Abstract
Background
Understanding the genetic mechanisms underlying diseases in ancestrally diverse populations is an important step towards development of targeted treatments. African and African admixed populations enable mapping of complex traits, because of their genetic diversity, extensive population substructure, and distinct linkage disequilibrium patterns. We aimed to do a comprehensive genome-wide assessment in African and African admixed individuals to better understand the genetic architecture of Parkinson’s disease in these underserved populations.
Methods
We performed a genome-wide association study in people of African and African admixed ancestry with and without Parkinson’s disease. We characterised ancestry-specific risk, differential haplotype structure and admixture, coding and structural genetic variation, and enzymatic activity.
Findings
We included 197 918 individuals (1488 cases; 196 430 controls) in our genome-wide analysis. We identified a novel common risk factor for PD and age at onset at the GBA1 locus (risk, rs3115534-G; OR=1.58, 95% CI = 1.37 – 1.80, P=2.397E-14; age at onset, BETA =−2.004, SE =0.57, P = 0.0005), that was found to be rare in non-African or non-African admixed populations. Downstream short- and long-read whole genome sequencing analyses did not reveal any coding or structural variant underlying the GWAS signal. However, we identified that this signal is associated with decreased glucocerebrosidase activity levels.
Interpretation
The present study identifies a novel genetic risk factor in GBA1 in people of African ancestry, which could be a major mechanistic basis of PD in this population. This striking result contrasts to previous work in Northern European populations, both in terms of mechanism and attributable risk. This finding highlights the importance of understanding ancestry-specific genetic risk in complex diseases, a particularly crucial point as the field moves toward targeted treatments in PD clinical trials and while recognizing the need for equitable inclusion of ancestrally diverse groups in such trials. Given the distinctive genetics of these underrepresented populations, their inclusion represents a valuable step towards insights into novel genetic determinants underlying PD etiology. This opens new avenues towards RNA-based and other therapeutic strategies aimed at reducing lifetime risk.
Keywords: genetics, Parkinson’s disease, genome-wide association study, African, African Admixed, GBA1, expression quantitative trait locus, therapeutic interventions
Introduction
Genome-wide association studies (GWAS) have been instrumental for identifying common variants associated with complex diseases like Parkinson’s disease (PD), unraveling the genetics and heritability of PD in European populations. The largest published GWAS meta-analysis of PD risk to date was performed on individuals of European ancestry and identified 90 independent genome-wide significant risk signals that explain 16–22% of the heritable risk of PD1,2. However, very little is known about the genetics of PD in non-European populations. There has been considerable ethnic variability in the distribution of monogenic causes and genetic risk variants documented across populations. For instance, the relatively common LRRK2 p.G2019S mutation remains unreported in some sub-Saharan African populations, despite being most commonly associated with familial and sporadic PD in Zambia and Northern Africa3–6.
African and African admixed populations offer unique opportunities for studying the genetics of both monogenic and complex diseases because they contain the largest portion of the within-population genetic variability in the world, shorter linkage disequilibrium (LD) blocks, and abundant alleles that are private to these populations7. In addition to promoting scientific equity to address health disparities, diverse representation provides a platform for replication studies to explore the strength and relevance of findings reported from other populations. Additionally, studying diverse populations have the potential to facilitate the identification of novel or unique loci and investigate genotype-phenotype correlations that can further expand our understanding of pathological and pathogenetic disease mechanisms in PD.
This study provides the first GWAS-based insights into the genetics of PD in the African and African admixed populations. Here we performed a comprehensive genome-wide assessment of PD risk and age at onset, characterizing ancestry-specific risk, haplotype structure, and genetic admixture. Leveraging this unique population genetic structure, our analyses identified a novel association signal in GBA1, the gene encoding the lysosomal enzyme glucocerebrosidase (GCase).
Methods
Study Design and Participants
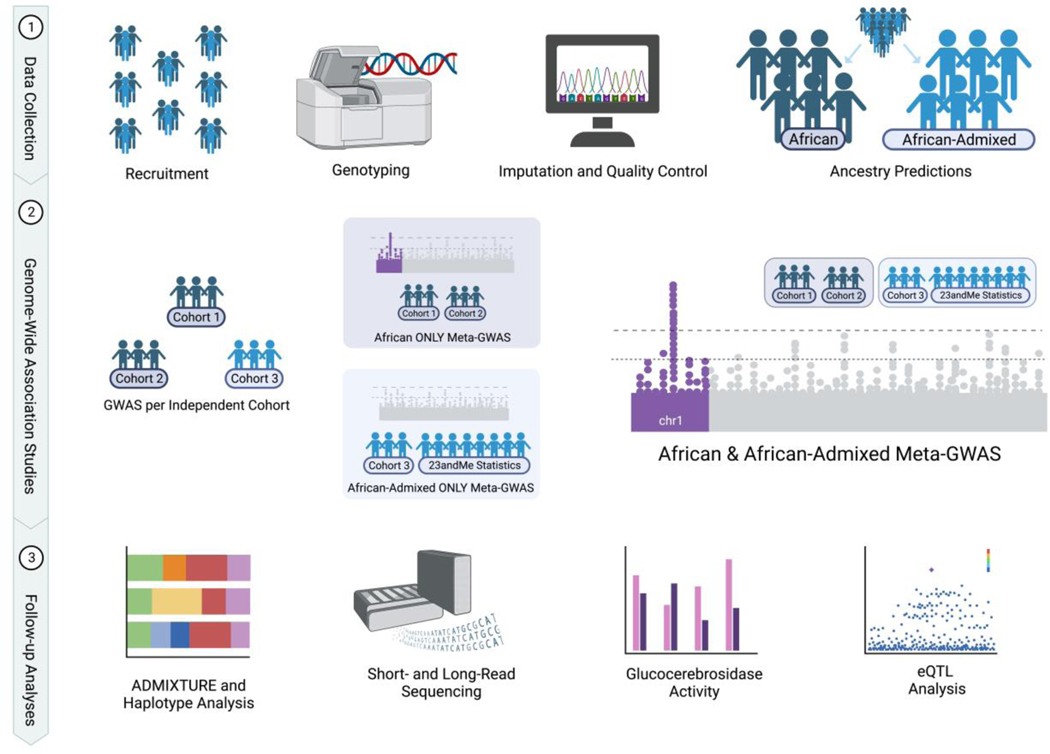
An overview of the study design can be found in Figure 1. Three sources of data were included in this study: Individual level data from the International Parkinson’s Disease Genomics Consortium - Africa (IPDGCAN) and the Global Parkinson’s Disease Genetics Program (GP2), and GWAS summary statistics from 23andMe, Inc. PD cases provided from efforts in Africa are predominantly from West Africa, specifically Nigeria, and therefore unlikely to be representative of the entirety of Africa. However, the majority of controls in this study come from global efforts and have higher percentages of admixture. Some of the individuals predicted to be of African descent cannot with certainty be defined as from Nigeria, but nonetheless unmistakably African (Supplementary Figure 3). Additionally, we define African admixed as individuals ancestrally similar to the following 1000 Genomes ancestry labels: African ancestry in Southwest United States of America (abbreviated as ASW in the 1000 Genomes project) and African Caribbean in Barbados (abbreviated as ACB in the 1000 Genomes project).
