Abstract
Background
Glutamatergic projection neurons of the lateral habenula (LHb) drive behavioral state modulation by regulating the activity of midbrain monoaminergic neurons. Identifying circuit mechanisms that modulate LHb output is of interest for understanding control of motivated behaviors.
Methods
A small population of neurons within the medial subnucleus of the mouse LHb express the GABAergic (gamma-aminobutyric acidergic)-synthesizing enzyme GAD2, and they can inhibit nearby LHb projection neurons; however, these neurons lack markers of classic inhibitory interneurons, and they coexpress the vesicular glutamate transporter VGLUT2. To determine the molecular phenotype of these neurons, we genetically tagged the nuclei of GAD2-positive cells and used fluorescence-activated nuclear sorting to isolate and enrich these nuclei for single-nucleus RNA sequencing.
Results
Our data confirm that GAD2+/VGLUT2+ neurons intrinsic to the LHb coexpress markers of both glutamatergic and GABAergic transmission and that they are transcriptionally distinct from either GABAergic interneurons or habenular glutamatergic neurons. We identify gene expression programs within these cells that show sex-specific differences in expression and that are implicated in major depressive disorder, which has been linked to LHb hyperactivity. Finally, we identify the Ntng2 gene encoding the cell adhesion protein netrin-G2 as a marker of LHb GAD2+/VGLUT2+ neurons and a gene product that may contribute to their target projections.
Conclusions
These data show the value of using genetic enrichment of rare cell types for transcriptome studies, and they advance understanding of the molecular composition of a functionally important class of GAD2+ neurons in the LHb.
Keywords: Depression, GAD2, Lateral habenula, Neurotransmitter phenotype, snRNAseq, Transcriptome
The habenula is a hub for anatomical connections that link the limbic forebrain with midbrain monoaminergic nuclei (1). The habenula comprises 2 major subregions: the medial habenula (MHb) and lateral habenula (LHb) (1). The LHb receives inputs from the hypothalamus, cortex, ventral tegmental area, dorsal raphe nuclei (DRN), and locus coeruleus, and sends projections to modulate the firing of monoaminergic neurons in the ventral tegmental area, DRN, and locus coeruleus (2). Dysfunction of the LHb is thought to contribute to psychiatric disorders, including drug addiction and depression (2).
The LHb is composed predominantly of glutamatergic neurons that project to downstream target regions in the midbrain (3, 4, 5). Cellular and synaptic mechanisms that promote activity of these excitatory projections have been associated with depressive-like behaviors (6, 7, 8, 9), whereas inhibition of LHb activity has been suggested as a therapeutic option for major depressive disorder (MDD) (10). These observations have driven interest in identifying sources of inhibition that reduce the firing of LHb projection neurons. In addition to GABAergic (gamma-aminobutyric acidergic) afferents that project to the LHb (11), inhibition of LHb efferents could be mediated by local GABAergic neurons (12). A small population of neurons expressing the GABA-synthesizing enzyme GAD2 are found in the medial subnucleus of the LHb, though these cells do not express VGAT or GAD1, and they coexpress the vesicular glutamate transporter VGLUT2 (13). Using Gad2-Cre mice in combination with Cre-dependent AAV-ChR2 expression in the LHb, optogenetic stimulation of these neurons resulted in inhibitory currents recorded from locally connected neurons within the LHb (14) but excitatory currents recorded from targets in the mesopontine tegmentum (13). The molecular mechanisms that allow this GAD2+ cell population to have properties of both local inhibitory interneurons and excitatory projection neurons have remained unknown.
Single-cell sequencing has revolutionized neuronal classification by reducing traditional reliance on categorical marker genes and facilitating holistic analysis of gene sets that define cellular function (15). Cellular diversity has recently been characterized within the MHb and LHb, yielding insights into the gene expression programs that characterize downstream projection patterns (3,4,16,17). Although a small number of Gad2-expressing cells were detected in these studies, there were insufficient numbers to drive these neurons into a cluster of their own, and thus, the transcriptome of this population remains uncharacterized. To overcome this limitation, we and others have been using transgenes to enrich single cells (18) or single nuclei (19) of rare cell types by fluorescence-activated sorting (fluorescence-activated cell sorting/fluorescence-activated nuclear sorting [FANS]) from heterogeneous brain tissues prior to sequencing. Here, we provide and analyze a comprehensive single-nucleus RNA sequencing (snRNA-seq) dataset of nuclei from GAD2+ neurons enriched from the LHb. By integrating our Gad2-enriched dataset with previous total single-cell RNA sequencing (scRNA-seq) data from the habenula, we describe the expression of key genes that define the transcriptional profiles of LHb GAD2+ neurons compared with other LHb neurons.
Methods and Materials
Animals
We used adult (>P60) female and male mice, and all experiments were conducted in accordance with an animal protocol approved by the Duke University Institutional Animal Care and Use Committee. For nuclear isolation, we crossed homozygous Gad2-IRES-Cre mice (Gad2tm2(cre)Zjh/J, RRID: IMSR_JAX:010802) (20) with homozygous mice expressing a Cre-inducible Sun1-myc-sfGFP transgene, also known as INTACT (B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J, RRID: IMSR_JAX: -21039) (21) to generate dual Gad2-Cre/INTACT heterozygotes.
Isolation and Sequence of Gad2+ Nuclei
LHb was dissected by punch biopsy bilaterally from Gad2-Cre/INTACT heterozygotes mice (n = 3 female, n = 7 male). Nuclei were isolated, pooled by sex, and incubated with multiplexing using lipid-tagged indices-sequencing lipid-modified oligonucleotides (22) prior to FANS, gating on DAPI and GFP (green fluorescent protein). We performed 10x Genomics 3′ Gene Expression (version 3 chemistry) library construction and sequenced all nuclei on a single 10x GEMwell. The count matrix was generated using CellRanger version 3.0.2 and used as input to Seurat version 417 (23) for downstream analysis. Differential expression (DE) was performed between clusters using a Wilcoxon rank-sum test using log fold change > 0.25 and p values < .05. Data are deposited at Gene Expression Omnibus datasets at GSE179198. Details of nuclear isolation and sequence analysis, including the integration of our sequencing data with a total LHb scRNA-seq dataset (3), can be found in the Supplement.
