Abstract
A key parameter of interest recovered from hyperpolarized (HP) MRI measurements is the apparent pyruvate-to-lactate exchange rate, , for measuring tumor metabolism. This manuscript presents an information-theory-based optimal experimental design approach that minimizes the uncertainty in the rate parameter, , recovered from HP-MRI measurements. Mutual information is employed to measure the information content of the HP measurements with respect to the first-order exchange kinetics of the pyruvate conversion to lactate. Flip angles of the pulse sequence acquisition are optimized with respect to the mutual information. A time-varying flip angle scheme leads to a higher parameter optimization that can further improve the quantitative value of mutual information over a constant flip angle scheme. However, the constant flip angle scheme, 35 and 28 degrees for pyruvate and lactate measurements, leads to an accuracy and precision comparable to the variable flip angle schemes obtained from our method. Combining the comparable performance and practical implementation, optimized pyruvate and lactate flip angles of 35 and 28 degrees, respectively, are recommended.
Subject terms: Functional magnetic resonance imaging, Computational science, Computational models, Image processing
Introduction
The potential of hyperpolarized (HP) C-Pyruvate magnetic resonance imaging (MRI) to characterize cancer biology, predict progression, and monitor early responses to treatment is well known (e.g.,1–9). Ongoing studies in prostate, brain, breast, liver, cervical, and ovarian cancer1, 3, 4 have shown that HP C-Pyruvate MRI is safe and effective. One of the central aspects of HP-MRI that make it appealing is the elevated chemical conversion of pyruvate to lactate, even in the presence of oxygen, via lactate dehydrogenase (LDH), also referred to as the Warburg effect10, 11. The higher production of lactate has been shown to correlate with disease presence, the aggressiveness of the disease (e.g., cancer and inflation), and response to therapy. The rate of pyruvate-to-lactate exchange () is a crucial parameter of interest in locating aggressive disease and assessing the biological state of the tissue. HP-MRI presents a unique opportunity to observe tumor metabolism in vivo2, 3, 5, 6 and use this information to make inferences about tumor aggressiveness and response to therapy. However, a recent white paper3 highlights the need for further development of spatial, temporal, and spectral encoding strategies that minimize uncertainty while maximizing the efficiency of HP-MRI. An example of uncertainty is the variability of the reported HP measurements in the literature12–14. In the present work, we develop an information-theory-based approach to determine the optimal MRI design parameters, such as flip angles, with a goal of reducing the uncertainty in the recovered rate parameter, .
The physics of the dynamic nuclear polarization (DNP) method that enables MR imaging of HP C-pyruvate is described in3, 5. The time history of pyruvate and lactate magnetization within the imaging voxels constitutes the data of interest. Together with a pharmacokinetic HP-MRI model, pyruvate and lactate magnetization data are employed to recover the pyruvate-to-lactate apparent exchange rate, ; e.g.,2, 15, 16. Accuracy and uncertainty in the recovered rate parameter depend on the data’s information content and the pharmacokinetic model’s fidelity. This work aims to determine the MRI design parameters that increase the information content in the data and reduce the uncertainty in the rate parameter. The mutual information (MI) between the data and critical model parameters is utilized toward this goal. Information theory and, specifically, mutual information provides a rigorous mathematical framework for optimizing acquisition protocols to improve reproducibility.
In this work, data to verify the reduction in uncertainty of recovered rate parameter when using optimal design parameters is generated synthetically from the pharmacokinetic model. Different signal-to-noise ratios are considered in generated noisy data to analyze the uncertainty in the recovered for various signal-to-noise ratios. The optimal design parameters is shown to reduce uncertainty in the pyruvate-to-lactate apparent exchange rate, . The codes and relevant files to reproduce the results will be publicly available in the following GitHub repository: https://github.com/prashjha/HyperpolarizedMRI.
Related works
Semi-quantitative metrics such as the ratio of the integrals of the lactate-to-pyruvate signals (area under the curve, AUC) are often preferred due to their simplicity. However, the lactate-to-pyruvate ratio is biased by HP pyruvate in blood vessels that are inaccessible to enzymes that mediate conversion from HP pyruvate to lactate. For example, if the vascular blood volume decreases by 10 percent in a region of a tumor, the lactate-to-pyruvate ratio could increase even if the true metabolic state of cells does not change. The lactate-to-pyruvate ratio is also affected by the excitation scheme. Small flip angles consume less pyruvate magnetization and permit the signal pool to remain longer for the conversion to lactate. The potential for variability is realized in the literature. In applications to brain cancer (glioma) Grist12 reports a lactate to pyruvate ratio of .25 ± .08 and .22 ± .06 in white matter and gray matter, respectively. Lactate-to-pyruvate ratios greater than 1.0 are reported in13 in gray matter. Lactate-to-pyruvate ratios in white matter of 0.43 ± 0.14 were reported in14.
Various strategies have been proposed to optimize the HP acquisition in terms of the variability and accuracy of the pyruvate to lactate conversion rate measurements, . Walker et al.15 utilized a two-species kinetic model and compared variable excitation angle acquisitions and conventional constant excitation angle acquisitions. The variable excitation scheme was obtained from a recursive relation that depletes the signal at the final acquisition17. Optimization strategies included maximization of the lactate signal or maintaining a constant signal. Either constant excitation angle or variable excitation angles that attempt to maximize total signal of the acquisition, as opposed to maintaining a constant signal level, were seen to produce the best recovery in terms of accuracy and repeatability. Larson et al.18 developed a novel kinetic modeling approach that did not require a manual input forcing function for the governing two-species kinetic model equations. The ‘inputless’ method required no assumptions regarding the input function and was compared to a tradition kinetic model that manually identified the known input signal. Assuming time-varying excitation angles, the ‘inputless’ method was seen to provide no loss in accuracy and precision over the classical method. Maidens et al.19 develop a Fisher information approach for determining the optimal time varying pulse sequence and demonstrate a decrease in uncertainty when compared with strategies that maximize the signal SNR (Signal-to-noise ratio). In20, the OED formulation for magnetic resonance fingerprinting based on the Cramér-Rao bound is presented. Cramér-Rao bound was used for optimal estimation of parameters in Bloch equation in21.
The information theoretic approach developed in this work is an extension of optimal experimental design approaches that use the Fisher information matrix and the Cramér-Rao bound as a lower bound on the variance of unbiased estimators16, 19–28. Indeed, the de Bruijn identity29 provides a direct connection between derivatives of our entropy calculations and the Fisher information matrix. However, optimizing the Fisher information matrix requires estimates of the unknown tissue parameters, such as relaxation losses and pyruvate-to-lactate conversion rate (parameter to be recovered from the data), to calculate the Fisher information. The Fisher information must be iteratively re-optimized as more accurate estimates of the tissue properties are obtained. In contrast, the present approach provides a mathematical framework to directly include the tissue parameter uncertainty in the mutual information and considers a range of tissue parameters (determined by the input probability distributions) to calculate and optimize the mutual information.
