetonitrile and added to the mixture. Following it, an aqueous solution of 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (0.75 eq) was added dropwise to the mixture. The reaction was stirred overnight at 55°C. The solvent was removed under reduced pressure, and the solid was dissolved in water. The solution was filtered through a pad of cyano-functionalized silica. The solution was then dialyzed against a 10,000 MWCO pretreated dialysis tubing in DI water for three days. The dialyzed solution was frozen at −80°C and lyophilized until dry.
Functionalization of alginate with B1-A51 amine:
1.5 g of UP-VLVG (1 equiv) was dissolved in 45 mL of water. Then 7.65 mmol of the B1-A51 (1 eq) amine was dissolved in 22.5 mL acetonitrile and added to the mixture. Following it, an aqueous solution of 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (0.75 eq) was added dropwise to the mixture. The reaction was stirred overnight at 55°C. The solvent was removed under reduced pressure, and the solid was dissolved in water. The solution was filtered through a pad of cyano-functionalized silica. The solution was then dialyzed against a 10,000 MWCO pretreated dialysis tubing in DI water for three days. The dialyzed solution was frozen at −80°C and lyophilized until dry.
Functionalization of alginate with Z1-A34 amine:
2g (1eq) of UP-VLVG (BP-1903–04; Novamatrix) was dissolved in water (75 mL). Then Z1-A34 small molecule (3.99 g, 10.20 mmol, 1eq) was dissolved in water under vortexing. The pH of the Z1-A34 solution is adjusted to 7.4 using HCl. Then Z1-A34 aqueous solution is slowly added to the UP-VLVG solution while stirring. Then a solution of (4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium chloride) (DMTMM, 0.5 eq.) was added dropwise to the mixture of UP-VLVG and Z1-A34. The reaction was heated to 55°C and stirred overnight. The solution was filtered through a pad of cyano-functionalized silica. The solution was then dialyzed against a 10,000 MWCO pretreated dialysis tubing in a beaker using saline (2 days) and mili-Q water (3 days). The dialyzed solution was frozen at −80°C and lyophilized until dry.
Gelation assay
The 211 alginate analogs were tested for their ability to efficiently crosslink in the presence of barium to form a hydrogel in a fluorophore retention assay. 100 μL of a 1% (w/v) solution of modified alginate polymers in 0.9% saline were dispensed into a 96-well plate. 1 μL of a 1% (w/v) solution of rhodamine B in DMSO was added to each well, followed by 50 μL of a 1 M barium chloride solution, then incubated for 10 mins on an orbital shaker. The wells were washed three times with deionized water, followed by fluorescence measurements (ex:540 nm/em:580 nm). A formerly synthesized UP-VLVG alginate was used as a positive control, and deionized water as a negative control.
Mechanical Testing
Mechanical testing of capsules made with lead hydrogel analogs (Z1-A3, Z4-A10, Z2-A19) and controls (unmodified alginate SLG20, previous lead Z1-A34, and negative control alginate analogs B1-A51 and Z1-A28) was performed with a Texture analyzer. Capsules were individually compressed using a cylindrical tip, and work-to-burst was calculated for each capsule group with n=20 capsules.
Permeability Test
Lead hydrogel analogs (Z1-A3, Z4-A10, Z2-A19) and controls (unmodified alginate SLG20, previous lead Z1-A34, and negative control alginate analogs B1-A51 and Z1-A28) were assessed for permeability to various size FITC-dextran molecules. Alginate solutions were prepared as a 70:30 mixture of the named alginate at 4 wt.% with 3 wt.% SLG100, except for SLG20 being 1.4 wt.% and having no SLG100. Saturated stock solutions of 25 mg/mL were made for four different molecular weights of FITC-dextran: 40 kDa (FD40S, Sigma), 70 kDa (FD70S, Sigma), 150 kDa (FD150S, Sigma), 500 kDa (FD500S, Sigma), and 2 MDa (FD2000S, Sigma). Capsules were made for each combination of FITC-dextran and alginate by mixing 950 uL of the alginate solution with 50 uL of the FITC-dextran and then electrospraying into 20 mM barium chloride crosslinker. The capsules were crosslinked for fifteen minutes, washed twice with HEPES buffer, and then placed in Krebs buffer on ice to minimize diffusion until all groups were prepared.
Capsules were aliquoted into black 96-well plates, washed with Krebs buffer to remove any external FITC-dextran, and finally incubated in 100 uL of Krebs buffer for the duration of the study. For each specified timepoint (5 min, 2 h, 4 h, 8 h, 24 h), n = 5 replicates were prepared for each group of capsules. A replicate here is a well with five capsules from a specific capsule group (e.g., five capsules consisting of 40 kDa-loaded SLG20 in 100 uL Krebs buffer). The plates were covered in aluminum foil and continuously shaken on a plate shaker at 400 rpm. At the specified timepoints, 50 uL of the incubation buffer was collected from all groups, with n = 5 replicates per group, and transferred to an empty black 96-well plate. A Tecan Infinite M Plex plate reader was then used to measure the fluorescence of the wells, with any samples that read as overflow being diluted until they were readable.
The saturated FITC-dextran stock solutions were serially diluted to obtain standard curve equations for relating the measured fluorescence to the concentration of FITC-dextran. Note that the different FITC-dextrans had differing fluorescent intensity levels at the same concentration and were therefore normalized to account for this when calculating concentrations from fluorescence. To calculate the percent retention of FITC-dextran in the various capsule groups over time, the theoretical maximum amount of FITC-dextran was calculated using the experimental capsule diameter (1.5 mm) and the known loading concentration. Percent released of the FITC-dextran at a given timepoint was then calculated as the ratio of measured concentration (converted from fluorescence with the standard curves) to theoretical maximum fluorescence within five capsules times 100. Percent retention was then calculated by one minus percent released.
Coating of catheters with Met-Z1-A3/Met-B2-A17/Met-Z1-A34 using surface plasma cleaning
Barium impregnated silicone rubber catheters (Codman® HOLTER® Atrial Distal catheter, Ref# 821670) were cut into identical length pieces using a surgical blade. To perform the chemical modifications, the catheters were plasma treated three times at 1 min intervals, rotated between exposures to cover all sides, using high RF frequency at 600–700 mbar pressure (Expanded Plasma Cleaner, Harrick Plasma, P/N PDC-001). The plasma-treated discs were immediately dropped into a 0.02 M solution of Met-Z1-A3/Met-B2-A17/Met-Z1-A34 in 5% DMSO in toluene. The reaction was kept under stirring for 2 hours, and the implants were thoroughly washed three times in methanol, three times in ethanol, three times with sterile grade water, and finally again with sterile grade ethanol. Finally, the implants were vacuum-dried overnight.