Results
Single-Nuclear Gene Expression Profiles Define Distinct Populations of LHb Gad2+ Neurons
We genetically tagged and enriched the nuclei of LHb Gad2+ neurons prior to performing snRNA-seq on the 10x Genomics platform (Figure 1A; Figure S1). We recovered transcriptomes from 3651 GFP+ cells (Figure S2) that fell into 11 clusters of cells. Almost all the clusters contained some nuclei with Gad2 messenger RNA (Figure S2C), and variation in Gad2 expression between cells in a cluster could reflect RNA dropout. However, when we assessed the expression of known cell-type marker genes, we found that many clusters consisted of non-neuronal cells (Figure S2B). This may reflect the expression of the Gad2-Cre transgene in non-neuronal cells (Figure S1B). To focus our analysis on neurons, we filtered clusters for the expression of the neuronal marker Rbfox3 (Figure S2D). This resulted in 5 clusters of 1491 cells (Figure 1B) with similar quality control features between clusters (Figure S3). The expression of activity-inducible genes was uniform across the neuronal clusters, supporting that these neurons clustered by cell type rather than activity state (Figure S4).
Figure 1.
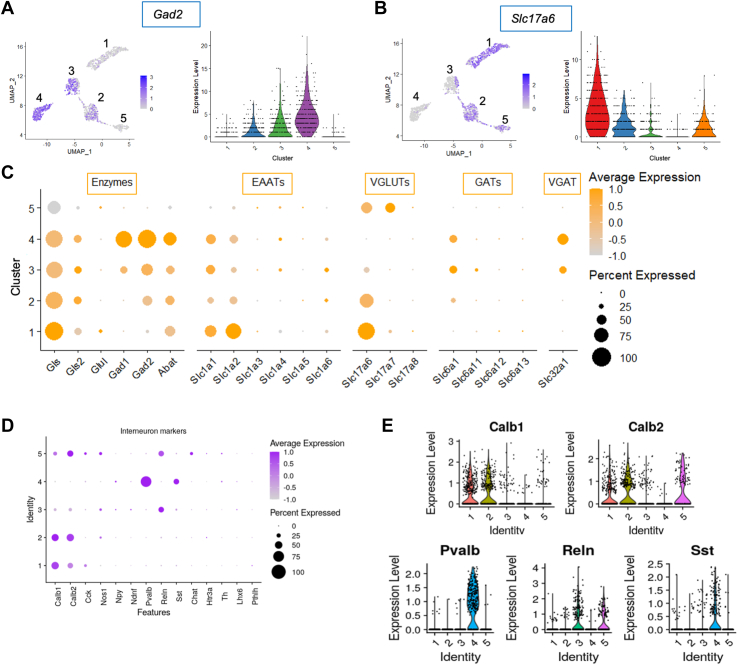
Isolation and single-nucleus RNA sequencing analysis of Gad2+ neurons from the LHb. (A) Schematic of the approach designed to enrich for Gad2+ neurons within the LHb and subsequent single-nucleus RNA sequencing. (B) UMAP clustering of the 5 neuronal clusters retrieved from single-nucleus RNA sequencing, filtered by expression of Rbfox3. (C) Heatmap showing differential expression of top 50 genes per cluster using a nonparametric Wilcoxon rank-sum test, adjusted p value ≤ 10 × 10−6 and log2 fold change ≥ |0.25|. Columns represent each cluster, with subcolumns representing single cells. Each row represents a different gene. (D) Dot plot showing scaled expression of representative top differential expression genes per cluster. (E) Spatial expression from the Allen Institute Brain Atlas showing Gad2 expression enriched in the medial portion of the LHb. A reference map is shown for orientation. (F) Representative top differential expression genes per cluster to highlight the respective anatomical origins. FACS, fluorescence-activated cell sorting; GFP, green fluorescent protein; LHb, lateral habenula; LMO, lipid-modified oligonucleotide; MHb, medial habenula; PVT, paraventricular nucleus of the thalamus; UMAP, Uniform Manifold Approximation and Projection.
We assessed the most differentially expressed genes (DEGs) between clusters (Figure 1C; Table S1) and used a subset of these DEGs (Figure 1D) in combination with the Allen Brain Atlas to verify the original anatomical origins of these neuronal clusters (Figure 1E). Clusters 2 and 4 are marked by expression of Megf11 and Pvalb, respectively, both of which have expression patterns in the LHb that have similarities to Gad2. Pvalb+ neurons are also found in the dorsal thalamus, which immediately borders the LHb on the lateral side. Some of the clusters appear to have originated from regions outside of but in close apposition to the LHb. Markers of clusters 1 (Ntng1) and 3 (Dab1) are enriched in the dorsal thalamus, whereas cluster 5 expressed high levels of Nwd2, which is highly expressed in the MHb.
Of the 5 neuronal clusters, clusters 2, 3, and 4 expressed the highest levels of Gad2 (Figure 2A). Given our evidence (Figure 1F) that clusters 2 and 4 are the most likely to represent Gad2+ neurons arising from the LHb, we focused on comparing gene expression between these 2 clusters. Unlike canonical cortical GABAergic interneurons (24), Gad2-expressing cells in the medial LHb are known for their coexpression of Slc17a6 encoding the excitatory glutamate vesicular transporter VGLUT2 (13). Cluster 2 but not cluster 4 expressed Slc17a6 as well as Gad2 (Figure 2B, C). When we graphed the levels of Gad2 expression against Slc17a6 expression for each single nucleus in each of the 5 clusters (Figure S5), we saw that whereas most nuclei in clusters 1 and 5 showed Slc17a6 expression and most nuclei in clusters 3 and 4 showed Gad2 expression, only cluster 2 had a significant number of cells that showed expression of both transporter genes.
Figure 2.
