Summary
The basolateral amygdala (BLA) projects widely across the macaque frontal cortex1-4, and amygdalo-frontal projections are critical for optimal emotional responding5 and decision-making6. While it is appreciated that single BLA neurons branch and project to multiple areas in frontal cortex, the organization and frequency of this branching has yet to be fully characterized. Here, we determined the projection patterns of over 3,000 macaque BLA neurons. We found that one-third of BLA neurons had two or more distinct projection targets in frontal cortex and subcortical structures. The patterns of single BLA neuron projections to multiple areas were organized into repeating motifs that targeted distinct sets of areas in medial and ventral frontal cortex indicative of separable BLA networks. Our findings begin to reveal the rich structure of single neuron connections in the non-human primate brain, providing a neuroanatomical basis for the role of BLA in coordinating brain-wide responses to valent stimuli7.
eTOC Blurb
Zeisler et al. utilize multiplexed analysis of projections by sequencing (MAPseq) to profile the anatomical connections of single neurons in primate basolateral amygdala. Analyzing the connections of over 3000 single neurons, they identify highly structured and repeating single neuron projection motifs that are indicative of separable BLA centered networks.
Introduction
Basolateral amygdala (BLA) is essential for adaptive emotional responding in humans and animals5,8,9. In humans, dysfunction within or damage to the circuits that connect through the amygdala are theorized to be the cause of numerous psychiatric disorders, including autism spectrum disorder, post-traumatic stress disorder, and schizophrenia10-12. Based on decades of tract-tracing studies in macaques, we know that the primate BLA projects widely across the brain, sending connections primarily to ventral and medial parts of the frontal lobe, as well as the temporal and occipital cortex, thalamus, and striatum1-4. These widespread connections, especially those to frontal lobe, are central to accounts of how BLA coordinates learning about and responding to different emotionally salient events13,14.
Despite the appreciation that the BLA plays a central role in coordinating activity across distributed networks in frontal cortex to guide emotional behavior, the anatomical organization of single neuron projections from this area to frontal cortex are largely unknown. Understanding these fine-grained patterns of connections is critical to understanding how BLA interacts with the rest of the brain. It is possible that single BLA neurons project to only one specific target, transmitting information to frontal areas in dedicated pathways. Such an organizing principle or connectional motif would align closely with the BLA’s known role in model-based behaviors and the processing of sensory-specific stimuli through interaction with distinct parts of frontal cortex15,16. An alternative is that single neurons in BLA branch and project to many different areas, such that activity can be efficiently coordinated across distributed networks of areas. This organization fits with the BLA’s role in more general aspects of motivation and response invigoration to approach or avoid salient stimuli that are more characteristic of model-free behaviors7.
These two potential projection motifs of single BLA neurons – specific vs branching – are not necessarily mutually exclusive. However, at present, the extent to which either motif best characterizes the projections of single BLA neurons is not known. Further, if single BLA neurons do branch and project to multiple areas, how are these projections organized? In part these details are unclear because gold-standard tract-tracing approaches are either too coarse to detect the projection patterns of individual neurons17,18 or because the available single neuron tracing techniques do not scale practically to non-human primates19.
To surmount these issues, we applied a high-throughput sequencing approach, multiplexed analysis of projections by sequencing (MAPseq20) to macaque monkeys. MAPseq uses barcoded mRNA technology21 to map the connections of individual neurons at scale and critically it permits the analysis of single neuron projections to many target areas, allowing the extent and organization of single-neuron branching to be discerned. Because of their potential importance in psychiatric disorders10, we focused on projections from BLA to frontal cortex, striatum, anterior temporal lobe, and mediodorsal nucleus of the thalamus (MD), a part of thalamus that receives input from both frontal cortex and amygdala1. Using this approach, we found that individual BLA neurons project widely in frontal cortex; about half of the neurons that leave amygdala project to more than one target area of those that we sampled. The pattern of these branching projections was not random and single neuron connections were organized into distinct and reproducible connectional motifs.
Results
MAPseq uses a sindbis virus vector to infect neurons with unique RNA barcodes. The approach works on the basis that single neurons are mostly likely to take up only a single virus particle coding for a unique RNA barcode. These RNA barcodes are conjugated to a non-functional presynaptic protein and thus are expressed in the cell body and transported down axons to synaptic terminals20. Then, by dissecting and sequencing samples from both injection sites and target sites, single-neuron projection patterns can be determined by comparing the list of barcodes found in each of the samples.
Validation of MAPseq in macaques
We made bilateral, MRI-guided stereotactic injections of barcoded sindbis virus into the BLA of two rhesus macaques (Figure 1A, see Methods and Supplemental Figure 1 for more detail). Following perfusion, brains were extracted, and the hemispheres separated and sectioned. The BLA and target areas in frontal cortex, striatum, entorhinal cortex, hippocampus, and MD were dissected according to gray/white matter boundaries as well as sulcal landmarks (Figure 1B). This set of areas was chosen because it encompasses the primary projection targets of BLA to frontal cortex1-4. It is important to note that because we selected a subset of possible target regions (for instance, we did not dissect targets from more caudal temporal cortex), we are not necessarily capturing the full extent of each sequenced neuron’s projections. Instead, all conclusions are based solely with reference to the set of target described here.
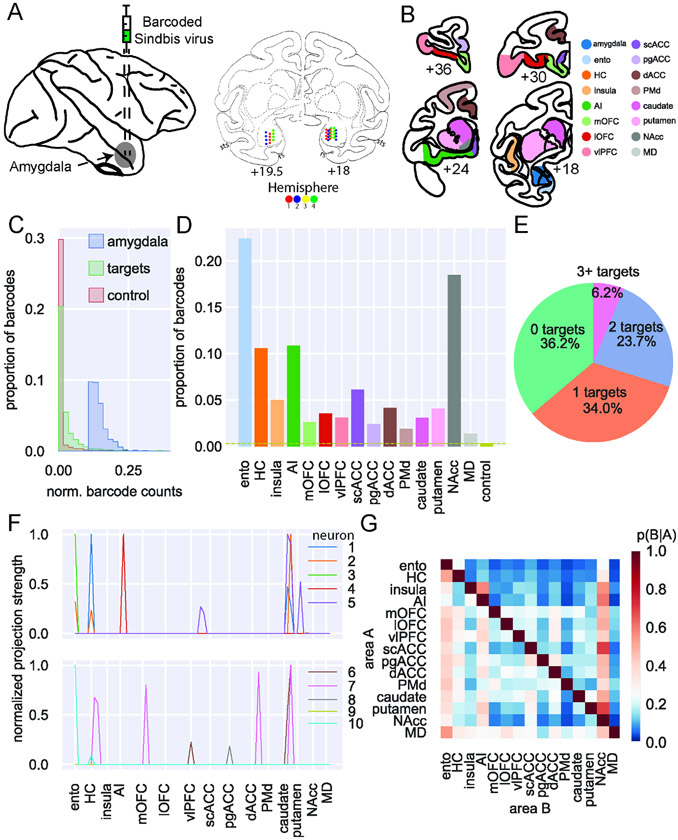
Figure 1: MAPseq of macaque BLA.
A) Left: schematic of injection approach. Right: actual injection sites plotted on coronal atlas sections (interaural distance in mm). In the anterior section (19.5 mm), injections were made in only one vertical track per animal; in the posterior section (18 mm), injections were made in two tracks per animal. Hemispheres 1 and 2 (red and blue, respectively) came from one animal, while hemispheres 3 and 4 came from the other (yellow and green, respectively). B) Example coronal sections showing dissection targets in striatum as well as frontal and temporal lobes. Ento, entorhinal; HC, hippocampus (not shown); AI, anterior insula cortex; mOFC, medial orbitofrontal cortex; lOFC, lateral orbitofrontal cortex; vlPFC, ventrolateral prefrontal cortex; scACC, subcallosal anterior cingulate cortex; pgACC, perigenual ACC; dACC, dorsal ACC; PMd, dorsal premotor cortex; NAcc, nucleus accumbens; MD, mediodorsal thalamus (also not shown). Anterior-posterior levels are in mm relative to the interaural plane. C) Distribution of barcode counts relative to max found in injection site across non-injection site amygdala samples (blue), all target structures (green; OLS regression vs amygdala: t(2) = 46.43, p < 0.0001), and control sites in cerebellum (red vs targets: t(2) = 4.77, p < 0.001). Significantly more barcode was expressed within the amygdala than distal target sites, and significantly more barcode was expressed in target sites than control tissue from cerebellum, to which the amygdala has a very low probability of projecting. D) Proportion of barcodes in each of the target and control areas; dashed line indicates the proportion of barcode found in control sites. E) Proportion of unique barcodes in no targets outside of amygdala (green), one (orange), two (blue), or more than two targets outside of amygdala (red). F) Normalized proportion of barcodes across all the potential target areas and control areas for 10 unique barcodes (colored lines). G) Conditional probability that a barcode in area A is also found in area B. Note that the order of indexing here alters the probability such that P(A∣B) can be greater than P(B∣A). See also Supplemental Figures 1-5.
We confirmed the integrity of the RNA in the tissue samples using Bioanalyzer and then used qPCR to confirm that barcode expression was robust compared to a housekeeping gene (Supplemental Figure 2). As expected, we recovered significantly more barcode in amygdala samples near the injection sites compared to samples from frontal cortex (OLS regression, t(2) = 46.43, p < 0.0001) and more in frontal cortex compared to control samples from cerebellum (t(2) = 4.77, p < 0.0001; Figure 1C). We specifically chose the cerebellum as a control site as amygdala is exceedingly unlikely to project to this part of the brain22. Thus, barcode mRNA was expressed in macaque neurons and was then transported to the axon terminals in frontal cortex and other brain areas.
High-throughput next-generation sequencing was then conducted on extracted mRNA from BLA and target areas. Counts of unique barcodes from the extracted RNA were normalized to a spike-in sequence (i.e. a known RNA sequence added to each sample to control for PCR efficiency across samples) and a threshold applied to control for spurious sequencing results20 (Supplemental Figure 3). The combined thresholded barcode counts from the four hemispheres were then analyzed together (see Supplemental Figures 4 and 5 for analyses of each hemisphere separately and comparisons between hemispheres, respectively).
In total, we recovered 3,115 unique barcodes in samples dissected from BLA across the four hemispheres. This yield of barcode counts is similar in level to that recovered in mice per injection21-24. This indicated that MAPseq was working as intended in the macaque brain. We also conducted an additional control analysis to assess whether in our data there was any evidence that a single neuron might take up two virus particles. MAPseq works on the basis that this is extremely unlikely20, but nevertheless we conducted an analysis of how often two barcodes had the exact same expression across all of our samples. Of the barcodes that met the threshold, only 10 out of 3,115 (0.32%) had the same pattern of expression. This indicates that the sindbis vector was likely only infecting individual neurons with a single barcode sequence and thus MAPseq is working similarly in macaques as it does in mice.
