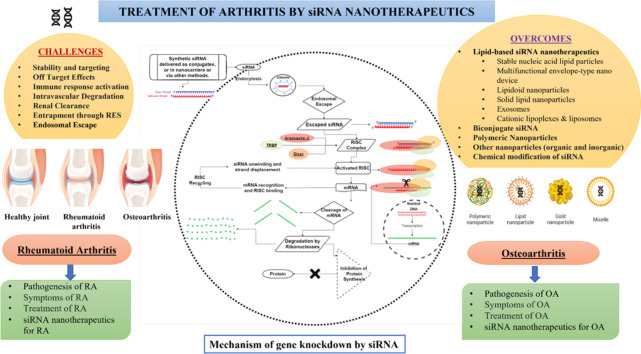
Abstract
RNA interference (RNAi) using small interfering RNA (siRNA) has shown potential as a therapeutic option for the treatment of arthritis by silencing specific genes. However, siRNA delivery faces several challenges, including stability, targeting, off-target effects, endosomal escape, immune response activation, intravascular degradation, and renal clearance. A variety of nanotherapeutics like lipidic nanoparticles, liposomes, polymeric nanoparticles, and solid lipid nanoparticles have been developed to improve siRNA cellular uptake, protect it from degradation, and enhance its therapeutic efficacy. Researchers are also investigating chemical modifications and bioconjugation to reduce its immunogenicity. This review discusses the potential of siRNA nanotherapeutics as a therapeutic option for various immune-mediated diseases, including rheumatoid arthritis, osteoarthritis, etc. siRNA nanotherapeutics have shown an upsurge of interest and the future looks promising for such interdisciplinary approach-based modalities that combine the principles of molecular biology, nanotechnology, and formulation sciences.
Keywords: RNAi, siRNA, Nanotherapeutics, Gene knockdown, Osteoarthritis, Rheumatoid arthritis
Graphical abstract
1. Introduction
Small interfering ribonucleic acid (siRNA) nanotherapeutics are promising therapeutic modalities that address the unmet medical needs not fulfilled by small molecules or antibodies in finding the cure for many deadly diseases. It is basically post-transcriptional gene silencing (PTGS) by delivering double-stranded RNA (dsRNA) molecules, viz. siRNA, microRNA (miRNA), short hairpin RNA (shRNA), etc. via carefully formulated nanocarriers (NCs) [1]. Through RNA-induced silencing, a wide range of genes may be targeted including splicing variants and undruggable proteins, making it extremely relevant for therapeutic usage in various types of diseases. The phenomenon is also known as RNA interference (RNAi) [2].
After 20 years of extensive research, there are some commercially available siRNA therapeutics which got approval by the US FDA (United States Food and Drug Administration). The first approved siRNA therapeutics was Onpattro (patisiran, 2018) for the treatment of polyneuropathy in patients suffering from familial transthyretin mediated amyloidosis [3]. This was followed by approval of Givlaari (givosiran, 2019) to treat acute hepatic porphyria [4,5] and Oxlumo (lumasiran, 2020) for treatment of primary hyperoxaluria type 1 (PH1) [6] and Amvuttra (vutrisiran, 2022) for the treatment of the rare disease hereditary transthyretin-mediated (hATTR) amyloidosis [5]. Alnylam Pharmaceuticals developed and marketed all these above listed siRNA therapeutics [7]. Leqvio (inclisiran, 2022) [8], was initially developed by Alnylam, but trialed and marketed by a subsidiary of Novartis. It is important to mention here that the above drugs’ mechanism and work are concentrated mainly on messenger RNA (mRNA) encoding in the liver and once the siRNA delivery limitations move past this, it can certainly have much wider applications. The most difficult challenge in siRNA therapeutics is getting siRNA delivered onto the target site. There are numerous siRNA delivery techniques, viz. lipidic NCs, chemical modification, dendrimers, lipidoids, exosomes, etc. Some of the major concerns in siRNA delivery are target site administration of the siRNA, prompt degradation by nucleases in the plasma, tissue and the cytoplasm, immune reactions, off-target effects and inability of RNA to cross the cellular membrane even after getting transported in the target tissue. Recent studies have used various methods to address the problems associated with target-specific delivery of siRNA. An interdisciplinary approach is needed to fully utilize the potential of siRNA. Formulation sciences help in the development of drug delivery vehicles for new treatments and medications. The utilization of nanotechnology principles in designing these drug delivery NCs, viz. solid lipid nanoparticles (SLNs), dendrimers, lipid nanoparticles, polymeric nanoparticles, lipidoids, etc. for siRNA delivery helped in the development of siRNA nanotherapeutics. Formulation of siRNA loaded NCs, conjugation of siRNA to a target ligand, development of ligand receptor pairs using cationic lipids as well as the combination of drugs and siRNA, chemical modification of siRNA followed by loading into the NCs are some of the strategies discussed herein [9].
Despite the significant progress made in utilizing nanomaterials for the treatment of arthritis, there are still gaps in our understanding of their therapeutic mechanisms and certain limitations hinder their widespread application. Currently, most research focuses on small molecules and monoclonal antibodies-based therapies working on symptomatic treatment. The potential strategy for treating arthritis could involve gene silencing by siRNA targeting specific pathways involved in the pathogenesis of rheumatoid arthritis (RA) and osteoarthritis (OA). This approach holds promise and may represent a new direction for the treatment of RA and OA in the future. This comprehensive review article highlights the current efforts for RNAi-mediated gene silencing using siRNA nanotherapeutics for the treatment of different types of arthritis. The technique's therapeutic potential and major challenges being faced by researchers in the delivery of siRNA are discussed. The recent research conducted on siRNA nanotherapeutics for RA and OA treatment have been exclusively reviewed and compiled for the readers.
2. siRNA mechanism of gene silencing
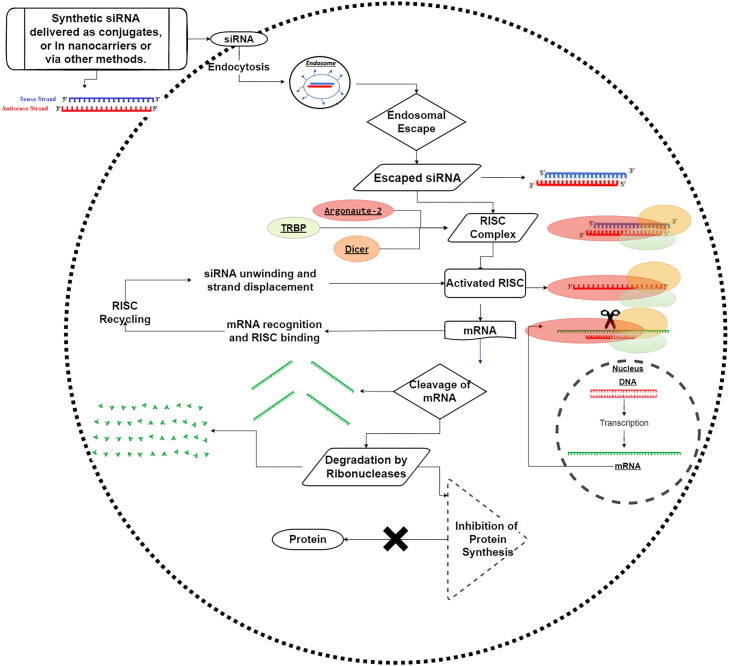
The siRNA is a double-stranded (ds) and non-coding RNA molecule. The ds siRNA molecule comprises of passenger strand and the guide strand [10]. The length of siRNA is approximately 7.5 nm and diameter 2 nm. Some of the properties of siRNA are listed in Table 1. siRNA is synthesized when the long dsRNA is cleaved with the help of introduction of ribonucleotide protein, also known as Dicer Enzymes (RNAse III endonuclease). This ds siRNA connects to a complex structure of proteins called RNA induced silencing complex (RISC). Further, Argonate-2 (Ago2) helps separate the passenger strand from the RISC-loading complex (RLC) followed by the connection of the guide strand to the target mRNA leading to cleavage into small mRNA fragments, thus silencing the gene expression. The gene silencing mechanism of siRNA is shown in Fig. 1. This ability of siRNA to silence genes has transformed or revolutionized the treatment of a variety of diseases [11].
Table 1.
Properties of siRNA.
| General Properties | Specific attribute |
|---|---|
| Primary mechanism of action | Translation Inhibition |
| Secondary mechanism of action | Cleavage of mRNA |
| Synthesis | Mostly Exogenous, Rarely Endogenous |
| Processing Enzymes | Dicer |
| Structure | 21–23 nucleotide RNA duplex with 2 nucleotide 3′ overhangs |
| Complementary | Fully complementary to mRNA |
| Synthesis Enzyme | RNA dependent-RNA Polymerase, Dicer |
| Gene regulation mechanism | Endonucleotide cleavage of mRNA |
| Function | Regulation of Protein-Coding Gene and Transposons |
| Clinical application | Therapeutic agent |
Fig. 1.
Mechanism of gene silencing or gene knockdown by siRNA.
3. Challenges in siRNA delivery
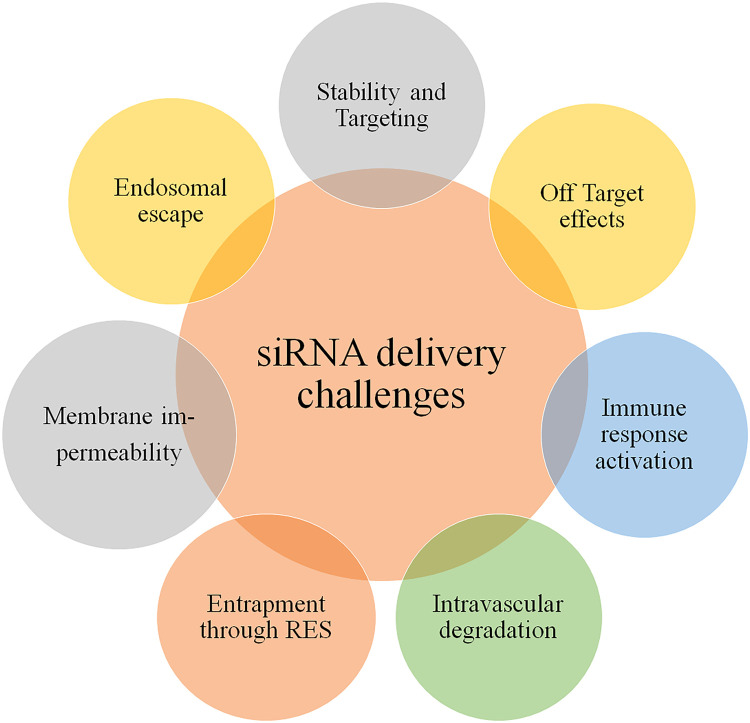
There are two main types of administration routes for delivery of siRNA, the first one is localized administration and the second one is systemic administration. In localized administration, local delivery of siRNA refers to the administration of siRNA directly to the target organ or tissues, offering several advantages, such as enhanced bioavailability at the desired site and bypassing systemic barriers. Whereas, the systemic administration of siRNA faces more challenges than localized delivery, when siRNA is administered intravenously, unmodified naked siRNA molecules are susceptible to degradation by endogenous enzymes [12]. Additionally, their small size (around 13 kDa) leads to rapid elimination through the kidneys. Moreover, due to their negatively charged nature, siRNA molecules face significant barriers in crossing biological membranes. As with every new advancement, there comes a new challenge related to its effective delivery. The siRNA delivery is also limited due to some major challenges as depicted in Fig. 2.
Fig. 2.
Challenges in siRNA delivery.
3.1. Stability and targeting of siRNA
siRNA is quickly degraded when it enters the cytoplasm, tissue as well as plasma. In serum, the naked siRNA half-life ranges from a couple of mins to 1 h [13]. Hence, accumulating target sites with quantities of therapeutical significance is a considerable challenge [14]. The small size of siRNA molecules [15] and the negative charge of siRNA stop them from entering the cell membrane, thus they can't accumulate intracellularly. Moreover, endocytosis-based siRNA delivery systems must account for endosomal escape. To prevent its destruction after entering the cell cytoplasm by intracellular RNA, siRNA must be efficiently identified and integrated into RISC [16]. To overcome these challenges, the stability and efficacy of siRNA complexes have been improved through the incorporation of locked nucleic acid modifications in their design. Studies have shown that sugar 2′-modifications are the most effective way to improve the effectiveness of siRNA. These modifications, which include 2′-O-methyl, 2′-fluoro, and 2′-methoxyethyl help improve the stability and binding affinity of siRNA duplexes. They also reduce the activation of the innate immune response [17]. These modifications have been successfully applied in the recently approved siRNA drug, Onpattro.
3.2. Off-Target effects
Apart from delivery issues, the RNAi paradigm of precise silencing showed practical problems. siRNA therapy can result in the silencing of genes that are not the desired gene targets, this is also known as off-target gene silencing [18]. Off-target gene silencing is undesirable as it may cause harmful gene expression mutations and unanticipated cell transformations. According to recent studies, homology with 6 to 7 nucleotides in the “seed region” of the siRNA sequence is what primarily causes off-target gene silencing. An even larger likelihood of a siRNA duplex matching unwanted targets might result from RISC's poor choice of guide strand over passenger strand. When creating siRNA-based treatments, off-target silencing cannot be disregarded, and all prospective siRNA sequences must undergo extensive testing. Researchers have turned to surface-ligand modifications for more specific targeted delivery. These modifications aim to overcome the off-target effects associated with the enhanced permeation and retention (EPR) effect, which is not significant enough and may potentially have harmful consequences in the long run. Recent advancements in siRNA formulation have incorporated antibody conjugation, chemical modifications, receptor-targeted delivery to improve targeting specificity and reduce off-target effects. Also, the use of antibody conjugation with siRNA (antibody-siRNA complex) has shown potential in reducing the off-target effects [19].