Methods
Hyperpolarized (HP) MRI model
Consider a tissue domain within which different constituents evolve depending on the local environment which includes, for example, extravascular (interstitial) and vascular. In this work, two spatial compartments, namely, extravascular and vascular, each containing HP pyruvate and lactate and complement of these two constituents, are considered. The model employed is based on2, 15 and accounts for relaxation loss, signal loss due to excitation at each scan, and pyruvate-to-lactate and lactate-to-pyruvate exchanges. We expect a dynamic, spatially, and spectrally localized data from the HP MR data acquisition such that the spatially invariant model may represent the data collection on a voxel-by-voxel basis.
Consider a tissue domain of volume in units of cm. Assuming is small enough that the spatial variation of hyperpolarized agents in can be ignored, we let and denote the relative density of HP pyruvate and lactate, respectively, at time t (in units of s). Here, the relative density of species is defined as the ratio of the density of species and the total density , i.e., . Then the total mass of HP pyruvate and lactate are , respectively, being the mass density (g/cm) of the continuum mixture. Discrete times at scans are denoted by , ; and are flip angles in scan whereas , , are repetition times and . With , the HP pyruvate and lactate available for measurement at the scan, , are given by15
| 1 |
where the matrix accounts for relaxation losses and pyruvate-lactate exchanges
| 2 |
Here denote the relaxation times (s), pyruvate-to-lactate and lactate-to-pyruvate exchange rates (s), vascular-tissue exchange rate (s), and the extravascular volume fraction. In (1), denotes the matrix that takes into account the loss of signal due to excitation at scan:
| 3 |
Lastly, is the vascular input function (dimensionless). Empirically determined blood flow through nonbranching vessels have been shown to correspond to a gamma variate input function30. Thus, is taken to be
| 4 |
where are constants, and denotes a gamma probability density function given by
| 5 |
The constant is associated with bolus arrival time and is treated as one of the uncertain model parameters. Constants and are dimensionless while is in units of seconds.
In (1), the parameters can be gathered in two different classes: 1) model parameters that depend on the tissue domain and specific voxel and includes , and 2) HP-MRI design parameters such as repetition times and flip angles . For simplicity, the parameters are assumed to be known and fixed so that . The default values of model and design parameters are provided in Tables 1and 2, respectively.
Table 1.
Default model parameters, , and remaining fixed model parameters used in initialization, optimization, and verification steps.
| Parameter | Default value | Description |
|---|---|---|
| 30 s, 25 s | Relaxation times31 | |
| 0.15 s | Pyruvate-to-lactate apparent exchange rate | |
| 0 s | Lactate-to-pyruvate apparent exchange rate | |
| 4 s | Bolus arrival time for HP pyruvate | |
| 100, 2.5, 4.5 s | Parameters in the gamma function (4) | |
| 0.05 s | Vascular-extravascular exchange rate | |
| 0.95 | Extravascular volume fraction |
Table 2.
Default design parameters, , used in initialization, optimization, and verification steps.
| Parameter | Default value | Range | Description |
|---|---|---|---|
| 30 | – | Number of HP-MRI scans | |
| 3 s | [1.5, 4.5] | Repetition times (for ) | |
| 20 degrees | [0, 35] | Flip angles for HP pyruvate (for all k) | |
| 30 degrees | [0, 35] | Flip angles for HP lactate (for all k) |
The total signal
The magnetization intensity of constituent , , at scan is assumed to be proportional to the mass of the constituent in , i.e., there is a constant such that and are the total magnetization of pyruvate and lactate, respectively. Without loss of generality, is such that . In this case, are the total magnetizations, and are the measured signal, and and are the signals remaining for the next measurements, respectively.
The signals measured at the scan are, see15,
| 6 |
i.e., one only measures the and fractions of the magnetization leaving the and fractions for the next measurement. The total signal is the sum of the individual signals at different scans and is given by
| 7 |
where the dependence of on the design parameters and model parameters is clear from (1) and (6). Note that the total signal depends on the entire history of the magnetization time evolution. The magnetization at the current acquisition depends on the HP signal available from the previous acquisition and the total signal is the sum over each time point. In general, the total signal is also proportional to the magnetization weighted by B1 sensitivity maps of the receive coils used for the acquisitions. Here, we are considering the HP data fits on a per pixel basis and assume that B1 variations are small across a given pixel.
Synthetic data
To verify the uncertainty reduction in using the optimal design parameters, HP-MRI experiment is synthetically simulated using the model in (1) with the optimal design parameters and the control design parameters (default values listed in Table 2). Suppose is the design parameter associated with some scenario ; correspond to the default design parameters, correspond to the optimized design parameters for the specific value.
Using the pharmacokinetic model (1), “ground truth” (signals at scans) is generated using design parameters and default model parameters listed in Table 1. The data (simulation results) is denoted by and takes the form:
where, denote the pyruvate and lactate signals at scan, see (6). A total of 25 samples of noisy data for different values of SNR are considered. Noisy samples are computed as follows:
| 8 |
where , given in (21), is the amount of noise in measured signals that depends on the assumed SNR, , and for each . Since SNR in the data, , is expected to vary with pixelwise location, a range of for different optimal design parameters is considered to comprehensively evaluate the accuracy and precision of the parameter recovery. The noise values correspond to the previous SNR range considered: for .
Mutual information based optimization of MR scan parameters
A major goal of this study is to formulate an optimization problem to determine the design parameters such that the MRI measurements reduce uncertainty in the rate parameter, . Treating total signal, defined in (7), as the data, and model parameters, , and data as random variables, an optimization problem of maximizing the mutual information (MI) between the data and model parameters is proposed. It is shown that uncertainty in recovered from synthetic noisy data is reduced when optimal design parameters are considered; see section "Results".
In what follows, after defining some preliminary quantities, the mutual information between data and the model parameters is defined. Let , , and , where are data, model parameter, and design parameter spaces, respectively. It is remarked that mutual information are intractable and suffer from the curse of dimensionality as the model complexity is increased to consider more variables. The curse of dimensionality appears from the nested integrals inherent to the mutual information calculation. Further, mutual information calculations in this work utilizes the spatially invariant model while incorporating spatial variations in the mutual information through the parameter variance. For example, spatial variations in the T1 relaxation times among biological compartments are expected and are represented by the variance in the T1 parameters of our model. Our Bayesian approach explicitly accounts for modeling uncertainty including differences among biological compartments through the parameter variance.