Coating of using MPC zwitterionic polymer
The MPC-coated catheters were prepared using a previously established method31. Pieces of unmodified catheter tubing were placed in a round bottom flask with a stir bar and were continuously purged with nitrogen. During this time, 2g of MPC monomer (6.8 mmol) was added to 12 mL of a 1:1 mixture of methanol and MilliQ water in a 25 mL round bottom flask. This flask was sealed with a rubber stopper before being placed on ice and degassed with nitrogen for 20 minutes. The ice bath was then removed before adding 9.8 mg of CuBr (0.069 mmol) and 21.2 mg of 2,2′-bipyridine while the mixture was continuously purged with nitrogen. The mixture was then charged into the catheter flask with a cannula. The reaction was performed at room temperature for three hours under continuous nitrogen flow. The coated catheters were thoroughly washed three times in methanol, three times in 10x PBS, three times in sterile grade water, and finally again with sterile grade ethanol. Finally, the implants were vacuum-dried overnight.
Scanning electron microscopy of coated and unmodified catheters
Unmodified and modified catheter samples were taped to an SEM pin mount, lightly treated with an air gun to remove particulates, and Au sputtered using a Denton Desk V Sputter system (Rice SEA). This was repeated for each coated catheter group. The sputtered catheters were imaged using a JEOL 6500F Scanning Electron Microscope.
XPS of unmodified and coated catheters
X-ray photoelectron spectroscopy (XPS) is a surface-sensitive spectroscopic method that quantitatively measures the elemental composition at the surface (within the 6 nm range) of any material when the sample is irradiated by mono-energetic X-rays causing emissions of photoelectrons from the material’s surface. The elemental compositions of uncoated, Met-Z1-A3 coated, Met-B2-A17 coated implants were analyzed by PHI Quantera XPS. A Survey technique at 1100eV, 200 μm spot size, 50W 15 kV ion gun neutralization was used for the analysis.
ToF-SIMS of unmodified and coated catheters
Positive and negative high mass resolution spectra were performed using a ToF-SIMS NCS instrument, which combines a TOF.SIMS5 instrument (ION-TOF GmbH, Münster, Germany) and an in-situ Scanning Probe Microscope (NanoScan, Switzerland) at Shared Equipment Authority from Rice University. A bunched 30 keV Bi3+ ions (with a measured current of 0.2 pA) was used as a primary probe for analyzing a field of view of 250 × 250 μm2, with a raster of 128 by 2128 pixels and by respecting the static limit of 1.1012 ions/cm2 not to damage the surface. A charge compensation with an electron flood gun has been applied during the analysis, and an adjustment of the charge effects has been operated using a surface potential. The cycle time was fixed to 100 μs (corresponding to m/z = 0 – 1102 a.m.u mass range).
Raman spectroscopy of unmodified and coated catheters (Pre-implant and post-explant)
Raman spectroscopy was performed with a Renishaw inVia Raman Microscope, a 785 nm laser for spectrum collection, and a 50x lens for focusing on the catheter surface. Unmodified and coated catheters were analyzed pre-implant, after the 4-week explant, and after the 8-week explant to assess the longevity of the small molecule coatings. The explanted catheters were washed with DI water and 1% SDS to remove attached proteins, cells, and lipids that could interfere with reading the catheter surface31.
Cell culture and expansion
Human umbilical vein endothelial cells (HUVECs, CC-2517, LONZA, MD, USA) from 20 different donors (Supplementary Table. 3) were cultured in VascuLife® VEGF medium complete kit (LL-0003, Lifeline Cell Technology, CA, USA). The HUVECs were sub-cultured for expansion and maintained in a humidified incubator at 37°C in a 5% CO2 atmosphere. The media was changed three times every week.
20 Donors’ SNP profiles
A 30-plex single nucleotide polymorphism (SNP) panel was designed to uniquely identify HUVEC donors through multiplex PCR reactions and next-generation sequencing (NGS). The SNP loci were downloaded from the 1000 Genomes database, and the genomic sequences were fetched from the National Center for Biotechnology Information (NCBI) website. The SNP were selected such that the minor allele’s population allele frequency was 10% and 90%, and their loci were evenly distributed across chromosomes 1–22.
Genomic DNA (gDNA) was extracted from 20 different donors of HUVECs using a DNeasy kit (Qiagen, catalog #69054). For each donor, 100 ng of gDNA was added to 50 μL of PCR reaction mix with a concentration of 50nM for each primer and Phusion Hot Start Flex 2X Master Mix (NEB, catalog #M0536L). The PCR reaction condition included an activation at 98°C for 30 sec, 20 cycles of denaturation at 98°C for 30 sec, annealing at 63°C for 2 min, and extension at 72°C for 1 min, and completed the reaction with incubation at 72°C for 5 min (shorten as 98°C: 30s- (98°C:10s-63°C:2min-72°C:1min) x 20 – 72°C:5min-4°C: hold). The PCR products were purified using Monarch PCR & DNA Cleanup Kit (5 μg) (NEB, catalog #T1030S). The purified PCR products were then prepared for NGS on the Illumina Miseq platform using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, catalog #E7645S) according to the manufacturer’s protocol. The library quantification and quality control were performed on Agilent 2100 Bioanalyzer. Then deep sequencing was performed at approximately 5000x depth to establish SNP profile for 20 different HUVEC donors.