Figure 1: Study design.
Figure created with BioRender.com. eQTL=expression quantitative trait locus. GWAS=genome-wide association study.
For the IPDGCAN and the GP2 cohorts, the diagnosis of PD was based on fulfillment of the United Kingdom PD Society Brain Bank criteria (excluding the requirement for not more than one affected relative)8. The respective ethical committees for medical research approved involvement in genetic studies, and participants gave informed written consent. All participants underwent a neurological examination conducted by a study neurologist to document clinical and neurological status. Controls were generally assessed to detect overall signs of neurological condition and samples presenting any clinical signs of neurodegenerative diseases were excluded from the control series.
For the 23andMe dataset, the diagnosis of PD was self-reported (see Supplementary Materials). Summary statistics for individuals with or without PD were provided through a collaborative agreement with 23andMe, Inc. Participants provided informed consent and volunteered to participate in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent (E&I) Review Services. As of 2022, E&I Review Services is part of Salus IRB (https://www.versiticlinicaltrials.org/salusirb; Supplementary Methods).
Statistical analyses
Genotype data generation, quality control, ancestry predictions, and imputation
The IPDGCAN and GP2 samples were genotyped using two different genotyping platforms (Table 1). The NeuroBooster array (v.1.0, Illumina, San Diego, CA) contains a backbone of 1,914,935 variants densely covering ancestry informative markers, markers for determination of identity by descent, X-chromosome SNPs for sex determination, and contains 96,517 customized variants. Samples collected as part of the GP2 initiative were genotyped on this array. Samples collected as part of the IPDGCAN initiative (Table 1) were genotyped using two different platforms; the Neurochip array, containing a backbone of 306,670 variants and customized content comprising 179,467 variants9, and the previously described NeuroBooster array.
Table 1:
Characteristics of cohorts in the study
| African predicted ancestry | African admixed predicted ancestry* | |||
|---|---|---|---|---|
| Nigerian origin (IPDGC cohort)† | African, broad unspecified origin (GP2 dataset)‡ | African admixed origin (GP2 dataset)§ | African admixed origin (23andMe dataset) | |
| Total participants | 589 | 1722 | 1334 | 194 273 |
| Recruited from Nigerian sites | 589 (100%) | 1330 (77%) | 50 (4%) | N/A |
| Cases | 304 | 711 | 185 | 288 |
| Recruited from Nigerian sites | 304 (100%) | 672 (95%) | 16 (9%) | N/A |
| Female | 80 (26%) | 206 (29%) | 80 (43%) | N/A |
| Male | 224 (74%) | 505 (71%) | 105 (57%) | N/A |
| Controls | 285 | 1011 | 1149 | 193 985 |
| Recruited from Nigerian sites | 285 (100%) | 658 (65%) | 34 (3%) | N/A |
| Female | 97 (34%) | 448 (44%) | 714 (62%) | N/A |
| Male | 188 (66%) | 563 (56%) | 435 (38%) | N/A |
| Case age at onset, years | 58·20 (9·67) | 59·31 (11·37) | 57·84 (14·69) | N/A |
| Control age at examination, years | 64·4 (7·56) | 65·09 (9·55) | 66·34 (8·71) | N/A |
| Array | NeuroChip | NeuroBooster | NeuroBooster | Omni Express & GSA & 550k |
Data are n, n (%) or mean (SD). N/A=not available.
African admixed defined as individuals ancestrally similar to the following 1000 Genomes project (https://www.internationalgenome.org/) ancestry labels: African ancestry in Southwest United States of America (abbreviated as ASW in the 1000 Genomes project) and African Caribbean in Barbados (abbreviated as ACB in the 1000 Genomes project).
See appendix (p 37) for a complete list of Nigerian hospitals and institutions contributing to this cohort.
GP2 cohorts with predicted African ancestry include Baylor College of Medicine (https://www.bcm.edu/), BioFIND (https://biofind.loni.usc.edu/), BLAAC PD (https://www.blaacpd.org/), Coriell (https://www.coriell.org/), Movement Disorders Genotypes and Phenotypes – King’s College London (MDGAP-KINGS; further details at https://gp2.org/the-components-of-gp2s-third-data-release/), PPMI (https://www.ppmi-info.org/), PAGE (https://www.pagestudy.org/), University of Maryland (https://umd.edu/), and IPDGCAF-NG (https://www.ipdgc-africa.com/).
GP2 cohorts with predicted African admixed ancestry include Baylor College of Medicine, BioFIND, BLAAC PD, Coriell, MDGAP-KINGS, PPMI, PAGE, University of Maryland, Systemic Synuclein Sampling Study (S4; https://pubmed.ncbi.nlm.nih.gov/28353371/), and IPDGCAF-NG.
Raw genotype data was passed through a custom ancestry prediction and pruning machine learning method as a part of the GenoTools pipeline (https://github.com/GP2code/GenoTools), as described elsewhere10. All samples underwent similar standardized quality control (QC). For additional information, please see the Supplementary Materials.