The overall proportion of barcode recovered in each target brain area (Figure 1D) mirrored known connections of BLA in macaques. The highest amount of barcode was recovered in entorhinal cortex and nucleus accumbens (NAcc), two areas which have previously been identified as receiving dense projections from BLA25. A lower amount of barcode was recovered from hippocampus and agranular insula (AI), which are also well-documented as receiving strong projections from BLA4,26; these areas were followed by subcallosal (scACC) and dorsal ACC (dACC), which were found to receive the strongest amygdala input among frontal areas in one of the few quantitative studies of amygdala projections4. Notably, relatively similar amounts of barcode were recovered from all other target areas in frontal cortex and striatum, which again matches known gradients of BLA connections across the frontal lobe4,27. Thus, the level of barcode mRNA recovered and the alignment between our findings and prior tract-tracing supports the validity of MAPseq for quantifying area-to-area projections in macaques.
To begin to establish the extent of projections from putative single neurons in BLA to multiple areas, we then determined whether each barcode found in BLA was present in any of the target brain areas after collapsing across all samples in a target area. Approximately one-third of the unique barcodes – which can be interpreted to represent single neurons – were found only in samples within the amygdala. We refer to these as having zero-targets. These cells are likely to be either intra-amygdala interneurons or cells that project to a target area that was not included in this analysis, such as parts of the anterior temporal lobes26. Another third projected to only one of the target areas we collected. We refer to these neurons as having specific projections (i.e., specific with respect to the set of target areas we assessed). Finally, the remaining third had two or more targets outside of amygdala. We refer to these neurons with multiple targets as having branching projections (Figure 1E). Thus, of the two-thirds of BLA neurons that we found that projected outside of amygdala, half of those branched to innervate multiple locations in frontal cortex. The likelihood of branching is significantly higher than prior estimates based on the tracing of single axons28, highlighting the unique contribution of the high throughput and multiplexed capability of MAPseq.
Quantitative analysis of branching connectional motifs of single BLA neurons
Prior analyses of the patterns of single neuron projections to multiple cortical areas have been limited in either the number of neurons that could be carefully traced28 or the number of targets that could be assessed29. MAPseq, however, overcomes this issue as it can reveal the connections of single neurons to multiple brain areas (Figure 1F). Thus, we next looked at the probability that a barcode found in one target structure would also be found in another target structure (Figure 1G). This approach allowed us to begin to ascertain the degree to which connections of single BLA neurons are either specific to a single area or branch to multiple areas. As can be seen in Figure 1G, BLA neurons often targeted multiple locations in frontal cortex. For example, BLA neurons that project to NAcc also have a high probability of projecting to all parts of frontal cortex, especially agranular insula cortex (AI). By contrast, single BLA neurons that project to AI are unlikely to project to either entorhinal cortex or hippocampus. Similar patterns of non-overlapping projections are also apparent in BLA projections to frontal cortex. For example, neurons that project to medial orbitofrontal cortex (mOFC) are unlikely to project to ventrolateral prefrontal cortex (vlPFC).
Notably, the pattern of these projections to two areas corresponds closely with the findings of two studies that investigated the patterns of BLA neuron branching to frontal cortex and thalamus. One by Sharma, Fudge and colleagues29 identified a subset of amygdala neurons that project to adjacent parts of medial frontal areas 25/14 (which correspond to our scACC and mOFC areas, respectively) and 32/24 (which correspond to our perigenual ACC [pgACC] and dACC areas, respectively). Our MAPseq results identified neurons that projected to both areas, although we identified a slightly higher proportion of neurons that projected to both areas (7-20%) compared to that prior work (7-10%). This difference likely reflects the fact that our dissected areas encompassed larger portions of medial frontal cortex than the relatively focused injections of retrograde tracers. We also found highly similar results to those reported by Timbie and Barbas30 who found two non-overlapping populations of amygdala neurons that project to either MD or posterior OFC/AI (Figure 1G). This lack of branching as discerned by our MAPseq analysis is an important finding. This is because it shows that this technique has the resolution to reveal both the expected pattern of branching projections to certain targets29, as well as expected pattern of specific projections30. Taken together these analyses provide evidence that BLA neurons often project to multiple targets in frontal cortex, arguing against specific or one-to-one targeting as the dominant connectivity principle.
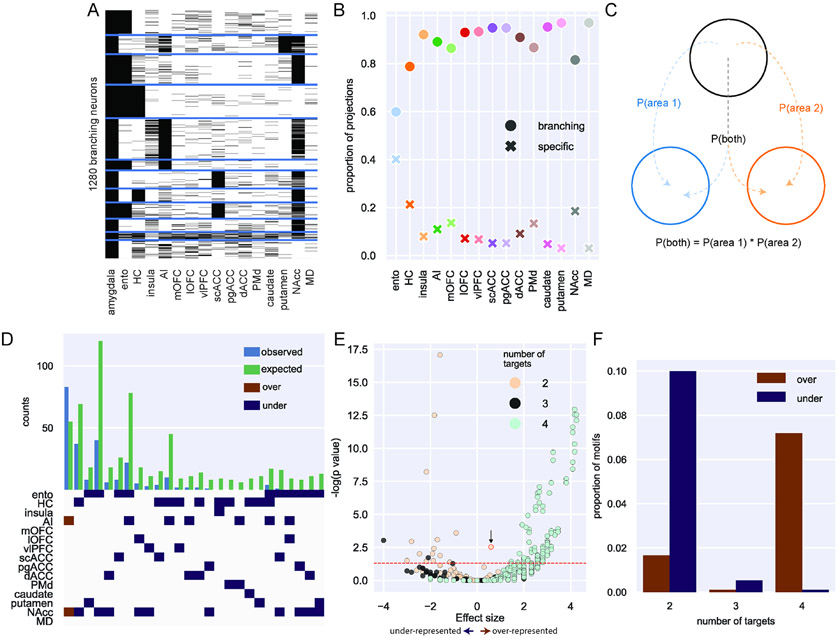
The prior analysis on the conditional probabilities of neuronal branching to two targets (Figure 1G), while revealing, does not capture the full set of connections that single BLA neurons make to multiple areas – one of the major strengths of MAPseq over standard tract-tracing approaches. Consequently, we focused our next analysis on the nearly 1,300 BLA neurons with projections that branch to multiple locations in frontal cortex, temporal cortex, striatum and thalamus (Figure 2A). While all target areas received the majority of their input from neurons that branched to multiple areas, entorhinal cortex received the highest proportion of projections from BLA that were specific and only targeted this area (Figure 2B, z-test for proportions: entorhinal vs hippocampus [next highest], z = 10.67, p < 0.0001). Such a difference could be related to this being the only cortical temporal lobe area that we assessed.
Figure 2: Single neuron analysis of branching projections from BLA.
A) K-means clustering of 1,300 neurons that project to more than one area outside of amygdala (k=12). B) Proportion of projections from BLA to frontal cortex areas, striatum, and mediodorsal thalamus that are from specific (crosses) or branching projections (filled circles). C) Logic of null distribution computation for branching motifs. D) Observed (blue) and expected (green) counts of neurons with projections to multiple areas (top). Specific over (red) or under-represented (blue) branching motifs by area (bottom). E) Volcano plot of probability of all possible branching motifs with 2 (cream), 3 (grey), and 4 (turquoise) target areas. Positive effect size indicates over-representation compared to the null distribution and negative effect size indicates under-representation. The red dashed line marks the level of statistical significance such that any points above it are significantly over- or under-represented after FDR correction. The point labeled with the black arrow and red outline refers to the bifurcating motif between NAcc and AI. F) Proportion of significantly over- (red) and under- (blue) represented 2-, 3- and 4- target area branching motifs. See also Supplemental Figure 6.
Next, we sought to determine whether any branching motifs were over- or under-represented compared to chance. First, we compared the frequency of these branching motifs to a null distribution based on a uniform likelihood of projecting to each area; assuming total independence for branching, the probability that a neuron projects to both areas A and B can be computed as the product of the independent probabilities of projecting to area A and area B24. Thus, with 120 possible unique BLA bifurcating motifs to two areas, the expected probability of each of those would be 0.0083; 560 unique trifurcating motifs result in an expected probability of 0.0018, and 43860 unique quadfurcations results in a probability of 0.000023. The results of this analysis are displayed in Supplemental Figure 6, and they indicate that motifs including areas that receive stronger input from BLA, like entorhinal cortex, are more likely to be found as over-represented. To better answer this question and account for the different projection strengths from BLA to each target area, we built another null distribution that was instead based on the overall distribution of barcodes across the target areas in our data (Figure 2C). Comparing the actual counts for each motif with the null distribution based on the relative proportion of barcodes, we identified only one over-represented bifurcating motif: neurons that project to both NAcc and AI (binomial test, p = 0.003; highlighted with the black arrow and red outline in Fig. 2E). This motif was found alongside predominantly under-represented bi- and trifurcations, many of which included projection motifs encompassing some combination of NAcc, entorhinal cortex, and hippocampus (Figure 2D and 2E). That branching to only two of these areas happens less frequently than expected is somewhat surprising considering these were the three areas most likely to receive projections from BLA in our data (Figure 1E). By contrast, single neurons that projected to four different target areas were far more likely to be over-represented compared to that same chance distribution (Figure 2F). In other words, there are fewer bifurcations than expected based solely on the proportions of barcodes found in each target area, while there were more BLA neurons that had quadfurcations than expected. Importantly, this analysis also revealed that the observed branching motifs are not simply a product of the distribution of barcodes (i.e., entorhinal cortex receives the highest proportion of projections from BLA, but projections to entorhinal are not the most likely to branch to multiple areas). Instead, branching of single BLA neurons appears to be highly structured.
BLA single neuron projection networks within frontal cortex
With the appreciation that the projections of single neurons in BLA are highly likely to branch to multiple areas we sought to understand how these projections are organized. Here we separately focused on the pattens of single BLA projections to the parts of frontal cortex that receive the highest density of connections, namely medial and ventral frontal cortex3,4,26,30. Furthermore, BLA projections to these areas are thought to be functionally distinct; interaction between BLA and medial frontal cortex is heavily linked to defensive threat conditioning in animals, aspects of social valuation31,32, and anxiety-related disorders such as PTSD in humans33. Projections from BLA to ventral frontal cortex are, by contrast, more frequently associated with reward-guided behaviors6,34-40 and dysfunction in these circuits is linked to addictive disorders41.
