The emergence of drug resistance complicating anti-human immunodeficiency virus type 1 (HIV-1) therapy remains a significant limitation in the clinical utility of inhibitors directed against specific HIV enzymatic targets.
Viral resistance is a direct consequence of genetic diversity. The inherent high error rate of reverse transcriptase (35, 56) and the high replication levels of virus in vivo (5, 33, 93) allow for the generation of many variants in the virus population. Most important is the rapid selection of resistant variants due to the selective pressure exerted by antiviral drugs when virus replication continues during therapy. Recombination may also play a significant role in the generation of HIV-1 genetic diversity (12, 13, 20, 80).
Early development of antiretroviral therapy focused on inhibitors of reverse transcriptase. Both nucleoside and nonnucleoside inhibitors of this enzyme showed significant antiviral activity (19). However, the clinical benefit of these drugs had been limited due to drug resistance, limited potency, and host cellular factors (78). Thus, inhibitors targeted against a second essential enzyme of HIV-1 were urgently needed.
In 1988, the protease enzyme of HIV-1 was crystallized and its three-dimensional structure was determined (67, 94), allowing for the rapid development of protease inhibitors. Initially, it was hypothesized that HIV-1 protease, unlike reverse transcriptase, would be unable to accommodate mutations leading to drug resistance. This is not the case, and to date more than 20 possible amino acid substitutions in the HIV-1 protease have been observed during treatment with the currently available protease inhibitors. The genetic pattern of mutations conferring resistance to these protease inhibitors is complex, and cross-resistance between structurally different compounds occurs.
In this review the structure and function of HIV-1 protease will be discussed. The clinically relevant protease inhibitors will be described separately with emphasis on the emergence both in vitro and in vivo of drug-resistant variants. Mechanisms including active-site and secondary amino acid substitutions as well as gag cleavage site mutations will be described. Finally, the crucial issues surrounding cross-resistance and sequential protease inhibitor therapy will be discussed.
HIV-1 PROTEASE STRUCTURE AND FUNCTION
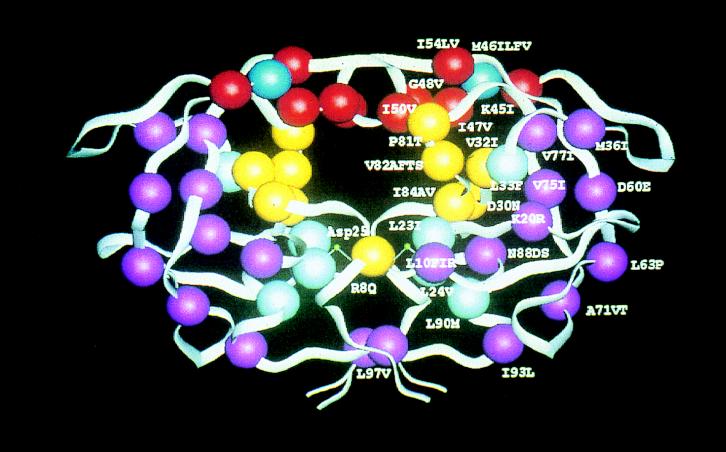
HIV-1 protease was classified as an aspartic proteinase on the basis of putative active-site homology (88), its inhibition by peptastin (77), and its crystal structure (67). The enzyme functions as a homodimer composed of two identical 99-amino-acid chains (18), with each chain containing the characteristic Asp-Thr-Gly active-site sequence at positions 25 to 27 (88). The crystal structure of HIV-1 protease reveals a dimer exhibiting exact twofold rotational C2 symmetry (67). The conserved active-site motifs are located in loops that approach the center of the dimer (Fig. 1). The two subunits are linked by a four-stranded antiparallel β-sheet involving both the amino and the carboxyl termini of each subunit. Upon binding, both subunits form a long cleft where the catalytically important aspartic acids are located in a coplanar configuration on the floor of the cleft. In addition, the enzyme contains a so-called “flap structure” in each subunit, an antiparallel β-hairpin with a β-turn that extends over the substrate binding site (34, 36). By convention, the peptide bond that is cleaved is referred to as the scissile bond that lies between P1 and P1′. The flanking amino acids going toward the amino terminus are named P1, P2, P3, etc., and those going toward the carboxyl terminus are referred to as P1′, P2′, P3′, etc. (39). The subsites of the enzyme interacting with the corresponding side chains of the polypeptide (substrate, inhibitor) are termed, starting from the central aspartates, S1, S2, S3, etc. and S1′, S2′, S3′, etc., respectively. It has been shown that HIV protease most efficiently cleaves peptide substrates seven amino acids long (P4-P3′) with the major processing subsites (S4-S3′) (17, 49, 54, 55, 81, 89). Substrate specificity of HIV-1 protease is significantly determined by subsites S2-S2′ (22).
FIG. 1.
Schematic structure of HIV-1 protease. Active-site residues are yellow; residues in the flap region are red; residue 46 and the flap hinge are dark blue; residues adjacent to the active site are light blue; residues distant from the active site of the enzyme are purple. Designations consist of the wild-type amino acid followed by the residue number and one or more described substitutions observed during protease inhibitor therapy; for example, I84V is a valine-for-isoleucine substitution at residue 84 in the protease monomer. (Courtesy of John Erickson.)
HIV protease processes gag (p55) and gag-pol (p160) polyprotein products into functional core proteins and viral enzymes (47, 50). During or immediately after budding, the polyproteins are cleaved by the enzyme at nine different cleavage sites to yield the structural proteins (p17, p24, p7, and p6) as well as the viral enzymes reverse transcriptase, integrase, and protease (75). An asparagine replacement for aspartic acid at active-site residue 25 results in the production of noninfectious viral particles with immature, defective cores (37, 44, 48, 74). Similarly, virus particles produced by infected cells treated with protease inhibitors contain unprocessed precursors and are noninfectious (15, 30, 45, 48, 74, 86). Unlike reverse transcriptase inhibitors, protease inhibitors block the production of infectious virus from chronically infected cells (53). Although the viral protease is a symmetric dimer, it binds inhibitors asymmetrically (23, 60). These findings together with the knowledge that amide bonds of proline residues are not susceptible to cleavage by mammalian endopeptidases gave rise to the first class of HIV-1 protease inhibitors based on the transition state mimetic concept, with the phenylalanine-proline cleavage site being the critical nonscissile bond (79).
SAQUINAVIR
Saquinavir, developed by Hoffmann-La Roche, was the first protease inhibitor to undergo clinical evaluation, demonstrating that HIV-1 protease was a valid target for the treatment of HIV infection (40). Saquinavir is a highly active peptidomimetic protease inhibitor with a 90% inhibitory concentration (IC90) of 6 nM (79). In vitro, saquinavir selects for variants with one or both of two amino acid substitutions in the HIV-1 protease gene, a valine-for-glycine substitution at position 48 (G48V), a methionine-for-leucine substitution at residue 90 (L90M), and the double substitution G48V-L90M (27, 41, 90). In most cases, G48V is the first mutation to appear, and continued selection results in highly resistant double-mutant variants. A substitution at either residue results in a 3- to 10-fold decreased susceptibility to the inhibitor, whereas the simultaneous occurrence of both substitutions causes a more severe loss of susceptibility of >100-fold (41).