HUVEC encapsulation within modified alginate
Capsule fabrication for mice implant:
Twenty different materials were tested for each round of screening, and a total of 149 materials were screened in C57BL/6J mice. Therefore, twenty different HUVECs donors were used to create corresponding cell barcodes for each material every round. Modified alginates were initially dissolved at 3–5% w/v in 0.8% saline and then blended with 3% w/v SLG100 (also dissolved in 0.8% saline) at a volume ratio of 70% modified alginate to 30% SLG100. Alginate solutions were sterilized by filtration through a 0.2-μm filter. Immediately before encapsulation, the cultured HUVECs were centrifuged at 250G for 5 minutes and washed with Ca-free Krebs buffer (4.7 mM KCl, 25 mM HEPES, 1.2 mM KH2PO4, 1.2 mM MgSO4∙7H2O, 135 mM NaCl). After washing, cells were centrifuged again, and all supernatants were aspirated. The cell pellet was then resuspended in alginate solution at the cell density of 5×106 cells per 0.5 mL alginate solution (~30,000 cells/capsule). Each HUVECs donor was encapsulated with their corresponding modified alginate solutions. Alginate capsules were made using an electro-spraying machine (Pump 11 Pico Plus, Harvard Apparatus, MA, USA). 18G blunt-tipped needle was attached to a 1-mL Luer-lock syringe containing the alginate solution, clipped to a syringe pump oriented vertically over a 150 mL of crosslinking solution bath (20mM BaCl2, 250mM D-Mannitol, 25mM HEPES with 0.01 v/v % tween 20). A voltage generator was attached to the needle tip and grounded to the crosslinking bath. The settings of the syringe pump were 5 mL/hr flow rate at 15~20 cm height. Cell density per capsule was maintained by adjusting a voltage between 5.5 and 7 kV. After the capsules were formed in the crosslinking bath, they were then collected and washed three times with HEPES buffer (25 mM HEPES (Gibco, Life Technologies, California, USA), 1.2 mM MgCl2×6H2O, 4.7 mM KCl, 132 mM NaCl). Capsules were washed three times with cell culture medium and cultured overnight in a 37°C incubator for transplantation. Ten capsules of each material were aliquoted and mixed into one 2 mL tube, and this mixture of 200 capsules was used for implantation. Immediately before implantation into the peritoneal cavity of mice, the capsules were washed an additional two times with 0.9% saline. All materials were observed under bright-field microscopy to verify the homogeneity of cell density and capsule size.
Microcapsule preparation and implantation:
Top 10 lead materials identified from mice screening and two controls (SLG20 and Z1-A34) were used for microcapsules implantation. Modified alginates were dissolved at 3~5% w/v in 0.8% saline and blended with 3% w/v SLG 100 at 70:30 ratio. SLG20 were dissolved at 1.4% w/v in 0.8 saline. Formulated alginate solutions were used to make 300~400 μm size capsules. Encapsulation procedures were the same with 1.5 mm size capsules, except that a 30G needle was used for microcapsules with a 200 μl/min flow rate. After washing, 400 μL of microcapsules were aliquoted and implanted into mice IP cavity space for two weeks.
Capsule fabrication for NHP implant:
By mixing two different HUVECs donors at specific ratios, 400 different donor combinations from twenty different donors can be created to tag the biomaterials. To confirm the feasibility of mixed donor identification, different ratios (1:2, 1:3, and 1:4) were tested, and determined what ratios are detectable via NGS. A total of nine combinations were made with the three cell lines (Supplementary Fig. 3a). Once the correct ratios were aliquoted in their respective test tubes, nine different ratio capsules were created using SLG20, followed by the encapsulation protocol mentioned in the previous section. Including 20 single HUVECs donors, a total of 400 combinations of cell barcodes can be generated, and 100 cell barcodes among 400 were used for the NHP study.
A total of 100 cell barcodes were created, and these mixtures of HUVECs were encapsulated with 100 different materials, respectively. Cell density per capsule was fixed at 60,000 cells/capsule, which included 20,000 cells of donor 1 and 40,000 cells of donor 2, following the same encapsulation method described in the previous section (mouse study). Thirty capsules per material were mixed into one T225 flask, and these mixed capsules were prepared for IP implantation. After the aliquoting desired number of capsules into flasks, these samples were shipped overnight for implantation in an NHP.
Human islet encapsulation with lead material
Human islets (from Prodo Labs) were cultured in PIM(S) media (Prodo Labs) for further use. The cultured islets were centrifuged at 1200 rpm for 3 mins and washed with Ca-free Krebs buffer. The islets were then centrifuged again. The islet pellet was then resuspended in a 5% solution of Z4-A10 (blended with 3% SLG100 at a 70:30 ratio) at an islet density of 5,000 islets per 1mL alginate solution. Capsules were crosslinked in BaCl2 solution, and their sizes were adjusted to 1.5 mm. After crosslinking, the capsules were washed with HEPES three times and washed again with media twice. As the islets had variable sizes (50 ~ 400 μm), the total number of encapsulated islets was recounted and converted into islets equivalents (IEQ) (Supplementary Fig. 9). The average IEQ of each capsule was ~ 10 IEQ/capsule (1x). To make high-density capsules (2x and 4x density), alginate volume was reduced to 0.5mL and 0.25mL, respectively, while keeping the same IEQ. Finally, ~20 IEQ/capsule (2x) and ~40 IEQ/capsule (4x) were fabricated with Z4-A10 modified alginate. SLG20 was used as a control material, followed by the same encapsulation method and islets density.
Optimization of DNA extraction (in vitro)
To increase the amount of extracted gDNA from a single capsule, different parameters were optimized using a DNesay kit (Qiagen, catalog #69054). Fresh capsules and flash-frozen capsules were compared with or without lysis conditions. 50 mM EDTA in 10 mM HEPES solution was applied to lyse capsules and then centrifuged at 250G for 5 mins. Only pelleted cells were used for the following DNA extraction process. Un-lysed capsules were also used after homogenization as a comparison. Second, the yield was tested against different elution temperature settings (RT vs. 56°C). Finally, different cell numbers per capsule (5,000, 10,000, 20,000, 40,000, and 80,000 cells per capsule) were compared to determine optimal cell density for in vivo study.
Optimization of NGS library preparation workflow
Because DNA extracted from capsules was at low concentration, and the input for constructing the NGS library is typically below 1 ng, the PCR reactions were more prone to primer-dimer, especially in multiplex PCR. Here, the library preparation workflow was optimized to reduce primer-dimer. The first PCR amplified SNPs with multiplex primers containing 5’-overhang sequences. The primer concentration, PCR cycles, and annealing time of the first PCR were adjusted to reduce primer-dimers that could arise from multiplex PCR. Following on-plate purification, the second PCR amended position barcodes by amplifying with row-specific and column-specific primers comprising hamming barcode sequence and sequence that annealed to the 5’-overhang region of SNP primers. The amplification cycles of the second PCR were also adjusted.
Implantation/Transplantation surgeries
IP implant of mixed capsules in C57BL/6J mice:
All mice experiments were approved by Rice University’s Institution Animal Care and Use Committee (IACUC). Immune-competent male C57BL/6J mice (Charles River Labs, 6~8 weeks old) were first weighed and anesthetized with 1–4% isoflurane in oxygen at a heating pad. Buprenorphine was administered subcutaneously based on their weight (0.5 mg/kg dose). Their abdomens were shaved and sterilized by scrubbing three times with betadine and isopropanol, respectively. A 0.5 – 10 cm midline incision through the skin was made using a sharp blade. The peritoneal wall was then grasped with forceps, and a 5 mm incision was made along the linea alba. A volume of 0.5 mL of capsules was then implanted into the peritoneal cavity. The abdominal muscle was closed using absorbable sutures. The skin was then closed with a suture.