Estimation of PD risk, age at onset and admixture
To estimate risk associated with PD, imputed dosages (meaning genotype probabilities for a variant to be A/A, A/B, or B/B from 0 to 2 that account for some uncertainty) were analyzed using a logistic regression model adjusted for sex, age, and the first ten PCs as covariates. The PCs were fit on the set of overlapping SNPs between the datasets and the reference panels before being transformed by UMAP to represent the population substructure (see Supplementary Materials). Age at onset (AAO) was used for cases and age at recruitment was used for controls. In instances where AAO was not available for cases, age at recruitment was used instead (less than 6% of individuals). For individuals who had no age information provided, average age was imputed (less than 5% and 2% of cases and controls, respectively). Summary statistics were generated using PLINK 1.9 and 2.011, and filtered for inclusion after meeting a minimum imputation quality of 0.30 and MAF > 5 %. To explore the influence of genetic variation on the AAO of PD cases, a linear regression model adjusted for the same covariates was performed. Here, AAO was defined as the self-reported date of first motor symptom. Additionally, we conducted linear regression analyses to explore how potential GWAS signals would correlate with admixture levels. All the analyses were performed on Terra (https://terra.bio/). GWAS was conducted on African and African admixed ancestries independently using PLINK and a Bonferroni threshold of 5E-8 prior to meta-analysis. We utilized fixed-effects meta-analyses as implemented in METAL12 to leverage summary statistics across all sources. Pairwise LD values were calculated using 1000 Genomes African population data through LD link (https://ldlink.nci.nih.gov/?tab=home).
Haplotype and fine-mapping analyses
Haplotype size was compared using individual level data across African, African admixed, and European PD cases. After standardizing the three datasets with the same genotyped SNPs passing identical QC steps, we determined the size of the haplotype blocks using default parameters in PLINK 1.9. This analysis estimates haplotype blocks by Haploview’s interpretation of the block definition. By default, only pairs of variants within 200 kilobases (kb) of each other were considered. Two variants are considered by this procedure to be in strong LD if the lower bound of the 90% D-prime confidence interval (CI) was >0.70, and the upper bound of the CI was at least 0.98. Fine-mapping analyses were conducted using the R package coloc (https://CRAN.R-project.org/package=coloc; Supplementary Methods).
Procedures
Short and Long read Whole Genome Sequencing
To further dissect the novel identified GWAS signal, we performed whole-genome sequencing (WGS) analyses in 206 individuals (141 cases and 65 controls) of which 39 individuals were GBA1 rs3115534-GG carriers, 69 were rs3115534-GT and 98 were rs3115534-TT carriers. Short-read WGS DNA sequencing was performed by Psomagen (detailed in Supplementary Methods).
Oxford Nanopore Technologies (ONT) long-read whole-genome sequencing data was generated for five GBA1 rs3115534-GG carriers, two heterozygotes and six GBA1 rs3115534-TT carriers. High molecular weight DNA was extracted from either frozen blood samples or cell-lines. Additional details are described in Supplementary Methods.
Glucocerebrosidase activity
Patient-derived lymphoblastoid cell lines (LCLs) were obtained from the Coriell repository (https://www.coriell.org/). LCLs were maintained as directed in suspension with RPMI 1640 (ThermoFisher Scientific, 11875093) containing 2mM Glutamax (ThermoFisher Scientific, 35050061), and 15% FBS (ThermoFisher Scientific, A3160501) at 37°C in 5% CO2. Protein was extracted from LCLs using a citrate-phosphate buffer (0.2 M Na2HPO4, 0.1 M citrate, protease inhibitor, pH 5.8, Millipore Sigma, 11836170001) that was activated with 0.25% Triton X-100. Cells were subjected to a 4-methylumbelliferone (4-MU, Sigma Aldrich, M1381) fluorometric glucocerebrosidase (GCase) activity assay in quadruplicate as previously reported in the literature13 with adjusted incubation time of 2.5 hours. A total of 5E6 cells were used per sample with protein concentrations normalized to 0.7 mg/ml via BCA Protein Assay (Thermo Fisher Scientific 23225).
Acknowledgements
We thank all the participants that contributed to this study. GP2 is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). Additional funding was provided by The Michael J. Fox Foundation for Parkinson’s Research through grant MJFF-009421/17483. The funders supported clinical data collection and genotyping data generation.
Results
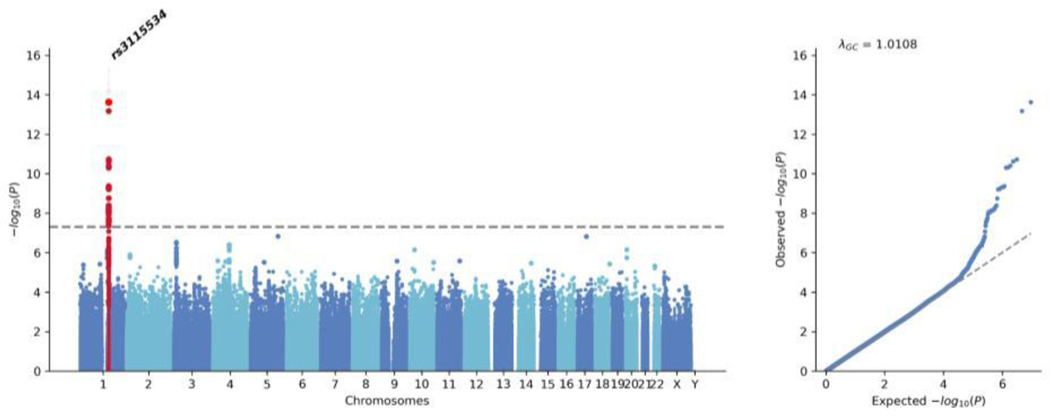
Here, we performed a GWAS meta-analysis (Figure 2) of the African (Supplementary Figure 4) and African admixed datasets (Supplementary Figure 5), totaling 1,488 cases and 196,430 controls. The demographic and clinical characteristics of the cohorts under study are provided in Table 1. This revealed that a total of 35 SNPs near the GBA1 gene were significantly associated with PD risk with consistent directionality of effect, the two most distant SNPs being 639,773 base pairs apart from each other (Supplementary Table 2). Conditional analyses on the top two SNPs suggested that there is only one causal signal driven by rs3115534 as the leading SNP. Of note, rs3115534-G is much more common in individuals of African or African admixed ancestry relative to other populations; allele frequency = 0.16 according to gnomAD (accessed February 2023)14 and allele frequency = 0.21 according to the African 1000 Genomes panel 15. The African and African admixed datasets used in this study yielded similar frequencies (African dataset; cohort MAF = 0.25, affected MAF = 0.33, unaffected MAF = 0.19), (African admixed datasets; cohort MAF = 0.14, affected MAF = 0.22, unaffected MAF = 0.13). Within our research cohorts, we found that rs3115534-G was more frequent in Nigerian populations (Supplementary Table 3). Linear regression analyses showed that the GBA1 rs3115534 variant was positively associated with a higher percentage of African ancestry (BETA = −0.001, SE= 0.0005, P= 0.011).