Gad2+ lateral habenula neurons in cluster 2 express both GABAergic and glutamatergic transmitter features. (A, B) Feature plots showing scaled expression and violin plots showing raw counts of Gad2(A) and Slc17a6(B) expression within the 5 neuronal Gad2+ clusters. (C) Dot plot showing scaled expression of genes related to synthesis, degradation, release, and reuptake of glutamate and GABA across the 5 neuronal Gad2+ clusters. (D) Dot plot and (E) violin plots showing scaled expression of GABAergic interneuron–associated markers in the 5 neuronal Gad2+ clusters. EAAT, excitatory amino acid transporter; GABA, gamma-aminobutyric acid; GAT, GABA transporter; UMAP, Uniform Manifold Approximation and Projection; VGAT, vesicular GAT; VGLUT, vesicular glutamate transporter.
Cluster 4 but not cluster 2 neurons expressed Gad1, encoding the GABA-synthesizing enzyme GAD67 (Figure 2C). Consistent with cluster 4 representing a subtype of GABAergic inhibitory interneurons, we found that this was the only cluster strongly expressing the interneuron markers Pvalb and Sst (Figure 2D, E) (25). By contrast, we observed little expression of any of the usual interneuron class markers in cluster 2, though these cells did express Calb1, encoding calbindin, and Calb2, encoding calretinin (Figure 2D, E). Consistent with prior studies of the Gad2+ neurons in the LHb, we found that neurons in cluster 2 did not express Slc32a1, encoding the vesicular GABA transporter VGAT, whereas this gene was strongly expressed in the Gad2+ neurons of cluster 4. Interestingly however, neurons in cluster 2 were seen to express Slc6a1, the gene encoding the plasma membrane GABA transporter GAT1 (Figure 2C). This is important because GAT1 function has been identified as a possible mechanism for the release of GABA by transporter reversal under membrane-depolarized conditions, even from neurons that lack VGAT to support vesicular GABA release (26,27).
To validate whether Gad2+ cells in the medial subnucleus of the LHb simultaneously coexpress both the glutamate transporter Slc17a6 and the plasmid membrane GABA transporter Slc6a1, we used RNAScope fluorescent in situ hybridization to quantify the expression and colocalization of these markers on brain sections (28) (Figure 3). Slc17a6 was expressed in a large percentage of cells across the MHb and LHb, consistent with the evidence that most neurons in this region are glutamatergic (Figure S6; Figure 3B, F). Slc6a1 was found in both MHb and LHb, though it was expressed at higher levels and in a greater percentage of cells in the LHb compared with the MHb (Figure S6; Figure 3C, F). Within the LHb, Slc17a6 and Slc6a1 were not limited to Gad2+ cells, but either one or both were coexpressed with Gad2 in the majority of Gad2+ cells (Figure 3D, E, G). These data confirm that cluster 2 represents the medial LHb Gad2+ cell cluster of functional interest for its coexpression of GABAergic and glutamatergic transmitter genes.
Figure 3.
Most Gad2+ neurons in the medial subnucleus of the LHb coexpress Slc17a6 and/or Slc6a1. (A–C) Representative RNAScope in situ hybridization images of Gad2(A), Slc17a6(B), and Slc6a1(C) in the habenula. DAPI, nuclei (blue). Dotted white line, boundary between the MHb (left) and LHb (right). (D) Overlap of Gad2 with Slc17a6 and Slc6a1. Dotted white box is centered on the medial subnucleus of the LHb and expanded in panel (E). Yellow circles show Gad2+ cells evaluated for overlap. (F) Quantification of the percent of positive nuclei for each individual brain section from thresholding in Figure S6. Forty cells/image/channel from 3 brains. (G) Percentage of the total of all Gad2+ positive nuclei from images of 3 brains that coexpress Slc17a6, Slc6a1, or both. A total of 51 cells were quantified on 3 brains. Scale bar = 10 μm. LHb, lateral habenula; MHb, medial habenula.
Gad2+/Slc17a6+ LHb Neurons Are Distinguished by Distinct Neurotransmitter Gene Expression
We identified the strongest DEGs between cluster 2 and the other 4 Gad2+ neuron clusters (Figure 4A; Table S2). Gene Ontology analysis of the DE genes identified glutamatergic synapse genes and other components of chemical synaptic transmission as defining categories (Figure 4B; Table S3). In cases where the same Gene Ontology category was significant in both of the genes that were significantly enriched and de-enriched in cluster 2, this is because different members of the same gene family were expressed in the different cell clusters. For example, in the molecular function category glutamate receptor activity (GO:0008066), Gria1, Gria2, and Grik4 were higher in cluster 2 relative to the other clusters, whereas Gria4, Grik2, and Grik3 were lower. Indeed, cluster 2 expressed a unique profile of genes for glutamatergic receptors in all the families, notably including low expression of genes for NMDA-type glutamate receptor subunits as well DE of subunits for AMPA-type, metabotropic, and kainate-type glutamate receptors (Figure 4C). Among the GABA receptor genes, neither clusters 2 nor 4 showed high expression especially compared with cluster 1, but cluster 2 displayed higher expression of Gabrg1, and cluster 4 had highest expression of Gabra3, again suggesting distinctions in the composition of GABA receptors among different classes of Gad2+ neurons (Figure 4D).
Figure 4.
Neurotransmitter and neuromodulator receptor profiles distinguish different Gad2+ neuronal populations in the lateral habenula. (A) Volcano plot highlighting top differential expression genes for cluster 2 compared with the other 4 neuronal clusters in the dataset using a nonparametric Wilcoxon rank-sum test. Dashed lines indicate cutoffs in the horizontal and vertical direction for log2 FC ≥ |0.25| and adj. p value ≤ 10 × 10−6, respectively. (B) GO analysis summary showing the top 5 GO terms in each category (molecular function, BP, and CC) for the up and downregulated differential expression genes, as determined from (A). (C) Heatmap displaying an expression of glutamate receptor subunits and (D) GABA receptor subunits per cluster. Each subcolumn within a cluster represents a single nucleus. Scaled expression is shown. (E) Dot plot showing expression of the following gene categories within each of the 5 Gad2+ neuronal clusters: serotonin receptors, acetylcholine receptors, dopamine receptors, peptides and their receptors, and adrenergic receptors. adj., adjusted; AMPAR, AMPA receptor; BP, biological process; CC, cellular component; FC, fold change; GABA, gamma-aminobutyric acid; GO, Gene Ontology; Kainate R, kainate-type glutamate receptor; MF, molecular function; mGluR, metabotropic glutamate receptor; NMDAR, NMDA receptor; NS, nonsignificant.