Amino acid residue 48 is in the flexible flap loop of the enzyme, while residue 90 is located outside the binding pocket of the enzyme. Substitutions at position 48 could result in less conformational freedom and greater rigidity of the flap (55). The L90M substitution may induce conformational perturbations in the enzyme altering binding of the inhibitor (3).
Ermolieff et al. reported that the inhibition constants Ki of the constructed mutants were significantly higher than those of wild-type virus: 3-fold for L90M, 13.5-fold for G48V, and 419-fold for G48V/L90M (29). Maschera et al. determined the affinity of the wild type and the three mutant proteases L90M, G48V, and L90M/G48V (59). The affinity values for saquinavir were 1/20, 1/160, and 1/1,000 of that of the wild type, respectively. The catalytic efficiencies (kcat/Km) of the mutant proteases G48V and L90M/G48V were markedly reduced, 1/10 to 1/20 of that of the wild-type protease. These findings document the deleterious effects of these mutations on enzyme function and significantly reduced binding of the inhibitor. Of note, the decreased catalytic efficiencies resulted primarily from increased Km values, as opposed to changes in kcat. This suggests that the enzyme will maintain function in the presence of substrate excess, explaining the viability of the mutant viruses.
In vivo saquinavir therapy appears to select almost exclusively for mutations at codons 90 and 48 (41, 42, 92). Saquinavir-resistant variants emerge in approximately 45% of patients after 1 year of monotherapy with 1,800 mg daily (14, 25, 41, 65). The frequency of genotypic resistance is lower (22%) in patients receiving combination therapy with zidovudine, zalcitabine, and saquinavir (6). In contrast to in vitro-selected virus, where the G48V mutation is the first step to resistance, the L90M exchange is the predominant mutation selected in vivo while the G48V (2%) or the double mutant (<2%) is rarely found (41). In another recent study of in vivo resistance during saquinavir monotherapy no patient was found to harbor a G48V mutant virus (38). Interestingly, Winters et al. (94) observed a higher frequency of the G48V mutation in patients receiving higher saquinavir doses as monotherapy. All patients (six of six) who initially developed G48V also acquired a V82A mutation either during saquinavir treatment or after switching to either indinavir or nelfinavir. An identical mutational pattern was found in another study during saquinavir monotherapy (26). Some residues represent sites of natural polymorphism of the HIV-1 protease (positions 10, 36, 63, and 71) and appear to be positively correlated to the L90M mutation (42). Another substitution, G73S, has been recently identified and may play a role in saquinavir resistance in vivo. Isolates from five patients with early saquinavir resistance and those from two patients with induced saquinavir resistance after a switch of therapy to indinavir carried the G73S and the L90M substitutions (24).
RITONAVIR
Ritonavir, developed by Abbott Laboratories, was the second HIV protease inhibitor to be licensed in the United States. Ritonavir is a potent and selective inhibitor of HIV protease that is derived from a C2-symmetric, peptidomimetic inhibitor (34). In vitro activity has been demonstrated against a variety of laboratory strains and clinical isolates of HIV-1 with IC90s of 70 to 200 nM (51).
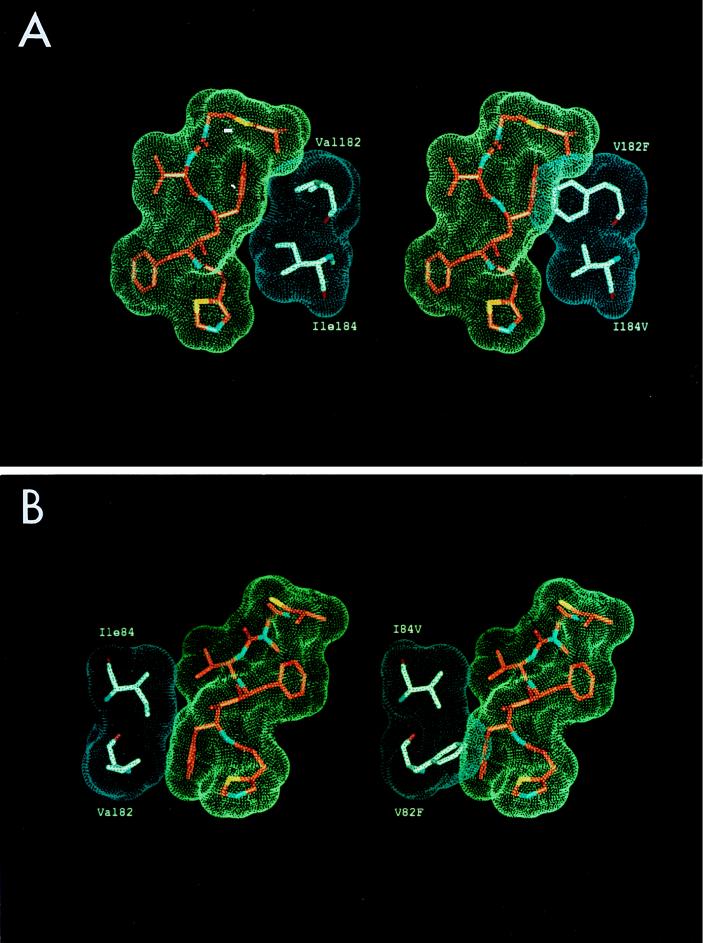
Phenotypically resistant virus generated by serial in vitro passages is associated with specific mutations at positions 84, 82, 71, 63, and 46 (58). The I84V substitution appeared to be the major determinant of resistance, resulting in a 10-fold reduction in sensitivity to ritonavir. Addition of the V82F mutation confers an even greater level of resistance, 10- to 20-fold. The substitutions M46I, L63P, and A71V, when introduced into the protease coding region of wild-type NL4-3, did not result in significant changes in drug sensitivity. Based on replication kinetics experiments, these changes are likely to be compensatory for active-site mutations, restoring the impaired replicative capacity of the combined V82F and I84V mutations. Computer modeling of ritonavir binding to the HIV-1 protease, based on the crystal structure of the related inhibitor A-78791 (36), demonstrated the structural effects of the V82F and I84V substitutions in HIV-1 protease on the interaction with ritonavir at the S1 and S1′ subsites (Fig. 2). Conformational adjustments due to these mutations result in decreases in enzyme-inhibitor interactions which are associated with loss of drug activity. Molecular dynamics simulation studies suggest that the M46I substitution results in a closed conformation of the flap domain relative to the wild-type enzyme, when a substrate or an inhibitor is bound (3, 7, 8). The role of the L63P and A71V substitutions is not obvious, since these mutations are away from the immediate vicinity of the active site and are not involved in substrate binding (96). The catalytic efficiencies (kcat/Km) for the mutant HIV proteases M46I, V82F, I84V, and M46I/V82F are reduced relative to wild-type enzyme by factors of 2.2, 3.8, 2.6, and 1.6, respectively (31). Schock et al. observed that the mutant with the double substitution M46I-L63P had a greater catalytic efficiency (per mole per second) than that of the wild-type enzyme for every substrate (110 to 360%), strongly suggesting that these changes likely play a compensatory role for other deleterious mutations (83).
FIG. 2.