Figure 2: African and African admixed GWAS meta-analysis assessing Parkinson’s disease risk.
Manhattan plot showing the significance of the association as −log10.(p value) against chromosomes at a genomic scale (Bonferroni correction highlighted in grey at 5 × 10 3). GWAS=genome-wide association study.
We tested whether the effect of the risk allele was additive by calculating the frequency of homozygotes for the risk allele and heterozygotes in cases versus controls (Supplementary Table 8). As a follow-up analysis, we assessed whether this GBA1 variant is associated with AAO. Linear regression analyses in 711 African ancestry cases and 185 African admixed ancestry cases showed that GBA1 rs3115534-G is also an AAO disease modifier (African ancestry: BETA =−2.004, SE = 0.57, P = 0.0005; African-admixed: BETA = −4.15, SE =0.58, P =0.015; Meta-analysis: BETA =−3.06, SE =0.40, P = 0.008) resulting in onset of PD three years earlier per risk allele (Supplementary Figure 7).
In an attempt to further dissect the novel signal identified in the GBA1 locus, we next compared effect estimates and directionality of effect leveraging summary statistics from the largest PD GWAS meta-analysis of PD in Europeans1, Latin American16, and East Asian populations17. The rs3115534-G allele is extremely rare in Europeans (allele frequency = 0.0015), East Asians (allele frequency = 0.0005), South Asians (allele frequency = 0.0017), and Ashkenazi Jewish populations (allele frequency = 0.0009) according to gnomAD.
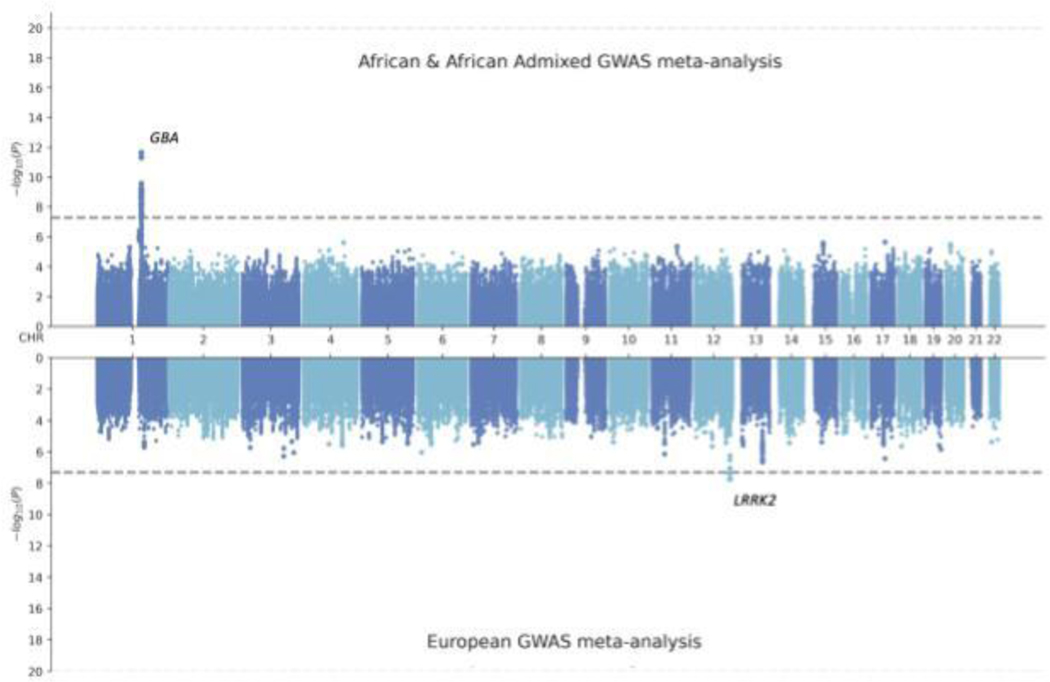
The GBA1 locus in African and African admixed populations differs substantially from Europeans (Figure 3; Supplementary Figure 8), whose association with disease risk is driven by two independent signals, including rs35749011 (GBA1-E326K) and rs76763715 (GBA1-N370S). These variants are very rare in individuals of African and African admixed ancestry (Supplementary Figure 14B). Similarly, the GBA1 locus considerably differs from the East Asian population, for which the rs3115534 variant was also not imputed in the largest East Asian GWAS meta-analysis17 (Supplementary Figure 14C). These differences are less noticeable when assessing the Amerindian/Latin American and indigenous populations, which harbor higher levels of African admixture (Supplementary Figure 14D); Loesch et al. GWAS16; rs3115534-G; OR = 1.13, 95% CI =0.41–1.86, P= 0.72; Amerindian/Latin American and indigenous 23andMe GWAS; rs3115534-G; OR = 1.56, 95% CI = 1.55–1.88, P= 0.01).
Figure 3: Miami plot comparing European versus African and African admixed GWAS meta-analyses of Parkinson’s disease risk.
Randomly sampled 1200 cases and 2445 controls were included in each of the GWAS meta-analyses. The grey horizontal lines indicate the significance threshold of 5 × 10 3. GWAS=genome-wide association study.
Interestingly, the larger sub-African population haplotypes spanning the rs3115534 variant were found in the Esan and the Yoruba in Ibadan (Nigerian) populations according to 1000 Genomes (Supplementary Figure 9), suggesting that this haplotype might have originated in these populations, given that founder effects result in decreased genetic admixture and therefore larger haplotype block sizes. Fine-mapping of this locus showed the lead SNP had a PP of 71.4% (rs3115534; Supplementary Table 4).
In an effort to identify a functional coding variant undetectable through genotyping or imputation that could explain the novel GWAS signal, we conducted short-read WGS analyses on a total of 206 individuals (141 cases and 65 controls) of which 39 individuals were GBA1 rs3115534-GG carriers, 69 were rs3115534-GT and 98 were rs3115534-TT carriers. A 96.6 % correlation was observed between short-read WGS and imputed genotyped data for rs3115534, validating the high quality of our imputed data. No differences in coding variation were observed between carriers and non-carriers of the GWAS signal (Table 2).