Prior, likelihood, and evidence
Suppose is the prior probability distribution function (PDF) of model parameters and is the PDF of the data . Then the joint PDF must satisfy
| 9 |
where is the conditional PDF of data conditioned on model parameters (also referred to as the likelihood function) and the conditional PDF of model parameters conditioned on data (posterior of ). The prior is taken as multi-variate Gaussian with mean and covariance matrix :
| 10 |
Here . Within this Bayesian setting, , is representative of biological variability. Within the scope of this manuscript, biological variability refers to potential patient specific differences in the pyruvate to lactate exchange rates, vascular tissue exchange rates, and bolus timing of the injected pyruvate.
To derive the expression for the likelihood function, first suppose that is the model prediction of data. Data and the model prediction are assumed to be related as follows:
| 11 |
where an additive model of noise is assumed and the noise, , is taken as Gaussian with a zero mean and standard deviation , i.e., . Here, we assume that the the phase of the real and imaginary MR data acquisition is constant over time such that the signal may be phase corrected. The MR data acquisition is, in general, complex valued with real and imaginary components that are independent and Gaussian. However, within our phase corrected approach, only the real component is non-zero and Gaussian noise for the real channel is appropriate.
Therefore, the likelihood function takes the Gaussian form:
| 12 |
Here denotes the Euclidean norm. Technically, is also conditioned on , but, for simplicity in notation, the dependence on is suppressed.
With and defined as above, the joint PDF can be computed using (9). Further, using (9), the PDF of data (evidence), , can be computed by marginalizing with respect to :
| 13 |
where is the parameter space.
Mutual information
Next, the statistical mutual information is defined and the optimization problem for design parameters is posed. Given HP-MRI data, the accurate inference of pyruvate-to-lactate exchange rate parameters (and other parameters in ) depends on the specific choice of control (design) parameters as selection of affects the information content in the measured data. The notion of mutual information29 provides a way to quantify the information content about the model parameters in the data . The mutual information between the two random variables z and with PDFs and and the joint PDF is defined as
| 14 |
Here, the mutual information I depends only on design parameters and the forward model (1).
Using Bayes theorem, , it can easily be shown that
| 15 |
or,
| 16 |
where the second term in the above equation is identified as information entropy and the first term as negative of the cross-information entropy, . That is
| 17 |
Optimization problem In order to maximize the reduction in the uncertainty in the model parameters (i.e. to have the most confident estimates of the parameters ), we propose to maximize the mutual information between the observation data and parameters of interest:
| 18 |
where is the design parameter for which I is maximum (assuming exist).
Given a Gaussian prior and conditional probability of the data with respect to the parameters, the entropy of the data conditioned on the model parameter, i.e., , is constant. Therefore, the optimization problem simplifies to
| 19 |
Approximation of mutual information and information entropy
Mutual information calculations are computationally demanding due to high-dimensional integration over the parameter and data spaces. Combinations of both quadrature and sampling methods have been employed for mutual information calculations, each of which is well-suited to certain function classes32–44. These methods include Monte Carlo and Quasi-Monte Carlo methods33–35, lattice rules36, adaptive subdivision37, 38, neural network approximations39 and numerical quadrature40. Here the problem structure is used to accelerate computations and facilitate tractable numerical integration. Following45, Gauss-Hermite quadrature is applied in each dimension of mutual information integrals defined in (16) to numerically integrate multi-variate Gaussian random variables. Quadrature order is increased until convergence is observed. Finite difference time stepping is used to solve the ODE-based model presented in Sect. “The total signal”.
Automatic differentiation accelerated optimization for OED calculations
For constant TR optimization, the auto-differentiation functions of MATLAB were used to calculate gradients of (16). In particular, design parameters and state variables were considered as optimization variables to minimize the objective function (16) with respect to the model constraints (1). Auto-differentiation provides the derivatives of the objective function and constraints with respect to this full space formulation. Given the derivatives in the full space formulation, the reduced space gradient of the objective function (16) with respect to the design parameters may be calculated using an adjoint solve. For varying TR optimization, adjoint gradients were calculated by hand.
Inverse problem to recover rate parameter
A MATLAB routine fmincon is used to solve the inverse problem of recovering model parameters in the model (1) from the data generated in Sect. “Synthetic data” As an objective function for the inverse problem, square of the difference between data and model prediction of signals is used. Similarly, derivatives from the automatic differentiation feature in the MATLAB are utilized for numerical optimization.
The Cramér-Rao bound is computed to provide a reference for the uncertainty observed in the recovered .
Here, J is the Fisher information matrix. Each time point of the HP signal evolution, Eq. (1), may be considered as an independent random variable, thus following46, the Cramér-Rao bound may be computed analytically with the Fisher information matrix given as follows.
Results
In this section, the main results of our analysis are presented. First, the optimal design parameters for different signal-to-noise ratios (SNRs) are shown. Optimal design parameters for both temporally constant and varying flip angles at each data acquisition are considered. Next, the reduction in uncertainty of when using optimal design parameters generated synthetic data is shown.
Optimized design parameters
As mentioned in Sect. “Mutual information based optimization of MR scan parameters”, multi-variate Gaussian is taken as a prior for uncertain model parameters, . The mean and diagonal covariance matrix are fixed to
| 20 |
For this choice of mean and variance, all quadrature points for a fifth order Gauss-Hermite quadrature approximation of numerical integration were positive. The remaining model parameters are fixed according to Table 1. Next, to fix the likelihood function, the Gaussian noise distribution, i.e., , is needed to be fixed. To consider reasonable values of , first the reference peak pyruvate signal is calculated using the solution of the model with the default model and design parameters in Tables 1 and 2; it is found to be . Then for different signal-to-noise ratios (), the noise (standard deviation) in the individual signals, , and the standard deviation of the total signal, , are computed as follows, for ,
| 21 |
where is the total number of scans. For each and corresponding in the above list, optimal design parameters are obtained by solving the optimization problem (19). For simplicity, let denote the optimized design parameter corresponding to and . The rationale behind considering different is that, in reality, data is expected to have varied signal-to-noise ratios and this is shown to impact the choice of optimal design parameters. The initial values of the design parameter are listed in Table 2.
Two cases of optimization as described below are considered:
Constant flip angle optimization. In this case, is fixed to 3 seconds for all k, and the flip angles are assumed to be same for all scans. As a result, the optimization problem involved only two variables, and .