All Gad2+ clusters expressed genes encoding serotonin and acetylcholine receptors, and they weakly expressed noradrenergic receptor subunits; however, we saw minimal expression of dopamine receptors in any of the clusters (Figure 4E). The most strikingly differential expression between clusters is among the acetylcholine receptors, with Chrm3 highly expressed in cluster 2 versus Chrm2 in cluster 4. Chrm3 was identified in a previous scRNA-seq study of the habenula as a marker of cells in both the lateral oval and medial subdivisions of the LHb (4). Given that Gad2 expression is restricted to the medial subdivision, our findings suggest that these are distinct Chrm3+ cell types in the 2 regions.
Gad2+/Slc17a6+ LHb Neurons Are Transcriptionally Distinct From Other Slc6a1+ LHb Neurons
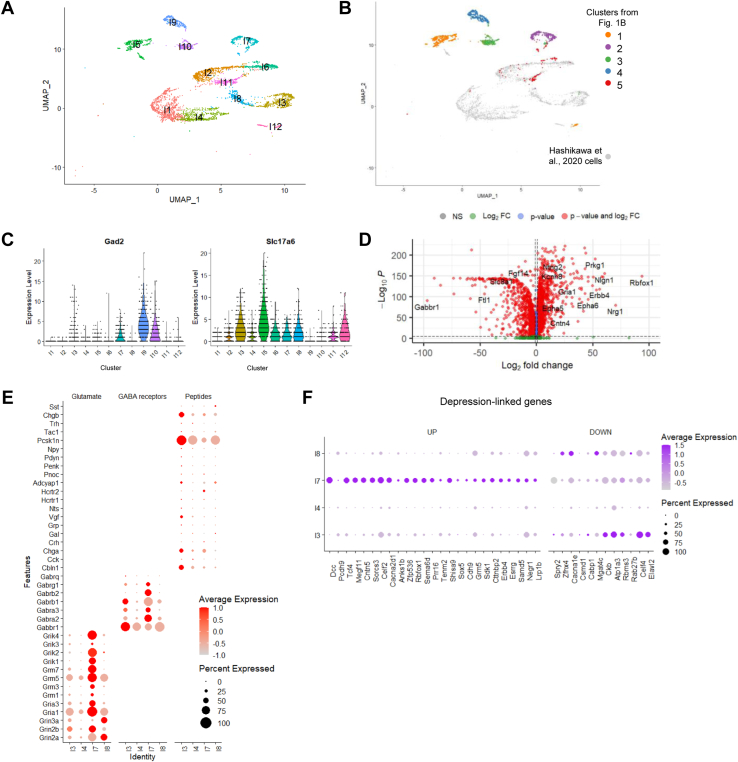
To compare the programs of gene expression that we observed in our Gad2+ clusters against those of Gad2− neurons of the LHb, we integrated our dataset with a previously published scRNA-seq dataset from the MHb and LHb (3). Clustering of the integrated datasets confirmed our identification of microglia, astrocytes, oligodendrocytes, and endothelial cells from our preliminary clusters (Figures S2 and S7). After filtering the integrated dataset for only Rbfox3+ neuron-containing clusters, we obtained 12 clusters. We call these clusters I1-I12 (I for Integrated) to distinguish them from the 5 Gad2+ clusters in our data from Figure 1. This clustering supports our conclusion that cluster 5 of our Gad2+ neurons (Figure 1B) contained cells from the MHb, as these neurons colocalized in clusters I2, I6, and I11 with MHb cells from the Hashikawa et al. (3) dataset (Figure 5A, B; Figure S8). As expected, the perihabenular Gad2+ cells from our clusters 1 and 3 failed to overlap with the habenular clusters. The highly Gad2+ cells in cluster 4 that we suggested were canonical interneurons (Figure 2) most closely cluster with perihabenular neurons in I9, suggesting either that they may not arise from the LHb, or that they may be too few in the larger dataset to permit them to cluster as LHb interneurons. The cells we identified as the Gad2/Slc17a6 dual-expressing cluster 2 from Figure 1B clustered on their own in the integrated dataset as integrated cluster I7 (Figure 5A, B). Cluster I7 was the only cluster that showed significant coexpression of Gad2 and Slc17a6 in the entire integrated dataset (Figure 5C).
Figure 5.
Gad2/Slc6a17 dual-positive neurons of the lateral habenula are transcriptomically distinct from other lateral habenula neurons. (A) UMAP clustering of all neurons after integration with Hashikawa et al. (3) dataset. (B) UMAP clustering of all neuron clusters with neurons from our dataset highlighted according to original cluster number from Figure 1B vs. neurons from Hashikawa et al. (3) dataset (gray). (C) Violin plot of expression of Gad2 and Slc17a6 across the 12 neuronal clusters in the integrated dataset. (D) Volcano plot highlighting top differential expression genes between the LHb clusters. Differential expression was run using a nonparametric Wilcoxon rank-sum test. Dashed lines indicate cutoffs in the horizontal and vertical direction for log2 FC ≥ |0.25| and adjusted p value ≤ 10e−32, respectively. (E) Dot plot showing expression of glutamate receptor subunits, GABA receptor subunits, and peptides between cluster I7 compared with I3, I4, and I8. Scaled expression is shown. (F) Dot plot showing the enriched (up) and de-enriched (down) genes in cluster I7 compared with clusters I3, I4, and I8 from cross-reference to a meta-analysis of 269 depression-linked genes. Scaled expression is shown. FC, fold change; GABA, gamma-aminobutyric acid; NS, nonsignificant; UMAP, Uniform Manifold Approximation and Projection.
We ran DE analysis between cluster I7 and the 3 clusters from the Hashikawa et al. (3) dataset that contain LHb neurons (clusters I3, I4, and I8) (Figure 5A; Figures S7, S8; Table S2) to determine which genes most strongly distinguish the Gad2/Slc17a6 double-positive neurons from Gad2− neurons in the LHb (Figure 5D). Prior studies have shown that Gad2+ cells of the medial subnucleus express orexin receptors (Hcrtr2) that mediate the modulatory effects of orexin on aggression (14). We confirmed the preferential expression of the orexin receptor Hcrtr2 in cluster I7 relative to other LHb neurons (Figure 5E), though we did not find that these neurons were particularly enriched for other peptide receptors (14). We confirmed the enrichment of Chrm3 in cluster I7 relative to the Gad2− LHb neurons and noted a relative paucity of neuromodulator receptors in any of the LHb neurons except the Gad2/Slc17a6 double-positive cells in cluster I7 (Figure S9).