Table 2:
Functional coding variants identified by short-read whole genome sequencing in carriers of the novel GBA1 rs3115534 variant
| Variant | Base change | Functional consequence | Genetic variant | Cases with functional variant (n) | Controls with functional variant (n) | rs3115534-GG carriers (n) | rs3115534-GT carriers (n) | rs3115534-TT carriers (n) |
|---|---|---|---|---|---|---|---|---|
| chr1:155236249:A:C | A→C | Non-synonymous SNV | Ile320Ser | 1 | 0 | 0 | 1 | 0 |
| rs149487315 | C→T | Non-synonymous SNV | Met313Ile | 1 | 0 | 0 | 0 | 1 |
| rs143222798 | C→T | Synonymous SNV | Gly277Gly | 6 | 3 | 0 | 6 | 3 |
| rs61748906 | A→G | Non-synonymous SNV | Trp136Arg | 1 | 0 | 1 | 0 | 0 |
| rs368786234 | G→T | Non-synonymous SNV | Ser77Arg | 1 | 0 | 0 | 1 | 0 |
| rs761621516 | GTA→deleted | Non-frameshift deletion | Trp75del (222_224del) | 1 | 0 | 0 | 1 | 0 |
| rs150466109 | T→C | Non-synonymous SNV | Lys13Arg | 12 | 8 | 0 | 10 | 10 |
Analyses were done in 141 cases and 65 controls. All variants were on chromosome 1, were exonic, and were heterozygous. SNV=single nucleotide variant.
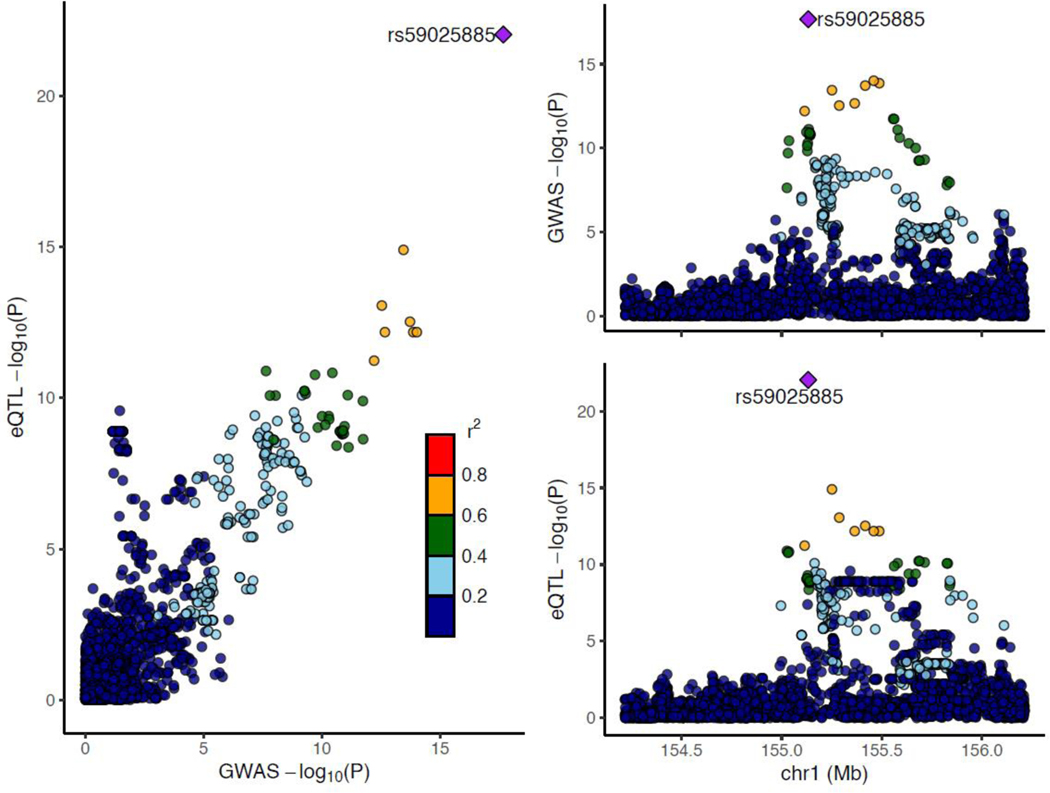
We leveraged existing whole blood expression quantitative trait locus (eQTL) summary statistics from Mak et al., 2021 based on RNA sequencing from 2,733 samples of predominantly African American and Indigenous American ancestries18. Of note, we identified a strong eQTL signal at rs3115534, located 8,821 bp from the canonical transcription start site (Figure 4; MAF = 0.15; P= 9.99E-25, BETA = 0.238, SE = 0.022). The rs3115534-G risk allele was found to be associated with increased GBA1 expression levels. We questioned whether this observation could be explained by the existence of multi-mapping reads between GBA1 and its pseudogene, GBAP1, which are often discarded in standard processing and do not contribute to gene-level quantification of expression in many publicly available datasets like GTEx (https://gtexportal.org/). Indeed, transcript diversity is a common and known biological phenomena19 that could explain the fact that rs3115534-G may increase the expression of a non-functional transcript that in turn would decrease the levels of the transcript responsible for optimal production of the protein isoform with GCase activity.
Figure 4. LocusZoom plot displaying African and African admixed Parkinson’s disease GWAS meta-analysis summary statistics versus African American eQTL summary statistics from blood.
eQTL=expression quantitative trait locus. GWAS=genome-wide association study.
Our data suggests a decreasing trend in GCase activity estimates when comparing rs3115534-GG homozygous risk allele (762.50 ± 273.50 U) versus rs3115534-GT heterozygous carriers (2743.76 ± 1960.83 U); (Welch Two Sample t-test - GG versus GT; t = −4.3138, df = 21.583, p-value = 0.00029) and rs3115534-TT homozygous non-risk allele carriers (1879.94 ± 1010.84 U) versus rs3115534-GG homozygous risk allele carrier; (Welch Two Sample t-test - GG versus TT; t = −4.7564, df = 18.363, p-value = 0.00014)(Supplementary Figures 11 and 12).
