Highlights
-
•
BTV-17 infection in sheep showed sustained viral RNA detection.
-
•
Infectious virus detection diminished during height of immune response.
-
•
BTV-17 causes increased expression of cytokines IFN-γ and CXCL10.
-
•
CD8+ t cells expand concurrently with proinflammatory cytokine expression.
Keywords: Bluetongue virus, Immune response, Sheep, BTV-17
Abstract
Bluetongue virus (BTV) is an economically important pathogen of ruminant species with worldwide prevalence. While many BTV infections are asymptomatic, animals with symptomatic presentation deteriorate quickly with the sickest succumbing to disease within one week. Animals that survive the infection often require months to recover. The immune response to BTV infection is thought to play a central role in controlling the disease. Key to understanding BTV disease is profiling vertebrate host immunological cellular and cytokine responses. Studies to characterize immune responses in ruminants have been limited by a lack of species-specific reagents and assay technology. Here we assess the longitudinal immunological response to experimental BTV-17-California (CA) infection in sheep using the most up to date assays. We infected a cohort of sheep with BTV-17-CA and longitudinally monitored each animal for clinical disease, viremia and specific immunological parameters (B cells, T cells, monocytes) by RT-qPCR, traditional flow cytometry and/or fluorescent based antibody arrays. BTV-inoculated sheep exhibited clinical signs characteristic of bluetongue virus disease. Circulating virus was demonstrated after 8 days post inoculation (DPI) and remained detectable for the remainder of the time course (24 DPI). A distinct lymphopenia was observed between 7 and 14 DPI that rebounded to mock-inoculated control levels at 17 DPI. In addition, we observed increased expression of pro-inflammatory cytokines after 8 DPI. Taken together, we have established a model of BTV infection in sheep and have successfully monitored the longitudinal vertebrate host immunological response and viral infection progression using a combination of traditional methods and cutting-edge technology.
Importance
Bluetongue virus, the causative agent of bluetongue disease, is an economically important pathogen of ruminant species. Although 36 serotypes are endemic in regions across the world, emergence of novel serotypes into new regions can have devastating impact on the agriculture economy and local wildlife. Predicting these novel emergences would be challenging, yet understanding the immune factors that drive positive outcomes of the disease will permit a more effective and rapid response when they occur. Outcomes of infection vary based on a multitude of factors including, the serotype of the virus, the host species that is infected, as well as the age and immunological status of the infected animal. Understanding the immune mechanisms that the host species employs to successfully combat and resolve BTV infections is critical for the development of novel therapeutics to help combat this persistent and ever emerging disease.
1. Introduction
Bluetongue disease in ruminant species is caused by bluetongue virus (BTV) infection and is spread by Culicoides biting midge (Diptera: Ceratopogonidae) (Mellor et al., 2000). The evolving emergence of novel BTV serotypes contributes to the detrimental impacts of this disease. The emergence of novel BTV serotypes in the U.S. and around the world is a growing concern as climates change and midges occupy new territory; i.e. the geographical distribution of BTV will inevitably expand and infect previously naïve ruminant populations (Gibbs et al., 2008). To date, up to 36 serotypes of BTV have been identified with variable geographic distribution and disease causing capacity (Ries et al., 2021). Co-infection with multiple serotypes of virus can occur in endemic regions, compounding the complications of disease (Hemadri et al., 2017; Veronesi et al., 2020). In 2006 BTV-8, normally endemic to the sub-Saharan region, emerged in northern European countries resulting in extensive damage to local and global trade including substantial economic loss (Velthuis et al., 2010). A successful vaccine strategy has been developed for several of the BTV serotypes, yet the prevalence of asymptomatic infections and the abundance of infected wild ruminants that act as a reservoir for virus ensure that BTV infections will continue to circulate throughout the world and impact vulnerable populations.
The host immunological response plays a key role in combating and controlling the disease yet is also necessary in perpetuating the viral lifecycle (Maclachlan et al., 2014; Schwartz-Cornil et al., 2008). BTV utilizes the local host immune cells at the bite site, such as lymphocytes, monocytes, dendritic cells and macrophages, to replicate and disseminate the virus to secondary sites of infection. Although these cells are necessary to promote infection, it has been shown that B and T cell lymphocytes are critical for controlling and eliminating the virus. The protection conveyed by T cells is illustrated by complete protection in animals that have received BTV specific T cell adoptive transfer (Jeggo et al., 1985). Additionally, B cells provide protection through the humoral response and are potently effective at preventing BTV disease in ruminants (Jeggo et al., 1983). Driving these cellular responses, cytokine production also plays a critical role in resolution and disease severity (Maclachlan et al., 2014; Schwartz-Cornil et al., 2008). It has been speculated that the differences in disease severity associated with different BTV serotypes is directly related to cytokine expression (Maclachlan et al., 2014). Although viral and immune cell interactions have been broadly characterized for BTV, these studies have been limited by lack of species-specific reagents and assay technology.
Here we report a characterization of sheep experimentally inoculated with BTV-17-CA monitored longitudinally over a 26 day time course. We evaluated clinical symptoms, viremia, serological response, cellular (i.e. T cells, B cells, CD14+ monocytes and neutrophils) and non-cellular (i.e. cytokine and chemokine) immune changes, and blood chemistry (i.e. liver enzymes).These data together will provide a comprehensive evaluation of viral and host changes over the course of BTV-17-CA infection in its natural host.
2. Materials & methods
2.1. Animal care and husbandry
Fifteen female Rambouillet sheep, ranging from 3 to 8 years of age. The range in age of sheep used for these studies was due to the necessity for stringent conditions to determine cohort inclusion. Prior to study start, ewes were screened for current and/or historic exposure to BTV by RT-qPCR and serology. The ewes were also screened for pregnancy to further reduce confounding variables to the study. At study start (Day 0), the animals were subcutaneously-inoculated with 1 mL media (mock-inoculated) or BTV-17 (USA1988/CA also known as BTV-17-CA, at a titer of 1.4 X 106 TCID50 mL−1). Female animals were chosen for housing purposes, and they have been shown to be more affected by BTV infection (Mahmoud et al., 2019). The animals were screened for presence of BTV prior to enrollment in the study and housed indoors where there was no access to wild insects. One mock-inoculated animal was cohoused with BTV-inoculated sheep and served as a negative contact transmission control. Animals were monitored for clinical signs throughout the experiment and a daily clinical score was recorded for each animal for the following criteria: (1) respiratory symptoms, (2) behavior changes (apathy, lethargy), (3) nasal/eye discharge, (4) ulcers (oral and/or nasal), (5) salivation, (6) facial edema. All signs were scored on a scale from 1 (very mild), 2 (moderate) to 3 (severe). Daily body temperatures were recorded for all animals. Animals were humanly euthanized at day 26 post inoculation. All experiments were carried out in accordance with the CSU IACUC and with approved animal protocols (IACUC #1400). EDTA blood and serum samples were taken from the jugular vein at regular intervals.
2.2. BTV virus isolate and propagation
The BTV-17 (Order Reovirales, family Reoviridae, genus Orbivirus, species Bluetongue virus) isolate used in this study is a characterized field isolate from California (GenBank, MT952971-MT952980) and has been used in previous studies (Kopanke et al., 2020). Briefly, it was isolated from whole blood harvested from a naturally occurring BTV infection in a clinically affected sheep. Prior to initiation of the current experiment, the BTV-17-CA isolate was expanded as previously described in BHK 21 cells (Clavijo et al., 2000). Use of the virus was limited to one passage post expansion and infectious titer was determined via 50 % tissue culture infectious dose (TCID50) via the Reed-Muench method.