Subcutaneous implant of catheter samples in C57BL/6J mice:
Unmodified and coated catheters were implanted in the subcutaneous space of C57BL/6J mice (Charles River Labs). Specifically, one incision site was made in the back of each mouse, and one separate subcutaneous pocket was made for each catheter implant. The incisions were suture closed.
IP implant of human islets capsules in STZ-induced diabetic C57BL/6J mice and blood glucose monitoring:
To create insulin-dependent diabetic mice, healthy C57BL/6J mice were treated with streptozotocin (STZ). For five consecutive days, STZ solution at 7.5mg/ml concentration (50mg/kg of STZ) was injected into the IP space. The blood glucose (BG) levels and weights of all the mice were measured after one hour of fasting. Only mice whose BG levels were above 350mg/dL for two consecutive days were considered diabetic and used for islets transplantation. Two hundred capsules containing human islets were implanted into diabetic mice (~2,000 IEQ per mouse) for the 1x group. Furthermore, 100 capsules at ~ 20 IEQ/capsule and 50 capsules at ~ 40 IEQ/capsule were implanted for 2x and 4x groups, keeping the same IEQ number per mouse (~2,000 IEQ per mouse). BG levels were monitored three times a week following the transplantation of islets containing Z4-A10, Z1-A34, and SLG20 capsules. Mice with BG levels below 250mg/dL were considered normoglycemic. Monitoring continued until all mice had returned to a hyperglycemic state, at which point they were euthanized, and the capsules were retrieved. Mice fasted for 4hrs before in vivo glucose tolerance test. Each mouse was given a bolus dose of 30% sterile glucose solution in saline at 1.5g/kg through a tail vein injection. Blood glucose levels were measured every 15 mins for 2 hrs after the glucose injection.
Implantation of capsules laparoscopically into IP space in NHP:
On the day of the scheduled surgery, the NHP (male Mauritian cynomolgus monkey) was sedated and anesthetized (as per approved animal protocol). The anterior abdomen was shaved and prepped from xiphoid to pubis. A small (2 cm) supraumbilical incision was performed, and a 5–12 mm trocar was inserted. Pneumoperitoneum was created with CO2 at a pressure of 10–14 mmHg. After warming up with heated saline, the camera was inserted into the peritoneal cavity through the trocar. Under the view of laparoscopy, another 2 small incisions (1–2 cm) were made (on the left and right flank), and 5–12 mm trocars were inserted into the peritoneal cavity. A 2 mL sterile pipette connected to the syringe by a silicone tube was inserted through the trocar to the abdominal cavity. The capsules were distributed evenly in the following spaces: perihepatic, retrogastric, perisplenic, left and colonflexium, omentum, and behind small bowels. The three small incisions were then sutured by layered closure using Vicryl 3–0 for muscle and 4–0 Vicryl for skin (subcuticular). The animal’s recovery was followed by the OR staff, veterinary staff, and technicians. The animal protocol involving the care and use of non-human primates in this study was reviewed and approved by the University of Illinois-Chicago (UIC) Institutional Animal Care and Use Committee (IACUC) prior to commencement. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of UIC.
Retrieval of materials
Capsule retrieval from mouse IP space:
Five mice from each round were euthanized under CO2 administration at a designated timepoint of implantation, followed by cervical dislocation. An incision was then made using forceps and scissors along the abdomen, skin, and peritoneal wall. Ca+ Krebs buffer was then used to wash out all capsules from the abdomen and transferred into Petri dishes for collection. After all the capsules were washed out or manually retrieved, they were transferred into 50 mL conical tubes. The capsules were washed several times with Krebs buffer until the washed buffer was void of blood and tissue traces. Then the mixed explanted capsules were processed for further imaging and selection.
Capsule retrieval from the IP cavity of NHPs via laparoscopic technique:
NHPs were sedated and anesthetized (as per approved protocol). The anterior abdomen was shaved and prepped from xiphoid to pubis. A 2 cm supraumbilical incision was performed, and a 5–12 mm trocar was inserted. Pneumoperitoneum was created with CO2 at a pressure of 10–14mmHg. After warming up with heated saline, the camera was inserted into the peritoneal cavity through the trocar. Under the view of laparoscopy, another incision (1–2 cm) was made (on the left or right flank), and a 5–12 mm trocar was inserted into the peritoneal cavity. Laparoscopic images and video was taken to record microcapsule distribution. 20–30cc of saline was used for flushing any free capsules laparoscopically that may be present in the pelvic space that may have leaked from the lesser sac. The animal protocol involving the care and use of non-human primates in this study was reviewed and approved by the University of Illinois-Chicago (UIC) Institutional Animal Care and Use Committee (IACUC) prior to commencement. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of UIC.
Retrieval, fixation, and histology of catheter explants:
Catheters were explanted at four weeks by carefully removing the catheters and attaching tissues. The subcutaneous catheter explants were firmly attached to the skin and were covered with a thin membrane that lightly adhered to the muscle. The tissue around the catheter was cut about 3 mm from the catheter, and the catheter with skin attached was removed from the mouse. Explants were fixed with 10% formalin (Sigma) for four days before being transferred to PBS. The Baylor Pathology and Histology Core did further processing, sectioning, and histology. Specifically, samples were paraffin-embedded, sectioned along the cross-sectional axis of the catheters, and stained with H&E staining.
Imaging and selection of the explanted capsules
Capsules were gently washed using Krebs buffer before phase-contrast imaging and were transferred into 35 mm Petri dishes for bright- and dark-field imaging using a Leica stereoscope. Based on the levels of fibrotic overgrowth on the capsule surface, we manually separated capsules into three different groups (L: Low, M: Medium, and H: High fibrosed groups). Selected clear capsules with low fibrosis (L group) were flash-frozen and used for DNA extraction; otherwise, left capsules were flash-frozen with liquid N2 and stored at −80 °C for further uses.
Live/dead cell staining
Fluorescent imaging of cells stained with live/dead assay (Invitrogen, catalog # L3224) was performed to check the viability of encapsulated HUVECs from either pre- or post-implant capsules. Five capsules of each material were washed with DPBS and stained with 2 μM calcein AM and 4 μM EthD-1 in complete media. Capsules were incubated for 30 minutes and imaged using an EVOS microscope with fluorescence filters. Live cells were imaged with a GFP filter as green, and dead cells were imaged with a Texas-Red filter as red color. In the case of explanted capsules from NSG mice, capsules were washed three times with Ca+ Krebs buffer and then incubated in a staining solution. Capsules from each mouse were transferred into a 35 mm Petri dish and washed twice with DPBS. They were imaged under 2X magnification, and acquired images were stitched to observe all explanted capsules in the entire dish.