Medial frontal cortex
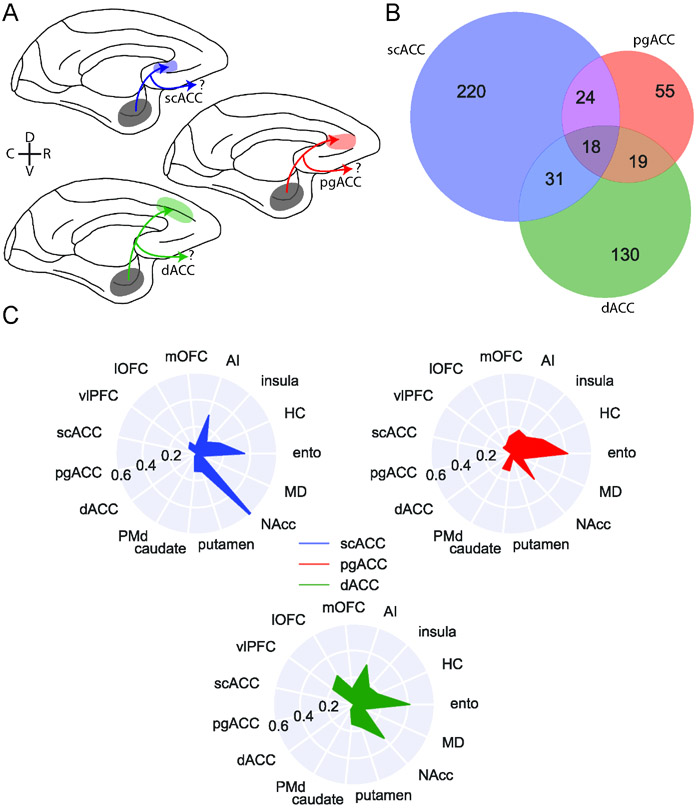
We identified 405 BLA neurons that project to either medial frontal areas scACC (area 25 as defined by Carmichael and Price42), pgACC (area 32), or dACC (area 24) (Figure 3A). Although these areas are densely interconnected17, we found marked differences in the organization of the BLA projections that they receive. First, over half of the BLA neurons that projected to medial frontal cortex exclusively targeted scACC (220/405), whereas a third targeted only dACC (130/405; z-test for proportions, z = 6.38, p < 0.0001); even fewer BLA projections to medial frontal cortex were specific to pgACC (55/405; z = 6.28, p < 0.0001) (Figure 3B). Indeed, the majority of BLA neurons targeting pgACC also projected to the other parts of medial frontal cortex, while a smaller proportion of dACC-projecting neurons branched within medial frontal cortex (z = 5.37, p < 0.0001). scACC-projecting neurons were least likely to branch and innervate other parts of medial frontal cortex (z = 3.17, p = 0.0015). Thus, within medial frontal cortex there is a hierarchy of specific versus branching BLA connections where pgACC receives the least specific BLA input and scACC receives the most.
Figure 3: BLA specific projections to medial frontal cortex.
A) Schematic showing the three populations of medial frontal cortex-projecting neurons analyzed here: BLA neurons targeting scACC (blue), pgACC (red), dACC (green). D and V refer to the dorsal and ventral directions, respectively; R and C designate rostral and caudal. B) Venn diagram illustrating projections to each medial frontal cortex area that also branch to the other two; pgACC-projecting neurons are more likely to branch than dACC-projecting neurons (z-test for proportions, z=3.17, p=0.0015), which are more likely to branch than scACC-projecting neurons (z=2.27, p=0.023). C) Likelihood of medial frontal cortex-projecting neurons projecting to non-medial frontal cortex targets. Populations of neurons here are non-overlapping, such that neurons which branch between scACC, pgACC, and dACC are excluded. See also Supplemental Figure 7.
Next, we focused only on the neurons that projected to one of the three medial frontal areas without branching between them. We did this in order to assess the structure of the specific projections that do not overlap. As we noted earlier, BLA neurons tend to have more projections that branch to multiple areas than projections to a single specific area (Figure 2C and E). Within this branching we found additional structure. Neurons that project to dACC were more likely to have four targets than neurons which projected to pgACC (z-test for proportions, proportion 4-targets: z = 2.47, p = 0.013), which were dominated by neurons that projected to two or three target areas (Supplemental Figure 7B). These results indicate that although much of the BLA input to pgACC is shared among other medial frontal areas, that same input is much less frequently broadcast to areas outside of medial frontal cortex, highlighting that a single cortical area can receive both highly shared and highly specific input at the same time.
Within the projections from BLA to medial frontal cortex there were notable differences in the areas that these single neurons sent branching projections to. Whereas the BLA neurons projecting to each medial area had similar proportions of bifurcations to hippocampus and entorhinal cortex (Figure 3C), neurons that projected to scACC were far more likely to also project to NAcc than the other areas (z-test for proportions: scACC vs pgACC: z = 5.85, p < 0.0001; scACC vs dACC: z = 6.33, p < 0.0001). This projection motif was evident at the level of individual neurons (Supplemental Figure 7A) and these cells were also highly likely to connect to AI. dACC-projecting BLA neurons, however, were more likely than other parts of medial frontal cortex to also project to lateral OFC (lOFC; dACC vs pgACC: z = 2.72, p = 0.0097; dACC vs scACC: z = 3.80, p < 0.001) and vlPFC (dACC vs pgACC: z = 2.04, p = 0.061; dACC vs scACC: z = 3.52, p < 0.001) on the ventral surface of the frontal lobe. This pattern of connections may be related to the role of this area in foraging and valuation which is thought to require interactions with ventral frontal areas6,43.
In summary, single BLA neurons targeting medial frontal cortex appeared to be split into distinct dorsal-ventral networks; those targeting the more ventral scACC were largely constrained to this area and primarily sent bifurcations to NAcc and AI in the posterior ventral frontal cortex. Given the known role for scACC and amygdala in regulating affect in general and mood in particular this branching projections could play a role in controlling affective states44. By contrast, BLA neurons projecting to more dorsal dACC also innervated parts of ventral frontal cortex that are central to accounts of value-based decision-making6,43 indicating that this network may be essential for adaptive foraging45,46.
Ventral Frontal Cortex
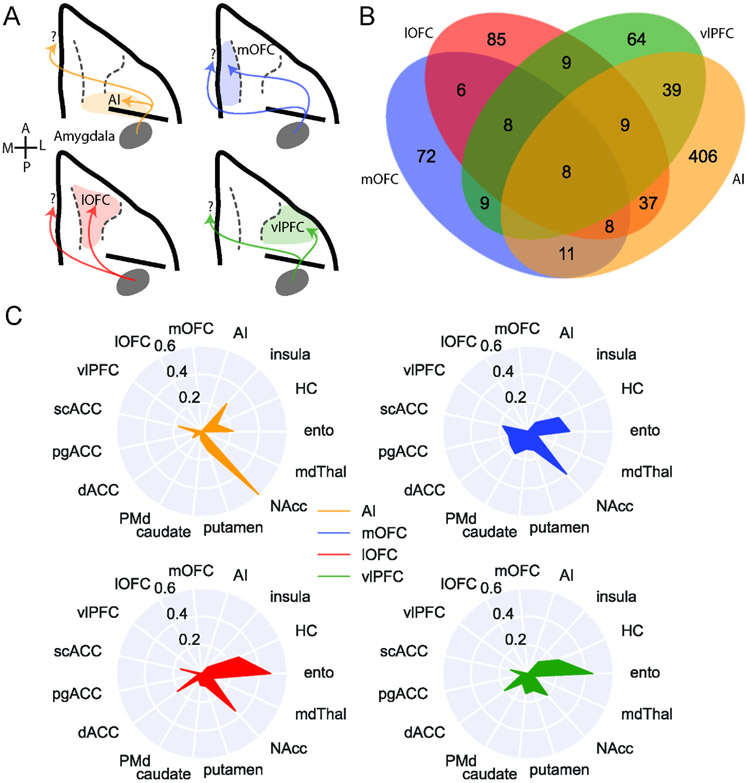
We conducted a similar analysis focused only on the 627 BLA neurons that projected to areas on the ventral surface of frontal cortex, including AI, mOFC, lOFC, and vlPFC (Figure 4A). Of those BLA neurons that projected to ventral frontal cortex, almost two thirds (65%) of BLA neurons solely targeted AI. By contrast, only 10% solely targeted vlPFC, meaning that the majority of BLA neurons projecting to vlPFC also targeted other parts of the ventral frontal cortex. Within ventral frontal cortex, these areas received the highest and lowest proportions of specific projections, respectively (z-test for proportions: z = 19.85, p < 0.0001, Figure 4B). BLA neurons projecting to mOFC and lOFC were evenly split between those that specifically projected to these areas or branched to other parts of ventral frontal cortex. Overall, of the BLA projections to ventral frontal cortex, those to AI are more specific compared to those directed to more anterior regions (AI vs mOFC: z = 4.43, p < 0.0001), with vlPFC receiving the fewest specific projections from BLA (vlPFC vs mOFC: z = 2.48, p = 0.013).
Figure 4: BLA specific projections to ventral frontal cortex.
A) Schematic showing the four populations of ventral frontal cortex-projecting neurons analyzed here: BLA neurons targeting AI (yellow), mOFC (blue), lOFC (red), and vlPFC (green). A and P designate anterior and posterior directions, respectively; M and L refer to medial and lateral. B) Venn diagram illustrating projections to each area that also branch to the other three; AI-projecting neurons were least likely to branch to the other areas (z-test for proportions, z=4.425, p<0.0001), while vlPFC-projecting neurons were most likely to branch within ventral FC (z=2.476, p=0.0133). C) Likelihood of ventral frontal cortex-projecting neurons projecting to non-ventral frontal cortex targets. Populations are non-overlapping, such that we are excluding neurons that branch between mOFC, lOFC, vlPFC, and AI. See also Supplemental Figure 7.
Similar to medial frontal cortex, BLA neurons projecting to ventral frontal cortex were more likely to send branching as opposed to specific projections. AI-, lOFC-, and vlPFC-projecting neurons were most likely to project to two areas (Supplemental Figure 7D; binomial test, vlPFC 2-targets vs null: p = 0.009), whereas mOFC-projecting neurons tended to innervate either one or four areas (mOFC 1-target or 4-targets vs null: p < 0.0001). AI- and vlPFC-projecting neurons were least likely to branch to four distinct areas, suggesting that these two areas receive more specific input from BLA.
When we limited our analyses to BLA neurons that only projected to one of the four ventral frontal areas without branching between them, BLA neurons that projected to AI exhibited the strongest projections to NAcc compared to other areas in ventral frontal cortex (Figure 4C and Supplemental Figure 7C; Fisher’s exact test: AI vs mOFC, p = 0.0057; AI vs lOFC, p = 0.00019; AI vs vlPFC, p < 0.0001). By contrast, vlPFC- and lOFC- projecting neurons were most likely to also project to entorhinal cortex compared to mOFC and AI (lOFC vs mOFC, p = 0.043; lOFC vs AI, p < 0.0001; vlPFC vs AI, p = 0.00051). Somewhat unexpectedly, BLA neurons with projections to mOFC were more likely to project to dorsal premotor cortex (PMd; mOFC vs AI, p < 0.00001; mOFC vs lOFC, p = 0.028) and other more medial areas in the frontal lobe such as the pgACC (mOFC vs AI, p = 0.019; mOFC vs lOFC, p = 0.038).