Computer-generated model of HIV-1 protease-ritonavir complexes of wild-type HIV-1NL4-3 and the V82F/I84V double mutant. (A) Interaction between ritonavir and the protease at the S1′ subsite; (B) the same interactions at the S1′ binding subsite. I84V decreases the interaction with the Cβ group of the benzyl side chain of ritonavir, whereas V82F results in a severe spatial overlap with the phenyl ring of the inhibitor at the P1′ site. Note the effects of the double mutant on the van der Waals interactions between the enzyme and the inhibitor.
Phase I/II clinical trials of ritonavir monotherapy demonstrated a rapid decline in plasma HIV RNA to ∼1% of baseline levels (16, 33, 57, 93). After 32 weeks of treatment, only patients receiving the highest dose of ritonavir (1,200 mg daily) maintained a mean HIV plasma RNA reduction >0.8 log unit (16). Genotypic analysis of the HIV protease of viral isolates from patients with a rebound in viremia revealed a stepwise, ordered accumulation of multiple mutations at nine different codons resulting in amino acid substitutions at residues 20, 33, 36, 46, 54, 71, 82, 84, and 90 (63). An initial mutation at position 82 was consistently observed and appeared to be necessary for the primary loss of antiviral effect. There was no patient with a rise in viral load without variation at residue 82, as either a single substitution or part of a complex mutational pattern (63). This observation suggests that the primary mechanism for resistance is the selection of preexisting V82 single mutants with incompletely suppressed replication in the presence of ritonavir (5). The substitution at position 82 is followed most frequently by mutations at positions 54, 71, and 36. In contrast to in vitro experiments with ritonavir, in vivo I84V emerged late and less frequently. The L90M mutation was also observed less frequently and was always accompanied by at least two additional mutations. Emergence of I54V was not predicted by in vitro selection. The coappearance of I54V with V82A/F in vivo suggests a possible compensatory role of I54V (82).
INDINAVIR
Indinavir, developed by Merck & Co., is the third HIV protease inhibitor licensed in the United States. Indinavir is a potent and selective inhibitor of HIV-1 and HIV-2 proteases with Ki values of 0.34 and 3.3 nM, respectively (91). The drug acts as peptidomimetic transition state analogue and belongs to the class of protease inhibitors known as HAPA (hydroxyaminopentane amide) compounds (91). Indinavir provides enhanced aqueous solubility and oral bioavailability and in cell culture exhibits an IC95 of 50 to 100 nM (28).
Despite early reports of a lack of in vitro resistance by selection with indinavir (91), Tisdale et al. (87) were able to obtain resistant variants during selection in MT-4 cells with substitutions at residues 32, 46, 71, and 82. At least four mutations were required to produce a significant loss of susceptibility (6.1-fold compared with the wild type). The mutation at position 71, described as compensatory (58), appeared to contribute phenotypic resistance and also to improve virus growth. Emini et al. (28) and Condra et al. (10) found by constructing mutant HIV-1 clones that at least three mutations at residues 46, 63, and 82 were required for the phenotypic manifestation of resistance with a fourfold loss of susceptibility.
Early dose-ranging phase I/II studies with indinavir demonstrated that resistance emerges rapidly at suboptimal doses of the drug (10). Detailed genotypic analysis of viral isolates from patients with evidence of indinavir resistance showed multiple substitutions among at least 11 protease amino acid residues (9). No single mutation was present in all resistant isolates. The occurrence of mutations at residues 10, 20, 24, 46, 54, 63, 64, 71, 82, 84, and 90 was correlated with the loss of viral susceptibility to indinavir in vivo. Substitutions at position 46 and/or 82 predicted resistance in all isolates. The number of substitutions was also correlated with the degree of resistance. A simple pathway to phenotypic resistance to indinavir could not be defined. The IC95s for constructed single and double mutants were identical to that for wild-type virus, indicating a cumulative evolution of resistance. Zhang et al. demonstrated similar IC50s for wild-type virus and the double mutant 46/82, confirming the finding of the previous study (97). However, their data suggest a common pathway for emergence of resistance in which the first mutations that occur are M46L/I and V82A followed by a mutation at either I54V or A71V/T.
NELFINAVIR
Nelfinavir, developed by Agouron Pharmaceuticals, is a selective, nonpeptidic HIV-1 protease inhibitor that was designed by protein structure-based techniques using iterative protein crystallographic analysis (1). In vitro, nelfinavir was found to be a potent inhibitor of HIV-1 protease with a Ki of 2.0 nM (43). The drug demonstrated antiviral activity against several laboratory and clinical HIV-1 and HIV-2 strains with 50% effective concentrations ranging from 9 to 60 nM (70). Nelfinavir exhibits additive-to-synergistic effects when combined with other antiretroviral drugs (69). Preclinical data showed high levels of the drug in mesenteric lymph nodes and the spleen and good oral bioavailability (84).
In vitro, following 22 serial passages of HIV-1NL4-3 in the presence of nelfinavir, a variant (P22) with a sevenfold reduced susceptibility was isolated. After an additional six passages a variant (P28) with a 30-fold-decreased susceptibility to nelfinavir was identified (71). Sequence analysis of the protease gene from these variants identified in decreasing frequency the substitutions D30N, A71V, and I84V for the P22 variant and mutations M46I, I84V/A, L63P, and A71V for the P28 variant. Antiviral susceptibility testing of recombinant mutant HIV-1NL4-3 containing various mutations resulted in a fivefold-increased 90% effective concentration for the I84V and D30N single mutants and the M46I/I84V double mutant, whereas no change in susceptibility was observed with M46I, L63P, or A71V alone (71).
In an early phase II monotherapy study with nelfinavir, viral isolates from 9 of 17 patients had evidence of phenotypic resistance to the inhibitor. Sequence analysis of these isolates revealed a D30N mutation in all phenotypically resistant variants. This substitution had appeared in vitro in the P22 isolates but not in later, more resistant isolates, suggesting the possibility of a negative impact on viral fitness. A subsequent study confirmed the importance of D30N. In 55 patients who developed nelfinavir resistance this mutation was always identified (72). Substitutions at positions 36, 46, 71, 88, and others were occasionally associated with the change at residue 30. However, critical primary-site mutations conferring resistance to the other protease inhibitors were not seen.
AMPRENAVIR (VX-478 OR 141W94)
Amprenavir is a novel protease inhibitor currently in phase III studies. Developed by Vertex Laboratories, it was designed from knowledge of the HIV-1 protease crystal structure (46). The drug belongs to the class of sulfonamide protease inhibitors and has been shown to be a potent inhibitor of HIV-1 and HIV-2, with IC50s of 80 and 340 nM, respectively. The mean IC50 for amprenavir against clinical viral isolates was 12 nM (85). HIV-1 variants 100-fold resistant to amprenavir have been selected by in vitro passage experiments (69). DNA sequence analysis of the protease of these variants revealed a sequential accumulation of point mutations resulting in amino acid substitutions L10F, M46I, I47V, and I50V. The key resistance mutation in the HIV-1 protease substrate binding site is I50V. As a single mutation it confers a two- to threefold decrease in susceptibility (69). The other substitutions did not result in reduced susceptibility when introduced as single mutations into an HIV-1 infectious clone (HXB2). However, a triple protease mutant clone containing the mutations M46I, I47V, and I50V was 20-fold less susceptible to amprenavir than wild-type virus. The I50V mutation has not been frequently reported in resistance studies with other HIV protease inhibitors. Kinetic characterization of these substitutions demonstrated an 80-fold reduction in the inhibition constant (Ki) for the I50V single-mutant protease and a 270-fold-reduced Ki for the triple mutant M46I/I47V/I50V, compared to the wild-type enzyme (73). The single mutants L10F, M46I, and I47V did not display reduced affinity for amprenavir. The catalytic efficiency (kcat/Km) of the I50V mutant was decreased up to 25-fold, while the triple mutant M46I/I47V/I50V had a 2-fold-higher processing efficiency than the I50V single mutant, confirming the compensatory role of the M46I-and-I47V mutation. The reduced catalytic efficiency (kcat/Km) for these mutants in processing peptides appeared to be due to both increased Km and decreased kcat values.