Variable flip angle and TR optimization. In this case, flip angles and TR values at all scans are optimized.
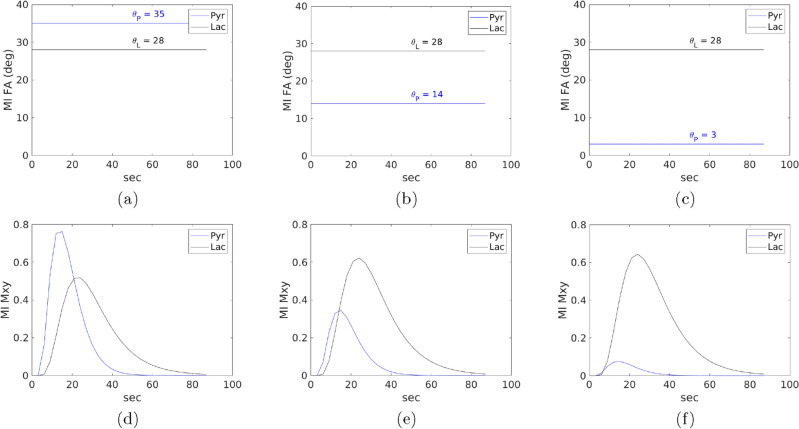
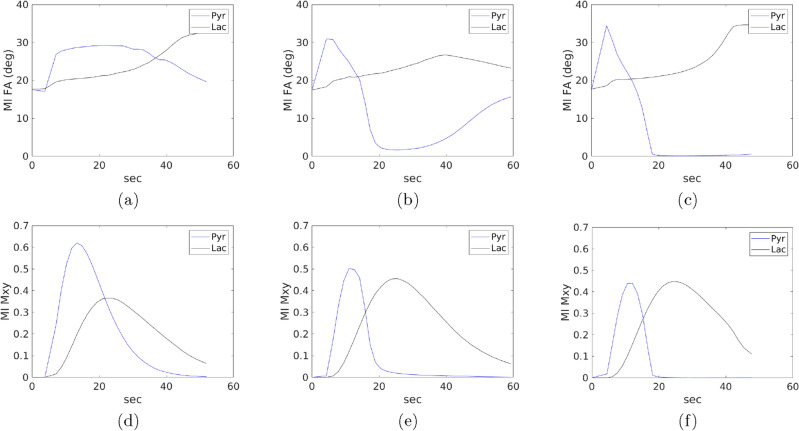
For the case of constant flip angle optimization, the optimized design parameters, , corresponding to the five SNRs are tabulated in Table 3. Figs. 1 and 2 represent the optimal design parameters, , for , , and for the two cases of optimization problems, respectively. Fig. 1 presents the optimal solution when considering a fixed repetition time of 3s and optimizing for pyruvate and lactate flip angles that are constant in time. Fig. 2 presents the optimal solution when allowing the repetition time and flip angles to vary at each acquisition for a fixed number of data acquisitions, . The optimal values of design parameters are shown in Fig. 1(a-c) and Fig. 2(a-c). The time varying optimized design parameters are significantly different from the constant value flip angle scheme. Fig. 1(d-f) and Fig. 2(d-f) show the transverse magnetizations for . For the case of variable design parameters, optimized values are shown in Fig. 3.
Table 3.
Optimized design parameters considering constant flip angles throughout the scan. Repetition time is fixed to s, for all k.
| (degrees) | (degrees) | (s) | (degrees) | (degrees) | (s) | ||
|---|---|---|---|---|---|---|---|
| 2 | 35 | 28 | 3 | 5 | 35 | 28 | 3 |
| 10 | 14 | 28 | 3 | 15 | 4 | 28 | 3 |
| 20 | 3 | 28 | 3 |
Figure 1.
Optimized design parameters along with the signals of pyruvate and lactate obtained from the solution of the forward model using optimal design parameters. In (a), for noise , i.e., for , the optimized flip angle scheme is shown for constant flip angles throughout the acquisition (optimal angles are degrees and degrees, for all ). The corresponding signal evolution of the transverse magnetizations () are shown in (d) for the constant flip angle case. Similarly, (b) and (c) present the optimized flip angles for and ; respectively. The corresponding signal evolution of the transverse magnetizations are shown in (e) and (f).
Figure 2.
Optimized design parameters along with the solution of the forward model. In (a), for noise , i.e., for , the optimizer considers jointly varying the flip angle and repetition time at each acquisition. The corresponding signal evolution of the transverse magnetizations () are shown in (d). Similarly, (b) and (c) present the optimized flip angles and repetition time for and , respectively. The corresponding signal evolution of the transverse magnetizations are shown in (e) and (f). Note here that x-axis in all plots is for time and therefore the plots implicitly include the values of optimized , .
Figure 3.
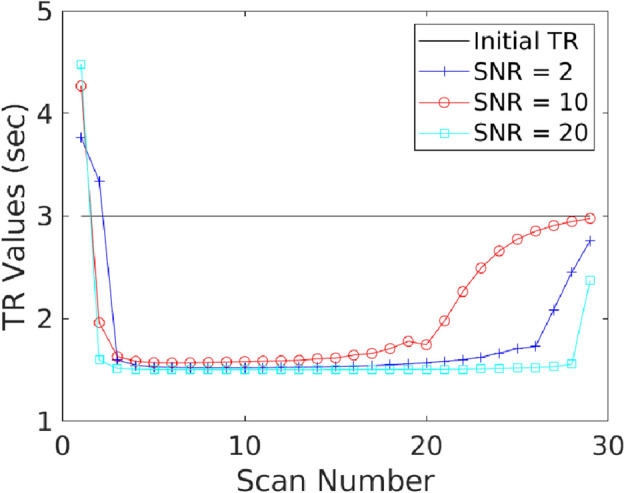
Plot of optimal repetition times for three cases of .
Validation of the uncertainty reduction using optimal design parameters
The basic workflow in verification includes generating the samples of noisy data associated with different design parameters following Sect. “Synthetic data” and then solving the inverse problem to recover uncertain model parameters for each sample of noisy data. From the recovered for different samples of data, the mean and the standard deviation is computed. Specifically, the standard deviation is used as a measure of the uncertainty in the recovered .