Further, the use of chemical modification with siRNA can also help inhibit off-target effects. Specifically, surface modifications with peptide ligands, such as cyclic arginyl-glycyl-aspartic acid (cRGD), facilitate the targeting of blood vessels which provides nutrients. cRGD, exhibits a strong preference for binding to avβ3 and avβ5 integrins found overexpressed in endothelial cells of angiogenic origin [20]. cRGD binds to integrin receptors with more affinity than linear RGD, making it a preferred option for surface modification. In a study utilizing functionalized chitosan nanoparticles that targeted PLXDC1 (a receptor abundantly expressed in tumor vasculature), the modified cRGD nanoparticles inhibited tumor growth by approximately 90%, exhibiting a 60% increase in efficacy than non-modified nanoparticles [21].
Finally, receptor targeted delivery focuses on targeting particular receptors that are articulated in the tissue type. By using GalNAc (N-acetyl galactosamine) conjugation and targeting the specific receptor, it confirms specific delivery to the desired tissue, diminishing the risk of unintentional effects on non-targeted tissues. Givosiran, an FDA-approved medication uses GalNAc conjugation to modify the 3′ terminus sense strand of siRNA. This conjugate binds to the asialoglycoprotein (ASGPR) receptor, via receptor binding with terminal galactose moieties and subsequent endocytosis [22]. In vivo studies have shown that this conjugate increases RNAi activity by approximately 5-fold. Targeting transthyretin (TTR) mRNA, considerable reduction of TTR mRNA levels has been reported in liver cells, specifically hepatocytes. Furthermore, the GalNAc conjugate with a phosphorothioate (PS) linkage modification improves protection against 5′-exonuclease degradation and requires meagre siRNA dose. Further investigations have suggested that both monovalent and tri-antennary GalNAc conjugations improve the binding affinity and gene silencing efficiency [23].
3.3. Immune response activation
The innate immune system provides a rapid, nonspecific response to protect from recognizing pathogens and eliminate them. As we have discussed earlier, siRNA (less than 30 nucleotides) avoids nonspecific stimulation of the interferon response by avoiding the immune system. However, further studies revealed that the production of cytokines by immune response elicited by siRNA both in vivo and in vitro [24]. This immune response can be triggered by the siRNA itself or by the vehicles used for in vivo siRNA delivery, such as cationic lipids [25]. Studies have shown that certain siRNA can stimulate the production of pro-inflammatory cytokines and interferon in immune cells through toll-like receptors (TLRs) and protein kinase R (PKR) pathways [26]. The immune response triggered by siRNA is often attributed to the presence of guanosine-cytosine (GC) rich sequences [27], which can activate various immune signaling pathways, including nuclear factor kappa B (NF-κB), interferon regulatory factors, and TLR-dependent or TLR-independent pathways (such as TLR7, TLR8 and TLR9). These pathways recognize siRNA as dsRNA and initiate inflammatory and antiviral responses. To avoid immune stimulation and enhance the therapeutic potential of siRNA-based treatments, various strategies have been explored as discussed below.
Chemical modifications of the ribose backbone have been employed to reduce the immune response in some studies. Modifying the 2′-OH group with 2′-H (2′-deoxy), 2′-F, or 2′-O-methyl (2′-O-Me) substitutions can alter the innate immune response and allow siRNA to evade immune detection while maintaining their intended activity [28]. Additionally, combining deoxyribonucleic acid (DNA) analogues, 2-F-modified RNAs and locked nucleic acids has shown promise in enhancing silencing efficiency while reducing immunostimulatory characteristics. Nucleobase structure modifications have also been investigated to mitigate the pro-inflammatory effects of siRNA delivery. Methylation of specific nitrogenous bases, such as 5-methyl-cytidine and 5-methyluridine, has demonstrated the potential to reduce the immunological response induced by siRNA [29]. By avoiding specific sequences, particularly those rich in uridine and guanosine, the immunological response can be reduced [30]. In the TLR-independent pathway, cytoplasmic RNA sensors like retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and PKR play crucial roles. RIG-I and MDA5 activate downstream signaling molecules that lead to the production of type I interferons and pro-inflammatory cytokines. PKR, on the other hand, is activated by binding to dsRNA, regardless of the sequence [31].
Therefore, inhibiting the TLR pathway or chemically modifying the siRNA duplex to prevent activation of the innate immune system can help mitigate unwanted immune responses. These approaches aim to enhance siRNA efficacy while minimizing immunostimulatory effects, thereby improving the therapeutic potential of siRNA-based treatments.
3.4. Intravascular degradation and renal clearance
The degradation of siRNA by plasma nuclease enzyme is the first biological obstacle after injection. The systemic circulation of naked siRNA is unstable and more sensitive to A-type nucleases which are found both extracellularly and intracellularly [32]. Additionally, siRNA has a relatively short half-life of 10 min to 1 h due to quick renal elimination [33]. The rapid breakdown of siRNA by nucleases presents in plasma or tissues within a short timeframe of mins to hours limits the potential applications of siRNA-based therapies [34]. This is due to the comparatively lesser molecular weight of siRNA, which is approximately 13 kDa and 7 nm in length, it is passed through the kidney easily [35]. Hence, the modification of siRNA, conjugation with polymer, siRNA-cationic polymer, and cationic comb-type copolymers (CCC) are needed to reduce its degradation via nucleases, improves the in vivo characteristics and encourage appropriate therapeutic action.
The modifications of sugar backbone oligoribonucleotide bases to enhance stability, prevent degradation in blood vessels and increased resistance of enzymatic degradation have been reported in the literature [36]. For example, base modification replacement of uridine with rF (2,4-difluorotoluyl ribonucleoside) substitutions has been found to improve resistance against degradation by nucleases present in the serum [37]. Modification of sugar, interchange of CH3 group or fluoride atom in the place of ribose 2′-OH group in both the sense and antisense strands of siRNA has been shown to enhance their resistance against endonucleases. This modification not only improves the stability of siRNA molecules but also enhances their therapeutic potency [38]. In the case of modification of backbone, replacing PS with morpholino oligomers has shown a more potent and longer half-life of siRNA duplex [39].
When siRNA was conjugated with cholesterol, its half-life increased to 90 min and plasma retention was increased owing to plasma protein binding [40]. This cholesterol-siRNA conjugation also increased nuclease resistance, improved serum stability and circulation time [41]. However, naked siRNA doesn't demonstrate any silencing activity within the cell. The addition of cholesterol to siRNA not only improves its stability and circulation but also facilitates its intracellular delivery and gene silencing efficacy [42]. On the other hand, conjugation with polyethylene glycol (PEG) polymer also increases the half-life of siRNA with improved pharmacokinetics profiles [43].
One more approach is siRNA cationic polymer, which has retained more than a 100-fold increase in concentration in blood as compared to naked siRNA. Besides, cationic CCC was found to improve siRNA stability against nucleases and plasma components. Complexing siRNA with CCC has a prolonged duration in blood circulation compared to naked siRNA or siRNA complexed with polyethyleneimine (PEI) modification [44].
3.5. Entrapment through reticuloendothelial system (RES)
RES poses the biggest threat to nucleic acid therapies due to phagocytosis [45]. In RES, the macrophages quickly remove siRNA-loaded nanoparticles that are being opsonized [2]. Once in circulation, siRNA therapies must be shielded from the mononuclear phagocyte system (MPS) phagocytic cells [46]. So, the siRNA nanoparticles in bloodstream get transported very quickly to RES organs. The sluggish processing and removal of these carriers, however, causes its prolonged retention in the organs. In cases where the target organ is one that is rich in RES [12], the process of RES filtration specifically based on siRNA treatments might be beneficial. RES absorption and biodistribution may be inhibited by a variety of elements, including carrier size, charge, and surface characteristics. Theoretical considerations suggest that carriers possessing a negative charge are more prone to elimination from the bloodstream compared to carriers with a positive or neutral charge [12].
The NCs for siRNA delivery can be tailored with hydrophilic polymers like PEG, polyethylene oxide and poloxamers to give them stealth properties, prolonging their presence in the bloodstream. This modification prevents protein adsorption, opsonization and ensures cargo stability. The stealth properties are most effective for carriers with sizes less than 200 nm. However, excessive PEGylation can compromise the carrier's ability to facilitate siRNA uptake into cells by neutralizing its positive surface charge [12]. Therefore, a specific PEGylation strategy is required to maintain both evasion of the RES and efficient cellular uptake of siRNA. Despite these advancements, further research is needed to optimize the size and molecular size of PEG for designing an ideal siRNA delivery system. Understanding these parameters will enhance the development of efficient and targeted siRNA therapies.
3.6. Impermeability of membrane
siRNA is unable to cross through the cell owing to its negative charge, size and excessive hydrophilic nature. As a result, to overcome this obstacle, effective delivery of siRNA requires modification. The siRNA, net negative charge can be hidden by complexing it with cationic polymers or lipids [47]. Additionally, these negatively charged cellular membranes and positively charged nanoparticles interact to cause internalization [48]. Alternative methods for effectively delivering siRNA include combining it with aptamers, ligands or immunoglobulins that recognize specific target cell antigens. These conjugates bind to the cells, where siRNA is picked up through endocytosis mediated by receptors. This process results in the creation of endosomes and subsequent transport of siRNA in the cytoplasm. Numerous carriers are utilized, including cationic polymers and peptides, because of the hydrophilic and negatively charged nature of siRNA. Due to the variety of their physiochemical characteristics and activities, peptides have drawn interest as they exhibit tremendous potential as siRNA carriers. Cell-penetrating peptides (CPPs) [49], non-covalent multifunctional peptide complexes and endosome-disrupting peptides are few kinds of peptides that can be employed depending on the purpose.
3.7. Endosomal escape
A significant barrier still exists after the siRNA has been internalized because it cannot escape endosomes. Therefore, for effective endosomal escape and siRNA gene silencing, a carrier or alteration that permits endosomal membrane breakdown is required. The transition from the extracellular to the endosomal milieu has been exploited by many modern delivery systems. The ability to traverse from the extracellular to the endosomal environment has facilitated the development of numerous delivery systems in recent times. Due to their large buffering capacity, it has been demonstrated that protonable cationic polymers may enhance the delivery efficiency of siRNA [50]. To evade the endosomal trap, siRNA carrier formulations can also contain versatile endosomolytic agents, viz. polymers, proteins, peptides, and small compounds like chloroquine [51]. These carriers elevate the concentrations of counter-ions, leading to osmotic swelling, endosomal membrane breach and subsequent release of siRNA into the cytosol [52].
Different approaches have been utilized to promote the efficient release of siRNA from endosomes, enhancing siRNA delivery systems. PEI acts as a “proton sponge,” facilitating siRNA release by inducing chloride ion influx and endosomal disruption. However, its cytotoxicity increases with size [53]. Poly (lactic-co-glycolic acid) (PLGA), a biodegradable copolymer, improves siRNA activity, provides sustained release and enhances cellular uptake. Combining PEI and PLGA in layer-by-layer complexes enhances endosomal escape [54]. Protonation of amino acids like arginine and lysine destabilizes cellular membranes, aiding siRNA release into the cytoplasm [55]. A PEG-based cleavable polymeric system, comprising lipid nanoparticles stabilized by PEG-conjugated vinyl ether lipids, allows for endosomal fusion and siRNA release in the acidic environment. Incorporating fusogenic molecules, such as dioleoylphosphatidylethanolamine (DOPE) [56] and fusogenic protein-based carriers like Hemagglutinin 2 (HA2 and glutamic-acid-alanine-leucine-alanine (GALA) peptides, promote endosomal escape by destabilizing the endosomal membrane. These approaches contribute to the development of promising platforms for siRNA delivery by improving endosomal release and enhancing siRNA therapeutic efficacy [57]. Some of the extracellular and intracellular barriers of siRNA effective delivery are depicted in Fig. 3 [58]. Hence, the delivery of siRNA has several difficulties, that need to be taken care of, while designing siRNA nanotherapeutics for the treatment of diseases.
Fig. 3.
Types of barriers for siRNA delivery.
4. Overcoming challenges of siRNA delivery using nanotherapeutics
The important attributes that are prerequisites for an effective siRNA delivery system are its non-immunogenicity, biocompatibility and degradable nature. For siRNA to be properly delivered to target cells or tissues, the delivery techniques should also protect active ds siRNA molecules against assault by serum nucleases. After systemic administration, specificity in target tissue must be ensured by the delivery techniques to prevent fast hepatic or renal clearance. Once siRNA is internalized into target cells through endocytosis, the delivery vehicle should facilitate its release from endosomes into the cytoplasm. This will allow siRNA and the endogenous RISC to interact [59]. Hence, there is a need to design effective NCs or delivery vehicles for siRNA that are non-viral to achieve cellular, tissue bioavailability and stability. Also, siRNA nanotherapeutics have been successfully developed by researchers in the last two decades. Some of these approaches to designing effective NCs for siRNA delivery are described below.
4.1. Lipid-based siRNA nanotherapeutics
Lipid-based nanoparticles (LNPs) possess significant potential for siRNA delivery owing to biocompatibility with limited toxicity as compared to their other counterparts like inorganic and synthetic nanoparticles. Because of their better pharmacokinetic profiles, high transfection efficiency into the mammalian cells and electrostatic interaction with nucleic acids, cationic lipids have become appealing raw material for siRNA delivery vehicles. The structure of lipid-based siRNA nanotherapeutics is depicted in Fig. 4.
Fig. 4.
Structure of lipid-based siRNA nanotherapeutics.