Dithizone staining
The explanted islets capsules were stained with dithizone (DTZ). 5mg of DTZ was dissolved in 1 mL of dimethyl sulfoxide (DMSO), thoroughly mixed, and incubated for 5 mins. 4mL of DPBS was added to the mixed solution and filtered through a 0.22 μm filter. Islets capsules were placed in a 35mm Petri dish and washed with PBS three times. Capsules were then incubated in DTZ solution for 5 mins and washed with DPBS three times to remove background staining. The stained capsules were imaged with a Leica microscope.
Immunofluorescence staining for confocal imaging
For immunofluorescence staining, retrieved microcapsules were washed with Krebs buffer and fixed in 4% paraformaldehyde overnight at 4°C. Samples were washed with PBS three times, and cells were permeabilized with 1% Triton X-100 for 15 mins at room temperature. After washing with PBS, samples were incubated in 1% bovine serum albumin (BSA) solution for 1 hour at room temperature for blocking and then incubated with staining solution containing the antibody cocktails (diluted at 1:200 Alexa Fluor 488 anti-mouse CD68 Antibody (Cat# 137012, BioLegend, CA, USA), 1:200 Anti-mouse α-Smooth Muscle-Cy3 (Cat# C6198, Sigma-Aldrich, MO, USA) in 1% BSA, and two drops/mL of DAPI (NucBlue Fixed Cell ReadyProbes Reagent, Cat# R37606, Invitrogen, CA, USA)) for 1 hour at room temperature. After washing with 0.1% tween 20 solution, samples were washed twice with PBS and transferred to a 50% glycerol solution in a glass-bottomed 24-well plate for imaging. Nikon A1-Rsi confocal microscope was used for immunofluorescence imaging.
Histological processing of retrieved catheters and histology-based assessment of tissue overgrowth on catheters
Stained sections of catheter explants were imaged with a Leica M165C light microscope. The dark band of purple tissue between the catheter and skin layer was measured using ImageJ software to determine the extent of tissue overgrowth on the catheters. This method is based on the findings of Xie et al., which showed the tissue band directly adjacent to the subcutaneous implant stains dark purple with H&E and blue with Trichrome23. Staining blue with Trichrome means that this tissue band is collagen-rich, suggesting this tissue is indicative of fibrotic overgrowth. Because of this correlation, the dark purple tissue was measured and used to assess the relative immune response to the differently coated catheters.
Specifically, the catheter section image was rotated to angle the adjacent skin tissue downwards. Three measurements, which were equally spaced along with the skin tissue, were taken of the dark purple band of tissue for each image and averaged to avoid arbitrarily measuring one spot. Measuring the length of the scale bar burned into the image was used to convert the ImageJ pixel measurement to millimeters. Some sections have large, lighter purple bands of tissue that run more perpendicular to the catheter. These were specifically avoided during measurements since they are part of scar tissue from the incision.
DNA/RNA extraction from explanted capsules and RT-qPCR
For DNA extraction from a single capsule, the optimized method was applied for this study. Encapsulated cells from pre- or post-implant capsules were lysed in 50 mM EDTA for 15 mins and centrifuged at 5000 rpm for 10 mins. The supernatant was aspirated, and the cell pellet was suspended in 200 μL of PBS. According to the manufacturer’s instructions, total gDNA from a single capsule was isolated using the DNeasy kit (Qiagen, catalog # 69504) with minor modifications under optimized conditions. Briefly, the cell suspension was lysed with proteinase K and RNase A for 5 minutes at RT and incubated with lysis buffer for 20 mins at 56 °C. After adding ethanol, the supernatant was transferred into a column and washed twice with a wash buffer. DNA was collected with elution buffer heated at 56°C and stored at −20°C for further uses. Total RNA was extracted from 100μl of retrieved microcapsules (300–400 μm size). Snap frozen capsules were thawed on ice, homogenized, and processed using RNeasy Mini Kit (Qiagen, catalog #74104) according to the manufacturer’s instructions. Extracted RNA was converted to cDNA for RT-qPCR using the high-capacity cDNA reverse transcription kit (Applied Biosystems, catalog #4368814). Real time qPCR was performed using 3 μL of gDNA/cDNA in a 10 μL reaction volume with SYBR Green (PowerUp SYBR Green Master Mix; Applied Biosystems, catalog # A25741) to quantify the PCR product. PCRs were carried out under the following conditions: 95°C for 10 s, 48°C for 20 s, 72°C for 30 s (40 cycles), 72°C for 5 min, 65°C for 5 s, and a final cycle at 95°C. All reactions were run in triplicates. Data were analyzed with the 2ΔΔCT method, and relative RNA levels were compared after normalization to mouse a-actin (ActB) and SLG20 (control). The primers used in this study are listed in the Supplementary Table. 5.
Next-Generation Sequencing (NGS)
NGS library preparation:
DNA content in individual capsules was semi-quantitatively evaluated using qPCR as a sample quality control procedure, with Ct values in negative correlation to extracted DNA content. Samples with amplifiable DNA content would undergo two PCR steps to amplify and barcode the target amplicons. With DNA from each capsule in individual wells of a 96-well PCR plate, the first PCR was performed using 30-plex primers targeting 30 non-pathogenic SNP whose collective genotype profile will uniquely identify a HUVEC donor (Supplementary Table. 6). The 30-plex primers all contained a 5’-overhang sequence to incorporate a universal binding domain to target amplicons. The reaction mixture comprised 30-plex SNP primers at a concentration of 50 nM each and Phusion Hot Start Flex 2X Master Mix at 1X. The reaction condition was an activation at 98°C for 30 s, seven cycles of denaturation at 98°C for 30 sec, annealing at 63°C for 5min, extension at 72°C for 1min, and complete the reaction with incubation at 72°C for 5min (shorten as 98°C:30s-(98°C:10s-63°C:5min-72°C:1min)x7–72°C:5min-4°C: hold). AMPure XP magnetic beads (Beckman Coulter, catalog #A63881) with a 1.2x volumetric ratio were added to the first PCR product. The suspension was incubated for 5min at room temperature, then placed on a magnetic stand to separate and discard the supernatant. The remaining magnetic beads were washed twice with 80% ethanol, and DNA content was eluted in water. The second PCR used primers carrying overhang Hamming code sequence to uniquely barcode capsule samples. A total of 12 barcoded column-specific forward primers and eight barcoded row-specific reverse primers were designed to have a distance of at least three from one another that allowed barcoding of 12 × 8 = 96 capsules and eased batch processing on a 96 well plate. The reaction condition was 98°C:30s - (98°C:10s −63°C:1min - 72°C:1min) x 20 −72°C:5min - 4°C:hold) with primer concentration of 400nM and Phusion Hot Start Flex 2X Master Mix at 1X. The row and column barcodes uniquely determined the sample position on the 96-well plate. Thus, barcoded amplicons could be pooled from all wells, forming a single library containing SNP information from 96 different capsule samples (Supplementary Table. 7). As previously described above, the products were purified using AMPure XP magnetic beads at a 1.0x volumetric ratio.