2.3. BTV competitive elisa
Sera collected from all experimental animals were tested for anti-BTV antibodies using a BTV-specific VP7 cELISA kit (Veterinary Medical Research and Diagnostics, Bluetongue Virus Antibody Test Kit, Inc., Pullman, US) and performed according to the instructions of the supplier. Positivity threshold was set at 60 % inhibition. BTV was not detected in environmental and negative mock-inoculated controls at any time point.
2.4. Longitudinal PCR viral detection in blood and terminal tissues (spleen and lung)
Whole blood (EDTA) was collected from each animal at regular intervals (daily post inoculation for 7 days, then every third day until the end of study). Detection of BTV viral RNA was performed as previously described (Ortega et al., 2010; Hofmann et al., 2008). Viral genomic material was isolated using MagMAX Pathogen RNA/DNA kit (Applied Biosystems, Foster City, CA, USA) per the manufacturer's instructions. BTV specific primers (BTV-Fwd:5′-TGGAYAAAGCRATGTCAAA −3′, BTV Rev: 5′-ACRTCATCACGAAACGCTTC-3′, BTV-Probe 5′/56-FAM/ARGCTGCAT/ZEN/TCGCATCGTACGC/3IABkFQ/3′) were used in an RT-qPCR reaction using SuperScript™ III Platinum One-step RT-qPCR kit (Invitrogen, Carlsbad, CA, USA). Samples were considered positive if the cycle to threshold (Ct) value was below 35. Infectious virus was determined by isolating virus from collected peripheral blood and CPE development when grown with BHK-21 cells, as previously described (Kopanke et al., 2020). Viral genomic detection from terminal tissues was performed by Qiagen kit isolation, and the RT-qPCR was performed using the same primers and methodology.
2.5. Flow cytometry, complete blood counts, and blood chemistry
The following anti-sheep antibodies were used according to manufacturer's specifications: CD8α (clone ST8, Cat# SHP2002, WSU), CD4 (clone S-17D, Cat #S-GT2002, WSU), CD72 (clone 2–104) (Young et al., 1997), CD14 (clone VPM65, Cat #MCA920GA, Bio-rad), FITC conjugated anti-mouse (Cat# 1071–02, Southern Bio), BV421 conjugated anti-mouse (Cat# 115–675–075, Jackson), & PE conjugated anti-mouse (Cat# Ab970024, Abcam). Sheep EDTA-blood was cleared of RBCs by red blood cell lysing buffer (ammonium chloride) for 3 min at room temperature and washed repeatedly with PBS. Terminal tissues were passed through a 100 μm cell strainer (Corning Life Sciences) and splenic tissues was cleared of RBCs by red blood cell lysing buffer (ammonium chloride 154.4 mM). The dissociated cells were incubated with the corresponding antibodies followed by incubation with a secondary antibody conjugated fluorophores for 30 min to 1 h at room temperature and washed with PBS. Samples were fixed in 2 % paraformaldehyde for 5 min, washed and resuspended in PBS. Samples were passed through a 35 μm cell strainer (Corning Life Sciences) immediately before analysis on an Aurora spectro-cytometer (Cytek) (Becton Dickinson). Data was analyzed using FlowJo (v.10) software.
Aliquots of EDTA-blood were submitted to the CSU Veterinary Diagnostic Laboratory for complete blood counts and blood chemistry. Analysis was sent as a report and data was collated by the researchers.
2.6. Cytokine array
The ovine cytokine array was performed on serum collected at indicated timepoints as per the manufacturer's guidelines (Raybiotech, Cat# QAO-CYT-1, Peachtree Corners, GA, USA). Fluorescent data scanning was performed by Raybiotech scanning services and raw data was provided for analysis and data collation. Serum cytokine biomarkers analyzed include CXCL10, IFN γ, and TNFα. Each sample was performed in at least triplicate (with four detection replicates per well). Concentrations were determined by a standard curve.
3. Results
3.1. Sheep experimentally inoculated with BTV-17-CA demonstrate detectable viremia and infectious virus throughout the study time course
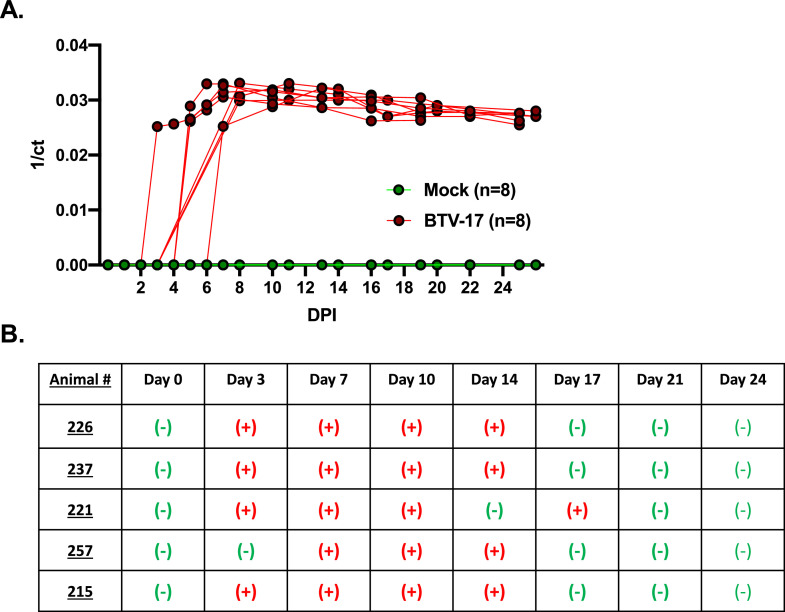
To evaluate the impact and response to BTV-17-CA infection, a cohort of sheep were experimentally inoculated with BTV-17 USA1988/CA, also named BTV-17-CA (hereafter referred to as BTV-17). A separate cohort of sheep was mock-inoculated to serve as negative controls. Peripheral blood was collected from the animals daily for the first week post inoculation, and every third day thereafter until study termination. BTV-17 viral genetic material was assessed by reverse transcription-quantitative PCR (RT-qPCR) using BTV specific primers (Fig. 1A). BTV-17 viral genetic material was detected in blood as early as 2 days post inoculation (DPI), with all animals exhibiting maximal viral levels by 8 DPI. Interestingly, the detection of BTV-17 viral genetic material in peripheral blood remained robust throughout the longitudinal study time course. As expected, none of the mock-inoculated animals showed positivity at any time point. To evaluate if the virus detected in the peripheral blood was infectious, we cultured BHK-21 cells with lysates of blood collected from the BTV-17 inoculated animals at 0, 3, 7, 10, 14, 17, 21, and 24 DPI (Fig. 1B). We found that RT-qPCR positive animals showed detectable viral plaques (infectious virus) as early as 3 DPI and a single positive animal as late at 17 DPI. We evaluated two mock-infected animals at the same time points and found no detectable plaques. Taken together, these data demonstrate active BTV-17 infection in sheep with robust infection over a twenty-six day time course.
Fig. 1.
