Key Points
Question
What is the performance of the novel multitarget stool RNA (mt-sRNA) test (ColoSense) for individuals 45 years and older undergoing colorectal cancer screening?
Findings
A pivotal prospective, cross-sectional clinical trial comprising 8920 eligible participants was used to evaluate the sensitivity and specificity of the mt-sRNA test compared with a colonoscopy. The sensitivity of the mt-sRNA test for detecting colorectal cancer was 94%, the sensitivity for detecting advanced adenomas was 46%, and the specificity for no lesions on colonoscopy was 88%.
Meaning
The mt-sRNA test exhibited strong performance across all age groups, including those aged 45 years and older. The mt-sRNA test demonstrated high sensitivity for detecting colorectal neoplasia and achieved a similar level of specificity compared with other molecular diagnostic tests.
Abstract
Importance
Noninvasive tests for colorectal cancer screening must include sensitive detection of colorectal cancer and precancerous lesions. These tests must be validated for the intended-use population, which includes average-risk individuals 45 years or older.
Objective
To evaluate the sensitivity and specificity of a noninvasive, multitarget stool RNA (mt-sRNA) test (ColoSense) test compared with results from a colonoscopy.
Design, Setting, and Participants
This phase 3 clinical trial (CRC-PREVENT) was a blinded, prospective, cross-sectional study to support a premarket approval application for a class III medical device. A total of 8920 participants were identified online using social media platforms and enrolled from June 2021 to June 2022 using a decentralized nurse call center. All participants completed the mt-sRNA test, which incorporated a commercially available fecal immunochemical test (FIT), concentration of 8 RNA transcripts, and participant-reported smoking status. Stool samples were collected prior to participants completing a colonoscopy at their local endoscopy center. The mt-sRNA test results (positive or negative) were compared with index lesions observed on colonoscopy. Over the course of 12 months, individuals 45 years and older were enrolled in the clinical trial using the decentralized recruitment strategy. Participants were enrolled from 49 US states and obtained colonoscopies at more than 3800 different endoscopy centers.
Main Outcomes and Measures
The primary outcomes included the sensitivity of the mt-sRNA test for detecting colorectal cancer and advanced adenomas and the specificity for no lesions on colonoscopy.
Results
The mean (range) age of participants was 55 (45-90) years, with 4% self-identified as Asian, 11% as Black, and 7% as Hispanic. Of the 8920 eligible participants, 36 (0.40%) had colorectal cancer and 606 (6.8%) had advanced adenomas. The mt-sRNA test sensitivity for detecting colorectal cancer was 94%, sensitivity for detecting advanced adenomas was 46%, and specificity for no lesions on colonoscopy was 88%. The mt-sRNA test showed significant improvement in sensitivity for colorectal cancer (94% vs 78%; McNemar P = .01) and advanced adenomas (46% vs 29%; McNemar P < .001) compared with results of the FIT.
Conclusions and Relevance
In individuals 45 years and older, the mt-sRNA test showed high sensitivity for colorectal neoplasia (colorectal cancer and advanced adenoma) with significant improvement in sensitivity relative to the FIT. Specificity for no lesions on colonoscopy was comparable to existing molecular diagnostic tests.
Trial Registration
ClinicalTrials.gov Identifier: NCT04739722
This article presents results from a prospective pivotal study in which individuals completed a multitarget stool RNA test prior to undergoing a colonoscopy to determine the sensitivity for detecting colorectal cancer and advanced ademonas.
Introduction
Colorectal cancer is the second deadliest cancer in the US, causing more than 52 000 deaths each year.1 Although most colorectal cancers occur in adults older than 50 years, approximately 12% of all cases are observed in individuals younger than 50 years.1 Screening for colorectal cancer can lower colorectal cancer mortality by detection of treatable early-stage disease or by detection of precancerous lesions, which can prevent colorectal cancer development if removed.2 The US Preventive Services Task Force (USPSTF) recommends both invasive and noninvasive approaches for colorectal cancer screening, with colonoscopy being the criterion standard for identifying and removing colorectal lesions, including polyps.3 Adherence rates for colonoscopies as a screening modality have remained consistently low at approximately 60%.4 Issues with low adherence have been compounded by recent guideline changes to include individuals aged 45 to 49 years in screening recommendations.5
Biomarker-based methods have critical advantages over classical screening modalities of imaging and endoscopy.6 Genomic biomarkers detected in either stool or blood have shown the potential to identify colorectal neoplasia with varying levels of sensitivity.7,8 Currently, the most sensitive US Food and Drug Administration (FDA)–approved or FDA-cleared noninvasive screening test for colorectal cancer is the mt-sDNA test (Cologuard). In patients 50 years and older, the mt-sDNA test has a reported 92% sensitivity for colorectal cancer and 42% sensitivity for advanced adenomas.7 Addendum data for the mt-sDNA test in those aged 45 to 49 years showed only 33% sensitivity for advanced adenomas, with no data on colorectal cancer sensitivity.9 Existing noninvasive tests play a crucial role in the colorectal cancer screening paradigm by increasing adherence and potentially identifying individuals who are more likely to have pathological findings on colonoscopy examinations,10,11 while simultaneously enabling endoscopic efforts to focus on higher risk-patients.12 Individuals often prefer noninvasive tests13 and are more likely to comply with colonoscopy after a positive noninvasive test result.14
There are currently no approved or cleared RNA-based molecular tests for the detection of colorectal cancer or advanced adenomas in average-risk individuals. Previous research has demonstrated that stool-derived eukaryotic RNA could serve as an attractive category of biomarkers for applications in stool-based colorectal cancer screening.15,16 This study presents the results from an 8920-participant prospective pivotal study, whereby individuals completed a multitarget stool RNA (mt-sRNA) test (ColoSense) prior to undergoing a colonoscopy. Data from this clinical trial will support a premarket approval application to the FDA.
Methods
Clinical Trial Design
From June 2021 through June 2022, participants were enrolled in a prospective cross-sectional study. The study was approved by an institutional review board (Advarra). All participants provided both written and oral informed consent to complete study requirements and provide medical records. Monitoring was completed by an independent third party (ARC Regulatory Consulting) at 3 points during the clinical trial. No serious adverse events occurred during trial execution. The clinical trial protocol is available in Supplement 1 and the statistical analysis plan is available in Supplement 2.