4.1.1. Stable nucleic acid lipid particles (SNALPs)
SNALPs comprise of an ionizable lipid of 1,2-dilinoleyloxy-N, N-dimethyl-3-aminopropane (DLin-DMA). It is an ether analog of ionizable lipid having oleic acid chains, viz. 1,2-dioleoyl-3- (N, N-dimethylamino) propane (DODAP). It has been reported that ionizable lipids with linoleic acid chains, such as DLin-DMA, exhibit superior siRNA initiation vis oleic acid chains like in DODAP [60,61]. Further studies revealed that the linker moiety and head of ionizable lipid in LNP fabricated with 2,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane (DLin-KC2-DMA) demonstrated better in vivo results. Further, the addition of 1,2-distearoyl-sn‑glycero-3-phosphocholine (DSPC), cholesterol PEG2000 derivatives are helper lipids that improve the stability of LNPs. Generally, the LNPs for nucleic acid delivery are fabricated by microfluidic mixing of lipids in ethanol and siRNA in citrate buffer. This was followed by dialysis to remove ethanol. This technique has shown optimum particle size, high efficiency of siRNA encapsulation and mass manufacture. The structure of LNPs reveals reverse micelles of a hydrophobic core made of DLin-KC2-DMA. The Cryogenic transmission electron microscopy (cryo-TEM) and small-angle X-ray techniques have shown that at pH 4.0, the siRNA/DLin-KC2-DMA complex is sandwiched within the lipid bilayer [62,63]. However, as the pH approaches neutral, the ionizable lipids form an amorphous oil phase in the middle of the LNP [64].
Cationic lipids of the 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) type are frequently employed in laboratories and can be purchased commercially. A liposomal siRNA/DOTAP/Cholesterol platform was developed by Kim et al. in 2007 for siRNA targeting hepatitis B virus (HBV) delivery to the liver [65]. Yagi et al. demonstrated the use of cationic DOTAP, egg phosphatidylcholine and PEG lipid in a 24:14.8 wt ratio to form a siRNA delivery complex [66]. For in vivo delivery and testing in several disease models, SNALPs have been produced by various researchers across the globe. For instance, a lipid bilayer composed of cationic lipids along with fusogenic acid contains siRNA in the center of SNALPs. To boost hydrophilicity and stability in the serum, PEG is applied to the surface of the SNALP. Compared to unformulated siRNA, siRNA-SNALP complex has a substantially longer half-life. Using a mouse replication model for HBV, an HBV targeted siRNA-SNALP (dose-3 mg/kg/d) caused a selective reduction in HBV mRNA [67]. Additionally, a single systemic dosage (2.5 mg/kg) of an Apolipoprotein B (ApoB) specific siRNA loaded in SNALPs demonstrated more than 90% silencing impact of ApoB mRNA in cynomolgus monkey's liver [61].
4.1.2. Multifunctional envelope-type nano device system for siRNA delivery
The idea of a multifunctional envelope-type nano-device (MEND) for nucleic acid delivery was developed to overcome different delivery barriers by incorporating functional moieties in one nanoparticle [68]. MEND consists of a lipid bilayer modified with peptides, encapsulating siRNA in its inner phase with diverse functions. One notable modification of MEND is the incorporation of octaarginine (R8), enabling cellular uptake through macropinocytosis and efficient release of nucleic acids into the cytoplasm [69]. The integration of fusible or pH-responsive lipids into the lipid membranes help in improving the intracellular dynamics. Harashima's group developed pH-responsive ionizable first-generation lipid (YSK05), second-generation (YSK13) and third generation lipid (CL4H6). YSK05, resembles the structure of DODAP, contains a tertiary amine, unsaturated fatty acid chains, and a pKa of 6.4, YSK05-MEND exhibits high membrane fusion and gene knockdown activity [70]. YSK13-MEND, demonstrated over a 4-fold decrease in the ED50 (0.015 mg/kg) for knockdown of blood-clotting factor VII (FVII) compared to YSK05-MEND [71]. Through systematic studies on ionizable lipid structures, CL4H6 was developed with a pKa of 6.25. The head group structure of ionizable lipids was identified as a primary determinant of pKa, crucial for the distribution and endosomal escape of LNPs. Interestingly, the hydrophobic tail structure was found to have no significant effect on the apparent pKa. Furthermore, intravenous injection of CL4H6 LNPs in mice resulted in an ED50 of 0.0025 mg/kg for FVII knockdown, demonstrating a substantial improvement in gene-silencing efficiency through the systematic study of ionizable lipid structure [72].
4.1.3. Lipidoid nanoparticles
The delivery of siRNA has also been studied using lipidoid nanoparticles composed of molecules resembling lipids. The flexibility of lipidoid structure allows for improved in vivo kinetics, effectiveness, and safety of lipidoids [73]. Lipidoids can be screened to see how changes in their partial structure affect their properties as lipidoids with minimal experimentation. Over the past decade, Anderson's group has conducted three screening studies on lipidoids for siRNA delivery. Each study evaluated the effectiveness of nanoparticles in achieving FVII knockdown via intravenous injection in mice. Love et al. developed C12–200 lipidoid nanoparticles, with an ED50 (0.01 mg/kg) of siRNA [74]. Whitehead and his team focused on enhancing biocompatibility and identified 304O13 nanoparticles, achieving gene knockdown with an ED50 (0.01 mg/kg) without severe cytokine induction or inflammation even at high siRNA doses (1 mg/kg) [75]. Dong et al. screened a peptide-based lipidoid library and discovered cKK-E12 nanoparticles with an impressive ED50 (0.002 mg/kg) for FVII knockdown, surpassing DLin-MC3-DMA-LNP [76].
During the screenings, lipidoid nanoparticles composition includes lipidoid, DSPC, cholesterol and DMG-mPEG2000 at specific molar ratios, with particle size of less than 90 nm. Like LNPs, it is found that lipidoid nanoparticles are coated with ApoE in the bloodstream and are taken up by hepatocytes through hepatic low-density lipoprotein (LDL) receptors. The small particle size likely facilitates their accumulation in the liver by passing through fenestrae in hepatic vessels. However, the exact delivery mechanism is not fully understood. Recent studies have traversed the use of lipidoid nanoparticles for siRNA delivery into the inflammation sites [77] and oral administration for intestinal diseases [78], expanding their potential applications beyond liver targeting in siRNA therapeutics [79].
4.1.4. Solid lipid nanoparticles (SLNs)
SLNs are composed of solid lipids gaining significant attention in drug delivery for therapeutic and cosmetic applications due to their high biocompatibility. SLNs have a solid lipid core surrounded by a lipid membrane, they can encapsulate lipophilic drugs and facilitate sustained drug release [80]. SLNs have been extensively studied for siRNA delivery. One approach involves incorporating cationic lipids into SLNs, forming electrostatic complexes with siRNA. These siRNA complexed SLNs have shown potential for treating cancer and liver diseases. Another method is the hydrophobic ion-pairing (HIP) technology to incorporate siRNA into the hydrophobic SLN core via ionic complexes with cationic lipids [81]. For example, siRNA/DOTAP encapsulated into a triolein core, followed by the addition of phosphatidylcholine and PEGylated lipids. Studies have shown that siRNA can be released from SLNs for up to 10 d in mice, with effective gene-silencing capabilities in vitro. Moreover, SLNs offer promising prospects for siRNA delivery, providing sustained release and efficient gene-silencing capabilities in various applications [82].
4.1.5. Exosomes
Exosomes are natural endogenous vesicles capable of carrying nucleic acids and proteins, have emerged as safe, efficient and promising siRNA delivery carriers [83,84]. Exosomes have shown tissue-specific accumulation and surface molecule variations depending on the cell type that produces them. To reduce safety concerns and immunogenicity, it is preferable to use exosomes produced from the recipient's own cells when delivering siRNA. Although exosomes are native siRNA delivery carriers, some studies have successfully encapsulated siRNA within exosomes obtained from human serum via electroporation. These exosomes have effectively delivered siRNA into human monocytes and lymphocytes, demonstrating the use of recipient-derived exosomes as siRNA carriers [85].
Alvarez-Erviti et al. reported the use of modified dendritic cells (DCs) to produce exosomes containing the rabies viral glycoprotein (RVG) peptide, which enables neuronal cell targeting. siRNA encapsulated in these RVG peptide-expressing exosomes successfully silenced the BACE1 gene in the brain, exposing their potential to cross the blood-brain barrier (BBB) and deliver therapeutic cargo to neuronal cells [86]. The exosome database ExoCarta (www.exocarta.org) provides details on the proteins, mRNAs, miRNAs, and lipids that make up exosomes [87]. Exosome investigations should lead to the development of artificial exosomes or exosome mimics that can carry siRNA to targeted tissues.
4.1.6. Cationic lipoplexes & liposomes
Cationic lipoplexes and liposomes have been extensively studied as transfection agents among all lipid-based systems due to their potential to show charge-based interactions with the cell membrane [88]. In the context of siRNA distribution, various cationic transfection agents such as Lipofectamine 2000, DOTAP, Lipofectamine RNAiMAX, HiPerfect and Cardiolipin analogues, have been investigated [89]. However, upon administration to mice, these carriers have demonstrated dose-dependent toxicity along with an increase in inflammation. The study showed that Lipofectamine, DOTAP and nonionic/anionic liposomes exhibited harmful effects in decreasing order [90].
Numerous formulation characteristics of liposomes enhance the effectiveness of siRNA delivery. There are several instances in the literature that demonstrate the use of both neutral and cationic lipids in creating siRNA delivery systems. The structure of cationic head, tail length of lipid and the type of linker, all have an impact on the transfection ability and toxicity associated with siRNA encapsulated by cationic liposomes. The development of such systems facilitates the endocytic uptake of siRNA enabling its intracellular release at the targeted site. It has been reported that the nitrogen/phosphorous molar ratio and concentration of nucleic acid are critical attributes to fabricate safe lipoplexes. Further, it has been reported that DOTAP or DC—Cholesterol associated with DOPE and complexation with PEG were developed as safe and effective lipoplexes [91].
4.2. Bioconjugate siRNA
Another strategy to improve the effectiveness of siRNA in vivo is using biomolecules that can be chemically modified or incorporated into nanoparticles for covalent coupling with siRNA. Various biomolecules, such as dendrimers, peptides, lipophilic molecules, ligands, cholesterol, aptamers, antibodies, and biopolymers can be conjugated to siRNA to improve its delivery. The cell targeting, cell-penetrating peptides or biofunctional peptides can be covalently linked to siRNA [92]. Covalently attaching drugs to the peripheral groups of PAMAM (poly amidoamine) dendrimers have been utilized to improve the effectiveness and solubility of therapeutics, minimize non-specific toxicity, and achieve a controlled and prolonged release of the drug [93]. The controlled modification of poly-amidoamine dendrimer surfaces with targeting ligands reduces cytotoxicity, improves transfection efficiency, and enhances targeting capability [94]. In a study by Choi et al. a self-crosslinked fusogenic KALA peptide and a cell-penetrating peptide, Hph1 were functionalized with siRNA conjugated with branching PEG to suppress gene expression in MDA-MB-435 in cell lines using a polyelectrolyte complex micelle [95]. Intratracheally administered siRNA against the p38 mitogen-activated protein kinase (MAPK) and the HIV TAT cell-penetrating peptide were used to try and inhibit innate immune responses in the lung [96].
When administered systemically, cholesterol-conjugated siRNA enhances intracellular activity and facilitates cellular uptake through LDL receptor-mediated endocytosis, taking advantage of the natural uptake of cholesterol by hepatocytes and lipoproteins. Target-specific siRNA conjugated with cholesterol has shown improved serum stability and increased suppression of liver apoB mRNA levels for apoB [97].
Antibodies can be directly linked to siRNA for targeted delivery to specific cell types or tissues. Antibody-based therapeutics such as pertuzumab, cetuximab and trastuzumab have been routinely administered with excellent results. Trastuzumab emtansine (T-DM1), an antibody-drug conjugate combining trastuzumab with the anti-microtubule agent DM1, is a notable example of antibody conjugation. Antibodies attached to small molecules of chemotherapy have also shown therapeutic effects [98]. Some laboratory and in vivo studies demonstrated superior siRNA targeting with KRAS2 (Kristen rat sarcoma viral oncogene homolog or alternatively Kristen murine sarcoma virus2 homolog) and an anti-EGFR (epidermal growth factor receptor) antibody [98,99].
Aptamers, nucleic acid-based targeting agents, can also be coupled with siRNA. These synthetic single-stranded (ss) DNA/RNA ligands are designed to bind to specific targets selectively and effectively [100]. Aptamers can be employed to deliver siRNA/shRNA molecules that specifically target retineic-acid-receptor-related orphan nuclear receptor gamma (RORγt). By replacing the targeted genes, such as GATA3, Tbet and signal transducer and activator of transcription (STAT) 3, with siRNA/shRNA, the CD4 aptamer offers a flexible approach to introduce gene silencing molecules into CD4+ T cells. This enables the manipulation of various subsets of Th cells. To assess the potential advantages in autoimmune diseases, additional animal and clinical trials involving CD4-AshR-RORγt chimeras are needed [101]. Aptamers targeting transmembrane receptors have gained attention as active targeting moieties, like siRNA. Hence, these strategies involving biomolecule conjugation with siRNA hold promise for improving siRNA delivery and enhancing its therapeutic efficacy.
4.3. Polymeric nanoparticles
To overcome the limitations of nucleic acid formulations, siRNA is often incorporated into nanoparticles, which offer improved serum stability, better distribution, and controlled release. Natural biodegradable nanoparticles have shown significant therapeutic effects. There are two types of polymers commonly used in siRNA delivery research, viz. natural polymers, and synthetic polymers. Examples of natural polymers include albumin, chitosan, cyclodextrin, gelatin, atelocollagen, etc. [102,103]. Synthetic polymers extensively studied for siRNA delivery include PLGA, PEI and PEG [104].
