Abstract
Evidence for the influence of chronic inflammation induced by microbial dysbiosis on aberrant DNA methylation supports a plausible connexion between disordered microbiota and precancerous lesions of gastric cancer (PLGC). Here, a comprehensive study including multi-omics data was performed to estimate the relationships amongst the gastric microbiome, inflammatory proteins and DNA methylation alterations and their roles in PLGC development. The results demonstrated that gastric dysbacteriosis increased the risk of PLGC and DNA methylation alterations in related tumour suppressor genes. Seven inflammatory biomarkers were identified for antrum and corpus tissues, respectively, amongst which the expression levels of several biomarkers were significantly correlated with the microbial dysbiosis index (MDI) and methylation status of specific tumour suppressor genes. Notably, mediation analysis revealed that microbial dysbiosis partially contributed to DNA methylation changes in the stomach via the inflammatory cytokines C–C motif chemokine 20 (CCL20) and tumour necrosis factor receptor superfamily member 9 (TNFRSF9). Overall, these results may provide new insights into the mechanisms that might link the gastric microbiome to PLGC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43657-023-00118-w.
Keywords: Gastric premalignant lesions, Microbiome dysbiosis, Inflammation, DNA methylation, Mediation analysis
Introduction
Chronic atrophic gastritis (CAG), intestinal metaplasia (IM) and dysplasia (DYS) are recognised as precancerous lesions of gastric cancer (PLGC). Accumulating evidence suggests that dysbiosis of the gastric microbiome contributes to the progression from PLGC to gastric cancer (Ndegwa et al. 2020; Zhang et al. 2021), but the mechanisms involved have not yet been well elucidated.
Growing evidence suggests that initiating and perpetuating chronic inflammation are one of the most important pathogenic mechanisms for gastric microflora dysregulation. Furthermore, given that inflammatory processes have been previously reported to be associated with abnormal alterations in methylation status (Abu-Remaileh et al. 2015; Ligthart et al. 2016), we speculated that inflammation may play a mediating role in the association between gastric dysbacteriosis and DNA methylation alterations. Hence, this population-based study aimed to evaluate the association between the gastric microbiome and DNA methylation changes in the development of PLGC and to explore the mediating effects of the inflammatory response on this relationship.
Materials and Methods
Study Population and Design
The participants in this study were recruited, in part, from a randomised controlled trial conducted in a high-risk area (Changle County in Fujian Province, China), which has been described in detail previously (Wong et al. 2004; Yan et al. 2022). We also recruited volunteers with good compliance residing in the same county. From August 2020 to July 2021, a total of 281 eligible patients were included in this study (Fig. S1).
16S rRNA Sequencing and Read Processing
Cytology brushing samples from the gastric antrum were analysed using 16S rRNA gene V3–V4 region sequencing with Illumina MiSeq. Raw sequence data were processed using the QIIME2 (version 2021.8). For downstream analyses, an R6 class object was created using the R package ‘microeco’ (version 4.1.0).
Methylation‑Specific PCR (MSP) of Candidate Genes
MSP was performed to detect the promoter methylation status of nine candidate genes associated with gastric cancer or its precancerous lesions using two sets of primers for methylated and unmethylated DNA. The primer sequences and corresponding annealing temperatures for each gene are listed in Table S1.
Determination of Inflammatory Biomarkers
To identify potential inflammatory biomarkers, the subjects were divided into discovery (n = 124) and validation (n = 156) sets. For biomarker discovery, the Proseek Multiplex Inflammation kit I (Olink Bioscience, Sweden), was used for detecting 92 inflammatory proteins in biopsy tissue specimens. For biomarker validation, C–C motif chemokine 20 (CCL20) and tumour necrosis factor receptor superfamily member 9 (TNFRSF9) were validated by specific ELISA kits (R&D Systems).
Statistical Analysis
The structure of the microbiome was analysed through alpha and beta diversity analyses. Linear discriminant analysis (LDA) effect size (LEfSe) analysis was used to identify differentially abundant genera between groups. The microbial dysbiosis index (MDI) was further calculated as the log of [total abundance in genera increased in PLGC] over [total abundance in genera decreased in PLGC] (Gunathilake et al. 2021). Differentially expressed protein markers of inflammation in the discovery set were identified using the ‘limma’ package. We further applied least absolute shrinkage and selection operator (LASSO) regression analysis to select the most predictive biomarkers for PLGC risk and then constructed a comprehensive inflammation scoring system (ISS) according to the corresponding coefficients. Correlations between the differentially abundant genera and protein markers were examined using Spearman rank-order correlation. Causal mediation analysis with parametric regression models was performed using the 'paramed' method in Stata 15.0 software.