Study Population
Participants were engaged using an online social media platform through a decentralized recruitment effort based on guidance from the FDA.17 Interested participants completed a self-reported online survey to screen for the intended use population and to obtain socioeconomic and demographic information (see eMethods in Supplement 3). Information on race and ethnicity was collected to ensure diverse representation in the clinical trial, per FDA guidance.18 Per the clinical trial protocol, a subset of non–average-risk participants (eg, family history of colorectal cancer or negative noninvasive screening test result) was permitted for inclusion, despite falling outside the intended use population. Participants were excluded if they reported a personal history of colorectal neoplasia, digestive cancer, a history of high-risk conditions (eg, hereditary cancer syndromes and inflammatory bowel disease), overt rectal bleeding within the last 30 days, undergoing a colonoscopy within the past 9 years, or having had a positive fecal immunochemical (FIT) test result (or immunochemical fecal occult blood test) in the past year or a positive mt-sDNA test result in the past 3 years (eMethods in Supplement 3).
Clinical Procedures
Enrolled participants were provided a stool sample collection kit before a colonoscopy. Samples were required to be collected before routine bowel preparation. All medical records associated with the procedure were obtained and reviewed using a centralized database. Participants were classified based on the most advanced colorectal epithelial lesion (ie, index lesion) (see eMethods in Supplement 3).
Laboratory Procedures
Samples were processed using previously defined methods (see eMethods in Supplement 3).16 Briefly, the FIT was assessed using the immunochemical fecal occult blood test OC Auto Micro 80 Analyzer (510(k), k041408). The stool sample was assessed using an automated nucleic acid extraction instrument (EMAG; bioMérieux) and Bio-Rad’s QXDx ddPCR System (510(k), k181661). A locked software component was used to generate mt-sRNA test results (eMethods in Supplement 3). A predetermined threshold value was used to determine whether the mt-sRNA test result was positive.15,16 The final output for each sample was generated prior to unblinding colonoscopy results.
Colonoscopy Procedures
After stool sample collection, all participants were navigated to complete a routine standard-care colonoscopy, which was prescribed by the participant’s health care professional and performed at a local endoscopy center. Reports from procedures were centrally reviewed by pathologists to determine participant classification (eMethods in Supplement 3).19
Statistical and Computational Analyses
Based on FDA guidance, acceptance criteria for primary objectives included sensitivity for colorectal cancer being at least 90% with a 2-sided lower bound 95% CI (Clopper-Pearson exact test) being at least 80%; sensitivity for advanced adenomas being at least 45% with a 2-sided lower-bound 95% CI being at least 40%; and specificity for all other findings being at least 80%. Ninety-five percent CIs were compared using Z scores and associated P values. A minimum study size of 8500 participants was required to power the study; this number was based on an expected colorectal neoplasia incidence of 8%. Receiver operating characteristics (ROC) area under the curve (AUC) was calculated by evaluating the binary colonoscopy outcome to the mt-sRNA score. Fisher exact and χ2 tests were used to identify significant differences in test performance between various subgroups. Z scores and McNemar tests were used to calculate differences between mt-sRNA performance and FIT performance. A random forest classification model was used to assess features impacting dropout. A multivariate logistic regression model, which leveraged inverse probability weighting, was used to analyze the impact of dropout on sensitivity and specificity of the mt-sRNA test (eMethods in Supplement 3).
Results
Enrollment
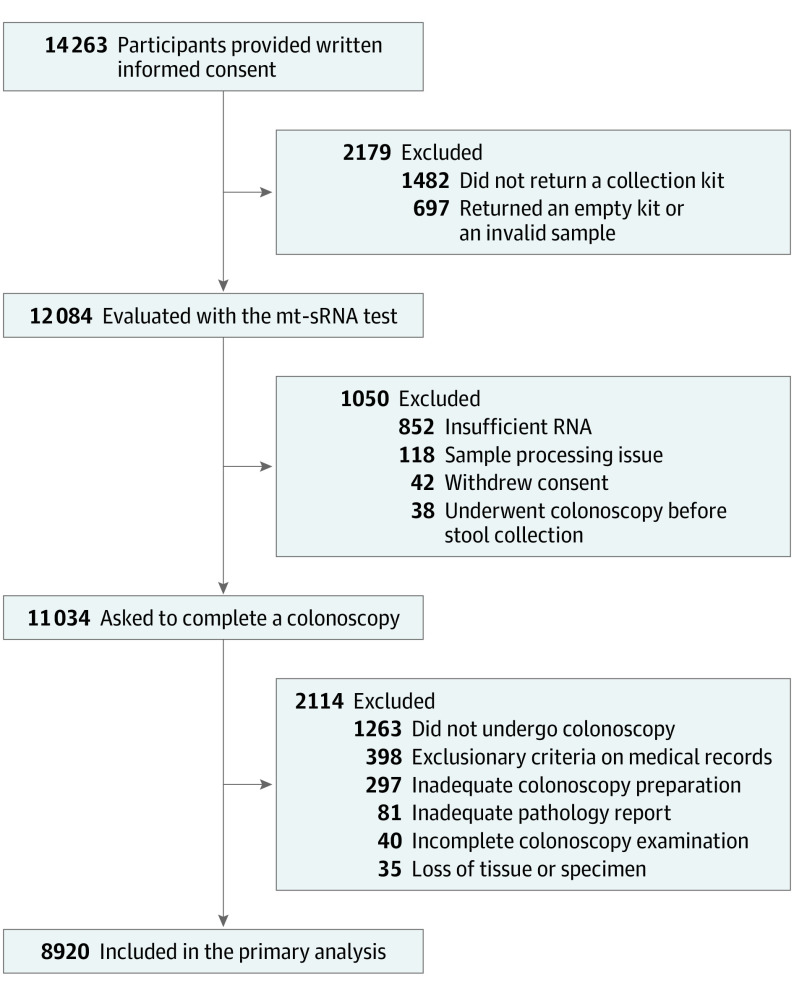
A total of 14 263 participants were enrolled in the CRC-PREVENT clinical trial (Figure 1). Among enrolled participants, 68% did not have a colonoscopy scheduled at survey completion and many required assistance with obtaining a colonoscopy appointment at a local endoscopy center. Additionally, 70% of participants had never completed a colonoscopy, with 32% of those individuals being older than 55 years (eTable 1 in Supplement 3). Across all enrolled participants, 2179 did not submit a valid stool sample (1482 did not return a collection kit and 697 returned an empty kit or invalid sample). The remaining 12 084 participants were evaluated with the mt-sRNA test, of whom 91% were eligible (852 had insufficient RNA, 118 had sample reception or processing issues, 42 withdrew consent, and 38 had a colonoscopy before stool collection). In total, 11 034 participants were asked to complete a colonoscopy, of whom 85% successfully completed the procedure (1263 did not complete a colonoscopy, 40 had incomplete colonoscopy examinations, and 297 had inadequate colonoscopy preparation). There were 398 participants who were withdrawn due to exclusionary criteria observed on medical records after study completion and 116 participants whose procedure or records did not permit adequate index lesion classification (eg, loss of tissue or inadequate pathology report). There were significant differences (P < .001) in sex, ethnicity, race, annual household income, insurance, and smoking status between participants who were withdrawn from the study relative to those who completed all study requirements (eTable 2 in Supplement 3). Higher dropout rates were observed in individuals who identified as male, Black or African American, or having low income (eFigure 1 in Supplement 3). A multivariate logistic regression model was employed on an expanded cohort (n = 13 785) to assess the impact of dropout on the mt-sRNA test with regards to sensitivity and specificity (see eTable 3 in Supplement 3).