The largest PD-GWAS and multi-ancestry GWAS meta-analyses to date identified a total of 104 independent significant PD risk variants 1,17,20. Out of the 104 variants, 91 variants passed QC, imputation filters, and were present in the African and African admixed GWAS meta-analysis (Supplementary Figure 15; Table 2). Out of the 91 variants, 16 variants were nominally significant (p < 0.05; Supplementary Table 5) in the African and African admixed meta-GWAS reported here.
Discussion
Although there have been a number of published studies exploring PD genetics in the African and African admixed populations21,22,23,24,25,26,27,28,6,29,30,31,32,33,34, in the present study, we have gathered the largest collection of PD patients and controls from African and African admixed ancestry populations to comprehensively assess the genetic architecture of PD on a genome-wide scale. Here, we identified a novel African-specific GWAS signal on the GBA1 locus, significantly associated with PD risk and AAO, to be the most important risk factor for PD in these African and African admixed populations. Remarkably, almost a four times larger sample size in cases was required to nominate GBA1 as one of the major PD risk factors in the European ancestry population through GWAS35, showing the power and benefit of using diverse ancestry data.
GBA1 is a classic pleomorphic locus, showing coding, structural, and non-coding variants that exert different degrees of risk36. Despite the large effect size driven by this signal, our study did not identify an association with any previously reported or new GBA1 coding or structural aberration that could explain this signal37,38,39,40.
Strikingly, by leveraging existing eQTL data predominantly of African American ancestry, we found the rs3115534-G risk allele to be associated with increased GBA1 expression levels in whole blood, but paradoxically linked with a trend towards decreased GCase activity, which may be due to challenges with RNA-seq in this locus. Future large scale single cell expression studies should investigate in which brain cell types these expression differences are most prominent. This potential novel mechanism opens new avenues towards efficient RNA-based therapeutic strategies, such as antisense oligonucleotides or short interfering RNAs aimed at reducing lifetime risk.
Interestingly, given the high population frequency of the identified signal and the phenotypic characteristics of the homozygous Africans and African admixed carriers, our study does not support the notion that this variant causes Gaucher disease. Furthermore, the rs3115534 variant has been found to be extremely rare in non-African/African admixed populations. These findings suggest an African founder effect, and reinforce that the genetic architecture of this locus and its influence in risk and onset is different across populations. Interestingly, rs3115534 was also found to be associated with PD AAO in our study.
Here, we produce crucial insights into targeted construction of African ancestral haplotypes and potential novel pathogenic mechanisms underlying PD etiology. The utility of genetically characterizing populations of African and African admixed ancestry is unquestionable. This study demonstrates the importance of haplotype substructure discoveries for future fine-mapping efforts, showing how leveraging unique populations can benefit our understanding of complex diseases.
Overall, addressing the genetic complexity underlying these underrepresented populations, our study represents a valuable resource for identifying and tracking GBA1 carriers that may prove relevant for enrollment in target-specific PD clinical trials. We envisage that these data generated under the Global Parkinson’s Genetics Program initiative will be key to shed light on the molecular mechanisms involved in the disease process and might pave the way for future clinical trials and therapeutic interventions.
Limitations
Although we have made progress in assessing genetic risk factors for PD in an African-specific manner, there are a number of limitations to our study. Unraveling additional susceptibility genetic risk and phenotypic relationships would have been possible if a larger cohort had been analyzed. Considering our limited sample size, we lacked statistical power to detect common genetic variants of smaller effect sizes (Supplemental Figure 13). Additionally, an important proportion of the genetic risk contributing to the missing heritability of PD in the African and African admixed populations might result from rare alleles and structural variants that have not been assessed in the present study. Due to the lack of well-powered and African or African admixed RNA sequencing datasets, the added complexity of multi-mapping reads between GBA and GBAP1 and the limited number of LCLs to explore GCase activity in a large scale manner, we assume the limitation that this potential novel functional mechanism merits further study. We are aware that although this represents the first PD GWAS in the African and African admixed populations, two-thirds of the cases are of Nigerian descent, therefore likely unrepresentative of the substantial genetic diversity across the continent.
Data Sharing
All GP2 data is hosted in collaboration with the Accelerating Medicines Partnership in Parkinson’s disease, and is available via application on the website (https://amp-pd.org/register-for-amp-pd). The GWAS summary statistics from this study, excluding 23andMe, are available as of GP2’s release 5. 23andMe summary statistics are available via application on the website (https://research.23andme.com/dataset-access/). Genotyping imputation, quality control, ancestry prediction, and processing was performed using GenoTools v1.0, publicly available on GitHub (https://github.com/GP2code/GenoTools). All scripts for analyses are publicly available on GitHub [https://github.com/GP2code/GP2-AFR-AAC-metaGWAS; 10.5281/zenodo.7888141].
Ethics Statement
All cohorts recruited to the GP2 initiative undergo a thorough review of the consent forms in the Operations and Compliance working group, ensuring that each contributing study abided by the ethics guidelines set out by their institutional review boards, and all participants gave informed consent for inclusion in both their initial cohorts and subsequent studies within local law constraints. All GP2 data is hosted in collaboration with the Accelerating Medicines Partnership in Parkinson’s disease, and is available via application on the website (https://amp-pd.org/register-for-amp-pd).
Summary statistics for individuals with or without PD were provided through a collaborative agreement with 23andMe, Inc. Participants provided informed consent and volunteered to participate in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent (E&I) Review Services. As of 2022, E&I Review Services is part of Salus IRB (https://www.versiticlinicaltrials.org/salusirb). 23andMe summary statistics are available via application on the website (https://research.23andme.com/dataset-access/).
Supplementary Material
Research in Context.
Evidence Before this Study
Our current understanding of Parkinson’s disease (PD) is disproportionately based on studying populations of European ancestry, leading to a significant gap in our knowledge about the genetics, clinical characteristics, and pathophysiology in underrepresented populations. This is particularly notable in individuals of African and African admixed ancestries. Over the last two decades, we have witnessed a revolution in the research area of complex genetic diseases. In the PD field, large-scale genome-wide association studies in the European, Asian, and Latin American populations have identified multiple risk loci associated with disease. These include 78 loci and 90 independent signals associated with PD risk in the European population, nine replicated loci and two novel ancestry-specific signals in the Asian population, and a total of 11 novel loci recently nominated through multi-ancestry GWAS efforts. Nevertheless, the African and African admixed populations remain completely unexplored in the context of PD genetics.