Results
The general characteristics were comparable between the PLGC and control groups (Table S2). Figure S2 shows a significant difference between the groups on beta diversity, but not on alpha diversity. Eleven genera were the dominant microbiota in the controls, whilst three genera were significantly enriched in cases with PLGC (Fig. 1a). Further, the PLGC group exhibited a significantly higher MDI value than the control group (Fig. 1b).
Fig. 1.
Microbial dysbiosis is associated with the risk of PLGC and DNA methylation alterations. a Histogram of the linear discriminant analysis (LDA) scores for differentially abundant genera with relative abundances higher than 0.01% between the PLGC and control groups. Significance obtained by LDA effect size (LEfSe) at p < 0.05 and LDA score > 2. b MDI comparison amongst the study groups. c, d Comparison of the MDI between subjects with different DNA methylation statuses of genes: c gastric antrum tissues and d gastric corpus tissues. The p values were determined with the Wilcoxon rank-sum test, *p < 0.05; **p < 0.01; ***p < 0.001
Table S3 shows that higher methylation rates of cadherin 1 (CDH1), protocadherin 10 (PCDH10), reprimo (RPRM), homeobox D10 (HOXD10) and runt-related transcription factor 3 (RUNX3) were associated with a significantly increased risk of PLGC. The MDI value of individuals with methylated CDH1, PCDH10, RPRM or HOXD10 in the gastric antrum was significantly higher than that of individuals with unmethylated genes (Fig. 1c). Additionally, a higher MDI value was observed in individuals with methylated CDH1, PCDH10, HOXD10 or RUNX3 in the gastric corpus than in those with unmethylated genes (Fig. 1d).
A total of 46 proteins were included in the analysis (Table S4). Differentially expressed proteins were identified between groups and are depicted in volcano plots (Fig. S3a and S3b). Using LASSO regression analysis, seven biomarkers were retained for antrum and corpus tissues respectively. There appeared to be a dose‒response increase in the risk of PLGC with higher tertiles of ISS (Fig. S3c). In addition, the values of area under the curve for ISSantrum and ISScorpus were 0.717 and 0.752, respectively (Fig. S3d).
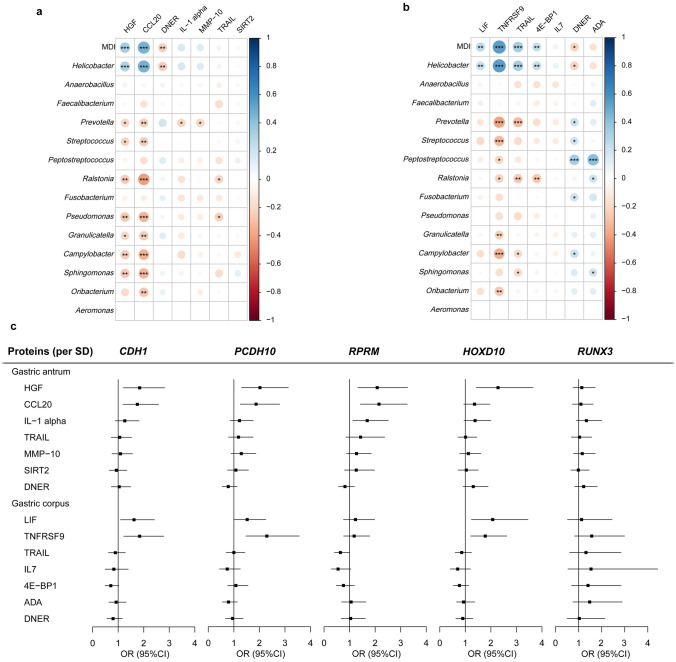
We next selected proteins that were associated with the MDI and methylated genes related to inflammatory markers for the mediation analysis (Fig. 2). As shown in Table 1, there was a significant mediation effect of CCL20 on the association between MDI and aberrant promoter methylation of CDH1 in the gastric antrum. Furthermore, significant indirect effects of TNFRSF9 on the relationships between MDI and aberrant promoter methylation of CDH1 and PCDH10 were observed in the gastric corpus. We further verified the role of key proteins CCL20 and TNFRSF9 in the validation group of 94 cases and 62 controls (Figs. S4–S5 and Table S5). Importantly, the mediating role of CCL20 (OR, 1.26, 95% CI 1.05–1.69, proportion mediated = 32.6%) and TNFRSF9 (OR, 1.24, 95% CI 1.03–1.62, proportion mediated = 68.3%) on the relationship between gastric dysbacteriosis and DNA methylation changes was confirmed (Table 1).
Fig. 2.