Sheep experimentally inoculated with BTV-17 exhibited detectable viremia and infectious virus during the experimental time course. (A) Detection of BTV-17 genomes by RT-qPCR. Peripheral blood was drawn and processed for BTV detection. BTV was not detected in environmental and negative mock-inoculated controls at any time point. Assay used a threshold of 0.04 and the controls were valid. Data is represented as 1 over the count to threshold (Ct). (B) Blood was processed for infectious BTV virus. Blood samples were lysed and virus was propagated on BHK-21 cells until clear CPE was observed. An additional round of propagation was performed to confirm BTV as the causative agent of the CPE. Negative (-) and positive (+) represent no clear CPE detected and CPE detectable respectively from samples taken at that time point.
3.2. Sheep experimentally infected with BTV-17 exhibit clinical signs and mount an acute humoral response to infection
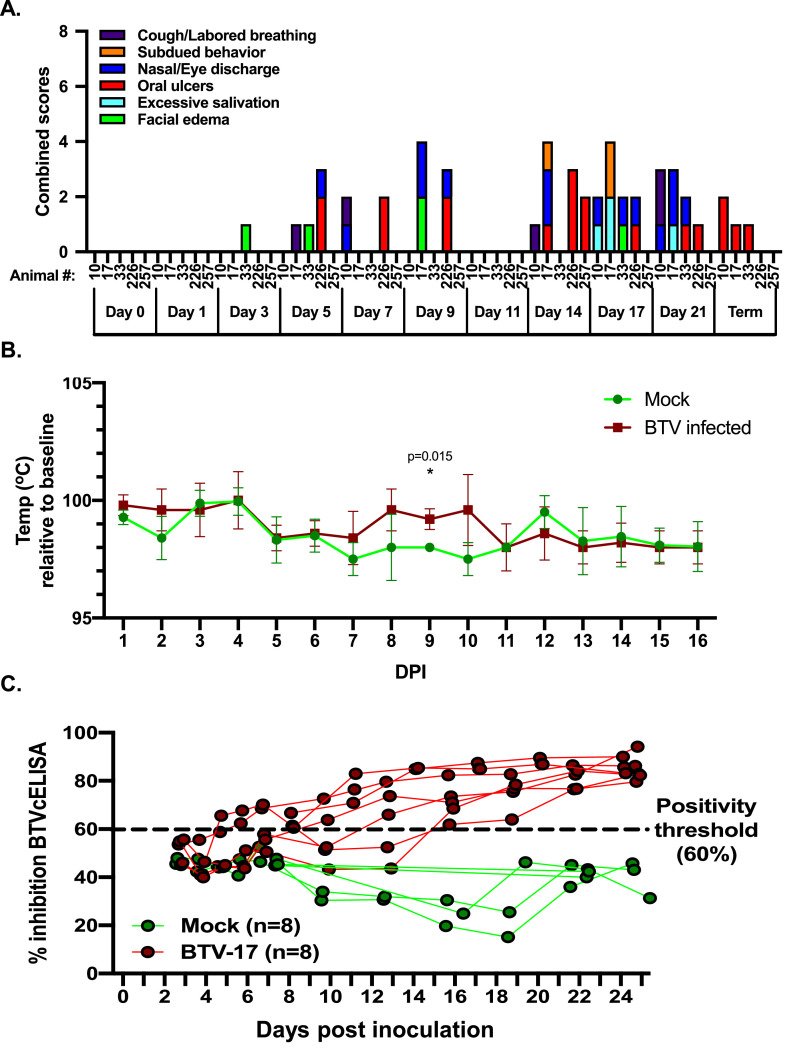
Animals were monitored and evaluated daily for signs of clinical disease (Fig. 2A & B). Five of the eight (5/8) RT-qPCR positive sheep exhibited clinical disease consistent with BTV infection. Modest facial edema was observed as early as 3 DPI. A discernable increase in body temperature was observed in the infected animals at 7 DPI and remained elevated for 3 days prior to returning to body temperatures recorded in mock-inoculated animals. The most pronounced clinical symptoms were observed 2 weeks post inoculation, with all animals exhibiting multiple symptoms, including excessive salivation, lethargy, eye and nasal discharge. While the majority of these symptoms resolved, oral ulcerations were observed until the end of the study.
Fig. 2.
Experimental sheep were infected with BTV-17 and revealed moderate clinical signs of infection. (A) Animals were evaluated for: respiratory symptoms, behavior changes (apathy, lethargy), nasal/eye discharge, ulcers (oral and/or nasal), salivation, and facial edema. All signs were scored on a scale from 1 (very mild), 2 (moderate) to 3 (severe). Values of each are represented as additive per animal at each time point. (B) Animal temperature was recorded using a rectal thermometer at time of blood collection. (C) Serum taken at blood draw time points was evaluated for detection of anti-BTV antibodies using a BTV specific competitive ELISA (cELISA). A sample was considered positive if it was above 60 % inhibition of signal threshold.
To evaluate if the infected sheep mounted an acute humoral response to the BTV-17 infection, a BTV inhibition competitive ELISA was performed using sheep serum (Fig. 2C). At 7 DPI, a clear separation began to emerge between BTV-17 and mock-inoculated animals, with all BTV-17 inoculated animals crossing the 60 % positivity threshold by 16 DPI. These data reveal that the BTV-17 inoculated sheep exhibited clear signs of BTV infection and mounted a humoral response to the infection.
3.3. Experimentally infected sheep exhibited dynamic cellular (CD4, CD8, CD14 and neutrophil) immune responses over time
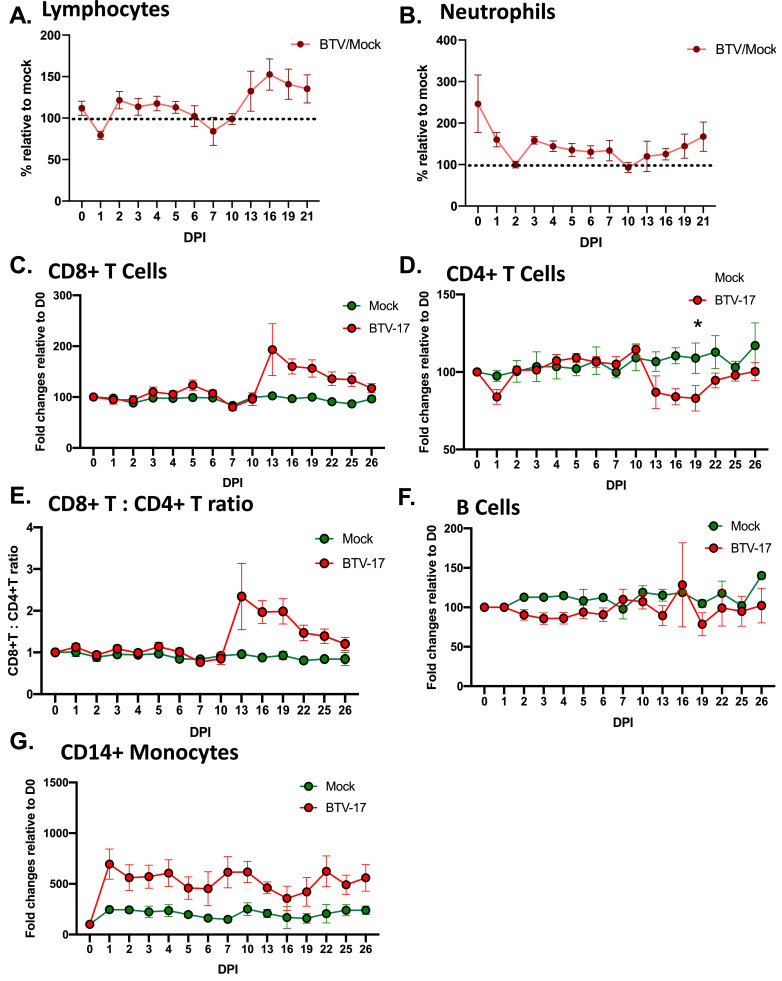
Previous studies have shown that BTV infections can result in leukopenia seven to ten days into the infection. Using flow cytometry and complete blood cell counts (CBCs), we monitored several immune populations that are known to be critical to infection. T cell responses have been cited as critical for clearing infection and producing sufficient memory response to protect animals via adoptive transfer. CBC analysis of broader cell populations revealed a stark lymphopenia one day after inoculation and a second lymphopenia at 7 DPI (Fig. 3A). A reduction in lymphocyte population during BTV infection has been previously described (Ellis et al., 1990). A decline in neutrophil cell population was observed in our study animals, however, due to high neutrophil cell counts at day 0 in the BTV-17 inoculated cohort, they appear elevated as compared to the mock-inoculated cohort (Fig. 3B). The decline in neutrophil counts occurred at 2 and 10 DPI, lagging behind the decline in lymphocyte cell counts. Interestingly, a rebound in neutrophil cell counts occurs at the time of lymphocyte recovery. We found that CD8+ T cells modestly declined by 7 DPI (Fig. 3C) with a robust rebound and expansion near the end of the study. Interestingly, CD4+ T cells