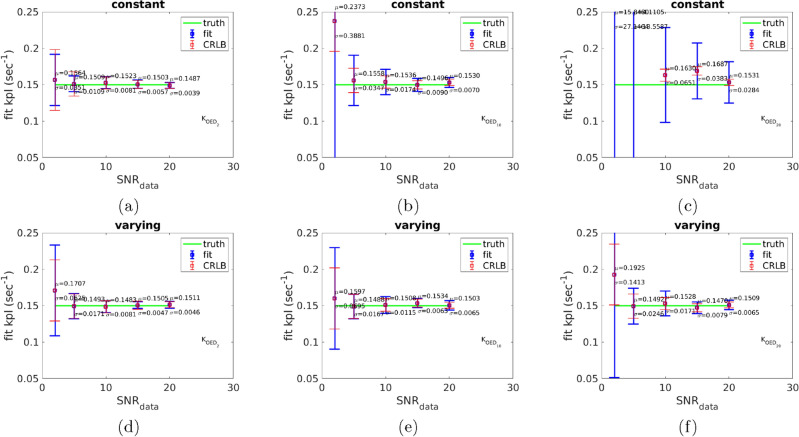
First using the model in (1), data (for ), corresponding to , is obtained. Then the uncertain parameters in the model are recovered from the 25 samples of noisy data. The remaining model parameters are drawn from the Table 1. In Fig. 4, the statistics (mean and standard deviation) of recovered corresponding to optimal design parameters shown in Figs. 1and 2 is presented. The SNR (SNR to generate noisy data) along the x-axis corresponds to the value of added to the data to generate noisy data; see (8) for the definition of noisy data. The y-axis represents the mean and standard deviation of the recovered from the inference. The known value of used to generate the noise corrupted data is shown as a horizontal line at for a reference. Fig. 4(a) and Fig. 4(d) correspond to for a constant and varying flip angle scheme; respectively. Fig. 4(b) and Fig. 4(e) correspond to for a constant and varying flip angle scheme; respectively. And finally Fig. 4(c) and Fig. 4(f) correspond to for a constant and varying flip angle scheme; respectively. The Cramér-Rao bound is provided as a reference for the lower bound of the variance in each. Fig. 5 provides a control for the accuracy and precision obtained for flip angles similar to those currently in use in our clinic47, 48, and . Similarly, the Cramér-Rao bound is provided as a reference for the lower bound of the variance. The results show that the uncertainties in recovered parameter using are comparable to the current clinical pulse sequence implementation. In fact, demonstrates improved performance for all except .
Figure 4.
Plot of inferred from 25 samples of noisy data based on the synthetic data from the solutions of the model in (1). The x-axis is the value of employed in computing noisy data. (a) and (d) correspond to for a constant and varying flip angle and TR scheme, respectively. (b) and (e) correspond to for a constant and varying flip angle and TR scheme, respectively. (c) and (f) correspond to for a constant and varying flip angle and TR scheme, respectively.
Figure 5.
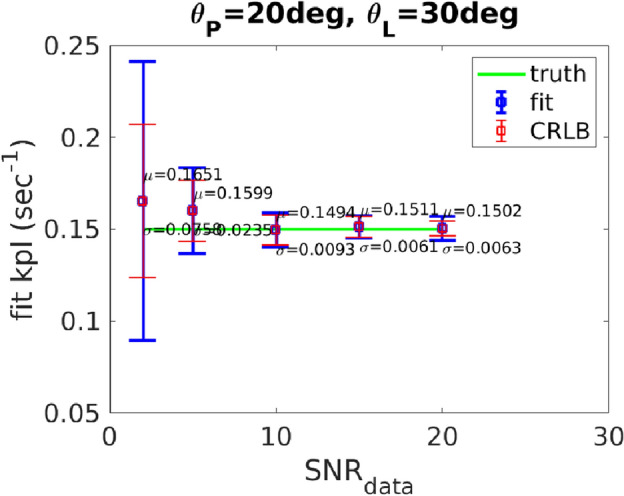
Plot of inferred from 25 samples of noisy data based on the synthetic data from the solutions of the model in (1). The x-axis is the value of employed in computing noisy data. Here, the accuracy and precision obtained for flip angles are similar to values currently used in our human studies, and is shown as a control for Fig. 4.
Generally, an improvement in the accuracy and precision of the recovered parameter is seen with increasing . A time-varying flip angle scheme leads to a higher parameter optimization that can further improve the quantitative value of mutual information over a constant flip angle scheme. However, the constant flip angle scheme, 35 and 28 degrees for pyruvate and lactate measurements, leads to the accuracy and precision comparable to the variable flip angle schemes obtained from our method.
Discussion
The evaluation of accuracy and precision as a function of pulse sequence design is effectively a bi-level optimization problem where the goal is to solve two nested optimization problems: (1) find the pulse sequence that produces the best accuracy and precision for the (2) best curve fit to the data. Direct numerical optimization of the bi-level cost functions(s) is challenging49. The mutual information objective function of this study as well as signal maximization and Fisher information in the literature15, 18, 19 are effectively proposing a surrogate objective function as a numerically tractable approximation to the bi-level optimization problem of interest. Within the mutual information based optimal experimental design formulation, a time varying flip angle and repetition time scheme is seen to provide significant differences in the pulse sequence as compared to the case when excitation angles are fixed to a constant value over time with a fixed repetition time of 3 s. Indeed, the varying flip angle and repetition time scheme leads to a higher dimensional parameter optimization that provides more degrees of freedom to further improve the quantitative value of mutual information over the constant flip angle scheme. However, as seen in Fig. 4, the constant flip angle scheme leads to comparable accuracy and precision when considering the inference from noise-corrupted data. The time varying scheme is seen to be more sensitive to noise corruption of the expected signal and is generally seen to have the higher variance in the parameter recovery at lower . The mutual information calculations are generally seen to achieve the Cramér-Rao bound with higher SNR.
The purely theoretical nature of these results is a limitation of the study. However, the two-compartment model analyzed in this work is utilized in numerous studies50–56 and, when parameterized with physically meaningful values of the parameters, good data agreement is shown within these studies. Results of this manuscript are thus relevant towards guiding future data collection.
The reduction in the recovered variance is seen to be correlated with the assumed noise value added to the data. Intuitively, less noise resulted in less variance in the parameter recovery. Less intuitively, the optimal MI solutions for flip angles are seen to vary with the noise value of the signal conditional probability model . The greatest reduction in measurement uncertainty is seen for the MI optimal solution corresponding to low SNR of the signal conditional probability model. Here, the lower flip angle is applied to the non-injected secondary substrate and higher pyruvate signal is maintained throughout the acquisition. This could be due to the system being so signal limited for the low SNR case that it is forced to leverage the pyruvate signal to extract any additional information about the metabolic exchange rate. The excitation angles optimized for the higher SNR condition reduce the pyruvate excitation angle to save magnetization for subsequent conversion while simultaneously increasing the lactate flip angle. Within the time varying optimization, the pyruvate excitation angle is reduced to zero after 20 s. For high SNR this suggests that the lactate signal is sufficient to accurately determine the metabolic exchange rate and measuring a large pyruvate signal after the initial bolus is less important.