Associations between the levels of inflammatory biomarkers and microbial dysbiosis and DNA methylation alterations in the discovery set. a, b Spearman correlation of inflammatory biomarker expression related to the MDI and differential microbiota: a gastric antrum tissues and b gastric corpus tissues. *p < 0.05; **p < 0.01; ***p < 0.001. c Forest plots for the relationship between inflammatory biomarker expression and DNA methylation changes in gastric antrum and corpus tissues. Proteins were analysed as a continuous variable per 1-standard deviation (SD) increase
Table 1.
Mediation analysis of the mediation effect of microbiota on DNA methylation via inflammatory factors
| Exposurea | Mediatora | Methylated genes | Marginal total effect | Natural direct effect | Natural indirect effect | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | Proportion mediated (%)b | |||
| Discovery group | |||||||||
| Gastric antrum site | |||||||||
| MDI | HGF | CDH1 | 3.99 (1.63–11.18) | < 0.01 | 3.56 (1.52–9.60) | < 0.01 | 1.12 (0.90–1.49) | 0.37 | |
| PCDH10 | 4.10 (1.83–10.05) | < 0.01 | 3.86 (1.73–9.11) | < 0.01 | 1.06 (0.96–1.32) | 0.40 | |||
| RPRM | 1.42 (0.65–3.38) | 0.38 | 1.30 (0.60–2.97) | 0.49 | 1.09 (0.93–1.34) | 0.38 | |||
| HOXD10 | 1.76 (0.76–3.69) | 0.15 | 1.61 (0.70–3.41) | 0.21 | 1.09 (0.92–1.40) | 0.38 | |||
| CCL20 | CDH1 | 3.79 (1.47–9.99) | < 0.01 | 2.45 (0.93–6.41) | 0.05 | 1.55 (1.19–2.34) | < 0.01 | 32.9 | |
| PCDH10 | 4.14 (1.85–11.08) | < 0.01 | 3.25 (1.33–7.83) | < 0.01 | 1.28 (0.99–1.82) | 0.10 | |||
| RPRM | 1.38 (0.63–3.21) | 0.40 | 1.07 (0.48–2.48) | 0.87 | 1.30 (1.06–1.77) | 0.03 | 81.5 | ||
| Gastric corpus site | |||||||||
| MDI | LIF | CDH1 | 1.76 (0.91–3.91) | 0.13 | 1.64 (0.79–3.39) | 0.18 | 1.07 (0.97–1.33) | 0.33 | |
| PCDH10 | 4.64 (2.16–10.35) | < 0.01 | 4.66 (2.11–10.91) | < 0.01 | 1.00 (0.82–1.15) | 0.95 | |||
| HOXD10 | 2.62 (1.28–6.50) | 0.02 | 2.31 (1.10–5.60) | 0.03 | 1.13 (1.00–1.44) | 0.15 | |||
| TNFRSF9 | CDH1 | 1.75 (0.85–3.86) | 0.14 | 1.27 (0.56–2.96) | 0.55 | 1.38 (1.06–2.12) | 0.03 | 57.6 | |
| PCDH10 | 4.81 (2.24–11.66) | < 0.01 | 3.54 (1.45–8.12) | < 0.01 | 1.36 (1.02–1.99) | 0.04 | 19.6 | ||
| HOXD10 | 2.57 (1.27–6.19) | 0.01 | 2.25 (0.95–5.82) | 0.05 | 1.15 (0.86–1.65) | 0.38 | |||
| TRAIL | RPRM | 1.24 (0.53–3.41) | 0.62 | 1.35 (0.57–3.71) | 0.51 | 0.92 (0.70–1.05) | 0.38 | ||
| Validation group | |||||||||
| Gastric antrum site | |||||||||
| MDI | CCL20 | CDH1 | 2.59 (1.34–4.87) | < 0.01 | 2.23 (1.10–4.26) | 0.02 | 1.16 (0.96–1.53) | 0.16 | |
| RPRM | 2.03 (1.01–4.21) | 0.04 | 1.61 (0.74–3.15) | 0.17 | 1.26 (1.05–1.69) | 0.04 | 32.6 | ||
| Gastric corpus site | |||||||||
| MDI | TNFRSF9 | CDH1 | 2.34 (1.27–4.80) | 0.01 | 1.99 (1.04–4.03) | 0.05 | 1.18 (0.97–1.55) | 0.13 | |
| PCDH10 | 1.37 (0.72–2.80) | 0.35 | 1.11 (0.56–2.23) | 0.77 | 1.24 (1.03–1.62) | 0.05 | 68.3 | ||
aContinuous variables were dichotomized by median values
bProportion mediated was calculated as log (natural indirect effect)/log (marginal total effect)
Discussion
It is widely accepted that methylation changes in specific genes are closely linked to Helicobacter pylori (H. pylori) infection. In the present study, we used the MDI instead of single H. pylori infection status, and found that subjects with methylated genes had a higher MDI value than those with unmethylated genes. The results of our study suggested that microbiome imbalance may also be associated with DNA methylation alterations.