The description of in vivo emergence of resistance to amprenavir is still preliminary. Ninety-two patients with no prior protease inhibitor treatment were treated with amprenavir alone or in combination with zidovudine-lamivudine. The mutations at residues 46, 47, and 50 selected in vitro were also observed in some patients in vivo. Additional substitutions associated with the I50V mutation included changes at residues 54 and 84. Mutations at positions 10, 20, 54, 82, and 84 also occurred independently of the I50V mutation (66).
The significance of these genotypic changes as they relate to phenotypic resistance and in vivo drug activity remains to be further elucidated.
PROTEASE INHIBITORS IN PRECLINICAL DEVELOPMENT
PNU-140690.
PNU-140690 (sulfonamide-containing 5,6-dihydro-4-hydroxy-2-pyrone) is a potent nonpeptidic HIV-1 protease inhibitor that was developed by structure-based design by Pharmacia & Upjohn. In H9 cell cultures, PNU-140690 inhibited acute infection with laboratory strain HIV-1IIIB at an average IC90 of 0.16 U (76). Enzymatic data underscore its potency and selectivity with inhibition constant values of Ki < 0.01 nM for the HIV-1 protease and Ki < 1 nM for the HIV-2 protease. PNU-140690 is active against HIV-1 isolates that are significantly resistant to ritonavir. An HIV-1NL4-3 passaged in the presence of ritonavir containing the substitutions L10I, M46I, L63P, and I84V/A conferred profound resistance (47- to >125-fold) to several structurally different protease inhibitors. However, this highly resistant isolate demonstrated only a sixfold increase in the IC90 of PNU-140690 versus wild-type HIV-1NL4-3 (76). The combination of PNU-140690 with ritonavir in vitro resulted in additive-to-synergistic effects, with even greater synergy demonstrated against ritonavir-resistant isolates (4). The specific pattern of in vitro resistance to PNU-140690 has yet to be identified.
ABT-378.
ABT-378 is a novel C2-symmetric protease inhibitor currently in phase I studies. The drug, developed by Abbott Laboratories, appears to be 10-fold more potent than its precursor, ritonavir, and demonstrates in vitro activity against ritonavir-resistant isolates (52). However, preliminary reports of amino acid substitutions selected by ABT-378 include L10F, V32I, M46I, I84V, and T91S (61).
CROSS-RESISTANCE AND SEQUENTIAL THERAPY
The issues surrounding cross-resistance to protease inhibitors and sequential therapy remain poorly defined and complex. Condra et al. (10) described patients undergoing therapy with indinavir. Five of the variants resistant to this inhibitor were cross-resistant to a panel of other protease inhibitors including saquinavir and amprenavir (10). Cross-resistance to all inhibitors required a minimum of four substitutions in the protease (M46I, L63P, V82T, and I84V). In a subsequent study 15 additional virus isolates were characterized (9). Resistance to indinavir was associated with a loss of susceptibility to ritonavir, whereas only a subset of indinavir-resistant variants exhibited decreased susceptibility to saquinavir (63%) and amprenavir (81%). The major contributors to cross-resistance to ritonavir appeared to be substitutions at either V82 or I84. The pattern of cross-resistance among indinavir, saquinavir, and amprenavir was not clearly discernible and seemed to be more complex. Tisdale et al. (86) analyzed variants individually selected for in vitro resistance to five different protease inhibitors. A total of 11 different mutations were selected on passage in the presence of the different compounds. The saquinavir-selected quadruple mutant (G48V, A71V, I84V, L90M) showed different levels of resistance to indinavir (3.8-fold), amprenavir (1.8-fold), and A-77003 (5.6-fold), the parent compound of ritonavir. The indinavir-selected quadruple variant (V32I, M46L, A71V, V82I) showed cross-resistance to A-77003 (9.8-fold) but had 5- to 6-fold-increased susceptibility to saquinavir and only 1.5-fold-increased resistance to amprenavir. The amprenavir-selected variant with the double mutation M46I/L-I50V exhibited a three- to fourfold-increased susceptibility to saquinavir and a two- to fourfold-enhanced susceptibility to indinavir compared to the wild type. These in vitro data suggest that combination protease inhibitor therapy may be a cogent strategy in protease-naive patients. During in vitro selection studies with the protease inhibitor ritonavir, three different viral variants resistant to ritonavir were assayed for cross-resistance to saquinavir and indinavir (58). The single mutants I84V and V82F and the quadruple mutant M46I/A71V/V82F/I84V displayed comparable levels of resistance to indinavir and ritonavir. In contrast, susceptibility to saquinavir was only moderately reduced, twofold for both single mutants and threefold for the quadruple mutant. Mo et al. (61) investigated cross-resistance of several HIV-1 variants to a panel of six structurally diverse protease inhibitors. They confirmed the previous finding of only discrete loss of susceptibility for single mutant I84V and multiple mutant M46I/L63P/A71V/V82F/I84V to saquinavir. Interestingly, the triple mutation M46I/L63P/I84A conferred a significant level of resistance, with IC90 increases of 80-fold for ritonavir, indinavir, and saquinavir and 125-fold for nelfinavir compared with the wild type. Molla et al. (62) examined the cross-resistance of mutant HIV-1 selected by ritonavir in vivo to the four protease inhibitors saquinavir, indinavir, nelfinavir, and amprenavir. Cross-resistance to indinavir and nelfinavir was observed in some of the patient isolates with multiple mutations, with 3- to 8-fold and 4- to 14-fold losses of susceptibility, respectively. No significant cross-resistance to either saquinavir or amprenavir was detected. Susceptibility testing of molecular mutant clones revealed a similar pattern of cross-resistance. The double mutant V82T-I54V and the highly ritonavir-resistant multiple mutants displayed significant cross-resistance to indinavir and nelfinavir but retained wild-type sensitivity to saquinavir and amprenavir.