Finally, because the LHb has been implicated in MDD and antidepressant action (2,6,9,29), we sought to determine whether there were any significant DEGs preferentially expressed in cluster I7 versus other neurons in the LHb that were genetically associated with MDD. A recent meta-analysis (30) yielded a list of 269 genes with significant genetic risk for MDD. We cross-referenced this list against the set of genes that were preferentially expressed in cluster I7 Gad2/Slc17a6+ neurons relative to Gad2− LHb neurons from clusters I3, I4, and I8 (Table S4). We identified 25 MDD-associated genes that were preferentially expressed in cluster I7 relative to other LHb neurons (Figure 5F). Only 12 of the 269 genes were more strongly enriched in any of the 3 other LHb clusters compared with I7. Of the I7-enriched genes, 10 were also significantly enriched in cluster 2 relative to other Gad2+ neurons from the LHb (Dcc, Tcf4, Megf11, Cacna2d1, Sorcs3, Erbb4, Chd9, Sema6d, Zfp536, and Sdk125) (Figure S10).
Sex-Specific Differences in LHb Gad2+ Neuron Gene Expression
A prior study has shown that Gad2/Slc17a6 double-positive LHb neurons express estrogen receptors and suggested that gonadal steroid-dependent modulation of these neurons might contribute to sex-specific and behaviorally relevant regulation of LHb activity (31). To determine whether Gad2+ LHb neurons show sex-specific differences in gene expression, we tagged the nuclei harvested from either female or male mice with distinct lipid-modified oligonucleotide barcodes (22). We confirmed that the Y-chromosome genes Eif2s3y, Uty, Kdm5d, and Ddx3y were expressed only in nuclei marked by the barcode we added to nuclei of male mice, whereas Xist and Tsix, which mediate X chromosome inactivation, were found exclusively in nuclei containing the female barcode (Figure 6A). After sex classification and filtering for expression of the neuronal marker Rbfox3, we unambiguously identified 107 female and 816 male nuclei.
Figure 6.
Sex bias in gene expression within Gad2+ populations from the lateral habenula. (A) Expression of known female and male-specific genes within nuclei tagged by lipid-modified oligonucleotide barcodes added to nuclei isolated from female or male mice, respectively. (B, C) DE of genes in Gad2+ lateral habenula nuclei from female or male mice: expression higher in females (B) and expression higher in males (C). (D) Table of comparison between sex-biased genes and dbSNP genes in the disease categories shown. The p values were calculated as described in the results section and were Bonferroni corrected for multiple hypothesis testing. Observed vs. expected overlap between sex-biased genes and dbSNP genes for depression (E) and Alzheimer’s disease (F). Dotted red line, observed; black line, expected. ADHD, attention-deficit/hyperactivity disorder; DE, differential expression.
Excluding the 6 genes we used for sex classification, we found 17 additional genes that were significantly differentially expressed between Gad2+ neurons of female and male mice (Table S5; Figure S11A). Sixteen are autosomal genes, 5 of which were expressed significantly more highly in Gad2+ nuclei from female mice (Figure 6B), whereas the other 11 were expressed more highly in Gad2+ nuclei from male mice (Figure 6C). We confirmed that the differences in gene expression did not arise from skewing of cell recovery across the 5 Rbfox3+ clusters and that female- and male-classified nuclei expressed similar levels of Gad2 (Figure S11). Because we pooled nuclei across female or male mice in this study, one limitation of our analysis is that we are not able to correct for individual variability when comparing differences in the expression of these genes between sexes (32). However, at least 6 of these genes (Nav1, Nos1ap, Pde1a, Plekha5, Ptprd, and Zfp804c) were also reported as sex biased in estrogen receptor–expressing neuronal populations from other brain regions (33).
We next asked whether the genes we identified as sex differentially expressed in Gad2+ neurons showed genetic association with psychiatric or neurological disorders. Using all genes with average expression above 0.25 from our LHb neuronal clusters as background, a one-sided Fisher’s exact test was used to determine if our sex-biased genes had significant enrichment within the dbSNP database of genes linked to a variety of neuronal phenotypes (34). We computed the overlap of our 16 autosomal sex-biased genes with 6 categories of neuronal phenotypes from the dbSNP database, which we also filtered for autosomal genes and calculated the statistical significance of this overlap. These analyses revealed a significant overlap of LHb Gad2+ sex-biased genes with depression and Alzheimer’s disease, both of which are known to have a female bias in the human population (Figure 6D, E) (35).
Ntng2 as a Marker of Gad2/Slc17a6 Double-Positive LHb Neurons
The gene for netrin-G2, Ntng2, appeared near the top of the DEG lists for upregulated genes when we compared these cells within our dataset of Gad2+ neurons (Figure 4A; Table S1), whereas Ntng1 was significantly downregulated. The netrins-Gs are known to mediate axonal guidance and thus raised our interest in the possibility that these molecules could define the projection targets of the Gad2/Slc17a6+ LHb neurons. Ntng2 was also significantly upregulated when compared with other LHb neurons from the Hashikawaet al. (3) dataset, suggesting that it could define these cells regionally (Figure 5D; Table S2).