declined at 10 DPI, as was observed for CD8+ T cells (Fig. 3D). The CD4+ T cell population remained significantly reduced for the remainder of the study. This is in stark contrast to the observed expansion of CD8+ T cells at roughly the same time of evaluation. We also evaluated CD8+ and CD4+ T cell populations as a ratio of each other (Fig 3E). We observed a pronounced increase in the ratio at 13 DPI, suggesting a strong CD8+ T cell expansion compared to the CD4+ T cell population. The other major cell class of lymphocytes in the blood, B cells, exhibited a modest reduction in cell numbers at the same timeframe as the T cell populations but remained not significantly changed throughout the infection time course (Fig. 3F). These data suggest CD8+ T cells are the most dynamically influenced lymphocyte population during BTV-17 infection in sheep.
Fig. 3.
Experimentally infected sheep exhibited dynamic immune responses over the study time course. Sheep peripheral blood was analyzed for specific cell subsets using flow cytometry and cell specific antibodies. Broader cell populations, as indicated by complete blood counts, show (A) lymphocytes, and (B) neutrophils cell counts. CBC data is shown as BTV-17 infected relative to mock (set to 100 %) for each timepoint. Specific cell populations were identified by flow cytometry (C) CD8+ T cells, (D) CD4+ T cells, the CD8+/CD4+ T cells ratio (E), (F) B cells, (G) and CD14+ monocytes were detected and evaluated. Data is shown as mock infected (green) and BTV-17 infected (red). To account for animal-to-animal variation, all data points were standardized to signal at day 0..
Although many myeloid cells play critical roles during BTV infection, monocytes have been shown to shuttle the virus throughout the animal from the bite site (Barratt-Boyes and MacLachlan 1994). We found CD14+ monocytes were significantly elevated after just 2 DPI and remained elevated for roughly two weeks post inoculation (Fig. 3G). This elevation in monocytes remained above that recorded for the mock-inoculated animals throughout the study time course. From our observations, the CD14+ monocytes did not undergo the population suppression observed in the other lymphocyte immune populations. Taken together, these data provide evidence for compelling trends of leukocyte populations modifications during BTV infection. Although the described leukopenia was observed in many cases, our findings show the population changes to be more dynamic when evaluated at the specific cell subset level. Furthermore, these data suggest that CD8+ T cell, CD14+ monocyte, and neutrophil immune cell populations may play a key role in controlling or perpetuating BTV-17 infection in sheep.
3.4. Blood chemistry and cytokine changes during BTV infection in sheep
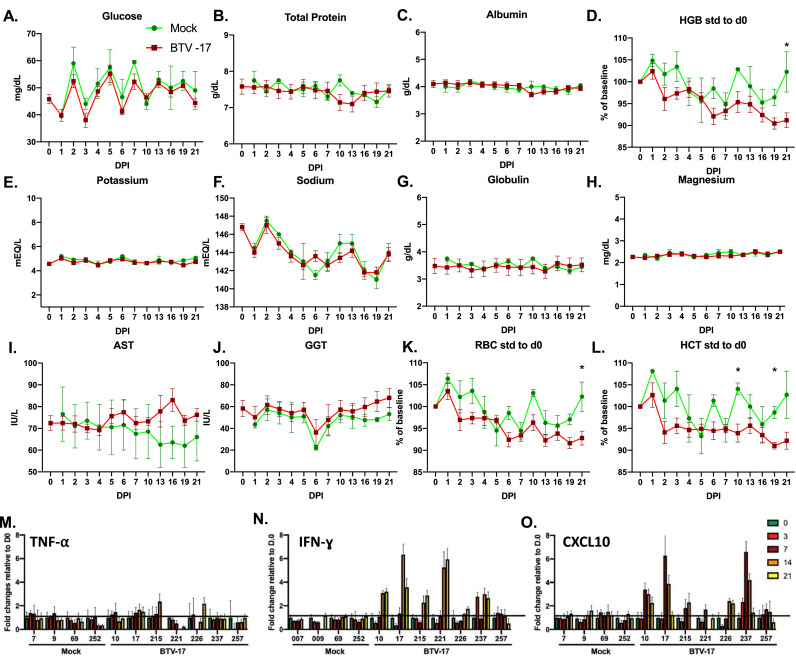
Blood chemistry panels revealed near identical profiles between the BTV- and mock-inoculated cohorts (Fig. 4A–G). Exceptions to this include liver enzymes (aspartate transaminase (AST), glutathione S-transferases (GGT), and components related to erythrocytes (hemoglobin (HGB), red blood cell count (RBC), and hematocrit (HCT)) (Fig. 4H–L). Liver enzymes exhibited a clear departure from the mock-inoculated animals at 10 to 13 DPI. HGB, RBC, and HCT revealed dynamic changes throughout the study. As the handling and timing of blood collections was consistent for all animals, differences between the BTV- and mock-inoculated cohorts are most likely infection related. A decline in HGB, RBC, and HCT values at 2 DPI remained below levels reported in mock-inoculated animals throughout the study. The most significant difference in these parameters was noted at 10 DPI, with levels in mock-inoculated animals returning to baseline while the levels in BTV-inoculated animal remained reduced.
Fig. 4.
Blood chemistry and cytokine changes during BTV infection in sheep. (A–L) Blood chemistry performed by the CSU-VDL was performed on peripheral blood taken at each time point. Data is shown as the average of BTV-17 inoculated animals relative to the average mock-infected animals. (M–O) Serum cytokines TNFα, IFN γ, and CXCL10 were evaluated from the peripheral blood of three negative and five positive animals at day 0, 3, 7, 14, 21, and 28 using the Raybiotech ovine sheep cytokine array. Data is shown as RFU with background signal removed.