Limited resistance data from sequential therapy with different protease inhibitors are slowly becoming available. Twenty-two patients initially treated with saquinavir were switched to indinavir due to a poor virological response. Eleven of the 22 did not respond to indinavir. Five patients had saquinavir resistance mutations including L90M (24). These mutations were maintained during indinavir therapy, and a limited number of other resistance substitutions were added, including L10I, M36I, L63P, A71V, and the previously uncharacterized substitution G73S. Viral isolates from two patients contained G48V, and the switch to indinavir resulted in the rapid selection of V82A. In the remaining four patients no mutations were noted at the time of the switch. However, typical substitutions of saquinavir resistance emerged under indinavir pressure. These findings strongly suggest that in these four patients saquinavir-resistant variants were present as a minority in the virus population, with indinavir exposure selecting for their rapid emergence. Despite the demonstration of relatively modest cross-resistance in vitro, saquinavir treatment followed by indinavir therapy resulted in the rapid selection of HIV-1 variants resistant to both saquinavir and indinavir. Nine of the 11 individuals with no response to indinavir failed saquinavir therapy with the typical saquinavir-resistant genotypes. Another study examined the genotypes of patients that failed long-term saquinavir therapy and were switched to either indinavir or nelfinavir (94). Three of nine patients who initially developed L90M on saquinavir therapy acquired the M36I or M46I substitution following nelfinavir or indinavir treatment. Only one patient with the L90M mutation showed evidence of the D30N mutation after nelfinavir failure. Patients without L90M or G48V following saquinavir therapy acquired L90M (four of seven) or D30N (one of seven) mutations on exposure to the second protease inhibitor. Some isolates of saquinavir-treated patients already had decreased in vitro susceptibility to nelfinavir and/or indinavir prior to therapy with these protease inhibitors. Moreover, after treatment with a second protease inhibitor, viruses emerged with reduced susceptibility to more than one inhibitor. These results indicate that initial therapy with one protease inhibitor may provide a genotypic foundation for the emergence of resistance mutations selected by exposure to a subsequent protease inhibitor.
Taken together, these data suggest that emergence of cross-resistance to protease inhibitors in vivo is a very complex and dynamic process which cannot be adequately predicted by standard viral genotypic and phenotypic assays (11). Novel phenotypic recombinant assays have been developed to rapidly determine drug susceptibility (32, 68). The clinical utility of these new technologies awaits clear definition.
CLEAVAGE SITE MUTATIONS
HIV-1 protease cleaves the gag and gag-pol precursor proteins at nine different cleavage sites (75). Cleavage sites are divided into two types: the “classical,” which are situated between p17/p24, p11 (protease)/p51, and the N terminus of protease, and the “nonclassical,” which include the remaining cleavage sites. Different cleavage sites are cleaved at different rates: p2/p7 and TF (transframe protein)/PR are the most rapidly processed cleavage sites, while p7/p1 and p1/p6 are the most slowly cleaved (17, 90). Doyon et al. (21) determined catalytic efficiency and growth kinetics of protease-resistant variants selected in vitro in the presence of two different substrate analog inhibitors, BILA 1906 BS and BILA 2185 BS. Sequence analysis of drug-resistant clones included not only typical mutations in the protease gene but also substitutions in gag precursor p1/p6 and/or p7/p1 cleavage sites. The p1/p6 mutation was found in 17 of 19 clones, whereas the p7/p1 mutation was observed only in highly resistant variants carrying seven or eight mutations in the protease gene. Growth kinetic studies revealed that variants containing mutations in both the protease gene and the cleavage sites grew significantly less well than wild-type virus but more efficiently than constructed chimeric variants lacking the cleavage site mutations. Analysis of the enzymatic activity of highly mutated variants towards peptides representing wild type or mutant cleavage sites with an L-to-F substitution in the P1′ position of the p1/p6 junction and/or a QA-to-RV change in the P3 and P2 residues of the p7/p1 junction indicated that the catalytic activity of the mutant enzyme was 2- to 10-fold increased when peptides with cleavage site mutations were used. However, wild-type peptides were more efficiently cleaved by wild-type HIV-1 protease than by mutated protease and mutated peptides. These data provide the first evidence of cleavage site mutations improving polyprotein processing of mutant HIV-1 protease, which permits compensation for impaired protease activity.
Zhang et al. analyzed sequences of the protease gene and cleavage sites of six patients with a rebound in plasma virus levels during antiretroviral therapy with indinavir (97). In addition to the sequential acquisition of protease mutations at residues 46, 54, 71, 82, 89, and 90, each patient had an identical mutation at position P2 in gag p7/p1. In one patient, an additional mutation at the gag p1/p6 cleavage site appeared. Recombinant HIV-1 variants with protease mutations at residues 46 and 82, but without mutations in p7/p1 and p1/p6, displayed a 68% reduction in replication rate compared to wild-type virus. Introduction of an additional mutation in the p7/p1 cleavage site resulted in a 41% increase in the replication rate compared to the latter variant. These findings confirm the in vitro results reported by Doyon et al.: cleavage site mutations compensate for the deleterious effect of a certain mutation in vivo and confer a significant growth advantage in the presence of the inhibitor (21).
CONCLUSIONS
HIV-1 protease inhibitors represent an expanding class of potent antiretroviral agents with superior in vivo antiviral activity. Given the results of in vitro selection and analysis of patient isolates for each protease inhibitor, it is evident that there is a striking overlap of resistance-conferring mutations among most of the available protease inhibitors (Table 1).
TABLE 1.
HIV-1 resistance to protease inhibitors
| Drug | Position(s) in protease
|
Reference(s) | |
|---|---|---|---|
| Critical substitutions | Secondary substitutionsa | ||
| Saquinavir | 48, 90 | 10, 36, 63, 71 | 43 |
| Ritonavir | 82, 84 | 20, 36, 46, 54, 63, 71, 90 | 59, 65 |
| Indinavir | 46, 82 | 10, 20, 24, 32, 54, 63, 71, 84, 90 | 10, 90 |
| Nelfinavir | 30 | 46, 63, 71, 88, 90 | 73, 74 |
| Amprenavir | 50 | 10, 46, 47 | 71 |
The role of secondary substitutions in the development of resistance is not completely understood. Some substitutions are known to be compensatory, enhancing the replicative efficiency of the virus. Other changes exist as naturally occurring polymorphisms in untreated patients.
The distinct pattern of viral resistance selected by individual drugs is an important element for the design of ideal combination therapies. Clearly, one criterion for selection of drugs used in combination therapy should be non-cross-resistance. Given the observation that initial pathways to resistance among protease inhibitors may differ, theoretically, a combination of protease inhibitors with different initial pathways to resistance may represent a rational strategy for efficient long-term suppression of viral replication. Studies using combinations of protease inhibitors with and without reverse transcriptase inhibitors are ongoing (2). The development of potent novel protease inhibitors with divergent mutational pathways hold promise for design of highly efficient and long-term suppressive antiretroviral combination therapy.
ACKNOWLEDGMENTS
We thank John Erickson for providing figures and David D. Ho for continued support.