Figure 1. Participants Enrolled in the CRC-PREVENT Clinical Trial .
Demographic Information
The 8920 eligible participants were enrolled from 49 US states across approximately 5400 different zip codes with colonoscopies completed at more than 3800 different endoscopy centers (eFigure 2 in Supplement 3). The mean (range) participant age was 55 (45-90) years and 5326 (60%) were women. Overall, 4% of participants self-identified as Asian, 11% as Black or African American, 7% as Hispanic or Latino, and 84% as White. In the study, 34% of participants had an annual household income less than $50 000 per year, 30% had public insurance or no insurance, and 5% were from rural areas. Additionally, 34% of participants reported primary exposure to tobacco products and 7% had a first-degree relative with a history of colorectal cancer (Table 1; eTable 4 in Supplement 3).
Table 1. Demographic and Socioeconomic Characteristics of Eligible Participants (N = 8920).
| Characteristic | No. (%) |
|---|---|
| Age, mean (SD), y | 55 (7.8) |
| Sexa | |
| Women | 5326 (59.7) |
| Men | 3584 (40.2) |
| Other | 10 (0.1) |
| Ethnicityb | n = 8743 |
| Hispanic or Latino | 588 (6.7) |
| Raceb | n = 8583 |
| American Indian or Alaska Native | 75 (0.9) |
| Asian | 343 (4.0) |
| Black or African American | 980 (11.4) |
| Native Hawaiian or Pacific Islander | 21 (0.2) |
| White | 7164 (83.5) |
| Annual household income, $b | n = 7996 |
| ≤29 999 | 1528 (19.1) |
| 30 000-49 999 | 1167 (14.6) |
| 50 000-74 999 | 1204 (15.1) |
| 75 000-99 999 | 1102 (13.8) |
| 100 000-149 999 | 1462 (18.3) |
| 150 000-199 999 | 725 (9.1) |
| ≥200 000 | 808 (10.1) |
| Insurancec | |
| Public insurance | 2549 (28.6) |
| Private insurance | 6117 (68.6) |
| Self-insured | 166 (1.9) |
| No insurance | 88 (1.0) |
| Locationd | |
| Urban or suburban | 8498 (95.3) |
| Rural | 422 (4.7) |
| Prior or current history of smokinge | |
| Yes | 3062 (34.3) |
| Family history of colorectal cancerf | |
| Yes | 580 (6.5) |
| Prior colonoscopyb | n = 6444 |
| Yes | 1874 (29.1) |
| Prior noninvasive testb | n = 6336 |
| mt-sDNA | 269 (4.2) |
| FIT/iFOBT | 451 (7.1) |
| No | 5616 (88.6) |
Abbreviations: FIT, fecal immunochemical test; iFOBT, immunochemical fecal occult blood test; mt-sDNA, multitarget stool DNA.
Sex was self-reported. Responses were mutually exclusive and included the option of “Other.”
Responses were self-reported. Responses were mutually exclusive and included the option of “Prefer not to answer.”
Insurance was self-reported. Responses were mutually exclusive. Public insurance included participants who selected Medicaid, Medicare, or Medicare Advantage.
Location was determined based on self-reported zip codes, and rural areas were defined as areas in which the local city center had a population of fewer than 10 000 inhabitants.
Smoking status was self-reported. Participants were asked to report any prior primary exposure to tobacco products.
Family history was self-reported. A positive family history was defined as a first-degree relative (sibling, parent, or child) with colorectal cancer.
Study Population
A total of 36 participants were found to have colorectal cancer (prevalence of 0.4%). A total of 606 participants were found to have advanced adenomas (prevalence of 6.8%). A total of 649 participants were found to have a medium-risk adenomas (MRAs; prevalence of 7.3%) and 2284 participants were found to have a low-risk adenomas (LRAs; prevalence of 25.6%). The adenoma detection rate across all eligible participants was 40.1%. A total of 5345 participants had either hyperplastic polyps less than 10 mm, other negative lesions (eg, lipoma, inflammatory polyp), or no lesions on colonoscopy (prevalence of 59.9%) (Table 2). Colonoscopy procedure withdrawal time for participants with no findings on colonoscopy was approximately 8.8 minutes (eTable 5 in Supplement 3).