This work considers uncertainty in the vascular-tissue exchange parameter, bolus arrival time, and rate constants modeled through a Gaussian prior. However, a more comprehensive evaluation of additional uncertain parameters would further evaluate the stability of our results. Additionally, the effect of alternative prior formulations such as uniform distributions for the prior parameters may also be investigated. The numerical computation in this work is also limited by the quadrature scheme for numerical integration of the mutual information integrals. Adding additional sources of uncertainty suffers from the well-known curse of dimensionality57 and alternative integration schemes such as Markov chain Monte Carlo may be more effective.
Further, the current approach considers the real component of the readout and assumes SNR such that Gaussian statistics is an appropriate noise model for the signal acquisition. Rician statistics58 is known to be more appropriate as the noise model for low SNR and the low SNR range is expected to be more important toward the end of the HP data acquisition as the signal decays. Rician statistics will be considered in future efforts to optimize acquisition parameters at low SNR or when considering both the real and imaginary components of the signal magnitude.
Alternative to the spatial-invariant model, high-fidelity models may also be considered to determine optimal design parameters and to recover model parameters from the data. However, for such an approach to work, a realistic high-fidelity model is needed keeping in mind the major factors in HP-MRI physics. Additional model fidelity may include permutations of lactate and pyruvate that are endogenous as well as hyperpolarized. Intravascular, extracellular, and intracellular species may also be considered. Nonlinear pyruvate-to-lactate conversion parameters such as from a Michaelis-Menten relationship may also be considered. Additional formulations may also consider the impact of blood flow in the simulations directly though Dirichlet boundary conditions, as convective transport through porous media59, or as a sophisticated 3D-1D coupling with vasculature treated as 1D curvilinear segments60, 61.
In summary, our results suggest that the constant flip angle scheme corresponding to is the best choice in terms of accuracy and precision of the parameter recovery. Results at , and , are comparable to the current clinical pulse sequence implementations, and , and demonstrate an improved performance at low . Further, the constant flip angle scheme may represent a practical choice for implementation on the pulse sequence hardware.
Acknowledgements
We acknowledge Reshmi Patel and Collin J. Harlan for sharing the insights from their work. The work of PKJ and JTO was supported by the U.S. Department of Energy, Office of Science, USA, Office of Advanced Scientific Computing Research, Mathematical Multifaceted Integrated Capability Centers (MMICCS), under Award Number DE-SC0019303. DF, JB, and DS are supported under R21CA249373A1. This work was supported as one of the pilot projects by the MDACC-Oden Institute-TACC joint initiative. JB, DF, and PKJ acknowledge this support.
Author contributions
D.F. and P.K.J. worked on developing computational methods and numerical simulations. J.B., D.S., and C.W. provided necessary insights into hyperpolarized MRI experimentation and helped with introduction and modeling. D.F., P.K.J., D.M., D.S., and C.W. contributed to software development. J.T.O. and J.B. offered valuable insights into the optimal experimental design problem. All authors contributed to the preparation and review of the manuscript and helped in analyzing numerical results.
Data availability
The codes and relevant data files to reproduce the results will be publicly available in the following GitHub repository: https://github.com/prashjha/HyperpolarizedMRI.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Prashant K. Jha, Email: prashant.jha@austin.utexas.edu
David T. Fuentes, Email: dtfuentes@mdanderson.org
References
- 1.Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstark AL, Ferrone M, Criekinge MV, Chang JW, Bok R, Park I, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13c] pyruvate. Sci. Transl. Med. 2013;5(198):198ra108–198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bankson JA, Walker CM, Ramirez MS, Stefan W, Fuentes D, Merritt ME, Lee J, Sandulache VC, Chen Y, Phan L, Chou P-C, Rao A, Yeung S-CJ, Lee M-H, Schellingerhout D, Conrad CA, Malloy C, Sherry AD, Lai SY, Hazle JD. Kinetic modeling and constrained reconstruction of hyperpolarized [1-sup13/supC]-pyruvate offers improved metabolic imaging of tumors. Cancer Res. 2015;75(22):4708–4718. doi: 10.1158/0008-5472.CAN-15-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, Bankson JA, Brindle K, Cunningham CH, Gallagher FA, Keshari KR, Kjaer A, Laustsen C, et al. Hyperpolarized 13c MRI: Path to clinical translation in oncology. Neoplasia. 2019;21(1):1–16. doi: 10.1016/j.neo.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granlund KL, Tee S-S, Vargas HA, Lyashchenko SK, Reznik ED, Fine S, Laudone V, Eastham JA, Touijer KA, Reuter VE, et al. Hyperpolarized MRI of human prostate cancer reveals increased lactate with tumor grade driven by monocarboxylate transporter 1. Cell Metab. 2020;31(1):105–114. doi: 10.1016/j.cmet.2019.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miloushev VZ, Keshari KR, Holodny AI. Hyperpolarization MRI: Preclinical models and potential applications in neuroradiology. Topics Magnetic Reson. Imag. TMRI. 2016;25(1):31. doi: 10.1097/RMR.0000000000000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, De Angelis LM, Mellinghoff IK, Brennan CW, Tabar V, Yang TJ, Holodny AI, et al. Metabolic imaging of the human brain with hyperpolarized 13c pyruvate demonstrates 13c lactate production in brain tumor patients. Cancer Res. 2018;78(14):3755–3760. doi: 10.1158/0008-5472.CAN-18-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal R, Vigneron DB, Kurhanewicz J. Hyperpolarized 1-[13c]-pyruvate magnetic resonance imaging detects an early metabolic response to androgen ablation therapy in prostate cancer. Eur. Urol. 2017;72(6):1028. doi: 10.1016/j.eururo.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher FA, Woitek R, McLean MA, Gill AB, Garcia RM, Provenzano E, Riemer F, Kaggie J, Chhabra A, Ursprung S, et al. Imaging breast cancer using hyperpolarized carbon-13 MRI. Proc. Natl. Acad. Sci. 2020;117(4):2092–2098. doi: 10.1073/pnas.1913841117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woitek R, McLean MA, Gill AB, Grist JT, Provenzano E, Patterson AJ, Ursprung S, Torheim T, Zaccagna F, Locke M, et al. Hyperpolarized 13c MRI of tumor metabolism demonstrates early metabolic response to neoadjuvant chemotherapy in breast cancer. Radiol. Imag. Cancer. 2020;2(4):e200017. doi: 10.1148/rycan.2020200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg Otto. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 11.Heiden MGV, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grist JT, McLean MA, Riemer F, Schulte RF, Deen SS, Zaccagna F, Woitek R, Daniels CJ, Kaggie JD, Matys T, et al. Quantifying normal human brain metabolism using hyperpolarized [1-13c] pyruvate and magnetic resonance imaging. NeuroImage. 2019;189:171–179. doi: 10.1016/j.neuroimage.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CY, Soliman H, Geraghty BJ, Chen AP, Connelly KA, Endre R, Perks WJ, Heyn C, Black SE, Cunningham CH. Lactate topography of the human brain using hyperpolarized 13c-MRI. NeuroImage. 2020;204:116202. doi: 10.1016/j.neuroimage.2019.116202. [DOI] [PubMed] [Google Scholar]
- 14.Autry AW, Gordon JW, Carvajal L, Mareyam A, Chen HY, Park I, Mammoli D, Vareth M, Chang SM, Wald LL, et al. Comparison between 8-and 32-channel phased-array receive coils for in vivo hyperpolarized 13c imaging of the human brain. Magnetic Reson. Med. 2019;82(2):833–841. doi: 10.1002/mrm.27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker CM, Fuentes D, Larson PEZ, Kundra V, Vigneron DB, Bankson JA. Effects of excitation angle strategy on quantitative analysis of hyperpolarized pyruvate. Magnetic Reson. Med. 2019;81(6):3754–3762. doi: 10.1002/mrm.27687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maidens, J., Larson, P. E. Z., & Arcak, M. Optimal experiment design for physiological parameter estimation using hyperpolarized carbon-13 magnetic resonance imaging. In Proc. 2015 American Control Conference (ACC), 5770–5775. IEEE, (2015).