Cytokines play a key role in the progression and ultimate outcome of disease (Maclachlan et al., 2014; Schwartz-Cornil et al., 2008). It has been suggested that variation in cytokine induction by BTV serotypes may contribute to the divergent disease severity outcomes observed in BTV infections. Vasoactive cytokines, such as TNFα and IL-1, are increased dependent upon the BTV serotype responsible for infection and are suspected to be responsible for increased vascular permeability resulting in tissue injury (Sánchez‐Cordón et al., 2013). Disease resolution has been associated with high expression of cytokines that assist cell mediated immunity, including IFN-γ, and IL-2 (Maclachlan et al., 2014; Schwartz-Cornil et al., 2008; Bitew et al., 2019). Using a commercially available ovine serum cytokine kit, we evaluated 9 cytokines associated with infection. We evaluated 4 mock- and 7 BTV-inoculated sheep at 0, 3, 7, 14, and 21 DPI. Interestingly, the majority of cytokines were not significantly elevated above their mock-inoculated counterparts. A modest induction of TNFα response was detected at 14 DPI (Fig. 4M). The most robust change observed was in the proinflammatory cytokines IFN-γ and CXCL10 in most (5/6) of the BTV-inoculated animals (Fig. 4N-O). Both cytokines showed significant induction at 14 DPI. This is the time point at which CD8+ T cells demonstrated a marked expansion. Taken together, both the blood chemistry and cytokine profiles exhibit clear differences particularly at the time of CD8+ T cell expansion, suggesting their potential role in establishing control of the infection.
3.5. Evaluation of terminal tissues harvested from BTV infected sheep
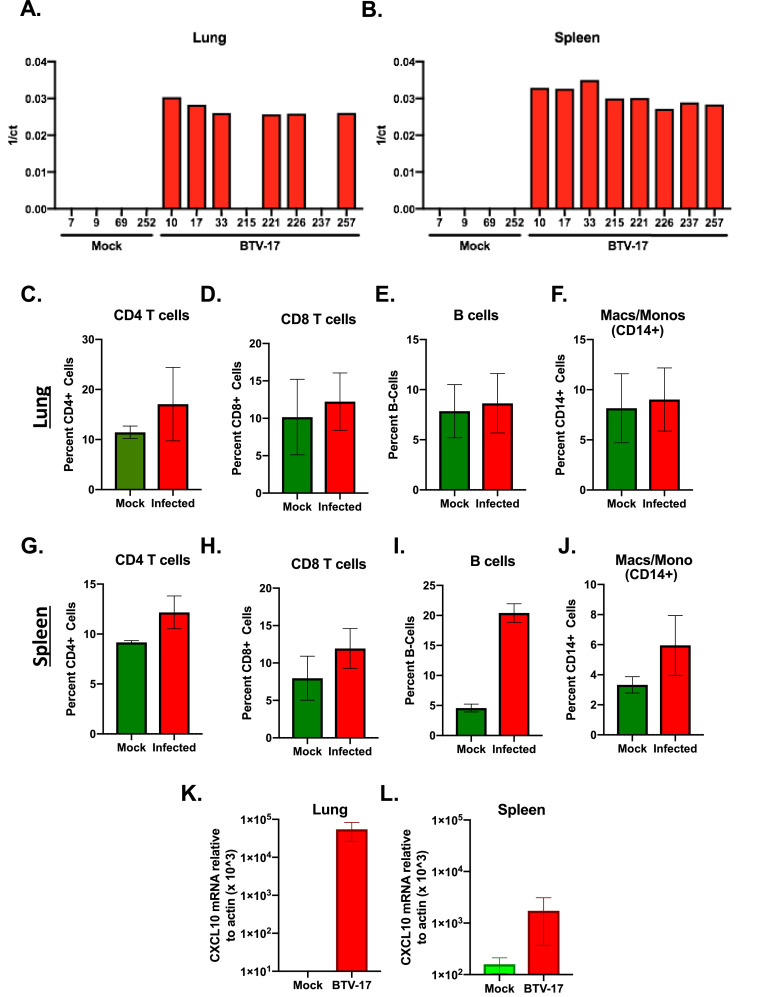
At the conclusion of the study, the animals were euthanized, and several tissues were evaluated. BTV viral detection was still highly detectable in the blood at the time of sacrifice (Fig. 1A). Samples of lung and spleen, tissues previously described to harbor BTV (Saminathan et al. 2020a), were evaluated for BTV genomic detection by RT-qPCR (Fig. 5A & B). Although all positive animals exhibited robust BTV detection in splenic tissues, 2 of the positive animals showed no detectable BTV-17 RNA in their sampled lung tissue. No detection was observed in mock-inoculated animals. To evaluate immune cell populations in these tissues, we analyzed lung and spleen tissues by flow cytometry using the same panel of antibodies used to detect blood immune cell populations (Fig. 3). We found the lung exhibited modest increases in all immune cell types but not to a significant degree (Fig. 5C–F). In spleen harvested from BTV-infected animals we observed significant increases in CD4+ T cells, B cells, and CD14+ cells populations in BTV-infected animals over mock controls (Fig. 5G–J). As CXCL10 was significantly elevated in the blood of infected animals, we also evaluated CXCL10 transcript levels in lung and splenic tissues (Fig. 5K & L). We observed a robust induction of CXCL10 transcript in both tissues, correlating with our observations in the blood. These data revealed terminal tissues, specifically lung and spleen, have robust BTV viral genome detection, exhibit increased cellular immune populations, and show an increase in CXCL10 production. Taken together, at the conclusion of our study, the BTV infected animals are still in the process of combating the infection.
Fig. 5.
Evaluation of terminal tissues of BTV infected sheep. RT-qPCR was performed on terminal lung (A) and spleen (B) tissue samples of three positively infected animals using BTV specific primers. Immune populations (CD8+ T, CD4+ T, B cells, and CD14+ monocytes were evaluated in lung (C–F), and spleen (G–J) of terminal collected tissues. (K, L) CXCL10 expression in terminal lung and spleen tissue of both mock and BTV-17 infected animals was determined by RT-qPCR using ovine CXCL10 specific primers.
4. Discussion
Although bluetongue virus has been recognized as the causative agent for bluetongue disease for over a hundred years, encroachments of novel serotypes into previously naïve territory represents a significant concern for both domestic and wild ruminant populations. These encroachments are becoming more frequent as the Culicoides vector habitable zone is expanding as a result of climate change. Between 2006 and 2009, BTV-8, normally endemic to the Saharan region emerged in northern Europe (Szmaragd et al., 2010; Bessell et al., 2016). This resulted in extensive economic losses to the agricultural industry as thousands of farms in prior naïve regions were impacted. Prevention of the movement of existing and novel BTV serotypes into new territories may not be possible. Thus, understanding the mechanisms of BTV disease and immune cells critical to perpetuation or control of disease is necessary for the development of effective therapeutics. In this study we evaluated cohorts of mock- and BTV-17-inoculated sheep longitudinally over 26 days. Here we characterize the longitudinal progression of BTV-17 infection in sheep across a spectrum of immunological, virological, and pathological metrics. We observed clinical signs associated with the disease and detected BTV genome burden in peripheral blood collected from all BTV-inoculated animals by RT-qPCR starting as early as 2 DPI that was maintained until the end of the study. The prolonged detection of BTV viral RNA is not unexpected as BTV viral RNA was shown to be detectable as late as 222 DPI (Bonneau et al., 2002). Interestingly, robust virus detection was demonstrated in animals that exhibited few clinical signs, suggesting BTV infections with low viral burdens. We were also able to detect the presence of infectious virus in the peripheral blood of most infected animals (Fig. 1B) for up to 17 DPI. While we were unable to identify infectious virus after this point, viral genome positivity remained consistent to study termination, suggesting an interesting disconnect between viral genomes and infectious virus in the peripheral blood. Potentially contributing to this disparity, an effective neutralized humoral response against BTV was observed as early as 5 DPI, with all animals showing 60 % neutralization by day 16. An additional curious finding was that two of the BTV-17 inoculated sheep exhibited no detectable BTV viral genomes in their lungs (Fig. 5A), despite robust detection in both their peripheral blood and spleen (Fig. 1A & 5B). Although these sheep did not differ in terms of clinical presentation, immune cell populations or cytokine levels, it is possible that at this stage of infection BTV was highly localized and the tissue we evaluated was not harboring the virus. An alternative explanation is that BTV was being cleared from the tissue at our terminal time point, and we captured the initial stages of recovery. The former is more likely, as BTV has been shown to be harbored in both blood and tissues for extended periods of time (Worwa et al., 2010).