Cyclodextrin polymer (CDP) was the first nanoparticle delivery technology used in clinical trials for siRNA, where it was utilized as the basic material. CDP is derived from the breakdown of cellulose by bacteria. It is a polycationic oligosaccharide used by pharmaceutical companies to deliver small compounds [105]. Self-assembled CPD containing human transferrin (Tf) and PEG, referred to as CALAA-01, has shown better capacity to target cells from siRNA delivery formulation [106]. This method was then put to the test in a clinical phase experiment. Chitosan-based systems are another type of polymeric nanoparticles commonly used in nanomedicine. Chitosan is a polysaccharide derived from chitin, composed of N-acetyl-D-glucosamine (deacetylated unit) and – (1–4)-linked D-glucosamine (acetylated unit). Due to their positive charge, chitosan nanoparticles can easily interact with negatively charged siRNA through electrostatic interactions, facilitating the formulation process [107]. When atelocollagen/siRNA complexes were injected intratumorally, tumor development of an orthotopic xenograft cancer model was inhibited [108]. Natural polymers are advantageous for siRNA administration because they are biodegradable, have little immunological stimulation, and are simple to condense with nucleic acids. Despite having higher transfection efficiency, macromolecules containing cationic lipids may still cause cytotoxicity and an immunological response. Its adaptability for loading siRNA, biological compatibility, and the combined administration of small molecule treatments with siRNA, is due to the polymer's ability to change a chemical bond [109]. Several natural polymers, including gelatin, chitosan, sodium alginate, albumin and lectins, are employed for the delivery of therapeutic siRNA [110].
Poly(Ɛ-caprolactone), PLGA, polycaprolactone and polyglutamic acid are examples of synthetic polymers used for the transport of siRNA [111]. Polymeric NCs have shown to be more adaptable to surface and chemical alterations, integration of small functionalities is one of the unique features of polymeric nanoparticles for the effective delivery of siRNA.
4.4. Other nanoparticle delivery systems for therapeutic siRNA
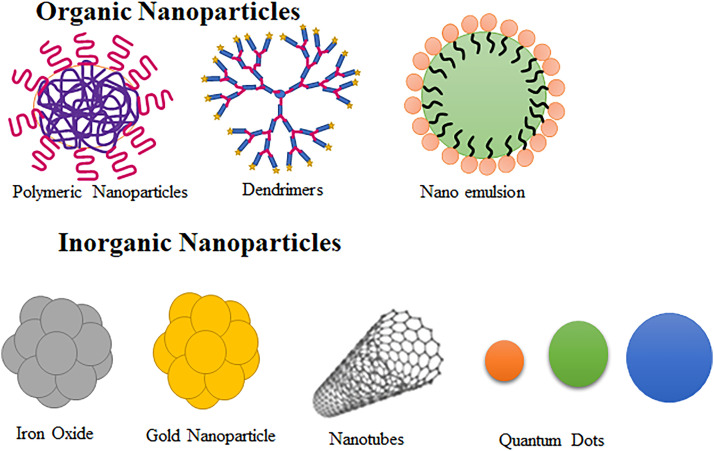
The two main categories of nanoparticles used for siRNA delivery are organic or soft nanoparticles and inorganic or hard nanoparticles. In the case of hard or inorganic nanoparticles, they are often coated with polymers to enhance their solubility.
4.4.1. Organic or soft nanoparticles
Organic nanoparticles are made from natural organic materials or synthetic organic materials such as self-aggregating surfactants or polymers. Common examples of organic nanoparticles include dendrimers, polymer nanoparticles, nanoemulsions and liposomes. Liposomes, composed of organic lipid molecules in a bilayer structure, can have various charges. Although neutral liposomes are commonly preferred, their limited entrapment efficiency is a drawback. To address this, the zwitterionic compound 1,2-dioleoyl-sn‑glycero-3-phosphatidylcholine (DOPC) is often used in the preparation of liposomes [46]. The resemblance of liposomes to natural biological membranes is the unique advantage of this NC making it the most widely used biocompatible NC. The liposome like particles can carry both hydrophilic and lipophilic drugs and are ideal NCs for siRNA delivery.
4.4.2. Inorganic or hard nanoparticles
These nanoparticles fall into the category of non-biodegradable, bio-persistent inorganic and insoluble nanoparticles. They include metals and their oxides, carbon-based compounds such as nanotubes, fullerenes, and fibers [112]. Magnetic nanoparticles, specifically super paramagnetic iron oxide nanoparticles (SPION), are a type of inorganic nanoparticle [113] and gold nanoparticles are adaptable enough to be employed as both medicinal agents and delivery systems. It has been observed that the magnetic nanomaterials relevant for nucleic acid delivery are either positively charged particles alone or the formulations including positively charged lipids or polymers containing both positively and negatively charged particles. Quantum dots are a relatively new type of nanocrystal made of colloidal semiconductors, known for their exceptional electrical and optical properties, making them efficient carriers for siRNA. Nanodiamonds (NDs) are among the most advanced delivery technologies being investigated for siRNA therapies. In a study, a fluorescent cationic coated ND vector was synthesized for siRNA delivery into Ewing sarcoma cells in culture. The coatings were done using polyallylamine hydrochloride (PAH) and PEI. This vector depicted larger efficiency in inhibiting the gene expression, and lesser toxicity. A larger adsorption affinity of siRNA was observed with PAH-coated NDs and ND-PEI carriers exhibit lesser toxicity. Also, slower dissociation of the siRNA:ND-PAH complex than of the siRNA:ND-PEI ones and hence a lower siRNA-associated biological activity [114]. Carbon nanotubes, particularly those containing nanoneedles, are extensively researched due to their potential to induce cell death [46]. Fig. 5 depicts the structure of organic and inorganic nanoparticles.
Fig. 5.
Structure of organic and inorganic nanoparticles for siRNA delivery.
4.5. Chemical modification of siRNA
The modification geometries of siRNA have been attempted to increase its stability for sustained circulation in vivo. Several modifications have been applied to different locations within the siRNA duplex to enhance nuclease tolerance. A popular technique is substituting the phosphodiester (PO4) group with a PS group at the 3′ end [115]. Additional modifications such as adding a fluoro group (2′-F), O-methyl group (2′-O-Me), or 2-methoxyethyl group (2′-O-MOE) have extended RNAi effects and half-lives in cultured cells and plasma [16]. Other methods include using 2′,5′-phosphodiester links, a 2′-O-alkyl alteration combined with 4′-thiolation and the locked nucleic acid substitution with the 2′- and 4′-positions connected by a methylene bond [116]. Small compounds like 2,4-dinitrophenol (DNP) have also been used to modify siRNA, increasing its nuclease resistance and membrane permeability.
Chemically modified siRNA demonstrated in vivo absorption and targeted downregulation of endogenous proteins [42]. However, the breakdown of synthetic compounds used in the modifications can lead to the production of toxic metabolites. Concerns such as off-target effects, decreased RNAi activity and decreased therapeutic index may arise from chemical modifications [117]. Additionally, the breakdown of a modified siRNA into non-naturally occurring chemicals in the body raises safety concerns regarding the potential toxicity of these metabolites.
5. Potential of siRNA delivery in the treatment of arthritis
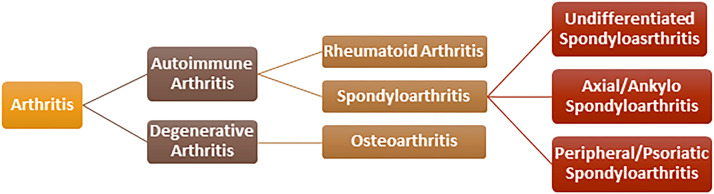
The word “arthritis” comes from the Greek language, meaning “disease of the joints”. It is generally characterized by joint inflammation (acute or persistent) with severe pain and structural damage to bone and cartilage degradation. The most noticeable signs include stiffness, pain, reduced range of motion and joint abnormalities. More than 100 different forms of arthritis have been identified like OA, RA, ankylosing spondylitis, psoriatic arthritis, etc. The classification of arthritis is depicted in Fig. 6. This review article is mainly focused on siRNA therapeutics under the research and development pipeline in OA and RA.
Fig. 6.
Classification of arthritis.
5.1. Rheumatoid arthritis
RA is a chronic systemic inflammatory disease of autoimmune origin characterized by inflammation of the synovial tissue, leading to stiffness, swelling and pain in the joints that ultimately leads to cartilage damage. There are multiple factors involved, viz. smoking, genetics, environmental factors, etc. that increase the predisposition of an individual towards RA [118]. According to the Arthritis Foundation (AF), the number of adults diagnosed with arthritis is close to 60 million, whereas approximately 300,000 children are affected by juvenile arthritis. Women are more prone to RA as compared to men and account for 75% of the total cases. Although RA may afflict persons of any age, it often appears in those between the ages of 30 and 50.
5.1.1. Pathogenesis of RA
An increased immunological response of T-cells is a defining feature of RA. Inflammatory cytokines, such as interleukin-1 (IL-1), IL-6, IL-17 and tumor necrosis factor-α (TNF-α) are produced by T-cells, synovial fibroblasts and macrophages. These cytokines can destroy bones by stimulating osteoclasts. Subsets of T-helper (Th) cells are Th1, Th2 and Th17 cells. Through the action of IL-1, IL-6, IL-21 and transforming growth factor (TGF), T cells can become Th17 cells. IL-17 produced by Th17 cells stimulates osteoclasts by causing the receptor activator of NF-κB ligand (RANKL) to be produced in synovial fibroblasts. This is achieved by influencing several immune cells and inflammatory mediators. Chondrocytes, synovial fibroblasts, and synovial macrophages all generate matrix metallopeptidase (MMP) and a disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS), which cause damage of cartilage [119]. Several signaling pathways of the kinase inhibitors such as MAPK, phosphoinositide 3-kinase/protein kinase B (PI3K/AKT), NF-κB and janus kinase/signal transduction and transcription activation protein (JAK/STAT), among others, have been shown to be effective therapeutic alternatives in RA. Fig. 7 depicts the pathogenesis of RA in detail.
Fig. 7.
Pathogenesis of rheumatoid arthritis.
5.1.2. Symptoms of RA
Multiple joints can be affected by RA. Patients with RA may also notice that the same joints on both sides of their body are affected. For instance, both knees and wrists can be affected. RA typically manifests itself in small joints such as finger bones or wrist. Additionally, it can also affect the lungs, eyes and skin. The major symptoms of RA include joint pain and stiffness, redness of joints, loss of motion, weight loss, weakness and fever.
5.1.3. Treatment of RA
At present there is no permanent treatment available for RA. Therapeutic approaches used in treatment include non-steroidal anti-inflammatory drugs (NSAIDs), corticosteroids, disease modifying anti-rheumatic drugs (DMARDs), biological-disease modifying anti-rheumatic drugs (bDMARDs), IL-6 inhibitors, TNF-α inhibitors, etc. These treatments can help in partially treating the symptoms associated with RA. However, they also have some side effects and may not help in prolonging survival [119]. All the above treatments provide some disease modification and symptomatic relief from pain and inflammation but don't offer any permanent solution to the problem of RA. The gene silencing potential of siRNA has shown promising results in reversing the disease progression of RA if effective delivery of siRNA is achieved. siRNA has several unique characteristics like rapid design of sequences for gene knockdown and its synthesis not requiring specific complex cellular expression. These characteristics have increased the interest of researchers and the pharmaceutical industry in developing siRNA therapeutics for RA treatment. The combination of drugs and siRNA in therapy holds promise for the treatment of RA. There are numerous ways to overcome the challenges faced during delivery of siRNA to the cells which have already been discussed above in Section 4.
5.1.4. Limitations of nanoparticles or nanomaterials in the context of RA
Despite the significant progress made in utilizing nanomaterials/nanoparticles for the treatment of RA, there are still gaps in our understanding of their uptake mechanisms and certain limitations hinder their widespread application. Further exploration and research are necessary to investigate the biocompatibility of NCs and their metabolic pathways within the body. Additionally, achieving precise control over drug release rates and retention times should be a key focus for future advancements [120]. The major limitations include poor water solubility, poor hydrophobicity and limited bioaccumulation. However, these limitations can be overcome by selection of suitable polymers.
As nanotechnology continues to advance, we anticipate that advanced nanomaterials will play a pivotal role in the treatment of RA. These materials offer unique properties and capabilities that can be harnessed to overcome current challenges. By optimizing the design of NCs, researchers can improve their compatibility with biological systems, enhance drug delivery efficiency and minimize adverse effects. The precise control of drug release rates and retention times is a critical aspect that requires further investigation. By tailoring the design and composition of NCs, it may be possible to achieve controlled and sustained drug release, leading to prolonged therapeutic effects and reduced dosing frequencies. This approach can enhance patient compliance and minimize side effects. Furthermore, a deeper understanding of the interactions between nanomaterials and the immune system is crucial. This knowledge will aid in the development of NCs that can evade immune surveillance and effectively target specific sites of inflammation. Additionally, elucidating the metabolic pathways of nanomaterials within the body will contribute to their safe and efficient utilization. Currently, most research focuses on single-targeted therapies. However, a potential strategy for treating arthritis could involve targeting multiple pathways simultaneously. By using multiple drugs that synergistically block various pathways involved in the pathogenesis of RA, we may be able to alleviate the disease progress and enhance therapeutic outcomes. This approach holds promise and may represent a new direction for the treatment of RA in the future.
In the end, while there are still uncertainties and limitations surrounding the use of nanomaterials for RA treatment, the future holds great promise. Through continued research and development, we can advance our understanding of nanomaterial biocompatibility, metabolic pathways, and therapeutic mechanisms. By targeting multiple pathways simultaneously and achieving precise control over drug release, nanomaterials have the potential to deliver molecules like siRNA or mRNA for specific targeting of signaling pathways involved in the pathogenesis of RA.
5.1.5. Some recent work on siRNA nanotherapeutics related to RA
Interdisciplinary approach-based modalities, such as siRNA nanotherapeutics, are emerging as a promising area of research for the treatment of RA worldwide. Table 2 provides a brief overview of recent studies on siRNA nanotherapeutics in the context of RA as per the timeline.