Illumina sequencing adapters were amended by ligation-based method, and the library index was added by PCR using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, catalog #E7645S) according to the manufacturer’s protocol. The library quantification and quality control were performed on Agilent 2100 Bioanalyzer.
Single donor sample analysis:
Encapsulating materials were barcoded by the co-encapsulated HUVEC donor cells, and thus determining HUVEC donor identities through sequencing data analysis could reveal material information. NGS fastq data were demultiplexed by row and column barcodes to re-group sequences amplified from the same DNA input. Sequence alignment to target amplicons was conducted in bowtie2. Then for each amplicon sequence, a grep function was applied to search the dominant and minor alleles to calculate variant allele frequency (VAF) for each SNP locus. Since capsules implanted in mice contained only one HUVEC donor, the HUVEC donor with the highest match rate to the analyte sample was identified as the barcoding cell.
Dual donors sample analysis:
Log-likelihood was employed to analyze explanted samples that encapsulated one or two HUVEC donors. Specifically, SNP VAF profiles were calculated for all possible compositions of donor cells. in equation (1) represents the expected VAF of the kth SNP when donor i and donor j were mixed at a ratio of 1:R. Here, all possible combinations of 20 different HUVEC donors had 20×20=400 different compositions. For each possible composition, the observed VAF of each SNP, depending on how close or far the observed value was from the VAF in the composition, a probability, was calculated from Gaussian distribution. The overall log-likelihood of each composition, , is obtained from summing the log-likelihood of all SNPs, and the composition with the highest overall log-likelihood is determined as the barcoding cell composition.
| (1) |
| (2) |
| (3) |
Statistical analysis
Statistical analysis for material screening used a customized MATLAB script. The retrieved percentage for low-fibrosed capsules was averaged among biological replicates (the number of mice N = 5). An error bar of 95% confidence interval was calculated for each material from a binomial distribution. Any two materials with a non-overlapping error bar were identified as statistically significant (p < 0.05). For statistical analysis of other graphs, one-way or two-way ANOVA with Bonferroni multiple-comparison correction was used (****P < 0.0001, ***P < 0.002).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw sequencing data generated in this study can be accessed via Figshare at https://figshare.com/projects/In_vivo_screening_of_hydrogel_library_using_cellular_barcoding_identifies_biomaterials_that_mitigate_host_immune_responses_and_fibrosis/144375. The datasets generated during and/or analysed during the study are available for research purpose from the corresponding author on reasonable request.
Extended Data
ED Fig. 1. Schematic diagram for the synthesis of 211 alginate analogs.
Left of the scheme are two of the previous triazole-containing lead alginates (B1-A21 and Z1-A34) that prevent fibrosis in mouse and NHP models. A total of 211 new alginate analogs were synthesized by varying the lead hydrophilic linker (Azido PEG-amine) and hydrophobic linker (iodobenzylamine) combined with a set of alkyne classes highlighted in blue colors.
ED Fig. 2. Capsule optimization.
a, Gelation assay of alginate analogs using Rhodamine B entrapment. b, Representative images of capsule formation using the alginate analogs. Scale bar, 2mm. c, After initial characterization studies, including purity, solubility, and gel-forming ability, 149 alginate polymers were used for the screening test. d, Mechanical testing (work to burst) with different alginate formulations. One-way ANOVA with Bonferroni correction was used for statistical analysis; ***P = 0.0007 (Z1-A34), ***P = 0.0009 (B1-A51), ns; non significant for comparisons with SLG20. All error bars denote mean ± s.e.m of n=20 replicates. e-i, FITC-dextran permeability test with different modified alginates. All error bars denote mean ± s.e.m of n=5 replicates.
ED Fig. 3. Optimization of NGS library preparation workflow.
The library preparation involved a multiplex PCR step in amplifying the SNP loci, a barcoding PCR step to add position barcode to each sample, and a ligation-based sequencing adapter amendment procedure. a, Before optimization, the library on-target rate was <10%, with primer-dimers and non-specific PCR products contributing to the majority of reads. b, After optimization, the on-target rate was increased to >80% regardless of low DNA input (<1ng) in the starting material.
ED Fig. 4. Bioinformatic pipeline for determining material identity/composition from NGS sequencing data.
Fastq NGS data was demultiplexed by row and column barcodes to re-group sequences amplified from the same DNA input. Then for each amplicon sequence, the grep function was applied to search the dominant and variant alleles to calculate variant allele frequency (VAF) for each SNP locus. If the encapsulated cells comprised only one donor, the VAF profile was compared against profiles of the 20 pre-screened HUVEC donors. The donor with the highest match rate was identified as the encapsulated donor cell. When one or two donors were used as encapsulated cells, the log-likelihood of all possible donor compositions was calculated. The composition with the highest overall log-likelihood was determined as the cell composition (Quality control for log-likelihood analysis: 1) at least 25/30 SNP loci had sequencing coverage >50; and 2) overall log-likelihood higher than −200, and 3) goodness measurement higher than 10 where goodness is defined as the difference of log-likelihood between the most likely and the second most likely donor pairs). The material corresponding to the identified donor cell or cell composition would be the material encapsulating cells.
ED Fig. 5. Dual donor barcoding identification in C57BL/6J mice.
a, Three different materials were tested; UP-VLVG (control), B1-A51 (one of the negative materials), and Z1-A34 (one of the positive materials). b, Schematic workflow of three materials screening containing mixed dual donors. c-d, After two weeks of implant, capsules were retrieved from each mouse (M1-M3) and were separated into three groups depending on fibrosis levels. e, Representative heatmap result of identified donor pair. f, 39 mapped to Z1-A34 (positive control material), coded by H16:H14 at 1:2 ratio; 4 mapped to UP-VLVG coded by H6:H8 at 1:2 ratio; 0 sample mapped to B1-A51. Overalled 43/45 samples were mapped from the 400 donor SNP profile. Proportions of each material corresponded to donor pairs were plotted, and Z1-A34 showed the highest value indicating the best immune-protective properties.