Added Value of this Study
To address the lack of diversity in our research field, this study aimed to conduct the first genome-wide assessment of PD genetics in the African and African admixed populations. Here, we identified a genetic risk factor linked to PD etiology, dissected ancestry-specific differences in risk and age at onset, characterized known genetic risk factors, and highlighted the utility of the African and African admixed risk haplotype substructure for future fine-mapping efforts.
Implications of all the Available Evidence
We nominate a novel signal impacting GBA1 as the major genetic risk factor for PD in the African and African admixed populations. The present study could inform future GBA1 clinical trials, improving patient stratification. In this regard, genetic testing can help to design trials likely to provide meaningful and actionable answers. We identified a novel disease mechanism via expression changes consistent with decreased GBA1 activity levels. This novel mechanism may hold promise for future efficient RNA-based therapeutic strategies such as antisense oligonucleotid es or short interfering RNAs aimed at preventing and decreasing disease risk. This work represents a valuable resource in an underserved population, supporting pioneering research within the Global Parkinson’s Genetics Program (GP2) and beyond. Deciphering causal and genetic risk factors in all these ancestries will help determine whether interventions, potential targets for disease modifying treatment, and prevention strategies that are being studied in the European populations are relevant to the African and African admixed populations.
Funding:
Data used in the preparation of this article were obtained from Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). For a complete list of GP2 members see https://gp2.org. Additional funding was provided by The Michael J. Fox Foundation for Parkinson’s Research through grant MJFF-009421/17483.
Declaration of Interests
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA), National Institutes of Health, Department of Health and Human Services; project number ZO1 AG000535 and ZIA AG000949, as well as the National Institute of Neurological Disorders and Stroke (NINDS) and the National Human Genome Research Institute (NHGRI).
This work was supported in part by the Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Against Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). Additional funding was provided by The Michael J. Fox Foundation for Parkinson’s Research through grant MJFF-009421/17483. For a complete list of GP2 members see https://gp2.org
D.V., H.I., H.L.L., K.L. and M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research.
K.H. and members of the 23andMe Research Team are employed by and hold stock or stock options in 23andMe, Inc. M.A.N. also currently serves on the scientific advisory board for Character Biosciences Inc and Neuron 23 Inc.
DGS is a member of the faculty of the University of Alabama at Birmingham and is supported by endowment and University funds, is an investigator in studies funded by Abbvie, Inc., the American Parkinson Disease Association, the Michael J. Fox Foundation for Parkinson Research, The National Parkinson Foundation, Alabama Department of Commerce, Alabama Innovation Fund, Genentech, the Department of Defense, and NIH grants P50NS108675 and R25NS079188 and has a clinical practice and is compensated for these activities through the University of Alabama Health Services Foundation. He serves as Deputy Editor for the journal Movement Disorders and is compensated for this role by the International Parkinson and Movement Disorders Society. In addition, since January 1, 2022 he has served as a consultant for or received honoraria from Abbvie Inc., Curium Pharma, Appello, Theravance, Sanofi-Aventis, Alnylam Pharmaceutics, Coave Therapeutics, BlueRock Therapeutics and F. Hoffman-La Roche.
We thank the research participants and employees of 23andMe. The following members of the 23andMe Research Team contributed to this study:
Stella Aslibekyan, Adam Auton, Elizabeth Babalola, Robert K. Bell, Jessica Bielenberg, Katarzyna Bryc, Emily Bullis, Paul Cannon, Daniella Coker, Gabriel Cuellar Partida, Devika Dhamija, Sayantan Das, Sarah L. Elson, Nicholas Eriksson, Teresa Filshtein, Alison Fitch, Kipper Fletez-Brant, Pierre Fontanillas, Will Freyman, Julie M. Granka, Alejandro Hernandez, Barry Hicks, David A. Hinds, Ethan M. Jewett, Yunxuan Jiang, Katelyn Kukar, Alan Kwong, Keng-Han Lin, Bianca A. Llamas, Maya Lowe, Jey C. McCreight, Matthew H. McIntyre, Steven J. Micheletti, Meghan E. Moreno, Priyanka Nandakumar, Dominique T. Nguyen, Elizabeth S. Noblin, Jared O’Connell, Aaron A. Petrakovitz, G. David Poznik, Alexandra Reynoso, Madeleine Schloetter, Morgan Schumacher, Anjali J. Shastri, Janie F. Shelton, Jingchunzi Shi, Suyash Shringarpure, Qiaojuan Jane Su, Susana A. Tat, Christophe Toukam Tchakouté, Vinh Tran, Joyce Y. Tung, Xin Wang, Wei Wang, Catherine H. Weldon, Peter Wilton, Corinna D. Wong
We thank Cynthia J. Casaceli, Debbie Baker and Christi Alessi-Fox from the University of Rochester Clinical Trials Coordination Center for its contribution to the coordination of the BLAAC PD Study.
We thank Lisa Shulman for her contribution to the design of the protocol for BLAAC PD clinical assessments.
We thank the Biowulf team, as this study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://hpc.nih.gov).
Footnotes
Figure 1 was generated on www.biorender.com.