Aberrant DNA methylation has also been reported to occur in chronic inflammatory conditions (Jammula et al. 2020; Xu et al. 2022), which suggests that inflammation is an important contributor to DNA methylation changes. This is further supported by findings from previous epigenome-wide association studies (Myte et al. 2019). In this study, significant mediating effects of CCL20 and TNFRSF9 between microbial dysbiosis and DNA methylation changes were found and further verified. These findings may provide new insights into the mechanisms that link gastric dysbiosis to PLGC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank all the participants who provided precious samples for this study and the staff of the Department of Gastroenterology at Affiliated Union Hospital of Fujian Medical University who performed gastroscopies.
Abbreviations
- CAG
Chronic atrophic gastritis
- CCL20
C–C motif chemokine 20
- CDH1
Cadherin 1
- DYS
Dysplasia
- H. pylori
Helicobacter pylori
- HOXD10
Homeobox D10
- IM
Intestinal metaplasia
- ISS
Inflammation scoring system
- LASSO
Least absolute shrinkage and selection operator
- LEfSe
Linear discriminant analysis (LDA) effect size
- MDI
Microbial dysbiosis index
- MSP
Methylation-specific PCR
- PCDH10
Protocadherin 10
- PLGC
Precancerous lesions of gastric cancer
- RPRM
Reprimo
- RUNX3
Runt-related transcription factor 3
- SG
Superficial gastritis
- TNFRSF9
Tumour necrosis factor receptor superfamily member 9
Authors’ Contributions
Concept and design: WY; Acquisition, analysis, or interpretation of data: LY, JW and YC; Experimental design and operation: LY and WL; Drafting of the manuscript: LY and WL; Critical revision of the manuscript for important intellectual content: LY, WL and WY; Statistical analysis: LY; Obtained funding: WY; Administrative, technical, or material support: FC, JC and WY; Supervision: WY.
Funding
This study was funded by Grant of Science and Technology of Fujian, China (2019L3006), Special Funds of Fujian Finance Department (2020czbz01), and High-level Talents Research Start-up Project of Fujian Medical University (XRCZX2017035 and XRCZX2020034).
Data Availability
The data generated in this study are available upon request from the corresponding author.
Declarations
Conflict of Interest
WY is the Editorial Board Member of Phenomics, and he was not involved in reviewing this paper.
Ethical Approval
This study was approved by the Institutional Review Board (IRB) of Fujian Medical University.
Consent to Participate
All participants signed written informed consent forms.
Consent for Publication
All authors have approved the publication of this work.
Footnotes
Lingjun Yan and Wanxin Li contributed equally to this work.
References
- Abu-Remaileh M, Bender S, Raddatz G, et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015;75(10):2120–2130. doi: 10.1158/0008-5472.Can-14-3295. [DOI] [PubMed] [Google Scholar]
- Gunathilake M, Lee JH, Choi IJ, et al. Effect of the interaction between dietary patterns and the gastric microbiome on the risk of gastric cancer. Nutrients. 2021;13(8):2692. doi: 10.3390/nu13082692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammula S, Katz-Summercorn AC, Li X, et al. Identification of subtypes of barrett's esophagus and esophageal adenocarcinoma based on DNA methylation profiles and integration of transcriptome and genome data. Gastroenterology. 2020;158(6):1682–1697. doi: 10.1053/j.gastro.2020.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligthart S, Marzi C, Aslibekyan S, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17(1):255. doi: 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myte R, Sundkvist A, Van Guelpen B, et al. Circulating levels of inflammatory markers and DNA methylation, an analysis of repeated samples from a population based cohort. Epigenetics. 2019;14(7):649–659. doi: 10.1080/15592294.2019.1603962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndegwa N, Ploner A, Andersson AF, et al. Gastric microbiota in a low-helicobacter pylori prevalence general population and their associations with gastric lesions. Clin Translat Gastroenterol. 2020;11(7):e00191. doi: 10.14309/ctg.0000000000000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong BC, Lam SK, Wong WM, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291(2):187–194. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- Xu J, Xu HM, Yang MF, et al. new insights into the epigenetic regulation of inflammatory bowel disease. Front Pharmacol. 2022;13:813659. doi: 10.3389/fphar.2022.813659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Chen Y, Chen F, et al. Effect of Helicobacter pylori eradication on gastric cancer prevention: updated report from a randomized controlled trial with 26.5 years of follow-up. Gastroenterology. 2022;163(1):154–162. doi: 10.1053/j.gastro.2022.03.039. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li C, Cao W, et al. Alterations of Gastric Microbiota in Gastric Cancer and Precancerous Stages. Front Cell Infect Microbiol. 2021;11:559148. doi: 10.3389/fcimb.2021.559148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.