These patterns of projections indicate the BLA neurons targeting the ventral frontal cortex have reproducible patterns of connections and innervate separable networks from frontal cortex areas; those targeting AI are less likely also to project to other ventral frontal areas. When they do branch to other areas, they primarily innervate other areas in the posterior frontal lobe such as scACC and NAcc. By contrast, BLA projections to more anterior parts of ventral frontal cortex, especially vlPFC, branch more both within ventral frontal cortex and also to other areas, including dACC, scACC, and NAcc. This finding that vlPFC-projecting BLA neurons connect so broadly aligns closely with the known role of the BLA-vlPFC circuit in representing and updating model-free stimulus-outcome associations47, information that is likely to be shared among other ventral frontal areas to inform reward-guided choice behavior48.
Taken together, the patterns of projections we found reveal the existence of BLA neurons that target distinct parts in the medial and ventral frontal cortex, indicative of separable multi-region BLA networks. Those BLA neurons that target scACC and AI in the posterior frontal cortex preferentially innervate NAcc. By contrast, BLA neurons that project to more anterior medial and ventral frontal areas appear to be part of more distributed networks of areas.
Retrograde tracing of specific and branching BLA projections to ventral frontal cortex
Next, we sought to assess the most distinct BLA projection patterns revealed by MAPseq using standard retrograde viral tracing. We note, however, that by making discrete injections of retro-AAV into subregions of the areas that we analyzed with MAPseq, that our two approaches are complimentary, not equivalent. For the MAPseq experiments, we collected large samples from each of our target areas in order to assess collateralization at an area-by-area level by combining dissected samples across A/P levels. Retrograde tracing, by contrast, provides much more specific insight into the origins of the neurons that project to a specific subregion of a target area. These two approaches, when utilized in concert, are essential to provide evidence for single-cell collateralization to two areas, as well as to identify particular subregions or even cell layers wherein collateralization might be more or less common.
In a single long-tailed macaque, MRI-guided stereotactic injections of retro-AAV2 coding for mCherry and EGFP fluorophores were injected into NAcc (mCherry) and a lateral subregion of AI (EGFP) in one hemisphere. Injections were targeted to posterior-lateral lOFC (EGFP, area 13m as defined by Carmichael and Price42) and posterior-medial vlPFC (mCherry, area 12o) in the other hemisphere (Figure 5A and B). As cross-hemispheric BLA connections are negligible27, we were able to analyze the two hemispheres separately. We then conducted unbiased stereological counting of neurons in BLA that were either single or double labeled with each fluorophore49. While we observe dense labeling of cell bodies in the cortex immediately ventral to our injection site in NAcc, our injection was indeed into NAcc as opposed to the caudal-medial agranular cortex. This is because based on prior work, if this part of the caudal cortex had been injected we would have expected retrograde labeling in the putamen, lateral caudate nucleus, and AI lateral to the injection50, which we did not observe. Instead, we found an overall pattern of retrograde labeling across the ventromedial frontal cortex indicative of the previously published connections of NAcc and medial striatum51 and relatively diffuse labeling of processes around the injection site as would be expected.
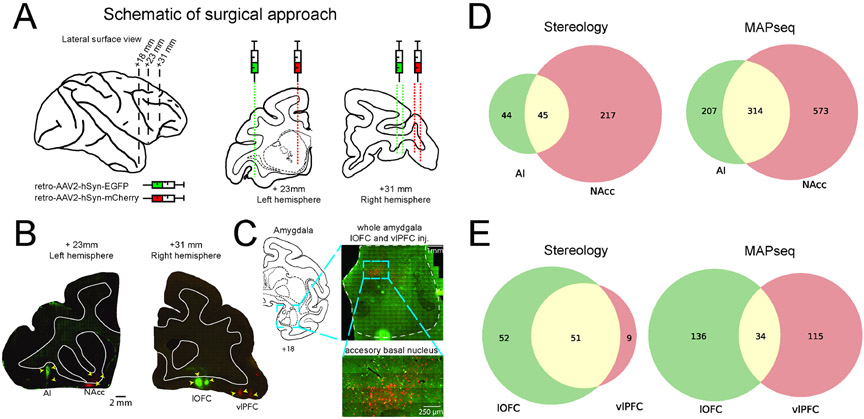
Figure 5: Tract-tracing confirms branching motifs identified by MAPseq.
A) Left: lateral surface of macaque brain; dotted lines indicate sections shown with intraaural distance in mm. Right: Schematic of injection targets plotted on atlas sections. B) Photomicrographs depicting actual injection sites: AI (EGFP) and NAcc (mCherry) in the left hemisphere and lOFC (EGFP) and vlPFC (mCherry) in the right hemisphere. Intraaural distance in mm is displayed beneath each photomicrograph along with a 2mm scale bar; atlas and photomicrographs are both to scale. C) Example retrograde labelling in amygdala. Left: Atlas section and intraaural distance in mm. Right, top: Representative photomicrograph (left hemisphere) depicting entire amygdala with retrograde labelling; scale bar is 1mm. Right, bottom: higher magnification depicting double-labelled cells in accessory basal nucleus of the amygdala; scale bar is 250μm. D) Left: stereological counts of both single- and double-labelled cells for AI/NAcc hemisphere. Right: the same comparison based on MAPseq results. E) Same as D for lOFC/vlPFC hemisphere.
Comparison of the stereology and MAPseq results found broadly similar patterns of connectivity although with expected differences. For BLA neurons projecting to AI, we found a high degree of correspondence in the proportion of branching neurons between stereology and MAPseq estimates (z-test for proportions p > 0.05, Figure 5D). For NAcc-projecting neurons, however, our MAPseq and stereology results were less consistent; stereological estimates of the proportion of projections that were specific to a single area were higher than we found with MAPseq (p < 0.0001). For BLA neurons projecting to lOFC and/or vlPFC the proportion of neurons that specifically projected to only one of these areas or branched to both areas was also different to what we found with MAPseq (p < 0.001, Figure 5E). This was primarily because stereological estimates of BLA projections to vlPFC almost entirely overlapped with those targeting lOFC. Thus, this standard tract-tracing approach confirmed our prior MAPseq findings that vlPFC connections from BLA are highly overlapping with other parts of ventral frontal cortex (Figure 4B).
As we noted earlier, the difference in estimates of specific versus branching BLA projections between retrograde tracing and MAPseq results is not unexpected22. This difference between techniques is, however, revealing as it indicates that projections to this subregion of area 12 are even less specific than indicated by the MAPseq analysis of the connectivity of the larger vlPFC.
Discussion
We used MAPseq to characterize the single-neuron connectivity of individual neurons in macaque BLA. Establishing this approach in macaques opens new avenues of research into the organization of single-cell connections in non-human primates. Using this approach, we successfully determined the projection patterns of over 3,000 single neurons from BLA to frontal cortex, striatum, parts of the temporal lobe, and MD. Notably, the bulk patterns of connections identified here are in close alignment with previous reports of BLA projections based on traditional tract-tracing techniques1-3,25,26,30,52 (Figures 1 and 2). We then revealed that single BLA neurons are highly likely to branch and project to multiple areas, with branching to four distinct targets areas being the most likely (Figure 2). Further, we found that the projection patterns of single neurons were not random but had distinct and reproducible connectional motifs; BLA projections to posterior parts of the medial and ventral frontal cortex, including AI and scACC, were highly specific and less likely to branch to other areas that we sampled from. By contrast, projections to more anterior or dorsal areas were much more likely to branch and project to multiple other parts of frontal cortex (Figures 3 and 4). This difference between the projection patterns of BLA neurons to frontal cortex is potentially indicative of separable BLA-centered networks. Additional experiments using gold-standard tract-tracing methods corroborated the patterns of BLA projections that we observed using MAPseq (Figure 5) showing the validity of our results. Taken together, our findings begin to reveal the unique patterns of projections of single BLA neurons to frontal cortex and show that these projections are organized into distinct networks.
Specific and branching pattern of single amygdala neurons and their relation to function
Prior studies have shown that BLA projections to frontal cortex exhibit an anterior-posterior gradient whereby projections are more dense to posterior regions and weaker to more anterior regions2,4, a finding that we successfully replicated using MAPseq (Figure 1D). On top of this we report that in medial frontal cortex, single BLA neurons projecting to anterior areas were the most likely to branch and project to multiple areas. For instance, BLA neurons projecting to pgACC showed the highest degree of branching as these neurons also projected to other parts for medial frontal cortex (Figure 3B). By contrast, neurons projecting to the more posterior scACC were the least likely to branch to other parts of medial frontal cortex that we took samples from. This difference means that BLA signals directed to pgACC are broadcast widely across medial frontal cortex, whereas those to scACC are specifically targeted to this part of medial frontal cortex.
The specific and segregated inputs to scACC and pgACC identified here potentially provide a neuroanatomical basis for their opposing roles in affective responding that has been identified in non-human primates44. Variation in the patterns of projections from BLA to medial frontal cortex could also help to explain circuit-based treatments for psychiatric disorders; stimulation targets that are close together can have profoundly different effects on brain-wide networks and clinical symptoms53-55. Specifically, deep brain stimulation of the white matter tracts near to the scACC is an emerging therapy for treatment resistant depression56, but this therapy is most clinically effective when targeted to specific locations that likely involve the BLA projections to scACC57, often determined by patient-specific fiber tractography58.
In ventral frontal cortex we also observed a distinction in the organization of projections from BLA, with more anterior areas receiving projections that branched to more posterior areas (Figure 4). The notable exception to this was the most lateral part of ventral frontal cortex, vlPFC. This area received the highest degree of projections that branched to other areas in ventral frontal cortex (Figure 4). The widespread projections from BLA to vlPFC may relate to the role of this area in model-free as opposed to model-based behaviors43. Notably, lesions of vlPFC do not impact reinforcer devaluation that depends on specific sensory model-based information about an outcome47. By contrast, they do impact probabilistic learning that depends more on model-free associations that rely on the presence or absence of a reward. Thus, the patterns of specific and branching connections that we identified from single BLA neurons to ventral frontal cortex may help to shape the functional roles of these areas in cognition and behavior.
We note, however, that while anatomical and functional connectivity are often closely aligned59, functional relationships can exist in the absence of a direct anatomical connection60, sometimes through oscillatory coupling that may not rely on direct single cell projections32,61. Thus, while we have speculated about the functional significance of the projection motifs that we identified here using MAPseq, such speculation does not preclude the existence of other functional interactions between BLA and areas in which we found little or no barcodes.
Interpretational limitations
As this is the first time that MAPseq has successfully been applied to macaques, we took a fairly coarse approach with our dissections, relying on sulci to distinguish parts of cortex from one another. Using these large dissections, we were able to quantitatively assess the extent of single neuron branching to multiple areas at a scale that is largely beyond the reach of standard anatomical techniques. Our tract-tracing results (Figure 5), however, revealed that connectional motifs to small subregions of a particular brain area may be subtly different from those that target the entire area under investigation. This highlights that MAPseq is a complementary approach to be used alongside traditional anatomical approaches to determine the projection patterns of single neurons from one area to another.