- 17.Nagashima Kaz. Optimum pulse flip angles for multi-scan acquisition of hyperpolarized NMR and MRI. J. Magnetic Reson. 2008;190(2):183–188. doi: 10.1016/j.jmr.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Larson PEZ, Chen H, Gordon JW, Korn N, Maidens J, Arcak M, Tang S, Criekinge M, Carvajal L, Mammoli D, et al. Investigation of analysis methods for hyperpolarized 13c-pyruvate metabolic MRI in prostate cancer patients. NMR Biomed. 2018;31(11):e3997. doi: 10.1002/nbm.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidens J, Gordon JW, Arcak M, Larson PEZ. Optimizing flip angles for metabolic rate estimation in hyperpolarized carbon-13 MRI. IEEE Trans. Med. Imag. 2016;35(11):2403–2412. doi: 10.1109/TMI.2016.2574240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao, B., Haldar, J. P., Setsompop, K., & Wald, L. L . Optimal experiment design for magnetic resonance fingerprinting. In Proc. 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 453–456. IEEE, (2016). [DOI] [PMC free article] [PubMed]
- 21.Assländer J, Lattanzi R, Sodickson DK, Cloos MA. Optimized quantification of spin relaxation times in the hybrid state. Magnetic Reson. Med. 2019;82(4):1385–1397. doi: 10.1002/mrm.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marseille GJ, De Beer R, Fuderer M, Mehlkopf AF, van Ormondt D. Maximum Entropy and Bayesian Methods. Springer; 1996. Bayesian estimation of MR images from incomplete raw data; pp. 13–22. [Google Scholar]
- 23.Brihuega-Moreno O, Heese FP, Hall LD. Optimization of diffusion measurements using Cramer-Rao lower bound theory and its application to articular cartilage. Magnetic Reson. Med. 2003;50(5):1069–1076. doi: 10.1002/mrm.10628. [DOI] [PubMed] [Google Scholar]
- 24.Poot DHJ, den Dekker AJ, Achten E, Verhoye M, Sijbers J. Optimal experimental design for diffusion kurtosis imaging. IEEE Trans. Med. Imag. 2010;29(3):819–829. doi: 10.1109/TMI.2009.2037915. [DOI] [PubMed] [Google Scholar]
- 25.Cercignani M, Alexander DC. Optimal acquisition schemes for in vivo quantitative magnetization transfer MRI. Magnetic Reson. Med. 2006;56(4):803–810. doi: 10.1002/mrm.21003. [DOI] [PubMed] [Google Scholar]
- 26.Reeves SJ, Zhe Z. Sequential algorithms for observation selection. IEEE Trans. Signal Process. 1999;47(1):123–132. [Google Scholar]
- 27.Ji S, Xue Y, Carin L. Bayesian compressive sensing. IEEE Trans. Signal Process. 2008;56(6):2346–2356. [Google Scholar]
- 28.Seeger M, Nickisch H, Pohmann R, Schölkopf B. Optimization of k-space trajectories for compressed sensing by Bayesian experimental design. Magnetic Reson. Med. 2010;63(1):116–126. doi: 10.1002/mrm.22180. [DOI] [PubMed] [Google Scholar]
- 29.Cover TM, Thomas JA. Elements of Information Theory. Wiley; 2012. [Google Scholar]
- 30.Davenport R. The derivation of the gamma-variate relationship for tracer dilution curves. J. Nucl. Med. 1983;24(10):945–948. [PubMed] [Google Scholar]
- 31.Mammoli D, Gordon J, Autry A, Larson PEZ, Li Y, Chen H-Y, Chung B, Shin P, Criekinge MV, Carvajal L, et al. Kinetic modeling of hyperpolarized carbon-13 pyruvate metabolism in the human brain. IEEE Trans. Med. Imag. 2019;39(2):320–327. doi: 10.1109/TMI.2019.2926437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerstner T, Griebel M. Numerical integration using sparse grids. Numer. Algorithms. 1998;18:209–232. [Google Scholar]
- 33.Niederreiter H. Random Number Generation and Monte Carlo Methods. SIAM; 1992. [Google Scholar]
- 34.Robert C, Casella G. Monte Carlo Statistical Methods. Springer; 2013. [Google Scholar]
- 35.Gordon S, McBride BJ. Computer Program for Calculation of Complex Chemical Equilibrium Compositions and Applications. CiteSeer; 1996. [Google Scholar]
- 36.Sloan IH, Joe S, Joe SLMS. Lattice Methods for Multiple Integration. Oxford science publications; 1994. [Google Scholar]
- 37.Genz A. A Package for Testing Multiple Integration Subroutines. Springer; 1987. pp. 337–340. [Google Scholar]
- 38.van Dooren P, de Ridder L. An adaptive algorithm for numerical integration over an n-dimensional cube. J. Comput. Appl. Math. 1976;2(3):207–217. [Google Scholar]
- 39.Barron AR. Approximation and estimation bounds for artificial neural networks. Mach. Learn. 1994;14(1):115–133. [Google Scholar]
- 40.Cavagnaro DR, Myung JI, Pitt MA, Kujala JV. Adaptive design optimization: a mutual information-based approach to model discrimination in cognitive science. Neural Comput. 2010;22(1956):1–15. doi: 10.1162/neco.2009.02-09-959. [DOI] [PubMed] [Google Scholar]
- 41.Drovandi CC, McGree JM, Pettitt AN. A sequential Monte Carlo algorithm to incorporate model uncertainty in Bayesian sequential design. J. Comput. Graph. Stat. 2014;23(1):3–24. [Google Scholar]
- 42.Ryan EG, Drovandi CC, Thompson MH, Pettitt AN. Towards Bayesian experimental design for nonlinear models that require a large number of sampling times. Comput. Stat. Data Anal. 2014;70:45–60. [Google Scholar]
- 43.Ryan KJ. Estimating expected information gains for experimental designs with application to the random fatigue-limit model. J. Comput. Graph. Stat. 2003;12(3):585–603. [Google Scholar]