REFERENCES
- 1.Appelt K R, Bacquet J, Bartlett C, Booth C L J, Freer S T, Fuhry M M, Gehring M R, Herrmann S M, Howland E F, Janson C A, Jones T R, Kan C C, Kathardekar V, Lewis K K, Marzoni G P, Mathews D A, Mohr C, Moomaw E W, Morse C A, Oatley S J, Ogden R C, Reddy M R, Reich S H, Schoettlin W S, Smith W W, Varney M D, Villafranca J E, Ward R W, Webber S, Webber S E, Welsh K M, White J. Design of enzyme inhibitors using iterative protein crystallographic analysis. J Med Chem. 1991;34:1925–1928. doi: 10.1021/jm00111a001. [DOI] [PubMed] [Google Scholar]
- 2.Cameron, D. W., A. J. Japour, Y. Xu, A. Hsu, C. Cohen, C. Farthing, S. Follansbee, M. Markowitz, J. Mellors, D. Poretz, J. B. Angel, D. Ho, D. McMahon, V. Devanarayan, R. Rode, M. Salgo, D. Kempf, R. Granneman, J. M. Leonard, and E. Sun. Ritonavir and saquinavir combination therapy for the treatment of HIV infection. Submitted for publication. [DOI] [PubMed]
- 3.Chen Z, Li Y, Hall D, Chen E, Kuo L C. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J Biol Chem. 1995;270:21433–21436. doi: 10.1074/jbc.270.37.21433. [DOI] [PubMed] [Google Scholar]
- 4.Chong K-T, Pagano P J. In vitro combination of PNU-140690, a human immunodeficiency virus protease inhibitor, with ritonavir against ritonavir-sensitive and -resistant clinical isolates. Antimicrob Agents Chemother. 1997;41:2367–2373. doi: 10.1128/aac.41.11.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J M. HIV population in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 6.Collier A C, Coombs R, Schoenfeld D A, Bassett R L, Joseph Timpone M S, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe V J, Friedman H M, Merigan T C, Reichmann R C, Hooper C, Corey L. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. N Engl J Med. 1996;334:1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 7.Collins J R, Burt S K, Erickson J W. Flap opening in HIV-1 protease simulated by activated molecular dynamics. Nat Struct Biol. 1995;2:334–338. doi: 10.1038/nsb0495-334. [DOI] [PubMed] [Google Scholar]
- 8.Collins J R, Burt S K, Erickson J W. Activated dynamics of flap opening in HIV-1 protease. Adv Exp Med Biol. 1995;362:455–460. doi: 10.1007/978-1-4615-1871-6_59. [DOI] [PubMed] [Google Scholar]
- 9.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 11.Condra J H, Holder D J, Graham D J, Shivaprakash M, Laird D T, Schleif W A, Chodakewitz J A, Emini E A. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, Fla. 1997. Genotypic or phenotypic susceptibility testing may not predict clinical responses to indinavir, abstr. 47; p. 31. [Google Scholar]
- 12.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J the UNAIDS Network for HIV Isolation and Characterization. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:8209–8212. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelissen M, van den Burg R, Zorgdrager F, Lukashov V, Goudsmit J. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol. 1997;71:6348–6358. doi: 10.1128/jvi.71.9.6348-6358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig I C, Duncan I B, Roberts N A, Whittaker L. Abstracts of the 9th International Conference on AIDS, Berlin, Germany. 1993. Ro 31-8959, an inhibitor of HIV proteinase, appears relatively refractory to the generation of virus mutants with reduced sensitivity, abstr. PO-A26-0694; p. 243. [Google Scholar]
- 15.Crawford S, Goff S P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danner S A, Carr A, Leonard J M, Lehman L M, Guidiol F, Gonzales J, Raventos A, Rubio R, Bouza E, Pintado V, Aguado A G, De Lomas J G, Delgado R, Borleffs J C C, Hsu A, Valdes J M, Boucher C A B, Cooper D A. A short-term study of the safety, pharmacokinetics, and efficacy of ritonavir, an inhibitor of HIV-1 protease. N Engl J Med. 1995;333:1528–1533. doi: 10.1056/NEJM199512073332303. [DOI] [PubMed] [Google Scholar]
- 17.Darke P L, Nutt R F, Brady S F, Garsky V M, Ciccarone T M, Leu C T, Lumma P K, Freidinger R M, Veber D F, Sigal I S. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem Biophys Res Commun. 1988;156:297–303. doi: 10.1016/s0006-291x(88)80839-8. [DOI] [PubMed] [Google Scholar]
- 18.Debouck C, Navia M A, Fitzgerald P M D, McKeever B M, Leu C-T, Heimbach J C, Herber W K, Sigal I S, Darke P L, Springer J P. Human immunodeficiency virus protease expressed in Escherichia coli exhibits autoprocessing and specific maturation of the gag precursor. Proc Natl Acad Sci USA. 1987;84:8903–8906. doi: 10.1073/pnas.84.24.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Clerq E. HIV inhibitors targeted at the reverse transcriptase. AIDS Res Hum Retroviruses. 1992;8:119–134. doi: 10.1089/aid.1992.8.119. [DOI] [PubMed] [Google Scholar]
- 20.Diaz R S, Sabino E C, Mayer A, Mosley J W, Bush M P The Transfusion Safety Study Group. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J Virol. 1995;69:3273–3281. doi: 10.1128/jvi.69.6.3273-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreyer G B, Lambert D M, Meek T D, Carr T J, Tomaszek T A, Jr, Fernandez A V, Bartus H, Cacciavillani E, Hassell A M, Minnich M. Hydroxyethylene isostere inhibitors of human immunodeficiency virus-1 protease: structure-activity analysis using enzyme kinetics, X-ray crystallography, and infected T-cell assays. Biochemistry. 1992;31:6646–6659. doi: 10.1021/bi00144a004. [DOI] [PubMed] [Google Scholar]
- 23.Dreyer G B, Boehm J C, Chenera B, DesJarlais R L, Hassell A M, Meek T D, Tomaszek T A J, Lewis M. A symmetric inhibitor binds HIV-1 protease asymmetrically. Biochemistry. 1993;32:937–947. doi: 10.1021/bi00054a027. [DOI] [PubMed] [Google Scholar]
- 24.Dulioust A, Paulous S, Guillemot L, Bouè F, Galanaud P, Clavel F. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, Fla. 1997. Selection of saquinavir resistant mutants by indinavir following a switch from saquinavir, abstr. 16; p. 11. [Google Scholar]
- 25.Duncan I B, Jacobsen H, Owen S, Roberts N A. Abstracts of the 3rd Conference of Retroviruses and Opportunistic Infections, Washington, D.C. 1996. Reduced HIV sensitivity during treatment with proteinase inhibitor saquinavir, abstr. 155. [Google Scholar]
- 26.Eastman P S, Duncan I B, Gee C, Race E. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, Fla. 1997. Acquisition of genotypic mutations associated with reduced susceptibility to protease inhibitors during saquinavir monotherapy, abstr. 30; p. 19. [Google Scholar]
- 27.Eberle J, Bechowsky B, Rose D, Hauser U, vonder Helm K, Guertler L, Nitschko H. Resistance of HIV type 1 to proteinase inhibitor Ro 31-8959. AIDS Res Hum Retroviruses. 1995;11:671–676. doi: 10.1089/aid.1995.11.671. [DOI] [PubMed] [Google Scholar]
- 28.Emini E A, Schleif W A, Deutsch P, Condra J H. In vivo resistance of HIV-1 variants with reduced susceptibility to the protease inhibitor L-735,524 and related compounds. Antiviral Chemother. 1996;4:327–331. doi: 10.1007/978-1-4757-9209-6_30. [DOI] [PubMed] [Google Scholar]
- 29.Ermolieff J, Hong L, Lin X, Foundling S, Hartsuck J A, Tang J. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, Fla. 1997. Kinetic and structural basis of saquinavir resistance of HIV-1 protease mutants, abstr. 14; p. 9. [Google Scholar]