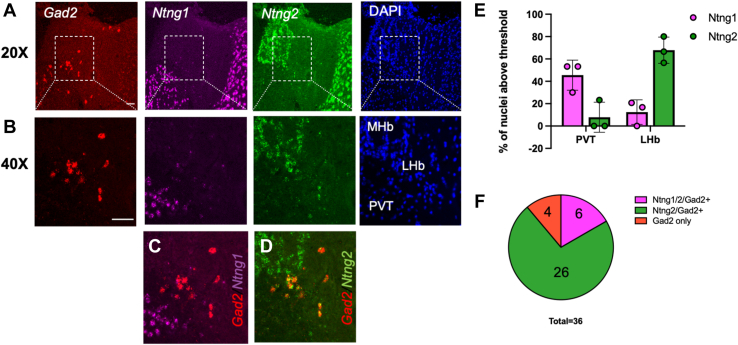
We performed in situ labeling using RNAScope fluorescent in situ hybridization probes for Gad2, Ntng1, and Ntng2 to determine the anatomical expression in LHb tissue and confirm the expression of Ntng2 within Gad2+ neurons (Figure 7; Figure S12). We found distinct patterns of Ntng1 and Ntng2 expression in and around the habenular complex, with Ntng1 expressed predominantly in the surrounding regions, including the paraventricular nucleus of the thalamus (Figure 7A–C, E). Interestingly, though Ntng2 was densely expressed in the MHb, it showed a scattered distribution into the medial part of the LHb (Figure 7A, B, D, E) where nearly all the Gad2+ neurons were observed to coexpress Ntng2 (Figure 7D, F). These findings validate the results from the snRNA-seq study and provide a new marker for this cell type within the LHb.
Figure 7.
Ntng2 is coexpressed in Gad2-positive neurons in the medial subnucleus of the LHb. (A) Representative RNAScope fluorescent in situ hybridization) images for Gad2, Ntng1, and Ntng2 as well as DAPI at 20×. The medial subnucleus of the LHb on each image is centered within the dotted white box, which is shown at 40× in panel (B). The regions that correspond to the MHb, LHb, and PVT are labeled on the DAPI image. (C) Non-overlap of Gad2 with Ntng1 and (D) overlap with Ntng2 in the LHb. (E) Quantification of the percent of positive nuclei for each individual brain section from thresholding in Figure S12. Thirty cells/image/channel from 3 brains. (F) Percentage of the total of all Gad2+ positive nuclei from images of 3 brains that coexpress Ntng1, Ntng2, or both. Scale bar = 40 μm. LHb, lateral habenula; MHb, medial habenula; PVT, paraventricular nucleus of the thalamus.
Discussion
Single-cell sequencing methods have advanced knowledge about brain complexity. However, rare cell types remain challenging to characterize because they are found in too low abundance to drive subclusters with sufficient power for DEG analysis. One approach to this limitation is to genetically enrich rare cells prior to single-cell sequencing (18,19). Here, we used transgenic expression of Sun1-GFP to purify nuclei of Gad2+ neurons from the LHb for snRNA-seq. GFP+ nuclei comprised only approximately 3% of all the DAPI+ nuclei in our samples. We recovered <700 of these cells from the LHb of every single mouse, which is consistent with Gad2+ cells comprising a few percent of the estimated 13,000 total LHb cells per mouse (36). This low abundance may explain why they failed to be detected as a distinct cluster in either of 2 prior scRNA-seq datasets from mouse habenula (3,4).
Our analysis resolved 5 clusters of FANS-enriched neurons. Gad2 messenger RNA was only weakly expressed within some of these clusters (clusters 1 and 5; Figure S5); however, this was presumably sufficient to drive enough Sun1-GFP expression from the Cre-dependent transgene to allow these cells to be sorted by FANS (Figure S1C). When we compared expression of the top DEGs in each cluster with in situ from the Allen Brain Atlas, only cluster 2 markers strongly overlapped the distribution of the Gad2+/Slc17a6+ population in the medial subnucleus of the LHb (Figure 1F; Figure S1B). Clusters 1 and 3 appear to be derived primarily from Gad2-driven Sun1-GFP transgene expression (Figure S1A) in the dorsal thalamic regions immediately flanking the LHb. Cluster 5 contains marker genes that are widely expressed in the MHb as well as in neurons scattered through the LHb. Given that the MHb shows no detectable Gad2 expression (Figure 1E) or Gad2-driven Sun1-GFP transgene expression (Figure S1A), cluster 5 is likely to come from cells in the LHb that share some similarities in gene expression with MHb neurons. Finally, cluster 4’s expression of canonical GABAergic genes suggests that this is a local inhibitory interneuron population likely to arise from the lateral LHb, where the inhibitory functions of Pvalb+ neurons have been previously characterized (12).
Neurons that coexpress markers of more than one neurotransmitter system have now been found in many regions of the brain (37). Among our purified Gad2+ LHb neurons, the cells in cluster 2 were unique for their expression of Hcrtr2, the gene encoding the orexin receptor ORXR2, which supports these neurons as the same cells determined by Flanigan et al. (14) to be locally inhibitory within the LHb. We observed both by single-cell sequencing (Figure S5) analysis of cluster 2 and by in situ hybridization (Figure 3) that Gad2+ in the medial subnucleus of the LHb coexpress the vesicular glutamatergic transporter Slc17a6, encoding VGLUT2. We confirmed previous reports that these cells fail to coexpress the vesicular GABA transporter VGAT, encoded by Slc32a1 (4,13), but we did find colocalization of Gad2/+Slc17a6+ cells in the LHb with Slc6a1, encoding the plasma membrane GABA transporter (Figure 3).
If LHb GAD2+ neurons release GABA, one possibility is that the reversal of this plasma membrane GABA transporter could be used for nonvesicular GABA release, explaining how these neurons can drive inhibition (38). Alternatively, other transporters could package GABA in vesicles; for example, midbrain dopamine neurons use Slc18a2, encoding VMAT2, to package GABA into synaptic vesicles for release (39). In some of the dual GABA/glutamate-releasing neurons, GABA and glutamate transporters are segregated into separate populations of vesicles within a single terminal (37). By contrast, the local inhibitory (14) and distal excitatory (13) currents measured upon activation of the dual Gad2/Slc17a6+ LHb neurons studied here might suggest that these neurons release GABA and glutamate from distinct projections.