Table 2.
siRNA based nanotherapeutics for treatment of RA.
| Strategy | Formulation | Target pathways | Characterizations | Animal model | Route | Outcomes | Ref. |
|---|---|---|---|---|---|---|---|
| CH nanoparticles | CH/ siRNA nanoparticles |
TNF-α siRNA | Particle size: 350 and 450 nm |
CIA | Intraperitoneal injection |
Use 2′-O-Me-modified Dicer substrate siRNA shown least severe arthritic scores, reduced activation of type I interferon, minimal cartilage destruction and inflammatory cell infiltration. | [121] |
| DCs | Ag-loaded DCs, siRNA | Knockdown of CD40, CD80, and CD86 | – | CIA | Intravenously injected | Gene knockdown of CD40, CD80 and CD8, Downregulation of IL-2, IFN-γ, TNF-α and IL-17 and increased FoxP3+ cells with regulatory activity. | [122] |
| Encapsulation | RGD peptide PLGA nanoparticles, siRNA | STAT1 siRNA | Particle size: 250.5 nm, Zeta potential: −0.13 mV | Mouse model | Intradermally injected | Protected siRNA from serum degradation by nanoparticles, inhibition of macrophage and DC activation by decrease level of STAT1 mRNA and increase level of mannose receptor 1 (Mrc-1) and IL-10 mRNA | [123] |
| Co-delivery PLGA nanoparticles | siRNA and dexamethasone loaded PLGA nanoparticles | COX-2 siRNA-complexed, TNF-α, iNOS |
Particle size: 90 nm | – | – | Inhibit the expression of certain genes and proteins improve arthritis features in C28/I2 cells. | [124] |
| PEGylation on cationic bilayer | siRNA/ wrapsome |
TNFα /ws | Particle size: 100 nm | CIA mice | Intravenously injected | Decrease level of CD11b cells present in macrophages and neutrophils in the inflamed synovium. |
[125] |
| Polymeric nanoparticles | G4 and G7 PAMAM dendrimers as well as dextran nanogels | TNF-α | – | Rat model | – | Dextran nanogels and PAMAM dendrimers: safe and efficient siRNA delivery systems for high gene silencing, low toxicity and reduced off-target. | [126] |
| Encapsulation cyclic cationic head lipid-polymer hybrid NCs (CyLiPns) | anti-TNF siRNA (siTNF) Capsaicin (Cap)-encapsulated | TNF-α, IL-17, IL-23, NF-κB and Ki-67 | Particle size: 163 ± 9 nm Zeta Potential: 35.14 ± 8.23 mV |
Psoriasis like mouse mode | Intra-articular injection | Inhibit gene expression of TNFα, NF-κB, IL-17, IL-23 and Ki-67. | [127] |
| – | VE-cadherin siRNA | SAINT-C18 based liposomes (SAINT-O-Somes |
Particle size: 106 ± 48 nm Zeta potential: 3.8 ± 5 mV |
Acute systemic inflammation mice | Intravenously injected | Surface-modified with antibodies, E-selectin or VCAM-1, downregulation of both the target gene mRNA and protein without exerting cellular toxicity. | [128] |
| tGC polymer | Poly siRNA-tGC- nanoparticles | TNF-α | Particle size: 370 nm Zeta Potential: 4.01 ± 0.72 mV |
CIA | Intravenously injected | Rapid cellular uptake of psi-tGC- nanoparticles and TNF-α gene silencing efficacy microcomputed tomography and specific nanoprobe show targeted, accumulation at joint site. |
[129] |
| Magnetic targeting with nanocarrier | PEI-SPIONP (45 nm) siRNA |
IL-2/IL-15Rβ siRNA | Particle size: 161.5 ± 3.5 nm Zeta potential: +26.32 ± 5.34 mV PDI: 0.262 |
Rat arthritis model | Intravenously Injected | PEI-SPIOs effectively targeted macrophages and T cells while delivering IL-2/IL-15Rb siRNA to inflamed joints. In CIA treatment, combining magnetic field exposure increased the anti-inflammatory effect. | [130] |
| tGC nanoparticles | tGC loading siRNA nanoparticles | Notch1 TNF-α | Particle size: 230 nm Zeta Potential: 6.29 ± 0.512 mV |
CIA | Intradermallyi njected |
Inhibit Notch1, slow down the inflammation, bone erosion and cartilage damage. | [131] |
| Peptide modified polymeric micelle | NF-κB subunit siRNA (siRelA) | NF-κBp65 subunit RelA | Particle size: 50 nm | CIA | Intradermally Injected |
Peptide modified micelles with arginine-histidine improved siRNA uptake and decreased inflammatory cytokine level (TNF-α, IL-6 and IL-12) in synoviocytes as well as accumulation of siRNA in the arthritic paws, effectively suppressing RelA mRNA expression, inflammatory cytokines and improving clinical symptoms. | [132] |
| Biodegradable Cationic Polymer PDAPEI | TNF-α shRNA (siRNA) | TNF-α | Particle size: 80 nm Zeta Potential: 57 mV (pH 5.4) and −9 mV (pH 9.4) |
CIA | Intravenously injected | PDAPEI effectively delivered TNF-α shRNA to macrophages, reducing TNF-α expression with lower cytotoxicity and higher transfection efficiency. | [133] |
| Co-delivery copolymers of PCL-PEI and PCL-PEG | glucocorticoid dexamethasone and siRNA | NF-κB | Particle size: 98 nm Zeta potential: Neutral |
CIA | Intradermally injected | Inhibit conversion of macrophages (M1) to the anti-inflammatory M2 state, targeting in inflamed joints, reducing inflammation without harmful effect of kidney or liver function. Inhibition of activation of NF-κB in inflammatory tissue. | [134] |
| Ligand | Modified CH using PEG, folic acid and DEAE | Silencing TNF-α expression | Particles size: 259 ± 3 nm, Zeta potential: 28.3 ± 0.8 mV |
murine CIA | Intraperitoneal injection | Folate-PEG-CH-DEAE15/siRNA lower TNF-α protein concentrations and decreased inflammation. | [135] |
| PEGylation | SNPs that are PEGylated and acid sensitive (AS) sheddable | TNF-α siRNA | Particle size: 118 ± 7 nm Zeta potential: −13.8 ± 5.8 mV. |
CAIA model | Intravenously injected | AS-TNF-α-siRNA-SLNs show high encapsulation efficiency with less burst release and improve siRNA delivery to inflammation sites. The therapeutic potential was demonstrated by reduced paw thickness, bone loss and histopathological scores. | [136] |
| Hybrid nanoparticles | Calcium phosphate/liposome, siRNA and MTX | NF-κB -targeted | Particle size: 45 nm Zeta potential: −23.6 mV. |
CIA | Intradermally injected | Inhibit NF-κB pathways, reducing pro-inflammatory cytokine expression and avoiding adverse effects of MTX. | [137] |
| Polymeric nanoparticles | Targeted ligand of folate-PEG-PLGA and DOTAP/siRNA | MCL-1, Bcl-2, macrophages | Particle size: 142.6 ± 0.61 nm, Zeta potential: 3.6 ± 0.43 mV |
AIA rat model | Intravenously injected | Facilitated escape from the RES, accumulation in inflamed tissues, reduced pro-inflammatory cytokine levels, minimal fibroplasia and mild inflammatory cell infiltration. | [138] |
| Lipidoid-polymer hybrid nanoparticles | Lipidoid and PLGA loaded siRNA | TNF-α, Th1 response | Particle size: 210.2 ± 9.1 Zeta potential: 12.5 ± 6.5 mV |
Murine experimental arthritis model | Intra-articulari njection |
Inhibit pro-inflammatory cytokine and reduce inflammation. | [139] |
| Co-delivery In situ hydrogel |
Indomethacin, MTX, siRNA | MMP-9, inflammatory cytokines | Particle size: 80 nm Zeta potential: 53.50 mV |
CIA | Intra-articular injected | Reduced joint swelling and inhibited gene expression (TNF-α, IL-6 and MMP-9) in plasma and joints. | [140] |
| Encapsulated nanoparticles | Rituximab, siRNA | BAFF-R siRNA | – | Intravenously injected | Reduced arthritis score, ankle diameter, serum anti-collagen IgG level as well as increased collagen type II and osteocalcin expression. Decreased B cell percentage and pro-inflammatory cytokine production. | [141] | |
| Co-delivery of nanoparticles encapsulation | HA-coated pH-responsive nanoparticles, dexamethasone siRNA | MCL-1 | Particle size: 117.07 ± 2.21 nm Zeta potential: 5.53 ± 1.06 mV |
AIA rat model | Intravenously injected | Showed superior inhibition of RA compared to individual drug-loaded nanoparticles, combining MCL-1 siRNA and Dex proved more effective therapies. | [142] |
| Microspheres loaded with HA CH nanoparticles | PLGA loaded and CADK | siRNA MCL-1 | Particle size: 113.6 nm PDI: 0.154 ± 0.013 Zeta potential: 15.0 ± 0.38 mV EE: 90.02% ± 3.02% |
SD rats | Intravenously injected /Intramuscularly injected | Sustained release of nanoparticles in microsphere, protected siRNA from nuclease degradation and readily cross the cellular membrane. | [143] |
| SWCNTs | HiPco- and carboxyl-SWCNT | siRNA Notch1 (siRNA/MTX loaded nanotubes) | Particle size: 407.67±120.21 PDI: 0.087 |
Mice | Intraperitoneally injected | Accumulation in inflamed joints, improve targeting specificity to neutrophils. No impact on B cells and monocytes. | [144] |
| PEGylation | siRNA incorporated PLGA nanoparticles | TNF-α siRNA Mcl-1 siRNA | Particle size: 192 ± 12 nm, Zeta potential: −26.7 ± 2.3 mV |
CIA | Intravenously injected | Inhibited macrophage-based cytokine release and anti-inflammatory effect. | [145] |
CH: Chitosan; CIA: collagen-induced arthritis; MCL-1: myeloid cell leukemia-I; COX2: Cyclooxygenase-2; iNOS: inducible nitric oxide; Ki-67: Kiel-67 (monoclonal antibody); SAINT-C18: Cationic lipid 1-methyl-4-(cis-9-dioleyl)methyl-pyridinium-chloride; VCAM-1: Vascular cell adhesion molecule 1; tGC: Thiolated glycol chitosan; PDAPEI: Dopamine and PEI copolymerized nanodots; PCL: Poly-ε-caprolactone; DEAE: Diethylethylamine; CAIA: Collagen-Antibody Induced Arthritis; MTX: Methotrexate; AIA: Adjuvant-induced arthritis; BAFF-R: B-cell activating factor receptor; HA: Hyaluronic acid; LPCE:low-molecular-weight-polyethylenimine-cholesterol–polyethylene-glycol; PCADK: poly (cyclohexane-1,4-diyl acetone dimethylene ketal); SWCNTs: single-walled carbon nanotubes.
5.2. Osteoarthritis
OA is a multifaceted and age-related, distinct form of arthritis that is linked to the breakdown of articular cartilage. It is also a degenerative joint condition. Some of the symptoms of this chronic condition are pain, localized tissue damage and cartilage damage. OA has become a concerning chronic disease due to the global increase in life expectancy and more elderly population [146]. It is generally recognized that reactive oxygen species (ROS) play a crucial role in cartilage degradation and chondrocyte mortality [147].
5.2.1. Pathogenesis of OA
In the early stages of articular cartilage degeneration, hypertrophic chondrocytes play a significant role. These cells release MMPs and ADAMTS contributing to OA development. The Runt-related transcription factor 2 (Runx2), regulated by hedgehog signaling, is a crucial transcription factor in chondrocyte hypertrophy. On the other side, parathyroid hormone (PTH) inhibits chondrocyte hypertrophy. Various factors including HIF-2 (hypoxia-inducible factor-2), TLR4 signaling Notch 1, NF-κB and Hes1 encourage the release of different degrading enzymes, such as MMPs & ADAMTS. However, miR-140 helps preserve cartilage by inhibiting the production of ADAMTS-5. Cartilage specific protein, carminerin is involved in chondrocyte calcification. All these changes are involved in the progression and pathogenesis of OA related degeneration of joints [119]. Fig. 8 depicts the pathogenesis of OA.
Fig. 8.
Pathogenesis of osteoarthritis.
5.2.2. Symptoms of OA
OA joint pain is commonly aggravated by activity and relieved by rest. The key signs indicating an OA diagnosis include pain, reduced function, stiffness, joint instability and buckling or giving way. Patients may also report decreased mobility, deformity, edema and crepitus, as per age (OA is rare before the age of 40) without systemic symptoms like fever. In addition, persistent pain might cause psychological discomfort [148].
5.2.3. Treatment of OA
Generally, OA treatments primarily address pain and inflammation symptoms using steroidal or NSAIDs drugs. However, as our understanding of OA underlying mechanisms has improved, new treatment targets called disease-modifying OA drugs (DMOADs) have emerged. They help in reducing the resulting structural damage to avoid permanent impairment. The goal of the DMOADs currently undergoing clinical trials is to re-establish the stability of matrix metabolism. DMOADs efficiently manage the degenerative changes in osteoarthritic cartilage by concentrating on matrix-degrading enzymes, inflammatory cytokines, the Wnt pathway and OA-related pain [146]. Numerous DMOADs are in phase II/III clinical trials and many repurposed drugs are under investigation. In 2010, the FDA approved the antidepressant duloxetine hydrochloride (CymbaltaⓇ) to alleviate OA discomfort, including lower back pain. This has been a significant breakthrough for those who are unable to handle NSAIDs or other medications.
5.2.4. Some recent work on siRNA nanotherapeutics related to OA
siRNA nanotherapeutics are emerging as a promising area of research for the treatment of OA worldwide. Table 3 provides a brief overview of recent studies on siRNA nanotherapeutics in the context of OA as per the timeline.