Table 2. Sensitivity and Specificity of the Multitarget Stool RNA (mt-sRNA) Test and Fecal Immunochemical Test (FIT) .
| Outcome | mt-sRNA test (RNA and FIT) | Fecal immunochemical test (FIT) | ||
|---|---|---|---|---|
| Positive findings (sensitivity) | No./total No. | Sensitivity (95% CI), % | No./total No. | Sensitivity (95% CI), % |
| Colorectal cancer (CRC) | 34/36 | 94.4 (81-99) | 28/36 | 77.8 (61-90) |
| Stage I | 14/14 | 100 (77-100) | 10/14 | 71.4 (42-92) |
| Stage II | 10/12 | 83.3 (52-98) | 9/12 | 75.0 (43-95) |
| Stage III | 9/9 | 100 (66-100) | 8/9 | 88.9 (52-100) |
| Stage IV | ||||
| Unknown | 1/1 | 100 (0-100) | 1/1 | 100 (0-100) |
| Advanced adenomas | 278/606 | 45.9 (42-50) | 175/606 | 28.9 (25-33) |
| HGD or ≥10 adenomas (any size) | 30/46 | 65.2 (50-79) | 22/46 | 47.8 (33-63) |
| TVA (any size) | 78/164 | 47.6 (40-55) | 53/164 | 32.3 (25-40) |
| TSA or TA ≥10 mm | 170/396 | 42.9 (38-48) | 100/396 | 25.3 (21-31) |
| Negative findings (specificity) | No./total No. | Specificity (95% CI), % | No./total No. | Specificity (95% CI), % |
| Medium-risk adenomas | 502/649 | 77.3 (74-81) | 590/649 | 90.9 (88-93) |
| HP or SSL ≥10 mm | 159/192 | 82.8 (77-88) | 180/192 | 93.8 (89-97) |
| 5-9 adenomas (TA and SSL) <10 mm | 82/110 | 74.5 (65-82) | 98/110 | 89.1 (82-94) |
| 3-4 adenomas (TA and SSL) <10 mm | 261/347 | 75.2 (70-80) | 312/347 | 89.9 (86-93) |
| Low-risk adenomas | 1925/2284 | 84.7 (83-86) | 2163/2284 | 94.7 (94-96) |
| 1-2 adenomas (TA and SSL) 5-9 mm | 497/617 | 80.6 (77-84) | 570/617 | 92.4 (90-94) |
| 1-2 adenomas (TA and SSL) <5 mm | 1428/1667 | 85.7 (84-87) | 1593/1667 | 95.6 (95-97) |
| No findings | 4647/5345 | 86.9 (86-88) | 5100/5345 | 95.4 (95-96) |
| HP <10 mm or other negative lesions | 1343/1585 | 84.7 (83-86) | 1501/1585 | 94.7 (93-96) |
| No lesions on colonoscopy | 3304/3760 | 87.9 (87-89) | 3599/3760 | 95.7 (95-96) |
Abbreviations: HGD, high-grade dysplasia; HP, hyperplastic polyp; SSL, sessile serrated lesion; TA, tubular adenoma; TSA, traditional serrated adenoma; TVA, tubulovillous adenoma.
Multitarget Stool RNA Test Profile
The mt-sRNA test detected 34 of 36 participants with colorectal cancer (94.4% sensitivity [95% CI, 81%-99%]), with 100% sensitivity (n = 14) for stage I colorectal cancer (Table 2). The mt-sRNA test detected 278 of 606 participants with advanced adenomas (45.9% sensitivity [95% CI, 42%-50%]) (Table 2). Positive predictive value for colorectal neoplasia (colorectal cancer or advanced adenomas) was 20.6% (eTable 6 in Supplement 3). The diagnostic likelihood ratio for detecting colorectal cancer was 5.7 and the diagnostic likelihood ratio for detecting colorectal neoplasia (colorectal cancer or advanced adenomas) was 3.3 (eTable 7 in Supplement 3).
Within the colorectal cancer category, mt-sRNA test sensitivity for stage I/II colorectal cancer was not significantly different from sensitivity for stage III/IV colorectal cancer (92.3% vs 100.0%; P = .39). Within the advanced adenomas category, mt-sRNA test sensitivity for participants with at least 10 adenomas or lesions with high-grade dysplasia was 65.2% (n = 46), sensitivity for tubulovillous adenomas (any size) was 47.6% (n = 164), and sensitivity for traditional serrated adenomas or tubular adenomas (≥10 mm) was 42.9% (n = 396) (Table 2). The mt-sRNA test detected lesions with the highest malignant transformation rates at a higher sensitivity relative to those with lower malignant transformation rates (65.2% vs 42.9%; P = .002). Additionally, the mt-sRNA test sensitivity was proportional to maximum lesion size, such that sensitivity for advanced adenomas greater than 20 mm was 50.0%, whereas sensitivity for advanced adenomas less than 10 mm was 35.5% (P = .03). Sensitivity for advanced adenomas was significantly higher for distal lesions relative to proximal lesions (53.2% vs 37.0%; P <.001) (eTable 8 in Supplement 3).
Relative to the FIT, the mt-sRNA test showed a significant increase in sensitivity for colorectal cancer (94.4% vs 77.8%; McNemar P = .01) and advanced adenomas (45.9% vs 28.9%; McNemar P < .001) (Table 2; eTable 9 in Supplement 3). Across all advanced adenoma subcategories, the mt-sRNA test sensitivity was significantly elevated relative to the FIT (eTable 8 in Supplement 3).
The mt-sRNA test specificity for all other findings (MRA, LRA, and no findings; n = 8278) was 85.5% (Table 2). Within this category, specificity for no lesions on colonoscopy (n = 3760) was 87.9% (Table 2). Relative to the FIT, the mt-sRNA test showed a significant decrease in the specificity for no lesions on colonoscopy (95.7% vs 87.9%; P < .001) (Table 2).
ROC curves were generated for colorectal cancer compared with all other findings (MRA, LRA, and no findings) and for advanced adenomas compared with all other findings (MRA, LRA, and no findings). ROC AUC was 0.92 for colorectal cancer compared with all other findings and 0.68 for advanced adenomas compared with all other findings (Figure 2). When isolating the mt-sRNA molecular component (ie, no FIT component), ROC AUC was 0.62 for colorectal cancer compared with all other findings and 0.58 for advanced adenomas compared with all other findings (eFigure 3 in Supplement 3).