- 30.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulnik S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 32.Hertogs K, deBethune M-P, Miller V, Ivens T, Schel P, van Cauwenberge A, van den Eynde C, van Gerwen V, Azijn H, van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 34.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, Vander-Pol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 36.Hosur M V, Bhat T N, Kempf D J, Baldwin E T, Liu B, Gulnik S, Wideburg N E, Norbeck D W, Appelt K, Erickson J W. Influence of stereochemistry on activity and binding modes for C2 symmetry-based-inhibitors of HIV-1 protease. J Am Chem Soc. 1994;116:847–855. [Google Scholar]
- 37.Huff J R. HIV protease: a novel chemotherapeutic target for AIDS. J Med Chem. 1991;34:2305–2314. doi: 10.1021/jm00112a001. [DOI] [PubMed] [Google Scholar]
- 38.Ives K J, Jacobsen H, Galpin S A, Garaev M M, Dorrell L, Mous J, Bragman K, Weber J N. Emergence of resistant variants of HIV in vivo during monotherapy with the proteinase inhibitor saquinavir. J Antimicrob Chemother. 1997;39:771–779. doi: 10.1093/jac/39.6.771. [DOI] [PubMed] [Google Scholar]
- 39.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1998;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 40.Jacobsen H, Brun-Vezinet F, Duncan I, Hanggi M, Ott M, Vella S, Weber J, Mous J. Genotypic characterization of HIV-1 from patients after prolonged treatment with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobsen H, Yasargil K, Winslow D L, Craig J C, Kroehn A, Duncan I B, Mous J. Characterization of human immunodeficiency virus type 1 mutant with decreased sensitivity to proteinase inhibitor Ro 31-8959. Virology. 1995;206:527–534. doi: 10.1016/s0042-6822(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 42.Jacobsen H, Hangi M, Ott M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 43.Kaldor S W, Kalish V J, Davies J F, Shetty B V, Fritz J E, Appelt K, Burgess J A, Campanale K M, Chirgadze N Y, Clawson D K, Dressman B A, Hatch S D, Khalil D A, Kosa M B, Lubbehusen P P, Muesing M A, Patick A K, Reich S H, Su K S, Tatlock J H. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J Med Chem. 1997;40:3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan A H, Zack J A, Knigge M, Paul D A, Kempf D J, Norbeck D W, Swanstrom R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol. 1993;67:4050–4055. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh I, Yoshinaka Y, Rein A, Shibuya M, Odaka T, Oroszlan S. Murine leukemia virus maturation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology. 1985;145:280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- 46.Kim E E, Baker C T, Dwyer M D, Murcko M A, Rao B G, Tung R D, Navia M A. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J Am Chem Soc. 1995;117:1181–1182. [Google Scholar]
- 47.Kohl N E, Diehl R E, Rands E, Davis L J, Hanobik M G, Wolanski B, Dixon R A. Expression of active human immunodeficiency virus type 1 protease by noninfectious chimeric virus particles. J Virol. 1991;65:3007–3014. doi: 10.1128/jvi.65.6.3007-3014.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohl N E, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A, Scolnik E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotler M, Katz R A, Danho W, Leis J, Skalka A M. Synthetic peptides as substrates and inhibitors of a retroviral protease. Proc Natl Acad Sci USA. 1988;85:4185–4189. doi: 10.1073/pnas.85.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kramer R A, Schaber M D, Skalka A M, Ganguly K, Wong-Staal F, Reddy E P. HTLV-III gag protein is processed in yeast cells by the virus pol-protease. Science. 1986;231:1580–1584. doi: 10.1126/science.2420008. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda M J, El-Farrash M A, Cloudhury S, Harada S. Impaired infectivity of HIV-1 after a single point mutation in the pol gene to escape the effect of a protease inhibitor in vitro. Virology. 1995;210:212–216. doi: 10.1006/viro.1995.1334. [DOI] [PubMed] [Google Scholar]
- 52.Lal R, Hsu A, Granneman G R, El-Shoubargy T, Johnson M, Lam W, Manning L, Japour A Sun E. Abbott Laboratories. Abstracts of the 5th Conference on Retrovirus and Opportunistic Infections, Chicago, Ill. 1998. Multiple dose safety, tolerability and pharmacokinetics of ABT-378 in combination with ritonavir, abstr. 647; p. 201. [Google Scholar]
- 53.Lambert D M, Petteway S R, Jr, McDanal C E, Hart T K, Leary J J, Dreyer G B, Meek T D, Bugelski P J, Bolognesi D P, Metcalf B W, Matthews T J. Human immunodeficiency virus type 1 protease inhibitors irreversibly block infectivity of purified virions from chronically infected cells. Antimicrob Agents Chemother. 1992;36:982–988. doi: 10.1128/aac.36.5.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lillehoj E P, Salazar F H R, Mervis R J, Raum M G, Chan H W, Ahmad N, Venkatesan S. Purification and structural characterization of the putative gag-pol protease of human immunodeficiency virus. J Virol. 1988;62:3053–3058. doi: 10.1128/jvi.62.8.3053-3058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y, Lin X, Hong L, Foundling S, Heinrikson R L, Thaisrivongs S, Leelamanit W, Raterman D, Shah M, Dunn B M, Tang J. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type 1 protease: relationship to drug resistance. Biochemistry. 1995;34:1143–1152. doi: 10.1021/bi00004a007. [DOI] [PubMed] [Google Scholar]
- 56.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1996;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markowitz M, Saag M, Powderly W G, Hurley A M, Hsu A, Valdes J M, Henry D, Sattler F, La Marca A, Leonard J M, Ho D D. A preliminary study of ritonavir, an inhibitor of HIV-1 protease, to treat HIV-1 infection. N Engl J Med. 1995;333:1534–1539. doi: 10.1056/NEJM199512073332204. [DOI] [PubMed] [Google Scholar]
- 58.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maschera B, Darby G, Palú G, Wright L L, Tisdale M, Meyers R, Blair E D, Furfine E S. Human immunodeficiency virus. Mutations in the viral protease that confers resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J Biol Chem. 1996;271:33231–33235. doi: 10.1074/jbc.271.52.33231. [DOI] [PubMed] [Google Scholar]
- 60.Miller M, Schneider J, Sathyanarayana B K, Toth M V, Marshall G R, Clawson L, Selk L, Kent S B, Wlodawer A. Structure of complex of synthetic HIV-1 protease with a substrate base inhibitor at 2.3 Å resolution. Science. 1989;246:1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 61.Mo H, Markowitz M, Ho D D. Abstracts of the 2nd National Conference on Human Retroviruses and Related Infections, Washington, D.C. 1995. Pattern of specific mutations that confer resistance to a panel of protease inhibitors, abstr. 188; p. 89. [Google Scholar]
- 62.Molla A, Korneyeva M, Chernyavskiy T, Colgrove R, Chung P, Japour A, Mellors J, Xu Y, Rode R, Hsu A, Granneman G R, Kempf J, Leonard J the M96-462 Study Team. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, Fla. 1998. Characterization of HIV-1 protease mutations, compliance and drug concentrations in patients who have an HIV RNA rebound on ritonavir-saquinavir, abstr. 83; p. 54. [Google Scholar]
- 63.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H-M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A B, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 64.Molla, A. Personal communication.