ED Fig. 6. Diabetic reversal study with lead material.
a, Capsules containing human islets were fabricated at final cell density with 4,000, 8,000, and 16,000 IEQ/alginate volume (mL). The final IEQ values in each capsule were 10, 20, and 40 IEQ per capsule, respectively. In each group, 500μL, 250μL, and 125μL of capsules were implanted in IP space, containing total 2,000 IEQ per mouse. All error bars denote mean ± s.e.m of n=20 biological replicates. b, Representative images of pre-implant Z1-A34 capsules. Dithizone staining indicates viable islets within the capsule matrix. After encapsulation, islets show good viability (live: green, dead: red).
ED Fig. 7. Material characterization and evaluation of catheters coated with lead molecules.
a-b, The total intensity of two main peaks analyzed by ToF-SIMS (a, CN- and b, Br-) were plotted to compare with the unmodified catheter. c, XPS data for unmodified, Met-Z1A3, and Met-B2-A17 modified catheters showing wt. % of small molecule-specific atoms, indicating successful coating. d, Representative SEM images of unmodified and coated catheters. e-g, Surface characteristic of catheters coated with lead molecules using Raman spectroscopy. Pre-implant or post-explant catheters (4 or 8 weeks) were analyzed to confirm the presence of coated molecules (e, Met-Z1-A3, f, Met-B2-A17, g, Met-Z1-A34 catheters). All error bars denote mean ± s.e.m of n=2 replicates.
Supplementary Material
Acknowledgements
This work was supported by the US National Institute of Health (R01 DK120459) to O.V. and D. Y. Z. The authors acknowledge the support of the National Science Foundation for access to the ToF-SIMS, supported through CBET1626418. ToF-SIMS analyses and spin coating were carried out with support provided by the Shared Equipment Authority at Rice University. The authors would like to thank Rice University animal resource facility staff for their assistance with animal research.
Funding
O.V. and D.Y.Z. disclose support for the research described in this study from the US National Institute of Health (grant R01DK120459).
Footnotes
Competing interests
O.V. is a member of the Scientific Advisory Board of Sigilon Therapeutics and holds equity in Avenge Bio and Pana Bio. D.Y.Z. owns significant equity in and receives consulting income from NuProbe Global and Torus Biosystems and owns significant equity in Pana Bio.
Additional information [please do not modify this block of text]
Extended data is available for this paper at https://doi.org/10.1038/s41551-02X-XXXX-X.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-02X-XXXX-X.
Peer review information Nature Biomedical Engineering thanks Reviewer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
References
- 1.Anderson JM, Rodriguez A. & Chang DT Foreign body reaction to biomaterials. Semin Immunol 20, 86–100, doi: 10.1016/j.smim.2007.11.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wick G. et al. The immunology of fibrosis. Annu Rev Immunol 31, 107–135, doi: 10.1146/annurev-immunol-032712-095937 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA & Ramalingam TR Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18, 1028–1040, doi: 10.1038/nm.2807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veiseh O. & Vegas AJ Domesticating the foreign body response: Recent advances and applications. Adv Drug Deliv Rev 144, 148–161, doi: 10.1016/j.addr.2019.08.010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenneth Ward W. A review of the foreign-body response to subcutaneously-implanted devices: the role of macrophages and cytokines in biofouling and fibrosis. J Diabetes Sci Technol 2, 768–777, doi: 10.1177/193229680800200504 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vegas AJ et al. Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat Biotechnol 34, 345–352, doi: 10.1038/nbt.3462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veiseh O. et al. Size- and shape-dependent foreign body immune response to materials implanted in rodents and non-human primates. Nat Mater 14, 643–651, doi: 10.1038/nmat4290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell M. & Mooney DJ Leveraging advances in biology to design biomaterials. Nature Materials 16, 1178–1185, doi: 10.1038/nmat4991 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Peters A, Brey DM & Burdick JA High-Throughput and Combinatorial Technologies for Tissue Engineering Applications. Tissue Engineering Part B: Reviews 15, 225–239, doi: 10.1089/ten.teb.2009.0049 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Grainger DW All charged up about implanted biomaterials. Nature Biotechnology 31, 507–509, doi: 10.1038/nbt.2600 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Zhang L. et al. Zwitterionic hydrogels implanted in mice resist the foreign-body reaction. Nat Biotechnol 31, 553–556, doi: 10.1038/nbt.2580 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Witherel CE, Abebayehu D, Barker TH & Spiller KL Macrophage and Fibroblast Interactions in Biomaterial-Mediated Fibrosis. Adv Healthc Mater 8, e1801451, doi: 10.1002/adhm.201801451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratner BD Biomaterials: Been There, Done That, and Evolving into the Future. Annu Rev Biomed Eng 21, 171–191, doi: 10.1146/annurev-bioeng-062117-120940 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Bank RA Limiting biomaterial fibrosis. Nat Mater 18, 781, doi: 10.1038/s41563-019-0428-y (2019). [DOI] [PubMed] [Google Scholar]
- 15.Noskovicova N. et al. Suppression of the fibrotic encapsulation of silicone implants by inhibiting the mechanical activation of pro-fibrotic TGF-β. Nature Biomedical Engineering, doi: 10.1038/s41551-021-00722-z (2021). [DOI] [PubMed] [Google Scholar]
- 16.Ernst AU, Wang L-H & Ma M. Islet encapsulation. Journal of Materials Chemistry B 6, 6705–6722, doi: 10.1039/C8TB02020E (2018). [DOI] [PubMed] [Google Scholar]
- 17.Kearney CJ & Mooney DJ Macroscale delivery systems for molecular and cellular payloads. Nat Mater 12, 1004–1017, doi: 10.1038/nmat3758 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Bochenek MA et al. Alginate encapsulation as long-term immune protection of allogeneic pancreatic islet cells transplanted into the omental bursa of macaques. Nat Biomed Eng 2, 810–821, doi: 10.1038/s41551-018-0275-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vegas AJ et al. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice (vol 22, pg 306, 2016). Nat Med 22, 446–446, doi:DOI 10.1038/nm0416-446e (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidarra SJ, Barrias CC & Granja PL Injectable alginate hydrogels for cell delivery in tissue engineering. Acta Biomaterialia 10, 1646–1662, doi: 10.1016/j.actbio.2013.12.006 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Weir MD & Xu HHK An injectable calcium phosphate-alginate hydrogel-umbilical cord mesenchymal stem cell paste for bone tissue engineering. Biomaterials 31, 6502–6510, doi: 10.1016/j.biomaterials.2010.05.017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X. et al. A nanofibrous encapsulation device for safe delivery of insulin-producing cells to treat type 1 diabetes. Science Translational Medicine 13, eabb4601, doi:doi: 10.1126/scitranslmed.abb4601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X. et al. Reduction of measurement noise in a continuous glucose monitor by coating the sensor with a zwitterionic polymer. Nat Biomed Eng 2, 894–906, doi: 10.1038/s41551-018-0273-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs-Tulleneers-Thevissen D. et al. Sustained function of alginate-encapsulated human islet cell implants in the peritoneal cavity of mice leading to a pilot study in a type 1 diabetic patient. Diabetologia 56, 1605–1614, doi: 10.1007/s00125-013-2906-0 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Robitaille R. et al. Inflammatory response to peritoneal implantation of alginate-poly-L-lysine microcapsules. Biomaterials 26, 4119–4127, doi: 10.1016/j.biomaterials.2004.10.028 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Rokstad AM et al. Alginate microbeads are complement compatible, in contrast to polycation containing microcapsules, as revealed in a human whole blood model. Acta Biomater 7, 2566–2578, doi: 10.1016/j.actbio.2011.03.011 (2011). [DOI] [PubMed] [Google Scholar]