Figure 2. Sensitivity and Specificity of the Multitarget Stool RNA (mt-sRNA) Test and the Fecal Immunochemical Test (FIT) for Index Lesions Observed on Colonoscopy.
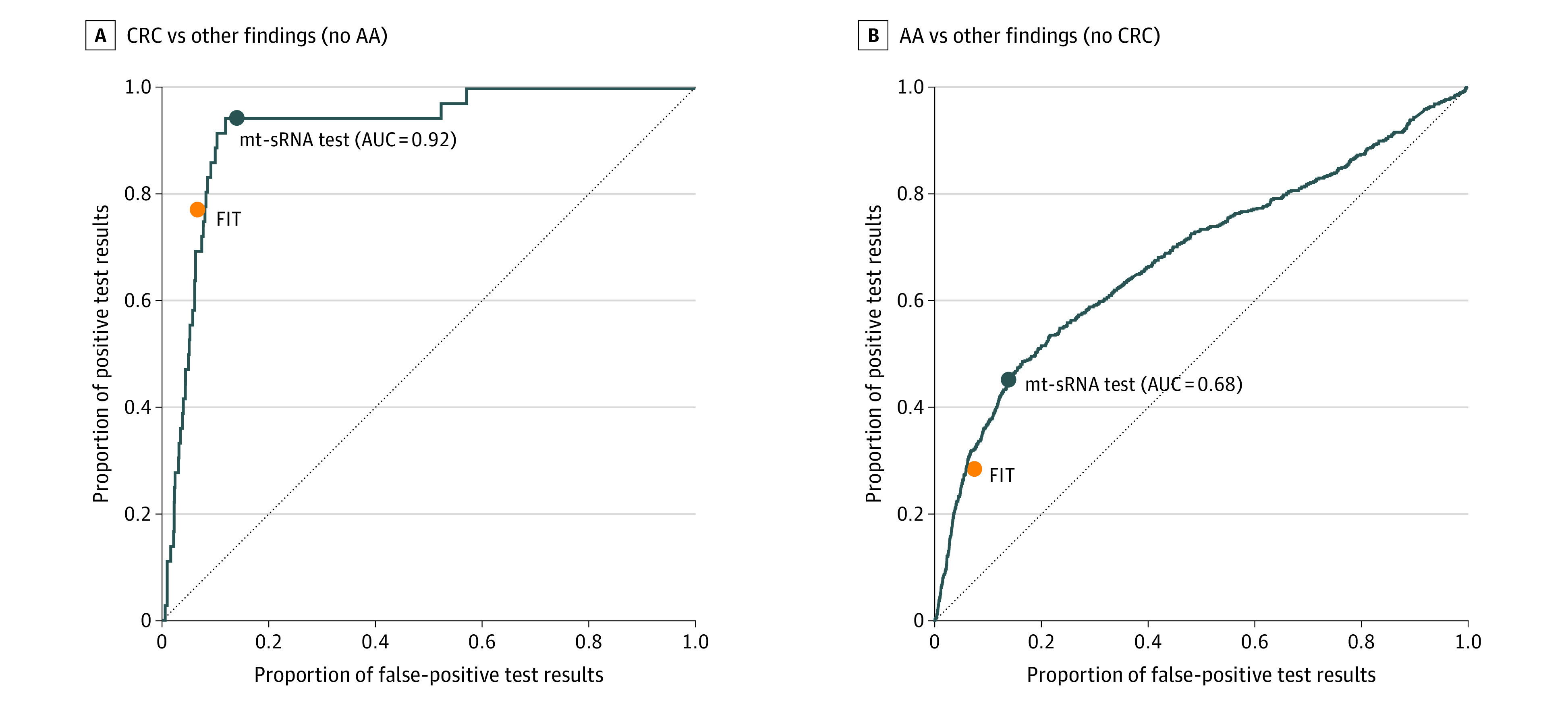
The mt-sRNA test receiver operating characteristic area under the curve (AUC) evaluated colorectal cancer (CRC) and advanced adenoma (AA) findings compared with all other findings (medium-risk adenomas, low-risk adenomas, and no findings).
An analysis was performed to assess the mt-sRNA test performance by age. In total, 23% of participants were aged 45 to 49 years and 33% were aged 50 to 54 years (eTable 4 in Supplement 3). Of all participants with colorectal cancer, 47% were 54 years or younger. In those aged 45 to 49 years, mt-sRNA detection of colorectal cancer was 100% (n = 17) and mt-sRNA detection of advanced adenomas was 45% (n = 103) (eTable 10 in Supplement 3).
An additional risk-based subset analysis showed significant differences in sensitivity for advanced adenomas (eTable 11 in Supplement 3). Sensitivity for advanced adenomas was significantly elevated in participants with low income (<$50 000 annual household income) relative to high income (≥$150 000 annual household income) (54.9% vs 35.5%; P = .001) and participants with public insurance relative to private insurance (55.8% vs 39.5%; P < .001). Larger lesions with higher malignant transformation rates were observed more frequently in cohorts with higher sensitivity for advanced adenoma.
Although this study did not complete a head-to-head performance comparison with other noninvasive molecular tests, a total of 323 participants reported a negative noninvasive screening test result (mt-sDNA in the last 3 years or FIT in the last year) prior to completing the mt-sRNA test and subsequent colonoscopy. Among this cohort with prior negative test results, the mt-sRNA test had 100% sensitivity for colorectal cancer (n = 2) and 63.2% sensitivity for advanced adenomas (n = 19), with an 84.1% specificity for all other findings (n = 302) (eTable 12 in Supplement 3). There were 398 participants who were withdrawn from analysis due to having exclusionary criteria on medical records (eTable 13 in Supplement 3). For these participants, the mt-sRNA test had 100% sensitivity for colorectal cancer (n = 1) and 61.7% sensitivity for advanced adenomas (n = 47), with a specificity of 76.6% for all other findings (n = 350).