Several studies have evaluated aspects of the naïve host immune response and pathogenic course of infection to BTV. BTV-1 and BTV-8 are the most commonly studied in these models (Bitew et al., 2019; Ruscanu et al., 2012; Sánchez‐Cordón et al. 2015; Melzi et al., 2016; Rojas et al., 2017; Ratinier et al., 2016; Drolet et al., 2015; Breard et al., 2015), however BTV-2, 4, 9, 10, 12, 16, and 23 have also been evaluated using systems including human cell culture, mouse models and sheep infections (Bitew et al., 2019; Ruscanu et al., 2012; Breard et al., 2015; Putty et al., 2019; Yang et al., 2020b, Dhanasekaran et al., 2013). Understanding BTV infections has been complicated by the use of several model systems and BTV serotypes, making comprehensive conclusions about the impact of BTV immune responses challenging. A single study from 1990 that evaluated the cellular immune response to BTV-17 in a small cohort of sheep observed reduction of T lymphocytes, mild clinical presentation and consistent viremia detection in agreement with our findings (Ellis et al., 1990). Studies evaluating several BTV serotypes in sheep have also shown consistent trends in cellular population modifications. Leukopenia has been reported in sheep experimentally infected with BTV-1, 8, and 9 (Sánchez‐Cordón et al. 2013, Chatzinasiou et al., 2017). Interestingly, BTV-related leukopenia has also been reported in interferon deficient mouse models of infection (Saminathan et al. 2020b; Saminathan et al., 2021). This immune suppression is thought to be driven by follicular dendritic cells as was observed in a sheep model of BTV-8 infection (Melzi et al., 2016). We observed dynamic changes in the CD14+ cell population in our BTV-17 inoculated animals (Fig. 3G). Interestingly, monocytes have long been known to be a target cell population for BTV infection (Whetter et al., 1989), with some data suggesting that activation of monocytes may be, in part responsible for vascular injury (Drew et al., 2010). Future studies to determine the BTV viral load in CD14+ cell populations would be of interest. A narrowing of T cell repertoire has been shown in sheep infected with BTV-8 (Rojas et al., 2017), exemplifying further broad changes to the adaptive immune cell compartment. We also observed lymphopenia, with CD4+ T cells showing the most dynamic changes over the disease course (Fig. 3D). And while less dramatic, in our study CD8+ T cells also exhibited a decline in population (Fig. 3C) that rebounded and continued to expand throughout the study. A previous study (Bitew et al., 2019) had shown expansion of immune cell populations in response to vaccination across several BTV serotypes (BTV-1, −2, −10, −16, and −23) suggesting that these cell populations are key to establishing disease protection. Our data collectively shows that specific immune cell subsets are more responsive to BTV mediated leukopenia, and that the expansion of T cell populations, specifically CD8+ T cells, may be necessary to combat BTV infection.
Several studies have also been performed to evaluate the role of non-cellular immune factors (cytokines and chemokines) during BTV infection. Interleukin 4 (IL-4) promotes the differentiation of naïve T cells to the T helper type 2 (Th2) cells. The Th2 response has been shown to be induced upon BTV infection (Sánchez‐Cordón et al. 2013; Yang et al., 2020b; Rodríguez-Martín et al., 2021; Kopanke et al., 2020). IL-4 has been shown to be specifically induced by the BTV viral protein VP2 (Yang et al., 2020b) suggesting a deliberate mechanism where BTV drives the immune response away from an antiviral Th1 response. The Th2 response is critical for supporting a humoral immune response and IL-4 induction in BTV infections has been shown to increase throughout recovery (Sánchez‐Cordón et al. 2013). This may ultimately help the animal clear the infection over time and establish potent immunity against reinfection. Our cytokine array analysis reveals that the most consistently induced include IFN-γ and CXCL10. Although IFN-γ is typically associated with a robust cellular immune response to BTV (Bitew et al., 2019), to date no studies have evaluated how CXCL10 may shape the expanded immune response following the lymphopenia we and others have observed. Two studies evaluating whole transcriptome changes with BTV-1 and BTV-8, have noted a marked increase in CXCL10 expression (Rojas et al., 2017; Lu et al., 2022). Given the conserved induction of CXCL10 across serotype, it would be tempting to speculate that it is involved in BTV spread throughout the host. However, at least at the initial stages of infection, when combined with the infectious bite of a Culicoides midge, CXCL10 does not appear to be induced (Pages et al., 2014). Therefore, given the timing of the CXCL10 induction in our system, it may play a role in controlling the BTV infection. Interestingly, the induction of these cytokines occurs just prior to the expansion of CD8+ and to a lesser degree CD4+ T cells populations. Given that these cytokines play a critical role in helping initiate T cell responses, it would be interesting to evaluate their role in the expansion of the suppressed immune populations and the role they play in the overall outcomes of these infections in the sheep. To further understand the impact of CXCL10, identifying the source (or sources) of the CXCL10 response will be important. Several immune cell types including neutrophils, eosinophils, monocytes, as well as non-immune cells such as epithelial, endothelial, stromal and keratinocytes can secrete CXCL10 (Liu et al., 2011), many of which are known to harbor BTV infection. Given the role of CXCL10 as promoting T cell and macrophage migration and adhesion to endothelial cells, a principal target of BTV, it is a promising factor to evaluate as a potential signal that ultimately allows for the host immune response too rebound and combat the BTV infection.
5. Conclusion
In summary, here we show that both the cellular and non-cellular immune responses are critical to combat BTV-17 infections. Specifically, the increased expression of CXCL10 and IFN-γ preceding the expansion of CD8+ T cells, suggest key immune responses needed to control the infection. The establishment of our native host sheep BTV infection system permits further exploration of the interconnection between virus and host factors important to the development of therapeutics and control of BTV infections in ruminant populations impacted by this disease.
Compliance with ethics guidelines
This study was carried out in accordance with the CSU IACUC and with approved animal protocols (IACUC #1400).
CRediT authorship contribution statement
Joseph A Westrich: Investigation, Methodology, Writing – original draft, Conceptualization. Erin E McNulty: Investigation, Methodology, Writing – review & editing, Resources. Molly Carpenter: Investigation, Writing – review & editing. Mollie Burton: Investigation, Writing – review & editing. Kirsten Reed: Investigation, Writing – review & editing, Resources, Formal analysis. Amy Nalls: Writing – review & editing, Resources. Audrey Sandoval: Investigation, Writing – review & editing. Christie Mayo: Funding acquisition, Project administration, Supervision, Writing – review & editing, Conceptualization. Candace K Mathiason: Funding acquisition, Project administration, Supervision, Writing – review & editing, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by the USDA-NIFA AFRI (Grant Number 2019–67015–28982) as part of the joint USDA-NSF-NIH-BBSRC-BSF Ecology and Evolution of Infectious Diseases program, and from the Colorado State University College of Veterinary Medicine and Biomedical Sciences College Research Council/ USDA Animal Health and Disease (AHD) (project number COLV 2021–02). They authors have no conflicts of interest to declare.