We also acknowledge that the over/under-represented analysis approach taken in Figure 2 relies on a number of assumptions about the rules that govern the patterns of BLA single neuron branching. Because quantitative data on which to build our null distribution for the probability of neuron branching are not available, we had to rely on the bulk projection patterns that we identified (Figure 1D). As more data on the degree of branching becomes available, future studies will be able to characterize more carefully the extent to which certain motifs are more or less common than expected. It is also important to note that the proportions on which these null models we built were restricted to the areas from which we collected samples. Thus, we are likely not capturing the full extent of the branching projections of single BLA neurons. For example, the large proportion of single neurons that we identified that had no targets outside of amygdala could project to areas in the temporal lobe or subcortical areas that we did not collect samples from. Determining how single BLA neurons connect to temporal and subcortical areas will be a key avenue for future research.
Summary and relevance to mental health
Appreciating the unique features of BLA projections to frontal cortex is potentially useful for understanding the basis of a number of amygdala-linked psychiatric disorders10. For example, obsessive-compulsive disorder is associated with dysfunction in basal ganglia-OFC circuitry regulating valuation as well as basal ganglia-dACC circuits involved in action selection62. Here we identified a population of dACC-projecting BLA neurons that also preferentially targeted parts of ventral frontal cortex including lOFC (Figure 3). Our findings therefore provide a potential anatomical basis through which dysfunction in a single population of BLA neurons could influence a distributed network of areas. Further, we also provide additional evidence at the level of single neurons for the rich interconnected circuit encompassing BLA, ventral striatum and insula51,63 through both our MAPseq results (Figure 4) as well as our tract-tracing results (Figure 5).
Determining the distinct functions of the different BLA projection motifs identified here as well as their molecular signatures23,64 is a key next step, coupled with careful tract-tracing studies to identify subregions of interest for more precise study than the large brain regions assessed here. Further, recent work in humans has shown that differences in amygdala-NAcc functional connectivity are highly correlated with mental health outcomes65; careful profiling of individual differences in behavior in macaques and relating this to projection patterns could lend key insight to the potential neural basis of these difference connectivity profiles in humans. Such work also has the potential to bring network-level understanding to basic and translational neuroscience and might provide a more biologically-realistic basis for the construction of neural network architectures66.
STAR Methods
Resource Availability
Lead Contact:
Further information and requests for resources and reagents should be directed to the lead contact, Peter Rudebeck (peter.rudebeck@mssm.edu).
Materials Availability:
This study did not generate new unique reagents.
Data and Code Availability:
Analysis code and pre-processed data are archived on Zenodo under the DOI listed in the key resources table. Sequencing data are available at the NIH NCBI Sequencing Read Archive under accession numbers SRR22578455, SRR22578456, and SRR22578457. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| n/a | ||
| Bacterial and virus strains | ||
| Sindbis Virus for MAPseq (20 million barcode diversity) | Cold Spring Harbor MAPseq Core | https://www.cshl.edu/research/core-facilities/mapseq/ |
| pAAV2-hSyn-mCherry (2 x 1013 GC/ml) | Addgene | #114472 |
| pAAV2-hSyn-eGFP (2.2 x 1013 GC/ml) | Addgene | #50465 |
| Biological samples | ||
| n/a | ||
| Chemicals, peptides, and recombinant proteins | ||
| n/a | ||
| Critical commercial assays | ||
| n/a | ||
| Deposited data | ||
| Sequencing Data | NIH NCBI Sequencing Read Archive | SRR22578455, SRR22578456, SRR22578457 |
| Pre-processed connectivity matrix | Zenodo | 10.5281/zenodo.8319819 |
| Experimental models: Cell lines | ||
| n/a | ||
| Experimental models: Organisms/strains | ||
| Rhesus macaque (Macaca mullatta) | National Institute of Mental Health | https://www.nimh.nih.gov/ |
| Long-tailed macaque (Macaca fascicularis) | National Institute of Mental Health | https://www.nimh.nih.gov/ |
| Oligonucleotides | ||
| n/a | ||
| Recombinant DNA | ||
| n/a | ||
| Software and algorithms | ||
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| Python 3.9 | Python Software Foundation | https://www.python.org |
| StereoInvestigator | MBF Bioscience | https://www.mbfbioscience.com/products/stereo-investigator |
| Original analysis code | Zenodo | 10.5281/zenodo.8319819 |
| Other | ||
| n/a | ||
Experimental Model and Study Participant Details
Two male rhesus monkeys (Macaca mulatta) and one male long-tailed macaque (Macaca fascicularis), 8-9 years of age, were used for our experiments, weighing between 10 and 15 kg. The two rhesus macaques were used in MAPseq experiments whereas the long-tailed macaque was used in retro-AAV tracing experiment. All animals were housed individually and kept on a 12-hour light/dark cycle. Food was provided daily with water ad libitum. Environmental enrichment was provided daily, in the form of play objects or small food items. All procedures were approved by the Icahn School of Medicine IACUC and were carried out in accordance with NIH standards for work involving non-human primates.
Method Details
Virus prep
Modified sindbis virus for MAPseq was obtained from the MAPseq Core Facility at Cold Spring Harbor Laboratory20. The viral library used in this study had a diversity of 20,000,000 unique barcodes. Retro-AAV2 coding for mCherry (pAAV2-hSyn-mCherry, Addgene #114472, 2 x 1013 GC/ml) and green fluorescent protein (pAAV2-hSyn-eGFP, Addgene #50465, 2.2 x 1013 GC/ml) under the human synapsin promoter were obtained from Addgene. All viruses were stored at −80°C and aliquots were thawed over wet ice immediately prior to injection.
Sindbis virus injections
For each animal, T1-weighted MRIs were obtained on a Skyra 3T scanner (Siemens) for surgical targeting. Animals were anesthetized with 10 mg/kg ketamine and 0.015 mg/kg dexmedetomidine; anesthesia was maintained with isoflurane (1-3%) as needed for the duration of the scan. Animals were scanned using an MRI-compatible stereotaxic frame (Jerry-Rig, Inc.). 3-4 images were obtained per scan, which were subsequently averaged together using the AFNI software suite (NIH)67. Then, stereotactic coordinates for the BLA were computed using ImageJ (NIH)68. Two injection tracks were planned to target the middle of the BLA’s anterior/posterior extent, equally spaced in the medial/lateral plane. One anterior injection track was also planned 1.5 mm in front of the middle of the BLA. Within each track, 4-5 injections 1.5 mm apart were planned in the dorsal/ventral plane to cover the entire extent of the basal and accessory amygdala nuclei.
After allowing approximately one week to elapse after the MRI, anesthesia was induced using ketamine and dexmedetomidine and maintained with isoflurane as described above. Surgery was performed under aseptic conditions, using the toothmarker method69 to place the animals in the same 3D position as the MRI. The skin, fascia, and muscles were retracted, and holes were drilled in the skull at each injection location using a surgical drill, widened if necessary, using rongeurs. Small dural incisions permitted Hamilton syringes access to the brain surface. 0.4 μl of virus was delivered at each injection site at a rate of 0.2 μl per minute, after which the needle was left in place for at least 3 minutes before moving to the next site. Injections proceeded from the deepest site to the most superficial. The post-injection wait period was extended to at least 5 minutes at the top of each injection track before removing needles from the brain and proceeding to the next injection track.
After all injections were completed, the muscles, fascia, and skin were closed in anatomical layers. Following surgery, the animal was closely monitored in the home cage until normal behavior resumed. Postoperative treatment included buprenorphine (0.01 mg/kg, i.m., every 8 h) and meloxicam (0.2 mg/kg, i.m., every 24 h), based on attending veterinary guidance, as well as cefazolin (25 mg/kg, i.m., every 24 h) and dexamethasone sodium phosphate (0.4 −1 mg/kg, every 12–24 h) on a descending dose schedule.
Perfusion took place 67-72 hours after the final injection series with RNAase-free solutions. All tools were cleaned with RNAseZap solution (Fisher), and all solutions were prepared using RNAse-free reagents Animals were first anesthetized with ketamine /dexmedetomidine (10 mg/kg and 0.125 mg/kg, respectively) and then given sodium pentobarbital (100 mg/kg). The animal was perfused transcardially with ice-cold 1% paraformaldehyde (PFA; Electron Microscopy Science) in phosphate-buffered saline (PBS; Invitrogen) for approximately two minutes, followed by 4% PFA in PBS for approximately 18 minutes. Breathing was supplemented by manual ventilation until access to the heart was obtained.
Following brain extraction, the brain was placed in 4% PFA briefly before dissection and blocking as follows. After the cerebellum was removed, the brain was separated into hemispheres, the temporal lobes were dissected, and the remaining brain was cut into two blocks using a cryostat blade: one coronal cut was performed posterior to the central sulcus, separating the frontal and anterior parietal lobes from the remaining caudal portions of the brain – ensuring that the thalamus remained in the anterior block. Brain blocks were then post-fixed in 4% PFA for 48 hours. After post-fix, blocks were frozen slowly over dry ice before being stored at −80°C until sectioning.
Prior to the experiments presented here, we conducted a series of surgeries to determine the optimal incubation time for sindbis virus in macaques. We found that injections of sindbis virus either 120 or 96 hours before perfusion resulted in poor barcode RNA recovery, and other post-fixation protocols led to poor RNA quality in general. Supplemental Figure 2 shows the quality control results used to conclude that MAPseq was working successfully when animals were perfused <72 hours after injections of sindbis virus and extracted brain blocks were post-fixed for 48 hours.
Retro-AAV2 injections
In another, separate experiment, using the same MRI-guided stereotactic approach as above we targeted retro-AAV injections to the NAcc and AI in the left hemisphere and lOFC and vlPFC in the right hemisphere of a single long-tailed macaque. This is a separate animal to those used for the MAPseq experiments described above. Again, prior to surgery the animal was anesthetized, scanned at 3 T to obtain structural images, and injection targets were planned based on these scans. On the day of surgery, the animal was deeply anesthetized as described above and injections were made during an aseptic neurosurgery. The skin, fascia, and muscles were opened in anatomical layers, burr holes were drilled over target locations and the dura opened. Injection syringes were then lowered into the brain and injections of retro-AAV2 were then made into the targets. At each location we injected 10 μl of virus, at a rate of 0.5 μl per minute. At the conclusion of each set of injections the needle was left in place for at least 5 minutes to allow the virus to diffuse. After all injections were completed, the muscles, fascia, and skin were closed in anatomical layers. Following surgery, the animal was closely monitored in their home cage until normal behavior resumed. Postoperative treatment included buprenorphine (0.01 mg/kg, i.m., every 8 h) and meloxicam (0.2 mg/kg, i.m., every 24 h), based on attending veterinary guidance, as well as cefazolin (25 mg/kg, i.m., every 24 h) and dexamethasone sodium phosphate (0.4 −1 mg/kg, every 12–24 h) on a descending dose schedule.