Table 3.
siRNA based nanotherapeutics for treatment of OA.
| Carrier | Drug/ Formulation |
Targeted Pathways | Characteristics | Animal Model | Administration Route | Outcomes | Ref |
|---|---|---|---|---|---|---|---|
| Adenoviral vector | Ad-siRNA NF-κBp65 | NF-κBp65 | Optical density: 260 | SD male rats | Intra-articular injection | Decrease NF-κBp65 expression and mitigate synovial inflammation by inhibition of f IL-1β, TNF-α and cartilage degradation. | [149] |
| – | siRNA with phenotypic features of OA Ob. | Inhibitors of leptin signaling TGF-β1 | Final concentration: 100 µg/ml | Human subchondral Ob cell culture | – | Blocking leptin signaling via siRNA decreased the levels of OC, ALP, TGF-β1, Ob Rb in OA Ob expression. Inhibition of leptin production in OA Ob improved the phenotypic expression of Ob. | [150] |
| – | SDF-1, CXCR4 siRNA, or CXCR4 antibody | Signaling between SDF-1 and CXCR4 with AMD3100 | 5 µg pU6RNAi-CXCR4 vector transferred into chondrocyte | Guinea pig OA model. | Small subcutaneous pockets of Alzet mini osmotic pump (44.44 mg/ml of AMD3100) | SDF-1 entered the cartilage, leading to reduced proteoglycan staining. The blocked of SDF-1, CXCR4 signaling reduced the levels of SDF-1, MMPs, GAG and IL-1b in synovial fluid showing promising therapeutics option to decreased cartilage degeneration in OA. | [151] |
| – | Lentivirus-mediated siRNA | ADAMTS-5 knockdown | – | Rat model | Intra-articular injection | Downregulation of ADAMTS-5 protein expression and prevented the degradation of articular cartilage. | [152] |
| Chondrocyte‐homing peptide/PEI nanoparticles | HIF-2α, siRNA | HIF-2α, ADAMTS-4, MMP-13, and MMP-9, VEGF, NF-κB and collagen type X | – | OA-affected mice | Intra-articular injection | Downregulation of catabolic factors, (HIF-2α, MMP-13 and MMP-9, ADAMTS-4, VEGF, collagen type X and NF-κB). Decreased IL-1β levels and cartilage integrity maintained. |
[153] |
| Surgically | MMP-13 or ADAMTS-5 siRNA | IL-1β stimulation, MMP13 mRNA expression in FLS | – | DMM Mouse Model | Intra-articular injection | siRNA-treated all three groups showed significant improvement in histological scores compared to the control siRNA group. | [154] |
| – | siRNA for NR1D1 or BMAL1 | TGF-β pathway. circadian rhythm pathway |
– | Mouse model | – | Knock down of NR1D1 that increased BMAL1 expression, while knock down BMAL1 resulted in decreased NR1D1 levels. These changes also affected the TGF-α signaling pathway. | [155] |
| – | Lipofectamine 2000, siRNA | NLRP1 and NLRP3 siRNA | – | – | – | Blocking NLRP1 and NLRP3, decreased the production of LPS induced pyroptosis and its related cytokines. | [156] |
| – | Lorecivivint | CLK2 and DYRK1A for Wnt pathway inhibition, siRNA, NF-κB and STAT | – | MIA-induced OA rats | Intra-articular injection | Early chondrogenesis due to decreased CLK2. Enhanced mature chondrocyte function by DYRK1A, inhibition of NF-κB and STAT3 via lorecivivint reduced inflammation, cytokines levels and cartilage-degrading enzymes. All the above finding improves cartilage degradation and weight bearing function of joints. | [157] |
| Photothermal-triggered NO (650 nm NIR laser irradiation) nanogenerators | NHsPP nanoparticles, NO, siRNA or PTT | Pro-inflammatory cytokines, macrophage | Particle size: 200 nm |
OA mice model | In situ injections | Inhibits the inflammatory response effectively by reducing the level of pro-inflammatory cytokines and the macrophage response and also prevents cartilage erosion efficiently. | [158] |
| – | YAP siRNA | IL-1β, Hippo/YAP signaling pathway | – | Surgery-induced OA animal models. | Intra-articular injection | Knockdown of IL-1β and prevent cartilage degradation. | [159] |
| Encapsulate PLGA nanoparticles | p66shc- siRNA-loaded nanoparticles | ROS-associated proteins p-p66shc expression levels | Particle Size: 183.7 ± 72.21 nm Zeta potential: 41.1 ± 4.81 mV |
MIA-induced OA rats | Intra-articular injection | Blocking p66shc phosphorylation reduced ROS in chondrocytes, alleviated pain, cartilage damage and inflammation. | [160] |
| PLGA nanoparticles | siRNA p47phox | Oxidative stress and ROS | Particle size: 126 ± 55 nm, Zeta potential: −23 ± 2 mV |
MIA-induced OA rats | Intra-articular injection | Inhibition of ROS generation, chondrocyte cell death can be reduced as well as decrease cartilage degradation | [147] |
| – | siRNA into chondrocyte | Mitofusin 2 (MFN2) is Parkin receptor |
– | Rats with OA | – | Silencing MFN2 using siRNA reversed age-related metabolic alterations, reduced inflammation and increase Parkin level. | [161] |
| Functionalized nanoparticles antibody | MMP13, siRNA Methylprednisolone |
MMP13, type II collagen | Particle size:100 nm Zeta potential: neutral |
Post-traumatic OA. | Intra-articular injection | Gene knockdown of MMP-13 decrease gene clusters linked to tissue remodeling, angiogenesis, immune responses and proteolysis. Reduced disease progression compared to single or weekly methylprednisolone injections. | [162] |
| – | GPER, siRNA silencing Piezo1 |
AP and ARHGAP29, and the YAP nuclear localization, RhoA/LIMK/coflin pathway | – | Rat OA model | Intra-articular injection | GPER suppressed the RhoA/LIMK/coflin pathway, actin polymerization and Piezo1 by upregulating YAP and ARHGAP29 leading to reduced cartilage degeneration. | [163] |
Ad-siRNANF-kBp65: Adenoviral vector-mediated NF-κB; Ob: Osteoblast; OC: Osteocalcin release; ALP: Alkaline phosphatase activity;; SDF-1: Stromal cell-derived factor-1; CXCR4: C-X-C chemokine receptor type 4; GAG: glycosaminoglycans; FLS: Fibroblast-like synoviocyte; DMM: Disproportionate Micromelia; NLRP1 and NLRP3: Inflammasomes; CLK2: CDC-like kinase 2; DYRK1A: Dual-specificity tyrosine phosphorylation- regulated kinase 1A; MIA: Monoiodoacetate; PTT: Photothermal Therapy; NO: Nitric oxide; NHsPP: NO—Hb@siRNA@PLGA-PEG; YAP: Yes-Associated Protein; GPER: G protein coupled oestrogen receptor; ARHGAP29: YAP/Rho GTPase activating protein 29.
6. Conclusions
The potential of siRNA nanotherapeutics in the treatment of arthritis has shown an upsurge of interest and is very promising in finding the cure for disease progression in arthritis. The difficulty of effectively delivering siRNA to specific diseased sites in vivo is well documented in the literature and has been discussed at length in this review. The inefficient transport of therapeutic drugs to the local chondrocytes in avascular/articular cartilage presents a significant obstacle to the successful development of arthritis therapy. The use of versatile NCs in improving the delivery of siRNA to the desired target is the upcoming area of research in the successful development of siRNA nanotherapeutics. In the case of arthritis, the gene-silencing potential of customized siRNA specific to a particular pathway involved in pathogenesis has shown a new hope for its treatment. Knocking down/silencing of various pathogenic pathways, such as STAT1, Notch1, NF-kB, TNF-α, JAK-STAT and MMPs, etc. has been explored as a potential approach. In preclinical research, various nanostructures have been utilized as carriers for siRNA delivery. Self-assembling nanostructures enable synchronized release of siRNA and can achieve endosomal escape while also preventing the oxidation of siRNA in endosomes and the bloodstream. The combination of drugs and siRNA in therapy holds promise for enhancing treatment outcomes, minimizing side effects have opened new therapeutic horizons for the treatment of arthritis. Demand for cutting-edge therapeutic approaches is increasing and a revolutionary approach to treat the disease is made possible by the RNAi phenomenon and its success has led to FDA approval of five siRNA therapeutics in the last five years. The chemical modifications of siRNA and customized NCs have helped achieve long-term therapeutic potential at much lesser doses of siRNA in various diseases. However, none of the approved agents were used for the treatment of arthritis but the technology has opened the doors for researchers to explore the pathways involved in immune-mediated disease like arthritis. The short duration of siRNA research and development time as compared to small molecules, and monoclonal antibodies holds great promise in the future of this revolutionary technology. Further, these modalities need to be administered quarterly or half yearly and this will in turn help in improving patient compliance. With more advancements in the chemical modification of siRNA geometries and tailored NCs, the siRNA nanotherapeutics will revolutionize the precise and personalized treatment of inflammatory diseases like RA and OA.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y., Zhang C., Liang W. Development of RNAi technology for targeted therapy–a track of siRNA-based agents to RNAi therapeutics. J Control Release. 2014;193:270–281. doi: 10.1016/j.jconrel.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 3.Hoy S.M. Patisiran: first global approval. Drugs. 2018;78(15):1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 4.Scott L.J. Givosiran: first approval. Drugs. 2020;80(3):335–339. doi: 10.1007/s40265-020-01269-0. [DOI] [PubMed] [Google Scholar]
- 5.Habtemariam B.A., Karsten V., Attarwala H., Goel V., Melch M., Clausen V.A., et al. Single-dose pharmacokinetics and pharmacodynamics of transthyretin targeting N-acetylgalactosamine-small interfering ribonucleic acid conjugate, vutrisiran, in healthy subjects. Clin Pharmacol Ther. 2021;109(2):372–382. doi: 10.1002/cpt.1974. [DOI] [PubMed] [Google Scholar]
- 6.Scott L.J., Keam S.J. Lumasiran: first approval. Drugs. 2021;81(2):277–282. doi: 10.1007/s40265-020-01463-0. [DOI] [PubMed] [Google Scholar]
- 7.Rossi J.J., Rossi D.J. siRNA drugs: here to stay. Mol Ther. 2021;29(2):431–432. doi: 10.1016/j.ymthe.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray K.K., Wright R.S., Kallend D., Koenig W., Leiter L.A., Raal F.J., et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med. 2020;382(16):1507–1519. doi: 10.1056/NEJMoa1912387. [DOI] [PubMed] [Google Scholar]
- 9.Zeinali M., Abbaspour-Ravasjani S., Ghorbani M., Babazadeh A., Soltanfam T., Santos A.C., et al. Nanovehicles for co-delivery of anticancer agents. Drug Discov Today. 2020;25(8):1416–1430. doi: 10.1016/j.drudis.2020.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behlke M.A. Progress towards in vivo use of siRNAs. Mol. Ther. 2006;13(4):644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo P., Coban O., Snead N.M., Trebley J., Hoeprich S., Guo S., et al. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev. 2010;62(6):650–666. doi: 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C., Wang J. Delivery systems for siRNA drug development in cancer therapy. Asian J Pharm Sci. 2015;10(1):1–12. [Google Scholar]
- 16.Gavrilov K., Saltzman W.M. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85(2):187–200. [PMC free article] [PubMed] [Google Scholar]
- 17.Paul A., Muralidharan A., Biswas A., Kamath B.V., Joseph A., Alex A.T. siRNA therapeutics and its challenges: recent advances in effective delivery for cancer therapy. OpenNano. 2022;7 [Google Scholar]
- 18.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 19.Song E., Zhu P., Lee S.K., Chowdhury D., Kussman S., Dykxhoorn D.M., et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 20.Leng Q., Woodle M.C., Mixson A.J. Targeted delivery of siRNA therapeutics to malignant tumors. J Drug Deliv. 2017;2017 doi: 10.1155/2017/6971297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han H.D., Mangala L.S., Lee J.W., Shahzad M.M., Kim H.S., Shen D., et al. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clin Cancer Res. 2010;16(15):3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda S., Keiser K., Nair J.K., Charisse K., Manoharan R.M., Kretschmer P., et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem Biol. 2015;10(5):1181–1187. doi: 10.1021/cb501028c. [DOI] [PubMed] [Google Scholar]
- 23.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc. 2014;136(49):16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 24.Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 25.Robbins M., Judge A., MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19(2):89–101. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 26.Judge A., MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19(2):111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 27.Forsbach A., Nemorin J.G., Montino C., Müller C., Samulowitz U., Vicari A.P., et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180(6):3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 28.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348(5):1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Meng Z., Lu M. RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 32.Shegokar R., Al Shaal L., Mishra P.R. SiRNA delivery: challenges and role of carrier systems. Pharmazie. 2011;66(5):313–318. [PubMed] [Google Scholar]
- 33.Zahir-Jouzdani F., Mottaghitalab F., Dinarvand M., Atyabi F. siRNA delivery for treatment of degenerative diseases, new hopes and challenges. J Drug Deliv Sci Technol. 2018;45:428–441. [Google Scholar]