- 65.Mous J, Brun-Vezinet F, Duncan I B, Haenggi M, Jacobsen H, Vella S. Abstracts of the 10th International Conference on AIDS, Yokohama, Japan. 1994. Characterization of in vivo selected HIV-1 variants with reduced sensitivity to proteinase inhibitor saquinavir, abstr. 515B; p. 90. [Google Scholar]
- 66.Murphy R, Degruttola V, Gulick R, D’Aquila R, Eron J, Sommadossi J P, Smeaton L, Currier J, Tung R, Kuritzkes D. Abstracts of the 5th Conference on Retrovirus and Opportunistic Infections, Chicago, Ill. 1998. 141W94 with or without zidovudine/3TC in patients with no prior protease inhibitor or 3TC therapy-ACTG 347, abstr. 512; p. 175. [Google Scholar]
- 67.Navia M A, Fitzgerald P M D, McKeever B M, Leu C-T, Heimbach J C, Herber W K, Sigal I S, Darke P L, Springer J P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989;337:615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- 68.Parkin N, Whitcomb J, Smith D, Tian T, Limoli K, Lie Y S, Winslow G, Wrin T, Capon D, Petropoulos C J. Program addendum of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Toronto, Canada: American Society for Microbiology; 1997. The use of a rapid phenotypic HIV-1 drug resistance and susceptibility assay in analyzing the emergence of drug-resistant virus during triple combination therapy, abstr. LB-1; p. 7. [Google Scholar]
- 69.Partaledis J A, Yamaguchi K, Tisdale M, Blair E E, Falcione C, Maschera B, Myers R E, Pazhanisamy S, Futer O, Cullinan A B, Stuver C M, Byrn R A, Livingston D J. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69:5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patick A K, Boritzki T J, Bloom L A. Activities of the human immunodeficiency virus type 1 (HIV-1) protease inhibitor nelfinavir mesylate in combination with reverse transcriptase and protease inhibitors against acute HIV-1 infection in vitro. Antimicrob Agents Chemother. 1997;41:2159–2164. doi: 10.1128/aac.41.10.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patick A K, Ho H, Markowitz M, Appelt K, Wu B, Musick L, Kaldor S, Reich S, Ho D, Webber S. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob Agents Chemother. 1996;40:292–297. doi: 10.1128/aac.40.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. Kuritzkes, D. D. Ho, and M. Markowitz. Genotypic analysis of HIV-1 variants isolated from patients treated with the protease inhibitor, nelfinavir. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 73.Pazhanisamy S, Stuver C M, Cullinan A B, Margolin N, Rao B G. Kinetic characterization of human immunodeficiency virus type 1 protease resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 74.Peng C, Ho B K, Chang T W, Chang N T. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pettit S C, Michael S F, Swanstrom R. The specificity of the HIV-1 protease. Perspect Drug Discov Des. 1993;1:69–83. [Google Scholar]
- 76.Poppe S M, Slade D E, Chong K-T, Hinshaw R R, Pagano P J, Markowitz M, Ho D D, Mo H, Gorman III R R, Dueweke T J, Thaisrivongs S, Tarpley W G. Antiviral activity of the dihydropyrone PNU-140690, a new nonpeptidic human immunodeficiency virus protease inhibitor. Antimicrob Agents Chemother. 1997;41:1058–1063. doi: 10.1128/aac.41.5.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richards A D, Roberts R, Dunn B M, Graves M C, Kay J. Effective blocking of HIV-1 proteinase activity by characteristic inhibitors of aspartic proteinases. FEBS Lett. 1989;247:113. doi: 10.1016/0014-5793(89)81251-7. [DOI] [PubMed] [Google Scholar]
- 78.Richman D D. HIV drug resistance. Annu Rev Pharmacol Toxicol. 1993;32:149–164. doi: 10.1146/annurev.pa.33.040193.001053. [DOI] [PubMed] [Google Scholar]
- 79.Roberts N A, Martin J A, Kinchington D, Broadhurst A V, Craig J C, Duncan I B, Galpin S A, Handa B K, Kay J, Kröhn A, Lambert R W, Merett J H, Mills J S, Parkes K E B, Redshaw S, Ritchie A J, Taylor D L, Thomas G J, Machin P J. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 80.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 81.Schechter I, Berger A. On the size of the active side in proteases. Biochem Biophys Res Commun. 1967;27:157. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 82.Schmit J-C, Ruiz L, Clotet B, Raventos A, Tor J, Leonard J, Desmyter J, De Clerq E, Vandamme A M. Resistance-related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538) AIDS. 1996;10:995–999. doi: 10.1097/00002030-199610090-00010. [DOI] [PubMed] [Google Scholar]
- 83.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 84.Shetty B V, Kosa M B, Khalil D A, Webber S. Preclinical pharmacokinetics and distribution to tissue of AG1343, an inhibitor of human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 1996;40:110–114. doi: 10.1128/aac.40.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.St. Clair M H S, Millard J, Rooney J, Tisdale M, Parry N, Sadler B M, Blum M R, Painter G. In vitro activity of 141W94 (VX-478) in combination with other antiretroviral agents. Antiviral Res. 1996;29:53–56. doi: 10.1016/0166-3542(95)00916-7. [DOI] [PubMed] [Google Scholar]
- 86.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Blair E D. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toh H, Ono M, Saigo K, Miyata T. Retroviral protease-like sequence in the yeast transposon Ty 1. Nature. 1985;315:691. [Google Scholar]
- 89.Tozser J, Gustchina A, Weber I T, Blaha I, Wondrak E M, Oroszlan S. Studies on the role of the S4 substrate binding site of HIV proteinases. FEBS Lett. 1991;279:356–360. doi: 10.1016/0014-5793(91)80186-7. [DOI] [PubMed] [Google Scholar]
- 90.Turriziani O, Antonelli G, Jacobsen H, Mous J, Riva E, Pistello M, Dianzani F. Identification of an amino acid substitution involved in the reduction of sensitivity of HIV-1 to an inhibitor of viral proteinase. Acta Virol. 1994;38:297–298. [PubMed] [Google Scholar]
- 91.Vacca J P, Dorsey B D, Schleif W A, Levin R B, McDaniel S L, Darke P L, Zugay J, Quintero J C, Blahy O M, Roth E, Sardana V V, Schlabach A J, Graham P I, Condra J H, Gotlib L, Holloway M K, Lin J, Chen I-W, Vastag K, Ostovich D, Anderson P S, Emini E A, Huff J R. L-735,524, an orally bioavailable human immunodeficiency virus type 1 protease inhibitor. Proc Natl Acad Sci USA. 1994;91:4096–4100. doi: 10.1073/pnas.91.9.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vella S, Galluzzo C, Giannini G, Pirillo M F, Duncan I, Jacobsen H, Andreoni M, Sarmati L, Ercoli L. Saquinavir/zidovudine combination in patients with advanced HIV infection and no prior antiretroviral therapy: CD4+ lymphocyte/plasma RNA changes, and emergence of HIV strains with reduced phenotypic sensitivity. Antiviral Res. 1996;29:91–93. doi: 10.1016/0166-3542(95)00926-4. [DOI] [PubMed] [Google Scholar]
- 93.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G S. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 94.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, Fla. 1997. Genotypic and phenotypic analysis of the protease gene in HIV-1 infected patients that failed long-term saquinavir therapy and switched to other protease inhibitors, abstr. 17; p. 11. [Google Scholar]
- 95.Wlodawer A, Erickson J W. Structure based inhibitors of HIV-1 protease. Annu Rev Biochem. 1993;62:543–580. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 96.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana B K, Baldwin E, Weber I T, Selk L M, Clawson L, Schneider J, Kent S B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]