Eight weeks after surgery, the animal was perfused transcardially using the methods described above with ice-cold 1% PFA (Electron Microscopy Sciences) in PBS (Invitrogen) for approximately two minutes, followed by 4% PFA in PBS for approximately 18 minutes. The brain was then extracted, postfixed for 24 h in 4% PFA at 4°C and cryoprotected in 10% glycerol in PBS for 24 h, followed by 20% glycerol in PBS for another 24 h. The brain was then blocked with one coronal cut posterior to the thalamus, and the two resulting blocks were frozen in isopentane before storage at −80°C. The brain was then sectioned in the coronal plane on a freezing stage sliding microtome (Leica) at 40 μm in a 1:8 series. Tissue sets were stored either in PBS with 0.1% sodium azide (Sigma) at 4°C or in cryoprotectant comprised of glycerol, ethylene glycol, PBS, and distilled water (30/30/10/30 v/v/v/v, respectively) at −80°C.
Sectioning and dissection
From brains injected with sindbis virus, tissue was sectioned at 200 μm on a Leica 3050S cryostat that had been cleaned with RNAseZap prior to use. Sections were collected over dry ice and stored at −80°C prior to dissection. Cortical areas were then dissected according to sulcal landmarks over dry ice. The areas that were collected and our operational definitions of their boundaries can be found in Table 1.
Table 1. Definitions of brain regions.
Here, we provide the names, abbreviations, and anatomical features used to determine the brain regions collected for the MAPseq experiments.
| Brain Region | Abbreviation | Approximate Walker’s Areas |
A/P boundaries | M/L boundaries |
|---|---|---|---|---|
| Amygdala | n/a | All nuclei across entire A/P extent | ||
| Entorhinal cortex | ento | From the anterior tip to a point adjacent to the mid-hippocampus. | Midline to fundus of rhinal sulcus | |
| Hippocampus | HC | Anterior tip to the middle of hippocampus | ||
| Medial orbitofrontal cortex | mOFC | 11m, 13a/b, 14 | From emergence of medial orbital sulcus to its disappearance | Between rostral sulcus and medial orbital sulcus |
| Lateral orbitofrontal cortex | lOFC | 11l, 12m/r, 13m/l | From emergence of lateral orbital sulcus to its disappearance | Between fundus of medial sulcus and fundus of lateral orbital sulcus |
| Ventrolateral prefrontal cortex | vlPFC | 45, 12o, 12l | From emergence of lateral orbital sulcus to its disappearance | Between lateral orbital sulcus and lateral convexity of ventral surface |
| Dorsal anterior cingulate cortex | dACC | 24 | From emergence of cingulate sulcus until area 25 disappears from medial wall/emergence of septum. | Dorsal and ventral banks of cingulate sulcus, and cortex immediately dorsal to corpus callosum |
| Subcallosal anterior cingulate cortex | scACC | 25 | Medial surface, ventral to corpus callosum, medial/dorsal to medial orbital sulcus | |
| Perigenual anterior cingulate cortex | pgACC | 32/24 | Anterior to corpus callosum | Cortex on medial surface of the frontal lobe between the rostral sulcus and the cingulate sulcus |
| Dorsal premotor cortex | PMd | 6 | Posterior to emergence of dorsal arcuate sulcus to emergence of arcuate spur | Dorsal convexity to arcuate spur |
| Caudate | n/a | |||
| Putamen | n/a | |||
| Nucleus Accumbens | NAcc | From presence of internal capsule to the full separation of caudate nucleus and putamen | Ventromedial portion of striatum | |
| Agranular Insula | AI | Orbital surface after disappearance of medial and lateral orbital sulci | From medial corner of orbital surface to approximate location of fundus of lateral sulcus | |
| Insula | n/a | Posterior to frontal/temporal junction to midpoint of hippocampus | Most medial vertically-oriented cortex in depth of lateral fissure | |
| Medial dorsal thalamus | MD | From the optic tract joining the cerebral white matter to the lateral habenula | Dorsal and medial thalamic nuclei |
Samples from each area were combined across 3 sections in the anterior/posterior plane into 1.5-ml Eppendorf tubes, which were stored at −80°C prior to shipping frozen on dry ice for sequencing.
mRNA sequencing and preprocessing
Sequencing of MAPseq projections was performed by the MAPseq Core Facility at CSHL as described in Kebschull et al. 201620. Briefly, sections were homogenized and treated with protease before RNA extraction. Total RNA was extracted using an established Trizol-based protocol. RNA quality was verified on Bioanalyzer and bulk amounts of barcodes were examined by qPCR and compared with the amount of beta actin prior to sequencing (as described in Kebschull et al. 201620). Barcode RNA was reverse transcribed into cDNA, and a known amount of RNA (spike-in sequence) was added to each sample during reverse transcription. Barcode cDNA was then double-stranded, PCR amplified to produce a sequencing library, and the purified barcode library was then submitted for a paired-end36 run on the Illumina NextSeq platform.
Preprocessing was performed by CSHL using MATLAB (Mathworks) to create a barcode matrix with size (n samples x n barcodes). Corresponding barcode sequences and spike-in counts were also extracted to allow for normalization and assessment of duplicates.
Filtering and analysis
Filtering was performed in Python 3.970-73 using a modification of the publicly available normBCmat.m script20; specific analyses can be found in the code available on Zenodo. Briefly, for each animal, barcodes are filtered, then the raw barcode counts are divided by the spike-in counts for normalization; barcodes survive filtering if the max barcode amount is found in the injection sites (any amygdala sample), the max count is greater than 20, and if any other sample (whether within amygdala or outside) has greater than 5 barcode counts. This adaptation from the original filtering allows barcodes which send their strongest projections only within amygdala to survive filtering.
To determine how many barcode expression patterns were unique, as a proxy to the rate of single neurons being infected by multiple virus particles and thus expressing multiple barcodes, we used np.unique. Of the over 3000 barcodes that survived thresholding, only 10 barcodes (0.32%) were found to have exactly the same expression pattern across samples. Thus, while it is possible that single neurons were expressing two barcodes, after filtering, that proportion was very small.
The resulting barcode matrix was then binarized and collapsed within brain regions: if a barcode was found in any of the multiple samples for each brain area, then we counted that neuron as projecting to that brain area. This collapsing and binarization allowed us to combine the results from the four sequencing runs: each of the two hemispheres from one animal, and one hemisphere sequenced twice from the other animal. Only unique barcodes from the resequencing were included in analyses; duplicates were removed based on their barcode sequences. This filtered, binarized, and collapsed matrix combined across the four hemispheres was the basis for all analyses in this study.
Population summary
Projection proportions were calculated by summing the barcodes found in one brain area and dividing by the total number of barcodes in the dataset; chance level was defined as the barcode proportion found in control sites collected from cerebellum. The number of targets for each neuron is calculated by summing the number of areas in which individual barcodes were found and subtracting one, to count the number of non-amygdala targets for each neuron; thus, a neuron which projects from only one amygdala sample to another was defined as having zero targets.
Conditional probabilities
To calculate the conditional probability of a neuron projecting to two areas, A and B, we first found all of the cells that projected to area A (irrespective of their other targets). Then, we found the subset of those cells which also project to area B. Thus, the conditional probability of B∣A is the number of cells that project to both A and B divided by the number of cells that project to A. This analysis was not restricted to neurons with only two targets.
K-means clustering
We performed k-means clustering using scikit-learn Python library. The optimal number of clusters was determined using the elbow method, in which we plotted the number of clusters against the within-cluster sum of squares. We found k= 12 to be optimal. We then sorted the projection matrices by k-means cluster.
Over- and under-represented motifs
To assess whether any branching motifs were under- or over- represented compared to chance, we first constructed two null distributions: a uniform distribution where a given neuron is equally likely to project to any target area, and another based on the overall proportion of barcode found in each area (Figure 1D). In the uniform distribution, with 120 possible unique bifurcating motifs, the expected probability of each of those would be 0.0083; 560 unique bifurcating motifs result in an expected probability of 0.0018, and 43860 quadfurcations results in a probability of 0.000023. In the latter distribution, we seek to control for the confound that areas that received more barcodes might receive more branching input (Supplemental Figure 6). Then, we assumed that the probability of a neuron branching to two areas, A and B, would be the product of the independent probabilities of projecting to A and B: p(A&B) = p(A) * p(B). This approach was taken for bi-, tri-, and quadrifurcations, with different null distributions being built for each branching degree (e.g., the null likelihood of a neuron branching and projecting to entorhinal cortex and hippocampus would be the product of the proportion of two-target neurons that were found in each area). We did not pursue analysis of neurons with five or more targets, as there were fewer than 100 neurons with each number of targets, leading to insufficient sample size on which to perform count-based statistics.
Then, we compared our actual barcode counts to the expected null distribution using a binomial probability test (scipy.stats.binomtest). Resulting p-values were FDR-corrected within each branching degree, and effect sizes were calculated as .
Network analysis
To compare projections within the medial and ventral frontal cortex, we first isolated projections to only one, not multiple, of the areas. We defined these networks by comparing areas on the orbital surface (mOFC, lOFC, vlPFC, AI) to one another and areas on the medial surface (dACC, pgACC, scACC). First, we computed the degree of overlap within these networks by preparing Venn diagrams of projections specific to the areas and which branched between them. The proportion of branching within each area was compared using z-tests for proportions; p-values were adjusted using FDR correction.
We then focused on the projections which were specific while excluding any neurons which projected to multiple areas in the same network. Projection strength to other areas were calculated as described above and compared using pairwise z-tests for proportions and Fisher’s exact tests, corrected for multiple comparisons. Degrees of branching were calculated as before, and branching was compared between brain areas using a permutation test in which area labels were shuffled 1000x to generate a null distribution; distributions were then compared using Chi-squared tests. Clustered projection heatmaps were constructed using seaborn.clustermap with Ward’s distance.
Comparison of neurons across populations
To understand whether projection motifs were reproducible across hemispheres and across animals, we compared each of our independent sequencing runs to simulated neurons from a uniform distribution 22, that is, neurons which are equally likely to project to any target area we collected (Supplemental Figure 5). 250 neurons were randomly sampled from each of the sequencing runs and compared to other runs and to the simulated neurons using cosine distance.
Stereology
One series of tissue for stereology was mounted from PBS onto gelatin coated slides and mounted and coverslipped using Vectashield Vibrance Antifade aqueous mounting medium (Vector Labs). Slides were stored in a lightproof slide box at 4°C to prevent fluorophore fading during analysis.
An adjacent series was histochemically stained for acetylcholinesterase (AChE) for more reliable identification of amygdala boundaries. Tissue was incubated overnight at 4°C in a solution containing 0.68% sodium acetate (Thermo Scientific), 0.1% copper (II) sulfate (Thermo Scientific), 0.12% glycine (Thermo Scientific), 0.12% acetylthiocholine iodide (TCI America), and 0.003% ethopropazine (Sigma Aldrich). The following morning, sections were rinsed 3x in PBS for 5 minutes each, transferred to a solution of 0.1 M acetic acid (LabChem) with 1% sodium sulfide (Thermo Scientific) for 1-2 minutes, and then rinsed again 3x with PBS before mounting as above.