- 34.Rana T.M. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8(1):23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 35.Kim S.S., Garg H., Joshi A., Manjunath N. Strategies for targeted nonviral delivery of siRNAs in vivo. Trends Mol Med. 2009;15(11):491–500. doi: 10.1016/j.molmed.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czauderna F., Fechtner M., Dames S., Aygün H., Klippel A., Pronk G.J., et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31(11):2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia J., Noronha A., Toudjarska I., Li F., Akinc A., Braich R., et al. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem Biol. 2006;1(3):176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 38.Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342(3):919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 39.Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31(2):589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25(10):1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 41.Chernikov I.V., Gladkikh D.V., Meschaninova M.I., Ven'yaminova A.G., Zenkova M.A., Vlassov V.V., et al. Cholesterol-containing nuclease-resistant siRNA accumulates in tumors in a carrier-free mode and silences MDR1 gene. Mol Ther Nucleic Acids. 2017;6:209–220. doi: 10.1016/j.omtn.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 43.Gaziova Z., Baumann V., Winkler A.M., Winkler J. Chemically defined polyethylene glycol siRNA conjugates with enhanced gene silencing effect. Bioorg Med Chem. 2014;22(7):2320–2326. doi: 10.1016/j.bmc.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato A., Choi S.W., Hirai M., Yamayoshi A., Moriyama R., Yamano T., et al. Polymer brush-stabilized polyplex for a siRNA carrier with long circulatory half-life. J Control Release. 2007;122(3):209–216. doi: 10.1016/j.jconrel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 45.Peer D., Lieberman J. Special delivery: targeted therapy with small RNAs. Gene Ther. 2011;18(12):1127–1133. doi: 10.1038/gt.2011.56. [DOI] [PubMed] [Google Scholar]
- 46.Tatiparti K., Sau S., Kashaw S.K., Iyer A.K. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials. 2017;7(4):77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musacchio T., Torchilin V.P. siRNA delivery: from basics to therapeutic applications. Front Biosci. 2013;18(1):58–79. doi: 10.2741/4087. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Song Z., Zheng N., Nagasaka K., Yin L., Cheng J. Systemic siRNA delivery to tumors by cell-penetrating α-helical polypeptide-based metastable nanoparticles. Nanoscale. 2018;10(32):15339–15349. doi: 10.1039/c8nr03976c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cummings J.C., Zhang H., Jakymiw A. Peptide carriers to the rescue: overcoming the barriers to siRNA delivery for cancer treatment. Transl Res. 2019;214:92–104. doi: 10.1016/j.trsl.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mainini F., Eccles M.R. Lipid and Polymer-based nanoparticle siRNA delivery systems for cancer therapy. Molecules. 2020;25(11):2692. doi: 10.3390/molecules25112692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151(3):220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Pei D., Buyanova M. Overcoming endosomal entrapment in drug delivery. Bioconjug Chem. 2019;30(2):273–283. doi: 10.1021/acs.bioconjchem.8b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu S., Morris V.B., Labhasetwar V. Effectiveness of small interfering RNA delivery via arginine-rich polyethylenimine-based polyplex in metastatic and doxorubicin-resistant breast cancer cells. J Pharmacol Exp Ther. 2019;370(3):902–910. doi: 10.1124/jpet.119.256909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonawane N.D., Szoka F.C., Jr, Verkman A.S. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 55.Huang H., Yuan S., Ma Z., Ji P., Ma X., Wu Z., et al. Genetic recombination of poly(l-lysine) functionalized apoferritin nanocages that resemble viral capsid nanometer-sized platforms for gene therapy. Biomater Sci. 2020;8(6):1759–1770. doi: 10.1039/c9bm01822k. [DOI] [PubMed] [Google Scholar]
- 56.Shin J., Shum P., Thompson D.H. Acid-triggered release via dePEGylation of DOPE liposomes containing acid-labile vinyl ether PEG-lipids. J Control Release. 2003;91(1–2):187–200. doi: 10.1016/s0168-3659(03)00232-3. [DOI] [PubMed] [Google Scholar]
- 57.Subhan M.A., Torchilin V.P. siRNA based drug design, quality, delivery and clinical translation. Nanomedicine. 2020;29 doi: 10.1016/j.nano.2020.102239. [DOI] [PubMed] [Google Scholar]
- 58.Sajid M.I., Moazzam M., Kato S., Yeseom Cho K., Tiwari R.K. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals (Basel) 2020;13(10):294. doi: 10.3390/ph13100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juliano R., Alam M.R., Dixit V., Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36(12):4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107(2):276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 61.Zimmermann T.S., Lee A.C., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 62.Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., et al. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12(5):4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 63.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc Chem Res. 2019;52(9):2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 64.Leung A.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured Core. J Phys Chem C Nanomater Interfaces. 2012;116(34):18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S.I., Shin D., Choi T.H., Lee J.C., Cheon G.J., Kim K.Y., et al. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol Ther. 2007;15(6):1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- 66.Yagi N., Manabe I., Tottori T., Ishihara A., Ogata F., Kim J.H., et al. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Res. 2009;69(16):6531–6538. doi: 10.1158/0008-5472.CAN-08-3945. [DOI] [PubMed] [Google Scholar]
- 67.Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 68.Kogure K., Akita H., Yamada Y., Harashima H. Multifunctional envelope-type nano device (MEND) as a non-viral gene delivery system. Adv Drug Deliv Rev. 2008;60(4–5):559–571. doi: 10.1016/j.addr.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 69.Nakamura Y., Kogure K., Futaki S., Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. J Control Release. 2007;119(3):360–367. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 70.Sato Y., Hatakeyama H., Sakurai Y., Hyodo M., Akita H., Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163(3):267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto N., Sato Y., Munakata T., Kakuni M., Tateno C., Sanada T., et al. Novel pH-sensitive multifunctional envelope-type nanodevice for siRNA-based treatments for chronic HBV infection. J Hepatol. 2016;64(3):547–555. doi: 10.1016/j.jhep.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 72.Sato Y., Hashiba K., Sasaki K., Maeki M., Tokeshi M., Harashima H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J Control Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 73.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love K.T., Mahon K.P., Levins C.G., Whitehead K.A., Querbes W., Dorkin J.R., et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A. 2014;111(11):3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasiewicz L.N., Whitehead K.A. Lipid nanoparticles silence tumor necrosis factor α to improve wound healing in diabetic mice. Bioeng Transl Med. 2018;4(1):75–82. doi: 10.1002/btm2.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ball R.L., Bajaj P., Whitehead K.A. Oral delivery of siRNA lipid nanoparticles: fate in the GI tract. Sci Rep. 2018;8(1):2178. doi: 10.1038/s41598-018-20632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yonezawa S., Koide H., Asai T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv Drug Deliv Rev. 2020;154-155:64–78. doi: 10.1016/j.addr.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martins S., Costa-Lima S., Carneiro T., Cordeiro-da-Silva A., Souto E.B., Ferreira D.C. Solid lipid nanoparticles as intracellular drug transporters: an investigation of the uptake mechanism and pathway. Int J Pharm. 2012;430(1–2):216–227. doi: 10.1016/j.ijpharm.2012.03.032. [DOI] [PubMed] [Google Scholar]
- 81.Lee S., Yang S.C., Kao C.Y., Pierce R.H., Murthy N. Solid polymeric microparticles enhance the delivery of siRNA to macrophages in vivo. Nucleic Acids Res. 2009;37(22):e145. doi: 10.1093/nar/gkp758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobson G.B., Gonzalez-Gonzalez E., Spitler R., Shinde R., Leake D., Kaspar R.L., et al. Biodegradable nanoparticles with sustained release of functional siRNA in skin. J Pharm Sci. 2010;99(10):4261–4266. doi: 10.1002/jps.22147. [DOI] [PubMed] [Google Scholar]
- 83.Lai R.C., Yeo R.W., Tan K.H., Lim S.K. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31(5):543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 84.EL Andaloussi S., Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 85.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 87.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., et al. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428(4):688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tseng Y.C., Mozumdar S., Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61(9):721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin Q., Chen J., Zhang Z., Zheng G. Lipid-based nanoparticles in the systemic delivery of siRNA. Nanomedicine (Lond) 2014;9(1):105–120. doi: 10.2217/nnm.13.192. [DOI] [PubMed] [Google Scholar]
- 90.Dokka S., Toledo D., Shi X., Castranova V., Rojanasakul Y. Oxygen radical-mediated pulmonary toxicity induced by some cationic liposomes. Pharm Res. 2000;17(5):521–525. doi: 10.1023/a:1007504613351. [DOI] [PubMed] [Google Scholar]
- 91.Wang J., Lu Z., Wientjes M.G., Au J.L. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tai W. Current aspects of siRNA bioconjugate for in vitro and in vivo delivery. Molecules. 2019;24(12):2211. doi: 10.3390/molecules24122211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abedi-Gaballu F., Dehghan G., Ghaffari M., Yekta R., Abbaspour-Ravasjani S., Baradaran B., et al. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl Mater Today. 2018;12:177–190. doi: 10.1016/j.apmt.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abedi Gaballu F., Cho W.C., Dehghan G., Zarebkohan A., Baradaran B., Mansoori B., et al. Silencing of HMGA2 by siRNA loaded methotrexate functionalized polyamidoamine dendrimer for human breast cancer cell therapy. Genes (Basel) 2021;12(7):1102. doi: 10.3390/genes12071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Choi S.W., Lee S.H., Mok H., Park T.G. Multifunctional siRNA delivery system: polyelectrolyte complex micelles of six-arm PEG conjugate of siRNA and cell penetrating peptide with crosslinked fusogenic peptide. Biotechnol Prog. 2010;26(1):57–63. doi: 10.1002/btpr.310. [DOI] [PubMed] [Google Scholar]
- 96.Moschos S.A., Jones S.W., Perry M.M., Williams A.E., Erjefalt J.S., Turner J.J., et al. Lung delivery studies using siRNA conjugated to TAT (48-60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjug Chem. 2007;18(5):1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Osborn M.F., Khvorova A. Improving siRNA delivery in vivo through lipid conjugation. Nucleic Acid Ther. 2018;28(3):128–136. doi: 10.1089/nat.2018.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong D.J., Hurvitz S.A. Recent advances in the development of anti-HER2 antibodies and antibody-drug conjugates. Ann Transl Med. 2014;2(12):122. doi: 10.3978/j.issn.2305-5839.2014.08.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bäumer S., Bäumer N., Appel N., Terheyden L., Fremerey J., Schelhaas S., et al. Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clin Cancer Res. 2015;21(6):1383–1394. doi: 10.1158/1078-0432.CCR-13-2017. [DOI] [PubMed] [Google Scholar]
- 100.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin H., Song P., Zhao Y., Xue L.J., Liu Y., Chu C.Q. Targeting Th17 cells with small molecules and small interference RNA. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/290657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S.J., Yhee J.Y., Kim S.H., Kwon I.C., Kim K. Biocompatible gelatin nanoparticles for tumor-targeted delivery of polymerized siRNA in tumor-bearing mice. J Control Release. 2013;172(1):358–366. doi: 10.1016/j.jconrel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 103.Yhee J.Y., Lee S.J., Lee S., Song S., Min H.S., Kang S.W., et al. Tumor-targeting transferrin nanoparticles for systemic polymerized siRNA delivery in tumor-bearing mice. Bioconjug Chem. 2013;24(11):1850–1860. doi: 10.1021/bc400226b. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y., Li Z., Han Y., Liang L.H., Ji A. Nanoparticle-based delivery system for application of siRNA in vivo. Curr Drug Metab. 2010;11(2):182–196. doi: 10.2174/138920010791110863. [DOI] [PubMed] [Google Scholar]
- 105.Davis M.E. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6(3):659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 106.Davis M.E., Brewster M.E. Cyclodextrin-based pharmaceutics: past, present and future. Nat Rev Drug Discov. 2004;3(12):1023–1035. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 107.Han H.D., Mora E.M., Roh J.W., Nishimura M., Lee S.J., Stone R.L., et al. Chitosan hydrogel for localized gene silencing. Cancer Biol Ther. 2011;11(9):839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Inaba S., Nagahara S., Makita N., Tarumi Y., Ishimoto T., Matsuo S., et al. Atelocollagen-mediated systemic delivery prevents immunostimulatory adverse effects of siRNA in mammals. Mol Ther. 2012;20(2):356–366. doi: 10.1038/mt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pack D.W., Hoffman A.S., Pun S., Stayton P.S. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 110.Zhao M.D., Cheng J.L., Yan J.J., Chen F.Y., Sheng J.Z., Sun D.L., et al. Hyaluronic acid reagent functional chitosan-PEI conjugate with AQP2-siRNA suppressed endometriotic lesion formation. Int J Nanomedicine. 2016;11:1323–1336. doi: 10.2147/IJN.S99692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singha K., Namgung R., Kim W.J. Polymers in small-interfering RNA delivery. Nucleic Acid Ther. 2011;21(3):133–147. doi: 10.1089/nat.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim T., Hyeon T. Applications of inorganic nanoparticles as therapeutic agents. Nanotechnology. 2014;25(1) doi: 10.1088/0957-4484/25/1/012001. [DOI] [PubMed] [Google Scholar]
- 113.Mody V.V., Cox A., Shah S., Singh A., Bevins W., Parihar H. Magnetic nanoparticle drug delivery systems for targeting tumor. Appl Nanosci (Switzerland) 2014;4(4):385–392. [Google Scholar]
- 114.Alhaddad A., Adam M.P., Botsoa J., Dantelle G., Perruchas S., Gacoin T., et al. Nanodiamond as a vector for siRNA delivery to Ewing sarcoma cells. Small. 2011;7(21):3087–3095. doi: 10.1002/smll.201101193. [DOI] [PubMed] [Google Scholar]