References
- 1.Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 2019; 18: 1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol 2020; 19: 170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okubadejo N, Britton A, Crews C, et al. Analysis of Nigerians with apparently sporadic Parkinson disease for mutations in LRRK2, PRKN and ATXN3. PLoS One 2008; 3: e3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cilia R, Sironi F, Akpalu A, et al. Screening LRRK2 gene mutations in patients with Parkinson’s disease in Ghana. J Neurol 2012; 259: 569–70. [DOI] [PubMed] [Google Scholar]
- 5.Okubadejo NU, Rizig M, Ojo OO, et al. Leucine rich repeat kinase 2 (LRRK2) GLY2019SER mutation is absent in a second cohort of Nigerian Africans with Parkinson disease. PLoS One 2018; 13: e0207984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonova-Doing E, Atadzhanov M, Quadri M, et al. Analysis of LRRK2, SNCA, Parkin, PINK1, and DJ-1 in Zambian patients with Parkinson’s disease. Parkinsonism Relat Disord 2012; 18: 567–71. [DOI] [PubMed] [Google Scholar]
- 7.Choudhury A, Aron S, Botigué LR, et al. High-depth African genomes inform human migration and health. Nature 2020; 586: 741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992; 55: 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blauwendraat C, Faghri F, Pihlstrom L, et al. NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol Aging 2017; 57: 247.e9–247.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koretsky MJ, Alvarado C, Makarious MB, et al. Genetic risk factor clustering within and across neurodegenerative diseases. bioRxiv. 2022; published online Dec 3. DOI: 10.1101/2022.12.01.22282945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters SP, Lee RE, Glew RH. A microassay for Gaucher’s disease. Clin Chim Acta 1975; 60: 391–6. [DOI] [PubMed] [Google Scholar]
- 14.Karczewski KJ, Francioli LC, Tiao G, et al. Author Correction: The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2021; 590: E53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loesch DP, Horimoto ARVR, Heilbron K, et al. Characterizing the Genetic Architecture of Parkinson’s Disease in Latinos. Ann Neurol 2021; 90: 353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foo JN, Chew EGY, Chung SJ, et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans: A Genome-Wide Association Study. JAMA Neurol 2020; 77: 746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mak ACY, Kachuri L, Hu D, et al. Gene expression in African Americans and Latinos reveals ancestry-specific patterns of genetic architecture. bioRxiv. 2021; : 2021.08.19.456901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustavsson EK, Sethi S, Gao Y, et al. The annotation and function of the Parkinson’s and Gaucher disease-linked gene GBA1 has been concealed by its protein-coding pseudogene GBAP1. bioRxiv. 2023; : 2022.10.21.513169. [Google Scholar]
- 20.Kim JJ, Vitale D, Véliz Otani D, et al. Multi-ancestry genome-wide meta-analysis in Parkinson’s disease. bioRxiv. 2022; published online Aug 6. DOI: 10.1101/2022.08.04.22278432. [DOI] [Google Scholar]
- 21.Ross OA, Wilhoite GJ, Bacon JA, et al. LRRK2 variation and Parkinson’s disease in African Americans. Mov Disord 2010; 25: 1973–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark LN, Levy G, Tang M-X, et al. The Saitohin ‘Q7R’ polymorphism and tau haplotype in multiethnic Alzheimer disease and Parkinson’s disease cohorts. Neuroscience Letters. 2003; 347: 17–20. [DOI] [PubMed] [Google Scholar]
- 23.Gwinn-Hardy K, Singleton A, O’Suilleabhain P, et al. Spinocerebellar ataxia type 3 phenotypically resembling parkinson disease in a black family. Arch Neurol 2001; 58. DOI: 10.1001/archneur.58.2.296. [DOI] [PubMed] [Google Scholar]
- 24.Okubadejo NU, Okunoye O, Ojo OO, et al. APOE E4 is associated with impaired self-declared cognition but not disease risk or age of onset in Nigerians with Parkinson’s disease. npj Parkinson’s Disease 2022; 8: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milanowski LM, Oshinaike O, Walton RL, et al. Screening of GBA Mutations in Nigerian Patients with Parkinson’s Disease. Mov Disord 2021; 36: 2971–3. [DOI] [PubMed] [Google Scholar]
- 26.Nishioka K, Ross OA, Vilariño-Güell C, et al. Glucocerebrosidase mutations in diffuse Lewy body disease. Parkinsonism Relat Disord 2011; 17: 55–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardien S, Keyser R, Yako Y, Lombard D, Carr J. Molecular analysis of the parkin gene in South African patients diagnosed with Parkinson’s disease. Parkinsonism Relat Disord 2009; 15: 116–21. [DOI] [PubMed] [Google Scholar]
- 28.Hashad DI, Abou-Zeid AA, Achmawy GA, Allah HMOS, Saad MA. G2019S mutation of the leucinerich repeat kinase 2 gene in a cohort of Egyptian patients with Parkinson’s disease. Genet Test Mol Biomarkers 2011; 15: 861–6. [DOI] [PubMed] [Google Scholar]
- 29.Keyser RJ, Lombard D, Veikondis R, Carr J, Bardien S. Analysis of exon dosage using MLPA in South African Parkinson’s disease patients. Neurogenetics 2010; 11: 305–12. [DOI] [PubMed] [Google Scholar]
- 30.Hulihan MM, Ishihara-Paul L, Kachergus J, et al. LRRK2 Gly2019Ser penetrance in Arab-Berber patients from Tunisia: a case-control genetic study. Lancet Neurol 2008; 7: 591–4. [DOI] [PubMed] [Google Scholar]
- 31.Ishihara-Paul L, Hulihan MM, Kachergus J, et al. PINK1 mutations and parkinsonism. Neurology 2008; 71: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouhouche A, Tesson C, Regragui W, et al. Mutation Analysis of Consanguineous Moroccan Patients with Parkinson’s Disease Combining Microarray and Gene Panel. Front Neurol 2017; 8: 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trinh J, Gustavsson EK, Vilariño-Güell C, et al. DNM3 and genetic modifiers of age of onset in LRRK2 Gly2019Ser parkinsonism: a genome-wide linkage and association study. Lancet Neurol 2016; 15: 1248–56. [DOI] [PubMed] [Google Scholar]
- 34.Okunoye O, Ojo O, Abiodun O, et al. allele and haplotype frequencies in Nigerian Africans: population distribution and association with Parkinson’s disease risk and age at onset. medRxiv 2023; published online March 24. DOI: 10.1101/2023.03.24.23287684. [DOI] [PubMed] [Google Scholar]
- 35.Simón-Sánchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 2009; 41: 1308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Hum Mol Genet 2011; 20: R158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toffoli M, Chen X, Sedlazeck FJ, et al. Comprehensive short and long read sequencing analysis for the Gaucher and Parkinson’s disease-associated GBA gene. Commun Biol 2022; 5: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JK, Koprivica V, Andrews DQ, et al. Glucocerebrosidase mutations among African-American patients with type 1 Gaucher disease. Am J Med Genet 2001; 99: 147–51. [DOI] [PubMed] [Google Scholar]
- 39.Tayebi N, Park J, Madike V, Sidransky E. Gene rearranagement on 1q21 introducing a duplication of the glucocerebrosidase pseudogene and a metaxin fusion gene. Hum Genet 2000; 107: 400–3. [DOI] [PubMed] [Google Scholar]
- 40.Mahungu AC, Anderson DG, Rossouw AC, et al. Screening of the glucocerebrosidase (GBA) gene in South Africans of African ancestry with Parkinson’s disease. Neurobiol Aging 2020; 88: 156.e11–156.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.