Data availability
Data will be made available upon request.
References
- Barratt-Boyes S.M., MacLachlan N.J. Dynamics of viral spread in bluetongue virus infected calves. Vet. Microbiol. 1994;40(3–4):361–371. doi: 10.1016/0378-1135(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Bessell P.R., Searle K.R., Auty H.K., Handel I.G., Purse B.V., Bronsvoort B.M. Assessing the potential for Bluetongue virus 8 to spread and vaccination strategies in Scotland. Sci. Rep. 2016;6(1):1–3. doi: 10.1038/srep38940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitew M., Ravishankar C., Chakravarti S., Kumar Sharma G., Nandi S. Comparative evaluation of T-cell immune response to BTV infection in sheep vaccinated with pentavalent btv vaccine when compared to un-vaccinated animals. Vet. Med. Int. 2019;2019 doi: 10.1155/2019/8762780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau K.R., DeMaula C.D., Mullens B.A., MacLachlan N.J. Duration of viraemia infectious to Culicoides sonorensis in bluetongue virus-infected cattle and sheep. Vet. Microbiol. 2002;88(2):115–125. doi: 10.1016/s0378-1135(02)00106-2. [DOI] [PubMed] [Google Scholar]
- Breard E., Belbis G., Viarouge C., Nomikou K., Haegeman A., De Clercq K., Hudelet P., Hamers C., Moreau F., Lilin T., Durand B. Evaluation of adaptive immune responses and heterologous protection induced by inactivated bluetongue virus vaccines. Vaccine. 2015;33(4):512–518. doi: 10.1016/j.vaccine.2014.11.053. [DOI] [PubMed] [Google Scholar]
- Chatzinasiou E., Chaintoutis S.C., Dovas C.I., Papanastassopoulou M., Papadopoulos O. Immunosuppression in sheep induced by cyclophosphamide, bluetongue virus and their combination: effect on clinical reaction and viremia. Microb. Pathog. 2017;104:318–327. doi: 10.1016/j.micpath.2017.01.048. [DOI] [PubMed] [Google Scholar]
- Clavijo A., Heckert R.A., Dulac G.C., Afshar A. Isolation and identification of bluetongue virus. J. Virol. Methods. 2000;87(1–2):13–23. doi: 10.1016/s0166-0934(00)00150-6. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran S., Vignesh A.R., Raj G.D., Reddy Y.K., Raja A., Tirumurugaan K.G. Comparative analysis of innate immune response following in vitro stimulation of sheep and goat peripheral blood mononuclear cells with bluetongue virus–serotype 23. Vet. Res. Commun. 2013;37:319–327. doi: 10.1007/s11259-013-9579-5. [DOI] [PubMed] [Google Scholar]
- Drew C.P., Heller M.C., Mayo C., Watson J.L., MacLachlan N.J. Bluetongue virus infection activates bovine monocyte-derived macrophages and pulmonary artery endothelial cells. Vet. Immunol. Immunopathol. 2010;136(3–4):292–296. doi: 10.1016/j.vetimm.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet B.S., Reister L.M., Lehiy C.J., Van Rijn P.A., Bowen R.A. Effect of culicoides sonorensis salivary proteins on clinical disease outcome in experimental Bleutongue virus serotype 8 infection of Dorset sheep. Vet. Ital. 2015;51(4):379–384. doi: 10.12834/VetIt.496.2398.1. [DOI] [PubMed] [Google Scholar]
- Ellis J.A., Luedke A.J., Davis W.C., Wechsler S.J., Mecham J.O., Pratt D.L., Elliott J.D. T lymphocyte subset alterations following bluetongue virus infection in sheep and cattle. Vet. Immunol. Immunopathol. 1990;24(1):49–67. doi: 10.1016/0165-2427(90)90077-6. [DOI] [PubMed] [Google Scholar]
- Gibbs E.P., Tabachnick W.J., Holt T.J., Stallknecht D.E. US concerns over bluetongue. Science. 2008;320(5878):872. doi: 10.1126/science.320.5878.872a. -NEW YORK THEN WASHINGTON. [DOI] [PubMed] [Google Scholar]
- Hemadri D., Maan S., Chanda M.M., Rao P.P., Putty K., Krishnajyothi Y., Reddy G.H., Kumar V., Batra K., Reddy Y.V., Maan N.S. Dual infection with bluetongue virus serotypes and first-time isolation of serotype 5 in India. Transbound Emerg. Dis. 2017;64(6):1912–1917. doi: 10.1111/tbed.12589. [DOI] [PubMed] [Google Scholar]
- Hofmann M., Renzullo S., Mader M., Chaignat V., Worwa G., Thuer B. Genetic characterization of toggenburg orbivirus, a new bluetongue virus, from goats, Switzerland. Emerg. Infect. Dis. 2008;14:1855–1861. doi: 10.3201/eid1412.080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo M.H., Gumm I.D., Taylor W.P. Clinical and serological response of sheep to serial challenge with different bluetongue virus types. Res. Vet. Sci. 1983;34(2):205–211. [PubMed] [Google Scholar]
- Jeggo M.H., Wardley R.C., Brownlie J. Importance of ovine cytotoxic T cells in protection against bluetongue virus infection. Prog. Clin. Biol. Res. 1985;178:477–487. [PubMed] [Google Scholar]
- Kopanke J.H., Lee J.S., Stenglein M.D., Mayo C.E. The genetic diversification of a single bluetongue virus strain using an in vitro model of alternating-host transmission. Viruses. 2020;12(9):1038. doi: 10.3390/v12091038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Guo S., Stiles J.K. The emerging role of CXCL10 in cancer. Oncol. Lett. 2011;2(4):583–589. doi: 10.3892/ol.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Li Z., Zhu P., Yang Z., Yang H., Li Z., Li H., Li Z. Whole-transcriptome analyses of sheep embryonic testicular cells infected with the bluetongue virus. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1053059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclachlan N.J., Henderson C., Schwartz-Cornil I., Zientara S. The immune response of ruminant livestock to bluetongue virus: from type I interferon to antibody. Virus Res. 2014;182:71–77. doi: 10.1016/j.virusres.2013.09.040. [DOI] [PubMed] [Google Scholar]
- Mahmoud A.S., Savini G., Spedicato M., Monaco F., Carmine I., Lorusso A., Francesco T., Mazzei M., Forzan M., Eldaghayes I., Dayhum A. Exploiting serological data to understand the epidemiology of bluetongue virus serotypes circulating in Libya. Vet. Med. Sci. 2019;5(1):79–86. doi: 10.1002/vms3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor P.S., Boorman J., Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu. Rev. Entomol. 2000;45(1):307–340. doi: 10.1146/annurev.ento.45.1.307. [DOI] [PubMed] [Google Scholar]
- Melzi E., Caporale M., Rocchi M., Martín V., Gamino V., di Provvido A., Marruchella G., Entrican G., Sevilla N., Palmarini M. Follicular dendritic cell disruption as a novel mechanism of virus-induced immunosuppression. Proc. Natl Acad. Sci. 2016;113(41):E6238–E6247. doi: 10.1073/pnas.1610012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J., Crossley B., Dechant J.E., Drew C.P., James MacLachlan N. Fatal bluetongue virus infection in an alpaca (Vicugna pacos) in California. J. Vet. Diagn. Investig. 2010;22:134–136. doi: 10.1177/104063871002200129. [DOI] [PubMed] [Google Scholar]