Stereology was performed using a Zeiss Apotome.2 microscope equipped with a Q-Imaging digital camera, motorized stage, and Stereo Investigator software (MBF Bioscience). A total of 12 sections were used for stereology, evenly distributed to cover the anterior-posterior extent of the BLA. The borders of the amygdala were identified using a 5x objective on the AChE-stained sections, then ROI contours were realigned with the unstained sections for stereological analysis. EGFP-, mCherry-, and double-labelled cells were counted based on the soma as the counting target; the optical fractionator probe was used for stereological estimation as described in West et al., 199149. Neurons were counted under a 10x objective, with a counting frame of size 150 x 150 x 15 μm; a 5-μm guard was applied to the dorsal aspect of each section and a 20-μm guard to the ventral side. Counting frames were arranged in a 670.8 x 670.8 μm grid for systematic-random sampling. Results of the stereological analysis were compared to MAPseq results using z-tests for proportions.
Quantification and Statistical Analysis
All statistical tests used are described as they occur throughout the manuscript. The associated test statistics, p-values, and, where appropriate, degrees of freedom are all presented. In general, ordinary least squares (OLS) regressions were used to compare multi-level data, especially related to quantitative sequencing results (Figure 1C). For the analysis of the barcode matrix, we used z-tests for proportions to compare the likelihood that a single neuron would branch and project to many areas (Figure 2B). Binomial tests were then used to determine whether motifs were over- or under-represented compared to our null distributions, and FDR correction for multiple comparisons was employed when appropriate (Figure 2D, 2E, 2F). For the network analyses for single neuron BLA projections to medial and ventral frontal cortex (Figures 3 and 4), we again used z-tests for proportions to compare the likelihood of a single neuron branching within ventral or medial frontal cortex, as well as when comparing the frequency of the number of areas that each single neuron projected to. In medial frontal cortex, we again used z-tests for proportions to determine whether populations of neurons were more or less likely to also project to other areas; in ventral frontal cortex, we instead used Fisher’s exact test to perform these comparisons because of the smaller populations of neurons particularly in mOFC and lOFC. Finally, when we analyzed the results of the stereology analysis (Figure 5D, 5E), we used z-tests for proportions to compare the likelihood of branching between MAPseq and the counts from stereology.
Supplementary Material
Highlights.
Single basolateral amygdala (BLA) neurons branch to multiple parts of frontal cortex
The branching patterns of single BLA neurons are organized into structured motifs
BLA neurons that project to frontal cortex very rarely branch to thalamus as well
BLA projection patterns to ventral and medial frontal cortex suggest separable networks
Acknowledgements:
ZRZ, RLC, PRH, and PHR are supported by a grant from the BRAIN Initiative (R34NS122050). We thank Allison Sowa of the ISMMS Microscopy and Advanced Bioimaging CoRE for help with tissue preparation, members of the Rudebeck Lab for assistance with surgical procedures, and the MAPseq Core at CSHL for assistance in tissue processing and sequencing. We also thank Anthony Zador and Alex Vaughan for advice and encouragement and Elisabeth Murray for comments on an earlier version of the manuscript.
Inclusion and Diversity:
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors have no disclosures to report.
References
- 1.Aggleton JP, and Mishkin M (1984). Projections of the amygdala to the thalamus in the cynomolgus monkey. J. Comp. Neurol 222, 56–68. 10.1002/cne.902220106. [DOI] [PubMed] [Google Scholar]
- 2.Amaral DG, and Price JL (1984). Amygdalo-cortical projections in the monkey (Macaca fascicularis). Journal of Comparative Neurology 230, 465–496. 10.1002/cne.902300402. [DOI] [PubMed] [Google Scholar]
- 3.Ghashghaei HT, and Barbas H (2002). Pathways for emotion: interactions of prefrontal and anterior temporal pathways in the amygdala of the rhesus monkey. Neuroscience 115, 1261–1279. 10.1016/S0306-4522(02)00446-3. [DOI] [PubMed] [Google Scholar]
- 4.Ghashghaei HT, Hilgetag CC, and Barbas H (2007). Sequence of information processing for emotions based on the anatomic dialogue between prefrontal cortex and amygdala. NeuroImage 34, 905–923. 10.1016/j.neuroimage.2006.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucy PC, and Klüver H (1955). An anatomical investigation of the temporal lobe in the monkey (Macaca mulatta). Journal of Comparative Neurology 103, 151–251. 10.1002/cne.901030202. [DOI] [PubMed] [Google Scholar]
- 6.Rudebeck PH, Ripple JA, Mitz AR, Averbeck BB, and Murray EA (2017). Amygdala Contributions to Stimulus–Reward Encoding in the Macaque Medial and Orbital Frontal Cortex during Learning. J. Neurosci 37, 2186–2202. 10.1523/JNEUROSCI.0933-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M, and Whalen PJ (2001). The amygdala: vigilance and emotion. Mol Psychiatry 6, 13–34. 10.1038/sj.mp.4000812. [DOI] [PubMed] [Google Scholar]
- 8.Horvath FE (1963). Effects of basolateral amygdalectomy on three types of avoidance behavior in cats. Journal of Comparative and Physiological Psychology 56, 380–389. 10.1037/h0041108. [DOI] [PubMed] [Google Scholar]
- 9.Sengupta A, Yau JOY, Jean-Richard-Dit-Bressel P, Liu Y, Millan EZ, Power JM, and McNally GP (2018). Basolateral Amygdala Neurons Maintain Aversive Emotional Salience. J. Neurosci 38, 3001–3012. 10.1523/JNEUROSCI.2460-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumann CM, Bauman MD, and Amaral DG (2011). Abnormal structure or function of the amygdala is a common component of neurodevelopmental disorders. Neuropsychologia 49, 745–759. 10.1016/j.neuropsychologia.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady RO, Masters GA, Mathew IT, Margolis A, Cohen BM, Öngür D, and Keshavan M (2016). State dependent cortico-amygdala circuit dysfunction in bipolar disorder. Journal of Affective Disorders 201, 79–87. 10.1016/j.jad.2016.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marusak HA, Thomason ME, Peters C, Zundel C, Elrahal F, and Rabinak CA (2016). You say ‘prefrontal cortex’ and I say ‘anterior cingulate’: meta-analysis of spatial overlap in amygdala-to-prefrontal connectivity and internalizing symptomology. Transl Psychiatry 6, e944–e944. 10.1038/tp.2016.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin S, Liu Y, Petro NM, Keil A, and Ding M (2018). Amygdala Adaptation and Temporal Dynamics of the Salience Network in Conditioned Fear: A Single-Trial fMRI Study. eNeuro 5. 10.1523/ENEURO.0445-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray EA, and Fellows LK (2022). Prefrontal cortex interactions with the amygdala in primates. Neuropsychopharmacol. 47, 163–179. 10.1038/s41386-021-01128-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AW, Gallagher M, and Holland PC (2009). The Basolateral Amygdala Is Critical to the Expression of Pavlovian and Instrumental Outcome-Specific Reinforcer Devaluation Effects. J. Neurosci 29, 696–704. 10.1523/JNEUROSCI.3758-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes SEV, and Murray EA (2013). Differential Effects of Amygdala, Orbital Prefrontal Cortex, and Prelimbic Cortex Lesions on Goal-Directed Behavior in Rhesus Macaques. J. Neurosci 33, 3380–3389. 10.1523/JNEUROSCI.4374-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmichael ST, and Price JL (1996). Connectional networks within the orbital and medial prefrontal cortex of macaque monkeys. Journal of Comparative Neurology 371, 179–207. . [DOI] [PubMed] [Google Scholar]
- 18.Rockland KS (2018). Axon Collaterals and Brain States. Front. Syst. Neurosci 12. 10.3389/fnsys.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockland KS (2020). What we can learn from the complex architecture of single axons. Brain Struct Funct 225, 1327–1347. 10.1007/s00429-019-02023-3. [DOI] [PubMed] [Google Scholar]
- 20.Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, and Zador AM (2016). High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987. 10.1016/j.neuron.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kebschull JM, and Zador AM (2018). Cellular barcoding: lineage tracing, screening and beyond. Nat Methods 15, 871–879. 10.1038/s41592-018-0185-x. [DOI] [PubMed] [Google Scholar]
- 22.Han Y, Kebschull JM, Campbell RAA, Cowan D, Imhof F, Zador AM, and Mrsic-Flogel TD (2018). The logic of single-cell projections from visual cortex. Nature 556, 51–56. 10.1038/nature26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Sun Y-C, Zhan H, Kebschull JM, Fischer S, Matho K, Huang ZJ, Gillis J, and Zador AM (2019). High-Throughput Mapping of Long-Range Neuronal Projection Using In Situ Sequencing. Cell 179, 772–786.e19. 10.1016/j.cell.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gergues MM, Han KJ, Choi HS, Brown B, Clausing KJ, Turner VS, Vainchtein ID, Molofsky AV, and Kheirbek MA (2020). Circuit and molecular architecture of a ventral hippocampal network. Nature Neuroscience, 1–9. 10.1038/s41593-020-0705-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russchen FT, Bakst I, Amaral DG, and Price JL (1985). The amygdalostriatal projections in the monkey. An anterograde tracing study. Brain Research 329, 241–257. 10.1016/0006-8993(85)90530-X. [DOI] [PubMed] [Google Scholar]
- 26.Höistad M, and Barbas H (2008). Sequence of information processing for emotions through pathways linking temporal and insular cortices with the amygdala. Neuroimage 40, 1016–1033. 10.1016/j.neuroimage.2007.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aggleton JP, Wright NF, Rosene DL, and Saunders RC (2015). Complementary Patterns of Direct Amygdala and Hippocampal Projections to the Macaque Prefrontal Cortex. Cereb Cortex 25, 4351–4373. 10.1093/cercor/bhv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rockland KS (2013). Collateral branching of long-distance cortical projections in monkey. Journal of Comparative Neurology 521, 4112–4123. 10.1002/cne.23414. [DOI] [PubMed] [Google Scholar]
- 29.Sharma KK, Kelly EA, Pfeifer CW, and Fudge JL (2019). Translating Fear Circuitry: Amygdala Projections to Subgenual and Perigenual Anterior Cingulate in the Macaque. Cerebral Cortex, bhz106. 10.1093/cercor/bhz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timbie C, and Barbas H (2015). Pathways for Emotions: Specializations in the Amygdalar, Mediodorsal Thalamic, and Posterior Orbitofrontal Network. J. Neurosci 35, 11976–11987. 10.1523/JNEUROSCI.2157-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pujara MS, Ciesinski NK, Reyelts JF, Rhodes SEV, and Murray EA (2022). Selective Prefrontal–Amygdala Circuit Interactions Underlie Social and Nonsocial Valuation in Rhesus Macaques. J. Neurosci 42, 5593–5604. 10.1523/JNEUROSCI.0794-21.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dal Monte O, Chu CCJ, Fagan NA, and Chang SWC (2020). Specialized medial prefrontal–amygdala coordination in other-regarding decision preference. Nature Neuroscience, 1–10. 10.1038/s41593-020-0593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J, and Moghaddam B (2017). Impact of anxiety on prefrontal cortex encoding of cognitive flexibility. Neuroscience 345, 193–202. 10.1016/j.neuroscience.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenbaum G, Setlow B, Saddoris MP, and Gallagher M (2003). Encoding Predicted Outcome and Acquired Value in Orbitofrontal Cortex during Cue Sampling Depends upon Input from Basolateral Amygdala. Neuron 39, 855–867. 10.1016/S0896-6273(03)00474-4. [DOI] [PubMed] [Google Scholar]