- 44.Ryan EG, Drovandi CC, Mcgree JM, Pettitt AN. A review of modern computational algorithms for Bayesian optimal design. Int. Stat. Rev. 2016;84(1):128–154. [Google Scholar]
- 45.Mitchell DP, Hwang K-P, Bankson JA, Stafford RJ, Banerjee S, Takei N, Fuentes D. An information theory model for optimizing quantitative magnetic resonance imaging acquisitions. Phys. Med. Biol. 2020;65(22):225008. doi: 10.1088/1361-6560/abb9f6. [DOI] [PubMed] [Google Scholar]
- 46.Porat B, Friedlander B. Computation of the exact information matrix of gaussian time series with stationary random components. IEEE Trans. Acoust. Speech Signal Process. 1986;34(1):118–130. [Google Scholar]
- 47.Tang S, Meng MV, Slater JB, Gordon JW, Vigneron DB, Stohr BA, Larson PEZ, Wang ZJ. Metabolic imaging with hyperpolarized 13c pyruvate magnetic resonance imaging in patients with renal tumors initial experience. Cancer. 2021;127(15):2693–2704. doi: 10.1002/cncr.33554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schellingerhout, D. Hyperpolarized carbon c 13 pyruvate in diagnosing glioma in patients with brain tumors, https://clinicaltrials.gov/study/NCT03830151 (2022).
- 49.Sinha A, Malo P, Deb K. A review on bilevel optimization: From classical to evolutionary approaches and applications. IEEE Trans. Evol. Comput. 2017;22(2):276–295. [Google Scholar]
- 50.Lee PM, Chen H-Y, Gordon JW, Wang ZJ, Bok R, Hashoian R, Kim Y, Liu X, Nickles T, Cheung K, et al. Whole-abdomen metabolic imaging of healthy volunteers using hyperpolarized [1-13c] pyruvate MRI. J. Magnetic Reson. Imag. 2022;56(6):1792–1806. doi: 10.1002/jmri.28196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joergensen SH, Hansen ESS, Bøgh N, Bertelsen LB, Staehr PB, Schulte RF, Malloy C, Wiggers H, Laustsen C. Detection of increased pyruvate dehydrogenase flux in the human heart during adenosine stress test using hyperpolarized [1-13c] pyruvate cardiovascular magnetic resonance imaging. J. Cardiovasc. Magnetic Reson. 2022;24(1):1–9. doi: 10.1186/s12968-022-00860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu JY, Kim Y, Autry AW, Frost MM, Bok RA, Villanueva-Meyer JE, Xu D, Li Y, Larson PEZ, Vigneron DB, et al. Kinetic analysis of multi-resolution hyperpolarized 13c human brain MRI to study cerebral metabolism. Magnetic Reson. Med. 2022;88(5):2190–2197. doi: 10.1002/mrm.29354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsieh C-Y, Sung C-H, Shen Y-L, Lai Y-C, Kuan-Ying L, Lin G. Developing a method to estimate the downstream metabolite signals from hyperpolarized [1-13c] pyruvate. Sensors. 2022;22(15):5480. doi: 10.3390/s22155480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reynolds S, Kazan SM, Anton A, Alizadeh T, Gunn RN, Paley MN, Tozer GM, Cunningham VJ. Kinetic modelling of dissolution dynamic nuclear polarisation 13c magnetic resonance spectroscopy data for analysis of pyruvate delivery and fate in tumours. NMR Biomed. 2022;35(5):e4650. doi: 10.1002/nbm.4650. [DOI] [PubMed] [Google Scholar]
- 55.Grashei M, Biechl P, Schilling F, Otto AM. Conversion of hyperpolarized [1-13c] pyruvate in breast cancer cells depends on their malignancy, metabolic program and nutrient microenvironment. Cancers. 2022;14(7):1845. doi: 10.3390/cancers14071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Kouchkovsky I, Chen H-Y, Ohliger MA, Wang ZJ, Bok RA, Gordon JW, Larson PEZ, Frost M, Okamoto K, Cooperberg MR, et al. Hyperpolarized 1-[13 c]-pyruvate magnetic resonance imaging detects an early metabolic response to immune checkpoint inhibitor therapy in prostate cancer. Eur. Urol. 2022;81(2):219–221. doi: 10.1016/j.eururo.2021.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berchtold,S., Böhm, C., & Kriegal, H.-P. The pyramid-technique: Towards breaking the curse of dimensionality. In Proc. of the 1998 ACM SIGMOD international conference on Management of data, 142–153, (1998).
- 58.Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magnetic Reson. Med. 1995;34(6):910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuentes D, Thompson E, Jacobsen M, Crouch AC, Layman RR, Riviere B, Cressman E. Imaging-based characterization of convective tissue properties. Int. J. Hyperth. 2020;37(3):155–163. doi: 10.1080/02656736.2020.1845403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fritz M, Jha PK, Köppl T, Oden JT, Wohlmuth B. Analysis of a new multispecies tumor growth model coupling 3d phase-fields with a 1d vascular network. Nonlinear Anal. Real World Appl. 2021;61:103331. [Google Scholar]
- 61.Fritz M, Jha PK, Köppl T, Oden JT, Wagner A, Wohlmuth B. Modeling and simulation of vascular tumors embedded in evolving capillary networks. Comput. Methods Appl. Mech. Eng. 2021;384:113975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The codes and relevant data files to reproduce the results will be publicly available in the following GitHub repository: https://github.com/prashjha/HyperpolarizedMRI.