- Pages N., Breard E., Urien C., Talavera S., Viarouge C., Lorca-Oro C., Jouneau L., Charley B., Zientara S., Bensaid A., Solanes D. Culicoides midge bites modulate the host response and impact on bluetongue virus infection in sheep. PLoS One. 2014;9(1):e83683. doi: 10.1371/journal.pone.0083683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putty K., Shaik A.M., Peera S.J., Reddy Y.N., Rao P.P., Patil S.R., Reddy M.S., Susmitha B., Jyothi J.S. Infection kinetics and antibody responses in Deccani sheep during experimental infection and superinfection with bluetongue virus serotypes 4 and 16. Vet World. 2019;12(1):41. doi: 10.14202/vetworld.2019.41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratinier M., Shaw A.E., Barry G., Gu Q., Di Gialleonardo L., Janowicz A., Varela M., Randall R.E., Caporale M., Palmarini M. Bluetongue virus NS4 protein is an interferon antagonist and a determinant of virus virulence. J. Virol. 2016;90(11):5427–5439. doi: 10.1128/JVI.00422-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries C., Vögtlin A., Hüssy D., Jandt T., Gobet H., Hilbe M., Burgener C., Schweizer L., Häfliger-Speiser S., Beer M., Hoffmann B. Putative novel atypical BTV serotype ‘36’identified in small ruminants in Switzerland. Viruses. 2021;13(5):721. doi: 10.3390/v13050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martín D., Louloudes-Lázaro A., Avia M., Martín V., Rojas J.M., Sevilla N. The interplay between bluetongue virus infections and adaptive immunity. Viruses. 2021;13(8):1511. doi: 10.3390/v13081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas J.M., Rodríguez-Calvo T., Sevilla N. Recall T cell responses to bluetongue virus produce a narrowing of the T cell repertoire. Vet. Res. 2017;48:1–2. doi: 10.1186/s13567-017-0444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscanu S., Pascale F., Bourge M., Hemati B., Elhmouzi-Younes J., Urien C., Bonneau M., Takamatsu H., Hope J., Mertens P., Meyer G. The double-stranded RNA bluetongue virus induces type I interferon in plasmacytoid dendritic cells via a MYD88-dependent TLR7/8-independent signaling pathway. J. Virol. 2012;86(10):5817–5828. doi: 10.1128/JVI.06716-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saminathan M., Singh K.P., Khorajiya J.H., Dinesh M., Vineetha S., Maity M., Rahman A.F., Misri J., Malik Y.S., Gupta V.K., Singh R.K. An updated review on bluetongue virus: epidemiology, pathobiology, and advances in diagnosis and control with special reference to India. Vet. Q. 2020;40(1):258–321. doi: 10.1080/01652176.2020.1831708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saminathan M., Singh K.P., Vineetha S., Maity M., Biswas S.K., Manjunathareddy G.B., Chauhan H.C., Milton A.A., Ramakrishnan M.A., Maan S., Maan N.S. Virological, immunological and pathological findings of transplacentally transmitted bluetongue virus serotype 1 in IFNAR1-blocked mice during early and mid gestation. Sci. Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-58268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saminathan M., Singh K.P., Maity M., Vineetha S., Manjunathareddy G.B., Dhama K., Malik Y.S., Ramakrishnan M.A., Misri J., Gupta V.K. Pathological and immunological characterization of bluetongue virus serotype 1 infection in type I interferons blocked immunocompetent adult mice. J. Adv. Res. 2021;31:137–153. doi: 10.1016/j.jare.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Cordón P.J., Pedrera M., Risalde M.A., Molina V., Rodríguez-Sánchez B., Nunez A., Sánchez-Vizcaíno J.M., Gómez-Villamandos J.C. Potential role of proinflammatory cytokines in the pathogenetic mechanisms of vascular lesions in goats naturally infected with bluetongue virus serotype 1. Transbound Emerg. Dis. 2013;60(3):252–262. doi: 10.1111/j.1865-1682.2012.01343.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cordon P.J., de Diego A.P., Gomez-Villamandos J.C., Sanchez-Vizcaino J.M., Pleguezuelos F.J., Garfia B., Del Carmen P., Pedrera M. Comparative analysis of cellular immune responses and cytokine levels in sheep experimentally infected with bluetongue virus serotype 1 and 8. Vet. Microbiol. 2015;177(1–2):95–105. doi: 10.1016/j.vetmic.2015.02.022. [DOI] [PubMed] [Google Scholar]
- Schwartz-Cornil I., Mertens P., Contreras V., Hemati B., Pascale F., Bréard E., Mellor P., MacLachlan N., Zientara S. Bluetongue virus: virology, pathogenesis and immunity. Vet. Res. 2008;39(5):1. doi: 10.1051/vetres:2008023. [DOI] [PubMed] [Google Scholar]
- Szmaragd C., Wilson A.J., Carpenter S., Wood J.L., Mellor P.S., Gubbins S. The spread of bluetongue virus serotype 8 in Great Britain and its control by vaccination. PLoS One. 2010;5(2):e9353. doi: 10.1371/journal.pone.0009353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velthuis A.G., Saatkamp H.W., Mourits M.C., De Koeijer A.A., Elbers A.R. Financial consequences of the Dutch bluetongue serotype 8 epidemics of 2006 and 2007. Prev. Vet. Med. 2010;93(4):294–304. doi: 10.1016/j.prevetmed.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Veronesi E., Darpel K., Gubbins S., Batten C., Nomikou K., Mertens P., Carpenter S. Diversity of transmission outcomes following co-infection of sheep with strains of bluetongue virus serotype 1 and 8. Microorganisms. 2020;8(6):851. doi: 10.3390/microorganisms8060851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetter L.E., Maclachlan* N.J., Gebhard D.H., Heidner H.W., Moore P.F. Bluetongue virus infection of bovine monocytes. J. Gen. Virol. 1989;70(7):1663–1676. doi: 10.1099/0022-1317-70-7-1663. [DOI] [PubMed] [Google Scholar]
- Worwa G., Hilbe M., Chaignat V., Hofmann M.A., Griot C., Ehrensperger F., Doherr M.G., Thür B. Virological and pathological findings in Bluetongue virus serotype 8 infected sheep. Vet. Microbiol. 2010;144(3–4):264–273. doi: 10.1016/j.vetmic.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Yang J.L., Chang C.Y., Sheng C.S., Wang C.C., Wang F.I. The tip region on VP2 protein of bluetongue virus contains potential IL-4-inducing amino acid peptide segments. Pathogens. 2020;10(1):3. doi: 10.3390/pathogens10010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.J., Marston W.L., Dessing M., Dudler L., Hein W.R. Distinct recirculating and non-recirculating B-lymphocyte pools in the peripheral blood are defined by coordinated expression of CD21 and L-selectin. Blood. 1997;90(12):4865–4875. , The Journal of the American Society of Hematology. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available upon request.