- 35.Sharpe MJ, and Schoenbaum G (2016). Back to basics: Making predictions in the orbitofrontal–amygdala circuit. Neurobiology of Learning and Memory 131, 201–206. 10.1016/j.nlm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray EA, and Rudebeck PH (2018). Specializations for reward-guided decision-making in the primate ventral prefrontal cortex. Nat Rev Neurosci 19, 404–417. 10.1038/s41583-018-0013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard JD, Gottfried JA, Tobler PN, and Kahnt T (2015). Identity-specific coding of future rewards in the human orbitofrontal cortex. Proceedings of the National Academy of Sciences 112, 5195–5200. 10.1073/pnas.1503550112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiuzat EC, Rhodes SEV, and Murray EA (2017). The Role of Orbitofrontal–Amygdala Interactions in Updating Action–Outcome Valuations in Macaques. J. Neurosci 37, 2463–2470. 10.1523/JNEUROSCI.1839-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izquierdo A, and Murray EA (2010). Functional Interaction of Medial Mediodorsal Thalamic Nucleus But Not Nucleus Accumbens with Amygdala and Orbital Prefrontal Cortex Is Essential for Adaptive Response Selection after Reinforcer Devaluation. J. Neurosci 30, 661–669. 10.1523/JNEUROSCI.3795-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaffan D, and Murray EA (1990). Amygdalar interaction with the mediodorsal nucleus of the thalamus and the ventromedial prefrontal cortex in stimulus-reward associative learning in the monkey. J. Neurosci 10, 3479–3493. 10.1523/JNEUROSCI.10-11-03479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldstein RZ, and Volkow ND (2002). Drug Addiction and Its Underlying Neurobiological Basis: Neuroimaging Evidence for the Involvement of the Frontal Cortex. AJP 159, 1642–1652. 10.1176/appi.ajp.159.10.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmichael ST, and Price JL (1994). Architectonic subdivision of the orbital and medial prefrontal cortex in the macaque monkey. Journal of Comparative Neurology 346, 366–402. 10.1002/cne.903460305. [DOI] [PubMed] [Google Scholar]
- 43.Monosov IE, and Rushworth MFS (2022). Interactions between ventrolateral prefrontal and anterior cingulate cortex during learning and behavioural change. Neuropsychopharmacol. 47, 196–210. 10.1038/s41386-021-01079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallis CU, Cardinal RN, Alexander L, Roberts AC, and Clarke HF (2017). Opposing roles of primate areas 25 and 32 and their putative rodent homologs in the regulation of negative emotion. PNAS 114, E4075–E4084. 10.1073/pnas.1620115114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudebeck PH, and Izquierdo A (2022). Foraging with the frontal cortex: A cross-species evaluation of reward-guided behavior. Neuropsychopharmacol. 47, 134–146. 10.1038/s41386-021-01140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balewski ZZ, Elston TW, Knudsen EB, and Wallis JD (2023). Value dynamics affect choice preparation during decision-making. Nat Neurosci, 1–9. 10.1038/s41593-023-01407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudebeck PH, Saunders RC, Lundgren DA, and Murray EA (2017). Specialized Representations of Value in the Orbital and Ventrolateral Prefrontal Cortex: Desirability versus Availability of Outcomes. Neuron 95, 1208–1220.e5. 10.1016/j.neuron.2017.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padoa-Schioppa C, and Conen KE (2017). Orbitofrontal Cortex: A Neural Circuit for Economic Decisions. Neuron 96, 736–754. 10.1016/j.neuron.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West MJ, Slomianka L, and Gundersen HJG (1991). Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. The Anatomical Record 231, 482–497. 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- 50.Ferry AT, Öngür D, An X, and Price JL (2000). Prefrontal cortical projections to the striatum in macaque monkeys: Evidence for an organization related to prefrontal networks. Journal of Comparative Neurology 425, 447–470. . [DOI] [PubMed] [Google Scholar]
- 51.Haber SN, Kim K-S, Mailly P, and Calzavara R (2006). Reward-Related Cortical Inputs Define a Large Striatal Region in Primates That Interface with Associative Cortical Connections, Providing a Substrate for Incentive-Based Learning. J. Neurosci 26, 8368–8376. 10.1523/JNEUROSCI.0271-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avendaño C, Price JL, and Amaral DG (1983). Evidence for an amygdaloid projection to premotor cortex but not to motor cortex in the monkey. Brain Research 264, 111–117. 10.1016/0006-8993(83)91126-5. [DOI] [PubMed] [Google Scholar]
- 53.Riva-Posse P, Choi KS, Holtzheimer PE, Crowell AL, Garlow SJ, Rajendra JK, McIntyre CC, Gross RE, and Mayberg HS (2018). A connectomic approach for subcallosal cingulate deep brain stimulation surgery: prospective targeting in treatment-resistant depression. Mol Psychiatry 23, 843–849. 10.1038/mp.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haber SN, Yendiki A, and Jbabdi S (2021). Four Deep Brain Stimulation Targets for Obsessive-Compulsive Disorder: Are They Different? Biological Psychiatry 90, 667–677. 10.1016/j.biopsych.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sui Y, Tian Y, Ko WKD, Wang Z, Jia F, Horn A, De Ridder D, Choi KS, Bari AA, Wang S, et al. (2021). Deep Brain Stimulation Initiative: Toward Innovative Technology, New Disease Indications, and Approaches to Current and Future Clinical Challenges in Neuromodulation Therapy. Frontiers in Neurology 11. 10.3389/fneur.2020.597451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheth SA, and Mayberg HS (2023). Deep Brain Stimulation for Obsessive-Compulsive Disorder and Depression. Annu. Rev. Neurosci 46, annurev-neuro–110122-110434. 10.1146/annurev-neuro-110122-110434. [DOI] [PubMed] [Google Scholar]
- 57.Hamani C, Mayberg H, Stone S, Laxton A, Haber S, and Lozano AM (2011). The Subcallosal Cingulate Gyrus in the Context of Major Depression. Biological Psychiatry 69, 301–308. 10.1016/j.biopsych.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 58.Zhu Z, Hubbard E, Guo X, Barbosa DAN, Popal AM, Cai C, Jiang H, Zheng Z, Lin J, Gao W, et al. (2021). A connectomic analysis of deep brain stimulation for treatment-resistant depression. Brain Stimulation 14, 1226–1233. 10.1016/j.brs.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Trambaiolli LR, Peng X, Lehman JF, Linn G, Russ BE, Schroeder CE, Liu H, and Haber SN (2022). Anatomical and functional connectivity support the existence of a salience network node within the caudal ventrolateral prefrontal cortex. eLife 11, e76334. 10.7554/eLife.76334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, and Hagmann P (2009). Predicting human resting-state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences 106, 2035–2040. 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taub AH, Perets R, Kahana E, and Paz R (2018). Oscillations Synchronize Amygdala-to-Prefrontal Primate Circuits during Aversive Learning. Neuron 97, 291–298.e3. 10.1016/j.neuron.2017.11.042. [DOI] [PubMed] [Google Scholar]
- 62.Ahmari SE, and Dougherty DD (2015). Dissecting Ocd Circuits: From Animal Models to Targeted Treatments. Depression and Anxiety 32, 550–562. 10.1002/da.22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho YT, Ernst M, and Fudge JL (2013). Cortico–Amygdala–Striatal Circuits Are Organized as Hierarchical Subsystems through the Primate Amygdala. J. Neurosci 33, 14017–14030. 10.1523/JNEUROSCI.0170-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Y-C, Chen X, Fischer S, Lu S, Zhan H, Gillis J, and Zador AM (2021). Integrating barcoded neuroanatomy with spatial transcriptional profiling enables identification of gene correlates of projections. Nature Neuroscience, 1–13. 10.1038/s41593-021-00842-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein-Flügge MC, Jensen DEA, Takagi Y, Priestley L, Verhagen L, Smith SM, and Rushworth MFS (2022). Relationship between nuclei-specific amygdala connectivity and mental health dimensions in humans. Nat Hum Behav 6, 1705–1722. 10.1038/s41562-022-01434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zador AM (2019). A critique of pure learning and what artificial neural networks can learn from animal brains. Nature Communications 10, 3770. 10.1038/s41467-019-11786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox RW (1996). AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages. Computers and Biomedical Research 29, 162–173. 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 68.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saunders RC, Aigner TG, and Frank JA (1990). Magnetic resonance imaging of the rhesus monkey brain: use for stereotactic neurosurgery. Exp Brain Res 81, 443–446. 10.1007/BF00228139. [DOI] [PubMed] [Google Scholar]
- 70.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, et al. (2020). Array programming with NumPy. Nature 585, 357–362. 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 12, 2825–2830. [Google Scholar]
- 72.Hunter JD (2007). Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering 9, 90–95. 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 73.Waskom ML (2021). seaborn: statistical data visualization. Journal of Open Source Software 6, 3021. 10.21105/joss.03021. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Analysis code and pre-processed data are archived on Zenodo under the DOI listed in the key resources table. Sequencing data are available at the NIH NCBI Sequencing Read Archive under accession numbers SRR22578455, SRR22578456, and SRR22578457. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| n/a | ||
| Bacterial and virus strains | ||
| Sindbis Virus for MAPseq (20 million barcode diversity) | Cold Spring Harbor MAPseq Core | https://www.cshl.edu/research/core-facilities/mapseq/ |
| pAAV2-hSyn-mCherry (2 x 1013 GC/ml) | Addgene | #114472 |
| pAAV2-hSyn-eGFP (2.2 x 1013 GC/ml) | Addgene | #50465 |
| Biological samples | ||
| n/a | ||
| Chemicals, peptides, and recombinant proteins | ||
| n/a | ||
| Critical commercial assays | ||
| n/a | ||
| Deposited data | ||
| Sequencing Data | NIH NCBI Sequencing Read Archive | SRR22578455, SRR22578456, SRR22578457 |
| Pre-processed connectivity matrix | Zenodo | 10.5281/zenodo.8319819 |
| Experimental models: Cell lines | ||
| n/a | ||
| Experimental models: Organisms/strains | ||
| Rhesus macaque (Macaca mullatta) | National Institute of Mental Health | https://www.nimh.nih.gov/ |
| Long-tailed macaque (Macaca fascicularis) | National Institute of Mental Health | https://www.nimh.nih.gov/ |
| Oligonucleotides | ||
| n/a | ||
| Recombinant DNA | ||
| n/a | ||
| Software and algorithms | ||
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| Python 3.9 | Python Software Foundation | https://www.python.org |
| StereoInvestigator | MBF Bioscience | https://www.mbfbioscience.com/products/stereo-investigator |
| Original analysis code | Zenodo | 10.5281/zenodo.8319819 |
| Other | ||
| n/a | ||