Discussion
This study demonstrates that the novel mt-sRNA test can accurately detect individuals with colorectal cancer (94% sensitivity) and advanced adenomas (46% sensitivity) among those aged 45 years or older. The mt-sRNA test demonstrated 100% sensitivity for stage I cancer (n = 14) and 65% sensitivity for high-grade dysplasia or 10 or more adenomas. Further, the mt-sRNA test maintained performance in individuals aged 45 to 49 years (100% colorectal cancer sensitivity and 45% advanced adenoma sensitivity), for whom screening is now recommended per the USPSTF.
This study represents the first known instance of leveraging a decentralized real-world, cohesive colorectal cancer screening program for clinical validation. Decentralized recruitment outside of endoscopy practices provided unique engagement with participants who were not yet actively participating in colorectal cancer screening and required navigation to colonoscopy at a local endoscopy center. This is in direct contrast to previously conducted clinical trials in which participant recruitment occurred at endoscopy centers for individuals who already had a colonoscopy scheduled.7 Although remuneration might have played a role in adherence (see eMethods in Supplement 3), high participation was observed relative to other noninvasive testing platforms.20,21 Given that noninvasive colorectal cancer screening depends on the completion of colonoscopy for individuals with a positive screening test result, employing colonoscopy navigation methods described in this study could be used to improve colonoscopy compliance after a positive mt-sRNA test result.22
Colorectal cancer rates are increasing in younger populations,23 resulting in the recent recommendation to reduce the screening age from 50 to 45 years.5,24,25 In this study, the adenoma detection rate in the youngest age bracket (45-49 years) was 35% and colorectal cancer incidence was 0.24%. Despite increasing rates of disease in younger patient populations, minimal data have been reported to demonstrate performance characteristics of noninvasive screening tests in this cohort. Previous clinical trials for the mt-sDNA test have shown no data on colorectal cancer sensitivity in the age group of 45 to 49 years, with a significant reduction in advanced adenomas sensitivity from 42% (age ≥50 years) to 33% (age 45-49 years).26 Additionally, in the mt-sDNA study, 10% of all participants and 25% of participants with colorectal cancer were in age categories for which the USPSTF does not recommend screening (>75 years).3 In the trial presented here, 99% of all participants, and 100% of participants with colorectal cancer, were within age ranges that would be endorsed by the USPSTF with an A or B rating. Importantly, 23% of participants were age 45 to 49 years, with preserved sensitivity for colorectal cancer (100%) and advanced adenoma (45%) compared with other age groups. This may be attributable to the use of RNA biomarkers, which are not subject to age-related methylation patterns that can impact test results across different age groups.7,27,28,29,30
Participants in this study were classified based on the risk of developing metachronous advanced neoplasia after lesion observation on colonoscopy findings,31,32,33,34 with updates to reflect pertinent guidelines.9,35 Specifically, sessile serrated lesions (SSLs) greater than or equal to 10 mm were labeled as MRAs due to the recent redefinition whereby the crypt burden required for SSL classification was reduced from 2 to 3 crypts to 1 crypt.36 Due to this change, more than double the reported incidence of large SSLs relative to previous studies was observed,7 many of which would have been classified as hyperplastic polyps with previous guidelines.37,38 Although it is known that SSLs of 10 mm or more with the 2 to 3 crypt requirement resulted in a significantly higher risk of developing colorectal cancer compared with no adenoma,39,40,41,42 further research needs to be completed to understand if SSLs with only 1 crypt maintain the same increase in risk for developing colorectal cancer.
The adenoma detection rate is a critical quality metric for gastroenterologists.43,44,45 Recommended adenoma detection rates on screening colonoscopy are 25%, whereby endoscopy centers with higher adenoma detection rates report improved clinical outcomes for individuals.46 In this study, the total adenoma detection rate across all participants was 40%. For participants with a positive mt-sRNA test result, adenoma detection rate was 54%. Given the mt-sRNA test’s higher sensitivity in detecting advanced adenomas, it could be used as a tool to identify individuals more likely to have precancerous lesions, thus enriching the population undergoing colonoscopy.10,11
The mt-sRNA test showed comparable levels of sensitivity for colorectal cancer (94%) and advanced adenomas (46%) relative to the mt-sDNA test (92% for colorectal cancer and 42% for advanced adenomas) for individuals 50 years or older, and a significant increase in sensitivity for colorectal neoplasia for individuals younger than 50 years (47% [n = 108] vs 33% [n = 49]; P = .04). Although specificity of the mt-sRNA and mt-sDNA tests are reduced relative to FIT, increased sensitivity for colorectal neoplasia permits a 3-year screening interval, whereas FITs are being offered annually. Therefore the combined specificity for all 3 noninvasive diagnostics across a 3-year interval is comparable. Additional data from a blood-based test are pending publication, but is preliminarily showing reduced sensitivity for advanced adenomas (13%) and colorectal cancer (83%) relative to stool-based screening tests.47
Limitations
Study limitations were mostly attributable to use of a decentralized clinical trial for participant recruitment. First, center-to-center variations in colonoscopy quality metrics (eg, adenoma detection rate, withdrawal time), which are attributable to a physician’s experience, technique, and training, might have increased the false-positive and false-negative result rate of the mt-sRNA test. Second, there were differences in participant treatment (eg, colonoscopy scheduling, bowel preparation) and reporting practices in the colonoscopy and pathology reports (eg, nomenclature for SSLs, lesion sizing). These factors could have contributed to the variability of results. Third, the decentralized approach likely contributed to the high dropout rate.
Conclusions
This study leads to several conclusions to improve colorectal cancer screening. The mt-sRNA test can be an effective noninvasive test that is sensitive for colorectal cancer and advanced adenomas, with a comparable level of false-positive results compared with existing molecular screening tests. Moreover, the test characteristics in a younger cohort (45-49 years), now recommended for screening, is preserved. Also, the use of social media could potentially identify individuals not actively participating in colorectal cancer screening to potentially improve screening rates for this patient population.