One of the most powerful applications of scRNA-seq is linking molecular signatures of distinct cell types with the physiology assigned to a given brain region (40). Though many studies have focused on the importance of LHb projections in the control of dopamine systems in the brain, the Gad2+/Slc17a6+ neurons are thought to project primarily to serotonergic nuclei of the DRN and median raphe nuclei (13). We found that all the Gad2+ neurons we isolated not only express multiple serotonin receptors (Figure 4E) but also express them at higher levels compared with other types of LHb neurons (Figure S9). The LHb receives a dense serotonergic projection back from the DRN (41), which could suggest that these neurons engage in a bidirectional feedback loop that contributes to serotonergic regulation of LHb functions (42). Notably, we found a significant overrepresentation of genes associated with depression (30) in the Gad2+/Slc17a6+ population compared with other LHb neurons (Figure 5F), and genes that showed sex-DE in LHb Gad2+ cells were more likely than chance to be associated with depression and Alzheimer’s risk genes (Figure 6D, E). We also observed segregation in the LHb and surrounding regions between cells that express Ntng1 and Ntng2, encoding glycosylphosphatidylinositol-anchored netrin-G proteins. Ntng2 is highly expressed in glutamatergic neurons of the MHb, whereas within the LHb, it is selectively expressed in the GAD2+ population of the medial subnucleus. The netrin-Gs are axon guidance/cell adhesion molecules that play important functions in establishing specificity of excitatory synapse formation during development (43,44). What functions these proteins play in mature neurons and whether their expression in GAD2+ neurons of the LHb contributes to the neurological phenotypes seen in humans with NTNG2 mutations (45,46) will be interesting questions for the future.
Acknowledgments and Disclosures
This work was supported by National Institutes of Health (Grant No. R01DA047115 [to AEW]).
We thank Mariah Hazlett for critical reading of the manuscript and Melyssa Minto for support with the bioinformatics.
A previous version of this article was published as a preprint on bioRxiv: https://doi.org/10.1101/2023.01.09.523312.
The authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2023.04.004.
Supplementary Material
References
- 1.Hikosaka O. The habenula: From stress evasion to value-based decision-making. Nat Rev Neurosci. 2010;11:503–513. doi: 10.1038/nrn2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu H., Cui Y., Yang Y. Circuits and functions of the lateral habenula in health and in disease. Nat Rev Neurosci. 2020;21:277–295. doi: 10.1038/s41583-020-0292-4. [DOI] [PubMed] [Google Scholar]
- 3.Hashikawa Y., Hashikawa K., Rossi M.A., Basiri M.L., Liu Y., Johnston N.L., et al. Transcriptional and spatial resolution of cell types in the mammalian habenula. Neuron. 2020;106:743–758.e5. doi: 10.1016/j.neuron.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace M.L., Huang K.W., Hochbaum D., Hyun M., Radeljic G., Sabatini B.L. Anatomical and single-cell transcriptional profiling of the murine habenular complex. eLife. 2020;9 doi: 10.7554/eLife.51271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jhou T.C., Fields H.L., Baxter M.G., Saper C.B., Holland P.C. The rostromedial tegmental nucleus (RMTg), a GABAergic afferent to midbrain dopamine neurons, encodes aversive stimuli and inhibits motor responses. Neuron. 2009;61:786–800. doi: 10.1016/j.neuron.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B., Piriz J., Mirrione M., Chung C., Proulx C.D., Schulz D., et al. Synaptic potentiation onto habenula neurons in the learned helplessness model of depression. Nature. 2011;470:535–539. doi: 10.1038/nature09742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Cui Y., Sang K., Dong Y., Ni Z., Ma S., Hu H. Ketamine blocks bursting in the lateral habenula to rapidly relieve depression. Nature. 2018;554:317–322. doi: 10.1038/nature25509. [DOI] [PubMed] [Google Scholar]
- 8.Tchenio A., Lecca S., Valentinova K., Mameli M. Limiting habenular hyperactivity ameliorates maternal separation-driven depressive-like symptoms. Nat Commun. 2017;8:1135. doi: 10.1038/s41467-017-01192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shabel S.J., Proulx C.D., Piriz J., Malinow R. Mood regulation. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science. 2014;345:1494–1498. doi: 10.1126/science.1250469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sartorius A., Kiening K.L., Kirsch P., von Gall C.C., Haberkorn U., Unterberg A.W., et al. Remission of major depression under deep brain stimulation of the lateral habenula in a therapy-refractory patient. Biol Psychiatry. 2010;67:e9–e11. doi: 10.1016/j.biopsych.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Webster J.F., Lecca S., Wozny C. Inhibition within the lateral habenula-Implications for affective disorders. Front Behav Neurosci. 2021;15 doi: 10.3389/fnbeh.2021.786011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webster J.F., Vroman R., Balueva K., Wulff P., Sakata S., Wozny C. Disentangling neuronal inhibition and inhibitory pathways in the lateral habenula. Sci Rep. 2020;10:8490. doi: 10.1038/s41598-020-65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quina L.A., Walker A., Morton G., Han V., Turner E.E. GAD2 expression defines a class of excitatory lateral habenula neurons in mice that project to the raphe and pontine tegmentum. eNeuro. 2020;7 doi: 10.1523/ENEURO.0527-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanigan M.E., Aleyasin H., Li L., Burnett C.J., Chan K.L., LeClair K.B., et al. Orexin signaling in GABAergic lateral habenula neurons modulates aggressive behavior in male mice. Nat Neurosci. 2020;23:638–650. doi: 10.1038/s41593-020-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Z.J., Paul A. The diversity of GABAergic neurons and neural communication elements. Nat Rev Neurosci. 2019;20:563–572. doi: 10.1038/s41583-019-0195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey S., Shekhar K., Regev A., Schier A.F. Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Curr Biol. 2018;28:1052–1065.e7. doi: 10.1016/j.cub.2018.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerniauskas I., Winterer J., de Jong J.W., Lukacsovich D., Yang H., Khan F., et al. Chronic stress induces activity, synaptic, and transcriptional remodeling of the lateral habenula associated with deficits in motivated behaviors. Neuron. 2019;104:899–915.e8. doi: 10.1016/j.neuron.