- 27.King A, Sandler S. & Andersson A. The effect of host factors and capsule composition on the cellular overgrowth on implanted alginate capsules. J Biomed Mater Res 57, 374–383, doi: (2001). [DOI] [PubMed] [Google Scholar]
- 28.Manoury B, Caulet-Maugendre S, Guenon I, Lagente V. & Boichot E. TIMP-1 is a key factor of fibrogenic response to bleomycin in mouse lung. Int J Immunopathol Pharmacol 19, 471–487, doi: 10.1177/039463200601900303 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Desai T. & Shea LD Advances in islet encapsulation technologies. Nature Reviews Drug Discovery 16, 338–350, doi: 10.1038/nrd.2016.232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuch BE et al. Safety and Viability of Microencapsulated Human Islets Transplanted Into Diabetic Humans. Diabetes Care 32, 1887–1889, doi: 10.2337/dc09-0744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bose S. et al. A retrievable implant for the long-term encapsulation and survival of therapeutic xenogeneic cells. Nat Biomed Eng 4, 814–826, doi: 10.1038/s41551-020-0538-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farah S. et al. Long-term implant fibrosis prevention in rodents and non-human primates using crystallized drug formulations. Nat Mater 18, 892–904, doi: 10.1038/s41563-019-0377-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krutzik PO & Nolan GP Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods 3, 361–368, doi: 10.1038/nmeth872 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Strobel B. et al. High-throughput identification of synthetic riboswitches by barcode-free amplicon-sequencing in human cells. Nat Commun 11, 714, doi: 10.1038/s41467-020-14491-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyons E, Sheridan P, Tremmel G, Miyano S. & Sugano S. Large-scale DNA Barcode Library Generation for Biomolecule Identification in High-throughput Screens. Sci Rep 7, 13899, doi: 10.1038/s41598-017-12825-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binan L, Drobetsky EA & Costantino S. Exploiting Molecular Barcodes in High-Throughput Cellular Assays. SLAS Technol 24, 298–307, doi: 10.1177/2472630318824337 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Bykov YS et al. High-throughput ultrastructure screening using electron microscopy and fluorescent barcoding. J Cell Biol 218, 2797–2811, doi: 10.1083/jcb.201812081 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira MB & Mano JF High-throughput screening for integrative biomaterials design: exploring advances and new trends. Trends in Biotechnology 32, 627–636, doi: 10.1016/j.tibtech.2014.09.009 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Bratlie KM et al. Rapid Biocompatibility Analysis of Materials via In Vivo Fluorescence Imaging of Mouse Models. PLOS ONE 5, e10032, doi: 10.1371/journal.pone.0010032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee K. et al. The effect of 3D hydrogel scaffold modulus on osteoblast differentiation and mineralization revealed by combinatorial screening. Biomaterials 31, 5051–5062, doi: 10.1016/j.biomaterials.2010.03.024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vega SL et al. Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments. Nature Communications 9, 614, doi: 10.1038/s41467-018-03021-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreiuk B. et al. Fluorescent Polymer Nanoparticles for Cell Barcoding In Vitro and In Vivo. Small 13, 1701582, doi: 10.1002/smll.201701582 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Lokugamage MP, Sago CD & Dahlman JE Testing thousands of nanoparticles in vivo using DNA barcodes. Curr Opin Biomed Eng 7, 1–8, doi: 10.1016/j.cobme.2018.08.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paunovska K. et al. A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation. Nano Letters 18, 2148–2157, doi: 10.1021/acs.nanolett.8b00432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vorholt SM et al. High-Throughput Screening of Blood Donors for Twelve Human Platelet Antigen Systems Using Next-Generation Sequencing Reveals Detection of Rare Polymorphisms and Two Novel Protein-Changing Variants. Transfus Med Hemother 47, 33–44, doi: 10.1159/000504894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nyyssonen M. et al. Coupled high-throughput functional screening and next generation sequencing for identification of plant polymer decomposing enzymes in metagenomic libraries. Front Microbiol 4, 282, doi: 10.3389/fmicb.2013.00282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klaper R, Arndt D, Bozich J. & Dominguez G. Molecular interactions of nanomaterials and organisms: defining biomarkers for toxicity and high-throughput screening using traditional and next-generation sequencing approaches. Analyst 139, 882–895, doi: 10.1039/c3an01644g (2014). [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa F. et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood 106, 1565–1573, doi: 10.1182/blood-2005-02-0516 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shultz LD et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 174, 6477–6489, doi: 10.4049/jimmunol.174.10.6477 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Doloff JC et al. Colony stimulating factor-1 receptor is a central component of the foreign body response to biomaterial implants in rodents and non-human primates. Nature Materials 16, 671–680, doi: 10.1038/nmat4866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doloff JC et al. The surface topography of silicone breast implants mediates the foreign body response in mice, rabbits and humans. Nature Biomedical Engineering 5, 1115–1130, doi: 10.1038/s41551-021-00739-4 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Qi M. et al. Survival of human islets in microbeads containing high guluronic acid alginate crosslinked with Ca2+ and Ba2+. Xenotransplantation 19, 355–364, doi: 10.1111/xen.12009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw sequencing data generated in this study can be accessed via Figshare at https://figshare.com/projects/In_vivo_screening_of_hydrogel_library_using_cellular_barcoding_identifies_biomaterials_that_mitigate_host_immune_responses_and_fibrosis/144375. The datasets generated during and/or analysed during the study are available for research purpose from the corresponding author on reasonable request.