Trial protocol
Statistical analysis plan
eMethods
Data sharing statement
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17-48. doi: 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- 2.Cronin KA, Scott S, Firth A, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2022;128(24):4251-4284. doi: 10.1002/cncr.34479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudsen AB, Rutter CM, Peterse EFP, et al. Colorectal cancer screening: an updated modeling study for the US Preventive Services Task Force. JAMA. 2021;325(19):1998-2011. doi: 10.1001/jama.2021.5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joseph DA, King JB, Dowling NF, Thomas CC, Richardson LC. Vital signs: colorectal cancer screening test use: United States, 2018. MMWR Morb Mortal Wkly Rep. 2020;69(10):253-259. doi: 10.15585/mmwr.mm6910a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson KW, Barry MJ, Mangione CM, et al. ; US Preventive Services Task Force . Screening for colorectal cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2021;325(19):1965-1977. doi: 10.1001/jama.2021.6238 [DOI] [PubMed] [Google Scholar]
- 6.Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19(8):521-531. doi: 10.1038/s41575-022-00612-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287-1297. doi: 10.1056/NEJMoa1311194 [DOI] [PubMed] [Google Scholar]
- 8.Church TR, Wandell M, Lofton-Day C, et al. ; PRESEPT Clinical Study Steering Committee, Investigators and Study Team . Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317-325. doi: 10.1136/gutjnl-2012-304149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imperiale TF, Kisiel JB, Itzkowitz SH, et al. Specificity of the multi-target stool DNA test for colorectal cancer screening in average-risk 45-49 year-olds: a cross-sectional study. Cancer Prev Res (Phila). 2021;14(4):489-496. doi: 10.1158/1940-6207.CAPR-20-0294 [DOI] [PubMed] [Google Scholar]
- 10.Wong JCT, Chiu HM, Kim HS, et al. ; Asia-Pacific Working Group on Colorectal Cancer . Adenoma detection rates in colonoscopies for positive fecal immunochemical tests versus direct screening colonoscopies. Gastrointest Endosc. 2019;89(3):607-613. doi: 10.1016/j.gie.2018.11.014 [DOI] [PubMed] [Google Scholar]
- 11.Dominitz JA, et al. S252 screening with FIT-DNA: Impact on colonoscopy withdrawal time, adenoma detection and endoscopist’s recommendation for follow-up. Am J Gastroenterol. 2022;117:e177-e178. doi: 10.14309/01.ajg.0000857648.98703.d6 [DOI] [Google Scholar]
- 12.Denis B, Gendre I, Tuzin N, et al. Adenoma detection rate is enough to assess endoscopist performance: a population-based observational study of FIT-positive colonoscopies. Endosc Int Open. 2022;10(9):E1208-E1217. doi: 10.1055/a-1859-8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young GP, Chen G, Wilson CJ, et al. “Rescue” of nonparticipants in colorectal cancer screening: a randomized controlled trial of three noninvasive test options. Cancer Prev Res (Phila). 2021;14(8):803-810. doi: 10.1158/1940-6207.CAPR-21-0080 [DOI] [PubMed] [Google Scholar]
- 14.Cooper GS, Grimes A, Werner J, Cao S, Fu P, Stange KC. Barriers to follow-up colonoscopy after positive fit or multitarget stool DNA testing. J Am Board Fam Med. 2021;34(1):61-69. doi: 10.3122/jabfm.2021.01.200345 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Barnell EK, Kang Y, Wurtzler EM, Griffith M, Chaudhuri AA, Griffith OL; Geneoscopy Scientists . Noninvasive detection of high-risk adenomas using stool-derived eukaryotic RNA sequences as biomarkers. Gastroenterology. 2019;157(3):884-887. doi: 10.1053/j.gastro.2019.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnell EK, Kang Y, Barnell AR, et al. Multitarget stool RNA test for noninvasive detection of colorectal neoplasias in a multicenter, prospective, and retrospective cohort. Clin Transl Gastroenterol. 2021;12(5):e00360. doi: 10.14309/ctg.0000000000000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conduct of Clinical Trials of Medical Products During the COVID-19 Public Health Emergency: Guidance for Industry, Investigators, and Institutional Review Boards. US Department of Health and Human Services; 2021. Accessed September 18, 2023. https://collections.nlm.nih.gov/catalog/nlm:nlmuid-9918248910206676-pdf [Google Scholar]
- 18.Office of the Commissioner . Diversity plans to improve enrollment of participants from underrepresented racial and ethnic populations in clinical trials. US Food and Drug Administration. Published April 2022. Accessed September 18, 2023. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/diversity-plans-improve-enrollment-participants-underrepresented-racial-and-ethnic-populations
- 19.AJCC Cancer Staging Manual 8th Edition. National Cancer Institute; 2020 [Google Scholar]
- 20.Mohl JT, Ciemins EL, Miller-Wilson LA, Gillen A, Luo R, Colangelo F. Rates of follow-up colonoscopy after a positive stool-based screening test result for colorectal cancer among health care organizations in the US, 2017-2020. JAMA Netw Open. 2023;6(1):e2251384. doi: 10.1001/jamanetworkopen.2022.51384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selby K, Jensen CD, Zhao WK, et al. Strategies to improve follow-up after positive fecal immunochemical tests in a community-based setting: a mixed-methods study. Clin Transl Gastroenterol. 2019;10(2):e00010. doi: 10.14309/ctg.0000000000000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman D, Ladabaum U, Brill JV, et al. Reducing the burden of colorectal cancer: AGA position statements. Gastroenterology. 2022;163(2):520-526. doi: 10.1053/j.gastro.2022.05.011 [DOI] [PubMed] [Google Scholar]
- 23.Siegel, R. L., et al. Colorectal cancer incidence patterns in the United States, 1974-2013. J Natl Cancer Inst. 2017;109(8):djw322. doi: 10.1093/jnci/djw322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crockett SD, Ladabaum U. Potential effects of lowering colorectal cancer screening age to 45 years on colonoscopy demand, case mix, and adenoma detection rate. Gastroenterology. 2022;162(3):984-986. doi: 10.1053/j.gastro.2021.11.024 [DOI] [PubMed] [Google Scholar]