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz-Manchado A.B., Bengtsson Gonzales C., Zeisel A., Munguba H., Bekkouche B., Skene N.G., et al. Diversity of interneurons in the dorsal striatum revealed by single-cell RNA sequencing and PatchSeq. Cell Rep. 2018;24:2179–2190.e7. doi: 10.1016/j.celrep.2018.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallegos D.A., Minto M., Liu F., Hazlett M.F., Aryana Yousefzadeh S., Bartelt L.C., West A.E. Cell-type specific transcriptional adaptations of nucleus accumbens interneurons to amphetamine [published online Feb 16] Mol Psychiatry. 2022 doi: 10.1038/s41380-022-01466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo A., Mukamel E.A., Davis F.P., Luo C., Henry G.L., Picard S., et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinnis C.S., Patterson D.M., Winkler J., Conrad D.N., Hein M.Y., Srivastava V., et al. MULTI-seq: Sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. 2019;16:619–626. doi: 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson K.C., Anderson K.J., Courtois E.T., Gujar A.D., Barthel F.P., Varn F.S., et al. Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response. Nat Genet. 2021;53:1456–1468. doi: 10.1038/s41588-021-00926-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul A., Crow M., Raudales R., He M., Gillis J., Huang Z.J. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell. 2017;171:522–539.e20. doi: 10.1016/j.cell.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuste R., Hawrylycz M., Aalling N., Aguilar-Valles A., Arendt D., Armañanzas R.A., et al. A community-based transcriptomics classification and nomenclature of neocortical cell types. Nat Neurosci. 2020;23:1456–1468. doi: 10.1038/s41593-020-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Attwell D., Barbour B., Szatkowski M. Nonvesicular release of neurotransmitter. Neuron. 1993;11:401–407. doi: 10.1016/0896-6273(93)90145-h. [DOI] [PubMed] [Google Scholar]
- 27.Cammack J.N., Rakhilin S.V., Schwartz E.A. A GABA transporter operates asymmetrically and with variable stoichiometry. Neuron. 1994;13:949–960. doi: 10.1016/0896-6273(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 28.Wang X., Gallegos D.A., Pogorelov V.M., O’Hare J.K., Calakos N., Wetsel W.C., West A.E. Parvalbumin interneurons of the mouse nucleus accumbens are required for amphetamine-induced locomotor sensitization and conditioned place preference. Neuropsychopharmacology. 2018;43:953–963. doi: 10.1038/npp.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold P.W., Kadriu B. A major role for the lateral habenula in depressive illness: Physiologic and molecular mechanisms. Front Psychiatry. 2019;10:320. doi: 10.3389/fpsyt.2019.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard D.M., Adams M.J., Clarke T.K., Hafferty J.D., Gibson J., Shirali M., et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Hernández V.S., Swinny J.D., Verma A.K., Giesecke T., Emery A.C., et al. A GABAergic cell type in the lateral habenula links hypothalamic homeostatic and midbrain motivation circuits with sex steroid signaling. Transl Psychiatry. 2018;8:50. doi: 10.1038/s41398-018-0099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Squair J.W., Gautier M., Kathe C., Anderson M.A., James N.D., Hutson T.H., et al. Confronting false discoveries in single-cell differential expression. Nat Commun. 2021;12:5692. doi: 10.1038/s41467-021-25960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoedler J.R., Inoue S., Bayless D.W., Yang T., Tantry A., Davis C.H., et al. A functional cellular framework for sex and estrous cycle-dependent gene expression and behavior. Cell. 2022;185:654–671.e22. doi: 10.1016/j.cell.2021.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M.J., Wang P., Liu X., Lim E.L., Wang Z., Yeager M., et al. GWASdb: A database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012;40:D1047–D1054. doi: 10.1093/nar/gkr1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laws K.R., Irvine K., Gale T.M. Sex differences in Alzheimer’s disease. Curr Opin Psychiatry. 2018;31:133–139. doi: 10.1097/YCO.0000000000000401. [DOI] [PubMed] [Google Scholar]
- 36.Zhang R., Oorschot D.E. Total number of neurons in the habenular nuclei of the rat epithalamus: A stereological study. J Anat. 2006;208:577–585. doi: 10.1111/j.1469-7580.2006.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Root D.H., Zhang S., Barker D.J., Miranda-Barrientos J., Liu B., Wang H.L., Morales M. Selective brain distribution and distinctive synaptic architecture of dual glutamatergic-GABAergic neurons. Cell Rep. 2018;23:3465–3479. doi: 10.1016/j.celrep.2018.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richerson G.B., Wu Y. Dynamic equilibrium of neurotransmitter transporters: Not just for reuptake anymore. J Neurophysiol. 2003;90:1363–1374. doi: 10.1152/jn.00317.2003. [DOI] [PubMed] [Google Scholar]
- 39.Tritsch N.X., Ding J.B., Sabatini B.L. Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature. 2012;490:262–266. doi: 10.1038/nature11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armand E.J., Li J., Xie F., Luo C., Mukamel E.A. Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron. 2021;109:11–26. doi: 10.1016/j.neuron.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metzger M., Souza R., Lima L.B., Bueno D., Gonçalves L., Sego C., et al. Habenular connections with the dopaminergic and serotonergic system and their role in stress-related psychiatric disorders. Eur J Neurosci. 2021;53:65–88. doi: 10.1111/ejn.14647. [DOI] [PubMed] [Google Scholar]
- 42.Tchenio A., Valentinova K., Mameli M. Can the lateral habenula crack the serotonin code? Front Synaptic Neurosci. 2016;8:34. doi: 10.3389/fnsyn.2016.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S., Burette A., Chung H.S., Kwon S.K., Woo J., Lee H.W., et al. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 44.Matsukawa H., Akiyoshi-Nishimura S., Zhang Q., Luján R., Yamaguchi K., Goto H., et al. Netrin-G/NGL complexes encode functional synaptic diversification. J Neurosci. 2014;34:15779–15792. doi: 10.1523/JNEUROSCI.1141-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heimer G., van Woerden G.M., Barel O., Marek-Yagel D., Kol N., Munting J.B., et al. Netrin-G2 dysfunction causes a Rett-like phenotype with areflexia. Hum Mutat. 2020;41:476–486. doi: 10.1002/humu.23945. [DOI] [PubMed] [Google Scholar]
- 46.Abu-Libdeh B., Ashhab M., Shahrour M., Daana M., Dudin A., Elpeleg O., et al. Homozygous frameshift variant in NTNG2, encoding a synaptic cell adhesion molecule, in individuals with developmental delay, hypotonia, and autistic features. Neurogenetics. 2019;20:209–213. doi: 10.1007/s10048-019-00583-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.