- 115.Selvam C., Mutisya D., Prakash S., Ranganna K., Thilagavathi R. Therapeutic potential of chemically modified siRNA: recent trends. Chem Biol Drug Des. 2017;90(5):665–678. doi: 10.1111/cbdd.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liao H., Wang J.H. Biomembrane-permeable and Ribonuclease-resistant siRNA with enhanced activity. Oligonucleotides. 2005;15(3):196–205. doi: 10.1089/oli.2005.15.196. [DOI] [PubMed] [Google Scholar]
- 117.Li L., Shen Y. Overcoming obstacles to develop effective and safe siRNA therapeutics. Expert Opin Biol Ther. 2009;9(5):609–619. doi: 10.1517/14712590902911420. [DOI] [PubMed] [Google Scholar]
- 118.Senthelal S., Li J., Ardeshirzadeh S., Arthritis Thomas MA. StatPearls Publishing; Treasure IslandFL: 2023. StatPearls.https://www.ncbi.nlm.nih.gov/books/NBK518992/ Available from. [Google Scholar]
- 119.Tateiwa D., Yoshikawa H., Kaito T. Cartilage and bone destruction in arthritis: pathogenesis and treatment strategy: a literature review. Cells. 2019;8(8):818. doi: 10.3390/cells8080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zheng M., Jia H., Wang H., Liu L., He Z., Zhang Z., et al. Application of nanomaterials in the treatment of rheumatoid arthritis. RSC Adv. 2021;11(13):7129–7137. doi: 10.1039/d1ra00328c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Howard K.A., Paludan S.R., Behlke M.A., Besenbacher F., Deleuran B., Kjems J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther. 2009;17(1):162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng X., Suzuki M., Ichim T.E., Zhang X., Sun H., Zhu F., et al. Treatment of autoimmune arthritis using RNA interference-modulated dendritic cells. J Immunol. 2010;184(11):6457–6464. doi: 10.4049/jimmunol.0901717. [DOI] [PubMed] [Google Scholar]
- 123.Scheinman R.I., Trivedi R., Vermillion S., Kompella U.B. Functionalized STAT1 siRNA nanoparticles regress rheumatoid arthritis in a mouse model. Nanomedicine (Lond) 2011;6(10):1669–1682. doi: 10.2217/nnm.11.90. [DOI] [PubMed] [Google Scholar]
- 124.Park J.S., Yang H.N., Jeon S.Y., Woo D.G., Kim M.S., Park K.H. The use of anti-COX2 siRNA coated onto PLGA nanoparticles loading dexamethasone in the treatment of rheumatoid arthritis. Biomaterials. 2012;33(33):8600–8612. doi: 10.1016/j.biomaterials.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 125.Komano Y., Yagi N., Onoue I., Kaneko K., Miyasaka N., Nanki T. Arthritic joint-targeting small interfering RNA-encapsulated liposome: implication for treatment strategy for rheumatoid arthritis. J Pharmacol Exp Ther. 2012;340(1):109–113. doi: 10.1124/jpet.111.185884. [DOI] [PubMed] [Google Scholar]
- 126.Jensen L.B., Griger J., Naeye B., Varkouhi A.K., Raemdonck K., Schiffelers R., et al. Comparison of polymeric siRNA nanocarriers in a murine LPS-activated macrophage cell line: gene silencing, toxicity and off-target gene expression. Pharm Res. 2012;29(3):669–682. doi: 10.1007/s11095-011-0589-0. [DOI] [PubMed] [Google Scholar]
- 127.Desai P.R., Marepally S., Patel A.R., Voshavar C., Chaudhuri A., Singh M. Topical delivery of anti-TNFα siRNA and capsaicin via novel lipid-polymer hybrid nanoparticles efficiently inhibits skin inflammation in vivo. J Control Release. 2013;170(1):51–63. doi: 10.1016/j.jconrel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kowalski P.S., Lintermans L.L., Morselt H.W., Leus N.G., Ruiters M.H., Molema G., et al. Anti-VCAM-1 and anti-E-selectin SAINT-O-Somes for selective delivery of siRNA into inflammation-activated primary endothelial cells. Mol Pharm. 2013;10(8):3033–3044. doi: 10.1021/mp4001124. [DOI] [PubMed] [Google Scholar]
- 129.Lee S.J., Lee A., Hwang S.R., Park J.S., Jang J., Huh M.S., et al. TNF-α gene silencing using polymerized siRNA/thiolated glycol chitosan nanoparticles for rheumatoid arthritis. Mol Ther. 2014;22(2):397–408. doi: 10.1038/mt.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duan J., Dong J., Zhang T., Su Z., Ding J., Zhang Y., et al. Polyethyleneimine-functionalized iron oxide nanoparticles for systemic siRNA delivery in experimental arthritis. Nanomedicine (Lond) 2014;9(6):789–801. doi: 10.2217/nnm.13.217. [DOI] [PubMed] [Google Scholar]
- 131.Kim M.J., Park J.S., Lee S.J., Jang J., Park J.S., Back S.H., et al. Notch1 targeting siRNA delivery nanoparticles for rheumatoid arthritis therapy. J Control Release. 2015;216:140–148. doi: 10.1016/j.jconrel.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 132.Kanazawa T., Endo T., Arima N., Ibaraki H., Takashima Y., Seta Y. Systemic delivery of small interfering RNA targeting nuclear factor κB in mice with collagen-induced arthritis using arginine-histidine-cysteine based oligopeptide-modified polymer nanomicelles. Int J Pharm. 2016;515(1–2):315–323. doi: 10.1016/j.ijpharm.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 133.Song J., Chen Y., Jiang S., Yang K., Li X., Zhao X., et al. Efficient and non-toxic biological response carrier delivering TNF-α shRNA for gene silencing in a murine model of rheumatoid arthritis. Front Immunol. 2016;7:305. doi: 10.3389/fimmu.2016.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Q., Jiang H., Li Y., Chen W., Li H., Peng K., et al. Targeting NF-kB signaling with polymeric hybrid micelles that co-deliver siRNA and dexamethasone for arthritis therapy. Biomaterials. 2017;122:10–22. doi: 10.1016/j.biomaterials.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 135.Shi Q., Rondon-Cavanzo E.P., Dalla Picola I.P., Tiera M.J., Zhang X., Dai K., et al. In vivo therapeutic efficacy of TNFα silencing by folate-PEG-chitosan-DEAE/siRNA nanoparticles in arthritic mice. Int J Nanomedicine. 2018;13:387–402. doi: 10.2147/IJN.S146942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aldayel A.M., O'Mary H.L., Valdes S.A., Li X., Thakkar S.G., Mustafa B.E., et al. Lipid nanoparticles with minimum burst release of TNF-α siRNA show strong activity against rheumatoid arthritis unresponsive to methotrexate. J Control Release. 2018;283:280–289. doi: 10.1016/j.jconrel.2018.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Duan W., Li H. Combination of NF-kB targeted siRNA and methotrexate in a hybrid nanocarrier towards the effective treatment in rheumatoid arthritis. J Nanobiotechnology. 2018;16(1):58. doi: 10.1186/s12951-018-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun X., Dong S., Li X., Yu K., Sun F., Lee R.J., et al. Delivery of siRNA using folate receptor-targeted pH-sensitive polymeric nanoparticles for rheumatoid arthritis therapy. Nanomedicine. 2019;20 doi: 10.1016/j.nano.2019.102017. [DOI] [PubMed] [Google Scholar]
- 139.Jansen M.A.A., Klausen L.H., Thanki K., Lyngsø J., Skov Pedersen J., Franzyk H., et al. Lipidoid-polymer hybrid nanoparticles loaded with TNF siRNA suppress inflammation after intra-articular administration in a murine experimental arthritis model. Eur J Pharm Biopharm. 2019;142:38–48. doi: 10.1016/j.ejpb.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 140.Yin N., Tan X., Liu H., He F., Ding N., Gou J., et al. A novel indomethacin/methotrexate/MMP-9 siRNA in situ hydrogel with dual effects of anti-inflammatory activity and reversal of cartilage disruption for the synergistic treatment of rheumatoid arthritis. Nanoscale. 2020;12(15):8546–8562. doi: 10.1039/d0nr00454e. [DOI] [PubMed] [Google Scholar]
- 141.Wu H., Su S., Wu Y., Wu Y., Zhang Z., Chen Q. Nanoparticle-facilitated delivery of BAFF-R siRNA for B cell intervention and rheumatoid arthritis therapy. Int Immunopharmacol. 2020;88 doi: 10.1016/j.intimp.2020.106933. [DOI] [PubMed] [Google Scholar]
- 142.Li X., Yu C., Meng X., Hou Y., Cui Y., Zhu T., et al. Study of double-targeting nanoparticles loaded with MCL-1 siRNA and dexamethasone for adjuvant-induced arthritis therapy. Eur J Pharm Biopharm. 2020;154:136–143. doi: 10.1016/j.ejpb.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 143.Zhao M., Zhu T., Chen J., Cui Y., Zhang X., Lee R.J., et al. PLGA/PCADK composite microspheres containing hyaluronic acid-chitosan siRNA nanoparticles: a rational design for rheumatoid arthritis therapy. Int J Pharm. 2021;596 doi: 10.1016/j.ijpharm.2021.120204. [DOI] [PubMed] [Google Scholar]
- 144.Andersen C.K., Khatri S., Hansen J., Slott S., Parvathaneni R.P., Mendes A.C., et al. Carbon nanotubes-potent carriers for targeted drug delivery in rheumatoid arthritis. Pharmaceutics. 2021;13(4):453. doi: 10.3390/pharmaceutics13040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen X., Zhou B., Gao Y., Wang K., Wu J., Shuai M., et al. Efficient treatment of rheumatoid arthritis by degradable LPCE nano-conjugate-delivered p65 siRNA. Pharmaceutics. 2022;14(1):162. doi: 10.3390/pharmaceutics14010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cho Y., Jeong S., Kim H., Kang D., Lee J., Kang S.B., et al. Disease-modifying therapeutic strategies in osteoarthritis: current status and future directions. Exp Mol Med. 2021;53(11):1689–1696. doi: 10.1038/s12276-021-00710-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shin H.J., Park H., Shin N., Kwon H.H., Yin Y., Hwang J.A., et al. p47phox siRNA-Loaded PLGA nanoparticles suppress ROS/oxidative stress-induced chondrocyte damage in osteoarthritis. Polymers (Basel) 2020;12(2):443. doi: 10.3390/polym12020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hunter D.J., McDougall J.J., Keefe F.J. The symptoms of osteoarthritis and the genesis of pain. Rheum Dis Clin North Am. 2008;34(3):623–643. doi: 10.1016/j.rdc.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen L.X., Lin L., Wang H.J., Wei X.L., Fu X., Zhang J.Y., et al. Suppression of early experimental osteoarthritis by in vivo delivery of the adenoviral vector-mediated NF-kappaBp65-specific siRNA. Osteoarthritis Cartilage. 2008;16(2):174–184. doi: 10.1016/j.joca.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 150.Mutabaruka M.S., Aissa M.A., Delalandre A., Lavigne M., Lajeunesse D. Local leptin production in osteoarthritis subchondral osteoblasts may be responsible for their abnormal phenotypic expression. Arthritis Res Ther. 2010;12(1):R20. doi: 10.1186/ar2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wei F., Moore D.C., Wei L., Li Y., Zhang G., Wei X., et al. Attenuation of osteoarthritis via blockade of the SDF-1/CXCR4 signaling pathway. Arthritis Res Ther. 2012;14(4):R177. doi: 10.1186/ar3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chu X., You H., Yuan X., Zhao W., Li W., Guo X. Protective effect of lentivirus-mediated siRNA targeting ADAMTS-5 on cartilage degradation in a rat model of osteoarthritis. Int J Mol Med. 2013;31(5):1222–1228. doi: 10.3892/ijmm.2013.1318. [DOI] [PubMed] [Google Scholar]
- 153.Pi Y., Zhang X., Shao Z., Zhao F., Hu X., Ao Y. Intra-articular delivery of anti-Hif-2α siRNA by chondrocyte-homing nanoparticles to prevent cartilage degeneration in arthritic mice. Gene Ther. 2015;22(6):439–448. doi: 10.1038/gt.2015.16. [DOI] [PubMed] [Google Scholar]
- 154.Hoshi H., Akagi R., Yamaguchi S., Muramatsu Y., Akatsu Y., Yamamoto Y., et al. Effect of inhibiting MMP13 and ADAMTS5 by intra-articular injection of small interfering RNA in a surgically induced osteoarthritis model of mice. Cell Tissue Res. 2017;368(2):379–387. doi: 10.1007/s00441-016-2563-y. [DOI] [PubMed] [Google Scholar]
- 155.Akagi R., Akatsu Y., Fisch K.M., Alvarez-Garcia O., Teramura T., Muramatsu Y., et al. Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF-β signaling in chondrocytes. Osteoarthritis Cartilage. 2017;25(6):943–951. doi: 10.1016/j.joca.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao L.R., Xing R.L., Wang P.M., Zhang N.S., Yin S.J., Li X.C., et al. NLRP1 and NLRP3 inflammasomes mediate LPS/ATPinduced pyroptosis in knee osteoarthritis. Mol Med Rep. 2018;17(4):5463–5469. doi: 10.3892/mmr.2018.8520. [DOI] [PubMed] [Google Scholar]
- 157.Deshmukh V., O'Green A.L., Bossard C., Seo T., Lamangan L., Ibanez M., et al. Modulation of the Wnt pathway through inhibition of CLK2 and DYRK1A by lorecivivint as a novel, potentially disease-modifying approach for knee osteoarthritis treatment. Osteoarthritis Cartilage. 2019;27(9):1347–1360. doi: 10.1016/j.joca.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 158.Chen X., Liu Y., Wen Y., Yu Q., Liu J., Zhao Y., et al. A photothermal-triggered nitric oxide nanogenerator combined with siRNA for precise therapy of osteoarthritis by suppressing macrophage inflammation. Nanoscale. 2019;11(14):6693–6709. doi: 10.1039/c8nr10013f. [DOI] [PubMed] [Google Scholar]
- 159.Gong Y., Li S.J., Liu R., Zhan J.F., Tan C., Fang Y.F., et al. Inhibition of YAP with siRNA prevents cartilage degradation and ameliorates osteoarthritis development. J Mol Med (Berl) 2019;97(1):103–114. doi: 10.1007/s00109-018-1705-y. [DOI] [PubMed] [Google Scholar]
- 160.Shin H.J., Park H., Shin N., Shin J., Gwon D.H., Kwon H.H., et al. p66shc siRNA nanoparticles ameliorate chondrocytic mitochondrial dysfunction in osteoarthritis. Int J Nanomedicine. 2020;15:2379–2390. doi: 10.2147/IJN.S234198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu L., Wu Z., He Y., Chen Z., Xu K., Yu W., et al. MFN2 contributes to metabolic disorders and inflammation in the aging of rat chondrocytes and osteoarthritis. Osteoarthritis Cartilage. 2020;28(8):1079–1091. doi: 10.1016/j.joca.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 162.Bedingfield S.K., Colazo J.M., Yu F., Liu D.D., Jackson M.A., Himmel L.E., et al. Amelioration of post-traumatic osteoarthritis via nanoparticle depots delivering small interfering RNA to damaged cartilage. Nat Biomed Eng. 2021;5(9):1069–1083. doi: 10.1038/s41551-021-00780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sun Y., Leng P., Guo P., Gao H., Liu Y., Li C., et al. G protein coupled estrogen receptor attenuates mechanical stress-mediated apoptosis of chondrocyte in osteoarthritis via suppression of Piezo1. Mol Med. 2021;27(1):96. doi: 10.1186/s10020-021-00360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]