- 25.Piscitello A, Edwards DK. Estimating the screening-eligible population size, ages 45-74, at average risk to develop colorectal cancer in the United States. Cancer Prev Res (Phila). 2020;13(5):443-448. doi: 10.1158/1940-6207.CAPR-19-0527 [DOI] [PubMed] [Google Scholar]
- 26.Imperiale TF, Kisiel JB, Itzkowitz SH, et al. Specificity of the multi-target stool DNA test for colorectal cancer screening in average-risk 45–49 year-olds: a cross-sectional study. Cancer Prev Res (Phila). 2021;14(4):489-496. doi: 10.1158/1940-6207.CAPR-20-0294 [DOI] [PubMed] [Google Scholar]
- 27.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371-384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 28.Busó EJ, Iborra M. Sequenom MassARRAY technology for the analysis of DNA methylation. García-Giménez JL, ed. Epigenetic Biomarkers and Diagnostics. Elsevier Academic Press; 2016: 137-153. [Google Scholar]
- 29.Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 2018;25(4):1066-1080. doi: 10.1016/j.celrep.2018.09.082 [DOI] [PubMed] [Google Scholar]
- 30.Cologuard: patients use. US Food and Drug Administration. Accessed September 18, 2023. https://www.accessdata.fda.gov/cdrh_docs/pdf13/P130017c.pdf
- 31.Gupta S, Lieberman D, Anderson JC, et al. Recommendations for follow-up after colonoscopy and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastrointest Endosc. 2020;91(3):463-485. doi: 10.1016/j.gie.2020.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart DB. Updated USPSTF guidelines for colorectal cancer screening: the earlier the better. JAMA Surg. 2021;156(8):708-709. doi: 10.1001/jamasurg.2021.1939 [DOI] [PubMed] [Google Scholar]
- 33.Crockett SD, Nagtegaal ID. Terminology, molecular features, epidemiology, and management of serrated colorectal neoplasia. Gastroenterology. 2019;157(4):949-966.e4. doi: 10.1053/j.gastro.2019.06.041 [DOI] [PubMed] [Google Scholar]
- 34.Shaukat A. Colorectal Polyps, An Issue of Gastrointestinal Endoscopy Clinics. Elsevier Health Sciences; 2022. [Google Scholar]
- 35.Nagtegaal ID, Odze RD, Klimstra D, et al. ; WHO Classification of Tumours Editorial Board . The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76(2):182-188. doi: 10.1111/his.13975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Kang GH. Evolving pathologic concepts of serrated lesions of the colorectum. J Pathol Transl Med. 2020;54(4):276-289. doi: 10.4132/jptm.2020.04.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bustamante-Balén M, Bernet L, Cano R, Morell L, López A. Assessing the reproducibility of the microscopic diagnosis of sessile serrated adenoma of the colon. Rev Esp Enferm Dig. 2009;101(4):258-264. doi: 10.4321/S1130-01082009000400004 [DOI] [PubMed] [Google Scholar]
- 38.Ensari A, Bilezikçi B, Carneiro F, et al. Serrated polyps of the colon: how reproducible is their classification? Virchows Arch. 2012;461(5):495-504. doi: 10.1007/s00428-012-1319-7 [DOI] [PubMed] [Google Scholar]
- 39.Erichsen R, et al. Increased risk of colorectal cancer development among patients with serrated polyps. Gastroenterology. 2016;150(4):895-902. doi: 10.1053/j.gastro.2015.11.046 [DOI] [PubMed] [Google Scholar]
- 40.Holme Ø, Bretthauer M, Eide TJ, et al. Long-term risk of colorectal cancer in individuals with serrated polyps. Gut. 2015;64(6):929-936. doi: 10.1136/gutjnl-2014-307793 [DOI] [PubMed] [Google Scholar]
- 41.Schreiner MA, Weiss DG, Lieberman DA. Proximal and large hyperplastic and nondysplastic serrated polyps detected by colonoscopy are associated with neoplasia. Gastroenterology. 2010;139(5):1497-1502. doi: 10.1053/j.gastro.2010.06.074 [DOI] [PubMed] [Google Scholar]
- 42.Anderson JC, Butterly LF, Robinson CM, Weiss JE, Amos C, Srivastava A. Risk of metachronous high-risk adenomas and large serrated polyps in individuals with serrated polyps on index colonoscopy: data from the New Hampshire Colonoscopy Registry. Gastroenterology. 2018;154(1):117-127.e2. doi: 10.1053/j.gastro.2017.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rex DK, Eid E. Considerations regarding the present and future roles of colonoscopy in colorectal cancer prevention. Clin Gastroenterol Hepatol. 2008;6(5):506-514. doi: 10.1016/j.cgh.2008.02.025 [DOI] [PubMed] [Google Scholar]
- 44.Khan R, Vaska M, Ruan Y, et al. Interventions to improve the quality of screening-related colonoscopy: protocol for a systematic review and network meta-analysis of randomised controlled trials. BMJ Open. 2022;12(11):e061855. doi: 10.1136/bmjopen-2022-061855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keswani RN, Crockett SD, Calderwood AH. AGA Clinical Practice Update on strategies to improve quality of screening and surveillance colonoscopy: expert review. Gastroenterology. 2021;161(2):701-711. doi: 10.1053/j.gastro.2021.05.041 [DOI] [PubMed] [Google Scholar]
- 46.Corte CJ, Leong RW. Improving the utility of colonoscopy: recent advances in practice. J Gastroenterol Hepatol. 2016;31(1):32-44. doi: 10.1111/jgh.13056 [DOI] [PubMed] [Google Scholar]
- 47.Chung DC, Gray DM II, Greenson JK, et al. Clinical validation of a cell-free DNA blood-based test for colorectal cancer screening in an average risk population. Presentation at Digestive Disease Week; May 7-9, 2023. Accessed September 18, 2023. https://guardanthealth.com/wp-content/uploads/ChungGrady_DDW_Abstract-913e_FINAL.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Chung DC, Gray DM II, Greenson JK, et al. Clinical validation of a cell-free DNA blood-based test for colorectal cancer screening in an average risk population. Presentation at Digestive Disease Week; May 7-9, 2023. Accessed September 18, 2023. https://guardanthealth.com/wp-content/uploads/ChungGrady_DDW_Abstract-913e_FINAL.pdf
Supplementary Materials
Trial protocol
Statistical analysis plan
eMethods
Data sharing statement